- 1Hunan Provincial Engineering Research Center for Potatoes, College of Horticulture, Hunan Agricultural University, Changsha, China
- 2Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
- 3Key Laboratory for Vegetable Biology of Hunan Province, Changsha, China
In various plant species, many transcription factors (TFs), such as MYB, bHLH, and WD40, have been identified as regulators of anthocyanin biosynthesis in underground organs. However, the regulatory elements of anthocyanin biosynthesis in the tuberous roots of sweet potato have not been elucidated yet. Here, we selected the purple-fleshed sweet potato cultivar “Zhezi1” (ZZP) and its spontaneous yellow-fleshed mutant “Xinli” (XLY) to investigate the regulatory mechanism of the anthocyanin biosynthesis in the tuberous roots of sweet potato. By analyzing the IbMYB1 genotype in ZZP and XLY, we found that the IbMYB1-2, a MYB TF involved in anthocyanin biosynthesis, was missing in the XLY genome, which might lead to an extreme decrease in anthocyanins in XLY. A comparative transcriptome analysis of ZZP and XLY was conducted to find the TFs involved in anthocyanin biosynthesis in ZZP and XLY. The anthocyanin structural genes were significantly enriched among the differentially expressed genes. Moreover, one MYB activator (IbMYB1), one bHLH (IbbHLH2), three WRKY activator candidates (IbWRKY21, IbWRKY24, and IbWRKY44), and two MYB repressors (IbMYB27 and IbMYBx-ZZ) were highly expressed in ZZP accompanied with anthocyanin structural genes. We also tested the expression of these TFs in six purple- and two orange-fleshed sweet potato cultivars. Interestingly, most of these TFs were significantly positively correlated with anthocyanin contents in these cultivars. The function of the anthocyanin biosynthesis repression of IbMYB27 and IbMYBx-ZZ was verified through transient co-transformation with IbMYB1 into tobacco leaves. Further functional verification of the above TFs was conducted by Y2H, BiFC, and dual-luciferase assays. These tests showed that the MYB-bHLH-WD40/MYB-bHLH-WD40-WRKY complex activated the promoter of anthocyanin structural gene IbDFR and promoters for IbWRKY44, IbMYB27, and IbMYBx-ZZ, indicating reinforcement and feedback regulation to maintain the level of anthocyanin accumulation in the tuberous roots of purple-fleshed sweet potato. These results may provide new insights into the regulatory mechanism of anthocyanin biosynthesis and accumulation in underground organs of sweet potatoes.
Introduction
Sweet potato (Ipomoea batatas [L.] Lam.) is the seventh-most important food crop in the world after wheat, rice, maize, potato, barley, and cassava. The global production of sweet potato is centered in Asia, and China is the largest producer, accounting for 56.05% of the total production (FAOSTAT, 2020). Sweet potatoes are rich in carbohydrates, dietary fibers, β-carotene, minerals, and other nutrients (Endrias et al., 2016). Moreover, cultivars with purple flesh are rich in anthocyanins (Mano et al., 2007).
In plants, structural genes encoding phenylalanine ammonia lyase (PAL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3’H), dihydroflavonol 4-reductase (DFR), anthocyanin synthase (ANS), and glutathione S-transferase (GST) are involved in anthocyanin biosynthesis (Holton and Cornish, 1995). These anthocyanin biosynthesis genes are activated by a transcriptional activation complex comprising basic helix–loop–helix (bHLH), R2R3-MYB, and WD-repeat (WDR) proteins (Chu et al., 2013; Albert et al., 2014). The R2R3-MYB factors in the complex are most crucial for the spatial and temporal localization of anthocyanins (Davies et al., 2012). For example, in the overground part, AtPAP1, AtMYB113, and AtMYB114 in Arabidopsis (Gonzalez et al., 2010) and PhAN2 in petunia (Quattrocchio et al., 1999) have been reported. In underground organs, DcMYB7 and DcMYB113 in carrot (Xu et al., 2019; Bæksted Holme et al., 2021) and AN1 in potato have been identified (Liu et al., 2016). However, two types of MYB repressors, i.e., R2R3-MYBs and R3-MYBs, which repress anthocyanin biosynthesis, are identified. For example, FaMYB1 (R2R3-MYB) in strawberry (Aharoni et al., 2001; Salvatierra et al., 2013), PhMYB27 (R2R3-MYB), and PhMYBx (R3-MYB) in petunia (Albert et al., 2011) have been reported. The MYB-bHLH-WD40 (MBW) activation complex and MYB repressors participate in anthocyanin biosynthesis in petunia leaf through hierarchical and feedback regulation to prevent inappropriate anthocyanin level (Albert et al., 2014).
Moreover, WRKY can promote anthocyanin biosynthesis by forming MYB-bHLH-WD40-WRKY (MBWW) complexes with MYB, bHLH, and WD40 (Verweij et al., 2016). In Arabidopsis thaliana, AtTTG2 (WRKY) can interact with AtTTG1 (WD40) to activate the accumulation of PAs in the seed coat (Gonzalez et al., 2016). In Petunia hybrida, PhPH3 can directly interact with the MBW complex to regulate the biosynthesis of anthocyanins (Verweij et al., 2016). WRKY proteins, as a large family of transcription factors (TFs) in plants, influence physiological processes, such as plant stress resistance, growth, and development (Alessandra et al., 2016). On the basis of the number of WRKY domains (WRKYGQ) and the characteristics of the zinc finger’s structure, WRKY proteins can be divided into three groups (Eulgem et al., 2000). However, reports on the regulation of anthocyanin biosynthesis by WRKY TFs in sweet potatoes are few.
Previous research suggested that anthocyanin biosynthesis in the storage roots of purple-fleshed sweet potato is regulated by an MYB activator, i.e., IbMYB1 (Mano et al., 2007). Two genotypes of the IbMYB1 gene, named IbMYB1-1 and IbMYB1-2, are found in the genome of a purple-fleshed sweet potato cultivar Ayamurasaki, whereas its spontaneous white-fleshed mutant and other white- or yellow-fleshed cultivars have only IbMYB1-1 (Tanaka et al., 2012). Although IbMYB1-1 and IbMYB1-2 have identical coding sequences, only IbMYB1-2 is identified to be responsible for anthocyanin accumulation in tuberous roots (Tanaka et al., 2012). In sweet potato, the molecular mechanism for the loss of anthocyanins in red young leaves gradually turning green is explored (Deng et al., 2020). Three MYB activator genes (i.e., IbMYB1, IbMYB2, and IbMYB3) are simultaneously expressed with four MYB repressor genes (i.e., IbMYB27, IbMYB4a, IbMYB4c, and IbMYBx) in the early stage of leaf development, and these MYB TFs are involved in the hierarchical and feedback regulation of anthocyanin accumulation (Deng et al., 2020). However, as underground organs, the tuberous roots of purple-fleshed sweet potato remain purple during the whole development stage. Therefore, the pattern of spatial and temporal localization of anthocyanins in underground organs may be different. Whether MYB repressors, MYB activators, and WRKYs collaboratively participate in the regulation of anthocyanin biosynthesis in underground organs remains to be clarified.
In this manuscript, a spontaneous yellow-fleshed mutant Xinli (XLY) is obtained from the purple-fleshed sweet potato cultivar “Zhezi1” (ZZP). The transcriptome analysis of two homologous materials ZZP and XLY is performed to elucidate the regulatory mechanism of the anthocyanin biosynthesis in the tuberous roots of purple-fleshed sweet potato. Candidate TFs are cloned, and their functions are verified. Our results suggest that MYB activators, bHLHs, WRKYs, and MYB repressors are collaboratively involved in the regulation of anthocyanin biosynthesis in the tuberous roots of purple-fleshed sweet potato to prevent the excessive accumulation of anthocyanins.
Materials and methods
Plant materials
A spontaneous yellow-fleshed mutant of ZZP was obtained from the experimental field of Hunan Agricultural University in 2012 and was named XLY. Eight recent Chinese sweet potato cultivars with different genetic backgrounds, including six purple- (i.e., XZ118, GZ11, GZ10, YZ7, NY76, and XZ7) and two orange-fleshed (i.e., X19 and G87) sweet potatoes, bred in different departments in different regions of China were grown in an experimental field of the Hunan Agricultural University in Changsha, Hunan in the autumn of 2019. Each sample replicate consisted of five tubers from different plants in the tuber mature stage, and three biological replicates were used. Cylinders with diameter of 1 cm from the middle parts of tuberous roots were stored in liquid nitrogen for transcriptome analysis and quantitative real-time (qRT)-PCR. Others were used in determining the content of anthocyanins. Tobacco (Nicotiana tabacum L., for transient assays and Nicotiana benthamiana L. for dual-luciferase assays) was grown in a pot with 16 h/8 h light/dark photocycle at 25°C.
Extraction and quantification of anthocyanins
Anthocyanins in tuberous roots of sweet potato were extracted and quantified in accordance with the methods described by Sun and Guo (2008). In brief, 0.5 g tuber tissues were immersed and extracted with methanol and 0.1% HCl. Anthocyanin levels from methanol extracts were estimated in accordance with equation: A = ([A520 × 50]/[98.2 × 0.5]) × 100. Absorbance was measured using the Multiskan spectrum device (Thermo Scientific Multiskan GO 1510, Finland).
RNA and DNA isolation and genomic PCR of ZZP and XLY
The total RNAs of experimental materials used in this paper were extracted using the RNA-prep Pure Plant Plus Kit (Polysaccharides and Polyphenolics-rich; Tiangen Biotech [Beijing] CO. LTD.). The RNA of each sample was reverse-transcribed into cDNA by the premix kit (Tiangen Biotech [Beijing] CO. LTD.). The total DNAs of experimental materials used in this manuscript were isolated using a plant genomic DNA kit (Tian gen, Beijing, China). Genomic PCR was conducted for IbMYB1 and structural genes using the PCR conditions and primers described by Mano et al. (2007). For the analysis of the IbMYB1 genotype in ZZP and XLY, IbMYB1-1 and IbMYB1-2 fragments were amplified using PCR conditions and primers (Supplementary Table 1) described by Tanaka et al. (2012).
Transcriptome analysis
Approximately 3 μg RNA per sample was used as input material for RNA sample preparations. The NEB Next® Ultra RNA library prep kit for Illumina® (NEB, MA, United States) was used to generate sequencing libraries. Index codes were added to attribute sequences to each sample.
