- 1School of Life Sciences, Guizhou Normal University, Guiyang, China
- 2Key Laboratory of Plant Physiology and Development Regulation, Guizhou Normal University, Guiyang, China
- 3Key Laboratory of National Forestry and Grassland Administration on Biodiversity Conservation in Karst Mountainous Areas of Southwestern China, Guizhou Normal University, Guiyang, China
- 4Sichuan Provincial Academy of Natural Resource Science, Chengdu, China
Fruit prickles are widely distributed on the pericarp and exhibit polymorphic traits at different developmental stages. Although they are multicellular appendages that are well-known for helping plants defend against biotic and abiotic stresses, their origination and molecular mechanism are still less known. Here, we studied the origination and molecular mechanism of fruit prickles in Rosa roxburghii. Using morphological and histological observations, we found that the fruit prickle primordium of R. roxburghii originated from the ground meristem that underwent cell division to form flagelliform prickles, continued to enlarge, and finally lignified to form mature fruit prickles. We amplified a homolog of candidate gene TRANSPARENT TESTA GLABRA1 (TTG1) from R. roxburghii, named RrTTG1. RrTTG1 harbored four conserved WD-repeat domains and was exclusively nuclear-localized. Using qRT-PCR and in situ hybridization, we found that RrTTG1 was constitutively expressed and highly expressed during the initiation and cell expansion phases of fruit prickles. Ectopic expression analysis in Arabidopsis proved that RrTTG1 substantially enhanced the number of trichome and pigmentation production and inhibited root hair formation. Besides, RrTTG1 complemented the phenotypes of the ttg1 mutant in Arabidopsis, thus indicating that RrTTG1 played pleiotropic roles akin to AtTTG1. We demonstrated that the RrTTG1 only interacted with RrEGL3, a homolog of ENHANCER OF GLABRA3 (EGL3), via yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays. Briefly, RrTTG1 might positively regulate the initiation of fruit prickle primordium and cell enlargement by forming the RrTTG1-RrEGL3-RrGL1 complex in R. roxburghii. Therefore, our results help characterize the RrTTG1 in R. roxburghii and also elucidate the establishment of the prickles regulatory system in the Rosaceae plants.
Introduction
Prickle is a sharply pointed emergence originating from multiple cellular divisions from the epidermis or cortex, which lack vasculature. It is a typical structure involved in thermoregulation and defense against multiple biotic and abiotic stresses (Kellogg et al., 2011). Rosa roxburghii Tratt, a Chinese perennial Rosaceae rosebush, is well-known for its high nutritional and medicinal values, but produces prickles on the surface of stem, sepals, petioles, and fruits (Wang D. J. et al., 2019; Yan et al., 2021). Its fruit prickles exhibit multiple morphological structures, including flagelliform, triangular, conical, and acicular glandular shapes at various stages (Wang D. J. et al., 2019; Li et al., 2022). However, the fruit prickles are considered an undesirable trait that damages the gardening management and injures workers. The fruit prickles of R. roxburghii were considered trichomes are large cells and oval in shape with large vacuoles and thin cell walls with large intercellular spaces containing abundant starch grains and plastids (Wang et al., 2021). Although the internal structure of fruit prickles has been available, precise histological observation is required for clearly deciphering the initiation and development of fruit prickles.
A fruit prickle is considered an extension or a lignified modification of the glandular trichomes, which are accompanied by deposition of phenolic compounds and development into sharp and hard appendages, thus resulting in non-glandular prickles at the late stage (Kellogg et al., 2011). Trichomes are unicellular or multicellular appendages that originate only from the protoderm of aerial epidermis cells and are diverse according to the location, morphology, and secretion of the individual plant (Serna and Martin, 2006; Wester et al., 2009). The underlying genetic and molecular mechanisms of trichome development are well understood in the model Arabidopsis thaliana. Furthermore, numerous genes have been previously identified through glabrous mutants and gene cloning (Walker et al., 1999; Payne et al., 2000; Zhang et al., 2003). TTG1 contains WD-40 motifs that interact with the bHLH transcription factor GLABRA3 (GL3) or ENHANCER OF GLABRA3 (EGL3) (Humphries et al., 2005). Subsequently, GL3/EGL3 interacts with the MYB transcription factor GLABRA1 (GL1) to form an MYB-bHLH-WD40 (MBW) chimera complex, which activates the downstream target gene GLABRA2 (GL2). The homeodomain-leucine zipper protein GL2 finally induces trichome initiation and growth (Balkunde et al., 2011). In addition, the TTG1 protein directly binds to various bHLH proteins, which further interact with a larger of multiple MYB proteins to form the TTG1-bHLHs-MYBs complex that plays different pleiotropic roles, such as root hair formation, flavonoid biosynthesis pathway, seed pigmentation, and flowering time (Zhang et al., 2003; Chen et al., 2015). The ttg1 mutant produced aborted trichomes and had a defective anthocyanin phenotype due to the failure of the MBW complex formation (Zhang et al., 2003). The single amino acid substitutions in the TTG1 protein could generate different phenotypes, including proanthocyanin accumulation, trichome, and root hair production in Arabidopsis. Besides, the amino acid changes in TTG1 altered the interaction of TTG1 with GL3 (Long and Schiefelbein, 2020).
Extensive studies have been performed mainly on stem prickles in the Rosaceae family for garden plants or cut flower production. Based on the ultrastructural observation of stem prickles of rose (Rosa hybrida), blackberry (Rubus rubus), and raspberry (Rubus idaeus), the well-accepted mainstream theory is that stem prickle is an outgrowth of the epidermis formed by multiple cellular divisions and it lacks vasculature thereby being biologically similarity to the trichome (Kellogg et al., 2011). Recently, using morphological studies of rose, stem prickles have been proposed to originate from multiple cells of the ground meristem beneath the protoderm that differs from the modified trichomes derived from the protoderm (Zhou et al., 2021). Based on the transcriptomic approach, homologs of the trichome candidate genes that regulate stem prickles’ development have been identified in the Rosaceae family. After screening the quantitative trait locus (QTL) region on Chr3 of a haploid rose line from Rosa chinensis, RC3G0244800 among them exhibited substantial similarity with TTG2, which acts genetically downstream of MBW while sharing the partial activity with GL2 to initiate trichome development (Hibrand Saint-Oyant et al., 2018). The high expression of RcGL2 triggered the differentiation of the epidermal cells to form an outgrowth of rose (Swarnkar et al., 2021). RNA-seq analysis found that the GL1 homolog R2R3-MYB gene was downregulated in the prickle-free raspberry (Khadgi and Weber, 2020). TTG1 expression significantly decreased in the glabrous cultivar of Rosa chinensis named “Basye’s Thornless” and increased in a densely prickled cultivar of Rosa rugosa (Feng et al., 2015; Zhou et al., 2020). The previous results were mainly based on gene expression or transcriptome analysis between prickle and prickle-less cultivars. Therefore, an in-depth investigation of the candidate gene that played in the development of fruit prickles should be made.
Some differences are observed between fruit and stem prickles in the rosaceous plants, despite similar morphology and function (Wang D. J. et al., 2019). The cells of fruit prickles contain granules, a high cytoplasm content, a range of plastids, and thin cell walls. In contrast, few organelles were observed in the cytoplasm of stem prickles cells with thick cell walls (Wang et al., 2021). Furthermore, the fruit prickles of R. roxburghii were distributed with spindle orientation, whereas binate prickles arose on the stem nodes (Wang D. J. et al., 2019). The potential functions and molecular mechanism of the BMW complex underlining fruit prickle are unknown. Previously, we found that RrGL1 from R. roxburghii was evolutionarily similar to GL1 and positively regulated the epidermal cell fate (Huang et al., 2019). RrEGL3 had significantly higher expression than RrGL3 in R. roxburghii (Yan et al., 2021). However, EGL3 in Arabidopsis had a much smaller effect than GL3 on trichome development (Bernhardt et al., 2003). Considering the ultrastructural and distributional divergence of stem and fruit prickles and the functional divergence of RrEGL3 and RrGL3, it is requested to demonstrate the function and molecular mechanisms of RrTTG1 in fruit prickles of R. roxburghii.
In this study, we elucidated their development from floral bud to mature fruit using the morphological and histological analyses of fruit prickles. Furthermore, we studied the homologous candidate gene TTG1 in R. roxburghii. qRT-PCR analysis and in situ hybridization revealed that RrTTG1 was constitutively expressed and highly expressed from fruit prickle primordium to young fruit prickles. Ectopic expression of RrTTG1 could promote trichomes formation, inhibit root hair development, and increase seed coat pigmentation and anthocyanin biosynthesis in Arabidopsis. The complex RrTTG1-RrEGL3-RrGL1 was detected by yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays. The results would lay the ground for further deciphering the regulatory mechanisms and genetic improvement of prickle-free R. roxburghii.
Materials and methods
Plant materials
R. roxburghii cultivar “Guinong 5” plants were planted in Guizhou Normal University (Guiyang, China, N 26°42.408′; E 106°67.353′). Stems, leaves, petioles, and sepals were collected before flowering in April. The reproductive tissues were collected from April to August, including seeds, receptacles, and fruits at 20, 40, 60, 80, and 100 days after pollination (DAP). Some of the materials for the paraffin section were stored in the FAA solution (50% ethanol, 4% formaldehyde, and 10% acetic acid). The others were frozen in liquid nitrogen immediately and stored in a freezing chamber at −80°C.
All seeds of Arabidopsis were in Columbia ecotype (Col-0) background. The plants were sterilized and plated on 1/2 MS (Murashige and Skoog) medium by keeping them in darkness at 4°C for 3 days and then transformed into an environment-controlled chamber under long-day conditions (16-h light/8-h dark) at 25°C.
Morphological and histological observation
Materials were observed by a stereomicroscope (Leica S8 APO, Germany). The tissues were fixed in the FAA solution and then subjected to vacuum infiltrations until the tissue pieces sank. The paraffin sections were performed as previously described (Huang et al., 2016). Finally, the tissues were embedded in wax blocks and sectioned at 8 μm. The sections were viewed with an Olympus BX53 microscope (Olympus, Japan) and imaged using a SPOT FLEXTM camera (SPOT, United States).
Gene amplification and phylogenic tree analysis
The cDNA regions were isolated by 5′ and 3′ RACE assay with the SMART™ RACE. Total RNA extracts as templates and degenerated primers of RrTTG1 (Supplementary Table 1) were used for SMART™ RACE cDNA amplification KIT (Clontech, United States). The final amplification products were cloned into the pMD18-T vector (TaKaRa, Japan) and sequenced. For phylogenic tree analysis, multiple sequence alignment was used by ClustalW1 with default parameters. The alignments were submitted to MEGA 102 to construct an unrooted phylogenic tree using a neighbor-joining statistical method and bootstrap analysis (1,000 replicates).
