- 1Zhengzhou Research Base, State Key Laboratory of Cotton Biology, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, China
- 2Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
- 3College of Agriculture, Shanxi Agricultural University, Taigu, China
Dwarfism is a beneficial trait in many crops. Dwarf crops hold certain advantages over taller crops in lodging resistance, fertilizer tolerance, and yield. Overexpression of CBF/DREB transcription factors can lead to dwarfing in many plant species, but the molecular mechanism of plant dwarfing caused by overexpression of CBF/DREB in upland cotton (Gossypium hirsutum) remains unclear. In this study, we observed that overexpression of the Ammopiptanthus mongolicus AmCBF1 transcription factor in upland cotton R15 reduced plant height, whereas virus-induced gene silencing of AmCBF1 in the derived dwarf lines L28 and L30 partially restored plant height. Five protein phosphatase (PP2C) genes (GhPP2C1 to GhPP2C5) in cotton were identified by RNA-sequencing among genes differentially expressed in L28 or L30 in comparison with R15 and thus may play an important role in AmCBF1-regulated dwarfing in cotton. Gene expression analysis showed that the GhPP2C genes were down-regulated significantly in L28 and L30, and silencing of GhPP2C1 or GhPP2C2 in R15 inhibited the growth of cotton seedlings. Subcellular localization assays revealed that GhPP2C1 was localized to the cell membrane and nucleus, whereas GhPP2C2 was exclusively localized to the nucleus. Yeast one-hybrid and dual-luciferase assays showed that AmCBF1 was able to bind to the CRT/DRE elements of the upstream promoter of GhPP2C1 or GhPP2C2 and repress their expression. These findings provide insight into the mechanism of dwarfing and may contribute to the breeding of dwarf cultivars of upland cotton.
Introduction
Dwarfism is a valuable agronomic trait, as it helps to improve the light use efficiency of the population and increases the ability of plants to resist an adverse external environment (Khush, 1999). In agricultural and horticultural crops, breeding for dwarfism is particularly important. Some tall crops have weak stalks and are susceptible to lodging, whereas dwarf crops have strong stalks and are resistant to lodging, highly fertilizer tolerant, and produce significantly increased yields through reasonably dense planting. Dwarf and compact horticultural plants are increasingly popular with consumers because of their high ornamental value. In recent years, considerable progress has been made in screening and identifying genes associated with plant dwarfing. Phytohormone-related genes play an important role in causing dwarfing in plants (Hu et al., 2017; Li et al., 2018; Graeff et al., 2020). In addition, genes that affect plant cell wall synthesis, such as a mutation of the cellulose biosynthesis gene CESA, or regulation by transcription factors such as C-repeat binding factor/dehydration responsive element-binding factor (CBF/DREB) proteins, can cause dwarfing in plants (Xie et al., 2011). In cotton, the dwarf variety AS98 was detected in an introgressed line derived from an interspecific hybrid and exogenous application of gibberellic acid (GA3) restores the plant height phenotype (Zhang et al., 2011; Jia et al., 2016). In the cotton pag1 mutant, cell growth is disrupted, resulting in a dwarf phenotype and reduced fiber quality (Yang et al., 2014). The contents of indoleacetic acid and abscisic acid (ABA) are reduced in the Asian cotton dwarf sda mutant, and genetic analysis indicates that the dwarfism trait is controlled by a pair of recessive genes (Wu et al., 2009). The aforementioned studies indicate that utilization of a variety of dwarfing resources provides a means to increase cotton yield by changing the plant architecture.
The CBF proteins comprise a subfamily of the APETALA 2/ETHYLENE-RESPONSIVE FACTOR (AP2/ERF) family of transcription factors, also termed DREBs, which are involved in a range of processes during the plant life cycle (Agarwal et al., 2006). The CBF protein binds to the C-repeat/dehydration-responsive element (CRT/DRE) sequence in the promoter of downstream genes to regulate their expression, and enhances plant resistance to a variety of stresses, such as drought, freezing, and salinity (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994; Dong et al., 2006). Overexpression of CBF transcription factors in diverse plants affects plant growth and leads to dwarfing, while affecting plant stress resistance. For example, overexpression of AtDREB1A or OsDREB1A/2A in Arabidopsis improves cold and drought tolerance, but growth of transgenic Arabidopsis is severely inhibited (Liu et al., 1998; Dubouzet et al., 2003). Similarly, overexpression of OsDREB1A/1B in rice (Oryza sativa) results in dwarfing (Ito et al., 2006). Interestingly, application of exogenous gibberellin (GA) partially restores the height of plants overexpressing CBF, suggesting that dwarfing caused by CBF overexpression may be due to GA deficiency (Hsieh et al., 2002). Overexpression of DgDREB1B in Arabidopsis leads to reduction in plant height and significant up-regulation of GA2ox7 and GA-inactivating enzymes, suggesting that DgDREB1B may be involved in GA-mediated development in plants (Tong et al., 2009). Twenty-one CBF genes have been identified in the upland cotton (Gossypium hirsutum) genome and are categorized into four groups: GhCBF I, GhCBF II, GhCBF III, and GhCBF IV (Ma et al., 2014). Overexpression of GhDREB1 in tobacco (Nicotiana tabacum) causes retarded growth and delays flowering (Shan et al., 2007). Recently, it was shown that overexpression of GhDREB1B in upland cotton results in significant reduction in plant height, shorter branch lengths, and reduced branch angles (Ji et al., 2021). These results confirm that overexpression of CBF/DREB transcription factors can lead to plant dwarfing, but the specific mechanism of dwarfing remains to be studied.
Post-translational modification of proteins plays an important role in plant developmental processes. Among these modifications, protein phosphorylation/dephosphorylation plays a role at various stages of plant growth and development by regulating protein activity and participating in the transmission of a number of signals in plants (Schweighofer et al., 2007; Fuchs et al., 2013; Singh et al., 2016). This reversible protein phosphorylation is achieved through the activities of the enzyme pair protein kinase and protein phosphatase. Depending on the catalytic substrate, protein phosphatases are mainly divided into protein tyrosine phosphatases and protein serine/threonine phosphatases (Xin et al., 2021). Protein phosphatase 2C (PP2C) comprises a class of protein serine/threonine phosphatases and is a monomeric enzyme (Zhang et al., 2022). In plants, PP2C is less conserved at the N terminus and more conserved at the C terminus, which confers PP2C with functional diversity (Ingebritsen and Cohen, 1983; Hubbard and Cohen, 1993; Bork et al., 1996). A recognized mechanism of action is that PP2C participates in the ABA signaling pathway. In this signaling mechanism, PP2Cs bind to SnRK2, resulting in the loss of SnRK2 phosphokinase activity and the inability of downstream substrates to be phosphorylated (Soon et al., 2012). In the presence of ABA, the ABA receptor PYR/PYL/RCAR binds to PP2C and SnRK2 is released, phosphorylating downstream transcription factors, such as ABA-responsive element-binding factors, and further activating the expression of certain downstream genes (Miyazono et al., 2009; Cutler et al., 2010; Hubbard et al., 2010; Rodrigues et al., 2013; Zhu, 2016).
The function of PP2C genes is closely associated with processes such as signal transduction, growth and development, hormone regulation, and oxidative stress (Gaits et al., 1997; Rodriguez, 1998; Kerk et al., 2002; Schweighofer et al., 2004). Nine and 10 subgroup A PP2C members have been identified in Arabidopsis and rice, respectively, of which most play a negative regulatory role in ABA signaling (Saez et al., 2004; Kuhn et al., 2006; Nishimura et al., 2007; Bhaskara et al., 2012; He et al., 2014; Née et al., 2017; Miao et al., 2018). Overexpression of OsPP108 in Arabidopsis enhances plant tolerance to salt and drought stress, and reduces ABA sensitivity (Singh et al., 2015). Overexpression of OsPP2C09 in rice results in rapid growth and increases individual plant yield, whereas its mutant reduces plant height and yield (Miao et al., 2020). A lack of SlPP2C3 expression in tomato (Solanum lycopersicum) not only affects plant growth, but also accelerates the onset of fruit ripening and affects fruit gloss by altering the outer epidermal structure (Liang et al., 2021). Genome-wide identification of PP2C family members detected 87, 99, 147, and 181 PP2C genes in Gossypium arboreum, Gossypium raimondii, Gossypium barbadense, and G. hirsutum, respectively. Among these genes, the 181 PP2Cs in G. hirsutum were roughly divided into 12 subgroups; the transcription of some of these PP2C genes was induced by high temperature, low temperature, drought, or high salt stress separately, suggesting that PP2C genes play a crucial role in abiotic stress response in cotton (Shazadee et al., 2019). However, the relationship between PP2C genes and a dwarf phenotype in upland cotton has not been examined.
Cotton, which is among the world’s most important oilseed and textile crops, plays an important role in global agricultural and industrial production. With technological developments, mechanized harvesting of cotton is increasingly important and dwarf cotton is a major breeding target on account of the compact plant height and suitability for machine harvesting. Although dwarf cotton varieties have various advantages, there are still relatively few dwarf cotton varieties on the market today. Therefore, development of new cotton varieties showing stable inheritance of dwarfism is a matter of urgency. Although a number of dwarfing-related genes have been identified, little information is available on their biological functions, especially the molecular mechanisms that regulate cotton growth and development.
In our previous work, Ammopiptanthus mongolicus AmCBF1 was overexpressed in the upland cotton line R15 to evaluate its function in abiotic stress response. We observed that overexpression of AmCBF1 increased the resistance of cotton to low temperature and drought stress, and decreased the plant height. In the present study, we further confirmed the relationship between AmCBF1 and the dwarf phenotype in cotton. Five down-regulated GhPP2C (GhPP2C1-GhPP2C5) genes were identified among differentially expressed genes by RNA-Seq and AmCBF1 transcription factors could negatively regulate the expression of GhPP2C1 or GhPP2C2. These results will help to understand the molecular mechanism of cotton dwarf and provide theoretical basis for cotton dwarf breeding.
Materials and Methods
Plant Materials and Growing Conditions
Transgenic dwarf cotton lines (L28 and L30), transgene recipient (R15), and tobacco (Nicotiana benthamiana) plants were grown in pots containing nutrient soil in a greenhouse (25 ± 3°C, 50% relative humidity, and 16 h light/8 h dark cycle). In addition, R15, L28, and L30 were grown in the field (39°36′10.3”N,116°36′07.3″E). Cotton and tobacco plants at different growth stages were selected for subsequent experiments.