A qualified complete cDNA library was obtained using the PacBio sequel platform and sequenced in a cell. The Agilent Bioanalyzer2100 sequencing platform (Illumina) was used to sequence second-generation libraries. Following the manufacturer’s instructions, the clustering of index-coded samples was performed on a cBot cluster generation system with the TruSeq PE cluster kit v3-cBot-HS (Illumina). Briefly, mRNA was enriched by Oligo (dT) beads. The enriched mRNA was fragmented into short fragments with fragmentation buffer and reverse-transcribed into cDNA with random primers. The second-strand cDNA was synthesized by DNA polymerase I, RNase H, dNTP, and buffer. Then, cDNA fragments were purified, end repaired, poly (A) added, and ligated to Illumina sequencing adapters. Ligation products were selected by size through agarose gel electrophoresis, PCR amplified, and sequenced using the AMPure XP system. High-quality clean reads were first aligned to de novo cognate assemblies of “ZZP” and “XLY” and mapped to the reference genome by the HISAT v2.1.0 after the release of the genomic sequence of the sweet potato cultivar “Taizhong 6” (Yang et al., 2017). No fundamental difference was displayed in the comprehensive analysis of the categories of expressed genes. However, subsequent analysis with the reference genome for specific gene sequences showed considerable incompleteness and inaccuracy that hindered further investigation. Alignment to de novo cognate assemblies was used as the main data to bypass these issues, whereas alignment to the “Taizhong 6” reference genome was used as auxiliary data for the subsequent homologous cloning of the promoters together with genomes of I. nil (Hoshino et al., 2016), I. trifida, and I. triloba (Wu et al., 2018). Differential expression gene analysis was performed on ZZP and XLY with the DESeq2 R package (1.22.2). q-value was < 0.05. For the anthocyanin biosynthesis genes, differentially expressed genes (DEGs) were recruited by log2 (fold change) > 2, and the q-value was < 0.01. Unigenes were functionally annotated by searching against NCBI nucleotide sequences (Nt), NCBI non-redundant protein, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), euKaryotic Ortholog Groups, Protein family, and Swiss-Prot databases, and the E-value was 10–5.
qRT-PCR
The primers used for qRT-PCR analysis were designed using the Primer 5 (Supplementary Table 1). qRT-PCR was performed using the Roche LightCycler® 480II system with the TSINKE2 × T5 Fast qPCR Mix (SYBR Green I) (TSINKE, China) under the following conditions: 95°C for 60 s, 40 cycles of 95°C for 10 s, and 60°C for 15 s. The expression levels of genes were normalized to the level of constitutive IbACTIN expression. The 2–ΔCT method was used in analyzing the qRT-PCR results for eight varieties of sweet potato and tobacco leaves, whereas the 2– ΔΔCT method was used for ZZP and XLY.
Phylogenetic analysis for the anthocyanin-related WRKY and MYB
Phylogenetic trees were constructed by MEGA X (Tamura et al., 2011). Parameters for the NJ tree were set as p-distance model and pairwise deletion with the bootstrap value as 1,000. According to results, the TF clusters of MYB and WRKY were used in protein sequence alignment analysis through the GenDoc.
Gene function assays
For the gene function analysis, the full-length coding sequences of IbMYB1, IbMYB27, and IbMYBx-ZZ were cloned using the Planta Super-Fidelity DNA Polymerase (Abm, BC, Canada) and inserted into a p2300 vector under the control of a 35S promoter. Primer sequences are listed in Supplementary Table 1. The transformation of recombinant vectors into GV3101 strains of Agrobacterium tumefaciens was performed in accordance with the methods described by Zhou et al. (2019).
Agrobacterium cells harboring IbMYB1, IbMYBx-ZZ, IbMYB27, and empty vector (CK) were mixed in different ratios and used in testing the repressing effects of IbMYB27 and IbMYBx-ZZ. The methods of mixing Agrobacterium cells and transforming into N. tabacum leaves were performed in accordance with the methods described by Zhou et al. (2019). Plants were incubated in the dark for 24 h and moved to a greenhouse with artificial irradiance (16 h day). Photographs were taken 7 days after injection, and the total RNA was collected for extraction when necessary.
Yeast two-hybrid assays
The open reading frames (ORFs) of IbMYB1, IbMYB27, IbMYBx-ZZ, IbWRKY44, and IbWD40 were cloned into the pGADT7 plasmid to generate IbMYB1-AD, IbMYB27-AD, IbMYBx-ZZ-AD, IbWRKY44-AD, and IbWD40-AD, respectively. The ORFs of IbMYB1, IbbHLH1, IbbHLH2, IbWD40, and IbMYB27 were inserted into the pGBKT7 plasmid to generate IbMYB1-BD, IbbHLH1-BD, IbbHLH2-BD, IbWD40-BD, and IbMYB27-BD, respectively. The primers used to clone these genes are listed in Supplementary Table 1. Recombinant vectors were transformed into Y187 yeast (Clontech, CA, United States) by PEG3350-mediated transformation. The transformed yeast cells were cultured in -T/-L (-tryptophan/-leucine) and -T/-L/-H/-A (-tryptophan/-leucine/-histidine/-adenine) media for 3 days to observe yeast growth.
Bimolecular fluorescence complementation assays
The ORFs of IbMYB1, IbMYB27, IbMYBx-ZZ, IbbHLH1, IbbHLH2, IbWD40, and IbWRKY44 were inserted into pSPYNE-35S/pUC-SPYNE (provided by our own laboratory) or pSPYCE-35S/pUC-SPYCE plasmids to generate IbbHLH1-YFPC, IbbHLH2-YFPC, IbWD40-YFPC and IbMYB1-YFPN, IbWRKY44-YFPN, IbMYB27-YFPN, IbMYBx-ZZ-YFPN, respectively. Recombinant vectors were transformed into GV3101 strains of Agrobacterium tumefaciens in accordance with the methods described in Section “Gene function assays.” A. tumefaciens mixtures (ratio = 1:1) were infiltrated into N. benthamiana leaves and cultured in a greenhouse for 2 days before sample collection. Fluorescence was detected using a confocal laser scanning microscope (TCIT SP2, Leica).
Dual-luciferase assays
The ORFs of IbMYB1, IbbHLH1, IbbHLH2, IbWD40, IbWRKY44, IbMYBx-ZZ, and IbMYB27 were cloned into the pGreen62-SK plasmid (provided by our own laboratory) to generate IbMYB1-pGreen62-SK, IbbHLH1-pGreen62-SK, IbbHLH2-pGreen62-SK, IbWD40-pGreen62-SK, IbWRKY44-pGreen62-SK, IbMYBx-ZZ-pGreen62-SK, and IbMYB27-pGreen62-SK, respectively. The promoter sequences of IbDFR, IbMYB27, IbMYBx-ZZ, and IbWRKY44 were inserted into the pGreen0800-LUC plasmid to generate pIbDFR-LUC, pIbMYB27-LUC, pIbMYBx-ZZ-LUC, and pIbWRKY44-LUC, respectively. The primers used to clone these gene promoters are listed in Supplementary Table 1. Recombinant vectors were transformed into the GV3101 strains of A. tumefaciens in accordance with the methods described in Section “Gene function assays.” A. tumefaciens mixtures (ratio = 1:1) were infiltrated into N. benthamiana leaves and cultured in a greenhouse for 2 days before sample collection. Afterward, 0.2 mM luciferin (Biyuntian, Jiangsu, China) was infiltrated into the same positions where A. tumefaciens was infiltrated and held in a dark room for 0.5 h. The luciferase activity was detected by live plant imaging system (Xenogen, Alameda, CA, United States, IVIS Spectrum). The LUC imaging was visualized using the Tanon gel imaging software. Promoter sequences are presented in Supplementary Table 3.
Statistical analysis
Samples were subjected to statistical analysis by using the IBM SPSS statistical software.1 Data were analyzed by one-way analysis of variance (ANOVA), and the t-test (P < 0.01) was used to separate the means in ZZP and XLY.
Results
Phenotypes of the ZZP and XLY mutant
XLY, a spontaneous tuberous root mutant of ZZP, was first found among the ZZP tuberous root when it was harvested in the field (Figure 1A). The skin color of the storage roots of XLY became white. The pigmentation on the stem of the plant, which first grew from the tuberous roots of the XLY, was slighter than that of ZZP (Figure 1B). No anthocyanin pigmentation was observed in stems, aerial roots, fiber roots, and storage roots of XLY plants growing from stem cuttings (Figures 1C–E). The anthocyanin contents in the skin and flesh of the storage roots of XLY were significantly (p < 0.01) reduced to about 1/9th and 1/7th, respectively, compared with those of ZZP in the tuber mature stage (Figure 1F). However, the color of the leaves seemed unchanged (Figures 1B,C). No other evident phenotypic difference was observed on plants between XLY and ZZP except for anthocyanin accumulation.
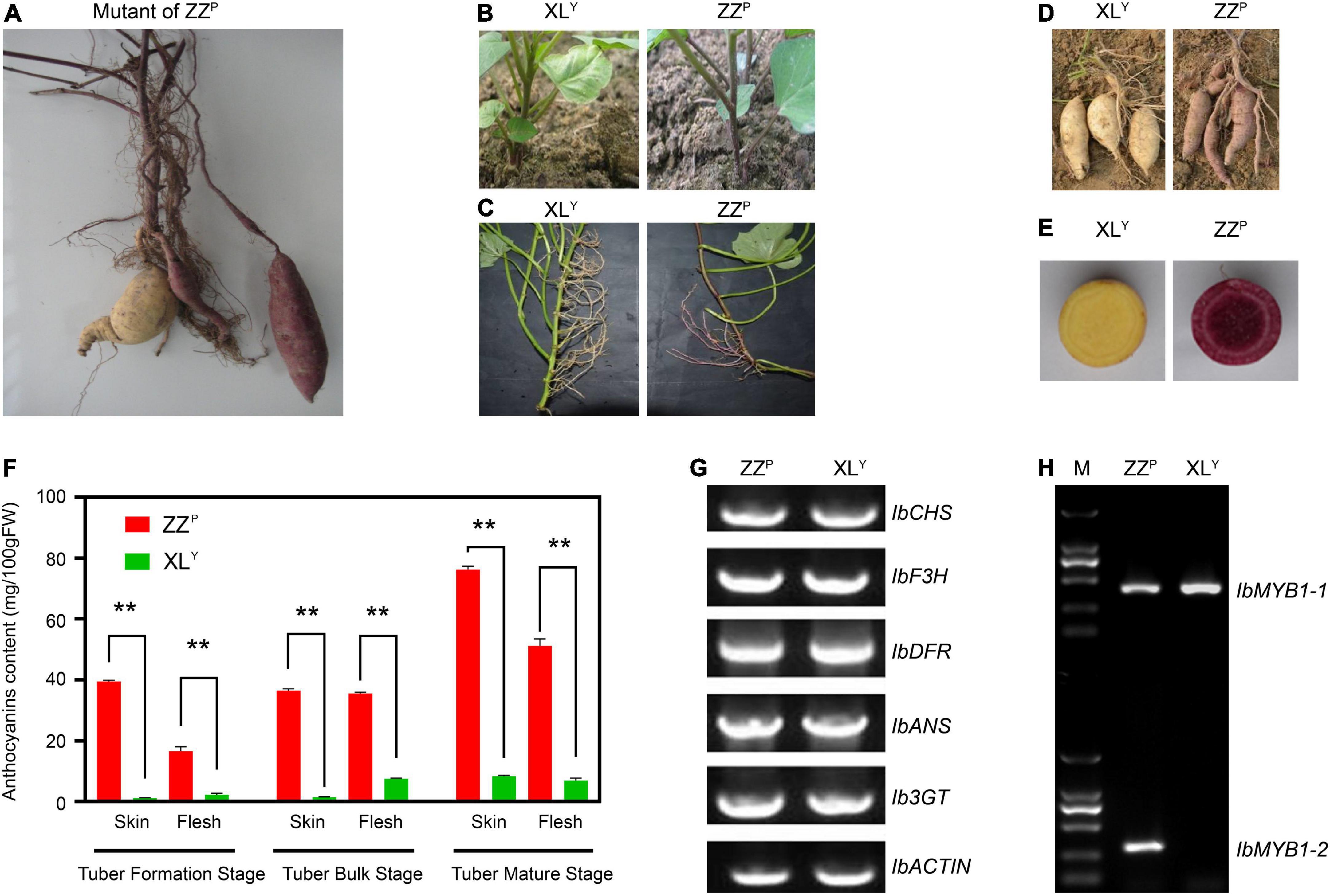
Figure 1. Phenotype and genotype of the Zhezi.1 (ZZP) and Xinli (XLY) mutant. (A) First appearance of ZZP mutant. (B) Plants of XLY (Left) and ZZP (Right) grown from tuberous roots. (C) Stems, (D) tuberous roots, and (E) cross-section of tuberous roots of XLY (Left) and ZZP (Right). (F) Anthocyanin content in tuberous roots of ZZP and XLY in different parts and stages. Error bars indicate the standard deviation from three biological replicates. Asterisks represent statistically significant differences (**P < 0.01), analyzed using Student’s t-test. (G) Genomic PCR for full CDS of structural genes involved in anthocyanin biosynthesis on ZZP and XLY. (H) Genotype of IbMYB1 in ZZP and XLY mutant. M, Marker of 2000.
