
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 07 July 2022
Sec. Plant Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.924417
 Yongmei Zhou1,2
Yongmei Zhou1,2 Wenqin Fang1,2
Wenqin Fang1,2 Ziqin Pang1,2
Ziqin Pang1,2 Li-Yu Chen1,2
Li-Yu Chen1,2 Hanyang Cai1,2
Hanyang Cai1,2 Noor-Ul-Ain1,2
Noor-Ul-Ain1,2 Men-Chi Chang3
Men-Chi Chang3 Ray Ming4*
Ray Ming4*During sexual reproduction in flowering plants, haploid spores are formed from meiosis of spore mother cells. The spores then undergo mitosis, develop into female and male gametophytes, and give rise to seeds after fertilization. We identified a female sterile mutant ap1g2-4 from EMS mutagenesis, and analyses of two T-DNA insertion mutants, ap1g2-1+/− and ap1g2-3−/−, and detected a partial female and male sterility. The ap1g2 mutant gametophyte development was arrested at one nuclear stage. A complementation test using a genomic sequence of AP1G2 with its native promoter restored the function in the three ap1g2 mutant lines. Transcriptome profiling of ap1g2 ovules revealed that four genes encoding clathrin assembly proteins PICALM5A/B and PICALM9A/B, which were involved in endocytosis, were downregulated, which were confirmed to interact with AP1G2 through yeast two-hybrid assays and BIFC analysis. Our result also demonstrated that RALFL4-8-15-19-26 CML16 and several calcium-dependent protein kinases, including CPK14-16-17, were all downregulated in the ovules of ap1g2-1+/−. Moreover, Ca2+ concentration was low in impaired gametophytes. Therefore, we proposed that through interaction with PICALM5A/B and PICALM9A/B, AP1G2 may mediate gametogenesis accompanied by Ca2+ signaling in Arabidopsis. Our findings revealed a crucial role of AP1G2 in female and male gametogenesis in Arabidopsis and enhanced our understanding of the molecular mechanisms underpinning sexual reproduction in flowering plants.
Plants have a life cycle variation between haploid and diploid phases, resulting in an alternation of generations. Angiosperms have evolved to produce a female gametophyte (FG; embryo sac) and a male gametophyte (MG; pollen). The female gametophyte plays an essential role in plant reproduction, including guiding pollen tube growth, fertilization, and seed development. In Arabidopsis, maize, and rice, genetic analyses have revealed that mutant defects were found at all stages of female gametophyte development, and analysis of the mutants began to reveal the characteristics of female gametophyte developmental programs (Yadegari and Drews, 2004; Drews and Koltunow, 2011; Bonnemaison et al., 2014). These studies improved our understanding of the regulatory network in the development of female gametophytes (Yadegari and Drews, 2004).
Large-scale screening for female sterile mutants has identified hundreds of female gametophyte mutants, while some mutants had defects in male gametophytes as well. Most of the characterized genes mediate essential functions (Yadegari and Drews, 2004; Pagnussat et al., 2005; Drews and Koltunow, 2011). Screening for female sterile mutants is a challenging task as it requires one additional generation to identify female sterile mutants. It also takes one additional generation to make crosses because such mutants can only maintain in heterozygous genotype. Our objective is to build a collection of female sterile mutants, ranging from the inception of carpel primordia, abortion of female reproduction organ, to pre- and post-meiosis mutations affection female gametophyte development to explore gene network controlling sex determination in male flowers.
The gene AP1G2 was analyzed by whole-genome resequencing, and, apparently, it may encode a large subunit of adaptor protein complex-1 (AP-1). Adaptor protein (AP) complexes, the predominant coat proteins that link the membrane proteins with clathrin molecules that form the coat of a lipid vesicle, have been characterized in various eukaryotic cells. They interact with membrane proteins, such as different classes of cargo receptors in the process of generating a clathrin-coated vesicle (CCV). The structure of AP complexes is highly conserved across all eukaryotes and comprises two large subunits (α/γ/δ/ε and β), one medium μ subunit, and one small σ subunit, and the lack of any single APs subunit impairs the function of APs (Teh et al., 2013; Bonnemaison et al., 2015; Navarro Negredo et al., 2017). Among the APs, AP-1 plays a role in insoluble enzyme composition and some membrane proteins from trans-Golgi network (TGN) to endosomes and lysosomal transport (Bonnemaison et al., 2014; Wang et al., 2014, 2017). The γ subunit of AP-1 is encoded by AP1G1 and AP1G2 (Park et al., 2013). Earlier studies demonstrated that AP1G is crucial for synergid-controlled pollen tube reception and pollen development through mediating vacuolar remodeling (Feng et al., 2017; Wang et al., 2017). Here, we report new functions for AP1G2 in gametogenesis. In this study, we characterized AP1G2 in female and male gametogenesis. We identified three loci, At1g22410, At1g22730, and At1g23900 after mapping the causal mutation of a female sterile mutant obtained by EMS mutagenesis, and then screening and annotation of candidate SNPs showed that suppression of recombination occurred in all non-reference alleles (Ozias-Akins and Van Dijk, 2007). Confirmation of candidate genes was performed by studying single mutants of each candidate gene, and we found the T-DNA insertion lines of At1g23900 (AP1G2) showed mitosis malfunction during gametogenesis, which appeared in the sterile EMS mutant. We transformed the construct ProAP1G2:AP1G2 into the EMS mutant, and the transgenic plants showed partial restoration infertility. Then, we did transcriptome profiling of ap1g2 ovules, which revealed four downregulation of four genes encoding clathrin assembly proteins PICALM5A/B and PICALM9A/B, which have been reported to mediate endocytosis. Additional characterization by BIFC and yeast two-hybrid assays comfirmed their interaction in vivo and in vitro. Additionally, RALFL genes, CML16, and several calcium-dependent protein kinases, including CPK14-16-17, were all downregulated in the ovules of ap1g2. And Ca2+ concentration was low in impaired gametophytes. Therefore, we propose that, through interaction with PICALM5A/B and PICALM9A/B, AP1G2 mediates female and male gametogenesis by virtue of its regulation role during Ca2+ signaling in Arabidopsis.
The T-DNA insertion lines of AP1G2, SALK_032500 (ap1g2-1), SALK_137129 (ap1g2-3), and lines of At1g22410 and At1g22730 in Supplementary Table 2 were obtained from the Arabidopsis Biological Resource Center (ABRC). The pAKV:H2B-YFP marker line was kindly provided by W.C. Yang at Academician of the Chinese Academy of Sciences. All the seeds were sterilized with 75% ethanol, cold-treated at 4°C overnight, and germinated in Murashige and Skoog (MS) mediums. Seedlings were planted into the soil containing [(peat moss:perlite, 2:1 (v/v)] in plastic pots and put in an air-conditioned room under 22°C, 70% relative humidity, and a 16:8 (light:dark; L:D) photoperiod (Chen et al., 2017).
The wild type and mutants used in the experiment were all from the Col-0 ecotype. Wild-type seeds were mutagenized with 40-m EMS for 8 h, and mutants induced by EMS were identified by screening plants from the second generation. The identified female-sterile mutants were backcrossed to the wild type to generate BC1 progeny and propagated by self-pollination to generate the BC1F2 segregating population.
Genomic DNA extracted from leaves of mutant and non-mutant plants was pooled. The libraries of both pools were constructed and sequenced by an Illumina HiSeq ™2500 platform at Novogene Corporation. The average sequencing depth was about 32-× coverage for both mutant and non-mutant pools. The reads we obtained from mutant and non-mutant pools were aligned with the Col-0 reference genome (Arabidopsis thaliana.TAIR10.21) by the software BWA, and SAMtools-mpileup was used to identify potential SNPs as described by Zhu et al. (2012). Single nucleotide polymorphisms (SNPs) of mutants were detected among BC1F2 segregants. Each candidate SNP was confirmed by PCR and sequencing of at least 12 mutants separately in BC2 progeny using primers as listed in Supplementary Table 1.
Molecular identification of genotypes of the T-DNA insertion lines was performed as described by Pagnussat et al. (2007). The primers used for identification were designed using the online tool (http://signal.salk.edu/tdnaprimers.2.html). For all T-DNA insertion mutants, T-DNA-specific primer LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′) was used, and the genomic sequence-specific primers were included in Supplementary Table 2. INO (INNER NO OUTER) is a key gene regulating the development of the outer integument (Meister et al., 2004; Sieber et al., 2004). To examine whether INO expression changed in EMS mutants, we collected the carpels from WT, wild-type plants from the BC1F2 population (ap1g2−4+/+ and ap1g2−4+/−), ap1g2−4−/−, ap1g2−4−/−/Pro:AP1G:AP1G2, ap1g2-1+/−, ap1g2-3−/−, ap1g2-1+/−/Pro:AP1G:AP1G2, and ap1g2-3−/−/Pro:AP1G:AP1G2 to extract total RNA and perform qRT-PCR. Total RNA was extracted using Total RNA Kit I (OMEGA). The first-strand cDNA synthesis for real-time PCR and the latter were, respectively, performed using PrimeScript® RT reagent Kit and SYBR Premix Ex Taq™ (Takara). The paired primers used for qRT-PCR were INO-F(5′ CAATGGTGGTGACTGTGAG 3′) and INO-R (5′ GCTTCTCAGGTGGTTTATT 3′) for INO, and Actin2F (5′TCCCTCAGCACATTCCAGCAGAT3′) and Actin2R (5′AACGATTCCTGGACCTGCCTCATC3′) for the reference gene ACTIN2. A t-test was used to analyze the differences between every two groups of samples. And qRT-PCR was performed for confirmation of mutation by T-DNA on target sites in T-DNA insertion lines using the primers AP1G2q-F (5′CTCCTGGCAAGCGGTAGT3′), AP1G2q-R (5′GCGAGGGAAGTTGCTGAC3′) for AP1G2 with Actin2F and Actin2R for the reference gene ACTIN2. The relative expression levels in different tissues were calculated with the cycle threshold methodology 2-ΔCt.
To analyze the seed set of the wild type and mutants, the total number of ovules and seeds was counted (n = 10), as described Alvarez and Smyth (1999). The seed set in each silique was the percentage of seeds to the total number of ovules. Statistical significance of the values was accessed using one-way ANOVA, followed by a least significant difference (LSD) test using a 95% confidence interval. For megagametophyte analysis, ovules were excised from different-sized pistils previously fixed in FAA (70% ethanol: acetic acid: 30% methylaldehyde, 9:0.5:0.5). They were cleared with chloral hydrate solution (chloral hydrate: glycerol: sterilized water, 8:1:2) and examined with the Olympus BX63 microscope equipped with DIC and phase-contrast optics.
The genomic region-2kb before the start codon ATG corresponding to the putative AP1G2 promoter was amplified by PCR from wild-type genomic DNA using proAP1G2-F (5′-CACCAATACATGAGGGAAAGGTGAGA-3′) in combination with the reverse primer proAP1G2-R (5′-TTGGTCCACCGGCAACTTTA-3′). For the molecular complementation test, the 7,912 bp of the genomic fragment containing the promoter and the gene of AP1G2 was amplified by PCR using the forward primer proAP1G2-F, in combination with the reverse primer AP1G2-R (5′-CAACCCGCGAGGGAAGTTG-3′) upstream of the stop codon. All PCR products were cloned in the pENTR/D/TOPO vector (Invitrogen). The generated entry vectors were subsequently used for generating the corresponding expression vectors PGWB533-GUS. Arabidopsis plants were transformed with agrobacterium tumefaciens strains GV3101 using the floral dip method (Clough and Bent, 1998). The presence of the transgene in T1 plants was confirmed by PCR using forward primer pro-F (5′AGTAGAGTAGGTAGCGTCAGAA-3′) for transgenic lines with promoter and gene-F (5′-ACGGAAAAGATGTATTAGAGG-3′) for complementation lines, and combined with the reverse primer GUS-R (5′-CGGCGAAATTCCATACCTG-3′).
For ap1g2 phenotypic analysis, the whole inflorescences from wild type and ap1g2 mutants were fixed in FAA fixative solution overnight, and transferred to sterilized water for 2 min. The ovaries were dissected on a slide and cleared in chloral hydrate solution and finally photographed using an Olympus BX63 microscope equipped with differential interference contrast (DIC) and phase-contrast optics. For β-Glucuronidase (GUS) staining, a GUS histochemical assays kit (Real-Time, China) was used following the manufacturer's protocol. Inflorescences and developing seedlings were incubated in a GUS staining buffer at 37°C overnight. Ovules were then washed three times in 75% ethanol for 7 h, followed by examination and photographs. To test the viability of the pollen, the anthers were collected at the anthesis stage and evaluated with the Alexander Red stain (Alexander, 1969). The development stage of pollen in wild type and mutants was analyzed for each subsequent bud in the same inflorescence from an open flower to the next 10 unopened buds. Anthers were placed on slides with DAPI staining solution (0.02-M citric acid, 0.16-M Na2HPO4, 0.2 μgml−1 DAPI (D'ippolito et al., 2017), and photographed using an Olympus BX63 microscope.
To confirm that megagametogenesis was affected in ap1g2 ovules, ap1g2-1+/− and ap1g2-3−/− were crossed with pAKV:H2B-YFP marker lines. We analyzed five progenies with an ap1g2-1 and ap1g2-3, which showed partial sterility. The ovules at different flower stages were observed using a Leica TCS SP8X confocal microscope at an excitation wavelength of 488 nm. Emissions were collected at 500–530 nm to visualize yellow fluorescence protein (YFP). Anthers for each successive bud mounted into slides with DAPI staining solution were also observed using the confocal microscope at an excitation wavelength of 405 nm, and emissions were collected at 415–500 nm.
To obtain the expression profiles during female gametogenesis of the wild type and ap1g2 mutants, we collected the ovules under a microscope from the wild type and the three ap1g2 mutants at different stages for RNA-Seq. The RNA-seq data reads were mapped against the Arabidopsis thaliana reference genome by Hisat2 (Pertea et al., 2016). We used StringTie to calculate expression levels, which were normalized by fragment per kb of transcript per million fragments mapped (FPKM). Differentially expressed genes (DEGs) among samples were defined using fold change values by the expression of assembled transcripts. DEGs were selected on the criteria, having an adjusted P < 0.05 and fold change value > 2 by DESeq2.R (Anders and Huber, 2010). We visualized the ratios of each DEG of the three mutants by Ternary Plot Visualization (Bulgarelli et al., 2012; Pang et al., 2021). We used online software (https://www.omicshare.com/tools/Home/Soft/gogsea) to perform the GO enrichment analysis. We identified 20 gene clusters definedby Fuzzy c-means clustering using the expression profiles of the ovules from wild type and ap1g2 mutant lines at the MMC, FG1, and FG3 stages (Gao et al., 2017).
