
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 June 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.924645
Recent studies have shown that reprogramming of gene expression in a genome can induce the production of proteins enabling yield increase. The transcription activator-like effectors (TALEs) from several species of bacterial Xanthomonas have been extensively studied, and a series of research tools, such as genome editing tool TALENs and gene expression activators, have been developed based on the specific protein–nucleic acid recognition and binding mechanisms of TALEs. In this proof-of-principle study, we designed and constructed a designer TALE (dTALE), designated as dTALE-NOG1, to specifically target the promoter of OsNOG1 gene in rice, and demonstrated that this dTALE can be used as a new type of plant growth regulator for better crop growth and harvest. In doing so, the dTALE-NOG1 was transferred into the non-pathogenic Xanthomonas oryzae pv. oryzae (Xoo) strain PH to generate a genetically engineered bacteria (GEB) strain called PH-dtNOG1. Functional verification showed that dTALE-NOG1 could significantly induce the expression of OsNOG1. By spraying cell suspension of PH-dtNOG1 on the rice plants during the tillering stage, the transcription level of OsNOG1 was highly enhanced, the grain number of rice plants was increased by more than 11.40%, and the grain yield per plant increased by more than 11.08%, demonstrating that the dTALE-NOG1 was highly effective in enhancing rice yield. This work provided a new strategy for manipulating agronomical traits by reprogramming gene expression in a crop genome.
Food security has always been a major issue for human beings, which has become even more prominent during the COVID-19 pandemic (Stark, 2021). Over the past century, crop breeding and commercialization of new varieties have brought about a tremendous increase in food productivity. However, the yield of major crops seems to be approaching a ceiling in the recent decade (Mann, 1999; Zhang et al., 2014). As such, feeding the huge and ever-growing human population has become a bigger challenge due to the global climate change along with frequent emergences of pests/pathogens and other environmental hazards.
Basically, crop breeding relies on genetic recombination to introduce useful genes/alleles into elite germplasms, which is not only time-consuming but also expensive. Particularly, in recent times and future, it is getting harder and harder to generate and select a significantly improved genotype of major crops because favorable alleles for most simply inherited traits are already prevalent and often fixed in elite germplasms (Varshney et al., 2012; Ansari et al., 2017). On the other hand, transgenic technology of plants, especially food crops, is still strictly limited in the application of agricultural production. Therefore, new technologies that can genetically manipulate the agronomic performance of current crop varieties without the need to change their genome are highly desired; we call such technology crop agronomic performance manipulation epigenetically (CAPME). Recently, the RNA viral transfection (RVT)-based CAPME technology has been developed, which can fine-turn the crop agronomic traits by reprogramming the plant genome through transient viral induction that was delivered to the crop through aerial spray (Torti et al., 2021).
The grain yield of rice (Oryza sativa L.), a staple crop feeding nearly half of the world’s population (Birla et al., 2017), is a complex agronomic trait composed of several factors, including effective tiller number, grain number per panicle, and grain weight, and among them panicle formation is the key factor (Khush, 1995). Studies have revealed that OsNOG1 gene encodes an enoyl-CoA hydratase/isomerase protein that enhances the activity of panicle meristem and increases the number of spikelets by decreasing the jasmonic acid (JA) content in rice (Huo et al., 2017). Consistent with this, the overexpression of OsNOG1 significantly increases rice yield via the increase of grain number per panicle (Huo et al., 2017). Therefore, we speculate that induction of OsNOG1 expression has the potential to increase rice yield.
Transcription activator-like effectors (TALEs) are a class of proteins identified in plant pathogenic Xanthomonas spp., including Xanthomonas oryzae pv. oryzae (Xoo), the causal agent of bacterial blight, a worldwide disease of rice (Boch and Bonas, 2010). The Xanthomonas pathogens inject their TALEs through the type-III secretion system (T3SS) into plant cells primarily to activate the expression of host genes that contribute to pathogen growth (Boch and Bonas, 2010), but they occasionally trigger the executor gene-mediated disease resistance in rice (Ji et al., 2022). TALEs are structurally conserved and function as eukaryotic transcription factors in inducing gene expression (Cornelis, 2006). A typical TALE in nature usually contains a secretion domain (SD), a chaperone binding domain (CBD), a DNA binding domain (DBD), multiple nuclear localization signals (NLS), and an acid transcription activation domain (TAD). The DBD consists of a variable number of 33–35 amino acid repeats that are nearly identical, except for the two variable amino acids at positions 12 and 13, designated as repeat variable di-residues (RVDs). Each RVD specifically recognizes one nucleotide, and the DNA target sequence of a given TALE is determined by the combination of a number and composition of the RVDs in the repeats (Boch et al., 2009; Moscou and Bogdanove, 2009). Based on the codes between RVDs and their recognized nucleotides (Boch et al., 2009), the central repeat region or DNA-binding domain of a TALE can be designed and modularly assembled, and this has allowed for broad applications in genome and epigenome engineering, including genome editing, transcriptional activation, transcriptional repression, and live visualization of chromatin dynamics (Anderson et al., 2020).
Here, we report the development of another CAPME technology based on the designer TALE (dTALE). A dTALE targeting the rice OsNOG1 gene, designated as dTALE-NOG1, was constructed and transferred into the non-pathogenic Xoo strain PH (Ji et al., 2016) to generate the genetically engineered bacteria (GEB) strain, designated as PH-dtNOG1, that can deliver the dTALE-NOG1 into rice cells. We showed that the expression of OsNOG1 was successfully induced by PH-dtNOG1, and rice yield was increased by simply spraying rice plants with cell suspension of PH-dtNOG1. This CAPME strategy does not involve any transgene in plants, and so it is safer and more reliable. This work provides a new strategy for promoting crop growth and increasing yield by a CAPME technology, with a significance for agricultural production and global food security.
The rice plants of japonica varieties Nipponbare (Nip), Jinjing 818 (JJ818), and indica varieties Guichao No.2 (GC2), R007 were planted in the screenhouse of Institute of Crop Science of Chinese Academy of Agricultural Sciences, Beijing, China. The growth season is from May to October, and the rice plants were managed with regular water and fertilizer management and natural lighting. The Nicotiana benthamiana plants were planted in phytotron with 14 h light (20,000 Lux) and 10 h dark, 25°C, relative humidity 70%, and watered with fertilizer once a week.
The dTALE targets in promoters of OsHYR (LOC4331412), OsSUT2 (LOC9266530), and OsNOG1 (LOC4324637) were selected by considering the number of repeat units (Boch and Bonas, 2010; Zheng et al., 2014), distance to the start codon (Grau et al., 2013), distance to the transcription start site (TSS; Antony et al., 2010), sequence specificity in the rice genome, and coverage with other transcription regulatory elements (Brückner et al., 2015). One target was selected for each gene.
