- 1Institute of Wetland Agriculture and Ecology, Shandong Academy of Agricultural Sciences, Jinan, China
- 2Institute of Crop Germplasm Resources, Shandong Academy of Agricultural Sciences, Jinan, China
Asian cultivated rice consists of two main subspecies, xian/indica (XI) and geng/japonica (GJ), and GJ accessions have significantly lower nitrogen-use efficiency (NUE) than XI accessions. In order to facilitate genetic improvement of NUE in GJ accessions, we conducted haplotype analysis of 14 cloned NUE genes using 36 rice germplasm accessions with high-quality reference genome and developed 18 intragenic markers for elite haplotypes, which were then used to evaluate NUE genes in another 41 genetically diverse germplasm accessions from 12 countries and 71 approved GJ cultivars from northern provinces of China. Our results show that elite haplotypes of 12 NUE genes are mainly existed in XI accessions, but few is distributed in GJ accessions. The number of elite haplotypes carried by an XI accession can reach 10, while that carried by a GJ accession is less than 3. Surprisingly, the elite haplotype of gene DEP1 is nearly fixed in approved GJ cultivars, and elite haplotypes of gene MYB61 and NGR5 have been introduced into some approved GJ cultivars. The developed intragenic markers for NUE genes and evaluated 77 genetically diverse rice accessions could be of great use in the improvement of NUE in GJ cultivars.
Introduction
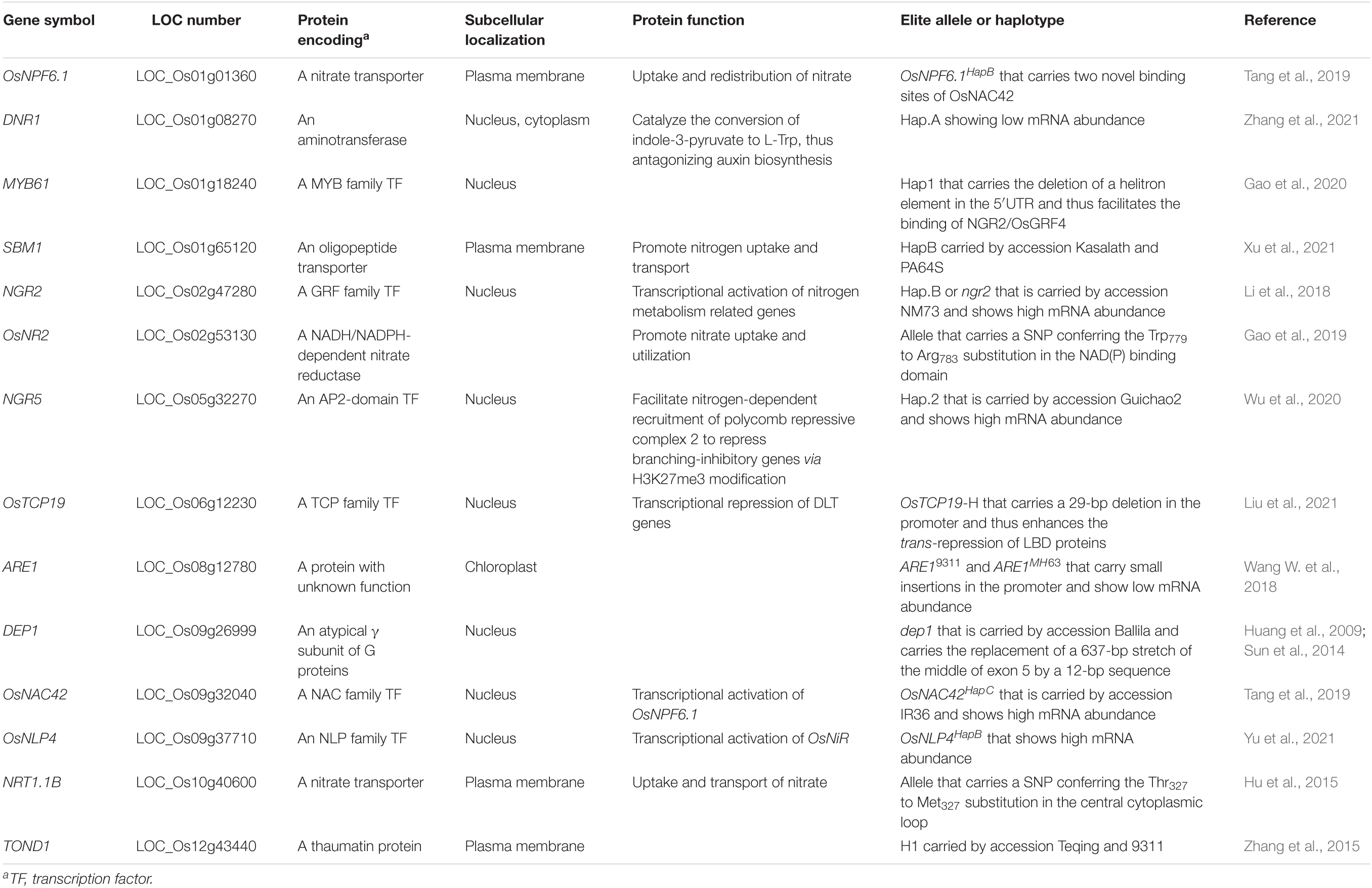
Rice is a staple food that feeds more than half of the world population. Nitrogen is a macronutrient that is essential to the growth and grain yield of rice plants. The adoption of semidwarf rice varieties and nitrogen fertilizer has strikingly boosted rice yield since 1960s (Khush, 1999). However, the excess input of nitrogen fertilizer not only contributes little to the increase in grain yield, but also brings great threat to environment in recent years. Therefore, adoption of rice cultivars with high nitrogen-use efficiency (NUE) and reduction of nitrogen input is a promising way to achieve sustainable agriculture. Asian