- 1Institute of Crops and Nuclear Technology Utilization, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2Yongding Agriculture and Rural Bureau of Longyan, Longyan, China
- 3Huzhou Agricultural Science and Technology Development Center/Huzhou Academy of Agricultural Sciences, Huzhou, China
- 4College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
Colorful flowers of rapeseed (Brassica napus L.) have been a hotspot for researchers, but the underlying mechanisms of pigment formation still need to be clarified. In this study, two stages of unopened rapeseed petals with red, white, and yellow colors were selected to identify the metabolites and genes involved in red pigment formation. Metabolomic analysis showed that flavonoids enriched the most co-differentially accumulated metabolites among all categories, and showed higher accumulation in red petal rapeseed than in white and yellow petal ones. RNA-seq analysis showed that among co-differentially expressed genes involved in red pigment formation, genes involved in anthocyanin (belonging to flavonoids) biosynthesis pathway were largely regulated by ANS, DFR, and UF3GT. The expression of those genes was higher in red petals of rapeseed than in white and yellow petals ones as well. Results of RNA interference of BnaA03.ANS in red rapeseed altered petal colors from raspberry red to beige red and zinc yellow under different interference levels, with the contents of pelargonidin, cyanidin, lutein, neoxanthin, β-carotene, and lycopene significantly decreased. However, overexpression of BnaA03.ANS in yellow rapeseed petals did not change the color of yellow petals. This study confirmed the important function of flavonoids, especially anthocyanins on red pigment formation, and for the first time, identified the irreplaceable role of BnaA03.ANS on red-flowered rapeseed.
Introduction
Brassica napus L. (AACC, 2n = 38), an allopolyploid species derived from the hybridization of Brassica rapa (AA, 2n = 20) and Brassica oleracea (CC, 2n = 18), is an important cash crop not only showing edible and industrial oil purpose but also exhibiting great ornamental value (Chalhoub et al., 2014; Fu et al., 2016). For example, Hubei Province, China, owns over 25.68 million hectares of rapeseed, the tourism of which caused by rapeseed “flower sea” has contributed to more than half of the county's GDP in 2011 (Fu et al., 2016).
Flower color is critical for ornamental and landscaping utilization by providing aesthetically pleasing scenes. The traditional flower color of rapeseed is yellow and some mutants with light to dark yellow, and with the exploitation of the ornamental values, various petal colors were bred, such as white, orange, pink, red, and purple (Yin et al., 2019). However, the mechanisms for regulating different petal colorations are still unclear.
Carotenoids, flavonoids (namely, anthocyanins, flavones, and flavonols), and betalain biosynthesis are reported as the three most important pathways which produce secondary metabolites that contribute to natural petal color display. For instance, carotenoid and betalain pathways contributed to yellow and red, while orange, yellow, red, and blue were attributed to the anthocyanin pathway (Tanaka et al., 2008). More than that, petal tissue structure, epidermal cell shapes, and pH were also reported to influence the formation of petal colors (Vignolini et al., 2015; Zhao and Tao, 2015). Anthocyanins are the main group of flavonoids, and play a crucial role in plant color development, ranging from pink to blue and purple. Over 100 anthocyanins have been identified, primarily originating from six common types, namely, pelargonidin, cyanidin, delphinidin, peonidin, petunidin, and malvidin (Veitch and Grayer, 2008). Previous studies showed that the color differences are highly correlated to different anthocyanin contents and components. By measuring the anthocyanin content of a series of butterfly pea petals, Kazuma et al. (2003) found that the content of anthocyanins was significantly higher in blue petals than in other colors while no anthocyanins were identified in white color petals. As for cineraria, delphinidin and cyanidin mainly contributed to blue and red flower colors, pink flowers were mainly determined by cyanidin and pelargonidin, while delphinidin and cyanidin were the core anthocyanins in purple flowers (Sun et al., 2009). Moreover, Zhang et al. (2011) found that the main pigment deposited in Lagenaria red petals was cyanidins, and pelargonidins primarily leaned toward scarlet.
Based on the irreplaceable role, anthocyanin biosynthesis has risen to be a hotspot of research for plant secondary metabolism and the key genes that participated in its biosynthetic pathway in plants have been clarified (Cheynier et al., 2013). CHALCONE SYNTHASE (CHS) is the key gene encoding the first enzyme in the anthocyanin biosynthesis pathway, hence influencing the coloration of flower petals. Ectopic expression of CHS1 gene from Freesia hybrid in petunia altered flower color from white to pink (Sun et al., 2015), and transgenic tobacco plants with CHS gene from Malus crabapple showed higher anthocyanin accumulation and a deep red color than the wild-type (Tai et al., 2014). CHALCONE ISOMERASE (CHI) encodes the second enzyme in anthocyanin biosynthesis and catalyzes the formation of chalcone. The decrease of CHI expression level in the petal of asters, tobacco, and carnations was found to lead to a greater accumulation of chalcone and turn the color into yellow (Nishihara et al., 2005). DIHYDROFLAVONOL 4-REDUCTASE (DFR) is another key gene encoding an enzyme that transfers three types of dihydroflavonols to their corresponding colorless anthocyanins with NADPH. Zhao et al. (2012) found that in different herbaceous peony organs, the highest expression level of DFR was observed in petals which accumulated the most anthocyanins, and similar results were also reported in Asian lily and gentian, suggesting the important role of DFR in flower color formation (Nakatsuka et al., 2003, 2005). ANTHOCYANIDIN SYNTHASE (ANS) is a key gene in the late stage of anthocyanin biosynthesis, which catalyzes the leucoanthocyanin to colored anthocyanidin (Heller et al., 1985). It was identified that ANS is a small gene family and these genes have been successfully cloned from several ornamental plants, such as Forsythia supensa, herbaceous peony, and gerbera (Rosati et al., 1999; Wellmann et al., 2006; Zhao et al., 2012). Rosati et al. (1999) found that null expression of ANS in Forsythia suspensa resulted in little anthocyanins accumulation in petals, and the absence of expression of ANS was found to be the underlying reason for the color change in lisianthus flowers (Shimizu et al., 2011). To date, the cloning and functional verification of genes that control petal color have paved for gene engineering of plants to obtain colorful exhibitions. For example, in Brassica napus, Liu et al. (2020) reported that silencing of BnaA09.Zep and BnaC09.Zep through Crispr/Cas9 confers orange color in B. napus petals. By ectopic overexpression of the OvPAP2 (Orychophragmus violaceus) gene, Fu et al. (2018) successfully produced red anthers and petals in yellow oilseed rape. Zhang et al. (2015) reported that disruption of BnaC03.CCD4 gene enhanced the accumulation of carotenoids leading to the transfer of petal color from white to yellow.
However, how the anthocyanin pathway regulates rapeseed petal color formation and the detailed molecular mechanisms remain unclear, and few studies have reported color-related genes or elaborated on the molecular regulation mechanism underlying anthocyanin-based variation in oilseed rape petal colors (Sagawa et al., 2016; Nikolov, 2019). The function of ANS, the key genes in anthocyanin biosynthesis, on flower coloration in Brassica napus is not reported in comparison with others. Here, we performed a metabolomics and RNA-seq study on two different stages of unopened petals of red, pure white, and yellow petal rapeseed lines, aiming at elaborating the pigment formation and development mechanisms in B. napus. BnaA03.ANS was identified as a high expression co-differentially expressed gene (co-DEG) according to RNA-seq and qRT-PCR. Disruption of BnaA03.ANS converted rapeseed petal color from raspberry red to beige red or zinc yellow, while overexpression of BnaA03.ANS made no change in petal color, but anthocyanins and carotenoids contents showed large differences. This study first comprehensively elaborated on the pigment formation and development mechanisms from genes to metabolites which contributed a lot to the foundation for colorful rapeseed breeding and the function of BnaA03.ANS on red color formation was first identified in B. napus.
