
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 27 June 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.924044
This article is part of the Research Topic Artificial Polyploidy in Plants, Volume II View all 15 articles
Polyploid breeding is an effective approach to improve plant biomass and quality. Both fast growth and dwarf types of in vitro or ex vitro plants are produced after polyploidization. However, little is known regarding the dwarf type mechanism in polyploids grown in vitro. In this study, the morphological and cytological characteristics were measured in tetraploid and diploid hybrid sweetgum (Liquidambar styraciflua × L. formosana) with the same genetic background. RNA sequencing (RNA-Seq) was used to analyse shoot and root variations between tetraploid and diploid plants; important metabolites were validated. The results showed that the shoot and root lengths were significantly shorter in tetraploids than in diploids after 25 d of culture. Most tetraploid root cells were wider and more irregular, and the length of the meristematic zone was shorter, while tetraploid cells were significantly larger than diploid cells. Differentially expressed genes (DEGs) were significantly enriched in the plant growth and organ elongation pathways, such as plant hormone biosynthesis and signal transduction, sugar and starch metabolism, and cell cycles. Hormone biosynthesis and signal transduction genes, such as YUCCA, TAA1, GH3, SAUR, CPS, KO, KAO, GA20ox, GA3ox, BAS1 and CYCD3, which help to regulate organ elongation, were generally downregulated. The auxin, gibberellin, and brassinolide (BL) contents in roots and stems were significantly lower in tetraploids than in diploids, which may greatly contribute to slow growth in the roots and stems of tetraploid regenerated plants. Exogenous gibberellic acid (GA3) and indole-3-acetic acid (IAA), which induced plant cell elongation, could significantly promote growth in the stems and roots of tetraploids. In summary, comparative transcriptomics and metabolite analysis showed that the slow growth of regenerated tetraploid hybrid sweetgum was strongly related to auxin and gibberellin deficiency. Our findings provide insights into the molecular mechanisms that underlie dwarfism in allopolyploid hybrid sweetgum.
Polyploidy or genome doubling results from the merging of more than two full sets of chromosomes (Comai, 2005; Song and Chen, 2015); polyploidization substantially contributes to plant evolution, new species formation, and plant genetic improvement (Leitch and Leitch, 2008; Jiao et al., 2011). After chromosome doubling, polyploid plants often exhibit multiple variations and generally develop large organs, strong resistance, high crop yield, wood quality, and ornamental value (Zhu et al., 1995; Leitch and Leitch, 2008; Tiku et al., 2014). However, after polyploidization, the changes in plant height differ among species; there is usually a fast growing type (Ni et al., 2009; Miller et al., 2012; Dai et al., 2015; Liao et al., 2016) and a dwarf type (Mu et al., 2012; Tokumoto et al., 2016; Xue et al., 2017). Similar to fast growth, dwarfing is a common phenomenon in tetraploid plants, especially woody plants. It remains unclear why specific species exhibit fast or slow growth after chromosome doubling. Dwarf tetraploid plants have a high ornamental value (Ye et al., 2010); they are often favoured in courtyard and urban landscapes where space may be limited. Such plants often exhibit strong resistance to abiotic and biotic stresses (Tan et al., 2015) and can be used as parents for triploid production.
In addition, fast growth (Gantait et al., 2011; Salma et al., 2018) and slow growth (Gu et al., 2005; Rao et al., 2019) polyploids have been observed in vitro. Regenerated plants of tetraploid Lagerstroemia indica (Tong, 2007), Ziziphus jujube (Gu et al., 2005), Punica granatum (Shao et al., 2003), and Lycium ruthenicum (Rao et al., 2019) exhibit dwarfing or shortened roots, which can affect regeneration efficiency. The transcriptional expression levels of fast and slow-growing polyploids may substantially change after polyploidization (Cheng et al., 2015; Ma et al., 2016). However, there have been few reports (Ma et al., 2016) concerning the molecular mechanism that underlies the in vitro dwarf polyploid type. Moreover, to promote stem and root elongation by improving culture conditions or medium formulation, there is a need to understand which genes and metabolites substantially change in tetraploid plants.
Organ elongation is an important process in plant tissue culture. The degree of difficulty in modifying organ length varies among species (Bizet et al., 2015). Light intensity, agar concentration, growth regulators, and nutrients can affect the lengths of adventitious shoots, stem segments, and roots in tissue culture. Plant hormones have critical effects on organ lengths in vitro; for example, auxin, gibberellin, and brassinolide (BL) can promote root or stem elongation (Su et al., 2011). Tong (2007) found that a diploid medium was unsuitable for in vitro tetraploid plants. After polyploidization, plants exhibit altered hormone synthesis and changes in the levels of metabolites related to signal transduction genes (Mu et al., 2012; Ma et al., 2016).
Forest trees of Liquidambar spp. have important economic and ecological value (Harlow et al., 1996; Merkle and Battle, 2000; Zheng et al., 2015). Chinese sweetgum (Liquidambar formosana Hance) is mainly used for ornamental and medicinal purposes (Chen et al., 2020). American sweetgum (Liquidambar styraciflua L.) is an important feedstock for the timber and paper industries in South America. In a previous study, several clones of hybrid sweetgum demonstrated obvious heterosis in terms of growth rate and wood density (Merkle et al., 2010). For allotetraploid species, the advantages of some traits have been linked to heterosis (Fuentes et al., 2014). Tetraploid hybrid sweetgum (L. styraciflua × L. formosana; Zhang et al., 2017) has been reported to show dwarf traits (Unpublished). Some new varieties of sweetgum, such as the dwarfing cultivar ‘Gumball’, have a shrubby appearance (maximum height of 4.5 m) (Sommer et al., 1999).
What are the key contributors to the dwarf resulting from genome doubling in hybrid sweetgum? Whether the roots and stems of tetraploid hybrid sweetgum have mutated; whether endogenous hormones in plants, the most important factors affecting plant growth, change their regulatory mechanism after polyploidy; whether the growth of dwarf tetraploid can be promoted by application of exogenous plant growth regulators. To answer those questions, in this study, in vitro tetraploid hybrid sweetgum was compared with diploid hybrid sweetgum. Morphological changes in stems and roots were observed at various growth stages; organ variations of tetraploid hybrid sweetgum were analysed at the phenotypic, cytological, and molecular levels. Histological analysis, phytohormone analysis and transcriptome sequencing were also performed to identify mechanisms responsible for the variation.
Three genotypes (Z1, Z2, and Z3) of diploid hybrid sweetgum (D1, D2, and D3) and their tetraploid counterparts (T1, T2, and T3; same genetic background) were used as research material in the present study. In vitro diploid and tetraploid families were obtained by hybridization (L. styraciflua × L. formosana). Artificial tetraploids were acquired from in vitro shoot regeneration via colchicine treatment, as previously described (Zhang et al., 2017). To acquire sufficient clones for each genotype, all materials were derived from tissue culture propagation. Plantlets were rooted in half-strength woody plant medium supplemented with 2.0 mg/L indole butyric acid, 0.1 mg/L naphthaleneacetic acid, 2.0 g/L agar, 4.0 g/L polygel, and 30 g/L sucrose, called primitive rooting medium. The pH was adjusted to 5.8 prior to autoclaving. Vigorously growing leaves of all genotypes were collected; the ploidy level was analysed using a Cyflow Ploidy Analyser (Partec, Görlitz, SN, Germany). Ploidy level was determined in accordance with the methods described by Zhang et al. (2017).
To maintain consistency, adventitious shoots at the same regeneration time were excised and cut to approximately 1.5 cm for rooting. The rooting medium contained half-strength woody plant medium supplemented with 2.0 mg/L indole butyric acid and 0.1 mg/L naphthaleneacetic acid; it was placed in a culture bottle with 50 ml medium. The phenotypes of the plantlets were measured after 0, 25, 50, and 70 d of subculture: 0–25 d was stage I, 25–50 d was stage II, and 50–70 d was stage III. In addition to plant height, ground diameter, number of stem nodes, root length, number of roots, leaf area, petiole length, and number of leaves were measured. The primary root diameter was measured by ImageJ.1 Three biological replicates were performed; each consisted of three genotypes, including three to five plantlets for each genotype.
Leaves (second or third leaf position), roots, stems, and shoot tips of diploid and tetraploid plants at 25, 50, and 70 d were prepared as paraffin sections. Materials were immobilised in formaldehyde alcohol acetic acid solution (5 ml 38% formaldehyde +5 ml glacial acetic acid +90 ml 50% ethanol, 1:1:18, by vol.) for 24 h. The materials were dehydrated in ethanol and pellucidum in xylene, then embedded in paraffin. The slices had a thickness of 8 mm; they were stained with saffron and solid green. All materials were observed under a biomicroscope (BX43, Olympus, Tokyo, Japan) and photographed by an image acquisition and analysis system (DP73, Olympus). The following items were observed: the thicknesses of leaves, spongy tissues, palisade tissues, and main veins; stem diameter and the thicknesses of epidermis, cortex, and vascular column; root diameter, epidermis cortex radius, and central column diameter; upper column diameter and cell transection area of leaf xylem, as well as cell transection areas of the lower epidermis, spongy tissue, and palisade tissue; cell transection areas of xylem, cortex, and phloem, as well as stem pith; and cell transection areas of xylem, epidermis, and root cortex. Three biological replicates were obtained. Each section was randomly measured for 20–30 cells, with three to five replicate measurements. All measurements were performed using ImageJ software.
Four types of RNA were extracted (each containing three genotypes): diploid stem (DS), tetraploid stem (TS), diploid root (DR), and tetraploid root (TR). Three biological replicates were used for total RNA extraction. The 12 samples were named DS-1, DS-2, DS-3, TS-1, TS-2, TS-3, DR-1, DR-2, DR-3, TR-1, TR-2, and TR-3, respectively. All materials were collected after 50 d of subculture, immediately frozen in liquid nitrogen, and stored at −80°C until RNA extraction. RNA extraction was performed in accordance with the methods described by Zhang et al. (2019). RNA purity was checked using a NanoPhotometer® spectrophotometer (IMPLEN, CA, United States). RNA concentrations were measured using a Qubit® RNA Assay Kit in a Qubit®2.0 fluorometer (Life Technologies, Carlsbad, CA, United States).
The construction and sequencing of a cDNA library were performed by Shanghai OE Biotech. Co., Ltd. (China); fragments with lengths of 150–200 bp were selected using an AMPure XP system (Beckman Coulter, Beverly, MA, United States). Library quality was assessed on a bioanalyzer (2100, Agilent, Santa Clara, CA, United States). Nine cDNA libraries were sequenced on a Hiseq 2000 platform (Illumina, CA, United States). High-quality reads were assembled de novo using Trinity software (Grabherr et al., 2011). The final unique consensus sequences were regarded as unigenes. For gene annotation, BLAST was used to align the unigenes to the National Center for Biotechnology Information (NCBI) nonredundant protein database (Nr), NCBI non-redundant nucleotide database (Nt), Swiss-Prot protein database (Swiss-Prot), protein families database (Pfam), Kyoto Encyclopaedia of Genes and Genomes (KEGG), and Clusters of Orthologous Groups of proteins (COG). Gene Ontology (GO)2 annotation was performed using Blast2GO software (version 2.5; Gotz et al., 2008) based on the results of Nr and Pfam annotation. Fragments per kilobase per million base pairs (FPKM) values were used to evaluate the expression patterns of unigenes with the thresholds of |log2Ratio| ≥ 1.0 and false discovery rate ≤ 0.001.
GO enrichment analysis (corrected p < 0.05) of the DEGs was performed with the GOseq R package after the data had been evaluated using the Kolmogorov–Smirnov test. KEGG pathways were assigned to the unigenes and pathway maps using the online KEGG Automatic Annotation Server3 with the bidirectional best-hit method (Moriya et al., 2007). KOBAS software (Mao et al., 2005) was used to determine DEG enrichment in KEGG pathways.
Heat maps were used to visualise gene expression; the maps were constructed using log2-transformed FPKM values in MeV v4.8.1 (Saeed et al., 2003), with Pearson’s correlation as a similarity metric.
Quantitative real-time polymerase chain reaction (qPCR) was conducted in accordance with the methods described by Zhang et al. (2019). Primer sequences are listed in Supplementary Table S1. 18s rRNA was used as the endogenous reference gene; relative expression levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Soluble sugar content was determined by the anthrone colorimetry method (Fukao et al., 2006). Fructose (Fructose assay kit A085-1-1), sucrose (Sucrose measurement kit A099-1-1), glucose (Glucose kit a154-1-1), and starch (Starch content kit A148-1-1) contents were measured using an assay kit (Nanjing Jiancheng Biotechnology Company, Nanjing, China). Units were expressed as mg metabolites per g of plant tissue. The experiment was conducted with three biological and three technical replicates.
