- 1Department of Applied Biosciences, Graduate School, Kyungpook National University, Daegu, South Korea
- 2Crop Breeding Division, National Institute of Crop Science, Rural Development Administration, Wanju, South Korea
Climate change leads to soil salinization, and the dynamic scarcity of freshwater has negatively affected crop production worldwide, especially Oryza sativa. The association among ion uptake, gene expression, antioxidant, biomass, and root and shoot development under different salt stress are not fully understood. Many studies are related to the effect of NaCl only. This study used two salts (CaCl2 and MgCl2) along with NaCl and analyzed their effects on mineral uptake (macronutrients and micronutrients), gene expression, seed germination, antioxidants, plant growth, and biomass in different rice genotypes. CaCl2 (up to 200 mM) slightly increased the germination percentage and seedling growth, whereas, 150 mM MgCl2 in the soil increased the root, shoot length, and fresh and dry weight in cultivars IR 28 and Cheongcheong. All agronomic traits among rice genotypes were drastically reduced by NaCl stress compared to other salts. Different salt stress differentially regulated ion uptake in the roots and shoots among different rice genotypes. Under different salt stress, a consistent decrease in Ca2+, Mn2+, and Fe2+ ions was observed in the roots of Cheongcheong, Nagdong, and IR 28. Similarly, under different salts, the stress in the shoots of Cheongcheong (Ca2+, Na+, and Zn2+) and Nagdong (Ca2+, Mg2+, Na+, and Zn2+) and the shoots of IR 28 (Ca2+ and Mg2+) consistently increased. Under different salts, a salt stress-related gene was expressed differentially in the roots of rice genotypes. However, after 6 and 12 h, there was consistent OsHKT1, OsNHX1, and OsSOS1 gene upregulation in the shoots of Nagdong and roots and shoots of the salt-tolerant cultivar Pokkali. Under different salt stress, glutathione (GSH) content increased in the shoot of IR 28 and Nagdong by NaCl, and MgCl2 salt, whereas, POD activity increased significantly by CaCl2 and MgCl2 in cultivar Cheongcheong and IR 28 shoot. Therefore, this study suggested that Pokkali responded well to NaCl stress only, whereas, the plant molecular breeding lab cultivar Nagdong showed more salt tolerance to different salts (NaCl, CaCl2, and MgCl2). This can potentially be used by agriculturists to develop the new salt-tolerant cultivar “Nagdong”-like Pokkali.
Introduction
Rice (Oryza sativa L.) is the main cereal crop worldwide with respect to cultivated area and total production. It provides almost 20% of the world’s dietary energy supply. Salinity is a crucial abiotic ecological factor that decreases rice and other plant growth and productivity worldwide. Soil salinity affects agriculture productivity in several regions globally, including Australia, Argentina, China, Egypt, Iran, Iraq, Pakistan, Thailand, and the United States [i.e., an area of >800 million ha; Rengasamy (2010)]. An estimated 5% or 3.85 million hectares of the total cultivated area in the world is affected by salinity (Sheng et al., 2008). By 2050, salt-affected soil is predicted to be increased up to 16.2 million ha, resulting in food insecurity for the world’s population (Yadav et al., 2017).
Soil is considered saline when it exhibits an apparent electrical conductivity of >4 ds m–1 (Daliakopoulos et al., 2016). Naturally, soil contains various kinds of soluble salts. Increasing salt concentration imposes ionic and osmotic stress on plants, causing several morphological and physiological changes. High salt content, particularly Cl– and Na+ sulfates, alters plant growth by modifying their anatomical, morphological, and physiological traits (Muscolo et al., 2003). Many studies reported that salt stress leads to a decrease in xylem development (Hilal et al., 1998). Some studies suggested that salt stress negatively decreases the root and shoot system’s fresh and dry weight by changing their concentration (Taffouo et al., 2010). The major cations found from soluble salts in saline soils consist of Na+, Ca2+, and Mg2+, whereas the most common anions are chloride, sulfates, and carbonates (including bicarbonates). Among various salts, sodium chloride (NaCl) is the most familiar worldwide. Most research studying plant physiological responses to salinity depends on experiments on NaCl stress only (Chinnusamy et al., 2005).
Salt stress activates the expression of various osmoresponsive genes and proteins in rice tissue (Chourey et al., 2003). The response of rice to salinity varies with their growth phases. Therefore, most regularly cultivated rice cultivar seedlings are very sensitive to salinity (Lutts et al., 1995; Zeng and Shannon, 2000). Thus, it can cause reductions in final germination percentage, germination energy percentage, and germination speed and lead to reduced root/shoot growth and dry matter (Ologundudu et al., 2014). Rice is considered the most sensitive crop for salinity stress at various growth stages. Salinity stress significantly damages the root surface area. Normal root growth needs essential elements, including Ca2+, Mg2+, Fe2+, and Zn2+, which are affected during salt stress. Thus, the reduction in root growth might influence Ca2+, Mg2+, Fe2+, and Zn2+ uptake (Robin et al., 2016). Quinoa seeds demonstrated decreased Ca2+, Mg2+, Zn2+, and Mn2+ contents in response to saline-sodic soil [e.g., in Larissa, Greece croplands; Karyotis et al. (2003)]. Some studies reported that salinity stress decreases Mn levels in the shoots of corn (Rahman et al., 1993). In contrast, some studies suggested that salt stress had no effect (Al-Harbi, 1995) or increased Mn content in the shoot or leaf tissue (Niazi and Ahmed, 1984). Most studies reported that salinity stress enhances Zn2+ concentrations in the shoots of maize (Rahman et al., 1993), citrus (Ruiz et al., 1997), and tomato (Knight et al., 1992). However, it was decreased in cucumber leaves (Al-Harbi, 1995). Some studies showed that salinity stress has an antagonistic effect on Ca2+, K2+, Fe2+, Mn2+, P, and Zn2+ but has a synergistic effect on Mg and N in rice (Jung et al., 2009; García et al., 2010).
After the salt overly sensitive (SOS) pathway proteins, another high-affinity K+ transporter (HKT) family of monovalent ion transporters is known as a relevant component in the plant defense against increased salinity levels (Rus et al., 2001, 2006). However, the main function of plant ion transporters in the HKT family remains unspecific. HKT1 was first confined and described in wheat (Triticum aestivum) as an HKT with Na+/K+ content carrier (Rubio et al., 1995). However, in Arabidopsis thaliana, the HKT1 functions as selective Na+ transporter (Uozumi et al., 2000). Salinity stress-induced cellular Ca2+ is considered a starting point in the SOS pathway for Na+ efflux facilitation. Furthermore, it forms a complex between a calcium-binding protein, SOS3 (CBL4), and a protein kinase, SOS2. Phosphorylation can occur between the SOS3/SOS2 complex and thus activate the Na+/H+ antiporter SOS1, which enhances Na+ export from cells. The Na+ export is not restricted only to the root surface, but their redistribution is more important to the whole body of the plant (Shi et al., 2000, 2003; Qiu et al., 2002; Oh et al., 2009). The Na+/H+ antiporter AtNHX family is present in A. thaliana, which plays a positive role in Na+ compartmentation (Blumwald, 2000). The Na+/H+ antiporter genes OsNHX1 to OsNHX5 have also been identified in rice (O. sativa L.; Fukuda et al., 2011). Across cell membranes, these genes play a pivotal role in catalysis to exchange Na+ for H+ that regulates cell volume, internal pH, and Na+ levels in the cytoplasm. Antiporter genes are also found in bacteria, yeast, animals, and plants with localization mainly in the cell membrane (Orlowski and Grinstein, 1997) and other organelles along with the perivacuolar compartment (Nass and Rao, 1998).
Salt stress, increases the formation of ROS within plant cells, and its high accumulation leads in oxidative damage of membrane lipids, protein, and nucleic acids (Ali et al., 2017; Sahin et al., 2018). There are some Enzymatic and non-enzymatic antioxidant which play a crucial role to minimize high ROS levels, an efficient plant system (Karuppanapandian et al., 2011; Chawla et al., 2013). Glutathione (GSH; c-glutamyl-cysteinyl-glycine) is a small intracellular thiol molecule which is considered as a strong non-enzymatic whereas peroxidase (POD) is enzymatic antioxidant (Foyer and Noctor, 2011; Hasanuzzaman et al., 2017). Higher GSH levels retain osmotic balance by induction of osmoprotectants and they prevent damage to, amino acids, lipids, and polysaccharides, thereby preventing damage to membranes, mitochondria, photosynthetic pigments, and other organelles which ultimately show tolerance to abiotic stresses (Nahar et al., 2015a,b,c). PODs function not only concern in scavenging H2O2 but also in plant growth, development, lignification, suberization, and cross-linking of cell wall compounds (Passardi et al., 2005). Higher activity of POD and some other antioxidants has been observed in salt tolerant cultivars of Calendula, Jatropha, pea, and tomato (Hernandez et al., 2000; Chaparzadeh et al., 2004; Mittova et al., 2004; Gao et al., 2008).
The O. sativa cultivar Nipponbare genome consists of nine HKT, five OsNHX1, and three genes of the SOS1 family. These genes play a crucial role in ion balance during salinity stress. Therefore, one gene from each family was selected in this study, and their relative gene expression was checked at different time points under different salt stress. However, different species varieties or cultivars have different salinity tolerance mechanisms. This study tested the two-parent cultivars, Cheongcheong and Nagdong of Cheongcheong/Nagdong Doubled haploid population (CNDH), with salt-sensitive cultivar IR 28 and salt-tolerant cultivar Pokkali. Most studies focused only on NaCl stress to check the plant physiological response against NaCl. However, this study investigated the effect of different salts, such as NaCl, CaCl2, and MgCl2, on the uptake of macronutrients and micronutrients, growth index, and gene expression in relation to salinity in four different rice cultivars at the early vegetative stage. The findings could help determine which salt most seriously affected growth attributes, ion uptake, and gene expression among different rice cultivars. Furthermore, the findings could help develop new salt-tolerant or salt-sensitive cultivars in the future for plant molecular breeding in the laboratory using genome editing techniques.
Materials and Methods
Plant Material and Preparation of Brine Solution
Seeds of four different rice cultivars, Cheongcheong, Nagdong, IR 28, and Pokkali, were used in the experiment. IR 28 is a salt-sensitive cultivar, and Pokkali is a salt-tolerant cultivar used worldwide, whereas cultivars Cheongcheong and Nagdong are two famous cultivars of plant molecular breeding laboratories in South Korea. Three different salts NaCl, CaCl2, and MgCl2 with concentrations of 100, 150, 200, and 250 mM were used for seed germination and seedling analysis of four different rice genotypes.
Seed Germination and Seedling Analysis
Before germination, seeds of four rice genotypes were surface sterilized with 70% ethanol for 1 min, followed by 5% sodium hypochlorite for 20 min. Afterward, seeds were washed five times with double-distilled water and dried for 40 min using autoclaved filter paper inside a clean bench. Small Petri dishes (∼9 cm in diameter) containing autoclaved filter paper were used for germination tests; 45 seeds were placed in a single Petri dish to give three replicates for each cultivar and treatment. Different salt treatments (NaCl, CaCl2, and MgCl2) were made at concentrations of 100, 150, 200, and 250 mM. The seeds were kept for a 14 h light/10 h dark photoperiod at 28°C in the light and dark. Relative humidity was maintained at 60% inside the growth room. The different salt stress treatments lasted for 2 weeks. Seeds were considered to have germinated when the radical was protruding from the seed coat. Germinated seeds were counted after each week. Finally, after 14 days, germination rate and seedling growth were recorded phenotypically.
Seedling Analysis in Soil Using Different Salt Treatments
Four rice genotypes were pregerminated and grown in small water cups of 500 mL (Σ100 mm in diameter and 120 mm in height) containing soil for 4 weeks in the greenhouse. Before salinity stress, the pH and EC of the soil were measured (4.5–5.5 and 2.0, respectively). Each water cup contains 10–12 germinated seedlings and makes three replicates of each salt treatment for four rice cultivars. Therefore, when rice seedlings reach the four- or five-leaf stage, 150-mM NaCl, CaCl2, and MgCl2 salts (200 mL) were applied twice daily for 7 days. Afterward, phenotypic differences were observed among rice cultivars, and the root and shoot length of the rice seedlings were measured.