cultivated rice consists of two main subspecies, xian/indica (XI) and geng/japonica (GJ), which display significant difference in various traits, including NUE (Hu et al., 2015; Gao et al., 2019). GJ cultivars, favored by consumers in Japan, South Korea, and the northern provinces of China, have low NUE, and massive input of nitrogen fertilizer is the way selected by farmers to improve rice yield. Therefore, genetic improvement of NUE in GJ cultivars is of great necessity to achieve sustainable agriculture. NUE involves nitrogen uptake, translocation, and assimilation, and results in changes in plant height, tiller number, and grain yield in rice (Li et al., 2017). Thus, it is a complex trait controlled by quantitative trait loci (QTL) (Zhou et al., 2017; Tang et al., 2019; Shen et al., 2021). Up to now, 14 genes conferring natural variation in NUE have been cloned and characterized (Table 1). Introgression of elite alleles or haplotypes of these cloned genes into the background of GJ cultivars could be a promising way to improve NUE of GJ cultivars. Marker-assisted selection has been proved to be an effective method in genetic improvement of rice cultivars, of which the premise is the development of effective molecular markers for target genes (Wang et al., 2017; Jiang et al., 2019; Mao et al., 2021).
Objective
Our aim was to develop effective intragenic markers for the 14 cloned NUE genes in rice, and then to evaluate NUE genes of some germplasm accessions and approved GJ cultivars, which could guide the following genetic improvement. Haplotype analysis of the 14 NUE genes from 36 rice accessions with high-quality reference genome provided many useful sequence variations for marker development, and developed intragenic markers facilitated the evaluation of NUE genes in germplasm accessions and approved cultivars.
Materials and Methods
Rice Accessions and Cultivars
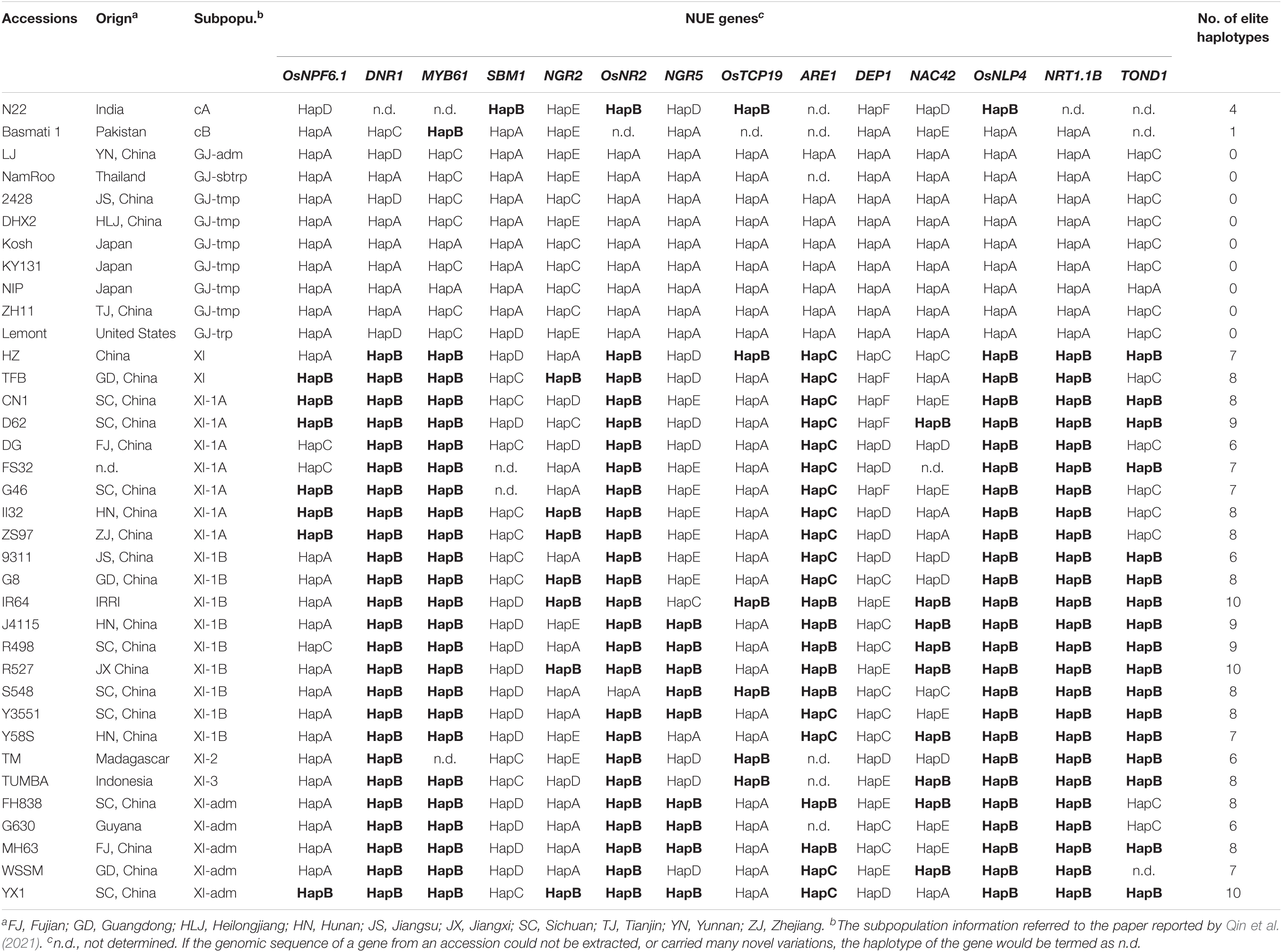