Endogenous IAA, GA3, and BL were measured by liquid chromatography-mass spectrometry. Tetraploid and diploid stems and roots (leaves removed) were collected after 50 d of growth, then frozen in liquid nitrogen. IAA and GA3 detection methods were adopted from the work by Chen et al. (2012); the BL detection method was adopted from the work by Ding et al. (2013). Quantum triple quadrupole liquid chromatography-mass spectrometry (Thermo Fisher Scientific, Waltham, MA, United States) was used for plant hormone detection; three biological replicates were included in this experiment.
Adventitious shoots of tetraploid hybrid sweetgum were incubated in three types of new media for 0, 30, or 40 d. Each new medium contained one hormone: 1.5 mg/L IAA, 0.5 mg/L GA3, or 0.1 mg/L epibrassinolide. After determination of the optimal replacement time, the adventitious shoots were placed in rooting media that contained different hormones; eight combinations were used (Supplementary Table S2). There were three replicates in each treatment; each replicate contained 10 plantlets, including five genotypes. Each adventitious shoot was cut to a length of approximately 1.5 cm. After 60–70 d of growth (i.e., when the plant stopped growing), the plant height, ground diameter, internode number, main root number, and root length were measured.
Significant differences among treatments were evaluated by Duncan’s multiple range test using a threshold of p < 0.05, after a homogeneity test had been conducted; all statistical analyses were performed using SPSS version 18.0 (SPSS Inc., Chicago, IL, United States). Histograms and line graphs were constructed using Microsoft Excel 2010 software (Microsoft Corp., Redmond, WA, United States).
The ploidy levels of three genotypes were determined by flow cytometry before morphological and molecular examinations (Figure 1A). The phenotypes of tetraploid and diploid plants were measured after they had been cultured in rooting medium for 0, 25, 50, and 70 d (Table 1; Figure 1B). The results showed that there were no significant differences in ploidy among the plants.
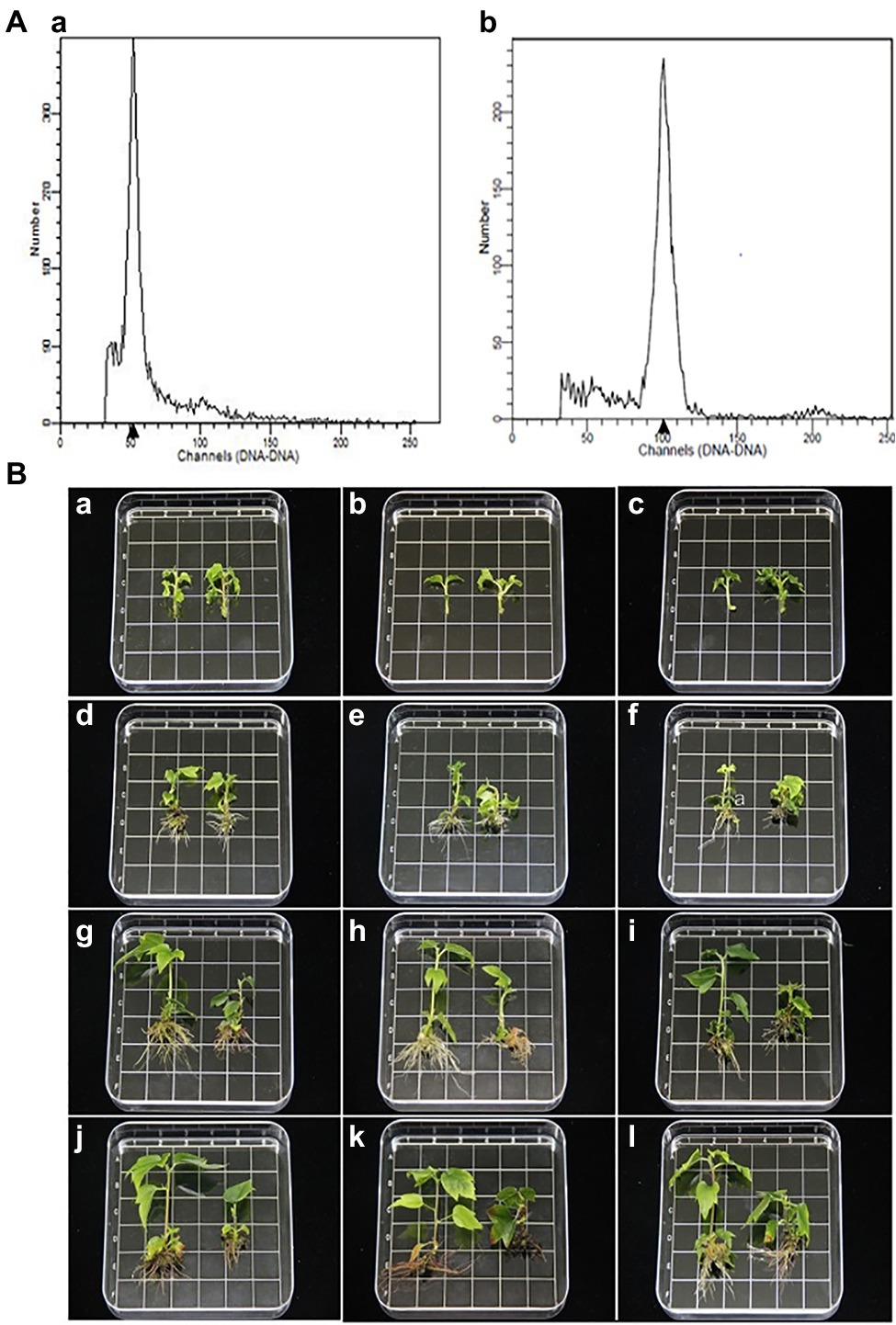
Figure 1. (A) Histograms of flow cytometry findings for Liquidambar styraciflua × Liquidambar formosana: a diploid plant (control); b tetraploid plant. (B) Morphological comparison of diploid and tetraploid hybrid sweetgum plants at various developmental stages of in vitro growth. a,d,g,j Z1; b,e,h,k Z2; c,f,i,l Z3. a–c: 0 d; d–f: 25 d; g–i: 50 d; j–l 70 d. a–l tetraploid on the right and diploid on the left in each picture. Area of each square: 1 × 1 = 1 cm2.
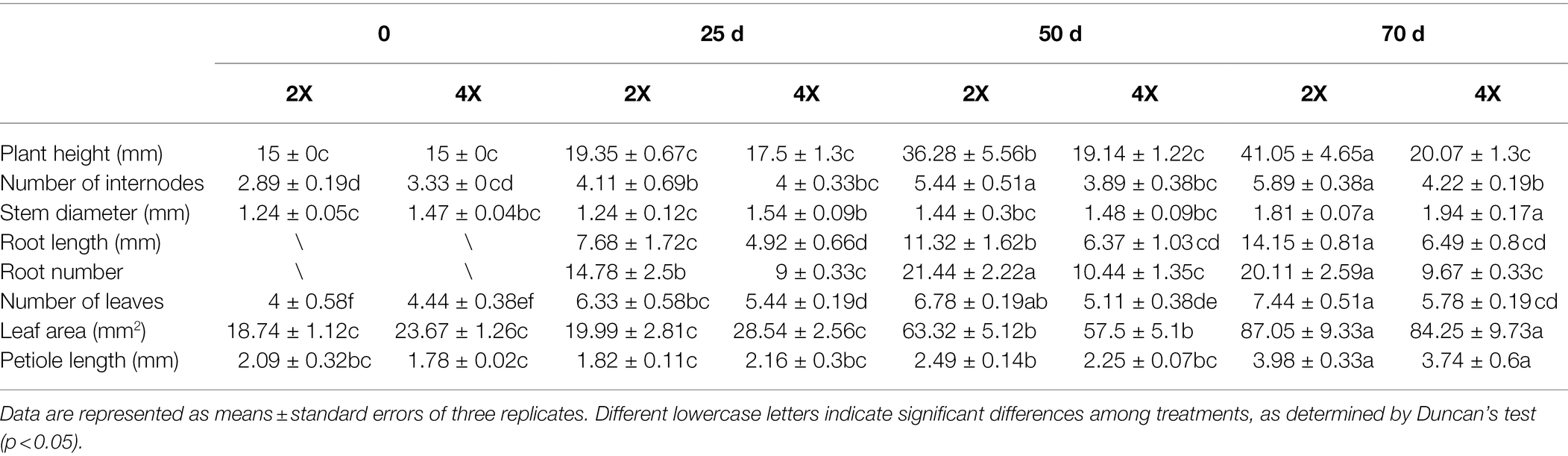
Table 1. In vitro growth statistics of diploid and tetraploid hybrid sweetgum plants at various developmental stages.
In stage I, the height of tetraploids only increased by 1.93 mm, while the diploids grew rapidly; in stage II, a significant difference was apparent (Figure 1B). In stage III, the plant growth rates of both tetraploid and diploid plants were less than the rates in stage II (increases of 0.93 and 4.77 mm, respectively). At 50 d, the plant height and number of internodes were significantly shorter and fewer, respectively, in tetraploids than in diploids. In all three stages, the stem diameter was always larger in tetraploids than in diploids, but the difference was not statistically significant at 50 or 70 d. There were significantly fewer tetraploid leaves than diploid leaves at 25 d, and the area of tetraploid leaves was less than the area of diploid leaves at 50 d; however, there was no significant difference in petiole length at any developmental stage. The rooting times of tetraploid and diploid plants were similar; the root number and length were significantly fewer and shorter, respectively, in tetraploids than in diploids (Table 1; Figure 1B). These results showed that diploid growth was rapid in stage II, while tetraploid growth was not. Regenerated tetraploid plants were shorter and could grow fewer new buds after upper stem segments had been cut.
Observation of the second or third leaf positions of tetraploid and diploid plants showed that tetraploid leaves were significantly thicker than diploid leaves (Figure 2A; Supplementary Table S3). Tetraploid veins were also thicker than diploid veins, but this difference was not statistically significant at stage III. Palisade tissue and spongy tissue were also thicker in tetraploids than in diploids. The areas of xylem transverse cells, upper epidermal cells, lower epidermal cells, spongy tissue cells, and palisade tissue cells were larger in tetraploids than in diploids (Supplementary Table S4).
