
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 09 August 2022
Sec. Plant Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.932767
This article is part of the Research TopicThe Role of Nitrate in Plant Response to Biotic and Abiotic StressView all 8 articles
 Binbin Wen1,2
Binbin Wen1,2 Xingyao Gong1,2
Xingyao Gong1,2 Wenpeng Deng1,2
Wenpeng Deng1,2 Xiude Chen1,2,3
Xiude Chen1,2,3 Dongmei Li1,2,3
Dongmei Li1,2,3 Xiling Fu1,2,3
Xiling Fu1,2,3 Ling Li1,2,3*
Ling Li1,2,3* Qiuping Tan1,2,3*
Qiuping Tan1,2,3*The regulation of plant gene expression by nitrate is a complex regulatory process. Here, we identified 90 GARP family genes in apples by genome-wide analysis. As a member of the GARP gene family, the expression of MdHHO3 (Malus domestica HYPERSENSITIVITY TO LOW PHOSPHATE-ELICITED PRIMARY ROOT SHORTENING1 HOMOLOG 3) is upregulated under N (nitrogen) supply. The results of DNA-binding site analysis and electrophoretic mobility shift assays (EMSA) showed that MdHHO3 binds to the motif-containing GAATC. Furthermore, MdHHO3 binds to its promoter sequence and inhibits its activity. In addition, the overexpression of MdHHO3 in apple calli resulted in less accumulation of nitrate in 35S:MdHHO3-GFP calli and downregulated the expression of the nitrate transport-related genes but upregulated the expression of the nitrate assimilation-related genes. Similarly, the expression of the nitrate transport-related genes was downregulated and the expression of the nitrate assimilation-related genes was upregulated in MdHHO3 overexpression Arabidopsis and tobacco plants. Interaction experiments showed that MdHHO3 could bind to the promoter MdNRT2.1 (NITRATE TRANSPORTER 2.1) and negatively regulate its expression. Moreover, the exposure of MdHHO3-overexpressing Arabidopsis and tobacco to nitrate deficiency resulted in an early senescence phenotype as compared to the WT plants. These results show that MdHHO3 can not only negatively regulate nitrate accumulation in response to nitrate but also promote early leaf senescence under nitrate deficiency. This information may be useful to further reveal the mechanism of the nitrate response and demonstrates that nitrate deficiency induces leaf senescence in apples.
The growth and reproduction of plants are closely related to changes in the environment. Therefore, plants can only grow and reproduce normally once they adapt to changes in the external environment. The soil nutrient status is one of the key environmental factors affecting plant growth and reproduction (Gallardo et al., 2006). In natural environments, plants often face N starvation conditions (Jackson and Caldwell, 1993; Miller and Cramer, 2004).
N is often a factor limiting plant growth and development. The regulation of plant gene expression by N is very complex, and some genes involved in the adaptive regulation of N starvation have been identified. Nitrate is the main N source absorbed by plants. When plants face a nitrate-deficient environment, the two families of nitrate transporters (NRT1 and NRT2) play important roles in high-affinity nitrate transport uptake (Nacry et al., 2013; Krapp et al., 2014). In the presence of a low external nitrate concentration, the dual-affinity transporter NRT1.1 is phosphorylated into a high-affinity transporter to participate in the absorption of a low concentration of nitrate (Miller et al., 2007; Ho et al., 2009). The NRT2 gene family members, NRT2.1, NRT2.2, NRT2.4, and NRT2.5, are involved in the absorption of nitrate at a low concentration, and among these, only NRT2.1 showed a significant correlation between transcript abundance and high-affinity nitrate influx after nitrate supply to nitrate-deficient plants (Lezhneva et al., 2014; Kiba and Krapp, 2016). In addition, NRT2.1 plays a leading role in root nitrate absorption, and the activity of Arabidopsis high-affinity transport system (HATs) was decreased by 63–75% when NRT2.1 was mutated (Li et al., 2007). However, overexpression of NRT2.1 resulted in a significant increase in plant biomass and a 38% increase in yield (Orsel et al., 2019). The role of NRT2.1 in the HATs supports its management of nitrate uptake in natural conditions, but the transcriptional repressors that regulate NRT2 expression in a time- and nitrate concentration-dependent manner remain to be identified. Rice NIGT1 (NITRATE-INDUCIBLE GARP-TYPE TRANSCRIPTIONAL REPRESSOR1) has been demonstrated to be involved in the nitrate response by encoding a high-affinity transporter gene (Sawaki et al., 2013). Further characterization of the NIGT1 gene revealed that it encodes a transcriptional repressor of the regulatory network underlying the nitrate response (Sawaki et al., 2013). In addition, the binding region of NIGT1 is conserved in the promoter region of Arabidopsis NRT2.1 (Maeda et al., 2018), which suggests that NIGT1 and its homologous genes may play similar roles in the N response.
N is an essential nutrient for plant growth and development and is the main component of amino acids, nucleotides, and chlorophyll, among other compounds. Therefore, N availability has a profound impact on plant productivity and growth. Nitrate in the soil is the main source of N that is absorbed by most plants, and its deficiency induces leaf senescence (Park et al., 2018). Plants adapt to N deficiency by regulating gene expression and metabolism to efficiently acquire and utilize the available N (Nacry et al., 2013). NLA (NITROGEN LIMITATION ADAPTATION) plays a key role in the process of leaf senescence induced by N deficiency (Kant et al., 2011). In addition, NLA protein can interact with ORE1 protein, the key gene for leaf senescence, to determine the leaf senescence process by regulating the stability of ORE1 proteins (Park et al., 2018). NRT1.5 plays an essential role in perceiving nitrate deficiency signals and inhibits leaf senescence induced by nitrate deficiency (Chen et al., 2021). NIGT1 hampers nitrate-induced positive regulation and reduces chlorophyll content in the NIGT1 overexpressors (Sawaki et al., 2013). Since N is the main component of chlorophyll, NIGT1 may be an important regulator to inhibit the absorption of nitrate and lead to the decline of chlorophyll. Chlorophyll degradation is the first important sign of leaf senescence. As a perennial deciduous fruit tree, the time of leaf senescence directly affects the yield and quality of the fruit (Woo et al., 2013; An et al., 2018b). Therefore, it is important to investigate the molecular mechanism of NIGT1 in N starvation-induced leaf senescence for the maintenance of apple leaf function.
To reveal the complex molecular mechanism of gene regulation in the nitrate supply and nitrate starvation, we identified the function of the MdHHO3 in this study. The supply of nitrate first rapidly induced the expression of MdHHO3, but longer exposure to nitrate limited the expression of this gene. Under nitrate-free conditions, the MdHHO3-overexpressing plants showed an early senescence phenotype. These results indicate that nitrate acts not only as a nutrient but also as a signal molecule and nitrate deficiency may generate signals to regulate the senescence of apple leaves. More significantly, the identification of the function of the MdHHO3 gene clarifies the molecular mechanism of nitrate uptake and transport in apples and the adaptation of apples to a low-N environment, which provides a direction for the future breeding of this plant.
The tissue culture plantlets of Malus × domestica “GL3” were grown on an MS medium containing 0.5 mg/L GA3, 0.5 mg/L 6-BA, and 0.1 mg/L NAA, and subcultured every 30 days. The plantlets subcultured for 30 days were selected to root in 1/2 MS medium containing 0.2 mg/L IAA. Then, the rooted apple plantlets were transferred to nursery pots for cultivation. In the hydroponic experiment, seedlings (n = 100) with the same growth parameters were planted on foam plates with 20 holes in each plate. Single Malus × domestica “GL3” plantlet was planted in each well and cultured with 1/2 Hoagland solution (Hoagland and Arnon, 1950) nutrient solution for 7 days. The nutrient solution was then replaced every 2 days with Hoagland nutrient solution. Once the plantlet grew 10 true leaves and to a height of approximately 12 cm, they were treated with distilled water for 7 days. The apple plantlets were grown at 25°C/23°C under long-day conditions (14 h of light:10 h of dark) with a relative humidity of 75% and a light intensity of 2,000 lx and were treated with various compounds. The apple calli (Malus domestica, Orin) were grown at 24°C in the dark and subcultured every 15 days. The Arabidopsis thaliana used in the study was Col-0, and the tobacco was Nicotiana benthamiana; both of these plants were grown at 25°C/23°C under long-day conditions (14 h of light:10 h of dark).
In this study, the Arabidopsis GARP-type protein sequences were used as a reference to construct the phylogenetic tree of apple GARP-type protein. The GARP-type gene family sequences from apple and Arabidopsis were downloaded from the Plant TF Database v. 4.01 (Jin et al., 2017). All candidate GARP-type proteins were confirmed using the Conserved Domain Database2 and SMART.3 The confirmed GARP-type protein sequences from apple and Arabidopsis were used to construct a phylogenetic tree using MEGA-X software with the NJ method, a bootstrap value of 1,000, and other default parameters. We then used an online webserver named EvolView4 to clean up the phylogenetic tree. The protein sequences are listed in Supplementary Table 1. Multiple sequence alignments of the HHO3 protein of different species were performed using DNAMAN 6.0.3.99, and the HHO3 protein sequences of these different species are listed in Supplementary Table 2.
Total RNA was extracted from apple, calli, Arabidopsis, and tobacco using an RNAprep Pure Kit (DP441, Tiangen, Beijing, China). First-strand cDNA was obtained using a cDNA reverse transcription kit (RR047A, Takara, Dalian, China). The primers used for qRT-PCR were designed with NCBI and synthesized by the Sangon Biotech Technology (Shanghai, China). The primers used are listed in Supplementary Table 3. SuperReal PreMix Plus (FP205, Tiangen, Beijing, China) was used for qRT-PCR. Three biological replicates and three technical replicates were analyzed using a CFX Connect™ Real-time System (Bio-Rad, Hercules, CA, United States). The qRT-PCR conditions were as follows: 95°C for 15 min and 40 cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 30 s. At the end of the reaction, a melting curve was obtained for each sample, and a transcription-level analysis was performed using the comparative Ct (2–ΔΔ Ct) method. The primers used for qRT-PCR are shown in Supplementary Table 3.
The chlorophyll content was determined as described above (Wen et al., 2019). In short, the leaves were beaten into small pieces with a punch and then submersed in 95% ethanol for 24 h until the leaves turned white. The absorbance was measured at wavelengths of 470, 649, and 665 nm using a spectrophotometer (UV-2600, Shimadzu, Shanghai, China).
The outer skin of the onion was removed, and the inner skin of the onion was peeled with a scalpel on a super clean workbench, cut into small 1-cm2 pieces, placed on MS solid medium, and cultured in the dark at 28°C for 24 h. The primers used for subcellular localization are shown in Supplementary Table 3. The complete sequence of MdHHO3 without a stop codon was cloned and inserted into the pZP211 vector. The plasmids harboring 35S:MdHHO3-GFP and 35S:GFP were transformed into the Agrobacterium tumefaciens strain LBA4404 and infected onion epidermal cells. GFP fluorescence was observed under a Zeiss LSM880 microscope (Carl Zeiss, Oberkochen, Germany) and analyzed using the ZEN lite software.5 The subcellular localization analysis was repeated three times, and the results were similar.
MdHHO3 was cloned into the pRI101-AN vector containing the GFP tag sequence to form an overexpression plasmid. The primer is shown in Supplementary Table 3. The recombinant plasmid was transformed into A. tumefaciens GV3101 and LBA4404, and the recombinant vector was then transformed into calli, Arabidopsis, and tobacco using A. tumefaciens cells. Transgenic apple calli, Arabidopsis, and tobacco were obtained as described previously (Horsch et al., 1985; Clough and Bent, 1998; An et al., 2018a). Three transgenic lines were verified as MdHHO3-overexpressing plants.
