
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 03 August 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.937473
This article is part of the Research Topic Genetics of Plant-Nematode Interactions View all 8 articles
The migratory plant-parasitic nematode Bursaphelenchus xylophilus is the pathogen of the pine wilt disease (PWD), causing serious damage to pine forests in China. During the process of plant resistance to multiple pathogens, plant immunity plays a key role. In this current study, the pathogen-associated molecular pattern (PAMP) BxCDP1 in B. xylophilus has been identified, but the host target protein of BxCDP1 and its key amino acid region inducing the plant immunity have yet to be elucidated. We found that BxCDP1 could trigger superoxide production, H2O2 production, and callose deposits. A RING-H2 finger protein 1 (RHF1) of Pinus thunbergii was screened and characterized as a target protein of BxCDP1 by yeast two-hybrid and co-immunoprecipitation (Co-IP). Moreover, two peptides (namely M9 and M16) proved to be key regions of BxCDP1 to induce PAMP-triggered immunity (PTI) in Nicotiana benthamiana, which also induced the expression of pathogenesis-related (PR) genes (PtPR-3, PtPR-4, and PtPR-5) in P. thunbergii and enhanced the resistance of the host to B. xylophilus. These results indicate that BxCDP1 plays a critical role in the interaction between B. xylophilus and P. thunbergii, and both peptides M9 and M16 have the potential to be developed and utilized as immune inducers of pines against B. xylophilus in future.
The pine wood nematode (PWN) Bursaphelenchus xylophilus, as an important migratory plant-parasitic nematode (PPN), causes the pine wilt disease (PWD). At present, PWD has become the most serious pine forest disease in China (Zhou et al., 2017; Huang L. et al., 2019; Wang et al., 2022). Due to the complexity of the occurrence and development of PWD, the exact pathogenic mechanism of B. xylophilus is not completely clear, making PWD difficult to predict and manage.
During the interaction of pathogens and plants, plants evolve a sophisticated basal immune system, and pathogens must overcome or escape plant immunity to colonize (Jones and Dangl, 2006). GPlants have two layers of the innate immune system. In the first tier, pathogen or microbe-associated molecular patterns (PAMPs or MAMPs, respectively) are recognized by plant plasma membrane-bound receptors (pattern recognition receptors [PRRs]) to induce innate immunity (PAMP-triggered immunity [PTI]) (Yang et al., 2017). In turn, the pathogen secretes effectors to suppress PTI in order to promote infection. In the second tier, plants employ the nucleotide-binding and leucine-rich repeat (NB-LRR) proteins encoded by disease resistance (R) genes to recognize effectors and trigger further innate immunity (effector-triggered immunity [ETI]) (Chisholm et al., 2006; Jones and Dangl, 2006). Many studies showed that PAMPs and effectors of pathogens have a key role in the interaction of pathogens and plants, which have become research hotspots (Jing et al., 2016; Chen et al., 2017; Kettles et al., 2017; Du et al., 2021).
Nowadays, research on PAMPs and effectors of B. xylophilus has attracted much attention. For example, effectors were broadly predicted and screened by transcriptomic analysis (Espada et al., 2016; Tsai et al., 2016; Margarida et al., 2018; Tanaka et al., 2019; Hu et al., 2021). Effectors BxSapB1, BxSapB2, and BxSapB3 were shown to trigger cell death and contribute to virulence (Hu et al., 2019; Huang X. et al., 2019; Zhao et al., 2020). A PAMP BxCDP1 was characterized in B. xylophilus (Hu et al., 2020). An effector Bx-FAR-1 inhibits plant immunity by mediating the jasmonic acid pathway in pines (Li et al., 2020; Wen et al., 2022). An effector BxSCD1 suppresses PAMP BxCDP1-triggered immunity and interacts with a host target protein PtACO1 (Wen et al., 2021).
In nature, oxygen molecules are in a less active oxidation state. However, when plants are subjected to biological stress such as pathogen infection, the dynamic balance of electron transfer and redox in plants is destroyed, and the electrons generated in cells can easily convert oxidized oxygen into reduced oxygen, producing superoxide [containing superoxide anion (O2–), hydroxyl free radical (OH–), and hydrogen peroxide (H2O2)]. These substances are collectively referred to as reactive oxygen species (ROS) (Wojtaszek, 1997). Plant immunity is stimulated by the accumulation of ROS, deposition of callose, induction of pathogenesis-related (PR) genes, and regulation of hormone signaling (Dodds and Rathjen, 2010; Tsuda and Katagiri, 2010). In our previous study, a PAMP of B. xylophilus BxCDP1 was identified, which triggered cell death and plant immunity in Nicotiana benthamiana and enhanced defense responses of P. thunbergii (Hu et al., 2020). However, it remains unclear whether BxCDP1 can activate the burst of ROS by increasing the content of superoxide and H2O2 and whether it can improve the deposition of callose in the plant.
The function of a protein is usually determined by its amino acid composition and protein structure (Li et al., 2018). The key amino acid regions of triggered or inhibited plant immunity of several PAMPs or effectors have been characterized. For example, flg22 peptides, a highly conserved region at the N-terminal of bacterial PAMP flagellin, can induce plant immunity and act on SA, MAPK, and other signal pathways, thus having an important impact on plant disease resistance, stomatal movement, growth, and development (Zipfel et al., 2004). The SCRE11–179 from Ustilaginoidea virens effector SCRE1 could suppress pattern-triggered rice immunity, and the 45CPARS49 motif has proven to be crucial for its suppression activity (Zhang et al., 2020).
As an important post-translational modification (PTM), protein ubiquitination is widely involved in various life activities, such as plant growth and development, biological and abiotic stress response, cycle regulation, and signal transduction by mediating the degradation of specific proteins (Stone et al., 2005; Liu and Stone, 2011). The ubiquitination system is a complex reaction process including three enzymes, namely E1 ubiquitin-activating enzyme, E2 ubiquitin-binding enzyme, and E3 ubiquitin ligase (Marino et al., 2012). E3 ubiquitin ligase can specifically recognize target proteins and has been proved important in multiple stages of plant immunity (Nagels et al., 2016). E3 ubiquitin ligases are divided into HECT, U-box, RING, and CRLs4 according to their domains and binding modes with substrates. The RING domain is a Cys-rich region made up of 40∼60 amino acids and contains eight spatially conserved Cys and His residues as metal ligands to chelate two zinc ions to realize the transfer of ubiquitin to the target protein (Marino et al., 2012). Many RING-type E3 ligase proteins have been identified as plant immune modulators. For example, RING-H2 finger protein EL5 in response to rice n-acetylchitosan oligosaccharide elicitor and ATL gene family members of RING zinc finger E3 ubiquitin ligase activated by elicitor treatment in Arabidopsis have been proven to play essential roles in plant defense in response to pathogen infection (Durrant et al., 2000). Constitutive expression of ATL2 in Arabidopsis induces defense-related gene expression (Serrano and Guzmán, 2004). In pepper, encoding RING-type protein CaRING1 is an active E3 ligase, which is necessary for hypersensitive response (HR) and resistance response of pepper to the infection of avirulent Xanthomonas campestris pv vesicatoria (Lee et al., 2011).
In this study, the target protein of BxCDP1 was identified to explore the function of BxCDP1 in host pine by yeast two-hybrid screening assay and co-immunoprecipitation (Co-IP). In order to develop potential immune inducers of pines against B. xylophilus, the key peptide regions of the BxCDP1-triggered plant immunity were identified and the roles of these peptides in promoting the defense ability of the host were also explored. Moreover, the content of superoxide and H2O2 by BxCDP1 triggered in N. benthamiana was detected by the NBT and DAB staining, respectively. The callose deposits were also detected in N. benthamiana to further confirm that the plant immunity was stimulated by BxCDP1.
Botrytis cinerea was cultured on potato dextrose agar (PDA) plates at 25°C for 7 days. Then, AMA3, a B. xylophilus highly virulent strain, was transferred into the B. cinerea plates under the same culture conditions above. The nematodes were extracted using the Baermann funnel technique (Qiu et al., 2013). N. benthamiana was grown in a glasshouse at 25°C with a relative humidity of 60% under 16:8-h light:dark conditions, and the leaves of 4–5-week-old N. benthamiana plants were used in the infiltration assay. The 3-year-old P. thunbergii seedlings were cultivated at temperatures ranging from 28 to 32°C with the relative humidity ranging from 65 to 75%.
BxCDP1 and its 19 deletion mutants were cloned using B. xylophilus cDNA. These amplified fragments of BxCDP1 and its 19 deletion mutants were ligated into pBINRFP (pCAM1300-RFP) using the Clone Express II One Step Cloning Kit (Vazyme, Nanjing, China). The amplified fragments of BxCDP1 were also ligated into PVX (pGR107-3*HA). The interacting protein PtRHF1 of BxCDP1 was cloned using Pinus thunbergii cDNA. These amplified fragments of PtRHF1 were also ligated into pBINGFP [a plasmid containing green fluorescent protein (GFP)]. Individual colonies of each construct were examined by using PCR, and the positive clones were verified by sequencing. The primers we used are listed in Supplementary Table 1.
The A. tumefaciens infiltration assays were performed according to previous operating instructions (Hu et al., 2019). Briefly, constructs were transformed into A. tumefaciens strain GV3101 by electroporation, and the cells were cultured on Luria Bertani (LB) agar plates with kanamycin and rifampicin. After culturing at 30°C with a rotation speed of 200 rpm for 12 h, the recombinant A. tumefaciens cells were resuspended in a wash buffer (10 mM MgCl2 and MES, and 100 μM acetosyringone, pH 5.6) and diluted to a final optical density (OD) at 600 nm of 0.4. The A. tumefaciens suspensions were infiltrated into the N. benthamiana leaves using a needleless syringe. After 7 days, the symptom development of infiltration points was observed and photographed. In the A. tumefaciens infiltration experiment of screening the key amino acid regions of BxCDP1, the empty vector pBINRFP was used as a control. The pBINGFP was used as the negative control when protein interaction verification was performed in N. benthamiana.