Three panels of rice materials were exploited in this study. The first panel consisted of 36 rice accessions with reference genome (Table 2), including GJ accession Nipponbare (NIP) (International Rice Genome Sequencing Project, 2005), 31 genetically diverse rice accessions (Qin et al., 2021), XI accessions Huazhan and Tianfeng (Zhang et al., 2022), and XI accessions Minghui63 and Zhenshan97 (Song et al., 2021). The second panel consisted of 41 genetically diverse rice accessions from 12 countries (Supplementary Table S16). The third panel consisted of 71 GJ cultivars approved in the northern provinces of China, including 2 from Anhui, 1 from Henan, 9 from Jiangsu, 39 from Shandong, 1 from Tianjin, and 19 approved in several provinces (Supplementary Table S17). The subpopulation of accessions in the first panel and second panel referred to a previous study (Wang W. et al., 2018).
Haplotype Analysis
The genomic sequences of 14 cloned NUE genes from 36 rice accessions with reference genome were extracted using the software BioEdit (Hall, 1999) and software SeqBuilder from the Lasergene package (DNASTAR, lnc., Madison, USA), including 2-kb region upstream of the start codon termed as 5′ untranslated region (5′UTR), the coding region, and 1-kb region downstream of the stop codon termed as 3′UTR. Elite alleles of NGR2, NGR5, DEP1, and IR36 were reported to be carried by a very few accessions not included in the 36 accessions, and their genomic sequences could not be found in related databases and papers. Therefore, genomic sequences of allele ngr2 from accession NM73, allele NGR5 from accession Guichao2, allele dep1 from accession Ballila, and allele NAC42 from accession IR36 were resequenced using the Sanger sequencing method and assembled using the software SeqMan from the Lasergene package, and related primers were listed in Supplementary Table S1. Sequence alignment for each gene was conducted using the software MEGA 7. Haplotype analysis of each gene was conducted, based on a combination of functional variations reported in previous studies and sequence variations identified in this study. For each NUE gene, the haplotype of accession NIP was defined as HapA, and reported elite haplotype was defined as HapB (Table 2 and Supplementary Tables S2–S15).
