- 1CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, CAS Center for Excellence in Molecular Plant Sciences, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3School of Life Sciences, University of Science and Technology of China, Hefei, China
- 4Soybean Research Institute, Jilin Academy of Agricultural Sciences, Changchun, China
- 5Yunnan Key Laboratory of Plant Reproductive Adaptation and Evolutionary Ecology and Institute of Biodiversity, School of Ecology and Environmental Science, Yunnan University, Kunming, China
Soybean is one of the most important legume crops that can provide the rich source of protein and oil for human beings and livestock. In the twenty-one century, the total production of soybean is seriously behind the needs of a growing world population. Cultivated soybean [Glycine max (L.) Merr.] was domesticated from wild soybean (G. soja Sieb. and Zucc.) with the significant morphology and organ size changes in China around 5,000 years ago, including twisted stems to erect stems, small seeds to large seeds. Then it was spread worldwide to become one of the most popular and important crops. The release of the reference soybean genome and omics data provides powerful tools for researchers and breeders to dissect the functional genes and apply the germplasm in their work. Here, we summarized the function genes related to yield traits and organ size in soybean, including stem growth habit, leaf size and shape, seed size and weight. In addition, we also summarized the selection of organ traits during soybean domestication. In the end, we also discussed the application of new technology including the gene editing on the basic research and breeding of soybean, and the challenges and research hotspots in the future.
Introduction
With the growth of the world food crisis, the plant as the main source of food is particularly important. To increase the crop yield and biomass, breeders used the artificial hybridization approach to select the later generation with significant morphology and organ size changes (Yin et al., 2020). The organ size is also an important parameter for describing organ morphology and characterizing key function genes. However, the molecular mechanism on regulation of plant organ size is also a fundamental question in the field of developmental biology (Yin et al., 2020). Stems, leaves and seeds are three types of important organs which determine the production of plants. All of them are also important agronomic traits during domestication, and cultivars in the same region will be domesticated with similar phenotypes to increase yields or adapt to the environment (Lenser and Theißen, 2013). Increasing the size of plant organs, especially seeds, is one of the most important ways to improve crop yields.
Soybean [Glycine max (L.) Merr.] is a major crop for protein and oil production, and is widely grown worldwide. It can provide the rich source of protein and oil for human being and livestock that accounts for approximately 56% of global oilseed production (Zhou et al., 2015; Lu et al., 2016). Recently research showed that global crop production must be doubled in 2050 to meet the growing demands (Foley et al., 2011; Tilman et al., 2011). However, the soybean production is seriously behind the demand of the growing world population, which just has only 1.3% rate of increase per year (Ray et al., 2013). Therefore, increasing yield is an important part of soybean breeding. With the release of soybean reference genome, rapid progresses on functional genes of soybean have been made in the past decade (Zhang et al., 2021). Here, we reviewed functional genes related to yield traits and organ size in soybean, including stem growth habit, leaf size and shape, seed size and weight. Furthermore, we focused on the evolution of three important yield-related traits in soybean during the long-term domestication. In addition, we also discussed the application of new technology including the gene editing on the basic research and breeding of soybean, and the challenges and research hotspots in the future.
Stem Growth Habit
Stem growth habit can affect plant height and node number, thus determines grain yield. However, the increase of plant height will incur costs in the construction and maintenance of the stem (Falster and Westoby, 2003). Soybean stem is determined by plant height (PH), node number (NN) and internode length (IL). PH was significantly and positively correlated with NN and IL (Chang et al., 2018; Kou et al., 2021). Both PH and NN affect seed yield by influence on lodging and adaptability of soybean (Assefa et al., 2019). The structure of soybean stems can be divided into three main categories, usually referred to as determinate, semi-determinate and indeterminate (Bernard, 1972). Dt1 and Dt2 are two major loci which control the stem growth habit of soybean (Bernard, 1972; Heatherly and Smith, 2004). In the Dt1Dt1 genetic background, the Dt2Dt2 genotypes produce semi-determinate phenotypes, but dt2dt2 genotypes produce indeterminate phenotypes. In the dt1dt1 genetic background, the phenotype is determinate. These results suggest that the dt1 allele has an epistatic effect on the expression of the Dt2/dt2 locus (Liu B. et al., 2010; Ping et al., 2014). Dt1 encodes the GmTFL1b protein which is an ortholog of Arabidopsis TERMINAL FLOWER1. PH and IL in the dt1 mutants were severely reduced under long-day conditions, but there were not obvious variations under short-day conditions (Liu B. et al., 2010). Dt2 is a gain-of-function of MADS-domain factor which directly binds to the promoter region of Dt1 to inhibit its transcription (Ping et al., 2014; Liu et al., 2016). SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) encodes a MIKCc-type MADS-box transcription factor which is involved in regulating the flowering time (Lee and Lee, 2010). Loss of function of GmSOC1 in soybean exhibited delaying the flowering time and increased the PH under both short-day and long-day conditions (Kou et al., 2022). There are two SOC1 homologs present in soybean (SOC1a and SOC1b). The GmSOC1a and GmSOC1b complex interacts with Dt2 to repress the transcription of Dt1 through binding to the Dt1 promoter, indicating the role of SOC1 in soybean stem growth habit (Figure 1; Liu et al., 2016; Kou et al., 2022). Moreover, Phytochromes E3 and E4 promote PH and NN by inducing Dt1 transcription indirectly (Figure 1; Xu et al., 2013).