DEGs in the top significant enriched GO terms that occurred in ap1g2-1+/− were selected for STRING analysis (https://string-db.org). DEGs in the top significant enriched GO terms identified in ap1g2-1+/− were selected for STRING analysis (https://string-db.org). The DEGs that occurred in Cluster 13 were also used for STRING analysis.
Interaction between AP1G2 and the four PICALM proteins was tested in a yeast two-hybrid assay using a DUAL membrane pairwise interaction kit (DualsystemsBiotech). AP1G2 was cloned into the bait vector pBT3-N-AP1G2, and the four PICALM proteins were cloned into the prey vector pPR3-N (pPR3-N-PICALM5A, pPR3-N-PICALM5B, pPR3-N-PICALM9A, pPR3-N-PICALM9B). pBT3-N-AP1G2 and pOst1-NubI were co-transformed into the yeast strain NMY51 to test AP1G2 expression. pBT3-N-AP1G2 and pPR3-N, pTSU2-APP and pPR3-N-PICALM5A, pTSU2-APP and pPR3-N-PICALM5B, pTSU2-APP and pPR3-N-PICALM9A, and pTSU2-APP and pPR3-N-PICALM9B were co-transformed into NMY51 as a negative control. pTSU2-APP and pNubG-Fe65 were co-transformed into NMY51 as a positive control. Interactions of pairs pBT3-N-AP1G2 and pPR3-N-PICALM5A, pBT3-N-AP1G2 and pPR3-N-PICALM5B, pBT3-N-AP1G2 and pPR3-N-PICALM9A, and pBT3-N-AP1G2 and pPR3-N-PICALM9B were assayed. All transformants were grown on synthetic dropout SD-Trp-Leu agar plates and SD-Trp-Leu-His-Ade agar plates for 4 days at 30°C.
The full-length AP1G2 and the four PICALM genes were amplified without stop codons, and cloned to YN and YC, respectively. Plasmids were transformed into Agrobacterium strain GV3101. Agrobacterium strains carrying the BiFC constructs were infiltrated into leaves of 5- to 6-week-old Nicotianabenthamiana plants. The YFP signals were examined by confocal microscopy (Leica TCS SP8X) in the leaf epidermis 24 h after the infiltration.
A calcium ion detection kit was used to observe the changes in calcium concentration during female gametogenesis in the wild type and ap1g2−1+/−, which adopted the BBcellProbe F03 intracellular calcium ion concentration fluorescent probe to detect the change of calcium ion concentration. BBcellProbe F03 has very high cell permeability, and, after, it can combine with calcium ion in living cells and produce strong fluorescence. According to the instruction of this kit, we diluted the probe loading concentration by 1,000 times, using HBSS (Hank's Balanced Salt Solution; components: 140 mg/L CaCl2, 100 mg/L MgCl2.6H2O, 100 mg/L MgSO4.7H2O, 400 mg/L KCI, 60 mg/L KH2PO4, 350 mg/L NaHCO3, 8,000 mg/L NaCl, 48 mg/L Na2HPO, 1,000 mg/L D-Glucose). After washing the isolated ovules at different developing stages with a PBS buffer on slides, we added 25 ul F03 solution and incubated the slides at 22°C for 30 min. Approximately, a 25-ul PBS buffer was added instead of the F03 solution before incubation as negative control to determine the false positive staining reaction. The fluorescence signal of the ovules was detected using a Leica TCS SP8X confocal microscope at an excitation wavelength of 488 nm. Emissions were collected at 515–530 nm. To compare the Ca2+ level in wild-type and aborted embryo sacs, the software ImageJ (Schneider et al., 2012) was used to measure the mean fluorescent intensity.
The fresh inflorescences of the wild type and ap1g2-1+/− were fixed in 2% glutaraldehyde (v/v) in a.1-M KH2PO4 buffer (pH 7.8), containing 1% potassium pyroantimonate (K2H2Sb2O7·4H2O) for overnight in 4°C (Qiu et al., 2005). The inflorescences were then washed three times with the same buffer containing 1% potassium pyroantimonate at 25°C and immersed for 4 h at 25°C in the same buffer, containing 1% (w/v) Osmium Tetraoxide. Inflorescences were then dehydrated using an acetone series [30, 50, 70, 85, 95, and 100% (v/v)]. Anthers and pistils at different developmental stages were finally embedded in Spurr's resin, and sections were observed with a transmission electron microscope (H-7650, HITACHI). We also fixed the wild-type inflorescences in the 2% glutaraldehyde, which was diluted with a.1-M KH2PO4 buffer (pH 7.8) and made sections as control. We examined the abundance of calcium precipitation in embryo sacs using imaging software (Simple PCI Version 6.6). Since calcium precipitates were not found in the central vacuoles, the central vacuoles were excluded when considering the areas of embryo sacs. So the abundance of calcium precipitates in wild-type embryo sacs only represented the average abundance in nuclear and cytoplasm. And, for the microspores, we only examined the abundance of young microspores with one nucleus centrally positioned, because, at this stage, the mutant microspores of ap1g2-1+/− began to degenerate. The number of calcium precipitates per μm2 each of the two groups was analyzed using t-test.
We obtained a sterile mutant from an ethyl methanesulfonate (EMS) mutagenesis screening, which showed shorter siliques with no seed set (Figure 1A). Cytological observations showed that the mutant had defects in the outer integument and embryo sac development (Figures 1B,C). About 56.3% of ovules were arrested at Stage FG1 (Christensen et al., 1998) in which the functional megaspore either persisted or degenerated after this stage. Approximately, 43.70% of ovules could undergo three mitoses and develop into mature embryo sac (Figure 1C).
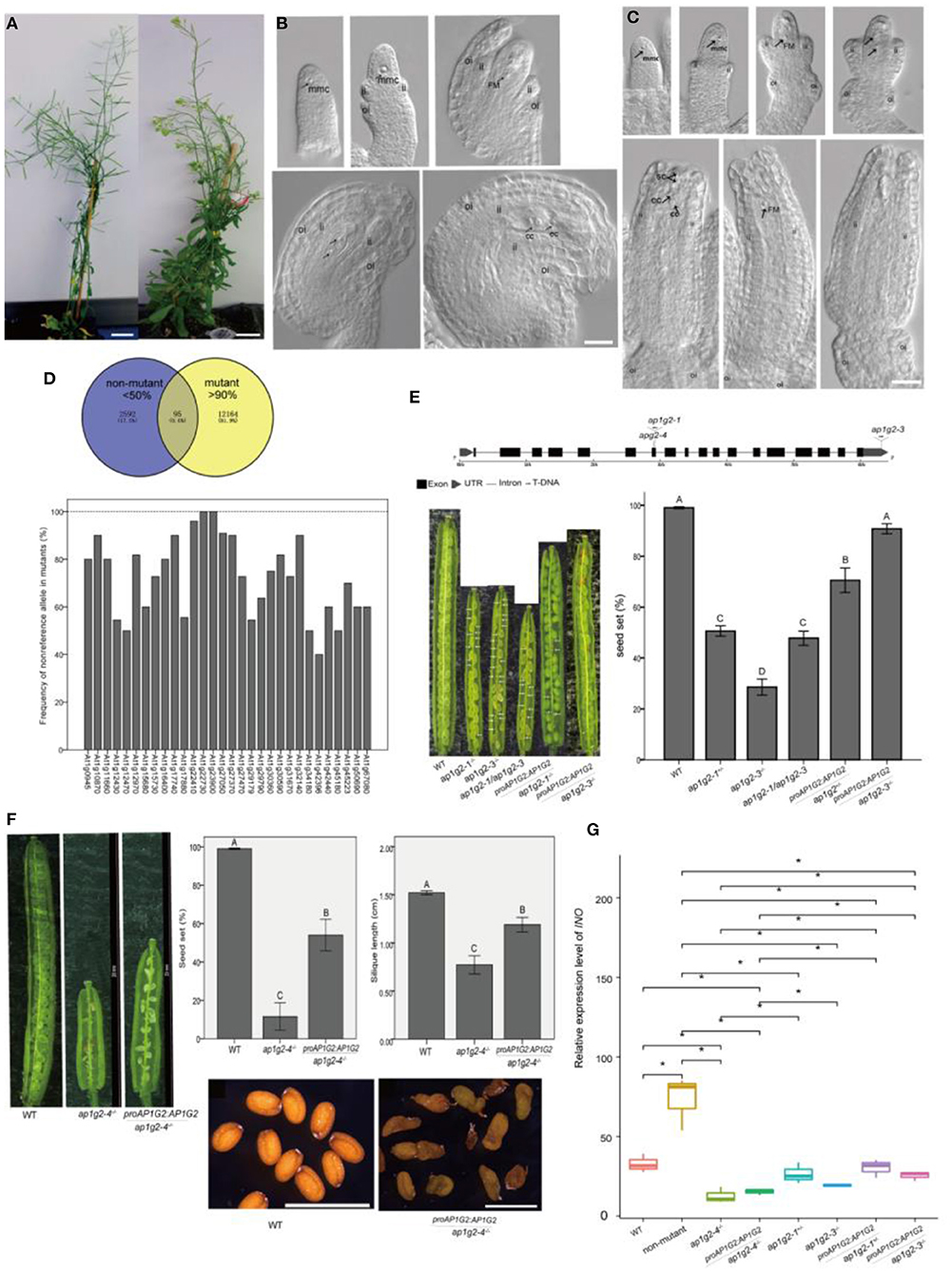
Figure 1. (A–C) Phenotypes of the EMS-induced sterile mutant. (B,C) Ovule development in WT and the EMS-induced sterile mutant. A scale bar = 20 mm in (A). A scale bar = 20 μm in (B,C), ii, inner integuments; oi, outer integuments; mmc, megaspore mother cell; FM, functional megaspore; cc, central cell; ec, egg cell; sc, synergid cells. (D) Ninety-five candidate SNPs with the non-reference allele >90% frequency in the mutant pool and <50% in the non-mutant pool, and frequency of the non-reference allele, which changed amino acid in homozygous mutants of BC2 progeny. (E) Schematic (Hu et al., 2015) representation of the gene AP1G2, with the positions of the various T-DNA insertions (ap1g2-1 and ap1g2-3) and the EMS mutant harboring a G-to-A base change at nucleotide position 8444076 (ap1g2-4). The seed set of ap1g2-1+/−, ap1g2-3−/−, apg2-1/apg2-3 mutants was significantly lower than that of wild-type plants and the genetic complementation lines of ap1g2-1+/− and ap1g2-3−/−. (F) AP1G2 driven by its native promoter could partially complement the infertility phenotype of ap1g2-4, but the seeds of transgenic plants had no heavy seed coat. Error bars indicate standard error. The capital letters above the bars mean very significant difference with the wild type (p < 0.01). (G) Quantitative qRT-PCR analysis of the INO expression. Each result was the average of three independent biological replicates. Error bars indicate standard error. *means significant difference (p < 0.05).
After backcross to wild-type plants, the self-pollinated BC1F2 plants were analyzed. The segregation ratio of wild type to mutants fits the expected 3:1 ratio (chi-square = 0.140, df = 1, p = 0.708). After backcross to the wild type, BC1 individuals were self-pollinated, and DNA of BC1F2 plants of mutants (n = 40) and non-mutants (n = 40) was pooled separately using the whole genome sequence as described by Nordstrom et al. (2013). We compared a causative mutation based on the frequency of the non-reference allele of a single nucleotide polymorphism (SNP) in the mutant and the non-mutant pools. If the non-reference allele of an SNP is the causal mutation, its frequency in the mutant pool should be 100% and about 33% in the non-mutant pool, and the SNPs associated with the causal gene should also display the high frequency of non-reference alleles in the mutant pool (Zhu et al., 2012). We selected 95 candidate SNPs (0.6% of total SNPs) with a frequency higher than 90% and lower than 50% in the mutant and non-mutant pools, respectively (Figure 1D). Among the 95 candidate SNPs, 81.05% of them were on Chromosome I, 0.07% on chromosome IV, 0.06% on Chromosome II, 0.02% on each chromosome III and chromosome V. To better understand the impact of SNPs on Chromosome I, we selected SNPs (30 associated genes) in coding regions that caused non-synonymous mutations or located in UTR (untranslated regions) for further analysis in the backcross BC2 progeny. As recombination events of the SNPs linked to the causal gene, each SNP was confirmed by PCR, and least 12 mutants were separately sequenced in the BC2 progeny. We found that At1g22730 and At1g23900 had 100% frequency of a non-reference allele in mutants, and At1g22410 had a frequency of 96%, making At1g22730, At1g23900, and At1g22410 candidate genes of the sterile mutant (Figure 1D).
To determine which causal gene is associated with the sterile mutant, we studied several mutant lines with T-DNA insertion in each candidate gene. Among the T-DNA insertion lines, only two lines with insertions in AP1G2 (At1g23900) showed the phenotype of a reduced seed set. ap1g2-1 (SALK_032500) had T-DNA insertion in the 7th exon, and heterozygous plants ap1g2-1+/− showed a 51.9% seed set (Figure 1E). For ap1g2-3 (SALK_137129) with T-DNA insertion in 3′ UTR, 56 bp upstream from the poly A tail of the mRNA, we obtained homozygous plants (ap1g2-3−/−), with 23.27% of the seed set (Figure 1E). To understand whether there were other T-DNA insertions, genomic DNA of ap1g2-1+/− was resequenced at a coverage rate of 24-× coverage of its whole genome, and we applied the software MaSuRCA 3.1.3 to assembly contigs (Zimin et al., 2013). Using the boundaries of the inserted fragment to align onto the assembled contigs, only one T-DNA insertion in the 7th exon of AP1G2 was uncovered. Pollen grains from ap1g2-3−/− were used as male parents to pollinate ap1g2-1+/−, by which heteroallelic homozygous mutants ap1g2-1+/−/ap1g2-3+/−were obtained with siliques containing 47.75% normal seeds (Figure 1E).
