- Department of Pomology, College of Agronomy, Jiangxi Agricultural University, Nanchang, China
Drought stress often occurred in citrus to limit its growth, distribution, and fruit quality. Cuticular waxes play an important role in regulating plant tolerance to drought stress. Plant enoyl-CoA reductase (ECR) is involved in the biosynthesis of cuticular waxes and catalyzes the last step of very long-chain fatty acids (VLCFAs) elongation. In this study, a putative ECR gene, named CsECR, was cloned from “Newhall” navel orange. CsECR protein has high identities with other plant ECR proteins and contained a conserved NADP/NAD-binding motif and three conserved functional sites. The highest expression of CsECR was observed in leaves, followed by stems, flavedos, ovaries, juice sacs, stigmas, stamens, albedos, and petals. Besides, the expression of CsECR was significantly induced by PEG6000 and ABA treatments. Ectopic overexpression of CsECR increased the contents of total waxes and aliphatic wax fractions (n-fatty acids, unsaturated fatty acids, n-alkanes, alkenes, iso-, and anteiso-alkanes) in the leaves and fruits of the transgenic tomato. Furthermore, ectopic overexpression of CsECR reduced the cuticle permeability in the leaves and fruits of the transgenic tomato and increased its tolerance to drought stress. Taken together, our results revealed that CsECR plays an important role in plant response to drought stresses by regulating cuticular wax biosynthesis.
Introduction
Cuticular waxes cover plant surfaces and serve as a hydrophobic barrier against biotic and abiotic stresses. In plants, cuticular waxes are mainly composed of very long-chain fatty acids (VLCFAs) and their derivatives with carbon chain lengths ranging from C20 to C34, such as alkanes, primary alcohols, secondary alcohols, aldehydes, esters, and ketones. In addition, cyclic compounds including triterpenoids, alkaloids, and phenylpropanoids are also detected in plant cuticular waxes. The biosynthesis of cuticular waxes started with the de novo synthesis of C16 and C18 acyl chains in the plastid of plant epidermal cells. Afterward, the C16 and C18 fatty acids are released from the acyl carrier protein (ACP) and esterified to CoA to produce C16 and C18 fatty acyl CoAs. The C16 and C18 fatty acyl CoAs are further extended to VLCFAs in the endoplasmic reticulum (ER) and generate cuticular wax components by the decarbonylation and acyl reduction pathways (Lee and Suh, 2015a).
The elongation of C16 and C18 fatty acids to VLCFAs in plants includes a series of reaction cycles catalyzed by the multienzyme fatty acid elongase (FAE) complex. Two carbons are added to the acyl chain per cycle. Each cycle consists of four consecutive reactions: biosynthesis of 3-ketoacyl-CoA by condensation of malonyl-CoA with a long chain acyl CoA catalyzed by a 3-ketoacyl-CoA synthase (KCS); reduction of 3-ketoacyl-CoA to 3-hydroxyacyl-CoA catalyzed by a 3-ketoacyl-CoA reductase (KCR); dehydration of 3-hydroxyacyl-CoA to trans-2-enoyl-CoA catalyzed by a 3-hydroxyacyl-CoA dehydratase (HCD); and reduction of trans-2-enoyl-CoA to acyl-CoA catalyzed by an enoyl-CoA reductase (ECR) (Haslam and Kunst, 2013).
To date, a number of genes involved in VLCFA biosynthesis have been reported in plants. For example, the KCSs are suggested to harbor fatty acid substrate specificity and determine the chain lengths of the final VLCFA product. A total of 21 KCS genes have been identified in the Arabidopsis genome. Among these genes, KCS1, KCS2/DAISY, KCS6/CUT1/CER6, KCS9, KCS10, KCS13, and KCS20 are proved to be involved in cuticular wax synthesis (Millar and Kunst, 1997; Millar et al., 1999; Todd et al., 1999; Lee et al., 2009; Kim et al., 2013). In addition to KCS, the other three enzymes involved in VLCFA biosynthesis have no substrate specificities and take part in the formation of VLCFAs with different carbon chain lengths. There are two KCR genes in the Arabidopsis genome, of which only KCR1 is involved in VLCFA biosynthesis (Beaudoin et al., 2009). However, two KCR genes (GL8A and GL8B) in maize participate in the biosynthesis of VLCFAs (Dietrich et al., 2005). In Arabidopsis, a single HCD gene named PASTICCINO2 (PAS2) is involved in VLCFA biosynthesis (Bach et al., 2008).
The first ECR gene, named ScTSC13, was identified from Saccharomyces cerevisiae. The silence of ScTSC13 decreased the formation of VLCFAs in yeast (Kohlwein et al., 2001). A single ECR candidate gene At3g55360 was identified in the Arabidopsis genome by sequence similarity search with ScTSC13. Further research suggested that the Arabidopsis cer10 mutant was disrupted in the At3g55360 gene. Expression of At3g55360 in the yeast tsc13 mutant and the Arabidopsis cer10 mutant revealed that CER10 has functional ECR activity. Furthermore, the biosynthesis of VLCFAs in Arabidopsis cer10 mutant was interrupted by the ECR defect and led to a sharp decrease in cuticular wax load, suggesting that the Arabidopsis AtECR gene is involved in VLCFA biosynthesis (Gable et al., 2004; Zheng et al., 2005). Interestingly, the interactions between HCD and other FAE proteins such as KCR and ECR were detected in Arabidopsis, which indicated that HCD is a molecular scaffold for the FAE (Roudier et al., 2010). Further study revealed that the expression of AtECR was positively regulated by AtMYB30 and AtMYB94, but negatively regulated by AtDEWAX (Raffaele et al., 2008; Go et al., 2014; Lee and Suh, 2015b). In addition to Arabidopsis, the identification of ECR homologs was reported in tobacco (Park et al., 2005) and cotton (Song et al., 2009; Mustafa et al., 2017).
Citrus is one of the most important fruit trees in the world. The growth, development, and fruit quality of citrus were severely limited by drought stress. The cuticular waxes limit non-stomatal water loss and play an important role in citrus tolerance to drought stress. To date, the cuticular wax morphology and chemical composition have been studied on many kinds of citrus, such as clementine mandarin (Baker et al., 1975), grapefruit (Nordby and McDonald, 1995), navel orange (Cajuste et al., 2010; Wang et al., 2014; Liu et al., 2015), sweet orange (Ding et al., 2018), satsuma mandarin (Ding et al., 2020), and lemon (Zhou et al., 2021). The molecular mechanism of cuticular wax biosynthesis in citrus has been studied in several reports. For example, the genes involved in the regulation of cuticular wax biosynthesis during fruit development in “Newhall” navel orange were revealed by transcriptomic and metabolomic analyses (Wang et al., 2015). In the past, we reported a navel orange variety, named “Ganqi 3,” originated from a bud mutation of “Newhall” navel orange (Citrus sinensis [L.] Osbeck cv. Newhall). Further research showed that many wax-related genes were downregulated on the surfaces of “Ganqi 3” fruits, resulting in a significant decrease in cuticular wax amounts and wax crystal numbers, and finally leading to the glossy surface of “Ganqi 3” fruits (Liu et al., 2012, 2015, 2016). Our recent reports revealed that the expression levels of many wax-related genes changed during cold storage, leading to the alteration of cuticular wax morphology and chemical composition, and finally changed the navel orange tolerances to postharvest decay (Liu et al., 2021). Interestingly, the transcriptomic and metabolomics analyses of another wax-deficient mutant “Gannan NO.1” derived from “Newhall” navel orange revealed that many genes belonging to the VLCFAs elongation and cuticular wax biosynthesis pathways were decreased in “Gannan NO.1” fruits (He et al., 2018). Our group cloned a 3-Ketoacyl-CoA synthase (KCS) gene CsKCS6 from “Newhall” navel orange. Ectopic overexpression of CsKCS6 increased the biosynthesis of VLCFAs and enhanced the drought and high salt tolerance of Arabidopsis (Guo et al., 2020). Similarly, ectopic overexpression of CsMYB30 and CsMYB96 from Citrus sinensis enhanced plant drought resistance by promoting cuticular wax accumulation (Wen et al., 2021; Zhang et al., 2022).
However, previous research focused on screening wax-related genes in citrus by high-throughput sequencing. There are few reports concerned with the function identification of specific wax-related genes in citrus. Under drought stress, plants usually close their stomata to reduce transpiration. However, residual stomatal transpiration even minimal stomatal conductance can interfere with the study of the contribution of plant cuticular waxes to cuticular permeability (Kerstiens, 1996). The astomatous tomato fruits possess a continuous cuticular wax layer relatively impervious to water loss, making it an ideal model for functional characterization of cuticular waxes from plants with defined genetic modifications and therefore revealing the impact of cuticular waxes on plant cuticular permeability and drought tolerance. A series of reports confirmed the important role of the tomato as a suitable model plant to relate cuticular waxes to plant cuticular permeability and drought tolerance. For example, fruits of tomato lecer6 and positional sterile (ps) mutants showed a significant increase in cuticular permeability, which was caused by a decline in n-alkanes and an increase in triterpenoids (Vogg et al., 2004; Leide et al., 2007, 2011). Overexpression of SlSHN1 reduced cuticular permeability and enhanced drought tolerance in tomatoes by increasing cuticular wax production (Al-Abdallat et al., 2014). SlCER1s and SlCER3s tomatoes are involved in the biosynthesis of n-alkanes, which are the major cuticular wax components in both tomato leaves and fruits. Overexpression of SlCER1–1 increases n-alkane accumulation in tomato leaves and fruits, therefore enhancing its drought tolerance and fruit storability (Wu et al., 2022). In the present study, a novel cuticular wax biosynthesis gene encoding enoyl-CoA reductase, named CsECR, was cloned from “Newhall” navel orange. Moreover, transgenic tomato plants' ectopic overexpressing CsECR were produced to determine whether CsECR is involved in cuticular wax biosynthesis and plant response to drought stress. The results of this study provide new insights into the molecular regulation of cuticular wax biosynthesis and drought resistance in citrus.
Materials and Methods
Plant Materials, Growth Conditions, and Treatments
All citrus tissues were sampled from 8-year-old trees of “Newhall” navel orange (Citrus sinensis [L.] Osbeck cv. Newhall) cultivated in an orchard of Xinfeng county, Ganzhou city, Jiangxi province of China. Yong leaves were used for CsECR cloning. Mature leaves, stems, flavedos, ovaries, juice sacs, stigmas, stamens, albedos, and petals were used for tissue-specific expression analysis.
For expression analysis after drought and ABA treatments, shoots of “Newhall” navel orange were grafted on trifoliate orange (Poncirus trifoliata (L.) Raf.) to produce grafted seedlings. After growing to the height of 30 ± 2 cm in soil culture, the grafted seedlings were transferred to Hoagland's solution in a growth chamber under normal growth conditions (25°C, 16 h light/8 h dark in a day, and 80% relative humidity) for 7 days and then subjected to the Hoagland's solution containing 20% PEG6000 and 100 μM ABA, respectively, for the drought and ABA treatments. For each treatment per biological replicate, the leaves were collected from 15 seedlings at 0, 1, 3, 6, 12, and 24 h. Three biological replicates were performed for each time point of each treatment.
