- 1Research Institute of Subtropical Forestry, Chinese Academy of Forestry, Hangzhou, China
- 2Zhejiang Provincial Key Laboratory of Tree Breeding, Hangzhou, China
Xyloglucan endotransglucosylase/hydrolase (XTH), belonging to glycoside hydrolase family 16, is one of the key enzymes in plant cell wall remodeling. Schima superba is an important timber and fireproof tree species in southern China. However, little is known about XTHs in S. superba. In the present study, a total of 34 SsuXTHs were obtained, which were classified into three subfamilies based on the phylogenetic relationship and unevenly distributed on 18 chromosomes. Furthermore, the intron–exon structure and conserved motif composition of them supported the classification and the members belonging to the same subfamily shared similar gene structures. Segmental and tandem duplication events did not lead to SsuXTH gene family expansion, and strong purifying selection pressures during evolution led to similar structure and function of SsuXTH gene family. The interaction network and cis-acting regulatory elements analysis revealed the SsuXTH expression might be regulated by multiple hormones, abiotic stresses and transcription factors. Finally, expression profiles and GO enrichment analysis showed most of the tandem repeat genes were mainly expressed in the phloem and xylem and they mainly participated in glycoside metabolic processes through the transfer and hydrolysis of xyloglucan in the cell wall and then regulated fiber elongation.
Introduction
Xyloglucan endotransglucosylases/hydrolases (XETs/XEHs, also named XTHs) are xyloglucan-modifying enzymes and belong to glycoside hydrolase family 16. The enzymes have two distinct catalytic activities, XET and XEH, and participate in cell wall relaxation, synthesis and degradation (Campbell and Braam, 1999; Rose et al., 2002; Baumann et al., 2007). Multigene families of XTHs have been identified in a variety of plant species, including Arabidopsis thaliana (33), Oryza sativa (29), Triticum aestivum (5), Sorghum bicolor (35), Nicotiana tabacum (56), Glycine max (61), Solanum lycopersicum (25), Actinidia deliciosa (14), Malus sieversii (11), Gossypium hirsutum (23), Brassica rapa (53), Brassica oleracea (38), B. juncea (74), and Populus spp. (41; Yokoyama and Nishitani, 2001; Yokoyama et al., 2004; Geisler-Lee et al., 2006; Saladié et al., 2006; Liu et al., 2007; Atkinson et al., 2009; Lee et al., 2010; Rai et al., 2016; Nawaz et al., 2017; Song et al., 2018; Wang et al., 2018; Li et al., 2020; Wu et al., 2020; Cheng et al., 2021). Based on their phylogenetic relationships, the XTH genes are normally classified into three major groups, Group I, Group II, and Group III, and one minor ancestral group exists differently in species (Yokoyama and Nishitani, 2001; Baumann et al., 2007; Michailidis et al., 2009; Wang et al., 2018; Li et al., 2020; Wu et al., 2020). In some species, groups I and II cannot be separated, such as OsXTHs in O. sativa, which are clustered into groups I/II and III (Yokoyama et al., 2004). In addition, the members in group III could be subclustered into groups IIIA and IIIB (Baumann et al., 2007). Studies have shown that XTH genes in groups I, II and IIIB mainly possess XET activity, whereas group IIIA has XEH activity (Campbell and Braam, 1999; Rose et al., 2002; Baumann et al., 2007).
XTHs have spatiotemporal expression specificity and play an important role in plant growth and development. For example, in A. thaliana, five genes are expressed in green siliques, two genes are expressed in stems, and at least 10 genes are significantly expressed in the root differentiation zone, promoting cell elongation, expansion, and cell wall formation (Yokoyama and Nishitani, 2001; Maris et al., 2009). Research has revealed that XTH remolds the structure and ductility of the cell wall through bond breaking and glycoside rearrangement in xyloglucan chains (Seale, 2020). At present, several XTH genes have been identified to participate in cell wall remodeling in woody plants (Bourquin et al., 2002; Mellerowicz and Sundberg, 2008; Nishikubo et al., 2011; Sundell et al., 2017; Ghosh Dasgupta et al., 2021). For example, PttXET16A is involved in the formation of secondary vascular tissue in poplar (Populus tremula × Populus tremuloides; Bourquin et al., 2002). Overexpression of PttXET16-34 increases xyloglucan levels in primary-walled xylem and promotes vascular growth and development but decreases in secondary-walled xylem (Nishikubo et al., 2011). XTH activity is positively associated with the Runkel ratio (2 × fiber cell wall thickness/lumen diameter) in Eucalyptus grandis (Ghosh Dasgupta et al., 2021). In addition, researchers have found that the higher the XTH activity during the cell elongation stage in cotton, the longer the fiber becomes (Michailidis et al., 2009; Lee et al., 2010; Shao et al., 2011).
XTH genes also participate in the response to biotic and abiotic stresses by cell wall remodeling. The Capsicum annuum XTH homologous genes CaXTH1, CaXTH2 and CaXTH3 are upregulated under drought, high salinity and cold temperature, and overexpression of CaXTH3 enhances tolerance to salt and drought stresses in A. thaliana and Lycopersicon esculentum (Cho et al., 2006; Choi et al., 2011). PeXTH (Populus euphratica) could enhance salt tolerance in tobacco plants (Han et al., 2013). AtXTH31 improves flood resistance in G. max (Song et al., 2018). AtXTH19 also enhances freeze tolerance after cold and subzero acclimation in A. thaliana (Takahashi et al., 2021). The promoter activities and expression of CsXTH1 and CsXTH3 induced by wounding in Cucumis sativus reveal that external mechanical factors play an important role in the regulation of the expression of these genes (Malinowski et al., 2018).
Schima superba, belonging to Theaceae, is an evergreen broad-leaved tree species in southern China (Yang et al., 2017; Zhou et al., 2020a). Because of its superior vascular anatomical structures, such as longer fiber and vessel length and thickened secondary cell walls, its timber has great rigidity and toughness and is valued commercially (Li, 2014). S. superba grows fast and has a strong ability to resist drought and barrenness and adapt to various environments. Therefore, these characteristics give this species a strong understory regeneration ability with functions in fire prevention. However, little is known about XTHs in S. superba. In this study, we performed a genome-wide analysis of the XTH genes in S. superba. From phylogenetic and gene duplication analyses, we revealed the origin and evolution of the SsuXTH family; from motif and structural analyses, we demonstrated the conservation of gene structure and function; and from the analysis of cis-acting regulatory elements, expression patterns and functional interactions, we predicted the possible functions and regulatory factors of these genes. This is the first time to perform gene family analysis at the genome level of S. superba, and it has been found that XTH genes may regulate wood fiber elongation. Here, we provide a new insight into the function of SsuXTH and lay the foundation for further exploration of the decisive genes of fiber development.
Materials and Methods
Genome-Wide Identification of XTH Genes
Schima superba, its related species Camellia sinensis, and the important broad-leaved timber tree species, Populus and E. grandis, genome sequences were recovered from the S. superba genome database (our laboratory, unpublished), Tea Plant Information Archive (TPIA),1 Populus trichocarpa genome database (version 4.1),2 and E. grandis genome database (version 2.0).3 We downloaded XTH protein sequences of A. thaliana from The Arabidopsis Information Resource (TAIR)4 and used them as the query sequences to scan the Populus, E. grandis, C. sinensis, and S. superba genome sequences using the Protein Basic Logical Alignment Search Tool (BLASTP, United States National Library of Medicine) with an e-value (≤1e−5) and an identity match (≥50%) as thresholds (Cui et al., 2019). To further confirm the presence of domains, we also conducted a hidden Markov model (HMM) search for sequence homologs using the HMMER 3.0 program (Mistry et al., 2021),5 and XET_C (PF06955.12) and Glyco_hydro_16 (PF00722.21) were used as baits. The BLASTP and HMM search results were then integrated to identify candidate XTH genes. Their sequences were then submitted to the NCBI conserved domains database (CDD; Lu et al., 2020)6 to verify the presence of the XET and GH16 domains (Zhu et al., 2020).
Sequence Analysis of XTH Proteins
The physicochemical properties of proteins encoded by XTH proteins in Populus, E. grandis, C. sinensis and S. superba, including molecular weight (MW), isoelectric point (PI), instability index, grand average of hydropathy (GRAVY) values and so on, were calculated using the ExPASy online tool (Wilkins et al., 1999).7 Finally, the subcellular localization of XTH proteins was predicted using Euk-mPLoc 2.0 (Chou and Shen, 2010).8
Phylogenetic Analysis of XTH Proteins
The XTH peptide sequences from P. trichocarpa (Ptri), E. grandis (Egr), C. sinensis (Cs), S. superba (Ssu) and A. thaliana (At) were aligned by using ClustalW (Larkin et al., 2007) with default parameters. The aligned sequences were then used to generate the phylogenetic tree using MEGA7.0 software (Kumar et al., 2016). The tree was constructed using the neighbor-joining (NJ) algorithm with default parameters. The reliability of the phylogenetic tree was analyzed by the bootstrap method, and replicates were set to 1,000. The resulting tree was visualized using EvolView v3 (Subramanian et al., 2019).9
Conserved Motif and Gene Structure Analyses
In order to identify the conserved motifs, the MEME (Bailey et al., 2015)10 suite was used with default parameters. Exons and introns of each XTH were identified using the gff3 file of the S. superba genome, which contained information regarding gene structure, and visualized using TBtools (Chen et al., 2020).
Chromosomal Localization and Ka/Ks Calculation
The chromosomal location information of the XTH gene family and the length of each chromosome were extracted using the annotated files of the S. superba genome. The gene positions on chromosomes were drafted by using TBtools (Chen et al., 2020).
And paralogous genes were determined by aligning and phylogenetically analyzing XTH proteins (He et al., 2016; Zhou et al., 2020b). Genes with a distribution on the chromosome in the range of 100 kb and separated by less than five genes were considered to be tandem duplicates according to the same standard as in rice (TIGR Rice Genome Annotation).11
Furthermore, the Ka/Ks ratios were evaluated using TBtools software (Chen et al., 2020) to assess the synonymous and nonsynonymous groups. The Ks values represent the divergence time of duplication events, and the Ka/Ks values represent the selective pressure of duplicate genes. Divergence time (T) was calculated by Ks/2r × 10−6 million years ago (Mya; Khan et al., 2018), where the r is the rate of divergence. For Theaceae plants, r = 5.62 × 10−9(Xia et al., 2020). In general, Ka/Ks < 1.0 represents purifying or negative selection, Ka/Ks = 1.0 represents neutral selection, and Ka/Ks > 1.0 represents positive selection (Zhang et al., 2006).
cis-Regulatory Element Analyses
The promoter sequences of 2,000 bp of the XTH genes were retrieved from the S. superba genome database (unpublished) to analyze the cis-acting regulatory elements (CAREs). PlantCARE 10 (Lescot et al., 2002)12 was used for identifying and analyzing the CAREs.
