- 1Chinese Academy of Agricultural Sciences, Tobacco Research Institute, Qingdao, China
- 2Graduate School of Chinese Academy of Agricultural Sciences, Beijing, China
- 3Technology Center, China Tobacco Hunan Industrial Co., Ltd., Changsha, China
- 4QuJing Tobacco Company, Qujing, China
Leaf senescence is a highly coordinated process and has a significant impact on agriculture. Plant peptides are known to act as important cell-to-cell communication signals that are involved in multiple biological processes such as development and stress responses. However, very limited number of peptides has been reported to be associated with leaf senescence. Here, we report the characterization of the INFLORESCENCE DEFICIENT IN ABSCISSION-LIKE6 (IDL6) peptide as a regulator of leaf senescence. The expression of IDL6 was up-regulated in senescing leaves. Exogenous application of synthetic IDL6 peptides accelerated the process of leaf senescence. The idl6 mutant plants showed delayed natural leaf senescence as well as senescence included by darkness, indicating a regulatory role of IDL6 peptides in leaf senescence. The role of IDL6 as a positive regulator of leaf senescence was further supported by the results of overexpression analysis and complementation test. Transcriptome analysis revealed differential expression of phytohormone-responsive genes in idl6 mutant plants. Further analysis indicated that altered expression of IDL6 led to changes in leaf senescence phenotypes induced by ABA and ethylene treatments. The results from this study suggest that the IDL6 peptide positively regulates leaf senescence in Arabidopsis thaliana.
Introduction
As a vital part in plants’ life cycle, leaf senescence is a type of post-mitotic senescence which involves a strictly programmed cell death process (Gan, 2003). During leaf senescence, cellular organelles and macromolecules are degraded and nutrients are remobilized to reproductive organs and new tissues (Li et al., 2017; Guo et al., 2021). In agricultural applications, artificially accelerating or delaying leaf senescence in crop plants could achieve higher yields and better quality (Gan, 2014; Guo and Gan, 2014; Woo et al., 2019). During the past two decades, a large number of stay-green loci and senescence regulators have been identified from model plant systems which can be potentially used in manipulating leaf senescence for crop improvement (Vadez et al., 2011; Guo et al., 2015; Del Pozo et al., 2016).
Leaf senescence is an extremely complicated process and can be triggered by diverse factors including internal and external factors (Guo and Gan, 2005). The former includes phytohormones and reproduction growth (Lim et al., 2007). For instance, abscisic acid (ABA; He et al., 2005), ethylene (Iqbal et al., 2017), Jasmonate (JA; Hu et al., 2017) and salicylic acid (SA; Abreu and Munné-Bosch, 2008) accelerate leaf senescence, while cytokinin (CK; Hwang et al., 2012), auxin (IAA; Lim et al., 2010) and gibberellins (GA; Rodrigues et al., 2012) are negative regulators of senescence. External senescence-regulating factors are complex and generally associated with biotic/abiotic stresses, such as darkness, salinity, drought, extreme temperature and pathogen infection. Interestingly, some genes involved in biotic/abiotic stresses have also been reported to regulate leaf senescence. WRKY55 was reported to be involved in both leaf senescence and disease resistance by regulating the accumulation of reactive oxygen species (ROS) and SA (Wang et al., 2020). In rice, ONAC106 can be induced by salt stress, and functions as a negative regulator of leaf senescence (Sakuraba et al., 2015).
As an important part of cell-to-cell interaction in higher plants, peptide signals have been characterized to be involved in various aspects of plants’ life cycle including meristem organization (Guo et al., 2015), self-incompatibility (Liu et al., 2021), reproduction (Kim et al., 2021), organ abscission (Jinn et al., 2000), root growth (Huang et al., 2006), stress responses (Yamaguchi et al., 2010), hormone signaling (Estornell et al., 2013), nodule development (Mohd-Radzman et al., 2016) and RNA metabolism (Matsubayashi and Sakagami, 2006; Zhang et al., 2020). INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) and IDA-Like (IDL) peptides are a small subgroup of plant peptides with 9 members in Arabidopsis, which possess an N-terminal signal peptide and a C-terminal extended PIP (EPIP) domain (Stenvik et al., 2008a,b). Some IDL peptides have been characterized for their roles in cell separation and stress responses. The IDA peptide was identified to control floral organ abscission and lateral root emergence via interacting with its receptors HAESA (HAE) and HAESA-LIKE2 (Stenvik et al., 2008b; Kumpf et al., 2013; Liu et al., 2013). The IDA-HAE/HSL2 signaling module functions in activating the mitogen-activated protein (MAP) kinase cascades, which in turn regulate the expression of cell wall-modifying and hydrolytic enzymes (Kumpf et al., 2013; Meng et al., 2016). In addition, SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 (SERK1) has been shown to act as a co-receptor of the IDA peptide in regulating flower abscission (Santiago et al., 2016). The coding genes of IDL6 and IDL7 peptides were shown to be induced rapidly by various stresses and have been suggested to be negative modulators of stress-induced ROS signaling (Vie et al., 2015, 2017). A recent study showed that the IDL6-HAE/HSL2 signaling module functions in facilitating infection of Pseudomonas syringae pv. tomato (Pst) DC3000 by promoting pectin degradation in Arabidopsis leaves (Wang et al., 2017).
A recent study reported that the CLAVATA3/ESR-RELATED 14 (CLE14) peptide serves as a senescence-regulating signal in Arabidopsis (Zhang et al., 2021), raising the possibility of more peptide signals involved in leaf senescence. Here we describe the characterization of the IDL6 peptide in regulating leaf senescence of Arabidopsis. The expression of IDL6 was detected to be up-regulated in senescing leaves. The loss-of-function idl6 mutant displayed a delayed senescence phenotype, and this phenotype could be rescued by the IDL6 gene. Overexpressing IDL6 or exogenous application of synthetic IDL6 peptides accelerated leaf senescence. Transcriptome analysis showed differential expression of phytohormones-responsive genes in idl6 mutant plants. Further test of ABA and ethylene-induced senescence on detached leaves suggest that IDL6 might function via affecting ABA and ethylene signaling. Taken together, the results from this study indicate that IDL6 is a positive modulator of leaf senescence in Arabidopsis.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis Columbia ecotype (Col-0) was used as the wild type in this study. The idl6 mutant (SALK_074245) was obtained from the Arabidopsis Biological Resource Center (ABRC) and genotyped via PCR. The transcript abundance of the IDL6 gene in idl6 and Col-0 plants was detected by quantitative real-time PCR (qRT-PCR).
Arabidopsis seeds were surface sterilized with 70% (v/v) ethanol for 5 min and spread evenly in half-strength Murashige and Skoog (1/2 MS) media. Then the media were cultivated at 23°C with continuous light after being placed in a 4°C refrigerator for 2 days. Two weeks later, seedlings were transplanted to soil mixture, and kept in a growth chamber at 23°C with continuous light (Li et al., 2021; Zhang et al., 2021). Leaves at different development stages were selected for gene expression analysis. The fifth and sixth rosette leaves at different developmental stages were collected to explore the expression pattern of IDL6 (YL, young leaves, 4-week-old; NS, non-senescence, 4.5-week-old; ES, early senescence, 5.5-week-old, LS, late senescence, 6.5-week-old). To this end, leaf samples of three biological replicates were collected and frozen in liquid nitrogen for RNA extraction.
Generation of Constructs and Transgenic Plants
For overexpression analysis, IDL6 (AT5G05300) CDS was PCR amplified from cDNA of Col-0 leaves. The PCR product was purified and inserted into the enzyme digested pCHF3 vector with Sac I by Infusion (Clontech, Beijing, China). The promoter sequence of IDL6 was amplified from genomic DNA of Col-0 leaves and the PCR product was purified and inserted into the enzyme digested pBI121a vector (modified from pBI121) with Sac I by Infusion. Similarly, the promoter plus CDS fragment of IDL6 was cloned into pZP211 for complementation test. All constructs were confirmed by Sanger sequencing and transformed into Agrobacterium competent cells (GV3101), which were used to transform Arabidopsis by Agrobacterium-mediated floral dip method (Zhang et al., 2006). The positive transgenic plants were screened on 1/2 MS medium containing 50 mg/l kanamycin and T3 homozygous lines were used for further study.
RNA Extraction and qRT-PCR
Total RNAs from each sample were extracted by using the Ultrapure RNA Kit (cwbiotech, Beijing, China). Reverse transcriptions were performed using the Evo M-MLV Mix Kit with gDNA Clean for qPCR (Accurate Biotechnology, Changsha, China). qRT-PCR was performed using a Roche LightCycler 480 Real-Time PCR instrument with SYBR® Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology, Changsha, China). ACT2 was used as an internal control and all experimental data were obtained with three technical repetitions. The resulted Data were analyzed via the 2–ΔΔCt method (Livak and Schmittgen, 2001). All primers used in this study are listed in Supplementary Table 1.
Determination of Fv/Fm, Chlorophyll Content and Ion Leakage
The chlorophyll fluorescence Fv/Fm of individual leaves was determined using the IMAGING-PAM Mseries Chlorophyll Fluorescence System (LI-6400-40 LCF, Walz, Effeltrich, Germany) according to the manufacturer’s instructions (Rossel et al., 2006). For determination of chlorophyll content, 100% methanol was used in dissolving chlorophyll from leaves. After chlorophyll was completely released, absorbance at 666 and 653 nm was obtained with a spectrophotometer (ClarioSTAR, BMG LABTECH, Offenburg, Germany), chlorophyll content was calculated as previously described (Lightenthaler, 1987). For ion leakage measurement, leaves were immersed in deionized distilled water, shaken at 25°C for 30 min, and the beginning conductivity was measured using a digital conductivity meter (Thermo Fisher Scientific Traceable, Hampton, NH, United States of America). The samples were then boiled for 15 min and then the second conductivity was measured. The percentage of the first measurement over the second measurement was used as the membrane leakage indicator (Zhao et al., 2018).