Arabidopsis and tobacco seeds were germinated on a 0.8% agar medium containing Murashige and Skoog salts and 30 g⋅L–1 sucrose. To induce leaf senescence in Arabidopsis seedlings in response to nitrate deficiency, 2-week-old Arabidopsis seedlings were grown on a nitrate-free medium (nitrate concentration: 0%) under long-day conditions (14 h of light:10 h of dark) at 24°C/22°C for 4 days. To induce leaf senescence in tobacco seedlings in response to nitrate deficiency, 4-week-old tobacco seedlings were grown in either nitrate-sufficient or a nitrate-free nutrient solution prepared with modified Hoagland’s solution (pH 5.8) for 3 weeks. After observing and recording the phenotype, the leaves of these plants were used to determine the chlorophyll content and the expression of senescence-related genes.
The MdNRT2.1 promoter fragment (sequence of the fragment containing cis-acting elements) was inserted into the pAbAi vector to construct the reporter gene pBait-AbAi vector, and the full-length CDS of MdHHO3 was cloned into the pGADT7 vector. The primers used in yeast one-hybrid assays are shown in Supplementary Table 3. To determine the minimum concentration of AbA (Aureobasidin A) that inhibits bait strain growth, the pAbAi plasmid was linearized by the restriction enzyme Bsp1191 (FD0124, Thermo Fisher Scientific™, Shanghai, China) and then transformed into the Y1H yeast strain, and the transformants were screened on SD/-Ura medium containing different concentrations of AbA. Single colonies were selected, and Matchmaker Insert Check PCR Mix 1 (Clontech) was used for colony PCR to verify the positive colonies. The minimum AbA concentration that inhibits the growth of the bait strain was then determined. The linearized pAbAi-MdNRT2.1 plasmid was co-transfected with the pGADT7-MdHHO3 plasmid into the Y1H yeast strain, and the yeast transformation product was then tested on medium (SD/-Ura/-Leu) containing an optimal concentration of AbA.
The full-length CDS of MdHHO3 was cloned into the overexpression vector pGEX-4T-1 to obtain a recombinant plasmid. The MdHHO3 recombinant plasmid was expressed in Escherichia coli strain BL21 (Tiangen, Beijing, China) and purified using Glutathione-Sepharose Resin (CWbiotech, Beijing, China). The synthesis and labeling of the probe were performed by Sangon Biotech Co., Ltd. (Shanghai, China). The probes used for EMSA are shown in Supplementary Table 3. The purified fusion protein and biotin-labeled probes were incubated in a binding buffer at room temperature under dark conditions for 30 min. After incubation, a protein loading buffer was added, and the proteins were separated by polyacrylamide gel electrophoresis (Thermo Fisher Scientific, Shanghai, China).
The promoter fragment of the MdNRT2.1 gene (including the cis-acting site sequence) was cloned into the pGreenII 0800-LUC vector to generate a reporter vector. The full-length CDS of MdHHO3 was inserted into the pGreenII 62-SK vector to construct the effector vector. The recombinant plasmid was transformed into the Agrobacterium strain GV3101. Tobacco leaves were injected with the Agrobacterium strain, cultivated in the dark for 12 h, and then placed in a light incubator. After 48 h, the injected tobacco leaves were collected and sprayed with D-Luciferin sodium salt (Sangon Biotech), and the fluorescence was then observed using an in vivo imaging system (Xenogen, Alameda, CA, United States).
GraphPad Prism 6 software (GraphPad Software, La Jolla, CA, United States) was used for the drawing, and SPSS 19.0 software (SPSS, Chicago, IL, United States) was used for the statistical analyses. The significance of the differences was tested by t-tests (*P < 0.05, significant; **P < 0.01, extremely significant).
To study the evolutionary relationship between the MdHHOs and AtHHOs genes, we constructed a rootless phylogenetic tree of apple GARP-type proteins using the Arabidopsis GARP-type protein sequences as a reference (Figure 1). A total of 90 GARP-type proteins were identified in the apple genome. The GARP-type gene family was classed into five groups (I–V) based on evolutionary relationships. Groups I–V contain 29, 16, 9, 23, and 13 genes, respectively. Among them, MdHHO3 was classified in the V group. In addition, the phylogenetic relationship between GARP-type proteins in apple and Arabidopsis suggests that apple MdHHO3 (MdGARP40) is the closest homolog of Arabidopsis AtHHO3/AtNIGT1.1 (AtGARP17) and AtHHO2/AtNIGT1.2 (AtGARP20).
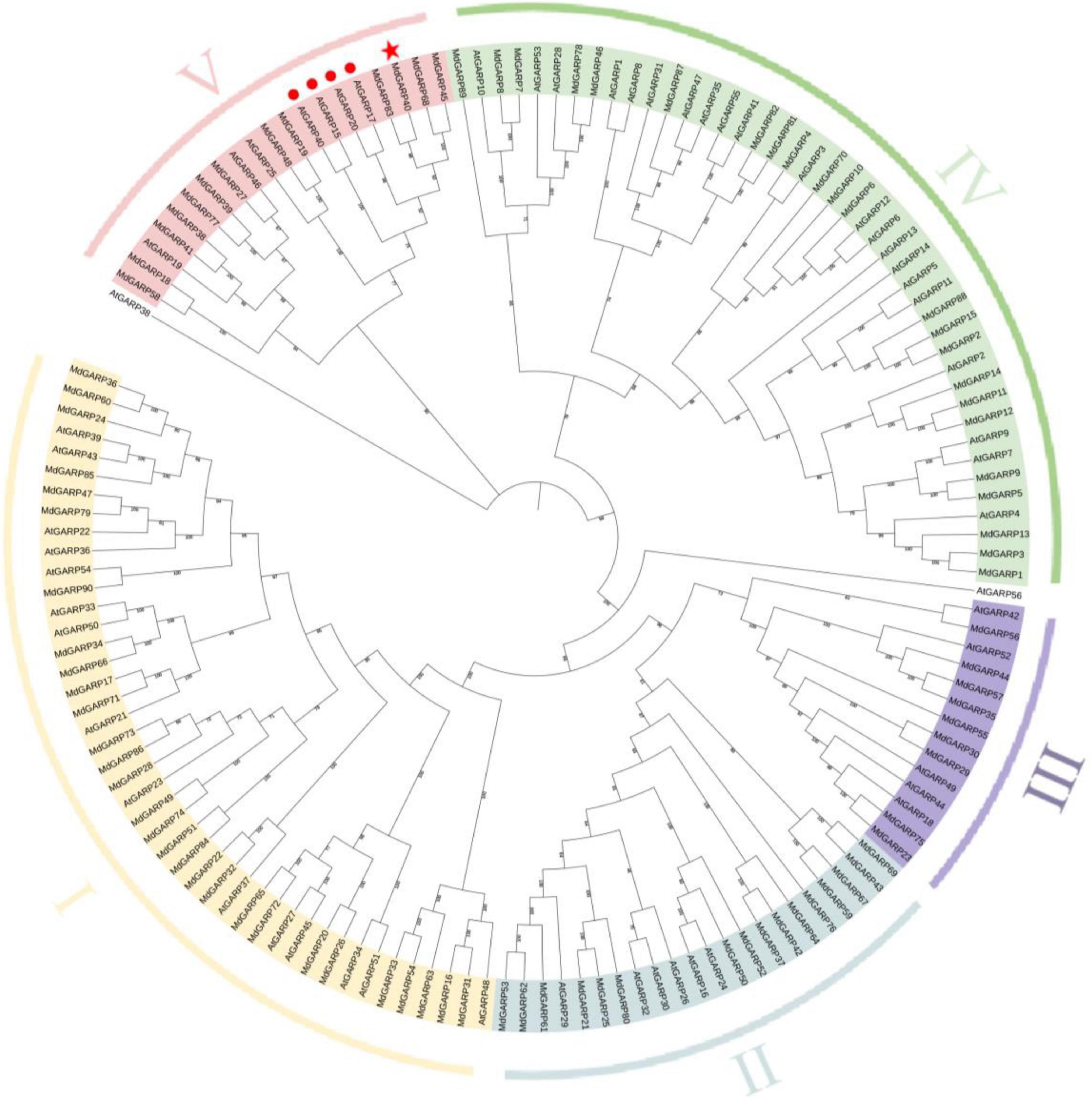
Figure 1. Phylogenetic tree of GARP-type proteins from apple and Arabidopsis. The phylogenetic tree was constructed using the NJ (neighbor-joining) algorithm. GARP proteins were clustered into five classes, and the five subfamilies are distinguished by different colors. The confirmed GARP-type protein sequences from apple and Arabidopsis were used to construct a phylogenetic tree using the MEGA software with the NJ method, a bootstrap value of 1,000, and other default parameters. The red star indicates MdHHO3(MdGARP40). The red circles indicate Arabidopsis four NIGT1s (NIGT1.1-1.4), among which AtGARP17, AtGARP20, AtGARP40, and AtGARP15 indicate AtHHO3/AtNIGT1.1, AtHHO2/AtNIGT1.2, AtHHO1/AtNIGT1.3, and AtHRS1/AtNIGT1.4, respectively. The protein sequences are listed in Supplementary Table 1.
Phylogenetic tree analyses showed that MdHHO3 was most closely related to PbEFM (Figure 2A). In addition, MdHHO3 is phylogenetically close to AtHHO3/NIGT1.1, OsNIGT1, and ZmNIGT1. Generally, phylogenetically closer orthologs may have similar functions. Sequence analysis showed that MdHHO3 contained 376 amino acids with a putative molecular weight of 41.44 kDa and a theoretical isoelectric point of 6.66. The multiple sequence alignment analyses showed that MdHHO3 had a myb-like DNA-binding domain and was similar to those of PbEFM (EARLY FLOWERING MYB PROTEIN) and PpEFM (Figure 2B). We constructed a 35S:MdHHO3-GFP vector and used the vector to transform onion epidermal cells. The subcellular localization of the 35S:MdHHO3-GFP fusion protein and the 35S:GFP protein was detected by fluorescence microscopy. The results showed that 35S:MdHHO3-GFP was located in the nucleus, whereas 35S:GFP was distributed throughout the whole cell (Figure 2C). These results suggest that MdHHO3, as a nuclear protein, plays a role in the response to nitrate.
