- National Key Laboratory of Crop Genetics and Germplasm Enhancement, Key Laboratory for Biology and Genetic Improvement of Soybean (General, Ministry of Agriculture), National Center for Soybean Improvement, Jiangsu Collaborative Innovation Center for Modern Crop Production, Nanjing Agricultural University, Nanjing, China
Alkaline soil has a high pH due to carbonate salts and usually causes more detrimental effects on crop growth than saline soil. Sodium hydrogen exchangers (NHXs) are pivotal regulators of cellular Na+/K+ and pH homeostasis, which is essential for salt tolerance; however, their role in alkaline salt tolerance is largely unknown. Therefore, in this study, we investigated the function of a soybean NHX gene, GmNHX6, in plant response to alkaline salt stress. GmNHX6 encodes a Golgi-localized sodium/hydrogen exchanger, and its transcript abundance is more upregulated in alkaline salt tolerant soybean variety in response to NaHCO3 stress. Ectopic expression of GmNHX6 in Arabidopsis enhanced alkaline salt tolerance by maintaining high K+ content and low Na+/K+ ratio. Overexpression of GmNHX6 also improved soybean tolerance to alkaline salt stress. A single nucleotide polymorphism in the promoter region of NHX6 is associated with the alkaline salt tolerance in soybean germplasm. A superior promoter of GmNHX6 was isolated from an alkaline salt tolerant soybean variety, which showed stronger activity than the promoter from an alkaline salt sensitive soybean variety in response to alkali stress, by luciferase transient expression assays. Our results suggested soybean NHX6 gene plays an important role in plant tolerance to alkaline salt stress.
Introduction
Saline-alkali soils currently account for 20% of irrigated land (Bouras et al., 2022). Land alkalization has become a major and increasingly serious problem in the world. Compared with neutral salt stress, the high pH environment in alkaline soil destroys soil structure and affects the absorption of essential elements such as phosphorus and iron by plants (Shi and Wang, 2005). Therefore, alkaline salt causes more serious effects than neutral salt stress on plants. Soil alkalization can reduce soil osmotic potential, and cause ion imbalance, disrupt physiological processes, inhibit the growth and development of plants, leading to a serious decline in the yield and quality, and even the death of plants (Yang et al., 2011). Saline-alkali stress leads to leaf wilting and chlorosis (Sun et al., 2021), fewer pods, seeds and lower 100-seed weight (He et al., 2020), inhibited nodule development (He et al., 2021) and other phenomena (Chen et al., 2018) of soybean, resulting in soybean growth retardation, and eventually lead to a serious decline in soybean yield (He et al., 2020). Plants can effectively reduce the damage caused by salt-alkali stress through ion selective absorption, compartmentalization and scavenging reactive oxygen species, etc. (Zhu, 2001; Zhang et al., 2019a). Sodium hydrogen exchanger (NHXs) are integral membrane proteins residing in the plasma membrane, endosome (Dragwidge et al., 2019), and vacuole (Sellamuthu et al., 2020), which belong to the monovalent cation/proton antiporter family (Bassil et al., 2011b). NHXs play important roles in regulating of cellular ion homeostasis (Bassil et al., 2019), pH (Reguera et al., 2015), vesicle trafficking (Bassil et al., 2019), protein transport (Dragwidge et al., 2019), auxin transport (Zhang et al., 2020a), salt tolerance (Long et al., 2020), cell turgor and expansion (Walker et al., 1996), as well as growth and development (Bassil et al., 2019) in many species ranging from bacteria to human. In plants, NHX utilizes an H+ electrochemical gradient established by (H+)-ATPase (in the plasma membrane) and (H+)-PPase (in vacuoles) to allow Na+ or K+ exchange for H+, to maintain pH and ion homeostasis (Rodríguez-Rosales et al., 2009; Bassil et al., 2012).
In Arabidopsis thaliana, the NHX exchanger family contains eight members which are divided into three subclasses based on their subcellular localizations: vacuolar (AtNHX1-AtNHX4), endosomal (AtNHX5 and AtNHX6), and plasma membrane (AtNHX7/SOS1and AtNHX8) localized exchangers (Reguera et al., 2014; Qiu, 2016a). Plasma membrane NHXs (AtNHX7/SOS1and AtNHX8) are required for Na+, K+, and pH homeostasis, and play an important role in salt tolerance (An et al., 2007; Oh et al., 2010). Vacuolar membrane NHXs (AtNHX1-AtNHX4) are critical for vacuolar pH and K+ homeostasis (Barragán et al., 2012), salt and drought stress responses (Moghaieb et al., 2014; Zhang et al., 2019b), osmotic adjustment (Quintero et al., 2000), flower development (Bassil et al., 2011b), as well as plant growth and development (Qiu, 2016a). AtNHX5 and AtNHX6 localize to the Golgi and trans-Golgi network (TGN), and are important for plant growth and response to salt stress (Bassil et al., 2011b), maintaining ion and pH homeostasis (Wang et al., 2015), protein transport (Qiu, 2016b), plant growth and development (Lv et al., 2020), and seedling growth (Zhang et al., 2020b). The Arabidopsis nhx5 nhx6 double mutant showed reduced growth, smaller and fewer cells, smaller rosettes and shorter seedlings, late flowering, and increased sensitivity to salinity (Bassil et al., 2011a). The nhx5 nhx6 syp22 triple mutant had short siliques and low seed setting rate, but larger seeds. In addition, the triple mutant had numerous smaller protein storage vacuoles (PSVs) and accumulated precursors of seed storage proteins, suggesting that AtNHX5 and AtNHX6 play important roles in seed production, protein trafficking and PSV biogenesis (Wu et al., 2016). A recent report suggests that NHX5 and NHX6 might regulate auxin transport through endoplasmic reticulum (Fan et al., 2018).
There are also few studies on NHX5 or NHX6 from other plant species in addition to Arabidopsis. Overexpression of PdNHX6 from Phoenix dactylifera in Arabidopsis plants enhanced salt tolerance, retained higher chlorophyll and water content, maintained a balanced Na+/K+ ratio, and increased seed germination under salinity when compared to the wild-type plants (Al-Harrasi et al., 2020). AoNHX6 from Avicennia officinalis showed high expression levels in the roots, and complementation with AoNHX6 improved the tolerance of yeast mutants and Arabidopsis mutants to both NaCl and KCl stress (Krishnamurthy et al., 2019). The ectopic expression of endosomal-type MnNHX6 from Morus notabilis in Arabidopsis and nhx1 yeast mutant can greatly enhance their salt tolerance compared with vacuolar-type MnNHXs (Cao et al., 2020). Up to now, the role of NHX genes in soybean tolerance to salt-alkali stress is largely unknown, except that GmNHX5 is found regulating salt tolerance in a recent study (Sun et al., 2021). Alkaline soils cause damage to plants not only through salt stress, but also through high pH (Shi and Wang, 2005), therefore, alkali stress is more serious than salt stress, while the role of NHX genes in alkali tolerance of soybean remains unclear.
In our previous transcriptomic study (unpublished) using an alkaline salt tolerant wild soybean variety, we identified an alkali-responsive gene (corresponding to Glyma.09G018200 in the reference genome of soybean variety Williams 82), which encodes a sodium/hydrogen exchanger, and is designated as GmNHX6. Here we investigated the role of GmNHX6 in alkali stress by comparing the relative expression level of GmNHX6 gene in alkaline salt tolerant and sensitive soybean varieties, and study the function of this gene in transgenic soybean composite plants and Arabidopsis. The sequence variation in GmNHX6 and its association with alkaline salt tolerance was also analyzed, to gain a better understanding of its role in alkaline salt tolerance. The possible molecular mechanism of GmNHX6 in response to alkaline salt stress was investigated by the content of Na+, K+, the Na+/K+ ratio, and promoter luciferase (LUC) assay. This study aims to provide new insights into the role of GmNHX6 in soybean tolerance to alkaline salt stress and the possible underlying mechanisms.
Materials and methods
Soybean accessions and sodic tolerance rating
A total of 60 soybean accessions (Supplementary Table 1), were obtained from the National Center for Soybean Improvement (Nanjing, China). Twelve seeds of each soybean accession were germinated in plastic pots (φ10 × 8 cm) filled with sterile nutrient soil, and irrigated with 1/2 Hoagland nutrient solution form bottom, at 28°C (day)/24°C (night) with 14 h (light)/10 h (dark) photoperiod. Fifteen pots were placed in a plastic tray (55 cm × 36 cm × 8 cm) containing 2 L fresh 1/2 Hoagland nutrient solution (pH ≈ 6.5), and the solution was changed every 2 days. After emergence, the seedings were thinned to four plants in each pot. When the second trifoliolate leaves appeared (12-day-old soybean seedlings), plants were treated with 1/2 Hoagland solution containing 0 or 90 mM NaHCO3. Sixteen days after treatment, the alkaline salt tolerance was determined by Sodic Tolerance Rating (STR) according to the previously published method (Tuyen et al., 2010).
RNA extraction and gene expression analysis
RNA-seq data from different soybean tissues were downloaded from Soybase.1 To experimentally determine the expression of GmNHX6, tissues were collected from soybean plants grown in vermiculite in greenhouse with controlled temperature (day/night, 28°C/24°C) and light cycle (day/night, 14/10 h). Arabidopsis thaliana plants were grown under controlled temperatures of day/night as 22/22°C and light cycle of day/night for 16/8 h. Fresh tissues were ground and extracted for total RNA using an RNAprep Pure Plant Kit (Tiangen Biotech, China). The gene specific primers were designed using the Primer Premier 5 software2 and listed in Supplementary Table 2. The cDNA was synthesized by the PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa, Japan). The qRT-PCR was conducted using SYBR Premix ExTaq™ II Mix (TaKaRa, Japan) on a Roche 480 Real-time detection system (Roche Diagnostics, Switzerland) according to the manufacturers’ instructions. Each experiment was performed in triplicates. Transcript levels in soybean plants were calculated in relative to the reference gene GmUKN1 (Guan et al., 2014), using the 2-△△CT methods (Livak and Schmittgen, 2001). The expression of GmUKN1 is stable across all samples in this study (Supplementary Table 3). The qRT-PCR of GmNHX6 gene in transgenic A. thaliana was conducted using the OE-2 line (with lowest expression level of GmNHX6) as the control (relative expression = 1) and AtACTIN7 (Miao et al., 2020) as the reference gene. Data was collected from three biological replicates. The amplification efficiencies (E) of primer pairs were estimated (Supplementary Table 2) by qRT-PCR using 1:10, 1:20, 1:40, 1:80, and 1:160 dilutions of cDNA templates, according to the equation: E = [10−1/slope] − 1 (Pfaffl, 2001).
Sequence alignment, phylogenetic analysis, and sequence analysis
The full sequences of GmNHX6 and other NHXs proteins obtained from Phytozome,3 were used for multiple sequence alignments by ClustalW2.4 The unrooted phylogenetic tree was then constructed using MEGA 6.0 (Tamura et al., 2013) based on the Maximum Likelihood (ML) algorithm with 1,000 bootstraps. The putative promoter sequence of 2000-bp upstream GmNHX6 was analyzed using the PlantCARE.5 Protein transmembrane topology and signal peptides were predicted from amino acid sequences using Protter database.6
Subcellular localization of GmNHX6
For subcellular localization of GmNHX6 protein, the open reading frame (ORF) of GmNHX6 was amplified by PCR using specific primers (Supplementary Table 2). The PCR product was then introduced into the pAN580 vector, which contains a green florescence protein (GFP) reporter gene, under the drive of CaMV 35S promoter for N-terminal GFP fusion (Zhang et al., 2016), using the ClonExpress II One Step Cloning Kit (Vazyme, China) according to the manufacturer’s protocol. To further determine the specific subcellular localization of GmNHX6, the fluorescent marker protein, Man1-mCherry, which is characteristic for the cis-Golgi (Tse et al., 2004), was co-expressed with GmNHX6. The transient expression of above proteins in Arabidopsis mesophyll protoplasts was performed following the published methods (Wu et al., 2009). Confocal imaging analysis was performed using a laser scanning microscope (Zeiss LSM780 META, Jena, Germany).