“Micro-Tom” tomato (Solanum lycopersicum L. cv. Micro-Tom) was used to produce transgenic plants ectopic overexpressing CsECR. For soil culture, seeds of the wild-type “Micro-Tom” tomato (WT) and the transgenic tomato lines were germinated on filter paper in Petri dishes at 25°C in darkness. Germinated seeds were transferred to pots filled with nutritional soil and placed in a growth chamber (25°C, 16 h light/8 h dark in a day, and 80% relative humidity). For in vitro culture, tomato plants were grown on a modified MS basal medium at 25°C under a photoperiod of 16 h light/8 h dark. The details of in vitro culture were carried out as described in the report by Dan et al. (2006).
Total RNA Extraction and First-Strand CDNA Synthesis
Total RNA was extracted from different tissues of “Newhall” navel orange, leaves, and fruit peels of “Micro Tom” tomato using the MiniBEST Plant RNA Extraction Kit (TaKaRa, Dalian, China). Trace genomic DNA was removed by DNase I (Promega, Madison, USA). The RNA yield and quality were detected according to our previous report (Liu et al., 2015). The first-strand cDNA was synthesized by PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) and used for gene clone and expression analysis.
Cloning and Bioinformatics Analysis of CsECR
The homologous cDNA sequence (Cs7g29210) of CsECR was obtained from the genomic database of sweet orange (http://citrus.hzau.edu.cn). According to the cDNA sequence of Cs7g29210, a pair of gene-specific primers (sense: 5′ AGAAATAACGAGGGAAGGAT 3′; antisense: 5′ AAAGGCTGCTAGATTACACA 3′) were designed to clone CsECR by PCR. The PCR for CsECR cloning was performed by 50 μL volume containing 25 μL PrimeSTAR Max Premix (TaKaRa, Dalian, China), 1 μL sense primer, 1 μL antisense primer, 1 μL cDNA, and 22 μL ddH2O. The PCR protocol was 5 min at 94°C, 35 cycles of 30 s at 94°C, 30 s at 55 °C, 1.5 min at 72 °C, and finally 10 min at 72°C. The PCR product was cloned into pMD18T vector (Takara, Dalian, China), transformed into Escherichia coli DH5α, and sequenced at Shanghai Sangon Biotech as described in our previous report (Liu et al., 2010).
The ExPASy ProtParam tool (http://us.expasy.org/tools/protparam.html) was used to analyze the theoretical isoelectric point and mass values of CsECR protein. The ECR proteins from other plants were obtained by BLAST search in the National Center for Biotechnology Information Server (http://www.ncbi.nlm.nih.gov/). The ClustalW program was used to conduct the multiple sequence alignment. The phylogenetic tree construction was performed by the Minimum-Evolution algorithm of Mega X. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches.
Expression Analysis of CsECR in Navel Orange
Expression profiles of CsECR in different tissues of navel orange and after drought and ABA treatments were performed by real-time quantitative PCR (qRT-PCR). The CsECR-specific primers (sense: 5′ CGCTTCAGTCATGCAACGTC 3′; antisense: 5′ GGGGTATATCGTGGGTGGTTC 3′) were designed for qRT-PCR analysis. The qRT-PCR was performed on the CFX96 Touch Real-Time PCR Detection cycler (Bio-Rad, Hercules, USA) using the SYBR®Premix Ex Taq™ kit (Takara, Dalian, China). The qRT-PCR was performed by 20 μL volume containing 10 μL SYBR®Premix Ex Taq, 0.5 μL sense primer, 0.5 μL antisense primer, 1 μL cDNA, and 8 μL ddH2O. The qRT-PCR program was 30 s at 95°C for pre-denaturation, 40 cycles of 5 s at 95°C for denaturation, and 30 s at 60°C for annealing. Melting curve analysis of the qRT-PCR products was performed to confirm the amplicon specificity. The relative gene expression levels were normalized relative to the expression of the Citrus β-actin gene (sense: 5′ ACTGCTCATTCTCAGC CTATG 3′; antisense: 5′ TGCACCCTGTTCTTCTTACTG 3′) and calculated by the 2−ΔΔCT method.
Generation of “Micro Tom” Tomato Ectopic Overexpressing CsECR
A pair of CsECR-specific primers with Nco I and Bgl II restriction sites (sense: 5′ CGACCATGGAT GAAGGTGACTGTAA 3′; antisense: 5′ TTTAGATCTCTACAGGAATGGGGGC 3′) were used to clone the coding sequence of CsECR by RT-PCR. The PCR product was digested by Nco I and Bgl II and ligated into the pCAMBIA1301 vector to produce the expression vector pCAMBIA1301-CsECR under the control of the CaMV35S promoter. The freeze-thaw method was used to introduce this overexpression vector into Agrobacterium tumefaciens strain LBA4404. “Micro Tom” tomato ectopic overexpressing CsECR was generated by Agrobacterium-mediated cotyledon transformation according to the report of Dan et al. (2006). The regeneration of transgenic plants was performed according to the method described by Dan et al. (2006). The positive T2 transgenic tomato lines were screened by hygromycin and further confirmed by genomic PCR with a pair of primers (CaMV35S sense primer: 5′ GCCGTAAAGACTGGCGAACA 3′; CsECR antisense: 5′ ATTGGCGACGTTGCA TGACT 3′). The genomic PCR was performed by a TransDirect® Plant Tissue PCR Kit (Transgen, Beijing, China). The expression levels of CsECR in leaves and fruit peels of the WT and T2 transgenic lines were examined by qRT-PCR with the CsECR-specific primers as described in the above Section Expression Analysis of CsECR in Navel Orange. Tomato actin gene (sense: 5′ GAAATAGCATAAGATGGCAGACG 3′; antisense: 5′ ATACCCACCATCACACCA GTAT 3′) was used as the endogenous control to normalize the expression levels between different samples.
Scanning Electron Microscopy
The cuticular wax morphology on the surfaces of the fully expanded leaves and red ripe fruits in the WT and transgenic tomato lines was observed by SEM. The samples were prepared as described in our previous report (Liu et al., 2015). In brief, fruit peel and leaf disks with a diameter of 0.5 cm were excised from the equatorial zones of the fruits and the leaves. After transferring to aluminum holders, the disks were frozen in liquid N2, freeze-dried, and sputter-coated with gold film in a sputter coater (SBC-12, KYKY, Beijing, China). The SEM (Zeiss DSM 962, Carl Zeiss, Oberkochen, Germany) was used to observe the coated disks at ×1,000 and ×5,000 magnification.
Extraction and Chemical Analysis of Cuticular Waxes
Three biological replicates were carried out for cuticular wax extraction and chemical analysis. A total of 20 fully expanded leaves and 15 red ripe fruits in each line per replicate were used for cuticular wax analysis. The leaves and fruits were immersed three times in chloroform at room temperature for 1 min each time to extract the cuticular waxes. Then, 100 μg of n-tetracosane (C24) was added to the cuticular wax solution as an internal standard. The cuticular wax solution was filtered, concentrated by a rotary evaporator at 35°C under reduced pressure, and dried under a gentle stream of nitrogen. Prior to the gas chromatography–mass spectrometry (GC–MS) experiment, the samples were treated with bis-N,N-(trimethylsilyl)-trifluoroacetamide (BSTFA) in pyridine for 40 min at 70°C to transform the carboxyl and hydroxyl compounds to their corresponding trimethylsilyl ethers and esters. After derivatization, the residual BSTFA was removed under a gentle stream of nitrogen and dissolved in 1 ml of chloroform for GC-MS analysis.
The GC-MS analysis was carried out according to the method of Vogg et al. (2004). In brief, 1 μL of the wax sample was analyzed using Agilent 6890N GC equipped with an HP-5 MS capillary column (30 m × 0.25 mm i.d. × 0.25 μm, Agilent, Palo Alto, CA, USA) and combined with an Agilent 5973N mass spectrometry detector (Palo Alto, CA, USA). Helium was used as the carrier gas with a constant low rate of 2 ml·min−1. GC was performed with an on-column injection temperature of 50°C, held for 2 min at 50°C, increased by 40°C·min−1 to 200°C, held for 2 min at 200°C, increased by 3°C·min−1 to 320 °C, and held for 30 min at 320°C. The MS was performed in positive electron ionization mode at 70 eV, obtaining spectra with an m/z range of 50–850. The wax components were identified by matching their electron ionization mass spectra with those from the NIST 14 MS library. The quantitative analysis of the wax components was performed by the same GC system equipped with a name ionization detector (FID). The same chromatographic conditions as above were used to quantify the wax components, but with H2 as carrier gas. The wax components were quantified by peak area comparisons with the internal standard. The leaf area was determined using a LI-3100C area meter (Li-Cor, Lincoln, NE, USA). Tomato Analyzer version 3.0 was used to assess the fruit surface area (Brewer et al., 2006). The amounts of wax constituents were expressed per unit of leaf or fruit surface area.
Measurement of Water Loss Rate and Chlorophyll Leaching Rate
Leaf and fruit non-stomatal water loss rates were investigated as described by Kosma et al. (2009). Briefly, tomato plants were first transferred to dark for 12 h to make sure that stomata were fully closed. Then, 30 fully expanded leaves and 30 red ripe fruits of the same size were detached from tomato plants in each line per replicate. The excised leaves and fruits were placed at room temperature in the dark for 6 h and 10 days, respectively, and weighed on an analytical balance every 1 h for leaves and every 2 days for fruits. Water loss was calculated as a percentage of the initial fresh weight of leaves or fruits at each time point.
For measurement of chlorophyll leaching rate, 20 fully expanded leaves per biological replicate in each line were sampled and soaked in 80% ethanol under a dark environment. After 15, 30, 60, 90, 120, 150, and 180 min of initial immersion, 2 ml of the solution was used to determine the absorbance of the extract at 647 and 664 nm wavelength by a UV-2600 spectrophotometer (Shimadzu, Kyoto, Japan). The leaf chlorophyll concentration was calculated using the following equation: total micromoles chlorophyll = 7.93 (A664) + 19.53 (A647) (Lolle et al., 1998). Chlorophyll leaching rate was expressed as a percentage of the chlorophyll concentration at each time point in total chlorophyll extracted after 24 h. Three biological replicates were used for water loss and chlorophyll leaching analysis.
Analyses of Phenotype, Ion Leakage, and Survival Rates in WT Plants and Transgenic Lines Under Drought Stress
The 60-day-old seedlings grown in soil were used to compare the difference in phenotype, ion leakage, and survival rates between the WT and two transgenic lines. For drought treatment, the seedlings of the two genotypes were deprived of water for 15 days and then re-watered for 5 days. The control plants of the two genotypes were well-watered during treatment. For measurement of leaf ion leakage, 30 fully expanded leaves in each line per replicate were sampled from each line per replicate after drought treatments. The ion leakage was measured according to the report of Huang et al. (2010). Meanwhile, 30 seedlings in each line per replicate were used to calculate the survival rates under drought stresses. The survival rates were expressed by the percentage of live plants after drought treatment. Three biological replicates were used for the investigation of ion leakage and survival rates.
Statistical Analysis
All data in this paper are shown as the means ± standard deviations of three biological replicates. Statistical analyses were carried out using Student's t-test (for two independent samples) and Duncan's multiple range tests (for one-way ANOVA) in SPSS version 22.
