- 1Key Laboratory of Plant Resource Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), College of Agricultural Sciences, Guizhou University, Guiyang, China
- 2Center of Applied Biotechnology, Wuhan University of Bioengineering, Wuhan, China
Melatonin plays an important role in plant resistance to biotic and abiotic stresses. However, whether melatonin is involved in the regulation of plant architecture, such as the formation of axillary bud outgrowth or tillering, in rice remains unknown. Here, we found that different concentrations of melatonin influenced axillary bud outgrowth in rice, and moderate melatonin concentrations also alleviated the inhibition of axillary bud outgrowth in the presence of high concentrations of basic amino acids lysine and arginine. Furthermore, transcriptome analysis demonstrated that genes involved in nitrogen metabolism and phytohormone signal transduction pathways may affect axillary bud outgrowth, which is regulated by melatonin. We determined that the differentially expressed genes glutamine synthetase OsGS2 and amino acid transporter OsAAP14, which are involved in nitrogen metabolism and are regulated by melatonin and basic amino acids, were the key regulators of axillary bud outgrowth in rice. In addition, we validated the functions of OsGS2 and OsAAP14 using rice transgenic plants with altered axillary bud outgrowth and tillers. Taken together, these results suggest that melatonin mediates axillary bud outgrowth by improving nitrogen assimilation and transport in rice.
Introduction
Melatonin (N-acetyl-5-methoxytryptamine) is an indolic compound first reported in bovine pineal glands (Lerner et al., 1958). Recently, melatonin has been widely used as a plant biostimulator to regulate growth and enhance plant resistance to both biotic and abiotic stresses. On the one hand, melatonin is involved in multiple plant biological processes, such as leaf senescence (Zhang et al., 2017), photosynthesis (Tan et al., 2019), flowering (Arnao and Hernández-Ruiz, 2020), fruit ripening (Onik et al., 2021), root induction (Yang et al., 2021), and other growth and developmental processes. On the other hand, melatonin confers tolerance to plants against several abiotic stresses, including cold (Bajwa et al., 2014), high temperatures (Larkindale and Huang, 2005), heavy metals (Posmyk et al., 2008), salinity (Vafadar et al., 2020), oxidative stress (Bahcesular et al., 2020), and drought (Wang et al., 2021), and various biotic stressors, including bacteria (Lee et al., 2014), fungi (Mandal et al., 2018), and viruses (Sofy et al., 2021). Nonetheless, whether melatonin is involved in the regulation of plant architecture, such as the formation of axillary bud outgrowth or tillering, in rice still remains unclear.
Tillering is an important agronomic trait that contributes to grain yield, and also a model system for the study of branching in monocotyledonous plants. Tillering occurs in a two-stage process: the formation of an axillary bud at each leaf axil and its subsequent outgrowth (Li et al., 2003). Tillering is regulated by several phytohormones and external environmental factors. The phytohormone auxin affects the formation of tillering buds, and the auxin transporter genes OsPIN1 (Xu et al., 2005), OsPIN2 (Chen et al., 2012), OsPIN5b (Lu et al., 2015), and OsPIN9 (Hou et al., 2021b) regulate rice tillering by altering the local distribution of auxin. Strigolactones (SLs) regulate rice tillering by inhibiting the elongation of rice axillary buds (Umehara et al., 2008). The synthetic genes D27 (Lin et al., 2009), D17 (Zou et al., 2005), D10 (Arite et al., 2007), receptor gene D14 (Arite et al., 2009), and downstream signaling genes D53 and FC1 (Jiang et al., 2013; Zhou et al., 2013) in the SL pathway are involved in the regulation of rice tillering. Recent evidence has revealed abscisic acid (ABA)-repression of lateral bud outgrowth in Arabidopsis (Yao and Finlayson, 2015). A high concentration of ABA acts as a growth inhibitor to induce axillary bud dormancy under adverse environmental conditions (Gonzalez-Grandio et al., 2017). In the ABA signaling pathway, studies have shown that OsNCED1 plays important roles in ABA biosynthesis and inhibition of bud outgrowth in rice (Luo et al., 2019). In addition, ABA can also interact with other phytohormones to affect plant growth and development. SLs promote ABA biosynthesis in shoot basal part, whereas ABA inhibits SL biosynthesis, revealing SL and ABA biosynthesis are closely integrated to coordinately repress tillering in rice (Liu et al., 2020). Besides, ABA and GA antagonistically regulate the growth of rice root and the increase of tillering (Lin et al., 2020). However, cytokinins (CKs) promote the elongation of axillary buds (Duan et al., 2019), but the rice CK oxidase genes OsCKX2 (Yeh et al., 2015), OsCKX4 (Gao et al., 2014), and OsCKX9 (Duan et al., 2019) negatively regulate tillering. Recently, brassinosteroids (BRs) have also been shown to play significant roles in the regulation of tillering, with OsBZR1–DLT–RLA1 signaling complex required for axillary bud outgrowth (Fang et al., 2020).
Nitrogen is an important external environmental factor required for axillary bud outgrowth, which influences tillering and grain yield in rice. Nitrate, ammonium, and several amino acids can be utilized by plants after being converted into other organic nitrogen forms by various processes. Reports suggest that elevated levels of nitrate or ammonium ions in the soil can promote rice tillering (Sasaki et al., 2002). Recently, it is reported that different nitrogen concentrations significantly influence the outgrowth of axillary buds for rice tillering (Wang et al., 2020). The lengths of axillary buds were shorter under lower nitrogen concentrations 0.5 and 1.0 mm than those under the optimal nitrogen concentration (2.0 mm), whereas the lengths of axillary buds were longer under 5.0 and 10.0 mm nitrogen concentrations. Remarkably, the axillary buds were shorter when the nitrogen concentration reached 15.0 mm (Wang et al., 2020). Nitrogen displays a significant promoting effect on tiller development by regulating nitrogen-metabolism and endogenous hormone levels (Liu et al., 2011). Farmers usually apply nitrogen fertilizers to enhance the number of tillers, and a sufficient application of N could increase rice tillers and panicles (Zhang et al., 2013). Therefore, increasing nitrogen application rate in rice with high tillering capacity could compensate for the yield reduction of low planting density (Huang et al., 2013).
Furthermore, NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) genes OsNPF7.1, OsNPF7.2, OsNPF7.3, OsNPF7.7, and OsNPF8.20 positively regulate rice tiller number, whereas OsNPF7.4 negatively regulates rice tillering (Fang et al., 2013, 2017; Huang et al., 2018, 2019; Wang et al., 2018). Importantly, OsNPF6.5 (OsNRT1.1B) is a key gene controlling nitrogen uptake and utilization, and influence grain yield by regulating rice tillering (Hu et al., 2015). Recently, it was reported that OsNPF5.16, a nitrate transporter gene with natural variation in its promoter sequence, is essential for rice tillering and yield (Wang et al., 2022b). In addition, the glutamine synthetase is a key point in nitrogen assimilation where ammonium is incorporated into glutamine, providing the precursor for production of all amino acids, nucleic acids, and chlorophylls (Kissen et al., 2010). In rice, there are three genes that encode cytosolic GS1 (OsGS1;1, OsGS1;2 and OsGS1;3), and one gene encodes chloroplastic GS2 (OsGS2; James et al., 2018). Mutants of cytosolic glutamine synthetase 1;2 reduce yield by reducing tillering and panicle number (Funayama et al., 2013). Omics analysis was then performed in the basal part of the seedlings and showed that the reduction in the number of tillering in OsGS1;2 mutants is due to a change in metabolic balance and is independent of the regulation of tillering by strigolactones (Ohashi et al., 2015). Besides, deletion of OsGS1;2 reduces IPT4 gene expression at the base, which is shown to reduce the amount of tZ-type cytokinins, resulting in reduced tiller number (Ohashi et al., 2017), and deletion of OsGS1;2 also reduces expression of the fructose-1,6-bisphosphatase gene (Ohashi et al., 2018). Furthermore, it has been shown that OsGS2 regulate axillary bud growth and tiller number in rice (Wang et al., 2020).
Amino acid transporters also play important roles in rice tillering. OsAAP1 primarily mediates neutral amino acids, such as proline, alanine, and tyrosine, and positively regulates rice tillering (Ji et al., 2020). OsAAP3 and OsAAP5 predominantly transport lysine (Lys) and arginine (Arg), and negatively regulate rice tillering (Lu et al., 2018; Wang et al., 2019). OsAAP4 regulates the transport of neutral amino acids valine and proline, and plants over-expressing OsAAP4 exhibit increased axillary bud elongation under different neutral amino acid concentrations (Fang et al., 2021). OsAAP6 facilitates the uptake and transport of threonine, serine, glycine, alanine, proline, and acidic amino acids by rice roots (Peng et al., 2014). In rice, low concentrations of Lys and Arg promote the elongation of axillary buds, whereas slightly higher concentrations inhibit axillary bud elongation (Lu et al., 2018). However, the molecular mechanism underlying the inhibition of rice axillary bud elongation by high concentrations of Lys and Arg remains to be elucidated. Therefore, this study aimed to determine the effects of different concentrations of melatonin, Lys, and Arg on the growth of axillary buds in rice. We further identify the genes involved in the regulation of rice axillary bud elongation and validate their functions using transgenic plants.
Materials and Methods
Plant Material and Growth Conditions
The germinated seeds of wild-type (WT) rice cultivar ZH11 (Oryza sativa L. ssp. japonica) were grown in basic rice nutrient solution for 14 days. The basic nutrient solution was composed of 1.0 mm NH4NO3, 0.32 mm NaH2PO4, 0.51 mm K2SO4, 1.0 mm CaCl2, 1.65 mm MgSO4, 8.9 μm MnSO4, 0.5 μm Na2MoO4, 18.4 μm H3BO3, 0.14 μm ZnSO4, 0.16 μm CuSO4, and 40.0 μm FeSO4. The seedlings were then transferred in a greenhouse and treated with basic nutrient solutions supplemented with different concentrations of melatonin (0 μm, 0.1 μm, 1 μm, 10 μm, 100 μm, and 1,000 μm), 1.5 mm Lys + 0.5 mm Arg, or 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin for 21 days. The solutions were renewed every 3 days, and the greenhouse maintained a 16 h-light (32°C)/8 h-dark (25°C) photoperiod. At 35 days after germination, the length of first and second axillary buds of ZH11 plants were measured. Simultaneously, axillary buds and basal parts from each treatment were excised and immediately frozen in liquid N2 for total RNA extraction. Thereafter, total RNA was extracted using TRIzol reagent (Vazyme, Nanjing, China) for RNA-Seq. The raw data collected from RNA-seq are available at the National Center for Biotechnology Information (NCBI): https://dataview.ncbi.nlm.nih.gov/object/PRJNA820302?reviewer=rlhbj4ujtu845m90qfffvupk0p.
Library Preparation for RNA-Seq and Data Processing
A total of 1 μg RNA per sample was used for constructing cDNA libraries. cDNA library preparation and sequencing were carried out by Beijing Nuohe Zhiyuan Technology Co. Ltd. (Nuohe, Beijing, China) following manufacturer’s recommendations. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina, San Diego, United States) according to the manufacturer’s instructions. After cluster generation, the libraries were sequenced on an Illumina Novaseq platform and 150-bp paired-end reads were generated.
Raw reads in the FASTQ format were first processed using in-house Perl scripts, and then processed reads were obtained by removing reads containing adapter and poly-N and low-quality reads from the raw data. Subsequently, the clean reads were aligned to the Nipponbare reference genome (Ensemble_37) using Hisat2 v2.0.5. To determine the expression levels of genes in the samples, fragments per kilobase of transcript per million reads (FPKM) were calculated using FeatureCounts v1.5.0-p3. Then, principal component analysis was performed online,1 and expression correlations between samples were calculated using the cor function in R software.
Gene Ontology and KEGG Enrichment Analysis of Differentially Expressed Genes
Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs) was performed using the cluster Profiler R package, wherein gene length bias was corrected. GO terms with corrected value of p < 0.05 were considered significantly enriched. We used the clusterProfiler R package to test the statistical enrichment of DEGs in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted using Trizol reagent according to the manufacturer’s instructions (Vazyme, Nanjing, China). First-strand cDNA were synthesized using 3 μg of total RNA extracted from each sample using MLV reverse transcriptase (Vazyme, Nanjing, China). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed in a 20 μl reaction volume containing 10 μl 2 × SYBR Green Mix (Vazyme, Nanjing, China), 1 μl cDNA solution, and 1 μl gene-specific primers (10 μm) under the following conditions: 94°C for 2 min (1 cycle), 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s (40 cycles), and 72°C for 1 min (1 cycle) using 7,500 RT qPCR system (Applied Biosystems, Foster City, United States). The primers used for qPCR are listed in Supplementary Table 1.
