
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 01 June 2022
Sec. Plant Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.910228
This article is part of the Research TopicRising Stars: Plant Cell Biology 2022View all 5 articles
Plant trichomes, protrusions formed from specialized aboveground epidermal cells, provide protection against various biotic and abiotic stresses. Trichomes can be unicellular, bicellular or multicellular, with multiple branches or no branches at all. Unicellular trichomes are generally not secretory, whereas multicellular trichomes include both secretory and non-secretory hairs. The secretory trichomes release secondary metabolites such as artemisinin, which is valuable as an antimalarial agent. Cotton trichomes, also known as cotton fibers, are an important natural product for the textile industry. In recent years, much progress has been made in unraveling the molecular mechanisms of trichome formation in Arabidopsis thaliana, Gossypium hirsutum, Oryza sativa, Cucumis sativus, Solanum lycopersicum, Nicotiana tabacum, and Artemisia annua. Here, we review current knowledge of the molecular mechanisms underlying fate determination and initiation, elongation, and maturation of unicellular, bicellular and multicellular trichomes in several representative plants. We emphasize the regulatory roles of plant hormones, transcription factors, the cell cycle and epigenetic modifications in different stages of trichome development. Finally, we identify the obstacles and key points for future research on plant trichome development, and speculated the development relationship between the salt glands of halophytes and the trichomes of non-halophytes, which provides a reference for future studying the development of plant epidermal cells.
Trichomes are visible on leaves, stems and flower organs of many terrestrial plants (Szymanski et al., 2000; Yang and Ye, 2013; Fei et al., 2020). These hair-like organs derive from a proliferation of epidermal cells, which undergo cell division, differentiation, and growth to produce tissues that extend from the surface of the epidermis. The resulting structures play a variety of roles in plant growth and stress tolerance (reviewed by Balkunde et al., 2010; see also Fei et al., 2020; Mirnezami et al., 2020). For example, trichomes provide a barrier that must be broken before any successful pathogen or herbivore attack; as such, they constitute the first line of plant defense. Trichomes are found in most angiosperms and some gymnosperms and bryophytes (Schuurink and Tissier, 2020; Spirina et al., 2020). Trichome variation is the result of natural selection (Mauricio, 1998; Sletvold et al., 2010; Züst et al., 2012). During evolution, plants have evolved different epidermal structures and their derived structures, such as trichomes and thorn-like leaves, in order to resist various biotic and abiotic stresses (Han et al., 2016). Compared with individuals without trichomes, individuals with trichomes have advantages, particularly in herbivore-rich environments and arid areas (Han et al., 2016; Ying et al., 2018).
Trichomes come in different shapes, sizes and densities. Morphologically, trichomes can be unicellular, bicellular, or multicellular (Andrade et al., 2017; Fei et al., 2020). They may be glandular or non-glandular, and branched or non-branched (Wang et al., 2013b; Shang et al., 2020). Some trichomes are obvious, such as the multicellular trichomes in nettles or tomatoes (Solanum lycopersicum), and cause the appearance of a “hairy plant,” while others are small, such as the shield coat of mint species (Turner et al., 2000; Deore et al., 2021).
Unicellular trichomes have a simple structure and usually do not have glands. Non-glandular trichomes are present in most angiosperms, some gymnosperms and bryophytes. However, multicellular trichomes are usually complex. For example, the trichomes of cucumber (Cucumis sativus), tobacco (Nicotiana tabacum), tomato and snapdragon (Antirrhinum majus) are multicellular (Yang and Ye, 2013; Tan et al., 2020; Zhang et al., 2021f). Some multicellular trichomes are secretory and some are not (Koudounas et al., 2015; Xue et al., 2019; Konarska and Łotocka, 2020).
Although such non-glandular hairs do not have secretory or metabolic functions (Yuan et al., 2020), they play an important role in resisting extreme environments, inducing pollination, preventing ultraviolet radiation, resisting drought, adapting to high salinity, absorbing heavy metals and preventing biological invasion and mechanical damage (Riddick and Simmons, 2014; Wang et al., 2020b; Zhang et al., 2021b). For example, cotton (Gossypium hirsutum) trichomes (cotton fibers) play an important role in resisting pests such as cotton boll weevil (Ye et al., 2020). Non-glandular hairs can affect herbivorous arthropods and prevent them from climbing over the leaf surface. The hooked tip of non-glandular hairs sometimes hinders their movement, interfering with their feeding or trapping them (Giuliani et al., 2020). After salt treatment, especially moderate and severe salt treatment, Schizonepeta tenuifolia increased its density of total trichomes on both sides of leaves to alleviate the effect of salt concentration (Ying et al., 2018). At a low temperature, the protoplast flow of Saintpaulia ionantha trichomes stopped immediately, and the aggregation of chloroplasts in cells slowed down. After heating to 20°C, the flow state recovered slowly (Saltveit and Hepler, 2004).
Secretory trichomes, also known as glandular hairs, are usually composed of three parts: the base of the trichome, the gland, and the head of the trichome. Only one cell in the base is tightly connected to the epidermis. The gland is composed of 1–5 short or long cylinder cells, which lack chloroplasts in the cytoplasm. The secretory process of the plant epidermis is closely related to the physiological activities and interactions of various organelles in the cell (Stratmann and Bequette, 2016). Functionally, glandular hairs have the ability to synthesize, store and secrete many specialized metabolites, many of which are of commercial importance in food additives, drugs, flavors, and natural pesticides (reviewed by Feng et al., 2021b). Many are also signal molecules, regulating plant growth and development (Rodziewicz et al., 2019). For example, the chemicals secreted by glandular hairs of crops can repel or even trap insects and mites, resulting in their death due to drying or starvation (Saltveit and Hepler, 2004; Feng et al., 2021b).
Bicellular trichomes are present in plants such as rice and maize (Angeles-Shim et al., 2012; Kong et al., 2021). They are arranged in a cylindrical shape between the stem veins of the upper and lower epidermis of the leaves (Angeles-Shim et al., 2012). Rice trichomes can be subdivided into long hairs, hook hairs, thorn hairs, slender hairs and serrated hairs. Bicellular trichomes also play an important role in plant biotic and abiotic stress responses.
Plant trichome development is influenced by genetic factors and environmental factors such as hormones, water and light (reviewed by Khan et al., 2021). The effects of hormones, water, and light on trichome development have been demonstrated in detail in previous publications (Ning et al., 2016; Li et al., 2021b; Yu et al., 2021b). The trichome pattern formation process has been well studied, mainly because trichomes can easily be observed and used in experiments (Han et al., 2016). Trichomes are an excellent model system for studying cell differentiation, cell cycle regulation, cell polarity, and cell expansion (Stratmann and Bequette, 2016). Therefore, trichomes are often analyzed at the genetic, genomic, and cellular level (Schellmann and Hülskamp, 2005; Xiao et al., 2021). Various mutants have been used to study trichome cell cycle regulation and cell morphogenesis (Aashima et al., 2014; Chen et al., 2021). Plant trichome development generally includes three stages: (1) fate determination and initiation, (2) branching, and (3) elongation and maturation. Many studies have confirmed that different transcription factor families, such as HD-ZIP type proteins (Henriksson et al., 2005; Xie et al., 2021a; Zhang et al., 2021c), C2H2 zinc finger proteins (Gan et al., 2007; Liao et al., 2021), basic helix-loop-helix (bHLH) type proteins (Payne et al., 2000; Liu et al., 2021), and v-myb avian myeloblastosis viral oncogene homolog (MYB) family proteins (Larkin et al., 1994; Khan et al., 2021), all play a key role in plant trichome development. In addition, trichome development is strictly regulated by a variety of plant hormones (Chang et al., 2018; Du et al., 2020; Han et al., 2020). Hormone signaling regulates the formation of trichomes by regulating the expression of downstream genes. Epigenetic modifications also play an important role in plant trichome development (Zhang et al., 2017b; Wu et al., 2020a).
In this paper, we review the molecular regulatory mechanisms of unicellular, bicellular and multicellular trichome development in different plants. We also propose future research directions.
Arabidopsis has both unicellular and non-glandular trichomes, which have been systematically studied as models for epidermal cell differentiation. The cell walls of Arabidopsis trichomes gradually become thinner from top to bottom. Upon invasion by external organisms or abiotic stress, the highly sensitive base quickly initiates a defense response (Han et al., 2016). In addition, Arabidopsis mutants with more trichomes showed less sensitivity to UV compared to wild-type plants, while mutants with fewer trichomes are more sensitive to UV, highlighting that trichomes have an important shielding effect against UV radiation (Riddick and Simmons, 2014; Karabourniotis et al., 2020).
In Arabidopsis, trichomes are present in most aerial organs, such as rosette leaves, stems, stem leaves and sepals, but not on hypocotyls and cotyledons (Akhtar et al., 2017). Most studies on the development of trichomes focus on the rosette leaves. Trichomes are single and branched cells covering the whole leaf surface. The trichomes of Arabidopsis are developed from a single protoepidermal cell at the leaf base. They are usually separated by three to four epidermis cells. As leaves grow, new trichomes form at the base of leaves, while the existing trichomes separate due to the division of intermediate epidermal cells (Banyar et al., 2015). The morphological photographs of Arabidopsis trichomes were shown in Supplementary Figure 1A (Breuer et al., 2009). The occurrence of trichomes is the result of external signals and endogenous transcriptional regulation (Hülskamp et al., 1994; Pesch and Hülskamp, 2009; Maes et al., 2011). As early as the 1990s, more than 70 Arabidopsis trichome mutants have been isolated; they can be divided into the following six types: with trichomes, without trichome, trichome ribbons, trichome reduction, trichome distortion, and vitreous trichomes (Hülskamp et al., 1994). Over the past few decades, researchers have focused on the molecular mechanisms of a series of trichome mutants such as gl1, gl2, and ttg1 (Oppenheimer et al., 1991; Rerie et al., 1994; Walker et al., 1999). With the discovery of novel genes regulating trichome development, new mutants at various stages of trichome development, such as gis, kak, and etc1 were complemented in the mutant library (Kirik et al., 2004; Yan et al., 2012; Bensussan et al., 2015). Many important genes that regulate the development of trichomes have been discovered, most of which are transcription factors (reviewed by Schwab et al., 2000; Han et al., 2021).
Based on the study of the hair development process in Arabidopsis, we divided the development of Arabidopsis trichomes into three stages: fate determination and initiation, trichome branching, and trichome elongation and maturation (reviewed by Payne et al., 1999; see also Hülskamp, 2004; Schellmann and Hülskamp, 2005). Morphogenesis can be divided into six stages, beginning which the trichome cell expanding rapidly relative to the surrounding normal epidermal cells and ending with a circle of trichome-supporting cells surrounding the trichome (Szymanski and Marks, 1998).
In the fate determination and initiation stage, positive regulators, such as R2R3-MYB transcription factors, WD40 (WD40 repeat) proteins, bHLH transcription factors and C2H2 zinc finger proteins, and negative regulators, such as R3-MYB transcription factors, play a key role in this process (Zang et al., 2015; Han et al., 2021).
C2H2-type zinc finger proteins play a key role in upstream trichome fate determination and initiation in Arabidopsis. GLABROUS INFLORESCENCE STEMS (GIS), GIS2, GIS3, ZINC FINGER PROTEIN 1 (ZFP1), ZFP5, ZFP6, and ZFP8 all encode C2H2 zinc finger proteins that play important roles in trichome development (Gan et al., 2007; Zhou et al., 2011, 2012, 2013; Sun et al., 2015a; Liu et al., 2017; Zhang et al., 2020; Han et al., 2021). GIS functions upstream of the trichome initiation complex GLABRA1 (GL1)-GLABRA3 (GL3)-TRANSPARENT TESTA GLABRA 1 (TTG1) (GL1-GL3-TTG1) and downstream of the gibberellin (GA) signal repressor SPINDLY (SPY). It responds to GA and promotes inflorescence trichome initiation. This pathway is negatively regulated by SPY and the DELLA repressor GIBBERELLIC ACID INSENSITIVE (GAI) (Guan et al., 2007). GIS3 receives GA and cytokinin (CTK) signal transduction and directly targets the downstream genes GIS and GIS2 to regulate the production of trichomes (Sun et al., 2015a). ZFP6 responds to GA and CTK signaling and plays a role upstream of ZFP5 (Zhou et al., 2013). ZFP5 acts upstream of the GIS gene family and key trichome initiation regulators, participates in GA signal transduction and controls the development of trichomes. ZFP8 is also a direct target gene of ZFP5 (Zhou et al., 2011, 2012). In addition, the function of the protein encoded by ZFP5 in controlling trichome initiation is equivalent to that of GIS and GIS2. They all play critical roles upstream of the trichome initiation complex GL1-GL3-TTG1. CTK induces the expression of ZFP1, thereby increasing the expression of GL3 and finally promoting trichome initiation in Arabidopsis (Sun et al., 2015a; Xie et al., 2019; Zhang et al., 2020). TRICHOME-RELATED PROTEIN (TRP) is also a recently identified C2H2 zinc finger protein, which negatively regulates trichome initiation-related transcription factors through GA signaling. TRP can interact with ZFP5, preventing ZFP5 from binding to the ZFP8 promoter and inhibiting the occurrence of superficial trichomes (Kim et al., 2018).
Downstream of C2H2 zinc finger proteins, GL1 was the first factor found by researchers to control the development of plant trichomes (Oppenheimer et al., 1991). It encodes an MYB-like protein, and knockout of this protein will produce hairless leaves (Ram et al., 2015). GL3 encodes a bHLH transcription factor and has redundancy with ENHANCER GLABRA3 (EGL3) (Payne et al., 2000; Zhang et al., 2003; Hung et al., 2020). A double mutation of GL3 and EGL3 leads to a trichome defect. TRANSPARENT TESTA GLABRA (TTG) has been studied since the 1990s (Larkin et al., 1994, 1999). TTG1 encodes a protein with 4–5 repeating WD-40 motifs, interacts with GL3, and ultimately promotes trichome differentiation (Walker et al., 1999; Champagne and Boutry, 2017). The semi deletion alleles of GL1 and TTG1 produce aborted trichomes, indicating that GL1 and TTG1 function as complexes and dual regulators in trichome development (Jose et al., 2018; Hung et al., 2020). SUPER SENSITIVE TO ABA AND DROUGHT2 (SAD2) encodes an importin β-domain protein that has the same function as GL1, GL2 and GL3 in regulating trichome development (Gao et al., 2008; Du et al., 2018). SAD2 can mediate the function of GL3 and regulate the expression of GL1, TTG1 and GL2 (Gao et al., 2008). TRIPTYCHON (TRY) and CAPRICE (CPC) negatively regulate the development of trichomes; they mainly encode R3-MYB transcription factors (Wada et al., 1997; Szymanski and Marks, 1998; Champagne and Boutry, 2017). They can competitively bind to the N-terminal of GL3 and EGL3, damage the function of GL3- and EGL3-related complexes, and interfere with the differentiation and development of trichomes (Feng et al., 2017). The R3-MYB transcription factors ENHANCER OF TRY AND CPC 1 (ETC1) inhibit the fate of Arabidopsis aboveground trichomes and non-hairy root epidermal cells (Victor et al., 2004). etc1 mutants have no distinct phenotype but enhance the roles of cpc and try in trichome development. etc1 try cpc triple mutants produce more trichomes on the upper epidermis of leaves and hypocotyl than the wild type, etc1, try, cpc, etc1 try double mutant, etc1 cpc double mutant and try cpc double mutant (Victor et al., 2004). SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) gene is also a negative regulator of trichome development. SPL temporally controls trichome distribution during flowering. Increasing SPL transcription levels were associated with progressive loss of stem hair cells. SPL9 directly activates its expression by binding to TCL1 and TRY promoters, which is independent of GL1; GIS-dependent pathways do not affect SPL9 regulation of TCL1 and TRY (Yu et al., 2010). SPL is targeted by microRNA 156 (miR156); overexpression of miR156 results in ectopic trichomes on the stem and flower organs of plants, while an increased SPL expression level in plants results in less trichomes than that in the wild type (Yu et al., 2010; Wei et al., 2012). TTG2, the first WRKY transcription factor associated with the mutant phenotype, regulates the development of trichomes, seed coats and root hairs (Johnson et al., 2002). Analysis of trichome and root hair mutants showed that the expression of TTG2 is regulated by MYB and bHLH genes. TTG2 mutations lead to phenotypic defects in trichome development (reviewed by Ishida et al., 2007). Most leaf surface hairs in ttg2 mutants are unbranched (> 95%), and the number of trichomes formed per leaf is approximately halved, and even glassy and twisted trichomes appear (Johnson et al., 2002). GL2, a homeobox family gene that encodes the HD-ZIP IV transcription factor, plays an essential role in and is expressed throughout trichome development (Rerie et al., 1994; Bryant et al., 2016). The upstream pathway of GL2 regulating the differentiation and development of plant epidermal cells is relatively clear. The R2R3-MYB transcription factor GL1/MYB23, WD40 protein TTG1 and bHLH transcription factor GL3/EGL3 together form a trimeric complex activator, which directly acts on the downstream GL2/TTG2 to regulate trichome development. However, the regulatory pathway downstream of GL2 is not clear. GL2, as a transcription factor, may affect cell differentiation and development by regulating the expression of downstream target genes. Studies have shown that knockdown of the bHLH-type transcription factor AtMYC1 reduces the density of Arabidopsis trichomes (Symonds et al., 2011). Further studies showed that MYC1 could regulate the intracellular localization of GL1 and TRY, suggesting that MYC1 may inhibit the activity of CPC and TRY, thereby regulating the number of trichomes (Pesch et al., 2013).
In the branching stage of trichomes, branching is regulated by microtubules, and the arrangement direction of microtubules controls the growth direction and branching ability of trichomes (Tominaga-Wada et al., 2012). The DNA in trichome nuclei needs four replications to form a branching structure. SIAMESE (SIM), CONSTITUTIVE EXPRESSION OF PR GENES 5 (CPR5) and RETINOBLASTOMA-RELATED (RBR) negatively regulate trichome branching by controlling nuclear replication (Kirik et al., 2001; Wang et al., 2021c). SIM is a cell cycle mitotic inhibitor of endonuclear replication (Walker et al., 2000). The sim mutant has fewer branched trichomes than the wild type because the transition of trichome cells from mitosis to intranuclear replication is inhibited (reviewed by Walker et al., 2000; see also Churchman et al., 2006; Grebe, 2012). SIM is likely a direct target for GL3 to control intracellular replication. In a chromatin immunoprecipitation (ChIP) experiment, GL1 and GL3 bound to the promoter region of SIM (Walker et al., 2000). CELL CYCLE SWITCH PROTEIN 52 A1 (CCS52A1) and SIM cooperate to inhibit the accumulation of mitotic cyclin to establish the inner loop of trichomes (Remmy et al., 2010). MYB5 and MYB23 are members of the R2R3 MYB family that regulate trichome extension and branching. The myb5 mutant showed little change in trichome morphology, whereas the myb23 mutant produced more small trichomes and two branched trichomes. The myb5 myb23 double mutant had shorter trichomes and more double-branched trichomes on rosette leaves than the single mutant (Li et al., 2009). In addition, the R2R3-MYB transcription factor MIXTA gene AtMYB106 negatively regulates the SIM gene (Jakoby et al., 2008; Tian and Zhang, 2021). CPR5 encodes a protein of unknown function that might be a membrane-bound protein. Compared with wild type, cpr5 mutants have smaller trichomes, less branching, and lower nuclear DNA content. In cpr5 mutant trichomes, the endonuclear replication cycle stops after two rounds instead of four, and the trichome cells are less branched than normal. The cpr5 mutant also has altered cell walls and reduced cellulose content in leaves and trichomes (El Refy et al., 2003; Brininstool et al., 2008). DA3-encoded UBIQUITIN-SPECIFIC PROTEASE 14 (UBP14) acts upstream of CYCLIN-DEPENDENT KINASE B1;1 (CDKB1;1), affecting endonuclear replication and cell growth in Arabidopsis. The da3-1 inhibitor SUPPRESSOR OF DA3-1 6; SUD6, which encodes CYCLIN-DEPENDENT KINASE G2 (CDKG2), promotes nuclear replication and cell growth and ultimately increases the number of trichome branches (Jiang et al., 2022). Studies have shown that TRY and ETC1 genes cooperate to regulate trichome branches. The trichomes of try etc1 double mutants have more branches than wild type and try single mutants. ETC1 can enhance the function of TRY to regulate the number of trichome branches (Tominaga-Wada and Nukumizu, 2012). In cpl3 mutants, the level of inscribed replication in the epidermis increases, resulting in a reduction of trichome branches (Tominaga-Wada and Nukumizu, 2012). Furthermore, GIS plays an inhibitory role in trichome branching, acting downstream of the key regulators STICHEL (STI) and SIM (Yan et al., 2014). Class I TCP transcription factors TCP14 and TCP15 regulate hyperdivided trichomes and increase stratum corneum permeability in Arabidopsis. TCP14 inhibits trichome branching in Arabidopsis leaves and inflorescence stems by direct transcriptional activation of GIS (Vadde et al., 2018). The protein encoded by AtTCP15 binds directly to the promoter regions of CYCLIN A 2;3 (CYCA2;3) and RBR genes and plays a key role in endonuclear replication (Li et al., 2012b). ANGUSTIFOLIA (AN) enriches high concentrations of microtubules at the top of trichome cells (Kim et al., 2002; Hülskamp, 2004; Hashida et al., 2020); ZWICHEL (ZWI) is involved in the initiation of trichome branching (Reddy et al., 2004; Chen et al., 2016b); STI and TUBULIN FOLDING COFACTOR A/C (TFCA/C) are involved in the formation of trichome branching (Victor et al., 2002); and FASS/TONNEAU2 (TON2) and SPIKE are involved in trichome branching by regulating microtubule tissue (Traas et al., 1995; Qiu et al., 2002; Deeks and Hussey, 2003; Ilgenfritz et al., 2003). INHIBITOR/INTERACTOR OF CDK 1 (ICK1)/KIP RELATED PROTEIN 1 (KRP1) positively regulates trichome branching. Overexpression of ICK1/KRP1 in Arabidopsis reduces intranuclear replication and cell size and induces cell death (Ilgenfritz et al., 2003; Schnittger et al., 2003; Wang et al., 2021c). ROOT HAIRLESS 2 (RHL2) and HYPOCOTYL6 (HYP6) encode subunits of DNA topoisomerase VI, indicating the existence of DNA replication pathways specific to internally replicating cells (Sugimoto-Shirasu et al., 2002). In rhl2, hyp6, and spindly (spy) mutants, trichomes are smaller, defective, branched, and have lower nuclear DNA content than in the wild type (Jacobsen and Olszewski, 1993; Sugimoto-Shirasu et al., 2002). In kaktus (kak) mutants, trichome branching increases (El Refy et al., 2003; Schellmann and Hülskamp, 2005).
