- 1Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding/Key Laboratory of Plant Functional Genomics of the Ministry of Education/Jiangsu Key Laboratory of Crop Genetics and Physiology/Jiangsu Co-Innovation Center for Modern Production Technology of Grain Crops, College of Agriculture, Yangzhou University, Yangzhou, China
- 2National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4Innovation Academy for Seed Design, Chinese Academy of Sciences, Beijing, China
Grain size and the endosperm starch content determine grain yield and quality in rice. Although these yield components have been intensively studied, their regulatory mechanisms are still largely unknown. In this study, we show that loss-of-function of OsNAC129, a member of the NAC transcription factor gene family that has its highest expression in the immature seed, greatly increased grain length, grain weight, apparent amylose content (AAC), and plant height. Overexpression of OsNAC129 had the opposite effect, significantly decreasing grain width, grain weight, AAC, and plant height. Cytological observation of the outer epidermal cells of the lemma using a scanning electron microscope (SEM) revealed that increased grain length in the osnac129 mutant was due to increased cell length compared with wild-type (WT) plants. The expression of OsPGL1 and OsPGL2, two positive grain-size regulators that control cell elongation, was consistently upregulated in osnac129 mutant plants but downregulated in OsNAC129 overexpression plants. Furthermore, we also found that several starch synthase-encoding genes, including OsGBSSI, were upregulated in the osnac129 mutant and downregulated in the overexpression plants compared with WT plants, implying a negative regulatory role for OsNAC129 both in grain size and starch biosynthesis. Additionally, we found that the expression of OsNAC129 was induced exclusively by abscisic acid (ABA) in seedlings, but OsNAC129-overexpressing plants displayed reduced sensitivity to exogenous brassinolide (BR). Therefore, the results of our study demonstrate that OsNAC129 negatively regulates seed development and plant growth, and further suggest that OsNAC129 participates in the BR signaling pathway.
Introduction
Rice is one of the most important cereal crops, providing a staple food for more than half of the world’s population. Thus, grain yield and grain quality are the most two critical issues in rice production. Grain yield is generally determined by tiller number (also referred as panicle number per plant), grain number per panicle, and grain size. Grain size, in turn, is determined by a combination of grain length, grain width, and grain thickness. Grain size is a complex quantitative trait controlled by multiple genes. To date, at least 280 grain size-related genes (TO:0000397) have been reported and accessed in the China Rice Data Center.1 These cloned genes have provided considerable insight into the individual molecular basis of grain size and shape regulation. Based on the results of previous studies, grain size is regulated by multiple pathways including the G-protein signaling pathway, the ubiquitin-proteasome pathway, the IKU pathway, the MAPK signaling pathway, the phytohormone signaling pathway, and the transcriptional regulatory pathway (Zuo and Li, 2014; Li and Li, 2016; Li et al., 2019). In any case, these pathways ultimately control seed size by influencing cell proliferation or/and cell expansion in maternal tissues or endosperm growth.
Among these pathways, the brassinosteroid (BR) pathway and transcriptional regulatory pathway are the most widely studied and well-characterized pathways. BRs are a class of polyhydroxysteroid phytohormones that are essential for the proper regulation of multiple physiological processes during plant growth and development (Brosa, 1999; Müssig, 2005; Gudesblat and Russinova, 2011; Ackerman-Lavert and Savaldi-Goldstein, 2020). Many studies have shown that BRs also participate in grain size regulation. For example, the BR-deficient mutants dwarf11 (d11) and dwarf2 (d2) and the BR-insensitive mutants dwarf61 (d61/bri1) and 08sg2 (bak1) all produce small and short grains, whereas overexpression of OsBZR1 increases grain length, grain width, and grain weight (Hong et al., 2005; Tanabe et al., 2005; Morinaka et al., 2006; Zhu et al., 2015; Wu et al., 2016; Yuan et al., 2017). In addition to these direct BR components, other indirect components such as GL3.1, SLG, SG1, GSE5, BU1, GS5, and DLT all participate in the regulation of grain size through the BR signaling pathway (Tanaka et al., 2009; Li et al., 2011; Hu et al., 2012; Nakagawa et al., 2012; Qi et al., 2012; Tong et al., 2012; Zhang et al., 2012; Feng et al., 2016; Duan et al., 2017; Liu et al., 2017; Gao et al., 2019). Transcriptional regulation of grain size is another important pathway because many of the identified grain-size-related genes encode transcription factors. For example, it has been reported that GLW7 (OsSPL13) and GW8 (OsSPL16), genes that encode the plant-specific transcription factor (TF) SQUAMOSA PROMOTER BINDING PROTEIN-LIKE, positively regulate grain length and yield by promoting cell expansion and cell division in the grain hull, respectively (Wang et al., 2012a; Si et al., 2016). Elevated expression of GL7 (GW7/SLG7), a gene that encodes a TON1 RECRUITING MOTIF (TRM)-containing protein, leads to slender grains, but further studies are needed to clarify whether GL7 (GW7/SLG7) controls grain size through cell proliferation or cell expansion (Wang et al., 2015a,b; Zhou et al., 2015). GS2 encodes a plant-specific transcription factor called GROWTH-REGULATING FACTOR 4 (OsGRF4), which regulates grain size mainly by increasing cell expansion and slightly promoting cell proliferation in the spikelet hull (Che et al., 2015; Duan et al., 2015; Hu et al., 2015; Li et al., 2016; Sun et al., 2016).
Additionally, these pathways do not always act in isolation, but interact with one another to shape and fine-tune grain size. For example, two atypical bHLH domain transcription factors without DNA-binding activity are encoded by OsPGL1 and OsPGL2/OsBUL1 and function as inhibitors of a typical DNA-binding bHLH TF OsAPG through heterodimerization (Heang and Sassa, 2012a,b; Jang et al., 2017; Jang and Li, 2017). Overexpression of both of these genes leads to increased grain length caused by increased cell length. Further study revealed that OsPGL1 and OsPGL2/OsBUL1 are BR-related genes, even though OsPGL1 is not BR-inducible (Heang and Sassa, 2012a,b; Jang et al., 2017; Jang and Li, 2017). Thus, OsPGL1 and OsPGL2/OsBUL1 function in the transcriptional regulation and BR pathways. Recently, it was reported that the MAPK signaling pathway component OsMKK4 and its substrate OsMAPK6 in the OsMKKK10-OsMKK4-OsMPK6 cascade positively regulate grain size by promoting cell division of the spikelet hulls (Guo et al., 2018; Xu et al., 2018). Moreover, both of these proteins affect BR responses and the expression of BR-related genes, indicating that there is crosstalk between the BR and MAPK pathways.
Storage starch, a mixture of two glucose polymers called amylose and amylopectin, accounts for 90% of the endosperm dry weight (Keeling and Myers, 2010; Zeeman et al., 2010), and thus has a very large effect on grain yield and quality. The starch biosynthesis pathway in the endosperm of cereal species is highly conserved. After decades of investigation, the key enzymes including ADP-glucose pyrophosphorylase (AGPase), granule bound starch synthases (GBSSs), soluble starch synthases (SSs), starch branching enzymes (SBEs), debranching enzymes (DBEs), and starch phosphorylases (PHOs) involved in this pathway have been fully characterized and are well reviewed in the literature (Jeon et al., 2010; Zhu et al., 2020; Huang et al., 2021). Among these enzymes, AGPase is an allosterically rate-limited enzyme responsible for synthesis of the starch substrate ADP-glucose (ADPG); GBSSs, especially GBSSI, are the only enzymes responsible for amylose synthesis; SSs, SBEs, and DBEs are cooperatively responsible for the synthesis of amylopectin; PHOs are thought to function in starch initiator synthesis (Jeon et al., 2010; Zhu et al., 2020; Huang et al., 2021). When these synthases involved in starch synthesis are dysfunctional, seed development and the filling process are disturbed, leading to abnormal seed phenotypes. In addition, the direct or indirect transcriptional regulators of starch synthase encoding genes also have significant influence on grain starch content and seed development. To date, however, only a few starch biosynthesis regulators have been functionally identified. For example, bZIP TF members including OsbZIP58, OsbZIP33, OsbZIP76, O2, ZmbZIP22, and ZmbZIP91 are all reported to directly bind and activate/suppress one or more starch synthase-coding genes (Yang et al., 2001; Wang et al., 2013; Chen et al., 2016; Zhang et al., 2016; Dong et al., 2019; Niu et al., 2020). In addition to the bZIP TFs, recently, a number of NAC TFs such as OsNAC20, OsNAC26, OsNAC127, OsNAC129, ZmNAC36, ZmNAC128, ZmNAC130, and TaNAC019 have been shown to regulate starch biosynthesis and/or storage protein synthesis (Zhang et al., 2014, 2019b; Peng et al., 2019; Wang et al., 2020; Gao et al., 2021; Ren et al., 2021).
NAM, ATAF1/2, and CUC2 (NAC) TFs comprise one of the largest plant-specific TF protein families and are charactered by the presence of a conserved NAC domain at the N-terminus and a highly variable transcriptional regulation region in the C-terminus (Olsen et al., 2005; Nuruzzaman et al., 2010, 2013). The NAC domain is further separated into five sub-domains (A–E), each of which has a specific biological function: sub-domain A plays a role in dimer formation with other TFs; sub-domains B and E are relatively divergent and may be related to NAC protein functional diversity; sub-domains C and D are responsible for binding to DNA (Ernst et al., 2004; Olsen et al., 2005; Jensen et al., 2010; Chen et al., 2011; Puranik et al., 2012). NAC TFs play multiple vital roles in plant growth and development, the stress response, plant hormone signal transduction, and leaf senescence (Singh et al., 2021). It has been reported that there are at least 151 NAC TF genes in the rice genome, and that nine of them are seed-specific, indicating a wider range of functions for NAC TFs in seed development (Fang et al., 2008; Nie et al., 2013).
