- 1State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, South China Agricultural University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Protein Function and Regulation in Agricultural Organisms, College of Life Sciences, South China Agricultural University, Guangzhou, China
- 3Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China
- 4State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Sciences and Technology, Guangxi University, Nanning, China
Eukaryotic translation initiation factor 4E1 (eIF4E1) is required for the initiation of protein synthesis. The biological function of eIF4E1 in plant–potyvirus interactions has been extensively studied. However, the role of eIF4E1 in Arabidopsis development remains unclear. In this study, we show that eIF4E1 is highly expressed in the embryo and root apical meristem. In addition, eIF4E1 expression is induced by auxin. eIF4E1 mutants show embryonic cell division defects and short primary roots, a result of reduced cell divisions. Furthermore, our results show that mutation in eIF4E1 severely reduces the accumulation of PIN-FORMED (PIN) proteins and decreases auxin-responsive gene expression at the root tip. Yeast two-hybrid assays identified that eIF4E1 interacts with an RAC/ROP GTPase activator, RopGEF7, which has been previously reported to be involved in the maintenance of the root apical meristem. The interaction between eIF4E1 and RopGEF7 is confirmed by protein pull-down and bimolecular fluorescent complementation assays in plant cells. Taken together, our results demonstrated that eIF4E1 is important for auxin-regulated embryo development and root growth. The eIF4E1–RopGEF7 interaction suggests that eIF4E1 may act through ROP signaling to regulate auxin transport, thus regulating auxin-dependent patterning.
Introduction
RAC/ROP GTPases are multi-functional signaling molecules which regulate a wide range of processes during plant growth and development, including pollen tube growth, leaf epidermal cell morphogenesis, root hair development, embryonic development, seedling growth, stress tolerance, and hormone response (Yang and Fu, 2007; Wu et al., 2011; Craddock et al., 2012; Bloch and Yalovsky, 2013; Li et al., 2021). RAC/ROPs undergo exchange reactions between an active GTP-bound form and an inactive GDP-bound form (Nibau et al., 2013; Akamatsu et al., 2015). The cycling of GTP- and GDP-bound forms is regulated by GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs). In short, GAPs catalyze GTP hydrolysis and convert them back to GDP-bound forms. Oppositely, GEFs activate RAC/ROPs by stimulating the exchange of GDP- to GTP-bound forms (Yalovsky, 2015). The plant-specific family of GEFs (i.e., RopGEFs) is utilized predominantly by RAC/ROPs to activate the signaling pathway to regulate plant development (Berken et al., 2005; Chen et al., 2011; Huang et al., 2018).
Arabidopsis possesses a distinct small GTPase family of RopGEFs (Gu et al., 2006; Liu et al., 2017). There are 14 RopGEF genes in Arabidopsis, namely, RopGEF1 to RopGEF14, which are versatile signaling molecules in plant growth and development, as well as responses to various stresses (Wu et al., 2011; Nibau et al., 2013; Liu et al., 2017; Ashraf and Rahman, 2019). The transient expression of RopGEF1 and RopGEF12 suggested that these genes mediate polarized pollen tube growth (Gu et al., 2006; Zhang and McCormick, 2007; Chang et al., 2013). In addition, studies of ropgef1 ropgef4 double mutants and overexpression transgenic lines showed that RopGEF1 and RopGEF4 are involved in ABA-mediated stomatal closure in response to drought stress (Li and Liu, 2012). A recent study reported that RopGEF1 participates in the ABA-mediated inhibition of lateral root growth via ABA-mediated protein degradation (Li et al., 2016). Moreover, ABA-induced trafficking and degradation of RopGEF1 are disrupted in a cpk3/4/6/11 quadruple mutant of calcium-dependent protein kinase (CPK) genes in Arabidopsis, suggesting that RopGEF1 cooperates with CPKs and plays crucial roles in root hair polar growth and seedling growth (Li Z. et al., 2018). In our previous study, we have shown that RopGEF1 plays essential roles in the early embryonic and seedling development through regulating polar auxin transport and cell polarity (Liu et al., 2017). Another study reported that phytochrome B (phyB) acts genetically upstream of RopGEF2 and RopGEF4 to activate RopGEF2 regulating stomatal opening (Wang et al., 2017). Our group reported that RopGEF7 participates in regulating auxin-dependent PLETHORA1 (PLT1)- and PLT2-mediated maintenance of root stem cell niches (Chen et al., 2011). In addition, we found that ROP3, which interacts with RopGEF7 (Chen et al., 2011), plays a critical role in maintaining the polarity of auxin efflux proteins (PINs) at the plasma membrane (PM), and whereby regulates embryonic development and postembryonic growth (Huang et al., 2014). To further investigate the role of RopGEF7, we used RopGEF7 as a bait in the screen of Arabidopsis seedling cDNA library and identified a potential interaction protein, a translation initiation factor, eIF4E1.
Eukaryotic translation initiation factor 4E (eIF4E) was initially named as the “cap-binding protein” since it interacts specifically with the 5′-terminal cap of mRNA to initiate mRNA translation, a rate-limiting step of protein synthesis (Jackson et al., 2010; Wang and Krishnaswamy, 2012). In Arabidopsis, eIF4E comprises a small multiple gene family, including eIF4E1 (At4g18040, also known as the canonical eIF4E), eIF4E2 (At1g29590, also known as eIF4E1C), eIF4E3 (At1g29550, also known as eIF4E1B), eIF4E1 isoform eIF(iso)4E (At5g35620), and eIF4E isoforms nCBP-1 and nCBP-2 (Patrick et al., 2014; Kropiwnicka et al., 2015; Tajima et al., 2017; Gomez et al., 2019). Published microarray and RNA sequencing data indicate that eIF4E1 and eIF(iso)4E are broadly expressed in various plant tissues at high levels (Zimmermann et al., 2004; Winter et al., 2007), whereas eIF4E2 and eIF4E3 are mainly restricted to reproductive tissues and developing embryos at low levels (Zimmermann et al., 2004; Winter et al., 2007; Patrick et al., 2014). Thus, eIF4E1 and eIF(iso)4E show the major activity of this small gene family.
Extensive studies have shown that eIF4E genes play an essential role in plant virus infection. Natural mutations in eIF4E1 and eIF(iso)4E enhance the resistance against plant viruses in diverse plant species (Li G. et al., 2018; Liu and Goss, 2018). ncbp-1/ncbp-2, a double mutant of the cassava eIF4E isoforms nCBP-1 and nCBP-2, displayed delayed and attenuated cassava brown streak disease symptom (Gomez et al., 2019). However, how exactly eIF4E mediates plant growth and development has not been well-studied. Downregulation of both eIF4E1 and eIF(iso)4E in antisense tobacco plants caused a semi-dwarf phenotype (Combe et al., 2005). Similarly, silencing of both eIF4E1 and eIF4E2 with RNAi strategy in tomato resulted in dwarf plants with fewer seeds and smaller fruits (Mazier et al., 2011). A more pronounced phenotype was observed in the tomato transgenic lines silenced for eIF4E1, eIF4E2, and eIF(iso)4E (Mazier et al., 2011). The knockout of eIF(iso)4E and overexpression of transgenic plants were associated with phosphate-regulated root growth (Martínez-Silva et al., 2012). Arabidopsis eif4e1 knockout mutants displayed a consistent 7-day bolting delay compared with the wild-type Columbia (Bastet et al., 2018). In this article, we report a specific role for eIF4E1 in embryo development and root growth through interactions with RopGEF7 in the regulation of the auxin pathway.
Results
eIF4E1 Is Expressed in Embryos and Seedlings and Is Induced by Auxin
To investigate the expression patterns of eIF4E1, we generated the transgenic plants of eIF4E1pro:YFP-eIF4E1 with a YFP reporter, and eIF4E1pro:GUS with a β-glucuronidase (GUS) reporter. Analysis of several independent eIF4E1pro:YFP-eIF4E1 transgenic lines showed that eIF4E1pro:YFP-eIF4E1 was ubiquitously expressed in both embryo proper and suspensor during embryogenesis. In detail, the YFP signal was detected in 8-cell, 16-cell, 32-cell, globular, triangle, early heart, and heart and mature embryo stages (Figure 1, A1–9), compared to the wild-type embryos which did not show any GFP fluorescence (Supplementary Figure S1). Similarly, eIF4E1pro:GUS transgenic lines displayed that GUS activities were expressed in embryogenesis. The GUS activities were detected in early globular, globular, early heart, and heart and mature embryo stages (Supplementary Figure S2). In postembryonic stages, eIF4E1 was expressed in primary roots (Figure 1, B1, 2; Supplementary Figure S3), lateral root primordia and lateral roots (Figure 1, B3, 4), and anthers (Figure 1, B5, 6), especially with strong expression in pollens (Figure 1, B6). To investigate how eIF4E1 expression is regulated, we treated the roots of 7-day-old eIF4E1pro:GUS seedlings with exogenous auxin NAA. The results indicated that GUS activity was markedly induced by NAA treatment, especially in the meristem region. Compared to untreated control, NAA enlarged the expression area of eIF4E1 and enhanced GUS activity in the elongation zone of roots (Figure 1C). To confirm such observations, we analyzed the eIF4E1 relative expression levels by quantitative real-time PCR (qRT-PCR) in 7-day-old wild-type seedling roots. eIF4E1 expression showed an increase following NAA treatment to up to 24 h; 24-h NAA treatment induced over a two-fold increase in eIF4E1 relative expression (Figures 1D,E).