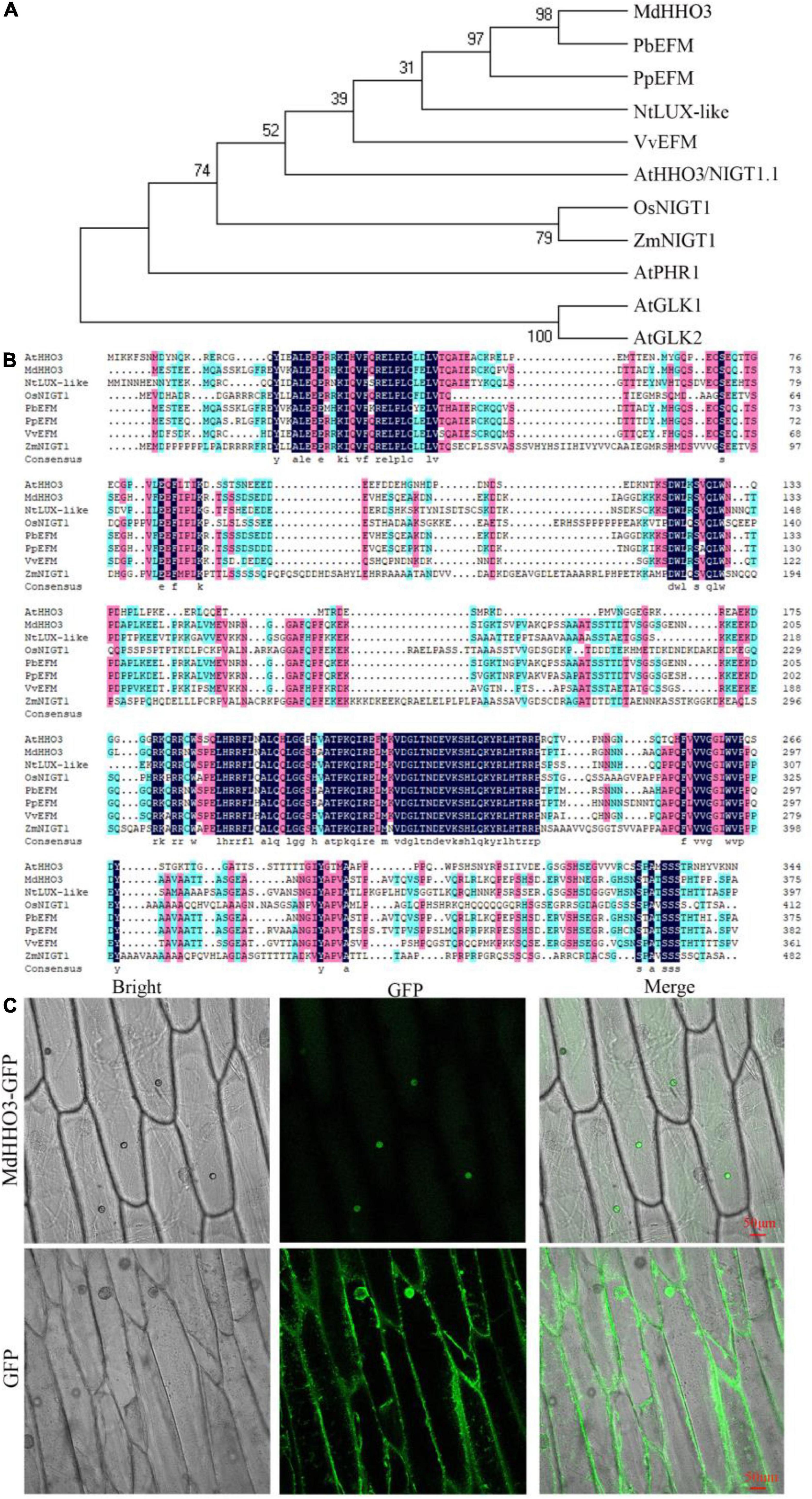
Figure 2. Phylogenetic analysis and subcellular localization of MdHHO3. (A) Phylogenetic tree analysis of HHOs of different species. The protein sequences of HHOs are listed in Supplementary Table 2. (B) Sequence alignment of 8 HHOs proteins. PbEFM (XP_009336613.1) in white pear (Pyrus bretschneideri), PpEFM (XP_007223224.2) in peach (Prunus persica), NtLUX-like (LUX ARRHYTHMO-like XP_016469710.1) in tobacco (Nicotiana tabacum), VvHHO3-like (AUZ62361.1) in grape (Vitis vinifera), AtHHO3/NIGT1.1 (NP_564236.1) in Arabidopsis thaliana, OsNIGT1 (XP_015623779.1) in rice (Oryza sativa), and ZmNIGT1 (PWZ44894.1) in maize (Zea mays) were used in the phylogenetic tree and sequence alignment analyses. (C) Subcellular localization of the 35S:MdHHO3-GFP fusion protein in onion; 35S: GFP was used as the control. GFP represents a green fluorescent protein.
To verify the temporal pattern of MdHHO3 expression induced by nitrate supply, we supplied nitrate to apple seedlings treated with N starvation and performed the quantitative real-time polymerase chain reaction (qRT-PCR) analysis using RNA obtained from roots collected at different times. During nitrate treatment, the expression of MdHHO3 was induced within 1 h, reached a peak approximately 6 h later, and then decreased, as shown in Figure 3A. To further verify that MdHHO3 expression specifically responds to nitrate, we supplied ammonium, phosphorus, glutamate, and trans-zeatin and found that none of these compounds could induce the expression of MdHHO3 (Figure 3B). We also found that the expression of MdHHO3 could be induced by a low concentration of nitrate (Figure 3C), and the expression of MdHHO3 was induced not only in the roots but also in the stems, leaves, and petioles (Figure 3D). These results indicate that MdHHO3 is a TF that is induced by nitrate and probably plays a role in nitrate-specific regulation in N response.
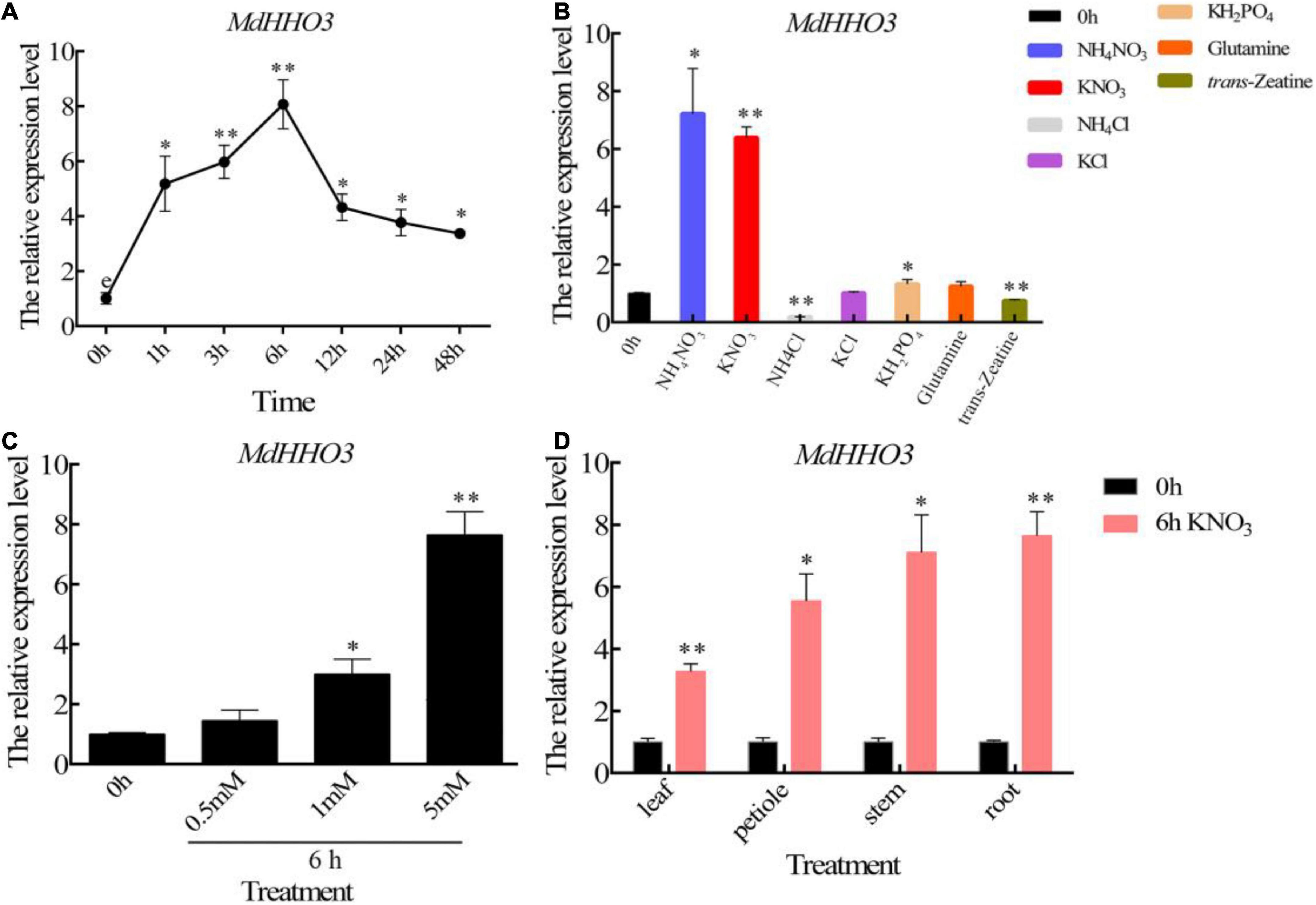
Figure 3. Nitrate specifically induces the expression of MdHHO3. (A) Time-dependent expression patterns of the MdHHO3 gene after the supply of 5 mM KNO3. (B) Expression pattern of the MdHHO3 gene after different N treatments. RNA was prepared from the roots of apple seedlings treated for 6 h with 5 mM NH4NO3, KNO3, NH4Cl, KCl, KH2PO4, and glutamine but not trans-zeatin (5 μM). (C) Expression of the MdHHO3 gene induced by different nitrate concentrations. RNA was prepared from the roots of apple seedlings treated with 0, 0.5, 1, and 5 mM KNO3 for 6 h. (D) Nitrate induced the expression of MdHHO3 in different plant tissues. Leaves, petioles, stems, and roots were collected from apple seedlings treated with 5 mM KNO3 for 6 h. Malus × domestica “GL3” seedlings grown for 1 week with distilled water were used as controls. Three biological replicates of each sample were included, and the data are expressed as the means ± SDs (n = 3). Significant differences were detected by a t-test: *P < 0.05 and **P < 0.01.
The nitrate supply experiment revealed that the expression of MdHHO3 showed a decreasing trend with increases in the duration of nitrate supply. This finding suggests that MdHHO3 may be a transcription inhibitor that inhibits its expression by binding to several sites of its own promoter. Previous studies have revealed the specific binding sequences of a few GARP family proteins (Waters et al., 2009). The DNA sequence selected in this study is GAATC. A promoter sequence analysis of MdHHO3 identified a total of eight possible binding sites (GAATC) and these sites were named P1, P2, P3, P4, and P5 based on the distances between them (Figures 4A,B). To confirm that MdHHO3 can bind to its own promoter, we performed EMSA experiments. As shown in Figure 4C, MdHHO3-GST fusion protein could bind to these sites. Furthermore, we used P1 as the binding probe according to the binding strength. The complex still bound to compete for probes at different concentrations but did not bind to mutant probes (5′-CAATG-3′) (Figure 4D). These results indicate that MdHHO3 can bind to the specific sequence of its own promoter.