Vector Construction and Transgenic Plant Generation
To construct OsGS2 and OsAAP14 promoter-GUS vectors, 2,238-bp and 2,300-bp fragments upstream of the start codon (ATG) of OsGS2 and OsAAP14, respectively, were inserted into the pCAMBIA1391Z vector using HindIII and NcoI restriction enzymes. To construct OsGS2- and OsAAP14-overexpression (OE) vectors, 1,287-bp OsGS2 (LOC_Os04g56400) and 1,410-bp OsAAP14 (LOC_Os04g56470) cDNA sequences were inserted downstream of the CaMV 35S promoter in the pCAMBIA1306 vector using KpnI and XbaI restriction enzymes. To generate the OsGS2-RNAi vector, two 217-bp fragments of OsGS2 cDNA were inserted downstream of the Ubi-1 promoter in the rice RNAi vector pTCK303 using BamHI/KpnI and SpeI/SacI, respectively. The OsAAP14 CRISPR plasmid was constructed using the CRISPR/Cas9-based multiplex genome editing system for monocot and dicot plants (Ma et al., 2015). These vectors were then transformed into Agrobacterium tumefaciens strain EHA105 and introduced into the calli of ZH11, following which T0 plants were selected using 50 mg l−1 hygromycin. Homologous T2 lines were used for all experiments.
Gus Signal Analysis of the OsAAP14 and OsGS2 Promoter
To analyze OsGS2 and OsAAP14 expression levels in the presence of different melatonin concentrations, three-week-old OsGS2 and OsAAP14 promoter-GUS transgenic plants were treated with basic solutions supplemented with different concentrations of melatonin (0 μm, 0.1 μm, 1 μm, and 10 μm), 1.5 mm Lys + 0.5 mm Arg, or 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin for 2 h and 24 h. GUS staining was performed according to a previously described histochemical staining method (Fang et al., 2017). The stained samples were then observed using a stereomicroscope (OLYMPUS SZX16, Tokyo, Japan). To make sections, the stained tissues were rinsed and fixed in FAA at 4°C for 24 h, gradually dehydrated with ethanol for 15 min each time, and washed twice with 100% ethanol for 30 min each time. Finally, the samples were embedded in Spurr resin, and ultramicrotome sections (2–8 μm) were applied onto poly-l-lysine-coated slides with glass knives. The sections were observed using Zeiss Axio Imager M2 (Carl Zeiss AG, Oberkochen, Germany).
FITC-Labeled Amino Acid Uptake Assay
Amino acids with FITC (Arg-FITC, Lys-FITC) were synthesized by Yuan Peptide Biotechnology Company, Nanjing, China. Then rice etiolated seedlings were prepared from ZH11, OsGS2, and OsAAP14 transgenic plants. Fluorescence was detected after culturing seedlings of OsAAP14 and OsGS2 transgenic plants with FITC-labeled amino acids Arg for 2 h and 6 h, and Lys for 2 h and 6 h. Finally, fluorescence was observed using a fluorescence analyzer (Qinxiang, Shanghai, China).
Determination of Free Amino Acids and Chlorophyll Content in Rice
To analyze the effects of OsGS2 and OsAAP14 expression levels on nitrogen metabolism and photosynthetic pathway, five-week-old OsGS2 and OsAAP14 transgenic plants were taken for total free amino acids and total chlorophyll content. Total free amino acid content was measured by the ninhydrin method (Fang et al., 2013). Total chlorophyll content was determined according to Luo et al. (2022).
Statistical Analyses
The statistical chart were performed using GraphPad Prism 7, and the heatmaps were performed using TBtools. For multiple comparisons, Duncan’s multiple range test was performed using SPSS software, indicating significant difference at p < 0.05.
Results
Exogenous Low Concentration Melatonin Promoting Axillary Bud Elongation
To investigate the effects of melatonin on tillering in rice, two-week-old ZH11 seedlings were cultured in basic nutrient solution supplemented with different concentrations of melatonin for 21 days, and the length of first and second axillary buds was measured. The results showed that low concentrations of melatonin (0–1 μm) promoted the elongation of the first and second axillary buds in rice, but higher concentrations (10–1,000 μm) inhibited bud elongation (Figures 1A–C). Furthermore, 1.5 mm Lys + 0.5 mm Arg inhibited the elongation of both first and second axillary buds (Figure 1). However, when these two amino acids were added with 1 μm melatonin, the inhibitory effect of the amino acids on rice axillary buds was not only alleviated but also the elongation of axillary buds increased. The effect of 1.5 mm Lys, 0.5 mm Arg and 1 μm melatonin on axillary bud elongation was stronger than that of 1 μm melatonin alone (Figure 1). These results demonstrated that melatonin plays an important role in axillary bud outgrowth in rice, and can alleviate the inhibition caused by high concentrations of basic amino acids on axillary bud outgrowth.
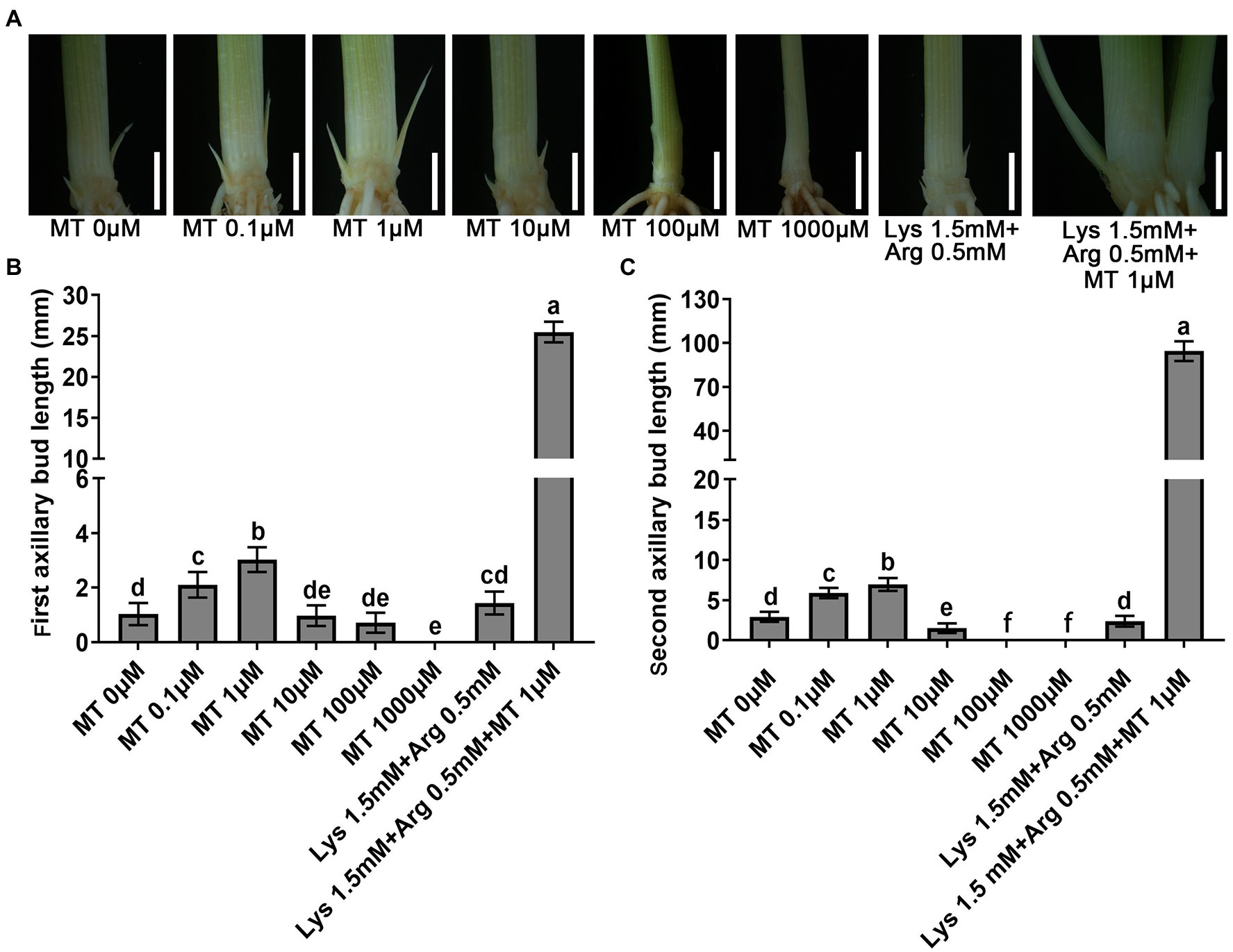
Figure 1. Phenotypes of outgrowth axillary bud under different concentrations of melatonin and amino acids treatments. (A) Phenotypes of outgrowth axillary buds under basic nutrient solutions supplemented with different concentrations of melatonin (0 μm, 0.1 μm, 1 μm, 10 μm, 100 μm, and 1,000 μm), 1.5 mm Lys + 0.5 mm Arg, or 1.5 mM Lys + 0.5 mM Arg + 1 μm melatonin. Statistical analysis of the first (B) and second (C) axillary bud length in (A). MT represents melatonin, Lys represents lysine, and Arg represents arginine. Values are means ± SD (n > 20), and the significance levels of different lowercase letters were as follows, p < 0.05. Scale bar = 3 mm.
Transcriptome Profiles of Axillary Buds and Basal Parts of Plants Grown Under Different Melatonin and Amino Acid Concentrations
To investigate the regulatory mechanisms underlying rice axillary bud outgrowth by melatonin, we sampled a mixture of the first and second axillary buds and basal parts of 35-day-old rice seedlings grown under different concentrations of melatonin for RNA-Seq analysis. We identified 4,480 DEGs in the axillary buds of plants treated with 0.1 μm melatonin compared with those treated with 0 μm melatonin, and 4,612 DEGs in the axillary buds of plants treated with 1 μm melatonin compared with those treated with 0.1 μm melatonin (Supplementary Figure S1A). Furthermore, 7,047 and 1,519 DEGs were detected in 10 μm melatonin-treated axillary buds compared with 1 μm and 0.1 μm melatonin-treated axillary buds, respectively (Supplementary Figure S1A). In addition, 2086 upregulated and 2,526 downregulated DEGs were detected in the axillary buds of plants treated with 0.1 μm melatonin compared with those treated with 1 μm melatonin (Supplementary Figure S2A). Compared to 10 μm melatonin treatment, 3,803 upregulated and 3,244 downregulated DEGs were detected in the axillary buds of plants treated with 1 μm melatonin (Supplementary Figure S2B). However, more DEGs were identified in the axillary buds than basal parts when exposed to the above treatments (Supplementary Figure S1B). For basal parts, 698 upregulated and 813 downregulated DEGs were detected upon treatment with 0.1 μm melatonin compared to the treatment with 1 μm melatonin (Supplementary Figure S3A), and 1,066 upregulated and 946 downregulated DEGs were detected upon treatment with 1 μm melatonin compared to the treatment with 10 μm melatonin (Supplementary Figure S3B), indicating that melatonin has a greater effect on gene expression in rice axillary buds than that in the basal part for axillary buds outgrowth.
Additionally, 2,365 DEGs were upregulated whereas 2,149 DEG were downregulated in the axillary buds of plants exposed to 1.5 mm Lys + 0.5 mm Arg treatment compared to 0 μm melatonin treatment (Supplementary Figures S4A, S5A). Compared to 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatment, 2,107 DEGs were upregulated whereas 2,705 DEGs were downregulated in the axillary buds of plants exposed to 1.5 mm Lys + 0.5 mm Arg treatment (Supplementary Figures S4A, S5B). In the basal parts, 1913 (1,074 upregulated and 839 downregulated) and 465 (298 upregulated and 167 downregulated) DEGs were detected upon 1.5 mm Lys + 0.5 mm Arg treatment compared to 0 μm melatonin (Supplementary Figures S4B, S6A) and 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatments (Supplementary Figures S4B, S6B), respectively, indicating that DEGs in axillary bud was higher than that in basal part under amino acids Lys + Arg treatments.
DEGs in the Axillary Buds and Basal Parts of Plants Grown Under Different Melatonin and Amino Acid Concentrations
To investigate the processes associated with axillary bud outgrowth under different melatonin concentrations, we conducted GO enrichment analysis for each treatment. The genes responsive to 0.1–1 μm melatonin in the axillary buds (Figure 2A) and basal parts (Supplementary Figure S7A) were primarily involved in cellular carbohydrate metabolism and oxidative stress responses. In addition, KEGG analysis indicated that these DEGs were mainly enriched in nitrogen metabolism, plant–pathogen interactions, and phytohormone signal transduction pathways in axillary buds (Figure 2B) and basal parts (Supplementary Figure S7B) of plants exposed to 0.1 μm and 1 μm melatonin treatments. The genes responsive to 1–10 μm melatonin in axillary buds (Figure 2C) and basal parts (Supplementary Figure S7C) were primarily involved in cellular nitrogen compound catabolism, cellular amide metabolism, and amide biosynthesis, and these DEGs were mainly enriched in nitrogen metabolism and phytohormone signal transduction in axillary buds (Figure 2D) and carbon metabolism and amino acid biosynthesis in basal parts (Supplementary Figure S7D). These results indicate that nitrogen metabolism and phytohormone signal transduction pathways play important roles in response to melatonin to promote or inhibit axillary bud outgrowth.
