- 1The New Zealand Institute for Plant and Food Research Limited, Palmerston North, New Zealand
- 2School of Biological Sciences, The University of Auckland, Auckland, New Zealand
- 3The New Zealand Institute for Plant and Food Research Limited, Auckland, New Zealand
- 4Department of Arctic and Marine Biology, UiT The Arctic University of Norway, Tromsø, Norway
- 5Norwegian Institute of Bioeconomy Research (NIBIO), Tromsø, Norway
Vaccinium berries are regarded as “superfoods” owing to their high concentrations of anthocyanins, flavonoid metabolites that provide pigmentation and positively affect human health. Anthocyanin localization differs between the fruit of cultivated highbush blueberry (V. corymbosum) and wild bilberry (V. myrtillus), with the latter having deep red flesh coloration. Analysis of comparative transcriptomics across a developmental series of blueberry and bilberry fruit skin and flesh identified candidate anthocyanin regulators responsible for this distinction. This included multiple activator and repressor transcription factors (TFs) that correlated strongly with anthocyanin production and had minimal expression in blueberry (non-pigmented) flesh. R2R3 MYB TFs appeared key to the presence and absence of anthocyanin-based pigmentation; MYBA1 and MYBPA1.1 co-activated the pathway while MYBC2.1 repressed it. Transient overexpression of MYBA1 in Nicotiana benthamiana strongly induced anthocyanins, but this was substantially reduced when co-infiltrated with MYBC2.1. Co-infiltration of MYBC2.1 with MYBA1 also reduced activation of DFR and UFGT, key anthocyanin biosynthesis genes, in promoter activation studies. We demonstrated that these TFs operate within a regulatory hierarchy where MYBA1 activated the promoters of MYBC2.1 and bHLH2. Stable overexpression of VcMYBA1 in blueberry elevated anthocyanin content in transgenic plants, indicating that MYBA1 is sufficient to upregulate the TF module and activate the pathway. Our findings identify TF activators and repressors that are hierarchically regulated by SG6 MYBA1, and fine-tune anthocyanin production in Vaccinium. The lack of this TF module in blueberry flesh results in an absence of anthocyanins.
Introduction
Anthocyanins are plant-specific compounds, which are usually responsible for the blue, purple and red colors found in plants. They are some of the most abundant secondary metabolites in Vaccinium fruit and contribute to their health promoting properties (Khoo et al., 2017), leading to Vaccinium berries being described as “superfoods”. Blueberries (Vaccinium spp., mainly Vaccinium corymbosum and Vaccinium virgatum) are one of the most well-known and commercially cultivated Vaccinium species, while there are numerous species such as bilberry (Vaccinium myrtillus), which are harvested from the wild. The anthocyanin localization in the fruit of these species differs, with blueberries having a pale, colorless flesh while bilberries have a red, anthocyanin-rich flesh (Lafferty et al., 2022).
In Vaccinium, proanthocyanidins (PAs) are the most abundant secondary metabolite during early fruit development (Zifkin et al., 2012; Karppinen et al., 2016). Proanthocyanidins confer the astringent and bitter taste associated with unripe fruit, which deters consumption before seed maturity, while anthocyanin accumulation signals fruit ripening (Rauf et al., 2019). Both PAs and anthocyanins are produced via the flavonoid pathway, sharing many common biosynthetic steps. This includes the conversion of leucoanthocyanidin into anthocyanidin by a 2-oxoglutarate dependent dioxygenase known as anthocyanidin synthase (ANS) or leucoanthocyanidin dioxygenase (LDOX). While some species have a single ANS/LDOX gene, others have multiple gene family members that can be phylogenetically separated into distinct clades, which are referred to as ANS (anthocyanins) or LDOX (proanthocyanidins; Jun et al., 2018; Wang et al., 2021). The PA specific biosynthetic steps occur when anthocyanin reductase (ANR) converts anthocyanidins to epicatechin or when leucoanthocyanidin reductase (LAR) converts leucoanthocyanidins to catechin. As Vaccinium fruit ripen the PA concentrations decrease and anthocyanins begin to accumulate, seen as pigmentation development during ripening (Zifkin et al., 2012; Karppinen et al., 2016). The comitted step for anthocyanin biosynthesis involves the glycosylation of anthocyanidins by uridine diphosphate (UDP)-glucose: flavonoid-O-glycosyltransferase (UFGT).
The flavonoid pathway is regulated at the transcriptional level, with an MBW complex promoting transcription of biosynthetic genes, consisting of R2R3 MYB, bHLH and WDR proteins (Baudry et al., 2004; Albert and Allan, 2021). The MYB transcription factor (TF) determines the specificity of the complex for the different biosynthetic gene promoters. Subgroup (SG) 5 MYBs typically regulate PA biosynthesis, and have been characterized in a number of fruiting species, including blueberry, bilberry, grape and strawberry (Terrier et al., 2009; Schaart et al., 2013; Karppinen et al., 2021; Lafferty et al., 2022). These MYBs drive MBW activation of the PA specific ANR and LAR genes. SG6 MYBs are involved in anthocyanin regulation, upregulating flavonoid biosynthetic genes and the anthocyanin specific UFGT. These are key regulators for controlling anthocyanin production in fruit (Albert and Allan, 2021), including blueberry and bilberry (Plunkett et al., 2018; Karppinen et al., 2021). An additional class of MYBs, MYBPA1-type, are dual regulators of PA and anthocyanin biosynthesis in fruit and are required for full activation of both pathways (Karppinen et al., 2021; Lafferty et al., 2022). These activate genes common to PA and anthocyanin biosynthesis, but are not sufficient to activate these pathways, and require the SG5 and SG6 MYBs to activate the additional pathway-specific genes for metabolite production. Arabidopsis lacks a MYBPA1 gene, but these MYBs appear to be important for species with more complex flavonoid profiles, such as are commonly found in fruit.
MYB repressors also contribute to PA and anthocyanin regulation. R3 MYB and SG4 R2R3 MYBs are two well-characterized classes of MYB repressors with distinct functions. R3 MYBs are small proteins that lack activation and repression motifs, yet have retained their bHLH binding domain (Zimmermann et al., 2004). When R3 MYBs bind bHLH proteins, this limits the formation of activator MBW complexes, referred to as passive repression (Zhang et al., 2009; Albert et al., 2014). While these repressors are best characterized for their roles that affect flower color and patterning (petunia, Mimulus; Albert et al., 2014; Ding et al., 2020), R3 MYBs have also been characterized and shown to inhibit anthocyanin production in fruiting species such as tomato (Colanero et al., 2018). Two R3 MYBs have been identified in bilberry and bog bilberry (V. uligonosum), but have yet to be functionally characterized (Primetta et al., 2015; Zorenc et al., 2017). The SG4 R2R3 MYBs function through active repression, as they have an ethylene-responsive element binding factor (ERF) associated amphiphilic repression (EAR) motif (Albert et al., 2014). This is recognized by TOPLESS (TPL), which recruits chromatin remodeling factors to the promoter region and inhibits transcription at the epigenetic level (Kagale and Rozwadowski, 2011). SG4 MYBs often have TLLLFR motifs that contribute to repressive activity (Matsui et al., 2008). Inhibition of the production of both PAs and anthocyanins by SG4 MYB repressors has been reported in a number of fruiting species, including apple, grape, peach and strawberry (Aharoni et al., 2001; Gao et al., 2011; Pérez-Díaz et al., 2016; Zhou et al., 2019; Zhu et al., 2019).
Hierarchical regulation, where a TF activates the expression of additional TFs, is important for flavonoid regulation in some species, including fruit systems. This has been observed for MYB repressors, where activator MYBs promoted their expression as part of a feedback repression loop, controlling the extent of activation for each pathway (Albert et al., 2014). This has also been reported for SG4 MYBs in petunia, poplar, Medicago truncatula and citrus (Albert et al., 2014; Jun et al., 2015; Yoshida et al., 2015; Huang et al., 2019) and for R3 MYBs in petunia and tomato (Albert et al., 2014; Yan et al., 2020). Hierarchy between activator TFs also occurs, with activator MYBs upregulating bHLH genes in Arabidopsis, kiwifruit, clover, Antirrhinum, tobacco and petunia (Baudry et al., 2006; Xu et al., 2013; Albert et al., 2014, 2021; Albert, 2015; Montefiori et al., 2015; Liu et al., 2021). Recently, the hierarchical regulation of blueberry MYBPA1.1 by both SG5 and SG6 MYB activators was reported (Lafferty et al., 2022). This was the first report of a SG6 MYB regulating MYBPA1-type MYBs, while their regulation by SG5 MYBs had been seen in apple, grape and poplar (Terrier et al., 2009; James et al., 2017; Wang et al., 2018). It is not currently known how this aspect of flavonoid regulation fits within the wider module of anthocyanin regulators, or whether any hierarchical regulation between members exists. Furthermore, flavonoid repressors have not yet been functionally characterized in Vaccinium spp.
The flavonoid profile of Vaccinium fruit varies during both fruit development and ripening. We hypothesize that coordinated expression of multiple TFs is required for full activation of the PA and anthocyanin pathways. Knowledge of these processes may help resolve the causal regulatory changes in bilberry that confer red flesh. To study this, we performed differential expression analysis on an RNA-sequencing (RNA-seq) dataset to identify candidate anthocyanin regulators. This dataset included both a blueberry and bilberry fruit developmental series, with skin and flesh tissues separated. Candidate genes were functionally characterized and hierarchical regulation was investigated. Finally, stable overexpression of the SG6 VcMYBA1 in blueberry was carried out to determine whether expression of this TF was sufficient for full activation of the anthocyanin pathway via activation of secondary transcriptional regulators.
Materials and Methods
Plant Material and Sampling
Northern Highbush blueberry (V. corymbosum) ‘Nui’ fruit and wild bilberry fruit were obtained as described previously, representing a developmental series from stage 4 to 8 (Günther et al., 2020; Lafferty et al., 2022). Additionally, stage 1–8 whole berry samples from “Nui” blueberry were sampled for qRT-PCR (Lafferty et al., 2022).
RNA-Sequencing, Differential Expression and Correlation Analysis
The raw RNAseq datasets used for this study were described previously for blueberry (Günther et al., 2020) and bilberry (Wu et al., 2021) and were deposited on NCBI under PRJNA591663 and PRJNA739815, respectively. The analysis of raw sequence datasets was performed and mapped to a V. corymbosum “Draper” genome (Colle et al., 2019). Raw read counts were normalized via the median of ratios method (Love et al., 2014).
