
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 September 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.927001
This article is part of the Research Topic Interspecific Hybridization in Plant Biology View all 16 articles
 Jianke Du1,2†
Jianke Du1,2† Chunfeng Ge1,3†
Chunfeng Ge1,3† Tao Wang1†
Tao Wang1† Jing Wang1
Jing Wang1 Zhiyou Ni1
Zhiyou Ni1 Shiwei Xiao1
Shiwei Xiao1 Fengli Zhao1
Fengli Zhao1 Mizhen Zhao4*
Mizhen Zhao4* Yushan Qiao1,4*
Yushan Qiao1,4*Fragaria viridis exhibits S-RNase-based gametophytic self-incompatibility, in which S-RNase is the major factor inhibiting pollen tube growth. However, the pathways involved in and the immediate causes of the inhibition of pollen tube growth remain unknown. Here, interactive RNA sequencing and proteome analysis revealed changes in the transcriptomic and proteomic profiles of F. viridis styles harvested at 0 and 24 h after self-pollination. A total of 2,181 differentially expressed genes and 200 differentially abundant proteins were identified during the pollen development stage of self-pollination. Differentially expressed genes and differentially abundant proteins associated with self-incompatible pollination were further mined, and multiple pathways were found to be involved. Interestingly, the expression pattern of the transcription factor FviYABBY1, which is linked to polar growth, differed from those of other genes within the same family. Specifically, FviYABBY1 expression was extremely high in pollen, and its expression trend in self-pollinated styles was consistent with that of S-RNase. Furthermore, FviYABBY1 interacted with S-RNase in a non-S haplotype way. Therefore, FviYABBY1 affects the expression of polar growth-related genes in self-pollen tubes and is positively regulated by S-RNase.
Self-incompatibility (SI) is a widespread mating system that avoids inbreeding and promotes outcrossing in angiosperms (Allen and Hiscock, 2008; Vieira et al., 2019; Kumar et al., 2021). In the family Rosaceae, the mechanisms of S-RNase-based gametophytic self-incompatibility (GSI) are diverse (Franklin-Tong and Franklin, 2003; McClure and Franklin-Tong, 2006; Sassa et al., 2010; Wu et al., 2013). In particular, studies on self- and non-self-recognition genera belonging, respectively, to tribes Amygdaleae (e.g., Prunus) and Maleae (e.g., Malus and Pyrus) have offered key insights into SI mating systems in the subfamily Amygdaloideae (Kubo et al., 2010; Iwano and Takayama, 2012; Fujii et al., 2016; Chen et al., 2020; Muñoz-Sanz et al., 2020; Harkness and Brandvain, 2021; Vekemans and Castric, 2021). In addition, an increasing number of modifiers have been explored, enabling comprehensive analyses of mechanisms underlying S-RNase-based SI (Wu et al., 2013; Claessen et al., 2019; Muñoz-Sanz et al., 2020). Within the genus Fragaria of the subfamily Rosoideae (family Rosaceae), some diploid species, such as Fragaria viridis, F. nubicola, F. pentaphylla, and F. nipponica, exhibit self-incompatibility as the major pollination barrier (Evans and Jones, 1967; Hancock, 1999; Sargent et al., 2004; Staudt, 2009; Bošković et al., 2010). The germplasm of low-ploidy wild strawberries contains abundant genetic resources controlling valuable traits, and it is a potential resource for improving cultivated strawberries (Hancock, 1999; Marta et al., 2004; Sargent et al., 2004; Staudt, 2009; Liston et al., 2014). However, as a pollination barrier between styles and pollen, SI has considerably hampered strawberry breeding (Evans and Jones, 1967; Li et al., 2000; Marta et al., 2004; Sargent et al., 2004; Bošković et al., 2010; Liston et al., 2014). As such, SI greatly limits the exploitation of valuable traits in diploid Fragaria species, such as enhanced disease resistance, stress tolerance, aromaticity, and solid content, for breeding (Evans, 1982; Hancock and Luby, 1993; Maas et al., 1997; Hancock et al., 2002; Ulrich et al., 2007). Therefore, elucidating molecular mechanisms underlying SI is expected to aid the development of strategies aimed at improving molecular breeding in strawberries. The mechanisms of SI in Fragaria have been studied for decades (Evans and Jones, 1967; Bošković et al., 2010; Aguiar et al., 2015). Specifically, GSI in Fragaria is known to be mediated by S-RNase (Du et al., 2021), similar to that in Rosa (Vieira et al., 2021); however, molecular mechanisms underlying the S-RNase-mediated inhibition of self-pollen tubes remain unclear.
High-throughput sequencing technologies, such as RNA sequencing (RNA-Seq) and proteomic analysis, have been used as efficient tools for studying SI mechanisms. In Petunia inflate, pollens of haplotypes S2 and S3 were analyzed using de novo transcriptome sequencing, and 17 pollen SLF determinants were identified (Williams et al., 2014; Wu et al., 2020). Moreover, comparative transcriptomics among unpollinated, self-pollinated, and cross-pollinated pistils of “Xiangshui” lemon revealed a series of genes related to pollen tube growth, programmed cell death (PCD), signal transduction, and transcription. In addition, a novel putative self-pollen rejection-associated S-RNase was detected, building the foundation for further research on SI mechanisms and the breeding of seedless varieties in lemons (Zhang et al., 2015). Additionally, proteomic analysis, as a tool for transcriptomics, has been widely applied in studies exploring SI mechanisms. Specifically, seven S-RNase-associated proteins have been identified in Japanese pears using two-dimensional gel electrophoresis (2-DE) (Ishimizu et al., 1996). Similarly, using 2-DE, Wang et al. (2013) discovered 22 proteins, some of which may be related to pollen maturation, fertility, germination, growth, and cell death. In a study exploring SI mechanisms in Solanum pennellii, proteomic analysis with isobaric tags for relative and absolute quantitation (iTRAQ) revealed that S-RNase, HT-A, cell wall-loosening proteins, and defense response-related proteins play key roles in an interspecific reproductive barrier between wild and domesticated species (Chalivendra et al., 2012). Overall, these cutting-edge techniques can not only directly identify potential pollen or style S-determinants but also reveal large-scale changes at the transcriptomic and proteomic levels in various SI-related biological processes during pollination.
Through ABCF transport proteins, S-RNase enters the pollen tube and functions non-specifically (Meng et al., 2014a,b). S-RNase is cytotoxic to self-pollen tubes, while its cytotoxicity is attenuated in non-self-pollen tubes (Claessen et al., 2019). Initially, the cytotoxicity of S-RNase was attributed to RNA degradation, and S-RNase was believed to enter the self-pollen tube and degrade RNA, explaining S-RNase-based GSI (Franklin-Tong and Franklin, 2003; Kubo et al., 2010; Tao and Iezzoni, 2010; Wu et al., 2013). S-RNase can break the gradient of reactive oxygen species (ROS) and calcium ions at the tip of the pollen tube, inducing a series of physiological and biochemical changes and PCD responses. Multiple signal transduction cascades are involved in S-RNase-based GSI (Wang et al., 2009, 2010; Wu et al., 2013, 2021; Li et al., 2018; Chen et al., 2020). Based on the evidence, the cytotoxicity of S-RNase in self-pollen tubes can be attributed to a multitude of reactions and signals, and the S-RNase-mediated degradation of RNA in pollen tubes may be the result of SI response or RNA homeostasis. The pollen tube is a classic model of polar growth, and the polarity of the self-pollen tube breaks following SI (Feijó, 2010; Wu et al., 2013; Del Duca et al., 2014). The YABBY gene family belongs to the zinc finger superfamily and plays pivotal roles in the development of vegetative and reproductive tissues in angiosperms (Kumaran et al., 2002; Bartholmes et al., 2012; Finet et al., 2016; Filyushin et al., 2017). As transcription factors, an important function of YABBYs is related to the establishment of polarity (Kumaran et al., 2002; Stahle et al., 2009); however, their link to SI and their involvement in breaking the polar growth of incompatible pollen tubes remain unknown.
The green strawberry F. viridis is a self-incompatible diploid (2n = 2x = 14) species. The fruits have an apple-like aroma, and the plants exhibit potent cold hardness and high soil pH tolerance (Labokas and Bagdonaitë, 2005; Staudt, 2009; Gruner et al., 2017). F. viridis is the germplasm for strawberry breeding (Gruner et al., 2017), and this species is a suitable model for further research on SI in the genus Fragaria (Bošković et al., 2010). In our experiment, at 24 h after self-pollination, most of the F. viridis pollen tubes stopped growing at 2/3 of the style. Thus, the receptacle with gynoecium from F. viridis line #10-42 (SI) at 0 and 24 h after self-pollination was selected for subsequent proteome sequencing. Integrative analysis with previous RNA-Seq data provided valuable information on the development of self-pollen tubes in the styles of F. viridis. In addition, the interaction between FviYABBY1 and S-RNase in F. viridis implicated polar growth regulators in self-incompatibility.
Styles of F. viridis were self-pollinated and harvested at 1, 6, 12, 18, 24, and 48 h after pollination (HAP). The styles were fixed in FAA solution (5% formalin, 5% acetic acid, and 65% ethanol) for 2 h at 65°C. Next, the styles were washed with distilled water three times, treated with 4 M NaOH for 2 h at 65°C, and washed with distilled water three times before staining with 0.1% (w/v) aniline blue for 12 h at 4°C in the dark. Self-pollinated pistils of Fragaria vesca after 24 h were used as controls. The stained styles of F. viridis were placed on a glass slide and mounted in a drop of glycerol for observation under a fluorescence microscope (BX53; Olympus, Tokyo, Japan) at a wavelength of 356 μm.
The self-pollinated receptacles with gynoecium from F. viridis lines #10–42 were harvested after 0 and 24 h and stored in liquid nitrogen for transcriptome sequencing and data analysis. Total RNA extraction, cDNA preparation, Illumina sequencing, and de novo assembly were performed as previously described (Du et al., 2021). The obtained non-redundant unigenes were annotated against the Nr, Nt, Swiss-Prot, COG, and KEGG databases using BLAST (Altschul et al., 1990), GO annotation was performed using Blast2GO, and InterPro annotation was performed using InterProScan5. ESTScan (Iseli et al., 1999) was used to predict the directions of unigenes that could not be annotated.
Unigene expression was estimated as fragments per kilobase of transcript per million fragments mapped reads (FPKM) (Mortazavi et al., 2008). Differentially expressed genes (DEGs) were detected using DEseq2 (Love et al., 2014), and p-values related to DEGs were modulated for multiple testing using the Benjamini–Hochberg false discovery rate (FDR) method (Benjamini and Hochberg, 1995). Unigenes deemed differently expressed were screened with an FDR-modulated p-value of ≤ 0.05 and |log2 FC| of ≥1. DEGs were subjected to GO functional and KEGG pathway analyses.
iTRAQ analysis was performed at the Beijing Genomics Institute (BGI, Shenzhen, China). Plant material was the same as that for the RNA-Seq analysis described earlier. Two biological replicates were set per sample. Proteins were extracted using the trichloroacetic acid (TCA)/acetone method (Gu et al., 2013). Protein concentration was measured using the Bradford method. For each sample, 100 μg protein was digested with Trypsin Gold (Promega, Madison, WI, USA) at a protein-to-trypsin ratio of 20:1 for 4 h at 37°C. Following trypsin digestion, the peptides were vacuum centrifuged to dryness and dissolved in 0.5 M TEAB. iTRAQ labeling of peptide samples was performed using the iTRAQ® Reagents-8plex Applications Kit according to the manufacturer's protocol. After labeling, the samples were combined and lyophilized, and the fractions were then analyzed using LC-ESI-MS/MS.