The complete sequences of OsNOG1 in all rice varieties used in this study were amplified using 2 × Rapid Taq Master Mix (P222, Vazyme) with primers No. 1–10 (Supplementary Table 1) and sequenced. The alignment was generated by SnapGene software (from Insightful Science1) using MUSCLE method with default parameter settings. Promoters of OsHYR, OsSUT2, and OsNOG1 were analyzed by New PLACE.2
Three vectors for expressing dTALEs targeting promoters of the above three genes were assembled using an improved unit assembly method (Huang et al., 2011; Zheng et al., 2014). A, T, C, and G were respectively identified by RVDs NI, NG, HD, and NN. Based on the one-unit vectors (pTALE-A, pTALE-T, pTALE-C, and pTALE-G), the pTALE-NOG1, including 20 RVDs repeats coding fragments was generated by several rounds of digestion and ligation. The digestions were performed by using FastDigest NheI (FD0973, Thermo Scientific) and FastDigest BcuI (FD1254, Thermo Scientific) with FastDigest HindIII (FD0504, Thermo Scientific), respectively. The ligation was achieved by using T4 DNA Ligase (2011A, Takara). The assembled DNA fragment coding the designed RVDs repeats was cut-off from pTALE-NOG1 by BcuI and NheI and inserted into the NheI site of pKSS-AvrXaΔRepeat, an expression vector containing the N-segment and C-segment including DYKDDDDK tag and TAD of TALE, resulting in a vector designated as pdTALE-NOG1. The pdTALE-NOG1 was linearized and ligated into the HindIII site of the cosmid vector pHM1 (Gu et al., 2005; Figure 1). The final dTALE transformation vector pHM1–dTALE-NOG1 (Figure 1) was transferred into the Xoo strain PH, a PXO99A none TALEs mutant, by electroporation, resulting in the engineered Xoo strain PH-dtNOG1 (Figure 2C). The other two dTALEs were constructed by the same process. Every step of the vector construction process was validated by Sanger sequencing. Since pHM1 contains spectinomycin adenylyltransferase and pKSS-AvrXaΔRepeat contains β-lactamase (Figure 1), the PH-dt strains, containing the corresponding dTALE, were cultured with NA medium (1.0% sucrose, 0.5% polypeptone, 0.1% yeast extract, 0.3% beef extract, pH 6.8) with ampicillin and spectinomycin added.
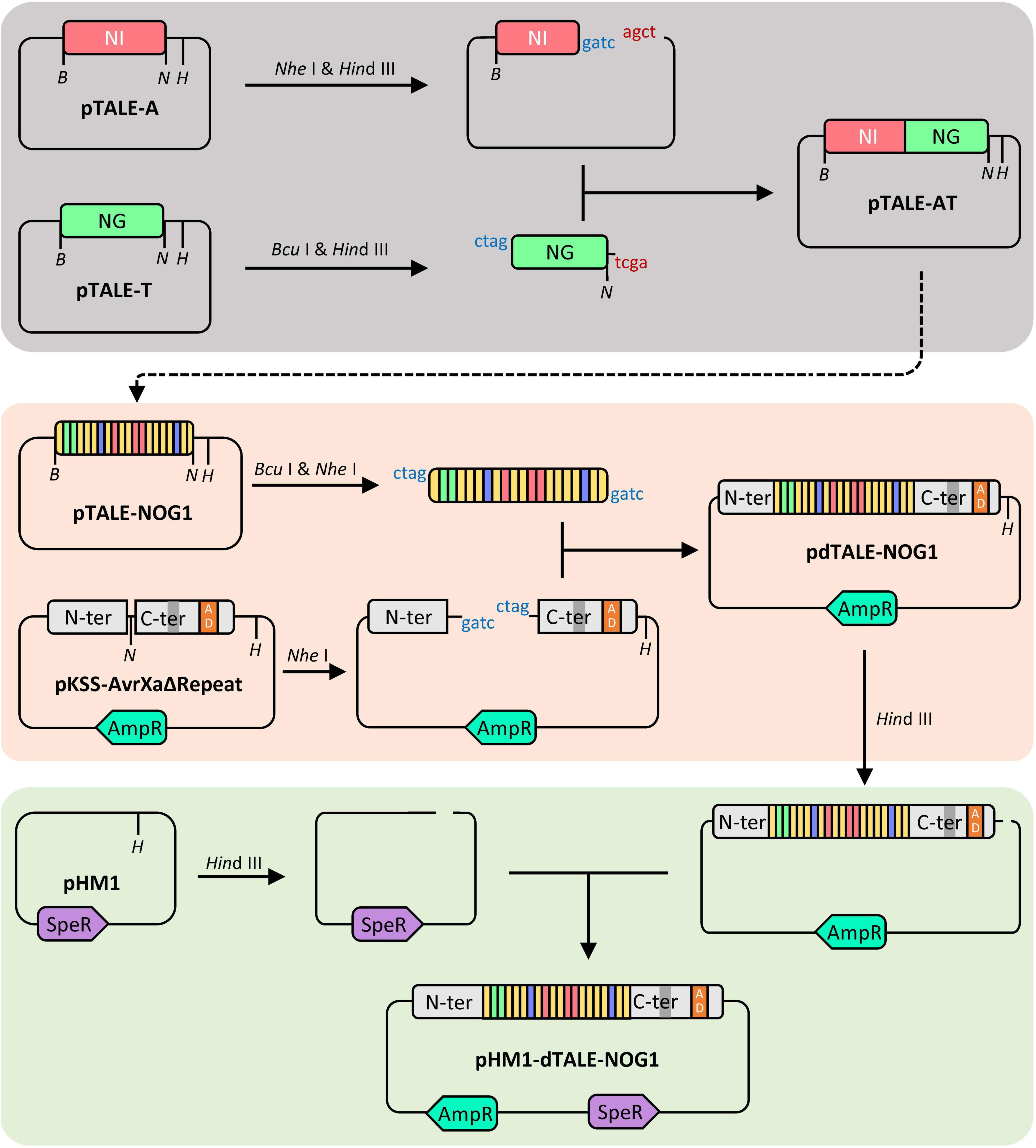
Figure 1. Schematic diagram of dTALE vector construction. The process of dTALE vector assembly. Based on 4 pTALE unit vectors (e.g., pTALE-A), the pTALE-NOG1 was generated by several rounds of digestion and ligation. The RVDs repeats coding region from pTALE-NOG1 was cut-off and fused into pKSS-AvrXaΔRepeat, resulting in pdTALE-NOG1. The ligation of pHM1 and pdTALE-NOG1 generated the final dTALE transformation vector pHM1-dTALE-NOG1. Restriction enzyme cutting sites of BcuI, NheI, and HindIII are represented by the letter B, N, and H, respectively. AmpR codes β-lactamase for ampicillin resistance and SpeR codes spectinomycin adenylytransferase for spectinomycin resistance.