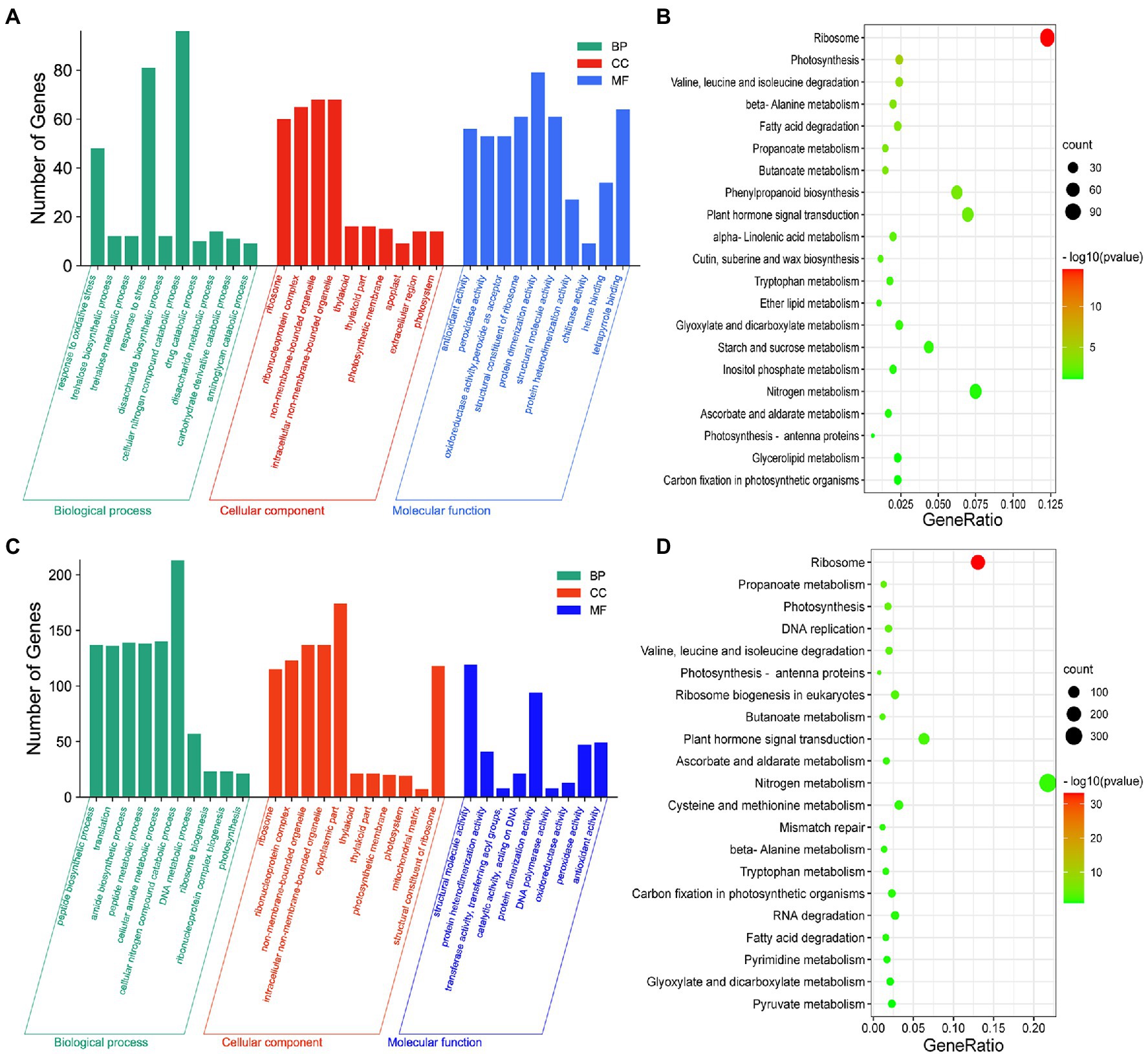
Figure 2. Analysis of differential expression genes (DEGs) with different expression patterns in axillary bud under different concentrations of melatonin treatments. The GO enrichment (A) and KEGG pathway enrichment (B) analysis of DEGs at 0.1 μm melatonin compared to 1 μm melatonin. The GO enrichment (C) and KEGG pathway enrichment (D) analysis of DEGs at 1 μm melatonin compared to 10 μm melatonin. Y-axis indicates the number of GO annotated genes, and X-axis indicates the processes/components in different biological processes, cellular components, and molecular functions (A,C). Y-axis indicates KEGG pathway, and X-axis indicates the ratio of the number of enriched genes to the number of annotated genes in the pathway (B,D). The color of the dot represents value of p, and the size of the dot represents the number of DEGs mapped to the referent pathway.
To investigate the mechanism underlying the inhibition of axillary bud elongation by Lys and Arg in rice, we conducted GO enrichment analysis of axillary buds of plants exposed to 1.5 mm Lys + 0.5 mm Arg and the control. DEGs in axillary buds (Figure 3A) and basal parts (Supplementary Figure S8A) of plants exposed to 1.5 mm Lys + 0.5 mm Arg treatment were mainly involved in cellular carbohydrate metabolism and amide biosynthesis, and were mainly enriched in nitrogen metabolism, plant–pathogen interactions, and phytohormone signal transduction pathways (Figure 3B; Supplementary Figure S8B). In addition, compared to 1.5 mm Lys + 0.5 mm Arg treatment, DEGs in the axillary buds (Figure 3C) and basal parts (Supplementary Figure S8C) of plants exposed to 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin were primarily involved in cellular nitrogen compound catabolism, carbohydrate metabolism, and stress responses, and were enriched in nitrogen metabolism, plant–pathogen interactions, photosynthesis, and phytohormone signal transduction pathways in axillary buds (Figure 3D) and photosynthesis, nitrogen metabolism, and carbon metabolism in basal parts (Supplementary Figure S8D). These results indicate that nitrogen metabolism, plant–pathogen interactions, and phytohormone signal transduction pathways play important roles in response to amino acids Lys + Arg alone or amino acids Lys + Arg together with melatonin to promote or inhibit axillary bud outgrowth.
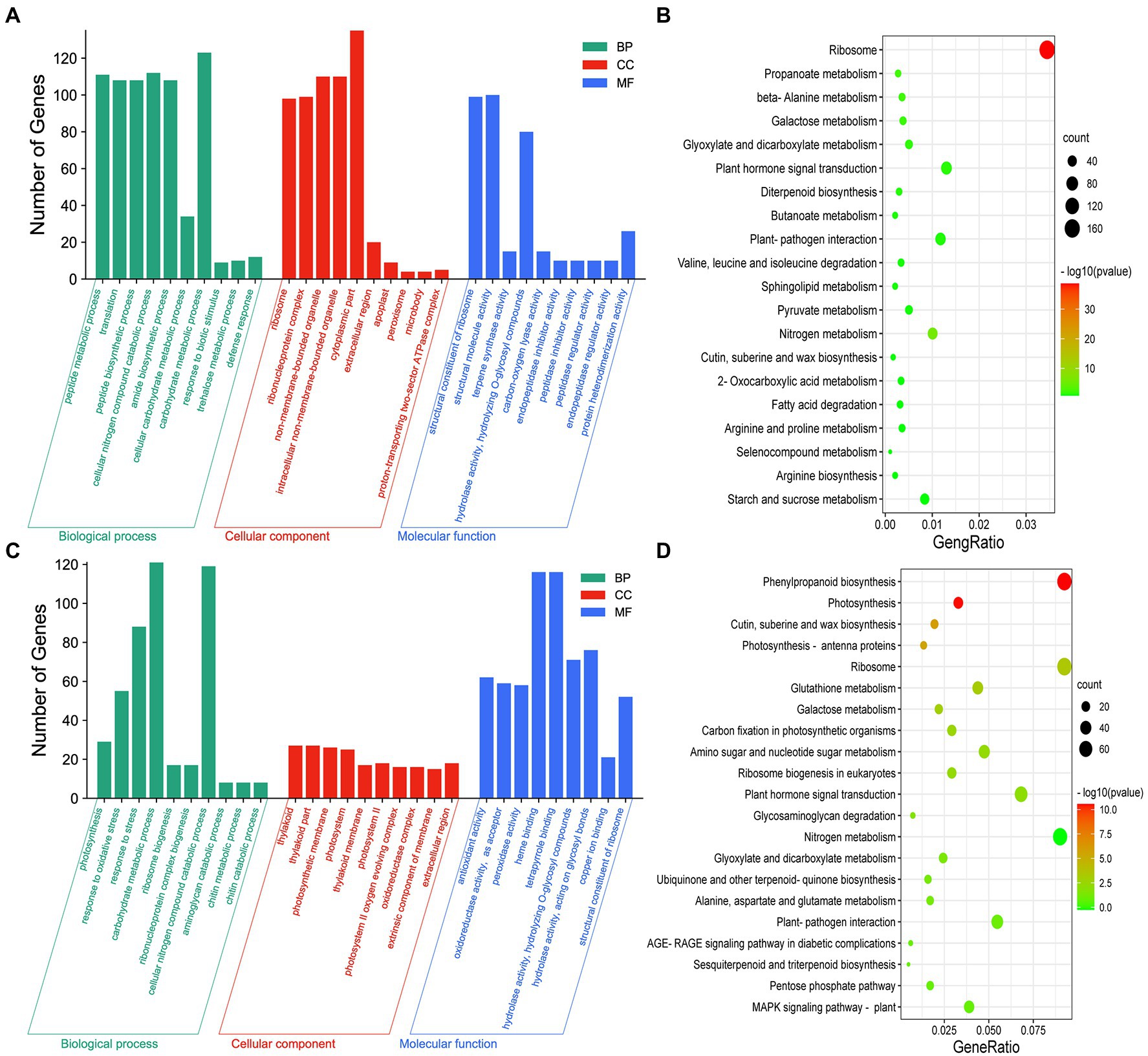
Figure 3. Analysis of differential expression genes (DEGs) with different expression patterns in axillary bud under amino acids related treatments. The GO enrichment (A) and KEGG pathway enrichment (B) analysis of DEGs at 0 μm melatonin compared to 1.5 mm Lys + 0.5 mm Arg. The GO enrichment (C) and KEGG pathway enrichment (D) analysis of DEGs at 1.5 mm Lys + 0.5 mm Arg compared to 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin. Y-axis indicates the number of GO annotated genes, and X-axis indicates the processes/components in different biological processes, cellular components, and molecular functions (A,C). Y-axis indicates KEGG pathway, and X-axis indicates the ratio of the number of enriched genes to the number of annotated genes in the pathway (B,D). The color of the dot represents value of p, and the size of the dot represents the number of DEGs mapped to the referent pathway.
Expression Profiles of Genes Related to Nitrogen Metabolism, Photosynthesis, and Stress Response Pathways
Based on the enriched processes associated with nitrogen metabolism, photosynthesis, stress response, and phytohormone pathways in the axillary buds and basal parts, we inferred that these processes predominantly determined the growth rate of axillary buds. Therefore, we analyzed the expression profiles of genes involved in nitrate, ammonium, and amino acid transport and assimilation in axillary buds using a heatmap (Figures 4A,B). Glutamine synthetase gene OsGS2 (Figure 4A) and amino acid transporter gene OsAAP14 (Figure 4B) were highly expressed upon treatment with 1 μm melatonin and 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin, indicating that nitrogen assimilation and amino acid transport were activated by 1 μm melatonin to promote axillary bud growth. The expression of OsGS2 and OsAAP14 was strongly inhibited by 10 μm melatonin and 1.5 mm Lys + 0.5 mm Arg (Figures 4A,B). Besides, the expression of OsCAT11 was also inhibited by 10 μm melatonin (Figure 4B). In addition, the expression of OsNPF5.4 was increased under low and moderate concentrations of melatonin, whereas OsNPF6.5 was significantly expressed only when the amino acids were complemented with melatonin (Figure 4A), indicating that melatonin could greatly promote nitrogen transport and assimilation under the high concentration nitrogen stress. However, the ammonium transporter gene was not remarkably expressed upon melatonin treatment (Figure 4A). In addition, in the basal parts, there were no significant differences in the expression of the genes involved in nitrogen metabolism among different melatonin and amino acid treatments (Supplementary Figure S9). These results suggest that the induction of nitrogen assimilation gene OsGS2 and amino acid transporter gene OsAAP14 by melatonin was critical for melatonin to promote axillary bud elongation.
