- 1Department of Plant Pathology, University of California, Davis, Davis, CA, United States
- 2United States Department of Agriculture, Agricultural Research Service, Salinas, CA, United States
Currently, Fusarium oxysporum f. sp. apii (Foa) race 4 in celery and F. oxysporum f. sp. coriandrii (Foci) in coriander have the characteristics of emerging infectious plant diseases in coastal southern California: the pathogens are spreading, yield losses can be severe, and there are currently no economical solutions for their control. Celery, and possibly coriander, production in these regions is are likely to have more severe disease from projected warmer conditions in the historically cool, coastal regions. Experimental evidence shows that Foa race 4 causes much higher disease severity when temperatures exceed 21°C. A phylogenomic analysis indicated that Foa race 4, an older, less virulent, and uncommon Foa race 3, and two Foci are closely related in their conserved genomes. These closely related genotypes are somatically compatible. Foa race 4 can also cause disease in coriander and the two organisms readily form “hetero” conidial anastomosis tubes (CAT), further increasing the likelihood of parasexual recombination and the generation of novel pathotypes. A horizontal chromosome transfer event likely accounts for the difference in host range between Foci versus Foa races 4 and 3 because they differ primarily in one or two accessory chromosomes. How Foa race 4 evolved its hyper-virulence is unknown. Although the accessory chromosomes of Foa races 3 and 4 are highly similar, there is no evidence that Foa race 4 evolved directly from race 3, and races 3 and 4 probably only have a common ancestor. Foa race 2, which is in a different clade within the Fusarium oxysporum species complex (FOSC) than the other Foa, did not contribute to the evolution of race 4, and does not form CATs with Foa race 4; consequently, while inter-isolate CAT formation is genetically less restrictive than somatic compatibility, it might be more restricted between FOSC clades than currently known. Other relatively new F. oxysporum in coastal California include F. oxysporum f. sp. fragariae on strawberry (Fof). Curiously, Fof “yellows-fragariae” isolates also have similar core genomes to Foa races 4 and 3 and Foci, perhaps suggesting that there may be core genome factors in this lineage that favor establishment in these soils.
Introduction
Fusarium oxysporum f. sp. apii (Foa) race 4 is a pathogen of celery (Apium graveolens var. dulce) that causes an emerging disease in California, United States. Foa race 4 apparently arose in approximately 2013 (Epstein et al., 2017), is highly aggressive at temperatures above 22°C (Kaur et al., 2022), has been spreading in its geographic distribution within California, and currently cannot be controlled via either host resistance or economical methods that reduce the pathogen abundance in infested soil. Although F. oxysporum f. sp. apii have been long-recognized, first by Wollenweber in 1917, (Snyder and Hansen, 1940; Armstrong and Armstrong, 1981; Edel-Hermann and Lecomte, 2019), Foa are not currently as well known by the scientific community as forma speciales (ff. spp.) such as lycopersici on tomato, cubense on banana, conglutinans on Arabidopsis and cabbage, etc. (Husaini et al., 2018; Yang et al., 2020). Whole genome sequencing and analysis was recently conducted for Foa races 2, 3, and 4, and F. oxysporum f. sp. coriandrii (Foci), a pathogen of coriander (synonym, cilantro; Coriandrum sativum; Henry et al., 2020). In this review, we highlight aspects of the biology of Foa and its host–pathogen interactions that are potentially relevant for a broader understanding of other Fusarium pathosystems.
History
What we now call Foa race 1, causal agent of Fusarium yellows of celery, is polymorphic (i.e., highly variable in their two-locus haplotypes), and apparently endemic in European and United States soils (Nelson et al., 1937; O’Donnell et al., 2009). In California, Foa race 1 strains are only virulent on heirloom, “yellow” celery (Apium graveolens var. dulce) cultivars that have not been commercially important since the 1950’s (Epstein et al., 2017). Foa race 2 was first observed in approximately 1959 on the “green” Pascal-type celery cultivars, e.g., Tall Utah, in California (Otto et al., 1976) (Figure 1); thereafter, race 2 was distributed, presumably on farm equipment, into all celery production areas in California. After Foa race 2 was established in California, it was apparently transported to other states (Subbarao and Elmer, 2002), Canada (Cerkauskas and Chiba, 1991), Japan (Akanuma and Shimizu, 1994), and Argentina (Lori et al., 2016), presumably on infested seed. If celery seed is produced in a field with Foa inoculum in the soil, a presumably low percentage of seed can be contaminated by wind-blown soil particles. Population sampling suggests that Foa race 2 is a monomorphic clonal lineage in the Fusarium oxysporum species complex (FOSC) clade 3 (Epstein et al., 2017). Largely based on somatic compatibility groups, Foa race 3 (Puhalla, 1984) was also present at least for some time in California, but is not highly virulent and has never been an important pathogen. More recently, Foa race 3 was identified in Costa Rica (Retana et al., 2018). After introgression of gene(s) for resistance from celeriac (Apium graveolens var. rapaceum; Orton et al., 1984b), Foa race 2 (and any race 3) were adequately controlled by resistant cultivars, e.g., cv. Challenger (Figure 1). Foa race 4 apparently first appeared in California in Camarillo, Ventura County in ca. 2013. Within a decade after its first sighting, race 4 had been moved, most likely on farm equipment, first to all celery production areas within Ventura county on the south coast, and more recently to Monterey county on the central coast. Foa race 1 haplotypes, Foa race 3, and Foa race 4 are in FOSC clade 2, which means that the newly emerged race 4 is only more distantly related in its core genome to Foa race 2.

Figure 1. Virulence of three F. oxysporum f. sp. apii (Foa) races on celery cultivars Tall Utah and Challenger. Two-month-old celery cultivars were transplanted into uninfested soil (mock) or soil infested with a Foa race. After 49 days, the median plant (n = 20) in height was photographed. Tall Utah 5270R-Improved is susceptible to the three races and Challenger is resistant to Foa races 2 and 3 but susceptible to Foa race 4. The image is a portion of a figure from Henry et al. (2020). BMC Genomics 21, No. 730 and is reprinted in accordance with BMC Genomics.
Foci was considered an insufficiently documented forma specialis (Edel-Hermann and Lecomte, 2019), but our recent work should change that. The disease was first reported in California in Santa Barbara County (Koike and Gordon, 2005). In Ventura county, during the time period in which Foa race 4 apparently arose, there was also an increase in Fusarium wilt in coriander, caused by Foci; similar to Foa race 4, the Foci are highly virulent on their host. Interestingly, Foa race 4 can cause modest disease in coriander, although Foci are more virulent on coriander than Foa race 4; both pathogens can co-infect coriander (Henry et al., 2020). The Foci are avirulent on celery. Both celery and coriander are in the family Apiaceae, and the genomic similarities between Foa race 4 and Foci suggest there may be common susceptibility factors in celery and coriander.
Symptoms
All Foa induce celery to produce a vascular discoloration in infected roots and crown; in young transplants, the discoloration is orangish-brown (Figure 2A), and in older plants, the discoloration is a darker brown (Figure 2B). In severe cases, particularly with Foa race 4, the discoloration can extend into the base of the petioles. Stunting is strongly associated with vascular discoloration on more than 1/4 of the circumference of the vascular ring in the crown (Kaur et al., 2022). The extent of stunting and vascular discoloration is also associated with more Foa biomass in the celery crown (Kaur et al., 2022). Both Foa races 2 and 4 can cause some chlorosis, frequently in a lower leaf, but chlorosis (Figure 2C), is typically more noticeable in a field with race 2 than race 4. Both Foa races 2 and 4 cause stunting and a loss of yield and quality. However, at similar inoculum levels, particularly in soil temperatures above 21°C, Foa race 4 causes far more severe disease than Foa race 2 (Figure 1). In addition to severe stunting, Foa race 4 can cause death (Figure 2D), particularly on young plants; typically, the progression to death begins with stunting, sometimes with lower leaf chlorosis and proceeds to wilting of the entire plant and death. In the field, particularly in wet conditions, race 4 infections can result in root necrosis/sloughing off of the roots and/or a crown rot.
