
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 July 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.903808
 Chenguang Zhang1†
Chenguang Zhang1† XIaoshuang Zhang1†
XIaoshuang Zhang1† Bo Cheng2†
Bo Cheng2† Junkai Wu1
Junkai Wu1 Libin Zhang1
Libin Zhang1 Xiao Xiao1
Xiao Xiao1 Dong Zhang2
Dong Zhang2 Caiping Zhao2
Caiping Zhao2 Na An2*
Na An2* Mingyu Han2*
Mingyu Han2* Libo Xing2*
Libo Xing2*Flowering-related problems in “Fuji” apple have severely restricted the development of China’s apple industry. Nuclear pore complexes (NPCs) control nucleoplasmic transport and play an important role in the regulation of plant growth and development. However, the effects of NPCs on apple flowering have not been reported. Here, we analysed the expression and function of MdNup54, a component of apple NPC. MdNup54 expression was the highest in flower buds and maintained during 30–70 days after flowering. MdNup54-overexpressing (OE) Arabidopsis lines displayed significantly earlier flowering than that of the wild type. We further confirmed that MdNup54 interacts with MdHSP70, MdMYB11, and MdKNAT4/6. Consistent with these observations, flowering time of MdHSP70-OE Arabidopsis lines was also significantly earlier. Therefore, our findings suggest a possible interaction of MdNup54 with MdHSP70 to mediate its nuclear and cytoplasmic transport and to regulate apple flowering. The results enhance the understanding of the flowering mechanism in apple and propose a novel strategy to study nucleoporins.
The nucleus is the main site for the regulation of genetic processes and metabolism in eukaryotes. The nuclear pore complex (NPC) constitutes the nuclear pore, which is the main channel for communication between the nucleus and cytoplasm, and it controls the flow of macromolecules such as RNA and proteins in and out of the nucleus. NPCs are essential for the maintenance of normal cellular functions and play a vital role in growth and development (D’Angelo and Hetzer, 2008; Zhang A. et al., 2020). NPCs are composed of various nucleoporins (Nups). Till date, more than 30 Nups have been identified in Arabidopsis and apple (Tamura et al., 2010; Zhang C. et al., 2020).
Nups play an important role in plant growth, development, and stress response pathways (Xu and Meier, 2008; Greco et al., 2012; Yang et al., 2017). For example, the resistance of nup96 mutants against infection by pathogens such as Peronospora parasitica 4 (RPP4), Pseudomonas syringae pv. maculicola (RPM1), and Pseudomonas syringae 4 (RPS4) was decreased (Zhang and Li, 2005). Nucleoporin Nup160 and Seh1 regulate the levels of Enhanced disease susceptibility1 (EDS1) and are involved in plant immunity (Roth and Wiermer, 2012). Moreover, Arabidopsis Nup62, Nup96, Nup160, and NUCLEAR PORE ANCHOR (TPR/NUA) are involved in auxin signalling pathways (Parry et al., 2006; Boeglin et al., 2016), and HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1) and CONSTITUTIVE EXPRESSOR OF PATHOGENESIS-RELATED GENES 5 (CPR5) regulate ethylene signalling (Lee and Seo, 2015; Wang et al., 2017). Furthermore, HOS1 and Nup160 are involved in regulating low-temperature stress response (Dong C. et al., 2006; Dong C. H. et al., 2006), while CPR5 is involved in high-temperature stress response (Wang et al., 2012). In addition, Nup85 mediates the regulation of plant salt tolerance by interacting with MEDIATOR SUBUNIT 18 (MED18) (Zhu et al., 2017).
Nups play an important role in regulating flowering. Nup136 is a plant-specific nucleopore protein, and its mutation leads to a significant acceleration of flowering in Arabidopsis (Tamura et al., 2010). HOS1 regulates the binding of certain nuclear genes to FLOWERING LOCUS C (FLC) chromatin at low temperatures and reduces the transcriptional inhibition of FLC by HISTONE DEACETYLASE 6 (HDA6), thereby inhibiting plant flowering (Jung et al., 2013). Compared with the wild type (WT), the nup54, nup58, nup62, and nup160 deletion mutants show significantly early flowering, whereas the nup85, nup98A, and seh1 deletion mutants show no significant difference in flowering time (Parry, 2014). It was further found that nup98A and nup98B single mutants show no obvious abnormal phenotype compared to WT, while the nup98A/nup98B double mutants show an early flowering phenotype (Jiang et al., 2019). In Arabidopsis thaliana, Nup96 and HOS1 mutually enhance the stability of one another. HOS1 binds to and degrades CONSTANS (CO), thereby promoting FLC transcription and leading to delayed flowering, while Nup96 maintains this regulatory pathway to control flowering time (Lazaro et al., 2015; Cheng et al., 2020).
Heat shock proteins (HSPs) are a class of chaperones that play a crucial role in protein folding, assembly, translocation, and degradation. They stabilise proteins and cell membranes and assist in protein refolding under stress conditions (Wang et al., 2004). According to their molecular weight, HSPs are divided into five subfamilies: HSP100, HSP90, HSP70, HSP60, and small HSP (Vierling, 1991). HSPs were first studied for their involvement in heat tolerance in plants (Lindquist, 1986). In Arabidopsis, HSP70 inhibits the activities of HEAT SHOCK FACTOR A1 and B1 (HSFA1 and HSFB1), whereas HSP90 stimulates the DNA-binding activity of HSFB1. HSP90 is also involved in regulating the rate of synthesis of HSFA2 by controlling the degradation of HSFA2 transcripts, thus affecting the abundance of HSFA2 and HSFB1 (Hahn et al., 2011). In cotton, the overexpression of AsHSP70 in plants significantly enhances heat tolerance (Batcho et al., 2021).
In addition to heat tolerance, HSPs have been reported to participate in other stress response pathways such as salt and drought stress response (Zou et al., 2012; Wang et al., 2019; He et al., 2021). Further, the function of HSPs in flowering has been reported. HSP90 plays an important role in floral induction and flower development in Arabidopsis (Margaritopoulou et al., 2016). HSP101 promotes flowering under non-stressed conditions, in association with FLC and SHORT VEGETATIVE PHASE (SVP) (Qin et al., 2021).
Apple (Malus domestica) is a globally important fruit grown predominantly in temperate regions. The apple variety “Fuji” covers the largest cultivated area in China. However, difficulty in flowering and alternate bearing has restricted the development of the apple industry in China. Therefore, it is particularly important to strengthen research on floral transition in apple. In this study, we analysed the expression of MdNup54 and developed early flowering MdNup54-overexpression (OE) lines in A. thaliana for functional characterisation. In addition, four interacting proteins [MdMYB11, HOMEOBOX PROTEIN KNOTTED-1 LIKE 4/6 (MdKNAT4/6), and MdHSP70] of MdNup54 were screened and validated. MdHSP70-OE Arabidopsis was shown to promote early flowering. Our findings provide a reference for further research on the various functions of MdNup54.
Multiple sequence alignment of Nup54 homologs of 10 plant species (A. thaliana, Malus domestica, Populus trichocarpa, Oryza sativa, Rosa chinensis, Pyrus communis, Ananas comosus, Vitis vinifera, Zea mays, and Prunus persica) revealed a universally conserved Nup54 domain, while eight of them also showed a conserved Nucleoporin_FG domain (Figure 1A), suggesting that Nup54 is conserved across these plants. Tissue-specific expression analysis showed that the expression level of MdNup54 was the highest in flower buds and the lowest in mature flowers (Figure 1B). Moreover, the expression of MdNup54 in the terminal bud of short shoots increased during the initial stages of development, peaked between 30 and 70 days after flowering, and showed a decreasing trend thereafter (Figure 1C). These results indicate that MdNup54 is highly expressed during the physiological differentiation of floral buds, suggesting a possible involvement in floral development in apple.