Measurement of Fresh and Dry Weights
After salinity stress with 150-mM NaCl, CaCl2, and MgCl2, roots, and shoots from rice cultivars were collected, and the fresh weight was measured. Furthermore, the samples were dried at 65°C for 5 days, and the root and shoot dry weight was determined.
Ion Determination
Roots and shoots of four rice cultivars were used for ion analysis, as described by Rus et al. (2001) with slight modification. Briefly, the samples were dried at 65°C for 7 days and ground immediately in liquid nitrogen. Furthermore, 100-mg powder was extracted with 10 mL of 0.1 N HNO3 for 30 min. Furthermore, the samples were diluted with 2% HNO3, and macronutrients and micronutrients were analyzed using an ICP spectrometer (I) (Optima 7300DV & Avio500; Perkin-Elmer).
Quantitative Real-Time Polymerase Chain Reaction Analysis
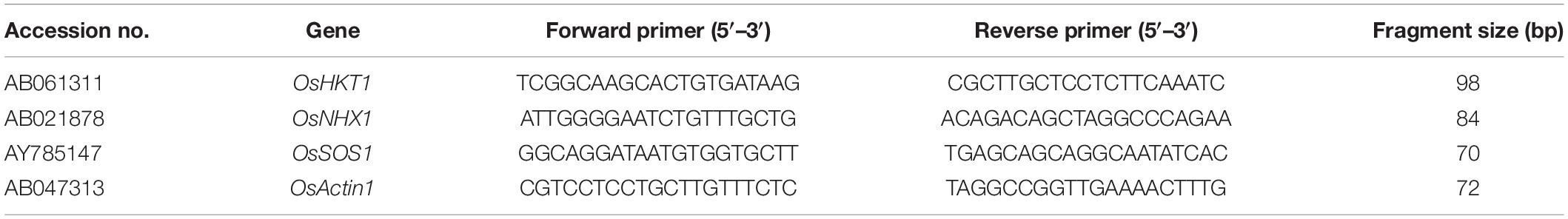
Pregerminating seeds of each rice genotype were grown for 3 weeks in a water cup containing soil up to the three- to four-leaf stage. These plants were treated with 150 mM (NaCl, CaCl2, and MgCl2), and RNA was extracted from their roots and shoots after 0, 6, and 12 h using a RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. A NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, United States) was used to measure RNA concentrations. For first-strand cDNA synthesis, the qPCRBIO cDNA Synthesis Kit and 600 ng total RNA were used. For quantitative real-time polymerase chain reaction (qRT-PCR), this study used the StepOnePlus™ Real-Time PCR System, Life Technologies Holdings Pte Ltd. (Singapore), BioFACT™ 2X Real-Time PCR Master Mix (including SYBR® Green I; www.bio-ft.com; South Korea), and primers specific to the selected genes (Table 1). OsActin1 (accession no. AB047313) was used as an internal reference gene for normalization.
Glutathione and Peroxidase
Rice genotypes were grown for 3 weeks in a water cup containing soil up to the three- to four-leaf stage. To determine the activity of GSH and POD these plants were treated with 150 mM (NaCl, CaCl2, and MgCl2), and the leaf samples were collected after 24 h. Two hundred milligrams (FW) of frozen tissues were ground immediately in liquid nitrogen. Briefly, 200 mg samples were homogenized in 2 mL of 10% trichloroacetic acid (TCA), followed by centrifugation at 10,000 × g for 15 min. The 0.1 mL supernatant was transferred to 3 mL of 150 mM monosodium phosphate buffer, and 0.5 mL Ellman’s reagent, followed by incubation at 30oC for 5 min. Afterward, the absorbance was measured spectrophotometrically at 420 nm. The POD value was measured according to the method described by Khan et al. (2019). Briefly, 500 mg plant samples were ground immediately in liquid nitrogen. Afterward, 0.1 M potassium phosphate buffer with (6.8 pH) was added to the samples and centrifuged at 4oC for 15 min at 5,000 r.p.m. A reaction mixture contained 0.1 M potassium phosphate buffer (pH 6.8), 50 μl pyrogallol (50 μM), and 50 μl H2O2 (50 μM), were mixed with 100 μl of the sample crude extract, and the reaction mixture was incubated at 25°C for 5 min. After incubation 5% H2SO4 (v/v) was added to stop the enzymatic reaction. The resulting absorbance was measured spectrophotometrically at 420 nm. One unit of POD was directly measured by an increase of 0.1 units of absorbance.
Statistical Analyses
Statistical analysis was performed for three replicates, where each replication was considered a block and arranged in different Petri dishes or pots in the control growth room conditions. The experiment was repeated three times. Differences among treatment means were evaluated using Duncan’s multiple range test with the significance set at p < 0.05. Data analysis was conducted in SPSS (IBM SPSS Statistics version 22). Figures were produced using GraphPad Prism version 9.0 (Graph Pad Software, Inc., San Diego, CA, United States).
Results
Effects of Different Salts on Seed Germination and Seedling Growth
The germination rate of four rice genotypes was observed after 7 and 14 days under three different salt stresses (NaCl, CaCl2, and MgCl2) at concentrations of 100, 150, 200, and 250 mM (Figure 1). After 7 days of different salt stresses, a uniform nature of seeds germination was observed for cultivar Pokkali and IR 28 up to 150- and 200-mM, whereas Nagdong showed a high germination rate compared to other cultivars (Figures 1A–C). In contrast, the Cheongcheong germination rate was lower than other cultivars. Under CaCl2 stress, their germination was slightly higher than that of NaCl and MgCl2 stresses (Figure 1B). Finally, after 14 days, up to 250 mM NaCl and CaCl2 stress seeds of Pokkali showed >60 and >20% germination rates, respectively. However, Pokkali seeds can germinate only up to 150 mM MgCl2 stress (Figures 1D–F). IR 28 seeds up to 250 mM germinated 40% in CaCl2 (Figure 1E) stress and <40% in NaCl stress; similarly, up to 150 mM, it can germinate to <40% in MgCl2 (Figures 1D,F). Under different salt stress, the highest germination rate was recorded in Nagdong compared to other cultivars and lowest in Cheongcheong. Nagdong seeds germinated 60 and 40% up to 250 mM NaCl and CaCl2 stress, respectively, whereas, >30% of Nagdong seeds germinated up to 200 mM MgCl2 stress (Figures 1D–F). Maximum seedling growth up to 200 mM under different salt stress was observed in Nagdong compared to other cultivars. MgCl2 stress from 150 to 250 mM significantly inhibited seed germination and seedling growth in all cultivars, except Nagdong (Figure 1H). Similarly, under 200 mM CaCl2, Cheongcheong, and salt-sensitive IR 28 seedlings responded well-compared to NaCl and MgCl2 (Figures 1C,G,I). In contrast, among different salt stress, Pokkali seedlings responded well to NaCl only (Figure 1J).
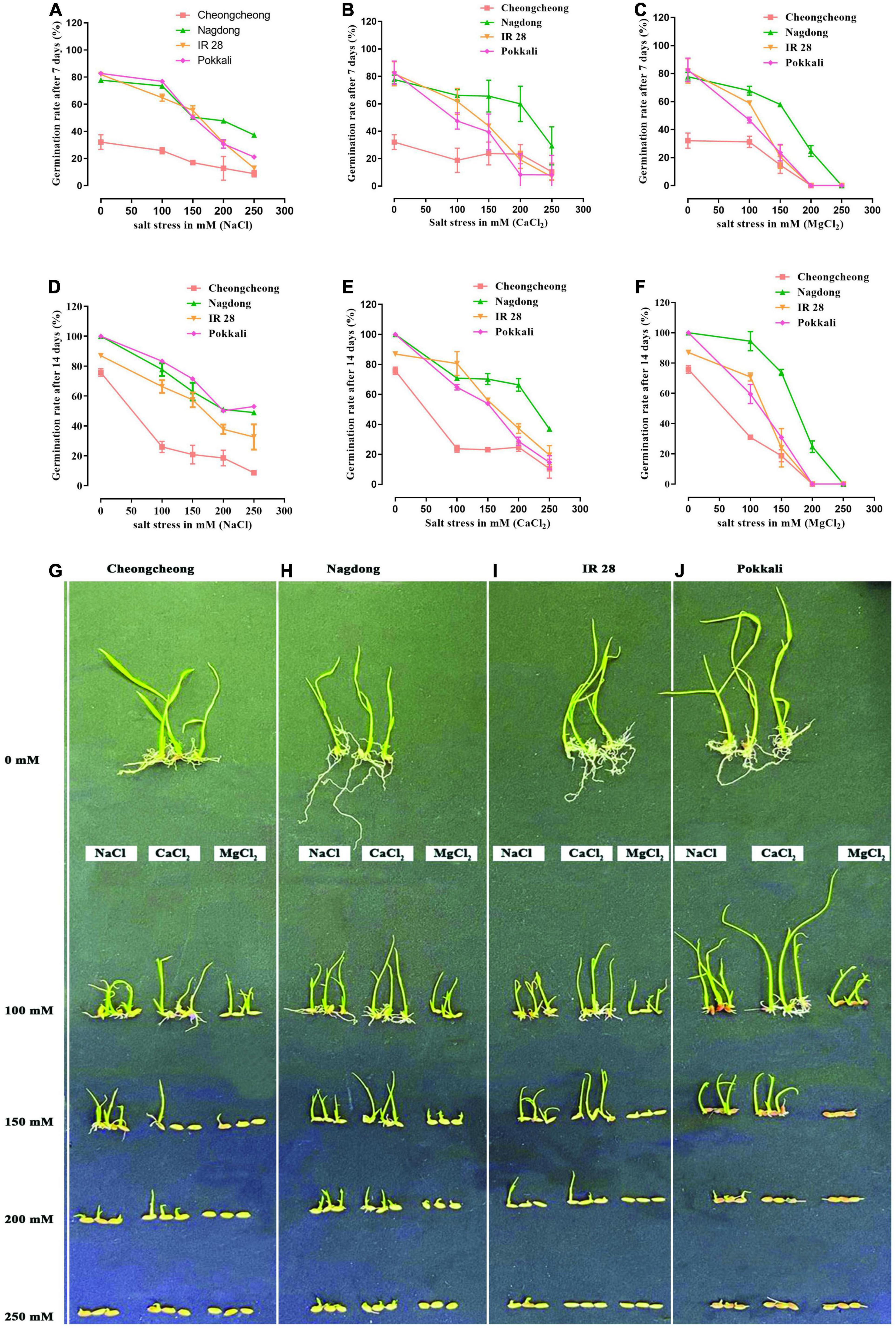
Figure 1. Seed germination test among rice genotypes under different salt stress. Different concentrations of different salts (NaCl, CaCl2, and MgCl2) were used to evaluate the seed germination and seedling growth (A–J).
Growth Determination in Soil Under Different Salt Stress
Seedling growth of four rice cultivars was determined after 7 days of 150-mM continuous stress of different salts (NaCl, CaCl2, and MgCl2). A phenotypical difference was observed among rice genotypes after 0, 3, 5, and 7 days (Figure 2A). After 3 days of stress, the most growth reduction in Cheongcheong and IR 28 was examined. After 5 days of continuous salt stress, the leaf apex and base of the tillers started burning and became dry. Finally, at 7 days, the growth rate was drastically reduced in all rice genotypes compared to control. However, Cheongcheong and IR 28 responded well to MgCl2 stress in soil compared to NaCl and CaCl2 stress. In contrast, Nagdong and Pokkali seedlings showed more vigor under NaCl and CaCl2 stress than MgCl2 (Figure 2A). Similarly, the root and shoot length was measured after 7 days of salt stress. Shoot length significantly decreased in all rice genotypes compared to their respective control. However, under different salt stress, Cheoncheong and IR 28 shoots were reduced in the same way as to control plants. The shoot length in MgCl2 was higher in Cheoncheong and IR 28 than NaCl and CaCl2 stress (Figure 2C). In contrast, Nagdong and Pokkali shoot length significantly decreased under MgCl2 stress compared to NaCl and CaCl2 stress. However, the shoot length of Nagdong and Pokkali was higher than that of Cheongcheong and IR 28 (Figure 2C). Under different salt stress, the root length of IR 28 significantly decreased compared to their respective control (Figure 2D). Similarly, NaCl stress significantly decreased the root length in cultivar Cheongcheong, whereas, the root length of Cheongcheong was non-significant on CaCl2 and MgCl2 stress compared to control plants (Figure 2D). In contrast, under different salt stress, there was no significant difference in the root length of Nagdong and Pokkali compared to control plants (Figure 2D).
