- Ecology and Evolution, Research School of Biology, The Australian National University, Canberra, ACT, Australia
The Orchidaceae is rivaled only by the Asteraceae as the largest plant family, with the estimated number of species exceeding 25,000 and encompassing more than 700 genera. To gain insights into the mechanisms driving species diversity across both global and local scales, well-supported phylogenies targeting different taxonomic groups and/or geographical regions will be crucial. High-throughput sequencing technologies have revolutionized the field of molecular phylogenetics by simplifying the process of obtaining genome-scale sequence data. Consequently, there has been an explosive growth of such data in public repositories. Here we took advantage of this unprecedented access to transcriptome data from predominantly non-phylogenetic studies to assess if it can be repurposed to gain rapid and accurate phylogenetic insights across the orchids. Exhaustive searches revealed transcriptomic data for more than 100 orchid species spanning 5 subfamilies, 13 tribes, 21 subtribes, and 50 genera that were amendable for exploratory phylotranscriptomic analysis. Next, we performed re-assembly of the transcriptomes before strategic selection of the final samples based on a gene completeness evaluation. Drawing on these data, we report phylogenetic analyses at both deep and shallow evolutionary scales via maximum likelihood and shortcut coalescent species tree methods. In this perspective, we discuss some key outcomes of this study and conclude by highlighting other complementary, albeit rarely explored, insights beyond phylogenetic analysis that repurposed multi-tissue transcriptome can offer.
Introduction
The Orchidaceae is rivaled only by the Asteraceae as the largest plant family, with an estimate of some 25,000 species (∼ 8% of plants) shared across at least 700 genera (Chase et al., 2015). Orchids can be found in a wide range of habitats across tropical, subtropical, and temperate regions (Swarts and Dixon, 2009), and hold great interest from an ecological and evolutionary standpoint. They are particularly well known for their diverse morphological adaptations such as epiphytism, with at least 18,000 species adopting this lifestyle (Gravendeel et al., 2004), and their dependence on mycorrhizal fungi interactions for germination (Phillips et al., 2020). Additionally, many orchids employ highly specialized or unusual pollination strategies (Jersáková et al., 2006; Schiestl and Schlüter, 2009; Bohman et al., 2016; Wong et al., 2017b; Trunschke et al., 2021). These characteristics make orchids ideal targets for exploring diverse ecological and evolutionary questions (Peakall, 2007).
Plastome and Mitochondrial Genome Phylogenomics in Orchids
To gain insights into the mechanisms driving the species diversity of the Orchidaceae across both global and local scales, investigations targeting different taxonomic and geographic scales (e.g., across specific tribe to genus) and a well-supported phylogeny are crucial. Rapid advancements in high-throughput sequencing and bioinformatics methods have enabled the use of genome (nuclear, plastid, and mitochondrial) and transcriptome sequences (including coding and non-coding regions) amendable for phylogenetic inference (McKain et al., 2018). Currently, the newfound ease of sequencing whole plastomes makes cpDNA the most widely used target for both small- and broad-scale orchid phylogenetic studies in terms of size (i.e., from tens to hundreds of species) and taxonomic breadth (i.e., within a genus or across five subfamilies) (Barrett et al., 2014; Givnish et al., 2015; Li Y. X. et al., 2019; Kim et al., 2020; Liu et al., 2020; Smidt et al., 2020; Serna-Sánchez et al., 2021). Notably, the first broad-scale plastome phylogeny for the Orchidaceae was based on maximum-likelihood (ML) analysis of 75 cpDNA coding-genes across 39 genera representing all five subfamilies and 16 (of 17) tribes (Givnish et al., 2015). Many previously underrepresented genera are now included in a cpDNA study of 78 genes spanning 264 species, 117 genera, 18 tribes, and 28 subtribes (Serna-Sánchez et al., 2021).
Phylotranscriptomics in Orchids
Notwithstanding the extraordinary insight from plastome studies gained in the last decade, phylogenetic studies leveraging transcriptome datasets to identify putative single-copy genes (or orthogroups) are now emerging as a new tool to aid plant molecular systematics. Compared to the linked genes in cpDNA which collectively represent just one evolutionary history, nuclear genes offer several advantages including their biparental inheritance, higher substitution rates, and access to thousands of unlinked genes each with the potential to represent independent evolutionary histories, among others (Zhang et al., 2012; Smith, 2015).
To date, several broad-scale transcriptome-based phylogenetic analyses of the Orchidaceae have been performed, but with limited taxon sampling compared to the available plastome-based phylogenies (Deng et al., 2015; Zhang et al., 2017; Guo et al., 2018; Unruh et al., 2018; Piñeiro Fernández et al., 2019; Wong et al., 2019). Nonetheless, one study incorporated the transcriptomes of 13 orchid species representing five subfamilies and 10 phylogenetic informative non-orchid outgroups to build a high-confidence phylogenetic tree of the Orchidaceae from >700 single-copy orthogroups using both concatenation and coalescence-based summary methods (Unruh et al., 2018). This analysis firmly placed the genus Cypripedium sister to the rest of the slipper orchid genera (e.g., Phragmipedium, Mexipedium, Paphiopedilum, Selenipedium), a previously uncertain relationship. The effectiveness of using transcriptomes to resolve relationships at shallower scales have also been demonstrated. For example, previously uncertain phylogenetic relationships among closely related orchid species (e.g., 8 Cypripedium, 4 Ophrys, and 5 Gymnadenia spp.) have also been clarified using hundreds of putative single-copy genes (Guo et al., 2018; Piñeiro Fernández et al., 2019).
Target Sequence Capture Phylogenomics in Orchids
Putative single-copy genes are already being used to design target enrichment probe sets capable of spanning broad evolutionary scales. Two universal probe sets that provide orchid coverage are: Angiosperms353 (Johnson et al., 2019) and Orchidaceae963 (Eserman et al., 2021). For example, a recent phylogenomic analysis of 75 species representing 69 genera, 16 tribes, and 24 subtribes used the Angiosperms353 universal probe set to target 294 low-copy nuclear genes (Pérez-Escobar et al., 2021). This study revealed that higher-level phylogenetic relationships were robust and largely congruent with earlier plastome- and mitochondrial genome-based phylogenies (Li Y. X. et al., 2019; Serna-Sánchez et al., 2021). However, several instances of strongly supported discordances in both shallow and deep time were also revealed. The adaptability of universal probe sets, such as Angiosperms353, for resolving within-genus relationships of some Epidendrum (Granados Mendoza et al., 2020) and Lepanthes (Bogarín et al., 2018) orchids, has also been demonstrated. Nonetheless, the use of more targeted probe sets may be necessary to provide greater phylogenetic resolution at the subtribe to subspecific levels, as exemplified by a study of 30 genera spanning the tribe Diurideae and 24 closely related Caladenia species (Peakall et al., 2021).
Opportunities for Repurposing Transcriptomes for Phylogenetic Analysis in Orchids
In this perspective, we demonstrate that rapid and accurate phylogenetic insights across the Orchidaceae can be gained from repurposed multi-tissue transcriptomes that were sequenced primarily for non-phylogenetic studies. We conclude by discussing some feasible avenues of research beyond phylogenetic analysis that repurposed multi-tissue transcriptome can offer. In 2021, we conducted an exhaustive search for orchid transcriptome datasets in public repositories and the literature. Our search revealed transciptomes were available for more than 100 orchid species spanning 5 subfamilies, 13 tribes, and 20 subtribes, and drawn from over 50 published studies (Supplementary Table 1).