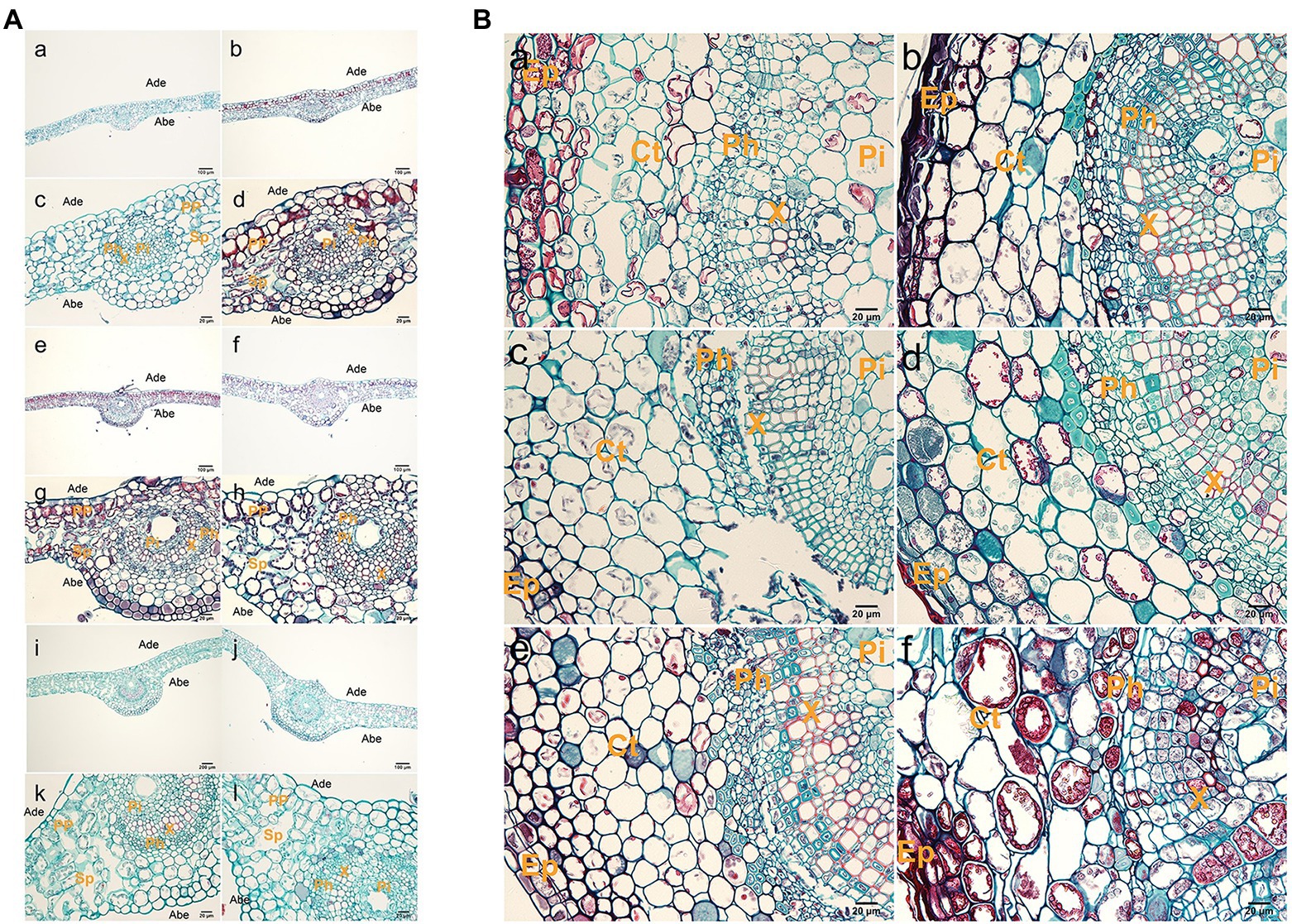
Figure 2. Histological observations of leaves and stems. (A) Anatomy of a leaf cross section from hybrid sweetgum. a,c,e,g,i,k Cross-section of a diploid leaf; b,d,f,h,g,l cross section of a tetraploid leaf. a–d 25 d; e–h 50 d; i–l: 70 d. Ade: adaxial epidermis; Abe: abaxial epidermis; PP: palisade parenchyma; Sp: spongy parenchyma; Pi: pith; Ph: phloem; X: xylem. Bars are 200 μm in a,b,e,f,i,j; they are 20 μm in c,d,g,h,k,l. (B) Anatomy of a stem cross section from hybrid sweetgum. a,c,e Cross section of a diploid shoot; b,d,f cross-section of a tetraploid shoot. a,b 25 d; c,d 50 d; e,f 70 d. Ct: cortex; Ep: epidermis; Pi: pith; Ph: phloem; X: xylem. Bars are 200 μm in a,b,e,f; they are 20 μm in c,d.
The shoot tissue thickness and cell size were greater in tetraploids than in diploids. The tissue thickness and cell area increased with as the number of culture days increased (Figure 2B; Supplementary Tables S5 and S6). The shoot diameter, epidermal thickness, stem cortex thickness, and vascular column thickness were not significantly different between tetraploids and diploids, although epidermal thickness differed at 70 d. There were significant differences between tetraploids and diploids in terms of xylem cells at all three stages, cortex cells at 70 d, and phloem cells at both 50 and 70 d. Figure 2B shows that the apical meristem cells in diploid and tetraploid plants divided vigorously at 25 d; most diploid apical meristem cells continued to vigorously divide at 50 and 70 d (Figures 3Ac,e). However, most tetraploids exhibited a dormant bud apex at 50 d (Figure 3Ad). At 70 d, the apical uds were completely dormant and the bud scale structure was present (Figure 3A f). Therefore, the slow growth rate of the apical meristem may have contributed to shorter plant height in tetraploids than in diploids.
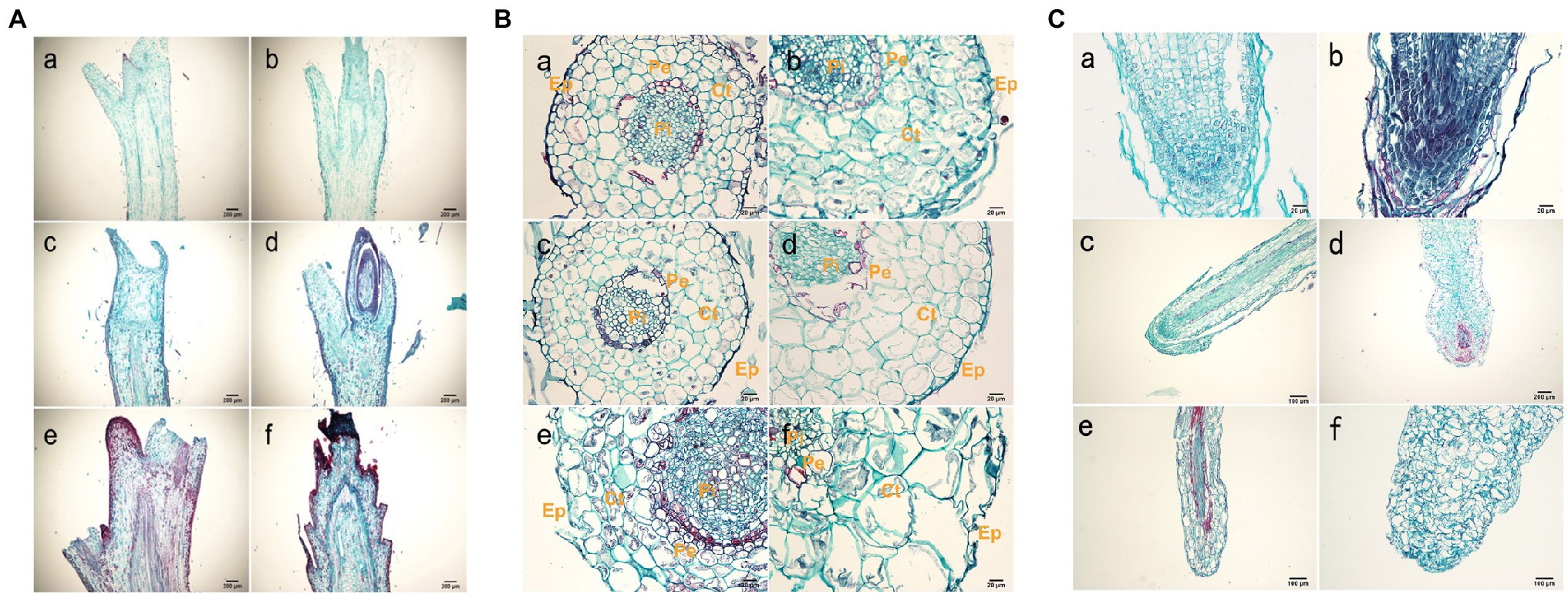
Figure 3. Histological observations of roots. (A) Anatomy of a shoot tip longitudinal-section from hybrid sweetgum. a, c, e Cross-section of a diploid shoot tip; b, d, f crosssection of a tetraploid shoot tip. a, b 25 d; c, d 50 d; e,f 70 d. Each bar is 200 μm. (B) Anatomy of a typical root cross section from hybrid sweetgum. a, c, e Cross-section of a diploid root; b, d, f cross-section of tetraploid roots. a, b 25 d; c, d 50 d; e, f 70 d. Ep: epidermis; Ct: cortex; Pi: pith; CC: central cylinder; Pe: pericycle. Bars are 100 μm in a,b,e,f; they are 20 μm in c, d. (C) Anatomy of a typical root tip longitudinal-section from hybrid sweetgum. a, c, e Cross-section of diploid root tips; b, d, f cross-section of tetraploid roof tips. a, b 25 d; c, d 50 d; e,f 70 d. Each bar is 200 μm.
The root diameter and root tissue thickness at 25 and 50 d were larger in tetraploids than in diploids, with the exception of cylinder diameter at 70 d. The root diameter at 50 and 70 d, and the thicknesses of the epidermis and cortex at all three stages, significantly differed between tetraploids and diploids (Figure 3B; Supplementary Tables S7 and S8). The areas of xylem cells, epidermal cells, and cortical cells were significantly greater in tetraploids at all three stages. The apical meristematic cells of new diploid and tetraploid roots divided vigorously at 25 d of rooting culture (Figures 3Ca,b); the apical meristematic cells of diploid roots continued to vigorously divide at 50 d. However, most tetraploids exhibited increased root cell width and irregular shapes at 50 d (Figures 3Cc,d); the length of the root meristematic zone also decreased in tetraploids. The proportion of malformed roots was 81.33%. Complete coleoptile structure was not detected. These results indicated that root cell shape and root structure began to change after stage II in tetraploids; their longitudinal elongation ability significantly decreased.
Twelve samples were analysed in this study. The original quality score (Q30) of each sample was 93.45–95.07%, the effective data were 6.26–6.90 G, the mean guanine-cytosine (GC) content was 46.50%, and 6.81–7.48 G raw bases were obtained by sequencing (Supplementary Table S9). In total, 4 526,576 to 4 677,234 clean reads (Supplementary Table S9) were obtained after low-quality sequences had been filtered out. Furthermore, 60,187 Unigenes were assembled, with a total length of 5 200,279 bp and mean length of 917 bp (Supplementary Table S10). Of these sequences, 0.24–63.82% were annotated in seven databases: Nr, Swiss-Prot, KEGG, COG, evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG), GO, and Pfam (Supplementary Table S11).
Statistical analysis of FPKM values in each group is shown in Supplementary Table S12 and Figure 4A. Overall, 14,095 and 13,117 DEGs were identified in tetraploid stems and roots, respectively, compared with diploid plants (Figure 4B). The qPCR results on selected genes were similar to the RNA-Seq data, which indicated that the results of RNA-Seq were reliable (Figure 5). In stems, 7,425 and 6,670 DEGs were upregulated and downregulated, respectively; in roots, 4,160 and 8,957 DEGs were upregulated and downregulated, respectively—these values constituted 31.71 and 68.29% of all DEGs, respectively (Figure 4C). Thus, gene expression was generally downregulated in the tetraploid roots of hybrid sweetgum.
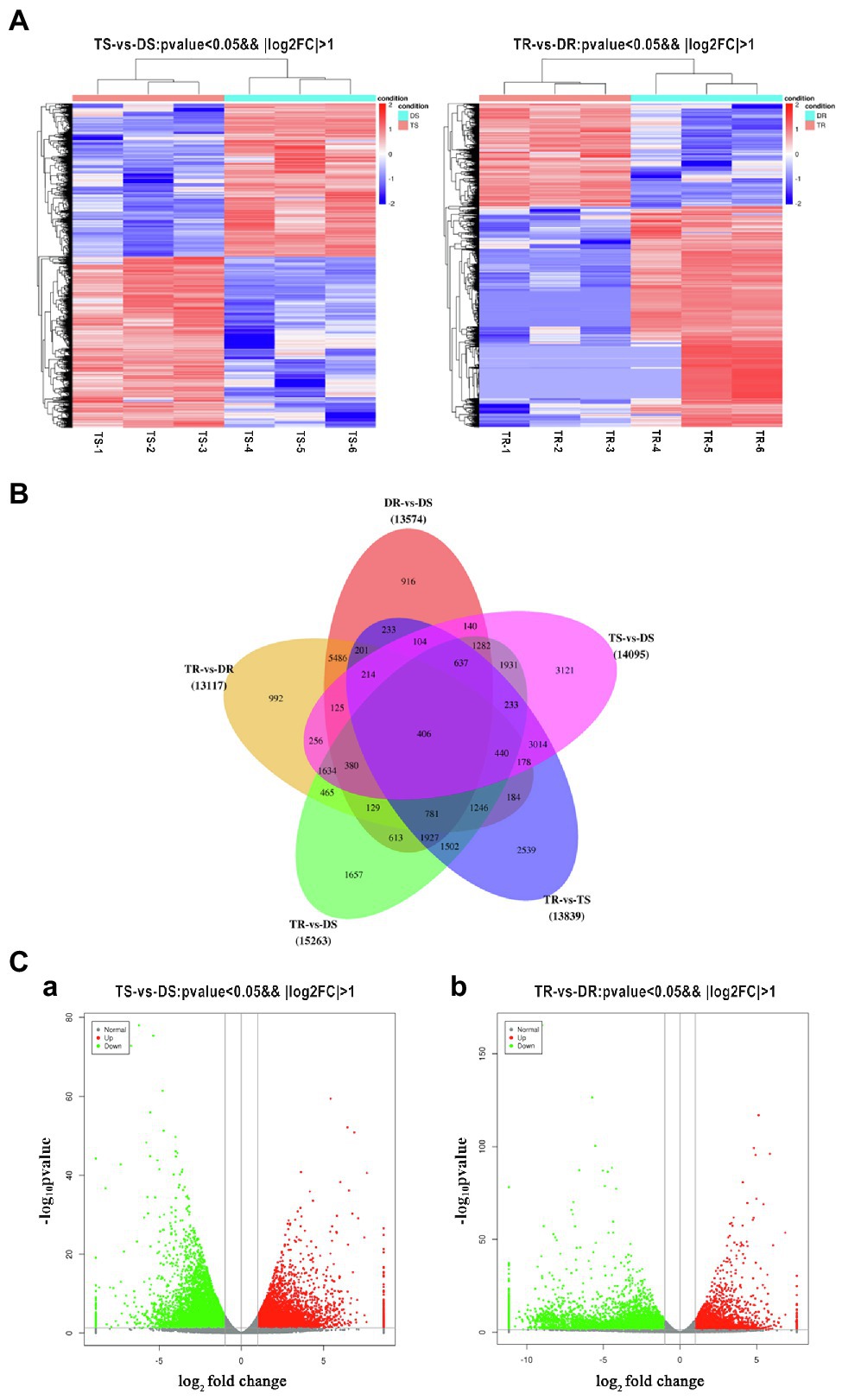
Figure 4. (A) Heatmap of DEG expression profiles (RNA-Seq) in tetraploid vs. diploid shoots. Left: DEGs were derived from TS-1_TS-2_ TS-3_vs DS-1_ DS-2_ DS-3. Right: DEGs were derived from TR-1_ TR-2_ TR-3_vs DR-1_ DR-2_ DR-3. Thresholds for DEG selection in the heatmap are p < 0.05 and |log2FoldChange| > 2. (B) Venn diagram analysis comparing five groups of DEGs. (C) Volcano plot of DEGs. a, b Represent the number of DEGs that were up- or downregulated for TS vs. DS and TR vs. DR, respectively. Grey indicates unigenes with similar expression patterns, red indicates significantly upregulated unigenes, and green indicates significantly downregulated unigenes. The x-axis is the log2FoldChange, and the y-axis is the log10P-value.