Functional Interaction Analyses
The protein interaction network was generated from the STRING database (Szklarczyk et al., 2019)13 based on an Arabidopsis association model with default parameters. The Arabidopsis model had to be employed due to the absence of the S. superba database in the STRING server. And the homologous proteins of Arabidopsis were designated STRING proteins and were selected based on high bit scores in the BLAST results (Chatterjee et al., 2020).
Structural Prediction of XTH Proteins
TMHMM Sever v2.0 (Krogh et al., 2001)14 was used to predict transmembrane helices (TMHs), SignalP-5.0 (Armenteros et al., 2019)15 was used to predict signal peptides, and NetPhos-3.1 (Blom et al., 1999)16 was used to predict phosphorylation sites in the amino acid sequence of each SsuXTH protein. We used the online software SOPMA (Levin et al., 1986; Levin and Garnier, 1988)17 and SWISS-MODEL (Waterhouse et al., 2018)18 to predict the secondary and three-dimensional (3D) structures of XTH proteins.
Gene Expression Profile Analysis and Quantitative Real-Time PCR
In August 2020, different tissues were collected for tissue-specific expression profile analysis from the germplasm bank of S. superba clones (119°06′E, 28°03’N) in Zhejiang Longquan. The tissues, including xylem, phloem, mature leaves, buds, fruits, and roots of tissue culture seedlings sub-cultured for 60 days (Yang et al., 2021). Each sample was set up in biological triplicates, and all samples were frozen with liquid nitrogen and stored at −80°C.
Total RNA was extracted from the different tissues of S. superba using the Easy Plant RNA Kit (Polysaccharides & Polyphenolics-rich; Code No. DR0407050, Easy-Do, China). Then, RNA samples with A260/A280 ratios between 1.8 and 2.1 were used to synthesize first-strand cDNA with PrimeScriptTM RT master mix (Perfect Real Time; Code No. RR036A, Takara, Japan). Quantitative real-time PCR (qRT–PCR) was performed using 2X TB Green Premix Ex Taq II (Tli RNase H Plus; Code No. RR820A, TaKaRa, Japan) and Applied Biosystems Q7 (United States). We used a previously described reaction system and qRT–PCR procedure, and SsuACT (Ssu18G01981) was used as an internal reference gene to normalize the gene expression level (Yang et al., 2021). The primers are listed in Supplementary Table 1.
Transcriptome Data and GO Classification Analysis
For the identification of cell wall remodeling- and fiber development-related XTH genes, we used RNA-seq data from three materials, which were secondary xylem, phloem and cambium of three 12-year-old half-sib progenies (S1, S2 and S3) from SC25 family in S. superba with different vascular characteristics. Specifically, the fiber length of S2 was significantly greater than that of S1 and S3, and there was no significant difference between those of S1 and S3 (p > 0.01). The cell wall thickness of S2 was significantly greater than that of S3 and was not significantly different from that of S1 (p > 0.01). The RNA-seq data in this study were deposited in the NCBI Sequence Read Archive (SRA) database under numbers SRR18212748 ~ SRR18212756.
Fragments per kilobase of transcript per million mapped reads (FPKM) values of SsuXTHs were used to evaluate transcript abundance. DESeq2 (Love et al., 2014) was used to conduct differential gene expression analysis, and gene ontology (GO) classification analysis was performed based on the differential gene expression analysis by the clusterProfiler R package (Wu et al., 2021). Finally, heatmaps of SsuXTH expression patterns were drawn by TBtools software (Chen et al., 2020).
Statistical Analyses
The average Ct value was calculated from three biological replicates and three technical replicates. Relative gene expression levels were calculated using the 2−△△Ct method (Livak and Schmittgen, 2001). Significance differences were determined by one-way analysis of variance with SPSS (Statistical Package for the Social Sciences) version 17.0 (Chicago, IL, United States).
Results
Identification and Sequence Analysis of XTH Genes
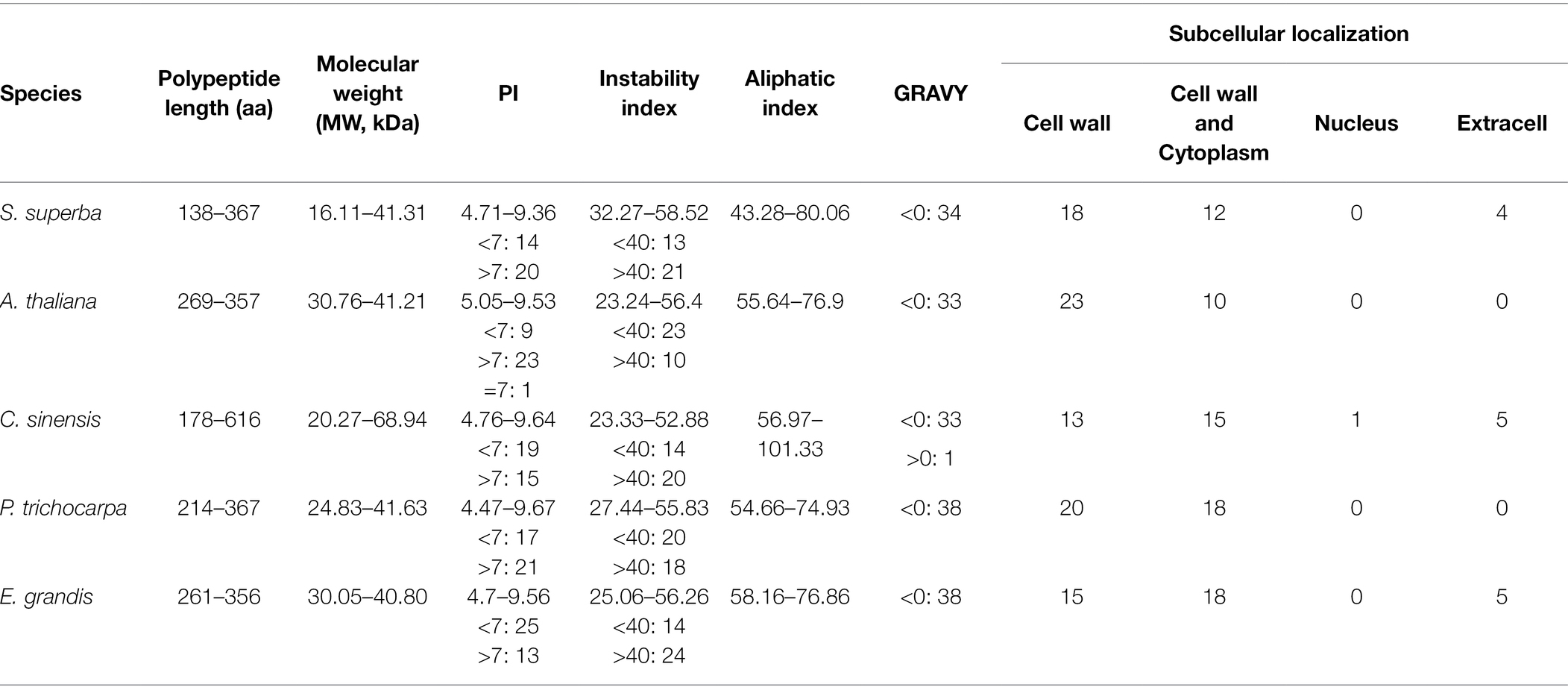
In our study, XTH genes were identified from the S. superba, C. sinensis, P. trichocarpa and E. grandis genomes. Compared with A. thaliana (33), P. trichocarpa (38), E. grandis (38) and C. sinensis (34), the XTH family in S. superba (34) was not obviously contracted or expanded (Supplementary Tables 2, 3).
The sequences of XTH proteins in the five plant species were similar (Table 1; Supplementary Table 4). Acidic proteins (PI < 7) accounted for 47.46%, alkaline proteins (PI > 7) accounted for 51.98%, and the remaining 0.56% were neutral proteins (PI = 7; AT4G37800.1_Atha). Stable (<40) and unstable (>40) proteins accounted for 47.46 and 52.54%, respectively. Most were hydrophilic in nature (99.44%), and only one gene corresponded to a hydrophobic protein (TEA007187.1_Csin). The aliphatic index of the peptides ranged from 43.28 (SsuXTH13) to 101.33 (TEA007187.1_Csin).
In S. superba, the polypeptide lengths and molecular weights of SsuXTH were short and low, especially those of SsuXTH22, compared with those of the other four plants. The number of alkaline proteins was greater than that of acidic proteins, consistent with results in A. thaliana and P. trichocarpa. The number of stable proteins was less than that of unstable proteins, consistent with results in C. sinensis and E. grandis.
Furthermore, the subcellular localization results of the XTH proteins in the five plant species were similar, and most were localized in the cell wall (Table 1; Supplementary Table 4). All XTH proteins of A. thaliana and P. trichocarpa were localized in the cell wall. However, 30, 28 and 33 XTH proteins were localized in the cell walls of S. superba, C. sinensis and E. grandis, respectively. This indicated that most XTH proteins in the five plants played an important role in cell wall remodeling.
Phylogenetic Analysis of XTH Proteins
According to the topological structure and sequence homology of the phylogenetic tree, the 177 XTH proteins were divided into three evolutionary branches, named Clade I (green), Clade II (pink) and Clade III (blue; Figure 1). From the tree, we found that Clade I had the largest number of proteins (86 out of 177), and the correlation between protein clustering and species was weak, indicating that the XTH proteins in the branch was primitive and less conserved, or the gene origin occurred before the differentiation of the five plants; the XTH proteins in Clade II (83 out of 177) were mostly clustered by species, indicating that the XTH proteins in this branch evolved late and were highly conserved; Clade III had the fewest members of the XTH proteins family, with only 8 proteins. In addition, it could be seen that two woody plants of Theaceae, S. superba and C. sinensis, often gathered in the same branch, while poplar and E. grandis were relatively closely related. It should be noted that the ancestral group included AtXTH1, AtXTH2, AtXTH3, AtXTH11, POPTR_0002s24570.1_Ptri, XP_010026021.1_Egr, XP_010039133.1_Egr, XP_010069639.1_Egr, SsuXTH20 and SsuXTH22, according to previous research (Baumann et al., 2007), and it was located in Clade I.