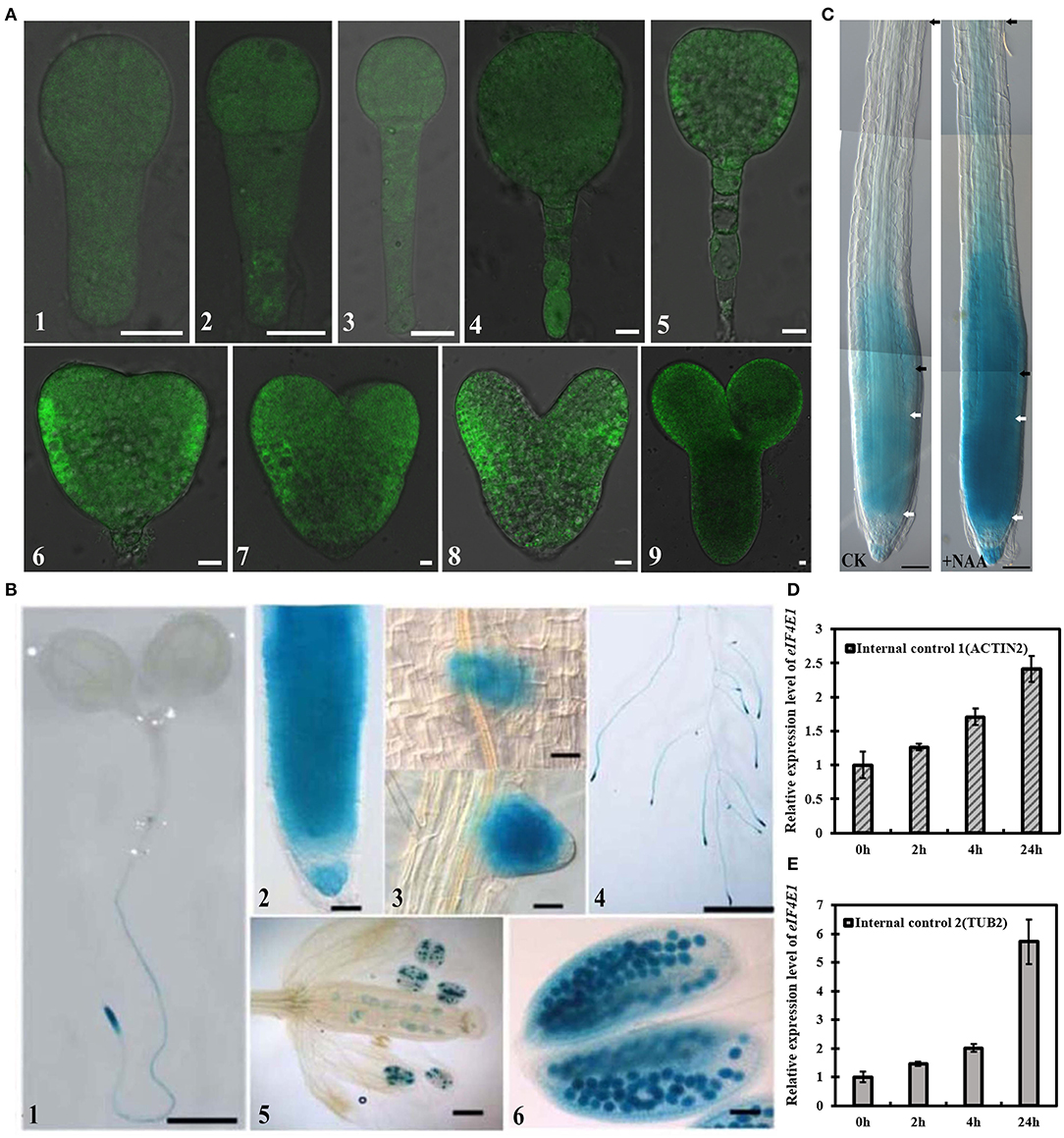
Figure 1. Expression profiles of eIF4E1 in Arabidopsis. (A) eIF4E1pro:YFP-eIF4E1 was expressed in embryos. The confocal images were merged by the DIC and the GFP channels. 8-cell (A1), 16-cell (A2), 32-cell (A3), globular (A4), triangle (A5), early heart (A6, 7), heart (A8), and mature (A9) embryo stages. (B) Detection of GUS activity in the primary roots (B1, 2) of 7-day-old eIF4E1pro:GUS seedlings, the early stage (up panel) and the late stage (down panel) of lateral root primordia (B3), and lateral roots (B4), as well as in anthers and pollens (B5, 6). (C) Detection of GUS activity under exogenous 10 μM NAA treatment. To display the meristem zone and entire elongation zone, (C) is generated by three field photographs (using the Photoshop6 software overlap function). The region between two white or two black arrowheads indicates the meristem or elongation zone, respectively. CK, a control without NAA treatment. (D,E) Expression level of eIF4E1 was significantly induced in 7-day-old wild-type seedlings by 10 μM NAA treatment. Transcription levels were normalized to ACTIN 2 (D) or TUBULIN 2 (E), respectively. Data are presented as mean values of three biological repeats. Scale bars: (A) 10 μm; (B1–4) 2 mm; (B5, 6; C) 50 μm.
Disruption of eIF4E1 Results in Developmental Defects During Embryogenesis and Seedling Growth
The expression patterns of eIF4E1 suggested that eIF4E1 may be functionally important for embryogenesis. To examine more preciously the role of eIF4E1, eif4e1-1 carrying a single base replacement mutation which caused a nonsense mutation from Trp (TGG) to a stop codon (TGA), and a T-DNA insertion mutant eif4e1-2 were used (Supplementary Figure S4A). The relative expression of eIF4E1 was undetectable in eif4e1-1 or reduced to <20% in eif4e1-2, respectively, compared to those of wild-type Col-0 (Supplementary Figures S4B,C).
Embryonic development defects were observed in eif4e1-1 and eif4e1-2 from the 8-cell stage. From the 8-cell to early heart stages, eif4e1-1 and eif4e1-2 embryos display abnormal divisions in contrast to the wild type (Figure 2, A1–4). eif4e1 mutants showed abnormal divisions in embryo proper and suspensor, which failed to form a clear boundary between apical and basal regions (Figure 2, A5, 6, 10), as well as an organized root pole (Figure 2, A7, 8, 12). In addition, from the 8-cell to early heart stages, mutant embryos showed aberrant cell divisions in suspensor in eif4e1-2 (Figure 2, A9, 11); 9.92% of eif4e1-1 (n = 252) and 8.88% of eif4e1-2 (n = 259) embryos displayed embryo developmental defects. The globular stages have the highest defect percentage with 15.15% (n = 99) in eif4e1-1 and 11.02% (n = 118) in eif4e1-2, respectively, in contrast to wild type, which only had very low percentage of abnormal embryos (1.20%, n = 167), as shown in Supplementary Table S2. Taken together, eIF4E1 is important for embryonic development in Arabidopsis, even though only ~10% of embryos showed a defect.
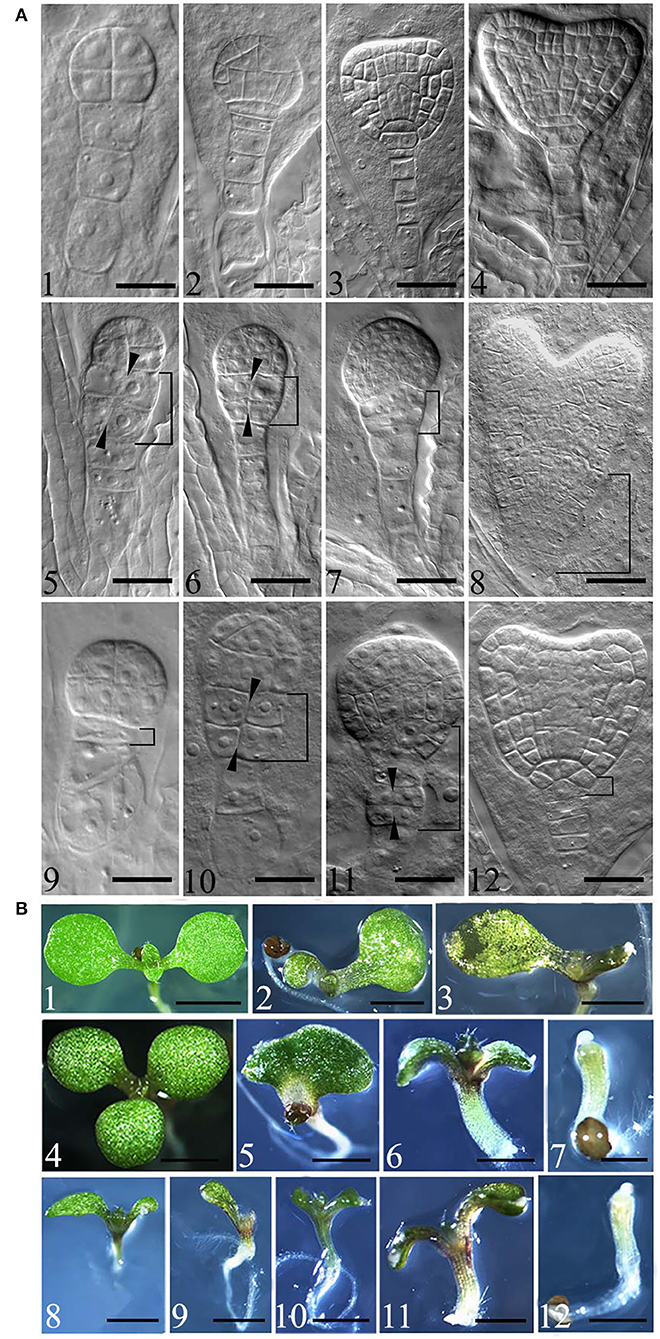
Figure 2. Mutation of eIF4E1 induces developmental defects in embryo and seedlings. (A) Embryo defects in eif4e1 mutants. Embryos at 8-cell to heart stages from wild type (A1–4), eif4e1-1 (A5–8), and eif4e1-2 (A9–12). The black arrowheads show the position of the aberrant cell division plate. The bracketed area indicates cell division defects region. (B) Seedling phenotypes of eif4e1 mutants. Seven-day-old seedlings of wild type (B1), eif4e1-1 (B2–7), and eif4e1-2 (B8–12) mutants. Scale bars: (A) 50 μm; (B) 2 mm.
To gain further insights into how eIF4E1 functions in plant development, we investigated the postembryonic development using 7-day-old seedlings of eif4e1-1 and eif4e1-2. Under normal growth conditions, the wild type has two symmetrical cotyledons and two true leaves (Figure 2, B1). We found that eif4e1-1 and eif4e1-2 showed similar phenotypes, for example, developmental defects of cotyledons (Figure 2, B2, 8), deterioration in advance of cotyledons (Figure 2, B3, 9, 10), mutant seedlings with three cotyledons (Figure 2, B4), single cotyledon (Figure 2, B5), extremely short roots (Figure 2, B6, 7, 11, 12), and cotyledon missing (Figure 2, B7, 12). Some of eif4e1 seedling phenotypes are very similar to auxin mutants (Friml et al., 2003; Blilou et al., 2005), whereas our observations showed that eif4e1-1 and eif4e1-2 had 7.74% (n = 168) and 6.8% (n = 146) developmental defect rates of cotyledon, respectively, as shown in Supplementary Table S3. eIF4E1 mutants appear to have a similar level of defects (~7%) on seedling cotyledon development in Arabidopsis.
eIF4E1 Is Necessary for Primary Root Growth
To investigate the roles of eIF4E1 in plant root development, we used eif4e1-1 and eif4e1-2 to analyze the root growth. The results indicated that the length of primary root of both mutants is remarkably shorter than that of the wild type (Figures 3A,C). To confirm that these phenotypes were caused by the mutation of eIF4E1, we generated the complementation transgenic lines by crossing eif4e1-1 and eIF4E1pro:YFP-eIF4E1. The complementary line rescued the shorter primary root phenotype (Figures 3A,C), suggesting that root growth defects were due to loss of eIF4E1. To further investigate how eIF4E1 mutation affects root growth, we measured the length of the root meristem and elongation zones and quantified their cell numbers. The length of root meristem (RM) and elongation zone of the eif4e1 mutants were significantly shorter than those of the wild type. However, complementary line was able to rescue the short root phenotype of eif4e1-1 (Figures 3B,D; Supplementary Figure S5). The cell numbers in the meristem and elongation zones of mutants were less than those of the wild type but recovered in the complementary transgenic line (Figure 3E; Supplementary Figure S5). We could not, however, see any difference in the cell size of the root apical meristem between eif4e1 mutants and wild type (Figure 3F), suggesting that eIF4E1 may regulate root growth by affecting cell division of root cells.