Quantitative Real-Time PCR
Total RNA was extracted from cotton leaves using an RNA extraction kit (Tiangen Biotech, Beijing, China). The RNA concentration was measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). Purified RNA (>1 μg) treated with gDNase was reverse transcribed with HiScript III RT SuperMix for qPCR (+gDNA wiper; Vazyme Biotech, Nanjing, China) to generate the first-strand cDNA. Gene expression levels were analyzed with an ABI ViiA™ 7 Real-Time PCR System (Applied Biosystems, Waltham, MA, United States) and ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech). The quantitative real-time PCR (qRT-PCR) protocol was as follows: 40 cycles at 95°C for 30 s, 95°C for 10 s, 60°C for 30 s, and 72°C for 15 s. Relative expression levels were calculated using the 2−ΔΔCt method. Primers were designed using PRIMER PREMIER 5 (Premier Biosoft, Palo Alto, CA, United States). The cotton ubq gene (Gh_D13G1489) was used as an internal control.
Virus-Induced Gene Silencing
Virus-induced gene silencing (VIGS) is an approach widely used to study gene function and functional genomics in a variety of plants (Becker and Lange, 2010). We performed a VIGS experiment to test if plant height was partly restored after silencing AmCBF1 in L28 or L30. The AmCBF1 fragment was amplified and inserted into the pTRV2 vector digested with the XbaI and KpnI restriction enzymes. The plasmids pTRV1, pTRV2, pTRV2-AmCBF1, and pTRV2-CLA1 were transferred to Agrobacterium tumefaciens strain GV3101 receptor cells by electroshock and incubated overnight in LB liquid medium supplemented with 50 mg L−1 kanamycin, 50 mg L−1 rifampicin, 10 mM MES, and 20 mM acetosyringone at 28°C on a shaker at 200 rpm. The Agrobacterium cells were collected by centrifugation and suspended in permeate [containing 10 mM MgCl2, 10 mM MES (pH 5.6), and 20 μM acetyleugenone] to optical density (OD600) = 1.2. After incubation at room temperature for 3 h in the dark, the suspensions containing the pTRV2, pTRV2-AmCBF1, or pTRV2-CLA1 plasmids were mixed with pTRV1 suspension (1:1, v/v) respectively. Cotton seedlings with well-grown, fully expanded cotyledons were selected and a small wound was made on the lower surface of the cotyledons with a syringe needle. Silencing of the CLA1 gene in R15 was used as a positive control to produce an albino phenotype in cotton. The injected cotton plants were cultured in the dark for 24 h, then for 4 weeks under a 16 h light/8 h dark cycle at 23°C and 50% relative humidity. Total RNA was extracted from young cotton leaves and qRT-PCR was performed to detect the relative expression level of AmCBF1. In addition, based on sequence alignment of GhPP2C1, GhPP2C2, and GhPP2C3, we designed specific VIGS fragments and qRT-PCR detection fragments (Supplementary Figure 3). The GhPP2C1, GhPP2C2, or GhPP2C3 genes were silenced in R15 by VIGS to investigate whether the genes affect the height of cotton plants. Fragments for silencing GhPP2C1, GhPP2C2, and GhPP2C3 were amplified from R15 cotton cDNA using specific primers and inserted into the XbaI and KpnI double-cleaved pTRV2 vector using seamless cloning. The specific experimental method was based on the gene silencing analysis of AmCBF1. The primers used for VIGS vector construction and qRT-PCR detection are listed in Supplementary Table 1.
Scanning Electron Microscopy
To investigate whether overexpression of AmCBF1 affects the morphology and size of cotton leaf cells, the upper epidermis of cotton seedling leaves was observed by scanning electron microscopy. The fourth true leaf of cotton seedlings was washed three times with deionized water and fixed with 2.5% glutaraldehyde at 4°C for more than 8 h. After dehydration, the fixed samples were dried in a CO2 critical-point desiccator for approximately 2 h. The samples were observed under a scanning electron microscope (Hitachi SU-8010, Tokyo, Japan) and photographed.
RNA-Sequencing Analysis
Total RNA extracted from fresh seedlings of R15, L28, and L30 was subjected to transcriptome sequencing by Libaijia Biologicals (Beijing, China). Two biological replicates of each material were designated R15R1, R15R2, L28R1, L28R2, L30R1, and L30R2. Quantification of gene expression levels was expressed as fragments per kilobase of transcript per million fragments mapped (FPKM). The fold change is the ratio of expression between the two samples, and the false discovery rate (FDR) is a value obtained by correcting for the difference in significance p-value (p-value). Fold change ≥2 and FDR < 0.01 were used as the screening criteria. The Benjamini–Hochberg correction method was used to correct the p-value obtained from the original hypothesis test to avoid the problem of false positives. Finally, the FDR was adopted as the crucial index for screening differentially expressed genes (DEGs). Twelve DEGs involved in different metabolic pathways were randomly selected to verify the transcriptome data. The selected genes, designated Q1–Q12, are listed in Supplementary Table 2.
Phylogenetic Analysis
The amino acid sequence of all PP2C proteins of upland cotton were downloaded from the Cotton Functional Genomics Database (cottonFGD).1 Sequences of the PP2C proteins of Arabidopsis were downloaded from The Arabidopsis Information Resource database (TAIR).2 Phylogenetic analyses were conducted using the neighbor-joining method implemented in the MEGA 7.0 software.
Subcellular Localization
The full-length coding regions (without the termination codon) of GhPP2C1, GhPP2C2, and GhPP2C3 were inserted into the pCambia1302 vector carrying the green fluorescent protein (GFP) gene. The plasmids 1302, 1302-GhPP2C1, 1302-GhPP2C2, and 1302-GhPP2C3 were transferred into A. tumefaciens strain EHA105 using the freeze–thaw method, and incubated in LB liquid medium supplemented with 50 mg L−1 kanamycin and 50 mg L−1 rifampicin for 16 h at 28°C on a shaker at 200 rpm. The Agrobacterium cells were centrifuged at 3,000 × g using a High-speed Micro Centrifuge and the supernatant was discarded. The sediment was resuspended in MMA solution [containing 10 mM MgCl2, 10 mM MES (pH 5.6), and 150 μM acetyleugenone] to OD600 = 1.2 and incubated at room temperature for 3 h in the dark. Healthy tobacco leaves were selected and the suspension was gently injected into the lower surface of the leaves with a syringe. Injection of GFP alone was used as a control, and H2B-mCherry was used as a nucleolus marker. After incubation for 16 h in the dark at 21°C, the leaves were incubated in the light for 8 h. Two days after injection, the fluorescence signal was observed using a LSM 700 confocal microscope (Zeiss, Oberkochen, Germany). The primers used for construction of the transient expression vectors are listed in Supplementary Table 1.
Yeast One-Hybrid assay
Three or two predicted CRT/DRE (A/GCCC/GAC) elements were identified in the 3 kb promoter regions upstream of the GhPP2C1 or GhPP2C2 genes, and designated P1, P2, P3, P4, and P5, respectively. A Y1H analysis was conducted to verify whether AmCBF1 is capable of interacting with the corresponding cis-elements in the upstream promoters of GhPP2C1 and GhPP2C2. The full-length coding sequence of AmCBF1 was amplified using specific primers and inserted into the pGADT7 vector digested with EcoRI and BamHI to generate the prey plasmid. The 2,049 bp or 882 bp promoter region containing all CRT/DRE elements of GhPP2C1 or GhPP2C2 was separately cloned and inserted into the pAbAi vector digested with KpnI and SalI to generate the bait plasmid. In addition, separate forward and reverse complementary oligonucleotide strands were synthesized for P1, P2, P3, P4, and P5, annealed to generate promoter DNA fragments containing the CRT/DRE elements, and inserted into the pAbAi bait vector. The bait plasmid was transformed into the yeast strain Y1H Gold by BstbI digestion using the EX-Yeast Transformation Kit (Zhuangmeng Bio, Beijing, China). The linearized bait plasmid was homologously integrated with the yeast Y1H Gold genome to generate a decoy reporter yeast strain, which was tested using an aureobasidin A (AbA) inhibition assay and Matchmaker Insert Check PCR Mix 1 (Clontech Laboratories, Inc., Palo Alto, CA, United States) to identify positive strains. The prey plasmids were then transformed into the bait reporter strains to verify DNA–protein interactions. The transformed cells were cultured on SD/−Leu/−Ura medium incubated for 3 days at 30°C. Positive strains were identified using the pGADT7 universal primer. Subsequently, yeast cells at different dilutions (1:10, 1:100, and 1:1,000) were cultured on SD/−Leu/−Ura medium supplemented with different concentration of AbA (0, 200, or 400 ng ml−1). The pGADT7 vector was used as a negative control. The primers used in this experiment are listed in Supplementary Table 1.
Dual-Luciferase Analysis
The full-length coding sequence of AmCBF1 was inserted by seamless cloning into the NcoI monozyme-cleaved pCambia1302 vector to generate the effector plasmids. A short truncated fragment of the promoter containing different numbers of CRT/DRE elements upstream of the ATG start codon for GhPP2C1 or GhPP2C2 was inserted by seamless cloning into the pGreenII 0800-LUC vector to generate GhPP2C1 Pro:LUC, C1:LUC, C2:LUC, GhPP2C2 Pro:LUC, and C3:LUC reporter plasmids. The recombinant plasmid and the negative control plasmid were introduced into Agrobacterium EHA105 (pSoup-p19) competent cells using the freeze–thaw method, inoculated in LB liquid medium supplemented with 50 mg L−1 kanamycin and 50 mg L−1 rifampicin, and incubated at 28°C on a shaker at 200 rpm to OD600 = 1.0. The Agrobacterium cells were centrifuged at 3,000 × g for 10 min. The supernatant was discarded and the Agrobacterium cells were suspended in permeate [containing 10 mM MgCl2, 10 mM MES (pH 5.6), and 150 μM acetosyringone] to OD600 = 1.2 and incubated for 3 h at room temperature in the dark. The suspensions were mixed (1:1, v/v) and injected into the lower surface of the tobacco leaves using a needleless syringe and incubated for 48–60 h under a 16 h light/8 h dark cycle at 23°C and 50% relative humidity. D-Luciferin potassium salt (10 μM) was sprayed onto the tobacco leaves and then imaged using the LB985 Night SHADE fluorescence imaging system (Berthold Technologies, Bad Wildbad, Germany). The dual LUC activity of the different samples was determined using a GloMax 20/20 luminometer (Promega, Madison, WI, United States) and the Dual-Luciferase Reporter Assay System (Promega) in accordance with the manufacturer’s instructions. The primers used in this experiment are listed in Supplementary Table 1.