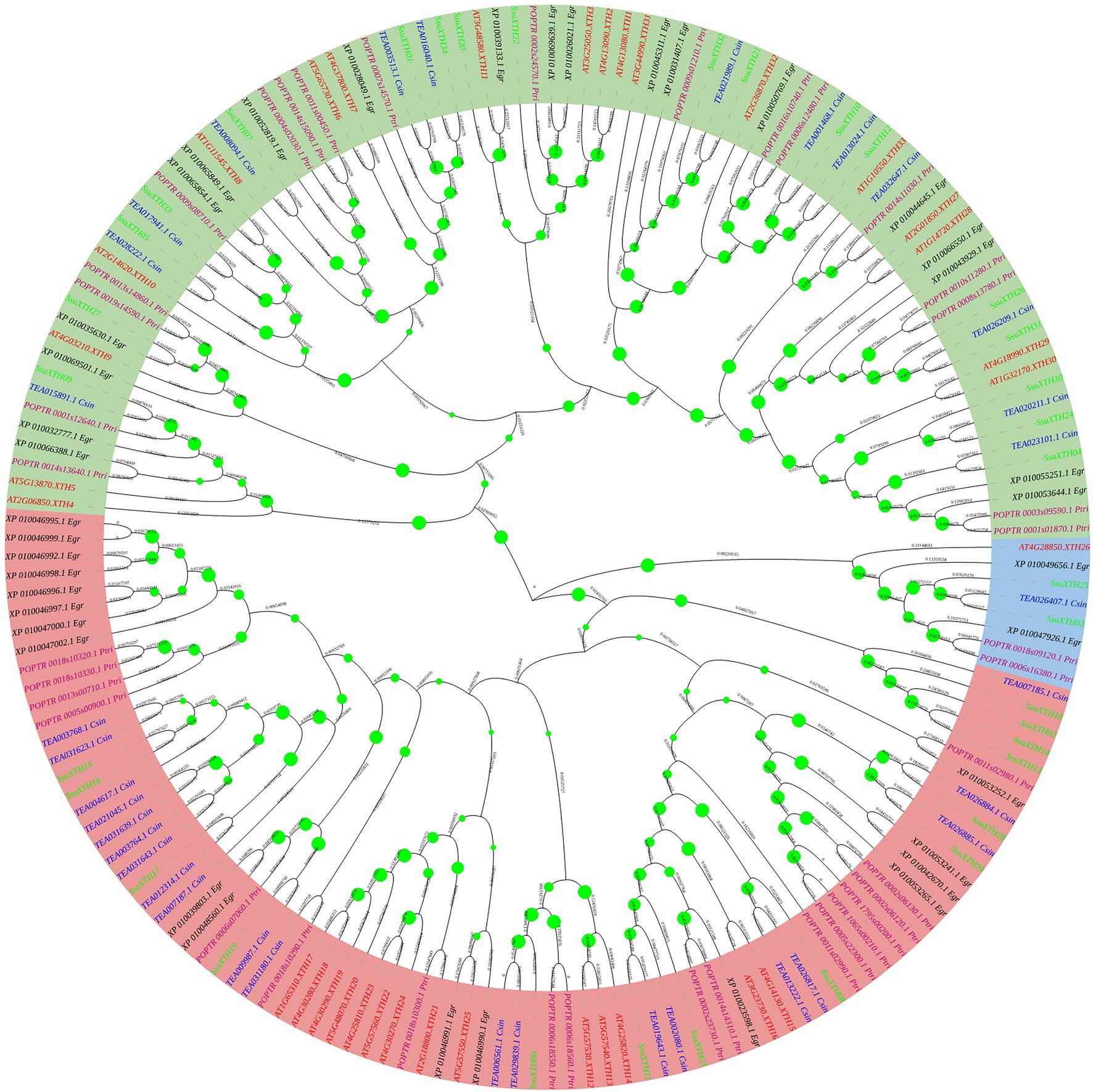
Figure 1. Phylogenetic tree of XTH proteins based on the full-length protein sequences using the neighbor-joining method. The three different groups are indicated by different background colors. The proteins of S. superba, tea plant, poplar, A. thaliana and E. grandis are indicated by different font colors.
Then, we divided the SsuXTHs into three groups according to the clustering branches of the phylogenetic tree (Ancestral group, Group I/II and Group III; Figure 2A), and group III was also divided into two branches (Group IIIA and Group IIIB). Then, the branches showed a direct relationship with the subcellular localization. For example, the ancestral group had only 2 proteins (SsuXTH20 and SsuXTH22) localized in extracellular spaces; Group III had 9 proteins localized in the cell wall; and Group I/II had 23 proteins localized in the cell wall (9 out of 34), extracellular space (2 out of 34) and cell wall and cytoplasm (12 out of 34).
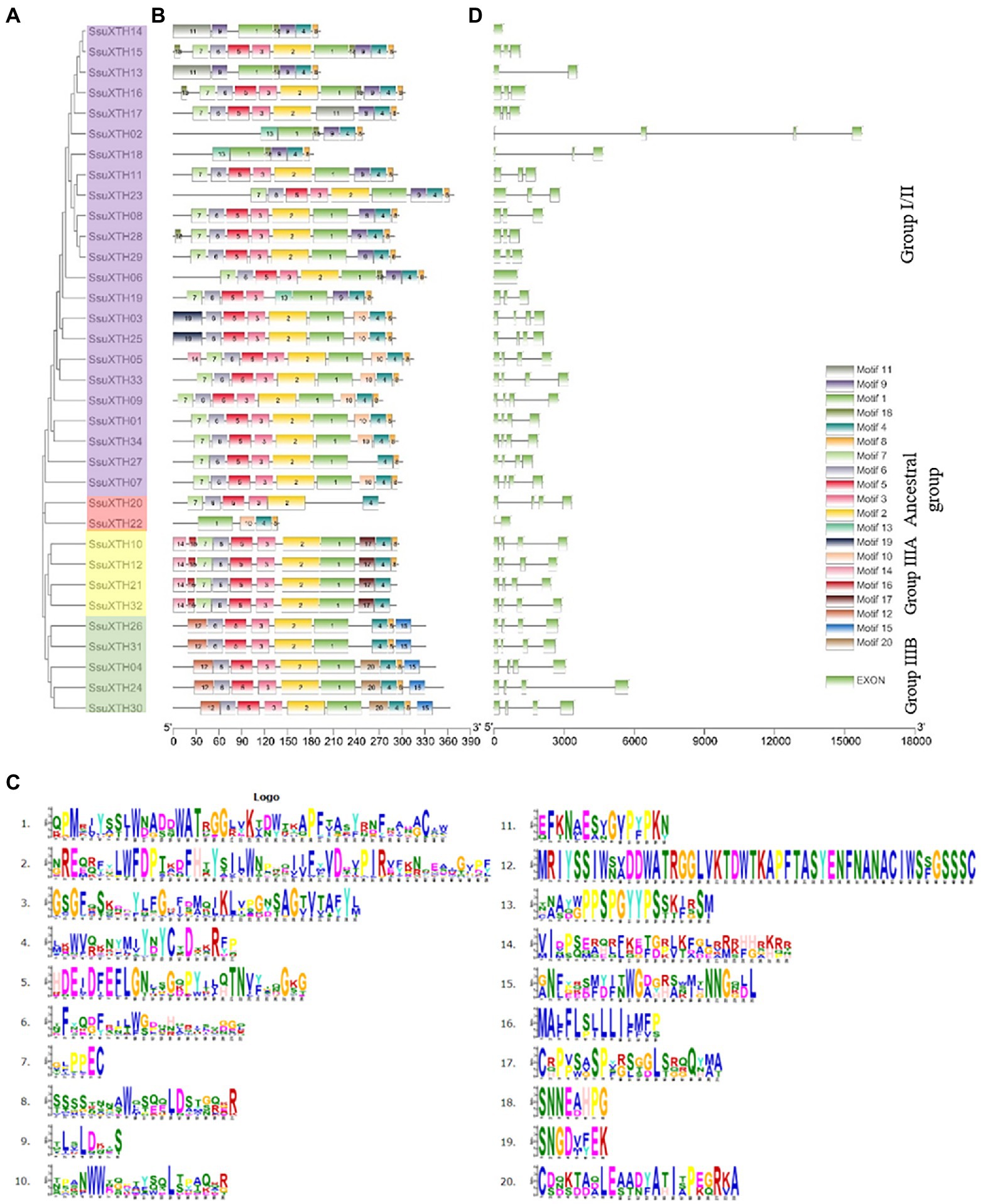
Figure 2. Evolutionary analysis, motif compositions, and gene structures of 34 SsuXTHs. (A) The phylogenetic tree of SsuXTHs, and four different groups are indicated by different background colors. (B) The motif location in SsuXTHs. (C) The gene structures of SsuXTHs. (D) The legend depicting the protein sequence of the corresponding motif.
Conserved Motif and Gene Structural Analyses
We detected 20 conserved motifs among SsuXTHs, and each motif appeared in 2–34 proteins (Figures 2B,C). Notably, motif 5 widely existed in Groups I/II and III, and DEIDFEFLG was the conserved motif that catalyzed the enzymatic reaction of XET and the characteristic motif of this family. Motif 4 existed in all SsuXTH proteins, indicating that this core conserved motif was also very important for XTH proteins, which meant that SsuXTH proteins had similar or the same structure and function. In addition, we also found that motifs 12, 14, 15, 16, 17 and 20 existed only in Group III, indicating that these motifs might have evolutionary specificity in the members of their group. In general, the motifs were highly conserved, which indicated that the functions of SsuXTH proteins were relatively similar.
To identify the structural characteristics of the SsuXTHs, the intron/exon architecture of the genes was analyzed using TBtools (Figure 2D). Analyzing these intron arrangements provides significant information regarding the evolution, regulation, and function of the XTHs (Liu et al., 2015). All the genes of Group III contained three introns and four exons. However, the numbers of introns and exons in the ancestral group and Group I/II were inconsistent.
Chromosomal Localization and Ka/Ks Calculation
The 34 SsuXTHs were unevenly mapped in the 18 chromosomes of S. superba (Figure 3). Chromosomes 12 and 16 did not carry any XTH genes. Among the rest, chromosome 7 contained the largest number (7; 20.59%) of SsuXTH genes, followed by chromosomes 1, 3 and 13, which contained three members (8.82%). Chromosomes 2, 4, 8, 9, 10 and 14 contained two members (5.88%), and the other chromosomes only contained one member. In addition, there was no evidence suggesting a positive correlation between the gene number and chromosome length.

Figure 3. The chromosomal distribution of SsuXTH genes. The left scale represents the length of the chromosomes.