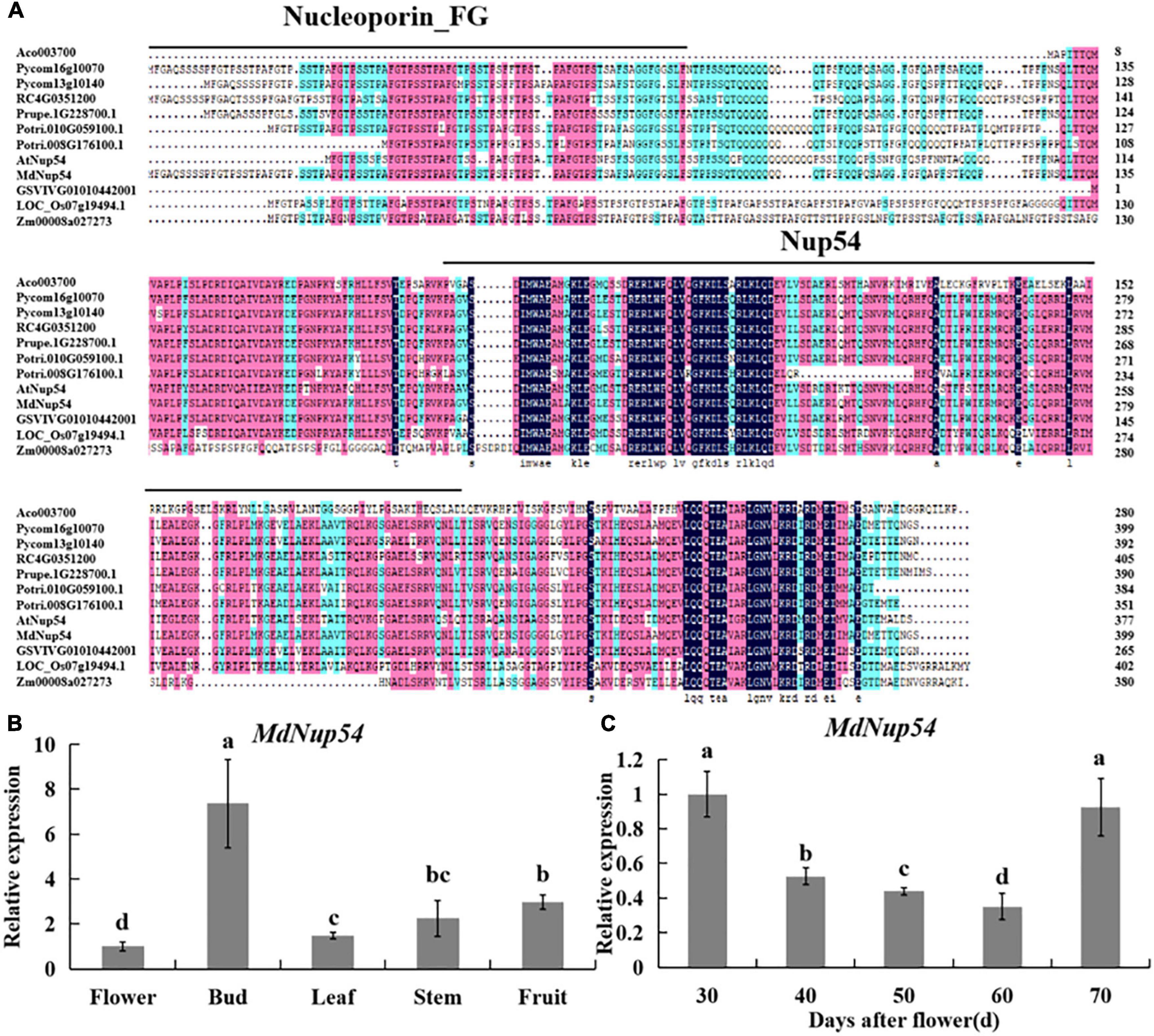
Figure 1. Bioinformatics and expression analysis of MdNup54. (A) The conservative domain of Nup54 in 10 plant species (Arabidopsis thaliana, Malus domestica, Populus trichocarpa, Oryza sativa, Rosa chinensis, Pyrus communis, Ananas comosus, Vitis vinifera, Zea mays, and Prunus persica). (B,C) Analyses of MdNup54 expression levels (B) in diverse “Nagafu No. 2” apple tissues and (C) in different flower bud developmental stages of “Nagafu No. 2.”
To analyse the subcellular localisation of MdNup54, the 35S:MdNup54-GFP construct was introduced into tobacco leaves. Tobacco leaves infiltrated with the empty vector were used as the control, which showed an instantaneous conversion of 35S:GFP. In tobacco leaves expressing 35S:MdNup54-GFP, the GFP signal was observed specifically in the nucleus and cytoplasm, whereas it was detected throughout the cells of the control tobacco leaves (Figure 2). These observations indicate that apple MdNup54 is localised in both nucleus and cytoplasm.
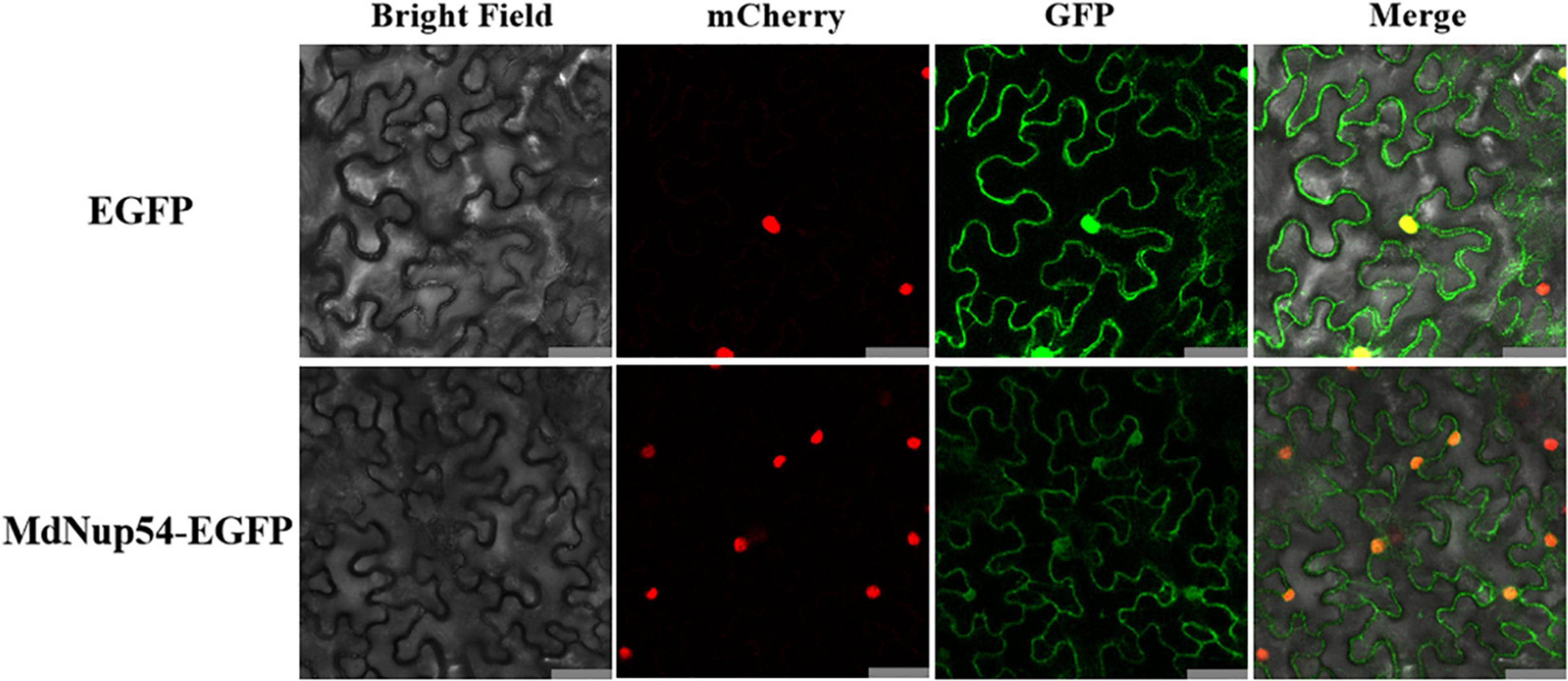
Figure 2. Subcellular localisation of MdNup54. The upper panel shows 35S:EGFP, and the lower panel shows 35S:MdNup54-EGFP. Bar = 50 μm.