During the elongation and maturation stage, trichome lengths increase to several times their previous length and expand in a polarized manner. The actin cytoskeleton determines the direction of trichomes cell expansion. Genes such as BRICK1 (BRK1), DISTORTED (DIS) and ROP are involved in trichome cell actin cytoskeleton expansion (Hülskamp et al., 1994; Yang, 2002). The mutation of Arabidopsis BRK1 gene leads to the morphological defects in trichome cells and related changes in the F-actin cytoskeleton. BRK1 is necessary for SCAR protein accumulation in vivo, which may explain the important role of BRK1 in ARP2/3 complex function (Frank and Smith, 2002). ARP2/3 is a large complex that promotes actin formation (Dipanwita et al., 2004; Papalazarou and Machesky, 2021). The following genes, known as DIS genes, encode components of the ARP2/3 complex: WRM, CRK, DIS2, DIS1, PIR, KLK, GRL and ITB1. Mutants of these genes have altered trichime elongation and growth direction; they have a phenotype of epidermal torsion, with some areas of cells bulging and displaying dysplasia (Hülskamp et al., 1994; Yang, 2002). A small GTPase called ROP controls downstream genes to regulate actin configuration in trichome cells. ROP binds to the ARP2/3 complex to release PIR121, NAP125, and ABI2, resulting in the activation of HSPC300 and SCAR/WAVE (Dipanwita et al., 2004).
Environmental and endogenous signals closely regulate trichome development. Several plant hormones, including those mentioned above (GA and CTK) are involved in trichome differentiation in Arabidopsis. Application of jasmonic acid (JA) leads to significantly more leaf trichomes (Qi et al., 2011). Salicylic acid (SA) can reduce the number of trichomes (Traw and Bergelson, 2003). CTK and GA stimulate the development of trichomes on inflorescence stems (Fan et al., 2020; Papalazarou and Machesky, 2021). In addition, plant hormones can also play an important signaling role in trichome development by mediating downstream genes (reviewed by Markus, 2012). GAI and SPY are a GA biosynthesis factor and a GA signal inhibitor, respectively. Mutations in these have a significant impact on epidermal hair development. There is a positive relationship between GA level and trichome development. GAs may rely on GL1 and TTG to promote trichome development (An et al., 2012). TEM1 and TEM2 encode members of ABI3 and VP1 transcription factor families. TEM1 and TEM2 play a negative regulatory role in trichome development, which depends on the GA pathway. TEMs not only regulate GA content but also the transportation and distribution of GA in mesophyll, which mediates trichome development. This indicates that subepidermal cells play a role in trichome initiation (Fiehn et al., 2000). A ChIP experiment showed that TEMs directly bind to the promoters of GL1, GL2, GIS2, and ZFP8, which may regulate and transcriptionally inhibit some transcription factors of coding genes in the initial stage of trichome development. As an important inhibitor of the JA signaling pathway, JASMONATE ZIM-DOMAIN PROTEIN 1 (JAZ1) can degrade under the action of JA, release MYB-bHLH-WD40 activity and promote trichome development. NUCLEOREDOXIN 2 (NRX2) promotes JA-mediated trichome formation in Arabidopsis; trichome formation is significantly reduced in the nrx2 mutant compared to in the wild type. JMJ29 is a histone demethylase-containing JMJC domain belonging to the JHDM2/KDM3 group. It participates in epidermal hair development by directly regulating GL3 expression (Fuyu et al., 2020). The cell cycle regulator CPR5 is involved in the SA signal pathway. Trichome length and number of branches are lower in cpr5 mutants than in the wild type (Kirik et al., 2001). These mutants also contain high SA content, suggesting that CPR5 may also be involved in SA biosynthesis (Peng et al., 2020).
Epigenetic modification is an important regulation mode, it is involved in a variety of protein post-translational modifications, such as ubiquitination, acetylation, methylation, glycosylation, and DNA methylation (Scoville et al., 2011). In addition, non-coding RNA, as an epigenetic regulator, also plays an important role in plant growth. Recent studies suggest that the multifunctional histone acetyltransferase AtGCN5 may have a positive effect on trichome branching by regulating TRY (Hülskamp et al., 1994). GCN5 also participates in the regulation of epidermal initiation through histone acetylation acting on the promoters of GL1, GL2, GL3, and CPC. Free degradation and mutant analysis of plants showed that UBIQUITIN PROTEIN LIGASE 3 (UPL3) promotes GL3 and EGL3 degradation, which in turn inhibits epidermal branching. As a histone chaperone, CAF-1 participates in trichome development through a pathway independent of internal duplication (reviewed by Hussey et al., 2006). In eukaryotes, the modification of mRNA is related to cell development and differentiation, in which N6-methyladenosine (m6A) is the most common genomic marker. Similar to DNA methylation, known regulatory genes can express m6A, and factors of different functions can also write, clear and read m6A (Wu et al., 2020a). ECT2 is an interpretation protein of m6A. It binds and stabilizes key transcripts related to trichomes, such as TTG1, affecting trichome development. This provides the groundwork for future research on post transcriptional modification as a key factor in trichome development (Wei et al., 2018b; Arribas-Hernández et al., 2021).
A model of trichome fate determination and initiation, branching, and elongation and maturation in Arabidopsis is shown in Figure 1.
Trichomes on cotton seed coats are usually called cotton fiber. Morphological photographs of cotton fibers were shown in Supplementary Figure 1B (Guan et al., 2014b). Cotton fiber is the main harvest of cotton and an important raw material for the textile industry. Cotton fiber development includes the following four main stages: initiation, elongation, secondary cell wall deposition and maturation (Hao et al., 2018).
Regulation of cotton fiber initiation [2–5 days after anthesis (DPA)] is similar to that of Arabidopsis trichomes, with several similar transcription factors and hormone signals.
Several cotton genes encoding MYB-type transcription factors that are homologous with Arabidopsis genes have been cloned and identified. GhMYB2 has a similar sequence to Arabidopsis GL1 and controls cotton fiber cell fate determination (Wang et al., 2004; Yong-Mei and Yu-Xian, 2011; Andreas et al., 2014; Zhao et al., 2020). The GhMYB2 gene promotes cotton fiber development and has functional homology with Arabidopsis GL1 in terms of trichome formation. Allotetraploid cotton containing GhMYB2A and GhMYB2D homologs is currently the most widely grown cotton variety. Cotton accumulates less GhMYB2A mRNA than GhMYB2D during the fiber initiation stage. GhMYB2D mRNA is targeted by miR828 and miR858, resulting in trans-siRNAs (ta-siRNAs) in the TRANS-ACTING SIRNA GENE 4 (TAS4) family. Four families of genes encoding ta-siRNA were found in Arabidopsis, namely TASl, TAS2, TAS3, and TAS4. TAS4 was discovered when a special algorithm clustered 21-base fragments within the genome. After the primary transcript of TAS4 was recognized by miR828, it was cleaved to generate the corresponding ta-siRNA. miR828 directs the cleavage of RNA derived from TAS4 and initiates the production of phasic small interfering RNA (siRNA) dependent on RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) (Bonar et al., 2018). Overexpression of GhMYB2A but not GhMYB2D complements gl1 phenotype. Mutation of the miR828 binding site or replacement of its downstream sequence can eliminate the production of ta-siRNA and restore trichome development in the gl1 mutant. In addition, blocking the biogenesis gene DICER-LIKE 4 (DCL4) or RDR6 of ta-siRNAs in gl1 GhMYB2D overexpressing body can restore trichome development (Voinnet, 2009). The functional difference between GhMYB2A and GhMYB2D homologous genes is caused by ta-siRNA mediated by miR828, which indicates the unique role of microRNA in the functional difference of target homologous genes. It also indicates that the evolution and selection of morphological characteristics of target homologous genes are vital. Small RNAs, including miRNAs and ta-siRNAs, are involved in the regulation of gene expression and development in plants and animals. In Arabidopsis, ta-siRNA biogenesis is usually triggered by miRNA cleavage at the TAS site (Allen et al., 2005). RNA-induced silencing complexes associated with the ARGONAUTE proteins AGO1 and AGO7 are assembled to target sites to cleave transcripts (Montgomery et al., 2008). Cleaved mRNA fragments serve as templates for RNA-dependent RNA polymerase 6 (RDR6), producing double-stranded RNAs (Peragine et al., 2004; Vazquez et al., 2004), which can be recognized by DCL4 and processed into 21-nt siRNAs or ta-siRNAs20. ta-siRNA regulates the development of trichomes in Arabidopsis leaves and the development of cotton fibers. GhMYB3 is a functional homolog of GL1 and GhMYB2. GhMYB3 interacts with Arabidopsis GL3 protein to regulate Arabidopsis trichome development. Ectopic expression of GhMYB3 rescues the hairless phenotype of the Arabidopsis gl1 mutant, produces more ectopic trichomes in the stems and flower organs of the inflorescence, and has an orthologous function in the development of plant trichomes (Shangguan et al., 2021). GhMYB25 and GhMYB25-like also play a positive regulatory role in fiber initiation. GhMYB25-like plays a role upstream of GhMYB25 and GhMYB109 (Wang et al., 2021a). GhMYB109 was found to be involved in the initiation and differentiation of fiber by RNAi experiments (Suo et al., 2003). GhMYB109 is structurally similar to GL1 and WER; which control Arabidopsis trichome initiation and have strong similarity to the R2R3 domain of GL1. GhMYB109 plays a positive regulatory role in the initiation and elongation of cotton fiber and affects the expression of downstream genes including GhACO1, GhACO2, GhTUB1, and GhACT1 (Suo et al., 2003). The gene encoding 1-aminocyclopropane-carboxylate oxidase (ACO), the last rate-limiting enzyme in ethylene (ETH) biosynthesis, plays a role in fiber development and is positively correlated with fiber elongation, maintaining ETH during fiber development (Wang et al., 2011; Cong et al., 2017; Li et al., 2021c). The MYB transcription factor GhCPC is homologous to Arabidopsis CPC and negatively regulates cotton fiber elongation. Transgenic evidence suggests that overexpression of GhCPC results in a delay in fiber initiation and a reduction in fiber length (Prakash et al., 2020). A yeast two-hybrid analysis showed that GhCPC can interact with the bHLH-type protein GhMYC1, and GhMYC1 can also interact with GhTTG1 and GhTTG4. GhCPC negatively regulates early fiber initiation and elongation through the CPC-MYC1-TTG1/4 complex (Liu et al., 2015; Zhang et al., 2021d). In the 35S:GhCPC transgenic line, the transcription levels of downstream genes GhHOX3 and GhRDL1 decreased significantly compared to in the wild type (Deng et al., 2012a). The promoter of the cotton dehydration-responsive gene RDL1 contains homeodomain-binding and MYB-binding motifs, which can specifically express trichomes of Arabidopsis (Wang et al., 2004; Chen et al., 2015).
HD-ZIP-type transcription factors also play an important regulatory role in cotton fiber initiation. PROTODERMAL FACTOR1 (GbPDF1) encodes the homologous framework HD-ZIP protein, which plays a role in fiber initiation and early elongation by interacting with PPIP1, PPIP2, and PPIP3. Knockout of GbPDF1 can result in delayed fiber initiation, fiber shortening, and decreased lint percentage, indicating that GbPDF1 plays an important role in fiber development (Jiang et al., 2012; Wang et al., 2019d). GbPDF1 is also involved in the homeostatic regulation of H2O2 during fiber development. Inhibiting the expression of GbPDF1 results in a substantial accumulation of H2O2 and delays the development of cotton fibers (Deng et al., 2012a). The HD-ZIP IV family transcription factor GOSSYPIUM BARBADENSE MERISTEM LAYER 1 (GbML1) interacts with GhMYB25 and specifically binds to the L1-box of the dehydration-inducing protein GbRDL1 promoter (Zhang et al., 2010). GaHOX1 is a class IV HD-ZIP transcription factor. When expressed under the control of the GL2 promoter, GaHOX1 rescues the trichome development of gl2-2 Arabidopsis hairless mutants. This indicates that GaHOX1 is a functional homolog of GL2 in the development of plant trichomes (Humphries et al., 2005; Guan et al., 2008; Zhang et al., 2010). The HD-Zip protein GhHD1 positively regulates cotton fiber formation by regulating reactive oxygen species (ROS) and ETH accumulation (Walford et al., 2012).
bHLH-type transcription factors are also involved in cotton fiber initiation. GhMYC1 is a bHLH-type transcription factor homologous to GL3; it can bind to the E-box sequence of the GhHOX1 promoter, indicating that GhHOX1 may be located downstream of GhMYC1 in fiber development regulation (Hu et al., 2018). In addition, the ectopic expression of the bHLH transcription factor GhDEL65 increases trichome density; it likely regulates cotton fiber development by interacting with GhMYB2 and GhMYB3 (Shangguan et al., 2016; Sun et al., 2020).
GhWRKY16 is a WRKY transcription factor; it can directly bind to the promoters of GhHOX3, GhMYB109, GhCesA6D-D11, and GhMYB25 to induce the expression of these genes, thereby promoting fiber initiation and elongation (Wang et al., 2021b).
The expression of VACUOLAR INVERTASE (VIN) is necessary for cotton fiber initiation, and RNAi-mediated inhibition of GhVIN1 leads to a significant decrease in VIN activity, thereby forming a fiber-free seed phenotype in a dose-dependent manner (Wang et al., 2014). GhVIN1-mediated hexose signaling acts upstream of GhMYB25-like, GhMYB25, and GhMYB109 transcription factors (Prakash et al., 2020).
Plant hormone signaling pathways also play an important role in the regulation of cotton fiber initiation. Auxin (AUX) biosynthesis and transport plays a major role in the accumulation of AUX in the ovule and fiber. The AUX efflux mediated by the IAA efflux transporter (PIN) in the ovule participates in the accumulation of AUX and the development of cotton fiber (Zhang et al., 2017b; Ma et al., 2019). Excessive GhPIN3 transcripts in the epidermis of the ovule promote the accumulation of fiber-specific AUX, thereby promoting fiber initiation, which indicates that all AUX signal transduction pathway components divide labor and play an important role in fiber development (Li et al., 2007; Zhang et al., 2017a). The AUX response factor ARF is expressed in multiple cotton tissues. GhARF2b, the Arabidopsis AtARF2 homolog, is preferentially expressed in developing ovules and fibers (Liu et al., 2020a; He et al., 2021). Overexpression of GhARF2b inhibits cotton fiber cell elongation but promotes fiber initiation. However, RNAi lines of GhARF2b resulted in fiber reduction but lengthening compared with that in the wild type. GhARF2b directly interacts with GhHOX3 and inhibits the transcriptional activity of GhHOX3 on target genes (Zhang et al., 2021d). GhARF2-1 and GhARF18-1 are only expressed in trichomes, and overexpression of these two genes in Arabidopsis enhances trichome initiation (Xiao et al., 2018). Similar to in Arabidopsis, GhJAZ2 protein is a negative regulatory protein in the JA pathway. GhJAZ2 interacts with GhMYB25-like, GhGL1, GhMYC2, GhWD40, and GhJI1 as a mediator of the JA signaling pathway and negatively regulates fiber development in cotton (Guan et al., 2014a). During cotton fiber initiation, JA regulates the downstream gene GhMYB25-like through JAZ2, while GhCPC prevents the expression of TTG1/MYC1, promotes the expression of GhHOX1, and initiates the formation of cotton fiber (Zhou et al., 2015). Exogenous ETH can promote the accumulation of H2O2 and play a positive regulatory function in fiber development, which indicates that ETH has a synergistic effect with the ROS pathway in fiber (Kim et al., 2017). PAGODA1 (PAG1) encodes CYP734A1, which degrades brassinosteroid (BR) through C26 hydroxylation and negatively regulates fiber development. As an acidic protein, Gh14-3-3 not only interacts with GhBZR1, but also regulates BR signals, thereby promoting fiber initiation and elongation (Li et al., 2013).
Transcription factors and hormone signaling pathways are also involved in the cotton trichome elongation stage (3–20 DPA).
The MYB transcription factor GhMYB212 is an important factor that regulates the transport of sucrose from the ovule to the fiber. GhMYB212 also controls expression of the sucrose transporter gene GhSWEET12, mediates the transport of sucrose and glucose, and mediates fiber development (Kai et al., 2017). GhMYB212 promotes the expression of GhSWEET12, and then GhPDF1 promotes the accumulation of ROS, which regulates cotton fiber elongation. During the transition from the initiation to the elongation stage, ETH and ROS mutually mediate cotton fiber initiation and elongation (Sun et al., 2019).
The bHLH transcription factor gene GhHOX3 is located in the 12th homologous chromosome of allotetraploid cotton varieties and is related to the quantitative trait locus (QTL) of fiber length (Shan et al., 2014). GhHOX3 can bind to the promoters of the cotton cell wall relaxin genes GhRDL1 and GhEXPA1 and activate their expression to promote cotton fiber elongation. The plant hormone GA can modulate the activity of GhHOX3 (Shan et al., 2014; Shangguan et al., 2021). The GhHOX3–GhHD1 interaction increases the transcriptional activity of GhHOX3 and its role in fiber elongation. The cotton bHLH transcription factor GhFP1 activates BR biosynthesis and signal transduction to positively regulate fiber elongation (Liu et al., 2020b). GhbHLH13 is a bHLH transcription factor that is upregulated during fiber elongation. GhPEL76 causes shortening of cotton fibers after virus-induced gene silencing (VIGS). Yeast one-hybrid and transient dual-luciferase assays showed that GhbHLH13 can activate GhPEL76 and regulate cotton fiber elongation by binding to the G-box in the promoter region of GhPEL76 (Sun et al., 2020).
Phospholipids and structural proteins are also involved in cotton fiber elongation. Phosphatidylinositol (PtdIns) is an important structural phospholipid, and exogenous PtdIns application can promote cotton fiber elongation (Long et al., 2018). In transgenic cotton plants, the expression of the cotton PtdIns synthase gene GhPIS is controlled by the fiber-specific promoter element, resulting in the specific up-regulation of GhPIS during cotton fiber elongation (Long et al., 2018). ARABINOGALACTAN PROTEINs (AGPs) are extracellular proteoglycans that play an important role in intercellular communication during cotton fiber elongation and secondary cell wall formation. Four AGP genes (GhAGP2, GhAGP3, GhAGP4, and GhFLA1) have been cloned from cotton fiber (Liu et al., 2008). GhAGP4 RNAi lines have markedly lower expression levels of GhAGP4 and partially down-regulated expression of GhAGP2, GhAGP3, and GhFLA1 compared to that in the wild type. As a result, fiber elongation is inhibited, reducing fiber quality (Li et al., 2010b). Transcription profile analysis showed that nine beta-tubulin (TUB) genes were highly expressed in elongated fiber cells compared with ovules without villi mutants (Li et al., 2002; He et al., 2008). Northern blot analysis showed that the GhTUB1 gene is specifically expressed in cotton fiber cells (Li et al., 2003).
Ethylene also plays a vital and active role in cotton fiber elongation. Several genes related to ETH biosynthesis are positively related to cotton fiber development, such as MAT (GA_Ed0052D12f), which is a homolog of GhMAT4 (Yu et al., 2021a). Overexpression of the Arabidopsis gene ANKYRIN REPEAT PROTEIN 2A (AKR2A) promotes cotton fiber elongation by increasing ETH biosynthesis and synergistic action with AUX accumulation (Hu et al., 2020). The plant hormones ABA and CTK inhibit fiber growth, and ETH may negatively regulate their abundance to promote fiber development (Yu et al., 2021a). GhDET2 encodes a rate-limiting enzyme 5α-reductase for BR biosynthesis, which promotes the density and length of cotton fibers (Luo et al., 2007). Gh14-3-3 interacts with GhBZR1 to regulate BR signaling and promote cotton fiber elongation (Zhou et al., 2015). Cotton GhPAG1 encodes a homolog of Arabidopsis CPY7341. The ghpag1 mutant exhibits a typical phenotype lacking BR. GhPAG1 potentially regulates cotton fiber by participating in the BR pathway. GhPAG1 is highly expressed in 15 DPA fibers and regulates fiber elongation by controlling the level of endogenous biologically active BRs (Yang et al., 2014b; Li et al., 2021b). Cotton DELLA protein GhSLR1 interferes with the stability of the GhHOX3-GhHD1 complex, inhibiting the transcription of downstream target genes and subsequent fiber elongation (Xia et al., 2018). GhMADS11 is a MADS transcription factor that can accumulate in cotton fibers and promote cell elongation (reviewed by Wang et al., 2019d). Heterologous overexpression of GhMAD14 can reduce GA content and shorten the length of Arabidopsis hypocotyls compared with that in the wild type (Zhou et al., 2014).
In the fibrous secondary cell wall deposition stage (16–40 DPA), a set of enzymes synthesize secondary cell walls (SCWs). The NAC-MYB-CESAs module defines the central pathway of SCW cellulose biosynthesis.
NAM, ATAF and CUC (NAC) transcription factors act as the main switch in the NAC-MYB-cellulose synthases (CESAs) pathway (Taylor-Teeples et al., 2015; Cao et al., 2020). In Gossypium hirsutum, GhFSN1 is a NAC transcription factor; it regulates SCW biosynthesis in cotton fibers. GhFSN1 positively regulates the formation of cotton fiber SCWs by regulating the expression of its downstream genes GhIRX12, GhMYB1, GhGUT1, and GhDUF231L1. Overexpression of GhFSN1 by the 35S promoter increases cotton fiber SCW thickness and slightly reduces fiber length (Jie et al., 2018; Zhang et al., 2018).
MYB-type transcription factors play an important role in the deposition of fibrous SCWs. Two MYBs, GhMYB46_D13 and GhMYB46_D9, are highly expressed in 20 DPA fibers. Both GhMYB46_D13 and GhMYB46_D9 can bind the promoter of the cotton fiber SCW cellulose synthase gene, enhancing its expression and promoting fiber SCW thickening (Huang et al., 2019a). In addition, two cotton R2R3 MYB transcription factors, GhMYB7 and GhMYBL1, play a role in the formation of fiber SCW. GhMYB7 regulates cotton fiber SCW cellulose synthase by directly binding to three different cis-elements in the GhCesA4, GhCesA7, and GhCesA8 promoters. Overexpression of GhMYB7 increases cell wall thickness (Huang et al., 2016, 2021b). In GhMYBL1 transgenic plants, the biosynthesis of cellulose and lignin is enhanced compared to in the wild type (Sun et al., 2015b). Therefore, GhMYB7 and GhMYBL1 may be involved in the regulation of cotton fiber SCW biosynthesis and deposition. The lipase/hydrolase gene GhGDSL from Gossypium hirsutum is expressed during SCW biosynthesis (19–25 DPA), and GhMYB1 is specifically expressed in 19 DPA fibers. A yeast one-hybrid assay showed that GhMYB1 binds to the GhGDSL promoter, indicating that GhMYB1 can regulate GhGDSL during fiber development (Yadav et al., 2017).