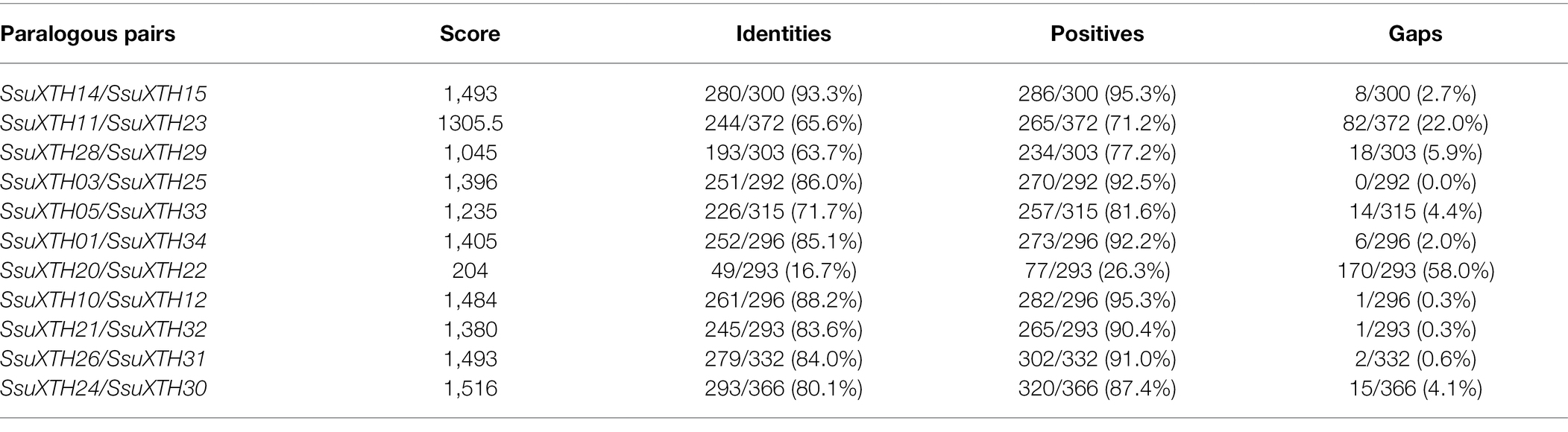
Moreover, we found two gene clusters (Figure 3). One was located on chromosome 7, including five XTH genes (SsuXTH14-18). The other was on chromosome 13, including two XTH genes (SsuXTH28-29). Thus, tandem repeats may be one of the important mechanisms of SsuXTH gene expansion. Gene duplication has long been considered one of the main forces in the evolution and expansion of the gene family (Crow and Wagner, 2006). Therefore, we investigated the different types of gene duplications in the SsuXTH gene family. According to phylogenetic analysis, we found 11 XTH paralogous gene pairs (Figure 2), with a proportion of 64.5%. We inferred that segmental gene duplication may be another important mechanism of SsuXTH gene expansion. Additionally, analyzing the amino acid sequence identity of paralogous gene pairs found that 10 paralogous pairs of SsuXTHs displayed high amino acid sequence identities (63.7–93.3%). On the flip side, the sequence identity of SsuXTH20/SsuXTH22 displayed lower levels (16.7%; Table 2); of course, their gene structures and conserved motifs were significantly different.
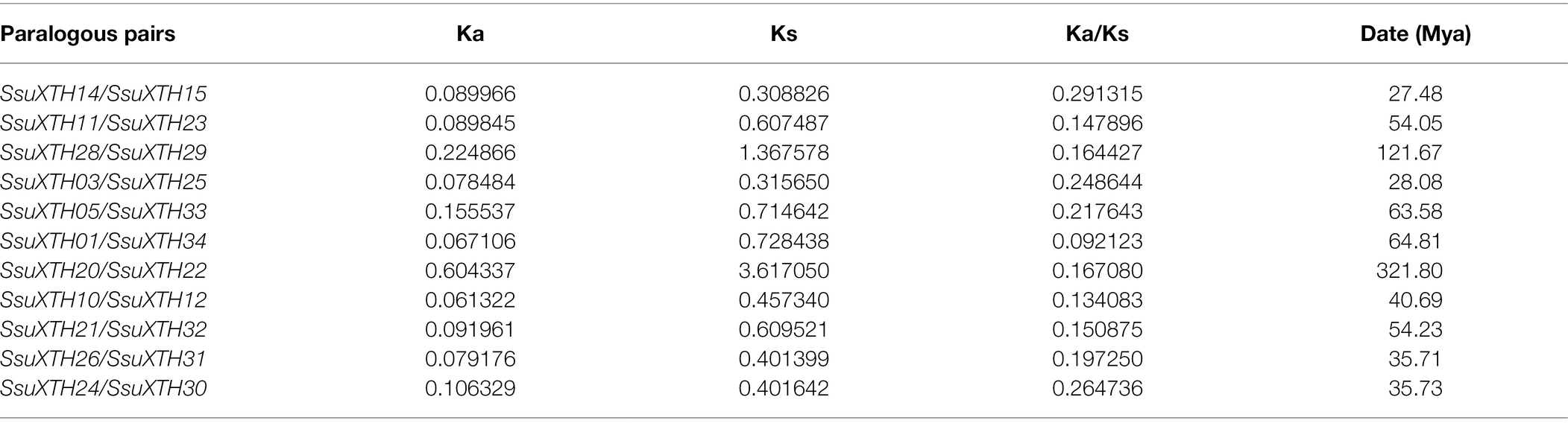
Additionally, the Ka/Ks ratios were calculated to understand the evolutionary pressure and gene divergence mechanism. The Ka/Ks ratio helps determine whether Darwinian selection pressures are involved in duplication events (Tian et al., 2017; Chatterjee et al., 2020). If the value of the Ka/Ks ratio is >1, it implies positive or Darwinian selection, and the gene is more prone to nonsynonymous mutation. If the ratio is equal to 1, it implies neutral selection, and if the ratio is <1, it determines purifying selection, and the gene is more prone to synonymous mutation (Bowers et al., 2003; Liu et al., 2014). The results showed that the Ka/Ks ratios of all gene pairs were less than 1 (Table 3), indicating negative or purifying selection.
Finally, we estimated the replication time of S. superba XTH paralogous gene pairs according to the Ks value (Table 3), in which the replication time of SsuXTH20/SsuXTH22 was 321.80 Mya, which was close to the origin time of gymnosperms, and the resulting times were very ancient. Interestingly, the gene pairs were also located in the ancestral group by phylogenetic analysis. The replication time of the SsuXTH28/SsuXTH29 gene pair was 121.67 Mya, which was in the Early Cretaceous. It was reported that the Schima genus possibly first appeared by the Late Cretaceous and continued to differentiate and expand during the Tertiary and probably first appeared in Asia (its modern distribution area) by the Late Oligocene (Shi et al., 2017). Therefore, the replication time of most gene pairs occurred between 27.48 and 64.81 Mya.
Analysis of cis-Acting Regulatory Elements
Various cis-acting regulatory elements were detected in the promoter regions of SsuXTH genes (Supplementary Tables 5, 6; Figure 4). Phytohormone responsive elements, such as ABREs (abscisic acid responsive elements), AuxREs (auxin responsive elements), GAREs (gibberellin responsive elements), MeJAREs (MeJA responsive elements) and SAREs (salicylic acid responsive elements), were included in the promoter regions, suggesting that the expression of SsuXTH genes might be regulated by multiple phytohormones. Additionally, some stress-related cis-acting elements, such as DSREs (drought and stress responsive elements) and LTREs (low-temperature responsive elements), were also found in the SsuXTH promoter regions, and these results indicated that SsuXTH genes might be closely related to the responses to multiple abiotic stresses. Moreover, each SsuXTH contained multiple copies of LREs (light responsive elements). Notably, the promoter region of SsuXTH genes contained some transcription factor-binding sites, such as those of MYBs, MYCs, HD-Zips, and GATAs. In particular, multiple MYB binding sites appeared in all SsuXTHs (Figure 4).
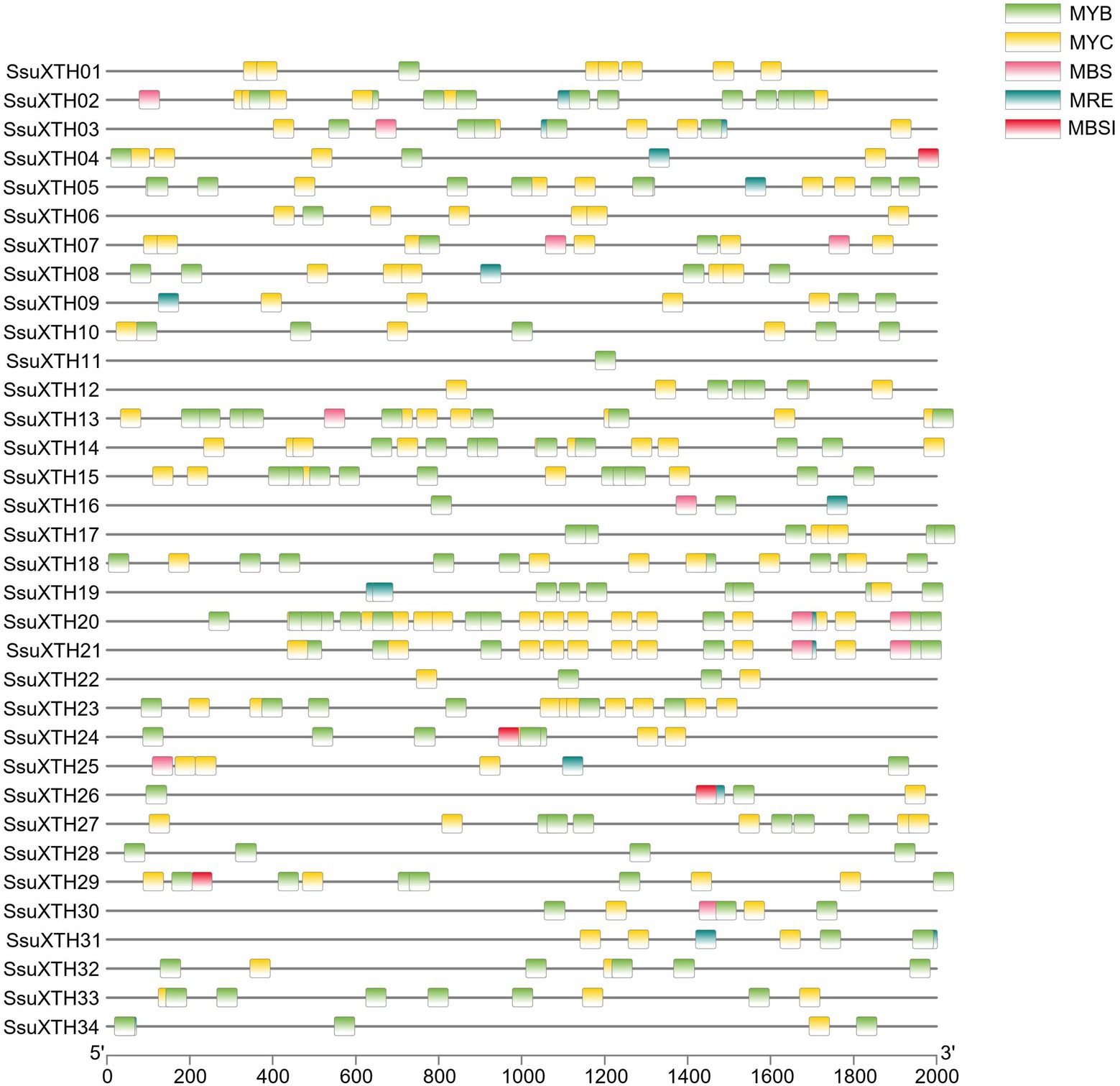
Figure 4. Analysis of the MYB and MYC binding sites in the promoter regions of SsuXTH genes. The 2-kb sequences of SsuXTH gene promoter regions were extracted and analyzed, and the different binding sites were color-coded. MBS: MYB binding site involved in the drought response; MRE: MYB binding site involved in light responsiveness; MBSI: MYB binding site involved in flavonoid biosynthetic gene regulation.
