- 1College of Life Sciences, Yantai University, Yantai, China
- 2Institute of Grain and Oil Crops, Yantai Academy of Agricultural Sciences, Yantai, China
- 3Dezhou Agricultural Technology Extension and Seed Industry Center, Dezhou, China
- 4Crop Research Institute, Shandong Academy of Agricultural Sciences, Jinan, China
Powdery mildew of wheat, caused by Blumeria graminis f. sp. tritici (Bgt), is a devastating disease that seriously reduces yield and quality worldwide. Utilization of plant resistance genes is an attractive and effective strategy for controlling this disease. Among the reported powdery mildew (Pm) resistance genes, Pm2 exhibits a diverse resistance spectrum among its multiple alleles. It has been widely used in China for resistance breeding for powdery mildew. To mine more Pm2 alleles and clarify their distribution, we screened 33 wheat cultivars/breeding lines carrying Pm2 alleles from 641 wheat genotypes using diagnostic and Pm2-linked markers. To further investigate the relationships within the Pm2 alleles, we compared their resistance spectra, polymorphism of marker alleles and gene sequences, and found that they have identical marker alleles and gene sequences but diverse resistance spectra. In addition, the diagnostic kompetitive allele-specific PCR (KASP) marker, YTU-KASP-Pm2, was developed and was shown to detect all the Pm2 alleles in the different genetic backgrounds. These findings provide valuable information for the distribution and rational use of Pm2 alleles, push forward their marker-assisted breeding (MAS), and hence improve the control of wheat powdery mildew.
Introduction
Common wheat (Triticum aestivum L.) is an important grain crop that provides 20% of the world’s food energy and 20% of its protein. Consequently, it is a major contributor to global food security (Hickey et al., 2019). However, wheat yield and quality are seriously affected by a variety of diseases, including powdery mildew caused by Blumeria graminis f. sp. tritici (Bgt; Costanzo and Bàrberi, 2014 Li et al., 2019). This disease can typically decrease wheat yield by 10%–15%, and up to 50% in serious cases (Morgounov et al., 2012; Xu et al., 2015). Several measures have been taken to control this disease, including the spraying of chemical agents and the use of resistance genes, with the latter being considered to be the most effective and the most environmentally acceptable (Zhang et al., 2019). However, to achieve control using resistance genes, both abundant powdery mildew resistance (Pm) genes/alleles and diversified donors are the perquisites for developing elite cultivars with high and/or durable resistance to powdery mildew (Laroche et al., 2019).
To date, more than 100 Pm genes/alleles (Pm1–Pm68, Pm8 is allelic to Pm17, Pm18 = Pm1c, Pm22 = Pm1e, Pm23 = Pm4c, and Pm31 = Pm21) have been identified at 63 loci from common wheat and its relatives, showing that there are abundant genetic resources for controlling wheat powdery mildew (McIntosh et al., 2020; He et al., 2021). However, most of these documented Pm genes/alleles cannot be directly used in wheat breeding due to observable linkage drag, adverse pleiotropism, and competition lag. For example, when the gene Pm16, derived from Triticum dicoccoides and which exhibits broad spectrum resistance to different Bgt isolates, was introgressed into wheat backgrounds, it caused up to 15% yield loss during production (Summers and Brown, 2013). Similar yield reductions have been shown for other Pm genes derived from wheat relatives and landraces (Xu et al., 2015).
Even for the genes with no linkage drag, evolving Bgt variants are another challenge (An et al., 2019). For example, Pm8 has been a widely used gene in breeding for resistance to powdery mildew. It is also an example of an extremely successful introgression of an elite alien gene from rye into common wheat (Kaur et al., 2017). Unfortunately, Pm8 has successively lost its resistance due to the co-evolution of pathogen virulence with host resistance (Chai et al., 2005). Clearly, the breeding value of the Pm genes depends not only on their effectiveness at disease control, but also on the agronomic performance of their donor (Ma et al., 2015a,b, 2018). Identification and utilization of Pm genes with no linkage drag offer an attractive prospect for the rapid improvement of resistance to powdery mildew in wheat.
Conventional breeding has made enormous contributions to resistance breeding in the past (Li et al., 2019, 2022), but marker-assisted selection (MAS) is currently considered to be the most effective way to accurately transfer targeted genes/loci (Li et al., 2019). To improve powdery mildew resistance, MAS has been used with more than 30 Pm genes in wheat breeding (Shah et al., 2018). In MAS, the key point is the development and screening of molecular markers that can efficiently trace the targeted genes. Various kinds of markers have been used in MAS, such as expressed sequence tags (EST), sequence-tagged sites (STS), and simple sequence repeat (SSR) markers based on gel electrophoresis detection (Li et al., 2019). With the development of high throughput detection platforms, kompetitive allele-specific PCR (KASP) markers have begun to be used in MAS (Makhoul et al., 2020). KASP was developed based on SNPs in alleles, and enables high-throughput, gel-free screening of markers.
The Pm2 gene, derived from Aegilops tauschii, has been used in resistance breeding for powdery mildew worldwide (Pugsley and Carter, 1953; Li et al., 2011). In our lab, we have identified a series of Pm2 alleles from different genotypes, including various wheat cultivars/breeding lines, indicating that Pm2 is a promising Pm gene (Gao et al., 2022). Although several Pm2 alleles no longer exhibit resistance to some Bgt isolates, there are still other Pm2 alleles that confer high resistance to powdery mildew in specific genotypes (Ma et al., 2015a,b, 2018). To maximize the effectiveness of Pm2 in resistance breeding, in this study we aimed to: (1) mine more Pm2 alleles and survey their distribution in wheat cultivars/breeding lines; (2) evaluate the resistance of different Pm2 alleles for their rational utilization in different genetic backgrounds and wheat production regions; and (3) develop diagnostic KASP markers to accelerate the transfer of Pm2 alleles into breeding lines.
Materials and Methods
Plant Materials
Six hundred and thirty-nine Chinese wheat cultivars/breeding lines and two wheat cultivars from New Zealand (Supplementary Table S1) were inoculated with Bgt isolate E09 for screening resistant genotypes. Twenty-two wheat cultivars/breeding lines (Shimai 22, Hanmai 13, Shixin 633, Taimai 1918, Tainong 18, Shannong 15,381, Taitianmai 118, Zhongxin 7503, Jimai 52, Jimai 61, Yannong 21, Youxuan 134, GY13029, Xinshiji 156, Shi 6609, Jinhe 13–205, Huixianhong, Mingxian 169, 12CA49, GY16022, GY16011, and 12CA49), which are susceptible to all the Bgt isolates tested were used as susceptible parents to cross with the resistant genotypes screened from the 641 wheat genotypes (Supplementary Table S1), to conduct F2 and F2:3 segregating populations for genetic analysis and molecular detection of their Pm genes. Susceptible cultivar Huixianhong was used as the susceptible control for phenotypic assessment. Ulka/8*Cc, which carries the known Pm2a gene (Sun et al., 2015a,b), was used as the resistant control.
Reactions to Different Bgt Isolates
At the seedling stage, the Bgt isolate E09, a dominant Bgt isolate in North China, was used to inoculate the 641 wheat cultivars/breeding line (Supplementary Table S1). Additionally, the Pm2 donors along with Ulka/8*Cc and Huixianhong were tested for their seedling reaction patterns to eight other Bgt isolates (A3, A10, A45, E15-1, E18, E20, E21, and E32) with different avirulence/virulence patterns and from different wheat production regions of China. The susceptible seedlings inoculated with an individual isolate were put in independent glass tubes to avoid cross infection. Five seeds of each genotype were sown in a 72-cell rectangular tray and put in an independent growth chamber to be infected with a Bgt isolate. When the seedlings had grown to the one-leaf stage, they were inoculated with fresh conidiospores previously cultivated on Huixianhong seedlings. At this time, the growth chambers were set at 100% humidity at 18°C for 24 h, after which the growing condition was set at 14 h light at 22°C and 10 h of darkness at 18°C. Inoculations were repeated twice in the following 2 days to ensure full infection. Infection types (ITs) were recorded when the spores were fully developed on the first leaves of Huixianhong seedlings based on the standard described by An et al. (2019), where ITs 0, 0; 1, 2, 3, and 4 are regarded as immune, hypersensitive, highly resistant, moderately resistant, moderately susceptible, and highly susceptible, respectively.