Results
Isolation and Sequence Analysis of CsECR
Based on the in silico cloning and RT-PCR strategy, a novel gene CsECR was isolated from the “Newhall” navel orange and deposited in GenBank under accession No. ON146445. The cDNA sequence of CsECR is 1,524 bp in length with a 933 bp open reading frame (ORF). The CsECR protein contained 310 amino acids with a predicted molecular mass of 36.11 kDa and an isoelectric point of 9.73.
BLAST analysis revealed that CsECR protein shared high identities with ECR proteins from other plant species, such as Citrus clementina CcECR (100%, GenBank accession number XP_006435990.1), Populus trichocarpa PtECR (89%, XP_002311166.1), Gossypium hirsutum GhECR (88%, NP_001314306.1), Malus domestica MdECR (87%, NP_001315672.1), Prunus avium PaECR (87%, XP_021813755.1), Glycine max GmECR (87%, NP_001276149.2), Solanum lycopersicum SlECR (85%, XP_010321381.1), Prunus persica PpECR (87%, XP_007218716.1), and Arabidopsis thaliana AtECR (83%, NP_191096.1). Multiple sequence alignment showed that CsECR contained a conserved NADP/NAD-binding motif (225-GSGGYQIPRG-234) and three conserved functional sites (Q89, K144, R145) (Figure 1A). To investigate the relationship between CsECR and other plant ECRs, a phylogenetic tree was constructed using the Minimum-Evolution algorithm of the MEGA X version. The investigated ECR proteins could be divided into three groups. CsECR was first classified with Citrus clementina CcECR, and then clustered with Ricinus communis RcECR, Vernicia fordii VfECR, Jatropha curcas JcECR, Manihot esculenta MeECR, Herrania umbratical HuECR, Theobroma cacao TcECR, Gossypium arboretum GaECR, Gossypium hirsutum GhECR, Durio zibethinus DzECR, Populus trichocarpa PtECR, Populus euphratica PeECR, Quercus sube QsECR and Juglans regia JrECR, Eucalyptus grandis EgECR, Vitis vinifera VvECR, Ziziphus jujuba ZjECR, Malus domestica MdECR, Prunus avium PaECR, Prunus persica PpECR, Glycine max GmECR, Mucuna pruriens MpECR, Vigna unguiculata VuECR, Cicer arietinum CaECR, Arachis ipaensis AiECR, Lupinus angustifolius LaECR, Cucurbita maxima CmECR, and Momordica charantia McECR, to form the group I. The group II consisted of Dianthus caryophyllus DcECR, Spinacia oleracea SoECR, Chenopodium quinoa CqECR, and Arabidopsis thaliana AtECR. The group III included Solanum tuberosum StECR, Solanum lycopersicum SlECR, Capsicum baccatum CbECR, Oryza sativa OsECR, Sesamum indicum SiECR, and Coffea eugenioides CeECR (Figure 1B).
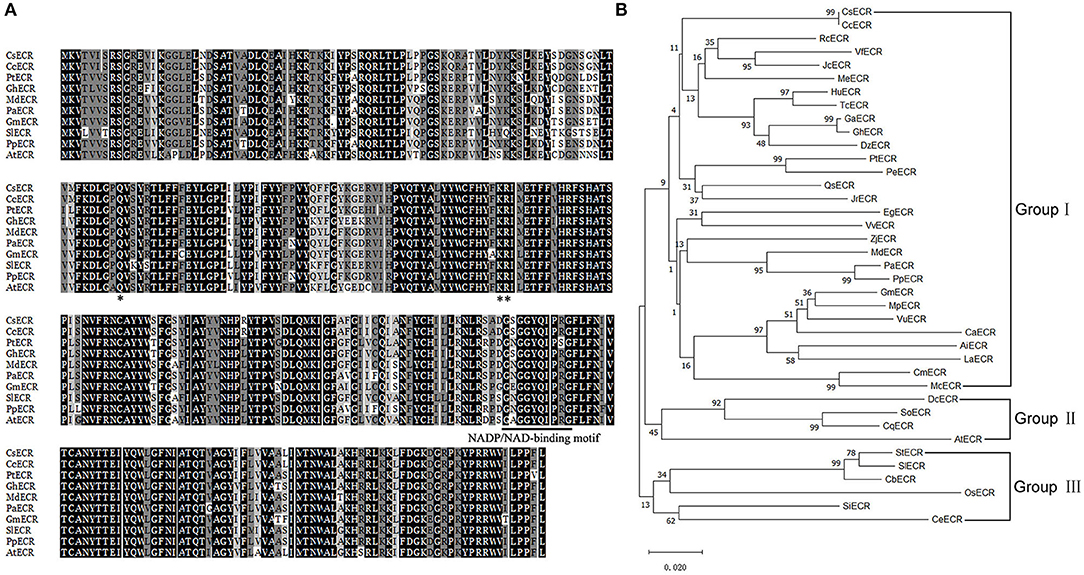
Figure 1. Multiple sequence alignment (A) and phylogenetic analysis (B) of CsECR with its homologous proteins from different plant species, including Citrus clementina CcECR (XP_006435990.1), Ricinus communis RcECR (XP_002530850.1), Vernicia fordii VfECR (AHI86054.1), Jatropha curcas JcECR (XP_012089325.1), Manihot esculenta MeECR (XP_021618477.1), Herrania umbratical HuECR (XP_021295484.1), Theobroma cacao TcECR (XP_007009348.1), Gossypium arboretum GaECR (GenBank accession number XP_017620211.1), Gossypium hirsutum GhECR (NP_001314306.1), Durio zibethinus DzECR (XP_022765174.1), Populus trichocarpa PtECR (XP_002311166.1), Populus euphratica PeECR (XP_011027192.1), Quercus sube QsECR (XP_023916079.1), Juglans regia JrECR (XP_018814522.1), Eucalyptus grandis EgECR (XP_010067405.1), Vitis vinifera VvECR (XP_010658577.1), Ziziphus jujuba ZjECR (XP_015876925.1), Malus domestica MdECR (NP_001315672.1), Prunus avium PaECR (XP_021813755.1), Prunus persica PpECR (XP_007218716.1), Glycine max GmECR (NP_001276149.2), Mucuna pruriens MpECR (RDX83442.1), Vigna unguiculata VuECR (XP_027941677.1), Cicer arietinum CaECR (NP_001265884.1), Arachis ipaensis AiECR (XP_016166715.1), Lupinus angustifolius LaECR (XP_019455882.1), Cucurbita maxima CmECR (XP_022992762.1), Momordica charantia McECR (XP_022134103.1), Dianthus caryophyllus DcECR (BAN15750.1), Spinacia oleracea SoECR (XP_021855497.1), Chenopodium quinoa CqECR (XP_021749534.1), Arabidopsis thaliana AtECR (NP_191096.1), Solanum tuberosum StECR (XP_006345960.1), Solanum lycopersicum SlECR (XP_010321381.1), Capsicum baccatum CbECR (PHT49178.1), Oryza sativa OsECR (XP_015621959.1), Sesamum indicum SiECR (XP_011091470.1), and Coffea eugenioides CeECR (XP_027160771.1). The NADP/NAD-binding motif was marked by solid bar. The conserved amino acid residues were marked by asterisk. Construction of the phylogenetic tree was performed with the Neighbor-Joining algorithm of MEGA 4 version.
Expression Analysis of CsECR in “Newhall” Navel Orange
The expression of CsECR was detected in all of the tested tissues in “Newhall” navel orange. The highest expression was detected in leaves, followed by stems, flavedos, ovaries, juice sacs, stigmas, stamens, albedos, and petals (Figure 2A).
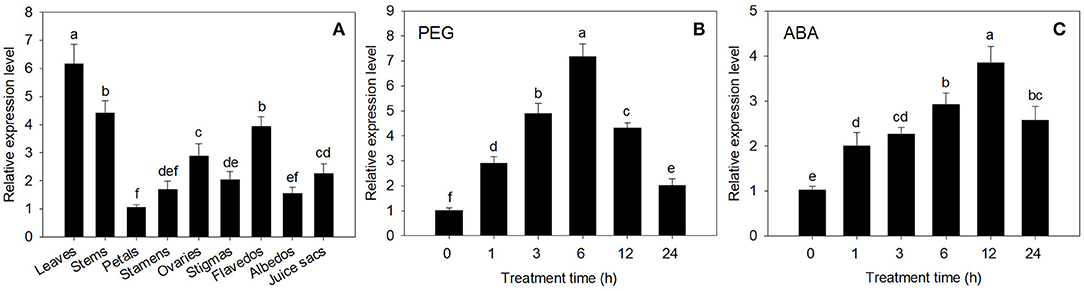
Figure 2. The expression analysis of CsECR in “Newhall” navel orange. (A) The expression of CsECR in different organs. (B,C) The expression patterns of CsECR in leaves of “Newhall” navel orange after drought (20% PEG 6000) and ABA (100 μM) treatments. The y-axis records the relative gene expression levels as calculated by the 2−ΔΔCT method with β-actin as the endogenous reference. Vertical bars represent standard deviations of the means (n = 3). Different small letters represent significant differences at P < 0.05 according to Duncan's multiple range tests.
The expression patterns of CsECR in leaves of “Newhall” navel orange after PEG6000 and ABA treatments were detected using qRT-PCR. The expression level of CsECR was significantly increased at 1 h after treatment with 20% PEG6000 for drought simulation, peaked at 6 h, and then continuously decreased from 6 to 24 h (Figure 2B). Under ABA (100 μM) stress, the CsECR expression was significantly increased at 1 h, reached the maximum level at 12 h, and then decreased from 12 to 24 h. These results suggested that the transcript levels of CsECR were strongly induced by drought and ABA treatments (Figure 2C).
Identification of Positive Transgenic Tomato Lines Ectopic Overexpressing CsECR
To analyze the physiological function of CsECR, we generated transgenic tomato plants that ectopic overexpressed CsECR under the control of CAMV 35S promoter. A total of nine positive T2 transgenic tomato lines were confirmed by kanamycin screening and by PCR at the DNA level (Figure 3A). The qRT-PCR results revealed that high transcript levels of CsECR were detected in leaves and fruit peels of all positive transgenic tomato lines, but no transcripts could be observed in the WT plants. Two T2 transgenic tomato lines (CsECR-OE2 and CsECR-OE6) with the highest expression of CsECR in leaves and fruit peels were selected for further research (Figure 3B). The growth behavior of the two transgenic lines was similar to the WT plants under normal growth conditions (Figure 3C). In addition, the leaves and fruits of the two genotypes exhibited similar morphological characteristics under normal growth conditions (Figures 3D–F).
