- 1Provincial Key Laboratory of Agrobiology, Institute of Food Crops, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 2College of Life Sciences, Nanjing Agricultural University, Nanjing, China
Salinity has become a crucial environmental factor seriously restricting maize (Zea mays L.) growth, development and productivity. However, how plants respond to salt stress is still poorly understood. In this study, we report that a maize brassinosteroid-signaling kinase gene ZmBSK1 plays a significant role in salt stress response. Expression pattern analysis revealed that the transcript level of ZmBSK1 was upregulated by NaCl treatment both in maize leaves, roots, and stems. Phenotypic and physiological analysis showed that overexpression of ZmBSK1 in maize improved salt tolerance by reducing the malondialdehyde (MDA) content, the percentage of electrolyte leakage, O2− and H2O2 accumulation under salt stress, relying on the increases of antioxidant defense enzyme activities and proline content. qRT-PCR analysis showed that overexpression of ZmBSK1 also positively modulated the expression levels of reactive oxygen species (ROS)-scavenging and proline biosynthesis-related genes under salt stress. Moreover, immunoprecipitation-mass spectrometry (IP-MS) assay and firefly luciferase complementation imaging (LCI) assay showed that ZmBSK1 could associate with heat shock protein ZmHSP8 and 14-3-3-like protein ZmGF14-6, and their gene expression levels could be significantly induced by NaCl treatment in different maize tissues. Our findings unravel the new function of ZmBSK1 in salt stress response, which provides the theoretical bases for the improvement of maize salt resistance.
Introduction
In past decades, plants have been frequently suffered from numerous environmental stimuli during their growth and development, including the biotic stresses resulted from pests and pathogens and the abiotic stresses resulted from salinity, drought, extreme temperature, heavy metals, etc (Atkinson and Urwin, 2012; Suzuki et al., 2014). Among them, salt stress is a major abiotic stress that can induce ionic stress, osmotic stress and secondary stresses such as oxidative stress caused by accumulation of excess reactive oxygen species (ROS; superoxide anion [O2−], hydroxyl radicals [OH•], singlet oxygen [1O2], and hydrogen peroxide [H2O2]) in plant cells, thus severely limiting plant growth and crop yield (Yang and Guo, 2018). Many genes, liking protein kinases and transcription factors, have been reported to play important roles in regulating plant tolerance to salt stress by affecting the downstream genes (Yan et al., 2014; Zhang et al., 2014; Yao et al., 2018; Qin et al., 2020).
Brassinosteroid (BR)-signaling kinase (BSK) is a kind of plant-specific protein kinase, which contains a kinase domain in N-terminus and a tetratricopeptide repeat (TPR) domain in C-terminus (Shiu et al., 2004; Kim and Wang, 2010). It belongs to the receptor-like cytoplasmic kinases (RLCKs) subfamily XII (Li et al., 2019). Until now, 12 BSKs in Arabidopsis and five BSKs in rice have been identified (Tang et al., 2008; Wang et al., 2017). Many studies have identified the different functions in Arabidopsis and rice. For example, AtBSK1, AtBSK2, AtBSK3, and AtBSK5 have been recognized as BR-responsive proteins in BR signaling pathway in Arabidopsis (Tang et al., 2008). AtBSK1 and Oryza sativa BSK3 (OsBSK3) can directly interact with and be phosphorylated by Arabidopsis BR insensitive 1 (AtBRI1) and rice BRI1 (OsBRI1), then the activated BSKs positively regulate BR signaling (Tang et al., 2008; Zhang et al., 2016). Sreeramulu et al. (2013) reveals that AtBSK3, AtBSK4, AtBSK6, AtBSK7, and AtBSK8 play a partial overlapping role in plant growth. However, only AtBSK3 has been found to be a scaffold protein to function in BR-mediated root growth, shoot growth, and organ separation (Ren et al., 2019a). Moreover, AtBSK1, AtBSK5, or AtBSK8 associates with the immune receptors to play crucial roles in activating the pathogen-associated molecular pattern (PAMP)-triggered immunity (Qi et al., 2011; Shi et al., 2013; Yan et al., 2018; Majhi et al., 2019). In rice, OsBSK1-2, an ortholog of AtBSK1, positively regulates flg22- and chitin-triggered defense responses (Wang et al., 2017).
Increasing evidences suggest that BSKs are also involved in responses of plants to abiotic stresses. For example, salinity, alkali (NaHCO3), drought, cold, phytohormones BR, and abscisic acid (ABA) can obviously upregulate the transcript levels of BSKs gene in many species including Arabidopsis, barley (Hordeum spontaneum L.), Populus tomentosa Carr., hemp (Cannabis sativa L.), potato (Solanum tuberosum L.), and kentucky bluegrass (Poa pratensis L.) (Li et al., 2012; Chen et al., 2019a,b; Yang et al., 2019; Jiang et al., 2021; Kang et al., 2021). In Arabidopsis, loss-of-function mutant bsk5 exhibits sensitivity to salinity and ABA, and further analysis shows that AtBSK5 is required for the tolerance of plants to salt stress and ABA-mediated drought stress (Li et al., 2012). Recently, there are nine BSKs have been identified in maize (Li et al., 2019). Only Zea mays BSK1 (ZmBSK1), an ortholog of AtBSK1, has been characterized to enhance drought tolerance (Liu et al., 2021). However, little is known about the biological function of BSKs in maize, especially under salt stress conditions.
In this study, we report that the gene expression of ZmBSK1 can be induced by NaCl and overexpression of ZmBSK1 enhances salt tolerance in maize. Further, we unravel the mechanism of ZmBSK1 in salt tolerance.
Materials and Methods
Plant Materials, Growth Conditions, and Salt Stress Treatment
Maize (Zea mays L.) inbred line B73 (from Nanjing Agricultural University, China) and tobacco (Nicotiana benthamiana L., from Nanjing Agricultural University, China) were used in this study. Maize and tobacco seeds were sown on pots containing soil mixture (soil: vermiculite, 1: 1, v/v) or grown in modified 1/2Hoagland nutrient solution (Coolaber, China) in an artificial climate chamber at a temperature of 28°C, photoperiod 14 h light: 10 h dark and 60% relative humidity (RH), and were watered daily. For tissue-specific expression analysis, roots, stems, and leaves were collected in three-leaf stage of maize, and pollens as well as pistils were collected in flowering stage of maize. For salt stress treatment, 10-day-old maize seedlings grown in nutrient solution were treated with or without 200 mM NaCl for various times, and then the second leaves were harvested and used for further analysis.
Generation of Transgenic Maize Plants
The full-length coding sequence of ZmBSK1 was cloned into pCUN-NHF expression vector driven by ubiquitin promoter with a 3 × Flag tag at the N terminus. The maize inbred line B73 was used as the plant receptor. The Agrobacterium-mediated maize transformation was performed as described by Liu et al. (2015). Positive transformants were selected by 75 mg L−1 herbicide Basta (Sangon Biotech, China) and were further confirmed by PCR. The primers are listed in Supplementary Table 1. The homozygous T3 lines were obtained for analysis of phenotypes, gene expressions, and physiological indexes.
RNA Extraction and qRT-PCR Analysis
Total RNA were isolated from different maize tissues using RNAiso Plus Kit (Takara, China) following the manufacturer’s protocol, and the cDNA was synthesized using M5 Super plus qPCR RT Kit (Mei5bio, China). Quantitative RT-PCR was performed on a CFX96 Touch System (Bio-Rad, United States) using 2 × M5 HiPer Realtime PCR Super Mix (Mei5bio, China) according to the manufacturer’s protocol. The gene expression levels were calculated by the 2-ΔΔCT method and were normalized against ZmActin2 gene. The specific primers used for qRT-PCR are listed in Supplementary Table 1.
Western Blot Analysis
Total proteins were extracted from maize leaves as described previously (Ma et al., 2012), and their content was quantified using Bradford Protein Assay Kit (Beyotime, China) according to the manufacturer’s protocol. Extracted proteins were separated by 12% SDS-PAGE and were transferred to a polyvinylidene fluoride (PVDF) membrane (Merck Millipore, United States). The membrane was blocked in PBST buffer containing 5% [w/v] skimmed milk powder for 2 h at 25°C, and was then incubated with primary antibody including anti-Flag antibody (1:5,000, Abmart) or anti-actin antibody (1:5,000, Biodragon, China). The secondary antibody, horseradish peroxidase (HRP)-conjugated anti-mouse antibody (Abmart), was used at 1:5,000 dilution. Chemifluorescent signal generated by BeyoECL plus (Beyotime, China) western blotting detection reagents was captured by a camera (Tanon 5200 Multi, China).
Phenotype and Oxidative Damage Analysis
For the root phenotype, maize seeds were spread on the paper containing 200 mM NaCl solution and kept for 4 days, after which the root length were measured. For the growth phenotype, 10-day-old maize seedlings were treated with 200 mM NaCl for 14 days. After recovery by rewatering for 5 days, the survival rate was calculated. For oxidative damage analysis, 10-day-old maize seedlings were treated with 200 mM NaCl for 2 days, the malondialdehyde (MDA) content and the percentage of electrolyte leakage were measured as described by Zhu et al. (2016).
Detection of H2O2 and O2−
About 10-day-old maize seedlings were treated with 200 mM NaCl for 2 days, and the second leaves were harvested for the subsequent analysis. For H2O2 staining and O2− staining, the leaves were incubated in 1 mg ml−1 3,3′-diaminobenzidine (DAB) solution (pH 3.8, Coolaber, China) and 0.5 mg ml−1 nitroblue tetrazolium chloride (NBT) solution (Solarbio, China) for 8 h in dark at room temperature, respectively. After staining, the leaves were boiled in 95% ethanol for 10 min to decolorize, and then photographed. For quantification, the contents of H2O2 and O2− were determined using H2O2 Detection Kit (Leagene, China) and Micro Superoxide Anion Assay Kit (Solarbio, China) according to the manufacturer’s protocol, respectively.
