
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 August 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.941026
This article is part of the Research Topic Abiotic Stress Adaptation and Tolerance Mechanisms in Crop Plants View all 40 articles
NAC proteins constitute one of the largest transcription factor families and are involved in regulation of plant development and stress responses. Our previous transcriptome analyses of tobacco revealed a significant increase in the expression of NtNAC028 during leaf yellowing. In this study, we found that NtNAC028 was rapidly upregulated in response to high salinity, dehydration, and abscisic acid (ABA) stresses, suggesting a vital role of this gene in abiotic stress response. NtNAC028 loss-of-function tobacco plants generated via CRISPR-Cas9 showed delayed leaf senescence and increased tolerance to drought and salt stresses. Meanwhile NtNAC028 overexpression led to precocious leaf senescence and hypersensitivity to abiotic stresses in Arabidopsis, indicating that NtNAC028 functions as a positive regulator of natural leaf senescence and a negative regulator of stress tolerance. Furthermore, NtNAC028-overexpressing Arabidopsis plants showed lower antioxidant enzyme activities, higher reactive oxygen species (ROS), and H2O2 accumulation under high salinity, resulted in more severe oxidative damage after salt stress treatments. On the other hand, NtNAC028 mutation in tobacco resulted in upregulated expression of ROS-scavenging and abiotic stress-related genes, higher antioxidant enzyme activities, and enhanced tolerance against abiotic stresses, suggesting that NtNAC028 might act as a vital regulator for plant stress response likely by mediating ROS scavenging ability. Collectively, our results indicated that the NtNAC028 plays a key regulatory role in leaf senescence and response to multiple abiotic stresses.
Plants are sessile organisms that must withstand and cope with environmental pressures, including biotic and abiotic stresses. Adverse changes in environmental conditions usually cause loss of crop yield, even the death of plant tissues (Liliane and Charles, 2020). Abiotic stresses, including drought and salinity, pose threats to food security worldwide by causing significant loss of crop production (Moghaddam et al., 2021). These stresses often lead to excessive accumulation of reactive oxygen species (ROS), which disrupts cellular redox homeostasis and causes oxidative damage of plant cells (Suzuki et al., 2012). In order to survive adverse environments, plants have developed a series of biochemical and physiological strategies during evolution (Haak et al., 2017). Gene regulation at the transcriptional level is the most critical and effective way for plants’ adaptation to stresses and DNA-binding transcription factors (TFs) play a vital role in this process (Lata et al., 2011; Le Hénanff et al., 2013). To date, extensive studies have revealed a wide range of TFs including MYB, bZIP, WRKY, and NAC (NAM, ATAF1/2, and CUC2) that are involved in regulation of abiotic stress response (Guo and Gan, 2012; Pascual et al., 2018; Zhang et al., 2018).
NAC (NAM, ATAF1/2, and CUC2) proteins are one of the largest families of plant-specific TFs, most of which harbor a highly conserved DNA-binding domain at the N-terminal region and a variable transcriptional regulation C-terminal domain (Diao et al., 2020). NAC TFs have been implicated in multiple aspects of plant development. For example, CUP-SHAPED COTYLEDON1 (CUC1) and NAC WITH TRANSMEMBRANE MOTIF1 (NTM1) are involved in shoot and lateral root development. NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1) and VASCULAR-RELATED NAC DOMAIN6 (VND6) are master regulators of secondary cell wall formation, ORESARA9 (ORE9) and NAC-LIKE, ACTIVATED BY AP3/PI (AtNAP) play important role in regulating leaf senescence (Takada et al., 2001; Guo and Gan, 2006; Kim et al., 2006; Zhong et al., 2010; Breeze et al., 2011; Diao et al., 2020; Liu et al., 2022). Besides, NAC proteins have also been reported to be implicated in different abiotic stress response (Nakashima et al., 2012). A number of NAC proteins involved in regulating stress response have been characterized and manipulated to develop crop varieties with enhanced stress tolerance. In Arabidopsis, expression of ANAC019, ANAC072/RD26, ANAC053/NTL4, or ANAC055 could improve salt and drought tolerance of transgenic plants (Tran et al., 2004; Lee et al., 2014). anac096 mutant is drought stress sensitive, indicating that ANAC096 functions as a positive regulator in dehydration stress response (Xu et al., 2013). Besides, ANAC016 could also positively regulate drought stress tolerance through suppressing AREB1 expression (Sakuraba et al., 2015). In contrast, ORE1 knockout mutants are more tolerant to salt stress, while overexpression lines show an opposite phenotype, suggesting that ORE1/ANAC092 functions as a negative regulator in salt stress response (Balazadeh et al., 2010). In rice, overexpression of OsNAC5, OsNAC6/SNAC2, OsNAC45, OsNAC52, or OsNAC066 significantly enhanced the tolerance to drought and salt stresses in transgenic rice (Nakashima et al., 2007; Sperotto et al., 2009; Gao et al., 2010; Yuan et al., 2019; Zhang et al., 2020). For wheat, TaNAC2, TaNAC67, and TaNAC29 have been reported to participate in abiotic stress responses. Overexpression of these genes in Arabidopsis and wheat improved plants’ tolerance to low temperature, high salinity, and drought by activating stress-responsive genes (Mao et al., 2012, 2014, 2016; Huang et al., 2015). Similarly, overexpression of several maize NACs, including ZmSNAC1, ZmNAC33, ZmNAC49, and ZmNAC55, could confer tolerance to drought stress in transgenic plants (Lu et al., 2012; Mao et al., 2016; Liu et al., 2019; Xiang et al., 2021). Interestingly, many stress-related genes might also be involved in plant senescence process. For instance, OsNAC2, known to negatively regulate many abiotic stresses tolerances, can also promote rice leaf senescence via ABA biosynthesis (Shen et al., 2017). TaNAC29 has been reported to play crucial roles in senescence and abiotic stress response (Huang et al., 2015; Xu et al., 2015). It has been hypothesized that some of these genes integrate the different signaling pathways and play important roles in the crosstalk between stress responses and age-dependent senescence (He et al., 2005; Guo and Gan, 2006).
Compared with Arabidopsis thaliana, Oryza sativa, and other plant species, few studies have been conducted on the functional characteristics of tobacco NAC genes. NtNAC-R1 has been reported to be involved in nicotine synthesis and lateral root development (Fu et al., 2013). Besides, overexpression of NtNAC2 or NtNAC053 could improve drought and/or salt stress tolerance in tobacco plants (Xu et al., 2018; Li et al., 2022). Our previous study has shown that overexpression of NtNAC080 promotes leaf senescence in Arabidopsis, suggesting that it acts as a positive regulator of natural leaf senescence in tobacco (Li et al., 2018). A recent study has indicated that NaNAC29 may participate in the defense responses to Alternaria alternata and promote leaf senescence in Nicotiana attenuata (Ma et al., 2020). To our knowledge, no further studies have been conducted on stress-related NAC genes in tobacco.
We previously identified a NAC TF, NtNAC028 (accession number XM_016636746), as a senescence upregulated gene. In this work, we found that NtNAC028 was also dramatically induced by high salinity, dehydration, and ABA treatments, suggesting that this gene might have multiple functions in different biological processes. Then we performed a detailed functional characterization of NtNAC028 in abiotic stress responses using transgenic tobacco and Arabidopsis. Phenotypic analysis showed that NtNAC028 played a negative role in response to drought and salt stress. Further study revealed that the enhanced tolerance of ntnac028 tobacco mutant obtained via CRISPR-Cas9 strategy was potentially achieved by activation of antioxidant enzymes in ROS scavenging and upregulation of stress-responsive genes. In addition to being involved in abiotic stress responses, NtNAC028 could also positively regulate leaf senescence in tobacco. Our findings demonstrated the role of NtNAC028 in leaf senescence, drought and salt stress responses and revealed the likely molecular mechanisms underlying abiotic stress response in tobacco.
Common tobacco (Nicotiana tabacum L. Cv. K326) seeds were sterilized by 70% ethanol for 1 min and 15% w/v H2O2 for 12 min, then rinsed in sterile water 4–5 times. The seeds were germinated and the plants were grown on Murashige and Skoog (MS) medium in a plant growth chamber (Conviron, Canada) at 25°C under long-day conditions (16 h light/8 dark). Then 4-week-old seedlings were removed from MS plates and treated in MS solution in 250 ml beakers with or without 200 mM NaCl, 150 μM abscisic acid (ABA), 150 μM salicylic acid (SA), or 150 μM jasmonic acid (JA) for 0–36 h. For drought treatments, 4-week-old seedlings were removed from MS medium, placed on Whatman No.5 filter paper, and dehydrated at room temperature for 0, 1, 3, 6, and 12 h. Whole plants were harvested, immediately frozen in liquid nitrogen, and stored at-80°C before further use. Three biological replicates were applied for each treatment.
Arabidopsis (Col-0) plants were grown on soil or on plates in a growth chamber at 22°C with hours a 16-h-light/8-h-dark cycle. For growth on plates, seeds were surface sterilized with 70% ethanol for 5 min, washed 4–5 times with sterile water, and then stored in the dark at 4°C for 3 days before the plates were transferred to growth chamber.
Total RNA was extracted using the Ultrapure RNA kit (CW biotech, Beijing, China), and first-strand cDNAs were synthesized using the HiScript III All-in-one RT SuperMix (Vazyme, NanJing, China). Then the cDNA samples were used for qRT-PCR analysis, which was performed on a Light Cycler96 real-time PCR system (Roche, Germany). NtActin and AtActin were used as internal reference genes in tobacco and Arabidopsis, respectively. Three technical replicates were used for all reactions and three biological replicates were performed for each experiment. The relative expression values were calculated using the 2-△△Ct method (Rao et al., 2013). The relevant primers are listed in Supplementary Table 1.