Expression and purification of the recombinant BxCDP1 and GFP were performed as described (Hu et al., 2020; Wen et al., 2021). Briefly, the ORFs for BxCDP1 and GFP were amplified using specific primers (Supplementary Table 1). The purified PCR products were inserted into the linearized vector pPICZαA (containing a 6 × His tag). After transforming into Escherichia coli TOP10, validating the transformants, and sequencing, the pPICZαA vector containing BxCDP1-6 × His and GFP-6 × His was transformed into the Pichia pastoris KM71H (Invitrogen). Positive clones were grown in a yeast extract–peptone–dextrose (YPD) medium containing 100 μg/ml of zeocin at 30°C for 3 days. The cultures were centrifuged to procure the supernatant containing BxCDP1-6 × His and GFP-6 × His after shaking in the BMGY (buffered glycerol complex medium) and BMMY (buffered methanol complex medium) media for 4 days. The expressed proteins were validated by SDS-PAGE. Purification of the recombinant protein BxCDP1-6 × His and GFP-6 × His from the culture supernatant was performed by affinity chromatography using Ni-NTA Superflow resin (Qiagen). To further detect the induction of superoxide, H2O2, and callose deposits, 1 μM purified BxCDP1 protein was infiltrated into leaves of N. benthamiana, and at 3 h after injection, the infiltrated into N. benthamiana leaves were stained using sodium azide, 3,3′-diaminobenzidine (DAB), or aniline blue as described (Dong et al., 2011). Meanwhile, the infiltrated leaves of N. benthamiana with 1 μM purified BxCDP1 protein were immersed in 40 ml of decolorizing solution (absolute ethanol:water:lactic acid:glycerin:phenol = 8:1:1:1:1) after injection of 3 h, and soaked in vacuum for 30 min. The samples were bathed in a 60°C water bath for 30 min to remove chlorophyll. After washing with water, the leaves without chlorophyll were immersed in 0.01% (w/v) aniline blue staining solution for 2–4 h under dark conditions. Then, the deposition of callose was observed under a laser confocal microscope (excitation light wavelength 377 nm and emission light wavelength 447 nm). The N. benthamiana leaves infiltrated with purified GFP protein were used as a control. The staining area of DAB or NBT and the number of callose deposition in N. benthamiana were quantified using ImageJ software, respectively. The staining area of DAB or NBT and the number of callose deposition by purified GFP protein treated were both set as 1. The relative staining area of DAB or NBT and the relative number of callose deposits by purified BxCDP1 protein treated were calculated, respectively. The experiment was performed three times, and three different plants with three infiltrated leaves were used for each assay.
About 3000 B. xylophilus virulent strains of AMA3 were inoculated to 3-year-old P. thunbergii. The stem segments of P. thunbergii about 3 cm near the inoculation site were cut into small segments at 12 h, 5 days, and 10 days after B. xylophilus infection. The samples were then mixed with the stem segments without B. xylophilus inoculation (0 h), followed by wrapping with tin foil and quick-frozen by using liquid nitrogen, and sent to Shanghai Ouyi Biomedical Co., Ltd. for pine yeast library construction.
BxCDP1 (without signal peptide) was cloned into the pGBKT7 bait vector and then transformed into yeast strain Y2H Gold. The B. xylophilus-infected pine cDNA library was screened following the Clontech protocols. Then, pGBKT7::BxCDP1 and potential interaction target proteins were performed in a yeast two-hybrid assay. In the Co-IP assay, BxCDP1 and PtRHF1 were cloned into PVX and pBINGFP and introduced into A. tumefaciens GV3101. The A. tumefaciens suspensions of PVX::BxCDP1, pBINGFP::PtRHF1, and their mixed bacterial suspensions were infiltrated into N. benthamiana. At 48 h after infiltration, the proteins were extracted and Co-IP assays were performed as described previously (Li et al., 2018).
The sequences of three deletion mutants of BxCDP1 (namely M9, M15, and M16), which triggered cell death in N. benthamiana, were synthesized by GenScript Biotech Corporation (Nanjing, China). The sequence of a deletion mutant of BxCDP1 that did not trigger cell death was also synthesized as a control (namely M14).
Agroinfiltrated N. benthamiana leaves were collected at 48-h post-inoculation (hpi). Total protein extraction and immunoblotting were performed according to a previous report (Yin et al., 2013). In brief, total proteins were separated by 12% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane (Bio-Rad, Hercules, CA, United States). The membranes were blocked with 5% (w/v) non-fat dry milk for 1 h at room temperature and then they were washed three times with PBS containing 0.1% Tween-20. Transient protein expression was assessed by incubating the membrane with a 1:5000 dilution of a primary mouse anti-HA antibody (Abmart, Berkeley Heights, NJ, United States) or an anti-RFP antibody (Abcam, Cambridge, United Kingdom) or an anti-GFP antibody (Abmart, Berkeley Heights, NJ, United States), followed by incubation with a goat anti-mouse secondary antibody at a 1:10,000 dilution (IRDye 800, 926-32210; LI-COR Biosciences, Lincoln, NE, United States). The four deletion mutants of BxCDP1 (M9, M15, M16, and M14) were analyzed by incubating the membrane with a 1:5000 dilution of a primary rabbit anti-RFP polyclonal antibody, followed by incubation with a peroxidase-conjugated goat anti-rabbit IgG secondary antibody at a 1:10,000 dilution (Zhongshan Bio-technique, Beijing, China). The results of protein expressions were observed by using an Odyssey® LI-COR imaging system. The Ponceau S staining was used to confirm the protein loading.
In the inoculation assay, we cut a small wound at the stem of the 3-year-old P. thunbergii seedling about 20 cm from the ground with a sterile scalpel, and the wound was as deep as the xylem of the pine tree. Then, the sealing film was wrapped around the wound to make a funnel, and 1 mL of synthesized peptides (M9, M15, and M16) and M14 (final concentration 50 μg/ml) (Xu et al., 2013) were added to the funnels. At 4 hpi, approximately 1,500 nematodes (a mixture of juveniles and adults) were inoculated into those previous funnels containing synthesized peptides, respectively. The morbidity degree of the P. thunbergii seedlings was classified into five different grades according to the previous study (Yu et al., 2012): 0, all needles of pines are green; I, a quarter of needles have turned yellow; II, half of the needles have turned yellow or brown; III, three-fourths of the needles have turned brown; and IV, the whole seedling withered. The formulas (Rempel and Hall, 1996) for calculating the infection ratio and disease severity index (DSI) of pine seedlings are as follows:
The infection levels of B. xylophilus were detected, and the infection ratio and DSI P. thunbergii seedlings were calculated. The infection assay of B. xylophilus was performed three times and a total of 18 individual P. thunbergii seedlings for each treatment were used. In addition, some 3-year-old P. thunbergii seedlings were inoculated with B. xylophilus for 2.5 h, 6 h, 12 h, 24 h, and 3 days, respectively. Some 3-year-old P. thunbergii seedlings were, respectively, inoculated with 1 mL of purified BxCDP1 and GFP proteins (with a final concentration of 50 μg/ml) (Xu et al., 2013) as described above. Briefly, 1 mL of purified BxCDP1 and GFP protein solutions was added to the funnels made of sealing film. Subsequently, the top of the funnel was sealed with another sealing film to prevent the rapid volatilization of the protein solutions.
The total RNA from each sample of N. benthamiana and P. thunbergii was separately extracted using the Plant Total RNA Kit (ZP405) (Zoman Biotech, Beijing, China), containing DNA digestive enzymes. The concentration of the total RNA was detected using an Ultraspec TM 2100 Pro UV/visible spectrophotometer, and the integrity of the total RNA was detected using 1% agarose gels.
The total RNA of each sample was reverse transcribed into cDNA using HiScript II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China) according to the manufacturer’s instructions. The protocol consists of two steps. First, gDNA of the total RNA was removed when the reaction mixture was incubated at 42°C for 2 min. Then, reverse transcription of total RNA was performed by following the procedure at 50°C for 15 min and 85°C for 5 s.
RT-qPCR assays were carried out using ChamQ SYBR qPCR Master Mix (Low ROX Premixed) (Vazyme, Nanjing, China) according to the manufacturer’s instructions. The RT-qPCR assays were kept at 95°C for 30 s and at 40 cycles of 95°C for 10 s, 60°C for 34 s, and 95°C for 15 s. Three 300 nM synthesized peptides (M9, M15, and M16) were, respectively, infiltrated into N. benthamiana for 3 h; then, the relative expressions of three PTI marker genes (NbAcre31, NbPTI5, and NbCyp71D20) were detected to evaluate the effects of the three peptides on them. N. benthamiana infiltrated with 300 nM synthesized peptide M14 was used as a control. Each assay was replicated three times, and the NbEF1α of N. benthamiana was used as a constitutively expressed endogenous control gene (Ma et al., 2015).
Relative transcript levels of PR genes (PtPR-3, PtPR-4, and PtPR-5) of 3-year-old P. thunbergii seedlings were detected after inoculating B. xylophilus with 50 μg/mL of synthesized peptides M9, M15, and M16, and the P. thunbergii seedlings inoculated with 1500 B. xylophilus (a mixture of juveniles and adults) with M14 were used as the control. The relative expressions of PtRHF1 were measured when P. thunbergii were infected with B. xylophilus for 2.5 h, 6 h, 12 h, 24 h, and 3 days, respectively. Moreover, the relative expressions of PtRHF1 were also measured when P. thunbergii were inoculated with BxCDP1 and GFP purified proteins. The assay was performed three times, and the PtEF1α of P. thunbergii was used as an endogenous control gene (Hirao et al., 2012).
