
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 31 May 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.905111
This article is part of the Research Topic The Plant Cell Wall: Advances and Current Perspectives View all 14 articles
 Zhenhua Chen1†
Zhenhua Chen1† Shouzhen Teng1†
Shouzhen Teng1† Di Liu1†
Di Liu1† Yuan Chang1
Yuan Chang1 Liying Zhang1
Liying Zhang1 Xuean Cui1
Xuean Cui1 Jinxia Wu1
Jinxia Wu1 Pengfei Ai2
Pengfei Ai2 Xuehui Sun1*
Xuehui Sun1* Tiegang Lu1*
Tiegang Lu1* Zhiguo Zhang1*
Zhiguo Zhang1*Leaf morphology is an important component of rice ideal plant type. To date, many regulatory genes influencing leaf morphology in rice have been cloned, and their underlying molecular regulatory mechanism has been preliminarily clarified. However, the fine regulation relationship of leaf morphogenesis and plant type remains largely elusive. In this study, a rolling-leaf mutant, named rlm1-D, was obtained and controlled by a pair of dominant nuclear genes. Cytological observations revealed that the rlm1 was mainly caused by abnormal deposition of secondary cell walls. Molecular evidence showed ectopic expression of a MYB-type transcription factor LOC_Os05g46610 was responsible for the phenotype of rlm1-D. A series of experiments, including the transcription factor-centered technology, DNA-binding assay, and electrophoretic mobility shift assay, verified that RLM1 can bind to the promoter of OsCAD2, a key gene responsible for lignin biosynthesis in rice. An interacting partner of RLM1, OsMAPK10, was identified. Multiple biochemical assays confirmed that OsMAPK10 interacted with RLM1. OsMAPK10 positively regulated the lignin content in the leaves and stems of rice. Moreover, OsMAPK10 contributes to RLM1 activation of downstream target genes. In particular, RLM1 is exclusively expressed in the stems at the mature plant stage. The yield of RLM1 knockdown lines increased by over 11% without other adverse agricultural trait penalties, indicating great practical application value. A MAPK-MYB-OsCAD2 genetic regulatory network controlling SCW was proposed, providing a theoretical significance and practical value for shaping the ideal plant type and improving rice yield.
Leaf morphology plays an important role in rice ideal plant type. Leaf morphology improvement focuses on erect leaves and moderately rolled leaves. Moderate rolling can keep rice leaves upright with no drooping, improving the light reception conditions at the base of the canopy at the middle and later stages, increase the light energy use rate, and enhancing root activity to improve lodging resistance (Zhang et al., 2009; Zou et al., 2011). Some rolling-leaf cultivars, such as Liangyoupeiyou and Miyang, have become popular with growers (Wang et al., 2003). Therefore, the study of leaf morphogenesis has become the focus of breeding for high-yield rice.
Leaf morphogenesis is a complex developmental process that is influenced by many factors. In Arabidopsis, the genetic regulatory network of leaf morphogenesis is relatively well known. HD-Zip III, YABBY, and KANADI are involved in the establishment and development of leaf polarity (Emery et al., 2003; Ha et al., 2010). By regulating the expression of HD-Zip III and ARF3/ARF4, miRNA165/166 and ta-siRNA, respectively, are also involved in the establishment and development of leaf polarity (Hunter et al., 2006; Merelo et al., 2016). To date, more than 10 regulatory genes controlling leaf morphogenesis have been cloned in rice, and their underlying molecular regulatory mechanisms have been preliminarily studied. The genes involved mainly include LC2 (Zhao et al., 2010), OsHB1 (Itoh et al., 2008), ACL1/ACL2 (Li et al., 2010), Brd1 (Hong et al., 2002), CLD1/SRL1 (Li et al., 2017); Yab1 (Dai et al., 2007), OsCSLD4/NRL1 (Hu et al., 2010), OsAGO7 (Shi et al., 2007), RL14 (Fang et al., 2012), CFL1 (Wu et al., 2011), SLL1 (Zhang et al., 2009), ROC5 (Zou et al., 2011), ROC8 (Sun et al., 2020), and PSL1 (Zhang et al., 2021). These genes mainly play a role in regulating the number and size of bulliform cells. RL14 and PSL1 positively regulate the development of bulliform cells, while Brd1, Yab1, RL14, LC2, ROC5, ROC8, OSHb1, and ACL1/ACL2 negatively regulate the development of these cells. In addition to the size and numbers of bulliform cells that cause leaf rolling in rice, the distribution of mesophyll cells, the formation of sclerenchyma cells, and the development of leaf cuticles can affect leaf rolling. SLL1 encodes a KANADI family MYB transcription factor. Mutation of SLL1 leads to a disruption of the programmed death process of abaxial mesophyll cells and inhibits the normal formation of abaxial sclerenchyma. CFL1 encodes an unknown protein with a WW domain and promotes leaf rolling. CFL1 interacts with HDG1 (HD-ZIP IV) proteins and negatively regulates the development of cuticles by affecting the expression of BDG and FDH genes related to cuticle development. Totally, leaf morphology underlying molecular regulatory mechanism has been preliminarily clarified. However, the fine regulation relationship of leaf morphogenesis and plant type remains largely elusive.
To better understand the molecular mechanism of leaf morphogenesis and plant type, the rolling-leaf mutant rlm1 was studied in depth. The rlm1 is controlled by a pair of dominant nuclear genes. Cytological observations revealed that RLM1 directly affects secondary cell wall (SCW) deposition, indirectly regulates the development of bulliform cells, and promotes leaf rolling. RLM1 was cloned by a map-based method and found to encode an R2R3 MYB transcription factor. The rlm1-D mutant phenotype was caused by the ectopic expression of the R2R3 MYB transcription factor. Combining biochemical, cytological, and genetic strategies, we proposed a new molecular regulatory network based on MAPK-MYB-OsCAD2. This study improves our understanding of the relationship between leaf morphogenesis and plant type and provides genetic resources for cultivating ideal plant-type rice materials.
The rlm1-D, WT, overexpression lines RLM1-OE1 and RLM1-OE2, mutant lines RLM1-m1 and RLM1-m2, and plants used for mapping populations were grown in the greenhouse and at a field station of the Chinese Academy of Agriculture Science. All the plants tested at the seedling stage were grown in a growth chamber with 10 h daytime/14 h night-time photoperiod at 28°C, under a light intensity of 150–200 μmol/m2/s and under 60–70% relative humidity (RH).
A mapping population was generated by the crossing of rlm1-D heterozygous plants with the Dular cultivar (Indica). Ten WT leaf samples were collected for preliminary mapping. Nearly 500 plants were used for fine mapping, and the location of RLM1 locus between Indel2670 and Indel2672 was narrowed. A T-DNA insertion was found in the promoter region of LOC_Os05g46610.
RLM1-pCAMBIA1302 vectors containing the full RLM1 ORF were constructed to reproduce the phenotype. Two Cas9 targets were selected via CRISPR-gene editing (GE),1 and RLM1-pP1C.7 was constructed. The full-length OsMAPK10 CDS was cloned into pCAMBIA1302, and two Cas9 targets were selected via CRISPR-GE to generate the OsMAPK10-pP1C.7 knockdown plasmids. All the plasmids were then transformed into the Agrobacterium AGL1 strain, which were then transformed into Nipponbare calli.
RNA samples were isolated from WT plants at the seedling and heading stages. The cDNAs were synthesized via FastKing gDNA Dispelling RT SuperMix (KR118, TIANGEN, Beijing). qRT-PCR was subsequently performed with SYBR qPCR Mix (ZF501, ZOMANBIO, Beijing) according to the manufacturer’s instructions. The housekeeping gene OsActin1 (LOC_Os03g0718100) was used as an internal control to normalize gene expression.
ProRLM1-1391Z vectors were used to determine the expression pattern, and the 2,000 bp sequence upstream was amplified from the RLM1 codon start site. The recombinant pRLM1-1391Z Agrobacterium strain was subsequently transformed into Nipponbare calli. At the seedling and heading stage, GUS staining was conducted, with 2 mM X-Gluc applied to different tissues.
The full-length CDS of RLM1 was cloned, amplified, and inserted into pAN580. The resulting RLM1-pAN580 constructs were transformed into Nipponbare protoplasts, and mCherry was selected as a nuclear marker. The signals were detected using a laser scanning microscope (LSM980, Zesis, Germany).
Stem samples from leaves or the 2nd internode were collected from rice at the heading stage. The tissues were cut to 0.5 cm and fixed in 2.5% glutaraldehyde for 12 h. The samples were prepared according to the methods of Wang et al. (2022) and then imaged with a scanning electron microscope (H-7500, Hitachi, Japan).
Samples of leaves and stems from WT, rlm1-D, RLM1-OE, RLM1-m, OsMAPK10-m, and OsMAPK10-OE plants at the heading stage were collected. The lignin and cellulose contents of 2 g samples were quantified by Wuhan Metwell Biotechnology Co., Ltd. (Wuhan, China).
Leaves from rlm1-D and WT plants were collected and fixed in Carnoy’s solution (ethanol/glacial acetic acid = 3/1) overnight. The samples were then treated according to the methods of Sun et al. (2020) and observed as previously described (Cui et al., 2019).
A BLAST search of Arabidopsis and Zea mays database was conducted, with the RLM1 protein sequence used as a query sequence. A phylogenetic tree was generated by MEGA XI software; the unrooted tree was constructed according to the neighbor-joining method.
Total RNA was extracted from WT and rlm1-D plants in the 3rd-leaf stage. Seqhealth Technology Co., Ltd. (Wuhan, China) prepared the libraries and performed high-throughput sequencing. The reference genome sequence was downloaded from the NCBI database (PRJDB1747). KEGG enrichment analysis of differentially expressed genes was implemented by KOBAS software (version 2.1.1) with a corrected p-value cut-off of 0.05 to judge statistically significant enrichment.
The full-length CDSs of RLM1 and OsMAPK10 were inserted into the EcoRI and BamHI sites of pGBKT7 (bait) and pGADT7 (prey), respectively, by an In-Fusion kit (Clontech, Japan). All the resulting constructs were verified by sequencing before being transformed into yeast strain AH109. The bait and prey plasmids were cotransformed into AH109 cells by an Ex-Yeast Transformation Kit (ZC135; ZOMANBIO, Beijing, China). The yeast colonies were transferred to plates containing SD/-Leu/-Trp and SD/-His/-Trp/-Leu/60 mM 3-AT media and then allowed to grow at 28°C for 3 days.
