
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 01 July 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.893508
This article is part of the Research Topic Spotlight on Solanaceae Metabolism: Biotechnological Application, Volume II View all 5 articles
 Gaobo Yu1*†
Gaobo Yu1*† Qiusen Chen1†
Qiusen Chen1† Fengqiong Chen2
Fengqiong Chen2 Hanlin Liu1
Hanlin Liu1 Jiaxin Lin1
Jiaxin Lin1 Runan Chen1,3
Runan Chen1,3 Chunyuan Ren2
Chunyuan Ren2 Jinpeng Wei2,4
Jinpeng Wei2,4 Yuxian Zhang2
Yuxian Zhang2 Fengjun Yang1
Fengjun Yang1 Yunyan Sheng1
Yunyan Sheng1Reduced glutathione (GSH) is a key antioxidant, which plays a crucial role in the detoxification of xenobiotics in plants. In the present study, glutathione could reduce chlorothalonil (CHT) residues in tomatoes by inducing the expression of the UDP-glycosyltransferase (UGT) gene. In plants, UGT is an important glycosylation catalyst, which can respond to stresses in time by activating plant hormones and defense compounds. Given the importance of plant growth and development, the genome-wipe analyses of Arabidopsis and soybean samples have been carried out, though not on the tomato, which is a vital vegetable crop. In this study, we identified 143 UGT genes in the tomato that were unevenly distributed on 12 chromosomes and divided into 16 subgroups and found that a variety of plant hormones and stress response cis-elements were discovered in the promoter region of the SlUGT genes, indicating that the UGT genes were involved in several aspects of the tomato stress response. Transcriptome analysis and results of qRT-PCR showed that most SlUGT genes could be induced by CHT, and the expression of these genes was regulated by glutathione. In addition, we found that SlUGT genes could participate in plant detoxification through interaction with transcription factors. These findings further clarify the potential function of the UGT gene family in the detoxification of exogenous substances in tomatoes and provide valuable information for the future study of functional genomics of tomatoes.
In modern agriculture, pesticides have become an important factor to prevent crop losses and increase yield. However, excessive exposure to pesticides can lead to disruption of endocrine, mitochondrial (dys) function, and genetic and epigenetic modifications (Mostafalou and Abdollahi, 2013). Pesticides, therefore, pose a direct threat to human health. Moreover, they are phytotoxic to plants, potentially inhibiting plant growth, photosynthesis, and the synthesis of proteins, DNA, and pigments (Liu et al., 2021; Shahid et al., 2021). In addition, it is worth noting that pesticide residues may remain in plant food organs. Pesticide residues, seriously affect food safety and crop quality and even threaten human health and the environment, which is a global problem (Carvalho, 2017). Plant have effective antioxidant mechanisms to respond to the adverse environments and toxicity of xenobiotics (Peco et al., 2020). At the same time, plants could utilize a three-phase detoxification system to transform pesticides, which typically involves conversion, conjugation, and compartmentalization processes (Su et al., 2019). In the first stage, pesticides are transformed into less toxic products by catalyzing oxidation, reduction, and hydrolysis catalyzed through cytochrome P450 monooxygenase and peroxidase. The second stage involves coupling pesticide metabolites to sugars, amino acids, or glutathione by glutathione-S-transferase (GST) and uridine diphosphate UDP-glycosyltransferase (UGT), which is less toxic and more soluble in water. In the third stage, these metabolites are transported from the cytoplasm to the vacuoles or blastocysts in plants (Sun et al., 2018). Glutathione (GSH) is a small peptide consisting of three amino acids, which is an important antioxidant and free radical scavenger in plants. At the same time, it can be used as the substrate of GST to participate in the two-phase detoxification in plants, to effectively alleviate the stress of toxic xenobiotics (Geu-Flores et al., 2011).
UGT is an enzyme that catalyzes glycosylation, which generally catalyzes the transfer of the glycosyl group from nucleoside diphosphate-activated sugars to a wide range of substrates (Barvkar et al., 2012). It exists widely in plants and plays an important role in response to abiotic stress (Chen T. et al., 2020). The plant UGT has a conserved sequence of 44 amino acids at the C-terminus, which is the plant’s secondary product glycosyltransferase box (PSPG box), the PSPG box is responsible for glycosyl binding (Caputi et al., 2012). N-terminal sequences vary widely and are responsible for identifying different receptor molecules (Vogt and Jones, 2000).
The UGT has been extensively studied in various plant species. In Arabidopsis thaliana, Zea mays, Glycine max, and Citrus grandis, over 100UGTs have been identified and classified into 14–17 groups based on the conservation of their amino acid sequences (Li et al., 2001, 2014; Mamoon Rehman et al., 2016; Wu et al., 2020). The functions of many UGT genes have been identified in plants and are related to different pathways. In particular, great progress has been made in the study of stress-related UGT. For example, AtUGT73B3 and AtUGT73B5 could respond to pathogens, thereby increasing the resistance of Arabidopsis to biotic stress (Langlois-Meurinne et al., 2005). Overexpression of UGT85A5 could effectively enhance the salt tolerance of tobacco (Sun et al., 2013). UGT91Q2 in tea plants plays a key role in regulating cold tolerance (Zhao et al., 2020).
Although the UGT gene plays an important role in the resistance of many plants to abiotic stresses, the UGT gene family in tomatoes is still not well understood. Therefore, in this study, genome-wide identification of the UGT gene family of tomato was characterized. The potential function of the UGT gene family in response to abiotic stresses in tomatoes was further clarified by phylogenetic relationship, chromosome location, expression profile, and protein structure analysis.
The seeds of the tomato cultivar “Zheza205” were grown on mixed soil in pots. Once six true leaves were fully expanded, the plants were pretreated with ddH2O, 1 mM BSO, or 5 mM GSSG, respectively, and then sprayed with 11.2 mM CHT after 24 h of pretreatment. The leaves were sampled at 24 h after CHT treatment for physiological indexes and qRT-PCR analysis, while the samples were taken on the 7th day after CHT treatment for the determination of pesticide residues. Triplicates were maintained for each treatment and control sample. The samples were immediately frozen in liquid nitrogen and stored at −80°C until further analyses.
The tomato sample (5 g) was grounded with 20 ml methanol. The extracted supernatant was used for analysis by UPLC. The residue of CHT in tomato was determined according to the method of Jongen et al. (1991).
The content of NPT in tomato was determined according to the method of Pal et al. (2019). The 0.2 g sample was homogenized in 3 ml of 5-sulfosalicylic acid (6.67%) and centrifuged at 13,000 × g at 4°C for 10 min. Diluted with Ellman reagent (1:9), the supernatant was taken after 15 min and the absorbance was recorded at 412 nm.
One gram (FW) tomato leaves were homogenized in 5 ml of 100 mM sodium phosphate buffer (pH 7.5) to determine the antioxidant enzymes. The determination of superoxide dismutase (SOD) activity by measuring the inhibition of NBT reduction at 560 nm (Beauchamp and Fridovich, 1971). The determination of catalase (CAT) activity by monitoring the decomposition of H2O2 at 240 nm (Aebi, 1984). The determination of ascorbate peroxidase (APX) activity was measured by spectrophotometric monitoring is based on ascorbic acid oxidation (Nakano and Asada, 1981). The determination of glutathione peroxidase (GPX) activity was measured by monitoring the non-enzymatic oxidation of NADPH (Halliwell and Foyer, 1978).
The content of ascorbic acid (AsA) in tomato leaves was estimated by homogenizing 0.5 g of sample tissue at 5% (w/v) metaphosphoric acid (Mukherjee and Choudhuri, 1983). The reduction level glutathione (GSH) in tomato leaves was estimated by homogenizing fresh sample tissue in 5% (w/v) of sulpho-salicylic acid (Smith, 1985).
The method of Sinha (1972) was used to determine the concentration of hydrogen peroxide (H2O2) in tomato leaves. Homogenizing 0.2 g of tomato leaves in ice-cold 0.1 M of PBS (pH 7.0). After centrifugation, the absorbance value was measured at 570 nm based on H2O2 mediated Fe2+ oxidation. According to the method of Chen et al. (2019), 0.2 g fresh leaves were homogenized in ice-cold 50 mM of PBS (pH 7.8) for the determination of superoxide radicals (O2−) concentration in tomato leaves.
One hundred twelve known IDs of A. thaliana UGT genes were put into the Arabidopsis genome database (TAIR)1 for protein sequences. Using these Arabidopsis UGT protein sequences as probes, the 146 candidate members of the Solanum lycopersicum UGT family were searched using BLASTP. On the SMART database,2 the domain of Pfam (PF00201) was identified and screened in the phytozome database. Subsequently, the duplicate UGT gene was deleted by DNAMAN (version 7.212, Lynnon Corp., Quebec, Canada), and genes with related amino acid sequence <200 were removed. Finally, 143 SlUGT genes were obtained and named SlUGT1–SlUGT143 according to their chromosomal positions. The grand average of hydropathicity (GRAVY), the number of amino acids, theoretical isoelectric points (pI), and secondary structure of all predicted SlUGTs were then determined by ExPASy, and predicting the subcellular localization using Plant-mPLoc (Wilkins et al., 1999; Chou and Shen, 2010).
The SlUGT protein sequence were obtained from the NCBI protein database.3 Use Gblocks4 to remove fuzzy areas in the alignment, and use conserved areas to construct phylogenetic trees (Castresana, 2000). Use MUSCLE to create multiple sequence alignment of all SlUGTs (Zhang et al., 2021). A neighbor-joining (NJ) phylogenetic tree of full-length sequences of AtUGTs and SlUGTs was constructed with 1,000 bootstrap replicates, using MEGA 7.0 (Morgulis et al., 2008). Identification of putative homologous chromosomal regions between tomato genome, Zea genome, and A. thaliana genomes genome using MCScanX toolkit, and then visualized with TBtools (Yu et al., 2017).
The Gene Structure Display Server (GSDS v2.0)5 is used to determine the exon-intron sequences of SlUGT genes by comparing the coding sequence of each SlUGT gene with its genome sequence (Hu et al., 2015). Search for the novel motifs of SlUGTs using MEME 5.0.36 (Bailey et al., 2009).
To identify the potential cis-acting sequences responsible for stress response in the promoter region of the UGTs, the cis-elements of SlUGT promoter regions were detected using the plant cis-acting regulatory element (Plant CARE) database with the default parameters (Rombauts et al., 1999). The promoter region of SlUGT is the genomic DNA sequence of 2000 bp upstream of the initiation codon (ATG).
UGT genes expression data in different tissues were obtained from the online tomato eFP database.
Tomato seedlings with six leaves were pretreated with ddH2O, 1 mM BSO, or 5 mM GSSG, respectively, after 24 h pretreatment, which was treated with 11.2 mM of chlorothalonil (CHT). After 24 h exposure to CHT, the leaves were sampled and analyzed by transcriptome sequencing. The expression profiles of the differentially expressed genes were presented. Amazing HeatMap software (Chen C. et al., 2020) was used to generate a heatmap.
TRIzol® reagent (Invitrogen, Carlsbad, CA, United States) were used to extract total RNA. One microgram per RNA sample was used as the template for synthesis of the first-strand cDNA, using the ReverTra Ace™ qPCR RT Master Mix with gDNA Remover (TOYOBO Co., Osaka, Japan). Next, using SYBR® Select Master Mix RT-PCR System (Takara) performed qRT-PCR on an optical 96-well plate. Actin was used as an internal reference. All the primers used for gene expression analysis were shown in Supplementary Table S1. The relative expression level was calculated with the formula 2−ΔΔCT (Livak and Schmittgen, 2001; Cui et al., 2019). Three independent biological replicates were analyzed.