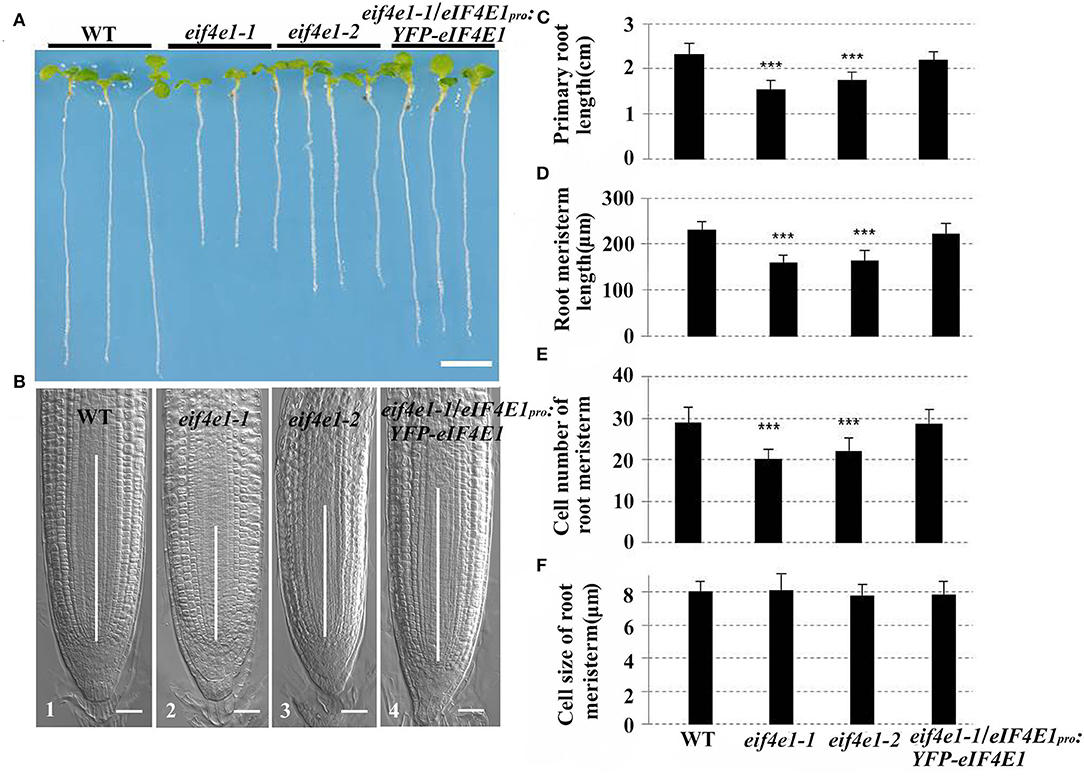
Figure 3. eIF4E1 mutation affects primary root growth. (A) Six-day-old seedlings of wild-type, eif4e1-1, eif4e1-2, and complementary line (transgenic plants harboring eIF4E1pro:YFP-eIF4E1 vector in the eif4e1-1 background). (B) Comparison of RM region of 6-day-old Arabidopsis among wild type (B1), eif4e1-1 (B2), eif4e1-2 (B3), and eif4e1-1/eIF4E1pro:YFP-eIF4E1 (B4). White lines in the middle of root tips indicate the length of RM. (C–F) Root analysis of 6-day-old seedlings. Length of primary root (C) and RM (D), cell number of RM (E), and cell size of RM (F). Data are presented as mean values with SD, n > 60. The asterisks indicate significant difference by Student's t-test (***P < 0.001). Scale bars: (A) 4 mm; (B) 20 μm.
eIF4E1 Regulates Auxin-Responsive Report Gene Expression and the Accumulation of PIN3 and PIN7 Proteins
The developmental phenotypes of the embryo and seedling of eif4e1 mutants (Figures 2A,B), and the expression levels of eIF4E1 were induced by exogenous NAA application (Figures 1D,E), suggesting that eIF4E1 may participate in the regulation and/or expression of auxin-mediated processes. To test our speculation, we examined whether auxin reporter DR5rev:GFP and auxin efflux carrier reporters were altered in eif4e1 mutants. First, we crossed DR5rev:GFP transgenic plants with eif4e1-1 mutants. We then examined DR5 activity during root growth in eif4e1-1 andcompared with that of the wild type. Two independent lines, L1 (Figure 4B) and L2 (Figure 4C), showed markedly reduction in GFP fluorescent signals in contrast to the wild type (Figure 4A), suggesting that eIF4E1 participates in auxin-regulated root growth.
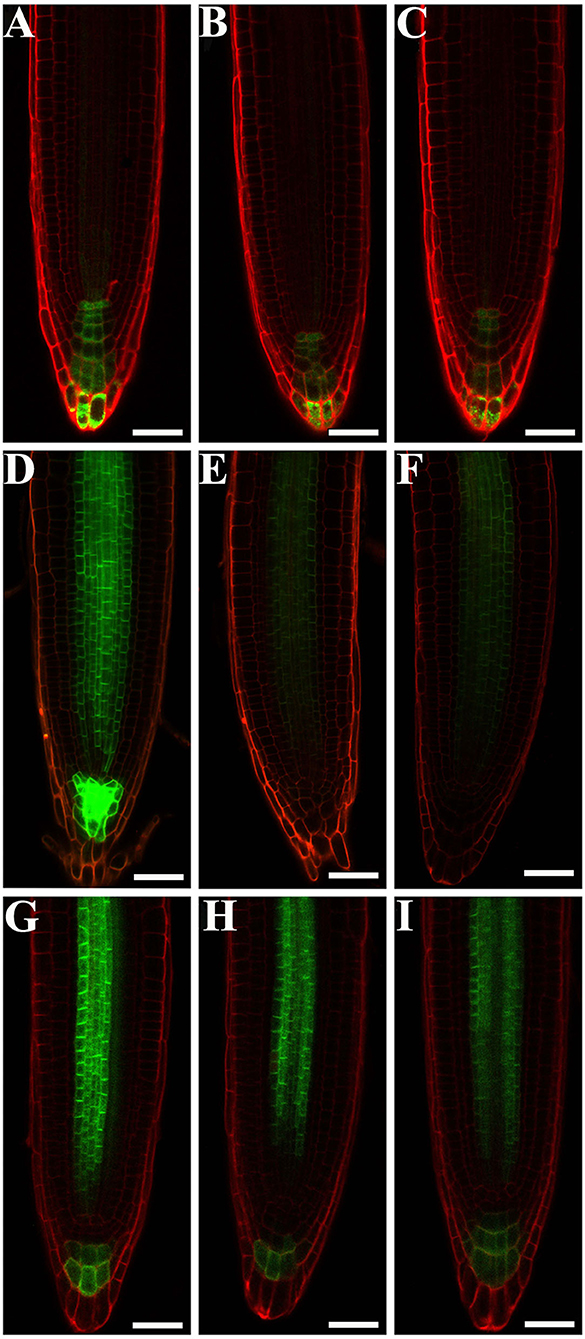
Figure 4. eIF4E1 regulates the expression of auxin-responsive report gene and the accumulation of PIN3 and PIN7 proteins. (A–C) DR5rev:GFP is expressed in roots of 4-day-old wild type (A), eif4e1-1 (B,C). “L1” and “L2” designate two lines of eif4e1-1 crossed by DR5rev:GFP. (D–F) PIN3pro:PIN3-GFP is expressed in roots of 4-day-old wild type (D), eif4e1-1 (E,F). “L1” and “L2” designate two lines of eif4e1-1 crossed by PIN3pro:PIN3-GFP. (G-I) PIN7pro:PIN7-GFP is expressed in roots of 4-day-old wild type (G), eif4e1-1 (H,I). “L1” and “L2” designate two lines of eif4e1-1 crossed by PIN7pro:PIN7-GFP. Scale bars: 20 μm.
Auxin efflux carrier PIN proteins are major players to generate auxin gradients in root tips (Adamowski and Friml, 2015; Sauer and Kleine-Vehn, 2019). To investigate the PIN expression in eif4e1 mutants, we crossed the PIN3pro:PIN3-GFP and PIN7pro:PIN7-GFP into eif4e1-1, respectively. We next analyzed the expression of PIN3pro:PIN3-GFP and PIN7pro:PIN7-GFP in the roots of the eif4e1-1 mutant. In wild-type roots, PIN3-GFP is localized in root stele cells and the root cap, while the PIN3-GFP signal was strongly reduced in eif4e1-1 L1 (Figure 4E) and L2 (Figure 4F) roots. Compared with the wild type, PIN3-GFP was even absent in the root cap (Figure 4D). We also found that PIN7-GFP was reduced in the roots of two independent eif4e1-1 lines (Figures 4H,I) compared to the wild type (Figure 4G), which further supports the importance of PIN3 and PIN7 in forming auxin gradients. We also examined the transcription level of PIN3 and PIN7 in eif4e1 mutants, and a slight elevation in the mutant background was observed compared to the wild type (Supplementary Figure S6). However, the location and accumulation of PIN1-GFP (Supplementary Figures S7B,C), PIN2-GFP (Supplementary Figures S7E,F), and AUX1-YFP (Supplementary Figures S7H,I) were not affected in eif4e1-1 in contrast to the wild type (Supplementary Figures S7A,D,G). Taken together, the loss-of-function mutation of eIF4E1 obviously reduced the expression of DR5rev:GFP and the abundance of PIN3 and PIN7 proteins but has no effect on the localization and accumulation of PIN1, PIN2, and AUX1.
