- 1Amity Institute of Biotechnology, Amity University Jharkhand, Ranchi, India
- 2Department of Botany, Shivaji College, University of Delhi, New Delhi, India
- 3School of Biotechnology, Gautam Buddha University, Greater Noida, India
Introduction
Modern single-celled, long, and spinnable cotton fibers are the result of allopolyploidy between A- and D-genome diploid species of cotton and millennia of selective breeding under domestication (Figure 1). Gossypium hirsutum is one of the domesticated allopolyploid lineages and is prevalent in cotton crop fields worldwide (Wendel and Cronn, 2003). Generally, cotton fibers are elongated and thickened epidermal cells of developing ovules, which undergo four tightly regulated stages during their development, namely, initiation, elongation/extension, secondary cell wall synthesis, and maturation (Haigler et al., 2012). The comparative transcriptome sequencing of wild and domesticated species reveals that the development of fiber initiation and elongation is an extraordinarily dynamic and complex process in which more than half of the genome is expressed in any particular stage of fiber development (Yoo and Wendel, 2014). Evidently, the morpho-evolution of fiber cells in domesticated cotton species is characterized by large transcriptomic biases, mostly in response to hormone signaling genes, antioxidant genes, and cell wall-modifying (CWM) genes (Chaudhary et al., 2008). The latter class of genes is responsible for (de)-polymerization of actin during fiber initiation and elongation, which is mediated by transcriptionally hyperactive CWM profilin (PRF) genes (Bao et al., 2011). PRFs are extremely conserved and ancient proteins present in viruses, flagellated prokaryotes, cyanobacteria, bacteria, and animalia and plantae kingdoms (Pandey and Chaudhary, 2017). Apparently, the GhACT1 gene responsible for constructing the actin cytoskeleton network is abundantly expressed in developing fiber cells, in particular during fiber elongation (Li et al., 2005). Mutation in the GhACT1 gene leads to formation of disrupted F-actin filaments and disordered cytoskeleton in elongating fiber cells (Thyssen et al., 2017). Additionally, the transcriptional dynamics and functional attributes of different CWM genes have also been investigated, which includes Ca2+dependent phospholipid-binding annexin genes (GhAnn2, AnxGb6, and GhFAnnxA) regulating the rate of Ca2+ flux and signaling mechanisms, rate of fiber cell polar extension and secondary cell wall synthesis through its interaction with actin filaments (Wang et al., 2010; Zhou et al., 2011; Tang et al., 2014; Zhang et al., 2016); β-tubulin gene (GhTub1) involved in the synthesis and rearrangement of microtubules required for cytoskeletal dynamicity during fiber formation (Li et al., 2003; Chen et al., 2021); expansin (GhEXPA8) gene stimulating cell wall loosening and extension during fiber cell elongation (Bajwa et al., 2015; Lv et al., 2020); and Fasciclin-Like Arabinogalactan (GhFLA1) protein responsible for the activation of primary cell wall biosynthesis genes in fiber initiation and elongation processes (Huang G. Q. et al., 2013). As a result of transcriptional loss of the domestication-driven CWM-associated profilin structural gene (GhPRF1), the cotton plant experiences severe developmental abnormalities in floral organs, predominantly due to disruption of coordinated gene expression profiles of CWM-gene clusters (triads) in the cellular milieu rather than single target gene silencing (Pandey and Chaudhary, 2021). This article presents a novel perspective on the synchronized gene expression evolution of CWM genes during floral, fiber, and boll development in cotton.