Pull-Down assays were performed with GST-RLM1 and His-OsMAPK10. The RLM1 and OsMAPK10 coding sequences were cloned into pGEX6p-1 or pACYCDuet1 and purified with GST-Tag or His-Tag magnetic beads, respectively. GST-RLM1 and His-MAPK10 were subsequently incubated in binding buffer [20 mM sodium dihydrogen phosphate, 300 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT; pH 7.4)] for 2 h, after which binding buffer was used to wash the beads 5 times. Then, elution buffer [20 mM sodium dihydrogen phosphate, 300 mM NaCl, 500 mM imidazole (pH 7.4)] was applied to elute the proteins for subsequent western blot assays. Anti-RLM1 and anti-GST antibodies were utilized to detect the pulled down RLM1, and anti-His antibodies were utilized to detect the input.
The RLM1 CDS was cloned into pCAMBIA1300-nLUC by an In-Fusion kit (Clontech, Japan), and the OsMAPK10 CDS was cloned into pCAMBIA1300-cLUC. The plasmid was sequenced for verification and then transformed into Agrobacterium strain GV3101. Four-week-old N. benthamiana plants were infiltrated by the transformed Agrobacterium cells and incubated at 23°C under dark conditions for 2 days; pCAMBIA1300-nLUC with pCAMBIA1330-cLUC served as a blank control. The leaves were sprayed with D-luciferin potassium salt (Yeasen, Shanghai, China), and the fluorescence was detected by an IVIS Lumina LT series III instrument (PerkinElmer).
The full-length CDS of RLM1 was inserted into the EcoRI and BamHI sites of pGADT7 by an In-Fusion kit (Clontech). The MYB1AT motif from the OsCAD2 promoter was inserted (as three tandem repeats) into pHis2 at both the EcoRI and SacI cloning sites. The plasmids were subsequently transformed into yeast strain Y187 by an Ex-Yeast Transformation Kit. The yeast colonies were transferred to plates containing SD/-Trp/-Leu and SD/-His/-Trp/-Leu media supplemented with 60 mM 3-AT and allowed to grow at 28°C for 3 days.
The direct binding of RLM1 to the OsCAD2 promoter was detected using an EMSA kit (GS009, Beyotime, Shanghai, China), following the manufacturer’s protocol, in conjunction with probes. The CDS of RLM1 was cloned into the Escherichia coli expression vector pGEX6p-1 with a GST tag at the BamHI and EcoRI sites. The resulting plasmid was then transformed into the E. coli Rosetta strain and induced with 0.8 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Recombinant GST-RLM1 was purified using GST purification magnetic beads (abs9902, Absin, Shanghai China) according to the manufacturer’s protocol. The biotin-labeled oligonucleotide probes were synthesized and labeled by Shanghai Sangon Company (Shanghai, China).
A 7 bp random motif library from yeast strain Y187 was purchased from Nanjing Ruiyuan Biotechnology Co., Ltd. (Nanjing, China). A TF-centered Y1H assay was performed as previously described (Ji et al., 2018). The yeast motif library was incubated overnight, after which an Ex-Yeast Transformation Kit was utilized to generate competent cells. Then, 35 μg of pGADT7-RLM1 was transformed into competent yeast library cells. The yeast was transferred to plates of SD/-His/-Trp/-Leu media supplemented with 60 mM 3-AT and allowed to grow at 28°C for 5 days. Monoclonal colonies were selected for sequencing, and random motif sequences between “GGG” and “CCC” (the SmaI site) were screened. The insertion sequences were analyzed using PLACE2 and PlantCARE3 to identify whether they were known motifs.
VP64 (encoding an exogenous activation domain) or EAR4 (encoding an exogenous repressor domain) was fused to the CDS of RLM1, which was then inserted into pGBKT7, and an Ex-Yeast Transformation Kit was used to transform these vectors into yeast strain AH109; the resulting yeast were then cultured on SD/-Trp media. Then, single colonies were selected and cultured on SD/-Ade-Trp-His media at 28°C for 3 days. The α-gal enzyme activity was assayed according to the Yeast Protocols Handbook (No. PT3024-1, Clontech). To finely map the active domain of OsRLM1, we divided the protein into different parts, as shown in Figure 1, and measured the α-gal enzyme activity.
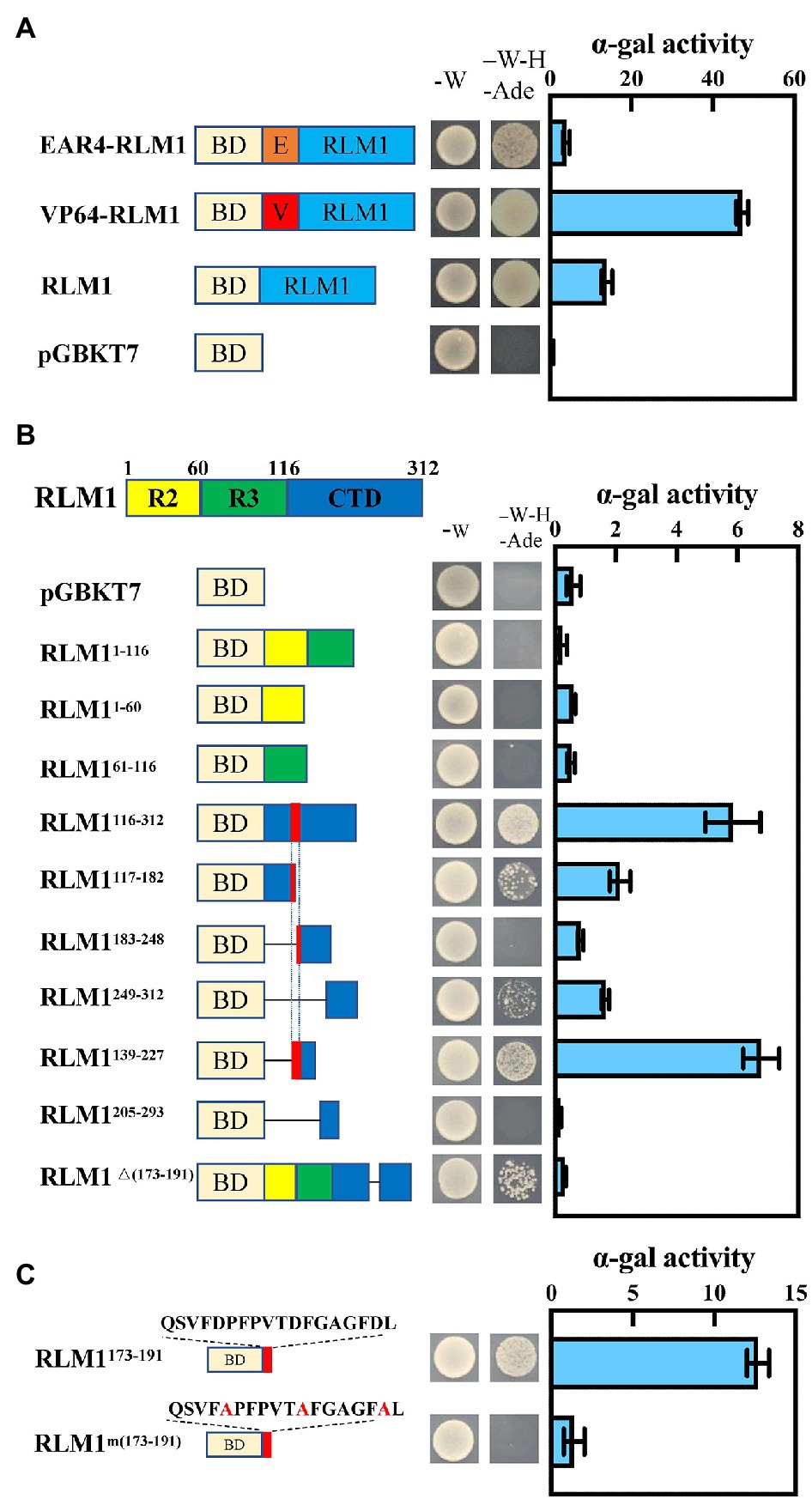
Figure 1. RLM1 acts as a transcriptional activator. (A) Transactivation analysis of RLM1, VP64-RLM1, and EAR4-RLM1 via a yeast assay. VP64 and EAR4 are an exogenous activation domain and an exogenous repression module, respectively. pGBKT7 alone was used as a negative control. Plate auxotroph and α-galactosidase assay showing the transcriptional activation of each protein. The bars represent the means ± SDs of three independent experiments. α-gal, α-galactosidase; -W, SD/-Trp; -W-H-Ade, SD/-Trp/-His/-Ade. (B) Mapping of the transactivation motif of RLM1 via a yeast assay. R2, aa 1–60; R3, aa 61–116; CTD, aa 117–312. Plate auxotroph and α-galactosidase assay showing the transcriptional activation of each protein. The bars represent the means ± SDs of three independent experiments. α-gal, α-galactosidase; pGBKT7, GAL4 DNA-binding domain; -W, SD; -W-H-Ade, SD/-Trp/-His/-Ade. (C) Transactivation analysis of the C-terminal motif QSVFDPFPVTDFGAGFDL via a yeast assay. BD represents the GAL4 DNA-binding domain. Mutations introduced into the C-terminal motif are indicated by the red font. Plate auxotroph and α-galactosidase assay showing the transcriptional activation by each protein. The bars represent the means ± SDs of three independent experiments. α-gal, α-galactosidase; -W, SD/-Trp; -W-H-Ade, SD/-Trp/-His/-Ade.
The promoter of OsCAD2, which was defined as the region 2 kb upstream from the transcription start site, was cloned and inserted into pGreenII-0800-Luc via the In-Fusion strategy to generate the corresponding reporter vector proOsCAD2-0800. The RLM1 and OsMAPK10 ORFs were cloned and inserted into pCAMBIA1307 to generate effector constructs. The resulting plasmids were transformed into rice protoplasts and cultured for 16 h. pCAMBIA1307 with pOsCAD2-0800 was used as a control. The LUC/REN ratio was determined by a Dual-Luciferase Reporter Assay System (Vazyme, DL101-01) according to the manufacturer’s instructions.
RLM1 phosphorylation by OsMAPK10 was performed with purified GST-RLM1 and His-OsMAPK10. The proteins were incubated in kinase buffer [50 mM hydroxyethyl piperazineethanesulfonic acid (HEPES; pH 7.4), 10 mM MnCl2, 1 mM DTT, 30 μM ATP] at 30°C for 30 min. The reaction was stopped, electrophoresis was performed on an SDS-PAGE gel (6% SDS, 5 μM Phos-tag, 10 μM MnCl2), and RLM1 was detected with an anti-GST antibody.
The experiments were repeated at least three times. The means with standard deviation are shown in the figures. Significant differences were determined by Student’s t test, and are marked with asterisks (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
The primers used are shown in Supplementary Table S3.