Subcellular localization assay
The CDS of RrTTG1 was cloned in-frame into the XbaI-digested pM999-eGFP plasmid, which contains an enhancer green fluorescent protein (eGFP) tag driven by CaMV 35S promoter. The constructed and empty vector plasmids were independently transformed into suspension protoplasts of Arabidopsis by PEG (polyethylene glycol) transformation (Yan et al., 2021). The fusion protein localization was monitored in the next 24 h. The nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI; Roche, Switzerland) at 0.1 μg/mL for 10 min. Meanwhile, the cell membrane was labeled by chloromethyl-benzamidodialkyl carbocyanine (CM-Dil, Thermo Fisher Scientific, United States) at 5 μg/mL for 15 min. The fluorescence was visualized using a laser confocal scanning microscope (SP8, Leica, Germany).
The real-time quantitative PCR
Total RNA from the above tissues was isolated using TRIzol reagent (TaKaRa) with DNase I treatment according to the manufacturer’s instructions. After DNase treatment, RT-PCR Kit® (TaKaRa) was performed according to the suppliers’ instructions using 2 μg of RNA and oligo dT-adaptor primer. The real-time quantitative PCR of RrTTG1 was performed in a LightCycler480 instrument (Roche, Switzerland). Each reaction contained 10 μL of SYBR green PCR master mix (TaKaRa), 1.0 μL cDNA, and 200 nM primers to the final volume of 20 μL. Amplification was performed at 95°C for 5 min, 40 cycles with 95°C for 20 s, 60°C for 30 s, and 72°C for 1 min. The expression levels relative to the RrActin were estimated by calculating △△Ct and subsequently analyzed using 2–△ △ Ct method. All samples were performed for three biological replicates.
In situ hybridization
The gene-specific fragments were amplified by PCR with the ISH-F/R primer (Supplementary Table 1) and cloned into the pGM-T vector (Tiangen, China) following the manufacturer’s instructions for a DIG Northern Starter kit (Roche, Switzerland), and digoxigenin-labeled sense and antisense RNA probes were synthesized using SP6 and T7 RNA polymerase, respectively. Then, prehybridization, hybridization, washing, and detecting were performed as previously described (Yan et al., 2021).
Arabidopsis transformation and phenotypic observation
RrTTG1 was cloned into pBI121 to construct an overexpression vector via XbaI/BamHI. Then, the constructed vector was transformed into Agrobacterium tumefaciens strain GV3101. The 6-week-old seedlings of Arabidopsis were transformed following the floral dipping method (Zhang et al., 2006). The harvested T1 seeds of transformed plants were sown on 1/2 MS medium containing 50 mg/L kanamycin. After kanamycin selection and RT-PCR confirmation (the primers were listed in Supplementary Table 1), the positive lines were selected, and phenotypes of transgenic plants were observed. For anthocyanin observation, 5-day-old seedlings and seeds were imaged using a Leica S8 APO stereomicroscope coupled to a Leica DC300 camera. For hair density, the number of hairs on the first 5 mm from the primary root tip of at least 30 plants per genotype was counted. Meanwhile, the number of trichomes on the first rosette leaves of 1-week-old seedlings was counted.
Extraction and quantification of anthocyanin evaluation
The 4-week-old Arabidopsis seedlings were ground thoroughly in liquid nitrogen and dissolved in the extraction solution (75% H2O, 24% methanol, and 1% formic acid) at 4°C for 12 h to extract anthocyanins. Then, the extracts were subjected to a 10 min centrifugation at 12,000 rpm. After centrifugation, the supernatant was filtered using a 0.22 μm sterile organic filter membrane. Dried delphinidin 3,5-diglucoside chloride (Sigma-Aldrich, United States) was used to make regression equations to measure the content of anthocyanin in samples. A C18 column (4.6 × 200 mm, 5 μm, Waters Corporation, United States) was used. The mobile phase consisted of solvent A: 5% (v/v) formic acid in water and solvent B: 5% (v/v) formic acid in acetonitrile. The anthocyanin was separated by starting with 100% A with a linear gradient to 25% B over 30 min, ramping to 80% B over 2 and 3 min to re-equilibrate to initial in acetonitrile (Pourcel et al., 2010). The absorbance of the mixtures was measured at 525 nm. The anthocyanin contents were expressed as μg/100 mg of fresh weight.
Yeast two-hybrid assay
For the Y2H analysis, the CDS of AtGL3, AtEGL3, RrGL3, and RrEGL3 was amplified and ligated into pGADT7 (AD) through BamHI and SacI double digestion sites. Full-length CDS of RrTTG1, AtTTG1, and RrGL1 was amplified and inserted into pGBKT7 (BD) at EcoRI and SmaI sites. Each combination of AD and BD plasmids was co-transformed into yeast strain AH109 separately using the LiAC/PEG method (Gietz and Schiestl, 2007). pGADT7-T was co-transformed with pGBKT7-53 as a positive control. pGADT7-T and pGBKT7-Lam were transformed as the negative controls (Supplementary Figure 1). All transformants were grown on SD-Trp-Leu (SD-LW), SD-Trp-Leu-Ade-His (SD-LWAH), and SD-LWAH-X-α-gal medium to detect the α-galactosidase activity of the yeast strains. The images were photographed at 5 day after incubation.
Bimolecular fluorescence complementation assay
Full-length CDS of AtGL3, AtEGL3, RrGL3, and RrEGL3 was amplified and ligated into pXY105 (cYFP) by BamHI and SalI sites for BiFC. Then, CDS of RrTTG1was amplified and inserted into pXY106 (nYFP) by BamHI and XbaI sites. These vectors were separately transformed into Agrobacterium EHA105. Transformants were harvested when OD600 reached 0.8 and resuspended in 1/2 MS medium (supplied with 50 μM acetosyringone). The agrobacterial cells containing the cYFP fusion vector were mixed with agrobacterial cells containing the nYFP fusion vector at a 1:1 ratio. The mixture was transiently expressed in tobacco (Nicotiana benthamiana) leave as reported (Lee et al., 2008). The YFP fluorescence signal was visualized and detected after 48 h by using a Leica SP5X confocal microscope (Leica Co., Germany).
Results
The origination and development of fruit prickles in Rosa roxburghii
The mature fruit prickles are big, conical, branchless, and non-glandular in R. roxburghii. They usually have two types: flagelliform and acicular (Wang D. J. et al., 2019). To investigate their origination and development in R. roxburghii, we carried out a detailed morphological and histological investigation from the floral bud to mature fruit (Figure 1).
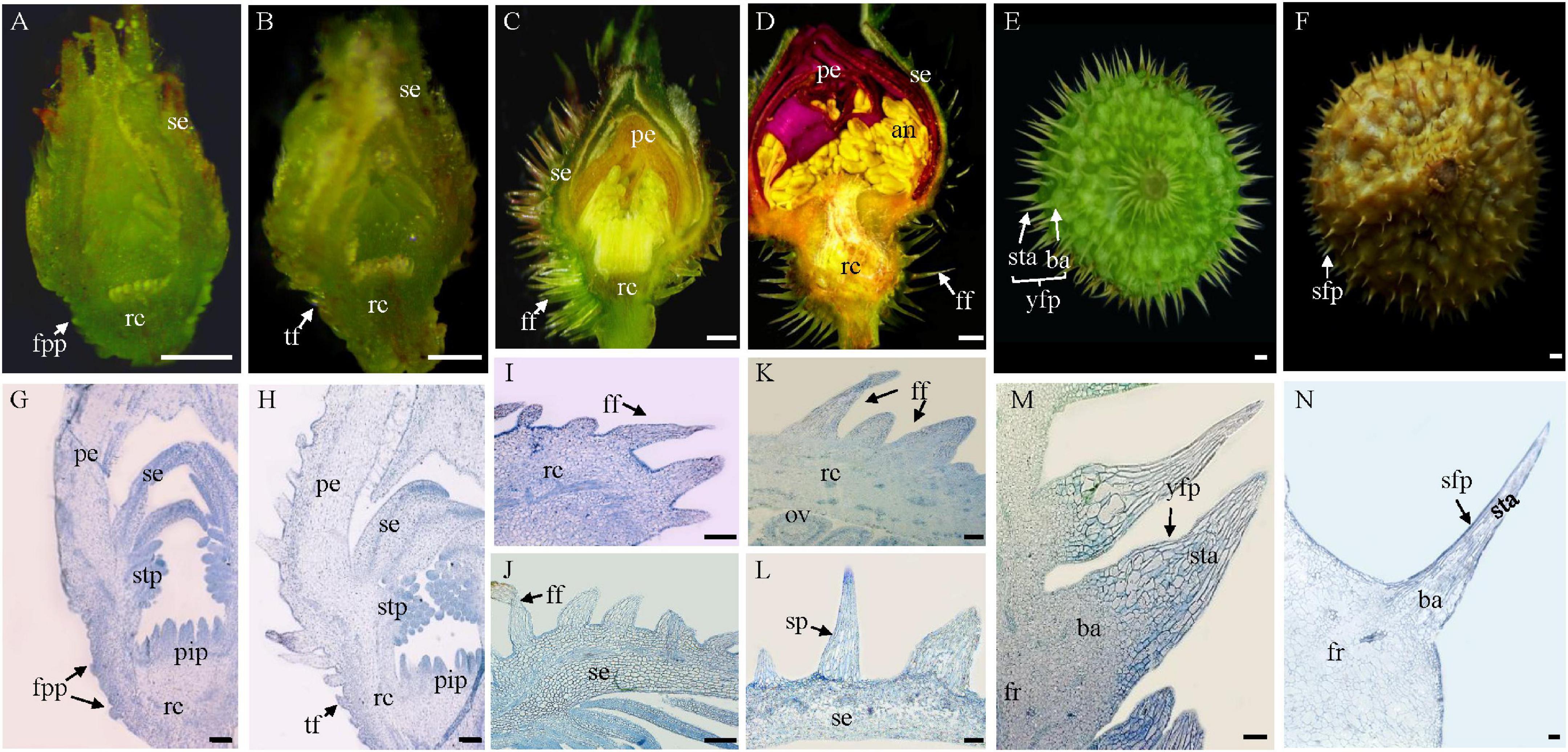
Figure 1. Origination and development of fruit prickles in R. roxburghii. (A–F) The macroscopic depiction of the development of fruit prickles during flower development (A–D) and fruit development (E,F). Anatomy of fruit prickle primordium (fpp) (G), triangular-shaped fpp (H), flagelliform fpp of sepal (I,J), flagelliform fpp of the receptacle (K,L), young fruit prickle on 20 DAP fruit (M), and sharp fruit prickle on 80 DAP fruit (N). se, sepal; rc, receptacle; pe, petal; stp, stamen primordium; pip, pistil primordium; fpp, fruit prickle primordium; tf, triangular fpp; ff, flagelliform fpp; ov, ovule; an, anther; fr, fruit; sta, stalk; ba, base; yfp, young fruit prickle; sfp, sharp fruit prickle. (A–F), bar = 500 μm; (G–N), bar = 100 μm.