For the functional characterisation of MdNup54, A. thaliana was transformed with the MdNup54 overexpression construct to obtained two transgenic lines (MdNup54-L1 and MdNup54-L2), and their flowering phenotypes were verified. Figure 3A shows that both MdNup54-OE A. thaliana lines flowered considerably earlier than WT plants did. The MdNup54-OE lines had fewer rosette leaves than those of WT (Figure 3B). Genotyping and expression analysis confirmed the presence of MdNup54 in transgenic plants (Figure 3C). Consistent with this, the expression levels of flowering genes A. thaliana SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 and LEAFY (AtSOC1 and AtLFY) in transgenic A. thaliana were significantly higher than those in WT (Figure 3D). These results demonstrate that MdNup54 promotes early flowering in plants.
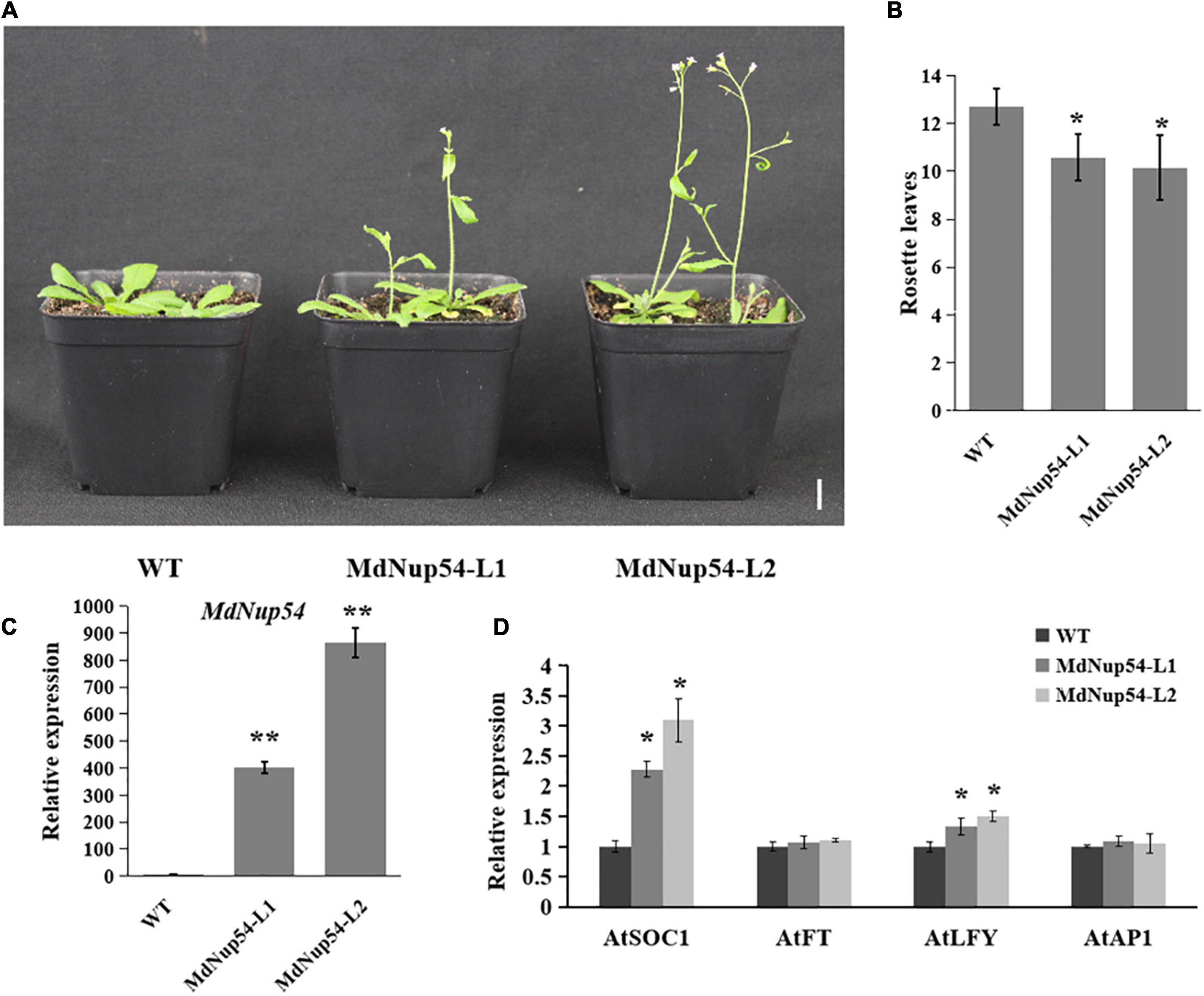
Figure 3. MdNup54 promotes flowering in Arabidopsis. (A) Phenotype of the MdNup54-OE Arabidopsis lines for flowering time. Bar = 2 cm. (B) Statistical analysis of rosette leaves of Arabidopsis thaliana during bolting (n = 15 plants per line). (*P < 0.05). (C) qRT-PCR analysis of MdNup54 expression in Arabidopsis samples. (**P < 0.01). (D) Relative expression levels of flowering genes (AtFT, AtLFY, AtSOC1, and AtAP1) in WT and MdNup54-OE lines. (*P < 0.05).
To investigate the mechanism through which MdNup54 is associated with the flowering pathway, a cDNA library representing the apple flower bud was screened using a yeast two-hybrid system to identify the interacting proteins. The interactions between MdNup54 and 10 proteins were verified, and MdNup54 was found to interacte with MdHSP70 and MdMYB11 (Figure 4). In addition, since yeast interactions between MdNup54 and MdKNAT4 and MdKNAT6 have been previously reported (Zhang C. et al., 2020), the interactions between MdNup54 and these four proteins were further validated using a split-LUC complementation assay. The combination of NLUC plus CLUC, NLUC plus MdNup54-CLUC, and MdHSP70/MdKNAT4/6/MdMYB11-NLUC plus CLUC displayed no fluorescence signal, whereas the combination of MdHSP70/MdKNAT4/6/MdMYB11-NLUC plus NUP54-CLUC showed obvious fluorescence signals (Figures 5A,B). These results confirm the interaction of MdNup54 with MdHSP70, MdKNAT4/6, and MdMYB11.