Differential gene expression analysis was performed with the “DESeq2” R package (Love et al., 2014). Transcripts with an adjusted P-value (padj) less than 0.01 and log2-fold change greater than 2 were determined to be highly differentially expressed genes (HDEG). Multiple DESeq2 comparisons were performed and overlaid with each other using the “VennDiagram” R package (Chen and Boutros, 2011). HDEGs can be found in Supplementary Table S1.
Highly differentially expressed genes were functionally annotated with Mapman bin numbers using Mercator4 V2.0, and flavonoid biosynthetic genes were visualized using the MapMan version 3.5.1R2 software (Schwacke et al., 2019), displaying the log-fold change between S7 blueberry and bilberry flesh. A table displaying the HDEGs annotated as TFs was produced in R using the “data.table” and “formattable” R packages. The closest Arabidopsis homolog was obtained by a BLAST of the gene ID against the Araport11 protein sequences dataset using TAIR BLAST 2.9.0 +.
Heatmaps were constructed using the “gplots” R package (Warnes et al., 2009). The blueberry genome includes all four haplotypes, and therefore the same gene may be represented by four gene IDs in the analysis. When this occurred, the gene ID with the greatest count data was chosen. Expression was displayed as the Z-score, which represents the number of standard deviations below (negative score) or above (positive score) the data mean for the given gene in each tissue, across development. The blueberry genome gene IDs for flavonoid biosynthetic and regulatory genes analyzed in this study are provided in Supplementary Table S2.
Correlation analysis was performed for each tissue type by calculating Spearman rank correlations on normalized read counts using R version 4.0.5 and the “Hmisc” package (Harrell and Dupont, 2017). The output was visualized using the “corrplot” package (Wei and Simko, 2017).
Cloning of TFs and Promoters
Cloning of GUS, PpbHLH3, VcMYBA1, VcMYBPA1.1 and VcMYBPA2.2 is described previously (Lafferty et al., 2022). Promoter cloning of V. virgatum DFR and UFGT and V. corymbosum ANR, into the pGreenII 0800-LUC vector was described previously (Lafferty et al., 2022). The VcMYBR3.1 sequence was identified in the blueberry reference transcriptome (V. corymbosum RefTrans V1), from the Genome Database for Vaccinium, based on its similarity to VuMYBR3 (KT186105.1). The VcMYBC2.1 gene ID was identified from the blueberry genome based on the differential expression analysis (Supplementary Table S5). Overexpression constructs for VcMYBR3.1 and VcMYBC2.1 and promoter constructs for VcMYBR3.1pro, VcMYBC2.1pro and VcbHLH2pro were made as described in Lafferty et al. (2022) and gene-specific primers used are provided in Supplementary Table S3.
Transient Transformation of Tobacco for Anthocyanin and Proanthocyanidin Production
Agrobacterium strains containing effector (35S:TF) constructs were prepared for infiltration as previously described (Lafferty et al., 2022). Agrobacterium mixtures, containing 1/3 volume of up to three Agrobacterium strains were prepared. This contained the activator MYB TF in combination with PpbHLH3 (Zhou et al., 2018) and either the repressor MYB TF or 35S:GUS (negative control). Approximately 500 μl of the mixture was infiltrated into the abaxial surface of young Nicotiana benthamiana ‘Northern Territory’ leaves using a 1 ml needleless syringe. For each treatment three biological replicates were performed, consisting of an infiltrated leaf on separate plants (Supplementary Figure S5). N. benthamiana growing conditions and Agrobacterium infiltration were as described previously (Lafferty et al., 2022). Leaves were photographed 5 days post-infiltration.
Promoter Activation Assays
For promoter activation assays, Agrobacterium cultures were prepared with 1/10 volume of promoter construct and 3/10 volume of up to three effector constructs, harboring effector TFs or the GUS negative controls. Mixtures were infiltrated into young N. benthamiana (“LAB” strain) leaves. Three leaves were infiltrated per treatment, as biological replicates. Dual luciferase assays were performed with DLAR-2B reagents (Targeting Systems), following the manufacturer’s instructions, using a Tecan Spark® 20 M multimode microplate reader. Values and error bars represent the mean of three biological replicates and ± standard error, respectively. Letters indicate significant differences assessed by one-way ANOVA and post hoc Tukey’s LSD tests (P < 0.05) performed on log-transformed data.
Stable Transformation of Blueberry
The 35Spro:MYBA1:OCS cassette from pKES8 (Plunkett et al., 2018) was cloned into pART27 binary vector (pKES10) and transformed into Agrobacterium tumefaciens GV3101 by electroporation. Agrobacteria were grown overnight in lysogeny broth to A600 = 0.6–0.8, harvested by centrifugation and resuspended in AB vir gene pre-induction medium (Gelvin and Liu, 1994) containing 200 μm acetosyringone to A600 = 0.2. Agrobacterium suspensions were then cultured for 4 h at 28°C and 250 rpm in an orbital shaker.
In vitro stock cultures of V. corymbosum “Draper” × “Legacy” were established from shoot tip and single-node cuttings of glasshouse plants were grown on “micropropagation” medium. Media compositions are provided in Supplementary Table S6. Leaf blade explants were cut transversely several times to produce more sites for callus and shoot regeneration, and pre-cultured for 2 days on ‘co-cultivation’ medium. Pre-cultured leaf blade explants were inoculated with agrobacteria by swirling the AB culture in a 50 ml Falcon tube for 10 min in an orbital shaker. Explants were transferred to filter paper on “co-cultivation medium” and cultured at 22°C for 6 days in the dark. After co-cultivation, explants were washed three times by shaking with 40 ml of liquid “regeneration” medium (“selection” medium without antibiotics or agar) in a 50 ml Falcon tube, rinsed with liquid “regeneration” medium containing 500 mg L−1 cefotaxime, and transferred to solid ‘selection’ medium without filter paper, and kept in the dark for 2 weeks at 25°C. Explants were transferred to new ‘selection’ media every 3 weeks for 5–6 months. Red calli were visible on transformed explants and green shoots on regeneration controls by 4 months after the experiment was set up. As shoots regenerated on explants, they were labeled with explant origin and transferred to “shoot proliferation” medium. Roots were initiated by transferring shoots onto “rooting” medium.
cDNA Synthesis and qRT-PCR Analysis for “Nui” Fruit Developmental Series and 35S:MYBA1 Blueberry Transformants
Vaccinium corymbosum “Nui” fruit from stages 1 to 3 were collected between November and December 2020 to supplement the existing fruit development series. Leaf and stem tissues of regenerating transgenic shoots were obtained from three independent transgenic lines and two regeneration controls, with three biological replicates. Total RNA was extracted from approximately 50 mg of frozen, ground leaf tissue, using the Spectrum™ Plant total RNA kit (Sigma-Aldrich, United States of America) with minor modification, as described previously (Günther et al., 2020). cDNA synthesis and qRT-PCR was performed with three biological replicates, normalized to the geometric mean of GAPDH and Actin (Lafferty et al., 2022). Primer efficiencies were verified using serial dilution with the gene-specific primers listed in Supplementary Table S4.
Results
Differential Expression Analysis of Blueberry and Bilberry Fruit Developmental Series Reveals Candidate Anthocyanin Regulators
A developmental series of berries was obtained as described previously (Lafferty et al., 2022). This consisted of blueberry (V. corymbosum) and bilberry (V. myrtillus) fruit, ranging from immature, small, green fruit (S4) to fully ripened blue fruit (S8). Anthocyanin pigmentation was restricted to the skin of blueberry and was present in both bilberry skin and flesh, developing at S5 and increasing as fruit ripened (Lafferty et al., 2022). The skin and flesh were separated and these samples were used for RNA sequencing (RNA-seq) analysis. RNA-seq reads were mapped against a tetraploid blueberry genome assembly, which included all four haplotypes (Colle et al., 2019).
Highly differentially expressed genes (HDEGs) had a log2-fold change greater than 2, with an adjusted P-value < 0.01. Comparisons were performed between ripe (S7) and unripe (S4) blueberry skin and bilberry skin and flesh, ripe (S7) blueberry skin and flesh, and ripe (S7) blueberry and bilberry flesh (Figure 1A). These comparisons were structured to filter out genes that were involved in ripening- and tissue-specific processes. In total, 839 genes were identified as being HDEGs (Supplementary Table S1). Their deduced amino acid sequences were extracted from the blueberry genome and annotated using Mercator4 V2.0 (Schwacke et al., 2019). The majority of HDEGs with annotation were predicted to have roles in either secondary metabolism, RNA biosynthesis or solute transport (Figure 1B), and 403 HDEGs were unannotated.
