- Key Laboratory of Grassland Resources of the Ministry of Education, College of Grassland Resources and Environment, Inner Mongolia Agricultural University, Hohhot, China
Abiotic stresses affect plant growth and productivity. The outstanding stress resistance of Medicago ruthenica makes it a desirable gene resource to improve the stress tolerance of other plants. The roles of three differently expressed genes [(DEGs) (MrERF, MrbZIP, and MrSURNod)] from M. ruthenica in stress resistance have not been fully elucidated. Therefore, we constructed their expression vectors, transformed them into tobacco, and subjected transgenic lines to abiotic stresses. Through comprehensive bioinformatics, transcriptomic, morphological, and physiological analyses of transgenic lines, we have revealed the critical role of these three DEGs in plant growth and abiotic stress response. The upregulation of genes enhanced the germination rate, biomass, root length number, etc. Additionally, the accumulation of osmolytes increased the activity of antioxidant enzymes. These genes are also associated with improved seed yield, increased branching, and early flowering, thereby shortening the growth period. Potentially, this is one of the ways for tobacco to cope with stress. Furthermore, the resistance of transgenic tobacco expressing MrERF or MrbZIP was better than that with MrSURNod. MrERF and MrbZIP can improve drought and salt tolerance of plants, whereas MrSURNod is beneficial in improving drought and cold resistance. Moreover, MrERF or MrbZIP can promote root elongation and increase the root number, whereas MrSURNod mainly promotes root elongation. This may be the reason why stress resistance conferred by MrSURNod is weaker than that associated with the other two genes. Overall, MrERF, MrbZIP, and MrSURNod positively modulate plant growth and stress tolerance.
Introduction
Grassland ecological environment is extremely complex, and forages are often exposed to cold, arid, and other stress environments not conducive to plant growth and high yield. Hence, it is necessary to develop new forage varieties with strong tolerance and high yield and protein. With the development of molecular technologies, scientists use genetic engineering to transfer stress-resistance genes to plants with a high yield but weak resistance to improve their stress resistance, which has become a new direction for plant resistance research.
Medicago ruthenica, a forage legume, is widely distributed in alpine and desertification grasslands in northern China, having the advantages of cold, drought, salinity, barren, and trampling resistance. Therefore, this species can grow well in cold and arid areas (Yang et al., 2011) and has tremendous potential for grassland improvement, ecological management, and grassland industry development (Zhao et al., 2013). Although it belongs to the genus Medicago as well as alfalfa (Medicago sativa), it has higher nutrient-use efficiency than alfalfa (Campbell et al., 1997) and can be used as a high-quality protein feed. In addition, domestic and foreign scholars agree that its outstanding resistance to drought, cold, salinity, and alkalinity stresses makes it not only a high-quality parent in breeding, but it can also be used as a gene resource to improve the stress resistance of alfalfa and other forages. It is thought that the new varieties of alfalfa with high yield and high stress resistance can be obtained by transferring the genes from M. ruthenica to alfalfa using transgenic technology (Campbell, 2003). Therefore, on the basis of previous morphological and physiological studies, transcriptome sequencing of M. ruthenica under drought was carried out to screen drought resistance-related genes.
In order to cope with various environmental stresses, plants have evolved a series of regulatory mechanisms. Plants perceive different stress signals from the outside and respond accordingly, and various hormonal and metabolic signaling pathways are involved in the processes. Notably, plant hormone signal transduction pathway plays a key role in resisting abiotic stress (Depuydt and Hardtke, 2011). Among them, ABA regulates not only plant growth and development but also plays an important role in plant responses to abiotic stress (Gupta and Rashotte, 2014; Vishwakarma et al., 2017). Numerous studies have shown that bZIP and AP2/EREBP transcription factors are involved in ABA signal transduction and stress responses in plants. Here, we studied three DEGs from M. ruthenica identified under drought, including two transcription factors (MrERF, MrbZIP) and one unknown gene (MrSURNod). MrERF belongs to AP2/EREBP family and MrbZIP to bZIP family. It is well documented that bZIP family genes are involved in plant adaptation to drought stress by regulating the expression of drought resistance-related genes (Wang et al., 2017). For example, overexpression of SlbZIP1 (Zhu et al., 2018), AtABF3 (Wang et al., 2016) or CaDILZ1 (Lim et al., 2018) enhanced drought tolerance of transgenic lines. However, they can also have an opposite effect, e.g., SlbZIP38 in tomato, enhancing plant sensitivity to drought (Pan et al., 2017). AP2/EREBP is involved in regulating abiotic stress responses as well as plant hormones (e.g., ABA) (Dossa et al., 2016). Studies have proved that overexpression of ERF in rice, tobacco, and other plants can improve plant tolerance to drought or high salinity. For example, the overexpression of OsERF48 (Jung et al., 2017), OsERF71 (Lee et al., 2016), and PpERF023 (Bielsa et al., 2018) enhanced drought resistance of transgenic plants. The overexpression of Arabidopsis ERF1 (Cheng et al., 2013) and wheat TaERF3 (Rong et al., 2014) improved the drought and salt tolerance of transgenic lines. The overexpression of MfERF1 in alfalfa improved the cold tolerance of transformed plants (Zhuo et al., 2018), and ERF105 (Bolt et al., 2017), ERF102, and ERF103 (Illgen et al., 2020) were confirmed to play important roles in cold acclimation of Arabidopsis thaliana.
In addition to bZIP and AP2/EREBP, an unknown gene MrSURNod with a high homology to Medicago truncatula was also identified in our study. The above genes were up-regulated under drought stress and may play an important role in the adaptation of M. ruthenica to drought. Therefore, the transcription factors MrbZIP and MrERF involved in ABA signal transduction pathways and an unknown gene MrSURNod were selected to construct plant expression vectors and transform them into tobacco. Three resistant tobacco plants were treated with drought, low temperature, and NaCl, and the expression pattern of the genes under stress was detected by qRT-PCR. We characterized the differences in phenotype and physiological and biochemical indices between WT and over-expression plants. Our research put forward new ideas for improving the abiotic stress of plant and laid the foundation for breeding forages for stress resistance.
Materials and Methods
Plant Materials and Growth Conditions
M. ruthenica (L.) cv. Zhilixing was selected as the experimental material. The seeds of this variety were collected in September 2008 from the experimental fields of Inner Mongolia Agricultural University located in Hohhot, Inner Mongolia, China. All seeds were soaked in concentrated H2SO4 for 5–8 min in order to break the hard coat and were then germinated on 1/2 MS medium until the seedlings grew 6–8 leaves. Subsequently, leaf samples were snap-frozen in liquid nitrogen and stored at −80°C for RNA extraction. Tobacco (Nicotiana benthamiana) was used for the generation of overexpression transgenic plants. Tobacco seeds came from the seed storage of Inner Mongolia Agricultural University. M. ruthenica and tobacco plants were grown in a greenhouse under controlled conditions (25°C, 16-h light/8-h dark, 50% humidity).
Gene Cloning, Sequencing, and Bioinformatics Analysis
Total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s protocol. A UEIris II RT-PCR System for first-strand cDNA synthesis (with dsDNase) (US Everbright, Suzhou, China) was used to synthesize cDNA. Polymerase chain reaction (PCR) was performed according to the manufacturer’s recommended protocol. After detection by agarose gel electrophoresis, the PCR product bands were recovered using a gel recovery kit (Omega, United States) and sequenced. Bioinformatics analysis was conducted through the following websites: ExPASy tools,1 Wolfpsort,2 TMHMM Server v.2.0,3 and CDD.4 The phylogenetic tree was constructed through neighbor-joining analysis using MEGA 7.0 (bootstrap 1000).
Plasmid Construction and Plant Transformation
The CDSs of MrERF, MrbZIP, and MrSURNod were ligated into the pCAMBIA2300-GFP vector using a homologous recombination system. The primer sequences used for vector construction are listed in Supplementary Table 1. The recombinant pCAMBIA2300—MrERF—GFP, pCAM BIA2300—MrbZIP—GFP, pCAMBIA2300—MrSURNod—GFP, and pCAMBIA2300—GFP plasmids were transformed into Agrobacterium tumefaciens (GV3101) for plant transformation. A stable transformation of tobacco was performed using a leaf disk co-cultivation protocol. The positive transgenic tobacco lines were selected by 100 mg/L kanamycin and confirmed by genomic DNA PCR (Plant Genome DNA Extraction and Amplification Kit (Biomed, Beijing, China), following the manufacturer’s protocol). After full development, the gene expression in the positive transgenic tobacco leaves was confirmed by qRT-PCR. The leaves with successful expression were selected for subculturing until roots formed, and seedlings were then transferred to soil for subsequent study.
Expression Pattern of MrbZIP, MrERF, and MrSURNod
The methods of RNA extraction and cDNA synthesis were described earlier. The qRT-PCR assays were performed using Fast Super EvaGreen® qPCR Master Mix (US Everbright, Suzhou, China) in an ABI 7500 system (Applied Biosystems, United States) with the following program: 95°C for 2 min; 45 cycles of 95°C for 10 s, 55°C for 10 s, 72°C for 30 s, 95°C for 15 s, and 60°C for 60 s. The primers used for the amplification of MrERF, MrbZIP, and MrSURNod (Supplementary Table 2) were designed according to the CDS of MrERF (Cluster-20905.1), MrbZIP (Cluster-60183.76174), and MrSURNod (Cluster-60183.75597). All the primers were synthesized by Invitrogen (Beijing, China). A 20 μL reaction mix was set up containing 10 μL of 2 × Fast Super EvaGreen® Master Mix, 1 μL of 10 × ROX, 5.5 μL of ddH2O, 3 μL of cDNA, and 0.5 μg of each primer. The relative expression changes of the endogenous reference and tested genes were analyzed by the 2–ΔΔCT method.
Abiotic Stress Treatment
The Resistance of Seeds at Germination Stage
Drought and salt treatment: The seeds of the transgenic (MrERF, MrbZIP, and MrSURNod) and WT lines were germinated on 1/2 MS medium containing mannitol (100, 200, and 300 mM) or NaCl (100, 150, and 200 mM) (Petri dish diameter 10 cm). Medium without additives was used as control (25°C, 16-h light/8-h dark, 50% humidity). Cold stress: The seeds were placed on 1/2 MS medium and placed in an incubator at 4°C. Three biological replicates were included in each treatment. Two weeks later, we scored the seed germination rate of transgenic and WT tobacco lines under different conditions.
The Resistance Identification at Seedling Stage
Seedlings were grown in pots (diameter 58 mm, depth 110 mm), and the planting substrate was organic nutrient soil that was purchased from Zhongshun Garden Machinery Co., Ltd. For the drought treatment, 5-week-old transgenic tobacco lines were subjected to no watering for 9 days. For the NaCl stress treatment, 5-week-old plants of the MrERF, MrbZIP, MrSURNod, and WT tobacco lines were watered with 200 mM NaCl for 9 days. For cold stress, seedlings were moved into an incubator set at 4°C and 16-h light/8-h dark. Leaves from tobacco lines were collected after 3, 6, and 9 days of stress. Three replicates per treatment were tested. All samples were collected at 10:00 am, snap-frozen in liquid nitrogen, and stored at −80°C. The gene expression of MrERF, MrbZIP, and MrSURNod in treated seedlings was detected after 9 days of growth under stress. Root length and number, plant height, and biomass at different time points during treatment were recorded. Soluble sugars, proline, MDA content, and SOD and POD under the treatment of drought, NaCl, and cold were measured. In addition, seed yield per plant was recorded.