Measurement of Antioxidant Defense Enzyme Activity and Proline Content
The harvest maize leaves were homogenized in extraction buffer containing 50 mM potassium phosphate (pH 7.0), 1 mM EDTA and 1% (w/v) polyvinylpyrrolidone 40. The homogenates were centrifuged at 12,000 × g for 30 min at 4°C, and the supernatants were immediately used for the subsequent antioxidant defense enzyme assays. Total activities of antioxidant defense enzymes were measured as described previously (Zhang et al., 2010). The proline content was determined using Proline Assay Kit (Leagene, China) according to the manufacturer’s instructions.
Immunoprecipitation-Mass Spectrometry
About 10-day-old OE-ZmBSK1 transgenic lines were treated with 200 mM NaCl for 0 or 10 h, and then total proteins from the leaves were extracted using lysis buffer [10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% (v/v) Triton X-100, 0.1% (w/v) SDS, 0.5 mM DTT, and protease inhibitor cocktail]. Protein extracts were immunoprecipitated with anti-Flag antibody (1:300, Abmart) bound to protein A/G agarose beads in PBS buffer containing protease inhibitor cocktail for 8 h at 4°C. The beads were washed for three times with PBS buffer and were boiled in SDS loading buffer (Beyotime, China). After centrifugation, the supernatants were processed according to a previous publication (Wang et al., 2015), and were analyzed using EASY-nLC 1200 ultra-high performance liquid system (Thermo Scientific, United States) and Q Exactive HF mass spectrometry system (Thermo Scientific, United States). For protein identification, the MS data were searched against maize database in UniProtKB using the Paragon algorithm in ProteinPilot ™ Software (v5.0.2, SCIEX). For the identified proteins, selected certain filtering criteria and peptides with an unused score > 1.3 were considered as credible peptides, and proteins containing at least one unique peptide were retained.
Firefly Luciferase Complementation Imaging Assay
The full-length coding sequence of ZmBSK1 was cloned into vector pCAMBIA1300-nLUC to express ZmBSK1-nLUC fusion protein. The full-length coding sequences of ZmHSP8 and ZmGF14-6 were cloned into vector pCAMBIA1300-cLUC to express cLUC-ZmHSP8 and cLUC-ZmGF14-6 fusion proteins, respectively. The empty vectors were used as negative control. Agrobacterium tumefaciens strain GV3101 containing each construct was transiently transfected into 4-week-old tobacco leaves. After 3 days of infiltration, 1 mM D-luciferin (PerkinElmer, United States) was sprayed onto the leaves and kept for 20 min in dark, and then the luminescence was captured by a low-light cooled charge coupled device camera (Tanon 5200 Multi, China).
Statistical Analysis
Statistical analysis was performed using the software SPSS (v16.0).1 One-way or two-way ANOVA corrected with Duncan’s multiple range test was used to determine statistical significance. Differences were considered significant at p < 0.05.
Accession Numbers
Sequence data from this article can be found in MaizeGDB database under the following accession numbers: ZmBSK1, Zm00001d048345; ZmActin2, Zm00001d013873; ZmcAPX, Zm00001d007234; ZmCAT1, Zm00001d014818; ZmCSD5, Zm00001d022505; ZmMSD2, Zm00001d009990; ZmP5CS1, Zm00001d012391; ZmP5CS2, Zm00001d010056; ZmHSP8, Zm00001d031325; and ZmGF14-6, Zm00001d003401.
Results
Expression Patterns of ZmBSK1
To determine the tissue-specific expression level of ZmBSK1, qRT-PCR was used to detect the transcript levels of ZmBSK1 in different organ tissues of maize, including roots, stems, leaves, pollens, and pistils. The results showed that the expression level of ZmBSK1 was higher in the leaves and pistils, and was lower in the roots and stems (Figure 1A), implying that ZmBSK1 may function in the process of ear development. To explore the effect of salt stress on the expression level of ZmBSK1 gene, qRT-PCR was conducted after maize plants were treated with or without 200 mM NaCl for various times. We found that the expression level of ZmBSK1 was gradually induced to a maximum value (5-fold in leaves, 2-fold in roots, and 2.3-fold in stems) within 9 h and then declined after treatment with 200 mM NaCl (Figures 1B–D). The results suggest that ZmBSK1 may play a positive role in plants response to salt stress.
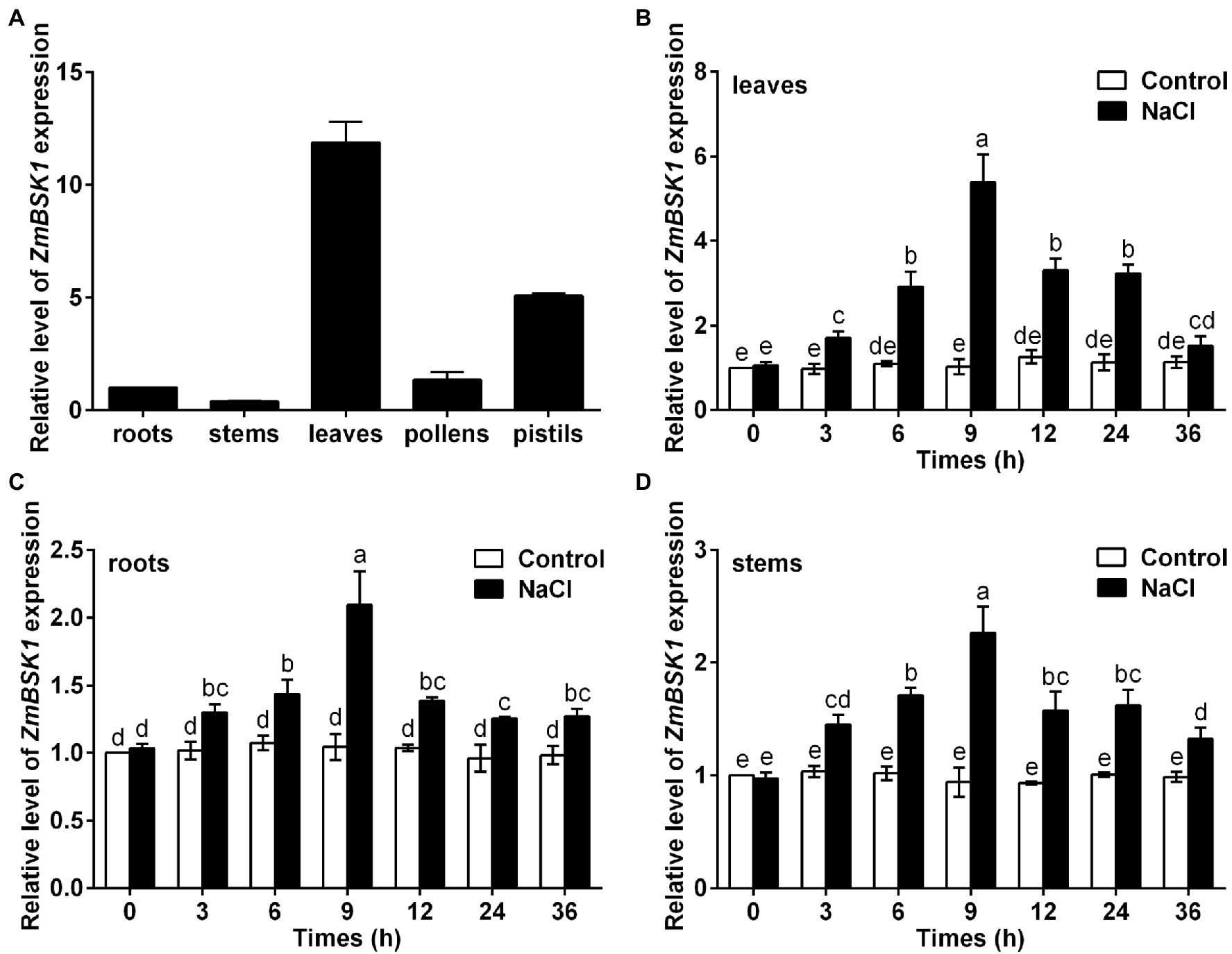
Figure 1. Expression patterns of ZmBSK1 gene in maize. (A) qRT-PCR analysis of the expression levels of ZmBSK1 in different maize tissues (roots, stems, leaves, pollens, and pistils). (B-D) The expression levels of ZmBSK1 in maize leaves (B), roots (C), and stems (D) exposed to salt stress. About 10-day-old wild type seedlings were treated with 200 mM NaCl for indicated times, and the expression was measured by qRT-PCR. ZmActin2 served as an internal control. Error bars represent ±SD (n = 3). Different letters indicate significant differences at p < 0.05 according to two-way ANOVA (Duncan’s multiple range test).
Overexpression of ZmBSK1 Enhances Salt Tolerance in Maize
To further investigate the biological function of ZmBSK1 in maize under salt stress, two independent ZmBSK1-overexpressing lines (OE-ZmBSK1#11 and OE-ZmBSK1#17) were generated, then the expressions of ZmBSK1 at transcript and protein levels were confirmed by qRT-PCR and immunobloting assays, respectively (Figures 2A,B). Wild type (WT) and OE-ZmBSK1 transgenic maize seeds were soaked in 200 mM NaCl solution at the germination stage, and then the root length was measured. As shown in Figures 2C,D, both two OE-ZmBSK1 maize seeds exhibited slightly longer root length than WT under normal conditions. However, these two transgenic maize seeds developed obviously longer roots than WT when exposed to salt treatment. Subsequently, 10-day-old WT and OE-ZmBSK1 transgenic seedlings were treated with or without 200 mM NaCl. Under normal conditions, there was no significant difference in the growth phenotypes between WT and transgenic lines (Figure 3A). Under salt stress, OE-ZmBSK1 lines presented less wilting and chlorosis than WT (Figure 3A). After recovery by rewatering, all transgenic lines had higher survival rates compared with WT plants (Figure 3B). These results indicate that ZmBSK1 positively regulates salt stress tolerance in maize.
