- State Key Laboratory of Environmental Criteria and Risk Assessment, Chinese Research Academy of Environmental Sciences, Beijing, China
Biological invasions have become one of the greatest threats to global biodiversity and ecosystem conservation. Most previous studies have revealed how successful invasive species adapt to new environments and climate change through phenotypic and genetic evolution. Some researchers suggested that understanding unsuccessful or less successful biological invasions might be important for understanding the relationships between invasion adaptability and climate factors. We compared the sexual reproduction ability, genetic diversity, and gene × environment interaction in two intentionally introduced alien species in China (Spartina anglica and Spartina alterniflora) based on restriction site-associated DNA (RAD) sequencing. After more than 50 years, the distribution of S. alterniflora has rapidly expanded, while S. anglica has experienced extreme dieback. A total of 212,939 single nucleotide polymorphisms (SNPs) for the two Spartina species were used for analysis. The multilocus genotype (MLG) analysis revealed that clonal reproduction was the prevalent mode of reproduction in both species, indicating that a change in the mode of reproduction was not the key factor enabling successful invasion by Spartina. All genetic diversity indicators (He, Ho, π) in S. alterniflora populations were at least two times higher than those in S. anglica populations, respectively (p < 0.001). Furthermore, the population genetic structure and stronger patterns of climate-associated loci provided support for rapid adaptive evolution in the populations of S. alterniflora in China. Altogether, our results highlight the importance of genetic diversity and local adaptation, which were driven by multiple source populations, in increasing the invasiveness of S. alterniflora.
Introduction
Biological invasions have become one of the greatest threats to global biodiversity and ecosystem conservation (Clavero and García-Berthou, 2005; Simberloff et al., 2013; Early et al., 2016). Understanding the mechanisms of invasion would contribute to methodologies for its prevention and control. In recent years, many researchers have explored invasion mechanisms by examining the genetic backgrounds of invasive alien species, especially the links between genetic diversity, bottlenecks, founder events, adaptive evolution, and invasion success (Dlugosch and Parker, 2008; Prentis et al., 2008; Estoup et al., 2016). Although the genetic diversity of invasive populations may be reduced due to founder events and genetic bottlenecks, most successful invasive species can overcome these negative effects through multiple introductions, phenotypic plasticity, asexual reproduction and hybridization. For instance, Ambrosia artemisiifolia invaded European, Australian and Chinese areas with high levels of genetic diversity due to multiple introductions (Genton et al., 2005; Gaudeul et al., 2011; van Boheemen et al., 2017; Li et al., 2019b). Phenotypic plasticity rather than genetic adaptation helps Alternanthera philoxeroides (Geng et al., 2007; Roman and Darling, 2007) and Spartina densiflora (Castillo et al., 2018) colonize a wide range of habitats. Some populations of Eichhornia crassipes in China can successfully invade without genetic variation via clonal reproduction in the introduced range (Ren et al., 2005; Roman and Darling, 2007). Spartina anglica successfully invaded the British Isles (Raybould et al., 1991) through interspecies hybridization. Therefore, disadvantages associated with founder events might be overstated (Estoup et al., 2016), and genetic diversity does not necessarily predict invasion success (Roman and Darling, 2007).
Some researchers hypothesized that understanding unsuccessful or less successful biological invasions is also important for determining the role of genetic diversity and bottlenecks in invasion success (Fridley et al., 2007; Roman and Darling, 2007; Zenni and Nuñez, 2013). Comparing genetic and environmental characteristics between successful and unsuccessful populations can also help reveal mechanisms producing fitness variations in different environments, which are valuable in studies of genotype-by-environment interactions in the introduced range (Lee, 2002). Wellband et al. (2017) showed that allelic diversity reduction was associated with lower invasion success by comparing highly successful and less successful invasive species of gobies and tunicates. Following this study, the authors used functional genetic markers to test the link between genetic selection and the invasion success of two pairs of invasive goby species. They proposed that increased evolutionary potential in invaded ranges may be associated with invasion success (Wellband et al., 2018). However, studies on unsuccessful or less successful invasions of different biological groups remain limited.
Spartina anglica and Spartina alterniflora are perennial grasses native to British estuaries and the Atlantic and Gulf Coast estuaries of North America, respectively. S. anglica originated by natural chromosome doubling of an infertile hybrid (S. × townsendii) between the European native species S. maritima and the alien species S. alterniflora (Groves and Groves, 1880). The new fertile dodecaploid species has rapidly expanded in salt marshes in several European countries (Gray and Raybould, 1991). As S. anglica and S. alterniflora are “ecological engineers,” their histories of introduction in China were recorded in detail. While S. anglica populations expanded rapidly on the European continent, they experienced dieback in coastal China. The species was introduced to China from England in 1963 (Chung, 1993), with planting sites as far as Dongow, Liaoning, in the north, and Hepu, Guangxi, in the south and a total planting area of almost 36,000 ha in 1981. S. alterniflora was introduced to China later from North Carolina, Georgia, and Florida of North America in 1979 (Chung, 1993). However, after more than 50 years, there were only three sites with S. anglica in Jiangsu and Zhejiang in 2000, with the area shrunken to 50 ha, whereas the area of S. alterniflora had expanded to 112,000 ha, as reported by An et al. (2007).
Most studies have proposed that the coexistence of various intraspecific hybrids, and mixtures of the three ecotypes sampled from their native ranges is the main reason for the wide spread of S. alterniflora (Xia et al., 2015), even though the genetic diversity of populations in China was lower than that in native populations (Deng et al., 2007; Xia et al., 2015; Bernik et al., 2016). Such genetic admixture within or between species in the invaded range may allow them to adapt to local conditions via increases in heterozygote frequency and the production of novel genotypes (Ellstrand and Schierenbeck, 2006; Schierenbeck and Ellstrand, 2008; Rius and Darling, 2014). On the basis of chloroplast sequence analysis and reciprocal transplant experiments, Qiao et al. (2019) reported that genetic admixture has facilitated the evolution of super competitive genotypes of S. alterniflora in China and that the super competitive genotypes could overcome the negative correlation between plant height and shoot regeneration in the invasive range. In addition, some studies have indicated that a lack of nitrogen in soil (Li et al., 2007), high plant density (Li et al., 2009), and unsuitable soil texture (Liu et al., 2016a) can cause dieback in S. anglica. One important reason for the failure of S. anglica in China is weak sexual propagation, while S. alterniflora had a strong capacity for both sexual and asexual reproduction (Chung, 1985; Xu and Zhuo, 1985; Xiao et al., 2011). Li et al. (2008) studied the breeding system of the species in one field site and a greenhouse, and the results implied that protogyny, poor pollen quality, and abnormal pollen grains and pollen tubes were the main causes of low seed production in S. anglica in coastal China. However, no studies have compared the genetic characteristics and genotype-by-environment interactions of the successful invader S. alterniflora and the less successful invader S. anglica.
Moreover, the distribution areas of S. anglica have become unclear in recent years. As reported by An et al. (2007), there were no populations of Spartina in Liaoning. Based on geographical information systems (GISs), global positioning systems (GPSs), and on-site investigations in 2007, Zuo et al. (2012) found 1 ha of S. anglica in Liaoning, 10 ha in Hebei, 3 ha in Shandong, 1 ha in Jiangsu, and 1 ha in Guangdong. The authors found that populations of S. alterniflora were also distributed in these provinces and occupied larger areas than S. anglica. Zhang et al. (2017) also reported one population of S. alterniflora in Liaoning but no S. anglica. In 2019, we found and identified S. anglica in Dandong and Huludao cities of Liaoning Province. We chose four populations of S. alterniflora, which are distributed from north to south in China, for comparison with the two populations of S. anglica. We used restriction site-associated DNA (RAD) sequencing (Baird et al., 2008) to improve the resolution to detect finer-scale spatial patterns of genetic structure and diversity. This sequencing method allows concurrent single nucleotide polymorphism (SNP) identification and genotyping via high-throughput sequencing of flanking regions of restriction enzyme digestion sites dispersed throughout the genome. Here, we addressed three main questions: (1) Did low sexual reproduction cause the dieback of S. anglica in coastal China? (2) Does the successful invasive species S. alterniflora have higher genetic diversity than the less successful invasive species S. anglica? (3) Do the highly successful invasive species S. alterniflora and less successful invasive species S. anglica differ in their levels of genetic adaptation? We also expected to provide evidence to clarify the role of genetic diversity in the colonization or invasion failure of species.
Materials and Methods
Sample Collection and Species Confirmation
We collected leaf samples from 88 individuals of Spartina from six sites (14–15 leaf samples per site) along the coastline of China in October 2018 (Supplementary Figure 1). From north to south, the six populations were distributed in Dandong (DD), Huludao (HLD), Tanggu (TG), Cixi (CX), Quanzhou (QZ), and Fangchenggang (FCG). Information on the numbers of individuals and locations of populations is listed in Table 1. Within each population, the distance between each pair of sampled individuals was approximately 10 m. We also obtained three pieces of soil samples from each site, and the depth of the soil samples was 0–25 cm.
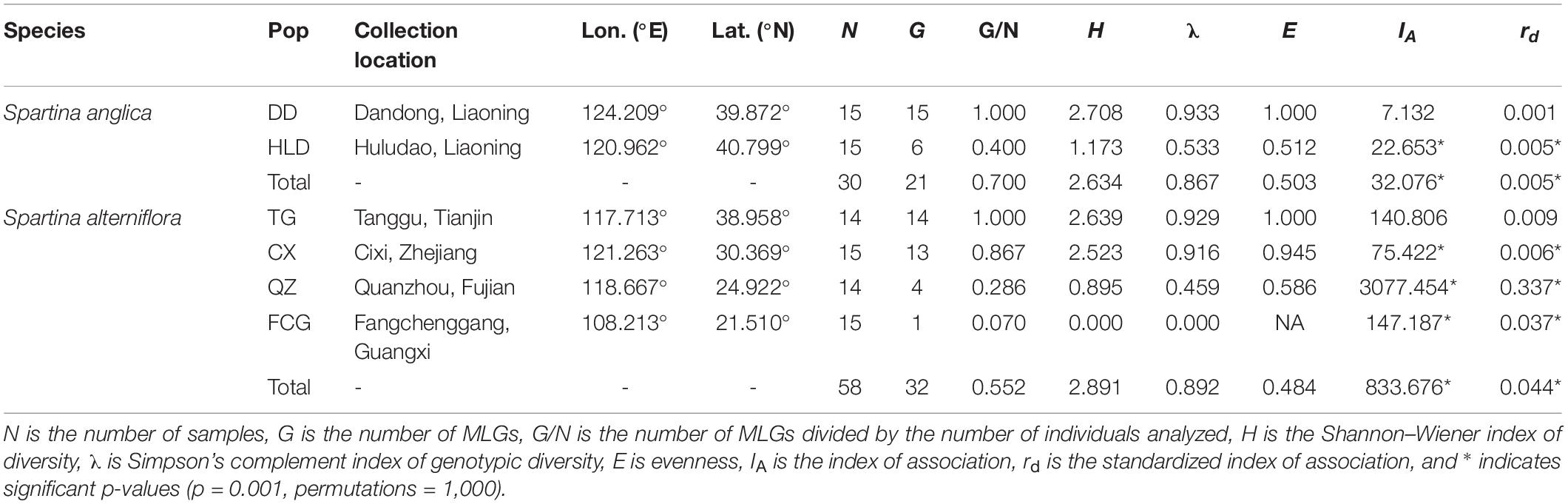
Table 1. Location information, number of sampled individuals, and genotypic diversity in populations of Spartina anglica and Spartina alterniflora.
