- 1Yunnan Key Laboratory for Rice Genetic Improvement, Food Crops Research Institute, Yunnan Academy of Agricultural Sciences (YAAS), Kunming, China
- 2Institute of Plant Resources, Yunnan University, Kunming, China
Intraspecific hybrid sterility is a common form of postzygotic reproductive isolation in Asian cultivated rice, which is also the major obstacle to utilize the strong heterosis in the rice breeding program. Here, we review recent progress in classification and hybrid sterility in Asian cultivated rice. A genome-wide analysis of numerous wild relatives of rice and Asian cultivated rice has provided insights into the origin and differentiation of Asian cultivated rice, and divided Asian cultivated rice into five subgroups. More than 40 conserved and specific loci were identified to be responsible for the hybrid sterility between subgroup crosses by genetic mapping, which also contributed to the divergence of Asian cultivated rice. Most of the studies are focused on the sterile barriers between indica and japonica crosses, ignoring hybrid sterility among other subgroups, leading to neither a systematical understanding of the nature of hybrid sterility and subgroup divergence, nor effectively utilizing strong heterosis between the subgroups in Asian cultivated rice. Future studies will aim at identifying and characterizing genes for hybrid sterility and segregation distortion, comparing and understanding the molecular mechanism of hybrid sterility, and drawing a blueprint for intraspecific hybrid sterility loci derived from cross combinations among the five subgroups. These studies would provide scientific and accurate guidelines to overcome the intraspecific hybrid sterility according to the parent subgroup type identification, allowing the utilization of heterosis among subgroups, also helping us unlock the mysterious relationship between hybrid sterility and Asian cultivated rice divergence.
Introduction
Biological species form when the gene flow between two divergent populations has mostly or completely limited, and the striking characteristic of species is that species can actually or potentially interbreed, but are prevented from producing the fertile progeny with other species (Paterson, 1985). Reproductive isolation is critical for the formation and maintenance of species between different populations (Oka, 1988). It could occur at different developmental stages, including prezygotic and postzygotic reproductive isolation, the former prevents mating or formation of hybrid and the latter reduces the fitness of hybrid (Seehausen et al., 2014). Hybrid sterility is a major form of postzygotic reproductive isolation in rice and has been a good example of genetic studies of population speciation as an intrinsic genetic factor (McDermott and Noor, 2010; Ouyang et al., 2010; Ouyang and Zhang, 2013).
Genus Oryza contains 23 wild species and two cultivated rice species with 11 genome types. Of the 11 genome types, 6 are diploid, including AA, BB, CC, EE, FF, and GG, and five are polyploidy, including BBCC, CCDD, HHJJ, HHKK, and KKLL (Khush, 1997; Stein et al., 2018). Six wild species of Oryza nivara, Oryza rufipogon, Oryza barthii, Oryza glumaepatula, Oryza longistaminata, and Oryza meridionalis, as well as the two cultivated species of Oryza sativa and Oryza glaberrima were classified into the AA genome (Khush, 1997). Thus, AA genome species were considered as the natural germplasm pool for rice genetic improvement (Vaughan, 1994). However, hybrid sterility occurs widely in intraspecific and interspecific hybrids in rice, which is the major barrier to utilize heterosis and make genetic improvement difficult by restricting gene exchange in the Asian cultivated rice, African cultivated rice, and between the two cultivated species and the wild relatives (Ouyang and Zhang, 2013; Li et al., 2020).
This review summarized recent progress in understanding the genetic differentiation in Asian cultivated rice, expatiated on the genetic basis of hybrid sterility between different subpopulations, discussed the role of hybrid sterility on the population divergence, as well as tried to outline the future efforts to be done in intraspecific hybrid sterility study.
Genetic Differentiation of the Asian Cultivated Rice
Asian cultivated rice (O. sativa L.) is widely cultivated around the world and mainly in Asia. Abundant genetic diversity and significant genetic differentiation due to the different geographic or ecological adaptation, as well as domestication, had been reported (Huang et al., 2011). The two distinguishable groups were recognized as “hsien” and “Keng” in Chinese ancient records from the Han dynasty (Ting, 1949a,b,c). These two different groups of O. sativa subs. japonica (Keng/Geng) and indica (hsien/Xian) were first named based on their morphological and serological characteristics, together with hybrid fertility differences (Kato et al., 1928), consistent with the classification by the ancient Chinese people. Then, distinct classification results emerged in view of different criteria. The three types, A, B, and C, were used to classify O. sativa according to the different geographical distribution, later regarded as japonica, javanica, and indica (Matsuo, 1952; Morinaga, 1954). Further studies indicated that Asian cultivated rice should be classified into three types and five subgroups according to hybrid affinity ability and geographic distributions. Japanese rice belonged to the japonica type, aman and tjereh subgroup belonged to indica type because of the low affinity of the Japanese subgroup, whereas aus together with bulu subgroup could be reconsidered as neither japonica nor indica, because they showed considerable high hybrid fertility to both types, so they were called the intermediate type. Thus, O. sativa might be grouped into three types: japonica, indica, and intermediate types (Morinaga and Kuriyama, 1955, 1958). A fast and feasible criterion for distinguishing Keng (japonica) from hsien (indica) based on some classification traits was established, such as apiculus hair length, grain size, hull color at heading, phenol reaction, and other traits, some uncertain intermediate varieties were assessed followed by traits of leaf hair and 1–2 internode length of the main panicle. This method, called the “Cheng's index,” was effective in practice (Cheng et al., 1984). Isozyme was also used to study the differentiation of O. sativa at the biochemical level. One thousand six hundred and eighty-eight Asian varieties were detected by 15 isozyme markers at eight loci, the result indicated that Asian cultivated rice was divided into six different groups: I, II, III, IV, V, and VI, respectively, corresponding indica, aus, deepwater rice in Bangladesh, deepwater rice in Northeast India, basmati, and japonica (Glaszmann, 1987). Reviewing the previous results, nearly all the researchers agreed with the opinion that Asian cultivated rice contained two major groups: japonica and indica, but the classification of intermediate groups was ambiguous. The limited number or geographic distribution of varieties, evaluated by a few traits and genetic markers, ignoring the role of origin, domestication, and selection on the divergence of O. sativa, resulted in the un-uniformed classification of O. sativa.
Progress in the development of a great number of molecular markers and genome sequencing provided a new insight into understanding the differentiation of O. sativa at the molecular level. Two hundred and thirty-four rice varieties representing a wide geographic range of O. sativa were genotyped using 169 nuclear SSRs and two chloroplast loci; the results indicated that five distinct groups were detected, corresponding to temperate japonica, tropical japonica, indica, aus, and aromatic rice (Garris et al., 2005). Twenty diverse varieties and landraces by genotyping 160,000 non-redundant SNPs not only fitted well with five groups of O. sativa, but also demonstrated genomic introgression from one varietal group into another under strong artificial selection for production or environment adaptation demands (McNally et al., 2009). The analysis of the genetic variation of 3,010 Asian cultivated rice accessions by genome-wide SNP supported the classification of the five major groups in O. sativa, but also revealed seven unreported and geographic distribution-related subgroups (Wang et al., 2018). Population genetics of 5,152 rice accessions analyzed by resequencing and genetic diversity analysis of 60 rice varieties using 190 SNPs had clearly demonstrated the genetic difference among temperate japonica, tropical japonica, indica, aus, and aromatic rice (Kishor et al., 2020; Yan et al., 2020). Taken together, the population structure and genetic diversity of the worldwide rice gave insights into the five distinct subgroups; this classification of O. sativa was widely recognized by rice scientists.
Basmati rice is the typical representative of the aromatic type, and the fragrance trait can be found in basmati (Dom-sufid from Iran), indica (KDM105 from Thailand), and tropical japonica (Azucena from Philippines) (Bradbury et al., 2005; Bourgis et al., 2008; Kovach et al., 2009; Myint et al., 2012); therefore, the word “aromatic” representing one of the five subgroups is easily confused. Here, we use basmati to be the japonica-clinous rice from Myanmar to South Asia and Iran so as to discriminate aromatic rice from indica, tropical japonica, temperate japonica, and aus.
In addition, a high-throughput genome sequence, coupled with archaeobotanical, paleoclimate, and geographic data, revealed the origin and domestication history of the five subgroups of O. sativa, thus supporting multiple origins and at least one domestication in Asian cultivated rice, which was accepted by scientists (Zhao, 2011; Huang et al., 2012; Gross and Zhao, 2014; Civan et al., 2015; Travis et al., 2015; Stein et al., 2018; Choi et al., 2020; Gutaker et al., 2020; Figure 1), but this classification was not proved by reproductive isolation.
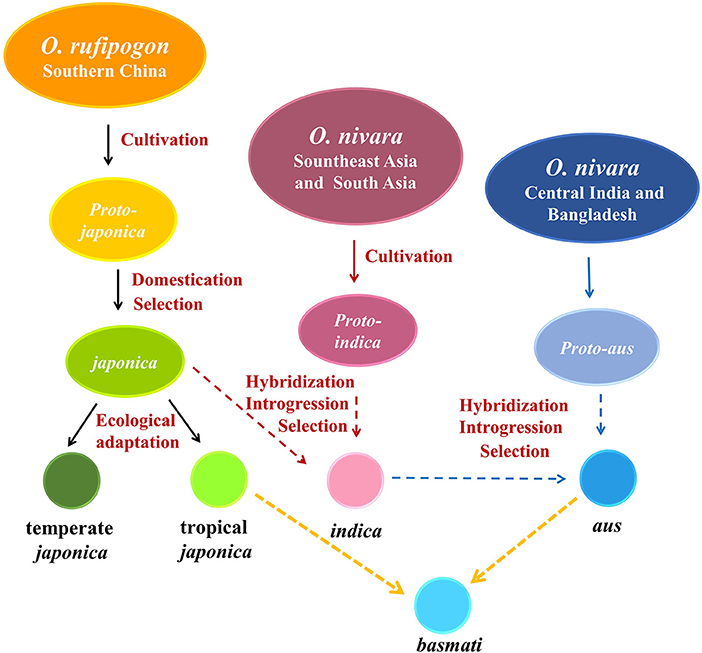
Figure 1. Origin and domestication model of Asian cultivated rice. Archaeological and genetic evidence indicated that japonica rice originated from a O. rufipogon population around the middle area of the Pear River and began to be cultivated about 9,000 years BP in the Yangtze Valley in southern China; japonica spread to South Asia about 4,000 years BP leading to the introgression of domestication genes into pro-indica derived from local O. nivara in Southeast Asia and South Asia, and indica was then formed to be cultivated in the lower Ganges valley (Zhao, 2011; Huang et al., 2012; Gross and Zhao, 2014; Stein et al., 2018). Tropical japonica and temperate japonica differed genetically due to a global reduction in temperature and humidity; it was the result of strong pressure to adapt to different climatic conditions (Kovach et al., 2007; Gutaker et al., 2020). Recently, two additional subspecies of Asian cultivated rice, aus and basmati, were recognized to differentiate distinctly from japonica and indica. O. nivara in central India and Bangladesh as the gene pool crossed with indica and selected to adapt to rainfed conditions, including rainfed upland, rainfed lowland, and deep-water, gave rise to aus group, whereas aromatic was the hybrid of aus and tropical japonica and was popular in Pakistan, northern India (basmati), and Iran (sadri) (Khush et al., 2013; Civan et al., 2015; Travis et al., 2015; Choi et al., 2020).