To use this large volume of data effectively, it was necessary to first re-assemble the transcriptomes from the publically accessible raw data. In total, the transcriptomes of 133 target orchid species were reassembled from a total of ∼ 8.5 billion high-quality reads using Trinity (Haas et al., 2013). Next, the outcomes of a gene completeness evaluation using BUSCOs (Waterhouse et al., 2018) was used to gauge assembly quality and to cull samples with poor scores (i.e., < 60%). Finally, generic level representation was restricted to a maximum of two (when available) species per genus (Supplementary Figure 1 and Text). Thus, our final high-quality orchid-wide phylotranscriptome data set contained 69 orchids (48 genera) spanning 5 subfamilies, 13 tribes, and 21 subtribes. Four non-orchid Asparagales species (i.e., Molineria capitulata, Hypoxis hemerocallidea, Lanaria larata, Borya sphaerocephala) were included as outgroups. Single-copy (strict and relaxed) phylogenetic-informed orthogroups were then identified using OrthoFinder (Emms and Kelly, 2019) and concatenated alignments of protein sequences were evaluated by maximum-likelihood and shortcut-coalescent phylogenetic analysis using IQTREE (Minh et al., 2020) and ASTRAL (Zhang et al., 2018), respectively (see Supplementary Text and Methods for additional details).
Taxonomic Representation Across the Phylotranscriptome Data Set
Despite originating from public databases which constrains the choice of samples available, the final phylotranscriptome data set of 69 species achieved wide taxonomic coverage across the Orchidaceae and outgroups (Figure 1). This included all known genera of the Apostasioideae and Cypripedioideae subfamilies, while only the tribe Pogonieae lacked representation within the Vanilloideae. Tribes spanning both the lower and higher Epidendroideae were also well covered, with a total of 16 genera, 9 tribes, and 10 subtribes represented. Finally, despite a trend of the Orchidoideae being previously under-represented, this gap has been filled in this study. In particular, genome-scale phylogenetic studies (based on whole plastome, transcriptome, or mitochondria genome) have had poor representation of the diverse Australasian tribe the Diurideae (Givnish et al., 2015; Li Y. X. et al., 2019; Kim et al., 2020; Smidt et al., 2020). Here our phylogenetic analysis included 18 Diurideae species from 10 genera and 8 subtribes. Consequently, this phylotranscriptome dataset provides one of the most comprehensive genera and subtribe representations for the Orchidoideae in phylogenomic studies to date, with a total number of genera, tribes, and subtribes of 22, 3, and 11, respectively (Figure 1).
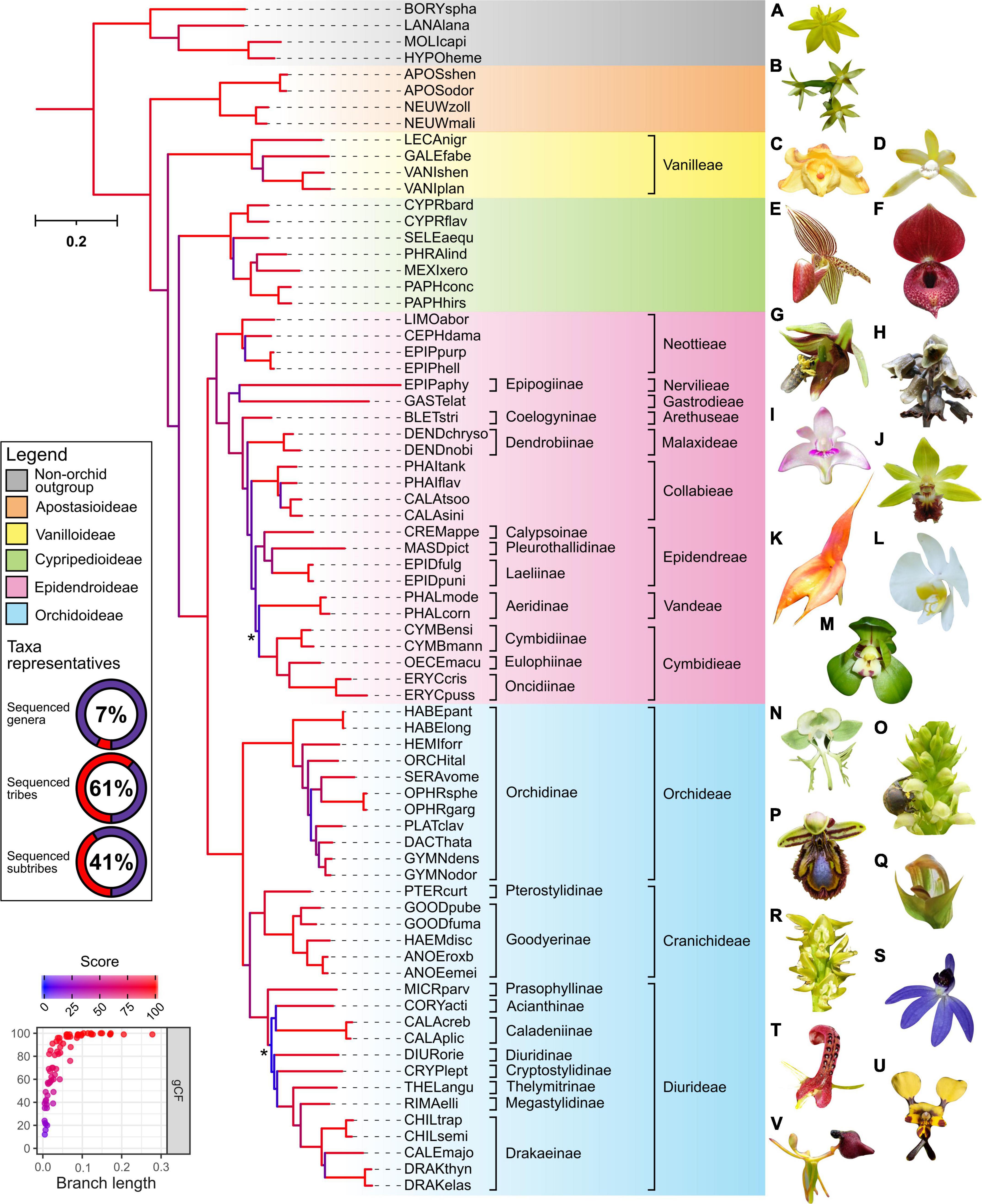
Figure 1. Outcomes of phylogenetic analysis across the Orchidaceae from repurposed multi-tissue transcriptomes. The maximum-likelihood IQ-TREE phylogeny of 69 orchids (spanning 5 subfamilies, 13 tribes, 21 subtribes, and 48 genera) and four non-orchid species as outgroup was based on partition analysis of 633 amino-acid alignments with a total of 317,221 (139,913 parsimony-informative) sites (see Supplementary Methods for details). The abbreviated species name is shown (see Supplementary Table 1 for details). Background color indicates subfamily grouping. The donut chart depict the taxonomic representativeness of included orchid genera, subtribe, and tribe (see relevant placeholder in the phylogeny for details). Asterisk indicate ultrafast bootstrap (UFboot) or SH-aLRT branch support <95 and <80%, respectively. Branch length indicates the total number of substitutions per site and branch colors correspond to gene concordance factors (gCF)–the proportion of gene trees that decisively support the given bifurcation. Inset (bottom left) shows a strong positive relationship of gCF scores with branch length. See Supplementary Figure 2 for additional details on the corresponding shortcut coalescent ASTRAL phylogeny. Representative species of the outgroup (A) Hypoxis hirsuta and subfamilies across the Orchidaceae are indicated (B) Apostasia nipponica–Apostasioideae; (C) Galeola lindleyana and (D) Lecanorchis suginoana–Vanilloideae; (E) Paphiopedilum rothschildianum and (F) Cypripedium lichiangense–Cypripedioideae; (G) Epipactis veratrifolia, (H) Gastrodia fontinalis, (I) Dendrobium kingianum, (J) Calanthe tricarinata, (K) Masdevallia veitchiana, (L) Phalaenopsis amabilis, and (M) Cymbidium goeringii–Epidendroideae; (N) Habenaria limprichtii, (O) Satyrium microrrhynchum, (P) Ophrys speculum, (Q) Pterostylis curta, (R) Prasophyllum elatum, (S) Cyanicula caerulea, (T) Cryptostylis leptochila, (U) Diuris pardina, and (V) Drakaea glyptodon–Orchidoideae. All images have been reproduced with permission from the respective copyright holders. Please refer to the section “Acknowledgments” for image credits.
