- 1Hunan Research Center of Engineering Technology for Utilization of Environmental and Resources Plant, Central South University of Forestry and Technology, Changsha, China
- 2Central South Inventory and Planning Institute of National Forestry and Grassland Administration, Changsha, China
- 3College of Forestry, Northwest A&F University, Yangling, China
Flavonoids are important secondary metabolites involved in plant development and environmental responses. Sex differences in flavonoids are common in plants. Broussonetia papyrifera is a dioecious plant that is rich in flavonoids. However, few studies have been done on its molecular mechanism, especially sex differences. In the present study, we performed an integrated transcriptomics and metabolomics analysis of the sex differences in the accumulation of flavonoids in B. papyrifera leaves at different developmental stages. In general, flavonoids accumulated gradually with developmental time, and the content in female plants was higher than that in male plants. The composition of flavonoids in female and male plants was similar, and 16 kinds of flavonoids accumulated after flowering. Correspondingly, a significant enrichment of differentially expressed genes and metabolites was observed in the flavonoid biosynthesis pathway. WGCNA and qRT-PCR analyses identified several key genes regulating the accumulation of flavonoids, such as those encoding CHS, CHI and DFR. In addition, 8 TFs were found to regulate flavonoid biosynthesis by promoting the expression of multiple structural genes. These findings provide insight into flavonoid biosynthesis in B. papyrifera associated molecular regulation.
Introduction
Dioecious plants are an important part of terrestrial ecosystems and play a positive role in protecting species diversity and maintaining ecosystem stability. From an evolutionary perspective, dioeciousness increases genetic diversity, allowing plants to exhibit a variety of morphologies and traits (Al-Dossary et al., 2021). On the other hand, environmental stress or human selection can further contribute to differences in plant traits between sexes. Actually, sex differences in metabolites are common in plants. In Rhamnus alpinus, male plants have higher concentrations of defense-related compounds (anthraquinones) in the leaves than female plants, thus indicating increased resistance to biotic stress in male plants (Banuelos and Obeso, 2004). In Populus cathayana, under normal conditions, the sucrose concentration of male leaves was higher than that of female leaves. Nitrogen or potassium deficiency significantly enhanced leaf sucrose in females, but had less effect on males (Yang et al., 2015; Wu et al., 2021). Until now, most reports have focused on the physiological basis of dioecious plants, but few studies have been done on molecular biology, which is a crucial question. Because changes in physiological traits may be due to a genetic basis or in combination with other factors such as allocation. It is necessary to distinguish temporary changes in stress from stable genetic features.
Flavonoids, as a class of polyphenolic compounds with a C6–C3–C6 double aromatic ring, are important secondary metabolites in plants development. Flavonoids can improve the adaptability of plants to the various terrestrial environments, and participate in plant ecological defense, such as promoting plant growth (Stafford, 1991), resisting diseases (Agati et al., 2012), resisting the damage of ultraviolet radiation (Harborne and Williams, 2000), scavenging oxygen free radicals (Agati et al., 2009, 2011; Lillo et al., 2010; Tattini et al., 2010), driving away foragers, and preventing the invasion of harmful microorganisms (Falcone Ferreyra et al., 2012). Therefore, flavonoids may be a secondary metabolite in response to the environment. In addition to being beneficial to plants themselves, flavonoids have also been developed and utilized for their medicinal effects. More and more evidences suggest that flavonoids and related compounds can provide beneficial effects on cancer and chronic diseases, including cardiovascular disease, type II diabetes and non-alcoholic fatty liver disease (NAFLD), due to their immunomodulatory, anti-inflammatory and antioxidant properties (González-Gallego et al., 2010; Ying et al., 2013; Pisonero-Vaquero et al., 2014). Sex differentiation of flavonoid content has recently been emphasized because it not only reflects the survival strategies of different sexual plants, but also affects the development and utilization. For instances, the content of flavonoids in female Hippophae rhamnoides leaves is slightly lower than that of male plants (Lu et al., 2018). Under limited phosphorus conditions, the synthesis of flavonoids and condensed tannins in leaves, as well as the synthesis of phenolic acids in stems and roots in females P. tremula, were greater than in males (Randriamanana et al., 2014). In Ginkgo biloba, more phenolics were extracted from leaves of male trees than from female trees (Koczka et al., 2015).
Broussonetia papyrifera (paper mulberry) is a dioecious plant belonging to the Moraceae family. Once an important source of forage, the distribution of B. papyrifera has expanded with human migration (Chang et al., 2015). It is widely distributed in Asia and Pacific countries, and is an important economic plant in most areas (Zheng et al., 2002). In recent year, due to its easily digestible crude fiber and high protein content, B. papyrifera is still used as forage to address the shortage of feedstuff. At the same time, the tree has the characteristics of strong germination, fast growth, resistance to salt stress, heavy metals and air pollution, and has been used as a pioneer plant in polluted areas (Li et al., 2012; Zhang et al., 2013; Zhao et al., 2014). Compared with other woody plants, B. papyrifera is rich in flavonoids, polyphenols, and fructose (Feng et al., 2019), which may be the reason why B. papyrifera has been used as a traditional Chinese medicine for a long history (Feng et al., 2008). For instance, prenylflavone derivatives from B. papyrifera have an inhibitory effect on cancer cells (Guo et al., 2013), and polyphenols from B. papyrifera can inhibit coronavirus protease (Park et al., 2017). Broussochalcone A, papyriflavonol A, 3′-(3-methylbut-2-enyl)-3′,4′,7-trihydroxyflavane, and broussoflavan A from B. papyrifera have been proved to be potent the main protease (Mpro) inhibitors. There are more effective than the two repurposed drugs (lopinavir and darunavir) and may serve as promising anti-COVID-19 drugs (Ghosh et al., 2021). B. papyrifera leaves and roots are rich in flavonoids. The high flavonoid content may result from the significant expansion of gene family in the flavonoid synthesis pathway (Peng et al., 2019). On the other hand, previous studies have shown that there are sex differences in the accumulation of flavonoids in B. papyrifera (Zhao, 2011), but the molecular mechanism is still unclear. Understanding synthesis mechanism of flavonoids in B. papyrifera leaves of different sexes will not only help us to understand the survival strategies of B. papyrifera in the wild, but also help us to select superior varieties.
At present, metabolomic and transcriptomic analysis have been widely used to study the response mechanisms of metabolite accumulation under different conditions (Zhang et al., 2021). In our study, we focus on the molecular mechanisms of flavonoid biosynthesis in B. papyrifera leaves, especially their sex differences. Our work aims to: (I) elucidate the accumulation of flavonoids in B. papyrifera leaves of different sexes, and (II) screen the key genes for the synthesis of B. papyrifera flavonoids. The results of this work may provide insight into flavonoid biosynthesis in B. papyrifera associated molecular regulation.
Materials and Methods
Plant Materials
Broussonetia papyrifera leaves were collected from April 2019 to October 2019 on the campus of Central South University of Forestry and Technology (28°6′25.48″ N, 112°59′37.68″ E), China. Three adult female (F) and three adult male (M) B. papyrifera were selected as experimental samples. They were divided into three groups, each consisting of a male and a female, and they were grown together. The distance between each group was about 200 meters. The six samples were the same age and well-grown. Subsequently, three typical phenological periods of B. papyrifera were selected for sampling. The first was 30 days after flowering (the flowering stage, F30/M30). At this point, the gender could be clearly distinguished, and the leaves were fully expanded. The second was 120 days after flowering (the fruiting stage, F120/M120). It was the peak fruiting period of B papyrifera, with green and red fruits accounting for about half. The third was 180 days after flowering (the defoliation stages, F180/M180). At this point, the leaves began to fall (Supplementary Figure S1). Every treatment was repeated three times. These leaves were immediately frozen in liquid nitrogen and stored at −80°C for metabolite extraction, transcriptome sequencing, and qRT-PCR analysis.
Total Flavonoids Analysis
The content of total flavonoids was determined by the aluminum nitrate chromogenic method (Yang, 2018). In brief, 1.00 g freeze-dried leaf powder mixed with 5 ml 70% (v/v) ethanol, and then extracted at 50°C for 1.5 h. The supernatant was collected by centrifugation at 8,000 r/min for 10 min at room temperature, and then diluted to 25 ml with 70% (v/v) ethanol. The content of total flavonoids was measured by spectrophotometer UV-1200 (Mapada, China) at 510 nm using rutin as the standard substance.
Flavonoid Identification and Quantification
100 mg of vacuum freeze-dried leaf samples were dissolved in 1.0 ml 70% (v/v) methanol. The supernatant was collected by centrifugation at 10,000 g for 10 min. The test solution was obtained by filtration with a microporous membrane (0.22 μm Pore Size). Flavonoids were analyzed by Metware Biotechnology Co., Ltd. (Wuhan, China) using an LC-ESI-MS/MS System (UPLC, Shim-Pack UFLC SHIMADZU CBM30A System, www.shimadzu.com.cn/; MS, Applied Biosystems 4000 Q TRAP, www.appliedbiosystems.com.cn/). Metabolite quantification was performed using a scheduled multiple reaction monitoring (MRM) method, which has been previously described (Chen et al., 2013). Setting |Log2 (Fold Change)| ≥2 and |Log2 (Fold Change)| ≤ 0.5, and P < 0.05 as thresholds for DAFs.