We carried out a complementation test using native promoter ProAP1G2, driving the wild-type allele. Five out of nineteen independent lines that were heterozygous for ap1g2-1 and carried the transgene showed a significantly higher seed set. For ap1g2-3−/−, twenty-six independent lines were obtained, and eight lines complemented the ap1g2-3−/− phenotype. The seed set of the ap1g2-3−/−mutant carrying the ProAP1G2:AP1G2 constructwas −90.81%, very close to the vaule shown by the WT (99.04%) (Figure 1E). We also complemented the EMS mutant, which partially recovered its severely reduced fertility with an increase of 40% (Figure 1F). The ovules of the transgenic genotype still had no outer integuments, which led to the loss of episperm in seeds. Because INO (INNER NO OUTER) is a key gene regulating the development of the ovule outer integument and the defects of outer integument in ap1g2-4−/− were very similar to those of ino mutants, so we doubted that the expression of this gene were affected in ap1g2-4−/− (Meister et al., 2004; Sieber et al., 2004). The INO expression level was tested in the carpels from the wild-type plants, ap1g2 mutants, and complementation lines, which showed INO was only regulated in ap1g2-4−/− and ap1g2-4−/−/ProAP1G2:AP1G2 (Figure 1G). These results revealed that the absence of the outer ovule integument was not caused by the mutation in AP1G2 of this EMS-induced mutant.
To understand at which stage of the megagametophyte development might be affected in the ap1g2 mutants, we observed the ovules from WT, ap1g2-1+/−, and ap1g2-3−/− at different stages of development. The results showed that these plants were all able to produce functional megaspore cells (Figures 2A,F). Thereafter at the FG3 stage, wild-type plants contained a two-nucleate embryo sac, and continued to develop, producing four-nucleate embryo sacs and then mature embryo sacs (Figures 2B–E). About 50% of the ovules from ap1g2-1+/− were arrested at Stage FG1, with the functional megaspore persisting or degenerating during development (Figures 2G–J). Ovules were observed (n = 989) in the ap1g2-1+/− mutant, and the aborted ovules accounted for half of the total number (1:1, chi-square = 0.171, p = 0.679). While, in plants ap1g2-3−/−, most of the ovules contained a single cell in the nucellus or degenerated gradually (Figures 2G–J).
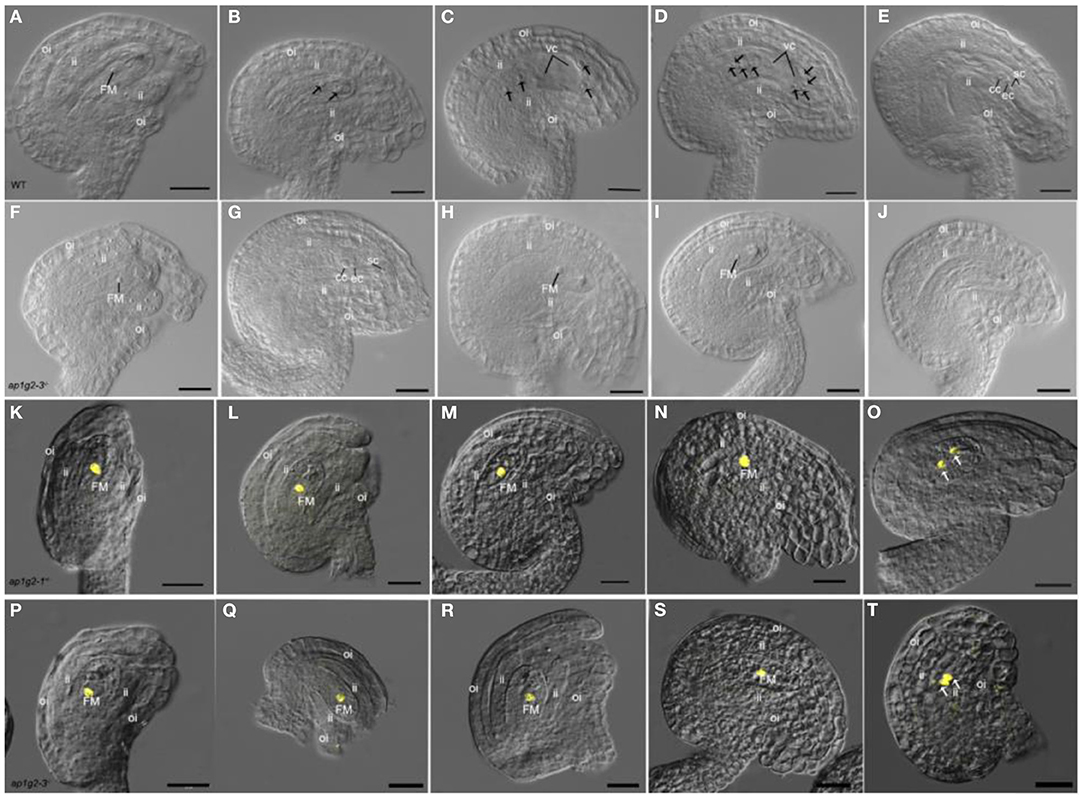
Figure 2. Female gametogenesis is impaired in ovules from ap1g2-1+/− and ap1g2-3−/− plants. (A–E) Ovule development in the wild-type, Stage FG1 (A); Stage FG3 (B); Stage FG4 (C); Stage FG5 (D); Stage FG7 (E). (F–J) Ovules in ap1g2-3−/−, a normal ovule at Stage FG1 (F); a mautrue ovule at FG7 aborted ovules arrested at FG1 (H,I); (J) Aborted ovules without functional megaspore degenerated. (K–O) Analysis of H2B-YFP expression under the control of the AKV promoter in ap1g2-1+/−. (L–N) Aborted ovules arrested at FG1 in ap1g2-1+/−. (O) YFP signals in the wild-type ovule at the FG2 stage. (P–T) Analysis of H2B-YFP expression under the control of the AKV promoter in ap1g2-3−/−. (Q–S) Aborted ovules arrested at FG1 in ap1g2-1+/−. (T) The wild-type ovule at the FG2 stage. A scale bar = 20 μm, ii, inner integuments; oi, outer integuments; FM, functional megaspore; cc, central cell; ec, egg cell; sc, synergid cells.
The ANTI-KEVORKIAN (AKV) cell-identity marker during megagametogenesis was used to confirm that the identity of the cell in the abortive ovules was the functional megaspore (Schmidt et al., 2011). The promoter pAKV is a gametophyte-specific promoter; therefore, the pAKV:H2B-YFP marker was specifically expressed in the nuclei of the functional megaspore and the developing gametophyte before cellularization (Su et al., 2017). We crossed ap1g2-1+/− and ap1g2-3−/− plants with pAKV:H2B-YFP marker lines and observed the ovules from the F1 plants with ap1g2-1+/− allele and F2 plants with ap1g2-3−/− allele. The YFP was expressed in the nuclei of the ovules at FG1 and wild-type ovules of ap1g2-1+/− and ap1g2-3−/−,(Figures 2K,O,P,T) as well as the aborted ovules arrested at the mononucleate stage (Figures 2L–N,Q–S).
The female gametophyte development within a pistil is generally synchronous, with a relatively narrow range of variation in WT (Christensen et al., 1997; Shi et al., 2005). To investigate the developmental synchrony of female gametophytes in the pistils of ap1g2 mutants, we emasculated flowers at the Stage 12 of development (Christensen et al., 1998), and, after 48–72 h, we fixed pistils from flowers of the wild type and mutants at different developmental stages. Compared with wild-type pistils, the developmental synchrony of female gametophytes in ap1g2 was disturbed (Table 1, Supplementary Table 3). In ap1g2-1+/− pistils, about half of the female gametophytes in each mutant pistil (P9–P14) either persisted at FG1 or degraded. While, in the ap1g2-3−/−, around 77.75% of all female gametophytes failed to undergo nuclear division (P9–P14).
The development of the pollen grain begins with the expansion of the microspore (Figures 3A,B) and a large vacuole produced, companied by the migration of the microspore nuclear to a peripheral position against the cell wall. The microspore then undergoes a first asymmetric cell division (PMI)which results in a bicellular pollen gains (Figure 3C). Afterward, the smaller germ cell continues through a further round of mitosis (PMII), to produce twin sperm cells (Figure 3D). To analyze whether ap1g2 led to pollen abortion, the viability of pollen grains was tested using Alexander Red stain. About 46.04% (n = 2,096) non-viable pollen was detected in mature anthers in ap1g2-1+/− plants (Figures 3E–H), and 49.71% non-viable pollen was obtained for ap1g2-3−/− (n = 1,750). However, the transgenic ap1g2-1+/−, carrying proAP1G2-AP1G2-GUS, had a 71.22% (n = 300) viability rate, and the transgenic ap1g2-3−/− plants had a similar-level viability rate as the wild type (98.70%, n = 230), resulting in 1.64% (n = 600) aborted pollen.
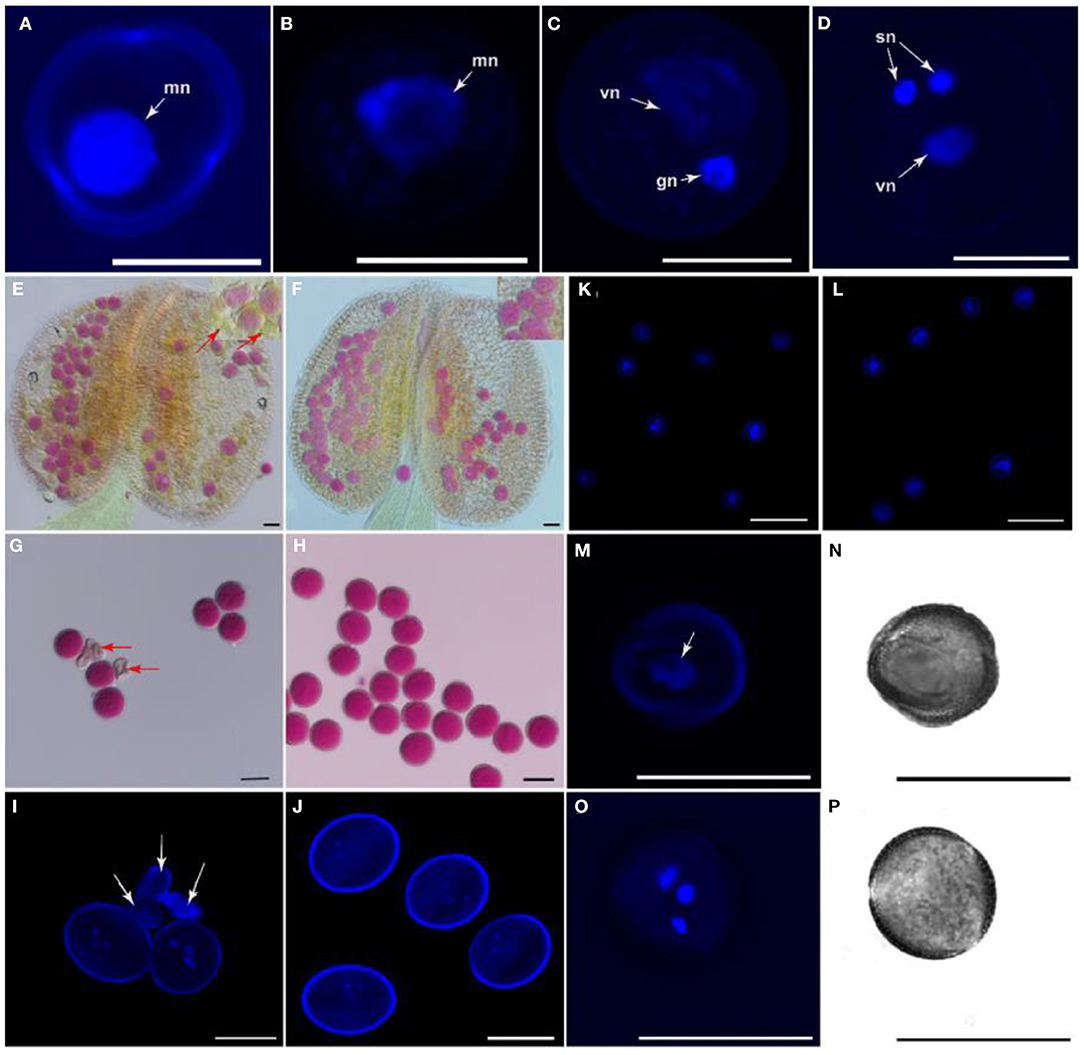
Figure 3. Morphology and viability of apg2-1+/− mutant pollen grains. (A–D) The nuclear constitution of WT pollen grains through microgametogenesis developmental stages. Nuclei are indicated by arrows; mn, microspore nucleus; vn, vegetative nucleus; gn, generative nucleus; sn, sperm neclei. Bars = 10 μm. (E,F) Pollen viability by means of Alexander's staining in anthers from WT and ap1g2-1+/−, respectively. Arrows indicate impaired pollen grains. (G,H) Alexander's staining of mature pollen grains from WT and ap1g2-1+/−, respectively. Arrows indicate impaired pollen grains. (I,J) 40,6-Diamidino-2-phenylindole (DAPI) staining of pollen grains from buds at Stage 13 in WT and ap1g2-1+/−, respectively. Arrows indicate impaired pollen grains. (K,L) DAPI staining of microspores from buds at Stage 9 in WT and ap1g2-1+/−, respectively. (M,O) Nuclear constitution by means of DAPI staining in pollen grains in buds at Stage 12 from WT and ap1g2-1+/−, respectively. Arrows indicate nuclei. (N,P) Bright field images of (M,O), respectively. Bars = 20 μm in (E–P).
We used 4′,6-diamidino-2-phenylindole (DAPI) staining to trace the pollen development in wild-type plants and ap1g2-1+/−, which was a single T-DNA insertion mutant. The normal mature pollen grains from wild type and ap1g2-1+/− contained a vegetative nucleus and two generative nuclei (Figures 3I,J), while about half of the pollen grains from ap1g2-1+/− did not possess a nuclear fluorescence signal as the pollen was abnormal and shriveled (Figure 3J). Although at the microspore stage, pollen grains in both WT and ap1g2-1+/− showed normal single nucleus fluorescence (Figures 3K,L), nearly half of the microspores of ap1g2-1+/−did not undergo nuclear polarization before pollen mitosis but still showed unicellular and shriveled microspores (Figures 3O,P) when tricellular pollen grains had formed in the wild type (Figures 3M,N).