Stage I corresponds to fruit prickle initiation. During the floral differentiation (Figures 1A–C), fruit prickle primordia were first observed on the surface of the receptacle after the formation of the stamen and pistil primordia (Figures 1A,G). The fruit prickle primordium was a global bulge with 10–20 small ground meristematic cells (Figure 1G). Then, with the increasing number of fruit prickle primordia, their top cells gradually formed a triangular-shaped fruit prickle (Figures 1B,H). However, the initiation of the sepal prickle was a little earlier than the fruit prickle of the receptacle (Figures 1A,B,G,H).
Fruit prickle primordia exhibited continuous growth and color development during a period of rapid cell division in stage II (Figures 1C,D). The triangular-shaped prickle underwent constant cell division to form the flagelliform fruit prickle, which had no obvious base until the flower bloomed (Figures 1I–K), whereas the sepal prickle cells stopped dividing, began enlarging, and finally lignified when the flower bloomed (Figures 1J–L). Thus, the cell proliferation ability of the flagelliform fruit prickle on the receptacle may determine the width of the fruit prickle base in stage III.
After pollination, the receptacle gradually enlarged with the fruit prickle entering stage III, when it began to lignify and gradually harden (Figures 1E,F). The base cells of the fruit prickle kept dividing, with the top cells started expanding during this process to form the young fruit prickle. Each comprised a base and a stalk (Figure 1E). Their bases were composed of hundreds of spherical cells smaller than those of the stalk cells that comprised tinny cylindrical-shaped cells (Figure 1M). The top cells lignified when the fruits ripened, leading to a hard and sharp fruit prickle (Figures 1F,N). Therefore, these results indicated that the fruit prickle primordia originated from parenchyma cells of the receptacle during floral budding and subsequently transformed into hard and sharp prickles postfertilization.
A WD40 protein RrTTG1 is closely related to FvTTG1 and localized in the nucleus
Since the WD40-repeat protein TTG1 functions as a critical positive regulator of trichome determined in Arabidopsis, a detailed analysis of TTG1 homolog in R. roxburghii might help us to understand the molecular mechanism of fruit prickles formation. The full-length cDNA of RrTTG1 contains an open reading frame (ORF) of 1,041 bp that encoded a polypeptide of 346 amino acids. RrTTG1 harbored four conserved WD40 repeats, with each repeat containing nearly 44 amino acid tandems (sites at 70–113 aa, 121–166 aa, 169–207 aa, and 258–298 aa). RrTTG1 aligned with its TTG1 orthologs in woodland strawberry (Fragaria vesca), rose (Rosa rugosa), peach (Prunnus persica), and Arabidopsis (Figure 2A). RrTTG1 showed over 95% similarity with its homologs of woodland strawberry and rose. However, we observed slightly less similarity to TTG1 which showed 79% identity.
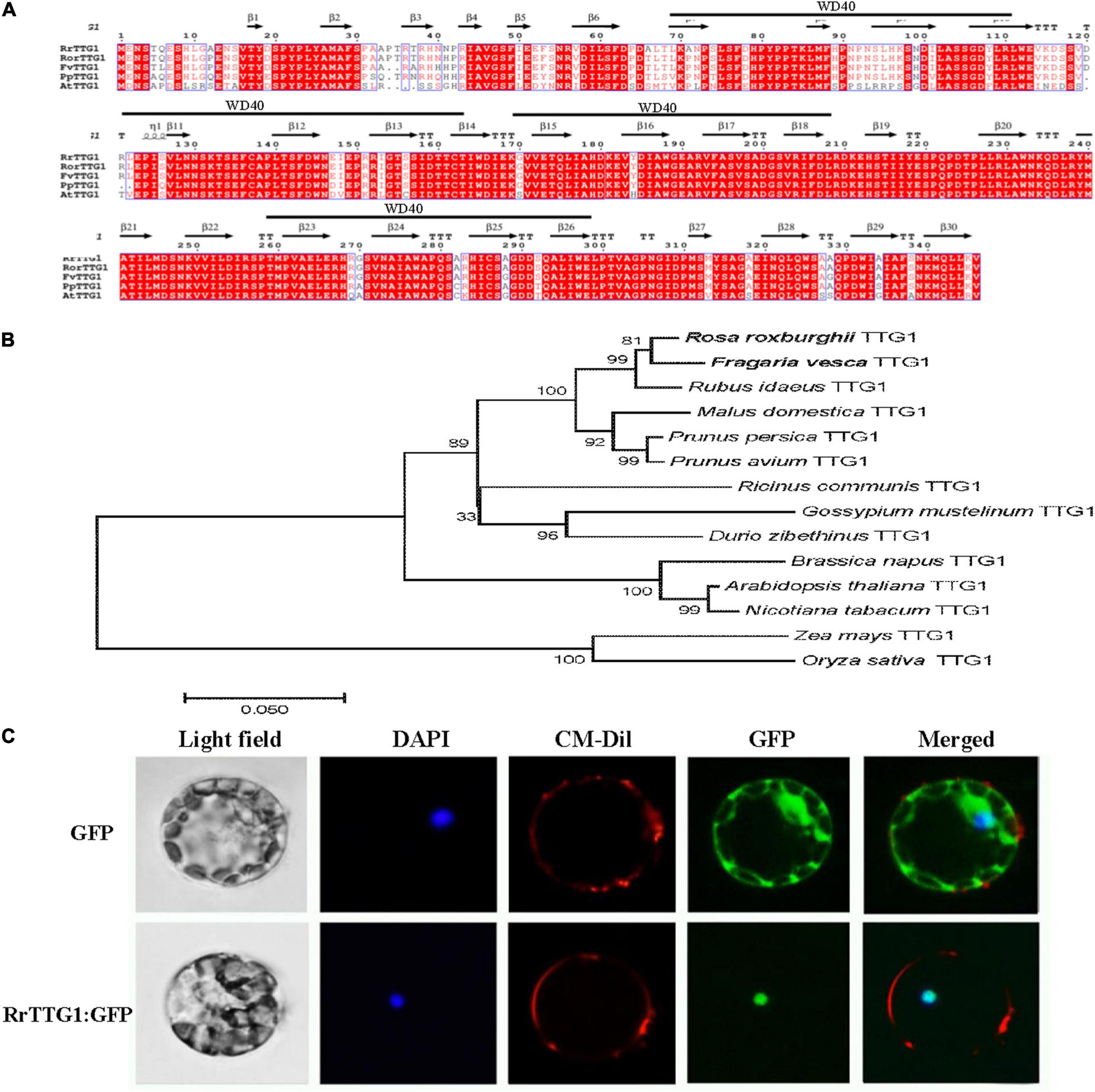
Figure 2. Alignment, phylogenetic, and subcellular analysis of RrTTG1. (A) The deduced amino acid sequence of RrTTG1 compared to TTG1 homologs from Rosa rugosa, Fragaria vesca, Prunnus persica, and Arabidopsis. Alignments were performed using Clustal W. The secondary structure was displayed above the sequences. Red shading indicated identical amino acids. The above black line represents the WD40 domain. (B) Phylogenetic analysis of TTG1 homologs in R. roxburghii and the other plants. The scale bar indicates the number of amino acid substitutions per site. GenBank accession numbers: Fragaria vesca TTG1 (XP_004307911.1), Rosa rugosa TTG1 (QCI162276.1), Rubus idaeus TTG1 (AEI55401.1), Malus domestica TTG1 (XP_008376821.1), Prunus persica TTG1 (ACQ65867.1), Prunus avium TTG1 (XP_021805418.1), Ricinus communis TTG1 (XP_025015182.1), Gossypium mustelinum TTG1 (TYI69075.1), Durio zibethinus TTG1 (XP_022773722.1), Brassica napus TTG1 (NP_001303154.1), Arabidopsis thaliana TTG1 (CAC10524.1), Nicotiana tabacum TTG1 (ACJ06978.1), Zea mays TTG1 (NP_001310302.1), and Oryza sativa TTG1 (XP_015626852.1). (C) RrTTG1 was localized in the nucleus. Protoplasts from Arabidopsis were transiently transformed with an 35S:RrTTG1-GFP or an empty vector. The empty 35S:GFP vector was used as the control. GFP, green fluorescence protein; DAPI, 4′, 6-diamino-2-phenylindol; CM-Dil, chloromethyl-benzamidodialkyl carbocyanine. Bars = 5 μm.
Phylogenetic analysis confirmed that RrTTG1 clusters together with the homologs from the Rosaceae family, thus displaying the closest relationship with TTG1 homologs from woodland strawberry and rose, and we also identified additional closely related sequences in homologs of raspberry and apple (Malus domestica) (Figure 2B). Furthermore, we also found that homologs of dicotyledonous plants, including castor beans (Ricinus communis), cotton (Gossypium mustelinum), and durian (Durio zibethinus), were also clustered. However, those of the homologous monocot sequences ZmTTG1and OsTTG1 were more distantly related (Figure 2B). To examine the detailed subcellular localization of RrTTG1, the fused protein RrTTG1: GFP was transformed into Arabidopsis protoplasts. DAPI and CM-Dil staining showed exclusively nuclear and cell membrane distributions, respectively (Figure 2C). Under the induction of 405 nm light, the protoplasts transformed with 35S:GFP presented the green fluorescent signals in the whole cell, including the membrane, cytoplasm, and nucleus. When the protoplast was transformed with 35S:RrTTG1:GFP, the GFP signals were only observed in the nucleus, similarly at the same site as DAPI, inferring that RrTTG1 was localized in the nucleus and may function as a transcription factor.
RrTTG1 was highly expressed during the division and expansion phases of fruits prickles
We selected various tissues to examine the expression patterns of RrTTG1 and found that it was constitutively expressed in young leaf, stem, petiole, sepal, and receptacle, whereas being low in seeds in R. roxburghii (Figure 3A). Moreover, the expression during fruit development was detected. We found that RrTTG1 was highly expressed in the fruits at the early stages and finally peaked at 40 DAP (Figure 3B). The transcript level was significantly reduced at 80 DAP when fruits were in the turning stage, and it reached the lowest at 100 DAP when fruits fully ripened (Figure 3B). Thus, these results mainly demonstrated that RrTTG1 was essential in the early stage of fruit prickle development.