RNA-seq Analysis
The total RNA was extracted from frozen leaf samples using an EASYspin Plus Plant RNA Kit by Biomarker Technologies Co., Ltd. (Beijing, China). A total amount of 1 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext®Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer's recommendations and transcriptome sequencing were performed by Biomarker Technologies Co., Ltd. (Beijing, China) using an Illumina Hiseq 2000 platform. Clean data were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. Transcriptome assembly was accomplished using Trinity (Grabherr et al., 2011) by default. The assembled transcript was mapped to the clean reads to obtain the mapped reads. Gene expression levels were estimated by RNA-seq by Expectation-Maximization (RSEM) (Li and Dewey, 2011), and fragments per kilobase of transcript per million mapped reads (FPKM) was used for gene/transcript level quantification. Differential expression analysis of two samples was performed using the EBSeq R package. P value was adjusted using q value. q value < 0.005 and |log2 (Fold Change) |>1 was set as the threshold for significantly differential expression. Gene functions were annotated based on the following databases: NR (NCBI non-redundant protein sequences), Pfam (Protein family), KOG/COG/eggNOG (Clusters of Orthologous Groups of proteins), Swiss-Prot (A manually annotated and reviewed protein sequence database), KEGG (Kyoto Encyclopedia of Genes and Genomes), GO (Gene Ontology). We used KOBAS (Chen et al., 2011) software to test the statistical enrichment of differential expression genes in the KEGG pathways.
Co-expression Network Analysis for Construction of Modules
To identify key regulatory genes in the flavonoid biosynthesis pathway, WGCNA was performed using the WGCNA R package (v 1.70). The adjacency matrix between different genes was constructed with a soft power of 26 and a dynamic tree cut procedure (merge cut height = 0.25, min module size = 30) was used to screen similar modules in the hierarchical tree. The module eigengene was defined as the first principal component of a given module, and then used to represent the expression profile of module genes in each sample. The Pearson correlations between the eigengenes of each module and the abundance of flavonoids were performed using OriginPro 2016. Genes associated with flavonoid biosynthesis pathways were identified by KEGG annotation (ko00941, ko00942, ko00943 and ko00944) (Guo et al., 2020). Genes related to flavonoid biosynthesis and TFs in flavonoid biosynthesis highly correlated modules were constructed co-expression networks with edge weights >0.3, and visualized the candidate target genes with Cytoscape software (Version 3.9.0) (Ding et al., 2021). Genes with high degrees in the network were selected as hub genes.
qRT-PCR Analysis
Total RNA was extracted from leaf samples according to the instructions of Biofit kit (Tsingke, China). The strand cDNA was synthesized using Goldenstar RT6 cDNA synthesis kit (Tsingke, China). The expression levels of representative unigenes and selected key genes in the flavonoid biosynthesis pathway were analyzed by qRT-PCR using FQD-96A (Bioer Technology, China). The 20 μl reaction mixture contained 10 μl SYBR® Green Real-time PCR Master Mix (CWBIO), 1 μl cDNA template, 0.8 μM of each forward and reverse primer, and 7.4 μl ddH2O. The following cycling parameters were applied for amplification: 95°C for 1 min followed by 40 cycles of 95°C for 15 s, 60°C for 15 s, 72°C for 30 s, then followed by 95°C for 5 s, 60°C for 1 min, 0.11°C / s to 95°C, 50°C for 30 s for plate reading. The primers list is shown in Supplementary Table S1. The actin gene was selected as an internal reference (Xu et al., 2019). Three replicates were performed for each sample. Quantitative data were analyzed using the 2−ΔΔCT method.
Statistical Analysis
Differences between samples were determined by one-way analysis of variance (ANOVA) and significant differences were calculated by the least significant difference (LSD) test at P < 0.05.
Results
Total Flavonoids in Broussonetia papyrifera Leaves
The content of flavonoids in leaves of female B. papyrifera was higher than that of male at three developmental stages. There were significant differences between flowering and defoliation stages (P < 0.05). Accumulation of flavonoids increased with leaf development. F180 and M180 had the highest content at 1.961 ± 0.191 mg g−1 and 1.571 ± 0.213 mg g−1, respectively (Figure 1).
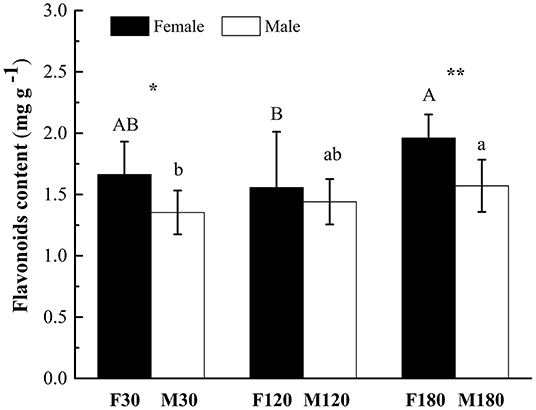
Figure 1. Total flavonoid contents in developing Broussonetia papyrifera leaves. Asterisks indicate statistically significant differences between female and male plants: no symbol P > 0.05; *P < 0.05; **P < 0.01. Capital letters indicate statistically significant differences (P < 0.05) among the three development stages of female plants. Lowercase letters indicate statistically significant differences (P < 0.05) among the three development stages of male plants.
Identification of Flavonoids in Broussonetia papyrifera Leaves
A total of 192 kinds of flavonoids substances were detected in B. papyrifera leaves, including 85 flavones, 46 flavonols, 23 flavonoid carbonoside, 10 flavan-3-ols, 9 anthocyanins, 6 dihydroflavone, 7 dihydroflavonol, 4 isoflavones and 2 chalcones (Supplementary Table S2). 170 kinds of flavonoids were identified in F30 or M30, and 188 kinds of flavonoids were identified in F120, M120, F180 or M180 (Figure 2A). 163 kinds of flavonoids were shared among 6 leaf samples. Besides, 16 kinds of flavonoids were found in the fruiting and defoliation stage but not in flowering stage (Figure 2B). Comparing the sex differences of each flavonoid, there were 50 DAFs in flowering stage, 49 DAFs in fruiting stage and 72 DAFs in defoliation stage. Among this DAFs, the content of most flavonoids in female B. papyrifera was higher than that in male (Supplementary Figure S2). Cluster analysis of 192 kinds of flavonoids showed F30 and M30 were located in the same branch, and F120, M120, F180 and M180 were located in the same branch. At the same time, these flavonoids were divided into 3 clade, clade I had high content substances in the F30 and M30, clade II high content substances in the M30 and F120, and clade III high content substances in the F180 and M120 (Figure 3A).
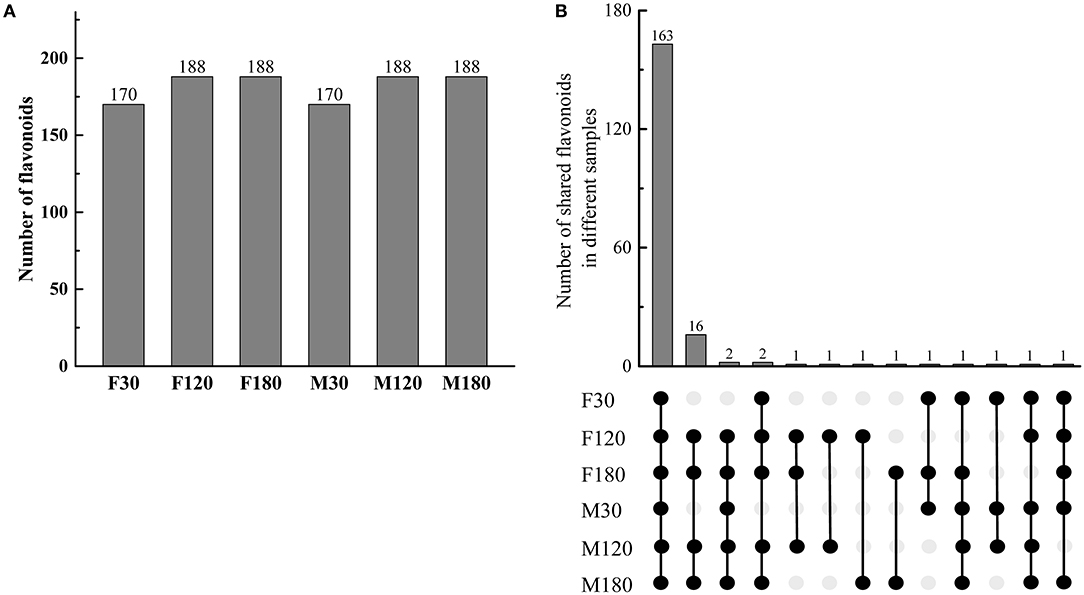
Figure 2. Gender differences in the types and numbers of flavonoid metabolites in Broussonetia papyrifera leaves. (A) Numbers of flavonoids in different samples. (B) Numbers of shared flavonoids in different samples. The black solid circle indicated the presence of compounds in samples, and the gray solid circle indicated the absence of compounds in samples.