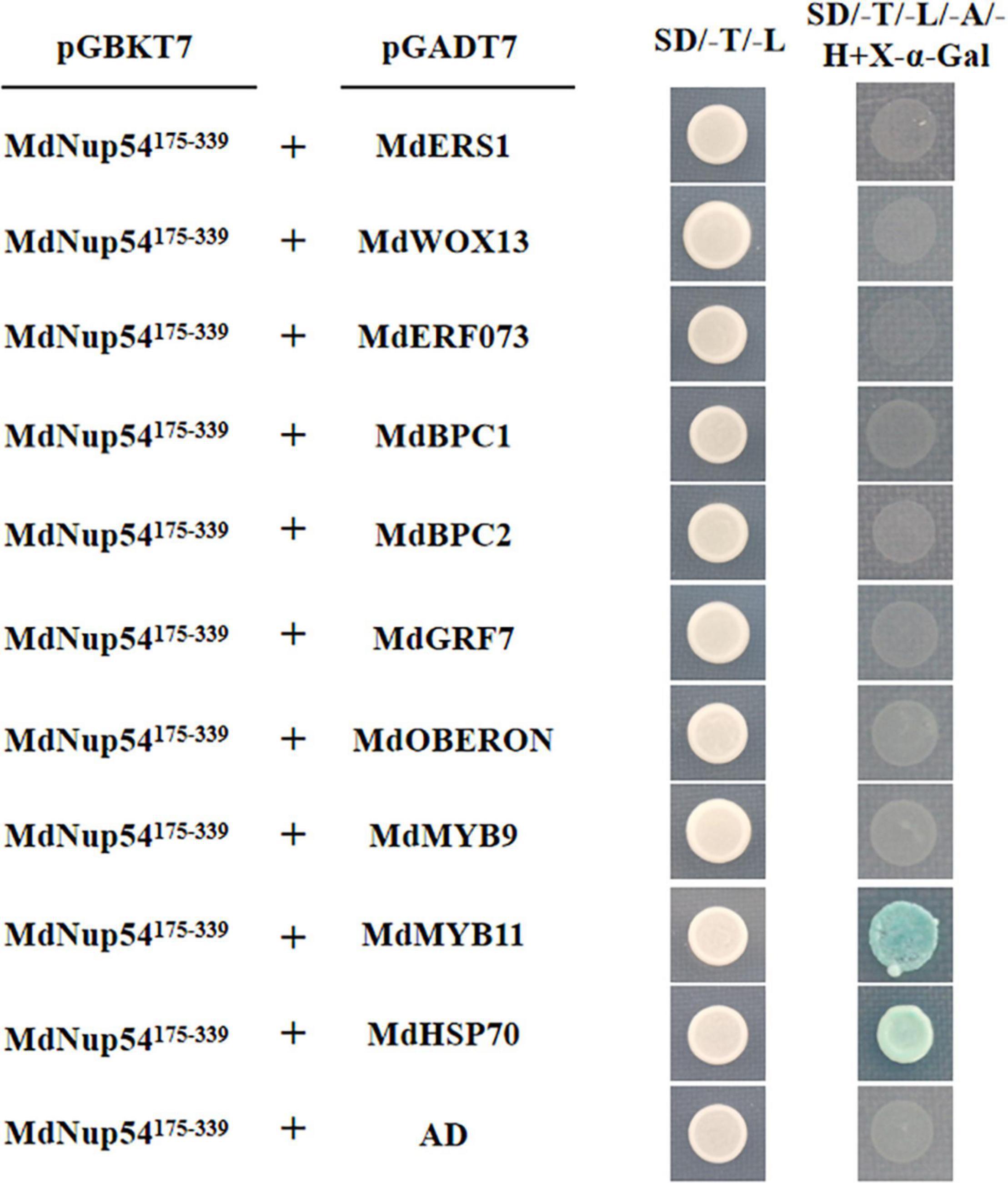
Figure 4. Yeast two-hybrid validation of MdNup54 interacting proteins. The MdNup54175– 339 truncated sequence was cloned into pGBKT7, whereas others were cloned independently into the pGADT7 vector. Empty pGADT7 plus MdNup54175– 339-pGBKT7 was used as the control.
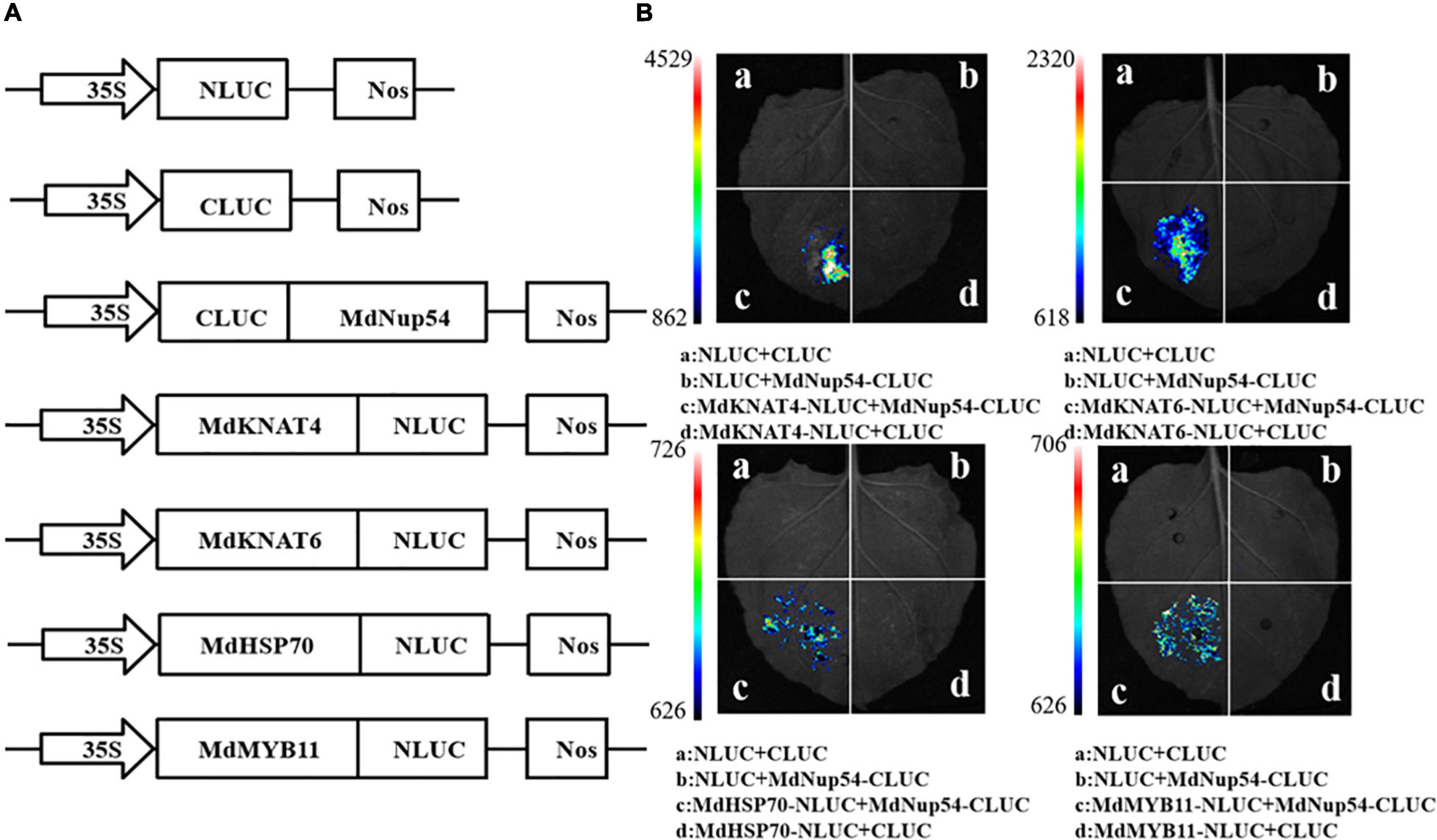
Figure 5. Luciferase (LUC) complementation validation of MdNup54 interacting proteins. (A) Carrier construction information of NLUC and CLUC. (B) Fluorescent signal in different combination.