We used PlantRegMap (Internet; Tian et al., 2020)7 to predict transcription factors associated with SlUGT. Meanwhile, the transcription factors (TFs) network was visualized using Origin 8.0.
To evaluate the effect of glutathione on CHT accumulation in tomato leaves, the content of CHT in the leaves of tomatoes was analyzed under different treatments. Significant CHT accumulation was observed in the leaves of plants treated with CHT (Figure 1A). Interestingly, the application of BSO (Glutathione synthesis inhibitor) or GSSG on CHT-treated plants significantly increased the content of CHT in leaves by 26.0% and 46.1%, respectively, compared with that in only CHT-treated plants. In addition, we observed that exposure to CHT significantly increased the NPT content in leaves by 16.8%, while NPT content was inhibited with the application of BSO and GSSG (Figure 1B), suggesting that glutathione improved CHT detoxification in the tomato plant.
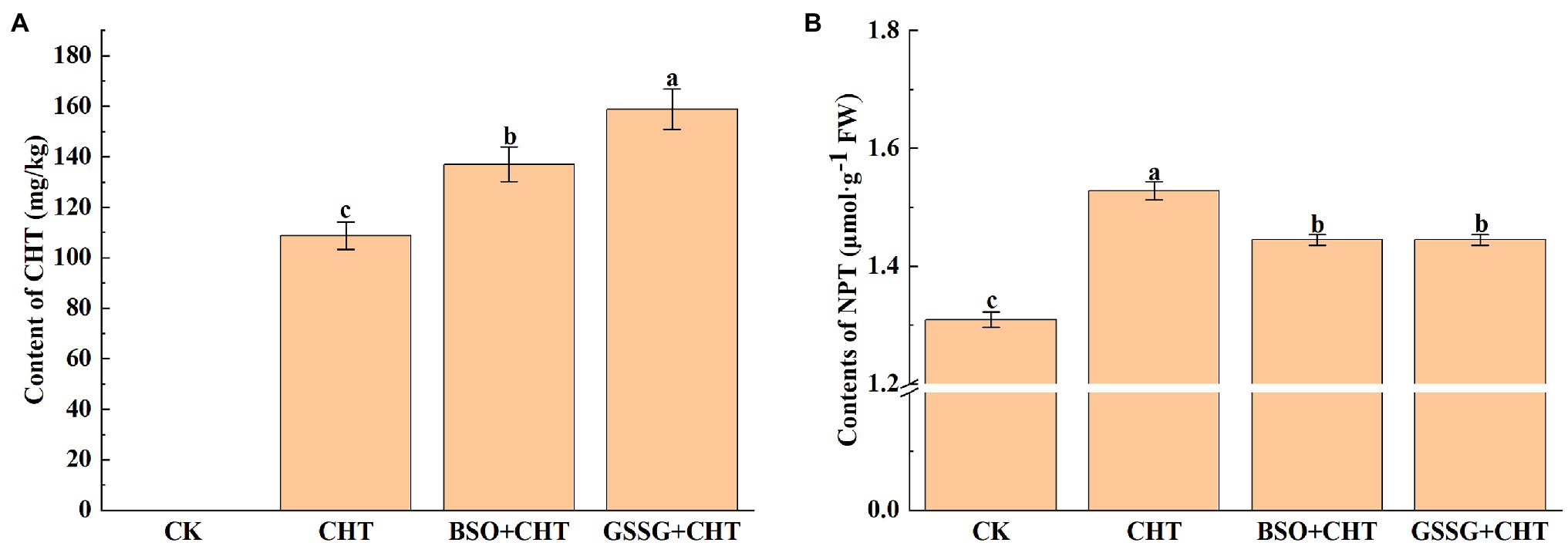
Figure 1. Pesticide residue (A) and NPT content (B) under exogenous treatment in tomato. NPT, non-protein thiols; CHT, chlorothalonil; BSO, L-buthionine-sulfoximine; and GSSG, glutathione (oxidized). Error bars represented the SD (n = 3). According to Duncan’s multiple test, bars with different letters were significantly different (p < 0.05).
To assess the CHT-caused injury to tomatoes, we analyzed ROS accumulation in tomatoes. The biochemical assay of ROS showed that CHT treatment significantly increased the H2O2 and O2− levels by 34.4% and 6.3%, respectively, compared to CK (Figures 2A,B). Meanwhile, a further increase was observed in the level of both H2O2 and O2− in tomatoes under CHT stress with BSO application, while only an increase in the content of O2− was found in tomatoes with GSSG treatment and no significant change in the production of H2O2.
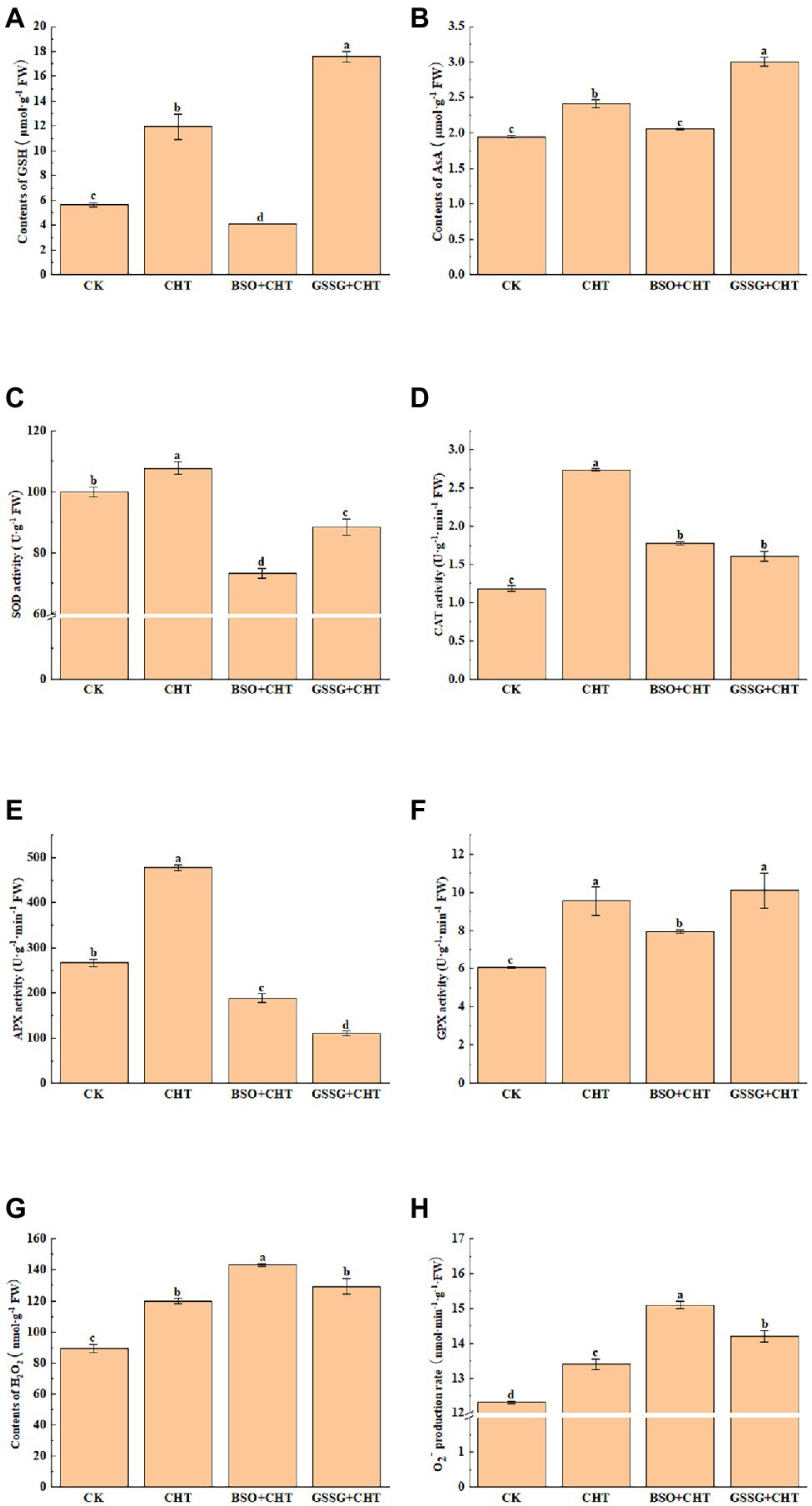
Figure 2. Effects of exogenous treatment on antioxidant system in tomato. (A) GSH content. (B) AsA content. (C) SOD activity. (D) CAT activity. (E) APX activity. (F) GPX activity. (G) H2O2 content. (H) O2− content. GSH, glutathione; AsA, ascorbic acid; SOD, superoxide dismutase; CAT, catalase; APX, ascorbate peroxidase; H2O2, hydrogen peroxide; O2−, superoxide anion; CHT, chlorothalonil; BSO, L-buthionine-sulfoximine; and GSSG, glutathione (oxidized). Error bars represented the SD (n = 3). According to Duncan’s multiple test, bars with different letters were significantly different (p < 0.05).
To unveil how glutathione alleviates CHT-induced oxidative stress, we measured the activity of antioxidant enzymes such as SOD, CAT, APX, and GPX. The CHT treatment significantly increased the activity of SOD, CAT, APX, and GPX by 7.8%, 30.3%, 79.2%, and 57.7%, respectively, compared with the control (Figures 2C–F). However, BSO administration substantially inhibited the activity of SOD, CAT, APX, and GPX by 32.0%, 35.0%, 60.5%, and 16.9%, respectively, compared with the only CHT treatment. Similarly, GSSG treatment also decreased the activity of antioxidant enzymes except for GPX, compared with the only CHT treatment.
To evaluate the remediation role of glutathione in ROS stress, we also investigated the non-enzymatic substances exposed to CHT. AsA and GSH were two common plant antioxidants. The CHT treatment significantly promoted the content of AsA and GSH by 23.6% and 111.0%, respectively, compared with the CK (Figures 2G,H). However, the application of BSO on CHT-treated plants significantly decreased the content of AsA and GSH in leaves by 26.0% and 46.1%, respectively, compared with that in only CHT-treated plants. These results suggested that glutathione alleviated CHT-induced oxidative stress by the antioxidant enzymes and non-enzymatic substances in tomatoes. Interestingly, GSSG application increased the levels of GSH and AsA, compared with that in only CHT-treated plants. This might be caused by the change in glutathione redox homeostasis.
P450, GST, UGT, and ABC play an important role in xenobiotic detoxification in plants. To further study the effect of glutathione on CHT detoxification as influenced, the gene expression of P450, GST, UGT and ABC were evaluated in tomato leaves. The results showed that the expression of P450, GST, UGT, and ABC increased after CHT treatment compared with the control. However, BSO administration substantially inhibited the expression of all these genes compared with the only CHT treatment. Similarly, GSSG treatment also reduced the expression of all these genes compared with the only CHT treatment (Figure 3), suggesting a role for glutathione in CHT detoxification in tomatoes.