Materials and Methods
Plant Materials and Sampling
Three B. napus lines with contrasting petal colors were used, which were Zhehuhong (red, abbreviated as V1), Zhehubai (white, abbreviated as V2), and Zheyou 50 (yellow, abbreviated as V3). The transgenic plants with overexpression and RNAi were obtained from Zheyou 50 and Zhehuhong, respectively. The rapeseed plants including the transgenic lines were planted in a glass solar greenhouse with three replications in a completely randomized block design at the Zhejiang Academy of Agricultural Science, China.
At the flowering stage, full plump unopened buds from the main inflorescence with <5 flowers were selected, then all the petals in each bud were separated out, and the developmental stages of unopened petals were defined as follows: stage 1, petals with 5 mm length and colors close to pale; stage 2, petals with 6–7 mm length; stage 3, petals with 8–9 mm length; and stage 4, petals with 10 mm length and were strongly pigmented. The petals in stage 1 and stage 4 of V1, V2, and V3 were utilized for RNA-seq and metabolomic analysis with 3 biological replicates. Petals at all stages were utilized for stereoscopic imaging, and petals at stage 4 were used for carotenoids and anthocyanins quantification. Petal colors were compared according to the RAL method (Long et al., 2011).
Metabolomic Analysis
Petals at stage 1 and stage 4 of V1, V2, and V3 were prepared, named V1-1, V1-4, V2-1, V2-4, V3-1, and V3-4, respectively, and 3 biological replicates were set. About 50 mg sample was weighed out for supernatant preparation. Agilent 1290 Infinity II UHPLC system coupled to an Agilent 6545 UHD and Accurate-Mass Q-TOF/MS was used for liquid chromatography-mass spectrometry (LC-MS) analysis. The chromatographic column used was Waters XSelect HSS T3 (2.5 μm, 100 mm × 2.1 mm). Raw data were converted to common (mz.data) using Agilent MassHunter Qualitative Analysis B.08.00 software (Agilent Technologies, USA). Then all data went through internal standard normalization and weight normalization. Visualization matrices containing sample name, m/z-RT pair, and peak area were obtained. After editing, the data matrices were imported into SIMCA-P 14.1 (Umetrics, Umea, Sweden), mean-centered and scaled to Pareto variance. Then, a multivariate analysis was conducted. |log2FoldChange|>1 and p ≤ 0.05 were determined as differentially accumulated metabolites (DAMs). DAMs Venn diagram, principal component analysis (PCA), Gene Ontology (GO), and KEGG enrichment (Kyoto Encyclopedia of Genes and Genomes, KEGG) were also conducted for further analysis.
RNA-seq Analysis
The preparation of samples was the same as metabolomic analysis. Total RNA was extracted using a polysaccharide and polyphenol total RNA isolation kit (Bioteke, Beijing, China). HISAT2 was used to align all clean reads against the reference genome Brassicanapus.Annotation_v4.1. FPKM values were used to calculate the expression level of genes. |log2FoldChange >1 and p ≤ 0.05 were determined as differentially expressed genes (DEGs). Venn diagram of DEGs, PCA, GO, and KEGG was also conducted for further analysis.
Quantification of Carotenoids and Anthocyanins Contents
Contents of carotenoids and anthocyanins in 9 rapeseed petals (V1-4, V2-4, V3-4, and each containing 3 biological replicates) were determined through HPLC methods. Carotenoids were performed according to Cao et al. (2012) and the contents of each carotenoid were determined as previously described (Morris et al., 2004).
The components of anthocyanins were determined according to Sun et al. (2014) with several modifications. Approximately 100 mg petal samples were weighed out and 800 μl methanol was added and vortex for 1 min, then shock under 4°C for 30 min. A total of 600 μl supernatant was prepared and concentrated to dry, then 200 μl methanol was added to redissolve the samples. The supernatants were used for LC-MS/MS analysis after centrifuging under 12,000 rpm 4°C for 10 min. The MS parameters were: ESI ion source, 35 arb curtain gas, 7 arb collision gas, 4,500 V ion spray voltage, 450°C temperature, 55 arb ion source gas1, and 55 arb ion source gas2. Multiple reaction monitoring was used for parameter acquisition and MultiQuant software was used for data calculation.
Plasmid Construction and Transformation
Petal-specific expression promoter XY355 and the open reading frame of BnaA03.ANS was amplified and cloned into the Pme1-Pac1 and Pac1-Asc1 sites of pMDC83, respectively, to construct overexpression plasmid pXY355::BnaA03.ANS. The plasmid was introduced into the Agrobacterium tumefaciens strain GV3101 and transformed into yellow-flowered rapeseed V3.
The target gene fragments were amplified using BnaA03.ANS RNAi primers (Supplementary Table 1), and then ligated into the pNC-Cam1304-RNAi vector using the Nimble Cloning kit (Chinese Academy of Tropical Agricultural Sciences) to construct an ANS interference vector. The constructed vector was transformed into Agrobacterium tumefaciens GV3101, and the genetic transformation of rapeseed hypocotyl mediated by Agrobacterium was used to transform it into red rapeseed V1. Both Nimble Cloning kit and pNC-Cam1304-RNAi vector were donated by Mr. Yan Pu (Chinese Academy of Tropical Agricultural Sciences). The genetic transformation methods were referred to by Zhou et al. (2012).
qRT-PCR Analysis
Total RNA was extracted from petal samples using a polysaccharide and polyphenol total RNA isolation kit (Bioteke, Beijing, China) and qRT-PCR was performed according to Xu et al. (2017). All primers used in the study were listed in Supplementary Table 1.
Statistics
Data analysis was performed using IBM SPSS v.22.0 statistical software. Duncan's multiple range test was used to evaluate significant treatment effects at the significance level of p ≤ 0.05. MultiQuant software was used to integrate the curves and calculate the contents of anthocyanins and carotenoids according to each standard curve.
Results
Phenotype Characterization of all B. napus Materials
In this experiment, the phenotype of V1, V2, V3, RNAi, and the overexpression lines were recorded (Figure 1). The opened petal color of V1, V2, and V3 matched RAL3017 (rose), RAL9016 (traffic white), and RAL1016 (sulfur yellow), respectively. Different from the edge of the V1 petal, the color of the center matched RAL3027 (raspberry red). While the unopened petals were deeper than those opened petals to some extent, the colors of petals at stage 4 matched RAL4004 (claret violet), RAL1001 (beige), and RAL1018 (zinc yellow), respectively, in V1, V2, and V3.

Figure 1. Inflorescence individuals and unopened petals phenotypes of Brassica napus with red (Zhehuhong, V1), white (Zhehubai, V2), yellow (Zheyou 50, V3) color, and the transgenic lines (ans#8 and ans#12 indicated RNAi-1 and RNAi-2 plants; ANS-OE indicated overexpression plants). Numbers on the top of unopened petals in V1 represented 4 development stages. R numbers represent colors alignment to RAL methods. Bars at the left bottom represent 1 cm and bars at the right bottom represent 0.5 cm.
Petal's color became shallow to some degree in BnaA03.ANSRNAi lines, with the color matched to RAL3012 (beige red, count 62.5% of all RNAi lines) and RAL1018 (zinc yellow, count 37.5% of all RNAi lines), while the color of unopened petals at T4 stage matched RAL3004 (purple red) and RAL1012 (lemon yellow), respectively. Overexpression of BnaA03.ANS did not alter the unopened petal color with the opened petal color match to RAL1026 (luminous yellow).
Identification and Functional Analysis of Differentially Accumulated Metabolites in V1, V2, and V3 at Stage 1 and Stage 4
The result of PCA showed that V1-1 and V1-4 separated significantly from other groups (Figures 2A,B). Under positive mode, a total of 163 and 170 co-DAMs were identified between V1 vs. V2 and V1 vs. V3 at stage 1 and stage 4 (Co-DAMs-T1 and Co-DAMs-T4), respectively. Among them, 46+2+103 metabolites were also co-differentially accumulated in stage 1 and stage 4 of V1, which were determined as Co-DAMs-red metabolites functioning on the red color formation (Figure 2C). The accumulation level of each sample group was shown as a heatmap after normalization processing (Figure 3A), and the top 3 classes were carboxylic acids and derivatives (12 DAMs), fatty acyls (11 DAMs), and benzene and substituted derivatives (8 DAMs). Furthermore, KEGG analysis was performed aiming at these 46+2+103 DAMs, and 10 KEGG pathways were enriched significantly (Figure 3C).