Genetic and Molecular Basis of Hybrid Sterility in Asian Cultivated Rice
Main Features of Hybrid Sterility in Asian Cultivated Rice
Hybrid sterility is widely observed in rice, causing reduced gamete viability of hybrids between divergent subspecies. Hybrid sterility between japonica and indica was first reported when Asian cultivated rice was classified into the two subspecies using the criterion of hybrid fertility (Kato et al., 1928). Since then, the genetic basis of hybrid sterility had been investigated and scientists found some distinguished features in hybrid sterility: (1) Cytoplasm had almost no effect on hybrid fertility because reciprocal crosses showed no significant difference in male and female gametes fertility of F1 plants (Kato, 1930; Oka, 1957); (2) F1 hybrids between indica and japonica were usually sterile, the hybrid fertility varied dependent on the number of hybrid sterility genes and genetic distance between the two parents (Oka, 1988; Zhang et al., 1989, 1994); (3) Sterility could be explained by hybrid sterility genes instead of chromosome disturbance (Kato, 1930); (4) The F1 and F2 fertilities are not correlated (Oka and Doida, 1962); (5) Hybrid sterility between japonica and indica could be observed due to male gamete abortion, female gamete abortion, and embryo abortion (Teng et al., 1996; Zhu et al., 1996; Liu et al., 2004); (6) Hybrid gene interactions, including both allelic interaction at the same locus and non-allelic interactions at the different loci, were responsible for varying degrees of hybrid sterility in crosses between subgroups of O. sativa (Zhang and Lu, 1993; Zhang et al., 1993; Kubo et al., 2008).
Identification, Cloning, and Molecular Characterization of Hybrid Sterility Loci
By now, more than 40 loci responsible for hybrid sterility in the crosses between the different subgroups are identified by genetic mapping in rice (Supplementary Table 1). Surprisingly, three loci, Ef1, Pf3, and Sf3, were found in the cross between ZS97 and Minghui 63, which were famous indica parents in hybrid rice breeding. It is suggested that hybrid sterility also happened within the indica cross (Supplementary Table 1). But it was notable that ZS97 and Minghui 63 were not traditional varieties, they were the product of modern breeding incorporating other genome introgression fragments from other subgroups by multiple round crosses (McNally et al., 2009); therefore, cloning and gene sequence analysis of three loci will answer whether those loci were responsible for hybrid sterility in indica × indica cross or not. Moreover, there is no gene or QTL identified in the cross between basmati and other subgroups, further study will pay more attention to identifying, cloning, and characterizing the molecular mechanism of hybrid sterile genes from basmati rice.
Until now, six loci for intraspecific hybrid sterility were cloned and characterized. Sa was the first characterized hybrid male sterility loci between indica and japonica. SaM encoded a small ubiquitin-like modifier E3 ligase, and SaF encoded an F-box protein. Three divergent genetic components: SaF+ (the indica allele), SaM+ (the indica allele), and SaM− (the japonica allele) consisted of a killer system to selectively abort the male gametes harboring SaM− in the indica×japonica hybrid (Long et al., 2008). S5 locus conferring female hybrid sterility consisted of three tightly linked genes, ORF3, ORF4, and ORF5, which acted in a killer–protector system. ORF3, ORF4, and ORF5 encoded an HSP70 heat shock protein, a transmembrane protein, and a putative aspartic protease, respectively. During female development, endoplasmic reticulum (ER) stress is caused by the action of ORF5+ (killer) and ORF4+ (partner), and ORF3+ (protector) could prevent this ER stress, but ORF3– could not prevent ER stress, triggering premature programmed cell death, and leading to embryo-sac abortion (Chen et al., 2008; Yang et al., 2012). A tetratricopeptide repeat domain-containing protein encoded by S7 was required for hybrid female sterility in the crosses between aus and indica or japonica (Yu et al., 2016). Two tightly linked genes, HSA1a and HSA1b, were responsible for hybrid female sterility in a cross between temperate japonica and indica. HSA1a encoded an unknown function protein with a highly conserved plant-specific domain, whereas HSA1b, encoded an uncharacterized protein with the nucleotide-binding domain. Heterozygous HSA1a with homozygous HSA1b allele from indica conferred hybrid sterility due to selective abortion of HSA1a-is gamete in the hybrid (Kubo et al., 2016a). Allelic suppression mediated by Sc contributed to hybrid male sterility between temperate japonica and indica. The japonica allele, Sc-j, encoded a DUF1618-domain protein; the indica allele, Sc-i, consisted of two or three tandem-duplicated Sc-j homolog. The selective abortion of Sc-j pollen leading to the transmission ratio distortion resulted from the expression of Sc-j suppressed by high expression of Sc-i in Sc-j/Sc-i hybrids (Shen et al., 2017). Hybrid pollen germination caused by reciprocal gene DPL1/DPL2 was isolated from japonica and aus cross. DPL encoded a plant-specific small protein. The pollen from Nipponbare possessed DPL1-N+/ DPL2-N− genotype, whereas pollen from Kasalath carried DPL1-K−/ DPL2-K+ genotype. Pollen harboring two nonfunctional alleles DPL1-K− and DPL2-N− failed to germinate and was not transmitted to the next generation (Mizuta et al., 2010). According to the above information, it is easy to find that the hybrid sterility of the different genetic materials was controlled by diverse genes or QTLs, and allelic interaction at each locus and non-allelic interaction at the different loci from two parents contribute to the intraspecific hybrid sterility. These genes or QTLs identified in the different populations will lay the foundation for elucidating the genetic and molecular mechanisms of hybrid sterility in Asian cultivated rice.
Two genetic models were popular for explaining hybrid sterility. One model was the one-locus sporo-gametophytic interaction model (Kitamura, 1962), and another was a duplicate gametic lethal model interpreting two independent loci involved in the hybrid sterility (Oka, 1957, 1974). S5, S7, Sa, Sc, and HSA1 fitted in with the one-locus sporo-gametophytic interaction model, and DPL1/DPL2 followed the duplicate gametic lethal model (Figure 2; Long et al., 2008; Mizuta et al., 2010; Yang et al., 2012; Kubo et al., 2016a; Yu et al., 2016; Shen et al., 2017).
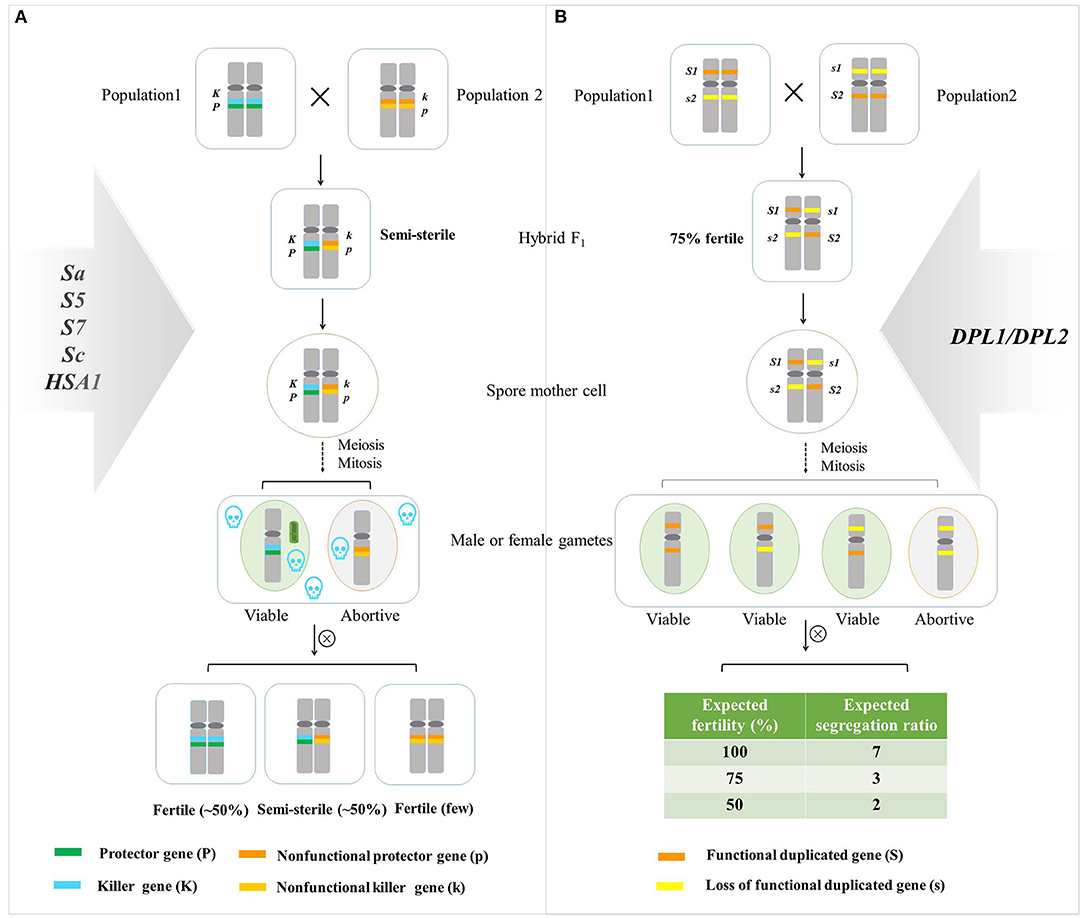
Figure 2. A genetic model of hybrid sterility. (A) Modified one-locus sporophytic–gametophytic allelic interaction model (Ikehashi and Araki, 1986). A hybrid sterility system consisted of at least two tightly linked genes: protector gene (P) and killer gene (K) in one population, nonfunctional protector gene (p), and nonfunctional killer gene (k) in another population. PP and KK constitute a functional killer system that triggers a sterility signal in the sporophytic stage, which selectively eliminates gametes without the PP gene, PP could protect the gametes in a gametophytic manner, leading to semi-sterility in the hybrid. Thus, the percentage of hybrid sterility plants in F1 hybrid accounted for 50%. The fertile plants and semi-sterile plants fit in with the ratio of 1:1 in the F2 population. S5, S7, Sa, Sc, and HSA1 fitted in with this model (Chen et al., 2008; Long et al., 2008; Yang et al., 2012; Kubo et al., 2016a; Yu et al., 2016; Shen et al., 2017). (B) Duplicate gametic lethal model interpreting two independent loci involved in the hybrid sterility. When the gametes carrying S1S1/s2s2 or s1s1/S2S2 developed normally, whereas two recessive alleles at both loci, s1s1/s2s2, conferred deleterious interaction, and about 25% of gametes were sterile (Oka, 1957, 1974). The ratio of the fertile plants, partial fertile plants, and semi-sterile plants conforms to the theoretical ratio of 7:3:2 in the progeny. DPL1/DPL2 followed this model (Mizuta et al., 2010).