Structural Prediction of SsuXTH Proteins
To study the functions of SsuXTH proteins, we predicted their transmembrane helices (TMHs), signal peptides and phosphorylation sites (Supplementary Table 7). Seventeen SsuXTH proteins predicted by TMHMM had only one TMH, and the TMHs might be a signal peptide. Signalp-5.0 predicted that 24 SsuXTH proteins had signal peptides, and the probability of signal peptides in 16 genes was greater than 0.9. The results predicted by the two tools were not completely consistent, possible because of the different algorithms that they employed.
Netphos-3.1 prediction results (Supplementary Table 7) showed that the number of phosphorylation sites of 34 SsuXTH proteins varied from 18 to 81, among which the number of SsuXTH22 was the lowest, and that of SsuXTH30 was the highest. In addition, except for SsuXTH09, SsuXTH22 and SsuXTH34, the numbers of serine residue sites were significantly greater than those of the other two amino acid residues of the other proteins.
The secondary structures of SsuXTH proteins were rich, including alpha helix, beta turn, extended strand and random coil structures (Supplementary Table 8). Among them, the random coil was the most common (46.39% ~ 57.29%), and the beta turn was the least common (3.93% ~ 7.58%). The quantitative order was random coil > extended strand/alpha helix > beta turn for the secondary structures in all proteins, indicating that the structures and functions of the SsuXTH proteins were conserved.
3D proteins could be effectively used for understanding the structure and mode of action of XTH enzymes, which will help reveal the functions of SsuXTHs. The structural properties of all SsuXTHs were displayed in homology-based tertiary (3D) protein models (Supplementary Table 9), which were predicted via the SWISS-MODEL website. Predicted models were based on the reported templates to heuristically maximize the alignment coverage, percentage identity, and confidence score for the tested sequences. All 3D protein models were constructed with 38–91% seq-identity, and the residue coverage varied from 63 to 99%, suggesting that structure prediction of SsuXTH proteins was highly reliable. Most SsuXTH proteins contained similar structures, especially those from the same branch, implying that SsuXTH proteins may evolved from same ancestor sequence and/or under purification selection force to keep stabilization during long-term acclimation after the initially divergent (Zhu et al., 2019). For example, the 3D structure of proteins from Group IIIA were same. In addition, the 3D structure of SsuXTH13, SsuXTH18 and SsuXTH22 were quite different from that of other proteins, and they were in different groups, but their polypeptide lengths and molecular weights were shortest and lowest. These predicted 3D models provided an important basis for the functional analysis of the XTH proteins.
Tissue-Specific Expression Profiles of SsuXTHs
The SsuXTHs had different expression patterns in different tissues (Figure 5A), which indicated that the SsuXTHs gene family might have different functions in developmental processes. The results showed that most SsuXTHs (21, 62%) were mainly expressed in leaves. This was followed by the phloem (6, 18%) and buds (5, 14%). Interestingly, the tandem repeat genes (SsuXTH14, SsuXTH15, SsuXTH16, SsuXTH17 and SsuXTH29) were mainly expressed in the phloem and xylem. And the expression of almost all genes in roots and fruits were very low.
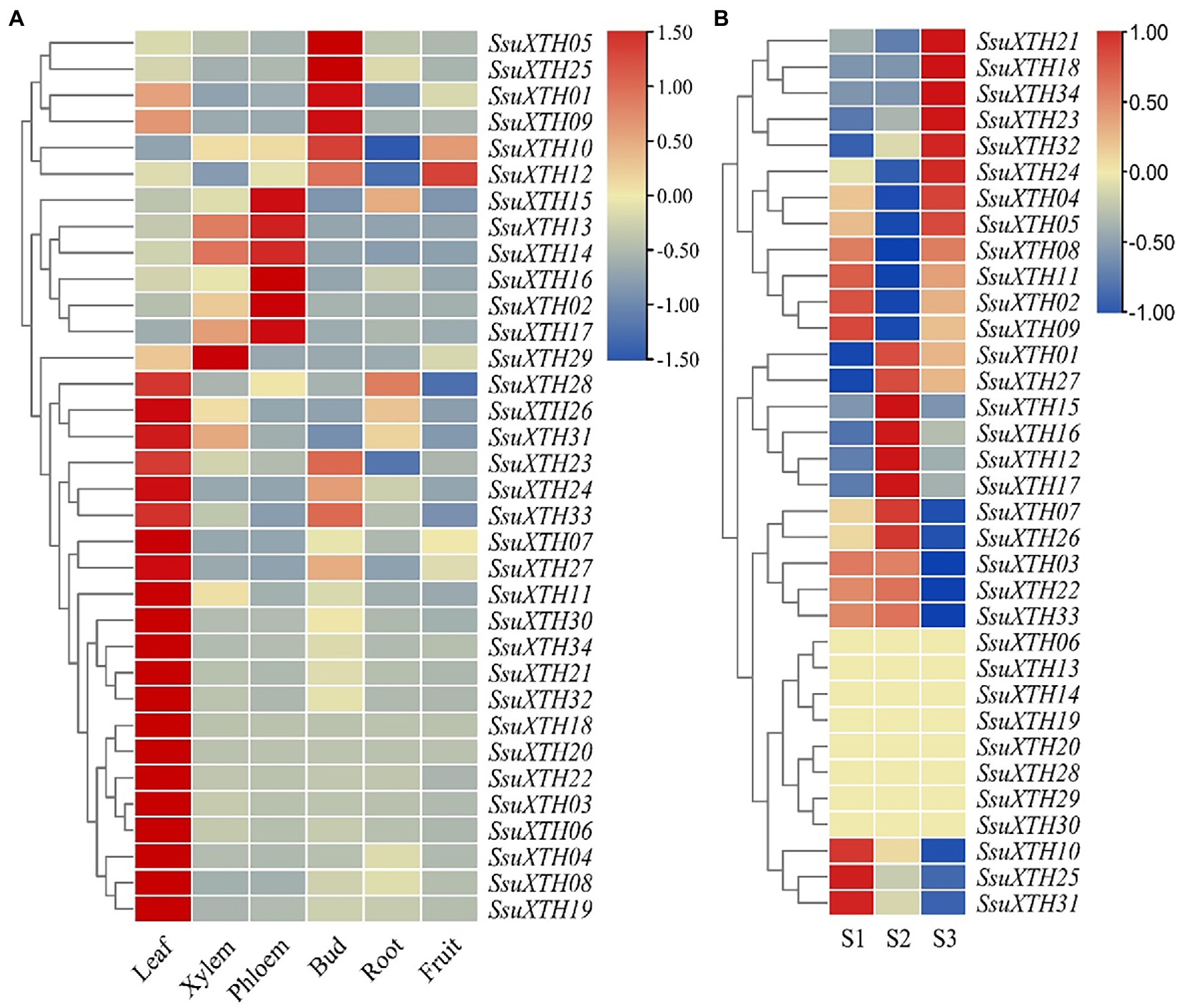
Figure 5. Expression profiles of SsuXTHs in different tissues and materials with different vascular characteristics. (A) The leaves, phloem, xylem, buds, roots and fruits were collected and then used for qRT-PCR. The expression level was calculated according to the 2−∆∆Ct method. Relative mRNA abundance of each gene was normalized with SsuACT gene. (B) RNA-seq data of three half-sib progenies (S1, S2 and S3) from SC25 family in S. superba with different vascular characteristics were used for expression profiles analysis. The expression level was determined by the FPKM calculated by StringTie v2.1.2 tool and log2 transformation was used for normalization.
Differential Expression of SsuXTHs Between Materials With Different Vascular Characteristics
The expression levels of 34 SsuXTHs in S1, S2 and S3 were retrieved from transcriptome databases (SRR18212748 ~ SRR18212756). The results revealed that only 14 differentially expressed SsuXTH genes were highly expressed among different materials (Figure 5B; Supplementary Table 10). Among, 10 SsuXTH genes were differentially expressed in S2 vs. S1, 12 genes were differentially expressed in S2 vs. S3, and 8 genes were differentially expressed in S1 vs. S3. Only 5 SsuXTH genes (SsuXTH10, SsuXTH16, SsuXTH17, SsuXTH23 and SsuXTH31) were differentially expressed in three comparison combinations. In addition, the expression of SsuXTH12, SsuXTH15, SsuXTH16 and SsuXTH17 in S2 was significantly greater than that in S1 and S3, which was consistent with the fiber length difference, indicating that these genes might positively regulate fiber elongation.
The GO enrichment analysis was conducted on the 14 differentially expressed genes, and it was found that they were involved in 16 GO terms (Supplementary Table 11), including 5 biological processes (BPs), 5 cell components (CCs) and 6 molecular functions (MFs). The BPs mainly included carbohydrate metabolism, polysaccharide metabolism and glucan metabolism, and the MFs mainly included transferase activity and hydrolase activity. The CCs mainly included the cell wall, extracellular region and apoplast, which were basically consistent with the predicted subcellular localizations.
Functional Interaction Network of SsuXTH Proteins
To understand and explore the interaction patterns of XTH genes in S. superba, a protein interaction network was constructed using the STRING server based on an Arabidopsis association model (Figure 6). The Arabidopsis model had to be employed due to the absence of the S. superba database in the STRING server. The interaction network was therefore constructed mainly based on XTR6 (AT4G25810) and TCH4 (AT5G57560) in A. thaliana, the homologs of SsuXTH15, SsuXTH16 and SsuXTH17. The results showed that various transcription factors specifically interacted with SsuXTH16 (Figure 6), such as ethylene-responsive transcription factors (ERF012 (AT1G21910), ERF104 (AT5G61600) and ERF105 (AT5G51190)), MYB77 (AT3G50060), and WRKY40 (AT1G80840). They interacted separately with the phosphate-responsive 1 family proteins PHI-1/EXL1 (AT1G35140) and EXO (AT4G08950). The PHI-1/EXL1 and EXO genes were identified as potential mediators of brassinosteroid (BR)-promoted growth (Schröder et al., 2009), and BR could promote cell expansion by increasing the expression of AtXTH22 and AtXTH24 (He et al., 2003). Notably, SsuXTH15/SsuXTH17 and SsuXTH16 could interact with each other.