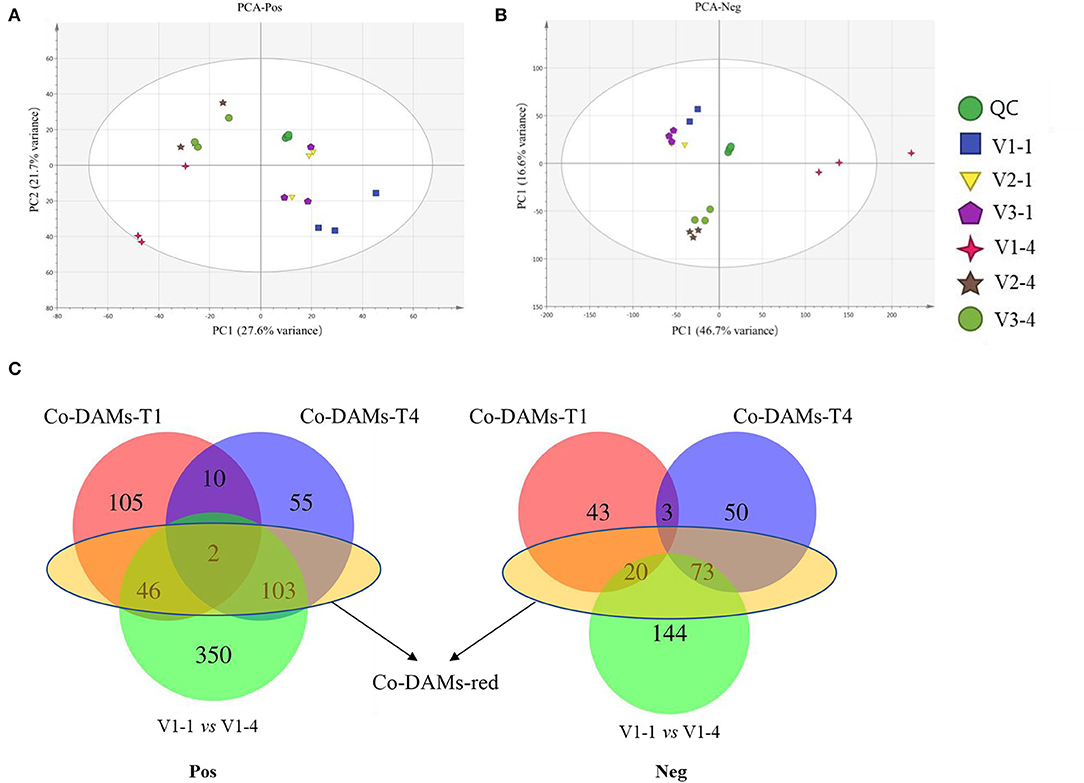
Figure 2. PCA analysis and Venn diagrams of metabolomic data. (A) PCA analysis of metabolomic data under positive mode. (B) PCA analysis of metabolomic data under negative mode. (C) Venn diagram analysis of differentially accumulated metabolites. V1-1, V2-1, and V3-4 represent the unopened petals of V1, V2, and V3 at stage 1. V1-4, V2-4, and V3-4 represent the unopened petals at stage 4. Co-DAMs-T1 represents the common differentially accumulated metabolites between V1 vs. V2 and V1 vs. V3 at stage 1. Co-DAMs-T4 represents the common differentially accumulated metabolites between V1 vs. V2 and V1 vs. V3 at stage 4. Co-DAMs-red represents the metabolites that were considered key metabolites functioning in red color. Pos represents the positive mode and Neg represents the negative mode.
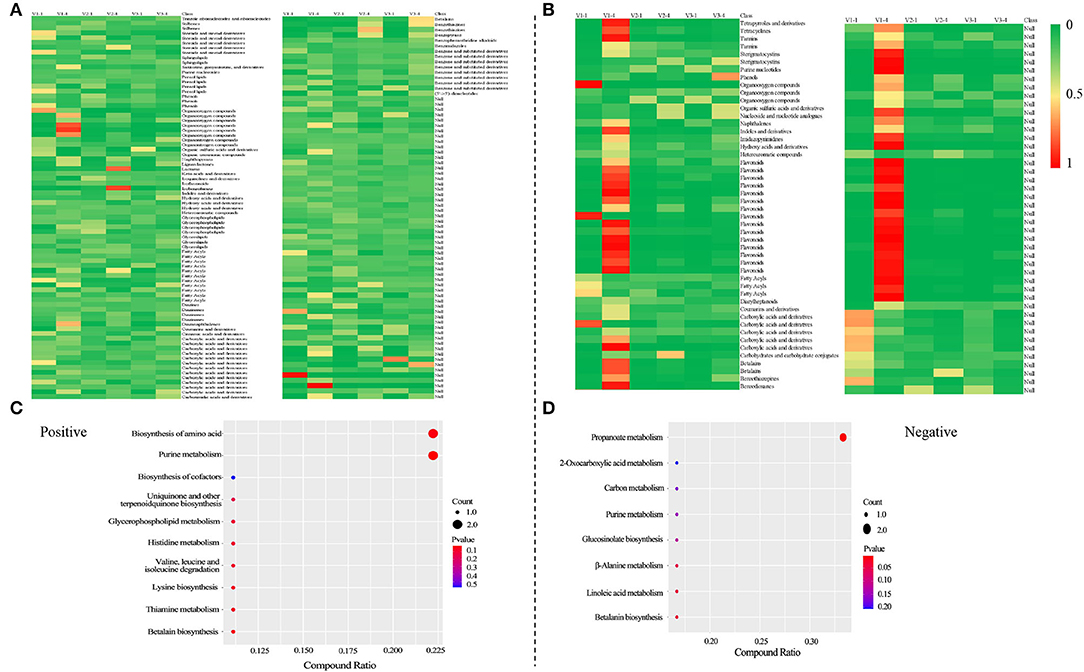
Figure 3. Heatmaps of the accumulation level of differentially accumulated metabolites after normalization and KEGG analysis of Co-DAMs-red. Heatmaps were drawn after normalization, that is take the sum of each metabolite accumulation value in all 6 samples as 1, and calculate the ratio in the sum. (A,C) were at positive mode; (B,D) were at negative mode.
Under negative mode, a total of 66 and 126 co-DAMs were identified between V1 vs. V2 and V1 vs. V3 at stage 1 and stage 4, respectively. Among them, 20+73 metabolites were also co-differentially accumulated at stage 1 and stage 4 of V1 (Figure 2C). The accumulation level of each sample group was shown as a heatmap after normalization processing (Figure 3B), with flavonoids (15 DAMs) being the most class. Furthermore, KEGG analysis was performed aiming at these 20+73 DAMs, and 8 KEGG pathways were enriched significantly (Figure 3D).
Identification and Functional Analysis of Differentially Expressed Genes in V1, V2, and V3 at Stage 1 and Stage 4
For RNA-seq analysis, raw reads, clean reads, total genes, and sequenced genes of each sample were shown in Supplementary Table 2. PCA results showed that these 6 group samples had a distinct separation except for V2 and V3 at stage 4 (Figure 4A). A total of 21 genes were selected randomly to validate the reliability of RNA-seq data by qRT-PCR analysis. Results showed that the relative coefficients of the Log10Value of the 21 genes between RNA-seq and qRT-PCR results at group V1-1 vs. V1-4, V2-1 vs. V2-4, and V3-1 vs. V3-4 were 0.7392, 0.7457, and 0.7229, which showed the high reliability of the RNA-seq data (Figures 4B–D).