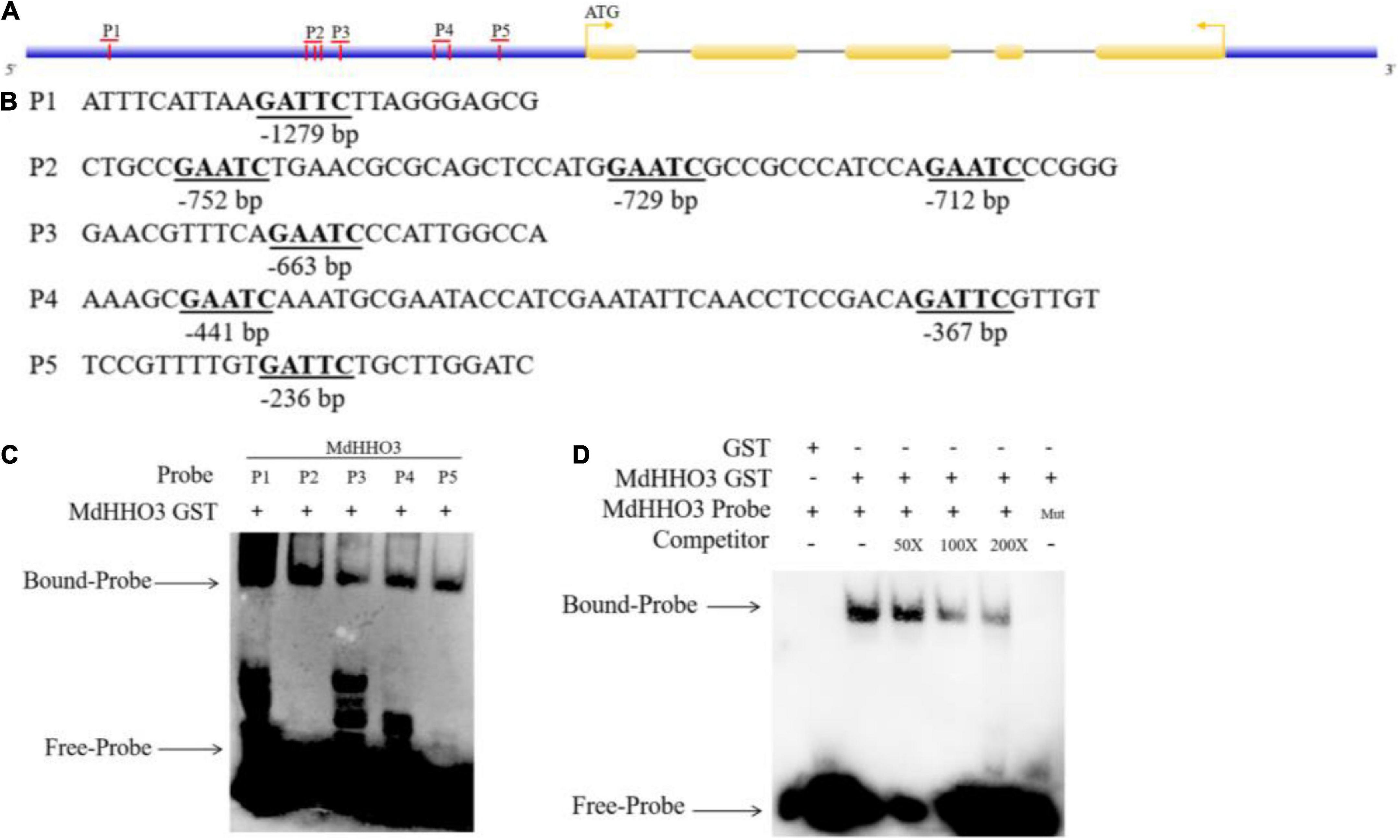
Figure 4. MdHHO3 binds to a specific sequence of its own promoter. (A) Diagram of the MdHHO3 gene promoter region. A vertical bar represents the potential sites to which MdHHO3 may bind. (B) GAATC and GATTC motifs in the promoter region selected in the DNA-binding site. (C) The electrophoretic mobility shift assay (EMSA) showed that the MdHHO3-GST fusion protein binds to the labeled DNA probe. (D) EMSA results showed that MdHHO3 binds to the labeled probe of the MdHHO3 promoter. The MdHHO3-GST fusion protein was incubated with a labeled or mutated probe DNA fragment. The labeled probe used in this study is p1 (5′-ATTTCATTAAGATTCTTAGGGAGCG-3′). The unlabeled probes were used as a competitor. 50×, 100×, and 200× represent the rates of the competitor. “Mut” representing the mutated probe in which the 5′-GAATC-3′ motif was replaced by 5′-CAATG-3′. The – symbol indicates absence, and the + symbol indicates presence.
MdHHO3 is an important regulator that regulates the response to nitrate. To verify the role of MdHHO3 in the processes of nitrate transport and assimilation, overexpressed MdHHO3 apple calli were obtained (Supplementary Figure 1). The transgenic and wild-type (WT) apple calli were cultured under normal conditions for 15 days, and the growth and fresh weight of the apple calli were then observed. The results revealed no significant difference in growth between the transgenic calli (MdHHO3) and the WT control (Figure 5). Because MdHHO3 can be specifically induced by nitrate, MdHHO3 may affect the accumulation of nitrate. Furthermore, we determined the content of nitrate in apple calli. As expected, the nitrate content in the overexpressed MdHHO3 transgenic calli was significantly lower than that in the WT calli (Figure 5). These results indicate that MdHHO3 may act as a negative regulator of nitrate accumulation.
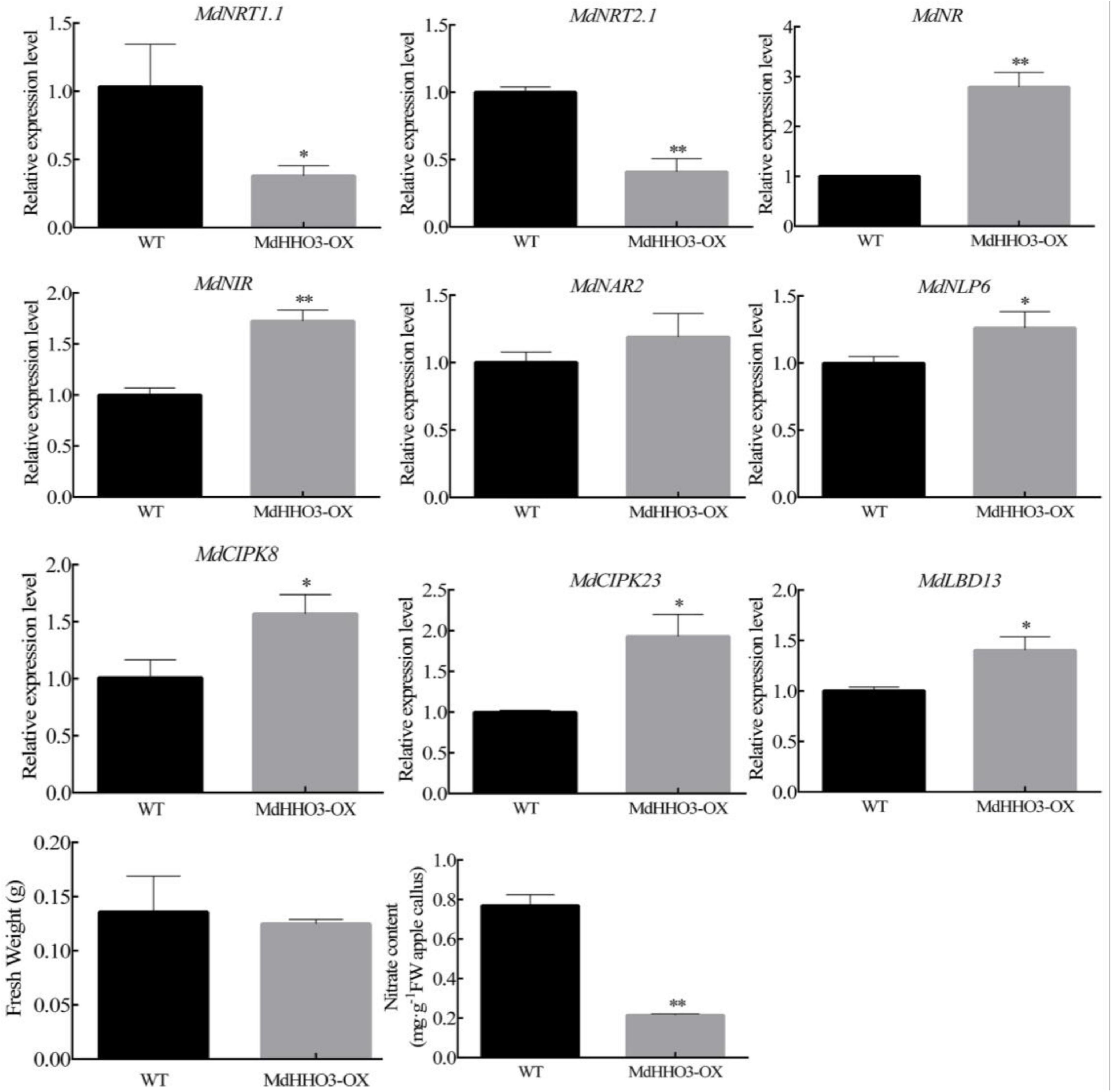
Figure 5. Expression of the nitrate regulatory genes (MdNRT1.1, MdNRT2.1, MdNR, MdNIR, MdCIPK8, MdCIPK23, MdNAR2, MdNLP6, and MdLBD13) in 2-week-old apple calli (WT, wild-type; MdHHO3-OX, with the MdHHO3 overexpression construct). The value for the WT control was set to 1. Three biological replicates of each sample were included, and the data are expressed as the means ± SDs (n = 3). Significant differences were detected by a t-test: *P < 0.05 and **P < 0.01.
The expression of nitrate transporter genes is usually used as a marker for nitrate uptake. To explore how MdHHO3 regulates the uptake and accumulation of nitrate, the expression of these genes related to nitrate transportation and assimilation were examined. Surprisingly, the expression of the nitrate transport-related genes (MdNRT1.1 and MdNRT2.1) was downregulated, whereas that of the nitrate assimilation-related genes (MdNR and MdNIR) were upregulated. In addition, the expression of five genes (MdCIPK8, MdCIPK23, MdNAR2, MdNLP6, and MdLBD13) that respond to nitrate was found to be upregulated (Figure 5). Interestingly, the expression of both the double-affinity transporter (NRT1.1) and high-affinity transporter (NRT2.1) was downregulated in the MdHHO3 transgenic calli. These results indicate that MdHHO3 may inhibit the absorption of nitrate by inhibiting the expression of high-affinity transporters.
To further verify the role of MdHHO3 in nitrate transport and accumulation, the MdHHO3 overexpression construct was generated and transformed individually into Arabidopsis and tobacco (Supplementary Figures 2, 3). The expression of the nitrate response-related genes AtNRT1.1 and AtNRT2.1 and the nitrate assimilation-related genes AtNR and AtNIR were then examined in Arabidopsis thaliana. In the overexpression lines, the expression of the tested nitrate response-related genes was downregulated, and the expression of the nitrate assimilation-related genes was upregulated (Figures 6A–D). Tobacco overexpressing MdHHO3 showed a phenotype similar to that of MdHHO3-overexpressing Arabidopsis. Correspondingly, the expression of genes related to the nitrate response was downregulated, whereas that of genes related to nitrate assimilation was upregulated (Figures 6E–H). These results indicate that MdHHO3 may act as a negative regulator of nitrate transport by inhibiting the accumulation of nitrate but improving enzyme activity in the nitrate assimilation pathway.