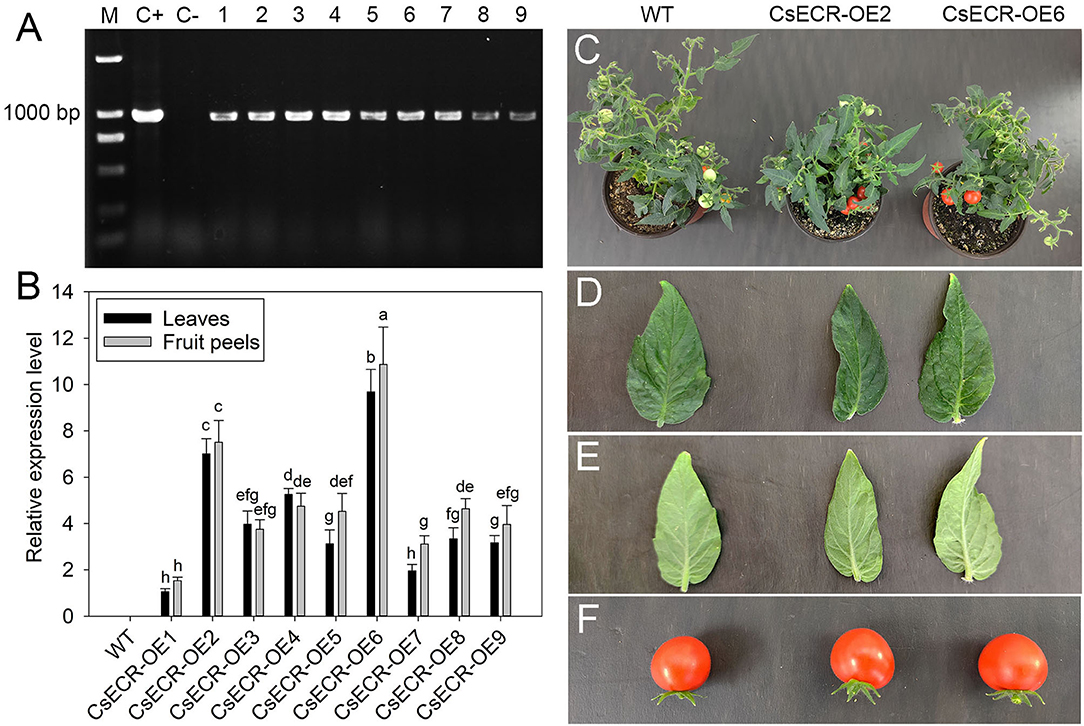
Figure 3. Identification and phenotype analysis of positive transgenic tomato lines. (A) Identification of positive transgenic tomato lines by PCR using CaMV35S sense primer and CsECR antisense primer. M: standard DNA maker; C+: positive control (plasmid contained recombinant expression vector pCAMBIA1301-CsECR); C–: DNA from the wild-type (WT) tomato leaves. Number 1–9: positive transgenic tomato lines. (B) Expression levels of CsECR in leaves and fruit peels of the WT and positive transgenic tomato lines (CsECR-OE1 to CsECR-OE9) were investigated by qRT-PCR. (C) Phenotype of the WT and two positive transgenic tomato lines. (D, E) Phenotype of the adaxial and abaxial sides of leaves, and (F) fruits of the WT and two positive transgenic tomato lines (CsECR-OE2 and CsECR-OE6). Vertical bars represent standard deviations of the means (n = 3). Different small letters represent significant differences at P < 0.05 according to Duncan's multiple range tests.
Ectopic Overexpression of CsECR Increases Cuticular Wax Accumulation on the Surfaces of Leaves and Fruits of the Transgenic Tomato Plants
SEM results showed that no significant difference in cuticular wax morphology was observed between the abaxial leaf surfaces of the WT and transgenic plants (Figures 4A–D). However, the wax crystal density on the fruit surfaces of the transgenic plants was much higher than that on the WT fruit surfaces (Figures 4E–H).
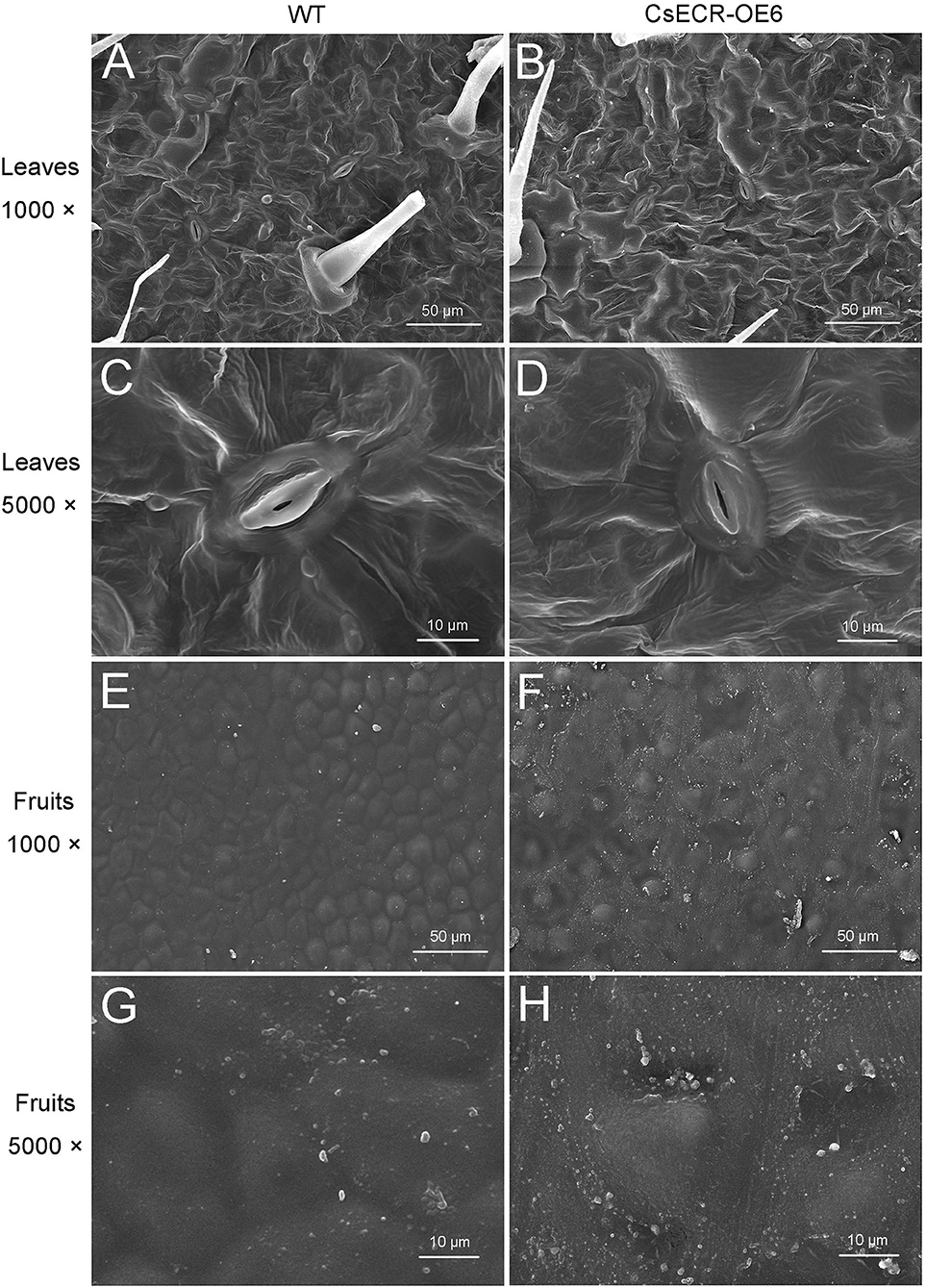
Figure 4. Scanning electron micrographs of cuticular waxes on the surfaces of leaves and fruits in the WT and transgenic tomato (CsECR-OE6). (A,B) The abaxial surfaces of leaves in the WT and transgenic tomato line were imaged at ×1,000 magnification. (C,D) The abaxial surfaces of leaves in the WT and transgenic tomato line were imaged at ×5,000 magnification. (E,F) The surfaces of fruits in the WT and transgenic tomato line were imaged at ×1,000 magnification. (G,H) The surfaces of fruits in the WT and transgenic tomato line were imaged at ×5,000 magnification.
The total wax loads in the leaves of the two transgenic tomato lines were 9.64 and 10.91 μg·cm−2, which increased by 49.46 and 69.15%, respectively, in comparison to the WT leaves (6.45 μg·cm−2) (Figure 5A). The most abundant cuticular wax fraction in the WT leaves was n-alkanes (3.39 μg·cm−2, accounting for 52.56% of total wax load), followed by triterpenoids (1.39 μg·cm−2, 21.55%), n-fatty acids (0.75 μg·cm−2, 11.63%), n-aldehydes (0.35 μg·cm−2, 5.43%), unsaturated fatty acids (0.29 μg·cm−2, 4.50%), iso-, and anteiso-alkanes (0.20 μg·cm−2, 3.10%), unidentified wax component (0.043 μg·cm−2, 0.67%), alkenes (0.018 μg·cm−2, 0.28%), sterols (0.0089 μg·cm−2, 0.14%), and n-primary alcohols (0.0059 μg·cm−2, 0.092%). The amounts of n-alkanes increased by 66.67% (5.65 μg·cm−2) and 89.38% (6.42 μg·cm−2), respectively, in the two transgenic lines compared to the WT leaves. Notably, the increases in n-alkane amounts accounted for 70.85 and 67.94% increases in total wax loads, respectively, in the two transgenic line leaves. Compared to the WT leaves, the amount of n-fatty acids increased by 26.67% (0.95 μg·cm−2) and 49.33% (1.12 μg·cm−2), respectively, in the two transgenic line leaves. Similarly, the content of unsaturated fatty acids increased by 106.90% (0.60 μg·cm−2) and 100.00% (0.58 μg·cm−2), the alkenes increased by 155.56% (0.046 μg·cm−2) and 211.11% (0.056 μg·cm−2), and the iso- and anteiso-alkanes increased by 60.00% (0.32 μg·cm−2) and 85.00% (0.37 μg·cm−2) in the two transgenic line leaves compared with the WT leaves. However, no significant differences were observed in the amounts of n-aldehydes, n-primary alcohols, triterpenoids, and unidentified wax components between the leaves of the two genotypes (Figures 5B, 6A). The n-alkanes in both genotype leaves were made of C22, C23, C25, C27, and C28 to C33 homologs. The amounts of C22:0, C23:0, C25:0, C27:0, C28:0, and C30:0 alkanes in the two transgenic line leaves were significantly higher than those in the WT leaves. The n-fatty acids in the leaves of both genotypes were composed of a series of homologs with even-numbered chain lengths from C16 to C28 and C23. Compared to the WT leaves (0.023 μg·cm−2), ectopic overexpression of CsECR increased the amounts of C26:0 fatty acid by 278.26% (0.087 μg·cm−2) and 595.65% (0.16 μg·cm−2), respectively, in the two transgenic line leaves. The unsaturated fatty acids consisted of C18:2 and C18:3 fatty acids in the leaves of both genotypes. The amounts of C18:2 fatty acid in the two transgenic line leaves increased by 136.36% (0.26 μg·cm−2) and 172.73% (0.30 μg·cm−2), respectively, in comparison to the WT leaves (0.11 μg·cm−2). Alkene was composed of C27:1, C29:1, and C33:1 alkene in the leaves of both genotypes. The amounts of C33:1 alkene increased by 218.18% (0.028 μg·cm−2) and 320.45% (0.037 μg·cm−2) in the two transgenic line leaves compared with the WT leaves (0.0088 μg·cm−2). A total of three iso-alkanes (C31–C33) and one anteiso-alkane (C32) were observed in the leaves of both genotypes. The amounts of C31 and C32 iso-alkanes in the two transgenic line leaves were significantly higher than those in the WT leaves. However, no significant changes were observed for the amounts of other wax components (C24:0 aldehyde, C18:0 primary alcohol, stigmasterol, α-amyrin, β-Amyrin, γ-Amyrin, and unidentified wax component) in the two transgenic line leaves (Figure 5C).