The relative expression levels of genes above were analyzed and calculated using the ABI Prism 7500 software (Applied Biosystems, Foster City, CA, United States) and the 2–ΔΔCt method, respectively. The RT-qPCR was conducted with three biological replicates and three technical replicates. All primer sequences are provided in Supplementary Table 1.
All experiments were repeated at least three times with similar results. The statistical significance was determined by one-way ANOVA followed by Duncan’s multiple range test using SPSS ver. 19 (SPSS Japan Inc., Tokyo, Japan).
Nitroblue tetrazolium (NBT) can react with O2– produced in plants to form a dark blue insoluble complex (Doke, 1983). Under dark conditions, DAB can react with hydrogen peroxide (H2O2) to produce dark brown precipitates visible to the naked eye (Thordal-Christensen et al., 1997). To determine whether BxCDP1 can induce ROS accumulation, the leaves of N. benthamiana injected with BxCDP1 and GFP pure protein were treated with NBT and DAB, respectively. The results showed that there were more dark blue areas and more dark brown sediments on leaves injected with purified BxCDP1 protein (Figures 1A,B) than those injected with purified GFP protein in the N. benthamiana (Figures 1C,D), respectively. The staining area of DAB or NBT by the former treated was calculated, which was compared with that of the latter treated. It showed that the relative staining area of DAB or NBT by BxCDP1 treated was both significantly bigger than GFP treated, which showed that the production of superoxide and H2O2 by BxCDP1 triggered was significantly higher than the GFP triggered (Figure 1G). These results indicated that BxCDP1 could induce ROS accumulation.
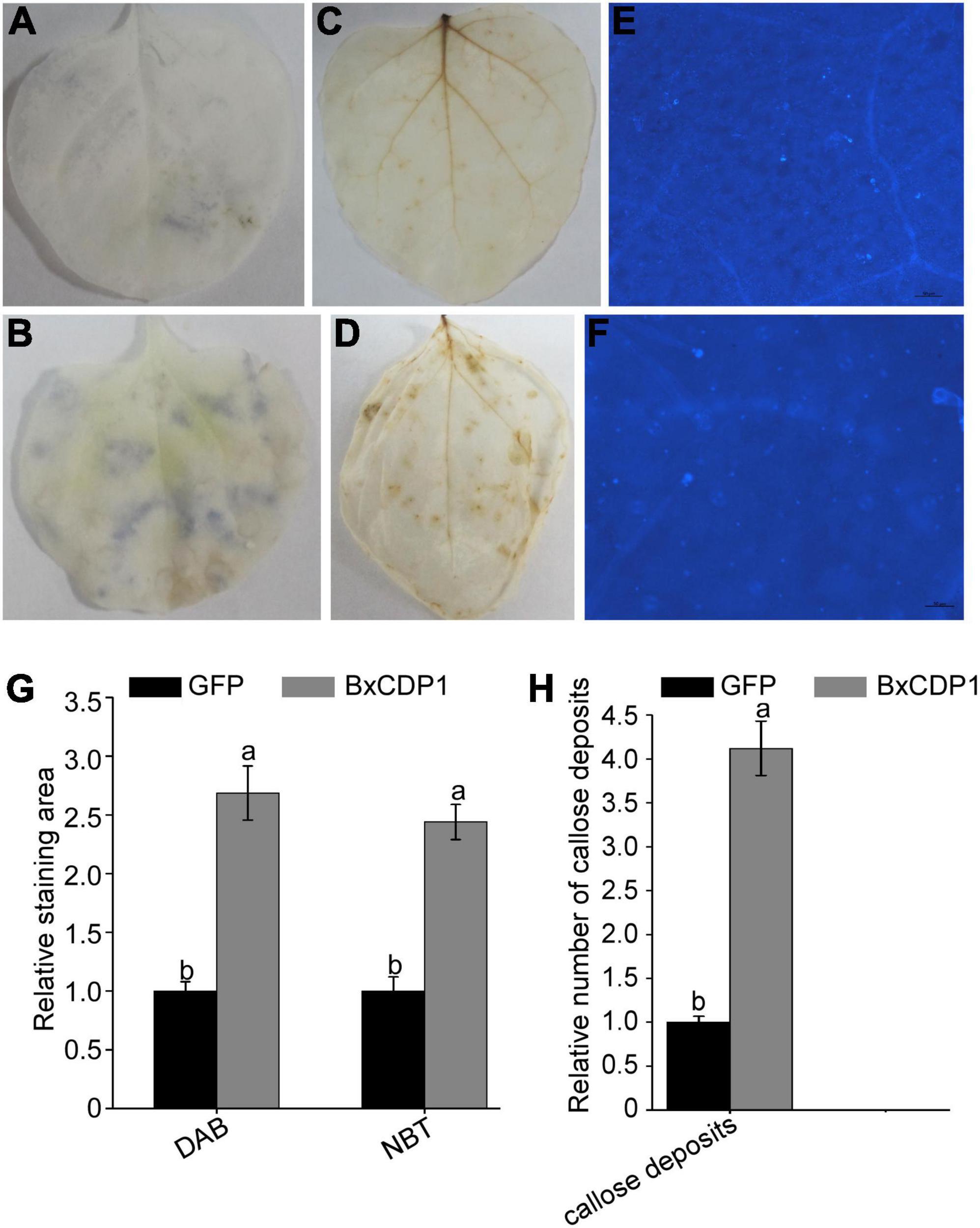
Figure 1. BxCDP1 induces the accumulation of superoxide and hydrogen peroxide and callose deposits in Nicotiana benthamiana. Representative N. benthamiana leaves stained using sodium azide after 3 h inoculation with purified GFP protein (A) and BxCDP1 protein (B). Representative N. benthamiana leaves stained using 3,3’-diaminobenzidine (DAB) after 3 h inoculation with purified GFP protein (C) and BxCDP1 protein (D). Representative N. benthamiana leaves stained using aniline blue after 3 h inoculation with purified GFP protein (E) and BxCDP1 protein (F). These infiltration assays were, respectively, performed three times, and three different plants with three inoculated leaves were used in each assay. (G) Quantification using ImageJ software. The relative staining area of DAB or NBT by purified BxCDP1 protein treated in N. benthamiana was calculated. The staining area of DAB or NBT by GFP treated was set as 1. (H) The number of callose deposits per microscopic field was quantitated using ImageJ software. The relative number of callose deposits by purified BxCDP1 protein treated in N. benthamiana was counted. The number of callose deposits by GFP treated was set as 1. Data are the means, and the error bars represent ± SD from three biological replicates. Different letters on top of the bars indicate statistically significant differences (p < 0.05, t-test) as measured by Duncan’s multiple range test.
Similarly, more callose was accumulated in N. benthamiana leaves injected with purified BxCDP1 protein than that injected with GFP (Figures 1E,F). The relative number of callose deposits by BxCDP1 triggered was also significantly more than the GFP triggered (Figure 1H). The result further verified that BxCDP1 could induce plant immunity.
To explore the avirulence function of BxCDP1 in host pine, a yeast two-hybrid screening assay was performed to identify potential host targets of BxCDP1. The result showed that BxCDP1 was self-activated in yeast (Supplementary Figure 1) and SD/-Trp/-Leu/-His/-Ade/+X-α-Gal+AbA plates with 1 mM 3-AT could effectively inhibit BxCDP1 self-activation (Supplementary Figure 2). The bait plasmid PGBKT7::BxCDP1 was co-transferred with yeast library plasmids of pine into yeast competent cell Y2H. Sixty-six positive transformants were screened on SD/-Trp/-Leu/-His/-Ade/+X-α-Gal+AbA plates with a 1-mM 3-AT plate. Then, the 66 positive transformants were sequenced and BLAST compared to a reference genome of Pinus taeda (Neale et al., 2014), and we found that most of them were not annotated (Supplementary Table 2). Among them, a RING-H2 finger protein 1 (RHF1) contains a RING-H2 domain, and some proteins containing this domain have been proved to participate in the immune response triggered by PAMP, the plant MAPK signal pathway, and apoptosis (Sinha et al., 2011; Park et al., 2012). At the same time, BxCDP1 proved to be a PAMP of B. xylophilus (Hu et al., 2020), so the PtRHF1, in which we were most interested in, was selected for subsequent one-to-one verification. The complete coding sequence of RHF1 from P. thunbergii was acquired according to the specific primer of P. taeda (Supplementary Table 1). The interaction between the full-length sequences of BxCDP1 and PtRHF1 was confirmed by one-to-one validation in yeast (Figure 2A). As expected, Co-IP experiments proved that BxCDP1 interacted with PtRHF1 in plants when they were co-expressed in N. benthamiana (Figure 2B). The results showed that PtRHF1, indeed, interacts with BxCDP1.