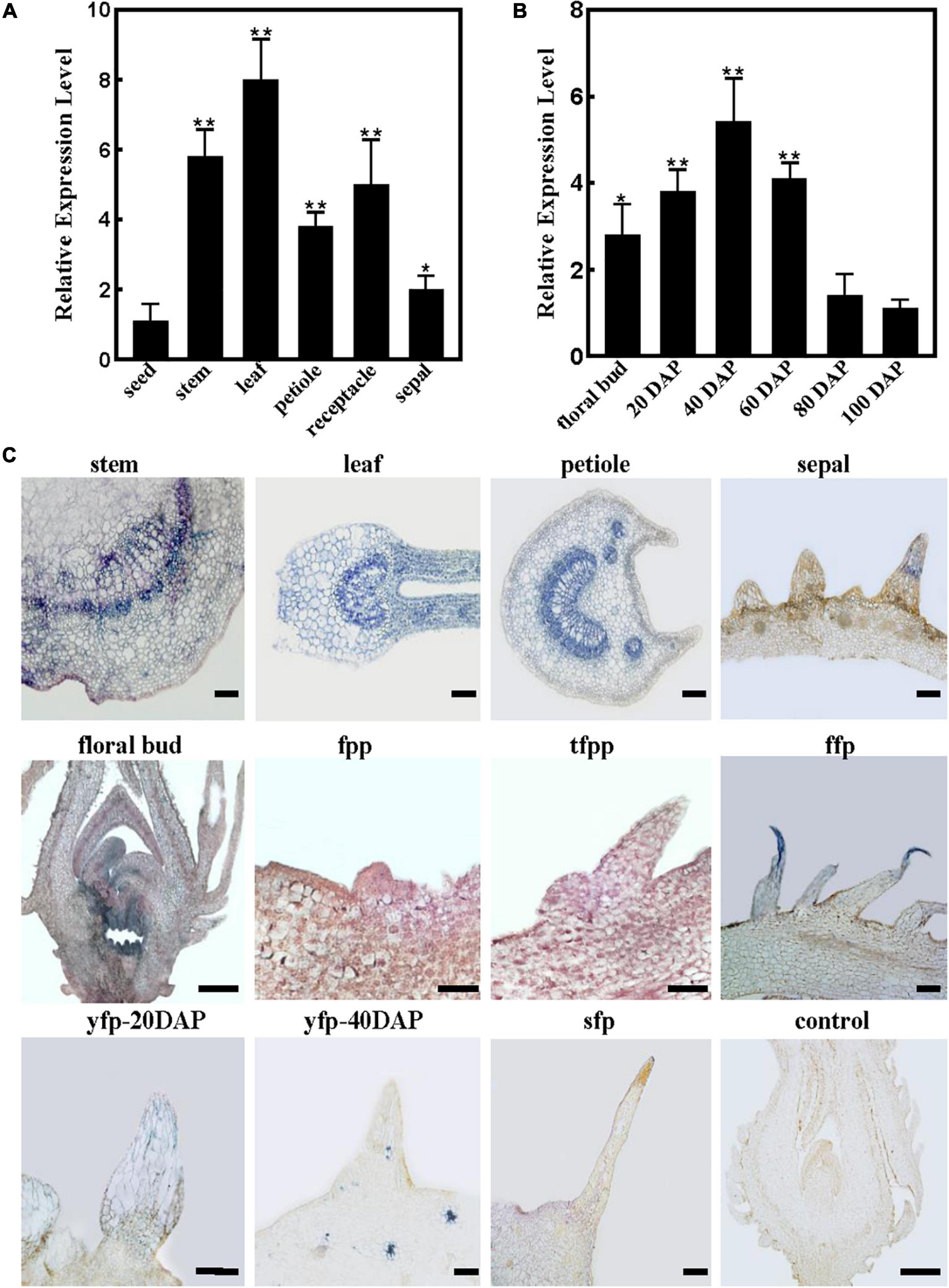
Figure 3. RrTTG1 was expressed constitutively and highly at the early stage of fruit prickles formation. (A,B) qRT-PCR analysis of RrTTG1 expression in various tissues (A) and fruits at different development stages (B). Values are presented as means ± SD (n = 3) and analyzed using one-way ANOVA (**p < 0.01, *p < 0.05). Primers are listed in Supplementary Table 1. (C) In situ hybridization analysis of the expression pattern of RrTTG1. The expression of RrTTG1 in the young stem, young leaf, petiole, sepal, floral bud, fruit prickle primordium (fpp), triangular-shaped fpp (tfpp), flagelliform fruit prickle (ffp), young fruit prickle on 20 DAP fruit (yfp-20 DAP), 40 DAP fruit (yfp-40 DAP), and 80 DAP fruit (sfp) was detected. The paraffin sections with sense RNA probes of RrTTG1 were used as the control. Bar = 200 μm.
To further identify the spatial and temporal expression patterns of RrTTG1 during fruit prickle development, in situ hybridization was performed (Figure 3C). The sense probe showed no detectable signal as the control. Despite the different arrangements of xylem and phloem cell types in the various organs, the cells in vascular bundles, cambium, and parenchyma cells of the young stem, young leaf, and petiole showed a much higher RrTTG1 transcript signal, which was consistent with the qRT-PCR results (Figure 3C). We also detected strong visible signals in the fruit prickle primordia and triangular-shaped fruit prickle primordia, especially the top cells at the flagelliform fruit prickle, which indicated the highly abundant transcript of RrTTG1 being involved in the prickle initiation (Figure 3C). RrTTG1 continued to be expressed highly in the parenchymal cells of fruit prickles on 20 DAP fruit. In the 40 DAP fruit, despite the high expression of RrTTG1 in the vascular bundles of fruit fresh, its expression decreased in the cells of fruit prickle. Corresponding with the low RrTTG1 expression in fruits at 80 DAP, there was no obvious signal in the sharp fruit prickle (Figure 3C). Therefore, the results indicated that RrTTG1 likely enhanced fruit prickle differentiation and enlargement.
RrTTG1 decreased the number of root hairs and enhanced the anthocyanin content
Due to no feasible transformation system for R. roxburghii, we ectopically expressed RrTTG1 in the Arabidopsis plants to explore its function. The transcript of RrTTG1 was detectable in the seedlings of transgenic plants, including three RrTTG1-ox lines and RrTTG1-ox/ttg1 plants, inferring that RrTTG1 was expressed in Arabidopsis (Figure 4A). The colors of hypocotyl and cotyledon of the seedlings in the RrTTG1-ox plants were more intense than wild-type (WT) (Figure 4B). The results of anthocyanin quantification showed a significant increase in RrTTG1-ox plants. The anthocyanin contents of three transgenic plants increased by 48.19, 53.64, and 57.45% compared to the WT, respectively (Figure 4C). Moreover, pigmentation of the seed coat endothelium was more condensed in RrTTG1-ox plants, while its color was less brown in WT, thereby suggesting that RrTTG1 could enhance the anthocyanin accumulation. Consistently, ectopic expression of RrTTG1 could rescue the pigmentation phenotype of the ttg1 mutant in Arabidopsis (Figure 4B).
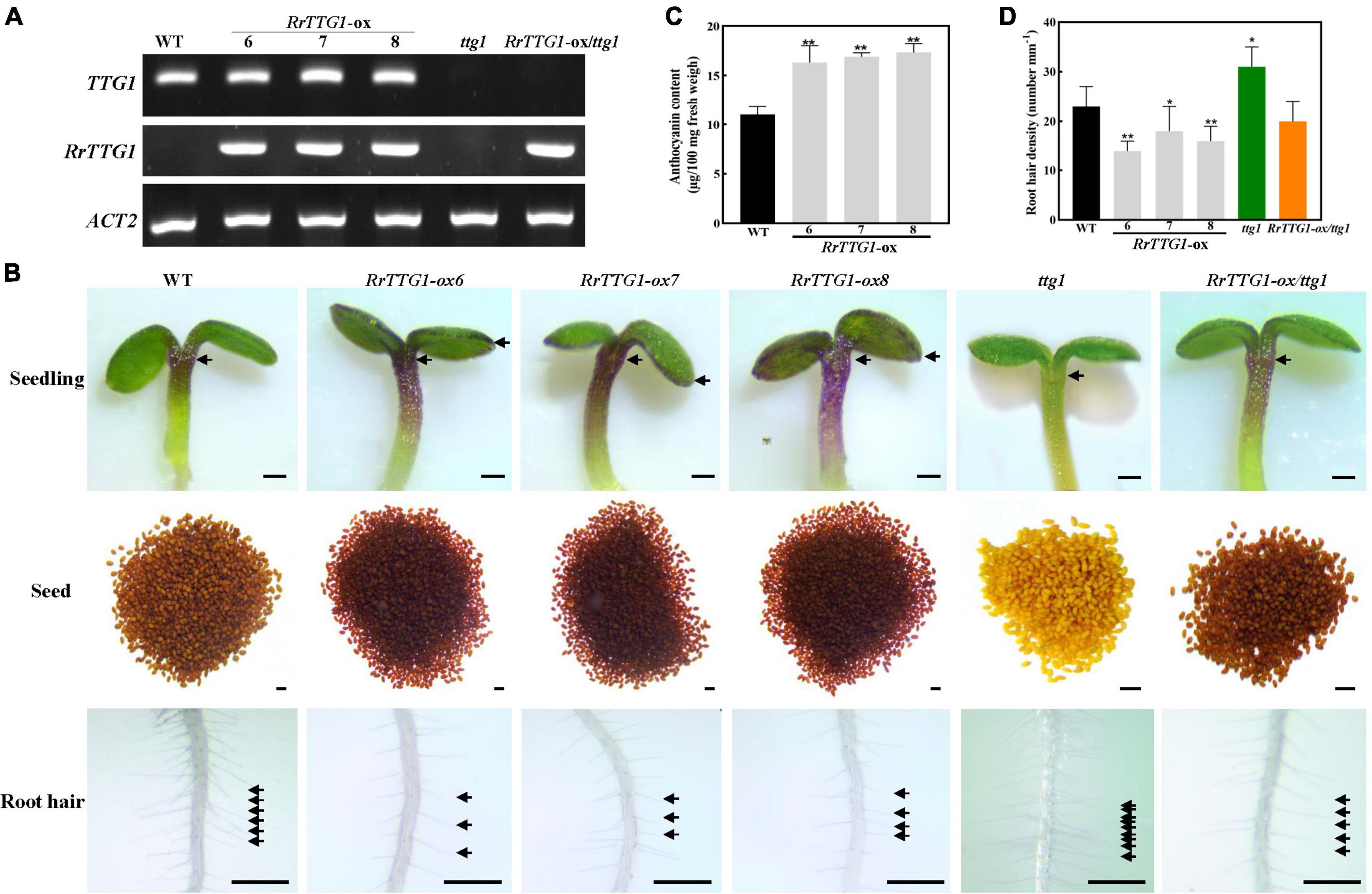
Figure 4. RrTTG1 promoted anthocyanin content and inhibited root hair formation. (A) Expression of TTG1 and RrTTG1 in WT (Col-0), ttg1 mutant, and transgenic plants (RrTTG1-ox and RrTTG1-ox/ttg1). RT-PCR was used to detect the expression of TTG1 and RrTTG1, and ACT2 was used as a control. Primers are listed in Supplementary Table 1. (B) Ectopic expression of RrTTG1 could promote anthocyanin content and inhibit root hair formation in Arabidopsis. The seeds and 5-day-old seedlings were collected for anthocyanidin observation. Root hair located 5 mm above the root tip was observed, bar = 1 mm. (C) Ectopic expression of RrTTG1 could promote anthocyanin content. The anthocyanin contents of 4-week-old WT and RrTTG1-ox transgenic plants were measured by HPLC analysis. (D) Ectopic expression of RrTTG1 could reduce the root hair density. The number of root hairs per mm (root hair density) of WT, ttg1 mutant, and transgenic plants (RrTTG1-ox and RrTTG1-ox/ttg1) was counted (n = 30). Data are mean ± SD and analyzed using one-way ANOVA (**p < 0.01, *p < 0.05).