The CELLULOSE SYNTHASE A (GhCESA) genes in cotton are divided into six subclasses, of which GhCESA4, GhCESA7, and GhCESA8 are mainly involved in SCW development (Yuan et al., 2015a; Wang et al., 2019b; Zhang et al., 2021e). GhCESA4 positively regulates cellulose biosynthesis during cotton fiber development (reviewed by Xu et al., 2021). As the developing cotton fiber produces SCW cellulose, the transcription level of GhCESA4 is significantly up-regulated (Kim et al., 2011).
GhTCP4 contains a conserved non-standard bHLH domain. During fiber development, transcriptome and promoter activity analysis showed that the overexpression of GhTCP4 up-regulates and accelerates SCW biosynthesis in fiber cells. The activation of the pathway leads to shorter fibers, different lengths, and thicker walls (Cao et al., 2020; Huang et al., 2021a). GhTCP4 interacts with GhHOX3 and inhibits its transcriptional activity. GhTCP4 and GhHOX3 antagonistically regulate cell elongation, thereby establishing the time control of the transition of fibroblasts to SCW (Ye et al., 2020; Huang et al., 2021a). GhTCP14 is also a key gene in the AUX pathway (Wang et al., 2013a) and promotes the biosynthesis of GA (Hao et al., 2012). The E3 ligase GhHUB2 can control fiber development by triggering the accumulation of GhKNL1. Overexpression of GhHUB2 can increase cotton fiber length and secondary cell wall thickness. The transcription inhibitor GhKNL1 is mainly expressed in developing fibers and can be degraded by GhHUB2 through the ubiquitin-26S proteasome pathway, eliminating the inhibitory effect of GhKNL1 on the accumulation of cotton fiber SCWs (Feng et al., 2018; Hao et al., 2018). GhKNL1 regulates fiber elongation and SCW formation, and can directly bind to the promoters of GhCesA4-2/4-4/8-2 and GhMYB46 to regulate cellulose biosynthesis during SCW deposition. GhKNL1 can also inhibit the expression of GhXPA2D/4A-1/4D-1/13A by combining with the promoter of GhXPA2D/4A-1/4D-1/13A, thereby regulating the fiber elongation of cotton (Wang et al., 2021d).
During the cotton fiber maturation stage (40–50 DPA), overexpression of the marshmallow sucrose synthase gene GhSusA1 can significantly increase the length and thickness of mature fibers. Overexpression of GhSusA1 can enhance secondary cell wall thickening and fiber quality compared to that in the wild type; whereas inhibition of GhSusA1 in transgenic cotton reduces fiber quality. Exogenous application of bioactive GA can promote the expression of GhSusA1 in cultured fibers and cotton hypocotyls (Jiang et al., 2012; Bai et al., 2014; Wang et al., 2020a). GhBRI1 is a BR receptor that regulates cellulose deposition during the SCW deposition and fiber maturation stages (Li et al., 2011; Sun et al., 2015c).
A regulation model of cotton trichome (fiber) initiation, elongation, secondary cell wall deposition and maturation are shown in Figure 2.
Rice (Oryza sativa) leaf trichomes are bicellular. They are arranged in a cylindrical shape between the stem veins of the upper and lower epidermis of the leaves, the morphological photographs of rice leaf trichomes were shown in Supplementary Figure 2 (Wang et al., 2013b). Rice trichomes commonly occur on the surface of leaves and glumes. They can be classified as macrohairs, micro hairs and glandular hairs (Wang et al., 2013b). Macrohairs and micro hairs are ubiquitous in plants. Macrohairs are mainly distributed on siliceous cells of cell vascular bundles, while micro hairs and glandular hairs are mainly distributed in the vicinity of stomata or motile cells (Wang et al., 2013b). Rice light-leaf mutants have filamentous glandular hairs that release secretions to the epidermal surface (Li et al., 2010a). Rice trichome development is a very complex process. Although some rice trichome-related genes have been located and cloned, the molecular mechanisms of rice trichome development are still largely unknown. At present, few genes involved in rice trichome development have been identified in the fate determination, initiation and elongation stages.
Several genes encoding different types of proteins and transcription factors are involved in the fate determination and initiation stage.
ESTRICTION FRAGMENT LENGTH POLYMORPHISM (RFLP) marker was used to preliminarily map GL-1, which controls the light-leaf trait of tropical japonica rice, on chromosome 5, which was linked to RG182 and RG403 (Yu et al., 1995). Researchers constructed the near isogenic line (NIL) of light-leaf rice using the American rice variety Rico No. 1 and light-leaf rice variety Jia64 (Li et al., 2012a). The light-leaf gene GLABROUS RICE1 (GLR1) was located within 21 kb between chromosome 5 markers M6 and M7. In a complementary experiment, weakening the expression of GLR1 significantly reduced or completely removed the number of trichomes, and the constitutive expression of GLR1 in the knockout line NILglr1 rescued the hairless phenotype of T0 generation transgenic lines (Li et al., 2012a). Complementary tests showed that the genomic fragments covering the open reading frame of DEGENERATIVE PALEA (DEP) can restore the formation of bristle-type trichomes on the leaves and glumes of hairless rice, indicating that DEP can regulate trichome formation on leaves and glumes (Angeles-Shim et al., 2012). GLR1, WUSCHEL-LIKE HOMEOBOX 3B (OsWOX3B), DEP and NUDA encode a homologous domain protein similar to that encoded by WUSCHEL (WUS); these genes are located at the same site on chromosome 5 and regulate rice trichome formation (Shang et al., 2020; Li et al., 2021a). The NUDA and GL-1 genes are alleles, and RNAi and complementary transgene studies have shown that this locus encodes the WUSCHEL-like homeobox gene OsWOX3B (Zhang et al., 2012). GLR2 has been screened from the tissue seedlings of Zhonghua 11, a light-leaf mutant. Genetic analysis revealed that glr2 mutants are controlled by a single-site recessive gene. A segregating population was used to locate glr2 on the first chromosome markers RM12124 and RM12136. There are 12 predicted genes in the 84.7 kb interval between the two genes, and the expression of three genes changed significantly, of which the expression of two genes (LOC_Os01g70020 and LOC_Os01g70090) was induced in the light-leaf mutant and that of one gene (LOC_Os01g70100) was repressed in the optica mutant. Therefore, LOC_Os01g70100 is a candidate gene for GLR2 (Wang et al., 2013b).
The recently reported SQUAMOSA PROMOTER BINDING PROTEIN BOX (SBP Box) family gene SQUAMOSA PROMOTER BINDING PROTEIN-LIKE10 (OsSPL10) also controls trichome initiation and positively regulates rice trichome formation (Tao et al., 2019). Knockout OsSPL10 rice displayed glabrous leaves and glumes, whereas overexpression mutants exhibited the opposite phenotype (Tao et al., 2019). At the same time, the researchers also found that OsSPL10 transcript levels increased when treated with the auxin analog 1-naphthylacetic acid (NAA) (Li et al., 2021a). OsTCL1 is a homolog of Arabidopsis AtTCL1, and OsTCL1 transgenic lines in Arabidopsis inhibit the formation of trichomes through direct interaction with GL3. However, there is no phenotypic difference in OsTCL1 overexpressing rice plants (Zheng et al., 2016). HAIRY LEAF6 (OsHL6) encodes APETALA2/ETHYLENE RESPONSE FACTOR type transcription factor. OsHL6 protein can interact with OsWOX3B, and enhance the binding ability of OsHL6 to the downstream AUX-related gene OsYUCCA5 (Sun et al., 2017).
OsGL6 localizes to the lower 79-kb region of chromosome 6. OsGL6 is a natural allele of OsHL6, which can stimulate the initiation of trichomes. In OsGL6 overexpressing rice, OsGL6 can promote trichome initiation but does not affect trichome elongation (Xie et al., 2020b). Furthermore, OsGL6 can interact with OSK3 and CSN5 proteins, and OsGL6 may regulate the formation of trichomes by forming a complex with OSK3 and CSN5 (Xie et al., 2020b).
In the rice trichome elongation stage, OsHL6 can not only promote trichome initiation but also affect rice trichome elongation. Suwangwanger (SWWR) is an indica rice variety from Sri Lanka with hairy leaves. HL6SWWR is a novel allele of the reported HL6 locus that regulates trichome formation in rice. Transgene complementation and knockout experiments confirmed that HL6SWWR regulates rice trichome elongation (Fei et al., 2020).
A regulation model of rice trichome initiation and elongation is shown in Figure 3.
Cucumber (Cucumis sativus) is a common vegetable worldwide. From stems and leaves to flowers, branches, fruits, and tendrils, their surface is covered with trichomes. The trichomes on its fruit are called thorns, and they have a multi-layered tubercule underneath (Pan et al., 2015). Both the tubercule and the thorns are collectively referred to as cucumber fruit wart. In cucumber, thorns combine with tubercule to form a verrucous fruit trait, which is important for fruit quality (reviewed by Liu et al., 2016).
Cucumber fruit spines are multicellular trichomes, and their development and regulation mechanisms are different from those of unicellular trichomes. According to the structure and morphology of cucumber fruit trichomes, the researchers divided cucumber fruit trichomes into eight types, the morphological photographs of cucumber trichomes were shown in Supplementary Figure 3 (Xue et al., 2019). Of these, type I and VI trichomes are glandular hairs (Xue et al., 2019). Except for two hairless mutants (csgl1 and csgl3), type I trichomes exist on the fruits of all cucumber varieties. Type I trichomes consist of a single short stalk with 3–5 cells and a 4–8-cell head region with glandular functions (Samuels et al., 1993). Type II trichomes are larger, without glands, and consist of a base and long stems. During non-glandular hair type II trichome development, precursor cells expand perpendicular to the surface of the epidermis and then divide around the cells to form a stalk composed of 5–7 rectangular cells and a pointed apical cell (Li et al., 2015). The stalk of type III trichomes consists of 3–6 cells and is much shorter than the stalk of type II trichomes. A particularly notable feature of type III trichomes is that the base cells divide to form a conical structure. Type IV trichomes undergo initiation and division to form multicellular stalks, and the base cells divide into multicellular structures similar to type II trichomes but smaller. Type V trichomes have a unique pyramid structure without an obvious slender stem and without an enlarged base (Xue et al., 2019). Type VI glandular hairs are rarely observed and have similar four-cell or five-cell glands on each head, but their stalk cells are longer than those of type I trichomes. Type VII and VIII trichomes are only found on the hairless mutant csgl1; they are invisible to the naked eye (Chen et al., 2014; Zhao et al., 2015). Type VII trichomes begin with the expansion of a hairy cell. Then, after several cell divisions, multiple cells are stacked together to form a blunt tip. The development process of type VIII trichomes is basically similar to that of type VII trichomes, but type VIII trichomes have extra branch formation in the later stage (Zhao et al., 2015). All non-glandular trichomes have gone through a stage of senescence, that is, the tip first becomes white or brown and then the other trichomes lose their green color from top to bottom and become white or brown (Zhang et al., 2016). The trichomes of cucumber cotyledons and fruits have relatively synchronized developmental trajectories and can be divided into five stages according to their morphogenesis: initiation, first division, formation of pointed head (non-glandular trichomes)/glandular head transformation (glandular trichomes), elongation (non-glandular trichomes)/glandular head formation (glandular trichomes) and multicellular base formation (non-glandular trichomes)/active metabolic processes (glandular trichomes) (Dong et al., 2022).
HD- ZIP-, MYB-, and C2H2-type transcription factors as well as WD-repeat proteins are involved in the cucumber trichome fate determination and initiation stage.
TRICHOME-LESS (Tril) and CsGL3 are two alleles of the HD-ZIP IV family; they play an important role in the fate determination and initiation of cucumber glandular hairs. Mutations in these two genes present a completely hairless phenotype on leaves, stems, flowers, sepals, and fruits (Pan et al., 2015; Wang et al., 2016). Tril expression controlled by its own promoter partially rescues the mutant phenotype of tril and csgl3 (Du et al., 2020). The hairless phenotype of csgl3 exhibited due to the loss of function of CsGL3 is due to the insertion of an autonomous, active class I transposable element in CsGL3 (Pan et al., 2015). It is speculated that Tril and CsGL3 may directly bind to CsTBH, CsMICT, and CsGL1 to activate downstream transcriptional activators such as CsMYB6, CsWIN1, and CsGL2, and then promote the initiation of trichomes in leaves, stems and fruits (Pan et al., 2015; Shvachko et al., 2020). The csgl2 mutant showed few trichomes or nodules on tendrils, calyx, ovary and fruit, but glabrous stems and leaves (Cui et al., 2016).
GLABROUS 1 (CsGL1), TINY BRANCHED HAIR (CsTBH), and MICRO-TRICHOME (CsMICT) genes of the HD-ZIP I family are involved in the formation of cucumber trichomes. Under scanning electron microscopy (SEM), many papillae can be observed on the epidermis of csgl1 mutant leaves. The density of papillae is similar to that of wild type trichomes, indicating that CsGL1 may be involved in the development of trichomes of leaves but not in fate determination and initiation (Pan et al., 2015). Studies have shown that CsGL1 may indirectly regulate the expression of CsMYB6 and GA20ox1 (Li et al., 2015). The other two allelic mutants of csgl1, tbh, and mict have similar phenotypes to csgl1. CsTBH is preferentially expressed in the multicellular trichomes of cucumber fruits. Overexpression of CsTBH in cstbh mutants restores the fruit trichome phenotype, while the silence of CsTBH in wild-type plants leads to spine dysplasia; however, CsTBH does not participate in the spine initiation (Chen et al., 2014; Xue et al., 2019). CsTBH can directly bind to the promoter of the cucumber 1-aminocyclopropane-1-carboxylic acid synthase (CsACS) gene and regulate its expression, thereby affecting the development of multicellular fruit trichomes (Zhang et al., 2021f). Therefore, CsTBH regulates cucumber fruit trichomes through the ETH pathway (Chen et al., 2014; Xue et al., 2019). CsMICT is expressed in trichome cells, and all leaf and fruit trichomes in its mutants are small and stunted (Zhao et al., 2015; Pan et al., 2021). Phenotypic analysis of mutants demonstrated that CsTBH, CsMICT and CsGL1 are involved in regulating the morphogenesis of glandular hairs rather than fate determination and initiation (Liu et al., 2016; Feng et al., 2021b).
The MYB-type transcription factors CsMYB6 (Csa3G824850) and CsTRY (Csa5G139610) negatively regulate cucumber fruit spine initiation (Zhang et al., 2019b). Transformation of CsTRY (Csa015371) into Arabidopsis can significantly inhibit the development of leaf trichomes. Researchers speculate that the CsTRY gene inhibits the development of cucumber trichomes, and the CsMYB6 gene, which is similar to CsTRY, negatively regulates the formation of cucumber trichomes (Yang et al., 2018, 2021).
The C2H2 zinc finger protein gene Tu is specifically expressed in cucumber tumor cells (Yang et al., 2014a). Tu is a key factor in controlling fruit tumor formation (Cui et al., 2016). CsGL1 has an epistatic effect on Tu (reviewed by Che and Zhang, 2019). The formation of nodules is related to high CTK content, and the expression of Tu affects the expression of Csa5M644580 and Csa5M224130 genes, which are homologous to CTK hydroxylase, thereby accelerating the biosynthesis of CTK and promoting the formation of fruit nodules (Yang et al., 2014a). Tu was present in all 38 warty (Wty) lines and completely absent in all 56 non-wart (nWty) lines. Moreover, Tu was required for the Wty fruit phenotype in Tu transgenic cucumber plants (Yang et al., 2014a).
The WD-repeat protein gene CsTTG1 (Csa4Gm097650) plays a key role in cucumber flowering and trichome and fruit tumor formation. Silencing of CsTTG1 inhibits the formation of fruit thorns (Guo et al., 2020). Molecular and genetic analysis showed that CsTTG1 and the key trichome forming factors CsMICT and CsGL1 have similar roles in regulating the initial development of trichomes (Chen et al., 2016a).
The bHLH-type transcription factor HECATE2 (CsHEC2) is highly expressed in cucumber pericarp (including tubercule and thorns). A mutant of the CsHEC2 gene obtained using the CRISPR/Cas9 system had a significantly lower fruit thorn and tumor density and lower accumulation of CTK compared to the wild type. Conversely, overexpression of CsHEC2 led to an increase in thorn and tumor density and CTK level. CsHEC2 directly binds to the promoter of the cytokinin hydroxylase gene CsCHL1, activating its expression. In addition, CsHEC2 directly interacted with CsGL3 and Tu, further enhancing its positive regulation of CsCHL1 expression (Wang et al., 2021e).
The plant hormone signaling pathway is also involved in cucumber trichomes occurrence. 6-benzylaminopurine (6-BA) and GA3 effect epidermal differentiation and fruit thorn development. They affect the number of trichomes in each fruit. Both 6-Benzylaminopurine (6-BAP) and GA stimulate the formation of trichomes in cucumber fruit in a concentration-dependent manner (Zhao et al., 2015; Xue et al., 2019). GA oxidase (GAoxs) is a key enzyme in the GA biosynthesis pathway. Overexpression of CsGA20ox1 reduces the length of cucumber fruit spines (Li et al., 2015; Sun et al., 2018). The NUMEROUS SPINES (NS) (Csa2G264590) gene encodes AUX transporter-like protein 3, which is a key negative regulator gene that determines the thorn density of cucumber fruit (Du et al., 2020). Through expression pattern analysis, it was found that the upstream genes of the AUX signaling pathway in two cucumber cultivars (NCG122 and NCG121), including NS, were down-regulated, while AUX signaling pathway downstream genes were up-regulated. This implies that NS is a negative regulator that determines the density of cucumber fruit thorns and may regulate the development of fruit thorns by regulating the AUX signaling pathway (Xie et al., 2018).
A regulation model of cucumber trichome development was shown in Figure 4.
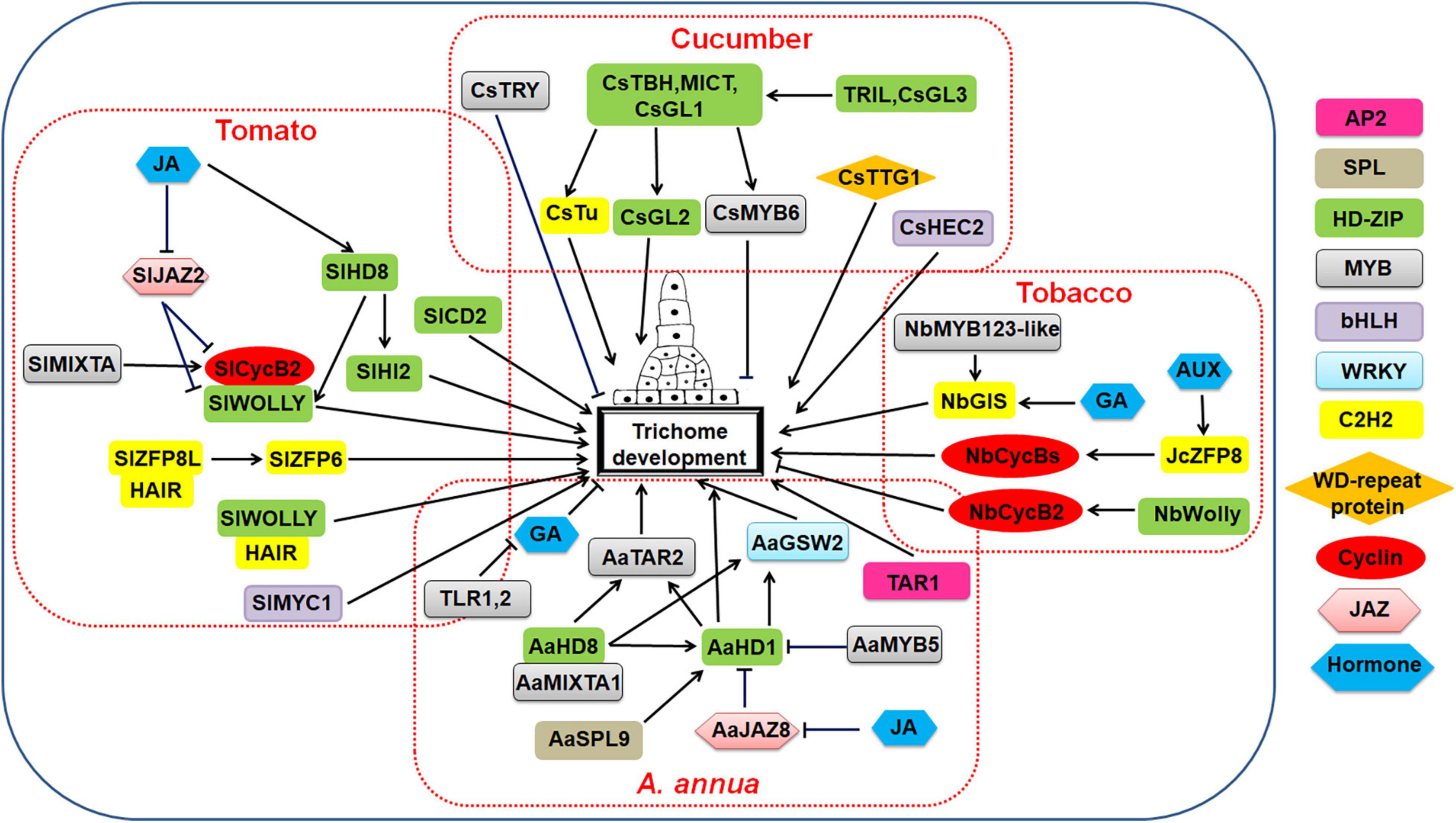
Figure 4. Regulatory network model for multicellular trichome development in cucumber, tomato, tobacco, and Artemisia annua.
Tobacco (Nicotiana tabacum L.), including Nicotiana benthamiana L., is commonly used as a heterogeneous production platform as an important model plant for basic biological research. It is a multicellular hairy plant that is widely cultivated worldwide. Most studies on multicellular trichomes in tobacco have primarily focused on morphological observations and identification of their secretions; few studies have investigated the molecular mechanisms of multicellular trichome differentiation in tobacco. Tobacco has three types of trichomes: long-stalked glandular trichomes (LGT), short-stalked glandular trichomes (SGT) and non-glandular trichomes (NGT) (Wang et al., 2021f; Zhang et al., 2021a). The glandular trichomes consist of one basal cell, 1–5 stalk cells and 1–12 head cells. They are mainly unbranched head-shaped glandular hairs composed of a multicellular stalk and a single or multicellular head, the morphological photographs of tobacco trichomes were shown in Supplementary Figure 4 (Amme et al., 2005; Liu et al., 2018a).