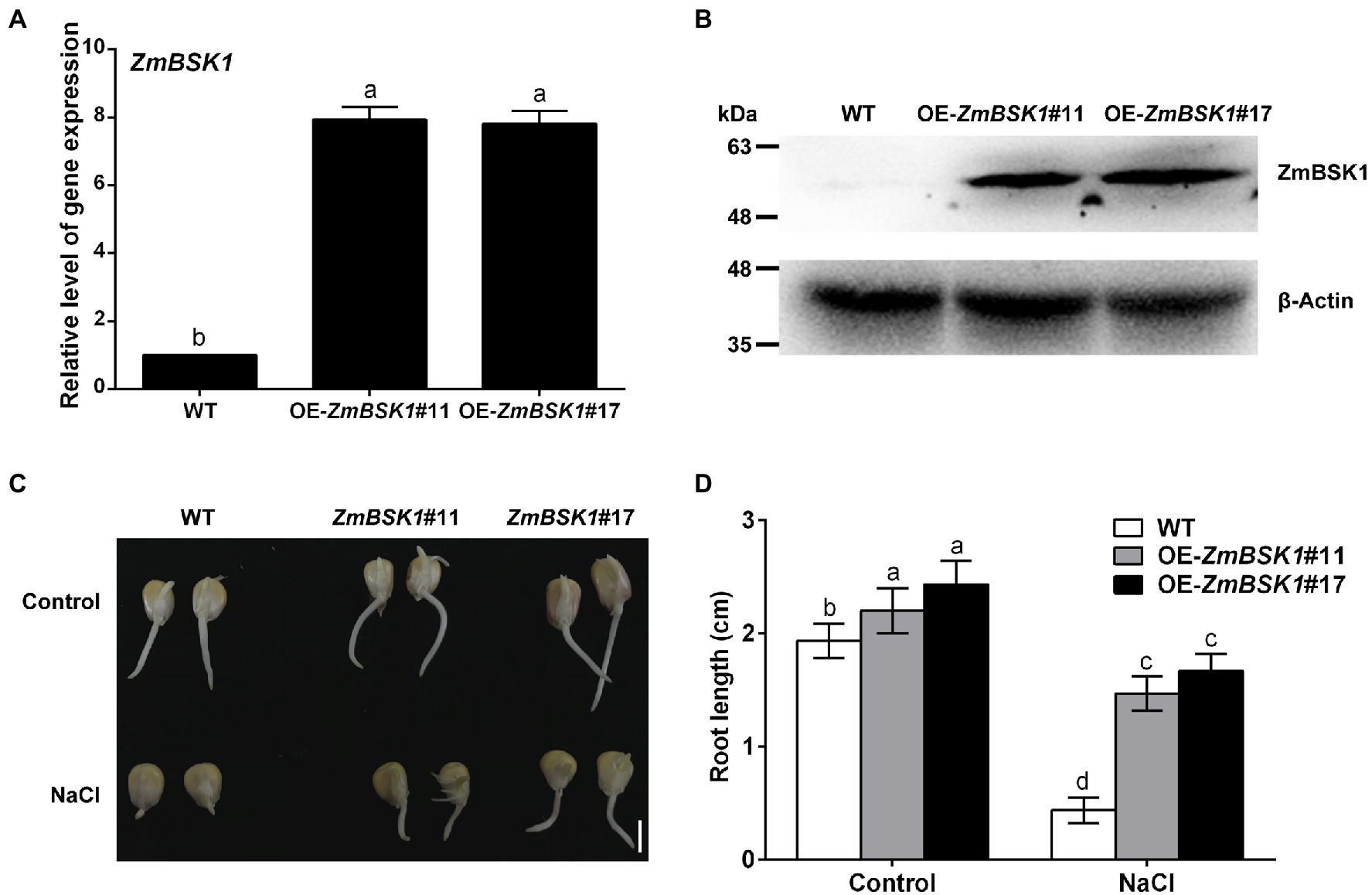
Figure 2. Effect of ZmBSK1 on root growth of maize seeds under salt stress. (A) The expression levels of ZmBSK1 in OE-ZmBSK1 and wild type (WT) plants. The expression of ZmBSK1 was measured by qRT-PCR, and ZmActin2 served as an internal control. (B) The protein levels of ZmBSK1 in OE-ZmBSK1 and WT plants. Total proteins extracted from the leaves were used for immunoblotting with anti-Flag antibody. β-actin was used as a loading control. (C) The root growth phenotypes of OE-ZmBSK1 and WT maize seeds under salt stress. The maize seeds were spread on paper containing 200 mM NaCl for 4 days during germination. Scale bar = 1 cm. (D) Statistical analysis of root length in (C). Error bars in (A,D) represent ±SD (n = 3). Different letters indicate significant differences at p < 0.05 according to one-way or two-way ANOVA (Duncan’s multiple range test).
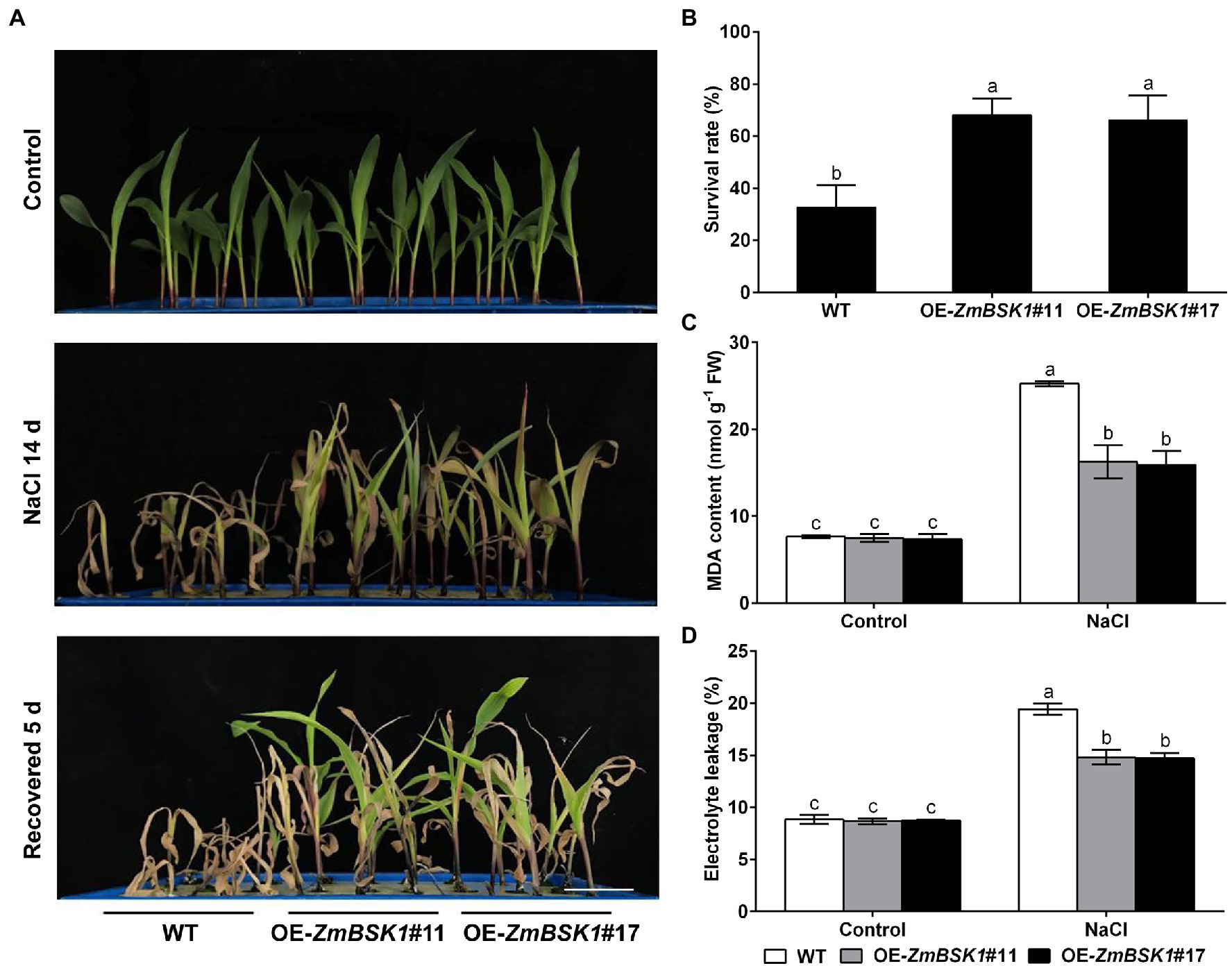
Figure 3. Overexpression of ZmBSK1 improves salt tolerance in maize. (A) The phenotypes of OE-ZmBSK1 and WT maize plants under salt stress. About 10-day-old maize seedlings were treated with 200 mM NaCl for 14 days, and then recovered for 5 days. Scale bar = 5 cm. (B) Survival rate (%) of maize plants in (A). At least 45 seedlings of each line per replicate were used for survival rate analysis. The malondialdehyde (MDA) content (C) and the percentage of electrolyte leakage (D) in leaves of OE-ZmBSK1 and WT maize plants under salt stress. About 10-day-old maize seedlings were treated with 200 mM NaCl for 2 days, and then the physiological indexes as indicated were measured. Error bars in (B–D) represent ±SD (n = 3). Different letters indicate significant differences at p < 0.05 according to one-way or two-way ANOVA (Duncan’s multiple range test).