To explore the regulatory factors controlling leaf morphogenesis in rice, a rolling-leaf mutant, named rlm1-D, was obtained from a rice T-DNA mutant library (Wan et al., 2009). The rlm1-D presented dwarfism, rolling leaves, and a dark leaf color at the tillering stage (Figures 2A–C). In particular, owing to the excessive rolling of its leaves, the rlm1-D mutant did not produce seeds normally. There were three phenotypes of the rlm1-D heterozygous offspring: excessive-rolling, half-rolling, and wild-type like (WT-like) phenotypes, with which the corresponding segregation ratio was 1/2/1 (Supplementary Figure S1C). This suggests that rlm1-D may be controlled by a pair of dominant nuclear genes and that the rlm1-D gene may have a dose-dependent effect on leaf rolling.
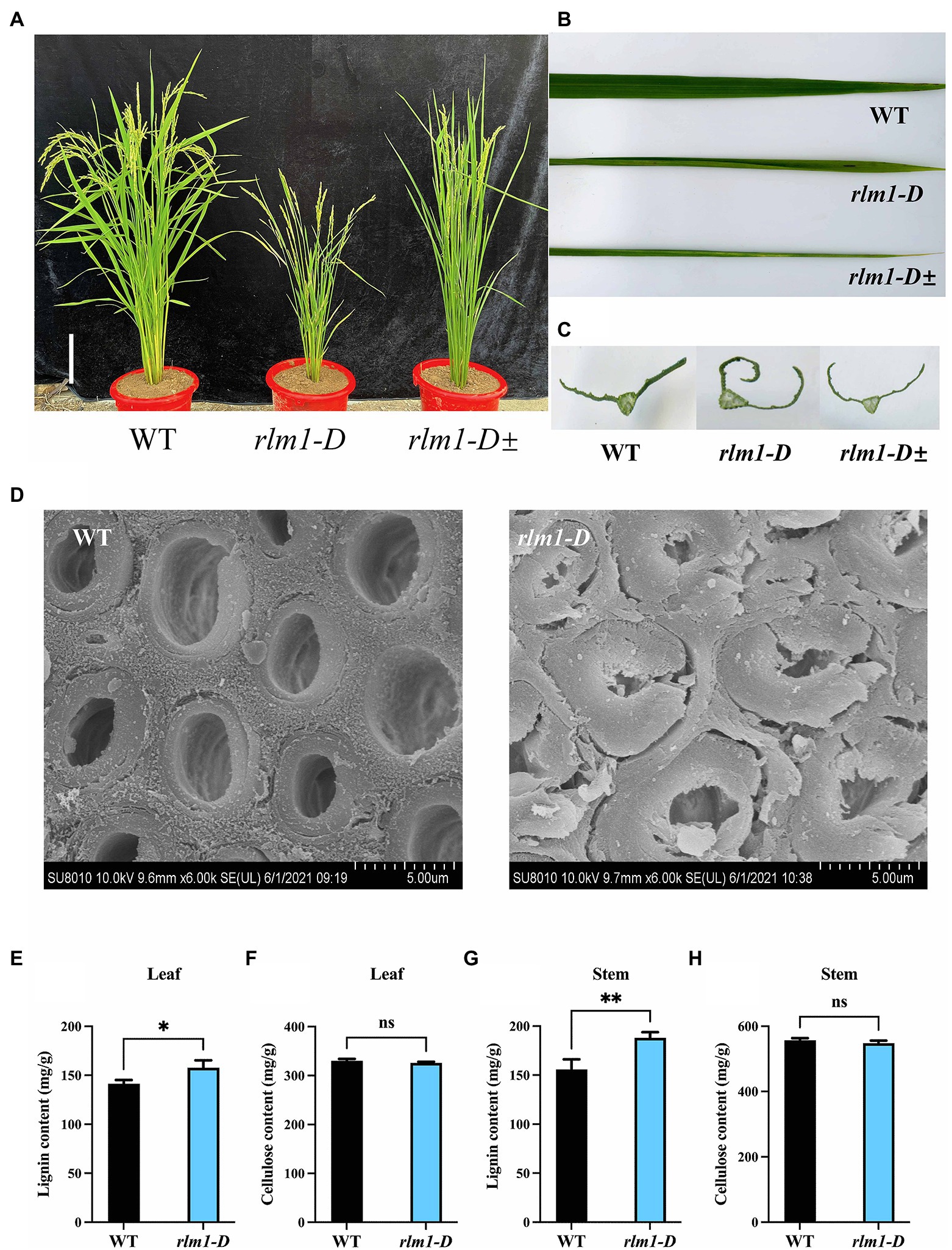
Figure 2. Clarification of the rlm1-D phenotype. (A) Phenotype comparisons of WT, rlm1-D, and rlm1 heterozygous (rlm1-D±) plants at the heading stage. Bar = 5 cm. (B) Flag leaf comparison of the plants shown (A). (C) Flag leaf cross-section comparisons of the plants shown in (A). (D) SEM observations of sclerenchyma cell walls in the internodes from the 3-month-old WT and rlm1-D plants. Bar = 5 μm. (E) Lignin content measurements in WT and rlm1-D leaf tissue. (F) Cellulose content measurements in WT and rlm1-D leaf tissue. (G) Lignin content measurements in the internodes of the 3-month-old WT and rlm1-D plants. (H) Cellulose content measurements in secondary stem tissues of WT and rlm1-D plants. *p < 0.05, **p < 0.01 (Student’s t-test).
To observe the cytological changes of rlm1-D, paraffin sectioning demonstrated that the size and number of bulliform cells in rlm1-D were significantly reduced compared to those in the WT (Supplementary Figures S1D,E). Since the rlm1-D plants exhibited a rolling-leaf phenotype accompanied by a dwarf-type phenotype, the features of rlm1-D were similar to those of the roc8 and rl14 mutants (Fang et al., 2012; Sun et al., 2020). Thus, we determined the leaf structure of rlm1-D and the WT using scanning electron microscopy (SEM). SEM results revealed that sclerenchyma cell wall (SCW) deposition was abnormal in rlm1-D internode tissues compared to WT tissues (Figure 2D). Therefore, we measured the contents of lignin and cellulose in rlm1-D and WT leaf and stem tissues. The results showed that the lignin contents of the stems and leaves of rlm1-D were significantly higher than those of the WT (Figures 2E,G). However, the cellulose content in rlm1-D was comparable to that in the WT (Figures 2F,H). The results of the abovementioned experiments indicated that abnormal SCWs in rlm1-D were perhaps the main factor responsible for leaf rolling. The abnormal SCWs in rlm1-D perhaps influenced water transport and indirectly regulated the size of bulliform cells, which promoted leaf rolling in rlm1-D, similar as rl14 mutant (Fang et al., 2012).
To clone RLM1, a genetic mapping population was generated by a cross between rlm1-D heterozygous plants and the rice indica cultivar Dular. RLM1 was preliminarily mapped to a 25.2–27.72 cM range on chromosome 5. Further fine mapping narrowed RLM1 to a 100 kb region (Figure 3A). Sequence analysis revealed a T-DNA insertion 750 bp upstream of the start codon (ATG) of LOC_Os05g46610 (Figure 3B). We speculated that the CaMV35S promoter in the T-DNA region activated LOC_Os05g46610 expression. To confirm this hypothesis, a quantitative reverse transcription PCR (qRT-PCR) experiment was performed to judge the expression levels of LOC_Os05g46570, LOC_Os05g46580, LOC_Os05g46610, LOC_Os05g46620, and LOC_Os05g46630 in rlm1-D, rlm1-D heterozygous plants and WT plants. The results demonstrated that the expression levels of the abovementioned genes (except for LOC_Os05g46610) in rlm1-D were comparable to those in the WT. The LOC_Os05g46610 expression level in rlm1-D increased by ~1,000-fold. Additionally, the expression level of LOC_Os05g46610 in the rlm1-D heterozygous plant increased to a level that was ~800-fold greater (Figures 3C–G). Overall, the map-based cloning and qRT-PCR analysis indicated that LOC_Os05g46610 was a candidate gene of RLM1.
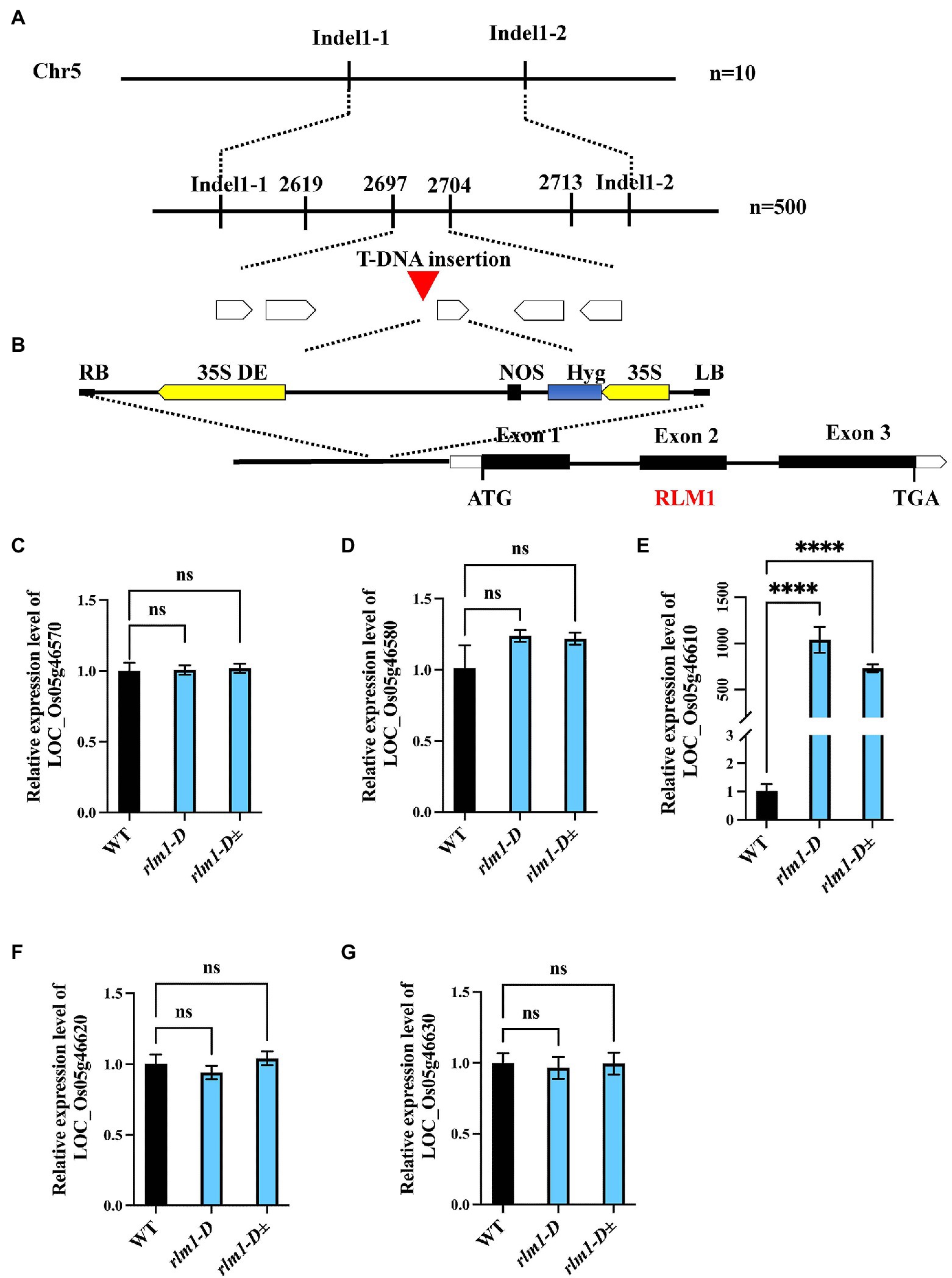
Figure 3. Map-based cloning of RLM1. (A) The RLM1 locus was mapped to a 2.5 cM interval on chromosome 5, between Indel markers Indel1-1 and Indel1-2. The location of the RLM1 locus was narrowed to a 10 kb region between insertion–deletion (InDel) markers 2,697 and 2,704 via 500 F2 WT-like plants. (B) The T-DNA insert site is located upstream of LOC_Os05g46610. The structure of RLM1 is shown. ATG and TGA, start and stop codons, respectively. Boxes, exons; lines between black boxes, introns. (C–G) Expression analysis of LOC_Os05g46570, LOC_Os05g46580, LOC_Os05g46570610, LOC_Os05g46620, and LOC_Os05g46630 in WT, rlm1-D, and heterozygous (rlm1-D±) plants. ****p < 0.0001 (Student’s t-test).