Analysis of the IbMYB1 genotype in ZZP and XLY mutant
Genomic PCR was conducted for structural gene coding sequence in accordance with the method described by previous research (Mano et al., 2007) to explore whether gene deletion in XLY resulted in a remarkable decrease in anthocyanin accumulation. All tested genes, including IbCHS, IbF3H, IbDFR, IbANS, and Ib3GT, were detected in ZZP and XLY (Figure 1G). Results implied that there was no missing of these genes in ZZP and XLY.
Previous research suggested that two variants of IbMYB1, namely, IbMYB1-1 and IbMYB1-2, are found in the genomes of purple-fleshed cultivar Ayamurasaki and that IbMYB1-2 is lost in its white-fleshed mutant cultivar AYM96 (Tanaka et al., 2012). The identification of the IbMYB1 genotype was conducted on ZZP and XLY in accordance with a previously described method (Tanaka et al., 2012). The IbMYB1-1 variant was detected in ZZP and XLY, but the IbMYB1-2 variant was only detected in ZZP (Figure 1H). These results implied that the missing IbMYB1-2 in XLY might lead to remarkable downregulation of IbMYB1, eventually leading to a dramatic reduction in anthocyanin accumulation in the XLY mutant.
Transcriptome analyses of ZZP and XLY mutant
RNA-seq was performed on tuberous roots of ZZP and XLY to screen DEGs in sweet potato tubers. The sequencing and assembly results implied the reliability of unigenes data (Supplementary Table 2). A total of 49.87 and 50.33 million raw reads were obtained, and the average Q30 of the raw reads were 91.09 and 91.17% in ZZP and XLY, respectively (Supplementary Table 2). A total of 3,876 DEGs, including 1,792 downregulated and 2,084 upregulated genes, were identified in XLY compared with ZZP (Supplementary Figure 1A). In the GO term, DEGs were further classified into 3,288 functional subcategories. In the “biological process” category, the “anthocyanin-containing compound metabolic process” (43 unigenes, 1.92%), “hemicellulose metabolic process” (37 unigenes, 1.65%), and “anthocyanin-containing compound biosynthetic process” (35 unigenes, 1.56%) were the most common assignments (Figure 2A). DEGs were assigned to 128 KEGG pathways. The “metabolic pathways,” “biosynthesis of secondary metabolites,” “phenylpropanoid biosynthesis,” and “flavonoid biosynthesis” categories were the top four enriched processes in DEGs (Supplementary Figure 1B). Hence, these results showed that the downregulated genes involved in flavonoid and anthocyanin biosyntheses might be the cause of the dramatic reduction in anthocyanin accumulation in XLY mutant.
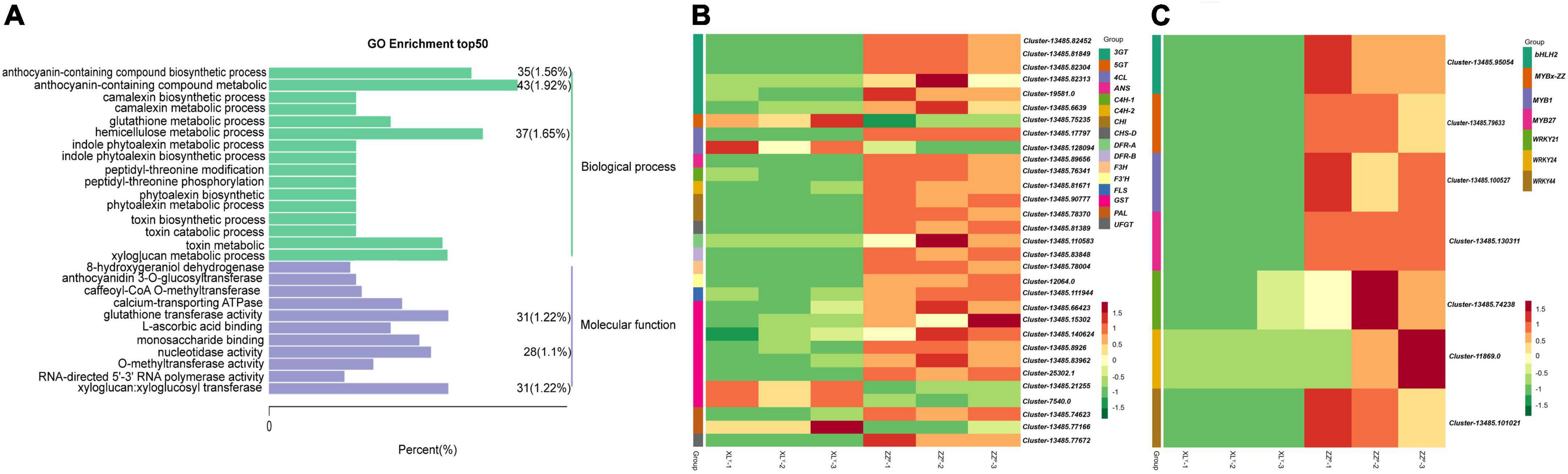
Figure 2. Differentially expressed genes (DEGs) in tuberous roots of ZZP and XLY. (A) Top 50 pathways with the most significant enrichment in GO term between ZZP and XLY. The X-axis indicates the number and percentage of DEGs under each functional classification, whereas the Y-axis represents the enriched GO functional classification, which is divided into two categories: biological process and molecular function. (B) Heat map of differentially expressed structural genes relative to anthocyanin biosynthesis pathway between ZZP and XLY. (C) Heat map of differentially expressed TFs relative to the anthocyanin biosynthesis pathway between ZZP and XLY. Each row represents a gene, and each column represents a sample (XLY-1, XLY-2, and XLY-3 represent three replicates of XLY sample. ZZP-1, ZZP-2, and ZZP-3 represent three replicates of ZZP sample). Different colors in the clustering heat map represent the different expression levels of genes. Red and green colors indicate high and low expression levels, respectively.
Identification of the structural genes and transcription factors involved in flavonoid and anthocyanin biosyntheses in ZZP
We focused on the structural genes and TFs involved in anthocyanin biosynthesis. The DEGs involved in the biosyntheses of anthocyanins and flavonoids were screened (Figures 2B,C). Among the differentially expressed structural genes involved in the biosyntheses of flavonoids and anthocyanins, 1 4CL, 1 CHS, 2 CHI, 1 F3’H, 1 F3H, 2 DFR, 1 ANS, 6 3GT, 1 UFGT, 1 PAL, 2 C4H, 1 FLS, and 6 GST genes in XLY were significantly downregulated compared with those in ZZP (Figure 2B). These results were consistent with the remarkable differences in anthocyanin accumulation in the storage roots of ZZP and XLY. Moreover, 260 TF genes, including 127 downregulated and 133 upregulated genes, were determined in XLY. Among these TF genes, 3 MYB genes, i.e., IbMYB1 (Cluster-13485.100527), IbMYB27 (Cluster-13485.130311), IbMYBx-ZZ (Cluster-13485.79633); 3 WRKY genes, i.e., IbWRKY21 (Cluster-13485.74238), IbWRKY44 (Cluster-13485.101021), IbWRKY24 (Cluster-11869.0); and 1 bHLH gene IbbHLH2 (Cluster-13485.95054) in XLY were markedly downregulated compared with those in ZZP (Figure 2C).
RNA-seq data were further confirmed by selecting 15 related anthocyanin biosynthesis genes for RT-qPCR (Figure 3). Results indicated that the expression patterns from qRT-PCR assay correlated well with sequencing results.

Figure 3. Verification of the relative expression level of DEGs in the tuberous roots of sweet potato by qRT-PCR. Expression levels were standardized to GADPH, and results in the ZZP were set to 1. Expression patterns of 15 DEGs related to anthocyanin biosynthesis pathway including (A–G) TFs and (H–O) structural genes. The red bar shows relative gene expression level measured by qRT-PCR, and the green bar shows the FPKM of relative gene in RNA-seq. Error bars indicate the standard deviation from three biological replicates. Asterisks represent statistically significant differences (****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05), as analyzed using Student’s t-test.
Functional verification of activator IbWRKY44 and two repressors IbMYBx-ZZ and IbMYB27
A phylogenetic tree of the WRKY family was constructed using the method described by Li et al. (2020). Phylogenetic analysis results indicated that the protein sequences of IbWRKY21, IbWRKY24, and IbWRKY44 were homologous to those of anthocyanin-related genes, including MdWRKY11, PhPH3, PbWRKY75, StWRKY13, VvWRKY26, and AtWRKY53 (Figure 4A). The derived polypeptide alignment of IbWRKY24, IbWRKY44, and IbWRKY21 was similar to that of other WRKY TFs involved in anthocyanin biosynthesis, and all of these WRKY TFs contained WRKY (β2-β3-β4) and zinc-finger-like (C-x5-C-x23-H-x-H) motifs (Figure 4B). IbWRKY44 and IbWRKY24 had two WRKY motifs, and IbWRKY21 had only one WRKY motif. However, the Leu zipper motif, which existed in MdWRKY40 and AtWRKY40 and consisted of a series of modules of seven residues, was not identified in IbWRKY24, IbWRKY44, and IbWRKY21.