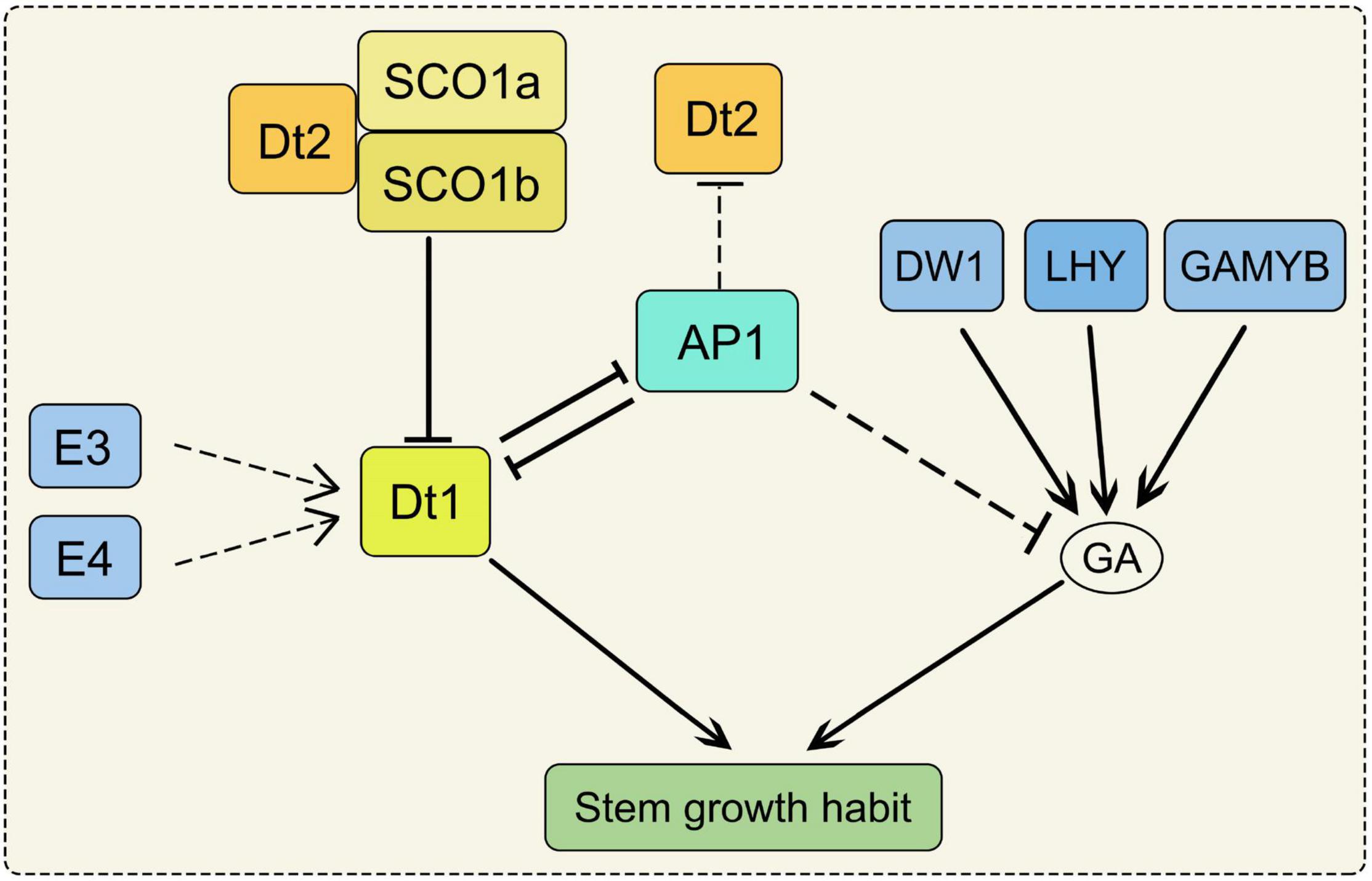
Figure 1. The regulation pathway of stem growth habit in soybean. SOC1a and SOC1b complex interacts with Dt2 to repress the transcription of Dt1. AP1 can inhibit Dt2 expression. Dt1 and AP1 form a suppressive regulatory feedback loop. AP1 may negatively regulate the expression of the GA metabolic pathway-related genes. Phytochromes E3 and E4 induce Dt1 transcription indirectly. Both of the SOC1, AP1, E3, E4 and Dt2 impact on the stem growth habit through Dt1 in soybean; DW1, LHY, GAMYB could regulate the stem growth habit through GA biosynthetic pathway. SOC1, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1; AP1, APETALA1; E3 and E4, phytochromes E3 and E4; DW1, a dwarf gene encodes an ent-kaurene synthase; LHY, LATE ELONGATED HYPOCOTYL; GA, gibberellin.
APETALA1 (AP1) encodes a member of MADS-box transcription factors family, and it belongs to the class A gene in the ABC model for floral organ development (Chen et al., 2020). Under short-day conditions, NN and IL were increased in the gmap1 quadruple mutant, resulting in the increased plant height (Chen et al., 2020). Dt1 and AP1 form a suppressive regulatory feedback loop, and AP1 can inhibit Dt2 expression (Yue et al., 2021). Moreover, AP1 may negatively regulate the expression of the GA metabolic pathway-related genes (Chen et al., 2020). As a member of FLOWERING LOCUS T (FT) family, GmFT5a may repress the expression of Dt1 by inducing GmAP1 transcription (Takeshima et al., 2019). Overexpression of GmFT5a in wild-type soybean reduced PH, and the same phenotype was obtained by overexpression GmFT5a in DT1/dt2 soybean, implying that GmFT5a can affect stem growth independently on the Dt2 (Takeshima et al., 2019). The dwarf gene GmDW1 encodes an ent-kaurene synthase (KS) which regulates the development of plant height in soybean. The gmdw1 is a gibberellin (GA) biosynthesis-deficient mutant, and the dwarf phenotype is due to the longitudinally decreased cell length (Figure 1; Li et al., 2018). As a MYB transcription factor, LATE ELONGATED HYPOCOTYL (GmLHY) also impacts PH through the GA biosynthetic pathway. The quadruple mutant of GmLHY causes dwarf plants due to defective GA biosynthesis (Figure 1; Cheng et al., 2019; Lu et al., 2020). As a R2R3 MYB transcription factor, overexpression of GmGAMYB increases PH in transgenic soybean by up-regulating the expression of GmGA20ox (Figure 1; Yang X. et al., 2021). However, another MYB transcription factor showed an opposite function, overexpression of GmMYB14 reduced PH and IL in soybean (Chen et al., 2021). Moreover, a small ubiquitin-related modifier E3 ligase GmSIZ1 positively regulates the vegetative growth of soybean. GmSIZ1-RNAi plants showed a phenotype of reduced leaf size and PH (Cai et al., 2017).