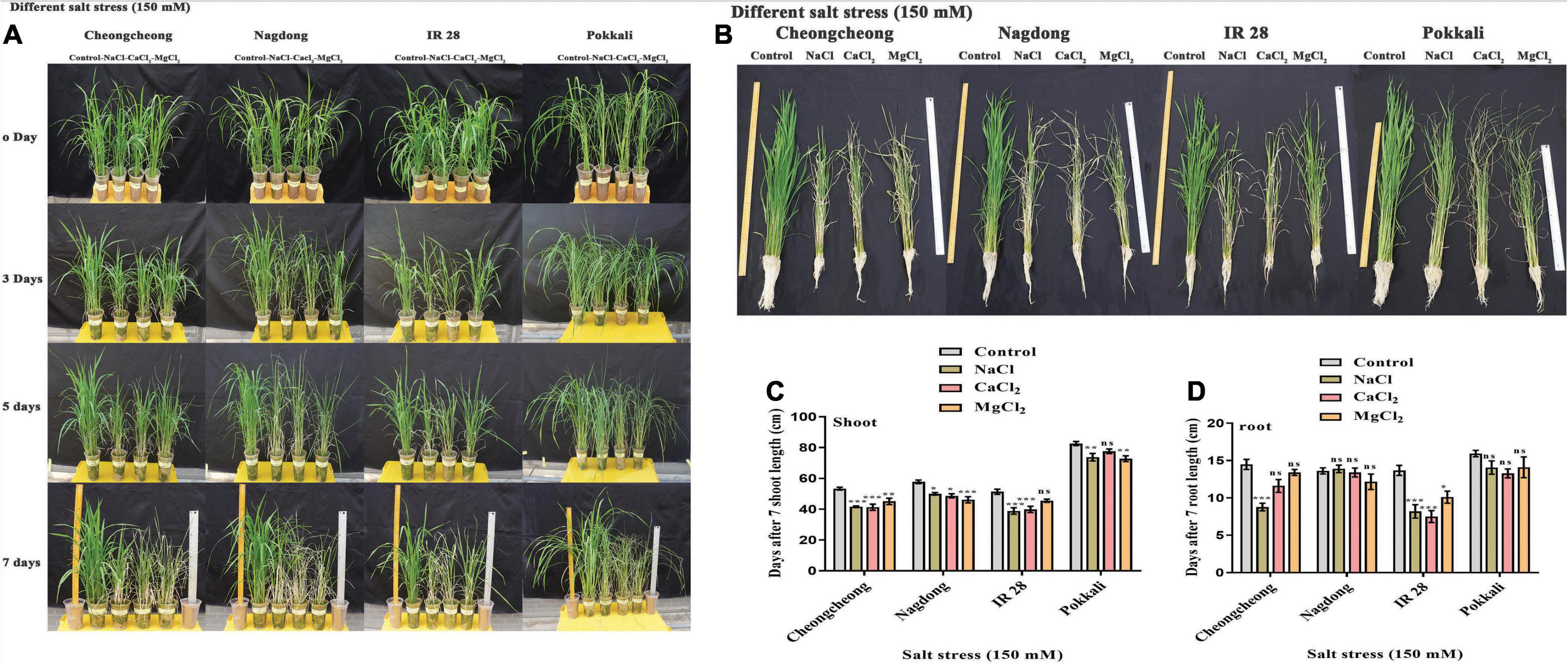
Figure 2. Phenotypic evaluation of rice genotypes under 150 mM of different salts (NaCl, CaCl2, and MgCl2) in a greenhouse. (A) Effects of different salts after 0, 3, 5, and 7 days. (B) Measurement of the growth index of each rice cultivar. (C,D) Shoot and root length determination. Statistical analysis was calculated by the Bonferroni posttest two-way repeated-measures analysis of variance. Asterisks in the vertical bar indicate a significant difference at *p < 0.05, **p < 0.01, ***p < 0.00, compared to control.
Effects of Salt Stress on the Fresh and Dry Weight of Rice
In soil, salinity stress significantly reduced the fresh and dry weight of the tested genotypes compared to control plants. However, Pokkali and Nagdong exhibited the highest root/shoot fresh and dry weight when exposed to different salt stress compared to the rest of the cultivars. Nagdong and Pokkali responded well under NaCl and CaCl2, whereas, MgCl2 stress significantly reduced the root/shoot fresh and dry weight (Figures 3A,B). However, under MgCl2 stress, Cheongcheong and IR 28 responded well and showed the highest root/shoot fresh and dry weight compared to NaCl and CaCl2 stress (Figures 3A,B).
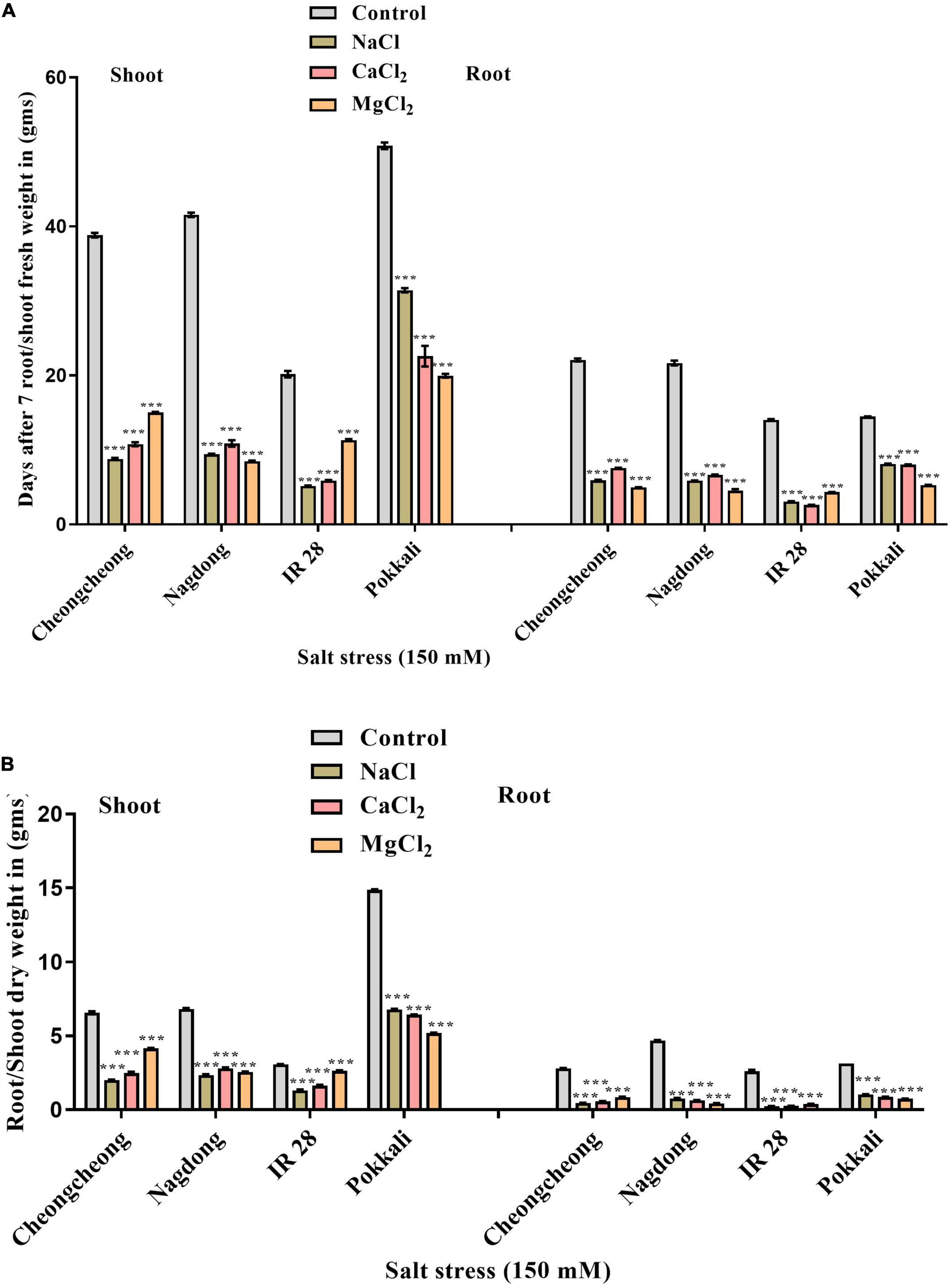
Figure 3. Effect of 150 mM of different salts on the shoot and root fresh and dry weight. (A) Fresh weight of shoots and roots. (B) Shoot and root dry weight. Statistical analysis was calculated by the Bonferroni posttest two-way repeated-measures analysis of variance. Asterisks in the vertical bar indicate a significant difference at ***p < 0.00, compared to control.