To further verify that LOC_Os05g46610 is responsible for the rlm1-D phenotype, an overexpression experiment was conducted. Agrobacterium tumefaciens strain AGL1 containing the recombinant construct (LOC_Os05g46610-pCambia1302) was used to infect the Nipponbare calli. Fifteen lines among the T1 progeny revealed rolling-leaf and dwarf-type phenotypes (Figures 4A–C). qRT-PCR analysis showed that the degree of the leaf rolling among the overexpression lines was positively correlated with the expression level of LOC_Os05g46610 (Figure 4D). SEM experiments also demonstrated that SCW deposition was abnormal in RLM1-OE1 and RLM1-OE2 (Figure 4E). Lignin and cellulose content measurements also verified the increased lignin content and normal cellulose content in RLM1-OE1 and RLM1-OE2, the contents of which were similar to those of the rlm1-D mutant (Figures 4F–I). The results of the abovementioned experiments confirmed that ectopic expression of LOC_Os05g46610 was responsible for the rolling-leaf phenotype of rlm1-D.
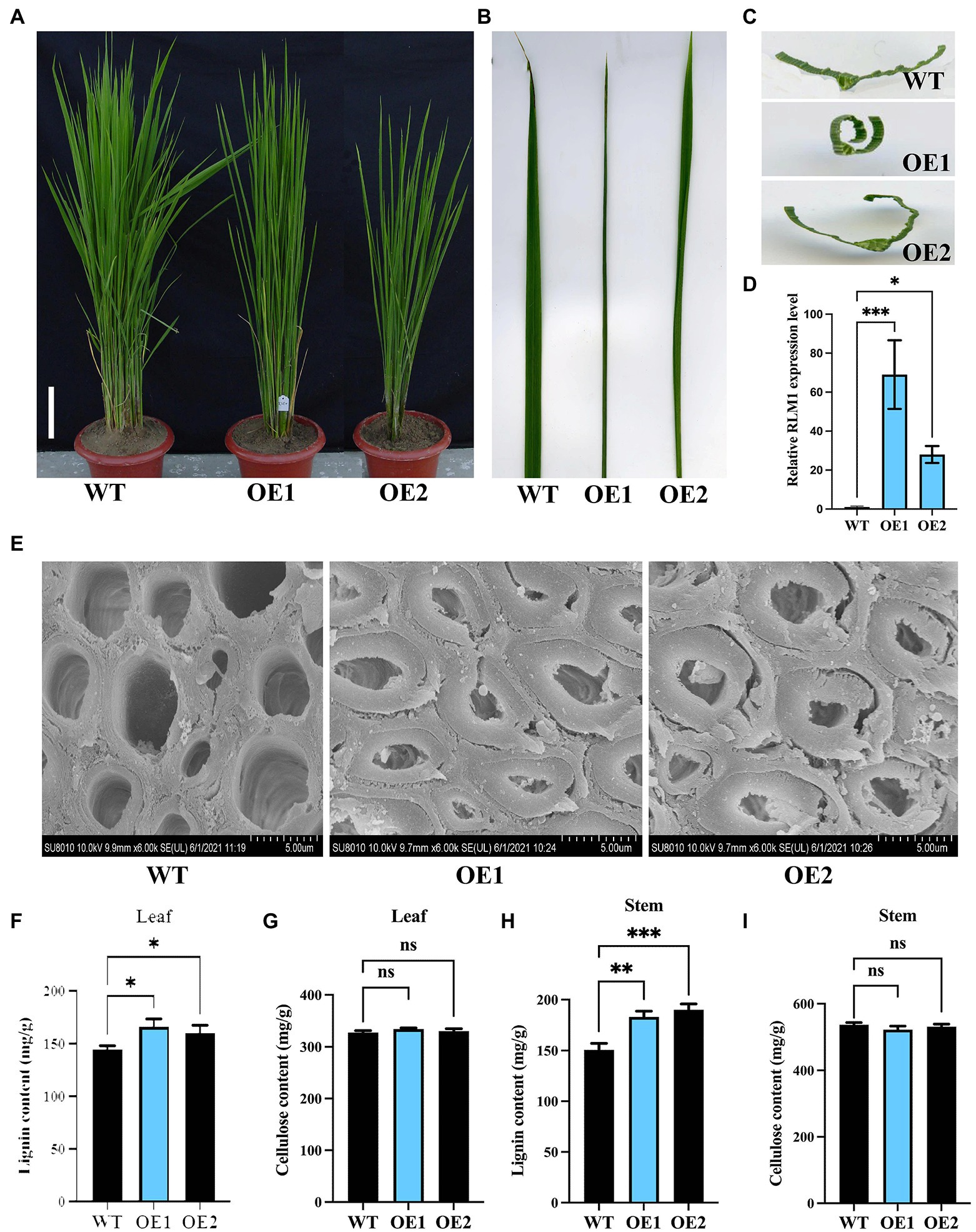
Figure 4. Analysis of RLM1 overexpression in the WT background. (A) Comparison of WT and RLM1 overexpression plants (OE1, OE2) at the late-tiller stage. Bar = 5 cm. (B) Flag leaf comparisons of the plants shown in (A). (C) Flag leaf cross-section comparisons of the plants shown in (A). (D). RLM1 expression level analysis of WT and RLM1-overexpressing plants (OE1, OE2). (E) SEM observations of sclerenchyma cell walls in the internodes from the 3-month-old WT, OE1, and OE2 plants. Bar = 5 μm. (F) Lignin content measurements in WT, OE1, and OE2 leaf tissues. (G) Cellulose content measurements in WT, OE1, and OE2 leaf tissue. (H) Lignin content measurements in the second internode tissues from 3-month-old WT, OE1, and OE2 plants. (I) Cellulose content measurements in the second internode tissues from 3-month-old WT, OE1, and OE2 plants. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t-test).
BLAST analysis demonstrated that RLM1 encoded an R2R3 MYB transcription factor. R2R3 MYB transcription factors are major transcriptional regulators that mediate a variety of plant biological processes, including seed germination, drought tolerance, stomatal conductance, lateral root development, hormone biosynthesis, anthocyanin accumulation, and cuticle wax biosynthesis (Jiang and Rao, 2020). The function of RLM1, which encodes an R2R3 MYB transcription factor, is unknown. RLM1 contains three exons and two introns, and its predicted ORF was 936 bp and encoded 312 amino acids (aa). Protein motif analysis demonstrated that RLM1 contained an N-terminal domain (R2R3) and a C-terminal domain (CTD). The N-terminus likely mainly interacts with other proteins, and the C-terminus activates or inhibits downstream target genes (Meng et al., 2019).
The amino acid sequence of RLM1 is 55% homologous with that of AtMYB4 from Arabidopsis (Supplementary Figure S2). In Arabidopsis, AtMYB4 acts as a transcriptional repressor (Jin et al., 2000). To confirm whether RLM1 has transcriptional inhibitory activity, the constructs (RLM1-pGBKT7, VP64-RLM1-pGBKT7, EAR4-RLM1-pGBKT7) were expressed in yeast, respectively. The yeast colony status on the selected media (SD/−Ade-Trp-His) showed that the activation activity of VP64-RLM1-pGBKT7, RLM1-pGBKT7, pGBKT7, and EAR4-RLM1-pGBKT7 decreased successively (Figure 1A). The above results showed that, in contrast to AtMYB4, RLM1 mainly had a transcriptional activation activity.
To further study which region of RLM1 had activation activity, the full-length RLM1 sequence was divided into three regions: R2 (RLM11–60), R3 (RLM161–116), and CTD (RLM1116–312). Then, three different constructs (R2-pGBKT7, R3-pGBKT7 and CTD-pGBKT7) were expressed in yeast, respectively. Yeast activation assays revealed that R2-pGBKT7 and R3-pGBKT7 had no activation effect in yeast growing on selective media (SD/-Ade-Trp-His). However, CTD- pGBKT7 had a stronger activation effect on yeast growing on the selective medium (SD/-Ade-Trp-His; Figure 1B).
Next, we tested the activation activities of different CTD deletion constructs. Yeast activation experiments revealed that an 18 amino acid (aa) sequence (QSVFDPFPVTDFGAGFDL) was very important for the activation activity of RLM1. Partial mutation or full deletion of these 18 aa had a great impact on the activation activity of RLM1. Moreover, the 18 aa motif alone had strong transcriptional activation activity, similar to that of the full RLM1-pGBKT7. This 18 aa motif contains some acidic amino acids, and after these acidic amino acids were mutated (Glu/Asp replaced by Ala), the activation activity was no longer detected, indicating that the acidic amino acids of these 18 aa were very important for the activation activity of RLM1 (Figure 1C).