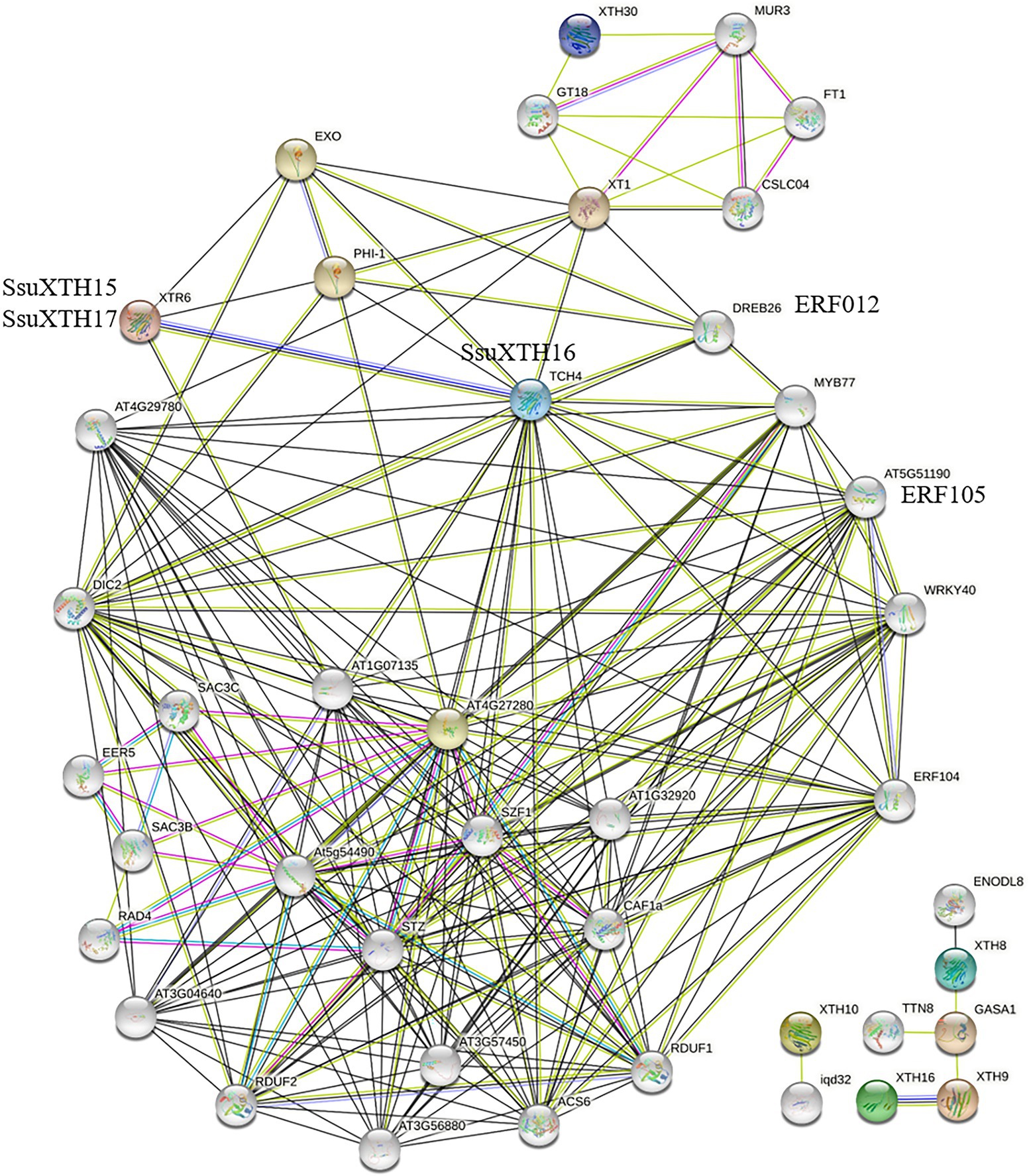
Figure 6. Prediction of the functional interaction network of SsuXTH based on the orthologs in A. thaliana. The colored nodes: query proteins and first shell of interactors; White nodes: second shell of interactors; Light blue edges: known interactions from curated databases; Pink edges: known interactions experimentally determined; Green edges: predicted interactions with gene neighborhood; Red edges: predicted interactions with gene fusions; Blue edges: predicted interactions with gene co-occurrence; Blue edges: text mining; Black edges: coexpression; Purple edges: protein homology.
Discussion
XTH gene families have already been identified and functionally characterized in several plants, including A. thaliana, O. sativa, G. hirsutum, B. rapa, B. oleracea, B. juncea and so on (Yokoyama and Nishitani, 2001; Yokoyama et al., 2004; Lee et al., 2010; Li et al., 2020; Wu et al., 2020). However, the XTH genes in S. superba have not yet been studied. In this study, we analyzed the gene structure, phylogenetic relationship, genomic distribution, and expression of XTH genes in S. superba at the genomic level. The results showed that a total of 34 XTH genes were in S. superba, and they were divided into three groups, which had different gene structures and subcellular locations (Figure 2A). For example, the genes from the ancestral group and Group III were localized in the extracellular space and cell wall, respectively, whereas genes in Group I/II were mainly in the cell wall, cytoplasm, and extracellular space. These findings were similar to those reported in pineapple and poplar (Li et al., 2019; Cheng et al., 2021). Moreover, all the genes in Group III contained three introns and four exons, and the types and numbers of conserved motifs within the group were similar.
Compared with those in A. thaliana, P. trichocarpa, E. grandis and C. sinensis, the XTH families in S. superba were not obviously contracted or expanded (Supplementary Table 3). Segmental and tandem duplication events did not lead to XTH gene family expansion but might increase functional divergence, which was an essential factor in adapting to environmental changes (Conant and Wolfe, 2008). Of course, segmental and tandem duplication events also are found among poplar XTH family genes (Cheng et al., 2021). Furthermore, the phylogenetic analysis of XTH encoded proteins among the five plants revealed that S. superba and C. sinensis, all belonging to Theaceae, had many more genetic relationships, while Populus and E. grandis had much closer relationships (Figure 1). In addition, the Ka/Ks ratio indicates different selection pressures and divergence times on genes throughout evolutionary changes. However, in S. superba, the Ka/Ks ratios indicated that strong purifying selection pressures had occurred during evolution, thereby enabling a number of different environmental factors to regulate the XTHs in the S. superba genome. This might also be the reason why most SsuXTH proteins contained similar structures (Zhu et al., 2019). The replication time of most SsuXTH gene pairs was between 27.48 and 64.81 Mya, which was consistent with the origin and expansion time of the Schima genus (Shi et al., 2017).
Cis-acting regulatory elements play the role of key molecular switches in the transcriptional regulation of gene expression (Ding et al., 2018; Verma et al., 2019). LeXTH2 could be enhanced by GA and inhibited by auxin in tomato (Catala et al., 2001). AtXTH23 is also upregulated by ABA in Arabidopsis (Yokoyama and Nishitani, 2001). These studies show that hormones could regulate XTH expression, and the cis-acting element in the promoter region of XTH is an important factor in the regulatory mechanism (Vissenberg et al., 2005). In this study, we found various phytohormone regulatory elements in the promoter regions of SsuXTH genes, including ABREs, AuxREs, GAREs, MeJAREs and SAREs. SsuXTH16 also interacted with ethylene-responsive transcription factors (ERFs) in protein interaction network, and AtXTH was upregulated by ethylene in root hairs (Vissenberg et al., 2001). Notably, the promoter region of SsuXTH genes contained MYB, MYC, HD-Zip and GATA transcription factor-binding sites, and MYB was fully covered in all SsuXTHs (Figure 4). The protein interaction analysis also proved that SsuXTH could be regulated by MYB, as SsuXTH16 interacted with AtMYB77 (Figure 6). Smit et al. (2020) shows that AtMYB77 is a key and core transcription factor in the vascular development transcriptional network and can switch on LBD4 (LATERAL ORGAN BOUNDARIES DOMAIN4), which regulates vascular cell number and organization in Arabidopsis. However, there are few reports on transcription factors regulated XTH expression. Only ANAC071 is bound to the AtXTH19 and AtXTH20 promoters to induce their expression by auxin in the distal part of an incised stem and their involvement in cell proliferation in the tissue reunion process (Pitaksaringkarn et al., 2014). In addition, each SsuXTH contained multiple copies of LREs, which were involved in light responsiveness, and S. superba had strong light-seeking ability in the early stage (Yao et al., 2018), suggesting that SsuXTH genes were an important component of the light response in S. superba. Finally, the existence of ARE and WRE suggested that the expression of SsuXTH genes might be induced by anoxic conditions and wounds, and this result was consistent with those of Oogawara et al. (2005). S. superba could quickly recover vitality after forest fire (Zhou et al., 2020a), and we speculated that anoxic conditions and burns induced the expression of SsuXTH genes and then promoted plant regeneration. All these cis-acting regulatory elements of SsuXTH implied that they had important functions in plant growth, development and stress resistance.
The SsuXTH genes had different expression patterns among tissues in S. superba (Figure 5A). This indicated that the XTH gene family provided opportunities to break the functional constraints of the original gene during the course of evolution. The same patterns were also found in barley and tobacco (Wang et al., 2018; Fu et al., 2019). And 5 SsuXTH genes (SsuXTH10, SsuXTH16, SsuXTH17, SsuXTH23 and SsuXTH31) were differentially expressed in three comparison combinations among different materials (Figure 5B), but only SsuXTH16 and SsuXTH17 were mainly expressed in the phloem and xylem. It was interesting that we found that most of the tandem repeat genes were highly expressed in the phloem and xylem, and from the phenotype correlation results, SsuXTH15, SsuXTH16 and SsuXTH17 might positively regulate fiber elongation (Figure 5B). The GO enrichment analysis showed that 14 differentially expressed genes were involved in carbohydrate metabolic processes and possessed transferase and hydrolase activity (Supplementary Table 11), which indicated that the SsuXTH enzyme mainly participated in glycoside metabolic processes by transferring and hydrolyzing xyloglucan in the cell wall and then regulating fiber elongation.