Effect of Different Salts on Nutrients Uptake
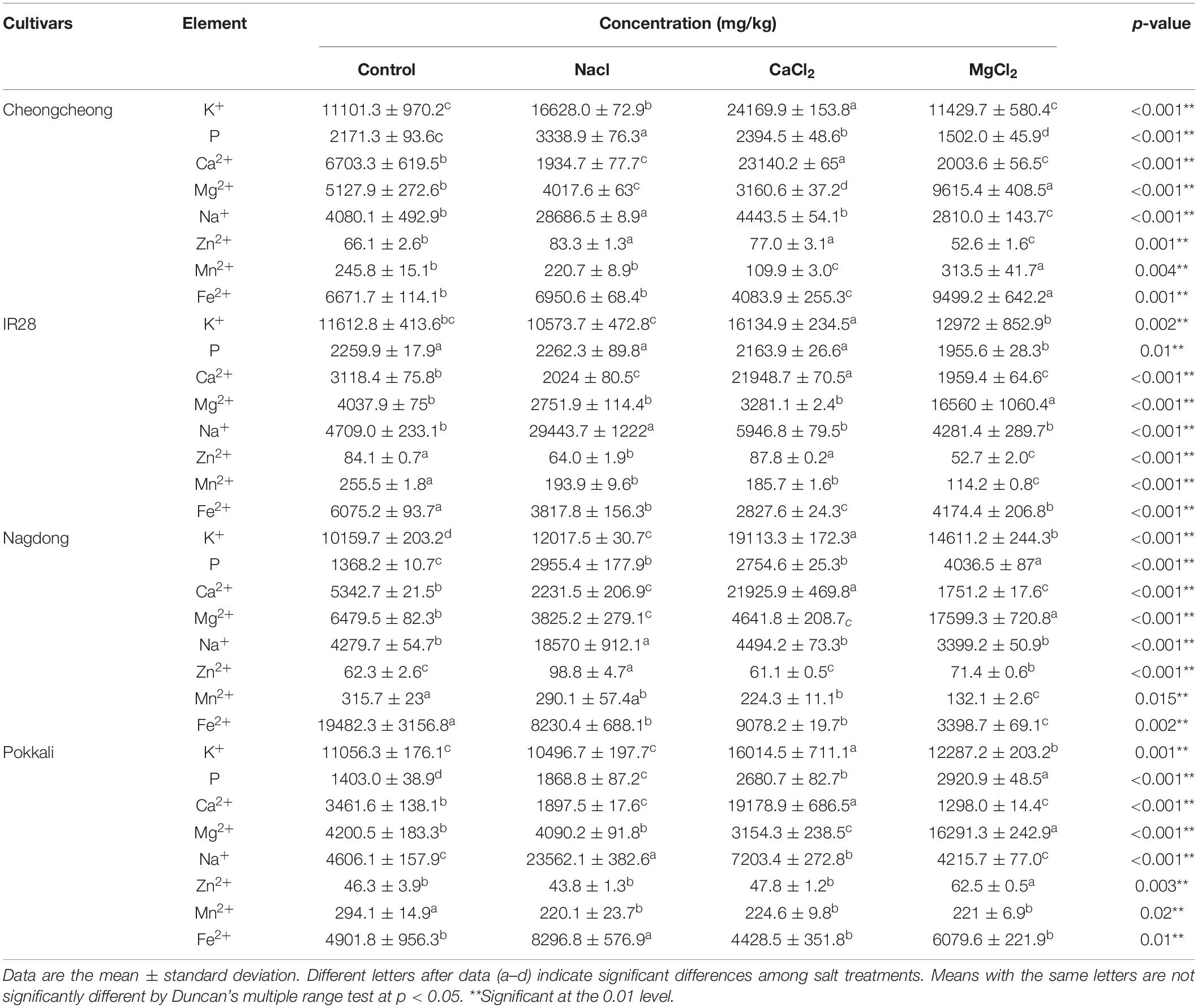
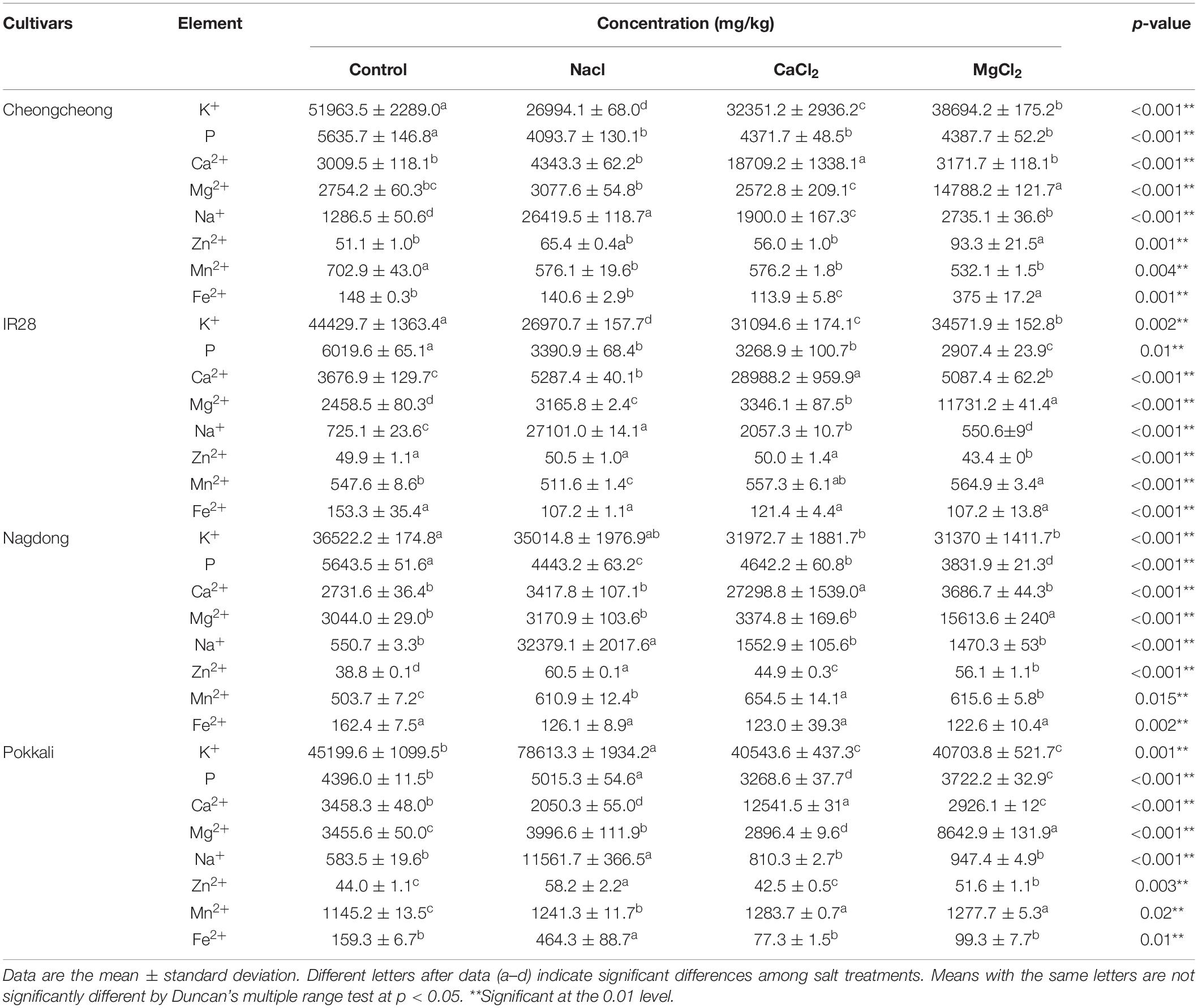
Under different salt stress, eight elements (namely K+, P, Ca2+, Mg2+, Na+, Zn2+, Mn2+, and Fe2+) were determined in the roots and shoots of control and treated rice plants (Tables 2, 3). Different salts stress differentially regulated the nutrients and their uptake in all rice genotypes. After 7 days of 150 mM of different salt stress, there was a high uptake of K+ and P ions was observed in the roots of Nagdong and Cheongcheong; however, P uptake in the roots of Cheongcheong under MgCl2 stress significantly decreased by 45.9-fold compared to control plants. The K+ and P uptake levels in the shoots of Cheongcheong, IR 28, and Nagdong significantly decreased when subjected to different salt stress. Under different salts stress, roots of IR 28 show significantly a high uptake of K+, but P uptake decreased significantly under CaCl2 and MgCl2 salts. However, significantly a high uptake of P was observed in the roots of Pokkali under three different salt stress compared to control plants. Similarly, under CaCl2 and MgCl2 stress, a maximum uptake of K+ and P was observed in the roots of Pokkali. Although CaCl2 and MgCl2 stress decreased significantly the uptake of K+ and P in the shoot of Pokkali, whereas in the shoot region NaCl stress significantly increased the uptake of K+ and P. Interestingly among all rice cultivars roots and shoots we observed that both CaCl2 and MgCl2 stress, significantly increased their respective ions compared to control plants e.g., CaCl2 salt increase Ca2+ ion and MgCl2 increase Mg2+ ion. However, different salt stresses significantly increase the uptake of Ca2+ and Mg2+ ions in the shoot of Cheongcheong, IR 28, and Nagdong except for the cultivar Pokkali (Tables 2, 3).
Tables 2, 3 show that the micronutrient Na+ significantly increased by NaCl stress in the roots and shoots of all rice genotypes compared to control plants under different salt stress. Both NaCl and CaCl2 salts significantly increase the Zn2+ uptake in the roots of Cheongcheong and shoot of IR 28. However, among all rice cultivars under NaCl stress, a remarkable uptake of Zn2+ was observed in the roots of Nagdong (Table 2). Under different salt stress, a maximum uptake of Mn2+ was observed in the shoots of Nagdong, Pokkali, and IR 28, whereas in the root of Cheongcheong MgCl2 significantly increased the uptake of Mn2+. However, under different salt stress, Mn2+ uptake significantly decreases in the roots of all rice cultivars. Different salt stress significantly decreased the uptake of total Fe2+ content in the roots and shoots of all rice genotypes, but Fe2+ content significantly increased under MgCl2 and NaCl stress in the roots and shoots of Cheongcheong and Pokkali (Tables 2, 3).
Ion Transport-Related Gene Expression Under Different Salt Stress Among Rice Cultivars
Under different salt stress at different time points, the expression of three very important ion transport-related genes, namely OsHKT1, OsNHX1, and OsSOS1, was examined (Figure 4). Gene regulation under different salt stress differed in roots and shoots among rice genotypes according to StepOnePlus™ Real-Time PCR System. After 6 h of different salt stress, NaCl and CaCl2 upregulated OsHKT1 and OsSOS1 in the roots of Cheongcheong, whereas these genes were downregulated after 12 h; OsSOS1 was 40% upregulated by MgCl2 in the roots of Cheongcheong (Figure 4A). In contrast, after 6 h of different salt stress, OsNHX1 and OsSOS1 were upregulated by CaCl2 and MgCl2 in the shoots of Cheongcheong. In addition, after 12 h of salt stress, CaCl2 and MgCl2 highly expressed OsHKT1, whereas OsNHX1 and OsSOS1 were upregulated by NaCl stress (Figure 4B). Similarly, IR 28 roots exhibited absolute expression of genes OsHKT1 and OsSOS1 after 6 h exposure to 150 mM salt concentrations of NaCl, CaCl2, and MgCl2, whereas after 12 h of stress, these genes were almost 90% upregulated by NaCl and MgCl2 (Figure 4C). Similarly, after 6 h of stress, NaCl and MgCl2 upregulated OsHKT1 and OsNHX1 in the shoots; however, these genes were significantly downregulated after 12 h of stress, except for OsSOS1, which was almost 40% upregulated by MgCl2 (Figure 4D). After 6 and 12 h of stress, NaCl and CaCl2 showed the most OsNHX1 and OsHKT1 upregulation in the roots of Nagdong, although MgCl2 up to 80% also induced OsNHX1. In contrast, NaCl, CaCl2, and MgCl2 downregulated OsSOS1 in the roots of Nagdong (Figure 4E). After 6 h of stress, NaCl, CalCl2, and MgCl2 upregulated OsHKT1 and OsNHX1 in Nagdong shoots; however, after 12 h of stress, NaCl, CaCl2, and MgCl2 upregulated OsHKT1 and OsSOS1 (Figure 4F). Pokkali roots after 6 h of stress showed very unique results; all ion transport-related genes, such as OsHKT1, OsNHX1, and OsSOS1, were 100% upregulated by NaCl, CaCl2, and MgCl2, except for OsHKT1, which was downregulated by NaCl (Figure 4G). In addition, after 12 h of stress, the maximum OsHKT1 upregulation was under NaCl and MgCl2 stress in the roots. Similarly, in the shoots of Pokkali after 6 h of stress, NaCl, CaCl2, and MgCl2 upregulated OsHKT1 and OsSOS1, whereas NaCl and CaCl2 100% upregulated OsNHX1 and OsSOS1 after 12 h of stress (Figure 4H). In contrast, after 6 and 12 h of stress, MgCl2 upregulated OsNHX1 in the shoots of Pokkali (Figure 4H).
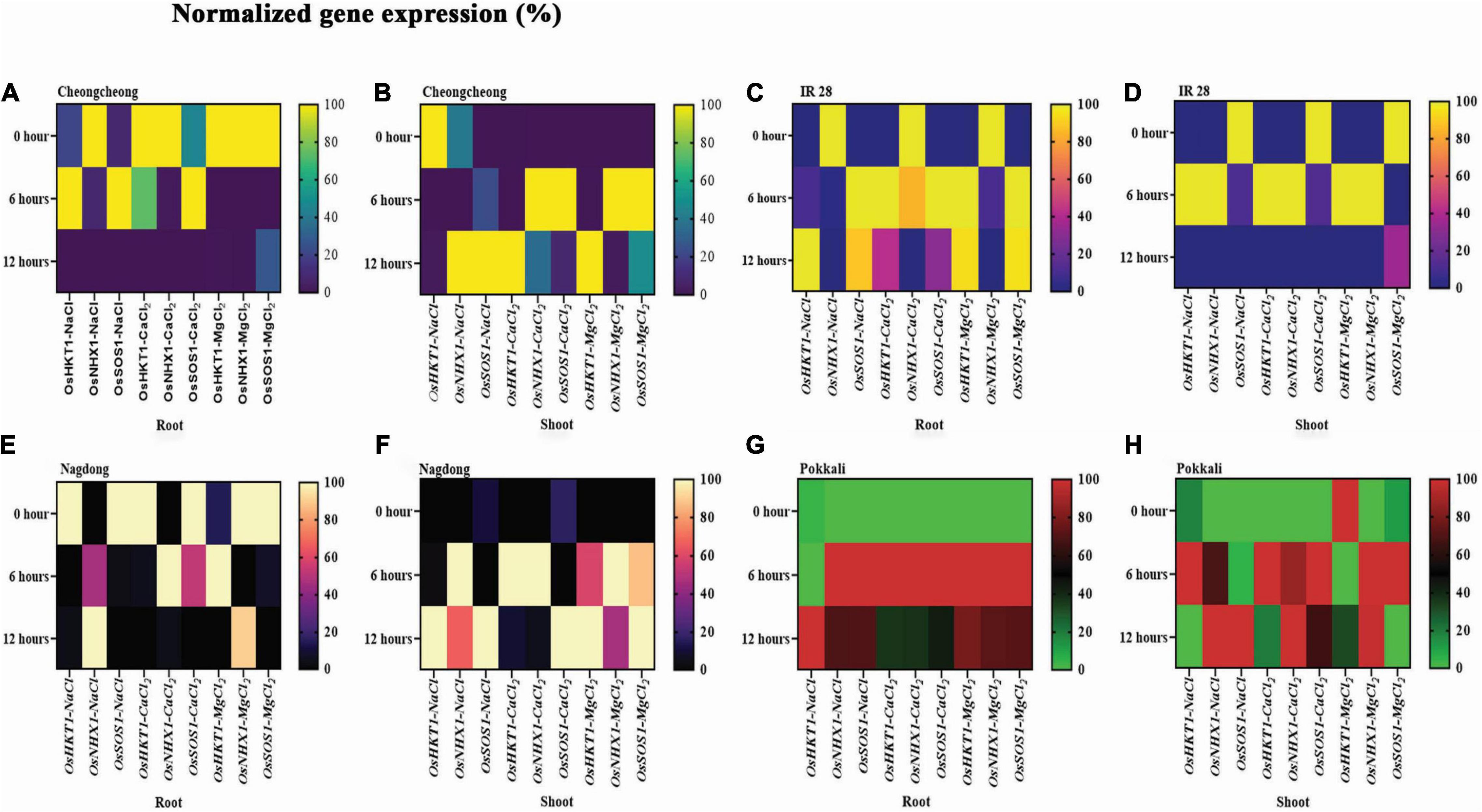
Figure 4. Under 150 mM of different salt stress, quantitative real-time polymerase chain reaction expression analysis of OsHKT1, OsNHX1, and OsSOS1 relative to salinity in the roots and shoots of rice genotypes. The expression pattern in the heatmap (A–H) was determined in normalized gene expression (%) after 0, 6, and 12 h of salt stress.