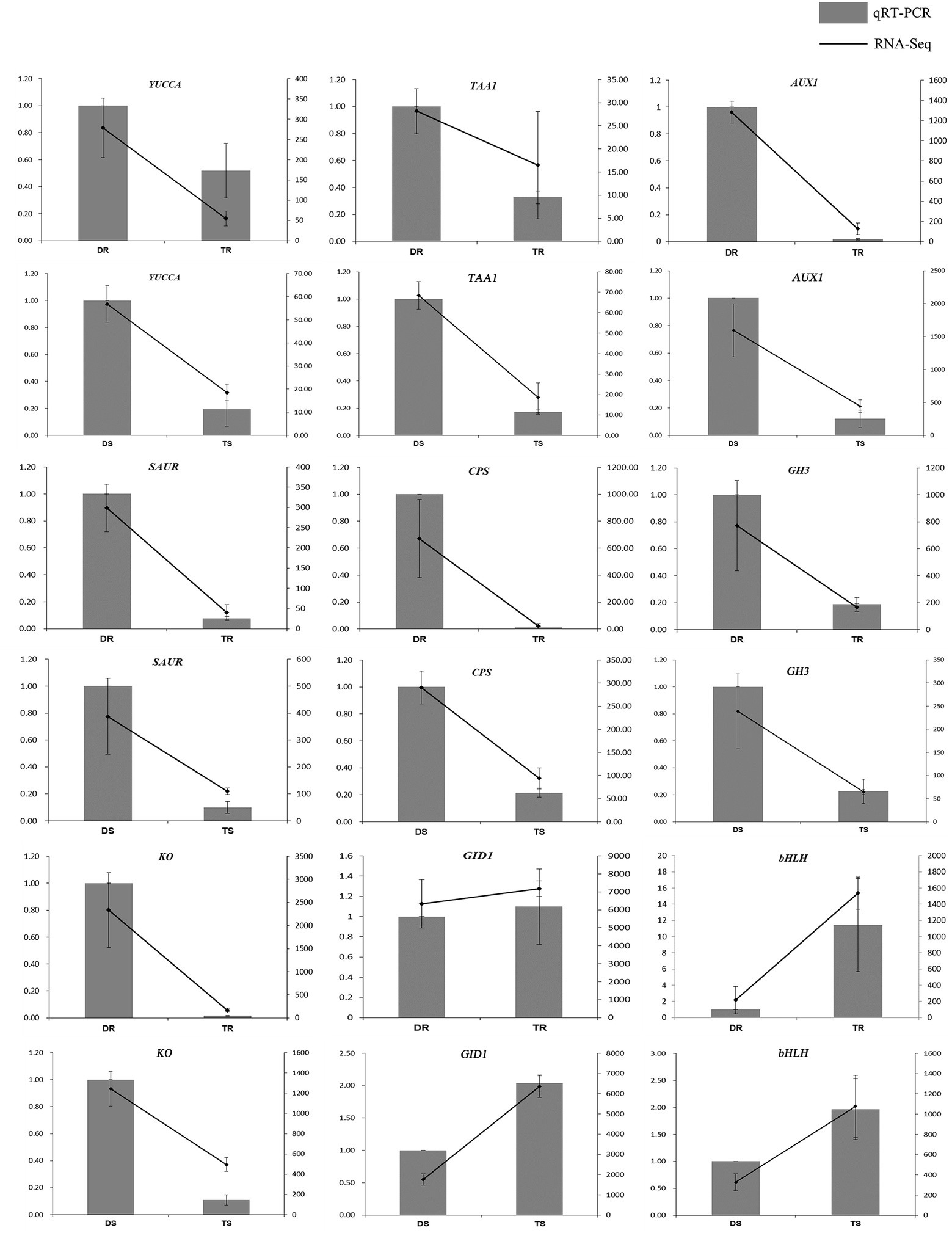
Figure 5. Comparison of expression profiles of nine representative genes as measured by RNA-seq and qRT-PCR. Columns represent expression determined by qRT-PCR (left y-axis), while lines represent expression by RNA-seq in FPKM values (right y-axis). The x-axis in each chart represents different plant materials. For qRT-PCR assay, the mean was calculated from three biological replicates each with three technical replicates (n = 9). For RNA-seq, each point is the mean of three biological replicates.
Gene ontology was used to analyse the DEGs of tetraploid and diploid stems and roots from three perspectives: biological process (BP), cell component (CC), and molecular function (MF). For stems and roots, DEGs were enriched in biosynthesis and signal transduction pathways, cell cycle, and carbon metabolism. For shoots, upregulated DEGs were enriched in guanosine triphosphate metabolic process (GO:0046039), gibberellin catabolic process (GO:0045487), other hormone biosynthesis and metabolic pathways, secondary metabolite biosynthetic process (GO:0044550), starch catabolic process (GO:0005983), and alkaloid metabolic process (GO:0009820; Supplementary Table S13). For shoots, downregulated DEGs were mainly enriched in histone phosphorylation (GO:0016572), cellular response to far red light (GO:0071490), cellular response to red light (GO:0071491), and histone kinase activity (H3-S10 specific; GO:0035175; Supplementary Table S14). For roots, upregulated DEGs were significantly enriched in maintenance of seed dormancy by abscisic acid (GO:0098755), response to low light intensity stimulus (GO:0009645), cellular response to freezing (GO:0071497), priming of cellular response to stress (GO:0080136), cellular response to boron-containing substance deprivation (GO:0080169), other stress response pathways, and NADH dehydrogenase complex (plastoquinone) assembly (GO:0010258; Supplementary Table S15). For roots, downregulated DEGs were mainly involved in septin ring assembly (GO:0000921) and cell cycle (GO:0007089), (GO:0071931, GO:2000045, GO:2000134; Supplementary Table S16).
The KEGG database was used to identify biological pathways that were enriched in DEGs (Figure 6; Supplementary Tables S17 and S18). DEGs in tetraploid and diploid stems and roots were enriched in 47 and 72 biological pathways (p < 0.05), respectively. These pathways included gibberellin, auxin, brassinosteroids, salicylic acid, and ethylene pathways, such as plant hormone signal transduction (ko04075), diterpene biosynthesis (ko00904), brassinosteroids biosynthesis (ko00905) and carotenoid biosynthesis (ko00906); they also included secondary metabolite pathways, such as phenylpropionic acid synthesis (ko00940), flavonoid biosynthesis (ko00941), starch and sugar metabolism (ko00500), fatty acid synthesis and metabolism related to energy synthesis and metabolism (ko00071, ko01212, ko00061), cell cycle (ko04110), and apoptosis (ko04210).
Plant hormone biosynthesis and signal transduction pathways include auxin, gibberellin, cytokinin, BL, abscisic acid, ethylene, jasmonic acid and salicylic acid synthesis and transduction. Many important genes involved in auxin, gibberellin, and BL biosynthesis and signal transduction pathways were downregulated in tetraploids (Figure 7A). In shoots, the auxin biosynthesis genes YUCCA (Indole-3-pyruvate monooxygenase YUCCA) and TAA1 (Tryptophan aminotransferase of Arabidopsis1) and the auxin influx carrier AUX1 (Auxin1) were downregulated; SAUR (Small auxin up RNA), IAA (Indole-3-acetic acid), and GH3 (Gretchen hagan 3) were generally downregulated (Figure 7A). Multiple gibberellin biosynthesis genes were also downregulated, including CPS (Copalyl pyrophosphate synthase), KAO (Kaurenoic acid oxidase), KO (Kaurene oxidase), GA20ox (GA20-oxidase), GA2ox (GA2-oxidase), and GA3ox (GA3-oxidase). The GID1 (GA-insensitive dwarf1) gene, which contributes to positive regulation of gibberellin signal transduction, was upregulated; its negative regulation counterpart, DELLA, was generally downregulated. The BL biosynthesis and signal transduction genes BAS1 (phyB-4 activation-tagged suppressor 1) and CYCD3 (Cyclin D3;1) were downregulated. ABA, PYL (Pyrabactin resistance 1-like), the negative regulation SnRKs (SNF1-related protein kinases; TRINITY_DN36419_c0_g1_i1_3, TRINITY_ DN39068_c1_g1_i10_3, TRINITY_DN44648_c2_g3_i1_1, TRINITY_ DN46092 _ c1 _g1_i3_1)—which negatively regulates abscisic acid signal transduction—and PP2C (Protein phosphatase 2C; TRINITY_DN34392_c0_g1_i3_2 and TRINITY_DN37551_c0_g1 _i6_2) were downregulated. However, the positive regulation counterpart, ABF (Abscisic acid responsive elements-binding factor), was generally downregulated.
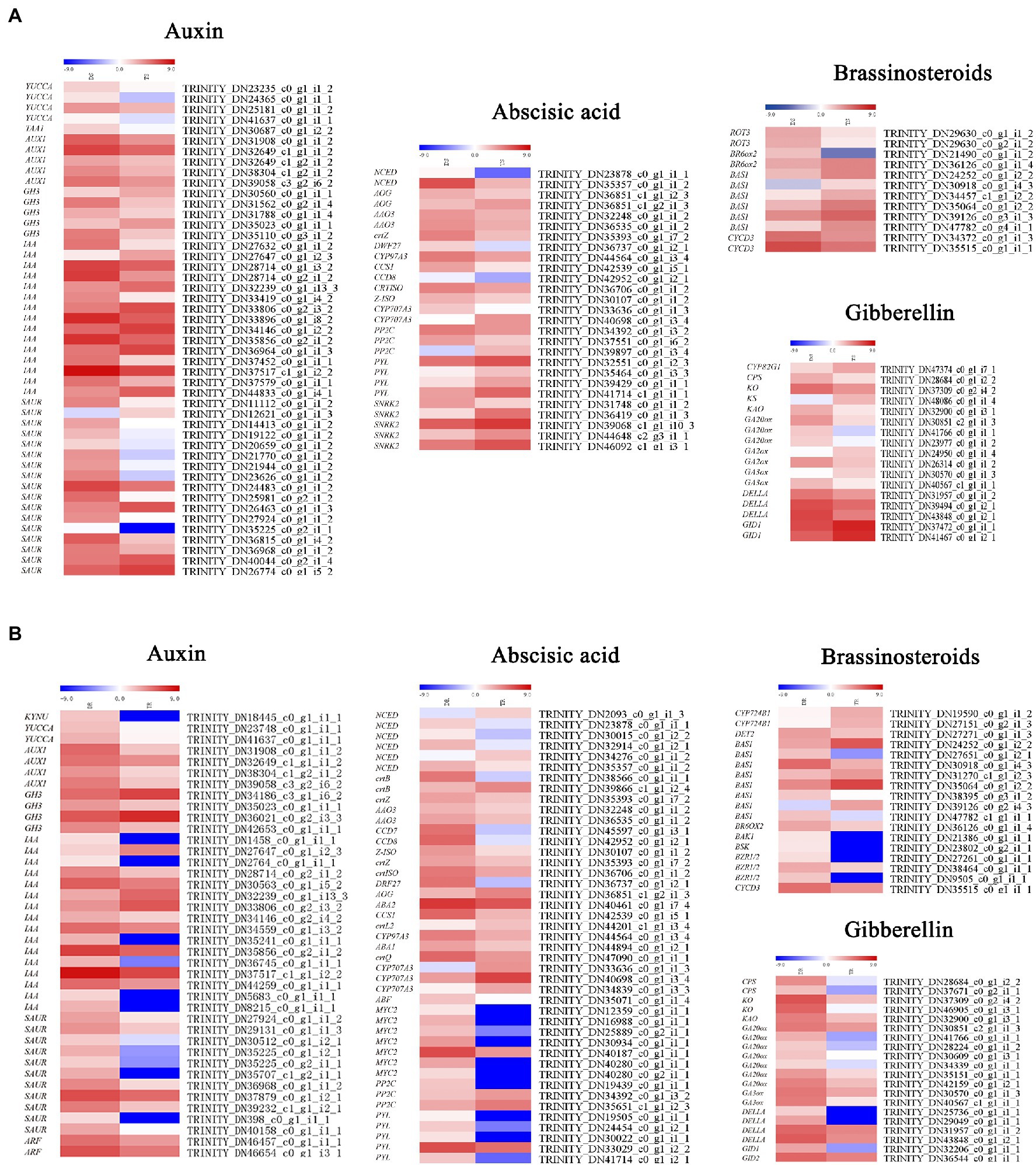
Figure 7. Heat maps depicting differential expression patterns of genes involved in plant hormone synthesis and signal transduction: (A) shoots, (B) roots.