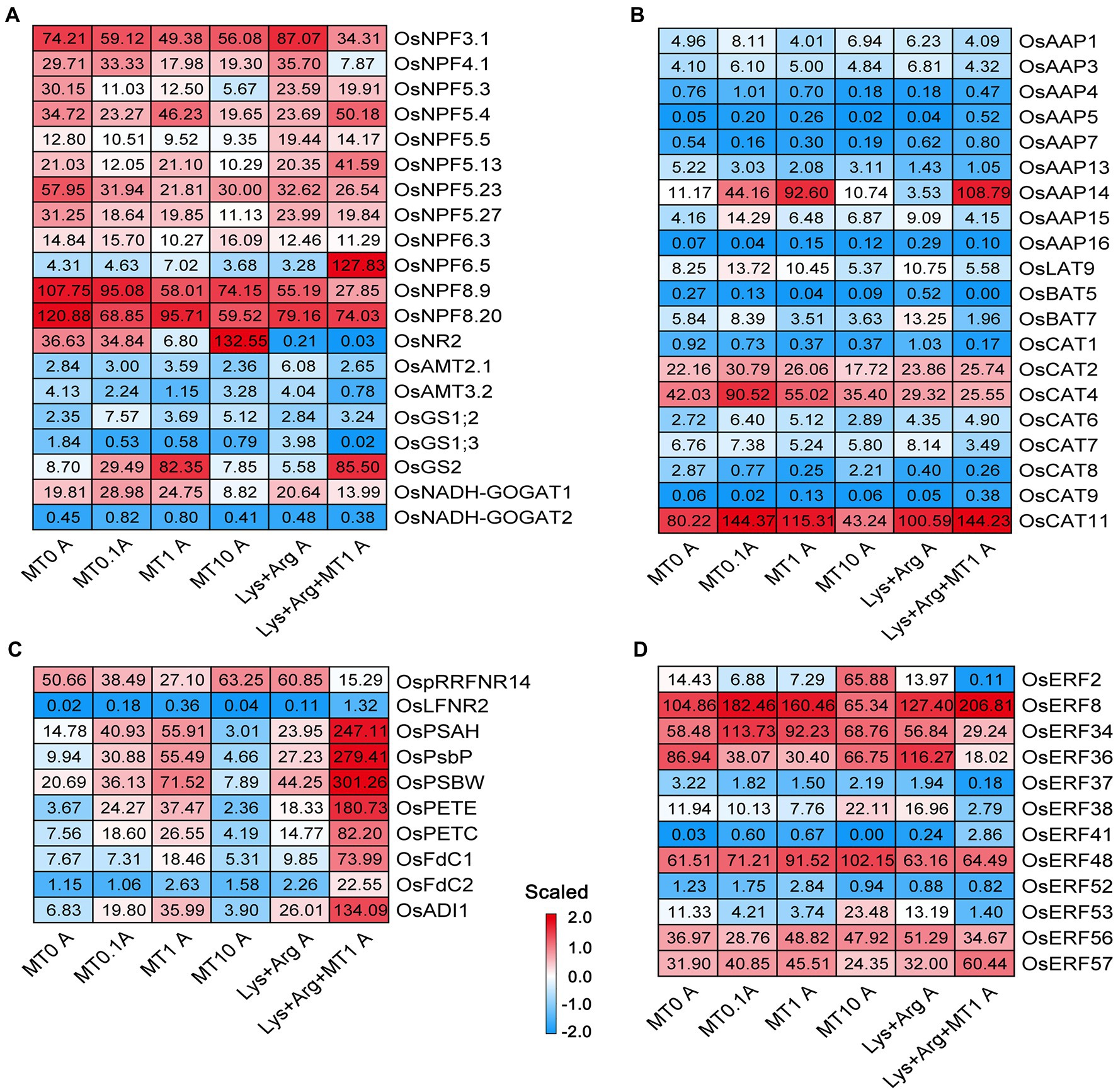
Figure 4. The expression of genes involved in nitrogen metabolism, photosynthesis and stress in axillary bud under different concentrations of melatonin and amino acids. Heatmaps display the expression patterns of genes involved in nitrate and ammonium transport and assimilation (A), amino acid transport (B), photosynthesis (C) and stress (D). Red and blue represent the highest and lowest level of expression. MT represents melatonin, Lys represents lysine, and Arg represents arginine. A at the bottom of each graph represents axillary bud, and B at the bottom of each graph represents basal part. MT0, MT0.1, MT1 and MT10 represents melatonin at 0 μm, 0.1 μm, 1 μm and 10 μm, respectively.
In the photosynthetic pathway, the expression of OsPSAH, OsPsbP, OsPSBW, OsPETE, OsPETC, OsFdC1, OsFdC2, and OsADI1 was higher in axillary buds upon treatment with 1 μm melatonin compared to treatment with 0 μm melatonin (Figure 4C). However, the expression of these genes was inhibited by 10 μm melatonin, significantly increased upon treatment with 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin (Figure 4C), and not inhibited upon treatment with 1.5 mm Lys + 0.5 mm Arg (Figure 4C). This suggests that ferredoxin-associated genes were involved in the regulation rice axillary bud elongation by melatonin, but not in elongation inhibition by Lys and Arg. Furthermore, the expression levels of some genes with unknown functions in the photosynthetic pathway were higher in axillary buds under 1 μm melatonin and 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatments compared to other treatments (Supplementary Figure S10A).
The expression of some genes with unknown functions in the carbon metabolic pathway was higher in axillary buds under 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatment compared to other treatments (Supplementary Figures S10B,C). However, there was no significant difference in the expression of these genes among different treatments in the basal parts (Supplementary Figures S11A–C). In the phenylpropanoid biosynthesis pathway, the expression of some genes was higher in axillary buds (Supplementary Figures S10D–F) and basal parts (Supplementary Figures S11D–F) under 10 μm melatonin treatment compared to other treatments. In the stress response pathway, some ethylene response factor genes were also differentially expressed; however, their expression was neither induced nor inhibited by melatonin (Figure 4D), indicating that melatonin did not directly regulate rice axillary bud elongation. Additionally, only the expression of OsCML10, OsCML15, and OsCML31 of the calmodulin family was higher in both axillary buds and basal parts under 1 μm melatonin treatment (Supplementary Figure S12).
Expression Profiles of Genes Related to Phytohormone Pathways
To determine the functions of phytohormones involved in the axillary bud outgrowth of plants exposed to different melatonin and amino acid treatments, we analyzed the expression profiles of genes associated with phytohormones [auxin, CKs, abscisic acid (ABA), BRs and SLs]. The expression of some auxin-responsive SMALL AUXIN UPREGULATED RNA and Aux/IAA gene family members increased when treated with 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin compared to other treatments (Figures 5A,B). The expression of AUXIN RESPONSE FACTOR (ARF) gene family members OsARF15 and OsARF25 increased in the plants exposed to 10 μm melatonin, whereas that of OsARF1 decreased in plants treated with 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin compared to other treatments (Figure 5C). Previous reports have shown that OsARF1 (Attia et al., 2008) and OsARF25 (Li et al., 2020) exhibit negative regulatory effects on rice tillers, which indicates that some ARF members involved in the auxin pathway are also involved in the regulation of axillary bud outgrowth by melatonin and amino acids. In the CK pathway, the expression of OsLOGL5 and OsLOGL10 increased upon treatment with 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin, whereas that of OsCKX4 was only induced upon treatment with 10 μm melatonin compared to other treatments (Figure 5D). Previous reports suggest that both OsLOGL5 (Hou et al., 2021a) and OsCKX4 (Wang et al., 2022a) regulate the elongation of axillary buds in rice.
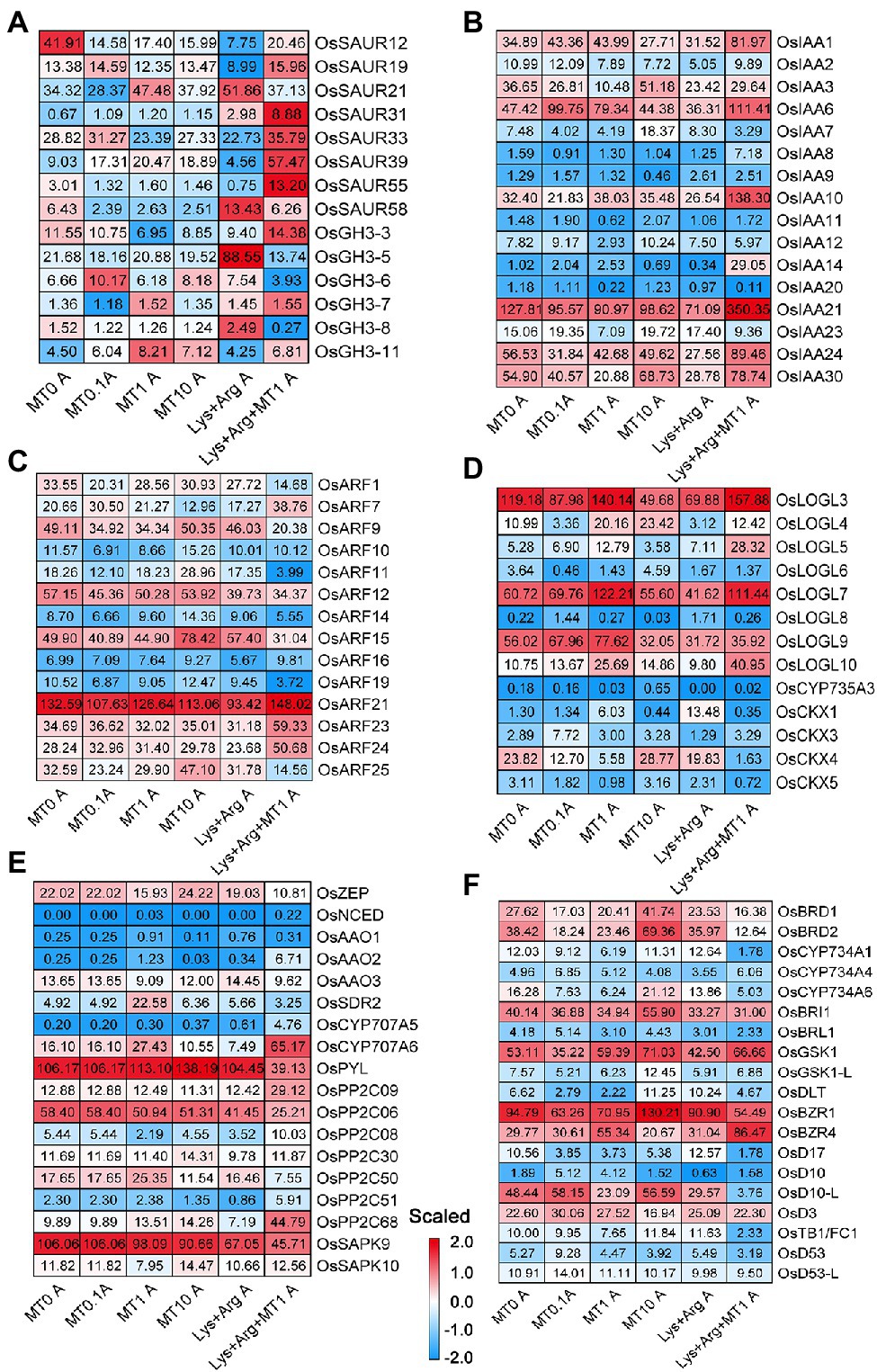
Figure 5. The expression of genes involved in plant hormone signal transduction in axillary bud under different concentrations of melatonin and amino acids. Heatmaps display the expression patterns of genes involved in auxin (A–C), cytokinin (D), abscisic acid (E), ethylene and strigolactone (F). Red and blue represent the highest and lowest level of expression. MT represents melatonin, Lys represents lysine, and Arg represents arginine. A at the bottom of each graph represents axillary bud, and B at the bottom of each graph represents basal part. MT0, MT0.1, MT1 and MT10 represents melatonin at 0 μm, 0.1 μm, 1 μm and 10 μm, respectively.
In the ABA pathway, only the expression of OsSDR2 and OsPP2C50 was induced in 1 μm melatonin treatment compared to other treatments (Figures 5E,F), whereas the expression of OsCYP707A6, OsPP2C09, OsPP2C68, and OsBZIP63 was higher in 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatment (Figures 5E,F). In bZIP transcription factor of ABA pathway, the expression of OsFBL30, and OsbZIP37 in axillary buds was higher under 1 μm Melatonin, and the expression of OsBZIP63 was higher in 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatment (Supplementary Figure S13). Reports suggest that OsPP2C09 regulates drought resistance, growth, and development in rice (Miao et al., 2020), indicating that ABA pathway may be involved in the regulation of axillary buds by melatonin and amino acids. In the BRs pathway, the expression of DEGs showed no obvious change under different treatments (Figure 5F). Furthermore, consistent with the role of SLs as inhibitors of axillary bud growth, the expression of SL biosynthesis-related gene OsD10-L and signaling-related gene OsTB1 was low under 1 μm melatonin and 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatments (Figure 5F). In addition, only the expression of OsLOGL3 in CK pathway was induced upon 1 μm melatonin and 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatments compared to other treatments in the basal parts (Supplementary Figure S14).
Melatonin and Basic Amino Acids Regulate the Expression of OsGS2 and OsAAP14 in Rice Roots
Since melatonin, Lys, and Arg, which are involved in the nitrogen metabolism pathway, affected the expression of OsGS2 and OsAAP14 (Figures 4A,B), OsGS2 and OsAAP14 promoter-GUS transgenic lines were perform for experiments. GUS staining and RT-qPCR showed that OsGS2 was highly expressed in root tip (Supplementary Figures S15A,H), lateral root (Supplementary Figures S15B,H) basal part (Supplementary Figures S15C,H), culm (Supplementary Figures S15D,H) and panicle (Supplementary Figures S15G,H), but was only slightly expressed in leaf sheath (Supplementary Figures S15E,H) and leaf blade (Supplementary Figures S15F,H). Furthermore, we found a similar expression pattern in OsAAP14 (Supplementary Figure S16). Besides, OsGS2 and OsAAP14 promoter-GUS transgenic lines were treated with exogenous melatonin, Lys, and Arg to determine whether they altered the expression of the above mentioned genes (Figure 6). Results showed that the root tip and lateral roots of OsGS2 (Figure 6A) and OsAAP14 (Figure 6B) promoter-GUS transgenic rice plants treated with 1 μm melatonin were deeply stained at both 2 h and 24 h compared to other concentrations of melatonin. With the increase in melatonin concentration (0–1 μm), GUS staining gradually deepened, but the intensity of the stain decreased when the concentration exceeded 1 μm (Figure 6). To systematically characterize the regulation of OsGS2 and OsAAP14 expression by basic amino acids or their combination with melatonin, GUS staining was performed after treatments with 1.5 mm Lys + 0.5 mm Arg or 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin. Weak and strong staining was observed at 2 h and 24 h upon 1.5 mm Lys + 0.5 mm Arg and 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatments, respectively (Figure 6). The above results were further determined by RT-qPCR (Supplementary Figure S17).