MS/MS raw data were converted to MGF files using the ProteoWizard tool (msConvert, http://proteowizard.sourceforge.net), and proteins were searched using the Mascot search engine (Matrix, Science, London, UK, vision2.3.02) against the de novo transcriptomic data of F. viridis style (Du et al., 2021). Proteins were quantified using IQuant (BGI, Shenzhen, China), as previously described (Wen et al., 2014). Permutation tests were used for statistical and protein quantification analyses. Proteins were identified and quantified separately for each biological replicate (Tolin et al., 2013). FDR correction was adopted with a threshold of <1% to reduce the false identification of peptides. Proteins with a |log2 FC| of ≥0.6 and a p-value of ≤0.05 were considered differentially expressed.
According to the hidden Markov model (HMM) of the YABBY domain, the hmmsearch program in HMMER (Finn et al., 2011) was used to search the whole-genome protein database of F. viridis and F. vesca for obtaining the YABBY gene family members. The selected proteins were input into the CDD database on NCBI (https://www.ncbi.nlm.nih.gov/cdd/?term) to verify the integrity of the YABBY domain, and proteins that did not contain the YABBY domain were filtered out. YABBY HMM (PF04690) can be downloaded directly from the Pfam database (http://pfam.xfam.org/). The protein database for F. viridis was derived and predicted based on the style transcript database after self-pollination (Du et al., 2021). The proteome database of F. vesca was downloaded from the Rosaceae Genome Database (https://www.rosaceae.org/), and F. vesca v4.0. a1 was selected.
Molecular weights and isoelectric points (pI) of YABBY gene family members were analyzed using ExPaSy (https://web.expasy.org/protparam/) (Artimo et al., 2012), and signal peptides were analyzed using SignalP (http://www.cbs.dtu.dk/services/SignalP/index.php) (Armenteros et al., 2019). The amino acid sequences of genes and proteins were compared using the alignment tool DNAMAN8.0 (Lynnon, Quebec, Canada). The online tool MEME (https://meme-suite.org/meme/tools/meme) was used for the motif analysis of YABBY family members, and TBtools was used for visualization. Using Arabidopsis and tomato as references (Bowman, 2000; Huang et al., 2013), multiple sequence alignment of YABBY protein sequences between F. viridis and F. vesca was performed with the MUSCLE program in MEGA 7.0 (Mello et al., 2017).
Receptacles with gynoecium from F. viridis were collected at 0, 6, 12, 24, 48, and 72 HAP. Total RNA extraction and cDNA synthesis were performed as previously described (Du et al., 2021). Based on reference unigene sequences, 21 primer pairs (Supplementary Table S1) were designed using an online real-time PCR tool (https://sg.idtdna.com/scitools/Applications/RealTimePCR/). EF-1α was used as the internal control (Du et al., 2021). In addition, for spatiotemporal expression analysis, pollen, leaves, peduncles, calyxes, and petals of F. viridis were collected for tissue-specific expression analysis of YABBY family genes. The SYBR Premix Ex Taq™ kit (TaKaRa) was used for RT-qPCR. The reactions were conducted with three biological replicates using the ABI 7300 Real-Time System (PE Applied Biosystems, CA, USA). The 2−ΔΔCT algorithm was used to analyze quantitative variation.
Primers were designed based on the open reading frame of the putative FviYABBY1 unigene (Unigene16298_All) (Du et al., 2021) (see Supplementary Table S1). rTaq was used for PCR. The reaction conditions were as follows: 94°C for 4 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and final extension at 72°C for 10 min. PCR products were evaluated on a 1% agarose gel. The target fragments were purified using the AxyPrep DNA gel extraction kit (Axygen, CA, USA), cloned into the pMD™ 19-T vector (Takara Bio, Dalian, China), and transformed into Escherichia coli DH5α competent cells. Positive clones were sequenced as described previously (Li et al., 2015).
The FviYABBY1 coding sequence (CDS) lacking stop codon was obtained from the plasmid containing FviYABBY1 using primers with Nco I–Spe I restriction sites. FviYABBY1 was ligated to the pCAMBIA1302-GFP expression vector using T4 ligase to construct the FviYAB1-GFP recombinant plasmid. Tobacco leaves were transiently transformed as described previously (Du et al., 2019). Following culture in the dark for ~60 h, the fluorescent signal of GFP in tobacco leaves was detected using a laser confocal fluorescence microscope.
The recombinant reporter vectors pGBKT7-Sa and pGBKT7-Sb were constructed, and yeast AH109 cells were transformed with the pGADT7 plasmid. To test the toxicity and self-activation of Sa-RNase and Sb-RNase, as well as to obtain the optimal 3-AT concentration for the background expression of eliminating reporter genes in yeast cells, pGADT7-largeT+pGBKT7-p53 and pGADT7-largeT+pGBKT7-laminC combinations were used as the positive and negative control, respectively. SD/-Trp/-Leu and SD/-Trp/-Leu/-His/Ade media with 3-AT concentration gradients of 0, 5, 10, and 15 mM were used, respectively.
The full-length CDSs of Sa-RNase and Sb-RNase were stored in the previously described vector as templates (Du et al., 2021), and primers with Nde I–Bam HI restriction sites were used to obtain the CDSs of mature Sa-RNase and Sb-RNase peptides. Enzyme digestion was used to construct the recombinant vectors pGBKT7-Sa and pGBKT7-Sb. The same method was used to recombine the full-length CDS of F. viridis YABBY1 into pGADT7 for obtaining pGADT7-FviYAB1 with Nde I–Bam HI restriction sites. The primers used are listed in Supplementary Table S1.
The recombinant plasmids pGBKT7-Sa (pGBKT7-Sb) and pGADT7-FviYAB1 were co-transformed into the yeast strain AH109. The yeast cells were spread on the SD/-Trp-Leu medium and cultured for 3–5 days at 28°C in an incubator. Single colonies were selected, transferred to SD/-Trp-Leu liquid medium, and shaken. After shaking, the yeast suspension was straked on SD/-Trp-Leu, SD/-Trp-Leu-His-Ade, and SD/-Trp-Leu-His-Ade/X-α-gal plates and cultivated at 28°C in an incubator for 3–5 days to observe colony growth. pGADT7-largeT+pGBKT7-p53 and pGADT7-largeT+pGBKT7-laminC were used as the positive and negative controls, respectively.
The target genes FviYABBY1 and S-RNase (Sa-RNase and Sb-RNase) from the pMD19 (Simple) T-vector containing the corresponding gene were amplified using primers with restriction sites for the gene-specific primers used (Supplementary Table S1). Using restriction digestion, the CDS of FviYABBY1 lacking the stop codon was cloned into the 35S:YCE vector with Asc I–Bam HI, and the full-length CDS of Sa-RNase (Sb-RNase) was cloned into the 35S:YNE vector with Asc I–Kpn I to obtain the recombinant vectors FviYABBY1-35S:YCE and Sa(Sb)-35S:YNE, respectively. Tobacco cells were transiently transfected with Agrobacterium following the same method as that for subcellular localization assay.
Aniline blue staining showed the growth of F. viridis pollen tubes in pistils at 1, 6, 12, 18, 24, and 48 HAP (Figures 1A–F). Pollen grains gradually attached to stigma papilla cells at 1 HAP, started germinating at 6 HAP (Figure 1B), and continued to elongate until reaching two-thirds of the pistil at 24 HAP (Figure 1E) refer to self compatible F. vesca (Figure 1G); no further distinct growth was observed at 48 HAP (Figure 1F). Thus, genes related to the inhibition of self-pollen tube growth may be expressed around 24 HAP based on the differential expression of pollen tube development. Accordingly, self-pollinated receptacles with gynoecia were harvested after 0 and 24 h and analyzed for differential expression.
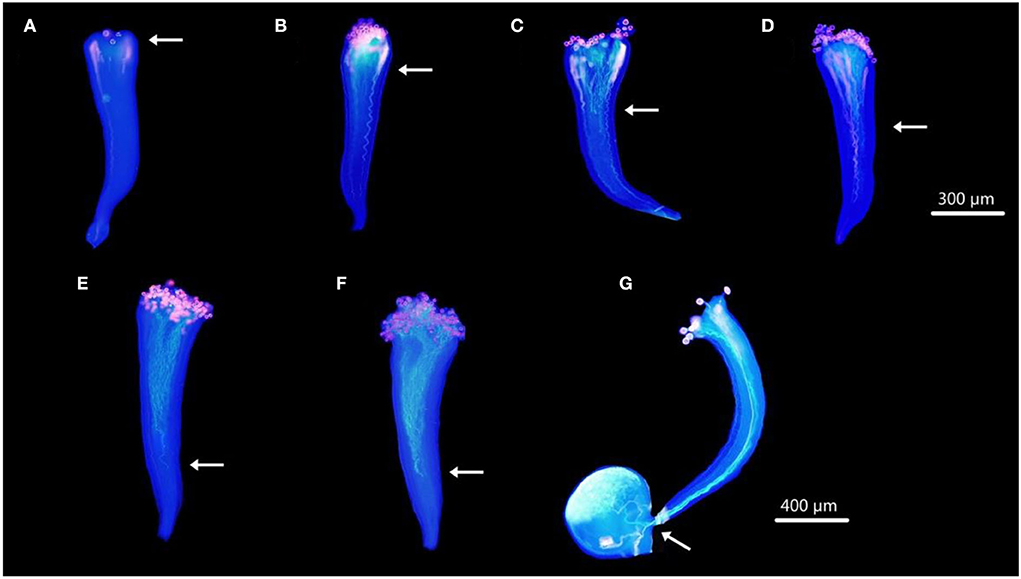
Figure 1. Dynamic development of pollen tubes in pistils under SP. (A–F) Styles of F. viridis at 1, 6, 12, 18, 24, and 48 HAP, respectively. (G) Pistil of F. vesca at 24 HAP. White arrows indicate the positions at which the majority of the pollen tubes were arrested in pistils. Bars (A–F) = 300 μm, (G) = 400 μm.
Six pools generated a total of 40.23 Gb of raw data (Supplementary Table S2). After removing low-quality, adaptor-polluted, and high-content unknown base (N) reads, 268.16 Mb of clean reads were generated (Q30 > 87.14%). A total of 71,756 unigenes were annotated using the Nr, Nt, Swiss-Prot, COG, KEGG, GO, and Interpro databases (Supplementary Figure S1A), with a mean length of 1,209 bp. The identified unigenes of F. viridis were highly homologous to those of F. vesca (85.5%), Prunus mume (1.59%), Prunus persica (1.31%), and Malus domestica (1.01%) (Supplementary Figure S1B, respectively). Global gene expression profiling between samples collected after 0 and 24 h revealed 2,181 DEGs, including 1,355 downregulated and 826 upregulated genes (Supplementary Table S3). These results indicate that many biological and molecular processes are altered during self-pollination. DEGs identified in the NR database were classified using GO functional analysis (Figures 2A–C), which is an important reference for pollen tube development in self-pollinated Fragaria species according to previous reports (Zhao et al., 2015; Zhang et al., 2016, 2017).