About half the pollen grains from ap1g2-3−/− were shown to be wrinkled shaped under scanning electron microscopy (SEM) (Supplementary Figures 2B,D), in contrast to those of wild type (Supplementary Figures 1A,C). Solid pollen germination mediums (Rodriguez Enriquez et al., 2013) were used to culture the pollen grains from WT and ap1g2-3−/−, and we obtained 72.22% (n = 180) and 30% (n = 180) germination, respectively (Supplementary Figures 1E,F).
The qRT-PCR analysis revealed that AP1G2 expression was found in a variety of tissues, including roots, leaves, stems, and flowers, where flowers revealed the highest relative expression, followed by stems and leaves (Figure 4A). For ap1g2-3−/− mutants with the T-DNA insertion in 3′ UTR, the expression levels were significantly downregulated as compared with the wild type using t-test (p < 0.01).
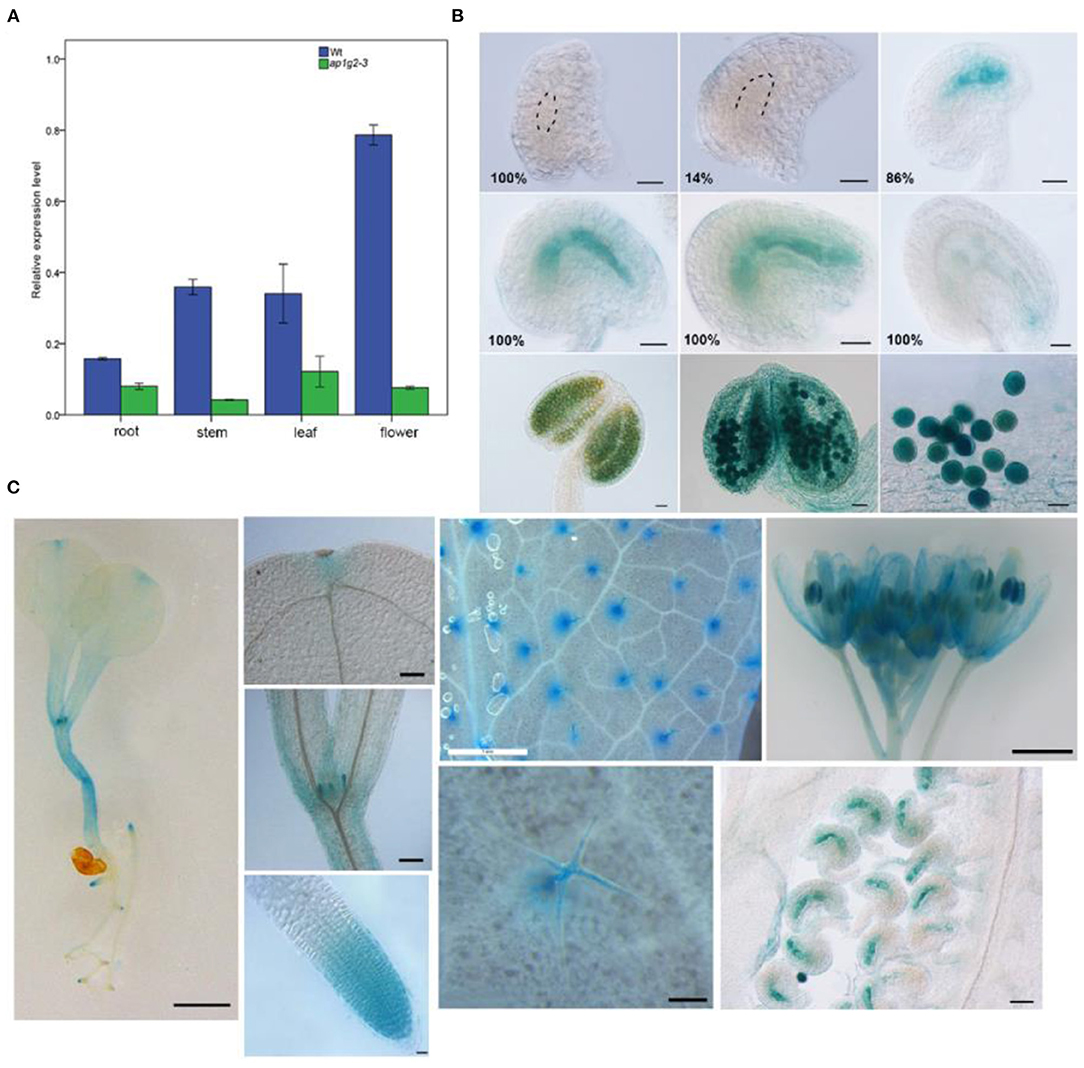
Figure 4. (A) Quantitative qRT-PCR analysis of the AP1G2 gene expression. Each result was the average of three independent biological replicates. Error bars indicate standard error. (B) Activity of AP1G2 in ovules and anthers. One hundred percent of ovules containing female gamete at Stage FG1 and 14% of ovules containing a female gametophyte at FG3 were not detected GUS expression. One hundred percent ovules expressed GUS strongly at FG4 to FG7 until the process of embryogenesis. GUS staining in anthers and pollen grains. Bars = 20 μm. (C) ProAP1G2:GUS expression in the seedlings, leaves, and flowers. Bar = 1 cm in the pictures of 8-day-old seeding, leaves, and inflorescence. Bar = 20 μm in in the pictures of leaf primordial, apical meristem, trichomes, and ovules.
Spatial and temporal GUS expression profiles of transgenic AP1G2 plants were observed. Twenty-three independent lines of the T2 generation were analyzed, of which 5 showed GUS expression in the female gametophyte, and GUS expression was detected after the big vacuole formed and remained until embryogenesis began; in later stages, GUS expression was reduced (Figure 4B), while the same pattern was observed in anthers. ProAP1G2:GUS was specifically expressed in the male gametophyte at maturation in all independent lines (Figure 4B). Additionally, ProAP1G2:GUS was expressed at the 8–10-day seedling stage, and expression was also detected in hypocotyle, leaves, leaf primordia, shoot apical meristem, flowers, anthers, filament, and pedicles. GUS expression was also observed in root tips, while strong GUS staining was noted in trichomes (Figure 4C).
To recover the mechanism that how AP1G2 mediated the process of female gametogenesis, we collected the ovules from the wild type and the three ap1g2 mutants at different stages for RNA-Seq analysis. Expression profiles were used for principal component analysis (PCA), which revealed that ap1g2-3−/− expressed at MMC and FG3 stages, which were significantly different from that of the wild type (Supplementary Figure 2A). For differentially expressed genes (DEGs), the number of upregulated DEGs of ap1g2-1+/− and ap1g2-1+/−/ap1g2-3+/− peaked at MMC stages and decreased significantly at FG1 and FG3 stages when the downregulated DEGs increased. The number of downregulated DEGs of ap1g2-3−/− at FG1 and FG7 stages also increased compared with the MMC stage (Supplementary Figure 2B). The total number of the DEGs at each stage of ap1g2-1+/−, ap1g2-1+/−/ap1g2-3+/−, and ap1g2-3−/− was 648, 636, and 837, respectively. Among them, there were 164 common DEGs at all phases of the three mutant lines (Supplementary Figure 2C).
To find out whether the DEGs identified belong to a particular mutant or are common in two or three mutants, all DEGs of the three mutant lines at each stage were divided into seven groups (Figure 10). For example, PHOSPHATIDYLINOSITOL BINDING CLATHRIN ASSEMBLY PROTEIN 5A (PICALM5A) was significantly upregulated in both ap1g2-1+/− and ap1g2-1+/−/ap1g2-3+/− at the MMC stage, and PICALM5B was upregulated in all ap1g2-1 at the MMC stage (Figure 5A).
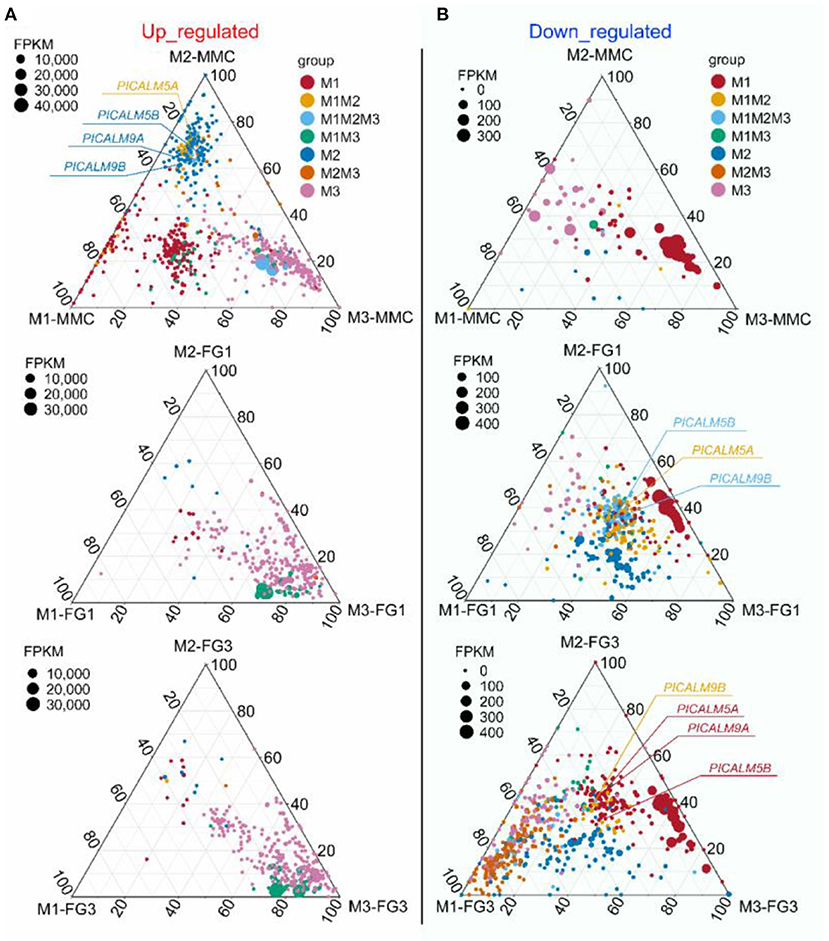
Figure 5. A ternary plot of all DEGs from ap1g2-1+/− (M1), ap1g2-1/ap1g2-3 (M2), and ap1g2-3−/−(M3) at MMC, FG1, and FG3 stages. (A) The upregulated DEGs of the three ap1g2 mutants at each stage; (B) The downregulated DEGs of the three ap1g2 mutants at each stage. Each circle represented one DEG. The size of each circle represented one DEG's FPKM. The position of each circle was determined by the contribution of the indicated compartments to the total FPKM. The dotted grid and numbers inside the plot indicated 20% increments of contribution from each gene. Red circles denoted the specific DEGs of ap1g2-1+/−. Yellow circles denoted the overlapping DEGs of ap1g2-1+/− and ap1g2-1+/−/ap1g2-3+/−. Light blue circles denoted the overlapping DEGs of ap1g2-1+/− and ap1g2-1+/−/ap1g2-3+/− and ap1g2-3−/−. Green circles represented the overlapping DEGs of ap1g2-1+/− and ap1g2-3−/−. Dark blue circles represented the specific DEGs of ap1g2-1+/−/ap1g2-3+/−. Orange circles represented the overlapping DEGs of ap1g2-1+/−/ap1g2-3+/− and ap1g2-3−/−, and pink circles represented the specific DEGs of ap1g2-3−/−.
From the MMC to the FG3 stage, there were 40, 66, and 21 common DEGs among the three mutant lines, respectively. At the FG1 stage, 66 DEGs were all downregulated in three mutant lines (Figure 5B). GO enrichment analysis revealed that downregulated DEGs of ap1g2-1+/− were enriched on pollination, cell wall organization, calcium-mediated signaling, and pectinesterase activity, calcium-dependent protein serine/threonine kinase activity, endocytosis, pectinesterase inhibitor activity, membrane, clathrin-coated vesicles, and vacuoles. These proteins, together with the proteins containing the domain of invertase/pectin methylesterase inhibitor, were used for STRING analysis (Figure 6A). Among the DEGs, PICLAM9B, and PICLAM5B were upregulated at the MMC stage but downregulated at the following stage when the development of partial ovules was arrested. The results showed that AP1G2, PICALM5A/B, and PICALM9A/B were located on the membrane and clathin-coated vesicles. And the four PICALM proteins, which have been reported to be involved in endocytosis, were predicted to interact with AP1G2 because the γ-adaptins and PICALM6 were found interacting in Musmusculus (Wang et al., 1995).