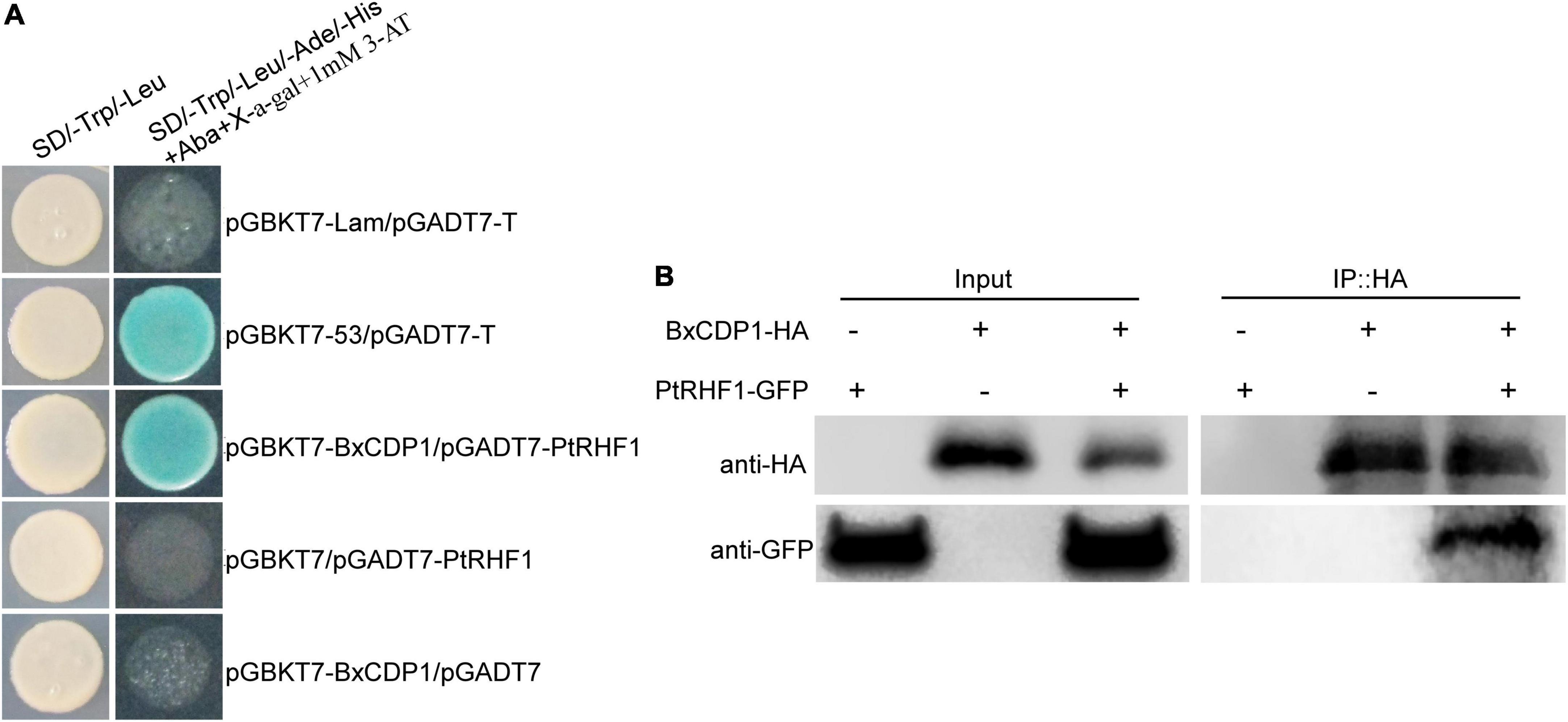
Figure 2. BxCDP1 interacts with Pinus thunbergii protein PtRHF1. (A) BxCDP1 interacts with PtRHF1 in yeast. Yeast strain Y2H Gold co-carrying pGBKT7-BxCDP1 and pGADT7-PtRHF1 was grown on SD/-Trp/-Leu and the selective medium SD/-Trp/-Leu/-Ade/-His+X-α-gal+AbA+1mM 3-AT. (B) BxCDP1 interacts with PtRHF1 in vivo. Co-immunoprecipitation (Co-IP) was performed on extracts of Nicotiana benthamiana leaves expressing both BxCDP1-HA and PtRHF1-GFP. BxCDP1-HA and PtRHF1-GFP were detected using anti-HA and anti-GFP antibodies by Western blot. The immune complexes were pulled down using anti-HA agarose beads.
To investigate whether PtRHF1 is involved in the interaction between pine and B. xylophilus, the expression of PtRHF1 was detected when P. thunbergii was inoculated with B. xylophilus for 0 h, 2.5 h, 6 h, 12 h, 24 h, and 3 days by RT-qPCR, respectively. The result showed that the expression of PtRHF1 was upregulated at 2.5 h after B. xylophilus infection and reached the peak at 12 h, thus showing a downward trend (Figure 3A). These results suggested that the expression of PtRHF1 was induced by the early infection of B. xylophilus and might be involved in the plant immune response.
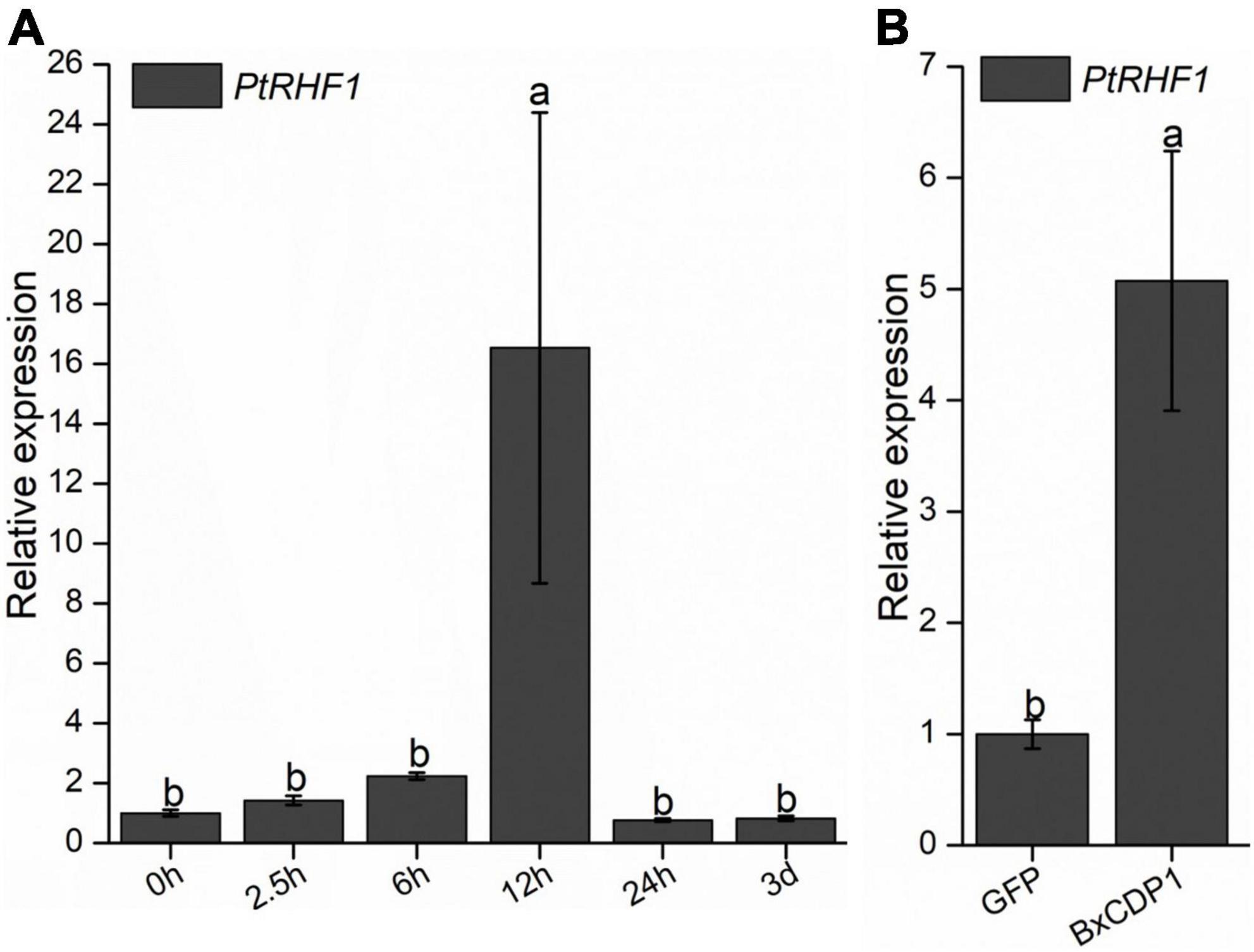
Figure 3. Relative transcript levels of PtRHF1 in Pinus thunbergii. (A) The PtRHF1 was upregulated at the early stages of Bursaphelenchus xylophilus infection. The P. thunbergii inoculated without B. xylophilus was used as a control. (B) The relative expression of PtRHF1 was induced when purified BxCDP1 protein was inoculated into P. thunbergii. The P. thunbergii inoculated with purified GFP protein was used as a control. Data are the means, and the error bars represent ± SD from three biological replicates. Different letters on top of the bars indicate statistically significant differences (p < 0.05, t-test) as measured by Duncan’s multiple range test.
In order to explore the relationship between BxCDP1 and PtRHF1, purified BxCDP1 protein was inoculated in 3-year-old P. thunbergii seedlings, and 6 h later, the relative expression levels of PtRHF1 were detected in pine seedlings. The results showed that the relative expression level of PtRHF1 in P. thunbergii inoculated with purified BxCDP1 protein was significantly higher than that of the control (Figure 3B), indicating that BxCDP1 can induce the upregulation of PtRHF1, suggesting that BxCDP1 may stimulate the defense response of P. thunbergii by inducing the upregulation of PtRHF1.
In order to determine the key amino acid region of BxCDP1 triggering cell death in N. benthamiana, 19 deletion mutants of BxCDP1 were constructed and finally transiently expressed these deletion mutants into N. benthamiana. It was found that the mutant lacking the N-terminal signal peptide could not cause cell death in N. benthamiana, indicating that the deletion of N-terminal signal peptide affected its function of causing death in N. benthamiana. However, 10 mutants from the remaining 18 BxCDP1 deletion mutants adding the N-terminal signal peptide could cause cell death in N. benthamiana, and three of them, namely M9, M15, and M16 (56–86aa, 107–136aa, and 153–173aa), were in the shortest region that could trigger cell death in N. benthamiana (Figure 4). The expression of the three deletion mutants of BxCDP1 was validated by Western blot analysis (Figure 5). In addition, we found that the degree of cell death induced by a deletion mutant M8 (including regions covered by M9, M15, and M16) was lower than that induced by M9, M15, and M16. We speculated that this could be due to two reasons. First, the protein has a spatial structure. In M8, the regions covered by M9, M15, and M16 may be partially wrapped in space, resulting in a lower degree of cell death. Second, the expression of different proteins mediated by A. tumefaciens may be different. That is to say, the expression of M8 mediated by A. tumefaciens may be lower than that of M9, M15, and M16 mediated by A. tumefaciens, resulting in a lower degree of cell death.

