
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 27 June 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.934877
This article is part of the Research TopicMulti-omics and Computational Biology in Horticultural Plants: From Genotype to PhenotypeView all 32 articles
Saline-alkali soils pose an increasingly serious global threat to plant growth and productivity. Much progress has been made in elucidating how plants adapt to salt stress by modulating ion homeostasis. Understanding the molecular mechanisms that affect salt tolerance and devising strategies to develop/breed salt-resilient crops have been the primary goals of plant salt stress signaling research over the past few decades. In this review, we reflect on recent major advances in our understanding of the cellular and physiological mechanisms underlying plant responses to salt stress, especially those involving temporally and spatially defined changes in signal perception, decoding, and transduction in specific organelles or cells.
Food production must increase by 70% worldwide by 2050 to meet the demands of the ever-increasing human population (High-Level Expert Forum, FAO, October 20091). Thus, attaining food security and developing strategies to improve crop productivity and quality have become urgent aims. A major obstacle to improving crop productivity is soil salinization and alkalization, as salt stress severely restricts the germination rates, growth, development, and biomass accumulation of plants. Crop loss due to soil salinity poses an increasing threat to modern agriculture (Munns and Tester, 2008; Zhu, 2016; Yang and Guo, 2018a). In addition to rising levels of groundwater with high salt levels and increased evaporation due to drought, the increase in soil salinization is also caused by overirrigation and climate change (Rengasamy, 2006). Approximately one-fifth of irrigated lands worldwide are affected by soil salinization (Morton et al., 2019). Thus, the sustainable use of saline land resources and improving the agricultural output of saline soils are of paramount importance for global food security.
In contrast to halophytes, which can grow at high salt concentrations (>200 mM NaCl) (Munns and Tester, 2008; Flowers and Colmer, 2015), glycophytes are sensitive to high salinity. Most crops are glycophytes and are not suitable for growth in saline lands. An effective strategy for increasing crop yields in salinized agricultural lands is to design and breed salt-tolerant crop varieties. To achieve this goal, we must better understand (1) how to reduce soil salinity via phytoremediation or by improving agricultural practices; (2) the mechanisms by which high salinity leads to reduced water availability; (3) the toxic effects of sodium and chlorine ions on plants. Recent achievements in deciphering the underlying salt stress sensing and tolerance mechanisms in plants will greatly facilitate the breeding of salt-tolerant crop varieties. In this review, we briefly summarize recent progress in our understanding of salt stress sensing and signal transduction, focusing on how plants sense salt stress and transmit and decode salt stress signals to alter the gene expression and/or protein stability/activity in response to salt stress.
The process of salt stress is divided into osmotic stress (early stage) and ionic toxicity (later stage) (Munns and Tester, 2008). To tolerate salt stress more effectively, plants have evolved various regulatory mechanisms that quickly perceive changes in Na+ concentrations and osmotic pressure caused by salt stress (Figure 1). NaCl or mannitol stimuli induce rapid increases in cytosolic Ca2+ levels within seconds (Knight et al., 1997), perhaps due to the strong coupling relationship between osmotic stress receptors and calcium channels. The increase in Ca2+ levels first occurs in roots (Tracy et al., 2008) and has been detected in several different cell types (Kiegle et al., 2000; Martí et al., 2013).
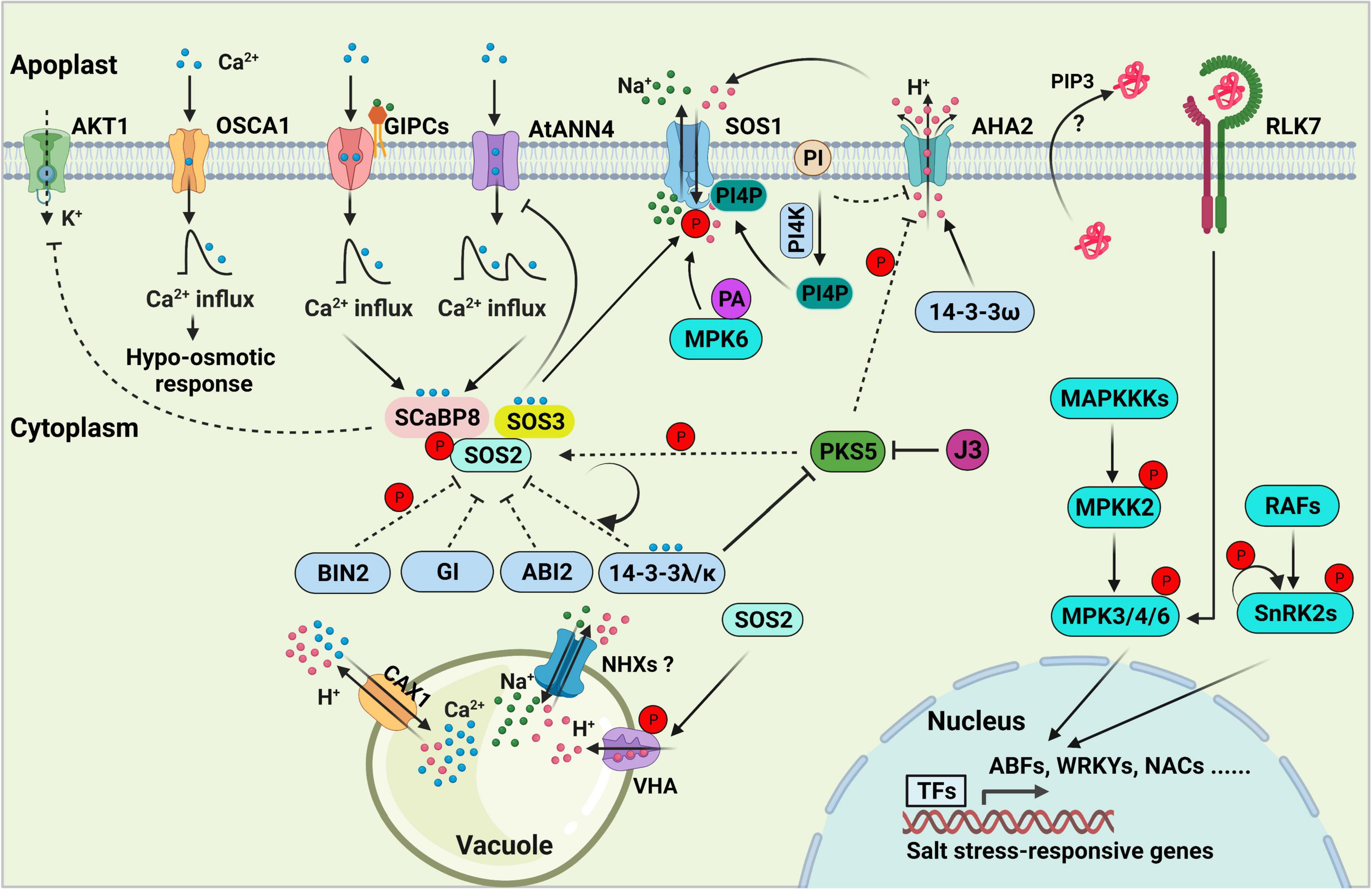
Figure 1. Salt stress signal transduction in plants. The SOS pathway, consisting of SOS3, SCaBP8, SOS2, and SOS1, is essential for decoding salt-induced calcium signals and maintaining ionic homeostasis in the plant cell. 14-3-3, GIGANTEA (GI), ABI2, and BIN2 proteins negatively regulate SOS pathway activity by directly interacting with SOS2 and repressing its kinase activity. PKS5-mediated phosphorylation of SOS2 enhances its interaction with 14-3-3, thereby inhibiting and maintaining basal levels of SOS2 activity under normal conditions. Arabidopsis K+ TRANSPORTER 1 (AKT1) activity is repressed by SCaBP8. GIPCs might function as monovalent-cation sensors that bind to Na+ and initiate calcium influx, which further activates the SOS pathway. AtANN4, a putative calcium-permeable transporter, might also generate calcium influx to activate the SOS pathway under salt stress. OSCA1 functions as an osmosensor to generate osmotic Ca2+ signaling in response to osmotic stress. Feedback regulation of AtANN4 by the SOS pathway is required to fine-tune the formation and duration of salt-induced calcium influx and long-term salt stress responses. Phosphatidylinositol (PI) directly binds to the C-terminus of the plasma membrane (PM) H+-ATPase AHA2 to repress its activity. PI is converted into phosphatidylinositol 4-phosphate (PI4P) to release the inhibition of AHA2 under salt stress. PI4P binds to and activates the PM Na+/H+ antiporter SOS1. PIP3 and RLK7 accumulate under salt stress, and the PIP3-RLK7 interaction contributes to the activation of RLK7, resulting in the activation of MPK3/6 to transduce stress signals. MAP kinase cascades are involved in regulating salt stress signal transduction. RAFs are required for the phosphorylation and activation of SnRK2s in response to salt-induced osmotic stress, and SnRK2 activity is amplified by auto-phosphorylation. In the nucleus, several specific transcription factors that are downstream targets of MPKs and SnRK2s bind to and activate the expression of salt stress-responsive genes. In the vacuole, NHXs, CAX1, the vacuolar Ca2+/H+ antiporter and vacuolar H+-ATPase (VHA) exclude Na+ from the cell. The dashed lines indicate regulatory roles under normal conditions.
Exogenous expression of Arabidopsis thaliana HISTIDINE KINASE 1 (AtHK1) suppressed the lethality of the temperature-sensitive and osmosensing-defective yeast mutation sln1-ts (Urao et al., 1999; Tran et al., 2007; Wohlbach et al., 2008), suggesting that AtHK1 functions in osmo-sensing. However, a subsequent study showed that the hk1 mutant exhibits significant physiological responses to osmotic stress (Kumar et al., 2013), indicating that some other unknown proteins must be responsible for sensing osmotic stress. Based on these findings, Kurusu et al. (2013) proposed that hyper-osmotic stress is sensed by a mechanical Ca2+ channel, which was described as a mechano-osmotic sensory modality (Kurusu et al., 2013). The osca1 (reduced hyperosmolality-induced [Ca2+]i increase 1) mutant was isolated in Arabidopsis using calcium imaging-based unbiased forward genetic screening. Similar to the osmosensor TRPV4 in animals (Liedtke et al., 2000), OSCA1 is involved in osmotic stress-induced rapid signal transduction, intermediate cellular responses, and prolonged growth and development (Yuan et al., 2014). OSCA1 was identified as a plasma membrane hyperosmolality-gated calcium-permeable channel and a putative osmosensor required for osmotic stress-induced Ca2+ signatures (Yuan et al., 2014; Zhang et al., 2020).
Using the same strategy, a genetic screen for mutants impaired in intracellular calcium concentration ([Ca2+]cyt) increases specifically induced by sodium (Na+) was performed in Arabidopsis. This led to the identification of the moca1 (monocation induced Ca2+ increases 1) mutant, which shows reduced cytosolic [Ca2+]cyt spikes in response to salt treatment (Jiang et al., 2019). The increases in [Ca2+]cyt in response to K+ or Li+ were also reduced in moca1, but it showed nearly wild type responses to H2O2, cold stress, and high external Ca2+ stimuli, indicating that moca1 is mainly sensitive to monovalent cations. Furthermore, moca1 only exhibits attenuated growth under Na+ stress, indicating that MOCA1 specifically regulates Na+ signaling (Jiang et al., 2019). MOCA1, an inositol phosphorylceramide glucuronosyltransferase also known as IPUT1, was also classified as PLANT GLYCOGENIN-LIKE STARCH INITIATION PROTEIN 6 (PGSIP6) in glucuronosyltransferase subfamily 8 (GT8) (Rennie et al., 2012, 2014). IPUT1 functions as an enzyme that transfers a glucuronic acid (GlcA) residue from UDP-GlcA to the precursor inositol phosphorylceramide (IPC) to form glycosyl inositol phosphorylceramide (GIPC) (Rennie et al., 2014). Indeed, moca1 plants contained higher levels of IPCs but lower levels of GIPCs than wild type plants. It appears that GIPC directly binds Na+ on the cell surface, thereby preventing the subsequent depolarization of the cell membrane potential that gates Ca2+ channels (Jiang et al., 2019).
Based on the mechano-osmotic sensory modality, salt stress and osmotic stress have long been regarded as mediating the mechanical properties of the cell wall, which then senses and transduces the salt stress signal. Accumulating evidence suggests that the receptor-like kinase FERONIA (FER) (Feng et al., 2018), ANNEXINs (Laohavisit et al., 2013; Ma et al., 2019), plastid K+ exchange antiporters (KEAs) (Stephan et al., 2016), the mechanosensitive ion channel MscS-like (MSL) (Hamilton et al., 2015a,b), MID1-COMPLEMENTING ACTIVITY (MCA) (Kurusu et al., 2012a,b, 2013), and two-pore calcium channel family proteins (Choi et al., 2014) sense osmotic stress or salt stress by mediating salt-induced Ca2+ signaling or by perceiving salt stress-induced turgor or changes in cell structure.
Feronia functions as a sensor of cell wall softening and induce cell-specific calcium transients to maintain cell wall integrity under salt stress (Feng et al., 2018). Salt stress induces the processing and secretion of mature RAPID ALKALINIZATION FACTOR 22/23 (RALF22/23), which in turn interact with the cell wall-localized leucine-rich repeat extensins LRX3, LRX4, and LRX5, together with FER, to sense and transduce salt stress signals by monitoring cell wall integrity (Zhao C. et al., 2018). AtANN1 is an important regulator of calcium signaling and adaptive root growth that mediates reaction oxygen species (ROS)-induced increases in cytosolic calcium concentration ([Ca2+]cyt) under salt stress. The atann1 mutant shows enhanced Na+ influx and K+ efflux at root epidermal cells and impaired root growth under saline conditions (Laohavisit et al., 2013). AtANN4, another calcium-dependent membrane-binding protein, and putative calcium-permeable transporter, is required for the salt stress response and salt-induced increases in [Ca2+]cyt and is also essential for salt overly sensitive (SOS) pathway activation (Ma et al., 2019).