Marker Development
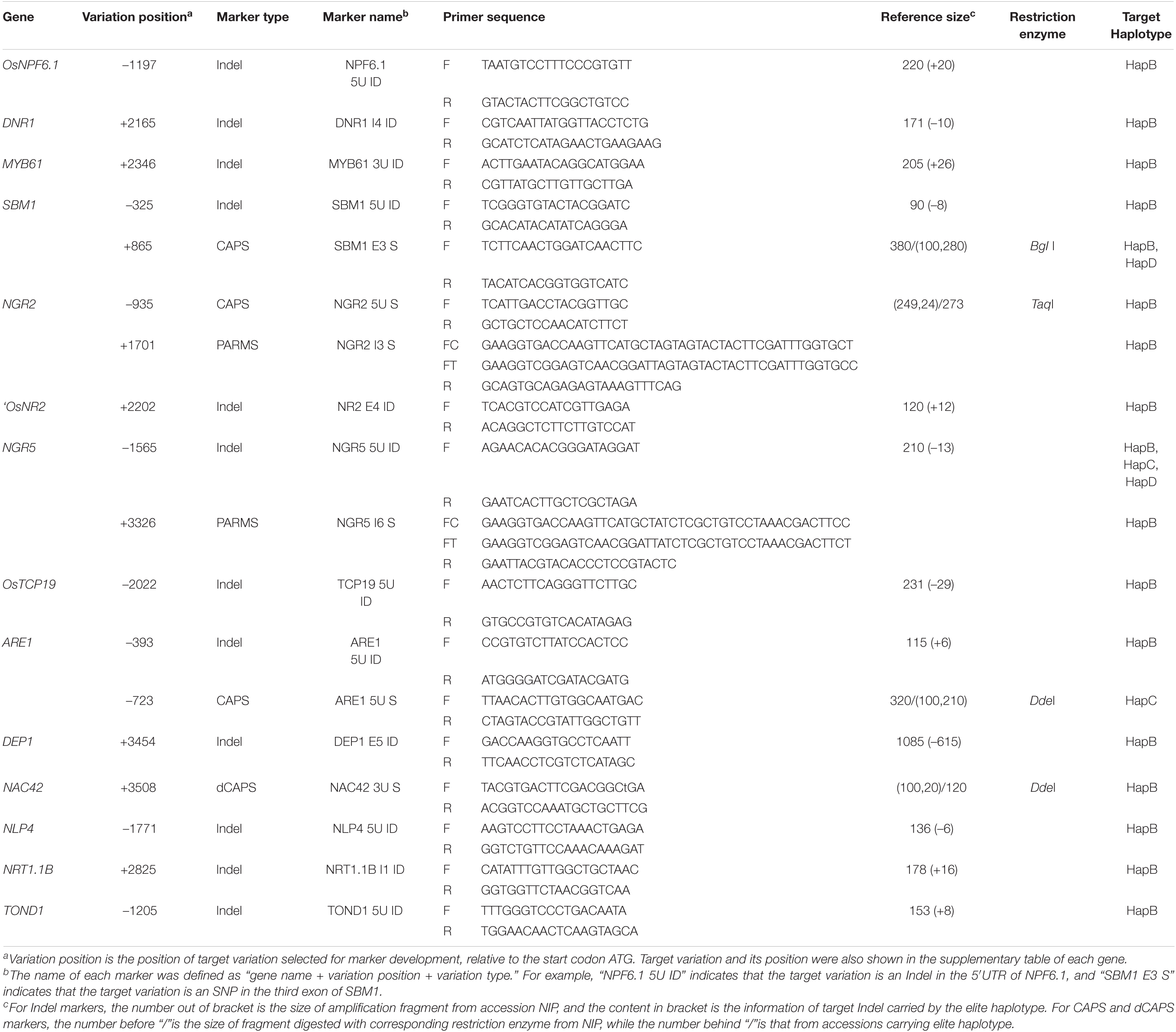
Several rules were followed during marker development. First of all, target variations should be unique to the elite haplotype of each gene. Secondly, an insertion/deletion (Indel) whose size was between 5 and 30 bp was given priority, and the Indel marker with amplification fragment between 100 and 300 bp was developed. Thirdly, if no Indel could be selected, a single-nucleotide polymorphism (SNP) that could be transformed into a common restriction enzyme site was taken into consideration, and then the cleaved amplified polymorphic sequences (CAPS) marker with amplification fragment between 100 and 500 bp or derived CAPS (dCAPS) marker with amplification fragment between 100 and 300 bp was developed. Finally, if no variation could meet the second and third rules, a penta-primer amplification-refractory mutation system (PARMS) marker was designed (Qing et al., 2018).
Indel markers were developed using the software Primer Premier 6 (PREMIER Biosoft, San Francisco, USA). CAPS markers and dCAPS markers were developed using the website dCAPS Finder 2.01 (Neff et al., 2002) and the software Primer Premier 6. PARMS markers were developed using the website SNPWay2.