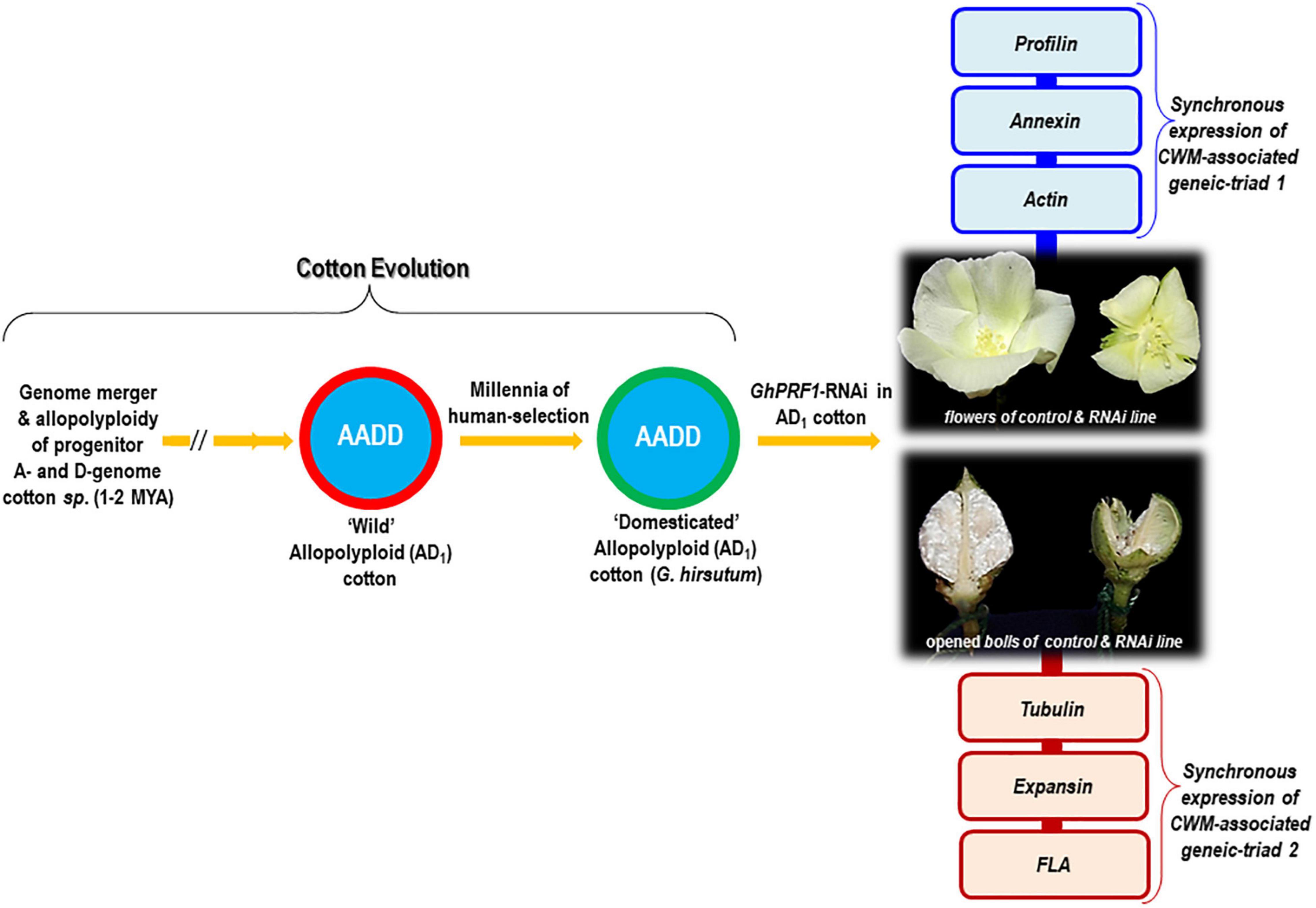
Figure 1. Summarized representation of cell wall-modifying (CMW) gene expression evolution accompanying the evolutionary trajectory of genomic merger and allopolyploid formation in cotton. RNAi of the GhPRF1 gene showed abnormal floral shapes and reduced fiber development in mature bolls. Relative temporal expression of the “GhPRF–GhAnnex–GhAct” and “Tub-Exp-FLA1” geneic-triads at the 5-, 10-, and 20-dpa stages of fiber development in the Coker 310 cultivar exhibits synchronous expression patterns and highlights their fine-scale coordination during floral and fiber development.