Obtaining and phenotyping of transgenic soybean composite plants
The coding sequence of GmNHX6 from soybean accession M8206 was cloned into pBinGFP4 vector under the control of CaMV 35S promoter. The construct containing 35S:GmNHX6 and the empty vector pBinGFP4, were separately transformed into Agrobacterium rhizogenes strain K599 (Kereszt et al., 2007), then used to infect soybean hypocotyls of an alkaline salt sensitive soybean variety Tianlong 1, to obtain transgenic soybean composite plants, according to the previously described method (Kereszt et al., 2007). After 20 days of plant growth, the green fluorescence signals of GFP in positive hairy roots were identified at 488 nm wavelength using a stereoscopic fluorescence microscope (Mshot, China), and the non-transgenic roots were cut-off. The transgenic soybean composite plants were treated with 1/2 Hoagland solution containing 0 or 90 mM NaHCO3 for 7 days, then the Soil and Plant Analysis Development (SPAD) value for chlorophyll content (Pérez-Patricio et al., 2018) and leaf relative water content (LRWC) (Zhang and Zhang, 2021) were measured. The SPAD values of the second, third and fourth trifoliolate leaves were recorded 10 times each by the Chlorophyll Meter SPAD-502 Plus (Konica Minolta, Tokyo, Japan), and the average value was calculated as the SPAD value of the plant. To measure LRWC, the fresh weight (FW) of the second, third and fourth trifoliolate leaves was recorded, and then the leaves were incubated in sterile water at room temperature for 12 h, and the turgid fresh weight (TFW) was measured. The fully turgid leaves were then dried at 80°C for 72 h and the dry weight (DW) was recorded. LRWC was measured according to the following equation: LRWC = (FW – DW)/(TFW – DW) × 100%. Three biological replications were performed and five independent transgenic plants (n = 3 × 5 = 15) were measured for each repeat.
Functional analysis of GmNHX6 in Arabidopsis thaliana
The coding sequence of GmNHX6 from soybean accession M8206 was cloned into pCAMBIA3301 vector under the control of CaMV 35S promoter. The pCAMBIA3301-35S:GmNHX6 vector was introduced into the A. tumefaciens strain EHA105 for A. thaliana transformation using Col-0 wild-type (WT) plants by the floral dipping method (Clough and Bent, 1998). Transgenic plants were screened based on 20 mg L−1 glufosinate (PhytoTech, United States) resistance (Ren et al., 2009). The homozygous T3 generation plants were subjected to GmNHX6 gene expression analyses and phenotypic evaluation. The Arabidopsis nhx6 T-DNA insertion mutant (stock, SALK-100042C, nhx6, Col-0 background) used in this study was obtained from the Arabidopsis Biological Resource Center.7
To measure the seed germination rate of A. thaliana, 36 stratified seeds for each of three independent GmNHX6-overexpressed lines (OE-1, OE-9, and OE-18), as well WT and nhx6 mutant, were plated on 1/2 MS agar medium, or the media supplemented with 150 mM NaCl (Zheng et al., 2021) or 8 mM NaHCO3 (Zhu et al., 2011), and then placed in a growth chamber under the long-day growth conditions (16 h light/8 h dark cycle at 22°C ± 2°C). The rates of cotyledon greening were measured as seed germination rates after 7 days. To measure the root length and fresh weight of Arabidopsis seedlings, the sterilized seeds of Arabidopsis were vertically grown on 1/2 MS agar medium for 7 days, and then the seedlings were transplanted to fresh 1/2 MS agar medium, or media supplemented with 150 mM NaCl (Zheng et al., 2021) or 8 mM NaHCO3 (Zhu et al., 2011) for another 10 days. The root length was analyzed using Image J software.8
Determination of ion content
Seven-day-old Arabidopsis seedlings were transplanted to fresh 1/2 MS agar medium, or media supplemented with 150 mM NaCl or 8 mM NaHCO3 for another 10 days. Then the seedlings were used for Na+ and K+ content determination. To determine the ion content, plants were rinsed thoroughly in deionized water, separated into roots and shoots, inactivated at 105°C for 2 h, and oven dried at 80°C until they attained a constant mass. The 0.1 g dry material was mineralized using 2 ml 10% HNO3. Then, the microwave digestion system ETHOS T (Milestone, Italy)was used for ion extraction, and the ion contents in the supernatants were analyzed using an Optima 8,000 ICP-OES DV Spectrometer (PerkinElmer, United States) according to the manufacturer’s instructions (Munns et al., 2012). Three biological replicates were performed.
Analysis of sequence variation and regional association study
Nucleotide sequences were downloaded from phytozome v10 (see Footnote 3), and the 2000-bp upstream of the start codon ATG was considered as the promoter region. The full-length coding sequences were amplified using soybean cDNA as template, and the promoter regions were amplified using soybean genomic DNA as template. The amplicons were sequenced at GenScript (Nanjing, China), and sequence variations among soybean accessions were analyzed. Regional association study was performed using TASSEL 5.0 software (Bradbury et al., 2007). The threshold for a significant association was determined using the previously published method (Yang et al., 2014; Wang et al., 2020), which is p < 3.3 × 10−2 (1/n, where n is the number of markers) in this study.
Promoter-LUC assays in tobacco leaves
The promoter activities were analyzed by promoter-LUC transient expression assays in tobacco (Nicotiana benthamiana) leaves, according to the procedures described previously (Jin et al., 2021). For vector constructs, the full-length 2-kb promoter regions of M8206 and ZY were amplified, respectively, and the amplicons were inserted into pGreenII0800-LUC to obtain different promoter-LUC vectors. Above constructs with LUC as a reporter, Pro(ZY)-LUC, Pro(M)-LUC, were transformed into A. tumefaciens strain GV3101 that carries pSoup-19, and introduced into tobacco leaves by infiltration. Then, the tobacco seedlings were transferred to 0 or 100 mM NaHCO3 for 16 h. LUC activity was observed with an in vivo plant imaging system (Berthold LB 985, Germany).
Statistical analyses
For qRT–PCR analyses, at least four individual plants were pooled per tissue sample, and at least three qRT–PCR reactions (technical replicates) with at least three biological replicates were performed. For phenotypic evaluation, at least 12 individual plants were analyzed per accession, and the exact numbers of individuals (n) are presented in all figure legends. Differences between groups or genotypes were analyzed using two-sided Wilcoxon test, Duncan’s multiple range test or Student’s t-test by R software and SAS 9.2 (SAS Institute Inc., Cary, NC, United States).
Results
GmNHX6 encodes a Golgi-localized sodium/hydrogen exchanger
GmNHX6 gene encodes a sodium/hydrogen exchanger consisting of 534 amino acids. Phylogenetic analyses of GmNHX6 with NHX proteins in Arabidopsis showed that GmNHX6 is closely related with AtNHX6 (Figure 1A). Prediction of GmNHX6 transmembrane topology displays 12 transmembrane domains (Supplementary Figure 1), sharing high similarities with AtNHX6. GmNHX6 contains four conserved acidic residues (Al-Harrasi et al., 2020) in transmembrane domains (Supplementary Figure 2), which were previously proven to be essential for K+ transport (Wang et al., 2015). To determine the subcellular localization of GmNHX6, the GmNHX6 gene was expressed in fusion with the GFP reporter gene, in the pAN580-GFP expression vector. The constructed vector and empty control vector were transferred to the protoplast of Arabidopsis, and the location of GFP or its fusion protein was observed. We found that, GFP itself was distributed evenly in the cytoplasm and the nucleus, whereas the GmNHX6–GFP fusion protein was mainly localized to the cytoplasm and to punctate compartments in the cytosol (Figure 1B). To determine the nature of these punctate compartments, we co-expressed GmNHX6–GFP and the fluorescent marker characteristic for the Golgi, Man1-mCherry (Tse et al., 2004). The co-expression results showed that GmNHX6-GFP fusion protein was mainly located in the Golgi (Figure 1C).
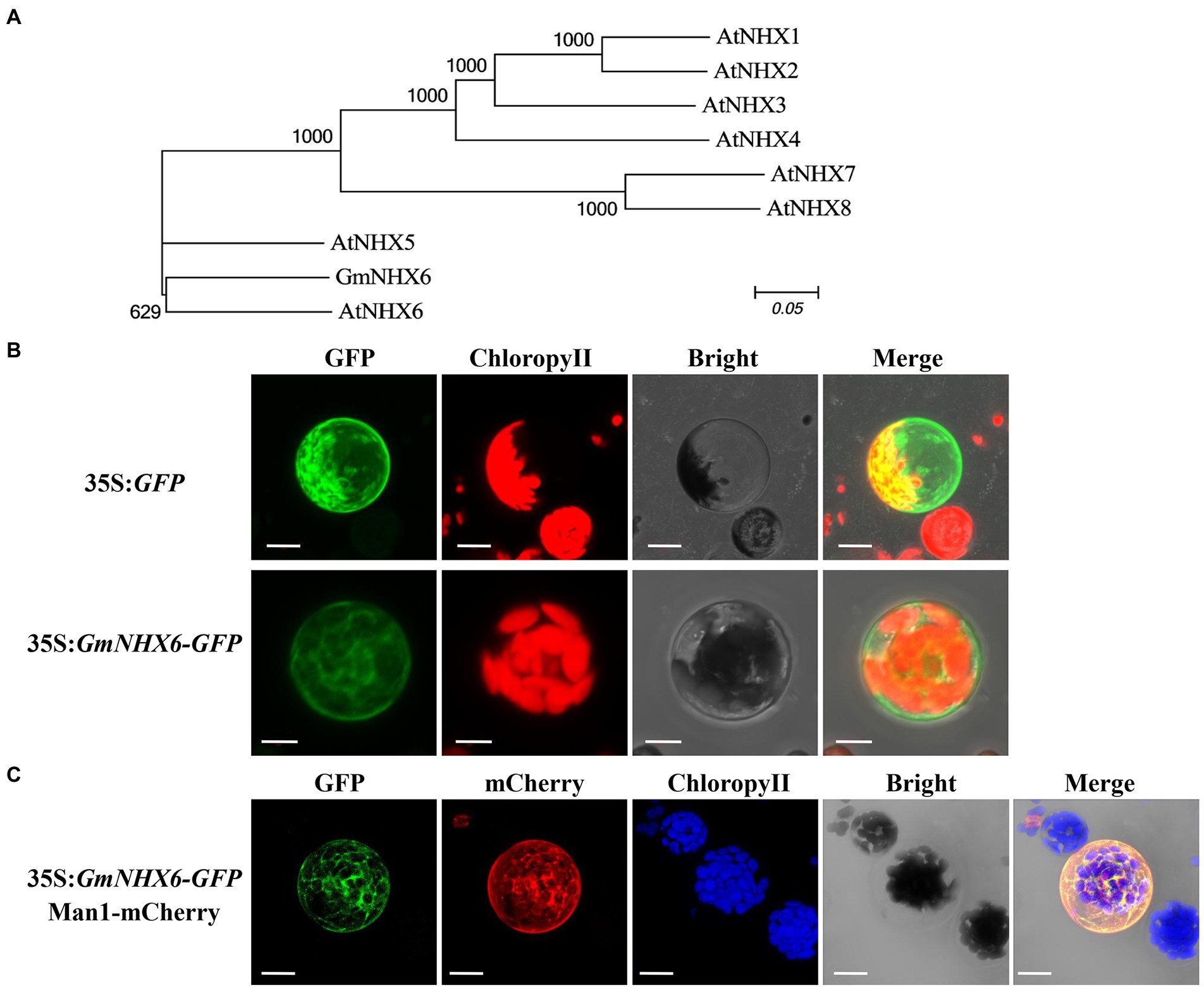
Figure 1. Phylogenetic analysis and subcellular localization of GmNHX6. (A) Phylogenetic analysis of GmNHX6 (XP_028181010.1) from soybean Glycine max and NHX proteins from Arabidopsis thaliana including AtNHX1 (NP_198067.1), AtNHX2 (NP_187154.1), AtNHX3 (NP_200358.1), AtNHX4 (NP_187288.2), AtNHX5 (NP_175839.2), AtNHX6 (NP_178079.2), AtNHX7 (NP_178307.2), AtNHX8 (NP_172918.2). The sequences of NHX proteins were downloaded from the Phytozome database. Unrooted phylogenetic tree was constructed using MEGA6.0 based on the Maximum Likelihood algorithm with 1,000 bootstraps. (B) Subcellular localization of GmNHX6. GFP protein and GmNHX6-GFP fusion protein expressed under the control of CaMV 35S promoter in Arabidopsis protoplasts were observed under a confocal microscope. Bar, 10 μm. (C) Images of Arabidopsis mesophyll protoplasts co-expressing GmNHX6-GFP fusion protein and a mCherry Golgi apparatus marker (Man1-mCherry). Bars = 10 μm.