Pioneering studies demonstrated that KEA1 and KEA2 are targeted to the inner envelope membrane of the chloroplast, whereas KEA3 is targeted to the thylakoid membrane, and that these transporters are required for plasmid ion homeostasis and osmoregulation (Kunz et al., 2014; Stephan et al., 2016). Mutation of KEA1, KEA2, or KEA3 leads to reduced rapid osmotic stress-induced calcium spikes, suggesting that Arabidopsis KEA1/2/3 function as osmotic sensors, endowing plants with the ability to sense the intensity of water limitation in response to osmotic stress (Stephan et al., 2016).
The rapidly reduced turgor pressure caused by high salinity is perceived by MSL and MCA family proteins. MSL10 is required for potentiating the [Ca2+]cyt increase and ROS accumulation in response to cell swelling (Basu and Haswell, 2020), suggesting that MSL10 might function as a membrane-based sensor that perceives cell swelling. Pollen-localized MSL8, a membrane tension-gated ion channel, is required for the survival of pollen and full male fertility under hypo-osmotic shock due to rehydration (Hamilton et al., 2015a), and MSL2 and MSL3 have been implicated in osmotic homeostasis in chloroplasts (Haswell and Meyerowitz, 2006; Wilson et al., 2011; Veley et al., 2012). MCA1 is also required for cell wall integrity (Denness et al., 2011; Wormit et al., 2012) and for promoting Ca2+ influx upon mechanical stimulation (Nakagawa et al., 2007). Plasma membrane-localized OsMCA1 regulates Ca2+ influx and ROS generation in rice (Oryza sativa) under hypo-osmotic stress conditions (Kurusu et al., 2012a).
High salinity stress rapidly triggers H2O2 bursts in plant cells, which function as important stress signals (Wang W. et al., 2020). The leucine-rich-repeat (LRR) receptor kinase HYDROGEN-PEROXIDE-INDUCED CA2+ INCREASES1 (HPCA1) functions as a hydrogen peroxide sensor that perceives the stress-induced extracellular H2O2 burst and generates increased [Ca2+]cyt under stress stimuli (Wu et al., 2020). The Arabidopsis Na+/H+ antiporter SOS1 may perform other activities in addition to its antiporter activity; its long cytoplasmic tail likely confers its salt stress sensing activity (Shi et al., 2002; Zhu, 2002). Plant harbors abundant proteins that can act as salt stress sensor(s) and sensing mechanisms operate in parallel will allow plants to decode the stress signals and adapt to salt stress more efficiently.
During the long evolutionary process, plants have evolved a series of physiological, biochemical, and molecular regulatory mechanisms to respond to and resist salt stress (Figure 1), including the selective absorption, accumulation, or excretion of ions, the regionalization of Na+ in the cytoplasm through the membrane system, and the induction of stress tolerance gene expression (Roy et al., 2014; Zhu et al., 2016; Yang and Guo, 2018a; van Zelm et al., 2020). The cell surface-localized receptors rapidly sense external environmental stimuli (hypersaline stress), and second messengers such as Ca2+, ROS, and phytohormones are generated in a spatiotemporal-specific manner. These signals are decoded by diverse Ca2+-dependent proteins, including calmodulins (CaM), calmodulin-like proteins (CMLs), calcium-dependent protein kinases (CDPKs), CBL-interacting protein kinases (CIPKs)/SOS2-like protein kinases (PKSs) and calcineurin B-like proteins (CBLs)/SOS3-like calcium-binding proteins (SCaBPs) (Dodd et al., 2010). Finally, the Ca2+ signatures are translated into protein-protein interactions, protein phosphorylation/de-phosphorylation, phospholipid metabolism, or gene expression (Hashimoto and Kudla, 2011).
In Arabidopsis, the evolutionarily conserved SOS pathway is essential for plant adaptation to salt stress by exporting excess Na+ (Zhu, 2016; Yang and Guo, 2018a,b; van Zelm et al., 2020; Zhao et al., 2020). The classical SOS pathway includes three major components: the Na+/H+ antiporter SOS1; the serine/threonine-protein kinase SOS2, which harbors an N-terminal kinase domain similar to that of SUCROSE NON-FERMENTING 1 (SNF1)/AMPK; the helix E-loop-helix F hand (EF-hand) calcium-binding proteins SOS3 and SCaBP8/CBL10 (Zhu et al., 1998; Halfter et al., 2000; Liu et al., 2000; Shi et al., 2000; Qiu et al., 2002; Quan et al., 2007; Lin et al., 2009). The calcium sensors SOS3 and SCaBP8 perceive the salt-induced [Ca2+]cyt to release the self-inhibition of SOS2 and promote its activity via interactions with its FISL motif (A, F, I, S, L, and F are absolutely conserved). These calcium sensors then recruit the activated SOS2 to the plasma membrane (Halfter et al., 2000; Guo et al., 2001; Quan et al., 2007). SOS3 primarily functions in roots, whereas SCaBP8 primarily functions in shoots in response to salt toxicity. The salt-hypersensitive phenotype of sos3 was partially rescued by ScaBP8 overexpression, but overexpressing SOS3 failed to complement the salt-sensitive phenotype of scabp8, indicating that the functions of SOS3 and ScaBP8 are only partially redundant, and each plays additional and unique roles in plant responses to salt stress (Quan et al., 2007). ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT-I (ESCRT-I) subunit VPS23A VACUOLAR PROTEIN SORTING 23A (VPS23A) interacts with and assists in targeting SOS2 to the plasma membrane, which enhances the SOS2-SOS3 interaction on the plasma membrane (Lou et al., 2020). The plasma membrane-localized SOS2, with high levels of kinase activity, further phosphorylates and activates the Na+/H+ antiporter SOS1 (Quan et al., 2007; Lou et al., 2020).
Salt overly sensitive 1 (SOS1) is a key systemic determinant of Na+ extrusion from the cytosol to the apoplast, and its functional mutant sos1-1 displays the exceptional sensitivity to salt stress (Shi et al., 2000, 2002; Qiu et al., 2002; Quintero et al., 2002, 2011). SOS1 forms a homodimer folded into an N-terminal transmembrane and a cytosolic, autoinhibitory C-terminal tail (Quintero et al., 2011; Núñez-Ramírez et al., 2012), which is activated when the amino acid residues (serine 1036 and serine 1038) at its C-terminus are phosphorylated by SOS2 (Quintero et al., 2011). Overexpression of SOS1 C terminus leads to increased salt tolerance by the sequestration of inhibitory 14-3-3 proteins (Duscha et al., 2020). SOS1 is also involved controlling long-distance Na+ transport from root to shoot (Shi et al., 2002; El Mahi et al., 2019). Besides SOS1, several Na+ transporters from different transporter protein families have been identified, for example, the HIGH-AFFINITY K+ TRANSPORTER 1 (HKT1) family transporter and the HAK-type Na+-selective ion transporter. HKT1 is responsible for promoting shoot Na+ exclusion by retrieving Na+ from xylem vessels (Ren et al., 2005; Møller et al., 2009). The AtHKT1 loss-of-function mutation was able to confer enhanced salt tolerance in both sos1, sos2, and sos3 mutants by limiting the accumulation of cytosolic Na+ and mitigating the root-shoot Na+ translocation, indicating the interplays between HKT1 and SOS pathway is essential for modulating Na+ homeostasis in plant cells (Rus et al., 2001, 2004; Pabuayon et al., 2021). The HAK Na+ transporter is responsible for modulating root-to-shoot Na+ translocation and root Na+ content (Zhang et al., 2019; Wang et al., 2020). Salt stress-induced Ca2+ binds to ZmNSA1 and triggers its degradation, then promotes the transcription of Maize PM-H+-ATPases (MEAs) to enhance root H+ efflux, thus promoting the pump activity of SOS1 in maize (Cao et al., 2020). Numerous studies have revealed how plants adjust Na+ transporters and SOS pathway activities to balance plant growth and salt tolerance to facilitate adaptation to the ever-changing environment.
In the absence of salt stress, the kinase activity of SOS2 is inhibited by several regulators, including BRASSINOSTEROID-INSENSITIVE 2 (BIN2; Li et al., 2020), SOS2-LIKE PROTEIN KINASE 5 (PKS5; Yang Z. et al., 2019), 14-3-3 proteins (Zhou et al., 2014) and GIGANTEA (GI; Kim et al., 2013), all of which interact with SOS2 to repress its kinase activity. The protein phosphatase 2C ABA INSENSITIVE 2 (ABI2) interacts with SOS2 and might also negatively regulate its kinase activity (Ohta et al., 2003). The protein phosphatase interaction (PPI) motif in SOS2, a protein domain of 37 amino acid residues, is sufficient and necessary for the ABI2-SOS2 interaction (Ohta et al., 2003).
GIGANTEA and 14-3-3 proteins physically interact with SOS2 and suppress its kinase activity under normal conditions. Under salt stress conditions, GI and 14-3-3 are degraded by the 26S proteasome degradation pathway, resulting in the release of SOS2 from SOS2-GI/14-3-3 complexes and the activation of SOS2 (Kim et al., 2013; Zhou et al., 2014). 14-3-3 proteins are a family of conserved regulators that are considered to be phosphoserine binding proteins due to their ability to bind to numerous, functionally diverse phosphorylated signaling proteins, including kinases, phosphatases, and transmembrane receptors (reviewed in Fu et al., 2000). Phosphorylation of the amino acid residue (serine 294) in SOS2 enhanced its binding affinity to 14-3-3. PKS5 is responsible for phosphorylating SOS2 at this amino acid residue to promote the SOS2-14-3-3 interaction, thereby limiting SOS2 activity to basal levels in the absence of salt stress (Yang Z. et al., 2019). Salt stress represses the kinase activity of PKS5 by promoting the PKS5-14-3-3 interaction, thereby releasing the inhibition of SOS2 (Yang Z. et al., 2019).
The glycogen synthase kinase 3 (GSK3)-like kinase BIN2 phosphorylates and inhibits SOS2 activity to negatively regulate plant salt tolerance (Li et al., 2020). During the rapid recovery phase after salt stress, SOS3 and SCaBP8 promote the membrane distribution of BIN2, where BIN2 phosphorylates SOS2 at threonine 172 (T172) to represses its kinase activity. Meanwhile, downstream targets such as BES1 (BRI1-EMS-SUPPRESSOR 1) and BZR1 (BRASSINAZOLE RESISTANT1) are released to promote plant growth, indicating that BIN2 functions as a molecular switch between salt tolerance and growth recovery (Li et al., 2020).
Upon salt stress, not only are negative regulators of the salt stress response de-activated but positive regulators of this process are also activated. The key regulatory step in the activation of the SOS pathway is the amplification of SOS2 kinase activity in response to salt stress. As mentioned above, AtANN4 plays a critical role in mediating Ca2+ transients, which are essential for the activation of SOS2 kinase in Arabidopsis (Ma et al., 2019). In addition, the feedback regulation of AtANN4 fine-tunes the formation and duration of salt stress-induced Ca2+ transients, thereby optimizing SOS2 kinase activity in response to long-term salt stress (Ma et al., 2019). The salt-induced Ca2+ signature is decoded by 14-3-3 proteins, resulting in the increased repression of PKS5 activity, reduced SOS2Ser294 phosphorylation, and thus reduced repression of SOS2 activity by 14-3-3 proteins (Yang Z. et al., 2019). The SCaBP1/CBL2-PKS5 module and the calcium sensor SCaBP3/CBL7 all negatively regulate plasma membrane H+-ATPase activity under normal conditions, and PKS5 activity is repressed to release the H+-ATPase activity under salt stress (Fuglsang et al., 2007; Yang et al., 2010; Yang Y. et al., 2019). Together, these findings indicate that the salt-induced calcium signature is decoded by 14-3-3 and SOS3/SCaBP8 to activate or suppress SOS2 and PKS5 to further mediate plasma membrane Na+/H+ antiporter and H+-ATPase activity, respectively (Yang et al., 2010; Yang Z. et al., 2019). In the presence of salt stress, BIN2 dissociates from the plasma membrane, whereas SOS2 accumulates on the plasma membrane, also leading to the release of SOS2 inhibition and the activation of SOS1 (Quan et al., 2007; Li et al., 2020).
Besides the activation of the SOS pathway, plants have evolved an array of strategies that help them withstand salt stress, including the accumulation of protective metabolites, such as polyols, betaine, trehalsose, ectoine, proline, soluble sugars, polyamines (PAs), free unsaturated fatty acids, phosphatidylinositol, phosphatidic acid, and Late Embryogenesis Abundant (LEA) proteins, that buffer the negative effects of toxic ions (Hasegawa et al., 2000; Thole and Nielsen, 2008; Liu et al., 2015; Yang et al., 2021). Overexpression of P5CS1, encoding the rate-limiting proline biosynthesis enzyme Δ1-pyrroline-5-carboxylate synthetase 1, increased proline contents and osmotolerance in transgenic plants (Yoshiba et al., 1999; Székely et al., 2008; Szabados and Savouré, 2010). The plasma membrane-localized L-type amino acid transporter LAT1 (also known as PUT3) exhibits a polyamine transport activity (Fujita et al., 2012; Fujita and Shinozaki, 2014; Shen et al., 2016). SOS2 and SOS1 physically and genetically interact with PUT3 to modulate its polyamine transport activity in response to high salt conditions (Chai et al., 2020).
Free unsaturated fatty acids play an essential role in salt stress tolerance (Zhang et al., 2012) by activating the plasma membrane H+-ATPase by directly binding to its C-terminus (Han et al., 2017). Phosphatidylinositol (PI) directly binds to the C-terminus of the plasma membrane H+-ATPase AHA2 and inhibits its activity. PI is converted into phosphatidylinositol 4-phosphate (PI4P) under salt stress to mediate the removal of AHA2 inhibition, while the accumulated PI4P positively regulates salt tolerance by interacting with and activating SOS1 (Yang et al., 2021). NaCl treatment leads to increased PA levels and the increased production and enzymatic activity of phospholipase D (Yu et al., 2010; Wang et al., 2019). In turn, PA activates MITOGEN-ACTIVATED PROTEIN KINASE 6 (MPK6), which phosphorylates and activates SOS1 to improve plant salt tolerance (Yu et al., 2010).