According to the manufacturer’s instructions, genomic DNA was extracted from the leaves using the EasyPure Plant Genomic DNA Kit (Beijing TransGen Biotech Co., Ltd., Beijing, China). Based on the published nrITS sequences of 145 species of Spartina available from GenBank,1 nrITS (KM010334 of S. anglica and KM010330 of S. alterniflora) was used to identify S. anglica and S. alterniflora (Peterson et al., 2014). We confirmed that the HLD and DD populations were S. anglica and the other four populations were all S. alterniflora through multiple sequence alignment and neighbor joining (NJ) tree construction with 1,000 bootstrap by MAFFT v. 7 (Katoh et al., 2019; Supplementary Figure 2).
Sequencing and Genotyping
All RAD libraries were created using 200 ng of genomic DNA from samples of Spartina. This DNA was double-digested with EcoRI and Mse l (New England Biolab, Beverly, MA, United States) and ligated to two-end adapters, allowing the resulting amplified fragments to bind to Illumina flow cells and uniquely identify the individual. All libraries with insert sizes of 300–500 bp were isolated by gel extraction and sequenced on the Illumina XTen platform using 150 nt paired-end reads (Jierui Biotech, Guangzhou, China). A total of ∼260 GB of data was obtained from Illumina sequencing of 88 samples from six populations. The data for each sample were split and filtered using the process_radtags module of Stacks 2.3e (Catchen et al., 2011). We removed reads with low quality, missing the restriction site or linked with incorrect barcodes. We combined each end of the retained reads from each sample. Here, we set two as the minimum depth of coverage (m) required to create a stack and three as the maximum distance (in nucleotides) allowed between stacks (M) within an individual. We then used the cstacks module in Stacks to build the catalogs for all these individuals, with two as the maximum number of mismatches allowed between individuals (n). We finally used the cstacks, ustacks, and populations modules in Stacks to obtain SNPs. We obtained the loci using the following limits in the populations module: 0.01 as the minor allele frequency, 75% as the percentage of individuals sharing the locus within the population, and appearance of the locus in all populations. Linkage disequilibrium (LD) between each pair was tested using VCFTOOLS v.0.1.15 (Danecek et al., 2011), and one marker was excluded from each pair with R2 > 0.8. The LD-excluded VCF data were further divided into S. anglica and S. alterniflora subsets for subsequent analysis by Tassel 5 (Bradbury et al., 2007), and the three datasets were converted into file formats necessary for analyses using PLINK v.1.9 (Purcell et al., 2007) and PGDSPIDER 2.0.9.0 (Lischer and Excoffier, 2012).
Environmental Data
To test for correlations between genetic and environmental variables of Spartina in China, we required environmental data at the sampling locations. We extracted the standard nineteen bioclimatic variables (Supplementary Table 1) from the WORLDCLIM dataset (Version 2; period 1970–2000) (Hijmans et al., 2005) for the set of georeferenced locations of available localities using DIVA-GIS software (Hijmans et al., 2004). The total organic nitrogen (TN) and total phosphorus (TP) of the soil samples were determined using the Kjeldahl method and the molybdenum-antimony antispectrophotometric method at Bowu Company, Beijing. Soil electrical conductivity (EC) was detected by a New Digital DDS-307 Conductivity-Salinity Meter Tester in a mixture of 1:5 soil:distilled water as an index of soil salinity (S). The average TN, TP, and S values of three samples and the standard nineteen bioclimatic variables at each site were all used as environmental variables (Supplementary Table 1).
Genetic Diversity and Clone Detection
Genetic diversity indices including the percentage of polymorphic loci (%poly), average nucleotide diversity (π), average observed heterozygosity per locus (Ho), and average expected heterozygosity per locus (He) were calculated using populations in Stacks version 2.3 based on the raw dataset (Catchen et al., 2011). Then, we used analysis of variation (ANOVA) to test for a difference in genetic diversity between S. anglica and S. alterniflora using the R 4.1.2 function “aov” (Chambers et al., 2017). The Tukey’s multiple comparisons test (TukeyHSD) was used for multiple comparisons (Yandell, 2017).
We referred to several analysis results to evaluate the reproductive systems of six populations. First, we calculated the average Wright’s inbreeding coefficient (FIS) using populations in Stacks version 2.3. The inbreeding coefficient (FIS) was often used to quantify the partial asexual reproduction population (Stoeckel and Masson, 2014). Negative FIS values may reflect excess heterozygosity associated with reproduction via clonality (Pantoja et al., 2017).
Second, the genetic clones of S. anglica and S. alterniflora subsets were calculated using the package “poppr v. 2.9.3” based on the putatively neutral dataset (Kamvar et al., 2014, 2015), which can identify multilocus genotypes (MLGs) in clonal, partially clonal, and/or sexual reproduction populations. Clonal membership to the same genet can be inferred through a shared MLG among multiple individuals within a population. The true number of MLGs was determined by the “mlg.filter” function with a Nei’s distance (Nei, 1978) threshold determined using the cutoff_predictor tool, which finds a gap in the distance distribution. We investigated additional evidence of clonal reproduction through the “poppr” function (Kamvar et al., 2014, 2015). Since this function cannot work with a large dataset, we used two stricter datasets that restricted data analysis to one random SNP per locus (8,314 SNPs for S. anglica and 21,029 SNPs for S. alterniflora) to calculate the index of association (IA), the standardized index of association (rd), and their p-values (Supplementary Table 2). We also ensured that the MLGs obtained from strict datasets were consistent with the MLGs obtained from non-strict datasets. The IA index determines individual’s recombination, which indicates panmixia, using the ratio of observed-to-expected variance in the number of loci (Agapow and Burt, 2001). The rd index is a modification of IA that removes sample size bias (Agapow and Burt, 2001). Clone correction was allowed, and the p-values of IA and rd were obtained with 999 permutations. We also constructed an unweighted pair group method with arithmetic mean (UPGMA) tree of all individuals, and minimum spanning networks (MSNs) of each MLG based on distance matrices.
We used “poppr” to calculate the clonal diversity of each population, including the Shannon–Wiener index (H) (Shannon, 1948), Simpson’s index (λ) (Simpson, 1949), and evenness (E). The parameter G/N was also estimated, and while it could be biased, it is frequently used (Saleh et al., 2012; Pantoja et al., 2017; Tsujimoto et al., 2020). G represents the number of MLGs and N is the number of samples.
Population Structure
To infer the genetic structure of Spartina populations, we used ADMIXTURE version 1.3.0 (Alexander et al., 2009; Zhou et al., 2011) to estimate individual ancestries with the combined datasets of the two species and subsets of each species. We designated K = 1 to K = 9 as ancestral modes, and the most likely number of K given the populations was estimated according to the lowest cross-validation error (Alexander et al., 2009). We created an output file for the most likely number of K and visualized the results in the R environment. Discriminant analysis of principal components (DAPC) (Jombart et al., 2010) was performed using the R package “adegenet.” This method is more appropriate than others for the analysis of invasive species because it does not make any assumptions about migration-drift equilibrium (Jombart et al., 2010). The number of clusters was assessed for both species and their combined dataset, ranging from 1 to 10. The optimal number of clusters was identified based on the Bayesian information criterion (BIC), as suggested by Jombart et al. (2010). We used the function find.clusters to transform the original data into principal components (PCs), retaining 100 PCs in the analysis. Discriminant analysis was performed using 50 PCs (>90% of variance explained), and one, four or five eigenvalues were retained and examined. The optimal number of PCs to be retained in the final analysis was identified by the optim.a.score function with twenty independent runs.
Moreover, a redundancy analysis (RDA) within the “vegan” package (v.2.5–7) (Oksanen et al., 2021) in R 4.1.2 was used to assess whether environmental variables influenced the genetic differentiation of S. alterniflora. A principal component analysis (PCA) was performed by TASSEL 5 (Glaubitz et al., 2014). Variance inflation factors (VIF) were excluded through the “vif.cca” function of “vegan” package (Oksanen et al., 2021). The important environmental variables were chosen by the “forward.sel” function of the “adespatial” package (Dray et al., 2018). Then we used the “rdacca.hp” package to estimate the percentage contribution of explained variation to each or the total important environmental variables among the genetic groups, and the significance of the RDA results was tested with a global permutation (999 permutations) (Lai et al., 2022).
Outlier Detection
The outlier loci were detected using two population differentiation (PD) analysis methods and two environment association (EA) analyses. The first PD method was implemented in ARLEQUIN (Excoffier et al., 2005), which identifies outliers by comparing genetic diversity and differentiation between populations. Both infinite island and hierarchical demographic models of this approach were used to estimate outliers, and we only chose the outliers identified by both models. The second PD method we used was in the “PCAdapt” package in R 4.1.2, which detects outlier loci based on principal component analysis (PCA). This approach overcomes the limitation of the traditional “outlier test” approach, which assumes an unrealistic island model of migration between populations. SNP with qvalue less than 0.01 was considered as an outlier (Capblancq et al., 2018).
For EA analysis, BAYENV identified outliers based on a correlation matrix of loci using SNPs, and 22 environmental variables and two geographical variables (longitude and latitude). A batch file estimated the BayesFactor with 100,000 MCMC steps. Regarding interpretation (Jeffreys, 1961), a BayesFactor more than 10 indicated strong supportive evidence for an association between an environmental parameter and a locus (Cullingham et al., 2014). Second, we used latent factor mixed models (LFMMs) implemented in the LEA package in R 4.1.2 (Frichot and François, 2015) to detect outliers correlated with climatic variables. The implementation of the LFMM is based on least-squares estimates (Caye et al., 2019). The prcomp function determines the number of latent factors (K). A fitted model was then used to perform association testing based on the ridge_lfmm function after ridge estimation. Finally, any obtained p-values were transformed into qvalues. The qvalues less than a cutoff of 0.01 were indicative of candidate loci. A Benjamini and Hochberg (1995) false discovery rate (FDR) correction of 5% was applied to p-values using the qvalue package in R (Storey et al., 2015). Binomial tests were used to assess differences in the number of outliers in the two species obtained by the four methods.
Single Nucleotide Polymorphism Annotation
To gain insights into the potential adaptive significance of outlier loci, we obtained the flanking sequence of outlier loci identified by at least two methods from the RAD library. Functional annotation was performed using the online Basic Local Alignment Search Tool (BLAST) database. We searched the Genbank database using BLASTn with >60% sequence similarity. If the sequence was related to a protein, we further searched for its molecular function from UniProt2 and identified its related functions in plants according to literature. Then, we calculated correlations between allele frequencies of outlier loci and climate variables to identify putative genes associated with local adaptation.
Results
Genome Sequencing
A total of 1680,866,670 paired-end (PE) reads (read length = 150 bp, 252.13 Gbp) were obtained from the ddRAD library constructed for 30 individuals of S. anglica and 58 individuals of S. alterniflora. We obtained a total of 1674,211,878 reads (read length = 135 bp, 226.02 Gbp) after demultiplexing and removing reads of low quality. The number of reads for the samples varied from 3,632,316 to 52,525,600, with the average number of reads per sample being 19,025,135. Our analysis with ustacks showed that the number of ddRAD tags recovered from these samples varied from 248,767 to 1,385,706. Using cstacks, we obtained 4,664,866 loci from the 30 S. anglica and 58 S. alterniflora samples. After the initial filtering steps, 212,939 SNPs were retained, followed by removing sites showing linkage disequilibrium, and 169,522 SNPs were retained for all samples. Based on the dataset with 169,522 SNPs, a total of 90,671 SNPs were identified in the S. alterniflora subset, and 10,542 SNPs were identified in the S. anglica subset, with a minimum allele frequency of 0.01 (Supplementary Table 2).
