
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 May 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.910408
This article is part of the Research Topic Multi-omics and Computational Biology in Horticultural Plants: From Genotype to Phenotype View all 32 articles
WRKY transcription factor participates in plant growth and development and response to biotic and abiotic stresses. Arachis duranensis, a turfgrass, has high drought tolerance, yet little is known about AdWRKYs response to drought stress in A. duranensis. In this study, RNA-seq identified five AdWRKYs, including AdWRKY18, AdWRKY40, AdWRKY42, AdWRKY56, and AdWRKY64, which were upregulated under drought stress. Orthologous relationships between AdWRKYs and Arabidopsis WRKY were determined to predict the regulatory networks of the five AdWRKYs based on AtWRKYs. Additionally, protein–protein interactions were predicted using differentially expressed proteins from RNA-seq. The quantitative real-time PCR (qRT-PCR) results showed that AdWRKY40 was upregulated, while AdWRKY42, AdWRKY56, and AdWRKY64 were downregulated at different time-points under drought stress. The predicted regulatory networks showed that AdWRKY40 activates COR47, RD21, and RD29A expression under drought stress. Besides, AdWRKY56 regulated CesA8 under drought stress. Aradu.YIQ80 (NAC019) interacted with AdWRKY40, AdWRKY42, AdWRKY56, and AdWRKY64, while Aradu.Z5H58 (NAC055) interacted with AdWRKY42 and AdWRKY64 under drought stress. This study used Arabidopsis to assess AdWRKYs function and regulatory networks, providing a basis for understanding drought tolerance in A. duranensis.
Drought severely impairs plant growth and development (Zhu, 2002, 2016). Therefore, plants have evolved complex adaptive strategies to cope with drought stress over time. Plants reduce water loss by regulating stomatal aperture and root development (Zhu, 2016; Ahammed et al., 2020). Briefly, accumulated abscisic acid (ABA) content changes the Ca2+ concentration of the guard cell, which activates Ca2+ signaling to increase water loss resulting in stomatal closure under drought stress (MacRobbie, 2006). High ABA content reduces the germination rate, root development, and plant growth (Finkelstein et al., 2002; Fujita et al., 2011). Similarly, accumulated ROS activates Ca2+ and K+ signaling, promoting stomatal closure under drought stress (Livak and Schmittgen, 2001; Sierla et al., 2016; Qi et al., 2018). Besides, plants also scavenge reactive oxygen species (ROS) to prevent harm (Zhu, 2016; Ahammed et al., 2020). Under drought stress, excess ROS content causes lipid peroxidation, protein degradation, nucleotide damage, cell death, and electron transport chain damage (Sierla et al., 2016; Xu et al., 2016; Qi et al., 2018). Oxidases (SOD, POD, and CAT) and non-oxidases (APX, GSH, and AsA) can scavenge ROS in plants (Sierla et al., 2016; Qi et al., 2018).
The WRKY transcription factor is involved in drought stress response (Rushton et al., 2010, 2012; Chen et al., 2017a). Besides the one conserved heptapeptide WRKYGQK motif located on the N-terminal WRKY domain (Eulgem et al., 2000), WRKY contains two types of zinc-finger structure, C-X4-5-C-X22-23-H-X-H (C2H2) and C-X5-8-C-X25-28-H-X1-2-C (C2HC; Eulgem et al., 2000; Rushton et al., 2010). WRKY can be classified into three groups based on WRKY domain number and zinc-finger structure type. Group I contains two WRKY domains and a C2H2; Group II contains one WRKY domain and a C2H2; and Group III contains one WRKY domain and a C2HC (Eulgem et al., 2000; Rushton et al., 2010). WRKYs have been identified at the genome level due to the development of sequencing technology: Arabidopsis thaliana (Eulgem et al., 2000), Oryza sativa (Ross et al., 2007), Medicago truncatula (Song and Nan, 2014), Triticum aestivum (Gupta et al., 2018), Glycine max (Song et al., 2016a), and Arachis species (Song et al., 2016b; Zhao et al., 2020). However, the role of WRKY has not been widely studied in Arachis.
WRKY controls gene expression by binding to the W-box element (C/TTGACT/C) of downstream genes (Eulgem et al., 2000; Ciolkowski et al., 2008; Rushton et al., 2010). Extensive studies have also demonstrated that WRKY improves drought stress response (Phukan et al., 2016; Jiang et al., 2017; Chen et al., 2017a, 2019). Moreover, AtWRKY11 and AtWRKY17 overexpression improves drought tolerance in Arabidopsis, promoting seed germination and root growth under drought stress (Ali et al., 2018). OsWRKY11 enhances drought tolerance in rice by upregulating HSP101 expression (Wu et al., 2009). WRKY regulates drought stress through abscisic acid (ABA) signaling (Rushton et al., 2012). AtWRKY57 improves drought stress in Arabidopsis by binding their W-box elements to activate the expressions of RD29A and NCED3 (Jiang et al., 2012). TaWRKY2 increases the drought and salt tolerance of wheat by binding to the W-box of STZ and RD29B (Niu et al., 2012). TaWRKY19 also increases drought and salt tolerance of wheat by binding to W-box of DREB2A and Cor6.6, thus activating DREB2A, RD29A, DR29B, and Cor6.6 (Niu et al., 2012). GmWRKY54 enhances drought stress of soybean by activating PYL8, SRK2A, CIPK11, and CPK3 (Wei et al., 2019). WRKY53 enhances drought stress of Pyrus betulaefolia by binding to the W-box of PbrNCED1 (Liu et al., 2019). Therefore, different plants regulate various downstream genes involved in drought stress tolerance.
Arachis duranensis, a turfgrass, has strong drought tolerance (Leal-Bertioli et al., 2012, 2018). The genome sequences of A. duranensis and drought-related RNA-seq datasets are available in the PeanutBase database (Brasileiro et al., 2015; Bertioli et al., 2016; Dash et al., 2016). A previous study identified 75 WRKYs (AdWRKYs) in A. duranensis (Song et al., 2016b), providing a basis for identifying AdWRKYs involved in drought stress response. In this RNA-seq based study, five AdWRKYs (AdWRKY18, AdWRKY40, AdWRKY42, AdWRKY56, and AdWRKY64) were differentially expressed under drought stress. The regulatory networks of AdWRKYs were then determined and verified. This work provides a theoretical basis for further analysis of the function of AdWRKYs.
The transcriptomes of A. duranensis under drought stress and normal growth conditions were sequenced and de novo assembled to detect differentially expressed genes (DEGs) under normal growth and drought stress conditions in 2015 (Brasileiro et al., 2015). In 2016, the RNA-seq was re-assembled using the A. duranensis genome as the reference, and the updated RNA-seq data were released in the PeanutBase database (Dash et al., 2016). The DEGs were identified between drought and control using the edgeR program (Robinson et al., 2010). Genes with log2(Foldchange) > 2 or <−2 at FDR < 0.05 were considered differentially expressed (Brasileiro et al., 2015; Dash et al., 2016).
A previous study identified WRKYs in A. duranensis (Song et al., 2016b). This study extracted the differentially expressed AdWRKY genes in the abovementioned RNA-seq datasets. The differentially expressed AdWRKY features and subcellular localization were predicted using ExPASy (Gasteiger et al., 2003) and Plant-mPLoc (Chou and Shen, 2010) with default parameters.
Phylogenetic trees were constructed using AdWRKYs and Arabidopsis WRKY (AtWRKY) to reveal their orthologous relationship. AtWRKYs were obtained from a public database (The Arabidopsis Information Resource, https://www.arabidopsis.org/index.jsp). Multiple sequence alignments were conducted using the MAFFT program (Katoh and Standley, 2013). The ProtTest program was used to estimate the best-fit model for constructing phylogenetic trees (Darriba et al., 2011). The maximum likelihood (ML) of the trees was determined using the IQ-tree program (Nguyen et al., 2015).
WRKY regulates downstream gene expression by binding to cis-acting elements, such as W-box (C/TTGACC/T), WT-box (GACTTT), WK-box (TTTTCCAC), PRE (TACTGCGCTTAGT), and SURE (TAAAGA TTACTAATAGGAA; Rinerson et al., 2015; Chen et al., 2019). Herein, the TBtools program was used to extract the 2-kb sequences upstream of the start codon of the predicted genes to identify AdWRKYs genes involved in regulation (Chen et al., 2020). PlantCARE (Rombauts et al., 1999) was used to predict the binding sites of the WRKY transcription factor.
WRKYs interact with other proteins involved in plant development and stress response (Hu et al., 2013; Chen et al., 2017b; Zhang et al., 2018). The protein interaction of AdWRKYs and their differentially expressed genes were predicted using the STRING public database1 with Arabidopsis protein sequences as the reference.
