
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 22 June 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.930592
This article is part of the Research Topic Frontiers in Global Regulatory Landscape of CRISPR-Edited Plants View all 13 articles
The efficiency of the CRISPR/Cas9 genome editing system remains limited in many crops. Utilizing strong promoters to boost the expression level of Cas9 are commonly used to improve the editing efficiency. However, these strategies also increase the risk of off-target mutation. Here, we developed a new strategy to utilize intron-mediated enhancement (IME)-assisted 35S promoter to drive Cas9 and sgRNA in a single transcript, which escalates the editing efficiency by moderately enhancing the expression of both Cas9 and sgRNA. In addition, we developed another strategy to enrich cells highly expressing Cas9/sgRNA by co-expressing the developmental regulator gene GRF5, which has been proved to ameliorate the transformation efficiency, and the transgenic plants from these cells also exhibited enhanced editing efficiency. This system elevated the genome editing efficiency from 14–28% to 54–81% on three targets tested in lettuce (Lactuca sativa) without increasing the off-target editing efficiency. Thus, we established a new genome editing system with highly improved on-target editing efficiency and without obvious increasement in off-target effects, which can be used to characterize genes of interest in lettuce and other crops.
The CRISPR/Cas9 system is a powerful genome editing tool that has been widely used in the past decade (Gao, 2021). With the complementary base pairing mechanism, the Cas9 endonuclease is guided to the specific DNA sequence by the guide RNA (gRNA), and generates double-stranded DNA breaks (DSBs) at the desired loci. Predominantly, the DSBs are repaired by the error-prone non-homologous end joining (NHEJ) pathway, which introduces insertions/deletions (indels) that range from one to hundreds of base pairs, that could lead to site-specific genetic alterations (Gao, 2021; Hassan et al., 2021). Until today, this technology has been successfully used to generate mutant plants and for agronomic trait enhancement in many crops. Nevertheless, the editing efficiency remains quite limited in several vegetable and crop plants.
Boosting the expression of Cas9 or sgRNA is the major method to improve the genome editing efficiency (Castel et al., 2019; Hassan et al., 2021). Several studies have utilized strong promoters, such as the RPS5A promoter (Tsutsui and Higashiyama, 2017; Castel et al., 2019; Ordon et al., 2020; Oh and Kim, 2021), the UBQ10 promoter (Wang and Chen, 2019; Wolabu et al., 2020), and the MAS promoter (An et al., 2021), to strengthen the expression level of Cas9, which leads to increasements in the genome editing efficiency. Also, the augmentation of sgRNA level, by using native U6/U3 promoters (Sun et al., 2015; Ren et al., 2021), or by using Pol II promoters such as the ubiquitin promoter (Ding et al., 2018), or the cestrum yellow leaf curling virus (CmYLCV) promoter (Cermak et al., 2017; Li et al., 2021), escalates genome editing efficiency. Also, the Cas9 with multiple introns efficiently generates more mutants than the conventional Cas9 (Grutzner et al., 2021). However, these strategies also increase the risk of off-target mutation, which might interfere phenotypic analysis of desired genes and more severely hinder deregulation and commercial release of genome-edited crops.
Intron-mediated enhancement (IME) is a well-known phenomenon to enhance homogeneous protein expression in plants and animals (Vain et al., 1996; Laxa, 2016). The introns located in the 5′-UTR region from several strong and constitutive genes, such the first intron of UBQ10, ACTIN, TRP1 (Rose, 2004; Jeong et al., 2009), have been proved to greatly improve the expression of downstream gene. For example, the first intron of maize ubiquitin 1 (ZmUbi1) located in the 5′-UTR region combines with CaMV 35S promoter leads to a over 90-fold increasement of gene expression in maize and bluegrass (Vain et al., 1996). Many works have attempted to identify the key cis-elements in this process, but the detailed mechanism is still not clear, since it has been found that the sequence and splicing process are not the key feathers of these introns (Rose and Beliakoff, 2000; Rose, 2004; Back and Walther, 2021). Thus, it’s promising to engineer these introns to enhance the strength of the promoters that drive the expression of the CRISPR/Cas9 system.
Several DR (DEVELPMENTAL REGULATOR) genes, such as the WUS (WUSCHEL), BBM (BABY BOOM) and GRFs (GROWTH-REGULATING FACTORs), have been proved to improve the transformation efficiency (Lowe et al., 2016; Debernardi et al., 2020; Kong et al., 2020; Qiu et al., 2022). Ectopic expression of the BBM gene, which is originally identified in Brassica napus, has diverse functions in plant cell proliferation, growth and development (Jha and Kumar, 2018). The co-expression of BBM with WUS greatly boosts the transformation efficiency of several monocot species, including rice, maize and sorghum (Lowe et al., 2016). Several plant-specific GRF transcription factors have successfully elevated the regeneration and transformation efficiency of crop plants, such as soybean, canola, and sunflower (Kong et al., 2020; Pan et al., 2022). The overexpression of a chimeric protein consisting of the GRF4 and GRF-interacting factor 1 (GIF1) proteins reinforce the regeneration efficiency and regeneration speed in wheat, triticale, rice and watermelon (Debernardi et al., 2020; Feng et al., 2021; Qiu et al., 2022). However, the effect of these DRs on the genome editing efficiency in the regenerated plants has not been investigated.