In this study, we characterized a seed-specific NAC TF gene OsNAC129 (LOC_Os11g31380). Loss-of-function of OsNAC129 led to increased grain length, grain weight, apparent amylose content (AAC), and plant height, while transgenic plants overexpressing OsNAC129 displayed essentially the opposite phenotypes. Further study revealed that OsNAC129 negatively regulates grain size and AAC by inhibiting the expression of genes related to grain size and starch synthesis. Moreover, we showed that OsNAC129 participates in the BR signaling pathways.
Materials and Methods
Plant Materials and Growth Conditions
For overexpression of OsNAC129, the full length DNA fragment including 2 kb of promoter sequence upstream of the start codon, the open reading frame (ORF) from the start codon to the stop codon, and 1 kb downstream of the stop codon was amplified from genomic DNA of the japonica rice (Oryza sativa L.) cultivar “Nipponbare” by PCR. The DNA fragment was then introduced into pCAMBIA 1300 by recombinase. For the promoter-driven GUS reporter, 2 kb of DNA sequence upstream of the start codon was amplified from genomic DNA of “Nipponbare” by PCR, and the DNA fragment was introduced into the pCAMBIA 1300-GN vector by recombinase. All of the recombinant plasmids were introduced into Agrobacterium tumefaciens strain EHA105 and used to infect callus of “Nipponbare” to obtain transgenic plants. The japonica rice cultivar “Dongjin” and the osnac129 T-DNA mutant (PFG_3A-60140) were obtained from Pohang University of Science and Technology, Korea (Jeong et al., 2002). Paddy field conditions: plants were grown at the experimental stations in Shanghai (121°24′ E, 31°00′ N) during the summer season, or at Lingshui (110°00′ E, 18°31′ N) during the winter season under natural conditions mainly for phenotypic analysis, detection of gene expression, and seed production. Plants were grown in the greenhouse at 28°C with an 11-h day/13-h night photoperiod mainly for seedling cultivation and for gene expression experiments.
DNA and RNA Isolation and qRT-PCR Analysis
Plant DNA and plasmid DNA isolation was performed using a plant DNA isolation kit (Tiangen Biotech) and a plasmid DNA extraction kit (Tiangen Biotech), respectively, following the manufacturer’s instructions. For RNA isolation, all tissues were collected fresh, flash-frozen in liquid nitrogen, and total RNA was extracted using a plant total RNA isolation kit (Tiangen Biotech) following the manufacturer’s instructions. First-strand cDNA was synthesized using a reverse transcription kit (TAKARA) as directed by the manufacturer. For qPCR, 2 μl of cDNA was used as template in 20 μl reaction volumes, and the qPCR amplifications were performed using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus; TAKARA) on a LightCycler® 480 instrument (Bio-Rad). The rice UBIQUITIN10 (OsUBQ10) or ACTIN1 genes were used as the internal controls for normalization of gene expression. All oligonucleotide primers used in this study are given in Supplementary Table 1.
Southern Blot Analysis
Southern blot analysis of the osnac129 mutant was performed as described previously (Liu et al., 2020).
GUS Assays
Histochemical analysis of GUS activity was performed as described in a previous study with slight modification (Edwards et al., 1990). Briefly, the tissues of transgenic plants expressing the OsNAC129 pro::GUS construct were collected and fixed in 90% acetone aqueous solution for 15 min on ice. The fixative solution was discarded and the tissues were washed twice with deionized water. Subsequently, the tissues were immersed in the GUS staining solution and allowed to stain overnight at 37°C. The GUS staining solution was discarded the following day and ethanol was added to decolorize the tissues so that the GUS staining signal could be observed and imaged.
Determination of Endosperm Starch Content
Total starch content (TSC) and apparent amylose content determination were performed as described previously (Wang et al., 2013).
Seed Phenotype Observation and Lemma Outer Epidermis Cell Measurement
Grain size of mature seeds harvested from transgenic plants was measured using the WSeen SC-G automatic seed testing system and thousand-grain weight analysis system (Hangzhou WSeen Detection Technology Co., Ltd., China) following the manufacturer’s operating instructions. There were three biological replicates per sample and each replicate contained more than 300 seeds. Lemma outer epidermis cell measurement was performed as previously described (Heang and Sassa, 2012a). Starch grain morphology observation was performed as described previously (Fu and Xue, 2010).
Plant Hormone Induction Assay
About 1-week-old “Nipponbare” seedlings were submerged in distilled water containing the various plant hormones, and distilled water without added plant hormones was used as the control. The hormone-treated samples were collected at different times and immediately frozen in liquid nitrogen. All the collected samples were then used in qRT-PCR assays to determine the relative levels of OsNAC129 expression. RNA isolation, reverse transcription, and qRT-PCR amplifications were performed as described above.
BL Sensitivity Assay
About 2-week-old “Nipponbare” and OsNAC129-OE seedlings were treated with brassinolide (BL) at three concentrations (1, 5, and 50 μM) for 24 h. Sterile water treatment was used as the control. The seedlings were then imaged and the leaf angles calculated by ImageJ following the manufacturer’s instructions.
Results
OsNAC129 Is Highly Expressed in Immature Rice Seeds
It was previously reported that nine NAC TFs gene show seed-specific expression patterns, and OsNAC129 was one of them (Fang et al., 2008; Nie et al., 2013). In order to comprehensively reveal the functions of OsNAC129, we determined its expression profile by qRT-PCR in various tissues and developmental stages in the japonica rice cultivar “Nipponbare” (NIP). The results showed that OsNAC129-specific mRNA was almost undetectable in vegetative tissues including roots and shoots at 7 days after germination, and in the flag leaves, leaf sheaths, and stems at heading (Figure 1A). Additionally, the expression level was also very low in panicles early in development (Figure 1A). Expression of OsNAC129 increased rapidly in immature seeds after fertilization and peaked at 7 days after fertilization (DAF), then gradually decreased with development (Figure 1A), which is consistent with the grain filling process. In addition, we confirmed that the highest expression in seeds was mainly detected in the endosperm (Figure 1A). We also found that OsNAC129 was highly expressed in the spikelet hull, seed coat, and embryo (Figure 1A), implying its potential functions in the regulation of seed development.
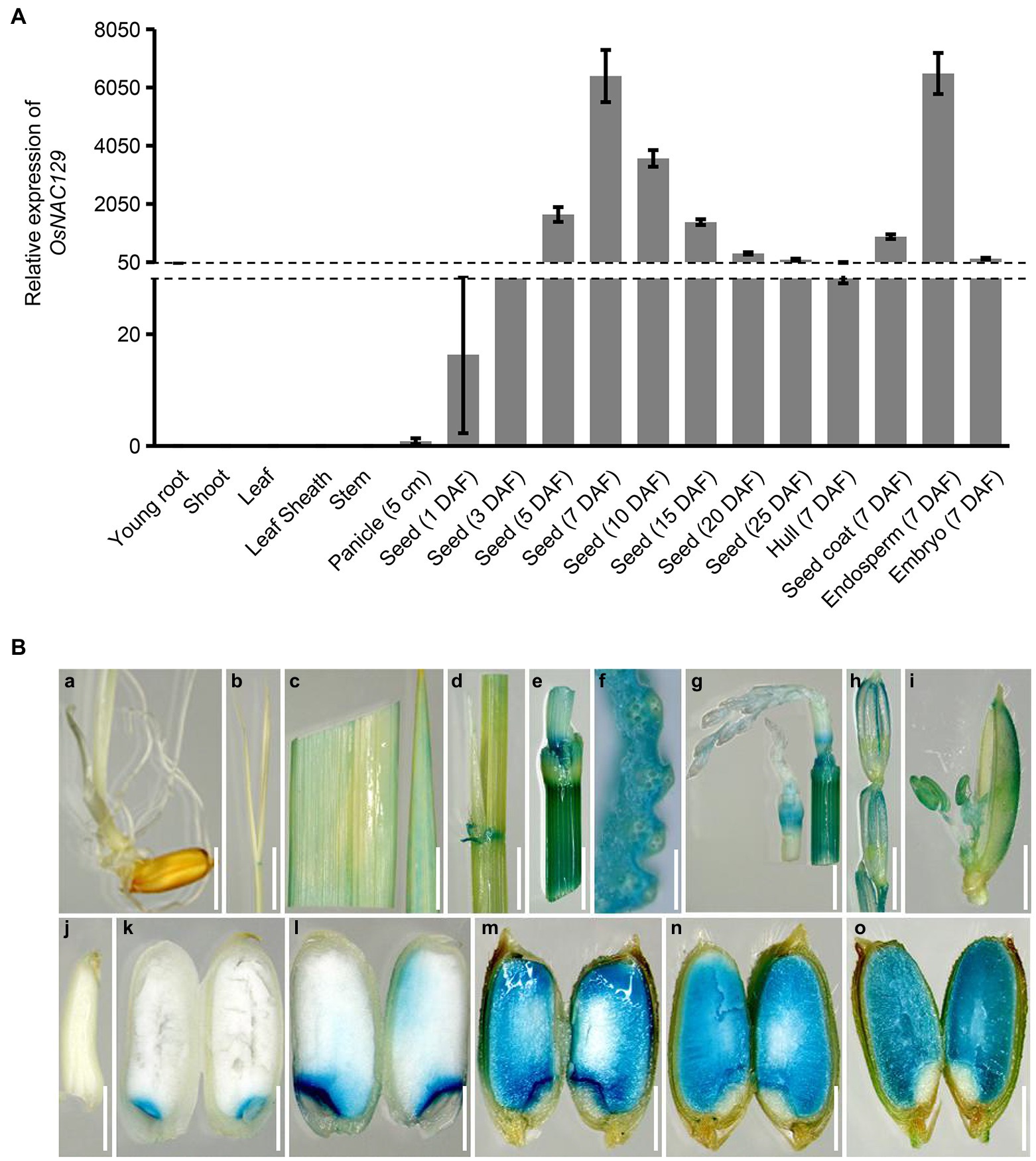
Figure 1. OsNAC129 is highly expressed in immature seeds. (A) Expression profiles of OsNAC129 in “Nipponbare.” Total RNA was extracted from roots and shoots at 7 days after germination, from leaves, leaf sheathes, stems, and young panicles before flowering, and from developing seeds from 1 to 25 days after fertilization (DAF). Actin1 was used as the internal control for normalization of gene expression. Data are means ± SD of four biological replicates. (B) OsNAC129 promoter-GUS expression patterns in transgenic plants. GUS expression in root (a) and shoot (b) at 7 days after germination, flag leaf (c), leaf sheath (d), stem (e), and cross section of stem (f) at heading date, younger panicle (g), older panicle (h), flowering spikelet (i), and endosperm at 3 DAF (j), 5 DAF (k), 7 DAF (l), 10 DAF (m), 15 DAF (n), and 25 DAF (o). Scale bars (a–e, g, and h) = 5 mm; scale bar (f) = 1 mm; scale bars (i–o) = 2.5 mm.