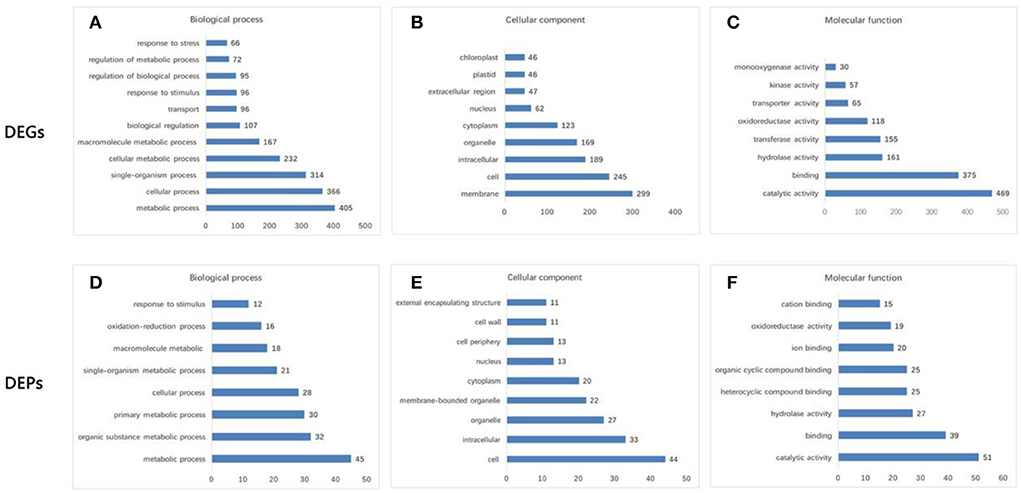
Figure 2. GO functional analysis of differentially expressed genes (DEGs) and differentially abundant proteins (DAPs). (A,D) Biological process. (B,E) Cellular component. (C,F) Molecular function. DEGs were mainly enriched in “metabolic process” (405 DEGs) and “cellular process” (366) for the biological process; “membrane” (299) and “cell” (245) for the cellular component; and “catalytic activity” (469) and “binding” (375) for the molecular function. The transcriptome data were significantly enriched in several GO categories, such as “cell wall organization and biogenesis,” “defense response,” “response to hormone,” “gametophyte development,” “pollen development,” “pollen germination,” and “pollination,” which are pollen–style interaction-associated biological processes. Moreover, GO terms annotation of DAPs revealed that “cell” (44) and “intracellular” (33) were the major cellular components; “catalytic activity” (51) and “binding” (39) were the major molecular functions, and “metabolic process” (45) and “cellular process” (28) were the major biological processes.
Furthermore, KEGG pathway analysis was performed to elucidate the biological pathways activated by pollen–style interaction. A total of 1,571 DEGs were annotated in 125 KEGG pathways (Supplementary Table S4). “Metabolic pathway” (ko01100), “biosynthesis of secondary metabolites” (ko01100), and “starch and sucrose metabolism” (ko00500) were the three major pathways. These results are consistent with the findings of transcriptome analysis of SI and SC pollination in tomatoes (Vieira et al., 2008). Interestingly, many genes were annotated to “plant–pathogen interaction” (ko04626), “phenylpropanoid biosynthesis” (ko00940), “plant hormone signal transduction” (ko04075), and “ubiquitin-mediated proteolysis” (ko04120), which are all notable pathways related to self-pollination (Elleman and Dickinson, 1999; Williams et al., 2015).
The quality of transcriptomic data was confirmed using RT-qPCR. Fourteen genes related to S-RNases, cell death, microtubule binding, pathogen interaction, hormone signaling, CDPK signaling, enzyme inhibitor activity, cell wall, pollen development, RNA degradation, and ubiquitin-mediated proteolysis were detected (Figures 3A–N). The results showed good concordance (R2 = 0.9409) between the RT-qPCR and RNA-Seq data (Figure 3O), confirming the reliability of our transcriptomic data. Consistent with previous reports, Sa-RNase and Sb-RNase showed an initial uptrend, followed by a peak at 12 HAP and then a downtrend. Interestingly, the expression trends of four genes encoding a PR protein, an auxin-induced protein, a Glu S.griseus protease inhibitor, and CAF1 were similar to that of S-RNase. The expression levels of these four genes (Pollen-specific LRX protein, CDPK34, PRK2, and SKP1) increased after self-pollination. Conversely, the expression levels of three genes (UBP1 associated protein, PE/PEI 17, GsSRK) first decreased and then increased.
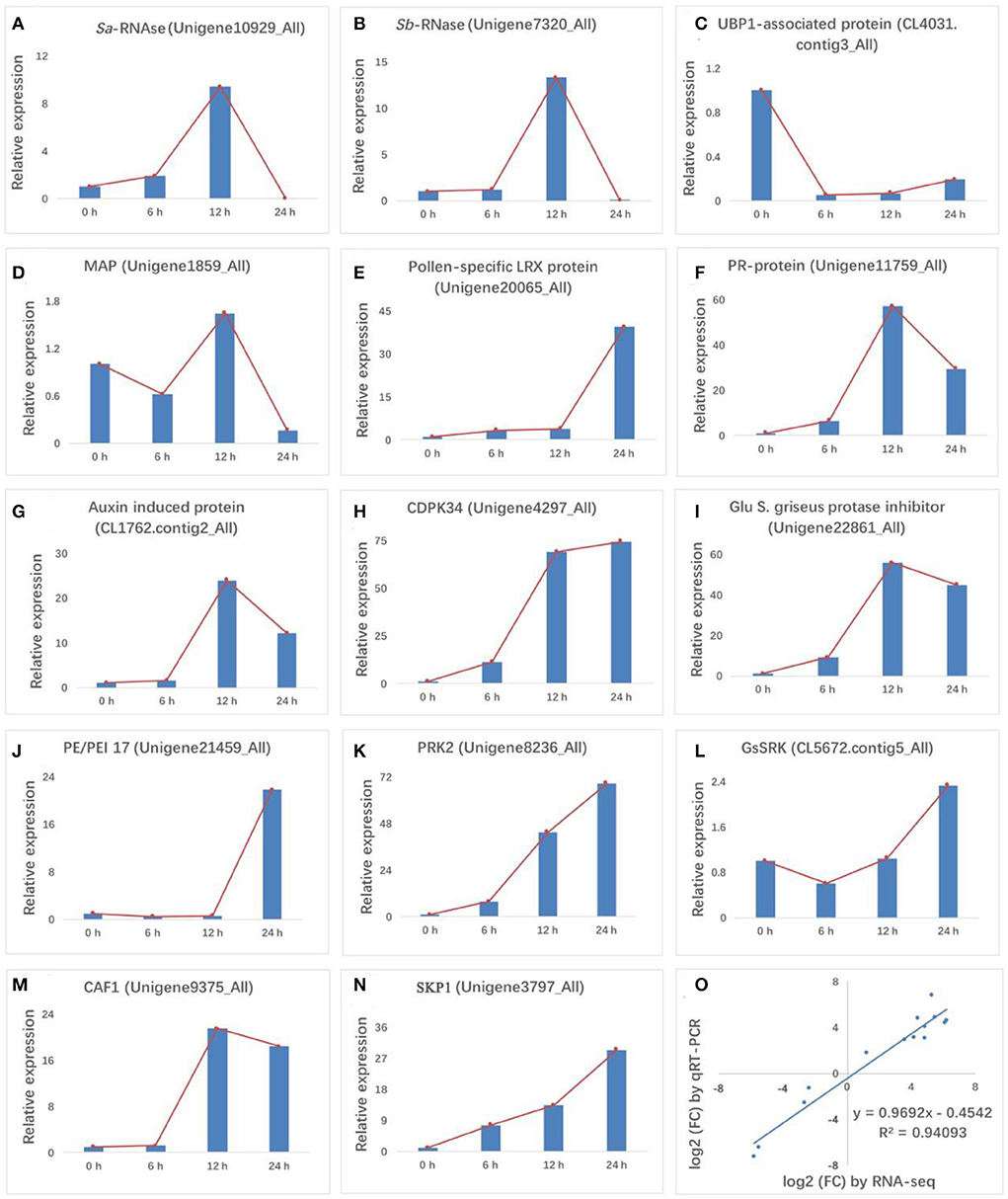
Figure 3. RT-qPCR validation of 14 differentially expressed genes (DEGs). (A,B) S-RNase, (C) cell death, (D) microtubule binding, (E) pathogen response, (F,G) hormone signaling, (H) CDPK signaling, (I) enzyme inhibitor activity, (J) cell wall, (K,L) pollen development, (M) RNA degradation, and (N) ubiquitin-mediated proteolysis were validated for gene expression levels at 0, 6, 12, and 24 HAP. (O) Comparison of fold-changes in gene expression (24 HAP/0 HAP) between RNA-Seq (x-axis) and RT-qPCR (y-axis) data. Both data were log2-transformed. In charts, red lines in (A–N) indicate the expression trend of each gene at the four pollination stages.
Comparative proteomic analysis between the two samples collected at 0 and 24 HAP was performed as a parallel complement to transcriptomic analysis. We identified 7,105 proteins from 385,662 spectra (data not shown), including 200 DAPs (FC > 1.5, p < 0.05), of which 109 were upregulated and 91 were downregulated (Supplementary Table S5). According to the transcriptomic data, the results of GO functional analysis of DAPs and DEGs were highly consistent during self-pollination (Figure 2). In addition, 151 DAPs were annotated to 74 KEGG pathways (Supplementary Table S6), and the majority of these pathways were related to pollen tube development.
Integrative analysis of DEGs and DAPs offered comprehensive information on key genes involved in pollen–style interactions. In the present study, we identified 3,273 genes (Supplementary Table S7) at both transcript and protein levels and used these for correlation analysis. The distributions of the corresponding transcript-to-protein ratios (colored plots) were presented as scatter plots of log2-transformed data (Figure 4). The expression of most genes (black, blue, and green plots) did not significantly vary at the transcript (|log2 FC| ≥ 1) and protein (|log2 FC| ≥ 0.6) levels. Next, we focused on genes (red plot) in four sections (A–D). Genes in sections B and C exhibited similar expression trends at the transcript and protein levels (both upregulated and downregulated), whereas genes in sections A and D showed negative expression trends. Twelve DAPs (|log2 FC| ≥ 1 and p ≤ 0.05) that were differentially expressed at the transcript level (|log2 FC| ≥ 1 and p ≤ 0.05) are listed in Supplementary Table S8. Eleven genes showed consistent trends at both levels, including eight upregulated and three downregulated genes. Only one gene (beta-glucosidase 3) showed a negative expression trend, being downregulated at the transcript level but upregulated at the protein level at 24 HAP compared with that at 0 HAP. Beta-glucosidase is involved in various biological processes, including the formation and degradation of cell walls and response to biotic and abiotic stresses. The difference in the transcript and protein levels of this gene may result from many factors rather than its involvement in the self-incompatibility reaction alone.
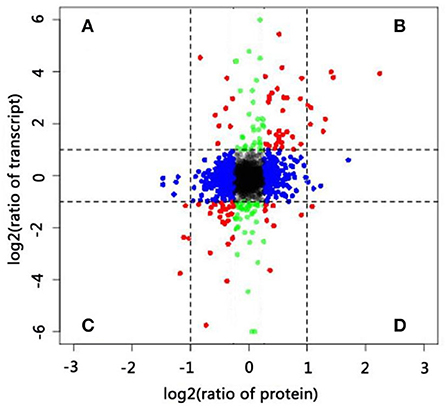
Figure 4. Correlations between protein and transcript levels. (A–D) Differentially expressed genes at both transcript and protein levels.
S-RNase-mediated GSI involves complex genetic mechanisms (Franklin-Tong and Franklin, 2003; Wu et al., 2013), and the integrity of SI depends on both the S locus (pistil S gene and pollen S gene) and the modifier genes (Sassa and Hirano, 2006; Wu et al., 2013; Claessen et al., 2019). These modifiers are closely related to PCD (Thomas and Franklin-Tong, 2004), plant-pathogen interactions (Elleman and Dickinson, 1999; Sanabria et al., 2008), Ca2+ signaling (Wu et al., 2013; Gu et al., 2015; Qu et al., 2016), ROS (Wang et al., 2010), hormone signaling (Shi et al., 2017), cell wall formation (Graaf et al., 2003), protein ubiquitination (McClure and Franklin-Tong, 2006), and pollen development (Zhang et al., 2016; Shi et al., 2017). Here, we clustered the SI-related DEGs and DAPs based on GO functional annotation, KEGG pathway classification, and Nr database annotation, as seen in Supplementary Table S9.