Lettuce is one of the most popular vegetable crops that is cultivated worldwide (Su et al., 2020; Assefa et al., 2021). The substantial amounts of ascorbic acid, vitamin A, carotenoids, folate, and other primary and secondary metabolites are beneficial to human health (Assefa et al., 2021). However, the candidate genes behinds these traits are poorly investigated. The CRISPR/Cas9 genome editing system, which is powerful and promising in generating the desired mutants and in crop breeding, has been utilized in the studies of lettuce in recent years (Bertier et al., 2018; Zhang et al., 2018; Luo et al., 2021). However, an improved and efficient genome editing has not been established for lettuce and is urgently needed.
In this work, we established an intron-mediated enhancement-based strategy to increase the expression of Cas9 and sgRNA, and also tested the effect of GRF5 on lettuce (Lactuca sativa) transformation and genome editing. These two methods successfully boosted the genome editing efficiency from 14–28% to 54–81% on three targets tested in lettuce without increasing the off-target editing efficiency.
First, we decided to moderately enhance the expression of both Cas9 and sgRNA through IME. Previous studies have successfully engineered the sgRNA expression cassette tRNA-sgRNA-tRNA into the first intron of ZmUbi1, thus the mature sgRNA can be generated by the endogenous tRNA-processing system (Xie et al., 2015; Zhong et al., 2020). We generated pZKD672 by inserting this engineered intron (Supplementary Table 1) into a 35S promoter-driven Cas9 expression cassette (Figure 1A). In this way, Cas9 and sgRNA were driven by an IME-assisted 35S promoter and co-expressed from a single transcriptional unit. We speculated that the expression levels of both Cas9 and sgRNA could be improved. The pKSE401 vector, in which Cas9 and sgRNA are respectively driven by 35S promoter and Arabidopsis U6-26 promoter (Xing et al., 2014), was used as the control.
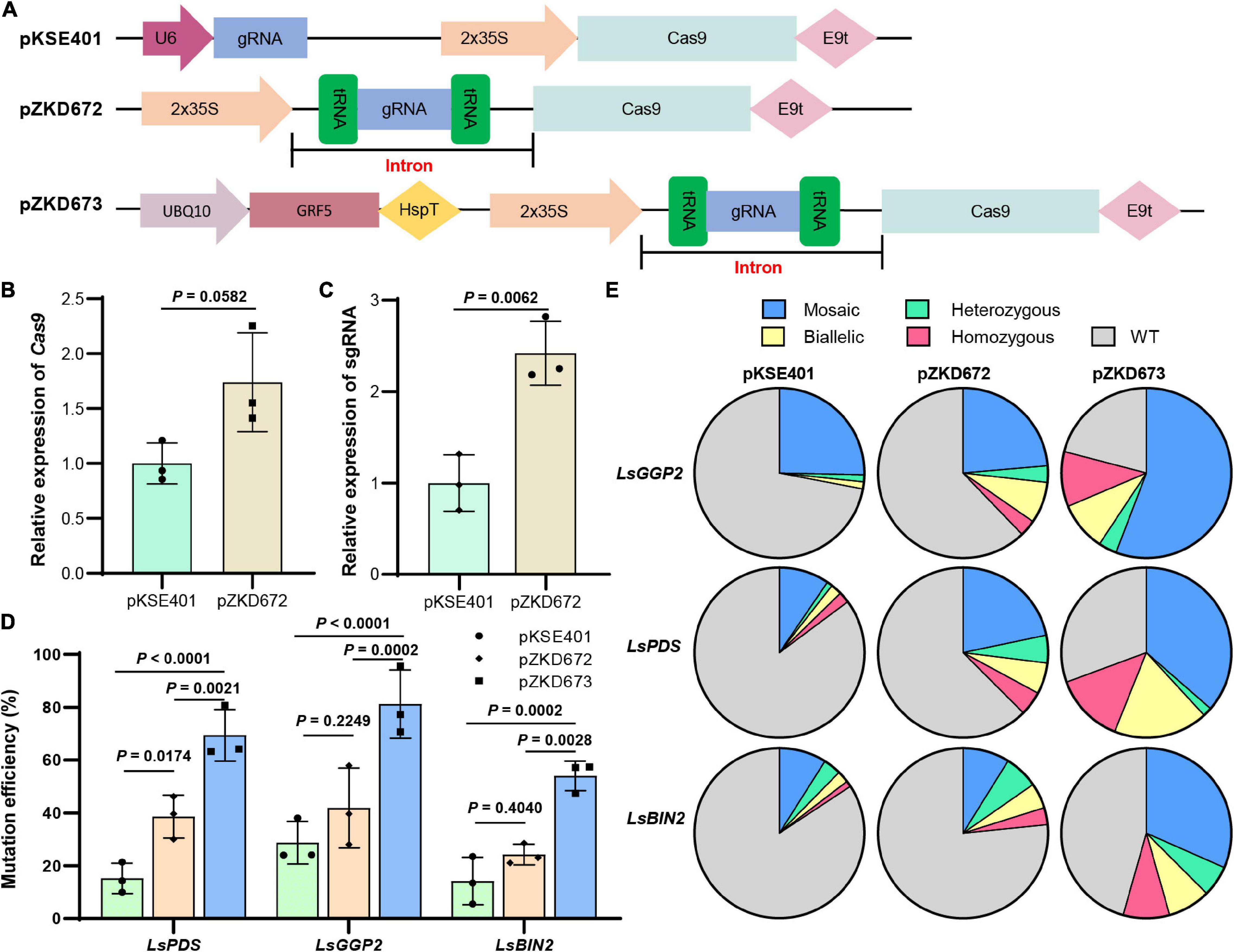