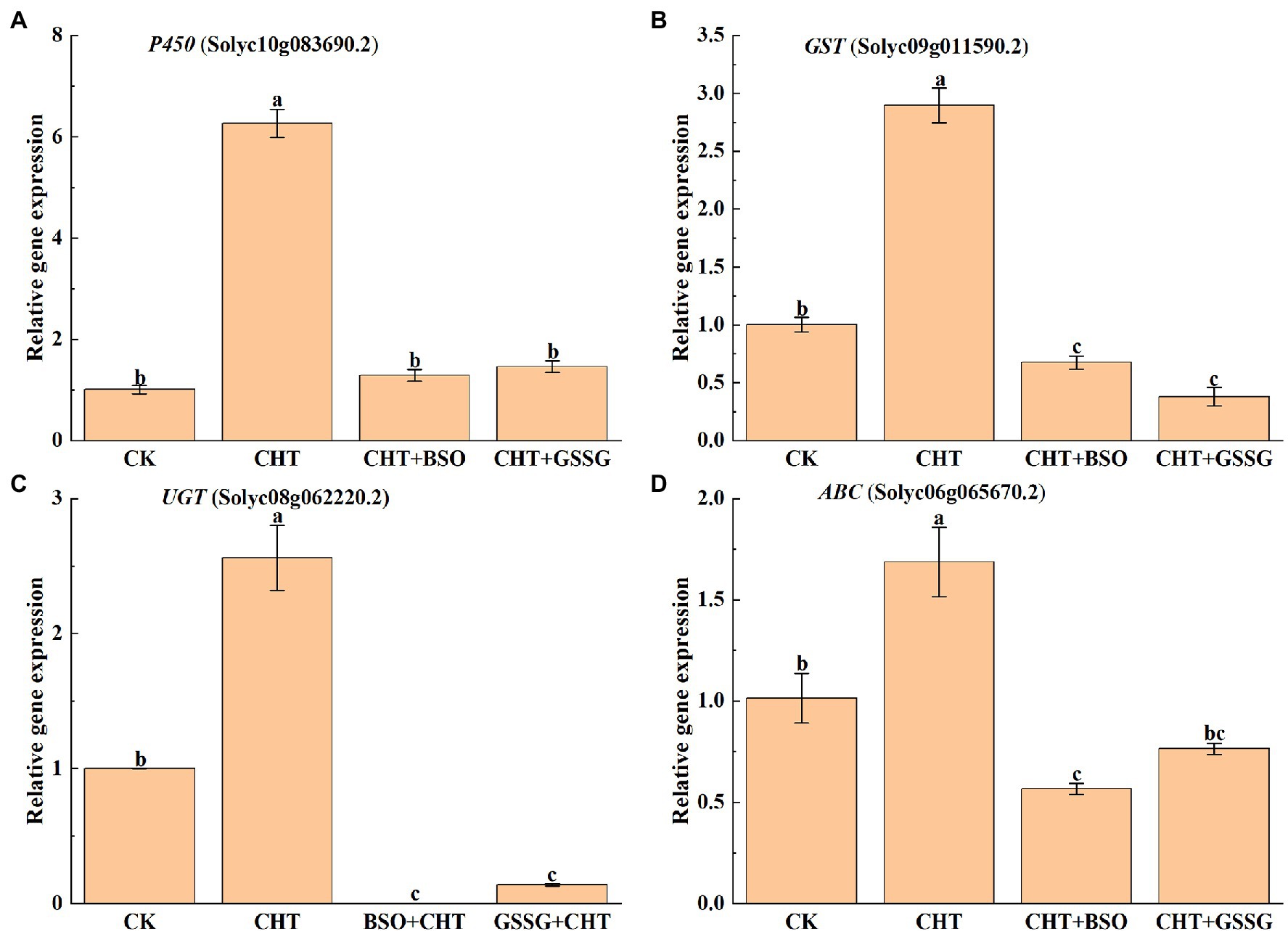
Figure 3. Effects of exogenous treatment on genes’ expression related to the detoxification system in tomato. (A) P450 expression. (B) GST expression. (C) UGT expression. (D) ABC expression. P450, cytochrome P450s; GST, glutathione S-transferase; UGT, UDP-glycosyltransferases; ABC, ATP-binding cassette transporters; CHT, chlorothalonil; BSO, L-buthionine-sulfoximine; and GSSG, glutathione (oxidized). Error bars represented the SD (n = 3). According to Duncan’s multiple test, bars with different letters were significantly different (p < 0.05).
Taking the A. thaliana UGT protein sequence as a reference, the candidate UGT protein was screened through a blast comparison of the tomato genome [SL2.50 (iTAG2.4)]. One hundred forty-three tomato UGT genes were screened for the conserved domain (PF00201) and de-redundancy. According to the position of the gene on the chromosome, the candidate gene was named SlUGT1 ~ SlUGT143 (Figure 4). The SlUGT protein sequence consisted of 292–952 amino acids, with an average length of 463 amino acids. Among them, 134 SlUGT proteins were acidic (with an isoelectric point <7), and only nine SlUGT proteins were alkaline (isoelectric point >7). The subcellular localization, as estimated by Plant-mPLoc, showed that SlUGT proteins were likely to be distributed in multiple organelles, including the peroxisome, mitochondria, and endoplasmic reticulum, though most SlUGTs were located in the chloroplast, cytoplasm, and nucleus. The total hydrophilic score of most SlUGT genes was negative, only three UGT genes (SlUGT29, SlUGT35, and SlUGT51) were positive in hydrophilicity, indicating that most SlUGT genes were hydrophilic (Supplementary Table S1).
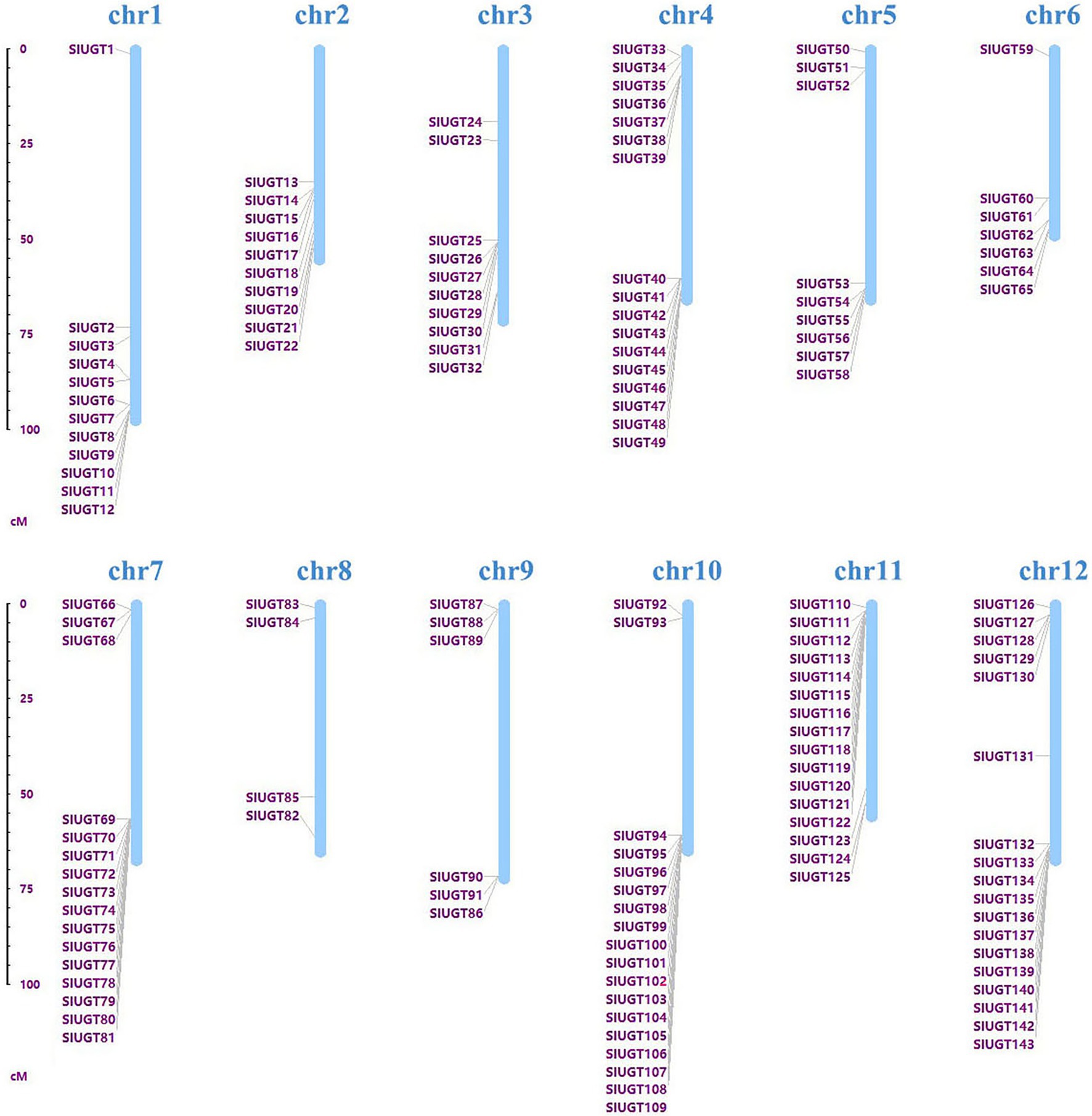
Figure 4. Chromosomal distribution of the 143 SlUGT genes identified in the present study. The chromosome number is indicated above each chromosome.
Multiple sequence alignment revealed that SlUGT proteins were highly conserved. The phylogenetic tree of UGT genes in the tomato, Z. mays, and A. thaliana was constructed by the neighbor-joining (NJ) method (One UGT gene was selected from each subfamily of Z. mays, and A. thaliana). The results indicated a classification of 16 major groups (A–P) in tomatoes, including two newly discovered groups (O and P), which were absent in Arabidopsis but present in higher plants like maize (Figure 5). These UGT groups (A–P) consisted 26, 2, 2, 18, 18, 2, 11, 5, 2, 1, 5, 18, 3, 1, 25, and 4 members, respectively. To clarify the evolutionary relationship of the SlUGT gene family, we constructed a collinear map between the tomato and one dicotyledon (A. thaliana) and one monocotyledon (maize)—12 collinear relationships were found between the UGT of tomato and dicotyledons and only two with monocotyledons (Figure 6).