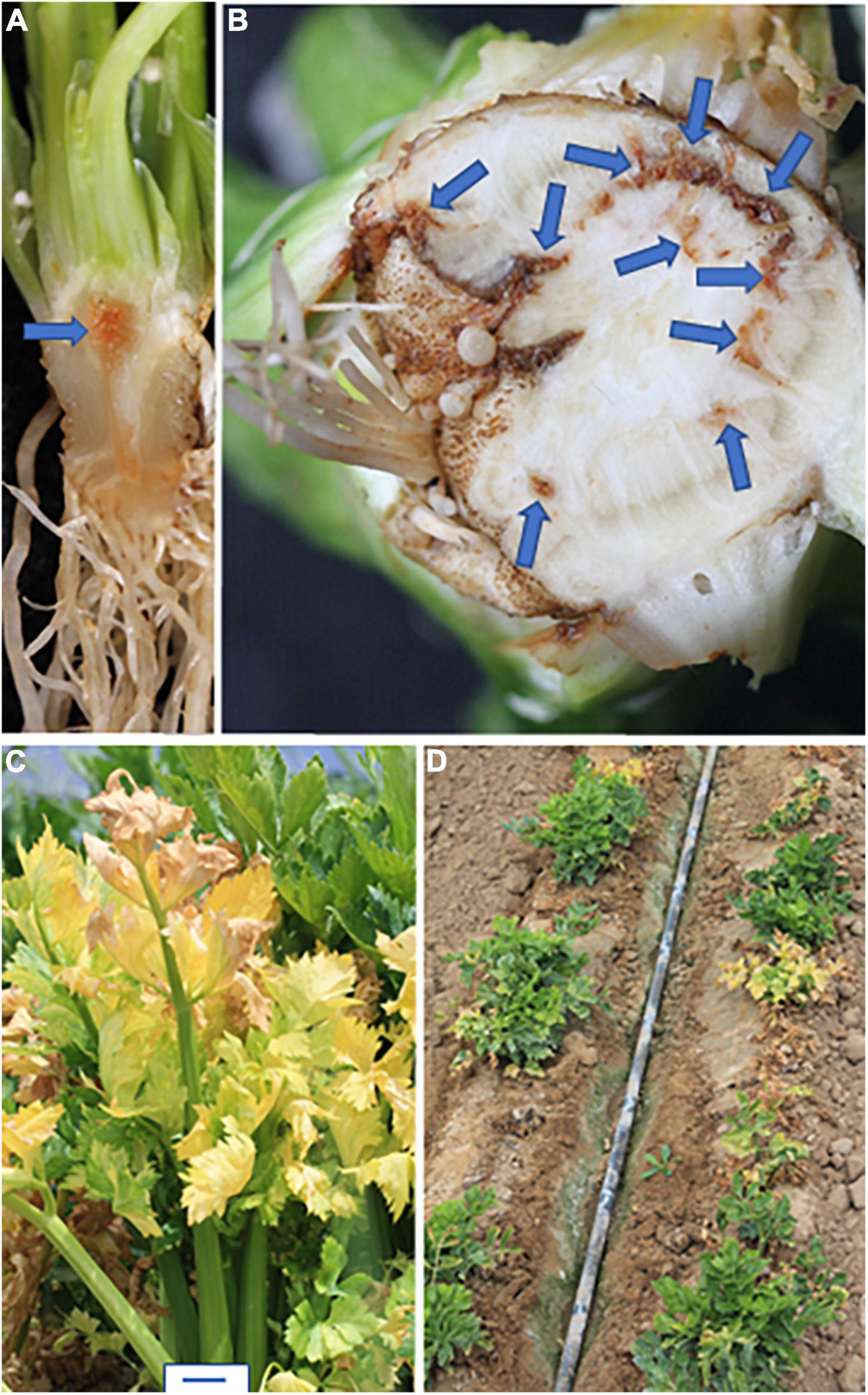
Figure 2. Symptoms of F. oxysporum f. sp. apii (Foa) in celery. (A,B) Vascular discoloration in the crown (shown here) and roots. These plants were infected with Foa race 4. (A) In young transplants, the discoloration is orangish-brown. (B) In older plants, the discoloration is darker brown. This field-grown celery plant had vascular discoloration on more than 25% of the circumference of the crown and was severely stunted. (C) Chlorosis of older leaves, here caused by Foa race 2. (D) Death and severe stunting of young plants caused by Foa race 4. Size bar = 2 cm.
In contrast to celery, which is transplanted into the field in California, coriander is direct-seeded. As a consequence, although Foci produce similar symptoms on coriander as Foa race 4 produces on celery, Foci-infected coriander can also have poor seedling emergence. In the field, Foci-infected plants are stunted, often with chlorotic lower foliage (Koike and Gordon, 2005); the vascular discoloration in the roots and crown can extend into the petioles. Particularly when younger plants are infected, typical disease progression includes sudden wilting and rapid death. Fibrous roots are sloughed off and the taproot is often rotted. With Foci in coriander, the vascular discoloration is reddish to light brown.
Lessons From Fusarium oxysporum f. sp. apii and Fusarium oxysporum f. sp. coriandrii in California
Genetic Plasticity Challenges Disease Management and Molecular Diagnostics
Few Genetic Differences Can Cause Major Differences in Host Specificity and Virulence
In individual strains of F. oxysporum, chromosomes can be classified as part of either the “core” genome that is conserved amongst strains or the “accessory” (i.e., dispensable) genome that is variable amongst strains (Yang et al., 2020). A landmark discovery by Ma et al. (2010) demonstrated that accessory chromosomes could be horizontally transferred between strains of F. oxysporum with the genes required for pathogenicity in a specific host. Specifically, they demonstrated the transfer of a chromosome from a tomato pathogen (Fol4287) to a previously non-pathogenic recipient (Fo47); the transformed Fo47+ was pathogenic on tomato. Since this discovery, multiple pathogenicity chromosomes in the accessory genome, and in some cases, just portions of a pathogenicity chromosome, have been identified that encode for all the genes necessary to transform a non-pathogen into a pathogen of a specific crop, including cucumber, melon, and cabbage (Vlaardingerbroek et al., 2016; van Dam et al., 2017; Li et al., 2020; Ayukawa et al., 2021). The work has revealed that relatively few genetic differences can have profound impacts on pathogenicity and host range.
Although there are profound differences in symptom severity and host range between Foa race 3, race 4, and Foci, they have similar genomes. In a comparison of 2,718 conserved, single copy orthologs in the core genome, we discovered that the low-virulence Foa race 3, highly virulent Foa race 4, and Foci isolates are tightly clustered in a monophyletic group (Figure 3; Henry et al., 2020). Given this close phylogenetic relationship, many genomic similarities are vertically inherited and identical by descent in these three groups. Figure 4 shows an analysis of the extent of conservation and lack thereof of both the core and accessory genomes between the three Foa and two Foci strains. Foa race 3 and race 4 are nearly identical in their core and their accessory genomes (Figure 4A), despite their profound differences in virulence (Henry et al., 2020). The two Foci, which were collected 12 years apart from two counties, have highly similar core and accessory genomes to each other (Figure 4B; Henry et al., 2020). In contrast, the two Foci differ from Foa race 3 and race 4 in approximately 1/3 of their accessory genome (Henry et al., 2020), i.e., probably in one or possibly two accessory chromosomes. The differences in host range between these Foa and Foci strains are likely to be caused by presence/absence variation in the divergent portion of the accessory genome.
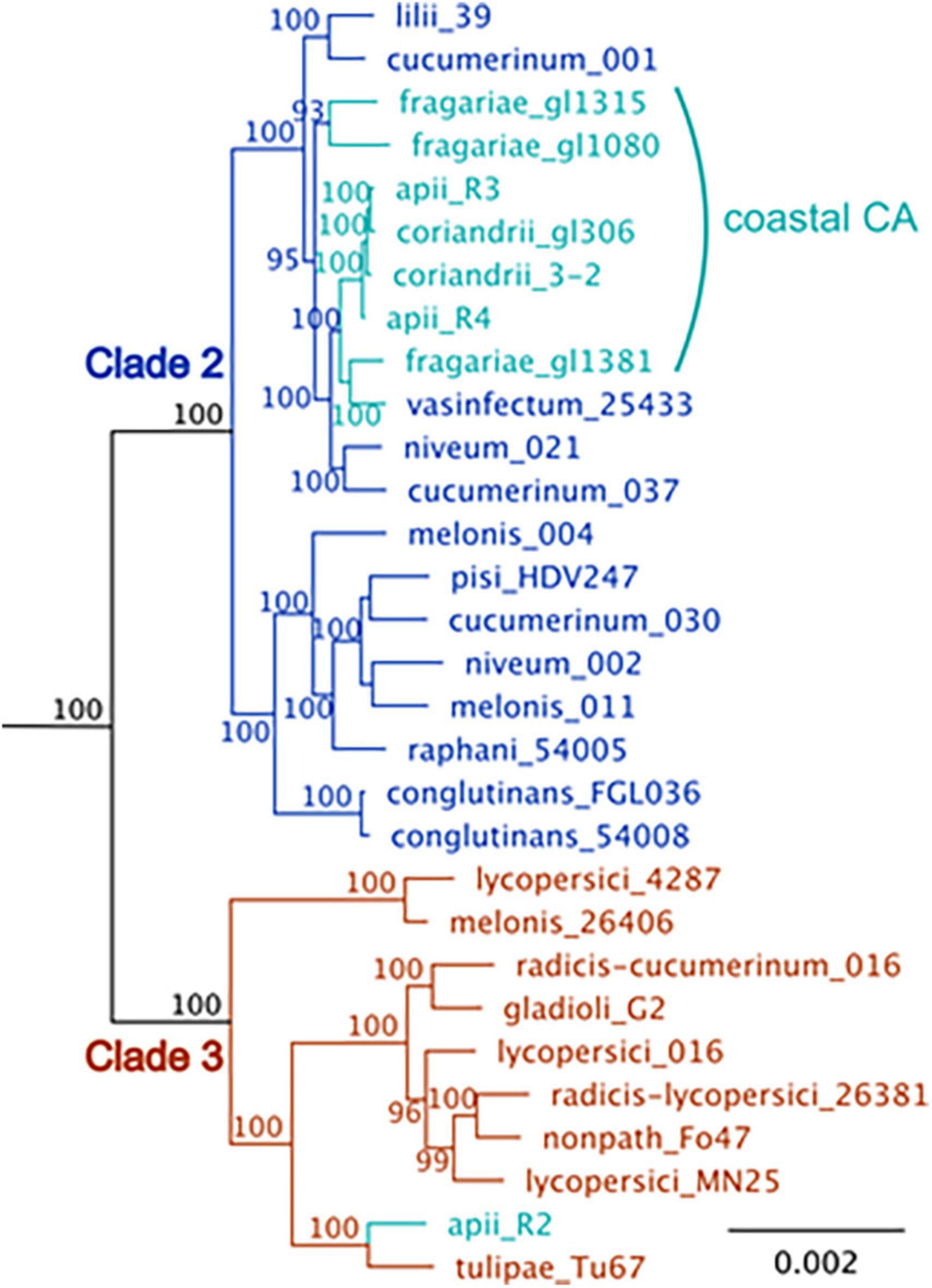
Figure 3. A maximum-likelihood phylogenetic tree of F. oxysporum isolates that cause disease in celery (apii), strawberry (fragariae), and coriander (coriandrii) in comparison to selected assemblies of F. oxysporum from NCBI. The figure is modified from Henry et al. (2020) with the addition of three F. oxysporum f. sp. fragariae sequences from Henry et al. (2021). BUSCO v. 2.0 (Benchmark of Unique Single Copy Orthologs) was used to identify 2,718 full-length, single-copy genes that were in all strains; these genes are primarily in the core genome. Sequences were aligned with MUSCLE and concatenated into a single ∼ 5.5 Mbp sequence. The tree was generated with RaxML with the general time-reversible evolutionary model, rooted with F. oxysporum f. sp. cubense tropical race 4, which is in Fusarium oxysporum species complex (FOSC) clade 1. Support for the tree is based on 1000 bootstrap replicates; bootstrap values below 70 are not shown. The coastal California isolates are shown in turquoise. F. oxysporum f. sp. apii race 2 (apii_R2), is in FOSC clade 3. The other F. oxysporum f. sp. apii (apii, R3, race 3) and (apii race 4, apii_R4) are in FOSC clade 2.
