
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 17 August 2022
Sec. Plant Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.938570
This article is part of the Research Topic Plant RNA Structure View all 7 articles
RNA molecules have the capacity to form a multitude of distinct secondary and tertiary structures, but only the most energetically favorable conformations are adopted at any given time. Formation of such structures strongly depends on the environment and consequently, these structures are highly dynamic and may refold as their surroundings change. Temperature is one of the most direct physical parameters that influence RNA structure dynamics, and in turn, thermosensitive RNA structures can be harnessed by a cell to perceive and respond to its temperature environment. Indeed, many thermosensitive RNA structures with biological function have been identified in prokaryotic organisms, but for a long time such structures remained elusive in eukaryotes. Recent discoveries, however, reveal that thermosensitive RNA structures are also found in plants, where they affect RNA stability, pre-mRNA splicing and translation efficiency in a temperature-dependent manner. In this minireview, we provide a short overview of thermosensitive RNA structures in prokaryotes and eukaryotes, highlight recent advances made in identifying such structures in plants and discuss their similarities and differences to established prokaryotic RNA thermosensors.
RNA molecules play vital roles in all extant forms of life: they serve as blueprints for protein synthesis, exert regulatory effects on DNA, other RNAs and proteins and even act as biocatalysts. Underpinning all these functions is their secondary and tertiary structure: inter- and intramolecular base pairing results in the formation of hairpins, apical and internal loops, three-way junctions and pseudo-knots (Cruz and Westhof, 2009). The critical role of these structures in non-coding RNAs has long been established, but over the years, it has become clear that they play important roles in transcription, processing, splicing, transport, translation as well as stability of mRNA (Jacobs et al., 2012; Lazzaretti and Bono, 2017; Soemedi et al., 2017; Bartys et al., 2019).
Thermodynamic free energy is a strong determinant of RNA secondary structure and thus, only the most energetically favorable will be adopted at any given time from a multitude of possible structures. While RNA folding is an intrinsic property of each RNA molecule, it is highly sensitive to the molecular environment and is influenced by RNA-binding proteins and ligands in addition to physical and chemical parameters. The free energy of RNA folding is directly dependent on temperature and therefore temperature changes can have profound and immediate consequences on RNA conformation. Transcriptome-wide RNA structure probing indeed revealed that shifts to higher temperature globally reduce RNA structure formation in several bacterial species (Righetti et al., 2016) as well as in yeast (Rouskin et al., 2014) and plants (Su et al., 2018).
Temperature affects every physical and biochemical process within the cell; it is thus vital for every organism to tightly monitor its temperature environment. Many organisms are known to undergo substantial physiological and developmental changes when their ambient temperature deviates from the desired optimum. As its structure is intrinsically thermosensitive, RNA represents an attractive candidate molecule to perceive temperature signals. While many conformational changes triggered by a rise or drop in temperature are likely to be subtle with limited functional consequences, some RNA structures undergo substantial rearrangements across a physiologically relevant temperature gradient, and it is these structures that can be harnessed by cells to monitor their temperature surroundings. Such RNA-based thermosensors are best described in bacteria, including plant symbionts and pathogens, where the so-called RNA thermometers control translation initiation in direct response to the temperature environment (Narberhaus et al., 2006).
However, thermosensory RNA elements in eukaryotes are not extensively explored to date. Previous studies on ectotherms such as Drosophila and Trypanosomes predict the possibility of mRNA 5′-UTR secondary structure mediated regulation in heat shock translation response (Ahmed and Duncan, 2004; David et al., 2010). Recent discoveries have shown that thermosensitive RNA structures that control gene expression in cis also exist in plants, although their mode of action differs from bacterial RNA thermometers (Chung et al., 2020). Temperate plants are exposed to a wide range of temperatures throughout the year and substantially change their growth habit and cellular composition in response to temperature changes. In the model plant Arabidopsis thaliana, warm ambient temperatures in the range of 25–30°C trigger increased elongation growth of stems and roots, reduced stomata formation and accelerated flowering (Nomoto et al., 2013; Song et al., 2018). These developmental adjustments largely aim at avoiding future heat and drought stress, but come at the cost of reduced immunity, particularly reduction in effector-triggered immune responses, at higher temperatures (Cheng et al., 2013; Menna et al., 2015; Huot et al., 2017; Qiu et al., 2022). Under severe heat stress above 40°C, growth instead ceases, and plants alter their physiology to minimize damage to membranes and proteins (Hayes et al., 2021). While these processes have been investigated for decades, studies have mainly focused on transcriptional or post-transcriptional mechanisms operating in trans, such as those mediated by miRNA, and on post-translational control, little attention has been paid to direct regulation of translation in cis by mRNAs (Megha et al., 2018).
Recently however, a genome-wide analysis of rice (Oryza sativa L.) seedlings reveals global structural changes in the transcriptome in response to high temperature due to unfolding of mRNA 5′ and 3′-untranslated regions (UTRs) (Su et al., 2018). This results in reduced mRNA stability and downregulation of global translation in rice seedlings. Temperature dependent splicing can be mediated by alterations of pre-mRNA secondary structures at intron-exon junctions thereby modulating the activity of spliceosome complex or that of splicing regulators (Shiina and Shimizu, 2020). Several examples of this mechanism are being identified, including alternative splicing of pre-mRNA encoding for an amino peptidase in Saccharomyces cerevisiae (Meyer et al., 2011) and more recently at the intron 2 of Heat shock factor (HsfA2) pre-mRNA from Solanum lycopersicum (Broft et al., 2022). Yet another study shows that thermosensitive RNA structures directly influence protein synthesis in cis in Arabidopsis, highlighting the importance of translational control in plant temperature responses (Chung et al., 2020). Likewise, increased RNA G-quadruplex (RG4) formation in 3′-UTRs of plant transcripts, in response to low temperatures, have been shown to enhance mRNA stability and adaptation to cold environment (Yang et al., 2022).
In this minireview, we compare recently identified thermosensitive RNA structures in plants with structures found in prokaryotic and other eukaryotic systems and contrast the different modes of action employed by these structures to control gene expression.
