- Department of Life Science (BK21 Program), Chung-Ang University, Seoul, South Korea
Abscisic acid (ABA) is a major phytohormone that regulates plant growth, development, and abiotic/biotic stress responses. Under stress, ABA is synthesized in various plant organs, and it plays roles in diverse adaptive processes, including seed dormancy, growth inhibition, and leaf senescence, by modulating stomatal closure and gene expression. ABA receptor, clade A protein phosphatase 2C (PP2C), and SNF1-related protein kinase 2 (SnRK2) proteins have been identified as core components of ABA signaling, which is initiated via perception of ABA with receptor and subsequent activation or inactivation by phosphorylation/dephosphorylation. The findings of several recent studies have established that the post-translational modification of these components, including phosphorylation and ubiquitination/deubiquitination, play important roles in regulating their activity and stability. In this review, we discuss the functions of the core components of ABA signaling and the regulation of their activities via post-translational modification under normal and stress conditions.
Introduction
Abscisic acid (ABA) is a major hormone that regulates growth, development, and responses to abiotic/biotic stress throughout the life cycle of plants. Under environmental conditions perceived as unsuitable for plant growth, ABA helps maintain seed dormancy and inhibits germination, whereas during the post-germination stage, this hormone inhibits growth to conserve energy (Chinnusamy et al., 2008; Raghavendra et al., 2010). In addition, ABA plays key roles in stomatal opening and closure, thereby contributing to the regulation of water loss under drought stress (Melotto et al., 2006; Hirayama and Shinozaki, 2007; Cutler et al., 2010; Finkelstein, 2013).
The core components of ABA signaling, along with their roles and regulatory mechanisms, are well known. As ABA receptors, pyrabactin resistance 1 (PYR1)/PYR1-like (PYL)/regulatory components of the ABA receptor (RCAR) family bind ABA in response to abiotic stress and positively regulate ABA signaling (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009; Gonzalez-Guzman et al., 2012; Antoni et al., 2013). Under normal conditions, clade A protein phosphatase 2Cs (PP2Cs) play roles as negative regulator of ABA signaling by binding to subclass III sucrose nonfermenting 1 (SNF1)-related protein kinase 2 (SnRK2) proteins, and subsequently these interactions inactivate downstream ABA signaling factors and inhibit ABA responses to facilitate growth and development (Mustilli et al., 2002; Yoshida et al., 2006a; Fujii et al., 2007; Umezawa et al., 2009; Vlad et al., 2009). Under stress conditions, ABA receptors interact with PP2Cs and SnRK2s are released from PP2Cs-mediated inhibition, leading to autophosphorylation or phosphorylation by other kinases, whereupon the activated SnRK2s phosphorylate downstream targets, including transcription factors and channel proteins, thereby facilitating abiotic stress responses (Fujii and Zhu, 2009; Geiger et al., 2009; Yin et al., 2009; Nishimura et al., 2010; Ng et al., 2011; Xue et al., 2011; Brandt et al., 2012; Soon et al., 2012).
The different components of ABA signaling are modulated to varying extents by post-translational modification (Table 1), with the ubiquitination/deubiquitination and phosphorylation of these components being generally well studied. Ubiquitination of ABA core components can influence ABA signaling and responses both positively and negatively (Coego et al., 2021). Deubiquitination mediated by members of the UBIQUITIN-SPECIFIC PROTEASE (UBP) protein family affects a range of plant physiological processes, including growth, development, immunity, and stress response (Zhou et al., 2017). However, the biochemical mechanisms underlying the involvement of UBPs in ABA signaling have yet to be sufficiently elucidated. A further major category of post-translational modification is phosphorylation; ABA receptors, PP2Cs, and SnRK2s can be positively or negatively regulated by phosphorylation. For example, SnRK2s can undergo autophosphorylation and activation (Yang et al., 2017; Chen et al., 2020). In this review, we focus on the roles of ABA, core ABA signaling pathways, and post-transcriptional regulation of ABA signaling components in plants.
The Role of ABA in Abiotic Stress Responses
Regulation of Stomatal Closure
In response to drought stress, ABA induces stomatal closure to reduce transpirational water loss (Bauer et al., 2013). ABA modulates stomatal movement via both Ca2+-dependent and -independent pathways. In the former, ABA induces Ca2+ channel opening to facilitate the influx of calcium ions, in response to which upregulated reactive oxygen species levels and inositol-1-4-5-triphosphate contribute to increases in cytosolic Ca2+ content (Lee et al., 1996; Pei et al., 2000; Murata et al., 2001; Mustilli et al., 2002), which in turn modulates multiple signal transduction. Calcium-dependent protein kinases (CPKs) 3/4/6/10/11 phosphorylate and activate slow-type anion efflux channels, including SLOW ANION CHANNEL-ASSOCIATED 1 (SLAC1) and SLAC1 HOMOLOG 3 (SLAH3; Mori et al., 2006; Zhu et al., 2007; Zou et al., 2010; Brandt et al., 2012), whereas CPK21 phosphorylates K+ outward rectifying channel GORK to promote the efflux of K+ ions; KAT1 and KAT2 are inactivated by CPK13 to inhibit K+ ion influx (Hosy et al., 2003; van Kleeff et al., 2018). In contrast, the Ca2+-independent pathway is associated with SnRK2.6 (Li et al., 2000), which activates SLAC1 and KUP6, the latter of which is a KUP/HAK/KT family K+ efflux transporter, thereby suppressing KAT1 activity via phosphorylation (Kwak et al., 2001; Geiger et al., 2009; Lee et al., 2009; Sato et al., 2009a; Osakabe et al., 2013). SnRK2.6 has also been shown to phosphorylate the ABA-responsive kinase substrate AKS1, which binds to the KAT1 promoter, thereby promoting the downregulated expression of KAT1 (Takahashi et al., 2013). Collectively, these Ca2+-dependent and Ca2+-independent pathways contribute to reductions in guard cell turgor, thereby leading to stomatal closure.
Regulation of Dormancy and Germination
As sessile organisms, plants are particularly responsive to environmental cues with respect to the initiation of germination. Under unfavorable environmental conditions, ABA contributes to the suppression of germination by promoting seed dormancy, as revealed by the reduced seed dormancy of ABA biosynthesis and signaling-impaired mutants (Koornneef et al., 1982; Leon-Kloosterziel et al., 1996; Lefebvre et al., 2006; Nakashima et al., 2009a; Zhao et al., 2018). For example, the Raf-like MAKKKs, Raf10 and Raf11, influence seed dormancy by phosphorylating SnRK2s and ABRE-binding factors (ABFs; Lee et al., 2015; Nguyen et al., 2019). ABA can also influence seed germination by modifying hormonal balance. In contrast to ABA, gibberellic acid (GA) is a phytohormone that promotes seed germination (Debeaujon and Koornneef, 2000; Ogawa et al., 2003), and the activities of these two hormones antagonistically regulate seed dormancy and germination in higher plants (Weiss and Ori, 2007; Golldack et al., 2013). Notably, compared with wild-type plants, ABA-deficient mutants are characterized by higher levels of GA, and it has accordingly been suggested that ABA plays a role in regulating the GA metabolic pathway (Seo et al., 2006). As a key repressor of GA signaling, REPRESSOR OF GA-LIKE 2 (RGL2) plays a negative role in seed germination, as indicated by the loss-of-function mutant rgl2, in which ABA concentrations are reduced and germination is promoted (Piskurewicz et al., 2008; Lee et al., 2010). RGL2 also promotes ABI5 expression levels, thereby inducing the ABA-mediated suppression of germination (Liu et al., 2016). Exogenous ABA activates the expression of RGL2 and ABI5 (Piskurewicz et al., 2008), and thus RGL2 is believed to modulate the balance between ABA and GA contents during seed germination.
Regulation of Drought-Responsive Gene Expression
On exposure to drought stress, ABA induces the expression of several genes in mature plants that play roles in the drought stress response. Representative ABA-responsive marker genes in this context include RESPONSIVE TO DESICCATION 29B (RD29B), RESPONSIVE TO ABA18 (RAB18), LATE EMBRYOGENESIS ABUNDANT 1 (EM1), and EM6, the expression of which is induced under conditions of water limitation (Yamaguchi-Shinozaki and Shinozaki, 1994; Mantyla et al., 1995; Uno et al., 2000; Carles et al., 2002; Umezawa et al., 2006; Wang et al., 2018b). The expression of these genes is regulated by a number ABA-inducible transcription factors, including ABA-responsive element (ABRE)-binding proteins (AREBs) and ABFs, which contribute to the regulation of ABA-dependent gene expression under drought stress conditions (Nakashima et al., 2009b; Yoshida et al., 2015). It has also been established that a number drought stress-induced genes are also responsive to exogenous ABA, which is mediated via promoter region ABREs (Nakashima et al., 2009b; Fujita et al., 2011). For example, plants overexpressing (OX) the AREB1/ABF2, AREB2/ABF4, ABF1, and ABF3 genes are characterized by enhanced ABA sensitivity and drought tolerance (Kang et al., 2002; Fujita et al., 2005). In contrast, areb1, areb2, and abf3 mutants show reduced drought tolerance and ABA sensitivity (Yoshida et al., 2010). The transcripts of other transcription factors, such as AtMYC2 and AtMYB2, have been found to accumulate in response to exogenous ABA, and in AtMYC2-OX and AtMYB2-OX plants, the ABA-inducible gene RESPONSIVE TO DESICCATION 22 (RD22) is upregulated, leading to ABA-sensitive and drought-tolerant phenotypes (Abe et al., 2003). Similarly, expression of the NAC transcription factor RD26 is induced by drought and ABA treatment, with RD26-OX plants being characterized by hypersensitivity to exogenous ABA, whereas contrastingly, RD26 dominant repressor plants exhibit an ABA-insensitive phenotype (Fujita et al., 2004). Collectively, these findings thus indicate that ABA induces ABA-responsive transcription factors, which in turn regulate the expression of ABA-responsive genes, thereby enhancing drought stress tolerance.