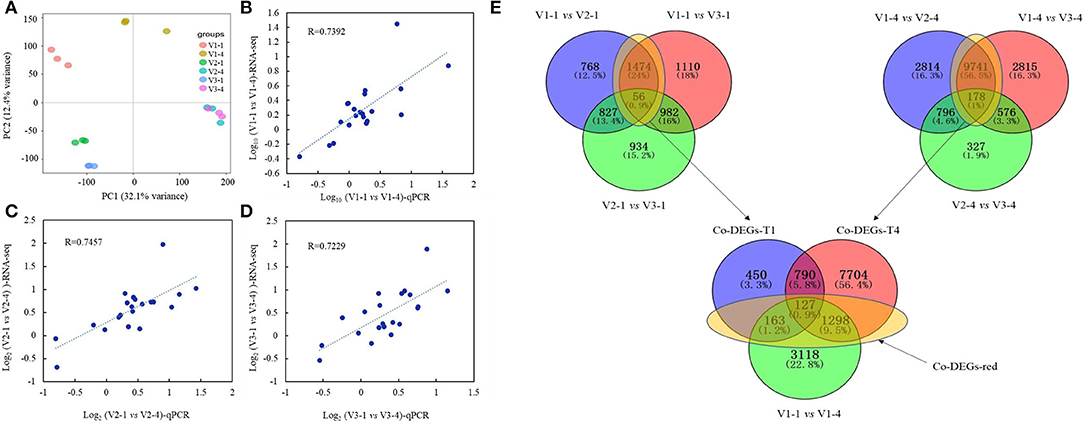
Figure 4. PCA analysis, qRT-PCR validation, and Venn diagrams of RNA-seq data. (A) PCA analysis of RNA-seq data; (B–D) represent the validation and correlation analysis of RNA-seq and qRT-PCR results of 21 selected genes. (B) V1-1 vs. V1-4; C, V2-1 vs. V2-4; D, V3-1 vs. V3-4. (E) Venn diagram of common differentially expressed genes (co-DEGs) between V1, V2, and V3 at stage 1 and stage 4. Co-DEGs-T1 represents the genes that are co-differentially expressed in V1 vs. V2 and V1 vs. V3 at stage 1. Co-DEGs-T4 represents the genes that are co-differentially expressed in V1 vs. V2 and V1 vs. V3 at stage 4. Co-DEGs-red represents the genes that were considered key genes that are functioning in red color formation.
The result of Venn diagrams showed that at stage 1, 1,530 co-DEGs (1,474+56, named Co-DEGs-T1) were identified between V1 vs. V2 and V1 vs. V3, and a total of 9,919 co-DEGs (9,471+178, named Co-DEGs-T4) were identified between V1 vs. V2 and V1 vs. V3 at stage 4. Among them, 163+127+1,298 co-DEGs were also differentially expressed between Co-DEGs-T1 and Co-DEGs-T4 with V1-1 vs. V1-4. These co-DEGs (163+127+1,298, named DEGs-red) were considered to function on the difference between red with white, red with yellow and light red with deep red petal color (Figure 4E).
Response of DEGs-Red Participating in Anthocyanin Biosynthesis Pathway, MYB and bHLH Transcription Factors
Aiming at DEGs-red, a total KEGG analysis was conducted and the results were shown in Supplementary Figure 1. Anthocyanin biosynthesis pathway was drawn and the heatmap of relative gene expression level was exhibited. Among them, ANS was significantly upregulated in five comparison groups (Figure 5). In addition, MYB, WD40, and bHLH transcription factor family are reported to be the three main transcription factors that affected petal color formation, and the heatmap of MYB and bHLH were shown in Supplementary Figure 2. What's more, qRT-PCR analysis was performed to further verify the expression level of genes relative to petal color (Supplementary Figure 3). Among them, ANS2, ANS4, PAP2, DFR1, DFR2, and UF3GT genes had significantly higher expression amounts in red petals than in white and yellow at both stage 1 and stage 4. The expression amount of CHS, CHI, F3H, F3'H, and MYB111 was significantly higher in V2 at stage 1 and was higher in V2 at stage 4 for PAP2 and MBY61 while it was higher in V3 at stage 1 for MYB5 and MYC1.
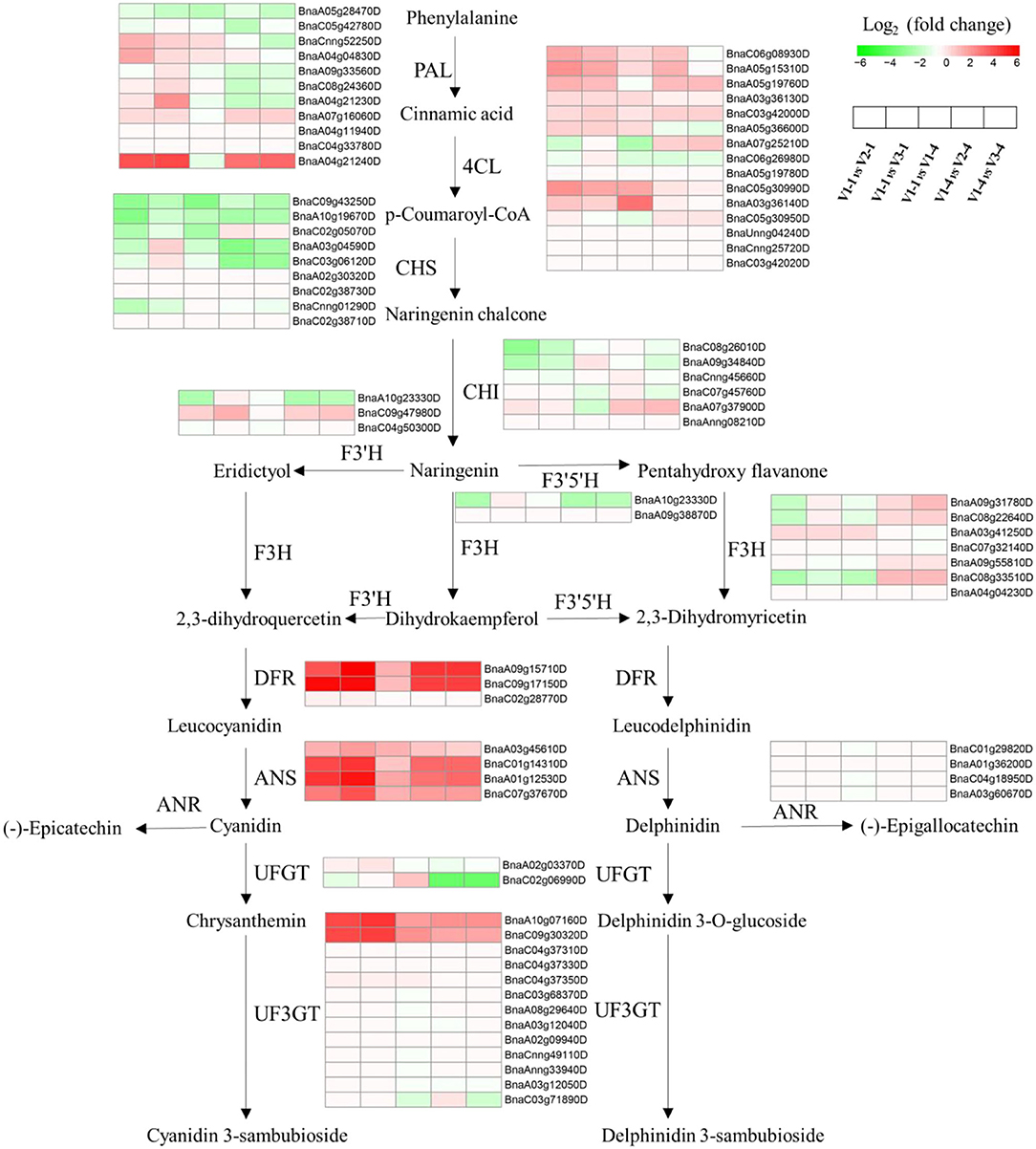
Figure 5. Overview of the transcriptional regulation of anthocyanin biosynthetic genes. Heatmaps represent the log2-fold change value of V1-1 vs. V2-1, V1-1 vs. V3-1, V1-1 vs. V1-4, V1-3 vs. V2-4, and V1-4 vs. V3-4, respectively. PAL, phenylalanine ammonia lyase; 4CL, 4-coumarate co ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3'H, flavonoid 3'-hydroxylase; F3H, flavanone 3-hydroxylase; F3'5'H, flavonoid-3'5'-hydroxylase; DFR, dihydroflavonol reductase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase; UFGT, uridine diphosphate glucose-flavonoid glucosyltransferase; UF3GT, UDP: flavonoid 3-O-glucosyltransferase.