The subcellular localisation of the four interacting partners of MdNup54 identified from the previous experiment were analysed. Since the nuclear localisation of MdMYB11 has already been reported (An et al., 2015), the subcellular localisation of MdHSP70 and MdKNAT4/6 was studied by introducing 35S:MdHSP70-GFP and 35S:MdKNAT4/6-GFP into tobacco leaves. Tobacco leaves infiltrated with empty vector, displaying an instantaneous expression of 35S:GFP, were used as controls. In the tobacco leaves expressing MdHSP70-GFP and MdKNAT6-GFP, fluorescence signals were observed in the nucleus and cytoplasm, and MdKNAT4-GFP was observed only in the nucleus. In contrast, the fluorescence signal was detected throughout the control tobacco leaf cells expressing 35S:GFP (Figure 6). These results indicate that MdHSP70 and MdKNAT6 were localised in the nucleus and cytoplasm, whereas MdKNAT4 was localised in the nucleus.
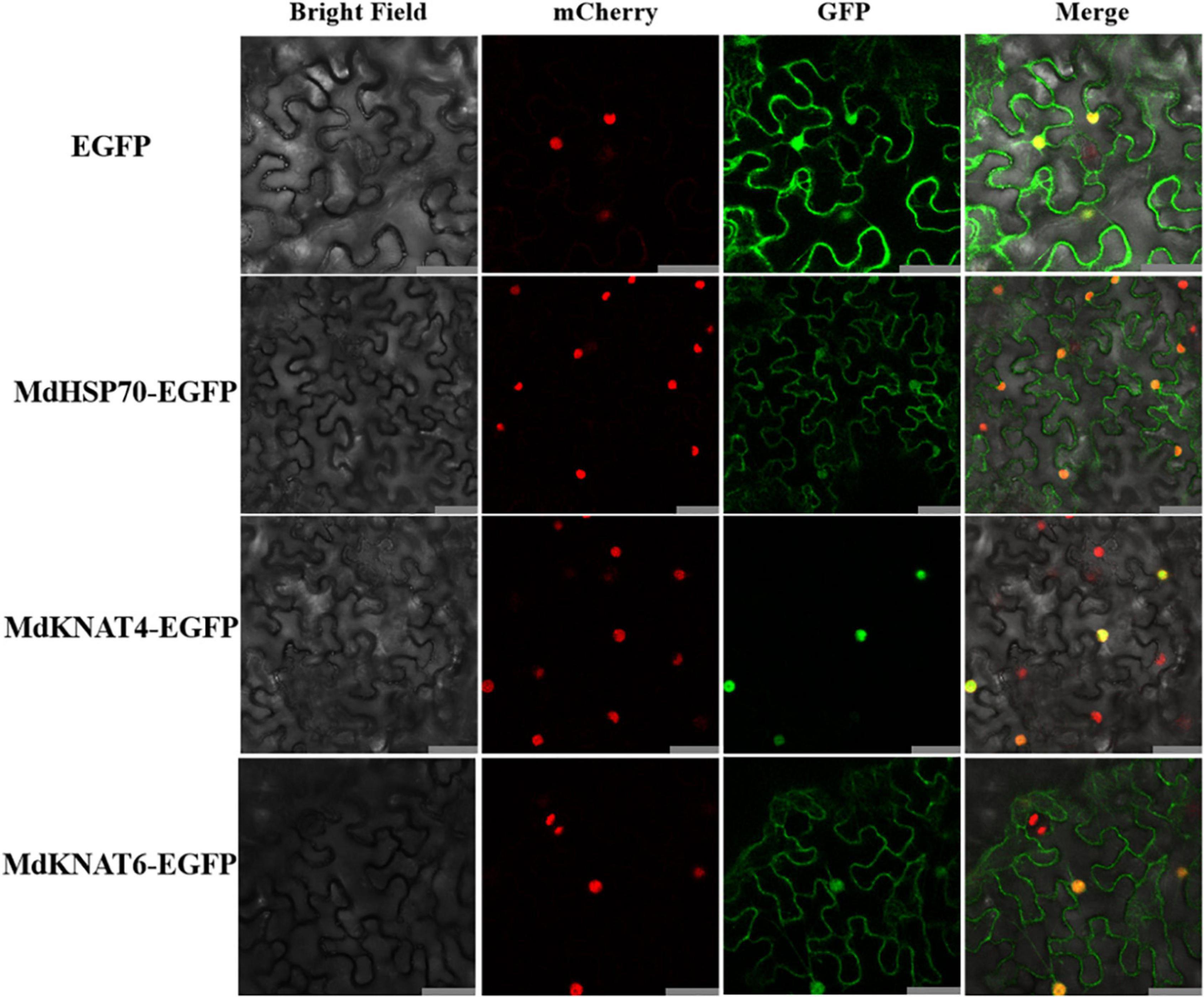
Figure 6. Subcellular localisation of MdHSP70 and MdKNAT4/6. The upper panel shows 35S:EGFP, the second is 35S:MdHSP70-EGFP, the third is 35S:MdKNAT4-EGFP, and the lower panel shows 35S: MdKNAT6-EGFP. Bar = 50 μm.
MdHSP70-OE transgenic A. thaliana lines were developed, and three lines (lines 1, 2, and 6) were selected to analyse flowering phenotypes. Genotyping and qRT-PCR-based expression analysis confirmed the presence of MdHSP70 in the transgenic plants. Figure 7A shows that all MdHSP70-OE plants flowered considerably earlier than the WT plants, and the number of rosette leaves was markedly lower than that of WT plants (Figure 7B). The expression of flowering genes (AtSOC1, AtFT, and AtLFY) were also increased in transgenic plants in comparison to WT (Figure 7C). These results clearly demonstrated that MdHSP70 promotes early flowering in plants.