The C2H2 transcription factor is involved in regulating the development of tobacco glandular trichomes. NbGIS has a positive regulatory effect on the development of tobacco glandular trichomes. In response to GA signals, it can control glandular trichome initiation and significantly affects the accumulation and expression of GA biosynthesis marker genes, which may lead to tobacco growth and maturity (Liu et al., 2018b). The Jatropha curcas C2H2 zinc finger protein gene, JcZFP8, regulates the development of transgenic tobacco trichomes (Li et al., 2016). JcZFP8 is related to NtZFP8, AtGIS and AtZFP8 during trichome development (Liu et al., 2017). Overexpression of JcZFP8 in tobacco increases the number of trichomes on flowers, and JcZFP8 induces trichome formation. The way JcZFP8 regulates trichome development is different from that of GIS. JcZFP8 regulates trichome development by inducing the expression of MYB and CycB related genes. This finding provided new insights into the regulatory mechanism of the C2H2 ZFP genes in trichome development (Shi et al., 2018).
MYB-type transcription factors play a role in tobacco trichome initiation (Schnittger et al., 2005). NbMYB123-like encodes an R2R3 MYB domain that regulates tobacco glandular trichome initiation by acting downstream of NbGIS (Liu et al., 2018b). The R2R3 MYB transcription factors MIXTA and MIXTA-like1 from snapdragon promote tobacco trichome development (Perez-Rodriguez et al., 2005).
The interaction between HD-ZIP transcription factors and B type cyclin genes regulates tobacco trichome development. The tobacco B type cyclin gene NbCycB2 is a negative regulator of multicellular trichome formation, and the NbWoolly (Nbwo) gene is an HD Zip IV transcription factor (Yang et al., 2015). NbWoV is a Nbwo gain-of-function allele (Wu et al., 2020b). The genomic sequences of Nbwo and NbWoV can be combined with the NbCycB2 promoter sequence to directly regulate the expression of NbCycB2, Nbwo and NbWoV. As a form of feedback regulation, at the protein level, NbCycB2 inhibits Nbwo activity to negatively regulate trichome formation. Mutation of the Nbwo wolly motif prevents NbCycB2 inhibition of NbWoV, resulting in a marked increase in the amount of active Nbwo protein, increasing trichome density and branching number (Wu et al., 2020b). Nbwo and NbWoV act on the L1-like box in the promoter region of NbCycB2 to affect the expression of NbCycB2, inhibit the activity of Nbwo, and regulate the level of proteins that form trichomes (Wu et al., 2020b). In addition, NbCycB2 can inhibit trichome initiation by binding to the LZ domain of NbWo66 (Wu et al., 2020b). Knocking out NtCycB2 (NtCycB2-KO) can promote the formation of LGT, while overexpression of NtCycB2 (NtCycB2-OE) can reduce LGT density (Wang et al., 2021f).
Various plant hormones are involved in tobacco trichome formation and branching (Di et al., 2015). In JcZFP8 transgenic tobacco, AUX may be involved in trichome initiation, while GA and JA may not be involved in this process, and SA is a negative regulator of trichome development (Traw and Bergelson, 2003; Han et al., 2021). The tobacco transcription repressor protein NtJAZ is potentially involved in abiotic stress responses and glandular hair development. NtJAZ-9 may play an important role in the induction of NtJAZs in glandular trichomes (Zhang et al., 2019a).
A regulation model of tobacco trichome development was shown in Figure 4.
Cultivated tomato (Solanum lycopersicum) and its wild relative (Lycopersicon esculentum) have seven different trichome types. Types II, III, and V are non-glandular hairs composed of a neck and base. They act as a physical barrier to function in resisting diseases and insects. Types I, IV, VI, and VII are glandular hairs, the morphological photographs of tomato trichomes were shown in Supplementary Figure 5 (Xu et al., 2018; Chalvin et al., 2020). In addition to the neck and base, they also have a gland top that can store and secrete various secondary metabolites (Chen et al., 2018). Type II and V tomato trichomes are the most abundant; they are the typical non-glandular hairs. Type II and V trichomes have similar morphology but are composed of different numbers of cells. Type II trichomes are mainly composed of 5–8 cells, and type V trichomes are mainly composed of three cells. In type II trichomes, cells are larger at the base and become smaller toward the top. A similar trend occurs in type V trichomes (Glas et al., 2012; Vendemiatti et al., 2017). Type III trichomes are shorter than type II trichomes. Most type I and VI trichomes are glandular. Type I trichomes have multicellular bases, multicellular stalks and glandular cells at the top, whereas type VI trichomes have shorter multicellular stalks and a pumpkin-shaped gland composed of four cells (Bergau et al., 2015). Type IV trichomes have glands similar to type I trichomes, with a single-cell base and two to three stalk cells, whereas type VII trichomes have shorter single-cell stalks and 4–8-cell glands (Vendemiatti et al., 2017).
Tomato trichomes are mainly regulated by R2R3-MYB, HD-ZIP IV, C2H2, and bHLH-type transcription factors (reviewed by Chezem and Clay, 2016; see also Chalvin et al., 2020).
The R2R3-MYB proteins SlMIXTA1 and SlMixta-like control tomato trichome initiation. After SlMixta-like silencing, many trichomes occur on leaves. SlMixta-like silence lines lead to abnormal trichome spacing and trichome aggregation (Ewas et al., 2017; Galdon-Armero et al., 2020). In SlMixta-like overexpression lines, the density of type I and IV trichomes was unaffected, while the density of type V and VI trichomes was significantly reduced compared to that in the wild type (Galdon-Armero et al., 2020).
HD-ZIP IV transcription factors are key regulators of tomato trichome development (Hua et al., 2021a). The SlWOOLLY (SlWo) gene encodes an HD-ZIP IV transcription factor that regulates tomato type I trichome formation mainly through heterodimer formation with the B-type cyclin SlCycB2 (Gao et al., 2015). SlWo also actively regulates mitosis in multicellular trichomes (Gao et al., 2017; Hua et al., 2021a). SlWo is mainly regulated by plant hormones such as JA, AUX and GA. Among them, JA has an obvious effect on inducing glandular hairs. The HD-ZIP IV transcription factor SlCD2 regulates type VI glandular trichome formation (Nadakuduti et al., 2012). A loss-of-function mutation in SlCD2 resulted in a sticky peel mutant phenotype in tomato, with less glandular hairs (especially type VI) than that in the wild type (Nadakuduti et al., 2012). Knockout of the HD Zip IV transcription factor HDZIPIV8 also distorts tomato trichomes (Xie et al., 2020a). A mutation of NCK-ASSOCIATED PROTEIN 1 named as Hairless-2 (Hl-2) in tomato caused serious distortions in all trichome types (Xie et al., 2020a). HDZIPIV8 regulates the expression of H1-2 by binding to the L1-Box of the H1-2 promoter region, thereby regulating the elongation and morphogenesis of tomato trichomes (Xie et al., 2020a). SlHZ45 is the HD-ZIP IV transcription factor with the highest homology to SlWo. Compared with a control group, the number of type I, IV and VI trichomes on the leaf margins of SlHZ45 overexpressing plants increased; these three trichome types are glandular hairs (Zhang et al., 2014). The HD-Zip transcription factor SlHD8 positively regulates tomato trichome elongation. Dual luciferase and ChIP experiments showed that SlHD8 regulates tomato trichome elongation by directly binding to a set of cell wall loose protein gene promoters and activating their transcription (Hua et al., 2021b).
C2H2 zinc finger proteins have been isolated and identified in tomato trichome formation. The C2H2 zinc finger protein gene is homologous with ZFP8 of Arabidopsis. It directly interacts with SlWo and regulates type I and VI trichomes (Chang et al., 2018). H overexpression lines can promote the elongation of type I, III and VI trichomes in tomato. H is a constitutively expressed gene, and lack of H may inhibit the function of SlWo (Tao et al., 2014). In addition, the similar regulatory effects of H gene homologs in the formation of trichomes indicate that these multicellular structures of solanaceous plants may be controlled by conservative molecular mechanisms. Recent studies have found that SlZFP8-like (SlZFP8L) directly interacts with SlWo to regulate tomato trichome initiation (Zheng et al., 2021). Overexpression of SlZFP8L will increase the length of type I, III, and VI trichomes and increase the density of type I, III, V, VI, and VII trichomes (Zheng et al., 2021). Researchers also found that H interacts with SlZFP8-like (SlZFP8L) to regulate trichome initiation and elongation by regulating SlZFP6 expression in tomato (Zheng et al., 2021).
Almost all genes identified so far that control glandular hair also affect non-glandular hair development; only one gene (SlMYC1) is involved in glandular hair regulation but not non-glandular hair regulation. SlMYC1 is a bHLH transcription factor that regulates the development of type VI glandular hairs in tomato (Xu et al., 2018). In mature gland cells of type VI trichomes, SlMYC1 can be recruited by SlWo to activate TPS gene expression for terpene biosynthesis. The SlWo-SlMYC1 functional module can be inhibited by SlJAZ2, and JA can relieve this inhibition. In addition to acting with SlWo, SlMYC1 plays a SlWo-independent role in glandular cell division and expansion (Xu et al., 2018). SlbHLH95 negatively regulates trichome initiation, and plants overexpressing SlbHLH95 have significantly less type I trichomes on stems (Chen et al., 2020). bHLH95 also regulates GA biosynthesis and trichome formation through GA20ox2 and KS5 (Chen et al., 2020).
Plant hormone signaling pathways are involved in regulating tomato trichome differentiation. The JAZ protein SlJAZ2 is a repressor protein of the tomato JA signaling pathway; it also negatively regulates the occurrence of glandular hairs. In SlJAZ2 overexpression plants, the expression of SlWo and SlCycB2 was suppressed, indicating that SlJAZ2 suppressed glandular hair initiation by suppressing the expression of SlWo and SlCycB2 (Yu et al., 2018). A yeast two-hybrid experiment found that SlJAZ2 interacts with CORONATINE INSENSITIVE1 (SlCOI1), which also positively regulates glandular hair development (Thines et al., 2007). AUX also plays an important role in regulating glandular hair growth. When the expression of the tomato AUX family gene SlIAA15 and the AUX response factor SlARF3 is down-regulated, the number of tomato type I, V, and VI trichomes decreases (Deng et al., 2012b). The negative regulator of trichome development JAZ4 is a key component of tomato trichome JA signal transduction. The HD-Zip transcription factor SlHD8, located downstream of the JA signal, positively regulates tomato trichome elongation and can interact with SlJAZ4 (Hua et al., 2021b).
A regulation model of tomato trichome development was shown in Figure 4.
In recent years, artemisinin has received widespread attention due to its antimalarial effects. The glandular hair of Artemisia annua is the only place where artemisinin is synthesized and stored. Artemisia annua has two types of trichomes, namely non-secretory glandular trichomes and glandular secretory trichomes (GSTs). Non-secretory glandular trichomes are also called T-shaped trichomes (TSTs) or T-type non-glandular trichome (TNGTs) because their appearance is similar to the letter “T.” The morphological photographs of Artemisia trichomes were shown in Supplementary Figure 6 (Chalvin et al., 2020).
The transcription factors involved in the regulation of glandular hair development in A. annua are similar to those in tomato, and most of them belong to the R2R3-MYB and HD-ZIP subfamilies (reviewed by Chezem and Clay, 2016; see also Chalvin et al., 2020).
AaHD1 and AaHD8 of the HD-ZIP IV family play a positive role in regulating the initiation of glandular hairs. Overexpression of AaHD1 or AaHD8 increases the density of glandular hairs, whereas inhibition of expression of either of them decreases the density of glandular hairs. SEM analysis found that the glandular and non-glandular hair densities of A. annua were significantly decreased in AaHD8 silenced lines and increased in AaHD8 overexpressing lines compared with the wild type. In addition, expression levels of AaHD1 followed changes in AaHD8 expression, which was significantly down-regulated in AaHD8 silencing and up-regulated in AaHD8 overexpressing A. annua lines (Yan et al., 2018).
The R2R3-MYB transcription factor AaMIXTA1 is a positive regulator of the initiation of glandular hair development and can directly activate the expression of genes related to the biosynthesis of the leaf stratum corneum (Matías-Hernández et al., 2017). Later research found that the transcription complex formed by AaHD8 and AaMIXTA1 can promote AaHD1 expression, which in turn promotes glandular hair initiation (Yan et al., 2018). Overexpression of AaMYB1 can increase the density of glandular hairs (Matías-Hernández et al., 2017). AaMYB1 is an ortholog of AtMYB61. Overexpression of both AaMYB1 and AtMYB61 affect Arabidopsis trichome initiation, root development, and stomatal pore size. Transgenic Arabidopsis overexpressing AaMYB1 have a higher density of trichomes on rosette leaves compared to the wild type, and a similar number of trichomes as atmyb61-2 mutant plants overexpressing AaMYB1 (myb61-2 35S:AaMYB1), indicating that AaMYB1 expression rescues the Atmyb61-2 mutant phenotype (Matías-Hernández et al., 2017). AaMYB16 positively regulates the initiation of glandular hairs. Compared with the wild type, AaMYB16-overexpressing transgenic lines had 28–45% higher GST density on the front surface of mature leaves, while AaMYB16-knockdown lines had 27–41% lower GST density (Xie et al., 2021a). SEM analysis showed that TNGT density was also higher in the overexpression lines and lower in the RNAi lines (Xie et al., 2021a). AaMYB5 has the opposite effect to AaMYB16 but neither can independently regulate GST formation. GST densities were higher in AaMYB5 knockdown lines and lower in AaMYB5 overexpression lines compared to the wild type. SEM analysis showed no difference in TST density between wild type and transgenic plants (Xie et al., 2021a). AaMYB5 and AaMYB16 compete by interacting and regulating the binding activity of the AaHD1 promoter (Xie et al., 2021a). R2R3 MYB transcription factor AaTAR2 is mainly expressed in young leaves of A. annua. Knockout and overexpression of AaTAR2 resulted in decreased and increased GST number and artemisinin content, respectively. In addition, the researchers also found that AaTAR2 can bind to the promoters of HD-ZIP transcription factors AaHD1 and AaHD8 to regulate the development of Artemisia trichomes (Zhou et al., 2020). R2R3 MYB transcription factors TRICHOME LESS REGULATOR 1 (TLR1) and TLR2 were found to negatively regulate trichome development in A. annua, the trichome density and artemisinin content were decreased in TLR1 and TLR2 overexpressing lines, and increased in TLR1-RNAi lines compared with the wild type. Studies of TLR1 and TLR2 in Arabidopsis and A. annua overexpression lines show that they negatively regulate trichome density by reducing gibberellin levels (Lv et al., 2022).
Overexpression of the GST-specific WRKY transcription factor AaGSW2 in A. annua significantly increases GST density. Knockout of AaGSW2 inhibits the initiation of GST in A. annua. Furthermore, AaHD1 and AaHD8 can bind to the L1-box on the promoter of AaGSW2 and positively regulate GST initiation (Xie et al., 2021b).
The AP2 transcription factor TRICHOME AND ARTEMISININ REGULATOR 1 (TAR1) regulates trichome development in A. annua. Compared to the wild type, TAR1 disturbed expression lines had altered trichome morphology and cuticle wax composition; significantly lower artemisinin content; higher permeability; less glandular hairs; and a collapsed glandular hair head phenotype (Tan et al., 2015). In addition, compared to the wild type, A. annua plants overexpressing the β-glucosidase gene (BGL1) had 20 and 66% more trichomes with 1.4 and 2.56% higher artemisinin content on leaves and inflorescences, respectively (Singh et al., 2016).
SQUAMOSA promoter-binding protein-like (SPL) is a kind of plant-specific transcription factor, studies have shown that overexpression of AaSPL9 increases the density of glandular hairs by 45–60% and the artemisinin content by 33–60%, indicating that AaSPL9 positively regulates the initiation of glandular hairs. Yeast one-hybrid, dual-luciferase and electrophoretic mobility shift assay (EMSA) demonstrated that AaSPL9 activates the expression of AaHD1 by directly binding the GTAC-box of the AaHD1 promoter to regulate the initiation of glandular hairs in A. annua (He et al., 2022).
The plant hormone signaling pathway is involved in A. annua trichome initiation. Trichome formation is mainly regulated by JA. AaJAZ8 is a repressor of the JA signaling pathway in A. annua. It can inhibit the activity of the positive regulator AaHD1 and reduce the density of glandular hairs (Yan et al., 2017). The zinc finger protein AaSAP1 can also respond to JA induction and positively regulate glandular hair development. However, the specific molecular mechanisms remain to be elucidated (Wang et al., 2019c).
A regulation model of A. annua trichome development was shown in Figure 4.
Three types of trichomes occur on adult maize (Zea mays) leaves: microscopic bicellular hairs, macrohairs, and prickle hairs (Kong et al., 2021). Overexpression of the maize HD-ZIP IV gene OCL4 in Arabidopsis resulted in a glabrous rosette leaf phenotype (Vernoud et al., 2009). The maize GLOSSY1 (ZmGL1) gene is a component of the pathway leading to maize seedling epidermal wax biosynthesis. zmgl1 mutants have altered maize epidermal development, including altered trichome size and impaired cuticle structure (Sturaro et al., 2005). The SQUAMOSA-promoter-binding protein-like (SPL) protein is a plant-specific transcription factor with SBP characteristics and a domain consisting of 76 amino acid residues (Wei et al., 2018a). Cytological analysis showed that the zmspl10/14/26 triple mutant was completely hairless (Feng et al., 2021a). Three homologous of the ZmSPL transcription factors, ZmSPL10, ZmSPL14, and ZmSPL26, act synergistically to promote trichome fate in maize leaves, possibly by regulating the expression of ZmWOX3A and AUX-related genes (Kong et al., 2021).
The prickles on rose plants are a specialized form of trichome. The TTG1 transcription factor gene RcTTG1 may be related to the development of rose bark, and the expression level of this gene significantly differs among different tissues (Feng et al., 2015).
Specialized trichomes of prickly pear (Rosa roxburghii) distributes on its leaves, stems, branches, sepals, pedicels and fruit, and the trichomes on the fruit are often called thorns; they affect the appearance and sensory quality of the plant (Wang et al., 2019a). The RrGL1 gene of prickly pear can functionally restore the formation of trichomes in Arabidopsis gl1 mutants, indicating that RrGL1 is involved in the development of prickly pear spines. Yeast hybridization assays indicate that RrGL1 may play a functional role in trichomes by forming an MYB-bHLH-WD40 complex. Thus, the formation of prickly pear spines is similar to that of Arabidopsis trichomes (Huang et al., 2019b). Moreover, there were significant differences in the expression levels of RrGL2 in stems and fruits of different stages, indicating that this gene may be closely related to fruit spine formation and development.
Overexpression of the MYB-like gene AmMIXTA from snapdragon (Antirrhinum majus) in tobacco resulted in more trichomes, suggesting that AmMIXTA regulates trichome development in snapdragon (Payne et al., 2000). Under the action of the CaMV35S promoter, there were more trichomes on snapdragon leaves, further proving that AmMIXTA has a role in regulating snapdragon trichome development (reviewed by Martin et al., 2002). The conserved domain of the snapdragon AmMYBML1 transcription factor is almost identical to that of the AmMIXTA transcription factor, and when overexpressed in tobacco, AmMYBML1 can promote trichome development in floral tissues (Glover et al., 2004).
Trichomes are small structures on plant arial parts. Their developmental regulation involves many hormones, transcription factors and metabolic pathways. The key genes in development of different trichome types are summarized in Supplementary Table 1. Although much research has been conducted on the fate determination, initiation, branching, elongation and maturation of trichomes in different species, there are still many knowledge gaps.
Many trichome development studies have focused on model plants such as Arabidopsis. Development of special forms of trichomes has been studies less, such as thorns on cucumber fruit and roses, which affect field management and picking efficiency. In addition, little research progress has been made on trichomes of monocotyledonous plants such as rice. Extensive research is needed on trichome development in various horticultural plants from multiple perspectives. Future research should focus on determining internal connections and interactions of trichomes with seeds and other organs; key regulatory factors related to trichome development; and how these regulatory factors change with environmental or developmental changes. This will provide new insights and genetic resources for plant development and crop improvement. Depending on the trichome-specific genes, corresponding promoters, such as FBP7 and E6, can also improve crop yield and quality. Therefore, the identification and application of tissue-specific promoters and specific genes are of great significance to the research of plant development and the advancement of crop genetics.
Many positive regulators and pathways in trichome development have been identified, but our understanding of negative regulatory mechanisms and key factors is still limited. For example, many C2H2 zinc finger protein transcription factors that positively regulate trichome development have been found in different species, but similar negative regulator have not been reported.
Plant hormones play an important role in trichome development. However, the regulatory mechanism of these plant hormones requires further research. AUX, BR, GA, and JA positively regulate cotton fiber initiation, whereas ABA and CTK negatively regulate cotton fiber initiation. The important roles of plant hormones in trichome development are well known, but the interactions among these hormones and their molecular mechanism requires further study. This will help identify and analyze the positive and negative factors in trichome development.
Glandular hairs development is closely related to their density on the plant surface and the types and contents of secondary metabolites. Studies in tomato, A. annua and cucumber have shown that C2H2 zinc finger proteins, and HD-ZIP- and MYB-type transcription factors play a key role in the glandular hair initiation and development. The biosynthesis of cytoskeleton and cutin also has an important role in the morphogenesis of glandular hairs. Interestingly, the reported glandular hair development genes often also affect the development of non-glandular hairs in plants. Therefore, it is necessary to further explore the relationship between glandular and non-glandular hair development and genes that independently regulate glandular hair development.
Similarities between trichomes and salt glands are also worthy of future study. Most plants have trichomes; however, the aerial part of the salt-secreting halophyte Limonium bicolor has no trichomes but has multicellular salt glands. The salt-secreting capacity of the salt glands significantly improves the salt tolerance of L. bicolor. The excess salt in the plant tissue can be excreted through salt glands to regulate the ion balance in the plant. In the epidermal structure of non-halophytes, such as Arabidopsis, tobacco and tomato, there are trichomes but not salt glands, while the leaf epidermal structure of L. bicolor has salt glands but not trichomes. The distribution of salt glands on L. bicolor leaves is similar to that of trichomes in Arabidopsis, they are both arranged in an interval pattern (Mishra et al., 2021). Both salt glands and trichomes differentiate earlier than stomata (Yuan et al., 2016b). In summary, salt glands and trichomes have similar temporal and spatial characteristics. In addition, transcriptome analysis of L. bicolor salt glands at different developmental stages showed that many differentially expressed genes in the early stage of salt gland development were homologous to the key genes in trichomes development in plants such as Arabidopsis (Yuan et al., 2015c). Furthermore, as an accessory structure of the plant epidermis with a secretory function, the trichomes of A. annua and tobacco have a strong secretory ability. Similarly, L. bicolor secretes salt through salt glands. Therefore, we speculate that there are certain similarities between salt-secreting glands and secretory trichomes. This raises many questions. Is there a homologous relationship between the development of trichomes and salt glands? Are the key genes that determine salt gland development homologous with the key genes in trichome development? What is the molecular evolutionary relationship between trichomes and salt glands? We propose the “common origin hypothesis” of salt glands and plant trichomes in non-halophytes, that is, there is a “common ancestor gene” that controls the differentiation of proepidermal cells, and separation may occur at a certain node in the long-term evolution of plants. One gene type controls the differentiation and development of proepidermal cells into salt glands to adapt to high salt environments, while another type controls the development into trichomes to survive under other biotic and abiotic stressors, such as arid and semiarid regions (Figure 5). To explore this hypothesis, we are currently researching salt gland development in our laboratory.