Mode of Reproduction
Using the clonal threshold of 0.124 identified by cutoff_predictor, we found 32 MLGs among 58 individuals of S. alterniflora (Supplementary Figure 3B), while 21 MLGs among 30 individuals of S. anglica were found with a threshold of 0.163 (Table 1 and Supplementary Figure 3D). Table 1 also shows genotypic (MLG) diversity calculated separately for each population. The FCG population of S. alterniflora had the lowest genotypic diversity, with a single MLG identified among 15 sampled individuals (G:N = 0.067). Other populations with low genotypic diversity were QZ (S. alterniflora) and HLD (S. anglica), which had G:N ratios of 28.6 and 40%, respectively. In contrast, populations DD (S. anglica) and TG (S. alterniflora) had G:N ratios of 100% (Table 1).
Both indices of association, IA and rd, demonstrated that the population of genotypes was significantly different (p = 0.001) from the expected products of panmixia for populations HLD (S. anglica), CX, QZ, and FCG (S. alterniflora) (Table 1 and Supplementary Figure 4), indicating clonal reproduction as the prevalent mode of reproduction at these sites. The p-values of IA and rd were not significant (p = 1) only for populations DD (S. anglica) and TG (S. alterniflora) (Table 1 and Supplementary Figure 4), indicating that the null hypothesis of sexual reproduction could not be rejected for these populations. The inbreeding coefficients (FIS) of all S. anglica populations (DD and HLD) and the QZ and FCG populations of S. alterniflora were negative, showing a clear trend of heterozygote excess (Table 2). In addition, the minimum spanning networks (MSNs) based on Euclidean distances of the clone-corrected datasets showed no sign of reticulation (Figure 1). MLG 54 at the center of the S. alterniflora network consisted of only one individual from the TG population, and MLGs 26 and 30, which consisted of 10 individuals from the TG population and all individuals from the FCG population, were near the center (Figure 1A). Ten individuals of HLD belonging to MLG 16 were at the center of the S. anglica network, and other MLGs were found in only one individual (Figure 1B).
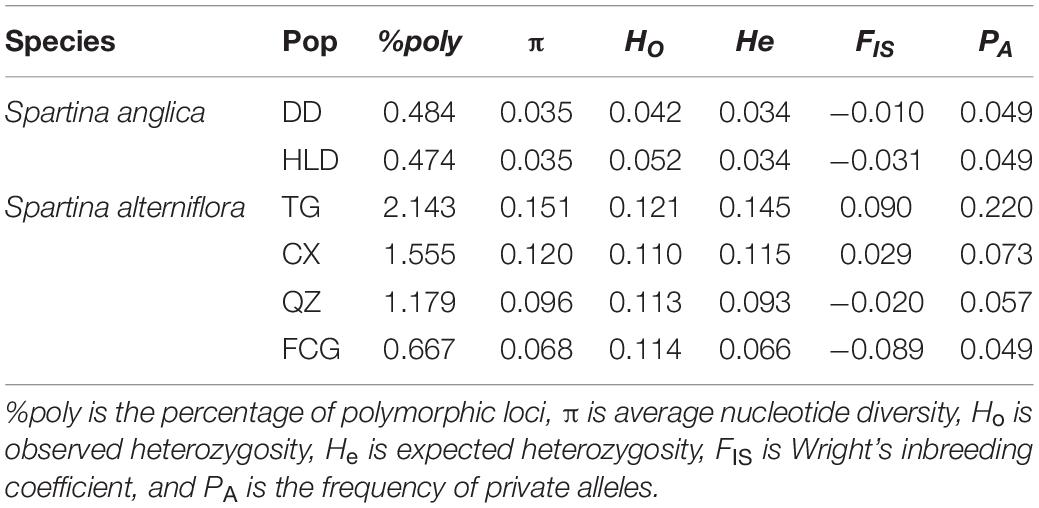
Table 2. Diversity indices of each sampled population of Spartina anglica and Spartina alterniflora.
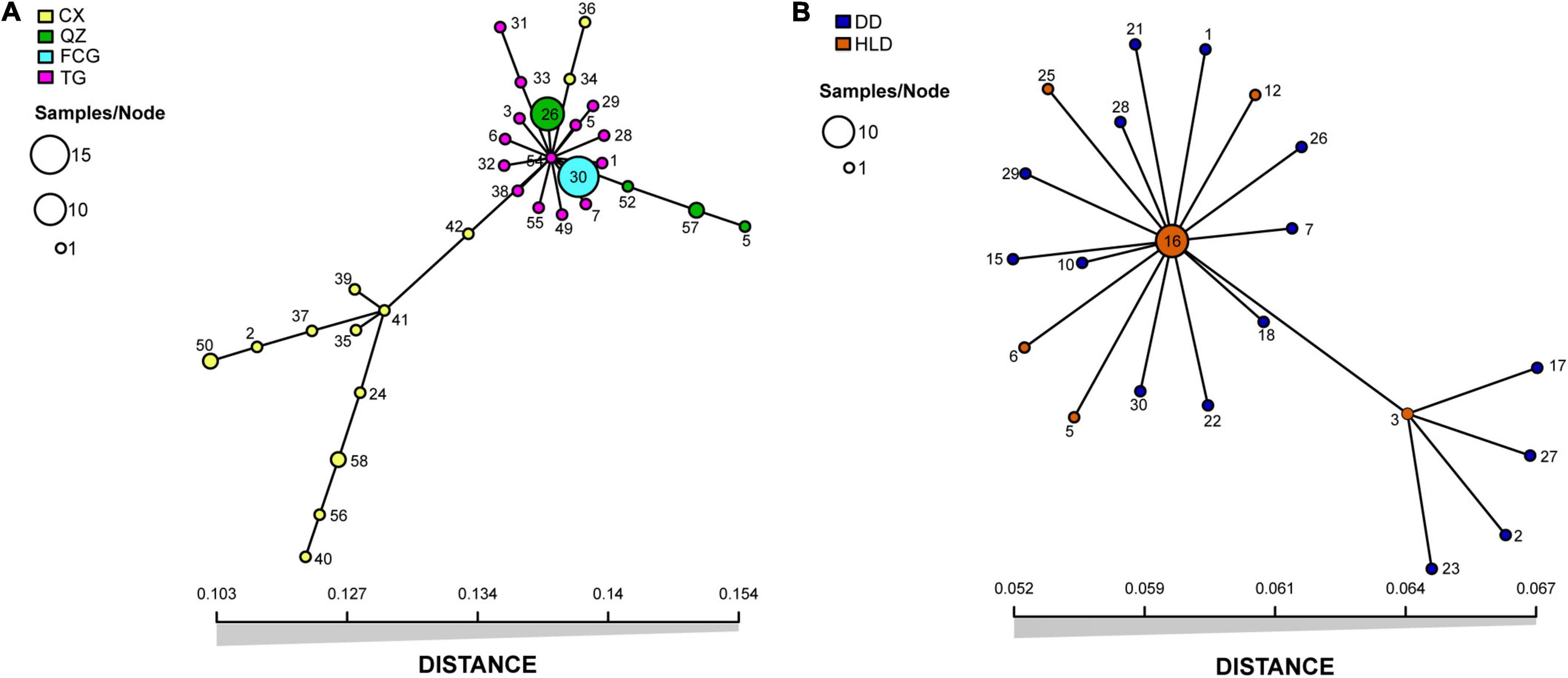
Figure 1. Minimum spanning networks (MSNs) of unique multilocus genotypes (MLGs) in populations of Spartina alterniflora (A) and Spartina anglica (B) collected from six fields in China. Each node represents a multilocus genotype (MLG), with size depending on the number of individuals with the MLG. The distance between the nodes represents the genetic distance between MLGs. Sampled fields are denoted by color, and the size of sample nodes represents either one or two isolates.
Genetic Diversity
There was higher genetic diversity in S. alterniflora populations than in S. anglica populations in terms of nucleotide diversity (π, 0.035 vs. 0.068-0.151, p < 0.001), observed heterozygosity (Ho, 0.042–0.052 vs. 0.110–0.121, p < 0.001) and expected heterozygosity (He, 0.034 vs. 0.145-0.066, p < 0.001) (Table 2 and Figures 2A–C). TukeyHSD also revealed significant differences among the six populations (p < 0.001), except He and π between HLD and DD (p = 0.976, 0.988), Ho between FCG and QZ (p = 0.699, Figures 2D–F and Supplementary Table 3). The TG population had the highest frequency of private alleles (21.98%), while the DD and FCG populations had the lowest frequencies (4.85 and 4.91%, respectively) (Table 2).
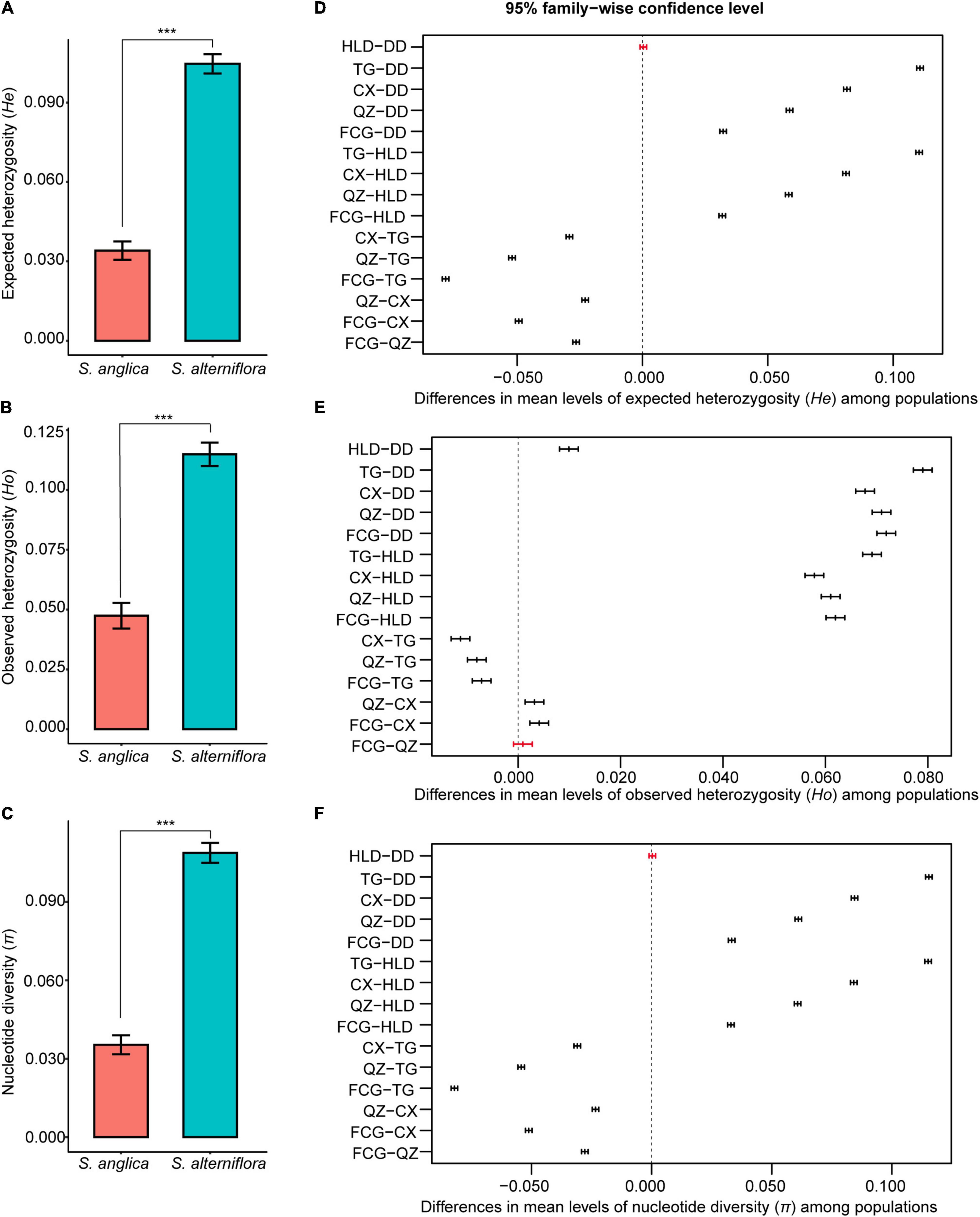
Figure 2. Comparison of expected heterozygosity (He), observed heterozygosity (Ho), and nucleotide diversity (π) between Spartina anglica and S. alterniflora (A–C) by the one-way ANOVA analysis, and among their populations (D–F) by Tukey’s multiple comparisons test. ***In panels (A–C) indicates p < 0.001. In panels (D–F), the bars represent differences in mean levels of genetic diversity indicators (He, Ho, π) between populations, p-values of black bars were less than 0.001, while those of red bars were more than 0.05.