The expression of GmNHX6 is more upregulated in alkaline salt tolerant soybean in response to NaHCO3 stress
The phenotypic difference between two soybean varieties of M8206 (alkaline salt tolerant) and ZY (alkaline salt sensitive) was obvious under 90 mM NaHCO3 stress (Figure 2A): ZY already showed wilted leaves at 24 h, but the leaves of M8206 were still normal. The leaves of ZY became yellow and dry out at 48 h and more severe at 72 h, while the leaves of M8206 did not show obvious wilting at 48 h until 72 h. The expression patterns of GmNHX6 in G. max (see Footnote 1) showed that the transcript abundance of GmNHX6 was higher in the young_leaf and root than the other organs (Supplementary Figure 3). The relative expression levels of GmNHX6 in response to alkaline salt stress (90 mM NaHCO3) in two soybean varieties were investigated using qRT-PCR. GmNHX6 gene expression was induced by 90 mM NaHCO3 treatment in the roots and leaves of both soybean varieties (Figures 2B,C). The relative expression level of GmNHX6 gene in the roots of M8206 (alkaline salt tolerant) showed significantly greater increase at 9, 12, 24, and 48 h after 90 mM NaHCO3 treatment than the alkaline salt sensitive soybean variety ZY (Figure 2B), and the relative expression level of GmNHX6 gene in the leaves of M8206 showed significantly greater increase at 9, 12, and 24 h in response to NaHCO3 treatment than ZY (Figure 2C). There was no significant difference in GmNHX6 gene expression between two soybean varieties under normal condition (Supplementary Figure 4). These results suggested that GmNHX6 might play an important role in soybean response to alkaline salt stress.
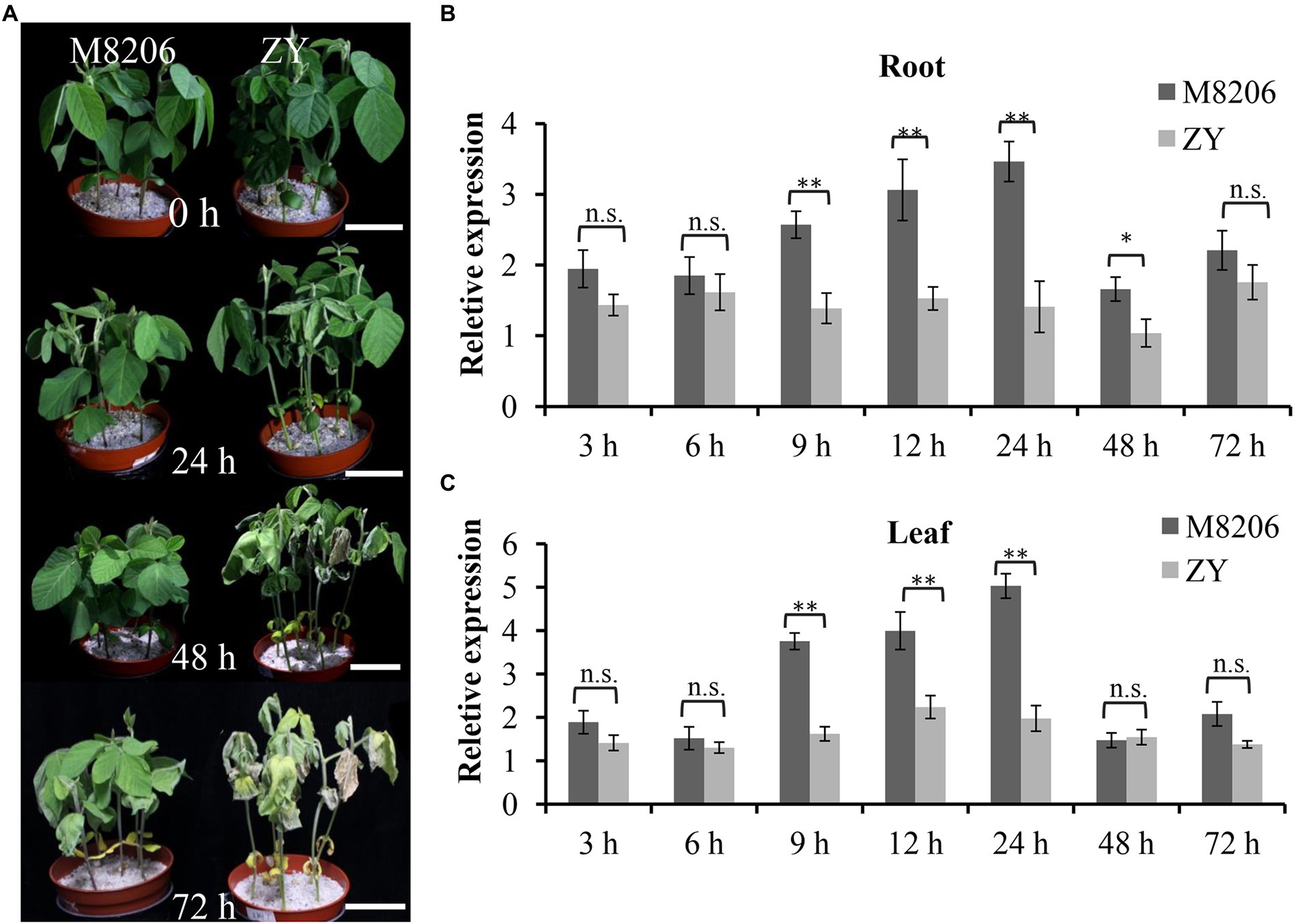
Figure 2. Phenotype and relative expression of GmNHX6 in two soybean varieties in response to alkali stress. (A) Phenotypes of two soybean varieties at 0, 24, 48, and 72 h after 90 mM NaHCO3 treatment. The 12-day-old seedlings were subjected to treatment. Bar = 5 cm. (B,C) Relative expression of GmNHX6 in response to alkali stress in roots and leaves, respectively. Soybean seedlings were treated with 0 or 90 mM NaHCO3 for 3, 6, 9, 12, 24, 48, and 72 h. Roots and leaves receiving 0 mM NaHCO3 treatment at each time point were used as controls. Data represents the mean ± standard deviation of three biological replications with three repeats within each replication (n = 3 × 3 = 9). Differences between two soybean varieties were evaluated using the two-tailed Student’s t-tests (n.s., not significant; *p < 0.05; **p < 0.01).
Overexpression of GmNHX6 improved soybean tolerance to NaHCO3 stress
Since a greater increase in the expression level of GmNHX6 was observed in alkaline salt tolerant soybean variety than the sensitive variety after NaHCO3 treatment, we next tested whether overexpression of GmNHX6 could improve soybean tolerance to alkaline salt stress or not. The coding region of GmNHX6 gene was expressed in fusion with GFP (while the empty vector 35S:GFP was used as control), and transformed into soybean hypocotyls to obtain transgenic composite plants, in the genetic background of an alkaline salt sensitive soybean variety Tianlong 1. The positive transgenic composite soybean plants were identified through GFP fluorescence signal in roots (Figure 3A). Under normal condition (0 mM NaHCO3), all soybean composite plants grew well, with no obvious difference (Figure 3B; Supplementary Figures 5A,E,I). When the transgenic soybean composite plants were treated with 90 mM NaHCO3 for 7 days, the 35S:GFP-transformed soybean composite plants showed obviously inhibited growth, leaf wilting and chlorosis, while the 35S:GmNHX6-GFP-transformed composite plants had much less damage (Figure 3B; Supplementary Figures 5A,E,I). The expression level of GmNHX6 gene was higher in 35S:GmNHX6-GFP-transformed soybean plants than the 35S:GFP-transformed plants either under 0 or 90 mM NaHCO3, and GmNHX6 gene expression was significantly upregulated at 24 h post 90 mM NaHCO3 treatment in 35S:GmNHX6-GFP-transformed soybean hairy roots (Figure 3C; Supplementary Figures 5B,F,J). The average SPAD value for chlorophyll content (Figure 3D; Supplementary Figures 5C,G,K) and the leaf relative water content (LRWC; Figure 3E; Supplementary Figures 5D,H,L) of 35S:GmNHX6-GFP-transformed soybean composite plants were significantly higher than those of the control plants transformed by the 35S:GFP under 90 mM NaHCO3 treatment. No significant differences were found in SPAD and LRWC between 35S:GmNHX6-GFP-transformed plants and 35S:GFP-transformed plants under normal condition (0 mM NaHCO3). These results demonstrated that overexpression of GmNHX6 reduced the damage of NaHCO3 treatment on soybean plants.