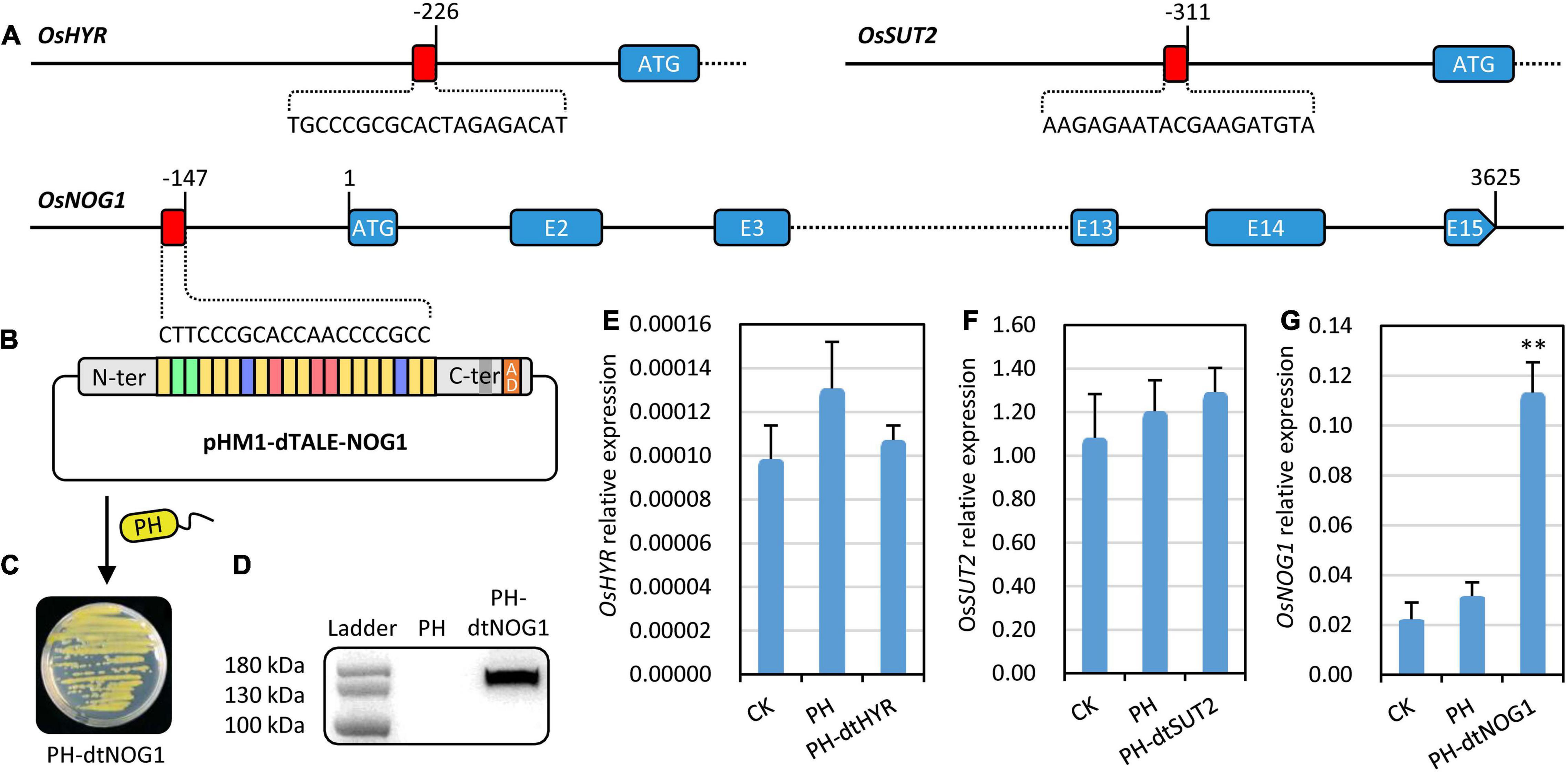
Figure 2. Targets of dTALEs and target gene responses to the genetically engineered bacteria (GEB) strains. (A) dTALE target sites in promoters of OsHYR, OsSUT2, and OsNOG1. The target sites were presented by red rectangle and the nucleotides were shown. The coding region or exons were shown by blue rectangles. (B) The schematic diagram of the construct pHM1–dTALE-NOG1, which was transferred into the non-pathogenic Xoo strain PH and resulted in GEB strain PH-dtNOG1. (C) PH-dtNOG1 grown on NA (Spe and Amp) plate. (D) Protein of dTALE-NOG1 expressed in PH-dtNOG1 detected by Western blot with PH as a negative control. (E–G) Expression of OsHYR, OsSUT2, and OsNOG1 in rice leaves injected with PH-dtHYR, PH-dtSUT2, and PH-dtNOG1, respectively. OsUbq (LOC4332169) was used as the reference gene for qRT-PCR. Data values were represented as mean with error bars representing standard deviation (n = 3). The significance of the difference was tested by t-test. **P < 0.01.
The activator, p1305-dTALE-NOG1, was generated by inserting the coding sequence of dTALE-NOG1 into NcoI and Eco72I sites of pCAMBIA1305. The reporter, pBI121-NOG1pro:GUS, was generated by inserting the promoter sequence of OsNOG1 of JJ818 and GC2 (amplified using primers No. 11–12, Supplementary Table 1) into HindIII and SmaI sites of pBI121 using ClonExpress® II One Step Cloning Kit (C112, Vazyme). These vectors were validated by Sanger sequencing.
A single colony of PH or PH-dtNOG1 was cultured in 3 mL of NA medium (spectinomycin and ampicillin added) at 200 RPM at 28°C for 24 h until turbidity. The bacteria culture was centrifuged at 12,000 RPM for 2 min to collect the bacteria and then 150 μL Solution I (25 mM pH 8.0 Tris–HCl, 10 mM EDTA, 50 mM Glucose) was added. After suspension, 30 μL lysozyme was added, mixed well, and let to stand for 5 min at room temperature. Then 200 μL of 1 × SDS loading buffer, which is precooled at 4°C, was added to the bacteria suspension, mixed well, and placed on ice. Then the suspension was for boiling water bath for 10 min, ice bath for 5 min, and centrifuged at 12,000 RPM for 1 min. Then 15 μL supernatant was used for SDS-PAGE electrophoresis and Western blot with ProteinFind® Anti-DYKDDDDK Mouse Monoclonal Antibody (HT201, TransGen), ProteinFind® Goat Anti-Mouse IgG (H + L), HRP Conjugate (HS201, TransGen), and Immobilon™ Western Chemiluminescent HRP Substrate (WBKL S0500, MILLIPORE) according to the product instructions. The above process was repeated independently at least three times.
The Agrobacterium tumefaciens strain GV3101, containing the corresponding plasmid, was cultured on LB medium (rifampicin and kanamycin added). Single clones were selected and transferred into 2 mL medium for culturing at 200 RPM at 28°C overnight. Then 100 μL bacterial suspension culture was transferred into 5 mL of LB medium (rifampicin and kanamycin added) with 2 μL 0.1 M acetosyringone (AS) and 100 μL 0.5 M 2-(N-morpholino) ethanesulfonic acid (MES) and cultured by shaking overnight. The bacteria were collected at 4,000 RPM and centrifuged for 5 min. The bacteria suspension OD600 value was adjusted to 1.0 with 10 mM MgCl2 solution (containing 0.1 mM AS and 10 mM MES). The bacterial solution was injected into the lower epidermis of N. benthamiana leaves on three plants using a 1 mL syringe without a needle.
Leaves of N. benthamiana were collected 2 days after injection and incubated in GUS staining buffer (0.1 M pH 7.0 sodium phosphate, 10 mM pH 8.0 EDTA, 0.1% Triton X-100, 1 mM K3Fe (CN)6, 2 mM X-Gluc in dimethylformamide; Jefferson, 1987) for 24 h at 37°C. Then the staining buffer was removed and the leaves were washed with several changes of ethanol (up to 2 h per wash) until the chlorophyll fades. The discolored leaves were stored in 75% ethanol for photography.
Leaves were collected from rice plants at the tillering stage or N. benthamiana 3 days after injection. RNA was extracted using KK Fast Plant Total RNA Kit (ZP405K, ZOMANBIO) according to the user manual. Total RNA of 2 μg was reversely transcribed into cDNA by using FastKing RT Kit (with gDNase; KR116, TIANGEN) according to the user manual. The cDNA product was diluted four times with ddH2O (RNase-free). A total of 20 μL volume, including 2 μL diluted cDNA was used in the qRT-PCR with Applied Biosystems 7500 Real-Time PCR System according to the user manual of Taq Pro Universal SYBR qPCR Master Mix (Q712, Vazyme). The OsHYR transcript was amplified using primers No. 13–14. The OsSUT2 transcript was amplified using primers No. 15–16. The OsNOG1 transcript was amplified using primers No. 17–18 (Supplementary Table 1). Each sample was amplified three times, and OsUbq (LOC4332169, primers No. 19–20, Supplementary Table 1) was used as a reference gene for rice. For N. benthamiana, the GUS gene was amplified using primers No. 21–22 (Supplementary Table 1). Each sample was amplified three times and NbEF-1α (Wang et al., 2018) (Nbv6.1trP76432, primers No. 23–24, Supplementary Table 1) was used as the reference gene. The results of qRT-PCR were analyzed by 2–ΔΔCt method (Livak and Schmittgen, 2001).