Regulation of Leaf Senescence
Exogenous ABA treatment has been widely demonstrated to induce leaf senescence (Gepstein and Thimann, 1980; Pourtau et al., 2004; Raab et al., 2009; Lee et al., 2011). Moreover, it has been observed that as leaves senesce, there is a corresponding increase in endogenous ABA levels (Gepstein and Thimann, 1980; Leon-Kloosterziel et al., 1996; Cheng et al., 2002; Breeze et al., 2011; Yang et al., 2014). In terms of gene expression, ABA signaling-associated genes have been found to be upregulated during leaf senescence and the expression of various senescence-associated genes can also be induced by the application of exogenous ABA (Tan et al., 2003; Parkash et al., 2014). An important factor associated with the induction of leaf senescence is chlorophyll degradation, which is mediated via genes such as STAY-GREEN 1 (SGR1), NON-YELLOW COLORING 1 (NYC1), PHEOPHYYTINASE (PPH), and PHEIDE a OXYGENASE (PaO) (Pruzinska et al., 2005; Ren et al., 2007; Hortensteiner, 2009; Sato et al., 2009b; Schelbert et al., 2009). In this regard, Arabidopsis thaliana NAC-LIKE, ACTIVATED BY AP3/PI (AtNAP) has been found to bind to the promoter of ABSCISIC ALDEHYDE OXIDASE 3 (AAO3) to enhance AAO3 transcription and ABA production, and this accumulation of ABA contributes to the induction of chlorophyll degradation-associated genes (Yang et al., 2014). ABA core components have also been established to be involved in chlorophyll degradation. For example, SnRK2s regulate the bZIP transcription factors ABI5 and ABF2/3/4, which directly activate NYC1, PAO, and SENESCENCE-ASSOCIATED GENE 12 (SAG12), which are considered senescence marker genes (Gao et al., 2016; Zhao et al., 2016b).
ABA Core Components
Under abiotic stress conditions, ABA mediates responses via a core signaling pathway. As ABA receptors, PYR1 and PYLs were initially identified based on the characterization of ABA-insensitive mutants, whereas RCARs were identified via yeast two-hybrid screens with clade A PP2Cs (Ma et al., 2009; Park et al., 2009). As positive regulators of ABA signaling, PYRs, PYLs, and RCARs (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009; Gonzalez-Guzman et al., 2012; Antoni et al., 2013) are characterized by star-related lipid-transfer domains that contain a conserved ligand-binding pocket and are localized to the nucleus, cytoplasm, and plasma membrane (Iyer et al., 2001; McConnell et al., 2001; Radauer et al., 2008). Arabidopsis also expresses a group of 13 genes that are similar to Pyr1, namely, Pyl1 to Pyl13 (PYR1-Like; Fujii et al., 2009; Li et al., 2013; Zhao et al., 2013), among which, Pyr1/Pyl1/pyl2/pyl4 mutant alleles show an ABA insensitive phenotype in terms of seed germination and root growth with diminished ABA-responsive gene expression and SnRK2.6 kinase activity (Park et al., 2009). Under normal conditions, PP2Cs negatively regulate ABA signaling by interacting with SnRK2s and inhibiting kinase activity, whereas under stress conditions, PYR1 and PYLs interact with group A PP2Cs, leading to the release and activation of SnRK2s. PYLs have different binding properties with respect to ABA and PP2Cs (Szostkiewicz et al., 2010; Hao et al., 2011; Antoni et al., 2012; Li et al., 2013; Zhao et al., 2013; Tischer et al., 2017), with two discrete binding types being identified, namely ABA-enhanced and ABA-dependent. In the former PYLs, including PYL4-6 and PYL8-10, bind with PP2Cs, whereas in the latter, dimeric PYLs, including PYR1 and PYL1-2, bind with PP2Cs (Hao et al., 2011).
In Arabidopsis, six of the nine identified clade A PP2Cs (ABI1, ABI2, HAB1, HAB2, AHG1, and PP2CA) function as ABA negative regulators (Merlot et al., 2001; Kuhn et al., 2006; Robert et al., 2006; Saez et al., 2006; Yoshida et al., 2006b; Nishimura et al., 2007; Rubio et al., 2009). Under normal conditions, PP2Cs bind to SnRK2s, thereby suppressing stress signal transmission, and thus facilitating appropriate plant growth. PP2Cs interact with C-terminal subdomain II of SnRK2s in an ABA-independent manner (Bhaskara et al., 2012) and repress ABA signaling via interaction with and dephosphorylation of SnRK2s (Mustilli et al., 2002; Yoshida et al., 2006a; Fujii et al., 2007; Lee et al., 2009; Umezawa et al., 2009; Vlad et al., 2009). In the presence of ABA, the different PP2Cs interact with specific ABA receptors and release SnRK2s, thereby inducing downstream signal transduction. Released subclass III SnRK2s are autophosphorylated and activated, with their activity being enhanced via phosphorylation by other protein kinases, including Raf kinases (Cai et al., 2014; Nguyen et al., 2019). In turn, these activated SnRK2s phosphorylate downstream transcription factors, including AREB and ABFs (Kobayashi et al., 2005; Furihata et al., 2006; Sirichandra et al., 2010).
Post-translational Modification of ABA Components
Ubiquitination and Deubiquitination
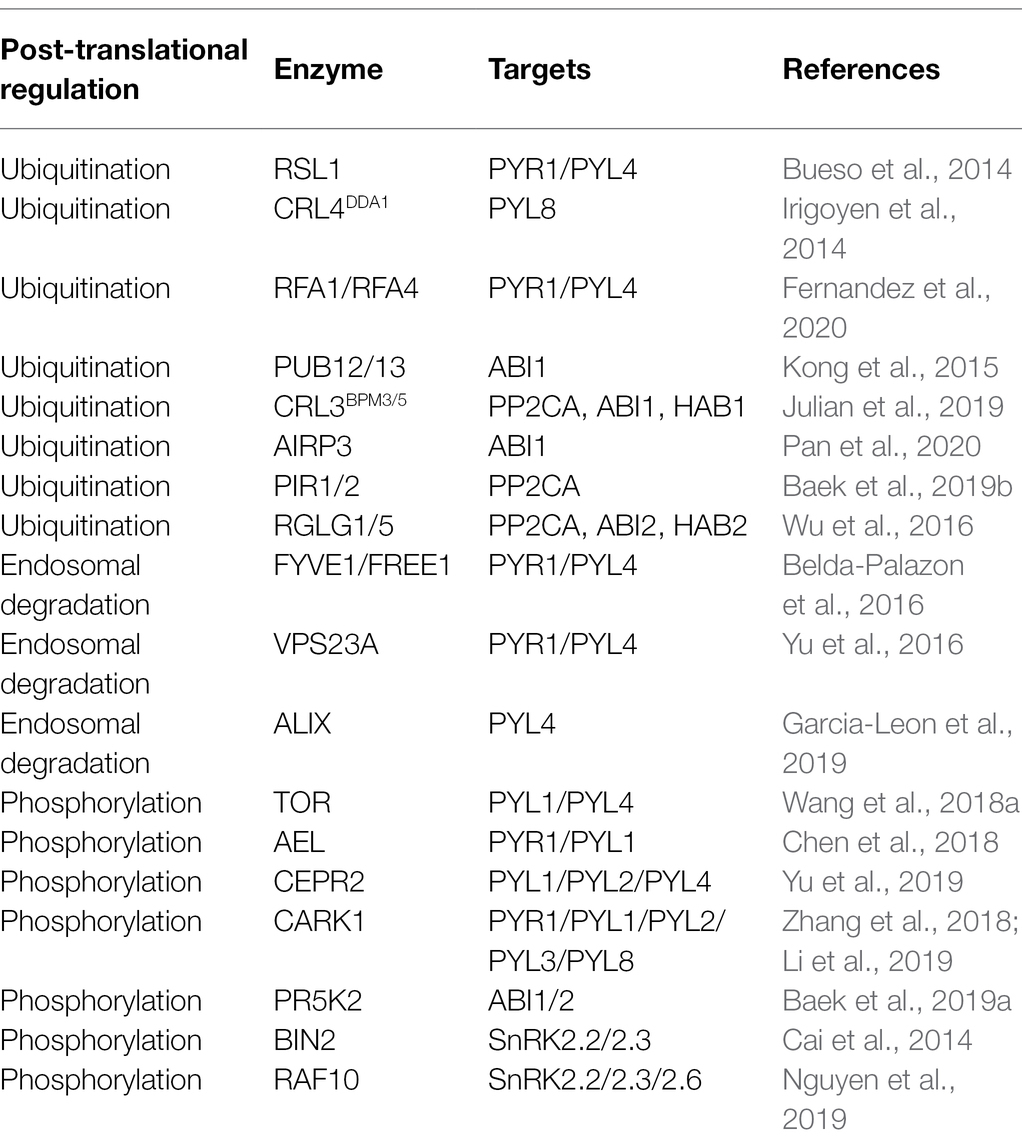
The core components of ABA signaling are modulated by different types of post-translational modification, among which, ubiquitination influences the responses to environmental factors. Ubiquitination is mediated by the activity of a series of enzymes, including ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and ubiquitin-protein ligase E3, and the substrate proteins thus ubiquitinated are subsequently degraded by 26S proteasome proteolysis (Stone, 2019). ABA components are regulated by E3 ligase complexes via successional steps (Figure 1). These complexes are classified into four groups, namely, really interesting new gene (RING), cullin-RING ligase (CRLs), U-box, and homologous to E6-AP carboxyl terminus (HECT; Vierstra, 2009), among which, RING, CRLs, and U-box E3 ligases are involved in ABA signaling.
