
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 May 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.906472
This article is part of the Research Topic Trends in Cotton Breeding: Meeting the Challenges of the 21st Century View all 22 articles
 Mukhtor M. Darmanov
Mukhtor M. Darmanov Abdusalom K. Makamov
Abdusalom K. Makamov Mirzakamol S. Ayubov*
Mirzakamol S. Ayubov* Naim N. Khusenov
Naim N. Khusenov Zabardast T. Buriev
Zabardast T. Buriev Shukhrat E. Shermatov
Shukhrat E. Shermatov Ilkhom B. Salakhutdinov
Ilkhom B. Salakhutdinov Khurshida A. Ubaydullaeva
Khurshida A. Ubaydullaeva Jurabek K. Norbekov
Jurabek K. Norbekov Maftuna M. Kholmuradova
Maftuna M. Kholmuradova Sardor E. Narmatov
Sardor E. Narmatov Ilyos S. Normamatov
Ilyos S. Normamatov Ibrokhim Y. Abdurakhmonov*
Ibrokhim Y. Abdurakhmonov*Marker-assisted selection (MAS) helps to shorten breeding time as well as reduce breeding resources and efforts. In our MAS program, we have targeted one of previously reported LD-blocks with its simple sequence repeat (SSR) marker(s), putatively associated with, at least, four different fibre quality QTLs such as fibre length, strength, micronaire and uniformity. In order to transfer targeted QTLs from a donor genotype to a cultivar of choice, we selected G. hirsutum donor genotypes L-141 and LN-1, possessing a fibre quality trait-associated LD-block from the chromosome 7/16. We crossed the donor lines with local elite G. hirsutum cultivars ‘Andijan-35’ and ‘Mekhnat’ as recipients. As a result, two segregating populations on LD-block of interest containing fibre QTLs were developed through backcrossing (BC) of F1 hybrids with their relative recipients (used as recurrent parents) up to five generations. In each BC and segregating BC1-5F1 populations, a transfer of targeted LD-block/QTLs was monitored using a highly polymorphic SSR marker, BNL1604 genotype. The homozygous cultivar genotypes with superior fibre quality and agronomic traits, bearing a targeted LD-block of interest, were individually selected from self-pollinated BC5F1 (BC5F2–5) population plants using the early-season PCR screening analysis of BNL1604 marker locus and the end-of-season fibre quality parameters. Only improved hybrids with superior fibre quality compared to original recipient parent were used for the next cycle of breeding. We successfully developed two novel MAS-derived cotton cultivars (named as ‘Ravnaq-1’ and ‘Ravnaq-2’) of BC5F5 generations. Both novel MAS cultivars possessed stronger and longer fibre as well as improved fibre uniformity and micronaire compared to the original recurrent parents, ‘Andijan-35’ and ‘Mekhnat’. Our efforts demonstrated a precise transfer of the same LD-block with, at least, four superior fibre QTLs in the two independent MAS breeding experiments exploiting different parental genotypes. Results exemplify the feasibility of MAS in cotton breeding.
The main goal of breeding programs is to mobilize genes from a donor genotype into an elite cultivar parent. Although traditional breeding methods showed efficiency in transferring a single gene/trait, it has limitations in the targeted-mobilization of multiple genes, regulating the complex quantitative traits (Abdurakhmonov et al., 2011). In particular, breeding of cotton cultivars with superior fibre quality is conventionally challenging because of multigenic regulation of quantitative trait loci (QTLs) for fibre quality as well as the existence of negative correlation among fibre quality traits, largely affected by a linkage drag (Lewis, 1962; Abdurakhmonov et al., 2008, 2009; Ijaz et al., 2019). In addition, the development of new cultivars using traditional selection methods is costly as well as time and resource-consuming task (Kushanov et al., 2021). To overcome these, DNA-based molecular markers are used in genetics and plant breeding to transfer traits of interest in a targeted manner, which is referred to as molecular breeding or marker-assisted selection (MAS; Abdurakhmonov, 2002; Collard and Mackill, 2008; Kushanov et al., 2021).
The molecular markers, based on the differences in the genetic material (DNA) sequence and closely located by a trait of interest, have evolved because of availability genetic sequence data. Molecular markers vary on types of genetic structure of DNA sequence and detection methods used to generate and record the polymorphisms between genotypes of interest (Collard et al., 2005; Amiteye., 2021). DNA markers associated with genomic regions of interest allow breeders to select plants early stage of plant growth based on a marker genotype rather than a phenotype that may get expressed later in plant vegetation. In particular, DNA markers are very useful and convenient for major QTLs, which are challenging task, if assessed by phenotypic evaluations during the course of breeding process. Therefore, DNA-based molecular markers and QTL-mapping results have become useful to find genome composition of interest (Tanksley et al., 1989). Usually, the targeted chromosomal segments of donor parents carry also undesirable traits along with a trait of interest. Because of power of trait-marker linkage association and linkage disequilibrium (LD) decay characteristics of neighbouring DNA markers over the genetic distance, DNA markers are the best instrumental for removing ‘undesired’ chunk of genetic material (linkage drag) coming from donor plant genome (Abdurakhmonov et al., 2008). In this case, DNA markers assist to minimize time to remove undesirable traits from F(n) generations after introducing QTL of interest into the recipient by a sexual crossing. Moreover, wild crops relatives have largely untapped source of desirable traits desirable alleles even with less expressed phenotypes (Migicovsky and Myles, 2017). Those can be identified by DNA markers and mobilized into elite germplasm to create new cultivars with superior phenotypes.
The huge number of QTL-mapping and genome-wide association studies in various cotton species and diverse sets of Upland cotton germplasms (He et al., 2007; Abdurakhmonov et al., 2008, 2009; Zeng et al., 2009; Zhang et al., 2012; Cai et al., 2014; Fang et al., 2014) have provided a portfolio of DNA markers, tightly linked with agronomically important cotton traits (Kushanov et al., 2021). Many QTLs for cotton fibre quality traits were identified for the last two decades through the mapping or association analyses of DNA markers in different populations or germplasm accessions in multiple environments (Sun et al., 2012; Liang et al., 2013; Fang et al., 2014; Shao et al., 2014; Zhiyuan et al., 2014; Said et al., 2015; Ma et al., 2019). However, a low level of functionality of many of these QTLs in different environments or breeding populations (Hugie et al., 2016) reduces their value in practical breeding (Fang et al., 2014). Loci that are detected in multiple environments are called stable and repeatable QTLs; therefore, they are favoured as reliable genetic loci for the MAS programs (Fang, 2018; Paudel et al., 2020). In this context, association studies, exploiting a large number of genotypes with simultaneous analyses of multiple alleles and majority of recombination events of genomic regions conditioning trait of interest, has become a powerful tool to mine stable QTLs for cotton breeding (Abdurakhmonov et al., 2008).
Sequencing efforts of several cotton genomes (diploid A and D, tetraploid AD; Pan et al., 2020) has provided a strong basis for accurate mapping of important QTLs useful for MAS programs. Moreover, genetic mapping in multiple populations using a large number of new generation molecular markers has led to reduce large gaps conventionally caused by the lack of polymorphism in certain complex genomic regions, helping to increase the number of mapped loci, confirm marker order, and increase marker coverage in cotton genome (Ulloa et al., 2017).
The competition for synthetic fibres, the variability of productivity from year to year, and the existence of new requirements for fibre quality due to technological developments in the textile industry continuously demand for novel cotton cultivars with superior fibre quality, fulfilling the industry requirements. The main fibre quality traits include fibre length (FL), fibre strength (FS), fibre uniformity (FU), fibre elongation (FE), and micronaire (FM) value. These are the most important characteristics that affect yarn quality (Yang et al., 2016). The application of MAS for the breeding of fibre quality traits along with yield potential is pivotal in cotton breeding (Li et al., 2016) although there is limited success of molecular breeding of fibre quality traits. To apply MAS tools for the improvement of fibre quality traits of commercially used Upland cotton of Uzbekistan, in this study, we exploited donor genotypes and fibre quality associated LD-block that was mapped using Simple Sequence Repeat (SSR) markers in our previous LD-based association study (Abdurakhmonov et al., 2009). As the result, two novel Upland cotton cultivars have been developed using MAS in a short period, with high fibre quality and agronomic potential. Our results should be helpful to accelerate cotton breeding programs, rapidly and precisely improving coarse fibred Upland cotton cultivars of Uzbekistan and timely responding the demand of national textile industry.
Twenty-six donor germplasm accessions bearing LD-block/QTLs of interest were identified in associated mapping studies of Upland cotton germplasm in our previous studies (Abdurakhmonov et al., 2008, 2009). For our MAS program, we have chosen L-141 and L-N1 lines as unique donor parental lines, bearing QTLs for the four important fibre quality traits such as FL, FS, FU, and FM. The local commercial G. hirsutum cotton cultivars ‘Andijan-35’ and ‘Mekhnat’, genetically polymorphic to the donor, were used as recipient parents. These are local cultivars, widely grown by farmers in Uzbekistan, but have coarse fibre compared to the donor parents. In addition, the G. hirsutum elite cultivar ‘Namangan-77’, widely used as a breeding standard in Uzbekistan (Abdukarimov et al., 2003), were used as the control genotype for comparisons of fibre quality improvement. The ‘Namangan-77’ cultivar was also used as the negative control (BC hybrid - lack of QTL allele) for the course of MAS breeding process.
Sexual crosses were performed between ‘Andijan-35’ × L-141 and ‘Mekhnat’ × L-N1. Further, F1 hybrids were backcrossed up to BC5 generations, creating segregating populations on targeted QTL regions. In each BC and segregating BC1-5F1 populations, the transfer of targeted LD-block/QTLs was monitored using the early-season PCR screening of SSR marker (see below section) and the end-of-season fibre quality parameters. Only improved hybrids with superior fibre quality compared to original recipient parent were used for the next cycle of breeding. Subsequently, BC5F1 genotypes of interest were self-pollinated and BC5F2 generation plant populations have been developed. Homozygous cultivar genotypes with advanced for fibre quality, morphological and agronomical traits and bearing a targeted QTL/LD-block of interest were individually selected from BC5F5 population plants based on marker state and trait improvement as described above. More than hundred genotypes, carrying homozygous QTLs, were planted separately in ten meters’ rows as separate families. Breeding was performed based on homozygous state of SSR marker genotype, fibre quality and yield to obtain new ‘Ravnaq-1′ and ‘Ravnaq-2′ cotton cultivars.