Peptide Synthesis and Detached Leaf Senescence Assay
The IDL6 peptide EPIP-domain (Supplementary Figure 1) sequence FGSLVLNALPKGSVPASGPSKRIN was synthesized by Genscript (Nanjing, China) at the purity of 95%. For detached leaf senescence assay, the seventh and eighth leaves of 8-week-old wild-type plants were excised and incubated in plates containing 1 μM IDL6 peptides. Leaf senescence phenotypes were recorded and chlorophyll contents were measured.
Dark- and Phytohormone-Induced Leaf Senescence
All plants were grown on soil in a growth chamber under continuous light at 23°C. For dark-induced leaf senescence analysis, the fifth, sixth and seventh leaves of 4.5-week-old wild-type, idl6 mutant, overexpression and rescued lines were excised and placed onto moistened filter paper inside foil-wrapped petri dishes as described previously (Li et al., 2016). Pictures were taken 0 d, 3 d, and 5 d after treatments. For hormone-induced senescence treatments, the seventh, eighth and nineth leaves of 6-week-old plants were incubated in liquid 1/2 MS media with or without 10 μM ABA or 50 μM ethephon, respectively, as described previously (Li et al., 2021). Pictures were taken at 0 d, 1 d, 3 d, and 5 d after treatments. For all measurements, three biological replicates were performed.
RNA-Sequencing Analysis
The sixth and seventh leaves of 4-week-old Col-0 and idl6 plants were collected and frozen in liquid nitrogen immediately. Each plant sample was represented by three biological replicates. The samples were entrusted to Shanghai OE Biotech for RNA-Seq. The quality of the sequencing data was scrutinized in terms of total raw reads, total clean reads, Q20 percentage, and GC percentage. DEGs were filtered using the following criteria: |Log2 (fold change)| > 2.0, p < 0.05. KEGG enrichment analysis was based on the path entries with the number of corresponding differential genes greater than 2, and sorted according to the corresponding -log10 p-value. Raw RNA-seq reads are available at the National Center for Biotechnology Information (BioProject ID: PRJNA821657).
Statistical Analysis
All data analyses in this study were performed based on at least three biological replicates. Statistically significant differences were determined using Student’s t-test (*p < 0.05, **p < 0.01, and ***p < 0.001). Values in graphs are the mean value ± SE of all replicates.
Results
ILD6 Is Up-Regulated During Leaf Senescence
The 5th and 6th rosette leaves of Arabidopsis (Col-0) plants were collected to explore the expression pattern of IDL6 at four different developmental stages, including young leaf (YL), non-senescence leaf (NS), early senescence leaf (ES) and late senescence leaf (LS; Figure 1A). Chlorophyll contents decreased from NS to LS stage, indicating progression of leaf senescence (Figure 1B). As expected (Gan and Amasino, 1995; Zhang et al., 2012; Guo et al., 2021), the rubisco small subunit encoding gene RBCS was down-regulated during leaf senescence (Figure 1C), while the senescence marker gene SAG12 was up-regulated from NS stage to LS stage, and was highly expressed at the late senescence stage (Figure 1D). The IDL6 transcripts were detected to be highly expressed in leaves at both early and late senescence stages (Figure 1E).
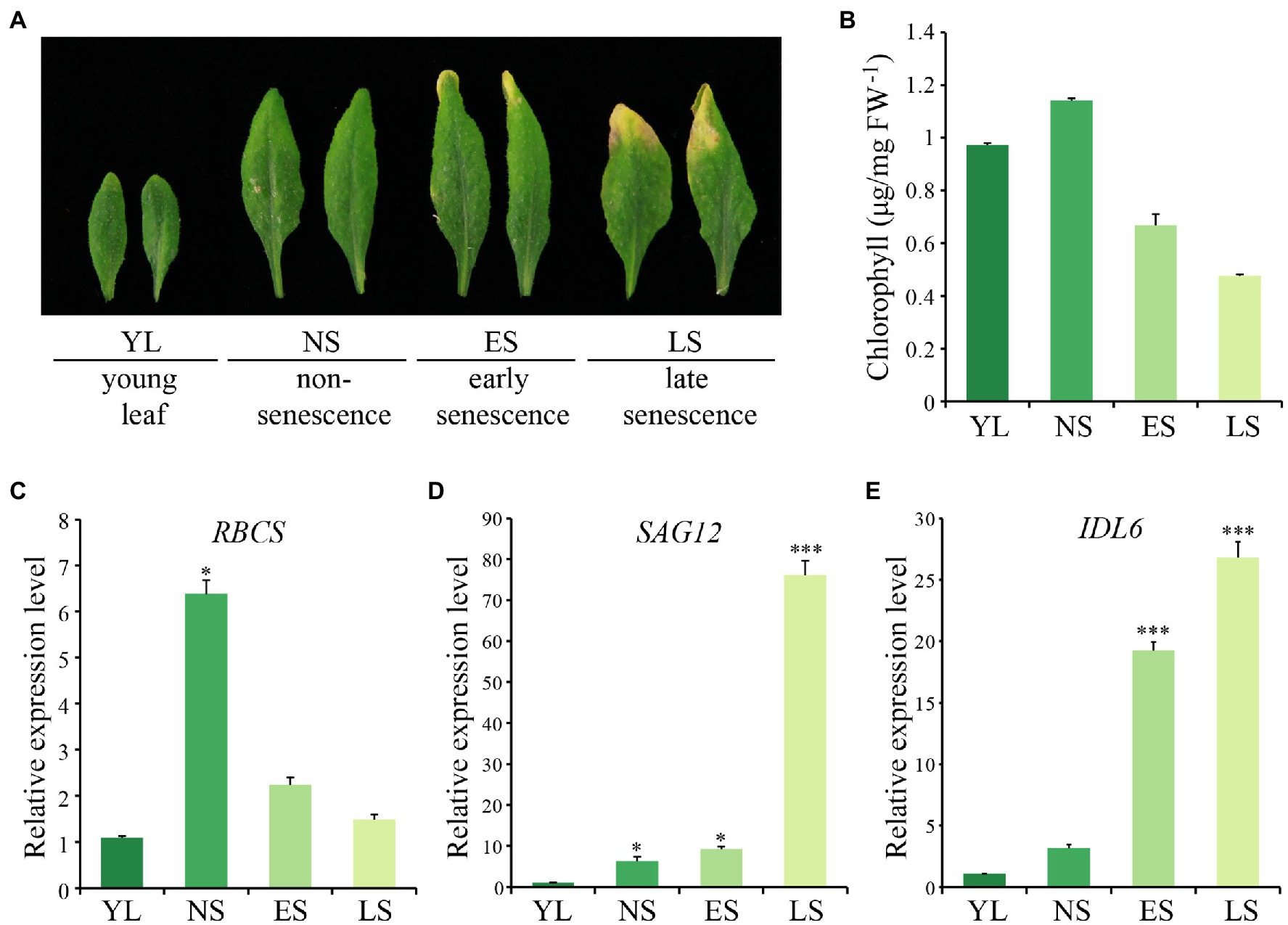
Figure 1. The expression pattern of IDL6 during leaf senescence. (A) The Arabidopsis wild-type leaves at different developmental stages. YL: young leaf, NS: non-senescence leaf, ES: early senescence leaf, LS: late senescence leaf. (B) The chlorophyll contents of leaves at different developmental stages. (C–E) The transcript abundance of RBCS, SAG12 and IDL6 during leaf senescence. The data are means ± SD of three biological repeats. Significant difference compared with the YL was determined by Student’s t-test (*p < 0.05 and ***p < 0.001).
Considering that natural senescence of a single leaf proceeds from the tip to the base part, an early senescence leaf from the 6th position of 8-week-old wild-type Arabidopsis was collected for analysis. From this leaf, three sections were isolated, including the tip, the middle and the base (Figure 2A). As expected, chlorophyll contents and photosynthetic rates declined gradually from tip to base (Figures 2B,C). Additionally, the transcript abundance of SAG12 and RBCS were correlated with the degree of leaf senescence, with the highest expression levels of RBCS and SAG12 detected in the base and the tip part, respectively, (Figures 2E,F). IDL6 gene was expressed similarly to SAG12, which increased continuously from the leaf base to the tip and was highly expressed in the senescent leaf tip (Figure 2G). Furthermore, the IDL6 promoter was cloned in front of the b-glucuronidase (GUS) gene and used for transforming wild-type plants. When ProIDL6::GUS leaves at the early senescence stage were used for GUS staining, more GUS signals were detected at the tip of leaves. Notably, strong GUS activity was observed at the base part of mechanically damaged leaves, suggesting that IDL6 was also induced by wounding (Figure 2D).