Deep and Shallow Phylotranscriptome Relationships Are Robust and Consistent With Other Phylogenomics Datasets
The outcomes of our phylotranscriptomic analysis (Figure 1) revealed robust and largely consistent findings with other phylogenomics datasets (Givnish et al., 2015; Kim et al., 2020; Liu et al., 2020; Eserman et al., 2021; Pérez-Escobar et al., 2021; Serna-Sánchez et al., 2021). As expected, the backbone phylogeny of the Orchidaceae revealed five distinct clades: Apostasioideae (4 species) was placed sister to the other four subfamilies, followed by Vanilloideae (4 species), and by Cypripedioideae (7 species) which is placed sister to Orchidoideae (30 species) and Epidendroideae (24 species). Within the Vanilloideae subfamily, Lecanorchis was placed sister to a clade formed by Galeola and Vanilla. Within the Cypripedioideae, Cypripedium is basal, followed by Selenipedium successively sister to a clade formed by Phragmipedium, Mexipedium, and Paphiopedilum species.
Within the Epidendroideae, for almost all branches leading to the relevant tribes except Vandeae, Epidendreae, and Cymbidieae, the topology was also consistent with other orchid-wide species trees inferred from genome-scale datasets. For example, clear separation between lower (i.e., tribes Neottieae, Gastrodieae, Nervilieae) and higher (i.e., in the order of tribes Arethuseae, Malaxideae, Collabieae, Epidendreae, Vandeae, and Cymbidieae) Epidendroid species were evident. The only case of poor branch support, low gCF and sCF (i.e., proportion of decisive gene trees and alignment sites supporting a given branch supporting a given branch, respectively), and local PP scores pertains to the placement of the tribe Vandeae (i.e., Phalaenopsis spp.) in the ML and ASTRAL species tree (Supplementary Figure 2 and Text for further information). This point of uncertainty is consistent with findings in other recent broad-scale nuclear orchid phylogenomic studies (Eserman et al., 2021; Pérez-Escobar et al., 2021). This now well characterized disparity is postulated to reflect widespread incomplete lineage sorting of nuclear genes during the rapid radiations at the early stages of the evolution of the Vandeae, Epidendreae, and Cymbideae tribes (Eserman et al., 2021).
Within the Orchidoideae, the tribe Orchideae (11 species) is placed sister to a clade formed by Diurideae (13 species) and Cranichideae (6 species) with robust support. Subtribe relationships within Cranichideae were resolved with the subtribe Goodyerinae forming a well-supported clade, sister to Pterostylis (subtribe Pterostylidinae). Relatively short branch lengths and low gCF/sCF scores along the backbone and some terminal taxa of the tribes Diurideae and Orchideae were also observed. Within Orchideae, the genus Habenaria was the outermost group followed by the genus Hemipilia and Orchis. Next, the genus Serapias forms a clade with Ophrys and is placed sister to a clade containing Platanthera, Dactylorhiza, and Gymnadenia species.
Concerning the tribe Diurideae, most subtribe and genus level relationships were congruent with those obtained from a recent phylogenomic study of this tribe using a multi-tiered sequence capture strategy (Peakall et al., 2021). Like the former study, poor support and very low gene concordance factors were observed on the very short but deep branch that separates the sole outermost genus, Microtis (Prasophyllinae) from the rest of the tribe. As in the Epidendroideae, such uncertainly likely reflects the incomplete lineage sorting during the rapid radiations underpinning the formation of the major subtribes (Peakall et al., 2021).
To ascertain if robust phylogenetic relationships can be attained at shallower evolutionary depths (i.e., within specific subfamily and genus) from repurposed transcriptomes, samples corresponding to the Cypripedioideae subfamily (19 species) and the genus Phalaenopsis (11 species) were targeted. Almost all nodes showed robust support and the topologies closely mirror those inferred using a few chloroplast and low-copy nuclear genes but with broader taxon sampling (Tsai et al., 2010; Guo et al., 2012). As already indicated by other phylotranscriptomic studies (Guo et al., 2018; Unruh et al., 2018; Piñeiro Fernández et al., 2019), our analyses re-confirm the potential for the phylogenetic analysis of transcriptomes to resolve relationships at relatively shallow scales.
The Prospects of Using Transcriptomic Datasets for Phylogenetic Inference and Beyond
While our study has convincingly demonstrated the potential of phylotranscriptomic studies across the Orchidaceae, there are still several major challenges posed across multiple levels. For example, despite our recent exhaustive search of orchid transcriptomes in public repositories, comprehensive coverage spanning: i. many genera, tribes, and subtribes (Li Y. X. et al., 2019; Kim et al., 2020; Serna-Sánchez et al., 2021) or ii. expanded coverage within a particular genus (Guo et al., 2021) is lacking when compared to those from recent plastome phylogenies. Thus, given the size of the Orchidaceae, we recommend targeted sequencing to help fill obvious gaps not yet available in the public datasets. Nonetheless, some phylotranscriptome studies demonstrate that past transcriptomic datasets can be a key resource for increasing taxonomic breadth, which when lacking is known to affect overall phylogenetic accuracy (Zwickl and Hillis, 2002). One example is the phylotranscriptome of Asterids where approximately 40% of the 365 species represented were largely repurposed (Zhang et al., 2020).
In the same way that transcriptomes can be utilized for phylogenetic studies, assembled whole genome sequences may be repurposed for combined phylogenomic/transcriptomic studies of the Orchidaceae. For example, high-depth whole-genome sequencing of 689 vascular plant species from a botanical garden (Liu et al., 2019) contained at least 10 orchid species spanning the genera Dendrobium, Renanthera, Vanda, Tropidia, and Hylophila, among others, may be used to complement the phylotranscriptomic analysis presented here.
In the Orchidaceae, mycoheterotrophy has evolved sporadically with estimates of some 200 almost fully mycoheterotrophic species, spanning 43 genera (Merckx, 2013). Partial mycoheterotrophy is also likely to be much more widespread across the orchids, than currently documented (Gebauer et al., 2016). Fully mycoheterotrophic orchids often exhibit severely reduced plastid genome, thus, they are often devoid of adequate plastid gene sequences for phylogenetic analysis (Feng et al., 2016). Partially mycoheterotrophic orchid species may also exhibit varying degrees of plastome degradation. In such cases, nuclear DNA sequences may provide the only reliable data for robust phylogenetic analysis (Cameron, 2004; Górniak et al., 2010; Freudenstein and Chase, 2015). Our findings from transcriptomic data show that the placement of Gastrodia elata is consistent with earlier orchid-wide phylotranscriptome studies (Deng et al., 2015) while the relationship of Epipogium aphyllum (tribe Nervilieae) being sister to G. elata (tribe Gastrodieae) appears to be a new finding from this present study. Interestingly, the exceptionally long branch lengths in the clades formed by the mycoheterotrophic G. elata and E. aphyllum also points to accelerated substitution rates of nuclear genes at a genome-wide scale, a general feature of many parasitic plants (Bromham et al., 2013).