The pollen tube grows smoothly in a compatible style, which is regulated by apical polarity (Feijó, 2010; Del Duca et al., 2014). A gene encoding a transcription factor related to the polar growth of the pollen tube, the axial regulator YABBY1 (Unigene16298_All), named FviYABBY1, was detected in the style at 0 HAP, and its expression was remarkably downregulated (log2 FC < −1) at 24 HAP. In the present study, FviYABBY1 was annotated to a GO term but not to a protein or a KEGG pathway. The tip of self-incompatible pollen tubes cannot extend normally, which limits their polar growth (Franklin-Tong and Franklin, 2003; McClure and Franklin-Tong, 2006; Wu et al., 2013; Claessen et al., 2019). Therefore, we focused only on the gene related to the polar growth of pollen tubes and explored its link to self-incompatibility through the following series of experiments.
The YABBY gene family comprises plant-specific transcription factors playing vital roles in many biological processes during plant growth and development. These genes are differentially expressed after self-pollination. To gain a comprehensive understanding of their roles and homologs in SI, the YABBY family genes expressed in the style and pollen interaction system of F. viridis were screened using F. vesca as the control. Through screening and verification, 13 YABBY family members were finally obtained—seven and six from the style and pollen interaction system of F. viridis and the whole genome of F. vesca, respectively. These genes were named according to their homology with the Arabidopsis YABBY family genes (Bowman, 2000), and their basic characteristics were analyzed (Supplementary Table S10). The identified YABBY family members harbor two conserved domains—a C2C2 zinc finger structure and a YABBY domain—and the degree of amino acid conservation in the YABBY domain is greater than that in the zinc finger structure (Supplementary Figure S2). In addition to the conserved motifs, the various members of the YABBY family contain unique motifs (Supplementary Figure S3), implying that these genes share certain commonalities and functional specificities.
Consistent with previous reports in Arabidopsis, tomato, rice, wheat, and pomegranate (Bowman, 2000; Toriba et al., 2007; Huang et al., 2013; Zeeshan et al., 2020; Zhao et al., 2020), the YABBY genes in F. viridis and F. vesca were divided into five subfamilies: INO, CRC, YAB2, YAB1/YAB3, and YAB5 (Figure 5A). Similar to those in F. vesca, INO, CRC, YAB2, and YAB5 in F. viridis comprise one member, while the remaining subfamily YAB1/YAB3 comprises three members, including one in YAB1 and two in YAB3, respectively. Meanwhile, in F. vesca, Arabidopsis (Bowman, 2000), and tomato (Huang et al., 2013), YAB1/YAB3 comprises two members, one each in YAB1 and YAB3.
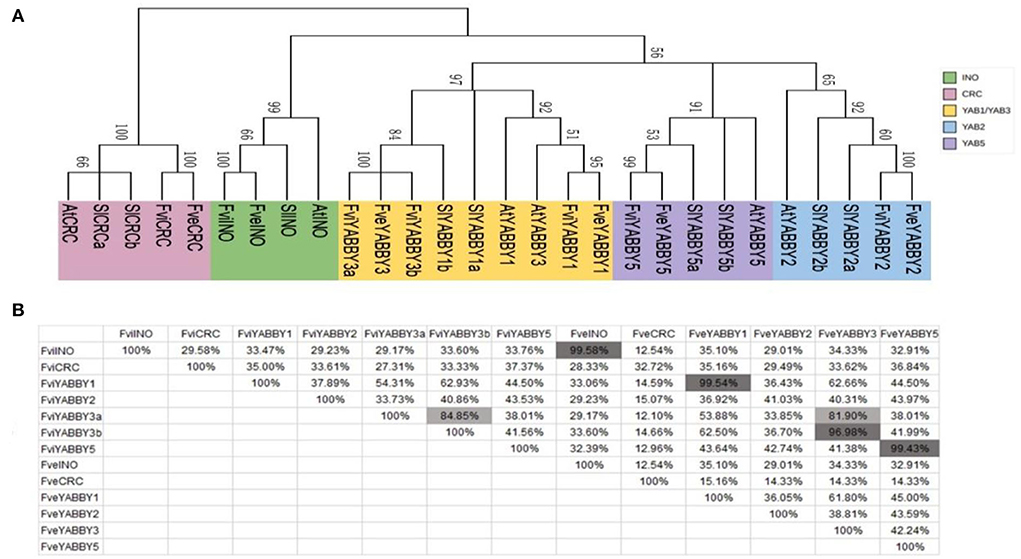
Figure 5. Homology analysis of YABBY family members in Fragaria viridis and Fragaria vesca. (A) Phylogenetic tree of YABBY family members in F. viridis and F. vesca. (B) Amino acid similarity between YABBY family members in F. viridis and F. vesca.
Homology analysis revealed that the functions of INO and YAB5 subfamily members are not differentiated between F. viridis and F. vesca, with similarity exceeding 99.43%; however, the YABBY2 and CRC subfamily members share a low similarity at 41.03 and 32.72%, respectively (Figure 5B). The YABBY1 and YABBY3 proteins in strawberries shared high homology, both within and between species, which explains the clustering of members in these two groups and their classification in the YAB1/3 subfamily. In addition, among the members of the strawberry YABBY family, only two YAB3 members (FviYABBY3a and FviYABBY3b) are present in F. viridis, both sharing high similarity, which suggests the occurrence of a gene duplication event in YAB3 throughout the evolution. The amino acid similarity was 99.54% between FviYABBY1 and FveYABBY1, 96.98% between FveYABBY3 and FviYABBY3a, and 81.90% between FveYABBY3 and FviYABBY3b, suggesting that FviYABBY3b has undergone functional diversification following the gene duplication event.
The expression levels of YABBY family genes in the pistil, pollen, leaves, peduncle, calyx, and petals of F. viridis were analyzed using RT-qPCR. As seen in Figure 6A, the expression levels of FviYABBY genes were relatively high in the pistil, suggesting their important roles in pistil development. Unlike other family members, FviYABBY1 showed the highest expression level in pollens; thus, the key function of FviYABBY1 may be in the pollen. We further analyzed the expression levels of these genes in pistils at different stages after self-pollination. As seen in Figure 6B, except for FviYABBY1, the other YABBY family genes reached their peak expression levels at 0 HAP. Further, while the expression of all FviYABBY genes significantly decreased at 6 h, their expression levels increased at 12 h, following another downward trend at 24 and 48 HAP. The YABBY family members were clustered on the phylogenetic tree and harbored the same conserved domains (Figure 5 and Supplementary Figure S3). FviYABBY3a and FviYABBY3b as well as FviYABBY2 and FviYABBY5 were highly similar in terms of their tissue-specific expression patterns and expression patterns in pistils at different stages after pollination, implying that they may serve similar functions. Although the expression level of FviYABBY1 in pistils decreased at 6 HAP, it reached the peak value at 12 HAP and then declined again. The expression trend of FviYABBY1 was consistent with that of S-RNase (Du et al., 2019), suggesting that FviYABBY1 is positively regulated by S-RNase.
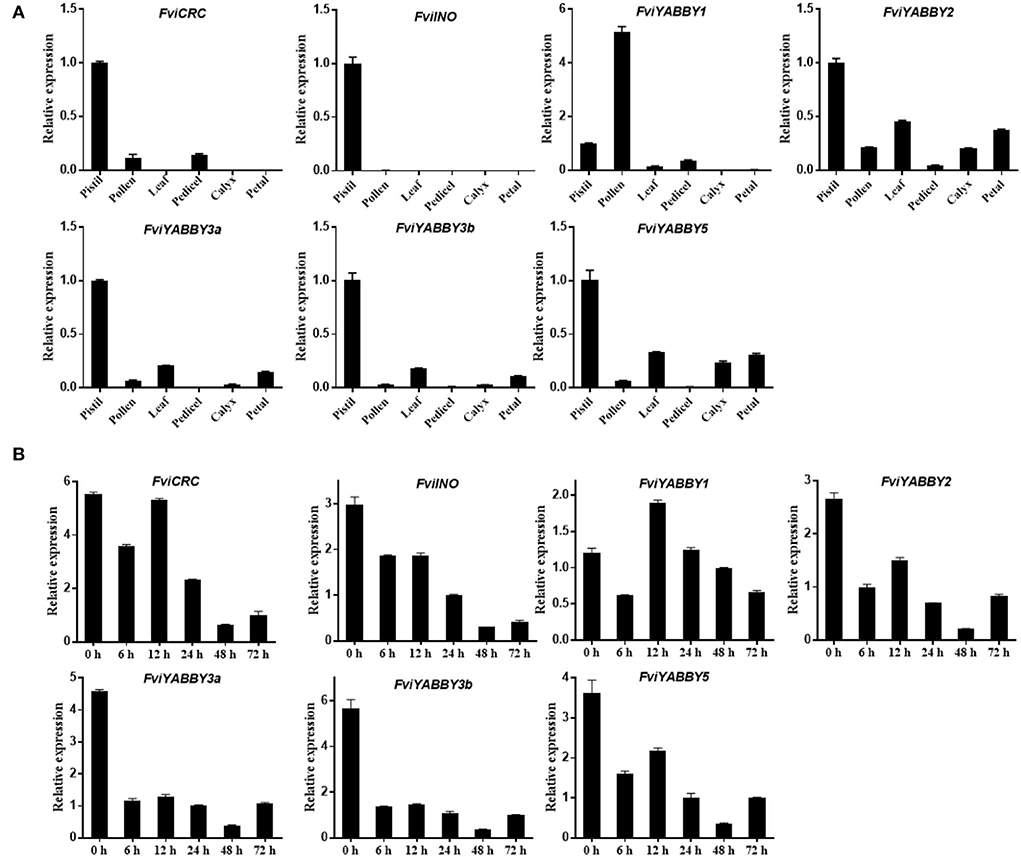
Figure 6. Analysis of gene expression pattern using RT-qPCR. (A) Tissue-specific expression patterns of YABBY family genes in Fragaria viridis. (B) Spatiotemporal expression after self-pollination of YABBY family genes in F. viridis.
Using F. viridis pistil cDNA as the template, the CDS-specific target band was obtained by PCR amplification (Figure 7A), with a length of 657 bp. Monoclonal sequencing results were consistent with the transcriptome data (Du et al., 2021). FviYABBY1 was cloned into pCAMBIA1302-GFP, and the recombinant vector was transferred into tobacco leaves through Agrobacterium-mediated transformation. The cellular distribution of FviYABBY1 protein was determined based on the GFP signal. Under 488 nm excitation wavelength, green fluorescent signals were observed in the nucleus and the cytoplasm (Figure 7B). Therefore, FviYABBY1 lacks a signal peptide or transmembrane structure (Supplementary Table S10) and is a non-secreted protein that cannot be transferred between cells.
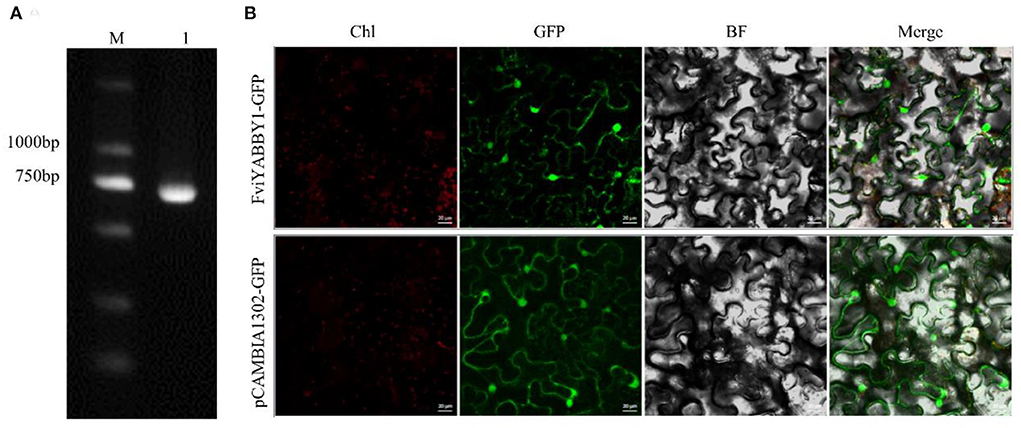
Figure 7. Gene cloning and subcellular localization of FviYABBY1. (A) FviYABBY1 cloning. M: 2000 DNA Marker; 1: Fragment of FviYABBY1 CDS. (B) Subcellular localization of FviYABBY1. Chl, chlorophyl II autofluorescence; GFP, green fluorescence protein; BF, bright field; Merge, merged image of Chl, GFP, and BF. Bar = 20 μm.