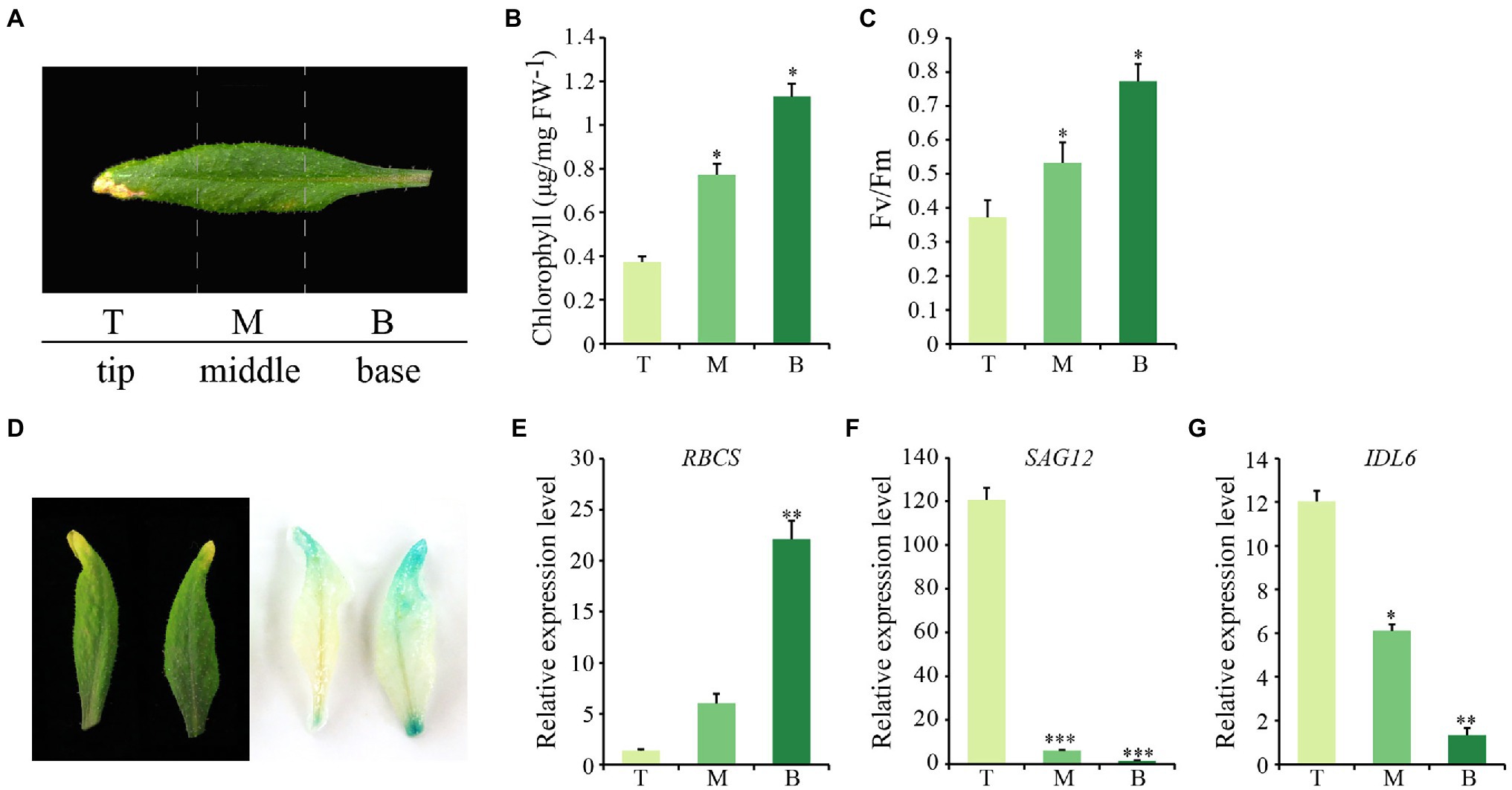
Figure 2. The expression of IDL6 in different parts of a senescing leaf. (A) Representative image of three sections of Arabidopsis wild-type leaves. T: leaf tip, M: middle part, B: leaf base. Chlorophyll contents (B) and photosynthetic rates (C) in different leaf sections. (D): GUS staining on 7th and 8th rosette leaves from 8-week-old ProIDL6::GUS transgenic line. (E-G) Transcript abundance of RBCS, SAG12 and IDL6 in different leaf sections. The data are means ± SD of three biological repeats. Significant differences compared with the tip part were determined by Student’s t-test (*p < 0.05, **p < 0.01, and ***p < 0.001).
Exogenous Application of Synthetic IDL6 Peptides Accelerates Leaf Senescence
As a special group of plant hormones, peptide ligands could be artificially synthesized to explore their function. In previous studies, the EPIP-domain (extended PIP) of AtIDA and MiIDL1 were synthesized in determining their roles in floral organ shedding (Stenvik et al., 2008b; Kim et al., 2018). The EPIP sequence of IDL6 peptide was confirmed by sequence alignment according to previous studies (Butenko et al., 2003; Stenvik et al., 2008b) and was artificially synthesized for treating detached leaves (Supplementary Figure 1). Leaf disks of the seventh and eighth leaves from 8-week-old wild-type plants were treated with 1 μM IDL6 EPIP peptides. Three and 4 days later, an early senescence phenotype can be observed on the leaf disks treated with IDL6 EPIP peptides compared to mock treatments (Figure 3A). In consistent with visible phenotypes, significant decline of chlorophyll content and Fv/Fm was detected in leaves treated with IDL6 EPIP peptides in comparison with mock treatments (Figures 3B,C). Besides, the RBCS expression level of mock treatments was higher than peptide treatments, while, the SAG12 expression level of mock treatments was lower than peptide treatments (Figures 3D,E). This result suggests that the IDL6 EPIP peptide functions in accelerating leaf senescence.
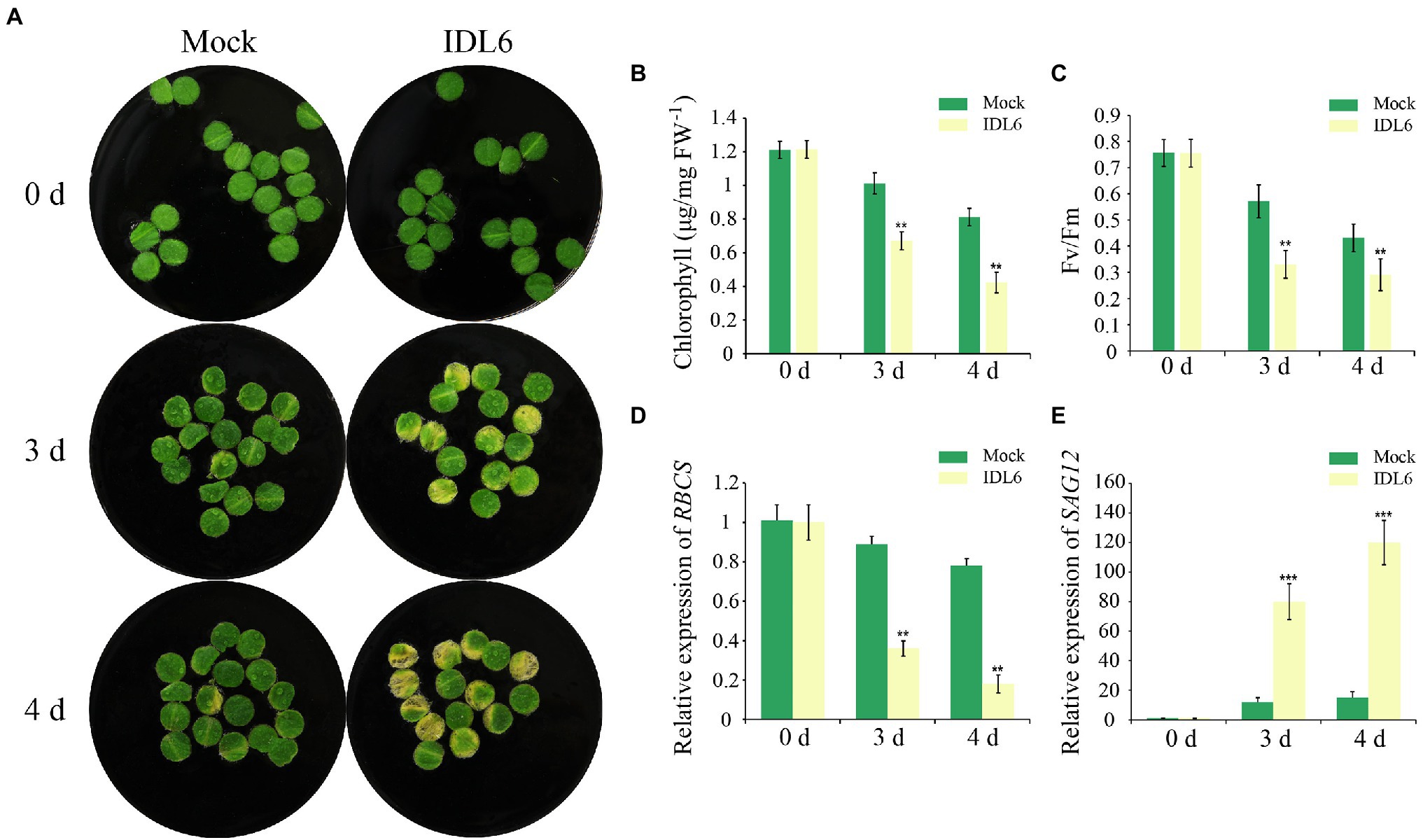
Figure 3. The senescence phenotypes of detached leaves treated with synthetic IDL6 peptides. (A) The detached leaves from wild type of Arabidopsis exhibited early senescence under IDL6 EPIP peptide treatments. Leaves were kept in a growth chamber at 23°C with continuous light. Changes in chlorophyll content (B) and photosynthetic rate (C) in the treated leaves. The relative expression of RBCS (A) and SAG12 (E) in leaves from each group. Error bars indicate the SE (n > 10). Significant difference compared with the Mock was determined by Student’s t-test (**p < 0.01 and ***p < 0.001).
Loss of IDL6 Function Delays Leaf Senescence
To confirm the role of IDL6 in regulating of leaf senescence, a mutant line SALK_074245 (idl6) with a T-DNA insertion before the start codon of the IDL6 gene was obtained (Figure 4A). PCR and Sanger sequencing were performed to identify homozygous idl6 mutant plants. IDL6 transcript was not detected in the late senescence leaf of idl6 plants, suggesting that is idl6 a null mutant (Figures 4B,C). Two rescue lines were obtained by transforming the IDL6 gene back to idl6 mutant plants (Figure 4C; Supplementary Figure 2).