Leaf Shape and Size
As the energy factory of plants, leaves absorb light energy through photosynthesis and convert it into bioenergy. Because photosynthesis mainly occurs in leaves, the size and shape of leaves have strong effects on the efficiency of photosynthesis, determining the final yield (Gonzalez et al., 2012). Leaves arise at the flank of the shoot apical meristem (SAM), and this process is regulated by multiple gene regulation networks (Efroni et al., 2010). In cultivated soybean, there is a classic phenomenon that narrow leaf cultivars tend to have more seeds per pod than broad leaf cultivars (Jeong et al., 2011). The narrow leaf phenotype is a single-gene recessive inheritance controlled by the Ln gene (Dinkins et al., 2002). Ln is a homologous gene of Arabidopsis JAGGED (JAG), which regulates lateral organ development and the number of seeds per pod (NSPP) (Jeong et al., 2013). Interestingly, ln widely exists in the high latitude soybean varieties, but it does not exist in the low latitude soybean varieties and is hardly found in wild soybean (Cai et al., 2021). GmJAG in low-latitude variety Huachun 6 was edited by CRISPR/Cas9-based genome-editing technology, the offspring showed the same phenotype as that of ln mutant which just exists in the high latitude soybean varieties. Furthermore, the yield of gmjag was higher than Huachun 6 (Cai et al., 2021). Another gene that can regulate the narrow-leaf phenotype of soybean is CTP (Chicken Toes-Like Leaf and Petalody Flower). CTP is a novel and critical pleiotropic regulator of leaf and flower development. The ctp mutant exhibits chicken toes-like leaf and petalody flower. As a novel gene, the function of CTP protein is unclear and valuable to explore in the future. Interestingly, the CTP homologous proteins are only found in the land plant, implying that the CTP gene could have played an important role during the evolution of land plants (Zhao et al., 2017). Moreover, overexpression of the flowering repressor gene E1 could result in small, curly unifoliolate leaves by directly repressing CINCINNATA (CIN)-like TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP) transcription factor genes (TCP14, TCP29) (Li Y. et al., 2021). GmKIX8-1 belongs to a family of KIX domain-containing protein, which regulates organ size in soybean. Loss of function of GmKIX8-1 in soybean leads to big leaf and seed by increased cell proliferation (Nguyen et al., 2021). YABBY genes play important roles in lateral organs such as leaves and floral organs. Overexpression of GmFILa, a soybean YABBY gene, causes curly and long-narrow leaves in Arabidopsis (Yang et al., 2019).
Seed Size and Weight
The seed is one of the most important organs in the plant because the size and number of seeds directly determine the final yield of crops. Seed size is critical to many aspects of evolution and influenced by several key transcription factors (Linkies et al., 2010). BIG SEEDS1 (BS1) encodes a member of group II of the TIFY family of transcription regulators, and it can affect both seed and leaf sizes in legume species (Ge et al., 2016). Down-regulation of soybean BS1 gene significantly increased the size and weight of seeds and leaves (Ge et al., 2016). SLB1/MIO1 encodes an Arabidopsis thaliana F-box protein SAP homologous in Medicago truncatula (Zhou et al., 2021). SLB1 interacts with BS1 to control organ size, and overexpression of SLB1 leads to the increased seed and leaf size in soybean (Yin et al., 2020). During the organogenesis of plant, both cell proliferation and cell growth together determine the size of the organ (Liu Z. et al., 2020). GmCIF1 is a cell wall invertase inhibitor in soybean, and GmCIF1-RNAi plant significantly improves seed weight and slightly increases seed size (Tang et al., 2017). GmFAD3, encoding an omega-3 fatty acid desaturase, can control the leaf and seed size in soybean. GmFAD3-silenced plant showed increased seed size and crinkled leaf, and it has been proved to increase seed yield without affecting seed protein or oil content in the greenhouse (Singh et al., 2011). Interestingly, overexpression of lesquerella (Physaria fendleri) FAD3 gene in soybean revealed that plant height, total seeds, and total seed weight were significantly increased (Yeom et al., 2020). These seem to imply that FAD3 is a key gene controlling seed size in soybean and its expression level needs to maintain a balance to control seed size. A phosphatase 2C-1 (PP2C-1) gene contributes to the increase in seed weight/size, and PP2C-1 may transduce the brassinosteroid (BR) signaling by promoting dephosphorylation of GmBZRs which is one of the key transcription factors in BR signaling. Moreover, GmBZR1 can promote seed weight/size in transgenic Arabidopsis for overexpression (Lu et al., 2017). Previous studies have revealed that SWEET proteins play important roles in sugar translocation to seeds, which in turn affects fruit set, and seed composition (Sosso et al., 2015). A knock-out GmSWEET10 (both GmSWEET10a and homolog GmSWEET10b) by CRISPR/Cas9 system, exhibited significantly decreased seed size. Inversely, in soybean lines with the overexpression of GmSWEET10, seeds are larger than Williams 82 control (Wang et al., 2020).