To determine the tissue specificity of RLM1, A. tumefaciens strain containing proRLM1-1391Z construct were used to infect Nipponbare calli. β-Glucuronidase staining of T1 progeny revealed that RLM1 is mainly expressed in young seedlings. In particular, RLM1 was also specifically expressed in the stem, possibly at the site of the intermediate meristem at the mature stage, and RLM1 was not expressed in the leaf, panicle, or root tissue at the mature stage (Supplementary Figures S3A–H). The qRT-PCR experiment showed that RLM1 was highly expressed at the young seedling stage and in stem tissue at the mature stage but was not expressed in the other tissues, which is generally consistent with the GUS staining assay results (Supplementary Figure S3I).
RLM1 encodes an R2R3-type MYB transcription factor and may localize to the nucleus. To judge whether RLM1 is localized in the nucleus, a RLM1-pAN580 construct was transiently transformed into rice protoplasts. Confocal observations revealed that RLM1 indeed localized to the nucleus and acted as a transcription factor (Supplementary Figures S4A–D).
To verify the regulatory pathways in which RLM1 is involved, RNA samples from 3rd-leaf stage WT and rlm1-D seedlings were collected. Then, high-throughput sequencing and analysis were performed by Seqhealth Technology Co., Ltd. (Wuhan, China). We obtained high-quality data (BioProject accession: PRJNA813008), with 97% of the reads mapped to the reference transcriptome. A total of 1,777 genes were differentially expressed in the rlm1-D plants, of which 973 genes were upregulated and 804 genes were downregulated, which is consistent with RLM1 acting as a transcriptional activator (at least three repeats, p ≤ 0.01; Supplementary Table S1). Kyoto Encyclopedia of Gene and Genomes (KEGG) analysis demonstrated that the upregulated genes were involved in the phenylpropanoid metabolic pathway and that the downregulated genes played a role in the biosynthesis of secondary metabolites (Supplementary Figures S5A,B). We selected 19 upregulated genes involved in the phenylpropanoid metabolic pathway. The gene expression levels were confirmed by qRT-PCR, and the results were generally consistent with the transcriptome data (Supplementary Figure S6A).
To identify the target sites that may be directly regulated by RLM1, using transcription factor-centered (TF-centered) yeast one-hybrid (Y1H) technology, we screened the motifs of RLM1-binding target sites. A pHis2 yeast library containing 7 bp random fragments was constructed and screened by using RLM1-AD according to the mentioned methods (Ji et al., 2014). A total of 122 motifs were obtained to perform enrichment analysis,4 and we found that 20 motifs possibly bind to RLM1 (Supplementary Table S2). Selection on yeast screening media (SD/-Trp-Leu-His with 60 mM 3-AT) revealed that all 20 motifs could be recognized by RLM1 (Supplementary Figure S7). These motifs are mainly composed of cis-acting elements whose core sequence is “MYB1AT.” However, other cis-acting elements, such as CAATBOX1 and GATABOX, were also found.
Combined with the transcriptome sequencing results, additional results showed that these motifs were present in the promoter of genes upregulated in the phenylpropanoid metabolic pathway. Because OsCCR1 and OsCAD2 showed higher expression in rlm1-D and its promoter contained an MYB1AT motif (Supplementary Figures S6B,C) and OsCAD2 was responsible for rice lignin synthesis (Ookawa et al., 2014). We selected OsCAD2 gene for further study.
Y1H assays confirmed that RLM1 could recognize and bind to the promoter of OsCAD2 (Figures 5A,B). A DNA-binding assay was performed to detect whether RLM1 can bind to the OsCAD2 promoter region in vitro. The results revealed that RLM1 fused to a glutathione S-transferase (GST) tag could bind to the MYB1AT motif, while the binding was significantly weakened in a mutant MYB1AT motif (TAACCATG to TAATTATA; Figure 5C). Next, EMSA experiment was used to judge the RLM1 binding to the promoter of OsCAD2. We incubated GST-RLM1 proteins together with a biotin-labeled probe. The protein-DNA complex showed an obvious band shift, but the mutant probe did not (Figure 5D). Thus, these results verified that RLM1 could bind to the OsCAD2 promoter containing the MYB1AT motif. In addition, a luciferase (LUC) activity assay was used to determine whether RLM1 can activate OsCAD2. In rice protoplasts cotransfected with the effector and reporter vectors, the ratio of firefly LUC to Renilla luciferase (REN) of the effector OsCAD2 pro-LUC was fourfold higher than that of the empty vector control (Figure 5E).
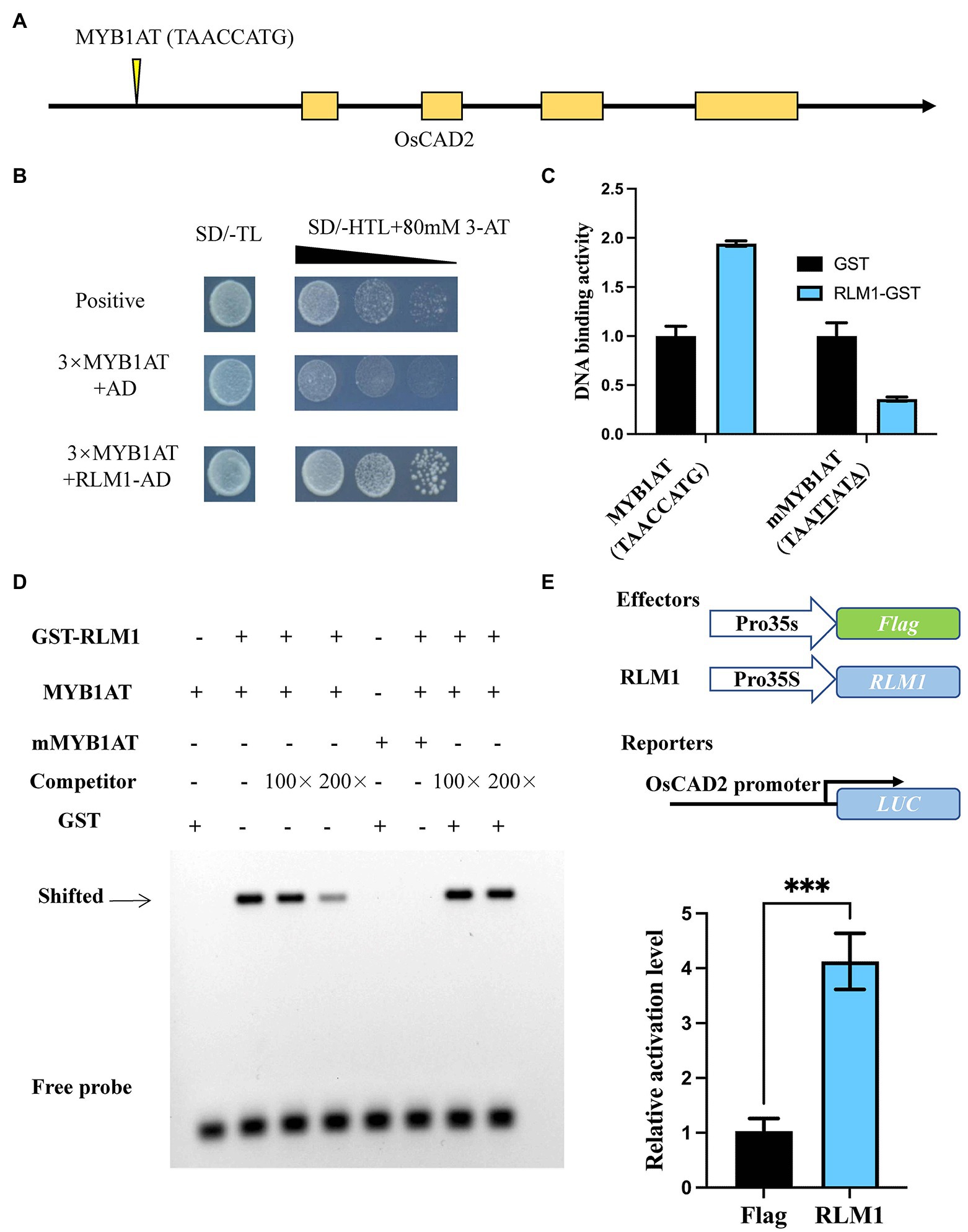
Figure 5. RLM1 binds to the motif of OsCAD2 and activates its expression. (A) The MYB1AT motif box is located approximately 1,931–1,938 bp upstream of the OsCAD2 start codon. (B) A Y1H assay was used to test RLM1 binding to the MYB1AT motif. An AD-only vector was used as a control. (C) DNA-protein binding assay revealing that RLM1 binds to the MYB1AT motif in the OsCAD2 promoter. MYB1AT, TAACCATG; mMYB1AT, TAATTATA. (A) Schematic diagram of OsCAD2. MYB1AT (TAACCATG) indicates the position in the OsCAD2 promoter. (B) Y1H assays show that RLM1 can bind to the MYB1AT motif, compared with the binding of the negative and positive controls. (C) DNA-binding activity of each group. GST was used as a negative control. The data are presented as the means ± SDs (n = 3). (D) EMSA shows that the GST-RLM1 recombinant protein directly binds to the biotin-labeled probe of the OsCAD2 promoter fragment. The signal in the RLM1 group and the MYB1AT motif in the OsCAD2 promoter shifted, but no signal shift was found in the RLM1 or mutant MYB1AT group. The shifted signal became increasingly weaker and eventually was no longer detected when unlabeled competitive probes were added. MYB1AT, TAACCATG; mMYB1AT, TAATTATA. (E) RLM1 induces proOsCAD2::LUC signaling. LUC activity was detected in rice protoplasts. The expression of REN was used as an internal control. The LUC/REN ratio indicates the expression level of the promoter (n = 3). ***p < 0.001 (Student’s t-test).
To clarify the regulatory network of RLM1, a yeast library screening assay was conducted. First, a self-activation test of RLM1-pGBKT7 was performed. We found that RLM1-pGBKT7 could not grow in selective media (SD/-Trp-His-Leu + 60 mM 3-AT). Then, the yeast screening library assay was carried out with a concentration of 60 mM 3-AT, and multiple clones were obtained by screening and sequencing. OsMAPK10 was repeated at least five times (Figure 6A). Through the RiceXPro database, we found that OsMAPK10 had a similar expression pattern as RLM1 (Supplementary Figures S8A,B). We subsequently carried out a follow-up study of OsMAPK10.