To better characterize the expression of OsNAC129, we also generated transgenic plants by transforming an OsNAC129 pro::GUS construct into the cultivar NIP. A histochemical GUS activity assay was then performed on tissues from the transgenic plants. The results showed that GUS staining signals could not be detected in roots and shoots at 7 days after germination except in the lamina joints (Figures 1Ba,b). Weak GUS staining was detected in leaves and leaf sheaths and relatively stronger GUS staining was detected in lamina joints before flowering (Figures 1Bc,d). Additionally, we detected strong GUS staining in the stem nodes and internodes (Figures 1Be,f). Moreover, as the panicles developed to the flowering stage, the GUS staining signals became stronger, and the stamen was also stained (Figures 1Bg–i). After flowering and fertilization, the embryo and endosperm begin to form. GUS staining was also undetectable until 5 DAF during the early stage of seed development (Figures 1Bj,k), and the staining was mainly concentrated at the junction of the embryo and endosperm (Figures 1Bk,l). Also, GUS staining became progressively stronger and was distributed throughout the entire endosperm after 7 DAF (Figures 1Bk–o). In conclusion, these results confirmed that OsNAC129 is a universally expressed gene with the highest level of expression detected in immature seeds, suggesting a key role in seed development and plant growth regulation.
Loss-of-Function of OsNAC129 Leads to Increases in Grain Size, Apparent Amylose Content, and Plant Height
Subsequently, we obtained a T-DNA insertion mutant called osnac129 (PFG_3A-60140) from the Rice T-DNA insertion sequence database (RISD) collection. According to description of this mutant, a T-DNA is inserted into the third exon of OsNAC129 (Supplementary Figure 1A). The T-DNA insertion site in the osnac129 mutant was verified by a PCR assay using gene- and T-DNA-specific primers (Supplementary Figures 1A,B). Homozygous mutant plants were further isolated from segregating progeny (Supplementary Figure 1B). Moreover, a Southern blot assay confirmed that there was only a single T-DNA insertion in this mutant (Supplementary Figure 1C), indicating a specific mutation in OsNAC129. The relative transcription of OsNAC129 was also found to be significantly reduced (Supplementary Figure 1D). Hence, osnac129 is a loss-of-function mutant. We next observed the phenotypes of osnac129 mature seeds. The results of this analysis showed that grain length and grain weight of osnac129 mutant seeds were increased compared with WT plants (Figures 2A,B,E), while grain width and grain thickness showed no obvious change (Figures 2A,C,D). These results further confirmed that OsNAC129 negatively regulates grain size, especially grain length. Likewise, the appearance of the endosperm in osnac129 mutant seeds was semi-transparent and non-chalky, indistinguishable from WT except for the slender seed shape (Figure 2F). Scanning electron microscope (SEM) images revealed that the starch granules observed in cross sections of osnac129 endosperm were closely packed and polyhedral, similar to WT (Figure 2F). We then measured the starch content of osnac129 and WT seeds, and found that the AAC was increased but that the TSC was reduced in osnac129 endosperm compared with WT (Figures 2G,H). Collectively, these results showed that OsNAC129 participates in the regulation of both grain shape and starch biosynthesis in rice.
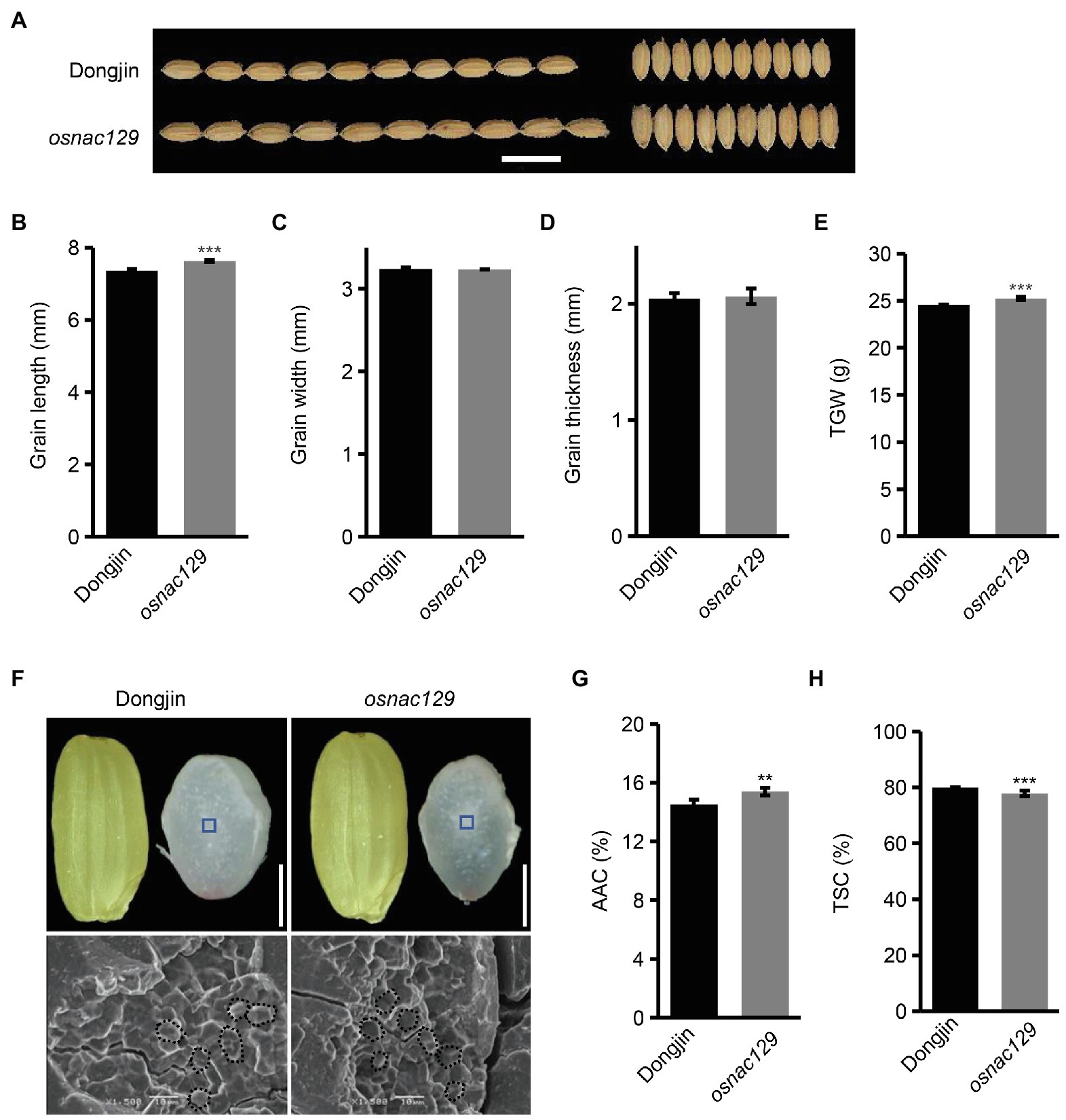
Figure 2. The osnac129 mutant has increased grain size and apparent amylose content. (A) Phenotypic observation of osnac129 grain at maturity. Scale bar = 10 mm. (B–E) Show the grain length, grain width, grain thickness, and 1,000-grain weight (TGW) of mature seeds, respectively. Data are means ± SD of three replicates. (F) Brown rice grains and cross sections of grains (upper panel, scale bars = 2.5 mm), and electron micrographs of starch granules (lower panel, scale bars = 10 μm) from the wild-type (WT) and osnac129 mutant. Blue boxes in the upper panels indicate the central area of the mature endosperm, where starch granules were analyzed by scanning electron microscope (SEM), and areas in the bottom panels delineated by dotted lines indicate starch grains. (G,H) Show the apparent amylose content (AAC) and total starch content (TSC) of grains from the WT and osnac129 mutant, respectively. Data are means ± SD of five replicates. **p < 0.01, ***p < 0.001 as determined by Student’s t-test.
Considering that OsNAC129 expression was also detected in stems, we sought to determine whether OsNAC129 plays any roles in vegetative growth and development. As a result, we found that the shoots of osnac129 mutant plants were longer than in WT plants at the seedling stage, while root lengths were reduced compared to WT (Supplementary Figures 2A–C). Moreover, plant height in the osnac129 mutant was also increased at maturity compared with WT (Supplementary Figures 2D,E), but tiller number was not affected. Therefore, these results indicate that OsNAC129 plays a negative role in regulating plant height in rice.