Statistical Analysis
Statistical analyses were performed using SPSS Statistics (version 21, IBM, Chicago, IL, United States) Excel 2016 (Microsoft, Washington, DC, United States). Three biological replicates were included in each experiment. The data are presented as the means ± SDs (n = 3). The differences among the tested lines were assessed using the Duncan multiple comparison test. The level of significance was set at p < 0.05.
Results
Bioinformatics Analysis of MrERF, MrbZIP, and MrSURNod
The fragments of MrERF, MrbZIP, and MrSURNod were amplified from M. ruthenica by RT-PCR. The coding regions (582, 1,308, and 1,029 bp) were cloned, sequenced, and submitted to the GenBank (accession numbers: MW811334, MW811335, and MW811336). The deduced amino acid sequences were 150 (MrERF), 427 (MrbZIP), and 340 amino acids (MrSURNod) (Supplementary Figure 1). The MrbZIP deduced protein has a bZIP domain of 54 amino acids comprising a leucine zipper. MrERF encodes a conserved AP2 domain (Figure 1). They have no transmembrane regions, whereas MrSURNod contained one SURNod domain and transmembrane regions (Supplementary Figure 2). Subsequently, phylogenetic trees were constructed using MEGA (version 7.0, Mega Limited, Auckland, New Zealand) (Figure 1). The amino acid sequences of MrERF and MrbZIP showed the highest similarity with the protein sequences from Arabidopsis thaliana (AtERF034 and AtbZIP36). The unknown gene MrSURNod had less than 40% homology to other sequences. The subcellular localization analysis implied the possible localization of MrERF and MrbZIP in the nucleus and MrSURNod in the chloroplast (Supplementary Figure 3). These findings demonstrated that MrERF, MrbZIP, and MrSURNod were sensitive to drought and were rapidly up-regulated, suggesting the roles of the ERF, bZIP, and MrSUDNod proteins in responses to drought.
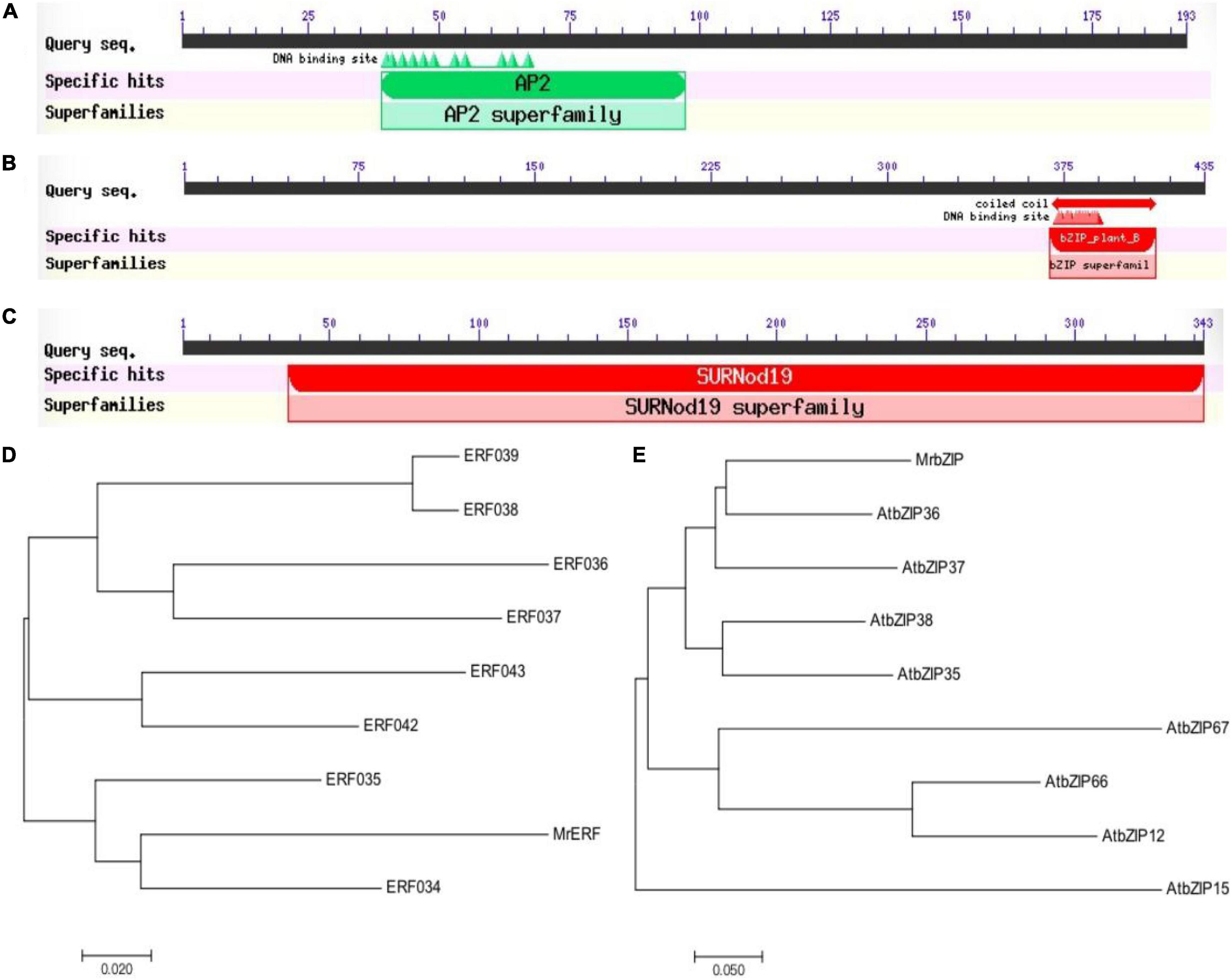
Figure 1. Prediction of the conserved domains, phylogenetic tree. (A) Conserved domain of MrERF; (B) conserved domain of MrbZIP; (C) conserved domain of MrSURNod; (D) phylogenetic tree of MrERF; and (E) phylogenetic tree of MrbZIP.
Vector Construction and Genetic Transformation
The target gene fragments of MrbZIP, MrERF, and MrSURNod were successfully ligated into the plant expression vector pCAMBIA2300-GFP, denoted as pCAMBIA2300-76174-GFP, pCAMBIA2300-20905.1-GFP, and pCAMBIA2300-75597-GFP, respectively (Supplementary Figures 4–6). Subsequently, the recombinant plasmids were transformed into agrobacterium. The results of colony PCR showed that the three vectors had been successfully transformed into agrobacterium strains and could be used later for plant transformation (Supplementary Figure 7). To investigate the biological functions of the three genes in response to abiotic stresses, tobacco (Nicotiana tabacum) leaves were transformed with above-mentioned fusion constructs via Agrobacterium-mediated transformation. Kanamycin-resistant T0 plants were evaluated with RT-PCR. The result showed that all transgenic plants had acquired the PCR product that was not present in WT (Figure 2). Positive transgenic plants containing MrbZIP, MrERF, or MrSURNod were cultured in water and transplanted to the soil a week later. Finally, the seeds of T0 generation were harvested.
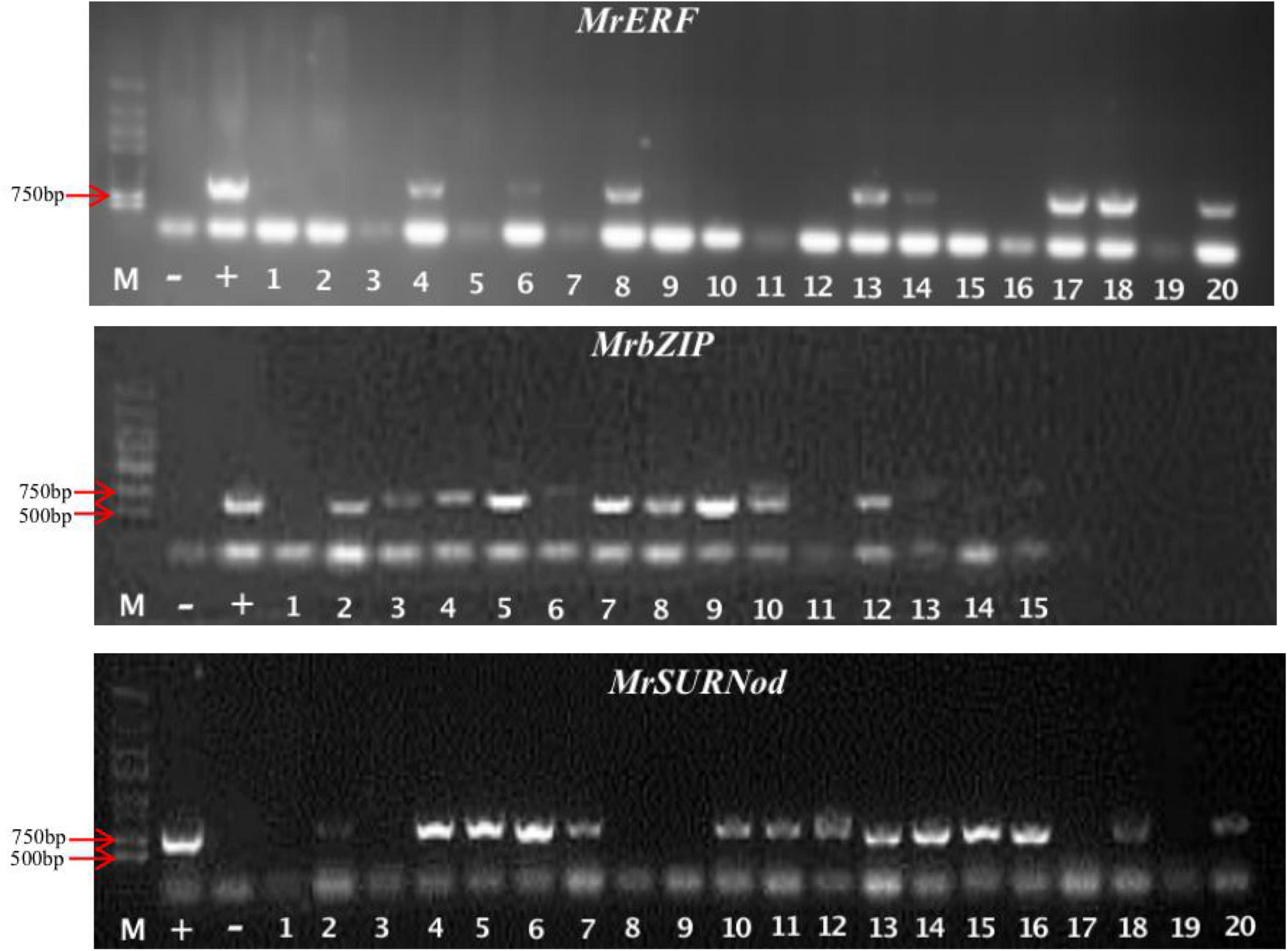
Figure 2. Identification of tobacco transgenic lines by PCR. “M”, Marker (DL2000); “–”, negative control; “+”, positive control, the rest were transgenic plants.