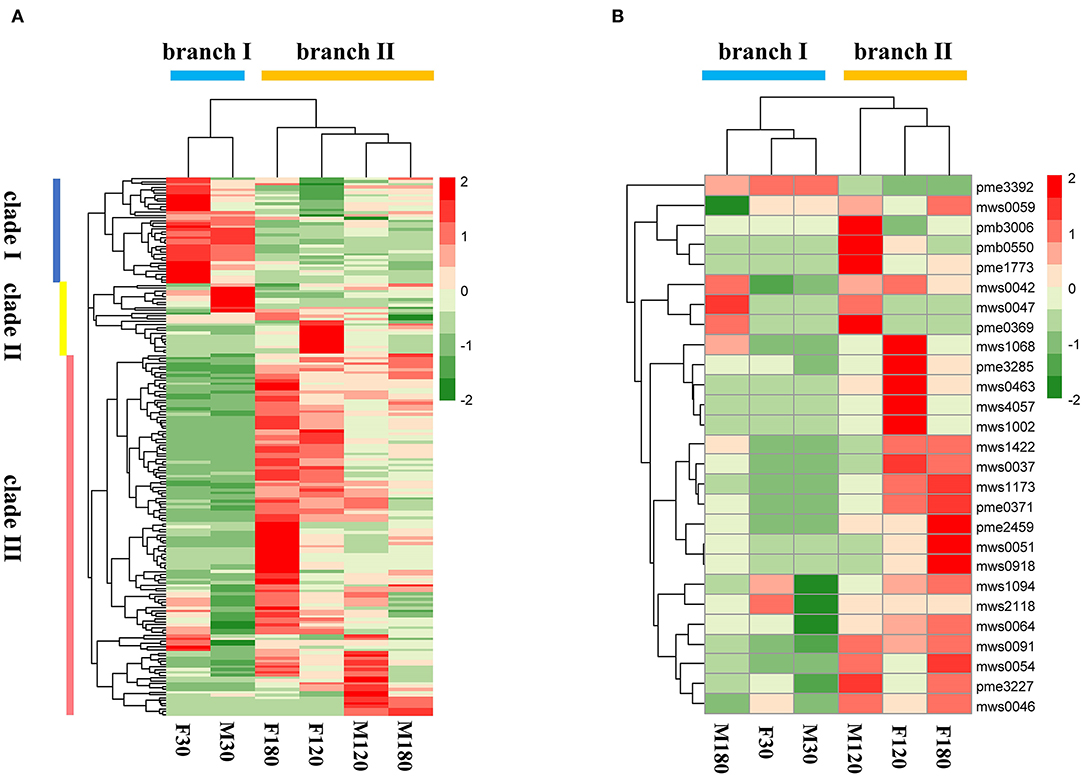
Figure 3. Cluster analysis of flavonoids in Broussonetia papyrifera leaves. (A) 192 flavonoids found in samples. (B) 27 flavonoids identified in the flavonoids biosynthesis pathway.
According to KEGG annotation results, 27 substances were identified in the flavonoids biosynthesis pathway. Remarkably, most substances were down-regulated in male samples compared with female samples (Supplementary Table S3). Cluster analysis of 27 flavonoids showed that F30, M30 and M180 were located in the same branch, while F120, M120 and F180 were located in the same branch. The number of flavonoids with high content in the former was lower than that in the latter (Figure 3B).
Transcriptome Analysis of Broussonetia papyrifera Leaves of Different Sexes
The throughput and sequencing quality were high enough to warrant further analysis. Each library received 22244561–22999380 clean reads and 17941528–21223800 mapped reads. The mapped ratio was in the range of 83.79–85.73%. Q30 percentages and GC percentages were 93.08–94.45% and 46.70–47.37%, respectively (Supplementary Table S4). During the development of B. papyrifera leaves, a total of 5,672 DEGs were obtained, including 1,475 at flowering stage, 4,391 at fruiting stage and 1,030 at defoliation stage (Supplementary Figure S3). Comparing the DEGs with the common databases, 4,887 differentially expressed genes were annotated, accounting for 86.16% (Supplementary Table S5).
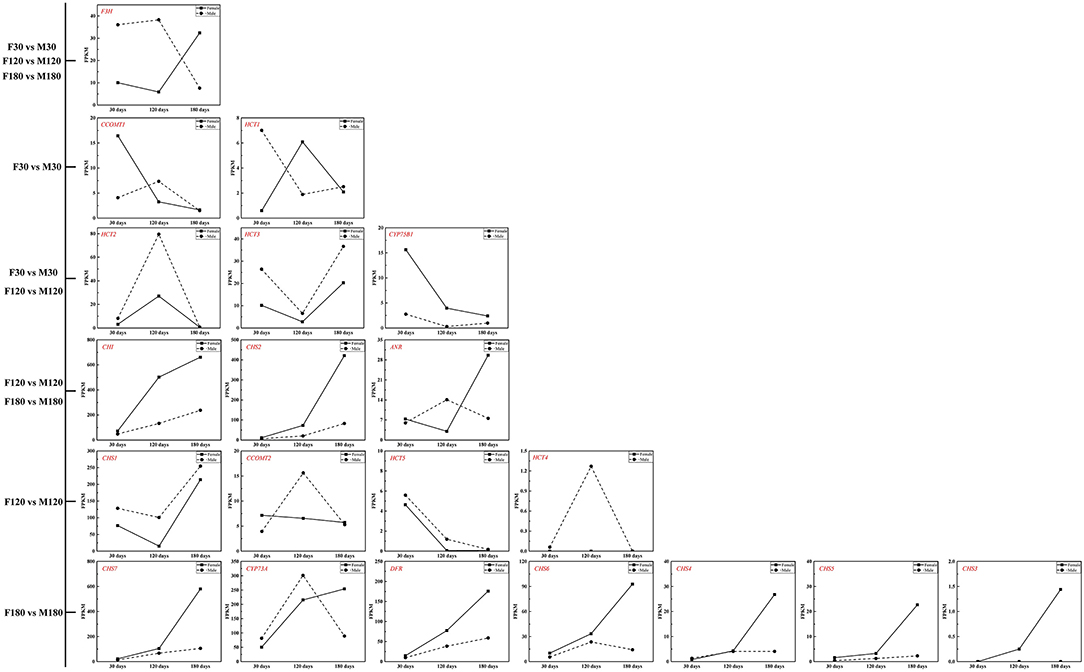
The KEGG enrichment analysis revealed that most genes were mainly enriched in “Ribosome,” “Plant-pathogen interaction,” and “Carbon metabolism” (Figure 4). Of note, there were 6 DEGs for F30 vs. M30, 11 DEGs for F120 vs. M120, and 11 DEGs for F180 vs. M180 in the flavonoid biosynthesis pathway. According to KEGG annotation results, 20 genes were identified in the flavonoids biosynthesis pathway (Supplementary Table S6). Among them, 14 genes were only highly expressed in the fruiting and defoliation stages, accounting for 70% (Figure 5; Supplementary Table S6). Among them, CHI, CHS2, CHS3, CHS4, CHS5, CHS6, CHS7, and DFR were highly expressed mainly in female plants. CHS1, HCT5 and HCT4 were highly expressed in male plants. ANR, CCOMT2 and CYP73A were highly expressed in male plants at fruiting stage, and highly expressed in female plants at defoliation stage (Figure 5). Notably, the expression of F3H was different in the three stages, the male plants were higher than the female plants in the flowering and fruiting stages, and the male plants were lower than the female plants in defoliation stage (Figure 5). Therefore, some structural genes may regulate the sex differentiation of flavonoid synthesis in B. papyrifera leaves, especially in the fruiting and defoliation stages.