The seeds of ‘Ravnaq-1’ and ‘Ravnaq-2’ were planted in one location with three replicates in a 90 × 15 × 1 plot scheme along with parental donor genotypes and control cotton cultivar ‘Namangan-77’ in three filed conditions with three replications during 2018 to 2020 years/seasons. Opened bolls from each replicate of all genotypes were individually harvested to analyse agronomic traits as harvesting materials. The traits such as 100 seed weight, and lint percentage were manually analysed in laboratory condition. The major fibre quality traits including FL, FS, FU and FM were measured using the High Volume Instrument (HVI; Uster Technologies, Inc., Knoxville, United States). Fibre quality traits of ‘Ravnaq-1’ and ‘Ravnaq-2’ cotton cultivars with their parent and control genotypes were studied over three years in three biological replications.
We tested several SSR markers from our association study (Abdurakhmonov et al., 2009) in the donor and recipient parental genotypes (Figure 1; Table 1) and have chosen one of highly polymorphic SSR marker - BNL1604 (Figure 1) to use as a molecular tool to monitor the transfer of LD-block associated with fibre QTLs of our interest.
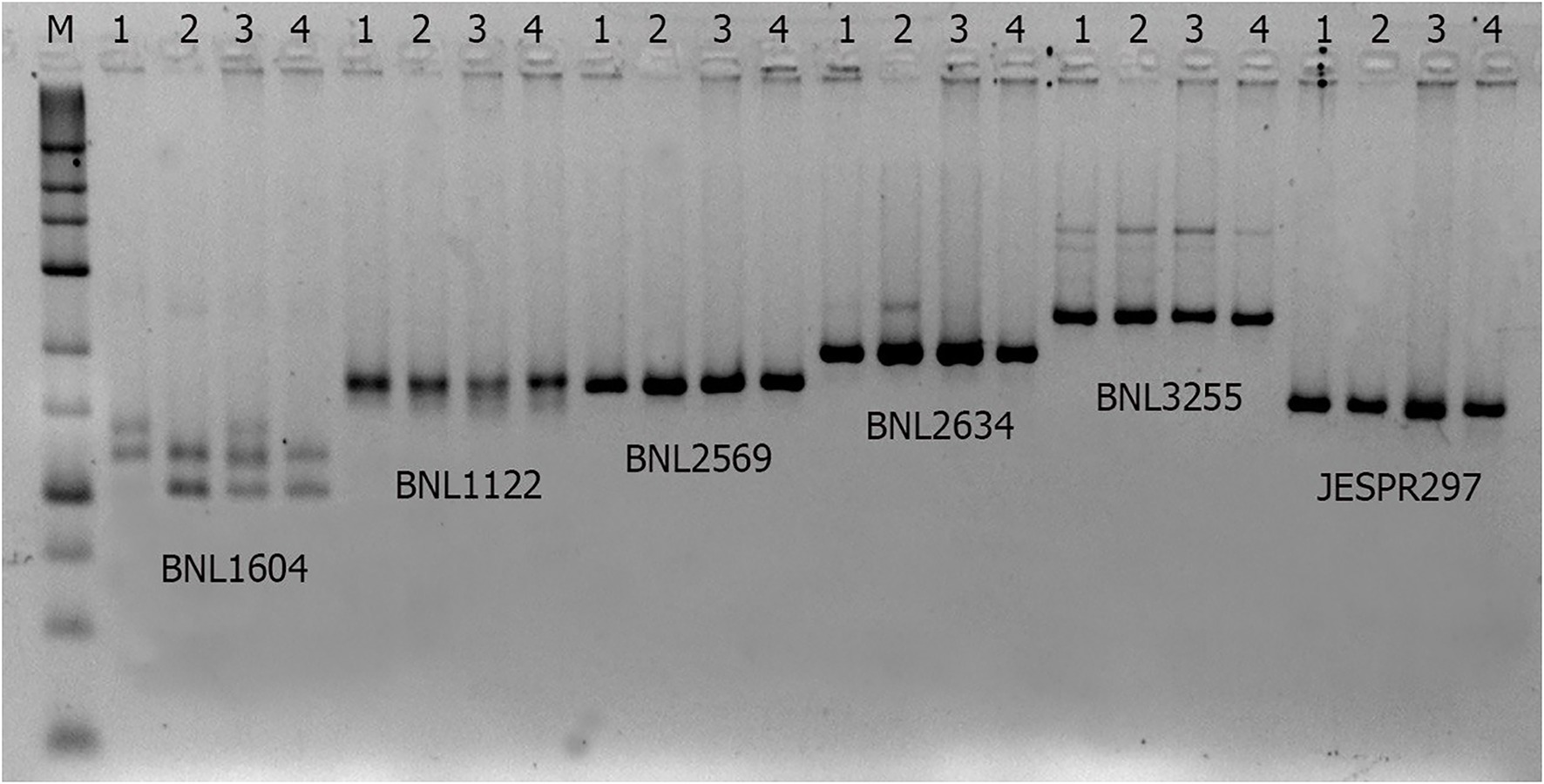
Figure 1. Agarose gel electropherogram of SSR markers in parental and hybrids. Parental genotypes (1 and 2), F1 hybrid (3) and BC generation hybrid (4).
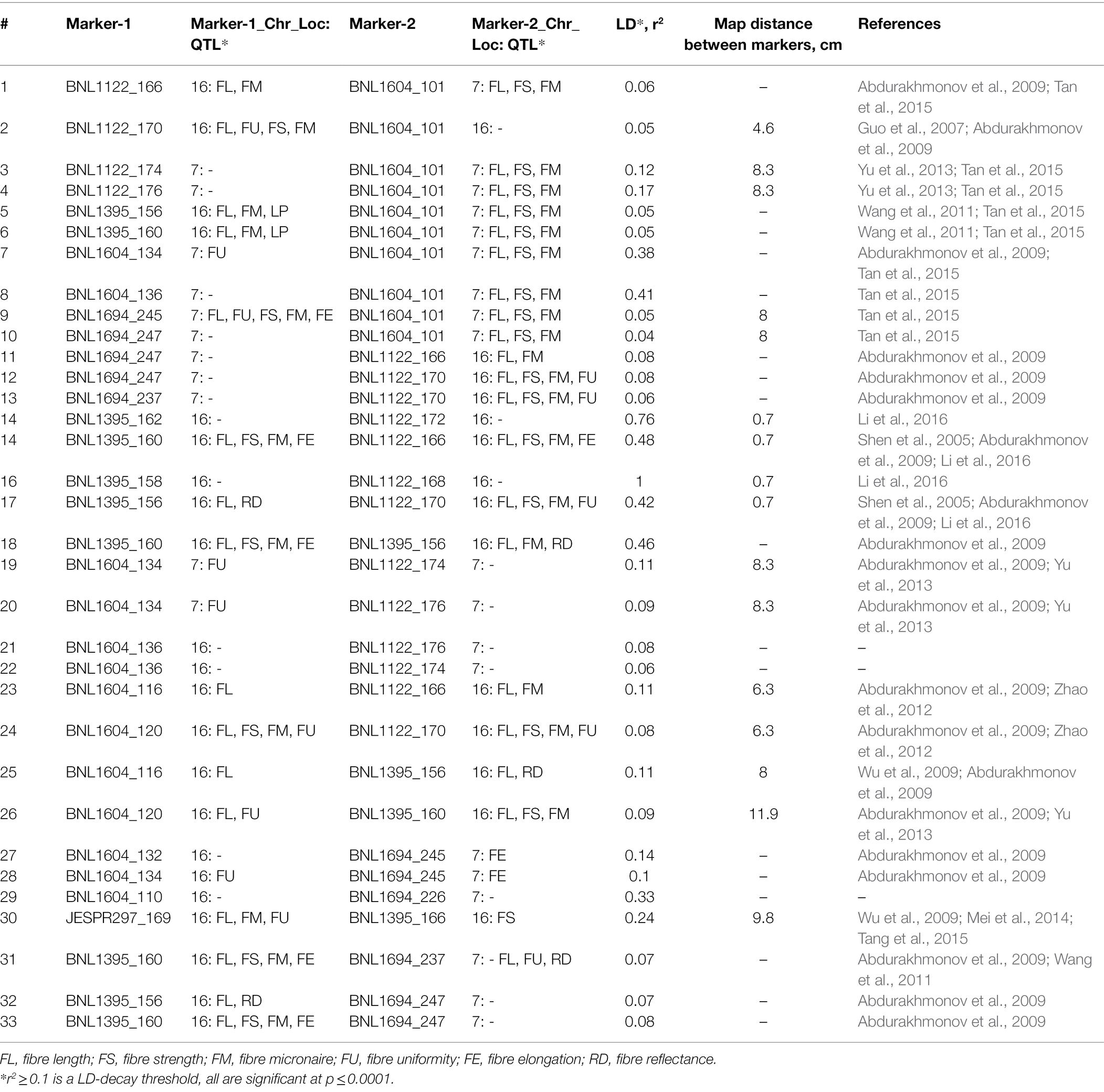
Table 1. Putative information on pairwise linkage, map distance, and QTL association of SSR markers in the targeted LD-block.
Leaf tissues were collected from all samples, and genomic DNAs were isolated from the frozen leaf tissues using the modified method of Dellaporta et al. (1983). Further, DNA concentrations were diluted in working solution (25 ng/μl) and stored in a refrigerator at −20°C. Amplification reactions were performed in 50 μl volumes containing 4.5 μl 10 × PCR buffer with 1.5 mM MgCl2, 1 μl BSA, 0.5 μl 25 mM of a dATP, dGTP, dTTP, and dCTP mix, 2.5 μl 50 ng/μl of microsatellite marker specific BNL1604 primers pairs (Supplementary Table 1) associated with fibre strength and fibre length, 1 μl of 25 ng/μl template DNA and 0.5 U Taq DNA polymerase (Sigma-Aldrich Chemie GmbH, Eschenstraße, Taufkirchen, Germany).