Overexpression of ZmBSK1 Alleviates Oxidative Damage Caused by Salt Stress in Maize
The MDA content and the percentage of electrolyte leakage are two major indicators of oxidative damage in plants (Jiang and Zhang, 2002; Zhang et al., 2011). Thus, these two physiological indicators were also detected. As shown in Figures 3C,D, without NaCl treatment, no significant difference was observed in the MDA content and the percentage of electrolyte leakage between WT plants and transgenic lines. Salt stress caused marked increases in the MDA content and the percentage of electrolyte leakage in WT plants compared with control conditions, which were further alleviated in OE-ZmBSK1 transgenic lines. These results suggest that overexpression of ZmBSK1 can largely protect maize plants from oxidative damage under salt stress.
ZmBSK1 Reduces the Accumulation of ROS by Enhancing Antioxidant Defense System Under Salt Stress in Maize
Salinity-induced ROS accumulation is responsible for oxidative damage (Foyer, 2018). The NBT and DAB staining were used to determine the levels of O2− and H2O2 in maize leaves. As shown in Figures 4A,B, the accumulation of O2− and H2O2 in OE-ZmBSK1 transgenic lines was obviously lower than that in WT plants exposed to salt stress. In agreement with the results of staining, the O2− production rate and the H2O2 content were lower in transgenic lines than that in WT plants under salt stress (Figures 4C,D). Under normal conditions, there was no obvious difference in the accumulation and the content of ROS in all lines (Figures 4A–D). These results show that overexpression of ZmBSK1 reduces ROS level in plants in response to salt stress.
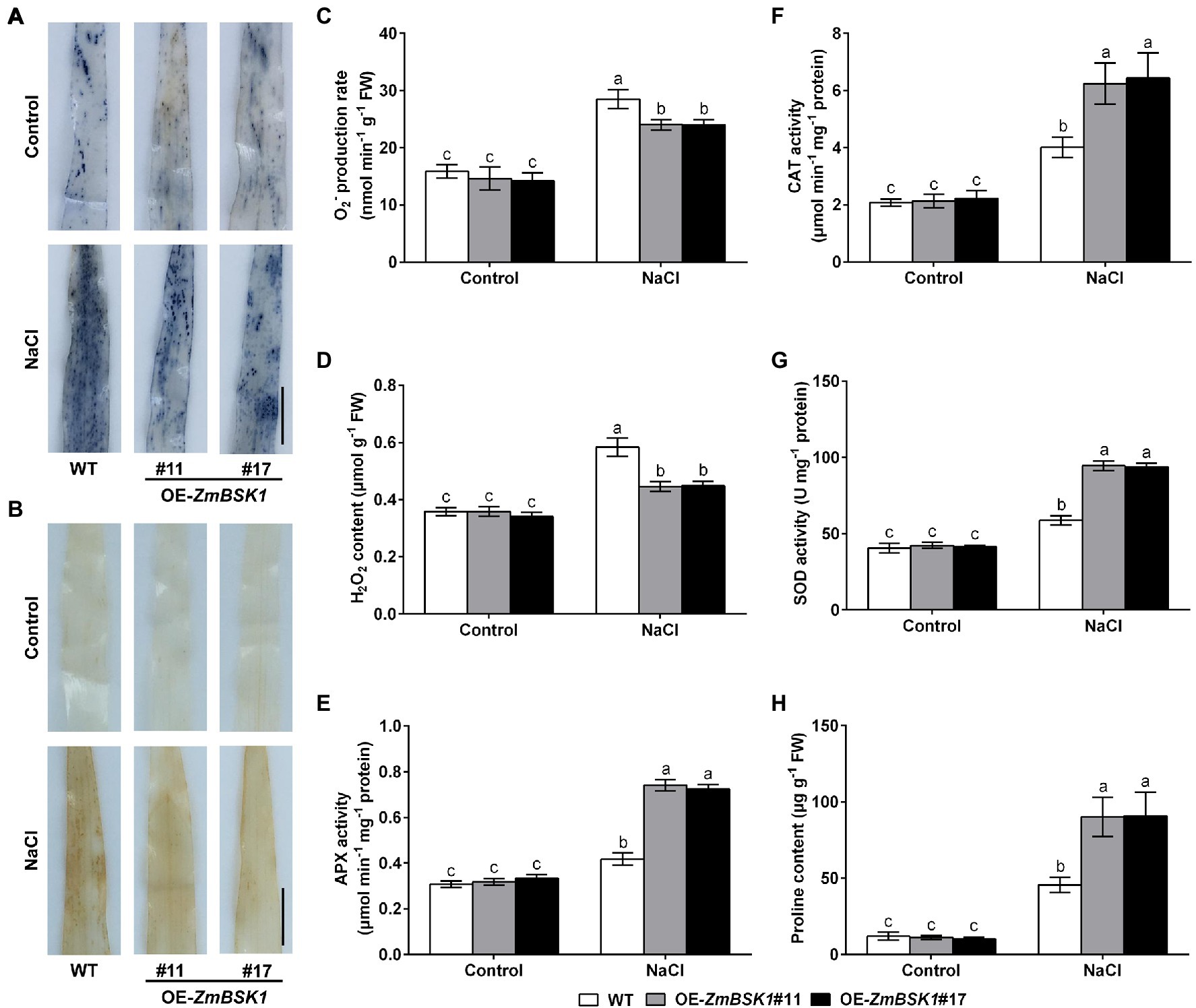
Figure 4. Effect of ZmBSK1 on reactive oxygen species (ROS) levels, antioxidant defense enzymes activity and proline content under salt stress. Nitroblue tetrazolium (NBT) staining of O2− (A) and 3,3′-diaminobenzidine (DAB) staining of H2O2 (B) in leaves of OE-ZmBSK1 and WT maize plants under salt stress. Scale bar = 1 cm. Statistical analysis of O2− production rates (C) and H2O2 content (D) in leaves of OE-ZmBSK1 and WT maize plants under salt stress. Ascorbate peroxidase (APX) activity (E), catalase (CAT) activity (F), superoxide dismutase (SOD) activity (G), and proline content (H) in leaves of OE-ZmBSK1 and WT maize plants under salt stress. About 10-day-old maize seedlings in (A–H) were treated with 200 mM NaCl for 2 days, and then the above physiological indexes as indicated were measured. Error bars in (C–H) represent ±SD (n = 3). Different letters indicate significant differences at p < 0.05 according to two-way ANOVA (Duncan’s multiple range test).
Antioxidant defense system is required for scavenging excess ROS produced by various abiotic stresses (Julkowska and Testerink, 2015; You and Chan, 2015; Hasanuzzaman et al., 2021). Accordingly, to explore the effect of ZmBSK1 on the antioxidant defense enzymes in maize in response to salt stress, the activities of three key enzymes ascorbate peroxidase (APX), catalase (CAT), and superoxide dismutase (SOD) were examined. Under normal conditions, both WT plants and OE-ZmBSK1 transgenic lines had the similar activities of APX, CAT, and SOD. Under salt stress, however, the activities of these three enzymes in transgenic lines increased more than those in WT (Figures 4E–G). In addition to the antioxidant defense enzymes, the content of the non-enzymatic antioxidants such as proline was also measured in maize. Consistent with the results of enzymes activities, the proline content was much higher in all transgenic lines than in WT plants when exposed to salt treatment (Figure 4H). Taken together, these results demonstrate that ZmBSK1 enhances the tolerance of maize plants to salt stress by inducing the activities of antioxidant defense enzymes and the accumulation of proline and alleviating ROS level.
ZmBSK1 Modulates the Expressions of Stress-Related Genes Under Salt Stress
To study the mechanism by which ZmBSK1 functioned in plant tolerance to salt stress, qRT-PCR was used to analyze the expression levels of several selected stress-related genes, including ROS-scavenging genes ZmcAPX (cytosolic APX), ZmCAT1, ZmCSD5 (Cu/Zn-SOD), and ZmMSD2 (Fe/Mn-SOD) as well as proline biosynthesis-related genes ZmP5CS1 and ZmP5CS2 (Wang et al., 2014). Under normal conditions, the expression levels of all genes were similar between WT and OE-ZmBSK1 transgenic lines (Figure 5). After salt treatment, compared with WT plants, ZmcAPX, ZmCAT1, ZmCSD5, ZmMSD2, and ZmP5CS2 genes displayed increased expression levels in the transgenic lines (Figures 5A–E). Nevertheless, among these genes, only ZmP5CS1 gene could not be induced in all lines (Figure 5F). This might be due to the fact that the expression of ZmP5CS1 is induced at a slower rate than that of ZmP5CS2 in the early stage of salt stress. In general, these data reveal that ZmBSK1 improves salt tolerance by modulating the expression levels of stress-related genes.