Figure 1. Boost genome editing efficiency through intron-mediated enhancement and GRF5 co-expression. (A) The schematic diagram of the vectors. In the conventional genome vector pKSE401, sgRNA is driven by the U6 promoter, and Cas9 is driven by the CaMV 35S promoter. In pZKD672, engineered intron contain sgRNA was used to boost Cas9 and sgRNA expression through the intron-mediated enhancement mechanisms. The endogenous tRNA-processing system was used to generate mature sgRNA. In pZKD673, the GRF5 driven by the UBQ10 promoter was added to the genome editing vector to facilitate the screening of plants with high expression levels. (B,C) The expression level of Cas9 (B) and sgRNA (C) in lettuce protoplasts using the pKSE401 or the pZKD672 vector. LsACT was used as the internal control. The P value was calculated with paired two-tailed Student’s t-test. (D) The mutation ratio of three tested target sites using the indicated vectors in transgenic lettuce plants in the T0 generation. The P value was calculated with Two-way ANOVA test. (E) The proportion of different mutation types among all the transgenic plants from three replicates.
To test our hypothesis, we utilized pKSE401 and pZDK672 to construct an sgRNA targeting LsPDS (PHYTOENE DESATURASE) in lettuce, and examined the expression of Cas9 and sgRNA in lettuce protoplasts. Compared with those from pKSE401, transcript levels of Cas9 and sgRNA from pZDK672 were increased by 0.74- and 1.42-fold, respectively (Figures 1B,C), proving the power of IME in moderately enhancing the expression of Cas9 and sgRNA.
To investigate whether the pZDK672 could elevate the editing efficiency in stable transgenic plants, we selected LsPDS and two additional target genes, LsBIN2 (BR-INSENSITIVE 2) and LsGGP2 (GDP-L-GALACTOSE PHOSPHORYLASE) to generate stable transgenic lettuces through Agrobacterium-mediated transformation (Zhang et al., 2018). The average mutation efficiencies by the pZKD672 were 38.59, 41.85, and 24.23% for LsPDS, LsGGP2, and LsBIN2 in 3 biological repeats, respectively, while only 15.24, 28.74, and 14.20% transgenic plants were mutated by pKSE401 (Figure 1D and Supplementary Tables 2, 3), with 1.53-, 0.80-, and 0.71-fold increasement. It suggests that the moderate magnification in the expression of Cas9 and sgRNA through IME resulted in a weak augmentation in editing efficiency.