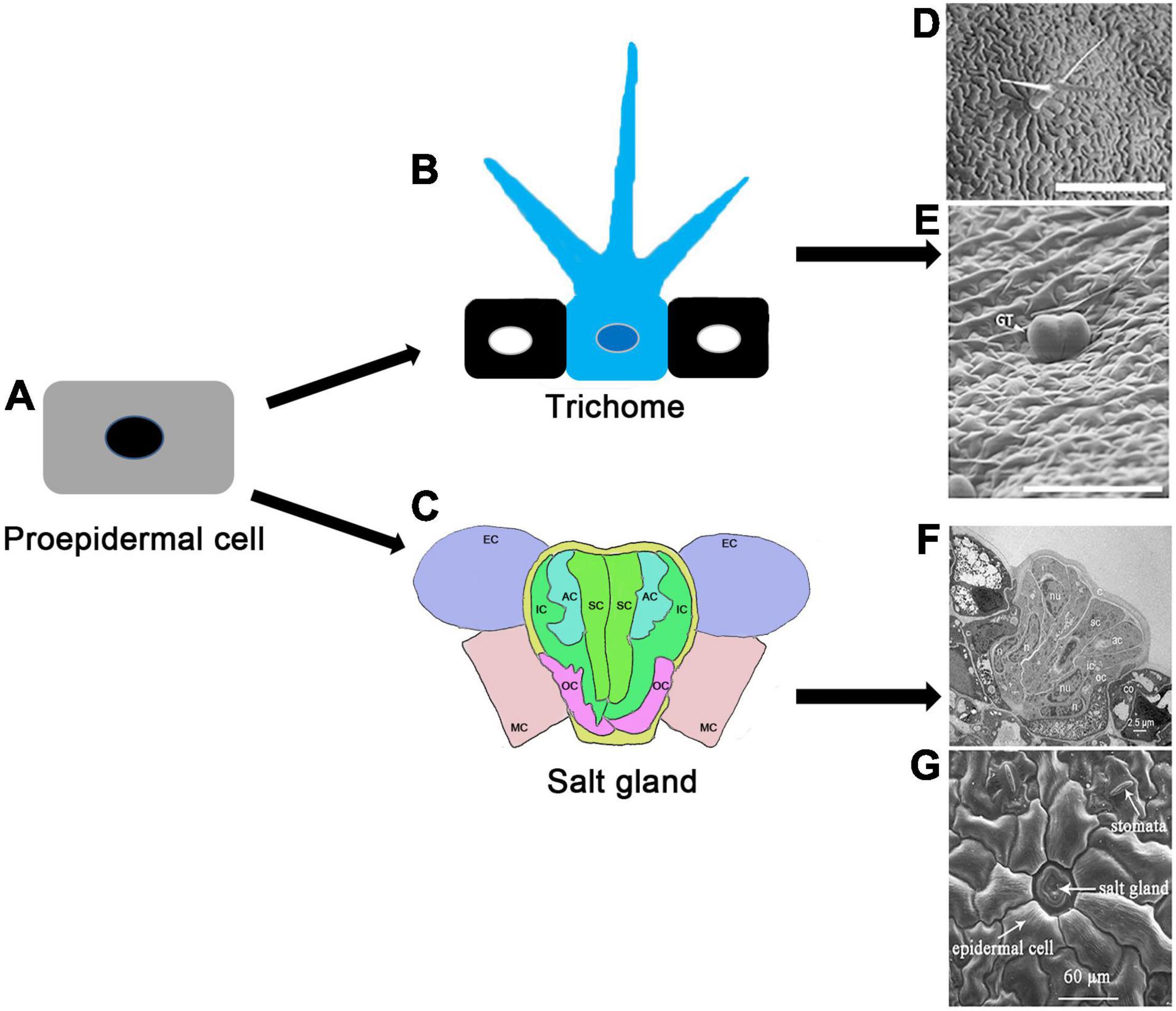
Figure 5. Possible evolutionary patterns in trichome and salt gland development (Breuer et al., 2009; Yuan et al., 2015b,2016a; Chalvin et al., 2020). (A) Schematic diagram of plant proepidermal cells; (B) schematic diagram of plant trichome; (C) schematic diagram of multicellular salt gland in Limonium bicolor. SC, secretory cell; AC, accessory cell; IC, inner cup cell; OC, outer cup cell; MC, mesophyll cell; EC, epidermal cell; (D) scanning electron microscope observation of Arabidopsis trichomes, 10-day-old wild-type [Columbia (Col)] on the first true leaf; (E) the multicellular glandular trichomes of A. annua leaf (adverse) were observed by scanning electron microscope. GT, glandular trichome. Scale bar = 100 μm. (F) Longitudinal section of salt glands in Limonium bicolor. Transmission electron microscope (TEM) image of salt glands of Limonium bicolor leaves prepared by high-pressure freezing (HPF) followed by freeze substitution (FS) and then embedded, sectioned, and stained. Scale bar = 25 μm; (G) morphologies of salt glands in Limonium bicolor leaves using environmental scanning electron microscopy (ESEM). Scale bar = 60 μm.
Innovation and application of cutting-edge science and technology and experimental methods will help future research on the molecular mechanisms of plant trichome development. These include chloroplast genetic engineering, nanoparticle bombardment transformation, the VIGS method, omics technology, single-cell sequencing and CRISPR-Cas9. With the rapid development and effective application of modern technologies in model plants, horticultural plants and crops, more genes and signaling pathways involved in trichome development will be identified.
GH and YL wrote this manuscript. ZY, CW, and YZ participated in the writing and modification of this manuscript. BW and GH conceptualized the idea. All authors read and approved the final manuscript.
This work was supported by National Natural Science Research Foundation of China (project nos. 32000209 and 32170301), Natural Science Research Foundation of Shandong Province (project no. ZR2020QC031), and China Postdoctoral Science Foundation (project no. 2020M672114).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.910228/full#supplementary-material
CGT, capitate glandular trichome; bHLH, basic helix-loop-helix; GA, gibberellin; CTK, cytokinin; DNA, deoxyribonucleic acid; JA, jasmonic acid; SA, salicylic acid; RNA, ribonucleic acid; mRNA, message RNA; m6A, N6-methyladenosine; DPA, days after anthesis; ta-siRNAs, trans-siRNAs; siRNA, small interfering RNA; miRNA, microRNA; ROS, reactive oxygen species; ETH, ethyne; AUX, auxin; IAA, indoleacetic acid; H2O2, hydrogen peroxide; QTL, quantitative trait locus; VIGS, virus-induced gene silencing; BR, brassinolide; RNAi, RNA interference; ABA, abscisic acid; SCW, secondary cell wall; SEM, scanning electron microscopy; Wty, warty; nWty, non-warty; 6-BA, 6-benzylaminopurine; GA3, gibberellin A3; LGT, long-stalked glandular trichomes; SGT, short-stalked glandular trichomes; NGT, non-glandular trichomes; ChIP, chromatin immunoprecipitation assay; GST, glandular secretory trichome; TST, T-shaped trichome; TNGT, T-type non-glandular trichome.
Aashima, K., Paper, J. M., Boehler, A. P., Bradley, A. M., Neumann, T. R., and Kathrin, S. (2014). HD-Zip Proteins GL2 and HDG11 Have Redundant Functions in Arabidopsis Trichomes, and GL2 Activates a Positive Feedback Loop via MYB23. Plant Cell 26, 2184–2200. doi: 10.1105/tpc.113.120360
Akhtar, M. Q., Qamar, N., Yadav, P., Kulkarni, P., Kumar, A., and Shasany, A. K. (2017). Comparative glandular trichome transcriptome-based gene characterization reveals reasons for differential (–)-menthol biosynthesis in Mentha species. Physiol. Plant. 160, 128–141. doi: 10.1111/ppl.12550
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). Microrna-Directed phasing during trans -acting siRNA Biogenesis in Plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Amme, S., Rutten, T., Melzer, M., Sonsmann, G., Vissers, J. P. C., Schlesier, B., et al. (2005). A proteome approach defines protective functions of tobacco leaf trichomes. Proteomics 5, 2508–2518. doi: 10.1002/pmic.200401274
An, L. J., Zhou, Z. J., Su, S., Yan, A., and Gan, Y. B. (2012). GLABROUS INFLORESCENCE STEMS (GIS) is required for trichome branching through gibberellic acid signaling in Arabidopsis. Plant. Cell Physiol. 53, 457–469. doi: 10.1093/pcp/pcr192
Andrade, M. C., Silva, A. A., Neiva, I. P., Oliveira, I. R. C., Castro, E. M., Francis, D. M., et al. (2017). Inheritance of type IV glandular trichome density and its association with whitefly resistance from Solanum galapagense accession LA1401. Euphytica 213:52.
Andreas, G., Kazuko, Y., Tran, L. T., and Peter, C. C. (2014). Characterization of an apple TT2-type R2R3 MYB transcription factor functionally similar to the poplar proanthocyanidin regulator PtMYB134. Planta 240, 497–511. doi: 10.1007/s00425-014-2098-y
Angeles-Shim, R. B., Asano, K., Takashi, T., Shim, J., Kuroha, T., Ayano, M., et al. (2012). A WUSCHEL-related homeobox 3B gene, depilous (dep), confers glabrousness of rice leaves and glumes. Rice (N Y) 5:28. doi: 10.1186/1939-8433-5-28
Arribas-Hernández, L., Rennie, S., Schon, M., Porcelli, C., Enugutti, B., Andersson, R., et al. (2021). The YTHDF proteins ECT2 and ECT3 bind largely overlapping target sets and influence target mRNA abundance, not alternative polyadenylation. Elife 10:e72377. doi: 10.7554/eLife.72377
Bai, W. Q., Xiao, Y. H., Zhao, J., Song, S. Q., Hu, L., Zeng, J. Y., et al. (2014). Gibberellin overproduction promotes sucrose synthase expression and secondary cell wall deposition in cotton fibers. PLoS One 9:e96537. doi: 10.1371/journal.pone.0096537
Balkunde, R., Pesch, M., and Hülskamp, M. (2010). Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr. Top Dev. Biol. 91, 299–321. doi: 10.1016/S0070-2153(10)91010-7
Banyar, A., Gruber, M. Y., Lisa, A., Khaled, O., Annick, B., and Abdelali, H. (2015). MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 13, 779–790. doi: 10.1111/pbi.12308
Bensussan, M., Lefebvre, V., Ducamp, A., Trouverie, J., Gineau, E., Fortabat, M. N., et al. (2015). Suppression of Dwarf and irregular xylem Phenotypes Generates Low-Acetylated Biomass Lines in Arabidopsis. Plant Physiol. 168, 452–463. doi: 10.1104/pp.15.00122
Bergau, N., Bennewitz, S., Syrowatka, F., Hause, G., and Tissier, A. (2015). The development of type VI glandular trichomes in the cultivated tomato Solanum lycopersicum and a related wild species S. habrochaites. BMC Plant Biol. 15:289. doi: 10.1186/s12870-015-0678-z
Bonar, N., Liney, M., Zhang, R., Austin, C., Dessoly, J., Davidson, D., et al. (2018). Potato miR828 Is Associated With Purple Tuber Skin and Flesh Color. Front. Plant Sci. 9:1742. doi: 10.3389/fpls.2018.01742
Breuer, C., Kawamura, A., Ichikawa, T., Tominaga-Wada, R., Wada, T., Kondou, Y., et al. (2009). The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell 21, 2307–2322. doi: 10.1105/tpc.109.068387
Brininstool, G., Kasili, R., Simmons, L. A., Kirik, V., Hülskamp, M., and Larkin, J. C. (2008). Constitutive Expressor Of Pathogenesis-related Genes5 affects cell wall biogenesis and trichome development. BMC Plant Biol 8:58. doi: 10.1186/1471-2229-8-58
Bryant, L., Patole, C., and Cramer, R. (2016). Proteomic analysis of the medicinal plant Artemisia annua : Data from leaf and trichome extracts. Data Brief 7, 325–331. doi: 10.1016/j.dib.2016.02.038
Cao, J. F., Zhao, B., Huang, C. C., Chen, Z. W., Zhao, T., Liu, H. R., et al. (2020). The miR319-Targeted GhTCP4 promotes the transition from cell elongation to wall thickening in cotton fiber. Mol. Plant 13, 1063–1077. doi: 10.1016/j.molp.2020.05.006
Chalvin, C., Drevensek, S., Dron, M., Bendahmane, A., and Boualem, A. (2020). Genetic control of glandular trichome development. Trends Plant Sci. 25, 477–487. doi: 10.1016/j.tplants.2019.12.025
Champagne, A., and Boutry, M. (2017). A comprehensive proteome map of glandular trichomes of hop (Humulus lupulus L.) female cones: identification of biosynthetic pathways of the major terpenoid-related compounds and possible transport proteins. Proteomics 17:8. doi: 10.1002/pmic.201600411
Chang, J., Yu, T., Yang, Q., Li, C., Xiong, C., Gao, S., et al. (2018). Hair, encoding a single C2H2 zinc-finger protein, regulates multicellular trichome formation in tomato. Plant J. 96, 90–102. doi: 10.1111/tpj.14018
Che, G., and Zhang, X. (2019). Molecular basis of cucumber fruit domestication. Curr. Opin. Plant Biol. 47, 38–46. doi: 10.1016/j.pbi.2018.08.006
Chen, C., Liu, M., Jiang, L., Liu, X., Zhao, J., Yan, S., et al. (2014). Transcriptome profiling reveals roles of meristem regulators and polarity genes during fruit trichome development in cucumber (Cucumis sativus L.). J. Exp. Bot. 65, 4943–4958. doi: 10.1093/jxb/eru258
Chen, L., Peng, Y., Tian, J., Wang, X., Kong, Z., Mao, T., et al. (2016b). TCS1, a Microtubule-Binding Protein. Interacts with KCBP/ZWICHEL to Regulate Trichome Cell Shape in Arabidopsis thaliana. PLoS Genet 12:e1006266. doi: 10.1371/journal.pgen.1006266
Chen, C., Yin, S., Liu, X., Liu, B., Yang, S., Xue, S., et al. (2016a). The WD-repeat protein CsTTG1 regulates fruit wart formation through interaction with the homeodomain-leucine zipper I protein Mict. Plant Physiol. 171, 1156–1168. doi: 10.1104/pp.16.00112
Chen, G., Klinkhamer, P. G. L., Escobar-Bravo, R., and Leiss, K. A. (2018). Type VI glandular trichome density and their derived volatiles are differently induced by jasmonic acid in developing and fully developed tomato leaves: implications for thrips resistance. Plant Sci. 276, 87–98. doi: 10.1016/j.plantsci.2018.08.007
Chen, Q., Wang, J., Danzeng, P., Danzeng, C., Song, S., Wang, L., et al. (2021). VvMYB114 mediated by miR828 negatively regulates trichome development of Arabidopsis. Plant Sci. 309:110936. doi: 10.1016/j.plantsci.2021.110936
Chen, T., Li, W., Hu, X., Guo, J., Liu, A., and Zhang, B. (2015). A cotton MYB transcription factor. GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 56, 917–929. doi: 10.1093/pcp/pcv019
Chen, Y., Su, D., Li, J., Ying, S., Deng, H., He, X., et al. (2020). Overexpression of bHLH95, a basic helix-loop-helix transcription factor family member, impacts trichome formation via regulating gibberellin biosynthesis in tomato. J. Exp. Bot. 71, 3450–3462. doi: 10.1093/jxb/eraa114
Chezem, W. R., and Clay, N. K. (2016). Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs. Phytochemistry 131, 26–43. doi: 10.1016/j.phytochem.2016.08.006
Churchman, M. L., Brown, M. L., Kato, N., Kirik, V., Hülskamp, M., Inzé, D., et al. (2006). SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18, 3145–3157. doi: 10.1105/tpc.106.044834
Cong, H., Xinhui, N., Chao, S., Chunyuan, Y., Wu, L., Wenxia, Z., et al. (2017). Population structure and genetic basis of the agronomic traits of upland cotton in china revealed by a genome-wide association study using high-density SNPs. Plant Biotechnol. J. 15, 1374–1386. doi: 10.1111/pbi.12722
Cui, J. Y., Miao, H., Ding, L. H., Wehner, T. C., Liu, P. N., Wang, Y., et al. (2016). A New Glabrous Gene (csgl3) Identified in Trichome Development in Cucumber (Cucumis sativus L.). PLoS One 11:e0148422. doi: 10.1371/journal.pone.0148422
Deeks, M. J., and Hussey, P. J. (2003). Arp2/3 and ‘The Shape of things to come’. Plant Biol. 6, 561–567. doi: 10.1016/j.pbi.2003.09.013
Deng, F., Tu, L., Tan, J., Li, Y., Nie, Y., and Zhang, X. (2012a). GbPDF1 is involved in cotton fiber initiation via the core CIS-Element HDZIP2ATATHB2. Plant Physiol. 158, 890–904. doi: 10.1104/pp.111.186742
Deng, W., Yang, Y., Ren, Z., Audran-Delalande, C., Mila, I., Wang, X., et al. (2012b). The tomato SlIAA15 is involved in trichome formation and axillary shoot development. New Phytol. 194, 379–390. doi: 10.1111/j.1469-8137.2012.04053.x
Deore, S. L., Ingole, S. R., and Baviskar, B. A. (2021). Comparative pharmacognostical phytochemical and biological evaluation of five ocimum species. Pharmacogn. J. 13, 463–474.
Di, W., Xiaobo, C., Zenglin, Z., Danmei, L., Gaoyuan, S., Xingchen, K., et al. (2015). A MADS-box gene NtSVP regulates pedicel elongation by directly suppressing a KNAT1-like KNOX gene NtBPL in tobacco (Nicotiana tabacum L.). J. Exp. Bot. 66, 6233–6244. doi: 10.1093/jxb/erv332
Dipanwita, B., El-Din El-Assal, S., Le, J., Mallery, E. L., and Szymanski, D. B. (2004). Interchangeable functions of Arabidopsis PIROGI and the human WAVE complex subunit SRA1 during leaf epidermal development. Development 131, 4345–4355. doi: 10.1242/dev.01307
Dong, M., Xue, S., Bartholomew, E. S., Zhai, X., Sun, L., Xu, S., et al. (2022). Transcriptomic and functional analysis provides molecular insights into multicellular trichome development. Plant Physiol. 16:kiac050. doi: 10.1093/plphys/kiac050
Du, H., Wang, G., Pan, J., Chen, Y., Xiao, T. T., Zhang, L. Y., et al. (2020). The HD-ZIP IV transcription factor Tril regulates fruit spine density through gene dosage effects in cucumber. J. Exp. Bot. 71, 6297–6310. doi: 10.1093/jxb/eraa344
Du, X., Huang, G., He, S., Yang, Z., Sun, G., Ma, X., et al. (2018). Resequencing of 243 diploid cotton accessions based on an updated a genome identifies the genetic basis of key agronomic traits. Nat. Genet. 50, 796–802. doi: 10.1038/s41588-018-0116-x
El Refy, A., Perazza, D., Zekraoui, L., Valay, J. G., Bechtold, N., Brown, S., et al. (2003). The Arabidopsis KAKTUS gene encodes a HECT protein and controls the number of endoreduplication cycles. Mol. Genet. Genom. 270, 403–414. doi: 10.1007/s00438-003-0932-1
Ewas, M., Gao, Y., Ali, F., Nishawy, E. M., Shahzad, R., Subthain, H., et al. (2017). RNA-seq reveals mechanisms of SlMX1 for enhanced carotenoids and terpenoids accumulation along with stress resistance in tomato. Sci. Bull. 62, 476–485.
Fan, D., Ran, L., Hu, J., Ye, X., Xu, D., Li, J., et al. (2020). miR319a/TCP module and DELLA protein regulate trichome initiation synergistically and improve insect defenses in Populus tomentosa. New Phytol. 227, 867–883. doi: 10.1111/nph.16585
Fei, S., Wenbin, M., Hao, W., Furong, X., Chunyan, X., and Jianfei, W. (2020). New allele of HL6 regulates trichome elongation in rice. Rice Sci. 27, 480–492.
Feng, Z., Bartholomew, E. S., Liu, Z., Cui, Y., Dong, Y., Li, S., et al. (2021b). Glandular trichomes: new focus on horticultural crops. Hortic. Res. 8:58. doi: 10.1038/s41438-021-00592-1
Feng, G., Han, J., Yang, Z., Liu, Q., Shuai, Y., Xu, X., et al. (2021a). Genome-wide identification, phylogenetic analysis, and expression analysis of the SPL gene family in orchardgrass (Dactylis glomerata L.). Genomics 113, 2413–2425. doi: 10.1016/j.ygeno.2021.05.032
Feng, H., Guo, L., Wang, G., Sun, J., Pan, Z., He, S., et al. (2017). The negative correlation between fiber color and quality traits revealed by QTL analysis. PLoS One 10:e0129490. doi: 10.1371/journal.pone.0129490
Feng, H., Li, X., Chen, H., Deng, J., Zhang, C., Liu, J., et al. (2018). GhHUB2, a ubiquitin ligase, is involved in cotton fiber development via the ubiquitin–26S proteasome pathway. J. Exp. Bot. 69, 5059–5075. doi: 10.1093/jxb/ery269
Feng, L.-G., Luan, X.-F., Wang, J., Xia, W., Wang, M., and Sheng, L.-X. (2015). Cloning and expression analysis of transcription factor RrTTG1 related to prickle development in rose (Rosa Rugosa). Arch. Biol. Sci. 67, 1219–1225.