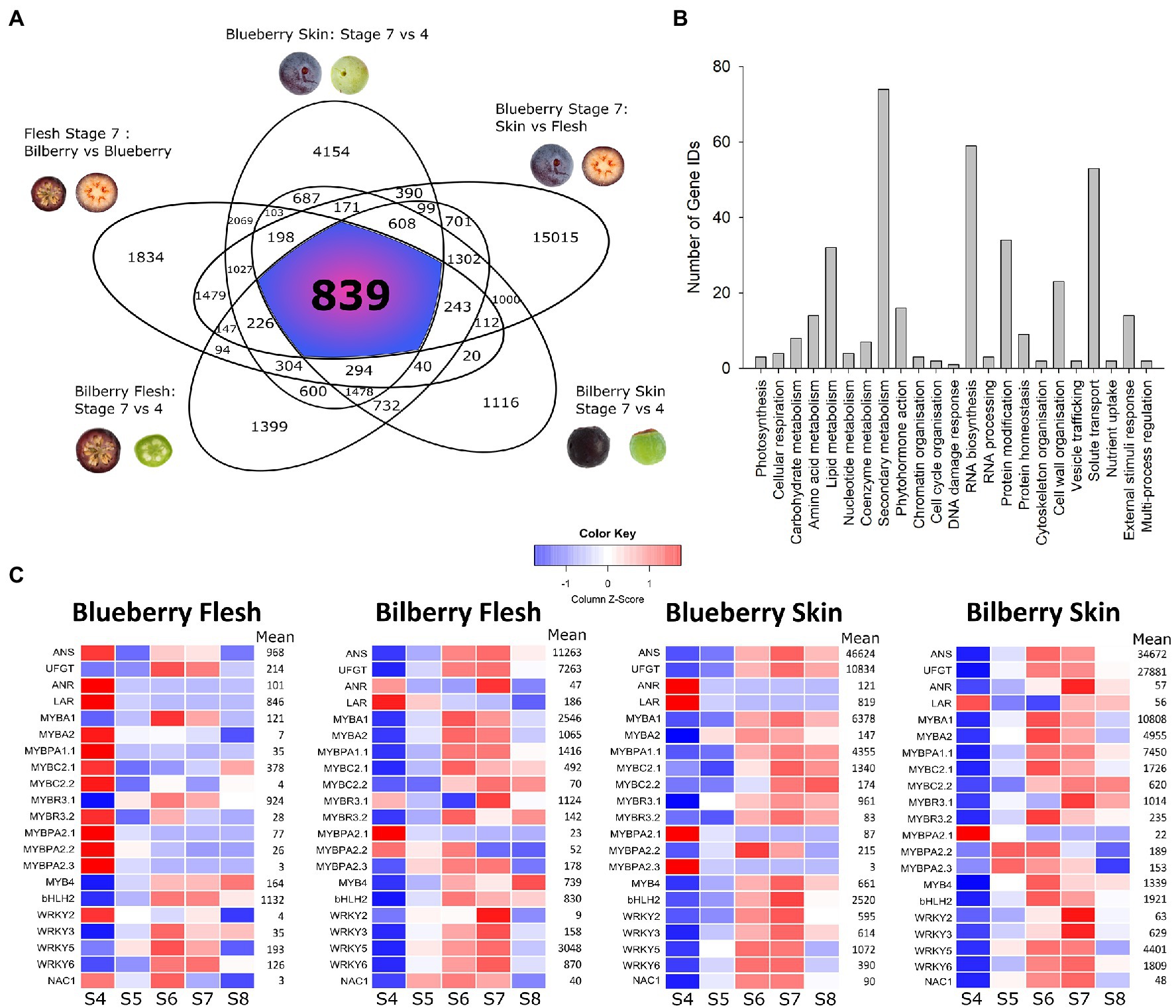
Figure 1. Differential expression analysis of the blueberry and bilberry RNA-seq dataset. (A) Highly differentially expressed genes (HDEGS) were identified by comparing high and low anthocyanin-producing tissues. HDEGs were described as having a log2-fold change greater than 2 in high-anthocyanin tissues, with an –adjusted p-value < 0.01. HDEGs consistently present across each comparison were identified and visualized as a Venn diagram. Comparisons were between blueberry skin S7 vs. S4, blueberry S7 skin vs. flesh, bilberry skin S7 vs. S4, bilberry flesh S7 vs. S4 and blueberry vs. bilberry S7 flesh. (B) HDEGs consistent across all comparisons were annotated using Mercator4 (V2.0), which assigned Mapman bin numbers. This was visualized as a bar graph in SigmaPlot (V14.0). (C) The developmental expression patterns of candidate anthocyanin transcription factors (TFs), SG5 MYBPA1-TFs and representative biosynthetic genes were visualized as a heatmap. ANS and UFGT represented anthocyanin biosynthesis while ANR and LAR represented proanthocyanidin (PA) biosynthesis. Raw counts were normalized via the median of ratios method. Count data were visualized as Z-scores, which calculated how many standard deviations a data point was from the mean of a gene across all samples in the given tissue, with blue and red representing negative and positive Z-scores, respectively. Mean represents the mean counts for each gene, in each tissue type, across development.
The expression profiles of general flavonoid and anthocyanin biosynthetic genes and transporters were compared between blueberry and bilberry ripe (S7) flesh. The log2-fold change of these genes was visualized (Supplementary Figure S1). Phenylpropanoid and flavonoid biosynthetic and transporter genes were highly upregulated in bilberry S7 flesh, with PAL, C4H, 4CL, CHS, F3′5′H, ANS, UFGT, GST, and MATE having approximately five log2-fold higher expression.
A number of HDEGs were annotated as being TFs (Supplementary Table S5). These were further annotated by finding the best BLAST hit to Arabidopsis. Up to four gene IDs may correspond to the same gene owing to all four haplotypes being represented in the tetraploid blueberry genome. Taking this into consideration, eight unique MYB genes were present in the analysis. Phylogenetic analysis identified the subgroups to which these R2R3 MYBs or R3 MYBs belonged (Supplementary Figure S2). This included the previously characterised SG6 VcMYBA1 and MYBPA1-type VcMYBPA1.1 (Zifkin et al., 2012; Plunkett et al., 2018). An additional SG6 MYB, named VcMYBA2, was present, which contained the [R/K]P[R/Q][P/R]RTF SG6 motif (Plunkett et al., 2018). Two SG4 MYBs were identified, named VcMYBC2.1 and VcMYBC2.2, and these contained both the [L/F]PDLN[L/F]x EAR and TLLLFR repression motifs (Ohta et al., 2001; Matsui et al., 2008). Additionally, there was a SG1 MYB, named VcMYB4 and an R3 MYB, named VcMYBR3.2. Furthermore, there were four unique WRKY TFs, VcWRKY2, VcWRKY3, VcWRKY5 and VcWRKY6, two bHLH TFs, and a NAC TF (Supplementary Table S5).
Expression and Correlation Analysis of Biosynthetic Genes and Candidate Anthocyanin Regulators
The expression profiles of flavonoid biosynthetic genes and candidate TFs, found by the differential expression analysis, were visualized (Figure 1C; Supplementary Figure S3). An anthocyanin related bHLH (bHLH2) and the SG5 R2R3 MYB PA regulators (MYBPA2.1–3), identified previously, were included (Günther et al., 2020; Lafferty et al., 2022). Additionally an R3 MYB, VcMYBR3.1, was included based on its sequence similarity to other R3 MYBs with established roles in anthocyanin regulation from other plants (Zhu et al., 2009; Albert et al., 2014; Primetta et al., 2015). The general flavonoid biosynthetic genes PAL, C4H, CHS, CHI, F3H, F3′H, F3′5′H, DFR, LDOX, and ANS, transporters GST and MATE8 and the anthocyanin specific UFGT all had similar expression profiles to each other (Figure 1C; Supplementary Figure S3). ANS and LDOX genes in blueberry were distinguished phylogenetically (Supplementary Figure S4). In blueberry skin, and bilberry skin and flesh, their transcript abundance increased at S6, peaked at S7 and then declined slightly at S8. This correlated with visible pigmentation (Lafferty et al., 2022), with this pattern described as being anthocyanin-related. In blueberry flesh, the expression of these genes was much lower, shown by the mean of the normalized count data. In blueberry flesh, transcript abundance for PAL, C4H, CHS, CHI, F3H, F3′5′H, DFR, LDOX, and MATE was highest in blueberry flesh at S4 (when PAs were probably still being synthesized), and declined as ripening proceeded. The PA specific biosynthetic genes ANR and LAR also shared this expression pattern. In both species, the mean count data were higher in the skin than the flesh and many genes, CHS, DFR, UFGT, and GST in particular, had higher count data in bilberry skin than in blueberry skin.
In blueberry skin and bilberry skin and flesh many candidate TFs also had an anthocyanin related expression pattern (Figure 1C). This included MYBA2, MYBPA1.1, MYBC2.1, MYBC2.2, MYBR3.2, WRKY2, and NAC1. Furthermore, MYBA1, MYBR3.1, MYB4, WRKY3, WRKY5, WKRY6, and bHLH2 had an anthocyanin related expression pattern in all tissues, including blueberry flesh. These TFs had much lower mean counts (normalized via the median of ratios method) in blueberry flesh, with the exception of MYBR3.1 and bHLH2, which had similar mean count data to bilberry flesh. The mean count data of these TFs were generally much higher in skin tissues, aligning with observations for the candidate genes. The SG5 MYBPA2.1 was most highly expressed at S4 in all tissues, corresponding to PA biosynthesis, while MYBPA2.3 showed this expression profile in blueberry skin and flesh. MYBPA2.2 and MYBPA2.3 had a more ripening-related expression pattern, but only in bilberry. The mean count data of the SG5 MYBs was relatively low in all tissues.
MYBA1 and MYBPA1.1 are key anthocyanin regulators in Vaccinium (Karppinen et al., 2021; Lafferty et al., 2022). Correlation analysis was performed to identify which candidate TFs strongly correlated with the expression of these TFs and of the anthocyanin related biosynthetic genes ANS and UFGT (Supplementary Figure S5). In blueberry skin and bilberry skin and flesh, bHLH2 consistently correlated with these genes, forming an anthocyanin-related cluster. In both bilberry tissues, this cluster also included MYBC2.1 and in bilberry skin MYBA2, MYB4, MYBR3.2 and WRKY5 were additionally present. In blueberry skin, the anthocyanin-related cluster contained MYB4, MYBR3.1, WRKY2, WRKY3, and NAC1. Blueberry flesh lacked many of the significant gene correlations seen in the other tissues, although significant correlations between UFGT, MYBA1, MYBR3.1, and bHLH2 were present.
SG4 and R3 MYB Repressors Inhibit Both the PA and Anthocyanin Biosynthesis Pathways
Two MYB repressors were analyzed further to elucidate their role in anthocyanin regulation. MYBC2.1 was identified in the differential expression and phylogenetic analysis as a candidate SG4 MYB repressor of anthocyanin biosynthesis (Figure 1C; Supplementary Figure S2). An R3 MYB repressor, MYBR3.1, was chosen as it was expressed in blueberry fruit flesh, where it may contribute to the lack of anthocyanins in this tissue. MYBR3.1 was not identified as an HDEG in the transcriptomic analysis due to its expression in blueberry flesh (Figure 1C). Although both MYBC2.2 and MYBR3.2 were identified as being HDEGs, they were not analyzed further due to their minimal expression in fruit tissues. Gene expression analysis, via qRT-PCR, across a full blueberry developmental series revealed that VcMYBC2.1 and VcMYBR3.1 were expressed highly during early development (S1–S3; Supplementary Figure S6). Because of this expression profile, it was hypothesized that these candidate TFs could also repress the PA pathway.
VcMYBC2.1 and VcMYBR3.1 were isolated from blueberry and cloned into overexpression vectors for functional analyses. The MYB activators VcMYBA1 and VcMYBPA2.2 regulate anthocyanin and proanthocyanidin biosynthesis, respectively, and function within MBW complexes (Karppinen et al., 2021; Lafferty et al., 2022). We were unable to include VcbHLH2 in these assays because despite amplifying bHLH2 cDNAs from blueberry (and bilberry), we found this sequence was unstable in plasmids, possibly due to a microsatellite repeat within exon six. Therefore, we used the orthologue from Prunus persica, PpbHLH3. Transient overexpression of either VcMYBA1 or VcMYBPA2.2 in N. benthamiana leaves, with PpbHLH3, resulted in strong anthocyanin and PA accumulation, respectively (Figure 2; Supplementary Figure S7). The presence of PAs was observed using p-dimethylaminocinnamaldehyde (DMACA) staining. Co-infiltration of either VcMYBC2.1 or VcMYBR3.1 with these MYB activators substantially reduced the amount of pigmentation and PA produced.