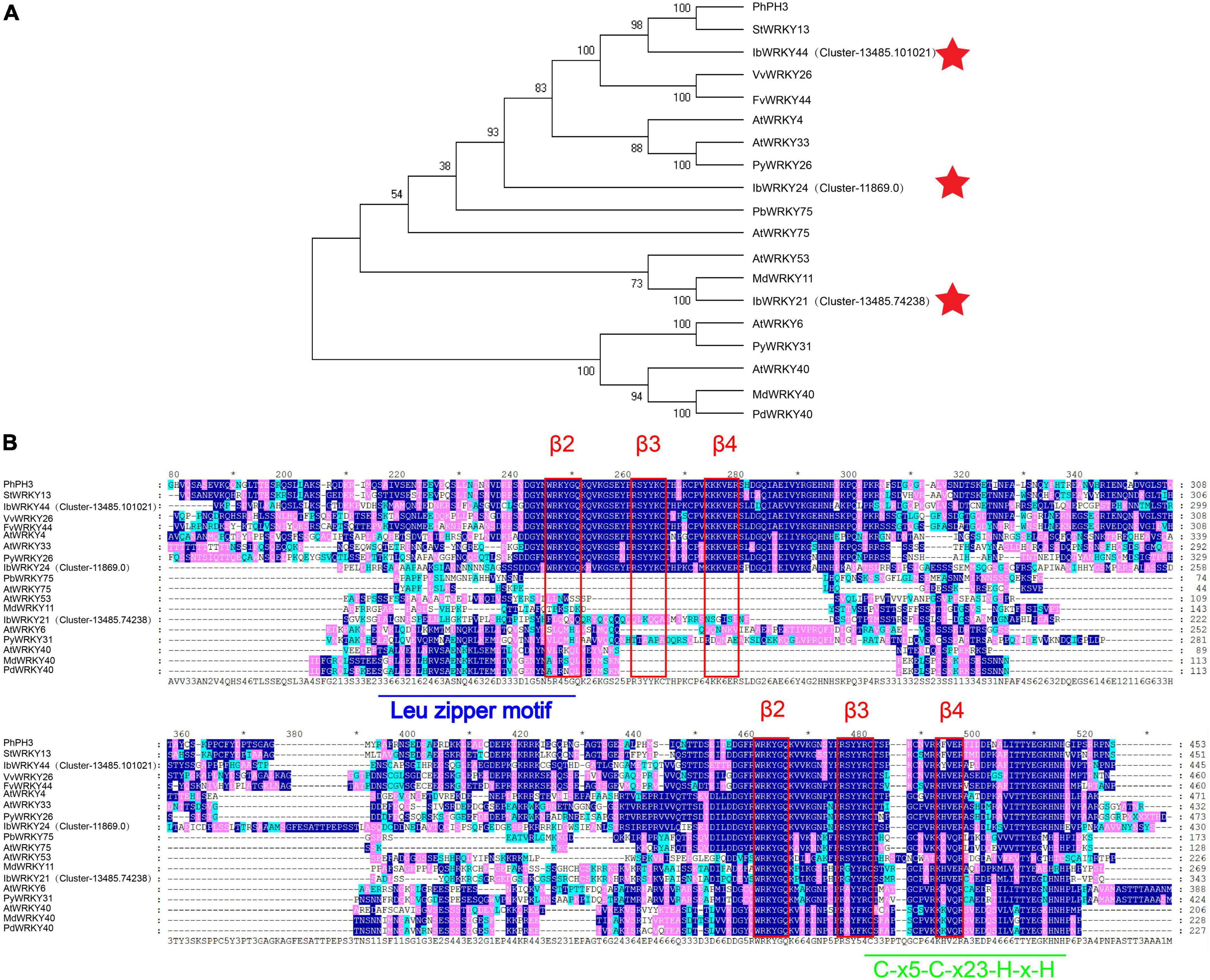
Figure 4. Phylogenetic analysis and protein sequence alignment for the putative WRKYs with function-verified homologous genes of other plant species. (A) Phylogenetic relationship of putative WRKYs in sweet potato and function-verified anthocyanin-related WRKYs in other species. Pentagram represents WRKYs screened from RNA-seq. (B) Protein sequence alignment for the putative MYBs with function-verified homologous genes of other plant species. Blue and green lines represent the Leu zipper motif and C-x5-C-x23-H-x-H motif, respectively. The red rectangle represents the β2-β3-β4 domains.
Three MYB genes screened by RNA-seq were aligned with classic anthocyanin MYBs in other plants, such as Arabidopsis, Petunia hybrida, Malus domestica, Fragaria × ananassa Duch, I. purpurea, and I. nil for the prediction of their functions (Supplementary Figure 2A). IbMYB1 and IbMYB27 were named after their homologous genes in I. batatas and clustered into MYB activator members and FaMYB1-like repressor members, respectively (Supplementary Figure 2A). IbMYBx-ZZ, which encoded a protein of 80 amino acids, had an identity of 78% and 60% with IbMYBx and PhMYBx, respectively, by protein blast and had been identified to repress anthocyanin biosynthesis in sweet potato and petunia (Albert et al., 2011; Deng et al., 2020; Supplementary Figure 2B). IbMYBx-ZZ was clustered into PhMYBx-like R3-MYB members (Supplementary Figure 2A).
To elucidate the repression functions of IbMYBx-ZZ and IbMYB27 in anthocyanin biosynthesis, we induced transient transformation in tobacco leaves in accordance with the method described by Liu et al. (2016). Agrobacterium cells harboring IbMYB1, IbMYBx-ZZ, IbMYB27, and empty vector (CK) were mixed in different ratios and coinfiltrated into tobacco leaves. As shown in Figure 6, the purple pigment was observed on the tobacco leaves when IbMYB1 was transformed in tobacco leaves, whereas no purple pigment was observed on patches when IbMYB27 or IbMYBx-ZZ was transformed alone in tobacco leaves. The lighter purple pigment was observed on leaves when IbMYB1 was cotransformed with IbMYB27 or IbMYBx-ZZ (Figures 5A,B). Moreover, qRT-PCR results showed that the expression levels of TFs and structural genes in tobacco leaves were significantly downregulated when IbMYB1 was cotransformed with IbMYB27 and IbMYBx-ZZ (Figures 5C,D). These results indicated that IbMYB27 and IbMYBx-ZZ could repress anthocyanin biosynthesis, which was induced by IbMYB1.
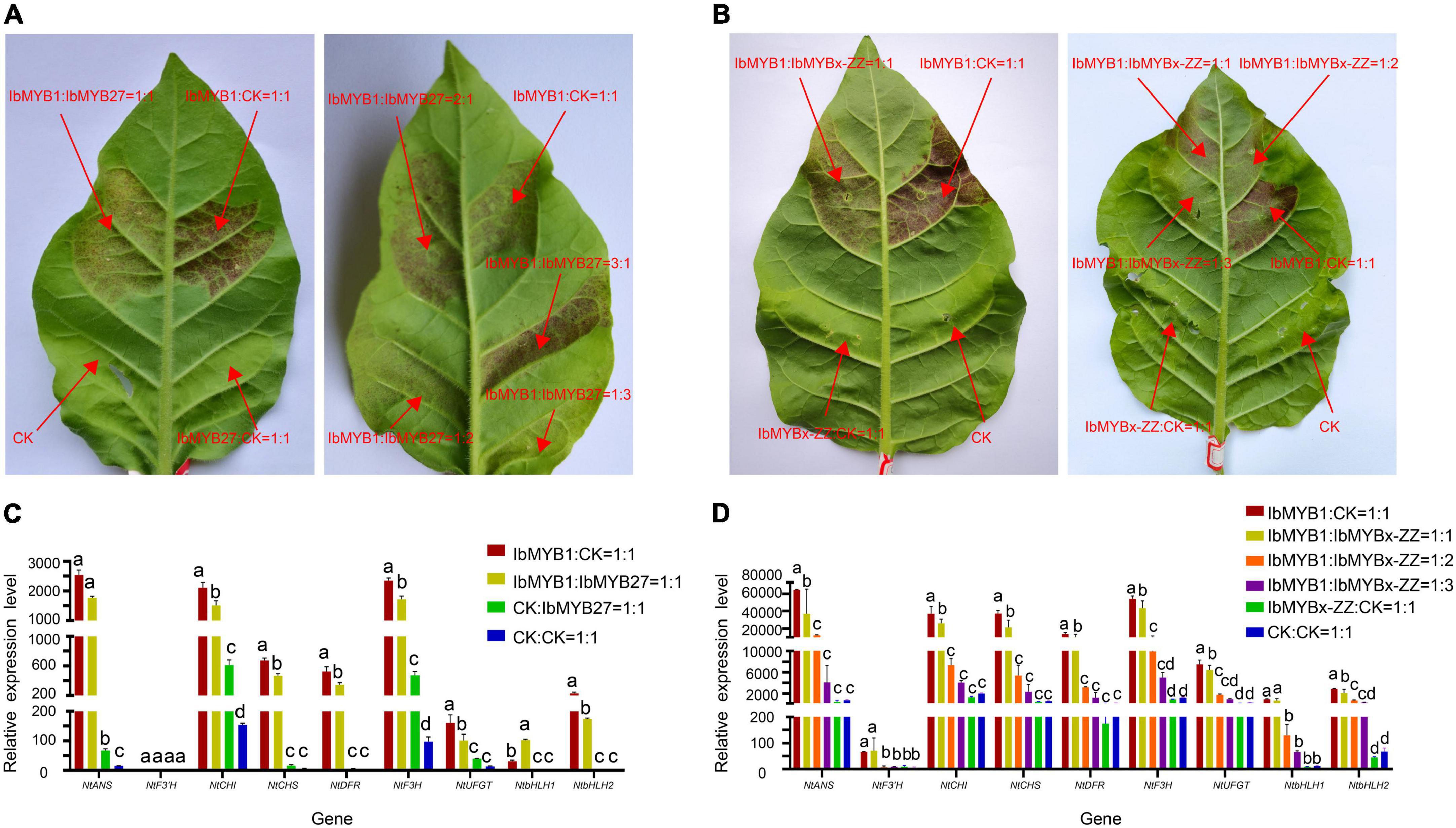
Figure 5. Functional analysis of the sweet potato IbMYB27 and IbMYBx-ZZ TFs. (A,B) Cotransformation of IbMYB1/IbMYB27 and IbMYB1/IbMYBx-ZZ-induced anthocyanin biosynthesis, as shown by transient expression assays in tobacco leaves. Tobacco leaves appeared 5 days after injection. Different ratios of IbMYB1, IbMYBx-ZZ, IbMYB27, or CK (empty vector) cotransformed into tobacco leaves. (C,D) Expression patterns of nine DEGs related to the anthocyanin biosynthesis pathway, including TFs and structural genes, after IbMYB1 and IbMYB27 or CK (empty vector) cotransformation into tobacco leaves. Leaf samples in panels (C,D) were cut out from the transformation part of panels (A,B). The X-axis represents the gene, and the Y-axis represents the relative expression level of the gene. Error bars indicate the standard deviation from three biological replicates. Statistical significance was determined by one-way ANOVA (P-values reported). Significant differences between means (Duncan, P = 0.05) were indicated by different letters above the bar.