Amplifications were carried out with an initial denaturation at 94°C for 3 min followed by 45 cycles of 94°C for 20 s, 55°C for 30 s, and 72°C for 50 s. A final 5 min extension at 72°C was then performed. The procedures of PCR cycles were modified intentionally according to amplicon sizes (Abdurakhmonov et al., 2009). For determining PCR product sizes, 3.5% high-resolution agarose (Affymetrix, Inc., Cleveland, Ohio, United States) gel electrophoresis was carried out in 0.5 × TBE buffer. Gels were stained with ethidium bromide. In addition, the samples were run on ABI 3130XL Capillary electrophoresis (Life technologies, Carlsbad, CA, United States) to know the polymorphism between Recurrent and Donor parents, as well as F1 hybrid and ‘Ravnaq’ cultivars using fluorescent labelled BNL1604 (PET)—SSR marker (Figure 2). Microsatellite marker genotyping method (recipient genotype—a donor genotype—b and heterosis genotype—h) developed by Reddy et al. (2001) was implemented.
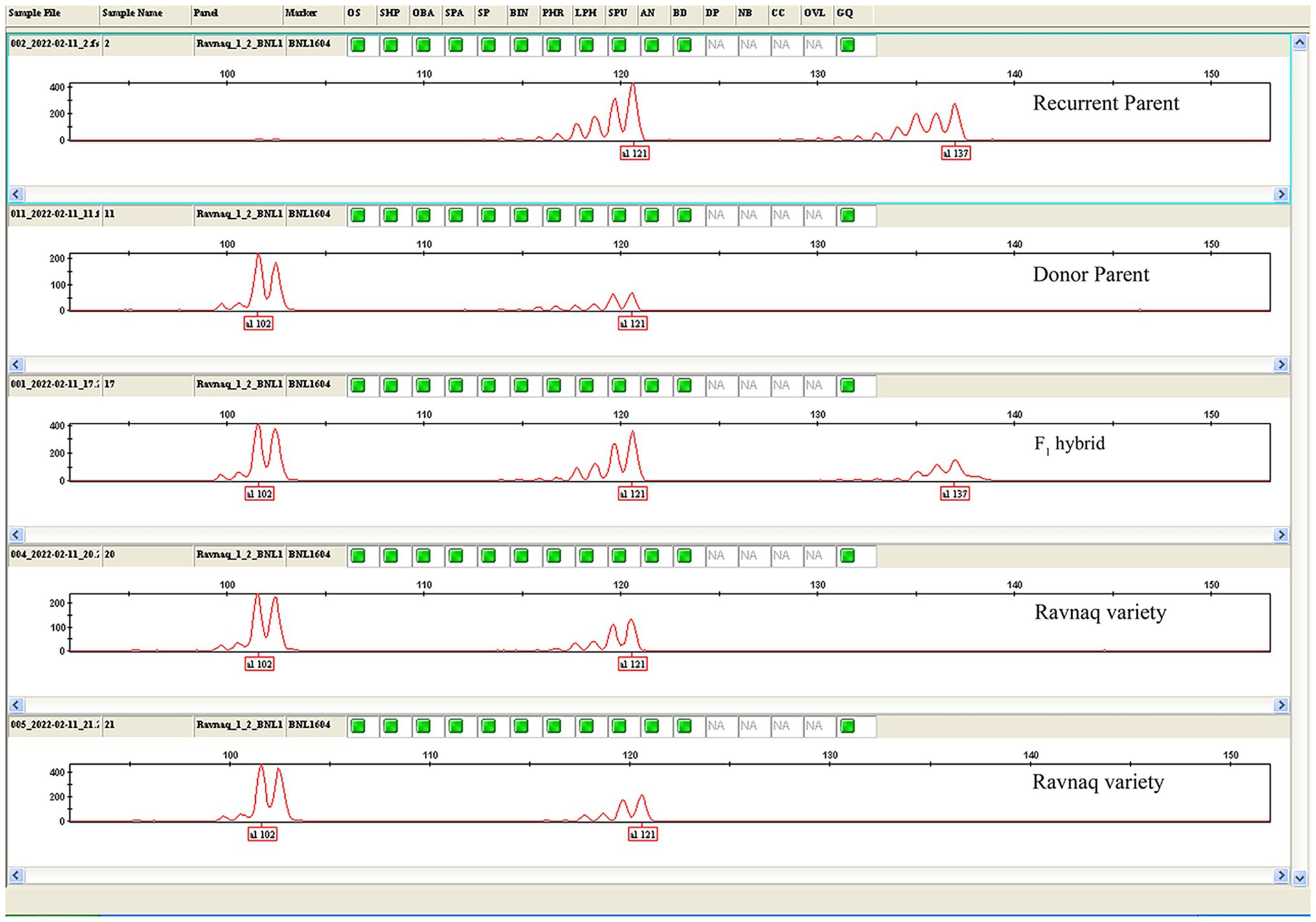
Figure 2. PCR amplicon separation for BNL1604 marker bands. ABI 3130XL Capillary electrophoresis results showing polymorphism between recurrent (with 121/137 amplicon) and donor [with 102/121 amplicon parents, F1 hybrid (102/121/137 amplicon)] and ‘Ravnaq’ cultivar (with 102/121 amplicon) for BNL1604–SSR marker PCR amplicons.
The variance and statistical analyses (Pearson’s Correlation, ANOVA, Two-Sample T-Test) were performed using the NCSS 2003 package software. The Kruskal–Wallis Multiple-Comparison Z-Value Test was performed to better identify the effects of the QTL allele introgressed by the MAS method. The HVI data obtained on fibre quality traits were analysed statistically according to the Pearson Correlation. The three-year data of fibre length and fibre strength characteristics obtained from each replication were analysed using the ANOVA and a comparative diagram was drawn in GraphPad Prism 8.01 (GraphPad Software, San Diego, CA, United States). Chi-square value (χ2) and broad sense heritability (H2) based on variations of FL and FS were analysed according to Petersen (1994) and Abdurakhmonov et al. (2005), respectively.
As highlighted above, our previous LD-based association mapping study (Abdurakhmonov et al., 2009) have revealed several important SSR markers, which were in pairwise linkage disequilibrium state on the homoeologous chromosomes 7 (At-genome) and 16 (Dt-genome) of the allotetraploid cotton genome. These markers were putatively linked to the important QTLs (Table 1) in various experimental mapping populations. The genetic position of SSR markers associated with, at least, four important fibre quality QTLs (FL, ST, FM and FU) was 8–8.3 cM in chromosome 7 or 0.7–11.9 cM in chromosome 16, which were in a range of the estimated genome-wide LD-decay (LD-block) for cotton (Abdurakhmonov et al., 2008, 2009). This suggested an opportunity to mobilize this LD-block of interest through MAS using SSR marker of choice(s).
Most of fibre QTL-associated SSRs in this LD-block have amplified polymorphic SSR marker bands between donor lines bearing QTL and local cultivars or their hybrids, which were not easily distinguishable (Figure 1) on the commonly used agarose gel electrophoresis. Therefore, we have selected a highly visible polymorphic BNL1604 marker, among others, for our MAS program as the marker of choice to mobilize the targeted LD-block of our interest. Here, it is important to mention that BNL1604 marker showed statistically significant pairwise LD (r2 > 0.1; p < 0.0001) with other fibre quality associated SSR markers in the targeted LD-block of our interest (Table 1). In parental genotypes used for MAS, the BNL1604 SSR primer pairs have amplified three different polymorphic bands of 137 bp, 121 bp, and 101/102 bp. Its 101/102 bp marker band was highly distinguishable polymorphic amplicon between donor (L-141 and L-N1 lines) and recipient (‘Andijan-35’ and ‘Mekhnat’) genotypes (Figures 1, 2).
BNL1604, with 25-times repeated AG dinucleotide motive (AG25) and chosen as a marker of choice to carry MAS in this study, was developed by Brookhaven National Laboratory (BNL)1 in 2000. This SSR locus was widely used for genetic mapping studies (Table 1; Supplementary Table 2) to identify important QTLs in many different cotton species. Marker bands of this SSR was mapped in homoeologous chromosomes 7 and 16 and revealed a putative genetic linkage with FS, FL (Tan et al., 2015; Fang et al., 2021), FM, and FU. In addition, its association with agronomic traits such as lint percentage (LP), fruit branches and wilt resistance (Zhang et al., 2015) were reported in the literature (Supplementary Table 2). Interestingly, in most of the studies, BNL1604 primer pairs amplified two loci with 100/101 bp and 115/116 bp lengths. In our study (Abdurakhmonov et al., 2009), we observed seven marker bands of 101, 116, 120, 132, 134, 140, and 142 bp in G. hirsutum cultivar germplasm accessions. It is worth mentioning that several independent studies have reported somewhat unconcordant results on the size of amplicon bands and their chromosomal locations of BNL1604. For example, Guo et al. (2007) and Zhao et al. (2012) have mapped BNL1604_115 to the chromosome 7, while Wu et al. (2009) observed only BNL1604_116 locus and have located it to the chromosome 16. BNL1604_100 was located to the chromosome 16 (Guo et al., 2007; He et al., 2008; Zhao et al., 2012) while Gutiérrez et al. (2009) located BNL1604_101 to the chromosome 7 (Table 1). Our in-silico PCR analysis, using reference allotetraploid cotton genome, has clearly revealed that BNL1604_102 marker is on chromosome A07 with the DNA position of 89,031,336–89,031,437 (102 bp; data not shown).