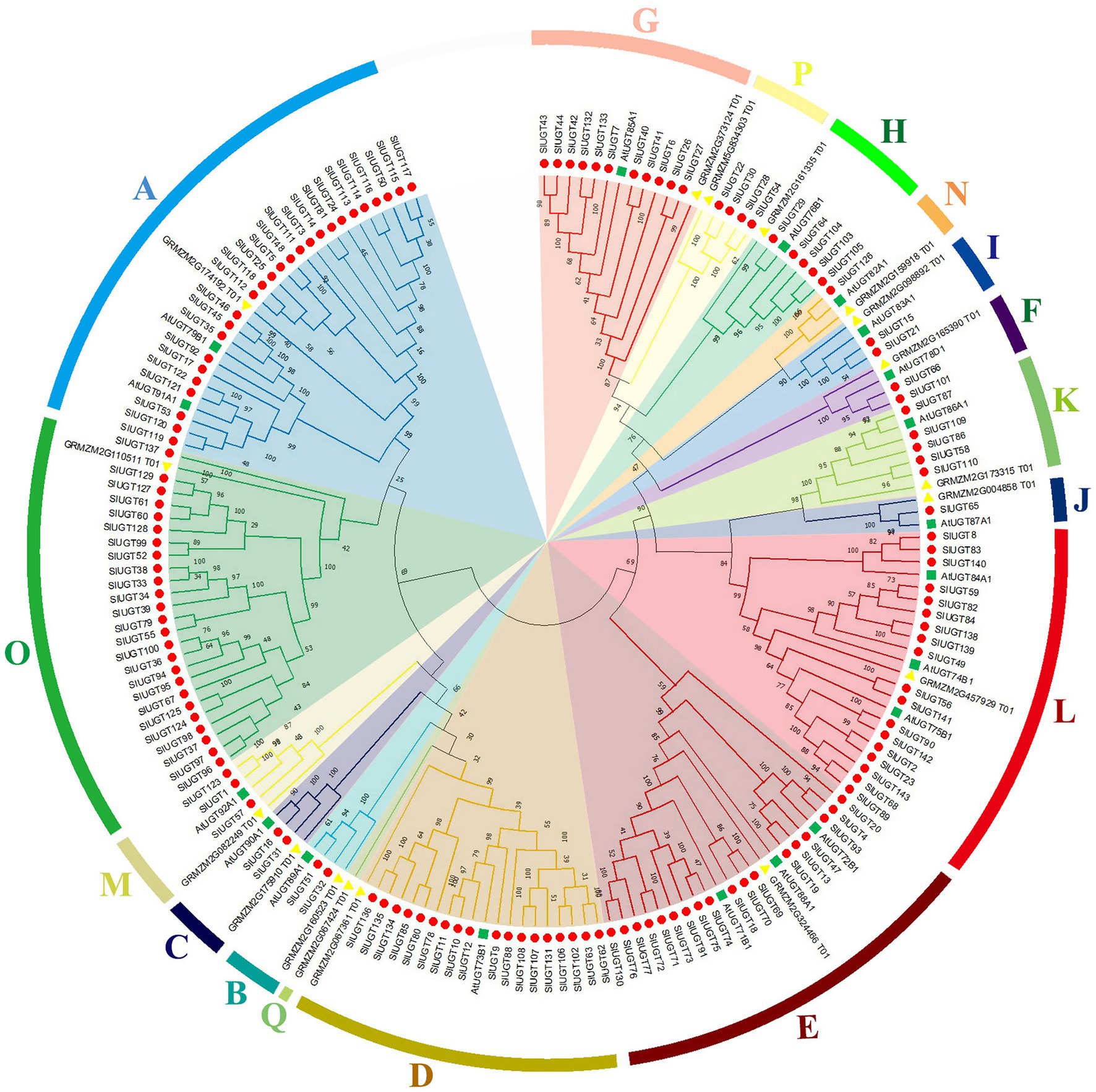
Figure 5. Phylogenetic analysis of UGT family genes from tomato. The tree was constructed using the neighbor-joining method by aligning the amino acid sequences of tomato UGTs, Zea mays UGTs, and Arabidopsis UGTs.
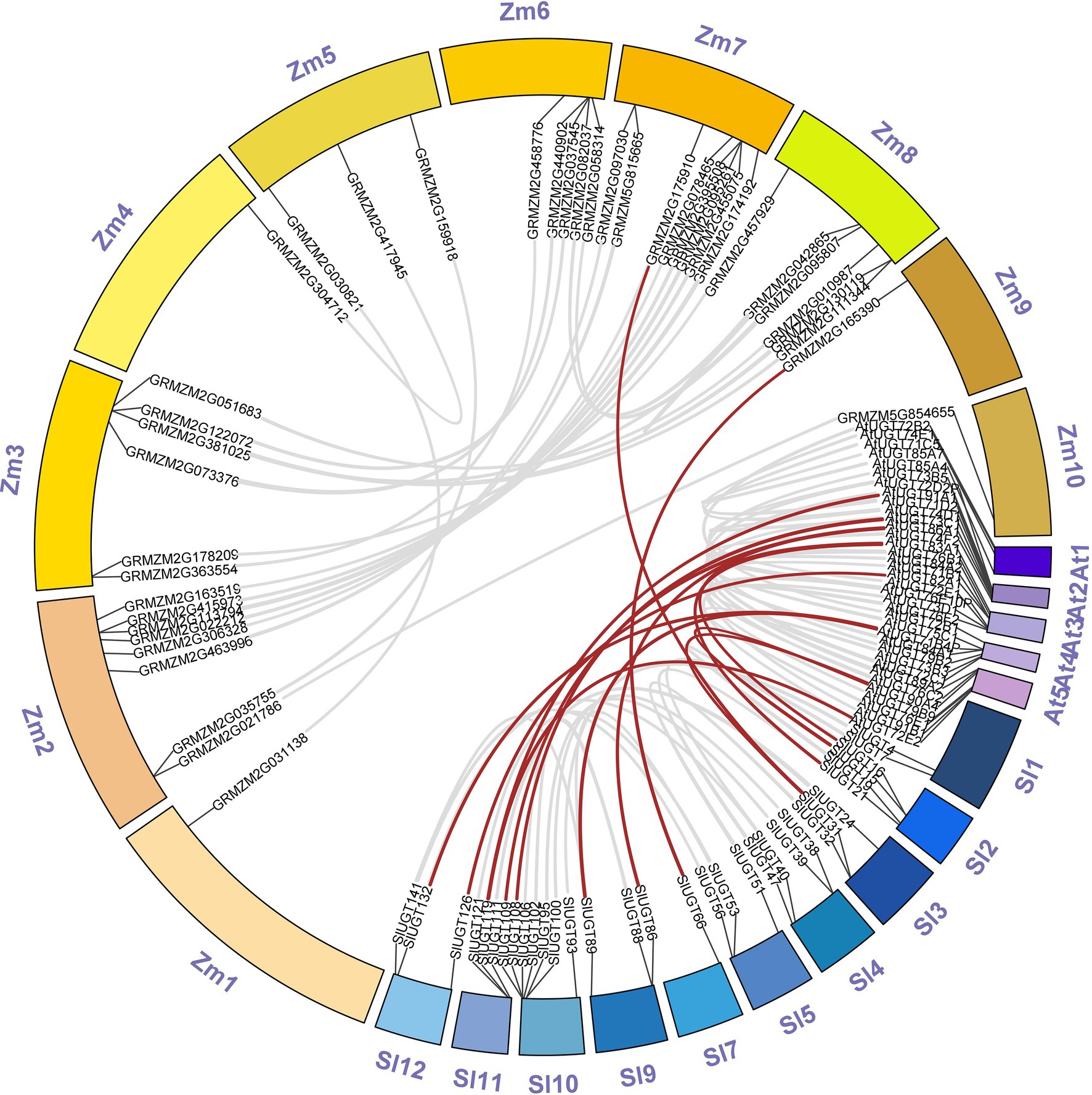
Figure 6. Synteny analysis of UGT genes between tomato and two representative plant species (Arabidopsis thaliana and Z. mays). “Sl” was on behalf of chromosomes of Solanum lycopersicum, “At” was on behalf of chromosomes of A. thaliana and “Zm” was on behalf of chromosomes of Z. mays.
To study the regulatory mechanism of the SlUGT gene, the sequence of the promoter codon 2000 bp upstream of (ATG) was scanned in the PlantCARE database, and a variety of cis-acting elements related to plant hormones and stress responses were obtained (Figure 7). Hormone response elements were mainly induced by Abscisic Acid (ABA), methyl jasmonate (MeJA), gibberellin (GA), indole acetic acid (IAA), and salicylic acid (SA). Among them, 108 SlUGT genes with ABA response elements were found, accounting for 75.5% of the total genes. In addition, 72 SlUGT genes containing MeJA response elements and 63 SlUGT genes containing GA elements were revealed. Furthermore, 47 SlUGT genes including IAA response elements and 40 SlUGT genes with at least one SA response element were discovered. Interestingly, the predicted stress response elements included MBS (drought induction), DRE (drought induction), LTR (low-temperature response), ARE (anaerobic induction), and defense and stress-responsive elements, among which, SlUGT genes containing ARE elements were the most frequent. Additionally, a large number of light response-regulatory elements (such as MRE), circadian response elements, and MYB (drought induction) binding sites (such as MBSI) were revealed in the promoter regions of several SlUGT genes. These results demonstrated that SlUGT genes were involved in response to a variety of stresses and hormones, to adapt to diverse environmental changes.
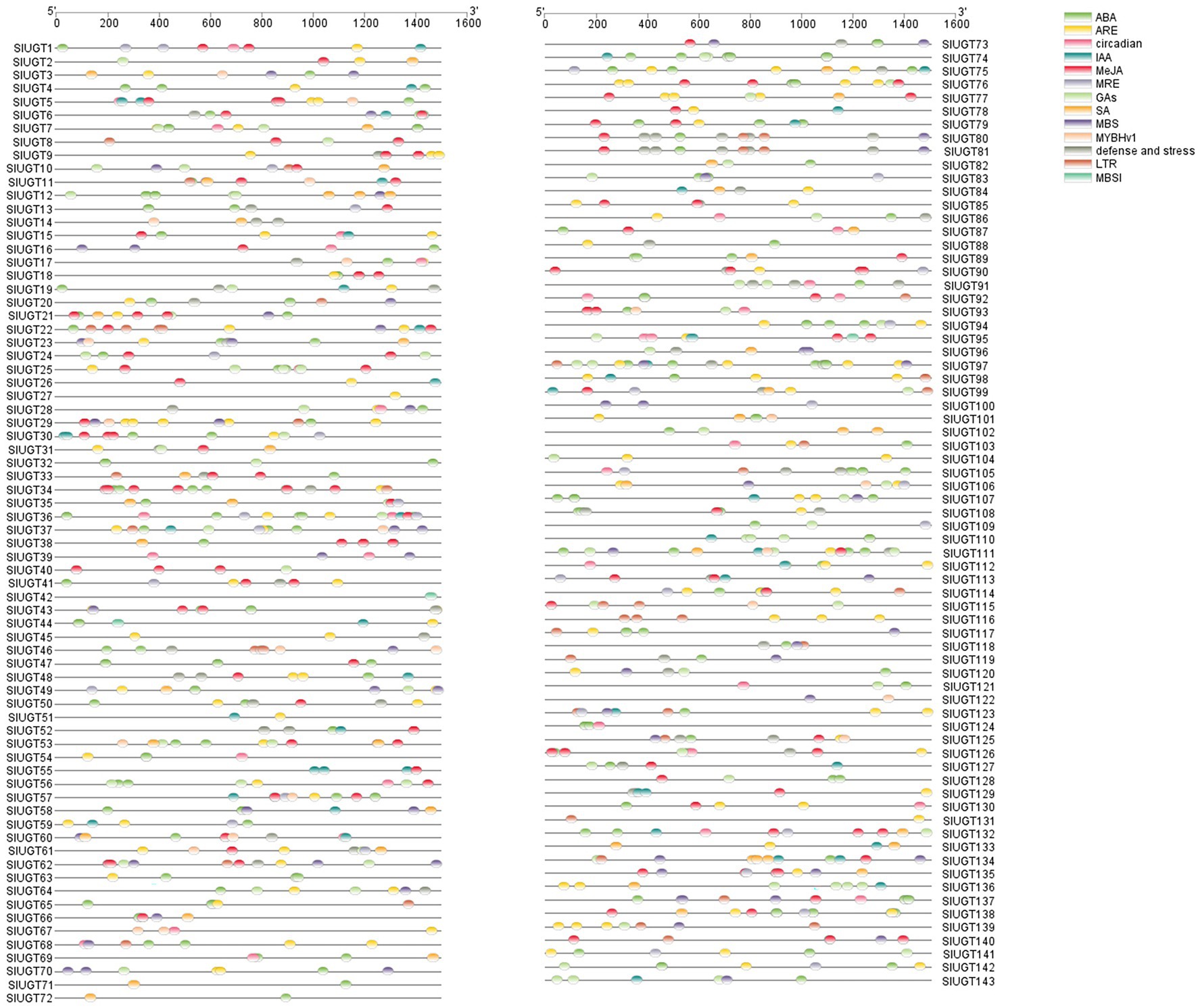
Figure 7. Analysis of the cis-acting elements in SlUGT genes. Different colors represented different cis-acting elements and their position in the SlUGT genes.