The PXO99A and PH were spread on the NA medium (with ampicillin and spectinomycin added for PH-dt strains) and cultured for 2 days at 30°C. The bacteria were eluted with sterile water, shaken well, and the OD600 value of the suspension was adjusted to 1.0. For the leaf-cutting inoculation, sterile scissors were used to soak the bacteria solution and the tip of 2 cm of rice fully expanded leaves were cut, with three leaves per plant and no less than three plants each for phenotypic identification. The lesions were measured and photographed after 2 weeks (Zhang et al., 2013). For the injection inoculation, a common method to observe the infection degree of Xanthomonas or the expression changes of related genes of rice leaves (Tariq et al., 2018), a sterile syringe without a needle was used to inject bacterial fluid from the undersurface of the fully expanded leaves of no less than three rice plants at about 10 o’clock in the morning. For the spraying treatment, the bacteria solution was sprayed evenly on both sides of rice leaves in the 10 × 10 plant-plots with a hand-held sprayer every 3 days at about 10 o’clock in the morning from tillering stage to the filling stage of rice.
The bacterial population density of PH-dtNOG1 was determined using the improved plate count method based on previous research (Bai et al., 2014). The whole leaves sprayed uniformly with the PH-dtNOG1 bacteria solution were collected. About 3 g leaves with 5 mL sterile water were ground into homogenate with sterilized mortar. A total of 50 μL homogenate was diluted 10, 100, and 1,000 times in sterile water, and then 50 μL was taken and coated on NA medium (ampicillin and spectinomycin added) solid plates and incubated at 30°C for 3 days. The plates with the number of colonies between 30 and 300 were selected for statistics. The final number of colony-forming unit per gram (CFU/g) was calculated according to this formula: CFU/g = A⋅F⋅100/W (A: counted colonies, F: dilution ratio, W: leaves weight). The final data was the mean of three experiments.
The agronomic traits of rice were investigated after the rice was fully mature. Plant height (the height of the ground part of the main tiller) and number of productive panicles (with more than 5 filled grains, NPP) were measured and recorded in the field. Panicle length, filled grain number per panicle (FGNPP), the number of empty grains per panicle, total grain number per panicle (TGNPP), setting rate, yield per plant, and 1,000-grain weight were measured after the fully mature panicles were harvested from the 6 × 6 plants area in the center of the 10 × 10 plants plot, with no less than 10 plants for each plot. The grain weight was measured after conventional drying of seeds. For JJ818, the agronomic traits were investigated independently in two growing seasons.
Rice growth or yield trait-related genes, OsHYR (Ambavaram et al., 2014), OsSUT2 (Eom et al., 2011), and OsNOG1 (Huo et al., 2017) were chosen as the target genes of dTALEs. The virulence-disarmed Xoo strain PH (Ji et al., 2016) was used as a device for delivering the dTALEs into rice plant cells. After analyzing the promoter sequences of the above-mentioned genes based on the reference genome of japonica rice variety Nip, along with considering factors including the number of repeat units of dTALEs to be assembled, the distance of target sites to the start codon and TSS of the target gene, target site sequence specificity in rice genome, and coverage with other transcription regulatory elements, three dTALEs targeting promoters of OsHYR, OsSUT2, and OsNOG1 were assembled, respectively (Figure 2A). Each dTALE was transferred into the mutant Xoo strain PH, resulting in GEB strains PH-dtHYR, PH-dtSUT2, and PH-dtNOG1, respectively (Figures 2B,C).
Western blotting confirmed the stable expression of dTALEs in the bacteria PH, where the expected dTALE-NOG1, dTALE-HYR, and dTALE-SUT2 proteins with DYKDDDDK tag (133.2, 132.9, and 129.2 kDa, respectively) were clearly detected (Figure 2D and Supplementary Figure 1). By injecting Nip leaves with cell suspensions of the three GEB strains, we found that the expression of OsNOG1 could be significantly induced by PH-dtNOG1, while the expression induction for the other two genes was not significant (Figures 2E–G). Therefore, OsNOG1 and PH-dtNOG1 were used for subsequent experiments.
The function of dTALE’s transcriptional activation is often affected by various factors (Schreiber and Tissier, 2017). To further confirm the functional feasibility of dTALE-NOG1 before field application, the GUS gene was used as a reporter by cloning it into vectors downstream of the OsNOG1 promotor from rice varieties GC2 or JJ818. The construct expressing dTALE-NOG1 was used as an activator (Figure 3A). The reporter and activator vectors were individually or co-transformed into N. benthamiana leaves by Agrobacterium-infiltration. GUS staining showed that the GUS expression, driven by OsNOG1 promoter from either GC2 or JJ818 was very weak, but the expression was significantly enhanced in the presence of dTALE-NOG1 (Figure 3B). qRT-PCR showed that the relative expression of GUS was more than 2.5 times increased by dTALE-NOG1 in N. benthamiana (Figure 3C). Therefore, the dTALE-NOG1 was functional for OsNOG1, and the PH-dtNOG1 could be tested in the field for exploring its effects on rice.

Figure 3. dTALE-NOG1 induced expression of GUS driven by OsNOG1 promoter in N. benthamiana. (A) The activator vector, p1305-dTALE-NOG1, was generated by inserting the coding sequence of dTALE-NOG1 into NcoI and Eco72I sites of pCAMBIA1305. The reporter vector, pBI121-NOG1pro:GUS, was generated by inserting the promoter sequence of OsNOG1 into HindIII and SmaI sites of pBI121. The dTALE target in OsNOG1 promoter was shown by a red rectangle. (B) GUS expression assays on a N. benthamiana leaf with treatments of I–VI, corresponding to Buffer, p1305-dTALE-NOG1 (dtNOG1), pBI121-GC2NOG1pro:GUS (GC2pro, the promoter fragment was from GC2), pBI121-818NOG1pro:GUS (818pro, the promoter fragment was from JJ818), GC2pro with dtNOG1, 818pro with dtNOG1, respectively, as shown in the sketch leaf. (C) Relative expression of GUS in N. benthamiana detected by qRT-PCR. NbEF-1α (Nbv6.1trP76432) was used as the reference gene. Data values were represented as mean with error bars representing standard deviation (n = 3). The significance of the difference was tested by t-test. **P < 0.01.
To choose the appropriate rice varieties to test the effects of PH-dtNOG1 in reprograming gene expression and relevant agronomic trait change, we confirmed the conservativeness of the target gene OsNOG1 in 4 varieties (Nip, GC2, JJ818, and R007) by sequencing. Nip was used as a sequence reference and control since it is the model variety of rice. GC2 is a high-yielding indica variety harboring the functional OsNOG1 (Huo et al., 2017). JJ818 is a high-quality japonica rice variety suitable for planting in single-season rice-growing areas in North China, with good compact plant type, which was selected as the main rice material in this study for its convenience for spraying GEB and the potential to improve yield. R007 was used as a representative of indica rice for the yield test. The full length of OsNOG1 in the above varieties was amplified and sequenced. Alignment analysis showed that the OsNOG1 from JJ818 contains 12 SNPs and 2 Indels in the introns, 1 SNP in Exon 2, which does not change its coding amino acid (Threonine), and a 3-bp (GAG, one glutamate added for protein) insertion in exon 14 (Supplementary Figure 2), which was the same as that in the reported cDNA from rice introgression line SIL176, not affecting the function of NOG1 protein (Huo et al., 2017).