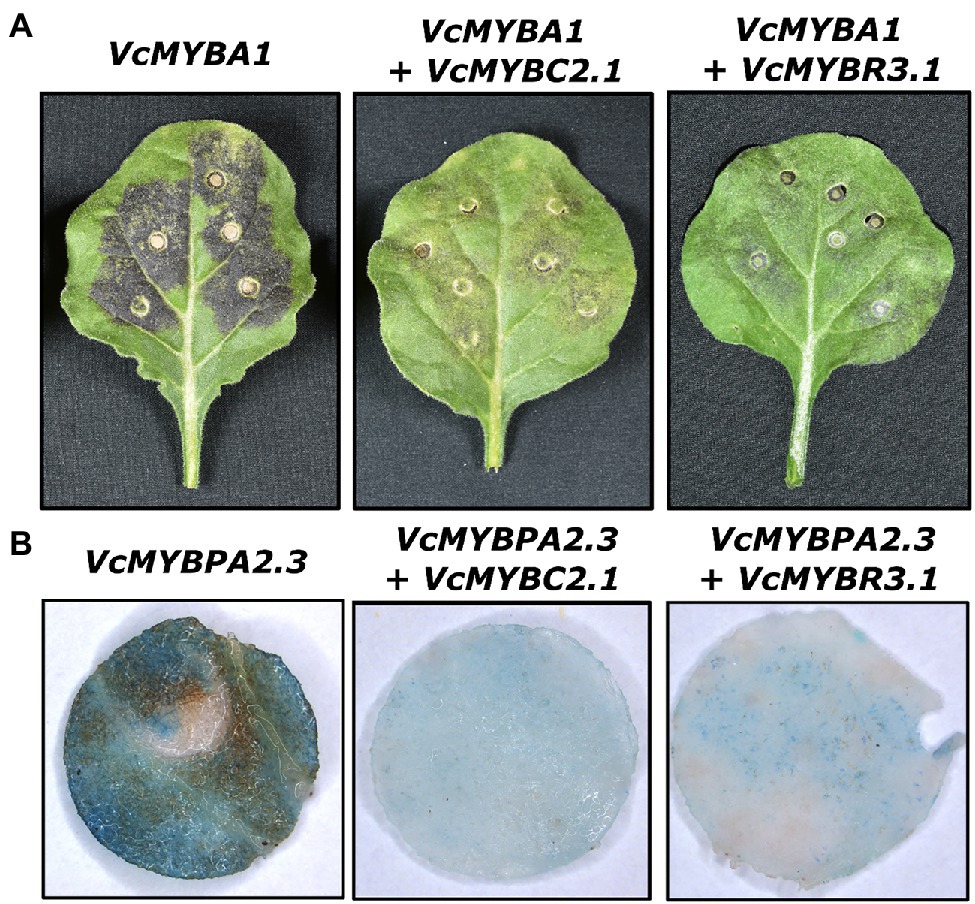
Figure 2. VcMYBC2.1 and VcMYBR3.1 inhibit anthocyanin and proanthocyanidin accumulation. Representative phenotypes of Agrobacterium infiltrated Nicotiana benthamiana leaves expressing MYB activators and PpbHLH3, and co-expressing the repressors VcMYBC2.1 or VcMYBR3.1. The SG6 MYB activator VcMYBA1 was used for anthocyanin accumulation assays (A) while the SG5 MYB activator VcMYBPA2.3 was used for proanthocyanidin accumulation assays (B) Leaves were photographed 7 days post infiltration. For proanthocyanidin accumulation studies, 1 cm diameter leaf discs were taken and DMACA stained before being photographed.
Promoter activation assays were performed on the Vaccinium DFR, ANR and UFGT promoters to assess the repression activity of these TFs (Figure 3). VcMYBA1 and VcMYBPA1.1 significantly activated DFRpro, VcMYBPA2.2 significantly activated ANRpro and VcMYBA1 significantly activated UFGTpro, either with and without PpbHLH3. The addition of VcMYBC2.1 in transient infiltrations significantly reduced activation of the DFRpro by VcMYBA1, VcMYBPA1.1 and VcMYBPA2.3 (all with PpbHLH3). Furthermore, there was significantly lower activation of UFGTpro when VcMYBC2.1 was co-infiltrated with VcMYBA1 and PpbHLH3. Co-infiltration of VcMYBPA2.3 and PpbHLH3 activated the ANRpro and addition VcMYBC2.1 had no effect on this activation. Combining VcMYBR3.1 with activator MYBs and PpbHLH3 had no effect on promoter activation. R3-MYB repressors are proposed to function by binding and titrating bHLH proteins (Albert et al., 2014), therefore the effects of altering the amount of supplied bHLH with VcMYBR3.3 was examined. Removal of PpbHLH3 from MYB activator and repressor co-infiltrations resulted in a significant drop in activation of all promoters, which was further reduced when the amount of VcMYBR3.1 co-infiltrated was doubled.
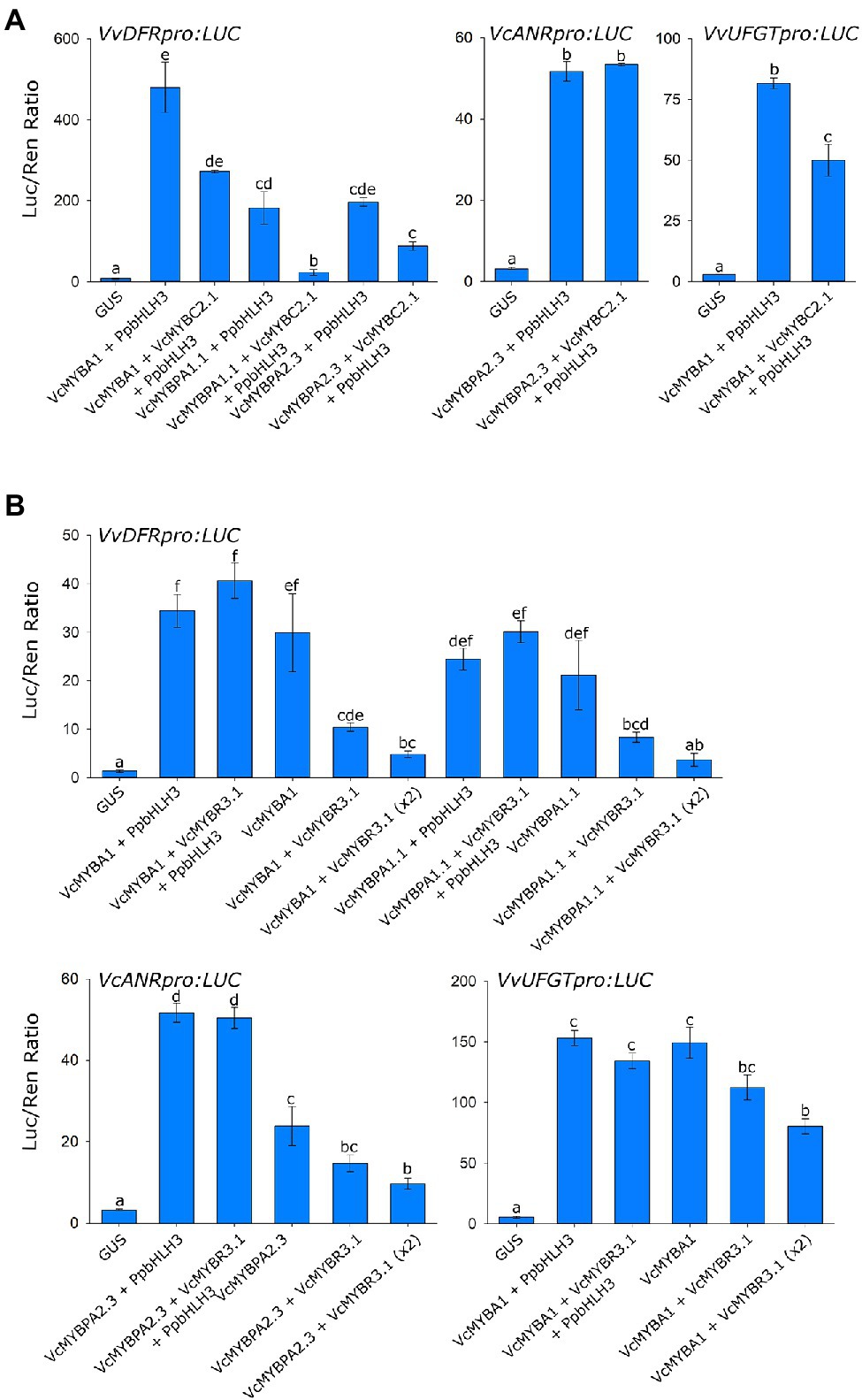
Figure 3. VcMYBC2.1 and VcMYBR3.1 repress anthocyanin and proanthocyanidin biosynthetic gene expression. Promoter activation assays were performed in Nicotiana benthamiana leaves via Agrobacterium-mediated co-infiltration of MYB activators and PpbHLH3, with and without either (A) VcMYBC2.1 or (B) VcMYBR3.1, against the DFR, ANR and UFGT promoter:Luciferase constructs. GUS was used as a negative control. Values represent the mean of three biological replicates. Error bars show ± SEM. Letters indicate significant differences between treatments, assessed by one-way ANOVA and post hoc Tukey’s test (P < 0.05) on log-transformed data.
Proanthocyanidin and Anthocyanin TFs Are Hierarchically Regulated
The correlation of MYBC2.1, MYBR3.1, and bHLH2 with the MYBA1 activator, in both blueberry and bilberry (Supplementary Figure S5), suggests they may be regulated by MYBA1 in an activator hierarchy. The three TFs are also involved in PA regulation and may also be regulated by MYBPA2.2 during early stages of fruit development. The promoters of the genes were isolated and cloned into dual luciferase vectors for promoter activation assays. All treatments included PpbHLH3, which was incapable of activating any promoters by itself. The VcMYBPA2.2, VcMYBA1, and VcMYBPA1.1 activators significantly activated VcMYBC2.1pro and VcbHLH2pro (Figure 4). No TF combinations assayed against the VcMYBR3.1pro were able to induce activity of this promoter.