Temperature-dependent changes in RNA secondary and tertiary structures are effectively utilized by many bacteria to modulate gene expression. These RNA thermometers form a powerful tool for rapid control of bacterial protein synthesis in response to cellular events such as entry into host, heat or cold stress (Narberhaus et al., 2006). Bacterial thermoregulatory RNAs are typically found at the 5′-UTR (5′-UTR) of the corresponding mRNA and encompass the Shine-Dalgarno (SD) sequence. As depicted in Figure 1, upon temperature dependent melting of these secondary structures, these non-coding mRNA regions are unmasked, providing accessibility for the 30S ribosomal subunits to interact with the SD sequence and identify the start codon, thereby resulting in translation initiation (Kortmann and Narberhaus, 2012; Somero, 2018).
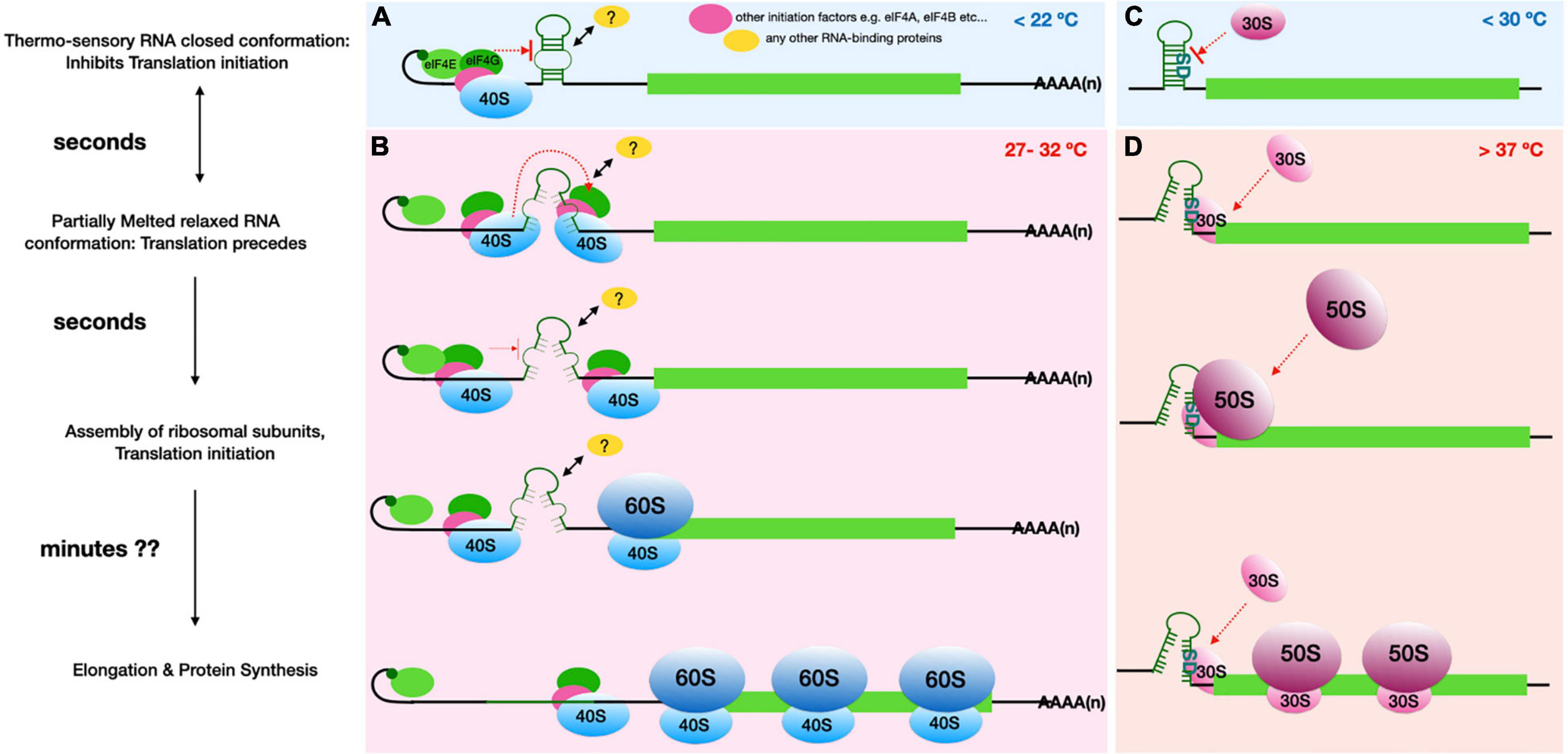
Figure 1. Comparison of the proposed plant RNA ThermoSwitch mechanism (A,B) with that of known Prokaryotic thermometers (C,D). (A) The plant RNA ThermoSwitch adopts a closed conformation at temperatures below 22°C, at which cap-dependent recruitment of 43S complex occurs, however further scanning and identification of the start codon is impeded by the ThermoSwitch. (B) When temperature reaches between 27 and 32°C, the ThermoSwitch RNA undergoes reversible partial melting, thereby adopting a relaxed conformation and allowing the 43S complex to precede scanning. The partially open conformation temporarily impedes further incoming scanning subunits until the preceding 43S complex encounters the start codon with sufficient initiation context, thereby facilitating 48S initiation complex formation followed by recruitment of the 60S ribosomal subunit. Thus, the switch from closed (<22°C) to relaxed conformation at 27-32°C may enhance translation whereby translation initiation rates are selectively increased at higher temperatures (Chung et al., 2020). The fully mature 80S ribosomes then enter the elongation stage of translation allowing protein synthesis, by which time (∼minutes) the ThermoSwitch RNA fully melts accompanied by the helicase activity of incoming scanning 43S complexes. It is also possible that the ThermoSwitch recruits an RNA binding protein, either at the stable conformation to further stabilise the ThermoSwitch at lower temperature (<22°C), or at the relaxed conformation to enhance 43S scanning. (C) The prokaryotic RNA thermometers operate by a distinct mechanism, since translation initiation here is dependent on the direct interaction of 16S rRNA within the 30S ribosomal subunit and the SD-sequence. The bacterial RNA thermosensors are typically located very close to the start codon and encompass the SD sequence preventing access to the initiating small ribosomal subunit. (D) At higher temperatures (>37°C), the RNA secondary structure either melts (e.g., ROSE element) or switches into an alternative structure (e.g., CspA cold shock transcript); both processes relieve the SD sequence from intra-molecular base pairing to allow interactions with the 30S ribosomal subunit and recruitment of 50S ribosomal subunit (Kortmann and Narberhaus, 2012). The fully assembled 70S ribosomes then proceed to elongation. Thus, translational induction mediated by prokaryotic RNA thermometers in cis is generally proportional to temperature: the higher the temperature, the less stable is the respective RNA secondary structure and therefore the more efficient is translation initiation. In both instances, translation initiation occurs within seconds of temperature rise while entry into elongation and complete protein synthesis may occur within minutes.