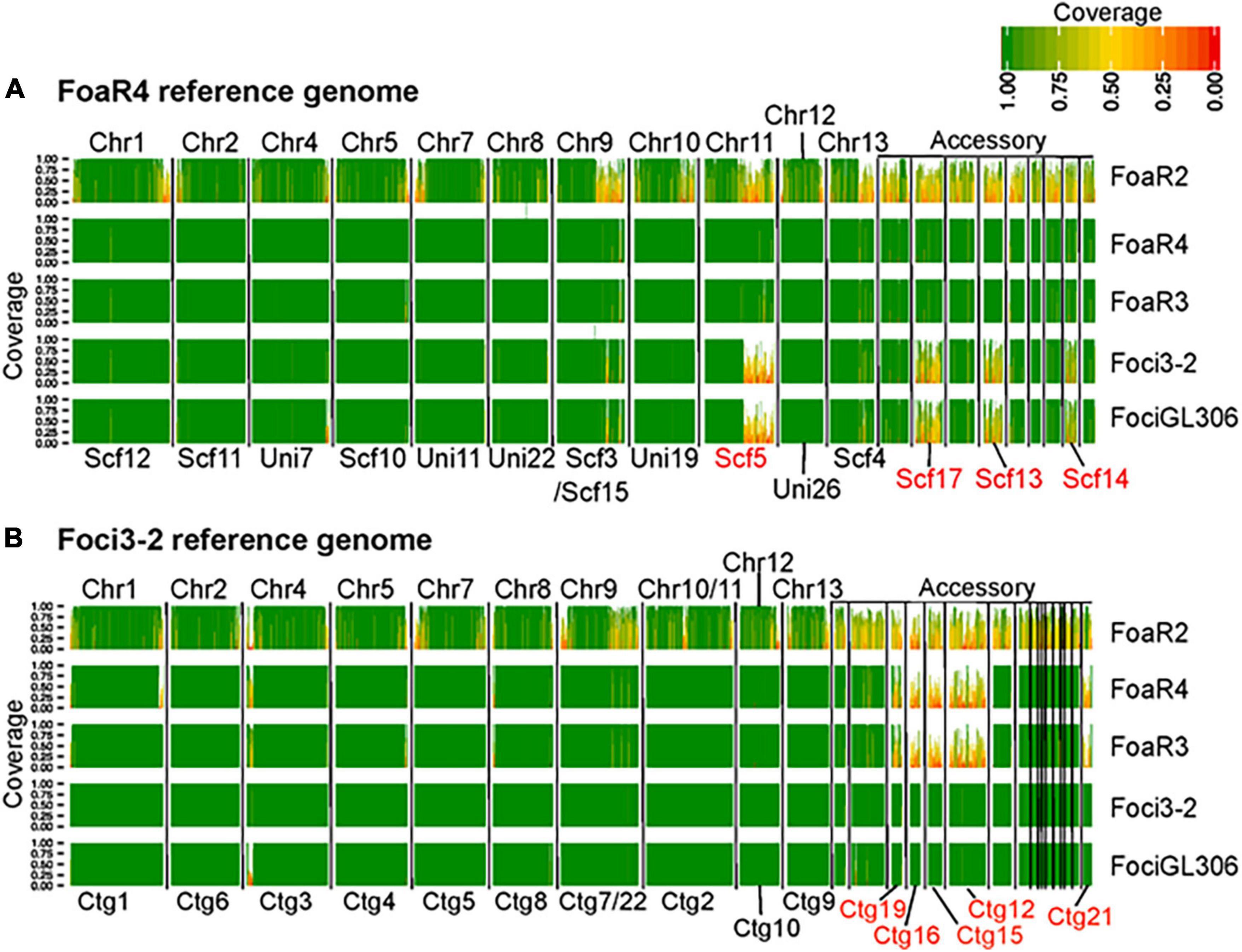
Figure 4. A comparative genomic analysis showing the extent of conservation in the core and accessory genomes between F. oxysporum f. sp. apii (Foa) race 4 and f. sp. apii races 2 and 3, and two strains of F. oxsporum f. sp. coriandrii (Foci). For this analysis, we used (A) Foa race 4 and (B) Foci 3–2 as reference assemblies, based on PacBio and Illumina sequencing. For each of the five strains, we mapped 6.5 Gbp (∼100× coverage) of quality-filtered Illumina reads onto each reference, and calculated the proportion of coverage of each 10 kbp window in the reference assemblies. Here, only contigs with a length greater than 150 kbp are shown and are separated by vertical black lines. The darkest green sections of the histogram have a 100% coverage and the reddest sections have coverage close to 0%. Coverage of 0.5 (yellow) indicates that only 5 kbp of the 10 kbp segment had Illumina coverage. The core chromosome (Chr) number of the F. oxysporum f. sp. lycopersici 4287 reference are labeled above each plot toward the left. The accessory portion of the reference genomes is indicated on the upper right of each figure. The IDs for contigs (Ctg) and scaffolds (Scf) that are associated with host specificity in our two references are lettered in red, below the plots. Each row shows the coverage from a single isolate, which is noted to the right of the graph (Foa race 2, FoaR2; Foa race 3, FoaR3). The image is a figure from Henry et al. (2020) (BMC Genomics 21, No. 730) and is reprinted in accordance with BMC Genomics.
The similarity between the accessory genomes of Foa race 4 and race 3 suggests that few genetic differences account for the hypervirulence in race 4. However, perhaps as because as small a difference as a single nucleotide polymorphism might account for the hyper-virulence, to date, we have not succeeded in generating a viable hypothesis of why Foa race 4 is hyper-virulent. None of the highly expressed putative effectors in Foa race 4, which would be prime candidates for disruption, are absent in Foa race 3. Furthermore, Foa race 3 produces comparatively little biomass in planta, making it extremely difficult to obtain sufficient RNAseq reads for Foa race 3 infection-specific transcriptome profiling (Kaur and Epstein, unpublished). We initially identified one potential avirulence effector in Foa race 3 in its accessory genome (Kaur, Epstein, Henry, and Stergiopolos, unpublished): the DNA (GenBank MT349842.1) is absent in Foa race 4, and based on reverse transcriptase real-time PCR, the gene is expressed in planta in race 3-infected celery but not in race 3 in vitro, or negative controls. However, after performing an Agrobacterium tumefaciens-mediated knock-out (Gold et al., 2017), the race 3 knock-outs have the same virulence as the wild type and ectopic recombinant controls, indicating that this gene is not an avirulence factor (data not shown).