Identification of Transgenic Tobacco Plants
The T0 generation tobacco seeds were screened on 1/2 MS medium containing 100 mg/L kanamycin (Figure 3A). The genomic DNA of transgenic plant leaves was extracted from healthy young leaves for PCR detection (Figure 3B). The results preliminarily proved that exogenous genes (MrERF, MrbZIP, and MrSURNod) had been successfully integrated into the tobacco genome. To detect the transcription expression of the genes, the transgenic plants with positive PCR detection were selected, and RNA was extracted. The results showed that exogenous genes in some transgenic plants were expressed, but the expression levels differed. After antibiotic screening, PCR detection, and qRT-PCR, T1 transgenic tobacco plants with stable integration of exogenous genes and successful expression were obtained (Figure 3C), including 68 plants with MrERF, 71 with MrbZIP, and 66 with MrSURNod; they were used for subsequent molecular, morphological and physiological stress resistance studies.
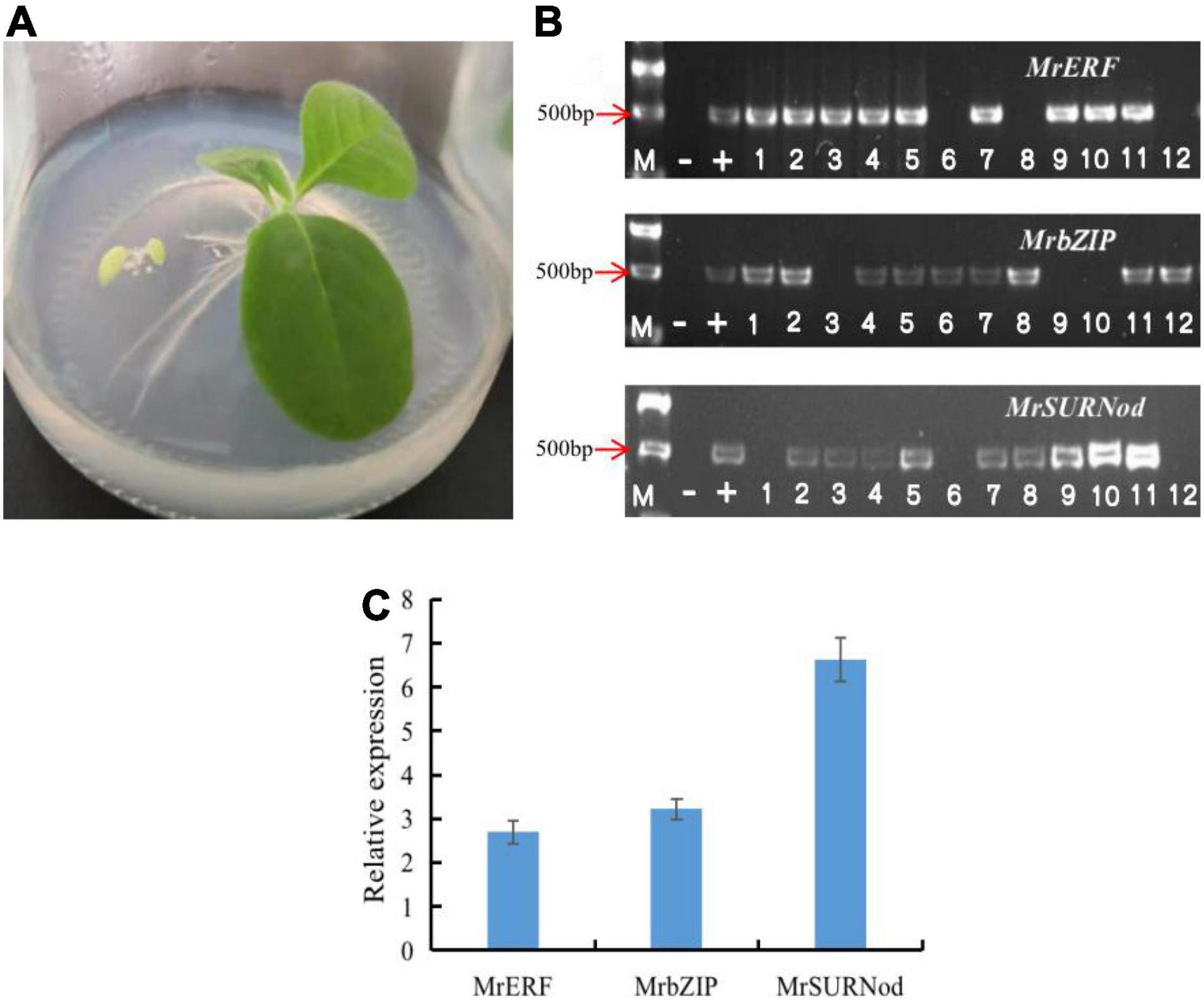
Figure 3. Transgenic plant identification. (A) Antibiotic selection; (B) identification of transgenic plants by PCR [“M”, Marker (DL2000); “–”, negative control; “+”, positive control, the rest were transgenic plants]; (C) qRT-PCR analysis of MrERF, MrbZIP, and MrSURNod in transgenic tobaccos.
Overexpression of MrERF, MrbZIP, and MrSURNod Affects Plant Growth and Development
To reveal whether the overexpression of MrERF, MrbZIP, or MrSURNod affects the flowering and fruiting of transgenic tobacco, the phenotypic characteristics at the mature stage under normal growth conditions were observed. The results showed that the over-expression transgenic lines grew better than WT, showing greater height, more branches, and earlier flowering (Figure 4C). In addition, the seed yield was significantly higher in transgenic tobacco than in WT (Figure 4A), being 14. 25-, 5. 56-, and 2.44-fold that of WT for MrERF, MrbZIP, and MrSURNod lines, respectively. The seed yield of the transgenic line (with MrERF) was significantly higher than others (P < 0.05). Transgenic plants also had longer and more numerous roots than WT (Figure 4B). This may contribute in improving the plant resistance. These results suggest that MrERF, MrbZIP, and MrSURNod are involved in plant growth. Their overexpression in tobacco-induced plant branching and earlier flowering and fruiting, thereby shortening the growth period.
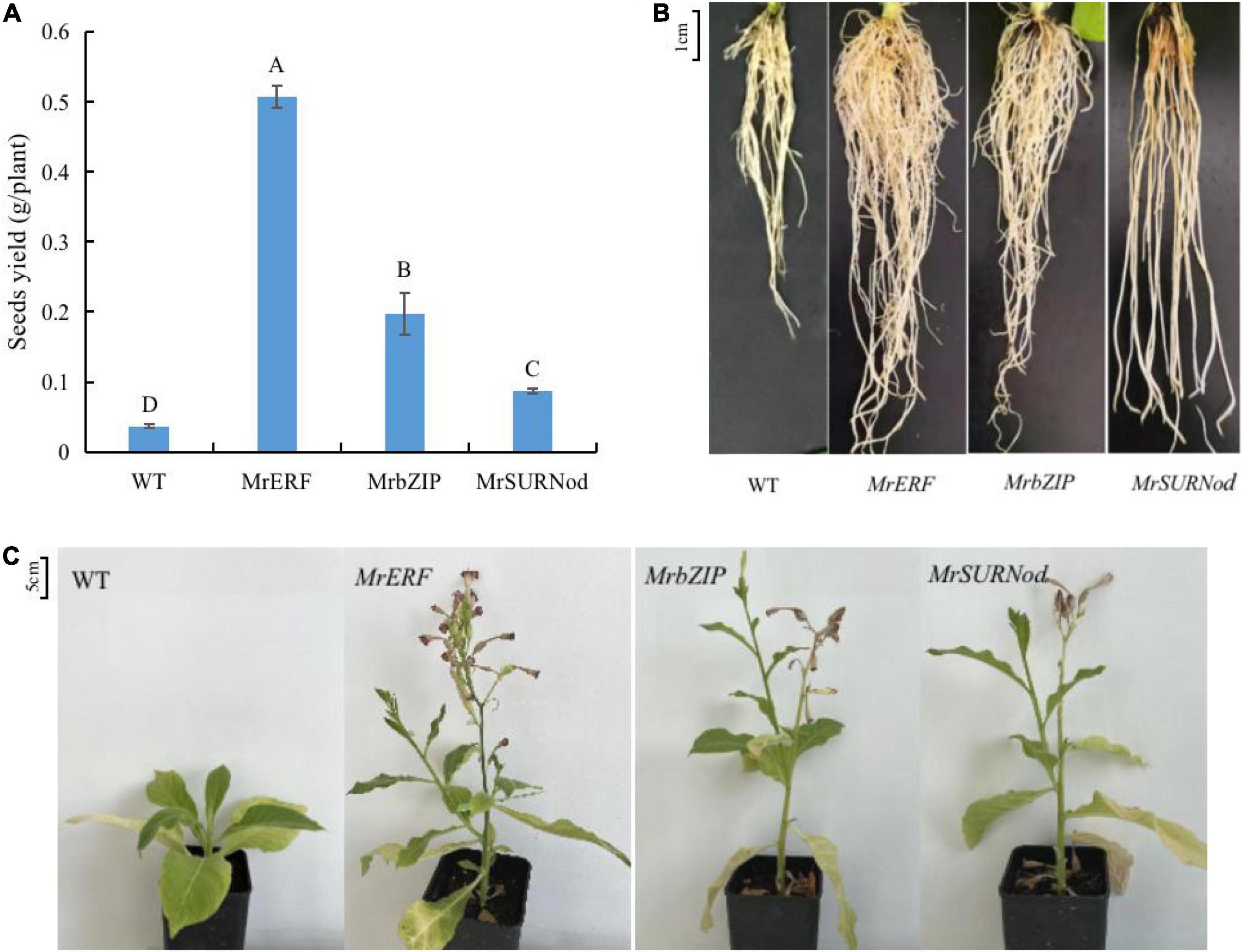
Figure 4. Effects of MrERF, MrbZIP, and MrSURNod on plant growth. (A) Seed yield of different tobacco lines (Different capital letters in the figure indicate significant differences at 0.05 level among different transgenic treatments). (B) Root morphology of different tobacco lines. (C) Morphological characteristics of aboveground parts of different tobacco lines.
Expression of MrERF, MrbZIP, and MrSURNod Under Abiotic Stress
To explore the expression of MrERF, MrbZIP, and MrSURNod in response to various stresses, we conducted qRT-PCR on seedlings of transgenic tobacco. The expression of MrERF, MrbZIP, and MrSURNod was induced strongly by water deficit, showing an overall upward trend with the duration of drought, and peaking (14. 77-, 15. 7-, and 4.9-fold higher than the non-stressed control, respectively) 9 days after the drought treatment started (Figure 5).