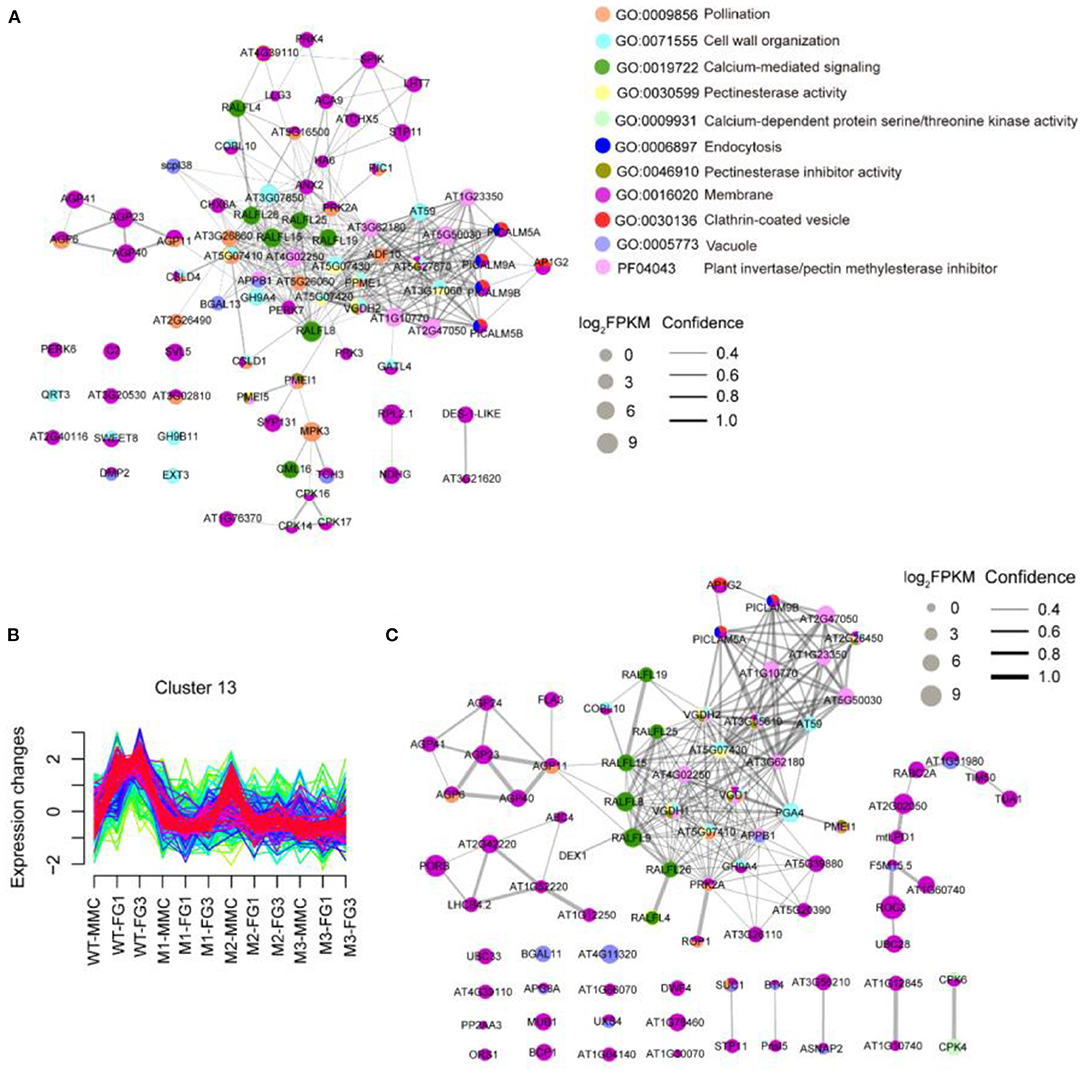
Figure 6. STRING protein-protein interaction network. (A) The STRING analysis downregulated DEGs of ap1g2-1+/−. (B) Cluster 13 showed an increase of the expression level at FG1 in wild-type ovules but repressed in the ap1g2 mutants at various stages. The y axis represented the standardized FPKM value of genes, and the x axis represented the different samples. (C) Genes of Cluster 13 that were enriched on the above GO terms in (A) were used for STRING analysis. Colored nodes from (A,C) indicated the proteins enriched on different GO terms. The size of each node indicated the normalized FPKM value. Lines between nodes represented both functional and physical protein associations. The line thickness indicated the confidence of association.
We also defined 20 gene clusters by Fuzzy c-means clustering (Supplementary Figure 3) using the expression profiles to show the expression trends of different genes at developing stages, in which Cluster 13 containing 270 genes showed decreased expression profiles of ap1g2 mutants at MMC and FG1 stages (Figure 6B). Eighty-three genes from Cluster 13 that were enriched on the above-mentioned GO terms or contained the domain of the invertase/pectin methylesterase inhibitor were selected for a STRING protein network map (Figure 6C). AP1G2 showed direct interactions with PICALM5A and PICALM9B (located in the periphery of the network diagram). Whereas, the phosphatidylinositol binding clathrin assembly proteins were predicted to have associations with those proteins that were putative pectin methylesterase/invertase inhibitors like AT2G47050, AT1G23350, AT1G10770, or other proteins involved in pectinesterase activity and cell wall organization, including AT5G07430 and VANGUARD 1 HOMOLOG 2 (VGDH2) (Figures 6A,C).
Some RALFL (Rapid Alkalinizationfactor-like) peptides, which induce a rapid increase in cytoplasmic calcium (Haruta et al., 2008; Somoza, 2021), also occurred in both network maps Calmodulin Like 16 (CML16), and calcium-dependent protein kinase genes, including CPK14, CPK16, and CPK17, which are calcium-binding genes, were downregulated in the ap1g2-1+/− mutant. In both network diagrams, members of the AGP (arabinogalactan protein) family appeared to be in connection with RALFL peptides (Figure 6).
PICALM5A/B and PICALM9A/B belong to the ANTH family. In the wild type, the four ANTH genes were barely expressed at the MMC stage in the wild type, but their expression levels dramatically increased at the FG1 stage (Figure 7), while PICALM5A and PICALM9A/B were upregulated at the MMC stage of ap1g2 mutants. PICALM5B and PICALM9B were downregulated in all mutants at Stage FG1, and PICALM5A was also downregulated in ap1g2-1+/− (Figures 6, 7A). At the FG7 stage of ap1g2-1+/−, the four genes showed lower expression than those of the wild type. Given the four genes may act redundantly, the total expression level of PICALM5A/B and PICALM9A/B at each stage of wild type; ap1g2 mutants were compared, which showed that the total expression level was significantly reduced at FG1 and FG7 stages in ap1g2 mutants. And qRT-PCR analysis also revealed that PICALM5A/B and PICALM9A/B were downregulated at developing ovules in ap1g2 (Supplementary Figure 4). In the ANTH family, only PICALM4A and PICALM9C have two ANTH domains, while the other members contained one. Among the ANTH family members, PICALM5A/B, PICALM1A/B, and PICALM2A have an ENTH domain, which is also a part of their ANTH domains. The genes of the ANTH family showed three expression patterns during the development of ovules in the wild type. PICALM4A, PICALM3, and PICALM1A/B maintained consistent expressions that might play very basic functions. Whereas members, including PICALM5A/B, PICALM9A/B, and PICALM10C, seemed to be involved in the megagametogenesis and pollination, since they showed low expressions before the FG1 stage, but kept relatively higher expression levels during the mitosis process of the female gametophyte and peaked at FG7 when the female gametophytes matured and were ready for double fertilization (Figure 7C). While PICALM4B, PICALM6, PICALM9C/D, PICALM2A/B, and PICALM10A/B showed extremely low transcriptional activity throughout the ovule development.
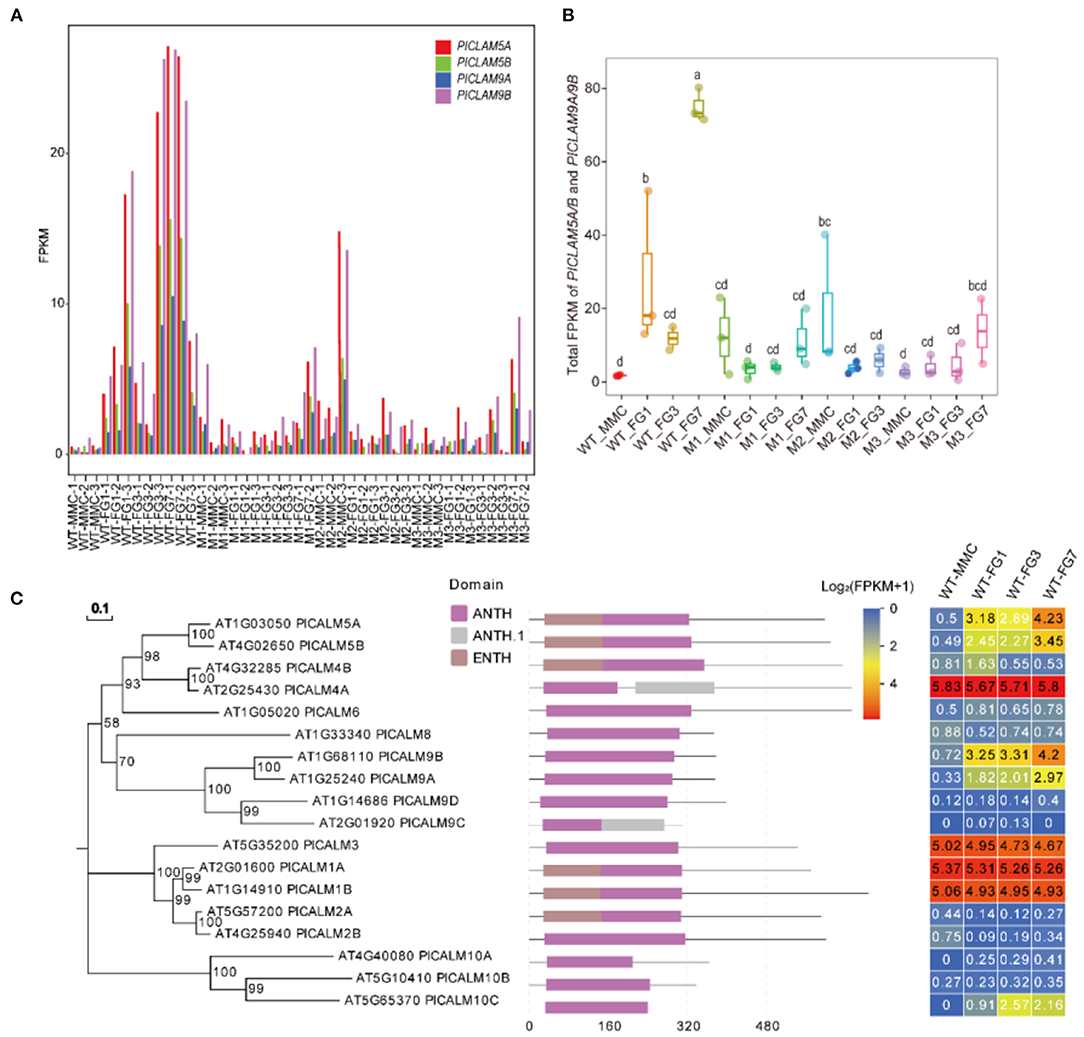
Figure 7. (A) FPKM value of PICALM5A/B and PICALM9A/B at each stage in WT, M1, M2, and M3. (B) The total FPKM values of the four PICALMs at each stage in WT, M1, M2, and M3. The lowercase letters above the bars mean significant difference (p < 0.05). (C) The expression profiles of each member of the ANTH family. Bootstraps (based on 1,000 replications) of the un-rooted phylogenetic tree are indicated at each node (bar = 0.1 amino acid substitutions per site).
To confirm the interactions between AP1G2 and the four candidate targets, we used split-ubiquitin yeast two-hybrid assays and a Bimolecular Fluorescence Complementation (BiFC) assay, which turned out that AP1G2 interacted with PICALM5A/B and PICALM9A/B in yeast and tobacco (Figures 8A–F). In tobacco leaf epidermal cells, we found the AP1G2, PICALM5A/B, and PICALM9A/B mainly localized and interact at the cellular plasma membrane and vesicles close to the plasma membrane (Figures 8C–F).
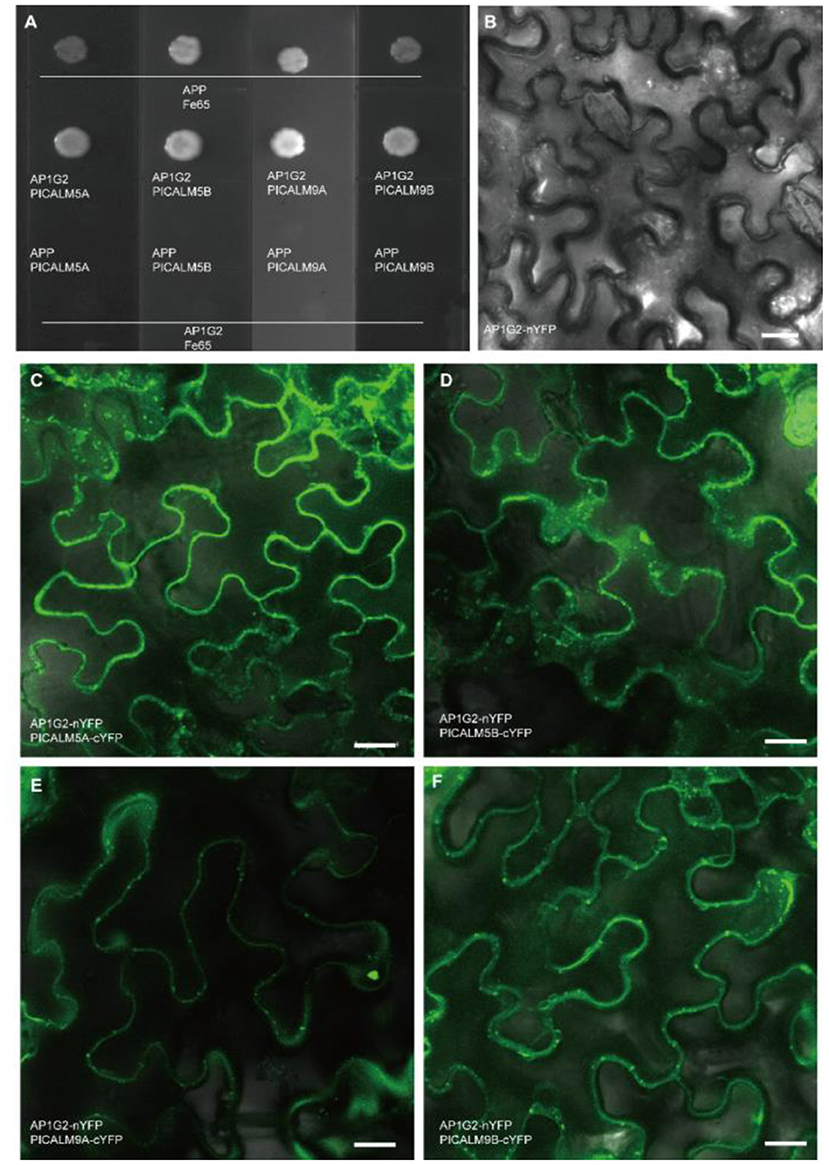
Figure 8. (A) The protein-protein interactions of AP1G2 and PICALM5A/B and PICALM9A/B in yeast. Vectors pTSU2-APP and pNubG-Fe65 were positive control bait and prey, respectively. (B–F) BiFC analysis of AP1G2 and PICALM6A/B and PICALM9A/B in tobacco leaf epidermis. (B) is a negative control. Confocal images showing YFP fluorescence indicated interaction.