Overexpression of OsNAC129 Leads to Reductions in Grain Size, Apparent Amylose Content, and Plant Height
To further verify the functions of OsNAC129 in rice seed development and plant growth, we also generated transgenic plants overexpressing the full length OsNAC129 gene (including ~2 kb promoter sequence upstream of the ATG, the ORF, and ~1 kb downstream of the stop codon) in the NIP background (hereafter referred as OE plants; Supplementary Figure 1E). The results of a phenotypic analysis of OE plants showed that, compared with WT, grain width rather than grain length was reduced in the OE plants, leading to a decrease in grain weight (Figures 3A–D). These results further confirmed that OsNAC129 plays a negative role in the regulation of grain size. Moreover, the AAC was reduced but TSC was unchanged in the seeds of OE plants compared with WT (Figures 3E,F). Above all, these results suggest that OsNAC129 simultaneously and negatively regulates grain size and starch biosynthesis, especially AAC.
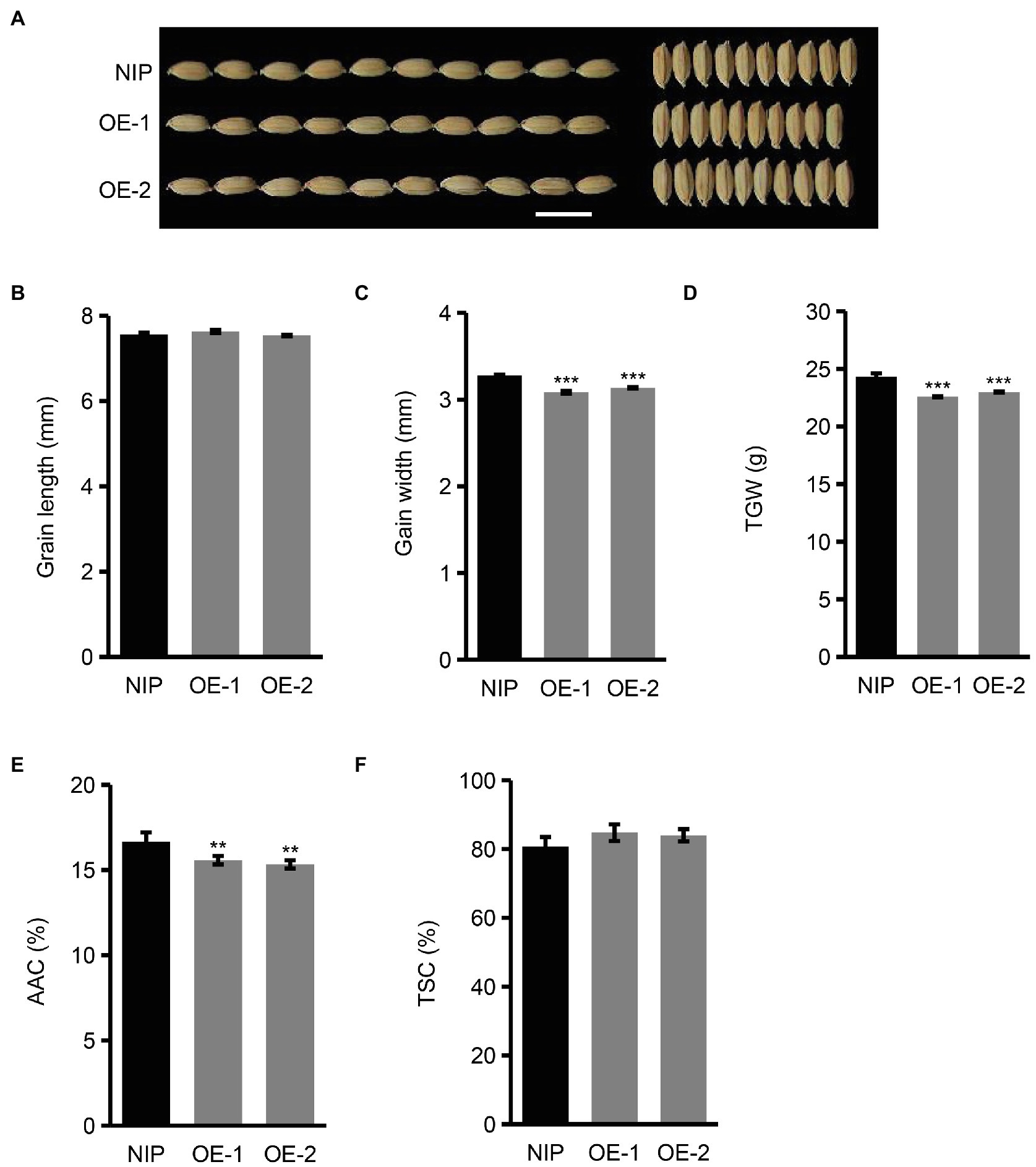
Figure 3. Overexpression of OsNAC129 leads to reductions in grain size and apparent amylose content. (A) Phenotypic observation of WT and OE plants mature seeds. Scale bar = 10 mm. (B–D) Show grain length, grain width, and TGW of mature seeds of NIP and two OsNAC129-OE lines, respectively. Data are means ± SD of three replicates. (E,F) Show the AAC and TSC of NIP and the two OsNAC129-OE lines, respectively. Data are means ± SD of five replicates. **p < 0.01, ***p < 0.001 as determined by Student’s t-test.
Additionally, we also checked the vegetative growth of OsNAC129-OE plants. In contrast to osnac129 mutant plants, shoots and roots of OE seedlings were shorter than in the WT (Supplementary Figures 3A–C). Moreover, plant height was also reduced at maturity in the OE plants (Supplementary Figures 3D–F). These results show conclusively that OsNAC129 plays negative roles in the regulation of grain size, starch synthesis, and plant height.
Expression of Grain Size and Starch Biosynthesis-Related Genes Is Altered in Both osnac129 Mutant and OsNAC129-OE Plants
The above results showed that OsNAC129 negatively regulates grain size (Figures 2A–E, 3A–D). In order to discover the regulatory mechanism, we observed cell differences in mature seeds of the osnac129 mutant and WT using SEM. The results showed that cell length rather than cell width was increased in the osnac129 mutant compared with WT, while the longitudinal cell number was reduced (Figures 4A–D), implying that OsNAC129 regulates grain length by controlling cell elongation and division. At present, several genes, such as OsSRS1, OsSRS3, OsSRS5, OsPGL1, and OsPGL2, have been shown to participate in the regulation of grain size by promoting cell expansion (Abe et al., 2010; Heang and Sassa, 2012a,b; Segami et al., 2012; Deng et al., 2015). Therefore, we asked whether OsNAC129-regulated grain size depends on one or more of these genes. We found that, of these genes, the expression levels of OsPGL1 and OsPGL2 were significantly upregulated in osnac129 seeds (Figure 4E), suggesting that the cell expansion promoted by OsNAC129 might partly depend on them. Consistent with this speculation, OsPGL1 and OsPGL2 were downregulated in the OsNAC129-OE plants (Figure 4F), implying that OsNAC129 negatively regulates OsPGL1 and OsPGL2 expression.
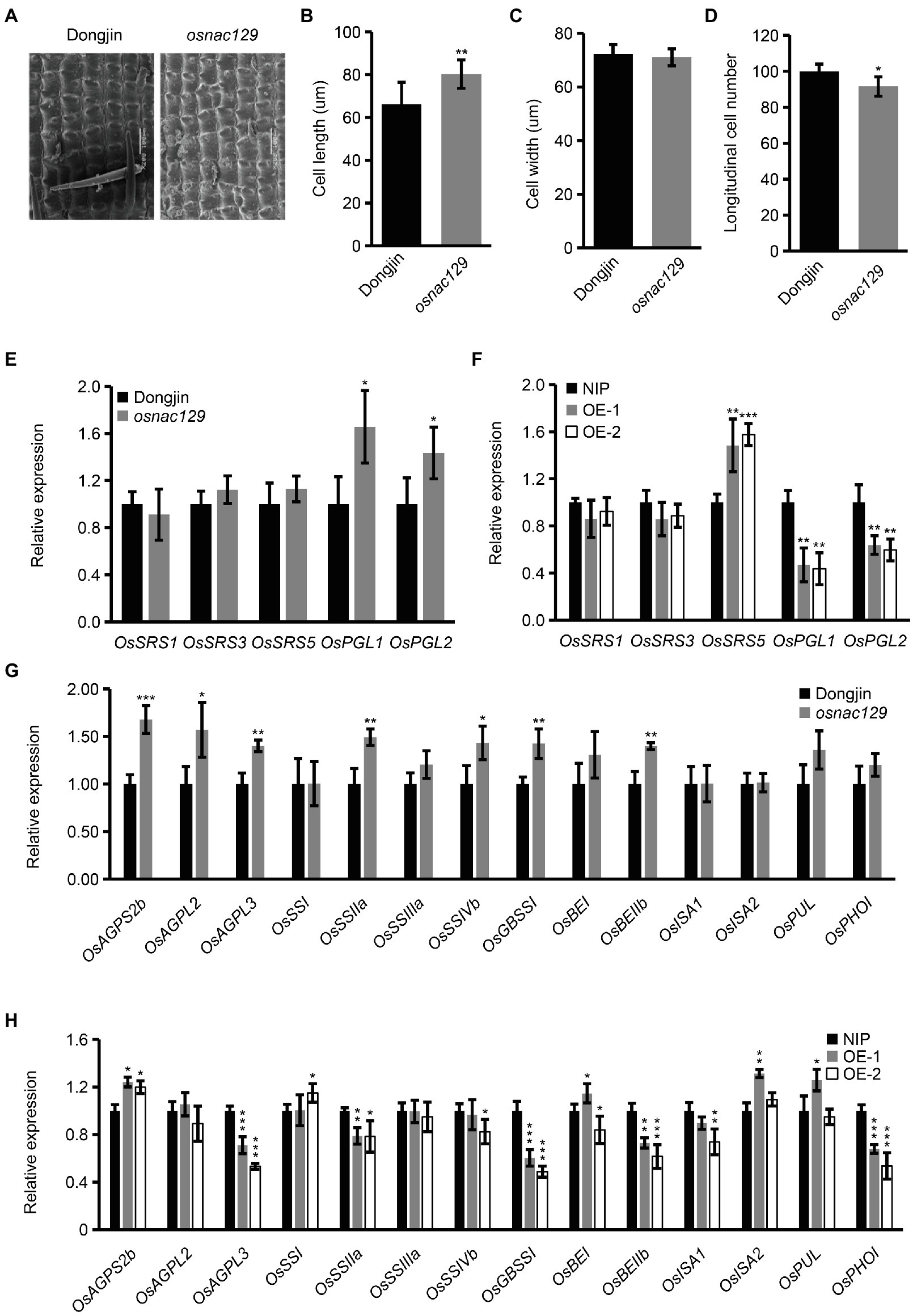
Figure 4. OsNAC129 simultaneously regulates cell elongation and starch biosynthesis in rice seeds. (A) Observation of the outer epidermal cells of WT and osnac129 mutant lemmas by SEM. Scale bars = 100 μm. (B–D) Show the cell length, cell width, and cell number measurement of the outer epidermal cells of the lemma. Data are means ± SD of three replicates (each replicate consisted of at least 30 cells) in (B,C), and four replicates in (D); **p < 0.01, ***p < 0.001 as determined by Student’s t-test. (E,F) Show the relative expression determined by qRT-PCR of genes reported to be related to grain size by controlling cell elongation in seeds from the WT, osnac129 mutant, and OE plants at 7 DAF. UBQ10 was used as the internal control for normalization of gene expression. Data are means ± SD of four replicates. (G,H) Show qRT-PCR determination of expression of starch synthase-encoding genes in seeds of the WT, osnac129, and OE plants at 7 DAF. UBQ10 was used as the internal control. Data are means ± SD of four replicates; *p < 0.05, **p < 0.01, and ***p < 0.001 as determined by Student’s t-test.