Similar to stems, the auxin biosynthesis genes YUCCA and KYNU (kynureninase), auxin signal transduction genes AUX/IAA (auxin/indole-3-acetic acid), SAUR, GH3 (TRINITY_DN35023 _c0_g1_i1_1, TRINITY_DN42653_c0_g1_i1_1) and IAA were downregulated in roots, while auxin influx carrier AUX1 was also similarly downregulated. The gibberellin biosynthesis biosynthetic genes (e.g., CPS, KO, KAO, GA20ox, and GA3ox) were all downregulated. However, GID1, which positively regulates gibberellin signal transduction, was upregulated; DELLA, its negative regulation counterpart, was downregulated. For ABA, except CYP707A3, the biosynthesis genes AAO3 and ABA2 were downregulated, while NCED (9-cis-epoxycarotenoid dioxygenase) was generally downregulated. In contrast to stems, with the exception of PP2C, the signal transduction genes PYL, SnRKs, and ABF were generally downregulated (Figure 7B).
In stems and roots, there were 42 and 65 DEGs related to starch and sugar metabolism, respectively (Figure 8); most were downregulated. In stems, sacA, SS (Starch synthase), glgA, otsB, GN1_2_3, GN4, GN5_6, SPS1 (Sucrose-phosphate synthase1), and SUS (Sucrose synthase) were downregulated; TPS (Trehalose 6-phosphate synthase) was upregulated. In roots, TPS, SS (TRINITY_DN 37738_c0_g1_i1_2, TRINITY_DN40072_c0_g1_i1_1, TRINITY_DN44913_c0_g2 _i1 _4), sac (TRINITY _DN 17363 _c 0_g1_i1_2, TRINITY_DN35727_c0_g1_i12_2, TRINITY_DN36819_c0_g1_i2_3, TRINITY_DN46169_c0_g1_i15_1), otsB, GN1_2 _3, and GN5_6 were downregulated.
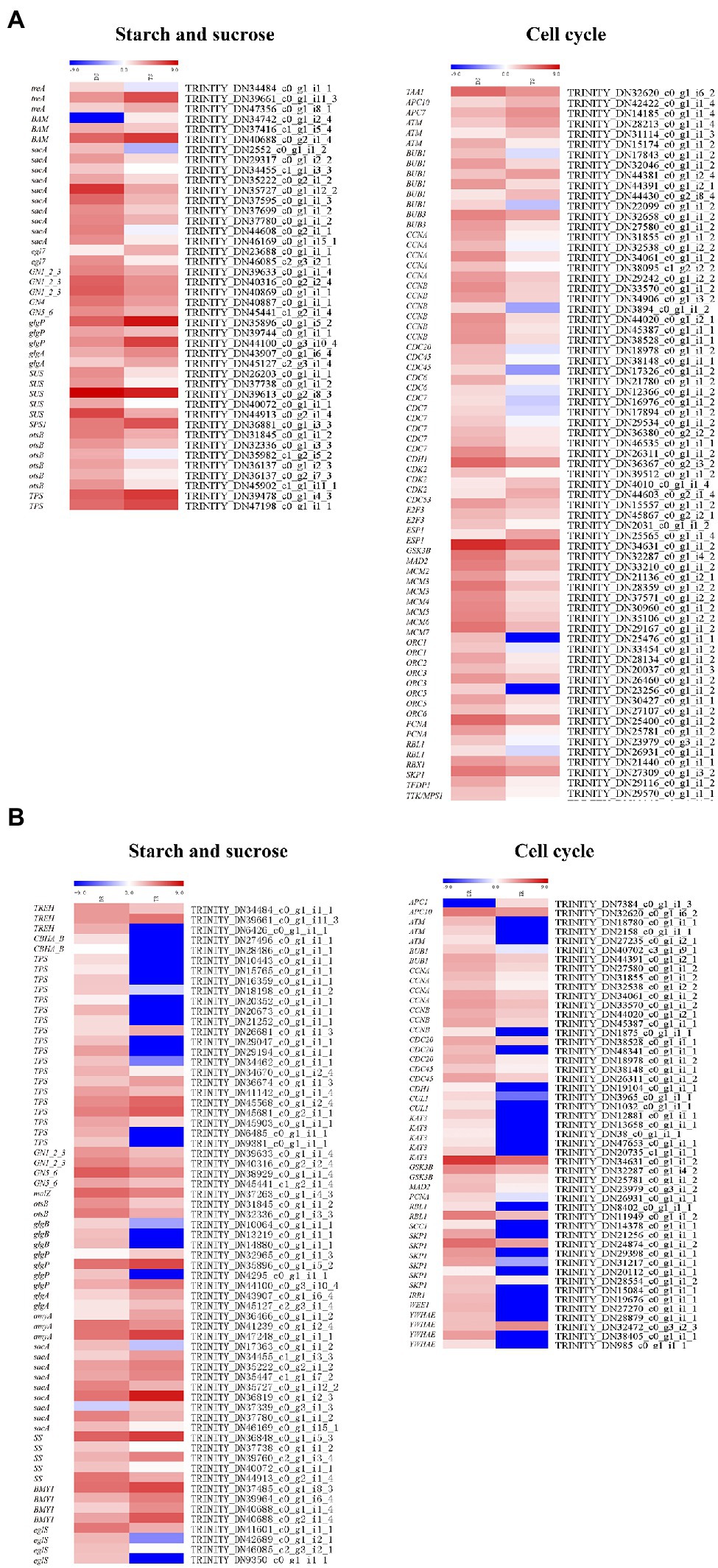
Figure 8. Heat maps depicting DEGs involved in starch and sucrose biosynthesis and metabolism and cell cycles: (A) shoots, (B) roots.
There were 76 and 49 genes involved in regulating cell cycles (ko04110) in stems and roots, respectively (Figure 8). In stems, these genes were generally downregulated, except ATM (Ataxia telangiectasia mutated), CDK2 (cyclin-dependent kinase 2), CDC6 (cell division control protein 6), CDC7, CDC20, CDC45, E2F3, Bub1, Bub3, CycA (Cyclin A), Orc1-Orc3, and Orc5; notably, Orc6—a contributor to cell proliferation and cell cycle regulation—was generally downregulated. In roots, except for two genes, the DEGs were downregulated in tetraploids, including ATM, BUB1, CDC20, CDC45, CUL1 (Cullin 1), SKP1 (S-phase kinase-associated protein 1), and GSK3B (glycogen synthase kinase 3 beta). These genes have important roles in regulating cell size and cell number. Therefore, the generally larger cells of tetraploid hybrid sweetgum, compared to diploid hybrid sweetgum, may be related to significant changes in cell cycle genes.
The IAA, GA3, and BL contents of diploid and tetraploid roots and stems cultured in rooting medium for 50 d were measured; the results are shown in Figure 9A. The endogenous auxin, gibberellin, and BL levels were significantly lower in tetraploids than in diploids, consistent with the results of the transcriptome analysis. Compared with diploids, only the glucose contents significantly differed in tetraploids (Figure 9B). The result showed that although starch and sugar biosynthesis genes were downregulated in tetraploid plants, the starch and soluble sugar contents did not significantly decrease. Therefore, gene expression and sugar accumulation were not synchronised at this stage.
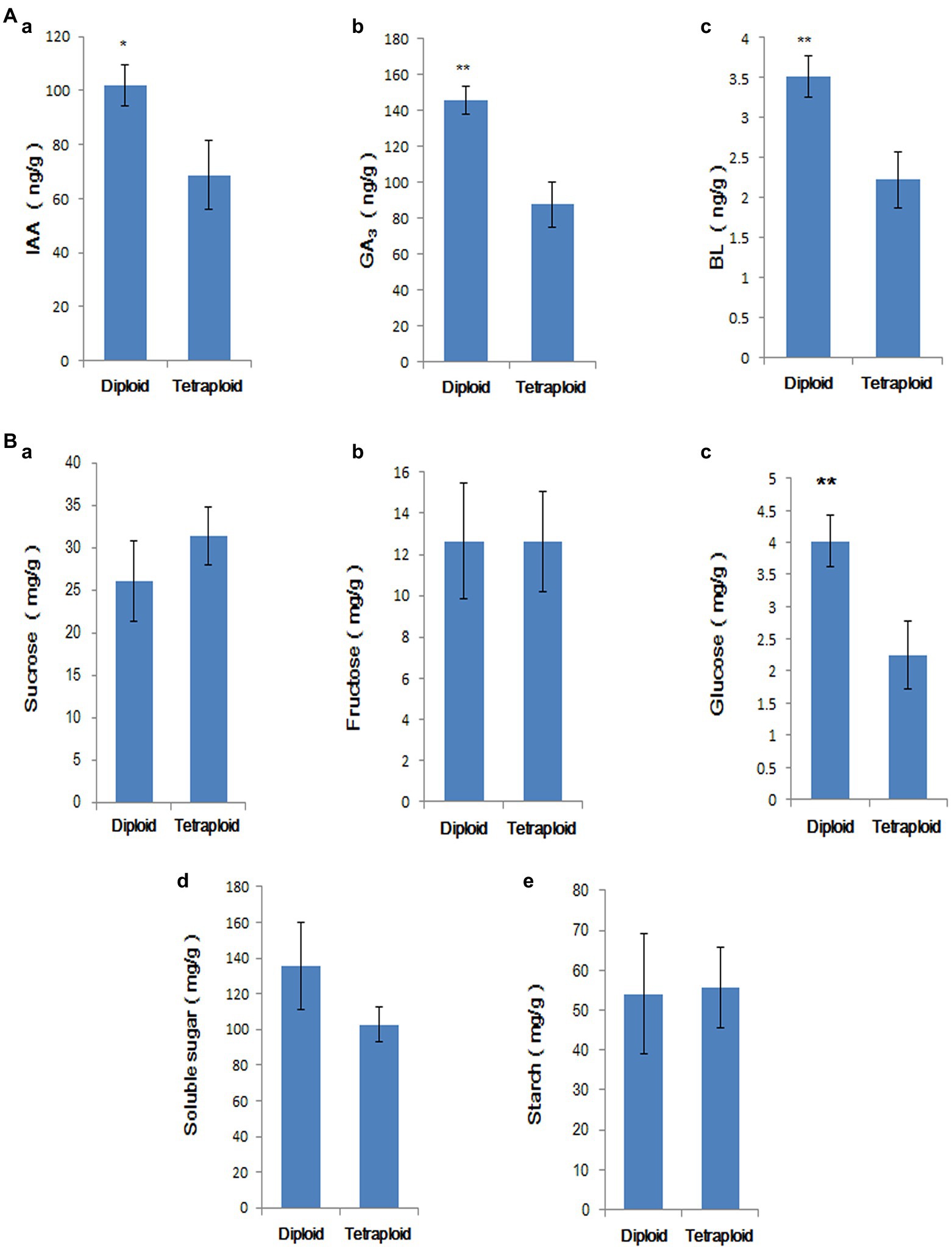
Figure 9. (A) Comparison of IAA (a), GA3 (b), and BR (c) contents in tetraploid and diploid stems and roots. ‘*’ indicates p < 0.05, ‘**’ indicates p < 0.01. (B) Comparison of sucrose (a), fructose (b), glucose (c), soluble sugar (d), and starch contents (e) in tetraploid and diploid stems and roots. ‘**’ indicates p < 0.01.