Marker Analysis
For Indel markers, CAPS markers and dCAPS markers, the PCR reaction was 3-μl genomic DNA with a concentration of 20 ng/μl, 0.5 μl of each primer with a concentration of 10 μM/L, 10-μl 2*Taq PCR MasterMix, and 6 μl of ddH2O. The PCR profile was 3 min at 94°C for denaturation, followed by 32 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min/kb, then by 3 min at 72°C for extension, and finally by 1 min at room temperature. The PCR products of Indel markers were run on 4% polyacrylamide gels with a voltage of 160 V for 80 min, and bands were then revealed using a silver staining procedure: gels were washed for 1 min with dH2O, followed by incubated for 10 min in staining solution (0.4 g AgNO3, 400 ml dH2O) with gentle shaking, then washed two times for 1min with dH2O, then incubated for 4 min in developing solution (6 g NaOH, 2 ml CH3CHO, 400 ml dH2O) with gentle shaking, and finally washed for 1 min with water. The PCR products of dCAPS markers and CAPS markers were digested with corresponding restriction enzymes purchased from Takara Biomedical Technology Co., LTD., Beijing, China, according to the recommended reaction and temperature. The digested products of dCAPS markers were analyzed as that of Indel markers described above. The digested products of CAPS markers were run 2% agarose gels containing GeneRed nuclelic acid dye with a voltage of 120 V for 20 min, and bands were developed in UVP EC3 Imaging System (Analytik Jena AG, Jena, Germany).
For PARMS markers, the PCR reaction was 2-μl genomic DNA with a concentration of 20 ng/μl, 0.15 μl of each forward primer and 0.4 μl of common reverse primer with a concentration of 10 μM/L, 5-μl 2*PARMS master mix containing two universal fluorescent primers, and 2.3 μl of ddH2O. The PCR profile was 15 min at 94°C for denaturation, followed by 10 cycles of 94°C for 20 s and 65°C (with each cycle minus 0.8°C) for 1 min, then by 28 cycles of 94°C for 20 s and 57°C for 1 min, and finally by 1 min at temperature. The PCR products were analyzed using ABI QuantStudio 5 fluorescent quantitative PCR system (Applied Biosystem, Foster, USA).
Results
Haplotype Analysis of 14 Nitrogen-Use Efficiency Genes
In order to have a closer examination of sequence variations in the 14 cloned NUE genes (Table 1), we extracted the genomic sequences from the 36 rice accessions with reference genome, including 2-kb 5′UTR, the coding region, and 1-kb 3′UTR. We also sequenced the genomic sequence of allele ngr2 from accession NM73, allele NGR5 from accession Guichao2, allele dep1 from accession Ballila, and allele NAC42 from accession IR36, which were reported elite alleles and whose sequences were not accessible. Subsequently, we performed sequence alignment and haplotype analysis of each gene, with the type of accession NIP termed as HapA and reported elite haplotype termed as HapB (Table 2).
For OsNPF6.1, four haplotypes were identified, including the two reported by Tang et al. (2019; Table 2 and Supplementary Table S2). Compared to HapA, both HapB and HapC carried two SNPs at positions −1041 and −443 that created two new binding sites for protein OsNAC42. In addition, HapB carried a non-synonymous SNP at position + 125 and HapC carried two non-synonymous SNPs at positions + 506 and + 535 in the first exon.
For DNR1, four haplotypes were identified, including the three types reported by Zhang et al. (2021), which were HapA (Hap.B-TEJ), HapB (Hap.A), HapC, and HapD (Hap.B-TRJ) (Table 2 and Supplementary Table S3). The four types were distributed in GJ-tmp, XI, cA/cB, and GJ-trp accessions, respectively (Table 2). Compared to HapA, HapB carried a 520-bp deletion, a 2-bp deletion and twenty SNPs in 5′UTR, five Indels and eighteen SNPs in the coding region, and three SNPs in the 3′UTR. In addition, both HapC and HapD carried the 520-bp deletion at position -1201.
For MYB61, three haplotypes were identified, in agreement with the three types reported by Gao et al. (2020), which were HapA (Hap3), HapB (Hap1), and HapC (Hap2) (Table 2 and Supplementary Table S4). HapC differed from HapA by an SNP G/A at position −323, of which both were distributed in GJ accessions. HapB, the type distributed in XI accessions, carried the deletion of 975-bp helitron at position −446 that was close to the binding site of NGR2/OsGRF4 in the 5′UTR, and a non-synonymous SNP at position + 1224 in the coding region relative to the other two types.
For SBM1, four haplotypes were identified, including the three types reported by Xu et al. (2021; Table 2 and Supplementary Table S5). HapA and HapB were distributed mainly in GJ and cA accessions, respectively, while both HapC and HapD were distributed mainly in XI accessions. Compared to HapA and HapC, HapB carried two Indels and fifteen SNPs in the 5′UTR, a 3-bp deletion and two SNPs in the coding region, and three SNPs in the 3′UTR. HapD carries a 1,204-bp deletion in the 5′UTR and a non-synonymous SNP at position + 2 which disrupted the translation start site, relative to the other three types (Supplementary Figure S1).