To identify the conserved structure of the SlUGT protein, 20 motifs were predicted by MEME motif analysis. The results revealed that most SlUGT proteins contained 11–17 motifs, while SlUGT14, SlUGT23, and SlUGT126 contained 10 motifs, SlUGT46 contained nine motifs, SlUGT77 and SlUGT134 contained only eight motifs (Figure 8). To understand the evolution of the SlUGT family, we visualized the exon/intron structure of the SlUGT gene (Figure 8). Most UGT genes contained 1–3 exons., and only UGT45 contained nine exons. Therefore, the gene structure and conserved motif analyses demonstrated that the SlUGTs were highly conserved.
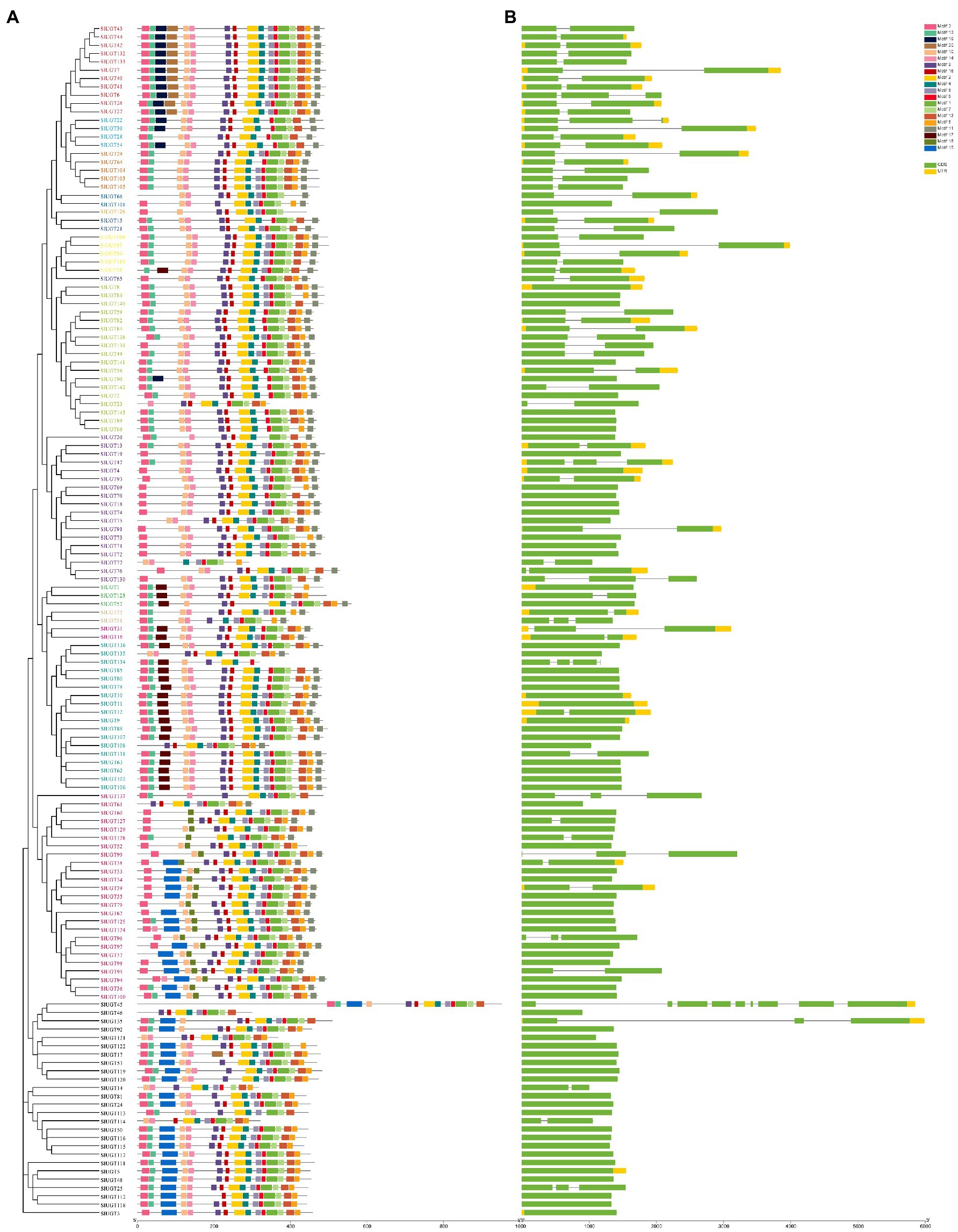
Figure 8. Gene structure and architecture of conserved motifs in SlUGT genes. (A) The motif composition of SlUGT proteins. Different colors represented different motifs. (B) Exon-intron structure and conserved domain of SlUGT genes.
To study the tissue-specific expression patterns of members of the SlUGT gene family, we analyzed the transcriptome data of UGTs present in roots, leaves, flowers, buds, and fruits in wild tomatoes (Solanum pimpinellifolium L.). As shown in Figure 9, most of the SlUGT genes, except for SlUGT23, SlUGT48, SlUGT72, and SlUGT113, could be expressed in tissue sites. Among them, 90 UGT genes were expressed in all tissues, accounting for 62.9% of the total UGT genes, while SlUGT3and SlUGT14, which were only expressed in buds, SlUGT53, SlUGT59, SlUGT110, and SlUGT112, which were only expressed in roots, and SlUGT2, SlUGT71 were only expressed in fruits. These results suggested that SlUGT was expressed in a tissue-specific manner in the tomato.
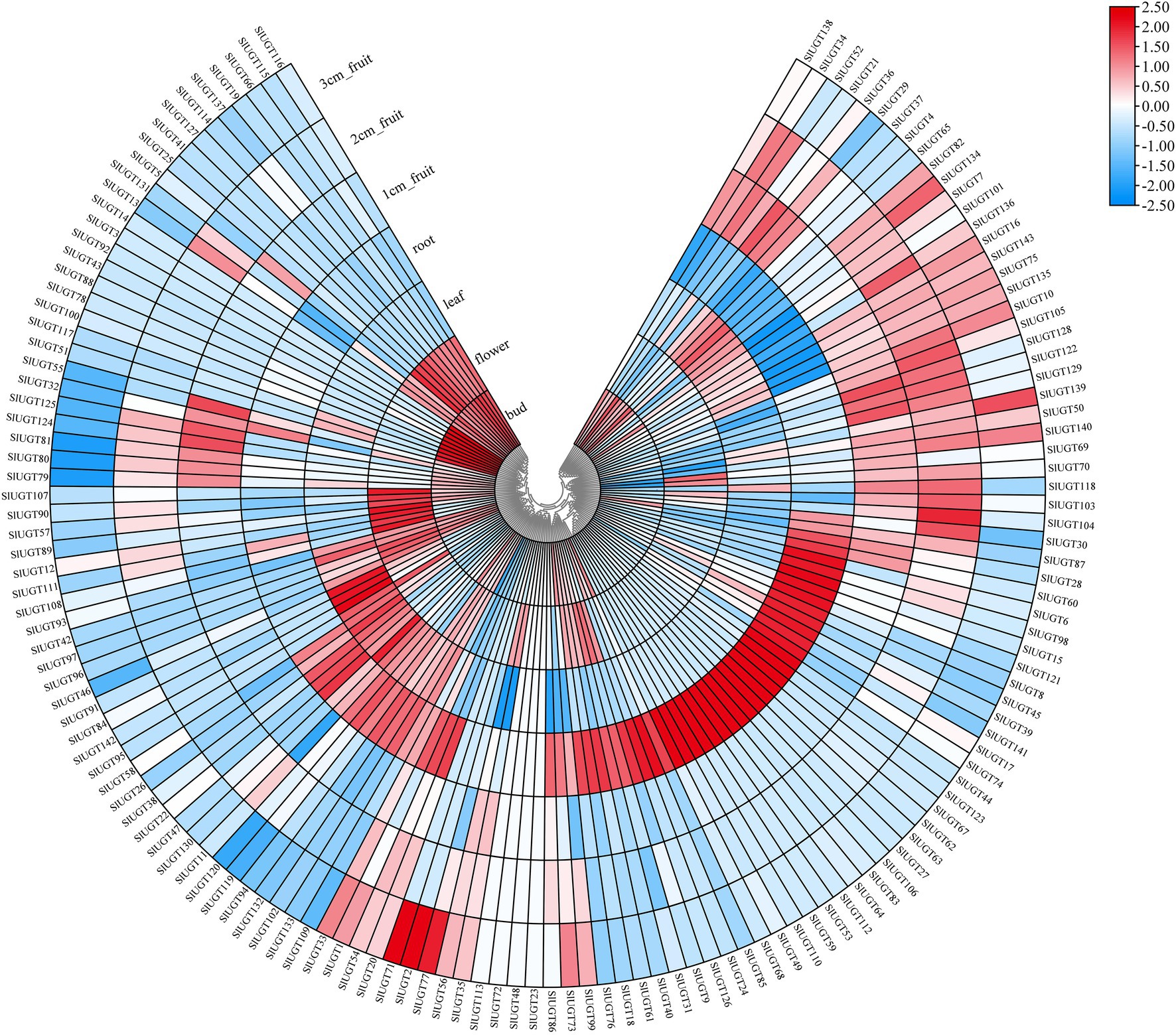
Figure 9. Expression profiles of SlUGT genes in different tissues. The expression levels were based on the transcriptome data. All gene expression levels were log scaled by row.
Based on the transcriptome data, we analyzed the SlUGT gene family members’ response to CHT and glutathione. The expression of 52 genes was upregulated, while that of 75 genes was downregulated after CHT treatment compared with the control (Figure 10). The transcription of 59 UGT genes was induced, however, that of 69 UGT genes was decreased by BSO treatment compared with the only CHT treatment. Similarly, the transcription of 67 UGT genes was induced, however, that of 61 UGT genes was decreased by GSSG treatment compared with the only CHT treatment (Figure 10). It showed that UGT genes responded to glutathione under CHT treatment.