Several other protein kinases are also involved in regulating salt stress responses. GEMINIVIRUS REP-INTERACTING KINASE 1 (GRIK1) and GRIK2 phosphorylate SOS2 at amino acid residue threonine 168 (T168), thereby increasing its kinase activity (Barajas-Lopez et al., 2018). The grik1-2 grik2-1 mutant is sensitive to both glucose and high salt, indicating that GRIKs are not only involved in sugar/energy-sensing but also in salinity signaling pathways (Barajas-Lopez et al., 2018). CIPK8, another CIPK protein family member and a close homolog of SOS2, interacts with CBL10 and activates SOS1 to form the CBL10-CIPK8-SOS1 module, which extrudes excess Na+ (Yin et al., 2020). In addition, OsMKK1 transcription and OsMKK1 kinase activity are markedly increased by salt treatment in rice. OsMKK1 then targets OsMPK4 to constitute a signaling pathway that positively regulates salt tolerance (Wang F. et al., 2014).
Arabidopsis MKK2 is activated by salt and cold stress, but not heat stress, hydrogen peroxide, or the flagellin-derived bacterial peptide elicitor flg22. Activated MKK2 specifically phosphorylates and activates MPK4 and MPK6, which induce the expression of cold- or salt stress-responsive genes (Teige et al., 2004). The mkk2 mutant is hypersensitive to cold and salt stress, whereas plants overexpressing MKK2 show increased salt and cold tolerance (Teige et al., 2004). MPK3/6 integrates cytokinin signaling by inducing the degradation of Arabidopsis RESPONSE REGULATOR 1 (ARR1), ARR10, and ARR12, thereby promoting salt tolerance (Yan et al., 2021).
By contrast, some MAP kinases play negative roles in salt stress tolerance. For example, the overexpression of OsMAPK33 and MKK9 led to increased salt sensitivity in rice and Arabidopsis, respectively (Xu et al., 2008; Lee et al., 2011). A T-DNA insertion mutant of MKK9 displayed insensitivity to 150 mM NaCl during seed germination, along with increased expression of the stress-related genes RESPONSIVE TO DEHYDRATION 22 (RD22) and RD29 (Alzwiy and Morris, 2007). Moreover, the Arabidopsis mapkkk20 knockout mutant showed enhanced tolerance to salt stress (Gao and Xiang, 2008), and the mpk9 mpk12 double mutant showed reduced water loss compared to the wild type (Jammes et al., 2009). In addition, five MPK genes (MPK9, MPK10, MPK11, MPK17, and MPK18), two MKK genes (MKK7 and MKK9), and four MEKK genes (MEKK3, MEKK5, MEKK6, and MEKK7) were induced by treatment with 200 mM NaCl (Moustafa et al., 2008), indicating that the MAP kinase signaling pathway plays a fundamental role in mediating plant salt tolerance. Raf-like protein kinases (RAFs) were classified as mitogen-activated protein kinase kinase kinases (MAPKKKs) in plants (Ichimura et al., 2002; Rao et al., 2010). Mutants of Raf-like kinase family members, such as ctr1, raf10, and raf11, are insensitive to ABA (Beaudoin et al., 2000; Lee et al., 2015), while the raf5 and sis8 mutants are hypersensitive to salt stress (Gao and Xiang, 2008). Plasma membrane-localized receptor-like kinases (RLKs) are also essential in sensing and transducing the salt stress signals (Li et al., 2014; Zhao X. et al., 2019; Zhou et al., 2022). The Lectin RLKs (LecRLKs) is reported to play critical roles in mediating plant salt stress and ABA responses (Vaid et al., 2013). The SALT INTOLERANCE 1 (SIT1) encodes a putative LecRLK and mediates salt sensitivity by activating MPK3/6 and promoting ethylene production and ROS accumulation in rice (Li et al., 2014). The salt-induced kinase activity of SIT1 is constrained by PP2A regulatory subunit B’κ-mediated dephosphorylation in its activation loop (Li et al., 2014; Zhao J. et al., 2019). In addition, the phosphorylation of RECEPTOR-LIKE KINASE 7 (RLK7) is enhanced with NaCl treatment, which positively regulates salt stress response by activating downstream MPK3/6 in Arabidopsis (Zhou et al., 2022). Understanding the specific mechanisms of salt stress signal transduction is essential for exploiting the potential of molecular and genetic markers/tools to create/breed salt-resilient crops.
Epigenetic regulation, including DNA methylation, histone modifications, histone variants, and some non-coding RNAs, plays essential roles in regulating plant adaptation to abiotic stress (Kinoshita and Seki, 2014; Chang et al., 2020; Wu et al., 2022). HKT1 is a salt tolerance determinant that mediates Na+ entry and high-affinity K+ uptake in roots (Rus et al., 2001). A putative small RNA target region of the HKT1 promoter is heavily methylated. The RNA-directed DNA Methylation (RdDM) component RDR2 is responsible for the methylation and expression of HKT1, indicating that the RdDM pathway functions in the salt stress response by negatively regulating HKT1 expression (Baek et al., 2011; Kumar et al., 2017). RDM16, another component of the RdDM pathway, regulates the overall methylation of transposable elements and the regions surrounding genes by influencing Pol V transcript levels (Huang et al., 2013). The rdm16 mutant is hypersensitive to salt stress and ABA, pointing to a tight connection between salt stress responses and DNA methylation (Huang et al., 2013).
In addition to DNA methylation, changes in histone modifications are also involved in regulating salt stress tolerance (Kim et al., 2015). Several ABA- and salt stress-responsive genes showed increased histone H3K9K14 acetylation and H3K4 trimethylation but decreased H3K9 dimethylation after ABA or salt treatment, indicating that the expression of stress-responsive genes is associated with changes in histone modifications (Chen et al., 2010). A higher-order mutant of HISTONE DEACETYLASE 6 (HDA6) showed decreased expression of ABA- and abiotic stress-responsive genes, thereby showing a salt-hypersensitive phenotype (Chen et al., 2010). The histone deacetylase HD2C interacts with HDA6 to repress the expression of ABI1 and ABI2 via histone modifications, and the hd2c mutant also showed increased sensitivity to ABA and salt stress (Luo et al., 2012).
The floral initiator SKB1 (SHK1 KINASE BINDING PROTEIN1) perceives salt stress and disassociates with chromatin to decrease H4R3sme2 (symmetric dimethylation of histone4 arginine3) levels to induce the transcription of FLOWERING LOCUS C, thereby regulating flowering time in Arabidopsis under salt stress (Zhang et al., 2011). Both transcriptomic changes and alternative mRNA splicing play vital roles in regulating the salt stress response (Barbazuk et al., 2008; Kornblihtt et al., 2013; Kong et al., 2014). The expression of linker histone variants is dependent on indicated environmental stimuli, for example, H1.S of tomato (Scippa et al., 2004) and HIS1-3 of Arabidopsis (Ascenzi and Gantt, 1997) could be induced by drought stress, and ABA treatment. However, the expression of HIS1-3 could be inhibited by salt stress (Wu et al., 2022). HIS1-3 negatively regulates plant salt stress response through the SOS pathway and the higher-order mutant of HIS1-3 confers plants’ enhanced salt tolerance in Arabidopsis (Wu et al., 2022). Arabidopsis SERRATE (SE) interacts with HYPONASTIC LEAVES 1 (HYL1) and CHR2 to facilitate microRNA biogenesis and fine-tune primary-microRNA processing (Lobbes et al., 2006; Yang et al., 2006; Wang Z. et al., 2018). High salinity represses the expression of SE at both the mRNA and protein levels, and the se mutant is hypersensitive to salt stress (Mou et al., 2021). SE positively regulates plant salt stress tolerance by modulating the pre-mRNA splicing of salt stress-responsive genes (Mou et al., 2021).
A study comparing the small RNA (sRNA) transcriptomes of the mangroves Bruguiera gymnorrhiza and Kandelia candel found that mangroves exhibit distinct sRNA expression patterns and regulatory networks that differ from those of glycophytes, indicating that changes in sRNA expression and sRNA-regulatory networks during evolution are essential for salt stress adaptation in plants (Wen et al., 2016). Salt stress decreases the accumulation of 24-nt siRNAs in Arabidopsis, thereby altering the expression of the transcription factor gene AtMYB74 to help the plant adapt to high salinity conditions (Xu et al., 2015). Indeed, overexpressing AtMYB47 led to hypersensitivity to salt stress during seed germination (Xu et al., 2015).
Phytohormones, including ABA, jasmonic acid (JA), gibberellin (GA), brassinosteroids (BRs), and ethylene are essential for plant growth and development and mediate biochemical and physiological responses to environmental stress (Figure 2), such as osmotic, salt, drought, cold and pathogen stress (Peleg and Blumwald, 2011; Yu et al., 2020). ABA, a 15-carbon sesquiterpenoid, plays important role in regulating plant growth, osmolyte accumulation, stomatal closure, leaf senescence, and root growth under abiotic stress. ABA also acts as an endogenous messenger involved in salt and drought stress signal transduction to initiate downstream gene expression (Finkelstein, 2013; Zhu, 2016).
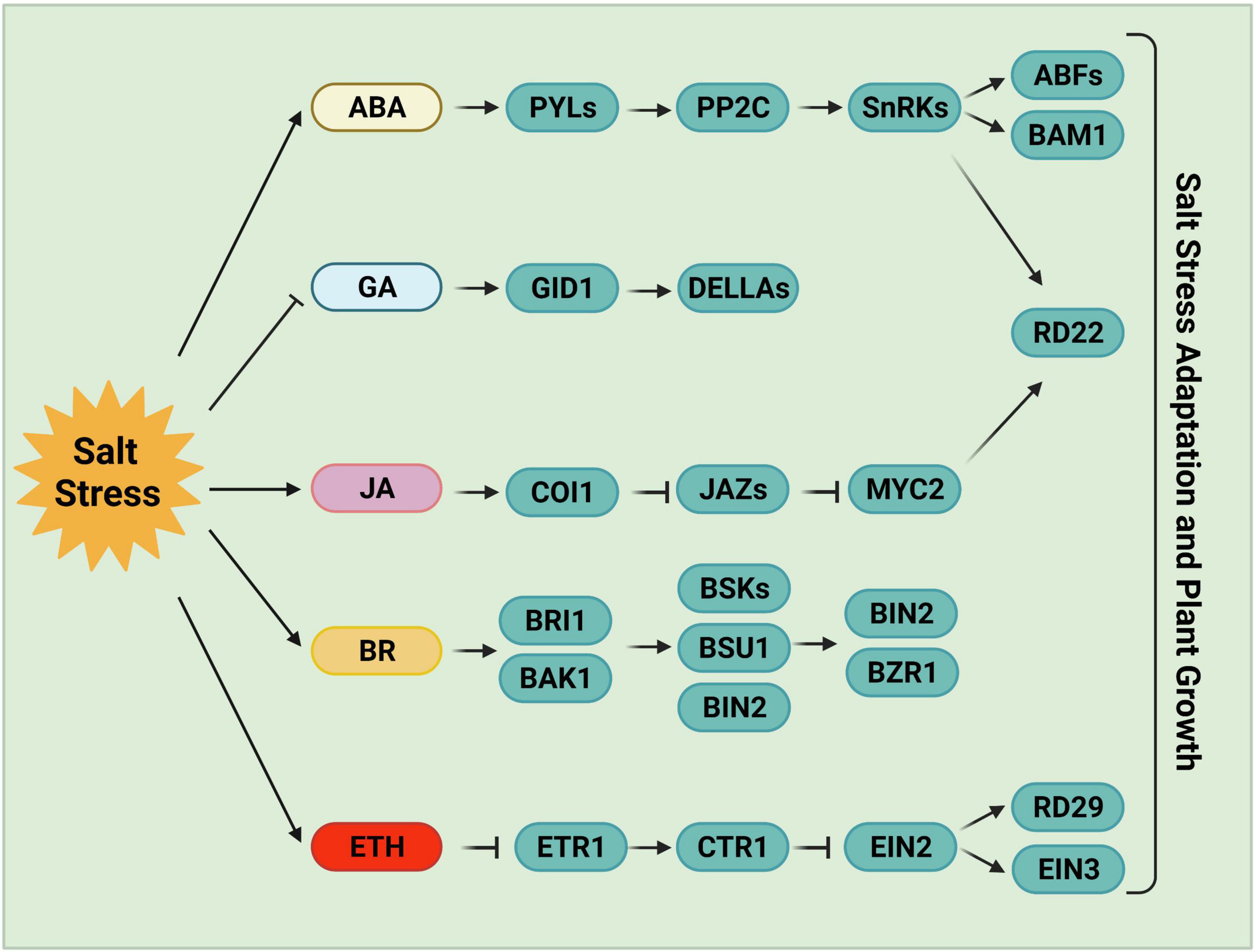
Figure 2. Phytohormone-mediated salt stress responses. ABA, one of the most important stress response hormones, plays a crucial role in salt stress tolerance. ABA-activated SnRK2s regulate stomatal closure, osmotic homeostasis, and gene expression. Salt stress negatively regulates the accumulation of bioactive GAs, and the reduced GAs levels or inactivated GAs promote plant salt tolerance following germination. JA levels increase and JA signaling is activated by high salinity stress. JA is required for the inhibition of primary root growth, which may be an adaptive strategy for survival under salt stress. BR, a growth-promoting phytohormone, accumulates upon salt stress to positively regulate plant salt tolerance. BR induces the formation of the BRI1-BAK1 heterodimer, which then initiates the phosphorylation relay cascades among BSKs, BSU1, and BIN2, ultimately remodeling gene expression via the regulation of BZR1 and BES1. Ethylene also accumulates under salt stress in plants. The components involved in ethylene homeostasis or the ethylene signaling pathway play either positive or negative roles in salt stress responses. ABA, abscisic acid; GA, gibberellin; JA, jasmonic acid; BR, brassinosteroid; ETH, ethylene.