In addition, the above results also showed that OsNAC129 participates in starch biosynthesis. Thus, we hypothesized that the expression levels of genes encoding starch synthase are affected in the osnac129 mutant and OE plants. qRT-PCR assays showed that several starch synthase genes such as OsAGPS2b, OsAGPL2, OsAGPL3, OsSIIa, OsSSIVb, OsGBSSI, and OsSBEIIb were upregulated in the osnac129 mutant (Figure 4G). However, we found that several starch synthase genes including OsAGPL3, OsSIIa, OsGBSSI, OsBEIIb, and OsPHOI were downregulated in OE endosperm, although OsAGPS2b was upregulated (Figure 4H). These results further confirm that OsNAC129 plays a negative role in regulation the expression of starch synthase genes. However, these results are contradictory in that they do not explain why the TSC decreased in osnac129 mutant seeds (Figure 2H) but was not changed in seeds from the OsNAC129-OE plants (Figure 3F). These results suggest that there might be more complex regulatory mechanisms that act at the post-transcriptional and translational levels in starch synthase-encoding genes.
OsNAC129 Expression Is Exclusively Induced by ABA, and OsNAC129 Participates in the BR Signaling Pathway
The above results suggest that the osnac129 mutant has the potential to improve grain size in rice due to its increased grain length and grain weight; thus, we further explored the types of upstream signals that allow OsNAC129 to participate in this process. Phytohormones are widely considered to be key regulatory signals that are involved in almost all aspects of plant growth and development. For example, indoleacetic acid (IAA) and BRs have been widely reported to positively regulate seed size in plants (Song, 2017; Figueiredo and Köhler, 2018; Cao et al., 2020). A very recent study showed that leaf-derived ABA regulates seed development by promoting the expression of starch synthesis genes (Qin et al., 2021). Therefore, we subsequently investigated the relationship between the expression of OsNAC129 and phytohormones, such as IAA, 6-benzylaminopurine (6-BA), gibberellin A3 (GA3), brassinolide (BL), and ABA. About 1-week-old WT seedlings were treated with these phytohormones (50 μM of each) and the water treatment group was used as the control. qRT-PCR assays showed that the expression level of OsNAC129 in seedlings was extremely low and almost undetectable, consistent with the expression profile determined previously (Figures 1A,B). However, after the hormone treatments, OsNAC129 was exclusively and significantly induced by ABA but not by the others (Figure 5A), indicating that ABA could be the upstream signal that allows OsNAC129 to participate in the corresponding regulation of a group of downstream genes.
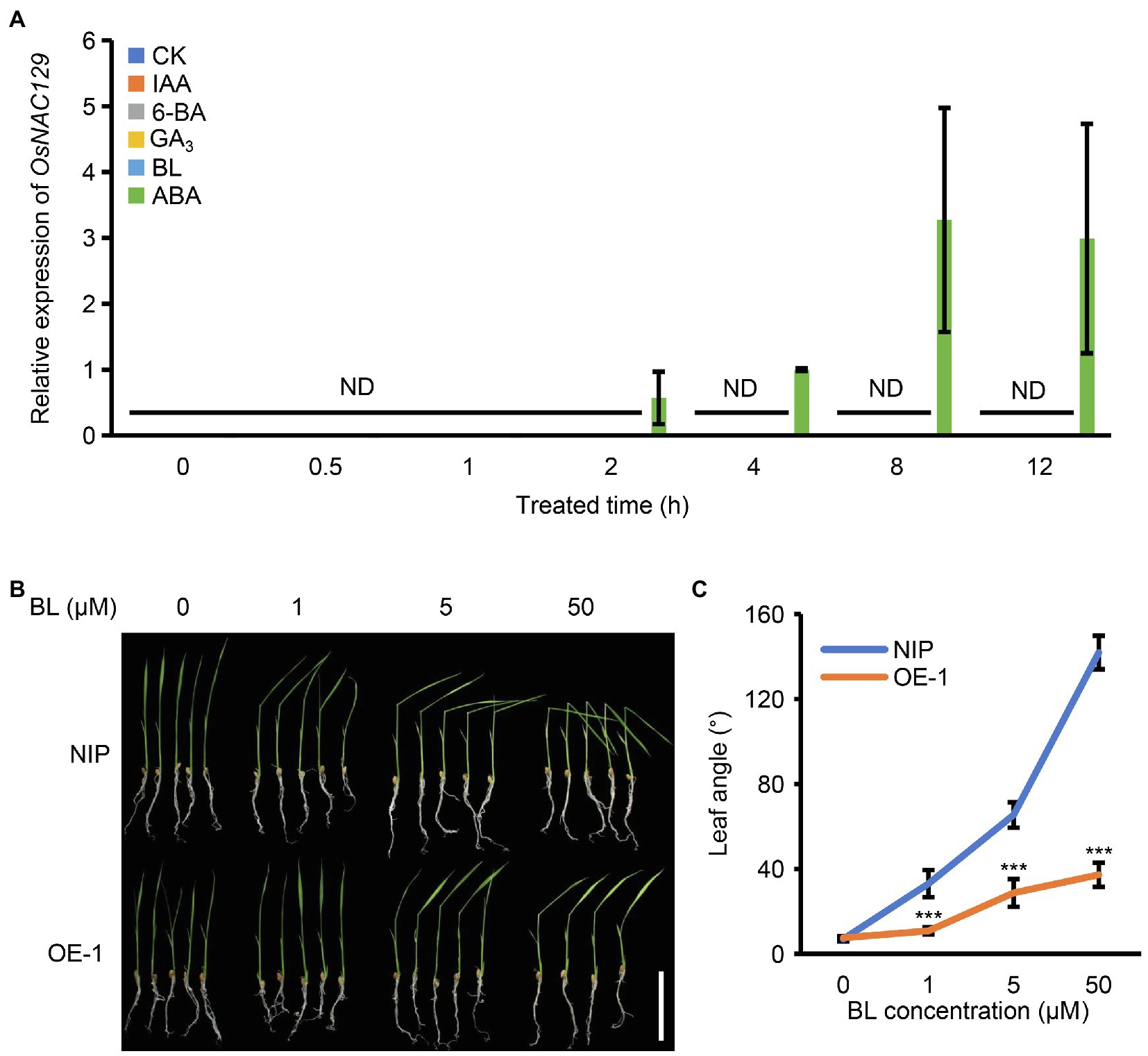
Figure 5. OsNAC129 is an abscisic acid (ABA)-inducible and brassinolide (BR)-related gene. (A) OsNAC129 was exclusively induced by ABA. About 1-week-old NIP seedlings were treated separately with IAA, 6-BA, GA3, BL, and ABA (50 μM of each phytohormone), and sterile water treatment was the control. Samples were collected, RNA was extracted, and the expression of OsNAC129 was determined by qRT-PCR. Data are means ± SD of four replicates. ND, not detected. (B) About 2-week-old seedlings of WT and the OsNAC129-OE-1 line were treated with three concentrations of BL for 24 h, after which the leaf angles were imaged. Scale bar = 5 cm. (C) Leaf angles of WT and OE-1 seedlings measured by Image J after BL treatment. Data are means ± SD of 10 replicates. ***p < 0.001 as determined by Student’s t-test.
Additionally, we showed that OsNAC129 negatively regulates grain length, and that this regulation is partially dependent on two other grain size regulator genes, OsPGL1 and OsPGL2. Both OsPGL1 and OsPGL2 were previously reported to be BR-related genes, even though OsPGL1 was not BR-inducible (Heang and Sassa, 2012a,b; Jang et al., 2017; Jang and Li, 2017). We also found that the expression of OsNAC129 could be detected in the lamina joints (Figures 1Bb,d), indicating a potential role in leaf bending/inclination regulation. Furthermore, promotion of leaf bending is representative of the BR response in cereal crops such as rice and maize. Thus, we wondered whether OsNAC129 is a BR-related gene even though it is not BR-inducible, similar to OsPGL1. We treated 2-week old WT and OsNAC129-OE seedlings with a variety of concentration of exogenous BL and the angle of leaf inclination was measured. The results of this experiment showed that at the seedling stage, leaf angle was indistinguishable between WT and OE seedlings without BL treatment. In seedlings treated with increasing BL concentrations, leaf angle in WT seedlings increased significantly, but the leaf angle increased much more slowly in the OE plants (Figures 5B,C), indicating significantly reduced sensitivity to BL and a negative role for OsNAC129 in the BR signaling pathway. Overall, these results suggest that transcription of OsNAC129 is induced by ABA and that it participates in the BR signaling pathway.