Sa-RNase and Sb-RNase are non-toxic and non-self-activated in yeast cells. Thus, yeast two-hybrid assays were conducted on SD/-Trp-Leu-His-Ade medium without 3-AT to verify whether other proteins interact with the target RNases (Supplementary Figure S4).
The recombinant plasmids FviYABBY1-pGADT7+Sa-pGBKT7 and FviYABBY1-pGADT7+Sb-pGBKT7 were co-transformed into the yeast strain AH109. The positive yeast strains on the SD/-Trp-Leu medium were multiplied and inoculated on prefabricated plates containing SD/-Trp-Leu, SD/-Ade-His-Trp-Leu, and SD/-Trp-Leu-His-Ade/X-α-gal media. As seen in Figure 8A, all yeast strains grew normally on SD/-Trp-Leu medium, indicating that yeast co-transformation was successful. Yeast strains containing FviYABBY1-pGADT7+Sa-pGBKT7 grew stably on thr SD/-Ade-His-Trp-Leu medium, while those containing FviYABBY1-pGADT7+Sb-pGBKT7 grew stably on the SD/-Trp-Leu-His-Ade medium and grew blue colonies on the SD/-Trp-Leu-His-Ade/X-α-gal medium. These results indicate the interaction of the FviYABBY1 protein with Sa-RNase and Sb-RNase without S-RNase haplotype specificity.
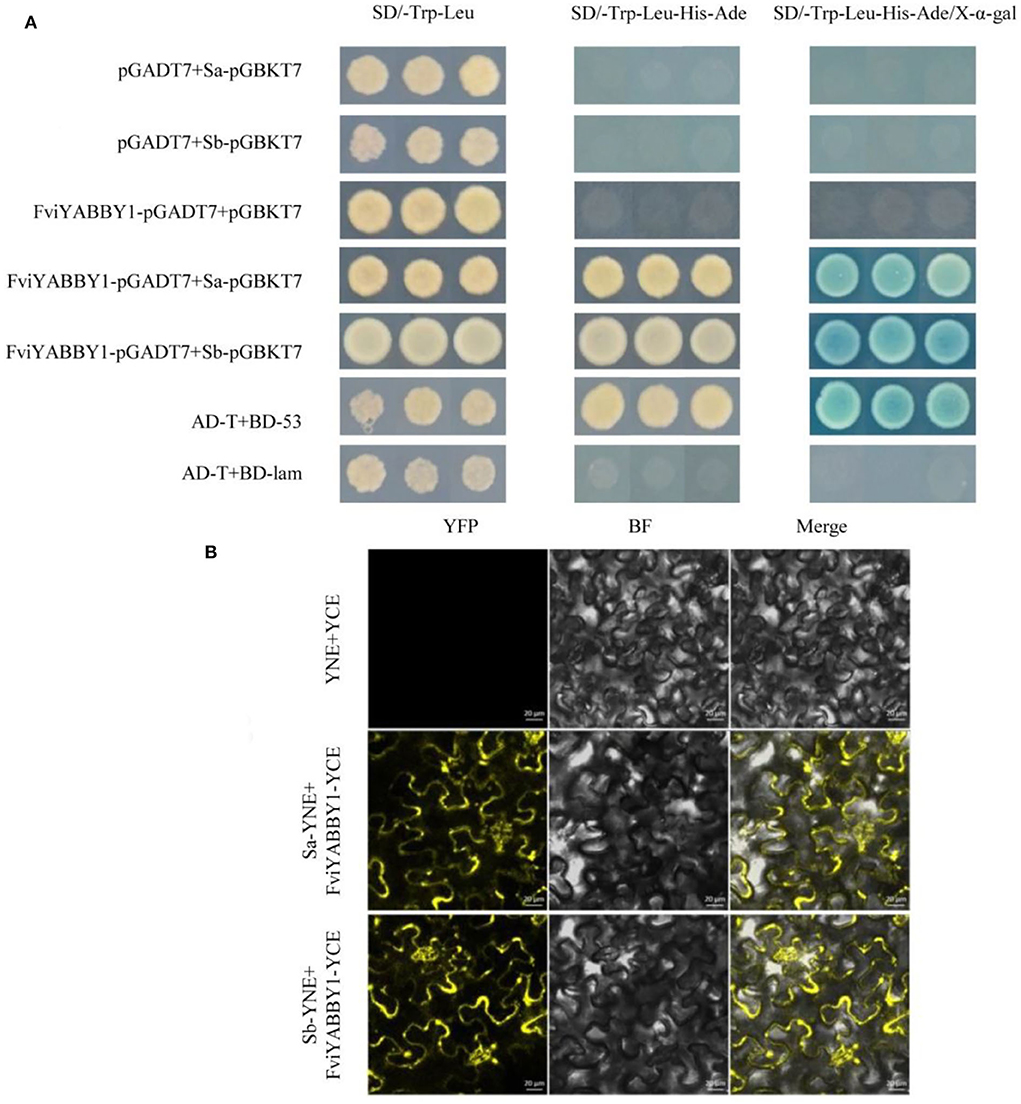
Figure 8. Interaction between FviYABBY1 and Sa-RNase/Sb-RNase. (A) Yeast two-hybrid assay. (B) Bimolecular fluorescence complementation (BiFC) assay. YFP, yellow fluorescence protein; BF, bright field; Merge, merged image of YFP and BF. Bar = 20 μm.
We further verified the protein interaction results of FviYABBY1 with Sa-RNase and Sb-RNase and the location of the interaction. The recombinant vectors were co-transferred into tobacco cells using Agrobacterium harboring FviYABBY1-35S:YCE+Sa-35S:YNE and FviYABBY1-35S:YCE+Sb-35S:YNE. As seen in Figure 8B, the tobacco cells containing FviYABBY1-35S:YCE+Sb-35S:YNE and FviYABBY1-35S:YCE+Sb-35S:YNE exhibited fluorescence, whereas control cells harboring YCE+35S:YNE did not. The results of the BiFC assay confirmed that FviYABBY1 interacts with Sa-RNase/Sb-RNase and that these interactions are not S-haplotype specific. Further, based on fluorescence signals, the cytoplasm is the main location of FviYABBY1–S-RNase interaction.
Fragaria viridis is a typical self-incompatible gametophyte. In this species, SI primarily manifests in the pollen tube, which is strongly inhibited in the styles (Evans and Jones, 1967). In a previous study, a pair of style determinants (Sa-RNase and Sb-RNase) was discovered based on transcriptomic data at 0 and 24 HAP, proving the occurrence of S-RNase-based GSI in Fragaria (Du et al., 2021). Consistent with previous reports, most self-incompatible pollen tubes eventually stop growing at 2/3 of the style in the genus Fragaria (Evans and Jones, 1967; Du et al., 2019, 2021). Contrary to that in other species, such as Solanum and Pyrus, the self-incompatible pollen tube in F. viridis showed a greater extension degree in styles, suggesting differences in the intermediate mechanisms or SI modifiers among self-incompatible species (Franklin-Tong and Franklin, 2003; Zhang et al., 2009; Baek et al., 2015; Claessen et al., 2019). Therefore, we further analyzed the previously obtained transcriptomic data and combined these with proteomic data to unveil the mechanisms of pollen tube development and pollen tube growth cessation due to SI in F. viridis. FviYABBY1, a putative transcription factor related to the polar growth of pollen tubes, has been a research hotspot, and its roles in F. viridis SI are discussed in this article.
In S-RNase-based GSI systems, a type of pollen–pistil interaction, pollen grains germinate and grow normally on the stigma, and the rejection of self-pollen tube is mainly determined by the style (Franklin-Tong and Franklin, 2003; Hiscock and Allen, 2008; Shi et al., 2017; Seth et al., 2019; Aloisi et al., 2020). In the present study, the majority of the F. viridis pollen tubes were arrested in the style at 24 HAP (Figure 1E), implying that certain time-specific genes (24 HAP) regulating pollen tube growth are the key targets of the Fragaria SI system. Successful manifestation of SI entails the involvement of modifiers other than the S-locus determinants (McClure et al., 2000; Sassa and Hirano, 2006). Here, we identified 2,181 genes and 200 proteins that were differentially expressed between 0 and 24 HAP in receptacles with gynoecia, and many SI-related genes were further predicted and classified (Supplementary Table S9). Interestingly, many additional DEGs and DAPs were enriched in GO terms, such as “pollen development,” “recognition of pollen,” “pollen germination,” and “pollen–style interaction.” Arabinogalactan proteins (AGPs) and the AGP extension hybrid glycoprotein TTS are closely linked to pollen development and control pollen tube elongation during S-RNase-based GSI (Majewska-Sawka and Nothnagel, 2000; Seifert and Roberts, 2007). Furthermore, genes encoding GDSL esterase/lipase protein, fasciclin-like arabinogalactan protein, stigma-specific stig1-like protein, and glycosyltransferase are involved in pollen development and germination (Goubet et al., 2003; Updegraff et al., 2009; Zhang et al., 2022). In addition, pollen receptor-like kinase (RLK) is a key factor in pollen–style recognition (Escobar-Restrepo et al., 2007), and RALF-like proteins play critical roles in the regulation of pollen tube growth (Covey et al., 2010; Murphy and De Smet, 2014; Zhang et al., 2020). All these genes were classified into the category “pollen development” and most of them were significantly upregulated at 24 HAP (Supplementary Table S9). These results suggest that the expression of the above genes is closely related to self-pollen development in styles. Pollen is a typical model of cell polar growth. Thus, a polar growth regulator of DEGs was further investigated according to functional annotation. Although the potential functions of these genes in Fragaria SI require further confirmation, our results indicate that some regulators related to pollen growth are involved in the SI response of F. viridis.
SI is an intricate biological process closely associated with ubiquitin-mediated proteolysis, which eventually leads to PCD and RNA degradation (Thomas and Franklin-Tong, 2004; McClure and Franklin-Tong, 2006; Zhang et al., 2009). In the present study, we found that SKP1 (Unigene3797_All), as a component of the ubiquitin–ligase complex, was gradually upregulated after self-pollination (FC > 29) at 24 h compared with the expression level at 0 h in the validation data (Figure 3N). Two DAPs (CL4737_Contig1_All and CL7114_Contig_All) and one DEG (Unigene8982_All) were annotated as E3 ubiquitin-protein ligases. A series of PCD-related DEGs and DAPs, such as MAPKKKs, 18.2 kDa class I heat shock protein (Unigene11193_All), and cytochrome P450, were identified (Supplementary Table S9), all of which are important regulators of PCD according to previous reports (Ludovico et al., 2002; del Pozo et al., 2004; Li et al., 2012). As the activity of S-RNase is protected in self-pollen tubes, S-RNase degrades cytoplasmic RNA, arrests protein synthesis, and finally inhibits pollen tube growth (Kao and Tsukamoto, 2004; Wang et al., 2009; Zhang et al., 2015). In the present study, 15 DEGs and 2 DAPs were enriched in the “RNA degradation” pathway (Supplementary Tables S4, S6), including one CCR4-associated factor (CAF), four NAC domain-containing proteins, three chaperonin 60 subunit beta 4, and two uncharacterized proteins (Supplementary Table S9). Many of these genes were annotated to “RNA degradation” in self-pollination for the first time in this work, and further research may reveal their necessity in the pollen–style interaction pathways.