Epistatic Interaction Between the Different Hybrid Sterility Loci
Hybrid sterility is a complex quantitative trait controlled by multiple loci, hence, it is conceived that gene–gene interaction is related to the sterility mechanism. Epistatic interaction between S35 and S24 regulated male sterility in rice intraspecific hybrid. S24 locus in the heterozygous plants caused abortion of male gametes from the japonica allele, independent of the S35 genotype, whereas male gametes from japonica alleles at the S35 were aborted when the homozygous genomic fragment harboring the S24 locus was introgressed into the heterozygous S35 background plants (Kubo et al., 2000, 2008). Further study confirmed that a locus INK tightly linked with S24, not S24, activated S35 to abort the male gametes from the japonica alleles at the S35 heterozygous locus. Homozygous EFS allele from indica can activate the S24 functioning in the pollen sterility, but the heterozygous EFS allele could eliminate the harmful effect of S24 on the male gametes in the hybrid (Kubo et al., 2016b). Another sample was that heterozygous HSA1a locus from indica × japonica cross caused the female gamete abortion in the homozygous HSA1b allele from indica, Conversely, the female gamete from japonica at heterozygous HSA1a locus was normally developed with homozygous HSA1b-j allele (Kubo et al., 2016a). Four S5-interacting QTLs, qSIG3.1, qSIG3.2, qSIG6.1, and qSIG12.1 were identified to interact with the S5 locus by genetic mapping (Rao et al., 2021). These evidences suggested that a genetic network with multiple genetic factors played a pivotal role in the intraspecific hybrid sterility in rice. However, to date, the gene–gene interactions among hybrid sterility loci are not clear; one reason might be this interaction depends on the special genetic background, and no effective investigation method of gene–gene interaction between the different loci might be another reason. In the future, improvements in the quantitative genetics methods coupled with a representative number of rice varieties could be needed to clarify this issue. In addition, in terms of the genetic model, there remain some substantial holes in explaining the epistasis effect on hybrid sterility, thus more comprehensive model will be established in light of the complex genetic basis of hybrid sterility.
Hybrid Sterility Loci and Divergence of Subgroups in Asian Cultivated Rice
Different Hybrid Sterility Loci Were Involved in the Divergence Between Two Subgroups
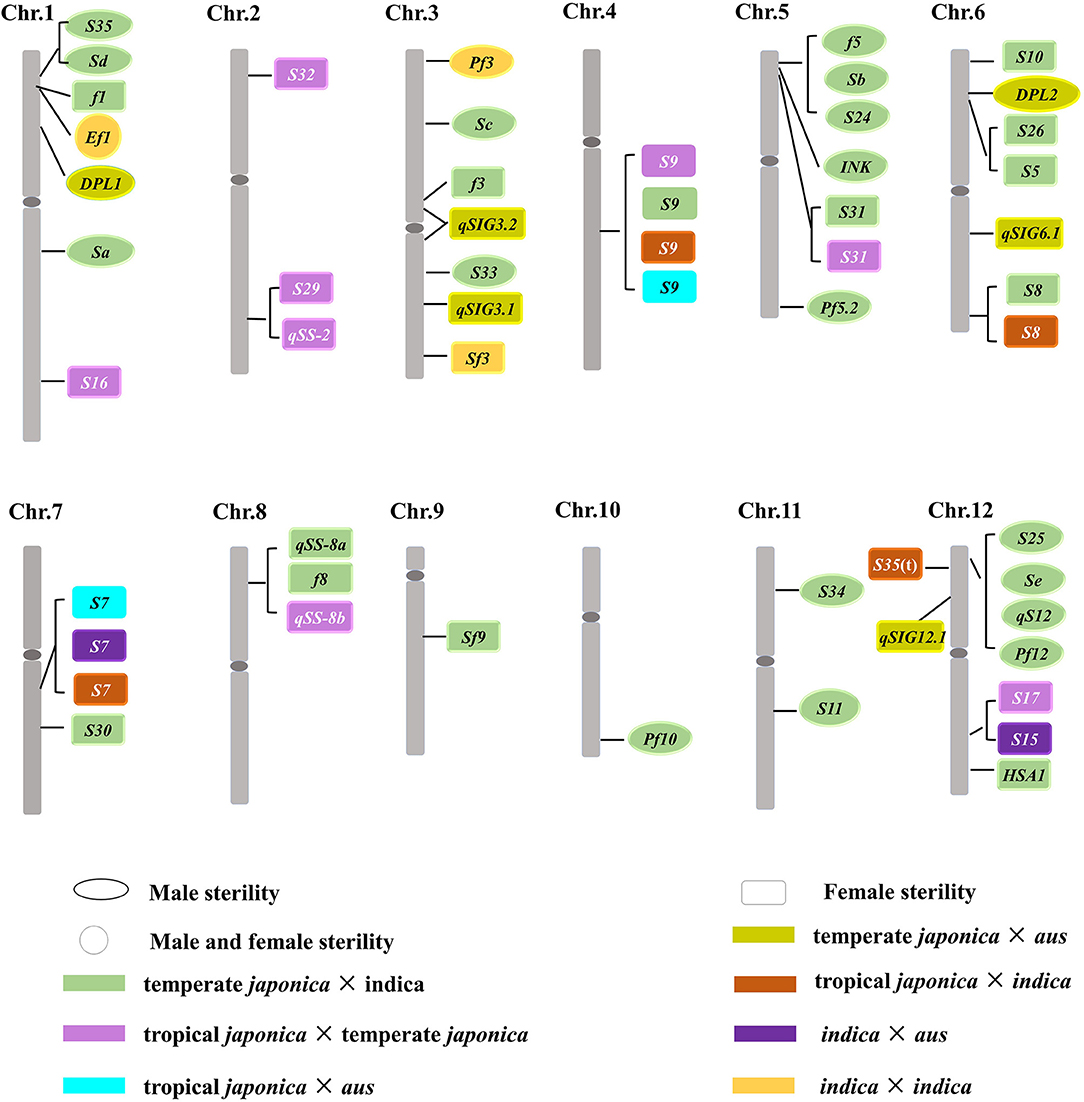
Twenty-two hybrid sterility loci had been described between indica and temperate japonica cultivars (Figure 3; Supplementary Table 1), including Sa, Sc, Sd, S11, S33, S34, S35, f5/Sb/S24, INK, Pf5.2, Pf10, and S25/Se/qS12/Pf12 for male gamete sterility (Sawamura and Sano, 1996; Kubo and Yoshimura, 2001; Yang et al., 2004; Li et al., 2006, 2008, 2017; Wang et al., 2006; Jing et al., 2007; Kubo et al., 2008; Long et al., 2008; Zhu et al., 2008; Zhang et al., 2011; Zhao et al., 2011; Shen et al., 2017) and f1, f3, S5, S8, S9, S10, S30, S31, S5/S26, qSS-8a/f8, Sf9, and HSA1 for female gamete sterility (Sano et al., 1994; Wan et al., 1996; Wang et al., 1998, 2005; Kubo and Yoshimura, 2001; Zhu et al., 2005b; Singh et al., 2006; Zhao et al., 2006; Chen et al., 2008; Kubo et al., 2016a; Li et al., 2017). It is suggested that those loci are also involved in the divergence between temperate japonica and indica subgroups. Low-hybrid fertility was also reported among other subgroups in previous studies (Morinaga, 1968; Engle et al., 1969), and then, hybrid sterility loci were identified by genetic mapping in succession (Figure 3; Supplementary Table 1). DPL1/DPL2 was responsible for hybrid pollen sterility (Mizuta et al., 2010), and qSIG3.1, qSIG3.2, qSIG6.1, as well as qSIG12.1 gave rise to female gamete abortion in the crosses between temperate japonica × aus (Rao et al., 2021). These results indicated that DPL1/DPL2, qSIG3.1, qSIG3.2, qSIG6.1, and qSIG12.1 were responsible for the divergence between temperate japonica and aus subgroups. S7 and S15 led to female gamete sterility in the hybrid between indica and aus (Wan et al., 1996; Yu et al., 2016), and it is suggested that both loci were involved in the reproductive isolation establishment between the two subgroups. Heterozygous loci at S9, S16, S17, S29, S31, S32, qSS-2, and qSS-8b resulted in gametes abortion in the hybrid between tropical japonica and temperate japonica (Wan and Ikehashi, 1995; Wan et al., 1998; Li et al., 2005, 2007; Wang et al., 2005; Zhu et al., 2005a; Zhao et al., 2007), which inferred that those loci acted in the genetic differentiation between tropical japonica and temperate japonica. In addition, S7, S8, S9, and S35(t) were responsible for the female sterility in the cross between tropical japonica and indica (Wan et al., 1993, 1996; Chen et al., 2012), it also indicated that those three loci promoted the differentiation between tropical japonica and indica. S7 and S9 controlled hybrid sterility and contributed to the divergence between tropical japonica and aus (Yanagihara et al., 1992; Wan et al., 1996). Taken together, the different combinations of distinct hybrid sterility loci could be the major driving force for Asian cultivated rice to differentiate into five subgroups, the type, and a number of loci involved in genetic differentiation varied with the subgroup types. The results supported the classification of five subgroups in Asian cultivated rice in terms of reproductive isolation.