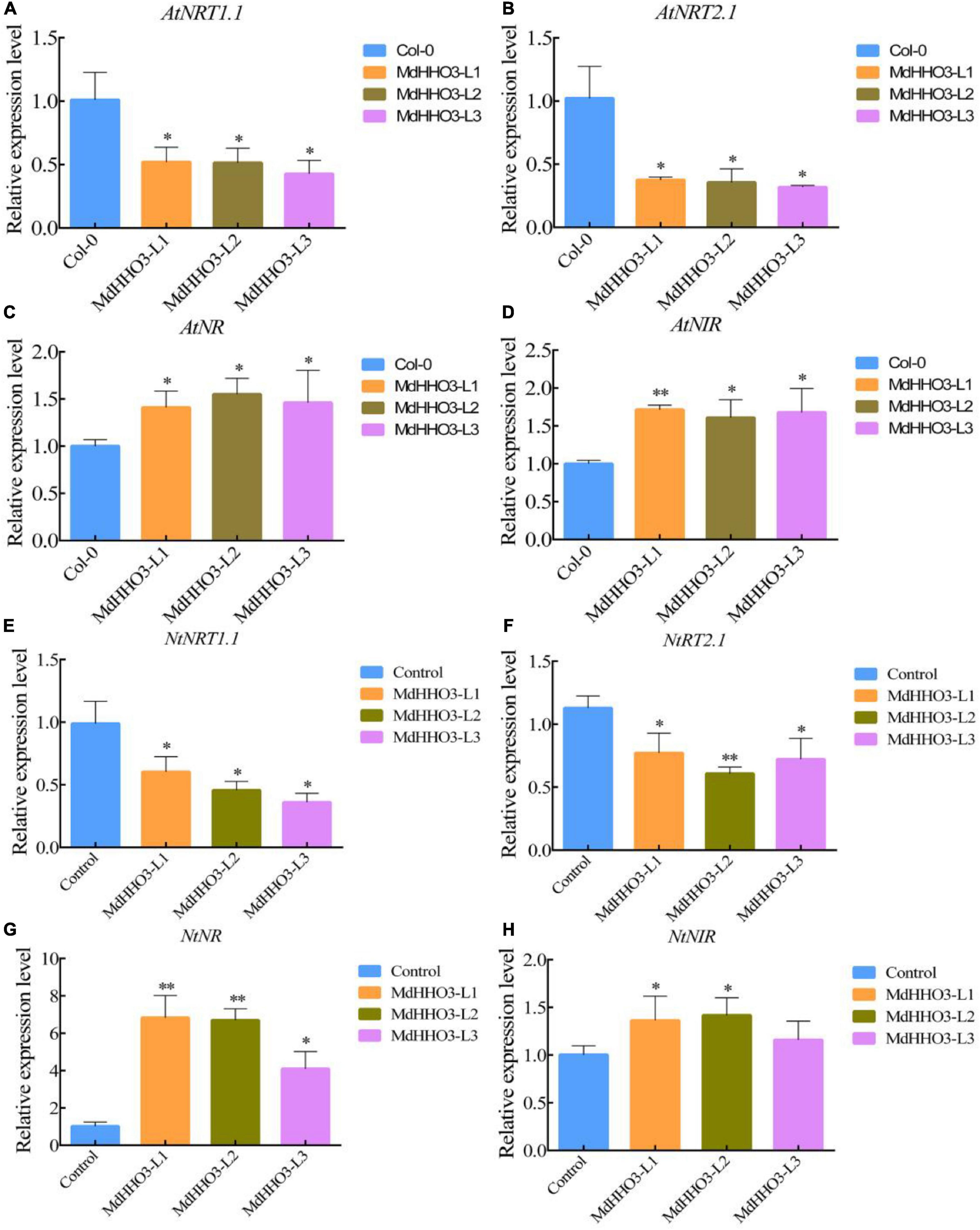
Figure 6. The overexpression of MdHHO3 downregulates the expression of the nitrate transport-related genes and upregulates the expression of the nitrate assimilation-related genes in Arabidopsis and tobacco. (A–D) Expression of genes related to nitrate transport (AtNRT1.1 and AtNRT2.1) and nitrate assimilation (AtNR and AtNIR) in the wild-type (Col-0) and MdHHO3-overexpressing Arabidopsis. (E–H) Expression of genes related to nitrate transport (NtNRT1.1 and NtNRT2.1) and nitrate assimilation (NtNR and NtNIR) in wild-type (Control) and MdHHO3-overexpressing tobacco. Three biological replicates of each sample were included, and the data are expressed as the means ± SDs (n = 3). Significant differences were detected by a t-test: *P < 0.05 and **P < 0.01.
As a high-affinity transporter, NRT2.1 plays an important role in nitrate transport. The expression of NRT2.1 in MdHHO3-overexpressing apple calli, Arabidopsis, and tobacco was inhibited (Figures 5, 6B,F), which suggests that a transcriptional repressor might regulate its expression. An analysis of the promoter sequence of NRT2.1 showed that it contains a sequence consistent with that of MdHHO3 (5′-GAATC-3′), and these sequences were named P1, P2, P3, P4, and P5 based on the distances between them (Figure 7A). To examine whether MdHHO3 regulates NRT2.1, an in vivo Y1H assay confirmed the interaction of the MdHHO3 protein and the MdNRT2.1 promoter. The full-length CDS of MdHHO3 and the NRT2.1 gene promoter fragment containing the 5′-GAATC-3′ motif were cloned to the PGADT7 and pAbAi vectors, respectively. The yeast strains co-transformed with pGADT7-MdHHO3 and pABAi-MdNRT2.1 grew normally in a selective medium, whereas the growth of the control yeast strain with the empty vector was inhibited, which indicated the existence of a direct interaction between the MdHHO3 and MdNRT2.1 (Figure 7C). To further examine the role of MdHHO3 in regulating the NRT2.1 expression, MdHHO3 sites in the NRT2.1 gene promoter were investigated in vitro using an EMSA with an integrated MdHHO3 protein. As shown in Supplementary Figure 4, five conserved motifs were putative MdHHO3-binding sites. According to the binding strength, we used P2 as the binding probe. In addition, the MdHHO3-GST fusion protein bound to the MdNRT2.1 probe (5′-GAATC-3′) but not to the mutated probe (5′-CAATG-3′), and the binding disappeared as the concentration of the competitive probe increased (Figure 7B).
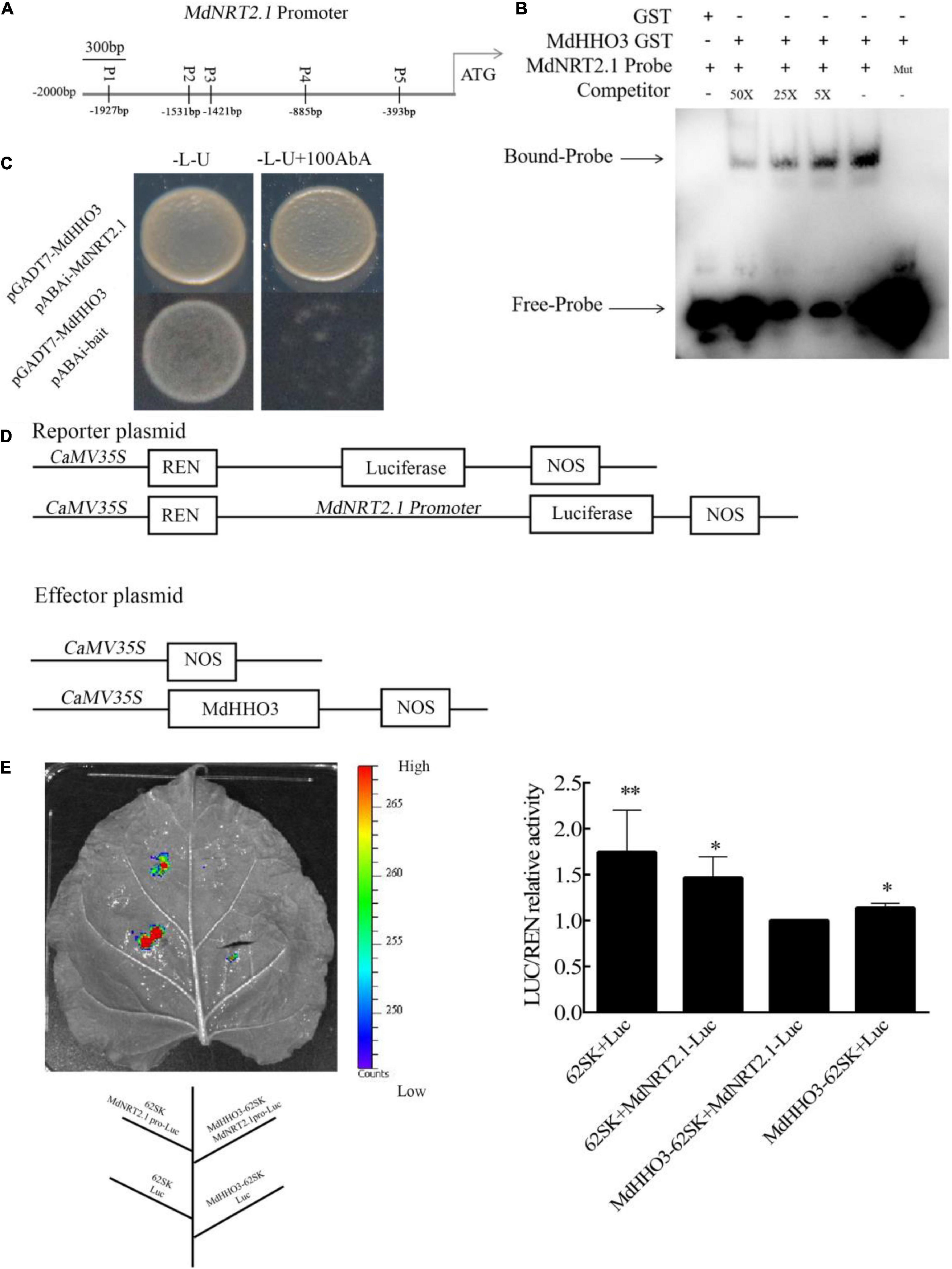
Figure 7. MdHHO3 inhibits the expression of MdNRT2.1. (A) Diagram of the MdNRT2.1 gene promoter region. p1–p5 represents the potential sites to which MdHHO3 may bind. (B) The electrophoretic mobility shift assay (EMSA) showed that the MdHHO3-GST fusion protein binds to the MdNRT2.1 promoter. The MdHHO3 fusion protein was incubated with a labeled or mutated probe DNA fragment. The labeled probe used in this study is p2 (5′-AACTCTTAACGAATCAATGGATGGA-3′). The unlabeled probes were used as a competitor. 50×, 100×, and 200× represent the rates of the competitor. “Mut” representing the mutated probe in which the 5′-GAATC-3′ motif was replaced by 5′-CAATG-3′. The – symbol indicates absence, and the + symbol indicates presence. (C) Yeast one-hybrid (Y1H) assay showing the interaction between MdHHO3 and the MdNRT2.1 promoter. The empty PGADT7 was used as a control. The fused yeast strains were grown on SD/-Leu/-Ura and SD/-Leu/-Ura + 100/200/400 mM AbA media. (D) Structure of the reporter and effector vectors used in the dual luciferase assays. (E) A transient expression assay of tobacco leaves showed that MdHHO3 inhibited the expression of MdNRT2.1. Quantitative analysis of LUC/REN activity. An empty vector was used as the control. The data are expressed as the means ± SDs (n = 3). Significant differences were detected by a t-test: *P < 0.05 and **P < 0.01.
Furthermore, to further confirm the direct regulation of MdNRT2.1 expression by MdHHO3, we constructed a dual-effect reporter system with MdHHO3 as the effector gene and MdNRT2.1 as the luciferase reporter gene (Figure 7D). The recombinant fusion plasmid of the MdHHO3 gene and the MdNRT2.1 promoter fragment containing the 5′-GAATC-3′ motif were injected into tobacco leaves. Clearly, the LUC activity of tobacco co-injected with the MdHHO3 and MdNRT2.1 promoters was lower than that of the control group (Figure 7E). These results indicate that MdHHO3 can bind to the promoter of the NRT2.1 gene and inhibit its expression.