Statistical Analysis
Statistical analyses were performed with the SPSS 22.0 software (IBM, Armonk, NY, United States). Values were the means ± SE deviations. Comparisons between groups were performed using one-way ANOVA followed by Duncan’s multiple comparisons. For all comparisons, significant at p < 0.05 and highly significant at p < 0.01 were considered statistically significant.
Results
AmCBF1 Is Closely Associated With Plant Height
The plant height of upland cotton overexpressing AmCBF1 was significantly reduced in both the greenhouse and the field. During the growth period, the plant height of L28 and L30 decreased by approximately 33 and 35% compared with that of R15 (Supplementary Figure 2). In the VIGS experiment, silencing of the cotton CLA1 gene in R15 was used as a positive control (Figure 1A). Silencing of AmCBF1 in L28 or L30 reduced the expression of AmCBF1 by approximately 75 and 85%, and the plant height was partly restored to 70.5 and 81.3% of that of R15, respectively (Figures 1A–C). The plant height of VIGS lines may be depending on the silencing efficiency of AmCBF1. These results indicated that overexpression of AmCBF1 was crucial for dwarfing in the cotton lines.
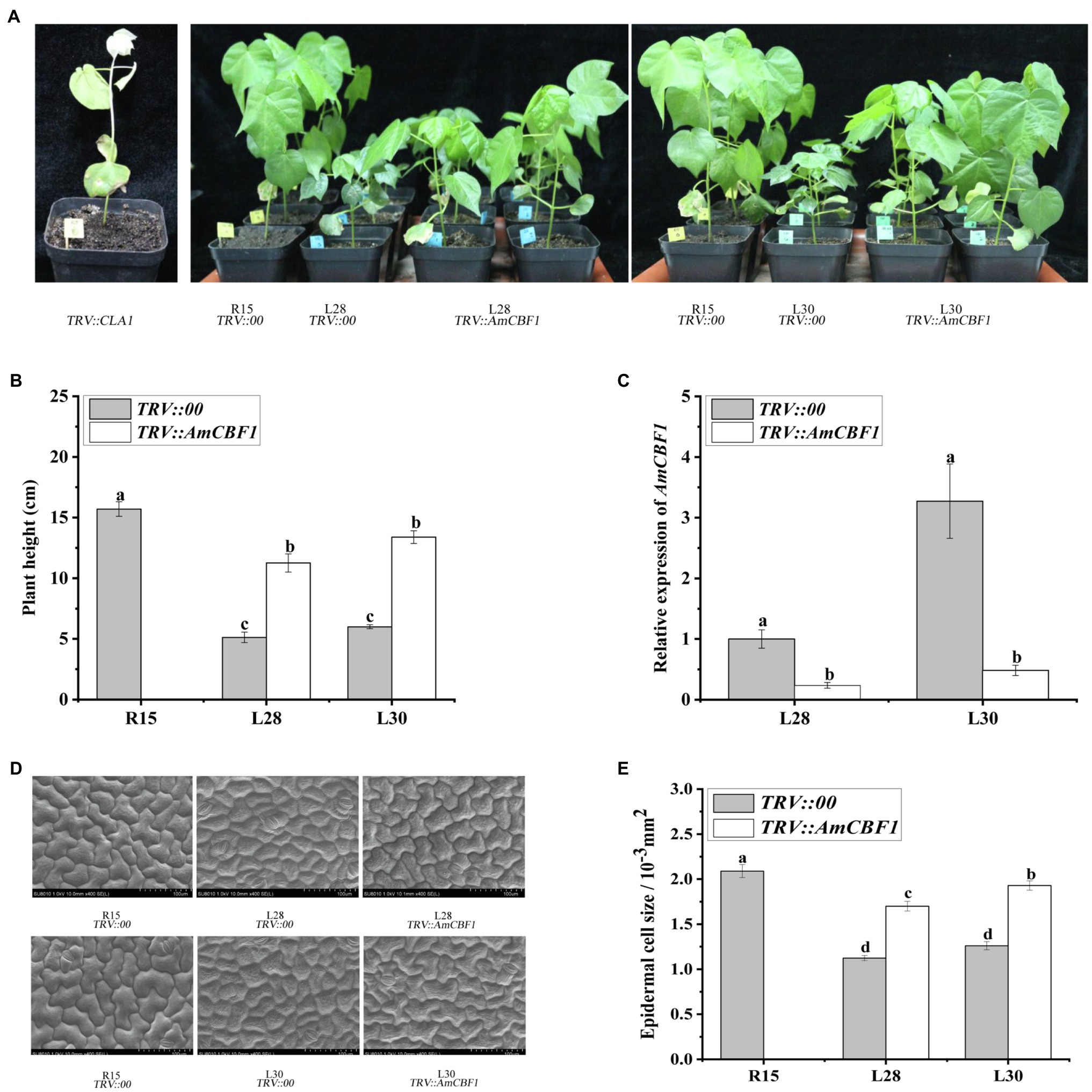
Figure 1. Cotton phenotypes and leaf epithelial cells observed before and after silencing of AmCBF1 by VIGS. (A) Phenotype of cotton after AmCBF1 silencing. TRV::CLA1, albino phenotype, positive control; TRV::AmCBF1, AmCBF1 silencing in L28 and L30. (B) Cotton plant height after AmCBF1 silencing. (C) Relative expression of AmCBF1 in cotton leaves after AmCBF1 silencing. (D,E) Scanning electron micrographs and size of the upper epidermal cells of true leaves. Error bars indicate the standard error (n = 3 biological replicates). Different lower-case letters above bars indicate a significant difference at p < 0.05 as determined using Duncan’s multiple range test.
AmCBF1 Negatively Regulates the Size of Cotton Leaf Epidermal Cells
The epidermal cells were observed with a scanning electron microscope. The epidermal cells of true leaves of L28 and L30 were smaller than those of R15, and the cells were more tightly packed. After silencing of AmCBF1 in L28 and L30, the epidermal cells of true leaves tended to enlarge, and partially recovered the size and morphology of the epidermal cells of R15 (Figure 1D). Statistical analysis indicated that the size of epidermal cells in L28 and L30 decreased by approximately 40 and 45% compared with that of R15, and the size of epidermal cells was recovered by approximately 25 and 35%, respectively, after silencing of AmCBF1 (Figure 1E). Overexpression of AmCBF1 led to reduction in the size of leaf epidermal cells, which suggested that the dwarfing mechanism in cotton may be regulated by AmCBF1.
RNA-Seq Analysis
To identify the regulatory pathway for AmCBF1-regulated plant dwarfing, we investigated the DEGs in R15, L28, and L30 in response to AmCBF1 overexpression by RNA-seq. Cluster analysis of the DEGs demonstrated that the number of down-regulated genes affected by overexpression of AmCBF1 was generally higher in L28 and L30 than in R15 (Figure 2A). A Venn diagram indicated that 780 DEGs were shared between R15 and L28, of which 309 were up-regulated and 471 were down-regulated in L28. There were 3,899 DEGs in common between R15 and L30, of which 1,424 were up-regulated and 2,475 were down-regulated in L30. Among these DEGs, 384 DEGs were common to L28 and L30 compared with R15 (Figure 2B). Analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment revealed that the DEGs were mainly enriched in phytohormone signaling, DNA replication, starch and sucrose metabolism, carbon metabolism, and amino acid biosynthesis (Figure 2C). Subsequently, 12 DEGs from different metabolic pathways were randomly selected to verify the results of transcriptome sequencing, which suggested the RNA-seq results were reliable (Figure 2D). Notably, five GhPP2C genes were down-regulated in the transcriptome data (Supplementary Figure 1). These genes are important intermediate receptors in ABA signal transduction.