Allelic Differentiation at Hybrid Sterility Locus and Genetic Differentiation in Asian Cultivated Rice
In recent years, more and more evidences showed that some loci responsible for hybrid sterility were identified in several subgroups crosses simultaneously (Supplementary Table 1). S7 contributed to the female sterility in the hybrid between tropical japonica or indica and aus, together with the tropical japonica and indica cross (Yanagihara et al., 1992; Wan et al., 1993; Yu et al., 2016), suggesting that the allelic differentiation at S7 locus contributed to the divergence among tropical japonica, indica, and aus subgroups. S8 locus conferring hybrid female sterility was reported in the different crosses between temperate japonica and indica, tropical japonica and indica (Wan et al., 1993; Singh et al., 2006), it indicated that S8 had a potential to lead the genetic differentiation between japonica and indica. A locus S9 for hybrid female sterility was found among the different subgroup crosses, including temperate japonica and tropical japonica, temperate japonica and indica, tropical japonica and aus, as well as tropical japonica and indica (Wan et al., 1996; Zhu et al., 2005a; Zhao et al., 2006), suggesting that allelic interaction at S9 locus was involved in the divergence of the above subgroups. Heterozygous S31 locus led to the hybrid female sterility in two kinds of populations, temperate japonica × indica and temperate japonica × tropical japonica (Li et al., 2005; Zhao et al., 2006), the same function had also been found in qSS-8a/qSS-8b/f8 (Wang et al., 1998, 2005), it indicated that the genetic differentiation between temperate japonica and tropical japonica or indica could partially resulted from the interaction of the different haplotypes at two loci. Evidence indicated that S15/S17 were mapped into the same region on chromosome 12, which might drove the divergence between indica and aus, temperate japonica and tropical japonica, respectively (Wan et al., 1996, 1998). Those six hot spots identified from different subgroup hybrids, with the same location region, as well as similar genetic behavior and phenotype, were probably orthologous genes and allelic to each other. It is suggested that these loci were conserved and contributed to hybrid sterility between different subgroups and allelic differentiation at hybrid sterility locus played important roles in triggering the divergence between distinct subgroups.
In a word, hybrid sterility is the major indication to distinguish the different subgroups in Asian cultivated rice at the subspecies level. A lot of genes or QTLs for male gamete abortion, female gamete abortion, and both male and female gamete abortions were identified in different populations. Some conserved loci governed the hybrid sterility among several subgroup crosses, whereas some loci were special for specific crosses; it suggested that multiple loci and allelic differentiation at hybrid sterility loci might be the important driving force of the divergence in Asian cultivated rice. However, the previous studies on hybrid sterility usually focused on the crosses between japonica and indica, with less progress on hybrid sterility between other subgroups. Future work should reinforce whole-genome-wide gene identification, and functional characterization of hybrid sterility loci in the hybrids between aus or basmati with other subgroups. It will help us to understand the genetic differentiation of subgroups and provide the guidelines for the heterosis strategy in rice breeding.
The Origin, The Evolution of Hybrid Sterility Loci, and the Divergence of Asian Cultivated Rice
Hybrid sterility resulted in the female or/and male gametes abortion in the hybrid, which hindered gene exchange and had substantial effects on the divergence between the different populations and speciation. How the hybrid sterility loci originated and evolved, fixed in the different populations, and lead to the population divergence finally was one of the hot spots in evolutionary biology.
Sa, S5, Sc, and HSA1 Are Involved in the Differentiation of Temperate Japonica From Indica
For Sa locus, SaF+/SaM+ haplotype was a major form presented in ancient Oryza species, SaF−/SaM+ and SaF−/SaM− haplotypes were generated by the sequential mutations in SaF+ and SaM+ in different accessions of O. rufipogon. And then, the increasing frequency of SaF+/SaM+ and SaF−/SaM+ fixed in indica populations, whereas the fixation of SaM−SaF− in japonica rice could be resulted from random genetic drift, together with geographical isolation or ecological adaptation (Long et al., 2008). In the light of SaF+, SaM+, and SaM consisted of killer system, it is supposed that the hybrid fertility should be observed in the hybrid between indica and O. rufipogon, indica and japonica, or within indica subspecies, and then it was confirmed that japonica haplotype at the Sa locus contributed to the pollen abortion in the crosses between temperate japonica and indica or wild rice species (Long et al., 2008). Accordingly, the reproductive isolation mediated by the Sa locus contributed to the genetic differentiation between temperate japonica and indica subspecies.
By a reciprocal BLASTN search using three genes sequence, it was postulated that the S5 complex could originated from Ospara3-5 by genome fragment duplication after Oryzeae tribe differentiation and produced a nascent loci ORF3+ORF4+ORF5+ with new introns and start codons. Most of the wild relatives in rice carried the functional ORF3+ORF4+ORF5+ allele. Mutations may occur in the partner gene by 11-bp deletion and killer gene by two SNP substitutions, leading to ORF3+ORF4–ORF5+ (the typical indica genotype), and ORF3+ORF4+ORF5– or ORF3+ORF4+ORF5n, respectively. And then, the protector ORF3+ mutated to ORF3– by 13bp deletion in the haplotypes of ORF3+ORF4+ORF5–, thus resulting in the typical japonica genotype of ORF3–ORF4+ORF5– in the pre-differentiated population. Subsequently, the increase of indica haplotype frequency was driven by natural selection, whereas the spread of japonica haplotype might be generated by the bottleneck during domestication. Once the frequencies of ORF3+ORF4–ORF5+ and ORF3–ORF4+ORF5– are fixed in the corresponding populations, indica and japonica subspecies would generate deleterious interactions upon hybridization, eventually resulting in reproductive isolation mediated by S5 locus (Du et al., 2011; Mi et al., 2020). In previous reports, 72% accession of O. rufipogon carried the ORF3+ORF4+ORF5+ or neutral haplotype at the S5 locus (Du et al., 2011; Mi et al., 2020) and this haplotype was compatible with the japonica or indica haplotypes. It suggested that it might be one of the reasons why hybrid sterility was universally observed in indica and japonica crosses, but almost never found in the crosses between O. rufipogon and O. sativa.
For the HSA1 locus, the ancestral population of O. rufipogon possessed the fertile HSA1a-j and the sterile HSA1b-is alleles. Mutation in HSA1a of the indica ancestry and in HSA1b of the japonica ancestry occurred separately, which generated indica haplotype (HSA1a-is/HSA1b-is) and temperate japonica haplotype (HSA1a-j/HSA1b-j). The differences in adaptive and domesticated evolution at the HSA1 locus accelerated the divergence between temperate japonica and indica subspecies (Kubo et al., 2016a). Sc-j and Sc-i haplotypes were found in the different O. rufipogon populations, suggesting that the origin of Sc occurred before the divergence between indica and japonica (Shen et al., 2017).
Therefore, hybrid sterility loci were diverged from the ancient haplotypes by a series of nucleotide variations and were further fixed in the japonica and indica subgroups, respectively, incompatible allelic interaction mediated by Sa, S5, HSA1, and Sc, activated the reproductive isolation system simultaneously, evenly restricting gene flow and resulting in the divergence of two subgroups.
DPL1/DPL2 Contributes to the Divergence Between Temperate Japonica and Aus
DPL originated from a small-scale genome duplication gene after the differentiation between Oryza and Brachypodium. Ancient DPL gene seemed to be functional by sequence analysis, but loss-of-function mutations of DPL1 genes fixed in indica and its wild ancestor, O. rufipogon. Especially, all the examined accessions of O. barthii and O. glaberrima had a deletion in the DPL1 exon, which suggested that the ancient DPL1 allele experienced the process of many times genome deletion, nucleotide substitution, and deletion, and gradually established non-functional haplotype in the different population, and the DPL2 gene defect was only found in the japonica cultivars (Mizuta et al., 2010). Based on the above data, hybrid pollen incompatibility governed by DPL had the potential to lead to reproductive isolation between japonica subspecies and O. barthii or O. glaberrima, but this phenomenon controlled by DPL has not been reported in interspecific hybridization population, there might be the function of DPL loci inhibited by an unknown factor during the stage of pollen germination. Furthermore, the origin and geographic distribution of DPL were investigated using 132 O. sativa, 296 accessions of O. nivara, and 388 accessions of O. rufipogon. DPL1− and DPL2− haplotypes of O. sativa separately emerged from an O. nivara population in India and O. rufipogon in South China, subsequently spread into other groups, especially aus and temperate japonica, respectively (Mizuta et al., 2010; Xu et al., 2021). Therefore, hybrid incompatibility established by DPL contributed to the genetic differentiation between aus and temperate japonica subspecies.