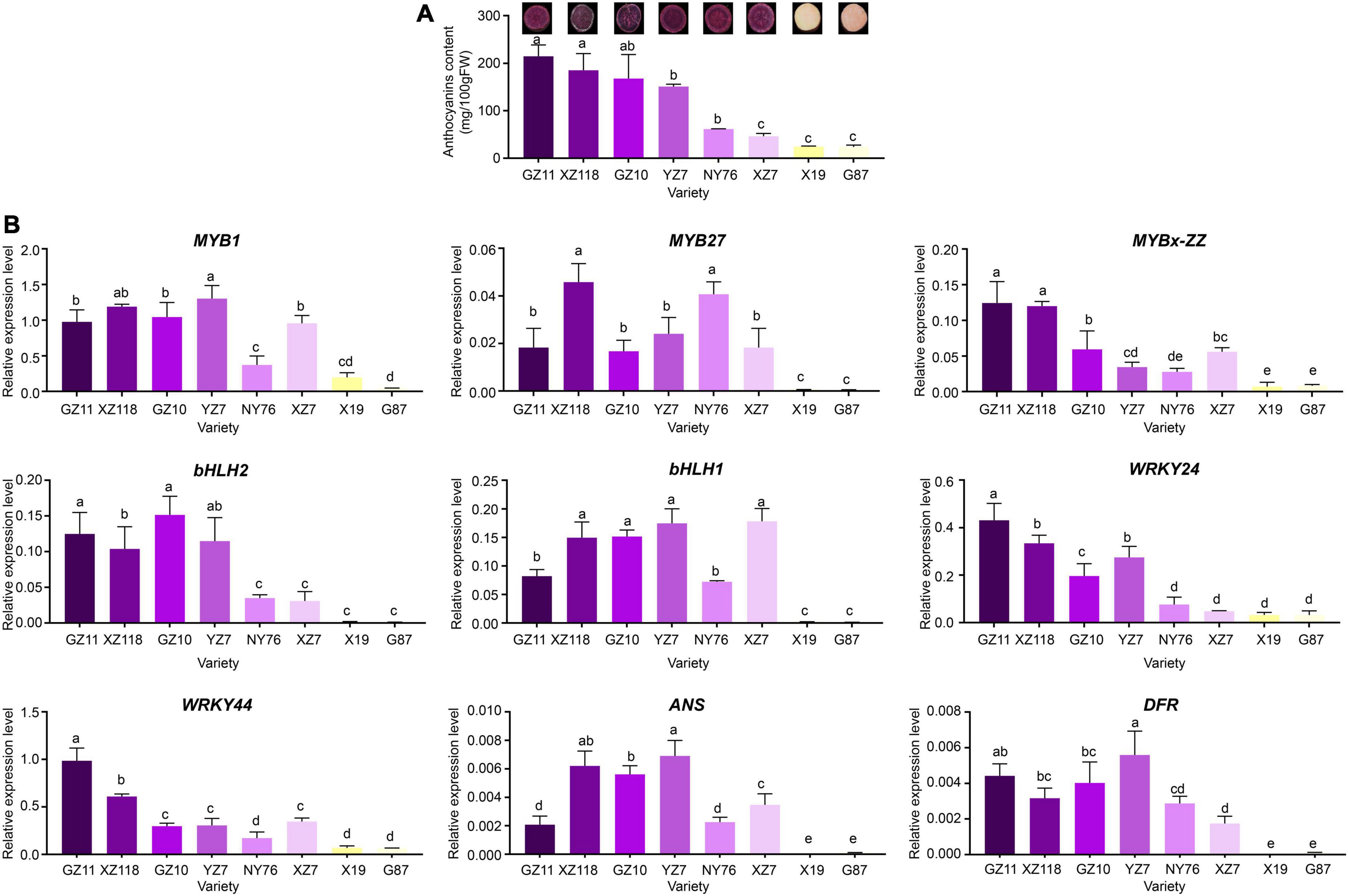
Figure 6. Total anthocyanin concentration and relative expression levels of key genes in various sweet potato accessions. (A) Cross-section of mature tuberous roots flesh and anthocyanin contents of the eight cultivars. The X-axis represents the cultivars, and the Y-axis represents the contents of anthocyanins. (B) Relative expression level of TFs and structural genes in the tuberous roots of the eight cultivars. The X-axis represents the gene, and the Y-axis represents the relative expression level of the gene. Error bars indicate the standard deviation from three biological replicates. Statistical significance was determined by one-way ANOVA (P-values reported). Significant differences between means (Duncan, P < 0.05) were indicated by different letters above the bar.
Expression levels of IbMYBs, IbbHLHs, IbWRKYs, and structural genes in various Chinese sweet potato cultivars
The expression levels of IbMYB1, IbMYB27, IbMYBx-ZZ, IbbHLH2, IbbHLH1, IbWRKY44, IbWRKY21, and IbWRKY24 and structural genes IbDFR and IbANS were analyzed in Chinese sweet potato cultivars, including 6 purple- and 2 orange-fleshed cultivars, to investigate whether the involvement of IbMYBs, IbbHLHs, and IbWRKYs in the regulation of anthocyanin accumulation was common in purple-fleshed sweet potato (Figure 6A). The expression levels of IbMYB1, IbMYB27, IbMYBx-ZZ, IbbHLH2, IbbHLH1, IbWRKY44, IbWRKY24, IbDFR, and IbANS in the tuberous roots of purple-fleshed sweet potato cultivars were higher than those of orange-fleshed sweet potato cultivars (Figure 6B). The expression levels of these genes and anthocyanin contents were significantly positively correlated (Supplementary Figure 3). These results further indicated that anthocyanin biosynthesis in the storage roots of purple-fleshed sweet potato might be regulated by activators, including IbMYB1, IbbHLH1, IbbHLH2, IbWRKY24, and IbWRKY44, and repressors, including IbMYB27 and IbMYBx-ZZ.
Functional verification of IbWRKY, MYB repressors, and MBW/MBWW complexes
Promoter activation tests were conducted by dual-luciferase assays to further confirm the activities of the screened TFs. Six effectors were generated by inserting 3 MYB (i.e., IbMYB1, IbMYB27, IbMYBx-ZZ), 1 bHLH (i.e., IbbHLH2), 1 WDR (i.e., IbWD40), and 1 WRKY (i.e., IbWRKY44) coding sequences into the pGreenII-62-SK vector. The promoters of IbMYB27, IbMYBx-ZZ, IbDFR, and IbWRKY44 were cloned into the pGreenII-0800-LUC vector to generate a reporter construct (Figure 7A). Vectors were separately infiltrated into N. benthamiana leaves. As shown in Figures 7B[1,2], the IbDFR promoter was activated by IbMYB1 alone or in combination with IbbHLH2 and IbWD40. This activation was repressed by IbMYB27 or IbMYBx-ZZ (Figures 7B[2,3]). IbMYB27 had a strong suppressive effect upon IbDFR promoter activation when coinfiltrated with IbbHLH2 and IbWD40 but only had a modestly suppressive effect upon activation by IbMYB1 (Figure 7B[3]). IbMYBx-ZZ also had a strong suppressive effect upon IbDFR promoter activation when coinfiltrated with IbbHLH2 and IbWD40 but only had a weakly suppressive effect upon activation by IbMYB1 (Figure 7B[3]). Besides, IbMYB27 showed a stronger suppression affect than IbMYBx-ZZ especially when they were coinfiltrated with IbMYB1 alone (Figure 7B[3]). At the same time, the promoters of IbMYBx-ZZ and IbMYB27 were activated by IbMYB1 alone or in combination with IbbHLH2 and IbWD40, and the activation effect of IbMYB1 together with IbbHLH2 and IbWD40 was stronger than that of IbMYB1 alone (Figure 7B[4]). In addition, the promoter of IbWRKY44 was activated by IbbHLH2 alone or in combination with either IbMYB1 or IbMYB1 and IbWD40 (Figure 7B[5]). The promoter of IbDFR was also activated by IbWRKY44 alone, but the activation effect of IbWRKY44 in combination with IbMYB1 was stronger than that of IbWRKY44 or IbMYB1 alone (Figure 7B[6]).
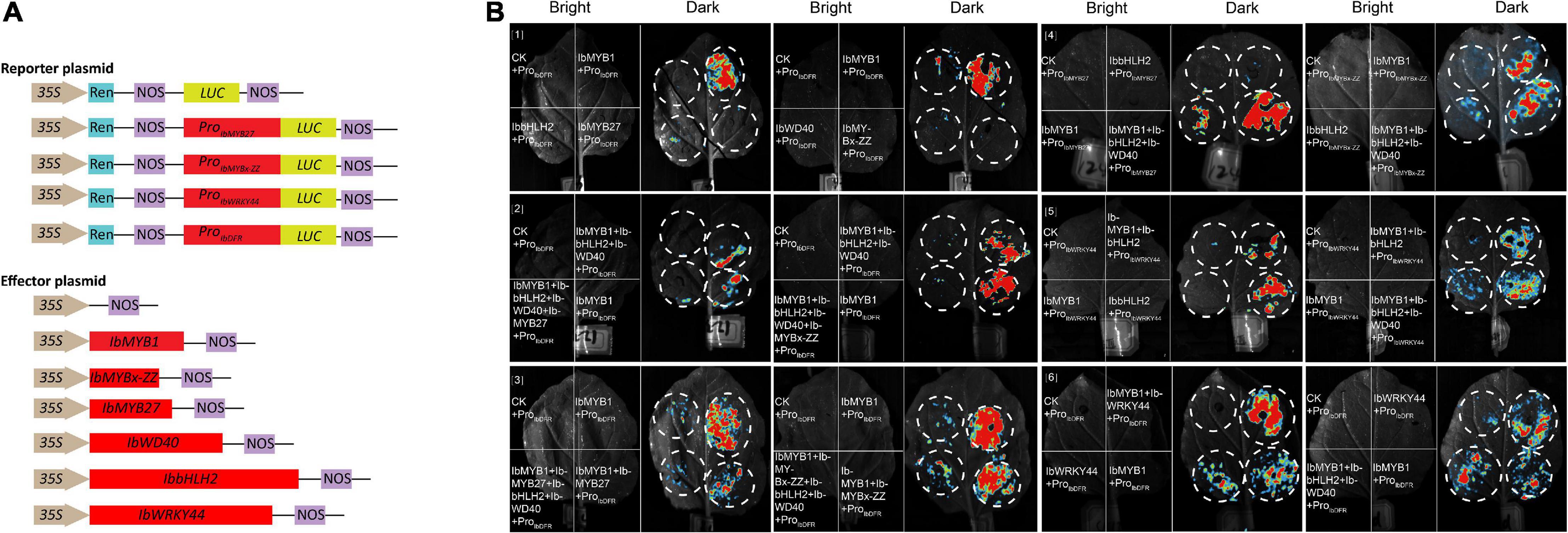
Figure 7. Identification of IbMYB1, IbMYB27, IbMYBx-ZZ, IbWRKY44, IbWD40, and IbbHLH2 transactivation activities of IbDFR, IbMYB27, IbMYBx-ZZ, and IbWRKY44 promoters by a dual-luciferase assay. (A) Schematic of reporter and effector constructs used in transient expression assay. (B) Transient expression assays showing that different TFs activates or represses the transcriptional function of promoters. Luminescence images of Nicotiana benthamiana leaves are shown 48 h after coinfiltration with the constructs indicated in the left. The red color represents strong signal, whereas the blue color represents weak signal.
Y2H and BiFC assays were performed to determine how TFs, including IbMYB1, IbMYB27, IbMYBx-ZZ, IbbHLH1, IbHLH2, and IbWRKY44, were involved in anthocyanin biosynthesis regulation. A summary of the interactions and their relative strengths is shown in Figure 8. The Y2H experiment result indicated that IbMYB1 interacted with IbbHLH1, IbbHLH2, or IbWD40 protein (Figure 8A). However, IbMYB27 interacted with IbWD40, and IbMYBx-ZZ interacted with IbbHLH1 or IbbHLH2 (Figure 8A). IbWRKY44 interacted with IbMYB1, IbbHLH1, or IbbHLH2. However, no interaction between IbWD40 and IbWRKY44, IbbHLH1, or IbbHLH2 was observed (Supplementary Figure 4A). These interaction results were confirmed by BiFC assays (Figures 8B; Supplementary Figures 4B,C).