Biological processes, such as “cell wall modification,” “accumulation extracellular calcium,” “synthesis of β-1,3 glucan callose,” and “presence of phenolic derivatives,” are common between SI and pathogen resistance (Zhou et al., 2014). In addition, enzymes, such as pectin lyase, cellulases, and glucanases, are typically secreted by pathogens for cell wall degradation (Wing et al., 1990; Yoder et al., 1990; Ham et al., 1997; Aich et al., 2017). Similar to that during pathogen defense (Wing et al., 1990; Marín-Rodríguez et al., 2002), cell wall modifiers, such as pectate lyase 5, beta-glucosidase 3, and glucan endo-1,3-beta-glucosidase, were downregulated at the transcript and protein levels (Supplementary Table S9). Ca2+ is vital in pathogen interactions, and the transient influx of Ca2+ across the plasma membrane plays a major role during the early stages of pathogen response (Zimmermann et al., 1997). In our transcriptome data, several Ca2+ signaling-related genes were markedly upregulated; as such, these genes were enriched in the KEGG pathway “plant–pathogen interaction” (Supplementary Figure S5 and Supplementary Table S9). As for pathogen response, hormones are essential for incompatibility response (Graaf et al., 2003; Robert-Seilaniantz et al., 2007; López et al., 2008; Song et al., 2013; Zhang et al., 2016). Here, we detected 56 DEGs and 8 DAPs that were enriched in the KEGG pathway “plant hormone signal transduction” (Supplementary Tables S4, S6). These findings support that the mechanism of plant–pathogen interactions are remarkably similar to that of pollen tube development and pollen–style recognition (Hodgkin et al., 1988; Thomas and Franklin-Tong, 2004).
YABBY family genes are seed-plant-specific transcription factors with critical roles in vegetative and reproductive development (Kumaran et al., 2002; Bartholmes et al., 2012; Finet et al., 2016; Filyushin et al., 2017; di Rienzo et al., 2021; Romanova et al., 2021; Luo et al., 2022). S-RNase-based SI occurs when pollen tubes stop growing and polar growth is hindered in self-styles (Wu et al., 2013; Claessen et al., 2019). Using transcriptomic data as clues, we focused on YABBY family genes, particularly YABBY1, which may be involved in the regulation of pollen tube polar growth. YABBY1 is primarily expressed in pollen and lacks a signal peptide, suggesting that it functions in the pollen. Yeast two-hybrid and BiFC assays confirmed that YABBY1 interacts with S-RNase, albeit without haplotype specificity. In species exhibiting SI, S-RNase enters the pollen tube non-specifically (Meng et al., 2014a,b) and exerts a toxic effect in the self-pollen tube; meanwhile, the toxicity of non-self S-RNase is eliminated (Wu et al., 2013; Sassa, 2016; Claessen et al., 2019). Therefore, self-S-RNase likely interacts with YABBY1 in the pollen tube, transmits the S-RNase cytotoxicity signal, and affects the polar growth of the pollen tube. In the non-self-pollen tube, the cytotoxicity of S-RNase is relieved by pollen determinants. In self-compatible F. vesca, given the loss of the S-RNase gene, YABBY1 executes the normal function of regulating the polar growth of the pollen tube tip. YABBY1 is positively correlated with S-RNase in an incompatible style–pollen interaction system after self-pollination. Therefore, S-RNase may regulate the expression of SI-related genes by positively regulating the expression of YABBY1.
SI is a system in which style interacts with pollen (Bedinger et al., 2017). In addition to YABBY1, six YABBY family members from F. viridis were highly expressed. Some YABBY family genes exhibited similar expression patterns and may be correlated to some extent. Genes in the same family may serve similar functions (Li et al., 2016; Lin et al., 2018); thus, other genes in the YABBY family may also be related to SI. However, whether they are involved in the SI response of F. viridis, similar to YABBY1, warrants further experimental exploration.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary materials.
JD, CG, and TW co-performed the experiment design, the major experiment operation, data handling, and manuscript writing. JW, ZN, SX, and FZ performed partial experiments. YQ and MZ proposed the research idea and experimental design and assisted with manuscript editing. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Nos. 32072540 and 31872056), the Fundamental Research Funds for the Central Universities (No. KYZZ2022004), Postgraduate Research & Practice Innovation Program of Jiangsu Province (No. KYCX21_0612), and the Jiangsu Province Agricultural Science and Technology Innovation Fund Projects [No. CX(21)2019].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.927001/full#supplementary-material
DEG, differentially expressed gene; DAP, differentially abundant protein; FC, fold change; GSI, gametophytic self-incompatibility; SLF, S-locus F-box protein; PCD, programmed cell death; SI, self-incompatibility; SC, self-incompatibility; iTRAQ, isobaric tags for relative and absolute quantitation; Nt, NCBI nucleotide sequences; Nr, NCBI non-redundant protein; COG, Cluster of Ortholog Genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; RNA-Seq, RNA sequencing; FDR, false discovery rate; 2-DE, two-dimensional gel electrophoresis; RT-qPCR, real-time quantitative PCR; FPKM, fragments per kilobase per million mapped reads; TCA, trichloroacetic acid; AGP, arabinogalactan proteins; CDPK, calcium-dependent protein kinase; ROS, reactive oxygen species.
Aguiar, B., Vieira, J., Cunha, A. E., Fonseca, N. A., Iezzoni, A., van Nocker, S., et al. (2015). Convergent evolution at the gametophytic self-incompatibility system in Malus and Prunus. PLoS ONE 10, e0126138. doi: 10.1371/journal.pone.0126138
Aich, S., Singh, R. K., Kundu, P., Pandey, S. P., and Datta, S. (2017). Genome-wide characterization of cellulases from the hemi-biotrophic plant pathogen, Bipolaris sorokiniana, reveals the presence of a highly stable GH7 endoglucanase. Biotechnol. Biofuels 10, 1–14. doi: 10.1186/s13068-017-0822-0
Allen, A. M., and Hiscock, S. J. (2008). “Evolution and phylogeny of self-incompatibility systems in angiosperms,” in Self Incompatibility in Flowering Plants. Evolution, Diversity, and Mechanisms, ed V. E. Tong (Berlin Heidelberg: Springer-Verlag), 73–101.
Aloisi, I., Distefano, G., Antognoni, F., Potente, G., Parrotta, L., Faleri, C., et al. (2020). Temperature-dependent compatible and incompatible pollen-style interactions in Citrus clementina Hort. ex Tan. show different transglutaminase features and polyamine pattern. Front. Plant Sci. 11, 1018. doi: 10.3389/fpls.2020.01018
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Armenteros, J. J. A., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Baek, Y. S., Covey, P. A., Petersen, J. J., Chetelat, R. T., McClure, B., and Bedinger, P. A. (2015). Testing the SI × SC rule: pollen-pistil interactions in interspecific crosses between members of the tomato clade (Solanum section Lycopersicon, Solanaceae). Am. J. Bot. 102, 302–311. doi: 10.3732/ajb.1400484
Bartholmes, C., Hidalgo, O., and Gleissberg, S. (2012). Evolution of the YABBY gene family with emphasis on the basal eudicot eschscholzia californica (Papaveraceae). Plant Biol. 14, 11–23. doi: 10.1111/j.1438-8677.2011.00486.x
Bedinger, P. A., Broz, A. K., Tovar-Mendez, A., and McClure, B. (2017). Pollen-pistil interactions and their role in mate selection. Plant Physiol. 173, 79–90. doi: 10.1104/pp.16.01286
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bošković, R. I., Sargent, D. J., and Tobutt, K. R. (2010). Genetic evidence that two independent S-loci control RNase-based self-incompatibility in diploid strawberry. J. Exp. Bot. 61, 755–763. doi: 10.1093/jxb/erp340
Bowman, J. L. (2000). The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 3, 17–22. doi: 10.1016/S1369-5266(99)00035-7
Chalivendra, S. C., Lopez-Casado, G., Kumar, A., Kassenbrock, A. R., Royer, S., Tovar-Mèndez, A., et al. (2012). Developmental onset of reproductive barriers and associated proteome changes in stigma/styles of Solanum pennellii. J. Exp. Bot. 64, 265–279. doi: 10.1093/jxb/ers324
Chen, X., Li, J., Cheng, T., Zhang, W., Liu, Y., Wu, P., et al. (2020). Molecular systematics of Rosoideae (Rosaceae). Plant Syst. Evol. 306, 1–12. doi: 10.1007/s00606-020-01629-z
Claessen, H., Keulemans, W., Van de Poel, B., and De Storme, N. (2019). Finding a compatible partner: self-incompatibility in European pear (Pyrus communis); molecular control, genetic determination, and impact on fertilization and fruit set. Front. Plant Sci. 10, 407. doi: 10.3389/fpls.2019.00407
Covey, P. A., Subbaiah, C. C., Parsons, R. L., Pearce, G., Lay, F. T., Anderson, M. A., et al. (2010). A pollen-specific RALF from tomato that regulates pollen tube elongation. Plant Physiol. 153, 703–715. doi: 10.1104/pp.110.155457
Del Duca, S., Serafini-Fracassini, D., and Cai, G. (2014). Senescence and programmed cell death in plants: polyamine action mediated by transglutaminase. Front. Plant Sci. 5, 120. doi: 10.3389/fpls.2014.00120
del Pozo, O., Pedley, K. F., and Martin, G. B. (2004). MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23, 3072–3082. doi: 10.1038/sj.emboj.7600283
di Rienzo, V., Imanifard, Z., Mascio, I., Gasser, C. S., Skinner, D. J., Pierri, C. L., et al. (2021). Functional conservation of the grapevine candidate gene INNER NO OUTER for ovule development and seed formation. Hortic Res. 8, 29. doi: 10.1038/s41438-021-00467-5
Du, J., Ge, C., Li, T., Wang, S., Gao, Z., Sassa, H., et al. (2021). Molecular characteristics of S-RNase alleles as the style determinant of self-incompatibility in Fragaria viridis. Hortic Res. 8, 185. doi: 10.1038/s41438-021-00623-x
Du, J., Lv, Y., Xiong, J., Ge, C., Iqbal, S., and Qiao, Y. (2019). Identifying genome-wide sequence variations and candidate genes implicated in self-incompatibility by resequencing Fragaria viridis. Int. J. Mol. Sci. 20, 1039. doi: 10.3390/ijms20051039
Elleman, C. J., and Dickinson, H. G. (1999). Commonalities between pollen/stigma and host/pathogen interactions: calcium accumulation during stigmatic penetration by Brassica oleracea pollen tubes. Sex. Plant Reprod. 12, 194–202. doi: 10.1007/s004970050192
Escobar-Restrepo, J. M., Huck, N., Kessler, S., Gagliardini, V., Gheyselinck, J., Yang, W. C., et al. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317, 656–660. doi: 10.1126/science.1143562
Evans, W. D. (1982). The production of multispecific octoploids from Fragaria species and the cultivated strawberry. Euphytica 31, 901–907. doi: 10.1007/BF00039230
Evans, W. D., and Jones, J. K. (1967). Incompatibility in Fragaria. Can. J. Genet. Cytol. 9, 831–836. doi: 10.1139/g67-088
Filyushin, M. A., Slugina, M. A., Shchennikova, A. V., and Kochieva, E. Z. (2017). YABBY3-orthologous genes in wild tomato species: structure, variability, and expression. Acta Naturae 9, 101–109. doi: 10.32607/20758251-2017-9-4-101-109
Finet, C., Floyd, S. K., Conway, S. J., Zhong, B., Scutt, C. P., and Bowman, J. L. (2016). Evolution of the YABBY gene family in seed plants. Evol. Dev. 18, 116–126. doi: 10.1111/ede.12173
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Franklin-Tong, N. V. E., and Franklin, F. C. H. (2003). Gametophytic self-incompatibility inhibits pollen tube growth using different mechanisms. Trends Plant Sci. 8, 598–605. doi: 10.1016/j.tplants.2003.10.008
Fujii, S., Kubo, K., and Takayama, S. (2016). Non-self-and self-recognition models in plant self-incompatibility. Nat. Plants 2, 16130. doi: 10.1038/nplants.2016.130
Goubet, F., Misrahi, A., Park, S. K., Zhang, Z., Twell, D., and Dupree, P. (2003). AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol. 131, 547–557. doi: 10.1104/pp.014555
Graaf, B. H., Knuiman, B. A., Derksen, J., and Mariani, C. (2003). Characterization and localization of the transmitting tissue-specific PELPIII proteins of Nicotiana tabacum. J. Exp. Bot. 54, 55–63. doi: 10.1093/jxb/erg002
Gruner, P., Ulrich, D., Neinhuis, C., and Olbricht, K. (2017). Fragaria viridis Weston: diversity and breeding potential of an underutilised strawberry species. Acta Hortic. 1156, 203–208. doi: 10.17660/ActaHortic.2017.1156.31
Gu, X., Gao, Z., Zhuang, W., Qiao, Y., Wang, X., Mi, L., et al. (2013). Comparative proteomic analysis of rd29A: RdreB1BI transgenic and non-transgenic strawberries exposed to low temperature. J. Plant Physiol. 170, 696–706. doi: 10.1016/j.jplph.2012.12.012
Gu, Z., Meng, D., Yang, Q., Yuan, H., Wang, A., Li, W., et al. (2015). A CBL gene, MdCBL5, controls the calcium signal and influences pollen tube growth in apple. Tree Genet. Genomes 11, 27. doi: 10.1007/s11295-015-0853-2
Ham, K. S., Wu, S. C., Darvill, A. G., and Albersheim, P. (1997). Fungal pathogens secrete an inhibitor protein that distinguishes isoforms of plant pathogenesis-related endo-β-1, 3-glucanases. Plant J. 11, 169–179. doi: 10.1046/j.1365-313X.1997.11020169.x
Hancock, J. F. (1999). “The strawberry species,” in Strawberries (Wallingford: CABI Publishing), 25–46.