Quantitative real-time PCR (qRT-PCR) analyses were used to verify the drought-tolerance function of the abovementioned genes. Briefly, the A. duranensis seeds were sterilized and germinated on wet filter paper at 28°C. The seedlings were then transferred to the Hoagland solution. Four-leaf plants were treated with 10% (w/v) PEG6000. The leaves were collected after 0, 6, 12, 24, 36, and 48 h of treatments. The control was sampled from 0 h. Three biological replicates were used.
Plant RNA Extraction Kit (TaKaRa, Dalian, China) was used to extract total RNA. The RNA (1 μg) was used for cDNAs synthesis via Reverse Transcriptase M-MLV System (TaKaRa, Dalian, China). The primers were designed by Beacon Designer 8 (Supplementary Table S1). The primers were specifically for amplification since WRKY sequences are conserved (Zhang et al., 2020a). qRT-PCR was performed using TB green premix ex Taq II (TaKaRa, Dalian, China) on the CFX96 real-time PCR machine (Bio-Rad, CA, United States) with UBI2 as the reference gene (Morgante et al., 2011). The PCR conditions included: 95°C denaturation for 30 s, followed by 40 cycles at 95°C for 5 s and 60°C for 45 s. A melting curve analysis was performed at the end of the PCR running end. The 2-ΔΔCt method was used for quantification (Livak and Schmittgen, 2001).
Five AdWRKYs were identified in A. duranensis RNA-seq datasets. These AdWRKYs (AdWRKY18, AdWRKY40, AdWRKY42, AdWRKY56, and AdWRKY64) had full-length sequences and were upregulated (Figure 1A). AdWRKY18 and AdWRKY56 had the highest (Log2foldchange = 4.9; Figure 1A), and lowest differential expression (Log2foldchange = 2.2, Figure 1A), respectively. The CDS length, DNA length, isoelectric point, and molecular weight were 471–1,623 bp, 783–2,328 bp, 6.59–9.67, and 18364.62–59830.41 Da (Table 1), respectively. The AdWRKYs were predicted in location in the nucleus (Table 1).
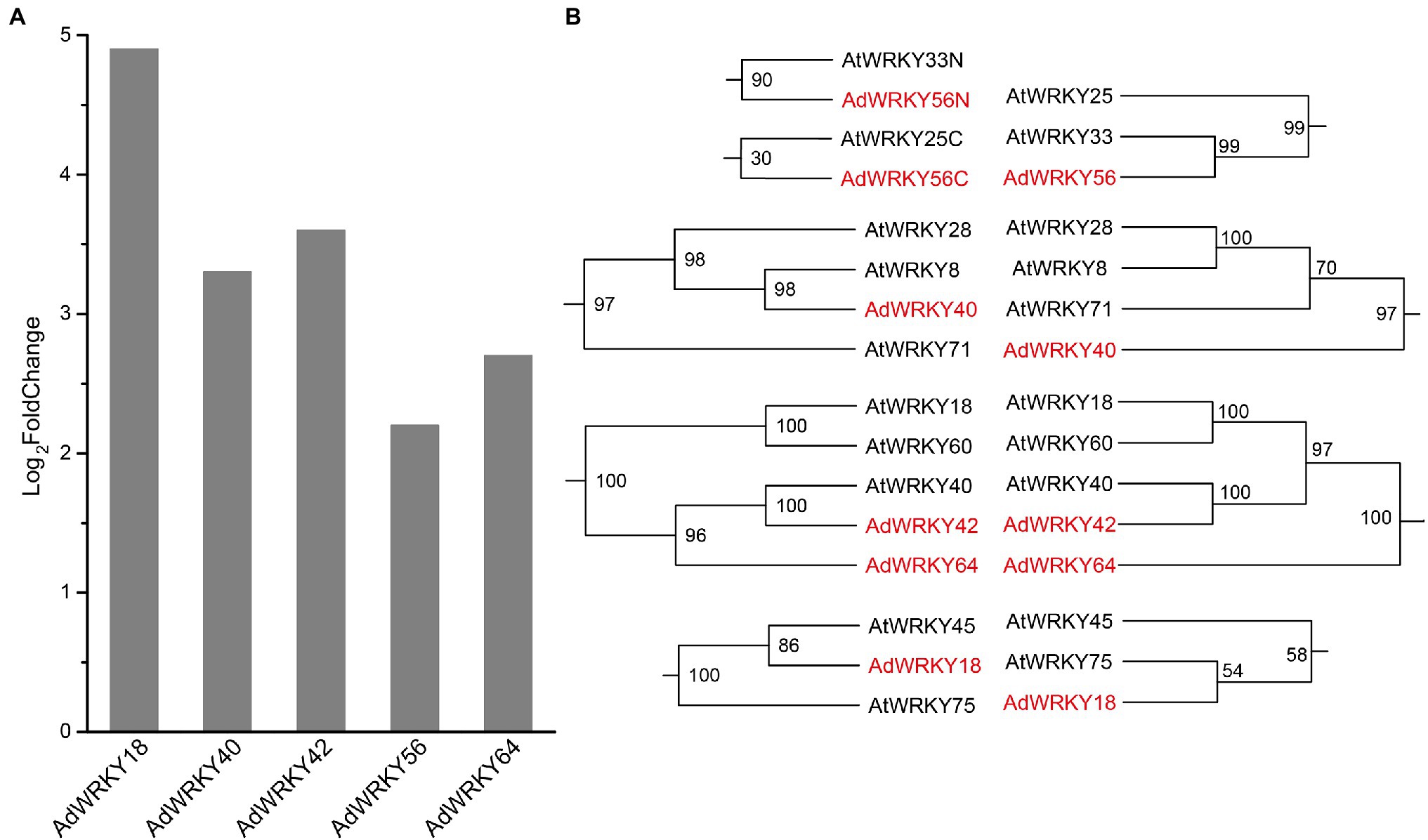
Figure 1. Differentially expressed AdWRKYs and phylogenetic trees. (A) The differential gene expression based on RNA-seq (Brasileiro et al., 2015). (B) The maximum likelihood trees construct using WRKY domains and full-length proteins.
The ML trees were constructed using WRKY domains and WRKY full-length proteins to reveal the orthologous relationships between AdWRKYs and AtWRKYs. The best-fit models were JTT + I + G and VT + I + G + F. The phylogenetic tree showed that AdWRKY42 had homology with AtWRKY40 (Figure 1B; Supplementary Figures S1, S2). The other four AdWRKYs had different topological structures between the WRKY domain tree and the WRKY full-length protein tree (Figure 1B; Supplementary Figures S1, S2). Altogether, the following homologous relationships were obtained based on two ML trees: AdWRKY18 with AtWRKY45 and AtWRKY75; AdWRKY40 with AtWRKY8, AtWRKY28, and AtWRKY71; AdWRKY42 with AtWRKY40; AdWRKY56 with AtWRKY33 and AtWRKY25; and AdWRKY64 with AdWRKY42, AtWRKY40, AtWRKY60, and AtWRKY18.
Experiments have shown the functions of many AtWRKYs. AtWRKY45 overexpression alleviates phosphate starvation and promotes leaf senescence in Arabidopsis (Wang et al., 2014; Chen et al., 2017b). Overexpression of AtWRKY75 promotes leaf senescence and flowering in Arabidopsis (Guo et al., 2017; Zhang et al., 2018). A recent study showed that PtrWRKY75, which is orthologous with AtWRKY75, improves drought tolerance in polar (Zhang et al., 2020b), indicating that AdWRKY18 may confer drought tolerance traits.
Overexpression of AtWRKY8 improves salt stress (Hu et al., 2013). Besides, AtWRKY28 and AtbHLH17 overexpression enhance drought and salt tolerance in Arabidopsis (Babitha et al., 2013). These results indicate that AdWRKY40 is tolerant to drought and salt stresses.
Drought stress induces AtWRKY40 expression (Chen et al., 2010), consistent with A. duranensis RNA-seq dataset results, which showed that AdWRKY42 is upregulated under drought stress. AtWRKY18, AtWRKY40, and AtWRKY60 are involved in the ABA-signaling pathway, but AtWRKY40 antagonizes AtWRKY18 and AtWRKY60 functions (Chen et al., 2010; Shang et al., 2010). Herein, AdWRKY64 had homology with AtWRKY18, AtWRKY40, and AtWRKY60, indicating that AdWRKY64 is involved in drought stress response.
Overexpression of AtWRKY33 enhances drought tolerance in Arabidopsis (Kim et al., 2013; Wang et al., 2013), indicating that drought stress induces AdWRKY56.
Therefore, AdWRKY18, AdWRKY40, AdWRKY42, AdWRKY56, and AdWRKY64 have potential functions in drought stress response based on the abovementioned homologous relationships.
AdWRKY18 has homology with PtrWRKY75. PtrWRKY75 directly regulates PHENYLALANINE AMMONIA LYASE 1 (PAL1; Figure 2), involved in the salicylic acid (SA) pathway, to scavenge ROS, thus improving drought stress (Zhang et al., 2020b).