We analyzed the effect of the transgene on root hair development. WT plants showed well-spaced root hairs, whereas the roots of RrTTG1-ox plants displayed sparsely spaced irregularly growing root hairs (Figure 4D). The RrTTG1-ox plants showed an average 30.43% decrease of root hair density compared to the WT. In addition, the ttg1 mutant produced approximately 1.35-fold of root hairs in WT root, while the RrTTG1-ox/ttg1 transgenic plants exhibited a near-normal root hair pattern (Figure 4D), thereby deducing that RrTTG1 substantially inhibited root hair formation.
RrTTG1 promoted the trichome number in Arabidopsis
To test the effect of RrTTG1 on trichome development, we observed the trichome phenotype in transgenic plants. We noticed that RrTTG1-ox Arabidopsis plants had a significantly greater number of trichomes per leaf than that of WT, and their trichome number increased by about half, finally reaching 52.38, 38.10, and 33.33%, respectively (Figures 5A,C). Undoubtedly, the trichome was absent in the ttg1 mutant. RrTTG1 was able to functionally complement the glabrous phenotype of the ttg1 mutant (Figures 5B,C). Our results indicated that RrTTG1 was functionally homologous to the TTG1 regulating trichome development, thus inferring that RrTTG1 might function as TTG1 homologs in the Rosaceae family to modulate the fruit prickles development of R. roxburghii.
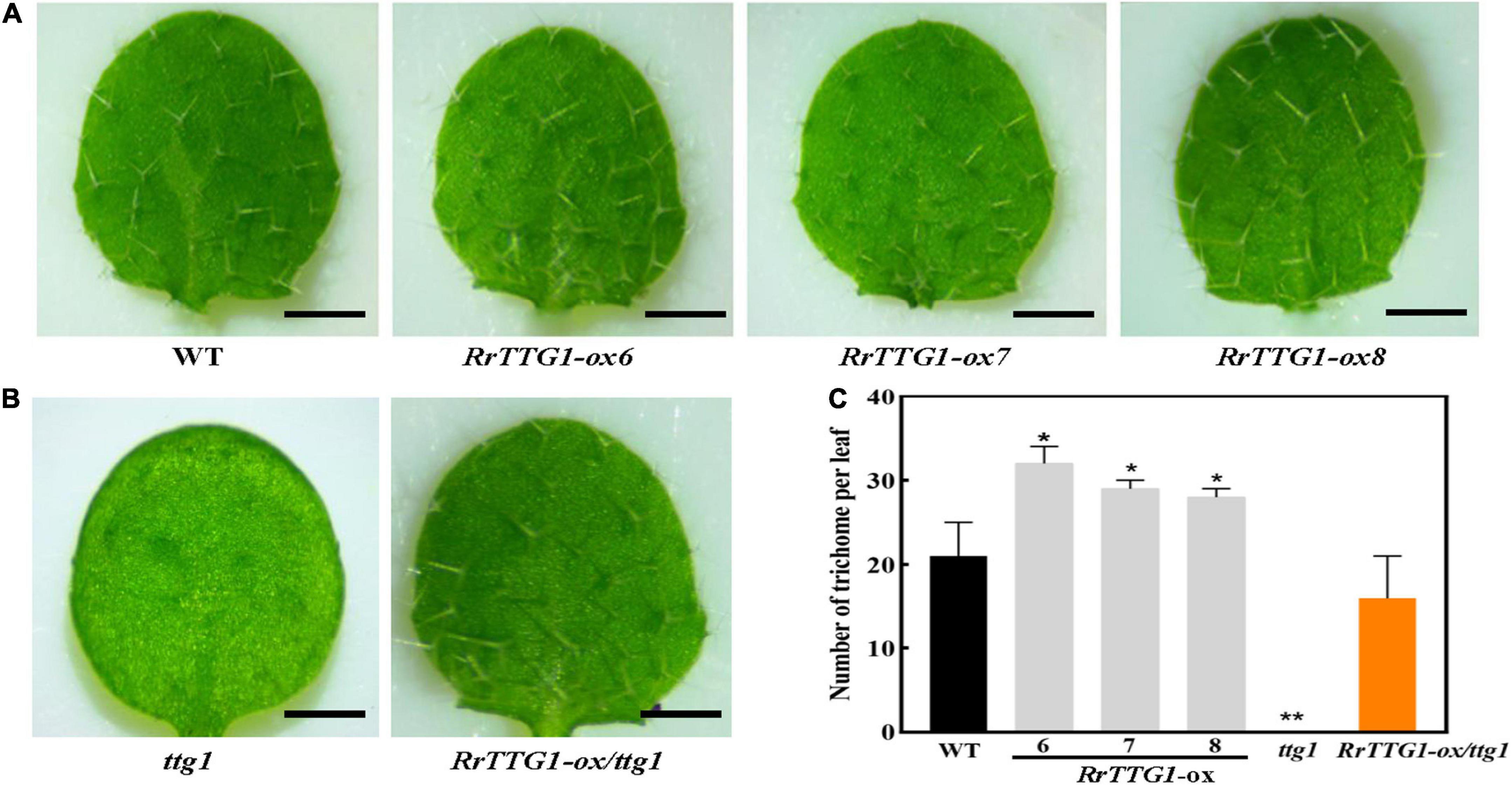
Figure 5. RrTTG1 promoted trichome formation in Arabidopsis. (A) Ectopic expression of RrTTG1 could increase the trichome number in Arabidopsis. Leaf trichome phenotypes from the first two rosette leaves of 1-week-old soil-grown seedlings of the WT (Col-0) and three RrTTG1-ox transgenic plants were compared, bar = 1 cm. (B) RrTTG1 complemented the glabrous phenotype of ttg1 mutant. Leaf trichome phenotypes from the first two rosette leaves of one-week-old soil-grown seedlings of the ttg1 and three RrTTG1-ox/ttg1 transgenic plants were compared, bar = 1 cm. (C) Comparison of the trichome number per leaf in the WT, ttg1 mutant, and transgenic plants (RrTTG1-ox and RrTTG1-ox/ttg1). Values are means ± SD (n = 30) and analyzed using one-way ANOVA (**p < 0.01, *p < 0.05).
The RrTTG1 only interacted with RrEGL3 to form an MYB-bHLH-WD40 complex in Rosa roxburghii
Since TTG1 interacted with the bHLH transcription factor GL3 or EGL3 to influence the trichome number and anthocyanin content in Arabidopsis, we performed the Y2H assay to detect the interaction partner proteins of RrTTG1. The combinations of empty vector (pGADT7 or pGBKT7) with all the tested proteins were also performed, and no self-activation was detected (Supplementary Figure 1). Y2H result showed that RrTTG1 could both directly interact with GL3 or EGL3 like TTG1 in yeast, thereby implying that RrTTG1 was evolutionarily similar to the TTG1. Notably, we found that RrTTG1 failed to bind to RrGL3 and only interacted with RrEGL3 (Figure 6A). Such interaction was also confirmed by the BiFC approach in Nicotiana benthamiana. The fluorescence signals revealed that RrTTG1 positively interacted with EGL3 or GL3 from Arabidopsis. Therefore, we also showed the positive interaction between RrTTG1 and RrEGL3. However, we did not detect any fluorescence signal from the combination of RrTTG1 and RrGL3, thus suggesting no direct interaction between RrGL3 and RrTTG1 (Figure 6B).