The expression profiles of RNA-Seq data showed that calcium-dependent protein kinases, i.e., CPK14, CPK17, and CPK36, were downregulated in ap1g2 mutants, and several RALFL peptides were also downregulated and grouped in Cluster 13 (Figures 6A,C); we examined calcium concentrations in the ovules and microspores of the wild type and ap1g2−1+/−. We used a calcium ion detection kit to observe the calcium dynamic changes during female gametogenesis in the wild type and ap1g2−1+/−, which adopted a fluorescent probe to detect the change of calcium ion concentration. The fluorescence signal was detected at the degenerated micropylar-most megaspores of ovules in both wild type (Figures 9A,B) and ap1g2-1+/− (Figures 9C,D). At later development stages, the wild-type embryo sacs showed a higher concentration of calcium ion than that of the aborted embryo sacs of ap1g2-1+/− (Figures 9E–O).
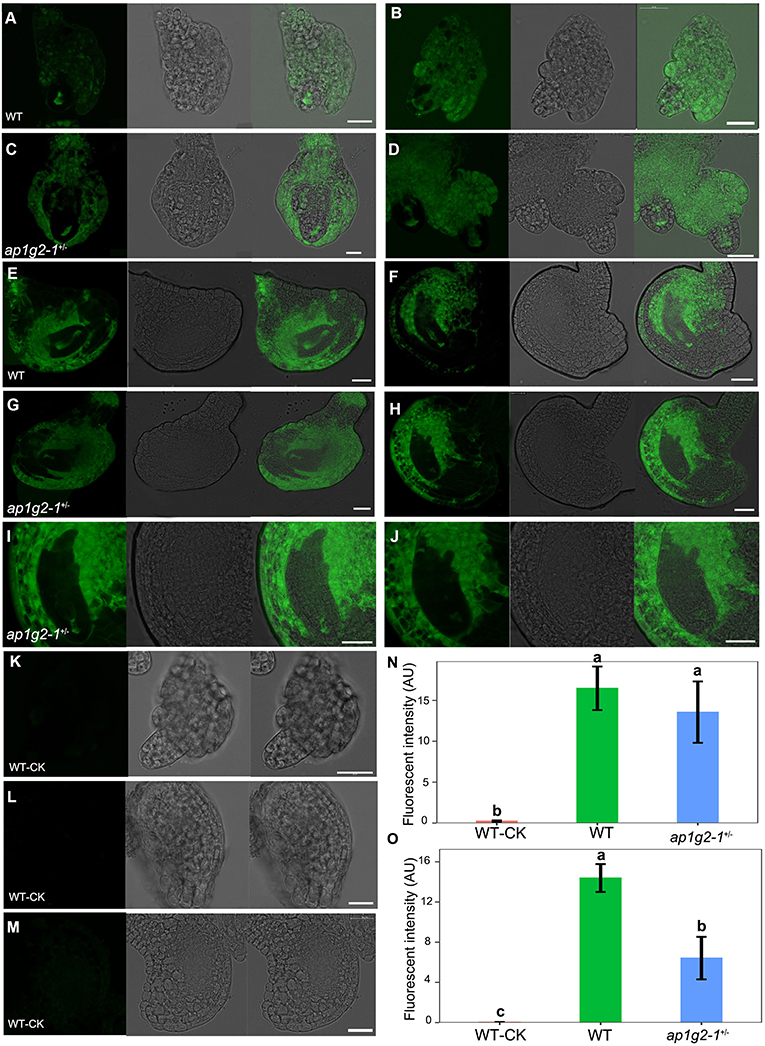
Figure 9. Calcium dynamics during female gametogenesis in WT and ap1g2-1+/−. (A,B) and (C,D) ovules at FG1 from WT and ap1g2-1+/− dyed by an intracellular calcium ion concentration fluorescent probe. (E,F) ovules around at FG3 and FG4 from WT, respectively. (G,H) Aborted embryo sacs from ap1g2-1+/−. (I,J) were magnification of (E,F). (K–M) were the ovules from the wild type without dyeing. (N) Quantification of the Ca2+ level in ovules of WT and ap1g2-1+/− at the FG1 stage. (O) Quantification of the Ca2+ level in ovules at later stages from WT and aborted ovules of ap1g2-1+/−. Bar = 20 um.
We also treated pistils and anthers at different stages from the wild type and ap1g2-1+/− with potassium pyroantimonate to localize loosely bound calcium, and observed the calcium precipitate through a transmission electron microscope (TEM). In the wild-type embryo sacs, there were many calcium precipitates that were distributed (Figures 10A,D,G,I, Supplementary Figure 4A), whereas ap1g2-1+/− abnormal embryo sacs contained fewer and smaller calcium precipitates (Figures 10C,E,F,H, Supplementary Figure 4A).
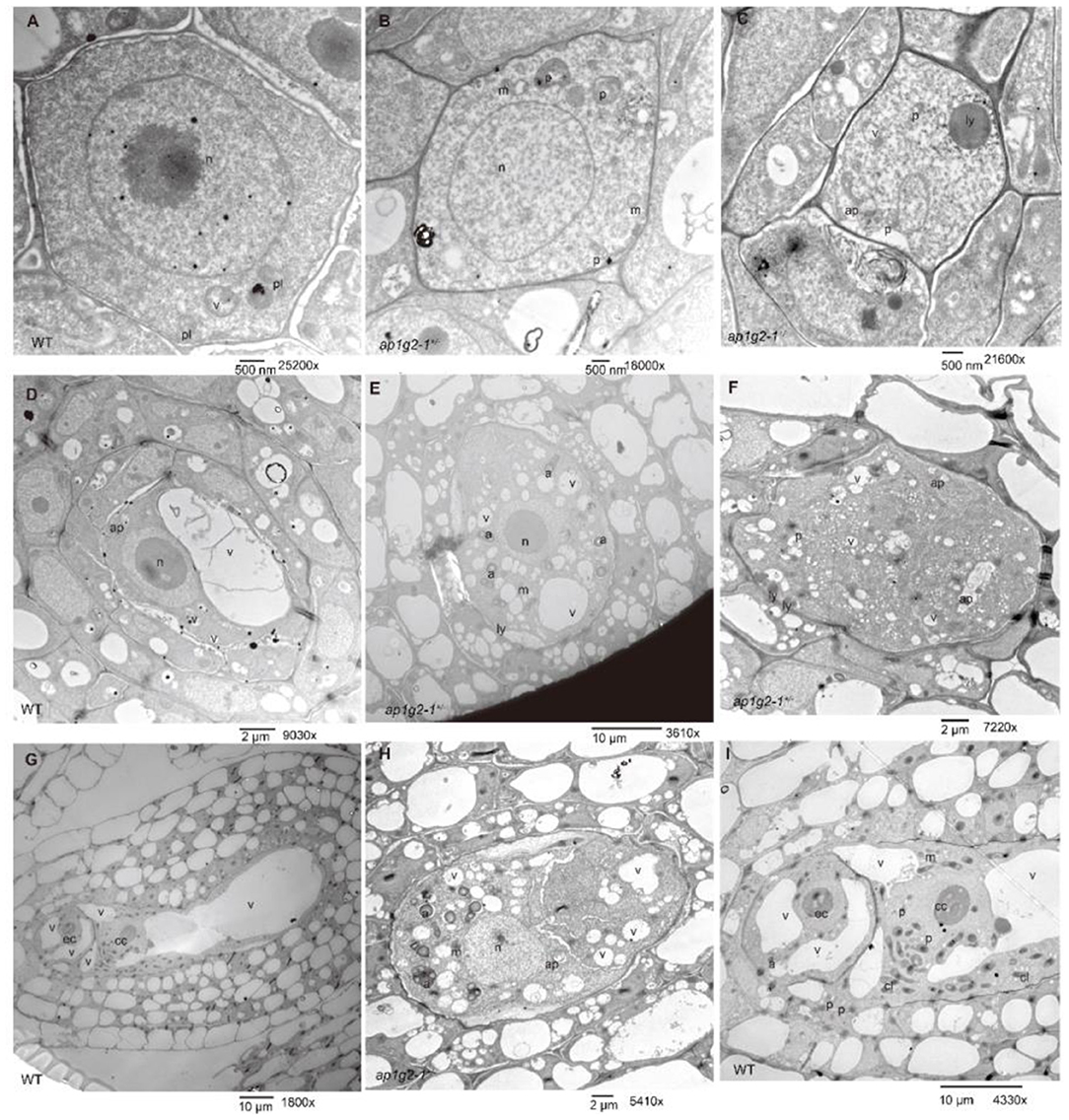
Figure 10. TEM micrographs of the pyroantimonate-labeled ovules from the wild type and ap1g2-1+/−. (A,D,G) Showed the ovules of the wild type at MMC, FG3, and FG7, respectively. (B,C) showed the ovules at the MMC stage from ap1g2-1+/−. (E,F,H) showed the aborted ovules from ap1g2-1+/−, while (I) was a magnification of (G). n, nucleus; p, proplastids; v,vacuoles; m, mitochondria; ap, autophagosomes; a, amyloplasts; ly, lysosomes; cl, chloroplasts; ec, egg cell; cc, central cell. The black dots were calcium precipitates.
Additionally, we found the aborted embryo sacs at FG1 that could not form a large central vacuole, which polarizes the two daughter nuclei in a wild-type embryo sac with a lot of randomly distributed small vacuoles. There were large amounts of amyloplasts that appeared in the abortive embryo sacs but very few in the wild-type ones (Figure 10).
To investigate the distribution of calcium precipitates in the abnormal developing microspores of ap1g2, anthers at different stages from the wild type and ap1g2-1+/− were treated with potassium pyroantimonate with non-treated wild-type anthers as control. Through TEM, very few black dots were observed in the control wild-type pollen grains (Figures 11A,E,I,M, Supplementary Figure 4B). A number of calcium precipitates appeared in the cytoplasm of well-developing microspores from the wild type (Figures 11B,F,J,N, Supplementary Figure 4A) and ap1g2-1+/− (Figures 11K,O,P), while, in the aborted microspores of ap1g2-1+/−, we observed few calcium precipitates (Figures 11C,D,G, Supplementary Figure 4A).
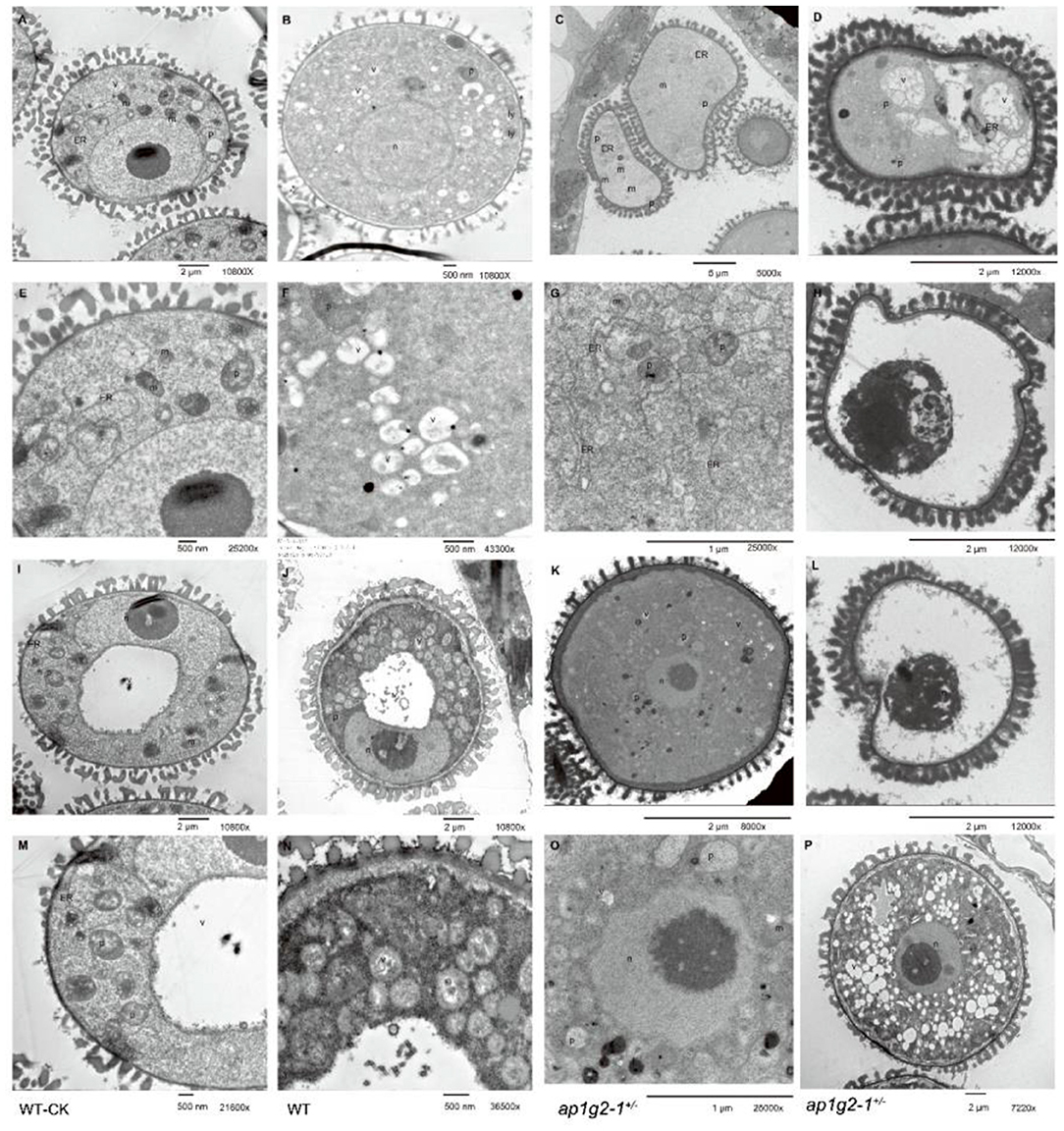
Figure 11. TEM of pyroantimonate-labeled microspores from the wild type and ap1g2-1+/−. (A,I) The microspores before pollen mitosis I from the wild type without potassium pyroantimonate treatment. (E,M) The magnifications of (A,I). (B,F,J) The microspores before PMI from the wild type with potassium pyroantimonate treatment. (N) The magnifications of (J). (C,D,G,H,L) Abnormal microspores failed to undergo pollen mitosis I from ap1g2-1+/−. (K,O,P) were normal biocellular pollens from ap1g2-1+/−. (G,O) were the magnifications of (C,K). n, nucleus; ER, endoplasmic reticulum; p, proplastids; v, vacuoles; m, mitochondria; ap, autophagosomes; a, amyloplasts; ly, lysosome.