Genetic Structure
Five genetic clusters (K = 5) were identified using ADMIXTURE based on the lowest cross-validation error (Supplementary Figure 5). The two Spartina species were clearly separated (Figure 3). All populations of S. alterniflora were divided into four clusters, and only four individuals of QZ belonged to the red cluster (Figures 3A,B). The two populations of S. anglica did not separate from each other (Figures 3A,B). DAPC performed on SNP datasets showed clear genetic structure in S. alterniflora populations but not in S. anglica (Figure 3C and Supplementary Figures 5B–D). All four populations of S. alterniflora were also separated from each other, and individuals of the FCG population were most distanced from other populations’ individuals (Figure 3D and Supplementary Figure 5C). The two populations of S. anglica were not separated based on the subdataset (Supplementary Figure 5). A UPGMA tree showed that individuals of S. alterniflora and S. anglica were divided into two major clusters with high bootstrap support. Individuals of four populations of S. alterniflora also separated into four subclusters with 100% bootstrap support (Figure 3E). The genetic distances among S. anglica individuals did not show clear separation (Figure 3E).
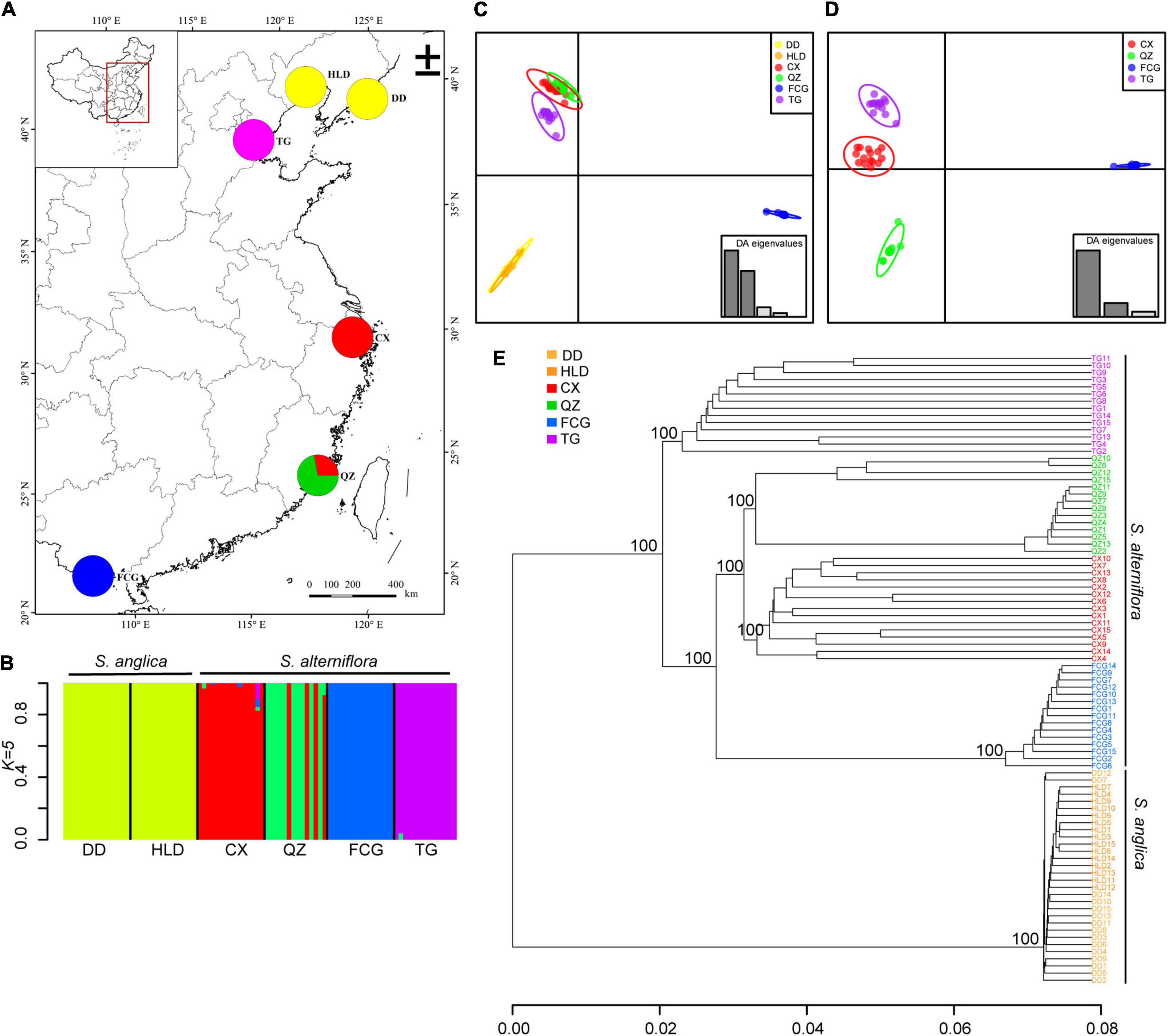
Figure 3. The genetic structure analysis of populations of Spartina anglica and Spartina alterniflora based on ADMIXTURE simulations, DAPC and UPGMA. (A) Map of collection sites for six populations. The pie chart shows the population cluster identified using ADMIXTURE when K = 5; (B) The ancestry composition of each individual of six populations. Each individual is represented by a vertical bar; (C) Scatter plots based on DAPC of all populations of Spartina, and (D) populations of S. alterniflora. Differently colored dots represent the individuals of different populations, and the insets indicate the eigenvalues from DAPC, with dark bars representing axes 1 and 2 of the plots. (E) Cluster analysis based on the genetic distance of six populations using a UPGMA tree with bootstrap support.
Three important environmental variables were kept through forward selection in the RDA analysis, including the minimum temperature of the coldest month (BIO6), the mean temperature of the wettest quarter (BIO8), and the precipitation of the warmest quarter (BIO18), which were significantly associated with genetic variation among populations of S. alterniflora (p = 0.001). The total explained variation was 0.784, and the precipitation of the warmest quarter (BIO18; 38.58% explained variation, p = 0.001) was the most important predictor (Supplementary Figure 6 and Supplementary Table 4).
Identification of Outliers and Analysis of Their Clines
A total of 7,868 and 238 outliers were identified in S. alterniflora and S. anglica, respectively, using four methods (Supplementary Figure 7). For the S. alterniflora subset, the two PD methods identified 4,791 outliers (4,145 from PCAdapt and 1,005 from ARLEQUIN), and the two EA methods identified 3,619 outliers (889 from BAYENV and 2,769 from LFMM). Only one outlier was identified by all four methods, 91 outliers were identified by three methods, and 848 outliers were identified by two methods (Supplementary Figure 7). For the S. anglica subset, the two PD methods identified 148 outliers (177 from PCAdapt and 93 from ARLEQUIN), and the two EA methods identified 31 outliers (12 from BAYENV and 19 from LFMM). None of the outliers were identified by all four methods, and only three outliers were identified by three methods; 60 outliers were identified by two methods (Supplementary Figure 7). The proportion of outliers obtained by the four methods was statistically different between the two species based on binomial tests (for PCAdapt, BAYENV and LFMM, p < 0.001; for ARLEQUIN, p < 0.05).
Functional annotation was not successful for any outlier loci of S. anglica. Fifteen loci obtained from the S. alterniflora subset were annotated (Supplementary Table 5). All of them were identified by at least two methods (11 from ARLEQUIN, 13 from PCAdapt, five from LFMM, and one from BAYENV). Two-thirds of the annotated outlier loci (10 outlier loci) were obtained with the two PD methods, while only one was obtained with the two EA methods. We identified 12 adjacent genes with a range of functions that were associated with plant growth and development, and four genes were associated with abiotic stress and pathogen attack. Based on the LFMM method, two (S10345, S199915) and four (S10345, S107946, S125087, S190589) genes identified were associated with precipitation (BIO11, 12, 13, 16, 18) and temperature (BIO1, 3, 4, 6, 7, 8, 9), respectively (Supplementary Table 6). The locus S199915 was identified by both EA methods, and was associated with precipitation and the TN content by the LFMM method, while it was associated with precipitation and temperature by BAYENV method. However, we only found strong associations between precipitation (Bio13, Bio16, and Bio18) and allele frequencies of two SNPs (S10345_125 and S199915_79), respectively (p < 0.05, Figure 4 and Supplementary Figure 8).