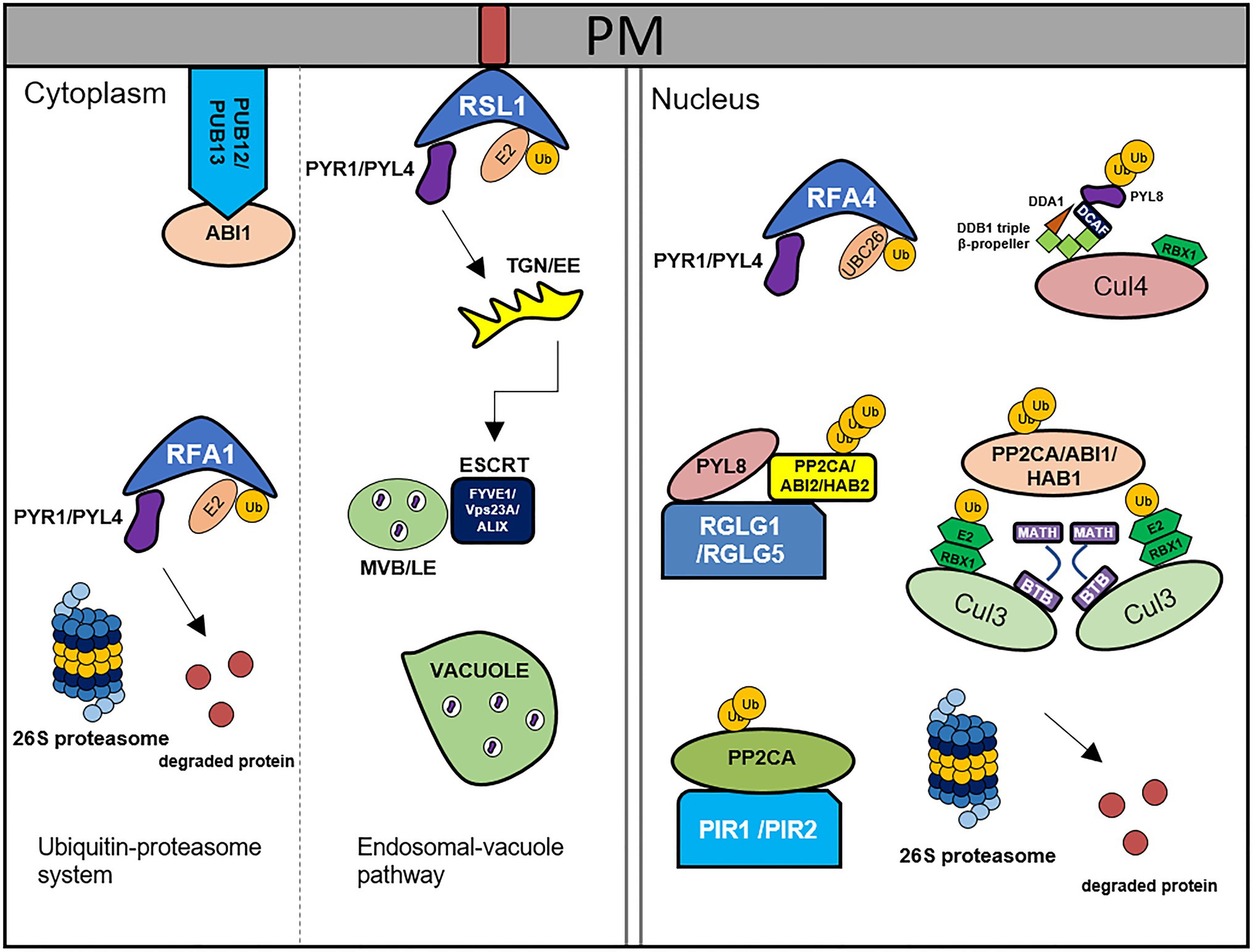
Figure 1. Degradation of ABA core components by the 26S ubiquitin-proteasome system and endosomal-vacuole pathway. PYR1/PYLs are degraded by RSL1, RFA1/RFA4, and CRL4DDA1 through ubiquitin-proteasome system. RSL1 ubiquitinates PYR1/PYL4 on the plasma membrane (PM), after that ubiquitinated substrates are degraded by ESCRT components, FIVE1/VPS23A/ALIX via endosomal-vacuole pathway. RFA1- and RFA4/CRL4DDA1-mediated ubiquitination processes occur in the cytosol and nucleus, respectively. PP2Cs are degraded by PUB12/13, CRL3BPM3/5(BTB-MATH), PIR1/2, and RGLG1/5 through 26S proteasome. PUB12/13 degrade ABI1 by ubiquitination in the PM. RGLG1/5 ubiquitinate PP2CA/ABI2/HAB2, CRL3BPM3/5(BTB-MATH) ubiquitinate PP2CA/ABI1/HAB1, and PIR1/2 ubiquitinates PP2CA in the nucleus. This figure has been modified from Coego et al. (2021).
Deubiquitination is catalyzed by the ubiquitin deconjugating enzymes (Chung and Baek, 1999). In contrast to E3 ligases, members of the deubiquitinase superfamily (DUB) proteins cleave ubiquitin from target proteins via the deconjugation or degradation of ubiquitin (Amerik and Hochstrasser, 2004). These processes contribute to enhancing target protein stability and activity in vivo (Taya et al., 1999; Lim et al., 2021b). There are five superfamilies among the DUBs, namely, ubiquitin-specific-processing proteases (UBPs), ubiquitin carboxy-terminal (UCH) proteases, the ovarian tumor proteases (OTUs), the Machado-Joseph disease protein domain proteases (MJDs), and JAB1/MPN+/MOV34 (JAMM) proteases. UBPs, UCHs, OTUs, and MJDs are cysteine proteases, whereas JAMMs are zinc metalloisopeptidases (Nijman et al., 2005). Of these five families, the UBPs are well studied in plants, and have been shown to play important roles in cellular processes, including signal transduction, protein degradation, and gene regulation (Yan et al., 2000; Liu et al., 2008).
Ubiquitination of ABA Receptors
CRL E3 ligase complexes and RING-type E3 ligases degrade PYR/PYLs via the 26S proteasome, and yeast two-hybrid screening has revealed that in this pathway, Cullin 4 (CUL4)/De-etiolated 1 (DET1)-damaged DNA binding protein 1 (DDB1)-associated 1 (DDA1) E3 ligase interacts with PYL4, PYL8, and PYL9 (Irigoyen et al., 2014). Furthermore, DDA1 promotes PYL8 protein degradation via the 26S proteasome and reduces ABA sensitivity in DDA1 overexpression mutants (Irigoyen et al., 2014), whereas in MG132-treated plants, accumulated PYL8 is polyubiquitinated and DDA1 promotes polyubiquitination, but ABA opposes this effect (Irigoyen et al., 2014). These findings provide evidence to indicate that ABA protects PYL8 from CRL4DDA1-mediated degradation, either by affecting the CDD-DDA1 complex or by promoting the formation of degradation-resistant PYL8-PP2C complexes (Irigoyen et al., 2014).
Ring finger of seed longevity 1 (RSL1), a member of the RBR-type E3 ligase family (Marin, 2010; Callis, 2014) interacts with PYR1 and PYL4 in the plasma membrane and subsequently polyubiquitinates these to promote their degradation via the 26S proteasome. Phenotypic analysis has revealed that RNA interference mutants of RSL1 are characterized by an ABA-hypersensitive phenotype, whereas an RSL1 overexpression line was found to show reduced ABA sensitivity during seed germination and early seedling growth (Bueso et al., 2014). These observations indicate that RSL1 plays a negative role in ABA signaling via the degradation of ABA receptors (Bueso et al., 2014). In the same gene family as RSL1, the RING finger ABA-related 1–9 (RFA1 to RFA9) proteins are similarly associated with ABA receptor degradation, among which, RFA1 and RFA4 are localized in the nucleus and promote the nuclear degradation of PYR1 and PYL4 via activity of the nuclear E2 enzyme UBC26 (Fernandez et al., 2020). Compared with wild-type plants, PYR1 and PYL4 were observed to show higher expression in an RFA1/RFA4 double knock-out mutant (Fernandez et al., 2020). The endosomal trafficking pathway functions as an important route for the modification of membrane proteins via the endosomal sorting complex required for transport (ESCRT) machinery. Ubiquitinated PYL4 is localized in the plasma membrane and degraded via a vacuolar degradation pathway mediated by the ESCRT components, in which ESCRT-I components FIVE1, VPS23A, and ALIX promote PYL4 degradation. Knock-down mutations of these ESCRT components are characterized a reduction in the degradation of PYL4 and enhanced ABA responses (Belda-Palazon et al., 2016; Yu et al., 2016; Garcia-Leon et al., 2019).