Expression Evolution of Cell Wall-Modifying-Associated “profilin” Genes During Fiber Development
Under cotton domestication, the selection of superior fiber traits in domesticated forms have led to the development of long and spinnable fiber phenotypes from the inferior wild short fuzz. Comparative temporal genomics of fiber development in wild and domesticated diploid and allopolyploid cotton species has identified CWM-associated cytoskeletal profilin (PRF) gene family members that are preferentially expressed (>400-fold) during emergence and extension of domesticated fiber initials (Bao et al., 2011). The functional characterization of PRF genes has demonstrated its direct role in the intricate process of fiber development and the regulation of floral development by activating various signaling pathways (Pandey and Chaudhary, 2016, 2021). The spatiotemporal expression analysis of PRF genes among vegetative, floral, and various stages of fiber development has identified PRF transcript abundance in developing fibers, particularly in fiber elongation stage (10 days post anthesis, dpa). Interestingly, increased PRF transcription exhibits proportional polymerization of F-actin levels in 10-dpa fiber tissues of different cotton cultivars, followed by 5- and 20-dpa fibers (Pandey and Chaudhary, 2019). This is attributed to the strong PRF genes’ expression-mediated F-actin polymerization and bundling during fiber elongation. Similarly to PRF genes, annexin, tubulin, and FLA genes are also expressed in developing fiber cells and are critical for fiber extension, secondary cell wall formation, and actin filament rearrangement (Huang Y. et al., 2013; Zhang et al., 2016). Hence, cell wall structural proteins, together with several glycoproteins and enzymes, form a rigid matrix of cellulose (Keegstra, 2010), and CWM genes contribute to maintaining a dynamic cell wall structure during fiber development.
RNAi of PRF Genes Exhibits Anomalous Floral Organ Shape and Reduced Fiber Elongation
The constitutive reduction in the transcription of domestication-driven PRF1 gene in cotton (GhPRF1) shows up to 40% less secondary branches and floral buds per transgenic plant compared to untransformed plants (Pandey and Chaudhary, 2021). Independent GhPRF1-RNAi lines exhibit floral organ abnormalities and shorter fiber lengths. Anthers are disoriented with shortened staminal tube, disorganized style protrusions, aberrant stigma tips, inadequate staminal tube growth, reduced pollen viability, and delayed and decreased fiber synthesis on the ovular surface. Most flowers fail to form seeds, and only a few produces underdeveloped bolls with stunted ovules and seeds. On the contrary, fiber-specific silencing of the GhPRF1 gene shows normal floral organ development but reduced emergence of fiber initials on the ovule surface. Together, the constitutive- and fiber-specific RNAi of PRF genes in cotton modulates the expression of actin and annexin genes, which also significantly influence the morpho-appearance of floral organs and fibers (Pandey and Chaudhary, 2019, 2021; Figure 1). These observations suggest that abnormal floral shapes among constitutive RNAi lines are precisely the effect of reduced PRF transcript levels and not a consequence of transgenesis. Moreover, the transcriptional loss of PRF gene(s) interfered with the synchronization of gene expression ratios in the “profilin–annexin–actin” (GhPRF1: Accession No. EF143832; GhAnnex3:Accession No. JX897059; and GhAct1: Accession No. AY305723) and “tubulin–expansin–FLA1” (GhTub1: Accession No. AF484959; GhExp1: Accession No. DQ204495; and GhFLA1: Accession No. EF672627) geneic triads, which may account for the observed phenotypic anomalies. Future experiments in this area would provide important insights into the strategic utilization of co-expressed CWM genes for enhanced agronomically important fiber-associated traits.