Influence of RNAi and Overexpression of BnaA03.ANS on Petal Colors and Pigment Contents
Semiquantitative RT-PCR was also conducted to identify the expression level of 4 copies of ANS genes in V1, V2, and V3. The results showed that BnaA03.ANS appeared evident difference in all lines, with the brighter band in V1 (Figure 6A). Hence, BnaA03.ANS was chosen to be the candidate gene for RNAi and overexpression validation.
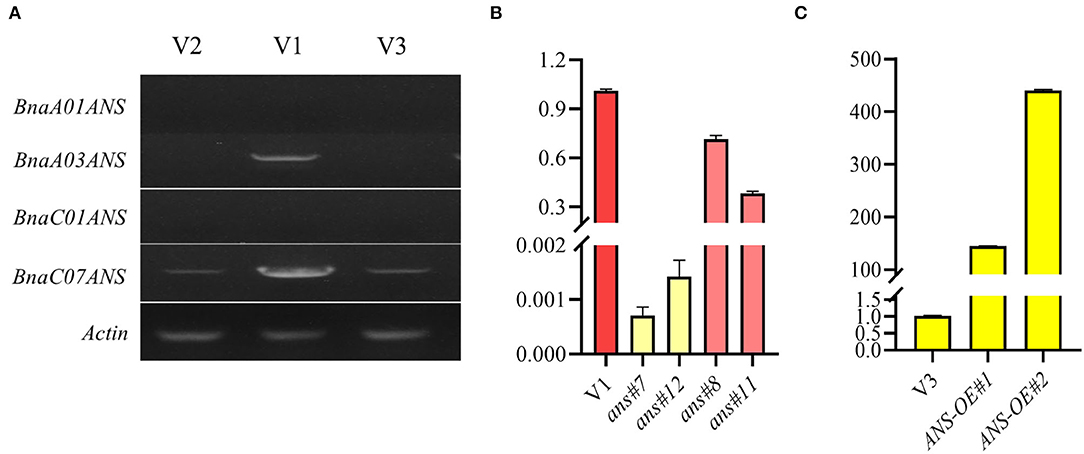
Figure 6. Identification of the expression level of 4 ANS copies, and the BnaA03.ANS expression validation in transgenic lines. (A) Identification of the expression level of 4 ANS copies by semi-RT-PCR; (B) qRT-PCR validation of BnaA03.ANS expression level in RNAi lines compared with the wild type (V1). (C) qRT-PCR validation of BnaA03.ANS expression level in overexpression lines compared with the wild type (V3).
RNA interference of BnaA03.ANS showed that, opened petal colors of V1 turned from rose and raspberry red to zinc yellow, with the expression level of BnaA03.ANS decreased by 700-fold −1,400-fold, and turn to beige red when the expression level of BnaA03.ANS was only 71.5% or 28.25% of V3 (Figure 6B).
Analysis of anthocyanin and carotenoid profiles indicated that pelargonidin and cyanidin were largely decreased by 97.0% and 37.5% in ans#7, and 97.8% and 62.1% in ans#8 compared with V1, respectively. As for carotenoids, lutein, neoxanthin, and β-carotene were also significantly decreased, with 82.5%, 76.2%, and 80.1% and 68.6%, 12.4%, and 60% lower in ans#7 and ans#8 than V1, respectively. It is interesting to note that no lycopene contents were detected when the expression of BnaA03.ANS was interferenced (Figure 7).
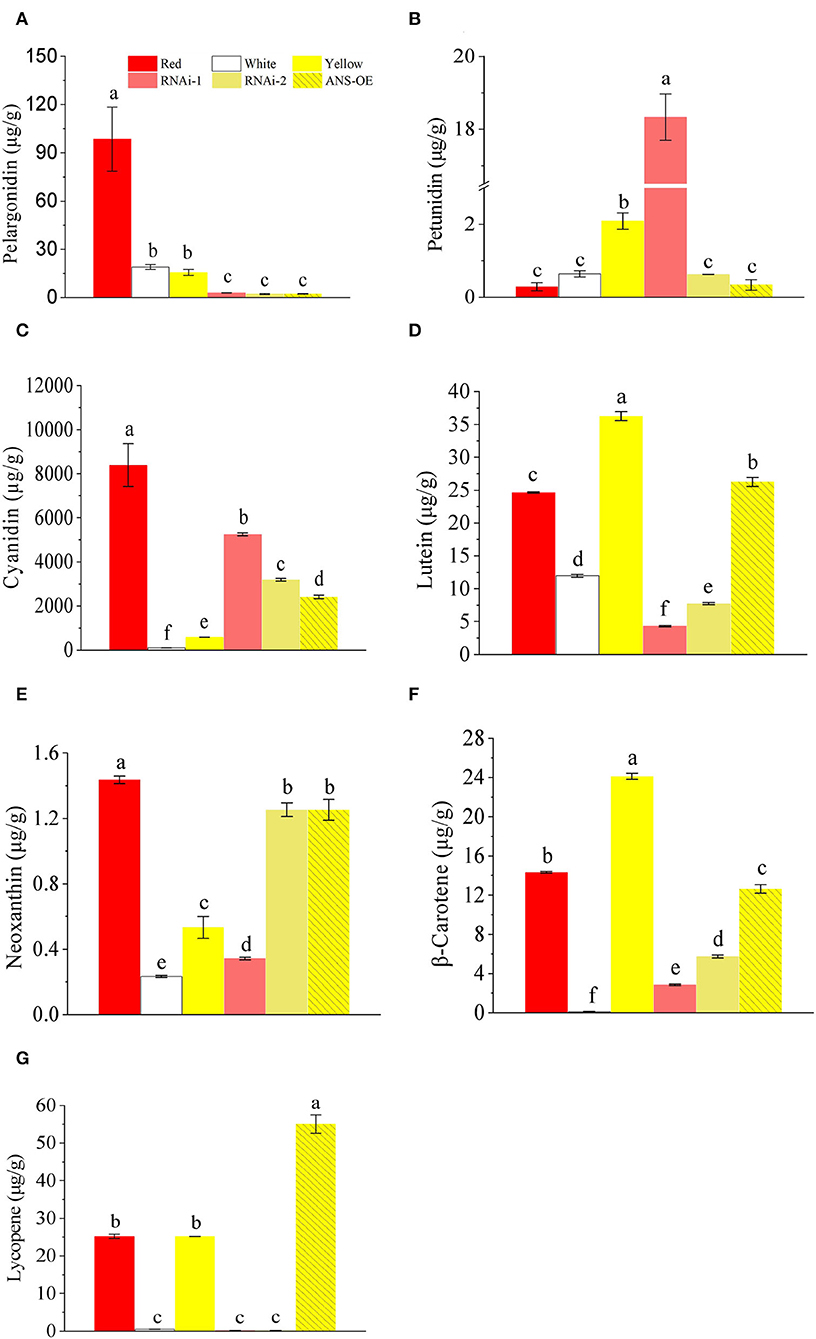
Figure 7. Comparison of anthocyanins and carotenoids contents in petals of red (V1), white (V2), yellow (V3), RNAi lines, and overexpression lines. (A) pelargonidin; (B) petunidin; (C) cyanidin; (D) lutein; (E) neoxanthin; (F) β-carotene; (G) lycopene. Letters above the columns represent a significant difference, p ≤ 0.05. Error bars represent SD values.
The full-length cDNA fragment of BnaA03.ANS was also overexpressed under the control of petal-specific promoter XY355 (Fu et al., 2018), but no obvious color change appeared with BnaA03.ANS was overexpressed by 144-fold or 440-fold (Figure 6C). Carotenoids and anthocyanin profiles showed that cyanidin (3.15-fold), neoxanthin (1.35-fold), and lycopene (1.18-fold) were high accumulated, while, pelargonidin (-85.7%), petunidin (-83.8%), lutein (-27.6%), and β-carotene (-47.7%) were lower in ANS-OE than in V3. However, the petal colors did not obviously differ from yellow (Figure 7).