Figure 4. Analysis of domain by constructing deletion mutants of BxCDP1. The representative Nicotiana benthamiana leaves at 7 days after agroinfiltration carrying BxCDP1 and its 19 deletion mutants. M9, M15, and M16 were the shortest regions that could trigger cell death in N. benthamiana. The infiltration assay was performed three times and three different N. benthamiana with three inoculated leaves were used in each assay.
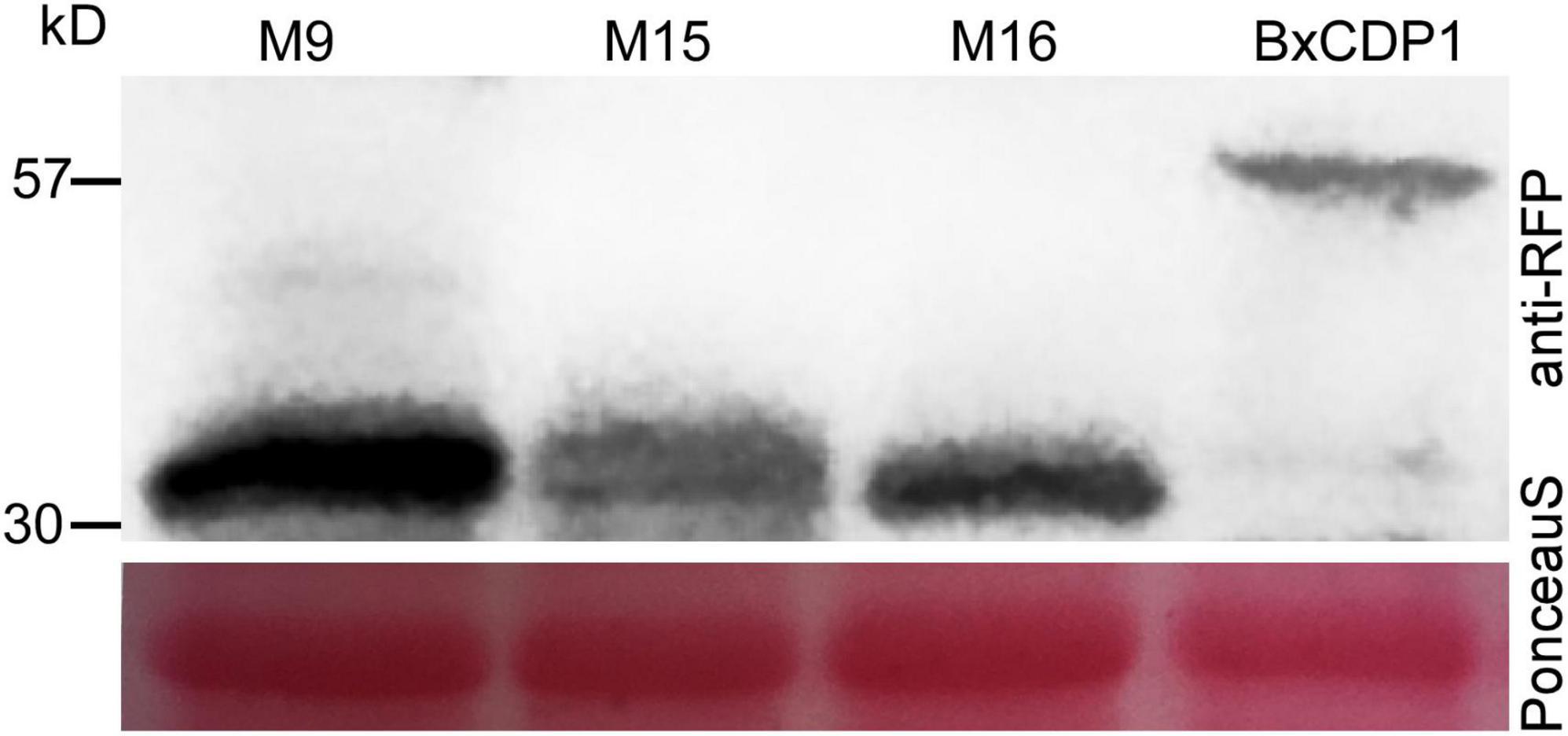
Figure 5. Western blot analysis of proteins from Nicotiana benthamiana leaves transiently expressing target proteins fused with anti-red fluorescent protein (anti-RFP) tags. Protein loading is shown by Ponceau S staining of RuBisCO.
In the previous study, BxCDP1 could significantly induce the expression of PTI marker genes NbAcre31, NbPTI5, and NbCyp71D20 (Hu et al., 2020). To determine whether the three regions (M9, M15, and M16) that triggered cell death can also induce immune responses in N. benthamiana, the sequences of M9, M15, and M16 and control M14 were synthesized. The 300 nM synthesized peptides M9, M15, and M16 and control M14 were infiltrated into leaves of N. benthamiana. The result showed that, compared with control M14, M9 could significantly induce the expressions of NbAcre31 and NbPTI5 (Figure 6A), M15 could significantly induce the expression of NbPTI5 (Figure 6B), and M16 could significantly induce the expression of NbCyp71D20 (Figure 6C). The result indicated that M9, M15, and M16 can significantly induce immune responses in N. benthamiana.
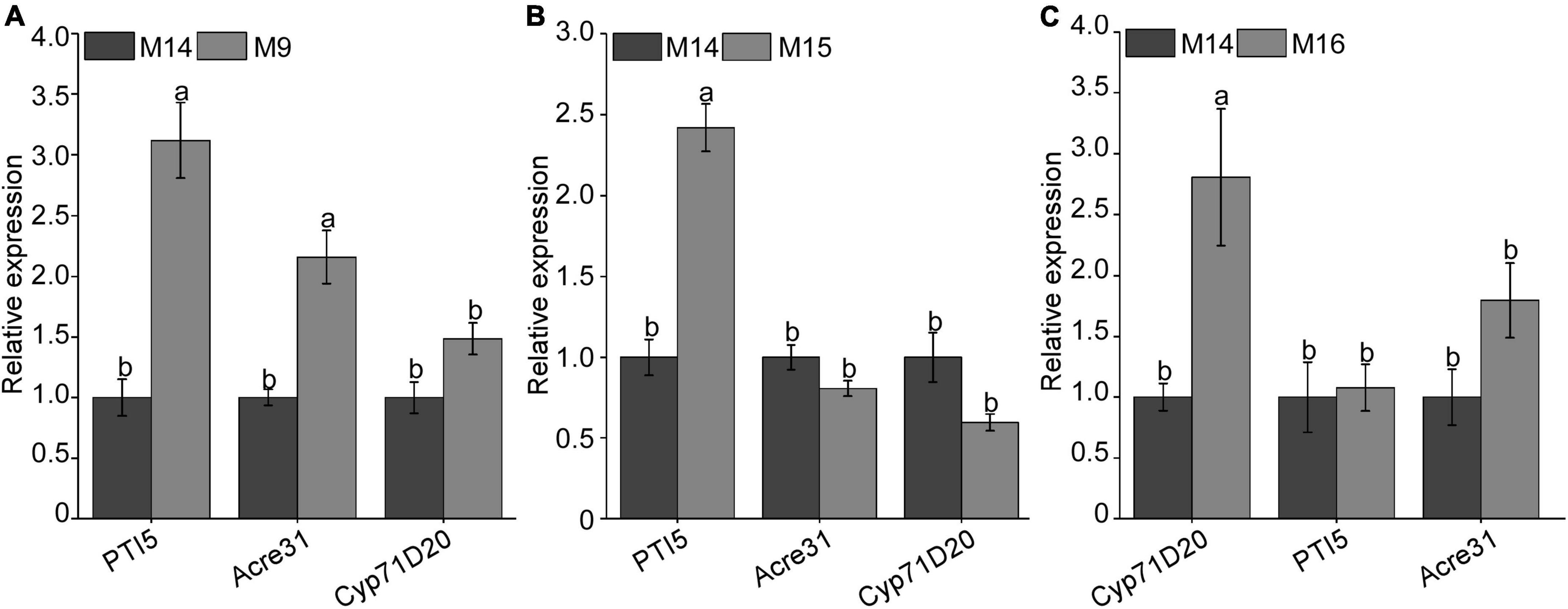
Figure 6. Detection of the relative expression of PAMP-triggered immunity (PTI) marker gene after injecting with three key peptides of BxCDP1 in Nicotiana benthamiana. (A–C) Transcriptional upregulation of N. benthamiana PTI marker genes triggered by 300 nM synthesized peptides M9, M15, and M16 in N. benthamiana. The N. benthamiana leaves infiltrated with synthesized peptide M14 were used as a control. The assay was conducted three times, and three different plants with three inoculated leaves were used in each assay. Data are the means, and the error bars represent ± SD from three biological replicates. Different letters on top of the bars indicate statistically significant differences (p < 0.05, t-test) as measured by Duncan’s multiple range test.