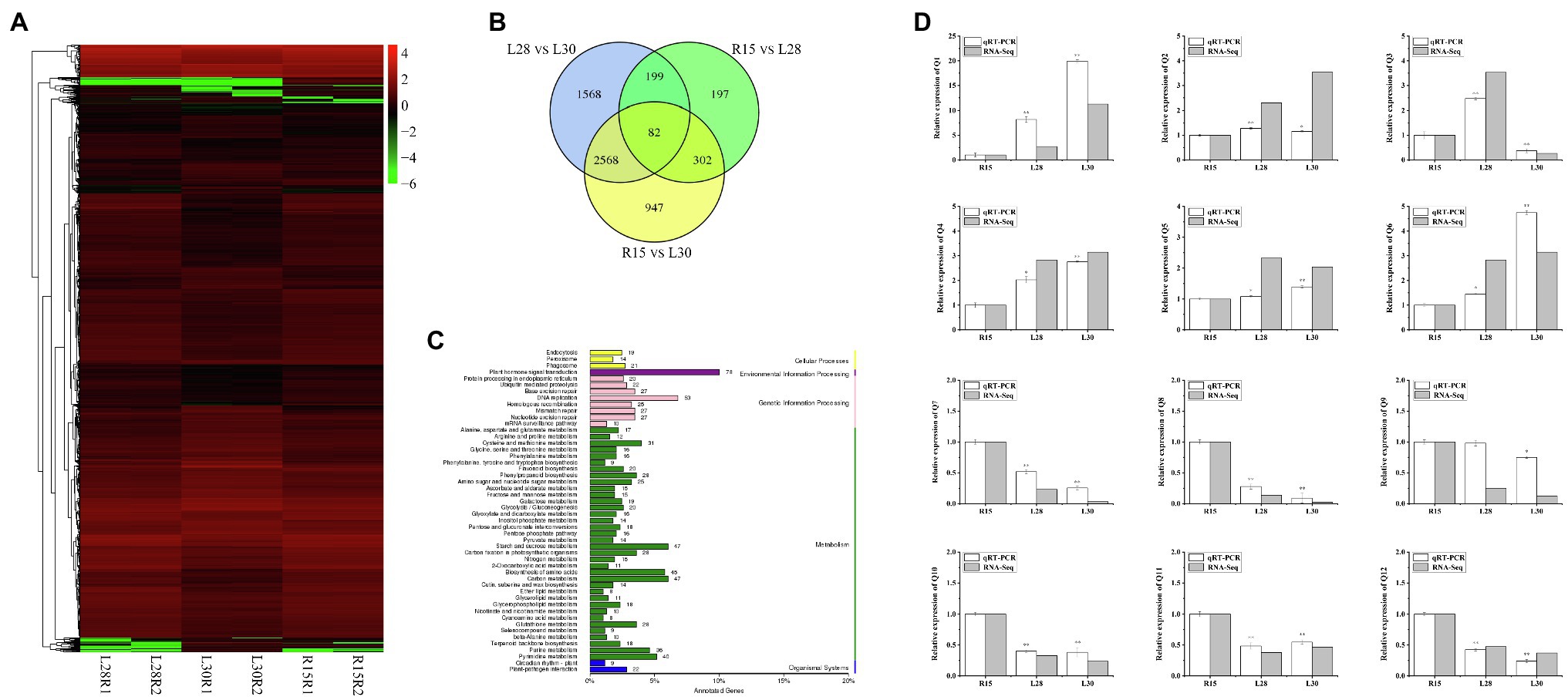
Figure 2. RNA-seq analysis of cotton R15, L28, and L30 seedlings. (A) Cluster diagram of expression patterns of all differentially expressed genes (DEGs). The horizontal axis represents the clustering results for the R15, L28, and L30 materials, and the vertical axis represents the clustering of the DEGs. Each column represents one sample of the plant materials and each row represents a different gene. The color indicates the log (FPKM +1) expression level of the gene in the sample. (B) Venn diagram of DEGs. Each circle in the plot represents a differential grouping, and the intersection of the circles represents the common DEGs, with the corresponding number of genes identified in the set. (C) KEGG classification plot of DEGs. The vertical axis is the name of the KEGG metabolic pathway, and the horizontal axis is the number of genes annotated to the pathway as a proportion of the total number of genes annotated. The value beside each bar is the number of genes annotated for each pathway. (D) Expression patterns of 12 genes in R15, L28, and L30 determined by RNA-seq and qRT-PCR analyses. Error bars indicate the standard error (n = 3 biological replicates). *p < 0.05, **p < 0.01 (Student’s t-test).
Phylogenetic Analyses of Upland Cotton PP2C Family Members
Eighty PP2C family members have been identified in Arabidopsis, which are classified into 12 subgroups (Fuchs et al., 2013). To clarify the phylogenetic relationships of PP2C family members in upland cotton, the keyword “protein phosphatase 2C” was used as the query term in a search of the cottonFGD database. A total of 265 PP2C family members from the upland cotton genome were retrieved. Phylogenetic analysis showed that the upland cotton PP2Cs could be divided into 12 subgroups (A–L; Figure 3A). Among these subgroups, 39 PP2C family members were present in subgroup A and may be involved in ABA signaling. Interestingly, four GhPP2C proteins (GhPP2C1–GhPP2C4) belonged to subgroup A of the PP2C family. Among these proteins, GhPP2C1, GhPP2C2, and GhPP2C3 were most closely related to AtHAI1/2/3 and AtAHG3 in Arabidopsis (Figure 3B).
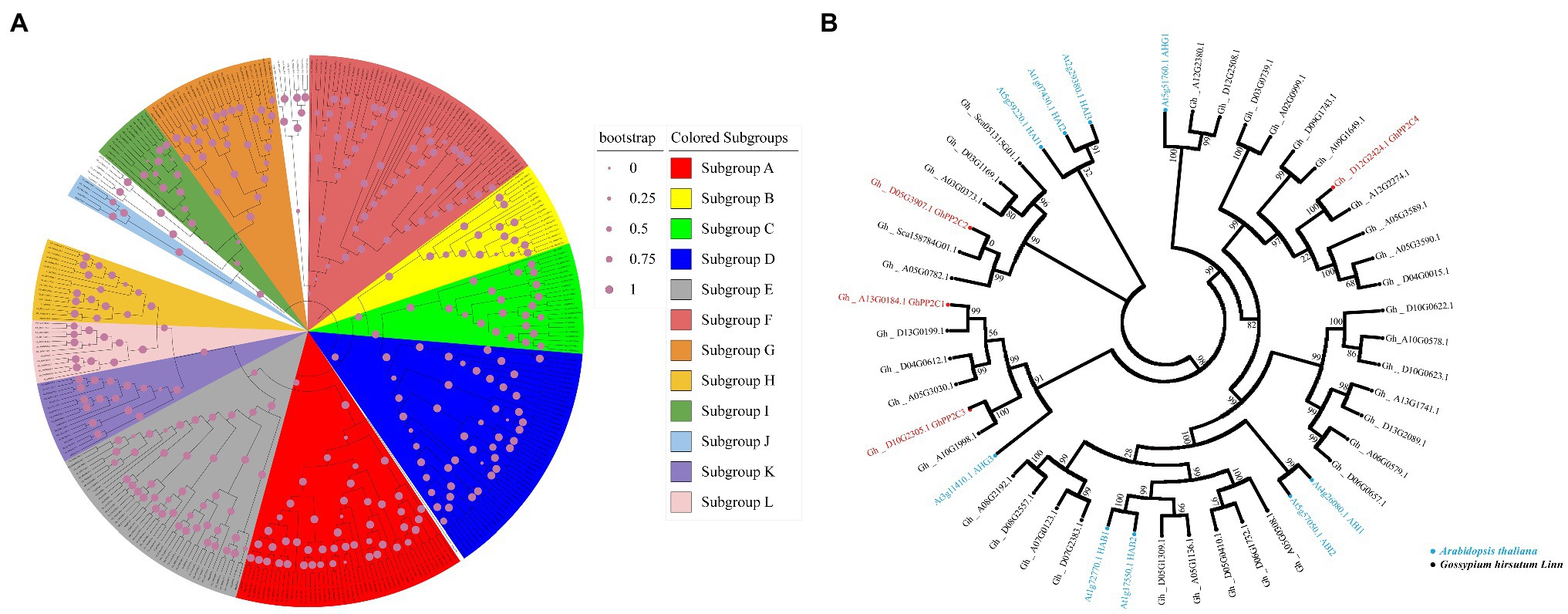
Figure 3. Phylogenetic relationships among PP2C family members. (A) Phylogenetic tree for PP2C family members of upland cotton and Arabidopsis. Sequences for PP2C family members of Arabidopsis were obtained from TAIR (http://www.arabidopsis.org). Subgroups A–L are as delimited in a previous study (Fuchs et al., 2013). Different subgroups are indicated by different colors. (B) Phylogenetic tree for subgroup A PP2C family members of upland cotton and Arabidopsis. Blue and black indicate Arabidopsis and upland cotton, respectively; red indicates the four GhPP2C genes detected in the transcriptome data. The evolutionary tree was constructed using the neighbor-joining method with MEGA7. The values at the nodes are bootstrap support percentages (n = 1,000 replicates).
Expression Analyses and Subcellular Localization of GhPP2Cs
The expression levels of GhPP2C1–GhPP2C4 were analyzed by qRT-PCR in three cotton materials (R15, L28, and L30). The expression levels were significantly lower in L28 compared with those in R15, and were extremely significantly decreased in L30. Compared with R15, the expression of these genes decreased by approximately 40–80% in L28 and 60–90% in L30 (Figure 4A). These results implied that the expression of these GhPP2C genes was negatively correlated with expression of AmCBF1.