The full-length NtNAC028 cDNA was amplified by PCR using primes (NtNAC028-35 s-F: 5′-CACTGT TGATACATATGGTTGGGAAAAATTGCTCCGAG-3′; NtNAC028-35 s-R: 5′-TGTTGATTCAGAATTCTCACTGGA ACTGAAAGGCTGGAT-3′) and cloned into linearized pRI101-AN vector (digested with NdeI and EcoRI) using the Infusion kit (Clontech, Bejing, China). The recombinant vector was transferred into Agrobacterium tumefaciens GV3101 and then transformed into Arabidopsis thaliana (Col-0) via the floral dip method (Clough and Bent, 1998). The transgenic plants were selected on 1/2 MS medium supplemented with 50 mg/ml kanamycin and confirmed by qRT-PCR. Homozygous T3 lines were used for senescence and stress response assays.
For CRISPR-Cas9 vector construction, an online software1 was used to design the guide RNA (gRNA) targeting the NtNAC028 coding region. The forward and reverse primers were annealed to form DNA double-strands and inserted into the pORE-Cas9 vector (Gao et al., 2015). The recombinant vector was introduced into Agrobacterium tumefaciens LBA4404 and transformed into common tobacco (K326) by Agrobacterium-mediated transformation (Horsch et al., 1989). The transgenic plants were screened by kanamycin (50 mg/ml) resistance and used for extraction of genomic DNA. For each transgenic plant, the region of the gRNA targeting site was amplified by PCR using primers (NtNAC028-TS-F: 5′-ATGGTTGGGAAAAATTGCT-3′; NtNAC028-TS-R: 5′-GATTGCTTTATCTGTGCCTGT-3′) and cloned into the pEASY-blunt Zero vector (Transgene, China). DNAs from single colonies were used for sequencing to detect the mutation type. T1 plants, derived from self-pollinated T0 mutant lines, were sequenced again to confirm mutations. The homozygous mutants were further analyzed.
The extraction and quantification of chlorophyll were performed as previously described (He and Gan, 2002). Briefly, 200 mg of fresh leaves was extracted with 5 ml of anhydrous ethanol in dark until the chlorophyll was completely dissolved. The supernatant was measured at 665 and 649 nm for absorbance. For determination of Fv/Fm, after 30 min of dark treatment, the leaves of individual plant were determined using a IMAGING-PAM M-series Chlorophyll Fluorescence System (LI-6400-40 LCF, Walz, Effeltrich, Germany). Chlorophyll contents and Fv/Fm were calculated and mean values from three biological replicates were obtained.
Leaf disks from 60-day-old plants of the ntnac028 mutant and WT were placed onto moistened filter paper inside Petri dishes, and kept in dark for 7 days. Three independent biological replicates of this experiment were carried out. For NaCl-induced senescence assay, leaf disks from 35-day-old plants were placed in dishes with 0 or 200 mM NaCl, and kept in light for 7 days. Similarly, the leaves of 4-week-old plants from NtNAC028-OE Arabidopsis lines and WT (Col-0) plants were placed in dishes with 0 or 200 mM NaCl, and kept in light for 3 days. Three independent biological replicates of this experiment were carried out.
For the drought tolerance assays, WT (Col-0) and NtNAC028-OE Arabidopsis lines were soil-grown for 4 weeks under well-watered conditions, then stressed by withholding water for 8 days. After that, the plants were re-watered and survival rate of each group of plants was calculated after 3 days. Similarly, 5-week-old plants of the ntnac028 mutant and WT (K326) were subject to dehydration by halting water for 12 days, and then re-watered for 2 days. After that, the survival rates of tobacco seedlings were calculated. The water loss rate was measured by calculating the weight change of detached leaves of different genotypes after dehydration for 2 h at room temperature.
For salt stress treatment, the surface-sterilized tobacco seeds grown on MS medium were placed vertically in a growth chamber for 2 weeks and transferred to MS medium supplemented with 0, 50, 100, 150, or 200 mM NaCl, respectively. The plants were then grown for 2 weeks before measuring the length of primary roots. Similarly, 4-day-old seedlings of different Arabidopsis lines were transferred into 1/2 MS medium supplemented with 100 or 150 mM NaCl, respectively. After 1 week, the length of the primary roots was measured.
For germination assay, surface-sterilized seeds of NtNAC028-OE and WT were sown on 1/2 MS medium supplemented with different concentrations of NaCl (0, 100, and 150 mM) or ABA (0.5 μM), and kept at 4°C for 3 days in the dark before being transferred to a growth chamber under normal growth conditions. After 1 week, the germination rates of each group were calculated based on the presence of open cotyledons. All the experiments were performed with three independent biological replicates.
The content of malondialdehyde (MDA) was determined as previously described (Draper et al., 1993). POD, SOD, and CAT enzymatic activities were measured using the detection kits provided by Beijing Solarbio Science & Technology Co., Ltd.
For NBT staining, the salt-treated leaves from NtNAC028-OE lines and WT were soaked in NBT staining buffer (10 mM potassium phosphate buffer, pH 7.2) for 12 h. After that, leaf chlorophyll was removed by a mixed solution (glycerol/acetic acid/ethanol, 1:1:3) and then stored in a solution consisting of ethanol/glycerol (4:1). The hydrogen peroxide content was determined using a hydrogen peroxide assay kit (Solarbio, China) according to the manufacturer’s instruction.
Our previous work showed that the expression of NtNAC028 increased significantly during tobacco leaf senescence, especially at the early stage of senescence (Li et al., 2018). In this study, qRT-PCR was performed to measure the expression of NtNAC028 in tobacco. Among the tested tissues, the transcript level of NtNAC028 was the highest in the senescent leaves and very low in young leaves and stems (Figure 1A). In addition, NtNAC028 showed high expression in roots, suggesting that NtNAC028 might have multiple biological functions. To understand whether NtNAC028 is also involved in abiotic stress response, we examined the expression patterns of NtNAC028 under salt and drought stress treatments. As shown in Figures 1B,C, the transcript level of NtNAC028 was induced significantly during the whole course (1–24 h) of these treatments. Meanwhile, the expression patterns of NtNAC028 after exogenous plant hormone treatment were analyzed. The expression of NtNAC028 increased continuously and rapidly after ABA treatments. However, no significant increase in NtNAC028 expression was observed after treatments with SA or JA (Figures 1D–F).
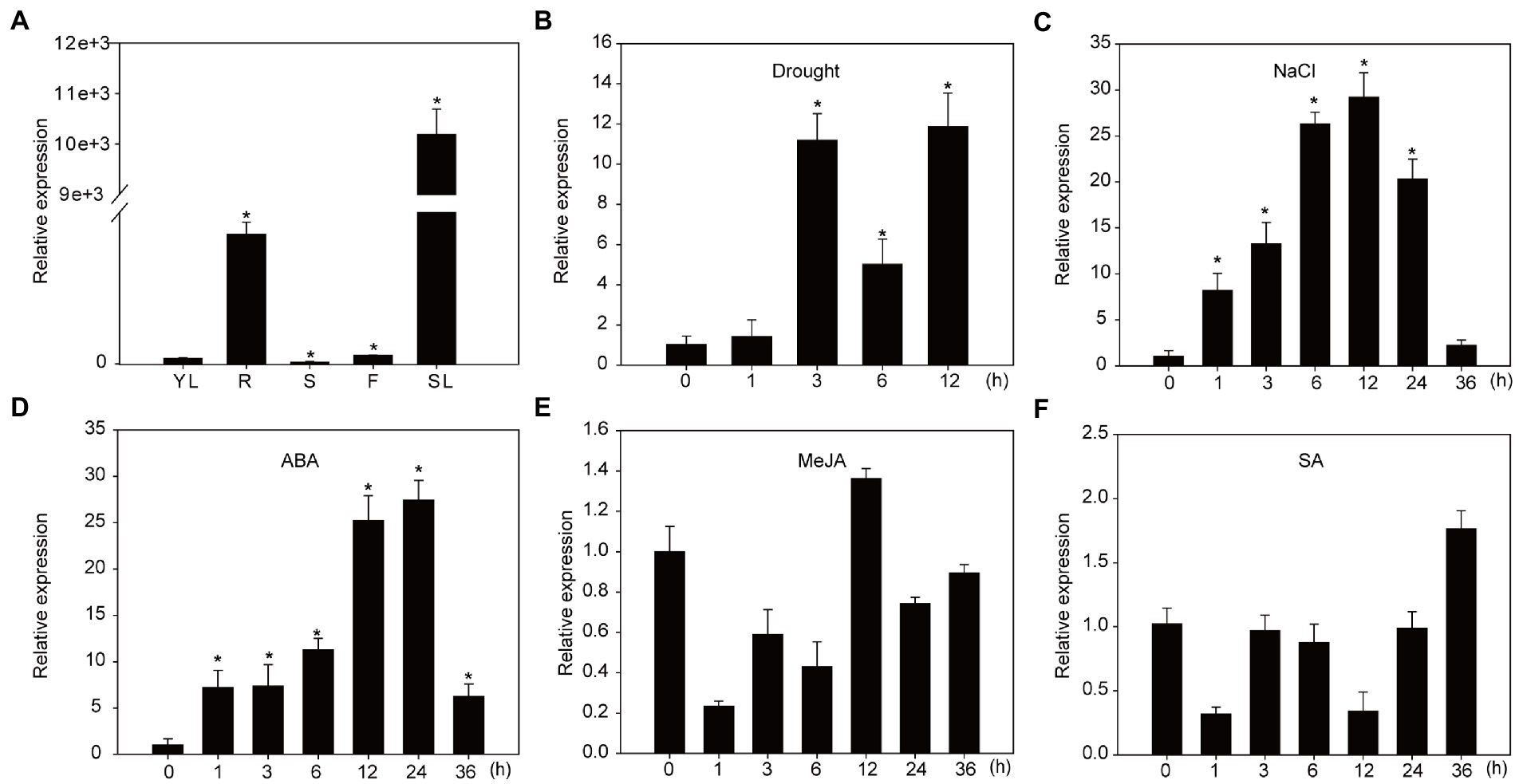
Figure 1. Expression profiling of NtNAC028. (A) The expression pattern of NtNAC028 in various organs of tobacco plants (K326). YL, young leaf; R, root; S, stem; F, flower; SL, senescence leaf. (B,C) NtNAC028 expression in response to different stress conditions. (D–F) The expression pattern of NtNAC028 under exogenous phytohormone treatments. ABA, abscisic acid; JA, jasmonic acid; and SA, salicylic acid. The bars are SD of three biological replicates. Asterisks indicate significant difference by Student’s t-test (*p < 0.05).