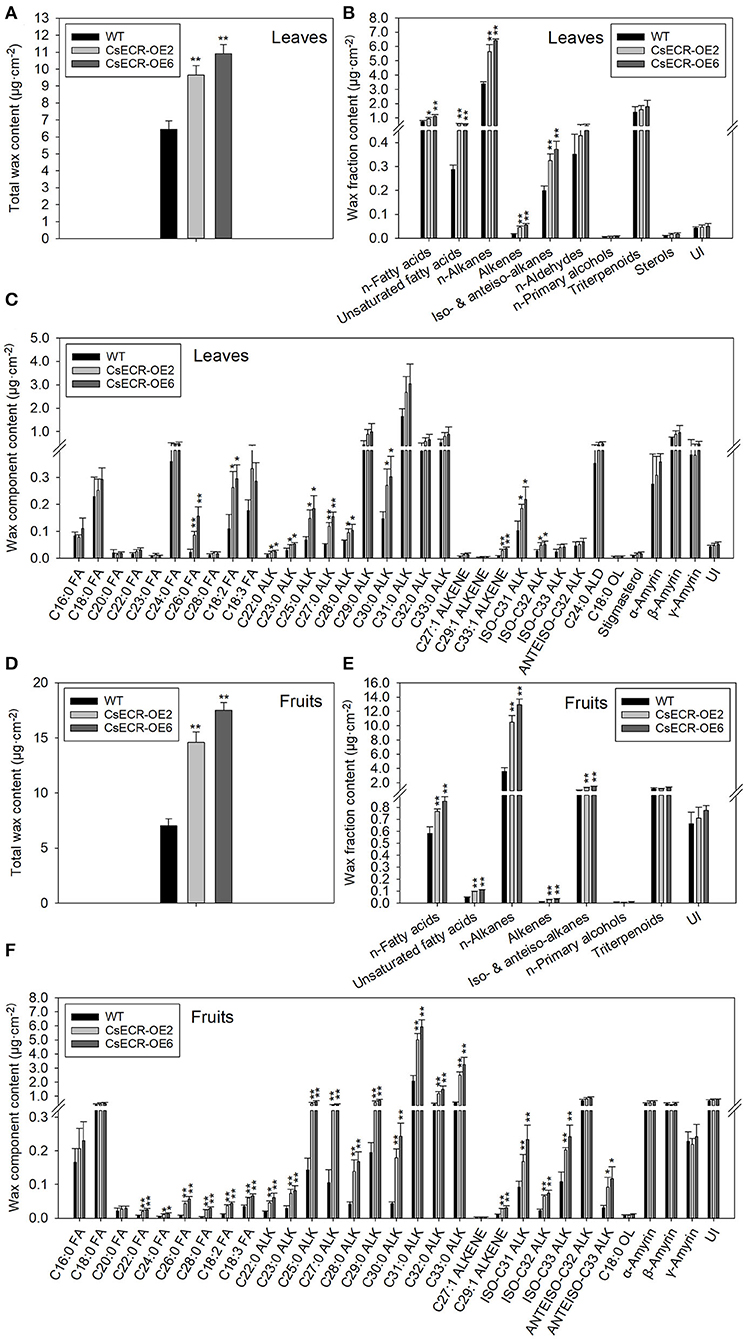
Figure 5. The contents of total waxes, wax fractions, and wax constituents of (A–C) leaves and (D–F) fruits in the WT and two transgenic tomato lines (CsECR-OE2 and CsECR-OE6). FA, fatty acid; ALK, alkane; ALD, aldehyde; OL, alcohol; UI, unidentified components. Vertical bars represent standard deviations of the means (n = 3). Significant differences between the WT and transgenic tomato lines at the P <0.05 and P <0.01 levels were indicated by single and double asterisks, respectively, according to Student's t-test.
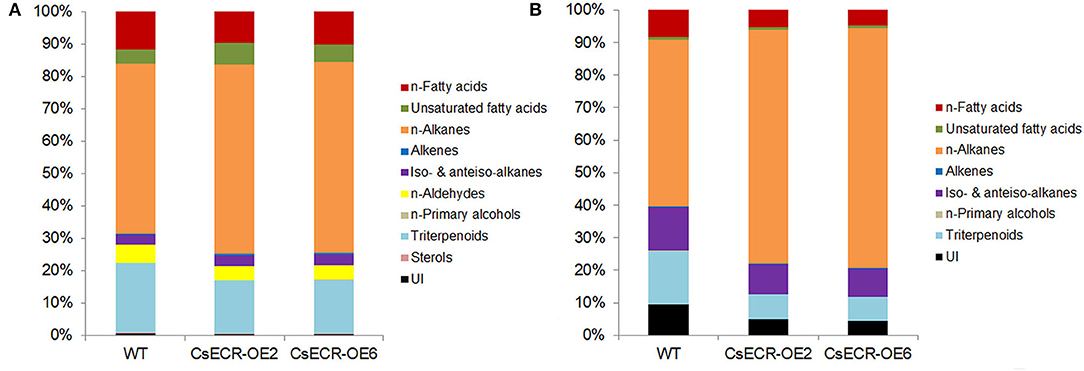
Figure 6. Relative content (%) of each wax fraction in (A) leaves and (B) fruits of the WT and two transgenic tomato lines (CsECR-OE2 and CsECR-OE6). Values are shown as the means of three biological replicates.
Compared to the WT fruits (7.01 μg·cm−2), the total wax loads increased by 108.43% (14.59 μg·cm−2) and 150.29% (17.52 μg·cm−2), respectively, on the surfaces of the two transgenic line fruits (Figure 5D). The most abundant wax fraction in the WT fruits was n-alkanes (3.60 μg·cm−2, accounting for 51.36% of total wax amounts), followed by triterpenoids (1.15 μg·cm−2, 16.41%), iso- and anteiso-alkanes (0.94 μg·cm−2, 13.41%), unidentified wax component (0.67 μg·cm−2, 9.56%), n-fatty acids (0.58 μg·cm−2, 8.27%), unsaturated fatty acids (0.048 μg·cm−2, 0.68%), alkenes (0.013 μg·cm−2, 0.19%), and n-primary alcohols (0.0085 μg·cm−2, 0.12%). Ectopic overexpression of CsECR increased the amounts of n-alkanes by 191.94% (10.51 μg·cm−2) and 259.17% (12.93 μg·cm−2), iso- and anteiso-alkanes by 42.55% (1.34 μg·cm−2) and 61.70% (1.52 μg·cm−2), n-fatty acids by 32.76% (0.77 μg·cm−2) and 46.55% (0.85 μg·cm−2), unsaturated fatty acids by 100.00% (0.096 μg·cm−2) and 129.17% (0.11 μg·cm−2), and alkenes by 130.77% (0.030 μg·cm−2) and 161.54% (0.034 μg·cm−2) in the fruits of two transgenic lines. Notably, 91.16 and 88.77% increases in the total wax loads in the two transgenic line fruit were attributed to the increases of n-alkanes (Figures 5E, 6B). A total of 10 n-alkane components (C22, C23, C25, and C27–C33) were observed in the fruits of both genotypes. The amounts of all n-alkane components in the two transgenic line fruit were significantly higher than those in the WT fruit. The most abundant wax component in the WT fruit was C31 alkane (2.07 μg·cm−2, accounting for 29.53% of total wax amounts), which increased by 142.03% (5.01 μg·cm−2) and 186.47% (5.93 μg·cm−2) in the two transgenic line fruits. Notably, the increases of C31 alkane in the two transgenic line fruit accounted for 38.79 and 36.73% increases in the total wax loads. A total of three iso-alkanes (C31–C33) and two anteiso-alkanes (C32, C33) were observed in the fruit of both genotypes. The amounts of all iso-alkanes and C33 anteiso-alkanes in the transgenic line fruit were significantly higher than those in the WT fruit. The n-fatty acids in the fruit of both genotypes consisted of series homologs with even chain lengths ranging from C16 to C28. The amounts of n-fatty acids with an even chain length larger than C22 significantly increased in the two transgenic line fruit compared to the WT fruits. Besides, the amounts of two unsaturated fatty acids (C18:2 and C18:3) and C29:1 alkene in the two transgenic line fruits were significantly higher than those in the WT fruits. However, other wax components including C27:1 alkene, C18:0 primary alcohol, α-amyrin, β-Amyrin, γ-Amyrin, and unidentified wax components shared similar amounts between the fruits of the two genotypes (Figure 5F).
Ectopic Overexpression of CsECR Reduces the Leaf and Fruit Cuticle Permeabilities and Enhances the Drought Tolerance of Transgenic Tomato Plants
Compared to the WT fruits, the fruits of two transgenic lines showed significantly lower water loss rates from 2 to 10 days after harvest (Figure 7A). Similarly, the water loss rates in the leaves of two transgenic lines were also much lower than those in the WT leaves after detachment from plants for 1–6 h (Figure 7B). Moreover, the two transgenic lines possessed much lower leaf chlorophyll leaching rates in comparison to the WT plants after 15–180 min alcohol treatment (Figure 7C). These results indicated that ectopic overexpression of CsECR reduces the cuticle permeabilities in the leaves and fruits of transgenic tomato plants.
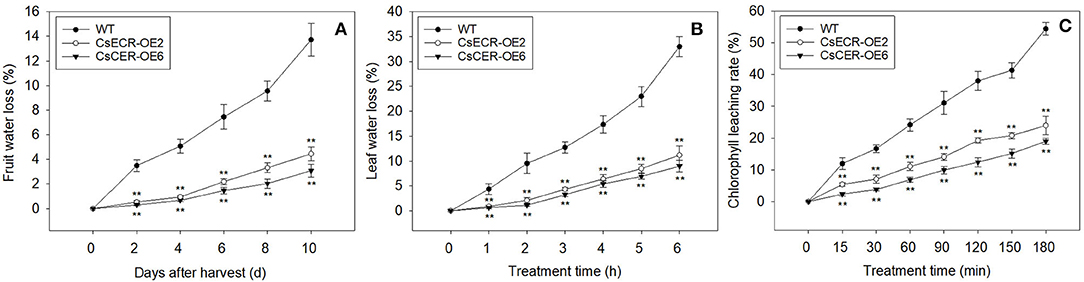
Figure 7. Analyses of cuticular permeability in the fruits and leaves from the WT and two transgenic tomato lines (CsECR-OE2 and CsECR-OE6). (A) The water loss of fruits and (B) leaves in the WT and two transgenic tomato lines. (C) The chlorophyll leaching rates in the leaves of the WT and two transgenic tomato lines. Vertical bars represent standard deviations of the means (n = 3). Significant differences between the WT and transgenic tomato lines at the P <0.01 levels were indicated by double asterisks according to Student's t-test.
After 15 days of drought treatment and 5 days of recovery, almost all leaves in the WT plants were severely wilted, while the two transgenic lines recovered well with only a few wilting leaves detected at the bottom (Figures 8A,B). The survival rates of the two transgenic lines were much higher than those in the WT plants after drought treatment (Figure 8C). In addition, the ion leakage in the leaves of the two transgenic lines was significantly decreased compared with that of the WT plants after drought treatment (Figure 8D). These results suggested that ectopic overexpression of CsECR increased the transgenic plant's resistance to drought stress.