Discussion
In this study, we characterized a NAC TF-encoding gene, OsNAC129, that is, highly expressed in seeds, to examine its potential roles in regulating seed development and plant growth. Comprehensive phenotypic analysis and comparison of a T-DNA insertion mutant and OsNAC129-OE plants with WT revealed that OsNAC129 simultaneously plays negative roles in regulating grain size, AAC, and plant height in rice. SEM observation of lemma outer epidermal cells indicated that OsNAC129 inhibits grain length mainly by regulating cell elongation, and that this regulation is partially dependent on OsPGL1 and OsPGL2. Furthermore, OsNAC129 was shown to be a BR signaling pathway-related gene that acts as a negative regulator, although it was not BR-inducible, being similar to OsPGL1 in that respect. Moreover, we also found that OsNAC129 expression was exclusively induced by ABA in seedlings. Taken together, these results strongly suggest that OsNAC129 regulates seed development and plant growth and participates in the BR signaling pathway.
OsNAC129 was previously reported to be a seed-specific gene (Fang et al., 2008). To date, several of these NAC family TF genes such as OsNAC20, OsNAC26, OsNAC127, and OsNAC129 had been confirmed to play important roles in the regulation of rice seed development (Wang et al., 2020; Ren et al., 2021). qRT-PCR assays confirmed that OsNAC129 is highly expressed in immature seeds, peaking at 7 DAF, after which the mRNA levels gradually decline (Figure 1A). This pattern was finely matched with the expression profiles of genes encoding starch synthases and the grain filling process, similar to the expression of OsNAC20, OsNAC26, OsNAC127, and OsNAC129 (Wang et al., 2020; Ren et al., 2021). However, histochemical GUS staining of transgenic plants expressing an OsNAC129 pro::GUS construct further showed that there was still some GUS staining present in a few regions of some vegetative tissues, such as the leaves, leaf sheaths, lamina joints, and stem nodes (Figure 1B). These expression patterns are consistent with those reported by Ren et al. (2021), and imply that OsNAC129 also functions in plant growth.
It is possible that the expression of OsNAC129 in the lamina joints is related to BR signaling, because promotion of leaf inclination is one of the typical physiological effects of BRs (Wang et al., 2012b, 2014; Li et al., 2020). Several BR-related components that control leaf angle also show lamina joint-specific expression patterns, such as OsBU1, OsPGL2/OsBUL1, and OsbHLH98 (Tanaka et al., 2009; Jang and Li, 2017; Guo et al., 2021). Furthermore, expression in the nodes and stems has been thought to relate to cell proliferation/expansion and nutrient transport. For example, our previous study reported that a spin-like gene, OsRRM, which modulates sugar transport, had a similar expression profile to OsNAC129 in stems, and the osrrm mutant displayed reduced plant height and grain size, similar to the phenotypes observed in OsNAC129-OE plants (Liu et al., 2020). Very recently, another group reported that OsNAC129 and its interactor OsNAC127 do indeed participate in grain filling regulation by directly targeting sugar transporter genes including OsMST6 and OsSWEET4 (Ren et al., 2021). Thus, in our study, we could not rule out the possibility that OsNAC129 might also participate in regulating sugar or other nutrient transport, and it would be worthwhile to further explore the mechanism. However, we did not observe the incomplete grain filling phenotype in osnac129 mutants and OsNAC129-OE plants in this study, probably due to the use of different cultivars [Ren et al. (2021) used ZH11, while we used “Dongjin” and “Nipponbare”], planting conditions, and phenotypes.
Ren et al. (2021) showed that OsNAC129 functions in grain filling. In our study, we showed that loss-of-function of OsNAC129 led to multiple phenotypes, including increased grain length, grain weight, AAC, and plant height, but reduced TSC (Figures 2A–H; Supplementary Figures 2A–F). Moreover, overexpression of OsNAC129 resulted in almost opposite phenotypes such as reduced grain width (instead of grain length), grain weight, AAC, and plant height, but slightly increased TSC (Figures 3A–F; Supplementary Figures 3A–F). These results provide evidence that OsNAC129 plays a negative role in regulating grain size, AAC, and plant height. Further observation of the cells in osnac129 and WT seeds revealed that the increase in grain length was caused by lemma cell elongation rather than cell proliferation (Figures 4A–D). qRT-PCR determination of the expression of several known genes associated with grain size through cell elongation control suggested that grain size regulation by OsNAC129 probably and depends partially on OsPGL1 and OsPGL2 (Figures 4E,F). OsPGL1 and OsPGL2 were previously reported to encode two atypical bHLH proteins that positively regulate grain length by heterodimerizing with a typical bHLH protein, APG, and inhibiting its activity. Overexpression of OsPGL1 and OsPGL2/OsBUL1 and knock-down of APG expression gave plants that produced longer seeds than WT, similar to the grains produced by the osnac129 mutant (Heang and Sassa, 2012a,b; Jang et al., 2017; Jang and Li, 2017). Moreover, OsPGL1 was shown to be a BR-related gene that functions as a positive regulator, because plants overexpressing OsPGL1 were hypersensitive to BL; however, its expression was not BL-inducible (Heang and Sassa, 2012a). The BR pathway is widely thought to be an important and classical pathway for the regulation of plant architecture (plant height, tiller number, and tiller angle), panicle morphology, and grain size (Müssig, 2005; Divi and Krishna, 2009; Fridman and Savaldi-Goldstein, 2013; Wei and Li, 2016; Zhang et al., 2019a; Ackerman-Lavert and Savaldi-Goldstein, 2020; Nolan et al., 2020). In this study, we found that OsNAC129 is expressed in the lamina joints, stem nodes, and the spikelet, and controls plant height and grain shape simultaneously, which is highly correlated with plant phenotypes that are regulated by BRs. However, the OE plants showed significantly reduced sensitivity to exogenous BL (Figures 5B,C). Thus, OsNAC129 seem to play a negative role in the BR pathway, antagonizing the functions of OsPGL1 and OsPGL2.
In addition, qRT-PCR assays of genes that encode starch synthases in osanc129 mutant and OsNAC129-OE plants revealed that OsNAC129 repressed the expression of several of them, including OsGBSSI (Wx; Figures 4G,H). OsGBSSI, encoded by the Wx gene, is the only enzyme responsible for amylose synthesis in endosperm (Wang et al., 1990, 1995). Thus, the AAC was increased in grains from the osnac129 mutant, but was reduced in OE plant grains compared to WT (Figures 2G, 3E). However, TSC was reduced in the osnac129 mutant (Figure 2H). There are two hypotheses that may explain this phenomenon: (1) OsNAC129 could also participate in regulating the synthesis of other storage molecules such as storage proteins; thus, loss-of-function of OsNAC129 increased the content of not only starch but also of other substances. For instance, several TF genes including OsNAC20, OsNAC26, ZmNAC128, ZmNAC130, and O2 have recently been reported to participate in regulating both starch and protein synthesis (Zhang et al., 2016, 2019b; Wang et al., 2020). (2) Starch synthases are regulated by complex mechanisms including the transcription level, post-transcription level, translation level, and protein interactions and modifications. For example, OsNAC20 and OsNAC26 can directly bind to the promoters and activate the expression of AGPS2b, AGPL2, and OsSBEI; however, the activities of AGPase and SBE were unchanged (Wang et al., 2020). Thus, it would be worthwhile to further explore the role of OsNAC129 in the regulation of other storage substances in seeds and relative changes in the activity of starch synthases. It has been reported that OsNAC129 directly and negatively regulates the expression of the sugar transporter encoding genes OsMST6 and OsSWEET4 during grain filling (Ren et al., 2021). Further analysis of whether starch-synthesis enzyme-coding genes are the direct target of OsNAC129 will help to clarify the molecular mechanism by which OsNAC129 regulates of starch synthesis.