As part of an investigation of potential effectors and virulence factors in Foa race 4, we quantified gene expression in planta within the crown compared to in culture. Two of the notable genes that were highly expressed and up-regulated in planta were transposons (Henry et al., 2020). Clearly, transposon activity in F. oxysporum contributes to isolate diversity, the potential for the evolution of new pathotypes, and potentially the loss of the informative markers used in molecular diagnostics.
Horizontal Chromosome Transfer Could Account for the Major Difference in the Accessory Genomes of Fusarium oxysporum f. sp. apii Race 4 and Fusarium oxysporum f. sp. coriandrii and for Their Differences in Host Range, but Not the Extreme Differences in Virulence Between Fusarium oxysporum f. sp. apii Races 3 and 4
Our work further supports the view that accessory chromosomes are present in a “mosaic” pattern between strains, where a range of presence/absence variation among similar chromosomes can produce new host specificities (Figure 4). Although our assemblies are fragmented in 50–75 contigs per genome, we can deduce that horizontal chromosome transfer must have occurred from an unknown source to either the Foci or to the progenitor of the Foa FOSC clade 2. This is the most parsimonious explanation for the presence/absence variation in approximately 1/3 of the accessory genome between these isolates that otherwise have nearly identical, vertically inherited genomes. In this scenario, the gain of one or more accessory chromosomes would have been accompanied with a concomitant loss of another accessory chromosome(s). This further substantiates the potential for these organisms and for a broader array of F. oxysporum to generate new combinations of accessory chromosomes that will contribute to its ability to evolve new pathogenic genotypes in the future.
In contrast, as illustrated in Figure 4, the hypervirulence of Foa race 4, in comparison to Foa race 3, was not caused by the acquisition of a large (> 0.5 Mbp) accessory chromosomal region from either any of the strains that we sequenced or from any other full genome-sequenced F. oxysporum (Henry et al., 2020). The Foa race 4 strain shares almost all (> 95%) of its accessory genome with Foa race 3.
Conidial Anastomosis Tube Formation and Somatic Compatibility Between Fusarium oxysporum f. sp. apii Race 4 and Fusarium oxysporum f. sp. coriandrii Increase the Potential for New Recombinants to Emerge
In general, somatic compatibility groups (SCGs), also known as vegetative compatibility groups or VCGs, have been a biologically useful way of indicating whether two F. oxysporum are closely related (Correll et al., 1986a). After our initial phylogenetic data indicated that an older Foa race 3, which we retrieved from a culture collection, and the emergent Foa race 4 were indistinguishable in our initial phylogenetic data (Epstein et al., 2017), we determined that Foa race 3 and 4 are in the same SCG, which indicates that they are closely related and share the requisite somatic compatibility gene alleles. Similarly, after we discovered the similarity in DNA sequence between Foci and Foa races 3 and 4, we demonstrated that Foci are also in the same SCG as Foa race 3 and race 4 (Henry et al., 2020). We are not aware of another example where organisms in two different formae speciales are in the same SCG. This somatic compatibility means that horizontal chromosome transfer could readily occur via hyphal fusion between co-localized Foci and Foa races 4 or 3. These strains can occur in the same fields and could be brought into close proximity by co-infection of root tissues.
Conidial anastomosis tubes are also an avenue for a horizontal chromosome transfer of an accessory/pathogenicity chromosome because nuclei can migrate across a CAT (Kurian et al., 2018), and importantly, CAT formation is a less restrictive barrier for nuclear entry than somatic/hyphal compatibility in F. oxysporum (Shahi et al., 2016), Colletotrichum lindemuthianum (Ishikawa et al., 2012), and Verticillium dahliae (Vangalis et al., 2021). Microconidia are frequently produced in planta in the xylem to facilitate systemic plant colonization. These conidia presumably could undergo CAT fusion. Because Foa race 4 and Foci can co-infect coriander and can form Foa race 4-Foci hetero-CATs (Figures 5A,B), coriander and, perhaps, celery may be a likely location for accessory chromosome transfer and evolution of new genotypes. We note that Foa race 4 and Foci form hetero-CATs (Figures 5C–F) as frequently as they form “homo” CATs (Henry et al., 2020). Thus, if Foa race 4 and Foci co-occur in conducive conditions for either somatic compatibility or CAT formation, they could undergo nuclear transfer and then horizontal chromosome transfer; such transfer could result in the evolution of a new genotype that could evade host recognition, increase virulence, and confound molecular detection of the new strain.
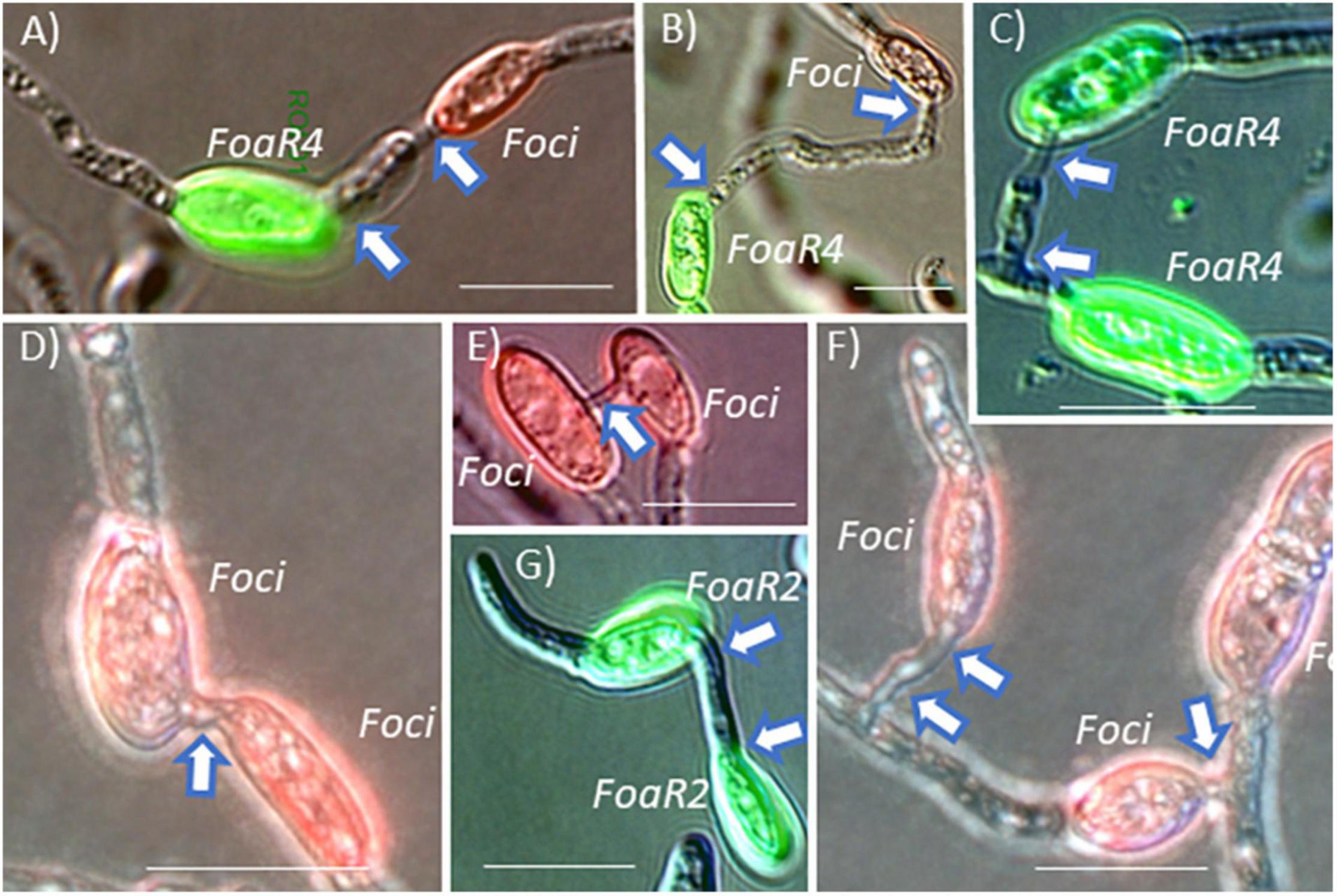
Figure 5. F. oxysporum microconidia were pre-stained with a fluorescent dye conjugate and then mixed with a differently labeled strain. After incubation on polystyrene in conditions conducive for conidial anastomosis tube (CAT) formation, “homo” and “hetero” CAT formation was enumerated (as shown in the original publication). (A,B) F. oxysporum f. sp. apii race 4 (FoaR4) and F. oxysporum f. sp. coriandrii 3-2 (Foci) formed hetero-CATs as frequently as each strain formed homo-CATs (C,D). (E) In compatible interactions, CATs were most commonly formed between two microconidia, but occasionally, CAT clusters with three microconidia (F) were observed. F. oxysporum f. sp. apii race 2 (FoaR2) also formed homo-CATs (G) but FoaR2 and FoaR4 were never observed to form a hetero-CAT. Short CATs are indicated with a single arrow and longer CATs are indicated with an arrow at each end of the CAT. Size bars = 10 μm. The image is a portion of a figure from Henry et al. (2020). (BMC Genomics 21, No. 730) and is reprinted in accordance with BMC Genomics.