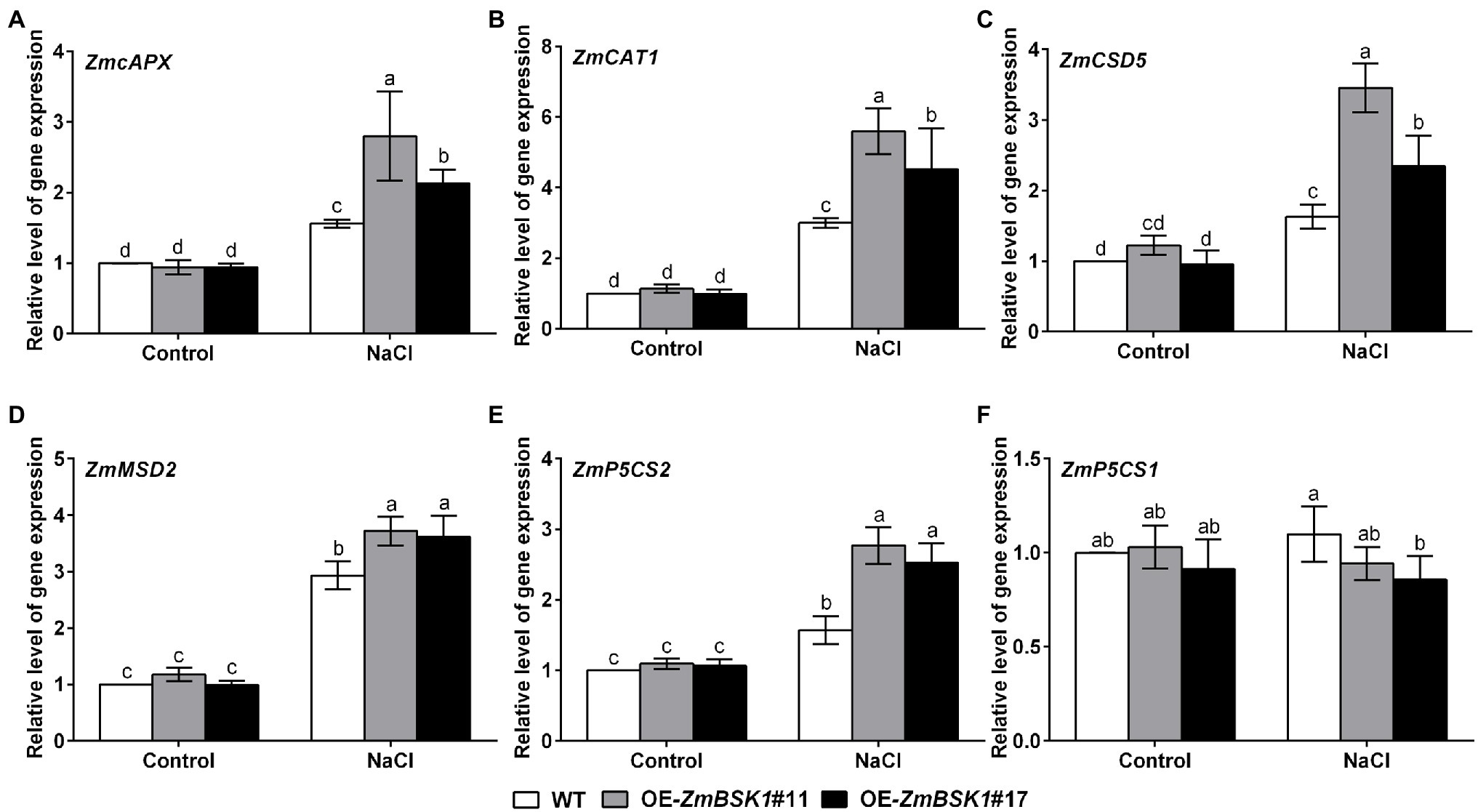
Figure 5. ZmBSK1 positively regulates the expression of stress-related genes in maize under salt stress. The expression levels of stress-related genes ZmcAPX (A), ZmCAT1 (B), ZmCSD5 (C), ZmMSD2 (D), ZmP5CS2 (E), and ZmP5CS1 (F) in OE-ZmBSK1 and WT maize plants exposed to salt stress. About 10-day-old maize seedlings were treated with 200 mM NaCl for 6 h, and the expressions were measured by qRT-PCR. ZmActin2 served as an internal control. Error bars in (A–F) represent ±SD (n = 3). Different letters indicate significant differences at p < 0.05 according to two-way ANOVA (Duncan’s multiple range test).
ZmBSK1 Interacts With ZmHSP8 and ZmGF14-6
Previous studies have demonstrated that BSKs can directly interact with several kinases and phosphatases, such as BRI1, BRI1-suppressor 1 (BSU1), ZmCCaMK, FLAGELLIN SENSING2 (FLS2), MEK Kinase5 (MAPKKK5), and RECEPTOR-LIKE KINASE 902 (RLK902) (Tang et al., 2008; Kim et al., 2009; Shi et al., 2013; Yan et al., 2018; Zhao et al., 2019; Liu et al., 2021). These findings suggest that BSKs require some different downstream components to regulate different development processes or stress responses. To further understand how ZmBSK1 functions in salt tolerance, immunoprecipitation-mass spectrometry (IP-MS) assay was carried out to identify the interacting proteins of ZmBSK1 in OE-Flag-ZmBSK1 transgenic lines using anti-Flag antibody under salt stress. Total 69 proteins were detected in this study (Figure 6A; Supplementary Table 2). Among these proteins, heat shock protein ZmHSP8 and 14-3-3-like protein ZmGF14-6 were chose to further confirm their interactions with ZmBSK1 using luciferase complementation imaging (LCI) assay (Figure 6B). ZmBSK1 was fused with the N-terminal fragment of luciferase (ZmBSK1-nLUC), meanwhile, ZmHSP8 and ZmGF14-6 were independently fused with the C-terminal fragment of luciferase (cLUC-ZmHSP8 and cLUC-ZmGF14-6). A strong luminescence signal was observed in tobacco leaves injected with ZmBSK1-nLUC and cLUC-ZmHSP8 (Figure 6C) as well as ZmBSK1-nLUC and cLUC-ZmGF14-6 (Figure 6D). These observations indicate that ZmBSK1 interacts with ZmHSP8 and ZmGF14-6, respectively.
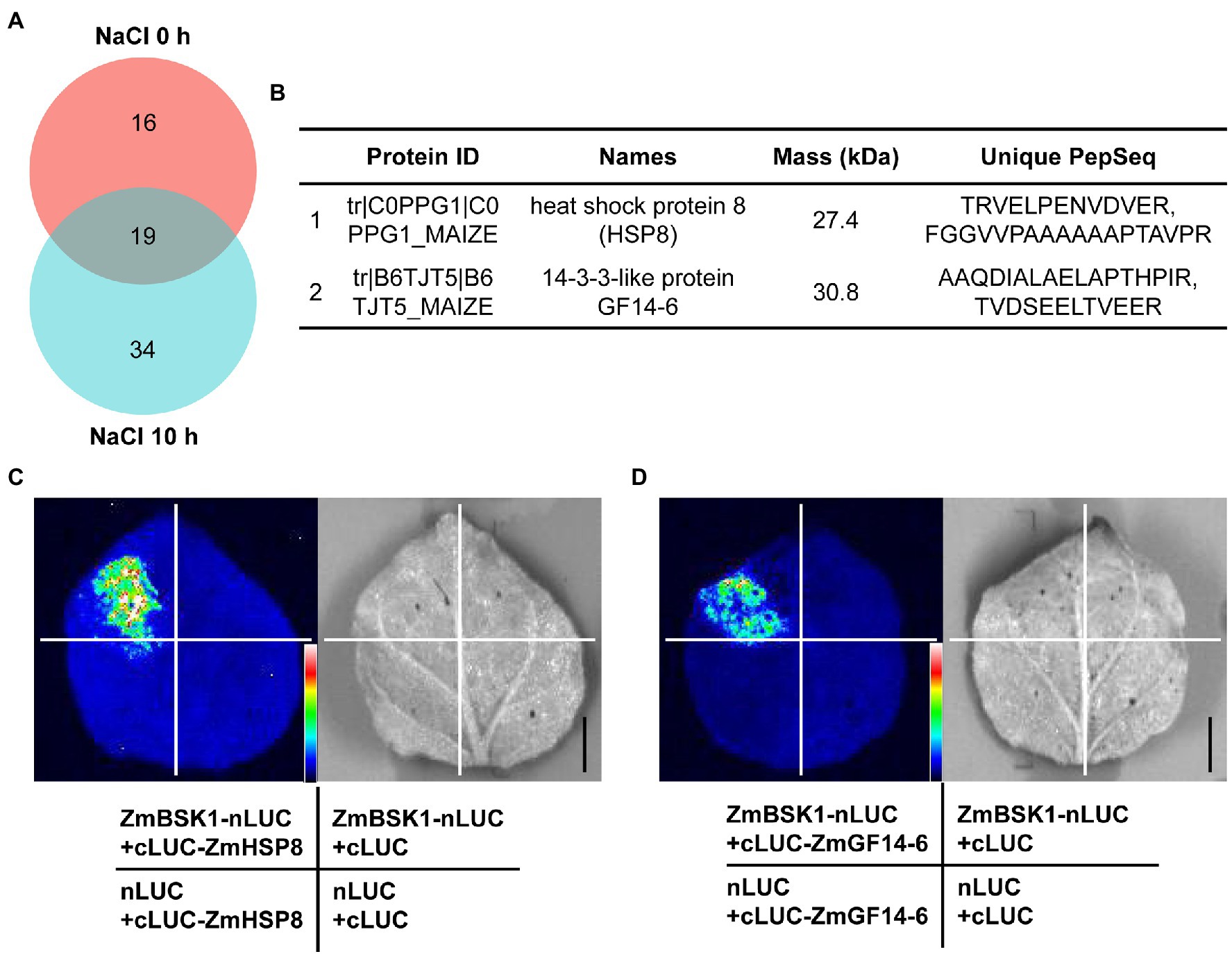
Figure 6. ZmBSK1 interacts with its target proteins ZmHSP8 and ZmGF14-6. (A) Venn diagram shows the number of ZmBSK1-interacting proteins in OE-ZmBSK1 maize plants with or without salt stress. About 10-day-old maize seedlings were treated with 200 mM NaCl for 0 or 10 h. Total proteins extracted from leaves were immunoprecipitated by anti-Flag antibody and the interacting proteins of ZmBSK1 were identified by immunoprecipitation-mass spectrometry (IP-MS). (B) The candidate target proteins of ZmBSK1 upon mass spectrometry analysis only after salt treatment. Firefly luciferase complementation imaging (LCI) assays confirm the interactions of ZmBSK1 with ZmHSP8 (C) and ZmGF14-6 (D). The combinations of ZmBSK1-nLUC and cLUC-ZmHSP8, ZmBSK1-nLUC and cLUC-ZmGF14-6 were co-transformed into tobacco leaves. The combinations of ZmBSK1-nLUC and cLUC, nLUC and cLUC-ZmHSP8, nLUC and cLUC-ZmGF14-6, nLUC and cLUC were used as negative controls. nLUC and cLUC represent the N-terminal and C-terminal fragments of firefly luciferase, respectively. Images were collected from the detached leaves after infiltration for 3 days. Scale bar = 1 cm.