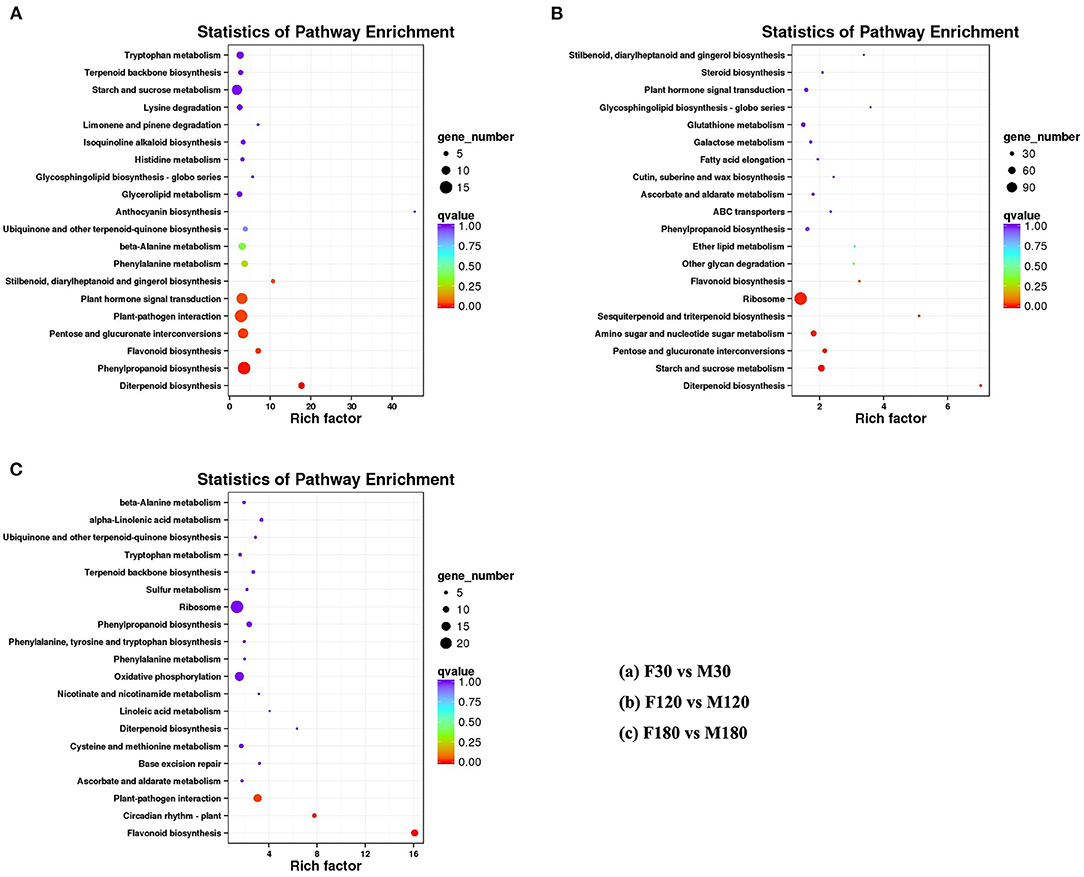
Figure 4. Significant pathways of metabolites in developing Broussonetia papyrifera leaves by KEGG functional enrichment. (A) F30 vs. M30. (B) F120 vs. M120. (C) F180 vs. M180.
Expression of Flavonoid Synthesis-Related Genes in Different Sexes
The profiles of transcriptional data and metabolic data during leaf development of B. papyrifera were exhibited in the flavonoid synthesis pathway (Figure 6). A total of 13 kinds of flavonoids were regulated by 12 genes in the pathway. Among these genes, the differences in gene expression of F3H, CHS1 and CYP75B1 had little effect on the content of flavonoids. High expression of CHS2, CHS3, CHS4, CHS5, CHS6, CHS7, DFR, ANR and CHI promoted the accumulation of flavonoids such as phlorizin, dihydrokaempferol, garbanzol, epigallocatechin, epiafzelechin, catechin, afzelechin, etc.
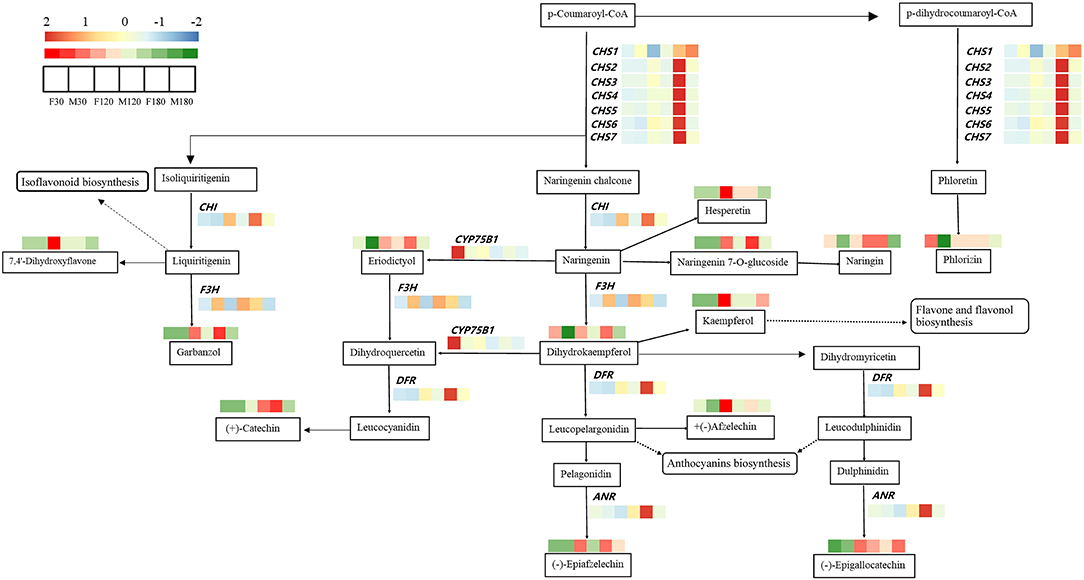
Figure 6. The transcriptional profile of structural genes and metabolites in the flavonoids biosynthesis pathway of Broussonetia papyrifera leaves.
Co-expression Analysis to Identify Differential Genes Related to Flavonoid Synthesis
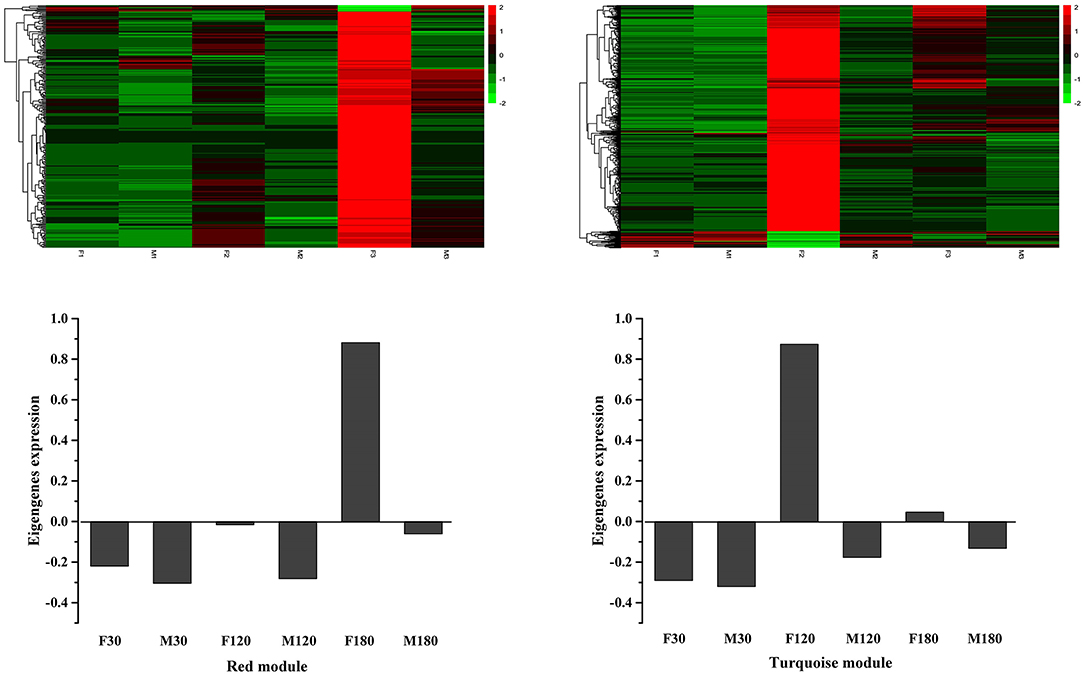
A gene cluster tree was constructed according to the correlation of expression levels between genes, and one branch of the tree corresponds to gene clusters whose expression levels were highly correlated (Figure 7A). The average adjacency coefficient of each gene was 196 (Supplementary Figure S4). 16 common expression modules were obtained (Figure 7B). Except for the gray module consisting of non-clustered genes, the turquoise module contained the largest number of genes (1,688). The midnightblue module contained the smallest number of genes (36). Correlation coefficients between modules and flavonoids showed that red and turquoise modules had the highest correlation with flavonoids synthesis (Supplementary Figure S5). The red module was significantly correlated with mws1173 (garbanzol) and pme0371 (naringenin 7-O-glucoside), while turquoise was correlated with mws0463 (hesperetin), mws1068 (kaempferol), mws4057 (7,4'-dihydroxyflavone) and pme3285 (afzelechin) (Supplementary Figure S5).
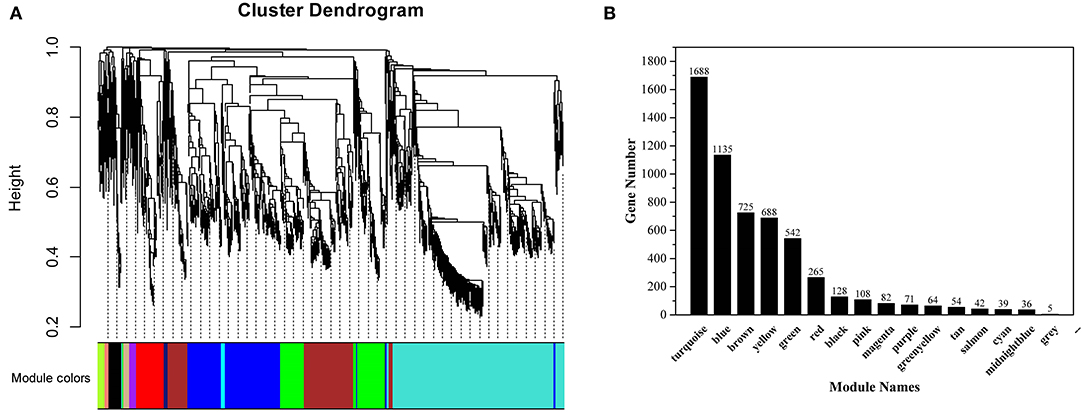
Figure 7. Co-expression network analysis of RNA-seq and physiological trait data. (A) Clustering dendrogram of differentially expressed genes and modules identified by the weighted gene co-expression network analysis. (B) The number of genes in each module.