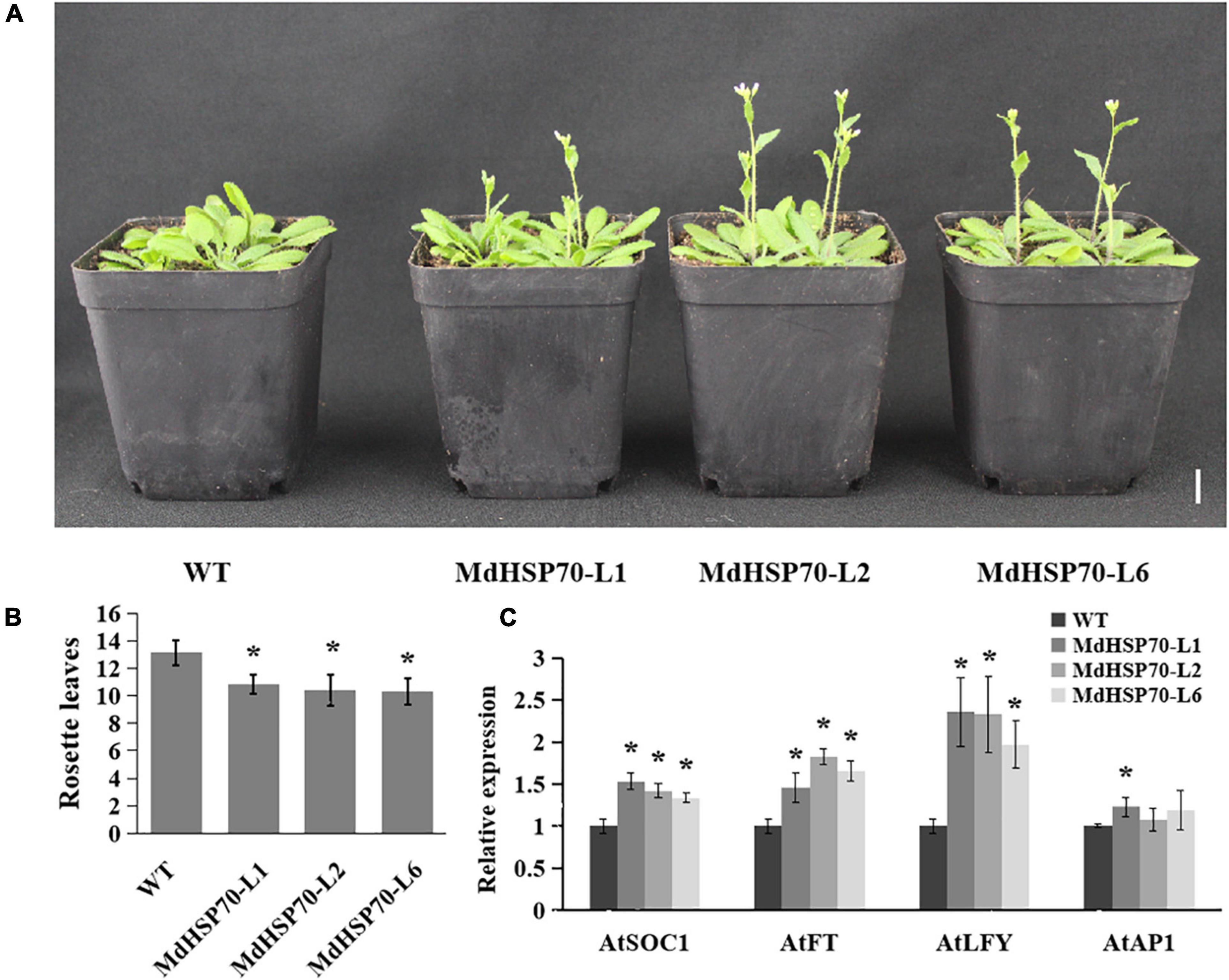
Figure 7. MdHSP70 promotes flowering in Arabidopsis. (A) Phenotype of the MdHSP70-OE Arabidopsis lines for flowering time. Bar = 2 cm. (B) Statistical analysis of rosette leaves of Arabidopsis thaliana during bolting (n = 15 plants per line). (*P < 0.05). (C) Relative expression levels of flowering genes (AtFT, AtLFY, AtSOC1, and AtAP1) in WT and MdNup54-OE lines. (*P < 0.05).
Nups play an important role in the regulation of crucial life processes in plants such as flowering. In the gene regulatory network of flowering, HOS1 not only weakens the transcriptional inhibition of FLC by HDA6 but also directly binds and degrades CO, thus promoting the expression of FLC and delaying Arabidopsis flowering (Jung et al., 2013). Moreover, Nup96 and HOS1 directly interact with each other to maintain the respective protein stability and flowering inhibition (Lazaro et al., 2015; Cheng et al., 2020). Mutations in Nup54, Nup58, Nup62, Nup136, and Nup160 significantly promote early flowering in Arabidopsis (Tamura et al., 2010; Parry, 2014). In this study, MdNup54 was highly expressed during the physiological differentiation of apple flower buds, and its heterologous overexpression in Arabidopsis significantly promoted early flowering. This suggested that the phenotype of MdNup54-OE in Arabidopsis was similar to the phenotype of the atnup54 deletion mutant in terms of flowering time. Interestingly, certain Nup overexpression and deletion mutants have been shown to display similar phenotypes, consistent with our findings. For instance, Arabidopsis HOS1-OE plants and hos1 deletion mutant both suppressed the response against cold stress (Ishitani et al., 1998; Dong C. et al., 2006). Moreover, AtNup62 showed a similar pattern in auxin response (Boeglin et al., 2016). These observations indicate that in case of nucleoproteins, overexpression does not result in gain-of-function, and instead a functional loss is noted yielding a phenotype similar to the mutant. Therefore, it is not surprising that the overexpression of MdNup54 in this study also resulted in loss-of-function. Nonetheless, the findings confirmed the involvement of MdNup54 in the flowering pathway.
To elaborate the mechanism of action of MdNup54, the interactions between MdNup54 and MdMYB11, MdKNAT4/6, and MdHSP70 were screened and confirmed. Nups play an important role in controlling the entry and exit of molecules in the nucleus (Zhang A. et al., 2020), and Nup54 plays a key role in nuclear transport owing to its localisation in the central part of the nuclear pore. Thus, apple MdNup54 may control the transport of MdMYB11, MdKNAT4/6, and MdHSP70 and participate in the corresponding biological processes. MYB11 is induced by methyl jasmonate (MeJA), and the overexpression of MYB11 significantly promotes the accumulation of anthocyanins and proanthocyanidins in apple calli (An et al., 2015). Furthermore, members of the KNOX family participate in plant hormone signalling (Jasinski et al., 2005; Sakamoto et al., 2006; Bolduc and Hake, 2009) and flower development (Khan et al., 2015). The MdKNAT4 and MdKNAT6 genes in apple are homologous to AtKNAT4 and AtKNAT6 genes in Arabidopsis, respectively. Previous studies have confirmed that AtKNAT4 affects seed dormancy (Chai et al., 2016), whereas AtKNAT6 plays an important role in maintaining meristem integrity and flowering (Khan et al., 2015). Moreover, MdHSP70 belongs to the HSP family and plays an important role in high-temperature stress tolerance (Hahn et al., 2011). Combined with these results, it is speculated that MdNup54 may be involved in colouration, flowering, seed germination, and response to high-temperature stress in apples.
Heat shock proteins have been reported to be involved in flowering (Margaritopoulou et al., 2016; Qin et al., 2021). HSP90 directly interacts with LFY, SOC1, and AGAMOUS-LIKE 24 (AGL24) to regulate flower differentiation, and the RNAi lines with suppressed HSP90 expression display severely impeded flowering (Margaritopoulou et al., 2016). The athsp101-knockout and AtHSP101-OE lines show delayed and advanced flowering, respectively. In addition, the late flowering phenotype can be restored by rescuing AtHSP101. Moreover, the promotion of flowering by AtHSP101 was found to be dependent on FLC and SVP (Qin et al., 2021). The MdHSP70-OE Arabidopsis lines obtained in this study significantly promoted early flowering. This suggested a possible role of MdHSP70 in apple flowering. As both MdNup54 and MdHSP70 are involved in the flowering process, we speculate that the mechanism through which MdNup54 influences flowering in apple involves the control of nuclear and cytoplasmic transport of HSP70.
Overall, the present study provides evidence regarding the role of a member of the NPC MdNup54 in regulating floral transition in apple. The MdNup54 gene is highly expressed in floral buds and during floral transition, and its heterologous overexpression in A. thaliana leads to early flowering. In addition, the MdNup54 protein was localised to both the nucleus and cytoplasm. Furthermore, the findings demonstrate that this function in floral transition is most likely mediated by interaction with several crucial genes involved in various developmental and stress response pathways, i.e., MdNup54, MdMYB11, MdKNAT4/6, and MdHSP70. MdHSP70 was also shown to accelerate flowering in A. thaliana.
The plant materials were 6-year-old apple trees (“Fuji”/T337/Malus robusta Rehd.) growing in the experimental orchard of the Horticulture College of Northwest A&F University (108°04′E, 34°16′N). The new shoots (2–3 mm in diameter) near the tips, fully expanded leaves near buds, flower buds, blooming flowers, and young fruit were collected for tissue specific expression analysis. And terminal bud of short shoots at 30, 40, 50, 60, and 70 days after flowering were collected. All the samples were immediately frozen in liquid nitrogen and stored at −80°C for later use.