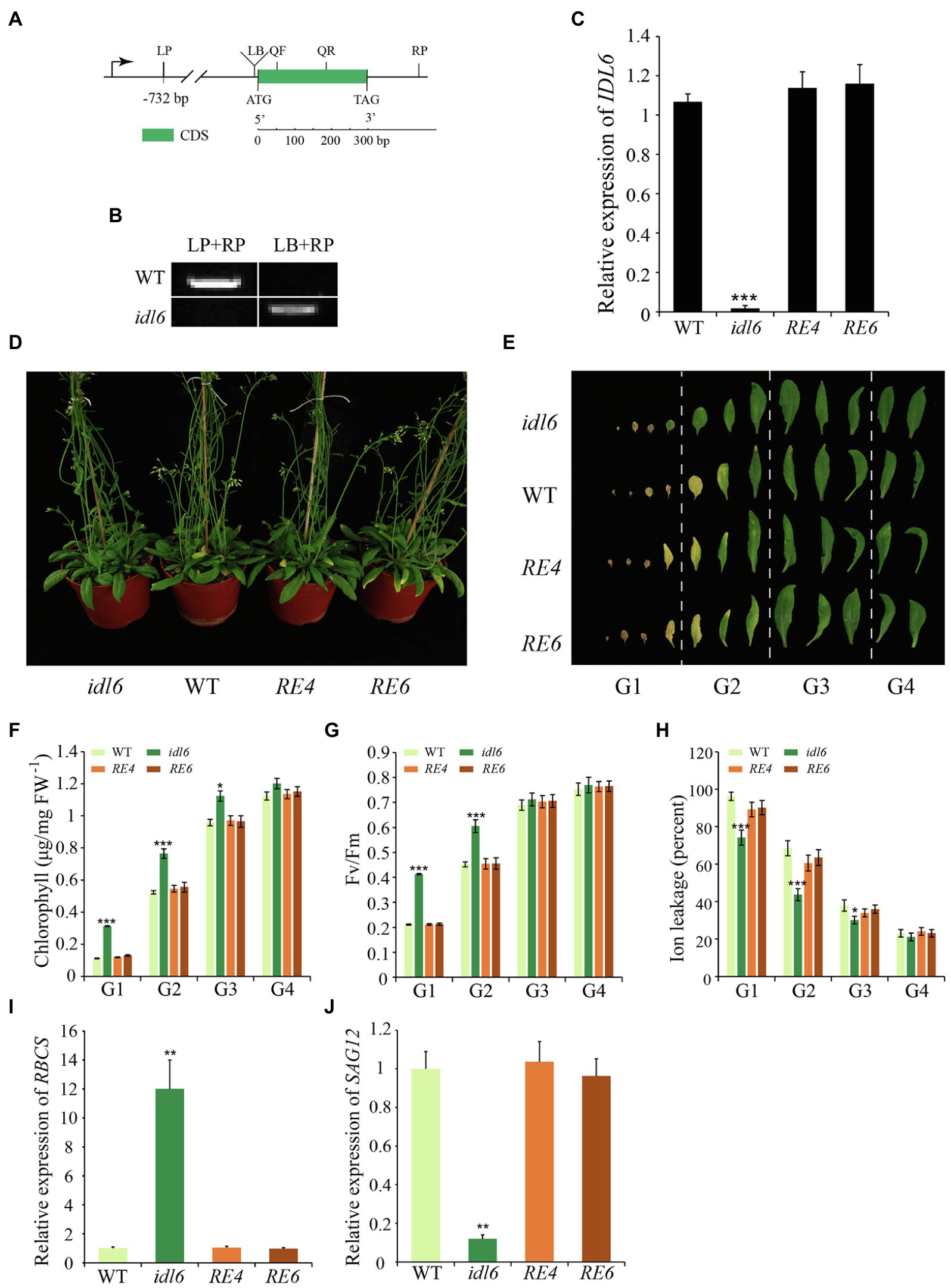
Figure 4. Leaf senescence phenotypes of IDL6 loss-of-function plants. (A) The gene structure of the IDL6 gene and location of the T-DNA insertion. (B) Homozygous mutant plants were identified by PCR. LP: left primer, RP: right primer, LB: T-DNA left border primer. (C) Quantification of IDL6 transcripts in wild-type, idl6 mutant and IDL6 rescue lines. (D) Leaf senescence phenotypes of 8-week-old wild-type, idl6 mutant and IDL6 rescue lines (RE4 and RE6). (E) Phenotypes of 12 detached rosette leaves from wild-type, idl6 mutant and IDL6 rescue lines. Quantification of chlorophyll contents (F), photosynthetic rates (G) and ion leakage (H) in leaves from each group. The relative expression of RBCS (I) and SAG12 (J) of group 2 leaves from each line. The data are means ± SD of three biological repeats. Significant differences (*p < 0.05, **p < 0.01, and ***p < 0.001) compared with the wild type in each group were determined by Student’s t-test.
No obvious phenotypic changes were found between the idl6 mutant line, wild-type and rescue lines at early developmental stages. At alter stage, when 8-week-old plants were compared, the loss-of-function idl6 mutant displayed a significant delay in leaf senescence. The delayed senescence phenotype of idl6 plants was rescued to wild type in plants of the rescue lines (Figures 4D,E). To further characterize the senescence phenotypes of the idl6 mutant, the 12 rosette leaves from plants of different genotypes were divided into four groups to collect physiological data (Figure 4E). As a result, the highest Fv/Fm ratio and chlorophyll contents were found in leaves of each group from the idl6 mutant (Figures 4F,G). Ion leakage is an important plasma membrane integrity indicator, and is considered one of the most important indicators of leaf senescence (Feller and Fischer, 1994). The leaves of the idl6 mutant showed significantly lower ion leakage rates compared with the wild-type and rescue lines (Figure 4H). As expected, the RBCS expression level of idl6 plants was higher than that of the wild-type and complementary lines, while the SAG12 expression level of the idl6 mutant was lower than wild-type and complementary lines (Figures 4I,J).
Overexpression of IDL6 Accelerates Leaf Senescence
To further explore the function of IDL6 in leaf senescence, the IDL6 gene was overexpressed using the 35S promoter. Two overexpression lines (namely OE-1 and OE-4) were selected to for further analysis. The results showed that leaves of these overexpression lines exhibit morphologies of smaller size, which is consistent with what Wang et al. (2017) reported. The IDL6-OE lines displayed an early leaf senescence phenotype compared with wild type (Figures 5A–C). The chlorophyll contents and Fv/Fm values in the overexpression lines were significantly lower than that of wild-type plants (Figures 5D,E). The high ion leakage in the overexpression lines also suggested that the leaf senescence progression was accelerated with enhanced IDL6 expression (Figure 5F). This was also demonstrated by the expression of RBCS and SAG12 in wild-type and overexpressed plants (Figures 5G,H).
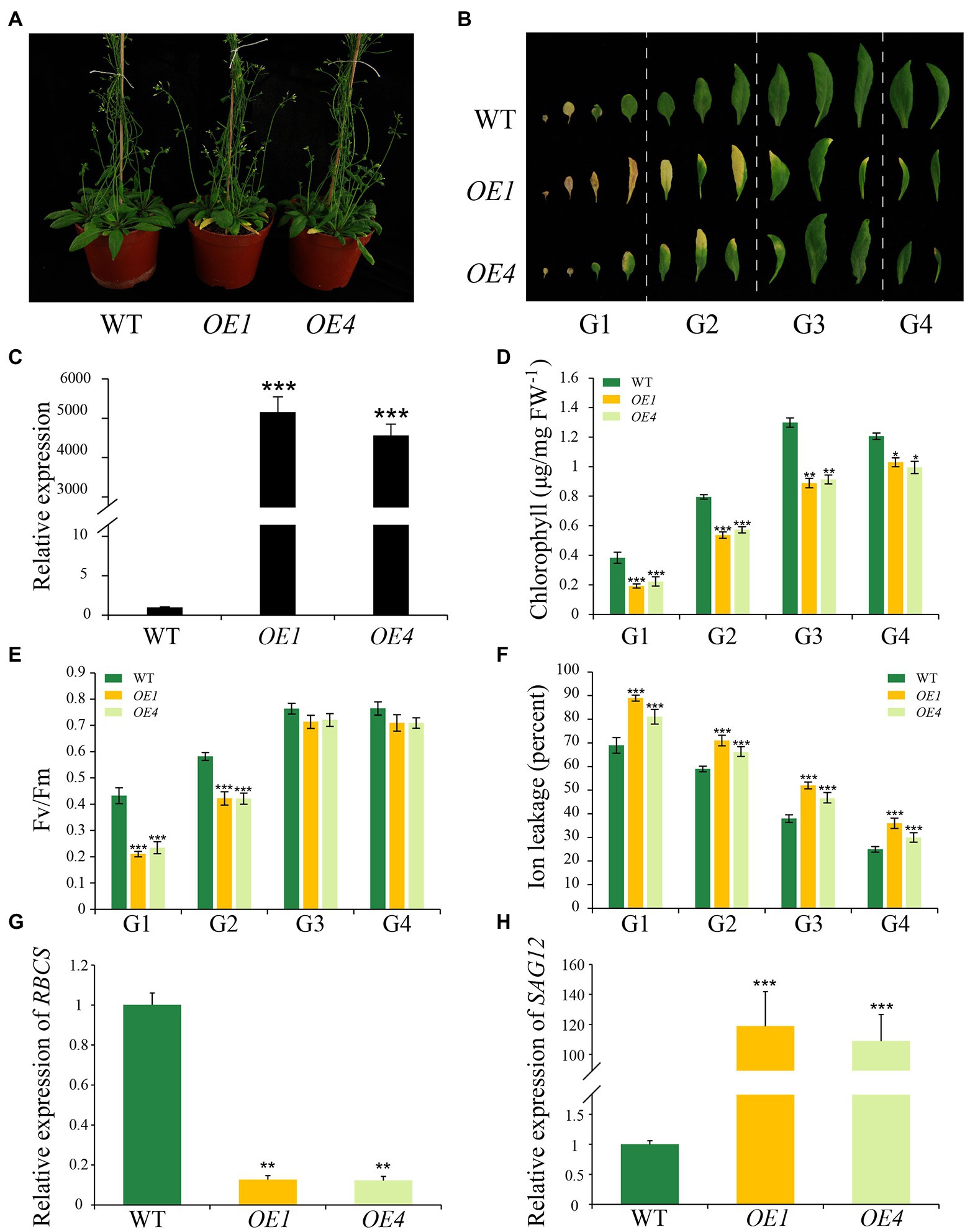
Figure 5. Leaf senescence phenotypes of IDL6 overexpression plants. (A) Leaf senescence phenotypes of 6-week-old wild-type and IDL6 overexpression lines (OE1 and OE4). (B) Phenotypes of 12 detached rosette leaves from wild-type and IDL6 overexpression lines. (C) Relative expression of the IDL6 gene in wild-type and IDL6 overexpression lines. Quantification of chlorophyll contents (D), photosynthetic rates (E) and ion leakage (F) in leaves from 4 groups of rosette leaves. The relative expression of RBCS (G) and SAG12 (H) of group 2 leaves from each line. The data are means ± SD of three biological repeats. Significant differences (*p < 0.05, **p < 0.01, and ***p < 0.001) compared with the wild type in each group were determined by Student’s t-test.