To understand the role of NtNAC028 in regulating tobacco leaf senescence, we generated a NtNAC028 loss-of-function line using the CRISPR-Cas9 system. A gRNA targeting the NAC domain of NtNAC028 was designed and used for Agrobacterium-mediated transformation of tobacco plants. T0 transgenic plants were obtained after kanamycin selection and sequenced to identify mutations. Then homozygous mutant plants were obtained from T1 generation for further phenotypic analysis (Figure 2A). Here, the ntnac028 mutant had a 1 bp insertion at the 3′ end of the gRNA sequence which introduced a stop codon after position P14 and led to a truncation of the DNA-binding NAC domain and loss of the transcriptional regulation domain at the C-terminus (Figure 2A). Compared with WT (K326), the ntnac028 mutant plants showed a delayed leaf senescence phenotype. The chlorophyll content of middle leaves from the ntnac028 mutant were higher than that in WT (Figures 2B,C). qRT-PCR showed that the transcript level of NtNAC028 in the mutant was significantly lower than WT (Figure 2D), possibly due to decreased stability of the truncated coding region. Furthermore, the transcript levels of senescence-related genes were determined by qRT-PCR. Compared to WT, the expression level of CYSTEINE PROTEINASE 1 (NtCP1, SENESCENCE-ASSOCIATED GENE 12/SAG12 homolog in tobacco) was lower and the RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN (NtRBCS) was higher in the ntnac028 mutant (Figures 2E,F). Likewise, we investigated the phenotype of the ntnac028 mutant leaves under dark treatments (Figure 2G). Detached leaves of WT and ntnac028 plants were covered with aluminum foil for 7 days. Leaves of the ntnac028 mutant showed a delayed senescence phenotype compared with those from WT. Consistent with this finding, the leaves of ntnac028 mutant had higher chlorophyll content compared with WT (Figure 2H). These data suggested that NtNAC028 could act as a positive regulator in tobacco leaf senescence.
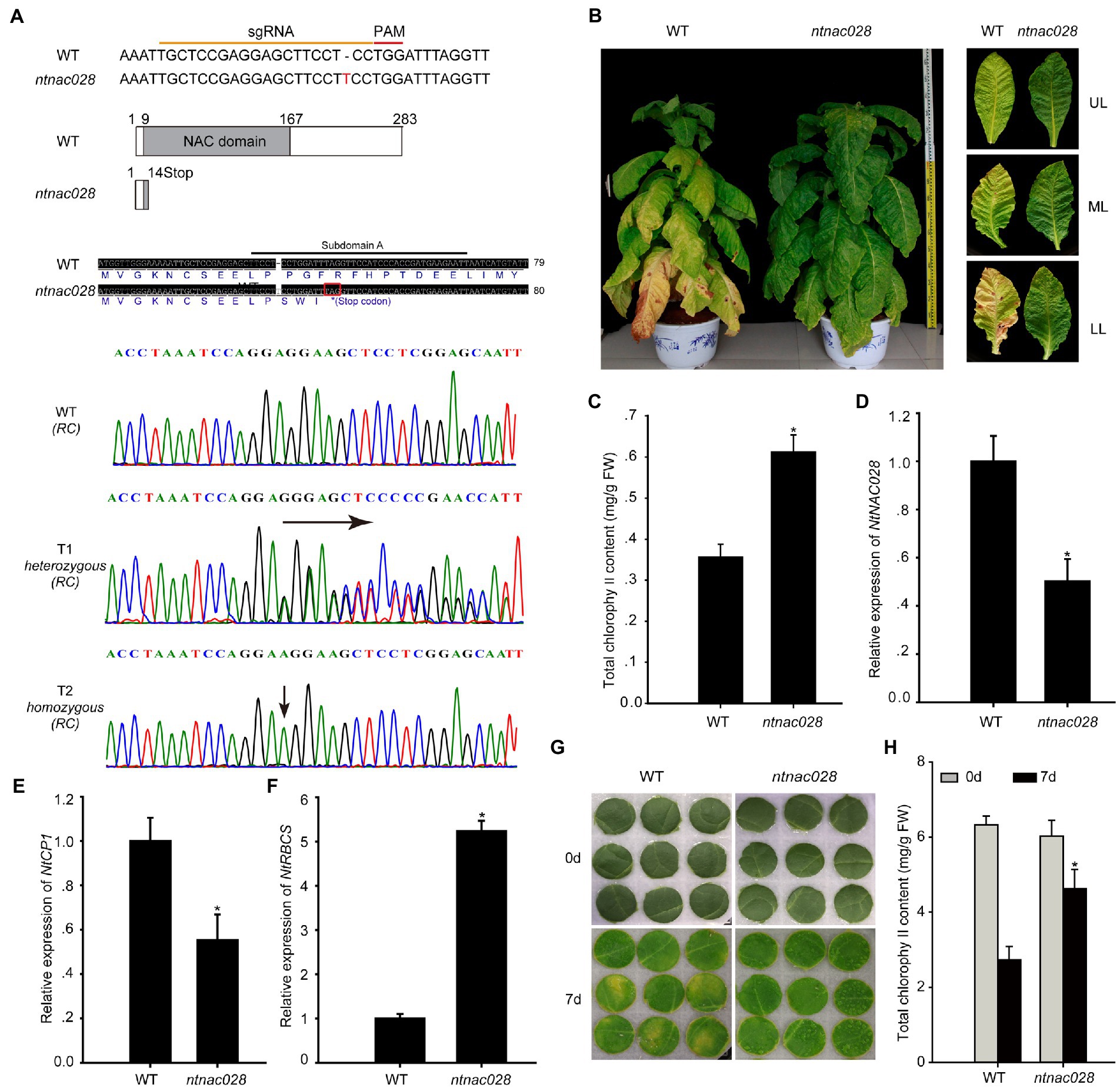
Figure 2. NtNAC028 positively regulates leaf senescence. (A) Construction of the NtNAC028 CRISPR-Cas9 vector. Sanger sequencing results showed an insert of 1 bp in the genomic DNA of ntnac028, compared to the WT. Red line represents the PAM sequence. (B) Leaf senescence phenotypes of ntnac028. (C) Chlorophyll content of middle leaves in age-matched wild-type (K326) and ntnac028 plants. UL (upper leaf), ML (middle leaf), and LL (low leaf). (D–F) Expression of NtNAC028, NtCP1, and NtRBCS in middle leaves of WT and ntnac028. (G) Phenotypes of detached leaves under dark conditions. (H) Chlorophyll content of detached leaves in WT and ntnac028 under darkness for 7 days. The bars are SD of three biological replicates. Asterisks indicate statistically significant difference from WT (*p < 0.05).
To further evaluate the role of NtNAC028 in regulating leaf senescence, we generated transgenic Arabidopsis plants overexpressing NtNAC028. 45 days after sowing, the overexpression transgenic plants, with expression of NtNAC028 confirmed by qRT-PCR, showed precocious leaf senescence (Supplementary Figures S1A,B). The chlorophyll contents and maximum quantum efficiency of PSII (Fv/Fm) of NtNAC028-OE plants (the fifth leaves) were lower than that of WT (Supplementary Figures S1C,D). Moreover, the premature senescence phenotype was also supported by the higher expression of AtSAG12 (a senescence marker gene) in the fifth leaves of the two NtNAC028 OE lines (Supplementary Figure S1E). These results also indicated that NtNAC028 could positively regulate leaf senescence.
In order to examine the biological role of NtNAC028 in response to drought stress, 5-week-old ntnac028 and WT tobacco plants were subject to drought stress treatments. As shown in Figure 3A, the ntnac028 plants displayed higher drought tolerance. Around 7 days after water withdraw, when most leaves of the WT plants were wilted, only a small portion of the ntnac028 leaves started wilting. Most of the WT plants became desiccated 12 days after drought treatment and died after re-watering, with a survival rate of 6.9%. The ntnac028 plants on the other hand, behaved much better at the end of the treatment with a survival rate of 96% after re-watering (Figures 3A,B). Detached leaves of the ntnac028 mutant and WT (K326) were subject to dehydration stress for 2 h and water loss rate was calculated at each time point. During the first 30 min of dehydration stress, there was no significant difference in relative water content between ntnac028 and WT plants. However, the relative water content of ntnac028 plants was significantly higher than that of WT after 1 h of dehydration stress. After 2 h dehydration stress, the water content of WT was less than 42%, while it was still more than 55% in the ntnac028 mutant (Figure 3C). This result was consistent with the enhanced drought tolerance in the ntnac028 mutant. Besides, we also analyzed the root length of ntnac028 and WT seedlings grown under treatments with different concentrations of NaCl. There was no significant difference in root length between ntnac028 plants and WT under normal conditions. However, in the presence of various concentrations of NaCl, the ntnac028 plants were more tolerant than WT. The root length of the ntnac028 plants was dramatically longer than that of WT (Figures 3D,E). For the germination assay, there was no significant difference between ntnac028 and WT plants in the absence of NaCl. However, the germination rates of ntnac028 seeds treated with 50 or 100 mM NaCl were significantly higher than that of WT plants, indicating that the seeds of ntnac028 mutant were insensitive to NaCl during the germination stage (Supplementary Figures S2A,B). Furthermore, detached leaves from 5-week-old plants of ntnac028 and WT were treated with 200 mM NaCl for 7 days. The leaves of WT senesced early with lower levels of chlorophyll content than ntnac028 leaves under NaCl stress conditions (Figures 3F,G). Taken together, these results indicated that the knockout of NtNAC028 led to elevated abiotic stress tolerance in tobacco.