Ubiquitination of Clade A PP2Cs
Plant U-box type E3 ubiquitin ligase (PUB)12/13, Ring domain ligase (RGLG)1/5, CUL3-RING-based E3 ligases, and other E3 ligases collectively play roles in the proteolytic degradation of PP2Cs. PUB12/13 interact with ABI1 and thereby promote the ubiquitination of ABI1 in response to exogenous ABA, which induces the production of the dimeric form of PYR1 or monomeric forms of PYL4/PYL9 (Kong et al., 2015). A pub12 pub13 double mutant was found to be characterized by a high accumulation of ABI1 and an ABA-insensitive phenotype, the latter of which was recovered following the crossing of double mutant plants with those containing an abi1-3 mutation. These findings thus indicate that PUB12 and PUB13 play roles in controlling ABI1 stability (Kong et al., 2015). RGLG1/5 E3 ligases contribute to the degradation of PP2CA. In vivo analysis has revealed that RGLG1 interacts with PP2CA, which is enhanced by ABA, and promotes ubiquitination. In vitro, however, it has been observed that RGLG1 ubiquitinates PP2CA in the absence of either ABA or PYL4 (Wu et al., 2016). In contrast to PP2CA, which is nuclear localized, myristoylated RGLG1 is localized in the plasma membrane. ABA prevents the myristoylation of RGLG1 and induces cycloheximide-insensitive translocation to the nucleus (Belda-Palazon et al., 2019). Thus, RGLG1 plays a positive role in the ABA response by repressing PP2CA (Wu et al., 2016). PP2CA has also been demonstrated to interact with BTB/POZ and MATH domain proteins (BPMs) in plants, based on co-immunoprecipitation analysis performed in conjunction with liquid chromatography–tandem mass spectrometry. Compared with wild-type plants, those overexpressing BPM3 and BPM5 were found to accumulate lower amounts of PP2CA protein following ABA treatment. Contrastingly, in response to ABA treatment, the levels of PP2CA accumulated in plants harboring a bpm3 bpm5 double knock mutation were observed to be higher than those in the wild-type plants. Consistently, BPM3 and BPM5 overexpressing plants are characterized by enhanced ABA sensitivity, whereas the bpm3 bpm5 mutant shows reduced ABA sensitivity. Consequently, CRL3BPM complexes appear to play vital roles in ABA responses and signaling (Julian et al., 2019).
By interacting with UBC27, AIRP3 contributes to ABI1 degradation via E3 ligase activity. The UBC27-AIRP3 ubiquitination complex modulates ABA responses by enhancing ABI1 protein stability. In ubc27 and airp3 mutant plants, the rate of ABI1 degradation is reduced, and with respect to cotyledon greening and stomatal movement, ubc27 and airp3 mutant plants are characterized by ABA-insensitive phenotypes (Pan et al., 2020). Among other RING proteins, PP2CA interacting ring finger protein (PIR)1 and PIR2 have been shown to interact with PP2CA, both in vitro and in vivo, thereby promoting its ubiquitination. pir1 pir2 double knock-out mutants were found to be characterized by higher PP2CA protein levels, thus indicating that PIR1 and PIR2 degrade PP2CA via the 26S proteasome (Baek et al., 2019b).
Regulation of SnRK2s Stability by E3 Ligases
Although E3 ligases have been established to contribute indirectly to the stability of SnRK2s, there has as yet been no conclusive evidence to indicate that E3 ligases directly target SnRK2s. To date, only AtPP2-B11, a component of the SKP1/Cullin/F-box complex, has been shown to interact directly with SnRK2.3, with cell-free degradation assays performed using AtPP2-B11-OE and amiR-AtPP2-B11 cell extracts indicating that AtPP2-B11 negatively influences SnRK2.3 stability. However, there is no evidence to indicate that AtPP2-B11 directly ubiquitinates SnRK2.3 (Cheng et al., 2017). In addition, the E3-ubiquitin ligase HOS15 has been found to interact with SnRK2.6, thereby influencing its stability, and compared with wild-type plants, those harboring a hos15-2 mutation were found to accumulate higher levels of SnRK2.6 (Ali et al., 2019). However, in common with AtPP2-B11, the mechanisms whereby HOS15 regulates SnRK2 stability have yet to be established.
Modulation of ABA Signaling by Ubiquitin-Specific Processing Proteases
With respect to ABA signaling, plant tolerance to high salinity and drought stresses is modulated by UBP24 via the regulation of stomatal closure. Knock-out of the UBP24 gene confers an ABA-hypersensitive phenotype during seedling growth, although contrastingly it reduces ABA sensitivity in the guard cells of mature plants. The latter effect results in a drought-sensitive phenotype, owing to a higher water loss compared with wild-type plants (Zhao et al., 2016a). In pepper plants, CaUBP12 has been observed to suppress the degradation of CaSnRK2.6 (Arabidopsis OST1 homolog; Lim et al., 2021b). CaUBP12 interacts with CaSnRK2.6 both in vitro and in vivo, and cell-free degradation analysis has revealed higher levels of CaSnRK2.6 degradation when incubated with crude extracts of the CaUBP12 knock-down pepper plants, although lower levels in the presence of crude extracts of CaUBP12 overexpressing Arabidopsis and tobacco plants (Lim et al., 2021a,b). These findings thus provide evidence to indicate that deubiquitinase-mediated post-transcriptional modification is implicated in the modulation of ABA signaling.
Phosphorylation of ABA Core Components
In response to the perception of stress, ABA signaling is regulated via the phosphorylation activity of a diverse range of kinases particularly with respect to ABA-induced responses under drought stress (Zhu, 2016). In the ABA core signaling pathway, SnRK2s phosphorylate downstream transcription factors (Merlot et al., 2001; Yoshida et al., 2002, 2006b; Saez et al., 2004, 2006; Kuhn et al., 2006; Robert et al., 2006; Rubio et al., 2009), whereas other kinases contribute to the phosphorylation of ABA receptor PYR/PYLs, PP2Cs, SnRK2s, and downstream transcription factors (Figure 2; Cai et al., 2014; Chen et al., 2018; Wang et al., 2018a; Zhang et al., 2018; Baek et al., 2019a; Li et al., 2019; Nguyen et al., 2019; Yu et al., 2019).
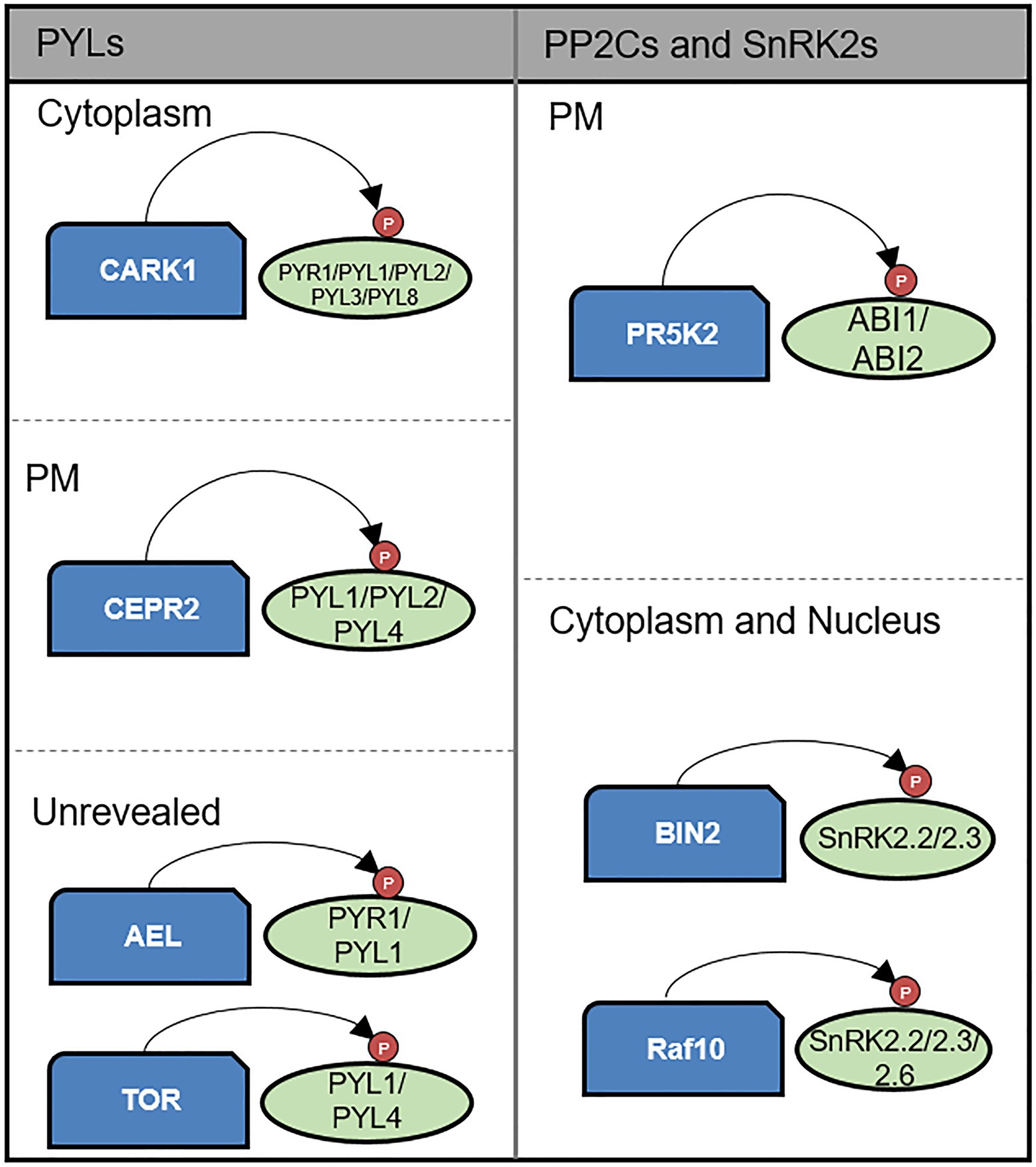
Figure 2. Kinase-mediated post-translational modification of core components of the ABA signaling pathway. CARK1, CEPR2, TOR kinase, and AEL phosphorylate PYLs. CARK1 positively regulates ABA signaling by enhancing PYLs activity on the cytoplasm. In contrast, CEPR2 and AEL negatively regulate ABA signaling by promoting degradation of PYLs. TOR kinase also plays negative role in ABA signaling by preventing PYL activation. CEPR2 acts on PM. ABI1 and ABI2 are phosphorylated and enhanced in their phosphatase activity by PR5K2 on the PM. BIN2 and RAF10 phosphorylate SnRK2s enhancing their kinase activity on the cytoplasm and nucleus.