Transcriptome Analysis Reveals the Importance of Hormone Signaling in IDL6-Mediated Leaf Senescence
Earlier studies suggested that 10–16% of all genes show differential expression during leaf senescence (Breeze et al., 2011; Woo et al., 2016). To explore gene expression changes caused by the idl6 mutation, the 6th and 7th leaves with similar degree of yellowing from wild-type and idl6 plants were collected to perform RNA-seq analysis.
The transcriptome analysis identified 2,618 differentially expressed genes (DEGs) in idl6 mutant compared with wild type (Figure 6A; Supplementary Table 2). Interestingly, The WRKY transcription factor WRKY53 (Miao et al., 2004) and Dof transcription factor CDF4 (Xu et al., 2020), which have been reported to be positive regulators of leaf senescence, were significantly down-regulated in the idl6 mutant (Figure 6C). Moreover, a number of stress responsive WRKY transcription factors, including WRKY38 and WRKY62 (Kim et al., 2008), were significantly down-regulated in idl6 leaves (Figure 6C), Also, the expression levels of these genes were confirmed in wild-type, idl6, rescue and overexpression lines with qRT-PCR (Supplementary Figure 3).
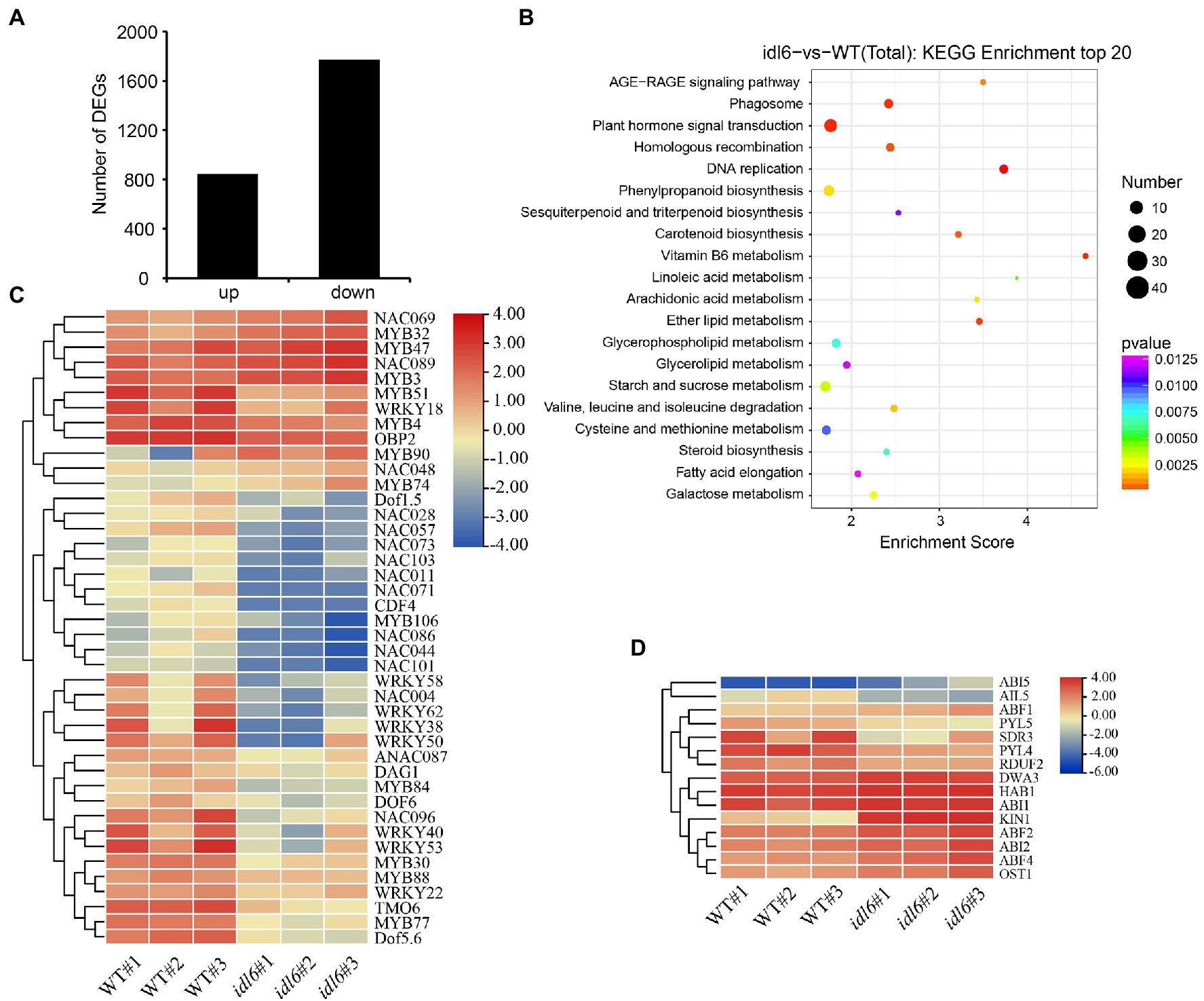
Figure 6. Genome-wide transcriptome analysis of idl6 mutant plants. (A) The number of up-regulated and down-regulated genes in rosette leaves of idl6 mutant vs. Col-0. (B) KEGG bubble diagram of the DEGs in rosette leaves of idl6 vs. Col-0. Bubble size represents numbers of DEGs, and bubble color represents p-values. The enrichment score represents the significance degree of DEGs in a certain signaling pathway. (C) Heatmap showing leaf senescence-related transcription factors in idl6 mutant leaves compared with Col-0. The log2 fold change scale is indicated on the right side of the heatmap. (D) The heatmap of ABA-related genes in idl6 mutant leaves compared to Col-0. The log2 fold change scale is indicated on the right side of the heatmap.
When KEGG analyses of DEGs were performed to compare wild-type and the idl6 mutant, more DEGs were enriched in plant hormone signaling pathways (Figure 6B). The ABA-INSENSITIVE1 (ABI1) protein phosphatase 2C and ABI2 were reported to be negative regulators of ABA signaling (Merlot et al., 2001). Both ABI1 and ABI2 were up-regulated in idl6 mutant (Figure 6D). In addition, a number of genes related to ABA signal transduction, including ABI5, PYL5, PYL4, OST1, ABF2 and ABF4 (Finkelstein and Lynch, 2000; Mustilli et al., 2002; Kim et al., 2004; Santiago et al., 2009) showed differential expression in idl6 plants (Figure 6D). These results suggest that IDL6 might be involved in hormonal-induced leaf senescence.
IDL6 Might Be Involved in ABA- and Ethylene-Induced Leaf Senescence
A growing body of evidence suggests that both ABA and ethylene are positive regulators of leaf senescence. In order to find out whether IDL6 is involved in leaf senescence induced by ABA or ethylene, the seventh, eighth and nineth leaves of wild-type, idl6, IDL6-OE1 and IDL6-RE4 plants were isolated and treated with 10 μm ABA or 50 μm ethylene. As expected, leaf senescence was accelerated by ABA and ethylene treatments. Significant leaf yellowing was observed on leaves from wild type 1 day after ABA treatments and 3 days after ethylene treatments. Leaves from the idl6 mutant showed delayed while leaves from the IDL6 overexpression line OE1 showed accelerated senescence compared with wild type. The IDL6 rescue-line showed the similar leaf senescence progress with wild type after both ABA and ethylene treatments (Figures 7A–C). Changes in photosynthetic rates and chlorophyll contents in the treated leaves were consistent with the leaf yellowing progression (Figures 7D,E). These results hint that IDL6 might be involved in ABA- and ethylene-induced leaf senescence.