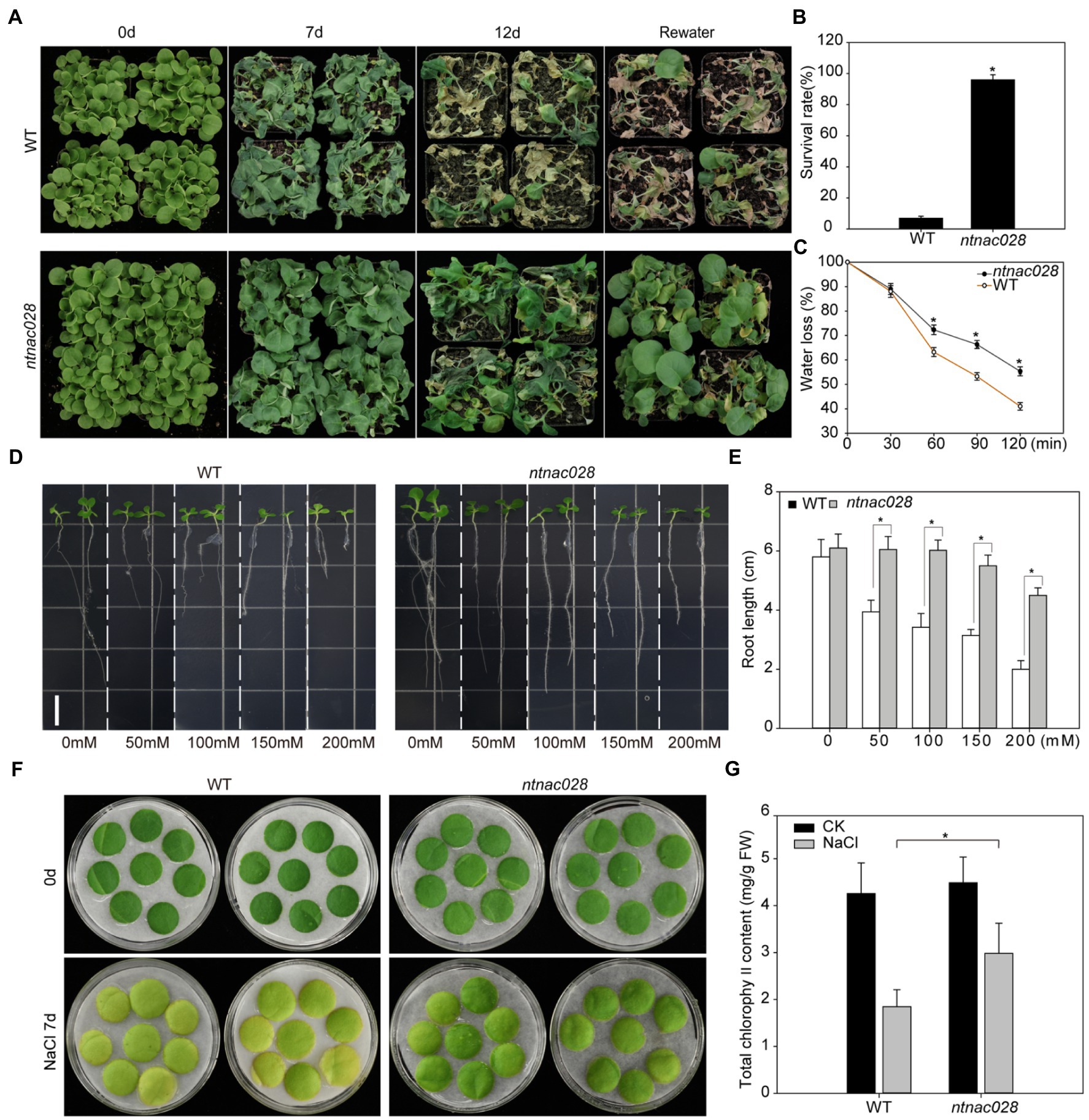
Figure 3. CRISPR/Cas9-mediated knockout of NtNAC028 enhances abiotic stress tolerance in tobacco. (A) Phenotypes of tobacco plants treated with drought stress for 12 days and recovered with water for 2 days. (B) Phenotypes of ntnac028 and wild-type tobacco under drought stress. (C) Water loss rates of detached leaves from WT and ntnac028. Water loss was determined as the percentage of initial fresh weight. Values are means from 10 plants for each of three independent experiments. (D,E) Root length of WT and ntnac028 under control and NaCl treatments. Seedlings were vertically cultivated on MS plates containing 0, 50, 100, 150, or 200 mM NaCl. Images were taken after 5 days. Bars = 1 cm. Data were calculated using 20 seedlings for each genotype. The bars are SDs of three biological replicates. Asterisks indicate statistically significant difference from WT (*p < 0.05). (F,G) The phenotypes and chlorophyll content of detached leaves from WT and ntnac028 under NaCl treatment for 7 days.
To evaluate the effect of NtNAC028 overexpression on drought stress tolerance, soil-grown Arabidopsis plants were subject to drought stress by withholding water. After the dehydration stress for 8 days, NtNAC028-OE6 and NtNAC028-OE17 plants exhibited severe leaf wilting phenotypes due to water deficiency, while no evident wilting symptom was observed for WT plants (Figure 4A). After re-watering for 3 days, less than 35% of the overexpression plants survived; whereas, the survival rates of WT plants were close to 100% (Figure 4B). Moreover, NtNAC028-OE lines lost more water than the WT plants at all time points in the water loss assay of detached leaves (Figure 4C). These results indicated that overexpression of NtNAC028 reduced drought tolerance in Arabidopsis.
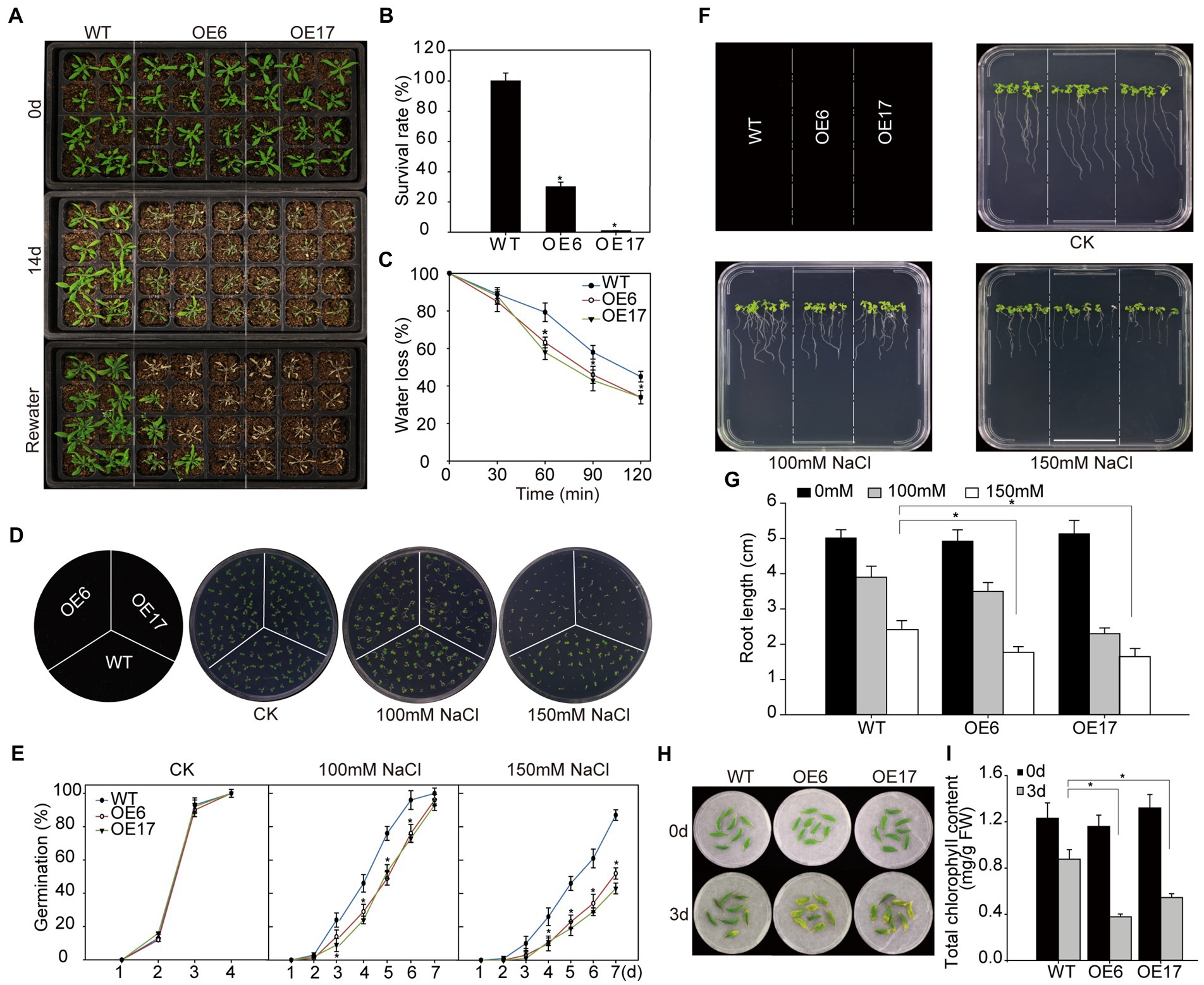
Figure 4. Overexpression of NtNAC028 confers hypersensitivity to abiotic stress in transgenic Arabidopsis. (A) Phenotypes of NtNAC028 overexpression lines (OE6 and OE17) and WT under drought stress treatments. Four-week-old NtNAC028 overexpression lines and WT were withheld from watering for 14 days, followed by re-watering and recovery for 3 days. (B) Survival rates of NtNAC028 overexpression lines and WT after drought stress. (C) Water loss rates of detached leaves from NtNAC028 overexpression lines and WT. (D,E) Germination of WT and NtNAC028 overexpression lines with different concentrations of NaCl as indicated. Representative images were taken after 7 days. (F,G) Root length of NtNAC028 overexpression lines and WT seedlings under NaCl treatments. Seedlings were vertically grown for 7 days on 1/2 MS plates containing 0, 100, or 150 mM NaCl. (H,I) The phenotypes and chlorophyll content of detached leaves from WT and NtNAC028 overexpression lines under NaCl treatments for 3 days. The bars are standard deviations (SD) of three biological replicates. Asterisks indicate statistically significant difference from WT (*p < 0.05).