Because of nitrate deficiency-induced leaf senescence, MdHHO3 negatively regulates the nitrate starvation response gene MdNRT2.1. Therefore, we speculate that MdHHO3 may play a key role in leaf senescence induced by nitrate deficiency. As expected, nitrate deficiency significantly induced leaf senescence in 2-week-old Arabidopsis after 4 days of treatment under long-day conditions (14 of light:10 h of dark) (Figure 8A). Correspondingly, the chlorophyll content of MdHHO3-overexpressing Arabidopsis was significantly lower than that of the WT control (Figure 8B). During the process of nitrate deficiency-induced leaf senescence, the chlorophyll catabolism-related genes AtSGR1 (Shimoda et al., 2016), AtNYC1 (Kusaba et al., 2007), and AtPAO (Pruzinská et al., 2005) were significantly upregulated, and the overexpression of MdHHO3 substantially increased the expression of these three genes (Figures 8C–E). However, under nitrate supply conditions, the overexpression of MdHHO3 did not significantly induce the expression of these genes, and no significant change in the chlorophyll contents was detected (Supplementary Figures 2C–G). These observations showed that MdHHO3 promotes the induction of leaf senescence in response to nitrate deficiency.
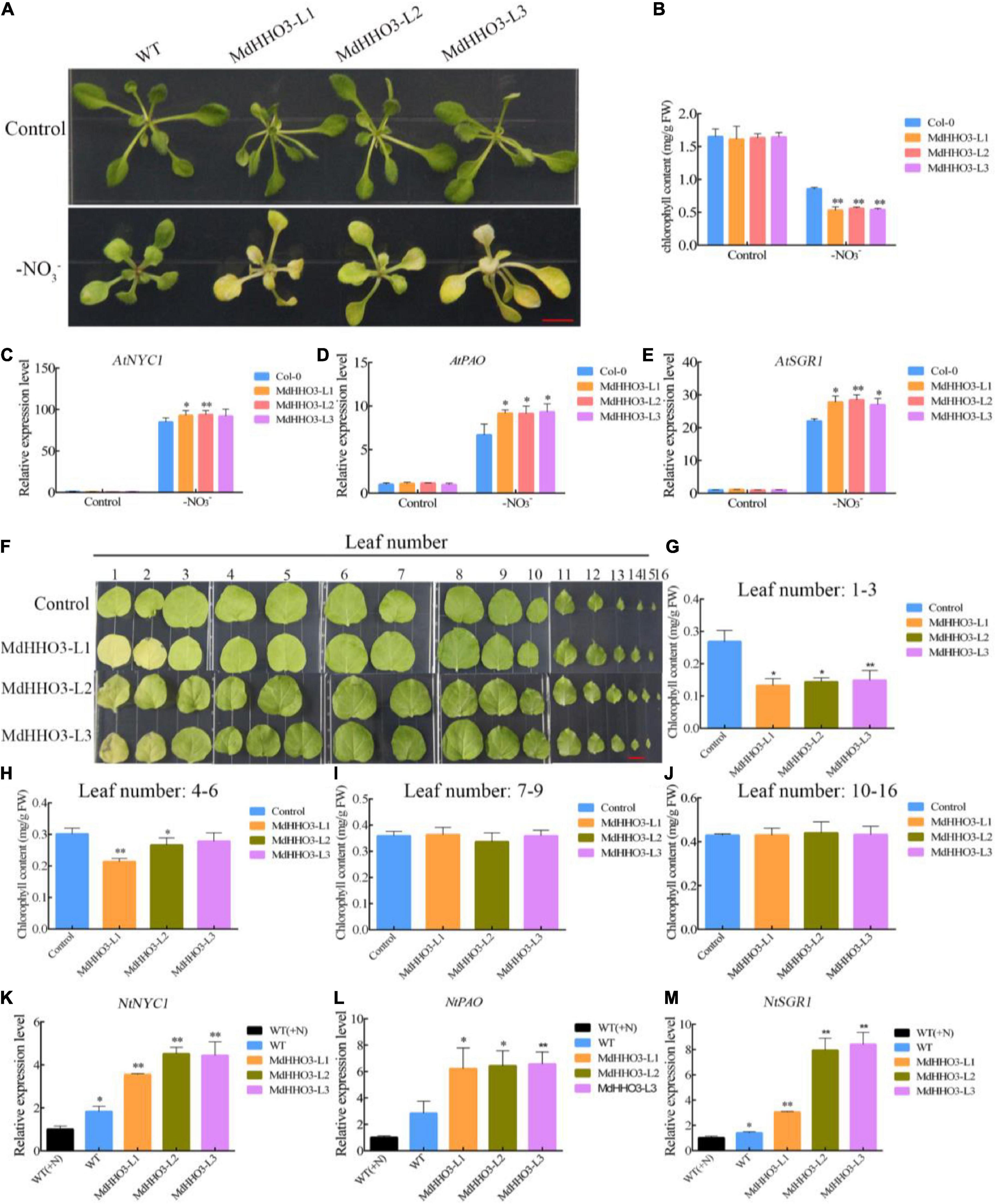
Figure 8. Overexpression of MdHHO3 causes early senescence in Arabidopsis and tobacco. (A) Phenotypes of wild-type (Control) and transgenic Arabidopsis plants (MdHHO3) after 4 days of nitrate deficiency treatment. (B) Total chlorophyll content in Arabidopsis leaves. (C–E) Expression of the senescence-related genes (AtSGR1, AtNYC1, and AtPAO) in Arabidopsis after exposure to nitrate deficiency for 4 days. (F) Phenotypes of wild-type (Control) and transgenic tobacco (MdHHO3) leaves after 30 days of nitrate deficiency treatment. (G–J) Total chlorophyll content in tobacco leaves 1–3, 4–6, 7–9, and 10–16. (K–M) Expression of the senescence-related genes (NtSGR1, NtNYC1, and NtPAO) in tobacco after exposure to nitrate deficiency for 30 days. The data are expressed as the means ± SDs (n = 3). Significant differences were detected by a t-test: *P < 0.05 and **P < 0.01.
Considering that Arabidopsis NIGT1 directly represses the expression of cytokinin synthase enzyme genes thereby leading to a semi-dwarf phenotype in the NIGT1.1 overexpressing Arabidopsis (Maeda et al., 2018). We examined the expression levels of the high-affinity nitrate transporter genes (NRT2.1, NRT2.4, and NRT2.5), as well as the cytokinin synthesis enzyme genes (CYP735A2, IP3, and ARR7) in the wild-type and MdHHO3 transgenic lines (Supplementary Figure 5). The expression of the high-affinity nitrate transporter genes (AtNRT2.1, AtNRT2.4, and AtNRT2.5) were downregulated in the MdHHO3-overexpressing Arabidopsis plants. Under nitrate-free conditions, the expression of the high-affinity transporter genes was upregulated in both the wild-type control and MdHHO3-overexpressing Arabidopsis plants. However, the expression level of these three genes in wild-type control was significantly higher than that in the MdHHO3-overexpressing Arabidopsis. This suggests that MdHHO3 inhibits the expression of the high-affinity transporter genes more significantly under N-deficient conditions. In addition, we examined the expression levels of genes (AtCYP735A2, AtIPT3, and AtARR7) related to cytokinin synthesis. Under N supply conditions, we did not find the effect of overexpression of MdHHO3 on the expression of the cytokinin synthesis-related genes. However, during N deficiency-induced leaf senescence, the expression levels of these three cytokinin synthesis-related genes decreased in both MdHHO3-overexpressing and wild-type control plants, but more significantly in MdHHO3-overexpressing plants. These results suggest that nitrate deficiency inhibits the expression of the cytokinin synthesis-related genes, and MdHHO3 may play an important role in cytokinin synthesis, but further studies are needed.
In a similar experiment, transgenic tobacco seedlings were generated with an MdHHO3 overexpression construct to further investigate the function of MdHHO3. As shown in Figure 8F, plants with different leaf numbers produced different senescence phenotypes after exposure to nitrate deficiency. We compared the chlorophyll content of leaves 1–3, 4–6, 7–9, and 10–16 between the overexpressed plants and the WT control. The changes in the chlorophyll content detected in leaves 1–3 and 4–6 were very similar, which showed that the overexpression of MdHHO3 resulted in lower chlorophyll content, but no significant difference in the chlorophyll content was found in leaves 7–9 and 10–16 (Figures 8G–J). During the process, we monitored the expression levels of the senescence-related genes NtSGR1, NtNYC1, and NtPAO, which may be the target genes of MdHHO3 under nitrate-free conditions (Figures 8K–M). The expression of these three target genes was induced by nitrate deficiency in the WT control. The induction level in the MdHHO3-overexpressing plants was 2–4-fold higher than that in the WT plants. The expression levels of these senescence-related genes were consistent with the leaf senescence phenotype. These results indicate that nitrate deficiency induces the expression of the senescence-related genes in MdHHO3-overexpressing plants and that MdHHO3 is a positive regulator of nitrate deficiency-induced leaf senescence.
Nitrate serves as a signaling molecule, and its regulation of plant gene expression is very complicated (Tsay et al., 2011). The supply of nitrate can induce the expression of genes related to nitrate transport and assimilation (Scheible et al., 2004). In addition, nitrate is an important N source that plants absorb from the soil, and N deficiency induces leaf senescence (Agüera et al., 2010; Krapp et al., 2014). Therefore, revealing the complex molecular mechanism of nitrate absorption by fruit trees and exploring the network through which nitrate deficiency regulates leaf senescence are of great significance to apple production.
GARPs are transcription factors involved in nutritional responses, and many GARPs have been identified in several plants (Safi et al., 2017). For example, 56 GARP family genes have been identified in Arabidopsis (Riechmann et al., 2000), 52 GARP family genes have been found in rice (Zeng et al., 2018), and 59 GARP family genes have been detected in maize (Liu et al., 2016). In this study, the GARP family in apple contains 90 genes and is divided into five groups based on their evolutionary relationship. MdHHO3 belongs to the V subfamily of the GARP gene family and is the closest evolutionary relationship with Arabidopsis At1g25550 (AtHHO3/AtNIGT1.1) and At1g68670 (AtHHO2/AtNIGT1.2). Previous studies have shown that Arabidopsis AtHHO3/AtNIGT1.1 and AtHHO2/AtNIGT1.2 are induced by nitrate treatment (Maeda et al., 2018). In Malus domestica, MdHHO3 was upregulated after nitrate treatment, but downregulated after prolonged nitrate treatment. These results suggested that metabolites downstream of the N metabolism pathway may trigger this negative feedback reaction (Loqué et al., 2003; Nazoa et al., 2003). However, this study revealed that the promoter sequence of the MdHHO3 gene contains its specific binding motif (5′-GAATC-3′). We speculate that over prolonged treatment, MdHHO3 may bind not only to the promoter of downstream genes but also to its own promoter motif, which results in autorepression. In addition, MdHHO3 was found to act as a potential negative regulator in the nitrate response by regulating its own transcription and the nitrate transporter genes (Figures 1, 3, 4). Previous studies have shown that conserved N-terminal CCD (coiled-coil domain) is necessary for NIGT1-NIGT1 interaction (Ueda et al., 2020). In addition, the NIGT1 protein form a dimer via the N-terminal CCD to recognize different types of DNA sequences (Ueda et al., 2020). However, in our study, no conserved CCD was found in MdHHO3 protein. This indicated that the mode of regulation of nutrient responses and target genes by MdHHO3 may differ from that of other NIGT1 family proteins. Therefore, we speculate that there may be additional amino acid residues on the surface of MdHHO3 protein that bind to GAATC or GATTC motifs and effectively repress MdHHO3 expression (Yanagisawa, 2013). These results indicate that the response of nitrate in plants involves not only the regulation of the upstream and downstream genes but also the self-regulation of genes. The regulatory network may be more complex than previously expected.
Subcellular localization analysis is an important method for understanding the specific location of fusion proteins in cells. In rice, the NIGT1-GFP fusion protein is predominantly located in the nucleus (Sawaki et al., 2013). Similarly, the NIGT1-GFP fusion protein is also localized in the nuclei of transgenic Arabidopsis plants overexpressing NIGT1-GFP, and its localization pattern is not altered by the availability of nitrate (Kiba et al., 2018). The subcellular localization analysis performed in this study indicated that the MdHHO3-GFP fusion protein is localized in the nucleus. This localization further indicates that HHO3, as a nuclear protein, is involved in the transcriptional regulation of nitrate.