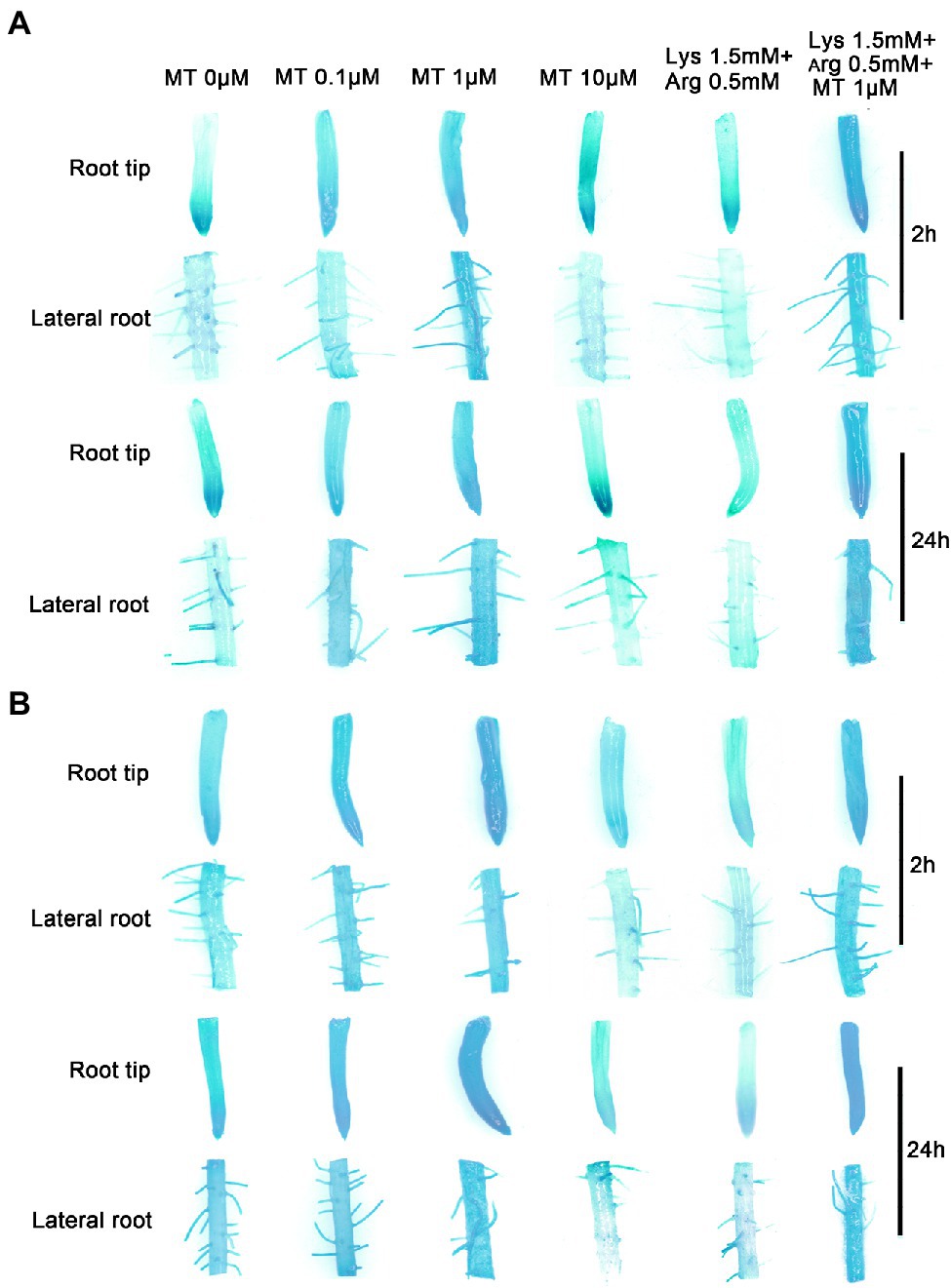
Figure 6. The expression of OsAAP14 and OsGS2 is regulated by melatonin and amino acids. Promoter-GUS transgenic seedlings were grown under basic nutrient solutions supplemented with different concentrations of melatonin (0 μm, 0.1 μm, 1 μm, 10 μm), 1.5 mm Lys + 0.5 mm Arg, or 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin for 2 h and 24 h, respectively. OsAAP14 promoter-GUS staining in the root tip and lateral root (A). OsGS2 promoter-GUS staining in the root tip and lateral root (B). MT represents melatonin, Lys represents lysine, and Arg represents arginine.
In addition, it showed that the axillary bud and basal part of OsAAP14 (Supplementary Figure S18A) and OsGS2 (Supplementary Figure S18B) promoter-GUS transgenic rice plants were deeply stained under 1 μm melatonin compared to other concentrations of melatonin. With the increase in melatonin concentration (0–1 μm), GUS staining gradually deepened, but the intensity of the stain decreased when the concentration exceeded 1 μm, and weak and strong staining was observed upon 1.5 mm Lys + 0.5 mm Arg and 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatments, respectively (Supplementary Figures S18A,B). The above results were further determined by RT-qPCR (Supplementary Figures S18C,D). Furthermore, to determine the tissue-specific expression pattern of OsAAP14 and OsGS2, sections of the GUS-stained organs were made (Figure 7). Sectioning confirmed that the expression of OsAAP14 was observed in the cortex parenchyma and vascular tissue of the root, and GUS activity was abundant in 1 μm melatonin or 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatments compared to other treatments (Figure 7A). Similarly, GUS activity of OsGS2 was abundant in the cortex parenchyma and vascular tissue of the root especially under 1 μm melatonin or 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatments compared to other treatments (Figure 7B). Overall, these observations show that OsGS2 and OsAAP14 were highly expressed in the roots and basal parts in response to melatonin and amino acids.
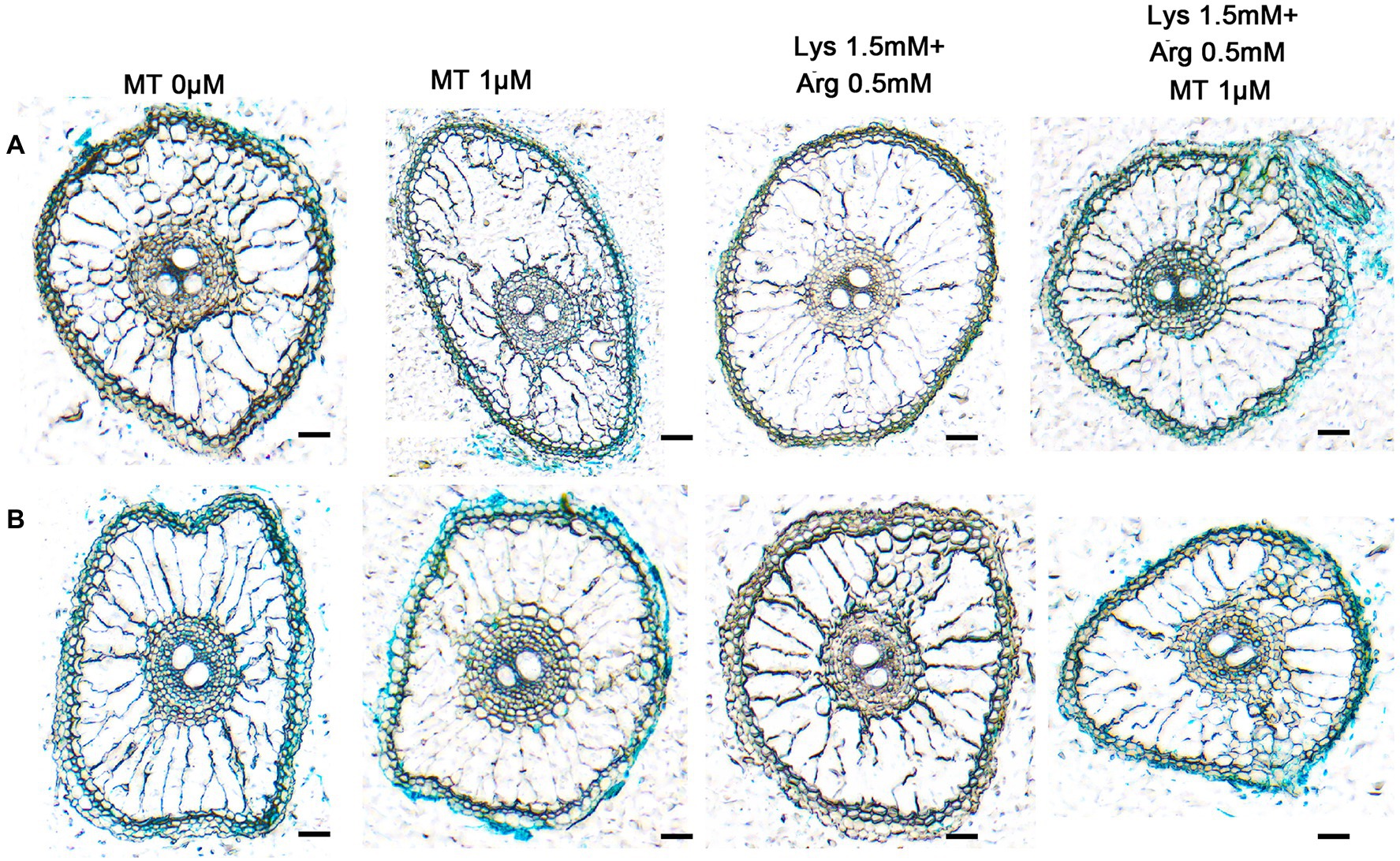
Figure 7. Section analysis of OsAAP14 and OsGS2 promoter-GUS transgenic plants regulating by melatonin and amino acids. Promoter-GUS transgenic seedlings were grown under basic nutrient solutions supplemented with different concentrations of melatonin (0 μm, 0.1 μm, 1 μm, 10 μm), 1.5 mm Lys + 0.5 mm Arg, or 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin. Paraffin sections of the stained roots of OsAAP14 (A) and OsGS2 (B) promoter-GUS transgenic plants. MT represents melatonin, Lys represents lysine, and Arg represents arginine. Scale bar = 40 μm.
Altered Expression of OsGS2 and OsAAP14 Influences Axillary Bud Outgrowth in Rice
To further determine the role of OsGS2 and OsAAP14 in regulating rice axillary bud outgrowth in response to melatonin and basic amino acid treatments, we generated OsGS2 OE and RNAi (Ri) lines and OsAAP14 OE (two splicing variants: OsAAP14a-OE and OsAAP14b-OE) and CRISPR lines in the japonica ZH11 background (Supplementary Figures S19, S20). We found that both the first and second axillary bud length in the OsGS2 OE line increased when grown in control solution compared to the Ri line and WT (Figures 8A,C,D). Furthermore, both the length of first and second axillary buds of the OE lines and WT significantly increased under 1 μm melatonin treatment, compared to their corresponding OE lines and WT grown in normal solution (Figures 8B,E,F). However, the axillary buds of the OsGS2 Ri line were short, and there was no difference in the length of axillary buds exposed to melatonin and normal solution (Figure 8). This indicated that the expression of OsGS2 in the Ri line induced by melatonin was lost, and that melatonin no longer influenced the elongation of axillary buds after the reduction in OsGS2 expression. Similarly, the first and second bud length in the OsAAP14a and OsAAP14b OE lines increased when grown in normal solution compared to that of the OsAAP14 CRISPR line and WT (Figures 9A,C,D). When treated with 1 μm melatonin, the axillary buds of OsAAP14a and OsAAP14b OE lines and WT further elongated, but the axillary bud length of the OsAAP14 CRISPR line did not change (Figure 9). Besides, we also found that both variants of OsAAP14 OE lines enhanced grain yield by increasing tiller and grain number per plant, while the OsAAP14 CRISPR line exhibited significantly reduced tiller number and grain yield (Supplementary Figure S21). In addition, it was showed that the contents of total free amino acids and total chlorophyll were significantly increased both in OsAAP14 OE lines and OsGS2 OE lines compared with WT, while significantly decreased in OsAAP14 CRISPR lines and OsGS2 Ri lines compared with WT (Supplementary Figure S22).