Meanwhile, NtNAC028-OE plants were examined for tolerance to salt stress. As shown in Figures 4D,E, no obvious difference in germination rate was observed without NaCl treatment. However, germination rates of NtNAC028-OE lines were significantly lower than that of WT in the presence of 100 or 150 mM NaCl. This result indicated that NaCl stress could severely inhibited seed germination of NtNAC028 overexpression lines compared to WT. Meanwhile, the germination assay was carried out to assess the ABA sensitivity of NtNAC028-OE lines. Interestingly, the NtNAC028-OE lines had significantly lower germination rates than WT under ABA treatments (Supplementary Figures S3A,B), suggesting that NtNAC028-OE seeds were more sensitive to ABA during germination. Besides, the sensitivity to salt in NtNAC028 overexpression lines was further assayed at the post-germination stage. Similarly, no significant difference in root length between NtNAC028-OE lines and WT was observed under the control conditions. In the presence of 100 or 150 mM NaCl, the roots of the NtNAC028-OE lines were significantly shorter than that of WT plants (Figures 4F,G). Furthermore, detached leaves from 4-week-old plants of NtNAC028-OE and WT were treated with 200 mM NaCl for 3 days. The leaves of NtNAC028-OE senesced early with lower levels of chlorophyll content than WT leaves under NaCl stress conditions (Figures 4H,I). Overall, these results suggested that overexpression of NtNAC028 reduced salt tolerance in Arabidopsis.
Drought or salt stress often leads to oxidative stress which involves excessive ROS generation in plant cells (Choudhury et al., 2017). To explore whether the reduced tolerance to drought and salt in NtNAC028-OE lines is associated with the alteration of ROS levels, we used NBT staining to visualize the levels of superoxide radical (O.2−) in leaves. Without NaCl treatment, the color of NBT staining in leaves showed no significant difference between NtNAC028-OE and WT plants. When stressed by 200 mM NaCl, the NtNAC028-OE lines accumulated significantly more O.2−, as densely stained with dark blue color in the leaves of NtNAC028-OE lines compared to WT (Figure 5A). Besides, we measured the endogenous H2O2 levels of plants with different genotypes. In the presence of NaCl stress, NtNAC028-OE plants accumulated significantly more H2O2 than that of WT (Figure 5B). These results indicated that NtNAC028 overexpression could enhance ROS production in transgenic Arabidopsis under salt stress treatments.
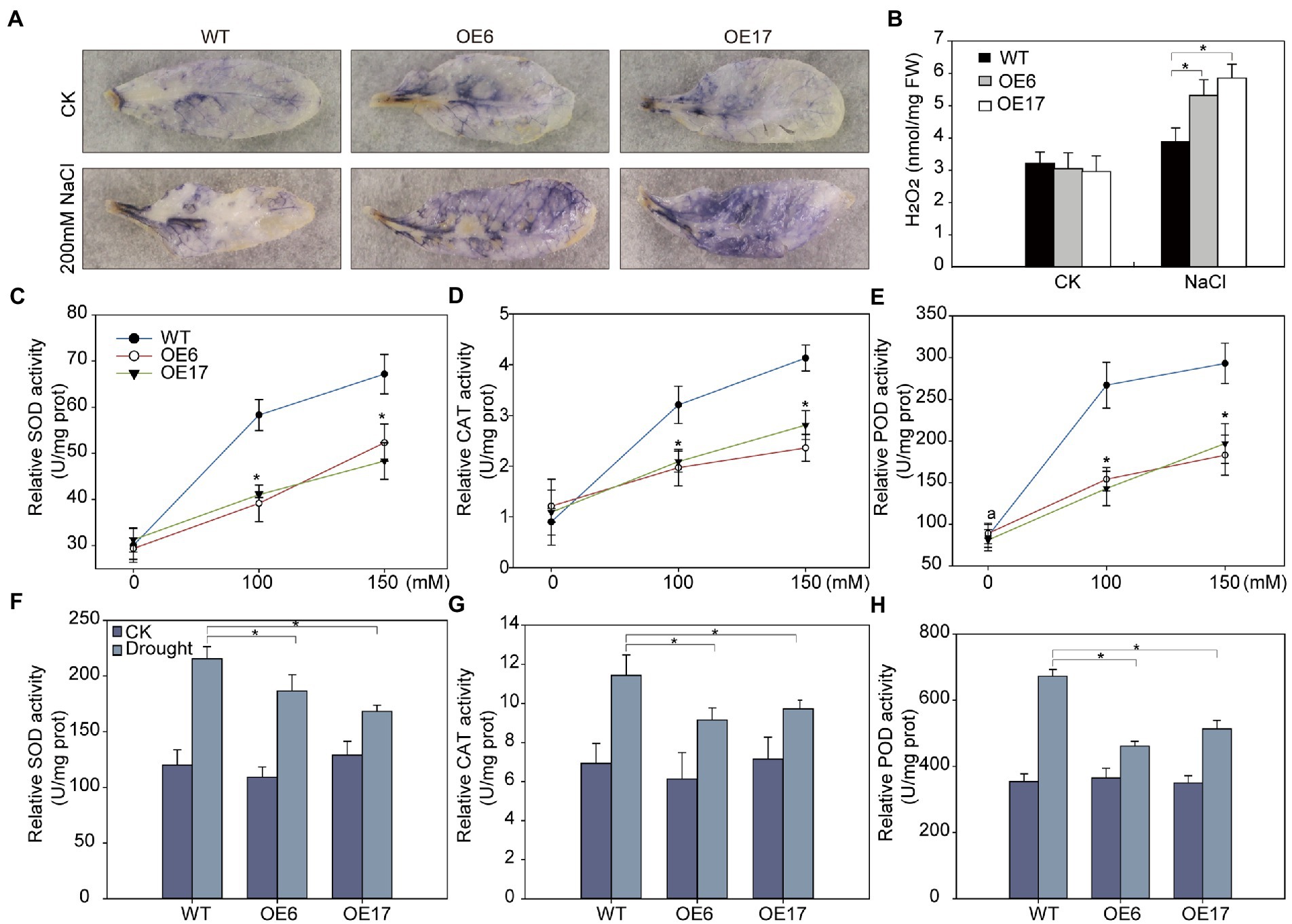
Figure 5. NtNAC028 promotes reactive oxygen species (ROS) accumulation under NaCl treatments. (A) NBT staining. The middle leaves of 4-week-old NtNAC028 overexpression lines and WT with or without NaCl treatments were used for NBT staining. (B) Measurement of accumulation of H2O2 in detached leaves of plants with different genotypes. (C–H) POD, SOD, and CAT activities of NtNAC028 overexpression lines and WT after salt and drought stress treatments. The bars are standard deviations (SD) of three biological replicates. Asterisks indicate statistically significant difference from WT (*p < 0.05).
Furtherly, we analyzed the activities of SOD, POD, and CAT, which are main enzymes responsible for scavenging ROS. No obvious difference was observed in the ROS scavenging activities under non-stress conditions. After salt and drought treatments, SOD, POD, and CAT activities of NtNAC028-OE lines and WT were all significantly increased. However, the increase in enzymatic activities of the NtNAC028-OE lines was significantly less than that of WT (Figures 5C–H). Furthermore, the expression levels of ROS-scavenging-related genes (SOD, POD, and CAT) in NtNAC028-OE lines were also lower than that of WT plants after salt and drought treatments (Supplementary Figures S4A–F). The expression patterns of stress-responsive genes including AtRD29A, AtDREB1B, AtRD26, and AtNCED3-2 were analyzed in NtNAC028-OE lines and WT plants before and after drought/salt treatments (Supplementary Figures S5A–H). Under no-stressed conditions, no significant difference in the transcript levels of these stress-related genes was observed between NtNAC028-OE and WT plants. However, the expression levels of these gene were significantly lower in NtNAC028-OE lines than in WT under drought or salt treatments. These results suggested that NtNAC028-OE could reduce the expression levels of ROS scavenging-related and stress-responsive genes upon abiotic stresses.
To ascertain whether the enhanced stress tolerance of ntnac028 plants was associated with the alteration of ROS scavenging capability, we examined the activities of SOD, POD, and CAT of ntnac028 and WT plants under normal or stressed conditions. No significant difference was observed in activities of the antioxidant enzymes under non-stress conditions. However, the activities of SOD, POD, and CAT were more dramatically increased in ntnac028 plants than that in WT after drought or salt stress treatments (Figures 6A–C,E). Consistently, MDA content of ntnac028 plants was significantly lower than that in WT following drought/salt stress treatments (Figures 6D,H). These results indicated that NtNAC028 mutation could enhance stress tolerance possibly through enhancing the activities of antioxidant enzymes.