Expression Analysis of ZmHSP8 and ZmGF14-6 Under Salt Stress
Since ZmHSP8 and ZmGF14-6 could directly interact with ZmBSK1, we wondered whether they functioned in salt response in maize. To test this, qRT-PCR was used to detect the transcript levels of ZmHSP8 and ZmGF14-6 genes in maize with or without NaCl treatment. As shown in Figure 7, under salt stress, the expression of ZmHSP8 gene was rapidly induced to reach the first peak value at 6 h and the second peak value at 36 h in leaves and was gradually induced to the peak value at 36 h in stems. Moreover, under salt stress, the expression of ZmGF14-6 gene gradually increased to its highest level within 12 h in leaves and 9 h in stems, whereas both ZmHSP8 and ZmGF14-6 were slightly upregulated in roots. These findings imply that ZmHSP8 and ZmGF14-6 may play a role in improving plant tolerance to salt stress.
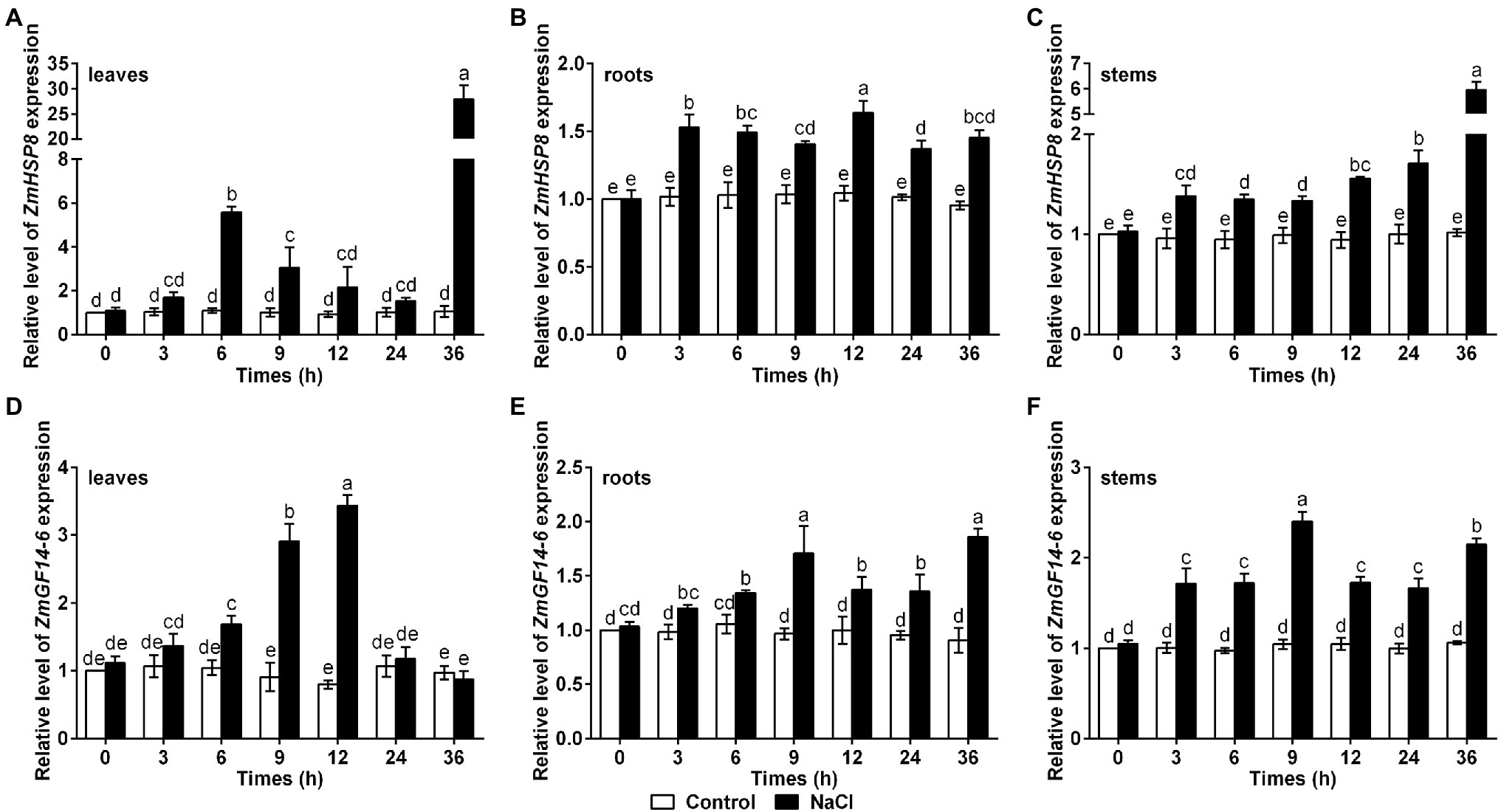
Figure 7. Gene expression patterns of ZmHSP8 and ZmGF14-6 under salt stress. (A–C) The expression levels of ZmHSP8 in maize leaves (A), roots (B), and stems (C) exposed to salt stress. (D–F) The expression levels of ZmGF14-6 in maize leaves (D), roots (E), and stems (F) exposed to salt stress. About 10-day-old wild type seedlings were treated with 200 mM NaCl for indicated times, and the expression was measured by qRT-PCR. ZmActin2 served as an internal control. Error bars represent ±SD (n = 3). Different letters indicate significant differences at p < 0.05 according to two-way ANOVA (Duncan’s multiple range test).
Discussion
Brassinosteroid (BR)-signaling kinases are first identified in Arabidopsis as a brassinosteroid-signaling kinase that is found to play a role in BR signaling pathway (Tang et al., 2008). Up to now, BSKs have been well characterized to play vital roles in many other biological processes such as plant growth and development as well as plant immunity (Sreeramulu et al., 2013; Yan et al., 2018). In addition, an increasing number of studies have demonstrated that BSKs are also involved in regulating plant responses to abiotic stresses (Li et al., 2012; Chen et al., 2019a; Yang et al., 2019; Liu et al., 2021). There are 9 BSKs have been found in maize (Li et al., 2019); however, the roles of ZmBSKs in response to abiotic stresses remain largely unknown.
In the present study, we revealed that ZmBSK1 acted as a positive regulator in plant salt tolerance, in agreement with AtBSK5 in a previous study (Li et al., 2012), based on the evidence that OE-ZmBSK1 transgenic lines displayed a better growth performance and a higher survival rate under salt stress (Figures 2, 3A,B). Moreover, changes of diverse physiological indicators appear to reflect the mechanisms by which transgenic lines cope with environmental stimuli. As previously reported, the expression of BSKs genes can be induced by different abiotic stresses (Yang et al., 2019; Jiang et al., 2021; Kang et al., 2021). And interestingly, numerous salinity stress-related cis-elements such as ARE were detected in the promoter region of ZmBSK1 (Supplementary Table 3), which might be responsible for the significant increase in ZmBSK1 gene expression under salt stress conditions (Figures 1B–D). The antioxidant defense system can be activated to mitigate oxidative damage caused by ROS under stress conditions (Sharma et al., 2012; Xiang et al., 2021; Yan et al., 2022). Our results showed that the MDA content and the percentage of electrolyte leakage in OE-ZmBSK1 transgenic lines were lower than those in WT plants (Figures 3C,D). Oxidative damage is tightly linked to the ROS accumulation (Gong et al., 2005; Li et al., 2008; Yang and Guo, 2018). Similarly, lower O2− and H2O2 levels were also observed in transgenic lines (Figures 4A–D), indicating that transgenic lines had lower ROS accumulation and oxidative damage degree under salt treatment. To protect plants from oxidative damage, the antioxidant defense system would be rapidly induced to scavenge excess ROS (Das and Roychoudhury, 2014; Moradbeygi et al., 2020). As expected, transgenic lines exhibited higher APX, CAT, and SOD activities and proline content (Figures 4E–H), contributing to maintain low ROS levels and the balance of osmotic pressure. The expressions of stress-responsive genes are crucial for enhancing plant tolerance to various stresses (Chini et al., 2004; Ren et al., 2019b). Indeed, a higher expressions of ROS-scavenging enzyme genes ZmcAPX, ZmCAT1, ZmCSD5, and ZmMSD2 were observed in transgenic lines (Figures 5A–D), which were consistent with the differences in the activities of ROS-scavenging enzymes between WT and transgenic lines under NaCl treatment. Furthermore, P5CS1/2 encodes a rate-limiting enzyme in the biosynthesis of proline (Wang et al., 2014), which can be upregulated under salt stress in maize (Wang et al., 2013). Likewise, in current study, the expression of ZmP5CS2 could be further induced in transgenic lines than that in WT plants under salt stress (Figure 5E). However, ZmP5CS1 showed the similar expressions in both transgenic lines and WT plants with or without NaCl treatment (Figure 5F). Due to the differences in NaCl processing time, one possible explanation is that ZmP5CS1 has not been induced during the initial stage of salt stress.