The ultrastructures of the microspores from the wild type and ap1g2−1+/− also confirmed that the infertile pollen grains were arrested before PMI (pollen mitosis I). After its microspores were released from tetrad, about half the pollen grains had abnormal shapes and failed to form a large vacuole (Figure 11D). In these sterile microspores, there was numerous endoplasmic reticulum (ER) distributed in the cytoplasm (Figure 11G), and then some abnormal vacuoles occurred inside the circles formed by ER (Figure 11D). The abnormal vacuolization led to the degradation of the cytoplasm compartments (Figures 11H,L).
In angiosperm, the gametophytes consist of fewer cells and are embedded within the sexual organs. The female gametophyte develops inside the ovule, while the male gametophyte, also called the pollen, develops in the anthers. Out of the −26,000 genes recognized in Arabidopsis, only a few thousand have been functionally defined (Bouche and Bouchez, 2001), while more than 30% of the expected Arabidopsis genes have not been studied with confidence. Earlier, various studies have been conducted related to gametophyte development. Here, we report the AP1G2 gene and its essential role in controlling female and male gametophyte development.
AP1G2 encodes one of the largest subunits of a heterotetrameric protein complex, which sorts proteins at the trans-Golgi network and endosomes (Park et al., 2013). The AP1G subunit is encoded by two homologs, AP1G1 and AP1G2, which are both constitutively expressed in male and female reproductive cells (Wang et al., 2017). We obtained two lines, ap1g2-1+/− and ap1g2-3−/−, which showed partial functional megaspore and microspores failed to undergo the first mitosis. A single T-DNA insertion in exon 7 of ap1g2-1+/− showed a nearly 50% seed set and no homozygous plants obtained in the self-pollinated progeny. While ap1g2-3 mutant with insertion in 3′ UTR (56 bp upstream from the poly A tail of mRNA), only the homozygous plants showed defective phenotypes in female and male gametophyte development, which were more severe than those of ap1g2-1+/−. The defects of ap1g2-3 were relatively weak compared with the ap1g2-1 mutant line, which was likely due to its rearward mutation position or other insertion mutations. The reason of caused-impaired function of AP1G2 might be that some miRNAs can decrease gene expression of mRNAs by binding to specific sites within the 3′ UTR, which leads to either inhibiting translation or directly causing degradation of the transcript (Barrett et al., 2012; Pichon et al., 2012). In this work, we found that insertion mutation in 3′ UTR of AP1G2 decreased its expression level in various organs. This result supports that the regulatory regions within the 3′ UTR could influence translation efficiency of the mRNA. The EMS mutant had additional integument defects, which were more likely caused by other mutations, which downregulated INO, the key regulatory gene of the outer integument because the two T-DNA mutants did not show the phynotype and decrease of INO expression. Moreover, the transgenic plants of ap1g2-4 carrying wild-type AP1G2 driven by the native promoter still lacked the outer integuments.
Female gametophyte mutations fall into two classes, including the female gametophyte-specific class and the general female gametophytic class (Drews et al., 1998). ap1g2 mutants belong to the second class, including male gametophyte developmental defects too. The ultrastructure of female gametophytes and male gametophytes from the wild type and ap1g2-1+/− showed the failure of some fundamental cellular processes of mutant gametophytes, such as mitosis, vacuole formation, cell expansion, and subcellular migration. ap1g2-impaired microspores were found without the formation of a large vacuole and an asymmetric cell division, but performing abnormal vacuolization, vacuole collapse, and cytoplasmic degradation. In the ap1g2-defect embryo sacs, neither the first mitosis nor the formation of a big vacuole that characterizes the FG3 stage was observed. These results suggested that ap1g2 mutations were responsible for the failure of mitosis and the normal large vacuole formation in both female and male gametophytes. The phenotypes of ap1g2 had a resemblance with those of the insertion lines in VACUOLELESS GAMETOPHYTES (VLG) as a DC1 domain-containing protein present in the endomembrane system (D'ippolito et al., 2017). VLG localized to plant prevacuolar compartments (PVCs) or multivesicular bodies (MVBs), which mediated protein trafficking to vacuoles in the secretory pathway and were also considered late endosomes in the endocytic pathways. The cytosolic adaptor protein-1 complex (AP-1) that was found localized on the trans-Golgi network (TGN)/endosomal membranes also played an essential role in protein trafficking between the TGN and endosomes by specific sorting signals (Bonnemaison et al., 2014; Wang et al., 2014, 2017). They suggested that the post-Golgi traffic pathway is crucial for gametophyte development.
Transcriptome analysis was carried out using the ovules at various stages from the wild type, ap1g2-1+/−, ap1g2-3−/−, and heterogenous homozygous mutant ap1g2-1+/−/ap1g2-3+/−. We used STRING analysis to predict the associations of the proteins of DEGs (Szklarczyk et al., 2019), which showed AP1G2 was predicted to have direct interaction with PICALM5A/B and PICALM9A/B, which were members of the PICALM (phosphatidylinositol binding clathrin assembly protein) subfamily. PICALM, is a clathrin adaptor protein containing functions in clathrin-coated vesicle formation (Ford et al., 2001; Meyerholz et al., 2005; Chen et al., 2011; De Craene et al., 2012; Muro et al., 2018). The PICALM subfamily has 18 group members (Muro et al., 2018), of which the members showed different expression patterns during the development of ovules in the wild type. PICLAM4A, PICLAM3, and PICLAM1A/B maintained consistent expression, indicating that they may provide basic and consistent functions, whereas members, including PICALM5A/B and PICALM9A/B, together with PICALM10C, seemed to be involved in the megagametogenesis and pollination because they showed a low expression level before the FG1 stage. However, they also maintained relatively higher transcriptional activity during the mitosis process of the female gametophyte and peaked when the matured female gametophytes were ready for the double fertilization. While the others showed extremely low transcriptional activity throughout the ovule development. These data demonstrated that PICALM proteins played roles during development by job division and coordination, and several members might act redundantly. It has been reported that via clathrin-mediated endocytosis, PICALM5A/B could recycle ANXUR kinases for the pollen tube integrity (Muro et al., 2018), and PICALM9B (EAP1) could antagonize REN4 by directly targeting and removing plasma membrane-localized REN4 (ROP1 Enhancer 4, AT2G26490) in pollen tubes (Li et al., 2018). The protein-protein interaction assays both in vitro and in vivo determined the four PICALM proteins directly interacted with AP1G2. Moreover, AP1G2 and the four PICALM proteins were found co-localized on the membrane. Thus, the complex formed with AP1G2 and four PICALM proteins might be required for gametophyte development via clathrin-mediated endocytosis.
AP complexes exhibit highly conserved roles in vesicular transport and act as a major hub of interactions in many organisms (Teh et al., 2013). There are five AP complexes (AP1–AP5) identified with conserved four heterotetramers structural compositions in the Arabidopsis genome (Teh et al., 2013; Wang et al., 2014). Reports for the AP-1 complex indicated that AP-1 is essential for viability in multicellular organisms. In mice, homozygous disruptions of the genes encoding γ1 or μ1A caused embryonic lethality (Zizioli et al., 1999; Meyer et al., 2000). In yeast, mutant cis1-1/apm1-1, an allele of the mammalian μ1A subunit of the AP-1 complex, was synthetically lethal, with a deletion of calcineurin, leading to pleiotropic defects in cellular processes, such as secretion, cytokinesis, vacuolar fusion, and cell wall integrity (Kita et al., 2004). In Arabidopsis, AP-1 has two putative copies of each adaptinβ (At4g11380 and At4g23460), γ (At1g60070 and At1g23900), μ (At1g60780 and At1g10730), and σ (At2g17380 and At1g35410). The medium subunit of AP1, redundant AP-1 μ-adaptins AP1M1 and AP1M2, were reported complexed with large subunits γ-adaptin of the heterotetrameric AP-1. The knockout mutation ap1m2 displayed impairing pollen function and arrested plant growth, and the ap1m1ap1m2 double mutant was nearly pollen-lethal (Park et al., 2013). Analysis of a double knockout ap1g1 g2/+ indicates AP1G was important to synergid-controlled pollen tube reception and pollen development by mediating vacuolar remodeling (Feng et al., 2017; Wang et al., 2017). Many DEGs of the three ap1g2 ovules were enriched on pollination, indicating AP1G2's role in pollen tube reception. And our study determined that the ap1g2 mutant had sterility defects in both female and male gametophyte development. Previous studies have indicated that AP-1 is required to execute somatic cytokinesis properly in root and shoot cells in Arabidopsis (Teh et al., 2013). AP1M1 promotes secretory and vacuolar trafficking that is required for cell division and growth during both pollen development and plant growth (Park et al., 2013). The expression of AP1G2 was also detected in the tissues of active cell division, for example, shoot apical meristems, and leaf primordial and root tips. Abnormal vacuolation occurred in both aborted female and male gametophytes, which failed to undergo mitosis. Therefore, we speculated that AP1G2 might have multiple functions and be involved in regulating the cell division cycle.
Calcium ion (Ca2+) signaling is essential for cells and is used for the crucial process by shifting a low concentration of Ca2+ to an increased level to activate or change some proteins that trigger downstream events. The importance of Ca2+ signaling in sexual reproduction has been widely reported, especially in pollen germination, guidance, and pollen tube growth (Iwano et al., 2012; Mahs et al., 2013; Steinhorst and Kudla, 2013; Lenartowski et al., 2015; Steinhorst et al., 2015; Wang et al., 2015; Zheng et al., 2019). Cytosolic calcium transient also occurred in the egg and central cells of female gametophytes to trigger pollen tube burst and ensure successful fertilization (Denninger et al., 2014; Hamamura et al., 2014). The changes in cytosolic free Ca2+ were detected during the processes of megasporogenesis and megaspore degeneration in lettuce (Qiu et al., 2005), but little is known about Ca2+ dynamics during megagametogenesis. In this study, we showed that the Ca2+ concentration was higher in normal developing female and male gametophytes than that in ap1g2-defective gametophytes, which revealed that free Ca2+ signaling was crucial for the process of gametogenesis. A genetic screen was carried out for mutations that were synthetically lethal, with a deletion of calcineurin that is a regulator of Ca2+ signaling in fission yeast, and apm1-1, an allele of the mammalian μ1A subunit of the AP-1 complex, was identified (Kita et al., 2004). Our transcriptomic analysis revealed that some RALFL genes, which lead to calcium-dependent signaling events through the transient increase of the cytoplasmic Ca2+ concentration, and CML16, a member of the calcium-binding EF-hand protein family, together with several calcium-dependent protein kinases, including CPK14, CPK16, and CPK17, were all downregulated in the ap1g2-1+/− mutant. And it has been reported that endocytosis is a calcium-dependent process. The Ca2+ channel could regulate clathrin-mediated endocytosis (Yao et al., 2017). Intracellular calcium may also play a role in the function of clathrin-coated vesicles (Nathke et al., 1990). Therefore, the Ca2+ dynamic might affect the clathrin-mediated endocytosis in the process of gametogenesis.
The clean data has been uploaded in SRA. The BioProjected ID: PRJNA849024: Arabidopsis ovule RNAseq data.
RM and YZ designed the research. YZ, WF, and HC conducted the research. YZ, ZP, and N-U-A analyzed the data. YZ wrote the manuscript. RM, L-YC, and M-CC guided the experiment and revised the manuscript. All authors approved the final version of the manuscript for publication.