Hancock, J. F., and Luby, J. J. (1993). Genetic resources at our doorstep: the wild strawberries. Bioscience 43, 141–147. doi: 10.2307/1312017
Hancock, J. F., Luby, J. J., Dale, A., Callow, P. W., Serce, S., and El-Shiek, A. (2002). Utilizing wild Fragaria virginiana in strawberry cultivar development: inheritance of photoperiod sensitivity, fruit size, gender, female fertility and disease resistance. Euphytica 126, 177–184. doi: 10.1023/A:1016309724998
Harkness, A., and Brandvain, Y. (2021). Non-self recognition-based self-incompatibility can alternatively promote or prevent introgression. New Phytol. 231, 1304–1307. doi: 10.1111/nph.17249
Hiscock, S. J., and Allen, A. M. (2008). Diverse cell signalling pathways regulate pollen-stigma interactions: the search for consensus. New Phytol. 179, 286–317. doi: 10.1111/j.1469-8137.2008.02457.x
Hodgkin, T., Lyon, G. D., and Dickinson, H. G. (1988). Recognition in flowering plants: a comparison of the Brassica self-incompatibility system and plant pathogen interactions. New Phytol. 110, 557–569. doi: 10.1111/j.1469-8137.1988.tb00296.x
Huang, Z., Van Houten, J., Gonzalez, G., Xiao, H., and van der Knaap, E. (2013). Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genom. 288, 111–129. doi: 10.1007/s00438-013-0733-0
Iseli, C., Jongeneel, C. V., and Bucher, P. (1999). ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. ISMB 99, 138–148.
Ishimizu, T., Sato, Y., Saito, T., Yoshimura, Y., Norioka, S., Nakanishi, T., et al. (1996). Identification and partial amino acid sequences of seven S-RNases associated with self-incompatibility of Japanese pear, Pyrus pyrifolia Nakai. J. Biochem. 120, 326–334. doi: 10.1093/oxfordjournals.jbchem.a021417
Iwano, M., and Takayama, S. (2012). Self/non-self discrimination in angiosperm self-incompatibility. Curr. Opin. Plant Biol. 15, 78–83. doi: 10.1016/j.pbi.2011.09.003
Kao, T., and Tsukamoto, T. (2004). The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell. 16, S72–S83. doi: 10.1105/tpc.016154
Kubo, K., Entani, T., Takara, A., Wang, N., Fields, A. M., Hua, Z., et al. (2010). Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330, 796–799. doi: 10.1126/science.1195243
Kumar, S., Deng, C. H., Hunt, M., Kirk, C., Wiedow, C., Rowan, D., et al. (2021). Homozygosity mapping reveals population history and trait architecture in self-incompatible pear (Pyrus spp.). Front. Plant Sci. 11, 590846. doi: 10.3389/fpls.2020.590846
Kumaran, M. K., Bowman, J. L., and Sundaresan, V. (2002). YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14, 2761–2770. doi: 10.1105/tpc.004911
Labokas, J., and Bagdonaitë, E. (2005). Phenotypic diversity of Fragaria vesca and F. viridis in Lithuania. Biologija 3, 19–22. Available online at: https://www.lmaleidykla.lt/ojs/index.php/biologija/article/view/583
Li, H., Yang, Q., Li, J., Gao, H., Li, P., and Zhou, H. (2015). The impact of temperature on microbial diversity and AOA activity in the Tengchong Geothermal Field China. Sci Rep. 5, 17056. doi: 10.1038/srep17056
Li, Q., Yan, W., Chen, H., Tan, C., Han, Z., Yao, W., et al. (2016). Duplication of OsHAP family genes and their association with heading date in rice. J. Exp. Bot. 67, 1759–1768. doi: 10.1093/jxb/erv566
Li, W., Meng, D., Gu, Z., Yang, Q., Yuan, H., Li, Y., et al. (2018). Apple S-RNase triggers inhibition of tRNA aminoacylation by interacting with a soluble inorganic pyrophosphatase in growing self-pollen tubes in vitro. New Phytol. 218, 579–593. doi: 10.1111/nph.15028
Li, Y., Hou, X., Lin, L., Jing, S., and Deng, M. (2000). Abnormal pollen germination and embryo abortion in the interspecific cross, Fragaria × ananassa × F. vesca, as related to cross-incompatibility. J. Jap. Soc. Hort. Sci. 69, 84–89. doi: 10.2503/jjshs.69.84
Li, Z., Yue, H., and Xing, D. (2012). MAP Kinase 6-mediated activation of vacuolar processing enzyme modulates heat shock-induced programmed cell death in Arabidopsis. New Phytol. 195, 85–96. doi: 10.1111/j.1469-8137.2012.04131.x
Lin, Y., Wang, K., Li, X., Sun, C., Yin, R., Wang, Y., et al. (2018). Evolution, functional differentiation, and co-expression of the RLK gene family revealed in Jilin ginseng, Panax ginseng CA Meyer. Mol. Genet. Genom. 293, 845–859. doi: 10.1007/s00438-018-1425-6
Liston, A., Cronn, R., and Ashman, T. L. (2014). Fragaria: a genus with deep historical roots and ripe for evolutionary and ecological insights. Am. J. Bot. 101, 1686–1699. doi: 10.3732/ajb.1400140
López, M. A., Bannenberg, G., and Castresana, C. (2008). Controlling hormone signaling is a plant and pathogen challenge for growth and survival. Curr. Opin. Plant Biol. 11, 420–427. doi: 10.1016/j.pbi.2008.05.002
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Ludovico, P., Rodrigues, F., Almeida, A., Silva, M. T., Barrientos, A., and Côrte-Real, M. (2002). Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol. Biol. Cell. 13, 2559–2976. doi: 10.1091/mbc.e01-12-0161
Luo, J., Zhou, Y., Pan, Q., Mu, Q., and Gu, T. (2022). Characterization of YABBY genes and the correlation between their transcript levels and histone modifications in strawberry. Sci. Hortic. 295, 110815. doi: 10.1016/j.scienta.2021.110815
Maas, J. L., Pooler, M. R., and Galletta, G. J. (1997). Bacterial angular leafspot disease strawberry: present status and prospects for control. Adv Strawb. 14, 18–24.
Majewska-Sawka, A., and Nothnagel, E. A. (2000). The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 122, 3–10. doi: 10.1104/pp.122.1.3
Marín-Rodríguez, M. C., Orchard, J., and Seymour, G. B. (2002). Pectate lyases, cell wall degradation and fruit softening. J. Exp. Bot. 53, 2115–2119. doi: 10.1093/jxb/erf089
Marta, A. E., Camadro, E. L., Díaz-Ricci, J. C., and Castagnaro, A. P. (2004). Breeding barriers between the cultivated strawberry, Fragaria × ananassa, and related wild germplasm. Euphytica 136, 139–150. doi: 10.1023/B:EUPH.0000030665.95757.76
McClure, B. A., Cruz-Garcia, F., Beecher, B., and Sulaman, W. (2000). Factors affecting inter-and intra-specific pollen rejection in Nicotiana. Ann. Bot. 85, 113–123. doi: 10.1006/anbo.1999.1061
McClure, B. A., and Franklin-Tong, V. (2006). Gametophytic self-incompatibility: understanding the cellular mechanisms involved in “self” pollen tube inhibition. Planta 224, 233–245. doi: 10.1007/s00425-006-0284-2
Mello, B., Tao, Q., Tamura, K., and Kumar, S. (2017). Fast and accurate estimates of divergence times from big data. Mol. Biol. Evol. 34, 45–50. doi: 10.1093/molbev/msw247
Meng, D., Gu, Z., Li, W., Wang, A., Yuan, H., Yang, Q., et al. (2014a). Apple MdABCF assists in the transportation of S-RNase into pollen tubes. Plant J. 78, 990–1002. doi: 10.1111/tpj.12524
Meng, D., Gu, Z., Yuan, H., Wang, A., Li, W., Yang, Q., et al. (2014b). The microtubule cytoskeleton and pollen tube Golgi vesicle system are required for in vitro S-RNase internalization and gametic self-incompatibility in apple. Plant Cell Physiol. 55, 977–989. doi: 10.1093/pcp/pcu031
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Muñoz-Sanz, J. V., Zuriaga, E., Cruz-García, F., McClure, B., and Romero, C. (2020). Self-(in)compatibility systems: target traits for crop-production, plant breeding, and biotechnology. Front. Plant Sci. 11, 195. doi: 10.3389/fpls.2020.00195
Murphy, E., and De Smet, I. (2014). Understanding the RALF family: a tale of many species. Trends Plant Sci. 19, 664–671. doi: 10.1016/j.tplants.2014.06.005
Qu, H., Zhang, Z., Wu, F., and Wang, Y. (2016). The role of Ca2+ and Ca2+ channels in the gametophytic self-incompatibility of Pyrus pyrifolia. Cell Calcium 60, 299–308. doi: 10.1016/j.ceca.2016.06.006
Robert-Seilaniantz, A., Navarro, L., Bari, R., and Jones, J. D. (2007). Pathological hormone imbalances. Curr. Opin. Plant Biol. 10, 372–379. doi: 10.1016/j.pbi.2007.06.003
Romanova, M. A., Maksimova, A. I., Pawlowski, K., and Voitsekhovskaja, O. V. (2021). YABBY genes in the development and evolution of land plants. Int. J. Mol. Sci. 22, 4139. doi: 10.3390/ijms22084139
Sanabria, N., Goring, D., Nürnberger, T., and Dubery, I. (2008). Self/nonself perception and recognition mechanisms in plants: a comparison of self-incompatibility and innate immunity. New Phytol. 178, 503–514. doi: 10.1111/j.1469-8137.2008.02403.x
Sargent, D. J., Geibel, M., Hawkins, J. A., Wilkinson, M. J., Battey, N. H., and Simpson, D. W. (2004). Quantitative and qualitative differences in morphological traits revealed between diploid Fragaria species. Ann. Bot. 94, 787–796. doi: 10.1093/aob/mch217
Sassa, H. (2016). Molecular mechanism of the S-RNase-based gametophytic self-incompatibility in fruit trees of Rosaceae. Breed. Sci. 66, 116–121. doi: 10.1270/jsbbs.66.116
Sassa, H., and Hirano, H. (2006). Identification of a new class of pistil-specific proteins of Petunia inflata that is structurally similar to, but functionally distinct from, the self-incompatibility factor HT. Mol. Genet. Genom. 275, 97–104. doi: 10.1007/s00438-005-0067-7
Sassa, H., Kakui, H., and Mai, M. (2010). Pollen-expressed F-box gene family and mechanism of S-RNase-based gametophytic self-incompatibility (GSI) in Rosaceae. Sex. Plant Reprod. 23, 39–43. doi: 10.1007/s00497-009-0111-6
Seifert, G. J., and Roberts, K. (2007). The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 58, 137–161. doi: 10.1146/annurev.arplant.58.032806.103801