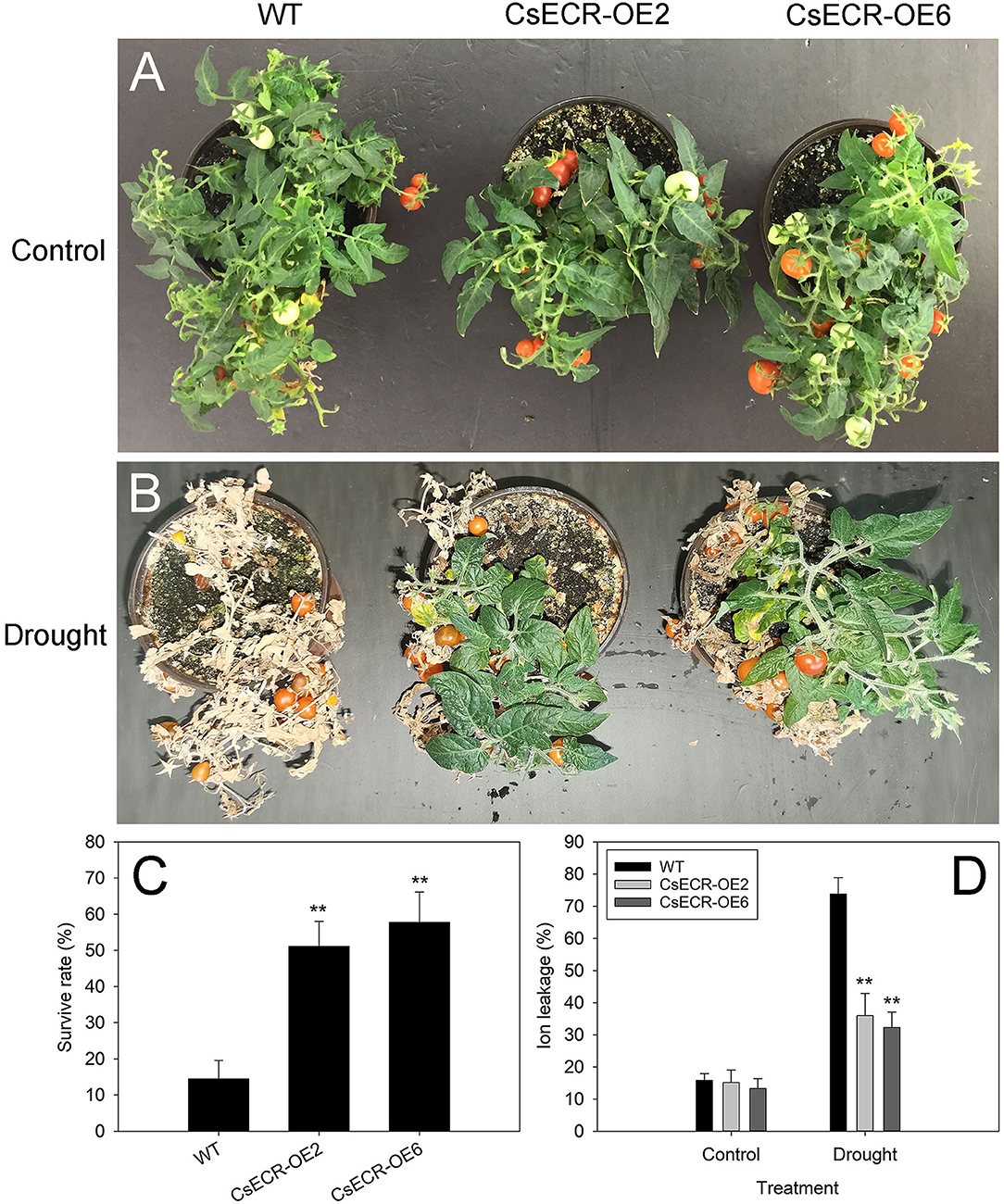
Figure 8. Phenotype (A,B), survival rate (C), and ion leakage (D) in the WT and two transgenic tomato lines (CsECR-OE2 and CsECR-OE6) under drought stress (without watering for 15 days before re-watering for 5 days). Vertical bars represent standard deviations of the means (n = 3). Significant differences between the WT and transgenic tomato lines at the P <0.01 levels were indicated by double asterisks according to Student's t-test.
Discussion
Drought is the main environmental stress that limits citrus growth and reduces its fruit quality and economic value (Rodríguez-Gamir et al., 2010). Our recent report revealed that cuticular waxes play an important role in limiting citrus non-stomatal water loss and regulating the citrus response to drought stress (Liang et al., 2022). However, the underlying molecular mechanism, especially the contribution of specific wax biosynthesis gene to drought tolerance is still unclear in citrus. VLCFAs are precursors and important components of plant cuticular waxes. An FAE system consists of four enzymes that catalyze a series of cyclic reactions to produce VLCFAs in plants. ECR has been shown to catalyze the final step of plant VLCFAs biosynthesis (Haslam and Kunst, 2013). Transcriptome and qRT-PCR analyses in our previous study showed that the expression level of CsECR was significantly downregulated in the fruit peels of “Ganqi 3,” a wax-deficient mutant derived from “Newhall” navel orange, suggesting that CsECR might be involved in cuticular wax biosynthesis and drought response in citrus (Liu et al., 2016). To prove this hypothesis, CsECR was cloned from “Newhall” navel orange and ectopic overexpressed in “Micro-Tom” tomato, a model plant suitable for functional analysis of cuticular waxes due to its astomatous fruit surfaces. CsECR showed high identities with other plant ECR proteins. In addition, all conserved motifs and functional sites of ECR were observed in CsECR. For example, the NADP/NAD-binding motif (225-GSGGYQIPRG-234) characterized in cotton GhECR and other plant ECR (Song et al., 2009), the functional site of K144, R145 in AtECR (Paul et al., 2007) and Q81 (Q89 in CsECR) in Saccharomyces cerevisiae (Kohlwein et al., 2001) were also conserved in CsECR (Figure 1A). The phylogenetic analysis showed that CsECR was classified with other plant ECR proteins, especially CcECR, RcECR, VfECR, JcECR, MeECR, HuECR, TcECR, GaECR, GhECR, and DzECR, which were closer to CsECR than other CsECR proteins (Figure 1B). The protein structural features and phylogenetic results suggested that CsECR was a member of the plant ECR family.
Previous reports showed that the expression levels of TaECR from wheat (Triticum aestivum), BnECR from oilseed rape (Brassica napus), MdCER10 (ECR homolog) from apple (Malus domestica) in leaves and stems were significantly higher than those in other tissues (Ni et al., 2011; Qi et al., 2019; Kong et al., 2020). Similarly, CsECR exhibited the highest expression level in leaves, followed by stems and flavedos (Figure 2A), suggesting that it might regulate cuticular wax biosynthesis mainly in these organs. Environmental stresses, such as drought, cold, and UV irradiation, have a great influence on the biosynthesis of plant cuticular waxes (Shepherd and Griffiths, 2006; Xue et al., 2017; Trivedi et al., 2019). It has been reported that many wax biosynthesis genes in plants are induced by drought stress. For example, the expression levels of AtKCS6 in Arabidopsis, PpCER1-2 in Poa pratensis, and CsKCS6 in “Newhall” navel orange increased under drought stress (Joubès et al., 2008; Guo et al., 2020; Wang et al., 2021). The ECR homolog MdCER10 from apple showed increased transcript levels after drought and salicylic acid treatments (Qi et al., 2019). Furthermore, the transcript levels of numerous wax biosynthesis genes increased after ABA treatment (Kosma et al., 2009; Seo et al., 2009; Wang et al., 2012). In addition, an Arabidopsis transcription factor MYB96 is proven to regulate cuticular wax biosynthesis by the ABA-mediated signaling pathway (Seo et al., 2011). In agreement with these reports, the expression of CsECR was induced by drought and ABA treatments (Figure 2B), suggesting that CsECR might be involved in citrus response to drought stress through the ABA-dependent pathway. Plant ECR genes play an important role in the biosynthesis of VLCFAs and other wax components. For example, virus-induced gene silencing of NbECR in tobacco reduced the amounts of VLCFAs (Park et al., 2005). Silencing of AtECR in Arabidopsis and TaECR in bread wheat decreased the amounts of total waxes, VLCFAs, and their derivatives such as alkanes, alcohols, aldehydes, and ketone (Zheng et al., 2005; Kong et al., 2020). In accordance with these reports, ectopic overexpression of CsECR significantly increased the amounts of total waxes, n-fatty acids, unsaturated fatty acids, n-alkanes, alkenes, iso-, and anteiso-alkanes in the leaves and fruits of transgenic tomato plants (Figures 5A–F), indicating that CsECR, like AtECR, NbECR, and TaECR, functions as an important gene responsible for the biosynthesis of VLCFAs and other aliphatic wax components. The cuticular wax composition detected in the WT leaves and fruits was similar to previous reports on “Micro Tom” tomato (Vogg et al., 2004; Leide et al., 2007). The n-alkanes dominated the culticular wax components in the leaves and fruit of “Micro Tom” tomato (Vogg et al., 2004; Leide et al., 2007). In this study, n-alkanes were also the most abundant wax fraction (accounting for >50% of total wax loads) and contributed to the major increases in total wax amounts (contributing to >67% increases in total wax loads in the leaves and fruits of transgenic tomatoes). Surprisingly, the n-fatty acids, which were the final product of ECR, increased much less than the n-alkanes in both leaves and fruits of the transgenic tomato plants (Figures 5B,E, 6A,B). This result suggested that a larger number of increased n-fatty acid components might be converted to n-alkanes and other aliphatic wax components in the leaves and fruits of transgenic tomatoes. A previous report revealed that the cuticular wax crystals on the fruit surfaces of tomatoes were mainly composed of aliphatic wax compounds (Vogg et al., 2004). Thus, the significant increases in the amounts of n-alkanes and other aliphatic wax components could explain the increases in cuticular wax crystal densities on the fruit surfaces of transgenic tomato plants (Figures 4E–H, 5E,F). To our surprise, the WT and transgenic tomato plants shared similar cuticular wax crystal morphology on the abaxial leaf surfaces, although the total waxes and aliphatic wax components increased significantly in transgenic tomato plant leaves (Figures 4A–D, 5B,C). It should be noted that the increases in total waxes (49.46 and 69.15%) and n-alkanes (66.67 and 89.38%) in leaves were much lower than those in fruits of two transgenic tomato lines (increased by 108.43 and 150.29% in total waxes, and 191.94 and 259.17% in n-alkanes) (Figures 5A,B,D,E). Thus, not like fruits, the increases of cuticular wax components might not be high enough to self-assemble into the cuticular wax crystals on the transgenic tomato leaf surfaces. Another explanation was that the increased cuticular waxes might cover the leaf surfaces of transgenic tomatoes with a morphology of thin film which could not be observed by SEM.