In the OsNOG1 promoter from JJ818, there were 33 irregularly distributed SNPs and 6 Indels compared with that from GC2. The 12-bp deletion at −2,209 bp upstream of ATG for JJ818 was consistent with that in the introgression line SIL176 described previously, which plays a key role in regulating OsNOG1 expression (Huo et al., 2017). The Japonica rice varieties JJ818 and Nip have 1 copy of the 12-bp Indel while the indica rice varieties GC2 and R007 have 2 copies of the 12-bp Indel. The 20 bp target site sequence (5′-CTTCCCGCACCAACCCCGCC-3′) of dTALE-NOG1 locates at -146 bp upstream of ATG was conserved in all the rice varieties tested (Figure 2B and Supplementary Figure 2). These results indicated that the OsNOG1 gene was conserved and functional in the tested rice varieties, and dTALE-NOG1 could be used to target the promoter of OsNOG1 in the rice varieties used in this study.
The PH is a TALE-free mutant strain of PXO99A that does not contain any TALE gene that plays key roles in rice disease (Ji et al., 2016) but still has a complete T3SS. This makes PH a good tool for dTALE delivery. Nevertheless, it is needed to determine the pathogenicity of the GEB PH-dtNOG1 before field application. Therefore, the wild-type Xoo strain PXO99A, the mutant strain PH, and the PH-dtNOG1 were inoculated on leaves of JJ818 and Nip by the cutting method (Zhang et al., 2013). After 2 weeks, the length of disease lesions was measured. Results showed that the disease lesion length caused by PXO99A on JJ818 was 14.3 cm on average, while the lesions caused by PH and PH-dtNOG1 were only 2.9 and 2.5 cm in length, respectively, significantly shorter than that of PXO99A (Figures 4A,B). Similar results were obtained on the Nip plants (Supplementary Figure 3). We further determined the pathogenicity of the Xoo strains by both injection and spraying methods, and results showed that no obvious disease phenotype was observed 4 days post-injection (Figure 4D) or spraying with PH or PH-dtNOG1 on JJ818 plants (Figure 4F). These observations confirmed that PH and PH-dtNOG1 had no obvious pathogenicity to JJ818 and Nip plants, and the mutant Xoo strain PH could be used to deliver dTALEs for regulating rice growth.
Figure 4. The response of rice variety JJ818 to Xoo strains PH and PH-dtNOG1. (A,B) PH and PH-dtNOG1 had no serious pathogenicity to JJ818 compared with the pathogenic strain PXO99A. The photos of leaves were taken 2 weeks post-inoculation. Scale, 2 cm. Data values were represented as the mean, with error bars representing standard deviation (n = 9). The significance of the difference was tested by t-test. **P < 0.01. (C) Effect of spraying PH-dtNOG1 on rice leaves. (D) Injection of PH and PH-dtNOG1 cells into leaves of JJ818, without pathogenicity. Photos were taken 4 days after the injection. (E) The retention of PH-dtNOG1 in JJ818 leaves after once spraying treatment. Data values were represented as mean with error bars representing standard deviation (n = 3). (F) The plants of JJ818 in the filling stage after multiple-spraying treatment with PH or PH-dtNOG1. (G) After multiple-spraying PH-dtNOG1, some thin spots appeared at the late growth stage of rice plants. Scale, 1 cm.
Under natural conditions, infection, colonization, and reproduction of Xanthomonas on host plants mainly depend on the activation of host susceptibility (S) genes by TALEs secreted by Xanthomonas, while other genes targeted by TALEs tend to be off-target (Nowack et al., 2021). Since PH has no TALE genes, it is unable to promote the expression of S genes, and this would affect its viability in rice leaves. To clarify this issue, the retention density of PH-dtNOG1 on rice JJ818 leaves was measured after once uniformly spraying the bacteria (Figure 4C). Results showed that the number of bacteria was kept maximum at 5 h post-spraying and decreased rapidly in the following 1–2 days and reached the lowest on the second day. However, from the third day, the number of bacteria of PH-dtNOG1 began to increase and remained at a relatively stable level at about 15 × 106 CFU/g, but still lower than the initial level (Figure 4E). These data showed that when the PH-dtNOG1 bacteria were transferred from the nutrient environment of the culture medium to the natural nutrient environment of leaves, a certain loss occurred, but the bacterial number would be recovered later. At the later stage of rice grain filling, some rice leaves with multiple-spraying treatment displayed tiny disease spots containing PH-dtNOG1, indicating that the GEB could be retained on rice leaves by spraying (Figure 4G).
During the tillering stage, the expression of OsNOG1 was detected in rice leaves of JJ818, Nip, and R007 treated with spraying PH-dtNOG1 once every 3 days until the late filling stage in the field. On the fifth day after the first spraying (1 day before the third spraying), there was no significant change in the relative expression of OsNOG1 in leaves sprayed with PH-dtNOG1 compared with the leaves sprayed with PH or without treatment (CK). The possible reason is that the first few days were colonization, recovery, and the reproduction stage of the GEB, when there was no enough bacteria to significantly affect the target gene OsNOG1. However, the relative expression of OsNOG1 was significantly increased 10 days after the first spraying (2 days before fifth spraying) and continued to 35 days post the first spraying (Figures 5A–C) for JJ818, Nip, and R007. These results indicate that the GEB PH-dtNOG1 can colonize on rice leaves, deliver the dTALE-NOG1 into rice cells and induce the expression of OsNOG1 when the plants were treated by simply repeatedly spraying the water solution of GEB without any additives.
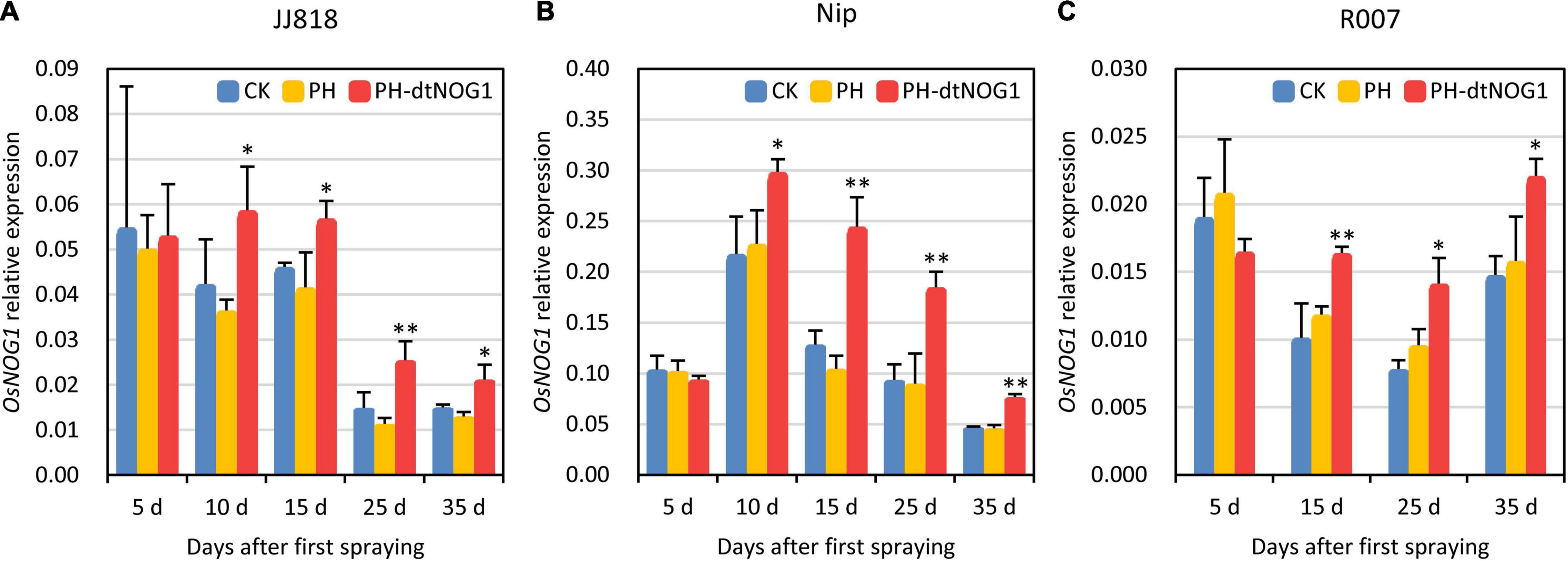
Figure 5. The expression of OsNOG1 was induced by spraying PH-dtNOG1. Panels (A–C) showed the expression of OsNOG1 in rice JJ818, Nip, and R007 untreated (CK, blue bars), treated with PH (yellow bars), and with PH-dtNOG1 (red bars), respectively, during the process of spraying treatments. OsUbq (LOC4332169) was used as the reference gene for qRT-PCR. Data values were represented as mean with error bars representing standard deviation (n = 3). The significance of the difference was tested by t-test. *0.01 < P < 0.05; **P < 0.01.