Nitrate is the main N source of most land plants (Sawaki et al., 2013). The regulation of gene expression induced by nitrate serves as the foundation for plant growth, but the regulatory mechanism of nitrate is largely unknown. Based on our findings, we propose that MdHHO3 may be a key regulator of nitrate-responsive transcription. As a regulator of nitrate-responsive transcription, HHO3 may play a critical role in determining the capacity of nitrate transport, assimilation, and accumulation. In this study, we found that the overexpression of MdHHO3 resulted in a decrease in the nitrate content in apple calli. Correspondingly, the expression of the nitrate transporter gene was significantly downregulated in the presence of the MdHHO3 overexpression construct compared with the level found in the WT control. In contrast, the expression of the nitrate assimilation gene was significantly upregulated in the presence of the MdHHO3 overexpression construct compared with that found with the WT control. Similarly, Arabidopsis and tobacco plants with MdHHO3 overexpression also showed downregulated and upregulated expression of the nitrate transport- and nitrate assimilation-related genes, respectively. Because the nitrate content could be a marker of N utilization efficiency (Chardon et al., 2010), the overexpression of MdHHO3 may negatively affect the N utilization efficiency by inhibiting the transport of nitrate and thus reducing the nitrate content. The content of nitrate directly affects the activity of N assimilating enzymes (Balotf et al., 2016). Previous studies have shown that under nitrate deficiency conditions, plants activate a high-affinity transport system, and thus increase nitrate reductase (NR) activity and mRNA expression (Tischner, 2000; Debouba et al., 2007). However, the expression of the NR and NIR genes was strongly induced by the overexpression of MdHHO3. It is possible that the overexpression of MdHHO3 inhibits the uptake and transport of nitrate, which is equivalent to the results observed in cells with an N-deficient environment.
Arabidopsis NRT2.1 is a typical nitrate-inducing gene that is essential for plant growth at low nitrate concentrations (Li et al., 2007). Several genes involved in the regulation of NRT2.1 expression have been identified. NIN-like protein 7 (NLP7) is involved in the response of plants to nitrate and can bind to the NRT2.1 promoter to enhance its expression (Marchive et al., 2013; Maeda et al., 2018). BT1/BT2 acts as a nitrate inhibitor to negatively regulate the expression of NRT2.1 and reduce the utilization efficiency of N under low nitrate concentrations (Araus et al., 2016). AtTCP20 could bind to the promoter of NRT2.1 and play a key role in the nitrate response (Guan et al., 2014), and TGA1 can directly bind to the promoter of NRT2.1 and promote its expression (Alvarez et al., 2014). In this study, a series of assays showed that MdHHO3 can also bind to the promoter of NRT2.1 and inhibit its expression (Figure 7). NRT2.1 is a typical nitrate-induced gene, which is indispensable for plant growth under low-nitrate concentration (Li et al., 2007). The expression of NRT2.1 was rapidly induced after supplying nitrate to nitrate deficient-cultivated plants, but was suppressed after a few hours (Zhuo et al., 1999), indicating that there was a transcriptional repress regulatory mechanism. Previous studies have shown that HHOs as important regulators of N starvation responses and play an important role in N starvation responses (Kiba et al., 2018). These results suggested that MdHHO3 may be a transcriptional repressor.
Leaf senescence is a complex regulatory process that serves as a comprehensive response to endogenous development and external environmental signals (Woo et al., 2004; Meng et al., 2016). Therefore, some genes involved in the response to environmental changes may be involved in leaf senescence. N is an important macronutrient absorbed by plants, and its deficiency induces leaf senescence (Schulte auf ’m Erley et al., 2007; Agüera et al., 2010). Some genes involved in N-deficiency-induced leaf senescence have been identified. Previous studies have shown that NRT1.7, a phloem nitrate transporter, delays N deficiency-induced leaf senescence (Fan et al., 2009). NLA, a N-limited adaptation regulator, regulates nitrate deficiency-induced leaf senescence by regulating the homeostasis of the ORE1 proteins (Park et al., 2018). The induction of leaf senescence by N deficiency is well-known, but its potential regulatory mechanism remains largely unknown. In our study, we found that the expression of MdHHO3 and the chlorophyll catabolism-related genes (MdNYC1, MdPAO, and MdSGR1) were significantly upregulated during N deficiency-induced leaf senescence (Supplementary Figure 6). Furthermore, under nitrate deficiency conditions, MdHHO3-overexpressing Arabidopsis and tobacco showed early senescence symptoms and a lower chlorophyll level than the WT controls, and a series of genes related to leaf senescence also showed higher expression levels in the overexpressing plants than in the WT controls. The results indicated that MdHHO3 may participate in nitrate deficiency-induced chlorophyll catabolism, but this hypothesis needs further research. In addition, another study found that N supply fails to induce early leaf senescence in Arabidopsis overexpressing MdHHO3 (Supplementary Figure 3). These results suggest that MdHHO3 may be a positive regulator of N deficiency-induced leaf senescence.
A working model was proposed based on the results of this study (Figure 9). Under nitrate supply, MdHHO3 binds to the promoter of MdNRT2.1 and inhibits its expression. With prolonged nitrate supply, MdHHO3 binds to its own promoter and inhibits its expression to reduce the nitrate content. Under nitrate deficiency conditions, MdHHO3 promoted leaf senescence induced by N deficiency and the expression of senescence related genes. These results might help us better understand the regulatory mechanisms of nitrate transport and nitrate deficiency-induced leaf senescence.
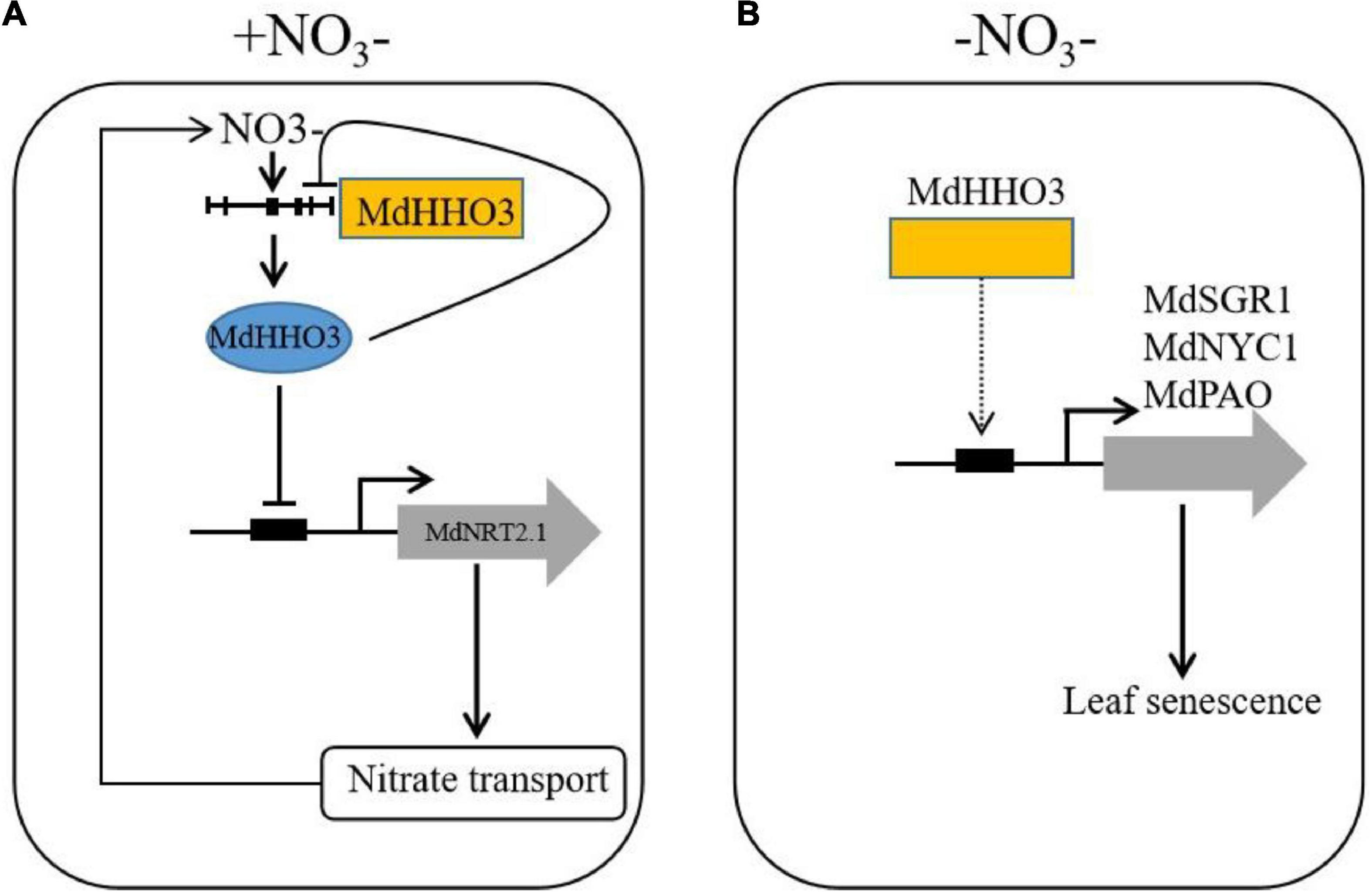
Figure 9. Working model of the function of MdHHO3 in promoting leaf senescence in response to nitrate. (A) Transcriptional regulatory model of the modulation of nitrate uptake. Under the condition of nitrate supply, nitrate activates the MdHHO3 clade gene promoters and is then repressed by the expression of MdHHO3. MdHHO3 binds to the MdNRT2.1 promoter and inhibits its expression. (B) Model of MdHHO3 regulating leaf senescence. In the absence of nitrate, MdHHO3 regulates the expression of the senescence-related genes and thus promotes leaf senescence.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
LL, QT, and BW designed the study. WD, XG, XC, DL, and XF performed the experiments and analyzed the data. BW wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was funded by the Shandong Province Major Science and Technology Innovation Project (2018CXGC0209), the Shandong Provincial Fruit Industry Technology System—Cultivation and Soil Fertilization Post (Grant No. SDAIT-06-04), and the Natural Science Foundation of Shandong Province (ZR2020ZD18).
We are grateful to the editor and reviewers for their valuable suggestions and comments, which improved our manuscript. We thank Futian Peng, Wei Xiao, and Hongyu Wu for their available suggestions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer D-GH declared a shared affiliation with the authors to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.932767/full#supplementary-material
Agüera, E., Cabello, P., and Haba, P. D. L. (2010). Induction of leaf senescence by low nitrogen nutrition in sunflower (Helianthus annuus) plants. Physiol. Plant. 138, 256–267. doi: 10.1111/j.1399-3054.2009.01336.x
Alvarez, J. M., Riveras, E., Vidal, E. A., Gras, D. E., and Gutiérrez, R. A. (2014). T approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 80, 1–13. doi: 10.1111/tpj.12618
An, J. P., Zhang, X. W., Bi, S. Q., You, C. X., Wang, X. F., and Hao, Y. J. (2018b). MdbHLH93, an apple activator regulating leaf senescence, is regulated by ABA and MdBT2 in antagonistic ways. New Phytol. 222, 735–751. doi: 10.1111/nph.15628
An, J. P., Yao, J. F., Xu, R. R., You, C. X., Wang, X. F., and Hao, Y. J. (2018a). Apple bZIP transcription factor MdbZIP44 regulates ABA-promoted anthocyanin accumulation. Plant Cell Environ. 2018, 2678–2692. doi: 10.1111/pce.13393
Araus, V., Vidal, E. A., Puelma, T., Alamos, S., Mieulet, D., Guiderdoni, E., et al. (2016). Members of BTB gene family regulate negatively nitrate uptake and nitrogen use efficiency in Arabidopsis thaliana and Oryza sativa. Plant Physiol. 171, 1523–1532.