Close Phylogenomic Relationships and the Potential for Recombination Complicate Efforts to Develop Molecular Diagnostics Specific to Fusarium oxysporum f. sp. apii Race 4 and Fusarium oxysporum f. sp. coriandrii
Using our whole genome shotgun sequences of Foa races 2, 3, and 4, and Foci, along with the NCBI GenBank wgs and selected SRA databases, we were able to design and validate PCR primers for detection of Foa races 2 and 4 and Foci either directly from plants or from culture (Henry et al., 2020). While empirical testing of a strain collection and in silico testing revealed comparatively few problems, not surprisingly, it is difficult to develop primers with no false positives. For example, in an in silico analysis of 437 F. oxysporum genomes in the Genbank wgs, and an additional 329 other Fusarium spp., the Foa race 4 primers FOAR4-447 would amplify 1.8% of the non-target F. oxysporum and one F. secorum.
Our preliminary results further suggest that recombination is occurring between strains of Foa race 4 and Foci in agricultural fields. After isolating from symptomatic coriander, we assembled a small collection of seven isolates that were bona fide Foci in that each caused disease on coriander but not on celery (Henry et al., 2020). In contrast to the other six Foci isolates, Foci10T was PCR-negative for two PCR primers on the host-specific Foci accessory genome (FOCI2-21 and FOCI-g_c31), and was PCR-positive for Foa race 4 primer R4-447, which is located an accessory region not shared by other, whole genome sequenced Foci isolates (Henry et al., 2020). This suggests that Foci10T lost regions of accessory chromosomes that are common to Foci and gained an Foa race 4-specific accessory chromosome region while retaining the avirulence-on-celery phenotype. This observation highlights the strong potential for recombination to confound efforts to distinguish these formae speciales solely with molecular markers.
There May Be a Barrier for Genetic Exchange Between Fusarium oxysporum f. sp. apii Race 4 and Race 2, and Consequently Between Other Members of Fusarium oxysporum Species Complex Clades 2 and 3
Fusarium oxysporum f. sp. apii (Foa) race 4 and Foa race 2 can be isolated from the same celery plant. When we initially observed co-occurring Foa races 2 and 4, and knew that the possibly extinct Foa race 3 was closely related to the new and highly aggressive Foa race 4, we postulated that Foa race 4 was perhaps the result of the acquisition of a Foa race 2 accessory chromosome in a Foa race 3 background. However, as illustrated in Figure 4A, the phylogenomic analyses indicate that Foa race 4 did not evolve from Foa race 2 and did not receive a portion of a chromosome from this extant pathogen of celery. Moreover, in our enumeration of CATs in assays with Foa race 2 and race 4 microconidia, we observed race 4-race 4 homo-CATs (Figure 5C) and race 2-race 2 homo-CATs (Figure 5G), but never a race 2-race 4 hetero-CAT. While one might argue that we would not have detected very low frequency events, nothing that we ever observed suggested that a Foa race 2 and a Foa race 4 might even start the process of CAT formation. Consequently, although CAT formation is a less restrictive conduit for nuclear entry than somatic/hyphal compatibility (Shahi et al., 2016), we postulate that there are more barriers than currently appreciated for inter-isolate CAT formation within the FOSC, and that barriers to CAT formation between FOSC clades may restrict nuclear entry and consequently accessory chromosome transfer. We note that the study on F. oxysporum CATs (Shahi et al., 2016) demonstrated transfer between two strains that are in FOSC clade 3: F. oxysporum f. sp. lycopersici 4287 and the biocontrol strain Fo47. Experimentally, using tagged pathogenicity chromosomes, Rep and colleagues have demonstrated that horizontal chromosome transfer can transform a non-pathogenic isolate (Fo47) into a pathogen of tomato (Ma et al., 2010), multiple cucurbits (van Dam et al., 2017), and melons (Li et al., 2020). However, those demonstrations, [with Fol4287, Forc016, and Fom001 (synonym, NRRL26406) and Fom005, respectively] are all within FOSC clade 3 (Figure 3), and the transfer between FOSC clades 2 and 3 has not been reported (van Dam et al., 2016; Henry et al., 2020). A critical question for future research in the broader Fusarium community is if two strains co-occur as microconidia within a host, are there limits to CAT formation between FOSC clades?
Climate Change Will Likely Exacerbate Fusarium Wilt Disease Severity and Incidence in Coastal California
The Disease Severity of Fusarium oxysporum f. sp. apii Race 4 in Celery and the Growth of Fusarium oxysporum f. sp. apii Race 4 in vivo Is Dramatically Increased as Temperatures Exceed 22°C
A major difference between Foa race 2 and Foa race 4 is that while both Foa race 2 and Foa race 4 are temperature-sensitive, the magnitude of the high temperature response in Foa race 4 is significantly greater (Kaur et al., 2022). In the celery cultivar Sonora, which is susceptible to both Foa race 2 and race 4, at 18°C, there were no significant (α = 0.05) differences between races in either stunting, i.e., the reduction in plant height, or in the concentration of the Foa in the crown, as estimated by qPCR. In contrast, at temperatures of 22, 24, and 26°C, there was significantly (α = 0.05) more Foa race 4 than Foa race 2 DNA in planta in crowns. Consequently, the predicted global warming in California (Oakely et al., 2019) will increase the Foa race 4 disease threat.
Global warming in California (Oakely et al., 2019) could have a large impact on the growth of multiple FOSC pathogens and consequently the production of high-quality crops in California. The FOSC typically have temperature optima from 25 to 28°C (Kaur et al., 2022). There are multiple examples of diseases caused by F. oxysporum formae speciales in which disease severity in susceptible varieties increases as temperatures increase from 22 to 28°C (e.g., f. sp. ciceris in chickpea, cepae in onions, conglutinans in cole crops, lactucae in lettuce, lycopersici in tomatoes, medicaginis in alfalfa, and melonis in melons; Bosland et al., 1988; Navas-Cortés et al., 2007; Scott et al., 2010; Jelínek et al., 2019). In the case of the diseases caused by f. sp. conglutinans on crucifers and ciceris on chickpeas, higher temperatures also decreased expression of resistance (Bosland et al., 1988; Landa et al., 2006). We note that celery, a cool weather crop, has a temperature optimum of 16 to 18°C to perhaps 21°C (Rubatzky et al., 1999; Smith, 2021), and that many of the crops in California that have an important F. oxysporum pathogen are also cool weather crops, including strawberry, chickpea, onions, cabbage, lettuce, alfalfa, spinach, caneberries, and garlic.
Fusarium oxysporum f. sp. apii and Fusarium oxysporum f. sp. coriandrii Are Only Two of the Fusarium oxysporum That Have Become More Important in Coastal California, United States Agriculture in the 21st Century
Based on disease reports, Foa races 2 and 4 appear to have originated in California fields. There are several other crops that are often grown in the same locales as celery and coriander in California that are infected by other F. oxysporum ff. spp. Here, we primarily mention pathogenic F. oxysporum on lettuce and strawberry. It is important to remember that F. oxysporum typically survive for many years in the absence of a plant host because although the host ranges for diseases are generally very narrow, F. oxysporum commonly infect and reproduce to some extent in root cortical cells of non-hosts (Henry et al., 2019).
Fusarium oxysporum f. sp. lactucae apparently originated in Japan and was then introduced into California presumably on contaminated lettuce seed in approximately 1990 (Paugh and Gordon, 2020). A single clonal lineage of f. sp. lactucae race 1 has now been transported, presumably on farm equipment, to all lettuce-growing areas in California, where the disease is problematic, particularly at higher temperatures (Scott et al., 2010; Paugh and Gordon, 2019). Based on SCGs, there has been some differentiation of isolates within the race 1 lineage (Paugh and Gordon, 2020). However, too few informative loci of f. sp. lactucae isolates have been sequenced to place the isolates in a robust phylogenetic tree.
Fusarium oxysporum f. sp. fragariae (Fof), a strawberry pathogen, was first reported in eastern Australia in 1962 and then in Japan in 1969, South Korea in 1974, and in multiple locales in the early 2000s including in California in 2006 (Henry et al., 2017). In California, the emergence of the chlorosis-inducing “yellows-fragariae” pathotype of Fof coincided with the phase-out of the soil fumigant methyl bromide, which essentially began in 1999 (Epstein, 2014). Currently, Fof is considered to be one of the three most important pathogens of strawberry in California. In contrast to Foa and f. sp. lactucae in California, which are in fewer clonal lineages, based on the conserved genome, Fof in California are variable, but fit into three distinct clonal lineages, of which all are in one subclade of FOSC clade 2 (Henry et al., 2021). Surprisingly, as we show in Figure 3, the three Fof from California are closely related to Foa races 3 and 4 and the Foci in their core genomes.