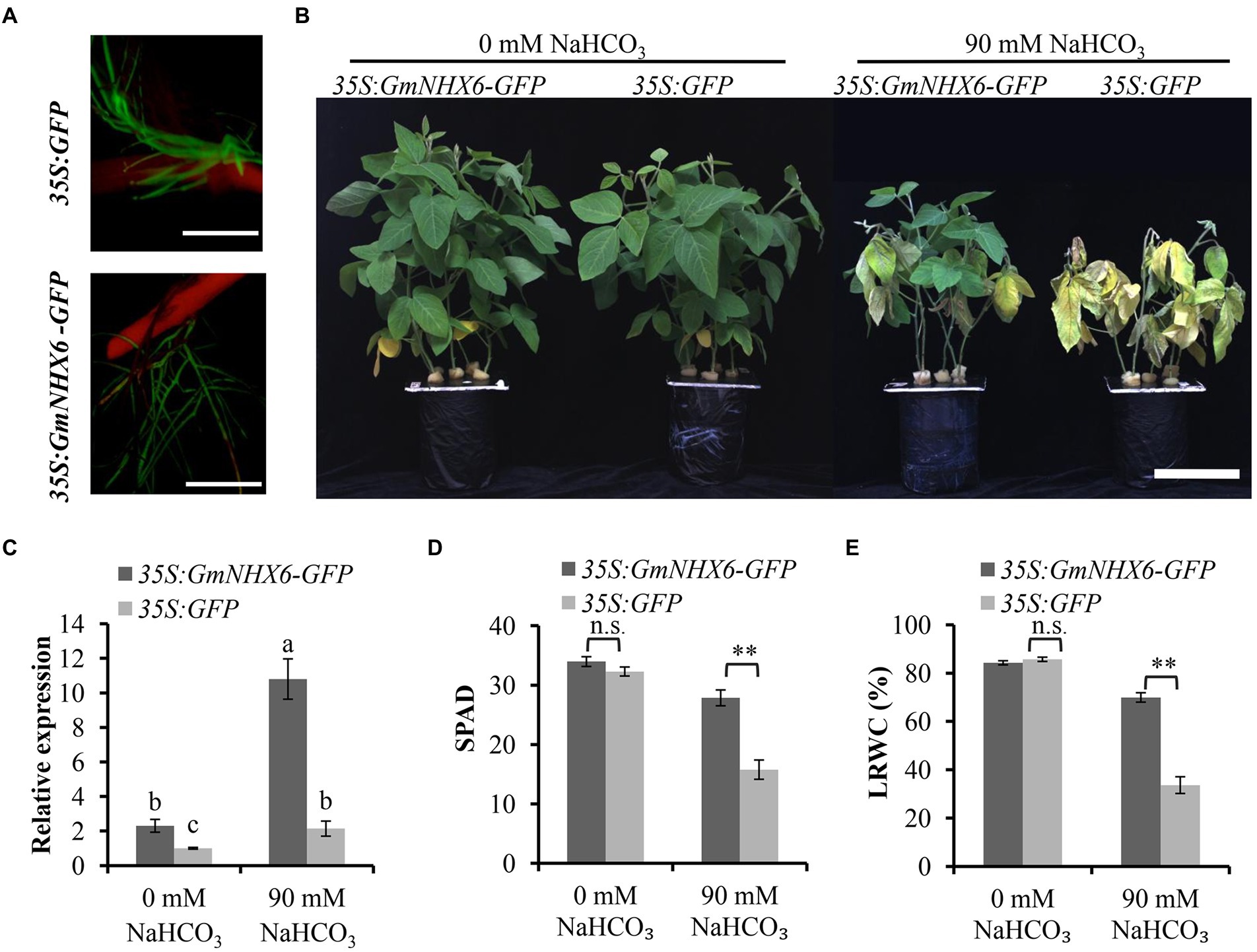
Figure 3. Alkaline salt tolerance analyses of transgenic soybean composite plants. (A) Identification of positive transgenic soybean lines by green fluorescence using a stereoscopic fluorescence microscope. Bar = 1 cm. (B) Representative phenotype of transgenic soybean composite plants (25-day-old) under 0 mM or 90 mM NaHCO3 for 7 days. Bar = 12 cm. (C) Relative expression of GmNHX6 gene in the roots of soybean composite plants at 24 h after 0 or 90 mM NaHCO3 treatment by qRT-PCR. For relative expression calculation, the sample from soybean composite plants with empty vector (35S:GFP) under 0 mM NaHCO3 was used as control and GmUKN1 was the reference gene. Data represents the mean ± standard deviation of three biological replications with three repeats within each replication (n = 3 × 3 = 9). Bars with the same letter in lowercase above bars indicate no significant difference according to Duncan’s multiple range test at 0.05 level. (D,E) The Soil and Plant Analysis Development (SPAD) value for chlorophyll content, and leaf relative water content (LRWC) of transgenic soybean composite plants under 0 mM or 90 mM NaHCO3 for 7 days, respectively. Data represents the mean ± standard deviation of three biological replications and each repeat contained five independent transgenic plants for each genotype (n = 3 × 5 = 15). Soybean variety of “TianLong1” was used. Differences were evaluated using the two-tailed Student’s t-tests (n.s., not significant; **p < 0.01).
Ectopic expression of GmNHX6 in Arabidopsis enhanced the alkaline salt tolerance by maintaining low Na+/K+ ratios
We further investigate the role of GmNHX6 in response to alkaline salt stress by transgenic Arabidopsis plants. GmNHX6 was overexpressed (OE) using the CaMV 35S promoter in Arabidopsis. Seven homozygous transgenic Arabidopsis lines containing 35S:GmNHX6 were obtained at T3 generation. Then three lines with higher GmNHX6 expression levels (OE-1, OE-18, and OE-9) were selected for further analyses (Supplementary Figure 6). The germination rates of Arabidopsis lines were compared in the absence or presence of NaHCO3, respectively. The results showed that there was no difference in germination rates between the wild type (WT) and GmNHX6 OE lines under normal condition without NaHCO3, but the germination rates of GmNHX6 OE lines were significantly higher than that of WT Arabidopsis under NaHCO3 stress (Figures 4A,B). To further test the effect of NaHCO3 stress on Arabidopsis, we measured the fresh weight and root length of Arabidopsis. Under normal condition, no significant difference was observed between GmNHX6 OE lines and WT (Figures 4C–E). After 10 days of 8 mM NaHCO3 treatment, GmNHX6 OE lines showed better growth than WT (Figure 4C): the average fresh weight and root length of GmNHX6 OE lines were significantly larger than those of WT Arabidopsis (Figures 4D,E). These results suggest that overexpression of GmNHX6 enhanced the alkaline salt tolerance in Arabidopsis.
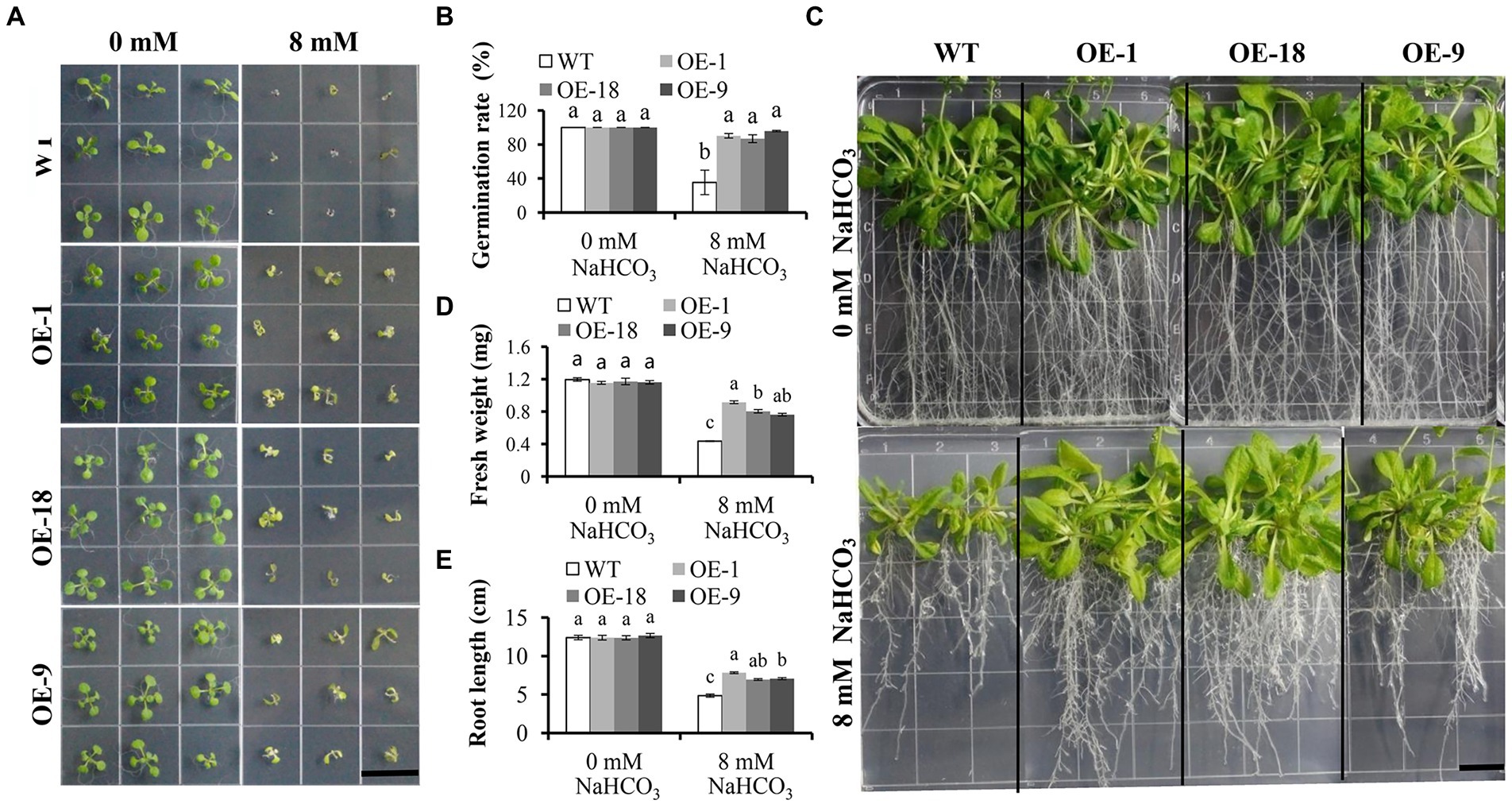
Figure 4. Effect of GmNHX6 overexpression on Arabidopsis tolerance to NaHCO3 treatment. (A) Seed germination assay of different Arabidopsis lines on 1/2 MS medium and 1/2 MS supplied with 8 mM NaHCO3. Photographs were taken 7 days after NaHCO3 treatment. WT: wild type; OE-1, OE-18, OE-9: Arabidopsis lines overexpressing GmNHX6. Bar = 1.5 cm. (B) Seed germination rates of different Arabidopsis lines as shown in A. Data represents the mean ± standard deviation of three biological replications and each repeat contained 36 seeds per line for each treatment (n = 36 × 3 = 108). (C) Phenotypes of Arabidopsis lines subjected to 0 and 8 mM NaHCO3 treatment, respectively. Seeds were grown vertically on 1/2 MS agar plates supplemented with 0 or 8 mM NaHCO3 for 10 days after seven days of normal growth. Bar = 1.5 cm. (D,E) Fresh weight and root length of Arabidopsis lines as shown in (C). Data represents the mean ± standard deviation of three biological replications and each repeat contained four plants per line for each treatment (n = 12). Bars with the same letter in lowercase above bars indicate no significant differences at the 0.05 level between lines under same treatment according to Duncan’s multiple range tests.
We also compared the content of Na+, K+, and the Na+/K+ ratio in the roots and leaves of GmNHX6 OE lines with WT Arabidopsis under NaHCO3 treatment and normal conditions (Figure 5). For Na+, under normal conditions, its content in leaf or root was similar between all lines. After NaHCO3 treatment, the leaf Na+ content in the GmNHX6 OE lines was significantly higher than that in the WT Arabidopsis, but the Na+ content in root showed the opposite pattern that OE lines had lower Na+ content than WT (Figure 5A). For K+, its content in both leaf and root of the GmNHX6 OE lines was significantly higher than that in the WT before and after NaHCO3 treatment (Figure 5B). For Na+/K+ ratios, there was no significant difference between GmNHX6 OE lines and WT under normal conditions in both leaves and roots. However, the Na+/K+ ratio of GmNHX6 OE lines was lower than that in the WT (Figure 5C). These results suggested that overexpression of GmNHX6 could improve the alkaline salt tolerance by keeping high K+ content and maintaining low Na+/K+ ratio in plants.