A well-studied class of bacterial RNA thermometers are the ROSE (repression of heat shock gene expression) elements which control the expression of small heat shock genes. Commonly found in Proteobacteria, these 5′-UTR hairpins undergo gradual melting in response to increasing temperature, which then leads to complete melting at high temperature (∼42°C) and full liberation of the SD and AUG start codon, allowing enhanced translation (Chowdhury et al., 2006; Waldminghaus et al., 2009). These mRNA regions often consist of up to 4 hairpins of varying stability. For example, the Escherichia coli ibpA RNA thermometer, which regulates expression of an inclusion-body associated protein, encompasses three RNA hairpins at the 5′-UTR. The two proximal hairpins were found to be stable at temperatures ranging from 20 to 50°C and may be involved in proper folding of the distal temperature sensitive hairpin encompassing the SD sequence and start codon (Waldminghaus et al., 2009).
RNA-based thermometers are effectively utilized by several plant pathogens and symbionts to regulate gene expression. Particularly well-studied in this context is the Rhizobiaceae family of plant symbiotic bacteria. In many rhizobial species, the expression of heat-shock genes is under the control of cis-acting ROSE elements. Structural and mutational studies on the small heat shock protein (HspA) transcript from Bradyrhizobium japonicum, a nitrogen-fixing root nodule bacterium, revealed a functional ROSE domain in the 5′-UTR that encompasses the transcript’s SD sequence. This domain forms an elaborate secondary structure consisting of four hairpins, which help in the ROSE at temperatures <30°C (Nocker et al., 2001a). Upon heat shock (∼42°C), the SD sequence gets exposed, allowing binding of the ribosomal 30S subunit.
Several non-canonical base pairs and transient hydrogen bonds are observed in all known ROSE-type thermometers in addition to a conserved short stretch of nucleotides UYGCU near the SD sequence. These secondary mRNA structure features are also observed in the heat-sensitive RNA structures of many other rhizobia such as Bradyrhizobium sp. (Parasponia), Mesorhizobium loti, and are functionally interchangeable among the species (Nocker et al., 2001a,b). High resolution NMR structures of the B. japonicum hspA 5′-UTR shows that the corresponding G83 nucleotide is involved in non-canonical base syn–anti base pairing with G94 in the SD region. In fact, the entire stretch of nucleotides complementary to the SD sequence are conserved in all known ROSE-like RNA elements in rhizobia (Chowdhury et al., 2006).
Similarly, translation of pAT-plasmid encoded small heat shock genes in Agrobacterium tumefaciens, plant pathogenic bacteria, is also mediated by ROSE-controlled expression. The heat shock gene transcripts hspAT1 and hspAT2 exhibit the characteristic features of ROSE-thermometers such as masked SD sequences and hairpin base pairing. In addition, the strictly conserved bulged G nucleotide opposite the SD sequence is observed in computer-aided secondary structure predictions of hspAT1 and hspAT2 transcripts (Balsiger et al., 2004). Thus, these RNA thermometers have evolved a conserved mechanism that is sufficient to control rapid synthesis of several heat-shock proteins in response to temperature.
In addition to the zipper-like RNA thermometers discussed so far, many bacteria contain a more switch-like thermoregulatory mechanism involving two mutually exclusive conformations. Examples of these are seen in bacterial cold shock responses such as in the expression of the CspA RNA chaperone in E. coli. The cspA transcript is unstable at 37°C but is highly stable when temperature drops to 10°C owing to reorganization of the mRNA thermosensory elements located at the 5′-UTR and extending a further 60 nucleotides downstream into the coding region. The rearrangements result in an alternate, mutually exclusive cspA mRNA conformation in response to cold shock, which in turn is efficiently translated due to the now highly exposed translation initiation sequence elements and low susceptibility to RNA degradation (Giuliodori et al., 2010; Phadtare and Severinov, 2010; Kortmann and Narberhaus, 2012).
Other well-characterized examples of bacterial RNA thermometers include those controlling virulence factor genes such as the mRNAs encoding Listeria monocytogenes virulence regulator PrfA (Johansson et al., 2002) and Yersinia virulence factor LcrF (Skurnik and Toivanen, 1992; Hoe and Goguen, 1993; Bohme et al., 2012; Pienkoss et al., 2021). In these pathogenic bacteria, a rapid temperature shift from low temperatures (<30°C) to 37°C, experienced upon host entry, triggers expression of the respective virulence factor genes. These protein factors in turn help evade immune responses and establish successful infection in the host. The 5′-UTR of Yersinia pestis lcrF controls access to the SD sequence via a small zipper-like RNA hairpin consisting of four consecutive uridines which base-pair with the SD sequence. Together with the Salmonella enterica small heat shock gene agsA mRNA 5′-UTR thermometer, this represents one of the founding members of the fourU type thermoregulatory RNA elements (Hoe and Goguen, 1993; Waldminghaus et al., 2007).
Interestingly, the L. monocytogenes prfA introduced above encompasses an elaborate zipper mechanism, which, in addition to encoding an elaborate temperature-controlled RNA secondary element in its 5′ leader, couples a truncated SAM riboswitch element (SreA), a trans-acting small RNA (sRNA). This sRNA in turn responds to elevated levels of S-adenosyl methionine (SAM) such as when inside the mammalian host, and functions in trans by binding to the prfA RNA thermosensor causing repression of PrfA synthesis. Thus, an additional feedback signal is integrated to regulate translation of the virulence regulator PrfA, thereby fine-tuning expression of several virulence genes and host immune modulators (Hoe and Goguen, 1993; Johansson et al., 2002; Bohme et al., 2012; Kortmann and Narberhaus, 2012).
In contrast to bacterial RNA thermometers, similar mechanisms in eukarya and archaea are only beginning to be understood. Some of the known or predicted eukaryotic thermoregulatory RNA mediates translation in response to temperature fluctuation, whether within physiological range, heat stress or cold shock. Rapid increase in temperature often leads to toxic accumulation of misfolded proteins which, if unchecked, can severely compromise cellular function even leading to apoptosis (Lindquist, 1986). A characteristic response to elevated temperatures involves the repression of normal cell metabolic processes and rapid production of large amounts of stress proteins known as heat shock proteins (HSPs) (Lindquist, 1981, 1986; Ahmed and Duncan, 2004).