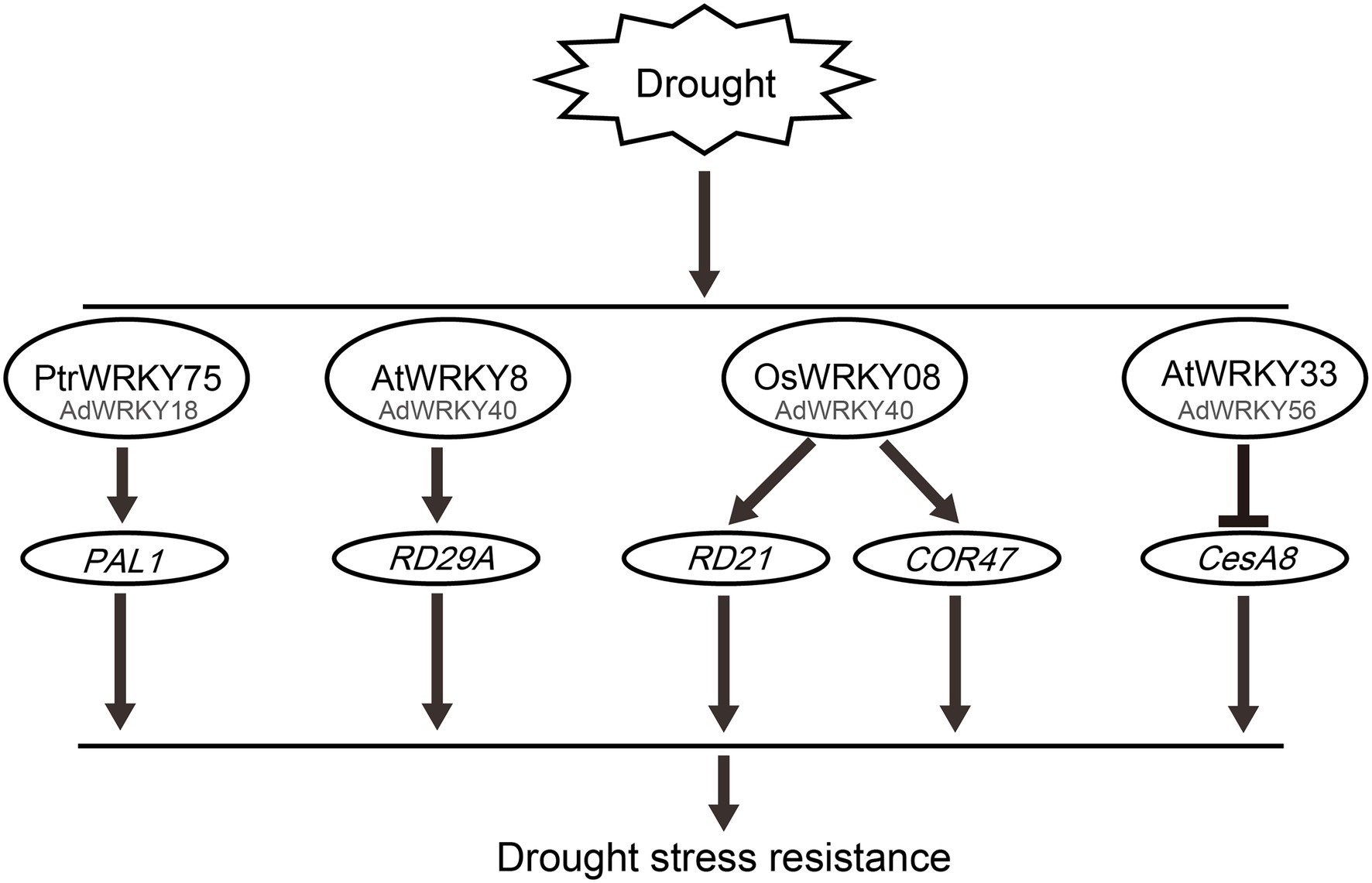
Figure 2. Potential roles of AdWRKYs regulate downstream genes under drought stress. The datasets are obtained from previous studies, including PtrWRKY75 (Zhang et al., 2020b), AtWRKY8 (Hu et al., 2013), OSWRKY8 (Song et al., 2010), and AtWRKY33 (Wang et al., 2013). The WRKY activates PAL1, RD29A, RD21, and COR47 genes expression, and represses CesA8 gene expression.
AdWRKY40 had homology with AtWRKY8, AtWRKY28, and AtWRKY71. AtWRKY8 enhances salt tolerance by activating AtRD29A expression (Hu et al., 2013). Drought, salt, and ABA induce RD29A expression (Narusaka et al., 2003; Hua et al., 2006). These results indicate that AtWRKY8 increases drought tolerance by activating AtRD29A. OsWRKY08 has homology with AtWRKY28 and AtWRKY71 (Song et al., 2010). OsWRKY08 improves drought stress by inducing RD21 and COR47 expression (Song et al., 2010).
Herein, AdWRKY42 had homology with AtWRKY40. Although drought stress induces AtWRKY40 expression (Chen et al., 2010), it is unknown how AtWRKY40 regulates drought stress. CRK5 improves drought tolerance (Lu et al., 2016). Although AtWRKY18, AtWRKY40, and AtWRKY60 triple mutants inhibit CRK5 expression, AtWRKY40 antagonizes AtWRKY18 and AtWRKY60 (Shang et al., 2010; Lu et al., 2016), indicating that only AtWRKY40 does not regulate drought stress through CRK5.
AtCesA8, a cellulose synthase catalytic subunit, plays a crucial role in cellulose synthesis in the secondary cell wall (Taylor et al., 2000, 2003). AtCesA8 mutant accumulates high ABA content, thus reducing the expression of stress-related genes (Chen et al., 2005). AtCesA8 negatively regulates drought stress (Chen et al., 2005; Wang et al., 2013). AtWRKY33 decreases gene expression by binding to CesA8 W-box element, thus increasing drought tolerance (Wang et al., 2013). Herein, AdWRKY56 had homology with AtWRKY33, indicating that AdWRKY56 can enhance drought tolerance by controlling CesA8. Therefore, these results suggest that AdWRKY18, AdWRKY40, and AdWRKY56 improve drought tolerance by regulating the expression of downstream genes.
WRKYs control downstream genes by binding to W-box, WT-box, WK-box, PRE, and SUR cis-acting elements (Sun et al., 2003; van Verk et al., 2008; Hu et al., 2013; Xiao et al., 2013; Machens et al., 2014; Rinerson et al., 2015; Chen et al., 2019). The AtPAL1, AtRD29A, AtRD21, AtCOR47, and AtCesA8 are orthologous with Aradu.NNP8F, Aradu.MEI7N, Aradu.08WSJ, Aradu.IF4XP, and Aradu.UPY7V in A. duranensis. Aradu.NNP8F, Aradu.MEI7N, and Aradu.08WSJ lack W-box element in the 2-kb promote region (Supplementary Table S2). AtWRKY33 inhibits CesA8 expression by binding to the distal W-box (~3-kb) of the CesA8 gene, thus increasing drought tolerance (Wang et al., 2013). The 3-kb promoter region of Aradu.NNP8F, Aradu.MEI7N, and Aradu.08WSJ also lack the W-box elements (Supplementary Table S2). These results indicate that AdWRKY18 cannot directly regulate Aradu.NNP8F (PAL1) under drought stress and AdWRKY40 cannot directly regulate Aradu.08WSJ (RD21) and Aradu.MEI7N (RD29A) under drought stress. Notably, RNA-seq showed that Aradu.IF4XP (COR47) gene was differentially expressed after drought stress, indicating that AdWRKY40 potentially regulates Aradu.IF4XP by binding the W-box element under drought stress.
Subcellular localization showed that AtPAL1 and Aradu.NNP8F are located in the cytoplasm, AtRD21 and Aradu.08WSJ are located in the vacuole, AtCesA8 and Aradu.UPY7V are located in the chloroplast, and others are located in the nucleus (Supplementary Table S2). A previous study revealed that AtWRKY40 moves from the nucleus to the cytoplasm to control downstream genes (Rushton et al., 2012). Similarly, AdWRKYs can potentially move from the nucleus to other organelles to regulate Aradu.NNP8F, Aradu.08WSJ, and Aradu.UPY7V is located outside the nucleus.
WRKYs can interact with other proteins involved in drought stress regulation (Perruc et al., 2004; Cheng et al., 2012; Hu et al., 2013). Herein, AdWRKY42 and AdWRKY64 mapped on the same Arabidopsis protein sequence, AT1G80840. AdWRKY18, AdWRKY40, AdWRKY42, AdWRKY56, and AdWRKY64 directly interacted with 17, 7, 57, 57, and 57 proteins, respectively (Figure 3A; Supplementary Table S3).