Discussion
Colorful flowers are one of the most important traits of many ornamental flowering plants and have gained more and more attention from breeders (Nishihara and Nakatsuka, 2011). Flower colors in rapeseed range from white, orange to yellow, and other rare colors such as pink, purple, and red also have been reported recently, which greatly enhanced its ornamental value (Fu et al., 2018). However, the underlying mechanisms of how genes and metabolites regulate color formation in red petals have not been elucidated thoroughly (Yin et al., 2019). In this study, the DAMs and DEGs in unopened small petals (stage 1) and unopened large petals (stage 4) of red color rapeseed (V1) compared with those in white (V2) and yellow (V3) were identified, aiming at understanding the molecular mechanism on the regulation of red petal color formation in rapeseed.
Metabolomic Analysis Revealed That Flavonoids May Contribute to Red Color Formation
The result of the principal component analysis of metabolomic research showed that two groups of red petals in rapeseed (V1-1 and V1-4) exhibited obvious separation from white and yellow ones (Figures 2A,B), which reflected that accumulation levels of metabolites in red petals have a huge difference in compared with white and yellow petals. Further analysis revealed that a total of 46+2+103 DAMs in positive mode and 20+73 DAMs in negative mode (Figure 2) were co-differentially accumulated in Co-DAMs-T1 (co-DAMs in V1 vs. V2 and V1 vs. V3 at stage 1), Co-DAMs-T4 (co-DAMs in V1 vs. V2 and V1 vs. V3 at stage 4), and group V1-1 vs. V1-4 (co-DAMs in V1 at stage 1 vs. V1 in stage 4), and these DAMs were considered as key metabolites that influence red petal formation and the differentiation from white and yellow petals. Among them, most DAMs were belonging to flavonoids (Figures 3A,B). Flavonoids, as the most important pigment, generate the widest spectrum of colors. Chen et al. (2012) reported that the flavonoid composition of white flower chrysanthemum only contained flavonols and flavones, while anthocyanins were detected in pink flowers. He et al. (2011) found that anthocyanins were presented in red and orange samples of lycoris longituba while no anthocyanins were detected within white and yellow. In this study, the heatmap of DAMs accumulation levels revealed that the accumulation level in red petals (V1-1 and V1-4) was higher than in white and yellow ones, with flavonoids being the most categories, which indicated the importance of flavonoids in red pigment formation compared with white and yellow.
Identification of RNA-seq Data Showed Anthocyanin Biosynthesis Pathway and MYB, bHLH Transcription Factors Were Largely Differentiated in V1, V2, and V3
Venn diagrams of RNA-seq revealed that a total of 1,588 (163+127+1,298) genes (Figure 4) were co-differentially expressed in group Co-DEGs-T1 (co-DEGs in V1 vs. V2 and V1 vs. V3 at stage 1), group Co-DEGs-T4 (co-DEGs in V1 vs. V2 and V1 vs. V3 at stage 4), and group V1-1 vs. V1-4 (co-DEGs in V1 at stage 1 vs. V1 at stage 4), and these co-DEGs were considered as key genes that influence the red petal color formation and the differentiation from white and yellow petal colors. Anthocyanins are formed by various sugars and anthocyanidins, and the diversity of colors is highly related to anthocyanin composition and content according to a previous study (Li et al., 2003). Genes involved in the anthocyanin biosynthesis pathway that function on petal colors have been well-reported, such as CHS in Malus crabapple (Sun et al., 2015), CHI in asters and cyclamen (Nishihara et al., 2005), F3H in carnation (Owens et al., 2008), DFR in Saussurea (Li et al., 2012), and ANS in lisianthus flowers (Shimizu et al., 2011), which demonstrated the importance of anthocyanin biosynthesis pathway on petal color formation. As a report, red color formation, as the result of anthocyanin accumulation, is controlled through the coordination of genes that encode the enzymes involved in the anthocyanin pathway (Lai et al., 2020). For example, in radish of Cruciferae, genes for the anthocyanin biosynthesis, namely, PAL, C4H, 4CL, CHS, CHI, F3H, DFR, and ANS, were identified by traditional manners and second-generation sequencing (Muleke et al., 2017; Sun et al., 2018). Among them, RsF3H, RsF3'H1, RsCHS3, RsANS, and particularly RsUFGT, were highly correlated with the anthocyanin contents in the flesh of red radish (Muleke et al., 2017). However, when both the skin and flesh were factored into account, only the expression of RsDFR and RsANS were correlated with the anthocyanin contents (Park et al., 2011). These results indicated that late structural genes involved in the flavonoid pathway are specifically involved in anthocyanin biosynthesis. According to our research, flavonoids were also highly accumulated from metabolomics data, and the genes and pathway involved in anthocyanin biosynthesis were drawn against these Co-DEGs-red (163+127+1,298). According to Figure 5, genes showed a diverse difference between red with white and yellow, while ANS, DFR, and UF3GT were greatly highly expressed in red petals than in white and yellow, and these results were further validated by qRT-PCR (Supplementary Figure 2).
In addition to these structural genes, transcription factors also play a role in petal color formation. MYB, bHLH, and WD40 families are reported to be the three major types of transcription factors that are involved in petal coloration and also regulate anthocyanin synthesis directly or indirectly (Zhang et al., 2003; Ramsay and Glover, 2005), for instance, the bleaching of gentian due to mutation of GtMYB3 (Nakatsuka et al., 2008), and also the positive regulation function of GhMYB10 and LhMYB6 on anthocyanin accumulation in gerbera and lily (Roosa et al., 2008; Yamagishi et al., 2010). Overexpression of an R2R3-MYB transcription factor, RsMYB1, resulted in the increasing production of red flowers in radish, and by ectopic overexpression of the OvPAP2 (Orychophragmus violaceus) gene, Fu et al. (2018) successfully produced red anthers and petals in yellow rapeseed (Lim et al., 2016; Fu et al., 2018). As for bHLH, in Arabidopsis, three bHLH transcription factors, GL3, TT8, and EGL3 were reported to participate in the biosynthesis of flavonoids (Nesi et al., 2000; Baudry et al., 2006). A recent report showed that the coexpression of RSMYB1 and RsTT8 in tobacco leaves remarkably increased the accumulation of anthocyanins, indicating the partnership of RsTT8 and RsMYB1 in anthocyanin biosynthesis (Lim et al., 2017). To unveil the mechanisms of red color formation, the two-most important regulators involved in anthocyanin biosynthesis were isolated, and the expression module of MYB and bHLH were drawn in Supplementary Figure 2, which showed huge differences in red petals than in white and yellow at stage 1 or stage 2, reflecting the importance of MYB and bHLH on red color formation in rapeseed.
RNA Interference of BnaA03.ANS Transferred Petal Color From Raspberry Red to Beige Red or Zinc Yellow, While no Obvious Petal Color Was Changed When BnaA03.ANS Was Overexpressed
According to metabolomics and RNA-seq data, aiming at flavonoids, we selected the 4 copies of ANS genes that played a key role in the anthocyanin biosynthesis pathway, and semi-RT-PCR was performed. The results showed that band of BnaA03.ANS in red petals exhibited an obvious difference from yellow and white petals compared with the other 3 copies, which indicates the importance of BnaA03.ANS. Disruption of BnaA03.ANS in red rapeseed led to converting the petal color from raspberry red to beige red when 28.5% or 71.7% of the expression level of BnaA03.ANS was suppressed, while the color changed to zinc yellow, when 99.93% or 99.86% of the expression level was suppressed. The result of anthocyanins and carotenoids contents analysis revealed that pelargonidin, cyanidin, lutein, neoxanthin, β-carotene, and lycopene contents were significantly inhibited (Figure 7), which indicated the importance of BnaA03.ANS on red petal formation. However, with the overexpression of BnaA03.ANS under XY355 promoter in yellow rapeseed, no change of petal color was detected, though the pigments showed a different change with cyanidin, neoxanthin, and lycopene content highly accumulated while pelargonidin, petunidin, lutein, and β-carotene were decreased in overexpression lines than control (yellow rapeseed). According to a previous report, even flowers of the same color exhibited distinct pigment profiles depending on the species and variety, for example, in yellow tomato flowers, neoxanthin and violaxanthin comprise the two major carotenoids, whereas 9-cis-violaxanthin is the predominant component in yellow rose flowers (Ariizumi et al., 2014; Wan et al., 2019; Liu et al., 2020). Therefore, further researches still need to be investigated.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA848086.