Our previous study found that BxCDP1 can induce the expression of pathogenesis-related marker genes PtPR-3, PtPR-4, and PtPR-5 of P. thunbergii, and P. thunbergii could sense the presence of xenobiotics and stimulate defense response (Hu et al., 2020). Therefore, in order to explore whether the identified peptides that can trigger cell death and immunity in N. benthamiana also have the ability to induce the expression of pathogenesis-related marker genes in P. thunbergii, M9, M15, M16, and control M14 were inoculated with 3-year-old P. thunbergii for 4 h, and then, about 1500 B. xylophilus strains were inoculated. The relative expressions of PtPR-3, PtPR-4, and PtPR-5 were detected by RT-qPCR. Compared with the expression levels in M15 and control M14-treated P. thunbergii, PtPR-3 was significantly upregulated both in the M9-treated and the M16-treated P. thunbergii (Figure 7A), PtPR-4 was significantly upregulated in the M16-treated samples (Figure 7B), and PtPR-5 was significantly upregulated in the M9-treated samples (Figure 7C).
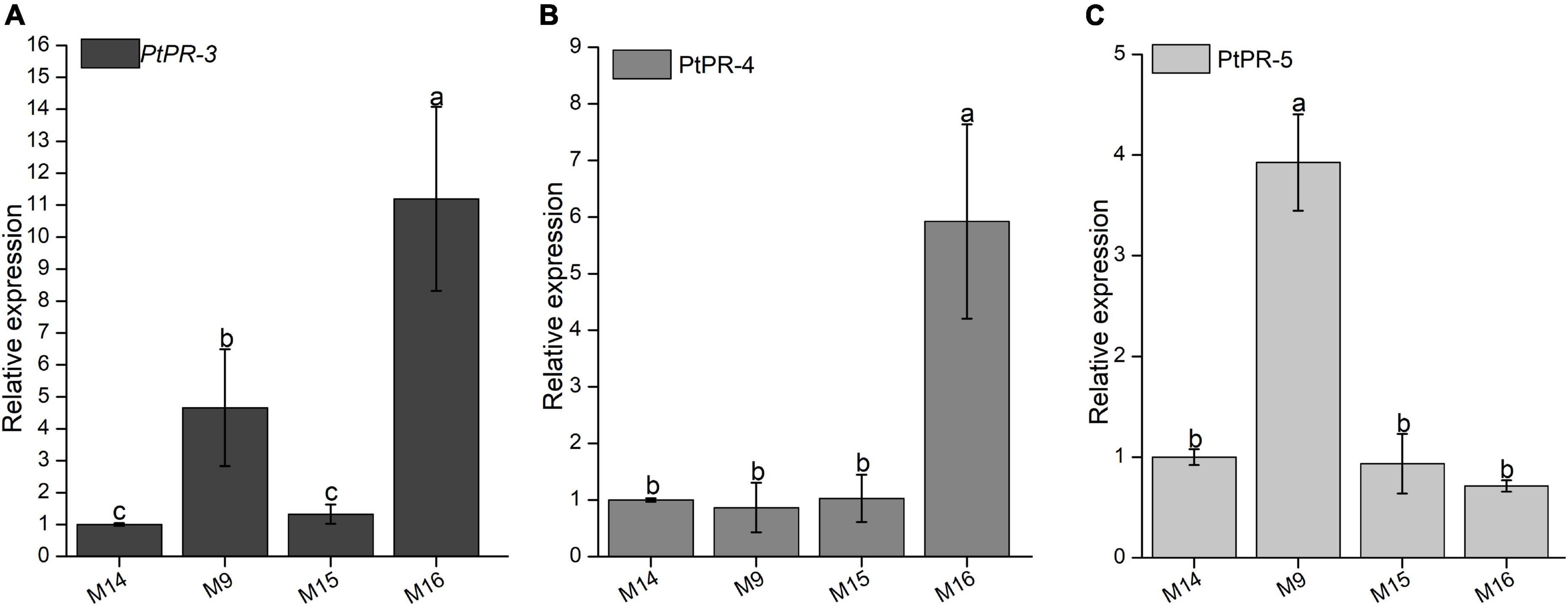
Figure 7. Relative transcript levels of pathogenesis-related (PR) genes in Pinus thunbergii inoculated with three synthesized peptides and Bursaphelenchus xylophilus. (A) The relative expression of PtPR-3 was detected by RT-qPCR. (B) The relative expression of PtPR-4 was detected by RT-qPCR. (C) The relative expression of PtPR-5 was detected by RT-qPCR. The P. thunbergii seedlings were treated with three synthesized peptides M9, M15, and M16 for 4 h and then inoculated with B. xylophilus. After 6 h, 2 cm stems in the length of P. thunbergii seedlings were used for RNA extraction, and the relative expressions of PtPR-3, PtPR-4, and PtPR-5 were detected. The P. thunbergii seedlings inoculated with B. xylophilus after inoculating with synthesized peptide M14 were used as the control. The inoculation assay was repeated three times, and in each assay, three different seedlings for each treatment were used. Data are the means, and the error bars represent ± SD from three independent experiments. Different letters on top of the bars indicate statistically significant differences (p < 0.05, t-test) as measured by Duncan’s multiple range test.
M9 and M16 could stimulate the expression of pathogenesis-related marker genes. However, it is unclear whether they can improve the resistance of P. thunbergii to B. xylophilus. Thus, M9, M15, and M16 and control M14 were inoculated into P. thunbergii for 4 h, and approximately 1,500 mixed-life-stage highly virulent strains of B. xylophilus AMA3 were inoculated at the inoculation wound of P. thunbergii. The pines infected with B. xylophilus would show early symptoms such as yellowing or browning or wilting needles. The result showed that this early symptom occurred later in seedlings inoculated with M9 and M16, compared with the ones inoculated with M14 and M15, whether on the 12 dpi or 15 dpi (Figure 8C). Moreover, the infection ratio and disease severity index of P. thunbergii seedlings inoculated with M9 and M16 were also significantly lower than the seedlings inoculated with M15 and control M14 at both 12 dpi and 15 dpi (Figures 8A,B).
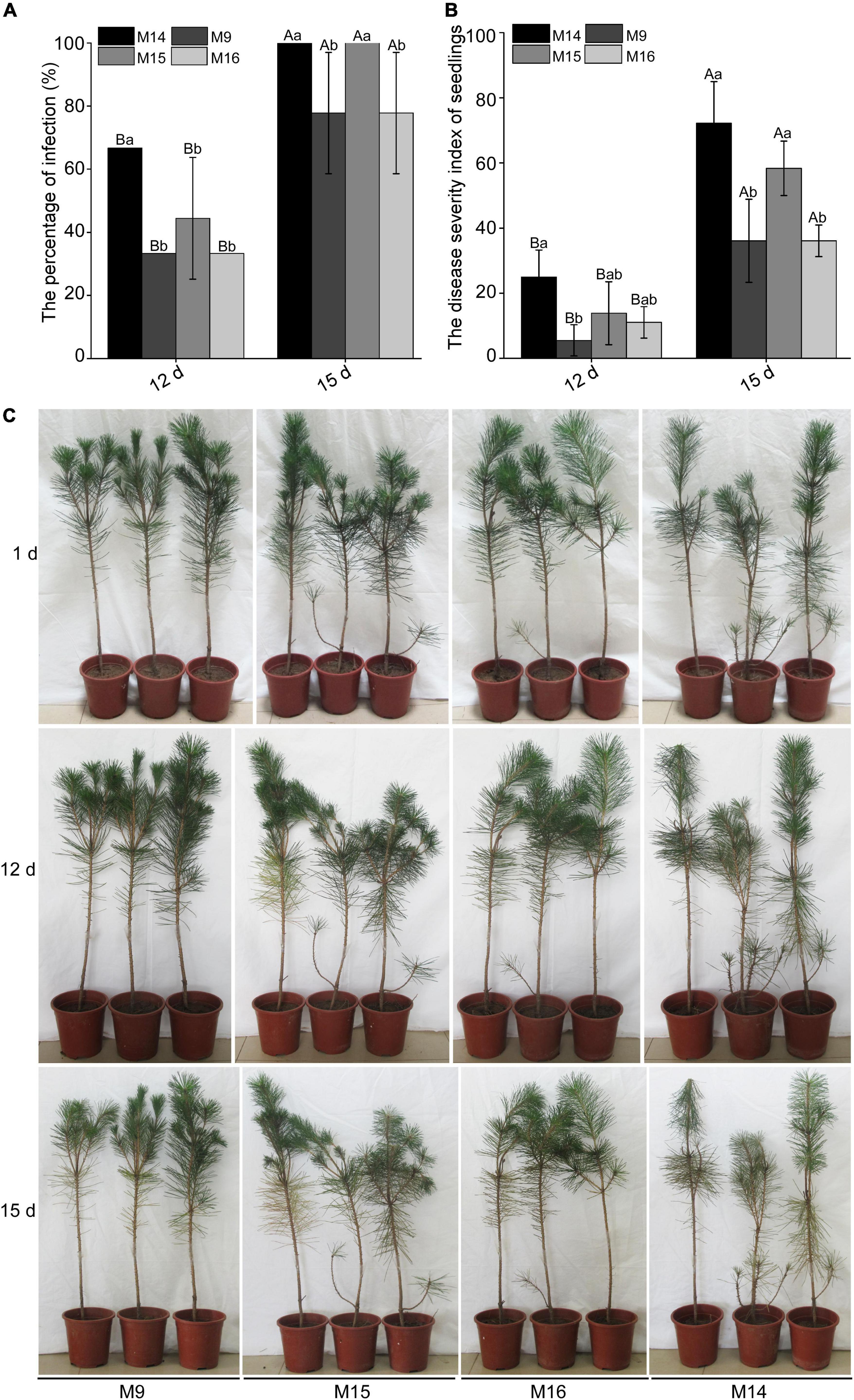
Figure 8. Peptides M9 and M16 of BxCDP1 can promote the resistance of Pinus thunbergii to Bursaphelenchus xylophilus. (A) The infection rates of P. thunbergii seedlings under four different treatments. (B) The disease severity index of P. thunbergii seedlings under four different treatments. The P. thunbergii seedlings were treated with three synthesized peptides M9, M15, and M16 for 4 h and then inoculated with 1500 B. xylophilus. The P. thunbergii seedlings inoculated with 1500 B. xylophilus after treating with synthesized peptide M14 for 4 h were used as control. Data are the means of three independent inoculation experiments and error bars (±SD) from three independent inoculation experiments. Different capital letters above the bars indicate significant differences between inoculation times in the same treatments (p < 0.05, t-test), as measured by Duncan’s multiple range test; different lowercase letters above the bars indicate significant differences between different treatments at the same inoculation time (p < 0.05, t-test), as measured by Duncan’s multiple range test. The infection experiment was repeated three times with three replicate P. thunbergii seedlings per treatment. (C) Representative photographs of P. thunbergii at 1 day, 12 days, and 15 days post-inoculation. The B. xylophilus infection assay was conducted three times.