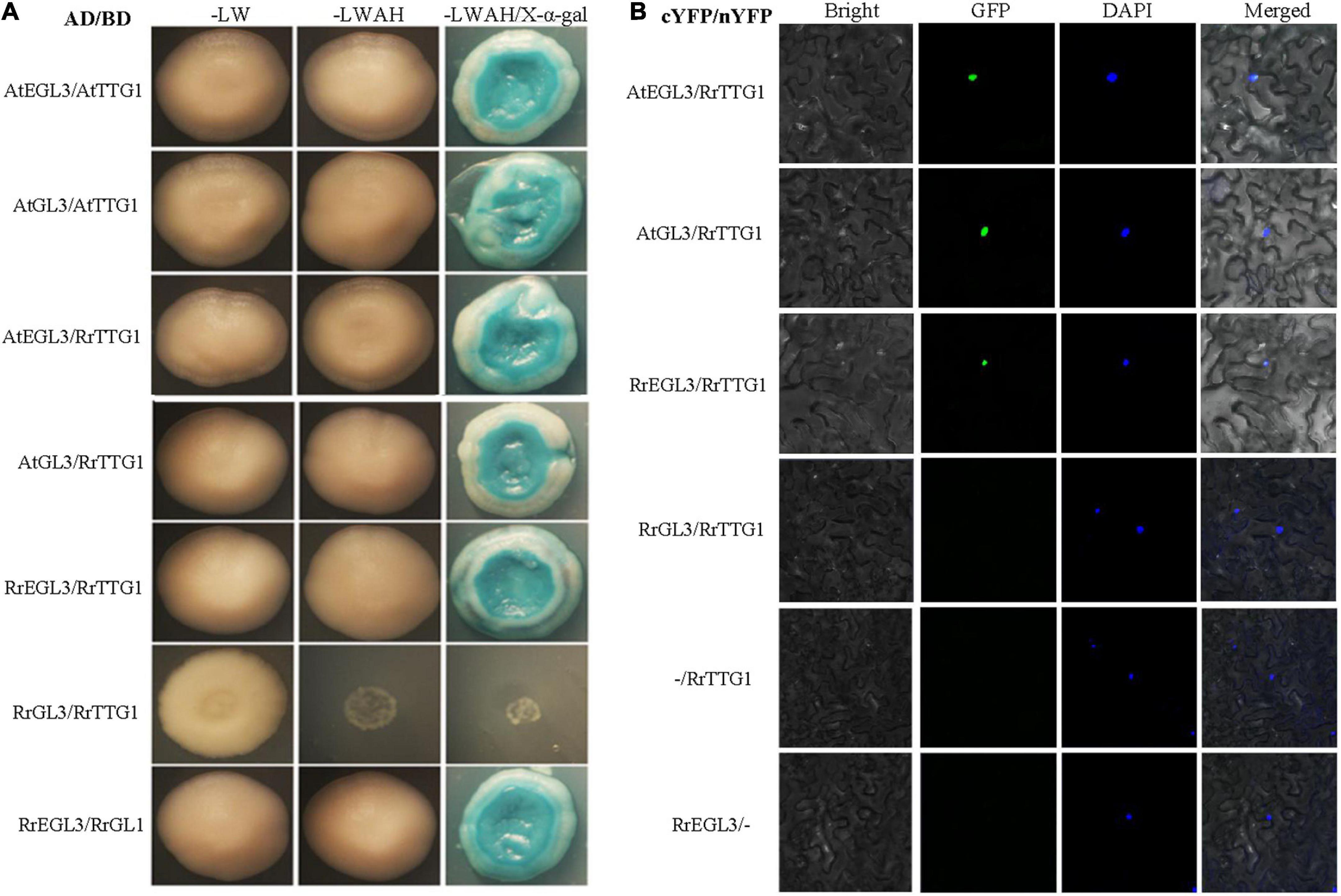
Figure 6. RrTTG1 only interacted with RrEGL3 to form the RrTTG1-RrEGL3-RrGL1 complex. (A) Y2H assay testing the interaction between RrTTG1 and GL3/EGL3/RrGL3/RrEGL3. Each combination of AD and BD plasmids was co-transformed into yeast strain AH109 separately, and all transformants were grown on SD-Trp-Leu (SD-LW), SD-Trp-Leu-Ade-His (SD-LWAH), and SD-LWAH-X-α-gal medium. The images were photographed at 5 day after incubation. All the controls were displayed in Supplementary Figure 1. (B) BiFC assay testing the interactions between RrTTG1 and GL3/EGL3/RrGL3/RrEGL3. Each combination of cYFP and nYFP was transiently expressed in Nicotiana benthamiana leave, and the YFP fluorescence signal was visualized and photographed after 48 h.
As RrTTG1 was highly expressed in the initiation of fruit prickle formation, to investigate whether its partner proteins have the same expression pattern, we explored the expressions of RrGL1, RrGL3, and RrEGL3 from fruit prickle primordia to young fruit prickle (Figure 7). The transcripts of RrGL1 and RrEGL3 having higher abundance were localized in the global bulges of fruit prickle primordia on the receptacle. RrGL1 and RrEGL3 were highly expressed in the flagelliform fruit prickle, especially RrEGL3 which was essential in the differentiation of fruit prickle primordia. Moreover, when the cells at the base began to enlarge, we found strong signals of both RrGL1 and RrEGL3 in the young fruit prickles that were derived from flagelliform fruit prickle, while no visible signal of RrGL3 was detected (Figure 7). These results showed that RrGL1 and RrEGL3, the partner proteins of RrTTG1, are also highly expressed at the early stage of fruit prickle formation. Therefore, such an expression pattern of the RrTTG1-RrEGL3-RrGL1 complex suggested it played a substantial role during the prickle primordium initiation and expansion of fruit prickles.
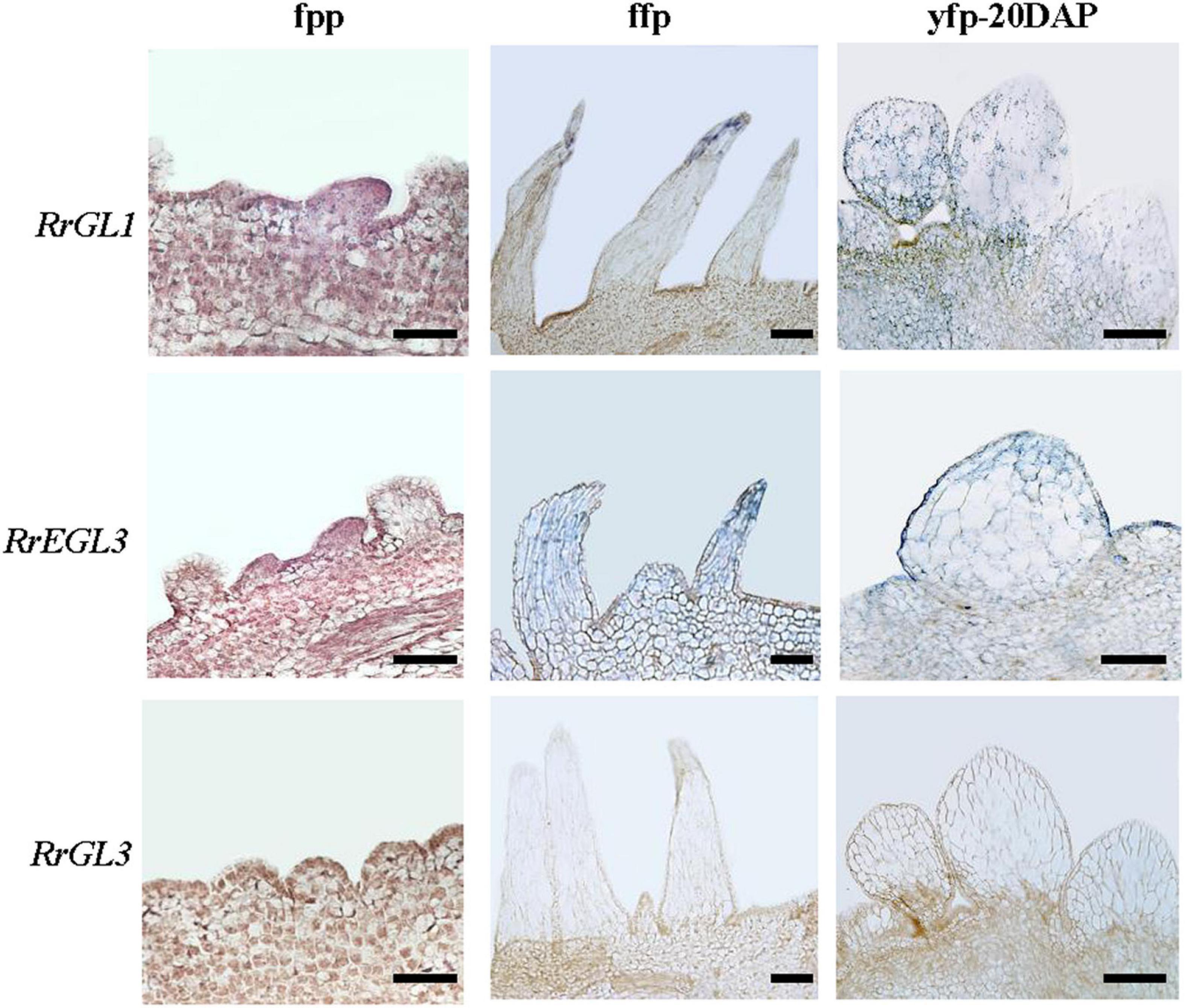
Figure 7. RrGL1 and RrEGL3 highly expressed at the early stage of fruit prickles formation. In situ hybridization testing the expression pattern of RrGL1, RrEGL3, and RrGL3 in fruit prickle primordium (ffp), flagelliform fruit prickle (ffp), and young fruit prickle on 20 DAP fruit (yfp-20 DAP). Bar = 200 μm.
Discussion
The fruit prickle was derived from the ground meristem beneath the protoderm of the receptacle
Prickles are previously proposed as extended and deformed trichomes originating from protodermal cells (Hibrand Saint-Oyant et al., 2018), while a recent study proved that stem prickle was not modified trichome due to its initiation from the ground meristem beneath the protoderm (Zhou et al., 2021). The fruit prickles are similar to the trichomes based on morphological and anatomical studies. Both lack vascular bundles and are effective deterrents against herbivores (Papatheodorou et al., 2020). Regarding differences, trichomes comprise non-glandular, branched, and unicellular cells originating from the protoderm (Serna and Martin, 2006). However, fruit prickles of R. roxburghii are derived from multiple cellular divisions along with lignification, thereby leading to sharp fruit prickles (Figure 1). Likewise, the prickles of eggplant were also shown to be multicellular and lignified (Zhang et al., 2021). Furthermore, through the histological analysis, the fruit prickle primordium of R. roxburghii with a slight global bulge of 40–70 spherical cells originated from the ground meristem beneath the protoderm of the receptacle, which is composed of parenchyma cells (Figure 1), supporting the divergence of fruit prickle and trichome. Understanding the histological aspect of prickles will benefit the fruit’s processing and harvesting that could be extended to other plant species.
Prickles are present on organs other than the fruit, such as stems, petioles, and pedicels. The prickles on the surface of fruit were found only in some species. Thus, little is known about the development of fruit prickles, whereas stem prickles have been made in numerous studies for cut rose production and gardening (Feng et al., 2015; Khadgi and Weber, 2020). The stem prickles of rose develop from multiple cells of the ground meristem beneath the protoderm of the stem apex. We found that fruit prickles also originated from the ground meristem beneath the protoderm of the receptacle, which is the axis of floral organs attached in botany. Nevertheless, we found a significantly divergent development between fruit and stem prickles. The base cells of stem prickles underwent one rapid proliferation and gradually stopped (Zhou et al., 2021). However, the cells of fruit prickle primordium underwent the first rapid proliferation to form flagelliform-shaped fruit prickle and stopped before flowering. The base cells of flagelliform-shaped fruit prickle underwent the second proliferation, and the top cells began to elongate after pollination (Figure 1). As we know, the cells of mature fruit prickles contain granules, a high cytoplasm content, a range of plastids, and thin cell walls. In contrast, few organelles were observed in the cytoplasm of stem prickles cells with thick cell walls (Wang et al., 2021). Thereby, the developmental discrepancy may underlie the divergence in shape, distribution, and internal structure between stem prickles and fruit prickles.