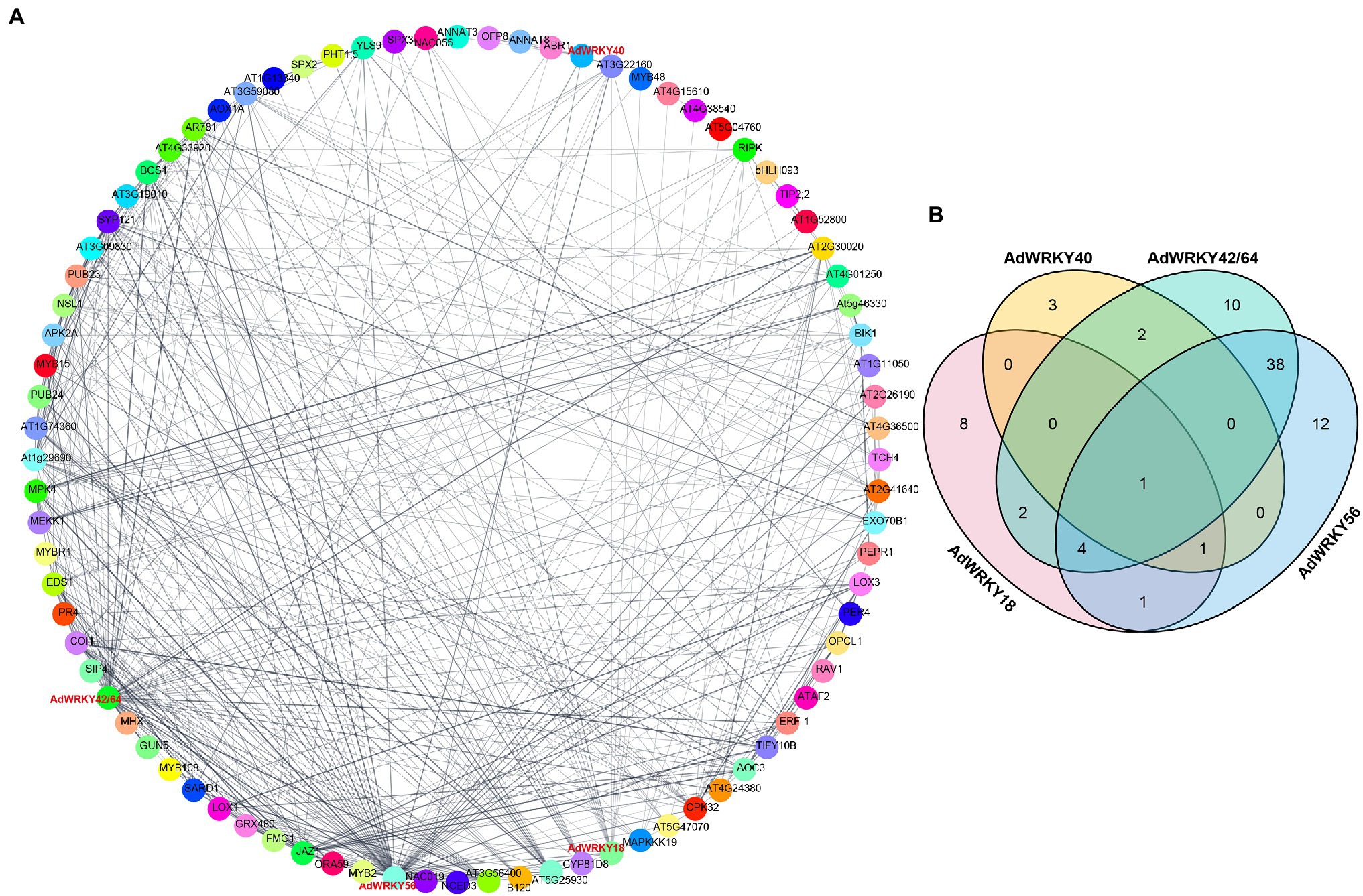
Figure 3. The interaction between AdWRKYs and other proteins. (A) The interaction between AdWRKYs and other proteins. (B) The number of other proteins interacting with AdWRKYs.
The five AdWRKYs interacted with common proteins and specific proteins (Figure 3B), of which four common proteins (NAC019, MYB2, NAC055, and ABR1) are involved in drought stress response. Aradu.YIQ80 (NAC019) interacted with the five AdWRKYs (Figure 4A; Supplementary Table S4) while Aradu.X7LBF (MYB2), Aradu.Z5H58 (NAC055), and Aradu.ME4LN (ABR1) interacted with at least two AdWRKYs (Figures 4B–D; Supplementary Table S4). AtNAC019 improves drought stress in Arabidopsis by activating ERD1 expression (Tran et al., 2004). AtWRKY1 enhances drought stress or ABA treatment by binding to the W-box cis-acting element of AtMYB2 (Qiao et al., 2016). AtNAC055 overexpression increases drought tolerance, and the AtNAC055 mutant has a decreased drought tolerance (Fu et al., 2018). Drought stress induces AtABR1 expression. However, mannitol stress can decrease AtABR1 expression, thus, reducing seed germination (Pandey et al., 2005).
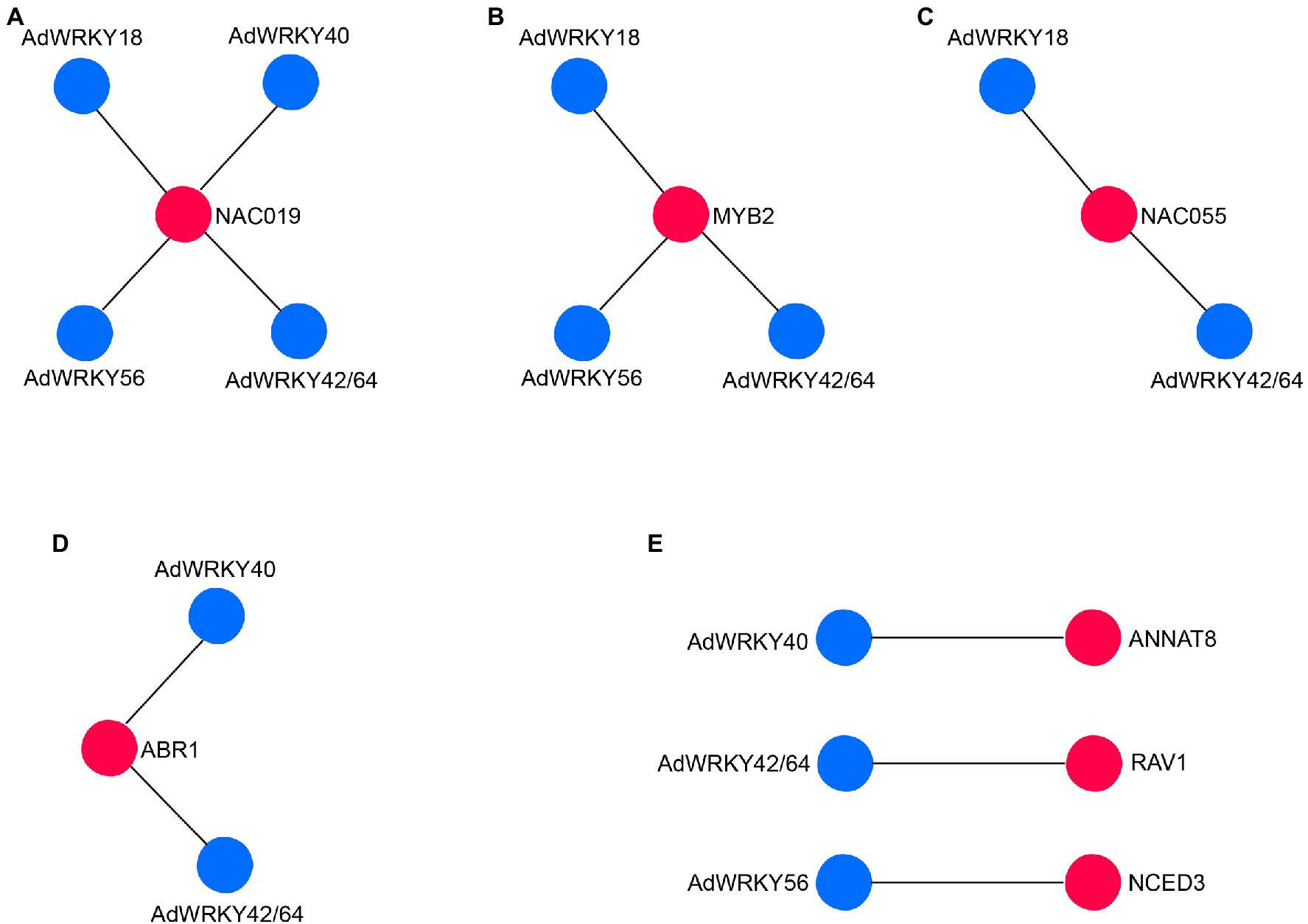
Figure 4. AdWRKYs interaction with proteins with drought-tolerance function based on Arabidopsis. (A) NAC019 interaction with AdWRKYs. (B) MYB2 interaction with AdWRKYs. (C) NAC055 interaction with AdWRKYs. (D) ABR1 interaction with AdWRKYs. (E) Interaction between AdWRKYs and other specific proteins.