To mobilize the LD-block of our interest into the widely-grown Uzbek cotton cultivars, we crossed G. hirsutum donor genotypes L-141 and LN-1 possessing fibre quality QTLs on the chromosome 7/16, with local elite G. hirsutum cultivars ‘Andijan-35’ and ‘Mekhnat’ as recipients. F1 hybrids were backcrossed with their relative recipient genotypes (as recurrent parents) up to five generations to develop segregating populations on targeted QTLs. In each BC and segregating BC1-5F1 populations, segregation of BNL1604 genotype was within range of 1:1 ratio (χ2 ≥ 0.4, p ≥ 0.2; Table 2) of homozygous as recipient versus heterozygous as F1 hybrid genotypes. For example, in the BC5F1 [(‘Andijan-35’ × L-141) × ‘Andijan-35’] and BC5F1 [(‘Mekhnat’ × L-N1) × ‘Mekhnat’] hybrid populations with 188 and 164 plants, the heterozygous versus homozygous marker loci ratios was 96:92 and 78:86, respectively (Figure 3; Table 2). Only improved hybrids with superior fibre quality compared to original recipient parent were used for the next cycle of breeding based on end-of-season fibre quality measurements.
Further, to get the homozygous cultivar genotypes, bearing the targeted LD-block/QTLs of interest, heterozygous BC5F1 plants in each combination were self-pollinated, resulting in BC5F2 segregating population for BNL1604 marker loci. In the BC5F2 [(‘Andijan-35’ × L-141) × ‘Andijan-35’] and BC5F2 [(‘Mekhnat’ × L-N1) × ‘Mekhnat’] combinations, 235 and 191 sample DNAs were screened by PCR. The results indicated the ration of 58:116:61 (1:2:1; χ2 = 0.115, p < 0.944) segregation in the combination of BC5F2 [(‘Andijan-35’ × L-141) × ‘Andijan-35’]. In that, 58 plants were homozygous (a) like the recipient parent, 116 samples were heterozygous (h) like the F1 hybrid and 61 samples were homozygous (b) like the donor parent. Similarly, in BC5F2 [(‘Mekhnat’ × LN1) × ‘Mekhnat’] combination, we observed monogenic marker segregation ratio of 44:99:48 (1:2:1; χ2 = 0.424, p < 0.809; Figure 3; Table 2). Genotyping analysis confirmed the stable monogenic Mendelian inheritance of the SSR marker band/LD-block in subsequent generations (1:2:1 segregation ratio with non-significant chi-square indices Table 2).
We also calculated the broad sense heritability (H2) based on variations of two major fibre quality trait data (FL and FS) of parental, BC5F1 and BC5F2 population genotypes of from above-mentioned two hybridization experiment (Table 3). Results revealed that in the cross of ‘Andijan-35’ and ‘Mekhnat’ as maternal parents and L-141 and L-N1 as paternal parents, 70 to 74% trait expression for FS was explained by the genetic variation, while 26–30%- trait variation was environmental. The broad sense heritability for FL was 0.53–0.54 in above combinations of parental lines and their hybrids, demonstrating 54% trait variation observed is explained by genetic loci transferred, while 46% trait variation is explained by environmental factors. Generally, a heritability value <20% is considered low, and a value >50% is considered high (Stanfield, 1983). As expected, most mapped QTLs corresponded to these characteristics with better genetic determination or stable heritability (Said et al., 2015). This showed suitability of the LD-block of our interest, exploited for fibre quality trait improvement using MAS.
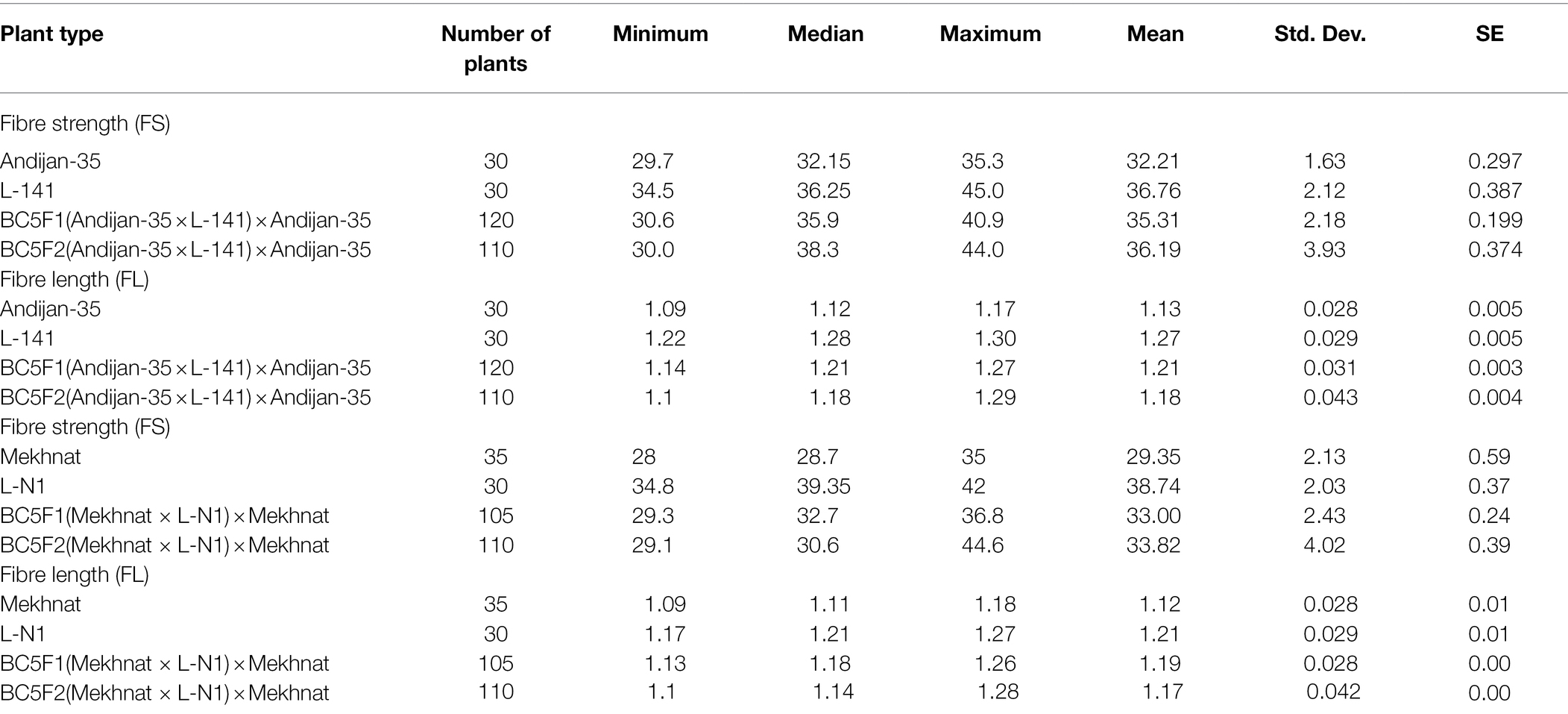
Table 3. Parameters for the fibre strength (FS) and fibre length (FL) traits in parental. BC5F1 and BC5F2 crosses.
Further, only homozygous samples for introgressed QTL alleles from the donor genotype were selected from the BC5F2 segregating population of above-described two independent hybridization experiment. Homozygous genotypes from each crossing combinations were bred in separate breeding nurseries, focusing on fibre strength, length and uniformity, as well as other agronomic traits until the BC5F5 generations. As a result, based on the early-season PCR screening of SSR marker locus and the end-season fibre quality parameters in each generation, individual homozygous plant genotypes bearing homozygous SSR marker of donor parent were selected for the next season field studies and consequent seed increasing. From final plant families, two highly homozygous novel lines of BC5F5 [(‘Andijan-35’ × L-141) × ‘Andijan-35’] and BC5F5 [(‘Mekhnat’ × LN1) × ‘Mekhnat’] were selected as the stable MAS-derived cultivars. These novel genotypes have been named as ‘Ravnaq-1’ and ‘Ravnaq-2’, respectively (Figures 4, 5; ‘Ravnaq’ translates as ‘flourishing or advancing’). These new cotton cultivars were then submitted to the State variety testing commission of the Republic of Uzbekistan in order to conduct field trials in different soil-climatic conditions, which are currently underway.
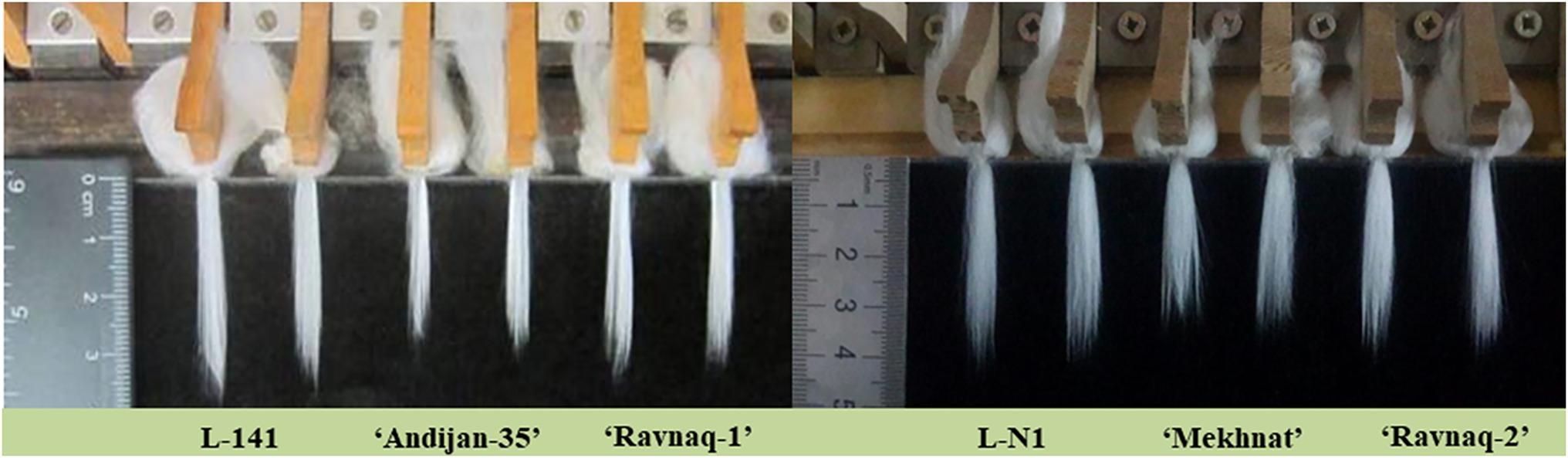
Figure 5. Fibre staple lengths (mm) of ‘Ravnaq-1’ and ‘Ravnaq-2’ cultivar compared to controls lines.