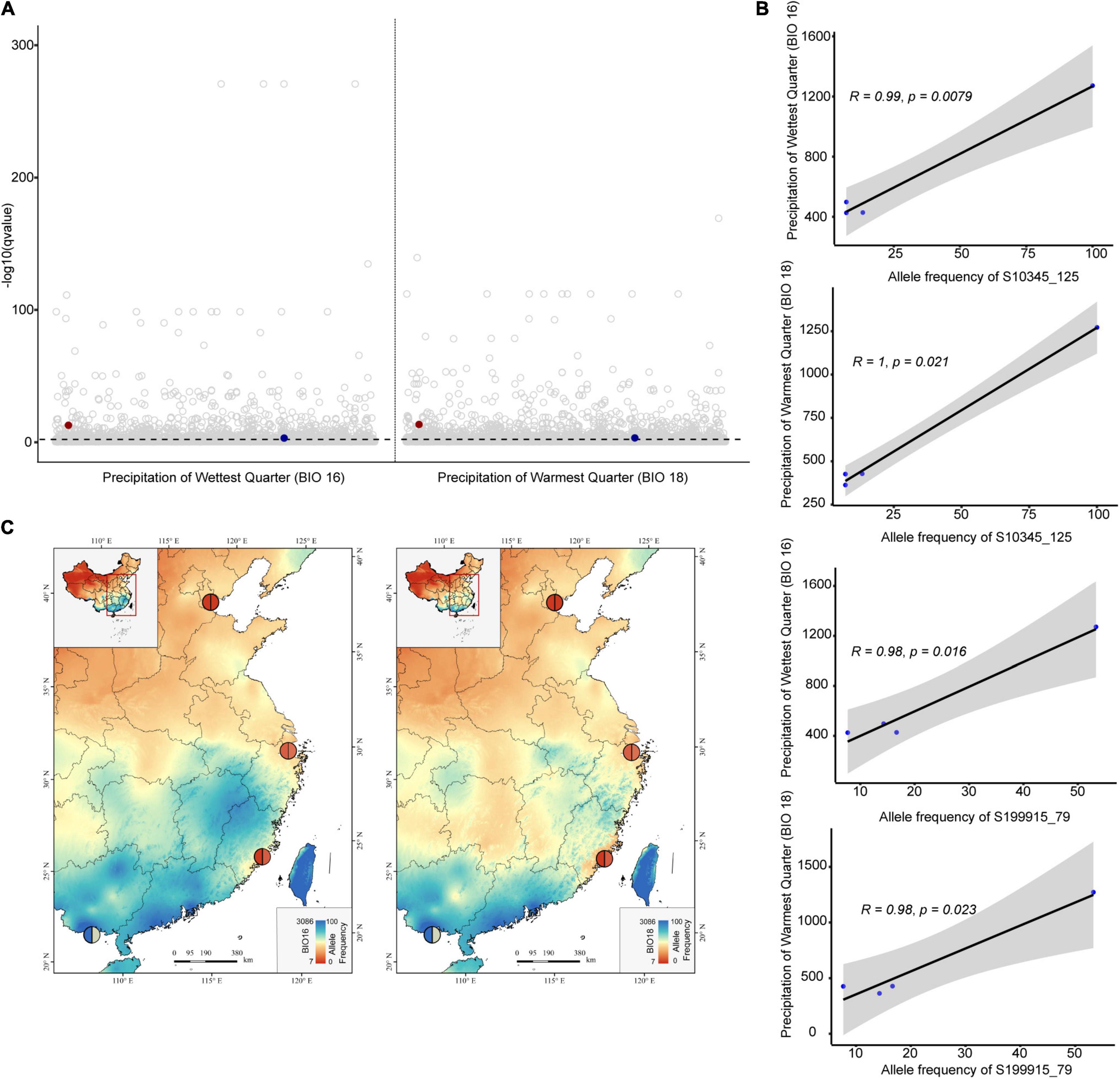
Figure 4. The outlier loci of Spartina alterniflora associated with climate variables based on the LFMM method. (A) Manhattan plots show the significance level for SNP associations with bioclimatic variables (BIO 16 and BIO18), the dark red points represent S10345_125, the dark blue points represent S199915_79, and the dashed line represents qvalue = 0.01. (B,C) Correlations between allele frequency and climate variables for S10345_125 and S199915_79 in populations of S. alterniflora. The colors of the pie chart on maps represent allele frequencies levels of S10345_125 (left half pie chart) and S199915_79 (right half pie chart).
Discussion
Our study confirmed that there are two species of Spartina along the coast of China. We compared the genetic backgrounds of the two species that were intentionally introduced to China decades ago. We used genome wide SNPs to analyze differences in modes of reproduction, genetic diversity, and gene × environment correlations between the two species in an effort to explain the reasons behind their differential invasion success. These analyses point to that high genetic diversity and genetic adaptation in response to the environment might promote S. alterniflora spreading on the coast of China.
Both High Clonality and Sexual Reproduction Were Revealed in the Two Spartina Species
Strong evidence of high clonal reproduction, such as a low number of genotypes, significant IA and rd (p < 0.001), and negative FIS, was found in populations of both S. alterniflora (FCG and QZ) and S. anglica (HLD). No reticulation of the minimum spanning networks (Figure 1) also provided an evidence of clonal spread (Maurice et al., 2019). However, sexual reproduction cannot be ignored in either species; population DD of S. anglica and population TG of S. alterniflora showed an extremely high G:N ratio. Although a slightly negative FIS (−0.010) was found in population DD of S. anglica, we still suspected that sexual reproduction rather than clonality was the main mode of reproduction in this population in consideration of the high number of genotypes, non-significant IA and rd, and lack of a large excess of heterozygotes. Negative FIS was usually interpreted as a signal of clonal reproduction in previous studies (Balloux et al., 2003; Halkett et al., 2005), while it was recently considered unreliable for evaluating reproduction. Both non-negative and negative FIS values can be frequently found in partially clonal populations, and the dynamics of FIS in (partially) clonal populations might be impacted by genetic drift (Reichel et al., 2016). Therefore, we focused more on the number of genotypes and the indices of association (IA and rd) when evaluating the mode of reproduction. Inconsistencies in FIS, IA, and rd were also found in population CX, which might be a partially clonal population with significant indices of association but a non-negative FIS (0.029). The results of reproduction analysis based on molecular genetic information in our study did not support the finding of previous studies that S. anglica mainly relies on asexual reproduction for population regeneration (Li et al., 2008). This may be due to population DD of S. anglica being located at a northern latitude (N 39.87°) with a lower annual mean temperature (9.3°C) and maximum temperature (28.88°C), while the population site in the study of Li et al. (2008) was located along the coast of the Yellow Sea in China (N 33.7°), with a higher annual mean temperature (15.0–15.6°C) and maximum temperature (39.0°C). The native distribution of S. anglica is in cool regions (the first record was in Lymington, Hampshire, England) (Gray and Raybould, 1991), and high temperatures might limit its seed production through less effective pollination or pollen germination (Li et al., 2008). Lower genotype diversities were found in populations FCG and QZ (latitudes below N 25°) than in TG and CX (latitudes above N 30°) (Table 1). This result was consistent with those of previous studies showing that sexual reproduction differed among populations of S. alterniflora in China and seed production decreased at low latitudes (Liu et al., 2016b).
High clonality is common in some notorious invasive alien species (Liu et al., 2006), such as Pueraria montana var. lobata (Bentley and Mauricio, 2016), Eichhornia crassipes (Barrett et al., 2008), Pistia stratiotes, and Eichhornia crassipes (Wang et al., 2016). In addition, most of these species are also capable of sexual propagation (Eckert, 2002) and plastic in their two methods of reproduction (Liu et al., 2006). Although low sexual reproduction may limit the local adaptation of alien species during an invasion due to little opportunity for genetic recombination, clonal reproduction can allow alien species to quickly establish populations regardless of whether the founder group sizes are small (Barrett et al., 2008). Our study revealed that the two Spartina species adopted both clonal and sexual reproduction strategies and might adjust seed production across latitudes. Therefore, the method of reproduction was not considered a major constraint on the dieback of S. anglica in China, even if this species may experience a reduced ability to produce seeds in low-latitude regions.
The Effect of High Genetic Diversity on Invasion Cannot Be Ignored
The newly introduced species suffered from bottlenecks, followed by reduced fitness and evolutionary potential, yet they often become invasive, and this dilemma was called the genetic paradox of invasion (Roman and Darling, 2007; Estoup et al., 2016). The genetic paradox of invasions has been thoroughly discussed by researchers in recent decades (Frankham, 2005; Stapley et al., 2015; Estoup et al., 2016; Schrieber and Lachmuth, 2017), and its importance among genetic mechanisms of invasions may be overestimated (Estoup et al., 2016). Many invaders do not exhibit the genetic paradox phenomenon because they have no loss of genetic diversity and no significant adaptive challenge in the invaded area compared to their native areas (Bossdorf et al., 2005; Roman and Darling, 2007). Some studies have already found that a negative founder effect with low genetic diversity can be overcome by high phenotypic variability, which can help Spartina species establish well in new environments (Castillo et al., 2018; Li et al., 2019a). In this study, the average nucleotide diversity (π) and observed heterozygosity (Ho) of each population reflected lower genetic diversity of the failed invader S. anglica (π: 0.035, Ho: 0.042–0.052) relative to the successful invader S. alterniflora. (π: 0.068–0.151, Ho: 0.110–0.121). Even though population FCG had extremely low genotype diversity, its genetic diversity was significantly higher than that in populations of S. anglica (p < 0.001, Figures 2D–F and Supplementary Table 3). This also suggests that differences in reproduction cannot explain the difference in genetic diversity observed here for S. anglica and S. alterniflora. Both species were intentionally introduced into China only once; the difference was that S. anglica came from a single source, while S. alterniflora came from multiple sources (Chung, 1993). The diverse genetic backgrounds resulting from multiple sources of S. alterniflora provided an opportunity for interbreeding and genetic recombination through sexual reproduction, whereas this was not possible for S. anglica with a single genetic source. Most previous studies also revealed that outcrossing of plants from the locations of origin (North Carolina, Georgia, and Florida in the US) of S. alterniflora generated a genetic admixture, and genetic variation remained high compared to those in the native populations (Bernik et al., 2016; Qiao et al., 2019; Xia et al., 2020). Moreover, only the TG and CX populations had higher expected heterozygosity (He, 0.145 and 0.115) than observed heterozygosity (Ho, 0.121 and 0.110), which may have been a reflection of earlier population bottlenecks (Rengefors et al., 2021), whereas, other populations did not show evidence of bottlenecks. Therefore, neither of the two Spartina species in China showed the genetic paradox of invasion. The low genetic diversity limiting population expansion of S. anglica might be due to inbreeding depression and loss of adaptive potential (Wellband et al., 2017, 2018). Significant differences in the levels of genetic diversity of successful and unsuccessful invasive species were consistent with those in a study of four aquatic invasive species (gobies and oysters) (Wellband et al., 2017). We also agree with the conclusion that the limitation of invasions via genetic diversity is species specific (Wellband et al., 2017).
Unlike most previous studies of S. alterniflora in China (Xia et al., 2015; Bernik et al., 2016; Qiao et al., 2019), ours revealed high genetic differentiation and low genetic admixture of populations based on structure analysis and DAPC, while the two populations of S. anglica in China could not be separated from each other. The findings of the current study do not support the previous findings of Wellband et al. (2017), who demonstrated greater divergence among populations of less-successful invasive species than among highly successful invasive species due to less gene flow among populations of the former. Population genetic structure in S. alterniflora in China was also found in the study of Xia et al. (2020), and they speculated that genetic divergence was mainly due to the low natural gene flow among populations. However, we believe that the existing genetic divergence is more likely a product of rapid adaptation, as Qiao et al. (2019) proved in their study. The significant association between environmental variables and genetic structure in this study also suggests that temperature (BIO6, 8) and precipitation (BIO18) are important drivers of genetic variation within S. alterniflora (Supplementary Figure 6). Therefore, high genetic variation promoted rapid genetic differentiation under environmental selection pressures is the possible explanation for why S. alterniflora in our study showed population genetic structure. Another reason is that RADseq technique afforded numerous SNPs with much more genetic information than other markers.
Genomic Signature Provides Support for Local Adaptation of Spartina alterniflora in China
Rapid local adaptation of invasive alien species usually includes two aspects: phenotypic plasticity and genetic evolution (Wellband and Heath, 2017; VanWallendael et al., 2018; Cao et al., 2021). Early studies on rapid adaptive evolution were mostly based on phenotypic changes between generations through common garden experiments or reciprocal transplant experiments (Turner et al., 2014; Ochocki and Miller, 2017). Recently, large genome-wide variation provided a more thorough understanding of the relationship between adaptive evolution and climate at the gene level (Zenni et al., 2014; Bay et al., 2018; Wellband et al., 2018). Based on the genomic scan, the performance of outlier test methods has been evaluated and used to provide evidence for local adaptive evolution in several studies (Nielsen et al., 2018; Gallego-Garcia et al., 2019; Cao et al., 2021). For example, Bay et al. (2018) identified the genomic regions which adapted across contemporary climate gradients in populations of yellow warbler (Setophaga petechia) by LFMM. To reveal signatures of adaptation within two montane bumble bee species, Jackson et al. (2020) detect outliers through several EA methods, including LFMM, Bayenv2, and RDA. All of these methods could lead to false positives and negatives, and it is hard to identify which outlier loci are false positives (Lotterhos and Whitlock, 2014), We try to avoid missing true positive outliers and adopt a cautious screening attitude to avoid false positives, the same as previous studies did (Cullingham et al., 2014; Wellband et al., 2018; Cao et al., 2021).