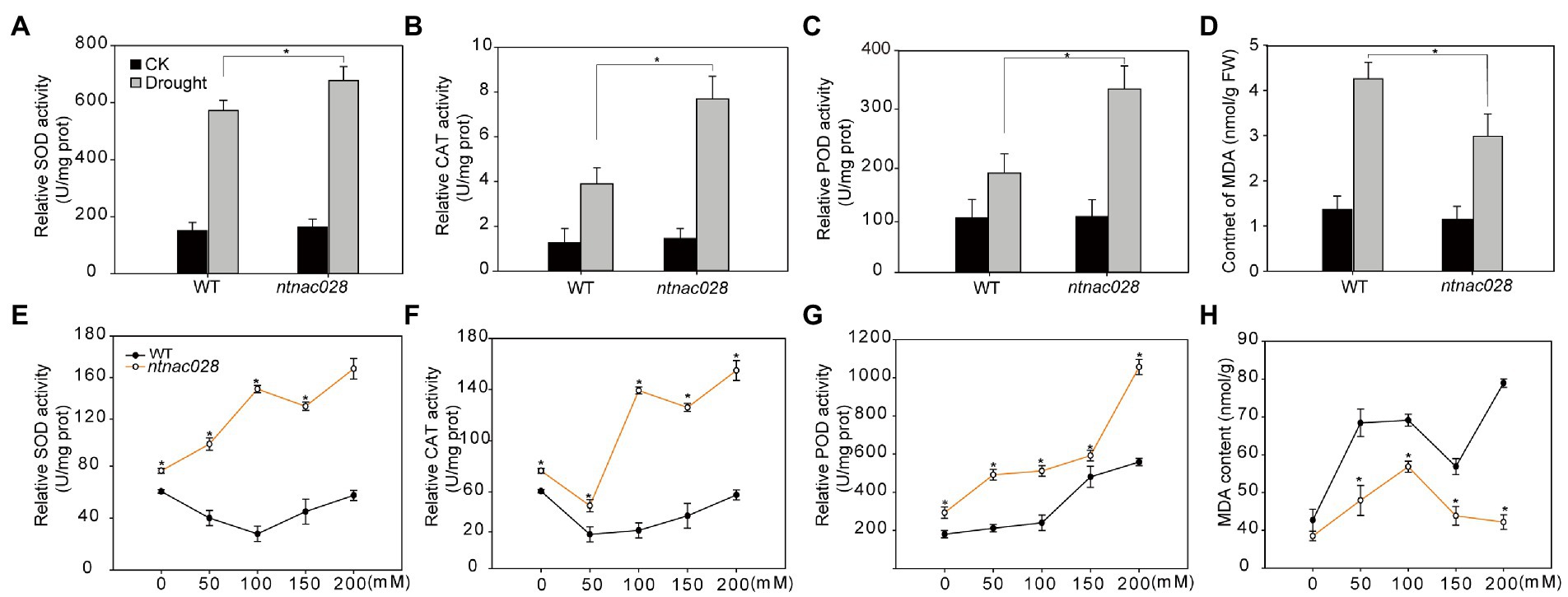
Figure 6. NtNAC028 mutation enhances the antioxidant activity of tobacco under abiotic stresses. (A–C) POD, SOD, and CAT activities of WT and ntnac028 plants with or without drought stress treatments. (E–G) POD, SOD, and CAT activities of WT and ntnac028 plants under various concentrations of NaCl. (D,H) MDA contents of WT and ntnac028 plants under drought and salt treatments. The bars are standard deviations (SD) of three biological replicates. Asterisks indicate statistically significant difference from WT (*p < 0.05).
To understand the mechanism of NtNAC028’s regulation of abiotic stress response, the expression patterns of stress-responsive genes, including NtRD29A, NtDREB1B, NtRD26, and NtNCED3-2 were analyzed in ntnac028 and WT plants before and after drought/salt treatments (Figures 7A–H). Under no-stressed conditions, no significant difference in the transcript levels of these stress-related genes was observed between ntnac028 and WT plants. However, the expression levels of these genes were significantly higher in ntnac028 than in WT plants under drought or salt treatments. Furthermore, salt and drought treatments strongly induced the expression of ROS-scavenging-related genes (SOD, POD, and CAT) in ntnac028 compared with WT (Supplementary Figures S6A–F). These results suggested that NtNAC028 knockout could enhance the expression levels of ROS scavenging-related and stress-responsive genes upon abiotic stresses.
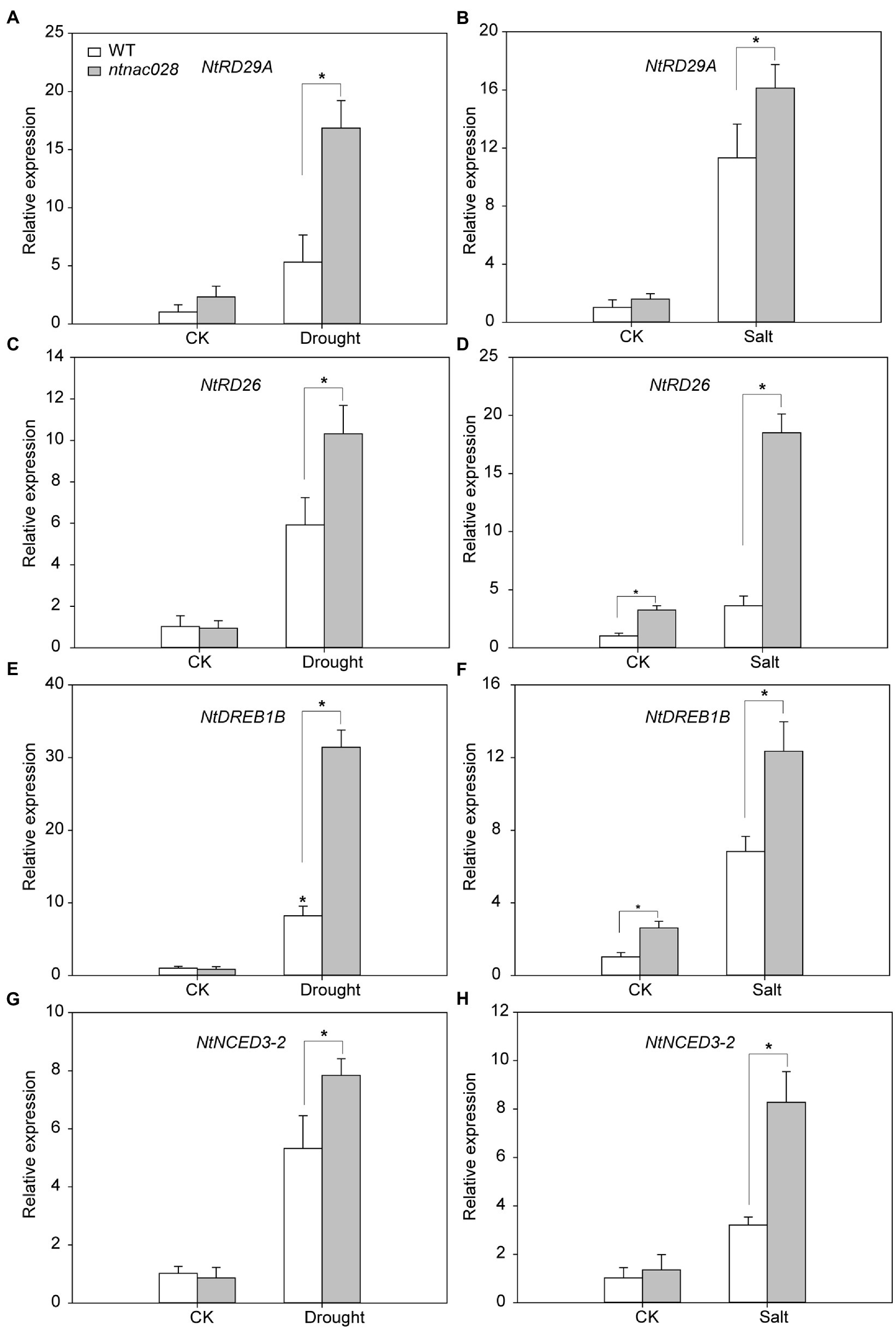
Figure 7. Expression patterns of stress-responsive genes in WT and ntnac028 plants under salt and drought treatments. Relative transcript levels of four stress-responsive genes (RD29A, RD26, DREB1B, and NCED3-2) in WT and ntnac028 plants before and after drought (A,C,E,G) and salt (B,D,F,H) treatments. The bars are standard deviations (SD) of three biological replicates. Asterisks indicate statistically significant difference from WT (*p < 0.05).
The NAC superfamily is one of the largest transcription factor families in plants, which plays important regulatory roles in plant development and stress response (Forlani et al., 2021). Previously, we identified a total of 154 NAC genes (NtNACs) in tobacco and demonstrated that NtNAC080 was a positive regulator of leaf senescence (Li et al., 2018). However, the functions of most of the remaining NtNACs remain unclear. In this study, we carried out a detailed functional characterization of NtNAC028, which belongs to the same phylogenetic subgroup as NtNAC080. The results demonstrated that NtNAC028 could also positively regulate leaf senescence, sharing a redundant role with NtNAC080. In addition, the NtNAC028 transcript was found to be upregulated rapidly and significantly under drought, salinity, and exogenous ABA treatments, suggesting it may be involved in abiotic stress response. Previous studies have revealed that some members of the NAC TFs are involved in senescence and also play vital roles in stress response (Guo and Gan, 2012; Zhang and Gan, 2012). For instance, overexpression of TaNAC29 from wheat can enhance salt and drought tolerance in transgenic Arabidopsis, which exhibits a delayed senescence phenotype (Huang et al., 2015). It was also showed that AtNAP negatively regulates tolerance to osmotic stress, while positively regulates leaf and fruit senescence (Guo and Gan, 2006; Xiaohong Kousup Sup, 2012). Interestingly, OsNAP, a rice homologue of AtNAP, functions as a positive regulator of leaf senescence and abiotic stress responses (high salinity and drought; Liang et al., 2014). In addition to having the similar function as positive regulators of leaf senescence, NAP homologues in different plant species seem to have more diversified functions in stress responses. In this study, several lines of evidence indicated that NtNAC028 plays an important role in response to drought and high salinity. As expected, the promoter sequence of NtNAC028 (about 3 kb upstream of the transcription start site) contains multiple cis-related elements of plant stress response, including seven ABA-responsive elements (ABREs) and three stress-related elements (TC-rich repeats: defense and stress responsiveness; LTR: low-temperature responsiveness; Supplementary Table 2). These results indicated that NtNAC028 might have broad functions in abiotic response.