The expression of ABA biosynthetic genes is strongly induced in certain tissues in response to salinity or water deficit, including ABA DEFICIENT genes (ABAs), ALDEHYDE OXIDASE 3, and NINE-CIS-EPOXYCAROTENOID DIOXYGENASE genes (NCEDs) (Barrero et al., 2006; Endo et al., 2008). NCED3, encoding a key enzyme for ABA biosynthesis, is induced in leaves under water-deficient stress (Iuchi et al., 2001; Tan et al., 2003; Endo et al., 2008). NCED5, which encodes a rate-limiting enzyme in ABA biosynthesis, is also rapidly induced under salt stress (Huang et al., 2019). The root-derived CLE25 (CLAVATA3/EMBRYO-SURROUNDING REGION-RELATED 25) protein transmits water-deficiency signals from the vascular system in roots to leaves, where it is sensed by BAM (BARELY ANY MERISTEM), indicating that the CLE-BAM module functions as a long-distance signaling pathway in response to dehydration stress (Takahashi et al., 2018). The application of CLE25 to roots induces NCED3 expression and the accumulation of ABA in leaves, leading to a level of stomatal closure similar to that induced by ABA application. Consistently, the CLE25 CRISPR-Cas9-derived knockout mutant cle25 and the bam1-5 bam3-3 double mutant are hypersensitive to both dehydration and salinity stress (Takahashi et al., 2018).
A very rapid and massive increase in ABA levels is observed in both roots and shoots under stress conditions (Jia et al., 2002; Fricke et al., 2004). This increase is sensed by the PYRABACTIN RESISTANCE 1 (PYR1)/PYR1-like (PYL)/REGULATORY COMPONENT OF ABA RECEPTORS (RCAR) family (PYLs) receptors (Ma et al., 2009; Park et al., 2009; Vlad et al., 2009). Once the PYLs bind to ABA, their conformation changes, allowing them to interact with protein phosphatase 2Cs (PP2Cs) and inhibit their activities, thus releasing SNF1-RELATED PROTEIN KINASE 2s (SnRK2s) from repression (Ma et al., 2009; Park et al., 2009). The released SnRK2s phosphorylate multiple downstream anion efflux channels and transcription factors, leading to decreased turgor pressure, stomatal closure, and gene expression reprogramming (Uno et al., 2000; Mustilli et al., 2002; Furihata et al., 2006; Fujii et al., 2007; Negi et al., 2008; Chen K. et al., 2020; Lin et al., 2020; Takahashi et al., 2020).
All 10 members of the SnRK2 family except SnRK2.9 are activated by osmotic stress (Boudsocq et al., 2004; Zhu, 2016), and the snrk2.1/2/3/4/5/6/7/8/9/10 decuple mutant is hypersensitive to osmotic stress (Fujii et al., 2011). The activation of SnRK2s induced by ABA (but not osmotic stress) is abolished in ABA insensitive 1 (abi1-1) or in higher-order mutants of PYR/PYL/RCAR ABA receptors (Vlad et al., 2010; Zhao Y. et al., 2018). Raf-like kinases (RAFs, especially B2, B3, and B4 RAFs) are required for the phosphorylation and activation of SnRK2s in response to early osmotic stress (Lin et al., 2020). The activated SnRK2s then trans-phosphorylate other SnRK2s to amplify the response (Lin et al., 2021). Higher-order mutants of RAFs are hypersensitive to osmotic stress induced by mannitol, NaCl, or polyethylene glycol treatment (Lin et al., 2020).
Abscisic acid-activated SnRK2s are also involved in regulating starch content in response to salt stress. SnRK2s phosphorylate and activate AREB/ABF transcription factors, which bind to the promoters of β-AMYLASE1 (BAM1) and α-AMYLASE3 (AMY3) and activate their expression (Thalmann et al., 2016). BAM1 and AMY3 mediate the degradation of starch to release sugar and sugar-derived osmolytes (Thalmann et al., 2016). The Arabidopsis amy3 bam1 double mutant is hypersensitive to osmotic and salinity stress (Thalmann et al., 2016). When environmental stress subsides or in the absence of stress conditions, the target of rapamycin (TOR) kinase phosphorylates PYLs to negative regulate their activity, resulting in the disassociation of the PYL-ABA-PP2C complex and the inactivation of ABA and stress signaling, which is sufficient to promote growth recovery (Wang P. et al., 2018). ABI2 is involved in modulating salt tolerance by suppressing the SOS pathway via the ABI2-SOS2 interaction (Ohta et al., 2003).
Emerging evidence points to the coordination between the JA pathway and salt stress signal transduction (Kazan, 2015; Delgado et al., 2021). Analysis of the root transcriptome of sweet potato revealed the upregulation of JA-biosynthesis genes under salt stress; the intensity of upregulation was much greater in a salt-tolerant variety vs. a salt-sensitive line. The JA signaling pathway is also involved in regulating the expression of salt stress-responsive genes. Therefore, the JA signaling pathway is essential for salt tolerance (Ma et al., 2006; Wang J. et al., 2020). A high-order mutant of LIPOXYGENASE3 (LOX3) exhibited salt hypersensitivity, which was rescued by treatment with methyl jasmonate, indicating that JA plays a positive role in salt tolerance (Ding et al., 2016).
The transcription factor MYC2, the regulatory hub of the JA pathway, also plays an essential role in salt tolerance (Abe et al., 2003; Chini et al., 2016; Valenzuela et al., 2016; Zander et al., 2020). MYC2 binds to the promoter of RD22 and activates its expression in response to NaCl or ABA treatment (Iwasaki et al., 1995; Abe et al., 2003). However, the salt stress-mediated activation of the JA signaling pathway inhibits cell elongation in the root elongation zone, and JA-related mutants (aos, col1, jaz3, myc2/3/4) exhibit longer primary roots than wild type plants under salt stress (Valenzuela et al., 2016). Finally, RICE SALT SENSITIVE3 (RSS3) interacts with Class-C bHLH transcription factors and JASMONATE ZIM-DOMAIN (JAZ) proteins (which negatively regulate JA signaling) to promote root cell elongation in rice in response to salt stress (Toda et al., 2013). Overexpressing OsJAZ9 and OsJAZ8 increased plant tolerance to soil salinity (Wu et al., 2015; Peethambaran et al., 2018). Therefore, the JA signaling pathway is required to inhibit root growth under high salinity conditions. Together, these findings suggest that JA has dual functions in plant responses to salt stress.
The phytohormone GA also plays an essential role in regulating plant growth under salt stress (Khan et al., 2012; Colebrook et al., 2014). Several GA-metabolism-related proteins positively regulate the salt stress response. Conversely, salt stress usually reduces bioactive GA levels and increases the accumulation of DELLA proteins, resulting in dwarfism and enhanced stress tolerance (Achard et al., 2006; Qin et al., 2011). For instance, microarray data indicate that several genes encoding GA deactivation enzymes are upregulated by high salinity, including GA2-oxidase 1 (GA2ox1), GA2ox2, GA2ox4, GA2ox6, and GA2ox8 (Kilian et al., 2007; Magome et al., 2008). GA2ox7, encoding a C20-GA deactivation enzyme, is also strongly induced by salt stress. A higher-order GA2ox mutant showed higher sensitivity to salt stress than the wild type (Magome et al., 2008). Consistently, a quadruple-DELLA mutant lacking GAI, RGA, RGL1, and RGL2 (Cheng et al., 2004) shows less salt-triggered inhibition of root growth and flowering than the wild type but is hypersensitive to salt-induced death (Achard et al., 2006), indicating that DELLA proteins play a central role in the trade-off between growth limitation and survival under salt stress.
Brassinosteroids are plant growth-promoting steroid hormones that play critical roles in plant growth, development, and stress responses (Nolan et al., 2019; Planas-Riverola et al., 2019). BR signaling is essential for plant salt tolerance (Cui et al., 2012; Zhao X. et al., 2019). BRs such as 24-epibrassinolide (eBL) binds to the receptor BR INSENSITIVE1 (BRI1) (Friedrichsen et al., 2000; He et al., 2000) or its homologs BRI1-LIKEs (BRLs) (Caño-Delgado et al., 2004; Kinoshita et al., 2005) and the coreceptor BAK1 to initiate the BR signaling pathway, leading to a phosphorylation relay cascade among BAK1, BRASSINOSTEROID-SIGNALING KINASEs (BSKs), BRI1 SUPPRESSOR 1 (BSU1) and BRASSINOSTEROID-INSENSITIVE 2 (BIN2) (Li and Nam, 2002; Russinova et al., 2004; Hohmann et al., 2018). BIN2 activity is inhibited by the activated BSU1, thereby promoting BR-induced gene expression and inhibiting BR-repressed gene expression via the regulation of the transcription factors BZR1, BES1, and other transcription factors or cofactors (Nolan et al., 2019; Planas-Riverola et al., 2019).
Under high salinity, the activity of the ethylene biosynthesis enzyme ACS (1-aminocyclopropane-1-carboxylate synthase) is enhanced by BR pretreatment, resulting in ethylene accumulation and better adaptation to salt stress (Tao et al., 2015; Zhu et al., 2016). Conversely, shutting down ethylene production represses BR-induced antioxidant enzyme activity and decreases salt tolerance (Tao et al., 2015; Zhu et al., 2016). High salinity strongly inhibits roots growth due to reduced accumulation of BZR1 in the nucleus and the repression of BR signaling (Geng et al., 2013). However, exogenous BR application partially rescued salt-induced growth inhibition (Zeng et al., 2010; Liu et al., 2014; Zhu et al., 2016). OsSERK2 localizes to the plasma membrane in rice and interacts with the BR receptor OsBRI1 to facilitate BR signaling. Notably, the CRISPR/Cas9 edited osserk2 mutant is impaired in BR signaling and shows hypersensitivity to salt stress (Dong et al., 2020). Finally, BIN2 is a negative regulator of the SOS pathway, which functions partially independently of the BR signaling pathway to balance plant growth and stress responses (Li et al., 2020). All of these findings demonstrate that the sensing and signaling of phytohormone pathways contribute to salt stress tolerance.
The plant cell wall consists of cellulose, pectins, hemicellulose, and various glycoproteins that help modulate cell wall extensibility. This property determines cell shape and size via the mechanical control of cell enlargement and expansion, thereby governing tissue and organ morphology (Cosgrove, 2005; Le Gall et al., 2015). Accumulating evidence demonstrates the important roles of the cell wall in plant responses to abiotic stress (Le Gall et al., 2015; Reviewed in Endler et al., 2015). The cell wall is a sensor of salt stress, and salt stress perception-to-signaling cascades mainly function at the cell wall-plasma membrane interface (Kacperska, 2004; Hou et al., 2005; Feng et al., 2018; Zhao C. et al., 2018). The nuclear-localized Agenet domain-containing protein SWO1 (SWOLLEN 1) functions together with importin α IMPA1 and IMPA2 to maintain cell integrity in Arabidopsis under salt stress (Wang et al., 2021). Several receptor-like protein kinases (RLKs) are regarded as sensors that perceive salt stress signals, such as the wall-associated kinases (WAKs) and FER (Baluska et al., 2005; Hou et al., 2005; Kohorn and Kohorn, 2012; Feng et al., 2018). WAK-LIKE KINASE 4 (WAKL4) but not WAK1 is highly induced under high salinity conditions (Hou et al., 2005), suggesting that WAKs play different roles in response to different stress conditions. The receptor-like kinase FER binds to RALF, senses salt-induced cell wall damage, and transduces the signals to downstream targets to maintain cell wall integrity under salt stress (Feng et al., 2018). The cell wall leucine-rich repeat extensins (LRX) 3/4/5 function together with the FER-RALF module to transduce cell wall signals to mediate plant growth and salt tolerance (Zhao C. et al., 2018).
In addition to sensing salt stress signals, the cell wall also plays a significant role in protecting the cell from salt stress-induced ionic toxicity. Several mutants with defective cell wall integrity are hypersensitive to salt. For instance, mutants of SOS6, encoding the cellulose synthase-like protein AtCSLD5, show reduced levels of arabinose, rhamnose, and galacturonic acid, which normally confer salt/osmotic stress tolerance. Thus, the sos6 mutant exhibits elevated ROS contents and a salt-hypersensitive phenotype under salt stress (Zhu et al., 2010). Membrane-localized cellulose synthase (CesA) complexes are required to synthesize cellulose, the main component of the cell wall (McFarlane et al., 2014). CESA1, CESA3, and CESA6 are required for primary cell wall synthesis (Arioli et al., 1998; Fagard et al., 2000), while CESA4, CESA6, and CESA8 are required for secondary cell wall synthesis (Taylor et al., 2003; Endler and Persson, 2011). The sustained cellulose synthesis conferred by CESA1, CESA6, and CELLULOSE SYNTHASE-INTERACTIVE PROTEIN 1 (CSI1) is important for salt tolerance in Arabidopsis (Kang et al., 2008; Gu et al., 2010; Li et al., 2012; Zhang et al., 2016), as knocking out either CESA1, CESA6 or CSI1 conferred increased sensitivity to salt stress. Two plant-specific components of the cellulose synthase complex, CC1 (COMPANION OF CELLULOSE SYNTHASE 1) and CC2 interact with CesA proteins and microtubules to promote the CesA activity and microtubule dynamics required for hypocotyl growth under salt stress (Endler et al., 2015).
The endoplasmic reticulum (ER) is the main site for the modification or folding of secretory and membrane proteins to achieve their native structures. In addition, the ER is a fundamental organelle involved in signal transduction that allows plants to adapt to diverse environmental stresses (Vitale and Boston, 2008; Liu and Howell, 2010; Jin and Daniell, 2014). The accumulation of unfolded or misfolded proteins is induced by various biotic and abiotic stresses, ER stress (Liu et al., 2007; Ye et al., 2011; Zhang and Wang, 2012). The cell has developed several important strategies to alleviate ER stress, including the accelerated degradation of misfolded proteins through the ER-associated degradation (ERAD) pathway (Vembar and Brodsky, 2008; Chen Q. et al., 2020). Numerous misfolded proteins accumulate in the ER under salt stress, which is recognized by the ERAD pathway and ubiquitinated and degraded through the ubiquitin/26S proteasome system (Liu et al., 2011), indicating that the ERAD pathway is essential for plant survival under high salinity conditions.