To determine the inheritance of powdery mildew resistance in the resistant genotypes, Bgt isolate E09 was selected to inoculate the resistant and susceptible parents and their F1, F2, and F2:3 progenies for genetic analysis. After phenotypic evaluation, the numbers of resistance and susceptible plants were counted, and then a goodness-of-fit assessment was performed to determine the resistant/susceptible ratio using a Chi-squared (χ2) test. The deviations of the observed phenotypic data from the theoretically expected segregation ratios were then evaluated using the SPSS 16.0 software (SPSS Inc., Chicago, United States) at p < 0.05.
Marker Analysis
Total genomic DNAs (gDNAs) of all the F2:3 families along with their parents were isolated after phenotypic evaluation using the TE-boiling method (He et al., 2017). For each population, equal amounts of gDNAs from 10 random homozygous resistant and 10 random homozygous susceptible F2:3 families were pooled to construct resistant and susceptible DNA bulks, respectively. The Pm2-linked marker Cfd81 (Ma et al., 2016) and the Pm2-diagnostic marker Pm2b-map-3 (Jin et al., 2021) were tested for polymorphisms between the resistant and susceptible parents and bulks. The polymorphic markers were genotyped on the corresponding F2:3 families. PCR amplifications and visualizations were as described by Ma et al. (2016).
Homology-Based Cloning of the Pm2 Alleles
Total RNA from each of the genotypes with Pm2 alleles was extracted using the Spectrum Plant Total RNA kit (Sigma-Aldrich, Shanghai, China) following the manufacturer’s recommendations. Then, they were quantified by measuring absorbance at the wavelengths of 260 and 280 nm using a Nano Drop 1000 spectrophotometer (Thermo Scientific, Shanghai, China). High quality RNA was treated by Promega DNase I and then used for cDNA synthesis using Invitrogen SuperScript-II reverse transcriptase following the manufacturer’s guidelines. Based on the report of the cloning of Pm2a (Sánchez-Martín et al., 2016), the primer pairs JS320 (Forward: 5′-3′: ACGATGATGTGAATCTTCCGTG) and JS305 (Reverse 5′-3′: AATGATAGCATGCATTTGGAG) were used to amplify the first exon of the Pm2 alleles identified in this study. Then, a nested PCR using the primer pairs JS314 (Forward: 5′-3′: TTTTCGCGGTATTGCTGGTG) and JS315 (Reverse 5′-3′: ACCTCCTGTCATCGGTTCAC) was performed to obtain the final sequence of the first exon. For amplifying the second and third predicted exons of the Pm2 alleles, primer pairs JS350 (Forward: 5′-3′: CCCTCCTCCTTGAAGAATCTGA) and JS313 (Reserve: 5′-3′: GCACAAACTCTACCCTGTTCC) were used. Finally, they were sequenced using Sanger sequencing and compared with that of the reported Pm2a (GenBank: LN999386.1; Sánchez-Martín et al., 2016).
Development of a Diagnostic KASP Marker for MAS
The sequences of the cloned Pm2 alleles were used to compare with the reference genome of Chinese Spring (v2.1, http://202.194.139.32/). Distinctive SNPs were identified after comparing the Pm2 sequences to the A, B, and D genomes of Chinese Spring, which does not carry Pm2 and is susceptible to powdery mildew. Sequences of 100 bp upstream and downstream of the distinctive SNPs were acquired and used for KASP development using both the Polymarker website1 and Premier 5 software.2 The amplification sequences of the primers were aligned once again with the reference genome of Chinese Spring in the Triticeae Multi-omics Center (http://202.194.139.32/) to ensure specificity of the sequences. The primers were then used to genotype all the F2:3 families carrying Pm2 alleles to confirm their polymorphisms. The diagnostic KASP marker was then used to genotype the breeding populations of Pm2 donors and susceptible cultivars. Combined with the phenotype against Bgt isolate E09, the diagnostic KASP marker was confirmed once again.
Genotyping using KASP primers was performed on a Bio-Rad CFX real-time PCR system (Bio-Rad Laboratories, Inc., CA, United States) with a final volume of 20 μl containing 6.00 μl of gDNA (~250 ng), 11.20 μl of 2 × KASP Master Mix (provided by LGC), 0.34 μl of primer mix (balanced mix of three pairs of primers for each marker), and 2.46 μl ddH2O. The amplification procedure was set as follows: 94°C for 15 min, followed by 10 touchdown cycles of 94°C for 20 s, 64°C to 58°C (decreasing 0.6°C per cycle), and 38 cycles of regular amplification (94°C for 20 s and 58°C for 60 s), and the final fluorescence was detected at 20°C using Bio-Rad CFX Manager 3.1 software (Bio-Rad Laboratories, Hercules, CA, United States).
Results
Screening of Resistant Genotypes and Inheritance Analysis
When inoculated with the Bgt isolate E09 at the seedling stage, 43 of the 641 accessions were resistant with ITs 0–2 (Table 1). The 43 resistant accessions were then crossed with susceptible cultivars to produce F1 hybrids, F2 populations, and F2:3 families (Table 1). The F1 plants of every hybridized combination all showed resistant phenotypes with ITs 0–2, suggesting dominant inheritance of the powdery mildew resistance in these accessions. Of their F2 populations, 40 fitted the expected ratios of 3:1 (resistant: susceptible individuals) for monogenic segregation using the same Bgt isolate, suggesting that a single dominant gene may be involved in the powdery mildew resistance of these accessions (Table 1). The 43 F2 populations were then transplanted in the field to produce F2:3 families to further confirm these results and validate the genotypes of the resistant F2 plants. The results showed that 37 F2:3 families fitted the expected ratios of 1:2:1 (Homozygous resistant: segregating: homozygous susceptible families; Table 1), confirming that a single dominant gene is involved in the powdery mildew resistance of these accessions. For three populations (Xinong 198 × Yannong 21, GQ17020 × Yannong 21, and GQ17014 × Yannong 21), although their F2 segregation ratio fitted the monogenic segregation of 3:1, their F2:3 segregation ratio did not fit the monogenic segregation of 1:2:1, suggesting that the powdery mildew resistance in these three accessions may not be controlled by a single dominant gene.
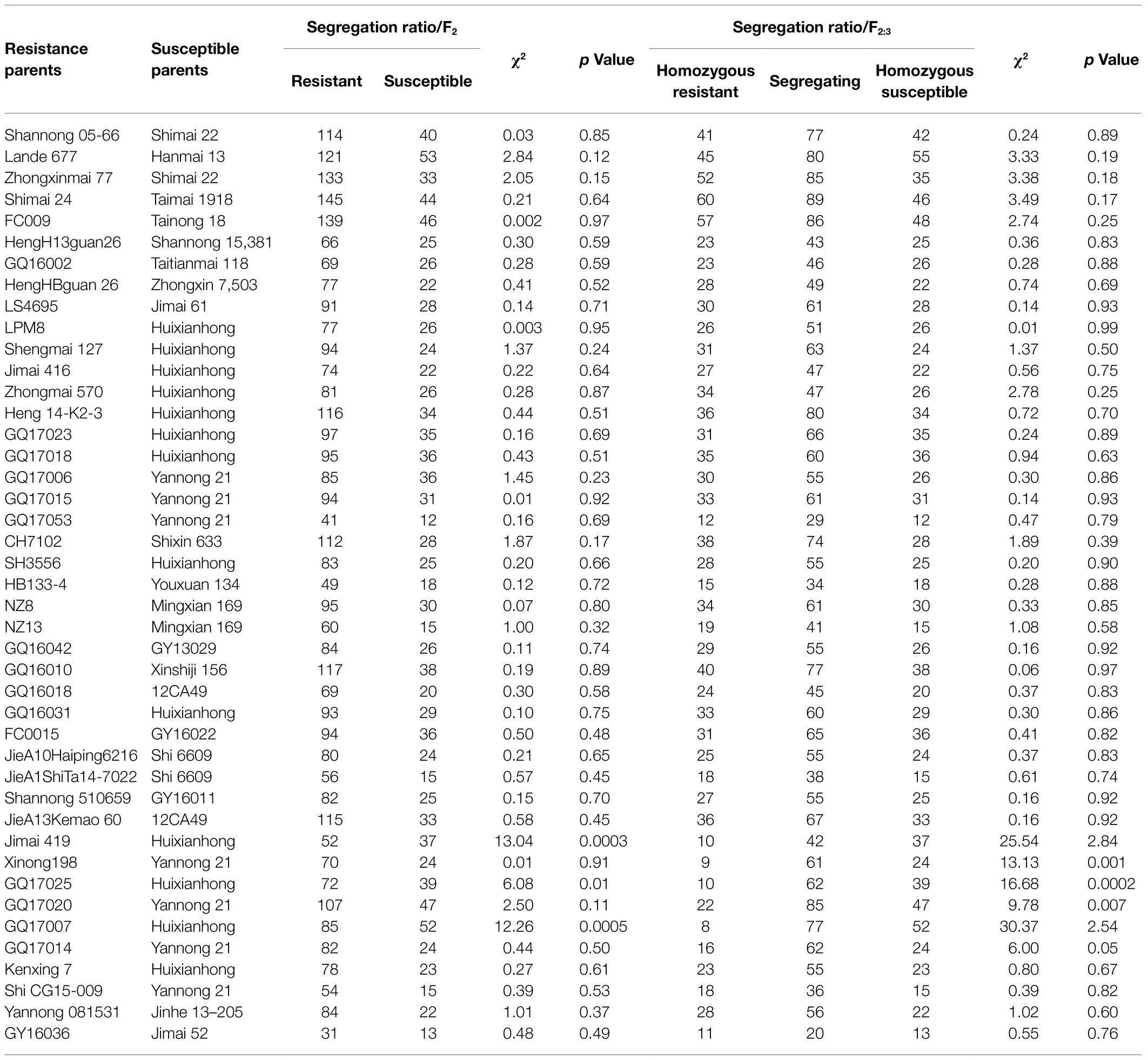
Table 1. Segregation ratios of F2 and F2:3 generations of resistant genotypes and different susceptible cultivars following inoculation with Blumeria graminis f. sp. tritici (Bgt) isolate E09 at the seedling stage.