AdWRKY40, AdWRKY42/64, and AdWRKY56-specific interaction with Aradu.MBZ2M (ANNAT8), Aradu.7AQ1B (RAV1), and Aradu.FJ7R7 (NCED3) are involved in drought stress response (Figure 4E; Supplementary Table S5). Heterologous expression of ANNAT8 enhances the response to drought and salt stresses in Arabidopsis and tobacco during the growth and development stages (Yadav et al., 2016). Overexpression of RAV1 promotes water loss by activating the expression of ABA-responsive genes (Fu et al., 2014), indicating that RAV1 negatively regulates drought stress. CED3 responds to water deficit through ABA synthesis (Sato et al., 2018; Baek et al., 2020).
This study identified five differentially expressed AdWRKYs using RNA-seq and their regulatory networks based on Arabidopsis. qRT-PCR was used to assess the drought stress response of the five AdWRKYs and their regulatory genes. AdWRKY42, AdWRKY56, and AdWRKY64 genes were downregulated at 6, 12, 24, 36, and 48 h (Figure 5). AdWRKY40 was upregulated at 6, 12, and 36 h, and downregulated at 48 h (Figure 5). AdWRKY18 was downregulated at 24 and 48 h and upregulated at 36 h (Figure 5). The five AdWRKYs were downregulated at 48 h. AdWRKY40 had similar qRT-PCR and RNA-seq results. The differential expression of AdWRKY42, AdWRKY56, and AdWRKY64 contradicted the RNA-seq results. The possible reason is different drought treatment and cultural environments between qRT-PCR and RNA-seq. The RNA-seq data were produced from the samples under natural drought and normal growth conditions (Brasileiro et al., 2015), while qRT-PCR was analyzed under five drought-stress time points using 10% (w/v) PEG6000 treatments.
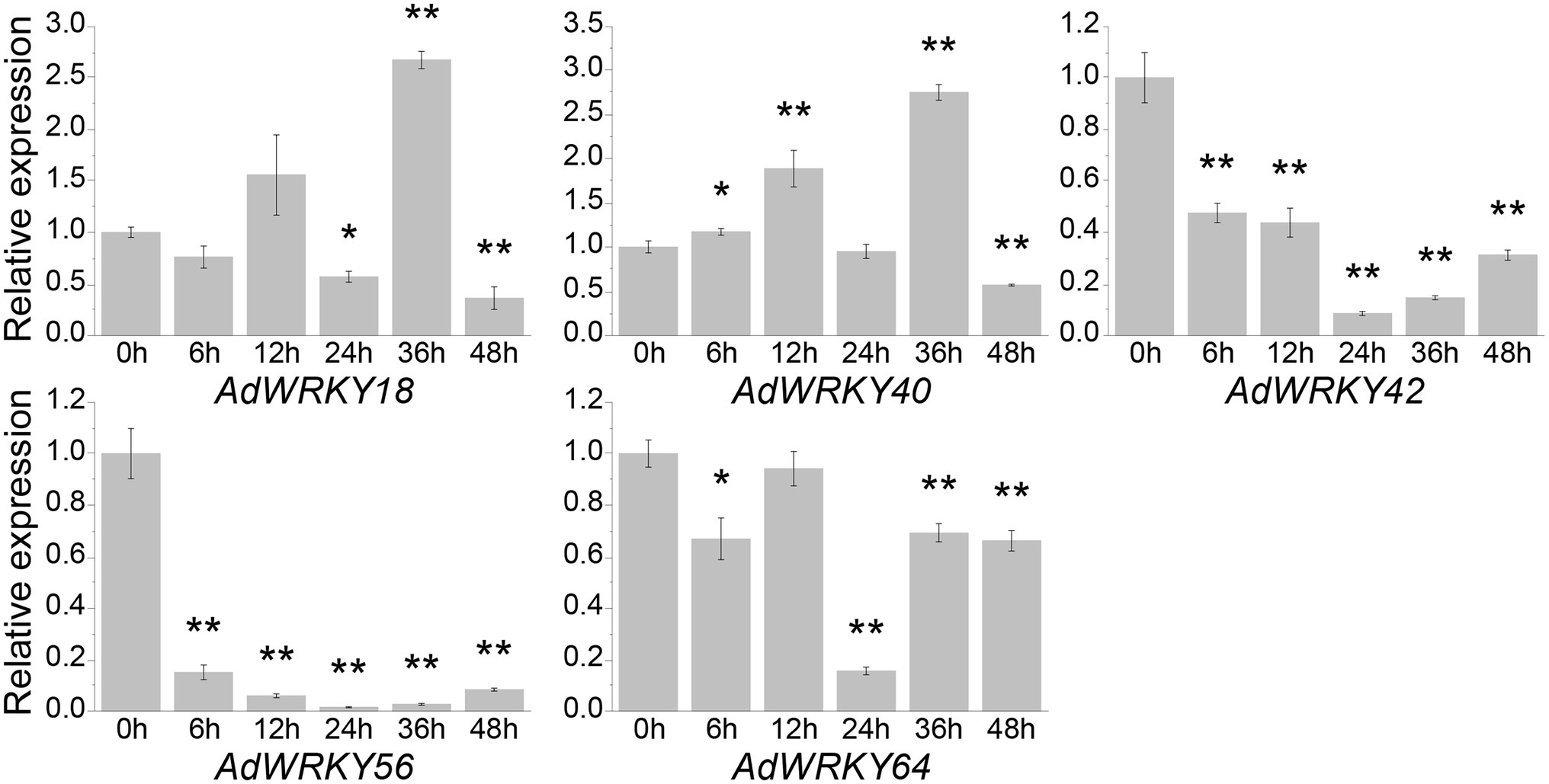
Figure 5. Differentially expressed AdWRKYs under drought stress detected by quantitative real-time PCR (qRT-PCR). Four-leaf plants were treated with 10% (w/v) PEG6000. The leaves were collected after 0, 6, 12, 24, 36, and 48 h of treatments. The control was sampled from 0 h. Three biological replicates were used. Asterisks * and ** indicate significant differences at 0.05 and 0.01 using t tests, respectively.
Quantitative real-time PCR showed that AdCOR47 and AdCesA8 genes were upregulated and downregulated, respectively, at five time-points (Figure 6). Similarly, previous studies showed that COR47 is positively regulated in Oryza sativa under drought stress, while CesA8 is negatively regulated in A. thaliana under drought stress (Song et al., 2010; Wang et al., 2013). Herein, AdPAL1 and AdRD29A were downregulated at 6, 12, 24, and 36 h (Figure 6). However, PtrPAL1 and AtRD29A are positively regulated under drought stress (Hu et al., 2013; Zhang et al., 2020b). WRKYs directly regulate PtrPAL1 and RD29A by binding to W-box elements. Although AdPAL1 and AdRD29A do not have W-box elements, they were upregulated at 48 h (Figure 6). AdRD21 also lacked the W-box element and was upregulated at five time-points. Similarly, a previous study showed that AtRD21 is positively regulated under drought stress (Narusaka et al., 2003; Hua et al., 2006; Song et al., 2010). Therefore, AdWRKY40 is involved in regulating different regulatory networks of downstream genes AdRD21 and AdRD29A.