Although cross-validation with multiple methods may increase the confidence of true positive outliers (Cullingham et al., 2014), we still consider the outlier loci, which can be successfully annotated and have significant correlations with environment variables, to be more reliable. As significant larger number of outliers in populations of S. alterniflora than in S. anglica, some of which were successfully annotated, provided evidence for rapid local adaptation in this study. It supports hypothesis of previous studies that rapid adaptive evolution is a key mechanism driving successful invasion of S. alterniflora along the coast of China through reciprocal transplant experiments (Qiao et al., 2019). Previous authors speculated that genetic admixture might promote the formation of super invasive genotypes with high competitive abilities (Qiao et al., 2019). We also cannot ignore the genetic admixture in our study; even though we chose four large geographically distant populations, genetic admixture was still found in population QZ. Therefore, a single introduction but with multiple sources could promote the adaptation of S. alterniflora to different environments in China even with the short time lag of invasion (Facon et al., 2006). Rapid adaptive evolution can promote phenotypic trait shifts to increase the fitness of alien species when they face new environments (Suarez and Tsutsui, 2008; Molina-Montenegro et al., 2018).
Previous studies revealed high phenotypic variation in S. alterniflora in China and proved the occurrence of latitudinal clines in vegetative and sexual traits of populations, especially a linear increase in seed set, which was the result of rapid evolution, through garden experiments at multiple latitudes (Liu et al., 2017, 2020). Moreover, some studies have also proven that seed traits are critical for dispersal syndromes and mechanisms allowing plants to cope with environmental stress (Baskin and Baskin, 1998; Moles et al., 2005), and adaptive evolution in an invasive plant can enhance the probability of seedling survival by increasing the thickness of the seed coat (Molina-Montenegro et al., 2018). Interestingly, the function of locus S50498 (Supplementary Table 5) in our study was related to grain size and shape in rice (Huang et al., 2017). In addition, we also found that locus S135470 (Supplementary Table 5) might play a crucial role in organelle biogenesis and seedling establishment (Rigas et al., 2009).
We further revealed two SNPs of locus S10345 and S199915 were strongly associated with precipitation variables. The highest allele frequencies at these SNPs both occurred in the Fangchengang city of Guangxi province, areas of the low latitude and high rainfall (Figure 4). The SNP on locus S10345 was related to diacylglycerol kinase 2 that belongs to the diacylglycerol kinase family (DGKs). DGKs phosphorylate diacylglycerol (DAG) to produce phosphatidic acid (PA), which is required for plant development and responses to both biotic and abiotic stress (Tan et al., 2018; Kue Foka et al., 2020), such as drought (Li et al., 2015), cold (Ruelland et al., 2002), and pathogen attack (Zhang and Xiao, 2015). The SNP on locus S199915 was the H + -pyrophosphatase gene, which is related to vacuolar membrane solute transport (Graus et al., 2018). Previous studies also proved up-regulation of this gene can enhance resistance to salt and drought stress in plants (Park et al., 2005; Brini et al., 2007). Nevertheless, there are clear limitations in this study to further confirm the role of these two genes in the adaptation of S. alterniflora to the environment, and we believe that these loci are worthy of further research and validation.
To conclude, we explored aspects of invasiveness from an evolutionary perspective through a genomic comparison of S. alterniflora and S. anglica, which have been introduced to China for several decades. Our results suggest that low sexual reproduction was not the reason for the dieback of S. anglica in coastal China. This further implies that the high genetic diversity caused by multiple introduction sources of S. alterniflora in China plays a role in the success of invasion. Moreover, comparisons of invading species and non-invasive related species might be more helpful for understanding the genetic paradox. Due to the limitations of the sampling strategy in our study (we only found two populations of S. anglica in China), we cannot directly demonstrate that rapid adaptive evolution was one of the main factors which promoted the success of S. alterniflora invasion. However, the evidence of the correlation between gene and environment of S. alterniflora revealed in this study might provide genetic support for previous research conclusion. Additionally, annotated outlier loci are worthwhile of further study in consideration that the functional protein-coding genes obtained from four populations of S. alterniflora might be critical for understanding processes influencing the range expansion of invasive species.
Data Availability Statement
The data presented in the study are deposited in the GenBank repository, accession numbers SRR18493548-SRR18493635.
Author Contributions
FL and CZ conceived the idea. FL, XL, JZ, and KG collected the samples in the field, performed the experiments, and collected the data. FL analyzed the data and wrote the original manuscript with edits completed by CZ. JL provided comments on the manuscript. All authors contributed to the final version before the manuscript was submitted.
Funding
This work was financially supported by grants from the National Key Research and Development Program of China (2020YFC1806300, 2016YFC1201100, and 2017YFC0506200), the special funded project of the Chinese Research Academy of Environmental Sciences in 2021 (2021-JY-27), and the Biodiversity Survey and Assessment Project of the Ministry of Ecology and Environment of China (2019HJ2096001006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Hongli Li (Beijing Forestry University) and Quanru Liu (Beijing Normal University) for their helpful suggestions of Spartina anglica in China.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.909429/full#supplementary-material
Footnotes
References
Agapow, P. M., and Burt, A. (2001). Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 1, 101–102. doi: 10.1046/j.1471-8278.2000.00014.x
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
An, S., Gu, B., Zhou, C., Wang, Z., Deng, Z., Zhi, Y., et al. (2007). Spartina invasion in China: implications for invasive species management and future research. Weed Res. 47, 183–191. doi: 10.1111/j.1365-3180.2007.00559.x
Baird, N. A., Etter, P. D., Atwood, T. S., Currey, M. C., Shiver, A. L., Lewis, Z. A., et al. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3:e3376. doi: 10.1371/journal.pone.0003376
Balloux, F., Lehmann, L., and de Meeûs, T. (2003). The population genetics of clonal and partially clonal diploids. Genetics 164, 1635–1644. doi: 10.1093/genetics/164.4.1635
Barrett, S. C., Colautti, R. I., and Eckert, C. G. (2008). Plant reproductive systems and evolution during biological invasion. Mol. Ecol. 17, 373–383. doi: 10.1111/j.1365-294X.2007.03503.x
Baskin, C. C., and Baskin, J. M. (1998). Seeds: Ecology, Biogeography, And, Evolution Of Dormancy And Germination. Elsevier: Amsterdam, doi: 10.1016/B978-0-12-080260-9.X5000-3
Bay, R. A., Harrigan, R. J., Underwood, V. L., Gibbs, H. L., Smith, T. B., and Ruegg, K. (2018). Genomic signals of selection predict climate-driven population declines in a migratory bird. Science 359, 83–86. doi: 10.1126/science.aan4380
Bentley, K. E., and Mauricio, R. (2016). High degree of clonal reproduction and lack of large-scale geographic patterning mark the introduced range of the invasive vine, kudzu (Pueraria montana var. lobata), in North America. Am. J. Bot. 103, 1499–1507. doi: 10.3732/ajb.1500434
Bernik, B. M., Li, H., and Blum, M. J. (2016). Genetic variation of Spartina alterniflora intentionally introduced to China. Biol. Invasions 18, 1485–1498. doi: 10.1007/s10530-016-1096-3
Bossdorf, O., Auge, H., Lafuma, L., Rogers, W. E., Siemann, E., and Prati, D. (2005). Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144, 1–11. doi: 10.1007/s00442-005-0070-z
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). Tassel: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Brini, F., Hanin, M., Mezghani, I., Berkowitz, G. A., and Masmoudi, K. (2007). Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt-and drought-stress tolerance in Arabidopsis thaliana plants. J. Exp. Bot. 58, 301–308. doi: 10.1093/jxb/erl251
Cao, L. J., Li, B. Y., Chen, J. C., Zhu, J. Y., Hoffmann, A. A., and Wei, S. J. (2021). Local climate adaptation and gene flow in the native range of two co-occurring fruit moths with contrasting invasiveness. Mol. Ecol. 30, 4204–4219. doi: 10.1111/mec.16055
Capblancq, T., Luu, K., Blum, M. G., and Bazin, E. (2018). Evaluation of redundancy analysis to identify signatures of local adaptation. Mol. Ecol. Resour. 18, 1223–1233. doi: 10.1111/1755-0998.12906
Castillo, J. M., Gallego-Tévar, B., Figueroa, E., Grewell, B. J., Vallet, D., Rousseau, H., et al. (2018). Low genetic diversity contrasts with high phenotypic variability in heptaploid Spartina densiflora populations invading the Pacific coast of North America. Ecol. Evol 8, 4992–5007. doi: 10.1002/ece3.4063
Catchen, J. M., Amores, A., Hohenlohe, P., Cresko, W., and Postlethwait, J. H. (2011). Stacks: building and genotyping loci de novo from short-read sequences. G3-Genes. Genom. Genet. 1, 171–182. doi: 10.1534/g3.111.000240
Caye, K., Jumentier, B., Lepeule, J., and François, O. (2019). LFMM 2: fast and accurate inference of gene-environment associations in genome-wide studies. Mol. Biol. Evol. 36, 852–860. doi: 10.1093/molbev/msz008
Chambers, J. M., Freeny, A. E., and Heiberger, R. M. (2017). “Analysis of variance,” in Statistical Models, ed Edn, ed. T. J. Hastie (Milton Park: Routledge), 145–193.
Chung, C. (1985). Brief History Of Spartina Anglica And Research Work Abroad. Nanjing: Nanjing University Press, 30.
Chung, C.-H. (1993). Thirty years of ecological engineering with Spartina plantations in China. Ecol. Eng. 2, 261–289. doi: 10.1016/0925-8574(93)90019-C
Clavero, M., and García-Berthou, E. (2005). Invasive species are a leading cause of animal extinctions. Trends Ecol. Evol. 20:110. doi: 10.1016/j.tree.2005.01.003
Cullingham, C. I., Cooke, J. E., and Coltman, D. W. (2014). Cross-species outlier detection reveals different evolutionary pressures between sister species. New Phytol. 204, 215–229. doi: 10.1111/nph.12896
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Deng, Z., An, S., Zhou, C., Wang, Z., Zhi, Y., Wang, Y., et al. (2007). Genetic structure and habitat selection of the tall form Spartina alterniflora loisel. in China. Hydrobiologia 583, 195–204. doi: 10.1007/s10750-006-0529-x
Dlugosch, K. M., and Parker, I. M. (2008). Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 17, 431–449. doi: 10.1111/j.1365-294X.2007.03538.x
Dray, S., Blanchet, G., Borcard, D., Guenard, G., Jombart, T., Larocque, G., et al. (2018). Package ‘Adespatial’. Available online at https://cran.microsoft.com/web/packages/adespatial/adespatial.pdf (accessed April 10, 2022).