Taken together, the origin and evolution of Sa could be explained by the sequential divergence model, the mode of S5 fitted well with the parallel-sequential divergence model, parallel divergence model could also be supported by the studies of HSA1 and DPL1/DPL2 (Ouyang and Zhang, 2018). Accordingly, these results supported the “multiple origins” hypothesis, in which the Asian cultivated rice has originated from the different wild relative groups. In the future, more and more cloned hybrid sterility loci would help understand the evolutional processes and history of Asian cultivated rice and find out the reasons for subspecies divergence governed by multiple hybrid sterility loci with complex mechanisms step by step. Moreover, the different haplotypes at each locus should be developed on the uniformed background as NILs to validate whether the haplotype combinations functioned in hybrid sterility or not, it will provide experimental evidence for the time and mechanism of reproductive isolation establishment.
Strategies for Overcoming Hybrid Sterility in an Intraspecific Hybrid Breeding Program
Genetic diversity representing genetic differences between parental lines is fundamentally important to utilize heterosis in rice. Current hybrid rice varieties mainly utilize the heterosis from intra-subspecific crosses, mostly between indica parental lines. Hybrid yields have reached a bottleneck due to the limited genetic diversity in rice. Five subgroups in Asian cultivated rice possess abundant genetic diversity, which is the basis of strong heterosis. For example, aus subgroup showed drought-tolerant, early-maturing traits, and 11 of 143 heterotic loci were contributed by the introgression from aus varieties (Lin et al., 2020); basmati varieties possess excellent quality traits such as slender and long grain, fluffy and soft texture after cooking, and special aroma. Therefore, basmati is designated as the most high priced group (Civan et al., 2019). However, high genetic diversity between subgroups might reinforce sterility barriers to maintain the independent gene pools. How to break hybrid sterility and breed intraspecific hybrids with strong heterosis has always been explored by scientists for many years. So far, three strategies have been developed to overcome hybrid sterility and enable breeders to utilize the strong hybrid vigor of O. sativa. The first strategy was to transfer of neutral alleles of hybrid sterility loci into the target parents to raise wide-compatibility lines for hybrid breeding (Kitamura, 1962). Neutral allele introgression, including S5-n, f5, and Sa-n, have been proved to enhance the pollen or spikelet fertility in the hybrid (Mi et al., 2016, 2019; Xie et al., 2017a; Ma et al., 2020). The second strategy is to develop indica-compatible japonica lines by introgression and pyramid of multiple hybrid sterility loci from indica into the japonica genetic background by using molecular marker assistant backcrossing methods (Guo et al., 2016). The third strategy is to create an artificial wide compatible line by genome editing technology, such as CRISPR/cas9 (Xie et al., 2017a,b, 2019), which is a feasible, effective, and safe method. However, hybrid sterility is a complex trait controlled by multiple loci dependent on the genetic background. If wide compatible lines eliminating the effect of the hybrid sterile loci were generated by CRISPR/cas9, the conditions are required as follows: knowing all the loci for hybrid sterility between two parental lines, clearing killer genes in the different sterility systems. Accordingly, it is necessary to utilize abundant germplasm and genome information, develop mutual intraspecific populations with five subgroups, and explore all the hybrid sterility loci by genome-wide approach, as well as characterize the molecular mechanism of the sterility loci, so as to find out or artificially create neutral allele at each locus special for target parental lines. Based on the comprehensive and systematic data, breeders could effectively and accurately utilize the stronger intraspecific heterosis by genomic breeding (Figure 4).
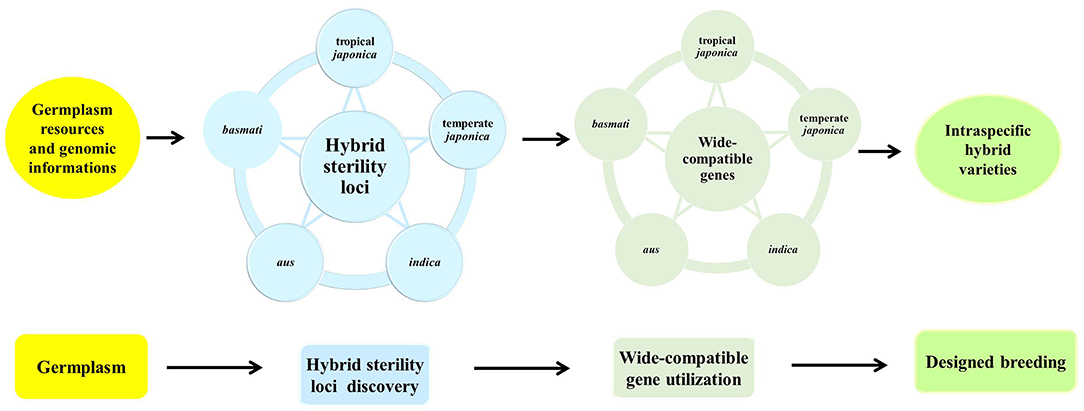
Figure 4. Strategies for developing intraspecific hybrid rice. The use of diverse rice accessions in five subgroups of Asian cultivated rice representing the abundant genetic diversity, together with plenty of genome information, the loci for intraspecific hybrid sterility among five subgroup crosses will be comprehensively and systematically explored, identified, and characterized by genome-wide genetic analysis, then natural neutral genes or wide-compatible genes will be created by CRISPR-Cas9 in light of special parental lines. Applying this information, breeders could design any intraspecific crosses using known wide-compatible genes, and will be able to breed fertile hybrid varieties with strong heterosis regardless of hybrid sterility.
Perspectives
Five subgroups of Asian cultivated rice have provided a natural model to investigate the hybrid sterility. The genetic basis and gene identification of hybrid sterility in Asian cultivated rice have been described by many researchers, especially in japonica × indica crosses (Ouyang and Zhang, 2013, 2018; Li et al., 2020). But it is still difficult to utilize the intraspecific heterosis in rice breeding; this issue has many reasons: first, identifying only a few sterility loci is not sufficient to reveal the whole process of hybrid sterility. Second, complex variation in hybrid sterility in the hybrids differs from one cross to another, and most of the studies focus on hybrid sterility derived from one or few combinations on one genetic background, we know little about the type, number, and genetic mode of hybrid sterility loci in other subgroups, except for japonica and indica. Scattered information about sterile barriers leads to the low efficiency of intraspecific varieties improvement. There is a long way to understand the genetic basis and regulatory network of hybrid sterility.
Comprehensive and Systematic Investigation of the Genetic Basis of Hybrid Sterility Based on Five Subgroups in Asian Cultivated Rice
To understand the genetic mechanism of hybrid sterility among the different subgroups comprehensively and systematically, the genetic population representing the typical five subgroups that crossed each other should be developed to elucidate the genetic basis of intraspecific hybrid sterility. Based on the amount of data in hybrid sterility special to parental lines, a blueprint for intraspecific hybrid sterility should be drawn in the distinct combinations derived from five subgroups, clearly marking the common or special features at gene types, gene numbers, conserved genes, special genes, major effect genes, minor effect genes, and gene–gene interaction, and so on. It would provide breeders with scientific and accurate guidelines to overcome the hybrid sterility special to target parental lines. Now, 20 varieties representing five subgroups were introgressed into a temperate japonica variety Dianjingyou 1 and an indica variety IR64 by multi-round backcrosses, respectively.
Moreover, basmati rice is considered a unique varietal group because of its aroma and superior grain quality (Ahuja et al., 1995; Siddiq et al., 2012; Civan et al., 2019) and it displays a different pattern of genetic diversity from other subgroups (Civan et al., 2019). Over the decades, less attention has been given to the hybrid sterility between basmati and other subgroups, though hybrid sterility was both observed in the hybrid between a basmati variety (Dom Sufid) and a japonica variety (Dianjingyou 1) or an indica variety (IR64) (data not shown). By now, none of the sterile loci was identified from the basmati variety, so, we should pay more attention to the hybrid sterility between the basmati group and other subgroups to utilize the genetic diversity and unique quality traits to breed intraspecific hybrid varieties.
Molecular Mechanism and Signal Network of Hybrid Sterility Loci
To date, only six loci involved in intraspecific hybrid sterility were cloned (Chen et al., 2008; Long et al., 2008; Mizuta et al., 2010; Yang et al., 2012; Kubo et al., 2016a; Yu et al., 2016; Shen et al., 2017). We only see a few separating points, which cannot be connected to obtain a clear genetic and regulatory pathway. Thus, it is necessary to clone more genes, elucidate the allelic and non-allelic interaction, and characterize the molecular mechanism of hybrid sterility. Recent evidence indicated the DUF1618 protein not only played an important role in hybrid sterility, but also contributed to gametophyte development (Kubo et al., 2016a; Shen et al., 2017). However, its molecular function was still unclear. It will provide a rapid and easy way to overcome hybrid sterility if we can find out the “key nodes” or “switch” in the crosstalk network of hybrid sterility.
Cloning and Characterization of Segregation Distortion Factors
Segregation distortion (SD) has substantial effects in population structure and fitness of the progenies, which would result in reproductive isolation and finally speciation. SD was observed widely in the cross populations, many loci for SD were identified by genetic analysis in rice (Figure 3; Kinoshita, 1991, 1993; Maekawa and Inukai, 1992; Rha et al., 1995; Xu et al., 1997; Harushima et al., 2001; Matsushita et al., 2003; Reflinur et al., 2014; Li et al., 2019; Xia and Ouyang, 2020). However, only one locus DPL1/DPL2 has been cloned and characterized (Mizuta et al., 2010), genetic and molecular mechanism of other loci remains to be unclear. Understanding the molecular events controlling SD will shed light on the reproductive isolation and speciation, as well as provide a potential approach to keep genetic diversity in populations. The above-mentioned studies will broaden our knowledge about hybrid sterility and divergence of Asian cultivated rice, and overcome the reproductive isolation so as to serve for rice intraspecific breeding.
Conclusion
In this article, we reviewed the comprehensive studies on the genetic differentiation and the origin of Asian cultivated rice, the comparative analysis of hybrid sterility loci among five subgroups, as well as an illustration of the relationships between hybrid sterility loci and the differentiation of subgroups in O. sativa. Clearly, Asian cultivated rice possesses abundant genetic diversity, which reflects the striking differences in traits, physiology, genome structure, geographic distribution, and ecological adaptation, accompanied by human civilization. Muti-dimensional discrepancies, including hybrid sterility, drive the genetic differentiation in rice. Conserved and special sterile loci contribute to the Asian cultivated rice divergence. Based on the genetic diversity of subgroups and information on the rice genome, it will facilitate gene flow by utilizing wide compatible genes for future rice breeding. Identifying loci conferring hybrid sterility between subgroups by genome-wide analysis, and elucidating the molecular mechanism and signal network will be easy to overcome the hybrid sterility by precise gene modification of key “killer genes” or “switch gene” in the sterility system.