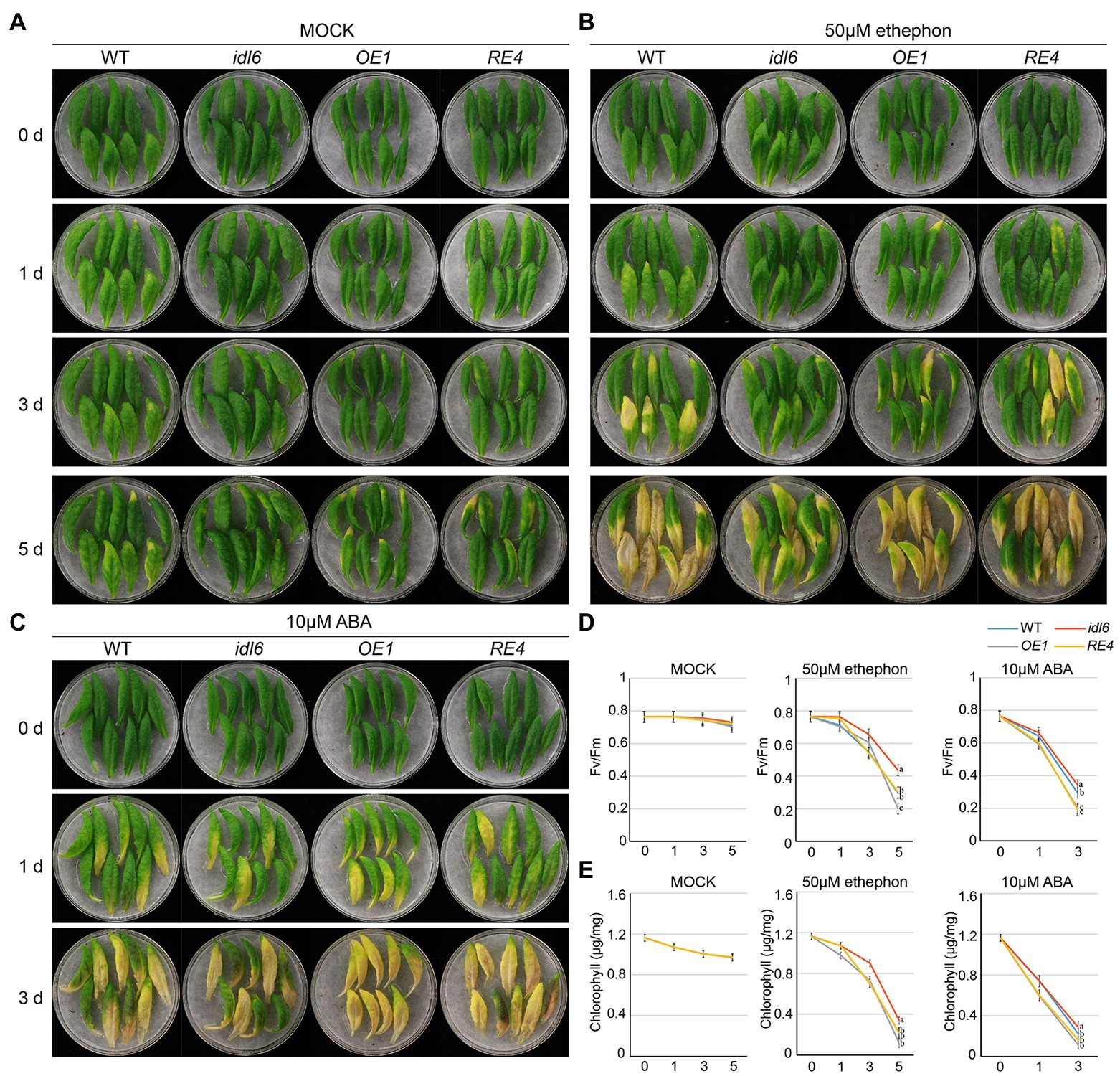
Figure 7. Senescence induced by ABA and ethylene on detached leaves of Arabidopsis. The 7th, 8th and 9th leaves of 6-week-old wild type, idl6 mutant, IDL6 OE plants and idl6 rescue plants were collected and treated by mock (A), 50 μM ethephon (B) and 10 μM ABA (C). The images of leaves were taken before and 1 day, 3 days, and 5 days after treatments. (D) The Fv/Fm and (E) chlorophyll concentrations in hormone treated and untreated leaves. For all conditions, statistically significant had been performed by two-way ANOVA analysis. All treatments were performed for three times.
IDL6 Functions in Promoting Dark-Induced Senescence
To further explore the role of IDL6 peptides in leaf senescence, dark treatments were performed to induce leaf senescence. The 5th, 6th, and 7th leaves were excised from 4.5-week-old plants (WT, idl6, OE1, OE4, RE4, and RE6), placed on moistened filter papers and incubated under darkness to induce senescence. Three days later, the chlorophyll levels of all leaves from the IDL6 OE lines were significantly reduced, while relatively less chlorophyll breakdown occurred in the leaves of idl6 mutant plants. After 5 days of dark treatment, the early senescence phenotype became more obvious on leaves from IDL6 OE lines (Figures 8A,B). Specifically, the chlorophyll contents in IDL6 OE lines were ~ 2.94, ~ 3.13, and 2.94 times lower than that of the wild-type, idl6 mutant and IDL6 RE lines, respectively. The changes in Fv/Fm also showed a similar trend (Figures 8B,C). Notably, overexpression of IDL6 could also affect RBCS and SAG12 expression in dark-induced senescence (Supplementary Figure 4). These results suggest that IDL6 is a positive regulator of dark-induced leaf senescence.
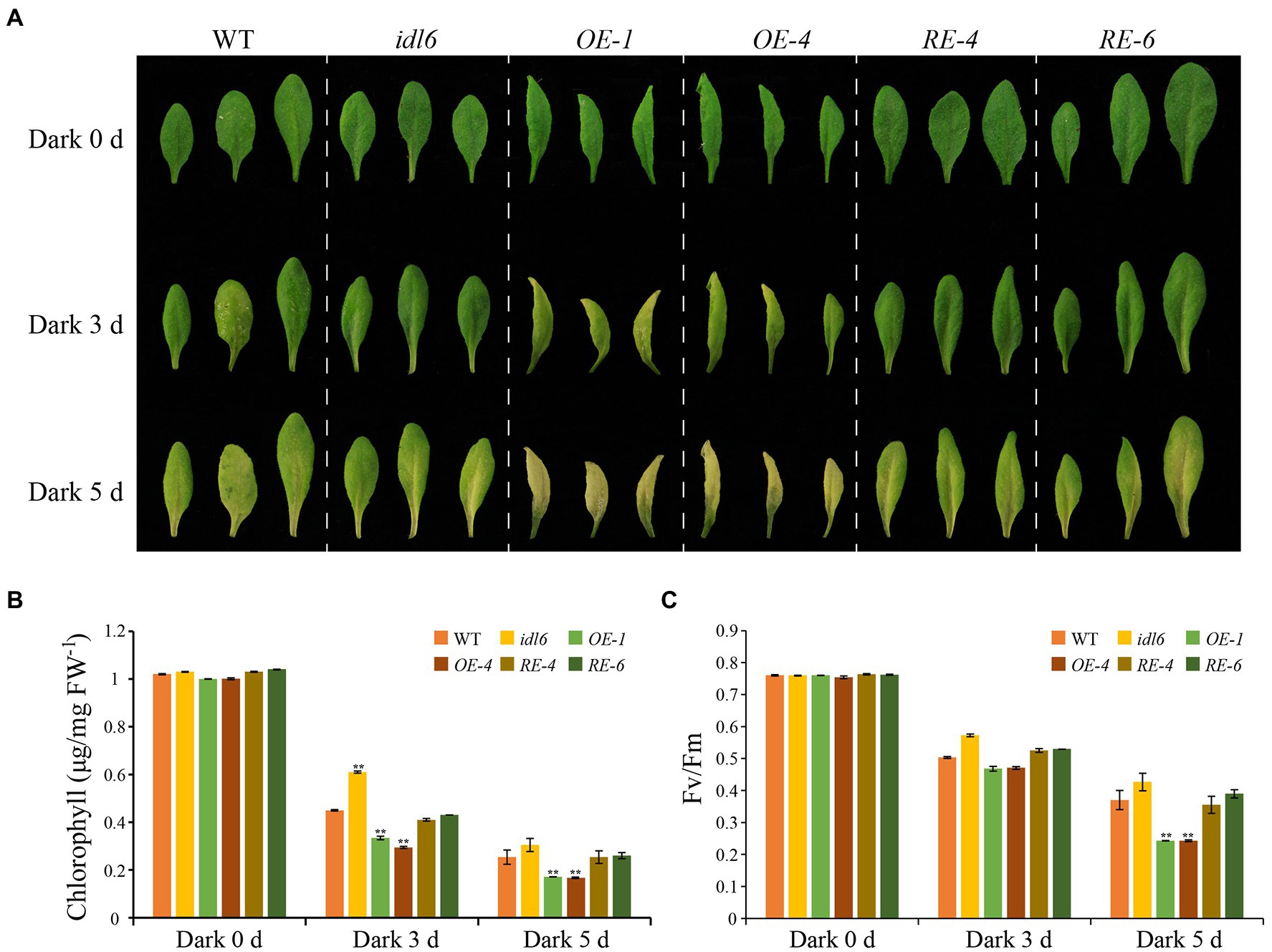
Figure 8. Dark-induced senescence in detached leaves with altered IDL6 expression. (A) The senescence phenotype s of the 5th, 6th, and 7th leaves from 4.5-week-old plants under dark treatment. Chlorophyll contents (B) and photosynthetic efficiency (C) of each leaf were measured after dark treatment for 0, 3 and 5 days. Error bars showed the SE (n = 3). Significant differences (**p < 0.01) compared with the wild type in each group were determined by Student’s t-test. Three independent experiments were carried out with similar results.
Discussion
Being stationary in nature, plants have evolved a series of sophisticated mechanisms for responding to unpredictable environmental stresses. Among them, cell-to-cell communication systems play a key role in growth and development. During the last decade, peptide ligands have emerged as vital mediators of cell-to-cell communications in plant growth, defense and stress responses in addition to the classical phytohormones (Matsubayashi and Sakagami, 2006). However, only one peptide, CLE14, has been functionally characterized and reported to be involved in leaf senescence (Zhang et al., 2021). In the current study, we found that IDL6, encoding a secreted peptide, was highly expressed in senescence leaves. Loss of IDL6 function mutation delayed leaf senescence while IDL6 overexpression and IDL6 peptide treatments caused precocious leaf senescence, supporting the role of IDL6 as a positive mediator of leaf senescence in Arabidopsis.
Comprising nine members (IDA and IDL1-8) in Arabidopsis, the IDL family members were detected to be expressed in floral organs, leaves, and roots. Among them, IDA has been shown to be important for flower abscission and lateral root emergence, while IDL6 and IDL7 have been reported to be negative regulators of genes associated with early responses to stresses (Vie et al., 2017). In a previous study, knockdown lines of IDL6 showed increased resistance to Pst DC3000 in Arabidopsis (Wang et al., 2017). In this study, IDL6 peptides were found to function in promoting age-dependent leaf senescence and senescence induced by darkness, ABA and ethylene treatments. Cross-talks between plant senescence and stress responses have been well recognized in earlier studies (Guo and Gan, 2012; Guo et al., 2021). IDL6 could be induced rapidly by various biotic and abiotic stresses, such as cold, salt, UV, P. syringae (Vie et al., 2015). It might act as a signaling hub where different pathways interconnect with each other. Notably, peptides from the same family are often found to be involved in similar biological processes (Matsubayashi and Sakagami, 2006). It will be no surprise if some of the other IDL family peptides are also found to be involved in plant senescence.