As a receptor-like protein kinase, BSKs are investigated to function in diverse biological processes by interacting with and phosphorylating its target proteins such as BSU1, MAPKKK5, and ZmCCaMK (Tang et al., 2008; Yan et al., 2018; Liu et al., 2021), which means that there are some unknown interactors of ZmBSK1 under salt stress conditions. Our IP-MS results revealed two novel interacting proteins of ZmBSK1 during NaCl treatment, ZmHSP8 and ZmGF14-6 (Figure 6). The molecular mechanisms underlying the functions of HSPs in biotic stress signaling, drought stress signaling, hormone signaling, and development have been extensively studied in many species (Jacob et al., 2017). Recently, transcriptomic analysis showed that ZmHSP8 was involved in response to drought and heat stresses in maize (Qian et al., 2019; Blein-Nicolas et al., 2020), whereas we found that NaCl treatment can obviously upregulate ZmHSP8 gene expression (Figures 7A–C), implying that ZmHSP8 may play a role in salt stress response. In animals, it is known that the phosphorylation of HSPs by stress kinase is one of the most important post-translational modifications, which functions in enhancing chaperone activities and its affinity for unfolded proteins during stress response (Späth et al., 2015). However, there are few reports on the roles of HSPs phosphorylation in plants. Recently, Zhao et al. (2021) finds that maize sHSP17.4 can interact with and be phosphorylated by ZmCDPK7 in regulating heat stress response. Furthermore, due to the lack of catalytic activity, 14-3-3 proteins usually function via physical interactions. Previous studies have reported that 14-3-3 proteins interact with various target proteins to regulate diverse biological processes, including metabolism, transcription, protein trafficking, and stress responses (Roberts, 2003; Zhou et al., 2014; Zhang et al., 2018). Additionally, the phosphorylation of 14-3-3 proteins also plays an important role in their functions. In Arabidopsis, a plasma membrane-localized kinase CRPK1 can phosphorylate 14-3-3 proteins, which followed by translocating from the cytoplasm into the nucleus to regulate the stability of CBFs in cold response (Liu et al., 2017). In maize, the gene expression of ZmGF14-6, encoding a 14-3-3-like protein, could be upregulated by salt stress (Campo et al., 2012), which was consistent with our findings (Figures 7D–F). Because ZmBSK1 is also a plasma membrane-anchored kinase, we wonder if it can recruit and phosphorylate ZmGF14-6 to further modulate salt tolerance. Here, our data provided the possibility of ZmBSK1 and ZmHSP8 or ZmGF14-6 in improving plant tolerance to salt stress through interacting with each other.
Based on these results, we provide a working model of ZmBSK1 in salt response. Salt stress rapidly induces the expression of ZmBSK1, thus upregulating the expressions of salt stress-responsive genes to enhance antioxidant defense enzyme activities and promote proline synthesis, improving the salt tolerance. In this process, the two interacting proteins of ZmBSK1, ZmHSP8, and ZmGF14-6, are identified and might function in salt response. Future work is needed to clarify the molecular mechanisms by which ZmBSK1 and its two interacting proteins positively regulate plant salt tolerance.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
JY, YC, and LL conceived the project and designed the experiments. LL performed most of the experiments, analyzed the data, and wrote the manuscript. YS performed the phenotype analysis. PD provided the resources of transgenic maize. YC, QM, and XW helped to analyze the data. JY and YC revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Postdoctoral Science Foundation of Jiangsu Province (2021K406C); the China Agriculture Research System (CARS-02); the Jiangsu Agriculture Science and Technology Innovation Fund [CX(21)3115]; and the earmarked fund for Jiangsu Agricultural Industry Technology System [JATS(2020)385].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to Jingwei Yan (Nanjing Agricultural University) for revising the manuscript, and we thank Aying Zhang (Nanjing Agricultural University) for providing the maize and tobacco seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.894710/full#supplementary-material
Footnotes
References
Atkinson, N. J., and Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. doi: 10.1093/jxb/ers100
Blein-Nicolas, M., Negro, S. S., Balliau, T., Welcker, C., Cabrera-Bosquet, L., Nicolas, S. D., et al. (2020). A systems genetics approach reveals environment-dependent associations between SNPs, protein coexpression, and drought-related traits in maize. Genome Res. 30, 1593–1604. doi: 10.1101/gr.255224.119
Campo, S., Peris-Peris, C., Montesinos, L., Peñas, G., Messeguer, J., and San Segundo, B. (2012). Expression of the maize ZmGF14-6 gene in rice confers tolerance to drought stress while enhancing susceptibility to pathogen infection. J. Exp. Bot. 63, 983–999. doi: 10.1093/jxb/err328
Chen, Y., Chen, Y., Shi, Z., Jin, Y., Sun, H., Xie, F., et al. (2019b). Biosynthesis and signal transduction of ABA, JA, and BRs in response to drought stress of kentucky bluegrass. Int. J. Mol. Sci. 20:1289. doi: 10.3390/ijms20061289
Chen, G., Wang, Y. Y., Wang, X. L., Yang, Q., Quan, X. Y., Zeng, J. B., et al. (2019a). Leaf epidermis transcriptome reveals drought-induced hormonal signaling for stomatal regulation in wild barley. Plant Growth Regul. 87, 39–54. doi: 10.1007/s10725-018-0450-0
Chini, A., Grant, J. J., Seki, M., Shinozaki, K., and Loake, G. J. (2004). Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J. 38, 810–822. doi: 10.1111/j.1365-313X.2004.02086.x
Das, K., and Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2:53. doi: 10.3389/fenvs.2014.00053
Foyer, C. H. (2018). Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ. Exp. Bot. 154, 134–142. doi: 10.1016/j.envexpbot.2018.05.003
Gong, H., Zhu, X., Chen, K., Wang, S., and Zhang, C. (2005). Silicon alleviates oxidative damage of wheat plants in pots under drought. Plant Sci. 169, 313–321. doi: 10.1016/j.plantsci.2005.02.023
Hasanuzzaman, M., Raihan, M. R. H., Masud, A. A. C., Rahman, K., Nowroz, F., Rahman, M., et al. (2021). Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int. J. Mol. Sci. 22:9326. doi: 10.3390/ijms22179326
Jacob, P., Hirt, H., and Bendahmane, A. (2017). The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 15, 405–414. doi: 10.1111/pbi.12659
Jiang, Y., Sun, Y., Zheng, D., Han, C., Cao, K., Xu, L., et al. (2021). Physiological and transcriptome analyses for assessing the effects of exogenous uniconazole on drought tolerance in hemp (Cannabis sativa L.). Sci. Rep. 11:14476. doi: 10.1038/s41598-021-93820-6
Jiang, M., and Zhang, J. (2002). Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 53, 2401–2410. doi: 10.1093/jxb/erf090
Julkowska, M. M., and Testerink, C. (2015). Tuning plant signaling and growth to survive salt. Trends Plant Sci. 20, 586–594. doi: 10.1016/j.tplants.2015.06.008
Kang, Y., Yang, X., Liu, Y., Shi, M., Zhang, W., Fan, Y., et al. (2021). Integration of mRNA and miRNA analysis reveals the molecular mechanism of potato (Solanum tuberosum L.) response to alkali stress. Int. J. Biol. Macromol. 182, 938–949. doi: 10.1016/j.ijbiomac.2021.04.094
Kim, T. W., Guan, S., Sun, Y., Deng, Z., Tang, W., Shang, J. X., et al. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11, 1254–1260. doi: 10.1038/ncb1970
Kim, T. W., and Wang, Z. Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61, 681–704. doi: 10.1146/annurev.arplant.043008.092057
Li, W. F., Shao, G., and Lam, H. M. (2008). Ectopic expression of GmPAP3 alleviates oxidative damage caused by salinity and osmotic stresses. New Phytol. 178, 80–91. doi: 10.1111/j.1469-8137.2007.02356.x
Li, Z., Shen, J., and Liang, J. (2019). Genome-wide identification, expression profile, and alternative splicing analysis of the brassinosteroid-signaling kinase (BSK) family genes in Arabidopsis. Int. J. Mol. Sci. 20:1138. doi: 10.3390/ijms20051138
Li, Z. Y., Xu, Z. S., He, G. Y., Yang, G. X., Chen, M., Li, L. C., et al. (2012). A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem. Biophys. Res. Commun. 426, 522–527. doi: 10.1016/j.bbrc.2012.08.118
Liu, Z. Y., Jia, Y. X., Ding, Y. L., Shi, Y. T., Li, Z., Guo, Y., et al. (2017). Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 66, 117–128. doi: 10.1016/j.molcel.2017.02.016
Liu, Y. B., Qin, L. J., Han, L. Z., Xiang, Y., and Zhao, D. G. (2015). Overexpression of maize SDD1 (ZmSDD1) improves drought resistance in Zea mays L. by reducing stomatal density. Plant Cell Tiss. Org. 122, 147–159. doi: 10.1007/s11240-015-0757-8