The cytochrome P450/CYP78A family plays an important role in regulating seed size (Adamski et al., 2009; Wang et al., 2015). Overexpression of GmCYP78A72 and GmCYP78A5 in soybean resulted in increased seed size and seed weight (Zhao et al., 2016; Du et al., 2017). Moreover, knock-out of GmCYP78A72 by CRISPR/Cas9 system does not reduce seed size, but simultaneous silencing of the GmCYP78A57, GmCYP78A70 and GmCYP78A72 genes reduced seed size. These results suggest that three GmCYP78A genes have redundant functions (Zhao et al., 2016). GmPDAT plays an important role in increasing seed size. Overexpression of GmPDAT results in increased seed size, while RNAi lines have the opposite effect (Liu J. Y. et al., 2020). GmNAP1 encodes a NCK-associated protein, and loss of function of GmNAP1 caused the reduced seed size and plant height in soybean (Campbell et al., 2016; Tang et al., 2020). A genome scan for loci involved in soybean domestication identified a semi-dominant locus, Seed Thickness 1 (ST1). ST1, encoding a UDP-D-glucuronate 4-epimerase, affects seed size through the pectin biosynthesis pathway. Loss of function of ST1 by CRISPR/Cas9-based genome-editing technology significantly decreased seed length, width and thickness in soybean (Li et al., 2022).
Organ Trait Selection During the Domestication Process of Soybean
Domesticated crops play important roles in human nutrition and agriculture. During the domestication of plants, wild populations respond to changing selection pressures and thus adapt to new cultivated ecological niches (Stitzer and Ross-Ibarra, 2018). Recently, some studies showed that cultivated soybeans [G. max (L.) Merr.] was domesticated from wild soybean (G. soja Sieb. and Zucc.) in China around 5,000 years ago, then it had been introduced to Korea and Japan. Cultivated soybean was introduced to the American continent between the 18th and 20th centuries (Zhou et al., 2015). Nowadays, soybean is widely grown around the world. Phylogenetic studies of whole-genome resequencing data demonstrate the monophyletic nature of domesticated soybean (Lam et al., 2010; Li et al., 2010; Zhou et al., 2015). However, there is a controversy about the domestication center of soybean, and it is mainly assumed that the domestication centers are the Yellow River of China (Li et al., 2010; Han et al., 2016; Wang et al., 2016). Furthermore, soybean populations are geographically structured, and some agronomic traits show geographic distribution patterns (Zhou et al., 2015). For example, soybean varieties from northern China tend to exhibit narrow leaf shapes, while varieties from southern China exhibit ovate leaf shapes (Zhou et al., 2015; Cai et al., 2021). The key regulatory gene responsible for this phenomenon may be Ln, which was not found in 383 re-sequenced accessions collected from low latitudes (Cai et al., 2021). Moreover, Dt1, a stem determinacy regulatory factor, showed a strong regional differentiation, as the frequency of mutant alleles gradually increased from northern to southern regions (Zhou et al., 2015).
During the soybean domestication process, the plant morphology has undergone tremendous changes. Wild soybean has much smaller seeds than cultivated soybean, and it also has procumbent and twining stems, breakable pods and impermeable seed coats (Chen and Nelson, 2004). These significant changes contribute to the growth adaptation and yield improvement of soybean. Therefore, some traits and their potential genes were selected and inherited during geographical differentiation or local breeding. Seed size as an important agronomic trait has been selected during crop domestication (Linkies et al., 2010; Ge et al., 2016). Compared to wild soybean, cultivated soybean seeds are longer, wider and thicker (Li et al., 2022). During the transition of soybean seeds from flat to round shape, ST1 as a key factor was strongly selected to increase seed size (Li et al., 2022). The SoyWRKY15a gene is differentially expressed in wild soybean and cultivated soybean, and this gene may be involved in the regulation of seed size and possibly in the domestication process of soybean (Gu et al., 2017). Recently, based on whole-genome resequencing data, GmSWEET10a was found to undergo a selection process during the soybean domestication (Wang et al., 2020). GmCYP78A10 has undergone significant selection during domestication. The GmCYP78A10a allele has been eliminated in cultivated soybean at the early stage of soybean breeding, while the GmCYP78A10b allele accumulated and became the dominant allele in cultivated soybean (Wang et al., 2015). However, some useful genetic loci may be lost during domestication in cultivated soybean. During the process of domestication, 70% of wild soybean genes were lost in cultivated soybean (Zhuang et al., 2022). Therefore, we can try to “pick up” the key function genes, which control good traits in wild soybean and lose in cultivated soybean, and apply them on molecular breeding to achieve the purpose of improving yield. Whole-genome sequencing analysis of wild and cultivated soybean uncovered 183 genomic rearrangements that affect important phenotypic traits such as flowering time, disease resistance, and stress tolerance. The genetic diversity of wild soybean could provide a resource for soybean breeding (Zhuang et al., 2022).
Concluding Remarks and Future Perspectives
Over the past 20 years, understanding of soybean organ development has grown, and the regulatory genes for many key traits have been identified in soybean (Table 1 and Figure 2), such as Dt1 and Dt2 for stem growth habit (Liu B. et al., 2010; Ping et al., 2014), Ln for narrow leaf (Jeong et al., 2013), and GmSWEET10 for seed size (Wang et al., 2020), etc. However, there are still many QTLs controlling organ development in soybean that have not been isolated and confirmed. In other species, the regulation mechanism of organ size has been studied in depth, but the research of functional genes in soybean has been fallen behind. The power of gene knock-out and RNAi-silence technologies has given a new way to study these functional genes in soybean. Targeted silencing of some possible key trait genes can explore their unknown functions in soybean. However, there are many redundant genes in soybean genome, so it is very difficult to study gene function by CRISPR/cas9 technology. Furthermore, Pan-genome sequencing and re-sequencing technologies can link genetic variation to candidate genes that determine important traits (Liu Y. et al., 2020).