Glutathione and Peroxidase Activity
After 24 h of different sat stress, GSH content was increased significantly in the leaves of IR 28 with 150 mM NaCl and MgCl2, and the highest GSH content was observed at MgCl2 salt compared to their respective control. Similarly, at 150 mM NaCl stress a higher GSH content was observed in the leaves of Nagdong. However, both CaCl2 and MgCl2 salts significantly decrease the GSH content in cultivar Pokkali whereas, in cultivar Cheongcheong CaCl2 significantly decreases the GSH content compared to the control group (Figure 5A). The second enzyme responsible for the deactivation of H2O2 is a peroxidase. POD activity increased significantly in both cultivar Cheongcheong and IR 28 at 150 mM MgCl2 and CaCl2 compared to Nagdong and Pokkali. However, under NaCl and CaCl2 salts the POD activity was found significantly higher in cultivar Cheongcheong compared to their respective control. In contrast, POD activity in IR 28 decreased significantly by NaCl and MgCl2 salts compared to the control. In the case of Cultivar Nagdong and Pokkali POD activity increased significantly under NaCl and MgCl2 salts however, MgCl2 salt decreased POD activity in cultivar Nagdong compared to the control group (Figure 5B).
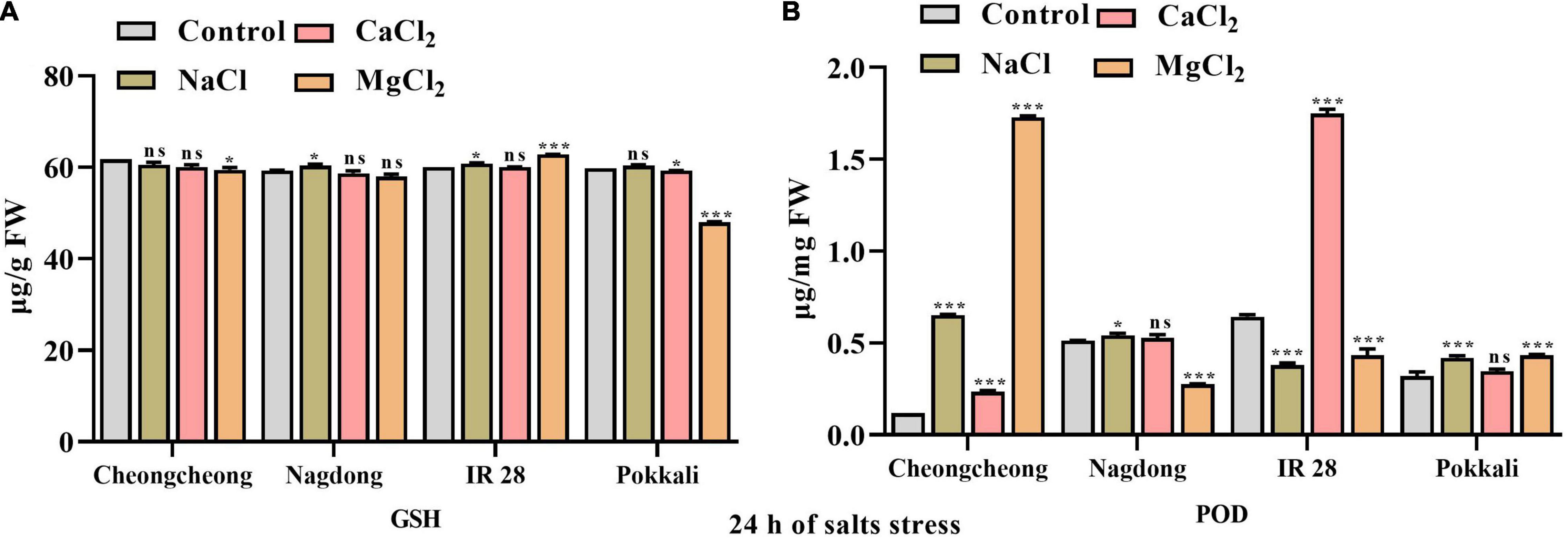
Figure 5. Effect of 150 mM of different salts on GSH and POD contents in the leaves of different rice genotypes. The GSH (A) and POD (B) activity was determined after 24 h of salt stress. Statistical analysis was calculated by the Bonferroni posttest one-way repeated-measures analysis of variance. Asterisks in the vertical bar indicate a significant difference at *p < 0.05, ***p < 0.00, compared to control.
Discussion
The response and adaptation of rice to salt stress is a very complex mechanism. Salt stress causes leaf rolling, root growth inhibition, and reduced plant height, tiller number, and spikelet sterility, leading to yield reduction (Ali et al., 2014; Razzaq et al., 2020). A previous study reported that salinity stress up to 20 ds m–1 dynamically inhibited rice seed germination, root and shoot length growth reduction, dry matter, and ultimately yield reduction (Hakim et al., 2010). However, another study demonstrated that rice seeds have a salt-tolerant ability up to some extent at germination and, in some cases, are not seriously affected by salts up to 16.3 ds m–1 (Khan et al., 1997). A previous study suggested that NaCl and its mixtures (MgSO4 + NaCl and Na2SO4 + CaCl2) significantly inhibited seed germination and root length in sugar beet cultivars with EC up to 8 and 4 ds/m, respectively (Jafarzadeh and Aliasgharzad, 2007). This study showed a higher reduction in seed germination and shoot and root length of rice cultivars by NaCl, along with CaCl2 and MgCl2, similar to findings for NaCl and Na2SO4 (Stewart, 1898). Bonilla et al. (2004) demonstrated high Ca content in moderate salinity levels up to 75 mM NaCl increased the proportion of germinated pea seeds and recovered the delay in germination. This study found a high germination percentage on Cheongcheong and IR 28; thus, this might be the role of Ca in CaCl2 (Figure 1E). A previous study reported that the germination percentage of sugar beet and cotton is significantly affected by NaCl, CaCl2, and Na2SO4, with a high extent or equal to 200 mM (Shonjani, 2002). In this study, under different salt stress, Cheongcheong and IR 28 significantly inhibited seed germination to ≥200 mm, contrary to Nagdong and Pokkali (Figures 1D–F). In current work under different salts stress cultivar Nagdong show significantly higher germination percentage and good seedlings growth compared to other cultivars. However, salt-sensitive cultivar IR 28, and Cheongcheong show high seeds germination under CaCl2 salt compared to the other two salts NaCl, and MgCl2. Thus, based on current work we believe that cultivar Nagdong has more salt-tolerant ability, and it should be used as a salt-tolerant cultivar for future studies.
Salinity stress reduced rice plant height, roots, and shoot dry matter at various growth phases (Puvanitha and Mahendran, 2017). NaCl treatment most prominently decreased the shoot growth in rice, sugar beet, and cotton compared to root growth (Shonjani, 2002). Another study demonstrated that salinity stress drastically reduced plant height, root and shoot dry weight, and yield of rice cultivars (Puvanitha and Mahendran, 2017). Many studies showed that the fresh and dry weights of the shoot and root system are affected negatively by changes in salinity concentration, depends on type of plant species, or type of salt present (Taffouo et al., 2010). In this study, different salt stress significantly affected rice plant height, root, and shoot dry matter compared to control. It might be due to the osmotic effect because in saline soil roots exclude all Na+ and Cl– ions while taking up water; the high concentration of these ions may lead to the reduction or death of the plant (Figures 2A–D). These findings agreed with Jamil et al. (2007). Growth reduction may be due to the toxic effects of NaCl or unbalanced nutrient uptake by the seedlings (Datta et al., 2009).
During salinity stress, a high concentration of Na+ and a higher Na+/K+ ratio disrupt ion homeostasis and decrease other nutrients (Simaei et al., 2012; Bose et al., 2014; Rahman et al., 2016b). Salt stress more seriously affects the ion balance of Na+, K+, and Cl– in old rice leaves compared to young leaves (Wang et al., 2012). In this study, the high accumulation of Na+/K+ ions among rice cultivars altered other macronutrients and micronutrients. Some studies reported that, as salinity level increases, Mn content increased in sunflower shoots compared to roots (Achakzai et al., 2010). The availability of most nutrients depends on the pH of the soil as well the nature of binding sites on organic and inorganic particle surfaces. In saline soils, the solubility of micronutrients such as Fe2+, Mn2+, Zn2+, and Cl– is particularly low, and plants grown in these soils often experience a deficiency in these elements (Pessarakli, 2019). Although the micronutrient concentration in plant shoots may increase, decrease, or have no effect depending on salinity, the salt tolerance of plant species, type of plant tissue, micronutrient concentration, environmental conditions, or sudden changes in the permeability of the plant cell membranes. Previous studies reported that salt-stress conditions decreased Mn content due to disrupted ion imbalance (Tunçturk et al., 2008; Wu and Wang, 2012). However our results indicated a high Mn content in the shoots of Nagdong and Pokkali, whereas varietal response was inconsistent; therefore, under different salt stress, cultivar Cheongcheong and IR 28 showed low Mn content in roots and shoots (Tables 2, 3). Thus it might be possible that the salt-tolerant ability of both cultivars Nagdong and Pokkali links with high Mn content instead of cultivar Cheongcheong and IR 28. Another study reported that exogenous application of Mn induced salt stress tolerance in rice seedlings (Rahman et al., 2016a). In present study under different salt stress, both cultivar Nagdong and Pokkali show high Mn content that might be induced salt stress tolerance in these cultivars compared to cultivars Cheongcheong and IR 28. A previous study reported that a high Na+ concentration has a contrasting effect on K+ ion, an essential plant nutrient for plant growth and development (Jung et al., 2009).
High Na+ in plant cells seriously damages membrane systems and organelles, resulting in abnormal plant growth and development and plant death (Quintero et al., 2007; Siringam et al., 2011). This study revealed the inconsistent uptake of Na+/K+ in the roots among rice cultivars under different salt stress but consistent increase and decrease between Na+/K+ ions in the shoots; thus, this unbalanced regulation of ions might be the cause of lower plant growth and fresh and dry matter. These findings agreed with Eker et al. (2006). Another study also reported the high uptake of Na+ over K+ ions in plant tissue of Butea monosperma (Hirpara et al., 2005). A previous study suggested that in a higher Na+/K+ ratio the K+ efflux activates non-selective cation channels (NSCCs) and guard cell outward rectifying potassium (GORK) channels, and K+ leakage also leads to a higher reactive oxygen species production that might decrease Mg2+, Mn2+, and Zn2+ contents (Demidchik et al., 2002). In this study, among different rice cultivars, NaCl and CaCl2 stress decreased Mg and Mn contents in the roots, whereas Zn and these elements differentially increased or decreased in the shoots of different genotypes (Tables 2, 3).