Arabidopsis plants (“Columbia”) growed under long-day conditions (16 h-light/8 h-dark) at 22°C. Measurements of flowering times were carried out when the plants were 4 weeks old after germination.
A protein sequence alignment of Nup54 from 10 plant species was performed using DNAMAN software. And the software was downloaded from LynnonBiosoft.1
The different tissues of apple trees (new shoots, leaves, flower buds, flowers, and young fruit), Arabidopsis seedlings leaves, and apple seedlings leaves samples were ground to powders in mortars under frozen conditions. Then, 100 milligram (mg) from them were taken, respectively, for total RNA extracting by using an RNA Plant Plus Reagent Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. The RNA was used as the template to synthesise cDNA with a PrimeScript RT Reagent Kit (Takara, Shiga, Japan). The qRT-PCR analysis was conducted on a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific, United States). The reaction solution contained 10 μL SYBR Green I Master Mix (CWBIO, Beijing, China), 0.5 μmol⋅L–1 primers (SANGON BIOTECH, Shanghai, China) (Supplementary Table 1), and 1 μL of each 1:5 diluted cDNA as a template in a total volume of 20 μL. The PCR programme was as follows: 95°C for 3 min; 40 cycles of 94°C for 15 s, 60°C for 20 s, and 72°C for 15 s. Apple MdActin (MD04G1127400) and Arabidopsis AtActin (AT2G37620) were used as reference genes, and the sequences can be found in GDR2 and TAIR3 database, respectively. All the samples were analysed with three biological replicates, each comprising three technical replicates. Relative gene expression levels were calculated in accordance with the 2–ΔΔCt method (Livak and Schmittgen, 2001).
The open reading frames (ORFs) of the MdNup54, MdHSP70, and MdKNAT4/6 genes were inserted independently into the pCAMBIA2300-EGFP vector to generate the 35S:MdNup54-EGFP, 35S:MdHSP70-EGFP, and 35S:MdKNAT4/6-EGFP recombinant plasmids, respectively. These recombinant plasmids were inserted independently into Agrobacterium tumefaciens strain GV3101 cells. Then they were infiltrated into tobacco leaves. GV3101 cells containing the pCAMBIA2300-EGFP vector (35S:EGFP) served as the control. After an additional 3 days of growth in the dark, green fluorescent protein (GFP) signals in transformed tobacco leaves were detected using a Leica TCS SP8 SR Laser Scanning Confocal Microscope (Leica, Germany). The primers used are shown in Supplementary Table 2.
The methods for transgenic Arabidopsis thaliana were according to published papers (Clough and Bent, 1998). The transgenic Arabidopsis lines were selected on MS plates supplemented with 50 mg⋅L–1kanamycin.
The MdNup54175–339 truncated sequence was cloned into the pGBKT7 vector. The other ORFs were cloned individually into the pGADT7 vector. The recombinant plasmids were inserted into Gold Yeast Two-Hybrid cells, which were then grown on a selective medium. The primers used are shown in Supplementary Table 2.
The full-length MdHSP70, MdKNAT4/6, and MdMYB11 coding sequences were cloned independently into the NLUC vector, while MdNup54 was cloned into the CLUC vector. The split-LUC complementation assay was performed with tobacco leaves. The reconstituted LUC activity was detected in the dark using a Princeton Lumazone Pylon 2048B cooling camera (Princeton, NJ, United States). The primers used are shown in Supplementary Table 2.
Statistical analyses were performed using SPSS software. Asterisks (*) indicate significant differences between treatments as assessed by Student’s t-test at P < 0.05 (*) and P < 0.01 (**). Different lowercase letters above the bars indicate significant differences (P < 0.05, Tukey’s test).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
DZ, CPZ, NA, MH, LX, and CGZ conceived and designed the experiment. CGZ, XZ, and BC performed the experiment. JW, LZ, XX, XZ, and CGZ analysed the data. CGZ and LX wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was financially supported by the National Natural Science Foundation of China (32072522), the China Postdoctoral Science Foundation (2018M631207 and 2019T120954), and the Key Research and Development Project of Ningxia Hui Autonomous Region (2021BBF02014).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.903808/full#supplementary-material
Supplementary Table 1 | Primers used for qRT-PCR.
Supplementary Table 2 | Primers used for plasmid construction.
Supplementary Figure 1 | Genomic PCR analyses of MdNup54 (a), MdHSP70 (b) and qRT-PCR analyses of MdHSP70 (c) in transgenic Arabidopsis lines.
An, X., Tian, Y., Chen, K., Liu, X., Liu, D., Xie, X. B., et al. (2015). MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 56, 650–662. doi: 10.1093/pcp/pcu205
Batcho, A. A., Sarwar, M. B., Rashid, B., Hassan, S., and Husnain, T. (2021). Heat shock protein gene identified from Agave sisalana (AsHSP70AsHSP70) confers heat stress tolerance in transgenic cotton (Gossypium hirsutum). Theor. Exp. Plant Physiol. 33, 141–156. doi: 10.1007/s40626-021-00200-6
Boeglin, M., Fuglsang, A. T., Luu, D., Sentenac, H., Gaillard, I., Chérel, I., et al. (2016). Reduced expression of AtNUP62 nucleoporin gene affects auxin response in Arabidopsis. BMC Plant Biol. 16::2. doi: 10.1186/s12870-015-0695-y
Bolduc, N., and Hake, S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism genega2ox1. Plant Cell 21, 1647–1658. doi: 10.1105/tpc.109.068221
Chai, M., Zhou, C., Molina, I., Fu, C., Nakashima, J., Li, G., et al. (2016). A class II KNOX gene, KNOX4, controls seed physical dormancy. Proc. Natl. Acad. Sci. 113, 6997–7002. doi: 10.1073/pnas.1601256113
Cheng, Z., Zhang, X., Huang, P., Huang, G., Zhu, J., Chen, F., et al. (2020). Nup96 and HOS1 are mutually stabilized and gate CONSTANS protein level, conferring Long-Day photoperiodic flowering regulation in arabidopsis. Plant Cell 32, 374–391. doi: 10.1105/tpc.19.00661
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
D’Angelo, M. A., and Hetzer, M. W. (2008). Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18, 456–466. doi: 10.1016/j.tcb.2008.07.009
Dong, C. H., Hu, X., Tang, W., Zheng, X., Kim, Y. S., Lee, B. H., et al. (2006). A putative arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol. 26, 9533–9543. doi: 10.1128/MCB.01063-06
Dong, C., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J. (2006). The negative regulator of plant cold responses, HOS1, is a RING e3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U.S.A. 103, 8281–8286. doi: 10.1073/pnas.0602874103
Greco, M., Chiappetta, A., Bruno, L., and Bitonti, M. B. (2012). In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 63, 695–709. doi: 10.1093/jxb/err313
Hahn, A., Bublak, D., Schleiff, E., and Scharf, K. (2011). Crosstalk between hsp90 and hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 23, 741–755. doi: 10.1105/tpc.110.076018