NAC TFs are known to be involved in environmental stresses, which in turn improve the ability of plants in tolerating these stresses (Nakashima et al., 2012). Previous studies have showed that abiotic stresses could induce premature leaf senescence, which accelerates dry matter accumulation in grains and plays a positive role in plant environmental adaptability (Sade et al., 2018). While the mutants with delayed leaf senescence often showed higher tolerance to stress treatments (Kang et al., 2019; Guo et al., 2021). Here, ntnac028 tobacco mutant generated by CRISPR/Cas9 showed a lower rate of water loss and enhanced salt and drought stress tolerance, while NtNAC028-OE Arabidopsis lines were more sensitive to these stress than WT. In addition, seed germination rates of NtNAC028-OE lines were much lower than that of WT under NaCl treatments. These results suggested that NtNAC028 might negatively regulate salt and drought stress responses. It is known that the responsiveness of plants to abiotic stresses was mediated by ABA-dependent and ABA-independent pathways. In the ntnac028 mutant, the ABA-dependent pathway genes NtRD26, NtRD29A, and NtNCED3-2, as well as the ABA-independent gene NtDREB1B, were expressed at a higher level under stress conditions. It has been reported that these genes are involved in the mediation of oxidative or osmotic damages induced by stresses (Wei et al., 2016; Huang et al., 2018; Jiang et al., 2019; Orbović et al., 2021). For example, RD26 acts as a transcriptional activator of ABA-inducible gene expression in plants under various abiotic stress conditions (Fujita et al., 2004). DREB1B is a member of the AP2/ERF family, which can activate a subset of downstream stress-responsive genes and plays an essential role in drought stress response (Wei et al., 2016). Thus, increased expression of these genes could help ntnac028 mutant plants in tolerating abiotic stresses. As being reported in our previous study, NtNAC028 is a homolog of AtNAP (Li et al., 2018). AtNAP has been documented to negatively regulate salt stress tolerance via repression of ABA-dependent stress-responsive genes (Seok et al., 2017), suggesting that NtNAC028 might perform similar regulatory function in abiotic stress response in tobacco.
It is well known that abiotic stresses could cause excessive accumulation of reactive oxygen species (ROS) in plants, which could lead to damage of cellular biomolecules, including proteins, lipids, and DNA (Parida and Das, 2005). In addition, the senescence process can also induce ROS generation, and the increase in ROS levels triggers and accelerates senescence (Maggiorani et al., 2017). Therefore, ROS may serve as a key signal in both senescence and stress responses. Besides, antioxidant enzymes, such as POD, SOD, and CAT are the enzymes responsible for scavenging ROS in plants (Gill and Tuteja, 2010; Miller et al., 2010). The genes encoding antioxidant enzymes are often upregulated to protect tissues under stresses (Jin et al., 2017). In the present study, NtNAC028-OE lines accumulated significantly higher levels of H2O2 than WT plants under salt stress conditions. The activities of POD, CAT, and SOD in NtNAC028-OE lines were also significantly lower than those of WT (Figure 5), indicating that the higher accumulation of ROS in NtNAC028-OE plants under stress conditions was due to their inability to remove ROS in time. In contrast, the transcript levels and activities of POD, SOD, and CAT in the ntnac028 mutant were significantly higher than that in WT (K326; Figures 6, 7). Consistently, MDA content of ntnac028 mutant plants was significantly lower than that of WT under stress conditions, indicating that the ntnac028 mutant plants produced less ROS. Thus, it is possible that a more efficient ROS scavenging system in the ntnac028 mutant is important for its abiotic stress tolerance.
In summary, we functionally characterized a tobacco NAC gene, NtNAC028, which was strongly induced by senescence and various abiotic stresses. NtNAC028 mutation induced by CRISPR/Cas9 led to delayed senescence and enhanced tolerance to drought and salinity in tobacco, while NtNAC028 overexpression transgenic Arabidopsis exhibited early senescence and hypersensitivity to high salinity and drought, indicating that NtNAC028 culd function as a negative regulator of leaf senescence and abiotic stress response in plants. Furthermore, our results revealed that the enhanced stress tolerance of the ntnac028 mutant might be due to enhanced scavenging capacity of ROS and increased expression of stress-responsive genes.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
YG and WL conceived the research. WL and LW generated the transgenic materials and performed all experiments, and drafted the manuscript. TL, ZD, and QW assisted in data collection and analysis. ZZ and WW designed the Crispr-Cas9 experiment and generated the constructs. All authors contributed to the article and approved the submitted version.
This work was financially supported by National Natural Science Foundation of China (No. 31970204), the Agricultural Science and Technology Innovation Program (ASTIP-TRIC02)and Funds for Special Projects of the Central Government in Guidance of Local Science and Technology Development (21-1-1-1-zyyd-nsh).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.941026/full#supplementary-material
Supplementary Figure S1 | Overexpression of NtNAC028 in Arabidopsis plants causes precocious senescence.
Supplementary Figure S2 | Seed germination under salt treatments.
Supplementary Figure S3 | Seed germination under ABA treatments.
Supplementary Figure S4 | Expression patterns of reactive oxidation species scavenging-related genes in WT and NtNAC028-OE plants in response to salt and drought treatments.
Supplementary Figure S5 | Expression patterns of stress responsive genes in WT and NtNAC028-OE plants under salt and drought treatments.
Supplementary Figure S6 | Expression patterns of ROS scavenging-related genes in WT and ntnac028 plants in response to salt and drought treatments.
Balazadeh, S., Siddiqui, H., Allu, A. D., Matallana Ramirez, L. P., Caldana, C., Mehrnia, M., et al. (2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62, 250–264. doi: 10.1111/j.1365-313X.2010.04151.x
Breeze, E., Elizabeth, H., Stuart, M., Linda, H., Richard, H., and Claire, H. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873–894. doi: 10.1105/tpc.111.083345
Choudhury, F. K., Rivero, R. M., Blumwald, E., and Mittler, R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867. doi: 10.1111/tpj.13299
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Diao, P., Chen, C., Zhang, Y., Meng, Q., Lv, W., and Ma, N. (2020). The role of NAC transcription factor in plant cold response. Plant Signal. Behav. 15:1785668. doi: 10.1080/15592324.2020.1785668
Draper, H. H., Squires, E. J., Mahmoodi, H., Wu, J., Agarwal, S., and Hadley, M. (1993). A comparative evaluation of thiobarbituric acid methods for the determination of malondialdehyde in biological materials. Free Radic. Biol. Med. 15, 353–363. doi: 10.1016/0891-5849(93)90035-s
Forlani, S., Mizzotti, C., and Masiero, S. (2021). The NAC side of the fruit: tuning of fruit development and maturation. BMC Plant Biol. 21, 238–214. doi: 10.1186/s12870-021-03029-y
Fu, Y., Guo, H., Cheng, Z., Wang, R., Li, G., Huo, G., et al. (2013). NtNAC-R1, a novel NAC transcription factor gene in tobacco roots, responds to mechanical damage of shoot meristem. Plant Physiol. Biochem. 69, 74–81. doi: 10.1016/j.plaphy.2013.05.004
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Gao, J., Wang, G., Ma, S., Xie, X., Wu, X., Zhang, X., et al. (2015). CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 87, 99–110. doi: 10.1007/s11103-014-0263-0
Gao, F., Xiong, A., Peng, R., Jin, X., Xu, J., Zhu, B., et al. (2010). OsNAC52, a rice NAC transcription factor, potentially responds to ABA and confers drought tolerance in transgenic plants. Plant Cell Tissue Organ Cult. 100, 255–262. doi: 10.1007/s11240-009-9640-9
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Guo, Y., and Gan, S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612. doi: 10.1111/j.1365-313X.2006.02723.x
Guo, Y., and Gan, S. S. (2012). Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 35, 644–655. doi: 10.1111/j.1365-3040.2011.02442.x
Guo, Y., Ren, G., Zhang, K., Li, Z., Miao, Y., and Guo, H. (2021). Leaf senescence: progression, regulation, and application. Mol. Hortic. 1, 1–25. doi: 10.1186/s43897-021-00006-9
Haak, D. C., Fukao, T., Grene, R., Hua, Z., Ivanov, R., Perrella, G., et al. (2017). Multilevel regulation of abiotic stress responses in plants. Front. Plant Sci. 8:1564. doi: 10.3389/fpls.2017.01564
He, Y., and Gan, S. (2002). A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 14, 805–815. doi: 10.1105/tpc.010422
He, X. J., Mu, R. L., Cao, W. H., Zhang, Z. G., Zhang, J. S., and Chen, S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916. doi: 10.1111/j.1365-313X.2005.02575.x
Horsch, R. B., Fry, J., Hoffmann, N., Neidermeyer, J., Rogers, S. G., and Fraley, R. T. (1989). Leaf disc transformation. Plant Molecular Biology Manual 7, 63–71. doi: 10.1007/978-94-009-0951-9_5
Huang, Y., Guo, Y., Liu, Y., Zhang, F., Wang, Z., Wang, H., et al. (2018). 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 9:162. doi: 10.3389/fpls.2018.00162
Huang, Q., Wang, Y., Li, B., Chang, J., Chen, M., Li, K., et al. (2015). TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 15, 268–215. doi: 10.1186/s12870-015-0644-9
Jiang, H., Tang, B., Xie, Z., Nolan, T., Ye, H., Song, G. Y., et al. (2019). GSK 3-like kinase BIN 2 phosphorylates RD 26 to potentiate drought signaling in Arabidopsis. Plant J. 100, 923–937. doi: 10.1111/tpj.14484
Jin, C., Li, K., Xu, X., Zhang, H., Chen, H., Chen, Y., et al. (2017). A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front. Plant Sci. 8:1049. doi: 10.3389/fpls.2017.01049
Kang, K., Shim, Y., Gi, E., An, G., and Paek, N.-C. (2019). Mutation of ONAC096 enhances grain yield by increasing panicle number and delaying leaf senescence during grain filling in rice. Int. J. Mol. Sci. 20:5241. doi: 10.3390/ijms20205241
Kim, Y., Sang-Gyu, K., Jung-Eun, P., Hye-Young, P., Mi-Hye, L., and Nam-Hai, C. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18, 3132–3144. doi: 10.1105/tpc.106.043018
Lata, C., Yadav, A., and Prasad, M. (2011). Role of plant transcription factors in abiotic stress tolerance. Abiotic Stress Response in Plants 10, 269–296. doi: 10.5772/23172
Le Hénanff, G., Profizi, C., Courteaux, B., Rabenoelina, F., Gérard, C., Clément, C., et al. (2013). Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. J. Exp. Bot. 64, 4877–4893. doi: 10.1093/jxb/ert277
Lee, S., Lee, H., Huh, S. U., Paek, K., Ha, J., and Park, C. M. (2014). The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci. 227, 76–83. doi: 10.1016/j.plantsci.2014.07.003
Li, W., Li, X., Chao, J., Zhang, Z., Wang, W., and Guo, Y. (2018). NAC family transcription factors in tobacco and their potential role in regulating leaf senescence. Front. Plant Sci. 9:1900. doi: 10.3389/fpls.2018.01900
Li, X., Wang, Q., Guo, C., Sun, J., Li, Z., Wang, Y., et al. (2022). NtNAC053, a novel NAC transcription factor, confers drought and salt tolerances in tobacco. Front. Plant Sci. 13:817106. doi: 10.3389/fpls.2022.817106
Liang, C., Wang, Y., Zhu, Y., Tang, J., Hu, B., Liu, L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. 111, 10013–10018. doi: 10.1073/pnas.1321568111
Liliane, T. N., and Charles, M. S. (2020). Factors Affecting Yield of Crops. Agronomy-Climate Change & Food Security. London, UK: IntechOpen, 9.