Liu, L., Xiang, Y., Yan, J., Di, P., Li, J., Sun, X., et al. (2021). Brassinosteroid-signaling kinase 1 phosphorylating calcium/calmodulin-dependent protein kinase functions in drought tolerance in maize. New Phytol. 231, 695–712. doi: 10.1111/nph.17403
Ma, F., Lu, R., Liu, H., Shi, B., Zhang, J., Tan, M., et al. (2012). Nitric oxide-activated calcium/calmodulin-dependent protein kinase regulates the abscisic acid-induced antioxidant defence in maize. J. Exp. Bot. 63, 4835–4847. doi: 10.1093/jxb/ers161
Majhi, B. B., Sreeramulu, S., and Sessa, G. (2019). Brassinosteroid-signaling kinase 5 associates with immune receptors and is required for immune responses. Plant Physiol. 180, 1166–1184. doi: 10.1104/pp.18.01492
Moradbeygi, H., Jamei, R., Heidari, R., and Darvishzadeh, R. (2020). Investigating the enzymatic and non-enzymatic antioxidant defense by applying iron oxide nanoparticles in Dracocephalum moldavica L. plant under salinity stress. Sci. Hortic. 272:109537. doi: 10.1016/j.scienta.2020.109537
Qi, Y., Tsuda, K., Glazebrook, J., and Katagiri, F. (2011). Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol. Plant Pathol. 12, 702–708. doi: 10.1111/j.1364-3703.2010.00704.x
Qian, Y. X., Ren, Q. Y., Zhang, J., and Chen, L. (2019). Transcriptomic analysis of the maize (Zea mays L.) inbred line B73 response to heat stress at the seedling stage. Gene 692, 68–78. doi: 10.1016/j.gene.2018.12.062
Qin, X., Duan, Z., Zheng, Y., Liu, W.-C., Guo, S., Botella, J. R., et al. (2020). ABC1K10a, an atypical kinase, functions in plant salt stress tolerance. BMC Plant Biol. 20, 1–13. doi: 10.1186/s12870-020-02467-4
Ren, H., Willige, B. C., Jaillais, Y., Geng, S., Park, M. Y., Gray, W. M., et al. (2019a). Brassinosteroid-signaling kinase 3, a plasma membrane-associated scaffold protein involved in early brassinosteroid signaling. PLoS Genet. 15:e1007904. doi: 10.1371/journal.pgen.1007904
Ren, Y. R., Yang, Y. Y., Zhang, R., You, C. X., Zhao, Q., and Hao, Y. J. (2019b). MdGRF11, an apple 14-3-3 protein, acts as a positive regulator of drought and salt tolerance. Plant Sci. 288:110219. doi: 10.1016/j.plantsci.2019.110219
Roberts, M. R. (2003). 14-3-3 proteins find new partners in plant cell signalling. Trends Plant Sci. 8, 218–223. doi: 10.1016/S1360-1385(03)00056-6
Sharma, P., Jha, A. B., Dubey, R. S., and Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. Aust. J. Bot. 2012, 1–26. doi: 10.1155/2012/217037
Shi, H., Shen, Q., Qi, Y., Yan, H., Nie, H., Chen, Y., et al. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25, 1143–1157. doi: 10.1105/tpc.112.107904
Shiu, S. H., Karlowski, W. M., Pan, R., Tzeng, Y. H., Mayer, K. F., and Li, W. H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. doi: 10.1105/tpc.020834
Späth, G. F., Drini, S., and Rachidi, N. (2015). A touch of Zen: post-translational regulation of the Leishmania stress response. Cell. Microbiol. 17, 632–638. doi: 10.1111/cmi.12440
Sreeramulu, S., Mostizky, Y., Sunitha, S., Shani, E., Nahum, H., Salomon, D., et al. (2013). BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 74, 905–919. doi: 10.1111/tpj.12175
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E., and Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytol. 203, 32–43. doi: 10.1111/nph.12797
Tang, W., Kim, T. W., Oses-Prieto, J. A., Sun, Y., Deng, Z., Zhu, S., et al. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321, 557–560. doi: 10.1126/science.1156973
Wang, P., Du, Y., Hou, Y. J., Zhao, Y., Hsu, C. C., Yuan, F., et al. (2015). Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. U. S. A. 112, 613–618. doi: 10.1073/pnas.1423481112
Wang, M., Liu, C., Li, S., Zhu, D., Zhao, Q., and Yu, J. (2013). Improved nutritive quality and salt resistance in transgenic maize by simultaneously overexpression of a natural lysine-rich protein gene, SBgLR, and an ERF transcription factor gene, TSRF1. Int. J. Mol. Sci. 14, 9459–9474. doi: 10.3390/ijms14059459
Wang, J., Shi, H., Zhou, L., Peng, C., Liu, D., Zhou, X., et al. (2017). OsBSK1-2, an orthologous of AtBSK1, is involved in rice immunity. Front. Plant Sci. 8:908. doi: 10.3389/fpls.2017.00908
Wang, G., Zhang, J., Wang, G., Fan, X., Sun, X., Qin, H., et al. (2014). Proline responding1 plays a critical role in regulating general protein synthesis and the cell cycle in maize. Plant Cell 26, 2582–2600. doi: 10.1105/tpc.114.125559
Xiang, Y., Bian, X., Wei, T., Yan, J., Sun, X., Han, T., et al. (2021). ZmMPK5 phosphorylates ZmNAC49 to enhance oxidative stress tolerance in maize. New Phytol. 232, 2400–2417. doi: 10.1111/nph.17761
Yan, H. R., Jia, H. H., Chen, X. B., Hao, L. L., An, H. L., and Guo, X. Q. (2014). The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 55, 2060–2076. doi: 10.1093/pcp/pcu133
Yan, J., Li, J., Zhang, H., Liu, Y., and Zhang, A. (2022). ZmWRKY104 positively regulates salt tolerance by modulating ZmSOD4 expression in maize. Crop J. 10, 555–564. doi: 10.1016/j.cj.2021.05.010
Yan, H., Zhao, Y., Shi, H., Li, J., Wang, Y., and Tang, D. (2018). Brassinosteroid-signaling kinase1 phosphorylates MAPKKK5 to regulate immunity in Arabidopsis. Plant Physiol. 176, 2991–3002. doi: 10.1104/pp.17.01757
Yang, Y., and Guo, Y. (2018). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804. doi: 10.1111/jipb.12689
Yang, X. Y., Zhao, T. Y., Rao, P., Gao, K., Yang, X., Chen, Z., et al. (2019). Transcriptome profiling of Populus tomentosa under cold stress. Ind. Crop. Prod. 135, 283–293. doi: 10.1016/j.indcrop.2019.04.056
Yao, W., Zhao, K., Cheng, Z., Li, X., Zhou, B., and Jiang, T. (2018). Transcriptome analysis of poplar under salt stress and over-expression of transcription factor NAC57 gene confers salt tolerance in transgenic Arabidopsis. Front. Plant Sci. 9:1121. doi: 10.3389/fpls.2018.01121
You, J., and Chan, Z. (2015). ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 6:1092. doi: 10.3389/fpls.2015.01092
Zhang, D., Jiang, S., Pan, J., Kong, X., Zhou, Y., Liu, Y., et al. (2014). The overexpression of a maize mitogen-activated protein kinase gene (ZmMPK5) confers salt stress tolerance and induces defence responses in tobacco. Plant Biol. 16, 558–570. doi: 10.1111/plb.12084
Zhang, B., Wang, X., Zhao, Z., Wang, R., Huang, X., Zhu, Y., et al. (2016). OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol. 170, 1149–1161. doi: 10.1104/pp.15.01668
Zhang, A., Zhang, J., Ye, N., Cao, J., Tan, M., Zhang, J., et al. (2010). ZmMPK5 is required for the NADPH oxidase-mediated self-propagation of apoplastic H2O2 in brassinosteroid-induced antioxidant defence in leaves of maize. J. Exp. Bot. 61, 4399–4411. doi: 10.1093/jxb/erq243
Zhang, A., Zhang, J., Zhang, J., Ye, N., Zhang, H., Tan, M., et al. (2011). Nitric oxide mediates brassinosteroid-induced ABA biosynthesis involved in oxidative stress tolerance in maize leaves. Plant Cell Physiol. 52, 181–192. doi: 10.1093/pcp/pcq187
Zhang, Y., Zhao, H., Zhou, S., He, Y., Luo, Q., Zhang, F., et al. (2018). Expression of TaGF14b, a 14-3-3 adaptor protein gene from wheat, enhances drought and salt tolerance in transgenic tobacco. Planta 248, 117–137. doi: 10.1007/s00425-018-2887-9
Zhao, Y., Du, H., Wang, Y., Wang, H., Yang, S., Li, C., et al. (2021). The calcium-dependent protein kinase ZmCDPK7 functions in heat-stress tolerance in maize. J. Integr. Plant Biol. 63, 510–527. doi: 10.1111/jipb.13056
Zhao, Y., Wu, G., Shi, H., and Tang, D. (2019). Receptor-like kinase 902 associates with and phosphorylates brassinosteroid-signaling kinase1 to regulate plant immunity. Mol. Plant 12, 59–70. doi: 10.1016/j.molp.2018.10.008
Zhou, H., Lin, H., Chen, S., Becker, K., Yang, Y., Zhao, J., et al. (2014). Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26, 1166–1182. doi: 10.1105/tpc.113.117069
Keywords: antioxidant defense enzyme, maize, protein interaction, reactive oxygen species, salt tolerance, ZmBSK1
Citation: Liu L, Sun Y, Di P, Cui Y, Meng Q, Wu X, Chen Y and Yuan J (2022) Overexpression of a Zea mays Brassinosteroid-Signaling Kinase Gene ZmBSK1 Confers Salt Stress Tolerance in Maize. Front. Plant Sci. 13:894710. doi: 10.3389/fpls.2022.894710
Edited by:
Yongfeng Guo, Tobacco Research Institute (CAAS), ChinaReviewed by:
Yuki Ohmuro-Matasuyama, Shimadzu Corporation, JapanÁgnes Szepesi, University of Szeged, Hungary
Copyright © 2022 Liu, Sun, Di, Cui, Meng, Wu, Chen and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanping Chen, Y2hlbnlwQGphYXMuYWMuY24=; Jianhua Yuan, eXVhbmpoMTEyM0AxNjMuY29t
 Lei Liu
Lei Liu Yanchao Sun1,2
Yanchao Sun1,2 Yanping Chen
Yanping Chen Jianhua Yuan
Jianhua Yuan