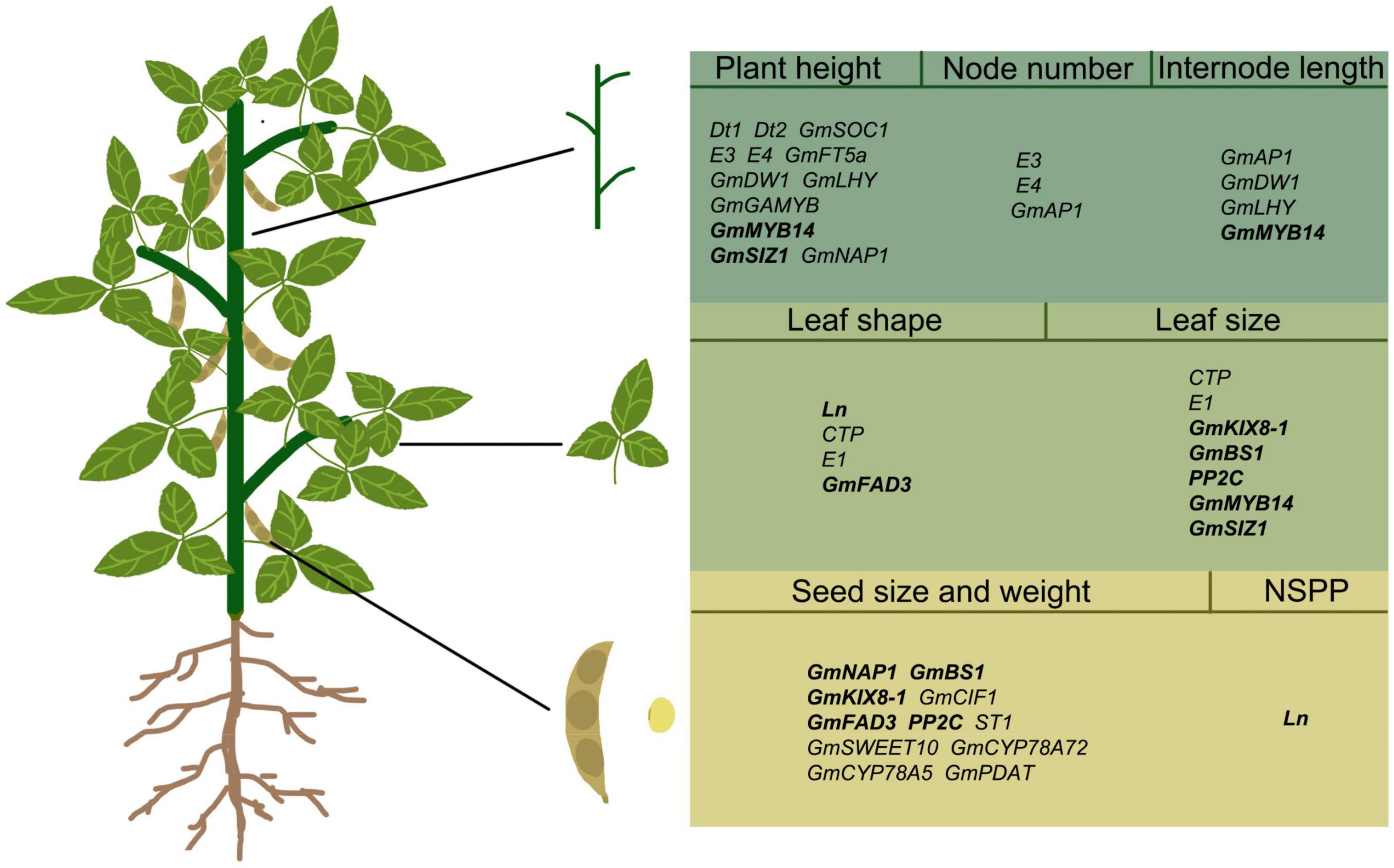
Figure 2. Regulatory genes in different organs of soybean. The words in bold refer to genes that simultaneously regulate the morphology of two soybean organs. Simultaneous regulation of stem and leaf: GmMYB14 and GmSIZ1; Simultaneous regulation of leaf and seed: Ln, GmKIX8-1, GmBS1, GmFAD3, PP2C; NSPP, number of seeds per pod.
In the future, soybean breeding is still an important but challenging work. Seed size is a key trait in breeding high-yielding soybeans, but there is a balance between seed size and yield. However, GmFAD3-silenced soybean showed both the larger seed and increased yield phenotype without affecting fat and protein content (Singh et al., 2011). It predicts that GmFAD3 may be a key factor in improving soybean yield. In addition, stem and leaf shape are also another key factors in improving soybean yield. Moderately increased planting densities have boosted crop yield. The vertical plant structure benefits from dense planting (Tian et al., 2019). The good morphology combined with proper planting density can effectively increase yield (Liu S. et al., 2020). Therefore, stem growth habit has been the focus of soybean domestication, and there are many QTLs for plant height waiting for our further identification (Yang Q. et al., 2021). Furthermore, resequencing data of wild and cultivated soybean revealed many QTLs related to agronomic traits, which provided the powerful database for the research of functional genes in the future (Lam et al., 2010; Zhou et al., 2015). The orderly integration of different superior traits with molecular tools will be one of the important challenges for soybean breeding in the future.