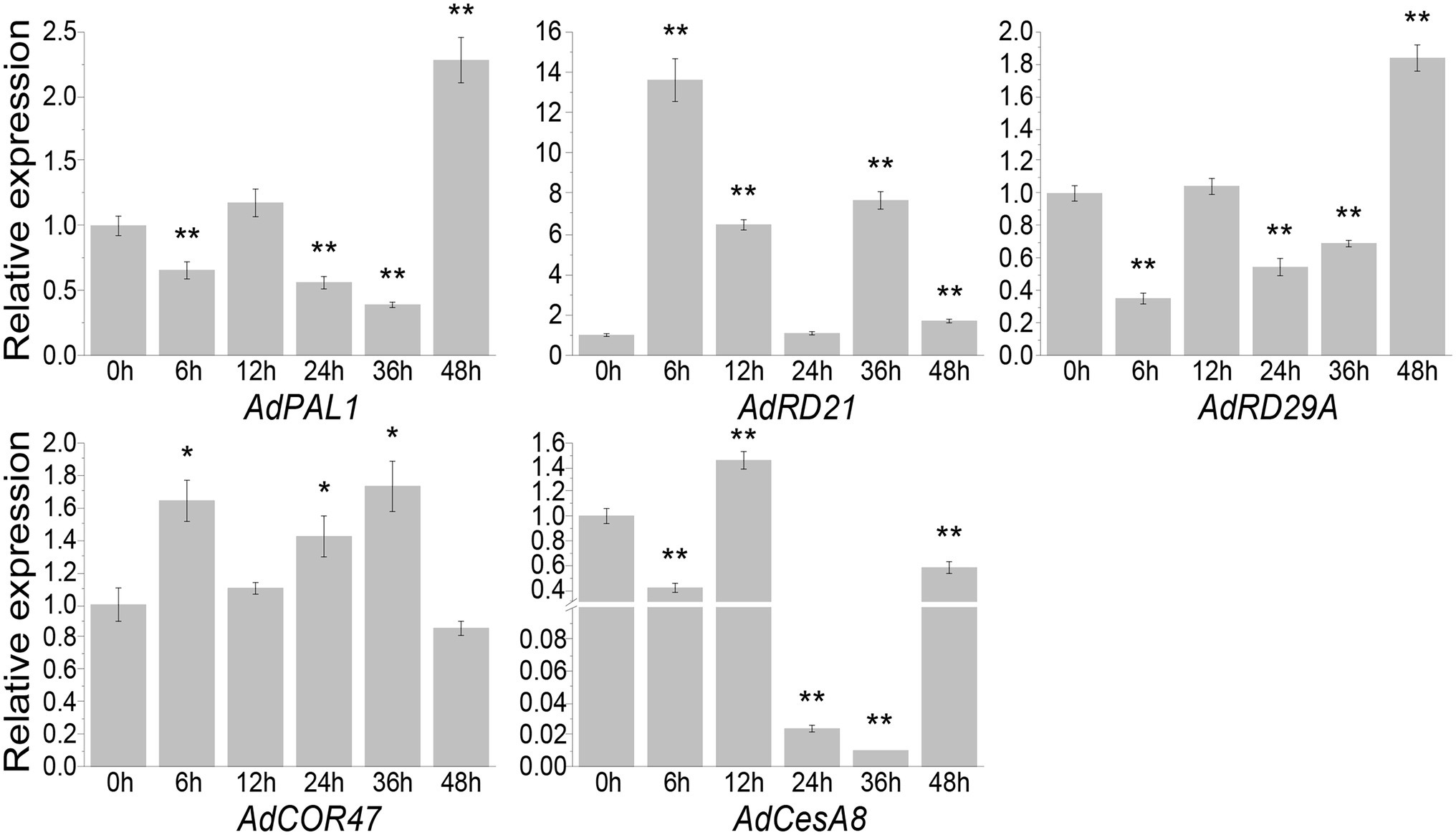
Figure 6. Differential expression levels of AdWRKYs regulating downstream genes under drought stress, assessed using quantitative real-time PCR. Four-leaf plants were treated with 10% (w/v) PEG6000. The leaves were collected after 0, 6, 12, 24, 36, and 48 h of treatments. The control was sampled from 0 h. Three biological replicates were used. Asterisks * and ** indicate significant differences at 0.05 and 0.01 using t tests, respectively.
Quantitative real-time PCR was used to assess the drought stress response of the genes that translated proteins-AdWRKYs interaction. This study found that AdNAC019 and AdNAC055 were differentially expressed after drought stress. However, other genes were not differentially expressed relative to the control group. AdNAC019 interacted with the five AdWRKYs, while AdNAC055 interacted with only AdWRKY42 and AdWRKY64. AdNAC019 was downregulated at the five time-points (Figure 6). AdNAC055 was upregulated at 6 and 36 h and downregulated at 12, 24, and 48 h (Figure 7). However, NAC019 and NAC055 positively regulate drought stress in Arabidopsis (Tran et al., 2004; Fu et al., 2018). Similarly, AdWRKY42, AdWRKY56, and AdWRKY64 were downregulated after drought stress, different from the expression patterns in Arabidopsis. These results indicate that Arachis and Arabidopsis have different regulatory networks under drought stress.

Figure 7. Differential expression levels of genes that are translated proteins interaction with AdWRKYs under drought stress detected via quantitative real-time PCR. Four-leaf plants were treated with 10% (w/v) PEG6000. The leaves were collected after 0, 6, 12, 24, 36, and 48 h of treatments. The control was sampled from 0 h. Three biological replicates were used. Asterisks * and ** indicate significant differences at 0.05 and 0.01 using t tests, respectively.
Herein, five AdWRKYs were identified under drought stress based on previous studies and RNA-seq (Brasileiro et al., 2015; Song et al., 2016b). Subsequently, orthologous relationships between AdWRKYs and AtWRKYs were constructed. The regulatory networks of the five AdWRKYs were determined based on AtWRKYs and verified using qRT-PCR. The results showed that AdWRKY40 positively regulated drought stress, while AdWRKY42, AdWRKY56, and AdWRKY64 negatively regulated drought stress. Moreover, AdWRKY40 was upregulated under drought stress, confirming the orthologous WRKYs from Arabidopsis and Oryza (Narusaka et al., 2003; Hua et al., 2006; Song et al., 2010; Hu et al., 2013). However, the orthologous AdWRKY42, AdWRKY56, and AdWRKY64 showed the opposite results in Arabidopsis under drought stress. Besides, qRT-PCR and RNA-seq results showed opposite results in A. duranensis. Additionally, Arabidopsis had different regulatory networks between AdWRKY40 and its orthologs. AtWRKYs activate COR47, RD21, and RD29A (Song et al., 2010; Hu et al., 2013). AdWRKY40 control COR47 by potential binding to the W-box element. AdWRKY40 indirectly regulates RD21 and RD29A because they lack the W-box element.
Protein–protein analyses are different from transcriptional regulation. To the best of our knowledge, no study has shown that the abovementioned proteins interact with WRKYs in Arabidopsis under drought stress. However, protein–protein interaction has been predicted based on Arabidopsis protein in A. duranensis. This study found that the NAC transcription factor interacts with WRKY involved in the drought stress response of A. duranensis. MaNAC5 interacts with MaWRKY1 and MaWRKY2 to activate PR1-1, PR2, PR10c, and CHIL1 expressions in response to Colletotrichum musae infection in bananas (Shan et al., 2016). Moreover, the CitNAC62-CitWRKY1 interaction enhances CitAco3 expression, thus decreasing citric acid content (Li et al., 2017). We hypothesize that AdWRKY40 and AdNAC interaction can control RD21 and RD29A expression. However, more experimental tests are needed to verify the hypothesis. Therefore, this study provides a model for studying gene function and a basis for revealing WRKY regulatory networks in A. duranensis under drought stress.
This study showed that AdWRKY18, AdWRKY40, AdWRKY42, AdWRKY56, and AdWRKY64 were differentially expressed under drought stress, but various regulatory networks were formed among the five AdWRKYs. AdWRKY18 was excluded from regulatory network analyses because it did not have the same differential expression pattern at two time-points. AdWRKY40 potentially regulates COR47 by binding the W-box element and indirectly regulates RD21 and RD29A under drought stress. AdWRKY56 controlled the CesA8 expression under drought stress (Figure 8). Protein–protein interaction results showed that AdNAC019 interacted with AdWRKY40, AdWRKY42, AdWRKY56, and AdWRKY64 under drought stress AdNAC055 interacted with AdWRKY42 and AdWRKY64 (Figure 8).
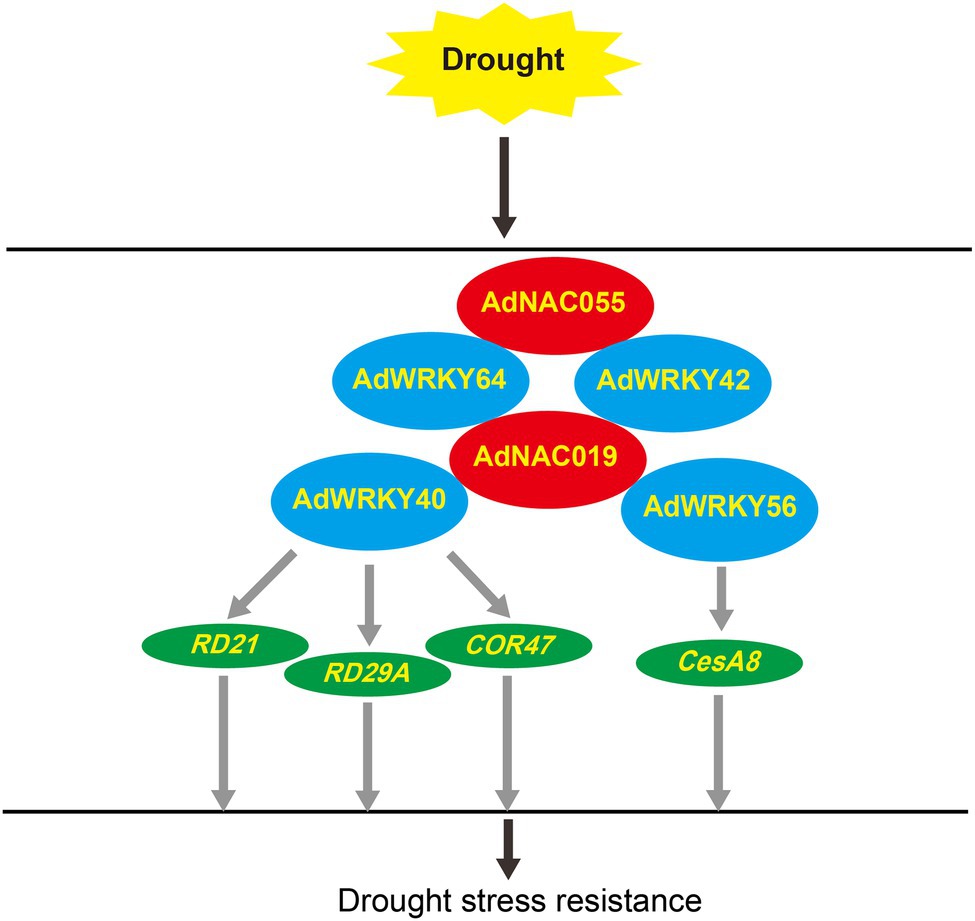
Figure 8. AdWRKYs regulatory networks under drought stress. The gray arrow indicates hypothesis but no experimental test.