For NGR2, five haplotypes were identified, including the three reported by Li et al. (2018) (Table 2 and Supplementary Table S6). HapB carried 14 SNPs at positions −1806, −1754, −1738, −1691, −1314, −935, −878, −844, −795, +511, +641, +654, +2275, and +3330 relative to the other four types, and was distributed in XI accessions. In addition, the two rare SNPs (+1187T > A and +1188C > A) preventing miR396-meidated cleavage of mRNA were not carried by all the 36 accessions.
For OsNR2, two haplotypes were identified, based on the functional SNP at position + 2692 reported by Gao et al. (2019; Table 2 and Supplementary Table S7). HapA and HapB were distributed in GJ and XI accessions, respectively. HapB differed from HapA by a 26-bp insertion and nine SNPs in the 5′UTR, a 12-bp insertion and three non-synonymous SNPs in the coding region, and four SNPs in the 3′UTR.
For NGR5, five haplotypes were identified, in agreement with the five reported by Wu et al. (2020), which were HapA (Hap1), HapB (Hap2), HapC (Hap3), HapD (Hap4), and HapE (Hap5) (Table 2 and Supplementary Table S8). Among those, HapB, HapC, and HapD shared many variations relative to the other two types, and differentiated from each other by an SNP, respectively. HapB carried an SNP at position + 3326 in the sixth intron, and HapC carried an SNP at position + 4020 in the eighth exon.
For OsTCP19, two haplotypes were identified, based on the functional 29-bp deletion at position −2022 reported by Liu et al. (2021; Table 2 and Supplementary Table S9).
For ARE1, three haplotypes were identified, in agreement with the three reported by Wang et al. (2018), which were HapA (ARE1NPB), HapB (ARE1MH63), and HapC (ARE19311) (Table 2 and Supplementary Table S10). Compared to HapA, HapB carried a 6-bp insertion and 10 SNPs in the 5′UTR, 11 SNPs in the coding region, and a 6-bp insertion and three SNPs in the 3′UTR. HapC carried a 2-bp deletion, a 7,808-bp insertion and eight SNPs in the 5′UTR, ten SNPs in the coding region, and a 6-bp insertion in the 3′UTR.
For DEP1, six haplotypes were identified (Table 2 and Supplementary Table S11). The functional variation – the replacement of a 637-bp segment of the middle of exon 5 by a 12-bp sequence, is carried by accession Ballila but not by all 36 accessions (Huang et al., 2009).
For OsNAC42, five haplotypes were identified (Table 2 and Supplementary Table S12). Compared to HapA, HapB carried eight SNPs in the 5′UTR, eleven SNPs in the coding region, and a 27-bp deletion and six SNPs in the 3′UTR.
For OsNLP4, two haplotypes were identified, in agreement with the two reported by Yu et al. (2021), which were distributed in GJ and XI accessions, respectively (Table 2 and Supplementary Table S13). Compared to HapA, HapB carried a 6-bp deletion and eight SNPs in the 5′UTR, five SNPs in the coding region, and five SNPs in the 3′UTR.
For NRT1.1B, two haplotypes were identified, based on the functional SNP at position + 3019 reported by Hu et al. (2015), which were distributed in GJ and XI accessions, respectively (Table 2 and Supplementary Table S14). Compared to HapA, HapB carried a 3-bp insertion and thirteen SNPs in the 5′UTR, five Indels and twenty-two SNPs in the coding region, and two Indels and nine SNPs in the 3′UTR.
For TOND1, three haplotypes were identified, including the elite type reported by Zhang et al. (2015), which was HapB (H1) (Table 2 and Supplementary Table S15). Compared to HapA and HapC, HapB carried an 8-bp insertion and three SNPs in the 5′UTR, and two non-synonymous SNPs in the coding region.
Development of Intragenic Markers for Nitrogen-Use Efficiency Genes
In order to develop effective markers for NUE genes, we focused on the variations only carried by HapB of each gene. As shown in Table 3, a total of 18 markers were developed for the 14 genes including twelve Indel markers, three CAPS makers, a dCAPS marker, and two PARMS markers. The effectiveness of 16 markers was evaluated by 20 germplasm accessions (Figure 1), and that of the two PARMS markers was evaluated by 57 germplasm accessions (Figure 2).