The possibility of a rapid-heat inducible RNA structural element was predicted in the Hsp90 transcripts in Drosophila (Ahmed and Duncan, 2004). Interestingly, the Drosophila Hsp90 mRNA 5′-UTR consists of extensive secondary structure regions in contrast to other HSP mRNAs (Hsp70 and Hsp22) (Ahmed and Duncan, 2004). Despite the importance of these rapid stress responders in cellular homeostasis, the detailed mechanism of the Hsp90 mRNA structure mediated regulation of this heat shock response in Drosophila and other eukaryotes is poorly understood.
The upregulation of HSPs plays a crucial role in ameliorating such lethal effects by assisting proper folding of damaged proteins, allowing repair, localization, and transport of proteins and re-establishing the equilibrium (Taipale et al., 2010). Under stress stimuli, the HSPs preferentially bind to aggregated misfolded proteins releasing HSFs (heat shock factors), allowing their translocation to the nucleus and trimerization into the active form, in turn inducing expression of HSPs. Once the stress induced protein damage to the cells is relieved, the HSPs negatively regulate their own synthesis by interacting with HSFs (Solis et al., 2016; Schopf et al., 2017). Recent studies have identified a small trans-acting RNA forming an essential regulatory component of the mammalian heat-shock-factor-1 (HSF-1) by assisting autoregulation of HSPs. This RNA element, termed heat-shock RNA1 (HSR1), undergoes temperature-dependent changes in conformation driving activation of HSF-1 leading to transcription of heat-shock protein genes. In addition to humans and rodents, these highly conserved HSR1 sequences were also identified in Drosophila, Caenorhabditis elegans, Arabidopsis, and bacteria (Shamovsky et al., 2006; Choi et al., 2015; Somero, 2018).
Eukaryotic heat shock responses can be affected by temperature-sensitive RNA structures, often found in the 3′-UTR of the corresponding transcripts. Examples of these type of control have been reported in Leishmania and rice (O. sativa L.), where they modulate RNA stability and consequently affect translation rates of the associated coding region.
A recent genome-wide study of heat shock response in rice shows that RNA secondary structure reprogramming broadly regulates protein synthesis at elevated temperatures (Su et al., 2018). In this study a dimethyl sulfate (DMS) based structure-seq analysis of rice (O. sativa L.) seedlings revealed global structural changes in the transcriptome at elevated temperature of 42°C due to RNA unfolding. However, a parallel Ribo-seq analysis showed no correlation between RNA unfolding and heat-induced changes in translation, while RNA-seq based transcript quantification indicates that the observed structural changes promote transcript degradation. Hence the authors conclude that the downregulation of global translation in rice, which reduces heat-stress induced damage, is facilitated by increased access of the RNA degradation machinery to unfolded mRNA 5′ and 3′-UTRs. Yet another study analyzed the nucleotide compositions of transcriptomes of 906 land plants (Yang et al., 2022) and found that plants growing in cold climates have guanine (G)-enriched transcripts. These transcripts in turn have high propensity to form RG4 structures. Further in vivo structure probing in Arabidopsis combined with immunofluorescence studies show that the RG4 formation in 3′-UTRs of plant transcripts, which are globally increased in response to low temperature (∼ 4°C), lead to enhanced mRNA stability and adaptation to cold (Yang et al., 2022).
Studies on Leishmania heat shock protein Hsp83 transcripts identified nucleotides 1–472 in the proximal 3′-UTR as having thermoregulatory function (Larreta et al., 2004; David et al., 2010). Translation regulation is effectively utilized by digenetic parasites like Leishmania for differential expression of genes during their life cycle. These parasites undergo differentiation from promastigote stage, in the alimentary tract of sandfly, to amastigotes once phagocytosed by macrophages of the mammalian host (Larreta et al., 2004). Adaptation to these distinct environments is achieved largely by controlling post-transcriptional events such as mRNA processing, stability and through translation regulation. Preferential translation of trypanosomatids in response to environmental cues such as temperature and pH, are heavily mediated by 3′-UTR of stage-specific transcripts (Zilka et al., 2001).
The regulatory element within this proximal 3′-UTR region consists of a long polypyrimidine tract (PPT) located between positions 312 and 341. Computer-based structure predictions showed that this region is positioned on a highly probable secondary structure. Thermal melting profiles at 260 nm and RNAase H assays of WT and mutated 3′-UTRs suggest that this regulatory region undergoes partial melting during a temperature shift from 26 to 37°C, indicating the presence of a thermosensory RNA sequence. The study also shows that Hsp83 preferential translation requires scanning of the Hsp83 5′-UTR, unlike cap-independent translation of Hsps of many higher eukaryotes at elevated temperatures (Hernandez et al., 2004; Duncan, 2008; David et al., 2010). While a detailed study of the molecular mechanism of this thermoregulatory PPT in the Leishmania Hsp83 transcript 3′-UTR is pending, it is hypothesized that the temperature-mediated exposure of this region results in binding of regulatory factors such as PPT binding protein (PTB), allowing interactions with the translation initiation complex. This may in turn lead to circularization of the mRNA by bridging of the 5′ and 3′-ends, thereby enhancing protein synthesis at elevated temperatures (Gingras et al., 1999; David et al., 2010; Mishra et al., 2020).
Plants have evolved various strategies for adaption to changes in surrounding temperatures. A recent study in the model plant Arabidopsis demonstrates, for the first time, the presence of plant RNA ThermoSwitches which directly control the expression of several transcripts in response to warm temperatures (Chung et al., 2020). This study thus illuminates the existence of an mRNA structure-mediated control mechanism in response to temperature changes during eukaryotic translation. The temperature-dependent translational enhancement in Arabidopsis has been investigated in detail for transcript encoding transcription factor PIF7 (PHYTOCHROME INTERACTING FACTOR 7): it is mediated by the formation an mRNA hairpin in the 5′-UTR, approximately 30 nucleotides upstream of the start codon. Further biophysical studies combined with in vitro translation experiments suggest that the hairpin remains stable at temperatures <22°C but switches to a partially melted open conformation at temperatures of 27–32°C. This structural change results in a direct increase in PIF7 protein synthesis, and high PIF7 protein levels in turn promote the transcription of several downstream genes, including those of the auxin biosynthetic pathway, thereby enhancing elongation growth during warm daytime (Chung et al., 2020).