Next, we hoped to optimize pZKD672 to further elevating its editing efficiency. The plant genetic transformation often generates populations with diverse gene expression levels. In the plant genome editing processes, the cells with higher expression levels of Cas9 and sgRNA, which leads to higher mutation rates, are the desired ones for regeneration. Direct enrichment of these cells or plants could also increase the mutation efficiency, such as the GLABRA2 mutation-based visible selection (GBVS) system which adds the GL2 target as a visible selection marker to identify plants with high mutation efficiency (Kong et al., 2021). However, mutation of a second gene might be a concern for crop breeding. This prompted us to explore novel strategies to enrich these cells. Several DRs have been proved to improve the transformation efficiency by promoting the somatic embryogenesis or regeneration rates (Lowe et al., 2016; Kong et al., 2020; Qiu et al., 2022). They have been widely used in plants recalcitrant to transformation (Kong et al., 2020; Pan et al., 2022; Qiu et al., 2022). However, the effect of these DRs on the editing efficiency of recipient plants during stable transformation had not yet been investigated. We surmised that the expression level of DRs should be correlated with that of Cas9 and sgRNA when they were constructed in a single T-DNA. The cells, highly expressing DRs, Cas9 and sgRNA, could gain an advantage over other cells to redifferentiation, hence co-expression of DRs could elevate mutation rate during stable transformation. To verify our speculation, we added an Arabidopsis GRF5 overexpression cassata (Supplementary Table 1) to pZKD672 to generate pZKD673 (Figure 1A). pZKD673 with corresponding target spacers were also transformed into lettuce. The mutation efficiencies of LsPDS, LsGGP2, and LsBIN2 by pZKD673 are 69.38, 81.22, and 54.00% in three biological replicates, respectively, and exhibit 0.80-, 0.94-, and 1.23-fold increase, compared with 38.59, 41.85, and 24.23% by pZKD672, respectively (Figure 1D and Supplementary Tables 2, 3). To further prove our postulate, we randomly selected about 24 transgenic plants for each vector and mixed them into 3 samples to check the expression level of Cas9. The result showed that the expression of Cas9 in pZKD673 transgenic plants is about 2.58-fold higher than the pZKD672 vector (Supplementary Figure 1). Our data indicate that co-expressing GRF5 could improve the editing efficiency of pZKD672.
Among all the T0 transgenic plants, the ratio of null mutants (homozygous and biallelic) was also increased. For example, the amount of lspds null mutants was raised from 4.23% for pKSE401 to 10.53% for pZKD672 and 31.21% for pZKD673 (Figure 1E and Supplementary Table 3). These results demonstrated that our new vectors could generate more null mutants, which are suitable for phenotyping or breeding in the offspring, and are labor- and time-saving.
Finally, to evaluate off-targeting efficiency, five predicted highly risky off-target sites for each target gene were identified through the CRISPOR program1 (Haeussler et al., 2016), and 20 on-target mutant lines for each plasmid were examined (except for the LsBIN2 by the pKSE401, with only 14 mutants obtained). No off-target mutation was detected, even at the off-target 1 (OT1) and OT2 of LsGGP2, and the OT1 of LsBIN2, which have 2 mismatches with the corresponding target sequence (Table 1 and Supplementary Table 4). These results indicated our newly established systems do not increase off-target efficiency.
In this study, we established a new genome editing system for creating mutations with high frequency in lettuce. With an intron expressing the sgRNA, and GRF5-mediated enrichment, we dramatically boosted the mutation efficiency compared with the commonly used vector in transgenic lettuce plants.
Successful engineering of introns to express sgRNA has been reported in other studies (Ding et al., 2018; Zhong et al., 2020). In these studies, the intron is inserted into the 5′-UTR or within the coding region of Cas9. All these experiments were conducted in rice, and compared to traditional genome editing vectors, these sgRNA containing introns didn’t significantly improved the editing efficiency (Ding et al., 2018; Zhong et al., 2020). This is probably because that the rice UBQ10 promoter or the maize Ubi promoter is used to drive Cas9. These Ubiquitin promoters itself contains the introns with IME. Therefore, additional adding of another IME introns probably doesn’t make functions. In this work, the genome editing is conducted in a dicot plant lettuce, and the most widely used 35S promoter is used to drive Cas9. It has been well proved that adding an IME intron could significantly boost the power of 35S promoter (Vain et al., 1996; Laxa, 2016). Our result proved that this modified intron could indeed improve the activity of the 35S promoter that expresses the Cas9. And it should be pointed out that the IME intron strategy might not be applicable to all the genome editing vectors, and not all the plant species.
In order to generate mature sgRNAs within the intron, the tRNA sequence was placed upstream and downstream of the spacer-sgRNA sequence. It has been well proved that these polycistronic gene can be processed by the endogenous tRNA-processing enzyme RNAse Z and RNAse P (Xie et al., 2015; Zhong et al., 2020). Successfully genome editing of endogenous targets in our experiments also confirmed these results. In addition to the tRNA-processing system, other sgRNA processing system, such as the dual HH-HDV ribozyme system (Gao and Zhao, 2014), or the sequence-specific RNase Csy4 (Przybilski et al., 2011), has also been used to express sgRNA in plants. In these experiments, the tRNA-processing system enables efficient sgRNA expression by the Pol II promoters, and efficient multiplex genome editing (Xie et al., 2015; Li et al., 2021). Also, the tRNA-processing system exhibited higher or comparable processing efficiency and mutation rates than the ribozyme system and the Csy4 system in these experiments (Tang et al., 2019; Hsieh-Feng and Yang, 2020; Zhong et al., 2020). Thus, our new vectors are promising in efficient multiplex genome editing in lettuce, and other dicot plants.