The phenomenon of increasing Ca2+ ions under mixed salt was also observed in New Zealand spinach (Tetragonia tetragonioides, Pall) and red orach (Atriplex hortensis L.) (Wilson et al., 2000). Another study demonstrate that an initial Ca2+ signal after salt stress was formed close to the root apex and then appeared to disperse to basal parts of this organ (Moore et al., 2002). In this study, CaCl2 stress mainly increased the Ca2+ level in the roots of different rice cultivars and the shoots of Pokkali. Several studies observed that the salt-induced Ca2+ rise originated within the roots (Kiegle et al., 2000; Tracy et al., 2008). However, under different salt stress, a high accumulation of Ca2+ ions was found in the shoots of Cheongcheong, Nagdong, and IR 28 compared to control (Table 3). Thus, these findings agreed with the above-mentioned statements that Ca2+ ions might be alleviate the toxic effects of Na+ ions because the Ca2+ ion concentration significantly decreased under NaCl and MgCl2 stress in Pokkali. A high salt level in rice decrease the Na-Ca selectivity (Zeng et al., 2003). Thus Na-Ca selectivity might be one salt tolerance component and effective selection criterion in screening for salt tolerance (Tables 2, 3). However, the exact phenomenon that Ca2+ ion alleviate the toxic effect of Na+ unknown, but recent findings put forward that the function of this second messenger in salt stress responses may be even more adaptable than so far appreciated. Indeed, accumulating evidence points to the involvement of a diverse array of Ca2+ sensor proteins in the various aspects of salinity tolerance. For example, CML9 [a member of the calmodulinlike (CML) gene family] was found to be up-regulated during salt stress, and cml9 loss-of-function mutants displayed hypersensitivity in germination assays on medium containing either NaCl or ABA, whereas adult cml9 plants show enhanced tolerance toward irrigation with salt water (Magnan et al., 2008).Thus, salinity stress either increases or decreases the uptake of micronutrients. High NaCl concentration generally increases Cu, Fe, and Zn contents (Alam, 1999; Víllora et al., 2000). Differences can be evaluated to plant species, plant tissue, environmental conditions, and salinity level. Another study reported that high salt stress decreases Ca2+, K+, Mg2+, P, Mn, and N contents (Talei et al., 2012). In saline soil micronutrients availability to plants is relatively low, and plants grown in such soil show nutrient deficiency in the plant body (Page, 1996). In this study, NaCl stress decreases the concentrations of all micronutrients (Fe2+, K+ Mn2+, Mg2+, P, Zn) in the roots and shoots of different rice cultivars. However, in the roots and shoots of different genotypes, these elements are differentially regulated by CaCl2 and MgCl2 (Tables 2, 3).
This study focused on OsHKT1, SOS1, and NHX1, three very important genes, and studied their expression pattern in the roots and shoots of rice cultivars with different time points. These genes are currently the most extensively studied mechanisms in controlling the salt stress response in plants. Thellungiella salsuginea comprised at least two HKT genes. TsHKT1;1 is expressed at very low levels, whereas TsHKT1;2 is transcriptionally mainly upregulated by salt stress (Ali et al., 2012). Another study demonstrated that, under NaCl stress, the OsHKT1 transcript was significantly downregulated in salt-tolerant cultivar. Pokkali but upregulated in salt-sensitive cv. BRRI Dhan29 shoot after 6 h intervals (Kader et al., 2006). However, in this study, NaCl stress after 6 and 12 h upregulated OsHKT1 in the roots and shoots of Pokkali and IR 28 (Figures 4C,D,G,H). Under NaCl stress, OsHKT1 was upregulated conversely in the roots of Cheongcheong and shoots of Nagdong at 6 and 12 h intervals (Figures 4A,F). In the same way, NaCl stress downregulated OsHKT1 in the roots of Nagdong and shoots of Cheongcheong (Figures 4B,E). We believe that under salt stress, OsHKT1 up-regulation might be closely link with Na+ concentration because in root the of Cheongcheong we observed high level of Na+ ion than that of cultivar Nagdong root. Conversely, in the shoot of Nagdong have a high Na+ ion compared to cheongcheong. Thus, it might possible that high accumulation of Na+ leads a high induction of OsHKT1. A previous study reported that, under high NaCl stress, high Na+ leads to K+ deficiency that might cause the induction of OsHKT1 (Maathuis and Amtmann, 1999). In the current study under NaCl stress, the Na+ concentration is relatively higher than K+ in the roots of different rice genotypes. However, K+ ions mainly higher in the shoot of different rice genotypes accept salt-sensitive cultivar IR 28 (Tables 2, 3). Thus, the high Na+ level in the root or shoot and high accumulation of K+ in the shoot might be the reason for OsHKT1 up-regulation. In this study, NaCl stress, along with other salts, such as CaCl2 and MgCl2, regulate OsHKT1 differentially among rice cultivars. After 6 and 12 h of stress, OsHKT1 was mostly upregulated by CaCl2 and MgCl2 in the roots and shoots of IR 28, Pokkali, Nagdong, and Cheongcheong; however, after 6 h of stress, CaCl2 upregulated OsHKT1 in the shoots of Nagdong, IR 28, and Pokkali. In the present study both CaCl2 and MgCl2 salt increase, K+ content compared to Na+ in the root and shoot of different rice genotypes. Thus, it might be the reason for OsHKT1 upregulation in the root and shoot region of rice genotypes. The previous study showed that, with an extended stress period, the salt-tolerant cultivar Pokkali starts to downregulate OsHKT1 expression in roots and shoots of rice (Kader et al., 2006). Our study demonstrated that NaCl stress downregulated OsHKT1 in the roots and shoots of cv. Pokkali after 6 and 12 h but in the roots of Pokkali after 12 h of NaCl stress it induced the expression of OsHKT1. Cultivar Pokkali is a salt-tolerant ability thus under salt stress initially it maintain internal Na+ ion in the root system. Therefore, it might show OsHKT1 downregulation in the root and shoot of Pokkali. On the other, hand prolong NaCl stress, increase the internal Na+ in the root of Pokkali that might cause a high induction of OsHKT1.
A previous study reported high expression of SOS1 and NHX under NaCl stress in the leaves and roots of Kochia scoparia after 48 h of stress (Fahmideh and Fooladvand, 2018). This study indicated that the expression pattern of OsNHX1 and OsSOS1 under different salt stress is different among rice genotypes. However, after 6 and 12 h of different salt stress, OsNHX1 and OsSOS1 were upregulated in the roots of cv. Pokkali and shoots of cv. Cheongcheong (Figures 4B,G). Thus, these findings agreed with Fahmideh and Fooladvand (2018). In the current study, under different salt stress, we observed that Na+ and K+ ions regulated inversely in the root and shoot of rice genotypes. The higher the Na+ level lower will be K+, and vice versa. Thus, in the root of Pokkali and shoot of Cheongcheong K+ ion higher over Na+ ion. Therefore, it might be the possible reason for OsNHX1 and OsSOS1 up-regulation. A previous study suggested that overexpression of AtNHX1 enhanced the salt tolerance ability of A. thaliana (Apse et al., 1999). In this study, there was a high expression of OsNHX1 in the roots and shoots of cv. Nagdong and Pokkali under NaCl stress (Figures 4E,H). Thus, the salt tolerance ability of Nogdong and Pokkali might be due to OsNHX1 upregulation. Another study reported that the Na+/H+ antiporter NHX is an important membrane protein that helps pump Na+ into the vacuoles to minimize Na+ toxicity and alleviate the serious effects of salt stress (Wu et al., 2004). In the SOS pathway, SOS1, in particular, leads to high sensitivity to NaCl in glycophytes and halophytes (Liu et al., 2000; Oh et al., 2009). Under salt stress, high gene expression of SOS1 was noted in the SOS3 or SOS2 mutant plant (Shi et al., 2000). In this study, under different salt stress, there was a high expression of OsSOS1 in the roots of IR 28, Pokkali, and Cheongcheong (Figures 4A,C,G). Similarly, after 6 and 12 h of salt stress, OsSOS1 upregulation was observed in the shoots of Cheongcheong, Nagdong, and Pokkali (Figures 4B,F,H). The ion homeostasis of the Na+/K+ ratio is a key factor see (Table 3) in the shoot of above mention cultivars that have high K+, compared to Na+. Thus, the upregulation of OsSOS1 might be linked to high K+ content. In the present study under different salt stress, cultivar Nagdong root shows high Na+ ion over K+, Thus, decreased K+ level in the root might be Consequences of down-regulate OsSOS1 gene. Conversely, high K+ ion the shoot of Nagdong show high induction of OsSOS1 gene.
Glutathione is a strong antioxidant; hence, it has a very important role in antioxidant defense. Under adverse environmental conditions, such as drought, salinity, etc., excessive amounts of ROS are formed, which leads to oxidative stress. Further, GHS helps to maintain the reduced state of constituents of the AsA-GSH pathway and thus detoxifies ROS. The DHAR enzyme uses GSH to convert oxidized ascorbic acid (DHA) to AsA (Foyer and Noctor, 2011). Another study suggested the function of GSH as an antioxidant that confers abiotic stress tolerance is well-established, but the mechanism underlying this defense response is unknown. Cross-communication of GSH with other signaling molecules helps in adaptation to abiotic stress (Ghanta and Chattopadhyay, 2011). Plants under stress conditions, strongly control the production, and elimination of ROSs by many enzymatic and non-enzymatic processes to the alleviate their damages (Jalali et al., 2020). In the experiments presented here, after 24 h of 150 mM NaCl, and MgCl2 salt we observed the high GSH content in the leaves of Nagdong and salt-sensitive cultivar IR 28. In the present work at 150 mM NaCl stress, cultivar Pokkali, and Cheongcheong did not affect GSH content (Figure 5A). Under NaCl stress, GSH contents in the chloroplasts of the cultivars Pokkali and Peta decrease significantly (Wang et al., 2014). However, a significant reduction was observed here under CaCl2 and MgCl2 in cultivars Pokkali, and Cheongcheong seedlings (Figure 5A). Therefore, it might be possible that a high GSH content in cultivars Nagdong under NaCl stress, and a high GSH content in IR 28 under MgCl2 stress help to maintain plant vigor under salinity stress (Figures 2A–C). Another study reported that POD activity during the seedling stage increased greatly in IR 28 and AT 353 grown at 4 and 6 dS/m compared to Pokkali (Safeena and Bandara, 2006). A previous study suggested that the POD activity of plants was shown to be up-regulated to reduce oxidative stress, cell membrane damage, and altered Ca2 + concentrations caused by ROS generated by salinity treatment (Sergio et al., 2012). In our study, CaCl2, and MgCl2 salt show the highest increase in POD activity in cultivars IR 28 and Cheongcheong. This increase is 2-fold higher than NaCl, as well as from their respective control. Therefore, it might be the possible reason that high POD activity under CaCl2 and MgCl2 stress reduces oxidative stress and ROS accumulation in cultivar Cheongcheong and IR 28 compared to NaCl treatment. Here we suggest that a high POD value might be involved to increase shoot growth, fresh and dry weight in cultivar Cheongcheong and IR 28 (Figures 2A–C, 3A,B).
Conclusion
The high worldwide population and industrialization lead to climate change that causes soil salinization and ultimately affects crop production, especially rice. This study tested four rice genotypes with three different salts (NaCl, CaCl2, and MgCl2). However, each cultivar responded differently to different salts, but NaCl stress most drastically reduced the agronomic traits among all rice cultivars. Cheongcheong and IR 28 had a mostly similar response to different salts. Nagdong and Pokkali had a similar response to different salts, but Pokkali showed more resistance to NaCl stress in the soil, whereas Nagdong showed more resistance to all salt. OsHKT1, OsNHX1, and OsSOS1 were upregulated in Nagdong and Pokkali. Under different sat stress a high induction of GSH was observed in cultivar Nagdong, and IR 28, whereas, higher POD activity was recorded in cultivar Cheoncheong and IR 28. Therefore, there is a need to develop a new overexpresser of Cheongcheong and a knockout of Nagdong in the near future using the CRISPR/Cas9 technique for plant molecular breeding laboratories in Korea.
Data Availability Statement
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
MF planned, designed, and performed the research study, analyzed the data, and wrote the findings. SA helped with data collection. Y-HJ and J-RP contributed to the ICP and statistical analysis. D-DZ and E-GK contributed to the experimental resources. K-MK edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2021M3E5E6022715).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Achakzai, A. K. K., Kayani, S. A., and Hanif, A. (2010). Effect of salinity on uptake of micronutrients in sunflower at early vegetative stage. Pak. J. Bot 42, 129–139.