Liu, G., Li, H., Grieson, D., and Fu, D. (2022). NAC transcription factor family regulation of fruit ripening and quality: a review. Cell 11:525. doi: 10.3390/cells11030525
Liu, W., Zhao, B., Chao, Q., Wang, B., Zhang, Q., Zhang, C., et al. (2019). Function analysis of ZmNAC33, a positive regulator in drought stress response in Arabidopsis. Plant Physiol. Biochem. 145, 174–183. doi: 10.1016/j.plaphy.2019.10.038
Lu, M., Ying, S., Zhang, D., Shi, Y., Song, Y., Wang, T., et al. (2012). A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep. 31, 1701–1711. doi: 10.1007/s00299-012-1284-2
Ma, L., Li, R., Ma, L., Song, N., Xu, Z., and Wu, J. (2020). Involvement of NAC transcription factor NaNAC29 in Alternaria alternata resistance and leaf senescence in Nicotiana attenuata. Plant Divers. 43, 502–509. doi: 10.1016/j.pld.2020.11.003
Maggiorani, D., Manzella, N., Edmondson, D. E., Mattevi, A., Parini, A., Binda, C., et al. (2017). Monoamine oxidases, oxidative stress, and altered mitochondrial dynamics in cardiac ageing. Oxidative Med. Cell. Longev. 2017, 1–8. doi: 10.1155/2017/3017947
Mao, X., Chen, S., Li, A., Zhai, C., and Jing, R. (2014). Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS One 9:e84359. doi: 10.1371/journal.pone.0084359
Mao, H., Yu, L., Han, R., Li, Z., and Liu, H. (2016). ZmNAC55, a maize stress-responsive NAC transcription factor, confers drought resistance in transgenic Arabidopsis. Plant Physiol. Biochem. 105, 55–66. doi: 10.1016/j.plaphy.2016.04.018
Mao, X., Zhang, H., Qian, X., Li, A., Zhao, G., and Jing, R. (2012). TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 63, 2933–2946. doi: 10.1093/jxb/err462
Miller, G., Suzuki, N., Ciftci Yilmaz, S., and Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Moghaddam, M. S. H., Safaie, N., Soltani, J., and Hagh-Doust, N. (2021). Desert-adapted fungal endophytes induce salinity and drought stress resistance in model crops. Plant Physiol. Biochem. 160, 225–238. doi: 10.1016/j.plaphy.2021.01.022
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. BBA Gene Regulat. Mechan. 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nakashima, K., Tran, L. S. P., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Orbović, V., Ravanfar, S. A., Acanda, Y., Narvaez, J., Merritt, B. A., Levy, A., et al. (2021). Stress-inducible Arabidopsis thaliana RD29A promoter constitutively drives Citrus sinensis APETALA1 and LEAFY expression and precocious flowering in transgenic citrus spp. Transgenic Res. 30, 687–699. doi: 10.1007/s11248-021-00260-z
Parida, A. K., and Das, A. B. (2005). Salt tolerance and salinity effects on plants: a review. Ecotoxicol. Environ. Saf. 60, 324–349. doi: 10.1016/j.ecoenv.2004.06.010
Pascual, M., Torre, F. D. L., Cañas, R. A., Cánovas, F. M., and Ávila, C. (2018). NAC transcription factors in woody plants. Progress in Botany 80, 195–222. doi: 10.1007/124_2018_19
Rao, X., Huang, X., Zhou, Z., and Xin, L. (2013). An improvement of the 2(−delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 3, 71–85.
Sade, N., del Mar Rubio-Wilhelmi, M., Umnajkitikorn, K., and Blumwald, E. (2018). Stress-induced senescence and plant tolerance to abiotic stress. J. Exp. Bot. 69, 845–853. doi: 10.1093/jxb/erx235
Sakuraba, Y., Kim, Y., Han, S., Lee, B., and Paek, N. (2015). The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27, 1771–1787. doi: 10.1105/tpc.15.00222
Seok, H., Woo, D., Nguyen, L. V., Tran, H. T., Tarte, V. N., Mehdi, S. M. M., et al. (2017). Arabidopsis AtNAP functions as a negative regulator via repression of AREB1 in salt stress response. Planta 245, 329–341. doi: 10.1007/s00425-016-2609-0
Shen, J., Lv, B., Luo, L., He, J., Mao, C., Xi, D., et al. (2017). The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci. Rep. 7, 1–16. doi: 10.1038/srep40641
Sperotto, R. A., Ricachenevsky, F. K., Duarte, G. L., Boff, T., Lopes, K. L., Sperb, E. R., et al. (2009). Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 230, 985–1002. doi: 10.1007/s00425-009-1000-9
Suzuki, N., Koussevitzky, S., Mittler, R., and Miller, G. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. doi: 10.1111/j.1365-3040.2011.02336.x
Takada, S., Ken-ichiro, H., Tetsuya, I., and Masao, T. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. doi: 10.1242/dev.128.7.1127
Tran, L. P., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Wei, T., Deng, K., Gao, Y., Liu, Y., Yang, M., Zhang, L., et al. (2016). Arabidopsis DREB1B in transgenic salvia miltiorrhiza increased tolerance to drought stress without stunting growth. Plant Physiol. Biochem. 104, 17–28. doi: 10.1016/j.plaphy.2016.03.003
Xiang, Y., Sun, X., Bian, X., Wei, T., Han, T., Yan, J., et al. (2021). The transcription factor ZmNAC49 reduces stomatal density and improves drought tolerance in maize. J. Exp. Bot. 72, 1399–1410. doi: 10.1093/jxb/eraa507
Xiaohong Kousup Sup, S. S. C. B. (2012). Arabidopsis AtNAP regulates fruit senescence. J. Exp. Bot. 63, 6139–6147. doi: 10.1093/jxb/ers266
Xu, Z., Kim, S. Y., Hyeon, D. Y., Kim, D. H., Dong, T., Park, Y., et al. (2013). The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25, 4708–4724. doi: 10.1105/tpc.113.119099
Xu, Z., Wang, C., Xue, F., Zhang, H., and Ji, W. (2015). Wheat NAC transcription factor TaNAC29 is involved in response to salt stress. Plant Physiol. Biochem. 96, 356–363. doi: 10.1016/j.plaphy.2015.08.013
Xu, X., Yao, X., Lu, L., and Zhao, D. (2018). Overexpression of the transcription factor NtNAC2 confers drought tolerance in tobacco. Plant Mol. Biol. Report. 36, 543–552. doi: 10.1371/journal.pone.0137098
Yuan, X., Wang, H., Cai, J., Bi, Y., Li, D., and Song, F. (2019). Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 19, 278–219. doi: 10.1186/s12870-019-1883-y
Zhang, K., and Gan, S. (2012). An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol. 158, 961–969. doi: 10.1104/pp.111.190876
Zhang, Y., Li, D., Wang, Y., Zhou, R., Wang, L., Zhang, Y., et al. (2018). Genome-wide identification and comprehensive analysis of the NAC transcription factor family in Sesamum indicum. PLoS One 13:e0199262. doi: 10.1371/journal.pone.0199262
Zhang, X., Long, Y., Huang, J., and Xia, J. (2020). OsNAC45 is involved in ABA response and salt tolerance in rice. Rice 13, 79–13. doi: 10.1186/s12284-020-00440-1
Keywords: tobacco, NtNAC028, abiotic stress, leaf senescence, ROS, Arabidopsis thaliana
Citation: Wen L, Liu T, Deng Z, Zhang Z, Wang Q, Wang W, Li W and Guo Y (2022) Characterization of NAC transcription factor NtNAC028 as a regulator of leaf senescence and stress responses. Front. Plant Sci. 13:941026. doi: 10.3389/fpls.2022.941026
Received: 11 May 2022; Accepted: 25 July 2022;
Published: 15 August 2022.
Edited by:
Ji-Hong Liu, Huazhong Agricultural University, ChinaReviewed by:
Massimo Galbiati, National Research Council, United StatesCopyright © 2022 Wen, Liu, Deng, Zhang, Wang, Wang, Li and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongfeng Guo, Z3VveW9uZ2ZlbmdAY2Fhcy5jbg==; Wei Li, bGl3ZWkwNkBjYWFzLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.