The difficulty in scaling up standard phylogenetic procedures to hundreds, if not thousands of species, will likely persist in future orchid phylotranscriptome studies. Therefore, we take the opportunity to highlight one promising approach using homologous k-mer (substring of sequence) blocks that bypass several arduous steps including annotation, ortholog detection, and alignment (Sanderson et al., 2017). Interestingly, Sanderson et al. (2017) recovered trees using homologous k-mer blocks and published ones were nearly identical in all seven genome-scale datasets tested containing tens to hundreds of species highlighting its robustness and scalability. Preliminary evaluation of this approach on target sequence capture data encompassing hundreds of loci and samples–67 samples spanning the tribe Diurideae and 72 samples involving 24 closely related Caladenia species–revealed its potential to recover species trees matching conventional workflows but also better group morphologically similar members of species complexes (Peakall et al., 2021).
Past and emerging transcriptome datasets can also be mined for the Angiosperms353 (Johnson et al., 2019) and Orchidaceae963 single-copy genes (Eserman et al., 2021) for downstream phylotranscriptomic analysis. This approach bypasses the need for the arduous ortholog inference that often scales poorly with large datasets spanning hundreds of species. However, it remains to be seen if these predefined single-copy genes will be effective in resolving shallow and/or complex evolutionary relationships (e.g., in recent and/or rapid radiating lineages). For the latter purpose, the use of thousands of single-copy genes unique to the study system enabled by targeted studies may be more effective as exemplified in this present study (Supplementary Figures 3, 4) and several others (Guo et al., 2018; Wu et al., 2018; Piñeiro Fernández et al., 2019; Züst et al., 2020; Peakall et al., 2021).
Benefits of Phylotranscriptomics for Gene Duplication and Selection
The prospects of using transcriptomic datasets to gain insights into whole-genome (WGD) or lineage-specific gene duplication events (Zhang et al., 2017; Unruh et al., 2018) and genome-wide detection of positively selected genes of specific evolutionary branches (Balao et al., 2017; Guo et al., 2018; Wong et al., 2019) are also promising avenues of research. For example, two studies have identified a single WGD event shared by all (extant) orchids using transcriptomes from a largely independent list of orchid and non-orchid species (Zhang et al., 2017; Unruh et al., 2018). Lineage-specific gene duplication events leading to the diversification of Cypripedioideae have also been inferred and mapped onto the phylogeny (Unruh et al., 2018).
Signatures of genes under positive selection that may have contributed to species diversification or adaptive evolution in specific evolutionary branches of orchids have also been revealed. For example, elevated evolutionary rates at several candidate volatile biosynthesis-related genes (e.g., fatty acid metabolism) have been independently observed in the unrelated genera of Chiloglottis and Cypripedium (Guo et al., 2018; Wong et al., 2019). In Chiloglottis, the genes encoding the β-ketoacyl-ACP synthase homologs implicated in the biosynthesis of the 2,5-dialkyl cyclohexane-1,3-dione volatiles (chiloglottones) that attract the specific male wasp pollinators (Peakall et al., 2010), show strong signatures of selection (Wong et al., 2019). In two Dactylorhiza orchids, positively selected genes were notably enriched with biotic defense-related pathways related to physical and chemical adaptations (Balao et al., 2017).
Conclusion
In this study, we showcase for the first time that a large set of publically available transcriptomes that were sequenced primarily for non-phylogenetic studies can be effectively repurposed to gain phylogenetic insights across broad evolutionary scales of the Orchidaceae. This strategy greatly increased the genus, tribe and subtribe representation of both small (e.g., Cypripedioideae) and larger subfamilies (e.g., Orchidoideae and Epidendroideae) and provided access to thousands of informative sequences (e.g., putative single-copy genes or orthogroups) amendable for phylogenetic inference. We predict that phylotranscriptomic studies will provide an additional platform for future molecular systematic studies and other investigations into the mechanisms driving the extraordinary species diversity of the Orchidaceae.
Data Availability Statement
The datasets presented in this study can be found in online repositories via NCBI Sequence Read Archive (SRA) accession from respective publications listed in Supplementary Table 1. When records of the original publication(s) are not available, the designated Bioproject accession (PRJNA#) is shown.
Author Contributions
DW designed the study and analyzed the data. RP and DW secured funding. DW wrote the manuscript with assistance from RP. Both authors have read and approved the manuscript.
Funding
This work was supported by the Australian Research Council projects DE190100249 to DW and DP150102762 to RP.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the plant research community for making a wide variety of high-throughput sequencing (especially RNA-seq) datasets publically available. Photographs in Figure 1 were taken by Zong-Xin Ren (a,c,f,g,j,n), Kenji Suetsugu (b,d,h), Jorun Tharaldsen (p), RP (e,k,l,m,o,r,t,u,v), and DW (i,q,s).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.910362/full#supplementary-material
References
Balao, F., Trucchi, E., Wolfe, T. M., Hao, B. H., Lorenzo, M. T., Baar, J., et al. (2017). Adaptive sequence evolution is driven by biotic stress in a pair of orchid species (Dactylorhiza) with distinct ecological optima. Mol. Ecol. 26, 3649–3662. doi: 10.1111/mec.14123
Barrett, C. F., Freudenstein, J. V., Li, J., Mayfield-Jones, D. R., Perez, L., Pires, J. C., et al. (2014). Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol. 31, 3095–3112. doi: 10.1093/molbev/msu252
Bogarín, D., Pérez-Escobar, O. A., Groenenberg, D., Holland, S. D., Karremans, A. P., Lemmon, E. M., et al. (2018). Anchored hybrid enrichment generated nuclear, plastid and mitochondrial markers resolve the Lepanthes horrida (Orchidaceae: Pleurothallidinae) species complex. Mol. Phylogenet. Evol. 129, 27–47. doi: 10.1016/j.ympev.2018.07.014
Bohman, B., Flematti, G. R., Barrow, R. A., Pichersky, E., and Peakall, R. (2016). Pollination by sexual deception - it takes chemistry to work. Curr. Opin. Plant Biol. 32, 37–46. doi: 10.1016/j.pbi.2016.06.004
Bromham, L., Cowman, P. F., and Lanfear, R. (2013). Parasitic plants have increased rates of molecular evolution across all three genomes. BMC Evol. Biol. 13:126. doi: 10.1186/1471-2148-13-126