Alam, S. M. (1999). “Nutrient uptake by plants under stress conditions,” in Handbook of Plant and Crop Stress, ed. P. Mohammad (New York: Marcel Dekker Press Inc.), 285–313. doi: 10.1201/9780824746728.ch12
Ali, M. N., Yeasmin, L., Gantait, S., Goswami, R., and Chakraborty, S. (2014). Screening of rice landraces for salinity tolerance at seedling stage through morphological and molecular markers. Physiol. Mol. Biol. Plants 20, 411–423. doi: 10.1007/s12298-014-0250-6
Ali, Q., Daud, M., Haider, M. Z., Ali, S., Rizwan, M., Aslam, N., et al. (2017). Seed priming by sodium nitroprusside improves salt tolerance in wheat (Triticum aestivum L.) by enhancing physiological and biochemical parameters. Plant Physiol. Biochem. 119, 50–58. doi: 10.1016/j.plaphy.2017.08.010
Ali, Z., Park, H. C., Ali, A., Oh, D.-H., Aman, R., Kropornicka, A., et al. (2012). TsHKT1; 2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiol. 158, 1463–1474. doi: 10.1104/pp.111.193110
Al-Harbi, A. R. (1995). Growth and nutrient composition of tomato and cucumber seedlings as affected by sodium chloride salinity and supplemental calcium. J. Plant Nutr. 18, 1403–1416. doi: 10.1080/01904169509364990
Apse, M. P., Aharon, G. S., Snedden, W. A., and Blumwald, E. (1999). Salt tolerance conferred by over expression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258. doi: 10.1126/science.285.5431.1256
Blumwald, E. (2000). Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 12, 431–434. doi: 10.1016/S0955-0674(00)00112-5
Bonilla, I., El-Hamdaoui, A., and Bolaños, L. (2004). Boron and calcium increase Pisum sativum seed germination and seedling development under salt stress. Plant Soil 267, 97–107. doi: 10.1007/s11104-005-4689-7
Bose, J., Rodrigo-Moreno, A., and Shabala, S. (2014). ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 65, 1241–1257. doi: 10.1093/jxb/ert430
Chaparzadeh, N., D’Amico, M. L., Khavari-Nejad, R.-A., Izzo, R., and Navari-Izzo, F. (2004). Antioxidative responses of Calendula officinalis under salinity conditions. Plant Physiol. Biochem. 42, 695–701. doi: 10.1016/j.plaphy.2004.07.001
Chawla, S., Jain, S., and Jain, V. (2013). Salinity induced oxidative stress and antioxidant system in salt-tolerant and salt-sensitive cultivars of rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 22, 27–34. doi: 10.1007/s13562-012-0107-4
Chinnusamy, V., Jagendorf, A., and Zhu, J. K. (2005). Understanding and improving salt tolerance in plants. Crop Sci. 45, 437–448. doi: 10.2135/cropsci2005.0437
Chourey, K., Ramani, S., and Apte, S. K. (2003). Accumulation of LEA proteins in salt (NaCl) stressed young seedlings of rice (Oryza sativaL.) cultivar Bura Rata and their degradation during recovery from salinity stress. J. Plant Physiol. 160, 1165–1174. doi: 10.1078/0176-1617-00909
Daliakopoulos, I., Tsanis, I., Koutroulis, A., Kourgialas, N., Varouchakis, A., Karatzas, G., et al. (2016). The threat of soil salinity: A European scale review. Sci. Total Environ. 573, 727–739. doi: 10.1016/j.scitotenv.2016.08.177
Datta, J., Nag, S., Banerjee, A., and Mondai, N. (2009). Impact of salt stress on five varieties of wheat (Triticum aestivum L.) cultivars under laboratory condition. J. Appl. Sci. Environ. Manag. 13, 93–97. doi: 10.4314/jasem.v13i3.55372
Demidchik, V., Davenport, R. J., and Tester, M. (2002). Nonselective cation channels in plants. Annu. Rev. Plant Biol. 53, 67–107. doi: 10.1146/annurev.arplant.53.091901.161540
Eker, S., Cömertpay, G., Konuşkan, Ö, Ülger, A. C., Öztürk, L., and Çakmak, I. (2006). Effect of salinity stress on dry matter production and ion accumulation in hybrid maize varieties. Turk. J. Agric. For. 30, 365–373.
Fahmideh, L., and Fooladvand, Z. (2018). Isolation and semi quantitative PCR of Na+/H+ antiporter (SOS1 and NHX) genes under salinity stress in Kochia scoparia. Biol. Proced. Online 20, 1–9. doi: 10.1186/s12575-018-0076-7
Foyer, C. H., and Noctor, G. (2011). Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 155, 2–18. doi: 10.1104/pp.110.167569
Fukuda, A., Nakamura, A., Hara, N., Toki, S., and Tanaka, Y. (2011). Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 233, 175–188. doi: 10.1007/s00425-010-1289-4
Gao, S., Ouyang, C., Wang, S., Xu, Y., Tang, L., and Chen, F. (2008). Effects of salt stress on growth, antioxidant enzyme and phenylalanine ammonia-lyase activities in Jatropha curcas L. seedlings. Plant Soil Environ. 54, 374–381. doi: 10.17221/410-PSE
García, M. J., Lucena, C., Romera, F. J., Alcántara, E., and Pérez-Vicente, R. (2010). Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J. Exp. Bot. 61, 3885–3899. doi: 10.1093/jxb/erq203
Ghanta, S., and Chattopadhyay, S. (2011). Glutathione as a signaling molecule-another challenge to pathogens: another challenge to pathogens. Plant Signal. Behav. 6, 783–788. doi: 10.4161/psb.6.6.15147
Hakim, M., Juraimi, A., Begum, M., Hanafi, M., Ismail, M. R., and Selamat, A. (2010). Effect of salt stress on germination and early seedling growth of rice (Oryza sativa L.). Afr. J. Biotechnol. 9, 1911–1918. doi: 10.5897/AJB09.1526
Hasanuzzaman, M., Nahar, K., Anee, T. I., and Fujita, M. (2017). Glutathione in plants: biosynthesis and physiological role in environmental stress tolerance. Physiol. Mol. Biol. Plants 23, 249–268. doi: 10.1007/s12298-017-0422-2
Hernandez, J. A., Jiménez, A., Mullineaux, P., and Sevilia, F. (2000). Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ. 23, 853–862. doi: 10.1046/j.1365-3040.2000.00602.x
Hilal, M., Zenoff, A. M., Ponessa, G., Moreno, H., and Massa, E. M. (1998). Saline stress alters the temporal patterns of xylem differentiation and alternative oxidase expression in developing soybean roots. Plant Physiol. 117, 695–701. doi: 10.1104/pp.117.2.695
Hirpara, K. D., Ramoliya, P. J., Patel, A. D., and Pandey, A. N. (2005). Effect of salinisation of soil on growth and macro-and micro-nutrient accumulation in seedlings of Butea monosperma (Fabaceae). Anal. Biol. 27, 3–14. doi: 10.1111/j.1744-7348.2004.tb00347.x
Jafarzadeh, A. A., and Aliasgharzad, N. (2007). Salinity and salt composition effects on seed germination and root length of four sugar beet cultivars. Biologia 62, 562–564. doi: 10.2478/s11756-007-0111-7
Jalali, P., Navabpour, S., Yamchi, A., Soltanloo, H., and Bagherikia, S. (2020). Differential responses of antioxidant system and expression profile of some genes of two rice genotypes in response to salinity stress. Biologia 75, 785–793. doi: 10.2478/s11756-019-00393-x
Jamil, M., Rehman, S., and Rha, E. (2007). Salinity effect on plant growth, PSII photochemistry and chlorophyll content in sugar beet (Beta vulgaris L.) and cabbage (Brassica oleracea capitata L.). Pak. J. Bot. 39, 753–760.
Jung, J.-Y., Shin, R., and Schachtman, D. P. (2009). Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21, 607–621. doi: 10.1105/tpc.108.063099
Kader, M. A., Seidel, T., Golldack, D., and Lindberg, S. (2006). Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa L.) cultivars. J. Exp. Bot. 57, 4257–4268. doi: 10.1093/jxb/erl199
Karuppanapandian, T., Moon, J.-C., Kim, C., Manoharan, K., and Kim, W. (2011). Reactive oxygen species in plants: their generation, signal transduction, and scavenging mechanisms. Aust. J. Crop Sci. 5, 709–725.
Karyotis, T., Iliadis, C., Noulas, C., and Mitsibonas, T. (2003). Preliminary research on seed production and nutrient content for certain quinoa varieties in a saline–sodic soil. J. Agron. Crop Sci. 189, 402–408. doi: 10.1046/j.0931-2250.2003.00063.x
Khan, M., Hamid, A., and Karim, M. (1997). Effect of sodium chloride on germination and seedling characters of different types of rice (Oryza sativa L.). J. Agron. Crop Sci. 179, 163–169. doi: 10.1111/j.1439-037X.1997.tb00512.x
Khan, M. A., Ullah, I., Waqas, M., Hamayun, M., Khan, A. L., Asaf, S., et al. (2019). Halo-tolerant rhizospheric Arthrobacter woluwensis AK1 mitigates salt stress and induces physio-hormonal changes and expression of GmST1 and GmLAX3 in soybean. Symbiosis 77, 9–21. doi: 10.1007/s13199-018-0562-3
Kiegle, E., Moore, C. A., Haseloff, J., Tester, M. A., and Knight, M. R. (2000). Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 23, 267–278. doi: 10.1046/j.1365-313x.2000.00786.x
Knight, S., Rogers, R., Smith, M., and Sporaer, L. (1992). Effects of NaCl salinity on miniature dwarf tomato ‘Micro-Tom’: I. Growth analyses and nutrient composition. J. Plant Nutr. 15, 2315–2327. doi: 10.1080/01904169209364476
Liu, J., Ishitani, M., Halfter, U., Kim, C.-S., and Zhu, J.-K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 97, 3730–3734. doi: 10.1073/pnas.97.7.3730
Lutts, S., Kinet, J., and Bouharmont, J. (1995). Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J. Exp. Bot. 46, 1843–1852. doi: 10.1093/jxb/46.12.1843
Maathuis, F. J., and Amtmann, A. (1999). K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann. Bot. 84, 123–133. doi: 10.1006/anbo.1999.0912
Magnan, F., Ranty, B., Charpenteau, M., Sotta, B., Galaud, J. P., and Aldon, D. (2008). Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 56, 575–589. doi: 10.1111/j.1365-313X.2008.03622.x
Mittova, V., Guy, M., Tal, M., and Volokita, M. (2004). Salinity up-regulates the antioxidative system in root mitochondria and peroxisomes of the wild salt-tolerant tomato species Lycopersicon pennellii. J. Exp. Bot. 55, 1105–1113. doi: 10.1093/jxb/erh113
Moore, C. A., Bowen, H. C., Scrase-Field, S., Knight, M. R., and White, P. J. (2002). The deposition of suberin lamellae determines the magnitude of cytosolic Ca2+ elevations in root endodermal cells subjected to cooling. Plant J. 30, 457–465. doi: 10.1046/j.1365-313X.2002.01306.x
Muscolo, A., Panuccio, M. R., and Sidari, M. (2003). Effects of salinity on growth, carbohydrate metabolism and nutritive properties of kikuyu grass (Pennisetum clandestinum Hochst). Plant Sci. 164, 1103–1110. doi: 10.1016/S0168-9452(03)00119-5
Nahar, K., Hasanuzzaman, M., Alam, M., and Fujita, M. (2015a). Glutathione-induced drought stress tolerance in mung bean: coordinated roles of the antioxidant defence and methylglyoxal detoxification systems. AoB Plants 7:lv069. doi: 10.1093/aobpla/plv069
Nahar, K., Hasanuzzaman, M., Alam, M., and Fujita, M. (2015b). Roles of exogenous glutathione in antioxidant defense system and methylglyoxal detoxification during salt stress in mung bean. Biol. Plant. 59, 745–756. doi: 10.1007/s10535-015-0542-x
Nahar, K., Hasanuzzaman, M., Alam, M. M., and Fujita, M. (2015c). Exogenous glutathione confers high temperature stress tolerance in mung bean (Vigna radiata L.) by modulating antioxidant defense and methylglyoxal detoxification system. Environ. Exp. Bot. 112, 44–54. doi: 10.1016/j.envexpbot.2014.12.001
Nass, R., and Rao, R. (1998). Novel localization of a Na+/H+ exchanger in a late endosomal compartment of yeast implications for vacuole biogenesis. J. Biol. Chem. 273, 21054–21060. doi: 10.1074/jbc.273.33.21054
Niazi, B., and Ahmed, T. (1984). Effect of sodium chloride and zinc on the growth of tomato. II. Uptake of ions. Geobios 11, 155–160.