Author Contributions
XZ, DW, YM, YZ, LZ, CZ, and JC drafted the manuscript. JC conceived to the article and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
We acknowledge the National Natural Science Foundation of China (U2102222 and 32070204), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB27030106), the Yunnan Fundamental Research Projects (202101AW070004), and Youth Innovation Promotion Association CAS (2021395) for providing support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adamski, N. M., Anastasiou, E., Eriksson, S., O’Neill, C. M., and Lenhard, M. (2009). Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 20115–20120. doi: 10.1073/pnas.0907024106
Assefa, T., Otyama, P. I., Brown, A. V., Kalberer, S. R., Kulkarni, R. S., and Cannon, S. B. (2019). Genome-wide associations and epistatic interactions for internode number, plant height, seed weight and seed yield in soybean. BMC Genomics 20:527. doi: 10.1186/s12864-019-5907-7
Bernard, R. L. (1972). Two genes affecting stem termination in soybeans 1. Crop Sci. 12, 235–239. doi: 10.2135/cropsci1972.0011183x001200020028x
Cai, B., Kong, X., Zhong, C., Sun, S., Zhou, X. F., Jin, Y. H., et al. (2017). SUMO E3 ligases GmSIZ1a and GmSIZ1b regulate vegetative growth in soybean. J. Integr. Plant Biol. 59, 2–14. doi: 10.1111/jipb.12504
Cai, Z., Xian, P., Cheng, Y., Ma, Q., Lian, T., Nian, H., et al. (2021). CRISPR/Cas9-mediated gene editing of GmJAGGED1 increased yield in the low-latitude soybean variety Huachun 6. Plant Biotechnol. J. 19, 1898–1900. doi: 10.1111/pbi.13673
Campbell, B. W., Hofstad, A. N., Sreekanta, S., Fu, F., Kono, T. J. Y., O’Rourke, J. A., et al. (2016). Fast neutron-induced structural rearrangements at a soybean NAP1 locus result in gnarled trichomes. Theor. Appl. Genet. 129, 1725–1738. doi: 10.1007/s00122-016-2735-x
Chang, F., Guo, C., Sun, F., Zhang, J., Wang, Z., Kong, J., et al. (2018). Genome-wide association studies for dynamic plant height and number of nodes on the main stem in summer sowing soybeans. Front. Plant Sci. 9:1184. doi: 10.3389/fpls.2018.01184
Chen, L., Nan, H., Kong, L., Yue, L., Yang, H., Zhao, Q., et al. (2020). Soybean AP1 homologs control flowering time and plant height. J. Integr. Plant Biol. 62, 1868–1879. doi: 10.1111/jipb.12988
Chen, L., Yang, H., Fang, Y., Guo, W., Chen, H., Zhang, X., et al. (2021). Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 19, 702–716. doi: 10.1111/pbi.13496
Chen, Y., and Nelson, R. L. (2004). Genetic variation and relationships among cultivated, wild, and semiwild soybean. Crop Sci. 44, 316–325. doi: 10.2135/cropsci2004.3160
Cheng, Q., Dong, L., Su, T., Li, T., Gan, Z., Nan, H., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol. 19:562. doi: 10.1186/s12870-019-2145-8
Dinkins, R. D., Keim, K. R., Farno, L., and Edwards, L. H. (2002). Expression of the narrow leaflet gene for yield and agronomic traits in soybean. J. Hered. 93, 346–351. doi: 10.1093/jhered/93.5.346
Du, J., Wang, S., He, C., Zhou, B., Ruan, Y. L., and Shou, H. (2017). Identifcation of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis. J. Exp. Bot. 68, 1955–1972. doi: 10.1093/jxb/erw460
Efroni, I., Eshed, Y., and Lifschitz, E. (2010). Morphogenesis of simple and compound leaves: a critical review. Plant Cell 22, 1019–1032. doi: 10.1105/tpc.109.073601
Falster, D. S., and Westoby, M. (2003). Plant height and evolutionary games. Trends Ecol. Evol. 18, 337–343. doi: 10.1016/S0169-5347(03)00061-2
Foley, J. A., Ramankutty, N., Brauman, K. A., Cassidy, E. S., Gerber, J. S., Johnston, M., et al. (2011). Solutions for a cultivated planet. Nature 478, 337–342. doi: 10.1038/nature10452
Ge, L., Yu, J., Wang, H., Luth, D., Bai, G., Wang, K., et al. (2016). Increasing seed size and quality by manipulating BIG SEEDS1 in legume species. Proc. Natl. Acad. Sci. U.S.A. 113, 12414–12419. doi: 10.1073/pnas.1611763113
Gonzalez, N., Vanhaeren, H., and Inzé, D. (2012). Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 17, 332–340. doi: 10.1016/j.tplants.2012.02.003
Gu, Y., Li, W., Jiang, H., Wang, Y., Gao, H., Liu, M., et al. (2017). Differential expression of a WRKY gene between wild and cultivated soybeans correlates to seed size. J. Exp. Bot. 68, 2717–2729. doi: 10.1093/jxb/erx147
Han, Y., Zhao, X., Liu, D., Li, Y., Lightfoot, D. A., Yang, Z., et al. (2016). Domestication footprints anchor genomic regions of agronomic importance in soybeans. New Phytol. 209, 871–884. doi: 10.1111/nph.13626
Heatherly, L. G., and Smith, J. R. (2004). Effect of soybean stem growth habit on height and node number after beginning bloom in the midsouthern USA. Crop Sci. 44, 1855–1858. doi: 10.2135/cropsci2004.1855
Jeong, N., Moon, J. K., Kim, H. S., Kim, C. G., and Jeong, S. C. (2011). Fine genetic mapping of the genomic region controlling leaflet shape and number of seeds per pod in the soybean. Theor. Appl. Genet. 122, 865–874. doi: 10.1007/s00122-010-1492-5
Jeong, N., Suh, S. J., Kim, M. H., Lee, S., Moon, J. K., Kim, H. S., et al. (2013). Ln is a key regulator of leaflet shape and number of seeds per pod in soybean. Plant Cell 24, 4807–4818. doi: 10.1105/tpc.112.104968