In addition to the strong evidence showing that OsNAC129 participates in the BR signaling pathway, we found that transcription of OsNAC129 is exclusively induced by ABA in seedlings, indicating a potential role for this gene in the ABA signaling pathway. Recently, it has been reported that leaf-derived ABA not only plays important roles in the stress response, senescence, and seed dormancy, but is also vital in promoting starch synthesis and grain filling (Qin et al., 2021). OsNAC129 is also a heat-stress responsive gene and both the osnac129 mutants and OE plants exhibit more sensitivity to heat stress (Ren et al., 2021). Therefore, we speculate that OsNAC129 probably coordinates BR and ABA signals to regulate diverse biological processes such as starch synthesis, grain filling, plant growth, and the heat stress response.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
J-PG and X-LC conceived the project and designed the study. S-KJ, M-QZ, Y-JL, L-NX, S-WJ, S-LW, TS, R-AW, Q-QY, and TT performed experiments. S-KJ, M-QZ, X-LC, and J-PG analyzed and interpreted the data. S-KJ and J-PG wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from Jiangsu Province Government [JBGS(2021)001], Guangdong Province Key Research and Development Program (2018B020202012), National Natural Science Foundation of China (31771754), The Independent Scientific Research Project funds of the Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding (PLR202101), Hainan Yazhou Bay Seed Lab (B21HJ0220-07), Natural Science Foundation of Shanghai (19ZR1466400), China Postdoctoral Science Foundation (2021M692723), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Xiao-Yan Gao, Zhi-Ping Zhang, and Ji-Qin Li (CAS Center for Excellence in Molecular Plant Sciences) for Scanning electron microscope observation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.905148/full#supplementary-material
Footnotes
References
Abe, Y., Mieda, K., Ando, T., Kono, I., Yano, M., Kitano, H., et al. (2010). The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice. Genes Genet. Syst. 85, 327–339. doi: 10.1266/ggs.85.327
Ackerman-Lavert, M., and Savaldi-Goldstein, S. (2020). Growth models from a brassinosteroid perspective. Curr. Opin. Plant Biol. 53, 90–97. doi: 10.1016/j.pbi.2019.10.008
Brosa, C. (1999). Biological effects of brassinosteroids. Crit. Rev. Biochem. Mol. Biol. 34, 339–358. doi: 10.1080/10409239991209345
Cao, J., Li, G., Qu, D., Li, X., and Wang, Y. (2020). Into the seed: auxin controls seed development and grain yield. Int. J. Mol. Sci. 21:1662. doi: 10.3390/ijms21051662
Che, R., Tong, H., Shi, B., Liu, Y., Fang, S., Liu, D., et al. (2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2:15195. doi: 10.1038/nplants.2015.195
Chen, Q., Wang, Q., Xiong, L., and Lou, Z. (2011). A structural view of the conserved domain of rice stress-responsive NAC1. Protein Cell 2, 55–63. doi: 10.1007/s13238-011-1010-9
Chen, J., Yi, Q., Cao, Y., Wei, B., Zheng, L., Xiao, Q., et al. (2016). ZmbZIP91 regulates expression of starch synthesis-related genes by binding to ACTCAT elements in their promoters. J. Exp. Bot. 67, 1327–1338. doi: 10.1093/jxb/erv527
Deng, Z. Y., Liu, L. T., Li, T., Yan, S., Kuang, B. J., Huang, S. J., et al. (2015). OsKinesin-13A is an active microtubule depolymerase involved in glume length regulation via affecting cell elongation. Sci. Rep. 5:9457. doi: 10.1038/srep09457
Divi, U. K., and Krishna, P. (2009). Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. New Biotechnol. 26, 131–136. doi: 10.1016/j.nbt.2009.07.006
Dong, Q., Xu, Q., Kong, J., Peng, X., Zhou, W., Chen, L., et al. (2019). Overexpression of ZmbZIP22 gene alters endosperm starch content and composition in maize and rice. Plant Sci. 283, 407–415. doi: 10.1016/j.plantsci.2019.03.001
Duan, P., Ni, S., Wang, J., Zhang, B., Xu, R., Wang, Y., et al. (2015). Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2:15203. doi: 10.1038/nplants.2015.203
Duan, P., Xu, J., Zeng, D., Zhang, B., Geng, M., Zhang, G., et al. (2017). Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 10, 685–694. doi: 10.1016/j.molp.2017.03.009
Edwards, J. W., Walker, E. L., and Coruzzi, G. M. (1990). Cell-specific expression in transgenic plants reveals nonoverlapping roles for chloroplast and cytosolic glutamine synthetase. Proc. Natl. Acad. Sci. U. S. A. 87, 3459–3463. doi: 10.1073/pnas.87.9.3459
Ernst, H. A., Olsen, A. N., Larsen, S., and Lo Leggio, L. (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303. doi: 10.1038/sj.embor.7400093
Fang, Y., You, J., Xie, K., Xie, W., and Xiong, L. (2008). Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Gen. Genomics. 280, 547–563. doi: 10.1007/s00438-008-0386-6
Feng, Z., Wu, C., Wang, C., Roh, J., Zhang, L., Chen, J., et al. (2016). SLG controls grain size and leaf angle by modulating brassinosteroid homeostasis in rice. J. Exp. Bot. 67, 4241–4253. doi: 10.1093/jxb/erw204
Figueiredo, D. D., and Köhler, C. (2018). Auxin: a molecular trigger of seed development. Genes Dev. 32, 479–490. doi: 10.1101/gad.312546.118
Fridman, Y., and Savaldi-Goldstein, S. (2013). Brassinosteroids in growth control: how, when and where. Plant Sci. 209, 24–31. doi: 10.1016/j.plantsci.2013.04.002
Fu, F. F., and Xue, H. W. (2010). Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 154, 927–938. doi: 10.1104/pp.110.159517
Gao, Y., An, K., Guo, W., Chen, Y., Zhang, R., Zhang, X., et al. (2021). The endosperm-specific transcription factor TaNAC019 regulates glutenin and starch accumulation and its elite allele improves wheat grain quality. Plant Cell 33, 603–622. doi: 10.1093/plcell/koaa040
Gao, X., Zhang, J. Q., Zhang, X., Zhou, J., Jiang, Z., Huang, P., et al. (2019). Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling. Plant Cell 31, 1077–1093. doi: 10.1105/tpc.18.00836
Gudesblat, G. E., and Russinova, E. (2011). Plants grow on brassinosteroids. Curr. Opin. Plant Biol. 14, 530–537. doi: 10.1016/j.pbi.2011.05.004
Guo, T., Chen, K., Dong, N. Q., Shi, C. L., Ye, W. W., Gao, J. P., et al. (2018). Grain size and number1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain NUMBER per panicle and grain size in rice. Plant Cell 30, 871–888. doi: 10.1105/tpc.17.00959
Guo, J., Li, W., Shang, L., Wang, Y., Yan, P., Bai, Y., et al. (2021). OsbHLH98 regulates leaf angle in rice through transcriptional repression of OsBUL1. New Phytol. 230, 1953–1966. doi: 10.1111/nph.17303
Heang, D., and Sassa, H. (2012a). Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS One 7:e31325. doi: 10.1371/journal.pone.0031325
Heang, D., and Sassa, H. (2012b). An atypical bHLH protein encoded by positive regulator of grain length 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breed. Sci. 62, 133–141. doi: 10.1270/jsbbs.62.133
Hong, Z., Ueguchi-Tanaka, M., Fujioka, S., Takatsuto, S., Yoshida, S., Hasegawa, Y., et al. (2005). The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17, 2243–2254. doi: 10.1105/tpc.105.030973
Hu, Z., He, H., Zhang, S., Sun, F., Xin, X., Wang, W., et al. (2012). A Kelch motif-containing serine/threonine protein phosphatase determines the large grain QTL trait in rice. J. Integr. Plant Biol. 54, 979–990. doi: 10.1111/jipb.12008
Hu, J., Wang, Y., Fang, Y., Zeng, L., Xu, J., Yu, H., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8, 1455–1465. doi: 10.1016/j.molp.2015.07.002
Huang, L., Tan, H., Zhang, C., Li, Q., and Liu, Q. (2021). Starch biosynthesis in cereal endosperms: an updated review over the last decade. Plant Commun. 2:100237. doi: 10.1016/j.xplc.2021.100237
Jang, S., An, G., and Li, H. Y. (2017). Rice leaf angle and grain size are affected by the OsBUL1 transcriptional activator complex. Plant Physiol. 173, 688–702. doi: 10.1104/pp.16.01653
Jang, S., and Li, H. Y. (2017). Oryza sativa brassinosteroid upregulated1 like1 induces the expression of a gene encoding a small leucine-rich-repeat protein to positively regulate lamina inclination and grain size in Rice. Front. Plant Sci. 8:1253. doi: 10.3389/fpls.2017.01253
Jensen, M. K., Kjaersgaard, T., Nielsen, M. M., Galberg, P., Petersen, K., O’Shea, C., et al. (2010). The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426, 183–196. doi: 10.1042/bj20091234
Jeon, J. S., Ryoo, N., Hahn, T. R., Walia, H., and Nakamura, Y. (2010). Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 48, 383–392. doi: 10.1016/j.plaphy.2010.03.006
Jeong, D. H., An, S., Kang, H. G., Moon, S., Han, J. J., Park, S., et al. (2002). T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 130, 1636–1644. doi: 10.1104/pp.014357
Keeling, P. L., and Myers, A. M. (2010). Biochemistry and genetics of starch synthesis. Annu. Rev. Food Sci. Technol. 1, 271–303. doi: 10.1146/annurev.food.102308.124214
Li, Y., Fan, C., Xing, Y., Jiang, Y., Luo, L., Sun, L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. doi: 10.1038/ng.977
Li, S., Gao, F., Xie, K., Zeng, X., Cao, Y., Zeng, J., et al. (2016). The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 14, 2134–2146. doi: 10.1111/pbi.12569
Li, N., and Li, Y. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. doi: 10.1016/j.pbi.2016.05.008
Li, X., Wu, P., Lu, Y., Guo, S., Zhong, Z., Shen, R., et al. (2020). Synergistic interaction of phytohormones in determining leaf angle in crops. Int. J. Mol. Sci. 21:5052. doi: 10.3390/ijms21145052
Li, N., Xu, R., and Li, Y. (2019). Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 70, 435–463. doi: 10.1146/annurev-arplant-050718-095851