Publicly available datasets were analyzed in this study. This data can be found at: https://www.ncbi.nlm.nih.gov/, JZ390113 to JZ390862.
HS conceived and designed this research, analyzed data, and wrote the manuscript. YZ and PD analyzed data. PD, FX, XZ, and HS evaluated and revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was funded by the Natural Science Foundation of Shandong Province, China (ZR2019QC017), Start-up Foundation for High Talents of Qingdao Agricultural University (no. 665/1120012), the first Class Grassland Science Discipline Program of Shandong Province, China, and Shandong Modern Agricultural Industrial and Technical System (SDAIT-23-01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.910408/full#supplementary-material
Supplementary Table S1 | Quantitative real-time PCR primers.
Supplementary Table S2 | W-box element and subcellular localization of downstream genes.
Supplementary Table S3 | Protein–protein interaction sequences.
Supplementary Table S4 | Common interaction between proteins and AdWRKYs.
Supplementary Table S5 | Specific interaction between proteins and AdWRKYs.
Supplementary Figure S1 | The maximum likelihood tree constructed using WRKY domains.
Supplementary Figure S2 | The maximum likelihood tree constructed using full-length WRKY proteins.
Ahammed, G. J., Li, X., Yang, Y., Liu, C., Zhou, G., Wan, H., et al. (2020). Tomato WRKY81 acts as a negtive regulator for drought tolerance by modulating guard cell H2O2-mediated stomatal closure. Environ. Exp. Bot. 171:103960. doi: 10.1016/j.envexpbot.2019.103960
Ali, M. A., Azeem, F., Nawaz, M. A., Acet, T., Abbas, A., Imran, Q. M., et al. (2018). Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J. Plant Physiol. 226, 12–21. doi: 10.1016/j.jplph.2018.04.007
Babitha, K. C., Ramu, S. V., Pruthvi, V., Mahesh, P., Nataraja, K. N., and Udayakumar, M. (2013). Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 22, 327–341. doi: 10.1007/s11248-012-9645-8
Baek, D., Kim, W. Y., Cha, J. Y., Park, H. J., Shin, G., Park, J., et al. (2020). The GIGANTEA-ENHANCED EM LEVEL complex enhances drought tolerance via regulation of abscisic acid synthesis. Plant Physiol. 184, 443–458. doi: 10.1104/pp.20.00779
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K. S., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446. doi: 10.1038/ng.3517
Brasileiro, A. C. M., Morgante, C. V., Araujo, A. C. G., Leal-Bertioli, S. C. M., Silva, A. K., Martins, A. C. Q., et al. (2015). Transcriptome profiling of wild Arachis from water-limited environments uncovers drought tolerance candidate genes. Plant Mol. Biol. Report. 33, 1876–1892. doi: 10.1007/s11105-015-0882-x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Z., Hong, X., Zhang, H., Wang, Y., Li, X., Zhu, J. K., et al. (2005). Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 43, 273–283. doi: 10.1111/j.1365-313X.2005.02452.x
Chen, F., Hu, Y., Vannozzi, A., Wu, K., Cai, H., Qin, Y., et al. (2017a). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36, 311–335. doi: 10.1080/07352689.2018.1441103
Chen, H., Lai, Z., Shi, J., Xiao, Y., Chen, Z., and Xu, X. (2010). Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10:281. doi: 10.1186/1471-2229-10-281
Chen, X., Li, C., Wang, H., and Guo, Z. (2019). WRKY transcription factors: evolution, binding, and action. Phytopathol. Res. 1:13. doi: 10.1186/s42483-019-0022-x
Chen, L., Xiang, S., Chen, Y., Li, D., and Yu, D. (2017b). Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant 10, 1174–1189. doi: 10.1016/j.molp.2017.07.008
Cheng, Y., Zhou, Y., Yang, Y., Chi, Y. J., Zhou, J., Chen, J. Y., et al. (2012). Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 159, 810–825. doi: 10.1104/pp.112.196816
Chou, K. C., and Shen, H. B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5:e11335. doi: 10.1371/journal.pone.0011335
Ciolkowski, I., Wanke, D., Birkenbihl, R., and Somssich, I. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92. doi: 10.1007/s11103-008-9353-1
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
Dash, S., Cannon, E. K. S., Kalberer, S. R., Farmer, A. D., and Cannon, S. B. (2016). “PeanutBase and other bioinformatic resources for peanut,” in Peanuts Genetics, Processing, and Utilization. eds. H. T. Stalker and R. F. Wilson (AOCS Press), 241–252.
Eulgem, T., Rushton, P., Robatzek, S., and Somssich, I. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Finkelstein, R. R., Gampala, S., and Rock, C. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14, S15–S45. doi: 10.1105/tpc.010441
Fu, M., Kang, H. K., Son, S. H., Kim, S. K., and Nam, K. H. (2014). A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol. 55, 1892–1904. doi: 10.1093/pcp/pcu118
Fu, Y., Ma, H., Chen, S., Gu, T., and Gong, J. (2018). Control of proline accumulation under drought via a novel pathway comprising the histone methylase CAU1 and the transcription factor ANAC055. J. Exp. Bot. 69, 579–588. doi: 10.1093/jxb/erx419
Fujita, Y., Fujita, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Marc Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Guo, P., Li, Z., Huang, P., Li, B., Fang, S., Chu, J., et al. (2017). A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29, 2854–2870. doi: 10.1105/tpc.17.00438
Gupta, S., Mishra, V. K., Kumari, S., Chand, R., and Varadwaj, P. K. (2018). Deciphering genome-wide WRKY gene family of Triticum aestivum L. and their functional role in response to abiotic stress. Genes Genomics 41, 79–94. doi: 10.1007/s13258-018-0742-9
Hu, Y., Chen, L., Wang, H., Zhang, L., Wang, F., and Yu, D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745. doi: 10.1111/tpj.12159
Hua, Z. M., Yang, X., and Fromm, M. E. (2006). Activation of the NaCl and drought-induced RD29A and RD29B promoters by constitutively active Arabidopsis MAPKK or MAPK proteins. Plant Cell Environ. 29, 1761–1770. doi: 10.1111/j.1365-3040.2006.01552.x
Jiang, Y. J., Liang, G., and Yu, D. Q. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 5, 1375–1388. doi: 10.1093/mp/sss080
Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J., and Zhang, J. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59, 86–101. doi: 10.1111/jipb.12513
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, W. C., Ko, J. H., Kim, J. Y., Kim, J., Bae, H. J., and Han, K. H. (2013). MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J. 73, 26–36. doi: 10.1111/j.1365-313x.2012.05124.x
Leal-Bertioli, S. C. M., Bertioli, D. J., Guimarães, P. M., Pereira, T. D., Galhardo, I., Silva, J. P., et al. (2012). The effect of tetraploidization of wild Arachis on leaf morphology and other drought-related traits. Environ. Exp. Bot. 84, 17–24. doi: 10.1016/j.envexpbot.2012.04.005
Leal-Bertioli, S. C. M., Godoy, I. J., Santos, J. F., Doyle, J. J., Guimarães, P. M., Abernathy, B. L., et al. (2018). Segmental allopolyploidy in action: increasing diversity through polyploid hybridization and homoeologous recombination. Am. J. Bot. 105, 1053–1066. doi: 10.1002/ajb2.1112
Li, S. J., Yin, X. R., Wang, W. L., Liu, X. F., Zhang, B., and Chen, K. S. (2017). Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up-regulation of CitAco3. J. Exp. Bot. 68, 3419–3426. doi: 10.1093/jxb/erx187
Liu, Y., Yang, T., Lin, Z., Gu, B., Xing, C., Zhao, L., et al. (2019). A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol. J. 17, 1770–1787. doi: 10.1111/pbi.13099
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene wxpression sata using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, K., Liang, S., Wu, Z., Bi, C., Yu, Y. T., Wang, X. F., et al. (2016). Overexpression of an Arabidopsis cysteine-rich receptor-like protein kinase, CRK5, enhances abscisic acid sensitivity and confers drought tolerance. J. Exp. Bot. 67, 5009–5027. doi: 10.1093/jxb/erw266