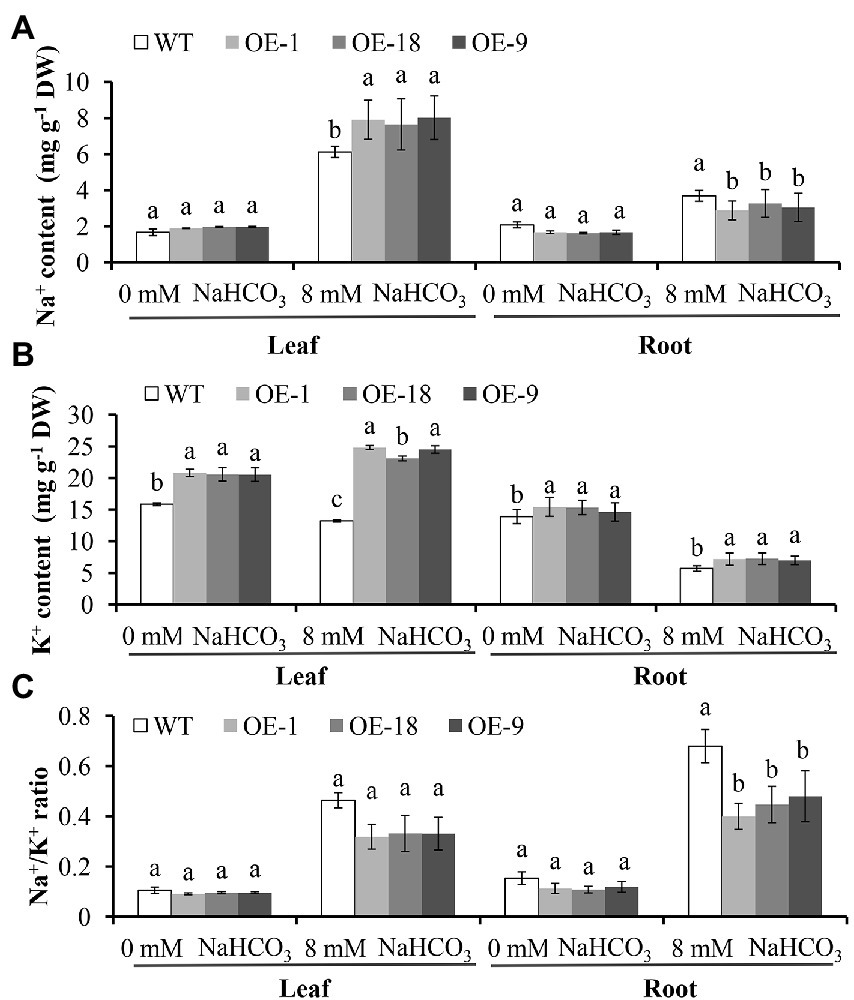
Figure 5. Na+ content (A), K+ content (B), and Na+/K + ratio (C) in the leaves and roots of Arabidopsis lines. Arabidopsis lines receiving 0 mM or 8 mM NaHCO3 treatment for 10 days, respectively. Data represents the mean ± standard deviation of three biological replications and each repeat contained 10 plants per line for each treatment (n = 30). Bars with the same letter in lowercase above bars indicate no significant differences at the 0.05 level between lines under same treatment according to Duncan’s multiple range tests.
Natural variation in the promoter region of soybean NHX6 is associated with alkaline salt tolerance
We sequenced the coding and promoter regions of soybean NHX6 from 30 wild soybean (15 alkaline salt tolerant and 15 alkaline salt sensitive soybean accessions) and 30 cultivated soybean (15 alkaline salt tolerant and 15 alkaline salt sensitive), respectively (Supplementary Table 1; Supplementary Figure 7). The results showed that only the NHX6 promoter region had sequence variation among 60 soybean accessions. A total of 30 SNPs were found in the 2-kb promoter region of soybean NHX6 (Supplementary Figure 8). Then, a regional association study was performed using the 30 SNPs in the 60 soybean accessions with extreme sodic tolerance rating (STR). Only one SNP (SNP−560, 560 bp upstream of the start codon) showed significant association with STR by mixed model in EMMAX (Supplementary Figure 9; Supplementary Table 4). Among the 60 soybean accessions, the majority (25 out of 30, 83%) of alkaline salt tolerant accessions had a base “C” at SNP−560 upstream of GmNHX6, while 70% (21 out of 30) alkaline salt sensitive accessions had a base “T” at SNP−560 (Supplementary Figure 8). There was a significant difference in alkaline salt tolerance between the two groups of SNP−560-C and SNP−560-T in 60 soybean accessions, as well as in wild soybean accessions and cultivated soybean accessions (Figure 6A; Supplementary Figure 10). In order to further compare the two types of promoters containing SNP−560-C and SNP−560-T, the representative promoters were cloned from two soybean varieties, M8206 and ZY, and designated as Pro-M8206 (containing SNP−560-C) and Pro-ZY (containing SNP−560-T), respectively. The promoter-LUC transient expression assays in tobacco leaves revealed that the activity of Pro-M was significantly stronger than that of Pro-ZY under alkali stress but not control condition (Figures 6B,C), suggesting that Pro-M is more responsive to alkali stress than Pro-ZY, therefore, leading to higher alkali induced expression level of GmNHX6 in the alkaline salt tolerant soybean accessions than sensitive accessions, thus resulting in enhanced alkaline salt tolerance.
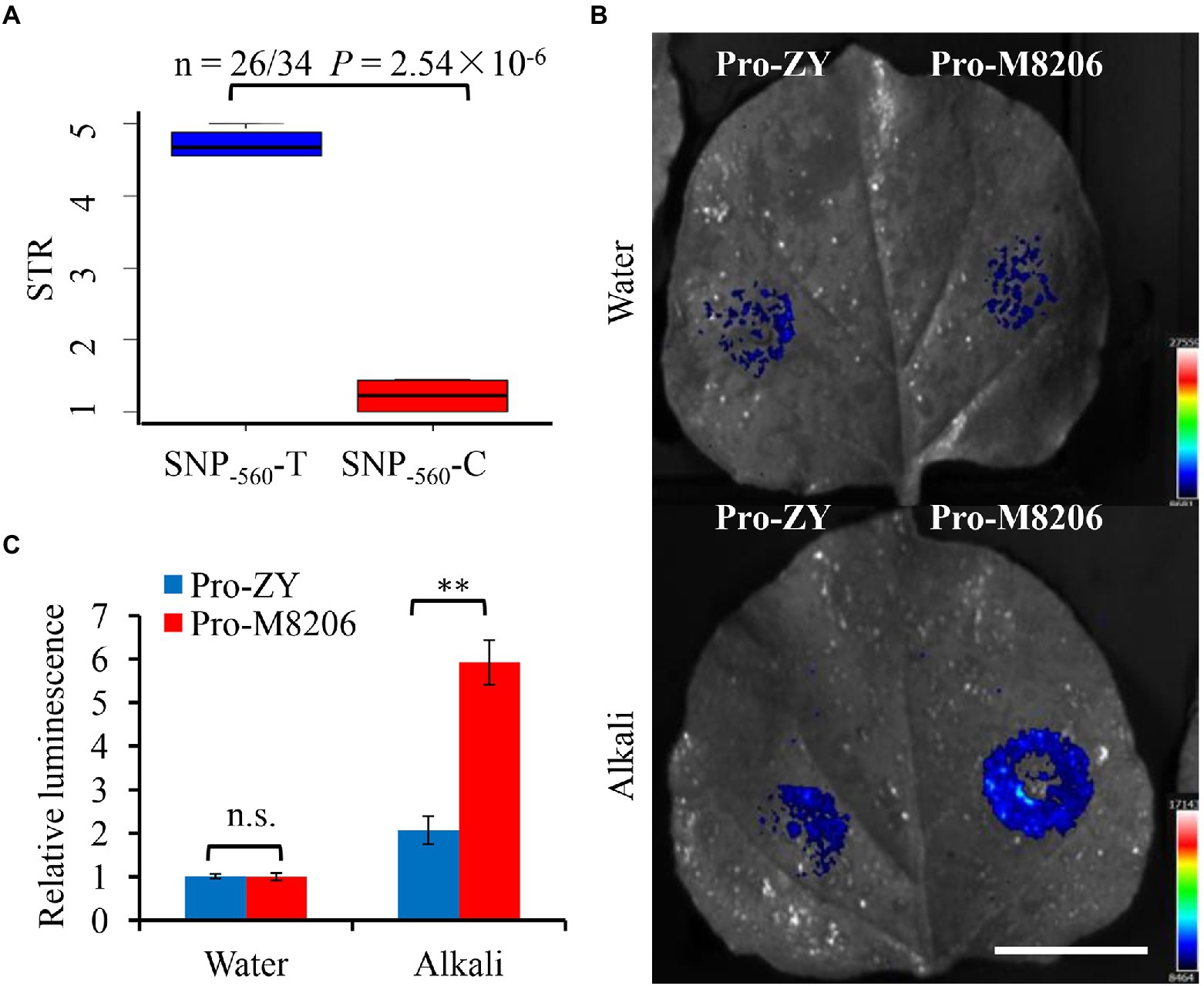
Figure 6. Natural variation and activity of soybean NHX6 promoter. (A) Boxplot of STR for two alleles of SNP−560 in GmNHX6 promoter among 60 soybean accessions. n denotes number of accessions. Statistical significance was detected by two-sided Wilcoxon test. The center bold line represents the median; box edges indicate the upper and lower quantiles; whiskers show the 1.5 × interquartile range. STR: sodic (alkaline salt) tolerance rating. (B,C) Promoter activities of two types of GmNHX6 promoters by luciferase (LUC) transient expression assays in tobacco leaves after alkaline salt (100 mM NaHCO3) treatment for 16 h. Pro-M8206 contains SNP−560-C and Pro-ZY contains SNP−560-T. The LUC reporter gene was driven by each type of promoter. The photos were taken using an in vivo plant imaging system. Bar = 3 cm. Data represents the mean ± standard deviation of four biological replications with two repeats within each replication (n = 4 × 2 = 8). Differences were evaluated using the two-tailed Student’s t-tests (**p < 0.01; n.s., not significant).
Discussion
Soil salinization and alkalization can reduce soil osmotic potential, and cause ion imbalance, disrupt physiological processes, inhibit growth and development of plants, leading to a serious decline in its yield and quality, and even the death of plants (Zhu, 2001; Yang et al., 2011). Soil salinization and alkalization frequently co-occur, but in the saline-alkaline land in mainland China, the soil salinization caused by alkali salts such as NaHCO3 and Na2CO3 is more serious than that caused by neutral salts such as NaCl and Na2SO4 (Yang et al., 2008, 2011). In general, the stress factors of neutral salts are mainly the ion stress of Na+ and osmotic stress of low water potential caused by high salt concentration, but for the alkaline salts, there is an added factor of high pH (Shi et al., 1998). The alkaline soil causes damage to plants not only through salt stress but also through high pH (Shi and Wang, 2005). Therefore, the deleterious effect of high pH stress or salinity alone is significantly less than that of the combined stress of high pH and salinity (Li et al., 2010). There have been lots of progress on plant tolerance to salt stress, while only few reports on alkali salt tolerance. Here, we investigated the role of soybean NHX6 gene in plant tolerance to alkaline salt (NaHCO3), which would broaden our knowledge on alkali salt tolerance.