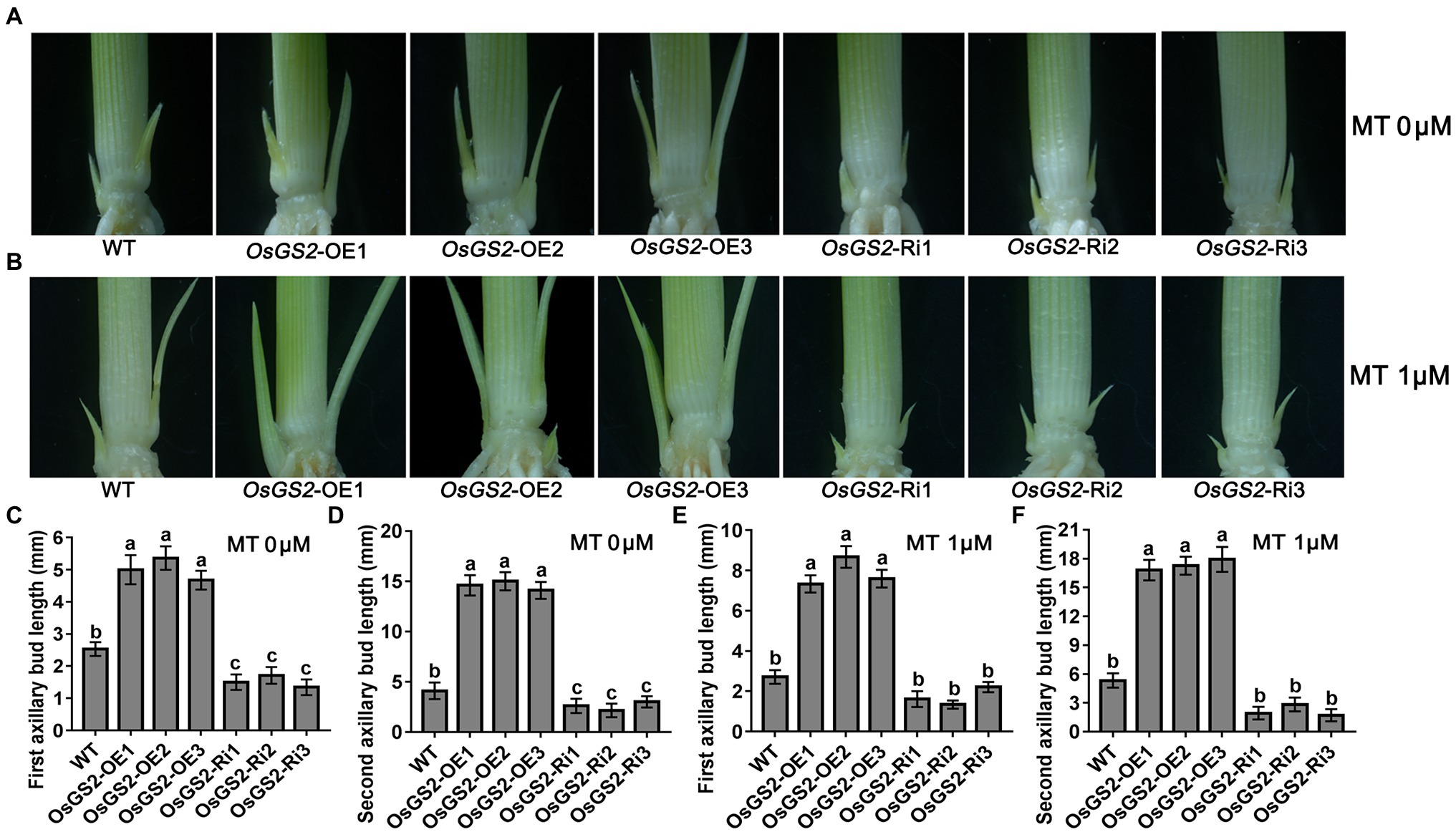
Figure 8. Effect of melatonin on outgrowth axillary bud of OsGS2 transgenic seedlings. Phenotypes of outgrowth axillary bud with altered expression of OsGS2 grown under 0 μm melatonin (A) and 1 μm melatonin (B). Statistical analysis of the first (C,E) and second (D,F) axillary bud length in (A,B). Values are means ± SD (n > 20), and the significance levels of different lowercase letters were as follows, p < 0.05. WT represents wild-type ZH11, OE1-OE3 represent three lines of OsGS2 over-expressing transgenic plants, and Ri1-Ri3 represent three lines of OsGS2 RNAi transgenic plants. MT represents melatonin.
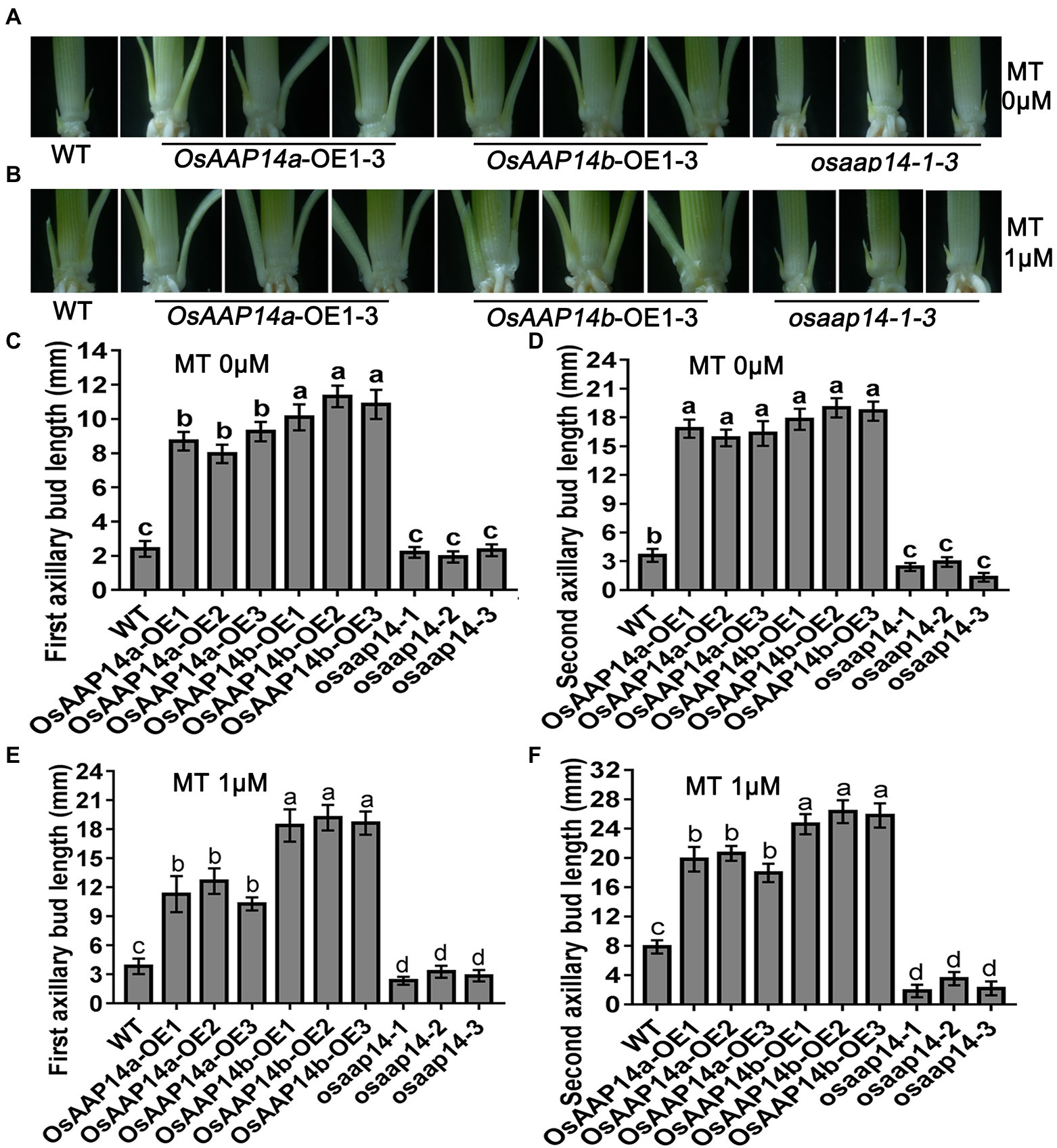
Figure 9. Effect of melatonin on outgrowth axillary bud of OsAAP14 transgenic seedlings. Phenotypes of outgrowth axillary bud with altered expression of OsAAP14 grown under 0 μm melatonin (A) and 1 μm melatonin (B). Statistical analysis of the first (C,E) and second (D,F) axillary bud length in (A,B). Values are means ± SD (n > 20), and the significance levels of different lowercase letters were as follows, p < 0.05. WT represents wild-type ZH11, OsAAP14a-OE1-OE3 represent three lines of OsAAP14 longer splicing variant over-expressing transgenic plants, OsAAP14b-OE1-OE3 represent three lines of OsAAP14 shorter splicing variant over-expressing transgenic plants, and osaap14-1-3 represent three lines of OsAAP14 CRISPR mutants. MT represents melatonin.
To investigate why the effect of 1.5 mm Lys, 0.5 mm Arg and 1 μm melatonin on axillary bud elongation was stronger than that of 1 μm melatonin alone, seedlings amino acid-uptake assay was performed under 1 μm melatonin. Stronger fluorescence signals were detected in the OsAAP14 OEb lines cultured with 0.5 mm Arg-FITC and 0.5 mm Lys-FITC for 2 h or 6 h than those of the ZH11 and OEa lines, and the FITC signal was weaker in Ri lines than in ZH11 (Supplementary Figures S23, S24). Similarly, Stronger fluorescence signals were detected in the OsGS2 OE lines cultured with 0.5 mm Arg-FITC, and 0.5 mm Lys-FITC for 2 h or 6 h than those of the ZH11, and the FITC signal was weaker in Ri lines than in ZH11 (Supplementary Figures S23, S24). These results suggest that OsGS2 and OsAAP14 play an important role in response to melatonin and the subsequent regulation of axillary bud outgrowth, tillering, and grain yield.
Discussion
Exogenous Low Melatonin Promotes Axillary Bud Outgrowth in Rice
Melatonin promotes coleoptile growth in four monocot species including canary grass, wheat, barley, and oat (Hernandez-Ruiz et al., 2005), and increases root growth in Brassica juncea (Chen et al., 2009). And it was found that melatonin might function as an auxin to promote vegetative growth (Kolář and Macháčková, 2005). Recently, the identification of the melatonin receptor CAND2/PMTR1 (Wei et al., 2018) established that the phytohormone melatonin plays an important role in plant growth, development, and stress resistance. However, whether melatonin affects axillary bud elongation and tillering in rice still remains unclear. In this study, we found that low concentrations of melatonin (0–1 μm) promoted the elongation of the first and second axillary buds in rice, but higher melatonin concentrations (10–1,000 μm) inhibited the elongation of these buds (Figure 1). The previous study reported that lateral root growth is stimulated at low melatonin levels, while lateral root growth is inhibited at higher melatonin levels (Park, 2011). Furthermore, a low concentration promotes adventitious root regeneration but a growth inhibitory effect at high concentrations, and the negative effect of the high melatonin concentration could be due to antagonism between melatonin and Ca2+-calmodulin (Sarropoulou et al., 2012). Thus, our results demonstrate that exogenous melatonin also promotes or inhibits axillary bud outgrowth in rice.
Similarly, previous study reported that 0.1 mm melatonin has a stimulatory effect on root growth of B. juncea, while 100 mm is inhibitory, and the effect on root growth and endogenous indole-3-acetic acid (IAA) levels determined (Chen et al., 2009). This study showed that the expression of some auxin-responsive and Aux/IAA gene family members increased when treated with 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin compared to other treatments (Figure 5). This indicates that there is a link between melatonin and auxin in growth regulation. Besides, the inhibitory effect of SLs was detectable at GR24 (a synthetic SL analogue) concentrations as low as 10 nM, and axillary bud outgrowth was approximately completely inhibited at 1 μm GR24 (Umehara et al., 2008). Furthermore, SLs inhibited axillary bud outgrowth in rice by altering the tillering inhibition pathway through increasing the expression of SL signaling-related gene OsTB1 (Fang et al., 2020). In our study, the expression of OsTB1 was low under 1 μm melatonin alone or 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin treatments (Figure 5). This suggests that melatonin may affect rice axillary buds through the expression of OsTB1 in SL pathway. In addition, BRs play a significant role in promoting rice tillering; 1 μm CS (a synthetic BR analogue) strongly promoted axillary bud outgrowth and significantly enhanced tillering in rice (Fang et al., 2020). However, different melatonin concentrations has no significant effect on the expression of BRs related genes (Figure 5). Therefore, how Auxin and SL pathways interact with melatonin in regulating rice axillary bud elongation, needs to be further studied.
Melatonin Relieves the Inhibition of Axillary Bud Outgrowth Under High Concentration of Lys and Arg
Amino acids are crucial for basal metabolism as they participate in protein synthesis and modulate plant growth and development (Yadav et al., 2015). It has been reported that Lys can inhibit mitotic activity in the root apical meristem, and that exogenous Lys can reduce the length of the main root of Arabidopsis (Yang et al., 2014). Recently, it has been shown that low concentrations of Lys and Arg promoted the elongation of rice buds, but their high concentrations inhibited bud elongation (Lu et al., 2018). In this study, the addition of high concentrations of Lys and Arg inhibited the elongation of first and second axillary buds in rice (Figure 1). One possible explanation is that Lys and Arg can influence antioxidant enzymes activities and photosynthetic pathway by initiating the ROS accumulation and nitric oxide signaling pathway (Wei et al., 2021). Our results also showed that nitrogen metabolism, stress, and photosynthetic pathways play important roles in response to amino acids Lys and Arg in axillary buds. However, the complementation of these amino acids were with 1 μm melatonin not only alleviated their inhibitory effect on rice axillary buds but also considerably promoted axillary bud elongation (Figure 1). This may be due to the dual role of melatonin in stress resistance and growth promotion. One previous study reported that melatonin enhances plant growth and abiotic stress tolerance in soybean plants (Wei et al., 2015). Transcriptome analysis revealed that melatonin may exert its functions mainly through regulation of photosynthesis, the cell cycle, DNA replication, starch/sucrose metabolism, and lipid biosynthesis (Wei et al., 2015).