The power of DRs in genetic transformation has been observed in many plant species. With the assistance of DRs such as GRF5, efficient and genotype-independent transformation can be achieved without obvious growth abnormities, and this system has been used to generate mutants by the CRISPR/Cas9 genome editing system (Debernardi et al., 2020; Pan et al., 2022). In our previous experiments, we demonstrated that GRF5 outperforms other DR genes, such as GRF4-GIF1, BBM and WUS, in the genetic transformation of watermelon (Citrullus lanatus) (Pan et al., 2022). However, the effect of GRF5 on the genome editing efficiency has not been investigated. In our work, we proved that overexpressing the GRF5 gene could dramatically elevate the genome editing efficiency. We suspect that this is a transgenic enrichment effect: as the GRF5 and Cas9 are constructed in a single T-DNA, efficient expression of Cas9 should co-relate with efficient expression of DRs, which facilitates the regeneration process. Thus, most of the transgenic plants we obtained with the pZKD673 vectors should have higher mutation efficiency. Our observation that the expression level of Cas9 is indeed higher in pZKD673 transgenic plants than the pZKD672 transgenic plants confirmed our hypothesis. Very recently, another group also observed the same mutation efficiency increasement by co-expressing WUS in sorghum (Che et al., 2022), but the detailed mechanism hasn’t been revealed. These works revealed the power of DRs, not only on genotype-independent genetic transformation, but also in efficient genome editing.
With great improvement in the genome editing efficiency, we can easily obtain large number of mutants. And the efficiency amplification also leads to higher ratio of homozygous and biallelic mutants in the T0 generation. What’s more, we didn’t observe significant increasement in the off-target mutation efficiency. Thus, these homozygous and biallelic mutants could directly be used for phenotype analysis and functional verification, which could save plenty of time and efforts.
In summary, we utilized two novel strategies, IME-mediated the moderate enhancement of Cas9/sgRNA expression and DR gene-associated transgene enrichment, to establish a highly efficient plant genome editing system without obvious off-targeting increase. These strategies could also be applied in other genome editing tools, such as base-editors and the prime-editors, and other crop species, to boost the editing efficiency.
The L. sativa L. var. capitata 101 was bought from Jingyan Yinong (Beijing) Seed Sci-Tech Co., Ltd. Plants were grown under a photoperiod of 16 h light (150 μmol m–2 s–1) and 8 h dark at 25°C.
The PTG sequence and codon optimized GRF5 coding sequence were synthesized at Sangon Biotech. The conventional CRISPR/Cas9 vector pKSE401 (Xing et al., 2014) were used as the control. The pKSE401 vector was first digested by HindIII, and the 14.5 kb backbone were ligated by T4 DNA ligase. The product was then digested by XbaI, and Gibson assembled (Sangon Biotech) with the PTG product amplified with the primer pair PTG-F/PTG-R, generating the pZKD672 vector.
The Arabidopsis UBQ10 promoter, the Arabidopsis Hsp terminator and codon optimized GRF5 coding sequence were amplified with UBQ10p-F/UBQ10p-R, HspT-F/HspT-R and GRF5-F/GRF5-R, respectively. The PCR products were then used as the template and amplified with UBQ10p-F/HspT-R. The 2.75 kb produce were then Gibson assembled with HindIII digested pZKD672, generating the pZKD673 vector.
The pKSE401, pZKD672, and pZKD673 vector were digested with BsaI, and ligated with annealed target oligos.
The primers were listed in Supplementary Table 5.
The lettuce protoplast preparation and transfection were performed according to a previous established method (Woo et al., 2015) with some modifications. Briefly, the heart of L. sativa L. var. capitata L 101 was sliced with double sides razor blades. The leaves were then digested with 1% Cellulase R10, 0.25% Macerozyme R10, 0.4 M Mannitol, 20 mM KCl, 20 mM MES pH 5.7, 20mM KCl for about 4 h. the enzyme solution was filtered with Miracloth (CALBIOCHEM), and collected by Centrifuged for 1 min at 100 g. The protoplasts were washed twice by 10 mL W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES pH 5.7. Then the protoplasts were suspended by 10 mL W5 solution and stand on ice for 30 min. The supernatant was removed and the MMG solution (0.4 M Mannitol, 15 mM MgCl2, 4 mM MES pH 5.7) was added to a final concentration of 2 × 104 to 2 × 105 cell/ml.
Ten microgram vector were mixed with 200 μL protoplast and mixed gently. Then 220 μL PEG/CaCl2 solution (40% PEG4000, 0.2 M Mannitol, 100 mM CaCl2) was added and mixed gently. The transfection was performed at room temperature for 15 min and stopped by adding 1.5 mL W5 solution and mixed gently. The protoplast was then washed by 1 mL W5 solution and risen in 200 μL. Then the protoplasts were kept at dark for 2 days at 24°C.