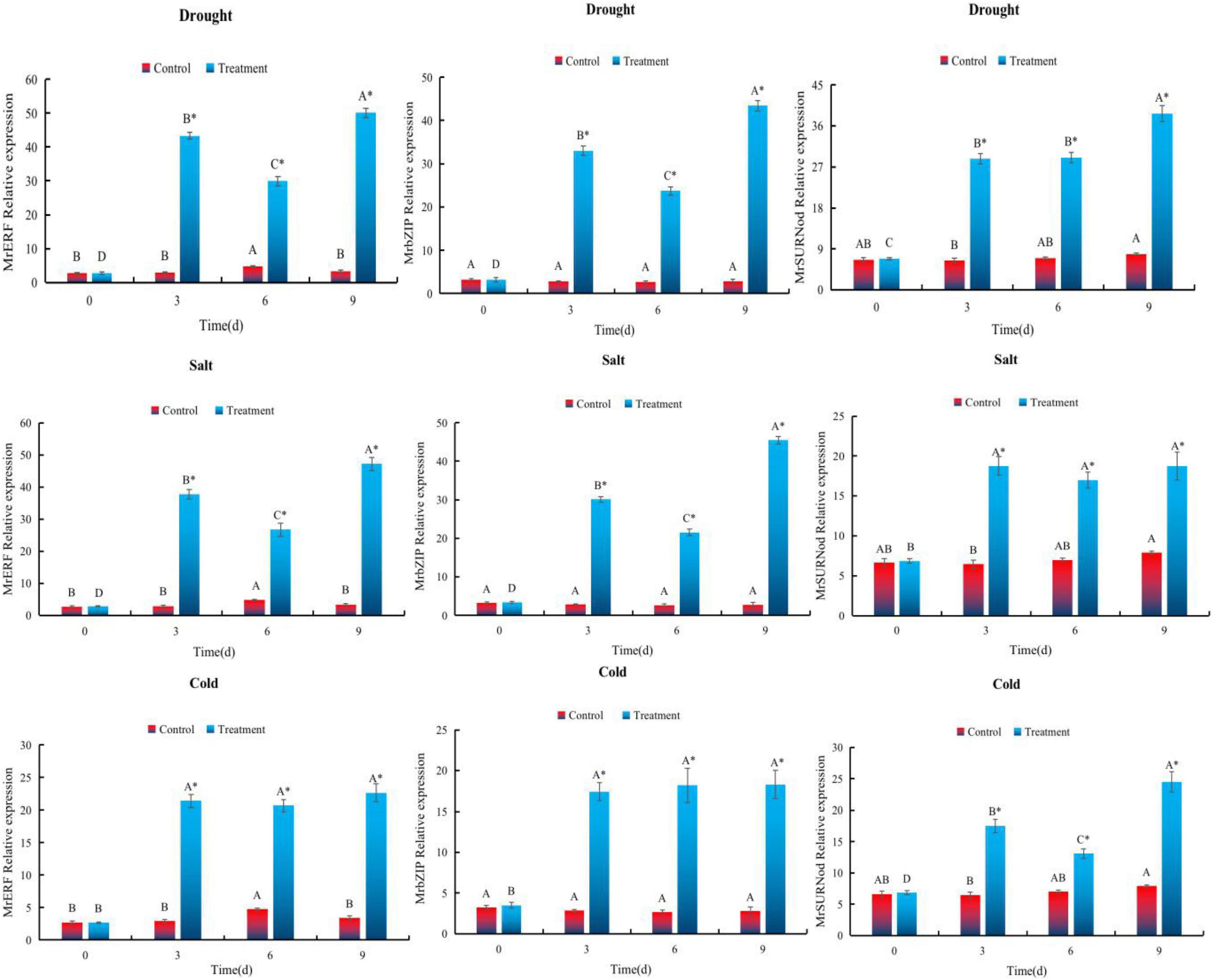
Figure 5. Expression patterns of MrERF, MrbZIP, and MrSURNod in leaf under abiotic stress. Different capital letters indicate significant differences under different treatments, and “*” indicates significant differences between control and treatment (P < 0.05).
In the NaCl treatment, expression patterns of MrERF and MrbZIP were consistent with those during drought stress. The MrERF and MrbZIP expression reached the maximum (around 13.92- and 16.45-fold of the control, respectively) on 9th day. By contrast, MrSURNod expression was 2.90 times higher than that of the control, but there was no significant change at three time points during the stress (Figure 5). Therefore, NaCl can induce the rapid expression of MrERF or MrbZIP in leaves, but its effect on MrSURNod is relatively weaker than the other two.
For cold stress, the expression of the genes was significantly higher than that of the control. The expression of MrERF and MrbZIP did not change with the duration of the stress, but MrSURNod showed a gradual upward trend, reaching the maximum (around 3.09-fold of the control) after 9 days. Hence, low temperature can strongly induce the expression of MrSURNod, but its effect on MrERF and MrbZIP is relatively weak.
In summary, it is preliminarily concluded that MrERF and MrbZIP mainly respond to salt and drought, while MrSURNod mainly responds to drought and cold stress.
Morphological and Physiological Changes in Transgenic Tobacco Under Abiotic Stress
Germination Rate of Transgenic Tobacco Seeds Under Abiotic Stress
The seed germination of transgenic tobacco lines was tested in this study (Figure 6). MrERF, MrbZIP, and MrSURNod are involved in the regulation of seed viability and can facilitate seed germination. Under normal conditions, the germination rate of WT or transgenic tobacco seeds was not significantly different. However, the leaf growth of three transgenic tobacco lines was better than that of WT, with MrERF having the best growth, followed by MrbZIP. The germination rate of the transgenic tobacco and WT seeds decreased with the increasing mannitol or NaCl concentration, but the germination rate of transgenic tobacco seeds was markedly higher than that of the WT seeds. The germination rate of transgenic tobacco lines with MrERF or MrbZIP was significantly higher than MrSURNod. Notably, transgenic tobacco with MrSURNod seeds did not germinate under 200 mM NaCl. Hence, MrERF and MrbZIP can improve the germination rate under osmotic stress or salt, whereas MrSURNod can improve seed germination rate under osmotic stress but only mild salt stress. In the cold treatment, WT and transgenic tobacco did not germinate.
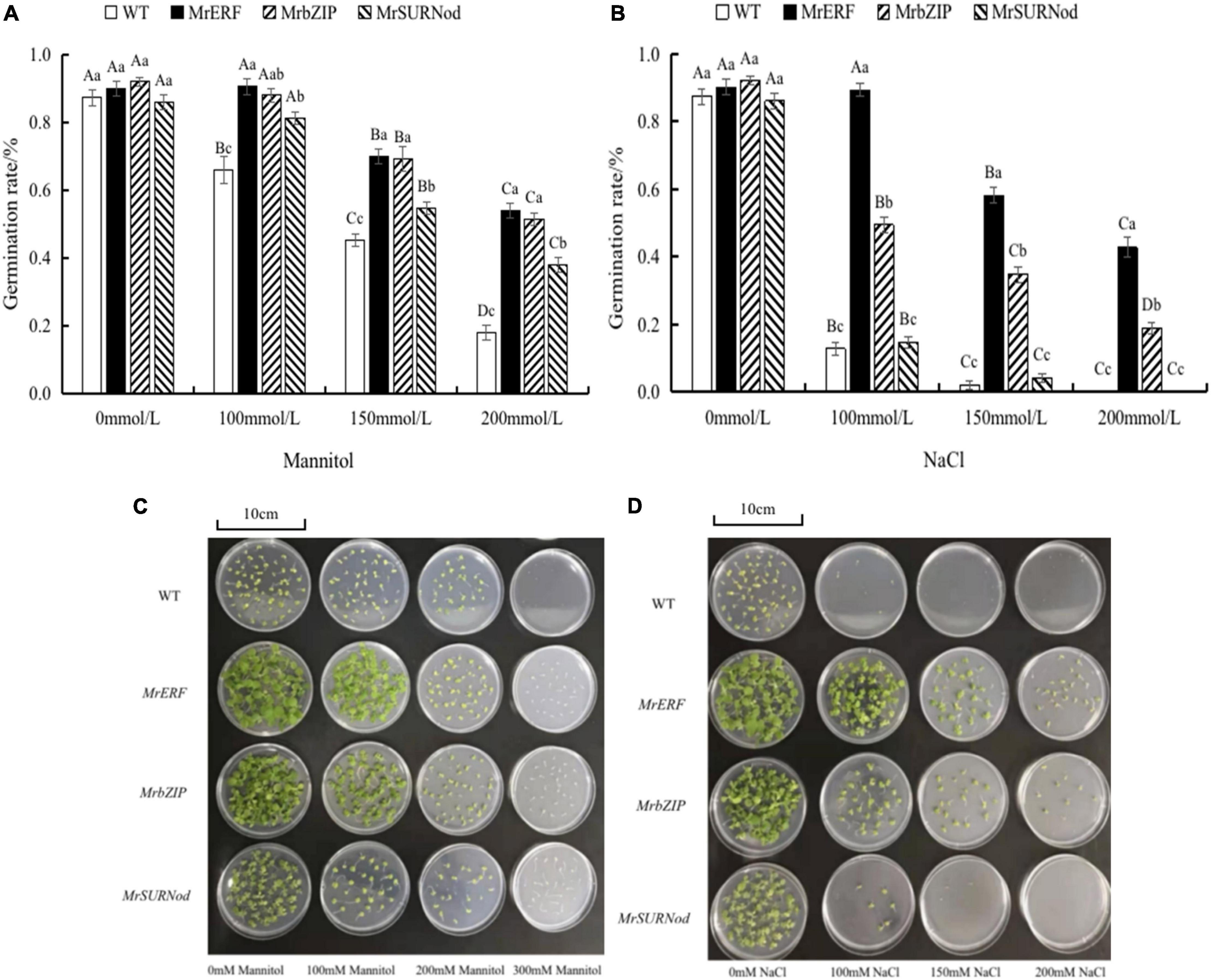
Figure 6. Germination of transgenic tobacco seeds treated with mannitol and NaCl. (A,C) Germination rate of transgenic tobacco seeds under mannitol treatment. (B,D) Germination rate of transgenic tobacco seeds under mannitol NaCl treatment. Different capital letters indicate significant differences under different treatments, and different lowercase letters indicate significant differences among different transgenic lines in the same treatment (P < 0.05).
Morphological Changes of Transgenic Tobacco Under Abiotic Stress
As shown in Figure 7, the growth retardation (plant height, biomass, and root/shoot ratio) under various abiotic stresses was lesser in transgenic tobacco than in WT. Under drought, with the duration of the stress plant height, biomass, and root/shoot increased, with the maximum on day 9. Under NaCl treatment, plant height, biomass, and root/shoot ratio of transgenic tobacco with MrERF or MrbZIP increased significantly, no significant change was noted for MrSURNod transgenic tobacco after day 6. However, in the low-temperature treatment, the contrary result was observed, i.e., plant height, biomass, and root/shoot ratio of MrSURNod transgenic lines increased obviously with the stress duration, while transgenic tobacco with MrERF or MrbZIP showed no obvious change from day 6.