Seth, R., Bhandawat, A., Parmar, R., Singh, P., Kumar, S., and Sharma, R. K. (2019). Global transcriptional insights of pollen-pistil interactions commencing self-incompatibility and fertilization in tea [Camellia sinensis (L.) O. Kuntze]. Int. J. Mol. Sci. 20, 539. doi: 10.3390/ijms20030539
Shi, D., Tang, C., Wang, R., Gu, C., Wu, X., Hu, S., et al. (2017). Transcriptome and phytohormone analysis reveals a comprehensive phytohormone and pathogen defence response in pear self-/cross-pollination. Plant Cell Rep. 36, 1785–1799. doi: 10.1007/s00299-017-2194-0
Song, S., Qi, T., Fan, M., Zhang, X., Gao, H., Huang, H., et al. (2013). The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 9, e1003653. doi: 10.1371/journal.pgen.1003653
Stahle, M. I., Kuehlich, J., Staron, L., von Arnim, A. G., and Golz, J. F. (2009). YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell. 21, 3105–3118. doi: 10.1105/tpc.109.070458
Staudt, G. (2009). Strawberry biogeography, genetics and systematics. Acta Hortic. 842, 71–84. doi: 10.17660/ActaHortic.2009.842.1
Tao, R., and Iezzoni, A. F. (2010). The S-RNase-based gametophytic self-incompatibility system in Prunus exhibits distinct genetic and molecular features. Sci. Hortic. 124, 423–433. doi: 10.1016/j.scienta.2010.01.025
Thomas, S. G., and Franklin-Tong, V. E. (2004). Self-incompatibility triggers programmed cell death in Papaver pollen. Nature 429, 305–309. doi: 10.1038/nature02540
Tolin, S., Arrigoni, G., Moscatiello, R., Masi, A., Navazio, L., Sablok, G., et al. (2013). Quantitative analysis of the naringenin-inducible proteome in Rhizobium leguminosarum by isobaric tagging and mass spectrometry. Proteomics 13, 1961–1972. doi: 10.1002/pmic.201200472
Toriba, T., Harada, K., Takamura, A., Nakamura, H., Ichikawa, H., Suzaki, T., et al. (2007). Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet. Genom. 277, 457–468 doi: 10.1007/s00438-006-0202-0
Ulrich, D., Komes, D., Olbricht, K., and Hoberg, E. (2007). Diversity of aroma patterns in wild and cultivated Fragaria accessions. Genet. Resour. Crop Evol. 54, 1185–1196. doi: 10.1007/s10722-006-9009-4
Updegraff, E. P., Zhao, F., and Preuss, D. (2009). The extracellular lipase EXL4 is required for efficient hydration of Arabidopsis pollen. Sex. Plant Reprod. 22, 197–204. doi: 10.1007/s00497-009-0104-5
Vekemans, X., and Castric, V. (2021). When the genetic architecture matters: evolutionary and ecological implications of self versus nonself recognition in plant self-incompatibility. New Phytol. 231, 1630–1643. doi: 10.1111/nph.17471
Vieira, J., Fonseca, N. A., and Vieira, C. P. (2008). An S-RNase-based gametophytic self-incompatibility system evolved only once in eudicots. J. Mol. Evol. 67, 179–190. doi: 10.1007/s00239-008-9137-x
Vieira, J., Pimenta, J., Gomes, A., Laia, J., Rocha, S., Heitzler, P., et al. (2021). The identification of the Rosa S-locus and implications on the evolution of the Rosaceae gametophytic self-incompatibility systems. Sci. Rep. 11, 3710. doi: 10.1038/s41598-021-83243-8
Vieira, J., Rocha, S., Vázquez, N., López-Fernández, H., Fdez-Riverola, F., Reboiro-Jato, M., et al. (2019). Predicting specificities under the non-self gametophytic self-incompatibility recognition model. Front. Plant Sci. 10, 879. doi: 10.3389/fpls.2019.00879
Wang, C., Wu, J., Xu, G., Gao, Y., Chen, G., Wu, J., et al. (2010). S-RNase disrupts tip-localized reactive oxygen species and induces nuclear DNA degradation in incompatible pollen tubes of Pyrus pyrifolia. J. Cell Sci. 123, 4301–4309. doi: 10.1242/jcs.075077
Wang, C., Xu, G., Jiang, X., Chen, G., Wu, J., Wu, H., et al. (2009). S-RNase triggers mitochondrial alteration and DNA degradation in the incompatible pollen tube of Pyrus pyrifolia in vitro. Plant J. 57, 220–229. doi: 10.1111/j.1365-313X.2008.03681.x
Wang, D., Skibbe, D. S., and Walbot, V. (2013). Maize Male sterile 8 (Ms8), a putative β-1, 3-galactosyltransferase, modulates cell division, expansion, and differentiation during early maize anther development. Plant Reprod. 26, 329–338. doi: 10.1007/s00497-013-0230-y
Wen, B., Zhou, R., Feng, Q., Wang, Q., Wang, J., and Liu, S. (2014). IQuant: an automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics 14, 2280–2285. doi: 10.1002/pmic.201300361
Williams, J. S., Der, J. P., dePamphili, C. W., and Kao, T.-h. (2014). Transcriptome analysis reveals the same 17 S-locus F-box genes in two haplotypes of the self-incompatibility locus of Petunia inflate. Plant Cell. 26, 2873–2888. doi: 10.1105/tpc.114.126920
Williams, J. S., Wu, L., Li, S., Sun, P., and Kao, T. H. (2015). Insight into S-RNase-based self-incompatibility in Petunia: recent findings and future directions. Front. Plant Sci. 6, 41. doi: 10.3389/fpls.2015.00041
Wing, R. A., Yamaguchi, J., Larabell, S. K., Ursin, V. M., and McCormick, S. (1990). Molecular and genetic characterization of two pollen-expressed genes that have sequence similarity to pectate lyases of the plant pathogen Erwinia. Plant Mol. Biol. 14, 17–28. doi: 10.1007/BF00015651
Wu, C., Gu, Z., Li, T., Yu, J., Liu, C., Fan, W., et al. (2021). The apple MdPTI1L kinase is phosphorylated by MdOXI1 during S-RNase-induced reactive oxygen species signaling in pollen tubes. Plant Sci. 305, 110824. doi: 10.1016/j.plantsci.2021.110824
Wu, J., Gu, C., Khan, M. A., Wu, J., Gao, Y., Wang, C., et al. (2013). Molecular determinants and mechanisms of gametophytic self-incompatibility in fruit trees of Rosaceae. Crit. Rev. Plant Sci. 32, 53–68. doi: 10.1080/07352689.2012.715986
Wu, L., Williams, J. S., Sun, L., and Kao, T. H. (2020). Sequence analysis of the Petunia inflata S-locus region containing 17 S-Locus F-Box genes and the S-RNase gene involved in self-incompatibility. Plant J. 104, 1348–1368. doi: 10.1111/tpj.15005
Yoder, M. D., DeChaine, D. A., and Jurnak, F. (1990). Preliminary crystallographic analysis of the plant pathogenic factor, pectate lyase C from Erwinia chrysanthemi. J. Biol. Chem. 265, 11429–11431. doi: 10.1016/S0021-9258(19)38415-7
Zeeshan, A. B., Yang, Y., Sharif, R., Wu, S. N., Xie, Y., and Wang, C. (2020). Genome wide identification, characterization, and expression analysis of YABBY-gene family in wheat (Triticum aestivum L.). Agronomy 10, 1189. doi: 10.3390/agronomy10081189
Zhang, C., Wang, L., Wei, K., Wu, L., Li, H., Zhang, F., et al. (2016). Transcriptome analysis reveals self-incompatibility in the tea plant (Camellia sinensis) might be under gametophytic control. BMC Genom. 17, 359. doi: 10.1186/s12864-016-2703-5
Zhang, H., Jing, X., Chen, Y., Liu, Z., Xin, Y., and Qiao, Y. (2020). The genome-wide analysis of RALF-Like genes in strawberry (wild and cultivated) and five other plant species (Rosaceae). Genes 11, 174. doi: 10.3390/genes11020174
Zhang, P., Zhang, H., Du, J., and Qiao, Y. (2022). Genome-wide identification and co-expression analysis of GDSL genes related to suberin formation during fruit russeting in pear. Hortic Plant J. 8, 153–170. doi: 10.1016/j.hpj.2021.11.010
Zhang, S., Ding, F., He, X., Luo, C., Huang, G., and Hu, Y. (2015). Characterization of the 'Xiangshui'lemon transcriptome by de novo assembly to discover genes associated with self-incompatibility. Mol. Genet. Genom. 290, 365–375. doi: 10.1007/s00438-014-0920-7
Zhang, S., Li, C., Cao, J., Zhang, Y., Zhang, S., Xia, Y., et al. (2009). Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3. 13 activation. Plant Physiol. 151, 1889–1901. doi: 10.1104/pp.109.146803
Zhang, T., Gao, C., Yue, Y., Liu, Z., Ma, C., Zhou, G., et al. (2017). Time-course transcriptome analysis of compatible and incompatible pollen-stigma interactions in Brassica napus L. Front. Plant Sci. 8, 682. doi: 10.3389/fpls.2017.00682
Zhao, P., Zhang, L., and Zhao, L. (2015). Dissection of the style's response to pollination using transcriptome profiling in self-compatible (Solanum pimpinellifolium) and self-incompatible (Solanum chilense) tomato species. BMC Plant Biol. 15, 119. doi: 10.1186/s12870-015-0492-7
Zhao, Y., Liu, C., Ge, D., Yan, M., Ren, Y., Huang, X., et al. (2020). Genome-wide identification and expression of YABBY genes family during flower development in Punica granatum L. Gene 752, 144784. doi: 10.1016/j.gene.2020.144784
Zhou, Q., Jia, J., Huang, X., Yan, X., Cheng, L., Chen, S., et al. (2014). The large-scale investigation of gene expression in Leymus chinensis stigmas provides a valuable resource for understanding the mechanisms of Poaceae self-incompatibility. BMC Genom. 15, 399. doi: 10.1186/1471-2164-15-399
Keywords: self-incompatibility, FviYABBY1, transcriptome, proteome, Fragaria viridis
Citation: Du J, Ge C, Wang T, Wang J, Ni Z, Xiao S, Zhao F, Zhao M and Qiao Y (2022) Combined transcriptomic and proteomic analysis reveals multiple pathways involved in self-pollen tube development and the potential roles of FviYABBY1 in self-incompatibility in Fragaria viridis. Front. Plant Sci. 13:927001. doi: 10.3389/fpls.2022.927001
Received: 23 April 2022; Accepted: 17 August 2022;
Published: 14 September 2022.
Edited by:
Dayun Tao, Yunnan Academy of Agricultural Sciences, ChinaReviewed by:
Igor Pacheco, Universidad de Chile, ChileCopyright © 2022 Du, Ge, Wang, Wang, Ni, Xiao, Zhao, Zhao and Qiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yushan Qiao, cWlhb3l1c2hhbkBuamF1LmVkdS5jbg==; Mizhen Zhao, bmp6aGFvbXpAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.