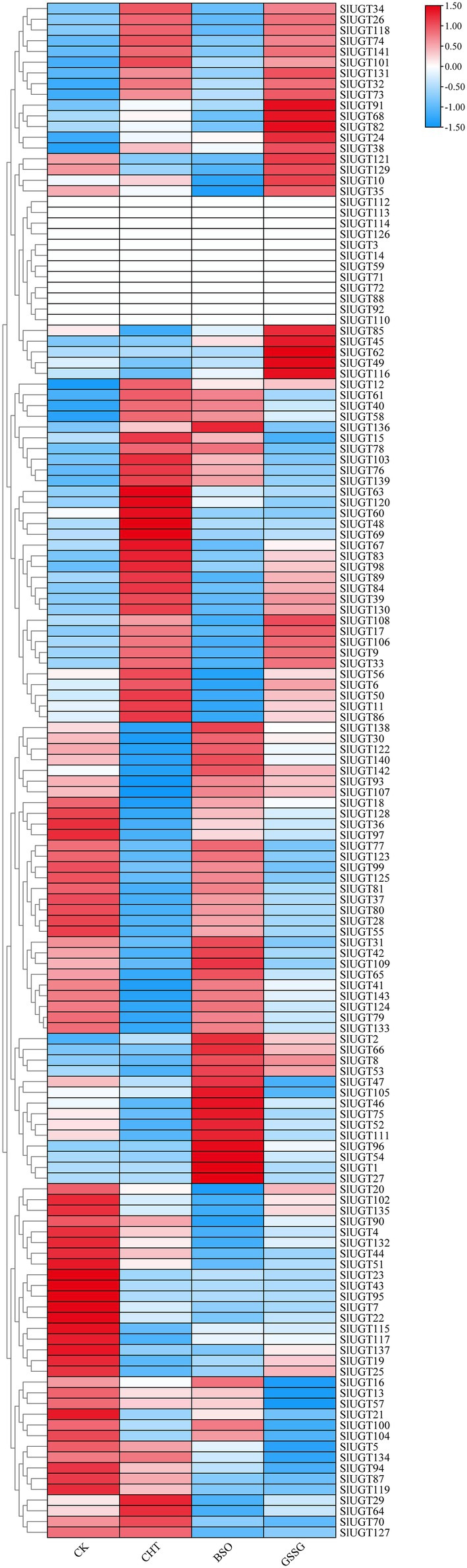
Figure 10. Expression profile of SlUGT genes in transcriptome data under different treatments. All gene expression levels were log scaled by row.
To confirm the effect of glutathione on SlUGT gene expression under CHT treatment, we selected several representative SlUGT genes and explored their genetic expression after treatment with BSO and GSSG under CHT treatment by qRT-PCR. After CHT treatment, the expression of eight UGT genes selected was significantly up-regulated compared with control, while after BSO and GSSG treatment, the expression of eight UGT genes was significantly inhibited compared with only CHT treatment (Figure 11), suggesting that they might be the key UGT genes induced by glutathione involved in the response to CHT stimulation in tomato.
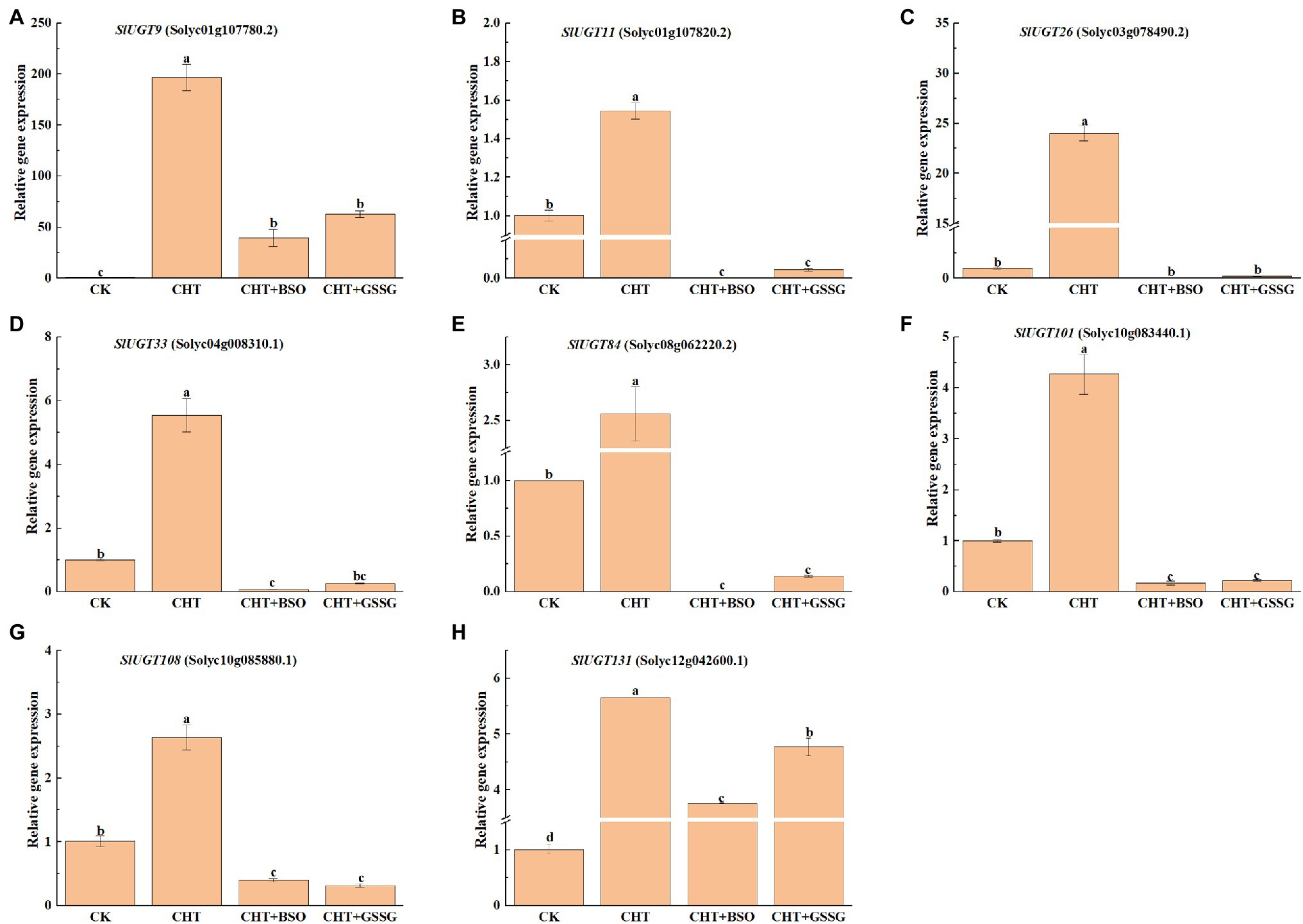
Figure 11. Expression profiles of eight representative SlUGT genes in response to different treatments. (A) SlUGT9 expression. (B) SlUGT11 expression. (C) SlUGT26 expression. (D) SlUGT33 expression. (E) SlUGT84 expression. (F) SlUGT101 expression. (G) SlUGT108 expression. (H) SlUGT131 expression. Error bars represented the SD (n = 3). According to Duncan’s multiple tests, bars with different letters were significantly different (p < 0.05).
To further explore the regulatory mechanism of the SlUGT gene, we predicted that a total of 12 transcription factor (TF) families interacted with the SlUGT gene (Figure 12). Among all SlUGT genes, nine transcription factors were predicted to interact with SlUGT73 and SlUGT75, with the largest number. While the transcription factor MYB interacted with 51 UGT genes, accounting for 35.7%. Interestingly, transcriptome data showed that each of the 12 transcription factor families contained genes that could be induced by CHT, while inhibited by BSO or GSSG treatment. The above results revealed that the UGT gene could participate in the regulation of plant growth and development and stress response through interaction with transcription factors in tomatoes, which meant that the SlUGT gene played an important role in plant detoxification through interaction with transcription factors.
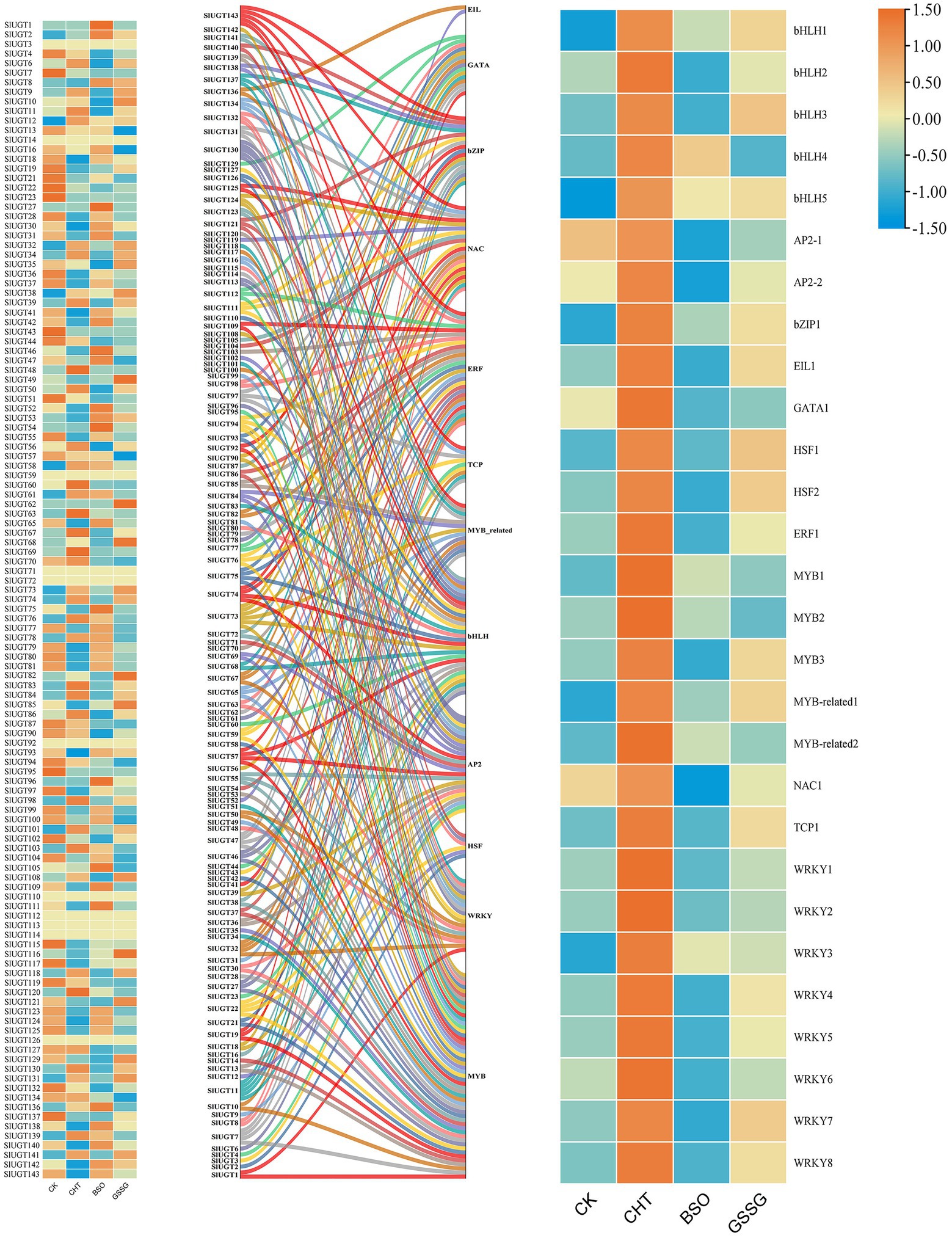
Figure 12. Network analysis of tomato UGT genes and transcription factors and Expression profiles of different SlUGT genes and transcription factors in response to different treatments. Different colors represent different SlUGT genes.
Fungicides are an essential part of intensive crop production, especially for economically important crop varieties (Alengebawy et al., 2021). It has been shown that approximately 50,000 plant pathogens can contribute to significant crop losses (Zhang, 2018). CHT is a widely used fungicide with an astonishingly high annual use (about 8,000 tons/year, such as in China; Wang et al., 2022). However, the massive use of pesticides will not only produce benefits but also cause excessive pesticide residues, which not only endangers the growth of plants and human health but also has a serious impact on the whole ecological environment (Ramel et al., 2012). It has been found that melatonin could promote BPA metabolism by improving glutathione-dependent detoxification of tomatoes (Kanwar et al., 2020). As a key antioxidant and redox buffer, GSH is widely distributed in all the cell compartments in plant (Noctor et al., 2012). In this study, we utilized BSO, a kind of glutathione inhibitor, and GSSG, Oxidized glutathione, to explore the effect of glutathione on the tolerance of CHT phytotoxicity in tomatoes. We found that BSO reduced NPT content under CHT stress, and improved CHT accumulation in tomatoes, suggesting that glutathione promoted CHT metabolism in tomato plants. Similarly, GSSG treatment could also increase the accumulation of chlorothalonil, although there was no significant difference in NPT content between GSSG treatment and pesticide treatment alone. Jung et al. (2021) also found that foliar application of glutathione could reduce cadmium-induced oxidative stress.