A phylogenomic analysis of an international collection of Fof (Henry et al., 2021) indicated that horizontal chromosome transfer of a pathogenicity/accessory chromosome was critical for Fof diversification of this forma specialis. Overall, in terms of core genome types and the pathogenicity chromosome, there are California Fof isolates in three groups: a group that includes isolates from western Australia that have a Y1 core and a T1080 accessory chromosome; a group that includes isolates from Japan with a Y2 core and a T1 accessory chromosome; and a group that includes isolates from Japan with a Y3 core and a T1381 accessory chromosome. Based on the international isolate collection, horizontal chromosome transfer of a pathogenicity chromosome has occurred at least four times. Thus, while the epiphytotic-causing strains of Foa and Foci may have originated in California, the problematic f. sp. lactucae and fragariae were introduced into California presumably on seed and on vegetative material, respectively (Okamoto et al., 1970; Nam et al., 2011; Pastrana et al., 2019). In the case of Fof, movement of the pathogen within California can occur on nursery-infected plants and/or on farm equipment.
As in Many Domesticated Crops, Celery Has a Narrow Genetic Base, and There Is a Need for a Broader Germplasm Collection With Potential Resistance Genes
Domesticated crops often have lost disease resistance genes that are present in wild progenitors (van de Wouw et al., 2010; Purugganan, 2019). The celery cultivar Ventura, which is susceptible to Foa races 2 and 4 (Kaur and Epstein, unpublished), has only 62 nucleotide binding site (NBS) disease resistance-type genes, all with leucine rich regions, including 10 with Toll/interleukin-1 receptors (TNL), 44 with coiled coil domains (CNL), and 8 with resistance to powdery mildew 8 (RPM8) (RNL) subtypes (Song et al., 2021).
Celery (A. graveolens var. dulce) can be crossed either with other celery, with celeriac (A. graveolens var. rapaceum), with “cutting celery” (A. graveolens var. secalinum), or with wild celery (A. graveolens var. graveolens). In the 1980s, UC Davis researchers introgressed resistance gene(s) to Foa race 2 (Orton et al., 1984a) from celeriac into celery, which ultimately resulted in the release of Foa race 2-resistant cultivars (Daugovish et al., 2008) such as Challenger and Sabroso. However, multiple celeriac accessions are resistant to Foa race 2 (Kaur and Epstein, unpublished). In contrast, very few A. graveolens accessions in the USDA and UC Davis Apium germplasm collections have resistance to Foa race 4 (Kaur and Epstein, unpublished). This reinforces the need for wild germplasm collections (Frese et al., 2018), particularly with wild Apium graveolens var. graveolens, which presumably have more resistance genes. Breeding programs would also benefit from wild germplasm that is adapted to warmer temperatures, i.e., above 21°C.
Interactions With and Relationship to Other Strains of Fusarium oxysporum
Celery Is an Excellent Host for Studying the Interaction of Endophytes and Saprophytic Strains With a Fusarium oxysporum f. sp. apii Pathogen
Endophytic F. oxysporum are readily isolated from internal tissue from celery crowns, from both healthy plants and from the margins of symptomatic tissue in infected plants (Epstein et al., 2017). Indeed, celery plants infected with a pathogenic race frequently contain at least one non-pathogenic F. oxysporum strain (Schneider, 1984; Correll et al., 1986b; Epstein et al., 2017). We assembled a collection of 174 F. oxysporum isolates from celery with symptoms of a Foa infection, primarily, so that we would be able to determine whether the pathogen population in California, and particularly Foa race 2, had changed between 1993 and 2014 (Epstein et al., 2017). Almost half (48%) of the isolates were non-pathogenic. Twenty-three of the non-pathogenic isolates were in the Foa race 2 elongation factor 1-α/intergenic spacer (EF-1α/IGS) two-locus sequence haplogroup and two were in one of two Foa race 1 haplogroups. Therefore, ∼30% of the 83 non-pathogenic strains may have a core genome that is compatible with pathogenicity in celery and some of those isolates may have lost their pathogenicity in storage. The remaining 58 non-pathogenic isolates represented 20 phylogenetically diverse two-locus haplotypes, with 18 lineages nested within FOSC clade 3 and two within clade 2. Thirteen of the putatively non-pathogenic lineages were represented by singletons. We selected seven non-pathogenic isolates, with six from within FOSC clade 3, and obtained 250 bp paired-end Illumina sequence to at least 10X coverage and examined SNPs and DNA sequence of 10 phylogenetically informative genes from the non-pathogenic isolates, nine pathogenic isolates from celery, and twelve F. oxysporum reference genomes. The results confirmed that in contrast to the highly clonal Foa race 2, the non-pathogenic isolates were diverse, but clustered with the FOSC clade 3 subclade with the F. oxysporum biocontrol Fo47 (synonym, NRRL 54002). In an earlier study of non-pathogenic F. oxysporum from apparently healthy celery in California (Correll et al., 1986b), 28 non-pathogenic isolates were shown to be in 14 SCGs. Using these 14 SCG as testers, only 27% of the remaining 82 isolates in their collection were in one of these SCGs. That is, even healthy celery is a host of a highly diverse population of F. oxysporum. We note that we have not observed a role of selected non-pathogenic F. oxysporum in disease caused by Foa race 4 (Kaur and Epstein, unpublished).
Some Recent Name Changes in Fusarium Will Create Chaos for the Fusarium Community
Because of the importance of Fusarium spp. in agricultural production and in disease of many hosts, the Fusarium community needs to be able to communicate essential information on a range of topics from the population genetics to the results of diagnostic assays to a diverse audience, including other scientists, plant pathologists, pest control advisors, growers, regulators, medical practitioners, and the public. As a consequence, the Fusarium community (Geiser et al., 2013, 2021; O’Donnell et al., 2020) has resolved that both the phylogeny and the collective knowledge and needs of the Fusarium community and the public should be considered before making taxonomic revisions. If changes, such as the division of F. oxysporum into 16 species (Lombard et al., 2019) were accepted, it’s unclear what all the organisms in this review should be called, our text would have a needless layer of confusing complexity and perhaps ambiguity, and no one would have a better understanding about either the biology or control of these organisms. While the polyphyletic origin of Foa and other forma speciales is admittedly taxonomically problematic, the ff. spp. designations encode much useful information (Edel-Hermann and Lecomte, 2019). As a community, let’s strive to only adopt taxonomic changes that are biologically informative and improve our ability to communicate about all of the incredibly diverse strains in F. oxysporum, and the still insufficiently defined potential that they do and do not have to transmit their genes.
Discussion and Conclusion
In 2021, 777 thousand tonnes of celery were produced in California,1 primarily in the south and central coasts (Daugovish et al, 2008). If the soil is infested with Foa race 4, the production of both celery, and to a lesser extent coriander, are threatened, particularly if the temperature exceeds 21°C. As with most F. oxysporum, Foa survive in the soil for many years. Full genome sequencing allowed the development of diagnostic PCR primers/probes that can be used to identify Foa races 2 and 4 and Foci in planta (Henry et al, 2020). Genome assembly of the Foa races and the race 4 relative, Foci, have allowed a view of both the core and accessory genomes of these pathogens; even though Foa race 2 and race 4 can co-infect celery, both the phylogenomics and experimental evidence indicate that horizontal chromosome transfer does not occur. In contrast, horizontal chromosome transfer could occur between Foa race 4 and Foci via either hypha or CATs, with the resultant formation of potentially new pathogen genotypes.
Within the last one to three decades, two other diseases caused by F. oxysporum have emerged in Coastal California agriculture: Fusarium wilt on lettuce, which is also more severe in warmer temperatures, and Fusarium wilt on strawberry. In contrast to Foa race 4 and Foci, which may be endemic, both f. sp. lactucae and f. sp. fragariae appear to have been introduced. Surprisingly, Fof is comparatively closely related to Foa race 4 and Foci (Figure 3); we do not know what this means in terms of the natural history of these organisms, but it seems unlikely that it’s a coincidence. We also do not know if Foa race 4 and Fof co-occur in conditions in which CATs form, but we do know that while Foa race 4 is not a strawberry pathogen, it can infect and survive in strawberry roots (Kaur and Epstein, unpublished). Regardless, the confluence of multiple factors indicate that coastal California will continue to be a hot spot for F. oxysporum diseases in the future: (1) F. oxysporum have an optimum of 25 to 28°C, the coastal climate is warming, the region will have more days in this near-optimum temperature range (Oakely et al., 2019), many of the intensive crops in this region, e.g., caneberries, celery, onions, cabbage, lettuce, spinach, and garlic, are better adapted for cooler temperatures, and disease in multiple ff. spp. is more severe at warmer temperatures; (2) the intensive agriculture, sometimes without crop rotation, allows production of high inoculum concentrations in the soil; (3) F. oxysporum inoculum is maintained in infested fields in multiple ways including, the pathogen can reproduce in non-host roots and debris at sufficient levels that populations persist despite rotation, and there is little knowledge of how to best manage crop residues so as to reduce inoculum concentration in soil; (4) F. oxysporum pathogens are routinely introduced into fields in soil and infected debris on harvesters and other equipment within and between counties, and on infected or infested seed and vegetative material that is part of global trade; (5) new pathotypes can arise via either CAT formation and horizontal chromosome transfer, or transposon activity, etc.; and (6) there are insufficient social factors that encourage growers to work toward long term disease control and multiple social factors that discourage it, including the economic pressure to farm for short-term profits and farming on leased land.