Similar hairpin sequences were identified in the 5′-UTRs of several other transcription factors such as WRKY22 and the heat shock regulator HSFA2, suggesting a conserved regulatory mechanism enabling plants to elicit rapid adaptive response at temperatures within physiological range (Chung et al., 2020). Computational analysis of these mRNA 5′-UTR regions shows high structural similarity consisting of a hairpin involving 28 nucleotides in the distal half of this region, despite the lack of sequence conservation.
These plant RNA ThermoSwitches appear to operate differently from bacterial RNA thermometers or from the mutually exclusive thermo-switch conformations seen in many bacterial cold shock protein transcripts (Breaker, 2010; Kortmann and Narberhaus, 2012), as depicted in Figure 1. In the bacterial system, translation initiation is dependent on the direct interaction of the 16S rRNA within the 30S ribosomal subunit and the SD-sequence, a purine-rich sequence approximately 5 nt upstream of the start codon on the messenger RNA (Steitz, 1969; Shine and Dalgarno, 1974; Steitz and Jakes, 1975). With the help of prokaryotic initiation factors IF1, IF2, and IF3, this interaction leads to the recruitment of the initiator met-tRNA to the start codon, followed by recruitment of the 50S ribosomal subunit to assemble the initiating 70S ribosome, placing the start codon:met-tRNA in the P-site of the 70S ribosome (Rodnina, 2018). The bacterial RNA thermometers are typically located very close to the start codon and encompass the SD sequence. At high temperature, the RNA secondary structure either melts (e.g., ROSE element) or switches into an alternative structure (e.g., CspA cold shock transcript); both processes relieve the SD sequence from intra-molecular base pairing to allow interactions with the 16S rRNA on the 30S ribosomal subunit (Kortmann and Narberhaus, 2012). Thus, translational induction mediated by these RNA thermometers in cis is generally proportional to temperature: the higher the temperature, the less stable is the respective RNA secondary structure and therefore the more efficient is translation initiation (Figure 1).
In contrast, hallmarks of canonical eukaryotic translation initiation involve scanning of the 43S complex through the 5′-UTR (Kozak, 1989; Jackson et al., 2010). Eukaryotic translation initiation is a highly involved multi-step process starting with recognition of the 5′ cap on the mRNA by the eIF4F complex, which leads to the recruitment of the 43S complex, comprising the 40S subunit and several initiating factors, including the eIF2-GTP-Met-tRNAMeti complex, to the 5′ end of the mRNA to begin the scanning process. Scanning by the 43S complex consists of two linked processes: movement of the 43S complex in a strictly 5′ to 3′ dependent manner and unwinding of secondary structures at the same time. Initiation then begins once the 43S complex encounters a start codon with sufficient initiation context, particularly with a purine at −3 and +4 position also known as Kozak consensus. The process is facilitated by eukaryotic initiation factors eIF1 and eIF1a and places the eIF2-GTP-Met-tRNAMeti with the start codon through base pairing with the tRNA anticodon followed by joining of the 60S ribosomal subunit. At higher temperatures the incoming scanning 43S complex is temporarily impeded by the relaxed, partially melted RNA ThermoSwitch, instead of interrupting the process of 48S Initiation complex formation facilitating recruitment of 60S ribosomal unit and assembly of the fully mature 80S ribosome to enter the elongation stage of translation (i.e., protein synthesis). Therefore, rather than providing accessibility for the ribosomal subunits to join the mRNA, the plant RNA ThermoSwitch is likely to regulate protein synthesis through the scanning process, possibly assuming a dual role: while its stable structure at temperatures less than 22°C may well impede movement of the scanning complex, its more relaxed structure at 27–32°C acts as a translational enhancer, increasing translation initiation rates selectively at these temperatures (Figure 1). This mode of action is evidenced by the fact that both disrupting and stabilizing the hairpin structure has a detrimental effect on translation rates and protein synthesis (Chung et al., 2020). The precise mechanism by which the ThermoSwitch enhances translation remains to be determined. RNA secondary structures are known to regulate RNA–RNA (Van Treeck and Parker, 2018) and RNA-protein interactions (Sanchez De Groot et al., 2019); such interactors may include RNAs or RNA-binding proteins that directly facilitate translation initiation, or proteins that affect RNA modification or localization and thereby indirectly influence the initiation process. In case of both prokaryotic and known eukaryotic thermoregulatory RNAs, the temperature mediated conformational change in RNA secondary structure and translation initiation happens within seconds of temperature rise while entry into elongation and complete protein synthesis may occur within minutes. Table 1 summarizes the known and predicted RNA structure mediated thermo-sensors discussed in this review.