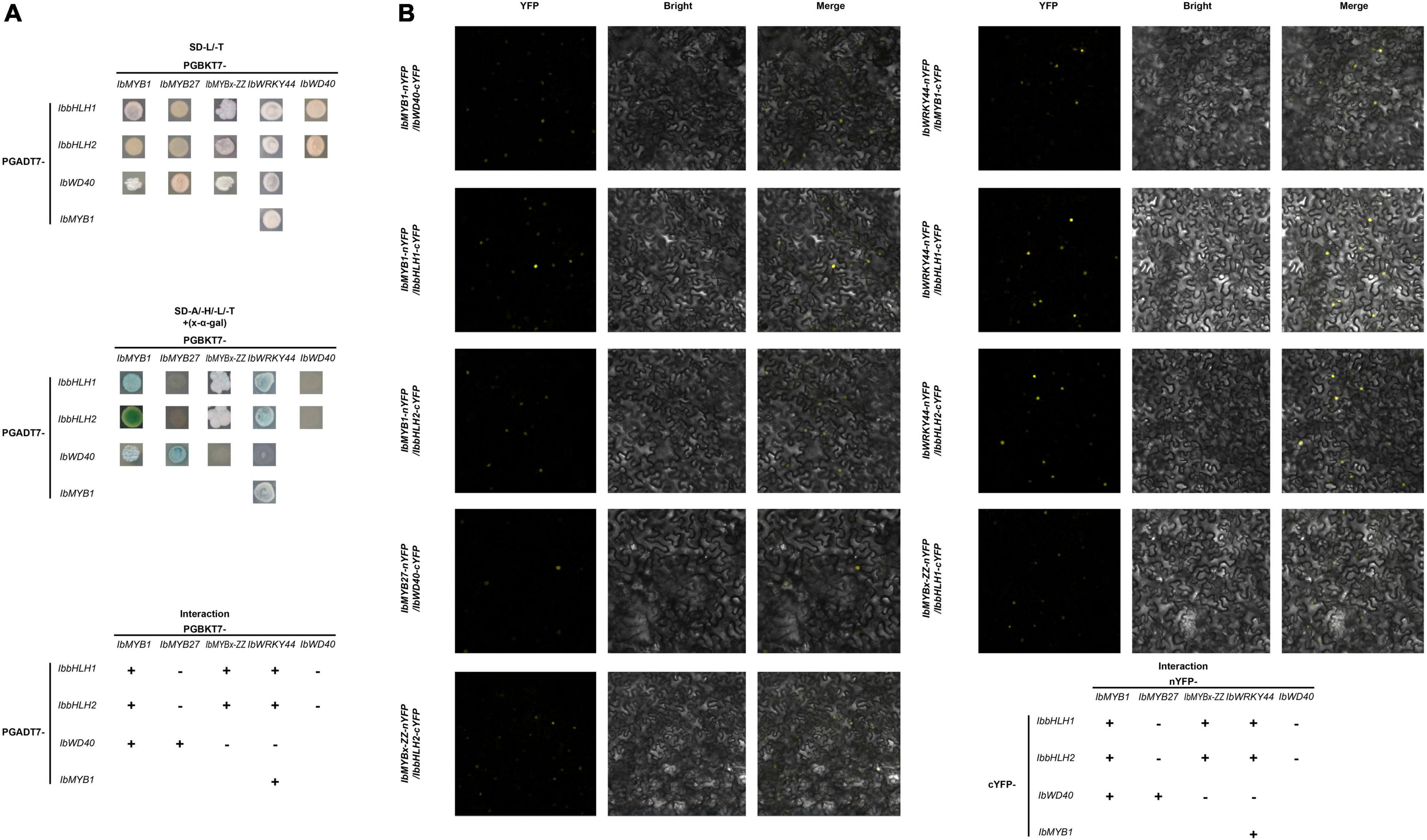
Figure 8. Interactions of IbMYB1, IbMYB27, IbMYBx-ZZ, and IbWRKY44 with IbWD40, IbbHLH1, or IbbHLH2 in vivo. (A) Interactions of IbMYB1, IbMYB27, IbMYBx-ZZ, and IbWRKY44 with IbWD40, IbbHLH1, or IbbHLH2 in vivo by a yeast two-hybrid (Y2H) assay. (B) Verification of the interaction of IbMYB1, IbMYB27, IbMYBx-ZZ, and IbWRKY44 with IbWD40, IbbHLH1, or IbbHLH2 in vivo by a bimolecular fluorescence complementation (BiFC) assay. Images show overlays of fluorescence and light views. “+” represents positive interaction between these two TFs, whereas “-” represents negative interaction.
Discussion
The MBW activation complex, which is composed of MYB, bHLH, and WD40 (WDR) protein, is conserved in multiple plant species to activate anthocyanin biosynthesis (Gonzalez et al., 2010; Albert et al., 2014; Li et al., 2021b). The R2R3-MYB factors in the complex are the most crucial for the spatial and temporal localization of anthocyanins (Davies et al., 2012). IbMYB1 controls anthocyanin biosynthesis in tuberous roots (Mano et al., 2007), and its expression is essential for the purple color of leaf and storage root in sweet potato (Zhang et al., 2021a). Based on the sequences of coding regions, IbMYB1 amplified products from a purple-fleshed sweet potato, Ayamurasaki, were classified into two genotypes, IbMYB1-1-type and IbMYB1-2-type, whereas IbMYB1 amplified product from its spontaneous mutant, AYM96, belonged to IbMYB1-1-type (Tanaka et al., 2012). Genome-wide analysis of expression quantitative trait loci (eQTLs) reveals one eQTL for IbMYB1-2 on chromosome 12 associated with flavonoid biosynthesis was the most promising candidate gene responsible for flesh color variation in sweet potato (Zhang et al., 2020). However, IbMYB1-1 and IbMYB1-2 shared the same coding sequences, only IbMYB1-2 is identified to be responsible for anthocyanin accumulation in tuberous roots (Tanaka et al., 2012). Further cloning and sequencing of the flanking regions of IbMYB1 sequences showed that the large differences in the flanking sequences, especially the differences in 5′ flanking sequences resulted in the functional difference (Tanaka et al., 2012). To determine the genotype of IbMYB1, fragments of different sizes were amplified from the 5′ flanking sequence of IbMYB1-1 and IbMYB1-2 using specific primers (Tanaka et al., 2012). In our research, the same PCR assays were performed on ZZP and its mutant, XLY, the missing IbMYB1-2 in the genome of XLY mutant caused evident pigmentation and anthocyanin accumulation losses in its tuberous roots (Figure 1). This finding was similar to that observed in a previous report (Tanaka et al., 2012). Besides, the expression levels of IbMYB1, IbbHLH2, and structural genes in XLY were significantly down-regulated compared with those in wild-type ZZP (Figures 2B,C). Moreover, IbMYB1 and IbbHLH2 were highly expressed in tuberous roots of six Chinese purple-fleshed sweet potato cultivars, and the expression levels of IbMYB1 (R2 = 0.56, p < 0.0001) and IbbHLH2 (R2 = 0.7873, p < 0.0001) showed significantly positive correlations with anthocyanin contents of eight Chinese sweet potato cultivars (Supplementary Figure 3). Although the expression of IbbHLH1 in XLY was not significantly down-regulated compared with that in ZZP by RNA-seq (data not shown), IbbHLH1 showed high expression level in all Chinese purple-fleshed sweet potato cultivars (Figure 6). Previous research showed that PhJAF13 (bHLH1) and PhAN1 (bHLH2) are involved in the anthocyanin accumulation in leaf and flower in petunia and that the R2R3-MYB activator, PhWDR (WD40), and PhJAF13 proteins form an MBW complex to activate the expression of PhAN1 (bHLH2). Then, the R2R3-MYB activator, PhWDR, and PhAN1 (bHLH2) protein form a core MBW complex to activate the expression of the anthocyanin biosynthesis genes and anthocyanin biosynthesis (Albert et al., 2014). DcMYB7 controls purple pigmentation in carrot roots by regulating its DcbHLH3 partner and the tested anthocyanin biosynthetic structural gene (Xu et al., 2019). Similarly, IbMYB1 interacted with IbbHLH1, IbbHLH2, or IbWD40 and might form a MYB1-bHLH1-WD40 or MYB1-bHLH2-WD40 complex in our results (Figure 8). Moreover, the MYB1-bHLH2-WD40 complex and IbMYB1 could activate the promoter of IbDFR (Figures 7B[1,2]). Besides the MBW complex, WRKY TFs are identified in some plants to regulate anthocyanin biosynthesis (Li et al., 2021a). For example, MdWRKY40, MdWRKY11, PyWRKY26, PbWRKY75, StWRKY13, and PhPH3 have been identified to promote anthocyanin accumulation (Verweij et al., 2016; Wang et al., 2018; An et al., 2019; Liu et al., 2019; Li et al., 2020; Zhai et al., 2021; Zhang et al., 2021b). In the present study, IbWRKY24, IbWRKY44, and IbWRKY21 homologous to MdWRKY11, PyWRKY26, StWRKY13, and PhPH3 (Figure 4) were found to be markedly downregulated in XLY (Figure 2C). Moreover, IbWRKY24 and IbWRKY44 were highly expressed in tuberous roots of all selected Chinese purple-fleshed sweet potato cultivars (Figure 6). The expression levels of IbWRKY24 (R2 = 0.7976, p < 0.0001) and IbWRKY44 (R2 = 0.6272, p < 0.0001) were significantly correlated with anthocyanin contents in the tuberous roots of Chinese sweet potato cultivars (Supplementary Figure 3). Additionally, IbWRKY44 could activate the promoter of IbDFR, and the coinfiltration of IbMYB1 and IbWRKY44 showed stronger activation than IbWRKY44 alone (Figure 7B[6]). Previous research reported that WRKY TFs are regulated by the MBW activation complex in Arabidopsis and Petunia and that the R2R3-MYB activator, WDR, bHLH, and WRKY protein form a MBWW complex to activate anthocyanin biosynthesis (Gonzalez et al., 2016; Verweij et al., 2016). The MBW activation complex containing bHLH2 activates the promoters of IbbHLH2 and PhAN1 (bHLH2) in sweet potato and petunia, and this activation serves as reinforcement regulation (Albert et al., 2014; Deng et al., 2020). In our results, the IbWRKY44 promoter was activated by MYB1-bHLH2-WD40 (Figure 7B[5]), and IbWRKY44 could interact with MYB1 and bHLH2, which were two members of the MYB1-bHLH2-WD40 activation complex. Thus, IbWRKY44 might form a MYB1-bHLH2-WD40-WRKY activation complex to activate anthocyanin biosynthesis in the tuberous root of purple-fleshed sweet potato. Moreover, the IbWRKY44 promoter was activated by bHLH2 and not by IbMYB1 (Figure 7B[5]).
Unlike MYB activators that induce anthocyanin accumulation, a fairly large number of MYB repressors that inhibit anthocyanin accumulation have been identified in the last decade (Lafountain and Yuan, 2021). In petunia, as a member of Convolvulaceae, repressors PhMYB27 (R2R3-MYB) and PhMYBx (R3-MYB) are identified to repress anthocyanin accumulation (Albert et al., 2014). Similarly, Deng et al. (2020) found that MYB repressor genes IbMYB27 and IbMYBx are highly expressed in young purple leaves, but their repression functions are not identified in their research. In the present paper, IbMYBx-ZZ, which coded a novel R3-MYB repressor, and IbMYB27 in the tuberous roots of ZZP were up-regulated compared with those in XLY, accompanied by the high expression of IbMYB1 (Figure 2C). IbMYB27 and IbMYBx-ZZ were highly expressed in the tuberous roots of all selected purple-fleshed sweet potato cultivars (Figure 6). The ectopic expression result showed that IbMYBx-ZZ and IbMYB27 repressed anthocyanin accumulation activated by IbMYB1 (Figure 5). Moreover, IbMYB27 and IbMYBx-ZZ repressed the activation of IbDFR promoter that was activated by IbMYB1 or MYB1-bHLH2-WD40 (Figures 7B[2,3]). At the same time, the promoters of IbMYB27 and IbMYBx-ZZ were activated by IbMYB1 or MYB1-bHLH2-WD40 (Figure 7B[4]), and these activations allowed for the feedback repression of anthocyanin biosynthesis. In petunia, PhMYB27 interacts with bHLH and is incorporated or binds to MBW complexes, thus changing the complex activity from activation to repression (Albert et al., 2014). MYBx and other R3-MYB, as competitive inhibitors, bind the bHLH that requires the formation of MBW complexes (Kroon, 2004; Pesch and Hülskamp, 2004; Wester et al., 2009). IbMYBx-ZZ interacted with IbbHLH1 and IbbHLH2 in our result (Figure 8). In petunia, MYB27 shows repression affected on activation of the DFR promoter when MYB27 is coinfiltrated with bHLH and DPL(MYB), and repression by MYB27 requires the ERF-associated amphiphilic repression (EAR) motif and the formation of the MBW complex. However, in their study, the effect of MYB27 repression on the activation of the DFR promoter by DPL alone is not determined. In our results, IbMYB27’s suppressive effect on the IbDFR promoter activation by IbMYB1 or by MBW activation complex was tested. IbMYB27 had a strong suppression to IbDFR promoter activation by the MBW activation complex and only a modest suppression on the activation by IbMYB1 (Figure 7B[3]). IbMYBx-ZZ also had a strong suppression to IbDFR promoter activation by MBW activation complex and a weak suppression on the activation by IbMYB1 (Figure 7B[3]). These results suggested that IbMYB27, similar to PnMYB27, contained the EAR motif and mediated transcriptional repression by binding to the promoter of target genes either directly itself or as part of a DNA-binding complex (Ohta et al., 2001; Kagale et al., 2010), just the later pattern showed a strong suppressive effect. However, IbMYB27 might repress anthocyanin biosynthesis in the tuberous root of purple-fleshed sweet potato by inhibiting the formation of active MBW complex via binding to WD40 (Figure 8). Thus, MYB27 had a potential double-lockdown mechanism for reducing anthocyanin production. However, IbMYBx-ZZ, a R3-MYB factor containing a single MYB repeat and lacking any repressive motif, asserted its repressive function only through competition for WD40 partner with IbMYB1 (Figure 7B[3]).