Our result showed that ‘Ravnaq-1’, developed by crossing of high-yielded but low fibre quality commercial Upland cultivar ‘Andijan-35’ (recipient) with donor G.hirsutun L-141 line bearing the superior fibre quality QTLs, have acquired new fibre quality QTLs located within the targeted LD-block of interest, which were successfully selected using single BNL1604 marker. The comparative fibre quality analysis of all genotypes in the same environmental growing conditions showed distinct fibre quality characteristics. The ‘Ravnaq-1’ have shown the mean fibre length—1.23 inches and strength—36.8 g/tex. These parameters were significantly (p < 0.0001) lower in the same environment-grown original recipient genotype ‘Andijan-35’, in which the mean fibre length was 1.13 inches and strength—32.7 g/tex. The donor genotype L-141 cotton line had the mean fibre length and strength of 1.25 inches and 40.2 g/tex in the same growing condition, respectively. The negative control ‘Namangan-77’ had the mean fibre length of 1.11inches and fibre strength of 32.0 g/tex. These suggested that in ‘Ravnaq-1’ cultivar development, molecular marker of our choice has effectively selected superior fibre quality loci of the donor. The transferred LD block/QTL region has significantly (p < 0.0001) improved FL by 8.8% and FS by 12.5% compared to its recurrent parent ‘Andijan-35’ (Table 4; Figure 6). Moreover, we observed significant (p < 0.0001) improvement on FM and FU, demonstrating superior quality of MAS cultivar compared to original recipient parent.
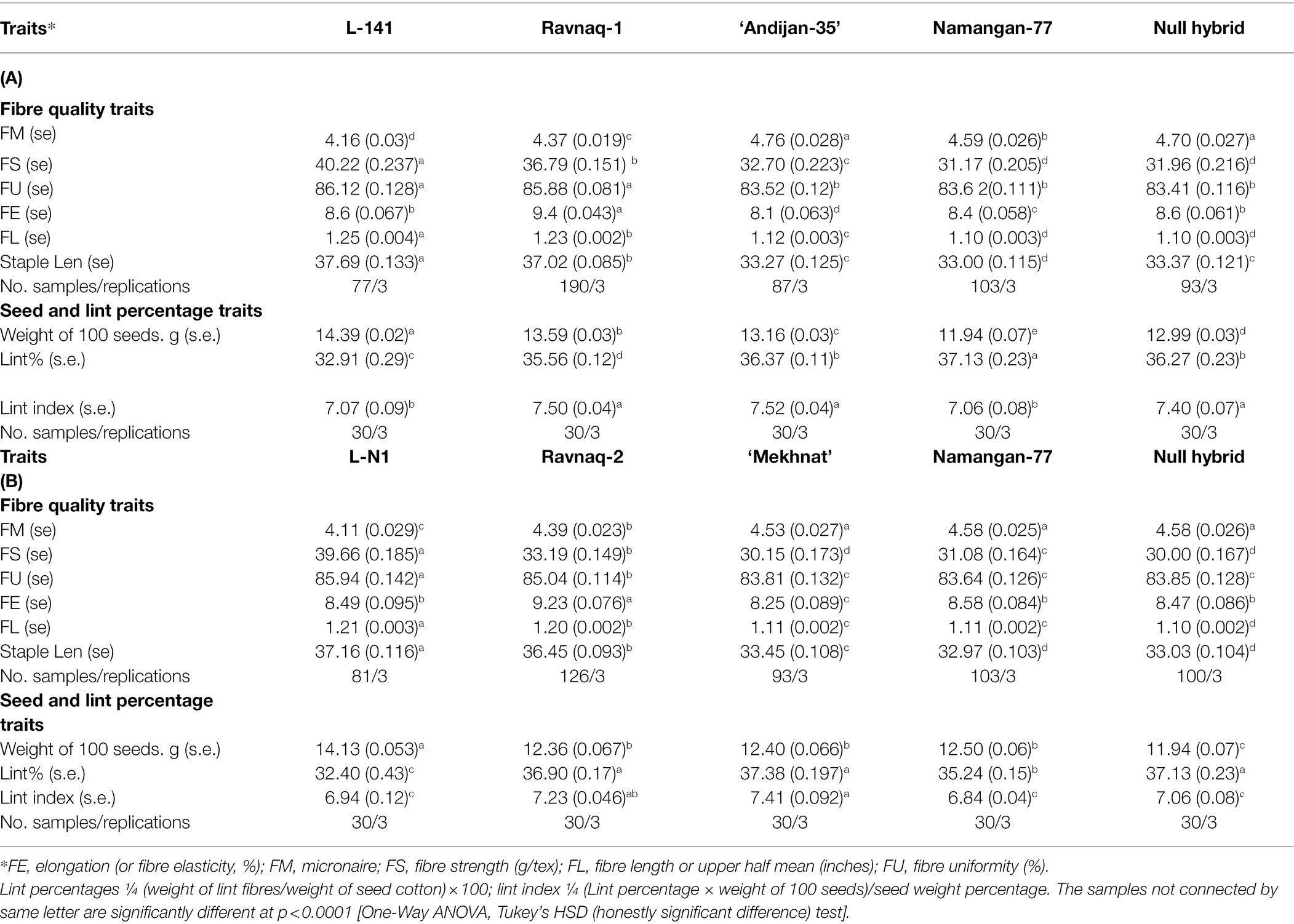
Table 4. Fibre quality and yield traits of ‘Ravnaq-1’ (A) and ‘Ravnaq-2’ (B) averaged from 2018 and 2020 growing environments.
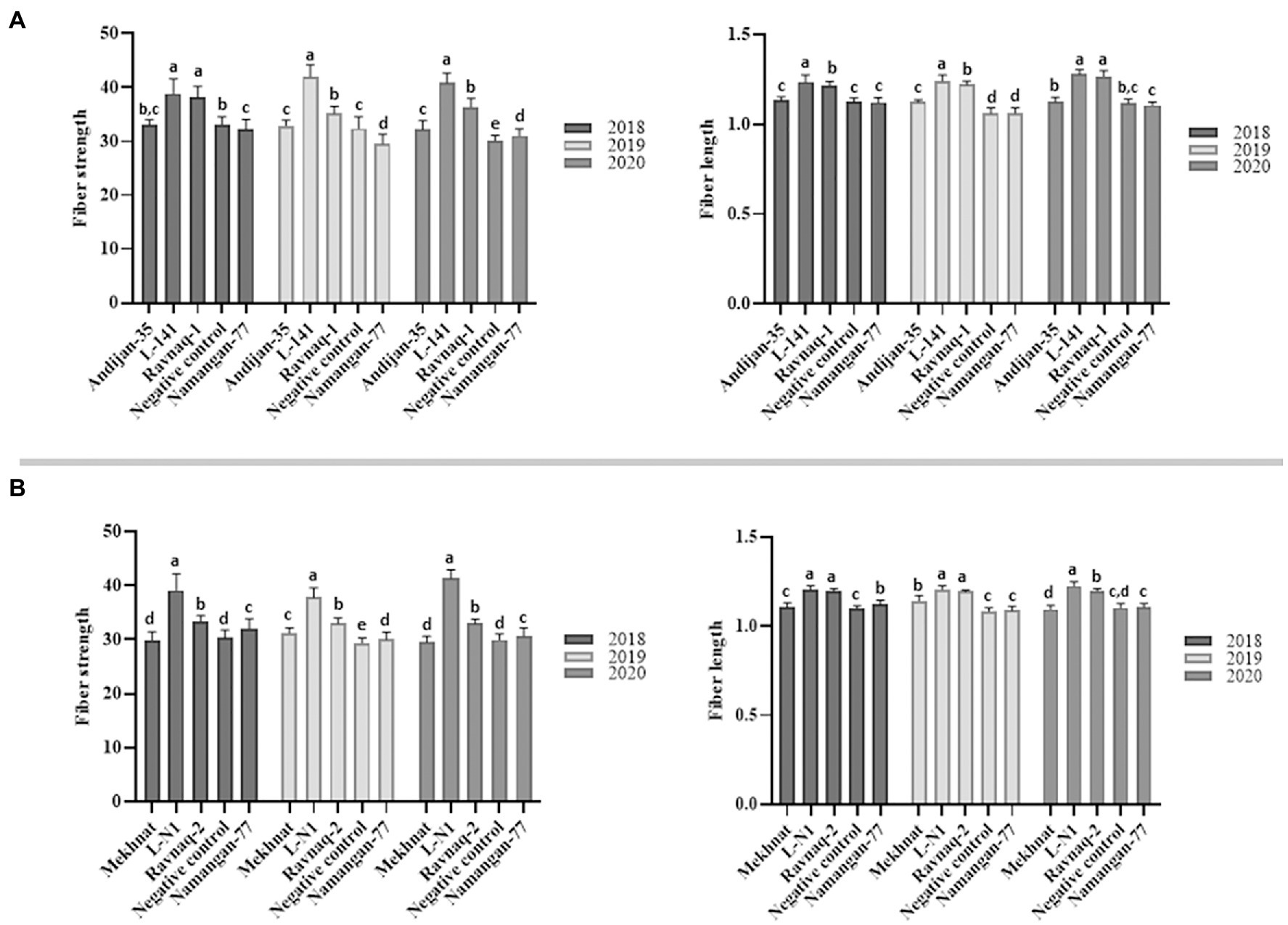
Figure 6. Comparative diagrams of fibre strength (FS) and length (FL) traits of ‘Ravnaq-1’ (A) and ‘Ravnaq-2’ (B) cultivars. The samples not connected by same letter are significantly different at p < 0.0001 [One-Way ANOVA, Tukey’s HSD (honestly significant difference) test].