Screening of Genotypes Carrying Pm2 Alleles
To identify Pm2 alleles in the 37 populations that fitted monogenic inheritance, the Pm2-linked marker Cfd81 and the Pm2-diagnostic marker Pm2b-map-3 were used to genotype all 37 segregating populations (Table 2). The results showed that these two markers can amplify consistent polymorphism between resistant parents and bulks in 33 populations. After genotyping the segregating populations, they were proved to be closely linked (Cfd81) and co-segregated (Pm2b-map-3) with the Pm genes in these resistant accessions (Figure 1), suggesting the existence Pm2 alleles in these resistant accessions. In four other populations (Kenxing 7 × Huixianhong, Shi CG15-009 × Yannong 21, Yannong 081531 × Jinhe 13–205, and GY16036 × Jimai 52), the markers Cfd81 and Pm2b-map-3 did not detect Pm2 alleles, suggesting these populations did not carry Pm2 alleles.
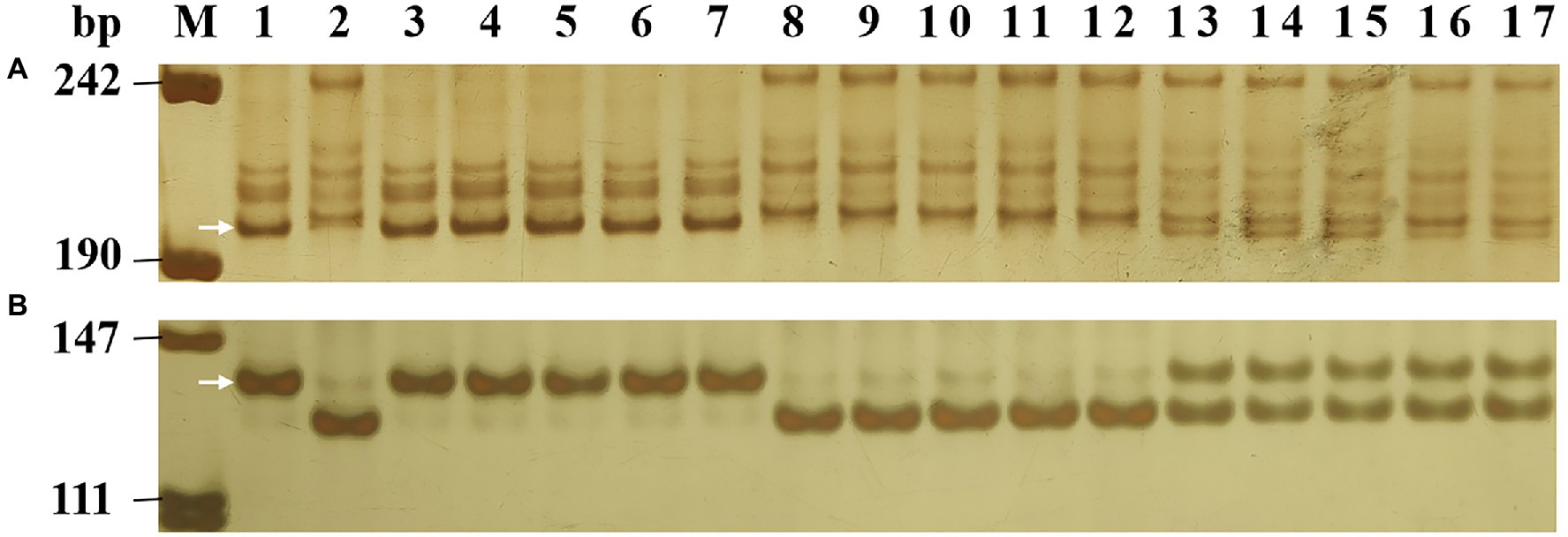
Figure 1. Amplification patterns of the Pm2-linked marker Cfd81 (A) and diagnostic marker Pm2b-map-3 (B) in genotyping Lande 677 (Pm2 donor), Hanmai 13 and random selected F2:3 families of Lande 677 × Hanmai 13. Lane M, pUC18 Msp I; lane 1, Lande 677; lane 2, Hanmai 13; lanes 3–7, homozygous resistant F2:3 families; lanes 8–12, heterozygous F2:3 families; and lanes 13–17, homozygous susceptible F2:3 families. The white arrows indicate the polymorphic bands in Lande 677.
To compare the Pm2 alleles, the 33 Pm2 donors were initially analyzed using the Pm2-linked marker Cfd81 and the Pm2-diagnostic marker Pm2b-map-3. The results showed that the 33 Pm2 donors have the same marker alleles, suggesting consistent genetic diversity in the Pm2 intervals of these donors (Figure 2). To further dissect the relationship of their Pm2 alleles, the sequences of the Pm2 alleles were analyzed after homology-based cloning based on the Pm2a sequence. The results indicated that all these Pm2 alleles were the Pm2a haplotype, suggesting that this haplotype has been widely used in resistance breeding for powdery mildew.
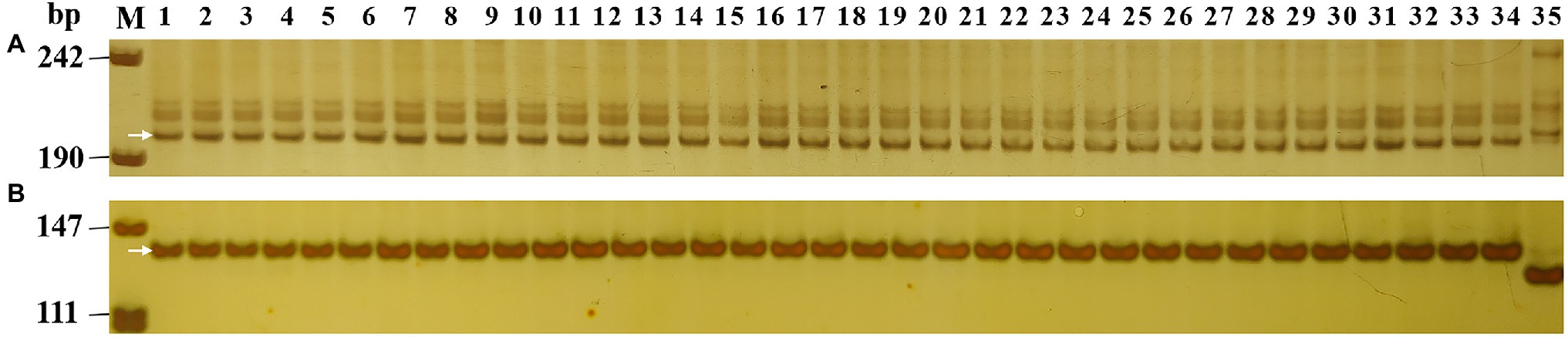
Figure 2. Marker analysis of 33 Pm2 donors using Pm2 linked marker Cfd81 (A) and diagnostic marker Pm2b-map-3 (B). Lane M, pUC18 Msp I; lane 1, Huixianhong (control without Pm2 allele); lane 2, Ulka/8*Cc (control with Pm2 allele); and lanes 3–35, the Pm2 donors identified in this study.