Phosphorylation of ABA Receptors
Several studies have provided evidence indicating the phosphorylation of ABA receptors. For example, TOR kinase, Arabidopsis Early flowering 1 (EL1)-like casein kinase (AEL), C-terminally encoded peptide receptor 2 (CEPR2), and cytosolic ABA receptor kinase 1 (CARK1) have been established to phosphorylate PYLs (Chen et al., 2018; Wang et al., 2018a; Zhang et al., 2018; Li et al., 2019; Yu et al., 2019). Among these, TOR kinase inactivates PYL1 and PYL4 by phosphorylating the Ser119 and Ser114 residues, respectively. Phosphomimic mutants of PYL1 and PYL4 are characterized by an inability to inhibit ABI1 phosphatase activity. Moreover, ABA has been shown to inhibit TOR kinase activity, thereby enhancing the ABA response. Thus, these observations indicate that TOR kinase functions as a negative regulator of ABA signaling by phosphorylating PYL1 and PYL4 (Wang et al., 2018a). AEL phosphorylates PYR1 and PYL1 at residues Ser109/152 and Ser136/182, respectively, and in doing so, promotes PYR1 and PYL1 degradation. Compared with wild-type plants, PYR1 and PYL1 proteins show greater stability in ael triple mutants. The ABA- and NaCl-sensitive phenotypes in knock-out mutants thus tend to indicate that AEL negatively regulates ABA signaling (Chen et al., 2018). CEPR2 has been found to interact with PYL4 in the plasma membrane, in which it phosphorylates Ser54, thus promoting an accelerated degradation of the PYL4 protein. Consistently, the levels of PYL4 protein in CEPR2 overexpressing and cepr2 mutant plants have been shown to be less and more stable, respectively (Yu et al., 2019). CARK1 phosphorylates PYR1 Thr78, PYL1 Thr105, PYL2 Ser81, and PYL3 Thr101, thereby enhancing ABA responses implicated in the inhibition of seed germination, root growth, and the expression of a number of ABA-responsive genes (Zhang et al., 2018; Li et al., 2019).
Phosphorylation of Clade A PP2Cs and Subclass III SnRK2s
PP2Cs are also phosphorylated by specific kinases. For example, ABI1 and ABI2 are phosphorylated by PR5 receptor-like kinase 2 (PR5K2), which phosphorylates the putative ABI1 phosphorylation site Ser 314 and ABI2 Ser304, thereby enhancing their respective phosphatase activities (Baek et al., 2019a). Plants overexpressing PR5K2 are characterized by ABA-insensitive and drought-sensitive phenotypes, whereas atpr5k2 mutant plants show the opposite phenotypes. SnRK2s are phosphorylated and activated by Raf-like MAKKKs, among which RAF10 phosphorylates SnRK2s at a site within C-terminal domain II, referred to as the ABA box. raf10 raf11 double-knock-out mutants are characterized by reduced levels of SnRK2 phosphorylation and germination is significantly delayed in the presence of ABA. In brassinosteroid (BR) signaling, BRASSINOSTEROID INSENSITIVE 2 (BIN2) plays a negative regulatory role by phosphorylating several transcription factors, which contrast with ABA signaling, wherein BIN2 plays the role of a positive regulator promoting ABA responses via the phosphorylation of SnRK2.2 and SnRK2.3, thus enhancing their kinase activities. These observations indicate that the modulation of stress responses in plants involves a certain degree of crosstalk between the ABA and BR signaling pathways (Cai et al., 2014).
Conclusions and Perspectives
Abscisic acid plays vital roles in plant growth, development, and biotic/abiotic stress responses, particularly with respect to seed dormancy/germination, stomatal opening and closure, and leaf senescence. Moreover, the ABA core signaling pathway and post-translational modification of pathway components have been extensively studied. Ubiquitination, catalyzed by E3 ligases, contributes to the 26S proteasome-mediated degradation of ABA receptor PYR/PYL and clade A PP2C proteins, and numerous E3 ligases have been found to regulate the stability of these target proteins, either via direct or indirect interactions. However, it has yet to be conclusively determined whether E3 ligases directly ubiquitinate SnRK2 proteins, which accordingly limits our current understanding of the E3 ligase-mediated modulation of SnRK2 protein stability. The regulation of target protein stability is also modulated by deubiquitination, although in Arabidopsis, the mechanisms underlying the deubiquitination of ABA signaling core components remain to be ascertained. The involvement of UBPs in stress responses and other developmental processes is well characterized; however, to date there have been few studies that have examined the mechanisms associated with protein deubiquitination. To determine whether target proteins are deubiquitinated by UBPs, it will be necessary to isolate either mono- or poly-ubiquitinated proteins. The phosphorylation of target proteins also contributes to their activity and/or stability. In this regard, several kinases that phosphorylate key ABA signaling factors have been shown to regulate drought stress responses, either positively or negatively. However, only a few kinases involved in the ABA core signaling-associated regulation of drought responses have been examined, and it can be assumed that there are numerous other functional kinases that await identification. Consequently, to gain a better understanding of the post-translational modification of ABA core components, it will be necessary to conduct further studies that focus on identifying and characterizing these undiscovered proteins, which will illustrate potentially novel routes for the regulation of ABA signaling and stress responses.