Treatment of CHX Can Mimic the Phenotypes of eif4e1
Considering that eIF4E1 is a eukaryotic translation initiation factor, we examined whether the treatment of translation inhibitor cycloheximide (CHX) can mimic the phenotypes of eif4e1 mutants. eif4e1 mutants and the wild type exhibit significant inhibition in root elongation in the presence of 50 nM CHX, compared to those without CHX treatment (Figures 5A,B), and eif4e1 mutants were less sensitive to CHX than the wild type (Figures 5A,B). In view of the decline of PIN3 and PIN7 protein abundance in eif4e1 mutants, we investigated whether the CHX treatment affects PIN accumulation. Seedlings harboring PIN3pro:PIN3-GFP or PIN7pro:PIN7-GFP display inhibited root growth similar to the wild type (Figures 5C,D). Moreover, the abundance of PIN3 and PIN7 proteins is reduced in the presence of CHX (Figures 5F,H), compared to that of CHX treatment (Figures 5E,G). Meanwhile, we examined the abundance of PIN1, PIN2, and AUX1 proteins after CHX treatment, and we also could see slight alterations on protein abundance compared to those without CHX treatment (Supplementary Figure S8). Taken together, our observations showed that the treatment of CHX can mimic the phenotypes of eif4e1, suggesting that root growth defects of eif4e1 mutants are attributed to altered polar auxin transport.
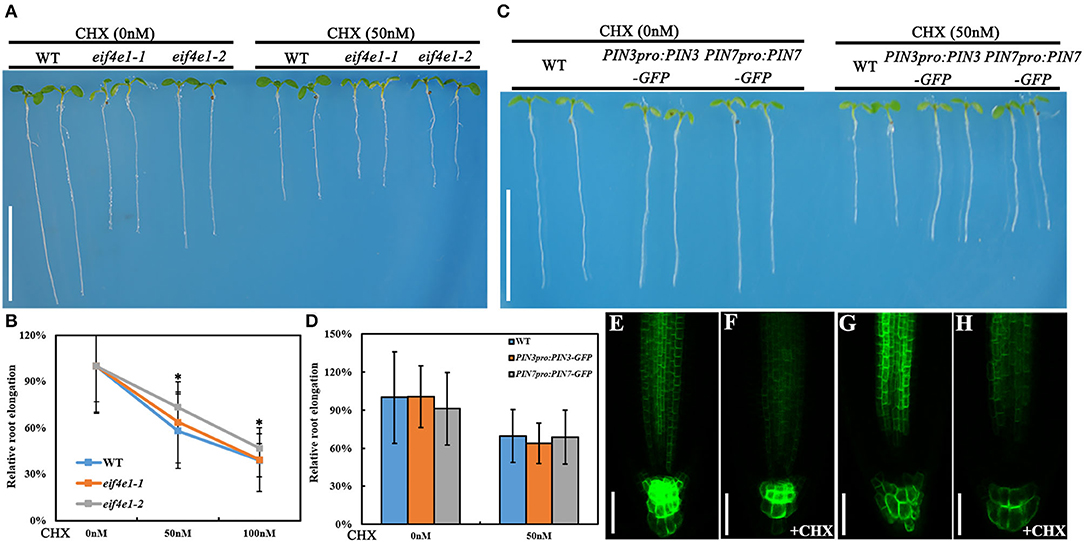
Figure 5. CHX treatment can mimic the eif4e1 phenotypes. (A) CHX treatment inhibits the root elongation. Four-day-old seedlings of wild type, eif4e1-1, and eif4e1-2 were transferred onto half-strength MS medium with or without 50 nM CHX for another 3-day growth. (B) Relative root elongation of wild type, eif4e1-1, and eif4e1-2 after treated with 50 or 100 nM CHX. (C) Four-day-old seedlings of wild type, PIN3pro:PIN3-GFP, and PIN7pro:PIN7-GFP were transferred onto half-strength MS medium with or without 50 nM CHX for another 3-day growth. (D) Relative root elongation of (C). (E–H) Confocal images showing the abundance of PIN3 and PIN7 proteins at the root cells of 4-day-old seedlings. Four-day-old seedlings of PIN3pro:PIN3-GFP and PIN7pro:PIN7-GFP were treated with (F,H) or without (E,G) CHX for another 3-day growth. The asterisks indicate significant difference by Student's t test (*P < 0.05). Scale bars: (A,C) 1 cm; (E–H) 50 μm.
eIF4E1 Interacts With RopGEF7
AtRopGEF7 is an activator of ROP GTPases, molecular switches function in multiple signaling pathways (Chen et al., 2011; Yalovsky, 2015). Previously, our group reported that AtRopGEF7 regulates the maintenance of the root apical meristem via affecting PIN-dependent polar auxin transport (Chen et al., 2011; Huang et al., 2014). Interestingly, we found a potential interacting eIF4E1, when we used RopGEF7 as bait in the screen of Arabidopsis seedling cDNA library. The cap-binding translation initiation factor, eIF4E1, could be involved in a potential RopGEF7 regulatory mechanism. To further confirm the interaction between eIF4E1 and RopGEF7, we performed the Y2H direct interaction assay. eIF4E1 strongly interacted with full-length protein of RopGEF7 (RopGEF7-CDS) and truncated proteins containing the PRONE domain (RopGEF7-ΔN and RopGEF7-PRONE), respectively, whereas eIF4E1 could not interact with the C terminus of RopGEF7 (RopGEF7-C) (Figures 6A,B), suggesting that the interaction requires a functional PRONE structure domain. Furthermore, we found eIF(iso)4E, a homolog of eIF4E1, also interacted with the PRONE domain of RopGEF7 but only weakly interacted with the full length of RopGEF7 in yeast cells (Figure 6C), suggesting this interaction may differ between these two forms of the cap-binding protein. Additionally, considering the conserved PRONE domain of RopGEFs, we tested whether the three homologs RopGEFs (RopGEF1, RopGEF4, and RopGEF6) closely related to RopGEF7 interact with eIF4E1 in yeast cells. However, eIF4E1 does not interact with them (Figure 6C), suggesting that eIF4E1 may predominantly interact with RopGEF7. The interaction between eIF4E1 and RopGEF7 was further confirmed by pull-down assays. In these experiments, the recombinant GST-tagged eIF4E1 protein and the YFP-RopGEF7 total plant protein were used as bait and prey, respectively. GFP antibody was used to detect the interaction between eIF4E1- and YFP-tagged RopGEF7 proteins. The results further support the interaction between eIF4E1 and RopGEF7 (Figure 6D). Moreover, to further investigate the relationship between RopGEF7 and eIF4E1, by using RopGEF7 antibody, the immunoblotting analysis showed that the abundance of RopGEF7 protein in eif4e1-1 and eif4e1-2 was markedly reduced (Figure 6F). On the contrary, the level of eIF4E1 protein in RopGEF7 RNAi transgenic lines was not affected (Figure 6E). To further verify the interactions between eIF4E1 and RopGEF7 in plant cells, we performed bimolecular fluorescence complementation (BiFC) assays in Arabidopsis protoplasts. The BiFC constructs of eIF4E1 in combination with RopGEF7 were co-transformed into Arabidopsis protoplasts, and the BiFC-generated yellow fluorescent protein (YFP) signal was observed in the cortical cytoplasm (Figure 7D), whereas the controls showed only the background signal (Figures 7A–C). We next observed the subcellular localization of RopGEF7 and eIF4E1. The results indicated that GFP-tagged eIF4E1 fusion protein was localized at the cytoplasm (Figure 8B), and mCherry-labeled RopGEF7 was localized at the cytoplasm and PM (Figure 8D), compared to their GFP or mCherry control vectors which are widely expressed at nucleus, cytoplasm, and PM (Figures 8A,C). Moreover, subcellular localization of YFP-eIF4E1 in roots of eIF4E1pro:YFP-eIF4E1 transgenic plants displayed that YFP-eIF4E1 is predominantly localized at the cytoplasm (Supplementary Figure S3). Meanwhile, co-transformation of 35S:eIF4E1-eGFP and 35S:mCherry-RopGEF7 in Arabidopsis protoplasts showed obviously merged signals in the cytoplasm (Figure 8E), indicating that eIF4E1 and RopGEF7 are colocalized at the cytoplasm. Taken together, these results show that eIF4E1 interacts with RopGEF7 and may act on the upstream of RopGEF7 to regulate RopGEF7 protein translation.
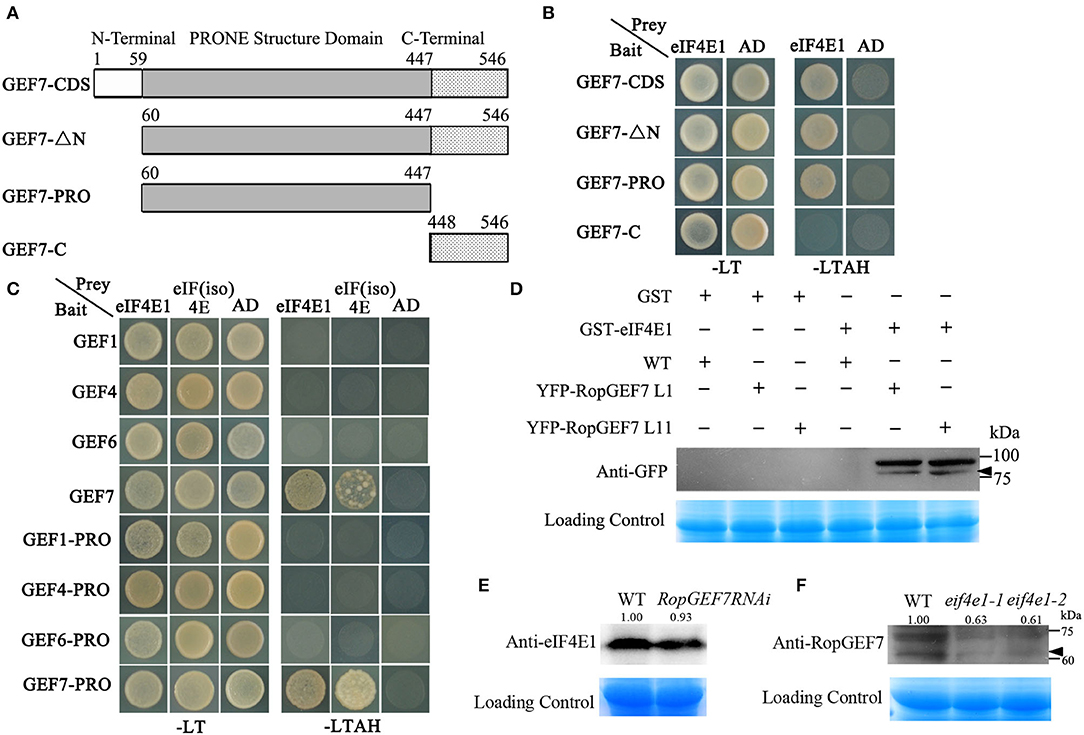
Figure 6. eIF4E1 interacts with RopGEF7. (A) Schematic representation of the different truncated versions of RopGEF7. (B) Yeast two-hybrid assay showed that eIF4E1 interacted with RopGEF7-CDS, -ΔN, and -PRONE, respectively. SD/-LT indicates SD/-Leu-Trp; SD/-LTAH indicates SD/-Leu-Trp-Ade-His. (C) Yeast two-hybrid assay showed that eIF4E1 and eIF(iso)4E interacted with RopGEF7-CDS and -PRONE, respectively. (D) Pull-down assays indicate the interactions between eIF4E1 and RopGEF7. 35Spro:YFP-RopGEF7 indicates 35Spro:YFP-RopGEF7 transgenic plants. L1 and L11 indicate two individual 35Spro:YFP-RopGEF7 transgenic lines. GFP antibody was used for detection. (E) Abundance of eIF4E1 protein in RopGEF7 RNAi transgenic line was detected by Western blotting. Anti-eIF4E1 was designated as eIF4E1 antibody. (F) Abundance of RopGEF7 protein in eif4e1-1 and eif4e1-2 was detected by Western blotting. Anti-RopGEF7 was designated as RopGEF7 antibody. The protein amount was quantified by its loading control for each sample, and the protein value of the wild type was set as one. The values are shown up the bands in (E,F). The black arrowheads indicate the aim protein bands in (D,F).