Resistance Spectra of the Pm2 Donors
Using nine Bgt isolates, including the four highly virulent isolates, E18, E20, E21, and E32, the resistance spectra of the 33 accessions carrying Pm2 alleles were evaluated. The results showed that the Pm2 alleles in different genetic backgrounds have different reaction patterns to the nine Bgt isolates (Figure 3; Table 3). Some Pm2 donors showed resistance to all the nine Bgt isolates tested, such as Shannong 05–66, GQ16002, GQ17023, and GQ16042. Some Pm2 donors have poor resistant spectrum, such as GQ16018 and GQ16031 which were susceptible to seven and six Bgt isolates. Other Pm2 donors were susceptible to Bgt isolates that were diversified from 1 to 5 ones. This may be related to diversity of genetic backgrounds and/or the interference of other related genes. These data provide a useful reference for breeders in different wheat production regions.

Figure 3. Reaction patterns of several Pm2 donors to four Blumeria graminis f. sp. tritici (Bgt) isolates, A3, A10, A45, and E15-1, collected from different regions of China using Huixianhong as susceptible control. 1: Huixianhong; 2: Lande 677; 3: Shimai 24; and 4: FC009.
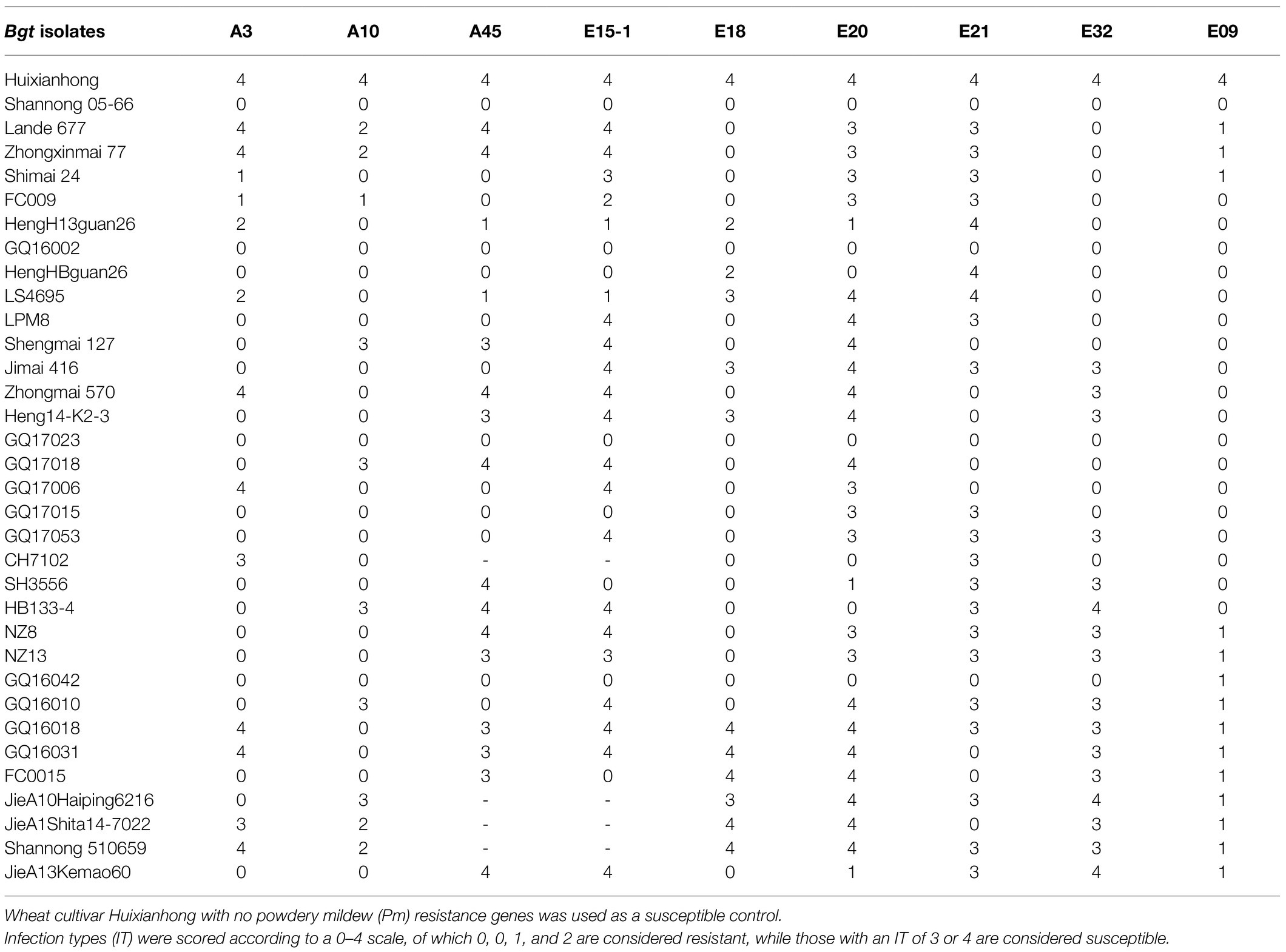
Table 3. Reaction patterns of the wheat genotypes with Pm2 alleles to nine Blumeria graminis f. sp. tritici (Bgt) isolates.
Evaluation of Markers for MAS
To transfer these Pm2 alleles to susceptible cultivars using MAS, the gel-based markers Cfd81 and Pm2b-map-3 were initially tested for their usefulness in MAS. The results demonstrated that both markers can detect polymorphic genotypes between the Pm2 donors and the 15 tested susceptible cultivars, suggesting that they can be used for MAS of Pm2 alleles when these are transferred into these susceptible cultivars by conventional hybridization (Figure 4; Table 4).
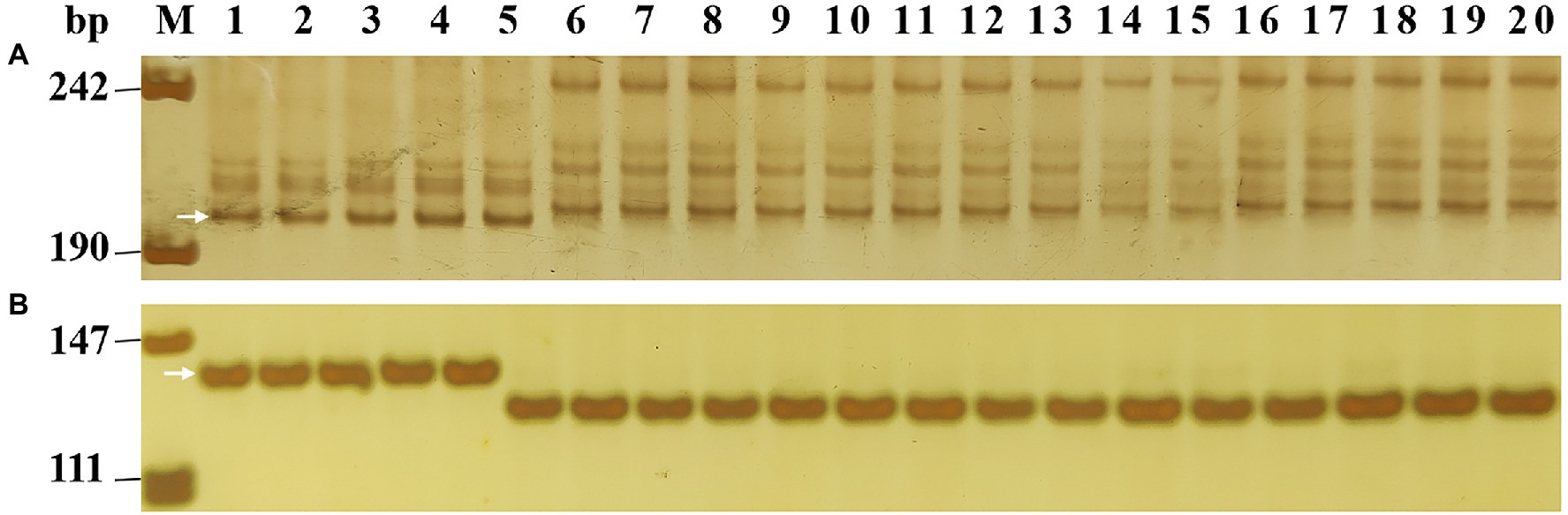
Figure 4. Amplification patterns of Pm2 linked marker Cfd81 (A) and diagnostic marker Pm2b-map-3 (B) in five Pm2 donors and 15 susceptible wheat cultivars/breeding lines. M, DNA marker pUC18 Msp I; lanes 1–5, five Pm2 donors with sequential order of Lande 677, LS4695, NZ8, NZ13, and GQ16031; lanes 6–20, wheat cultivars/breeding lines with sequential order of Shimai 22, Taimai 1918, Hanmai 13, Tainong 18, Shannong 15,381, Taitianmai 118, Zhongxin 7503, Jimai 61, Huixianhong, Yannong 21, Shixin 633, Mingxian 169, Xinshiji 156, 12CA49, and Shi 6609. The white arrows indicate the polymorphic bands in Pm2 donors.