Functional conserved RrTTG1 might modulate the formation of fruit prickle in Rosa roxburghii
The RrTTG1 protein has four WD40-repeat domains, which act as a scaffold for various protein–protein interactions for regulating the transcriptional processes (Stirnimann et al., 2010). We found that the TTG1 homologs from R. roxburghii and other dicotyledonous plants were clustered in the same branch (Figure 2). The nuclear localization and constitutive expression pattern of the RrTTG1 were consistent with that of TTG1, thereby suggesting that RrTTG1 may act as a transcription factor akin to TTG1 and have conserved functions (Zhao et al., 2008). TTG1 regulated epidermal cell fate specification, including trichome development in Arabidopsis (Payne et al., 2000; Lin and Schiefelbein, 2001). Trichome has served as an excellent model to study the molecular mechanism of prickle or other appendages of some plants lacking the genetic transformation system (Liu et al., 2016). Ectopic expression of RrTTG1 could increase the trichome number in Arabidopsis and rescue the glabrous phenotype in the ttg1 mutant (Figures 5, 6). This might be attributed to the conserved ability of RrTTG1 positively to interact with GL3 and EGL3 (Figure 6). Furthermore, RrTTG1 is highly expressed in the early stages of fruit prickles from fruit prickle primordium to young fruit prickles on 40 DAP fruits (Figure 3), which corresponds with TTG1 in the rosaceous plants is highly expressed in apical meristems or at the transition stages when cells differentiated to form stem prickles (Swarnkar et al., 2021; Zhong et al., 2021). Thus, the results demonstrated that RrTTG1 was homologous to the functions of TTG1 in Arabidopsis and might modulate the formation of fruit prickle.
RrTTG1 might function in fruit prickles formation through the RrTTG1-RrEGL3-RrGL1 complex in Rosa roxburghii
TTG1 is a central regulator by forming different MBW complexes to influence the development of appendages, including trichome, spine, prickle, and fiber (Zhang et al., 2003; Humphries et al., 2005). In Arabidopsis, TTG1 interacts with GL3 or EGL3 to regulate multiple processes in plants, such as trichome formation, anthocyanin production, and seed coat pigmentation, and negatively regulates root hair formation (Tian and Wang, 2020). Interestingly, GL3 plays a positive and critical role in trichome branching rather than EGL3 in Arabidopsis (Shen et al., 2006; Wang Z. et al., 2019). CsTTG1 is also an important regulator of fruit spine and wart formation in cucumber (Cucumis sativus) (Cui et al., 2016; Zhang et al., 2016). Regarding rice (Oryza sativa), only OsGL3B can interact with OsTTG1 to form the MBW complex to promote trichomes formation (Zheng et al., 2021). Similarly, SlTTG1 only interacts with SlEGL3 to regulate the trichomes formation in tomatoes (Solanum lycopersicum) (Tominaga-Wada et al., 2017). In our results, despite the conserved interaction of RrTTG1 with EGL3 or GL3, RrTTG1 only interacted with RrEGL3 and failed to bind RrGL3 (Figure 6). Furthermore, the expression of RrEGL3 was significantly higher than RrGL3 (Yan et al., 2021). Taken together, only RrEGL3 can interact with RrTTG1 to form an MBW complex in R. roxburghii.
The failure of RrTTG1 to interact with RrGL3 might attribute to the loss of function of RrTTG1 or RrGL3. The amino acid substitutions (S197F and L339F) of TTG1 could reduce its ability to interact with GL3 in Arabidopsis (Long and Schiefelbein, 2020). However, RrTTG1 shared the same amino acids with TTG1 at these two sites, indicting its conserved function of interacting with GL3 and fully restoring the deficient phenotype of the ttg1 mutant (Figures 4, 5). SlGL3 was found to lose its function of regulating trichome development (Wada et al., 2014). Similarly, a single amino acid substitution in the GL3 homologous gene MYC1 in Arabidopsis led to trichome pattern defects via the failure to interact with TTG1 (Zhao et al., 2012). The previous study showed that RrGL3 transcripts were absent during fruit development, and the ectopic expression of RrGL3 did not influence trichome patterning in Arabidopsis (Yan et al., 2021). Thus, the aborting of the RrTTG1-RrGL3 interaction is mainly attributed to the loss of function of RrGL3. By the way, the failure of the interaction of RrTTG1 with RrGL3 helped develop the non-branch fruit prickle in R. roxburghii.
A model of RrTTG1 function in the initiation and expansion of fruit prickles in Rosa roxburghii
The model of RrTTG1 function in the development of fruit prickles in R. roxburghii was shown (Figure 8). Fruit prickles’ formation undergoes three stages: initiation, enlargement, and lignification. The initiation stage corresponds to the occurrence of the fruit prickle primordium. The primordium derives from the meristem beneath the protoderm and forms the flagelliform trichome on the surfaces of the receptacle until the flower blooms. After pollination, with the receptacle gradually enlarging, the flagelliform prickles continue to grow via cell division and expansion. The lignification stage is defined as the mature stage when the hard and sharp fruit prickles have formed on the surface of the fruit. RrTTG1 only interacts with the bHLH transcription factor RrEGL3 to form an MBW complex (RrTTG1-RrEGL3-RrGL1) and fails to interact with RrGL3. The RrTTG1-RrEGL3-RrGL1 is highly expressed from fruit prickle primordia to young fruit prickle. Thus, RrTTG1-RrEGL3-RrGL1 might modulate the primordia initiation and enlargement of fruit prickles in R. roxburghii.
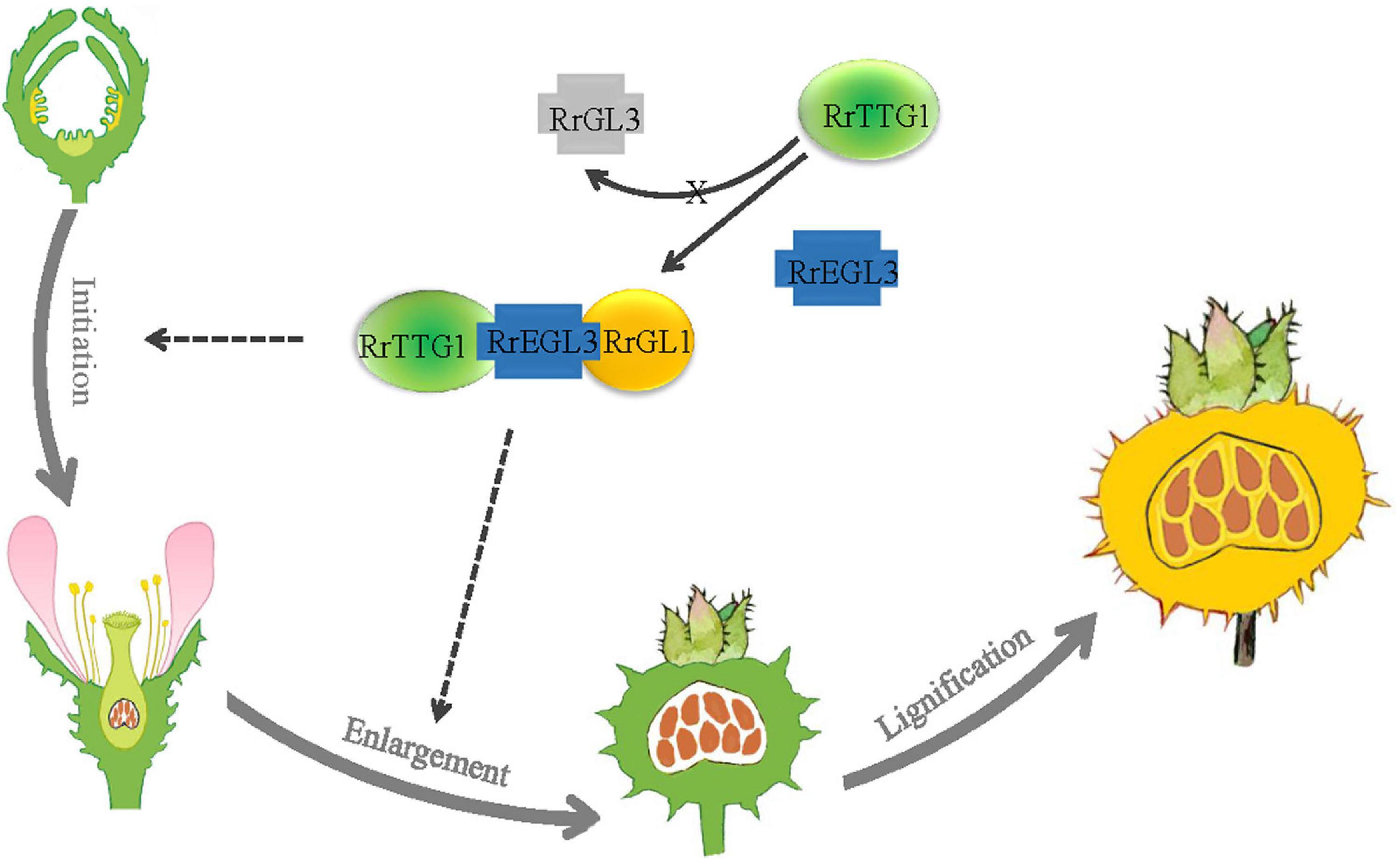
Figure 8. Model of RrTTG1 function in fruit prickles formation in R. roxburghii. Fruit prickles’ formation undergoes three stages: initiation, enlargement, and lignification. RrTTG1 only interacts with RrEGL3 to form the RrTTG1-RrEGL3-RrGL1 complex and fails to interact with RrGL3. The RrTTG1-RrEGL3-RrGL1 are highly expressed from fruit prickle primordia to young fruit prickle. Thus, RrTTG1-RrEGL3-RrGL1 might modulate the primordia initiation and enlargement of fruit prickles in R. roxburghii. The lines and dotted lines represent the results and speculations, respectively.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the NCBI repository, accession number: ON381945.
Author contributions
XH: writing, visualization, and editing. PY: formal analysis and investigation. YL, YJ, and QL: conceptualization and funding acquisition. YY and HY: project administration and supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (grant nos. 32060587 and 32160699), the Guizhou Province Natural Science Foundation (grant no. QKHJC-ZK[2022]ZD032), the Joint Fund of the National Natural Science Foundation of China, the Karst Science Research Center of Guizhou Province (grant no. U1812401), and the Sichuan Science and Technology (grant nos. 2021YJ0299 and 2021YFYZ0023).
Acknowledgments
We are very grateful for Prof. Tao Xia (Anhui Agricultural University) for providing the ttg1 mutant seeds.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.939270/full#supplementary-material
Supplementary Figure 1 | Control experiment of yeast two-hybrid assay. The combinations of pGADT7 and pGBKT7-Lam, as well as pGADT7 and pGBKT7-53, were used as negative and positive control, respectively. Meanwhile, the combinations of empty vector (pGADT7 or pGBKT7) with all the tested proteins were also performed, and no self-activation was detected.