Cai, J., Liu, X., Vanneste, K., Proost, S., Tsai, W.-C., Liu, K.-W., et al. (2015). The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 47, 65–72. doi: 10.1038/ng.3149
Cameron, K. M. (2004). Utility of plastid psaB gene sequences for investigating intrafamilial relationships within Orchidaceae. Mol. Phylogenet. Evol. 31, 1157–1180. doi: 10.1016/j.ympev.2003.10.010
Chao, Y. T., Chen, W. C., Chen, C. Y., Ho, H. Y., Yeh, C. H., Kuo, Y. T., et al. (2018). Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 16, 2027–2041. doi: 10.1111/pbi.12936
Chao, Y.-T., Yen, S.-H., Yeh, J.-H., Chen, W.-C., and Shih, M.-C. (2017). Orchidstra 2.0-A transcriptomics resource for the orchid family. Plant Cell Physiol. 58:e9. doi: 10.1093/pcp/pcw220
Chase, M. W., Cameron, K. M., Freudenstein, J. V., Pridgeon, A. M., Salazar, G., van den Berg, C., et al. (2015). An updated classification of Orchidaceae. Bot. J. Linn. Soc. 177, 151–174. doi: 10.1111/boj.12234
Chuang, Y. C., Hung, Y. C., Tsai, W. C., Chen, W. H., and Chen, H. H. (2018). PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids. J. Exp. Bot. 69, 4363–4377. doi: 10.1093/jxb/ery246
De Paolo, S., Salvemini, M., Gaudio, L., and Aceto, S. (2014). De novo transcriptome assembly from inflorescence of Orchis italica: analysis of coding and non-coding transcripts. PLoS One 9:e102155. doi: 10.1371/journal.pone.0102155
Deng, H., Zhang, G. Q., Lin, M., Wang, Y., and Liu, Z. J. (2015). Mining from transcriptomes: 315 single-copy orthologous genes concatenated for the phylogenetic analyses of Orchidaceae. Ecol. Evol. 5, 3800–3807. doi: 10.1002/ece3.1642
Dhiman, N., Sharma, N. K., Thapa, P., Sharma, I., Kumar Swarnkar, M., Chawla, A., et al. (2019). De novo transcriptome provides insights into the growth behaviour and resveratrol and trans-stilbenes biosynthesis in Dactylorhiza hatagirea - An endangered alpine terrestrial orchid of western Himalaya. Sci. Rep. 9:13133. doi: 10.1038/s41598-019-49446-w
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. doi: 10.1186/s13059-019-1832-y
Eserman, L. A., Thomas, S. K., Coffey, E. E. D., and Leebens-Mack, J. H. (2021). Target sequence capture in orchids: developing a kit to sequence hundreds of single-copy loci. Appl. Plant Sci. 9:e11416. doi: 10.1002/aps3.11416
Fang, L., Kong, X., Wen, Y., Li, J., Yin, Y., Li, L., et al. (2021). Characterization of embryo and protocorm development of Paphiopedilum spicerianum. Plant Physiol. Biochem. 167, 1024–1034. doi: 10.1016/j.plaphy.2021.09.001
Feng, Y. L., Wicke, S., Li, J. W., Han, Y., Lin, C. S., Li, D. Z., et al. (2016). Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol. Evol. 8, 2164–2175. doi: 10.1093/gbe/evw144
Fochi, V., Chitarra, W., Kohler, A., Voyron, S., Singan, V. R., Lindquist, E. A., et al. (2017). Fungal and plant gene expression in the Tulasnella calospora–Serapias vomeracea symbiosis provides clues about nitrogen pathways in orchid mycorrhizas. New Phytol. 213, 365–379. doi: 10.1111/nph.14279
Freudenstein, J. V., and Chase, M. W. (2015). Phylogenetic relationships in Epidendroideae (Orchidaceae), one of the great flowering plant radiations: progressive specialization and diversification. Ann. Bot. 115, 665–681. doi: 10.1093/aob/mcu253
Gebauer, G., Preiss, K., and Gebauer, A. C. (2016). Partial mycoheterotrophy is more widespread among orchids than previously assumed. New Phytol. 211, 11–15. doi: 10.1111/nph.13865
Givnish, T. J., Spalink, D., Ames, M., Lyon, S. P., Hunter, S. J., Zuluaga, A., et al. (2015). Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B Biol. Sci. 282:20151553. doi: 10.1098/rspb.2015.1553
Górniak, M., Paun, O., and Chase, M. W. (2010). Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, Xdh: congruence with organellar and nuclear ribosomal DNA results. Mol. Phylogenet. Evol. 56, 784–795. doi: 10.1016/j.ympev.2010.03.003
Granados Mendoza, C., Jost, M., Hágsater, E., Magallón, S., van den Berg, C., Lemmon, E. M., et al. (2020). Target nuclear and off-target plastid hybrid enrichment data inform a range of evolutionary depths in the orchid genus Epidendrum. Front. Plant Sci. 10:1761. doi: 10.3389/fpls.2019.01761
Gravendeel, B., Smithson, A., Slik, F. J. W., and Schuiteman, A. (2004). Epiphytism and pollinator specialization: drivers for orchid diversity? Philos. Trans. R. Soc. B Biol. Sci. 359, 1523–1535. doi: 10.1098/rstb.2004.1529
Guo, Y. Y., Luo, Y. B., Liu, Z. J., and Wang, X. Q. (2012). Evolution and biogeography of the slipper orchids: Eocene vicariance of the conduplicate genera in the old and new world tropics. PLoS One 7:e38788. doi: 10.1371/journal.pone.0038788
Guo, Y. Y., Yang, J. X., Bai, M. Z., Zhang, G. Q., and Liu, Z. J. (2021). The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC Plant Biol. 21:248. doi: 10.1186/s12870-021-03053-y
Guo, Y.-Y., Zhang, Y.-Q., Zhang, G.-Q., Huang, L.-Q., and Liu, Z.-J. (2018). Comparative transcriptomics provides insight into the molecular basis of species diversification of section Trigonopedia (Cypripedium) on the Qinghai-Tibetan Plateau. Sci. Rep. 8:11640. doi: 10.1038/s41598-018-30147-9
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hamabata, T., Kinoshita, G., Kurita, K., Cao, P. L., Ito, M., Murata, J., et al. (2019). Endangered island endemic plants have vulnerable genomes. Commun. Biol. 2:244. doi: 10.1038/s42003-019-0490-7
Heyduk, K., Hwang, M., Albert, V., Silvera, K., Lan, T., Farr, K., et al. (2019). Altered gene regulatory networks are associated with the transition from C3 to crassulacean acid metabolism in Erycina (Oncidiinae: Orchidaceae). Front. Plant Sci. 9:2000. doi: 10.3389/fpls.2018.02000
Hu, C., Yang, H., Jiang, K., Wang, L., Yang, B., Hsieh, T., et al. (2018). Development of polymorphic microsatellite markers by using de novo transcriptome assembly of Calanthe masuca and C. sinica (Orchidaceae). BMC Genomics 19:800. doi: 10.1186/s12864-018-5161-4
Hu, Y., Resende, M. F. R., Bombarely, A., Brym, M., Bassil, E., and Chambers, A. H. (2019). Genomics-based diversity analysis of Vanilla species using a Vanilla planifolia draft genome and genotyping-by-sequencing. Sci. Rep. 9:3416. doi: 10.1038/s41598-019-40144-1
Huang, H., Kuo, Y. W., Chuang, Y. C., Yang, Y. P., Huang, L. M., Jeng, M. F., et al. (2021). Terpene synthase-b and Terpene synthase-e/f genes produce monoterpenes for Phalaenopsis bellina floral scent. Front. Plant Sci. 12:700958. doi: 10.3389/fpls.2021.700958
Huolin, L., Yuan, T., Wenjing, Y., Liping, L., and Boyun, Y. (2019). De novo transcriptome assembly and analysis of the codon usage bias of the MADS-box gene family in Cymbidium kanran. Indian J. Genet. Plant Breed. 79, 485–493.