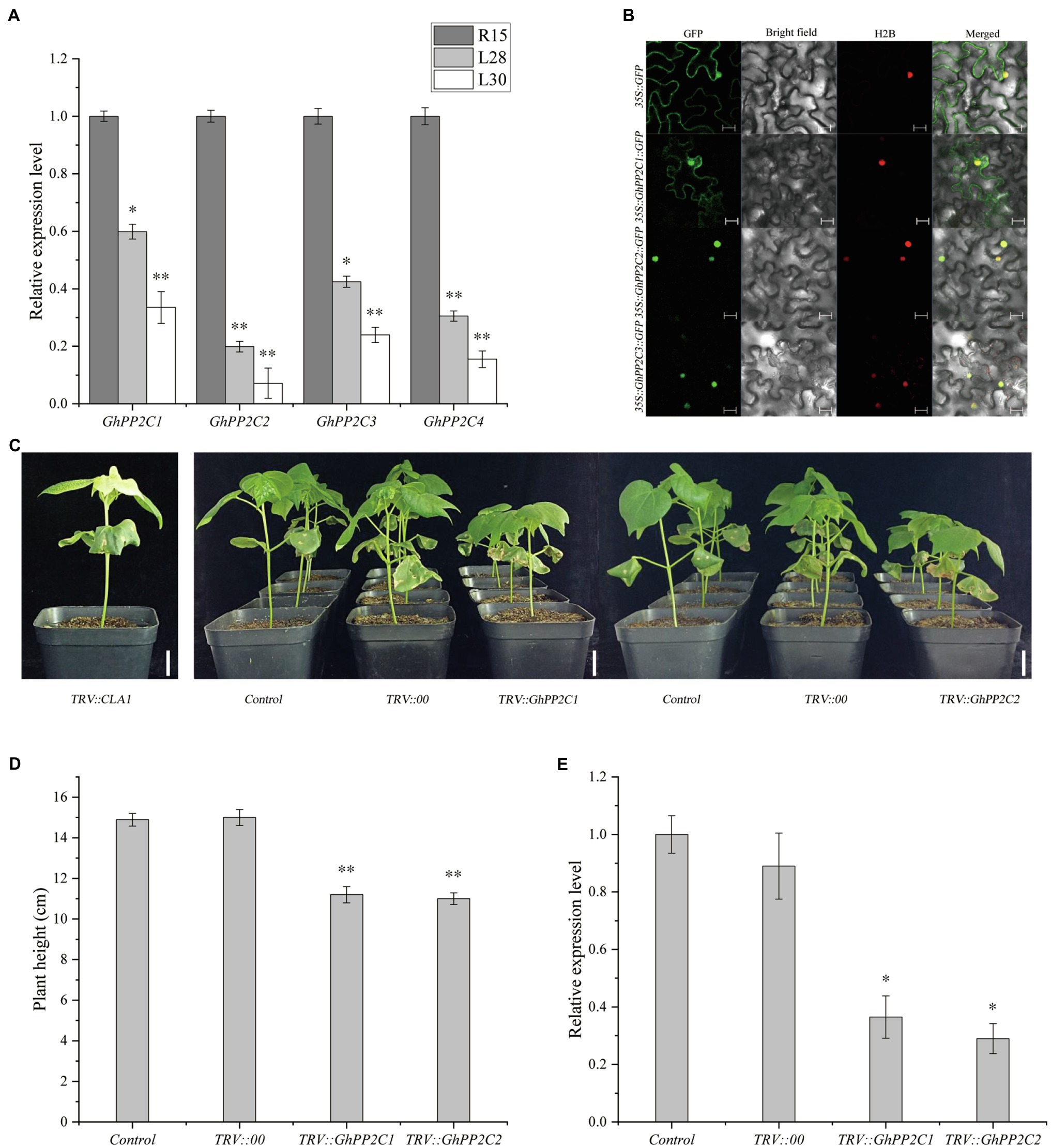
Figure 4. Preliminary analysis of GhPP2C gene functions. (A) Expression of four GhPP2C genes in cotton R15, L28, and L30 seedlings. (B) Subcellular localization of GhPP2C proteins. Fluorescence of GhPP2C1-GFP, GhPP2C2-GFP, and GhPP2C3-GFP fusion proteins and GFP (negative control) was detected at 488 nm by confocal microscopy with H2B-mCherry as the nucleolus marker; images were merged (Merged). Scale bar = 20 μm. (C) Plant phenotype after VIGS silencing of GhPP2C1 or GhPP2C2 in R15. TRV::CLA1, albino phenotype, positive control; Control, not treated; TRV::00, empty vector control. Scale bar = 2 cm. (D) Plant height after silencing of GhPP2C1 and GhPP2C2. (E) Gene expression assay after silencing of GhPP2C1 or GhPP2C2. Error bars indicate the standard error (n = 3 biological replicates). *p < 0.05, **p < 0.01 (Student’s t-test).
To investigate the subcellular localization of the GhPP2C proteins, we first predicted the subcellular localization using the CELLO 2.5 online tool.3 The results predicted that GhPP2C1–GhPP2C3 were localized in the nucleus, whereas GhPP2C4 was localized in the chloroplast. Combined with the phylogenetic analysis, the subcellular localization of GhPP2C1–GhPP2C3 was further verified by fusion expression with GFP. The fluorescence signal of the GhPP2C1-GFP fusion protein was localized to the cell membrane and nucleus, whereas the GFP signal of the GhPP2C2-GFP and GhPP2C3-GFP fusion proteins was localized to the nucleus (Figure 4B). The 1302-GFP protein was used as a control.
Silencing of GhPP2C Genes Reduces Plant Height
To further explore the function of GhPP2C1–GhPP2C3 in regulating plant height, we separately silenced these genes in R15 using VIGS and observed whether the plant phenotype was altered. Silencing of GhPP2C1 or GhPP2C2 resulted in a significant decrease in growth rate, and the plant height was reduced by approximately 25% compared with that of the control (Figures 4C,D). The qRT-PCR analysis indicated that the relative expression level of these two genes was significantly decreased (by approximately 65 and 70%) in gene-silenced plants than in the control (Figure 4E). These results suggested that GhPP2C played a vital role in regulating cotton plant height. Silencing of GhPP2C3 did not cause a significant change in phenotype, so no further studies were conducted on this gene (data not shown). TRV2::CLA1 was used as a positive control (Figure 4C).
AmCBF1 Binds Directly to the CRT/DRE Elements in the GhPP2C Promoter
Overexpression of AmCBF1 led to down-regulation in the expression level of GhPP2C genes, whereas silencing of the GhPP2C genes resulted in decreased plant height. These results suggested that there may be a regulatory relationship between AmCBF1 and GhPP2C proteins. The 3 kb promoter sequence upstream of GhPP2C1 or GhPP2C2 was further analyzed. Three or two CRT/DRE (A/GCCC/GAC) elements were identified and designated P1, P2, and P3 (upstream elements of GhPP2C1), and P4 and P5 (upstream elements of GhPP2C2; Figure 5A). A Y1H assay was conducted to verify whether AmCBF1 could bind to the promoter DNA fragments containing CRT/DRE elements. The yeast strains containing pGADT7 were unable to grow on SD/−Leu/−Ura medium supplemented with 400 ng ml−1 AbA, whereas the yeast strains harboring pGADT7-AmCBF1 were able to grow on this medium. Thus, AmCBF1 could directly bind to the CRT/DRE element in the promoter of GhPP2C1 (containing P1, P2, and P3) or GhPP2C2 (containing P4 and P5; Figure 5B). The binding properties between P1–P5 and AmCBF1 were also verified separately by means of a Y1H assay (Supplementary Figure 4A).
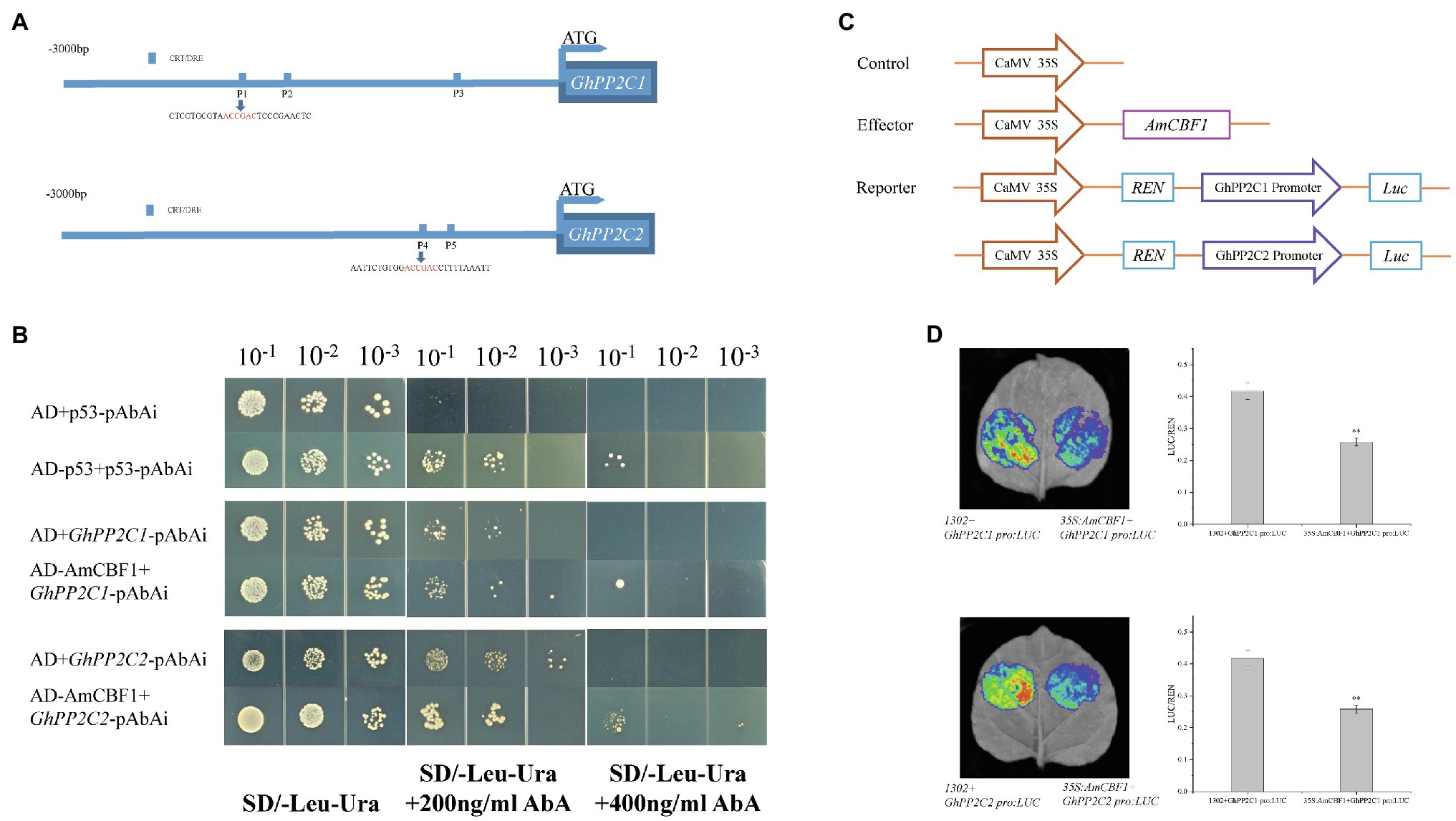
Figure 5. AmCBF1 transcription factor inhibition of the expression of GhPP2C1 and GhPP2C2. (A) Schematic diagram of the CRT/DRE binding element in the 3,000 bp promoter upstream of the ATG start codon of GhPP2C1 and GhPP2C2. (B) Yeast one-hybrid analysis showing binding of AmCBF1 to the promoter sequence of GhPP2C1 or GhPP2C2. Yeast cells were cultured at serial dilutions of 1:10, 1:100, and 1:1,000 on SD/−Leu/−Ura medium supplemented with or lacking 400 ng ml−1 aureobasidin A. The pGADT7 vector was used as a negative control. (C) Schematic diagram of vector construction of the reporter and effector plasmids for transient transcriptional activity assays in tobacco. REN, Renilla luciferase; LUC, firefly luciferase. (D) Tobacco leaf live-imaging observations. Red indicates the strongest fluorescence signal intensity. Luciferase activity was normalized to the respective REN activity and expressed as relative expression. Error bars indicate the standard error (n = 3 biological replicates). **p < 0.01 (Student’s t-test).