Since panicle differentiation has been completed in the late tillering stage (Xu et al., 2020), the spraying treatment must be carried out before panicle initiation to increase the grain number by changing the expression of OsNOG1. Meanwhile, to ensure a stable number of PH-dtNOG1 cells on the leaves, the early tillering stage was selected as the time for first spraying and continued to the late filling stage. The spraying treatment was carried out once every 3 days. The agronomic characters of JJ818 were investigated when the rice grains were fully mature. Results showed that the FGNPP, TGNPP, yield per plant of JJ818 were significantly increased by treatment with PH-dtNOG1, compared with the control plants sprayed with PH and plants without treatment (CK), while the plant height, NPP, panicle length, setting rate, and 1,000-grain weight did not change significantly compared with the controls (Figures 6A–H). The FGNPP increased by 11.40%, the TGNPP increased by 12.51% and the yield per plant increased by 15.09% compared with CK (Supplementary Table 3). Taken together, the effect of PH-dtNOG1 on grain number promoted the increase of rice yield, although sporadic and small disease spots appeared on some leaves at late growth stage of the rice plants (Figure 4G).
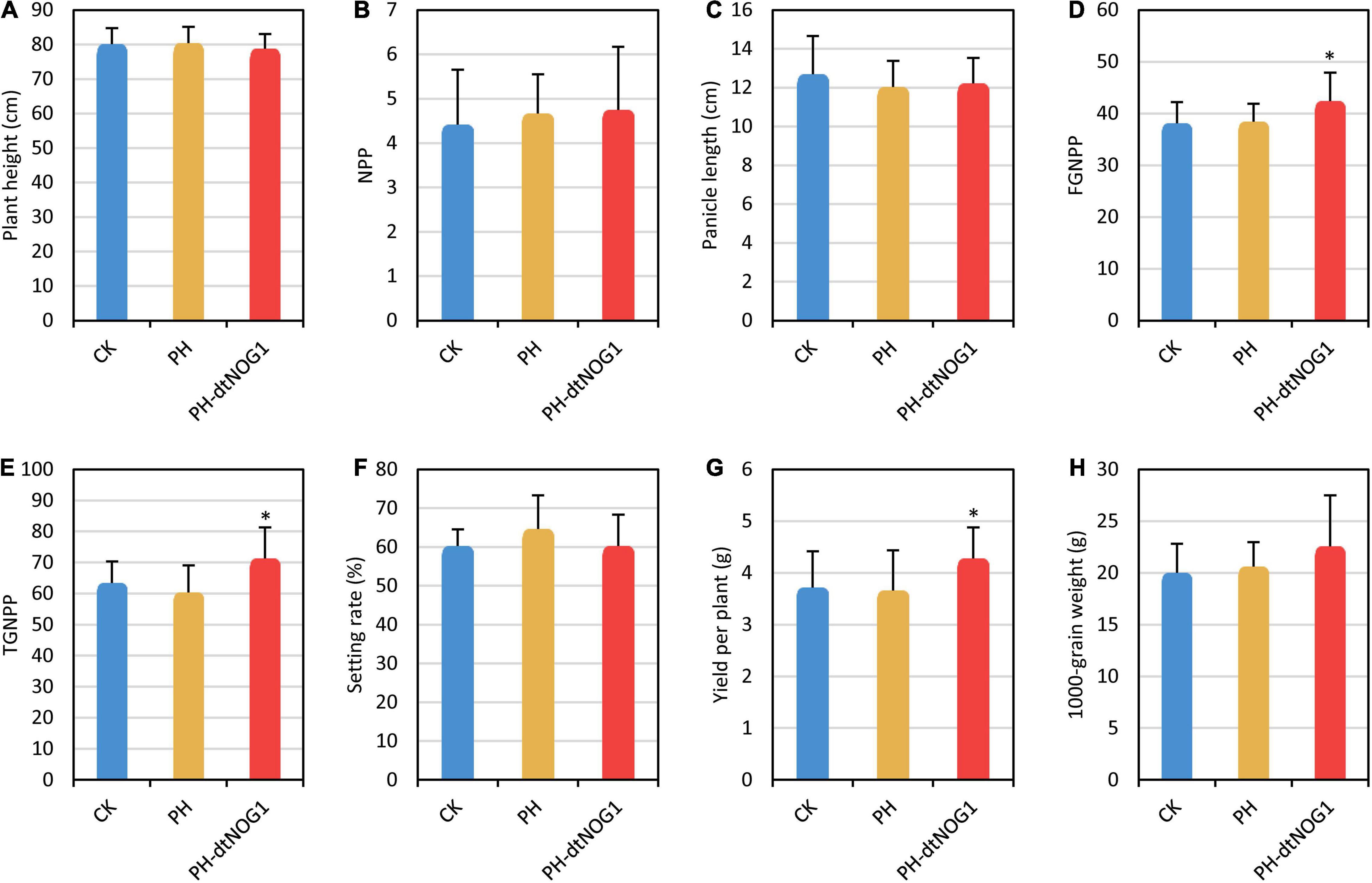
Figure 6. Rice yield was increased by spraying PH-dtNOG1. Panels (A–H) showed the plant height, number of productive panicles (NPP), panicle length, filled grain number per panicle (FGNPP), total grain number per panicle (TGNPP), setting rate, yield per plant and 1,000-grain weight of JJ818 untreated (CK, blue bars), and treated with PH (yellow bars) and PH-dtNOG1 (red bars), respectively. Data values were represented as mean with error bars representing standard deviation (n = 12). The significance of the difference was tested by t-test. *0.01 < P < 0.05.
Similar results have been obtained when other rice varieties were treated by spraying PH-dtNOG1. Compared with the plants without spraying, PH-dtNOG1 treated Nip showed no significant changes in plant height and NPP, but panicle length, FGNPP, TGNPP, and setting rate were significantly increased (Supplementary Figures 4A–H), among which FGNPP and TGNPP increased 17.21 and 12.10%, respectively. In addition, the yield per plant increased by 11.08%, and the 1,000-grain weight was also increased (Supplementary Table 3). Furthermore, PH-dtNOG1-treated R007, an indica variety, also showed a significant raise in TGNPP (Supplementary Figures 5C and Supplementary Table 3). These results demonstrated that regardless of the type of 1 or 2 copies of the 12-bp Indel in the OsNOG1 promoter, the number of grains and yield could be increased by PH-dtNOG1 via induced expression of OsNOG1. Therefore, the use of the GEB PH-dtNOG1 targeting OsNOG1 gene as the rice growth regulator can significantly promote rice yield traits without obvious pathogenicity or other side effects.