Table 4. Evaluation of Pm2-diagnostic and linked markers on Ulka/8*Cc (Pm2a donor) and 15 susceptible wheat cultivars/breeding lines in marker-assisted selection (MAS) breeding.
To transfer Pm2 alleles using a gel-free and high throughput genotyping platform, the diagnostic KASP marker YTU-KASP-Pm2 was developed to trace the Pm2 alleles based on the 609th base of the first exon of Pm2 (Figure 5; Table 2). Using this marker, different Pm2 donors, plants from the segregating populations and susceptible cultivars without Pm2 alleles all showed the required genotyping, suggesting YTU-KASP-Pm2 is an efficient diagnostic marker for Pm2 (Figure 6; Table 4). YTU-KASP-Pm2 was then used to genotype the 15 susceptible cultivars to evaluate its suitability for MAS. The result demonstrated that this marker can detect polymorphic genotypes between the Pm2 donor and each of the tested 15 susceptible cultivars (Figure 7). So, once the Pm2 allele is transferred into these susceptible genetic backgrounds through conventional hybridization, YTU-KASP-Pm2 can be used to trace it through the gel-free platform, thus providing a valuable supplement for Pm2 MAS.
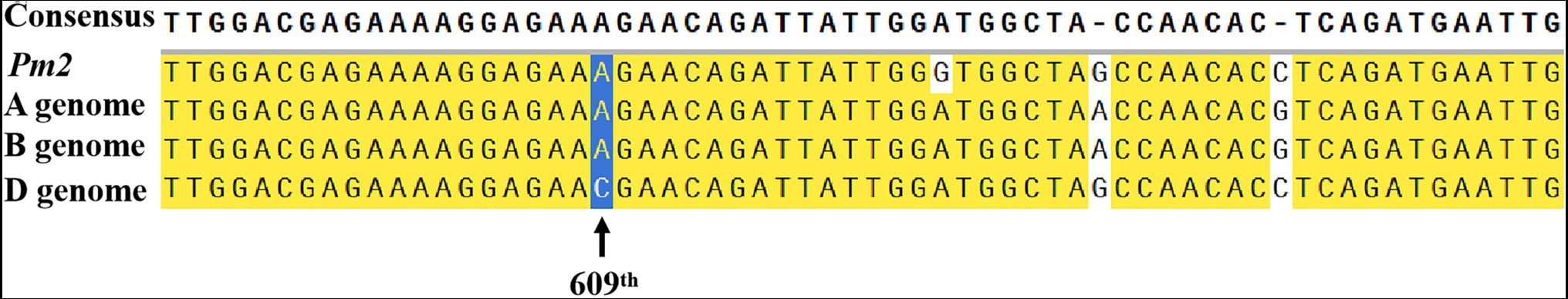
Figure 5. Sequence alignment of the first exon of Pm2a and its homologous sequences in the A, B, and D genomes of common wheat Chinese Spring to identify distinctive SNPs for development of diagnostic markers.
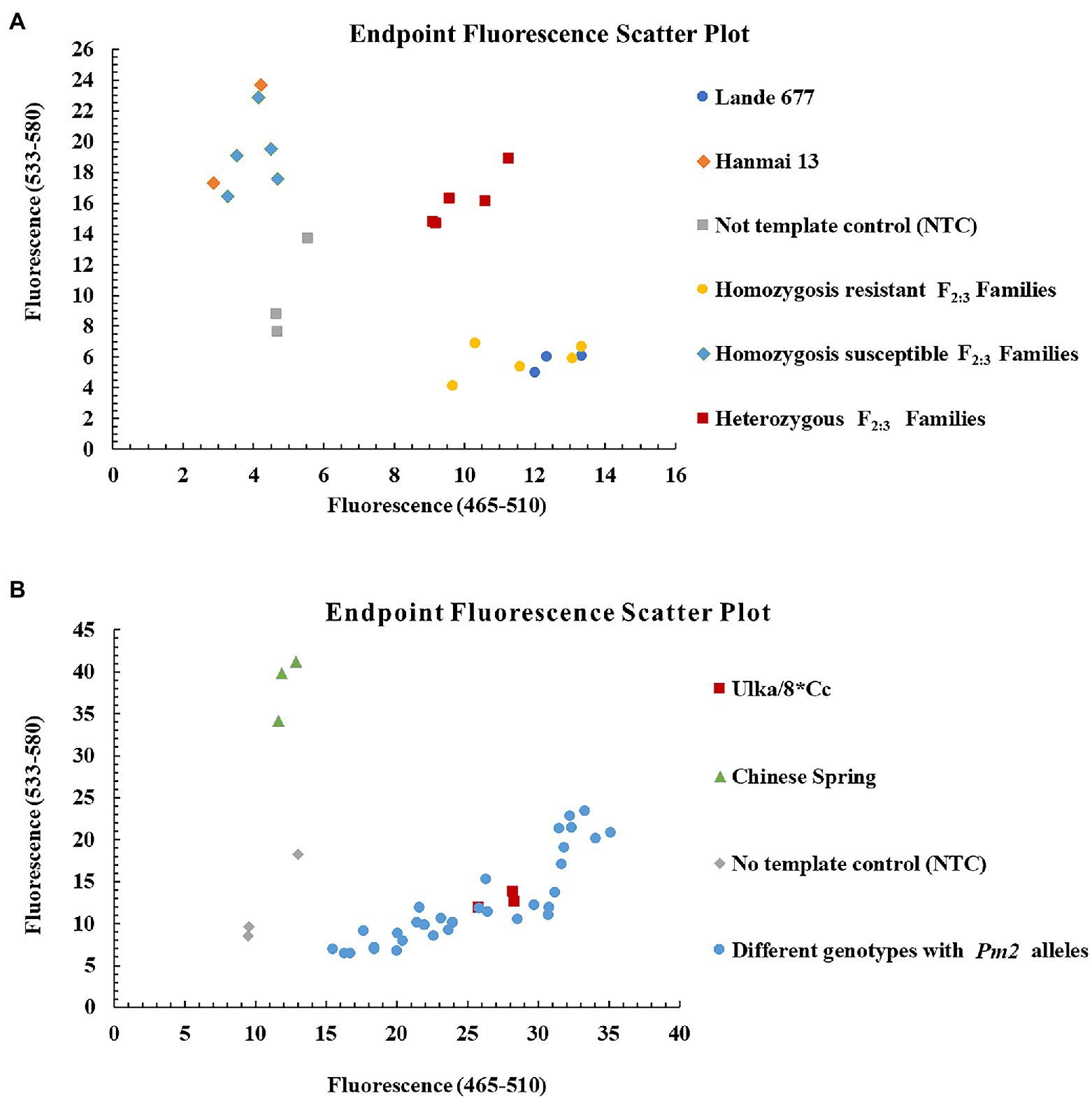
Figure 6. Genotyping results of the Pm2 diagnostic kompetitive allele-specific PCR (KASP) marker YTU-KASP-Pm2 for Lande 677 (Pm2 donor), Hanmai 13 and randomly selected F2:3 families of Lande 677 × Hanmai 13 (A) and different genotypes with Pm2 alleles (B).