Fiehn, O., Kopka, J., Dörmann, P., Altmann, T., Trethewey, R. N., and Willmitzer, L. (2000). Metabolite profiling for plant functional genomics. Nat. Biotechnol.: Sci. Bus. Biotechnol. 18, 1157–1161. doi: 10.1038/81137
Frank, M. J., and Smith, L. G. (2002). A small. novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr. Biol. 12, 849–853. doi: 10.1016/s0960-9822(02)00819-9
Fuyu, H., Jianhao, C., Yunru, F., Youcheng, L., Songguang, Y., and Keqiang, W. (2020). Arabidopsis JMJ29 is involved in trichome development by regulating the core trichome initiation gene GLABRA3. Plant J. 103, 1735–1743. doi: 10.1111/tpj.14858
Galdon-Armero, J., Arce-Rodriguez, L., Downie, M., Li, J., and Martin, C. (2020). A scanning electron micrograph-based resource for identification of loci involved in epidermal development in tomato: elucidation of a new function for the mixta-like transcription factor in leaves. Plant Cell 32, 1414–1433. doi: 10.1105/tpc.20.00127
Gan, Y., Liu, C., Yu, H., and Broun, P. (2007). Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134, 2073–2081. doi: 10.1242/dev.005017
Gao, S., Gao, Y., Xiong, C., Yu, G., Chang, J., Yang, Q., et al. (2017). The tomato B-type cyclin gene, SlCycB2, plays key roles in reproductive organ development, trichome initiation, terpenoids biosynthesis and Prodenia litura defense. Plant Sci. 262, 103–114. doi: 10.1016/j.plantsci.2017.05.006
Gao, Y., Gong, X., Cao, W., Zhao, J., Fu, L., Wang, X., et al. (2008). SAD2 in Arabidopsis functions in trichome initiation through mediating GL3 function and regulating GL1 TTG1 and GL2 expression. J. Integr. Plant Biol. 50, 906–917. doi: 10.1111/j.1744-7909.2008.00695.x
Gao, Y. N., Gao, S. H., Xiong, C., Yu, G., Chang, J., Ye, Z. B., et al. (2015). Comprehensive analysis and expression profile of the homeodomain leucine zipper IV transcription factor family in tomato. Plant Physiol. Biochem. 96, 141–153. doi: 10.1016/j.plaphy.2015.07.025
Giuliani, C., Giovanetti, M., Lupi, D., Mesiano, M. P., Barilli, R., Ascrizzi, R., et al. (2020). Tools to Tie: flower characteristics, VOC emission profile, and glandular trichomes of two mexican salvia species to attract bees. Plants (Basel) 9:1645. doi: 10.3390/plants9121645
Glas, J. J., Schimmel, B. C. J., Alba, J. M., Escobar-Bravo, R., Schuurink, R. C., and Kant, M. R. (2012). Plant glandular trichomes as targets for breeding or engineering of resistance to herbivores. Int. J. Mol. Sci. 13, 17077–17103. doi: 10.3390/ijms131217077
Glover, B. J., Bunnewell, S., and Martin, C. (2004). Convergent evolution within the genus Solanum: the specialised anther cone develops through alternative pathways. Gene 331, 1–7. doi: 10.1016/j.gene.2004.01.027
Grebe, M. (2012). The patterning of epidermal hairs in Arabidopsis - updated. Cur. Opin. Plant Biol. 15, 31–37. doi: 10.1016/j.pbi.2011.10.010
Guan, X., Song, Q., and Chen, Z. J. (2014b). Polyploidy and small RNA regulation of cotton fiber development. Trends Plant Sci. 19, 516–528. doi: 10.1016/j.tplants.2014.04.007
Guan, X., Pang, M., Nah, G., Shi, X., Ye, W., Stelly, D. M., et al. (2014a). miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat. Commun. 5:3050. doi: 10.1038/ncomms4050
Guan, X., Yu, N., Shangguan, X., Wang, S., Lu, S., Wang, L., et al. (2007). Arabidopsis trichome research sheds light on cotton fiber development mechanisms. Chin. Sci. Bull. 52, 1734–1741.
Guan, X. Y., Li, Q. J., Shan, C. M., Wang, S., Mao, Y. B., Wang, L. J., et al. (2008). The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiol. Plant 134, 174–182. doi: 10.1111/j.1399-3054.2008.01115.x
Guo, P., Chang, H., Li, Q., Wang, L., Ren, Z., Ren, H., et al. (2020). Transcriptome profiling reveals genes involved in spine development during CsTTG1-regulated pathway in cucumber (Cucumis sativus L.). Plant Sci. 291:110354. doi: 10.1016/j.plantsci.2019.110354
Han, G., Li, Y., Qiao, Z., Wang, C., Zhao, Y., Guo, J., et al. (2021). Advances in the regulation of epidermal cell development by C2H2 Zinc Finger Proteins in Plants. Front Plant Sci. 12:754512. doi: 10.3389/fpls.2021.754512
Han, G., Lu, C., Guo, J., Qiao, Z., Sui, N., Qiu, N., et al. (2020). C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front. Plant Sci. 11:115. doi: 10.3389/fpls.2020.00115
Han, L., Hong, Z. L., Jiaojiao, J., Shaobao, L., Zhanming, Z., Jian, L. T., et al. (2016). Gradient mechanical properties facilitate Arabidopsis trichome as mechanosensor. ACS Appl. Mater Interfaces 8, 9755–9761. doi: 10.1021/acsami.6b02253
Hao, F., Xin, L., Hong, C., Jie, D., Chaojun, Z., Ji, L., et al. (2018). GhHUB2, a ubiquitin ligase, is involved in cotton fiber development via the ubiquitin-26S proteasome pathway. J. Exp. Bot. 69, 5059–5075. doi: 10.1093/jxb/ery269
Hao, J., Tu, L., Hu, H., Tan, J., Deng, F., Tang, W., et al. (2012). GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 63, 6267–6281. doi: 10.1093/jxb/ers278
Hashida, Y., Takechi, K., Abiru, T., Yabe, N., Nagase, H., Hattori, K., et al. (2020). Two ANGUSTIFOLIA genes regulate gametophore and sporophyte development in Physcomitrella patens. Plant J. 101, 1318–1330. doi: 10.1111/tpj.14592
He, P., Zhang, Y., Li, H., Fu, X., Shang, H., Zou, C., et al. (2021). GhARF16-1 modulates leaf development by transcriptionally regulating theGhKNOX2-1gene in cotton. Plant Biotechnol. J. 19, 548–562. doi: 10.1111/pbi.13484
He, X.-C., Qin, Y.-M., Xu, Y., Hu, C.-Y., and Zhu, Y.-X. (2008). Molecular cloning, expression profiling, and yeast complementation of 19 beta-tubulin cDNAs from developing cotton ovules. J. Exp. Bot. 59, 2687–2695. doi: 10.1093/jxb/ern127
He, Y. L., Fu, X. Q., Li, L., Sun, X. F., Tang, K. X., and Zhao, J. Y. (2022). AaSPL9 affects glandular trichomes initiation by positively regulating expression of AaHD1 in Artemisia annua L. Plant Sci. 317:111172. doi: 10.1016/j.plantsci.2021.111172
Henriksson, E., Olsson, A. S., Johannesson, H., Johansson, H., Hanson, J., Engström, P., et al. (2005). Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 139, 509–518. doi: 10.1104/pp.105.063461
Hu, H., Wang, M., Ding, Y., Zhu, S., Zhao, G., Tu, L., et al. (2018). Transcriptomic repertoires depict the initiation of lint and fuzz fibres in cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 16, 1002–1012. doi: 10.1111/pbi.12844
Hu, W., Chen, L., Qiu, X., Wei, J., Lu, H., Sun, G., et al. (2020). AKR2A participates in the regulation of cotton fibre development by modulating biosynthesis of very-long-chain fatty acids. Plant Biotechnol. J. 18, 526–539. doi: 10.1111/pbi.13221
Hua, B., Chang, J., Wu, M., Xu, Z., Zhang, F., Yang, M., et al. (2021a). Mediation of JA signalling in glandular trichomes by thewoolly/SlMYC1regulatory module improves pest resistance in tomato. Plant Biotechnol. J. 19, 375–393. doi: 10.1111/pbi.13473
Hua, B., Chang, J., Xu, Z., Han, X., Xu, M., Yang, M., et al. (2021b). HOMEODOMAIN PROTEIN8 mediates jasmonate-triggered trichome elongation in tomato. New Phytol. 230, 1063–1077. doi: 10.1111/nph.17216
Huang, J., Chen, F., Guo, Y., Gan, X., Yang, M., Zeng, W., et al. (2021b). GhMYB7 promotes secondary wall cellulose deposition in cotton fibres by regulating GhCesA gene expression through three distinct cis-elements. New Phytol. 232, 1718–1737. doi: 10.1111/nph.17612
Huang, G., Huang, J.-Q., Chen, X.-Y., and Zhu, Y.-X. (2021a). Recent advances and future perspectives in cotton research. Annu. Rev. Plant. Biol. 72, 437–462. doi: 10.1146/annurev-arplant-080720-113241
Huang, J., Chen, F., Wu, S., Li, J., and Xu, W. (2016). Cotton GhMYB7 is predominantly expressed in developing fibers and regulates secondary cell wall biosynthesis in transgenic Arabidopsis. Sci. China Life Sci. 59, 194–205. doi: 10.1007/s11427-015-4991-4
Huang, J., Guo, Y., Sun, Q., Zeng, W., Li, J., Li, X., et al. (2019a). Genome-wide identification of R2R3-MYB transcription factors regulating secondary cell wall thickening in cotton fiber development. Plant Cell Physiol. 60, 687–701. doi: 10.1093/pcp/pcy238
Huang, X., Yan, H., Zhai, L., and Yi, Y. (2019b). GLABROUS1 from Rosa roxburghii Tratt regulates trichome formation by interacting with the GL3/EGL3 protein. Gene 692, 60–67. doi: 10.1016/j.gene.2018.12.071
Hülskamp, M. (2004). Plant trichomes: a model for cell differentiation. Nat. Rev. Mol. Cell. Biol 5, 471–480. doi: 10.1038/nrm1404
Hülskamp, M., Misŕa, S., and Jürgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. doi: 10.1016/0092-8674(94)90118-x
Humphries, J. A., Walker, A. R., Timmis, J. N., and Orford, S. J. (2005). Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol. Biol. 57, 67–81. doi: 10.1007/s11103-004-6768-1
Hung, F. Y., Chen, J. H., Feng, Y. R., Lai, Y. C., Yang, S. G., and Wu, K. Q. (2020). Arabidopsis JMJ29 is involved in trichome development by regulating the core trichome initiation geneGLABRA3. Plant J. 103, 1735–1743.
Hussey, P. J., Ketelaar, T., and Deeks, M. J. (2006). Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57, 109–125. doi: 10.1146/annurev.arplant.57.032905.105206
Ilgenfritz, H., Bouyer, D., Schnittger, A., Mathur, J., Kirik, V., Schwab, B., et al. (2003). The Arabidopsis STICHEL gene is a regulator of trichome branch number and encodes a novel protein. Plant Physiol. 131, 643–655. doi: 10.1104/pp.014209
Ishida, T., Hattori, S., Sano, R., Inoue, K., Shirano, Y., Hayashi, H., et al. (2007). Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19, 2531–2543. doi: 10.1105/tpc.107.052274
Jacobsen, S. E., and Olszewski, N. E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5, 887–896. doi: 10.1105/tpc.5.8.887
Jakoby, M. J., Falkenhan, D., Mader, M. T., Brininstool, G., Wischnitzki, E., Platz, N., et al. (2008). Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol. 148, 1583–1602. doi: 10.1104/pp.108.126979
Jiang, S., Wei, J., Li, N., Wang, Z., Zhang, Y., Xu, R., et al. (2022). The UBP14-CDKB1; 1-CDKG2 cascade controls endoreduplication and cell growth in Arabidopsis. Plant Cell 6:koac002. doi: 10.1093/plcell/koac002
Jiang, Y., Guo, W., Zhu, H., Ruan, Y. L., and Zhang, T. (2012). Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality. Plant Biotechnol. J. 10, 301–312. doi: 10.1111/j.1467-7652.2011.00662.x
Jie, Z., Geng-Qing, H., Dan, Z., Jing-Qiu, Y., Yang, L., Shan, H., et al. (2018). The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 217, 625–640.
Johnson, C. S., Kolevski, B., and Smyth, D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. doi: 10.1105/tpc.001404
Jose, D.-V., Caldu-Primo, J. L., Martinez-Garcia, J. C., and Alvarez-Buylla, E. R. (2018). Modeling the epigenetic landscape in plant development. Methods Mol. Biol. 1819, 357–383. doi: 10.1007/978-1-4939-8618-7_17
Kai, G., Lili, T., Yonghui, H., Jinwu, D., Maojun, W., Hui, H., et al. (2017). Interaction between calcium and potassium modulates elongation rate in cotton fiber cells. J. Exp. Bot. 68, 5161–5175. doi: 10.1093/jxb/erx346
Karabourniotis, G., Liakopoulos, G., Nikolopoulos, D., and Bresta, P. (2020). Protective and defensive roles of non-glandular trichomes against multiple stresses: structure–function coordination. J. For. Res. 31, 1–12.
Khan, R. A., Hurrah, I. M., Muzafar, S., Jan, S., and Abbas, N. (2021). Transcriptional regulation of trichome development in plants: an overview. Proc. Indian Natl,. Sci. Acad. U.S.A. 87, 36–47.
Kim, G. T., Shoda, K., Tsuge, T., Cho, K. H., Uchimiya, H., Yokoyama, R., et al. (2002). The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J. 21, 1267–1279. doi: 10.1093/emboj/21.6.1267
Kim, H. J., Hinchliffe, D. J., Triplett, B. A., Chen, Z. J., Stelly, D. M., Yeater, K. M., et al. (2017). Phytohormonal networks promote differentiation of fiber initials on pre-anthesis cotton ovules grown in vitro and in planta. PLoS One 10:e0125046. doi: 10.1371/journal.pone.0125046
Kim, H. J., Murai, N., Fang, D. D., and Triplett, B. A. (2011). Functional analysis of Gossypium hirsutum cellulose synthase catalytic subunit 4 promoter in transgenic Arabidopsis and cotton tissues. Plant Sci. 180, 323–332. doi: 10.1016/j.plantsci.2010.10.003
Kim, S. Y., Hyoung, S., So, W. M., and Shin, J. S. (2018). The novel transcription factor TRP interacts with ZFP5, a trichome initiation-related transcription factor, and negatively regulates trichome initiation through gibberellic acid signaling. Plant Mol. Biol. 96, 315–326. doi: 10.1007/s11103-018-0697-x
Kirik, V., Bouyer, D., Schöbinger, U., Bechtold, N., Herzog, M., Bonneville, J. M., et al. (2001). CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr. Biol. 11, 1891–1895. doi: 10.1016/s0960-9822(01)00590-5
Kirik, V., Simon, M., Huelskamp, M., and Schiefelbein, J. (2004). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268, 506–513.
Konarska, A., and Łotocka, B. (2020). Glandular trichomes of Robinia viscosa Vent. var. hartwigii (Koehne) Ashe (Faboideae, Fabaceae)-morphology, histochemistry and ultrastructure. Planta 252:102. doi: 10.1007/s00425-020-03513-z
Kong, D., Pan, X., Jing, Y., Zhao, Y., Duan, Y., Yang, J., et al. (2021). ZmSPL10/14/26 are required for epidermal hair cell fate specification on maize leaf. New Phytol. 230, 1533–1549. doi: 10.1111/nph.17293
Koudounas, K., Manioudaki, M. E., Kourti, A., Banilas, G., and Hatzopoulos, P. (2015). Transcriptional profiling unravels potential metabolic activities of the olive leaf non-glandular trichome. Front. Plant Sci. 6:633. doi: 10.3389/fpls.2015.00633
Larkin, J. C., Oppenheimer, D. G., Lloyd, A. M., Paparozzi, E. T., and Marks, M. D. (1994). Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA Genes in Arabidopsis trichome development. Plant Cell 6, 1065–1076. doi: 10.1105/tpc.6.8.1065
Larkin, J. C., Walker, J. D., Bolognesi-Winfield, A. C., Gray, J. C., and Walker, A. R. (1999). Allele-specific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics 151, 1591–1604. doi: 10.1093/genetics/151.4.1591
Li, B., Li, D.-D., Zhang, J., Xia, H., Wang, X.-L., Li, Y., et al. (2013). Cotton AnnGh3 encoding an annexin protein is preferentially expressed in fibers and promotes initiation and elongation of leaf trichomes in transgenic Arabidopsis. J. Integr. Plant Biol. 55, 902–916. doi: 10.1111/jipb.12063
Li, H. B., Qin, Y. M., Pang, Y., Song, W. Q., Mei, W. Q., and Zhu, Y. X. (2007). A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytol. 175, 462–471. doi: 10.1111/j.1469-8137.2007.02120.x
Li, J., Wang, X., Jiang, R., Dong, B., Fang, S., Li, Q., et al. (2021b). Phytohormone-Based Regulation of Trichome Development. Front. Plant Sci. 12:734776. doi: 10.3389/fpls.2021.734776
Li, X., Kong, X., Zhou, J., Luo, Z., Lu, H., Li, W., et al. (2021c). Seeding depth and seeding rate regulate apical hook formation by inducing GhHLS1 expression via ethylene during cotton emergence. Plant Physiol. Biochem. 164, 92–100. doi: 10.1016/j.plaphy.2021.04.030
Li, J., Tang, B., Li, Y., Li, C., Guo, M., Chen, H., et al. (2021a). Rice SPL10 positively regulates trichome development through expression of HL6 and auxin-related genes. J. Integr. Plant Biol. 63, 1521–1536. doi: 10.1111/jipb.13140
Li, Z. Y., Li, B., and Dong, A. W. (2012b). The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol. Plant 5, 270–280. doi: 10.1093/mp/ssr086
Li, J., Yuan, Y., Lu, Z., Yang, L., Gao, R., Lu, J., et al. (2012a). Glabrous Rice 1, encoding a homeodomain protein, regulates trichome development in rice. Rice (N Y) 5:32. doi: 10.1186/1939-8433-5-32
Li, L., Zhao, J., Zhao, Y., Lu, X., Zhou, Z., Zhao, C., et al. (2016). Comprehensive investigation of tobacco leaves during natural early senescence via multi-platform metabolomics analyses. Sci. Rep. 6:37976. doi: 10.1038/srep37976
Li, Q., Cao, C. X., Zhang, C. J., Zheng, S. S., Wang, Z. H., Wang, L. N., et al. (2015). The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of trichomes uncovers a novel function for the homeodomain-leucine zipper I gene. J. Exp. Bot. 66, 2515–2526. doi: 10.1093/jxb/erv046
Li, S. F., Milliken, O. N., Pham, H., Seyit, R., Napoli, R., Preston, J., et al. (2009). The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome Morphogenesis. Plant Cell 21, 72–89. doi: 10.1105/tpc.108.063503
Li, Y. J., Liu, D. Q., Tu, L. L., Zhang, X. L., Wang, L., Zhu, L. F., et al. (2010b). Suppression of GhAGP4 gene expression repressed the initiation and elongation of cotton fiber. Plant Cell Rep. 29, 193–202. doi: 10.1007/s00299-009-0812-1
Li, W., Wu, J., Weng, S., Zhang, D., Zhang, Y., and Shi, C. (2010a). Characterization and fine mapping of the glabrous leaf and hull mutants (gl1) in rice (Oryza sativa L.). Plant Cell Rep. 29, 617–627. doi: 10.1007/s00299-010-0848-2
Li, X. B., Cai, L., Cheng, N. H., and Liu, J. W. (2002). Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber. Plant Physiol. 130, 666–674. doi: 10.1104/pp.005538
Li, Y., Ning, H., Zhang, Z., Wu, Y., Jiang, J., Su, S., et al. (2011). A cotton gene encoding novel MADS-box protein is preferentially expressed in fibers and functions in cell elongation. Acta. Biochim. Biophys. Sin. 43:607. doi: 10.1093/abbs/gmr055
Li, Y. L., Sun, J., Li, C. H., Zhu, Y. Q., and Xia, G. X. (2003). Specific expression of a beta-tubulin gene (GhTub1) in developing cotton fibers. Sci. China Ser. C Life Sci. 46, 235–242. doi: 10.1360/03yc9025
Liao, X., Wang, L., Zhu, S., Zheng, F., and Yang, C. (2021). Identification, genomic organization, and expression profiles of single C2H2 zinc finger transcription factors in tomato (Solanum lycopersicum). J. Appl. Genet. 62, 1–15. doi: 10.1007/s13353-020-00587-z
Liu, B. L., Zhu, Y. C., and Zhang, T. Z. (2015). The R3-MYB Gene GhCPC negatively regulates cotton fiber elongation. PLoS One 10:e0116272. doi: 10.1371/journal.pone.0116272
Liu, D., Tu, L., Li, Y., Wang, L., Zhu, L., and Zhang, X. (2008). Genes encoding fasciclin-like arabinogalactan proteins are specifically expressed during cotton fiber development. Plant Mol. Biol. Rep. 26, 98–113.
Liu, N., Wu, S., Li, Z., Khan, A. Q., Hu, H., Zhang, X., et al. (2020a). Repression of microRNA 160 results in retarded seed integument growth and smaller final seed size in cotton. Crop J. 8, 602–612.
Liu, Z. H., Chen, Y., Wang, N. N., Chen, Y. H., Wei, N., Lu, R., et al. (2020b). A basic helix–loop–helix protein (GhFP1) promotes fibre elongation of cotton (Gossypium hirsutum) by modulating brassinosteroid biosynthesis and signalling. New Phytol. 225, 2439–2452. doi: 10.1111/nph.16301
Liu, R., Wang, Y., Tang, S., Cai, J., Liu, S., Zheng, P., et al. (2021). Genome-Wide Identification of Tea Plant bHLH transcription factor family and discovery of candidate regulator for trichome formation. Sci. Rep. 11:10764. doi: 10.1038/s41598-021-90205-7
Liu, X., Bartholomew, E., Cai, Y., and Ren, H. (2016). Trichome-related mutants provide a new perspective on multicellular trichome initiation and development in cucumber (Cucumis sativus L). Front. Plant Sci. 7:1187. doi: 10.3389/fpls.2016.01187
Liu, Y., Liu, D., Hu, R., Hua, C., Ali, I., Zhang, A., et al. (2017). AtGIS, a C2H2 zinc-finger transcription factor from Arabidopsis regulates glandular trichome development through GA signaling in tobacco. Biochem. Biophys. Res. Commun. 483, 209–215. doi: 10.1016/j.bbrc.2016.12.164
Liu, Y., Liu, D., Khan, A. R., Liu, B., Wu, M., Huang, L., et al. (2018a). NbGIS regulates glandular trichome initiation through GA signaling in tobacco. Plant Mol. Biol. 98, 153–167. doi: 10.1007/s11103-018-0772-3
Liu, Y. H., Liu, D. D., Khan, A. R., Liu, B. H., Wu, M. J., Huang, L. L., et al. (2018b). NbGIS regulates glandular trichome initiation through GA signaling in tobacco. Plant Mol. Biol. 98, 153–167.