Author Contributions
SH, HL, and LJ designed the experiment. PH, HL, BL, YR, and LH performed the experiment. PH and SH wrote the manuscript. All authors read and agreed to the final manuscript.
Funding
This work was supported by the earmarked fund for the China Agriculture Research System (CARS-12), the Zhejiang Natural Science Foundation (Y21C13014), the National Natural Science Foundation (32130076), the Zhejiang Science and Technology Major Program on Agricultural New Variety Breeding (2021C02064), and the Zhejiang Key Laboratory of Digital Dry Land Crops (2022E10012).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.940765/full#supplementary-material
Supplementary Figure 1. KEGG analysis of co-DEGs relative to red pigment formation.
Supplementary Figure 2. Heatmap of bHLH and MYB transcription factors among co-DEGs relative to red pigment formation.
Supplementary Figure 3. qRT-PCR identification of genes relative to anthocyanin biosynthesis.
Supplementary Table 1. Primers used in this research.
Supplementary Table 2. Basic results of RNA-seq data.
References
Ariizumi, T., Kishimoto, S., Kakami, R., Maoka, T., Hirakawa, H., Suzuki, Y., et al. (2014). Identification of the carotenoid modifying gene PALE YELLOW PETAL 1 as an essential factor in xanthophyll esterification and yellow flower pigmentation in tomato (Solanum lycopersicum). Plant J. 79, 453–465. doi: 10.1111/tpj.12570
Baudry, A., Caboche, M., and Lepiniec, L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46, 768–779. doi: 10.1111/j.1365-313X.2006.02733.x
Cao, H., Zhang, J., Xu, J., Ye, J., Yun, Z., Xu, Q., et al. (2012). Comprehending crystalline b-carotene accumulation by comparing engineered cell models and the natural carotenoid-rich system of citrus. J. Exp. Bot. 63, 4403–4417. doi: 10.1093/jxb/ers115
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, S. M., Li, C. H., Zhu, X. R., Deng, Y. M., Sun, W., and Wang, L. S. (2012). The identification of flavonoids and the expression of genes of anthocyanin biosynthesis in the chrysanthemum flowers. Biol. Plant. 56, 458–464. doi: 10.1007/s10535-012-0069-3
Cheynier, V., Comte, G., Davies, K. M., Lattanzio, V., and Martens, S. (2013). Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Bioch. 72, 1–20. doi: 10.1016/j.plaphy.2013.05.009
Fu, D. H., Jiang, L. Y., Masons, A. S., Xiao, M. L., Zhu, L. R., Li, L. Z., et al. (2016). Research progress and strategies for multifunctional rapeseed: a case study of China. J. Integ. Agric. 15, 1673–1684. doi: 10.1016/S2095-3119(16)61384-9
Fu, W. Q., Chen, D. Z., Pan, Q., Li, F. F., Zhao, Z. G., Ge, X. H., et al. (2018). Production of red-flowered oilseed rape via the ectopic expression of Orychophragmus violaceus OvPAP2. Plant Biotech. J. 16, 367–380. doi: 10.1111/pbi.12777
He, Q., Shen, Y., Wang, M., Huang, M., Yang, R., and Zhu, S. (2011). Natural variation in petal color in Lycoris longituba revealed by anthocyanin components. PLoS ONE 6, 22098. doi: 10.1371/journal.pone.0022098
Heller, W., Forkmann, G., Britsch, L., and Grisebach, H. (1985). Enzymatic reduction of (+)-dihydroflavonols to flavan-3 4-cis-diols with flower extracts from Matthiola incana and its role in anthocyanin biosynthesis. Planta 165, 284–287. doi: 10.1007/BF00395052
Kazuma, K., Noda, N., and Suzuki, M. (2003). Flavonoid composition related to petal color in different lines of Clitoria ternatea. Phytochemistry 64, 1133–1139. doi: 10.1016/S0031-9422(03)00504-1
Lai, B., Cheng, Y. Y., Liu, H., Wang, Q. X., Wang, Q., Wang, C. L., et al. (2020). Differential anthocyanin accumulation in radish taproot: importance of RsMYB1 gene structure. Plant Cell Rep. 39, 217–226. doi: 10.1007/s00299-019-02485-z
Li, H. H., Qiu, J., Chen, F. D., Lv, X. F., Fu, C. X., Zhao, D., et al. (2012). Molecular characterization and expression analysis of dihydroflavonol 4-reductase (DFR) gene in Saussurea medusa. Mol. Biol. Rep. 39, 2991–2999. doi: 10.1007/s11033-011-1061-2
Li, M. R., Chen, J. T., Sun, Z. J., Chen, Y. Z., and Li, H. Q. (2003). Advances in molecular breeding of ornamental plants. J. Trop. Subtrop. Bot. 11, 87–92. doi: 10.3969/j.issn.1005-3395.2003.01.017
Lim, S., Kim, D., Kim, J. K., Lee, J., and Ha, S. (2017). A radish basic helix-loop-helix transcription factor, RsTT8 acts a positive regulator for anthocyanin biosynthesis. Front. Plant Sci. 8, 1917. doi: 10.3389/fpls.2017.01917
Lim, S., Song, J., Kim, D., Kim, J. K., Lee, J., Kim, Y., et al. (2016). Activation of anthocyanin biosynthesis by expression of the radish R2R3-MYB transcription factor gene RsMYB1. Plant Cell Rep. 35, 641–653. doi: 10.1007/s00299-015-1909-3
Liu, Y. J., Ye, S. H., Yuan, G. G., Ma, X. W., Heng, S. P., Yi, B., et al. (2020). Gene silencing of BnaA09.ZEP and BnaC09.ZEP confers orange color in Brassica napus flowers. Plant J. 104, 932–949. doi: 10.1111/tpj.14970
Long, C. C., Flint, J. A., and Lepper, P. A. (2011). Insect attraction to wind turbines: does colour play a role? Eur. J. Wildlife Res. 57, 323–331. doi: 10.1007/s10344-010-0432-7
Morris, W., Ducreux, L., Griffiths, D., Stewart, D., Davies, H., and Taylor, M. (2004). Carotenogenesis during tuber development and storage in potato. J. Exp. Bot. 55, 975–982. doi: 10.1093/jxb/erh121
Muleke, E. M., Fan, L., Wang, Y., Xu, L., Zhu, X., Zhang, W., et al. (2017). Coordinated regulation of anthocyanin biosynthesis genes confers varied phenotypic and spatial-temporal anthocyanin accumulation in radish (Raphanus sativus L.). Front Plant Sci. 8, 1243. doi: 10.3389/fpls.2017.01243
Nakatsuka, A., Izumi, Y., and Yamagishi, M. (2003). Spatial and temporal expression of chalcone synthase and dihydroflavonol 4-reductase genes in the Asiatic hybrid lily. Plant Sci. 166, 759–767. doi: 10.1016/S0168-9452(03)00254-1
Nakatsuka, T., Haruta, K. S., Pitaksutheepong, C., Abe, Y., Kakizaki, Y., and Yamamoto, K. (2008). Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Physiol. 49, 1818–1829. doi: 10.1093/pcp/pcn163
Nakatsuka, T., Nishihara, M., Mishiba, K., and Yamamura, S. (2005). Temporal expression of flavonoid biosynthesis related genes regulates flower pigmentation in gentian plants. Plant Sci. 168, 1309–1318. doi: 10.1016/j.plantsci.2005.01.009
Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. doi: 10.1105/tpc.12.10.1863
Nikolov, L. A.. (2019). Brassicaceae flowers: diversity amid uniformity. J. Exp. Bot. 70, 2623–2635. doi: 10.1093/jxb/erz079
Nishihara, M., and Nakatsuka, T. (2011). Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 33, 433–441. doi: 10.1007/s10529-010-0461-z