Author Contributions
JL, CL, and SL: conceptualization and writing. JL and CL: data analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the Agriculture & Technology Development (project no. PJ01652101), the National Research Foundation of Korea (NRF) grant funded by the Korea Government (MSIT; 2021R1A2C2006338), and Rural Development Administration, Republic of Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Ali, A., Kim, J. K., Jan, M., Khan, H. A., Khan, I. U., Shen, M., et al. (2019). Rheostatic control of ABA signaling through HOS15-mediated OST1 degradation. Mol. Plant 12, 1447–1462. doi: 10.1016/j.molp.2019.08.005
Amerik, A. Y., and Hochstrasser, M. (2004). Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695, 189–207. doi: 10.1016/j.bbamcr.2004.10.003
Antoni, R., Gonzalez-Guzman, M., Rodriguez, L., Peirats-Llobet, M., Pizzio, G. A., Fernandez, M. A., et al. (2013). PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 161, 931–941. doi: 10.1104/pp.112.208678
Antoni, R., Gonzalez-Guzman, M., Rodriguez, L., Rodrigues, A., Pizzio, G. A., and Rodriguez, P. L. (2012). Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 158, 970–980. doi: 10.1104/pp.111.188623
Baek, D., Kim, M. C., Kumar, D., Park, B., Cheong, M. S., Choi, W., et al. (2019a). AtPR5K2, a PR5-Like receptor kinase, modulates plant responses to drought stress by phosphorylating protein phosphatase 2Cs. Front. Plant Sci. 10:1146. doi: 10.3389/fpls.2019.01146
Baek, W., Lim, C. W., Luan, S., and Lee, S. C. (2019b). The RING finger E3 ligases PIR1 and PIR2 mediate PP2CA degradation to enhance abscisic acid response in Arabidopsis. Plant J. 100, 473–486. doi: 10.1111/tpj.14507
Bauer, H., Ache, P., Lautner, S., Fromm, J., Hartung, W., Al-Rasheid, K. A., et al. (2013). The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr. Biol. 23, 53–57. doi: 10.1016/j.cub.2012.11.022
Belda-Palazon, B., Julian, J., Coego, A., Wu, Q., Zhang, X., Batistic, O., et al. (2019). ABA inhibits myristoylation and induces shuttling of the RGLG1 E3 ligase to promote nuclear degradation of PP2CA. Plant J. 98, 813–825. doi: 10.1111/tpj.14274
Belda-Palazon, B., Rodriguez, L., Fernandez, M. A., Castillo, M. C., Anderson, E. M., Gao, C., et al. (2016). FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. Plant Cell 28, 2291–2311. doi: 10.1105/tpc.16.00178
Bhaskara, G. B., Nguyen, T. T., and Verslues, P. E. (2012). Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 160, 379–395. doi: 10.1104/pp.112.202408
Brandt, B., Brodsky, D. E., Xue, S., Negi, J., Iba, K., Kangasjarvi, J., et al. (2012). Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. U. S. A. 109, 10593–10598. doi: 10.1073/pnas.1116590109
Breeze, E., Harrison, E., McHattie, S., Hughes, L., Hickman, R., Hill, C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873–894. doi: 10.1105/tpc.111.083345
Bueso, E., Rodriguez, L., Lorenzo-Orts, L., Gonzalez-Guzman, M., Sayas, E., Munoz-Bertomeu, J., et al. (2014). The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 80, 1057–1071. doi: 10.1111/tpj.12708
Cai, Z., Liu, J., Wang, H., Yang, C., Chen, Y., Li, Y., et al. (2014). GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 111, 9651–9656. doi: 10.1073/pnas.1316717111
Callis, J. (2014). The ubiquitination machinery of the ubiquitin system. Arabidopsis Book 12:e0174. doi: 10.1199/tab.0174
Carles, C., Bies-Etheve, N., Aspart, L., Léon-Kloosterziel, K. M., Koornneef, M., Echeverria, M., et al. (2002). Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J. 30, 373–383. doi: 10.1046/j.1365-313X.2002.01295.x
Chen, K., Li, G. J., Bressan, R. A., Song, C. P., Zhu, J. K., and Zhao, Y. (2020). Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 62, 25–54. doi: 10.1111/jipb.12899
Chen, H. H., Qu, L., Xu, Z. H., Zhu, J. K., and Xue, H. W. (2018). EL1-like casein kinases suppress ABA signaling and responses by phosphorylating and destabilizing the ABA receptors PYR/PYLs in Arabidopsis. Mol. Plant 11, 706–719. doi: 10.1016/j.molp.2018.02.012
Cheng, W. H., Endo, A., Zhou, L., Penney, J., Chen, H. C., Arroyo, A., et al. (2002). A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14, 2723–2743. doi: 10.1105/tpc.006494
Cheng, C., Wang, Z., Ren, Z., Zhi, L., Yao, B., Su, C., et al. (2017). SCFAtPP2-B11 modulates ABA signaling by facilitating SnRK2.3 degradation in Arabidopsis thaliana. PLoS Genet. 13:e1006947. doi: 10.1371/journal.pgen.1006947
Chinnusamy, V., Gong, Z., and Zhu, J. K. (2008). Abscisic acid-mediated epigenetic processes in plant development and stress responses. J. Integr. Plant Biol. 50, 1187–1195. doi: 10.1111/j.1744-7909.2008.00727.x
Chung, C. H., and Baek, S. H. (1999). Deubiquitinating enzymes: their diversity and emerging roles. Biochem. Biophys. Res. Commun. 266, 633–640. doi: 10.1006/bbrc.1999.1880
Coego, A., Julian, J., Lozano-Juste, J., Pizzio, G. A., Alrefaei, A. F., and Rodriguez, P. L. (2021). Ubiquitylation of ABA receptors and protein phosphatase 2C Coreceptors to modulate ABA signaling and stress response. Int. J. Mol. Sci. 22:7103. doi: 10.3390/ijms22137103
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Debeaujon, I., and Koornneef, M. (2000). Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 122, 415–424. doi: 10.1104/pp.122.2.415
Fernandez, M. A., Belda-Palazon, B., Julian, J., Coego, A., Lozano-Juste, J., Inigo, S., et al. (2020). RBR-type E3 ligases and the ubiquitin-conjugating enzyme UBC26 regulate Abscisic acid receptor levels and signaling. Plant Physiol. 182, 1723–1742. doi: 10.1104/pp.19.00898
Finkelstein, R. (2013). Abscisic acid synthesis and response. Arabidopsis Book 11:e0166. doi: 10.1199/tab.0166
Fujii, H., Chinnusamy, V., Rodrigues, A., Rubio, S., Antoni, R., Park, S. Y., et al. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. doi: 10.1038/nature08599
Fujii, H., Verslues, P. E., and Zhu, J. K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19, 485–494. doi: 10.1105/tpc.106.048538
Fujii, H., and Zhu, J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U. S. A. 106, 8380–8385. doi: 10.1073/pnas.0903144106
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488. doi: 10.1105/tpc.105.035659
Fujita, Y., Fujita, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Furihata, T., Maruyama, K., Fujita, Y., Umezawa, T., Yoshida, R., Shinozaki, K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U. S. A. 103, 1988–1993. doi: 10.1073/pnas.0505667103
Gao, S., Gao, J., Zhu, X., Song, Y., Li, Z., Ren, G., et al. (2016). ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant 9, 1272–1285. doi: 10.1016/j.molp.2016.06.006
Garcia-Leon, M., Cuyas, L., El-Moneim, D. A., Rodriguez, L., Belda-Palazon, B., Sanchez-Quant, E., et al. (2019). Arabidopsis ALIX regulates Stomatal aperture and turnover of Abscisic acid receptors. Plant Cell 31, 2411–2429. doi: 10.1105/tpc.19.00399
Geiger, D., Scherzer, S., Mumm, P., Stange, A., Marten, I., Bauer, H., et al. (2009). Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U. S. A. 106, 21425–21430. doi: 10.1073/pnas.0912021106
Gepstein, S., and Thimann, K. V. (1980). Changes in the abscisic acid content of oat leaves during senescence. Proc. Natl. Acad. Sci. U. S. A. 77, 2050–2053. doi: 10.1073/pnas.77.4.2050
Golldack, D., Li, C., Mohan, H., and Probst, N. (2013). Gibberellins and abscisic acid signal crosstalk: living and developing under unfavorable conditions. Plant Cell Rep. 32, 1007–1016. doi: 10.1007/s00299-013-1409-2
Gonzalez-Guzman, M., Pizzio, G. A., Antoni, R., Vera-Sirera, F., Merilo, E., Bassel, G. W., et al. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24, 2483–2496. doi: 10.1105/tpc.112.098574
Hao, Q., Yin, P., Li, W., Wang, L., Yan, C., Lin, Z., et al. (2011). The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol. Cell 42, 662–672. doi: 10.1016/j.molcel.2011.05.011
Hirayama, T., and Shinozaki, K. (2007). Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 12, 343–351. doi: 10.1016/j.tplants.2007.06.013
Hortensteiner, S. (2009). Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 14, 155–162. doi: 10.1016/j.tplants.2009.01.002
Hosy, E., Vavasseur, A., Mouline, K., Dreyer, I., Gaymard, F., Poree, F., et al. (2003). The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc. Natl. Acad. Sci. U. S. A. 100, 5549–5554. doi: 10.1073/pnas.0733970100
Irigoyen, M. L., Iniesto, E., Rodriguez, L., Puga, M. I., Yanagawa, Y., Pick, E., et al. (2014). Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 26, 712–728. doi: 10.1105/tpc.113.122234
Iyer, L. M., Koonin, E. V., and Aravind, L. (2001). Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins 43, 134–144. doi: 10.1002/1097-0134(20010501)43:2<134::AID-PROT1025>3.0.CO;2-I
Julian, J., Coego, A., Lozano-Juste, J., Lechner, E., Wu, Q., Zhang, X., et al. (2019). The MATH-BTB BPM3 and BPM5 subunits of Cullin3-RING E3 ubiquitin ligases target PP2CA and other clade A PP2Cs for degradation. Proc. Natl. Acad. Sci. U. S. A. 116, 15725–15734. doi: 10.1073/pnas.1908677116
Kang, J. Y., Choi, H. I., Im, M. Y., and Kim, S. Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. doi: 10.1105/tpc.010362