In plants, sodium hydrogen exchangers allow cation/H+ such as Na+/H+ to transmembrane transport, to regulate pH and maintain ion homeostasis to resist abiotic stress (Rodríguez-Rosales et al., 2009; Bassil et al., 2012). However, there is no report about its role in soybean tolerance to alkali stress. In this study, a sodium/hydrogen exchanger gene from the soybean, GmNHX6, was found to be involved in the alkaline salt tolerance of soybean. The expression of GmNHX6 was induced by NaHCO3 treatment and more upregulated in alkali salt tolerant soybean (Figure 2), suggesting it is involved in plant responses to alkaline stress. Transgenic soybean composite plants and Arabidopsis plants have been previously used to study gene functions in saline-alkaline tolerance (Al-Harrasi et al., 2020; Ketehouli et al., 2021). GmPKS4 overexpressing soybean composite plants and transgenic Arabidopsis plants had increased proline content as well as high antioxidant enzyme activities under salt and salt-alkali stress treatments, compared to the empty-vector-transformed or wild-type ones (Ketehouli et al., 2021). Then we employed transgenic soybean composite plants and Arabidopsis to investigate the role of GmNHX6 in alkaline salt tolerance (Figures 3–5; Supplementary Figure 5). We found that overexpression of GmNHX6 enhanced the alkaline salt tolerance of soybean and Arabidopsis.
Since NHX genes have been shown to play important roles in salt tolerance in a variety of plant species (Kobayashi et al., 2012; Al-Harrasi et al., 2020; Long et al., 2020; Sun et al., 2021), we further analyzed the function of GmNHX6 in salt tolerance. The germination rates of GmNHX6 OE lines were significantly higher than that of WT Arabidopsis under NaCl stress (Figures 7A,B). At seedling stage, overexpression of GmNHX6 in Arabidopsis increased the average fresh weight and root length under salt stress (Figures 7C–E). We also compared the contents of Na+ and K+ in the roots and leaves of Arabidopsis (Figures 7F–H). Under normal conditions, there is no significant difference in the content of Na+ content between WT and OE lines. However, under NaCl treatment, the Na+ content in the leaves of the GmNHX6 OE lines was significantly higher than that in the WT, but in the root Na+ content was just the opposite (Figure 7F). The K + content of GmNHX6 OE lines was significantly higher than that in the WT Arabidopsis in both leaves and roots (Figure 7G). The Na+/K+ ratios of the GmNHX6 OE lines were significantly lower than that in the WT Arabidopsis under NaCl treatment (Figure 7H). These results suggest that overexpression of GmNHX6 enhanced salt tolerance by maintaining Na+ and K+ homeostasis.
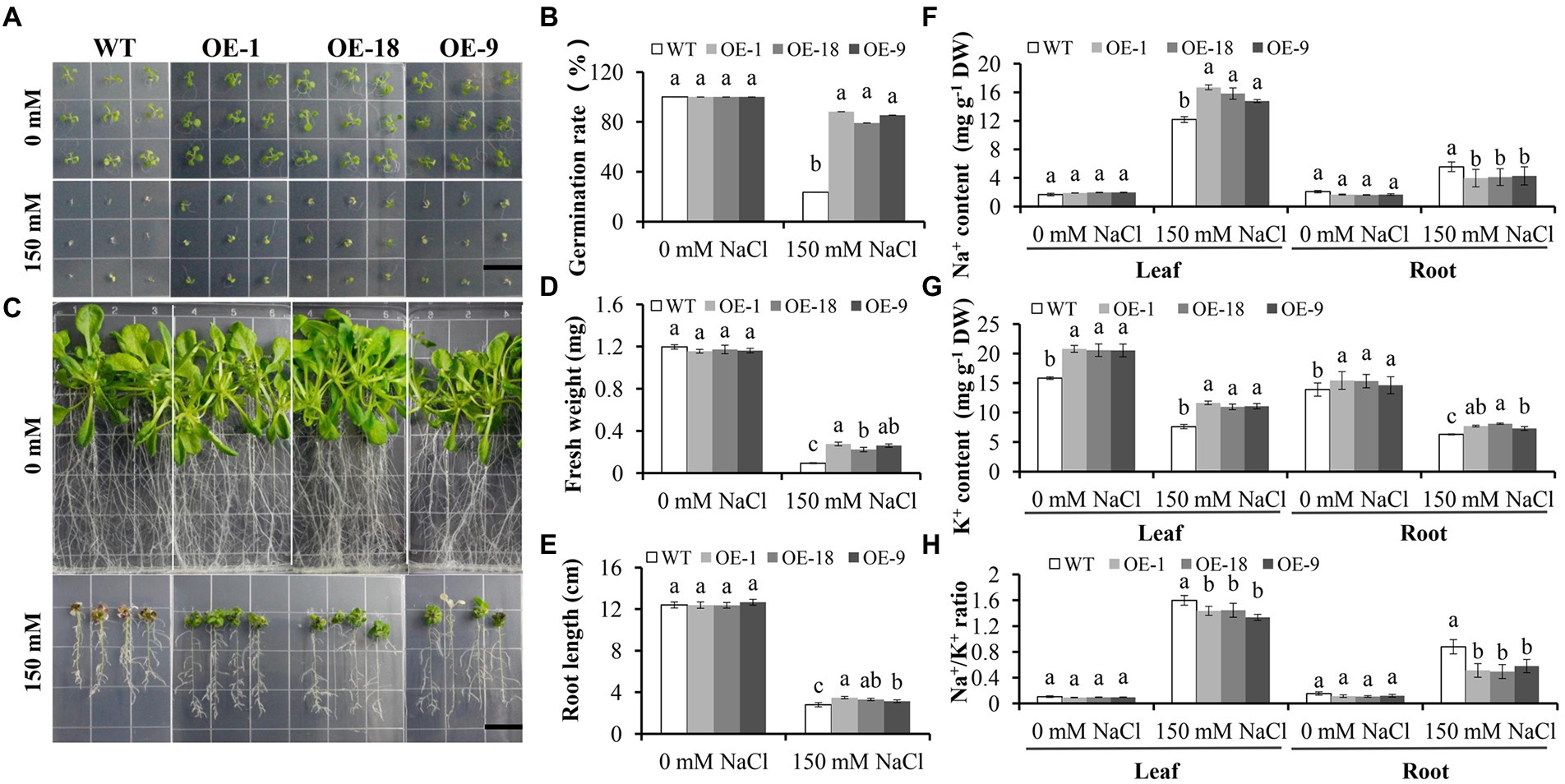
Figure 7. The effect of GmNHX6 overexpression on the salt tolerance of Arabidopsis. (A) Seed germination assay of different Arabidopsis lines on 1/2 MS medium supplied with 0 or 150 mM NaCl. Photographs were taken 7 days after treatment. WT: wild type; OE-1, OE-18, OE-9: Arabidopsis lines overexpressing GmNHX6. Bar = 1.5 cm. (B) Seed germination rates of different Arabidopsis lines as shown in A. Data represents the mean ± standard deviation of three biological replications and each repeat contained 36 seeds per line for each treatment (n = 36 × 3 = 108). (C) Arabidopsis seeds were grown vertically on 1/2 MS agar plates supplemented with 0 or 150 mM NaCl treatment for 10 days after 7 days of normal growth, respectively. Bar = 1.5 cm. (D,E) The fresh weight and root length of Arabidopsis lines shown in (C). Data represents the mean ± standard deviation of three biological replications and each repeat contained four plants per line for each treatment (n = 12). (F) Na+ content, (G) K + content, (H) Na+/K + ratio in the leaves and roots of Arabidopsis lines shown in (C). For (F–H), data represents the mean ± standard deviation of three biological replications and each replication has four repeats (n = 12), and 10 plants were pooled together for each sample. Bars with the same letter in lowercases above bars indicate no significant differences at the 0.05 level between lines under each condition according to Duncan’s multiple range test.
Different from salt stress, alkali stress caused by NaHCO3 and Na2CO3 leads to higher pH (Fang et al., 2021). According to the previous studies, the NHXs are important regulators of cellular pH and ion homeostasis (Rodríguez-Rosales et al., 2009; Bassil et al., 2012). NHX-type antiporters utilize the H+ electrochemical gradient to facilitate the exchange of H+ for cations such as Na+ or K+, thereby maintaining both pH and ion homeostasis (Sze and Chanroj, 2018). Studies also showed that plants can secrete large amounts of organic acids under alkali stress, which can play a buffer role and maintain intracellular pH stability (Fang et al., 2021). However, further experiments are needed to investigate the role of GmNHX6 in adjusting pH to alleviate alkaline salt stress.
K+ is an essential macronutrient for plant growth and the most abundant inorganic cation in plant cells. During plant growth and development, K+ is involved in the activation of more than 60 enzyme systems, photosynthesis, carbohydrate metabolism and protein synthesis, what’s more, K+ is a key modulator for cell homeostasis (Zhang et al., 2019a; Kumar et al., 2020). Na+ toxicity is one of the main harmful factors of saline-alkali stress (Apse and Blumwald, 2007). In root cells, Na+ is compartmentalized into vacuoles, radial transported to the stele cells, and loaded into the xylem, thus establishing the homeostatic control of Na+ in the cytosol (Apse and Blumwald, 2007). Plants can maintain high K+ content by increasing K+ absorption and reducing K+ loss (Zhu, 2003). Under saline-alkali stress, plant cells absorb a large amount of Na+, resulting in ion toxicity, which not only inhibits photosynthesis, but also inhibits the absorption of essential element such as K+ by plants, when Na+/K+ is too high, resulting in the change of ion homeostasis and metabolic disorder (Zhu, 2003). Therefore, maintaining a relatively high K+ content and low Na+/K+ ratio in plants under salt-alkali stress is a manifestation of salt-alkali tolerance. The NHXs play important roles in maintaining Na+ and K+ homeostasis have been well documented (Wang et al., 2015; Sun et al., 2021). Previous studies have shown that NHXs can compartmentalize Na+ into vacuoles under salt stress, thus maintaining the intracellular ion balance (Feng et al., 2021). Other studies found that vacuolar NHXs can simultaneously catalyze the exchange of Na+/H+ and K+/H+ to maintain intracellular ion balance (Sze and Chanroj, 2018). Under salt stress, NHX transporter, such as NHX1, transport sodium ions to and sequester them in the vacuole by vesicle transport, and release K+ from vacuolar into the cytoplasm (Barragán et al., 2012). In Arabidopsis, AtNHX5 and AtNHX6 are critical to K+ homeostasis in Arabidopsis (Wang et al., 2015). GmNHX5 positively regulates salt tolerance by maintaining higher K+/Na+ ratio in soybean (Sun et al., 2021). In this study, we found that overexpression of GmNHX6 enhanced Arabidopsis tolerance to both alkaline salt (NaHCO3) and salt (NaCl) stress, by maintaining higher K+ content and low Na+/K+ ratios.
We also obtained a homozygous mutant nhx6 (stock, SALK-100042C), a T-DNA insertion Arabidopsis mutant of AtNHX6 (Supplementary Figure 11). However, nhx6 mutant did not show alkaline salt tolerance or salt tolerance compared with the WT Arabidopsis (Supplementary Figure 12), which is consistent with the previous study by Bassil et al. (2011a) that the phenotype of nhx5 or nhx6 single-knockout line was not different from the WT, while the double mutant nhx5 nhx6 has reduced growth, smaller and fewer cells, and increased sensitivity to salinity. Through the EnsemblPlants database,11 we identified a total of 66 genes in the soybean NHX gene family, which might be functionally redundant in soybean tolerance to salt-alkali stress.