Nishihara, M., Nakatsuka, T., and Yamamura, S. (2005). Flavonoid components and flower color change in transgenic tobacco plants by suppression of chalcone isomerase gene. FEBS Lett. 579, 6074–6078. doi: 10.1016/j.febslet.2005.09.073
Owens, D. K., Crosby, K. C., Runac, J., Howard, B. A., and Winkel, B. S. (2008). Biochemical and genetic characterization of Arabidopsis flavanone 3β-hydroxylase. Plant Physiol. Biochem. 46, 833–843. doi: 10.1016/j.plaphy.2008.06.004
Park, N. I., Xu, H., Li, X., Jang, I. H., Park, S., Ahn, G. H., et al. (2011). Anthocyanin accumulation and expression of anthocyanin biosynthetic genes in radish (Raphanus sativus). J. Agric. Food Chem. 59, 6034–6039. doi: 10.1021/jf200824c
Ramsay, N. A., and Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Roosa, A. E., Laitinen, M., Ainasoja, M., Teeri, T. H., and Elomaa, P. (2008). Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida. J. Exp. Bot. 59, 3691–3703. doi: 10.1093/jxb/ern216
Rosati, C., Cadic, A., Duron, M., Ingouff, M., and Simoneaub, P. (1999). Molecular characterization of the anthocyanidin synthase gene in Forsythia x intermedia reveals organ-specific expression during flower development. Plant Sci. 149, 73–79. doi: 10.1016/S0168-9452(99)00146-6
Sagawa, J. M., Stanley, L. E., LaFountain, A. M., Frank, H. A., Liu, C., and Yuan, Y. W. (2016). An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol. 209, 1049–1057. doi: 10.1111/nph.13647
Shimizu, K., Ohnishi, N., Morikawa, N., Ishigami, A., Otake, S., Rabah, I. O., et al. (2011). A 94-bp deletion of anthocyanidin synthase gene in acyanic flower lines of lisianthus [Eustoma grandiflorum (Raf.) Shinn.]. J. Japan Soc. Hortic. Sci. 80, 434–442. doi: 10.2503/jjshs1.80.434
Sun, L. L., Gao, W., Zhang, M. M., Li, C., Wang, A. G., Su, Y. L., et al. (2014). Composition and antioxidant activity of the anthocyanins of the fruit of Berberis heteropoda Schrenk. Molecules 19, 19078–19096. doi: 10.3390/molecules191119078
Sun, W., Li, C., Wang, L., Dai, S., and Xu, Y. (2009). Anthocyanins present in flowers of Senecio cruentus with different colors. Acta Hortic. Sin. 36, 1775–1782.
Sun, W., Meng, X., Liang, L., Jiang, W., Huang, Y., He, J., et al. (2015). Molecular and biochemical analysis of chalcone synthase from Freesia hybrid in flavonoid biosynthetic pathway. PLoS ONE 10, e0119054. doi: 10.1371/journal.pone.0119054
Sun, Y., Wang, J., Qiu, Y., Liu, T., Song, J., and Li, X. (2018). Identification of ‘Xinlimei' radish candidate genes associated with anthocyanin biosynthesis based on a transcriptome analysis. Gene 657, 81–91. doi: 10.1016/j.gene.2018.03.001
Tai, D., Tian, J., Zhang, J., Song, T., and Yao, Y. (2014). A Malus crabapple chalcone synthase gene, McCHS, regulates red petal color and flavonoid biosynthesis. PLoS ONE 9, e110570. doi: 10.1371/journal.pone.0110570
Tanaka, Y., Sasaki, N., and Ohmiya, A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54, 733–749. doi: 10.1111/j.1365-313X.2008.03447.x
Veitch, N. C., and Grayer, R. J. (2008). Flavonoids and their glycosides including anthocyanins. Nat. Prod. Rep. 25, 555–611. doi: 10.1039/b718040n
Vignolini, S., Moyroud, E., Hingant, T., Banks, H., Rudall, P. J., Steiner, U., et al. (2015). The flower of Hibiscus trionum is both visibly and measurably iridescent. New Phytol. 205, 97–101. doi: 10.1111/nph.12958
Wan, H., Yu, C., Han, Y., Guo, X., Luo, L., Pan, H., et al. (2019). Determination of flavonoids and carotenoids and their contributions to various colors of rose cultivars (Rosa spp.). Front. Plant Sci. 10, 123. doi: 10.3389/fpls.2019.00123
Wellmann, F., Griesser, M., Schwab, W., Martens, S., Eisenreich, W., Matern, U., et al. (2006). Anthocyanidin synthase from Gerbera hybrida catalyzes the conversion of (+)-catechin to cyanidin and a novel procyanidin. FEBS Lett. 580, 1642–1648. doi: 10.1016/j.febslet.2006.02.004
Xu, P., Cao, S., Hu, K., Wang, X., Huang, W., Wang, G., et al. (2017). Trilocular phenotype in Brassica juncea L. resulted from interruption of CLAVATA1 gene homologue (BjMc1) transcription. Sci. Rep. 7, 3498. doi: 10.1038/s41598-017-03755-0
Yamagishi, M., Shimoyamada, Y., Nakatsuka, T., and Masuda, K. (2010). Two R2R3-MYB genes homologs of petunia AN2 regulate anthocyanin biosynthesis in flower tepals tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol. 51, 463–474. doi: 10.1093/pcp/pcq011
Yin, N. W., Wang, S. X., Jia, L. D., Zhu, M. C., Yang, J., Zhou, B. J., et al. (2019). Identification and characterization of major constituents in different-colored rapeseed petals by UPLC-HESI-MS/MS. J. Agric. Food Chem. 67, 11053–11065 doi: 10.1021/acs.jafc.9b05046
Zhang, B., Liu, C., Wang, Y. Q., Yao, X., Wang, F., Wu, J. S., et al. (2015). Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol. 206, 1513–1526. doi: 10.1111/nph.13335
Zhang, F., Gonzalez, A., Zhao, M. Z., Payne, C. T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. doi: 10.1242/dev.00681
Zhang, J., Wang, L., Gao, J., Li, S., Xu, Y., Li, C., et al. (2011). Identification of anthocyanins involving in petal coloration in Chaenomeles speciosa cultivars. Acta Hortic. Sin. 38, 527–534.
Zhao, D. Q., and Tao, J. (2015). Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 6, 261. doi: 10.3389/fpls.2015.00261
Zhao, D. Q., Tao, J., Han, C. X., and Ge, J. T. (2012). Flower color diversity revealed by differential expression of flavonoid biosynthetic genes and flavonoid accumulation in herbaceous peony (Paeonia lactiflora Pall.). Mol. Biol. Rep. 39, 11263–11275. doi: 10.1007/s11033-012-2036-7
Keywords: carotenoids, metabolomics, overexpression, RNA interference, RNA-seq
Citation: Hao P, Liu H, Lin B, Ren Y, Huang L, Jiang L and Hua S (2022) BnaA03.ANS Identified by Metabolomics and RNA-seq Partly Played Irreplaceable Role in Pigmentation of Red Rapeseed (Brassica napus) Petal. Front. Plant Sci. 13:940765. doi: 10.3389/fpls.2022.940765
Received: 10 May 2022; Accepted: 17 June 2022;
Published: 14 July 2022.
Edited by:
Sarvajeet Singh Gill, Maharshi Dayanand University, IndiaReviewed by:
Zhansheng Li, Insititute of Vegetables and Flowers (CAAS), ChinaCunmin Qu, Southwest University, China
Copyright © 2022 Hao, Liu, Lin, Ren, Huang, Jiang and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuijin Hua, c2podWExQDE2My5jb20=
†These authors have contributed equally to this work
 Pengfei Hao
Pengfei Hao Han Liu1,2†
Han Liu1,2† Baogang Lin
Baogang Lin Shuijin Hua
Shuijin Hua