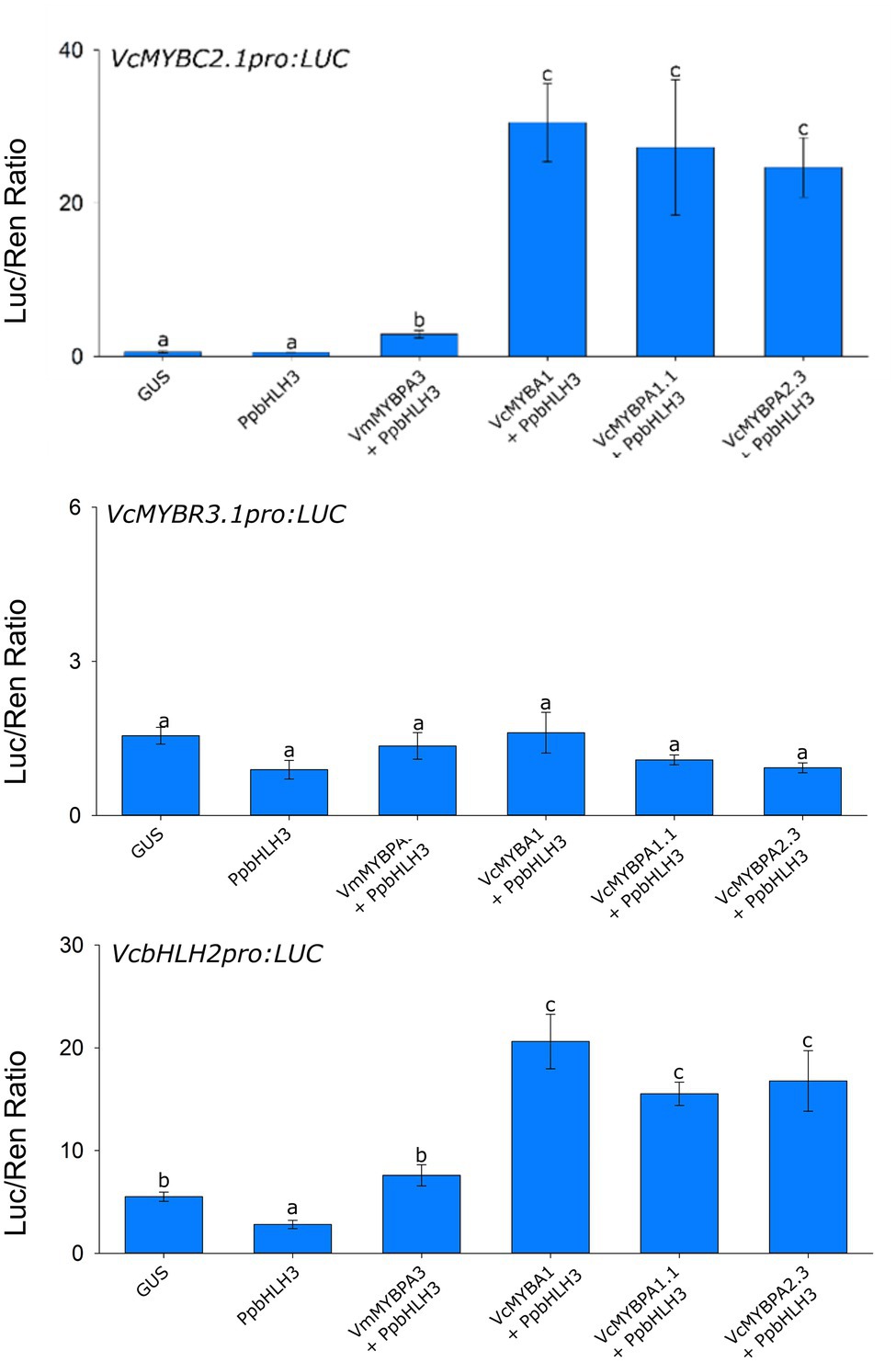
Figure 4. Hierarchical regulation of transcription factors by MYB activators. Promoter activation assays were performed in the leaves of N. benthamiana via Agrobacterium-mediated co-infiltration of SG5, SG6 and MYBPA1-type MYB activators, with PpbHLH3, against the VcMYBC2.1, VcMYBR3.1, VcMYBPA1.1 and VcbHLH2 promoter:Luciferase constructs. Values represent the mean of three biological replicates. Error bars are ± SEM. Letters indicate significant differences between treatments, assessed by one-way ANOVA and post hoc Tukey’s test (P < 0.05) on log-transformed data.
VcMYBA1 Overexpression in Blueberry Induces Anthocyanin Production
Evidence to date suggests that in addition to SG6/MYBA regulators, MYBPA1 and bHLH proteins are also required for regulating anthocyanins in bilberry and blueberry (Plunkett et al., 2018; Karppinen et al., 2021; Lafferty et al., 2022). To examine whether MYBA1 behaves as a high-level regulator within the hierarchy of TFs, we generated stable blueberry plants overexpressing VcMYBA1 from a CaMV35S promoter. The regenerating transformed callus developed deep red anthocyanin pigmentation and gave rise to plants with dark red leaves, stems and roots (Figure 5A; Supplementary Figure S8). Activation of the anthocyanin biosynthesis genes VcANS and VcUFGT in the 35S:MYBA1 lines was confirmed by qRT-PCR, with minimal expression detected in the regeneration controls (Figure 5B). In contrast, the PA biosynthetic gene ANR and the regulators MYBPA1.1 and MYBPA2.3 were highly expressed in the regeneration controls, suggesting PAs were accumulating in these tissues. This was confirmed by staining leaves with DMACA, which revealed a strong accumulation of PAs in the regeneration controls (Supplementary Figure S9). The presence of high concentrations of anthocyanins in 35S:MYBA1 plants obscured detection of PAs with DMACA staining, despite clearing tissues with acetic acid/ethanol. In two independent 35S:MYBA1 lines (1 and 3), MYBPA1.1 and MYBPA2.3 expression was moderately reduced with a commensurate reduction in ANR transcripts (Figure 5B). The expression of MYBC2.1 and bHLH2 were not greatly altered compared to controls.
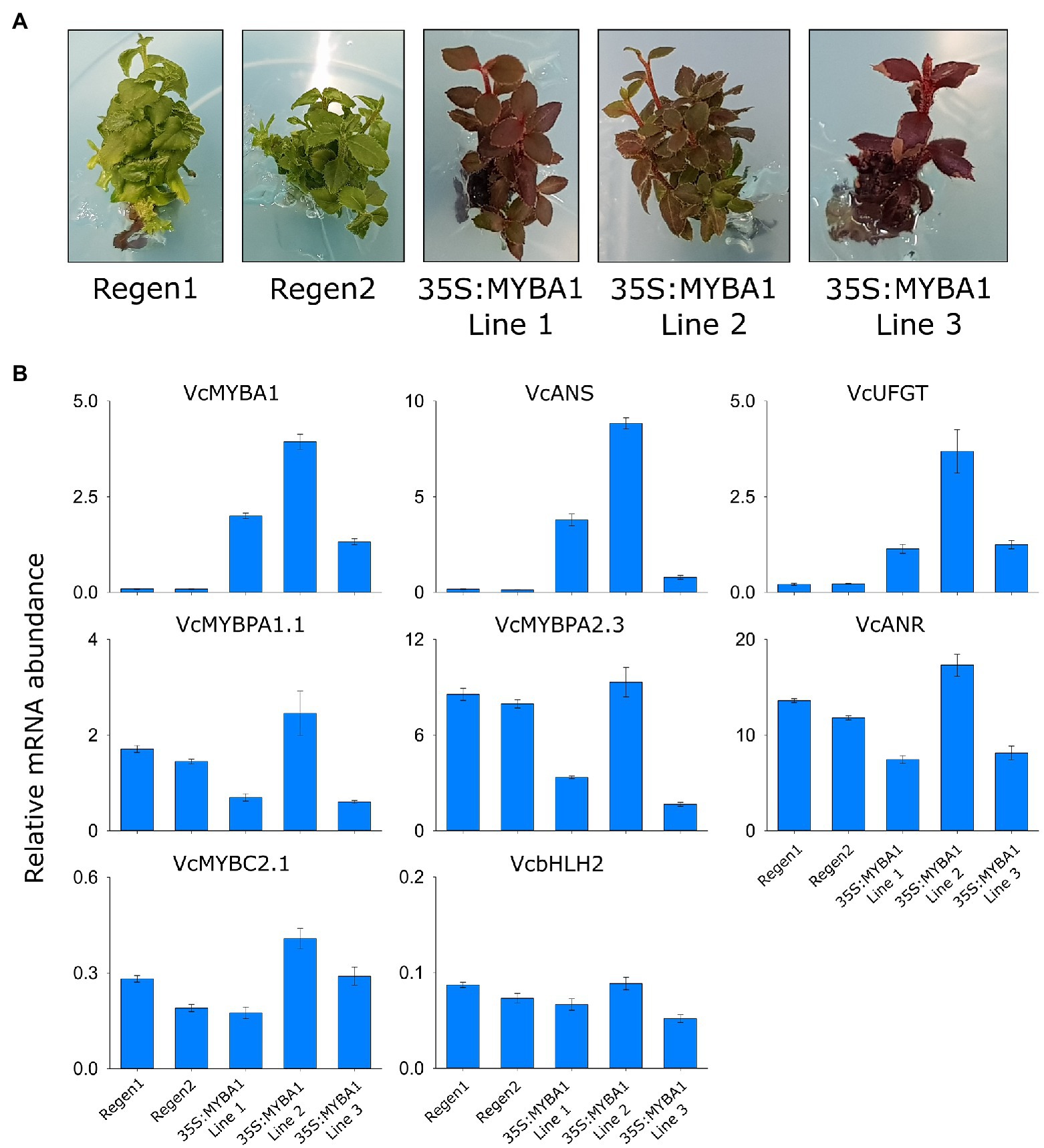
Figure 5. Stable overexpression of VcMYBA1 in blueberry was sufficient to enhance vegetative anthocyanin content. (A) Pigmentation phenotype of two regeneration lines, Regen 1 and 2, and three 35S:VcMYBA1 lines in tissue culture. (B) Expression analysis was performed on the regeneration controls and 35S:VcMYBA1 lines using qRT-PCR. Values represent means ± SEs of three biological replicates. All values are relative to the geometric mean of abundance of GAPDH and Actin reference gene transcripts.
Discussion
Anthocyanin and PA biosynthesis in Vaccinium is complex, involving several known activator and repressor TFs, and potentially additional uncharacterized TFs. This study aimed to unravel the regulation of these pathways and identify the conserved elements of anthocyanin and PA regulation. We used differential expression and correlation analysis of blueberry and bilberry fruit developmental series to identify candidate anthocyanin-related TFs, and to determine the expression patterns of the flavonoid-related biosynthetic and TF genes during berry development. Stable transformation and functional analyses were then used to reveal a regulatory hierarchy between candidate TFs that facilitates strong activation of both pathways at different stages of berry development.