KEGG enrichment analysis showed that in the red module, flavonoid biosynthesis, cysteine and methionine metabolism, and biosynthesis of amino acids were the top 3 in metabolism pathways (Supplementary Figure S6). Eight genes related to flavonoid synthesis, namely CHS2, CHS3, CH4, CH5, CHS6, CHS7, ANR, and DFR, were enriched in the flavonoid synthesis pathway (Supplementary Table S6). In the turquoise module, carbon metabolism, biosynthesis of amino acids, oxidative phosphorylation, glycolysis/gluconeogenesis, and purine metabolism were the top 5 in metabolism pathways (Supplementary Figure S6). In addition, a gene related to flavonoid synthesis, CHI, was enriched in the flavonoid synthesis pathway (Supplementary Table S6).
Using the FPKM values of genes to draw heat maps (Figure 8), the results showed that genes in the red module were highly expressed in F180 and significantly higher than in other samples, while genes in the turquoise module were highly expressed in F120. Although the red and turquoise modules were highly correlated with flavonoid synthesis, the genes in them exhibited distinct expression patterns and might perform distinct functions.
Identification of Key Genes for Flavonoids Synthesis
Key genes for flavonoids synthesis were screened using the gene expression levels of the major modules (Table 1). In the red module, CHS2, CH3, CHS4, CHS5, CHS6, CHS7, ANR and DFR were highly expressed at defoliation stage. In the turquoise module, CHI were highly expressed at fruiting stage and defoliation stage. CHS2, CH7, DFR and CHI had FPKM > 100 in our data, these genes may be key genes for flavonoid accumulation of B. papyrifera leaves.
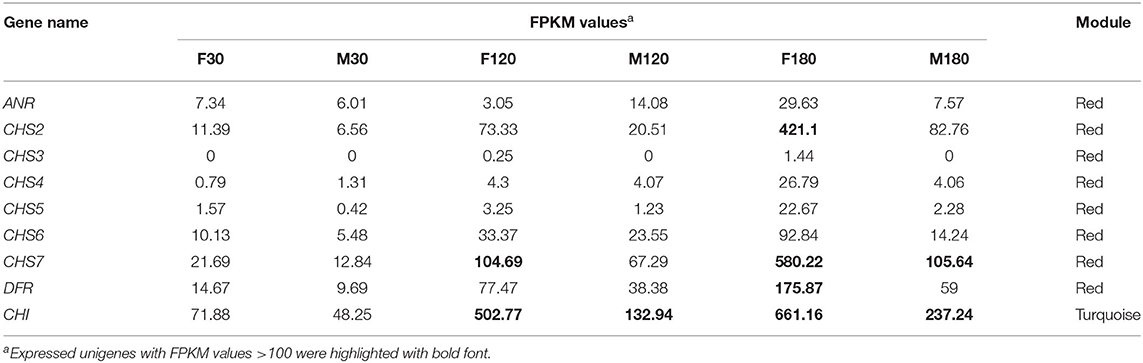
Table 1. Identification of flavonoid-related genes in the developing Broussonetia papyrifera leaves.
Identification of Transcription Factors for Flavonoids Synthesis
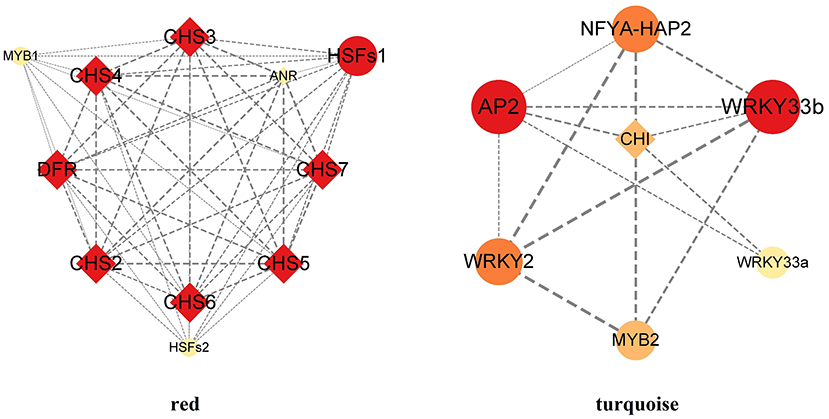
TFs were screened using the co-expression network of the major modules (Supplementary Table S7; Figure 9). The results showed that there were eight structural genes related to flavonoid synthesis, which were CHS2, CHS3, CH4, CH5, CHS6, CHS7, DFR, and CHI. There were eight TFs related to flavonoid synthesis. Among them, HSFs1, HSFs2, MYB1 were strongly associated with CHS and DFR. AP2, NFYA-HAP2, WRKY33a, WRKY33b and MYB2 were strongly associated with CHI.
qRT-PCR Analysis
To verify the stability of transcriptome, qRT-PCR verification on 10 representative unigenes in the flavonoid biosynthesis pathway was performed (Supplementary Table S1; Supplementary Figure S7). The correlation coefficient between qRT-PCR results and RNA-seq results reached 0.82. Therefore, our transcriptome sequencing data were generally accurate.
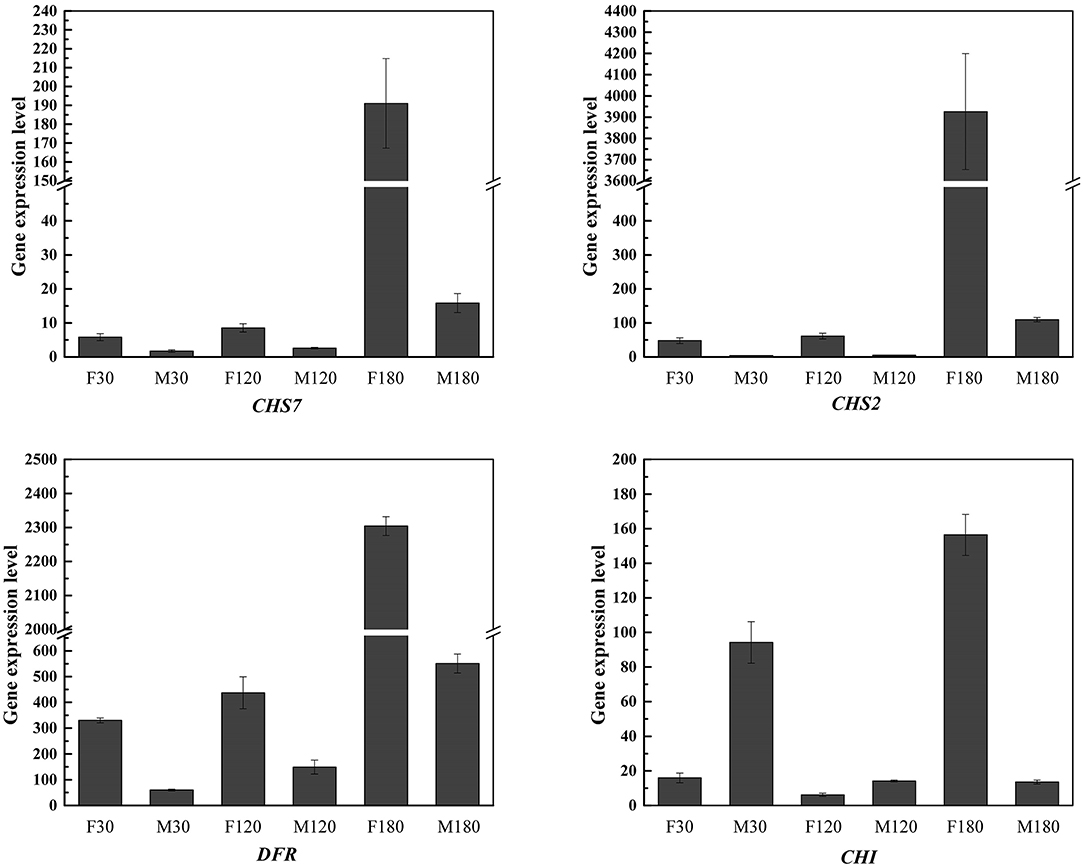
On the other hand, qRT-PCR verification on four key genes for flavonoid accumulation of B. papyrifera leaves was performed (Figure 10). The results showed that CHS2, CHS7, DFR and CHI were highly expressed in female plants at defoliation stage. By comparing the differences between female plants and male plants at different stages, CHS2 gene had the largest difference, reaching 35.77 times.