The burst of plant ROS, the upregulated expression of PR genes, and callose deposits are the indicators of plant immunity activation (Dodds and Rathjen, 2010). Our team identified a PAMP BxCDP1 in B. xylophilus, which can trigger cell death, and upregulated the expression of PTI marker genes and ROS outbreak in various plants, namely, N. benthamiana (Hu et al., 2020). In this study, the accumulation of superoxide, H2O2, and callose deposits in N. benthamiana treated with purified BxCDP1 protein was significantly higher than that in the control. It indicated that BxCDP1 promoted the outbreak of ROS by stimulating the production of more superoxide and H2O2 in N. benthamiana and callose deposits further verified that the immune response of N. benthamiana was activated by BxCDP1. These results indicated that BxCDP1 was a key molecular pattern to plant immunity activation.
Identifying the target proteins of PAMPs or effectors in the host can further reveal how PAMPs or effectors play a role in the infection of B. xylophilus. Many RING-type E3 ligase proteins have been identified as plant immune modulators. For example, the effector AvrPiz-t of Magnaporthe oryzae can be transported to rice cells and combined with the ubiquitin ligase APIP6 of rice RING E3 to make it unstable so as to inhibit the immunity induced by PAMP (Park et al., 2012). The expression of rice RING-type zinc finger protein OSRHC1 in Arabidopsis improves the resistance to highly toxic bacteria (Cheng et al., 2007). Moreover, the known homologous proteins containing the RING domain have been proven to be involved in ubiquitin-mediated protein hydrolysis, plant MAPK signal pathway, and apoptosis. Among them, plant MAPK and its signal transduction are considered to be important receptors and signal transduction processes involved in PAMPs signal transduction (Sinha et al., 2011). In this study, PtRHF1 was predicted to contain a RING-H2 domain and proved to be a host target protein interacting with PAMP BxCDP1 that activates plant PTI. Thus, we speculate that PtRHF1 may be a plant E3 ligase that participates in the ubiquitination process of BxCDP1 into plants so as to activate plant immunity. The relative expression of PtRHF1 started to increase at 2.5 h and reached the peak at 12 h after B. xylophilus infection, indicating that PtRHF1 played an important role in the early stage of B. xylophilus infection. In addition, the expression of PtRHF1 was significantly induced after purified BxCDP1 protein was inoculated into P. thunbergii. These results suggested that PtRHF1 was involved in the early immune response BxCDP1 triggered, which works by stimulating the defense response of P. thunbergii by inducing the upregulation of PtRHF1. However, because both the gene knockout and gene silencing system are not suitable for gymnosperm pine, we are restricted from further exploring the exact biological function of PtRHF1 in pine. Hence, in order to tackle the above speculation, multiple proofs of experiments need to be carried out in future. Next, we will further explore subcellular location and influence on nematodes (that can infect N. benthamiana, such as root-knot nematode) infection of PtRHF1 in N. benthamiana so as to indirectly reveal its function in the interaction between nematodes and pine trees. Moreover, a target protein PtACO1 of B. xylophilus effector BxSCD1 that can inhibit BxCDP1-triggered cell death and immunity was identified (Wen et al., 2021). Thus, whether there is a certain correlation between the interaction of BxCDP1 and PtRHF1 and the interaction of BxSCD1 and PtACO1, and the effect of their correlation on B. xylophilus infection need to be further studied.
At present, one of the common methods to screen and identify the key amino acid regions is to construct a protein deletion mutant and express it transiently in N. benthamiana. For example, the key amino acid regions of the effector SCRE1 of U. virens and the effectors PsAvh238 and PsAvh52 of Phytophthora sojae that inhibit plant immunity were identified through this method (Yang et al., 2017; Li et al., 2018). In this study, we successfully screened and identified three short amino acid regions of BxCDP1 named M9, M15, and M16 (31aa, 30aa, and 21aa, respectively), which can independently trigger cell death in N. benthamiana. Moreover, as the full length of BxCDP1, M9 and M16 could trigger the upregulated expression of PTI marker genes, indicating that they, as key regions of BxCDP1, indeed, triggered plant immunity. However, their ability to trigger the expression of the PTI marker gene was weaker than the full length of BxCDP1 (Hu et al., 2020). We speculated that the ability of BxCDP1 to trigger plant immunity is the superposition of the two regions.
BxCDP1 triggered the upregulated expressions of the host PR genes PtPR-3, PtPR-4, and PtPR-5 and consequently improved the pine resistance to PWD (Hu et al., 2020). In this study, we also found that synthesized peptides M9 could stimulate the upregulated expression of PtPR-3 and PtPR-5, and M16 could induce the upregulated expression of PtPR-3 and PtPR-4 in P. thunbergii. Meanwhile, pathogenicity tests showed that the early symptom of P. thunbergii seedlings inoculated with B. xylophilus after M9 and M16 occurred later than the control pines to some extent. Altogether, these results showed that both M9 and M16 could enhance the resistance of P. thunbergii seedlings to B. xylophilus by stimulating the expressions of host PR genes. Among all the synthesized peptides, M9 works the best in triggering cell death, inducing PTI Marker genes’ expression and improving pine resistance, followed by M16. In conclusion, both M9 and M16 have the potential to be developed and utilized as immune inducers against B. xylophilus in future.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
X-QW and L-JH were the leading investigators of this research program and planned and designed the research. L-JH performed the majority of experiments with the help of T-YW, Y-JQ, LR, and YZ. X-QW and J-RY contributed reagents, materials, and analysis tools. L-JH analyzed the data and wrote the manuscript with suggestions from X-QW and T-YW. All authors commented on the manuscript before submission and approved the submitted version.
This work was supported by the Natural Science Foundation of Jiangsu Province (BK20200774) and the National Natural Science Foundation of China (32001318).
We thank Prof. Yuanchao Wang (Nanjing Agricultural University) for providing the vector pGR107, pBinGFP, and pBinRFP and Nicotiana benthamiana seeds. We also thank Dr. Haiyang Li (Department of Plant Pathology, Henan Agricultural University, China) for his helpful suggestions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.937473/full#supplementary-material
Chen, J. S., Lin, B. R., Huang, Q. L., Hu, L. L., Zhuo, K., and Liao, J. L. (2017). A novel Meloidogyne graminicola effector, MgGPP, is secreted into host cells and undergoes glycosylation in concert with proteolysis to suppress plant defenses and promote parasitism. PLoS Pathog. 13:e1006301. doi: 10.1371/journal.ppat.1006301
Cheng, M. Y., Zeng, N. Y., Tong, S. W., Li, F. W. Y., Zhao, K. J., Zhang, Q., et al. (2007). Expression of a RING-HC protein from rice improves resistance to Pseudomonas syringae pv. tomato DC3000 in transgenic Arabidopsis thaliana. J. Exp. Bot. 58, 4147–4159. doi: 10.1093/jxb/erm272
Chisholm, S. T., Coaker, G., Day, B., and Staskawicz, B. J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124, 803–814.
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Doke, N. (1983). Involvement of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol. Phant Pathol. 23, 345–357.