Synchronized Transcriptional Dynamics of Cell Wall-Modifying Geneic Triads Regulates Floral Organ Shape and Fiber Architecture
Previously, temporal transcriptional biases in CWM-associated genes during vegetative and fiber development were investigated among diverse cotton cultivars, and interestingly, synchronous expression dynamicity and coordinated expression patterns were prominent among CWM-associated “profilin–annexin–actin” and “tubulin–expansin–FLA1” geneic-triads (Pandey and Chaudhary, 2019). Regardless of the cultivar’s genetic background, synchronous expression profiles of the GhPRF–GhAnnex–GhAct triad have been observed in root tissues, hypocotyl, cotyledons, cotyledonary callus, and leaf tissues. Simultaneously, transcriptional comparisons between GhPRFs-associated CWM-GhAnnex and -GhACT genes in developing fibers also demonstrate a highly significant association (correlation coefficient r = 0.95–1) across geneic comparisons. Similarly, fold-expression variations in actin-polymerizing GhPRFs in different stages of fiber development are correlated with GhACT bundle formation. In addition, RNAi-mediated constitutive suppression of domestication-driven GhPRF1 gene expression in cotton transgenics results in significant changes in CWM-GhAnnex and -GhACT gene expression profiles. At protein level, yeast two hybridization and BiFC based analyses indicate that the AnxGb6 homodimer interacts strongly with ACT protein resulting in enhanced F-actin accumulation in fibers and regulate fiber elongation process in cotton (Huang Y. et al., 2013). The transcriptional silencing of cotton PRF genes reveals a strong expression correlation among these genes, which has a direct impact on cotton organ development. In response to GhPRF1 expression reduction in transgenic cotton, GhAnnex3 and GhACT1 expression significantly increases. Such synchronized expression patterns are prominent in floral and fiber tissues. Notably, the GhAnnex3 and GhAct1 genes are significantly downregulated in deformed stamen tissues of GhPRF1-RNAi lines (Pandey and Chaudhary, 2019). Thus, GhPRF–GhAnnex–GhAct triad gene expression profiles are observed to be highly coordinated in their temporal patterns with intriguing interactions in various cellular contexts and cell types. Furthermore, the synchronized patterns of GhPRF–GhAnnex–GhAct triads coincide with the “tubulin (Tub)-expansin (Exp)-Fasciclin-Like Arabinogalactan-Protein (FLA1)” structural geneic triad during the emergence of cotton fibers. This triad mainly includes the Tub gene, which is responsible for enhanced and precise assembly of microtubules (Whittaker and Triplett, 1999; Mujahid et al., 2016); the Exp1 gene is responsible for fiber cell wall relaxation (effector) (Cosgrove, 2015); the FLA1 gene is responsible for cell wall integrity (Huang G. Q. et al., 2013). Hence, fine-scale and synchronized transcription of CWM geneic triads is essential for maintaining floral organ shapes and fiber development in cotton.
Conclusion
Modern forms of elongated and spinnable cotton fibers originated through the genomic hybridization between A- and D-genome progenitor cotton species, followed by the formation of allopolyploids and independent domestication of two polyploidy species. For such morphological transformation of wild fuzz into modern elongated fiber cells, various genes, and transcription factors, including CWM genes, have been recruited. Both diploid and polyploid species exhibit several hundred-fold expression evolution in actin (de)-polymerizing cytoskeletal profilin genes; and the RNAi of the PRF genes has profound effects on the appearance of floral organs as well as fiber architecture. A remarkable feature of the temporal expression profiles of the CWM-associated “profilin–annexin–actin” and “tubulin–expansin–FLA1” geneic triads in cotton is their coordination, with an array of fascinating interplay across a variety of cellular contexts and cell types. Both floral and fiber tissues demonstrate significant expression modulation of actin and annexin genes as a result of transcriptional loss of PRF genes. As a result, the concurrent transcriptional dynamics of cytoskeleton-associated structural genes in modern cotton fibers are very useful in understanding the evolutionary recruitment of CWM gene clusters for shaping the floral organs and determining fiber length (Figure 1).