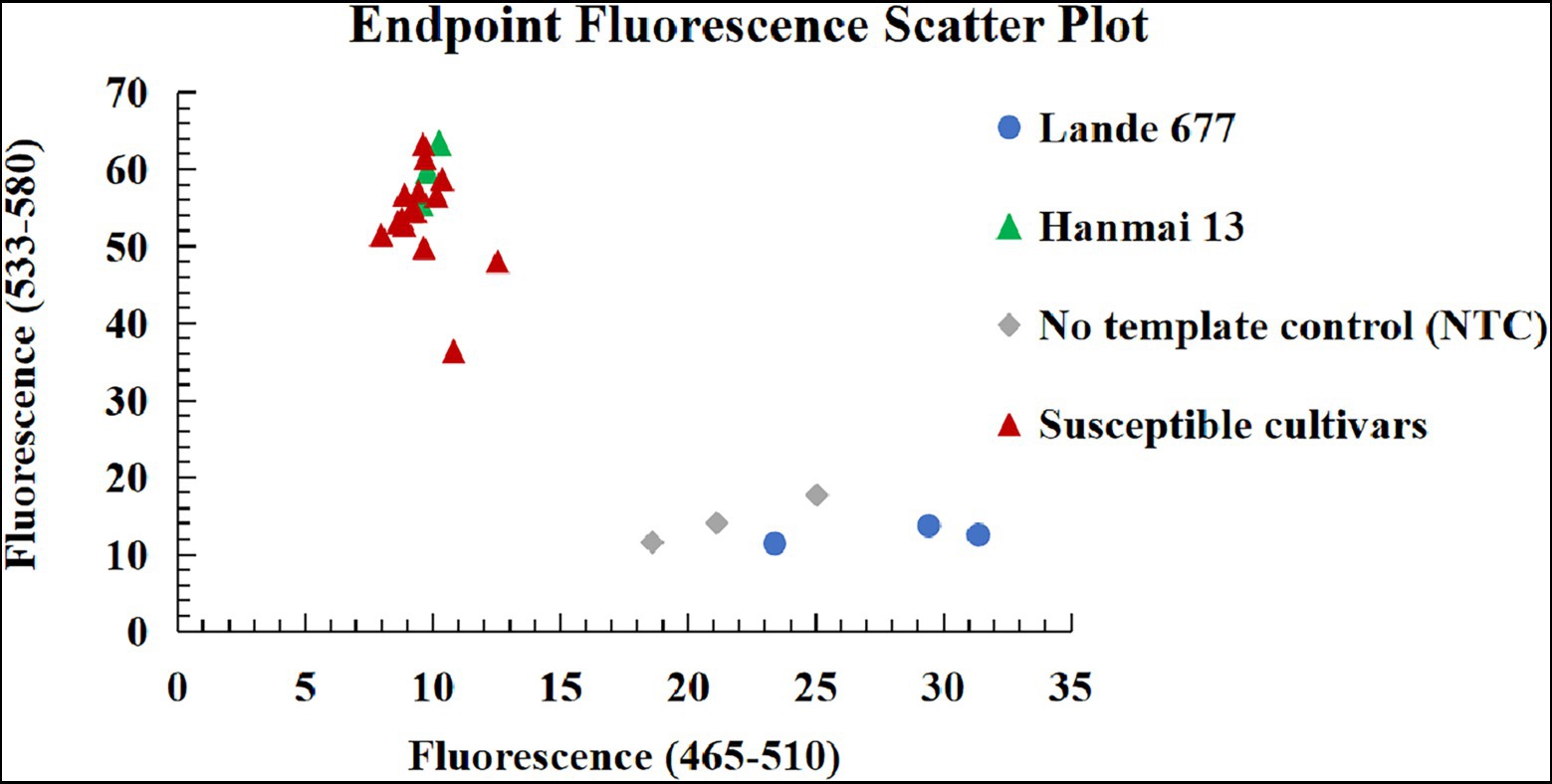
Figure 7. Genotyping results of the Pm2 KASP marker YTU-KASP-Pm2 for Lande 677, Hanmai 13 and 15 susceptible wheat cultivars/breeding lines Shimai 22, Taimai 1918, Hanmai 13, Tainong 18, Shannong 15381, Taitianmai 118, Zhongxin 7503, Jimai 61, Huixianhong, Yannong 21, Shixin 633, Mingxian 169, Xinshiji 156, 12CA49, and Shi 6609.
Discussion
Many Pm genes/alleles have been identified that confer resistance to wheat powdery mildew. Among them, only the Pm genes that are free of linkage drag and/or adverse pleiotropism have significant potential in resistance breeding. The gene, Pm2, was initially identified in the wheat landrace Ulka from the former Soviet Union in 1953 (Pugsley and Carter, 1953). In the 70 years of breeding history using Pm2, this gene has had exceptional performance in conferring resistance to powdery mildew. Many wheat cultivars carrying Pm2 have been bred and used in production, such as Liangxing 66 with PmLX66 (Huang et al., 2012), Jimai 22 and Jimai 23 with PmJM23 (Jia et al., 2020), Yingbo 700 with PmYB (Ma et al., 2015c), Zhongmai 155 with PmZ155 (Sun et al., 2015a), Nongda 399 with MlND399 (Li et al., 2013), Wennong 14 with PmW14 (Sun et al., 2015b), and Heng 4568 with PmH4568 (Gao et al., 2022). Apart from the cultivars in production, many breeding lines and landraces have also been shown to carry Pm2 alleles, including KM2939 with Pm2b (Ma et al., 2015a), Niaomai with Pm2c (Xu et al., 2015), Wangfengjian 34 with PmWFJ (Ma et al., 2015b), CH1357 with PmCH1357 (Chen et al., 2019), 10V-2 with Pm10V-2 (Ma et al., 2018), FG-1 with PmFG (Ma et al., 2016), Subtil with PmSub (Jin et al., 2018), and X3986-2 with PmX3986-2 (Ma et al., 2014). After 70 years in production, a number of Pm2 alleles continue to show high and broad-spectrum resistance in some genetic backgrounds, such as 10V-2, YingBo 700, KM2939, and Niaomai (Ma et al., 2015a,c, 2018; Xu et al., 2015). However, other alleles have reduced their ability to confer resistance (Ma et al., 2014, 2016).
It is necessary to identify more Pm2 donors from wheat cultivars and breeding lines and to also clarify their distribution in different wheat production regions, so that their use can be promoted in different regions. In this study, after wide screening, we identified a large number of Pm2 donors from 641 wheat cultivars and breeding lines collected from different wheat production regions. This is the first time that the existence of Pm2 alleles in current wheat breeding lines has been assessed and summarized, data which will contribute to realizing the synergistic improvement of resistance and other agronomic traits. The results revealed that Pm2 alleles accounted for a very high proportion of the resistance genes in resistant cultivars and breeding lines. This suggests more careful use of this gene in production is necessary, and the best strategy for its use may be in pyramiding it with other resistance genes to develop durable resistance.
Using mutant chromosome sequencing (MutChromSeq), the Pm2a allele was cloned (Sánchez-Martín et al., 2016). Furthermore, Chen et al. (2019) cloned PmCH1357 using map-based cloning and found that PmCH1357, Pm2c, PmLX66, and MlND399 all have identical sequences to Pm2a. However, Manser et al. (2021) identified seven new allelic variations of Pm2 from 28 Aegilops tauschii accessions. This implies that there may have been only one haplotype of Pm2 that is Pm2a, used in resistance breeding since it was first identified in 1953. To confirm this result, we identified additional Pm2 alleles in the 641 wheat cultivars and breeding lines and found that all of the Pm2 alleles were identical to haplotype Pm2a, further confirming the conservation of the Pm2a haplotype in wheat breeding. From this result, we speculate that the new Pm2 alleles identified in Aegilops tauschii could also become valuable contributors to resistance, just as Pm2a has been an elite allele used in breeding for more than 70 years and which still confers considerable resistance.
Nevertheless, there are still unsolved issues associated with the Pm2 locus. We, and others (Ma et al., 2014, 2018; Sun et al., 2015a,b; Jia et al., 2020), have now shown that different Pm2 donors have significantly different reaction patterns to different Bgt isolates, something that is hard to explain based solely on the different genetic backgrounds. We speculate that there may be other unknown associated genes in the different genetic backgrounds which, together, provide the powdery mildew resistance in specific genotypes. We also note that there is still no transgenic evidence to confirm the functionality of the cloned Pm2 gene, something that is required to clarify this locus.