The protoplasts were collected and total RNA were extracted with the Ultrapure RNA Kit (CWbio). The RNA was reverse transcript with the FastQuant RT Kit (With gDNase) (Tiangen) with some modification: 0.5 μL 10 μM qRT-LsPDS-R primer was added to a final 20 μL reverse transcription mixture. The quantitative real-time PCR were performed with CFX Opus real-time PCR system (BioRad) with the Talent qPCR PreMix (SYBR Green) (Tiangen). The corresponding primers were listed in supplementary Table 5.
The protocol for lettuce transfection was previously described (Zhang et al., 2018). In brief, surface sterilized lettuce seeds were placed on MS medium and incubated under a photoperiod of 16 h light (150 μmol m–2 s–1) and 8 h dark at 25°C. The cotyledons were excised from germinated seedlings and incubated for 10 min with the Agrobacterium (EHA105) suspension carrying the desired construct. The treated explants were placed on MS co-cultivation medium (MS supplemented with 30 g L–1 sucrose, 0.8% plant agar, 0.1 mg L–1 α-naphthalaneacetic acid, and 0.5 mg L–1 6-benzylaminopurine) and incubated at 25°C in dark for 48 h.
Afterward, explants were transferred to MS selection medium (MS supplemented with 30 g L–1 sucrose, 0.8% plant agar, 0.1 mg L–1 α-naphthalaneacetic acid, 0.5 mg L–1 6-benzylaminopurine, 40 mg/l kanamycin monosulfate, and 250 mg L–1 carbenicilin), and incubated under a 16 h light/8 h dark cycle at 25°C. After about 25 days, regenerated shoots were excised and transferred to MS rooting medium (1/2 MS supplemented with 15 g L–1 sucrose, 20 mg L–1 kanamycin monosulfate, and 250 mg L–1 carbenicilin) for root induction. The plantlets with well-developed shoot and root were transferred to soil and further examined.
The genomic DNA of regenerated lettuce plants was extracted with the CTAB method. Positive transgenic plants were examined with the Cas9-check-F2/Cas9-Check-R2 primer pair. The target regions were amplified with corresponding primer pairs, and the analyzed with Sanger sequencing. The sequencing chromatogram were decoded with the TIDE program2 (Brinkman et al., 2014).
The potential off-target editing sites were chosen through the CFD score in the CRISPOR program3(Haeussler et al., 2016). For each vector, 20 mutant plants were randomly chosen and the target regions were amplified with the corresponding primer pairs. The PCR products were analyzed with Sanger sequencing.
The primers were listed in Supplementary Table 5.
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
HZ conceived the study and agreed to serve as the author responsible for contact and ensures communication. HZ and DL supervised the research. WP and XL performed all experiments and analyzed the data with help from HZ and DL. All authors contributed to the article and approved the submitted version.
This work was supported by grants from Taishan Scholar Foundation of Shandong Province (tsqn202103160), Excellent Youth Foundation of Shandong Scientific Committee (ZR202103010168) for HZ, the China Postdoctoral Science Foundation (No. 2021T140017) for WP, and the Beijing Academy of Agriculture and Forestry Science (KJCX20220104) for DL.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Qijun Chen from China Agricultural University for sharing the pKSE401 plasmid.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.930592/full#supplementary-material
An, Y., Geng, Y., Yao, J., Wang, C., and Du, J. (2021). An Improved CRISPR/Cas9 System for Genome Editing in Populus by Using Mannopine Synthase (MAS) Promoter. Front. Plant Sci. 12:703546. doi: 10.3389/fpls.2021.703546
Assefa, A. D., Hur, O. S., Hahn, B. S., Kim, B., Ro, N. Y., and Rhee, J. H. (2021). Nutritional Metabolites of Red Pigmented Lettuce (Lactuca sativa) Germplasm and Correlations with Selected Phenotypic Characters. Foods 10:2504. doi: 10.3390/foods10102504
Back, G., and Walther, D. (2021). Identification of cis-regulatory motifs in first introns and the prediction of intron-mediated enhancement of gene expression in Arabidopsis thaliana. BMC Genomics 22:390. doi: 10.1186/s12864-021-07711-1
Bertier, L. D., Ron, M., Huo, H., Bradford, K. J., Britt, A. B., and Michelmore, R. W. (2018). High-Resolution Analysis of the Efficiency, Heritability, and Editing Outcomes of CRISPR/Cas9-Induced Modifications of NCED4 in Lettuce (Lactuca sativa). G3 8, 1513–1521. doi: 10.1534/g3.117.300396
Brinkman, E. K., Chen, T., Amendola, M., and Van Steensel, B. (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42:e168. doi: 10.1093/nar/gku936