Overall, through phenotype, genotype, and transcriptome analyses carried on the purple-fleshed sweet potato ZZP and its mutant XLY, the candidate TFs that involved anthocyanin accumulation in the tuberous root of purple-fleshed cultivars, including IbMYB1, IbbHLH2, IbWRKY24, IbWRKY44, IbWRKY21, IbMYB27, and IbMYBx-ZZ, were screened. Given that TFs shared a common expression pattern in various purple-fleshed sweet potato varieties, the anthocyanin biosynthesis of tuberous roots was likely regulated by IbMYB1, IbWD40, IbHLH1, IbbHLH2, IbWRKYs (i.e., IbWRKY24, IbWRKY44, IbWRKY21), IbMYB27, and IbMYBx-ZZ in sweet potato. Further functional verification of the above TFs was conducted by Y2H, BiFC, and dual-luciferase assays. These tests showed that IbMYB1 was the central determinant of the MBW complex and that IbbHLH2, IbWRKYs, and repressors IbMYB27 and IbMYBx-ZZ were activated by the MBW/MBWW activation complex (Figure 9). This regulatory network provides reinforcement and feedback regulation to maintain the level of anthocyanin accumulation in the tuberous roots of purple-fleshed sweet potato similar to other plants (Albert et al., 2014; Verweij et al., 2016; Peng et al., 2020).
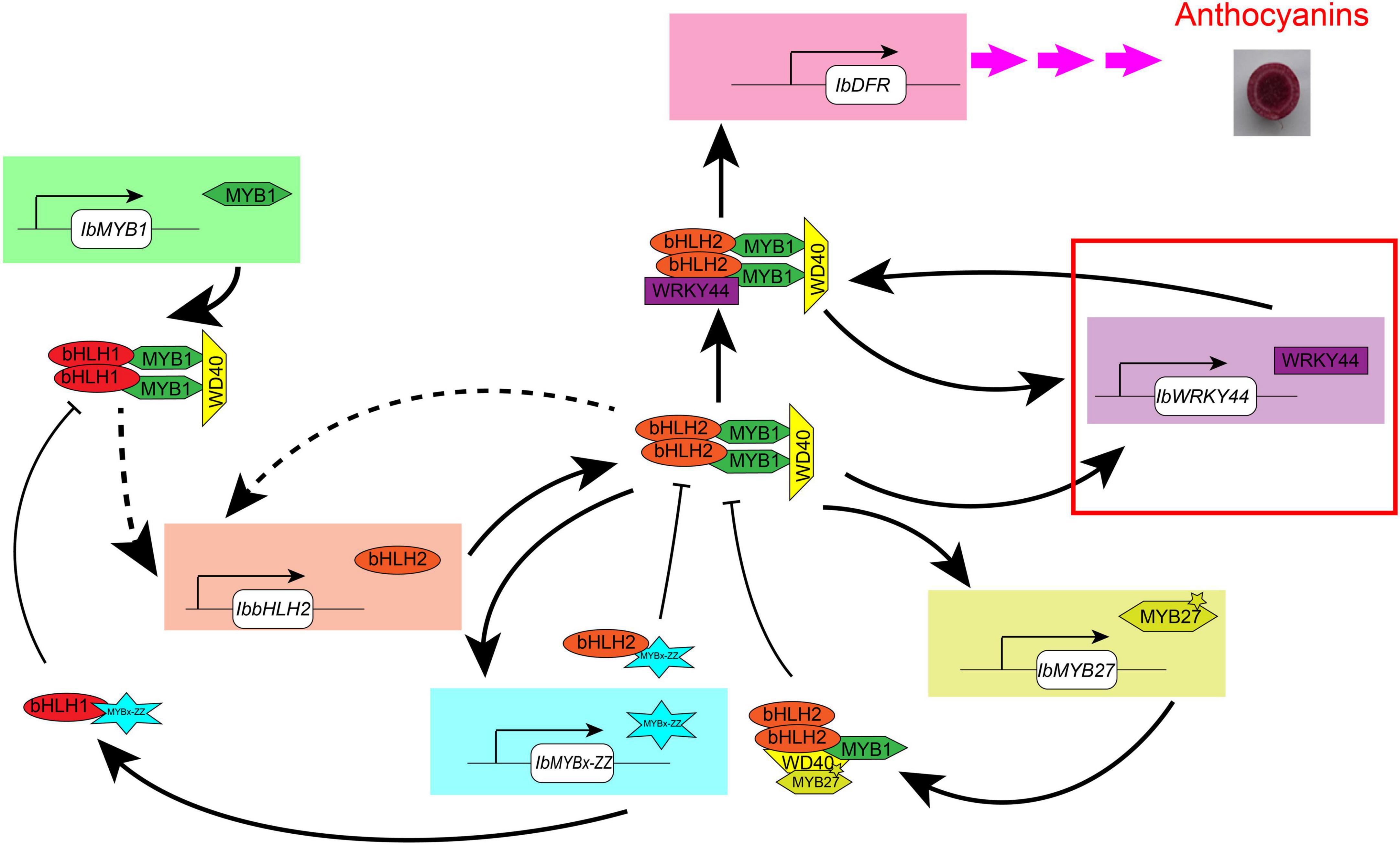
Figure 9. Model of the anthocyanin regulation network in tuberous roots of sweet potato. This model is modified in accordance with the method of Albert et al. (2014). IbMYB1 is expressed and then forms a MBW activation complex with bHLH1 and WD40, which activate the expression of IbbHLH2. Then, a core MBW activation complex containing bHLH2 forms combines with WRKY44 protein, forming a MBWW complex, and activates the expression of the anthocyanin biosynthesis genes, ultimately resulting in anthocyanin accumulation. The MBWW complex can reinforce the expression of IbWRKY44. The core MBW activation complex can also reinforce the expression of IbbHLH2 and activate the expression of IbWRKY44 and MYB repressors, like IbMYBx-ZZ and IbMYB27. Then, MYB repressors collaborate with bHLH and WD40 protein and inhibit the expression of anthocyanin biosynthesis genes. Arrow with solid line represents regulation proven in sweet potato leaves, and dotted arrow represents the regulation that has not been proven yet. The red rectangle represents novel TF that we found to be involved in anthocyanin biosynthesis in sweet potato.
Conclusion
The comparative transcriptome analysis of purple sweet potato and its yellow mutant were carried out. The missing IbMYB1-2 caused a dramatic downregulation of the expression of TFs IbMYB1, IbbHLH2, IbWRKY24, IbWRKY44, IbWRKY21, IbMYB27, and IbMYBx-ZZ and structural genes and ultimately resulted in a dramatic decrease in anthocyanin content. The functions of repressing anthocyanin accumulation of IbMYB27 and IbMYBx-ZZ were identified by transient transformation in tobacco leaves. The expression patterns of TFs and structural genes in the tuberous root of Chinese sweet potato cultivars showed that the expression levels of IbMYB1, IbbHLH1, IbbHLH2, IbWRKY24, IbWRKY44, IbMYB27, and IbMYBx-ZZ were significantly positively correlated with anthocyanin contents in tuberous roots. Further functional verification of the above TFs was conducted by Y2H, BiFC, and dual-luciferase assays. These tests showed that the MBW/MBWW complex activated the protomers of anthocyanin structural gene IbDFR and the promoters for IbWRKY44, IbMYB27, and IbMYBx-ZZ. These results indicated reinforcement and feedback regulation and provided valuable information for the regulation of anthocyanin biosynthesis in purple-fleshed sweet potato.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA837528.
Author contributions
WD, LT, and XH designed the study. WD, LT, and YP performed the experiments. YL and YQ analyzed the data. WD wrote the manuscript. XX and XH funded the research project. All authors read and approved the manuscript.
Funding
This research was funded by the Basic Research Project of Changsha Science and Technology Plan (KQ2004029) and the China Agriculture Research System Program under project number CARS-09-ES16 (awarded to XH).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.924379/full#supplementary-material
Supplementary Figure 1 | Volcano plots and KEGG classification of DEGs from ZZP and XLY. (A) Volcano plots displaying the upregulated (red), downregulated (green), and unregulated (blue) genes in XLY compared with those in ZZP. (B) KEGG enrichment of DEGs between ZZP and XLY. The X-axis represents the rich factor corresponding to the pathway, and the Y-axis represents the pathway name. The size of the q-value is represented by the dot color. A small q-value indicates close color to red. The number of DEGs contained in each pathway is represented by the dot size.
Supplementary Figure 2 | Phylogenetic analysis and protein sequence alignment for putative MYBs with function-verified homologous genes of other plant species. (A) Phylogenetic relationship of putative MYBs in sweet potato and function-verified anthocyanin-related MYBs in other species. Orange- and green-shaded areas indicate R2R3-MYB activators and repressors, respectively. The blue-shaded area shows R3-MYB repressors. The pentagram represents IbMYBx-ZZ. (B) Protein sequence alignment for the putative MYBs with function-verified homologous genes of other plant species. Black lines represent the R2 and R3 domains.
Supplementary Figure 3 | Correlation between contents of anthocyanins and relative expression level of nine DEGs in the tuberous roots of sweet potato. The X-axis represents the contents of anthocyanins, and the Y-axis represents the relative expression level of the gene.
Supplementary Figure 4 | Self-activated test of IbMYB1, IbMYB27, IbMYBx-ZZ, IbWRKY44, IbWD40, IbbHLH2, and IbbHLH1 in (A) Y2H and (B) BiFC experiments. (C) No fluorescence signal detected in IbMYBx-ZZ-IbWD40, IbbHLH1-IbWD40, IbbHLH2-IbWD40, IbWRKY44-IbWD40, IbMYB27-IbbHLH1, and IbMYB27-IbbHLH2 pairs.
Supplementary Table 1 | Primers used in this manuscript.
Supplementary Table 2 | Summary of RNA-seq data.
Supplementary Table 3 | Promoter sequences of IbMYB27, IbMYBx-ZZ, IbWRKY44, and IbDFR.
Supplementary Table 4 | Sequences of IbMYBx-ZZ, IbWRKY21, IbWRKY24, and IbWRKY44.
Supplementary Table 5 | Protein sequences used in Figure 4 and Supplementary Figure 2.