Machens, F., Becker, M., Umrath, F., and Hehl, R. (2014). Identification of a novel type of WRKY transcription factor binding site in elicitor-responsive cis-sequences from Arabidopsis thaliana. Plant Mol. Biol. 84, 371–385. doi: 10.1007/s11103-013-0136-y
MacRobbie, E. A. C. (2006). Control of volume and turgor in stomatal guard cells. J. Membr. Biol. 210, 131–142. doi: 10.1007/s00232-005-0851-7
Morgante, C. V., Guimarães, P. M., Martins, A. C. Q., Araújo, A. C. G., Leal-Bertioli, S. C. M., Bertioli, D. J., et al. (2011). Reference genes for quantitative reverse transcription-polymerase chain reaction expression studies in wild and cultivated peanut. BMC. Res. Notes 4:339. doi: 10.1186/1756-0500-4-339
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148. doi: 10.1046/j.1365-313X.2003.01708.x
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Niu, C. F., Wei, W., Zhou, Q. Y., Tian, A. G., Hao, Y. J., Zhang, W. K., et al. (2012). Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 35, 1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x
Pandey, G. K., Grant, J. J., Cheong, Y. H., Kim, B. G., Li, L., and Luan, S. (2005). ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 139, 1185–1193. doi: 10.1104/pp.105.066324
Perruc, E., Charpenteau, M., Ramirez, B. C., Jauneau, A., Galaud, J. P., Ranjeva, R., et al. (2004). A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J. 38, 410–420. doi: 10.1111/j.1365-313X.2004.02062.x
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Qi, J., Song, C. P., Wang, B., Zhou, J., Kangasjärvi, J., Zhu, J. K., et al. (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60, 805–826. doi: 10.1111/jipb.12654
Qiao, Z., Li, C. L., and Zhang, W. (2016). WRKY1 regulates stomatal movement in drought-stressed Arabidopsis thaliana. Plant Mol. Biol. 91, 53–65. doi: 10.1007/s11103-016-0441-3
Rinerson, C. I., Rabara, R. C., Tripathi, Q. J., Shen, P. J., and Rushton, P. J. (2015). The evolution of WRKY transcription factors. BMC Plant Biol. 15:66. doi: 10.1186/s12870-015-0456-y
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rombauts, S., Déhais, P., Van Montagu, M., and Rouzé, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Ross, C., Liu, Y., and Shen, Q. (2007). The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 49, 827–842. doi: 10.1111/j.1672-9072.2007.00504.x
Rushton, P., Somssich, I., Ringler, P., and Shen, Q. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Sato, H., Takasaki, H., Takahashi, F., Suzuki, T., Iuchi, S., Mitsuda, N., et al. (2018). Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. U. S. A. 115, E11178–E11187. doi: 10.1073/pnas.1811491115
Shan, W., Chen, J. Y., Kuang, J. F., and Lu, W. J. (2016). Banana fruit NAC transcription factor MaNAC5 cooperates with MaWRKYs to enhance the expression of pathogenesis-related genes against Colletotrichum musae. Mol. Plant Pathol. 17, 330–338. doi: 10.1111/mpp.12281
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to telieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Sierla, M., Waszczak, C., Vahisalu, T., and Kangasjärvi, J. (2016). Reactive oxygen species in the regulation of stomatal movements. Plant Physiol. 171, 1569–1580. doi: 10.1104/pp.16.00328
Song, Y., Jing, S., and Yu, D. (2010). Overexpression of the stress-induced OsWRKY08 improves osmotic stress tolerance in Arabidopsis. Chin. Sci. Bull. 54, 4671–4678. doi: 10.1007/s11434-009-0710-5
Song, H., and Nan, Z. (2014). Genome-wide indentification and analysis of WRKY transcription factors in Medicago truncatula. Hereditas 36, 152–168. doi: 10.3724/SP.J.1005.2014.0152
Song, H., Wang, P., Hou, L., Zhao, S., Zhao, C., Xia, H., et al. (2016a). Global analysis of WRKY genes and their response to dehydration and salt stress in soybean. Front. Plant Sci. 7:9. doi: 10.3389/fpls.2016.00009
Song, H., Wang, P., Lin, J. Y., Zhao, C., Bi, Y., and Wang, X. (2016b). Genome-wide identification and characterization of WRKY gene family in peanut. Front. Plant Sci. 7:534. doi: 10.3389/fpls.2016.00534
Sun, C., Palmqvist, S., Olsson, H., Borén, M., Ahlandsberg, S., and Jansson, C. (2003). A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15, 2076–2092. doi: 10.1105/tpc.014597
Taylor, N. G., Howells, R. M., Huttly, A. K., Vickers, K., and Turner, S. R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U. S. A. 100, 1450–1455. doi: 10.1073/pnas.0337628100
Taylor, N. G., Laurie, S., and Turner, S. R. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12, 2529–2539. doi: 10.1105/tpc.12.12.2529
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factor that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
van Verk, M. C., Pappaioannou, D., and Neeleman, L. (2008). A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 146, 1983–1995. doi: 10.1104/pp.107.112789
Wang, X., Du, B., Liu, M., Sun, N., and Qi, X. (2013). Arabidopsis transcription factor WRKY33 is involved in drought by directly regulating the expression of CesA8. Am. J. Plant Sci. 4, 21–27. doi: 10.4236/ajps.2013.46A004
Wang, H., Xu, Q., Kong, Y. H., Chen, Y., Duan, J. Y., Wu, W. H., et al. (2014). Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 164, 2020–2029. doi: 10.1104/pp.113.235077
Wei, W., Liang, D. W., Bian, X. H., Shen, M., Xiao, J. H., Zhang, W. K., et al. (2019). GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J. 100, 384–398. doi: 10.1111/tpj.14449
Wu, X. L., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xiao, J., Cheng, H., Li, X., Xiao, J., Xu, C., and Wang, S. (2013). Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiol. 163, 1868–1882. doi: 10.1104/pp.113.226019
Xu, Y., Burgess, P., Zhang, X. Z., and Huang, B. R. (2016). Enhancing cytokinin synthesis by overexpressing ipt alleviated drought inhibition of root growth through activating ROS-scavenging systems in Agrostis stolonifera. J. Exp. Bot. 67, 1979–1992. doi: 10.1093/jxb/erw019
Yadav, D., Ahmed, I., Shukla, P., Boyidi, P., and Kirti, P. B. (2016). Overexpression of Arabidopsis AnnAt8 alleviates abiotic stress in transgenic Arabidopsis and tobacco. Plan. Theory 5:18. doi: 10.3390/plants5020018
Zhang, L., Chen, L., and Yu, D. (2018). Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 176, 790–803. doi: 10.1104/pp.17.00657
Zhang, Y., Yin, D., and Song, H. (2020a). Genome-wide identification and characterization of gene families in Arachis: methods and strategies. Front. Genet. 11:525. doi: 10.3389/fgene.2020.00525
Zhang, Y., Zhou, Y., Zhang, D., Tang, X., Li, Z., Shen, C., et al. (2020b). PtrWRKY75 overexpression reduces stomatal aperture and improves drought tolerance by salicylic acid-induced reactive oxygen species accumulation in poplar. Environ. Exp. Bot. 176:104117. doi: 10.1016/j.envexpbot.2020.104117
Zhao, N., He, M., Li, L., Cui, S., Hou, M., Wang, L., et al. (2020). Identification and expression analysis of WRKY gene family under drought stress in peanut (Arachis hypogaea L.). PLoS One 15:e0231396. doi: 10.1371/journal.pone.0231396
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Keywords: Arachis duranensis, drought tolerance, protein–protein interaction, regulatory network, WRKY
Citation: Zhang Y, Du P, Xiong F, Zhang X and Song H (2022) WRKY Genes Improve Drought Tolerance in Arachis duranensis. Front. Plant Sci. 13:910408. doi: 10.3389/fpls.2022.910408
Received: 01 April 2022; Accepted: 05 May 2022;
Published: 26 May 2022.
Edited by:
Xiaoming Song, North China University of Science and Technology, ChinaReviewed by:
Lihu Wang, Hebei University of Engineering, ChinaCopyright © 2022 Zhang, Du, Xiong, Zhang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Song, Ymlvc29uZ2h1aUBvdXRsb29rLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.