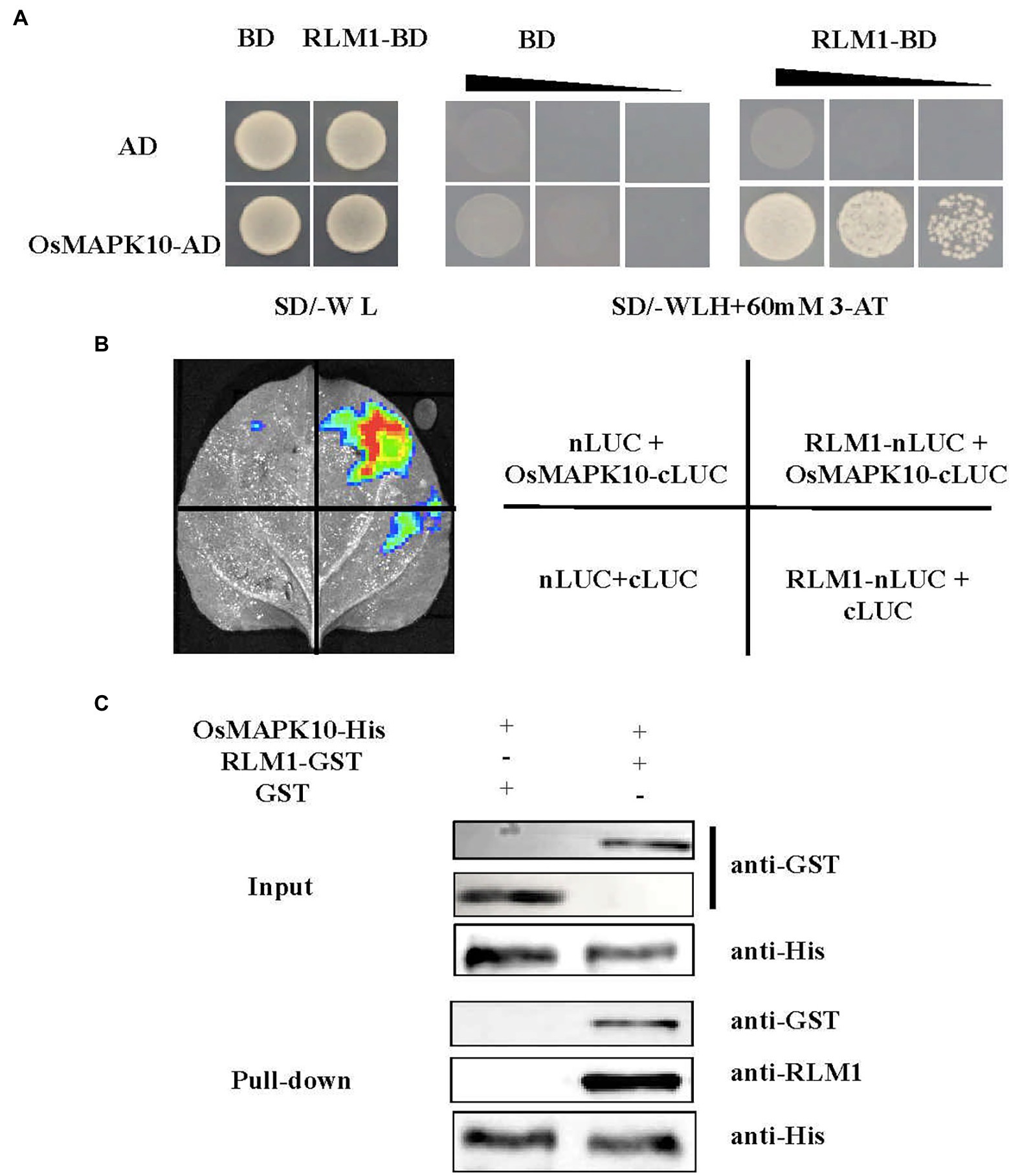
Figure 6. RLM1 physically interacts with OsMAPK10. (A) A Y2H interaction assay was performed between RLM1 and OsMAPK10. A pGADT7 vector (AD) was used as the negative control. RLM1 was fused to the pGBKT7 vector (RLM1-BD). OsMAPK10 proteins were fused to the pGADT7 vector (OsMAPK10-AD). (B) The full OsMAPK10 protein was fused to the pCAMBIA1300-cLUC vector (OsMAPK10-cLUC). The full RLM1 protein was fused to pCAMBIA1300-nLUC (RLM1-nLUC). Split-LUC complementation assay showing that RLM1 can interact with OsMAPK10 in cells of N. benthamiana leaves. The LUC signals were not detected in the corresponding negative controls. (C) Pull-Down assays of RLM1 with OsMAPK10 in vitro. RLM1-GST was incubated together with OsMAPK10-His beads, and GST proteins were used as controls. Western bolt was detected via anti-GST, anti-RLM1, and anti-his antibodies, respectively. Compared with the input and Pull-Down band results, the results showed that GST did not interact with OsMAPK10, but RLM1 interacted with OsMAPK10.
Three experiments were applied to verify the relationship between OsMAPK10 and RLM1. First, the full-length OsMAPK10 coding sequence was amplified and fused to pGADT7, and the yeast two-hybrid (Y2H) assay showed that OsMAPK10 interacted with RLM1-pGBKT7 in selective media (SD/-Trp-His-Leu + 60 mM 3AT; Figure 6A). Second, we used the split-LUC complementation experiment to judge the interaction of both. The pCAMBIA1300-nLUC (RLM1-nLUC), and pCAMBIA1300-cLUC (OsMAPK10-cLUC) were transformed into Agrobacterium GV3101, respectively. The different combinations were injected into tobacco, and LUC fluorescence was detected. The results suggested that RLM1-nLUC and MAPK10-cLUC could interact, causing LUC to be expressed and fluoresce (Figure 6B). Third, the pull-down assay also confirmed the interaction of OsMAPK10 and RLM1. The GST-RLM1 and GST proteins were incubated with beads bound to His-MAPK10, washed several times after incubation, denatured, detected via western blotting, and hybridized to anti-GST and anti-RLM1 antibodies. We found that GST-RLM1 proteins could be detected, but GST proteins could not be (Figure 6C). Taken together, the results of the three above mentioned experiments proved that RLM1 interacts with OsMAPK10.
To further study whether OsMAPK10 participates in leaf morphogenesis, OsMAPK10 knockout lines were created via gene-editing technology. The mutation of knockout line 1 (OsMAPK10-m1) occurred in the second exon, and 10 bp was deleted. The mutation of knockout line 2 (OsMAPK10-m2) occurred in the second exon, and 1 bp was deleted (Supplementary Figure S9A). Both mutations resulted in frameshift mutations (Supplementary Figure S9B). qRT-PCR analysis demonstrated that the expression level of OsMAPK10 reached a lower level than it did in the WT (Supplementary Figure S9D). The OsMAPK10-m1 and OsMAPK10-m2 lines were considered knockdown mutants. The OsMAPK10-m1 and OsMAPK10-m2 lines had a dwarf-type phenotype in the field (Supplementary Figure S9C). There were lower lignin contents in the leaves and stems in the OsMAPK10-m1 and OsMAPK10-m2 lines compared to the WT (Supplementary Figures S9E–H). Overexpression of OsMAPK10 driven by CaMV35S resulted in increased lignin contents in the leaves and stems of OE1 and OE2, similar to those of rlm1-D (Supplementary Figures S10A–G). The knockdown and overexpression experiments of OsMAPK10 showed that OsMAPK10 may directly participate in SCW deposition.
Three experiments were performed to study the relationship between OsMAPK10 and RLM1. MAPK family proteins mainly function by phosphorylating interacting partners. To judge whether OsMAPK10 phosphorylates RLM1, His-OsMAPK10 and GST-RLM1 fusion constructs were transformed into the E. coli Rosetta strain (DE3). Both fusion proteins were incubated and hybridized with GST antibody with or without the addition of a Phos-tag. The results demonstrated that when Phos-tag was added to the SDS-PAGE gel, the hybridization band was lagged without the addition of the Phos-tag (Supplementary Figure S11A). These results indicated that RLM1 could be phosphorylated by OsMAPK10. A LUC activity assay was applied to detect whether OsMAPK10 can facilitate RLM1 activation of downstream target genes. In rice protoplasts cotransfected with the effector and reporter vectors, the ratio of LUC to REN of the effector pOsCAD2-LUC was threefold higher than that of the empty vector control. While OsMAPK10 and RLM1 were transferred into rice protoplast together, the ratio of LUC to REN of the effector pOsCAD2-LUC was sixfold higher than that of the empty vector control (Supplementary Figure S11B). The above experiments preliminarily suggested that RLM1 is phosphorylated by OsMAPK10 and that phosphorylated RLM1 may activate the expression of downstream target genes.
To explore the function of RLM1, two RLM1-deletion mutant lines (rlm1-m1 and rlm1-m2) were generated using CRISPR editing technology. The rlm1-m1 line and rlm1-m2 line contained a 69 bp deletion in the second exon of RLM1 (from 199 to 267 bp) and a 1 bp deletion in the second exon of RLM1 (at 203 bp), respectively (Figures 7A,B). The qRT-PCR of rlm1-m1 and rlm1-m2 plants showed that the expression level of RLM1 was reduced significantly in the rlm1-m1 and rlm1-m2 plants, which indicated that both plants were knockdown mutants (Figure 7D). The leaf morphology phenotypes of the two knockdown lines were similar to that of the WT (Figure 7C). Because RLM1 was expressed in the stems only at maturity, SEM observation was applied, which revealed that the SCWs in the rlm1-m1 and rlm1-m2 stems were significantly thinner than those in the WT (Figure 7K). The result in the rlm1-m1 and rlm1-m2 stems was just contrary to the cytological observation of RLM1 overexpression lines. The lignin content of the rlm1-m1 and rlm1-m2 stems also decreased slightly compared to that of the WT (Figure 7G). However, the cellulose and lignin contents of the leaves of the rlm1-m1 and rlm1-m2 plants did not change significantly (Figures 7E,F,H). Moreover, the rlm1-m1 and rlm1-m2 had more grains per panicle (Figure 7I), and the yield per plant increased by 11% compared with that of the WT (Figure 7L), whereas the 1,000 grain weight in rlm1-m1 and rlm1-m2 were similar to WT (Figure 7J).