Footnotes
References
Aharoni, A., Vos, C., Wein, M., Sun, Z., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyaninsand flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/j.1365-313X.2001.01154.x
Albert, N., Davies, K., Lewis, D., Zhang, H., and Schwinn, K. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanins pigmentation in eudicots. Plant Cell 26, 962–980. doi: 10.1105/tpc.113.122069
Albert, N., Lewis, D., Zhang, H., Schwinn, K., Jameson, P., and Davies, K. (2011). Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65, 771–784. doi: 10.1111/j.1365-313X.2010.04465.x
Alessandra, A., Erika, C., Sara, Z., Finezzo, L., Maura, B., Benedetto, R., et al. (2016). A Grapevine TTG2-Like WRKY Transcription Factor Is Involved in Regulating Vacuolar Transport and Flavonoid Biosynthesis. Front. Plant Sci. 7:1979. doi: 10.3389/fpls.2016.01979
An, J., Zhang, X., You, C., Bi, S., Wang, X., and Hao, Y. (2019). Mdwrky40 promotes wounding-induced anthocyanins biosynthesis in association with MdMYB1 and undergoes MdBT2-mediated degradation. New Phytol. 224, 380–395. doi: 10.1111/nph.16008
Chu, H., Jeong, J., Kim, W., Chung, D., Jeon, H., Ahn, Y., et al. (2013). Expression of the sweetpotato R2R3-type IbMYB1a gene induces anthocyanin accumulation in Arabidopsis. Physiol. Planta. 148, 189–199. doi: 10.1111/j.1399-3054.2012.01706.x
Davies, K., Albert, N., and Schwinn, K. (2012). From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 39, 619–638. doi: 10.1071/FP12195
Deng, J., Wu, D., Shi, J., Balfour, K., Wang, H., Zhu, G., et al. (2020). Multiple MYB Activators and Repressors Collaboratively Regulate the Juvenile Red Fading in Leaves of Sweetpotato. Front. Plant Sci. 11:941. doi: 10.3389/fpls.2020.00941
Endrias, D., Negussie, R., and Gulelat, D. (2016). Comparison of Three Sweet Potato (Ipomoea Batatas (L.) Lam) varieties on Nutritional and Anti-Nutritional Factors. Glob. J. Sci. Front. Res. 16, 7–19.
Eulgem, T., Rushton, P., Robatzek, S., and Somssich, I. (2000). The wrky superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
FAOSTAT, (2020). Production of Crops. Available online at: http://www.fao.org/faostat/en/#home (accessed February 17, 2022).
Gonzalez, A., Brown, M., Hatlestad, G., Akhavan, N., Smith, T., Hembd, A., et al. (2016). TTG2 controls the developmental regulation of seed coat tannins in arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev. Biol. 419, 54–63. doi: 10.1016/j.ydbio.2016.03.031
Gonzalez, A., Zhao, M., Leavitt, J., and Lloyd, A. (2010). Regulation of the anthocyanins biosynthetic pathway by the TTG1/bHLH/MYB transcriptional complex in arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Bæksted Holme, I., Dionisio, G., and Brinch-Pedersen, H. (2021). A roadmap to modulated anthocyanin compositions in carrots. Plants 10:472. doi: 10.3390/plants10030472
Holton, T., and Cornish, E. (1995). Genetics and biochemistry of anthocyanins biosynthesis. Plant Cell 7, 1071–1083. doi: 10.1105/tpc.7.7.1071
Hoshino, A., Jayakumar, V., Nitasaka, E., Toyoda, A., Noguchi, H., Itoh, T., et al. (2016). Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat. Commun. 7:13295. doi: 10.1038/ncomms13295
Kagale, S., Links, M., and Rozwadowski, K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in. Plant Physiol. 152, 1109–1134. doi: 10.1104/pp.109.151704
Kroon, A. (2004). Transcription Regulation of the Anthocyanin Pathway in Petunia Hybrida. (Ph.D.thesis). Amsterdam: Vrije Universiteit.
Lafountain, A., and Yuan, Y. (2021). Repressors of anthocyanins biosynthesis. New Phytol. 231, 933–949. doi: 10.1111/nph.17397
Li, C., Wu, J., Hu, Ka, Wei, S., Sun, H., Hu, L., et al. (2020). PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanins biosynthesis and transport in red-skinned pears. Horticult. Res. 7:37. doi: 10.1038/s41438-020-0254-z
Li, L., Wei, Z., Zhou, Z., Zhao, D., Tang, J., Yang, F., et al. (2021b). A single amino acid mutant in the EAR motif of IbMYB44.2 reduced the inhibition of anthocyanins accumulation in the purple-fleshed sweetpotato. Plant Physiol. Biochem. 167, 410–419. doi: 10.1016/j.plaphy.2021.08.012
Li, D., Wang, Z., Sun, S., Xiao, K., Cao, M., Li, X., et al. (2021a). VvMYB15 and VvWRKY40 Positively Co-regulated Anthocyanins Biosynthesis in Grape Berries in Response to Root Restriction. Front. Plant Sci. 12:789002. doi: 10.3389/fpls.2021.789002
Liu, W., Wang, Y., Yu, L., Jiang, H., Guo, Z., Xu, H., et al. (2019). MdWRKY11 Participates in Anthocyanins Accumulation in Red-Fleshed Apples by Affecting MYB Transcription Factors and the Photoresponse Factor MdHY5. J. Agric. Food Chem. 67, 8783–8793. doi: 10.1021/acs.jafc.9b02920
Liu, Y., Wang, K., Espley, R., Wang, L., Yang, H., Yu, B., et al. (2016). Functional diversification of the potato R2R3 myb anthocyanins activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 67, 2159–2176. doi: 10.1093/jxb/erw014
Mano, H., Ogasawara, F., Sato, K., and Minobe, H. (2007). Isolation of a regulatory gene of anthocyanins biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol. 143, 1252–1268. doi: 10.1104/pp.106.094425
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class ii erf transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. doi: 10.1105/TPC.010127
Peng, Y., Thrimawithana, A., Cooney, J., Jensen, D., and Allan, A. (2020). The proanthocyanin-related transcription factors mybc1 and wrky44 regulate branch points in the kiwifruit anthocyanin pathway. Sci. Rep. 10:14161. doi: 10.1038/s41598-020-70977-0
Pesch, M., and Hülskamp, M. (2004). Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr. Opin. Genet. Dev. 14, 422–427. doi: 10.1016/j.gde.2004.06.007
Quattrocchio, F., Wing, J., Woude, K., Souer, E., Vetten, N., and Koes, M. (1999). Molecular analysis of the anthocyanins 2 gene of petunia and its role in the evolution of flower color. Plant Cell 11, 1433–1444. doi: 10.2134/agronj1997.00021962008900030003x
Salvatierra, A., Pimentel, P., Alejandra, M., and Herrera, R. (2013). Increased accumulation of anthocyanins in Fragaria chiloensis fruits bytransient suppression of FcMYB1 gene. Phytochemistry 90, 25–36. doi: 10.1016/j.phytochem.2013.02.016
Sun, R., and Guo, H. (2008). Comparision of Anthocyanins Content from Storage Roots of Seven Purple Sweet Potato Varieties. Crops 6, 56–59. doi: 10.1007/s00376-008-1009-1
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tanaka, M., Takahata, Y., Kurata, R., Nakayama, H., and Yoshinaga, M. (2012). Structural and functional characterization of IbMYB1 genes in recent japanese purple-fleshed sweetpotato cultivars. Mol. Breed. 29, 565–574. doi: 10.1007/s11032-011-9572-z
Verweij, W., Spelt, C., and Bliek, M. (2016). Functionally similar wrky proteins regulate vacuolar acidification in petunia and hair development in arabidopsis. Plant Cell 28, 786–803. doi: 10.1105/tpc.15.00608
Wang, N., Liu, W., Zhang, T., Jiang, S., Xu, H., Wang, Y., et al. (2018). Transcriptomic analysis of red-fleshed apples reveals the novel role of mdwrky11 in flavonoid and anthocyanins biosynthesis. J. Agric. Food Chem. 66, 7076–7086. doi: 10.1021/acs.jafc.8b01273
Wester, K., Digiuni, S., Geier, F., Timmer, J., Fleck, C., and Hülskamp, M. (2009). Functional diversity of R3 single-repeat genes in trichome development. Development 136, 1487–1496. doi: 10.1242/dev.021733
Wu, S., Lau, K., Cao, Q., Hamilton, J., Sun, H., Zhou, C., et al. (2018). Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat. Commun. 9:4580. doi: 10.1038/s41467-018-06983-8
Xu, Z., Yang, Q., Feng, K., and Xiong, A. (2019). Changing carrot color: insertions in dcmyb7 alter the regulation of anthocyanin biosynthesis and modification. Plant physiol. 181, 195–207. doi: 10.1104/pp.19.00523
Yang, J., Moeinzadeh, M., Kuhl, H., Helmuth, J., Xiao, P., Haas, S., et al. (2017). Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 3, 696–703. doi: 10.1038/s41477-017-0002-z
Zhai, R., Wang, Z., Sha, G., Zhang, S., Cong, L., Yang, C., et al. (2021). PbWRKY75 promotes anthocyanin synthesis by activating PbDFR, PbUFGT and PbMYB10b in pear. Physiol. Planta. 173, 1841–1849. doi: 10.1111/ppl.13525
Zhang, D., Tan, Y., Dong, F., Zhang, Y., Huang, Y., Zhou, Y., et al. (2021a). The Expression of IbMYB1 Is Essential to Maintain the Purple Color of Leaf and Storage Root in Sweet Potato [Ipomoea batatas (L.) Lam]. Front. Plant Sci. 12:688707. doi: 10.3389/fpls.2021.688707
Zhang, H., Zhang, Z., Zhao, Y., Guo, D., Zhao, X., Gao, W., et al. (2021b). StWRKY13 promotes anthocyanins biosynthesis in potato (Solanum tuberosum) tubers. Funct. Plant Biol. 49, 102–114. doi: 10.1071/FP21109
Zhang, L., Yu, Y., Shi, T., Kou, M., Sun, J., Xu, T., et al. (2020). Genome-wide analysis of expression quantitative trait loci (eQTLs) reveals the regulatory architecture of gene expression variation in the storage roots of sweet potato. Hortic. Res. 7:90. doi: 10.1038/s41438-020-0314-4
Keywords: sweet potato, tuberous root, anthocyanins, transcription factor, transcriptome analysis
Citation: Dong W, Tang L, Peng Y, Qin Y, Lin Y, Xiong X and Hu X (2022) Comparative transcriptome analysis of purple-fleshed sweet potato and its yellow-fleshed mutant provides insight into the transcription factors involved in anthocyanin biosynthesis in tuberous root. Front. Plant Sci. 13:924379. doi: 10.3389/fpls.2022.924379
Received: 20 April 2022; Accepted: 04 July 2022;
Published: 08 August 2022.
Edited by:
Wilfried Rozhon, Anhalt University of Applied Sciences, GermanyReviewed by:
Nick Albert, The New Zealand Institute for Plant and Food Research Ltd., New ZealandPablo Federico Cavagnaro, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina
Copyright © 2022 Dong, Tang, Peng, Qin, Lin, Xiong and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinxi Hu, aHVfeGlueGlAMTI2LmNvbQ==; Xingyao Xiong, eGlvbmd4aW5neWFvQGNhYXMuY24=
 Wen Dong
Wen Dong Linfei Tang1
Linfei Tang1 Yuan Lin
Yuan Lin Xingyao Xiong
Xingyao Xiong