The production of ROS production (H2O2 and O2−) in plant cells increased under stress. Studies have shown that glutathione can improve the salt tolerance of sorghum by scavenging ROS (Punia et al., 2021). In addition, a study on upland cotton plants under lead (Pb) stress proved the importance of GSH in HMS detoxification (Khan et al., 2016). Our results also show that exposure to the CHT leads to an increase of reactive oxygen species in tomatoes, suggesting that the CHT induces oxidative stress in tomatoes through the excessive accumulation of ROS. Consistent with the ROS content, an increased CHT accumulation increased the antioxidant enzymes activity and the content of non-enzymatic substances, indicating that ROS levels may exceed antioxidant elimination capacity, although plants can improve their scavenging capacity of ROS to resist again stress. Interestingly, when glutathione was inhibited, the content of ROS in tomatoes increased further, while the content of non-enzymatic antioxidants and the activity of antioxidant enzymes decreased significantly, revealing that glutathione enhanced antioxidant capacity to alleviate the CHT-induced oxidative stress in tomatoes, and then degraded the CHT residue in tomato.
In general, xenometabolism in both animals and higher plants consisted of three processes: transformation (phase I), binding (phase II), and compartmentalization (phase III; Yu et al., 2013). Glutathione (GSH) is a kind of low-molecular-weight thiol, which can be used as the substrate of GST to participate in phase II detoxification of plants (Geu-Flores et al., 2011). It has been found that ABC transporters can participate in the detoxification of CHT in tomatoes (Szabados and Savour’e, 2010). In addition, Hou et al. (2018) observed that the CHT degradation promoted by EBR was related to the increase of GST activity. In the present study, the expression of detoxification genes (including P450, GST, UGT, and ABC) increased after CHT treatment compared with the control. However, BSO and GSSG administration substantially inhibited the expression of all these genes compared with the only CHT treatment. It revealed that glutathione could degrade the residual CHT in tomatoes by inducing the expression of detoxification genes related to glutathione.
It is reported that UGT also plays a role in various abiotic stress and xenobiotic detoxification in plants (Mamoon Rehman et al., 2016). However, the role of UGT genes in tomatoes has not been studied. Previous studies have identified a total of 145 UGT genes in C. grandis,147 UGT genes in Z. mays, 179 UGT genes in Triticum aestivum, and 180 UGT genes in Oryza sativa (Caputi et al., 2012; Li et al., 2014; He et al., 2018; Wu et al., 2020). In the present study, a total of 143 UGT genes were identified in tomato, these UGT genes were divided into 16 groups, including 14 highly conserved groups (A–N) and two newly discovered groups O and P, which were absent in Arabidopsis, while Q Group only existed in maize, suggesting that this group might exist only in monocotyledons and could play a significant role in the UGT gene evolution of monocotyledons. In addition, Arabidopsis and tomato belong to dicotyledonous plants. There are a large number of collinear relationships between them, while there are only two collinear relationships with the monocotyledonous plant (maize). This result is consistent with the evolutionary relationship between dicotyledonous and monocotyledonous plants.
Introns are important components of genes. Although they are not involved in protein coding, the acquisition or loss of introns and the insertion position of introns relative to protein sequences are generally considered to be key clues to understanding the evolutionary diversity of gene families (Rogozin et al., 2000). Gene structure and motif analysis showed that the SlUGT protein was highly conserved, and most members of the SlUGT gene family contained a typical motif, and motif4 existed in all SlUGT genes. SlUGT genes of the same clade contained a similar number of exons/introns and motifs. In addition, some clades included a unique motif, such as, motif 20 was specific to group G, which might be the reason that these clades could perform specific functions.
Abiotic stress resistance is one of the important traits during tomato breed improvement (Cortina et al., 2005; Almaghamsi et al., 2021). In recent years, it has been found that UGT genes of Melilotus albus may participate in various defense responses against biotic and abiotic stresses (Duan et al., 2021). It has also been found that UGT genes could regulate tolerance under FHB (Fusarium head blight) in wheat (He et al., 2018). Here, we also found several plant hormone and stress response-regulatory elements in the promoter region of SlUGT genes. It showed that UGT genes in tomatoes were related to plant hormones and stress regulation. At the same time, transcriptome data revealed that most UGT genes could be induced or inhibited by CHT, and the expression of these genes was also changed after pretreatment with BSO or GSSG. This suggested that the SlUGT gene family played an important role in plant detoxification, which might be regulated by glutathione. QRT-PCR analysis further confirmed the above conclusions. Interestingly, most of the UGT genes induced by CHT belonged to subgroup D, indicating that this subgroup played a major role in tomato detoxification.
Transcription factors (TFs) are important proteins, which bind to specific DNA motifs, regulate gene expression and play an important role in plant stress response (Wu et al., 2022). The present study found that 121 tomato UGT genes interacted with 12 different transcription factors, 90% of which were regulated by glutathione under CHT stress. It could be seen that glutathione might promote the metabolism of pesticides in tomatoes by regulating the interaction between the UGT gene and transcription factors. In addition, among them, 54 UGT genes had complex relationship networks with MYB transcription factors, and the MYB transcription factor has been proved to be related to abiotic stress in plants (Dar et al., 2022). It has been reported that the UGT gene could work with transcription factors to affect the stress response of cassia seeds (Deng et al., 2018). Therefore, SlUGT genes could participate in the regulation of stress through the interaction with these transcription factors.
In summary, we found that CHT treatment-induced oxidative stress in tomatoes by producing ROS. However, glutathione increased the activity of antioxidative enzymes, the content of non-enzymatic substances, and the expression of detoxification genes to scavenge ROS and reduce the accumulation of CHT in tomato plants. In addition, we presented the first instance of comprehensive information about UGT family genes in tomatoes. The phylogenetic analysis showed that 143 SlUGT genes could be grouped into 16 subfamilies (Group A–P) with conserved domains and motifs. Most of the identified genes are expressed in different tissue types. The promoter region of SlUGT genes contained several plant hormone- and stress response-regulatory elements. Transcriptome data and qRT-PCR results concurred that most SlUGT genes could be induced by CHT, and the expression of these genes is regulated by glutathione. We also found SlUGT gene could participate in plant detoxification through interaction with transcription factors, which conferred a new direction for studies on the function of the SlUGT gene.
The data presented in the study are deposited in the NCBI repository, available at: https://dataview.ncbi.nlm.nih.gov/object/PRJNA835910?reviewer=kt60ukoga0butbtus3p7lk4nt.
GY: conceptualization, writing-review and editing, and funding acquisition. QC: methodology, formal analysis, writing-original draft. FC: investigation. HL: data curation. JL: software. RC: investigation. CR: data curation. JW: writing-review and editing. YZ: resources and funding acquisition. FY: resources. YS: resources. All authors have read and agreed to the published version of the manuscript.
This work was financially supported by the Postdoctoral Scientific Research Developmental Fund of Heilongjiang (LBH-Q20052), the Fund Program for Overseas Returnees, National Natural Science Foundation of China (31301769), and Heilongjiang Bayi Agricultural University PhD Research Initiation Fund, Heilongjiang Bayi Agricultural University Support Program for San Heng San Zong (TDJH202004), the Natural Science Foundation of Heilongjiang Province (QC2018023).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.893508/full#supplementary-material
1. ^http://www.arabidopsis.org/
2. ^http://smart.embl-heidelberg.de/
3. ^http://www.ncbi.nlm.nih.gov/protein/
4. ^http://phylogeny.lirmm.fr/phylo_cgi/one_task.cgi?task_type=gblocks
5. ^http://gsds.cbi.pku.edu.cn/
Alengebawy, A., Abdelkhalek, S. T., Qureshi, S. R., and Wang, M. Q. (2021). Heavy metals and pesticides toxicity in agricultural soil and plants: ecological risks and human health implications. Toxics 9:42. doi: 10.3390/toxics9030042
Almaghamsi, A., Nosarzewski, M., Kanayama, Y., and Archbold, D. D. (2021). Effects of abiotic stresses on sorbitol biosynthesis and metabolism in tomato (Solanum lycopersicum). Funct. Plant Biol. 48, 286–297. doi: 10.1071/FP20065
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Barvkar, V. T., Pardeshi, V. C., Kale, S. M., Kadoo, N. Y., and Gupta, V. S. (2012). Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genomics. 13:175. doi: 10.1186/1471-2164-13-175
Beauchamp, C., and Fridovich, I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287. doi: 10.1016/0003-2697(71)90370-8
Caputi, L., Malnoy, M., Goremykin, V., Nikiforova, S., and Martens, S. (2012). A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 69, 1030–1042. doi: 10.1111/j.1365-313X.2011.04853.x
Carvalho, F. P. (2017). Pesticides, environment, and food safety. Food Energy Secur. 6, 48–60. doi: 10.1002/fes3.108
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, T. T., Liu, F. F., Xiao, D. W., Jiang, X. Y., Li, P., Zhao, S. M., et al. (2020). The Arabidopsis UDP-glycosyltransferase75B1, conjugates abscisic acid and affects plant response to abiotic stresses. Plant Mol. Biol. 102, 389–401. doi: 10.1007/s11103-019-00953-4
Chen, Z. Y., Wang, Y. T., Pan, X. B., and Xi, Z. M. (2019). Amelioration of cold-induced oxidative stress by exogenous 24-epibrassinolide treatment in grapevine seedlings: toward regulating the ascorbate–glutathione cycle. Sci. Hortic. 244, 379–387. doi: 10.1016/j.scienta.2018.09.062
Chou, K. C., and Shen, H. B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5:11335. doi: 10.1371/journal.pone.0011335
Cortina, C., Francisco, A., and Culiáñez-Macià, F. A. (2005). Tomato abiotic stress enhanced tolerance by trehalose biosynthesis. Plant Sci. 169, 75–82. doi: 10.1007/s00425-019-03195-2