Soybean germplasm provides a wide range of saline-alkaline tolerance. To explore the natural variation in soybean NHX6 gene, we analyzed the sequence polymorphism of the 2-kb promoter region (Supplementary Figure 13) of NHX6 in 60 soybean accessions, including G. max and G. soja. Only one SNP (SNP−560) out of a total of 30 SNPs showed significant association with alkaline salt tolerance (STR) by mixed model in EMMAX (Supplementary Table 4). We analyzed the cis-elements of the 2-kb promoter region (Supplementary Figure 13) of GmNHX6 in two soybean accessions, M8206 and ZY. We found that in Pro-M8206 (containing SNP−560-C), the sequence forms four cis-acting elements around SNP−560, including Skn-1_motif (GTCAT, −562 to −558 bp), TCATTT element (−561 to −556 bp), TTGTCA motif (−564 to −559 bp), and TGTCAT motif (−563 to −559 bp). Skn-1_motif is responsive to abiotic stress and could be used in plant genetic engineering research on abiotic stress tolerance (De Silva et al., 2017; Zhang et al., 2018). TTGTCA element is involved in light induction (Mazouni et al., 2003). Previous studies showed that TCATTT element and TGTCAT element are the binding sites of downstream key functional genes. TCATTT-containing element acted as an enhancer (Perrotti et al., 1996), and was essential for inducible expression of the IL-5 gene (Miao et al., 2006). Tunicamycin (Tm)-activated sXBP1 bound to the TGTCAT element and suppressed XRCC2 expression to prevent tumor proliferation in vivo (Zhao et al., 2021). Interesting, promoter-LUC transient expression assays in tobacco leaves revealed that the promoter of GmNHX6 from the alkaline salt tolerant soybean variety, Pro-M8206, had a significantly stronger activity than Pro-ZY (from alkaline salt sensitive variety), under alkali stress (Figures 6B,C), suggesting that Pro-M8206 is more responsive to alkali stress than Pro-ZY. Therefore, it is likely that the SNP−560 might affect the promoter activity under alkali stress by interrupting the relevant cis-acting elements mentioned above. How these cis-acting elements in the promoter of GmNHX6 regulate gene expression in response to alkali stress needs further study.
Conclusion
In summary, GmNHX6 encodes a sodium/hydrogen exchanger. The expression of GmNHX6 was induced by NaHCO3 stress, and greater increase in its transcript abundance was observed in alkaline salt tolerant than in alkaline salt sensitive soybean variety. Overexpression of GmNHX6 enhanced the alkaline-salt tolerance of soybean composite plants and Arabidopsis. The GmNHX6 overexpressing Arabidopsis lines had higher K+ content and lower Na+/K+ ratio than the wild-type plants under NaHCO3 stress. Furthermore, a single nucleotide polymorphism in the promoter region of NHX6 is associated with the alkali tolerance in soybean. These findings would help to further understand the role of NHX6 and its regulatory mechanism in soybean tolerance to alkaline-salt stress.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
TJ and YL conceived and designed the experiments, interpreted the results, and wrote and revised the manuscript. TJ, JA, HX, JC, LP, RZ, and NW performed the experiments. TJ analyzed the data and generated the pictures. JG and YL contributed reagents and materials. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2021YFF1001204) and the Core Technology Development for Breeding Program of Jiangsu Province (JBGS-2021-014).
Acknowledgments
We appreciate Xinyuan Huang and Daolong Dou at Nanjing Agricultural University to provide us the vectors of pGreenII0800-LUC and pBinGFP4.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.938635/full#supplementary-material
Footnotes
2. ^http://www.premierbiosoft.com/primerdesign/
3. ^https://phytozome.jgi.doe.gov/pz/portal.html
4. ^https://www.ebi.ac.uk/Tools/msa/clustalw2/
5. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
6. ^https://wlab.ethz.ch/protter/start/
7. ^http://www.arabidopsis.org
References
Al-Harrasi, I., Jana, G. A., Patankar, H. V., Al-Yahyai, R., Rajappa, S., Kumar, P. P., et al. (2020). A novel tonoplast Na+/H+ antiporter gene from date palm (PdNHX6) confers enhanced salt tolerance response in Arabidopsis. Plant Cell Rep. 39, 1079–1093. doi: 10.1007/s00299-020-02549-5
An, R., Chen, Q. J., Chai, M. F., Lu, P. L., Su, Z., Qin, Z. X., et al. (2007). AtNHX8, a member of the monovalent cation: proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li+/H+ antiporter. Plant J. 49, 718–728. doi: 10.1111/j.1365-313X.2006.02990.x
Apse, M. P., and Blumwald, E. (2007). Na+ transport in plants. FEBS Lett. 581, 2247–2254. doi: 10.1016/j.febslet.2007.04.014
Barragán, V., Leidi, E. O., Andrés, Z., Rubio, L., De Luca, A., Fernández, J. A., et al. (2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24, 1127–1142. doi: 10.1105/tpc.111.095273
Bassil, E., Coku, A., and Blumwald, E. (2012). Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 63, 5727–5740. doi: 10.1093/jxb/ers250
Bassil, E., Ohto, M. A., Esumi, T., Tajima, H., Zhu, Z., Cagnac, O., et al. (2011a). The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23, 224–239. doi: 10.1105/tpc.110.079426
Bassil, E., Tajima, H., Liang, Y. C., Ohto, M. A., Ushijima, K., Nakano, R., et al. (2011b). The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23, 3482–3497. doi: 10.1105/tpc.111.089581
Bassil, E., Zhang, S., Gong, H., Tajima, H., and Blumwald, E. (2019). Cation specificity of vacuolar NHX-type cation/H+ antiporters. Plant Physiol. 179, 616–629. doi: 10.1104/pp.18.01103
Bouras, H., Choukr-Allah, R., Amouaouch, Y., Bouaziz, A., Devkota, K. P., El Mouttaqi, A., et al. (2022). How does quinoa (Chenopodium quinoa Willd.) respond to phosphorus fertilization and irrigation water salinity? Plant Theory 11:216. doi: 10.3390/plants11020216
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Cao, B., Xia, Z., and Liu, C. (2020). New insights into the structure-function relationship of the endosomal-type Na+, K+/H+ antiporter NHX6 from mulberry (Morus notabilis). Int. J. Mol. Sci. 21:428. doi: 10.3390/ijms21020428
Chen, C., Yu, Y., Ding, X., Liu, B., Duanmu, H., Zhu, D., et al. (2018). Genome-wide analysis and expression profiling of PP2C clade D under saline and alkali stresses in wild soybean and Arabidopsis. Protoplasma 255, 643–654. doi: 10.1007/s00709-017-1172-2
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
De Silva, W. S. I., Perera, M. M. N., Perera, K., Wickramasuriya, A. M., and Jayasekera, G. A. U. (2017). In silico analysis of osr40c1 promoter sequence isolated from Indica variety Pokkali. Rice Sci. 24, 228–234. doi: 10.1016/j.rsci.2016.11.002
Dragwidge, J. M., Scholl, S., and Schumacher, K. (2019). NHX-type Na+, K+/H+ antiporters are required for TGN/EE trafficking and endosomal ion homeostasis in Arabidopsis thaliana. J. Cell Sci. 132:jcs226472. doi: 10.1242/jcs.226472
Fan, L., Zhao, L., Hu, W., Li, W., Novák, O., Strnad, M., et al. (2018). Na+, K+ /H+ antiporters regulate the pH of endoplasmic reticulum and auxin-mediated development. Plant Cell Environ. 41, 850–864. doi: 10.1111/pce.13153
Fang, S., Hou, X., and Liang, X. (2021). Response mechanisms of plants under saline-alkali stress. Front. Plant Sci. 12:667458. doi: 10.3389/fpls.2021.667458
Feng, J., Ma, W., Ma, Z., Ren, Z., and Zhou, Y. (2021). GhNHX3D, a vacuolar-localized Na+/H+ antiporter, positively regulates salt response in upland cotton. Int. J. Mol. Sci. 22:4047. doi: 10.3390/ijms22084047
Guan, R., Qu, Y., Guo, Y., Yu, L., Liu, Y., Jiang, J., et al. (2014). Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 80, 937–950. doi: 10.1111/tpj.12695
He, Y., Dong, Y., Yang, X., Guo, D., Qian, X., Yan, F., et al. (2020). Functional activation of a novel R2R3-MYB protein gene, GmMYB68, confers salt-alkali resistance in soybean (Glycine max L.). Genome 63, 13–26. doi: 10.1139/gen-2018-0132
He, C., Gao, H., Wang, H., Guo, Y., He, M., Peng, Y., et al. (2021). GSK3-mediated stress signaling inhibits legume-rhizobium symbiosis by phosphorylating GmNSP1 in soybean. Mol. Plant 14, 488–502. doi: 10.1016/j.molp.2020.12.015
Jin, T., Sun, Y., Shan, Z., He, J., Wang, N., Gai, J., et al. (2021). Natural variation in the promoter of GsERD15B affects salt tolerance in soybean. Plant Biotechnol. J. 19, 1155–1169. doi: 10.1111/pbi.13536
Kereszt, A., Li, D., Indrasumunar, A., Nguyen, C. D., Nontachaiyapoom, S., Kinkema, M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2, 948–952. doi: 10.1038/nprot.2007.141
Ketehouli, T., Zhou, Y. G., Dai, S. Y., Carther, K. F. I., Sun, D. Q., Li, Y., et al. (2021). A soybean calcineurin B-like protein-interacting protein kinase, GmPKS4, regulates plant responses to salt and alkali stresses. J. Plant Physiol. 256:153331. doi: 10.1016/j.jplph.2020.153331
Kobayashi, S., Abe, N., Yoshida, K. T., Liu, S., and Takano, T. (2012). Molecular cloning and characterization of plasma membrane- and vacuolar-type Na+/H+ antiporters of an alkaline-salt-tolerant monocot, Puccinellia tenuiflora. J. Plant Res. 125, 587–594. doi: 10.1007/s10265-012-0475-9
Krishnamurthy, P., Vishal, B., Khoo, K., Rajappa, S., Loh, C. S., and Kumar, P. P. (2019). Expression of AoNHX1 increases salt tolerance of rice and Arabidopsis, and bHLH transcription factors regulate AtNHX1 and AtNHX6 in Arabidopsis. Plant Cell Rep. 38, 1299–1315. doi: 10.1007/s00299-019-02450-w
Kumar, P., Kumar, T., Singh, S., Tuteja, N., Prasad, R., and Singh, J. (2020). Potassium: a key modulator for cell homeostasis. J. Biotechnol. 324, 198–210. doi: 10.1016/j.jbiotec.2020.10.018
Li, R., Shi, F., and Fukuda, K. (2010). Interactive effects of various salt and alkali stresses on growth, organic solutes, and cation accumulation in a halophyte Spartina alterniflora (Poaceae). Environ. Exp. Bot. 68, 66–74. doi: 10.1016/j.envexpbot.2009.10.004
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Long, L., Zhao, J. R., Guo, D. D., Ma, X. N., Xu, F. C., Yang, W. W., et al. (2020). Identification of NHXs in Gossypium species and the positive role of GhNHX1 in salt tolerance. BMC Plant Biol. 20:147. doi: 10.1186/s12870-020-02345-z