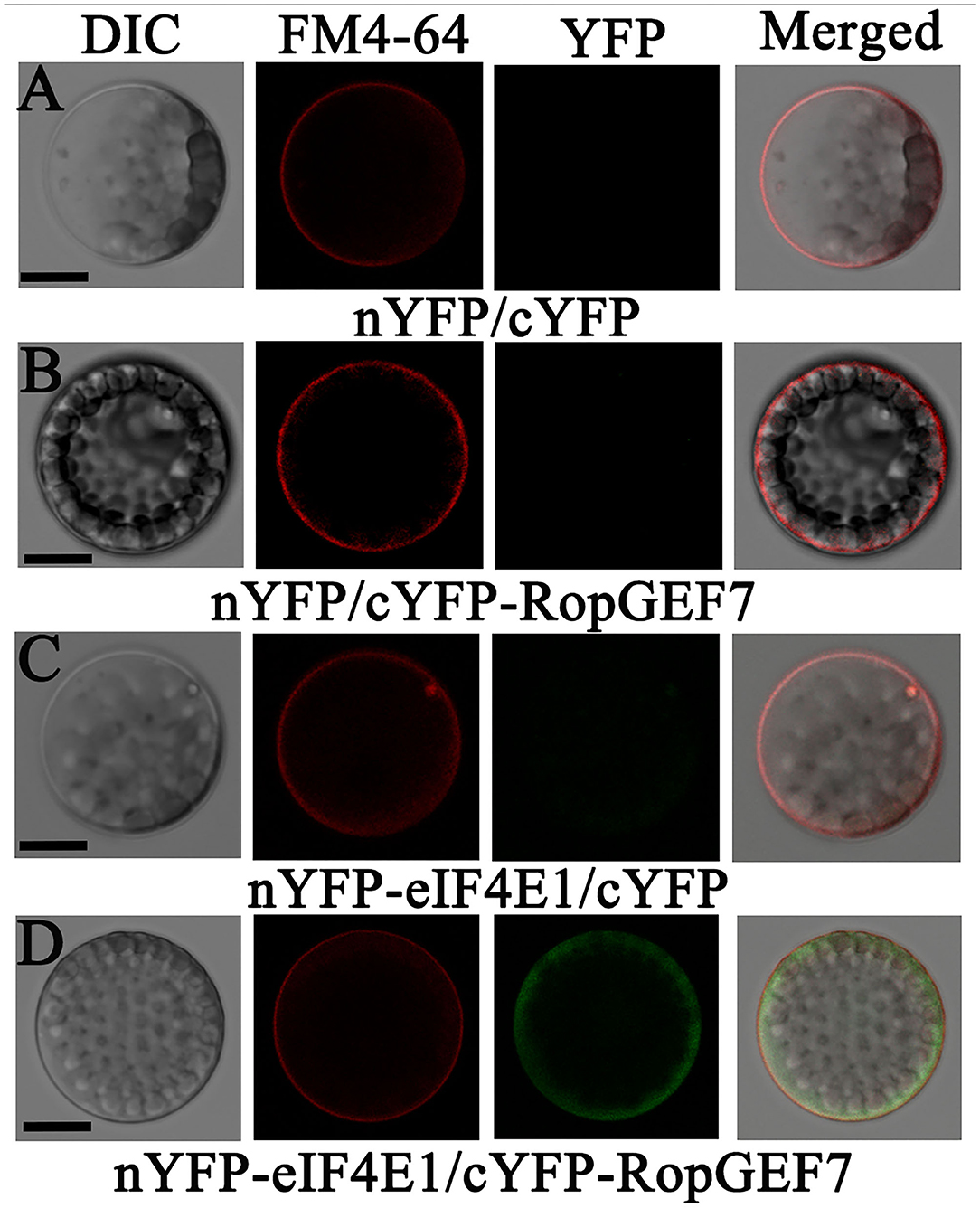
Figure 7. eIF4E1 interacts with RopGEF7 in Arabidopsis protoplasts. (A–D) Arabidopsis protoplasts were co-transfected with (A) N-terminal YFP and C-terminal YFP, (B) nYFP and cYFP-RopGEF7, (C) nYFP-eIF4E1 and cYFP, and (D) nYFP-eIF4E1 and cYFP-RopGEF7. The FM4-64 dye labels the plasma membrane. Scale bars: 20 μm.
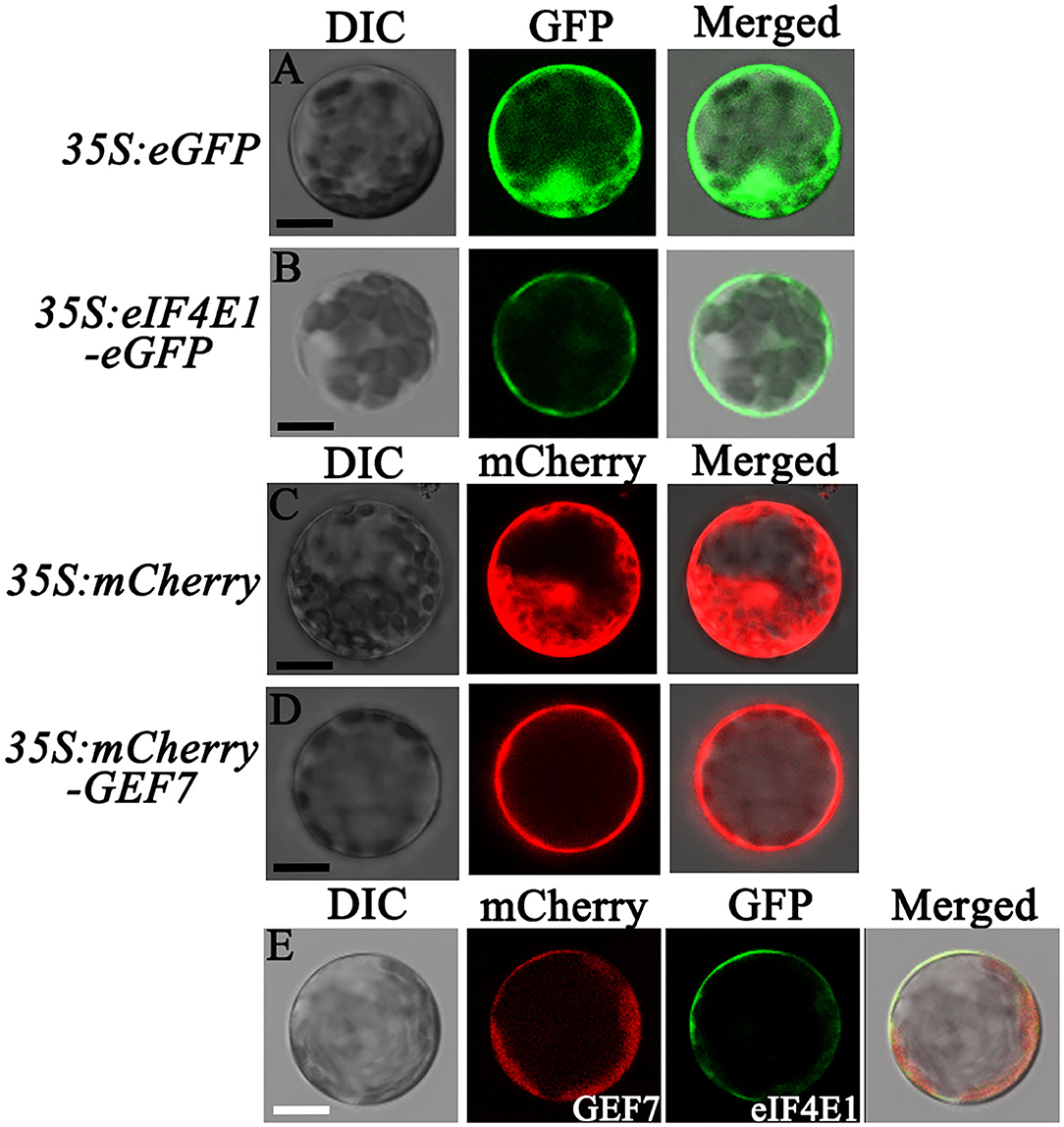
Figure 8. Subcellular localization of eIF4E1 and RopGEF7 in Arabidopsis protoplasts. (A-D) Subcellular localization of GFP from a cell harboring a 35S:eGFP empty vector (A), GFP-tagged eIF4E1 in a 35S:eIF4E1-eGFP expressed cell (B), mCherry in a cell expressed 35S:mCherry empty vector (C), and mCherry-labeled RopGEF7 in a 35S:mCherry-RopGEF7 expressed cell (D) in Arabidopsis protoplasts, respectively. (E) Co-localization of eIF4E1-eGFP and mCherry-RopGEF7 in Arabidopsis protoplasts. Scale bars: 20 μm.