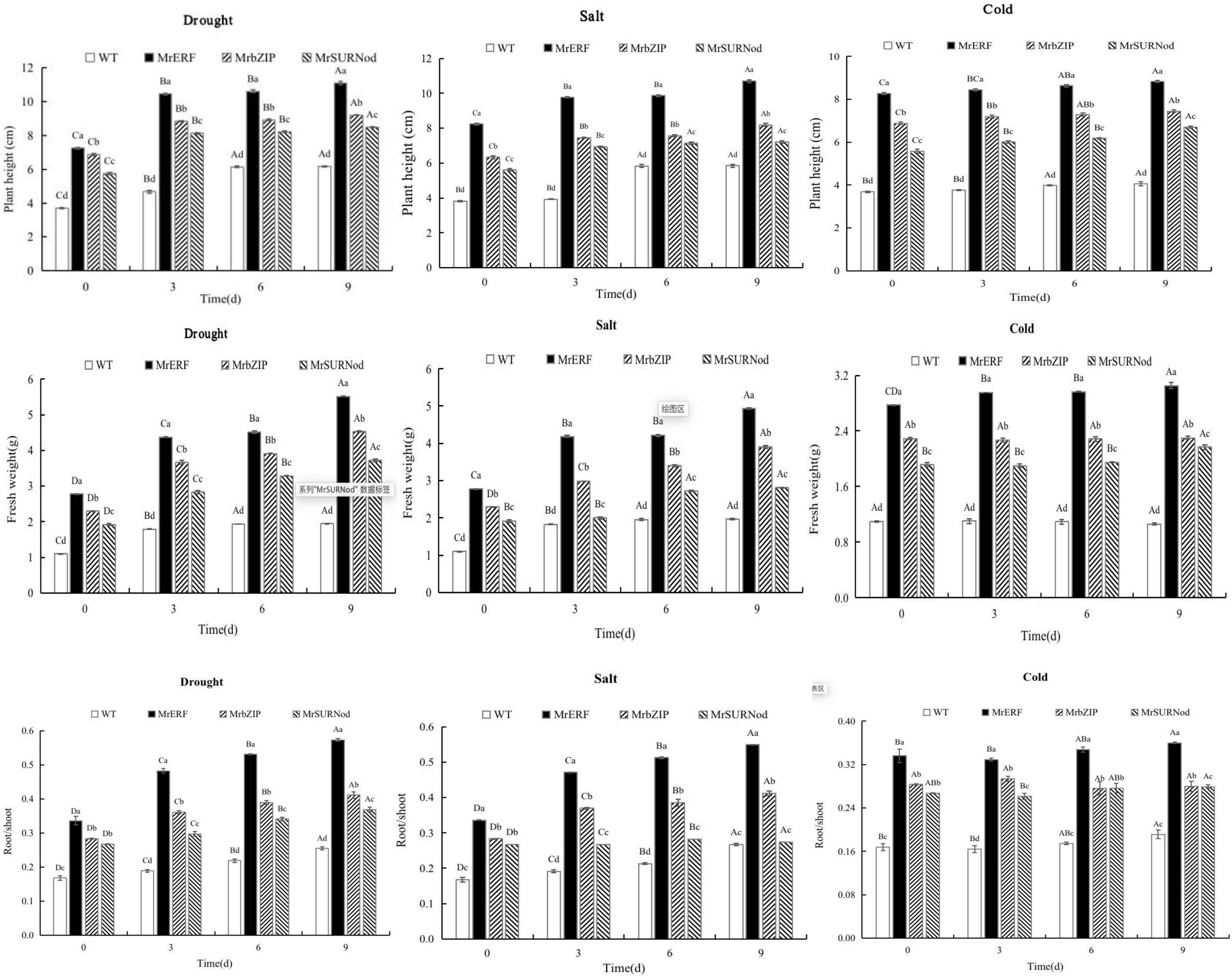
Figure 7. Plant height, biomass, and root/shoot of transgenic tobacco under abiotic stress. Different capital letters indicate significant differences under different treatments, and different lowercase letters indicate significant differences among different transgenic lines in the same treatment (P < 0.05).
We also noted that the root length and number were generally greater in transgenic plants than in WT (Figure 8). The transgenic plants with MrSURNod had the longest roots, followed by MrbZIP. With the duration of drought, the root length of MrERF, MrbZIP, and MrSURNod transgenic lines increased significantly, around 1. 48-, 1. 62-, and 1.73-fold of WT, respectively. Under salt stress, the root length of MrERF or MrbZIP transgenic lines increased, around 2.03- and 2.19-fold of WT, respectively, but the root length of MrSURNod transgenic plants increased only slightly. The root number increased gradually under drought or salt stress. The root number of transgenic tobacco with MrERF was the largest, around 2.76-fold of WT, followed by MrbZIP (2.09-fold increase), and MrSURNod was the smallest. In the cold treatment, the root length of MrSURNod transgenic tobacco increased significantly, but the other lines showed no significant change.
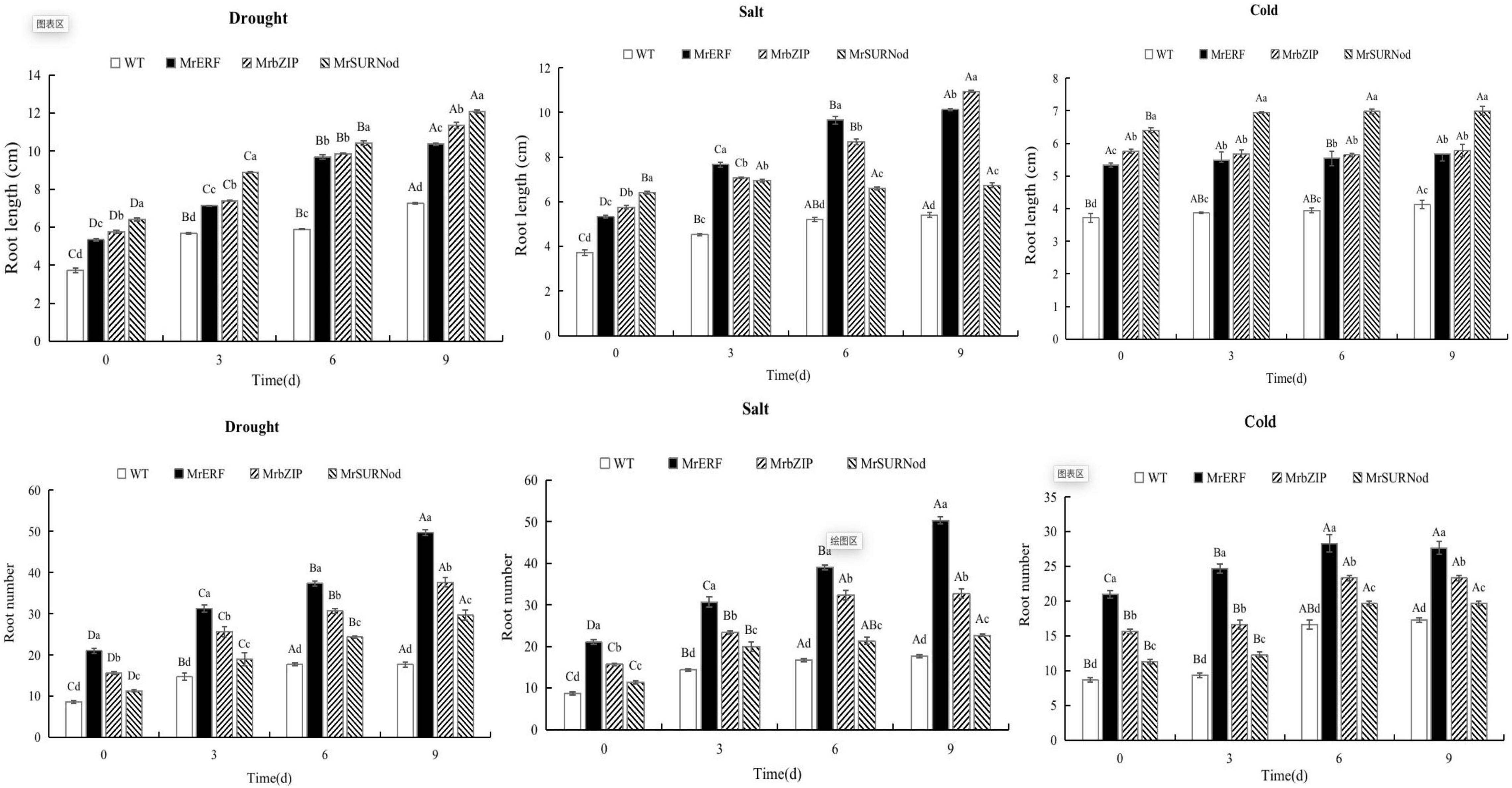
Figure 8. Effects of different stresses on root growth of tobacco. Different capital letters indicate significant differences under different treatments, and different lowercase letters indicate significant differences among different transgenic lines in the same treatment (P < 0.05).
Physiological Changes in Transgenic Tobacco Under Abiotic Stress
In addition to morphological changes, we also quantified the stress-related physiological and biochemical indicators, including osmoregulation substances (Figure 9), protective enzymes (Figure 10), and MDA (Figure 11). Significant increases in soluble sugars, proline content, SOD, and POD were observed under abiotic stress. With the prolonged drought and salt stress, the osmolyte content and antioxidant enzyme activities showed an upward trend and peaked after 9 days of stress. This finding indicates that the introduction of exogenous transcription factors can increase the accumulation of soluble sugars and proline and activity of the antioxidant enzyme in transgenic lines, thereby improving the resistance to drought and salt stress. These indices were highest in transgenic tobacco with MrERF, followed by MrbZIP and MrSURNod. However, with the duration of salt stress, the soluble sugar and proline contents and activities of SOD and POD in MrERF or MrbZIP transgenic tobacco leaves increased significantly, whereas in MrSURNod transgenic tobacco leaves, there was no significant increase after 6 days of stress. In the cold treatment, these indices first increased (peaking at day 6) and then decreased in MrERF or MrbZIP transgenic tobacco, but continued to increase significantly in MrSURNod transgenic tobacco. Overall, these results indicate that the salt tolerance of transgenic tobacco with MrERF or MrbZIP is stronger than that of MrSURNod, whereas the cold tolerance of the transgenic line with MrSURNod is the highest of the three lines. This was further verified by MDA content.