SG6 R2R3 MYBs are well-characterized anthocyanin regulators, and MYBA1 is a key activator of anthocyanin biosynthesis in both blueberry and bilberry (Plunkett et al., 2018; Karppinen et al., 2021). Recently, a MYBPA1-type TF (MYBPA1.1) was shown to co-regulate the pathway and to be regulated by MYBA1 (Karppinen et al., 2021; Lafferty et al., 2022). Both MYBA1 and MYBPA1.1 were strongly correlated with anthocyanin production and were expressed in bilberry red flesh, albeit at a lower concentration than in skin, while having minimal expression in pale blueberry flesh. These TFs correlated strongly with the anthocyanin specific biosynthetic genes ANS and UFGT in all anthocyanin-producing tissues (Supplementary Figure S4). Stable overexpression of VcMYBA1 in blueberry was sufficient to elevate the anthocyanin content in vegetative tissues, with ANS and UFGT being highly upregulated (Figure 5). Interestingly, the leaves of the regeneration controls also contained high amounts of PAs, shown by DMACA staining, and the PA-specific regulator MYBPA2.3 and biosynthesis gene ANR are highly expressed (Figure 5; Supplementary Figure S9). This finding was somewhat unexpected, but provides further support for the hierarchical regulation of TF genes (bHLH2, MYBPA1.1, and MYBC2.1) by MYBPA2.3 (Figure 4; Lafferty et al., 2022). However, this makes it very difficult to separate hierarchical regulation by MYBA1 overexpression from the endogenous MYBPA2.3 action. It is anticipated that the 35S:MYBA1 blueberry plants will have additional phenotypes when they have been exflasked and reached maturity, which may include more intensely colored flowers and fruit with altered flesh color.
bHLH proteins are required for the formation of MBW complexes, and are themselves regulated by these complexes via a mechanism referred to as hierarchical regulation (Baudry et al., 2004, 2006; Albert et al., 2014). bHLH regulation has been reported by SG5 MYBs in Arabidopsis (Baudry et al., 2006; Xu et al., 2013), by SG6 MYBs in Arabidopsis, petunia, Antirrhinum and kiwifruit (Baudry et al., 2006; Xu et al., 2013; Albert et al., 2014, 2021; Liu et al., 2021) and by MYBPA1-type MYBs in grape (Hichri et al., 2010). VcbHLH2, previously identified as a potential PA and anthocyanin regulator in blueberry (Günther et al., 2020; Lafferty et al., 2022), was found to correlate with anthocyanin production and biosynthetic gene activity in tissues containing high concentrations of anthocyanins (Figure 1C; Supplementary Figure S5). The VcbHLH2 promoter was activated by the SG5 VcMYBPA2.3, SG6 VcMYBA1 and MYBPA1-type VcMYBPA1.1 activators, revealing that bHLH2 was also hierarchically regulated in Vaccinium (Figure 4). Hierarchical regulation of bHLH2 by MYBPA2.3 explains why bHLH2 is expressed in wild type blueberry leaves (Figure 4); MYBPA2.3 is highly expressed and regulates PA biosynthesis, bHLH2 and other TF targets (see below). Interestingly, overexpression of MYBA1 was unable to increase VcbHLH2 expression any further (Figure 5).
MYB repressors have previously been proposed to fine-tune the anthocyanin and PA pathways, preventing over-accumulation of these metabolites (Albert et al., 2014; Albert, 2015). In this study, the R3 MYB, MYBR3.1, was expressed during early berry development, when PAs are accumulating, and at ripening, when anthocyanins are produced (Figure 2A), and functional assays demonstrate VcMYBR3.1 inhibits the production of both metabolites (Figure 2B). This suggests that R3-MYB repressors are also important for regulating these pathways in Vaccinium berries, as they are in other characterized systems (Schellmann et al., 2002; Zhu et al., 2009; Albert et al., 2014; Cao et al., 2017; Colanero et al., 2018; Ding et al., 2020).
MYBR3.1 has repressive activity upon anthocyanin and PA biosynthesis that is dependent on bHLH concentration. The addition of PpbHLH3 in promoter activation assays removed its repression activity, while doubling the amount of VcMYBR3.1 in infiltrations increased the inhibitory effect on activator MYBs (Figure 2C). This is consistent with the proposed role of R3 MYB repressors titrating bHLH proteins (Zhu et al., 2009; Albert et al., 2014). The activator MYBs assayed (MYBA, MYBPA1, MYBPA2) were probably able to function without the addition of PpbHLH3 because of the presence of endogenous N. benthamiana bHLHs (Montefiori et al., 2015). R3 MYB repressors can also be hierarchically regulated by activator MYBs, as reported in petunia and tomato (Albert et al., 2014; Yan et al., 2020). This was, however, not seen for the VcMYBR3.1 promoter assayed in this study. Additionally, MYBR3.1 had similar expression in blueberry and bilberry flesh, regardless of MYB activators having minimal expression in blueberry flesh. This indicated an additional (MYBA1-independent) regulatory mechanism regulates MYBR3.1 expression in blueberry flesh at ripening. This is further supported by the analysis of an albino bilberry mutant, in which VmMYBR3 had elevated expression (Zorenc et al., 2017), where VmMYBA1 expression is expected to be minimal.
VcMYBC2.1 encodes a SG4 MYB and was identified in the expression analyses that was expressed highly during early development and ripening, indicating it was a repressor of both PA and anthocyanin production. VcMYBC2.1 correlated strongly with anthocyanin biosynthesis and UFGT in anthocyanin rich tissues (Figure 1C; Supplementary Figure S5). The anthocyanin and PA accumulation and promoter activation assays confirmed that VcMYBC2.1 was a repressor of both pathways (Figure 3). In other plant species, expression of the SG4 MYB genes correlated with the pathways they repress, because they are themselves regulated by SG5 and/or SG6 MYBs (Albert et al., 2014; Jun et al., 2015; Yoshida et al., 2015; Huang et al., 2019). The hierarchical regulation by both SG5 and SG6 MYBs was found in Vaccinium (Figure 6), supporting this as a conserved aspect of anthocyanin and PA pathway regulation in different tissues across eudicots.
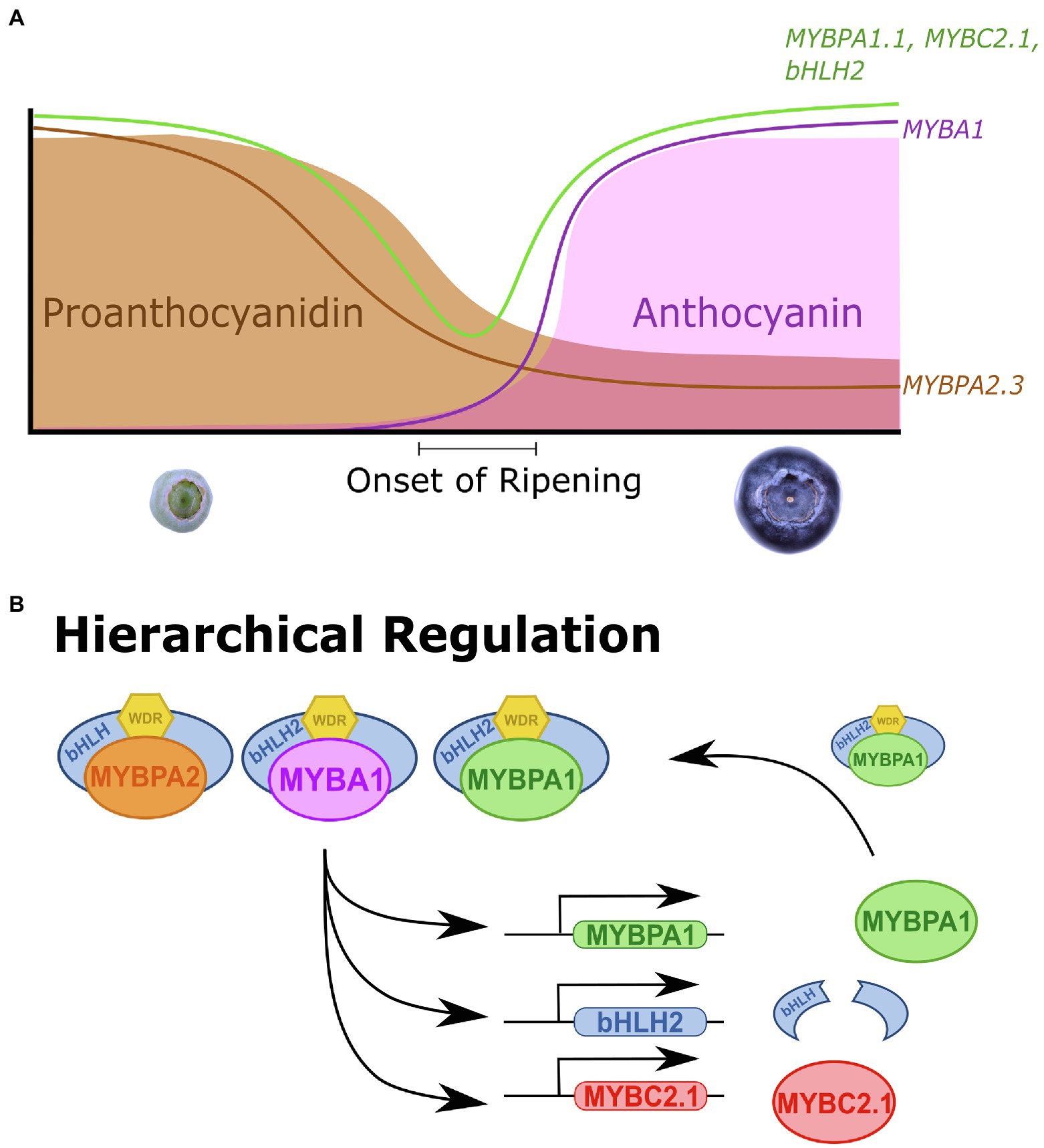
Figure 6. Vaccinium MYB activators and repressors are coordinately regulated for precise control of proanthocyanidin and anthocyanin biosynthesis. (A) During early stages of Vaccinium fruit development (S1–S3) proanthocyanidin (PA) concentrations are high, before declining as development proceeds. This correlates with the expression of the MYBPA2.3, a PA-specific SG5 MYB activator. At the onset of ripening (S5) anthocyanins begin accumulating and concentrations rise as ripening progresses, correlating with the anthocyanin-specific SG6 MYB, MYBA1. MYBPA1.1, MYBC2.1, MYBR3.1, and bHLH2 regulate both PA and anthocyanin biosynthesis and this was reflected in their biphasic expression pattern, being more strongly expressed during early development and again during ripening. (B) SG5 MYBPA2, SG6 MYBA1 and MYBPA1-type MYBPA1 MBW complexes hierarchically regulate MYBPA1, bHLH2, and MYBC2.1. This establishes feed forward activation for bHLH2 and MYBPA1 and a feedback repression loop for MYBC2.1.