Early, R., Bradley, B. A., Dukes, J. S., Lawler, J. J., Olden, J. D., Blumenthal, D. M., et al. (2016). Global threats from invasive alien species in the twenty-first century and national response capacities. Nat. Commun. 7, 1–9. doi: 10.1038/ncomms12485
Eckert, C. G. (2002). “The loss of sex in clonal plants,” in Ecology and Evolutionary Biology Of Clonal Plants, Dordrecht, edts Edn, eds J. F. Stuefer, B. Erschbamer, H. Huber, and J. I. Suzuki (New York:NY: Springer Press), 279–298. doi: 10.1007/978-94-017-1345-0_15
Ellstrand, N. C., and Schierenbeck, K. A. (2006). Hybridization as a stimulus for the evolution of invasiveness in plants. Euphytica 148, 35–46. doi: 10.1007/s10681-006-5939-3
Estoup, A., Ravigné, V., Hufbauer, R., Vitalis, R., Gautier, M., and Facon, B. (2016). Is there a genetic paradox of biological invasion? Annu. Rev. Ecol. Evol. Syst. 47, 51–72. doi: 10.1146/annurev-ecolsys-121415-032116
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinf. 1, 47–50.
Facon, B., Genton, B. J., Shykoff, J., Jarne, P., Estoup, A., and David, P. (2006). A general eco-evolutionary framework for understanding bioinvasions. Trends Ecol. Evol. 21, 130–135. doi: 10.1016/j.tree.2005.10.012
Frankham, R. (2005). Resolving the genetic paradox in invasive species. Heredity (Edinb) 94:385. doi: 10.1038/sj.hdy.6800634
Frichot, E., and François, O. (2015). LEA: An R package for landscape and ecological association studies. Methods Ecol. Evol. 6, 925–929. doi: 10.1111/2041-210X.12382
Fridley, J. D., Stachowicz, J., Naeem, S., Sax, D., Seabloom, E., Smith, M., et al. (2007). The invasion paradox: reconciling pattern and process in species invasions. Ecology 88, 3–17. doi: 10.1890/0012-9658(2007)88[3:tiprpa]2.0.co;2
Gallego-Garcia, N., Forero-Medina, G., Vargas-Ramirez, M., Caballero, S., and Shaffer, H. B. (2019). Landscape genomic signatures indicate reduced gene flow and forest-associated adaptive divergence in an endangered neotropical turtle. Mol. Ecol. 28, 2757–2771. doi: 10.1111/mec.15112
Gaudeul, M., Giraud, T., Kiss, L., and Shykoff, J. A. (2011). Nuclear and chloroplast microsatellites show multiple introductions in the worldwide invasion history of common ragweed, Ambrosia artemisiifolia. PLoS One 6:e17658. doi: 10.1371/journal.pone.0017658
Geng, Y.-P., Pan, X.-Y., Xu, C.-Y., Zhang, W.-J., Li, B., Chen, J.-K., et al. (2007). Phenotypic plasticity rather than locally adapted ecotypes allows the invasive alligator weed to colonize a wide range of habitats. Biol. Invasions 9, 245–256. doi: 10.1007/s10530-006-9029-1
Genton, B. J., Shykoff, J. A., and Giraud, T. (2005). High genetic diversity in French invasive populations of common ragweed, Ambrosia artemisiifolia, as a result of multiple sources of introduction. Mol. Ecol. 14, 4275–4285. doi: 10.1111/j.1365-294X.2005.02750.x
Glaubitz, J. C., Casstevens, T. M., Lu, F., Harriman, J., Elshire, R. J., Sun, Q., et al. (2014). TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9:e90346. doi: 10.1371/journal.pone.0090346
Graus, D., Konrad, K. R., Bemm, F., Patir Nebioglu, M. G., Lorey, C., Duscha, K., et al. (2018). High V-PPase activity is beneficial under high salt loads, but detrimental without salinity. New Phytol. 219, 1421–1432. doi: 10.1111/nph.15280
Gray, M., and Raybould, A. (1991). “A century of evolution in Spartina anglica,” in Advances in Ecological Research, Elsevier, eds A. MacFayden and E. D. Ford (Cambridge: Academic Press), 1–62. doi: 10.1016/s0065-2504(08)60096-3
Groves, H., and Groves, J. (1880). Spartina townsendii nobis. Rep. Bot. Soc. Exch. Club Br. Id. 1, 37.
Halkett, F., Simon, J.-C., and Balloux, F. (2005). Tackling the population genetics of clonal and partially clonal organisms. Trends in Ecol. Evol. 20, 194–201. doi: 10.1111/nph.15280
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G., and Jarvis, A. (2005). Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978. doi: 10.1016/j.tree.2005.01.001
Hijmans, R. J., Guarino, L., Bussink, C., Mathur, P., Cruz, M., Barrentes, I., et al. (2004). Diva-gis: Vsn. Available online at http://diva-gis.org (Accessed 6 December 2019)
Huang, K., Wang, D., Duan, P., Zhang, B., Xu, R., Li, N., et al. (2017). Wide and thick grain 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice. Plant J. 91, 849–860. doi: 10.1111/tpj.13613
Jackson, J. M., Pimsler, M. L., Oyen, K. J., Strange, J. P., Dillon, M. E., and Lozier, J. D. (2020). Local adaptation across a complex bioclimatic landscape in two montane bumble bee species. Mol. Ecol. 29, 920–939. doi: 10.1111/mec.15376
Jombart, T., Devillard, S., and Balloux, F. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94. doi: 10.1186/1471-2156-11-94
Kamvar, Z. N., Brooks, J. C., and Grünwald, N. J. (2015). Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 6:208. doi: 10.3389/fgene.2015.00208
Kamvar, Z. N., Tabima, J. F., and Grünwald, N. J. (2014). Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. Peer. J. 2:e281. doi: 10.7717/peerj.281
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kue Foka, I. C., Ketehouli, T., Zhou, Y., Li, X.-W., Wang, F.-W., and Li, H. (2020). The emerging roles of diacylglycerol kinase (DGK) in plant stress tolerance, growth, and development. Agronomy 10:1375. doi: 10.3390/agronomy10091375
Lai, J., Zou, Y., and Zhang, J. Peres-Neto, P. R. (2022). Generalizing hierarchical and variation partitioning in multiple regression and canonical analyses using the rdacca. hp R package. Methods Ecol. Evol. 13, 782–788. doi: 10.1111/2041-210X.13800
Lee, C. E. (2002). Evolutionary genetics of invasive species. Trends Ecol. Evol. 17, 386–391. doi: 10.1016/S0169-5347(02)02554-5
Li, F., van Kleunen, M., Li, J., Liu, X., Gao, K., Zhu, J., et al. (2019b). Patterns of genetic variation reflect multiple introductions and pre-admixture sources of common ragweed (Ambrosia artemisiifolia) in China. Biol. Invasions 21, 2191–2209. doi: 10.1007/s10530-019-01966-2
Li, F.-F., Gong, L., Li, J.-S., Liu, X.-Y., and Zhao, C.-Y. (2019a). Low genetic differentiation yet high phenotypic variation in the invasive populations of Spartina alterniflora in Guangxi China. PLoS One 14:e0222646. doi: 10.1371/journal.pone.0222646
Li, H., An, S., Zhi, Y., Yan, C., Zhao, L., Zhou, C., et al. (2008). Protogynous, pollen limitation and low seed production reasoned for the dieback of Spartina anglica in coastal China. Plant Sci. 174, 299–309. doi: 10.1016/j.plantsci.2007.11.015
Li, H., Zhi, Y., An, S., Zhao, L., Zhou, C., Deng, Z., et al. (2009). Density-dependent effects on the dieback of exotic species Spartina anglica in coastal China. Ecol. Eng. 35, 544–552. doi: 10.1016/j.ecoleng.2008.03.001
Li, H., Zhi, Y., Zhao, L., An, S., Deng, Z., Zhou, C., et al. (2007). Eco-physiological responses of the declining population Spartina anglica to N and P fertilizer addition. Sheng. Xuebao 27, 2725–2732. doi: 10.1016/s1872-2032(07)60060-2
Li, Y., Tan, Y., Shao, Y., Li, M., and Ma, F. (2015). Comprehensive genomic analysis and expression profiling of diacylglycerol kinase gene family in Malus prunifolia (Willd.) Borkh. Gene 561, 225–234. doi: 10.1016/j.gene.2015.02.029
Lischer, H. E., and Excoffier, L. (2012). PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28, 298–299. doi: 10.1093/bioinformatics/btr642
Liu, J., Dong, M., Miao, S. L., Li, Z. Y., Song, M. H., and Wang, R. Q. (2006). Invasive alien plants in China: role of clonality and geographical origin. Biol. Inv. 8, 1461–1470. doi: 10.1007/s10530-005-5838-x
Liu, L., An, S., Zhi, Y., Zhang, M., and Li, H. (2016a). Effects of different sediment type and burial depth on growth traits and biomass accumulation of Spartina anglica. Shengwu Duoyangxing 24, 1279–1287. doi: 10.17520/biods.2016024
Liu, W., Maung-Douglass, K., Strong, D. R., Pennings, S. C., and Zhang, Y. (2016b). Geographical variation in vegetative growth and sexual reproduction of the invasive Spartina alterniflora in China. J. Ecol. 104, 173–181. doi: 10.1111/1365-2745.12487
Liu, W., Strong, D. R., Pennings, S. C., and Zhang, Y. (2017). Provenance-by-environment interaction of reproductive traits in the invasion of Spartina alterniflora in China. Ecology 98, 1591–1599. doi: 10.1002/ecy
Liu, W., Zhang, Y., Chen, X., Maung-Douglass, K., Strong, D. R., and Pennings, S. C. (2020). Contrasting plant adaptation strategies to latitude in the native and invasive range of Spartina alterniflora. New Phytol. 226, 623–634. doi: 10.1111/nph.16371
Lotterhos, K. E., and Whitlock, M. C. (2014). Evaluation of demographic history and neutral parameterization on the performance of FST outlier tests. Mol. Ecol. 23, 2178–2192. doi: 10.1111/mec.12725
Maurice, S., Montes, M. S., Nielsen, B. J., Bodker, L., Martin, M. D., Jonck, C. G., et al. (2019). Population genomics of an outbreak of the potato late blight pathogen, Phytophthora infestans, reveals both clonality and high genotypic diversity. Mol. Plant Pathol. 20, 1134–1146. doi: 10.1111/mpp.12819
Moles, A. T., Ackerly, D. D., Webb, C. O., Tweddle, J. C., Dickie, J. B., Pitman, A. J., et al. (2005). Factors that shape seed mass evolution. Proc. Natl. Acad. Sci. 102, 10540–10544. doi: 10.1073/pnas.0501473102
Molina-Montenegro, M. A., Acuna-Rodriguez, I. S., Flores, T. S. M., Hereme, R., Lafon, A., Atala, C., et al. (2018). Is the success of plant invasions the result of rapid adaptive evolution in seed traits? Evidence from a latitudinal rainfall gradient. Front. Plant Sci. 9:208. doi: 10.3389/fpls.2018.00208
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89, 583–590. doi: 10.1093/genetics/89.3.583
Nielsen, E. S., Henriques, R., Toonen, R. J., Knapp, I. S. S., Guo, B., and Von Der Heyden, S. (2018). Complex signatures of genomic variation of two non-model marine species in a homogeneous environment. BMC Genomics 19:347. doi: 10.1186/s12864-018-4721-y
Ochocki, B. M., and Miller, T. E. X. (2017). Rapid evolution of dispersal ability makes biological invasions faster and more variable. Nat. Commun. 8, 1–8. doi: 10.1038/ncomms14315
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., Mcglinn, D., et al. (2021). Vegan: Community Ecology Package. R package version 2.5-7. 2020.Available online at https://CRAN.R-project.org/package=vegan. (accessed 10 February 2022).