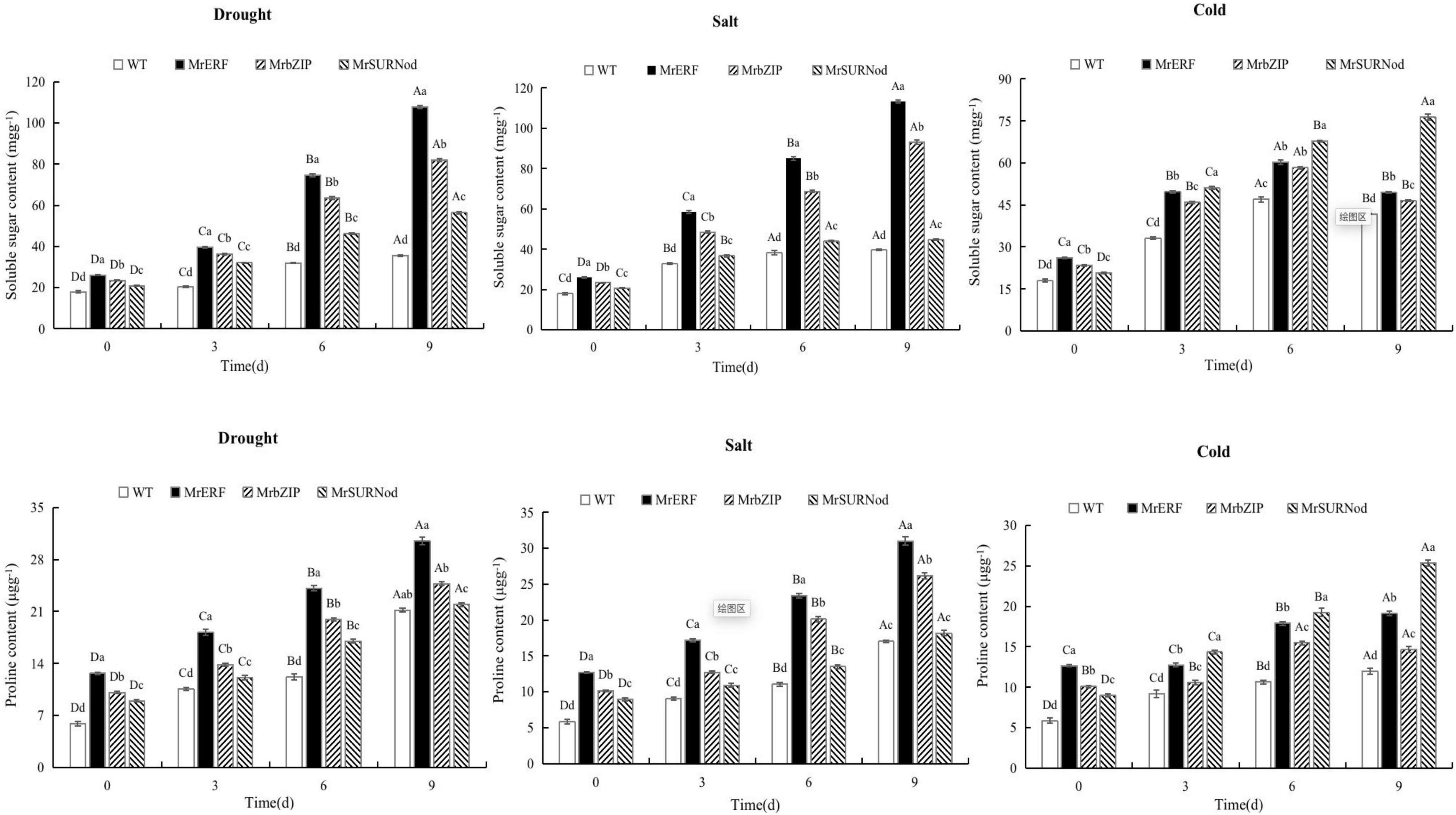
Figure 9. Effects of different stresses on osmotic adjustment substance of tobacco. Different capital letters indicate significant differences under different treatments, and different lowercase letters indicate significant differences among different transgenic lines in the same treatment (P < 0.05).
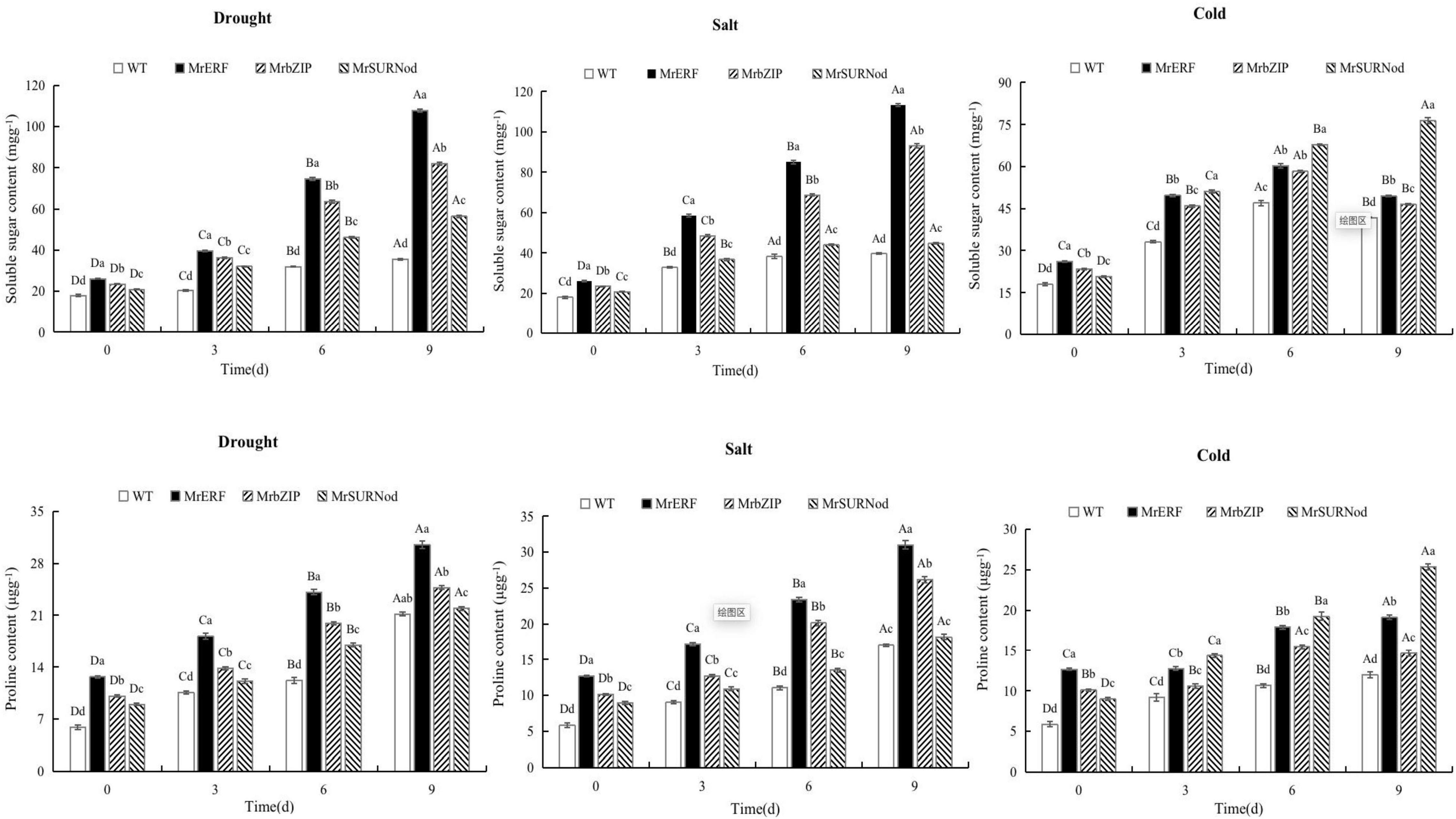
Figure 10. Effects of different stresses on antioxidant enzyme activity of tobacco. Different capital letters indicate significant differences under different treatments, and different lowercase letters indicate significant differences among different transgenic lines in the same treatment (P < 0.05).
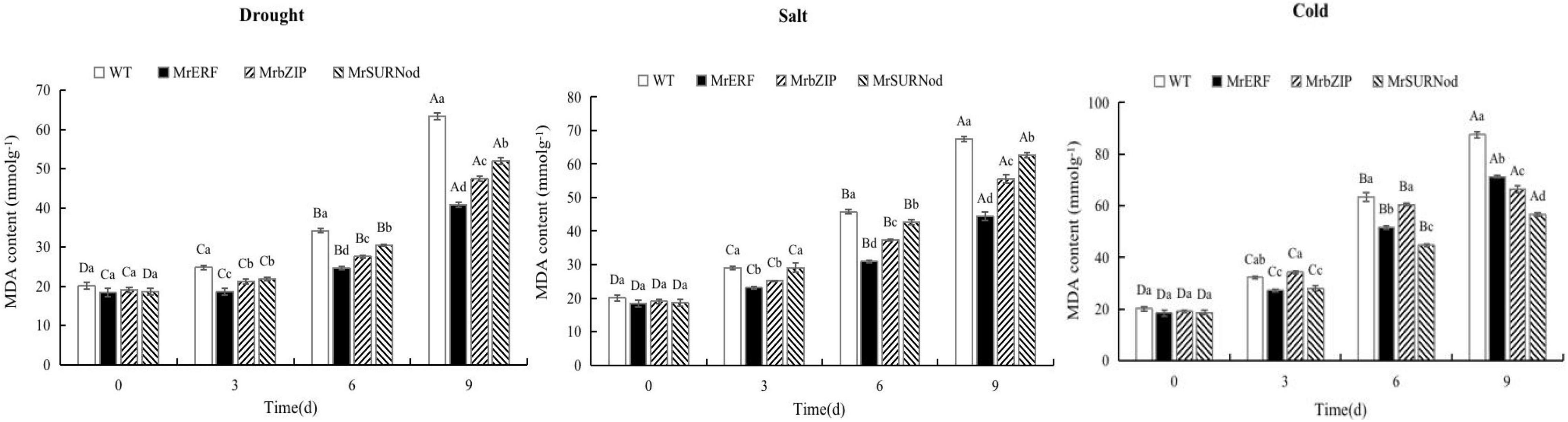
Figure 11. Effects of different stresses on MDA content of tobacco. Different capital letters indicate significant differences under different treatments, and different lowercase letters indicate significant differences among different transgenic lines in the same treatment (P < 0.05).
Under abiotic stress, the MDA content in WT exhibited a significant rise when compared with three transgenic tobacco lines, and reached the maximum after 9 days of stress, indicating that the severity of damage was lower in transgenic plants than in WT. Accumulation of MDA in MrERF transgenic plants was the lowest, followed by MrbZIP. Its accumulation in MrERF or MrbZIP transgenic lines under salt was relatively low, and the accumulation in MrSURNod was relatively high, indicating that the cell membrane damage in MrSURNod transgenic plants was more severe. However, the opposite result was found in the cold treatment, with a degree of damage lower in MrSURNod transgenic plants than MrERF or MrbZIP transgenic plants.