Previous reports showed that the increases of cuticular waxes could reduce the cuticle permeability and enhance the plant tolerance to drought stress. For example, Arabidopsis and rice plants overexpressing wheat TaCER1-1A gene exhibited significant increases of n-alkanes, decreases in cuticle permeability, and enhanced tolerance to drought stress (Li et al., 2019). Ectopic overexpression of navel orange CsKCS6 in Arabidopsis decreased the cuticle permeability and enhanced the transgenic plant tolerance to drought stress by promoting cuticular wax accumulation (Guo et al., 2020). In fact, various wax biosynthesis genes from different plant species have been reported to enhance plant tolerance to drought stress by increasing wax production and reducing the cuticle permeability, such as AtWXP1 and AtWXP2 from Arabidopsis, AhKCS1 from groundnut (Arachis hypogaea), MdSHINE2 and MdLACS2 from apple (Malus × domestica), PpCER1 from Poa pratensis, and so on (Zhang et al., 2007, 2019, 2020; Lokesh et al., 2019; Wang et al., 2021). The plant ECR genes have been reported to be involved in normal shoot development, cell expansion, membrane biogenesis, and fungal pathogen resistance (Park et al., 2005; Zheng et al., 2005; Kong et al., 2020). However, it is still unknown whether plant ECR genes are involved in plant response to drought stress. Our results showed that the increases in cuticular wax contents led to the decreases in cuticle permeability in the leaves and fruits of transgenic tomato plants. Further study revealed that the two transgenic tomato lines were much more tolerant to drought stress than the WT plants, as indicated by their fewer wilting leaves, significantly higher survival rates and much lower ion leakage under drought stress (Figures 8A–D). Our findings proved for the first time that CsECR, an ECR homologous gene encoding enoyl-CoA reductase in navel orange, has a function of enhancing plant drought tolerance by increasing cuticular wax accumulation. However, further study must be performed to investigate the CsECR function in citrus other than in model plants by gene overexpressing and gene silencing techniques. Moreover, the results of this study will prompt us to further investigate the detailed molecular mechanism of CsECR regulating the cuticular wax biosynthesis and drought tolerance, including analysis of CsECR promotor, identification of the upstream transcription factor, and interacting protein of CsECR.
Conclusion
In summary, a novel gene CsECR encoding enoyl-CoA reductase was cloned from “Newhall” navel orange. CsECR exhibited differential transcriptional levels in the tested tissues of “Newhall” navel orange, with the highest expression in leaves. The expression of CsECR was induced by PEG6000 and ABA. Further study revealed that ectopic overexpression of CsECR increased the amounts of n-fatty acids, unsaturated fatty acids, n-alkanes, alkenes, and iso- and anteiso-alkanes, decreased the cuticle permeability, and enhanced the transgenic tomato tolerance to drought stress. These results suggested that CsECR was involved in the biosynthesis of citrus cuticular waxes and plays an important role in regulating plant tolerance to drought stress.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI; ON146445.
Author Contributions
DL: performed major experimental works, writing the original draft, and acquiring financial funding. WG: performed some physiological experiments and visualization of data. XG: performed statistical analyses. LY: visualization of data, and writing—review and editing. WH: interpreting the data. LK: reviewing of data. YH: visualization of data. JX: writing—review and editing. YL: conception, design of the study, writing—review and editing, and financial funding. All authors reviewed and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (Grant No. 2019YFD1000100), the National Natural Science Foundation of China (Grant Nos. 31701896 and 31860544), the Natural Science Foundation of Jiangxi Province (Grant No. 20202BAB205001), and by the earmarked fund for Jiangxi Agriculture Research System (Grant No. JXARS-07-cultivation post).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Abdallat, A., Al-Debei, H., Ayad, J., and Hasan, S. (2014). Over-expression of SlSHN1 gene improves drought tolerance by increasing cuticular wax accumulation in tomato. Int. J. Mol. Sci. 15, 19499–19515. doi: 10.3390/ijms151119499
Bach, L., Michaelson, L. V., Haslam, R., Bellec, Y., Gissot, L., Marion, J., et al. (2008). The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. U. S. A. 105, 14727–14731. doi: 10.1073/pnas.0805089105
Baker, E. A., Procopiou, J., and Hunt, G. M. (1975). The cuticles of citrus species. Composition of leaf and fruit waxes. J. Sci. Food Agr. 26, 1093–1101. doi: 10.1002/jsfa.2740260913
Beaudoin, F., Wu, X., Li, F., Haslam, R. P., Markham, J. E., Zheng, H., et al. (2009). Functional characterization of the Arabidopsis thaliana β-ketoacyl-CoA reductase candidates of the fatty acid elongase. Plant Physiol. 150, 1174–1191. doi: 10.1104/pp.109.137497
Brewer, M. T., Lang, L. X., Fujimura, K., Dujmovic, N., Gray, S., and van der Knaap, E. (2006). Development of a controlled vocabulary and software application to analyze fruit shape variation in tomato and other plant species. Plant Physiol. 141, 15–25. doi: 10.2307/20205718
Cajuste, J. F., González-Candelas, L., Veyrat, A., García-Breijo, F. J., Reig-Armiñana, J., and Lafuente, M. T. (2010). Epicuticular wax content and morphology as related to ethylene and storage performance of ‘Navelate' orange fruit. Postharvest Biol. Technol. 55, 29–35. doi: 10.1016/j.postharvbio.2009.07.005
Dan, Y., Yan, H., Munyikwa, T., Dong, J., Zhang, Y., and Armstrong, C. L. (2006). MicroTom-a high-throughput model transformation system for functional genomics. Plant Cell Rep. 25, 432–441. doi: 10.1007/s00299-005-0084-3
Dietrich, C. R., Perera, M. A. D. N., Yandeau-Nelson, M. D., Meeley, R. B., Nikolau, B. J., and Schnable, P. S. (2005). Characterization of two GL8 paralogs reveals that the 3-ketoacyl reductase component of fatty acid elongase is essential for maize (Zea mays L.) development. Plant J. 42, 844–861. doi: 10.1111/j.1365-313x.2005.02418.x
Ding, S., Zhang, J., Wang, R., Ou, S., and Shan, Y. (2018). Changes in cuticle compositions and crystal structure of ‘Bingtang' sweet orange fruits (Citrus sinensis) during storage. Int. J. Food Prop. 21, 2411–2427. doi: 10.1080/10942912.2018.1528272
Ding, S., Zhang, J., Yang, L., Wang, X., Fu, F., Wang, R., et al. (2020). Changes in cuticle components and morphology of ‘Satsuma' Mandarin (Citrus unshiu) during ambient storage and their potential role on Penicillium digitatum infection. Molecules 25, 412. doi: 10.3390/molecules25020412
Gable, K., Garton, S., Napier, J. A., and Dunn, T. M. (2004). Functional characterization of the Arabidopsis thaliana orthologue of Tsc13p, the enoyl reductase of the yeast microsomal fatty acid elongating system. J. Exp. Bot. 55, 543–545. doi: 10.1093/jxb/erh061
Go, Y. S., Kim, H., Kim, H. J., and Suh, M. C. (2014). Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell 26, 1666–1680. doi: 10.1105/tpc.114.123307
Guo, W. F., Wu, Q., Yang, L., Hu, W., Liu, D. C., and Liu, Y. (2020). Ectopic expression of CsKCS6 from navel orange promotes the production of very-long-chain fatty acids (VLCFAs) and increases the abiotic stress tolerance of Arabidopsis thaliana. Front. Plant Sci. 11, 564656. doi: 10.3389/fpls.2020.564656
Haslam, T. M., and Kunst, L. (2013). Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210, 93–107. doi: 10.1016/j.plantsci.2013.05.008
He, Y., Han, J., Liu, R., Ding, Y., Wang, J., Sun, L., et al. (2018). Integrated transcriptomic and metabolomic analyses of a wax deficient citrus mutant exhibiting jasmonic acid-mediated defense against fungal pathogens. Hortic. Res. 5, 43. doi: 10.1038/s41438-018-0051-0
Huang, X. S., Liu, J. H., and Chen, X. J. (2010). Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC Plant Biol. 10, 230. doi: 10.1186/1471-2229-10-230
Joubès, J., Raffaele, S., Bourdenx, B., Garcia, C., and Laroche-Traineau, J. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67, 547. doi: 10.1007/s11103-008-9339-z
Kerstiens, G. (1996). Cuticular water permeability and its physiological significance. J. Exp. Bot. 47, 1813–1832.