This proof-of-principle study proved that the mutant Xoo strain PH can be used as a device to deliver dTALEs, and the GEB strain PH-dtNOG1, which contains the artificially designed dTALE-NOG1 targeting the promoter of rice OsNOG1 gene, had no obvious pathogenicity to rice, but could deliver the dTALE-NOG1 into rice cells and induce the expression of OsNOG1, and finally resulted in rice yield increase. Our experiments showed that the expression of OsNOG1 was significantly up-regulated by spraying cell suspension of PH-dtNOG1 on rice plants during the tillering stage. For the japonica rice JJ818, the grain number per panicle of rice was increased by 12.51% and the yield per plant was increased by 15.09% compared with CK control. Similar results have been achieved on japonica rice Nip as well as the indica rice R007. Based on these findings, we proposed a new CAPME strategy for rice, namely GEB-dTALE strategy (Figure 7), which contains four steps: (1) choose appropriate genes as the targets of dTALEs; (2) design and construct the gene-specific dTALE’s expression vector, and perform functional verification of dTALEs; (3) transfer the dTALE-expressing construct into suitable delivery bacteria to generate GEB; and (4) apply the GEB on crop plant through methods such as spraying (Figure 7). The dTALEs should be secreted into the host cell through T3SS and should induce the target gene expression, resulting in changes in plant traits, such as improving nutrient absorption by plant root, leaf photosynthesis, source-sink transformation, effects of microbiome, and eventually achieve the increase of harvest (Figure 7).
Figure 7. The GEB-dTALE strategy for yield increase by regulating crop plants growth. Four steps of GEB-dTALE strategy: (1) select target genes for dTALEs; (2) design and construct dTALE-expressing vectors; (3) transfer the dTALE construct into suitable delivery bacteria, resulting in genetically engineered bacteria (GEB); and (4) apply the GEB on crop plant through methods such as spraying. The dTALEs proteins (golden pentagram) are secreted by GEB and delivered into rice cells through T3SS of the GEB-dTALE cells. Afterward, dTALEs regulate the transcription of their target genes. Therefore, rice plants could obtain enhanced functions (red arrows) of root nutrient (circles of different colors) absorption, leaf photosynthesis, and carbon sequestration, source-sink transformation, effects with microbiome, etc., which may contribute greatly to rice yield increase. The function indicated by the dotted lines in the rice root needs further confirmation.
In recent decade, some new technologies for promoting plant growth have been developed and applied in agricultural production, such as microbial growth regulators (Wei and Jousset, 2017) and nano regulation technology (Gogos et al., 2012). A number of studies have shown that many bacteria from the genera Bacillus, Pseudomonas, Enterobacter, and Streptomyces can promote crop growth, yield increase, or resistance enhancement, and they have been developed as biofertilizers (Ngalimat et al., 2021). In principle, these biofertilizers regulate crop phenotypes through physiological metabolism, which is lacking in accuracy, and possibly cause physiological toxicity problems to plants (Chun et al., 2020). In contrast, the GEB-dTALE strategy could specifically regulate the target gene expression genetically and the associated agronomical traits by individual dTALE protein, which should be more accurate and do not cause physiological toxicity to plants. However, it is noteworthy that the GEB-dTALE strategy may face a pathogenic risk if a GEB strain is used for many years, and the usage of a genetically engineered microbe may lead to additional regulatory hurdles in some countries. Although the PH strain had no obvious pathogenic effect on cultivated rice varieties, small disease spots still appeared on some leaves at a later stage of spraying treatment (Figure 4G). It is needed to keep watch on the possibility of bacterial mutation in the field environment. Theoretically, bacteria do not cause disease, but with T3SS they can be used as dTALE-delivery tools. Therefore, other bacteria strains, such as endophytes, could be identified as better dTALE-delivery tools in future.
In the use of GEB-dTALE strategy, the selection of appropriate genes and target site sequences for dTALEs is particularly important. The OsNOG1 gene used in this study is the only known single gene that plays a significant role in regulating rice yield traits, while other genes associated with rice growth or yield traits are involved in complex regulatory pathways regulating the corresponding phenotypes (Li et al., 2021). This might explain why the genes OsHYR or OsSUT2 could not be induced by the constructed dTALEs (Figures 2E,F). However, studies have shown that a multiplex transcription activator-like effector activation (mTALE-Act) system can simultaneously activate the expression of four endogenous genes in plants by transgenic technology (Lowder et al., 2018). Therefore, it is theoretically possible to simultaneously up-regulate the expression of multiple genes through GEB-dTALE strategy. In addition, the dTALE can be modified by replacing the AD domain with transcription inhibitors (Bernstein et al., 2015) or just cascading a repression domain after dTALE (Mahfouz et al., 2012) or occupying key site of promoter (Sakono and Hayakawa, 2021) to inhibit or down-regulate gene expression. All of these provide a basis for using dTALE system to comprehensively regulate (induction or suppression or both) genes or gene networks, which makes the GEB-dTALE strategy have tremendous potential for further improvement. For a crop species, its gene resources are limited, and current crop breeding selection and cultivation approaches have almost pushed the crop yield to a plateau, which often accompanied by great time and material costs. In this context, the GEB-dTALE strategy provides a new idea and great potential for further exploration in the field of CAPME technology.
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
KZ, CW, and ZJ conceived and designed the research. YT, CW, FW, ML, and YF performed the experiments. YT, KZ, and ZJ wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by grants from the National Key R&D Program of China (2021YFD200501), the National Natural Science Foundation of China (32072412), the Beijing Municipal Natural Science Foundation (6202031), and the National Key Research and Development Program of China (2017YFD0201301).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Thanks to Jie Wang (Institute of Crop Sciences of Chinese Academy of Agricultural Sciences) for providing JJ818 seeds.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.924645/full#supplementary-material
Ambavaram, M. M., Basu, S., Krishnan, A., Ramegowda, V., Batlang, U., Rahman, L., et al. (2014). Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat. Commun. 5:5302. doi: 10.1038/ncomms6302
Anderson, J. T., Rogers, J. M., Barrera, L. A., and Bulyk, M. L. (2020). Context and number of noncanonical repeat variable diresidues impede the design of TALE proteins with improved DNA targeting. Protein Sci. 29, 606–616. doi: 10.1002/pro.3801
Ansari, A., Wang, C., Wang, J., Wang, F., Liu, P., Gao, Y., et al. (2017). Engineered dwarf male-sterile rice: a promising genetic tool for facilitating recurrent selection in rice. Front. Plant Sci. 8:2132. doi: 10.3389/fpls.2017.02132
Antony, G., Zhou, J., Huang, S., Li, T., Liu, B., White, F., et al. (2010). Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22, 3864–3876. doi: 10.1105/tpc.110.078964