Receptor-like kinases are indispensable sensors that contribute to intercellular communication, especially peptide ligand signaling (Diévart and Clark, 2003; Gish and Clark, 2011). At present, most receptors of peptides have been identified to be in the Leucine-rich repeat receptor-like kinases (LRR-RLK) family, such as RLK7, CEPR, BAM, and PEPR (Deyoung et al., 2006; Klauser et al., 2015; Zhou et al., 2022). As the receptors of the IDA peptide, HAE and HSL2 were reported to participate in flower abscission and lateral root emergence (Stenvik et al., 2008b; Kumpf et al., 2013). HAE and HSL2 also have been characterized as receptors of the IDL6 peptide in regulating plant disease resistance by activating cell wall synthesis genes (Wang et al., 2017). Peptide signals might be perceived by the same or different receptors when functioning in different biological processes (Zhang et al., 2021). Whether IDL6 functions through interacting with HAE and HSL2 in promoting leaf senescence remains to be elucidated.
The roles of phytohormones in leaf senescence have been well established (Guo and Gan, 2012). The transcriptome analysis in this study revealed multiple DEGs related to phytohormones in the idl6 mutant (Figures 6B,D). Further study indicated that IDL6 functioned in leaf senescence induced by ABA and ethylene (Figure 7). How peptide signals including CLE14 and IDL6 interact with known senescence-regulating phytohormones and other senescence-regulating signals will be the next questions to be addressed in this field.
Leaf senescence as a complex and orderly controlled physiological process, requires hierarchical but also coordinated regulation by multiple transcription factors (Guo et al., 2004). The transcriptomic analyses of leaves from different developmental stages have identified numerous TFs differentially expressed during leaf senescence and more and more studies had characterized these TFs’ functions in leaf senescence (Guo, 2013). In this study, we found that WRKY53 and CDF4 were down-regulated in the idl6 mutant (Figure 6C), while they showed higher expression in overexpression IDL6 gene line (Supplemental Figure 3). These results indicated that transcription factors WRKY53 and CDF4, both have been characterized as positive regulators of senescence (Miao et al., 2004; Xu et al., 2020), might function downstream of IDL6 in regulating leaf senescence. The specific regulatory mechanisms underlying the interactions between IDL6 and senescence-regulating transcription factors remain to be further studied.
Conclusion
In this study, we functionally characterized the IDL6 peptide, the encoding gene of which exhibited the highest expression level in naturally senescing leaves. Exogenous application of synthetic IDL6 EPIP peptides accelerated leaf senescence. Transgenic Arabidopsis plants with depleted or overexpressed IDL6 had delayed or accelerated leaf senescence, respectively, indicating a positive role of IDL6 peptides in regulating leaf senescence. Furthermore, IDL6 peptides induced leaf senescence under darkness and hormonal treatments. Several senescence-associated transcription factors were significantly down-regulated in the idl6 mutant, suggesting extensive cross talks between the IDL6 signal and known senescence-regulating pathways.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
CG and XL conducted the research and participated in drafting the manuscript. ZeZ, QW, ZhZ, LW, CL, ZD, YC, and TL assisted in data collection and analysis. YG conceived this research, designed the experiments, and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Agricultural Science and Technology Innovation Program (ASTIP-TRIC02), Funds for Special Projects of the Central Government in Guidance of Local Science and Technology Development (21-1-1-1-zyyd-nsh), and National Natural Science Foundation of China (31571494).
Conflict of Interest
XL was employed by China Tobacco Hunan Industrial Co., Ltd. CL was employed by QuJing Tobacco Company.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.909378/full#supplementary-material
Supplementary Figure 1 | Sequence information of the IDL6 peptide.
Supplementary Figure 2 | Growth of wild-type, idl6 mutant and a complementary line on 1/2 MS medium containing Kanamycin.
Supplementary Figure 3 | The relative expression of WRKY53, CDF4, WRKY38, and WRKY62 in wild-type, idl6 mutant, rescue and overexpression lines.
Supplementary Figure 4 | The expression levels of RBCS and SAG12 in wild-type and overexpression plants under dark treatments.
Supplementary Figure 5 | The senescence phenotypes of detached leaves treated with 1 μM and 10 μM IDL6 peptides. (A) The detached leaves from wild type of Arabidopsis exhibited early senescence under IDL6 peptide treatments. Leaves were kept in a growth chamber at 23 °C with continuous light. Changes in chlorophyll content (B) and photosynthetic rate (C) in the treated leaves. Significant differences compared with the Mock were determined by Student’s t-test (*p < 0.05, **p < 0.01).
References
Abreu, M. E., and Munné-Bosch, S. (2008). Salicylic acid may be involved in the regulation of drought-induced leaf senescence in perennials: a case study in field-grown Salvia officinalis L. plants. Env. Exp. Bot. 64, 105–112. doi: 10.1016/j.envexpbot.2007.12.016
Breeze, E., Harrison, E., Mchattie, S., Hughes, L., Hickman, R., Hill, C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873–894. doi: 10.1105/tpc.111.083345
Butenko, M. A., Patterson, S. E., Grini, P. E., Stenvik, G. E., Amundsen, S. S., Mandal, A., et al. (2003). Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15, 2296–2307. doi: 10.1105/tpc.014365
Del Pozo, A., Yáñez, A., Matus, I. A., Tapia, G., Castillo, D., Sanchez-Jardón, L., et al. (2016). Physiological traits associated with wheat yield potential and performance under water-stress in a Mediterranean environment. Front. Plant Sci. 7:987. doi: 10.3389/fpls.2016.00987
Deyoung, B. J., Bickle, K. L., Schrage, K. J., Muskett, P., Patel, K., and Clark, S. E. (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45, 1–16. doi: 10.1111/j.1365-313X.2005.02592.x
Diévart, A., and Clark, S. E. (2003). Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Curr. Opin. Plant Biol. 6, 507–516. doi: 10.1016/s1369-5266(03)00089-x
Estornell, L. H., Agustí, J., Merelo, P., Talón, M., and Tadeo, F. R. (2013). Elucidating mechanisms underlying organ abscission. Plant Sci. 199, 48–60. doi: 10.1016/j.plantsci.2012.10.008
Feller, U., and Fischer, A. (1994). Nitrogen metabolism in senescing leaves. Crit. Rev. Plant Sci. 13, 241–273. doi: 10.1080/07352689409701916
Finkelstein, R. R., and Lynch, T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. doi: 10.1105/tpc.12.4.599
Gan, S. (2003). Mitotic and postmitotic senescence in plants. Sci. Aging Knowl. Environ. 24, 2003–2041. doi: 10.1126/sageke.2003.38.re7
Gan, S. (2014). Leaf senescence as an important target for improving crop production. Adv Crop Sci. Technol. 2, 10–11. doi: 10.4172/2329-8863.1000e116
Gan, S., and Amasino, R. M. (1995). Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270, 1986–1988. doi: 10.1126/science.270.5244.1986
Gish, L. A., and Clark, S. E. (2011). The RLK/Pelle family of kinases. Plant J. 66, 117–127. doi: 10.1111/j.1365-313X.2011.04518.x
Guo, Y. (2013). Towards systems biological understanding of leaf senescence. Plant Mol. Biol. 82, 519–528. doi: 10.1007/s11103-012-9974-2
Guo, Y., Cai, Z., and Gan, S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27, 521–549. doi: 10.1111/j.1365-3040.2003.01158.x
Guo, Y., and Gan, S. (2005). Leaf senescence: signals, execution, and regulation. Curr. Top. Dev. Biol. 71, 83–112. doi: 10.1016/S0070-2153(05)71003-6
Guo, Y., and Gan, S. (2012). Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 35, 644–655. doi: 10.1111/j.1365-3040.2011.02442.x
Guo, Y., and Gan, S. (2014). Translational researches on leaf senescence for enhancing plant productivity and quality. J. Exp. Bot. 65, 3901–3913. doi: 10.1093/jxb/eru248
Guo, Y., Ren, G., Zhang, K., Li, Z., Miao, Y., and Guo, H. (2021). Leaf senescence: progression, regulation, and application. Mol. Hortic. 1, 1–25. doi: 10.1186/s43897-021-00006-9
Guo, H., Zhang, W., Tian, H., Zheng, K., Dai, X., Liu, S., et al. (2015). An auxin responsive CLE gene regulates shoot apical meristem development in Arabidopsis. Front. Plant Sci. 6:295. doi: 10.3389/fpls.2015.00295
He, P., Osaki, M., Takebe, M., Shinano, T., and Wasaki, J. (2005). Endogenous hormones and expression of senescence-related genes in different senescent types of maize. J. Exp. Bot. 56, 1117–1128. doi: 10.1093/jxb/eri103
Hu, Y., Jiang, Y., Han, X., Wang, H., Pan, J., and Yu, D. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68, 1361–1369. doi: 10.1093/jxb/erx0004
Huang, G., Dong, R., Allen, R., Davis, E. L., Baum, T. J., and Hussey, R. S. (2006). A root-knot nematode secretory peptide functions as a ligand for a plant transcription factor. Mol. Plant-Microbe Interact. 19, 463–470. doi: 10.1094/MPMI-19-0463
Hwang, I., Sheen, J., and Müller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380. doi: 10.1146/annurev-arplant-042811-105503
Iqbal, N., Khan, N. A., Ferrante, A., Trivellini, A., Francini, A., and Khan, M. (2017). Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front. Plant Sci. 8:475. doi: 10.3389/fpls.2017.00475