Hierarchical activation of MYBPA1 by SG5 MYB activators has been reported in grape, poplar, apple and blueberry (Terrier et al., 2009; James et al., 2017; Wang et al., 2018; Lafferty et al., 2022) and activation by SG6 MYB activators has recently been shown in blueberry (Lafferty et al., 2022). Furthermore, MYBPA1 proteins have been shown to activate the promoters of many general flavonoid biosynthetic genes leading to PA and anthocyanin production (Bogs et al., 2007; Ravaglia et al., 2013; Karppinen et al., 2021; Lafferty et al., 2022). However, the position of the different TF types in the wider context of flavonoid production and hierarchical regulation has not been elucidated. Here we show that VcMYBPA1.1 could activate the VcbHLH2 promoter, contributing to the feedforward activation loop. Additionally, VcMYBPA1.1 could activate the VcMYBC2.1 promoter as part of a feedback repression loop. This establishes MYBPA1 proteins as important hierarchical regulators in the wider PA and anthocyanin regulatory modules, expanding the models proposed by Albert et al. (2014) of hierarchical regulation within the MBW complex.
Our results underpin a proposed model that highlights the complex nature of Vaccinium anthocyanin and PA regulation (Figure 6). During early fruit development PA metabolites are produced in unripe fruit, deterring herbivory when seeds are not mature (Jaakola et al., 2002; Zifkin et al., 2012). The PA specific SG5 MYBPA2 activators are expressed, which hierarchically regulates the MYBPA1 (Lafferty et al., 2022) and bHLH2 activators. This functions as a feed forward activation loop, providing additional components for MBW complex formation. The activators also regulate the SG4 MYB repressor MYBC2.1, establishing a feedback repression loop (Albert et al., 2014), which limits the activation of the pathway. MYBR3.1 also represses the PA pathway, although how it fits into the regulatory system is unresolved. PA concentration and MYBPA2 expression fall as development proceeds. Anthocyanin content rises at the onset of ripening (Jaakola et al., 2002; Zifkin et al., 2012). This coincides with a rise in expression of the anthocyanin-specific SG6 MYBA1 TF. The expression of MYBPA1 and bHLH2 activators and the MYBC2.1 repressor also increase, concurrent with the feedforward activation and feedback repression loops regulating anthocyanin biosynthesis, along with MYBR3.1. The lack of anthocyanin content in blueberry flesh is probably due to the minimal expression of MYBA1. Target TF and biosynthetic genes also have minimal expression and the anthocyanin pathway was unable to be activated. It remains to be seen if MYBA1 is indeed the limiting factor for flesh color, but our transgenic results show strong anthocyanin accumulation in all vegetative tissues with the overexpression of MYBA and, based on data from other species, it is likely that this will extend to flesh colors (Espley et al., 2007; Lin-Wang et al., 2014; Rinaldo et al., 2015; Hijaz et al., 2018).
In conclusion, transcriptomic analysis in fruit skin and flesh samples across blueberry and bilberry development identified a number of TFs that strongly correlate with anthocyanin pigmentation in ripening blueberry skin and bilberry skin and flesh. These included two distinct MYB repressors, MYBC2.1 and MYBR3.1, which were functionally characterized. Promoter activation assays revealed MYBPA1 as an important hierarchical regulator for PA and anthocyanin regulation, being able to activate VcbHLH2 and VcMYBC2.1. These TFs were also activated by the SG5 MYBPA2 and SG6 MYBA1 proteins. This confirmed that regulation of PA and anthocyanin biosynthesis is complex, requiring coordinated expression of both activator and repressor TFs. Furthermore, the overexpression of MYBA1 was sufficient for strong activation of the anthocyanin pathway in transgenic blueberry plants. In the transcriptomic analyses MYBA1 was expressed in bilberry skin and flesh and blueberry skin, with minimal expression in blueberry flesh. Based on these results we conclude that the expression of MYBA1 in bilberry flesh is probably the crucial determinant of red flesh.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, with NCBI accessions PRJNA591663 and PRJNA739815.
Author Contributions
DJL: performed experimental work and data analysis; CHD and CSG: provided bioinformatic support; LJ and KK: providing berry samples; MB, LW, and HL: produced transgenic lines; NWA, APD, RVE, and ACA: conceptualization. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the New Zealand Ministry of Business, Innovation, and Employment contract C11X1704 “Filling the Void: boosting the nutritional content of New Zealand fruit.”
Conflict of Interest
Authors DJL, RVE, CHD, APD, CSG, MB, LW, ACA, and NWA were employed by the New Zealand Institute for Plant and Food Research Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Rebecca Kirk for coordination of the program, Ian King for care of Nicotiana plants, Andrew Mullan and Belinda Diepenheim for preparation of tissue culture media, and Steve Arathoon for lab support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.910155/full#supplementary-material
Abbreviations
PA, Proanthocyanidin; TF, Transcription factor; CHS, Chalcone synthase; F3′5′ H, Flavonoid 3′5′ -hydroxylase; DFR, Dihydroflavonol 4-reductase; ANS, Anthocyanidin synthase; UFGT, UDP-glucose:flavonoid 3-O-glucosyltransferase; GST, Glutathione S-transferase; MATE, Multidrug and Toxic Compound Extrusion; ANR, Anthocyanidin reductase; LAR, Leucoanthocyanidin reductase; FLS, Flavonol synthase; MYB, Myeloblastosis; bHLH, Basic helix loop helix.
References
Aharoni, A., De Vos, C. R., Wein, M., Sun, Z., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/j.1365-313X.2001.01154.x
Albert, N. W. (2015). Subspecialization of R2R3-MYB repressors for anthocyanin and proanthocyanidin regulation in forage legumes. Front. Plant Sci. 6:1165. doi: 10.3389/fpls.2015.01165
Albert, N. W., and Allan, A. C. (2021). MYB genes involved in domestication and crop improvement. Annu. Plant Rev. Online 4, 199–242. doi: 10.1002/9781119312994.apr0767
Albert, N. W., Butelli, E., Moss, S. M. A., Piazza, P., Waite, C. N., Schwinn, K. E., et al. (2021). Discrete bHLH transcription factors play functionally overlapping roles in pigmentation patterning in flowers of Antirrhinum majus. New Phytol. 231, 849–863. doi: 10.1111/nph.17142
Albert, N. W., Davies, K. M., Lewis, D. H., Zhang, H., Montefiori, M., Brendolise, C., et al. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26, 962–980. doi: 10.1105/tpc.113.122069
Baudry, A., Caboche, M., and Lepiniec, L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46, 768–779. doi: 10.1111/j.1365-313X.2006.02733.x
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Bogs, J., Jaffé, F. W., Takos, A. M., Walker, A. R., and Robinson, S. P. (2007). The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 143, 1347–1361. doi: 10.1104/pp.106.093203
Cao, X., Qiu, Z., Wang, X., Van Giang, T., Liu, X., Wang, J., et al. (2017). A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit. J. Exp. Bot. 68, 5745–5758. doi: 10.1093/jxb/erx382
Chen, H., and Boutros, P. C. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 12:35. doi: 10.1186/1471-2105-12-35
Colanero, S., Perata, P., and Gonzali, S. (2018). The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front. Plant Sci. 9:830. doi: 10.3389/fpls.2018.00830
Colle, M., Leisner, C. P., Wai, C. M., Ou, S., Bird, K. A., Wang, J., et al. (2019). Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. GigaScience 8:giz012. doi: 10.1093/gigascience/giz012
Ding, B., Patterson, E. L., Holalu, S. V., Li, J., Johnson, G. A., Stanley, L. E., et al. (2020). Two MYB proteins in a self-organizing activator-inhibitor system produce spotted pigmentation patterns. Curr. Biol. 30, 802–814.e8. doi: 10.1016/j.cub.2019.12.067
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Gao, J.-J., Shen, X.-F., Zhang, Z., Peng, R.-H., Xiong, A.-S., Xu, J., et al. (2011). The myb transcription factor MdMYB6 suppresses anthocyanin biosynthesis in transgenic Arabidopsis. Plant cell. Tissue Organ Cult. (PCTOC) 106, 235–242. doi: 10.1007/s11240-010-9912-4
Gelvin, S. B., and Liu, C.-N. (1994). “Genetic manipulation of Agrobacterium tumefaciens strains to improve transformation of recalcitrant plant species,” in Plant Molecular Biology Manual. eds. S. B. Gelvin and R. A. Schilperoort (Dordrecht: Springer), 85–97.