Discussion
In the present study, we investigated sex differences in flavonoid accumulation of B. papyrifera leaves. The results showed that flavonoids gradually accumulated with developmental time, and the content in female plants was higher than that in male plants (Figure 1; Supplementary Figure S2). Moreover, the composition of flavonoids in female and male plants was very similar, and 16 kinds of flavonoids accumulated after flowering (Figure 2). Xu et al. (2009) studied the seasonal changes of total flavonoids in different parts of B. papyrifera, and the results showed that flavonoids in B. papyrifera leaves increased with the developmental stage, which was consistent with our research. Generally speaking, the different selection pressures faced by female and male plants lead to different nutritional requirements. Female plants have a higher investment in reproduction and show stronger compensatory mechanisms (Delph and Herlihy, 2012). Studies have found that in resource-rich environments, the sex ratio tends to favor female plants (Ward et al., 2002), while in resource-poor environments, the sex ratio tends to favor male plants (Dawson and Ehleringer, 1993; Chen and Li, 2014; Li et al., 2017). Therefore, we hypothesize that the different survival strategies of male and female plants of B. papyrifera leaves lead to differences in flavonoids.
Combined with the results of WGCNA and qRT-PCR analysis, CHS2, CHS7, CHI and DFR may the key genes for flavonoid accumulation of B. papyrifera leaves, and the expression of these genes was affected by sex differences and developmental stages (Figures 5, 9, 10; Table 1). CHS directs the phenylpropane metabolic pathway to the synthesis of flavonoids. In B. papyrifera leaves, CHS enzymes code for proteins exhibiting polyketide synthase activity that accept either p-dihydrocoumaroyl-CoA or p-coumaroyl-CoA as starter CoA substrates, leading to production of phloretin, naringenin chalcone, and isoliquiritigenin (Figure 6), which supports the findings in apple (Yahyaa et al., 2017). CHI enzymes (EC 5.5.1.6) accept either naringenin chalcone or isoliquiritigenin as starter, leading to production of naringenin and liquiritigenin (Figure 6). DFR enzymes (EC 1.1.1.219) catalyz the NADPH-mediated and stereospecific reduction of dihydroflavonols to leucoanthocyanidins, which are the precursors for the formation of anthocyanins, catechins, and proanthocyanidins (Fischer et al., 2003). Specifically, it accepts either dihydrokaempferol, dihydroquercetin or dihdydromyricerin as starter, leading to production of leucopelargonidin, leucocyanidin and leucodelphinidin (Figure 6). However, these results are based on statistical method, and the function of these genes needs to be further verified. So far, little was known about the flavonoid-regulated genes in B. papyrifera leaves. Relevant molecular studies have only demonstrated that four key enzyme families (CHS, F3'H, I2'H, and DFR) in the flavonoid synthesis pathway exhibited significant expansion (Peng et al., 2014; Feng et al., 2019). The high flavonoid abundance and strong disease resistance of B. papyrifera may originate from these expanded gene families.
CHS2, CHS7, CHI and DFR were highly expressed in female B. papyrifera (Figures 5, 10). Most of them are upstream genes in flavonoids biosynthesis pathway. Correlation coefficients between modules and flavonoids showed that CHS2, CHS7 and DFR was significantly correlated with garbanzol and naringenin 7-O-glucoside. CHI was significantly correlated with hesperetin, kaempferol, 7,4'-dihydroxyflavone and afzelechin (Supplementary Figure S5). Therefore, we speculate that the key to affecting the accumulation of flavonoids in B. papyrifera leaves of different sexes is the expression of upstream genes in flavonoids biosynthesis pathway, and we can increase the accumulation of flavonoids by enhancing the expression of upstream genes.
TFs that may interact with flavonoid genes were screened from two key modules closely related to flavonoids, and 8 reliable TFs were found (Figure 9). Among them, HSFs and MYB TFs were strongly correlated with CHS. DFR. AP2, NFYA-HAP2, WRKY and MYB TFs were strongly correlated with CHI.
The HSFs family mediates the activation of genes responsive to diverse stresses. The N-terminal region of the HSF protein sequence contains a highly conserved DNA-binding domain, which can accurately identify and bind the heat shock cis-elements (HSE) of the downstream target gene promoter (Wang et al., 2020). Our results suggest that HSFs1 and HSFs2 genes in B. papyrifera may promote the expression of genes encoding related enzymes, such as CHS and DFR (Figure 9). The results are consistent with studies on other species. In soybean, HSFB2b can bind to the HSE cis-elements and promotes expression of GmCHS and GmFLS (Bian et al., 2020). In Malus domestica, MdHSFA8a can regulate the expression of MdDFR, MdFLS, and MdANS (Wang et al., 2020).
The MYB family is one of the largest families of regulatory proteins, and plays important roles in many secondary metabolic pathways (Zhang et al., 2018b). We found that the MYB1 gene was strongly associated with CHS and DFR, while the MYB2 gene was strongly associated with CHI (Figure 9). These genes may be involved in the flavonoid synthesis. In recent years, many MYB genes associated with the flavonoid biosynthesis have been isolated and characterized. Overexpression of GbMYBFL under the control of the CaMV35S promoter promoted the accumulation of flavonoids and anthocyanin in G. biloba (Zhang et al., 2018b). MYB6, a major regulator of flavonoid synthesis in P. tomentosa, up-regulated the expression of flavonoid biosynthetic genes (Wang et al., 2019).
The AP2/ERF TFs can directly or indirectly participate in multiple processes of plant development such as seed development, morphogenesis of organs such as flowers and fruits by responding to the regulation of ethylene, cytokinin and auxin; in addition to primary metabolism. AP2/ERF also has a significant effect on plant secondary metabolism, especially in regulating the synthesis of main medicinal active components (such as artemisinin, paclitaxel and lignin) (Zhang et al., 2018a). In citrus, the CitCHIL1 gene is transcriptionally activated by three AP2/ERF TFs, and significantly enhanced the accumulation of flavonoids (Zhao et al., 2021), and our results supported this point (Figure 9).
The WRKY family is involved in plant growth, development, abiotic stress and regulation of secondary metabolites (Dai et al., 2021). Our results suggested that WRKY33a and WRKY33b may regular the transcription of CHI, which is involved in the production of naringenin and liquiritigenin in B. papyrifera. In addition, the WRKY family is also involved in regulating other pathways of flavonoid synthesis. In apple calli, MdWRKY11 up-regulated the expression of F3H, FLS, DFR, ANS, and UFGT and increased the accumulation of flavonoids and anthocyanin (Wang et al., 2018). In grape, overexpression of VvWRKY26 possibly promoted the accumulation of proanthocyanidin biosynthesis (Amato et al., 2017).
Although there are few reports on the regulation of flavonoid synthesis by NFYA-HAP2 TFs, the NFYA-HAP2 gene may be involved in the stress resistance B. papyrifera. For example, NFYA-HAP2 showed significant responses to heat shock treatment and F. oxysporum infection in Brassica oleracea (Kim et al., 2015).
At present, based on the statistical method, we have screened out the key genes and TFs that may regulate the synthesis of B. papyrifera flavonoids. To provide more detailed information about the function of the genes and TFs in B. papyrifera, it is necessary to use biological techniques such as yeast two-hybrid and yeast one-hybrid methods to verify their transcriptional activities and interactions with the target genes promoter in the future study.
Conclusion
In this study, we presented the integrative metabolome and transcriptome analysis of flavonoid biosynthesis genes in B. papyrifera leaves from the perspective of sex differentiation. A total of 192 flavonoids substances were detected in B. papyrifera leaves. The accumulation law of flavonoids was that they accumulated gradually with the development time, and the content in female plants was higher than that in male plants. The composition of flavonoids in female and male plants was very similar, and 16 kinds of flavonoids accumulated after flowering. Four key structural genes involved in flavonoids accumulation during leaves development were identified. Most of them are upstream genes in the flavonoids biosynthesis pathway, and overexpression of these genes increased the accumulation of flavonoids, such as garbanzol, naringenin 7-O-glucoside, hesperetin, kaempferol, 7,4'-dihydroxyflavone and afzelechin. Furthermore, 8 reliable TFs (two HSFs, two MYBP, three AP2/ERF, one NFYA-HAP2, and two WRKY) were found. Based on statistical methods and previous findings, these TFs may interact with flavonoid genes CHI, CHS and DFR in B. papyrifera. This study will be useful for further research on flavonoid biosynthesis in B. papyrifera associated molecular regulation.
Data Availability Statement
The original contributions presented in the study are publicly available. The RNA-Seq data presented in the study are deposited in the NCBI's Short Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession number PRJNA819010.
Author Contributions
PJ: conceptualization, writing—original draft, and visualization. LC: investigation. ZY: supervision and funding acquisition. XZ: methodology and writing review and editing. ZW and DJ: data administration. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Key Projects of National Forestry and Grassland Bureau (201801), Postgraduate Scientific Research Innovation Project of Hunan Province (CX20200711), China Postdoctoral Science Foundation (2020M683592), and Natural Science Foundation of Shaanxi Province (2022JQ-202).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.900030/full#supplementary-material
Supplementary Figure S1. Phenotypes of Broussonetia papyrifera leaves of different sexes at three developmental stages.
Supplementary Figure S2. Differentially accumulated flavonoids in leaf samples.
Supplementary Figure S3. Differentially expressed genes in leaf samples.