We observed similar results of fibre quality improvement in the other crossing combination experiment, where MAS-derived ‘Ravnaq-2’ successfully received novel fibre QTLs from the donor G. hirsutum line L-N1 bearing the LD-block of our interest and expressing a superior fibre quality compared to the recipient Upland cultivar ‘Mekhnat’. The results of fibre quality analysis of ‘Ravnaq-2’ under the same environmental conditions with its donor and recipient genotypes have shown the mean fibre length of 1.20 inches and strength of 33.2 g/tex. These parameters were significantly lower in the recipient genotype ‘Mekhnat’, in which the mean fibre length was 1.11 inches and strength—30.2 g/tex in the same growing condition. The donor genotype L-N1 cotton line had the mean fibre length and strength of 1.21 inches and 39.7 g/tex, respectively. The negative control had the mean fibre length—1.10 inches, fibre strength - 30.0 g/tex (Table 4). The trait improvement (p < 0.0001) in MAS-derived ‘Ravnaq-2’ genotype, by mobilizing the LD-block of interest using BNL1604 SSR marker, was 8.1% for fibre length and 9.9% for fibre strength, showing the genetic power of the novel QTLs (Table 4). The other two important fibre quality traits such as FM (4.39, p < 0.0001) and FU (85.04, p < 0.0001) have also improved compared to recipient parent ‘Mekhnat (4.53 and 83.81).
Further, in both MAS cultivars, we observed no significant changes or subtle but statistically significant improvements on seed and lint percentage traits, mainly acquired from the recipient parent genomes (Table 4). These suggested that MAS cultivars having better fibre quality have also kept some of other key trait parameters of the original recipient cultivars, attributing to the yield and other agronomic performance.
To establish the effect of genetically related QTL allele on fibre length and strength traits in ‘Ravnaq-1’ cultivar., parent samples, hybrids without marker allele (negative control), and standard ‘Namangan-77’ fibre quality parameters were analysed by Kruskal–Wallis Multiple-Comparison Z-Value Test method. According to the results of the analysis, the characteristics of fibre length and strength in ‘Ravnaq-1’ cultivar were significantly different (α = 0.05) from the recipient - ‘Andijan-35’ cultivar., Negative control, and standard cultivar ‘Namangan-77’ (Table 5). Similarly, the Kruskal–Wallis multidimensional comparison analysis results showed that the fibre length and strength parameters of the ‘Ravnaq-2’ cultivar were significantly higher (α = 0.05) than the parent lines and control cultivar genotypes grown in the same environments and/or bread with similar approaches (Table 6). These results have suggested the genetic role of mobilized LD-block/QTLs from the donors in trait improvements of MAS cultivars. Results also demonstrated the feasibility of MAS in the tetraploid cotton genome with a total recombinational length of about 5,200 cM using single or few markers (Abdurakhmonov et al., 2008), provided selection of polymorphic DNA marker from an LD-block of interest.
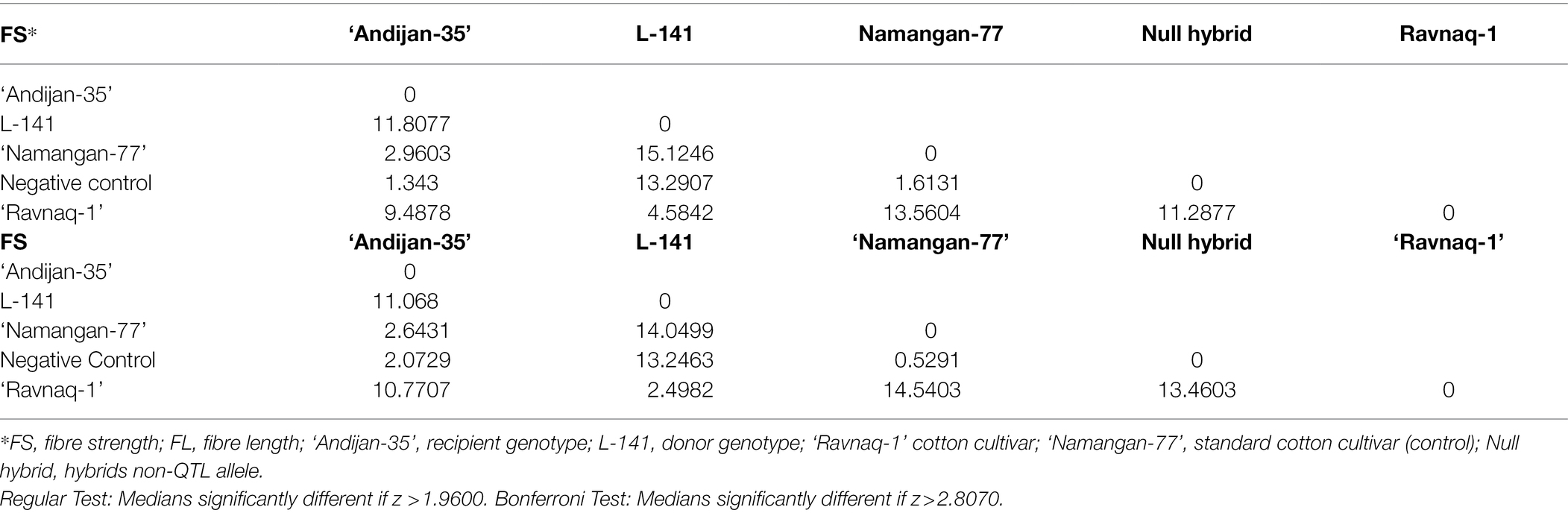
Table 5. Kruskal–Wallis Multiple-Comparison Z-Value Test method fibre length and strength in ‘Ravnaq-1’ cultivar.
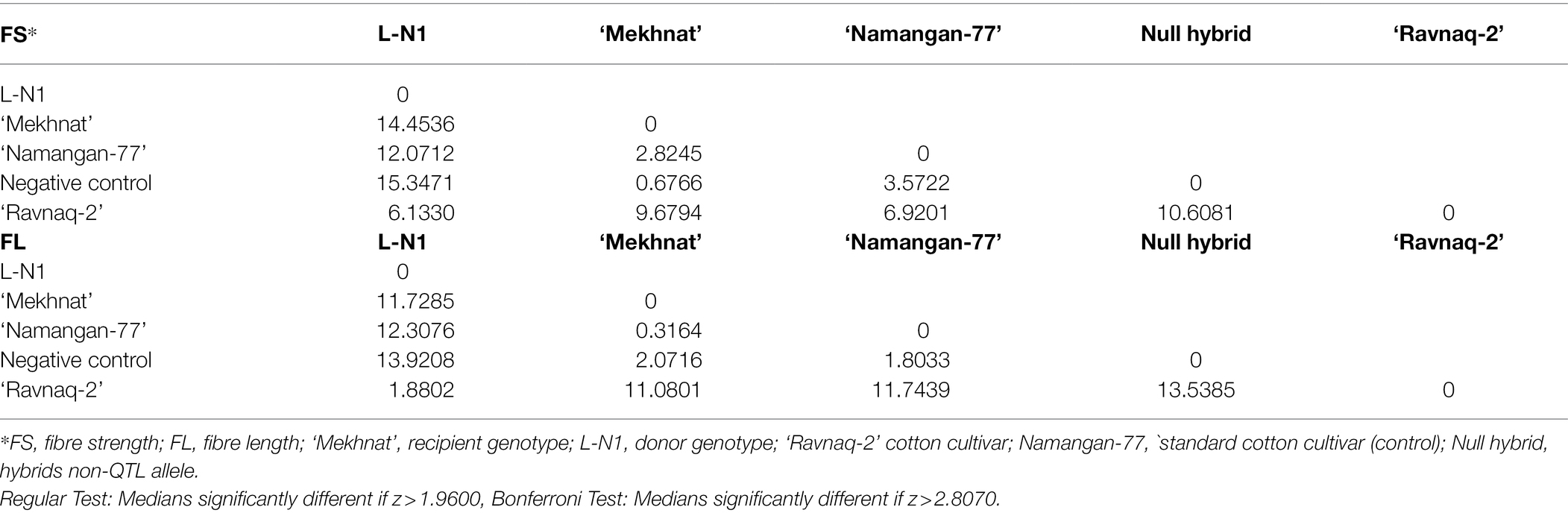
Table 6. Kruskal–Wallis Multiple-Comparison Z-Value Test method fibre length and strength in ‘Ravnaq-2’ cultivar.
Statistical analyses were performed using the Pearson method to study the genetic correlations among fibre quality parameters of MAS-derived genotypes of ‘Ravnaq-1’ and ‘Ravnaq-2’. In ‘Ravnaq-1’, a significant correlation (p < 0.001, r = 0.516) between the fibre strength and fibre length was obtained. Fibre strength (p < 0.001, r = 0.549) and fibre length (p < 0.001, r = 0.370) were also found to have a high and moderate positive correlation between fibre uniformity, and a weak positive or negative correlation between these two traits and other quality parameters were identified. Similar results were obtained by Wang X.Q. and others (2013) have shown a high positive correlation between fibre strength, fibre length (p < 0.01, r = 0.69**) and fibre uniformity (p < 0.01, r = 0.62**).
A low-value negative correlation was observed between FM and FL (p < 0.001, r = −0.248), FS (p < 0.001, r = −0.159), and FU (p < 0.001, r = −0.183). Genetic analysis of fibre quality parameters showed a significant high and moderate negative correlation between FM and FL (p < 0.001, −0.850) and FS (p < 0.029, −0.499) in the study of Yaqoob et al. (2016). According to the results of the Pearson correlation analysis, no opposite genetic correlation was observed between all fibre quality traits in the ‘Ravnaq-2’ cultivar at ≤0.05. For example, weak positive correlations were observed between FL and FS, FE and FU. In many other studies, a significant positive correlation between FL and FS and FE, as well as a negative correlation with FM were observed (Patil et al., 2014; Shang et al., 2016). A significant positive correlation was also observed between FS and FE, while a negative correlation was observed with FM. A similar positive correlation between FL and FS were observed in our previous exotic cotton germplasm studies (Abdurakhmonov et al., 2008). All these showed that fibre strength, elongation and uniformity become higher when fibre length is improved. Thus, it can be concluded that the correct correlation between the key fibre QTLs in the MAS-derived cultivars, described herein, is an additive effect of the QTLs and/or a joint effect of genes transferred from the donor genotypes to the cultivars. The positive correlation between lint percentage and FL in our MAS-derived cultivars can also be explained by the fact that the correct breeding approach was carried out in each backcrossed hybrid on FL and lint percentage parameters.