Dong, S. M., Yin, W. X., Kong, G. H., Yang, X. Y., Qutob, D., Chen, Q. H., et al. (2011). Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP-ribose pyrophosphorylase that modulates plant immunity. PLoS Pathog. 7:e1002353. doi: 10.1371/journal.ppat.1002353
Du, Y., Chen, X. K., Guo, Y. L., Zhang, X. J., Zhang, H. X., Li, F. F., et al. (2021). Phytophthora infestans RXLR effector PITG20303 targets a potato MKK1 protein to suppress plant immunity. N. Phytol. 229, 501–515. doi: 10.1111/nph.16861
Durrant, W. E., Rowland, O., Piedras, P., Hammond-Kosack, K. E., and Jones, J. D. (2000). cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell Rep. 12, 963–977. doi: 10.1105/tpc.12.6.963
Espada, M., Silva, A. C., Eves van den Akker, S., Cock, P. J., Mota, M., and Jones, J. T. (2016). Identification and characterization of parasitism genes from the pinewood nematode Bursaphelenchus xylophilus reveals a multilayered detoxification strategy. Mol. Plant Pathol. 17, 286–295. doi: 10.1111/mpp.12280
Hirao, T., Fukatsu, E., and Watanabe, A. (2012). Characterization of resistance to pine wood nematode infection in Pinus thunbergii using suppression subtractive hybridization. BMC Plant Biol. 12:13. doi: 10.1186/1471-2229-12-13
Hu, L. J., Wu, X. Q., Ding, X. L., and Ye, J. R. (2021). Comparative transcriptomic analysis of candidate effectors to explore the infection and survival strategy of Bursaphelenchus xylophilus during different interaction stages with pine trees. BMC Plant Biol. 1:224–237. doi: 10.1186/s12870-021-02993-9
Hu, L. J., Wu, X. Q., Li, H. Y., Wang, Y. C., Huang, X., Wang, Y., et al. (2020). BxCDP1 from the pine wood nematode Bursaphelenchus xylophilus is recognized as a novel molecular pattern. Mol. Plant Pathol. 21, 923–935. doi: 10.1111/mpp.12939
Hu, L. J., Wu, X. Q., Li, H. Y., Zhao, Q., Wang, Y. C., and Ye, J. R. (2019). An effector, BxSapB1, induces cell death and contributes to virulence in the pine wood nematode Bursaphelenchus xylophilus. Mol. Plant Microbe Interact. 32, 452–463. doi: 10.1094/MPMI-10-18-0275-R
Huang, L., Wang, P., Tian, M. Q., Zhu, L. H., and Ye, J. R. (2019). Major sperm protein BxMSP10 is required for reproduction and egg hatching in Bursaphelenchus xylophilus. Exp. Parasitol. 197, 51–56. doi: 10.1016/j.exppara.2019.01.004
Huang, X., Hu, L. J., and Wu, X. Q. (2019). Identication of a novel effector BxSapB3 that enhances the virulence of pine wood nematode Bursaphelenchus xylophilus. Acta Biochim. Biophys. Sin. 51, 1071–1078. doi: 10.1093/abbs/gmz100
Jing, M. F., Guo, B. D., Li, H. Y., Yang, B., Wang, H. N., Kong, G. H., et al. (2016). A Phytophthora sojae effector suppresses endoplasmic reticulum stress-mediated immunity by stabilizing plant binding immunoglobulin proteins. Nat. Commun. 7:11685. doi: 10.1038/ncomms11685
Kettles, G. J., Bayon, C., Canning, G., Rudd, J. J., and Kanyuka, K. (2017). Apoplastic recognition of multiple candidate effectors from the wheat pathogen Zymoseptoria tritici in the nonhost plant Nicotiana benthamiana. N. Phytol. 213, 338–350. doi: 10.1111/nph.14215
Lee, D. H., Choi, H. W., and Wang, B. K. (2011). The pepper E3 ubiquitin ligase RING1 gene, CaRING1, is required for cell death and the salicylic acid-dependent defense response. Plant Physiol. 156, 2011–2025. doi: 10.1104/pp.111.177568
Li, H. Y., Wang, H. N., Jing, M. F., Zhu, J. Y., Guo, B. D., Wang, Y., et al. (2018). A Phytophthora effector recruits a host cytoplasmic transacetylase into nuclear speckles to enhance plant susceptibility. eLife 7:e40039. doi: 10.7554/eLife.40039
Li, Y., Hu, L. J., Wu, X. Q., and Ye, J. R. (2020). A Bursaphelenchus xylophilus effector, Bx-FAR-1, suppresses plant defense and affects nematode infection of pine trees. Eur. J. Plant Pathol. 153, 637–650.
Liu, H., and Stone, S. L. (2011). E3 ubiquitin ligases and abscisic acid signaling. Plant Signal Behav. 6, 344–348.
Ma, Z. C., Song, T. Q., Zhu, L., Ye, W. W., Wang, Y., Shao, Y. Y., et al. (2015). A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant cell 27, 2057–2072. doi: 10.1105/tpc.15.00390
Margarida, E., Sebastian, E. V. D. A., and Tom, M. (2018). STATAWAARS: A promoter motif associated with spatial expression in the major effector-producing tissues of the plant-parasitic nematode Bursaphelenchus xylophilus. BMC Genom. 19:553. doi: 10.1186/s12864-018-4908-2
Marino, D., Peeters, N., and Rivas, S. (2012). Ubiquitination during plant immune signaling. Plant Physiol. 160, 15–27.
Nagels, D. A., Pauwels, L., and Goossens, A. (2016). The ubiquitin system and jasmonate signaling. Plants 5:6.
Neale, D. B., Wegrzyn, J. L., Stevens, K. A., Zimin, A. V., Puiu, D., Crepeau, M. W., et al. (2014). Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 15:R59. doi: 10.1186/gb-2014-15-3-r59
Park, C. H., Chen, S., and Shirsekar, G. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24, 4748–4762. doi: 10.1105/tpc.112.105429
Qiu, X. W., Wu, X. Q., Huang, L., Tian, M., and Ye, J. R. (2013). Specifically expressed genes of the nematode Bursaphelenchus xylophilus involved with early interactions with pine trees. PLoS One 8:e78063. doi: 10.1371/journal.pone.0078063
Rempel, C. B., and Hall, R. (1996). Comparison of disease measures for assessing resistance in canola (Brassica napus) to blackleg (Leptosphaeria maculans). Can. J. Bot. 74, 1930–1936.
Serrano, M., and Guzmán, P. (2004). Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genet. Mol. Res. 167, 919–929. doi: 10.1534/genetics.104.028043
Sinha, A. K., Jaggi, M., Raghuram, B., and Tuteja, N. (2011). Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 6, 196–203.
Stone, S. L., Herborg, H., and Andrew, T. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137, 13–30.
Tanaka, S. E., Dayi, M., Maeda, Y., Tsai, I. J., Tanaka, R., Bligh, M., et al. (2019). Stage-specific transcriptome of Bursaphelenchus xylophilus reveals temporal regulation of effector genes and roles of the dauer-like stages in the lifecycle. Sci. Rep. 9:6080. doi: 10.1038/s41598-019-42570-7
Thordal-Christensen, H., Zhang, Z., and Wei, Y. D. (1997). H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1104/pp.123.4.1289
Tsai, I. J., Tanaka, R., Kanzaki, N., Akiba, M., Yokoi, T., Espada, M., et al. (2016). Transcriptional and morphological changes in the transition from mycetophagous to phytophagous phase in the plant-parasitic nematode Bursaphelenchus xylophilus. Mol. Plant Pathol. 17, 77–83. doi: 10.1111/mpp.12261
Tsuda, K., and Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Wang, J. H., Liu, H. L., Guo, K., Yu, H. S., Ye, J. R., and Hu, J. F. (2022). Molecular characterization and functional analysis of Bxy-octr-1 in Bursaphelenchus xylophilus. Gene 823:146350. doi: 10.1016/j.gene.2022.146350
Wen, T. Y., Wu, X. Q., Hu, L. J., Qiu, Y. J., Rui, L., Zhang, Y., et al. (2021). A novel pine wood nematode effector, BxSCD1, suppresses plant immunity and interacts with an ethylene-forming enzyme in pine. Mol. Plant Pathol. 22, 1399–1412. doi: 10.1111/mpp.13121
Wen, T. Y., Wu, X. Q., Ye, J. R., Qiu, Y. J., Rui, L., and Zhang, Y. (2022). A Bursaphelenchus xylophilus pathogenic protein Bx-FAR -1, as potential control target, mediates jasmonic acid pathway in pines. Pest Manag. Sci. 78, 1870–1880. doi: 10.1002/ps.6805
Wojtaszek, P. (1997). Oxidative burst an early plant response to pathogen infection. Biochem. J. 322, 681–692.
Xu, Z. Q., Yu, J., Cui, L. M., Li, M., Li, R. G., and Guo, D. S. (2013). Effects of Pseudomonas fluorescens flagellin on physiological and biochemical characteristics in the suspension cells of Pinus thunbergii. Eur. J. Plant Pathol. 136, 729–736.
Yang, B., Wang, Q. Q., Jing, M. F., Guo, B. D., Wu, J. W., Wang, H. N., et al. (2017). Distinct regions of the Phytophthora essential effector Avh238 determine its function in cell death activation and plant immunity suppression. N. Phytol. 214, 361–375. doi: 10.1111/nph.14430
Yin, W. X., Dong, S. M., Zhai, L. C., Lin, Y. C., Zheng, X. B., and Wang, Y. C. (2013). The Phytophthora sojae Avr1d gene encodes an RxLR-dEER effector with presence and absence polymorphisms among pathogen strains. Mol. Plant Microbe Interact. 26, 958–968. doi: 10.1094/MPMI-02-13-0035-R
Yu, L. Z., Wu, X. Q., Ye, J. R., Zhang, S. N., and Wang, C. (2012). NOS-likemediated nitric oxide is involved in Pinus thunbergii response to the invasion of Bursaphelenchus xylophilus. Plant Cell Rep. 31, 1813–1821.
Zhang, N., Yang, J. Y., Fang, A. F., Wang, J. Y., Li, D. Y., Li, Y. J., et al. (2020). The essential effector SCRE1 in Ustilaginoidea virens suppresses rice immunity via a small peptide region. Mol. Plant Pathol. 21, 445–459. doi: 10.1111/mpp.12894
Zhao, Q., Hu, L. J., Wu, X. Q., and Wang, Y. C. (2020). A key effector, BxSapB2, plays a role in pathogenicity of pine wood nematode Bursaphelenchus xylophilus. For. Pathol. 50:e12600.
Zhou, L. F., Chen, F. M., Xie, L. Y., Pan, H. Y., and Ye, J. R. (2017). Genetic diversity of pine-parasitic nematodes Bursaphelenchus xylophilus and Bursaphelenchus mucronatus in China. For. Pathol. 47:e12334.
Keywords: Bursaphelenchus xylophilus, Pinus thunbergii, key peptides, immunity induction, protein interaction
Citation: Hu L-J, Wu X-Q, Wen T-Y, Ye J-R, Qiu Y-J, Rui L and Zhang Y (2022) The key molecular pattern BxCDP1 of Bursaphelenchus xylophilus induces plant immunity and enhances plant defense response via two small peptide regions. Front. Plant Sci. 13:937473. doi: 10.3389/fpls.2022.937473
Received: 06 May 2022; Accepted: 13 July 2022;
Published: 03 August 2022.
Edited by:
Carolina Escobar, University of Castilla-La Mancha, SpainReviewed by:
Margarida Espada, University of Évora, PortugalCopyright © 2022 Hu, Wu, Wen, Ye, Qiu, Rui and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Qin Wu, eHF3dUBuamZ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.