Irrespective of the unsolved issues associated with the Pm2 locus, this has not prevented its use in breeding in view of the advantages of this locus in conferring resistance. In wheat resistance breeding, MAS is a rapid and effective method to trace targeted genes in breeding programs, and development of molecular markers for MAS has been the key factor (William et al., 2007; Gupta et al., 2010; Li et al., 2019). Currently, there are two types of breeding markers (gel-based and gel-free), which can meet the needs of different labs in terms of throughput and which are applicable to different platforms (gel-based or gel-free) for marker analysis. Gel-based markers for Pm2 have been reported in previous studies (Ma et al., 2015a,b,c, 2016, 2018). We verified their suitability in identifying Pm2 donors in this study, including both linked and diagnostic markers.
With the development of high throughput genotyping platforms, gel-free KASP markers are now also widely used. Compared with the gel-based markers, KASP markers have the advantages of good stability and high throughput, are free of specific fluorescent probes and are low cost (Rasheed et al., 2016). However, no KASP markers suitable for MAS had been reported for the elite Pm2 gene. To efficiently and accurately transfer and trace Pm2 alleles using MAS, we developed the diagnostic KASP maker YTU-KASP-Pm2 based on the presence of a stable SNP between homologous genes in the susceptible cultivars and the Pm2 donors. This means that the Pm2 alleles can be detected through two platforms: the gel-based diagnostic marker Pm2b-map-3 can be used by breeders with only basic screening facilities, and the gel-free marker YTU-KASP-Pm2 is available to breeders with high-throughput genotyping platforms.
When an elite resistance gene is identified, its breeding value depends not only on its resistance but also on the agronomic performance of the gene donor. Gene donors with poor agronomic performance will greatly limit their utilization in breeding (Summers and Brown, 2013; Liu et al., 2019, 2020). In this study, all the Pm2 alleles identified were from wheat cultivars or breeding lines. Their donors all have elite yield and agronomic performance and, more importantly, they came from different wheat production regions, which can meet the requirements of different wheat production regions in corresponding resistance breeding. These Pm2 alleles can be individually used in corresponding ecoregions, and also can be used in pyramiding breeding with other resistance genes for durable resistance. The abundance of Pm2 donors provides the possibility of screening the optimum pyramiding model in breeding and realizing the dual improvement in both resistance and agronomic traits.
Conclusion
In conclusion, we have identified 37 Pm2 alleles from wheat cultivars and breeding lines collected from different wheat production regions and confirmed that they all have the Pm2a sequence, but have different reaction patterns to different Bgt isolates depending on their specific genetic background. Molecular markers available for MAS were screened and confirmed, and a diagnostic KASP marker YTU-KASP-Pm2 was identified for use in high-throughput genotyping platforms. Our study has provided valuable information on the distribution and rational use of Pm2 alleles and will contribute to the control of wheat powdery mildew.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
PM, LL, and NS conceived the research. ZY, LX, and FS performed the experiments. WL, YM, WZ, LW, and XL analyzed the data. FL, RH, NS, and LL performed phenotypic assessment. PM wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by the National Natural Science Foundation of China (32072053, 31971874, and 32001544) and Key Technology Research and Development Program of Shandong (2020CXGC010805 and 2021RKY06100).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Emerita Paula E. Jameson, University of Canterbury, financially supported by “Double Hundred” Plan for Foreign Experts in Shandong Province, China, for critically reviewing and editing this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.912589/full#supplementary-material
Footnotes
References
An, D. G., Ma, P. T., Zheng, Q., Fu, S. L., Li, L. H., Han, F. P., et al. (2019). Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor. Appl. Genet. 132, 257–272. doi: 10.1007/s00122-018-3214-3
Chai, J. F., Liu, X., and Jia, J. Z. (2005). Homoeologous cloning of omega-secalin gene family in a wheat 1BL/1RS translocation. Cell Res. 15, 658–664. doi: 10.1038/sj.cr.7290335
Chen, F., Jia, H. Y., Zhang, X. J., Qiao, L. Y., Li, X., Zheng, J., et al. (2019). Positional cloning of PmCH1357 reveals the origin and allelic variation of the Pm2 gene for powdery mildew resistance in wheat. Crop J. 7, 771–783. doi: 10.1016/j.cj.2019.08.004
Costanzo, A., and Bàrberi, P. (2014). Functional agrobiodiversity and agroecosystem services in sustainable wheat production. A review. Agron. Sustain. Dev. 34, 327–348. doi: 10.1007/s13593-013-0178-1
Gao, H. M., Xu, X. Z., Ai, P. F., Luo, F. Y., Guo, P., and Ma, P. T. (2022). Identification of the powdery mildew resistance in Chinese wheat cultivar Heng 4568 and its evaluation in marker-assisted selection. Front. Genet. 13:819844. doi: 10.3389/fgene.2022.819844
Gupta, P. K., Langridge, P., and Mir, R. R. (2010). Marker-assisted wheat breeding: present status and future possibilities. Mol. Breed. 26, 145–161. doi: 10.1007/s11032-009-9359-7
He, H. G., Ji, Y. Y., Zhu, S. Y., Li, B., Zhao, R. H., Jiang, Z. Z., et al. (2017). Genetic, physical and comparative mapping of the powdery mildew resistance gene Pm21 originating from Dasypyrum villosum. Front. Plant Sci. 8:1914. doi: 10.3389/fpls.2017.01914
He, H. G., Liu, R. K., Ma, P. T., Du, H. N., Zhang, H. H., Wu, Q. H., et al. (2021). Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor. Appl. Genet. 134, 53–62. doi: 10.1007/s00122-020-03681-2
Hickey, L. T., Hafeez, A. N., Robinson, H., Jackson, S. A., Leal-Bertioli, S. C. M., Tester, M., et al. (2019). Breeding crops to feed 10 billion. Nat. Biotechnol. 37, 744–754. doi: 10.1038/s41587-019-0152-9
Huang, J., Zhao, Z., Song, F. J., Wang, X. M., Xu, H. X., Huang, Y., et al. (2012). Molecular detection of a gene effective against powdery mildew in the wheat cultivar liangxing 66. Mol. Breed. 30, 1737–1745. doi: 10.1007/s11032-012-9757-0
Jia, M. S., Xu, H. X., Liu, C., Mao, R. X., Li, H. S., Liu, J. J., et al. (2020). Characterization of the powdery mildew resistance gene in the elite wheat cultivar Jimai 23 and its application in marker-assisted selection. Front. Genet. 11:241. doi: 10.3389/fgene.2020.00241
Jin, Y. L., Shi, F. Y., Liu, W. H., Fu, X. Y., Gu, T. T., Han, G. H., et al. (2021). Identification of resistant germplasm and detection of genes for resistance to powdery mildew and leaf rust from 2,978 wheat accessions. Plant Dis. 105, 3900–3908. doi: 10.1094/PDIS-03-21-0532-RE
Jin, Y. L., Xu, H. X., Ma, P. T., Fu, X. Y., Song, L. P., Xu, Y. F., et al. (2018). Characterization of a new Pm2 allele associated with broad-spectrum powdery mildew resistance in wheat line Subtil. Sci. Rep. 8:475. doi: 10.1038/s41598-017-18827-4
Kaur, R., Vyas, P., Sharma, P., Sheikh, I., Kumar, R., and Dhaliwal, H. S. (2017). “Marker-assisted breeding of recombinant 1RS.1BL chromosome for improvement of bread making quality and yield of wheat (Triticum aestivum L).” in Applications of Biotechnology for Sustainable Development. eds. K. Mukhopadhyay, A. Sachan, and M. Kumar (Singapore: Springer)