Discussion
Previous investigations revealed the important role of eIF4E1 in plant–potyvirus interactions (Wang, 2015). Little is known about its function in plant growth and development. In this work, we show that mutations in eIF4E1 resulted in embryo defects and reduced root growth (Figures 2, 3). The developmental abnormalities of eif4e1 might be attributed to altered polar auxin transport based on the auxin marker analysis (Figure 4). Furthermore, we discovered that eIF4E1 interacts directly with RopGEF7, an activator of ROP GTPases to regulate the abundance of RopGEF7 protein (Figure 6). Our results provide compelling evidence that eIF4E1 regulates auxin-dependent development and also establishes a link between protein translation and ROP signaling.
We found that eIF4E1 is broadly expressed in embryos from the early embryo stage to the mature embryo stage (Figure 1, A1–9), and the eif4e1 mutant embryos showed cell division defects at embryo proper, basal region, and suspensor from the 8-cell stage to the mature embryo stage (Figure 2A; Supplementary Table S2). RopGEF7 is also expressed in embryos, but its expression did not fully overlap with eIF4E1; it is rather restricted to the precursor cells of the quiescent center (QC) during the early heart stage to the mature stage of embryogenesis (Chen et al., 2011). The knockdown of RopGEF7 by RNA interference technique caused the defects in the root pole and suspensor (Chen et al., 2011). Interestingly, the expression pattern for RopGEF1, a close homolog of RopGEF7, is comparable to that of eIF4E1 in the embryo (Liu et al., 2017). Moreover, the phenotypes of the embryo of ropgef1 resemble to those of eif4e1 mutants (Liu et al., 2017).
To further understand the role of eIF4E1 in plant early development, we characterized a point mutation mutant eif4e1-1 (cum1) and a T-DNA insertion mutant eif4e1-2 (Supplementary Figure S4). Both mutant lines displayed embryo phenotypes and seedling cotyledon phenotypes (Figure 2), which partially originated from defective embryo development. These data indicates that eIF4E1 is important for embryo development. Further study to examine whether eIF(iso)4E, an isoform of eIF4E1, contributes to embryo development would provide more information. Several genetic studies failed to generate eif4e1 eif(iso)4e double mutants but only obtained plants homozygous for one locus and heterozygous for the other locus, due to embryo or gametophyte lethality (Callot and Gallois, 2014; Patrick et al., 2014).
The strong expression of eIF4E1 in the root apical meristem (Figure 1, B1; Supplementary Figure S3) supports its importance in root development. The expression pattern in the root apical meristem shown by eIF4E1pro:GUS reporter (Figure 1) is consistent with previous investigation by mRNA in situ hybridization (Rodriguez et al., 1998). eIF4E1 expression is induced by auxin (Figures 1C–E), suggesting that it might function in the auxin-regulated pathway. eif4e1 mutants developed shorter roots with small size of root apical meristems and elongation zones than those of the wild type (Figure 3; Supplementary Figure S5). Auxin maxima were reduced in the eif4e1 root tips (Figures 4A–C). Auxin efflux carrier PIN proteins are major contributors to establish the auxin maxima in the root tips. We also observed the abundance of PIN3 and PIN7 to be reduced in eif4e1 mutants (Figures 4D–I). However, the transcript level of PIN3 and PIN7 did not decrease in eif4e1 mutants (Supplementary Figure S6). We cannot exclude the possibility that eIF4E1 directly affects PIN protein translation. CHX treatment resulted in reduced root growth (Figures 5C,D) and a decreased abundance of PIN3 and PIN7 proteins (Figures 5E–H). Further study is required to decipher the precise mechanism how eIF4E1 affects the PIN accumulation and auxin-dependent development.
Our previous work showed that the knockdown of RopGEF7 by RNA interference technique caused defects in root growth and failure to maintain root stem cell niche (Chen et al., 2011). RopGEF7 could activate ROP GTPases, including ROP3, which is required for the localization of several PIN proteins in the plasma membrane of root cells (Chen et al., 2011; Huang et al., 2014). RopGEF7RNAi plants and ROP3-dominant negative plants showed reduced auxin maxima reflected by the DR5rev:GFP auxin reporter in the root tips (Chen et al., 2011; Huang et al., 2014). An ROP effector ICR1 is required for the recruitment of PIN proteins to the PM, and loss of function in ICR1 resulted in the reduced auxin maxima in the root tips and caused the collapse of the root apical meristem (Hazak et al., 2010). Interestingly, ICR1 induced by auxin is essential for auxin transport. When auxin reaches certain levels, ICR1 is degraded to result in auxin accumulation in the embryo hypophysis and the root tips (Hazak et al., 2014). In eif4e1 mutants, the perturbed auxin maxima in the root tips (Figures 4A–C) likely led to the retarded root growth. At present, it remains unclear how eIF4E1 acts through ROP signaling to affect auxin transport; additional work will be needed to elucidate how eIF4E1 regulates RopGEF7 to activate ROP in the plant cells. A recent study has shown that RALF1 peptide hormone promotes FERONIA receptor kinase-mediated phosphorylation of eIF4E1, which modulates mRNA translation and root hair polar growth (Zhu et al., 2020). FERONIA is also known to activate ROP signaling by interacting with several RopGEF1/4/7/10 to regulate auxin-dependent root hair development (Duan et al., 2010). Our recent study showed that FERONIA targeted PIN2 polarity to regulate lateral root growth and root gravitropic response (Dong et al., 2019). It is likely that FERONIA, eIF4E1, and RopGEF7 may act on the same pathway to regulate PIN-dependent auxin transport, which is essential for embryo and root development. Taken together, we provide evidence that eIF4E1 plays a critical role in embryo development and root growth in Arabidopsis. Mutations in eIF4E1 caused aberrant cell divisions in embryos and reduced cell numbers of the root apical meristem that might be attributed in part to the altered polar auxin transport. Moreover, eIF4E1 physically interacts with RopGEF7 (Figures 6B–D) to regulate the accumulation of RopGEF7 protein (Figure 6F) for targetingauxin-mediated embryo and root development in Arabidopsis.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis eif4e1-1 (CS6552) and eif4e1-2 (SALK_145583C) mutants were provided by ABRC (Ohio State University). eif4e1-1 carrying a single base replacement mutation caused a nonsense mutation from Trp (TGG) to a stop codon (TGA), and eif4e1-2 is a T-DNA insertion mutant. All Arabidopsis mutants and transgenic lines were derived from ecotype Col-0. Marker lines were used as previously described: DR5rev:GFP (Benková et al., 2003), PIN2pro:PIN2-GFP (Blilou et al., 2005), PIN3pro:PIN3-GFP (Blilou et al., 2005), PIN7pro:PIN7-GFP (Blilou et al., 2005), and AUX1pro:AUX1-YFP (Swarup et al., 2004). Hybridization lines were obtained by crossing eif4e1-1 with the aforementioned marker lines, respectively. Seeds were surface-sterilized and germinated on half-strength Murashige and Skoog (MS) medium supplemented with 1.0% sucrose and were treated at 4°C for 2–3 days in the dark for synchronization and transferred to growth chambers at 22°C under long-day conditions (16-h light/8-h dark cycle). Then 7-day-old seedlings were transferred to soil and grown under the same conditions (Huang et al., 2014; Liu et al., 2017).
Plasmid Construction and Plant Transformation
To construct a vector for the eIF4E1pro:GUS transgenic plants, a fragment of the 3,926-bp eIF4E1 promoter sequence was cloned into pBI101.1 at HindIII and BamHI sites. To generate 35Spro:YFP-RopGEF7 transgenic plants, a constitutive promoter from cauliflower mosaic virus (CaMV) was used, and two independently transgenic lines, 35Spro:YFP-RopGEF7 L1 and L11 with high levels of YFP-RopGEF7 expression, were obtained. eIF4E1pro:YFP-eIF4E1 vector contains a YFP coding sequence (CDS) fused in-frame with an eIF4E1 CDS under the control of the same 3,926-bp eIF4E1 promoter region.
The aforementioned constructs were introduced into Agrobacterium tumefaciens strain GV3101 and subsequently used to transform Arabidopsis (Col-0) by using the floral dip method (Clough and Bent, 1998). The transgenic plants were screened on half-strength MS agar medium containing 25 μg/mL kanamycin or 15 μg/mL hygromycin. The primers used for generating the constructs are listed in Supplementary Table S1.
Quantitative RT-PCR Analysis
For qRT-PCR analysis, total RNA was extracted from 7-day-old seedlings using the RNeasy plant mini kit (Qiagen), and the first-strand cDNA was synthesized using the Takara PrimeScript First-strand cDNA synthesis kit (TAKARA). Next, PCR was performed on an Illumine Eco (Illumina) System with an SYBR probe (TAKARA). Expression levels of eIF4E1 were normalized to the expression level of two reference genes (ACTIN 2 and TUBULIN 2), respectively. The primers used are listed in Supplementary Table S1. Results obtained from three biological repeats with standard deviation (SD).
Length Analysis of Primary Root, Root Meristem, and Elongation Zone
eif4e1-1 (CS6552), which was also reported as the cum1 mutant by Yoshii et al. (2004), and eif4e1-2 (SALK_145583C) mutants; the complementary line eif4e1-1/eIF4E1pro:YFP-eIF4E1, which was generated by the cross of eif4e1-1, eIF4E1pro:YFP-eIF4E1 transgenic line; and the wild-type seeds were surfaced sterilized and germinated on half-strength MS medium, and then grown under normal growth conditions. The length of the primary root of 6-day-old seedlings was measured. The length of RM and elongation zone were measured using 6-day-old seedlings. Data were presented as averages of over 60 seedlings from at least three biological replicates with SD.
Auxin Treatment
Wild-type (Col-0) seeds were surface-sterilized and germinated on the half-strength MS medium. The total RNA of 7-day-old wild-type seedlings was extracted after 0, 2, 4, and 24 h of treatment with exogenous 10 μM NAA. The expression levels of eIF4E1 were analyzed by qRT-PCR. In addition, 5-day-old eIF4E1pro:GUS transgenic seedlings were transferred to the 1/2 MS liquid medium with or without 10 μM NAA treatment for 12 h, and GUS staining samples were observed under an Olympus BX51 microscope.
CHX Treatment
Four-day-old seedlings were transferred onto half-strength MS medium containing 50 or 100 nM CHX (Cat: AC466, Genview) for another 3-day growth and then observed or took photos under a confocal microscope.
Marker Gene Analysis
Reporter lines DR5rev:GFP, PIN1pro:PIN1-GFP, PIN2pro:PIN2-GFP, PIN3pro:PIN3-GFP, PIN7pro:PIN7-GFP, and AUX1pro:AUX1-YFP were individually crossed with eif4e1-1. Homozygous plants were isolated from F2 populations. These homozygotes in F3 and later generations were used for analyses.