Conclusion
This study systematically identified and characterized the XTH family in S. superba. A total of 34 SsuXTHs were obtained, which were classified into three subfamilies based on the phylogenetic relationship and unevenly distributed on 18 chromosomes. Furthermore, the intron–exon structure and conserved motif composition of them supported the classification and the members belonging to the same subfamily shared similar gene structures. Segmental and tandem duplication events did not lead to SsuXTH gene family expansion, and strong purifying selection pressures during evolution led to similar structure and function of SsuXTH gene family. The interaction network and cis-acting regulatory elements analysis provided useful clues for revealing the SsuXTH regulation pathway. Finally, expression profiles and GO enrichment analysis showed most of the tandem repeat genes were mainly expressed in the phloem and xylem and they mainly participated in glycoside metabolic processes through the transfer and hydrolysis of xyloglucan in the cell wall and then regulated fiber elongation. Here, we provide a new insight into the function of SsuXTH and lay the foundation for further exploration of the decisive genes of fiber development. However, most of the analyses were primarily in silico, next we would identify the function of tandem repeat SsuXTH genes through genetic transformation and in vitro enzyme activity etc.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The RNA-seq data in this study were deposited in the NCBI Sequence Read Archive (SRA) database under numbers SRR18212748 ~ SRR18212756.
Author Contributions
ZY, RZ, and ZZ designed the research. ZY conducted the experiment, analyzed the data, and wrote the manuscript. All authors contributed to the discussion of the results, reviewed the manuscript, and approved the final article.
Funding
This study was supported by the Zhejiang Science and Technology Major Program on Agricultural New Variety Breeding (grant number 202102070–9), the Science and Technology Innovation Project in Jiangxi Province (grant number 2019–19), and the Sixth Stages of Planting and Seedling Science and Technology of Fujian Province (grant number 2019–06).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.911761/full#supplementary-material
Footnotes
2. ^https://phytozome-next.jgi.doe.gov/info/Ptrichocarpa_v4_1
3. ^https://phytozome-next.jgi.doe.gov/info/Egrandis_v2_0
4. ^https://www.arabidopsis.org/
6. ^http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
7. ^http://web.expasy.org/protparam
8. ^http://www.csbio.sjtu.edu.cn/bioinf/euk-multi2/
10. ^http://meme.nbcr.net/meme/cgi-bin/meme.cgi
12. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
13. ^https://string-db.org/?tdsourcetag=s_pctim_aiom
14. ^https://services.healthtech.dtu.dk/service.php?TMHMM-2.0
15. ^https://services.healthtech.dtu.dk/service.php?SignalP-5.0
16. ^https://services.healthtech.dtu.dk/service.php?NetPhos-3.1
17. ^https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html
References
Armenteros, J. J. A., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Atkinson, R. G., Johnston, S. L., Yauk, Y.-K., Sharma, N. N., and Schröder, R. (2009). Analysis of xyloglucan endotransglucosylase/hydrolase (XTH) gene families in kiwifruit and apple. Postharvest Biol Tec. 51, 149–157. doi: 10.1016/j.postharvbio.2008.06.014
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Baumann, M. J., Eklöf, J. M., Michel, G., Kallas, A. M., Teeri, T. T., Czjzek, M., et al. (2007). Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell 19, 1947–1963. doi: 10.1105/tpc.107.051391
Blom, N., Gammeltoft, S., and Brunak, S. (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362. doi: 10.1006/jmbi.1999.3310
Bourquin, V., Nishikubo, N., Abe, H., Brumer, H., Denman, S., Eklund, M., et al. (2002). Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14, 3073–3088. doi: 10.1105/tpc.007773
Bowers, J. E., Chapman, B. A., Rong, J., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521
Campbell, P., and Braam, J. (1999). Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 4, 361–366. doi: 10.1016/s1360-1385(99)01468-5
Catala, C., Rose, J. K., York, W. S., Albersheim, P., Darvill, A. G., and Bennett, A. B. (2001). Characterization of a tomato xyloglucan endotransglucosylase gene that is down-regulated by auxin in etiolated hypocotyls. Plant Physiol. 127, 1180–1192. doi: 10.1104/pp.010481
Chatterjee, A., Paul, A., Unnati, G. M., Rajput, R., Biswas, T., Kar, T., et al. (2020). MAPK cascade gene family in Camellia sinensis: in-silico identification, expression profiles and regulatory network analysis. BMC Genomics 21, 1–17. doi: 10.1186/s12864-020-07030-x
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, Z., Zhang, X., Yao, W., Gao, Y., Zhao, K., Guo, Q., et al. (2021). Genome-wide identification and expression analysis of the xyloglucan endotransglucosylase/hydrolase gene family in poplar. BMC Genomics 22, 1–13. doi: 10.1186/s12864-021-08134-8
Cho, S. K., Kim, J. E., Park, J.-A., Eom, T. J., and Kim, W. T. (2006). Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett. 580, 3136–3144. doi: 10.1016/j.febslet.2006.04.062
Choi, J. Y., Seo, Y. S., Kim, S. J., Kim, W. T., and Shin, J. S. (2011). Constitutive expression of CaXTH3, a hot pepper xyloglucan endotransglucosylase/hydrolase, enhanced tolerance to salt and drought stresses without phenotypic defects in tomato plants (Solanum lycopersicum cv. Dotaerang). Plant Cell Rep. 30, 867–877. doi: 10.1007/s00299-010-0989-3
Chou, K. C., and Shen, H. B. (2010). A new method for predicting the subcellular localization of eukaryotic proteins with Both single and multiple sites: Euk-mPLoc 2.0. PLoS One 5:e9931. doi: 10.1371/journal.pone.0009931
Conant, G. C., and Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi: 10.1038/nrg2482
Crow, K. D., and Wagner, G. P. (2006). What is the role of genome duplication in the evolution of complexity and diversity. Mol. Biol. Evol. 23, 887–892. doi: 10.1093/molbev/msj083
Cui, L. C., Yang, G., Yan, J. L., Pan, Y., and Nie, X. J. (2019). Genome-wide identification, expression profiles and regulatory network of MAPK cascade gene family in barley. BMC Genomics 20, 1–20. doi: 10.1186/s12864-019-6144-9
Ding, X., Li, J. H., Pan, Y., Zhang, Y., Ni, L., Wang, Y. L., et al. (2018). Genome-wide identification and expression analysis of the UGlcAE gene family in tomato. Int. J. Mol. Sci. 19:1583. doi: 10.3390/ijms19061583
Fu, M. M., Liu, C., and Wu, F. B. (2019). Genome-wide identification, characterization and expression analysis of Xyloglucan Endotransglucosylase/hydrolase genes family in barley (Hordeum vulgare). Molecules 24:1935. doi: 10.3390/molecules24101935
Geisler-Lee, J., Geisler, M., Coutinho, P. M., Segerman, B., Nishikubo, N., Takahashi, J., et al. (2006). Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol. 140, 946–962. doi: 10.1104/pp.105.072652
Ghosh Dasgupta, M., Abdul Bari, M. P., Shanmugavel, S., Dharanishanthi, V., Muthupandi, M., Kumar, N., et al. (2021). Targeted re-sequencing and genome-wide association analysis for wood property traits in breeding population of Eucalyptus tereticornis × E. grandis. Genomics 113, 4276–4292. doi: 10.1016/j.ygeno.2021.11.013
Han, Y., Wang, W., Sun, J., Ding, M., Zhao, R., Deng, S., et al. (2013). Populus euphratica XTH overexpression enhances salinity tolerance by the development of leaf succulence in transgenic tobacco plants. J. Exp. Bot. 64, 4225–4238. doi: 10.1093/jxb/ert229
He, J. X., Fujioka, S., Li, T., Kang, S. G., Seto, H., Takatsuto, S., et al. (2003). Sterols regulate development and gene expression in Arabidopsis. Plant Physiol. 131, 1258–1269. doi: 10.1104/pp.014605
He, Q. L., Jones, D. C., Li, W., Xie, F. L., Ma, J., Sun, R., et al. (2016). Genome-wide identification of R2R3-MYB genes and expression analyses During abiotic stress in Gossypium raimondii. Sci. Rep. 6:22980. doi: 10.1038/srep22980
Khan, N., Hu, C.-M., Khan, W. A., Wang, W., Ke, H., Huijie, D., et al. (2018). Genome-wide identification, classification, and expression pattern of Homeobox gene family in Brassica rapa under various stresses. Sci. Rep. 8:16265. doi: 10.1038/s41598-018-34448-x
Krogh, A., Larsson, B., Heijne, G. V., and Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, J. H., Burns, T. H., Light, G., Sun, Y., Fokar, M., Kasukabe, Y., et al. (2010). Xyloglucan endotransglycosylase/hydrolase genes in cotton and their role in fiber elongation. Planta 232, 1191–1205. doi: 10.1007/s00425-010-1246-2
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Peer, Y. V. D., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Levin, J. M., and Garnier, J. (1988). Improvements in a secondary structure prediction method based on a search for local sequence homologies and its use as a model building tool. Biochim. Biophys. Acta 955, 283–295. doi: 10.1016/0167-4838(88)90206-3
Levin, J. M., Robson, B., and Garnier, J. (1986). An algorithm for secondary structure determination in proteins based on sequence similarity. FEBS Lett. 205, 303–308. doi: 10.1016/0014-5793(86)80917-6
Li, Q. Y., Li, H. Y., Yin, C. Y., Wang, X. T., Jiang, Q., Zhang, R., et al. (2019). Genome-wide identification and characterization of Xyloglucan Endotransglycosylase/hydrolase in Ananas comosus during development. Genes. 10:537. doi: 10.3390/genes10070537
Li, M. Y., Xie, F. J., He, Q., Li, J., Liu, J. L., Sun, B., et al. (2020). Expression analysis of XTH in stem swelling of stem mustard and selection of reference genes. Genes. 11:113. doi: 10.3390/genes11010113
Liu, W., Li, W., He, Q. L., Daud, M. K., Chen, J. H., and Zhu, S. J. (2014). Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium raimondii. PLoS One 9:e98189. doi: 10.1371/journal.pone.0116352
Liu, Y., Liu, D. C., Zhang, H. Y., Gao, H. B., Guo, X. L., Wang, D. W., et al. (2007). The α- and β-expansin and xyloglucan endotransglucosylase/hydrolase gene families of wheat: molecular cloning, gene expression, and EST data mining. Genomics 90, 516–529. doi: 10.1016/j.ygeno.2007.06.012