Ja̧kalski, M., Minasiewicz, J., Caius, J., May, M., Selosse, M. A., and Delannoy, E. (2021). The genomic impact of mycoheterotrophy in orchids. Front. Plant Sci. 12:632033. doi: 10.3389/fpls.2021.632033
Jersáková, J., Johnson, S. D., and Kindlmann, P. (2006). Mechanisms and evolution of deceptive pollination in orchids. Biol. Rev. 81, 219–235. doi: 10.1017/S1464793105006986
Jiang, Y., Tian, M., Wang, C., and Zhang, Y. (2021). Transcriptome sequencing and differential gene expression analysis reveal the mechanisms involved in seed germination and protocorm development of Calanthe tsoongiana. Gene 772:145355. doi: 10.1016/j.gene.2020.145355
Johnson, M. G., Pokorny, L., Dodsworth, S., Botigué, L. R., Cowan, R. S., Devault, A., et al. (2019). A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Syst. Biol. 68, 594–606. doi: 10.1093/sysbio/syy086
Kim, Y. K., Jo, S., Cheon, S. H., Joo, M. J., Hong, J. R., Kwak, M., et al. (2020). Plastome evolution and phylogeny of Orchidaceae, with 24 new sequences. Front. Plant Sci. 11:22. doi: 10.3389/fpls.2020.00022
Lallemand, F., Martin-Magniette, M. L., Gilard, F., Gakière, B., Launay-Avon, A., Delannoy, É, et al. (2019). In situ transcriptomic and metabolomic study of the loss of photosynthesis in the leaves of mixotrophic plants exploiting fungi. Plant J. 98, 826–841. doi: 10.1111/tpj.14276
Leal, B. S. S., Brandão, M. M., Palma-Silva, C., and Pinheiro, F. (2020). Differential gene expression reveals mechanisms related to habitat divergence between hybridizing orchids from the Neotropical coastal plains. BMC Plant Biol. 20:554. doi: 10.1186/s12870-020-02757-x
Lee, Y. I., Chen, M. C., Lin, L., Chung, M. C., and Leu, W. M. (2018). Increased expression of 9-cis-epoxycarotenoid dioxygenase, PtNCED1, associated with inhibited seed germination in a terrestrial orchid, Phaius tankervilliae. Front. Plant Sci. 9:1043. doi: 10.3389/fpls.2018.01043
Leebens-Mack, J. H., Barker, M. S., Carpenter, E. J., Deyholos, M. K., Gitzendanner, M. A., Graham, S. W., et al. (2019). One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685. doi: 10.1038/s41586-019-1693-2
Li, D. M., Wu, W., Zhang, D., Liu, X. R., Liu, X. F., and Lin, Y. J. (2015). Floral transcriptome analyses of four Paphiopedilum orchids with distinct flowering behaviors and development of simple sequence repeat markers. Plant Mol. Biol. Report. 33, 1928–1952. doi: 10.1007/s11105-015-0886-6
Li, J. W., Zhang, S. B., Xi, H. P., Bradshaw, C. J. A., and Zhang, J. L. (2020). Processes controlling programmed cell death of root velamen radicum in an epiphytic orchid. Ann. Bot. 126, 261–275. doi: 10.1093/aob/mcaa077
Li, J., Xu, Y., and Wang, Z. (2019). Construction of a high-density genetic map by RNA sequencing and eQTL analysis for stem length and diameter in Dendrobium (Dendrobium nobile × Dendrobium wardianum). Ind. Crops Prod. 128, 48–54. doi: 10.1016/j.indcrop.2018.10.073
Li, X., Luo, J., Yan, T., Xiang, L., Jin, F., Qin, D., et al. (2013). Deep sequencing-based analysis of the Cymbidium ensifolium floral transcriptome. PLoS One 8:e85480. doi: 10.1371/journal.pone.0085480
Li, Y. X., Li, Z. H., Schuitman, A., Chase, M. W., Li, J. W., Huang, W. C., et al. (2019). Phylogenomics of Orchidaceae based on plastid and mitochondrial genomes. Mol. Phylogenet. Evol. 139:106540. doi: 10.1016/j.ympev.2019.106540
Lin, C. S., Chen, J. J. W., Chiu, C. C., Hsiao, H. C. W., Yang, C. J., Jin, X. H., et al. (2017). Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 90, 994–1006. doi: 10.1111/tpj.13525
Lin, C. S., Hsu, C. T., Liao, D. C., Chang, W. J., Chou, M. L., Huang, Y. T., et al. (2016). Transcriptome-wide analysis of the MADS-box gene family in the orchid Erycina pusilla. Plant Biotechnol. J. 14, 284–298. doi: 10.1111/pbi.12383
Liu, D. K., Tu, X., Zhao, Z., Zeng, M. Y., Zhang, S., Ma, L., et al. (2020). Plastid phylogenomic data yield new and robust insights into the phylogeny of Cleisostoma–Gastrochilus clades (Orchidaceae, Aeridinae). Mol. Phylogenet. Evol. 145:106729. doi: 10.1016/j.ympev.2019.106729
Liu, H., Wei, J., Yang, T., Mu, W., Song, B., Yang, T., et al. (2019). Molecular digitization of a botanical garden: high-depth whole-genome sequencing of 689 vascular plant species from the Ruili botanical garden. Gigascience 8:giz007. doi: 10.1093/gigascience/giz007
Liu, S. S., Chen, J., Li, S. C., Zeng, X., Meng, Z. X., and Guo, S. X. (2015). Comparative transcriptome analysis of genes involved in GA-GID1-DELLA regulatory module in symbiotic and asymbiotic seed germination of Anoectochilus roxburghii (Wall.) Lindl. (Orchidaceae). Int. J. Mol. Sci. 16, 30190–30203. doi: 10.3390/ijms161226224
Lv, X., Zhang, M., Li, X., Ye, R., and Wang, X. (2018). Transcriptome profiles reveal the crucial roles of auxin and cytokinin in the “shoot branching” of Cremastra appendiculata. Int. J. Mol. Sci. 19:3354. doi: 10.3390/ijms19113354
Ma, X., Yin, X., Tang, Z., Ito, H., Shao, C., Meng, Y., et al. (2020). The RNA degradome: a precious resource for deciphering RNA processing and regulation codes in plants. RNA Biol. 17, 1223–1227. doi: 10.1080/15476286.2020.1757898
McKain, M. R., Johnson, M. G., Uribe-Convers, S., Eaton, D., and Yang, Y. (2018). Practical considerations for plant phylogenomics. Appl. Plant Sci. 6:e1038. doi: 10.1002/aps3.1038
Meng, X., Li, G., Gu, L., Sun, Y., Li, Z., Liu, J., et al. (2020). Comparative metabolomic and transcriptome analysis reveal distinct flavonoid biosynthesis regulation between petals of white and purple Phalaenopsis amabilis. J. Plant Growth Regul. 39, 823–840. doi: 10.1007/s00344-019-10025-y
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Niu, S. C., Huang, J., Zhang, Y. Q., Li, P. X., Zhang, G. Q., Xu, Q., et al. (2017). Lack of S-RNase-based gametophytic self-incompatibility in orchids suggests that this system evolved after the monocot-eudicot split. Front. Plant Sci. 8:1106. doi: 10.3389/fpls.2017.01106
Peakall, R. (2007). Speciation in the Orchidaceae: confronting the challenges. Mol. Ecol. 16, 2834–2837. doi: 10.1111/j.1365-294X.2007.03311.x