Plants also alleviate salt stress by activating the expression of ER chaperones. HRD3A, the functional homolog of the yeast HRD1/HRD3 complex, is an active component of the ERAD complex in Arabidopsis. The hrd3a mutant exhibits an enhanced unfolded protein response under ER stress and increased sensitivity to salt stress (Liu et al., 2011). Accumulating evidence indicates that the ERAD complex plays a positive role in the salt stress response. For example, the ERAD complex mutants mns4/mns5, atos9, and hrd1a/hrd1b are hypersensitive to salt stress (Hüttner et al., 2012, 2014; Su et al., 2012). In addition, a defect in UBC32, a salt stress-induced functional ubiquitin conjugation enzyme (E2) required for the activation of the ERAD complex in Arabidopsis, confers increased tolerance to salt stress via a BR-dependent pathway (Cui et al., 2012). SES1 (SENSITIVE TO SALT 1), an ER-localized chaperone, protects plants from salt stress by alleviating salt-induced ER stress. The salt-sensitive phenotype of ses1 is due to the over-activation of the unfolded protein response. In addition, the ER stress sensor bZIP17 directly binds to the promoter of SES1 to activate its transcription under salt stress (Guan et al., 2018). Finally, a recent study demonstrated that the Arabidopsis receptor-like kinase SIMP1 (SALT-INDUCED MALECTIN-LIKE DOMAIN-CONTAINING PROTEIN 1) positively modulates plant salt tolerance by interacting with and stabilizing the putative proteasome maturation factor UMP1A, thus leading to enhanced proteasome maturation and ERAD efficiency, resulting in the mitigation of salt stress-induced ER stress (He et al., 2021).
Accumulating evidence points to a tight connection between the salt signaling pathway and other signal transduction pathways. For instance, light signaling plays a vital role in shaping the salt stress response. The localization of CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) is tightly controlled by light signals (von Arnim and Deng, 1994), whereas salt treatment promotes the translocation of COP1 to the cytosol (Yu et al., 2016). Light signaling is involved in regulating the salt-induced transcriptional memory response of P5CS1 to proline (Feng et al., 2016). In addition, PHYTOCHROME-INTERACTING FACTOR4 (PIF4), a negative regulator of photomorphogenesis, negatively regulates plant salt tolerance by directly modulating the expression of diverse salt-responsive genes (Leivar and Quail, 2011; Lee and Choi, 2017; Sakuraba et al., 2017). Moreover, the regulation of the salinity stress response depends on diurnal cycles and the circadian clock via modulating the expression of RD29A and SOS1. The abundance of SOS1 protein also appears to occur in a diurnal cycle (Park et al., 2016). Finally, low levels of NaCl in the soil severely inhibit shade-induced hypocotyl elongation via the BR and ABA signaling pathways (Hayes et al., 2019).
Genetic and molecular studies have implicated pattern recognition receptors (PRRs) in salt stress tolerance. Arabidopsis PROPEP3 functions in the plant immune system (Huffaker et al., 2006) and positively regulates the salt stress response (Nakaminami et al., 2018). PROPEP3 is highly induced under salinity stress conditions, and both PROPEP3 overexpression and Pep3 application enhance salt stress tolerance via PEP-RECEPTOR l (Nakaminami et al., 2018). Similarly, PAMP-INDUCED SECRETED PEPTIDE 3 (PIP3) functions together with RLK7, a leucine-rich repeat receptor-like kinase (LRR-RLK), to further activate MPK3/6, therefore conferring salt tolerance (Zhou et al., 2022). All of these findings suggest that the sensing and signaling of damage-associated molecular patterns contribute to salt stress tolerance.
Soil salinity severely threatens plant growth, crop productivity, and food security worldwide (Yang and Guo, 2018a,b; van Zelm et al., 2020). Identifying and characterizing the determinants and regulatory mechanisms of salt stress signaling represents the most effective way to breed salt-tolerant crops and improve agricultural development. In the last two decades, many new advances have been made in elucidating the key components of the salt stress response. In addition, several genetic loci involved in salt tolerance have been identified and the underlying genes cloned, representing candidate targets for designing the next generation of crop varieties. Studies of the SOS pathway have clearly demonstrated the mechanism of sodium ion (Na+) efflux and ion homeostasis. However, continuous efforts leading to substantial new discoveries are needed to better understand and further improve salt tolerance in plants.
Although GIPCs were characterized as potential monovalent-cation sensors (Jiang et al., 2019), the identification of other sodium sensors or receptors is still the most important goal of plant salt stress signaling research. As salt stress severely inhibits or destroys chloroplast development and photosynthesis, it is important to investigate whether salt sensors are present in different organelles, such as the chloroplast, ER, and vacuole. In addition, salt stress induces rapid calcium signaling in the cytosol (Knight et al., 1997; Zhu et al., 2016; Ma et al., 2019), and the long-distance transmission of Ca2+ waves from root to shoot is channeled through the cortex and endodermal cell layers, which is dependent on the vacuolar ion channel TPC1 (Choi et al., 2014). Therefore, it is critical to understand how the local salt stress signal in roots is sensed by different tissues and how plants integrate tissue-specific signals to confer stress tolerance throughout the plant.
The evolutionarily conserved SOS pathway primarily mediates Na+ homeostasis through the activation of the SOS3/SCaBP8-SOS2-SOS1 module under high salinity stress. Higher-order mutants of SOS2 and SOS1 exhibit severely reduced primary and lateral root growth under salt stress. As root system architecture is not only shaped by salt stress signals but also by plant nutrient status, whether and how the SOS pathway integrates different signal pathways (such as nutrient signaling and salt stress signaling) to balance plant root growth and stress tolerance needs to be explored in the future.
LM wrote the manuscript. XL and WL participated in writing and modification of the manuscript. LM and YY edited the manuscript. All authors have read and agreed to the published version of the manuscript.
This work was sponsored/funded by the China Postdoctoral Science Foundation (2019M660865).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We apologize to colleagues whose work could not be cited due to space limitations.
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. doi: 10.1126/science.1118642
Alzwiy, I. A., and Morris, P. C. (2007). A mutation in the Arabidopsis MAP kinase kinase 9 gene results in enhanced seedling stress tolerance. Plant Sci. 173, 302–308.
Arioli, T., Peng, L., Betzner, A. S., Burn, J., Wittke, W., Herth, W., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720.
Ascenzi, R., and Gantt, J. S. (1997). A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol. Biol. 34, 629–641. doi: 10.1023/a:1005886011722
Baek, D., Jiang, J., Chung, J. S., Wang, B., Chen, J., Xin, Z., et al. (2011). Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol. 52, 149–161. doi: 10.1093/pcp/pcq182
Baluska, F., Liners, F., Hlavacka, A., Schlicht, M., Van Cutsem, P., McCurdy, D. W., et al. (2005). Cell wall pectins and xyloglucans are internalized into dividing root cells and accumulate within cell plates during cytokinesis. Protoplasma 225, 141–155. doi: 10.1007/s00709-005-0095-5
Barajas-Lopez, J. D., Moreno, J. R., Gamez-Arjona, F. M., Pardo, J. M., Punkkinen, M., Zhu, J. K., et al. (2018). Upstream kinases of plant SnRKs are involved in salt stress tolerance. Plant J. 93, 107–118. doi: 10.1111/tpj.13761
Barbazuk, W. B., Fu, Y., and McGinnis, K. M. (2008). Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 18, 1381–1392. doi: 10.1101/gr.053678.106
Barrero, J. M., Rodríguez, P. L., Quesada, V., Piqueras, P., Ponce, M. R., and Micol, J. L. (2006). Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 29, 2000–2008. doi: 10.1111/j.1365-3040.2006.01576.x
Basu, D., and Haswell, E. S. (2020). The mechanosensitive ion channel MSL10 potentiates responses to cell swelling in Arabidopsis seedlings. Curr. Biol. 30, 2716–2728.e2716. doi: 10.1016/j.cub.2020.05.015
Beaudoin, N., Serizet, C., Gosti, F., and Giraudat, J. (2000). Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12, 1103–1116. doi: 10.1105/tpc.12.7.1103
Boudsocq, M., Barbier-Brygoo, H., and Laurière, C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279, 41758–41766. doi: 10.1074/jbc.M405259200
Caño-Delgado, A., Yin, Y., Yu, C., Vafeados, D., Mora-García, S., Cheng, J. C., et al. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131, 5341–5351. doi: 10.1242/dev.01403
Cao, Y., Zhang, M., Liang, X., Li, F., Shi, Y., Yang, X., et al. (2020). Natural variation of an EF-hand Ca2+-binding-protein coding gene confers saline-alkaline tolerance in maize. Nat. Commun. 11:186. doi: 10.1038/s41467-019-14027-y
Chai, H., Guo, J., Zhong, Y., and Hsu, C. C. (2020). The plasma-membrane polyamine transporter PUT3 is regulated by the Na+/H+ antiporter SOS1 and protein kinase SOS2. New Phytol. 226, 785–797. doi: 10.1111/nph.16407
Chang, Y. N., Zhu, C., Jiang, J., Zhang, H., Zhu, J. K., and Duan, C. G. (2020). Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 62, 563–580. doi: 10.1111/jipb.12901
Chen, K., Li, G. J., Bressan, R. A., Song, C. P., Zhu, J. K., and Zhao, Y. (2020). Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 62, 25–54. doi: 10.1111/jipb.12899
Chen, L. T., Luo, M., Wang, Y. Y., and Wu, K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. doi: 10.1093/jxb/erq154
Chen, Q., Yu, F., and Xie, Q. (2020). Insights into endoplasmic reticulum-associated degradation in plants. New Phytol. 226, 345–350. doi: 10.1111/nph.16369
Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D. E., Cao, D., et al. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064. doi: 10.1242/dev.00992
Chini, A., Gimenez-Ibanez, S., Goossens, A., and Solano, R. (2016). Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 33, 147–156. doi: 10.1016/j.pbi.2016.07.005
Choi, W. G., Toyota, M., Kim, S. H., Hilleary, R., and Gilroy, S. (2014). Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 111, 6497–6502. doi: 10.1073/pnas.1319955111
Colebrook, E. H., Thomas, S. G., Phillips, A. L., and Hedden, P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217, 67–75. doi: 10.1242/jeb.089938
Cui, F., Liu, L., Zhao, Q., Zhang, Z., Li, Q., Lin, B., et al. (2012). Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24, 233–244. doi: 10.1105/tpc.111.093062
Delgado, C., Mora-Poblete, F., Ahmar, S., Chen, J. T., and Figueroa, C. R. (2021). Jasmonates and plant salt stress: molecular players, physiological effects, and improving tolerance by using genome-associated tools. Int. J. Mol. Sci. 22:3082. doi: 10.3390/ijms22063082
Denness, L., McKenna, J. F., Segonzac, C., Wormit, A., Madhou, P., Bennett, M., et al. (2011). Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 156, 1364–1374. doi: 10.1104/pp.111.175737
Ding, H., Lai, J., Wu, Q., Zhang, S., Chen, L., Dai, Y. S., et al. (2016). Jasmonate complements the function of Arabidopsis lipoxygenase3 in salinity stress response. Plant Sci. 244, 1–7. doi: 10.1016/j.plantsci.2015.11.009
Dodd, A. N., Kudla, J., and Sanders, D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620. doi: 10.1146/annurev-arplant-070109-104628
Dong, N., Yin, W., Liu, D., Zhang, X., Yu, Z., Huang, W., et al. (2020). Regulation of brassinosteroid signaling and salt resistance by SERK2 and potential utilization for crop improvement in rice. Front. Plant Sci. 11:621859. doi: 10.3389/fpls.2020.621859
Duscha, K., Martins Rodrigues, C., Müller, M., Wartenberg, R., Fliegel, L., Deitmer, J. W., et al. (2020). 14-3-3 Proteins and other candidates form protein-protein interactions with the cytosolic C-terminal end of SOS1 affecting its transport activity. Int. J. Mol. Sci. 21:3334. doi: 10.3390/ijms21093334
El Mahi, H., Pérez-Hormaeche, J., De Luca, A., Villalta, I., Espartero, J., Gámez-Arjona, F., et al. (2019). A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 180, 1046–1065. doi: 10.1104/pp.19.00324
Endler, A., Kesten, C., Schneider, R., Zhang, Y., Ivakov, A., Froehlich, A., et al. (2015). A mechanism for sustained cellulose synthesis during salt stress. Cell 162, 1353–1364. doi: 10.1016/j.cell.2015.08.028
Endler, A., and Persson, S. (2011). Cellulose synthases and synthesis in Arabidopsis. Mol. Plant 4, 199–211.