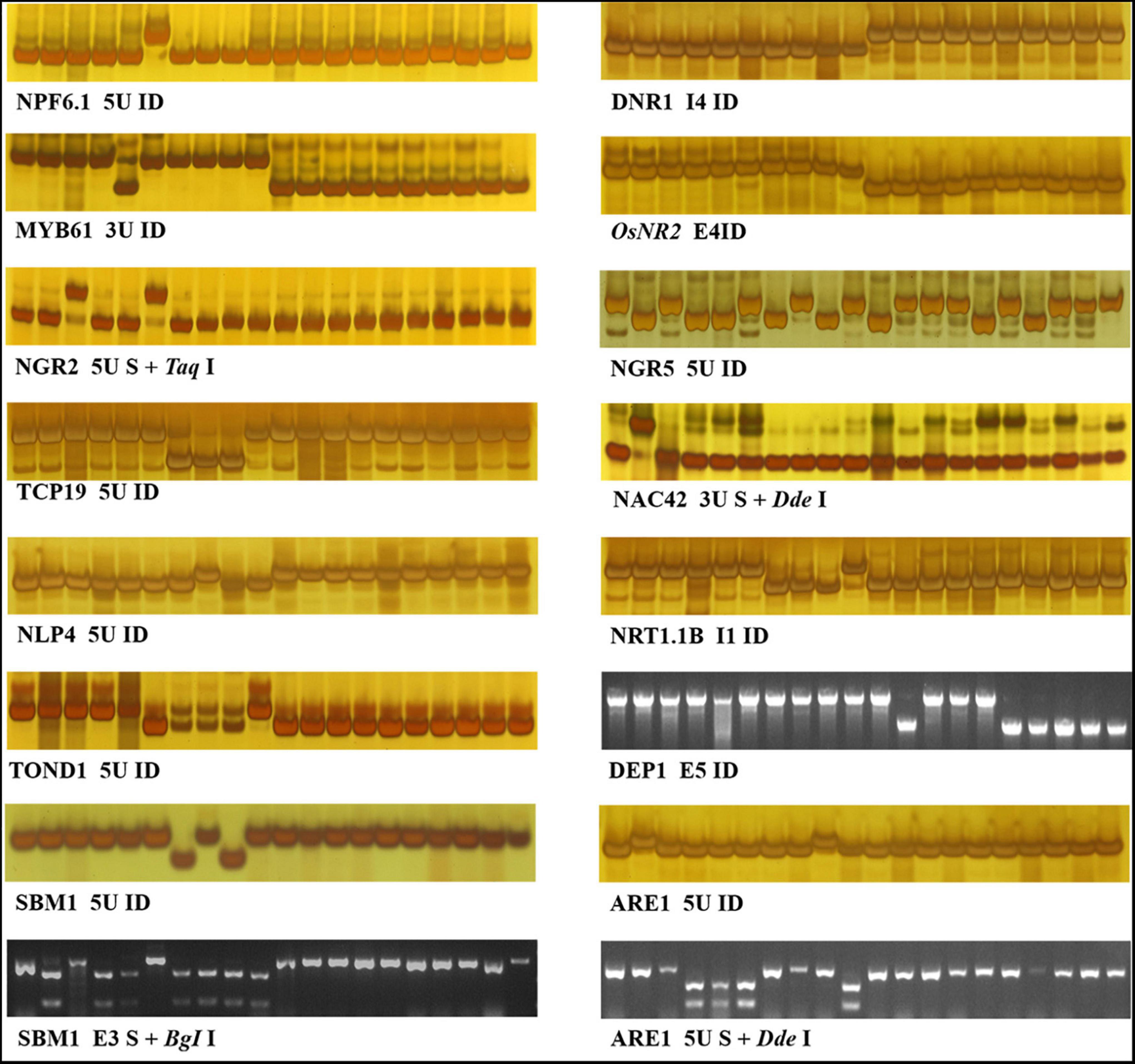
Figure 1. Indel, CAPS, and dCAPS markers for 14 NUE genes. The 20 accessions used for marker evaluation were 9311, IR36, NM73, Guicao 2, Huagengxian 74, ZS97, Kasalath, Basmati 370, Dular, NJ11, 02428, Ballila, NIP, Kosh, KY131, Runnong11, Suxiu 867, Huaidao 5, Xudao 3, and Huageng5 (here from left to right). The first 15 accessions were germplasm accessions, including donors of the 14 NUE genes, and the other 5 accessions were mainstay cultivars in the Huang–Huai rice area of China. In the figure of marker “TOND1 5U ID,” no band was amplified from accession Kasalath, Basmati 370, and Dular.