Author Contributions
BC, VK, and DP conceptualized this study. All authors have contributed in the writing and further approved this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are thankful to the Department of Biotechnology, and CSIR, Government of India for providing financial support to conduct this study.
References
Bajwa, K. S., Shahid, A. A., Rao, A. Q., Bashir, A., Aftab, A., and Husnain, T. (2015). Stable transformation and expression of GhEXPA8 fiber expansin gene to improve fiber length and micronaire value in cotton. Front. Plant Sci. 6:838. doi: 10.3389/FPLS.2015.00838
Bao, Y., Hu, G., Flagel, L. E., Salmon, A., Bezanilla, M., Paterson, A. H., et al. (2011). Parallel up-regulation of the profilin gene family following independent domestication of diploid and allopolyploid cotton (Gossypium). Proc. Natl. Acad. Sci. USA 108, 21152–21157. doi: 10.1073/pnas.1115926109
Chaudhary, B., Hovav, R., Rapp, R., Verma, N., Udall, J. A., and Wendel, J. F. (2008). Global analysis of gene expression in cotton fibers from wild and domesticated Gossypium barbadense. Evol. Dev. 10, 567–582. doi: 10.1111/j.1525-142X.2008.00272.x
Chen, B., Zhao, J., Fu, G., Pei, X., Pan, Z., Li, H., et al. (2021). Identification and expression analysis of Tubulin gene family in upland cotton. J. Cott. Res. 4, 1–10. doi: 10.1186/S42397-021-00097-1/FIGURES/6
Cosgrove, D. J. (2015). Plant expansins: diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 25, 162–172. doi: 10.1016/J.PBI.2015.05.014
Haigler, C. H., Betancur, L., Stiff, M. R., and Tuttle, J. R. (2012). Cotton fiber: A powerful single-cell model for cell wall and cellulose research. Front. Plant Sci. 3, 1–7. doi: 10.3389/fpls.2012.00104
Huang, G. Q., Gong, S. Y., Xu, W. L., Li, W., Li, P., Zhang, C. J., et al. (2013). A Fasciclin-Like Arabinogalactan Protein, GhFLA1, is involved in fiber initiation and elongation of cotton. Plant Physiol. 161, 1278–1290. doi: 10.1104/PP.112.203760
Huang, Y., Wang, J., Zhang, L., and Zuo, K. (2013). A Cotton Annexin Protein AnxGb6 Regulates fiber elongation through its interaction with Actin 1. PLoS One 8:e66160. doi: 10.1371/journal.pone.0066160
Li, X. B., Fan, X. P., Wang, X. L., Cai, L., and Yang, W. C. (2005). The Cotton ACTIN1 Gene Is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17:859. doi: 10.1105/TPC.104.029629
Li, Y., Sun, J., Li, C., Zhu, Y., and Xia, G. (2003). Specific expression of a β-tubulin gene (GhTub1) in developing cotton fibers. Sci. China Ser. C Life Sci. 463, 235–242. doi: 10.1360/03YC9025
Lv, L. M., Zuo, D. Y., Wang, X. F., Cheng, H. L., Zhang, Y. P., Wang, Q. L., et al. (2020). Genome-wide identification of the expansin gene family reveals that expansin genes are involved in fibre cell growth in cotton. BMC Plant Biol. 20:1–13. doi: 10.1186/S12870-020-02362-Y/FIGURES/6
Mujahid, H., Pendarvis, K., Reddy, J. S., Nallamilli, B. R. R., Reddy, K. R., Nanduri, B., et al. (2016). Comparative proteomic analysis of cotton fiber development and protein extraction method comparison in late stage fibers. Proteomes 4:7. doi: 10.3390/PROTEOMES4010007