Based on the results of morphological and physiological parameters, transgenic tobacco performed better than WT in all aspects, with MrERF transgenic plants performing best, followed by MrbZIP. Overall, it is speculated that transgenic plants with MrERF or MrbZIP have stronger drought and salt tolerance than MrSURNod, whereas transgenic tobacco with MrSURNod is more cold tolerant.
Discussion
It has been well documented that they are involved not only in plant growth and development by regulating plant hormones (e.g., ABA) but also involved in the response to abiotic stresses, e.g., drought, low temperature, and salinity. Compared with other cultivated medics (M. sativa, M. truncatula, etc.), the AP2/ERF and bZIP family genes in M. ruthenica were more abundant (Wang et al., 2021). In addition, the SURNod19 gene family is annotated in NCBI as up-regulated by stress. Although SURNod19 has been identified as a cold-induced gene by the gene-chip analysis (Nakamura et al., 2013), it has not been reported whether it responds to other abiotic stresses. Here, we have selected a gene from each of the three gene families mentioned above. Our study highlights the importance of MrERF, MrbZIP, and MrSURNod in plant growth and the responses to abiotic stresses.
In our study, the three genes (MrERF, MrbZIP, and MrSURNod) with homology to ERF, bZIP, and SURNod19, respectively, were among the DEGs in response to drought stress in M. ruthenica. The deduced proteins (MrERF and MrbZIP) showed high similarity with, respectively, AtERF034 and AtbZIP46 from A. thaliana. Subcellular localization analysis implied the possible localization of MrERF and MrbZIP in the nucleus and MrSURNod in the chloroplast. Up-regulation of these genes improved the drought tolerance of M. ruthenica. The expression patterns of the three genes in transgenic tobacco lines were consistent in showing an upward trend under drought, cold, and NaCl stress. This is similar to the expression patterns of other transcription factors related to plant stress resistance, such as OsERF65 (Zhao et al., 2021) and VrbZIP52 (Wang L. et al., 2018), indicating that the genes may be related to plant stress responses. Salt and drought stress significantly induced the expression of MrERF and MrbZIP in tobacco, suggesting an important role of these two genes in plant responses to salt and drought stresses. The MrSURNod was induced by drought or cold stress, suggesting a regulatory function in response to drought and low temperature.
MrERF, MrbZIP, and MrSURNod Are Involved in Plant Growth and Development
The ERF transcription factors respond to the ethylene (ETH) signal and play an important role in ethylene-dependent physiological activities (Bolt et al., 2017). For example, the tomato SlERF52 affects the abscission of pedicel (Nakano et al., 2014), overexpression of AtERF019 can slow down plant growth and delay senescence (Scarpeci et al., 2017), TINY causes growth retardation (Xie et al., 2019), and the rice OsEATB can inhibit internode elongation and thereby result in dwarfed phenotype (Qi et al., 2011). The bZIP transcription factors also regulate growth, senescence, flower development, and seed maturation, e.g., AtbZIP9 and AtbZIP46 are involved in regulating leaf development. However, there are few studies on the effect of the SURNod19 family genes on growth and development. In our study, MrERF, MrbZIP, and MrSURNod belong to the same families as the example genes listed above, but they have distinct functions. Under non-stress conditions, transgenic tobacco over-expressing one of the three genes showed different growth and development characteristics than WT. For example, the plant height, biomass, and seed yield of tobacco with MrERF, MrbZIP, or MrSURNod were higher than those of WT, and all the three transgenic lines flowered earlier and had a shorter growth period than WT, which is speculated to be one of the ways to cope with stress. These findings were similar to PpcERF5, whose overexpression was associated with earlier flowering in Arabidopsis (Gao et al., 2021). The three genes also promoted branching in transgenic tobacco. In addition, MrERF, MrbZIP, and MrSURNod were beneficial to root development. Both MrERF and MrbZIP increased root length and number, whereas MrSURNod mainly promoted root elongation. Enhanced root growth and development is conducive to increasing the shoot height and biomass. In summary, the three genes tested are involved in plant growth and development, and their regulatory mechanisms need to be explored further.
MrERF, MrbZIP, and MrSURNod Improved Abiotic Stress Resistance by Changing Plant Morphology
In addition to promoting the growth and development of plants, the MrERF, MrbZIP, and MrSURNod may also serve as regulators of abiotic stress responses by altering gene expression. Water and nutrient absorption is strongly correlated with root morphology and distribution in soil. Deep roots distributed widely and with more branches are beneficial for water absorption and utilization, i.e., the water absorption is increased by increasing the root contact area with soil particles (Jones and Dolan, 2012). It has been shown that root distribution down the soil profile is an important determinant of drought adaptability (Grieder et al., 2014). Moreover, the increase in root number also can improve the drought resistance of plants (Henry et al., 2011). In our study, the root length as well as number were significantly higher in transgenic plants (MrERF, MrbZIP, and MrSURNod) than in WT, indicating that MrERF, MrbZIP, and MrSURNod can improve the plant capacity to utilize soil moisture by increasing the number and length of roots in horizontal and vertical directions. However, the roots of transgenic tobacco over-expressing MrSURNod were longer in length but fewer in number, whereas transgenics over-expressing the other two genes showed the reverse results. Transgenic tobacco over-expressing MrERF had the largest number of roots, and its shoot height and biomass and root/shoot ratio were significantly higher than those of wild type and other transgenic tobacco lines. In summary, MrERF and MrbZIP promote root elongation and branching (root number), whereas MrSURNod mainly promotes root elongation, but all these responses are likely to enhance adaptation to abiotic stresses.
The root/shoot ratio reflects the environment adaptability of plants. Increased root/shoot ratio was associated with dehydration (Du et al., 2020). Similarly, in our study, the root/shoot ratio of tobacco lines over-expressing MrERF, MrbZIP, or MrSURNod was significantly higher than that of WT. This resource allocation in plants is a strategy associated with enhanced adaptation to drought.
Additionally, we found the three genes to be beneficial to seed germination under stress, except in cold environments. The over-expression of MrERF was linked to the highest germination rate, and its function appeared to be similar to PsnERF75 (Wang S. J. et al., 2018) and BoERF1 (Jiang et al., 2018).
MrERF, MrbZIP, and MrSURNod Improved Osmotic Adjustment and Antioxidant Capacity to Enhance Resistance to Abiotic Stresses
Previous studies have shown significantly higher root length and number in the three over-expression lines compared with WT under stress, indicating that MrERF, MrbZIP, and MrSURNod play important roles in resistance to abiotic stress. The physiological parameters of transgenic lines under abiotic stress confirmed that MrERF, MrbZIP, and MrSURNod play an important positive regulatory role in plant abiotic stress response. Regardless of the growth conditions, the content of osmoregulation substances and the activity of antioxidant enzymes were higher in transgenic plants than in WT, implying that the transgenic lines may have more efficient osmoregulation and antioxidant systems. MDA is an important product of lipid peroxidation in the cell membranes, and its concentration varies in response to biotic and abiotic stresses. Here, we found that less MDA was generated in transgenic than WT tobacco seedlings, suggesting that over-expression of the tested genes can significantly improve plant tolerance to stress.
The osmolyte content and antioxidant enzyme activity in tobacco lines over-expressing MrERF or MrbZIP were higher than in the line over-expressing MrSURNod. The stress damage in MrERF or MrbZIP transgenic plants under drought and salt stress was light, indicating strong resistance, which is similar to chrysanthemum CmERF053 (Nie et al., 2018), OsERF71 (Ahn et al., 2017), sweet potato IbRAP2-12 (Li et al., 2019), OsbZIP71 (Liu et al., 2018), StbZIP65 (Zhao et al., 2020), AtbZIP36, AtbZIP37, and AtbZIP38 (Takuya et al., 2010). MrERF performed better in drought and salt tolerance than MrbZIP. The SURNod 19 family has been identified in Brachypodium distachyon (Martin et al., 2018), but its role in the plant response to the stress is unclear, although it has been considered a cold-inducible gene in Bromus inermis Leyss (Nakamura et al., 2013). In our study, the expression of this gene responded mainly to drought and to a small extent to cold stress. Fewer roots in tobacco over-expressing this gene might have been the reason for their weak salt tolerance. Overall, the three genes from M. ruthenica can enhance the resistance of transgenic tobacco to stress, with MrERF and MrbZIP being most effective in improving the resistance of tobacco to stress.
Conclusion
Through comprehensive bioinformatics, morphological, physiological, and transcriptomic analyses of the transgenic tobacco plant under various abiotic stresses, we have revealed the critical role of MrERF, MrbZIP, and MrSURNod in plant growth and abiotic stress responses. In addition to plant height and biomass, the over-expression of MrERF, MrbZIP, and MrSURNod promoted the production of branches and improved the yield of tobacco seeds. Furthermore, these genes were associated with early flowering and shortened growth periods in tobacco, which is one of the ways for transgenic tobacco to cope with stress. MrERF, MrbZIP, and MrSURNod also enhanced root development, which is conducive to resisting stress. Also, under stress, different transgenic lines enhanced stress resistance by increasing the osmolyte content and antioxidant enzyme activity.
In summary, MrERF, MrbZIP, and MrSURNod positively modulate stress resistance. MrERF and MrbZIP can improve drought and salt tolerance of plants, whereas MrSURNod is beneficial to improving drought and cold resistance. Transgenic tobacco over-expressing MrERF or MrbZIP showed better resistance than transgenic tobacco over-expressing MrSURNod.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
FLS and RNW conceived the original screening and research plans. FLS supervised the experiments. RNW and BX designed and performed the experiments. BX analyzed the data. RNW conceived the project and wrote the article with the contributions of all the authors. RNW agreed to serve as the author responsible for contact and ensures communication. All authors reviewed and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (32160323) and Key Projects in Science and Technology of Inner Mongolia (2021ZD0031).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to express their gratitude to EditSprings for the expert linguistic services provided.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.907674/full#supplementary-material
Footnotes
- ^ http://web.expasy.org
- ^ http://wolfpsort.org/
- ^ http://www.cbs.dtu.dk/services/TMHMM/
- ^ http://smart.embl-heidelberg.de/
References
Ahn, H., Jung, I., Shin, S. J., Park, J., Rhee, S., Kim, J. K., et al. (2017). Transcriptional network analysis reveals drought resistance mechanisms of AP2/ERF transgenic rice. Front. Plant Sci. 8:1044. doi: 10.3389/fpls.2017.01044
Bielsa, B., Hewitt, S., Reyes-Chin-Wo, S., Dhingra, A., and Rubio-Cabetas, M. J. (2018). Identification of water use efficiency related genes in ‘garnem’ almond-peach rootstock using time-course transcriptome analysis. PLoS One 13:e0205493. doi: 10.1371/journal.pone.0205493
Bolt, S., Zuther, E., Zintl, S., Hincha, D. K., and Schmulling, T. (2017). ERF105 is a transcription factor gene of Arabidopsis thaliana required for freezing tolerance and cold acclimation. Plant Cell Environ. 40, 108–120. doi: 10.1111/pce.12838
Campbell, T. A. (2003). Investigation of variations in NBS motifs in alfalfa (Medicago sativa), M. edgeworthli, and M. ruthenica. Rev. Can. Phytotech. 83, 371–376.