Jinn, T. L., Stone, J. M., and Walker, J. C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14, 108–117. doi: 10.1101/gad.14.1.108
Kim, M. J., Jeon, B. W., Oh, E., Seo, P. J., and Kim, J. (2021). Peptide signaling during plant reproduction. Trends Plant Sci. 26, 822–835. doi: 10.1016/j.tplants.2021.02.008
Kim, S., Kang, J. Y., Cho, D. I., Park, J. H., and Kim, S. Y. (2004). ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40, 75–87. doi: 10.1111/j.1365-313X.2004.02192.x
Kim, K. C., Lai, Z., Fan, B., and Chen, Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20, 2357–2371. doi: 10.1105/tpc.107.055566
Kim, J., Yang, R., Chang, C., Park, Y., and Tucker, M. L. (2018). The root-knot nematode Meloidogyne incognita produces a functional mimic of the Arabidopsis INFLORESCENCE DEFICIENT IN ABSCISSION signaling peptide. J. Exp. Bot. 69, 3009–3021. doi: 10.1093/jxb/ery135
Klauser, D., Desurmont, G. A., Glauser, G., Vallat, A., Flury, P., Boller, T., et al. (2015). The Arabidopsis pep-PEPR system is induced by herbivore feeding and contributes to JA-mediated plant defence against herbivory. J. Exp. Bot. 66, 5327–5336. doi: 10.1093/jxb/erv250
Kumpf, R. P., Shi, C. L., Larrieu, A., Stø, I. M., Butenko, M. A., Péret, B., et al. (2013). Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. U. S. A. 110, 5235–5240. doi: 10.1073/pnas.1210835110
Li, L., Kubiszewski-Jakubiak, S., Radomiljac, J., Wang, Y., Law, S. R., Keech, O., et al. (2016). Characterization of a novel β-barrel protein (AtOM47) from the mitochondrial outer membrane of Arabidopsis thaliana. J. Exp. Bot. 67, 6061–6075. doi: 10.1093/jxb/erw366
Li, L., Li, K., Ali, A., and Guo, Y. (2021). AtWAKL10, a Cell Wall associated receptor-Like kinase, negatively regulates leaf senescence in Arabidopsis thaliana. Int. J. Mol. Sci. 22:4885. doi: 10.3390/ijms22094885
Li, W., Zhang, H., Li, X., Zhang, F., Liu, C., Du, Y., et al. (2017). Intergrative metabolomic and transcriptomic analyses unveil nutrient remobilization events in leaf senescence of tobacco. Sci. Rep. 7:12126. doi: 10.1038/s41598-017-11615-0
Lightenthaler, H. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Lim, P. O., Kim, H. J., and Gil Nam, H. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Lim, P. O., Lee, I. C., Kim, J., Kim, H. J., Ryu, J. S., Woo, H. R., et al. (2010). Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J. Exp. Bot. 61, 1419–1430. doi: 10.1093/jxb/erq010
Liu, B., Butenko, M. A., Shi, C. L., Bolivar, J. L., Winge, P., Stenvik, G. E., et al. (2013). NEVERSHED and INFLORESCENCE DEFICIENT IN ABSCISSION are differentially required for cell expansion and cell separation during floral organ abscission in Arabidopsis thaliana. J. Exp. Bot. 64, 5345–5357. doi: 10.1093/jxb/ert232
Liu, C., Shen, L., Xiao, Y., Vyshedsky, D., Peng, C., Sun, X., et al. (2021). Pollen PCP-B peptides unlock a stigma peptide–receptor kinase gating mechanism for pollination. Science 372, 171–175. doi: 10.1126/science.abc6107
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Matsubayashi, Y., and Sakagami, Y. (2006). Peptide hormones in plants. Annu. Rev. Plant Biol. 57, 649–674. doi: 10.1146/annurev.arplant.56.032604.144204
Meng, X., Zhou, J., Tang, J., Li, B., De Oliveira, M. V., Chai, J., et al. (2016). Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Rep. 14, 1330–1338. doi: 10.1016/j.celrep.2016.01.023
Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25, 295–303. doi: 10.1046/j.1365-313x.2001.00965.x
Miao, Y., Laun, T., Zimmermann, P., and Zentgraf, U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55, 853–867. doi: 10.1007/s11103-004-2142-6
Mohd-Radzman, N. A., Laffont, C., Ivanovici, A., Patel, N., Reid, D., Stougaard, J., et al. (2016). Different pathways act downstream of the CEP peptide receptor CRA2 to regulate lateral root and nodule development. Plant Physiol. 171, 2536–2548. doi: 10.1104/pp.16.00113
Mustilli, A. C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099. doi: 10.1105/tpc.007906
Rodrigues, C., Vandenberghe, L. P. D. S., De Oliveira, J., and Soccol, C. R. (2012). New perspectives of gibberellic acid production: a review. Crit. Rev. Biotechnol. 32, 263–273. doi: 10.3109/07388551.2011.615297
Rossel, J. B., Walter, P. B., Hendrickson, L., Chow, W. S., Poole, A., Mullineaux, P. M., et al. (2006). A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ. 29, 269–281. doi: 10.1111/j.1365-3040.2005.01419.x
Sakuraba, Y., Piao, W., Lim, J. H., Han, S. H., Kim, Y. S., An, G., et al. (2015). Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 56, 2325–2339. doi: 10.1093/pcp/pcv144
Santiago, J., Brandt, B., Wildhagen, M., Hohmann, U., Hothorn, L. A., Butenko, M. A., et al. (2016). Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. elife 5:e15075. doi: 10.7554/eLife.15075
Santiago, J., Rodrigues, A., Saez, A., Rubio, S., Antoni, R., Dupeux, F., et al. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60, 575–588. doi: 10.1111/j.1365-313X.2009.03981.x
Stenvik, G. E., Butenko, M. A., and Aalen, R. B. (2008a). Identification of a putative receptor-ligand pair controlling cell separation in plants. Plant Signal. Behav. 3, 1109–1110. doi: 10.4161/psb.3.12.7009
Stenvik, G. E., Tandstad, N. M., Guo, Y., Shi, C. L., Kristiansen, W., Holmgren, A., et al. (2008b). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20, 1805–1817. doi: 10.1105/tpc.108.059139
Vadez, V., Deshpande, S. P., Kholova, J., Hammer, G. L., Borrell, A. K., Talwar, H. S., et al. (2011). Stay-green quantitative trait loci’s effects on water extraction, transpiration efficiency and seed yield depend on recipient parent background. Funct. Plant Biol. 38, 553–566. doi: 10.1071/FP11073
Vie, A. K., Najafi, J., Liu, B., Winge, P., Butenko, M. A., Hornslien, K. S., et al. (2015). The IDA/IDA-LIKE and PIP/PIP-LIKE gene families in Arabidopsis: phylogenetic relationship, expression patterns, and transcriptional effect of the PIPL3 peptide. J. Exp. Bot. 66, 5351–5365. doi: 10.1093/jxb/erv285
Vie, A. K., Najafi, J., Winge, P., Cattan, E., Wrzaczek, M., Kangasjärvi, J., et al. (2017). The IDA-LIKE peptides IDL6 and IDL7 are negative modulators of stress responses in Arabidopsis thaliana. J. Exp. Bot. 68, 3557–3571. doi: 10.1093/jxb/erx168
Wang, Y., Cui, X., Yang, B., Xu, S., Wei, X., Zhao, P., et al. (2020). WRKY55 transcription factor positively regulates leaf senescence and the defense response by modulating the transcription of genes implicated in the biosynthesis of reactive oxygen species and salicylic acid in Arabidopsis. Development 147:dev189647. doi: 10.1242/dev.189647
Wang, X., Hou, S., Wu, Q., Lin, M., Acharya, B. R., Wu, D., et al. (2017). IDL6-HAE/HSL2 impacts pectin degradation and resistance to Pseudomonas syringae pv tomato DC 3000 in Arabidopsis leaves. Plant J. 89, 250–263. doi: 10.1111/tpj.13380
Woo, H. R., Kim, H. J., Lim, P. O., and Nam, H. G. (2019). Leaf senescence: systems and dynamics aspects. Annu. Rev. Plant Biol. 70, 347–376. doi: 10.1146/annurev-arplant-050718-095859
Woo, H. R., Koo, H. J., Kim, J., Jeong, H., Yang, J. O., Lee, I. H., et al. (2016). Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol. 171, 452–467. doi: 10.1104/pp.15.01929
Xu, P., Chen, H., and Cai, W. (2020). Transcription factor CDF4 promotes leaf senescence and floral organ abscission by regulating abscisic acid and reactive oxygen species pathways in Arabidopsis. EMBO Rep. 21:e48967. doi: 10.15252/embr.201948967
Yamaguchi, Y., Huffaker, A., Bryan, A. C., Tax, F. E., and Ryan, C. A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22, 508–522. doi: 10.1105/tpc.109.068874
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhang, Z., Liu, C., Li, K., Li, X., Xu, M., and Guo, Y. (2021). CLE14 functions as a “brake signal” suppressing age-dependent and stress-induced leaf senescence through promoting JUB1-mediated ROS scavenge in Arabidopsis. Mol. Plant 15, 179–188. doi: 10.1016/j.molp.2021.09.006
Zhang, K., Xia, X., Zhang, Y., and Gan, S. S. (2012). An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J. 69, 667–678. doi: 10.1111/j.1365-313X.2011.04821.x
Zhang, X., Yang, Z., Wu, D., and Yu, F. (2020). RALF–FERONIA signaling: linking plant immune response with cell growth. Plant Commun 1:100084. doi: 10.1016/j.xplc.2020.100084
Zhao, L., Xia, Y., Wu, X. Y., Schippers, J. H., and Jing, H. C. (2018). Phenotypic analysis and molecular markers of leaf senescence. Methods Mol. Biol. 1744, 35–48. doi: 10.1007/978-1-4939-7672-0_3
Keywords: leaf senescence, IDL6, plant peptide, Arabidopsis, transcriptome analysis, phytohormone
Citation: Guo C, Li X, Zhang Z, Wang Q, Zhang Z, Wen L, Liu C, Deng Z, Chu Y, Liu T and Guo Y (2022) The INFLORESCENCE DEFICIENT IN ABSCISSION-LIKE6 Peptide Functions as a Positive Modulator of Leaf Senescence in Arabidopsis thaliana. Front. Plant Sci. 13:909378. doi: 10.3389/fpls.2022.909378
Edited by:
Zhonghai Li, Beijing Forestry University, ChinaReviewed by:
Emily Breeze, University of Warwick, United KingdomGuodong Wang, Shaanxi Normal University, China
Copyright © 2022 Guo, Li, Zhang, Wang, Zhang, Wen, Liu, Deng, Chu, Liu and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongfeng Guo, Z3VveW9uZ2ZlbmdAY2Fhcy5jbg==
†These authors have contributed equally to this work
 Cun Guo
Cun Guo Xiaoxu Li
Xiaoxu Li Zenglin Zhang
Zenglin Zhang Qi Wang
Qi Wang Zhenbiao Zhang1,2
Zhenbiao Zhang1,2 Yongfeng Guo
Yongfeng Guo