Besides, it has been reported that the addition of melatonin lessens the amount of oxidative damage brought on by salinity, perhaps by directly scavenging H2O2 or enhancing the activities of antioxidative enzymes such as ascorbate peroxidase, catalase, and peroxidase (Li et al., 2012). And 0.1 μm melatonin significantly alleviates growth inhibition and enables plants to maintain an improved photosynthetic capacity in Malus hupehensis Rehd (Li et al., 2012). In addition, melatonin promotes seed germination under high salinity by regulating antioxidant systems, ABA and GA4 interaction in cucumber (Zhang et al., 2014). Our results showed that the expression of OsPSAH, OsPsbP, OsPSBW, OsPETE, OsPETC, OsFdC1, OsFdC2, and OsADI1 in photosynthetic pathway was higher in axillary buds upon treatment with 1.5 mm Lys + 0.5 mm Arg + 1 μm melatonin compared to other treatments (Figure 4). Therefore, it was demonstrated that melatonin promoted axillary bud outgrowth and tillering in rice exposed to high concentrations of basic amino acids, indicating that melatonin can alleviate the inhibition of rice axillary bud elongation under nitrogen stress, such as high concentrations of amino acids Lys and Arg.
Key Genes of Nitrogen Metabolism That Respond to Changes in Melatonin and Amino Acids to Regulate Tillering in Rice
Previously, pathway enrichment analysis indicated that eight pathways were over-represented among differentially expressed genes between control and melatonin-treated bermudagrass plants, including N metabolism, major carbohydrate metabolism, TCA/org transformation, transport, hormone metabolism, metal handling, redox, and secondary metabolism (Shi et al., 2015). In this study, we found that the expression of nitrate transporter gene OsNPF6.5, the glutamine synthetase gene OsGS2 and amino acid transporter gene OsAAP14 involved in nitrogen metabolism was regulated by different concentrations of melatonin, while the expression of these genes was inhibited by basic amino acid treatment (Figure 4). If melatonin was added with basic amino acids, the expression of these genes notably increased (Figure 4). It was reported that OsNPF6.5 (OsNRT1.1B) indica variant enhances nitrate uptake, tiller number and nitrogen use efficiency in rice (Hu et al., 2015). Recently, glutamine synthetase OsGS2 has been shown to respond to different nitrogen concentrations, and regulate the elongation of axillary buds and tiller number in rice (Wang et al., 2020). Besides, the previous study indicated that another glutamine synthetase gene OsGS1.2 could mediate axillary buds outgrowth and tiller number in rice by regulating CK pathway (Yamaya and Kusano, 2014). In addition, other amino acid transporter genes, such as OsAAP1 and OsAAP4, positively regulate rice axillary bud outgrowth and tillering by promoting the transport of neutral amino acids (Ji et al., 2020; Fang et al., 2021). Therefore, melatonin could greatly promote nitrogen transport and assimilation through relieving the high concentration nitrogen inhibition. We also found that the grain yield was enhanced in OsAAP14 OE lines by increasing tiller and grain number per plant compared with WT (Supplementary Figure S21), and the contents of total free amino acids and total chlorophyll were also significantly increased both in OsAAP14 OE lines and OsGS2 OE lines compared with WT (Supplementary Figure S22). This indicates that OsNPF6.5, OsGS2 and OsAAP14 are key melatonin- and basic amino acid-responsive genes of nitrogen metabolism that regulate axillary bud outgrowth in rice. Overall, the present study provides insights into axillary bud outgrowth in plants, and how melatonin can help improve rice grain yield, especially under nitrogen stress.
Concluding Remarks
In this study, we found that different concentrations of melatonin influenced axillary bud outgrowth in rice, and moderate melatonin concentrations alleviated the inhibition of axillary bud outgrowth under high concentrations of Lys and Arg. RNA-seq data indicated that the genes involved in nitrogen metabolism, phytohormone signal transduction, and phenylpropanoid biosynthesis were involved in the elongation of axillary bud outgrowth under different melatonin, Lys, and Arg concentrations. In addition, we indicated that the rice glutamine synthetase gene OsGS2 and amino acid transporter OsAAP14, involved in the nitrogen metabolism pathway, were involved in the regulation of axillary bud outgrowth under different concentration of melatonin, Lys, and Arg. These results elucidate the regulatory mechanism underlying the effect of melatonin on axillary bud outgrowth under different amino acid concentrations, and provide insights into improving rice tillering and grain yield using melatonin under amino acid-rich environments.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
ZF designed the study. GY and XW performed the experiments. ZF and GY drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Wuhan Science and Technology Project (2020020601012259), the Talent Project from Guizhou Education Department [Qian jiao he KY zi (2021) 024], the Key Cultivation Project of Guizhou University (201903).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.900262/full#supplementary-material
Footnotes
References
Arite, T., Iwata, H., Ohshima, K., Maekawa, M., Nakajima, M., Kojima, M., et al. (2007). DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51, 1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x
Arite, T., Umehara, M., Ishikawa, S., Hanada, A., Maekawa, M., Yamaguchi, S., et al. (2009). d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424. doi: 10.1093/pcp/pcp091
Arnao, M. B., and Hernández-Ruiz, J. (2020). Melatonin in flowering, fruit set and fruit ripening. Plant Reprod. 33, 77–87. doi: 10.1007/s00497-020-00388-8
Attia, A., Abdelkhalik, A. F., Ammar, M. H., Wei, C., Yang, J., Lightfoot, D. A., et al. (2008). Antisense phenotypes reveal a functional expression of OsARF1, an auxin response factor, in transgenic rice. Curr. Issues Mol. Biol. 11, i29–i34. doi: 10.21775/9781912530069.04
Bahcesular, B., Yildirim, E. D., Karaçocuk, M., Kulak, M., and Karaman, S. (2020). Seed priming with melatonin effects on growth, essential oil compounds and antioxidant activity of basil (Ocimum basilicum L.) under salinity stress. Ind. Crop. Prod. 146:112165. doi: 10.1016/j.indcrop.2020.112165
Bajwa, V. S., Shukla, M. R., Sherif, S. M., Murch, S. J., and Saxena, P. K. (2014). Role of melatonin in alleviating cold stress in Arabidopsis thaliana. J. Pineal Res. 56, 238–245. doi: 10.1111/jpi.12115
Chen, Y., Fan, X., Song, W., Zhang, Y., and Xu, G. (2012). Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 10, 139–149. doi: 10.1111/j.1467-7652.2011.00637.x
Chen, Q., Qi, W., Reiter, R. J., Wei, W., and Wang, B. (2009). Exogenously applied melatonin stimulates root growth and raises endogenous indoleacetic acid in roots of etiolated seedlings of Brassica juncea. J. Plant Physiol. 166, 324–328. doi: 10.1016/j.jplph.2008.06.002
Duan, J., Yu, H., Yuan, K., Liao, Z., Meng, X., Jing, Y., et al. (2019). Strigolactone promotes cytokinin degradation through transcriptional activation of CYTOKININ oxidase/dehydrogenase 9 in rice. Proc. Natl. Acad. Sci. U. S. A. 116, 14319–14324. doi: 10.1073/pnas.1810980116
Fang, Z., Bai, G., Huang, W., Wang, Z., Wang, X., and Zhang, M. (2017). The rice peptide transporter OsNPF7.3 is induced by organic nitrogen, and contributes to nitrogen allocation and grain yield. Front. Plant Sci. 8:1338. doi: 10.3389/fpls.2017.01338
Fang, Z., Ji, Y., Hu, J., Guo, R., Sun, S., and Wang, X. (2020). Strigolactones and brassinosteroids antagonistically regulate the stability of the D53-OsBZR1 complex to determine FC1 expression in rice tillering. Mol. Plant 13, 586–597. doi: 10.1016/j.molp.2019.12.005
Fang, Z., Wu, B., and Ji, Y. (2021). The amino acid transporter OsAAP4 contributes to rice tillering and grain yield by regulating neutral amino acid allocation through two splicing variants. Rice. 14:2. doi: 10.1186/s12284-020-00446-9
Fang, Z., Xia, K., Yang, X., Grotemeyer, M. S., Meier, S., Rentsch, D., et al. (2013). Altered expression of the PTR/NRT1 homologue OsPTR9 affects nitrogen utilization efficiency, growth and grain yield in rice. Plant Biotechnol. J. 11, 446–458. doi: 10.1111/pbi.12031
Funayama, K., Kojima, S., Tabuchi-Kobayashi, M., Sawa, Y., Nakayama, Y., Hayakawa, T., et al. (2013). Cytosolic glutamine synthetase1;2 is responsible for the primary assimilation of ammonium in rice roots. Plant Cell Physiol. 54, 934–943. doi: 10.1371/journal.pone.0095581
Gao, S., Fang, J., Xu, F., Wang, W., Sun, X., Chu, J., et al. (2014). CYTOKININ oxidase/DEHYDROGENASE4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiol. 165, 1035–1046. doi: 10.1104/pp.114.238584
Gonzalez-Grandio, E., Pajoro, A., Franco-Zorrilla, J. M., Tarancon, C., Immink, R. G., and Cubas, P. (2017). Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascad in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. U. S. A. 114, E245–E254. doi: 10.1073/pnas.1613199114
Hernandez-Ruiz, J., Cano, A., and Arnao, M. B. (2005). Melatonin acts as a growth-stimulating compound in some monocot species. J. Pineal Res. 39, 137–142. doi: 10.1111/j.1600-079x.2005.00226.x
Hou, M., Luo, F., Wu, D., Zhang, X., Lou, M., Shen, D., et al. (2021b). OsPIN9, an auxin efflux carrier, is required for the regulation of rice tiller bud outgrowth by ammonium. New Phytol. 229, 935–949. doi: 10.1111/nph.16901
Hou, M., Yu, M., Li, Z., Ai, Z., and Chen, J. (2021a). Molecular regulatory networks for improving nitrogen use efficiency in rice. Int. J. Mol. Sci. 22:9040. doi: 10.3390/ijms22169040
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838. doi: 10.1038/ng.3337
Huang, W., Bai, G., Wang, J., Zhu, W., Zeng, Q., Lu, K., et al. (2018). Two splicing variants of OsNPF7.7 regulate shoot branching and nitrogen utilization efficiency in rice. Front. Plant Sci. 9:300. doi: 10.3389/fpls.2018.00300
Huang, W., Nie, H., Feng, F., Wang, J., Lu, K., and Fang, Z. (2019). Altered expression of OsNPF7.1 and OsNPF7.4 differentially regulates tillering and grain yield in rice. Plant Sci. 283, 23–31. doi: 10.1016/j.plantsci.2019.01.019
Huang, M., Yang, C., Ji, Q., Jiang, L., Tan, J., and Li, Y. (2013). Tillering responses of rice to plant density and nitrogen rate in a subtropical environment of southern China. Field Crops Res. 149, 187–192. doi: 10.1016/j.fcr.2013.04.029
James, D., Borphukan, B., Fartyal, D., Ram, B., Singh, J., Manna, M., et al. (2018). Concurrent overexpression of OsGS1;1 and OsGS2 genes in transgenic rice (Oryza sativa L.): impact on tolerance to abiotic stresses. Front. Plant Sci. 9:786. doi: 10.3389/fpls.2018.00786
Ji, Y., Huang, W., Wu, B., Fang, Z., and Wang, X. (2020). The amino acid transporter AAP1 mediates growth and grain yield by regulating neutral amino acid uptake and reallocation in Oryza sativa. J. Exp. Bot. 71, 4763–4777. doi: 10.1093/jxb/eraa256
Jiang, L., Liu, X., Xiong, G., Liu, H., Chen, F., Wang, L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405. doi: 10.1038/nature12870
Kissen, R., Winge, P., Tran, D. H. T., Jørstad, T. S., Størseth, T. R., Christensen, T., et al. (2010). Transcriptional profiling of an Fd-GOGAT1/GLU1 mutant in Arabidopsis thaliana reveals a multiple stress response and extensive reprogramming of the transcriptome. BMC Genomics 11:190. doi: 10.1186/1471-2164-11-190
Kolář, J., and Macháčková, I. (2005). Melatonin in higher plants: occurrence and possible functions. J. Pineal Res. 39, 333–341. doi: 10.1111/j.1600-079X.2005.00276.x
Larkindale, J., and Huang, B. R. (2005). Effects of abscisic acid, salicylic acid, ethylene and hydrogen peroxide in thermotolerance and recovery for creeping bentgrass. Plant Growth Regul. 47, 17–28. doi: 10.1007/s10725-005-1536-z