Castel, B., Tomlinson, L., Locci, F., Yang, Y., and Jones, J. D. G. (2019). Optimization of T-DNA architecture for Cas9-mediated mutagenesis in Arabidopsis. PLoS One 14:e0204778. doi: 10.1371/journal.pone.0204778
Cermak, T., Curtin, S. J., Gil-Humanes, J., Cegan, R., Kono, T. J. Y., and Konecna, E. (2017). A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants. Plant Cell 29, 1196–1217. doi: 10.1105/tpc.16.00922
Che, P., Wu, E., Simon, M. K., Anand, A., Lowe, K., Gao, H., et al. (2022). Wuschel2 enables highly efficient CRISPR/Cas-targeted genome editing during rapid de novo shoot regeneration in sorghum. Commun. Biol. 5:344. doi: 10.1038/s42003-022-03308-w
Debernardi, J. M., Tricoli, D. M., Ercoli, M. F., Hayta, S., Ronald, P., Palatnik, J. F., et al. (2020). A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38, 1274–1279. doi: 10.1038/s41587-020-0703-0
Ding, D., Chen, K., Chen, Y., Li, H., and Xie, K. (2018). Engineering Introns to Express RNA Guides for Cas9- and Cpf1-Mediated Multiplex Genome Editing. Mol. Plant 11, 542–552. doi: 10.1016/j.molp.2018.02.005
Feng, Q., Xiao, L., He, Y., Liu, M., Wang, J., Tian, S., et al. (2021). Highly efficient, genotype-independent transformation and gene editing in watermelon (Citrullus lanatus) using a chimeric ClGRF4-GIF1 gene. J. Integr. Plant Biol. 63, 2038–2042. doi: 10.1111/jipb.13199
Gao, C. (2021). Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635. doi: 10.1016/j.cell.2021.01.005
Gao, Y., and Zhao, Y. (2014). Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 56, 343–349. doi: 10.1111/jipb.12152
Grutzner, R., Martin, P., Horn, C., Mortensen, S., Cram, E. J., Lee-Parsons, C. W. T., et al. (2021). High-efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns. Plant Commun. 2:100135. doi: 10.1016/j.xplc.2020.100135
Haeussler, M., Schonig, K., Eckert, H., Eschstruth, A., Mianne, J., Renaud, J. B., et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17:148. doi: 10.1186/s13059-016-1012-2
Hassan, M. M., Zhang, Y., Yuan, G., De, K., Chen, J. G., Muchero, W., et al. (2021). Construct design for CRISPR/Cas-based genome editing in plants. Trends Plant Sci. 26, 1133–1152. doi: 10.1016/j.tplants.2021.06.015
Hsieh-Feng, V., and Yang, Y. (2020). Efficient expression of multiple guide RNAs for CRISPR/Cas genome editing. aBIOTECH 1, 123–134.
Jeong, Y.-M., Jung, E.-J., Hwang, H.-J., Kim, H., Lee, S.-Y., and Kim, S.-G. (2009). Roles of the first intron on the expression of Arabidopsis (Arabidopsis thaliana) genes for actin and actin-binding proteins. Plant Sci. 176, 58–65.
Jha, P., and Kumar, V. (2018). BABY BOOM (BBM): a candidate transcription factor gene in plant biotechnology. Biotechnol. Lett. 40, 1467–1475. doi: 10.1007/s10529-018-2613-5
Kong, J., Martin-Ortigosa, S., Finer, J., Orchard, N., Gunadi, A., Batts, L. A., et al. (2020). Overexpression of the Transcription Factor GROWTH-REGULATING FACTOR5 Improves Transformation of Dicot and Monocot Species. Front. Plant Sci. 11:572319. doi: 10.3389/fpls.2020.572319
Kong, X., Pan, W., Sun, N., Zhang, T., Liu, L., and Zhang, H. (2021). GLABRA2-based selection efficiently enriches Cas9-generated nonchimeric mutants in the T1 generation. Plant Physiol. 187, 758–768. doi: 10.1093/plphys/kiab356
Laxa, M. (2016). Intron-Mediated Enhancement: A Tool for Heterologous Gene Expression in Plants? Front. Plant Sci. 7:1977. doi: 10.3389/fpls.2016.01977
Li, J., Zhang, S., Zhang, R., Gao, J., Qi, Y., Song, G., et al. (2021). Efficient multiplex genome editing by CRISPR/Cas9 in common wheat. Plant Biotechnol. J. 19, 427–429. doi: 10.1111/pbi.13508
Lowe, K., Wu, E., Wang, N., Hoerster, G., Hastings, C., Cho, M. J., et al. (2016). Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 28, 1998–2015. doi: 10.1105/tpc.16.00124
Luo, C., Wang, S., Ning, K., Chen, Z., Wang, Y., Yang, J., et al. (2021). LsAP2 regulates leaf morphology by inhibiting CIN-like TCP transcription factors and repressing LsKAN2 in lettuce. Hortic Res. 8:184. doi: 10.1038/s41438-021-00622-y
Oh, Y., and Kim, S. G. (2021). RPS5A Promoter-Driven Cas9 Produces Heritable Virus-Induced Genome Editing in Nicotiana attenuata. Mol. Cells 44, 911–919. doi: 10.14348/molcells.2021.0237
Ordon, J., Bressan, M., Kretschmer, C., Dall’osto, L., Marillonnet, S., Bassi, R., et al. (2020). Optimized Cas9 expression systems for highly efficient Arabidopsis genome editing facilitate isolation of complex alleles in a single generation. Funct. Integr. Genomics 20, 151–162.