Lv, S., Wang, L., Zhang, X., Li, X., Fan, L., Xu, Y., et al. (2020). Arabidopsis NHX5 and NHX6 regulate PIN6-mediated auxin homeostasis and growth. J. Plant Physiol. 255:153305. doi: 10.1016/j.jplph.2020.153305
Mazouni, K., Domain, F., Chauvat, F., and Cassier-Chauvat, C. (2003). Expression and regulation of the crucial plant-like ferredoxin of cyanobacteria. Mol. Microbiol. 49, 1019–1029. doi: 10.1046/j.1365-2958.2003.03609.x
Miao, Y., Yan, P. K., Kim, H., Hwang, I., and Jiang, L. (2006). Localization of green fluorescent protein fusions with the seven Arabidopsis vacuolar sorting receptors to prevacuolar compartments in tobacco BY-2 cells. Plant Physiol. 142, 945–962. doi: 10.1104/pp.106.083618
Miao, L., Yang, S., Zhang, K., He, J., Wu, C., Ren, Y., et al. (2020). Natural variation and selection in GmSWEET39 affect soybean seed oil content. New Phytol. 225, 1651–1666. doi: 10.1111/nph.16250
Moghaieb, R. E., Sharaf, A. N., Soliman, M. H., El-Arabi, N. I., and Momtaz, O. A. (2014). An efficient and reproducible protocol for the production of salt tolerant transgenic wheat plants expressing the Arabidopsis AtNHX1 gene. GM Crops Food 5, 132–138. doi: 10.4161/gmcr.28941
Munns, R., James, R. A., Xu, B., Athman, A., Conn, S. J., Jordans, C., et al. (2012). Wheat grain yield on saline soils is improved by an ancestral Na+transporter gene. Nat. Biotechnol. 30, 360–364. doi: 10.1038/nbt.2120
Oh, D. H., Lee, S. Y., Bressan, R. A., Yun, D. J., and Bohnert, H. J. (2010). Intracellular consequences of SOS1 deficiency during salt stress. J. Exp. Bot. 61, 1205–1213. doi: 10.1093/jxb/erp391
Pérez-Patricio, M., Camas-Anzueto, J. L., Sanchez-Alegría, A., Aguilar-González, A., and Gutiérrez-Miceli, F. (2018). Optical method for estimating the chlorophyll contents in plant leaves. Sensors 18:650. doi: 10.3390/s18020650
Perrotti, D., Bellón, T., Trotta, R., Martinez, R., and Calabretta, B. (1996). A cell proliferation-dependent multiprotein complex NC-3A positively regulates the CD34 promoter via a TCATTT-containing element. Blood 88, 3336–3348. doi: 10.1182/blood.V88.9.3336.bloodjournal8893336
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Qiu, Q. S. (2016a). Plant endosomal NHX antiporters: activity and function. Plant Signal. Behav. 11:e1147643. doi: 10.1080/15592324.2016.1147643
Qiu, Q. S. (2016b). AtNHX5 and AtNHX6: roles in protein transport. Plant Signal. Behav. 11:e1184810. doi: 10.1080/15592324.2016.1184810
Quintero, F. J., Blatt, M. R., and Pardo, J. M. (2000). Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett. 471, 224–228. doi: 10.1016/s0014-5793(00)01412-5
Reguera, M., Bassil, E., and Blumwald, E. (2014). Intracellular NHX-type cation/H+ antiporters in plants. Mol. Plant 7, 261–263. doi: 10.1093/mp/sst091
Reguera, M., Bassil, E., Tajima, H., Wimmer, M., Chanoca, A., Otegui, M. S., et al. (2015). pH regulation by NHX-type antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell 27, 1200–1217. doi: 10.1105/tpc.114.135699
Ren, Y., Lv, J., Wang, H., Li, L., Peng, Y., and Qu, L. J. (2009). A comparative proteomics approach to detect unintended effects in transgenic Arabidopsis. J. Genet. Genomics 36, 629–639. doi: 10.1016/s1673-8527(08)60155-1
Rodríguez-Rosales, M. P., Gálvez, F. J., Huertas, R., Aranda, M. N., Baghour, M., Cagnac, O., et al. (2009). Plant NHX cation/proton antiporters. Plant Signal. Behav. 4, 265–276. doi: 10.4161/psb.4.4.7919
Sellamuthu, G., Jegadeeson, V., Sajeevan, R. S., Rajakani, R., Parthasarathy, P., Raju, K., et al. (2020). Distinct evolutionary origins of intron retention splicing events in NHX1 antiporter transcripts relate to sequence specific distinctions in Oryza species. Front. Plant Sci. 11:267. doi: 10.3389/fpls.2020.00267
Shi, D. C., Sheng, Y. M., and Zhao, K. F. (1998). Stress effects of mixed salts with various salinities on the seedlings of Aneurolepidium chinense. Acta Bot. Sin. 40, 1136–1142.
Shi, D., and Wang, D. (2005). Effects of various salt-alkaline mixed stresses on Aneurolepidium chinense (Trin.) Kitag. Plant and Soil 271, 15–26. doi: 10.1007/s11104-004-1307-z
Sun, T., Ma, N., Wang, C., Fan, H., Wang, M., Zhang, J., et al. (2021). A golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K+/Na+ ratio in soybean. Front. Plant Sci. 12:638340. doi: 10.3389/fpls.2021.638340
Sze, H., and Chanroj, S. (2018). Plant endomembrane dynamics: studies of K+/H+ antiporters provide insights on the effects of pH and ion homeostasis. Plant Physiol. 177, 875–895. doi: 10.1104/pp.18.00142
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tse, Y. C., Mo, B., Hillmer, S., Zhao, M., Lo, S. W., Robinson, D. G., et al. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16, 672–693. doi: 10.1105/tpc.019703
Tuyen, D. D., Lal, S. K., and Xu, D. H. (2010). Identification of a major QTL allele from wild soybean (Glycine soja Sieb. & Zucc.) for increasing alkaline salt tolerance in soybean. Theor. Appl. Genet. 121, 229–236. doi: 10.1007/s00122-010-1304-y
Walker, D. J., Leigh, R. A., and Miller, A. J. (1996). Potassium homeostasis in vacuolate plant cells. Proc. Natl. Acad. Sci. U. S. A. 93, 10510–10514. doi: 10.1073/pnas.93.19.10510
Wang, L., Wu, X., Liu, Y., and Qiu, Q. S. (2015). AtNHX5 and AtNHX6 control cellular K+ and pH homeostasis in Arabidopsis: three conserved acidic residues are essential for K+ transport. PLoS One 10:e0144716. doi: 10.1371/journal.pone.0144716
Wang, L., Yang, Y., Zhang, S., Che, Z., Yuan, W., and Yu, D. (2020). GWAS reveals two novel loci for photosynthesis-related traits in soybean. Mol. Genet. Genomics 295, 705–716. doi: 10.1007/s00438-020-01661-1
Wu, X., Ebine, K., Ueda, T., and Qiu, Q. S. (2016). AtNHX5 and AtNHX6 are required for the subcellular localization of the SNARE complex that mediates the trafficking of seed storage proteins in Arabidopsis. PLoS One 11:e0151658. doi: 10.1371/journal.pone.0151658
Wu, F. H., Shen, S. C., Lee, L. Y., Lee, S. H., Chan, M. T., and Lin, C. S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5:16. doi: 10.1186/1746-4811-5-16
Yang, C. W., Wang, P., Li, C. Y., Shi, D. C., and Wang, D. L. (2008). Comparison of effects of salt and alkali stresses on the growth and photosynthesis of wheat. Photosynthetica 46, 107–114. doi: 10.1007/s11099-008-0018-8
Yang, J., Zaitlen, N. A., Goddard, M. E., Visscher, P. M., and Price, A. L. (2014). Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 46, 100–106. doi: 10.1038/ng.2876
Yang, J. Y., Zheng, W., Tian, Y., Wu, Y., and Zhou, D. W. (2011). Effects of various mixed salt-alkaline stresses on growth, photosynthesis, and photosynthetic pigment concentrations of Medicago ruthenica seedlings. Photosynthetica 49, 275–284. doi: 10.1007/s11099-011-0037-8
Zhang, Z., Cheng, Z. J., Gan, L., Zhang, H., Wu, F. Q., Lin, Q. B., et al. (2016). OsHSD1, a hydroxysteroid dehydrogenase, is involved in cuticle formation and lipid homeostasis in rice. Plant Sci. 249, 35–45. doi: 10.1016/j.plantsci.2016.05.005
Zhang, Y., Deng, G., Fan, W., Yuan, L., and Wang, H. (2019b). NHX1 and eIF4A1-stacked transgenic sweetpotato shows enhanced tolerance to drought stress. Plant Cell Rep. 38, 1427–1438. doi: 10.1007/s00299-019-02454-6
Zhang, X., Li, Z., Li, X., Xu, Y., Xie, H., and Qiu, Q. S. (2020b). CBL3 and CIPK18 are required for the function of NHX5 and NHX6 in mediating Li+ homeostasis in Arabidopsis. J. Plant Physiol. 255:153295. doi: 10.1016/j.jplph.2020.153295
Zhang, M., Liang, X., Wang, L., Cao, Y., Song, W., and Shi, J. (2019a). A HAK family Na+ transporter confers natural variation of salt tolerance in maize. Nat. Plants 5, 1297–1308. doi: 10.1038/s41477-019-0565-y
Zhang, S., Tajima, H., Nambara, E., and Blumwald, E. (2020a). Auxin homeostasis and distribution of the auxin efflux carrier PIN2 require vacuolar NHX-type cation/H+ antiporter activity. Plant Theory 9:1311. doi: 10.3390/plants9101311
Zhang, F., and Zhang, J. (2021). Bulk analysis by resequencing and RNA-seq identifies candidate genes for maintaining leaf water content under water deficit in maize. Physiol. Plant. 173, 1935–1945. doi: 10.1111/ppl.13537
Zhang, H., Zheng, J., Su, H., Xia, K., Jian, S., and Zhang, M. (2018). Molecular cloning and functional characterization of the Dehydrin (IpDHN) gene from Ipomoea pes-caprae. Front. Plant Sci. 9:1454. doi: 10.3389/fpls.2018.01454
Zhao, Z., He, K., Zhang, Y., Hua, X., Feng, M., Zhao, Z., et al. (2021). XRCC2 repairs mitochondrial DNA damage and fuels malignant behavior in hepatocellular carcinoma. Cancer Lett. 512, 1–14. doi: 10.1016/j.canlet.2021.04.026
Zheng, J., Lin, R., Pu, L., Wang, Z., Mei, Q., and Zhang, M. (2021). Ectopic expression of CrPIP2;3, a plasma membrane intrinsic protein gene from the halophyte Canavalia rosea, enhances drought and salt-alkali stress tolerance in Arabidopsis. Int. J. Mol. Sci. 22:565. doi: 10.3390/ijms22020565
Zhu, J. K. (2001). Plant salt tolerance. Trends Plant Sci. 6, 66–71. doi: 10.1016/S1360-1385(00)01838-0
Zhu, J. K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6, 441–445. doi: 10.1016/s1369-5266(03)00085-2
Keywords: abiotic stress, alkaline salt tolerance, natural variation, promoter, sodium bicarbonate, sodium hydrogen exchanger, soybean
Citation: Jin T, An J, Xu H, Chen J, Pan L, Zhao R, Wang N, Gai J and Li Y (2022) A soybean sodium/hydrogen exchanger GmNHX6 confers plant alkaline salt tolerance by regulating Na+/K+ homeostasis. Front. Plant Sci. 13:938635. doi: 10.3389/fpls.2022.938635
Edited by:
Rajeev K. Varshney, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), IndiaReviewed by:
Haoqiang Yu, Maize Research Institute of Sichuan Agricultural University, ChinaNeelam Mishra, St Joseph’s College (Autonomous), India
Copyright © 2022 Jin, An, Xu, Chen, Pan, Zhao, Wang, Gai and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Li, eWFubGkxQG5qYXUuZWR1LmNu
 Ting Jin
Ting Jin Jiaxin An
Jiaxin An Lang Pan
Lang Pan Ning Wang
Ning Wang Junyi Gai
Junyi Gai Yan Li
Yan Li