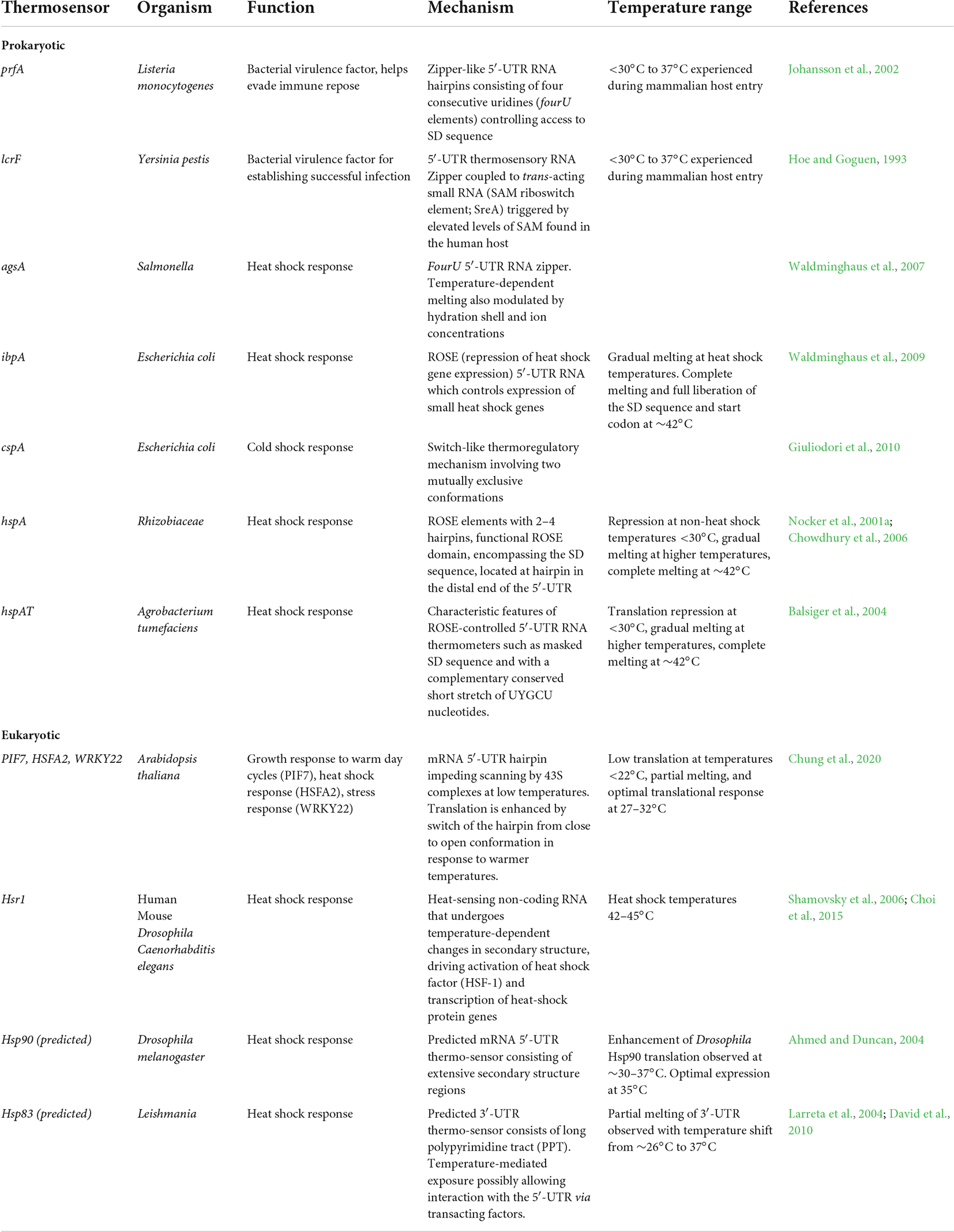
Table 1. Summary of known and predicted RNA structure mediated thermo-sensors discussed in this review.
The recent discovery of thermosensory RNA structures in plants emphasizes the important regulatory role RNA can play in the control of gene expression. Notably, we now know that RNA structures as part of thermosensory signaling modules are not confined to prokaryotes, but may be prevalent in eukaryotic systems, opening a new chapter of research into thermosensor research. It will be exciting to see whether similar thermosensory structures will be discovered in other eukaryotic species and how such structures have been harnessed by different organisms to tailor their development and physiology to their surrounding temperatures.
In addition, thermosensory RNA structures may provide a new means to manipulate gene expression to improve plant fitness and crop yield. It might become possible to tailor a gene’s expression level to specific temperature regimes by the addition of 5′-UTR thermoswitches or 3′-UTR G-quadruplexes. Temperature regulation of these structures is quick and, in case of thermoswitches, reversible, allowing for precise fine-tuning of temperature responses. The required structures could be introduced through gene editing techniques, avoiding the need to generate transgenic plants.
Likewise, plant RNA thermosensors can be utilized for advanced synthetic biology applications such as in inducible expression of plant pigments, phytochemicals and other biologicals of interest to biotechnological, pharmaceutical and food industry. Some recent studies involving the development of novel inducible riboswitches for metabolic engineering in green alga Chlamydomonas reinhardtii (Mehrshahi et al., 2020) and in tobacco (Nicotiana tabacum) plastids (Agrawal et al., 2022) are encouraging advances toward this aim. Further understanding of the essential features of these molecular thermometers and their precise functional mechanisms will facilitate future development of such biotechnological toolkits.
BC proposed the manuscript. BC and ST researched the content. All authors contributed to writing of the manuscript and approved the final version.
ST and BC are supported by the Medical Research Council Fellowship (MR/R021821/1). MB was supported by the Royal Society (University Research Fellowship: URF\R1\211672). BC lab was supported by the MRC (MR/R021821/1) and BBSRC project grants (BB/V017780/1, BB/V006096/1, and BB/W510609/1).
The authors would like to thank Tom Dever and Ivaylo Ivanov for discussions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agrawal, S., Karcher, D., Ruf, S., Erban, A., Hertle, A. P., Kopka, J., et al. (2022). Riboswitch-mediated inducible expression of an astaxanthin biosynthetic operon in plastids. Plant Physiol. 188, 637–652. doi: 10.1093/plphys/kiab428
Ahmed, R., and Duncan, R. F. (2004). Translational regulation of Hsp90 mRNA. AUG-proximal 5’-untranslated region elements essential for preferential heat shock translation. J. Biol. Chem. 279, 49919–49930. doi: 10.1074/jbc.M404681200
Balsiger, S., Ragaz, C., Baron, C., and Narberhaus, F. (2004). Replicon-specific regulation of small heat shock genes in Agrobacterium tumefaciens. J. Bacteriol. 186, 6824–6829. doi: 10.1128/JB.186.20.6824-6829.2004
Bartys, N., Kierzek, R., and Lisowiec-Wachnicka, J. (2019). The regulation properties of RNA secondary structure in alternative splicing. Biochim. Biophys. Acta Gene Regul. Mech. 1862:194401. doi: 10.1016/j.bbagrm.2019.07.002
Bohme, K., Steinmann, R., Kortmann, J., Seekircher, S., Heroven, A. K., and Berger, E. (2012). Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 8:e1002518. doi: 10.1371/journal.ppat.1002518
Breaker, R. R. (2010). RNA switches out in the cold. Mol. Cell. 37, 1–2. doi: 10.1016/j.molcel.2009.12.032
Broft, P., Rosenkranz, R., Schleiff, E., Hengesbach, M., and Schwalbe, H. (2022). Structural analysis of temperature-dependent alternative splicing of HsfA2 pre-mRNA from tomato plants. RNA Biol. 19, 266–278. doi: 10.1080/15476286.2021.2024034
Cheng, C., Gao, X., Feng, B., Sheen, J., Shan, L., and He, P. (2013). Plant immune response to pathogens differs with changing temperatures. Nat. Commun. 4:2530. doi: 10.1038/ncomms3530
Choi, D., Oh, H. J., Goh, C. J., Lee, K., and Hahn, Y. (2015). Heat Shock RNA 1, Known as a Eukaryotic Temperature-Sensing Noncoding RNA, Is of Bacterial Origin. J. Microbiol. Biotechnol. 25, 1234–1240. doi: 10.4014/jmb.1505.05014
Chowdhury, S., Maris, C., Allain, F. H., and Narberhaus, F. (2006). Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 25, 2487–2497. doi: 10.1038/sj.emboj.7601128
Chung, B. Y. W., Balcerowicz, M., Di Antonio, M., Jaeger, K. E., Geng, F., Franaszek, K., et al. (2020). An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat. Plants 6, 522–532. doi: 10.1038/s41477-020-0633-3
Cruz, J. A., and Westhof, E. (2009). The dynamic landscapes of RNA architecture. Cell 136, 604–609. doi: 10.1016/j.cell.2009.02.003