Supplementary Figure S4. Analysis of network topology for various soft-thresholding powers.
Supplementary Figure S5. Module-trait correlations. Each cell contains the corresponding correlation and p value. Red is positive correlation. Green is negative correlation.
Supplementary Figure S6. KEGG functional analysis of red and turquoise modules in metabolism pathways.
Supplementary Figure S7. Pearson correlation analysis between qRT-PCR results and RNA-seq results.
Supplementary Table S1. The primers list of genes for qRT-PCR analysis.
Supplementary Table S2. Types and relative contents of flavonoids in Broussonetia papyrifera leaves.
Supplementary Table S3. KEGG analysis of differential metabolites.
Supplementary Table S4. Transcriptome sequencing quality analysis.
Supplementary Table S5. Annotation of differentially expressed genes.
Supplementary Table S6. KEGG analysis of structural genes in the flavonoid biosynthesis pathway.
Supplementary Table S7. Candidate co-express genes related to flavonoid biosynthesis.
Abbreviations
ANR, Anthocyanin reductase; AP2/ERF, APETALA2/ethylene-responsive element binding factors; CHS, Chalcone synthase; CHI, Chalcone isomerase; DAFs Differentially accumulated flavonoids, ; DFR, Dihydroflavonol reductase; DEGs, Differentially expressed genes; F3H, Flavanone 3-hydroxylase; HSFs, Heat shock factors; MYB, v-myb avian myeloblastosis viral oncogene homolog; NFYA-HAP2, Nuclear transcription factor Y; qRT-PCR, Quantitative reverse transcription-polymerase chain reaction; TFs, Transcription factors; UPLC, Ultra performance liquid chromatography; WGCNA, Weighted gene co-expression network analysis.
References
Agati, G., Azzarello, E., Pollastri, S., and Tattini, M. (2012). Flavonoids as antioxidants in plants: location and functional significance. Plant Sci. 196, 67–76. doi: 10.1016/j.plantsci.2012.07.014
Agati, G., Biricolti, S., Guidi, L., Ferrini, F., Fini, A., and Tattini, M. (2011). The biosynthesis of flavonoids is enhanced similarly by UV radiation and root zone salinity in L. vulgare leaves. J. Plant Physiol. 168, 204–212. doi: 10.1016/j.jplph.2010.07.016
Agati, G., Stefano, G., Biricolti, S., and Tattini, M. (2009). Mesophyll distribution of 'antioxidant' flavonoid glycosides in Ligustrum vulgare leaves under contrasting sunlight irradiance. Ann. Bot. 104, 853–861. doi: 10.1093/aob/mcp177
Al-Dossary, O., Alsubaie, B., Kharabian-Masouleh, A., Al-Mssallem, I., Furtado, A., and Henry, R. J. (2021). The jojoba genome reveals wide divergence of the sex chromosomes in a dioecious plant. Plant J. 108, 1283–1294. doi: 10.1111/tpj.15509
Amato, A., Cavallini, E., Zenoni, S., Finezzo, L., Begheldo, M., Ruperti, B., et al. (2017). A grapevine TTG2-like WRKY transcription factor is involved in regulating vacuolar transport and flavonoid biosynthesis. Front. Plant Sci. 7, 1979. doi: 10.3389/fpls.2016.01979
Banuelos, M. J., and Obeso, J. R. (2004). Resource allocation in the dioecious shrub Rhamnus alpinus: the hidden costs of reproduction. Evol. Ecol. Res. 6, 397–413.
Bian, X.-H., Li, W., Niu, C.-F., Wei, W., Hu, Y., Han, J.-Q., et al. (2020). A class B heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol. 225, 268–283. doi: 10.1111/nph.16104
Chang, C.-S., Liu, H.-L., Moncada, X., Seelenfreund, A., Seelenfreund, D., and Chung, K.-F. (2015). A holistic picture of Austronesian migrations revealed by phylogeography of Pacific paper mulberry. Proc. Nat. Acad. Sci. 112, 13537–13542. doi: 10.1073/pnas.1503205112
Chen, J., and Li, C. (2014). Sex-specific responses to environmental stresses and sexual competition of dioecious plants. Chin. J. Appl. Environ. Biol. 2014, 743–750. doi: 10.3724/SP.J.1145.2014.06008
Chen, W., Gong, L., Guo, Z., Wang, W., Zhang, H., Liu, X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 6, 1769–1780. doi: 10.1093/mp/sst080
Chen, X., Mao, X., Huang, J., Yang, D., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucl. Acids Res. 39(suppl_2), w316–w322. doi: 10.1093/nar/gkr483
Dai, X., Liu, J., and Li, S. (2021). Bioinformatics analysis of WRKY gene family in Yulania biondii. J. Jiangsu For. Sci. Technol. 48, 1–9. doi: 10.3969/j.issn.1001?7380.2021.06.001
Dawson, T. E., and Ehleringer, J. R. (1993). Gender-specific physiology, carbon isotope discrimination, and habitat distribution in boxelder, Acer negundo. Ecology 74, 798–815. doi: 10.2307/1940807
Delph, L. F., and Herlihy, C. R. (2012). Sexual, fecundity, and viability selection on flower size and number in a sexually dimorphic plant. Evol. Int. J. Organ. Evol. 66, 1154–1166. doi: 10.1111/j.1558-5646.2011.01510.x
Ding, T., Zhang, R., Zhang, H., Zhou, Z., Liu, C., Wu, M., et al. (2021). Identification of gene co-expression networks and key genes regulating flavonoid accumulation in apple (Malus × domestica) fruit skin. Plant Sci. 304, 110747. doi: 10.1016/j.plantsci.2020.110747
Falcone Ferreyra, M. L., Rius, S., and Casati, P. (2012). Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3, 222. doi: 10.3389/fpls.2012.00222
Feng, J., Dong, P., Li, R., Li, C., Xie, X., and Shi, Q. (2019). Effects of wood fiber properties on mold resistance of wood polypropylene composites. Int. Biodeterior. Biodegr. 140, 152–159. doi: 10.1016/j.ibiod.2019.04.005
Feng, W., Li, H., and Zheng, X. (2008). Chemical constituents from the leaves of Broussonetia papyrifera. Acta Pharm. Sin. 43, 173–180. doi: 10.16438/j.0513-4870.2008.02.005
Fischer, T. C., Halbwirth, H., Meisel, B., Stich, K., and Forkmann, G. (2003). Molecular cloning, substrate specificity of the functionally expressed dihydroflavonol 4-reductases from Malus domestica and Pyrus communis cultivars and the consequences for flavonoid metabolism. Arch. Biochem. Biophys. 412, 223–230. doi: 10.1016/S0003-9861(03)00013-4
Ghosh, R., Chakraborty, A., Biswas, A., and Chowdhuri, S. (2021). Identification of polyphenols from Broussonetia papyrifera as SARS CoV-2 main protease inhibitors using in silico docking and molecular dynamics simulation approaches. J. Biomol. Struct. Dyn. 39, 6747–6760. doi: 10.1080/07391102.2020.1802347
González-Gallego, J., García-Mediavilla, M. V., Sánchez-Campos, S., and Tuñón, M. J. (2010). Fruit polyphenols, immunity and inflammation. Br. J. Nutr. 104, S15–S27. doi: 10.1017/S0007114510003910
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo, F., Feng, L., Huang, C., Ding, H., Zhang, X., Wang, Z., et al. (2013). Prenylflavone derivatives from Broussonetia papyrifera, inhibit the growth of breast cancer cells in vitro and in vivo. Phytochem. Lett. 6, 331–336. doi: 10.1016/j.phytol.2013.03.017
Guo, Y., Gao, C., Wang, M., Fu, F.-,f., El-Kassaby, Y. A., Wang, T., et al. (2020). Metabolome and transcriptome analyses reveal flavonoids biosynthesis differences in Ginkgo biloba associated with environmental conditions. Ind. Crops Prod. 158, 112963. doi: 10.1016/j.indcrop.2020.112963
Harborne, J. B., and Williams, C. A. (2000). Advances in flavonoid research since 1992. Phytochemistry 55, 481–504. doi: 10.1016/S0031-9422(00)00235-1
Kim, Y.-W., Jung, H.-J., Park, J.-I., Hur, Y., and Nou, I.-S. (2015). Response of NBS encoding resistance genes linked to both heat and fungal stress in Brassica oleracea. Plant Physiol. Biochem. 86, 130–136. doi: 10.1016/j.plaphy.2014.11.009
Koczka, N., Móczár, Z., Stefanovits-Bányai, É., and Ombódi, A. (2015). Differences in antioxidant properties of ginkgo leaves collected from male and female trees. Acta Pharm. 65, 99–104. doi: 10.1515/acph-2015-0001
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 12, 1–16. doi: 10.1186/1471-2105-12-323