This work was supported by National Science Foundation (NSF) Plant Genome Research Program Award DBI-1546890 to RM.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We greatly thank Syed Muhammad Azam for careful revision of this manuscript. We are thankful to Dr. Jane Wann-Neng from Academia Sinica for his support in the TEM sample preparation. We thank Prof. Taiyun Wei from Fujian Agriculture and Forestry University for kindly providing us the research platform to observe the ultrastructures of ovules and pollen grains. We thank the reviewers for their valuable and insightful comments and suggestions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.924417/full#supplementary-material
Alexander, M. P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122. doi: 10.3109/10520296909063335
Alvarez, J., and Smyth, D. R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386. doi: 10.1242/dev.126.11.2377
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. doi: 10.1186/gb-2010-11-10-r106
Barrett, L. W., Fletcher, S., and Wilton, S. D. (2012). Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell. Mol. Life Sci. 69, 3613–3634. doi: 10.1007/s00018-012-0990-9
Bonnemaison, M., Back, N., Lin, Y., Bonifacino, J. S., Mains, R., and Eipper, B. (2014). AP-1A controls secretory granule biogenesis and trafficking of membrane secretory granule proteins. Traffic 15, 1099–1121. doi: 10.1111/tra.12194
Bonnemaison, M. L., Back, N., Duffy, M. E., Ralle, M., Mains, R. E., and Eipper, B. A. (2015). Adaptor protein-1 complex affects the endocytic trafficking and function of peptidylglycine alpha-amidating monooxygenase, a luminal cuproenzyme. J. Biol. Chem. 290, 21264–21279. doi: 10.1074/jbc.M115.641027
Bouche, N., and Bouchez, D. (2001). Arabidopsis gene knockout: phenotypes wanted. Curr. Opin. Plant Biol. 4, 111–117. doi: 10.1016/S1369-5266(00)00145-X
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren Van Themaat, E., Ahmadinejad, N., Assenza, A., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Chen, P., Li, Y., Zhao, L., Hou, Z., Yan, M., Hu, B., et al. (2017). Genome-Wide identification and expression profiling of ATP-binding cassette (ABC) transporter gene family in pineapple (Ananas comosus (L.) Merr.) reveal the role of AcABCG38 in pollen development. Front. Plant Sci. 8, 2150. doi: 10.3389/fpls.2017.02150
Chen, X., Irani, N. G., and Friml, J. (2011). Clathrin-mediated endocytosis: the gateway into plant cells. Curr. Opin. Plant Biol. 14, 674–682. doi: 10.1016/j.pbi.2011.08.006
Christensen, C. A., King, E. J., Jordan, J. R., and Drews, G. N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10, 49–64. doi: 10.1007/s004970050067
Christensen, C. A., Subramanian, S., and Drews, G. N. (1998). Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202, 136–151. doi: 10.1006/dbio.1998.8980
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
De Craene, J. O., Ripp, R., Lecompte, O., Thompson, J. D., Poch, O., and Friant, S. (2012). Evolutionary analysis of the ENTH/ANTH/VHS protein superfamily reveals a coevolution between membrane trafficking and metabolism. BMC Genomics 13, 297. doi: 10.1186/1471-2164-13-297
Denninger, P., Bleckmann, A., Lausser, A., Vogler, F., Ott, T., Ehrhardt, D. W., et al. (2014). Male-female communication triggers calcium signatures during fertilization in Arabidopsis. Nat. Commun. 5, 4645. doi: 10.1038/ncomms5645
D'ippolito, S., Arias, L. A., Casalongue, C. A., Pagnussat, G. C., and Fiol, D. F. (2017). The DC1-domain protein VACUOLELESS GAMETOPHYTES is essential for development of female and male gametophytes in Arabidopsis. Plant J. 90, 261–275. doi: 10.1111/tpj.13486
Drews, G. N., and Koltunow, A. M. (2011). The female gametophyte. Arabidopsis Book 9, e0155. doi: 10.1199/tab.0155
Drews, G. N., Lee, D., and Christensen, C. A. (1998). Genetic analysis of female gametophyte development and function. Plant Cell 10, 5–17. doi: 10.1105/tpc.10.1.5
Feng, C., Wang, J. G., Liu, H. H., Li, S., and Zhang, Y. (2017). Arabidopsis adaptor protein 1G is critical for pollen development. J. Integr. Plant Biol. 59, 594–599. doi: 10.1111/jipb.12556
Ford, M. G., Pearse, B. M., Higgins, M. K., Vallis, Y., Owen, D. J., Gibson, A., et al. (2001). Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291, 1051–1055. doi: 10.1126/science.291.5506.1051
Gao, Y., Liu, X., Tang, B., Li, C., Kou, Z., Liu, W., et al. (2017). Protein expression landscape of mouse embryos during pre-implantation development. Cell Rep. 21, 3957–3969. doi: 10.1016/j.celrep.2017.11.111
Hamamura, Y., Nishimaki, M., Takeuchi, H., Geitmann, A., Kurihara, D., and Higashiyama, T. (2014). Live imaging of calcium spikes during double fertilization in Arabidopsis. Nat. Commun. 5, 4722. doi: 10.1038/ncomms5722
Haruta, M., Monshausen, G., Gilroy, S., and Sussman, M. R. (2008). A cytoplasmic Ca2+ functional assay for identifying and purifying endogenous cell signaling peptides in Arabidopsis seedlings: identification of AtRALF1 peptide. Biochemistry 47, 6311–6321. doi: 10.1021/bi8001488
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Iwano, M., Ngo, Q. A., Entani, T., Shiba, H., Nagai, T., et al. (2012). Cytoplasmic Ca2+ changes dynamically during the interaction of the pollen tube with synergid cells. Development 139, 4202–4209. doi: 10.1242/dev.081208
Kita, A., Sugiura, R., Shoji, H., He, Y., Deng, L., Lu, Y., et al. (2004). Loss of Apm1, the micro1 subunit of the clathrin-associated adaptor-protein-1 complex, causes distinct phenotypes and synthetic lethality with calcineurin deletion in fission yeast. Mol. Biol. Cell 15, 2920–2931. doi: 10.1091/mbc.e03-09-0659
Lenartowski, R., Suwinska, A., and Lenartowska, M. (2015). Calreticulin expression in relation to exchangeable Ca(2+) level that changes dynamically during anthesis, progamic phase, and double fertilization in Petunia. Planta 241, 209–227. doi: 10.1007/s00425-014-2178-z
Li, H., Luo, N., Wang, W., Liu, Z., Chen, J., Zhao, L., et al. (2018). The REN4 rheostat dynamically coordinates the apical and lateral domains of Arabidopsis pollen tubes. Nat. Commun. 9, 2573. doi: 10.1038/s41467-018-04838-w
Mahs, A., Steinhorst, L., Han, J. P., Shen, L. K., Wang, Y., and Kudla, J. (2013). The calcineurin B-like Ca2+ sensors CBL1 and CBL9 function in pollen germination and pollen tube growth in Arabidopsis. Mol. Plant 6, 1149–1162. doi: 10.1093/mp/sst095
Meister, R. J., Williams, L. A., Monfared, M. M., Gallagher, T. L., Kraft, E. A., Nelson, C. G., et al. (2004). Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. Plant J. 37, 426–438. doi: 10.1046/j.1365-313X.2003.01971.x
Meyer, C., Zizioli, D., Lausmann, S., Eskelinen, E. L., Hamann, J., Saftig, J., et al. (2000). mu1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 19, 2193–2203. doi: 10.1093/emboj/19.10.2193
Meyerholz, A., Hinrichsen, L., Groos, S., Esk, P. C., Brandes, G., and Ungewickell, E. J. (2005). Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic 6, 1225–1234. doi: 10.1111/j.1600-0854.2005.00355.x
Muro, K., Matsuura-Tokita, K., Tsukamoto, R., Kanaoka, M. M., Ebine, K., Higashiyama, T., et al. (2018). ANTH domain-containing proteins are required for the pollen tube plasma membrane integrity via recycling ANXUR kinases. Commun. Biol. 1, 152. doi: 10.1038/s42003-018-0158-8
Nathke, I., Hill, B. L., Parham, P., and Brodsky, F. M. (1990). The calcium-binding site of clathrin light chains. J. Biol. Chem. 265, 18621–18627. doi: 10.1016/S0021-9258(17)44797-1
Navarro Negredo, P., Edgar, J. R., Wrobel, A. G., Zaccai, N. R., Antrobus, R., Owen, D. J., et al. (2017). Contribution of the clathrin adaptor AP-1 subunit micro1 to acidic cluster protein sorting. J. Cell Biol. 216, 2927–2943. doi: 10.1083/jcb.201602058
Nordstrom, K. J., Albani, M. C., James, G. V., Gutjahr, C., Hartwig, B., Turck, F., et al. (2013). Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nat. Biotechnol. 31, 325–330. doi: 10.1038/nbt.2515
Ozias-Akins, P., and Van Dijk, P. J. (2007). Mendelian genetics of apomixis in plants. Annu. Rev. Genet. 41, 509–537. doi: 10.1146/annurev.genet.40.110405.090511
Pagnussat, G. C., Yu, H. J., Ngo, Q. A., Rajani, S., Mayalagu, S., Johnson, C. S., et al. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614. doi: 10.1242/dev.01595
Pagnussat, G. C., Yu, H. J., and Sundaresan, V. (2007). Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19, 3578–3592. doi: 10.1105/tpc.107.054890
Pang, Z., Dong, F., Liu, Q., Lin, W., Hu, C., and Yuan, Z. (2021). Soil metagenomics reveals effects of continuous sugarcane cropping on the structure and functional pathway of rhizospheric microbial community. Front. Microbiol. 12, 627569. doi: 10.3389/fmicb.2021.627569
Park, M., Song, K., Reichardt, I., Kim, H., Mayer, U., Hwang, I., et al. (2013). Arabidopsis μ-adaptin subunit AP1M of adaptor protein complex 1 mediates late secretory and vacuolar traffic and is required for growth. Proc. Nat. Acad. Sci. U.S.A. 110, 10318–10323. doi: 10.1073/pnas.1300460110
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Pichon, X., Wilson, L. A., Stoneley, M., Bastide, A., King, H. A., Somers, J., et al. (2012). RNA binding protein/RNA element interactions and the control of translation. Curr. Protein Pept. Sci. 13, 294–304. doi: 10.2174/138920312801619475
Qiu, Y. L., Liu, R. S., Xie, C. T., Yang, Y. H., Ge, L. L., Tian, H. Q., et al. (2005). The dynamic of calcium distribution during megasporegenesis of lettuce (Lactuca sativa L.). Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 31, 374–382. doi: 10.1360/aps040074
Rodriguez Enriquez, M. J., Mehdi, S., Dickinson, H. G., and Grant Downton, R. T. (2013). A novel method for efficient in vitro germination and tube growth of Arabidopsis thaliana pollen. New Phytol. 197, 668–679. doi: 10.1111/nph.12037
Schmidt, A., Wuest, S. E., Vijverberg, K., Baroux, C., Kleen, D., Grossniklaus, U., et al. (2011). Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS Biol. 9, e1001155. doi: 10.1371/journal.pbio.1001155
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9, 671–675. doi: 10.1038/nmeth.2089
Shi, D. Q., Liu, J., Xiang, Y. H., Ye, D., Sundaresan, V., and Yang, W. C. (2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17, 2340–2354. doi: 10.1105/tpc.105.033563
Sieber, P., Gheyselinck, J., Gross-Hardt, R., Laux, T., Grossniklaus, U., and Schneitz, K. (2004). Pattern formation during early ovule development in Arabidopsis thaliana. Dev. Biol. 273, 321–334. doi: 10.1016/j.ydbio.2004.05.037
Somoza, S. C., Sede, A. R., Boccardo, N. A., and Muschietti, J. P. (2021). Keeping up with the RALFs: how these small peptides control pollen-pistil interactions in Arabidopsis. New Phytologist. 229, 14–18. doi: 10.1111/nph.16817
Steinhorst, L., and Kudla, J. (2013). Calcium - a central regulator of pollen germination and tube growth. Biochim. Biophys. Acta 1833, 1573–1581. doi: 10.1016/j.bbamcr.2012.10.009
Steinhorst, L., Mahs, A., Ischebeck, T., Zhang, C., Zhang, X., Arendt, S., et al. (2015). Vacuolar CBL-CIPK12 Ca2+-sensor-kinase complexes are required for polarized pollen tube growth. Curr. Biol. 25, 1475–1482. doi: 10.1016/j.cub.2015.03.053
Su, Z., Zhao, L., Zhao, Y., Li, S., Won, S., Cai, H., et al. (2017). The THO complex non-cell-autonomously represses female germline specification through the TAS3-ARF3 module. Curr. Biol. 27, 1597–1609 doi: 10.1016/j.cub.2017.05.021
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, 607–613. doi: 10.1093/nar/gky1131
Teh, O. K., Shimono, Y., Shirakawa, M., Fukao, Y., Tamura, K., Shimada, T., et al. (2013). The AP-1 μ adaptin is required for KNOLLE localization at the cell plate to mediate cytokinesis in Arabidopsis. Plant Cell Physiol. 54, 838–847. doi: 10.1093/pcp/pct048
Wang, J. G., Feng, C., Liu, H. H., Feng, Q. N., Li, S., and Zhang, Y. (2017). AP1G mediates vacuolar acidification during synergid-controlled pollen tube reception. Proc. Natl. Acad. Sci. U.S.A. 114, 4877–4883. doi: 10.1073/pnas.1617967114
Wang, L. H., Sudhof, T. C., and Anderson, R. G. (1995). The appendage domain of alpha-adaptin is a high affinity binding site for dynamin. J. Biol. Chem. 270, 10079–10083. doi: 10.1074/jbc.270.17.10079
Wang, S. S., Diao, W. Z., Yang, X., Qiao, Z., Wang, M., Acharya, B., et al. (2015). Arabidopsis thaliana CML25 mediates the Ca2+ regulation of K+ transmembrane trafficking during pollen germination and tube elongation. Plant Cell Environ. 38, 2372–2386. doi: 10.1111/pce.12559
Wang, X., Cai, Y., Wang, H., Zeng, Y., Zhuang, X., Li, B., et al. (2014). Trans-Golgi network-located AP1 gamma adaptins mediate dileucine motif-directed vacuolar targeting in Arabidopsis. Plant Cell 26, 4102–4118. doi: 10.1105/tpc.114.129759
Yadegari, R., and Drews, G. N. (2004). Female gametophyte development. Plant Cell 16, 133–141. doi: 10.1105/tpc.018192
Yao, C. K., Liu, Y. T., Lee, I. C., Wang, Y. T., and Wu, P. Y. (2017). A Ca2+ channel differentially regulates clathrin-mediated and activity-dependent bulk endocytosis. PLoS Biol. 15, e2000931. doi: 10.1371/journal.pbio.2000931
Zheng, R. H., Su, S., Xiao, H., and Tian, H. Q. (2019). Calcium: a critical factor in pollen germination and tube elongation. Int. J. Mol. Sci. 20, 420. doi: 10.3390/ijms20020420
Zhu, Y., Mang, H.-G., Sun, Q., Qian, J., Hipps, A., and Hua, J. (2012). Gene discovery using mutagen-induced polymorphisms and deep sequencing: application to plant disease resistance. Genetics 192, 139–146. doi: 10.1534/genetics.112.141986
Zimin, A. V., Marcais, G., Puiu, D., Roberts, M., Salzberg, S. L., and Yorke, J. A. (2013). The MaSuRCA genome assembler. Bioinformatics 29, 2669–2677. doi: 10.1093/bioinformatics/btt476
Keywords: AP1G2, Arabidopsis, Ca2+ signaling, gametogenesis, PICALM proteins
Citation: Zhou Y, Fang W, Pang Z, Chen L-Y, Cai H, Ain N-U, Chang M-C and Ming R (2022) AP1G2 Affects Mitotic Cycles of Female and Male Gametophytes in Arabidopsis. Front. Plant Sci. 13:924417. doi: 10.3389/fpls.2022.924417
Received: 20 April 2022; Accepted: 26 May 2022;
Published: 07 July 2022.
Edited by:
Shanjin Huang, Tsinghua University, ChinaReviewed by:
Pablo Bolaños-Villegas, University of Costa Rica, Costa RicaCopyright © 2022 Zhou, Fang, Pang, Chen, Cai, Ain, Chang and Ming. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ray Ming, cmF5bWluZ0BpbGxpbm9pcy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.