Endo, A., Sawada, Y., Takahashi, H., Okamoto, M., Ikegami, K., Koiwai, H., et al. (2008). Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 147, 1984–1993. doi: 10.1104/pp.108.116632
Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., et al. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12, 2409–2424. doi: 10.1105/tpc.12.12.2409
Feng, X. J., Li, J. R., Qi, S. L., Lin, Q. F., Jin, J. B., and Hua, X. J. (2016). Light affects salt stress-induced transcriptional memory of P5CS1 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, E8335–E8343. doi: 10.1073/pnas.1610670114
Feng, W., Kita, D., Peaucelle, A., Cartwright, H. N., Doan, V., Duan, Q., et al. (2018). The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 28, 666–675.e665. doi: 10.1016/j.cub.2018.01.023
Flowers, T. J., and Colmer, T. D. (2015). Plant salt tolerance: adaptations in halophytes. Ann. Bot. 115, 327–331. doi: 10.1093/aob/mcu267
Fricke, W., Akhiyarova, G., Veselov, D., and Kudoyarova, G. (2004). Rapid and tissue-specific changes in ABA and in growth rate in response to salinity in barley leaves. J. Exp. Bot. 55, 1115–1123. doi: 10.1093/jxb/erh117
Friedrichsen, D. M., Joazeiro, C. A., Li, J., Hunter, T., and Chory, J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123, 1247–1256. doi: 10.1104/pp.123.4.1247
Fu, H., Subramanian, R. R., and Masters, S. C. (2000). 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40, 617–647. doi: 10.1146/annurev.pharmtox.40.1.617
Fuglsang, A. T., Guo, Y., Cuin, T. A., Qiu, Q., Song, C., Kristiansen, K. A., et al. (2007). Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19, 1617–1634. doi: 10.1105/tpc.105.035626
Fujii, H., Verslues, P. E., and Zhu, J. K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19, 485–494. doi: 10.1105/tpc.106.048538
Fujii, H., Verslues, P. E., and Zhu, J. K. (2011). Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 1717–1722. doi: 10.1073/pnas.1018367108
Fujita, M., Fujita, Y., Iuchi, S., Yamada, K., Kobayashi, Y., Urano, K., et al. (2012). Natural variation in a polyamine transporter determines paraquat tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 6343–6347. doi: 10.1073/pnas.1121406109
Fujita, M., and Shinozaki, K. (2014). Identification of polyamine transporters in plants: paraquat transport provides crucial clues. Plant Cell Physiol. 55, 855–861. doi: 10.1093/pcp/pcu032
Furihata, T., Maruyama, K., Fujita, Y., Umezawa, T., Yoshida, R., Shinozaki, K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 103, 1988–1993. doi: 10.1073/pnas.0505667103
Gao, L., and Xiang, C. B. (2008). The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in Arabidopsis. Plant. Mol. Biol. 67, 125–134. doi: 10.1007/s11103-008-9306-8
Geng, Y., Wu, R., Wee, C. W., Xie, F., Wei, X., Chan, P. M., et al. (2013). A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 25, 2132–2154. doi: 10.1105/tpc.113.112896
Gu, Y., Kaplinsky, N., Bringmann, M., Cobb, A., Carroll, A., Sampathkumar, A., et al. (2010). Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 12866–12871. doi: 10.1073/pnas.1007092107
Guan, P., Wang, J., Li, H., Xie, C., Zhang, S., Wu, C., et al. (2018). SENSITIVE TO SALT1, an endoplasmic reticulum-localized chaperone, positively regulates salt resistance. Plant Physiol. 178, 1390–1405. doi: 10.1104/pp.18.00840
Guo, Y., Halfter, U., Ishitani, M., and Zhu, J. K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13, 1383–1400. doi: 10.1105/tpc.13.6.1383
Halfter, U., Ishitani, M., and Zhu, J. K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. U.S.A. 97, 3735–3740. doi: 10.1073/pnas.97.7.3735
Hamilton, E. S., Jensen, G. S., Maksaev, G., Katims, A., Sherp, A. M., and Haswell, E. S. (2015a). Mechanosensitive channel MSL8 regulates osmotic forces during pollen hydration and germination. Science 350, 438–441. doi: 10.1126/science.aac6014
Hamilton, E. S., Schlegel, A. M., and Haswell, E. S. (2015b). United in diversity: mechanosensitive ion channels in plants. Annu. Rev. Plant Biol. 66, 113–137. doi: 10.1146/annurev-arplant-043014-114700
Han, X., Yang, Y., Wu, Y., Liu, X., Lei, X., and Guo, Y. (2017). A bioassay-guided fractionation system to identify endogenous small molecules that activate plasma membrane H+-ATPase activity in Arabidopsis. J. Exp. Bot. 68, 2951–2962. doi: 10.1093/jxb/erx156
Hasegawa, P. M., Bressan, R. A., Zhu, J. K., and Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. doi: 10.1146/annurev.arplant.51.1.463
Hashimoto, K., and Kudla, J. (2011). Calcium decoding mechanisms in plants. Biochimie 93, 2054–2059. doi: 10.1016/j.biochi.2011.05.019
Haswell, E. S., and Meyerowitz, E. M. (2006). MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr. Biol. 16, 1–11. doi: 10.1016/j.cub.2005.11.044
Hayes, S., Pantazopoulou, C. K., van Gelderen, K., Reinen, E., Tween, A. L., Sharma, A., et al. (2019). Soil salinity limits plant shade avoidance. Curr. Biol. 29, 1669–1676.e1664. doi: 10.1016/j.cub.2019.03.042
He, J., Zhuang, Y., Li, C., Sun, X., Zhao, S., Ma, C., et al. (2021). SIMP1 modulates salt tolerance by elevating ERAD efficiency through UMP1A-mediated proteasome maturation in plants. New Phytol. 232, 625–641. doi: 10.1111/nph.17628
He, Z., Wang, Z. Y., Li, J., Zhu, Q., Lamb, C., Ronald, P., et al. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288, 2360–2363. doi: 10.1126/science.288.5475.2360
Hohmann, U., Santiago, J., Nicolet, J., Olsson, V., Spiga, F. M., Hothorn, L. A., et al. (2018). Mechanistic basis for the activation of plant membrane receptor kinases by SERK-family coreceptors. Proc. Natl. Acad. Sci. U.S.A. 115, 3488–3493. doi: 10.1073/pnas.1714972115
Hou, X., Tong, H., Selby, J., Dewitt, J., Peng, X., and He, Z. H. (2005). Involvement of a cell wall-associated kinase, WAKL4, in Arabidopsis mineral responses. Plant Physiol. 139, 1704–1716. doi: 10.1104/pp.105.066910
Huang, C. F., Miki, D., Tang, K., Zhou, H. R., Zheng, Z., Chen, W., et al. (2013). A pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. PLoS Genet. 9:e1003779. doi: 10.1371/journal.pgen.1003779
Huang, Y., Jiao, Y., Xie, N., Guo, Y., Zhang, F., Xiang, Z., et al. (2019). OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 287, 110188. doi: 10.1016/j.plantsci.2019.110188
Huffaker, A., Pearce, G., and Ryan, C. A. (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 103, 10098–10103. doi: 10.1073/pnas.0603727103
Hüttner, S., Veit, C., Schoberer, J., Grass, J., and Strasser, R. (2012). Unraveling the function of Arabidopsis thaliana OS9 in the endoplasmic reticulum-associated degradation of glycoproteins. Plant Mol. Biol. 79, 21–33. doi: 10.1007/s11103-012-9891-4
Hüttner, S., Veit, C., Vavra, U., Schoberer, J., Liebminger, E., Maresch, D., et al. (2014). Arabidopsis class I α-mannosidases MNS4 and MNS5 are involved in endoplasmic reticulum-associated degradation of misfolded glycoproteins. Plant Cell 26, 1712–1728. doi: 10.1105/tpc.114.123216
Ichimura, K., Shinozaki, K., Tena, G., Sheen, J., Henry, Y., et al. (2002). Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7, 301–308. doi: 10.1016/s1360-1385(02)02302-6
Iuchi, S., Kobayashi, M., Taji, T., Naramoto, M., Seki, M., Kato, T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333. doi: 10.1046/j.1365-313x.2001.01096.x
Iwasaki, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1995). Identification of a cis-regulatory region of a gene in Arabidopsis thaliana whose induction by dehydration is mediated by abscisic acid and requires protein synthesis. Mol. Gen. Genet. 247, 391–398. doi: 10.1007/BF00293139
Jammes, F., Song, C., Shin, D., Munemasa, S., Takeda, K., Gu, D., et al. (2009). MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 20520–20525. doi: 10.1073/pnas.0907205106
Jia, W., Wang, Y., Zhang, S., and Zhang, J. (2002). Salt-stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J. Exp. Bot. 53, 2201–2206. doi: 10.1093/jxb/erf079
Jiang, Z., Zhou, X., Tao, M., Yuan, F., Liu, L., Wu, F., et al. (2019). Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572, 341–346. doi: 10.1038/s41586-019-1449-z
Jin, S., and Daniell, H. (2014). Expression of γ-tocopherol methyltransferase in chloroplasts results in massive proliferation of the inner envelope membrane and decreases susceptibility to salt and metal-induced oxidative stresses by reducing reactive oxygen species. Plant Biotechnol. J. 12, 1274–1285. doi: 10.1111/pbi.12224
Kacperska, A. (2004). Sensor types in signal transduction pathways in plant cells responding to abiotic stressors: do they depend on stress intensity? Physiol. Plant. 122, 159–168.
Kang, J. S., Frank, J., Kang, C. H., Kajiura, H., Vikram, M., Ueda, A., et al. (2008). Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. U.S.A. 105, 5933–5938. doi: 10.1073/pnas.0800237105
Kazan, K. (2015). Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20, 219–229. doi: 10.1016/j.tplants.2015.02.001
Khan, A. L., Hamayun, M., Kang, S. M., Kim, Y. H., Jung, H. Y., Lee, J. H., et al. (2012). Endophytic fungal association via gibberellins and indole acetic acid can improve plant growth under abiotic stress: an example of Paecilomyces formosus LHL10. BMC Microbiol. 12:3. doi: 10.1186/1471-2180-12-3
Kiegle, E., Moore, C. A., Haseloff, J., Tester, M. A., and Knight, M. R. (2000). Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 23, 267–278. doi: 10.1046/j.1365-313x.2000.00786.x
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363. doi: 10.1111/j.1365-313X.2007.03052.x
Kim, J. M., Sasaki, T., Ueda, M., Sako, K., and Seki, M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6:114. doi: 10.3389/fpls.2015.00114
Kim, W. Y., Ali, Z., Park, H. J., Park, S. J., Cha, J. Y., Perez-Hormaeche, J., et al. (2013). Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 4:1352. doi: 10.1038/ncomms2357
Kinoshita, T., Caño-Delgado, A., Seto, H., Hiranuma, S., Fujioka, S., Yoshida, S., et al. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433, 167–171. doi: 10.1038/nature03227
Kinoshita, T., and Seki, M. (2014). Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 55, 1859–1863. doi: 10.1093/pcp/pcu125
Knight, H., Trewavas, A. J., and Knight, M. R. (1997). Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 12, 1067–1078. doi: 10.1046/j.1365-313x.1997.12051067.x
Kohorn, B. D., and Kohorn, S. L. (2012). The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 3:88. doi: 10.3389/fpls.2012.00088
Kong, X., Ma, L., Yang, L., Chen, Q., Xiang, N., Yang, Y., et al. (2014). Quantitative proteomics analysis reveals that the nuclear cap-binding complex proteins Arabidopsis CBP20 and CBP80 modulate the salt stress response. J. Proteome Res. 13, 2495–2510. doi: 10.1021/pr4012624
Kornblihtt, A. R., Schor, I. E., Alló, M., Dujardin, G., Petrillo, E., and Muñoz, M. J. (2013). Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 14, 153–165. doi: 10.1038/nrm3525
Kumar, M. N., Jane, W. N., and Verslues, P. E. (2013). Role of the putative osmosensor Arabidopsis histidine kinase1 in dehydration avoidance and low-water-potential response. Plant Physiol. 161, 942–953. doi: 10.1104/pp.112.209791
Kumar, S., Beena, A. S., Awana, M., and Singh, A. (2017). Salt-induced tissue-specific cytosine methylation downregulates expression of HKT genes in contrasting wheat (Triticum aestivum L.) genotypes. DNA Cell Biol. 36, 283–294. doi: 10.1089/dna.2016.3505
Kunz, H. H., Gierth, M., Herdean, A., Satoh-Cruz, M., Kramer, D. M., Spetea, C., et al. (2014). Plastidial transporters KEA1, –2, and –3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 7480–7485. doi: 10.1073/pnas.1323899111
Kurusu, T., Kuchitsu, K., Nakano, M., Nakayama, Y., and Iida, H. (2013). Plant mechanosensing and Ca2+ transport. Trends Plant Sci. 18, 227–233. doi: 10.1016/j.tplants.2012.12.002
Kurusu, T., Nishikawa, D., Yamazaki, Y., Gotoh, M., Nakano, M., Hamada, H., et al. (2012a). Plasma membrane protein OsMCA1 is involved in regulation of hypo-osmotic shock-induced Ca2+ influx and modulates generation of reactive oxygen species in cultured rice cells. BMC Plant Biol. 12:11. doi: 10.1186/1471-2229-12-11
Kurusu, T., Yamanaka, T., Nakano, M., Takiguchi, A., Ogasawara, Y., Hayashi, T., et al. (2012b). Involvement of the putative Ca2+-permeable mechanosensitive channels, NtMCA1 and NtMCA2, in Ca2+ uptake, Ca2+-dependent cell proliferation and mechanical stress-induced gene expression in tobacco (Nicotiana tabacum) BY-2 cells. J. Plant Res. 125, 555–568. doi: 10.1007/s10265-011-0462-6
Laohavisit, A., Richards, S. L., Shabala, L., Chen, C., Colaco, R. D., Swarbreck, S. M., et al. (2013). Salinity-induced calcium signaling and root adaptation in Arabidopsis require the calcium regulatory protein annexin1. Plant Physiol. 163, 253–262. doi: 10.1104/pp.113.217810