Kobayashi, Y., Murata, M., Minami, H., Yamamoto, S., Kagaya, Y., Hobo, T., et al. (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44, 939–949. doi: 10.1111/j.1365-313X.2005.02583.x
Kong, L., Cheng, J., Zhu, Y., Ding, Y., Meng, J., Chen, Z., et al. (2015). Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 6:8630. doi: 10.1038/ncomms9630
Koornneef, M., Jorna, M. L., Brinkhorst-van der Swan, D. L., and Karssen, C. M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 61, 385–393. doi: 10.1007/BF00272861
Kuhn, J. M., Boisson-Dernier, A., Dizon, M. B., Maktabi, M. H., and Schroeder, J. I. (2006). The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140, 127–139. doi: 10.1104/pp.105.070318
Kwak, J. M., Murata, Y., Baizabal-Aguirre, V. M., Merrill, J., Wang, M., Kemper, A., et al. (2001). Dominant negative guard cell K+ channel mutants reduce inward-rectifying K+ currents and light-induced Stomatal opening in Arabidopsis. Plant Physiol. 127, 473–485. doi: 10.1104/pp.010428
Lee, Y., Choi, Y. B., Suh, S., Lee, J., Assmann, S. M., Joe, C. O., et al. (1996). Abscisic acid-induced phosphoinositide turnover in guard cell protoplasts of Vicia faba. Plant Physiol. 110, 987–996. doi: 10.1104/pp.110.3.987
Lee, I. C., Hong, S. W., Whang, S. S., Lim, P. O., Nam, H. G., and Koo, J. C. (2011). Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 52, 651–662. doi: 10.1093/pcp/pcr026
Lee, S. C., Lan, W., Buchanan, B. B., and Luan, S. (2009). A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. U. S. A. 106, 21419–21424. doi: 10.1073/pnas.0910601106
Lee, S. J., Lee, M. H., Kim, J. I., and Kim, S. Y. (2015). Arabidopsis putative MAP kinase kinase kinases Raf10 and Raf11 are positive regulators of seed dormancy and ABA response. Plant Cell Physiol. 56, 84–97. doi: 10.1093/pcp/pcu148
Lee, K. P., Piskurewicz, U., Tureckova, V., Strnad, M., and Lopez-Molina, L. (2010). A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc. Natl. Acad. Sci. U. S. A. 107, 19108–19113. doi: 10.1073/pnas.1012896107
Lefebvre, V., North, H., Frey, A., Sotta, B., Seo, M., Okamoto, M., et al. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45, 309–319. doi: 10.1111/j.1365-313X.2005.02622.x
Leon-Kloosterziel, K. M., Gil, M. A., Ruijs, G. J., Jacobsen, S. E., Olszewski, N. E., Schwartz, S. H., et al. (1996). Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 10, 655–661. doi: 10.1046/j.1365-313X.1996.10040655.x
Li, X., Kong, X., Huang, Q., Zhang, Q., Ge, H., Zhang, L., et al. (2019). CARK1 phosphorylates subfamily III members of ABA receptors. J. Exp. Bot. 70, 519–528. doi: 10.1093/jxb/ery374
Li, W., Wang, L., Sheng, X., Yan, C., Zhou, R., Hang, J., et al. (2013). Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13. Cell Res. 23, 1369–1379. doi: 10.1038/cr.2013.143
Li, J., Wang, X. Q., Watson, M. B., and Assmann, S. M. (2000). Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287, 300–303. doi: 10.1126/science.287.5451.300
Lim, C. W., Baek, W., and Lee, S. C. (2021a). Tobacco ubiquitin-specific protease 12 (NbUBP12) positively modulates drought resistance. Plant Signal. Behav. 16:1974725. doi: 10.1080/15592324.2021.1974725
Lim, C. W., Baek, W., Lim, J., Hong, E., and Lee, S. C. (2021b). Pepper ubiquitin-specific protease, CaUBP12, positively modulates dehydration resistance by enhancing CaSnRK2.6 stability. Plant J. 107, 1148–1165. doi: 10.1111/tpj.15374
Liu, X., Hu, P., Huang, M., Tang, Y., Li, Y., Li, L., et al. (2016). The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 7:12768. doi: 10.1038/ncomms12768
Liu, Y., Wang, F., Zhang, H., He, H., Ma, L., and Deng, X. W. (2008). Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J. 55, 844–856. doi: 10.1111/j.1365-313X.2008.03557.x
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Mantyla, E., Lang, V., and Palva, E. T. (1995). Role of abscisic acid in drought-induced freezing tolerance, cold acclimation, and accumulation of LT178 and RAB18 proteins in Arabidopsis thaliana. Plant Physiol. 107, 141–148. doi: 10.1104/pp.107.1.141
Marin, I. (2010). Diversification and specialization of plant RBR ubiquitin ligases. PLoS One 5:e11579. doi: 10.1371/journal.pone.0011579
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. doi: 10.1038/35079635
Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S. Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980. doi: 10.1016/j.cell.2006.06.054
Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25, 295–303. doi: 10.1046/j.1365-313x.2001.00965.x
Mori, I. C., Murata, Y., Yang, Y., Munemasa, S., Wang, Y. F., Andreoli, S., et al. (2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol. 4:e327. doi: 10.1371/journal.pbio.0040327
Murata, Y., Pei, Z. M., Mori, I. C., and Schroeder, J. (2001). Abscisic acid activation of plasma membrane Ca(2+) channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13, 2513–2523. doi: 10.1105/tpc.010210
Mustilli, A. C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099. doi: 10.1105/tpc.007906
Nakashima, K., Fujita, Y., Kanamori, N., Katagiri, T., Umezawa, T., Kidokoro, S., et al. (2009a). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50, 1345–1363. doi: 10.1093/pcp/pcp083
Nakashima, K., Ito, Y., and Yamaguchi-Shinozaki, K. (2009b). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. doi: 10.1104/pp.108.129791
Ng, L. M., Soon, F. F., Zhou, X. E., West, G. M., Kovach, A., Suino-Powell, K. M., et al. (2011). Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc. Natl. Acad. Sci. U. S. A. 108, 21259–21264. doi: 10.1073/pnas.1118651109
Nguyen, Q. T. C., Lee, S. J., Choi, S. W., Na, Y. J., Song, M. R., Hoang, Q. T. N., et al. (2019). Arabidopsis Raf-Like kinase Raf10 is a regulatory component of Core ABA signaling. Mol. Cell 42, 646–660. doi: 10.14348/molcells.2019.0173
Nijman, S. M., Luna-Vargas, M. P., Velds, A., Brummelkamp, T. R., Dirac, A. M., Sixma, T. K., et al. (2005). A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786. doi: 10.1016/j.cell.2005.11.007
Nishimura, N., Sarkeshik, A., Nito, K., Park, S. Y., Wang, A., Carvalho, P. C., et al. (2010). PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61, 290–299. doi: 10.1111/j.1365-313X.2009.04054.x
Nishimura, N., Yoshida, T., Kitahata, N., Asami, T., Shinozaki, K., and Hirayama, T. (2007). ABA-hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 50, 935–949. doi: 10.1111/j.1365-313X.2007.03107.x
Ogawa, M., Hanada, A., Yamauchi, Y., Kuwahara, A., Kamiya, Y., and Yamaguchi, S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15, 1591–1604. doi: 10.1105/tpc.011650
Osakabe, Y., Arinaga, N., Umezawa, T., Katsura, S., Nagamachi, K., Tanaka, H., et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25, 609–624. doi: 10.1105/tpc.112.105700
Pan, W., Lin, B., Yang, X., Liu, L., Xia, R., Li, J., et al. (2020). The UBC27-AIRP3 ubiquitination complex modulates ABA signaling by promoting the degradation of ABI1 in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 117, 27694–27702. doi: 10.1073/pnas.2007366117
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Parkash, J., Vaidya, T., Kirti, S., and Dutt, S. (2014). Translation initiation factor 5A in Picrorhiza is up-regulated during leaf senescence and in response to abscisic acid. Gene 542, 1–7. doi: 10.1016/j.gene.2014.03.032
Pei, Z. M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G. J., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734. doi: 10.1038/35021067
Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., and Lopez-Molina, L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745. doi: 10.1105/tpc.108.061515
Pourtau, N., Mares, M., Purdy, S., Quentin, N., Ruel, A., and Wingler, A. (2004). Interactions of abscisic acid and sugar signalling in the regulation of leaf senescence. Planta 219, 765–772. doi: 10.1007/s00425-004-1279-5
Pruzinska, A., Tanner, G., Aubry, S., Anders, I., Moser, S., Muller, T., et al. (2005). Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol. 139, 52–63. doi: 10.1104/pp.105.065870
Raab, S., Drechsel, G., Zarepour, M., Hartung, W., Koshiba, T., Bittner, F., et al. (2009). Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J. 59, 39–51. doi: 10.1111/j.1365-313X.2009.03846.x
Radauer, C., Lackner, P., and Breiteneder, H. (2008). The bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol. Biol. 8:286. doi: 10.1186/1471-2148-8-286
Raghavendra, A. S., Gonugunta, V. K., Christmann, A., and Grill, E. (2010). ABA perception and signalling. Trends Plant Sci. 15, 395–401. doi: 10.1016/j.tplants.2010.04.006
Ren, G., An, K., Liao, Y., Zhou, X., Cao, Y., Zhao, H., et al. (2007). Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 144, 1429–1441. doi: 10.1104/pp.107.100172
Robert, N., Merlot, S., N'Guyen, V., Boisson-Dernier, A., and Schroeder, J. I. (2006). A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett. 580, 4691–4696. doi: 10.1016/j.febslet.2006.07.047
Rubio, S., Rodrigues, A., Saez, A., Dizon, M. B., Galle, A., Kim, T. H., et al. (2009). Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 150, 1345–1355. doi: 10.1104/pp.109.137174