Pandey, D. K., and Chaudhary, B. (2016). Domestication-driven Gossypium profilin 1 (GhPRF1) gene transduces early flowering phenotype in tobacco by spatial alteration of apical/floral-meristem related gene expression. BMC Plant Biol. 16:201310. doi: 10.1186/s12870-016-0798-0
Pandey, D. K., and Chaudhary, B. (2017). Evolutionary expansion and structural functionalism of the ancient family of profilin proteins. Gene 626, 70–86. doi: 10.1016/j.gene.2017.05.024
Pandey, D. K., and Chaudhary, B. (2019). Synchronous transcription of cytoskeleton-associated genes is critical to cotton fiber elongation. J. Plant Growth Regul. 38, 1037–1061. doi: 10.1007/s00344-019-09913-0
Pandey, D. K., and Chaudhary, B. (2021). Transcriptional loss of domestication-driven cytoskeletal GhPRF1 gene causes defective floral and fiber development in cotton (Gossypium). Plant Mol. Biol. 107, 519–532. doi: 10.1007/s11103-021-01200-5
Tang, W., He, Y., Tu, L., Wang, M., Li, Y., Ruan, Y. L., et al. (2014). Down-regulating annexin gene GhAnn2 inhibits cotton fiber elongation and decreases Ca2+ influx at the cell apex. Plant Mol. Biol. 85, 613–625. doi: 10.1007/s11103-014-0208-7
Thyssen, G. N., Fang, D. D., Turley, R. B., Florane, C. B., Li, P., Mattison, C. P., et al. (2017). A Gly65Val substitution in an actin, GhACT_LI1, disrupts cell polarity and F-actin organization resulting in dwarf, lintless cotton plants. Plant J. 90, 111–121. doi: 10.1111/tpj.13477
Wang, L. K., Niu, X. W., Lv, Y. H., Zhang, T. Z., and Guo, W. Z. (2010). Molecular cloning and localization of a novel cotton annexin gene expressed preferentially during fiber development. Mol. Biol. Rep. 37, 3327–3334. doi: 10.1007/S11033-009-9919-2/FIGURES/4
Wendel, J. F., and Cronn, R. C. (2003). Polyploidy and the evolutionary history of cotton. Adv. Agron. 78, 139–186. doi: 10.1016/S0065-2113(02)78004-8
Whittaker, D. J., and Triplett, B. A. (1999). Gene-Specific Changes in α-tubulin transcript accumulation in developing cotton fibers. Plant Physiol. 121, 181–188. doi: 10.1104/PP.121.1.181
Yoo, M.-J., and Wendel, J. F. (2014). Comparative evolutionary and developmental dynamics of the cotton (gossypium hirsutum) fiber transcriptome. PLoS Genet. 10:e1004073. doi: 10.1371/journal.pgen.1004073
Zhang, F., Jin, X., Wang, L., Li, S., Wu, S., Cheng, C., et al. (2016). A cotton annexin affects fiber elongation and secondary cell wall biosynthesis associated with Ca2+ Influx, ROS Homeostasis, and actin filament reorganization. Plant Physiol. 171, 1750–1770. doi: 10.1104/PP.16.00597
Keywords: cotton, gene expression, evolution, cell wall genes, floral/fiber development
Citation: Pandey DK, Kumar V and Chaudhary B (2022) Concomitant Expression Evolution of Cell Wall Cytoskeletal Geneic Triad(s) Controls Floral Organ Shape and Fiber Emergence in Cotton (Gossypium). Front. Plant Sci. 13:900521. doi: 10.3389/fpls.2022.900521
Received: 20 March 2022; Accepted: 25 April 2022;
Published: 20 May 2022.
Edited by:
Hirokazu Tsukaya, The University of Tokyo, JapanReviewed by:
Youlu Yuan, Cotton Research Institute (CAAS), ChinaCopyright © 2022 Pandey, Kumar and Chaudhary. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bhupendra Chaudhary, Ymh1cGVuZHJhQGdidS5hYy5pbg==; orcid.org/0000-0001-9593-1016
 Dhananjay K. Pandey
Dhananjay K. Pandey Vijay Kumar
Vijay Kumar Bhupendra Chaudhary
Bhupendra Chaudhary