Long, Q., Yue, F., Liu, R. C., Song, S. Q., Li, X. B., Ding, B., et al. (2018). The phosphatidylinositol synthase gene (GhPIS) contributes to longer, stronger, and finer fibers in cotton. Mol. Genet. Genom. 293, 1139–1149. doi: 10.1007/s00438-018-1445-2
Luo, M., Xiao, Y., Li, X., Lu, X., Deng, W., Li, D., et al. (2007). GhDET2, a steroid 5α-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J. 51, 419–430. doi: 10.1111/j.1365-313X.2007.03144.x
Lv, Z., Li, J., Qiu, S., Qi, F., Su, H., Bu, Q., et al. (2022). The transcription factors TLR1 and TLR2 negatively regulate trichome density and artemisinin levels in Artemisia annua. J. Integr. Plant Biol. doi: 10.1111/jipb.13258
Ma, J., Pei, W., Ma, Q., Geng, Y., Liu, G., Liu, J., et al. (2019). QTL analysis and candidate gene identification for plant height in cotton based on an interspecific backcross inbred line population of Gossypium hirsutum x Gossypium barbadense. Theor. Appl. Genet. 132, 2663–2676. doi: 10.1007/s00122-019-03380-7
Maes, L., Nieuwerburgh, F. C. W. V., Zhang, Y., Reed, D. W., Pollier, J., Casteele, S. R. F. V., et al. (2011). Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytol. 189, 176–189. doi: 10.1111/j.1469-8137.2010.03466.x
Markus, G. (2012). The patterning of epidermal hairs in Arabidopsis–updated. Curr. opin. Plant Biol. 15, 31–37.
Martin, C., Bhatt, K., Baumann, K., Jin, H., Zachgo, S., Roberts, K., et al. (2002). The mechanics of cell fate determination in petals. Philos. Trans. R. Soc. Lond B Biol. Sci. 357, 809–813. doi: 10.1098/rstb.2002.1089
Matías-Hernández, L., Jiang, W., Yang, K., Tang, K., Brodelius, P. E., and Pelaz, S. (2017). Aa MYB 1 and its orthologue At MYB 61 affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana. Plant J. 90, 520–534.
Mauricio, R. (1998). Costs of resistance to natural enemies in field populations of the annual plant Arabidopsis thaliana. Am. Nat. 151, 20–28. doi: 10.1086/286099
Mirnezami, S. V., Young, T., Assefa, T., Prichard, S., Nagasubramanian, K., Sandhu, K., et al. (2020). Automated trichome counting in soybean using advanced image-processing techniques. Appl. Plant Sci. 8:e11375. doi: 10.1002/aps3.11375
Mishra, A., Gupta, P., Lal, R. K., and Dhawan, S. S. (2021). Assessing and integrating the transcriptome analysis with plant development, trichomes, and secondary metabolites yield potential in Mentha arvensis L. Plant Physiol. Biochem. 162, 517–530. doi: 10.1016/j.plaphy.2021.03.009
Montgomery, T. A., Howell, M. D., Cuperus, J. T., Li, D., Hansen, J. E., Alexander, A. L., et al. (2008). Specificity of ARGONAUTE7-miR390 Interaction and Dual Functionality in TAS3 Trans -Acting siRNA Formation. Cell 133, 128–141. doi: 10.1016/j.cell.2008.02.033
Nadakuduti, S. S., Pollard, M., Kosma, D. K., Allen, C. Jr., Ohlrogge, J. B., and Barry, C. S. (2012). Pleiotropic phenotypes of the sticky peel mutant provide new insight into the role of cutin deficient2 in epidermal cell function in tomato. Plant Physiol. 159, 945–960. doi: 10.1104/pp.112.198374
Ning, P., Wang, J., Zhou, Y., Gao, L., Wang, J., and Gong, C. (2016). Adaptional evolution of trichome in Caragana korshinskii to natural drought stress on the Loess Plateau, China. Ecol. Evol. 6, 3786–3795. doi: 10.1002/ece3.2157
Oppenheimer, D. G., Herman, P. L., Sivakumaran, S., Esch, J., and Marks, M. D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67, 483–493. doi: 10.1016/0092-8674(91)90523-2
Pan, J., Zhang, L., Chen, G., Wen, H., Chen, Y., Du, H., et al. (2021). Study of micro-trichome (mict) reveals novel connections between transcriptional regulation of multicellular trichome development and specific metabolism in cucumber. Hortic. Res. 8:21. doi: 10.1038/s41438-020-00456-0
Pan, Y. P., Bo, K. L., Cheng, Z. H., and Weng, Y. Q. (2015). The loss-of-function GLABROUS 3 mutation in cucumber is due to LTR-retrotransposon insertion in a class IV HD-ZIP transcription factor gene CsGL3 that is epistatic over CsGL1. BMC Plant Biol. 15:302. doi: 10.1186/s12870-015-0693-0
Papalazarou, V., and Machesky, L. M. (2021). The cell pushes back: the Arp2/3 complex is a key orchestrator of cellular responses to environmental forces. Curr. Opin. Cell Biol. 68, 37–44. doi: 10.1016/j.ceb.2020.08.012
Payne, C. T., Zhang, F., and Lloyd, A. M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362. doi: 10.1093/genetics/156.3.1349
Payne, T., Clement, J., Arnold, D., and Lloyd, A. (1999). Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum. Development 126, 671–682. doi: 10.1242/dev.126.4.671
Peng, S., Sun, K., Guo, Y., Liu, Y., and Wang, S. (2020). Arabidopsis nucleoporin CPR5 controls trichome cell death through the core cell cycle regulator CKI. Plant Biol. (Stuttg) 22, 337–345. doi: 10.1111/plb.13068
Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L., and Poethig, R. S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Cold Spring Harb. Lab. Press 18, 2368–2379. doi: 10.1101/gad.1231804
Perez-Rodriguez, M., Jaffe, F. W., Butelli, E., Glover, B. J., and Martin, C. (2005). Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development 132, 359–370. doi: 10.1242/dev.01584
Pesch, M., and Hülskamp, M. (2009). One, two, three.models for trichome patterning in Arabidopsis? Curr. Opin. Plant Biol. 12, 587–592. doi: 10.1016/j.pbi.2009.07.015
Pesch, M., Schultheiß, I., Digiuni, S., Uhrig, J. F., and Hülskamp, M. (2013). Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development 140, 3456–3467. doi: 10.1242/dev.094698
Prakash, P., Srivastava, R., Prasad, P., Tiwari, V. K., Kumar, A., Pandey, S., et al. (2020). Trajectories of cotton fiber initiation: a regulatory perspective. [Preprints] doi: 10.20944/preprints202011.0060.v1
Qi, T., Song, S., Ren, Q., Wu, D., Huang, H., Chen, Y., et al. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814. doi: 10.1105/tpc.111.083261
Qiu, J.-L., Jilk, R., Marks, M. D., and Szymanski, D. B. (2002). The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14, 101–118. doi: 10.1105/tpc.010346
Ram, B. D., Qing, W., Wolfgang, F., Bernhard, S., Sven, G., and Andreas, R. (2015). High resolution mass spectrometry imaging of plant tissues: towards a plant metabolite atlas. Analyst 140, 7696–7696. doi: 10.1039/c5an01065a
Reddy, V. S., Day, I. S., Thomas, T., and Reddy, A. S. N. (2004). KIC, a novel Ca2+ binding protein with one EF-hand motif, interacts with a microtubule motor protein and regulates trichome morphogenesis. Plant Cell 16, 185–200. doi: 10.1105/tpc.016600
Remmy, K., Walker, J. D., Simmons, L. A., Zhou, J., De Veylder, L., and Larkin, J. C. (2010). SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics 185, 257–268. doi: 10.1534/genetics.109.113274
Rerie, W. G., Feldmann, K. A., and Marks, M. D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8, 1388–1399. doi: 10.1101/gad.8.12.1388
Riddick, E. W., and Simmons, A. M. (2014). Plant trichomes have mixed impacts on predatory insects. Pest Manage. Sci. 70, 1668–1668. doi: 10.1002/ps.3811
Rodziewicz, P., Loroch, S., Marczak, Ł, Sickmann, A., and Kayser, O. (2019). Cannabinoid synthases and osmoprotective metabolites accumulate in the exudates of Cannabis sativa L. glandular trichomes. Plant Sci. 284, 108–116. doi: 10.1016/j.plantsci.2019.04.008
Saltveit, M. E., and Hepler, P. K. (2004). Effect of heat shock on the chilling sensitivity of trichomes and petioles of African violet (Saintpaulia ionantha). Physiol. Plant 121, 35–43. doi: 10.1111/j.0031-9317.2004.00288.x
Samuels, A., Glass, A., Ehret, D., and Menzies, J. (1993). The effects of silicon supplementation on cucumber fruit: changes in surface characteristics. Ann. Bot. 72, 433–440.
Schellmann, S., and Hülskamp, M. (2005). Epidermal differentiation: trichomes in Arabidopsis as a model system. Int. J. Dev. Biol. 49, 579–584. doi: 10.1387/ijdb.051983ss
Schnittger, A., Schöbinger, U., Stierhof, Y.-D., and Hülskamp, M. (2005). Ectopic B-Type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis Trichomes. Curr. Biol. 15:980. doi: 10.1016/s0960-9822(02)00693-0
Schnittger, A., Weinl, C., Bouyer, D., Schobinger, U., and Hulskamp, M. (2003). Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15, 303–315. doi: 10.1105/tpc.008342
Schuurink, R., and Tissier, A. (2020). Glandular trichomes: micro-organs with model status? New Phytol. 225, 2251–2266. doi: 10.1111/nph.16283
Schwab, B., Folkers, U., Ilgenfritz, H., and Hülskamp, M. (2000). Trichome morphogenesis in Arabidopsis. Philos. Trans. R. Soc. Lond B Biol. Sci. 355, 879–883.
Scoville, A. G., Barnett, L. L., Bodbyl-Roels, S., Kelly, J. K., and Hileman, L. C. (2011). Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol. 191, 251–263. doi: 10.1111/j.1469-8137.2011.03656.x
Shan, C. M., Shangguan, X. X., Zhao, B., Zhang, X. F., Chao, L. M., Yang, C. Q., et al. (2014). Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat. Commun. 5:5519. doi: 10.1038/ncomms6519
Shang, F., Mou, W., Wu, H., Xu, F., Xiang, C., and Wang, J. (2020). New Allele of HL6 regulates trichome elongation in rice. Rice Sci. 27, 480–492.
Shangguan, X., Yang, Q., Wu, X., and Cao, J. (2021). Function analysis of a cotton R2R3 MYB transcription factor GhMYB3 in regulating plant trichome development. Plant Biol. (Stuttg) 23, 1118–1127. doi: 10.1111/plb.13299
Shangguan, X. X., Yang, C. Q., Zhang, X. F., and Wang, L. J. (2016). Functional characterization of a basic helix-loop-helix (bHLH) transcription factor GhDEL65 from cotton (Gossypium hirsutum). Physiol. Plant 158, 200–212. doi: 10.1111/ppl.12450
Shi, X., Wu, Y., Dai, T., Gu, Y., Wang, L., Qin, X., et al. (2018). JcZFP8, a C2H2 zinc finger protein gene from Jatropha curcas, influences plant development in transgenic tobacco. Gene 34, 76–82.
Shvachko, N., Semilet, T., and Tikhonova, N. (2020). Trichomes of Higher Plants: homologous series in hereditary variability and molecular genetic mechanisms. Russ. J. Genet. 56, 1359–1370.
Singh, N. D., Kumar, S., and Daniell, H. (2016). Expression of β-glucosidase increases trichome density and artemisinin content in transgenic Artemisia annua plants. Plant Biotechnol. J. 14, 1034–1045. doi: 10.1111/pbi.12476
Sletvold, N., Huttunen, P., Handley, R., Karkkainen, K., and Agren, J. (2010). Cost of trichome production and resistance to a specialist insect herbivore in Arabidopsis lyrata. Evol. Ecol. 24, 1307–1319.
Spirina, U. N., Voronkova, T. V., and Ignatov, M. S. (2020). Are All Paraphyllia the Same? Front. Plant Sci. 11:858. doi: 10.3389/fpls.2020.00858
Stratmann, J. W., and Bequette, C. J. (2016). Hairless but no longer clueless: understanding glandular trichome development. J. Exp. Bot. 67, 5285–5287. doi: 10.1093/jxb/erw339
Sturaro, M., Hartings, H., Schmelzer, E., Velasco, R., Salamini, F., and Motto, M. (2005). Cloning and characterization of GLOSSY1, a maize gene involved in cuticle membrane and wax production. Plant Physiol. 138, 478–489. doi: 10.1104/pp.104.058164
Sugimoto-Shirasu, K., Stacey, N. J., Corsar, J., Roberts, K., and Mccann, M. C. (2002). DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 12, 1782–1786. doi: 10.1016/s0960-9822(02)01198-3
Sun, H., Hao, P., Gu, L., Cheng, S., Wang, H., Wu, A., et al. (2020). Pectate lyase-like Gene GhPEL76 regulates organ elongation in Arabidopsis and fiber elongation in cotton. Plant Sci. 293:110395. doi: 10.1016/j.plantsci.2019.110395
Sun, H., Pang, B. Y., Yan, J., Wang, T., Wang, L. N., Chen, C. H., et al. (2018). comprehensive analysis of cucumber gibberellin oxidase family genes and functional characterization of CsGA20ox1 in root development in Arabidopsis. Int. J. Mol. Sci. 19:3135.
Sun, L., Zhang, A., Zhou, Z., Zhao, Y., Yan, A., Bao, S., et al. (2015a). GLABROUS INFLORESCENCE STEMS3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol. 206, 220–230. doi: 10.1111/nph.13218
Sun, X., Gong, S. Y., Nie, X. Y., Li, Y., Li, W., Huang, G. Q., et al. (2015b). A R2R3-MYB transcription factor that is specifically expressed in cotton (Gossypium hirsutum) fibers affects secondary cell wall biosynthesis and deposition in transgenic Arabidopsis. Physiol. Plant 154, 420–432. doi: 10.1111/ppl.12317
Sun, Y., Veerabomma, S., Fokar, M., Abidi, N., Hequet, E., Payton, P., et al. (2015c). Brassinosteroid signaling affects secondary cell wall deposition in cotton fibers. Ind. Crop. Prod. 65, 334–342.
Sun, W., Gao, D., Xiong, Y., Tang, X., Xiao, X., Wang, C., et al. (2017). Hairy Leaf 6, an AP2/ERF Transcription Factor, Interacts with OsWOX3B and Regulates Trichome Formation in Rice. Mol. Plant 10, 1417–1433. doi: 10.1016/j.molp.2017.09.015
Sun, W., Gao, Z., Wang, J., Huang, Y., Chen, Y., Li, J., et al. (2019). Cotton fiber elongation requires the transcription factor Gh MYB 212 to regulate sucrose transportation into expanding fibers. New Phytol. 222, 864–881.
Suo, J. F., Liang, X. O., Pu, L., Zhang, Y. S., and Xue, Y. B. (2003). Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium Hirsutum L.). Biochim. Biophys. Acta 1630, 25–34. doi: 10.1016/j.bbaexp.2003.08.009
Symonds, V. V., Hatlestad, G., and Lloyd, A. M. (2011). Natural allelic variation defines a role for ATMYC1: trichome cell fate determination. PLoS Genet. 7:e1002069. doi: 10.1371/journal.pgen.1002069
Szymanski, D. B., Lloyd, A. M., and Marks, M. D. (2000). Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 5, 214–219. doi: 10.1016/s1360-1385(00)01597-1
Szymanski, D. B., and Marks, M. D. (1998). GLABROUS1 overexpression and TRIPTYCHON alter the cell cycle and trichome cell fate in Arabidopsis. Plant Cell 10, 2047–2062. doi: 10.1105/tpc.10.12.2047
Tan, H., Xiao, L., Gao, S., Li, Q., Chen, J., Xiao, Y., et al. (2015). TRICHOME AND ARTEMISININ REGULATOR 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua. Mol. Plant 8, 1396–1411.
Tan, Y., Barnbrook, M., Wilson, Y., Molnár, A., Bukys, A., and Hudson, A. (2020). Shared mutations in a novel glutaredoxin repressor of multicellular trichome fate underlie parallel evolution of Antirrhinum Species. Curr. Biol. 30, 1357–1366.e4. doi: 10.1016/j.cub.2020.01.060
Tao, L., Guangtao, Z., Junhong, Z., Xiangyang, X., Qinghui, Y., Zheng, Z., et al. (2014). Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 46, 1220–1226. doi: 10.1038/ng.3117
Tao, L., Yali, Z., Zilong, S., Shibo, Y., Haibing, S., Xiaoya, Z., et al. (2019). OsSPL10, a SBP-Box Gene, Plays a Dual Role in Salt Tolerance and Trichome Formation in Rice (Oryza sativa L.). G3 (Bethesda). 9, 4107–4114. doi: 10.1534/g3.119.400700
Taylor-Teeples, M., Lin, L., De Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665.
Tian, Y., and Zhang, T. (2021). MIXTAs and phytohormones orchestrate cotton fiber development. Curr. Opin. Plant Biol. 59, 101975. doi: 10.1016/j.pbi.2020.10.007
Tominaga-Wada, R., and Nukumizu, Y. (2012). The CAPRICE-LIKE MYB gene family cooperatively controls trichome branching and clustering in Arabidopsis. Plant Biotechnol. 29, 407–410.
Tominaga-Wada, R., Nukumizu, Y., Sato, S., Kato, T., Tabata, S., and Wada, T. (2012). Functional Divergence of MYB-Related Genes, WEREWOLF and AtMYB23 in Arabidopsis. Biosci. Biotechnol. Biochem. 76, 883–887. doi: 10.1271/bbb.110811
Traas, J., Bellini, C., Nacry, P., Kronenberger, J., Bouchez, D., and Caboche, M. (1995). Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 375, 676–677.
Traw, M. B., and Bergelson, J. (2003). Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol. 133, 1367–1375. doi: 10.1104/pp.103.027086
Turner, G. W., Gershenzon, J., and Croteau, R. B. (2000). Development of Peltate Glandular Trichomes of Peppermint. Plant Physiol. 124, 665–679. doi: 10.1104/pp.124.2.665
Vadde, B. V. L., Challa, K. R., and Nath, U. (2018). The TCP 4 transcription factor regulates trichome cell differentiation by directly activating GLABROUS INFLORESCENCE STEMS in Arabidopsis thaliana. Plant J. 93, 259–269. doi: 10.1111/tpj.13772
Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A. C., et al. (2004). Endogenous trans -Acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16, 69–79. doi: 10.1016/j.molcel.2004.09.028
Vendemiatti, E., Zsogon, A., Silva, G., De Jesus, F. A., Cutri, L., Figueiredo, C. R. F., et al. (2017). Loss of type-IV glandular trichomes is a heterochronic trait in tomato and can be reverted by promoting juvenility. Plant Sci. 259, 35–47. doi: 10.1016/j.plantsci.2017.03.006
Vernoud, V., Laigle, G., Rozier, F., Meeley, R. B., Perez, P., and Rogowsky, P. M. (2009). The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 59, 883–894. doi: 10.1111/j.1365-313X.2009.03916.x
Victor, K., Grini, P. E., Jaideep, M., Irene, K., Klaus, A., Nicole, B., et al. (2002). The Arabidopsis TUBULIN-FOLDING COFACTOR A gene is involved in the control of the alpha/beta-tubulin monomer balance. Plant Cell 14, 2265–2276. doi: 10.1105/tpc.003020
Victor, K., Marissa, S., Martin, H., and John, S. (2004). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268, 506–513.
Voinnet, O. (2009). Origin, biogenesis, and activity of plant MicroRNAs. Cell 136, 669–687. doi: 10.1016/j.cell.2009.01.046
Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. doi: 10.1126/science.277.5329.1113
Walford, S. A., Wu, Y., Llewellyn, D. J., and Dennis, E. S. (2012). Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. Plant J. 71, 464–478. doi: 10.1111/j.1365-313X.2012.05003.x
Walker, A. R., Davison, P. A., Bolognesi-Winfield, A. C., James, C. M., Srinivasan, N., Blundell, T. L., et al. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1350. doi: 10.1105/tpc.11.7.1337
Walker, J. D., Oppenheimer, D. G., Concienne, J., and Larkin, J. C. (2000). SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127, 3931–3940. doi: 10.1242/dev.127.18.3931
Wang, H., Mei, W., Qin, Y., and Zhu, Y. (2011). 1-Aminocyclopropane-1-carboxylic acid synthase 2 is phosphorylated by calcium-dependent protein kinase 1 during cotton fiber elongation. Acta Biochim. Biophys. Sin. 43, 654–661. doi: 10.1093/abbs/gmr056
Wang, L., Cook, A., Patrick, J. W., Chen, X. Y., and Ruan, Y. L. (2014). Silencing the vacuolar invertase gene GhVIN1 blocks cotton fiber initiation from the ovule epidermis, probably by suppressing a cohort of regulatory genes via sugar signaling. Plant J. 78, 686–696. doi: 10.1111/tpj.12512
Wang, Y., Zeng, J., Xia, X., Xu, Y., Sun, J., Gu, J., et al. (2020b). Comparative analysis of leaf trichomes, epidermal wax and defense enzymes activities in response to Puccinia horiana in Chrysanthemum and Ajania species. Hortic. Plant J. 6, 191–198.
Wang, L., Wang, G., Long, L., Altunok, S., Feng, Z., Wang, D., et al. (2020a). Understanding the role of phytohormones in cotton fiber development through omic approaches; recent advances and future directions. Int. J. Biol. Macromol. 163, 1301–1313.
Wang, Z., Yang, Z., and Li, F. (2019d). Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton. Plant Biotechnol. J. 17, 1706–1722. doi: 10.1111/pbi.13167
Wang, M., Tu, L., Yuan, D., Zhu, D., Shen, C., Li, J., et al. (2019b). Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 51, 224–229.
Wang, Y., Fu, X., Xie, L., Qin, W., Li, L., Sun, X., et al. (2019c). Stress associated protein 1 regulates the development of glandular trichomes in Artemisia annua. Plant Cell Tissue Organ Cult. 139, 249–259.
Wang, D.-J., Zeng, J.-W., Ma, W.-T., Lu, M., and An, H.-M. (2019a). Morphological and structural characters of trichomes on various organs of Rosa roxburghii. HortScience 54, 45–51.