To verify the effects of IAA, GA3, and BL on elongation in tetraploids, the effect of the timing of tetraploid transplantation into new medium was investigated. As shown in Supplementary Table S19, the growth of regenerated tetraploid plants was only significantly promoted at the initial stage when the rooting medium was supplemented with 0.5 mg/L GA3 or 1.5 mg/L IAA. These results showed that the timing of addition of exogenous hormones significantly affected the growth of regenerated plants. As time elapsed without replacement of the medium, the elongation effect gradually decreased.
After determination of the optimal medium replacement time, the effects of media containing different hormone ratios on tetraploid growth were investigated. The results showed that the greatest plant height was 4.42 cm in a rooting medium supplemented with 0.5 mg/L GA3; the plant height, ground diameter, root length, and leaf area were significantly increased compared with tetraploid hybrid sweetgum grown in primitive rooting medium, but the number of roots did not significantly differ (Figure 10; Supplementary Table S20). The plants placed in a medium with the addition of 0.5 mg/L GA3 combined with 1.5 mg/L IAA had the largest leaf area and greatest number of main roots. Therefore, GA3 was the hormone that most strongly supported stem elongation (Supplementary Table S20). The effects of BR alone or in combination with GA3 or IAA were weaker than the effects of other two hormones. These results indicated that the height of tetraploid plants could be significantly increased by adding an appropriate concentration of GA3 to the growth media. In addition, the stem segments used for subcultures could be rooted and elongated normally in the medium. Therefore, in vitro micro-cutting could be used to subculture and expand tetraploid plants. Additionally, 0.5 mg/L GA3 was added to the tetraploid adventitious bud elongation medium; it demonstrated a strong effect on adventitious bud elongation (Figure 10).

Figure 10. Effects of GA3 and IAA on shoot and root lengths of tetraploid hybrid sweetgum Z4 grown in vitro (A,B). (A) Control on the left (primitive rooting medium) and rooting medium supplemented with 0.5 mg/L GA3 on the right. (B) Control on the left (primitive rooting medium) and rooting medium supplemented with 0.5 mg/L GA3 and IAA on the right. Area of each square: 1 × 1 = 1 cm2. Effects of GA3 on adventitious bud length in tetraploid hybrid sweetgum (C,D). (C) Control. (D) Development of adventitious shoots in medium supplemented with 0.5 mg/L GA3.
Tetraploid hybrid sweetgum regenerated plants exhibited obvious morphological variations, with shorter stems and roots than diploid plants. Furthermore, the stem diameter and internode length were thicker and shorter, respectively, in tetraploids than in diploids. These observations were consistent with previous findings in some herbs, fruit trees, and timber trees (Nilanthi et al., 2009; Tan et al., 2015; Xu et al., 2018). However, with the exceptions of stems and roots, the other organs of dwarf tetraploids did not necessarily exhibit a growth disadvantage. The plant height was shorter in tetraploid apples than in diploid apples, but the leaf and flower organs were obviously larger in tetraploid apples (Xue et al., 2017). Thus, plant height is not necessarily positively related to the sizes of other organs. In addition, Corneillie et al. (2019) found that in Arabidopsis thaliana, the leaf area, stem length, and dry weight were greater in tetraploid plants than in plants with other ploidy levels; as the ploidy level increased, hexaploid and octoploid plants gradually exhibited less growth. The ploidy level at which optimal growth is reached differs among species.
The proliferation and enlargement rates of apical and intersegment meristematic cells have considerable effects on elongation and development in roots and stems (Takatsuka and Umeda, 2014). The relationships among organ size, cell size, and cell number remain unresolved (Tsukaya, 2008; Li et al., 2012). Previous studies have suggested that plant organ sizes are determined by both cell size and cell number (Krizek, 2009; Gonzalez et al., 2012). Generally, plant cell sizes significantly increase after chromosome doubling. Endoreduplication is considered an important cause of polyploid cell enlargement (Tsukaya, 2008). The numbers of cells vary among species and ploidy levels. Compared with diploids, triploid Populus plants showed a significant increase in cell area, but the number of cells did not decrease (Zhang et al., 2019). A similar observation was reported in a study of A. thaliana (Li et al., 2012). However, cell area was larger in octoploid Arabidopsis plants than in diploid plants, but the smaller number of cells resulted in a smaller leaf area (Levan, 1939). These results indicated that the cell size was greater in tetraploids than in diploids, but the tetraploid plants had dwarfing characteristics.
In the present study, tetraploid hybrid sweetgum root cells exhibited increased width and irregular shape, the tetraploid root meristematic zone length decreased, and no obvious coleoptile sheath or complete coleoptile structure were observed. Few observations of root malformation have been reported in tetraploid regenerated plants; however, in apples, the ratio of length to width of parenchyma cells is altered in the stem cortex (Ma et al., 2016). Compared with diploids, the apical bud entered dormancy earlier; it was difficult for most plants to exit dormancy and continue to elongate, which may explain why tetraploid regenerated plants were shorter than diploid plants. Sun et al. (2016) found that only a few individuals had autumn shoot growth, but none of these individuals were diploid. Changes in the content and proportion of endogenous hormones in plants are closely related to bud dormancy (Tanino, 2004; Ruttink et al., 2007), especially the ratio of gibberellin to abscisic acid (Rinne et al., 2011). In this study, the morphological variations of roots and stems may be closely related to hormone levels in tetraploid hybrid sweetgum; changes in the hormone ratio may lead to premature dormancy in tetraploid regenerated plants.
After polyploidization, growth conditions usually change in vitro; thus, the original medium for diploids is often unsuitable for tetraploid plant growth. For example, Lagerstroemia crassipes is difficult to root in an original culture medium (Tong, 2007). Root and stem elongation is an important process in plant tissue culture, but the elongation of adventitious shoots and plantlets is problematic in many species. Plant hormones (e.g., auxin, gibberellin, and BL) are the most important factors that affect the lengths of regenerated organs; such hormones can promote in vitro plant elongation (Sasaki, 2002; Zhang et al., 2013, 2018). The concentrations of auxin hormones (e.g., IAA and indole butyric acid) added to the medium have a significant effect on root length elongation (Nandagopal and Kumari, 2007). In this study, the rooting medium supplemented with IAA alone promoted roots of tetraploid hybrid sweetgum elongation; it also increased plant height. This result may have occurred because, although the polar transport of auxin occurs from top to bottom, the growth processes of underground and aboveground parts often interact. An increase in underground growth can provide more water, inorganic salts, organic substances, and hormones for the shoots (Agren and Franklin, 2003). In this study, the number of roots was significantly fewer in tetraploids than in diploids; a similar phenomenon has been reported, but in Gerbera jamesonii, the plant height was significantly greater in tetraploids than in diploids (Gantait et al., 2011). Therefore, tetraploid plant height may not be directly proportional to root number.
In the present study, gibberellin had effects on root and stem of tetraploid hybrid sweetgum elongation; however, the induction of root thinning by gibberellin may have been related to the gradient change in roots. Eiichi (2012) suggested that the gibberellin-mediated-concentration dependent-stimulation of elongation is important for regulating plant height and root length; GA3 could also significantly promote elongation in adventitious buds of sweetgum (Wang et al., 2018b).
Brassinolide also has important roles in promoting cell elongation, cell enlargement, and vascular tissue differentiation (Clouse, 2002). Previous studies have shown that BL can promote both lateral stem expansion and vertical stem growth (Sasse, 1997). Clouse and Sasse (1998) found that a low concentration of BL helped to accelerate cell division, whereas high concentrations had the opposite effect. In the present study, the addition of 0.1 mg/L BL did not have a significant effect on the lengths of roots and stems. Therefore, the concentration range is presumably unsuitable for tetraploid hybrid sweetgum elongation.
Previous studies have shown that reduced expression levels of genes with positive regulatory roles in gibberellin, auxin, and BL biosynthesis and signal transduction are important causes of plant dwarfing (Li et al., 2017; Ju et al., 2018; Wang et al., 2018a). Ma et al. (2016) suggested that changes in the auxin and BL contents in tetraploid apples might contribute to plant height variation. In the present study, the DEGs of tetraploid and diploid hybrid sweetgum stems and roots were also highly enriched in the hormone synthesis and signal transduction pathways, which were related to auxin, gibberellin, and BL levels.
Although the auxin synthesis pathway has not been fully analysed, an in-depth analysis of the indole-3-pyruvate pathway has been conducted. In the present study, the significant downregulation of YUCCA and TAA1 genes of tetraploid hybrid sweetgum had important effects on auxin biosynthesis. TAA participates in the transformation of tryptophan (Trp) into indole-3-pyruvate, while YUCs participate in the transformation of indole-3-pyruvate into IAA (Tao et al., 2008; Won et al., 2011; Zhao, 2012). YUC and TAA genes affect the growth of the yuc and tar mutants of A. thaliana, which have very short hypocotyls and roots (Won et al., 2011). Auxin signal transduction genes are classified into AUX/IAA, SAUR, and GH3 groups (Guilfoyle and Hagen, 2007), which have important roles in plant growth and development. AUX/IAA is an early auxin response gene, and its protein product can specifically bind to auxin response factor (ARF), thereby regulating the expression of auxin response genes; in the A. thaliana mutant AUX/IAA, the lengths of the stem and hypocotyl are altered; apical dominance and root formation are also modified (Liscum and Reed, 2002). In the present study, auxin signal transduction genes (e.g., AUX/IAA, GH3, and SAUR) and auxin influx carrier AUX1 (Swarup and Bhosale, 2019) were substantially downregulated in roots and stems.
Gibberellin biosynthesis genes also affect cell division and elongation, thereby influencing plant stem and root elongation. In this study, the CPS, KAO, KO, GA20ox, GA2ox, and GA3ox genes were generally downregulated in tetraploid hybrid sweetgum. These genes are involved in the three main steps of gibberellin synthesis. First, ent-kobaki pyrophosphate synthase (CPS) and ent-kaurene synthase catalyse the formation of ent-kaurene from yak’s geranyl pyrophosphate. Second, ent-kaurene catalyses the formation of GA12 and GA53 by ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO). Third, the oxidases GA20ox, GA2ox, and GA3ox eventually catalyse GA12 and GA53 to form GA1 and GA4 (Hedden and Phillips, 2000). Gibberellin and auxin are closely related and have significant interaction (Depuydt and Hardtke, 2011; Willige et al., 2011). Auxin reportedly can promote root growth in A. thaliana by acting on the gibberellin responsive protein, DELLA (Fu and Harberd, 2003). GID1 and DELLA have key roles in gibberellin signal transduction. After activation by gibberellin, the gibberellin receptor GID1 binds to DELLA via the E3 ubiquitination pathway, which leads to degradation of the DELLA protein and thus eliminates growth inhibition (Sun, 2010). However, in maize, DELLA deletion mutants showed dwarfing traits (Lawit et al., 2010). Therefore, although DELLA is a negative regulator, it is indispensable.
Changes in the expression patterns of BL biosynthesis and signal transduction genes also have significant effects on plant height and root length. DET2, ROT3, and BR6ox1 are important BL biosynthesis genes (Noguchi et al., 2000; He et al., 2005). A balanced BR signal is necessary to maintain the root meristem (Takatsuka and Umeda, 2014). The root meristem length of the bri1-116 mutant is decreased, but overexpression of the cell cycle gene CYCD3;1 inhibits this decrease (Gonzalez-Garcia et al., 2011). The overexpression of the mutant BZR1 significantly promoted hypocotyl elongation and increased plant height (Wang et al., 2002).
Previous studies have shown that epigenetic changes are important sources of transcriptional variation after polyploidization (Ni et al., 2009; Chen, 2013). The downregulation of the auxin signal transduction gene MdARF3 in tetraploid apples may be related to upregulation of the negative regulator MiR390; it may affect auxin signal transduction (Ma et al., 2016). Therefore, significant changes in the expression levels of hormone-related genes in tetraploid hybrid sweetgum may be related to the upregulation or downregulation of small RNA genes.
Based on our findings, we constructed a schematic of the effects of changes in endogenous hormone biosynthesis and signal transduction genes, as well as supplementation with exogenous hormones, on growth in tetraploid hybrid sweetgum (Figure 11). As discussed previously, reduced expression levels of genes in gibberellin, auxin, BL and other hormone biosynthesis and signal transduction are important causes of tetraploid hybrid sweetgum dwarfing. Furthermore, the down-regulated expression of important genes involved in cell cycle may also have an impact on tetraploid hybrid sweetgum. Exogenous GA3 and IAA, which induced plant cell elongation, could significantly promote growth in the stems and roots of tetraploids. It is currently unclear why some species exhibit two different phenotypes (i.e., fast growth or slow growth) after polyploidization. Species evolution and genome duplication induce gene redundancy. Epigenetic modification models differ among species; such models may influence the expression patterns of genes related to growth and development. This complex problem should be investigated in future studies.