Author Contributions
DT proposed the concept. YZ conceived and wrote the paper. JW, QP, YY, YL, JZ, JL, XD, and MW edited the manuscript. All the authors reviewed and approved the final manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (Grant Nos. 31991221, 31660380, 31201196, and 32160489), Yunnan Fundamental Research Projects (202001AS070003, 202101AS070036, 202101AT070193, and 530000210000000013809), the Yunnan Provincial Government (YNWR-QNBJ-2018-359), and the Yunnan Seed Industrialization Laboratory Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.908342/full#supplementary-material
References
Ahuja, S. C., Panwar, D. V. S., Uma, A., and Gupta, K. R. (1995). Basmati Rice: The Scented Pearl. Hisar: Directorate of Publications CCS Haryana Agricultural University).
Bourgis, F., Guyot, R., Gherbi, H., Tailliez, E., Amabile, I., Salse, J., et al. (2008). Characterization of the major fragance gene from an aromatic japonica rice and analysis of its diversity in Asian cultivated rice. Theor. Appl. Genet. 117, 353–368. doi: 10.1007/s00122-008-0780-9
Bradbury, L. M., Fitzgerald, T. L., Henry, R. J., Jin, Q., and Waters, D. L. (2005). The gene for fragrance in rice. Plant Biotechnol. J. 3, 363–370. doi: 10.1111/j.1467-7652.2005.00131.x
Chen, J., Ding, J., Ouyang, Y., Du, H., Yang, J., Cheng, K., et al. (2008). A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc. Natl. Acad. Sci. U.S.A. 105, 11436–11441. doi: 10.1073/pnas.0804761105
Chen, M., Zhao, Z., Jiang, L., and Wan, J. (2012). A new gene controlling hybrid sterility in rice (Oryza sativa L.). Euphytica 184, 15–22. doi: 10.1007/s10681-011-0467-1
Cheng, K., Wang, X., Zhou, J., Lu, Y., Lou, J., and Huang, H. (1984). Studies on indigenous rices in Yunnan and their utilization. 2. A revised classification of Asian cultivated rice (in Chinese). Acta Agron. Sin. 10, 271–280.
Choi, J., Lye, Z., Groen, S., Dai, X., Rughani, P., Zaaijer, S., et al. (2020). Nanopore sequencing-based genome assembly and evolutionary genomics of circum-basmati rice. Genome Biol. 21:21. doi: 10.1186/s13059-020-1938-2
Civan, P., Ali, S., Batista-Navarro, R., Drosou, K., Ihejieto, C., Chakraborty, D., et al. (2019). Origin of the aromatic group of cultivated rice (Oryza sativa L.) traced to the Indian subcontinent. Genome Biol. Evol. 11, 832–843. doi: 10.1093/gbe/evz039
Civan, P., Craig, H., Cox, C. J., and Brown, T. A. (2015). Three geographically separate domestications of Asian rice. Nat. Plants 1:15164. doi: 10.1038/nplants.2015.164
Du, H., Ouyang, Y., Zhang, C., and Zhang, Q. (2011). Complex evolution of S5, a major reproductive barrier regulator, in the cultivated rice Oryza sativa and its wild relatives. New Phytol. 191, 275–287. doi: 10.1111/j.1469-8137.2011.03691.x
Engle, L. M., Chang, T. T., and Ramirez, D. A. (1969). The cytogenetics of sterility in F1 hybrids of indica×indica and indica×javanica varieties of rice (Oryza sativa L.). Philipp. Agric. 53, 289–307.
Garris, A. J., Tai, T. H., Coburn, J., Kresovich, S., and Mccouch, S. (2005). Genetic structure and diversity in Oryza sativa L. Genetics 169, 1631–1638. doi: 10.1534/genetics.104.035642
Glaszmann, J. C. (1987). Isozymes and classification of Asian rice varieties. Theor. Appl. Genet. 74, 21–30. doi: 10.1007/BF00290078
Gross, B. L., and Zhao, Z. (2014). Archaeological and genetic insights into the origins of domesticated rice. Proc. Natl. Acad. Sci. U.S.A. 111, 6190–6197. doi: 10.1073/pnas.1308942110
Guo, J., Xu, X., Li, W., Zhu, W., Zhu, H., Liu, Z., et al. (2016). Overcoming inter-subspecific hybrid sterility in rice by developing indica-compatible japonica lines. Sci. Rep. 6:26878. doi: 10.1038/srep26878
Gutaker, R. M., Groen, S. C., Bellis, E. S., Choi, J. Y., Pires, I. S., Bocinsky, R. K., et al. (2020). Genomic history and ecology of the geographic spread of rice. Nat. Plants 6, 492–502. doi: 10.1038/s41477-020-0659-6
Harushima, Y., Nakagahra, M., Yano, M., Sasaki, T., and Kurata, N. (2001). A genome-wide survey of reproductive barriers in an intraspecific hybrid. Genetics 159, 883–892. doi: 10.1093/genetics/159.2.883
Huang, X., Kurata, N., Wei, X., Wang, Z. X., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Huang, X., Zhao, Y., Wei, X., Li, C., Wang, A., Zhao, Q., et al. (2011). Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32–39. doi: 10.1038/ng.1018
Ikehashi, H., and Araki, H. (1986). “Genetics of F1 sterility in remote crosses in rice,” in Rice Genetics Proceedings of the First Rice Genetics Symposium (Manila: IRRI), 119–130. doi: 10.1142/9789812814265_0011
Jing, W., Zhang, W., Jiang, L., Chen, L., Zhai, H., and Wan, J. (2007). Two novel loci for pollen sterility in hybrids between the weedy strain Ludao and the japonica variety Akihikari of rice (Oryza sativa L.). Theor. Appl. Genet. 114, 915–925. doi: 10.1007/s00122-006-0489-6
Kato, S. (1930). On the affinity of the cultivated varieties of rice plants, Oryza sativa L. J. Depart. Agric. Kyushu Imper. Univ. 2, 241–276. doi: 10.5109/22556
Kato, S., Kosaka, H., and Hara, S. (1928). On the affinity of rice varieties as shown by the fertility of hybrid plants. Bull. Sci. Fac. Agric. Kyushu Univ. 3, 132–147.
Khush, G. S. (1997). Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35, 25–34. doi: 10.1023/A:1005810616885
Khush, G. S., Brar, D. S., Virk, P. S., Tang, S., Malik, S. S., Busato, G. A., et al. (2013). Classifying Rice Germplasm by Isozyme Polymorphism and Origin of Cultivated Rice. IRRI discussion paper series No 46. Los Banos: International Rice research Institute, 11.
Kinoshita, T. (1991). Report of the committee on gene symbolization, nomenclature and linkage group. Rice Genet. Newslett. 8, 2–37.
Kinoshita, T. (1993). Report of the committee on gene symbolization, nomenclature and linkage group. Rice Genet. Newslett. 10, 7–39.
Kishor, D. S., Seo, J., Chin, J. H., and Koh, H. J. (2020). Evaluation of whole-genome Sequence, genetic diversity, and agronomic traits of Basmati rice (Oryza sativa L.). Front. Genet. 11:86. doi: 10.3389/fgene.2020.00086
Kitamura, E. (1962). Genetics studies on sterility observed in hybrids between distantly related varieties of rice (Oryza sativa L.). Bull. Chgoku Agric. Exp. Sta. Ser. A 8, 141–205.
Kovach, M. J., Calingacion, M. N., Fitzgerald, M. A., and Mccouch, S. R. (2009). The origin and evolution of fragrance in rice (Oryza sativa L.). Proc. Natl. Acad. Sci. U.S.A. 106, 14444–14449. doi: 10.1073/pnas.0904077106
Kovach, M. J., Sweeney, M. T., and Mccouch, S. R. (2007). New insights into the history of rice domestication. Trends Genet. 23, 578–587. doi: 10.1016/j.tig.2007.08.012
Kubo, T., Eguchi, M., and Yoshimura, A. (2000). A new gene for F1 pollen sterility in Japonica/Indica cross of rice. Rice Genet. Newslett. 17, 63–69.
Kubo, T., Takashi, T., Ashikari, M., Yoshimura, A., and Kurata, N. (2016a). Two tightly linked genes at the hsa1 locus cause both F1 and F2 hybrid sterility in rice. Mol. Plant 9 221–232. doi: 10.1016/j.molp.2015.09.014
Kubo, T., Yamagata, Y., Eguchi, M., and Yoshimura, A. (2008). A novel epistatic interaction at two loci causing hybrid male sterility in an inter-subspecific cross of rice (Oryza sativa L.). Genes Genet. Syst. 83, 443–453. doi: 10.1266/ggs.83.443
Kubo, T., and Yoshimura, A. (2001). Linkage analysis of an F1 sterility gene in japonica/indica cross of rice. Rice Genet. Newslett. 18, 52–53.
Kubo, T., Yoshimura, A., and Kurata, N. (2016b). Pollen killer gene S35 function requires interaction with an activator that maps close to S24, another pollen killer gene in rice. G3 6, 1459–1468. doi: 10.1534/g3.116.027573
Li, D., Chen, L., Jiang, L., Zhu, S., Zhao, Z., Liu, S., et al. (2007). Fine mapping of S32(t), a new gene causing hybrid embryo sac sterility in a Chinese landrace rice (Oryza sativa L.). Theor. Appl. Genet. 114, 515–524. doi: 10.1007/s00122-006-0450-8
Li, D., Zhu, S., Jiang, L., and Wan, J. (2005). Mapping for a new locus causing hybrid sterility in a China landrace (Oryza sativa L.). Rice Genet. Newslett. 22:20.