David, M., Gabdank, I., Ben-David, M., Zilka, A., Orr, I., Barash, D., et al. (2010). Preferential translation of Hsp83 in Leishmania requires a thermosensitive polypyrimidine-rich element in the 3’ UTR and involves scanning of the 5’ UTR. RNA 16, 364–374. doi: 10.1261/rna.1874710
Duncan, R. F. (2008). Rapamycin conditionally inhibits Hsp90 but not Hsp70 mRNA translation in Drosophila: implications for the mechanisms of Hsp mRNA translation. Cell Stress Chaperones 13, 143–155. doi: 10.1007/s12192-008-0024-6
Gingras, A. C., Raught, B., and Sonenberg, N. (1999). eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913–963. doi: 10.1146/annurev.biochem.68.1.913
Giuliodori, A. M., Di Pietro, F., Marzi, S., Masquida, B., Wagner, R., Romby, P., et al. (2010). The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell 37, 21–33. doi: 10.1016/j.molcel.2009.11.033
Hayes, S., Schachtschabel, J., Mishkind, M., Munnik, T., and Arisz, S. A. (2021). Hot topic: thermosensing in plants. Plant Cell Environ. 44, 2018–2033. doi: 10.1111/pce.13979
Hernandez, G., Vazquez-Pianzola, P., Sierra, J. M., and Rivera-Pomar, R. (2004). Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA 10, 1783–1797. doi: 10.1261/rna.7154104
Hoe, N. P., and Goguen, J. D. (1993). Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J. Bacteriol. 175, 7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993
Huot, B., Castroverde, C. D. M., Velasquez, A. C., Hubbard, E., Pulman, J. A., Yao, J., et al. (2017). Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nat. Commun. 8:1808. doi: 10.1038/s41467-017-01674-2
Jackson, R. J., Hellen, C. U., and Pestova, T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 11, 113–127. doi: 10.1038/nrm2838
Jacobs, E., Mills, J. D., and Janitz, M. (2012). The role of RNA structure in posttranscriptional regulation of gene expression. J. Genet. Genomics 39, 535–543. doi: 10.1016/j.jgg.2012.08.002
Johansson, J., Mandin, P., Renzoni, A., Chiaruttini, C., Springer, M., and Cossart, P. (2002). An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110, 551–561. doi: 10.1016/s0092-8674(02)00905-4
Kortmann, J., and Narberhaus, F. (2012). Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 10, 255–265. doi: 10.1038/nrmicro2730
Kozak, M. (1989). The scanning model for translation: an update. J. Cell Biol. 108, 229–241. doi: 10.1083/jcb.108.2.229
Larreta, R., Soto, M., Quijada, L., Folgueira, C., Abanades, D. R., Alonso, C., et al. (2004). The expression of HSP83 genes in Leishmania infantum is affected by temperature and by stage-differentiation and is regulated at the levels of mRNA stability and translation. BMC Mol. Biol. 5:3. doi: 10.1186/1471-2199-5-3
Lazzaretti, D., and Bono, F. (2017). mRNA localization in metazoans: a structural perspective. RNA Biol. 14, 1473–1484. doi: 10.1080/15476286.2017.1338231
Lindquist, S. (1981). Regulation of protein synthesis during heat shock. Nature 293, 311–314. doi: 10.1038/293311a0
Lindquist, S. (1986). The heat-shock response. Annu. Rev. Biochem. 55, 1151–1191. doi: 10.1098/rstb.2016.0525
Megha, S., Basu, U., and Kav, N. N. V. (2018). Regulation of low temperature stress in plants by microRNAs. Plant Cell Environ. 41, 1–15. doi: 10.1111/pce.12956
Mehrshahi, P., Nguyen, G., Gorchs Rovira, A., Sayer, A., Llavero-Pasquina, M., Lim Huei Sin, M., et al. (2020). Development of Novel Riboswitches for Synthetic Biology in the Green Alga Chlamydomonas. ACS Synth. Biol. 9, 1406–1417. doi: 10.1021/acssynbio.0c00082
Menna, A., Nguyen, D., Guttman, D. S., and Desveaux, D. (2015). Elevated Temperature Differentially Influences Effector-Triggered Immunity Outputs in Arabidopsis. Front. Plant Sci. 6:995. doi: 10.3389/fpls.2015.00995
Meyer, M., Plass, M., Perez-Valle, J., Eyras, E., and Vilardell, J. (2011). Deciphering 3’ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Mol. Cell 43, 1033–1039. doi: 10.1016/j.molcel.2011.07.030
Mishra, R. K., Datey, A., and Hussain, T. (2020). mRNA Recruiting eIF4 Factors Involved in Protein Synthesis and Its Regulation. Biochemistry 59, 34–46. doi: 10.1021/acs.biochem.9b00788
Narberhaus, F., Waldminghaus, T., and Chowdhury, S. (2006). RNA thermometers. FEMS Microbiol. Rev. 30, 3–16.
Nocker, A., Hausherr, T., Balsiger, S., Krstulovic, N. P., Hennecke, H., and Narberhaus, F. (2001a). A mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res. 29, 4800–4807. doi: 10.1093/nar/29.23.4800
Nocker, A., Krstulovic, N. P., Perret, X., and Narberhaus, F. (2001b). ROSE elements occur in disparate rhizobia and are functionally interchangeable between species. Arch. Microbiol. 176, 44–51. doi: 10.1007/s002030100294
Nomoto, Y., Kubozono, S., Miyachi, M., Yamashino, T., Nakamichi, N., and Mizuno, T. (2013). Circadian clock and PIF4-mediated external coincidence mechanism coordinately integrates both of the cues from seasonal changes in photoperiod and temperature to regulate plant growth in Arabidopsis thaliana. Plant Signal. Behav. 8:e22863. doi: 10.4161/psb.22863
Phadtare, S., and Severinov, K. (2010). RNA remodeling and gene regulation by cold shock proteins. RNA Biol. 7, 788–795.