Microscopy
To identify the phenotype of embryos and RM of eif4e1-1 and eif4e1-2, the ovules and seedling roots were cleared in modified Hoyer's solution and an HCG solution as previously described (Chen et al., 2011). The samples were observed by using an Olympus BX51 microscope connected to a Ritiga 2000R camera. Seedlings were observed under a Nikon SMZ1000 microscope equipped with a Nikon digital sight Ds-Fi1 camera.
For confocal microscopy, seedling roots were stained with 100 μg/mL propidium iodide (Sigma-Aldrich) and mounted in 5% glycerol. Images were taken using a Zeiss LSM 780 confocal laser scanning microscope with the following excitation (Ex) and emission (Em) wavelengths (Ex/Em): 488 nm/505–530 nm for GFP, 561 nm/591–635 nm for propidium iodide, and 543 nm/600 nm for FM4-64 as previously described (Huang et al., 2014).
Yeast Two-Hybrid Library Screening
Screening for interacting proteins of RopGEF7 was performed using a MatchmakerTM Library Construction & Screen Kit (Cat: 630445, Clontech). The cDNA library was generated by using 7-day-old Arabidopsis seedlings. The full-length coding sequence of RopGEF7 was constructed and used as a bait to screen the cDNA seedling library. The cDNA fragments coding for the RopGEF7 were amplified with primer pairs for RopGEF7-F and RopGEF7-R and then cloned into pGBKT7 (BD) to generate the bait of pGBKT7-GEF7. The primers used to generate the constructs are listed in Supplementary Table S1. Seedling cDNA was cloned with pGADT7 (AD) to generate the library. Next, the plasmids of pGBKT7-GEF7 and library were co-transformed into yeast (AH109) following the modified lithium acetate method as directed by the manufacturer. More than 2,000 transformants were screened, and the positive colonies were re-screened on the SD medium lacking Leu, Trp, Ade, and His. The selected clones were further tested on the same medium supplemented with 0 or 10 μM 3-amino-1,2,4-triazole. DNA was extracted from the positive clones and transformed into the Escherichia coli strain DH5α to amplify the cDNA inserts, and the plasmids were sequenced.
Direct Interaction Assays in Yeast Cells
The full-length cDNA, the N-terminal deleted fragment (GEF7-ΔN, aa 60-546), the PRONE domain (GEF7-PRONE, aa 60-447), and the C-terminal fragment (GEF7-C, aa 448-546) of RopGEF7 were cloned into vector pGBKT7, respectively. The cDNA of eIF4E1 was cloned into vector pGADT7, respectively. The primers used to generate the Y2H constructs are listed in Supplementary Table S1. The bait and prey constructs were co-transformed by pairs into the yeast AH109, and the transformants were selected on synthetic dextrose (SD) double dropout medium lacking tryptophan and leucine (SD/-Trp-Leu) following incubation at 28°C for 3 days. Single co-transformed yeast clones of double dropout plates were transferred to quadruple dropout medium (SD/-Trp-Leu-Ade-His). The positive (pGBKT7-53/pGADT7-T) and negative (pGBKT7-Lam/pGADT7-T) controls were used.
Pull-Down Assay
To generate the chimeric eIF4E1 protein, we cloned the full-length cDNA of eIF4E1 to the pGEX-4T-1 vector, which contains a GST tag before the multiple cloning sites (MCS). Next, the fused GST-eIF4E1 protein was expressed in E. coli and purified on glutathione affinity resin. The purified fusion protein GST-eIF4E1 was mixed with YFP-tagged RopGEF7 protein (extracted from 35Spro:YFP-RopGEF7 transgenic plants) and then incubated at 4°C for 2 h. The amino acid sequences of GFP and YFP differ only one amino acid, and GFP antibody (Abcam) was used as a substitute for the YFP antibody to detect the YFP-RopGEF7 plant protein. The primers used for plasmid construction are listed in Supplementary Table S1.
Western Blotting Assay
Plant total protein was extracted as previously described (Chen et al., 2011). Protein samples were separated using 12% sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membrane was incubated with GFP antibody (1:4,000 dilution, Abcam), eIF4E1 antibody (1:330 dilution, customized product at GenScript, USA), or RopGEF7 antibody (1:660 dilution, customized product at GenScript, USA) in TBS buffer (150 mM NaCl, 20 mM Tris–HCL PH 8.0, 0.05% Tween 20) with 5% non-fat dry milk, followed by rabbit horseradish peroxidase (HRP)-conjugated IgG secondary antibody (1:5,000 dilution, Boster Biological Technology Co. Ltd, USA). The signals were exposed to X-ray films using the chemiluminescence detection kit (Thermo-Scientific, USA). Coomassie brilliant blue-stained protein samples were used as loading controls.
Subcellular Localization and Colocalization of eIF4E1 and RopGEF7
For subcellular localization, the eIF4E1 coding sequence was inserted into a modified pCAMBIA-1300 vector (35S:eGFP) at the XbaI site by using a homologous recombination kit (Multis One Step Cloning Kit, VazymE). The RopGEF7 coding sequence was inserted into the pSAT6-mCherry-C1-B (pE3275) vector containing 35S:mCherry at the SalI and BamHI sites. Then the resulting constructs were transformed into Arabidopsis protoplasts, respectively. GFP and RFP were visualized by using a confocal microscope.
For subcellular colocalization, the aforementioned constructs of 35S:eIF4E1-GFP and 35S:mCherry-RopGEF7 were co-transformed into Arabidopsis protoplasts and then observed by using a confocal microscope.
Bimolecular Fluorescence Complement Analysis in Arabidopsis Protoplasts
To construct the fusion proteins of nYFP-eIF4E1 and cYFP-RopGEF7, the eIF4E1 coding sequence was inserted into the pSAT6-nEYFP-C1 vector, and the RopGEF7 coding sequence was inserted into the pSAT6-cEYFP-C1 vector (Chen et al., 2011). The resulting constructs were co-transformed into Arabidopsis protoplasts for BiFC assays. Protoplast isolation and transfection were performed as described (Tao et al., 2005; Chen et al., 2011). YFP was visualized by using an Olympus BX51 microscope using YFP filter sets as previously described (Chen et al., 2011; Huang et al., 2018). The FM4-64 dye labels the plasma membrane. The primers used for plasmid construction are listed in Supplementary Table S1.
Statistics
All the data were represented the average with SD from at least three biological replicates. A significant difference was determined by paired two-tailed Student's t-tests. P < 0.05 was considered significant.
Accession Numbers
The data that support the findings of this study are available in the Supplementary Data section of the online version of this article. Sequence data used in this work can be found in the Arabidopsis Genome initiative or GenBank/EMBL databases under the following accession numbers: eIF4E1 (At4g18040), eIF(iso)4E (At5g35620), RopGEF1 (At4g38430), RopGEF4 (At2g45890), RopGEF6 (At3g55660), RopGEF7 (At5g02010), PIN1 (At1g73590), PIN2 (At5g57090), PIN3 (At1g70940), PIN7 (At1g23080), AUX1 (At2g38120), ACTIN 2 (At3g18780), and TUBULIN 2 (AT5G62690).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
TL, QL, and L-ZT: designed the research. TL, QL, HL, JY, and L-ZT: analyzed the data. TL, QL, ZY, CW, HM, GL, RL, GP, and DC: performed the experiments. TL: wrote the manuscript. L-ZT: revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (32070320, and 31600217), the Natural Science Foundation of Guangdong Province (2019A1515011556 and 2018A030313931), and the Scientific Research Foundation of the State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (SKLCUSA-a201916).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the ABRC (Ohio State University) for providing the seeds used in this study. We thank Dr. Hao Wang (South China Agricultural University, Guangzhou, China) for helping with confocal imaging.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.938476/full#supplementary-material
References
Adamowski, M., and Friml, J. (2015). PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27, 20–32. doi: 10.1105/tpc.114.134874
Akamatsu, A., Uno, K., Kato, M., Wong, H. L., Shimamoto, K., and Kawano, Y. (2015). New insights into the dimerization of small GTPase Rac/ROP guanine nucleotide exchange factors in rice. Plant Signal. Behav. 10, e1044702. doi: 10.1080/15592324.2015.1044702
Ashraf, M. A., and Rahman, A. (2019). Cold stress response in Arabidopsis thaliana is mediated by GNOM ARF-GEF. Plant J. 97, 500–516. doi: 10.1111/tpj.14137
Bastet, A., Lederer, B., Giovinazzo, N., Arnoux, X., German-Retana, S., Reinbold, C., et al. (2018). Trans-species synthetic gene design allows resistance pyramiding and broad-spectrum engineering of virus resistance in plants. Plant Biotechnol J. 16, 1569–1581. doi: 10.1111/pbi.12896
Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/S0092-8674(03)00924-3
Berken, A., Thomas, C., and Wittinghofer, A. (2005). A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436, 1176–1180. doi: 10.1038/nature03883
Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., et al. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 43, 39–44. doi: 10.1038/nature03184
Bloch, D., and Yalovsky, S. (2013). Cell polarity signaling. Curr. Opin. Plant Biol. 16, 734–742. doi: 10.1016/j.pbi.2013.10.009
Callot, C., and Gallois, J. L. (2014). Pyramiding resistances based on translation initiation factors in Arabidopsis is impaired by male gametophyte lethality. Plant Signal. Behav. 9, e27940. doi: 10.4161/psb.27940
Chang, F., Gu, Y., Ma, H., and Yang, Z. (2013). AtPRK2 promotes ROP1 activation via RopGEFs in the control of polarized pollen tube growth. Mol. Plant 6, 1187–1201. doi: 10.1093/mp/sss103