Kou, K., Su, T., Wang, Y., Yang, H., Du, H., He, M., et al. (2021). Natural variation of the Dt2 promoter controls plant height and node number in semi-determinant soybean. Mol. Breed 41:40. doi: 10.1007/s11032-021-01235-y
Kou, K., Yang, H., Li, H., Fang, C., Chen, L., Yue, L., et al. (2022). A functionally divergent SOC1 homolog improves soybean yield and latitudinal adaptation. Curr. Biol. 32, 1728-1742.e6. doi: 10.1016/j.cub.2022.02.046
Lam, H. M., Xu, X., Liu, X., Chen, W., Yang, G., Wong, F. L., et al. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42, 1053–1059. doi: 10.1038/ng.715
Lee, J., and Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098
Lenser, T., and Theißen, G. (2013). Molecular mechanisms involved in convergent crop domestication. Trends Plant Sci. 18, 704–714. doi: 10.1016/j.tplants.2013.08.007
Li, J., Zhang, Y., Ma, R., Huang, W., Hou, J., Fang, C., et al. (2022). Identification of ST1 reveals a selection involving hitchhiking of seed morphology and oil content during soybean domestication. Plant Biotechnol. J 20, 1110–1121. doi: 10.1111/pbi.13791
Li, X., Fang, C., Yang, Y., Lv, T., Su, T., Chen, L., et al. (2021). Overcoming the genetic compensation response of soybean florigens to improve adaptation and yield at low latitudes. Curr. Biol 31, 3755–3767.e4. doi: 10.1016/j.cub.2021.06.037
Li, Y., Hou, Z., Li, W., Li, H., Lu, S., Gan, Z., et al. (2021). The legume-specific transcription factor E1 controls leaf morphology in soybean. BMC Plant Biol. 21:531. doi: 10.1186/s12870-021-03301-1
Li, Y. H., Li, W., Zhang, C., Yang, L., Chang, R. Z., Gaut, B. S., et al. (2010). Genetic diversity in domesticated soybean (Glycine max) and its wild progenitor (Glycine soja) for simple sequence repeat and single-nucleotide polymorphism loci. New Phytol. 188, 242–253. doi: 10.1111/j.1469-8137.2010.03344.x
Li, Z. F., Guo, Y., Ou, L., Hong, H., Wang, J., Liu, Z. X., et al. (2018). Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis. Theor. Appl. Genet 131, 1001–1016. doi: 10.1007/s00122-017-3044-8
Linkies, A., Graeber, K., Knight, C., and Leubner-Metzger, G. (2010). The evolution of seeds. New Phytol. 186, 817–831. doi: 10.1111/j.1469-8137.2010.03249.x
Liu, B., Watanabe, S., Uchiyama, T., Kong, F., Kanazawa, A., Xia, Z., et al. (2010). The soybean stem growth habit gene Dt1 is an ortholog of arabidopsis TERMINAL FLOWER1. Plant Physiol. 153, 198–210. doi: 10.1104/pp.109.150607
Liu, J. Y., Zhang, Y. W., Han, X., Zuo, J. F., Zhang, Z., Shang, H., et al. (2020). An evolutionary population structure model reveals pleiotropic effects of GmPDAT for traits related to seed size and oil content in soybean. J. Exp. Bot. 71, 6988–7002. doi: 10.1093/jxb/eraa426
Liu, S., Zhang, M., Feng, F., and Tian, Z. (2020). Toward a “green revolution” for Soybean. Mol. Plant 13, 688–697. doi: 10.1016/j.molp.2020.03.002
Liu, Y., Du, H., Li, P., Shen, Y., Peng, H., Liu, S., et al. (2020). Pan-genome of wild and cultivated soybeans. Cell 182, 162–176.e13. doi: 10.1016/j.cell.2020.05.023
Liu, Y., Zhang, D., Ping, J., Li, S., Chen, Z., and Ma, J. (2016). Innovation of a regulatory mechanism modulating semi-determinate stem growth through artificial selection in soybean. PLoS Genet. 12:e1005818. doi: 10.1371/journal.pgen.1005818
Liu, Z., Li, N., Zhang, Y., and Li, Y. (2020). Transcriptional repression of GIF1 by the KIX-PPD-MYC repressor complex controls seed size in Arabidopsis. Nat. Commun 11:1846. doi: 10.1038/s41467-020-15603-3
Lu, S., Dong, L., Fang, C., Liu, S., Kong, L., Cheng, Q., et al. (2020). Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52, 428–436. doi: 10.1038/s41588-020-0604-7
Lu, X., Li, Q. T., Xiong, Q., Li, W., Bi, Y. D., Lai, Y. C., et al. (2016). The transcriptomic signature of developing soybean seeds reveals the genetic basis of seed trait adaptation during domestication. Plant J. 86, 530–544. doi: 10.1111/tpj.13181
Lu, X., Xiong, Q., Cheng, T., Li, Q. T., Liu, X. L., Bi, Y. D., et al. (2017). A PP2C-1 allele underlying a quantitative trait locus enhances soybean 100-seed weight. Mol. Plant 10, 670–684. doi: 10.1016/j.molp.2017.03.006
Nguyen, C. X., Paddock, K. J., Zhang, Z., and Stacey, M. G. (2021). GmKIX8-1 regulates organ size in soybean and is the causative gene for the major seed weight QTL qSw17-1. New Phytol. 229, 920–934. doi: 10.1111/nph.16928
Ping, J., Liu, Y., Sun, L., Zhao, M., Li, Y., She, M., et al. (2014). Dt2 is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell 26, 2831–2842. doi: 10.1105/tpc.114.126938
Ray, D. K., Mueller, N. D., West, P. C., and Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PLoS One 8:e66428. doi: 10.1371/journal.pone.0066428
Singh, A. K., Fu, D. Q., El-Habbak, M., Navarre, D., Ghabrial, S., and Kachroo, A. (2011). Silencing genes encoding omega-3 fatty acid desaturase alters seed size and accumulation of Bean pod mottle virus in soybean. Mol. Plant Microbe Interact. 24, 506–515. doi: 10.1094/MPMI-09-10-0201
Sosso, D., Luo, D., Li, Q. B., Sasse, J., Yang, J., Gendrot, G., et al. (2015). Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 47, 1489–1493. doi: 10.1038/ng.3422