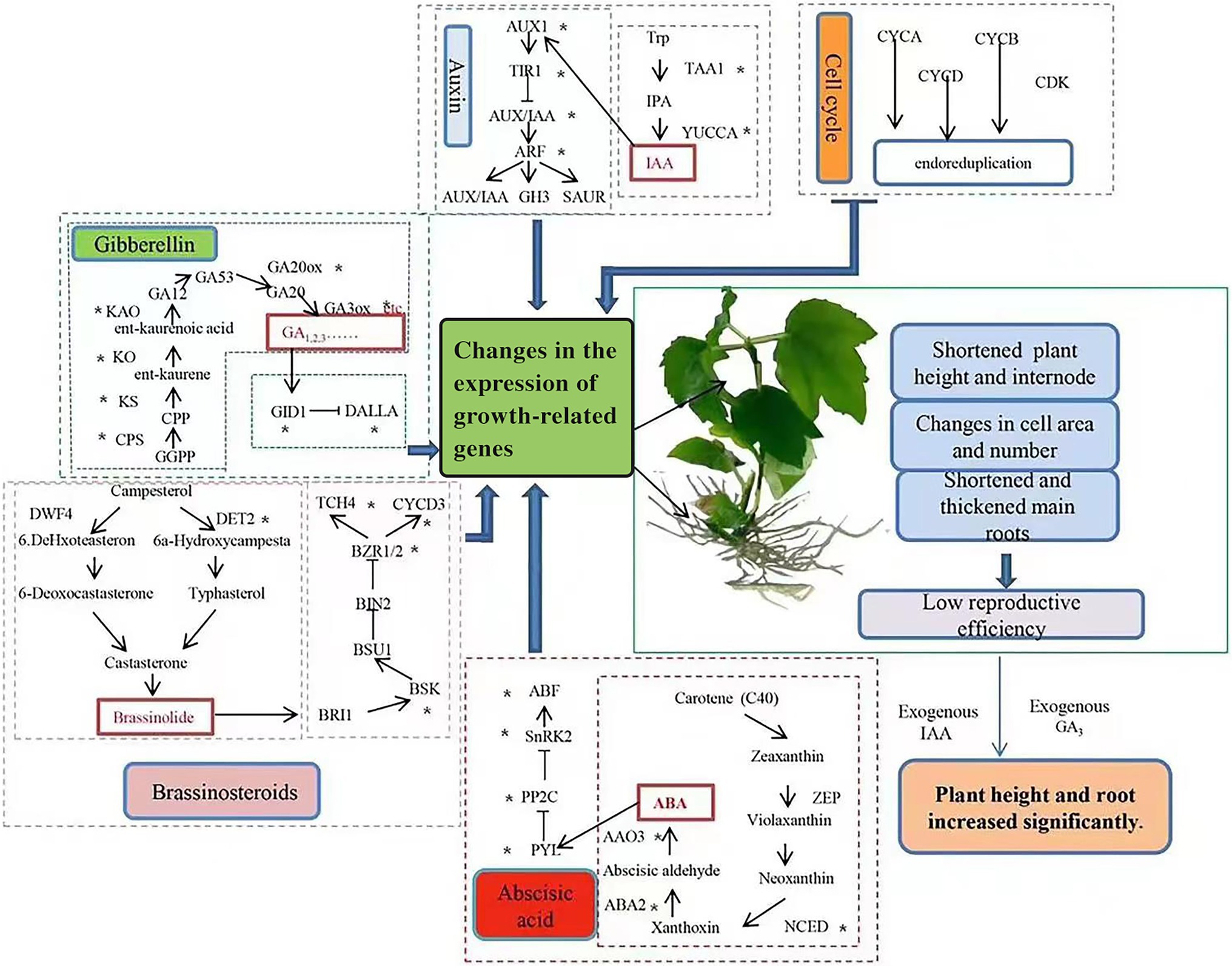
Figure 11. Effects of changes in endogenous hormone biosynthesis and signal transduction genes and supplementation with exogenous hormones on tetraploid hybrid sweetgum growth. ‘*’ indicates DEG enrichment in roots or stems.
To our knowledge, this is the first investigation of the polyploid molecular mechanism in the dwarf type of hybrid sweetgum grown in vitro. Genes that promote organ growth (e.g., auxin, gibberellin, and BL biosynthesis) and genes involved in signal transduction were downregulated. These changes may be the main cause of root and stem variations in tetraploids, which lead to slower growth in tetraploids than in diploids. Both IAA and GA3 can significantly promote shoot and root elongation in dwarf tetraploid hybrid sweetgum. Our findings provide insights into the molecular mechanisms that underlie dwarfism in allopolyploid hybrid sweetgum.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA831658.
JZhan and JZhao designed the study. YZ and SC conducted the experiments, analysed the data, and drafted the manuscript. SC, YZ, TZ, JZhao, and ZP critically reviewed and improved the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by National Forestry and Grassland Administration Promotion Project of China (2020133102); Scale-up Propagation Technology Promotion via Somatic Embryogenesis of Sweetgum Forestry Science and Technology Innovation Special Project of Jiangxi Province (2019-16); the Fundamental Research Funds for the Central Universities (2019ZY39); Major Science and Technology Special Project of Xuchang, Henan province, China (20170112006).
DZ and ZP are employed by Guangxi Bagui Forest and Flowers Seedlings Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.924044/full#supplementary-material
Agren, G. I., and Franklin, O. (2003). Root: shoot ratios, optimization and nitrogen productivity. Ann. Bot. 92, 795–800. doi: 10.1093/aob/mcg203
Bizet, F., Hummel, I., and Bogeat-Triboulot, M.-B. (2015). Length and activity of the root apical meristem revealed in vivo by infrared imaging. J. Exp. Bot. 66, 1387–1395. doi: 10.1093/jxb/eru488
Chen, Z. J. (2013). Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 14, 471–482. doi: 10.1038/nrg3503
Chen, S. Y., Dong, M. L., Zhang, Y., Qi, S. Z., Liu, X. Z., Zhang, J. F., et al. (2020). Development and characterization of simple sequence repeat markers for, and genetic diversity analysis of Liquidambar formosana. Forests 11:203. doi: 10.3390/f11020203
Chen, M.-L., Fu, X.-M., Liu, J.-Q., Ye, T.-T., Hou, S.-Y., Huang, Y.-Q., et al. (2012). Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J. Chromatogr. B-Analytical Technol. Biomed. Life Sci. 905, 67–74. doi: 10.1016/j.jchromb.2012.08.005
Cheng, S., Huang, Z., Li, Y., Liao, T., Suo, Y., Zhang, P., et al. (2015). Differential transcriptome analysis between Populus and its synthesized allotriploids driven by second-division restitution. J. Integr. Plant Biol. 57, 1031–1045. doi: 10.1111/jipb.12328
Clouse, S. D. (2002). Brassinosteroid signaling: novel downstream components emerge. Curr. Biol. 12, R485–R487. doi: 10.1016/s0960-9822(02)00964-8
Clouse, S. D., and Sasse, J. M. (1998). Brassinosteroids: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451. doi: 10.1146/annurev.arplant.49.1.427
Comai, L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846. doi: 10.1038/nrg1711
Corneillie, S., De Storme, N., Van Acker, R., Fangel, J. U., De Bruyne, M., De Rycke, R., et al. (2019). Polyploidy affects plant growth and alters Cell Wall composition. Plant Physiol. 179, 74–87. doi: 10.1104/pp.18.00967
Dai, F., Wang, Z., Luo, G., and Tang, C. (2015). Phenotypic and Transcriptomic analyses of Autotetraploid and diploid mulberry (Morus alba L.). Int. J. Mol. Sci. 16, 22938–22956. doi: 10.3390/ijms160922938
Depuydt, S., and Hardtke, C. S. (2011). Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21, R365–R373. doi: 10.1016/j.cub.2011.03.013
Ding, J., Mao, L.-J., Wang, S.-T., Yuan, B.-F., and Feng, Y.-Q. (2013). Determination of endogenous Brassinosteroids in plant tissues using solid-phase extraction with double layered cartridge followed by high-performance liquid chromatography-tandem mass spectrometry. Phytochem. Anal. 24, 386–394. doi: 10.1002/pca.2421
Eiichi, T. (2012). Tall or short? Slender or thick? A plant strategy for regulating elongation growth of roots by low concentrations of gibberellin. Ann. Bot. 110, 373–381. doi: 10.1093/aob/mcs049
Fu, X. D., and Harberd, N. P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421, 740–743. doi: 10.1038/nature01387
Fuentes, I., Stegemann, S., Golczyk, H., Karcher, D., and Bock, R. (2014). Horizontal genome transfer as an asexual path to the formation of new species. Nature 511, 232–235. doi: 10.1038/nature13291
Fukao, T., Xu, K. N., Ronald, P. C., and Bailey-Serres, J. (2006). A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice(W). Plant Cell, 18, 2021–2034. doi: 10.1105/tpc.106.043000
Gantait, S., Mandal, N., Bhattacharyya, S., and Das, P. K. (2011). Induction and identification of tetraploids using in vitro colchicine treatment of Gerbera jamesonii bolus cv. Sciella. Plant Cell Tissue Org. Cult. 106, 485–493. doi: 10.1007/s11240-011-9947-1
Gonzalez, N., Vanhaeren, H., and Inze, D. (2012). Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 17, 332–340. doi: 10.1016/j.tplants.2012.02.003
Gonzalez-Garcia, M.-P., Vilarrasa-Blasi, J., Zhiponova, M., Divol, F., Mora-Garcia, S., Russinova, E., et al. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138, 849–859. doi: 10.1242/dev.057331
Gotz, S., Garcia-Gomez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2go suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Gu, X. F., Yang, A. F., Meng, H., and Zhang, J. R. (2005). In vitro induction of tetraploid plants from diploid Zizyphus jujuba mill. cv. Zhanhua. Plant Cell Rep. 24, 671–676. doi: 10.1007/s00299-005-0017-1
Guilfoyle, T. J., and Hagen, G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460. doi: 10.1016/j.pbi.2007.08.014
Harlow, W., Harrar, E., Hardin, J. W., and White, F. M. (1996). Textbook of Dendrology. 8th Edn. New York, NY, United States: McGraw-Hill, 534.
He, J. X., Gendron, J. M., Sun, Y., Gampala, S. S. L., Gendron, N., Sun, C. Q., et al. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638. doi: 10.1126/science.1107580
Hedden, P., and Phillips, A. L. (2000). Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5, 523–530. doi: 10.1016/s1360-1385(00)01790-8
Jiao, Y., Wickett, N. J., Ayyampalayam, S., Chanderbali, A. S., Landherr, L., Ralph, P. E., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. doi: 10.1038/nature09916
Ju, Y., Feng, L., Wu, J., Ye, Y., Zheng, T., Cai, M., et al. (2018). Transcriptome analysis of the genes regulating phytohormone and cellular patterning in Lagerstroemia plant architecture. Sci. Rep. 8:15162. doi: 10.1038/s41598-018-33506-8
Krizek, B. A. (2009). Making bigger plants: key regulators of final organ size. Curr. Opin. Plant Biol. 12, 17–22. doi: 10.1016/j.pbi.2008.09.006
Lawit, S. J., Wych, H. M., Xu, D., Kundu, S., and Tomes, D. T. (2010). Maize DELLA proteins dwarf plant8 and dwarf plant9 as modulators of plant development. Plant Cell Physiol. 51, 1854–1868. doi: 10.1093/pcp/pcq153
Leitch, A. R., and Leitch, I. J. (2008). Perspective – genomic plasticity and the diversity of polyploid plants. Science 320, 481–483. doi: 10.1126/science.1153585
Levan, A. (1939). Tetraploidy and octaploidy induced by colchicines in diploid Petunia. Hereditas 25, 109–131. doi: 10.1111/j.1601-5223.1939.tb02689.x
Li, W., Katin-Grazzini, L., Gu, X., Wang, X., El-Tanbouly, R., Yer, H., et al. (2017). Transcriptome analysis reveals differential gene expression and a possible role of gibberellins in a shade-tolerant mutant of perennial ryegrass. Front. Plant Sci. 8:868. doi: 10.3389/fpls.2017.00868
Li, X., Yu, E., Fan, C., Zhang, C., Fu, T., and Zhou, Y. (2012). Developmental, cytological and transcriptional analysis of autotetraploid Arabidopsis. Planta 236, 579–596. doi: 10.1007/s00425-012-1629-7
Liao, T., Cheng, S., Zhu, X., Min, Y., and Kang, X. (2016). Effects of triploid status on growth, photosynthesis, and leaf area in Populus. Trees-Struct. Funct. 30, 1137–1147. doi: 10.1007/s00468-016-1352-2
Liscum, E., and Reed, J. W. (2002). Genetics of aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. doi: 10.1023/a:1015255030047
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Y., Xue, H., Zhang, L., Zhang, F., Ou, C., Wang, F., et al. (2016). Involvement of Auxin and Brassinosteroid in dwarfism of Autotetraploid apple (Malus x domestica). Sci. Rep. 6:26719. doi: 10.1038/srep26719
Mao, X. Z., Cai, T., Olyarchuk, J. G., and Wei, L. P. (2005). Automated genome annotation and pathway identification using the Kegg Orthology (Ko) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Merkle, S. A., and Battle, P. J. (2000). Enhancement of embryogenic culture initiation from tissues of mature sweetgum trees. Plant Cell Rep. 19, 268–273. doi: 10.1007/s002990050010
Merkle, S., Montello, P., Kormanik, T., and Le, H. (2010). Propagation of novel hybrid sweetgum phenotypes for ornamental use via somatic embryogenesis. Propag. Ornam. Plants 10, 220–226.