Lee, H. Y., Byeon, Y., and Back, K. (2014). Melatonin as a signal molecule triggering defense responses against pathogen attack in Arabidopsis and tobacco. J. Pineal Res. 57, 262–268. doi: 10.1111/jpi.12165
Lerner, A. B., Case, J. D., Takahashi, Y., Lee, T. H., and Mori, W. (1958). Isolation of melatonin, the pineal gland factor that lightens melanocyteS1. J. Am. Chem. Soc. 80:2587. doi: 10.1021/ja01543a060
Li, Y., Li, J., Chen, Z., Wei, Y., Qi, Y., and Wu, C. (2020). OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice. Plant Biotechnol. J. 18, 2015–2026. doi: 10.1111/pbi.13360
Li, X., Qian, Q., Fu, Z., Wang, Y., Xiong, G., Zeng, D., et al. (2003). Control of tillering in rice. Nature 422, 618–621. doi: 10.1038/nature01518
Li, C., Wang, P., Wei, Z., Liang, D., Liu, C., Yin, L., et al. (2012). The mitigation effects of exogenous melatonin on salinity-induced stress in Malus hupehensis. J. Pineal Res. 53, 298–306. doi: 10.1111/j.1600-079X.2012.00999.x
Lin, H., Wang, R., Qian, Q., Yan, M., Meng, X., Fu, Z., et al. (2009). DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21, 1512–1525. doi: 10.1105/tpc.109.065987
Lin, Q., Zhang, Z., Wu, F., Feng, M., Sun, Y., Chen, W., et al. (2020). The APC/CTE E3 ubiquitin ligase complex mediates the antagonistic regulation of root growth and tillering by ABA and GA. Plant Cell 32, 1973–1987. doi: 10.1105/tpc.20.00101
Liu, Y., Ding, Y., Wang, Q., Meng, D., and Wang, S. (2011). Effects of nitrogen and 6-Benzylaminopurine on rice tiller bud growth and changes in endogenous hormones and nitrogen. Crop Sci. 51, 786–792. doi: 10.2135/cropsci2010.04.0217
Liu, X., Hu, Q., Yan, J., Sun, K., Liang, Y., Jia, M., et al. (2020). ζ-Carotene isomerase suppresses tillering in rice through the coordinated biosynthesis of strigolactone and abscisic acid. Mol. Plant 13, 1784–1801. doi: 10.1016/j.molp.2020.10.001
Lu, G., Coneva, V., Casaretto, J. A., Ying, S., Mahmood, K., Liu, F., et al. (2015). OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 83, 913–925. doi: 10.1111/tpj.12939
Lu, K., Wu, B., Wang, J., Zhu, W., Nie, H., Qian, J., et al. (2018). Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechnol. J. 16, 1710–1722. doi: 10.1111/pbi.12907
Luo, J., Huang, W., Yan, J., Fang, Z., and Ren, M. (2022). The GzMYB-7D1 gene of Guizimai no. 1 wheat is essential for seed anthocyanins accumulation and yield regulation. Plant Sci. 320:111293. doi: 10.1016/j.plantsci.2022.111293
Luo, L., Takahashi, M., Kameoka, H., Qin, R., Shiga, T., Kanno, Y., et al. (2019). Developmental analysis of the early steps in strigolactone-mediated axillary bud dormancy in rice. Plant J. 97, 1006–1021. doi: 10.1111/tpj.14266
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Mandal, M. K., Suren, H., Ward, B., Boroujerdi, A., and Kousik, C. (2018). Differential roles of melatonin in plant-host resistance and pathogen suppression in cucurbits. J. Pineal Res. 65:e12505. doi: 10.1111/jpi.12505
Miao, J., Li, X., Li, X., Tan, W., You, A., Wu, S., et al. (2020). OsPP2C09, a negative regulatory factor in abscisic acid signalling, plays an essential role in balancing plant growth and drought tolerance in rice. New Phytol. 227, 1417–1433. doi: 10.1111/nph.16670
Ohashi, M., Ishiyama, K., Kojima, S., Kojima, M., Sakakibara, H., Yamaya, T., et al. (2017). Lack of cytosolic glutamine synthetase1;2 activity reduces nitrogen-dependent biosynthesis of cytokinin required for axillary bud outgrowth in rice seedlings. Plant Cell Physiol. 58, 679–690. doi: 10.1093/pcp/pcx022
Ohashi, M., Ishiyama, K., Kusano, M., Fukushima, A., Kojima, S., Hanada, A., et al. (2015). Lack of cytosolic glutamine synthetase1;2 in vascular tissues of axillary buds causes severe reduction in their outgrowth and disorder of metabolic balance in rice seedlings. Plant Cell Physiol. 81, 347–356. doi: 10.1111/tpj.12731
Ohashi, M., Ishiyama, K., Kusano, M., Fukushima, A., Kojima, S., Hayakawa, T., et al. (2018). Reduction in sucrose contents by downregulation of fructose-1,6-bisphosphatase2 caused cessation of tiller outgrowth of rice mutants lacking glutamine synthetase1;2 during the early growth stage. Rice 11:65. doi: 10.1186/s12284-018-0261-y
Onik, J. C., Wai, S. C., Li, A., Lin, Q., Sun, Q., Wang, Z., et al. (2021). Melatonin treatment reduces ethylene production and maintains fruit quality in apple during postharvest storage. Food Chem. 337:127753. doi: 10.1016/j.foodchem.2020.127753
Park, W. J. (2011). Melatonin as an endogenous plant regulatory signal: debates and perspectives. J. Plant Biol. 54, 143–149. doi: 10.1007/s12374-011-9159-6
Peng, B., Kong, H., Li, Y., Wang, L., Zhong, M., Sun, L., et al. (2014). OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat. Commun. 5:4847. doi: 10.1038/ncomms5847
Posmyk, M. M., Kuran, H., Marciniak, K., and Janas, K. M. (2008). Presowing seed treatment with melatonin protects red cabbage seedlings against toxic copper ion concentrations. J. Pineal Res. 45, 24–31. doi: 10.1111/j.1600-079X.2007.00552.x
Sarropoulou, V. N., Therios, I. N., and Dimassi-Theriou, K. N. (2012). Melatonin promotes adventitious root regeneration in in vitro shoot tip explants of the commercial sweet cherry rootstocks CAB-6P (Prunus cerasus L.), Gisela 6 (P. cerasus× P. canescens), and MxM 60 (P. avium× P. mahaleb). J. Pineal Res. 52, 38–46. doi: 10.1111/j.1600-079X.2011.00914.x
Sasaki, Y., Ando, H., and Kakuda, K. (2002). Relationship between ammonium nitrogen in soil solution and tiller number at early growth stage of rice. Soil Sci. Plant Nutr. 48, 57–63. doi: 10.1080/00380768.2002.10409171
Shi, H., Jiang, C., Ye, T., Tan, D., Reiter, R. J., Zhang, H., et al. (2015). Comparative physiological, metabolomic, and transcriptomic analyses reveal mechanisms of improved abiotic stress resistance in bermudagrass [Cynodon dactylon (L). Pers.] by exogenous melatonin. J. Exp. Bot. 66, 681–694. doi: 10.1093/jxb/eru373
Sofy, A. R., Sofy, M. R., Hmed, A. A., Dawoud, R. A., Refaey, E. E., Mohamed, H. I., et al. (2021). Molecular characterization of the alfalfa mosaic virus infecting Solanum melongena in Egypt and the control of its deleterious effects with melatonin and salicylic acid. Plants 10:459. doi: 10.3390/plants10030459
Tan, X. L., Fan, Z. Q., Kuang, J. F., Lu, W. J., Reiter, R. J., Lakshmanan, P., et al. (2019). Melatonin delays leaf senescence of Chinese flowering cabbage by suppressing ABFs-mediated abscisic acid biosynthesis and chlorophyll degradation. J. Pineal Res. 67:e12570. doi: 10.1111/jpi.12570
Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T., Takeda-Kamiya, N., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. doi: 10.1038/nature07272
Vafadar, F., Amooaghaie, R., Ehsanzadeh, P., Ghanati, F., and Sajedi, R. H. (2020). Crosstalk between melatonin and Ca2+/CaM evokes systemic salt tolerance in Dracocephalum kotschyi. J. Plant Physiol. 252:153237. doi: 10.1016/j.jplph.2020.153237
Wang, D., Chen, Q., Chen, W., Guo, Q., Xia, Y., Wang, S., et al. (2021). Physiological and transcription analyses reveal the regulatory mechanism of melatonin in inducing drought resistance in loquat (Eriobotrya japonica Lindl.) seedlings. Environ. Exp. Bot. 181:104291. doi: 10.1016/j.envexpbot.2020.104291
Wang, J., Lu, K., Nie, H., Zeng, Q., Wu, B., Qian, J., et al. (2018). Rice nitrate transporter OsNPF7.2 positively regulates tiller number and grain yield. Rice 11:12. doi: 10.1186/s12284-018-0205-6
Wang, R., Qian, J., Fang, Z., and Tang, J. (2020). Transcriptomic and physiological analyses of rice seedlings under different nitrogen supplies provide insight into the regulation involved in axillary bud outgrowth. BMC Plant Biol. 20:197. doi: 10.1186/s12870-020-02409-0
Wang, H., Tong, X., Tang, L., Wang, Y., Zhao, J., Li, Z., et al. (2022a). RLB (RICE LATERAL BRANCH) recruits PRC2-mediated H3K27 tri-methylation on OsCKX4 to regulate lateral branching. Plant Physiol. 188, 460–476. doi: 10.1093/plphys/kiab494
Wang, J., Wan, R., Nie, H., Xue, S., and Fang, Z. (2022b). OsNPF5.16, a nitrate transporter gene with natural variation, is essential for rice growth and yield. Crop J. 10, 397–406. doi: 10.1016/j.cj.2021.08.005
Wang, J., Wu, B., Lu, K., Wei, Q., Qian, J., Chen, Y., et al. (2019). The amino acid permease 5 (OsAAP5) regulates tiller number and grain yield in rice. Plant Physiol. 180, 1031–1045. doi: 10.1104/pp.19.00034
Wei, W., Li, Q., Chu, Y., Reiter, R., Yu, X., Zhu, D., et al. (2015). Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 66, 695–707. doi: 10.1093/jxb/eru392
Wei, J., Li, D. X., Zhang, J. R., Shan, C., Rengel, Z., Song, Z. B., et al. (2018). Phytomelatonin receptor PMTR1-mediated signaling regulates stomatal closure in Arabidopsis thaliana. J. Pineal Res. 65:e12500. doi: 10.1111/jpi.12500
Wei, Q., Yan, Z., Xiong, Y., and Fang, Z. (2021). Altered expression of OsAAP3 influences rice lesion mimic and leaf senescence by regulating arginine transport and nitric oxide pathway. Int. J. Mol. Sci. 22:2181. doi: 10.3390/ijms22042181
Xu, M., Zhu, L., Shou, H., and Wu, P. (2005). A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46, 1674–1681. doi: 10.1093/pcp/pci183
Yadav, U. P., Ayre, B. G., and Bush, D. R. (2015). Transgenic approaches to altering carbon and nitrogen partitioning in whole plants: assessing the potential to improve crop yields and nutritional quality. Front. Plant Sci. 6:275. doi: 10.3389/fpls.2015.00275
Yamaya, T., and Kusano, M. (2014). Evidence supporting distinct functions of three cytosolic glutamine synthetases and two NADH-glutamate synthases in rice. J. Exp. Bot. 65, 5519–5525. doi: 10.1093/jxb/eru103
Yang, H., Postel, S., Kemmerling, B., and Ludewig, U. (2014). Altered growth and improved resistance of Arabidopsis against pseudomonas syringae by overexpression of the basic amino acid transporter AtCAT1. Plant Cell Environ. 37, 1404–1414. doi: 10.1111/pce.12244
Yang, L., You, J., Li, J., Wang, Y., and Chan, Z. (2021). Melatonin promotes Arabidopsis primary root growth in an IAA-dependent manner. J. Exp. Bot. 72, 5599–5611. doi: 10.1093/jxb/erab196
Yao, C., and Finlayson, S. A. (2015). Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol. 169, 611–626. doi: 10.1104/pp.15.00682
Yeh, S. Y., Chen, H. W., Ng, C. Y., Lin, C. Y., Tseng, T. H., Li, W. H., et al. (2015). Down-regulation of cytokinin oxidase 2 expression increases tiller number and improves rice yield. Rice 8:36. doi: 10.1186/s12284-015-0070-5
Zhang, Z., Chu, G., Liu, L., Wang, Z., Wang, X., Zhang, H., et al. (2013). Mid-season nitrogen application strategies for rice varieties differing in panicle size. Field Crops Res. 150, 9–18. doi: 10.1016/j.fcr.2013.06.002
Zhang, J., Shi, Y., Zhang, X., Du, H., Xu, B., and Huang, B. (2017). Melatonin suppression of heat-induced leaf senescence involves changes in abscisic acid and cytokinin biosynthesis and signaling pathways in perennial ryegrass (Lolium perenne L.). Environ. Exp. Bot. 138, 36–45. doi: 10.1016/j.envexpbot.2017.02.012
Zhang, H., Zhang, N., Yang, R., Wang, L., Sun, Q., Li, D., et al. (2014). Melatonin promotes seed germination under high salinity by regulating antioxidant systems, ABA and GA4 interaction in cucumber (Cucumis sativus L.). J. Pineal Res. 57, 269–279. doi: 10.1111/jpi.12167
Zhou, F., Lin, Q., Zhu, L., Ren, Y., Zhou, K., Shabek, N., et al. (2013). D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. doi: 10.1038/nature16537
Keywords: melatonin, axillary bud, nitrogen, amino acid, OsAAP14, rice
Citation: Yang G, Wei X and Fang Z (2022) Melatonin Mediates Axillary Bud Outgrowth by Improving Nitrogen Assimilation and Transport in Rice. Front. Plant Sci. 13:900262. doi: 10.3389/fpls.2022.900262
Edited by:
Guillaume Pilot, Virginia Tech, United StatesCopyright © 2022 Yang, Wei and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongming Fang, em1mYW5nQGd6dS5lZHUuY24=
 Guo Yang
Guo Yang Xilin Wei1
Xilin Wei1 Zhongming Fang
Zhongming Fang