Author Contributions
LE wrote the manuscript with input from PMH and SK. All authors conducted the research and approved the manuscript.
Funding
We acknowledge the California Celery Research Advisory Board Grants EPS-19, EPS-20, EPS-21, and EPS-22 and the University of California Hansen Trust grant HF19-5830R.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Professor Radwan Barakat for conducting experiments on the conidial anastomosis tubes during his sabbatical, our colleagues and students who have worked on this research program at the University of California at Davis, and the Celery Research Advisory Board and the University of California Hansen Trust for their financial support.
Footnotes
References
Akanuma, R., and Shimizu, T. (1994). Epidemiology and control of Fusarium yellows of celery [Apium graveolens], 1: etiology of fusarium yellows of celery and occurrence of Fusarium oxysporum f. sp. apii race 2 in Nagano. Bullet. Nagano Veg. Ornam. Crops Exp. Stat. 8, 65–70.
Armstrong, G. M., and Armstrong, J. K. (1981). “Formae speciales and races of Fusarium oxysporum causing wilt diseases,” in Fusarium: Disease, Biology, and Taxonomy, eds P. E. Nelson, T. A. Toussoun, and R. James Cook (Pennsylvania: Pennsylvania State University Press), 391–399.
Ayukawa, Y., Asai, S., Gan, P., Tsushima, A., Ichihashi, Y., Shibata, A., et al. (2021). A pair of effectors encoded on a conditionally dispensable chromosome of Fusarium oxysporum suppress host-specific immunity. Commun. Biol. 4:707. doi: 10.1038/s42003-021-02245-4
Bosland, P. W., Williams, P. H., and Morrison, P. H. (1988). Influence of soil temperature on the expression of yellows and wilt of crucifers by Fusarium oxysporum. Plant Dis. 72, 777–780.
Cerkauskas, R. F., and Chiba, M. (1991). Soil densities of Fusarium oxysporum f. sp. apii race 2 in Ontario, and the association between celery cultivar resistance and photocarcinogenic furocoumarins. Can. J. Plant Pathol. 13, 305–314.
Correll, J. C., Puhalla, J. E., and Schneider, R. W. (1986a). Identification of Fusarium oxysporum f. sp. apii on the basis of colony size, virulence, and vegetative compatibility. Phytopathology 76, 396–400.
Correll, J. C., Puhalla, J. E., and Schneider, R. W. (1986b). Vegetative compatibility groups among nonpathogenic root-colonizing strains of Fusarium oxysporum. Can. J. Bot. 64, 2358–2361. doi: 10.1139/b86-310
Daugovish, O., Smith, R., Cahn, M., Koike, S., Smith, H., Aguiar, J., et al. (2008). Celery Production in California. Davis, CA: University of California, Agriculture and Natural Resources. doi: 10.3733/ucanr.7220
Edel-Hermann, V., and Lecomte, C. (2019). Current status of Fusarium oxysporum formae speciales and races. Phytopathology 109, 512–530. doi: 10.1094/PHYTO-08-18-0320-RVW
Epstein, L. (2014). Fifty years since silent spring. Annu. Rev. Phytopathol. 52, 377–402. doi: 10.1146/annurev-phyto-102313-045900
Epstein, L., Kaur, S., Chang, P. L., Carrasquilla-Garcia, N., Lyu, G., Cook, D. R., et al. (2017). Races of the celery pathogen Fusarium oxysporum f. sp. apii are polyphyletic. Phytopathology 107, 463–473. doi: 10.1094/PHYTO-04-16-0174-R
Frese, L., Bönisch, M., Nachtigall, M., and Schirmak, U. (2018). Patterns of genetic diversity and implications for in situ conservation of wild celery (Apium Graveolens L. ssp. graveolens). Agriculture 8:129. doi: 10.3390/agriculture8090129
Geiser, D. M., Al-Hatmi, A. M. S., Aoki, T., Arie, T., Balmas, V., Barnes, I., et al. (2021). Phylogenomic analysis of a 55.1-kb 19-gene dataset resolves a monophyletic Fusarium that includes the Fusarium Solani species complex. Phytopathology 111, 1064–1079. doi: 10.1094/PHYTO-08-20-0330-LE
Geiser, D. M., Aoki, T., Bacon, C. W., Baker, S. E., Bhattacharyya, M. K., Brandt, M. E., et al. (2013). One fungus, one name: defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology 103, 400–408. doi: 10.1094/PHYTO-07-12-0150-LE
Gold, S. E., Paz, Z., García-Pedrajas, M. D., and Glenn, A. E. (2017). Rapid deletion production in fungi via agrobacterium mediated transformation of OSCAR seletion xonstructs. J. Vis. Exp. 12:55239. doi: 10.3791/55239
Henry, P., Kaur, S., Pham, Q. A. T., Barakat, R., Brinker, S., Haensel, H., et al. (2020). Genomic differences between the new Fusarium oxysporum f. sp. apii (Foa) race 4 on celery, the less virulent Foa races 2 and 3, and the avirulent on celery f. sp. coriandrii. BMC Genomics 21:730. doi: 10.1186/s12864-020-07141-5
Henry, P. M., Kirkpatrick, S. C., Islas, C. M., Pastrana, A. M., Yoshisato, J. A., Koike, S. T., et al. (2017). The population of Fusarium oxysporum f. sp. fragariae, cause of Fusarium wilt of strawberry, in California. Plant Dis. 101, 550–556. doi: 10.1094/PDIS-07-16-1058-RE
Henry, P. M., Pastrana, A. M., Leveau, J. H. J., and Gordon, T. R. (2019). Persistence of Fusarium oxysporum f. sp. fragariae in soil through asymptomatic colonization of rotation crops. Phytopathology 109, 770–779. doi: 10.1094/PHYTO-11-18-0418-R
Henry, P. M., Pincot, D. D. A., Jenner, B. N., Borrero, C., Aviles, M., Nam, M.-H., et al. (2021). Horizontal chromosome transfer and independent evolution drive diversification in Fusarium oxysporum f. sp. fragariae. New Phytol. 230, 327–340. doi: 10.1111/nph.17141
Husaini, A. M., Sakina, A., and Cambay, S. R. (2018). Host–pathogen interaction in Fusarium oxysporum infections: where do we stand? MPMI. 31, 889–898. doi: 10.1094/MPMI-12-17-0302-CR
Ishikawa, F. H., Souza, E. A., Shoji, J., Connolly, L., Freitag, M., Read, N. D., et al. (2012). Heterokaryon incompatibility is suppressed following conidial anastomosis tube fusion in a fungal plant pathogen. PLoS One 7:e31175. doi: 10.1371/journal.pone.0031175
Jelínek, T., Koudela, M., Kofránková, V., and Salava, J. (2019). Effect of temperature on severity of Fusarium wilt of cabbage caused by Fusarium oxysporum f. sp. conglutinans. Eur. J. Plant Pathol. 155, 1277–1286. doi: 10.1007/s10658-019-01855-3
Kaur, S., Barakat, R., Kaur, J., and Epstein, L. (2022). The effect of temperature on disease severity and growth of Fusarium oxysporum f. sp. apii races 2 and 4 in celery. Phytopathology 112, 364–372. doi: 10.1094/PHYTO-11-20-0519-R
Kurian, S., Di Pietro, A., and Read, N. (2018). Live-cell imaging of conidial anastomosis tube fusion during colony initiation in Fusarium oxysporum. PLoS ONE 13, 1–32. doi: 10.1371/journal.pone.0195634
Koike, S. T., and Gordon, T. R. (2005). First report of Fusarium Wilt of cilantro caused by Fusarium oxysporum in California. Plant Dis. 89:1130. doi: 10.1094/PD-89-1130A
Landa, B. B., Navas-Cortés, J. A., Del Mar Jiménez-Gasco, M., Katan, J., Retig, B., and Jiménez-Díaz, R. M. (2006). Temperature response of chickpea cultivars to races of Fusarium oxysporum f. sp. ciceris, causal agent of Fusarium wilt. Plant Dis. 90, 365–374. doi: 10.1094/PD-90-0365
Li, J., Fokkens, L., van Dam, P., and Rep, M. (2020). Related mobile pathogenicity chromosomes in Fusarium oxysporum determine host range on cucurbits. Mol. Plant Pathol. 21, 761–776. doi: 10.1111/mpp.12927
Lombard, L., Sandoval-Denis, M., Lamprecht, S. C., and Crous, P. W. (2019). Epitypification of Fusarium oxysporum – clearing the taxonomic chaos. Persoonia 43, 1–47. doi: 10.3767/persoonia.2019.43.01
Lori, G. A., Birol, I., Mourelos, C. A., and Wolcan, S. M. (2016). First report of Fusarium oxysporum f. sp. apii race 2 causing Fusarium yellows on celery in Argentina. Plant Dis. 100:1020.