Footnotes
References
Balkunde, R., Bouyer, D., and Hulskamp, M. (2011). Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis. Development 138, 5039–5048. doi: 10.1242/dev.072454
Bernhardt, C., Lee, M. M., Gonzalez, A., Zhang, F., Lloyd, A., and Schiefelbein, J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130, 6431–6439. doi: 10.1242/dev.00880
Chen, M., Zhang, B., Li, C., Kulaveerasingam, H., Chew, F. T., and Yu, H. (2015). TRANSPARENT TESTA GLABRA1 regulates the accumulation of seed storage reserves in Arabidopsis. Plant Physiol. 169, 391–402. doi: 10.1104/pp.15.00943
Cui, J. Y., Miao, H., Ding, L. H., Wehner, T. C., Liu, P. N., Wang, Y., et al. (2016). A new glabrous gene (csgl3) identified in trichome development in cucumber (Cucumis sativus L.). PLoS One 11:e0148422. doi: 10.1371/journal.pone.0148422
Feng, L. G., Luan, X. F., Wang, J., Xia, W., Wang, M., and Sheng, L. X. (2015). Cloning and expression analysis of transcription factor RrTTG1 related to prickle development in rose (Rosa Rugosa). Arch. Biol. Sci. 67, 1219–1225.
Gietz, R. D., and Schiestl, R. H. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34.
Hibrand Saint-Oyant, L., Ruttink, T., Hamama, L., Kirov, I., Lakhwani, D., Zhou, N. N., et al. (2018). A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat. Plants 4, 473–484. doi: 10.1038/s41477-018-0166-1
Huang, X., Lu, Z., Wang, X., Ouyang, Y., Chen, W., Xie, K., et al. (2016). Imprinted gene OsFIE1 modulates rice seed development by influencing nutrient metabolism and modifying genome H3K27me3. Plant J. 87, 305–317. doi: 10.1111/tpj.13202
Huang, X., Yan, H., Zhai, L., and Yi, Y. (2019). GLABROUS1 from Rosa roxburghii Tratt regulates trichome formation by interacting with the GL3/EGL3 protein. Gene 692, 60–67. doi: 10.1016/j.gene.2018.12.071
Humphries, J. A., Walker, A. R., Timmis, J. N., and Orford, S. J. (2005). Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol. Biol. 57, 67–81. doi: 10.1007/s11103-004-6768-1
Kellogg, A. A., Branaman, T. J., Jones, N. M., Little, C. Z., and Swanson, J. D. (2011). Morphological studies of developing Rubus prickles suggest that they are modified glandular trichomes. Botany 89, 217–226.
Khadgi, A., and Weber, C. A. (2020). RNA-Seq analysis of prickled and prickle-free epidermis provides insight into the genetics of prickle development in red Raspberry (Rubus ideaus L.). Agronomy 10, 1904–1921.
Lee, L. Y., Fang, M. J., Kuang, L. Y., and Gelvin, S. B. (2008). Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4, 24. doi: 10.1186/1746-4811-4-24
Li, H., Fang, W., Wang, Z., and Chen, Y. (2022). Physicochemical, biological properties, and flavour profile of Rosa roxburghii Tratt, Pyracantha fortuneana, and Rosa laevigata Michx fruits: A comprehensive review. Food Chem. 366:130509. doi: 10.1016/j.foodchem.2021.130509
Lin, Y., and Schiefelbein, J. (2001). Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development 128, 3697–3705. doi: 10.1242/dev.128.19.3697
Liu, X., Bartholomew, E., Cai, Y., and Ren, H. (2016). Trichome-related mutants provide a new perspective on multicellular trichome initiation and development in cucumber (Cucumis sativus L). Front. Plant Sci. 7:1187. doi: 10.3389/fpls.2016.01187
Long, Y., and Schiefelbein, J. (2020). Novel ttg1 mutants modify root-hair pattern formation in Arabidopsis. Front. Plant Sci. 11:383. doi: 10.3389/fpls.2020.00383
Papatheodorou, I., Moreno, P., Manning, J., Fuentes, A. M., George, N., Fexova, S., et al. (2020). Expression atlas update: From tissues to single cells. Nucleic Acids Res. 48, D77–D83.
Payne, C. T., Zhang, F., and Lloyd, A. M. J. G. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362. doi: 10.1093/genetics/156.3.1349
Pourcel, L., Irani, N. G., Lu, Y., Riedl, K., Schwartz, S., and Grotewold, E. (2010). The formation of anthocyanic vacuolar inclusions in Arabidopsis thaliana and implications for the sequestration of anthocyanin pigments. Mol. Plant 3, 78–90. doi: 10.1093/mp/ssp071
Serna, L., and Martin, C. (2006). Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci. 11, 274–280. doi: 10.1016/j.tplants.2006.04.008
Shen, B., Sinkevicius, K. W., Selinger, D. A., and Tarczynski, M. C. (2006). The homeobox gene GLABRA2 affects seed oil content in Arabidopsis. Plant Mol. Biol. 60, 377–387. doi: 10.1007/s11103-005-4110-1
Stirnimann, C. U., Petsalaki, E., Russell, R. B., and Muller, C. W. (2010). WD40 proteins propel cellular networks. Trends Biochem. Sci. 35, 565–574.
Swarnkar, M. K., Kumar, P., Dogra, V., and Kumar, S. (2021). Prickle morphogenesis in rose is coupled with secondary metabolite accumulation and governed by canonical MBW transcriptional complex. Plant Direct 5:e00325. doi: 10.1002/pld3.325
Tian, H., and Wang, S. (2020). TRANSPARENT TESTA GLABRA1, a key regulator in plants with multiple roles and multiple function mechanisms. Int. J. Mol. Sci. 21, 4881–4895. doi: 10.3390/ijms21144881
Tominaga-Wada, R., Ota, K., Hayashi, N., Yamada, K., Sano, R., and Wada, T. (2017). Expression and protein localization analyses of Arabidopsis GLABRA3 (GL3) in tomato (Solanum lycopersicum) root epidermis. Plant Biotechnol. 34, 115–117. doi: 10.5511/plantbiotechnology.17.0418a
Wada, T., Kunihiro, A., and Tominaga-Wada, R. (2014). Arabidopsis CAPRICE (MYB) and GLABRA3 (bHLH) control tomato (Solanum lycopersicum) anthocyanin biosynthesis. PLoS One 9:e109093. doi: 10.1371/journal.pone.0109093
Walker, A. R., Davison, P. A., Bolognesi-Winfield, A. C., James, C. M., Srinivasan, N., Blundell, T. L., et al. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1350. doi: 10.1105/tpc.11.7.1337
Wang, D. J., Lu, M., Ludlow, R. A., Zeng, J. W., Ma, W. T., and An, H. M. (2021). Comparative ultrastructure of trichomes on various organs of Rosa roxburghii. Microsc. Res. Tech. 84, 2095–2103. doi: 10.1002/jemt.23765
Wang, D. J., Zeng, J. W., Ma, W. T., Lu, M., and An, H.-M. (2019). morphological and structural characters of trichomes on various organs of Rosa roxburghii. HortScience 54, 45–51.
Wang, Z., Yang, Z., and Li, F. (2019). Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton. Plant Biotechnol. J. 17, 1706–1722. doi: 10.1111/pbi.13167
Wester, K., Digiuni, S., Geier, F., Timmer, J., Fleck, C., and Hulskamp, M. (2009). Functional diversity of R3 single-repeat genes in trichome development. Development 136, 1487–1496. doi: 10.1242/dev.021733
Yan, H., Wu, Z., Liu, Y., Weng, Q., Yi, Y., and Huang, X. (2021). Functional divergence of RrGL3 and RrEGL3 from Rosa roxburghii in mediating trichome development. PCTOC 147, 313–324.
Zhang, F., Gonzalez, A., Zhao, M. Z., Payne, C. T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependentpathways of Arabidopsis. Development 130, 4859–4869.
Zhang, L., Sun, H., Xu, T., Shi, T., Li, Z., and Hou, W. (2021). Comparative transcriptome analysis reveals key genes and pathways involved in prickle development in eggplant. Genes 12, 341–355. doi: 10.3390/genes12030341
Zhang, S., Liu, S., Miao, H., Wang, M., Liu, P., Wehner, T. C., et al. (2016). Molecular mapping and candidate gene analysis for numerous spines on the fruit of cucumber. J. Hered. 107, 471–477.
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhao, H., Wang, X., Zhu, D., Cui, S., Li, X., Cao, Y., et al. (2012). A single amino acid substitution in IIIf subfamily of basic helix-loop-helix transcription factor AtMYC1 leads to trichome and root hair patterning defects by abolishing its interaction with partner proteins in Arabidopsis. J. Biol. Chem. 287, 14109–14121. doi: 10.1074/jbc.M111.280735
Zhao, M., Morohashi, K., Hatlestad, G., Grotewold, E., and Lloyd, A. J. D. (2008). The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135, 1991–1999. doi: 10.1242/dev.016873
Zheng, K., Wang, X., Wang, Y., and Wang, S. (2021). Conserved and non-conserved functions of the rice homologs of the Arabidopsis trichome initiation-regulating MBW complex proteins. BMC Plant Biol. 21:234. doi: 10.1186/s12870-021-03035-0
Zhong, M. C., Jiang, X. D., Yang, G. Q., Cui, W. H., Suo, Z. Q., Wang, W. J., et al. (2021). Rose without prickle: Genomic insights linked to moisture adaptation. Natl. Sci. Rev. 8, 92–103. doi: 10.1093/nsr/nwab092
Zhou, N., Simonneau, F., Thouroude, T., Oyant, L. H., and Foucher, F. (2021). Morphological studies of rose prickles provide new insights. Hortic. Res. 8:221. doi: 10.1038/s41438-021-00689-7
Keywords: Rosa roxburghii, RrTTG1, fruit prickle, trichome, MBW complex
Citation: Huang X, Yi P, Liu Y, Li Q, Jiang Y, Yi Y and Yan H (2022) RrTTG1 promotes fruit prickle development through an MBW complex in Rosa roxburghii. Front. Plant Sci. 13:939270. doi: 10.3389/fpls.2022.939270
Received: 09 May 2022; Accepted: 28 July 2022;
Published: 29 August 2022.
Edited by:
Ran Xu, Hainan University, ChinaReviewed by:
Xiang Gao, Northeast Normal University, ChinaJinzhi Zhang, Huazhong Agricultural University, China
Copyright © 2022 Huang, Yi, Liu, Li, Jiang, Yi and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiqing Yan, eWFuaHVpcWluZ0Bnem51LmVkdS5jbg==
†These authors have contributed equally to this work
‡ORCID: Huiqing Yan, orcid.org/0000-0001-9333-6336
 Xiaolong Huang
Xiaolong Huang Peipei Yi1,2,3†
Peipei Yi1,2,3† Qiaohong Li
Qiaohong Li Huiqing Yan
Huiqing Yan