Chen, M., Liu, H., Kong, J., Yang, Y., Zhang, N., Li, R., et al. (2011). RopGEF7 regulates PLETHORA-dependent maintenance of the root stem cell niche in Arabidopsis. Plant Cell 23, 2880–2894. doi: 10.1105/tpc.111.085514
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Combe, J. P., Petracek, M. E., van Eldik, G., Meulewaeter, F., and Twell, D. (2005). Translation initiation factors eIF4E and eIFiso4E are required for polysome formation and regulate plant growth in tobacco. Plant Mol. Biol. 57, 749–760. doi: 10.1007/s11103-005-3098-x
Craddock, C., Lavagi, I., and Yang, Z. (2012). New insights into Rho signaling from plant ROP/Rac GTPases. Trends Cell Biol. 22, 492–501. doi: 10.1016/j.tcb.2012.05.002
Dong, Q., Zhang, Z., Liu, Y., Tao, L. Z., and Liu, H. (2019). FERONIA regulates auxin-mediated lateral root development and primary root gravitropism. FEBS Lett. 593, 97–106. doi: 10.1002/1873-3468.13292
Duan, Q., Kita, D., Li, C., Cheung, A. Y., and Wu, H. M. (2010). FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. U. S. A. 107, 17821–17826. doi: 10.1073/pnas.1005366107
Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., et al. (2003). Efflflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153. doi: 10.1038/nature02085
Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., Hayden, L., Renninger, K., et al. (2019). Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17, 421–434. doi: 10.1111/pbi.12987
Gu, Y., Li, S., Lord, E. M., and Yang, Z. (2006). Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell 18, 366–381. doi: 10.1105/tpc.105.036434
Hazak, O., Bloch, D., Poraty, L., Sternberg, H., Zhang, J., Friml, J., et al. (2010). A rho scaffold integrates the secretory system with feedback mechanisms in regulation of auxin distribution. PLoS Biol. 8:e1000282. doi: 10.1371/journal.pbio.1000282
Hazak, O., Obolski, U., Prat, T., Friml, J., Hadany, L., and Yalovsky, S. (2014). Bimodal regulation of ICR1 levels generates self-organizing auxin distribution. Proc. Natl. Acad. Sci. U. S. A. 111, 5471–5479. doi: 10.1073/pnas.1413918111
Huang, J., Liu, H., Berberich, T., Liu, Y., Tao, L. Z., and Liu, T. (2018). Guanine Nucleotide Exchange Factor 7B (RopGEF7B) is involved in floral organ development in Oryza sativa. Rice 11, 42. doi: 10.1186/s12284-018-0235-0
Huang, J. B., Liu, H., Chen, M., Li, X., Wang, M., Yang, Y., et al. (2014). ROP3 GTPase contributes to polar auxin transport and auxin responses and is important for embryogenesis and seedling growth in Arabidopsis. Plant Cell 26, 3501–3518. doi: 10.1105/tpc.114.127902
Jackson, R. J., Hellen, C. U., and Pestova, T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. doi: 10.1038/nrm2838
Kropiwnicka, A., Kuchta, K., Lukaszewicz, M., Kowalska, J., Jemielity, J., Ginalski, K., et al. (2015). Five eIF4E isoforms from Arabidopsis thaliana are characterized by distinct features of cap analogs binding. Biochem. Biophys. Res. Commun. 456, 47–52. doi: 10.1016/j.bbrc.2014.11.032
Li, E., Zhang, Y. L., Shi, X., Li, H., Yuan, X., Li, S., et al. (2021). A positive feedback circuit for ROP-mediated polar growth. Mol. Plant 14, 395–410. doi: 10.1016/j.molp.2020.11.017
Li, G., Qian, W., Zhang, S., Zhang, S., Li, F., Zhang, H., et al. (2018). Variability in eukaryotic initiation factor iso4E in Brassica rapa influences interactions with the viral protein linked to the genome of Turnip mosaic virus. Sci. Rep. 8, 13588. doi: 10.1038/s41598-018-31739-1
Li, Z., and Liu, D. (2012). ROPGEF1 and ROPGEF4 are functional regulators of ROP11 GTPase in ABA-mediated stomatal closure in Arabidopsis. FEBS Lett. 586, 1253–1258. doi: 10.1016/j.febslet.2012.03.040
Li, Z., Takahashi, Y., Scavo, A., Brandt, B., Nguyen, D., Rieu, P., et al. (2018). Abscisic acid-induced degradation of Arabidopsis guanine nucleotide exchange factor requires calcium-dependent protein kinases. Proc. Natl. Acad. Sci. U. S. A. 115, 4522–4531. doi: 10.1073/pnas.1719659115
Li, Z., Waadt, R., and Schroeder, J. I. (2016). Release of GTP exchange factor mediated downregulation of abscisic acid signal transduction through ABA-induced rapid degradation of RopGEFs. PLoS Biol. 14:e1002461. doi: 10.1371/journal.pbio.1002461
Liu, Q., and Goss, D. J. (2018). The 3′ mRNA I-shaped structure of maize necrotic streak virus binds to eukaryotic translation factors for eIF4F-mediated translation initiation. J. Biol. Chem. 293, 9486–9495. doi: 10.1074/jbc.RA118.003377
Liu, Y., Dong, Q., Kita, D., Huang, J. B., Liu, G., Wu, X., et al. (2017). RopGEF1 plays a critical role in polar auxin transport in early development. Plant Physiol. 175, 157–171. doi: 10.1104/pp.17.00697
Martínez-Silva, A. V., Aguirre-Martínez, C., Flores-Tinoco, C. E., Alejandri-Ramírez, N. D., and Dinkova, T. D. (2012). Translation initiation factor AteIF(iso)4E is involved in selective mRNA translation in Arabidopsis thaliana seedlings. PLoS ONE 7:e31606. doi: 10.1371/journal.pone.0031606
Mazier, M., Flamain, F., Nicola,ï, M., Sarnette, V., and Caranta, C. (2011). Knock-down of both eIF4E1 and eIF4E2 genes confers broad-spectrum resistance against potyviruses in tomato. PLoS ONE 6:e29595. doi: 10.1371/journal.pone.0029595
Nibau, C., Tao, L., Levasseur, K., Wu, H. M., and Cheung, A. Y. (2013). The Arabidopsis small GTPase AtRAC7/ROP9 is a modulator of auxin and abscisic acid signaling. J. Exp. Bot. 64, 3425–3437. doi: 10.1093/jxb/ert179
Patrick, R. M., Mayberry, L. K., Choy, G., Woodard, L. E., Liu, J. S., White, A., et al. (2014). Two Arabidopsis loci encode novel eukaryotic initiation factor 4E isoforms that are functionally distinct from the conserved plant eukaryotic initiation factor 4E. Plant Physiol. 64, 1820–1830. doi: 10.1104/pp.113.227785
Rodriguez, C. M., Freire, M. A., Camilleri, C., and Robaglia, C. (1998). The Arabidopsis thaliana cDNAs coding for eIF4E and eIF(iso)4E are not functionally equivalent for yeast complementation and are differentially expressed during plant development. Plant J. 13, 465–473. doi: 10.1046/j.1365-313X.1998.00047.x
Sauer, M., and Kleine-Vehn, J. (2019). PIN-FORMED and PIN-LIKES auxin transport facilitators. Development 146, dev168088. doi: 10.1242/dev.168088
Swarup, R., Kargul, J., Marchant, A., Zadik, D., Rahman, A., Mills, R., et al. (2004). Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16, 3069–3083. doi: 10.1105/tpc.104.024737
Tajima, Y., Iwakawa, H. O., Hyodo, K., Kaido, M., Mise, K., and Okuno, T. (2017). Requirement for eukaryotic translation initiation factors in cap-independent translation differs between bipartite genomic RNAs of red clover necrotic mosaic virus. Virology 509, 152–158. doi: 10.1016/j.virol.2017.06.015
Tao, L. Z., Cheung, A. Y., Nibau, C., and Wu, H. M. (2005). RAC GTPases in tobacco and Arabidopsis mediate auxin-induced formation of proteolytically active nuclear protein bodies that contain AUX/IAA proteins. Plant Cell 17, 2369–2383. doi: 10.1105/tpc.105.032987
Wang, A. (2015). Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu. Rev. Phytopathol. 253, 45–66. doi: 10.1146/annurev-phyto-080614-120001
Wang, A., and Krishnaswamy, S. (2012). Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 13, 795–803. doi: 10.1111/j.1364-3703.2012.00791.x
Wang, W., Liu, Z., Bao, L. J., Zhang, S. S., Zhang, C. G., Li, X., et al. (2017). The RopGEF2-ROP7/ROP2 pathway activated by phyB suppresses red light-induced stomatal opening. Plant Physiol. 174, 717–731. doi: 10.1104/pp.16.01727
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718. doi: 10.1371/journal.pone.0000718
Wu, H. M., Hazak, O., Cheung, A. Y., and Yalovsky, S. (2011). RAC/ROP GTPases and auxin signaling. Plant Cell 23, 1208–1218. doi: 10.1105/tpc.111.083907
Yalovsky, S. (2015). Protein lipid modifications and the regulation of ROP GTPase function. J. Exp. Bot. 66, 1617–1624. doi: 10.1093/jxb/erv057
Yang, Z., and Fu, Y. (2007). ROP/RAC GTPase signaling. Curr. Opin. Plant Biol. 10, 490–494. doi: 10.1016/j.pbi.2007.07.005
Yoshii, M., Nishikiori, M., Tomita, K., Yoshioka, N., Kozuka, R., Naito, S., et al. (2004). The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 78, 6102–6111. doi: 10.1128/JVI.78.12.6102-6111.2004
Zhang, Y., and McCormick, S. (2007). A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 104, 18830–18835. doi: 10.1073/pnas.0705874104
Zhu, S., Estévez, J. M., Liao, H., Zhu, Y., Yang, T., Li, C., et al. (2020). The RALF1–FERONIA complex phosphorylates eIF4E1 to promote protein synthesis and polar root hair growth. Mol. Plant 13, 698–716. doi: 10.1016/j.molp.2019.12.014
Keywords: auxin, eIF4E1, RopGEF7, embryo development, RAC/ROP, root growth
Citation: Liu T, Liu Q, Yu Z, Wang C, Mai H, Liu G, Li R, Pang G, Chen D, Liu H, Yang J and Tao L-Z (2022) eIF4E1 Regulates Arabidopsis Embryo Development and Root Growth by Interacting With RopGEF7. Front. Plant Sci. 13:938476. doi: 10.3389/fpls.2022.938476
Received: 07 May 2022; Accepted: 06 June 2022;
Published: 30 June 2022.
Edited by:
Nobutoshi Yamaguchi, Nara Institute of Science and Technology (NAIST), JapanCopyright © 2022 Liu, Liu, Yu, Wang, Mai, Liu, Li, Pang, Chen, Liu, Yang and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taibo Liu, dGJsaXVAc2NhdS5lZHUuY24=; Li-Zhen Tao, bHp0YW8yMDA1QHNjYXUuZWR1LmNu
 Taibo Liu
Taibo Liu Qianyu Liu
Qianyu Liu Zhen Yu1,2,3
Zhen Yu1,2,3 Chunling Wang
Chunling Wang Huili Liu
Huili Liu Jiangyi Yang
Jiangyi Yang Li-Zhen Tao
Li-Zhen Tao