Li, G., Jin, J., Zhou, Y., Bai, X., Mao, D., Tan, C., et al. (2019). Genome-wide dissection of segregation distortion using multiple inter-subspecific crosses in rice. Sci. China Life Sci. 62, 507–516. doi: 10.1007/s11427-018-9452-8
Li, G., Li, X., Wang, Y., Mi, J., Xing, F., Zhang, D., et al. (2017). Three representative inter and intra-subspecific crosses reveal the genetic architecture of reproductive isolation in rice. Plant J. 92, 349–362. doi: 10.1111/tpj.13661
Li, L., Zhou, J., Zhang, Y., Yang, Y., Pu, Q., and Tao, D. (2020). New insights into the nature of interspecific hybrid sterility in rice. Front. Plant Sci. 11:555572. doi: 10.3389/fpls.2020.555572
Li, W., Zeng, R., Zhang, Z., Ding, X., and Zhang, G. (2006). Fine mapping of locus S-b for F1 pollen sterility in rice (Oryza sativa L.). Chinese Sci. Bull. 51, 675–680. doi: 10.1007/s11434-006-0675-6
Li, W., Zeng, R., Zhang, Z., Ding, X., and Zhang, G. (2008). Identification and fine mapping of S-d, a new locus conferring the partial pollen sterility of intersubspecific F1 hybrids in rice (Oryza sativa L.). Theor. Appl. Genet. 116, 915–922. doi: 10.1007/s00122-008-0723-5
Lin, Z., Qin, P., Zhang, X., Fu, C., Deng, H., Fu, X., et al. (2020). Divergent selection and genetic introgression shape the genome landscape of heterosis in hybrid rice. Proc. Natl. Acad. Sci. U.S.A. 117, 4623–4631. doi: 10.1073/pnas.1919086117
Liu, H., Xu, C., and Zhang, Q. (2004). Male and female gamete abortions, and reduced affinity between the uniting gametes as the causes for sterility in an indica/japonica hybrid in rice. Sex. Plant Reprod. 17, 55–62. doi: 10.1007/s00497-004-0214-z
Long, Y., Zhao, L., Niu, B., Su, J., Wu, H., Chen, Y., et al. (2008). Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc. Natl. Acad. Sci. U.S.A. 105, 18871–18876. doi: 10.1073/pnas.0810108105
Ma, X., Wei, X., Gao, G., and Jiang, H. (2020). Development and evaluation of improved lines based on an elite rice variety 9311 for overcoming hybrid sterility in rice. Mol. Breed. 40:102. doi: 10.1007/s11032-020-01180-2
Maekawa, M., and Inukai, T. (1992). Genes linked with d-2 in rice. Jpn. J. Breed. Sci. 42(Suppl 2), 212–213. doi: 10.1270/jsbbs1951.42.213
Matsuo, T. (1952). Genecological studies on cultivated rice (in Japanese). Bull. Nat. Inst. Agric. Sci. Jpn. D 3, 1–111.
Matsushita, S., Iseki, T., Fukuta, Y., Araki, E., Kobayashi, S., Osaki, M., et al. (2003). Characterization of segregation distortion on chromosome 3 induced in wide hybridization between indica and japonica type rice varieties. Euphytica 134, 27–32. doi: 10.1023/A:1026182312730
McDermott, S. R., and Noor, M. A. (2010). The role of meiotic drive in hybrid male sterility. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 1265–1272. doi: 10.1098/rstb.2009.0264
McNally, K. L., Childs, K. L., Bohnert, R., Davidson, R. M., Zhao, K., Ulat, V. J., et al. (2009). Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc. Natl. Acad. Sci. U.S.A. 106, 12273–12278. doi: 10.1073/pnas.0900992106
Mi, J., Li, G., Huang, J., Yu, H., Zhou, F., Zhang, Q., et al. (2016). Stacking S5-n and f5-n to overcome sterility in indica-japonica hybrid rice. Theor. Appl. Genet. 129, 563–575. doi: 10.1007/s00122-015-2648-0
Mi, J., Li, G., Xu, C., Yang, J., Yu, H., Wang, G., et al. (2020). Artificial selection in domestication and breeding prevents speciation in rice. Mol. Plant 13, 650–657. doi: 10.1016/j.molp.2020.01.005
Mi, J. Y., Kim, S. R., Prahalada, G. D., Ouyang, Y., and Mou, T. (2019). An effective strategy for fertility improvement of indica-japonica hybrid rice by pyramiding S5-n, f5-n, and pf12-j. Mol. Breed. 39:138. doi: 10.1007/s11032-019-1044-x
Mizuta, Y., Harushima, Y., and Kurata, N. (2010). Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc. Natl. Acad. Sci. U.S.A. 107, 20417–20422. doi: 10.1073/pnas.1003124107
Morinaga, T. (1954). “Classification of rice varieties on the basis of affinity,” in International Rice Commission, Working Party on Rice Breeding, Rep 5th Meeting, 1–14.
Morinaga, T. (1968). Origin and geographical distribution of Japanese rice. Jpn. Agric. Res. Q. 1–5.
Morinaga, T., and Kuriyama, H. (1955). Japonica type rice in the subcontinent of India and Java. Jpn. J. Breed. 5, 149–153. doi: 10.1270/jsbbs1951.5.149
Morinaga, T., and Kuriyama, H. (1958). Intermediate type of rice in the subcontinent of India and Java. Jpn. J. Breed. 7, 253–269. doi: 10.1270/jsbbs1951.7.253
Myint, K. M., Courtois, B., Risterucci, A. M., Frouin, J., Soe, K., Thet, K. M., et al. (2012). Specific patterns of genetic diversity among aromatic rice varieties in Myanmar. Rice 5:20. doi: 10.1186/1939-8433-5-20
Oka, H. I. (1957). Genic analysis for the sterility of hybrids between distantly related varieties of cultivated rice. J. Genet. 55, 397–409. doi: 10.1007/BF02984059
Oka, H. I. (1974). Analysis of genes controlling F1 sterility in rice by the use of isogenic lines. Jpn. J. Genet. 77, 521–534. doi: 10.1093/genetics/77.3.521
Oka, H. I., and Doida, Y. (1962). Phylogenetic differentiation of cultivated rice, XX. Analysis of the genetic basis of hybrid breakdown in rice. Jpn. J. Genet. 37, 24–35. doi: 10.1266/jjg.37.24
Ouyang, Y., Liu, Y. G., and Zhang, Q. (2010). Hybrid sterility in plant: stories from rice. Curr. Opin. Plant Biol. 13, 186–192. doi: 10.1016/j.pbi.2010.01.002
Ouyang, Y., and Zhang, Q. (2013). Understanding reproductive isolation based on the rice model. Annu. Rev. Plant Biol. 64, 111–135. doi: 10.1146/annurev-arplant-050312-120205
Ouyang, Y., and Zhang, Q. (2018). The molecular and evolutionary basis of reproductive isolation in plants. J. Genet. Genomics 45, 613–620. doi: 10.1016/j.jgg.2018.10.004
Paterson, H. E. H. (1985). “The recognition concept of species,” in Species and Speciation, ed E. Vrba (Pretoria: Transvaal Museum), 21–29.
Rao, J., Wang, X., Cai, Z., Fan, Y., and Yang, J. (2021). Genetic analysis of s5-interacting genes regulating hybrid sterility in rice. Rice 14:11. doi: 10.1186/s12284-020-00452-x
Reflinur, K. B., Jang, S. M., Chu, S. H., Bordiya, Y., Akter, M. B., et al. (2014). Analysis of segregation distortion and its relationship to hybrid barriers in rice. Rice 7:3. doi: 10.1186/s12284-014-0003-8
Rha, E. S., Wu, S. T., and Thseng, F. S. (1995). Distorted segregation of marker genes in wide crosses of rice (Oryza sativa L.). J. Agric. Ass. China 170, 104–123.
Sano, Y., Sano, R., Eiguchi, M., and Hirano, H. Y. (1994). Gamete eliminator adjacent to the wx locus as recealed by pollen analysis in rice. J. Hered. 85, 310–312. doi: 10.1093/oxfordjournals.jhered.a111465
Sawamura, N., and Sano, Y. (1996). Chromosomal location of gamete eliminator, S11(t), found in an Indica-Japonica hybrid. Rice Genet. Newslett. 13, 70–71.
Seehausen, O., Butlin, R. K., Keller, I., Wagner, C. E., Boughman, J. W., Hohenlohe, P. A., et al. (2014). Genomics and the origin of species. Nat. Rev. Genet. 15, 176–192. doi: 10.1038/nrg3644
Shen, R., Wang, L., Liu, X., Wu, J., Jin, W., Zhao, X., et al. (2017). Genomic structural variation-mediated allelic suppression causes hybrid male sterility in rice. Nat. Commun. 8:1310. doi: 10.1038/s41467-017-01400-y
Siddiq, E. A., Vemireddy, L. R., and Nagaraju, J. (2012). Basmati rices: genetics, breeding and trade. Agric. Res 1, 25–36. doi: 10.1007/s40003-011-0011-5
Singh, S. P., Sundaram, R. M., Biradar, S. K., Ahmed, M. I., Viraktamath, B. C., and Siddiq, E. A. (2006). Identification of simple sequence repeat markers for utilizing wide-compatibility genes in inter-subspecific hybrids in rice (Oryza sativa L.). Theor. Appl. Genet. 113, 509–517. doi: 10.1007/s00122-006-0316-0
Stein, J. C., Yu, Y., Copetti, D., Zwickl, D. J., Zhang, L., Zhang, C., et al. (2018). Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 50, 285–296. doi: 10.1038/s41588-018-0040-0
Teng, J., Xue, Q., and Wang, Y. (1996). Ultrastructural observations of pollen and anther wall developments between subspecies in rice (Oryza sativa L.) (in Chinese). J. Zhejiang Agric. Univ. 22, 467–473.
Ting, Y. (1949a). Origination of the rice cultivation in China. J. Coll. Agric. Sun Yat-Sen Univ. 7, 11–24.
Ting, Y. (1949b). Chronological studies of the cultivation and the distribution of rice varieties, Keng and Sen (in Chinese). Agr. Bull. Coll. Agric. Sun Yatsen Univ. 6, 1–32.