Wang, Y., Chen, W., Qin, P., Huang, Y., Ma, B., Ouyang, X., et al. (2013b). Characterization and fine mapping of Glabrous rice 2 in rice. J. Genet. Genom. 40, 579–582. doi: 10.1016/j.jgg.2013.06.001
Wang, M. Y., Zhao, P. M., Cheng, H. Q., Han, L. B., Wu, X. M., Gao, P., et al. (2013a). The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 162, 1669–1680. doi: 10.1104/pp.113.215673
Wang, X., Shen, C., Meng, P., Tan, G., and Lv, L. (2021c). Analysis and review of trichomes in plants. BMC Plant Biol 21:70. doi: 10.1186/s12870-021-02840-x
Wang, L., Kartika, D., and Ruan, Y. L. (2021a). Looking into ‘hair tonics’ for cotton fiber initiation. New Phytol. 229, 1844–1851. doi: 10.1111/nph.16898
Wang, N.-N., Li, Y., Chen, Y.-H., Lu, R., Zhou, L., Wang, Y., et al. (2021b). Phosphorylation of WRKY16 by MPK3-1 is essential for its transcriptional activity during fiber initiation and elongation in cotton (Gossypium hirsutum). Plant Cell 33, 2736–2752. doi: 10.1093/plcell/koab153
Wang, Y., Li, Y., Gong, S. Y., Qin, L. X., Nie, X. Y., Liu, D., et al. (2021d). GhKNL1 controls fiber elongation and secondary cell wall synthesis by repressing its downstream genes in cotton (Gossypium hirsutum). J. Integr. Plant Biol. 64, 39–55. doi: 10.1111/jipb.13192
Wang, Z., Wang, L., Han, L., Cheng, Z., Liu, X., Wang, S., et al. (2021e). HECATE2 acts with GLABROUS3 and Tu to boost cytokinin biosynthesis and regulate cucumber fruit wart formation. Plant Physiol. 187, 1619–1635. doi: 10.1093/plphys/kiab377
Wang, Z., Yan, X., Zhang, H., Meng, Y., Pan, Y., and Cui, H. (2021f). NtCycB2 negatively regulates tobacco glandular trichome formation, exudate accumulation, and aphid resistance. Plant Mol. Biol. 108, 65–76. doi: 10.1007/s11103-021-01222-z
Wang, S., Wang, J.-W., Yu, N., Li, C.-H., Luo, B., Gou, J.-Y., et al. (2004). Control of Plant Trichome Development by a Cotton Fiber MYB Gene. Plant Cell 16, 2323–2334. doi: 10.1105/tpc.104.024844
Wang, Y. L., Nie, J. T., Chen, H. M., Guo, C. L., Pan, J., He, H. L., et al. (2016). Identification and mapping of Tril, a homeodomain-leucine zipper gene involved in multicellular trichome initiation in Cucumis sativus. Theor. Appl. Genet. 129, 305–316. doi: 10.1007/s00122-015-2628-4
Wei, L.-H., Song, P., Wang, Y., Lu, Z., Tang, Q., Yu, Q., et al. (2018b). The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 30, 968–985. doi: 10.1105/tpc.17.00934
Wei, H., Zhao, Y., Xie, Y., and Wang, H. (2018a). Exploiting SPL genes to improve maize plant architecture tailored for high-density planting. J. Exp. Bot. 69, 4675–4688. doi: 10.1093/jxb/ery258
Wei, S., Gruber, M. Y., Yu, B., Gao, M. J., Khachatourians, G. G., Hegedus, D. D., et al. (2012). Arabidopsis mutant sk156 reveals complex regulation of SPL15 in a miR156-controlled gene network. BMC Plant Biol. 12:169. doi: 10.1186/1471-2229-12-169
Wu, J., Peled-Zehavi, H., and Galili, G. (2020a). The m6A reader ECT2 post-transcriptionally regulates proteasome activity in Arabidopsis. New Phytol. 228, 151–162. doi: 10.1111/nph.16660
Wu, M.-L., Cui, Y.-C., Ge, L., Cui, L.-P., Xu, Z.-C., Zhang, H.-Y., et al. (2020b). NbCycB2 represses Nbwo activity via a negative feedback loop in tobacco trichome development. J. Exp. Bot. 71, 1815–1827. doi: 10.1093/jxb/erz542
Xia, X. C., Hu, Q. Q., Li, W., Chen, Y., Han, L. H., Tao, M., et al. (2018). Cotton (Gossypium hirsutum) JAZ3 and SLR1 function in jasmonate and gibberellin mediated epidermal cell differentiation and elongation. Plant Cell Tissue Organ Cult. 133, 249–262.
Xiao, F., Gong, Q., Zhao, S., Lin, H., and Zhou, H. (2021). MYB30 and ETHYLENE INSENSITIVE3 antagonistically modulate root hair growth in Arabidopsis. Plant J. 106, 480–492. doi: 10.1111/tpj.15180
Xiao, G., He, P., Zhao, P., Liu, H., Zhang, L., Pang, C., et al. (2018). Genome-wide identification of the GhARF gene family reveals that GhARF2 and GhARF18 are involved in cotton fibre cell initiation. J. Exp. Bot. 69, 4323–4337. doi: 10.1093/jxb/ery219
Xie, L., Yan, T., Li, L., Chen, M., Hassani, D., Li, Y., et al. (2021a). An HD-ZIP-MYB complex regulates glandular secretory trichome initiation in Artemisia annua. New Phytol. 231, 2050–2064. doi: 10.1111/nph.17514
Xie, L., Yan, T., Li, L., Chen, M., Ma, Y., Hao, X., et al. (2021b). The WRKY transcription factor AaGSW2 promotes glandular trichome initiation in Artemisia annua. J. Exp. Bot. 72, 1691–1701. doi: 10.1093/jxb/eraa523
Xie, M., Sun, J., Gong, D., and Kong, Y. (2019). The Roles of Arabidopsis C1-2i Subclass of C2H2-type Zinc-Finger Transcription Factors. Genes 10:653. doi: 10.3390/genes10090653
Xie, Y. J., Yu, X. Z., Jiang, S. F., Xiao, K. Z., Wang, Y. P., Li, L. L., et al. (2020b). OsGL6, a conserved AP2 domain protein, promotes leaf trichome initiation in rice. Biochem. Biophys. Res. Commun. 522, 448–455. doi: 10.1016/j.bbrc.2019.11.125
Xie, Q., Gao, Y., Li, J., Yang, Q., Qu, X., Li, H., et al. (2020a). The HD-Zip IV transcription factor SlHDZIV8 controls multicellular trichome morphology by regulating the expression of Hairless-2. J. Exp. Bot. 71, 7132–7145. doi: 10.1093/jxb/eraa428
Xie, Q., Liu, P., Shi, L., Miao, H., Bo, K., Wang, Y., et al. (2018). Combined fine mapping, genetic diversity, and transcriptome profiling reveals that the auxin transporter gene ns plays an important role in cucumber fruit spine development. Theor. Appl. Genet. 131, 1239–1252. doi: 10.1007/s00122-018-3074-x
Xu, F., Chen, Q., Huang, L., and Luo, M. (2021). Advances about the roles of membranes in cotton fiber development. Membranes (Basel) 11:471. doi: 10.3390/membranes11070471
Xu, J., Van Herwijnen, Z. O., Drager, D. B., Sui, C., Haring, M. A., and Schuurink, R. C. (2018). SlMYC1 Regulates Type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant cell 30, 2988–3005. doi: 10.1105/tpc.18.00571
Xue, S., Dong, M., Liu, X., Xu, S., Pang, J., Zhang, W., et al. (2019). Classification of fruit trichomes in cucumber and effects of plant hormones on type II fruit trichome development. Planta 249, 407–416. doi: 10.1007/s00425-018-3004-9
Yadav, V. K., Yadav, V. K., Pant, P., Singh, S. P., Maurya, R., Sable, A., et al. (2017). GhMYB1 regulates SCW stage-specific expression of the GhGDSL promoter in the fibres of Gossypium hirsutum L. Plant Biotechnol. J. 15, 1163–1174. doi: 10.1111/pbi.12706
Yan, A., Pan, J., An, L., Gan, Y., and Feng, H. (2012). The responses of trichome mutants to enhanced ultraviolet-B radiation in Arabidopsis thaliana. J. Photochem. Photobiol. B 113, 29–35. doi: 10.1016/j.jphotobiol.2012.04.011
Yan, A., Wu, M., Zhao, Y., Zhang, A., Liu, B., Schiefelbein, J., et al. (2014). Involvement of C2H2 zinc finger proteins in the regulation of epidermal cell fate determination in Arabidopsis. J. Integr. Plant Biol. 56, 1112–1117. doi: 10.1111/jipb.12221
Yan, T., Chen, M., Shen, Q., Li, L., Fu, X., Pan, Q., et al. (2017). HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 213, 1145–1155. doi: 10.1111/nph.14205
Yan, T., Li, L., Xie, L., Chen, M., Shen, Q., Pan, Q., et al. (2018). A novel HD-ZIP IV/MIXTA complex promotes glandular trichome initiation and cuticle development in Artemisia annua. New Phytol. 218, 567–578. doi: 10.1111/nph.15005
Yang, C., Gao, Y., Gao, S., Yu, G., Xiong, C., Chang, J., et al. (2015). Transcriptome profile analysis of cell proliferation molecular processes during multicellular trichome formation induced by tomato Wov gene in tobacco. BMC Genomics 16:868. doi: 10.1016/j.gdata.2015.09.011
Yang, C., and Ye, Z. (2013). Trichomes as models for studying plant cell differentiation. Cell Mol. Life Sci. 70, 1937–1948. doi: 10.1007/s00018-012-1147-6
Yang, S., Cai, Y., Liu, X., Dong, M., Zhang, Y., Chen, S., et al. (2018). A CsMYB6-CsTRY module regulates fruit trichome initiation in cucumber. J. Exp. Bot. 69, 1887–1902. doi: 10.1093/jxb/ery047
Yang, Z., Zhang, C., Yang, X., Liu, K., Wu, Z., Zhang, X., et al. (2014b). PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 203, 437–448. doi: 10.1111/nph.12824
Yang, X., Zhang, W., He, H., Nie, J., Bie, B., Zhao, J., et al. (2014a). Tuberculate fruit gene Tu encodes a C2 H2 zinc finger protein that is required for the warty fruit phenotype in cucumber (Cucumis sativus L.). Plant J. 78, 1034–1046. doi: 10.1111/tpj.12531
Yang, Z., Song, M., Cheng, F., Zhang, M., Davoudi, M., Chen, J., et al. (2021). A SNP Mutation in Homeodomain-DDT (HD-DDT) Transcription Factor Results in Multiple Trichomes (mt) in Cucumber (Cucumis sativus L.). Genes (Basel) 12, 1478. doi: 10.3390/genes12101478
Yang, Z. B. (2002). Small GTPases: versatile signaling switches in plants. Plant Cell 14, S375–S388. doi: 10.1105/tpc.001065
Ye, Z., Qiao, L., Luo, X., Chen, X., Zhang, X., and Tu, L. (2020). Genome-wide identification of cotton GRAM family finds GhGRAM31 regulates fiber length. J. Exp. Bot. 72, 2477–2490. doi: 10.1093/jxb/eraa597
Ying, Z., Nanyu, T., Lijin, H., Yongjuan, Z., Xiaoqing, T., and Kangcai, W. (2018). Effects of Salt Stress on Plant Growth, Antioxidant Capacity, Glandular Trichome Density, and Volatile Exudates of Schizonepeta tenuifolia Briq. Int. J. Mol. Sci. 19:252. doi: 10.3390/ijms19010252
Yong-Mei, Q., and Yu-Xian, Z. (2011). How cotton fibers elongate: a tale of linear cell-growth mode. Curr. Opin. Plant Biol. 14, 106–111. doi: 10.1016/j.pbi.2010.09.010
Yu, X., Qi, X., Li, S., Fang, H., Bai, Y., Li, L., et al. (2021b). Transcriptome analysis of light-regulated monoterpenes biosynthesis in leaves of Mentha canadensis L. Plants (Basel) 10:930. doi: 10.3390/plants10050930
Yu, D., Li, X., Li, Y., Ali, F., Li, F., and Wang, Z. (2021a). Dynamic roles and intricate mechanisms of ethylene in epidermal hair development in Arabidopsis and cotton. New Phytol. 234, 375–391. doi: 10.1111/nph.17901
Yu, N., Cai, W. J., Wang, S., Shan, C. M., Wang, L. J., and Chen, X. Y. (2010). Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22, 2322–2335. doi: 10.1105/tpc.109.072579
Yu, X. H., Chen, G. P., Tang, B. Y., Zhang, J. L., Zhou, S. G., and Hu, Z. L. (2018). The Jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Science 267, 65–73. doi: 10.1016/j.plantsci.2017.11.008
Yu, Z. H., Mccouch, S. R., Tanksley, S. D., Kinoshita, T., and Sato, S. (1995). Association of morphological and RFLP markers in rice (Oryza sativa L.). Genome 38, 566–574. doi: 10.1139/g95-073
Yuan, D., Tang, Z., Wang, M., Gao, W., Tu, L., Jin, X., et al. (2015a). The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 5:17662. doi: 10.1038/srep17662
Yuan, F., Lyu, M. J. A., Leng, B. Y., Zheng, G. Y., Feng, Z. T., Li, P. H., et al. (2015c). Comparative transcriptome analysis of developmental stages of the L imonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 38, 1637–1657. doi: 10.1111/pce.12514
Yuan, F., Lyu, M. J., Leng, B. Y., Zheng, G. Y., Feng, Z. T., Li, P. H., et al. (2015b). Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 38, 1637–1657.
Yuan, F., Lyu, M. J., Leng, B. Y., Zhu, X. G., and Wang, B. S. (2016b). The transcriptome of NaCl-treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol. Biol. 91, 241–256. doi: 10.1007/s11103-016-0460-0
Yuan, F., Leng, B., and Wang, B. (2016a). Progress in studying salt secretion from the salt glands in recretohalophytes: how do plants secrete salt? Front. Plant Sci. 7:977. doi: 10.3389/fpls.2016.00977
Yuan, R., Cao, Y., Li, T., Yang, F., Yu, L., Qin, Y., et al. (2020). Comparative mapping and correlation analysis revealed differentiation in the genetic basis of stem trichome development between cultivated tetraploid cotton species. BMC Plant Biol. 21:115. doi: 10.21203/rs.3.rs-38572/v2
Zang, D., Wang, C., Ji, X., and Wang, Y. (2015). Tamarix hispida zinc finger protein ThZFP1 participates in salt and osmotic stress tolerance by increasing proline content and SOD and POD activities. Plant Sci. 235, 111–121. doi: 10.1016/j.plantsci.2015.02.016
Zhang, A., Liu, Y., Yu, C., Huang, L., Wu, M., Wu, J., et al. (2020). Zinc Finger Protein 1 (ZFP1) Is Involved in Trichome Initiation in Arabidopsis thaliana. Agricult.Basel 10:645.
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. doi: 10.1242/dev.00681
Zhang, F., Zuo, K., Zhang, J., Liu, X., Zhang, L., Sun, X., et al. (2010). An L1 box binding protein, GbML1, interacts with GbMYB25 to control cotton fibre development. J. Exp. Bot. 61, 3599–3613. doi: 10.1093/jxb/erq173
Zhang, Y., Shen, J., Bartholomew, E. S., Dong, M., Chen, S., Yin, S., et al. (2021f). TINY BRANCHED HAIR functions in multicellular trichome development through an ethylene pathway in Cucumis sativus L. Plant J. 106, 753–765. doi: 10.1111/tpj.15198
Zhang, H. N., Liu, P. H., Wang, B. S., and Yuan, F. (2021b). The roles of trichome development genes in stress resistance. Plant Growth Regul. 95, 137–148.
Zhang, L., Lv, D., Pan, J., Zhang, K., Wen, H., Chen, Y., et al. (2021c). A SNP of HD-ZIP I transcription factor leads to distortion of trichome morphology in cucumber (Cucumis sativus L.). BMC Plant Biol. 21:182. doi: 10.1186/s12870-021-02955-1
Zhang, X., Cao, J., Huang, C., Zheng, Z., Liu, X., Shangguan, X., et al. (2021d). Characterization of cotton ARF factors and the role of GhARF2b in fiber development. BMC Genom. 22:202. doi: 10.1186/s12864-021-07504-6
Zhang, X., Xue, Y., Guan, Z., Zhou, C., Nie, Y., Men, S., et al. (2021e). Structural insights into homotrimeric assembly of cellulose synthase CesA7 from Gossypium hirsutum. Plant Biotechnol. J. 19, 1579–1587. doi: 10.1111/pbi.13571
Zhang, H., Lu, X., Wang, Z., Yan, X., and Cui, H. (2021a). Excretion from long glandular trichomes contributes to alleviation of cadmium toxicity in Nicotiana tabacum. Environ. Pollut. 285:117184. doi: 10.1016/j.envpol.2021.117184
Zhang, L., Pan, J., Wang, G., Du, H., He, H., Pan, J., et al. (2019b). Cucumber CsTRY negatively regulates anthocyanin biosynthesis and trichome formation when expressed in tobacco. Front. Plant Sci. 10:1232. doi: 10.3389/fpls.2019.01232
Zhang, H., Li, W., Niu, D., Wang, Z., Yan, X., Yang, X., et al. (2019a). Tobacco transcription repressors NtJAZ: potential involvement in abiotic stress response and glandular trichome induction. Plant Physiol. Biochem. 141, 388–397. doi: 10.1016/j.plaphy.2019.06.021
Zhang, H., Wang, L., Zheng, S., Liu, Z., Wu, X., Gao, Z., et al. (2016). A fragment substitution in the promoter of CsHDZIV11/CsGL3 is responsible for fruit spine density in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 129, 1289–1301. doi: 10.1007/s00122-016-2703-5
Zhang, H., Wu, K., Wang, Y., Peng, Y., Hu, F., Wen, L., et al. (2012). A WUSCHEL-like homeobox gene, OsWOX3B responses to NUDA/GL-1 locus in rice. Rice (N Y) 5, 1–10. doi: 10.1186/1939-8433-5-30
Zhang, J., Huang, G. Q., Zou, D., Yan, J. Q., Li, Y., Hu, S., et al. (2018). The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 217, 625–640. doi: 10.1111/nph.14864
Zhang, M., Zeng, J.-Y., Long, H., Xiao, Y.-H., Yan, X.-Y., and Pei, Y. (2017b). Auxin regulates cotton fiber initiation via GhPIN-Mediated auxin transport. Plant Cell Physiol. 58, 385–397. doi: 10.1093/pcp/pcw203
Zhang, M., Xiao, Y., Zeng, J., and Pei, Y. (2017a). PIN-formed protein, a door to reveal the mechanism for auxin-triggered initiation of cotton fiber. Plant Signal. Behav. 12:e1319031. doi: 10.1080/15592324.2017.1319031
Zhang, Z., Chen, X., Guan, X., Liu, Y., Chen, H., Wang, T., et al. (2014). A genome-wide survey of homeodomain-leucine zipper genes and analysis of cold-responsive HD-Zip I members’ expression in tomato. Biosci. Biotechnol. Biochem. 78, 1337–1349. doi: 10.1080/09168451.2014.923292
Zhao, J. L., Pan, J. S., Guan, Y., Zhang, W. W., Bie, B. B., Wang, Y. L., et al. (2015). Micro-trichome as a class I homeodomain-leucine zipper gene regulates multicellular trichome development in Cucumis sativus. J. Integr. Plant Biol. 57, 925–935. doi: 10.1111/jipb.12345
Zhao, T., Tao, X., Li, M., Gao, M., Chen, J., Zhou, N., et al. (2020). Role of phasiRNAs from two distinct phasing frames of GhMYB2 loci in cis-gene regulation in the cotton genome. BMC Plant Biol 20:219. doi: 10.1186/s12870-020-02430-3
Zheng, F., Cui, L., Li, C., Xie, Q., Ai, G., Wang, J., et al. (2021). Hair (H) interacts with SlZFP8-like to regulate the initiation and elongation of trichomes by modulating SlZFP6 expression in tomato. J. Exp. Bot. 73, 228–244. doi: 10.1093/jxb/erab417
Zheng, K. J., Tian, H. N., Hu, Q. N., Guo, H. Y., Yang, L., Cai, L., et al. (2016). Ectopic expression of R3 MYB transcription factor gene OsTCL1 in Arabidopsis, but not rice, affects trichome and root hair formation. Sci. Rep. 6:19254. doi: 10.1038/srep19254
Zhou, Y., Li, B.-Y., Li, M., Li, X.-J., Zhang, Z.-T., Li, Y., et al. (2014). A MADS-box gene is specifically expressed in fibers of cotton (Gossypium hirsutum) and influences plant growth of transgenic Arabidopsis in a GA-dependent manner. Plant Physiol. Biochem. 75, 70–79. doi: 10.1016/j.plaphy.2013.12.003
Zhou, Y., Zhang, Z. T., Li, M., Wei, X. Z., Li, X. J., Li, B. Y., et al. (2015). Cotton (G ossypium hirsutum) 14-3-3 proteins participate in regulation of fibre initiation and elongation by modulating brassinosteroid signalling. Plant Biotechnol. J. 13, 269–280. doi: 10.1111/pbi.12275
Zhou, Z., An, L., Sun, L., and Gan, Y. (2012). ZFP5 encodes a functionally equivalent GIS protein to control trichome initiation. Plant signal. Behav. 7, 28–30. doi: 10.4161/psb.7.1.18404
Zhou, Z., An, L., Sun, L., Zhu, S., Xi, W., Broun, P., et al. (2011). Zinc Finger protein5 is required for the control of trichome initiation by acting upstream of zinc finger protein8 in Arabidopsis. Plant Physiol. 157, 673–682. doi: 10.1104/pp.111.180281
Zhou, Z., Sun, L., Zhao, Y., An, L., Yan, A., Meng, X., et al. (2013). Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol. 198, 699–708. doi: 10.1111/nph.12211
Zhou, Z., Tan, H. X., Li, Q., Li, Q., Wang, Y., Bu, Q. T., et al. (2020). TRICHOME AND ARTEMISININ REGULATOR 2positively regulates trichome development and artemisinin biosynthesis in Artemisia annua. New Phytol. 228, 932–945.
Keywords: plant, epidermal cell, trichome, development, molecular mechanism
Citation: Han G, Li Y, Yang Z, Wang C, Zhang Y and Wang B (2022) Molecular Mechanisms of Plant Trichome Development. Front. Plant Sci. 13:910228. doi: 10.3389/fpls.2022.910228
Received: 01 April 2022; Accepted: 13 May 2022;
Published: 01 June 2022.
Edited by:
Yuree Lee, Seoul National University, South KoreaReviewed by:
Yasuhiro Sato, University of Zurich, SwitzerlandCopyright © 2022 Han, Li, Yang, Wang, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoliang Han, Z2xfaGFuQHNkbnUuZWR1LmNu; YnN3YW5nQHNkbnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.