Liu, Z., Shi, L., Liu, Y., Tang, Q., Shen, L., Yang, S., et al. (2015). Genome wide identification and transcriptional expression analysis of mitogen-activated protein kinase and mitogen-activated protein kinase genes in Capsicum annuum. Front. Plant Sci. 6:780. doi: 10.3389/fpls.2015.00780
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, S. N., Wang, J. Y., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Malinowski, R., Fry, S. C., Zuzga, S., Wiśniewska, A., Godlewski, M., Noyszewski, A., et al. (2018). Developmental expression of the cucumber Cs-XTH1 and Cs-XTH3 genes, encoding xyloglucan endotransglucosylase/hydrolases, can be influenced by mechanical stimuli. Acta Physiol. Plant. 40, 130–140. doi: 10.1007/s11738-018-2707-7
Maris, A., Suslov, D., Fry, S. C., Verbelen, J. P., and Vissenberg, K. (2009). Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 60, 3959–3972. doi: 10.1093/jxb/erp229
Mellerowicz, E. J., and Sundberg, B. (2008). Wood cell walls: biosynthesis, developmental dynamics and their implications for wood properties. Curr. Opin. Plant Biol. 11, 293–300. doi: 10.1016/j.pbi.2008.03.003
Michailidis, G., Argiriou, A., Darzentas, N., and Tsaftaris, A. (2009). Analysis of xyloglucan endotransglycosylase/hydrolase (XTH) genes from allotetraploid (Gossypium hirsutum) cotton and its diploid progenitors expressed during fiber elongation. J. Plant Physiol. 166, 403–416. doi: 10.1016/j.jplph.2008.06.013
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Nawaz, M. A., Rehman, H. M., Imtiaz, M., Baloch, F. S., Lee, J. D., Yang, S. H., et al. (2017). Systems identification and characterization of cell wall reassembly and degradation related genes in Glycine max (L.) Merill, a bioenergy legume. Sci. Rep. 7:10862. doi: 10.1038/s41598-017-11495-4
Nishikubo, N., Takahashi, J., Roos, A. A., Derba-Maceluch, M., Piens, K., Brumer, H., et al. (2011). Xyloglucan endotransglycosylase-mediated xyloglucan rearrangements in developing wood of hybrid Aspen. Plant Physiol. 155, 399–413. doi: 10.1104/pp.110.166934
Oogawara, R., Satoh, S., Yoshioka, T., and Ishizawa, K. (2005). Expression of alpha-expansin and xyloglucan endotransglucosylase/hydrolase gene associated with shoot elongation enhanced by anoxia, ethylene and carbon dioxide in arrowhead (Sagittaria pygmaea Miq.) tubers. Ann. Bot. 96, 693–702. doi: 10.1093/aob/mci221
Pitaksaringkarn, W., Matsuoka, K., Asahina, M., Miura, K., Sage-Ono, K., Ono, M., et al. (2014). XTH20 and XTH19 regulated by ANAC071 under auxin flow are involved in cell proliferation in incised Arabidopsis inflorescence stems. Plant J. 80, 604–614. doi: 10.1111/tpj.12654
Rai, K. M., Thu, S. W., Balasubramanian, V. K., Cobos, C. J., Disasa, T., and Mendu, V. (2016). Identification, characterization, and expression analysis of cell wall related genes in Sorghum bicolor (L.) Moench, a food, fodder, and biofuel crop. Front. Plant Sci. 7:1287. doi: 10.3389/fpls.2016.01287
Rose, J. K. C., Braam, J., Fry, S. C., and Nishitani, K. (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol. 43, 1421–1435. doi: 10.1093/pcp/pcf171
Saladié, M., Rose, J. K. C., Cosgrove, D. J., and Catalá, C. (2006). Characterization of a new xyloglucan endotransglucosylase/hydrolase (XTH) from ripening tomato fruit and implications for the diverse modes of enzymic action. Plant J. 47, 282–295. doi: 10.1111/j.1365-313x.2006.02784.x
Schröder, F., Lisso, J., Lange, P., and Müssig, C. (2009). The extracellular EXO protein mediates cell expansion in Arabidopsis leaves. BMC Plant Biol. 9:20. doi: 10.1186/1471-2229-9-20
Seale, M. (2020). Cell Wall remodeling during wood development. Plant Physiol. 182, 1800–1801. doi: 10.1104/pp.20.00260
Shao, M. Y., Wang, X. D., Ni, M., Bibi, N., Yuan, S. N., Malik, W., et al. (2011). Regulation of cotton fiber elongation by xyloglucan endotransglycosylase/hydrolase genes. Genet. Mol. Res. 10, 3771–3782. doi: 10.4238/2011.October.27.1
Shi, X. G., Fu, Q. Y., Jin, J. H., and Quan, C. (2017). Mummified Oligocene fruits of Schima (Theaceae) and their systematic and biogeographic implications. Sci. Rep. 7:4009. doi: 10.1038/s41598-017-04349-6
Smit, M. E., McGregor, S. R., Sun, H., Gough, C., Bågman, A.-M., Soyars, C. L., et al. (2020). A PXY-mediated transcriptional network integrates signaling mechanisms to control vascular development in Arabidopsis. Plant Cell 32, 319–335. doi: 10.1105/tpc.19.00562
Song, L., Valliyodan, B., Prince, S., Wan, J., and Nguyen, H. T. (2018). Characterization of the XTH gene family: new insight to the roles in soybean flooding tolerance. Int. J. Mol. Sci. 19:2705. doi: 10.3390/ijms19092705
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., and Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Sundell, D., Street, N. R., Kumar, M., Mellerowicz, E. J., Kucukoglu, M., Johnsson, C., et al. (2017). AspWood: high-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 29, 1585–1604. doi: 10.1105/tpc.17.00153
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Cepas, J. H., et al. (2019). STRING v11: proteinprotein association networks with increased coverage, supporting functional discovery in genome wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Takahashi, D., Johnson, K. L., Hao, P., Tuong, T., Erban, A., Sampathkumar, A., et al. (2021). Cell wall modification by the xyloglucan endotransglucosylase/hydrolase XTH19 influences freezing tolerance after cold and sub-zero acclimation. Plant Cell Environ. 44, 915–930. doi: 10.1111/pce.13953
Tian, F. X., Chang, E., Li, Y., Sun, P., Hu, J., and Zhang, J. (2017). Expression and integrated network analyses revealed functional divergence of NHX-type Na+/H+ exchanger genes in poplar. Sci. Rep. 7:2607. doi: 10.1038/s41598-017-18149-5
Verma, D., Lakhanpal, N., and Singh, K. (2019). Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genomics 20:227. doi: 10.1186/s12864-019-5593-5
Vissenberg, K., Fry, S. C., and Verbelen, J. P. (2001). Root hair initiation is coupled to a highly localized increase of xyloglucan endotransglycosylase action in Arabidopsis roots. Plant Physiol. 127, 1125–1135. doi: 10.1104/pp.010295
Vissenberg, K., Ovama, M., Osato, Y., Yokoyama, R., Verbelen, J.-P., and Nishitani, K. (2005). Differential expression of AtXTH17, AtXTHl8, AtXTH19 and AtXTH20 genes in Arabidopsis roots. Physiological roles in specification in cell wall construction. Plant Cell Physiol. 46, 192–200. doi: 10.1093/pcp/pci013
Wang, M., Xu, Z. C., Ding, A. M., and Kong, Y. Z. (2018). Genome-wide identification and expression profiling analysis of the Xyloglucan Endotransglucosylase/hydrolase gene family in tobacco (Nicotiana tabacum L.). Genes. 9:273. doi: 10.3390/genes9060273
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2:100141. doi: 10.1016/j.xinn.2021.100141
Wu, D., Liu, A. Q., Qu, X. Y., Liang, J. Y., and Song, M. (2020). Genome-wide identification, and phylogenetic and expression profiling analyses, of XTH gene families in Brassica rapa L. and Brassica oleracea L. BMC Genomics 21:7153. doi: 10.1186/s12864-020-07153-1
Xia, E. H., Tong, W., Hou, Y., An, Y. L., Chen, L. B., Wu, Q., et al. (2020). The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol. Plant 13, 1013–1026. doi: 10.1016/j.molp.2020.04.010
Yang, H. B., Zhang, R., and Zhou, Z. C. (2017). Pollen dispersal, mating patterns and pollen contamination in an insect-pollinated seed orchard of Schima superba Gardn. Et champ. New Forest. 48, 431–444. doi: 10.1007/s11056-017-9568-6
Yang, Z. Y., Zhang, R., and Zhou, Z. C. (2021). Identification and validation of reference genes for gene expression analysis in Schima superba. Genes. 12:732. doi: 10.3390/genes12050732
Yao, J. B., Chu, X. L., Zhou, Z. C., Xu, H. B., and Zheng, X. J. (2018). Response of seedlings of three Schima superba provenances to different light environments when mixed planting with Cunninghamia lanceolata. For. Res. 31, 144–153. doi: 10.13275/j.cnki.Lykxyj.2018.06.020
Yokoyama, R., and Nishitani, K. (2001). A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 42, 1025–1033. doi: 10.1093/pcp/pce154
Yokoyama, R., Rose, J. K. C., and Nishitani, K. (2004). A surprising diversity and abundance of xyloglucan endotranglucosylase/hydrolases in rice classification and expression analysis. Plant Physiol. 134, 1088–1099. doi: 10.1104/pp.103.035261
Zhang, Z., Li, J., Zhao, X. Q., Wang, J., Wong, G. K. S., and Yu, J. (2006). KaKs_calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4, 259–263. doi: 10.1016/S1672-0229(07)60007-2
Zhou, Z. C., Zhang, R., Fan, H. H., and Chu, X. L. (2020a). Schima superba. Chinese Science Press: Beijing, China. pp. 1–9.
Zhou, W., Zhang, Q., Sun, Y., Yang, L., and Wang, Z. (2020b). Genome-wide identification and characterization of R2R3-MYB family in Hypericum perforatum under diverse abiotic stresses. Int. J. Biol. Macromol. 145, 341–354. doi: 10.1016/j.ijbiomac.2019.12.100
Zhu, J. H., Xia, D. N., Xu, J., Guo, D., Li, H. L., Wang, Y., et al. (2020). Identification of the bHLH gene family in Dracaena cambodiana reveals candidate genes involved in flavonoid biosynthesis. Ind. Crop. Prod. 150:112407. doi: 10.1016/j.indcrop.2020.112407
Keywords: XTH gene family, cell wall remodeling, Schima superba, genome-wide identification, expression analysis, functional interaction network
Citation: Yang Z, Zhang R and Zhou Z (2022) The XTH Gene Family in Schima superba: Genome-Wide Identification, Expression Profiles, and Functional Interaction Network Analysis. Front. Plant Sci. 13:911761. doi: 10.3389/fpls.2022.911761
Edited by:
Ashutosh Pandey, National Institute of Plant Genome Research (NIPGR), IndiaReviewed by:
Shivi Tyagi, National Institute of Plant Genome Research (NIPGR), IndiaGarima Bhatia, University of Pennsylvania, United States
Copyright © 2022 Yang, Zhang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Zhang, emhhbmdydWljYWZAY2FmLmFjLmNu; Zhichun Zhou, emN6aG91X3Jpc2ZAMTYzLmNvbQ==
 Zhongyi Yang
Zhongyi Yang Rui Zhang1,2*
Rui Zhang1,2*