Günther, C. S., Dare, A. P., Mcghie, T. K., Deng, C., Lafferty, D. J., Plunkett, B. J., et al. (2020). Spatiotemporal modulation of flavonoid metabolism in blueberries. Front. Plant Sci. 11:545. doi: 10.3389/fpls.2020.00545
Hichri, I., Heppel, S. C., Pillet, J., Léon, C., Czemmel, S., Delrot, S., et al. (2010). The basic Helix-loop-Helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 3, 509–523. doi: 10.1093/mp/ssp118
Hijaz, F., Nehela, Y., Jones, S. E., Dutt, M., Grosser, J. W., Manthey, J. A., et al. (2018). Metabolically engineered anthocyanin-producing lime provides additional nutritional value and antioxidant potential to juice. Plant Biotechnol. Rep. 12, 329–346. doi: 10.1007/s11816-018-0497-4
Huang, D., Tang, Z., Fu, J., Yuan, Y., Deng, X., and Xu, Q. (2019). CsMYB3 and CsRuby1 form an ‘activator-and-repressor’ loop for the regulation of anthocyanin biosynthesis in citrus. Plant Cell Physiol. 61, 318–330. doi: 10.1093/pcp/pcz198
Jaakola, L., Määttä, K., Pirttilä, A. M., Törrönen, R., Kärenlampi, S., and Hohtola, A. (2002). Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 130, 729–739. doi: 10.1104/pp.006957
James, A. M., Ma, D., Mellway, R., Gesell, A., Yoshida, K., Walker, V., et al. (2017). Poplar MYB115 and MYB134 transcription factors regulate proanthocyanidin synthesis and structure. Plant Physiol. 174, 154–171. doi: 10.1104/pp.16.01962
Jun, J. H., Liu, C., Xiao, X., and Dixon, R. A. (2015). The transcriptional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula. Plant Cell 27, 2860–2879. doi: 10.1105/tpc.15.00476
Jun, J. H., Xiao, X., Rao, X., and Dixon, R. A. (2018). Proanthocyanidin subunit composition determined by functionally diverged dioxygenases. Nat. Plants 4, 1034–1043. doi: 10.1038/s41477-018-0292-9
Kagale, S., and Rozwadowski, K. (2011). EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146. doi: 10.4161/epi.6.2.13627
Karppinen, K., Lafferty, D. J., Albert, N. W., Mikkola, N., Mcghie, T., Allan, A. C., et al. (2021). MYBA and MYBPA transcription factors co-regulate anthocyanin biosynthesis in blue-coloured berries. New Phytol. 232, 1350–1367. doi: 10.1111/nph.17669
Karppinen, K., Zoratti, L., Nguyenquynh, N., Häggman, H., and Jaakola, L. (2016). On the developmental and environmental regulation of secondary metabolism in Vaccinium spp. berries. Front. Plant Sci. 7:655. doi: 10.3389/fpls.2016.00655
Khoo, H. E., Azlan, A., Tang, S. T., and Lim, S. M. (2017). Anthocyanidins and anthocyanins: colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 61:1361779. doi: 10.1080/16546628.2017.1361779
Lafferty, D. J., Espley, R. V., Deng, C. H., Günther, C. S., Plunkett, B., Turner, J. L., et al. (2022). Hierarchical regulation of MYBPA1 by anthocyanin- and proanthocyanidin-related MYB proteins is conserved in Vaccinium species. J. Exp. Bot. 73, 1344–1356. doi: 10.1093/jxb/erab460
Lin-Wang, K., Mcghie, T. K., Wang, M., Liu, Y., Warren, B., Storey, R., et al. (2014). Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front. Plant Sci. 5:651. doi: 10.3389/fpls.2014.00651
Liu, Y., Ma, K., Qi, Y., Lv, G., Ren, X., Liu, Z., et al. (2021). Transcriptional regulation of anthocyanin synthesis by MYB-bHLH-WDR complexes in kiwifruit (Actinidia chinensis). J. Agric. Food Chem. 69, 3677–3691. doi: 10.1021/acs.jafc.0c07037
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Matsui, K., Umemura, Y., and Ohme-Takagi, M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55, 954–967. doi: 10.1111/j.1365-313X.2008.03565.x
Montefiori, M., Brendolise, C., Dare, A. P., Lin-Wang, K., Davies, K. M., Hellens, R. P., et al. (2015). In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 66, 1427–1436. doi: 10.1093/jxb/eru494
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. doi: 10.1105/tpc.010127
Pérez-Díaz, J. R., Pérez-Díaz, J., Madrid-Espinoza, J., González-Villanueva, E., Moreno, Y., and Ruiz-Lara, S. (2016). New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 90, 63–76. doi: 10.1007/s11103-015-0394-y
Plunkett, B. J., Espley, R. V., Dare, A. P., Warren, B. a. W., Grierson, E. R. P., Cordiner, S., et al. (2018). MYBA from blueberry (Vaccinium section cyanococcus) is a subgroup 6 type R2R3MYB transcription factor that activates anthocyanin production. Front. Plant Sci. 9:1300. doi: 10.3389/fpls.2018.01300
Primetta, A. K., Karppinen, K., Riihinen, K. R., and Jaakola, L. (2015). Metabolic and molecular analyses of white mutant Vaccinium berries show down-regulation of MYBPA1-type R2R3 MYB regulatory factor. Planta 242, 631–643. doi: 10.1007/s00425-015-2363-8
Rauf, A., Imran, M., Abu-Izneid, T., Iahtisham Ul, H., Patel, S., Pan, X., et al. (2019). Proanthocyanidins: a comprehensive review. Biomed. Pharmacother. 116:108999. doi: 10.1016/j.biopha.2019.108999
Ravaglia, D., Espley, R. V., Henry-Kirk, R. A., Andreotti, C., Ziosi, V., Hellens, R. P., et al. (2013). Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 13:14. doi: 10.1186/1471-2229-13-68
Rinaldo, A. R., Cavallini, E., Jia, Y., Moss, S. M. A., Mcdavid, D. a. J., Hooper, L. C., et al. (2015). A grapevine anthocyanin acyltransferase, transcriptionally regulated by VvMYBA, can produce Most acylated anthocyanins present in grape skins. Plant Physiol. 169, 1897–1916. doi: 10.1104/pp.15.01255
Schaart, J. G., Dubos, C., Romero De La Fuente, I., Van Houwelingen, A. M. M. L., De Vos, R. C. H., Jonker, H. H., et al. (2013). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 197, 454–467. doi: 10.1111/nph.12017
Schellmann, S., Schnittger, A., Kirik, V., Wada, T., Okada, K., Beermann, A., et al. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21, 5036–5046. doi: 10.1093/emboj/cdf524
Schwacke, R., Ponce-Soto, G. Y., Krause, K., Bolger, A. M., Arsova, B., Hallab, A., et al. (2019). MapMan4: a refined protein classification and annotation framework applicable to multi-omics data analysis. Mol. Plant 12, 879–892. doi: 10.1016/j.molp.2019.01.003
Terrier, N., Torregrosa, L., Ageorges, A., Vialet, S., Verriès, C., Cheynier, V., et al. (2009). Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149, 1028–1041. doi: 10.1104/pp.108.131862
Wang, N., Qu, C., Jiang, S., Chen, Z., Xu, H., Fang, H., et al. (2018). The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J. 96, 39–55. doi: 10.1111/tpj.14013
Wang, Y., Shi, Y., Li, K., Yang, D., Liu, N., Zhang, L., et al. (2021). Roles of the 2-Oxoglutarate-dependent Dioxygenase superfamily in the flavonoid pathway: A review of the functional diversity of F3H, FNS I, FLS, and LDOX/ANS. Molecules 26:6745. doi: 10.3390/molecules26216745
Warnes, G. R., Bolker, B., Bonebakker, L., Gentleman, R., Huber, W., Liaw, A., et al. (2009). Gplots: various R programming tools for plotting data. R package version 2, 1.
Wei, T., and Simko, V. (2017). R package “corrplot”: visualization of a correlation matrix (version 0.84). Available at: https://github.Com/taiyun/corrplot (Accessed September 2021).
Wu, C., Deng, C., Hilario, E., Albert, N. W., Lafferty, D., Grierson, E. R. P., et al. (2021). A chromosome-scale assembly of the bilberry genome identifies a complex locus controlling berry anthocyanin composition. Mol. Ecol. Resour. 22, 345–360. doi: 10.1111/1755-0998.13467
Xu, W., Grain, D., Le Gourrierec, J., Harscoët, E., Berger, A., Jauvion, V., et al. (2013). Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 198, 59–70. doi: 10.1111/nph.12142
Yan, S., Chen, N., Huang, Z., Li, D., Zhi, J., Yu, B., et al. (2020). Anthocyanin fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 225, 2048–2063. doi: 10.1111/nph.16272
Yoshida, K., Ma, D., and Constabel, C. P. (2015). The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 167, 693–710. doi: 10.1104/pp.114.253674
Zhang, W., Ning, G., Lv, H., Liao, L., and Bao, M. (2009). Single MYB-type transcription factor AtCAPRICE: A new efficient tool to engineer the production of anthocyanin in tobacco. Biochem. Biophys. Res. Commun. 388, 742–747. doi: 10.1016/j.bbrc.2009.08.092
Zhou, H., Liao, L., Xu, S., Ren, F., Zhao, J., Ogutu, C., et al. (2018). Two amino acid changes in the R3 repeat cause functional divergence of two clustered MYB10 genes in peach. Plant Mol. Biol. 98, 169–183. doi: 10.1007/s11103-018-0773-2
Zhou, H., Lin-Wang, K., Wang, F., Espley, R. V., Ren, F., Zhao, J., et al. (2019). Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 221, 1919–1934. doi: 10.1111/nph.15486
Zhu, H. F., Fitzsimmons, K., Khandelwal, A., and Kranz, R. G. (2009). CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2, 790–802. doi: 10.1093/mp/ssp030
Zhu, Z., Li, G., Liu, L., Zhang, Q., Han, Z., Chen, X., et al. (2019). A R2R3-MYB transcription factor, VvMYBC2L2, functions as a transcriptional repressor of anthocyanin biosynthesis in grapevine (Vitis vinifera L.). Molecules 24:92. doi: 10.3390/molecules24010092
Zifkin, M., Jin, A., Ozga, J. A., Zaharia, L. I., Schernthaner, J. P., Gesell, A., et al. (2012). Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol. 158, 200–224. doi: 10.1104/pp.111.180950
Zimmermann, I. M., Heim, M. A., Weisshaar, B., and Uhrig, J. F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40, 22–34. doi: 10.1111/j.1365-313X.2004.02183.x
Zorenc, Z., Veberic, R., Slatnar, A., Koron, D., Miosic, S., Chen, M.-H., et al. (2017). A wild ‘albino’ bilberry (Vaccinium myrtillus L.) from Slovenia shows three bottlenecks in the anthocyanin pathway and significant differences in the expression of several regulatory genes compared to the common blue berry type. PLoS One 12:e0190246. doi: 10.1371/journal.pone.0190246
Keywords: anthocyanin, proanthocyanidin, MYB, transcription factor, flavonoid, berry, repressor
Citation: Lafferty DJ, Espley RV, Deng CH, Dare AP, Günther CS, Jaakola L, Karppinen K, Boase MR, Wang L, Luo H, Allan AC and Albert NW (2022) The Coordinated Action of MYB Activators and Repressors Controls Proanthocyanidin and Anthocyanin Biosynthesis in Vaccinium. Front. Plant Sci. 13:910155. doi: 10.3389/fpls.2022.910155
Edited by:
Brian Farneti, Fondazione Edmund Mach, ItalyReviewed by:
Katia Petroni, University of Milan, ItalyHeidi Halbwirth, Vienna University of Technology, Austria
Copyright © 2022 Lafferty, Espley, Deng, Dare, Günther, Jaakola, Karppinen, Boase, Wang, Luo, Allan and Albert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nick W. Albert, bmljay5hbGJlcnRAcGxhbnRhbmRmb29kLmNvLm56
†Present address: Declan J. Lafferty, Boyce Thompson Institute, Ithaca, NY, United States
 Declan J. Lafferty
Declan J. Lafferty Richard V. Espley
Richard V. Espley Cecilia H. Deng
Cecilia H. Deng Andrew P. Dare
Andrew P. Dare Catrin S. Günther
Catrin S. Günther Laura Jaakola
Laura Jaakola Katja Karppinen
Katja Karppinen Murray R. Boase
Murray R. Boase Lei Wang1
Lei Wang1 Nick W. Albert
Nick W. Albert