The ‘Ravnaq-1’ and ‘Ravnaq-2’ cotton cultivars were first planted on 26 and 30 hectares of special seed increase farms in the Namangan region. In the cultivation of these cultivars, all agrotechnology measures were taken in a timely manner, which allowed farmers to get the seed cotton yield of 4.1 tons per hectare. In order to conduct field trails on larger areas in 2018, the ‘Ravnaq-1’ cultivar was planted on 500 hectares in the Namangan region, which allowed to obtain an average of more than 4.0 tons per hectare of seed cotton yield in the region. In 2019, the ‘Ravnaq-1’ cotton cultivar was planted in large areas in the Tashkent region, with an average yield of more than 3.6 tons per hectare of seed cotton yield in the region. Moreover, ‘Ravnaq-1’ has been planted in Surkhandaryo and Syrdaryo regions since 2021, and ‘Ravnaq-2’ cotton has been planted on farms and agriculture clusters in the Republic of Karakalpakstan, which is a relatively northern region of Uzbekistan. We observed high yield (3.8 tons per hectare of seed cotton yield) with high fibre quality in Karakalpakstan environment (data not shown). The seed cotton yield of MAS cultivars grown in the farmers field condition was competitive to those widely commercialized Upland cotton cultivars, including the recipient cultivars (average seed cotton yield of Uzbekistan cultivars is in range of 2.6 to 3.6 tons per hectare; data not shown). Some important characteristics of ‘Ravnaq’ cultivars are sown in Supplementary Figures 1, 2. These results demonstrated that novel MAS cultivars, in addition to newly acquired superior fibre quality traits, have competitive agronomic properties for sustainable farming in Uzbekistan.
Our research on the use of the first DNA-based MAS in Uzbekistan to improve one or more fibre quality traits in Upland cotton cultivars has proven to be a successful practice that led to the improvement of important fibre quality traits in widely-grown local Upland cultivars. Molecular marker BNL1604, chosen from the specific LD block associated with fibre QTLs along with donor genotypes, identified in our previous associative mapping studies using Upland cotton germplasm, were practically useful for our MAS program. We showed here that the mobilization of novel QTLs using SSR markers is effective to improve the key fibre quality traits such as FL, FS, FU, and FM. New cultivars, ‘Ravnaq-1/Ravnaq-2’, registered in the State Variety Testing Commission of Uzbekistan in 2014–2017 are the first generation of MAS-derived cotton cultivars in Uzbekistan. Our results highlight that LD-block of chromosome 7 with its mapped molecular marker(s) and donor genotypes, used herein, have efficiently helped to precisely and rapidly transfer superior fibre quality QTLs to the commercially grown Upland cotton cultivars, exemplifying the potential of MAS in cotton breeding.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
IA, MD, AM, MA, and ZB conceived and designed the study. MD, AM, KU, JN, MK, SN, and IN carried out field experiments. MD, AM, NK, SS, ZB, and IS performed data analyses. MA, AM, and IA drafted the manuscript. All authors contributed to manuscript revisions and approved the submitted version.
This research was funded by the Ministry of Innovative Development of Uzbekistan providing the research project (A6-T029, FA-F5-T030, IFA-2017-5-6, A-FA-2021-474).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge the Center of Genomics and Bioinformatics research team for helping with molecular and field experiments and data collection. Also, thanks to Akmal M. Asrorov for his help in manuscript preparation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.906472/full#supplementary-material
Supplementary Figure 1 | Some morphological phenotypes of MAS-derived ‘Ravnaq-1’ cultivar.
Supplementary Figure 2 | Some morphological phenotypes of MAS-derived ‘Ravnaq-2’ cultivar.
QTL, Quantitative trait loci; MAS, Marker-assisted selection; SSR, Simple sequence repeats; NIL, Near isogenic lines; BC, Back crossing; FS, Fibre strength; FL, Fibre length; FE, Fibre elongation; FM, Fibre micronaire; FU, Fibre uniformity; ‘Ravnaq’, Translates in English as flourishing or advancing.
Abdukarimov, A., Djataev, S., and Abdurakhmonov, I. Y. (2003). “Cotton research in Uzbekistan: Elite varieties and future of cotton breeding.” in Proceedings of World Cotton Research Conference ; March 9, 2003.
Abdurakhmonov, I. Y. (2002). Molecular cloning of new DNA-markers for marker-assisted selection of cotton. Dissertation’ Thesis. Tashkent, Uzbekistan: Institute of Genetics and plant experimental biology, academy of Uzbek sciences.
Abdurakhmonov, I. Y., Abdullaev, A. A., Saha, S., Buriev, Z. T., Arslanov, D., Kuryazov, Z., et al. (2005). Simple sequence repeat marker associated with a natural leaf defoliation trait in Tetraploid cotton. J. Hered. 96, 644–653. doi: 10.1093/jhered/esi097
Abdurakhmonov, I. Y., Buriev, Z. T., Shermatov, S. E., Kushanov, F. N., Makamov, A., Shopulatov, U., et al. (2011). “Utilization of natural diversity in Upland cotton (G. hirsutum) germplasm collection for pyramiding genes via marker-assisted selection program.” in Proceedings of 5th Meeting of Asian Cotton Research and Development Network ; February 23–25, 2011.
Abdurakhmonov, I. Y., Kohel, R. J., Yu, J. Z., Pepper, A. E., Abdullaev, A. A., Kushanov, F. N., et al. (2008). Molecular diversity and association mapping of fibre quality traits in exotic G. hirsutum L. germplasm. Genomics 98, 478–487. doi: 10.1016/j.ygeno.2008.07.013
Abdurakhmonov, I. Y., Saha, S., Jenkins, J. N., Buriev, Z. T., Shermatov, S. E., Scheffler, B. E., et al. (2009). Linkage disequilibrium-based association mapping of fibre quality traits in G. hirsutum L. variety germplasm. Genetica 136, 401–417. doi: 10.1007/s10709-008-9337-8
Amiteye,. (2021). Basic concepts and methodologies of DNA marker systems in plant molecular breeding. Heliyon 7:e08093. doi: 10.1016/j.heliyon.2021.e08093
Cai, C., Ye, W., Zhang, T., and Guo, W. (2014). Association analysis of fibre quality traits and exploration of elite alleles in upland cotton cultivars/accessions (Gossypium hirsutum L.). J. Integr. Plant Biol. 56, 51–62. doi: 10.1111/jipb.12124
Collard, B. C. Y., Jahufer, M. Z. Z., Brouwer, J. B., and Pang, E. C. K. (2005). An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142, 169–196. doi: 10.1007/s10681-005-1681-5
Collard, B. C., and Mackill, D. J. (2008). Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 363, 557–572. doi: 10.1098/rstb.2007.2170
Dellaporta, S. L., Wood, J., and Hicks, J. P. (1983). A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1, 19–21. doi: 10.1007/BF02712670
Fang, D. D. (ed.) (2018). “Cotton fibre genes and stable quantitative trait loci,” in Cotton Fibre: Physics, Chemistry and Biology. Cham, Switzerland: Springer-Nature Publishing Group, 151–178.