Le Gall, H., Philippe, F., Domon, J. M., Gillet, F., Pelloux, J., and Rayon, C. (2015). Cell wall metabolism in response to abiotic stress. Plants 4, 112–166. doi: 10.3390/plants4010112
Lee, N., and Choi, G. (2017). Phytochrome-interacting factor from Arabidopsis to liverwort. Curr. Opin. Plant Biol. 35, 54–60. doi: 10.1016/j.pbi.2016.11.004
Lee, S. J., Lee, M. H., Kim, J. I., and Kim, S. Y. (2015). Arabidopsis putative MAP kinase kinase kinases Raf10 and Raf11 are positive regulators of seed dormancy and ABA response. Plant Cell Physiol. 56, 84–97. doi: 10.1093/pcp/pcu148
Lee, S. K., Kim, B. G., Kwon, T. R., Jeong, M. J., Park, S. R., Lee, J. W., et al. (2011). Overexpression of the mitogen-activated protein kinase gene OsMAPK33 enhances sensitivity to salt stress in rice (Oryza sativa L.). J. Biosci. 36, 139–151. doi: 10.1007/s12038-011-9002-8
Leivar, P., and Quail, P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. doi: 10.1016/j.tplants.2010.08.003
Li, C., Wang, G., Zhao, J., Zhang, L., Ai, L., Han, Y., et al. (2014). The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 26, 2538–2553. doi: 10.1105/tpc.114.125187
Li, J., and Nam, K. H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295, 1299–1301. doi: 10.1126/science.1065769
Li, J., Zhou, H., Zhang, Y., Li, Z., Yang, Y., and Guo, Y. (2020). The GSK3-like kinase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev. Cell 55, 367–380.e366. doi: 10.1016/j.devcel.2020.08.005
Li, S., Lei, L., Somerville, C. R., and Gu, Y. (2012). Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc. Natl. Acad. Sci. U.S.A. 109, 185–190. doi: 10.1073/pnas.1118560109
Liedtke, W., Choe, Y., Martí-Renom, M. A., Bell, A. M., Denis, C. S., Sali, A., et al. (2000). Vanilloid receptor-related osmotically activated channel (VR-OAC), a candidate vertebrate osmoreceptor. Cell 103, 525–535. doi: 10.1016/s0092-8674(00)00143-4
Lin, H., Yang, Y., Quan, R., Mendoza, I., Wu, Y., Du, W., et al. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21, 1607–1619. doi: 10.1105/tpc.109.066217
Lin, Z., Li, Y., Wang, Y., Liu, X., Ma, L., Zhang, Z., et al. (2021). Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 12:2456. doi: 10.1038/s41467-021-22812-x
Lin, Z., Li, Y., Zhang, Z., Liu, X., Hsu, C. C., Du, Y., et al. (2020). A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun. 11:613. doi: 10.1038/s41467-020-14477-9
Liu, J., Gao, H., Wang, X., Zheng, Q., Wang, C., Wang, X., et al. (2014). Effects of 24-epibrassinolide on plant growth, osmotic regulation and ion homeostasis of salt-stressed canola. Plant Biol. 16, 440–450. doi: 10.1111/plb.12052
Liu, J., Ishitani, M., Halfter, U., Kim, C. S., and Zhu, J. K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 97, 3730–3734. doi: 10.1073/pnas.97.7.3730
Liu, J. H., Wang, W., Wu, H., Gong, X., and Moriguchi, T. (2015). Polyamines function in stress tolerance: from synthesis to regulation. Front. Plant Sci. 6:827. doi: 10.3389/fpls.2015.00827
Liu, J. X., and Howell, S. H. (2010). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22, 2930–2942. doi: 10.1105/tpc.110.078154
Liu, J. X., Srivastava, R., Che, P., and Howell, S. H. (2007). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51, 897–909. doi: 10.1111/j.1365-313X.2007.03195.x
Liu, L., Cui, F., Li, Q., Yin, B., Zhang, H., Lin, B., et al. (2011). The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 21, 957–969. doi: 10.1038/cr.2010.181
Lobbes, D., Rallapalli, G., Schmidt, D. D., Martin, C., and Clarke, J. (2006). SERRATE: a new player on the plant microRNA scene. EMBO Rep. 7, 1052–1058. doi: 10.1038/sj.embor.7400806
Lou, L., Yu, F., Tian, M., Liu, G., Wu, Y., Wu, Y., et al. (2020). ESCRT-I component VPS23A sustains salt tolerance by strengthening the SOS module in Arabidopsis. Mol. Plant 13, 1134–1148. doi: 10.1016/j.molp.2020.05.010
Luo, M., Wang, Y. Y., Liu, X., Yang, S., Lu, Q., Cui, Y., et al. (2012). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63, 3297–3306. doi: 10.1093/jxb/ers059
Ma, L., Ye, J., Yang, Y., Lin, H., Yue, L., Luo, J., et al. (2019). The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 48, 697–709.e695. doi: 10.1016/j.devcel.2019.02.010
Ma, S., Gong, Q., and Bohnert, H. J. (2006). Dissecting salt stress pathways. J. Exp. Bot. 57, 1097–1107. doi: 10.1093/jxb/erj098
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., and Oda, K. (2008). The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 56, 613–626. doi: 10.1111/j.1365-313X.2008.03627.x
Martí, M. C., Stancombe, M. A., and Webb, A. A. (2013). Cell- and stimulus type-specific intracellular free Ca2+ signals in Arabidopsis. Plant Physiol. 163, 625–634. doi: 10.1104/pp.113.222901
McFarlane, H. E., Döring, A., and Persson, S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65, 69–94. doi: 10.1146/annurev-arplant-050213-040240
Møller, I. S., Gilliham, M., Jha, D., Mayo, G. M., Roy, S. J., Coates, J. C., et al. (2009). Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21, 2163–2178. doi: 10.1105/tpc.108.064568
Morton, M. J. L., Awlia, M., Al-Tamimi, N., Saade, S., Pailles, Y., Negrão, S., et al. (2019). Salt stress under the scalpel – dissecting the genetics of salt tolerance. Plant J. 97, 148–163. doi: 10.1111/tpj.14189
Mou, M., Wang, Q., Chen, Y., Yu, D., and Chen, L. (2021). Functional characterization of the Arabidopsis SERRATE under salt stress. Plant Divers. 43, 71–77.
Moustafa, K., Lefebvre-De Vos, D., Leprince, A.-S., Savouré, A., and Laurière, C. (2008). Analysis of the Arabidopsis mitogen-activated protein kinase families: organ specificity and transcriptional regulation upon water stresses. Sch. Res. Exch. 2008:143656.
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681.
Mustilli, A. C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099. doi: 10.1105/tpc.007906
Nakagawa, Y., Katagiri, T., Shinozaki, K., Qi, Z., Tatsumi, H., Furuichi, T., et al. (2007). Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc. Natl. Acad. Sci. U.S.A. 104, 3639–3644. doi: 10.1073/pnas.0607703104
Nakaminami, K., Okamoto, M., Higuchi-Takeuchi, M., Yoshizumi, T., Yamaguchi, Y., Fukao, Y., et al. (2018). AtPep3 is a hormone-like peptide that plays a role in the salinity stress tolerance of plants. Proc. Natl. Acad. Sci. U.S.A. 115, 5810–5815. doi: 10.1073/pnas.1719491115
Negi, J., Matsuda, O., Nagasawa, T., Oba, Y., Takahashi, H., Kawai-Yamada, M., et al. (2008). CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452, 483–486. doi: 10.1038/nature06720
Nolan, T. M., Vukašinović, N., Liu, D., Russinova, E., and Yin, Y. (2019). Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32, 295–318. doi: 10.1105/tpc.19.00335
Núñez-Ramírez, R., Sánchez-Barrena, M. J., Villalta, I., Vega, J. F., Pardo, J. M., Quintero, F. J., et al. (2012). Structural insights on the plant salt-overly-sensitive 1 (SOS1) Na+/H+ antiporter. J. Mol. Biol. 424, 283–294. doi: 10.1016/j.jmb.2012.09.015
Ohta, M., Guo, Y., Halfter, U., and Zhu, J. K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. U.S.A. 100, 11771–11776. doi: 10.1073/pnas.2034853100
Pabuayon, I. C. M., Jiang, J., Qian, H., Chung, J.-S., and Shi, H. (2021). Gain-of-function mutations of AtNHX1 suppress sos1 salt sensitivity and improve salt tolerance in Arabidopsis. Stress Biol. 1:14.
Park, H. J., Qiang, Z., Kim, W.-Y., and Yun, D.-J. (2016). Diurnal and circadian regulation of salt tolerance in Arabidopsis. J. Plant Biol. 59, 569–578.
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Peethambaran, P. K., Glenz, R., Höninger, S., Shahinul Islam, S. M., Hummel, S., Harter, K., et al. (2018). Salt-inducible expression of OsJAZ8 improves resilience against salt-stress. BMC Plant Biol. 18:311. doi: 10.1186/s12870-018-1521-0
Peleg, Z., and Blumwald, E. (2011). Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 14, 290–295. doi: 10.1016/j.pbi.2011.02.001
Planas-Riverola, A., Gupta, A., Betegón-Putze, I., Bosch, N., Ibañes, M., and Caño-Delgado, A. I. (2019). Brassinosteroid signaling in plant development and adaptation to stress. Development 146:dev151894. doi: 10.1242/dev.151894
Qin, F., Kodaira, K. S., Maruyama, K., Mizoi, J., Tran, L. S., Fujita, Y., et al. (2011). SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 157, 1900–1913. doi: 10.1104/pp.111.187302
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Quan, R., Lin, H., Mendoza, I., Zhang, Y., Cao, W., Yang, Y., et al. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19, 1415–1431. doi: 10.1105/tpc.106.042291
Quintero, F. J., Martinez-Atienza, J., Villalta, I., Jiang, X., Kim, W. Y., Ali, Z., et al. (2011). Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 108, 2611–2616. doi: 10.1073/pnas.1018921108
Quintero, F. J., Ohta, M., Shi, H., Zhu, J. K., and Pardo, J. M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. U.S.A. 99, 9061–9066. doi: 10.1073/pnas.132092099
Rao, K. P., Richa, T., Kumar, K., Raghuram, B., and Sinha, A. K. (2010). In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res. 17, 139–153. doi: 10.1093/dnares/dsq011
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., Chao, D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146. doi: 10.1038/ng1643
Rengasamy, P. (2006). World salinization with emphasis on Australia. J. Exp. Bot. 57, 1017–1023. doi: 10.1093/jxb/erj108
Rennie, E. A., Ebert, B., Miles, G. P., Cahoon, R. E., Christiansen, K. M., Stonebloom, S., et al. (2014). Identification of a sphingolipid α-glucuronosyltransferase that is essential for pollen function in Arabidopsis. Plant Cell 26, 3314–3325. doi: 10.1105/tpc.114.129171
Rennie, E. A., Hansen, S. F., Baidoo, E. E., Hadi, M. Z., Keasling, J. D., and Scheller, H. V. (2012). Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases. Plant Physiol. 159, 1408–1417. doi: 10.1104/pp.112.200964
Roy, S. J., Negrão, S., and Tester, M. (2014). Salt resistant crop plants. Curr. Opin. Biotechnol. 26, 115–124. doi: 10.1016/j.copbio.2013.12.004
Rus, A., Lee, B. H., Muñoz-Mayor, A., Sharkhuu, A., Miura, K., Zhu, J. K., et al. (2004). AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 136, 2500–2511. doi: 10.1104/pp.104.042234
Rus, A., Yokoi, S., Sharkhuu, A., Reddy, M., Lee, B. H., Matsumoto, T. K., et al. (2001). AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc. Natl. Acad. Sci. U.S.A. 98, 14150–14155. doi: 10.1073/pnas.241501798
Russinova, E., Borst, J. W., Kwaaitaal, M., Caño-Delgado, A., Yin, Y., Chory, J., et al. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16, 3216–3229. doi: 10.1105/tpc.104.025387
Sakuraba, Y., Bulbul, S., Piao, W., Choi, G., and Paek, N. C. (2017). Arabidopsis EARLY FLOWERING3 increases salt tolerance by suppressing salt stress response pathways. Plant J. 92, 1106–1120. doi: 10.1111/tpj.13747
Scippa, G. S., Di Michele, M., Onelli, E., Patrignani, G., Chiatante, D., and Bray, E. A. (2004). The histone-like protein H1-S and the response of tomato leaves to water deficit. J. Exp. Bot. 55, 99–109. doi: 10.1093/jxb/erh022
Shen, Y., Ruan, Q., Chai, H., Yuan, Y., Yang, W., Chen, J., et al. (2016). The Arabidopsis polyamine transporter LHR1/PUT3 modulates heat responsive gene expression by enhancing mRNA stability. Plant J. 88, 1006–1021. doi: 10.1111/tpj.13310
Shi, H., Ishitani, M., Kim, C., and Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901. doi: 10.1073/pnas.120170197
Shi, H., Quintero, F. J., Pardo, J. M., and Zhu, J. K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14, 465–477. doi: 10.1105/tpc.010371
Stephan, A. B., Kunz, H. H., Yang, E., and Schroeder, J. I. (2016). Rapid hyperosmotic-induced Ca2+ responses in Arabidopsis thaliana exhibit sensory potentiation and involvement of plastidial KEA transporters. Proc. Natl. Acad. Sci. U.S.A. 113, E5242–E5249. doi: 10.1073/pnas.1519555113
Su, W., Liu, Y., Xia, Y., Hong, Z., and Li, J. (2012). The Arabidopsis homolog of the mammalian OS-9 protein plays a key role in the endoplasmic reticulum-associated degradation of misfolded receptor-like kinases. Mol. Plant 5, 929–940. doi: 10.1093/mp/sss042
Szabados, L., and Savouré, A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97.
Székely, G., Abrahám, E., Cséplo, A., Rigó, G., Zsigmond, L., Csiszár, J., et al. (2008). Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53, 11–28. doi: 10.1111/j.1365-313X.2007.03318.x
Takahashi, F., Suzuki, T., Osakabe, Y., Betsuyaku, S., Kondo, Y., Dohmae, N., et al. (2018). A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556, 235–238. doi: 10.1038/s41586-018-0009-2
Takahashi, Y., Zhang, J., Hsu, P. K., Ceciliato, P. H. O., Zhang, L., Dubeaux, G., et al. (2020). MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 11:12. doi: 10.1038/s41467-019-13875-y
Tan, B. C., Joseph, L. M., Deng, W. T., Liu, L., Li, Q. B., Cline, K., et al. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35, 44–56.