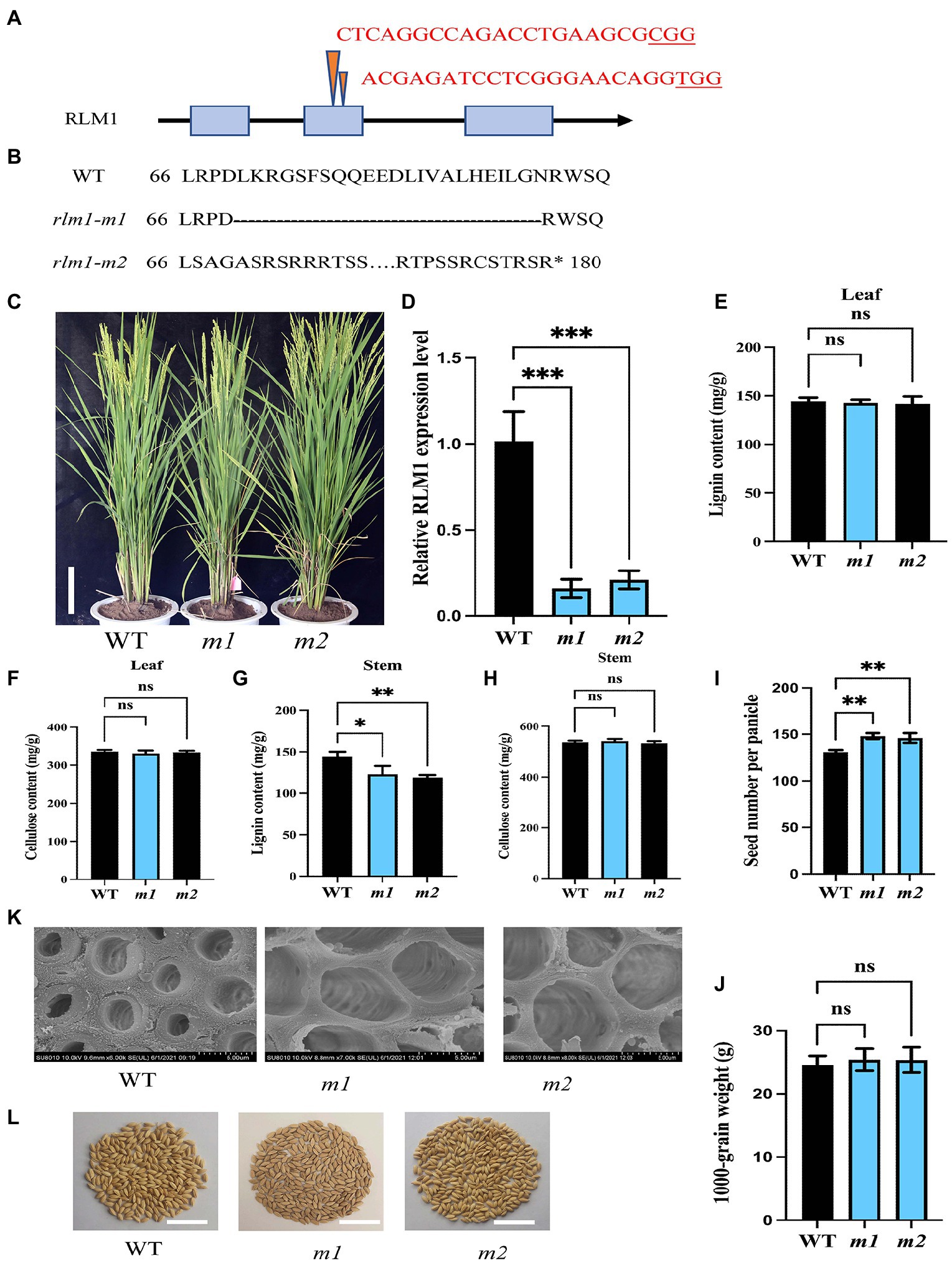
Figure 7. CRISPR-Cas9-induced mutations in RLM1. (A) Schematic map of the genomic region of RLM1 and the sgRNA target site. The arrow shows the sgRNA target site of the RLM1 genomic sequence, and the PAM motif (NGG) is shown in red. The blue boxes represent RLM1 exons, and the black lines indicate intron sequences. (B) Amino acid alignment surrounding the sgRNA target region showing the predicted peptide sequence of the WT and mutant alleles. The site of the predicted frame shifted sequence is underlined, and new stop codons are indicated by asterisks. (C) Phenotypes of the WT and homozygous mutants rlm1-m1 and indicated rlm1-m2 at the heading stage (Bar = 5 cm). (D) qRT-PCR analysis showing RLM1 expression levels in WT plants and rlm1-m1 and rlm1-m2 homozygous mutant plants. OsActin1 was selected as an internal reference. (E) Lignin content measurements in WT, rlm1-m1, and rlm1-m2 leaf tissue. (F) Cellulose content measurements in WT, rlm1-m1, and rlm1-m2 leaf tissue. (G) Lignin content measurements in the second internodes from 3-month-old WT, rlm1-m1, and rlm1-m2 plants. (H) Cellulose content measurements in the second internodes from 3-month-old WT, rlm1-m1, and rlm1-m2 plants. (I) Comparison of seed number per panicle among WT, rlm1-m1, and rlm1-m2 plants (n = 15). (K) SEM observations of sclerenchyma cell walls in the internodes from the 3-month-old WT, rlm1-m1, and rlm1-m2 plants. Bar = 5 μm. (J) Comparison of 1,000-grain weight among WT, rlm1-m1, and rlm1-m2 plants (n = 15). (L) Seeds per panicle of WT, rlm1-m1, and rlm1-m2. Bar = 5 cm. The bars represent the SDs of the means. The student’s t-test was performed to determine the significance: *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t-test).
Rice yield is mainly determined by leaf morphology and plant type. At present, many genes controlling leaf morphogenesis have been identified. However, how to understand the regulation relationship of leaf morphogenesis and plant type remains unclear. In this study, by screening a T-DNA insertion mutant population, we obtained a rolling-leaf mutant, rlm1-D, and cloned the RLM1 gene via the map-based cloning method. RLM1 encodes a typical R2R3 MYB transcription factor, and we proposed a preliminary a regulatory network based on cytological, biochemical, and genetic evidence. First, most of the reported genes, such as LC2 (Zhao et al., 2010), OsHB1 (Itoh et al., 2008), ACL1/ACL2 (Li et al., 2010), BRD1 (Hong et al., 2002), CLD1/SRL1 (Li et al., 2017), YAB1 (Dai et al., 2007), OsCSLD4/NRL1 (Hu et al., 2010), and OsAGO7 (Shi et al., 2007), directly regulate the development of bulliform cells and promote leaf rolling. A series of experiments, such as those involving paraffin sectioning, SEM observations, and measurements of cellulose and lignin contents, confirmed that the abnormal SCW disposition in rlm1-D plants was the direct cause of the rlm1-D rolling-leaf and dwarf-type phenotypes, and the reduction in the number and size of bulliform cells in rlm1-D was a secondary effect; these mechanisms were different from that governing the rolling-leaf phenotype. This mechanism controlling leaf morphology in rlm1-D was similar to that involving RL14 (Fang et al., 2012) and ROC8 (Sun et al., 2020). Second, multiple conclusions supported the rlm1-D was caused by the ectopic expression of RLM1. The expression level analysis showed that RLM1 is highly expressed in all parts of rlm1-D mutant (Supplementary Figure S12). Overexpression of RLM1 in Nipponbare derived from CaMV35S reproduces the rolling-leaf and dwarf-type phenotype, similar to the rlm1-D mutant phenotype. There was a dose-dependent effect on RLM1 expression due to heterologous activation of RLM1. Third, the biochemical analysis of RLM1 also confirmed that RLM1 had transcriptional activity, and this activity was determined by the CTD motif. Further experiments involving the deletion of the CTD motif determined that 18 aa was important for RLM1 transcriptional activity. Fourth, transcriptome analysis of rlm1-D and WT plants confirmed that RLM1 regulated the phenylpropanoid pathway. qRT-PCR, Y1H assay, and electrophoretic mobility shift assay (EMSA) experiments determined that RLM1 targets the promoter of OsCAD2 responsible for lignin synthesis by binding the MYB1AT motif. Fifth, Expression pattern analysis at the young seedling and mature stages suggested that RLM1 was mainly expressed in young seedlings and in mature stems, not in the other tissues of mature plants. The temporal and spatial expression specificity of RLM1 determines the role of RLM1 in leaf morphology and plant type. Totally, the above five aspects support that RLM1 influences SCW deposition by regulating lignin synthesis in leaves and stems in rice.
The components of the cell wall include cellulose, hemicellulose, and lignin, which provide support and defensive ability to plants. Together, the components of the cell wall also play the most important role in terms of biomass energy, degradation, and the transformation of straw. Therefore, the pyramidal hierarchy of SCW disposition regulation was determined in model plant species. NACs are the top-layer transcription factors for SCW deposition, and MYB transcription factors constitute the most important hub (Wang et al., 2021). Many MYBs in rice have been found to be involved in regulating the development of the SCW. OsMYB103L mediates cellulose biosynthesis and secondary wall formation in leaves and stems mainly by directly binding the promoters of CESA4, CESA7, CESA9, and BC1 and regulating their expression (Ye et al., 2015). NAC29/31 directly regulates OsMYB61, which in turn activates CESA expression (Ye et al., 2018). However, the function of RLM1 was different from that previously reported for OsMYB61 and OsMYB103L. We found that RLM1 regulates SCW deposition by regulating lignin synthesis. Lignin synthesis is specifically initiated in the cells that form SCWs, providing mechanical support for the upright growth of plants, helping to establish long-distance transport channels for water and materials, and strengthening the stress resistance barrier of plants. Notably, AtMYB4 is highly homologous to RLM1. AtMYB4 was shown to regulate the accumulation of the UV protectant compound sinapoyl malate through its ability to repress the transcription of the gene encoding the phenylpropanoid pathway enzyme cinnamate 4-hydroxylase (Jin et al., 2000). RLM1 acts as an activator, and AtMYB4 acts as a repressor. The opposite function of the two depends on 18 aa.
As components of complexes, MAPKs and MYBs participate in different biological development processes in plants. Mitogen-activated protein kinase 6 negatively regulates SCW biosynthesis by modulating MYB46 protein stability in Arabidopsis (Im et al., 2021). MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis (Li et al., 2016). However, the biological functions of MAPK and MYB complexes remain unknown in rice. OsMAPK10 regulates RLM1 based on the following considerations. Like that of RLM1, the expression of OsMAPK10 occurs predominantly in the stems. Y2H, split-LUC complementation, and pull-down assays confirmed the interaction between OsMAPK10 and RLM1. OsMAPK10 can phosphorylate RLM1. LUC activity assays revealed that OsMAPK10 facilitates the activation of downstream target genes by RLM1. We will study the specific phosphorylation sites in the future. Nevertheless, the knockdown lines of OsMAPK10 displayed a semi dwarf-type phenotype, and knockdown lines of RLM1 showed a normal-leaf phenotype, indicating that OsMAPK10 is perhaps involved in other plant development processes. The above mentioned findings suggest that a OsMAPK10-RLM1-OsCAD2 regulatory network controls leaf morphogenesis and plant type. However, the RLM1 phosphorylation sites modified by OsMAPK10 still need to be identified.