Pan, W., Cheng, Z., Han, Z., Yang, H., Zhang, W., and Zhang, H. (2022). Efficient genetic transformation and CRISPR/Cas9-mediated genome editing of watermelon assisted by genes encoding developmental regulators. J. Zhejiang Univ. Sci. B. 23, 339–344. doi: 10.1631/jzus.B2200119
Przybilski, R., Richter, C., Gristwood, T., Clulow, J. S., Vercoe, R. B., and Fineran, P. C. (2011). Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol. 8, 517–528. doi: 10.4161/rna.8.3.15190
Qiu, F., Xing, S., Xue, C., Liu, J., Chen, K., Chai, T., et al. (2022). Transient expression of a TaGRF4-TaGIF1 complex stimulates wheat regeneration and improves genome editing. Sci. China Life Sci. 65, 731–738. doi: 10.1007/s11427-021-1949-9
Ren, C., Liu, Y., Guo, Y., Duan, W., Fan, P., Li, S., et al. (2021). Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters. Hortic Res. 8:52. doi: 10.1038/s41438-021-00489-z
Rose, A. B. (2004). The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J. 40, 744–751. doi: 10.1111/j.1365-313X.2004.02247.x
Rose, A. B., and Beliakoff, J. A. (2000). Intron-mediated enhancement of gene expression independent of unique intron sequences and splicing. Plant Physiol. 122, 535–542. doi: 10.1104/pp.122.2.535
Su, W., Tao, R., Liu, W., Yu, C., Yue, Z., He, S., et al. (2020). Characterization of four polymorphic genes controlling red leaf colour in lettuce that have undergone disruptive selection since domestication. Plant Biotechnol. J. 18, 479–490. doi: 10.1111/pbi.13213
Sun, X., Hu, Z., Chen, R., Jiang, Q., Song, G., Zhang, H., et al. (2015). Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 5:10342. doi: 10.1038/srep10342
Tang, X., Ren, Q., Yang, L., Bao, Y., Zhong, Z., He, Y., et al. (2019). Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol. J. 17, 1431–1445. doi: 10.1111/pbi.13068
Tsutsui, H., and Higashiyama, T. (2017). pKAMA-ITACHI Vectors for Highly Efficient CRISPR/Cas9-Mediated Gene Knockout in Arabidopsis thaliana. Plant Cell Physiol. 58, 46–56.
Vain, P., Finer, K. R., Engler, D. E., Pratt, R. C., and Finer, J. J. (1996). Intron-mediated enhancement of gene expression in maize (Zea mays L.) and bluegrass (Poa pratensis L.). Plant Cell Rep. 15, 489–494. doi: 10.1007/BF00232980
Wang, J., and Chen, H. (2019). A novel CRISPR/Cas9 system for efficiently generating Cas9-free multiplex mutants in Arabidopsis. aBIOTECH 1, 6–14.
Wolabu, T. W., Park, J. J., Chen, M., Cong, L., Ge, Y., Jiang, Q., et al. (2020). Improving the genome editing efficiency of CRISPR/Cas9 in Arabidopsis and Medicago truncatula. Planta 252:15. doi: 10.1007/s00425-020-03415-0
Woo, J. W., Kim, J., Kwon, S. I., Corvalan, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164.
Xie, K., Minkenberg, B., and Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U. S. A. 112, 3570–3575. doi: 10.1073/pnas.1420294112
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14:327. doi: 10.1186/s12870-014-0327-y
Zhang, H., Si, X., Ji, X., Fan, R., Liu, J., Chen, K., et al. (2018). Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36, 894–898. doi: 10.1038/nbt.4202
Keywords: genome editing, CRISPR/Cas9, intron-mediated enhancement, GRF5, lettuce
Citation: Pan W, Liu X, Li D and Zhang H (2022) Establishment of an Efficient Genome Editing System in Lettuce Without Sacrificing Specificity. Front. Plant Sci. 13:930592. doi: 10.3389/fpls.2022.930592
Received: 28 April 2022; Accepted: 23 May 2022;
Published: 22 June 2022.
Edited by:
Kaijun Zhao, Institute of Crop Sciences (CAAS), ChinaReviewed by:
Peng-cheng Wei, Rice Research Institute, Anhui Academy of Agricultural Sciences, ChinaCopyright © 2022 Pan, Liu, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dayong Li, bGlkYXlvbmdAbmVyY3Yub3Jn; Huawei Zhang, aHVhd2VpLnpoYW5nQHBrdS1pYWFzLmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.