Miller, M., Zhang, C., and Chen, Z. J. (2012). Ploidy and hybridity effects on growth vigor and gene expression in Arabidopsis thaliana hybrids and their parents. G3 2, 505–513. doi: 10.1534/g3.112.002162
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
Mu, H.-Z., Liu, Z.-J., Lin, L., Li, H.-Y., Jiang, J., and Liu, G.-F. (2012). Transcriptomic analysis of phenotypic changes in birch (Betula platyphylla) Autotetraploids. Int. J. Mol. Sci. 13, 13012–13029. doi: 10.3390/ijms131013012
Nandagopal, S., and Kumari, B. R. (2007). Effectiveness of auxin induced in vitro root culture in chicory. J. Cent. Euro. Agricult. 8:8.
Ni, Z., Kim, E.-D., Ha, M., Lackey, E., Liu, J., Zhang, Y., et al. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457, 327–331. doi: 10.1038/nature07523
Nilanthi, D., Chen, X.-L., Zhao, F.-C., Yang, Y.-S., and Wu, H. (2009). Induction of Tetraploids from petiole explants through colchicine treatments in Echinacea purpurea L. J. Biomed. Biotechnol. 2009, 1–7. doi: 10.1155/2009/343485
Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Tax, F. E., Yoshida, S., et al. (2000). Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 124, 201–210. doi: 10.1104/pp.124.1.201
Rao, S., Kang, X., Li, J., and Chen, J. (2019). Induction, identification and characterization of tetraploidy in Lycium ruthenicum. Breed. Sci. 69, 160–168. doi: 10.1270/jsbbs.18144
Rinne, P. L. H., Welling, A., Vahala, J., Ripel, L., Ruonala, R., Kangasjarvi, J., et al. (2011). Chilling of dormant buds Hyperinduces flowering locus T and recruits GA-inducible 1,3-beta-Glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 23, 130–146. doi: 10.1105/tpc.110.081307
Ruttink, T., Arend, M., Morreel, K., Storme, V., Rombauts, S., Fromm, J., et al. (2007). A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19, 2370–2390. doi: 10.1105/tpc.107.052811
Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., et al. (2003). Tm4: a free, open-source system for microarray data management and analysis. BioTechniques 34, 374–378. doi: 10.2144/03342mt01
Salma, U., Kundu, S., Hazra, A. K., Ali, M. N., and Mandal, N. (2018). Augmentation of wedelolactone through in vitro tetraploid induction in Eclipta alba (L.) Hassk. Plant Cell Tissue Org. Cult. 133, 289–298. doi: 10.1007/s11240-018-1381-1
Sasaki, H. (2002). Brassinolide promotes adventitious shoot regeneration from cauliflower hypocotyl segments. PCTOC 71, 111–116. doi: 10.1023/A:1019913604202
Sasse, J. M. (1997). Recent progress in brassinosteroid research. J Physiologia Plantarum. 100, 696–701. doi: 10.1034/j.1399-3054.1997.1000333.x
Shao, J. Z., Chen, C. L., and Deng, X. X. (2003). In vitro induction of tetraploid in pomegranate (Punica granatum). Plant Cell Tissue Org. Cult. 75, 241–246. doi: 10.1023/a:1025871810813
Sommer, S. E., Wetzstein, S. Y., and Brown, S. L. (1999). Shoot growth and histological response of dwarf sweetgum to gibberellin. J. Horticult. Sci. Biotechnol. 74, 618–621. doi: 10.1080/14620316.1999.11511163
Song, Q., and Chen, Z. J. (2015). Epigenetic and developmental regulation in plant polyploids. Curr. Opin. Plant Biol. 24, 101–109. doi: 10.1016/j.pbi.2015.02.007
Su, Y.-H., Liu, Y.-B., and Zhang, X.-S. (2011). Auxin-Cytokinin interaction regulates meristem development. Mol. Plant 4, 616–625. doi: 10.1093/mp/ssr007
Sun, T.-P. (2010). Gibberellin-Gid1-Della: A pivotal regulatory module for plant growth and development. Plant Physiol. 154, 567–570. doi: 10.1104/pp.110.161554
Sun, R. X., Lin, F. R., Huang, P., and Zheng, Y. Q. (2016). Moderate genetic diversity and genetic differentiation in the relict tree liquidambar formosana hance revealed by genic simple sequence repeat markers. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01411
Swarup, R., and Bhosale, R. (2019). Developmental roles of AUX1/LAX Auxin influx carriers in plants. Front. Plant Sci. 10:1306. doi: 10.3389/fpls.2019.01306
Takatsuka, H., and Umeda, M. (2014). Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 65, 2633–2643. doi: 10.1093/jxb/ert485
Tan, F.-Q., Tu, H., Liang, W.-J., Long, J.-M., Wu, X.-M., Zhang, H.-Y., et al. (2015). Comparative metabolic and transcriptional analysis of a doubled diploid and its diploid citrus rootstock (C. junos cv. Ziyang Xiangcheng) suggests its potential value for stress resistance improvement. BMC Plant Biol. 15:89. doi: 10.1186/s12870-015-0450-4
Tanino, K. K. (2004). Hormones and endodormancy induction in woody plants. J. Crop Improv. 10, 157–199.
Tao, Y., Ferrer, J.-L., Ljung, K., Pojer, F., Hong, F., Long, J. A., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. doi: 10.1016/j.cell.2008.01.049
Tiku, A. R., Razdan, M. K., and Raina, S. N. (2014). Production of triploid plants from endosperm cultures of Phlox drummondii. Biol. Plant. 58, 153–158. doi: 10.1007/s10535-013-0372-7
Tokumoto, Y., Kajiura, H., Takeno, S., Harada, Y., Suzuki, N., Hosaka, T., et al. (2016). Induction of tetraploid hardy rubber tree, Eucommia ulmoides, and phenotypic differences from diploid. Plant Biotechnol. 33, 51–57. doi: 10.5511/plantbiotechnology.15.1219a
Tong, J. (2007). [D] Studies on Polyploid Induction Induction of Lagerstroemia indica by Colchicne. Wuhan: Huazhong Agricultural University.
Tsukaya, H. (2008). Controlling size in multicellular organs: focus on the leaf. PLoS Biol. 6, e174–e1376. doi: 10.1371/journal.pbio.0060174
Wang, T., Liu, L., Wang, X., Liang, L., Yue, J., and Li, L. (2018a). Comparative analyses of anatomical structure, Phytohormone levels, and gene expression profiles reveal potential dwarfing mechanisms in Shengyin bamboo (Phyllostachys edulis f. tubaeformis). Int. J. Mol. Sci. 19:1697. doi: 10.3390/ijms19061697
Wang, Z. Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., et al. (2002). Nuclear-localized Bzr1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513. doi: 10.1016/s1534-5807(02)00153-3
Wang, Z., Zhang, Y., Qi, S., Zhu, J., Yuan, W., Zhang, Y., et al. (2018b). Tissue culture regeneration of hybrid Liquidambar styraciflua×L. formosana. J. Beijing For. Univ. 40, 42–49.
Willige, B. C., Isono, E., Richter, R., Zourelidou, M., and Schwechheimer, C. (2011). Gibberellin regulates pin-formed abundance and is required for Auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell 23, 2184–2195. doi: 10.1105/tpc.111.086355
Won, C., Shen, X., Mashiguchi, K., Zheng, Z., Dai, X., Cheng, Y., et al. (2011). “Conversion of tryptophan to indole-3-acetic acid by tryptophan aminotransferase of Arabidopsis and YUCCAs in Arabidopsis.” Proceedings of the National Academy of Sciences of the United States of America 108, 18518–18523. doi: 10.1073/pnas.1108436108
Xu, C., Zhang, Y., Huang, Z., Yao, P., Li, Y., and Kang, X. (2018). Impact of the leaf cut callus development stages of Populus on the Tetraploid production rate by colchicine treatment. J. Plant Growth Regul. 37, 635–644. doi: 10.1007/s00344-017-9763-x
Xue, H., Zhang, B., Tian, J.-R., Chen, M.-M., Zhang, Y.-Y., Zhang, Z.-H., et al. (2017). Comparison of the morphology, growth and development of diploid and autotetraploid ‘Hanfu’ apple trees. Sci. Hortic. 225, 277–285. doi: 10.1016/j.scienta.2017.06.059
Ye, Y. M., Tong, J., Shi, X. P., Yuan, W., and Li, G. R. (2010). Morphological and cytological studies of diploid and colchicine-induced tetraploid lines of crape myrtle (Lagerstroemia indica L.). Sci. Hortic. 124, 95–101. doi: 10.1016/j.scienta.2009.12.016
Zhang, C., Fu, S., Tang, G., Hu, X., and Guo, J. (2013). Factors influencing direct shoot regeneration from mature leaves of Jatropha curcas, an important biofuel plant. In Vitro Cell. Dev. Biol. Plant 49, 529–540. doi: 10.1007/s11627-013-9530-z
Zhang, Y., Wang, B., Guo, L., Xu, W., Wang, Z., Li, B., et al. (2018). Factors influencing direct shoot regeneration from leaves, petioles, and plantlet roots of triploid hybrid Populus sect. Tacamahaca. J. For. Research 29, 1533–1545. doi: 10.1007/s11676-017-0559-4
Zhang, Y., Wang, B., Qi, S., Dong, M., Wang, Z., Li, Y., et al. (2019). Ploidy and hybridity effects on leaf size, cell size and related genes expression in triploids, diploids and their parents in Populus. Planta 249, 635–646. doi: 10.1007/s00425-018-3029-0
Zhang, Y., Wang, Z., Qi, S., Wang, X., Zhao, J., Zhang, J., et al. (2017). In vitro Tetraploid induction from leaf and petiole explants of hybrid Sweetgum (Liquidambar styraciflua x Liquidambar formosana). Forests 8:264. doi: 10.3390/f8080264
Zhao, Y. (2012). Auxin biosynthesis: a simple two-step pathway converts tryptophan to Indole-3-acetic acid in plants. Mol. Plant 5, 334–338. doi: 10.1093/mp/ssr104
Zheng, Y., Pan, B., and Itoh, T. (2015). Chemical induction of traumatic gum ducts in Chinese sweetgum, liquidambar formosana. IAWA J. 36, 58–68. doi: 10.1163/22941932-00000085
Keywords: polyploidization, RNA-seq, dwarf, plant hormone, hybrid sweetgum
Citation: Chen S, Zhang Y, Zhang T, Zhan D, Pang Z, Zhao J and Zhang J (2022) Comparative Transcriptomic, Anatomical and Phytohormone Analyses Provide New Insights Into Hormone-Mediated Tetraploid Dwarfing in Hybrid Sweetgum (Liquidambar styraciflua × L. formosana). Front. Plant Sci. 13:924044. doi: 10.3389/fpls.2022.924044
Received: 20 April 2022; Accepted: 24 May 2022;
Published: 27 June 2022.
Edited by:
Geoffrey Meru, University of Florida, United StatesReviewed by:
Wei Seng Ho, Universiti Malaysia Sarawak, MalaysiaCopyright © 2022 Chen, Zhang, Zhang, Zhan, Pang, Zhao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Zhao, emhhb2ppYW4wNzAzQGJqZnUuZWR1LmNu; Jinfeng Zhang, empmQGJqZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.