Temporal and spatial expression specificity of RLM1 indicates that RLM1 has potential application in production. In the RLM1 knockout lines, the morphological development of the leaves is not affected, but the SCW deposition of stems is modified. The aperture around stem SCW became bigger and was conducive to more water and mineral transport. Thus, there is reasonable to increase the productivity in RLM1 knockout lines. RLM1 perhaps coordinates the balance of C and N, which is very similar to OsMYB61 (Gao et al., 2020). RLM1 is highly expressed in the stems, and ectopic expression of RLM1 affects both leaf morphology and plant height, indicating that RLM1 perhaps participates in the gibberellin (GA) biosynthetic or signaling pathway. GA acts as the key hub between nitrogen and carbon (Gao et al., 2020), and RLM1 may coordinate the nitrogen and carbon balance in plants.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA813008.
ZC and ST conceived the main study. ZZ and YC obtained the mutant and constructed the genetic population. PA and JW constructed the genetic transformation. XC, XS, and DL constructed the expression analysis and sub-cell localization. LZ constructed the yeast interaction. ZZ, XC, and ZC contributed to the data analysis. ZZ and TL designed and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the National Key Research and Development Program of China (2020YFA0907603 and 2020YFE0202300), NSFC (32001532), NSFC-CGIAR (31861143006), the Agricultural Science and Technology Innovation Program for TL, XC, and ZZ support, and the Fundamental Research Funds for Central Non-Profit of Institute of Crop Sciences for TL, XC, and ZZ support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.905111/full#supplementary-material
Supplementary Table S1 | Up regulated genes and down regulated genes in WT and rlm1-D lists.
Supplementary Table S2 | List of RLM1 binding motifs.
2. ^http://www.dna.affrc.go.jp/PLACE/
3. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
Cui, X., Zhang, Z., Wang, Y., Wu, J., Han, X., Gu, X., et al. (2019). TWI1 regulates cell-to-cell movement of OSH15 to control leaf cell fate. New Phytol. 221, 326–340. doi: 10.1111/nph.15390
Dai, M., Hu, Y., Zhao, Y., Liu, H., and Zhou, D. X. (2007). A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 144, 380–390. doi: 10.1104/pp.107.095737
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A., et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. doi: 10.1016/j.cub.2003.09.035
Fang, L., Zhao, F., Cong, Y., Sang, X., Du, Q., Wang, D., et al. (2012). Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol. J. 10, 524–532. doi: 10.1111/j.1467-7652.2012.00679.x
Gao, Y., Xu, Z., Zhang, L., Li, S., Wang, S., Yang, H., et al. (2020). MYB61 is regulated by GRF4 and promotes nitrogen utilization and biomass production in rice. Nat. Commun. 11:5219. doi: 10.1038/s41467-020-19019-x
Ha, C. M., Jun, J. H., and Fletcher, J. C. (2010). Control of Arabidopsis leaf morphogenesis through regulation of the YABBY and KNOX families of transcription factors. Genetics 186, 197–206. doi: 10.1534/genetics.110.118703
Hong, Z., Ueguchi-Tanaka, M., Shimizu-Sato, S., Inukai, Y., Fujioka, S., Shimada, Y., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32, 495–508. doi: 10.1046/j.1365-313x.2002.01438.x
Hu, J., Zhu, L., Zeng, D., Gao, Z., Guo, L., Fang, Y., et al. (2010). Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating LEAF morphology and plant architecture in rice. Plant Mol. Biol. 73, 283–292. doi: 10.1007/s11103-010-9614-7
Hunter, C., Willmann, M. R., Wu, G., Yoshikawa, M., De La Luz Gutiérrez-Nava, M., and Poethig, S. R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133, 2973–2981. doi: 10.1242/dev.02491
Im, J. H., Ko, J. H., Kim, W. C., Crain, B., Keathley, D., and Han, K. H. (2021). Mitogen-activated protein kinase 6 negatively regulates secondary wall biosynthesis by modulating MYB46 protein stability in Arabidopsis thaliana. PLoS Genet. 17:e1009510. doi: 10.1371/journal.pgen.1009510
Itoh, J., Hibara, K., Sato, Y., and Nagato, Y. (2008). Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 147, 1960–1975. doi: 10.1104/pp.108.118679
Ji, X., Wang, L., Nie, X., He, L., Zang, D., Liu, Y., et al. (2014). A novel method to identify the DNA motifs recognized by a defined transcription factor. Plant Mol. Biol. 86, 367–380. doi: 10.1007/s11103-014-0234-5
Ji, X., Wang, L., Zang, D., and Wang, Y. (2018). “Transcription factor-centered yeast one-hybrid assay,” in Two-Hybrid Systems: Methods and Protocols. ed. L. Oñate-Sánchez (New York, NY: Springer New York).
Jiang, C. K., and Rao, G. Y. (2020). Insights into the diversification and evolution of R2R3-MYB transcription factors in plants. Plant Physiol. 183, 637–655. doi: 10.1104/pp.19.01082
Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Li, L., Shi, Z. Y., Li, L., Shen, G. Z., Wang, X. Q., An, L. S., et al. (2010). Overexpression of ACL1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol. Plant 3, 807–817. doi: 10.1093/mp/ssq022
Li, S., Wang, W., Gao, J., Yin, K., Wang, R., Wang, C., et al. (2016). MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28, 2866–2883. doi: 10.1105/tpc.16.00130
Li, W. Q., Zhang, M. J., Gan, P. F., Qiao, L., Yang, S. Q., Miao, H., et al. (2017). CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J. 92, 904–923. doi: 10.1111/tpj.13728
Meng, Y., Wang, Z., Wang, Y., Wang, C., Zhu, B., Liu, H., et al. (2019). The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40-1 to regulate carotenoid-derived flower pigmentation in Medicago truncatula. Plant Cell 31, 2751–2767. doi: 10.1105/tpc.19.00480
Merelo, P., Ram, H., Pia Caggiano, M., Ohno, C., Ott, F., Straub, D., et al. (2016). Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc. Natl. Acad. Sci. U. S. A. 113, 11973–11978. doi: 10.1073/pnas.1516110113
Ookawa, T., Inoue, K., Matsuoka, M., Ebitani, T., Takarada, T., Yamamoto, T., et al. (2014). Increased lodging resistance in long-culm, low-lignin gh2 rice for improved feed and bioenergy production. Sci. Rep. 4:6567. doi: 10.1038/srep06567
Shi, Z., Wang, J., Wan, X., Shen, G., Wang, X., and Zhang, J. (2007). Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226, 99–108. doi: 10.1007/s00425-006-0472-0
Sun, J., Cui, X., Teng, S., Kunnong, Z., Wang, Y., Chen, Z., et al. (2020). HD-ZIP IV gene Roc8 regulates the size of bulliform cells and lignin content in rice. Plant Biotechnol. J. 18, 2559–2572. doi: 10.1111/pbi.13435
Wan, S., Wu, J., Zhang, Z., Sun, X., Lv, Y., Gao, C., et al. (2009). Activation tagging, an efficient tool for functional analysis of the rice genome. Plant Mol. Biol. 69, 69–80. doi: 10.1007/s11103-008-9406-5
Wang, Y., Sun, J., Deng, C., Teng, S., Chen, G., Chen, Z., et al. (2022). Plasma membrane-localized SEM1 protein mediates sugar movement to sink rice tissues. Plant J. 109, 523–540. doi: 10.1111/tpj.15573
Wang, Y., Yu, W., Ran, L., Chen, Z., Wang, C., Dou, Y., et al. (2021). DELLA-NAC interactions mediate GA signaling to promote secondary Cell Wall formation in cotton stem. Front. Plant Sci. 12:655127. doi: 10.3389/fpls.2021.655127
Wang, R., Zhang, Y., Qian, L., and Yu, J. (2003). Photooxidation characteristics of super-hybrid rice “Liangyoupeijiu” and its parents. Ying Yong Sheng Tai Xue Bao 14, 1309–1312.
Wu, R., Li, S., He, S., Wassmann, F., Yu, C., Qin, G., et al. (2011). CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23, 3392–3411. doi: 10.1105/tpc.111.088625
Ye, Y., Liu, B., Zhao, M., Wu, K., Cheng, W., Chen, X., et al. (2015). CEF1/OsMYB103L is involved in GA-mediated regulation of secondary wall biosynthesis in rice. Plant Mol. Biol. 89, 385–401. doi: 10.1007/s11103-015-0376-0
Ye, Y., Wu, K., Chen, J., Liu, Q., Wu, Y., Liu, B., et al. (2018). OsSND2, a NAC family transcription factor, is involved in secondary cell wall biosynthesis through regulating MYBs expression in rice. Rice 11:36. doi: 10.1186/s12284-018-0228-z
Zhang, G., Hou, X., Wang, L., Xu, J., Chen, J., Fu, X., et al. (2021). PHOTO-SENSITIVE LEAF ROLLING 1 encodes a polygalacturonase that modifies cell wall structure and drought tolerance in rice. New Phytol. 229, 890–901. doi: 10.1111/nph.16899
Zhang, G. H., Xu, Q., Zhu, X. D., Qian, Q., and Xue, H. W. (2009). SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21, 719–735. doi: 10.1105/tpc.108.061457
Zhao, S. Q., Hu, J., Guo, L. B., Qian, Q., and Xue, H. W. (2010). Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res. 20, 935–947. doi: 10.1038/cr.2010.109
Keywords: rice, RLM1, R2R3 MYB transcription factor, secondary cell wall, yield
Citation: Chen Z, Teng S, Liu D, Chang Y, Zhang L, Cui X, Wu J, Ai P, Sun X, Lu T and Zhang Z (2022) RLM1, Encoding an R2R3 MYB Transcription Factor, Regulates the Development of Secondary Cell Wall in Rice. Front. Plant Sci. 13:905111. doi: 10.3389/fpls.2022.905111
Received: 26 March 2022; Accepted: 02 May 2022;
Published: 31 May 2022.
Edited by:
Wagner Rodrigo De Souza, Federal University of ABC, BrazilReviewed by:
Ruibo Hu, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), ChinaCopyright © 2022 Chen, Teng, Liu, Chang, Zhang, Cui, Wu, Ai, Sun, Lu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuehui Sun, c3VueHVlaHVpQGNhYXMuY24=; Tiegang Lu, bHV0aWVnYW5nQGNhYXMuY24=; Zhiguo Zhang, emhhbmd6aGlndW9AY2Fhcy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.