Campbell, T. A., Bao, G., and Xia, Z. L. (1997). Agronomic evaluation of Medicago ruthenica collected in Inner Mongolia. Crop Sci. 37, 599–604.
Cheng, M. C., Liao, P. M., Kuo, W. W., and Lin, T. P. (2013). The Arabidopsis ethylene response factor1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signal. Plant Physiol. 62, 1566–1582. doi: 10.1104/pp.113.221911
Depuydt, S., and Hardtke, C. S. (2011). Hormone signaling crosstalk in plant growth regulation. Curr. Biol. 21, R365–R373.
Dossa, K., Wei, X., Li, D., Fonceka, D., Zhang, Y., Wang, L., et al. (2016). Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant Biol. 16:171. doi: 10.1186/s12870-016-0859-4
Du, Y., Zhao, Q., Chen, L., Yao, X., Zhang, W., Zhang, B., et al. (2020). Effect of drought stress on sugar metabolism in leaves and roots of soybean seedlings. Plant Physiol. Biochem. 146, 1–12. doi: 10.1016/j.plaphy.2019.11.003
Gao, Y. D., Li, K., Zhu, Y. Y., Liu, X. L., Yin, Y. H., Wang, Y., et al. (2021). Cloning and functional analysis of ethylene responsive factor gene PpcERF5 from Chinese cherry (in China). Plant Physiol. J. 57, 59–68.
Grieder, C., Trachsel, S., and Hund, A. (2014). Early vertical distribution of roots and its association with drought tolerance in tropical maize. Plant Soil 377, 295–308.
Gupta, S., and Rashotte, A. M. (2014). Expression patterns and regulation of SlCRF3 and SlCRF5 in response to cytokinin and abiotic stresses in tomato (Solanum lycopersicum). J. Plant Physiol. 171, 349–358. doi: 10.1016/j.jplph.2013.09.003
Henry, A., Gowda, V. R. P., Torres, R. O., McNally, K. L., and Serraj, R. (2011). Variation in root system architecture and drought response in rice (Oryza sativa: phenotyping of the OryzaSNP panel in rainfed lowland fields. Field Crops Res. 120, 205–214.
Illgen, S., Zintl, S., Zuther, E., Hincha, D. K., and Schmulling, T. (2020). Characterisation of the ERF102 to ERF105 genes of Arabidopsis thaliana and their role in the response to cold stress. Plant Mol. Biol. 103, 303–320.
Jiang, M., Ye, Z. H., Zhang, H. J., and Miao, L. X. (2018). Broccoli plants over-expressing an ERF transcription factor gene BoERF1 facilitates both salt stress and Sclerotinia stem rot resistance. J. Plant Growth Regul. 38, 1–13.
Jones, V. A. S., and Dolan, L. (2012). The evolution of root hairs and rhizoids. Ann. Bot. 110, 205–212. doi: 10.1093/aob/mcs136
Jung, H., Chung, P. J., Park, S. H., Redillas, M. C. F. R., Kim, Y. S., Suh, J. W., et al. (2017). Overexpression of OsERF48 causes regulation of OsCML16, a calmodulin-like protein gene that enhances root growth and drought tolerance. Plant Biotechnol. J. 15, 1295–1308. doi: 10.1111/pbi.12716
Lee, D. K., Jung, H., Jang, G., Jeong, J. S., Kim, Y. S., Ha, S. H., et al. (2016). Overexpression of the OsERF71 transcription factor alters rice root structure and drought resistance. Plant Physiol. 172, 575–588. doi: 10.1104/pp.16.00379
Li, Y., Zhang, H., Zhang, Q., Liu, Q. C., Zhai, H., Zhao, N., et al. (2019). An AP2/ERF gene, IbRAP2-12, from sweetpotato is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Sci. 281, 19–30.
Lim, C. W., Baek, W., and Lee, S. C. (2018). Roles of pepper bZIP protein CaDILZ1 and its interacting partner RING-type E3 ligase CaDSR1 in modulation of drought tolerance. Plant J. 96, 452–467.
Liu, C., Mao, B., Ou, S., Wang, W., Liu, L., Wu, Y., et al. (2018). Correction to: OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 97, 467–468.
Martin, R. C., Vining, K., and Dombrowski, J. E. (2018). Genome-wide (ChIP-seq) identification of target genes regulated by BdbZIP10 during paraquat-induced oxidative stress. BMC Plant Biol. 18:58. doi: 10.1186/s12870-018-1275-8
Nakamura, T., Yazaki, J., Kishimoto, N., Kikuchi, S., Robertson, A. J., Gusta, L. V., et al. (2013). Comparison of long-term up-regulated genes during induction of freezing tolerance by cold and ABA in bromegrass cell cultures revealed by microarray analyses. Plant Growth Regul. 71, 113–136.
Nakano, T., Fujisawa, M., Shima, Y., and Ito, Y. (2014). The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. J. Exp. Bot. 65, 3111–3119. doi: 10.1093/jxb/eru154
Nie, J., Wen, C., Xi, L., Lv, S. H., Zhao, Q. C., Kou, Y. P., et al. (2018). The AP2/ERF transcription factor CmERF053 of chrysanthemum positively regulates shoot branching, lateral root, and drought tolerance. Plant Cell Rep. 37, 1049–1060. doi: 10.1007/s00299-018-2290-9
Pan, Y. L., Hu, X., Li, C. Y., Xu, X., Su, C. G., Li, J. H., et al. (2017). SlbZIP38, a tomato bZIP family gene downregulated by abscisic acid, is a negative regulator of drought and salt stress tolerance. Genes 8:402. doi: 10.3390/genes8120402
Qi, W. W., Sun, F., Wang, Q. J., Chen, M. L., Huang, Y. Q., Feng, Y. Q., et al. (2011). Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down – regulating agibberellin biosynthetic gene. Plant Physiol. 157, 216–228. doi: 10.1104/pp.111.179945
Rong, W., Qi, L., Wang, A., Du, L., Liang, H., Xin, Z., et al. (2014). The ERF transcription factor promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 12, 468–479. doi: 10.1111/pbi.12153
Scarpeci, T. E., Frea, V. S., Zanor, M. I., and Valle, E. M. (2017). Overexpression of AtERF019 delays plant growth and senescence, and improves drought tolerance in Arabidopsis. J. Exp. Bot. 68, 673–685. doi: 10.1093/jxb/erw429
Takuya, Y., Yasunari, F., Hiroko, S., Satoshi, K., Kyonoshin, M., Junya, M., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Vishwakarma, K., Upadhyay, N., Kumar, N., Yadav, G., Singh, J., Mishra, R. K., et al. (2017). Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 120:161. doi: 10.3389/fpls.2017.00161
Wang, C., Lu, G., Hao, Y., Guo, H., Guo, Y., Zhao, J., et al. (2017). ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta 246, 453–469. doi: 10.1007/s00425-017-2704-x
Wang, L., Zhu, J., Li, X., Wang, S., and Wu, J. (2018). Salt and drought stress and ABA responses related to bZIP genes from V. radiata and V. angularis. Gene 651, 152–160. doi: 10.1016/j.gene.2018.02.005
Wang, S. J., Zhou, B. R., Yao, W. J., and Jiang, T. (2018). PsnERF75 transcription factor from Populus simonii×P. nigra confers salt tolerance in transgenic Arabidopsis. J. Plant Biol. 61, 61–71.
Wang, T., Ren, L., Li, C., Zhang, D., Zhang, X., Zhou, G., et al. (2021). The genome of a wild Medicago species provides insights into the tolerant mechanisms of legume forage to environmental stress. BMC Biol. 19:96. doi: 10.1186/s12915-021-01033-0
Wang, Z., Su, G. X., Li, M., Ke, Q. B., Kim, S. Y., Li, H. B., et al. (2016). Over expressing Arabidopsis ABF3 increases tolerance to multiple abiotic stresses and reduces leaf size in alfalfa. Plant Physiol. Biochem. 109, 199–208. doi: 10.1016/j.plaphy.2016.09.020
Xie, Z. L., Nolan, T., Jiang, H., Tang, B. Y., Zang, M. C., Li, Z. H., et al. (2019). The AP2/ERF transcription factor TINY modulates brassinosteroid-regulated plant growth and drought responses in Arabidopsis. Plant Cell 31, 1788–1806. doi: 10.1105/tpc.18.00918
Yang, J. Y., Zheng, W., Tian, Y., Wu, Y., and Zhou, D. W. (2011). Effects of various mixed salt-alkaline stresses on growth, photosynthesis, and photosynthetic pigment concentrations of Medicago ruthenica seedlings. Photosynthetica 49, 275–284.
Zhao, H., Yang, S. Q., Xu, K., and Luo, L. J. (2021). Cloning, expression and trans-activation activity analysis of rice drought tolerance gene OsERF65 (in China). Mol. Plant Breed. 19, 361–369.
Zhao, L. P., Wu, G. L., and Shi, Z. H. (2013). Post-fire species recruitment in a semiarid perennial steppe on the Loess Plateau. Aust. J. Bot. 61, 29–35.
Zhao, P., Ye, M. G., Wang, R. Q., and Chen, Q. (2020). Systematic identification and functional analysis of potato (Solanum tuberosum L.) bZIP transcription factors and overexpression of potato bZIP transcription factor StbZIP-65 enhances salt tolerance. Int. J. Biol. Macromol. 161, 155–167. doi: 10.1016/j.ijbiomac.2020.06.032
Zhu, M. K., Meng, X. Q., Cai, J., Li, G., Dong, T. T., and Li, Z. Y. (2018). Basic leucine zipper transcription factor SlbZIP1 mediates salt and drought stress tolerance in tomato. BMC Plant Biol. 18:83. doi: 10.1186/s12870-018-1299-0
Zhuo, C. L., Liang, L., Zhao, Y. Q., Guo, Z. F., Lu, S. Y., and Zhou, C. L. (2018). A cold responsive ethylene responsive factor from Medicago falcata confers cold tolerance by up-regulation of polyamine turnover, antioxidant protection, and proline accumulation. Plant Cell Environ. 41, 2021–2032. doi: 10.1111/pce.13114
Keywords: Medicago ruthenica, abiotic stress, plant growth, transgenic tobacco, morphology, physiology
Citation: Wu RN, Xu B and Shi FL (2022) MrERF, MrbZIP, and MrSURNod of Medicago ruthenica Are Involved in Plant Growth and Abiotic Stress Response. Front. Plant Sci. 13:907674. doi: 10.3389/fpls.2022.907674
Received: 30 March 2022; Accepted: 05 May 2022;
Published: 02 June 2022.
Edited by:
Klára Kosová, Crop Research Institute (CRI), CzechiaReviewed by:
Tiejun Zhang, Beijing Forestry University, ChinaAlma Balestrazzi, University of Pavia, Italy
Copyright © 2022 Wu, Xu and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengling Shi, bm1jenlzZmxAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Rina Wu
Rina Wu Bo Xu†
Bo Xu†