Ma, L.-J., van der Does, H. C., Borkovich, K. A., Coleman, J. J., Daboussi, M.-J., Di Pietro, A., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373. doi: 10.1038/nature08850
Nam, M. H., Kang, Y. L., Kim, H. G., and Chun, C. (2011). Infection of daughter plants by Fusarium oxysporum f. sp. fragariae through runner propagation of strawberry. J. Hortic Sci. Technol. 29, 273–277.
Navas-Cortés, J. A., Landa, B. B., Méndez-Rodríguez, M. A., and Jiménez-Díaz, R. M. (2007). Quantitative modeling of the effects of temperature and inoculum density of Fusarium oxysporum f. sp. ciceris races 0 and 5 on development of Fusarium wilt in chickpea cultivars. Phytopathology 97, 564–573. doi: 10.1094/PHYTO-97-5-0564
Nelson, R., Coons, G. H., and Cochran, L. C. (1937). The Fusarium Yellows Disease of Celery (Apium graveolens L. var. dulce DC.). Technical Bulletin. East Lansing: Michigan Agricultural Experiment Station, 1–74.
Oakely, N. S., Hatchett, B. J., McEvoy, D. J., and Rodrigues, L. (2019). Projected Changes in Ventura County Climate. Reno, Nevada: Western Regional Climate Center, Desert Research Institute. Available online at: https://wrcc.dri.edu/Climate/reports.php (accessed May 29, 2022).
O’Donnell, K., Al-Hatmi, A. M. S., Aoki, T., Brankovics, B., Cano-Lira, J. F., Coleman, J. J., et al. (2020). No to Neocosmospora: phylogenomic and practical reasons for continued inclusion of the Fusarium solani species complex in the Genus Fusarium. Msphere 5, e810–e820. doi: 10.1128/mSphere.00810-20
O’Donnell, K., Gueidan, C., Sink, S., Johnston, P. R., Crous, P. W., Glenn, A., et al. (2009). A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet. Biol. 46, 936–948. doi: 10.1016/j.fgb.2009.08.006
Okamoto, H., Fujii, S., Kato, K., and Yoshioka, A. (1970). A new strawberry disease ‘Fusarium wilt.’. Plant Protection 24, 231–235.
Orton, T. J., Durgan, M. E., and Hulbert, S. H. (1984a). Studies on the inheritance of resistance to Fusarium oxysporum f. sp. apii in celery. Plant Dis. 68, 574–578.
Orton, T. J., Hulbert, S. H., Durgan, M. E., and Quiros, C. F. (1984b). UC1, Fusarium yellows-resistant celery breeding line. HortScience 19:594.
Otto, H. W., Paulus, A. O., Snyder, M. J., Endo, R. M., Hart, L. P., and Nelson, J. (1976). A crown rot of celery. Calif. Agric. 30, 10–11. doi: 10.1094/PD-90-0829A
Pastrana, A. M., Watson, D. C., and Gordon, T. R. (2019). Transmission of Fusarium oxysporum f. sp. fragariae through stolons in strawberry plants. Plant Dis. 103, 1249–1251. doi: 10.1094/PDIS-08-18-1353-RE
Paugh, K. R., and Gordon, T. R. (2019). Effect of planting date and inoculum density on severity of Fusarium Wilt of lettuce in California. Plant Dis. 103, 1498–1506. doi: 10.1094/PDIS-09-18-1614-RE
Paugh, K. R., and Gordon, T. R. (2020). The population of Fusarium oxysporum f. sp. lactucae in California and Arizona. Plant Dis. 104, 1811–1816. doi: 10.1094/PDIS-06-19-1228-RE
Puhalla, J. E. (1984). Races of Fusarium oxysporum f. sp. apii in California and their genetic interrelationships. Can. J. Bot. 62, 546–550. doi: 10.1139/b84-080
Purugganan, M. D. (2019). Evolutionary insights into the nature of plant domestication. Curr. Biol. 29, R705–R714. doi: 10.1016/j.cub.2019.05.053
Retana, K., Ramírez-Coché, J. A., Castro, O., and Blanco-Meneses, M. (2018). Caracterización morfológica y molecular de Fusarium oxysporum f. sp. apii asociado a la marchitez del apio en Costa Rica. Agron. Costarricense 42, 115–126. doi: 10.15517/rac.v42i1.32199
Rubatzky, V., Quiros, C., and Simon, P. (1999). Carrots and Related Vegetable Umbelliferae. New York, NY: CABI Pub.
Schneider, R. W. (1984). Effects of nonpathogenic strains of Fusarium oxysporum on celery root infection by F. oxysporum f. sp. apii and a novel use of the Lineweaver-Burk double reciprocal plot technique. Phytopathology 74:646. doi: 10.1094/Phyto-74-646
Scott, J. C., Gordon, T. R., Shaw, D. V., and Koike, S. T. (2010). Effect of temperature on severity of Fusarium wilt of lettuce caused by Fusarium oxysporum f. sp. lactucae. Plant Dis. 94, 13–17. doi: 10.1094/PDIS-94-1-0013
Shahi, S., Beerens, B., Bosch, M., Linmans, J., and Rep, M. (2016). Nuclear dynamics and genetic rearrangement in heterokaryotic colonies of Fusarium oxysporum. Fungal Genet. Biol. 91, 20–31. doi: 10.1016/j.fgb.2016.03.003
Smith, R. (2021). “Celery,” in Carrots and Related Apiaceae Crops, eds E. Geoffriau and P. W. Simon (Wallingford: CAB International), 272–282.
Snyder, W. C., and Hansen, H. N. (1940). The species concept in Fusarium. Am. J. Bot. 27, 64–67. doi: 10.2307/2436688
Song, X., Sun, P., Yuan, J., Gong, K., Li, N., Meng, F., et al. (2021). The celery genome sequence reveals sequential paleo-polyploidizations, karyotype evolution and resistance gene reduction in Apiales. Plant Biotechnol. J. 19, 731–744. doi: 10.1111/pbi.13499
Subbarao, K. V., and Elmer, W. H. (2002). “Fusarium yellows of celery,” in Compendium of Umbelliferous Crop Diseases, eds R. M. Davis and R. N. Raid (Saint Paul: American Phytopathological Society), 33–34.
van Dam, P., Fokkens, L., Ayukawa, Y., van der Gragt, M., Ter Hortst, A., Brankovics, B., et al. (2017). A mobile pathogenicity chromosome in Fusarium oxysporum for infection of multiple cucurbit species. Sci. Rep. 7:9042. doi: 10.1038/s41598-017-07995-y
van Dam, P., Fokkens, L., Schmidt, S. M., Linmans, J. H. J., Kistler, H. C., Ma, L.-J., et al. (2016). Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 18, 4087–4102. doi: 10.1111/1462-2920.13445
van de Wouw, M., Kik, C., van Hintum, T., van Treuren, R., and Visser, B. (2010). Genetic erosion in crops: concept, research results and challenges. Plant Genet. Res. 8, 1–15. doi: 10.1017/S1479262109990062
Vangalis, V., Knop, M., Typas, M. A., and Papaioannou, I. A. (2021). Establishment of conidial fusion in the asexual fungus Verticillium dahliae as a useful system for the study of non-sexual genetic interactions. Curr. Genet. 67, 471–485. doi: 10.1007/s00294-021-01157-4
Vlaardingerbroek, I., Beerens, B., Rose, L., Fokkens, L., Cornelissen, B. J. C., and Rep, M. (2016). Exchange of core chromosomes and horizontal transfer of lineage-specific chromosomes in Fusarium oxysporum. Environ. Microbiol. 18, 3702–3713. doi: 10.1111/1462-2920.13281
Keywords: Apium graveolens var. dulce, celery, cilantro, effectors, emerging pathogen, Fusarium oxysporum f. sp. apii, Fusarium oxysporum f. sp. coriandrii, Fusarium oxysporum species complex
Citation: Epstein L, Kaur S and Henry PM (2022) The Emergence of Fusarium oxysporum f. sp. apii Race 4 and Fusarium oxysporum f. sp. coriandrii Highlights Major Obstacles Facing Agricultural Production in Coastal California in a Warming Climate: A Case Study. Front. Plant Sci. 13:921516. doi: 10.3389/fpls.2022.921516
Received: 16 April 2022; Accepted: 13 May 2022;
Published: 13 June 2022.
Edited by:
Guixia Hao, Mycotoxin Prevention and Applied Microbiology Research, Agricultural Research Service (USDA), United StatesReviewed by:
Domingo Martinez-Soto, University of Massachusetts Amherst, United StatesFrancesco Tini, University of Perugia, Italy
Copyright © 2022 Epstein, Kaur and Henry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lynn Epstein, bGVwc3RlaW5AdWNkYXZpcy5lZHU=
 Lynn Epstein1*
Lynn Epstein1* Sukhwinder Kaur
Sukhwinder Kaur