Li, H., Li, T., Tu, B., Kou, Y., and Li, X. (2017). Host species shapes the co-occurrence patterns rather than diversity of stomach bacterial communities in pikas. Appl. Microbiol. Biotechnol. 101, 5519–5529. doi: 10.1007/s00253-017-8254-0
Li, M., Yan, L., and Li, H. (2012). Improvement of paper mulberry tolerance to abiotic stresses by ectopic expression of tall fescue FaDREB1. Tree Physiol. 32, 104–113. doi: 10.1093/treephys/tpr124
Lillo, C., Lea, U. S., and Ruoff, P. (2010). Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant Cell Environ. 31, 587–601. doi: 10.1111/j.1365-3040.2007.01748.x
Lu, W., Jianhua, T., Jucheng, W., Peng, S., Yicai, H., and Xuebin, G. (2018). Comparative analysis on total flavonoids content of Hippophae rhamnoides linn. leaf in Shanxi. South African J. Bot. 47, 33. doi: 10.1016/j.sajb.2012.11.005
Park, J.-Y., Yuk, H. J., Ryu, H. W., Lim, S. H., Kim, K. S., Park, K. H., et al. (2017). Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzyme Inhib. Med. Chem. 32, 504–512. doi: 10.1080/14756366.2016.1265519
Peng, X., Liu, H., Chen, P., Tang, F., Hu, Y., Wang, F., et al. (2019). A chromosome-scale genome assembly of paper mulberry (Broussonetia papyrifera) provides new insights into its forage and papermaking usage. Mol. Plant 12, 661–677. doi: 10.1016/j.molp.2019.01.021
Peng, X., Teng, L., Wang, X., Wang, Y., and Shen, S. (2014). De novo assembly of expressed transcripts and global transcriptomic analysis from seedlings of the paper mulberry (Broussonetia kazinoki x Broussonetia papyifera). PLoS ONE 9, e108613. doi: 10.1371/journal.pone.0097487
Pisonero-Vaquero, S., Garcia-Mediavilla, M. V., Jorquera, F., Majano, P. L., Benet, M., Jover, R., et al. (2014). Modulation of PI3K-LXRα-dependent lipogenesis mediated by oxidative/nitrosative stress contributes to inhibition of HCV replication by quercetin. Lab. Investig. 94, 262–274. doi: 10.1038/labinvest.2013.156
Randriamanana, T. R., Nybakken, L., Lavola, A., Aphalo, P. J., Nissinen, K., and Julkunen-Tiitto, R. (2014). Sex-related differences in growth and carbon allocation to defence in Populus tremula as explained by current plant defence theories. Tree Physiol. 34, 471–487. doi: 10.1093/treephys/tpu034
Stafford, H. A. (1991). Flavonoid evolution: an enzymic approach. Plant Physiol. 96, 680–685. doi: 10.1104/pp.96.3.680
Tattini, M., Galardi, C., Pinelli, P., Massai, R., and Agati, G. (2010). Differential accumulation of flavonoids and hydroxycinnamates in leaves of Ligustrum vulgare under excess light and drought stress. New Phytol. 163, 547–561. doi: 10.1111/j.1469-8137.2004.01126.x
Wang, L., Lu, W., Ran, L., Dou, L., Yao, S., Hu, J., et al. (2019). R2R3-MYB transcription factor MYB 6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 99, 733–751. doi: 10.1111/tpj.14364
Wang, N., Liu, W., Yu, L., Guo, Z., Chen, Z., Jiang, S., et al. (2020). HEAT SHOCK FACTOR A8a modulates flavonoid synthesis and drought tolerance. Plant Physiol. 184, 1273–1290. doi: 10.1104/pp.20.01106
Wang, N., Liu, W., Zhang, T., Jiang, S., Xu, H., Wang, Y., et al. (2018). Transcriptomic analysis of red-fleshed apples reveals the novel role of MdWRKY11 in flavonoid and anthocyanin biosynthesis. J. Agric. Food Chem. 66, 7076–7086. doi: 10.1021/acs.jafc.8b01273
Ward, J., Dawson, T., and Ehleringer, J. (2002). Responses of Acer negundo genders to interannual differences in water availability determined from carbon isotope ratios of tree ring cellulose. Tree Physiol. 22, 339–346. doi: 10.1093/treephys/22.5.339
Wu, X., Liu, J., Meng, Q., Fang, S., Kang, J., and Guo, Q. (2021). Differences in carbon and nitrogen metabolism between male and female Populus cathayana in response to deficient nitrogen. Tree Physiol. 41, 119–133. doi: 10.1093/treephys/tpaa108
Xu, Y., Guo, Z., Tan, L., Bu, X., and Long, L. (2009). Seasonal variation of total flavonoids contents for different parts of Broussonetia papyrifera. Res. Pract. Chin. Med. 23, 16–17. doi: 10.13728/j.1673-6427.2009.04.029
Xu, Z., Dong, M., Peng, X., Ku, W., Zhao, Y., and Yang, G. (2019). New insight into the molecular basis of cadmium stress responses of wild paper mulberry plant by transcriptome analysis. Ecotoxicol. Environ. Saf. 171, 301–312. doi: 10.1016/j.ecoenv.2018.12.084
Yahyaa, M., Ali, S., Davidovich-Rikanati, R., Ibdah, M., Shachtier, A., Eyal, Y., et al. (2017). Characterization of three chalcone synthase-like genes from apple (Malus x domestica Borkh.). Phytochemistry 140, 125–133. doi: 10.1016/j.phytochem.2017.04.022
Yang, Y. (2018). Study on Extraction, Purification and Antioxidant Activity of Total Flavonoids from Apocynum venetum. Master, South China University of Technology.
Yang, Y., Jiang, H., Wang, M., Korpelainen, H., and Li, C. (2015). Male poplars have a stronger ability to balance growth and carbohydrate accumulation than do females in response to a short-term potassium deficiency. Physiol. Plant. 155, 400–413. doi: 10.1111/ppl.12325
Ying, H.-Z., Liu, Y.-H., Yu, B., Wang, Z.-Y., Zang, J.-N., and Yu, C.-H. (2013). Dietary quercetin ameliorates nonalcoholic steatohepatitis induced by a high-fat diet in gerbils. Food Chem. Toxicol. 52, 53–60. doi: 10.1016/j.fct.2012.10.030
Zhang, L., Zhang, Z., Fang, S., Liu, Y., and Shang, X. (2021). Integrative analysis of metabolome and transcriptome reveals molecular regulatory mechanism of flavonoid biosynthesis in Cyclocarya paliurus under salt stress. Ind. Crops Prod. 170, 113823. doi: 10.1016/j.indcrop.2021.113823
Zhang, M., Fang, Y., Ji, Y., Jiang, Z., and Wang, L. (2013). Effects of salt stress on ion content, antioxidant enzymes and protein profile in different tissues of Broussonetia papyrifera. South Afric. J. Bot. 85, 1–9. doi: 10.1016/j.sajb.2012.11.005
Zhang, Q., Chen, J., Li, L., Zhao, M., Zhang, M., and Wang, Y. (2018a). Research progress on plant AP2/ERF transcription factor family. Biotechnol. Bull. 34, 1–7. doi: 10.13560/j.cnki.biotech.bull.1985.2017-1142
Zhang, W., Xu, F., Cheng, S., and Liao, Y. (2018b). Characterization and functional analysis of a MYB gene (GbMYBFL) related to flavonoid accumulation in Ginkgo biloba. Genes Genom. 40, 49–61. doi: 10.1007/s13258-017-0609-5
Zhao, C., Liu, X., Gong, Q., Cao, J., Shen, W., Yin, X., et al. (2021). Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol. J. 19, 671–688. doi: 10.1111/pbi.13494
Zhao, H. (2011). Quality Evaluation of Broussonetia papyrifera and the Intra-Specific Variation Under Heavy Metal Stress. Master, Fujian University of Traditional Chinese Medicine.
Zhao, X., Liu, J., Xia, X., Chu, J., Wei, Y., Shi, S., et al. (2014). The evaluation of heavy metal accumulation and application of a comprehensive bio-concentration index for woody species on contaminated sites in Hunan, China. Environ. Sci. Pollut. Res. 21, 5076–5085. doi: 10.1007/s11356-013-2393-3
Keywords: Broussonetia papyrifera, flavonoids, transcription factors, sex differentiation, WGCNA
Citation: Jiao P, Chaoyang L, Wenhan Z, Jingyi D, Yunlin Z and Zhenggang X (2022) Integrative Metabolome and Transcriptome Analysis of Flavonoid Biosynthesis Genes in Broussonetia papyrifera Leaves From the Perspective of Sex Differentiation. Front. Plant Sci. 13:900030. doi: 10.3389/fpls.2022.900030
Received: 19 March 2022; Accepted: 15 April 2022;
Published: 20 May 2022.
Edited by:
Da-Cheng Hao, Dalian Jiaotong University, ChinaReviewed by:
Feng Xu, Yangtze University, ChinaZou Xiaoxing, Fujian Agriculture and Forestry University, China
Chongde Sun, Zhejiang University, China
Copyright © 2022 Jiao, Chaoyang, Wenhan, Jingyi, Yunlin and Zhenggang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhao Yunlin, enlsODI5MTI5MEAxNjMuY29t; Xu Zhenggang, eHV6aGVuZ2dhbmdAbndhZnUuZWR1LmNu
 Peng Jiao
Peng Jiao Li Chaoyang
Li Chaoyang Zhai Wenhan3
Zhai Wenhan3 Xu Zhenggang
Xu Zhenggang