AmCBF1 Represses Transcription of GhPP2C1 or GhPP2C2
To verify how AmCBF1 regulates the transcription of GhPP2C1 or GhPP2C2, a dual-luciferase assay was performed in tobacco. The relevant vectors were constructed (Figure 5C; Supplementary Figure 4B). When the suspension containing the reporter plasmid GhPP2C1 Pro:LUC, C1:LUC, C2:LUC, GhPP2C2 Pro:LUC or C3:LUC were, respectively, mixed with the suspension containing the effector plasmid 35S:AmCBF1 to inject tobacco, the LUC luminescence intensity was significantly reduced and the relative LUC/Renilla luciferase (REN) activity was significantly lower than that of the control (decreased by approximately 30–45%; Figure 5D; Supplementary Figure 4C). These results further suggested that AmCBF1 may act as a transcriptional repressor and inhibits the expression of GhPP2C1 or GhPP2C2.
Discussion
Plant architecture strongly influences the fiber quality and yield of cotton. New cotton varieties with a dwarf and compact growth phenotype not only perform well in terms of lodging resistance and photosynthetic efficiency, but also are suitable for mechanical harvesting to reduce labor costs (Bednarz et al., 2005; Dai et al., 2015). Overexpression of CBF/DREB transcription factors in a variety of plant species, including Arabidopsis, tobacco, tomato, and cotton, can lead to dwarfing (Zhang et al., 2004; Zhou et al., 2014, 2017; Ji et al., 2021). In the present study, L28 and L30 overexpressing AmCBF1 both showed a dwarf phenotype in the greenhouse and in the field (Supplementary Figure 2). When AmCBF1 expression in L28 and L30 was inhibited by VIGS, the plant height was partially recovered (Figures 1A,B). This result was consistent with previous research that silencing of GhDREB1B in the AS98 cotton mutant exhibited partial restoration to normal plant height (Ji et al., 2021). At the cellular level, the upper epidermal cells were observed by scanning electron microscopy before and after AmCBF1 silencing. In addition to the change in size of epidermal cells, it was noteworthy that the epidermal cells were more tightly packed in L28 and L30 than in R15. The surface of the epidermal cells in L28 and L30 was less smooth than in R15, even after AmCBF1 silencing, and this phenotype was not visibly restored (Figure 1D). We speculated that AmCBF1 overexpression may affect the expression of genes associated with cell morphological structure, which leads to this change in cell morphology. Thus, the relationship between AmCBF1 and cell morphology requires further study.
To further investigate the genetic pathway of AmCBF1-regulated plant dwarfing, RNA-seq of R15, L28, and L30 seedlings was performed, and the genes differentially expressed in L28 or L30 in comparison with R15 were identified. Cluster analysis of the expression patterns of all DEGs revealed that most DEGs were significantly down-regulated in the overexpressed material, and more DEGs were down-regulated in L30 than in L28 (Figure 2A). A Venn diagram provided a visual representation of the number of DEGs common in the different materials (Figure 2B). However, the number of DEGs between R15 and L28 was 780, while 3,899 DEGs were found between R15 and L30. The possible reason for the huge differences in the number of DEGs may be due to the different insertion site of AmCBF1. A similar study revealed that in upland cotton overexpressing DREB1B, the number of up-regulated and down-regulated DEGs was approximately equal (Ji et al., 2021). We speculated that the difference in findings between these two studies was due to the different genes overexpressed in cotton or differences in the growth and development status of cotton seedlings used in transcriptome sequencing. The KEGG enrichment analysis of the DEGs revealed that most of the DEGs were enriched in phytohormone signaling pathways, and several genes were enriched in starch and sucrose metabolism, amino acid biosynthesis, carbon metabolism, and lignin biosynthesis pathways (Figure 2C). This result suggested that phytohormones may play an important role in plant dwarfing and was similar to the findings of previous reports (Chen et al., 2021; Sun et al., 2021).
Among the DEGs enriched in phytohormone signaling pathways, four GhPP2C genes were down-regulated. The function of PP2C genes has been widely studied in diverse plant species, such as Arabidopsis, rice, tomato, maize (Zea mays), and pepper (Capsicum annuum; Kerk et al., 2002; Singh et al., 2010; Kim et al., 2014; Wei and Pan, 2014; Liang et al., 2021). However, few studies have investigated the function of PP2C genes in cotton. In the present study, we analyzed the evolutionary relationships of PP2C family members in upland cotton and 265 putative PP2C genes were identified from the cottonFGD database. This result contrasted with a previous study in which 181 PP2C genes were identified in upland cotton (Shazadee et al., 2019). The difference might be due to the different cotton databases accessed or differences in retrieval methods. The 265 GhPP2C genes were classified into 12 subgroups of which 39 genes were classified in subgroup A. Interestingly, four of the five down-regulated GhPP2C genes identified by RNA-seq belonged to subgroup A. Among these genes, GhPP2C1–GhPP2C3 were closely related to AtHAI1/2/3 and AtAHG3 (Figure 3B). Subcellular localization analysis revealed that GhPP2C1 was localized to the nucleus and cell membrane, whereas GhPP2C2 and GhPP2C3 were localized to the nucleus (Figure 4B). It has previously been reported that PP2C proteins are localized in the cytoplasm (Amir Hossain et al., 2018). The aforementioned results indicate that PP2C proteins may be localized in different subcellular locations to perform different functions. In the present study, silencing of GhPP2C1 or GhPP2C2 resulted in dwarfing (Figure 4C). Their functions in regulating plant height may be similar to those of AtPP2CF1 (in Arabidopsis), OsPP2C09 (in rice), and SlPP2C3 (in tomato; Sugimoto et al., 2014).
Members of the CBF/DREB transcription factor subfamily interact with genes containing CRT/DRE elements in the promoter and either activate or repress gene expression (Magome et al., 2008; Tsutsui et al., 2009; Kuang et al., 2017; Zhou et al., 2017). The target genes of CBF/DREB transcription factors were significantly enriched in phytohormone signaling pathways, especially the ABA signaling pathway (Song et al., 2021). Whether GhPP2C genes are directly regulated by AmCBF1 and are involved in dwarfing in upland cotton has not been reported previously. In the present study, the upstream 3 kb promoter of GhPP2C1 or GhPP2C2 was analyzed. The Y1H assay verified that AmCBF1 was capable of binding to the CRT/DRE element in the promoter of GhPP2C1 or GhPP2C2 (Figure 5B; Supplementary Figure 4A). The dual-luciferase assay indicated that AmCBF1 directly inhibited transcription of GhPP2C1 or GhPP2C2 by binding to the CRT/DRE element in the promoter (Figure 5D; Supplementary Figures 4B,C). To our knowledge, the present study is the first to report that CBF-like transcription factors are involved in dwarfing of upland cotton by regulating the expression of GhPP2C genes.
There is increasing evidence that PP2C genes of subgroup A are crucial negative regulators of the ABA signaling pathway, and show distinct expression patterns and subcellular localization in plants (Saez et al., 2004; Nishimura et al., 2007; Umezawa et al., 2009; Zhang et al., 2012). Abscisic acid may exert various inhibitory effects in plant growth (Sharp and Lenoble, 2002; Chen et al., 2006; Zhao et al., 2015; Wang et al., 2018). The ABA–PYR/PYL/RCAR–PP2C complex is an important ternary complex that initiates signal transduction and activates the expression of downstream genes in the classical ABA signaling pathway (Hao et al., 2011; Antoni et al., 2012; Bhaskara et al., 2012; Chen et al., 2016). It is well known that GA and ABA have antagonistic effects in plants, jointly maintain the dynamic balance of plant hormones, and play roles in the growth and development of plants (Yoshida et al., 2014; Liu and Hou, 2018; Shu et al., 2018; Song et al., 2021; Tuan et al., 2021). Based on these findings and the present results, we hypothesized that AmCBF1 overexpression may affect ABA signaling by down-regulating the expression of GhPP2C1 or GhPP2C2, thereby leading to cotton dwarfing by disrupting the balance of ABA and GA.
Conclusion
Overexpression of the AmCBF1 transcription factor in upland cotton affected plant growth. GhPP2C1 and GhPP2C2 were identified among the DEGs detected by RNA-seq, and silencing of these two genes caused a dwarf phenotype in upland cotton. We verified that AmCBF1 could bind to CRT/DRE elements and negatively regulate the expression of GhPP2C genes, thereby regulating plant height. Based on these findings, we propose a molecular model for coordinated regulation of plant height by AmCBF1 and GhPP2C genes (Figure 6). The present findings expand our knowledge of the regulatory mechanism of dwarfing in cotton, and might contribute to the molecular breeding of cotton cultivars with a dwarf phenotype.
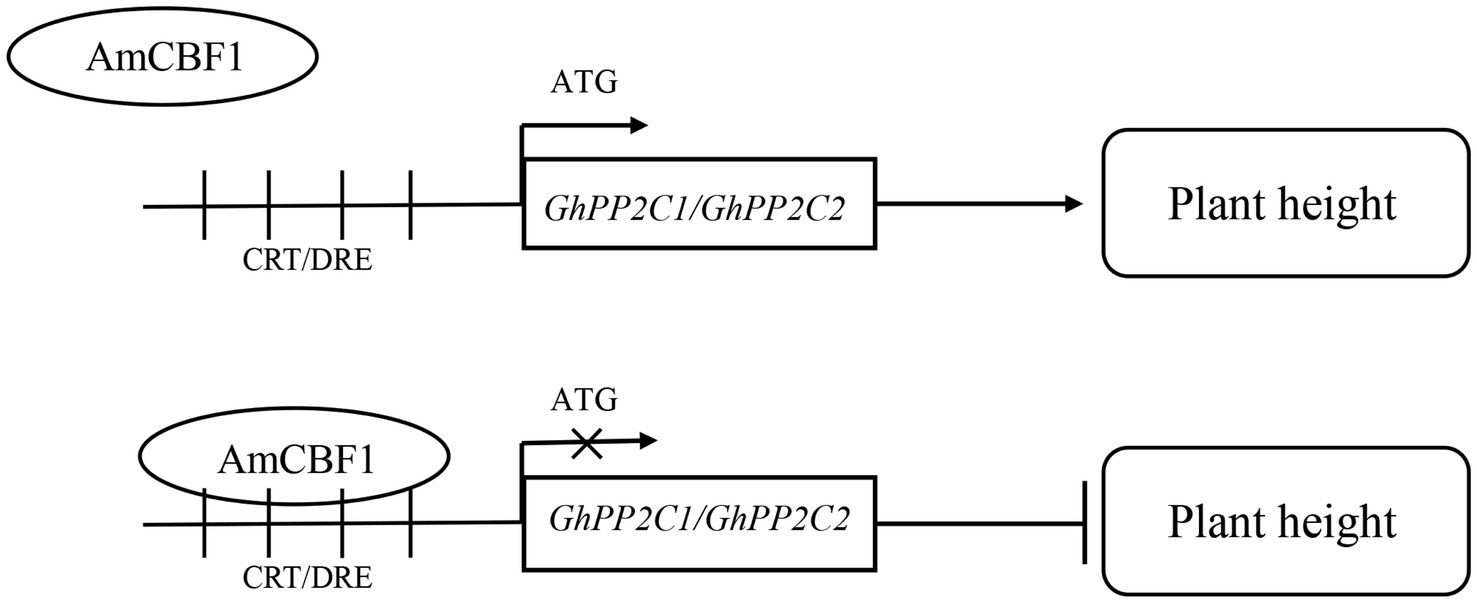
Figure 6. Schematic diagram of a working model for AmCBF1 regulation of GhPP2C genes. The transcription factor AmCBF1 inhibits expression of GhPP2C genes and negatively regulates the growth of upland cotton seedlings by binding to CRT/DRE elements. Bar, inhibition.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The transcriptome sequencing data can be found in the NCBI database under accession no. PRJNA791756.