Cui, J., Jiang, N., Meng, J., Yang, G., Liu, W., Zhou, X., et al. (2019). LncRNA33732-respiratory burst oxidase module associated with WRKY1 in tomato-Phytophthora infestans interactions. Plant J. 97, 933–946. doi: 10.1111/tpj.14173
Dar, N. A., Mir, M. A., Mir, J. I., Mansoor, S., Showkat, W., Parihar, T. J., et al. (2022). MYB-6 and LDOX-1 regulated accretion of anthocyanin response to cold stress in purple black carrot (Daucus carota L.). Mol. Biol. Rep. doi: 10.1007/s11033-021-07077-3 (Online ahead of print)
Deng, Y., Zheng, H., Yan, Z., Liao, D., Li, C., Zhou, J., et al. (2018). Full-length transcriptome survey and expression analysis of cassia obtusifolia to discover putative genes related to aurantio-obtusin biosynthesis, seed formation and development, and stress response. Int. J. Mol. Sci. 19:2476. doi: 10.3390/ijms19092476
Duan, Z., Yan, Q., Wu, F., Wang, Y., Wang, S., Zong, X., et al. (2021). Genome-wide analysis of the UDP-glycosyltransferase family reveals its roles in coumarin biosynthesis and abiotic stress in Melilotus albus. Int. J. Mol. Sci. 22:10826. doi: 10.3390/ijms221910826
Geu-Flores, F., Moldrup, M. E., Boettcher, C., Olsen, C. E., Scheel, D., and Halkier, B. A. (2011). Cytosolic gamma-glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in Arabidopsis. Plant Cell 23, 2456–2469. doi: 10.1105/tpc.111.083998
Halliwell, B., and Foyer, C. H. (1978). Properties and physiological function of a glutathione reductase purified from spinach leaves by affinity chromatography. Planta 139, 9–17. doi: 10.1007/BF00390803
He, Y., Ahmad, D., Zhang, X., Zhang, Y., Wu, L., Jiang, P., et al. (2018). Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 18:67. doi: 10.1186/s12870-018-1286-5
Hou, J., Zhang, Q., Zhou, Y., Ahammed, G. J., Zhou, Y., Yu, J., et al. (2018). Glutaredoxin GRXS16 mediates brassinosteroid-induced apoplastic H2O2 production to promote pesticide metabolism in tomato. Environ. Pollut. 240, 227–234. doi: 10.1016/j.envpol.2018.04.120
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jongen, M. J., Engel, R., and Leenheers, L. H. (1991). Determination of the pesticide chlorothalonil by HPLC and UV detection for occupational exposure assessment in greenhouse carnation culture. J. Anal. Toxicol. 15, 30–34. doi: 10.1093/jat/15.1.30
Jung, H. I., Lee, T. G., Lee, J., Chae, M. J., Lee, E. J., Kim, M. S., et al. (2021). Foliar-applied glutathione mitigates cadmium-induced oxidative stress by modulating antioxidant-scavenging, redox-regulating, and hormone-balancing systems in Brassica napus. Front. Plant Sci. 12:700413. doi: 10.3389/fpls.2021.700413
Kanwar, M. K., Xie, D., Yang, C., Ahammed, G. J., Qi, Z., Hasan, M. K., et al. (2020). Melatonin promotes metabolism of bisphenol A by enhancing glutathione-dependent detoxification in Solanum lycopersicum L. J. Hazard. Mater. 388:121727. doi: 10.1016/j.jhazmat.2019.121727
Khan, M., Daud, M. K., Basharat, A., Khan, M. J., Azizullah, A., Muhammad, N., et al. (2016). Alleviation of lead-induced physiological, metabolic, and ultramorphological changes in leaves of upland cotton through glutathione. Environ. Sci. Pollut. Res. Int. 23, 8431–8440. doi: 10.1007/s11356-015-5959-4
Langlois-Meurinne, M., Gachon, C. M., and Saindrenan, P. (2005). Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol. 139, 1890–1901. doi: 10.1104/pp.105.067223
Li, Y., Baldauf, S., Lim, E. K., and Bowles, D. J. (2001). Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 276, 4338–4343. doi: 10.1074/jbc.M007447200
Li, Y., Li, P., Wang, Y., Dong, R., Yu, H., and Hou, B. (2014). Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 239, 1265–1279. doi: 10.1007/s00425-014-2050-1
Liu, T., Li, T., Zhang, L., Li, H., Liu, S., Yang, S., et al. (2021). Exogenous salicylic acid alleviates the accumulation of pesticides and mitigates pesticide-induced oxidative stress in cucumber plants (Cucumis sativus L.). Ecotoxicol. Environ. Saf. 208:111654. doi: 10.1016/j.ecoenv.2020.111654
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mamoon Rehman, H., Amjad Nawaz, M., Bao, L., Hussain Shah, Z., Lee, J. M., Ahmad, M. Q., et al. (2016). Genome-wide analysis of family-1 UDP-glycosyltransferases in soybean confirms their abundance and varied expression during seed development. J. Plant Physiol. 206, 87–97. doi: 10.1016/j.jplph.2016.08.017
Morgulis, A., Coulouris, G., Raytselis, Y., Madden, T. L., Agarwala, R., and Schäffer, A. A. (2008). Database indexing for production MegaBLAST searches. Bioinformatics 24, 1757–1764. doi: 10.1093/bioinformatics/btn322
Mostafalou, S., and Abdollahi, M. (2013). Pesticides and human chronic diseases: evidences, mechanisms, and perspectives. Toxicol. Appl. Pharmacol. 268, 157–177. doi: 10.1016/j.taap.2013.01.025
Mukherjee, S. P., and Choudhuri, M. A. (1983). Implications of water stress-induced changes in the levels of endogenous ascorbic acid and hydrogen peroxide in Vigna seedlings. Physiol. Plant. 58, 166–170. doi: 10.1111/j.1399-3054.1983.tb04162.x
Nakano, Y., and Asada, K. (1981). Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 22, 867–880.
Noctor, G., Mhamdi, A., Chaouch, S., Han, Y., Neukermans, J., Marquez-Garcia, B., et al. (2012). Glutathione in plants: an integrated overview. Plant Cell Environ. 35, 454–484. doi: 10.1111/j.1365-3040.2011.02400.x
Pal, R., Kaur, R., Rajwar, D., and Narayan Rai, J. P. (2019). Induction of non-protein thiols and phytochelatins by cadmium in Eichhornia crassipes. Int. J. Phytoremediation 21, 790–798. doi: 10.1080/15226514.2019.1566881
Peco, J. D., Campos, J. A., Romero-Puertas, M. C., Olmedilla, A., Higueras, P., and Sandalio, L. M. (2020). Characterization of mechanisms involved in tolerance and accumulation of cd in Biscutella auriculata L. Ecotoxicol. Environ. Saf. 201:110784. doi: 10.1016/j.ecoenv.2020.110784
Punia, H., Tokas, J., Malik, A., Bajguz, A., El-Sheikh, M. A., and Ahmad, P. (2021). Ascorbate-glutathione oxidant scavengers, metabolome analysis and adaptation mechanisms of ion exclusion in sorghum under salt stress. Int. J. Mol. Sci. 22:13249. doi: 10.3390/ijms222413249
Ramel, F., Sulmon, C., Serra, A. A., Gouesbet, G., and Couée, I. (2012). Xenobiotic sensing and signalling in higher plants. J. Exp. Bot. 63, 3999–4014. doi: 10.1093/jxb/ers102
Rogozin, I. B., Lyonsweiler, J., and Koonin, E. V. (2000). Intron sliding in conserved gene families. Trends Genet. 16, 430–432. doi: 10.1016/S0168-9525(00)02096-5
Rombauts, S., Dehais, P., Van-Montagu, M., and Rouze, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Shahid, M., Khan, M. S., Ahmed, B., Syed, A., and Bahkali, A. H. (2021). Physiological disruption, structural deformation and low grain yield induced by neonicotinoid insecticides in chickpea: a long term phytotoxicity investigation. Chemosphere 262:128388. doi: 10.1016/j.chemosphere.2020.128388
Sinha, A. K. (1972). Colorimetric assay of catalase. Anal. Biochem. 47, 389–394. doi: 10.1016/0003-2697(72)90132-7
Smith, I. K. (1985). Stimulation of glutathione synthesis in photorespiring plants by catalase inhibitors. Plant Physiol. 79, 1044–1047. doi: 10.1104/pp.79.4.1044
Su, X. N., Zhang, J. J., Liu, J. T., Zhang, N., Ma, L. Y., Lu, F. F., et al. (2019). Biodegrading two pesticide residues in paddy plants and the environment by a genetically engineered approach. J. Agric. Food Chem. 67, 4947–4957. doi: 10.1021/acs.jafc.8b07251
Sun, Y. G., Wang, B., Jin, S. H., Qu, X. X., Li, Y. J., and Hou, B. K. (2013). Ectopic expression of Arabidopsis glycosyltransferase UGT85A5 enhances salt stress tolerance in tobacco. PLoS One 8:e59924. doi: 10.1371/journal.pone.0059924
Sun, L., Xua, H., Su, W., Xue, F., An, S., Lu, C., et al. (2018). The expression of detoxification genes in two maize cultivars by interaction of isoxadifen-ethyl and nicosulfuron. Plant Physiol. Biochem. 129, 101–108. doi: 10.1016/j.plaphy.2018.05.025
Szabados, L., and Savour’e, A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97. doi: 10.1016/j.tplants.2009.11.009
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J., and Gao, G. (2020). PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113. doi: 10.1093/nar/gkz1020
Vogt, T., and Jones, P. (2000). Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 5, 380–386. doi: 10.1016/S1360-1385(00)01720-9
Wang, Y., Zhang, T., Wang, J., Xu, S., and Shen, W. (2022). Regulation of chlorothalonil degradation by molecular hydrogen. J. Hazard. Mater. 424:127291. doi: 10.1016/j.jhazmat.2021.127402
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Wu, B., Liu, X., Xu, K., and Zhang, B. (2020). Genome-wide characterization, evolution and expression profiling of UDP-glycosyltransferase family in pomelo (Citrus grandis) fruit. BMC Plant Biol. 20:459. doi: 10.1186/s12870-020-02655-2
Wu, W., Zhu, S., Xu, L., Zhu, L., Wang, D., Liu, Y., et al. (2022). Genome-wide identification of the Liriodendron chinense WRKY gene family and its diverse roles in response to multiple abiotic stress. BMC Plant Biol. 22:25. doi: 10.1186/s12870-021-03371-1
Yu, J., Hu, F., Dossa, K., Wang, Z., and Ke, T. (2017). Genome-wide analysis of UDP-glycosyltransferase super family in Brassica rapa and Brassica oleracea reveals its evolutionary history and functional characterization. BMC Genomics 18:474. doi: 10.1186/s12864-017-3844-x
Yu, G. B., Zhang, Y., Ahammed, G. J., Xia, X. J., Mao, W. H., Shi, K., et al. (2013). Glutathione biosynthesis and regeneration play an important role in the metabolism of chlorothalonil in tomato. Chemosphere 90, 2563–2570. doi: 10.1016/j.chemosphere.2012.10.100
Zhang, W. (2018). Global pesticide use: profile, trend, cost/benefit and more. Proc. Int. Acad. Ecol. Environ. Sci. 8:1. doi: 10.13140/RG.2.2.28978.30405
Zhang, X., Gong, X., Li, D., Yue, H., Qin, Y., Liu, Z., et al. (2021). Genome-wide identification of PRP genes in apple genome and the role of MdPRP6 in response to heat stress. Int. J. Mol. Sci. 22:5942. doi: 10.3390/ijms22115942
Keywords: UDP-glycosyltransferase, tomato (Solanum lycopersicum), glutathione, chlorothalonil residues, gene expression
Citation: Yu G, Chen Q, Chen F, Liu H, Lin J, Chen R, Ren C, Wei J, Zhang Y, Yang F and Sheng Y (2022) Glutathione Promotes Degradation and Metabolism of Residual Fungicides by Inducing UDP-Glycosyltransferase Genes in Tomato. Front. Plant Sci. 13:893508. doi: 10.3389/fpls.2022.893508
Received: 17 March 2022; Accepted: 23 May 2022;
Published: 01 July 2022.
Edited by:
Amalia Barone, University of Naples Federico II, ItalyReviewed by:
Smita Kumar, King George's Medical University, IndiaCopyright © 2022 Yu, Chen, Chen, Liu, Lin, Chen, Ren, Wei, Zhang, Yang and Sheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gaobo Yu, eXVnYW9ibzgxQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.