Fang, D. D., Jenkins, J. N., Deng, D. D., McCarty, J. C., Li, P., and Wu, J. (2014). Quantitative trait loci analysis of Fier quality traits using a random-mated recombinant inbred population in upland cotton (Gossypium hirsutum L.). BMC Genomics 15:397. doi: 10.1186/1471-2164-15-397
Fang, D. D., Zeng, L., Thyssen, G. N., Delhom, C. D., Bechere, E., Jones, D. C., et al. (2021). Stability and transferability assessment of the cotton fibre strength QTL qFS-c7-1 on chromosome A07. Crop J. 9, 380–386. doi: 10.1016/j.cj.2020.06.016
Guo, W., Cai, C., Wang, C., Han, Z., Song, X., Wang, K., et al. (2007). A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium. Genetics 176, 527–541. doi: 10.1534/genetics.107.070375
Gutiérrez, O. A., Stelly, D. M., Saha, S., Jenkins, J. N., McCarty, J. M., Raska, D. A., et al. (2009). Integrative placement and orientation of non-redundant SSR loci in cotton linkage groups by deficiency analysis. Mol. Breeding 23, 693–707. doi: 10.1007/s11032-009-9266-y
He, Y., Guo, W., Shen, X., and Zhang, T. (2008). Molecular cloning and characterization of a cytosolic glutamine synthetase gene, a fibre strength-associated gene in cotton. Planta 228, 473–483. doi: 10.1007/s00425-008-0751-z
He, D. H., Lin, Z. X., Zhang, X. L., Nie, Y. C., Guo, X. P., Zhang, Y. X., et al. (2007). QTL mapping for economic traits based on a dense genetic map of cotton with PCR-based markers using the interspecific cross of Gossypium hirsutum × Gossypium barbadense. Euphytica 153, 181–197. doi: 10.1007/s10681-006-9254-9
Hugie, K. L., Fang, D. D., Smith, C. W., Li, P., Hinze, L. L., Hague, S. S., et al. (2016). Utility assessment of published microsatellite markers for fibre length and bundle strength QTL in a cotton breeding program. Crop Sci. 56, 2983–2995. doi: 10.2135/cropsci2016.02.0093
Ijaz, B., Zhao, N., Kong, J., and Hua, J. (2019). Fibre quality improvement in upland cotton (Gossypium hirsutum L.): quantitative trait loci mapping and marker assisted selection application. Front. Plant Sci. 10:1585. doi: 10.3389/fpls.2019.01585
Kushanov, F. N., Turaev, O. S., Ernazarova, D. K., Gapparov, B. M., Oripova, B. B., Kudratova, M. K., et al. (2021). Genetic diversity, QTL mapping, and marker assisted selection Technology in Cotton (Gossypium spp.). Front. Plant Sci. 12:779386. doi: 10.3389/fpls.2021.779386
Lewis, H. (1962). Catastrophic selection as a factor in speciation. Evolution 16, 257–271. doi: 10.1111/j.1558-5646.1962.tb03218.x
Li, C., Ai, N., Zhu, Y., Wang, Y., Chen, X., Li, F., et al. (2016). Association mapping and favourable allele exploration for plant architecture traits in upland cotton (Gossypium hirsutum L.) accessions. J. Agri. Sci. 154, 567–583. doi: 10.1017/S0021859615000428
Liang, Q., Hu, C., Hua, H., Li, Z., and Hua, J. (2013). Construction of a linkage map and QTL mapping for fibre quality traits in upland cotton (Gossypium hirsutum L.). Chin. Sci. Bull. 58, 3233–3243. doi: 10.1007/s11434-013-5807-1
Ma, L., Wang, Y., Ijaz, B., and Hua, J. (2019). Cumulative and different genetic effects contributed to yield heterosis using maternal and paternal testcross populations in upland cotton. Sci. Rep. 9:3984. doi: 10.1038/s41598-019-40611-9
Mei, H., Zhu, X., Guo, W., Cai, C., and Zhang, T. (2014). Exploitation of Chinese Upland Cotton Cultivar Germplasm Resources to Mine Favorable QTL Alleles Using Association Mapping. In I. Y. Abdurakhmonov, Ed., World Cotton Germplasm Resources. Intech, Rijeka, 55–85. doi: 10.5772/58587
Migicovsky, Z., and Myles, S. (2017). Exploiting wild relatives for genomics-assisted breeding of perennial crops. Front. Plant Sci. 8:460. doi: 10.3389/fpls.2017.00460
Pan, Y., Meng, F., and Wang, X. (2020). Sequencing multiple cotton genomes reveals complex structures and lays Foundation for Breeding. Front. Plant Sci. 11:560096. doi: 10.3389/fpls.2020.560096
Patil, M., Khadi, B. M., Basamma, K., and Katageri, I. S. (2014). Genetic variability and correlation analysis for fibre quality traits in diploid cotton (Gossypium spp.) American-Eurasian. J. Agric. Environ. Sci. 14, 392–395. doi: 10.5829/idosi.aejaes.2014.14.05.12322
Paudel, D., Dhakal, S., Parajuli, S., Adhikari, L., Peng, Z., Qian, Y., et al. (2020). “Use of quantitative trait loci to develop stress tolerance in plants,” in Plant Life Under Changing Environment. eds. D. K. Tripathi, V. P. Singh, D. K. Chauhan, and S. Sharma (United States: Academic Press), 917–965. doi: 10.5829/idosi.aejaes.2014.14.05.12322
Reddy, O. U. K., Pepper, A. E., Abdurakhmonov, I. Y., Saha, S., Jenkins, J. N., Brooks, T., et al. (2001). The identification of dinucleotide and trinucleotide microsatellite repeats loci from cotton G. hirsutum L. J. Cotton Sci. 5, 103–113.
Said, J. I., Song, M., Wang, H., Lin, Z., Zhang, X., Fang, D., et al. (2015). A comparative meta-analysis of QTL between intraspecifiс Gossypium hirsutum and interspecifi G. hirsutum × G. barbadense populations. Mol. Gen. Genomics. 290, 1003–1025. doi: 10.1007/s00438-014-0963-9
Shang, L., Ma, L., Wang, Y., Su, Y., Wang, X., Li, Y., et al. (2016). Main effect QTL with dominance determines Heterosis for dynamic plant height in upland cotton. G3 6, 3373–3379. doi: 10.1534/g3.116.034355
Shao, Q., Zhang, F., Tang, S., Liu, Y., Fang, X., Liu, D., et al. (2014). Identifying QTL for Fier quality traits with three upland cotton (Gossypium hirsutum L.) populations. Euphytica 198, 43–58. doi: 10.1007/s10681-014-1082-8
Shen, X., Guo, W., Zhu, X., Yuan, Y., Yu, J. Z., Kohel, R. J., et al. (2005). Molecular mapping of QTLs for fiber qualities in three diverse lines in Upland cotton using SSR markers. Mol. Breed. 15, 169–181. doi: 10.1007/s11032-004-4731-0
Stanfield, W. (1983). Theory and Problems of Genetics 2nd Schain’s Outline Series. New Delhi: Mc Grow Thill Book Co.
Sun, F. D., Zhang, J. H., Wang, S. F., Gong, W. K., Shi, Y. Z., and Liu, A. Y. (2012). QTL mapping for fibre quality traits across multiple generations and environments in upland cotton. Mol. Breeding 30, 569–582. doi: 10.1007/s11032-011-9645-z
Tan, Z., Fang, X., Tang, S., Zhang, J., Liu, D., Teng, Z., et al. (2015). Genetic map and QTL controlling fibre quality traits in upland cotton (Gossypium hirsutum L.). Euphytica 203, 615–628. doi: 10.1007/s10681-014-1288-9
Tang, S., Teng, Z., Zhai, T., Fang, X., Liu, F., Liu, D., et al. (2015). Construction of genetic map and QTL analysis of fiber quality traits for Upland cotton (Gossypium hirsutum L.). Euphytica 201, 195–213. doi: 10.1007/s10681-014-1189-y
Tanksley, S. D., Young, N. D., Paterson, A. H., and Bonierbale, M. W. (1989). RFLP mapping in plant breeding: new tools for an old science. Biotechnology 7, 257–264.
Ulloa, M., Hulse-Kemp, A. M., De Santiago, L. M., Stelly, D. M., and Burke, J. J. (2017). Insights into upland cotton (Gossypium hirsutum L.) genetic recombination based on 3 high-density single-nucleotide polymorphism and a consensus map developed independently With common parents. Genom. Insights. 10:1178631017735104. doi: 10.1177/1178631017735104
Wang, F., Gong, Y., Zhang, C., Liu, G., Wang, L., Xu, Z., et al. (2011). Genetic effects of introgression genomic components from Sea Island cotton (Gossypium barbadense L.) on fibre related traits in upland cotton (G. hirsutum L.). Euphytica 181, 41–53. doi: 10.1007/s10681-011-0378-1
Wu, J., Gutierrez, O. A., Jenkins, J. N., McCarty, J. C., and Zhu, J. (2009). Quantitative analysis and QTL mapping for agronomic and fibre traits in an RI population of upland cotton. Euphytica 165, 231–245. doi: 10.1007/s10681-008-9748-8
Yang, X., Wang, Y., Zhang, G., Wang, X., Wu, L., Ke, H., et al. (2016). Detection and validation of one stable fibre strength QTL on c9 in tetraploid cotton. Mol. Genet. Genomics 291, 1625–1638. doi: 10.1007/s00438-016-1206-z
Yaqoob, M., Fiaz, S., and Ijaz, B. (2016). Correlation analysis for yield and fibre quality traits in upland cotton. Commun. Plant Sci. 6, 55–60.
Yu, J., Zhang, K., Li, S., Yu, S., Zhai, H., Wu, M., et al. (2013). Mapping quantitative trait loci for lint yield and fibre quality across environments in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population. Theor. Appl. Genet. 126, 275–287. doi: 10.1007/s00122-012-1980-x
Zeng, L., Meredith, W. R., and Gutiérrez, O. A. (2009). Identification of associations between SSR markers and fiber traits in an exotic germplasm derived from multiple crosses among Gossypium tetraploid species. Theor. Appl. Genet. 119, 93–103. doi: 10.1007/s00122-009-1020-7
Zhang, J., Yu, J., Pei, W., Li, X., Said, J., Song, M., et al. (2015). Genetic analysis of Verticillium wilt resistance in a backcross inbred line population and a meta-analysis of quantitative trait loci for disease resistance in cotton. BMC Genomics 16: 577. doi: 10.1186/s12864-015-1682-2
Zhang, K., Zhang, J., Ma, J., Tang, S., Liu, D., Teng, Z., et al. (2012). Genetic mapping and quantitative trait locus analysis of fibre quality traits using a three-parent composite population in upland cotton (Gossypium hirsutum L.). Mol. Breeding. 29, 335–348. doi: 10.1007/s11032-011-9549-y
Zhao, L., Lv, Y., Cai, C., Tong, X., Chen, X., Zhang, W., et al. (2012). Toward allotetraploid cotton genome assembly: integration of a high-density molecular genetic linkage map with DNA sequence information. BMC Genomics 13:539. doi: 10.1186/1471-2164-13-539
Keywords: QTL, SSR markers, traits, fibre, Gossypium hirsutum, marker-assisted selection
Citation: Darmanov MM, Makamov AK, Ayubov MS, Khusenov NN, Buriev ZT, Shermatov SE, Salakhutdinov IB, Ubaydullaeva KA, Norbekov JK, Kholmuradova MM, Narmatov SE, Normamatov IS and Abdurakhmonov IY (2022) Development of Superior Fibre Quality Upland Cotton Cultivar Series ‘Ravnaq’ Using Marker-Assisted Selection. Front. Plant Sci. 13:906472. doi: 10.3389/fpls.2022.906472
Received: 28 March 2022; Accepted: 03 May 2022;
Published: 24 May 2022.
Edited by:
Linghe Zeng, United States Department of Agriculture (USDA), United StatesReviewed by:
Jinfa Zhang, New Mexico State University, United StatesCopyright © 2022 Darmanov, Makamov, Ayubov, Khusenov, Buriev, Shermatov, Salakhutdinov, Ubaydullaeva, Norbekov, Kholmuradova, Narmatov, Normamatov and Abdurakhmonov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ibrokhim Y. Abdurakhmonov, aS55LmFiZHVyYWtobW9ub3ZAZ21haWwuY29t; Mirzakamol S. Ayubov, bWlyem8uYXl1Ym92QGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.