Author Contributions
HG designed the experiments. JL and LW performed the experiments. JL and LW performed the data analysis. QZ, CM, XS, and HC provided valuable suggestions on the research design and improvement of the manuscript. JL and HG wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Central Public-interest Scientific Institution Basal Research Fund (nos. Y2022PT22 and Y2022ZK25).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Robert McKenzie, PhD, from Liwen Bianji (Edanz; www.liwenbianji.cn) for editing the English text of a draft of this manuscript. We thank the research team of Jianmin Wan, Institute of Crop Science, Chinese Academy of Agricultural Sciences, for providing the dual-luciferase plasmids. We thank the research team of Zhiying Ma, Hebei Agricultural University, for providing the VIGS related plasmids.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.914206/full#supplementary-material
Footnotes
References
Agarwal, P. K., Agarwal, P., Reddy, M. K., and Sopory, S. K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25, 1263–1274. doi: 10.1007/s00299-006-0204-8
Amir Hossain, M., Ahn, C.-H., Lee, I. S., Jeon, J.-S., An, G., and Park, P. B. (2018). Isolation of a novel protein phosphatase2C in rice and its response to gibberellin. Biochem. Biophys. Res. Commun. 503, 1987–1992. doi: 10.1016/j.bbrc.2018.07.146
Antoni, R., Gonzalez-Guzman, M., Rodriguez, L., Rodrigues, A., Pizzio, G. A., and Rodriguez, P. L. (2012). Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid aeceptors. Plant Physiol. 158, 970–980. doi: 10.1104/pp.111.188623
Baker, S. S., Wilhelm, K. S., and Thomashow, M. F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/BF00029852
Becker, A., and Lange, M. (2010). VIGS – genomics goes functional. Trends Plant Sci. 15, 1–4. doi: 10.1016/j.tplants.2009.09.002
Bednarz, C. W., Shurley, W. D., Anthony, W. S., and Nichols, R. L. (2005). Yield, quality, and profitability of cotton produced at varying plant densities. Agron. J. 97, 235–240. doi: 10.2134/agronj2005.0235a
Bhaskara, G. B., Nguyen, T. T., and Verslues, P. E. (2012). Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 160, 379–395. doi: 10.1104/pp.112.202408
Bork, P., Brown, N. P., Hegyi, H., and Schultz, J. (1996). The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci. 5, 1421–1425. doi: 10.1002/pro.5560050720
Chen, P., Sun, Y.-F., Kai, W.-B., Liang, B., Zhang, Y.-S., Zhai, X.-W., et al. (2016). Interactions of ABA signaling core components (SlPYLs, SlPP2Cs, and SlSnRK2s) in tomato (Solanum lycopersicon). J. Plant Physiol. 205, 67–74. doi: 10.1016/j.jplph.2016.07.016
Chen, L., Yang, H., Fang, Y., Guo, W., Chen, H., Zhang, X., et al. (2021). Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 19, 702–716. doi: 10.1111/pbi.13496
Chen, C.-W., Yang, Y.-W., Lur, H.-S., Tsai, Y.-G., and Chang, M.-C. (2006). A novel function of abscisic acid in the regulation of rice (Oryza sativa L.) root growth and development. Plant Cell Physiol. 47, 1–13. doi: 10.1093/pcp/pci216
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Dai, J., Li, W., Tang, W., Zhang, D., Li, Z., Lu, H., et al. (2015). Manipulation of dry matter accumulation and partitioning with plant density in relation to yield stability of cotton under intensive management. Field Crop Res. 180, 207–215. doi: 10.1016/j.fcr.2015.06.008
Dong, C.-H., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J.-K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U. S. A. 103, 8281–8286. doi: 10.1073/pnas.0602874103
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Fuchs, S., Grill, E., Meskiene, I., and Schweighofer, A. (2013). Type 2C protein phosphatases in plants. FEBS J. 280, 681–693. doi: 10.1111/j.1742-4658.2012.08670.x
Gaits, F., Shiozaki, K., and Russell, P. (1997). Protein phosphatase 2C acts independently of stress-activated kinase cascade to regulate the stress response in fission yeast. J. Biol. Chem. 272, 17873–17879. doi: 10.1074/jbc.272.28.17873
Graeff, M., Rana, S., Marhava, P., Moret, B., and Hardtke, C. S. (2020). Local and systemic effects of brassinosteroid perception in developing phloem. Curr. Biol. 30, 1626.e3–1638.e3. doi: 10.1016/j.cub.2020.02.029
Hao, Q., Yin, P., Li, W., Wang, L., Yan, C., Lin, Z., et al. (2011). The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol. Cell 42, 662–672. doi: 10.1016/j.molcel.2011.05.011
He, Y., Hao, Q., Li, W., Chuangye, Y., Yan, N., and Yin, P. (2014). Identification and characterization of ABA receptors in Oryza sativa. PLoS One 9:e95246. doi: 10.1371/journal.pone.0095246
Hsieh, T.-H., Lee, J.-T., Yang, P.-T., Chiu, L.-H., Charng, Y.-Y., Wang, Y.-C., et al. (2002). Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 129, 1086–1094. doi: 10.1104/pp.003442
Hu, Y.-X., Tao, Y.-B., and Xu, Z.-F. (2017). Overexpression of Jatropha gibberellin 2-oxidase 6 (JcGA2ox6) induces dwarfism and smaller leaves, flowers and fruits in Arabidopsis and Jatropha. Front. Plant Sci. 8:2103. doi: 10.3389/fpls.2017.02103
Hubbard, M. J., and Cohen, P. (1993). On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem. Sci. 18, 172–177. doi: 10.1016/0968-0004(93)90109-Z
Hubbard, K. E., Nishimura, N., Hitomi, K., Getzoff, E. D., and Schroeder, J. I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 24, 1695–1708. doi: 10.1101/gad.1953910
Ingebritsen, T. S., and Cohen, P. (1983). Protein phosphatases: properties and pole in cellular regulation. Science 221, 331–338. doi: 10.1126/science.6306765
Ito, Y., Katsura, K., Maruyama, K., Taji, T., Kobayashi, M., Seki, M., et al. (2006). Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47, 141–153. doi: 10.1093/pcp/pci230
Ji, G., Liang, C., Cai, Y., Pan, Z., Meng, Z., Li, Y., et al. (2021). A copy number variant at the HPDA-D12 locus confers compact plant architecture in cotton. New Phytol. 229, 2091–2103. doi: 10.1111/nph.17059
Jia, Y., Ding, Y., Shi, Y., Zhang, X., Gong, Z., and Yang, S. (2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212, 345–353. doi: 10.1111/nph.14088
Kerk, D., Bulgrien, J., Smith, D. W., Barsam, B., Veretnik, S., and Gribskov, M. (2002). The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 129, 908–925. doi: 10.1104/pp.004002
Khush, G. S. (1999). Green revolution: preparing for the 21st century. Genome 42, 646–655. doi: 10.1139/g99-044
Kim, S., Park, M., Yeom, S.-I., Kim, Y.-M., Lee, J. M., Lee, H.-A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Kuang, J.-F., Chen, J.-Y., Liu, X.-C., Han, Y.-C., Xiao, Y.-Y., Shan, W., et al. (2017). The transcriptional regulatory network mediated by banana (Musa acuminata) dehydration-responsive element binding (MaDREB) transcription factors in fruit ripening. New Phytol. 214, 762–781. doi: 10.1111/nph.14389
Kuhn, J. M., Boisson-Dernier, A. L., Dizon, M. B., Maktabi, M. H., and Schroeder, J. I. (2006). The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140, 127–139. doi: 10.1104/pp.105.070318
Li, Z., Zhang, X., Zhao, Y., Li, Y., Zhang, G., Peng, Z., et al. (2018). Enhancing auxin accumulation in maize root tips improves root growth and dwarfs plant height. Plant Biotechnol. J. 16, 86–99. doi: 10.1111/pbi.12751
Liang, B., Sun, Y., Wang, J., Zheng, Y., Zhang, W., Xu, Y., et al. (2021). Tomato protein phosphatase 2C influences the onset of fruit ripening and fruit glossiness. J. Exp. Bot. 72, 2403–2418. doi: 10.1093/jxb/eraa593
Liu, X., and Hou, X. (2018). Antagonistic regulation of ABA and GA in metabolism and signaling pathways. Front. Plant Sci. 9:251. doi: 10.3389/fpls.2018.00251
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Ma, L. F., Zhang, J. M., Huang, G. Q., Li, Y., Li, X. B., and Zheng, Y. (2014). Molecular characterization of cotton C-repeat/dehydration-responsive element binding factor genes that are involved in response to cold stress. Mol. Biol. Rep. 41, 4369–4379. doi: 10.1007/s11033-014-3308-1
Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., and Oda, K. (2008). The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 56, 613–626. doi: 10.1111/j.1365-313X.2008.03627.x
Miao, J., Li, X., Li, X., Tan, W., You, A., Wu, S., et al. (2020). OsPP2C09, a negative regulatory factor in abscisic acid signalling, plays an essential role in balancing plant growth and drought tolerance in rice. New Phytol. 227, 1417–1433. doi: 10.1111/nph.16670