He, Y., Yao, Y., Li, L., Li, Y., Gao, J., Fan, M., et al. (2021). A heat-shock 20 protein isolated from watermelon (ClHSP22.8) negatively regulates the response ofArabidopsis to salt stress via multiple signaling pathways. PeerJ. 9:e10524. doi: 10.7717/peerj.10524
Ishitani, M., Xiong, L., Lee, H., Stevenson, B., and Zhu, J. K. (1998). HOS1, a genetic locus involved in cold-responsive gene expression in arabidopsis. Plant Cell 10, 1151–1161. doi: 10.1105/tpc.10.7.1151
Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, I., et al. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15, 1560–1565. doi: 10.1016/j.cub.2005.07.023
Jiang, S., Xiao, L., Huang, P., Cheng, Z., Chen, F., Miao, Y., et al. (2019). Nucleoporin Nup98 participates in flowering regulation in a CONSTANS-independent mode. Plant Cell Rep. 38, 1263–1271. doi: 10.1007/s00299-019-02442-w
Jung, J. H., Park, J. H., Lee, S., To, T. K., Kim, J. M., and Seki, M. (2013). The cold signaling attenuator HIGH EXPRESSION of OSMOTICALLY RESPONSIVE GENE1 ActivatesFLOWERING LOCUS c transcription via chromatin remodeling under Short-Term cold stress in arabidopsis. Plant Cell 25, 4378–4390. doi: 10.1105/tpc.113.118364
Khan, M., Ragni, L., Tabb, P., Salasini, B. C., Chatfield, S., Datla, R., et al. (2015). Repression of lateral organ boundary genes by PENNYWISE and POUND-FOOLISH is essential for meristem maintenance and flowering in arabidopsis1[OPEN]. Plant Physiol. 169, 2166–2186. doi: 10.1104/pp.15.00915
Lazaro, A., Mouriz, A., Piñeiro, M., and Jarillo, J. A. (2015). Red Light-Mediated degradation of CONSTANS by the e3 ubiquitin ligase HOS1 regulates photoperiodic flowering in arabidopsis. The Plant Cell 27, 2437–2454. doi: 10.1105/tpc.15.00529
Lee, K., and Seo, P. J. (2015). The E3 ubiquitin ligase HOS1 is involved in ethylene regulation of leaf expansion in Arabidopsis. Plant Signal. Behav.Plant Signaling & Behavior 3, 1559–2324. doi: 10.1080/15592324.2014.1003755
Lindquist, S. (1986). Susan lindquist department of molecular genetics and cell biology, the university of chicago. Annu. Rev. Biochem.Annual Review of Biochemistry 55, 1151–1191.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Margaritopoulou, T., Kryovrysanaki, N., Megkoula, P., Prassinos, C., Samakovli, D., Milioni, D., et al. (2016). HSP90 canonical content organizes a molecular scaffold mechanism to progress flowering. The Plant J.ournal 87, 174–187. doi: 10.1111/tpj.13191
Parry, G. (2014). Components of the Arabidopsis nuclear pore complex play multiple diverse roles in control of plant growth. J. Exp. Bot.Journal of Experimental Botany 65, 6057–6067. doi: 10.1093/jxb/eru346
Parry, G., Ward, S., Cernac, A., Dharmasiri, S., and Estelle, M. (2006). The Arabidopsis SUPPRESSOR of AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18, 1590–1603. doi: 10.1105/tpc.106.041566
Qin, F., Yu, B., and Li, W. (2021). Heat shock protein 101 (HSP101) promotes flowering under nonstress conditions. Plant Physiology. * 186, 407–419. doi: 10.1093/plphys/kiab052
Roth, C., and Wiermer, M. (2012). Nucleoporins Nup160 and Seh1 are required for disease resistance in Arabidopsis. Plant Signal. Behav.Plant Signaling & Behavior 7, 1212–1214. doi: 10.4161/psb.21426
Sakamoto, T., Sakakibara, H., Kojima, M., Yamamoto, Y., Nagasaki, H., Inukai, Y., et al. (2006). Ectopic expression of KNOTTED1-Like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol.ogy 142, 54–62. doi: 10.1104/pp.106.085811
Tamura, K., Fukao, Y., Iwamoto, M., Haraguchi, T., and Hara-Nishimura, I. (2010). Identification and Characterization of Nuclear Pore Complex Components inArabidopsis thaliana. The Plant Cell 22, 4084–4097. doi: 10.1105/tpc.110.079947
Vierling, E. (1991). The roles of heat-shock proteins in plants. Annu. Rev. Plant Biol.Annual Review of Plant Biology 42, 579–620.
Wang, F., Wang, L., Qiao, L., Chen, J., Pappa, M. B., Pei, H., et al. (2017). Arabidopsis CPR5 regulates ethylene signaling via molecular association with the ETR1 receptor. J. Integr. Plant Biol. 59, 810–824. doi: 10.1111/jipb.12570
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends in Plant Sci.ence 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wang, X., Zhang, H., Shao, L., Yan, X., Peng, H., Ouyang, J. X., et al. (2019). Expression and function analysis of a rice OsHSP40 gene under salt stress. Genes & Genom.ics 41, 175–182. doi: 10.1007/s13258-018-0749-2
Wang, Y., Ye, Q., Zhang, M., and Yang, C. (2012). Involvement of Arabidopsis CPR5 in thermotolerance. Acta Physiol. Plant.Acta Physiologiae Plantarum 34, 2093–2103. doi: 10.1007/s11738-012-1008-9
Xu, X. M., and Meier, I. (2008). The nuclear pore comes to the fore. Trends Plant Sci.Trends in Plant Science 13, 20–27. doi: 10.1016/j.tplants.2007.12.001
Yang, Y., Wang, W., Chu, Z., Zhu, J., and Zhang, H. (2017). Roles of nuclear pores and nucleo-cytoplasmic trafficking in plant stress responses. Front. Plant Sci.Frontiers in Plant Science 8:574. doi: 10.3389/fpls.2017.00574
Zhang, A., Wang, S., Kim, J., Yan, J., Yan, X., Pang, Q., et al. (2020). Nuclear pore complex components have temperature-influenced roles in plant growth and immunity. Plant Cell Environ. 43, 1452–1466. doi: 10.1111/pce.13741
Zhang, C., An, N., Jia, P., Zhang, W., Liang, J., Zhang, X., et al. (2020). Genomic identification and expression analysis of nuclear pore proteins in Malus domestica. Sci. Rep.Scientific Reports 10:17426. doi: 10.1038/s41598-020-74171-0
Zhang, Y., and Li, X. (2005). A Putative Nucleoporin 96 is Required for Both Basal Defense and Constitutive Resistance Responses Mediated by suppressor of npr1-1, constitutive 1. The Plant Cell 17, 1306–1316. doi: 10.1105/tpc.104.029926
Zhu, Y., Wang, B., Tang, K., Hsu, C., Xie, S., Hai, D. U, et al. (2017). An Arabidopsis Nucleoporin NUP85 modulates plant responses to ABA and salt stress. PLoS Genet. PLoS Genetics 13:e1007124. doi: 10.1371/journal.pgen.1007124
Keywords: apple, MdNup54, interaction, MdHSP70, flowering
Citation: Zhang C, Zhang X, Cheng B, Wu J, Zhang L, Xiao X, Zhang D, Zhao C, An N, Han M and Xing L (2022) MdNup54 Interactions With MdHSP70 Involved in Flowering in Apple. Front. Plant Sci. 13:903808. doi: 10.3389/fpls.2022.903808
Received: 24 March 2022; Accepted: 07 June 2022;
Published: 05 July 2022.
Edited by:
Serena Varotto, University of Padua, ItalyReviewed by:
Xiao-Fei Wang, Shandong Agricultural University, ChinaCopyright © 2022 Zhang, Zhang, Cheng, Wu, Zhang, Xiao, Zhang, Zhao, An, Han and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Libo Xing, bGlib194aW5nQG53c3VhZi5lZHUuY24=, orcid.org/0000-0002-8918-7128; Na An, YW5uYTIwNkBud3N1YWYuZWR1LmNu; Mingyu Han, aGFubXlAbndzdWFmLmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.