Bai, Y., Zhi, D., Li, C., Liu, D., Zhang, J., Tian, J., et al. (2014). Infection and immune response in the nematode Caenorhabditis elegans elicited by the phytopathogen Xanthomonas. J. Microbiol. Biotechnol. 24, 1269–1279. doi: 10.4014/jmb.1401.01017
Bernstein, D. L., Le Lay, J. E., Ruano, E. G., and Kaestner, K. H. (2015). TALE-mediated epigenetic suppression of CDKN2A increases replication in human fibroblasts. J. Clin. Invest. 125, 1998–2006. doi: 10.1172/JCI77321
Birla, D. S., Malik, K., Sainger, M., Chaudhary, D., Jaiwal, R., and Jaiwal, P. K. (2017). Progress and challenges in improving the nutritional quality of rice (Oryza sativa L.). Crit. Rev. Food Sci. Nutr. 57, 2455–2481. doi: 10.1080/10408398.2015.1084992
Boch, J., and Bonas, U. (2010). Xanthomonas AvrBs3 family-Type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. doi: 10.1146/annurev-phyto-080508-081936
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., et al. (2009). Breaking the code of DNA binding specificity of TAL-Type III effectors. Science 326, 1509–1512. doi: 10.1126/science.1178811
Brückner, K., Schafer, P., Weber, E., Grutzner, R., Marillonnet, S., and Tissier, A. (2015). A library of synthetic transcription activator-like effector-activated promoters for coordinated orthogonal gene expression in plants. Plant J. 82, 707–716. doi: 10.1111/tpj.12843
Chun, S., Muthu, M., and Gopal, J. (2020). Nanotoxic impacts on staple food crops: there’s plenty of room for the unpredictables. Crit. Rev. Food Sci. Nutr. 60, 3725–3736. doi: 10.1080/10408398.2019.1707158
Cornelis, G. R. (2006). The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825. doi: 10.1038/nrmicro1526
Eom, J. S., Cho, J. I., Reinders, A., Lee, S. W., Yoo, Y., Tuan, P. Q., et al. (2011). Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 157, 109–119. doi: 10.1104/pp.111.176982
Gogos, A., Knauer, K., and Bucheli, T. D. (2012). Nanomaterials in plant protection and fertilization: current state, foreseen applications, and research priorities. J. Agric. Food Chem. 60, 9781–9792. doi: 10.1021/jf302154y
Grau, J., Wolf, A., Reschke, M., Bonas, U., Posch, S., and Boch, J. (2013). Computational predictions provide insights into the biology of TAL effector target sites. PLoS Comput. Biol. 9:e1002962. doi: 10.1371/journal.pcbi.1002962
Gu, K., Yang, B., Tian, D., Wu, L., Wang, D., Sreekala, C., et al. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125. doi: 10.1038/nature03630
Huang, P., Xiao, A., Zhou, M., Zhu, Z., Lin, S., and Zhang, B. (2011). Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 29, 699–700. doi: 10.1038/nbt.1939
Huo, X., Wu, S., Zhu, Z., Liu, F., Fu, Y., Cai, H., et al. (2017). NOG1 increases grain production in rice. Nat. Commun. 8:1497. doi: 10.1038/s41467-017-01501-8
Jefferson, R. A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. doi: 10.1007/BF02667740
Ji, Z., Guo, W., Chen, X., Wang, C., and Zhao, K. (2022). Plant executor genes. Int. J. Mol. Sci. 23:1524. doi: 10.3390/ijms23031524
Ji, Z., Ji, C., Liu, B., Zou, L., Chen, G., and Yang, B. (2016). Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat. Commun. 7:13435. doi: 10.1038/ncomms13435
Khush, G. S. (1995). Breaking the yield frontier of rice. GeoJournal 35, 329–332. doi: 10.1007/BF00989140
Li, G., Zhang, H., Li, J., Zhang, Z., and Li, Z. (2021). Genetic control of panicle architecture in rice. Crop J. 9, 590–597. doi: 10.1016/j.cj.2021.02.004
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lowder, L. G., Zhou, J., Zhang, Y., Malzahn, A., Zhong, Z., Hsieh, T. F., et al. (2018). Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-act systems. Mol. Plant 11, 245–256.
Mahfouz, M. M., Li, L., Piatek, M., Fang, X., Mansour, H., Bangarusamy, D. K., et al. (2012). Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein. Plant Mol. Biol. 78, 311–321.
Mann, C. C. (1999). Crop scientists seek a new revolution. Science 283, 310–314. doi: 10.1126/science.283.5400.310
Moscou, M. J., and Bogdanove, A. J. (2009). A Simple cipher governs DNA recognition by TAL effectors. Science 326:1501. doi: 10.1126/science.1178817
Ngalimat, M. S., Mohd Hata, E., Zulperi, D., Ismail, S. I., Ismail, M. R., Mohd Zainudin, N. A. I., et al. (2021). Plant growth-promoting bacteria as an emerging tool to manage bacterial rice pathogens. Microorganisms 9:682. doi: 10.3390/microorganisms9040682
Nowack, M. K., Holmes, D. R., and Lahaye, T. (2021). TALE-induced cell death executors: an origin outside immunity? Trends Plant Sci. S1360-1385, 309–305. doi: 10.1016/j.tplants.2021.11.003
Sakono, M., and Hayakawa, R. (2021). Repressor-like on-off regulation of protein expression by the DNA-binding transcription activator-like effector in T7 promoter-based cell-free protein synthesis. Chembiochem 22, 888–893. doi: 10.1002/cbic.202000591
Schreiber, T., and Tissier, A. (2017). Generation of dTALEs and libraries of synthetic TALE-activated promoters for engineering of gene regulatory networks in plants. Methods Mol. Biol. 1629, 185–204. doi: 10.1007/978-1-4939-7125-1_13
Stark, J. C. (2021). Food production, human health and planet health amid Covid-19. Explore 17, 179–180. doi: 10.1016/j.explore.2020.12.009
Tariq, R., Wang, C., Qin, T., Xu, F., Tang, Y., Gao, Y., et al. (2018). Comparative transcriptome profiling of rice near-isogenic line carrying Xa23 under Infection of Xanthomonas oryzae pv. oryzae. Int. J. Mol. Sci. 19:717. doi: 10.3390/ijms19030717
Torti, S., Schlesier, R., Thummler, A., Bartels, D., Romer, P., Koch, B., et al. (2021). Transient reprogramming of crop plants for agronomic performance. Nat. Plants 7, 159–171. doi: 10.1038/s41477-021-00851-y
Varshney, R. K., Ribaut, J. M., Buckler, E. S., Tuberosa, R., Rafalski, J. A., and Langridge, P. (2012). Can genomics boost productivity of orphan crops? Nat. Biotechnol. 30, 1172–1176. doi: 10.1038/nbt.2440
Wang, Y., Xu, Y., Sun, Y., Wang, H., Qi, J., Wan, B., et al. (2018). Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat. Commun. 9:594. doi: 10.1038/s41467-018-03010-8
Wei, Z., and Jousset, A. (2017). Plant breeding goes microbial. Trends Plant Sci. 22, 555–558. doi: 10.1016/j.tplants.2017.05.009
Xu, J., Henry, A., and Sreenivasulu, N. (2020). Rice yield formation under high day and night temperatures—a prerequisite to ensure future food security. Plant Cell Environ. 43, 1595–1608. doi: 10.1111/pce.13748
Zhang, T. Y., Yang, X. G., Wang, H. S., Li, Y., and Ye, Q. (2014). Climatic and technological ceilings for Chinese rice stagnation based on yield gaps and yield trend pattern analysis. Glob. Change Biol. 20, 1289–1298. doi: 10.1111/gcb.12428
Zhang, X. P., Wang, C. L., Zheng, C. K., Che, J. Y., Li, Y. Q., and Zhao, K. J. (2013). HrcQ is necessary for Xanthomonas oryzae pv. oryzae HR-induction in non-host tobacco and pathogenicity in host rice. Crop J. 1, 143–150. doi: 10.1016/j.cj.2013.07.009
Keywords: designer TALE, growth regulators, rice yield, OsNOG1, Xoo, genetically engineered bacteria
Citation: Tang Y, Wang C, Wang F, Li M, Fang Y, Ji Z and Zhao K (2022) Development of Designer Transcription Activator-Like Effector-Based Plant Growth Regulator for Higher Yield in Rice. Front. Plant Sci. 13:924645. doi: 10.3389/fpls.2022.924645
Received: 20 April 2022; Accepted: 19 May 2022;
Published: 14 June 2022.
Edited by:
Guotian Li, Huazhong Agricultural University, ChinaReviewed by:
Daniel Caddell, Nobell Foods, Inc., United StatesCopyright © 2022 Tang, Wang, Wang, Li, Fang, Ji and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyuan Ji, aml6aGl5dWFuQGNhYXMuY24=; Kaijun Zhao, emhhb2thaWp1bkBjYWFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.