Pienkoss, S., Javadi, S., Chaoprasid, P., Nolte, T., Twittenhoff, C., Dersch, P., et al. (2021). The gatekeeper of Yersinia type III secretion is under RNA thermometer control. PLoS Pathog. 17:e1009650. doi: 10.1371/journal.ppat.1009650
Qiu, J., Xie, J., Chen, Y., Shen, Z., Shi, H., Naqvi, N. I., et al. (2022). Warm temperature compromises JA-regulated basal resistance to enhance Magnaporthe oryzae infection in rice. Mol. Plant 15, 723–739. doi: 10.1016/j.molp.2022.02.014
Righetti, F., Nuss, A. M., Twittenhoff, C., Beele, S., Urban, K., Will, S., et al. (2016). Temperature-responsive in vitro RNA structurome of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 113, 7237–7242.
Rouskin, S., Zubradt, M., Washietl, S., Kellis, M., and Weissman, J. S. (2014). Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705. doi: 10.1038/nature12894
Sanchez De Groot, N., Armaos, A., Grana-Montes, R., Alriquet, M., Calloni, G., Vabulas, R. M., et al. (2019). RNA structure drives interaction with proteins. Nat. Commun. 10:3246.
Schopf, F. H., Biebl, M. M., and Buchner, J. (2017). The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 18, 345–360.
Shamovsky, I., Ivannikov, M., Kandel, E. S., Gershon, D., and Nudler, E. (2006). RNA-mediated response to heat shock in mammalian cells. Nature 440, 556–560.
Shiina, T., and Shimizu, Y. (2020). Temperature-Dependent Alternative Splicing of Precursor mRNAs and Its Biological Significance: a Review Focused on Post-Transcriptional Regulation of a Cold Shock Protein Gene in Hibernating Mammals. Int. J. Mol. Sci. 21:7599. doi: 10.3390/ijms21207599
Shine, J., and Dalgarno, L. (1974). The 3’-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U. S. A. 71, 1342–1346. doi: 10.1073/pnas.71.4.1342
Skurnik, M., and Toivanen, P. (1992). LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Bacteriol. 174, 2047–2051. doi: 10.1128/jb.174.6.2047-2051.1992
Soemedi, R., Cygan, K. J., Rhine, C. L., Glidden, D. T., Taggart, A. J., Lin, C. L., et al. (2017). The effects of structure on pre-mRNA processing and stability. Methods 125, 36–44.
Solis, E. J., Pandey, J. P., Zheng, X., Jin, D. X., Gupta, P. B., Airoldi, E. M., et al. (2016). Defining the Essential Function of Yeast Hsf1 Reveals a Compact Transcriptional Program for Maintaining Eukaryotic Proteostasis. Mol. Cell. 63, 60–71.
Somero, G. N. (2018). RNA thermosensors: how might animals exploit their regulatory potential? J. Exp. Biol. 221:jeb162842. doi: 10.1242/jeb.162842
Song, Y. H., Kubota, A., Kwon, M. S., Covington, M. F., Lee, N., Taagen, E. R., et al. (2018). Molecular basis of flowering under natural long-day conditions in Arabidopsis. Nat. Plants 4, 824–835. doi: 10.1038/s41477-018-0253-3
Steitz, J. A. (1969). Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature 224, 957–964.
Steitz, J. A., and Jakes, K. (1975). How ribosomes select initiator regions in mRNA: base pair formation between the 3’ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 72, 4734–4738. doi: 10.1073/pnas.72.12.4734
Su, Z., Tang, Y., Ritchey, L. E., Tack, D. C., Zhu, M., Bevilacqua, P. C., et al. (2018). Genome-wide RNA structurome reprogramming by acute heat shock globally regulates mRNA abundance. Proc. Natl. Acad. Sci. U. S. A. 115, 12170–12175. doi: 10.1073/pnas.1807988115
Taipale, M., Jarosz, D. F., and Lindquist, S. (2010). HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528. doi: 10.1038/nrm2918
Van Treeck, B., and Parker, R. (2018). Emerging Roles for Intermolecular RNA-RNA Interactions in RNP Assemblies. Cell 174, 791–802.
Waldminghaus, T., Gaubig, L. C., Klinkert, B., and Narberhaus, F. (2009). The Escherichia coli ibpA thermometer is comprised of stable and unstable structural elements. RNA Biol. 6, 455–463. doi: 10.4161/rna.6.4.9014
Waldminghaus, T., Heidrich, N., Brantl, S., and Narberhaus, F. (2007). FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 65, 413–424. doi: 10.1111/j.1365-2958.2007.05794.x
Yang, X., Yu, H., Duncan, S., Zhang, Y., Cheema, J., Miller, J. B., et al. (2022). RNA G-quadruplex structure contributes to cold adaptation in plants. bioRxiv [Preprint]. doi: 10.1101/2022.03.04.482910
Zilka, A., Garlapati, S., Dahan, E., Yaolsky, V., and Shapira, M. (2001). Developmental regulation of heat shock protein 83 in Leishmania. 3’ processing and mRNA stability control transcript abundance, and translation id directed by a determinant in the 3’-untranslated region. J. Biol. Chem. 276, 47922–47929. doi: 10.1074/jbc.M108271200
Keywords: RNA structure, plants, translation, protein synthesis, thermosensor, temperature
Citation: Thomas SE, Balcerowicz M and Chung BY-W (2022) RNA structure mediated thermoregulation: What can we learn from plants? Front. Plant Sci. 13:938570. doi: 10.3389/fpls.2022.938570
Received: 07 May 2022; Accepted: 27 June 2022;
Published: 17 August 2022.
Edited by:
William Zerges, Concordia University, CanadaReviewed by:
Steven Adriaan Arisz, University of Amsterdam, NetherlandsCopyright © 2022 Thomas, Balcerowicz and Chung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Betty Y.-W. Chung, YmN5MjNAY2FtLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.