Laroche, A., Frick, M., Graf, R. J., Larsen, J., and Laurie, J. D. (2019). Pyramiding disease resistance genes in elite winter wheat germplasm for Western Canada. Crop J. 7, 739–749. doi: 10.1016/j.cj.2019.08.005
Li, A. L., Hao, C. Y., Wang, Z. Y., Geng, S. F., Jia, M. L., Wang, F., et al. (2022). Wheat breeding history reveals synergistic selection of pleiotropic genomic sites for plant architecture and grain yield. Mol. Plant 15, 504–519. doi: 10.1016/j.molp.2022.01.004
Li, H. J., Wang, X. M., Song, F. J., Wu, C. P., Wu, X. F., Zhang, N., et al. (2011). Response to powdery mildew and detection of resistance genes in wheat cultivars from China. Acta Agron. Sin. 37, 943–954. doi: 10.1016/S1875-2780(11)60026-6
Li, D., Yuan, C. G., Wu, H. B., Zhang, D., Liang, Y., Wang, Z. Z., et al. (2013). SSR and AFLP-derived SCAR markers associated with the powdery mildew resistance gene in common wheat cultivar ND399. J. Plant Genet. Resour. 14, 104–108. doi: 10.3969/j.issn
Li, H. J., Zhou, Y., Xin, W. L., Wei, Y. Q., Zhang, J. L., and Guo, L. L. (2019). Wheat breeding in northern China: achievements and technical advances. Crop J. 7, 718–729. doi: 10.1016/j.cj.2019.09.003
Liu, C., Gong, W. P., Han, R., Guo, J., Li, G. R., Li, H. S., et al. (2019). Characterization, identification and evaluation of a set of wheat-Aegilops comosa chromosome lines. Sci. Rep. 9:4773. doi: 10.1038/s41598-019-41219-9
Liu, C., Han, R., Wang, X. L., Gong, W. P., Cheng, D. G., Cao, X. Y., et al. (2020). Research progress of wheat wild hybridization, disease resistance genes transfer and utilization. Agr. Sci. Sin. 53, 1287–1308. doi: 10.3864/j.issn.0578-1752.2020.07.001
Ma, P. T., Xu, H. X., Li, L. H., Zhang, H. X., Han, G. H., Xu, Y. F., et al. (2016). Characterization of a new Pm2 allele conferring powdery mildew resistance in the wheat germplasm line FG-1. Front. Plant Sci. 7:546. doi: 10.3389/fpls.2016.00546
Ma, P. T., Xu, H. X., Luo, Q. L., Qie, Y. M., Zhou, Y. L., Xu, Y. F., et al. (2014). Inheritance and genetic mapping of a gene for seedling resistance to powdery mildew in wheat line X3986-2. Euphytica 200, 149–157. doi: 10.1007/s10681-014-1178-1
Ma, P. T., Xu, H. X., Xu, Y. F., Li, L. H., Qie, Y. M., Luo, Q. L., et al. (2015a). Molecular mapping of a new powdery mildew resistance gene Pm2b in Chinese breeding line KM2939. Theor. Appl. Genet. 128, 613–622. doi: 10.1007/s00122-015-2457-5
Ma, P. T., Xu, H. X., Xu, Y. F., Song, L. P., Liang, S. S., Sheng, Y., et al. (2018). Characterization of a powdery mildew resistance gene in wheat breeding line 10V-2 and its application in marker-assisted selection. Plant Dis. 102, 925–931. doi: 10.1094/PDIS-02-17-0199-RE
Ma, P. T., Xu, H. X., Zhang, H. X., Li, L. H., Xu, Y. F., Zhang, X. T., et al. (2015b). The gene PmWFJ is a new member of the complex Pm2 locus conferring unique powdery mildew resistance in wheat breeding line Wanfengjian 34. Mol. Breed. 35:210. doi: 10.1007/s11032-015-0403-5
Ma, P. T., Zhang, H. X., Xu, H. X., Xu, Y. F., Cao, Y. W., Zhang, X. T., et al. (2015c). The gene PmYB confers broad-spectrum powdery mildew resistance in the multi-allelic Pm2 chromosome region of the Chinese wheat cultivar YingBo 700. Mol. Breed. 35:124. doi: 10.1007/s11032-015-0320-7
Makhoul, M., Rambla, C., Voss-Fels, K. P., Hickey, L. T., Snowdon, R. J., and Obermeier, C. (2020). Overcoming polyploidy pitfalls: a user guide for effective SNP conversion into KASP markers in wheat. Theor. Appl. Genet. 133, 2413–2430. doi: 10.1007/s00122-020-03608-x
Manser, B., Koller, T., Praz, C. R., Roulin, A. C., Zbinden, H., Arora, S., et al. (2021). Identification of specificity-defining amino acids of the wheat immune receptor Pm2 and powdery mildew effector AvrPm2. Plant J. 106, 993–1007. doi: 10.1111/tpj.15214
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Xia, X. C., and Raupp, W. J. (2020). “Catalogue of gene symbols for wheat: 2020 supplement,” in Annual Wheat Newsletter. ed. W. J. Raupp (Manhattan, USA), 98–113.
Morgounov, A., Tufan, H. A., Sharma, R., Akin, B., Bagci, A., Braun, H. J., et al. (2012). Global incidence of wheat rusts and powdery mildew during 1969–2010 and durability of resistance of winter wheat variety Bezostaya 1. Eur. J. Plant Pathol. 132, 323–340. doi: 10.1007/s10658-011-9879-y
Pugsley, A. T., and Carter, M. V. (1953). The resistance of twelve varieties of Triticum vulgare to Erysiphe graminis tritici. Aust. J. Biol. Sci. 6, 335–346. doi: 10.1071/BI9530335
Rasheed, A., Wen, W., Gao, F., Zhai, S., Jin, H., Liu, J., et al. (2016). Development and validation of kasp assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 129, 1843–1860. doi: 10.1007/s00122-016-2743-x
Sánchez-Martín, J., Steuernagel, B., Ghosh, S., Herren, G., Hurni, S., Adamski, N., et al. (2016). Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17:221. doi: 10.1186/s13059-016-1082-1
Shah, L., Rehman, S., Ali, A., Yahya, M., Riaz, M. W., Si, H. Q., et al. (2018). Genes responsible for powdery mildew resistance and improvement in wheat using molecular marker-assisted selection. J. Plant Dis. Protect. 125, 145–158. doi: 10.1007/s41348-017-0132-6
Summers, R. W., and Brown, J. K. M. (2013). Constraints on breeding for disease resistance in commercially competitive wheat cultivars. Plant Phothol. 62:115.
Sun, H. G., Song, W., Sun, Y. L., Chen, X. M., Liu, J. J., Zou, J. W., et al. (2015a). Resistance to powdery mildew in the wheat cultivar Zhongmai 155: effectiveness and molecular detection of the resistance gene. Crop Sci. 55, 1017–1025. doi: 10.2135/cropsci2014.05.0355
Sun, Y. L., Zou, J. W., Sun, H. G., Song, W., Wang, X. M., and Li, H. J. (2015b). PmLX66 and PmW14: new alleles of Pm2 for resistance to powdery mildew in the Chinese winter wheat cultivars Liangxing 66 and Wennong 14. Plant Dis. 99, 1118–1124. doi: 10.1094/PDIS-10-14-1079-RE
William, H. M., Trethowan, R., and Crosby-Galvan, E. M. (2007). Wheat breeding assisted by markers: CIMMYT’s experience. Euphytica 157, 307–319. doi: 10.1007/s10681-007-9405-7
Xu, H. X., Yi, Y. J., Ma, P. T., Qie, Y. M., Fu, X. Y., Xu, Y. F., et al. (2015). Molecular tagging of a new broad-spectrum powdery mildew resistance allele Pm2c in Chinese wheat landrace Niaomai. Theor. Appl. Genet. 128, 2077–2084. doi: 10.1007/s00122-015-2568-z
Zhang, D. Y., Zhu, K. Y., Dong, L. L., Liang, Y., Li, G. Q., Fang, T. L., et al. (2019). Wheat powdery mildew resistance gene Pm64 derived from wild emmer (Triticum turgidum var. dicoccoides) is tightly linked in repulsion with stripe rust resistance gene Yr5. Crop J. 7, 761–770. doi: 10.1016/j.cj.2019.03.003
Keywords: wheat, powdery mildew, Pm2, MAS, diagnostic KASP marker
Citation: Yu Z, Xiao L, Su F, Liu W, Luo F, Han R, Mu Y, Zhang W, Wu L, Liang X, Sun N, Li L and Ma P (2022) Mining of Wheat Pm2 Alleles for Goal-Oriented Marker-Assisted Breeding. Front. Plant Sci. 13:912589. doi: 10.3389/fpls.2022.912589
Edited by:
Handong Su, Huazhong Agricultural University, ChinaReviewed by:
Zhu-Qing Shao, Nanjing University, ChinaShunzong Ning, Sichuan Agricultural University, China
Copyright © 2022 Yu, Xiao, Su, Liu, Luo, Han, Mu, Zhang, Wu, Liang, Sun, Li and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengtao Ma, cHRtYUB5dHUuZWR1LmNu; Linzhi Li, bGluemhpbGkyMDAyQDE2My5jb20=; Nina Sun, c3VuMjAwNDM2QDE2My5jb20=
†These authors have contributed equally to this work
 Ziyang Yu1†
Ziyang Yu1† Ran Han
Ran Han Pengtao Ma
Pengtao Ma