Liu, J., Chen, J., Zheng, X., Wu, F., Lin, Q., Heng, Y., et al. (2017). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 3:17043. doi: 10.1038/nplants.2017.43
Liu, D., Xu, L., Wang, W., Jia, S., Jin, S., and Gao, J. (2020). OsRRM, an RNA-binding protein, modulates sugar transport in rice (Oryza sativa L.). Front. Plant Sci. 11:605276. doi: 10.3389/fpls.2020.605276
Morinaka, Y., Sakamoto, T., Inukai, Y., Agetsuma, M., Kitano, H., Ashikari, M., et al. (2006). Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 141, 924–931. doi: 10.1104/pp.106.077081
Müssig, C. (2005). Brassinosteroid-promoted growth. Plant Biol. 7, 110–117. doi: 10.1055/s-2005-837493
Nakagawa, H., Tanaka, A., Tanabata, T., Ohtake, M., Fujioka, S., Nakamura, H., et al. (2012). Short grain1 decreases organ elongation and brassinosteroid response in rice. Plant Physiol. 158, 1208–1219. doi: 10.1104/pp.111.187567
Nie, D. M., Ouyang, Y. D., Wang, X., Zhou, W., Hu, C. G., and Yao, J. (2013). Genome-wide analysis of endosperm-specific genes in rice. Gene 530, 236–247. doi: 10.1016/j.gene.2013.07.088
Niu, B., Deng, H., Li, T., Sharma, S., Yun, Q., Li, Q., et al. (2020). OsbZIP76 interacts with OsNF-YBs and regulates endosperm cellularization in rice (Oryza sativa). J. Integr. Plant Biol. 62, 1983–1996. doi: 10.1111/jipb.12989
Nolan, T. M., Vukašinović, N., Liu, D., Russinova, E., and Yin, Y. (2020). Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32, 295–318. doi: 10.1105/tpc.19.00335
Nuruzzaman, M., Manimekalai, R., Sharoni, A. M., Satoh, K., Kondoh, H., Ooka, H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44. doi: 10.1016/j.gene.2010.06.008
Nuruzzaman, M., Sharoni, A. M., and Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4:248. doi: 10.3389/fmicb.2013.00248
Olsen, A. N., Ernst, H. A., Leggio, L. L., and Skriver, K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Peng, X., Wang, Q., Wang, Y., Cheng, B., Zhao, Y., and Zhu, S. (2019). A maize NAC transcription factor, ZmNAC34, negatively regulates starch synthesis in rice. Plant Cell Rep. 38, 1473–1484. doi: 10.1007/s00299-019-02458-2
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Qi, P., Lin, Y. S., Song, X. J., Shen, J. B., Huang, W., Shan, J. X., et al. (2012). The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 22, 1666–1680. doi: 10.1038/cr.2012.151
Qin, P., Zhang, G., Hu, B., Wu, J., Chen, W., Ren, Z., et al. (2021). Leaf-derived ABA regulates rice seed development via a transporter-mediated and temperature-sensitive mechanism. Sci. Adv. 7:eabc8873. doi: 10.1126/sciadv.abc8873
Ren, Y., Huang, Z., Jiang, H., Wang, Z., Wu, F., Xiong, Y., et al. (2021). A heat stress responsive NAC transcription factor heterodimer plays key roles in rice grain filling. J. Exp. Bot. 72, 2947–2964. doi: 10.1093/jxb/erab027
Segami, S., Kono, I., Ando, T., Yano, M., Kitano, H., Miura, K., et al. (2012). Small and round seed 5 gene encodes alpha-tubulin regulating seed cell elongation in rice. Rice 5:4. doi: 10.1186/1939-8433-5-4
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Singh, S., Koyama, H., Bhati, K. K., and Alok, A. (2021). The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 134, 475–495. doi: 10.1007/s10265-021-01270-y
Song, X. J. (2017). Crop seed size: BR matters. Mol. Plant 10, 668–669. doi: 10.1016/j.molp.2017.04.007
Sun, P., Zhang, W., Wang, Y., He, Q., Shu, F., Liu, H., et al. (2016). OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Biol. 58, 836–847. doi: 10.1111/jipb.12473
Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S., Yano, M., et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17, 776–790. doi: 10.1105/tpc.104.024950
Tanaka, A., Nakagawa, H., Tomita, C., Shimatani, Z., Ohtake, M., and Nomura, T., et al. (2009). Brassinosteroid upregulated1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 151, 669–680. doi: 10.1104/pp.109.140806
Tong, H., Liu, L., Jin, Y., Du, L., Yin, Y., Qian, Q., et al. (2012). Dwarf and low-tillering acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24, 2562–2577. doi: 10.1105/tpc.112.097394
Wang, Z. Y., Bai, M. Y., Oh, E., and Zhu, J. Y. (2012b). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46, 701–724. doi: 10.1146/annurev-genet-102209-163450
Wang, W., Bai, M. Y., and Wang, Z. Y. (2014). The brassinosteroid signaling network-a paradigm of signal integration. Curr. Opin. Plant Biol. 21, 147–153. doi: 10.1016/j.pbi.2014.07.012
Wang, J., Chen, Z., Zhang, Q., Meng, S., and Wei, C. (2020). The NAC transcription factors OsNAC20 and OsNAC26 regulate starch and storage protein synthesis. Plant Physiol. 184, 1775–1791. doi: 10.1104/pp.20.00984
Wang, S., Li, S., Liu, Q., Wu, K., Zhang, J., Wang, S., et al. (2015a). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, Z. Y., Wu, Z. L., Xing, Y. Y., Zheng, F. G., Guo, X. L., Zhang, W. G., et al. (1990). Nucleotide sequence of rice waxy gene. Nucleic Acids Res. 18:5898. doi: 10.1093/nar/18.19.5898
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012a). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Wang, Y., Xiong, G., Hu, J., Jiang, L., Yu, H., Xu, J., et al. (2015b). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948. doi: 10.1038/ng.3346
Wang, J. C., Xu, H., Zhu, Y., Liu, Q. Q., and Cai, X. L. (2013). OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 64, 3453–3466. doi: 10.1093/jxb/ert187
Wang, Z. Y., Zheng, F. Q., Shen, G. Z., Gao, J. P., Snustad, D. P., Li, M. G., et al. (1995). The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 7, 613–622. doi: 10.1046/j.1365-313x.1995.7040613.x
Wei, Z., and Li, J. (2016). Brassinosteroids regulate root growth, development, and symbiosis. Mol. Plant 9, 86–100. doi: 10.1016/j.molp.2015.12.003
Wu, Y., Fu, Y., Zhao, S., Gu, P., Zhu, Z., Sun, C., et al. (2016). Clustered primary branch 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol. J. 14, 377–386. doi: 10.1111/pbi.12391
Xu, R., Duan, P., Yu, H., Zhou, Z., Zhang, B., Wang, R., et al. (2018). Control of grain size and weight by the OsMKKK10-OsMKK4-OsMAPK6 signaling pathway in rice. Mol. Plant 11, 860–873. doi: 10.1016/j.molp.2018.04.004
Yang, D., Wu, L., Hwang, Y. S., Chen, L., and Huang, N. (2001). Expression of the REB transcriptional activator in rice grains improves the yield of recombinant proteins whose genes are controlled by a Reb-responsive promoter. Proc. Natl. Acad. Sci. U. S. A. 98, 11438–11443. doi: 10.1073/pnas.201411298
Yuan, H., Fan, S., Huang, J., Zhan, S., Wang, S., Gao, P., et al. (2017). 08SG2/OsBAK1 regulates grain size and number, and functions differently in Indica and Japonica backgrounds in rice. Rice 10:25. doi: 10.1186/s12284-017-0165-2
Zeeman, S. C., Kossmann, J., and Smith, A. M. (2010). Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61, 209–234. doi: 10.1146/annurev-arplant-042809-112301
Zhang, J., Chen, J., Yi, Q., Hu, Y., Liu, H., Liu, Y., et al. (2014). Novel role of ZmaNAC36 in co-expression of starch synthetic genes in maize endosperm. Plant Mol. Biol. 84, 359–369. doi: 10.1007/s11103-013-0153-x
Zhang, Z., Dong, J., Ji, C., Wu, Y., and Messing, J. (2019b). NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc. Natl. Acad. Sci. U. S. A. 116, 11223–11228. doi: 10.1073/pnas.1904995116
Zhang, X., Wang, J., Huang, J., Lan, H., Wang, C., Yin, C., et al. (2012). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. U. S. A. 109, 21534–21539. doi: 10.1073/pnas.1219776110
Zhang, Z., Zheng, X., Yang, J., Messing, J., and Wu, Y. (2016). Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc. Natl. Acad. Sci. U. S. A. 113, 10842–10847. doi: 10.1073/pnas.1613721113
Zhang, W., Zhu, K., Wang, Z., Zhang, H., Gu, J., Liu, L., et al. (2019a). Brassinosteroids function in spikelet differentiation and degeneration in rice. J. Integr. Plant Biol. 61, 943–963. doi: 10.1111/jipb.12722
Zhou, Y., Miao, J., Gu, H., Peng, X., Leburu, M., Yuan, F., et al. (2015). Natural variations in SLG7 regulate grain shape in rice. Genetics 201, 1591–1599. doi: 10.1534/genetics.115.181115
Zhu, X., Liang, W., Cui, X., Chen, M., Yin, C., Luo, Z., et al. (2015). Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of carbon starved anther, a MYB domain protein. Plant J. 82, 570–581. doi: 10.1111/tpj.12820
Zhu, J., Yu, W., Zhang, C., Zhu, Y., Xu, J., Li, E., et al. (2020). New insights into amylose and amylopectin biosynthesis in rice endosperm. Carbohydr. Polym. 230:115656. doi: 10.1016/j.carbpol.2019.115656
Keywords: grain size, starch biosynthesis, NAC transcription factor, brassinosteroids, abscisic acid
Citation: Jin S-K, Zhang M-Q, Leng Y-J, Xu L-N, Jia S-W, Wang S-L, Song T, Wang R-A, Yang Q-Q, Tao T, Cai X-L and Gao J-P (2022) OsNAC129 Regulates Seed Development and Plant Growth and Participates in the Brassinosteroid Signaling Pathway. Front. Plant Sci. 13:905148. doi: 10.3389/fpls.2022.905148
Edited by:
Zhong-Nan Yang, Shanghai Normal University, ChinaReviewed by:
Huadong Zhan, Nanjing Agricultural University, ChinaShiyong Sun, Henan University, China
Copyright © 2022 Jin, Zhang, Leng, Xu, Jia, Wang, Song, Wang, Yang, Tao, Cai and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiu-Ling Cai, eGxjYWlAeXp1LmVkdS5jbg==; Ji-Ping Gao, anBnYW9AeXp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Su-Kui Jin1,2†
Su-Kui Jin1,2† Li-Na Xu
Li-Na Xu Shu-Wen Jia
Shu-Wen Jia Ji-Ping Gao
Ji-Ping Gao