Tao, J. J., Chen, H. W., Ma, B., Zhang, W. K., Chen, S. Y., and Zhang, J. S. (2015). The role of ethylene in plants under salinity stress. Front. Plant Sci. 6:1059. doi: 10.3389/fpls.2015.01059
Taylor, N. G., Howells, R. M., Huttly, A. K., Vickers, K., and Turner, S. R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 1450–1455. doi: 10.1073/pnas.0337628100
Teige, M., Scheikl, E., Eulgem, T., Dóczi, R., Ichimura, K., Shinozaki, K., et al. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15, 141–152. doi: 10.1016/j.molcel.2004.06.023
Thalmann, M., Pazmino, D., Seung, D., Horrer, D., Nigro, A., Meier, T., et al. (2016). Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 28, 1860–1878. doi: 10.1105/tpc.16.00143
Thole, J. M., and Nielsen, E. (2008). Phosphoinositides in plants: novel functions in membrane trafficking. Curr. Opin. Plant Biol. 11, 620–631. doi: 10.1016/j.pbi.2008.10.010
Toda, Y., Tanaka, M., Ogawa, D., Kurata, K., Kurotani, K., Habu, Y., et al. (2013). RICE SALT SENSITIVE3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell 25, 1709–1725. doi: 10.1105/tpc.113.112052
Tracy, F. E., Gilliham, M., Dodd, A. N., Webb, A. A., and Tester, M. (2008). NaCl-induced changes in cytosolic free Ca2+ in Arabidopsis thaliana are heterogeneous and modified by external ionic composition. Plant Cell Environ. 31, 1063–1073. doi: 10.1111/j.1365-3040.2008.01817.x
Tran, L. S., Urao, T., Qin, F., Maruyama, K., Kakimoto, T., Shinozaki, K., et al. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 20623–20628. doi: 10.1073/pnas.0706547105
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U.S.A. 97, 11632–11637. doi: 10.1073/pnas.190309197
Urao, T., Yakubov, B., Satoh, R., Yamaguchi-Shinozaki, K., Seki, M., Hirayama, T., et al. (1999). A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11, 1743–1754. doi: 10.1105/tpc.11.9.1743
Vaid, N., Macovei, A., and Tuteja, N. (2013). Knights in action: lectin receptor-like kinases in plant development and stress responses. Mol. Plant 6, 1405–1418. doi: 10.1093/mp/sst033
Valenzuela, C. E., Acevedo-Acevedo, O., Miranda, G. S., Vergara-Barros, P., Holuigue, L., Figueroa, C. R., et al. (2016). Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J. Exp. Bot. 67, 4209–4220. doi: 10.1093/jxb/erw202
van Zelm, E., Zhang, Y., and Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433.
Veley, K. M., Marshburn, S., Clure, C. E., and Haswell, E. S. (2012). Mechanosensitive channels protect plastids from hypoosmotic stress during normal plant growth. Curr. Biol. 22, 408–413. doi: 10.1016/j.cub.2012.01.027
Vembar, S. S., and Brodsky, J. L. (2008). One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944–957. doi: 10.1038/nrm2546
Vitale, A., and Boston, R. S. (2008). Endoplasmic reticulum quality control and the unfolded protein response: insights from plants. Traffic 9, 1581–1588. doi: 10.1111/j.1600-0854.2008.00780.x
Vlad, F., Droillard, M. J., Valot, B., Khafif, M., Rodrigues, A., Brault, M., et al. (2010). Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J. 63, 778–790. doi: 10.1111/j.1365-313X.2010.04281.x
Vlad, F., Rubio, S., Rodrigues, A., Sirichandra, C., Belin, C., Robert, N., et al. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21, 3170–3184. doi: 10.1105/tpc.109.069179
von Arnim, A. G., and Deng, X. W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79, 1035–1045. doi: 10.1016/0092-8674(94)90034-5
Wang, F., Jing, W., and Zhang, W. (2014). The mitogen-activated protein kinase cascade MKK1-MPK4 mediates salt signaling in rice. Plant Sci. 227, 181–189. doi: 10.1016/j.plantsci.2014.08.007
Wang, J., Song, L., Gong, X., Xu, J., and Li, M. (2020). Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 21:1446. doi: 10.3390/ijms21041446
Wang, P., Shen, L., Guo, J., Jing, W., Qu, Y., Li, W., et al. (2019). Phosphatidic Acid directly regulates PINOID-dependent phosphorylation and activation of the PIN-FORMED2 auxin efflux transporter in response to salt stress. Plant Cell 31, 250–271. doi: 10.1105/tpc.18.00528
Wang, P., Zhao, Y., Li, Z., Hsu, C. C., Liu, X., Fu, L., et al. (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 69, 100–112.e106. doi: 10.1016/j.molcel.2017.12.002
Wang, W., Xing, L., Xu, K., Ji, D., Xu, Y., Chen, C., et al. (2020). Salt stress-induced H2O2 and Ca2+ mediate K+/Na+ homeostasis in Pyropia haitanensis. J. Appl. Phycol. 32, 4199–4210.
Wang, Z., Hong, Y., Zhu, G., Li, Y., Niu, Q., Yao, J., et al. (2020). Loss of salt tolerance during tomato domestication conferred by variation in a Na+/K+ transporter. EMBO J. 39, e103256. doi: 10.15252/embj.2019103256
Wang, Z., Ma, Z., Castillo-González, C., Sun, D., Li, Y., Yu, B., et al. (2018). SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature 557, 516–521. doi: 10.1038/s41586-018-0135-x
Wang, Z., Wang, M., Yang, C., Zhao, L., Qin, G., Peng, L., et al. (2021). SWO1 modulates cell wall integrity under salt stress by interacting with importin α in Arabidopsis. Stress Biol. 1, 9.
Wen, M., Lin, X., Xie, M., Wang, Y., Xu, S., Liufu, Z., et al. (2016). Small RNA transcriptomes of mangroves evolve adaptively in extreme environments. Sci. Rep. 6:27551. doi: 10.1038/srep27551
Wilson, M. E., Jensen, G. S., and Haswell, E. S. (2011). Two mechanosensitive channel homologs influence division ring placement in Arabidopsis chloroplasts. Plant Cell 23, 2939–2949. doi: 10.1105/tpc.111.088112
Wohlbach, D. J., Quirino, B. F., and Sussman, M. R. (2008). Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 20, 1101–1117. doi: 10.1105/tpc.107.055871
Wormit, A., Butt, S. M., Chairam, I., McKenna, J. F., Nunes-Nesi, A., Kjaer, L., et al. (2012). Osmosensitive changes of carbohydrate metabolism in response to cellulose biosynthesis inhibition. Plant Physiol. 159, 105–117. doi: 10.1104/pp.112.195198
Wu, F., Chi, Y., Jiang, Z., Xu, Y., Xie, L., Huang, F., et al. (2020). Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 578, 577–581. doi: 10.1038/s41586-020-2032-3
Wu, H., Ye, H., Yao, R., Zhang, T., and Xiong, L. (2015). OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 232, 1–12. doi: 10.1016/j.plantsci.2014.12.010
Wu, X., Xu, J., Meng, X., Fang, X., Xia, M., Zhang, J., et al. (2022). Linker histone variant HIS1-3 and WRKY1 oppositely regulate salt stress tolerance in Arabidopsis. Plant Physiol doi: 10.1093/plphys/kiac174 [Epub ahead of print].
Xu, J., Li, Y., Wang, Y., Liu, H., Lei, L., Yang, H., et al. (2008). Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283, 26996–27006. doi: 10.1074/jbc.M801392200
Xu, R., Wang, Y., Zheng, H., Lu, W., Wu, C., Huang, J., et al. (2015). Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J. Exp. Bot. 66, 5997–6008. doi: 10.1093/jxb/erv312
Yan, Z., Wang, J., Wang, F., Xie, C., Lv, B., Yu, Z., et al. (2021). MPK3/6-induced degradation of ARR1/10/12 promotes salt tolerance in Arabidopsis. EMBO Rep. 22:e52457. doi: 10.15252/embr.202152457
Yang, L., Liu, Z., Lu, F., Dong, A., and Huang, H. (2006). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47, 841–850. doi: 10.1111/j.1365-313X.2006.02835.x
Yang, Y., and Guo, Y. (2018a). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, Y., and Guo, Y. (2018b). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804.
Yang, Y., Han, X., Ma, L., Wu, Y., Liu, X., Fu, H., et al. (2021). Dynamic changes of phosphatidylinositol and phosphatidylinositol 4-phosphate levels modulate H+-ATPase and Na+/H+ antiporter activities to maintain ion homeostasis in Arabidopsis under salt stress. Mol. Plant 14, 2000–2014. doi: 10.1016/j.molp.2021.07.020
Yang, Y., Qin, Y., Xie, C., Zhao, F., Zhao, J., Liu, D., et al. (2010). The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22, 1313–1332. doi: 10.1105/tpc.109.069609
Yang, Y., Wu, Y., Ma, L., Yang, Z., Dong, Q., Li, Q., et al. (2019). The Ca2+ sensor SCaBP3/CBL7 modulates plasma membrane H+-ATPase activity and promotes alkali tolerance in Arabidopsis. Plant Cell 31, 1367–1384.
Yang, Z., Wang, C., Xue, Y., Liu, X., Chen, S., Song, C., et al. (2019). Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 10:1199. doi: 10.1038/s41467-019-09181-2
Ye, C., Dickman, M. B., Whitham, S. A., Payton, M., and Verchot, J. (2011). The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol. 156, 741–755. doi: 10.1104/pp.111.174110
Yin, X., Xia, Y., Xie, Q., Cao, Y., Wang, Z., Hao, G., et al. (2020). The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance. New Phytol. 71, 1801–1814. doi: 10.1093/jxb/erz549
Yoshiba, Y., Nanjo, T., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Stress-responsive and developmental regulation of Delta(1)-pyrroline-5-carboxylate synthetase 1 (P5CS1) gene expression in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 261, 766–772. doi: 10.1006/bbrc.1999.1112
Yu, L., Nie, J., Cao, C., Jin, Y., Yan, M., Wang, F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773. doi: 10.1111/j.1469-8137.2010.03422.x
Yu, Y., Wang, J., Shi, H., Gu, J., Dong, J., Deng, X. W., et al. (2016). Salt stress and ethylene antagonistically regulate nucleocytoplasmic partitioning of COP1 to control seed germination. Plant Physiol. 170, 2340–2350. doi: 10.1104/pp.15.01724
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., and Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Yuan, F., Yang, H., Xue, Y., Kong, D., Ye, R., Li, C., et al. (2014). OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514, 367–371. doi: 10.1038/nature13593
Zander, M., Lewsey, M. G., Clark, N. M., Yin, L., Bartlett, A., Saldierna Guzmán, J. P., et al. (2020). Integrated multi-omics framework of the plant response to jasmonic acid. Nat. Plants 6, 290–302.
Zeng, H., Tang, Q., and Hua, X. (2010). Arabidopsis brassinosteroid mutants det2-1 and bin2-1 display altered salt tolerance. J. Plant Grow. Regul. 29, 44–52.
Zhang, L., and Wang, A. (2012). Virus-induced ER stress and the unfolded protein response. Front. Plant Sci. 3:293. doi: 10.3389/fpls.2012.00293
Zhang, M., Liang, X., Wang, L., Cao, Y., Song, W., Shi, J., et al. (2019). A HAK family Na+ transporter confers natural variation of salt tolerance in maize. Nat. Plants 5, 1297–1308. doi: 10.1038/s41477-019-0565-y
Zhang, Q., Lin, F., Mao, T., Nie, J., Yan, M., Yuan, M., et al. (2012). Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 24, 4555–4576. doi: 10.1105/tpc.112.104182
Zhang, S., Wu, Q. R., Liu, L. L., Zhang, H. M., Gao, J. W., and Pei, Z. M. (2020). Osmotic stress alters circadian cytosolic Ca2+ oscillations and OSCA1 is required in circadian gated stress adaptation. Plant Signal. Behav. 15:1836883. doi: 10.1080/15592324.2020.1836883
Zhang, S. S., Sun, L., Dong, X., Lu, S. J., Tian, W., and Liu, J. X. (2016). Cellulose synthesis genes CESA6 and CSI1 are important for salt stress tolerance in Arabidopsis. J. Integr. Plant Biol. 58, 623–626. doi: 10.1111/jipb.12442
Zhang, Z., Zhang, S., Zhang, Y., Wang, X., Li, D., Li, Q., et al. (2011). Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23, 396–411. doi: 10.1105/tpc.110.081356
Zhao, C., Zayed, O., Yu, Z., Jiang, W., Zhu, P., Hsu, C. C., et al. (2018). Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, 13123–13128.
Zhao, C., Zhang, H., Song, C., Zhu, J.-K., and Shabala, S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovation 1:100017. doi: 10.1016/j.xinn.2020.100017
Zhao, J., Zhang, L., Liu, N., Xu, S., Yue, Z., Zhang, L., et al. (2019). Mutual regulation of receptor-like kinase SIT1 and B’κ-PP2A shapes the early response of rice to salt stress. Plant Cell 31, 2131–2151. doi: 10.1105/tpc.18.00706
Zhao, X., Dou, L., Gong, Z., Wang, X., and Mao, T. (2019). BES1 hinders ABSCISIC ACID INSENSITIVE5 and promotes seed germination in Arabidopsis. New Phytol. 221, 908–918.
Zhao, Y., Zhang, Z., Gao, J., Wang, P., Hu, T., Wang, Z., et al. (2018). Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Rep 23, 3340–3351.e3345. doi: 10.1016/j.celrep.2018.05.044
Zhou, H., Lin, H., Chen, S., Becker, K., Yang, Y., Zhao, J., et al. (2014). Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26, 1166–1182. doi: 10.1105/tpc.113.117069
Zhou, H., Xiao, F., Zheng, Y., Liu, G., Zhuang, Y., Wang, Z., et al. (2022). Pamp-induced secreted peptide 3 modulates salt tolerance through receptor-like kinase 7 in plants. Plant Cell 34, 927–944. doi: 10.1093/plcell/koab292
Zhu, J., Lee, B. H., Dellinger, M., Cui, X., Zhang, C., Wu, S., et al. (2010). A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. Plant J. 63, 128–140. doi: 10.1111/j.1365-313X.2010.04227.x
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Zhu, J. K., Liu, J., and Xiong, L. (1998). Genetic analysis of salt tolerance in Arabidopsis. Evidence for a critical role of potassium nutrition. Plant Cell 10, 1181–1191.
Keywords: epigenetic regulation, hormonal regulation, salt stress, SOS pathway, signal transduction
Citation: Ma L, Liu X, Lv W and Yang Y (2022) Molecular Mechanisms of Plant Responses to Salt Stress. Front. Plant Sci. 13:934877. doi: 10.3389/fpls.2022.934877
Received: 03 May 2022; Accepted: 23 May 2022;
Published: 27 June 2022.
Edited by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaCopyright © 2022 Ma, Liu, Lv and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongqing Yang, eWFuZ3lvbmdxaW5nQGNhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.