Peakall, R., Ebert, D., Poldy, J., Barrow, R. A., Francke, W., Bower, C. C., et al. (2010). Pollinator specificity, floral odour chemistry and the phylogeny of Australian sexually deceptive Chiloglottis orchids: implications for pollinator-driven speciation. New Phytol. 188, 437–450. doi: 10.1111/j.1469-8137.2010.03308.x
Peakall, R., Wong, D. C. J., Phillips, R. D., Ruibal, M., Eyles, R., Rodriguez-Delgado, C., et al. (2021). A multitiered sequence capture strategy spanning broad evolutionary scales: application for phylogenetic and phylogeographic studies of orchids. Mol. Ecol. Resour. 21, 1118–1140. doi: 10.1111/1755-0998.13327
Pérez-Escobar, O. A., Dodsworth, S., Bogarín, D., Bellot, S., Balbuena, J. A., Schley, R. J., et al. (2021). Hundreds of nuclear and plastid loci yield novel insights into orchid relationships. Am. J. Bot. 108, 1166–1180. doi: 10.1002/ajb2.1702
Phillips, R. D., Reiter, N., and Peakall, R. (2020). Orchid conservation: from theory to practice. Ann. Bot. 126, 345–362. doi: 10.1093/aob/mcaa093
Piñeiro Fernández, L., Byers, K., Cai, J., Sedeek, K. E. M., Kellenberger, R. T., Russo, A., et al. (2019). A phylogenomic analysis of the floral transcriptomes of sexually deceptive and rewarding European orchids, Ophrys and Gymnadenia. Front. Plant Sci. 10:1553. doi: 10.3389/fpls.2019.01553
Ramya, M., Park, P. H., Chuang, Y. C., Kwon, O. K., An, H. R., Park, P. M., et al. (2019). RNA sequencing analysis of Cymbidium goeringii identifies floral scent biosynthesis related genes. BMC Plant Biol. 19:337. doi: 10.1186/s12870-019-1940-6
Sanderson, M. J., Nicolae, M., and McMahon, M. M. (2017). Homology-aware phylogenomics at gigabase scales. Syst. Biol. 66, 590–603. doi: 10.1093/sysbio/syw104
Schelkunov, M. I., Penin, A. A., and Logacheva, M. D. (2018). RNA-seq highlights parallel and contrasting patterns in the evolution of the nuclear genome of fully mycoheterotrophic plants. BMC Genomics 19:602. doi: 10.1186/s12864-018-4968-3
Schiestl, F. P., and Schlüter, P. M. (2009). Floral isolation, specialized pollination, and pollinator behavior in orchids. Annu. Rev. Entomol. 54, 425–446. doi: 10.1146/annurev.ento.54.110807.090603
Sedeek, K. E., Qi, W., Schauer, M. A., Gupta, A. K., Poveda, L., Xu, S., et al. (2013). Transcriptome and proteome data reveal candidate genes for pollinator attraction in sexually deceptive orchids. PLoS One 8:e64621. doi: 10.1371/journal.pone.0064621
Serna-Sánchez, M. A., Pérez-Escobar, O. A., Bogarín, D., Torres-Jimenez, M. F., Alvarez-Yela, A. C., Arcila-Galvis, J. E., et al. (2021). Plastid phylogenomics resolves ambiguous relationships within the orchid family and provides a solid timeframe for biogeography and macroevolution. Sci. Rep. 11:6858. doi: 10.1038/s41598-021-83664-5
Smidt, E. D. C., Páez, M. Z., Vieira, L. D. N., Viruel, J., De Baura, V. A., Balsanelli, E., et al. (2020). Characterization of sequence variability hotspots in Cranichideae plastomes (Orchidaceae, Orchidoideae). PLoS One 15:e0227991. doi: 10.1371/journal.pone.0227991
Smith, D. R. (2015). Mutation rates in plastid genomes: they are lower than you might think. Genome Biol. Evol. 7, 1227–1234. doi: 10.1093/gbe/evv069
Suetsugu, K., Yamato, M., Miura, C., Yamaguchi, K., Takahashi, K., Ida, Y., et al. (2017). Comparison of green and albino individuals of the partially mycoheterotrophic orchid Epipactis helleborine on molecular identities of mycorrhizal fungi, nutritional modes and gene expression in mycorrhizal roots. Mol. Ecol. 26, 1652–1669. doi: 10.1111/mec.14021
Sun, Y., Wang, G., Li, Y., Jiang, L., Yang, Y., and Guan, S. (2016). De novo transcriptome sequencing and comparative analysis to discover genes related to floral development in Cymbidium faberi Rolfe. Springerplus 5:1458. doi: 10.1186/s40064-016-3089-1
Swarts, N. D., and Dixon, K. W. (2009). Terrestrial orchid conservation in the age of extinction. Ann. Bot. 104, 543–556. doi: 10.1093/aob/mcp025
Trunschke, J., Lunau, K., Pyke, G. H., Ren, Z. X., and Wang, H. (2021). Flower color evolution and the evidence of pollinator-mediated selection. Front. Plant Sci. 12:617851. doi: 10.3389/fpls.2021.617851
Tsai, C. C., Chiang, Y. C., Huang, S. C., Chen, C. H., and Chou, C. H. (2010). Molecular phylogeny of Phalaenopsis Blume (Orchidaceae) on the basis of plastid and nuclear DNA. Plant Syst. Evol. 288, 77–98. doi: 10.1007/s00606-010-0314-1
Unruh, S. A., McKain, M. R., Lee, Y. I., Yukawa, T., McCormick, M. K., Shefferson, R. P., et al. (2018). Phylotranscriptomic analysis and genome evolution of the Cypripedioideae (Orchidaceae). Am. J. Bot. 105, 631–640. doi: 10.1002/ajb2.1047
Valadares, R. B. S., Marroni, F., Sillo, F., Oliveira, R. R. M., Balestrini, R., and Perotto, S. (2021). A transcriptomic approach provides insights on the mycorrhizal symbiosis of the Mediterranean orchid Limodorum abortivum in nature. Plants 10:251. doi: 10.3390/plants10020251
Valadares, R. B. S., Perotto, S., Lucheta, A. R., Santos, E. C., Oliveira, R. M., and Lambais, M. R. (2020). Proteomic and transcriptomic analyses indicate metabolic changes and reduced defense responses in mycorrhizal roots of Oeceoclades maculata (Orchidaceae) collected in nature. J. Fungi 6:148. doi: 10.3390/jof6030148
Waterhouse, R. M., Seppey, M., Simao, F. A., Manni, M., Ioannidis, P., Klioutchnikov, G., et al. (2018). BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543–548. doi: 10.1093/molbev/msx319
Wong, D. C. J., Amarasinghe, R., Falara, V., Pichersky, E., and Peakall, R. (2019). Duplication and selection in β-ketoacyl-ACP synthase gene lineages in the sexually deceptive Chiloglottis (Orchidaceace). Ann. Bot. 123, 1053–1066. doi: 10.1093/aob/mcz013
Wong, D. C. J., Amarasinghe, R., Pichersky, E., and Peakall, R. (2018). Evidence for the involvement of fatty acid biosynthesis and degradation in the formation of insect sex pheromone-mimicking chiloglottones in sexually deceptive Chiloglottis Orchids. Front. Plant Sci. 9:389. doi: 10.3389/fpls.2018.00839
Wong, D. C. J., Pichersky, E., and Peakall, R. (2017b). The biosynthesis of unusual floral volatiles and blends involved in orchid pollination by deception: current progress and future prospects. Front. Plant Sci. 8:1955. doi: 10.3389/fpls.2017.01955