Miao, C., Xiao, L., Hua, K., Zou, C., Zhao, Y., Bressan, R. A., et al. (2018). Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. U. S. A. 115, 6058–6063. doi: 10.1073/pnas.1804774115
Miyazono, K.-I., Miyakawa, T., Sawano, Y., Kubota, K., Kang, H.-J., Asano, A., et al. (2009). Structural basis of abscisic acid signalling. Nature 462, 609–614. doi: 10.1038/nature08583
Née, G., Kramer, K., Nakabayashi, K., Yuan, B., Xiang, Y., Miatton, E., et al. (2017). DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 8:72. doi: 10.1038/s41467-017-00113-6
Nishimura, N., Yoshida, T., Kitahata, N., Asami, T., Shinozaki, K., and Hirayama, T. (2007). ABA-hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 50, 935–949. doi: 10.1111/j.1365-313X.2007.03107.x
Rodrigues, A., Adamo, M., Crozet, P., Margalha, L., Confraria, A., Martinho, C., et al. (2013). ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell 25, 3871–3884. doi: 10.1105/tpc.113.114066
Rodriguez, P. L. (1998). Protein phosphatase 2C (PP2C) function in higher plants. Plant Mol. Biol. 38, 919–927. doi: 10.1023/A:1006054607850
Saez, A., Apostolova, N., Gonzalez-Guzman, M., Gonzalez-Garcia, M. P., Nicolas, C., Lorenzo, O., et al. (2004). Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 37, 354–369. doi: 10.1046/j.1365-313X.2003.01966.x
Schweighofer, A., Hirt, H., and Meskiene, I. (2004). Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 9, 236–243. doi: 10.1016/j.tplants.2004.03.007
Schweighofer, A., Kazanaviciute, V., Scheikl, E., Teige, M., Doczi, R., Hirt, H., et al. (2007). The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19, 2213–2224. doi: 10.1105/tpc.106.049585
Shan, D.-P., Huang, J.-G., Yang, Y.-T., Guo, Y.-H., Wu, C.-A., Yang, G.-D., et al. (2007). Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytol. 176, 70–81. doi: 10.1111/j.1469-8137.2007.02160.x
Sharp, R., and Lenoble, M. (2002). ABA, ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 53, 33–37. doi: 10.1093/jxb/53.366.33
Shazadee, H., Khan, N., Wang, J., Wang, C., Zeng, J., Huang, Z., et al. (2019). Identification and expression profiling of protein phosphatases (PP2C) gene family in Gossypium hirsutum L. Int. J. Mol. Sci. 20:1395. doi: 10.3390/ijms20061395
Shu, K., Zhou, W., and Yang, W. (2018). APETALA 2-domain-containing transcription factors: focusing on abscisic acid and gibberellins antagonism. New Phytol. 217, 977–983. doi: 10.1111/nph.14880
Singh, A., Giri, J., Kapoor, S., Tyagi, A. K., and Pandey, G. K. (2010). Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics 11:435. doi: 10.1186/1471-2164-11-435
Singh, A., Jha, S., Bagri, J., and Pandey, G. (2015). ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS One 10:e0125168. doi: 10.1371/journal.pone.0125168
Singh, A., Pandey, A., Srivastava, A. K., Tran, L.-S. P., and Pandey, G. K. (2016). Plant protein phosphatases 2C: from genomic diversity to functional multiplicity and importance in stress management. Crit. Rev. Biotechnol. 36, 1023–1035. doi: 10.3109/07388551.2015.1083941
Song, Y., Zhang, X., Li, M., Yang, H., Fu, D., Lv, J., et al. (2021). The direct targets of CBFs: in cold stress response and beyond. J. Integr. Plant Biol. 63, 1874–1887. doi: 10.1111/jipb.13161
Soon, F.-F., Ng, L.-M., Zhou, X. E., West Graham, M., Kovach, A., Tan, M. H. E., et al. (2012). Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335, 85–88. doi: 10.1126/science.1215106
Sugimoto, H., Kondo, S., Tanaka, T., Imamura, C., Muramoto, N., Hattori, E., et al. (2014). Overexpression of a novel Arabidopsis PP2C isoform, AtPP2CF1, enhances plant biomass production by increasing inflorescence stem growth. J. Exp. Bot. 65, 5385–5400. doi: 10.1093/jxb/eru297
Sun, Q., Xie, Y., Li, H., Liu, J., Geng, R., Wang, P., et al. (2021). Cotton GhBRC1 regulates branching, flowering, and growth by integrating multiple hormone pathways. Crop J. 10, 75–87. doi: 10.1016/j.cj.2021.01.007
Tong, Z., Hong, B., Yang, Y., Li, Q., Ma, N., Ma, C., et al. (2009). Overexpression of two chrysanthemum DgDREB1 group genes causing delayed flowering or dwarfism in Arabidopsis. Plant Mol. Biol. 71, 115–129. doi: 10.1007/s11103-009-9513-y
Tsutsui, T., Kato, W., Asada, Y., Sako, K., Sato, T., Sonoda, Y., et al. (2009). DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J. Plant Res. 122, 633–643. doi: 10.1007/s10265-009-0252-6
Tuan, P. A., Nguyen, T.-N., Jordan, M. C., and Ayele, B. T. (2021). A shift in abscisic acid/gibberellin balance underlies retention of dormancy induced by seed development temperature. Plant Cell Environ. 44, 2230–2244. doi: 10.1111/pce.13963
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 17588–17593. doi: 10.1073/pnas.0907095106
Wang, P., Zhao, Y., Li, Z., Hsu, C.-C., Liu, X., Fu, L., et al. (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 69, 100.e6–112.e6. doi: 10.1016/j.molcel.2017.12.002
Wei, K., and Pan, S. (2014). Maize protein phosphatase gene family: identification and molecular characterization. BMC Genomics 15:773. doi: 10.1186/1471-2164-15-773
Wu, C., Zhou, B., and Zhang, T. (2009). Isolation and characterization of a sterile-dwarf mutant in Asian cotton (Gossypium arboreum L.). J. Genet. Genomics 36, 343–353. doi: 10.1016/S1673-8527(08)60123-X
Xie, L., Yang, C., and Wang, X. (2011). Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. J. Exp. Bot. 62, 4495–4506. doi: 10.1093/jxb/err164
Xin, J., Li, C., Ning, K., Qin, Y., Shang, J.-X., and Sun, Y. (2021). AtPFA-DSP3, an atypical dual-specificity protein tyrosine phosphatase, affects salt stress response by modulating MPK3 and MPK6 activity. Plant Cell Environ. 44, 1534–1548. doi: 10.1111/pce.14002
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. doi: 10.1105/tpc.6.2.251
Yang, Z., Zhang, C., Yang, X., Liu, K., Wu, Z., Zhang, X., et al. (2014). PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 203, 437–448. doi: 10.1111/nph.12824
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Zhang, X., Fowler, S. G., Cheng, H., Lou, Y., Rhee, S. Y., Stockinger, E. J., et al. (2004). Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 39, 905–919. doi: 10.1111/j.1365-313X.2004.02176.x
Zhang, C., Sun, J.-L., Jia, Y.-H., Wang, J., Xu, Z.-J., and Du, X.-M. (2011). Morphological characters, inheritance and response to exogenous hormones of a cotton super-dwarf mutant of Gossypium hirsutum. Plant Breed. 130, 67–72. doi: 10.1111/j.1439-0523.2009.01759.x
Zhang, K., Xia, X., Zhang, Y., and Gan, S.-S. (2012). An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J. 69, 667–678. doi: 10.1111/j.1365-313X.2011.04821.x
Zhang, P., Yuan, Z., Wei, L., Qiu, X., Wang, G., Liu, Z., et al. (2022). Overexpression of ZmPP2C55 positively enhances tolerance to drought stress in transgenic maize plants. Plant Sci. 314:111127. doi: 10.1016/j.plantsci.2021.111127
Zhao, S., Wu, Y., He, Y., Wang, Y., Xiao, J., Li, L., et al. (2015). RopGEF2 is involved in ABA-suppression of seed germination and post-germination growth of Arabidopsis. Plant J. 84, 886–899. doi: 10.1111/tpj.13046
Zhou, M., Chen, H., Wei, D., Ma, H., and Lin, J. (2017). Arabidopsis CBF3 and DELLAs positively regulate each other in response to low temperature. Sci. Rep. 7:39819. doi: 10.1038/srep39819
Zhou, M., Xu, M., Wu, L., Shen, C., Ma, H., and Lin, J. (2014). CbCBF from Capsella bursa-pastoris enhances cold tolerance and restrains growth in Nicotiana tabacum by antagonizing with gibberellin and affecting cell cycle signaling. Plant Mol. Biol. 85, 259–275. doi: 10.1007/s11103-014-0181-1
Keywords: cotton, transcription factor, AmCBF1, GhPP2C, plant architecture and plant height
Citation: Lu J, Wang L, Zhang Q, Ma C, Su X, Cheng H and Guo H (2022) AmCBF1 Transcription Factor Regulates Plant Architecture by Repressing GhPP2C1 or GhPP2C2 in Gossypium hirsutum. Front. Plant Sci. 13:914206. doi: 10.3389/fpls.2022.914206
Edited by:
Weicong Qi, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaReviewed by:
Xiaoyang Ge, Cotton Institute of the Chinese Academy of Agricultural Sciences, ChinaZhikun Li, Hebei Agricultural University, China
Copyright © 2022 Lu, Wang, Zhang, Ma, Su, Cheng and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiming Guo, Z3VvaHVpbWluZ0BjYWFzLmNu
 Junchao Lu1,2
Junchao Lu1,2 Lihua Wang
Lihua Wang Xiaofeng Su
Xiaofeng Su Hongmei Cheng
Hongmei Cheng Huiming Guo
Huiming Guo