Balotf, S., Kavoosi, G., and Kholdebarin, B. (2016). Nitrate reductase, nitrite reductase, glutamine synthetase, and glutamate synthase expression and activity in response to different nitrogen sources in nitrogen-starved wheat seedlings. Biotechnol. Appl. Biochem. 63, 220–229. doi: 10.1002/bab.1362
Chardon, F., Barthélémy, J., Daniel-Vedele, F., and Masclaux-Daubresse, C. (2010). Natural variation of nitrate uptake and nitrogen use efficiency in Arabidopsis thaliana cultivated with limiting and ample nitrogen supply. J. Exp. Bot. 61, 2293–2302. doi: 10.1093/jxb/erq059
Chen, H., Zhang, Q., Wang, X. R., Zhang, J. H., Ismail, A. M., and Zhang, Z. H. (2021). Nitrogen form-mediated ethylene signal regulates root-to-shoot K+ translocation via NRT1.5. Plant Cell Environ. 44, 3576–3588. doi: 10.1111/pce.14182
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacteriummediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Debouba, M., Maâroufi-Dghimi, H., Suzuki, A., Ghorbel, M. H., and Gouia, H. (2007). Changes in Growth and Activity of Enzymes Involved in Nitrate Reduction and Ammonium Assimilation in Tomato Seedlings in Response to NaCl Stress. Ann. Bot. 99, 1143–1151. doi: 10.1093/aob/mcm050
Fan, S. C., Lin, C. S., Hsu, P. K., Lin, S. H., and Tsay, Y. F. (2009). The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21, 2750–2761. doi: 10.1105/tpc.109.067603
Gallardo, A., Parama, R., and Covelo, F. (2006). Differences between soil ammonium and nitrate spatial pattern in six plant communities. Simulated effect on plant populations. Plant Soil. 279, 333–346.
Guan, P., Wang, R., Nacry, P., Breton, G., Kay, S. A., Pruneda-Paz, J. L., et al. (2014). Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 111, 15267–15272. doi: 10.1073/pnas.1411375111
Ho, C. H., Lin, S. H., Hu, H. C., and Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194.
Hoagland, D. R., and Arnon, D. I. (1950). The water culture method for growing plants without soil. Calif. Agric. Exp. Stn. Circular 347, 4–32.
Horsch, R. B., Fry, J. E., Hoffman, N. L., Eichholtz, D., Rogers, S. A., and Fraley, R. T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1232.
Jackson, R. B., and Caldwell, M. M. (1993). The scale of nutrient heterogeneity around individual plants and its quantification with geostatistics. Ecology 74, 612–614.
Jin, J. P., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J. C., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Kant, S., Peng, M., and Rothstein, S. J. (2011). Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 7:e1002021. doi: 10.1371/journal.pgen.1002021
Kiba, T., Inaba, J., Kudo, T., Ueda, N., Konishi, M., Mitsudas, N., et al. (2018). Repression of Nitrogen-Starvation Responses by Members of the Arabidopsis GARP-Type Transcription Factor NIGT1/HRS1 Subfamily. Plant Cell 30, 925–945. doi: 10.1105/tpc.17.00810
Kiba, T., and Krapp, A. (2016). Plant nitrogen acquisition under low availability: regulation of uptake and root architecture. Plant Cell. Physiol. 57, 707–714.
Krapp, A., David, L. C., Chardin, C., Girin, T., Marmagne, A., Leprince, A. S., et al. (2014). Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 65, 789–798.
Kusaba, M., Ito, H., Morita, R., Iida, S., Sato, Y., Fujimoto, M., et al. (2007). Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell. 19, 1362–1375. doi: 10.1105/tpc.106.042911
Lezhneva, L., Kiba, T., Feriabourrellier, A. B., Lafouge, F., Boutetmercey, S., Zoufan, P., et al. (2014). The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 80, 230–241. doi: 10.1111/tpj.12626
Li, W. B., Wang, Y., Okamoto, M., Crawford, N. M., Yaeesh Siddiqi, M., and Glass, A. D. M. (2007). Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143, 425–433. doi: 10.1104/pp.106.091223
Liu, F., Xu, Y., Han, G., Zhou, L., Ali, A., Zhu, S., et al. (2016). Molecular evolution and genetic variation of G2-like transcription factor genes in maize. PLoS One 11:e0161763. doi: 10.1371/journal.pone.0161763
Loqué, D., Tillard, P., Gojon, A., and Lepetit, M. (2003). Gene expression of the NO3– transporter NRT1.1 and the nitrate reductase NIA1 is repressed in Arabidopsis roots by NO2–, the product of NO3– reduction. Plant Physiol. 132, 958–967. doi: 10.1104/pp.102.018523
Maeda, Y., Konishi, M., Kiba, T., Sakuraba, Y., Sawaki, N., Kurai, T., et al. (2018). A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 9:1376. doi: 10.1038/s41467-018-03832-6
Marchive, C., Roudier, F., Castaings, L., Bréhaut, V., Blondet, E., Colot, V., et al. (2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4:1713. doi: 10.1038/ncomms2650
Meng, S., Peng, J. S., He, Y. N., and Zhang, G. B. (2016). Arabidopsis NRT1.5 mediates the suppression of nitrate starvation-induced leaf senescence by modulating foliar potassium level. Mol. Plant. 9, 461–470. doi: 10.1016/j.molp.2015.12.015
Miller, A. J., and Cramer, M. D. (2004). Root nitrogen acquisition and assimilation. Plant Soil. 27, 1–36.
Miller, A. J., Fan, X., Orsel, M., Smith, S. J., and Wells, D. M. (2007). Nitrate transport and signalling. J. Exp. Bot. 58, 2297–2306.
Nacry, P., Bouguyon, E., and Gojon, A. (2013). Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil. 370, 1–29.
Nazoa, P., Vidmar, J. J., Tranbarger, T. J., Mouline, K., Damiani, I., Tillard, P., et al. (2003). Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: responses to nitrate, amino acids and developmental stage. Plant Mol. Biol. 52, 689–703. doi: 10.1023/a:1024899808018
Orsel, M., Eulenburg, K., Krapp, A., and Daniel-Vedele, F. (2019). Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 219, 714–721. doi: 10.1007/s00425-004-1266-x
Park, B. S., Yao, T., Seo, J. S., Wong, E. C. C., Mitsuda, N., Huang, C. H., et al. (2018). Arabidopsis NITROGEN LIMITATION ADAPTATION regulates ORE1 homeostasis during senescence induced by nitrogen deficiency. Nat. Plants 4, 898–903. doi: 10.1038/s41477-018-0269-8
Pruzinská, A., Tanner, G., Aubry, S., Anders, I., Moser, S., Müller, T., et al. (2005). Chlorophyll breakdown in senescent arabidopsis leaves. characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol. 139, 52–63. doi: 10.1104/pp.105.065870
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Safi, A., Medici, A., Szponarski, W., Ruffel, S., Lacombe, B., and Krouk, G. (2017). The world according to GARP transcription factors. Curr. Opin. Plant Biol. 39, 159–167. doi: 10.1016/j.pbi.2017.07.006
Sawaki, N., Tsujimoto, R., Shigyo, M., Konishi, M., Toki, S., Fujiwara, T., et al. (2013). A nitrate-inducible GARP family gene encodes an auto-repressible transcriptional repressor in rice. Plant Cell Physiol. 54, 506–517. doi: 10.1093/pcp/pct007
Scheible, W. R., Morcuende, R., Czechowski, T., Fritz, C., and Stitt, M. (2004). Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 136, 2483–2499. doi: 10.1104/pp.104.047019
Schulte auf ’m Erley, G., Begum, N., Worku, M., Bänziger, M., and Horst, W. J. (2007). Leaf senescence induced by nitrogen deficiency as indicator of genotypic differences in nitrogen efficiency in tropical maize. J. Plant Nutr. Soil Sci. 170, 106–114.
Shimoda, Y., Ito, H., and Tanaka, A. (2016). Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 28, 2147–2160. doi: 10.1105/tpc.16.00428
Tischner, R. (2000). Nitrate uptake and reduction in higher and lower plants. Plant Cell Environ. 23, 1005–1024.
Tsay, Y. F., Ho, C. H., Chen, H. Y., and Lin, S. H. (2011). Integration of nitrogen and potassium signaling. Annu. Rev. Plant Biol. 62, 207–226.
Ueda, Y., Nosaki, S., Sakuraba, Y., Miyakawa, T., Kiba, T., Tanokura, M., et al. (2020). NIGT1 family proteins exhibit dual mode DNA recognition to regulate nutrient response-associated genes in Arabidopsis. PLoS Genet. 16:e1009197. doi: 10.1371/journal.pgen.1009197
Waters, M. T., Wang, P., Korkaric, M., Capper, R. G., Saunders, N. J., and Langdale, J. A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21, 1109–1128. doi: 10.1105/tpc.108.065250
Wen, B. B., Li, C., Fu, X. L., Li, D. M., Li, L., Chen, X. D., et al. (2019). Effects of nitrate deficiency on nitrate assimilation and chlorophyll synthesis of detached apple leaves. Plant Physiol. Biochem. 142, 363–371. doi: 10.1016/j.plaphy.2019.07.007
Woo, H. R., Kim, J. H., Nam, H. J., and Lim, P. O. (2004). The delayed leaf senescence mutants of Arabidopsis, ore1, ore3, and ore9 are tolerant to oxidative stress. Plant Cell Physiol. 45, 923–932. doi: 10.1093/pcp/pch110
Woo, H. R., Kim, H. J., Nam, H. G., and Lim, P. O. (2013). Plant leaf senescence and deathregulation by multiple layers of control and implications for aging in general. J. Cell Sci. 126, 4823–4833. doi: 10.1242/jcs.109116
Yanagisawa, S. (2013). Characterization of a nitrate-inducible transcriptional repressor NIGT1 provides new insights into DNA recognition by the GARP family proteins. Plant Signal Behav. 8:e24447. doi: 10.4161/psb.24447
Zeng, L. P., Liu, X., Zhou, Z. Z., Li, D. Y., Zhao, X. F., Zhu, L. H., et al. (2018). Identification of a G2-like transcription factor, OsPHL3, functions as a negative regulator of flowering in rice by coexpression and reverse genetic analysis. BMC Plant Biol. 18:157. doi: 10.1186/s12870-018-1382-6
Keywords: apple, genome-wide, MdHHO3, nitrate response, nitrate deficiency, senescence
Citation: Wen B, Gong X, Deng W, Chen X, Li D, Fu X, Li L and Tan Q (2022) The apple GARP family gene MdHHO3 regulates the nitrate response and leaf senescence. Front. Plant Sci. 13:932767. doi: 10.3389/fpls.2022.932767
Received: 30 April 2022; Accepted: 19 July 2022;
Published: 09 August 2022.
Edited by:
Paul F. Devlin, Royal Holloway, University of London, United KingdomReviewed by:
Liangju Wang, Nanjing Agricultural University, ChinaCopyright © 2022 Wen, Gong, Deng, Chen, Li, Fu, Li and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Li, TGlsaW5nc2RhdUAxNjMuY29t; Qiuping Tan, ZHVuZHVuaGFwcHlAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.