Wong, D. C. J., Amarasinghe, R., Rodriguez-Delgado, C., Eyles, R., Pichersky, E., and Peakall, R. (2017a). Tissue-specific floral transcriptome analysis of the sexually deceptive orchid Chiloglottis trapeziformis provides insights into the biosynthesis and regulation of its unique UV-B dependent floral volatile, chiloglottone 1. Front. Plant Sci. 8:1260. doi: 10.3389/fpls.2017.01260
Wong, D. C. J., Perkins, J., and Peakall, R. (2022). Anthocyanin and flavonol glycoside metabolic pathways underpin floral color mimicry and contrast in a sexually deceptive orchid. Front. Plant Sci. 13:860997. doi: 10.3389/fpls.2022.860997
Wu, M., Kostyun, J. L., Hahn, M. W., and Moyle, L. C. (2018). Dissecting the basis of novel trait evolution in a radiation with widespread phylogenetic discordance. Mol. Ecol. 27, 3301–3316. doi: 10.1111/mec.14780
Xu, D., Chen, H., Aci, M., Pan, Y., Shangguan, Y., Ma, J., et al. (2018). De Novo assembly, characterization and development of EST-SSRs from Bletilla striata transcriptomes profiled throughout the whole growing period. PLoS One 13:e0205954. doi: 10.1371/journal.pone.0205954
Xu, H., Bohman, B., Wong, D. C. J., Rodriguez-Delgado, C., Scaffidi, A., Flematti, G. R., et al. (2017). Complex sexual deception in an orchid is achieved by co-opting two independent biosynthetic pathways for pollinator attraction. Curr. Biol. 27, 1867–1877. doi: 10.1016/j.cub.2017.05.065
Xu, Q., Niu, S.-C., Li, K.-L., Zheng, P.-J., Zhang, X.-J., Jia, Y., et al. (2022). Chromosome-scale assembly of the Dendrobium nobile genome provides insights into the molecular mechanism of the biosynthesis of the medicinal active ingredient of Dendrobium. Front. Genet. 13:844622. doi: 10.3389/fgene.2022.844622
Xu, Y., Jia, R., Zhou, Y., Cheng, H., Zhao, X., and Ge, H. (2018). Development and characterization of polymorphic EST-SSR markers for Paphiopedilum henryanum (Orchidaceae). Appl. Plant Sci. 6:e01152. doi: 10.1002/aps3.1152
Yu, J., Qiang, W., Qin-Qin, S., Bi-Ping, Z., and Jun-Rong, H. (2020). Transcriptome analysis reveals genes associated with leaf color mutants in Cymbidium longibracteatum. Tree Genet. Genomes 16:44. doi: 10.1007/s11295-020-01440-4
Yuan, Y., Jin, X., Liu, J., Zhao, X., Zhou, J., Wang, X., et al. (2018). The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 9:1615. doi: 10.1038/s41467-018-03423-5
Yuan, Y., Zhang, J., Kallman, J., Liu, X., Meng, M., and Lin, J. (2019). Polysaccharide biosynthetic pathway profiling and putative gene mining of Dendrobium moniliforme using RNA-Seq in different tissues. BMC Plant Biol. 19:521. doi: 10.1186/s12870-019-2138-7
Zhang, C., Rabiee, M., Sayyari, E., and Mirarab, S. (2018). ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19:153. doi: 10.1186/s12859-018-2129-y
Zhang, C., Zhang, T., Luebert, F., Xiang, Y., Huang, C.-H., Hu, Y., et al. (2020). Asterid phylogenomics/phylotranscriptomics uncover morphological evolutionary histories and support phylogenetic placement for numerous whole-genome duplications. Mol. Biol. Evol. 37, 3188–3210. doi: 10.1093/molbev/msaa160
Zhang, G. Q., Liu, K. W., Li, Z., Lohaus, R., Hsiao, Y. Y., Niu, S. C., et al. (2017). The Apostasia genome and the evolution of orchids. Nature 549, 379–383. doi: 10.1038/nature23897
Zhang, J., He, C., Wu, K., Teixeira da Silva, J. A., Zeng, S., Zhang, X., et al. (2016). Transcriptome analysis of Dendrobium officinale and its application to the identification of genes associated with polysaccharide synthesis. Front. Plant Sci. 7:5. doi: 10.3389/fpls.2016.00005
Zhang, J., Wu, K., Zeng, S., Teixeira da Silva, J. A., Zhao, X., Tian, C. E., et al. (2013). Transcriptome analysis of Cymbidium sinense and its application to the identification of genes associated with floral development. BMC Genomics 14:279. doi: 10.1186/1471-2164-14-279
Zhang, L., Chen, F., Zhang, G. Q., Zhang, Y. Q., Niu, S., Xiong, J. S., et al. (2016). Origin and mechanism of crassulacean acid metabolism in orchids as implied by comparative transcriptomics and genomics of the carbon fixation pathway. Plant J. 86, 175–185. doi: 10.1111/tpj.13159
Zhang, N., Zeng, L., Shan, H., and Ma, H. (2012). Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. New Phytol. 195, 923–937. doi: 10.1111/j.1469-8137.2012.04212.x
Zhang, Y. M., Su, Y., Dai, Z. W., Lu, M., Sun, W., Yang, W., et al. (2022). Integration of the metabolome and transcriptome reveals indigo biosynthesis in Phaius flavus flowers under freezing treatment. PeerJ 10:e13106. doi: 10.7717/peerj.13106
Zhang, Y., Zhang, G. Q., Zhang, D., Liu, X. D., Xu, X. Y., Sun, W. H., et al. (2021). Chromosome-scale assembly of the Dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic. Res. 8:183. doi: 10.1038/s41438-021-00621-z
Zhao, Y., Li, D., and Liu, T. (2019). Pollination-induced transcriptome and phylogenetic analysis in Cymbidium tortisepalum (Orchidaceae). Russ. J. Plant Physiol. 66, 618–627. doi: 10.1134/S1021443719040174
Zhou, Z., Ying, Z., Wu, Z., Yang, Y., Fu, S., Xu, W., et al. (2021). Anthocyanin genes involved in the flower coloration mechanisms of Cymbidium kanran. Front. Plant Sci. 12:737815. doi: 10.3389/fpls.2021.737815
Züst, T., Strickler, S. R., Powell, A. F., Mabry, M. E., An, H., Mirzaei, M., et al. (2020). Independent evolution of ancestral and novel defenses in a genus of toxic plants (Erysimum, Brassicaceae). Elife 9:e51712. doi: 10.7554/eLife.51712
Keywords: phylogeny, orchids, transcriptome, next-generation sequencing–NGS, plastome, target sequence capture sequencing, phylogenomics, phylotranscriptomics
Citation: Wong DCJ and Peakall R (2022) Orchid Phylotranscriptomics: The Prospects of Repurposing Multi-Tissue Transcriptomes for Phylogenetic Analysis and Beyond. Front. Plant Sci. 13:910362. doi: 10.3389/fpls.2022.910362
Received: 01 April 2022; Accepted: 21 April 2022;
Published: 27 May 2022.
Edited by:
Jen-Tsung Chen, National University of Kaohsiung, TaiwanReviewed by:
Tatiana Arias, Marie Selby Botanical Gardens, United StatesAli Raza, Fujian Agriculture and Forestry University, China
Copyright © 2022 Wong and Peakall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Darren C. J. Wong, ZGFycmVuLndvbmdAYW51LmVkdS5hdQ==; d29uZ2RjakBnbWFpbC5jb20=
 Darren C. J. Wong
Darren C. J. Wong Rod Peakall
Rod Peakall