Stitzer, M. C., and Ross-Ibarra, J. (2018). Maize domestication and gene interaction. New Phytol. 220, 395–408. doi: 10.1111/nph.15350
Takeshima, R., Nan, H., Harigai, K., Dong, L., Zhu, J., Lu, S., et al. (2019). Functional divergence between soybean FLOWERING LOCUS T orthologues FT2a and FT5a in post-flowering stem growth. J. Exp. Bot. 70, 3941–3953. doi: 10.1093/jxb/erz199
Tang, K., Yang, S., Feng, X., Wu, T., Leng, J., Zhou, H., et al. (2020). GmNAP1 is essential for trichome and leaf epidermal cell development in soybean. Plant Mol. Biol. 103, 609–621. doi: 10.1007/s11103-020-01013-y
Tang, X., Su, T., Han, M., Wei, L., Wang, W., Yu, Z., et al. (2017). Suppression of extracellular invertase inhibitor gene expression improves seed weight in soybean (Glycine max). J. Exp. Bot. 68, 469–482. doi: 10.1093/jxb/erw425
Tian, J., Wang, C., Xia, J., Wu, L., Xu, G., Wu, W., et al. (2019). Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365, 658–664. doi: 10.1126/science.aax5482
Tilman, D., Balzer, C., Hill, J., and Befort, B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U.S.A. 108, 20260–20264. doi: 10.1073/pnas.1116437108
Wang, J., Chu, S., Zhang, H., Zhu, Y., Cheng, H., and Yu, D. (2016). Development and application of a novel genome-wide SNP array reveals domestication history in soybean. Sci. Rep. 6:20728. doi: 10.1038/srep20728
Wang, S., Liu, S., Wang, J., Yokosho, K., Zhou, B., Yu, Y. C., et al. (2020). Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl. Sci. Rev. 7, 1776–1786. doi: 10.1093/nsr/nwaa110
Wang, X., Li, Y., Zhang, H., Sun, G., Zhang, W., and Qiu, L. (2015). Evolution and association analysis of GmCYP78A10 gene with seed size/weight and pod number in soybean. Mol. Biol. Rep. 42, 489–496. doi: 10.1007/s11033-014-3792-3
Xu, M., Xu, Z., Liu, B., Kong, F., Tsubokura, Y., Watanabe, S., et al. (2013). Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA-regulated post-flowering responses of soybean. BMC Plant Biol. 13:91. doi: 10.1186/1471-2229-13-91
Yang, H., Shi, G., Li, X., Hu, D., Cui, Y., Hou, J., et al. (2019). Overexpression of a soybean YABBY gene, GmFILa, causes leaf curling in Arabidopsis thaliana. BMC Plant Biol. 19:234. doi: 10.1186/s12870-019-1810-2
Yang, Q., Lin, G., Lv, H., Wang, C., Yang, Y., and Liao, H. (2021). Environmental and genetic regulation of plant height in soybean. BMC Plant Biol. 21:63. doi: 10.1186/s12870-021-02836-7
Yang, X., Li, X., Shan, J., Li, Y., Zhang, Y., Wang, Y., et al. (2021). Overexpression of GmGAMYB accelerates the transition to flowering and increases plant height in soybean. Front. Plant Sci. 12:667242. doi: 10.3389/fpls.2021.667242
Yeom, W. W., Kim, H. J., Lee, K. R., Cho, H. S., Kim, J. Y., Jung, H. W., et al. (2020). increased production of α-linolenic acid in soybean seeds by overexpression of Lesquerella FAD3-1. Front. Plant Sci. 10:1812. doi: 10.3389/fpls.2019.01812
Yin, P., Ma, Q., Wang, H., Feng, D., Wang, X., Pei, Y., et al. (2020). SMALL LEAF AND BUSHY1 controls organ size and lateral branching by modulating the stability of BIG SEEDS1 in Medicago truncatula. New Phytol. 226, 1399–1412. doi: 10.1111/nph.16449
Yue, L., Li, X., Fang, C., Chen, L., Yang, H., Yang, J., et al. (2021). FT5a interferes with the Dt1-AP1 feedback loop to control flowering time and shoot determinacy in soybean. J. Integr. Plant Biol. 63, 1004–1020. doi: 10.1111/jipb.13070
Zhang, M., Liu, S., Wang, Z., Yuan, Y., Zhang, Z., Liang, Q., et al. (2021). Progress in soybean functional genomics over the past decade. Plant Biotechnol. J. doi: 10.1111/pbi.13682
Zhao, B., Dai, A., Wei, H., Yang, S., Wang, B., Jiang, N., et al. (2016). Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 90, 33–47. doi: 10.1007/s11103-015-0392-0
Zhao, J., Chen, L., Zhao, T., and Gai, J. (2017). Chicken toes-like leaf and petalody flower (CTP) is a novel regulator that controls leaf and flower development in soybean. J. Exp. Bot. 68, 5565–5581. doi: 10.1093/jxb/erx358
Zhou, S., Yang, T., Mao, Y., Liu, Y., Guo, S., Wang, R., et al. (2021). The F-box protein MIO1/SLB1 regulates organ size and leaf movement in Medicago truncatula. J. Exp. Bot. 72, 2995–3011. doi: 10.1093/jxb/erab033
Zhou, Z., Jiang, Y., Wang, Z., Gou, Z., Lyu, J., Li, W., et al. (2015). Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 33, 408–414. doi: 10.1038/nbt.3096
Keywords: soybean, stem growth habit, leaf size and shape, seed size and weight, domestication
Citation: Zhou X, Wang D, Mao Y, Zhou Y, Zhao L, Zhang C, Liu Y and Chen J (2022) The Organ Size and Morphological Change During the Domestication Process of Soybean. Front. Plant Sci. 13:913238. doi: 10.3389/fpls.2022.913238
Received: 05 April 2022; Accepted: 24 May 2022;
Published: 09 June 2022.
Edited by:
Shengjun Li, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), ChinaReviewed by:
Chao Fang, Michigan State University, United StatesBianyun Yu, National Research Council Canada (NRC-CNRC), Canada
Copyright © 2022 Zhou, Wang, Mao, Zhou, Zhao, Zhang, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianghua Chen, amhjaGVuQHh0YmcuYWMuY24=
 Xuan Zhou
Xuan Zhou Dongfa Wang1,3
Dongfa Wang1,3 Yawen Mao
Yawen Mao Chunbao Zhang
Chunbao Zhang Jianghua Chen
Jianghua Chen