Saez, A., Apostolova, N., Gonzalez-Guzman, M., Gonzalez-Garcia, M. P., Nicolas, C., Lorenzo, O., et al. (2004). Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 37, 354–369. doi: 10.1046/j.1365-313X.2003.01966.x
Saez, A., Robert, N., Maktabi, M. H., Schroeder, J. I., Serrano, R., and Rodriguez, P. L. (2006). Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 141, 1389–1399. doi: 10.1104/pp.106.081018
Santiago, J., Rodrigues, A., Saez, A., Rubio, S., Antoni, R., Dupeux, F., et al. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60, 575–588. doi: 10.1111/j.1365-313X.2009.03981.x
Sato, Y., Morita, R., Katsuma, S., Nishimura, M., Tanaka, A., and Kusaba, M. (2009b). Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 57, 120–131. doi: 10.1111/j.1365-313X.2008.03670.x
Sato, A., Sato, Y., Fukao, Y., Fujiwara, M., Umezawa, T., Shinozaki, K., et al. (2009a). Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 424, 439–448. doi: 10.1042/BJ20091221
Schelbert, S., Aubry, S., Burla, B., Agne, B., Kessler, F., Krupinska, K., et al. (2009). Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21, 767–785. doi: 10.1105/tpc.108.064089
Seo, M., Hanada, A., Kuwahara, A., Endo, A., Okamoto, M., Yamauchi, Y., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48, 354–366. doi: 10.1111/j.1365-313X.2006.02881.x
Sirichandra, C., Davanture, M., Turk, B. E., Zivy, M., Valot, B., Leung, J., et al. (2010). The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS One 5:e13935. doi: 10.1371/journal.pone.0013935
Soon, F. F., Ng, L. M., Zhou, X. E., West, G. M., Kovach, A., Tan, M. H., et al. (2012). Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335, 85–88. doi: 10.1126/science.1215106
Stone, S. L. (2019). Role of the ubiquitin proteasome system in plant response to abiotic stress. Int. Rev. Cell Mol. Biol. 343, 65–110. doi: 10.1016/bs.ircmb.2018.05.012
Szostkiewicz, I., Richter, K., Kepka, M., Demmel, S., Ma, Y., Korte, A., et al. (2010). Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 61, 25–35. doi: 10.1111/j.1365-313X.2009.04025.x
Takahashi, Y., Ebisu, Y., Kinoshita, T., Doi, M., Okuma, E., Murata, Y., et al. (2013). bHLH transcription factors that facilitate K+ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci. Signal. 6:ra48. doi: 10.1126/scisignal.2003760
Tan, B. C., Joseph, L. M., Deng, W. T., Liu, L., Li, Q. B., Cline, K., et al. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35, 44–56. doi: 10.1046/j.1365-313X.2003.01786.x
Taya, S., Yamamoto, T., Kanai-Azuma, M., Wood, S. A., and Kaibuchi, K. (1999). The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin. Genes Cells 4, 757–767. doi: 10.1046/j.1365-2443.1999.00297.x
Tischer, S. V., Wunschel, C., Papacek, M., Kleigrewe, K., Hofmann, T., Christmann, A., et al. (2017). Combinatorial interaction network of abscisic acid receptors and coreceptors from Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 114, 10280–10285. doi: 10.1073/pnas.1706593114
Umezawa, T., Okamoto, M., Kushiro, T., Nambara, E., Oono, Y., Seki, M., et al. (2006). CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 46, 171–182. doi: 10.1111/j.1365-313X.2006.02683.x
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 17588–17593. doi: 10.1073/pnas.0907095106
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U. S. A. 97, 11632–11637. doi: 10.1073/pnas.190309197
van Kleeff, P. J. M., Gao, J., Mol, S., Zwart, N., Zhang, H., Li, K. W., et al. (2018). The Arabidopsis GORK K(+)-channel is phosphorylated by calcium-dependent protein kinase 21 (CPK21), which in turn is activated by 14-3-3 proteins. Plant Physiol. Biochem. 125, 219–231. doi: 10.1016/j.plaphy.2018.02.013
Vierstra, R. D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397. doi: 10.1038/nrm2688
Vlad, F., Rubio, S., Rodrigues, A., Sirichandra, C., Belin, C., Robert, N., et al. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21, 3170–3184. doi: 10.1105/tpc.109.069179
Wang, Z., Wang, F., Hong, Y., Yao, J., Ren, Z., Shi, H., et al. (2018b). The flowering repressor SVP confers drought resistance in Arabidopsis by regulating Abscisic acid catabolism. Mol. Plant 11, 1184–1197. doi: 10.1016/j.molp.2018.06.009
Wang, P., Zhao, Y., Li, Z., Hsu, C. C., Liu, X., Fu, L., et al. (2018a). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 69, 100–112.e6. doi: 10.1016/j.molcel.2017.12.002
Weiss, D., and Ori, N. (2007). Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 144, 1240–1246. doi: 10.1104/pp.107.100370
Wu, Q., Zhang, X., Peirats-Llobet, M., Belda-Palazon, B., Wang, X., Cui, S., et al. (2016). Ubiquitin ligases RGLG1 and RGLG5 regulate Abscisic acid signaling by controlling the turnover of phosphatase PP2CA. Plant Cell 28, 2178–2196. doi: 10.1105/tpc.16.00364
Xue, S., Hu, H., Ries, A., Merilo, E., Kollist, H., and Schroeder, J. I. (2011). Central functions of bicarbonate in S-type anion channel activation and OST1 protein kinase in CO2 signal transduction in guard cell. EMBO J. 30, 1645–1658. doi: 10.1038/emboj.2011.68
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. doi: 10.1105/tpc.6.2.251
Yan, N., Doelling, J. H., Falbel, T. G., Durski, A. M., and Vierstra, R. D. (2000). The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 124, 1828–1843. doi: 10.1104/pp.124.4.1828
Yang, J., Worley, E., and Udvardi, M. (2014). A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26, 4862–4874. doi: 10.1105/tpc.114.133769
Yang, W., Zhang, W., and Wang, X. (2017). Post-translational control of ABA signalling: the roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 15, 4–14. doi: 10.1111/pbi.12652
Yin, P., Fan, H., Hao, Q., Yuan, X., Wu, D., Pang, Y., et al. (2009). Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 16, 1230–1236. doi: 10.1038/nsmb.1730
Yoshida, T., Fujita, Y., Maruyama, K., Mogami, J., Todaka, D., Shinozaki, K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. doi: 10.1111/pce.12351
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Yoshida, R., Hobo, T., Ichimura, K., Mizoguchi, T., Takahashi, F., Aronso, J., et al. (2002). ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43, 1473–1483. doi: 10.1093/pcp/pcf188
Yoshida, T., Nishimura, N., Kitahata, N., Kuromori, T., Ito, T., Asami, T., et al. (2006b). ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 140, 115–126. doi: 10.1104/pp.105.070128
Yoshida, R., Umezawa, T., Mizoguchi, T., Takahashi, S., Takahashi, F., and Shinozaki, K. (2006a). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281, 5310–5318. doi: 10.1074/jbc.M509820200
Yu, F., Lou, L., Tian, M., Li, Q., Ding, Y., Cao, X., et al. (2016). ESCRT-I component VPS23A affects ABA signaling by recognizing ABA receptors for Endosomal degradation. Mol. Plant 9, 1570–1582. doi: 10.1016/j.molp.2016.11.002
Yu, Z., Zhang, D., Xu, Y., Jin, S., Zhang, L., Zhang, S., et al. (2019). CEPR2 phosphorylates and accelerates the degradation of PYR/PYLs in Arabidopsis. J. Exp. Bot. 70, 5457–5469. doi: 10.1093/jxb/erz302
Zhang, L., Li, X., Li, D., Sun, Y., Li, Y., Luo, Q., et al. (2018). CARK1 mediates ABA signaling by phosphorylation of ABA receptors. Cell Discov. 4:30. doi: 10.1038/s41421-018-0029-y
Zhao, Y., Chan, Z., Gao, J., Xing, L., Cao, M., Yu, C., et al. (2016b). ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. U. S. A. 113, 1949–1954. doi: 10.1073/pnas.1522840113
Zhao, Y., Chan, Z., Xing, L., Liu, X., Hou, Y. J., Chinnusamy, V., et al. (2013). The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Res. 23, 1380–1395. doi: 10.1038/cr.2013.149
Zhao, Y., Zhang, Z., Gao, J., Wang, P., Hu, T., Wang, Z., et al. (2018). Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Rep. 23, 3340.e5–3351.e5. doi: 10.1016/j.celrep.2018.05.044
Zhao, J., Zhou, H., Zhang, M., Gao, Y., Li, L., Gao, Y., et al. (2016a). Ubiquitin-specific protease 24 negatively regulates abscisic acid signalling in Arabidopsis thaliana. Plant Cell Environ. 39, 427–440. doi: 10.1111/pce.12628
Zhou, H., Zhao, J., Cai, J., and Patil, S. B. (2017). UBIQUITIN-SPECIFIC PROTEASES function in plant development and stress responses. Plant Mol. Biol. 94, 565–576. doi: 10.1007/s11103-017-0633-5
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Zhu, S. Y., Yu, X. C., Wang, X. J., Zhao, R., Li, Y., Fan, R. C., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19, 3019–3036. doi: 10.1105/tpc.107.050666
Keywords: abscisic acid, core component of ABA signaling, phosphorylation, post-translational modification, ubiquitination
Citation: Lim J, Lim CW and Lee SC (2022) Core Components of Abscisic Acid Signaling and Their Post-translational Modification. Front. Plant Sci. 13:895698. doi: 10.3389/fpls.2022.895698
Edited by:
Girdhar Kumar Pandey, University of Delhi, IndiaReviewed by:
Andreas Bachmair, University of Vienna, AustriaCopyright © 2022 Lim, Lim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sung Chul Lee, c2NsZWUxOTcyQGNhdS5hYy5rcg==
†These authors have contributed equally to this work
 Junsub Lim
Junsub Lim Chae Woo Lim
Chae Woo Lim Sung Chul Lee
Sung Chul Lee