Oh, D.-H., Leidi, E., Zhang, Q., Hwang, S.-M., Li, Y., Quintero, F. J., et al. (2009). Loss of halophytism by interference with SOS1 expression. Plant Physiol. 151, 210–222. doi: 10.1104/pp.109.137802
Ologundudu, A. F., Adelusi, A. A., and Akinwale, R. O. (2014). Effect of Salt Stress on Germination and Growth Parameters of Rice (Oryza sativa L.). Notulae Sci. Biol. 6, 237–243. doi: 10.15835/nsb629163
Orlowski, J., and Grinstein, S. (1997). Na+/H+ exchangers of mammalian cells. J. Biol. Chem. 272, 22373–22376. doi: 10.1074/jbc.272.36.22373
Page, A. (1996). “Deficiencies and toxicities of trace elements,” in Agricultural Salinity Assessment and Management (ASCE Manuals and Reports on Engineering Practice), eds W. W. Wesley and K. T. Kenneth (Reston: American Society of Civil Engineers), 138–160.
Passardi, F., Cosio, C., Penel, C., and Dunand, C. (2005). Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 24, 255–265. doi: 10.1007/s00299-005-0972-6
Pessarakli, M. (2019). Handbook of Plant and Crop Stress. Boca Raton: CRC press. doi: 10.1201/9781351104609
Puvanitha, S., and Mahendran, S. (2017). Effect of salinity on plant height, shoot and root dry weight of selected rice cultivars. Sch. J. Agric. Vet. Sci. 4, 126–131.
Qiu, Q.-S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J.-K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Quintero, J. M., Fournier, J. M., and Benlloch, M. (2007). Na+ accumulation in shoot is related to water transport in K+-starved sunflower plants but not in plants with a normal K+ status. J. Plant Physiol. 164, 60–67. doi: 10.1016/j.jplph.2005.10.010
Rahman, A., Nahar, K., Hasanuzzaman, M., and Fujita, M. (2016b). Calcium supplementation improves Na+/K+ ratio, antioxidant defense and glyoxalase systems in salt-stressed rice seedlings. Front. Plant Sci. 7:609. doi: 10.3389/fpls.2016.00609
Rahman, A., Hossain, M., Mahmud, J.-A., Nahar, K., Hasanuzzaman, M., and Fujita, M. (2016a). Manganese-induced salt stress tolerance in rice seedlings: regulation of ion homeostasis, antioxidant defense and glyoxalase systems. Physiol. Mol. Biol. Plants 22, 291–306. doi: 10.1007/s12298-016-0371-1
Rahman, S., Vance, G., and Munn, L. (1993). Salinity induced effects on the nutrient status of soil, corn leaves and kernels. Commun. Soil Sci. Plant Anal. 24, 2251–2269. doi: 10.1080/00103629309368953
Razzaq, A., Ali, A., Safdar, L. B., Zafar, M. M., Rui, Y., Shakeel, A., et al. (2020). Salt stress induces physiochemical alterations in rice grain composition and quality. J. Food Sci. 85, 14–20. doi: 10.1111/1750-3841.14983
Rengasamy, P. (2010). Soil processes affecting crop production in salt-affected soils. Funct. Plant Biol. 37, 613–620. doi: 10.1071/FP09249
Robin, A. H. K., Matthew, C., Uddin, M. J., and Bayazid, K. N. (2016). Salinity-induced reduction in root surface area and changes in major root and shoot traits at the phytomer level in wheat. J. Exp. Bot. 67, 3719–3729. doi: 10.1093/jxb/erw064
Rubio, F., Gassmann, W., and Schroeder, J. I. (1995). Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270, 1660–1663. doi: 10.1126/science.270.5242.1660
Ruiz, D., Martínez, V., and Cerdá, A. (1997). Citrus response to salinity: growth and nutrient uptake. Tree Physiol. 17, 141–150. doi: 10.1093/treephys/17.3.141
Rus, A., Baxter, I., Muthukumar, B., Gustin, J., Lahner, B., Yakubova, E., et al. (2006). Natural variants of At HKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2:e210. doi: 10.1371/journal.pgen.0020210
Rus, A., Yokoi, S., Sharkhuu, A., Reddy, M., Lee, B.-H., Matsumoto, T. K., et al. (2001). AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc. Natl. Acad. Sci. U.S.A. 98, 14150–14155. doi: 10.1073/pnas.241501798
Safeena, M., and Bandara, D. (2006). Antioxidant response of rice (Oryza sativa L.) varieties to salt stress at different growth stages. Trop. Agric. Res. 18, 1–12.
Sahin, U., Ekinci, M., Ors, S., Turan, M., Yildiz, S., and Yildirim, E. (2018). Effects of individual and combined effects of salinity and drought on physiological, nutritional and biochemical properties of cabbage (Brassica oleracea var. capitata). Sci. Hortic. 240, 196–204. doi: 10.1016/j.scienta.2018.06.016
Sergio, L., De Paola, A., Cantore, V., Pieralice, M., Cascarano, N. A., Bianco, V. V., et al. (2012). Effect of salt stress on growth parameters, enzymatic antioxidant system, and lipid peroxidation in wild chicory (Cichorium intybus L.). Acta Physiol. Plant. 34, 2349–2358. doi: 10.1007/s11738-012-1038-3
Sheng, M., Tang, M., Chen, H., Yang, B., Zhang, F., and Huang, Y. (2008). Influence of arbuscular mycorrhizae on photosynthesis and water status of maize plants under salt stress. Mycorrhiza 18, 287–296. doi: 10.1007/s00572-008-0180-7
Shi, H., Ishitani, M., Kim, C., and Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901. doi: 10.1073/pnas.120170197
Shi, H., Lee, B.-H., Wu, S.-J., and Zhu, J.-K. (2003). Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 21, 81–85. doi: 10.1038/nbt766
Shonjani, S. (2002). Salt Sensitivity of Rice, Maize, Sugar Beet, and Cotton During Germination and Early Vegetative Growth. [PhD thesis]. Giessen: Justus Leibig University.
Simaei, M., Khavari-Nejad, R., and Bernard, F. (2012). Exogenous application of salicylic acid and nitric oxide on the ionic contents and enzymatic activities in NaCl-stressed soybean plants. Am. J. Plant. Sci. 03, 1495–1503. doi: 10.4236/ajps.2012.310180
Siringam, K., Juntawong, N., Cha-Um, S., and Kirdmanee, C. (2011). Salt stress induced ion accumulation, ion homeostasis, membrane injury and sugar contents in salt-sensitive rice (Oryza sativa L. spp. indica) roots under isoosmotic conditions. Afr. J. Biotechnol. 10, 1340–1346.
Stewart, J. (1898). Effect of alkali on seed germination. Ann. Rept. Utah Agr. Col. Expt. St. 9, 26–35.
Taffouo, V., Wamba, O., Youmbi, E., Nono, G., and Akoa, A. (2010). Growth, yield, water status and ionic distribution response of three bambara groundnut (Vigna subterranea (L.) Verdc.) landraces grown under saline conditions. Int. J. Bot. 6, 53–58. doi: 10.3923/ijb.2010.53.58
Talei, D., Kadir, M. A., Yusop, M. K., Valdiani, A., and Abdullah, M. P. (2012). Salinity effects on macro and micronutrients uptake in medicinal plant King of Bitters (Andrographis paniculata Nees.). Plant Omics J. 5, 271–278.
Tracy, F. E., Gilliham, M., Dodd, A. N., Webb, A. A., and Tester, M. (2008). NaCl-induced changes in cytosolic free Ca2+ in Arabidopsis thaliana are heterogeneous and modified by external ionic composition. Plant Cell Environ. 31, 1063–1073. doi: 10.1111/j.1365-3040.2008.01817.x
Tunçturk, M., Tunçturk, R., and Yasar, F. (2008). Changes in micronutrients, dry weight and plant growth of soybean (Glycine max L. Merrill) cultivars under salt stress. Afr. J. Biotechnol. 7, 1650–1654. doi: 10.5897/AJB08.248
Uozumi, N., Kim, E. J., Rubio, F., Yamaguchi, T., Muto, S., Tsuboi, A., et al. (2000). The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 122, 1249–1260. doi: 10.1104/pp.122.4.1249
Víllora, G., Moreno, D. A., Pulgar, G., and Romero, L. (2000). Yield improvement in zucchini under salt stress: determining micronutrient balance. Sci. Hortic. 86, 175–183. doi: 10.1016/S0304-4238(00)00149-7
Wang, H., Zhang, M., Guo, R., Shi, D., Liu, B., Lin, X., et al. (2012). Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 12:194. doi: 10.1186/1471-2229-12-194
Wang, R., Liu, S., Zhou, F., and Ding, C. (2014). Exogenous ascorbic acid and glutathione alleviate oxidative stress induced by salt stress in the chloroplasts of Oryza sativa L. Zeitschrift für Naturforschung C 69, 226–236. doi: 10.5560/znc.2013-0117
Wilson, C., Lesch, S. M., and Grieve, C. M. (2000). Growth stage modulates salinity tolerance of New Zealand spinach (Tetragonia tetragonioides, Pall.) and red orach (Atriplex hortensis L.). Ann. Bot. 85, 501–509. doi: 10.1006/anbo.1999.1086
Wu, C.-A., Yang, G.-D., Meng, Q.-W., and Zheng, C.-C. (2004). The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant Cell Physiol. 45, 600–607. doi: 10.1093/pcp/pch071
Wu, G., and Wang, S. (2012). Calcium regulates K+/Na+ homeostasis in rice (Oryza sativa L.) under saline conditions. Plant Soil Environ. 58, 121–127. doi: 10.17221/374/2011-PSE
Yadav, R., Mahatma, M., Thirumalaisamy, P., Meena, H., Bhaduri, D., Arora, S., et al. (2017). “Arbuscular mycorrhizal fungi (AMF) for sustainable soil and plant health in salt-affected soils,” in Bioremediation of Salt Affected Soils: an Indian Perspective, eds A. Sanjay, K. S. Atul, and Y. P. Singh (Berlin: Springer), 133–156. doi: 10.1007/978-3-319-48257-6_7
Zeng, L., Poss, J. A., Wilson, C., Draz, A.-S. E., Gregorio, G. B., and Grieve, C. M. (2003). Evaluation of salt tolerance in rice genotypes by physiological characters. Euphytica 129, 281–292. doi: 10.1023/A:1022248522536
Keywords: OsHKT1, OsNHX1, OsSOS1, macronutrients, micronutrients, CaCl2, MgCl2
Citation: Farooq M, Asif S, Jang Y-H, Park J-R, Zhao D-D, Kim E-G and Kim K-M (2022) Effect of Different Salts on Nutrients Uptake, Gene Expression, Antioxidant, and Growth Pattern of Selected Rice Genotypes. Front. Plant Sci. 13:895282. doi: 10.3389/fpls.2022.895282
Received: 13 March 2022; Accepted: 16 May 2022;
Published: 16 June 2022.
Edited by:
Loredana F. Ciarmiello, University of Campania Luigi Vanvitelli, ItalyReviewed by:
Muhammad Manjurul Karim, University of Dhaka, BangladeshMirza Hasanuzzaman, Sher-e-Bangla Agricultural University, Bangladesh
Copyright © 2022 Farooq, Asif, Jang, Park, Zhao, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyung-Min Kim, a2ttQGtudS5hYy5rcg==
 Muhammad Farooq
Muhammad Farooq Saleem Asif
Saleem Asif Yoon-Hee Jang
Yoon-Hee Jang Jae-Ryoung Park
Jae-Ryoung Park Dan-Dan Zhao1
Dan-Dan Zhao1 Eun-Gyeong Kim
Eun-Gyeong Kim Kyung-Min Kim
Kyung-Min Kim