Kim, J., Jung, J. H., Lee, S. B., Go, Y. S., Kim, H. J., Cahoon, R., et al. (2013). Arabidopsis 3-ketoacyl-coenzyme A synthase 9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 162, 567–580. doi: 10.1104/pp.112.210450
Kohlwein, S. D., Eder, S., Oh, C. S., Martin, C. E., Gable, K., Bacikova, D., et al. (2001). Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell Biol. 21, 109–125. doi: 10.1128/MCB.21.1.109-125.2001
Kong, L., Zhi, P., Liu, J., Li, H., Zhang, X., Xu, J., et al. (2020). Epigenetic activation of enoyl-CoA reductase by an acetyltransferase complex triggers wheat wax biosynthesis. Plant Physiol. 183, 1250–1267. doi: 10.1104/pp.20.00603
Kosma, D. K., Bourdenx, B., Bernard, A., Parsons, E. P., Lu, S., Joubes, J., et al. (2009). The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 151, 1918–1929. doi: 10.1104/pp.109.141911
Lee, S. B., Jung, S. J., Go, Y. S., Kim, H. U., Kim, J. K., Cho, H. J., et al. (2009). Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but diferentially controlled by osmotic stress. Plant J. 60, 462–475. doi: 10.1111/j.1365-313X.2009.03973.x
Lee, S. B., and Suh, M. C. (2015a). Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 34, 557–572. doi: 10.1007/s00299-015-1772-2
Lee, S. B., and Suh, M. C. (2015b). Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 56, 48–60. doi: 10.1093/pcp/pcu142
Leide, J., Hildebrandt, U., Reussing, K., Riederer, M., and Vogg, G. (2007). The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: effects of a deficiency in a β-ketoacyl-coenzyme A synthase (LeCER6). Plant Physiol. 144, 1667–1679. doi: 10.1104/pp.107.099481
Leide, J., Hildebrandt, U., Vogg, G., and Riederer, M. (2011). The positional sterile (ps) mutation affects cuticular transpiration and wax biosynthesis of tomato fruits. J. Plant Physiol. 168, 871–877. doi: 10.1016/j.jplph.2010.11.014
Li, T., Sun, Y., Liu, T., Wu, H., An, P., Shui, Z., et al. (2019). TaCER1-1A is involved in cuticular wax alkane biosynthesis in hexaploid wheat and responds to plant abiotic stresses. Plant Cell Environ. 42, 3077–3091. doi: 10.1111/pce.13614
Liang, B., Wan, S., Ma, Q., Yang, L., Hu, W., Kuang, L., et al. (2022). Transcriptome and physiological analyses of a navel orange mutant with improved drought tolerance and water use efficiency caused by increases of cuticular wax accumulation and ROS scavenging capacity. Int. J. Mol. Sci. 23:5660. doi: 10.3390/ijms23105660
Liu, D. C., He, L. G., Wang, H. L., Xu, M., and Sun, Z. H. (2010). Molecular cloning, characterization and expression analysis of PtrHOS1, a novel gene of cold responses from trifoliate orange [Poncirus trifoliata (L.) Raf.]. Acta Physiol. Plant 32, 271–279. doi: 10.1007/s11738-009-0404-2
Liu, D. C., Ma, Q. L., Yang, L., Hu, W., Guo, W. F., Wang, M. L., et al. (2021). Comparative analysis of the cuticular waxes and related gene expression between ‘Newhall' and ‘Ganqi 3' navel orange during long-term cold storage. Plant Physiol. Biochem. 167, 1049–1060. doi: 10.1016/j.plaphy.2021.09.032
Liu, D. C., Yang, L., Wang, Y. C., Zhuang, X., Liu, C. F., Liu, S. B., et al. (2016). Transcriptome sequencing identified wax-related genes controlling the glossy phenotype formation of “Ganqi 3,” a bud mutant derived from wild-type “Newhall” navel orange. Tree Genet. Genomes 12, 55. doi: 10.1007/s11295-016-1017-8
Liu, D. C., Yang, L., Zheng, Q., Wang, Y. C., Wang, M. L., Zhuang, X., et al. (2015). Analysis of cuticular wax constituents and genes that contribute to the formation of ‘glossy Newhall', a spontaneous bud mutant from the wild-type ‘Newhall' navel orange. Plant Mol. Biol. 88, 573–590. doi: 10.1007/s11103-015-0343-9
Liu, D. C., Zeng, Q., Ji, Q. X., Liu, C. F., Liu, S. B., and Liu, Y. (2012). A comparison of the ultrastructure and composition of fruit' cuticular wax from the wild-type ‘Newhall' navel orange (Citrus sinensis [L.] Osbeck cv. Newhall) and its glossy mutant. Plant Cell Rep. 31, 2239–2246. doi: 10.1007/s00299-012-1333-x
Lokesh, U., Venkatesh, B., Kiranmai, K., Nareshkumar, A., Amarnathareddy, V., Rao, G. L., et al. (2019). Overexpression of ß-Ketoacyl Co-A synthase1 gene improves tolerance of drought susceptible groundnut (Arachis hypogaea L.) cultivar K-6 by increased leaf epicuticular wax accumulation. Front. Plant Sci. 9, 1869. doi: 10.3389/fpls.2018.01869
Lolle, S. J., Hsu, W., and Pruitt, R. E. (1998). Genetic analysis of organ fusion in Arabidopsis thaliana. Genetics 149, 607–619. doi: 10.1093/genetics/149.2.607
Millar, A. A., Clemens, S., Zachgo, S., Giblin, E. M., Taylor, D. C., and Kunst, L. (1999). CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11, 825–838. doi: 10.1105/tpc.11.5.825
Millar, A. A., and Kunst, L. (1997). Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12, 121–131. doi: 10.1046/j.1365-313x.1997.12010121.x
Mustafa, R., Hamza, M., Kamal, H., Mansoor, S., Scheffler, J., and Amin, I. (2017). Tobacco rattle virus-based silencing of Enoyl-CoA reductase gene and its role in resistance against cotton wilt disease. Mol. Biotechnol. 59, 241–250. doi: 10.1007/s12033-017-0014-y
Ni, Y., Zhang, F. C., Wang, Y. C., Pu, F., Wang, R., Chai, Y. R., et al. (2011). Cloning and functional analysis of enoyl-CoA reductase gene BnECR from oilseed rape (Brassica napus L.). Acta Agron. Sin. 37, 424–432. doi: 10.1016/S1875-2780(11)60012-6
Nordby, H. E., and McDonald, R. E. (1995). Variations in chilling injury and epicuticular wax composition of white grapefruit with canopy position and fruit development during the season. J. Agr. Food Chem. 43, 1828–1833. doi: 10.1021/jf00055a015
Park, J. A., Kim, T. W., Kim, S. K., Kim, W. T., and Pai, H. S. (2005). Silencing of NbECR encoding a putative enoyl-CoA reductase results in disorganized membrane structures and epidermal cell ablation in Nicotiana benthamiana. FEBS Lett. 579, 4459–4464. doi: 10.1016/j.febslet.2005.07.013
Paul, S., Gable, K., and Dunn, T. M. (2007). A six-membrane-spanning topology for yeast and Arabidopsis Tsc13p, the enoyl reductases of the microsomal fatty acid elongating system. J. Biol. Chem. 282, 19237–19246. doi: 10.1074/jbc.M701774200
Qi, C., Jiang, H., Zhao, X., Mao, K., Liu, H., Li, Y., et al. (2019). The characterization, authentication, and gene expression pattern of the MdCER family in Malus domestica. Hortic. Plant J. 5, 1–9. doi: 10.1016/j.hpj.2018.11.003
Raffaele, S., Vailleau, F., Léger, A., Joubès, J., Miersch, O., Huard, C., et al. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20, 752–767. doi: 10.1105/tpc.107.054858
Rodríguez-Gamir, J., Primo-Millo, E., Forner, J. B., and Forner-Giner, M. A. (2010). Citrus rootstock responses to water stress. Sci. Hortic. 126, 95–102. doi: 10.1016/j.scienta.2010.06.015
Roudier, F., Gissot, L., Beaudoin, F., Haslam, R., Michaelson, L., Marion, J., et al. (2010). Very-long-chain fatty acids are involved in polar auxin transport and developmental patterning in Arabidopsis. Plant Cell 22, 364–375. doi: 10.2307/25680057
Seo, P. J., Lee, S. B., Suh, M. C., Park, M. J., Go, Y. S., and Park, C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23, 1138–1152. doi: 10.1105/tpc.111.083485
Seo, P. J., Xiang, F., Qiao, M., Park, J. Y., Lee, Y. N., Kim, S. G., et al. (2009). The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289. doi: 10.2307/40537768
Shepherd, T., and Griffiths, D. W. (2006). The effects of stress on plant cuticular waxes. New Phytol. 171, 469–499. doi: 10.1111/j.1469-8137.2006.01826.x
Song, W. Q., Qin, Y. M., Saito, M., Shirai, T., Pujol, F. M., Kastaniotis, A. J., et al. (2009). Characterization of two cotton cDNAs encoding trans-2-enoyl-CoA reductase reveals a putative novel NADPH-binding motif. J. Exp. Bot. 60, 1839–1848. doi: 10.1093/jxb/erp057
Todd, J., Post-Beittenmiller, D., and Jaworski, J. G. (1999). KCS1 encodes a fatty acid elongase3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 17, 119–130. doi: 10.1046/j.1365-313X.1999.00352.x
Trivedi, P., Nguyen, N., Hykkerud, A. L., Häggman, H. M., Martinussen, I., Jaakola, L., et al. (2019). Developmental and environmental regulation of cuticular wax biosynthesis in fleshy fruits. Front. Plant Sci. 10, 431. doi: 10.3389/fpls.2019.00431
Vogg, G., Fischer, S., Leide, J., Emmanuel, E., Jetter, R., Levy, A. A., et al. (2004). Tomato fruit cuticular waxes and their effects on transpiration barrier properties: functional characterization of a mutant deficient in a very-long-chain fatty acid β-ketoacyl-CoA synthase. J. Exp. Bot. 55, 1401–1410. doi: 10.1093/jxb/erh149
Wang, D., Ni, Y., Liao, L., Xiao, Y., and Guo, Y. (2021). Poa pratensis ECERIFERUM1 (PpCER1) is involved in wax alkane biosynthesis and plant drought tolerance. Plant Physiol. Bioch. 159, 312–321. doi: 10.1016/j.plaphy.2020.12.032
Wang, J., Hao, H., Liu, R., Ma, Q., Xu, J., Chen, F., et al. (2014). Comparative analysis of surface wax in mature fruits between Satsuma mandarin (Citrus unshiu) and ‘Newhall' navel orange (Citrus sinensis) from the perspective of crystal morphology, chemical composition and key gene expression. Food Chem. 153, 177–185. doi: 10.1016/j.foodchem.2013.12.021
Wang, J., Sun, L., Xie, L., He, Y., Luo, T., Sheng, L., et al. (2015). Regulation of cuticle formation during fruit development and ripening in ‘Newhall' navel orange (Citrus sinensis Osbeck) revealed by transcriptomic and metabolomic profiling. Plant Science 243, 131–144. doi: 10.1016/j.plantsci.2015.12.010
Wang, Y, Wan, L., Zhang, L., Zhang, Z., Zhang, H., Quan, R., et al. (2012). An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 78, 275–288. doi: 10.1007/s11103-011-9861-2
Wen, X., Geng, F., Cheng, Y., and Wang, J. (2021). Ectopic expression of CsMYB30 from Citrus sinensis enhances salt and drought tolerance by regulating wax synthesis in Arabidopsis thaliana. Plant Physiol. Bioch. 166, 777–788. doi: 10.1016/j.plaphy.2021.06.045
Wu, H., Liu, L., Chen, Y., Liu, T., Jiang, Q., Wei, Z., et al. (2022). Tomato SlCER1–1 catalyzes the synthesis of wax alkanes, increasing drought tolerance and fruit storability. Hortic. Res. 9, uhac004. doi: 10.1093/hr/uhac004
Xue, D., Zhang, X., Lu, X., Chen, G., and Chen, Z. H. (2017). Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front. Plant Sci. 8, 621. doi: 10.3389/fpls.2017.00621
Zhang, C. L., Hu, X., Zhang, Y. L., Liu, Y., Wang, G. L., You, C. X., et al. (2020). An apple long-chain acyl-CoA synthetase 2 gene enhances plant resistance to abiotic stress by regulating the accumulation of cuticular wax. Tree Physiol. 40, 1450–1465. doi: 10.1093/treephys/tpaa079
Zhang, J. Y., Broeckling, C. D., Sumner, L. W., and Wang, Z. (2007). Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol. Biol. 64, 265–278. doi: 10.1007/s11103-007-9150-2
Zhang, M., Wang, J., Liu, R., Liu, H., Yang, H., Zhu, Z., et al. (2022). CsMYB96 confers water loss resistance in citrus fruit by simultaneous regulation of water transport and wax biosynthesis. J. Exp. Bot. 73, 953–966. doi: 10.1093/jxb/erab420
Zhang, Y. L., Zhang, C. L., Wang, G. L., Wang, Y. X., and Hao, Y. J. (2019). Apple AP2/EREBP transcription factor MdSHINE2 confers drought resistance by regulating wax biosynthesis. Planta 249, 1627–1643. doi: 10.1007/s00425-019-03115-4
Zheng, H., Rowland, O., and Kunst, L. (2005). Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17, 1467–1481. doi: 10.1105/tpc.104.030155
Keywords: navel orange, cuticular wax, drought, CsECR, transgenic tomato
Citation: Liu D, Guo W, Guo X, Yang L, Hu W, Kuang L, Huang Y, Xie J and Liu Y (2022) Ectopic Overexpression of CsECR From Navel Orange Increases Cuticular Wax Accumulation in Tomato and Enhances Its Tolerance to Drought Stress. Front. Plant Sci. 13:924552. doi: 10.3389/fpls.2022.924552
Received: 20 April 2022; Accepted: 10 June 2022;
Published: 05 July 2022.
Edited by:
Agnieszka Zienkiewicz, Nicolaus Copernicus University in Toruń, PolandReviewed by:
Adoración Zafra, Spanish National Research Council (CSIC), SpainNazim Hussain, Independent Researcher, Sharjah, United Arab Emirates
Copyright © 2022 Liu, Guo, Guo, Yang, Hu, Kuang, Huang, Xie and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Liu, bGl1eW9uZ2p4YXVAMTYzLmNvbQ==
 Dechun Liu
Dechun Liu Wenfang Guo
Wenfang Guo Yong Liu
Yong Liu