Pantoja, P. O., Simón-Porcar, V. I., Puzey, J. R., and Vallejo-Marín, M. (2017). Genetic variation and clonal diversity in introduced populations of Mimulus guttatus assessed by genotyping at 62 single nucleotide polymorphism loci. Plant Ecol. Divers. 10, 5–15. doi: 10.1080/17550874.2017.1287785
Park, S., Li, J., Pittman, J. K., Berkowitz, G. A., Yang, H., Undurraga, S., et al. (2005). Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc. Natl. Acad. Sci. 102, 18830–18835. doi: 10.1073/pnas.0509512102
Peterson, P. M., Romaschenko, K., Arrieta, Y. H., and Saarela, J. M. (2014). A molecular phylogeny and new subgeneric classification of Sporobolus (poaceae: chloridoideae: sporobolinae). Taxon 63, 1212–1243. doi: 10.12705/636.19
Prentis, P. J., Wilson, J. R., Dormontt, E. E., Richardson, D. M., and Lowe, A. J. (2008). Adaptive evolution in invasive species. Trends Plant Sci. 13, 288–294. doi: 10.1016/j.tplants.2008.03.004
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). Plink: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Qiao, H., Liu, W., Zhang, Y., Zhang, Y. Y., and Li, Q. Q. (2019). Genetic admixture accelerates invasion via provisioning rapid adaptive evolution. Mol. Ecol. 28, 4012–4027. doi: 10.1111/mec.15192
Raybould, A., Gray, A., Lawrence, M., and Marshall, D. (1991). The evolution of Spartina anglica CE Hubbard (Gramineae): origin and genetic variability. Biol. J. Linn. Soc. Lond. 43, 111–126. doi: 10.1111/j.1095-8312.1991.tb00588.x
Reichel, K., Masson, J.-P., Malrieu, F., Arnaud-Haond, S., and Stoeckel, S. (2016). Rare sex or out of reach equilibrium? BMC Genet. 17:76. doi: 10.1186/s12863-016-0388-z
Ren, M. X., Zhang, Q. G., and Zhang, D. Y. (2005). Random amplified polymorphic DNA markers reveal low genetic variation and a single dominant genotype in Eichhornia crassipes populations throughout China. Weed Res. 45, 236–244. doi: 10.1111/j.1365-3180.2005.00445.x
Rengefors, K., Gollnisch, R., Sassenhagen, I., Svensson, M., Lebret, K., Čertnerová, D., et al. (2021). Genome-wide SNP markers reveal population structure and dispersal direction of an expanding nuisance algal bloom species. Mol. Ecol. 30, 912–925. doi: 10.1111/mec.15787
Rigas, S., Daras, G., Laxa, M., Marathias, N., Fasseas, C., Sweetlove, L. J., et al. (2009). Role of Lon1 protease in post-germinative growth and maintenance of mitochondrial function in Arabidopsis thaliana. New Phytol. 181, 588–600. doi: 10.1111/j.1469-8137.2008.02701.x
Rius, M., and Darling, J. A. (2014). How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol. Evol. 29, 233–242. doi: 10.1016/j.tree.2014.02.003
Roman, J., and Darling, J. A. (2007). Paradox lost: genetic diversity and the success of aquatic invasions. Trends in Ecol. Evol. 22, 454–464. doi: 10.1016/j.tree.2007.07.002
Ruelland, E., Cantrel, C., Gawer, M., Kader, J.-C., and Zachowski, A. (2002). Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol. 130, 999–1007. doi: 10.1104/pp.006080
Saleh, D., Xu, P., Shen, Y., Li, C., Adreit, H., Milazzo, J., et al. (2012). Sex at the origin: an asian population of the rice blast fungus Magnaporthe oryzae reproduces sexually. Mol. Ecol. 21, 1330–1344. doi: 10.1111/j.1365-294X.2012.05469.x
Schierenbeck, K. A., and Ellstrand, N. C. (2008). Hybridization and the evolution of invasiveness in plants and other organisms. Biol. Invasions 11, 1093–1105. doi: 10.1007/s10530-008-9388-x
Schrieber, K., and Lachmuth, S. (2017). The genetic paradox of invasions revisited: the potential role of inbreeding x environment interactions in invasion success. Biol. Rev. Camb. Philos. Soc. 92, 939–952. doi: 10.1111/brv.12263
Shannon, C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Simberloff, D., Martin, J.-L., Genovesi, P., Maris, V., Wardle, D. A., Aronson, J., et al. (2013). Impacts of biological invasions: what’s what and the way forward. Trends Ecol. Evol. 28, 58–66. doi: 10.1016/j.tree.2012.07.013
Stapley, J., Santure, A. W., and Dennis, S. R. (2015). Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol. Ecol. 24, 2241–2252. doi: 10.1111/mec.13089
Stoeckel, S., and Masson, J. P. (2014). The exact distributions of F(IS) under partial asexuality in small finite populations with mutation. PLoS One 9:e85228. doi: 10.1371/journal.pone.0085228
Storey, J. D., Bass, A. J., Dabney, A., and Robinson, D. (2015). qvalue: Q-Value Estimation For False Discovery Rate Control. R package version 2.6.0.Available online at: github.com/jdstorey/qvalue. (accessed April 14, 2019).
Suarez, A. V., and Tsutsui, N. D. (2008). The evolutionary consequences of biological invasions. Mol. Ecol. 17, 351–360. doi: 10.1111/j.1365-294X.2007.03456.x
Tan, W.-J., Yang, Y.-C., Zhou, Y., Huang, L.-P., Xu, L., Chen, Q.-F., et al. (2018). Diacylglycerol acyltransferase and diacylglycerol kinase modulate triacylglycerol and phosphatidic acid production in the plant response to freezing stress. Plant Physiol. 177, 1303–1318. doi: 10.1104/pp.18.00402
Tsujimoto, M., Araki, K. S., Honjo, M. N., Yasugi, M., Nagano, A. J., Akama, S., et al. (2020). Genet assignment and population structure analysis in a clonal forest-floor herb. AOB Plants 12:lz080. doi: 10.1093/aobpla/plz080
Turner, K. G., Hufbauer, R. A., and Rieseberg, L. H. (2014). Rapid evolution of an invasive weed. New Phytol. 202, 309–321. doi: 10.1111/nph.12634
van Boheemen, L. A., Lombaert, E., Nurkowski, K. A., Gauffre, B., Rieseberg, L. H., and Hodgins, K. A. (2017). Multiple introductions, admixture and bridgehead invasion characterize the introduction history of ambrosia artemisiifolia in Europe and Australia. Mol. Ecol. 26, 5421–5434. doi: 10.1111/mec.14293
VanWallendael, A., Hamann, E., and Franks, S. J. (2018). Evidence for plasticity, but not local adaptation, in invasive Japanese knotweed (Reynoutria japonica) in North America. Ecol. Evol. 32, 395–410. doi: 10.1007/s10682-018-9942-7
Wang, P., Alpert, P., and Yu, F. H. (2016). Clonal integration increases relative competitive ability in an invasive aquatic plant. Am. J. Bot. 103, 2079–2086. doi: 10.3732/ajb.1600160
Wellband, K. W., and Heath, D. D. (2017). Plasticity in gene transcription explains the differential performance of two invasive fish species. Evol. Appl. 10, 563–576. doi: 10.1111/eva.12463
Wellband, K. W., Pettitt-Wade, H., Fisk, A. T., and Heath, D. D. (2017). Differential invasion success in aquatic invasive species: the role of within- and among-population genetic diversity. Biol. Inv. 19, 2609–2621. doi: 10.1007/s10530-017-1471-8
Wellband, K. W., Pettitt-Wade, H., Fisk, A. T., and Heath, D. D. (2018). Standing genetic diversity and selection at functional gene loci are associated with differential invasion success in two non-native fish species. Mol. Ecol. 27, 1572–1585. doi: 10.1111/mec.14557
Xia, L., Geng, Q., and An, S. (2020). Rapid genetic divergence of an invasive Species, Spartina alterniflora, in China. Front. Genet. 11:284. doi: 10.3389/fgene.2020.00284
Xia, L., Zhao, H., Yang, W., and An, S. (2015). Genetic diversity, ecotype hybrid, and mixture of invasive Spartina alterniflora Loisel in Coastal China. CLEAN–Soil Air Water 43, 1672–1681. doi: 10.1002/clen.201300882
Xiao, Y., Tang, J., Qing, H., Zhou, C., and An, S. (2011). Effects of salinity and clonal integration on growth and sexual reproduction of the invasive grass Spartina alterniflora. Flora Morphol. Distrib. Funct. Ecol. Plants. 206, 736–741. doi: 10.1016/j.flora.2010.12.003
Xu, G. W., and Zhuo, R. Z. (1985). Preliminary Studies Of Introduced Spartina alterniflora Loisel in China (I),” In Research Advances in Spartina: Achievements of Past 22 Years. Nanjing: Journal of Nanjing University, 212–215.
Yandell, B. S. (2017). Practical Data Analysis for Designed Experiments. New York:NY: Routledge Press.
Zenni, R. D., and Nuñez, M. A. (2013). The elephant in the room: the role of failed invasions in understanding invasion biology. Oikos 122, 801–815. doi: 10.1111/j.1600-0706.2012.00254.x
Zenni, R. D., Bailey, J. K., and Simberloff, D. (2014). Rapid evolution and range expansion of an invasive plant are driven by provenance-environment interactions. Ecol. Lett. 17, 727–735. doi: 10.1111/ele.12278
Zhang, D., Hu, Y., Liu, M., Chang, Y., Yan, X., Bu, R., et al. (2017). Introduction and spread of an exotic plant, spartina alterniflora, along coastal marshes of China. Wetlands 37, 1181–1193. doi: 10.1007/s13157-017-0950-0
Zhang, Q., and Xiao, S. (2015). Lipids in salicylic acid-mediated defense in plants: focusing on the roles of phosphatidic acid and phosphatidylinositol 4-phosphate. Front. Plant Sci. 6:387. doi: 10.3389/fpls.2015.00387
Zhou, H., Alexander, D., and Lange, K. (2011). A quasi-newton acceleration for high-dimensional optimization algorithms. Stat. Comput. 21, 261–273. doi: 10.1007/s11222-009-9166-3
Keywords: Spartina alterniflora, biological invasions, RAD, genetic paradox, environmental adaptation
Citation: Li F, Liu X, Zhu J, Li J, Gao K and Zhao C (2022) The Role of Genetic Factors in the Differential Invasion Success of Two Spartina Species in China. Front. Plant Sci. 13:909429. doi: 10.3389/fpls.2022.909429
Received: 31 March 2022; Accepted: 02 May 2022;
Published: 30 May 2022.
Edited by:
Francesco Sunseri, Mediterranea University of Reggio Calabria, ItalyReviewed by:
Jesús Manuel Castillo, Seville University, SpainNicole Kollars, Northeastern University, United States
Copyright © 2022 Li, Liu, Zhu, Li, Gao and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caiyun Zhao, emhhb2N5Y3JhZXNAMTI2LmNvbQ==, emhhb2N5QGNyYWVzLm9yZy5jbg==
 Feifei Li
Feifei Li Xiaoyan Liu
Xiaoyan Liu Jinfang Zhu
Jinfang Zhu Junsheng Li
Junsheng Li Kexiao Gao
Kexiao Gao Caiyun Zhao
Caiyun Zhao