Ting, Y. (1949c). A preliminary report on the cultivation and distribution of hsien and keng rices in ancient China and the classification of current cultivars (in Chinese). Mem. Coll. Agric. Sun Yatsen Univ. 6, 1–32.
Travis, A. J., Norton, G. J., Datta, S., Sarma, R., Dasgupta, T., Savio, F. L., et al. (2015). Assessing the genetic diversity of rice originating from Bangladesh, Assam and West Bengal. Rice 8:35. doi: 10.1186/s12284-015-0068-z
Vaughan, D. A. (1994). The Wild Relatives of Rice: A Genetic Resources Handbook. Manila: The International Rice Research Institute.
Wan, J., and Ikehashi, H. (1995). Identification of a new locus S-16 causing hybrid sterility in native rice varieties (Oryza sativa L.) from Tai-hu lake region and Yunnan Province, China. Breed. Sci. 45, 461–470. doi: 10.1270/jsbbs1951.45.461
Wan, J., Ikehashi, H., Sakai, M., Horisuei, H., and Imbe, T. (1998). Mapping of hybrid sterility gene S17 of rice (Oryza sativa L.) by isozyme and RFLP markers. Rice Genet. Newslett. 15, 151–154.
Wan, J., Yamaguchi, Y., Kato, H., and Ikehashi, H. (1996). Two new loci for hybrid sterility in cultivated rice (Oryza sativa L.). Theor. Appl. Genet. 92, 183–190. doi: 10.1007/BF00223375
Wan, J., Yanagihara, S., Kato, H., and Ikehashi, H. (1993). Multiple alleles at a new locus causing hybrid sterility between a Korean indica variety and a javanica variety in rice (Oryza sativa L.). Jpn. J. Breed. 43, 507–516. doi: 10.1270/jsbbs1951.43.507
Wang, C., Zhu, C., Zhai, H., and Wan, J. (2005). Mapping segregation distortion loci and quantitative trait loci for spikelet sterility in rice (Oryza sativa L.). Genet. Res. 86, 97–106. doi: 10.1017/S0016672305007779
Wang, G., He, Y., Xu, C., and Zhang, Q. (2006). Fine mapping of f5-Du, a gene conferring wide-compatibility for pollen fertility in inter-subspecific hybrids of rice (Oryza sativa L.). Theor. Appl. Genet. 112, 382–387. doi: 10.1007/s00122-005-0141-x
Wang, J., Liu, K., Xu, C., Li, X., and Zhang, Q. (1998). The high level of wide-compatibility of variety 'Dular' has a complex genetic basis. Theor. Appl. Genet. 97, 407–412. doi: 10.1007/s001220050910
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. doi: 10.1038/s41586-018-0063-9
Xia, F., and Ouyang, Y. (2020). Recurrent breakdown and rebalance of segregation distortion in the genomes: battle for the transmission advantage. aBIOTECH. 1, 246–254. doi: 10.1007/s42994-020-00023-0
Xie, Y., Niu, B., Long, Y., Li, G., Tang, J., Zhang, Y., et al. (2017a). Suppression or knockout of SaF/SaM overcomes the Sa-mediated hybrid male sterility in rice. J. Integr. Plant Biol. 59, 669–679. doi: 10.1111/jipb.12564
Xie, Y., Tang, J., Xie, X., Li, X., Huang, J., Fei, Y., et al. (2019). An asymmetric allelic interaction drives allele transmission bias in interspecific rice hybrids. Nat. Commun. 10:2501. doi: 10.1038/s41467-019-10488-3
Xie, Y., Xu, P., Huang, J., Ma, S., Xie, X., Tao, D., et al. (2017b). Interspecific hybrid sterility in rice is mediated by OgTPR1 at the S1 locus encoding a peptidase-like protein. Mol. Plant. 10, 1137–1140. doi: 10.1016/j.molp.2017.05.005
Xu, X., Ge, S., and Zhang, F. (2021). Genetic and geographic patterns of duplicate DPL genes causing genetic incompatibility within rice: implications for multiple domestication events in rice. Rice Sci. 28, 58–68. doi: 10.1016/j.rsci.2020.11.007
Xu, Y., Zhu, L., Xiao, J., Huang, N., and Mccouch, S. R. (1997). Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid, and recombinant inbred populations in rice (Oryza sativa L.). Mol. Gen. Genet. 253, 535–545. doi: 10.1007/s004380050355
Yan, J., Zou, D., Li, C., Zhang, Z., Song, S., and Wang, X. (2020). SR4R: an integrative SNP resource for genomic breeding and population research in rice. Genomics Proteomics Bioinformatics 18, 173–185. doi: 10.1016/j.gpb.2020.03.002
Yanagihara, S., Kato, H., and Ikehashi, H. (1992). A new locus for multiple alleles causing hybrid sterility between an Aus variety and javanica varieties in rice (Oryza sativa L.). Theor. Appl. Genet. 90, 182–188. doi: 10.1007/BF00222200
Yang, C., Chen, Z., Zhuang, C., Mei, M., and Liu, Y. (2004). Genetic and physical fine-mapping of the Sc locus conferring indica-japonica hybrid sterility in rice (Oryza sativa L.). Chinese Sci. Bull. 49, 1718–1721. doi: 10.1360/04wc0197
Yang, J., Zhao, X., Cheng, K., Du, H., Ouyang, Y., Chen, J., et al. (2012). A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337, 1336–1340. doi: 10.1126/science.1223702
Yu, Y., Zhao, Z., Shi, Y., Tian, H., Liu, L., Bian, X., et al. (2016). Hybrid sterility in rice (Oryza sativa L.) involves the tetratricopeptide repeat domain containing protein. Genetics 203, 1439–1451. doi: 10.1534/genetics.115.183848
Zhang, G., and Lu, Y. (1993). Genetic studies of the hybrid sterility in cultivated rice (Oryza sativa). II. Gene mode of F1 pollen sterility (in Chinese). Acta Genet. Sin. 20, 222–228.
Zhang, G., Lu, Y., Liu, G., Yang, J., and Zhang, H. (1989). Genetic studies of the hybrid sterility in culti vated rice (Oryza sativa). I. Diallel analysis of the hybrid sterility among isogenic F1 sterile lines (in Chinese). Chinese J. Rice Sci. 3, 97–101.
Zhang, G., Lu, Y., Liu, G., Yang, J., and Zhang, H. (1993). Genetic studies of the hybrid sterility in cultivated rice (Oryza sativa). III. Allele differentiation of F1 pollen sterility in different types of varieties (in Chinese). Acta Genet. Sin. 20, 541–551.
Zhang, G., Lu, Y., Zhang, H., Yang, J., and Liu, G. (1994). Genetic studies of hybrid sterility in cultivated rice (Oryza sativa), IV. Genotypes of pollen sterility genes (In Chinese). Acta Genet. Sin. 21, 31–41.
Zhang, H., Zhang, C., Sun, Z., Yu, W., Gu, M., Liu, Q., et al. (2011). A major locus qS12, located in a duplicated segment of chromosome 12, causes spikelet sterility in an indica-japonica rice hybrid. Theor. Appl. Genet. 123, 1247–1256. doi: 10.1007/s00122-011-1663-z
Zhao, Z. (2011). New archaeobotanic data for the study of the origins of Agriculture in China. Curr Anthropol S295–S306. doi: 10.1086/659308
Zhao, Z., Jiang, L., Zhang, W., Yu, C., Zhu, S., Xie, K., et al. (2007). Fine mapping of S31, a gene responsible for hybrid embryo-sac abortion in rice (Oryza sativa L.). Planta 226, 1087–1096. doi: 10.1007/s00425-007-0553-8
Zhao, Z., Wang, C., Jiang, L., Zhu, S., Ikehashi, H., and Wan, J. (2006). Identification of a new hybrid sterility gene in rice (Oryza sativa L.). Euphytica 151, 331–337. doi: 10.1007/s10681-006-9154-z
Zhao, Z., Zhu, S., Zhang, Y., Bian, X., Wang, Y., Jiang, L., et al. (2011). Molecular analysis of an additional case of hybrid sterility in rice (Oryza sativa L.). Planta 233, 485–494. doi: 10.1007/s00425-010-1313-8
Zhu, S., Jiang, L., and Wang, C. M. (2005b). The origin of weedy rice Ludao in China deduced by a genome wide analysis of its hybrid sterility genes. Breed. Sci. 55, 409–414. doi: 10.1270/jsbbs.55.409
Zhu, S., Wang, C., Zheng, T., Zhao, Z., Ikehashi, H., and Wan, J. (2005a). A new gene located on chromosome 2 causing hybrid sterility in a remote cross of rice. Plant Breed. 124, 440–445. doi: 10.1111/j.1439-0523.2005.01143.x
Zhu, W., Li, W., Ding, X., Zhang, Z., Zeng, R., Zhu, H., et al. (2008). Preliminary identification of F1 pollen sterility gene S-e in Oryza sativa (in Chinese). J. South China Agric. Univ. 29, 1–5.
Keywords: Oryza sativa L., hybrid sterility, subgroup, divergence, heterosis
Citation: Zhang Y, Wang J, Pu Q, Yang Y, Lv Y, Zhou J, Li J, Deng X, Wang M and Tao D (2022) Understanding the Nature of Hybrid Sterility and Divergence of Asian Cultivated Rice. Front. Plant Sci. 13:908342. doi: 10.3389/fpls.2022.908342
Received: 30 March 2022; Accepted: 30 May 2022;
Published: 27 June 2022.
Edited by:
Yongzhong Xing, Huazhong Agricultural University, ChinaCopyright © 2022 Zhang, Wang, Pu, Yang, Lv, Zhou, Li, Deng, Wang and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dayun Tao, dGFvZHkxMkBhbGl5dW4uY29t
 Yu Zhang
Yu Zhang Jie Wang
Jie Wang Qiuhong Pu
Qiuhong Pu Ying Yang
Ying Yang Yonggang Lv
Yonggang Lv Jiawu Zhou
Jiawu Zhou Jing Li
Jing Li Xianneng Deng
Xianneng Deng Min Wang1,2
Min Wang1,2 Dayun Tao
Dayun Tao