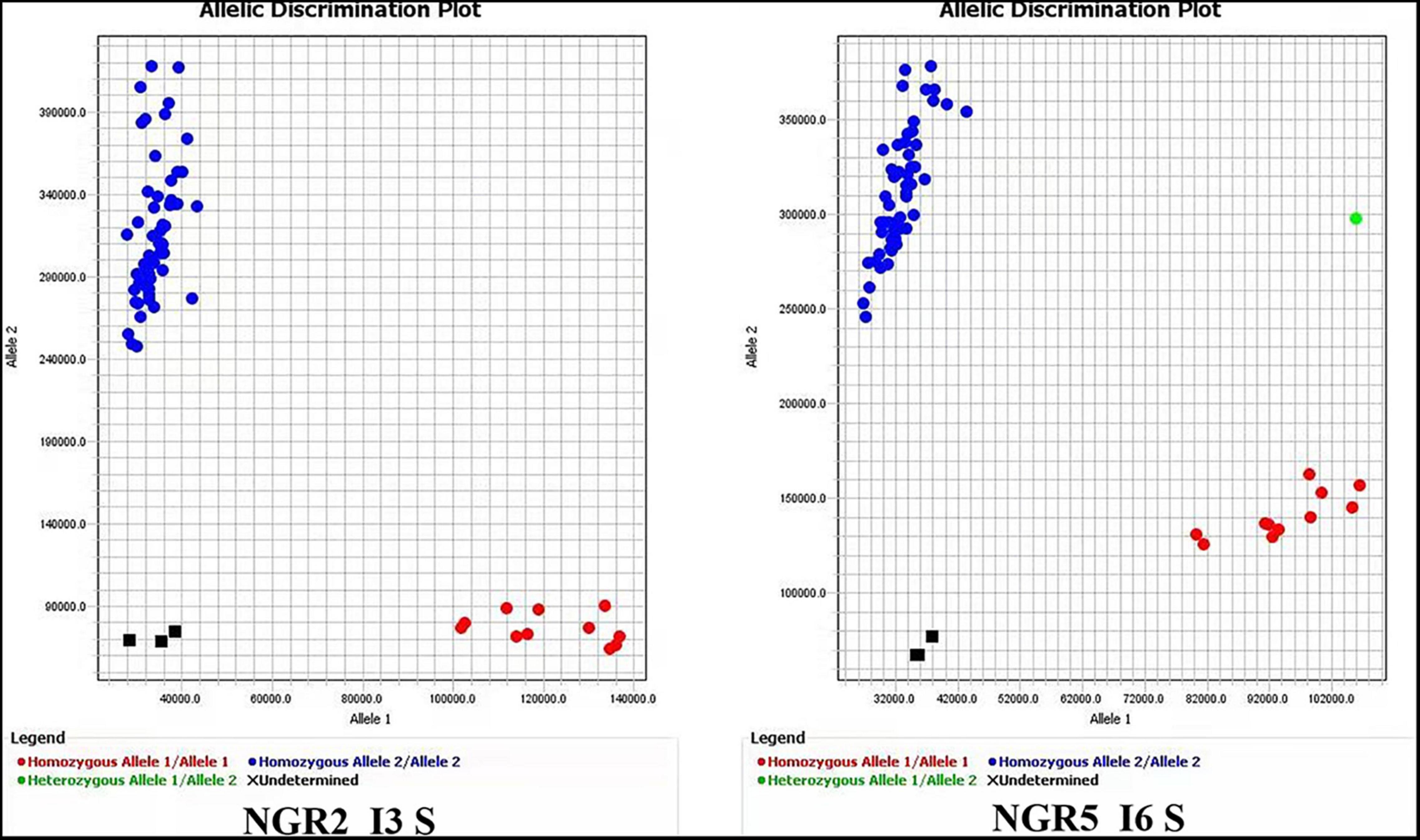
Figure 2. PAMRS markers for NGR2 and NGR5. The accessions used for marker evaluation were the 41 germplasm accessions in Supplementary Table S16 and the first 16 accessions in Figure 1.
SBM1 was reported as a negative regulator of NUE and grain yield, and the elite type HapB carried by accession Kasalath was a type with weak function (Xu et al., 2021). HapD carried a 1,204-bp deletion in the 5′UTR region and a non-synonymous SNP at position + 2 that would disrupt the translation start site, and was likely to have a better performance than HapB (Supplementary Table S5). Thus, the CAPS marker “SBM1 E3 S” together with restriction enzyme BgI I was developed to select HapB and HapD, which could also differentiate the two types together with the Indel marker “SBM1 5U ID.”
Two elite types of ARE1 were reported, which were carried by accession Minghui63 and 9311, respectively (Wang et al. 2018). However, the 6-bp insertion reported in the promoter region of accession 9311 was not observed in all the 36 accessions with reference genome, including the resequenced accession 9311. A 7,808-bp insertion in the HapC carried by 15 accessions was observed at position −466, which might reduce the expression of ARE1 and make it an elite type (Supplementary Table S10). Thus, the CAPS marker “ARE1 5U S” together with restriction enzyme DdeI was developed to select HapC.
NGR5 was classified into five haplotypes, among which HapB, HapC, and HapD differentiated from each other only by one SNP (Supplementary Table S8). HapB was reported to be superior to HapC and HapD, but the SNP in the sixth intron could not account for the difference. We then examined the 7-kb region upstream of the start codon, and found that no additional variation could further differentiate the three types (data not shown). Therefore, it was likely that all the three types were elite, and an Indel marker “NGR5 5U ID” was developed for the 13-bp deletion at position −1565, which was shared by the three types. In addition, a PARMS marker “NGR5 In6 S” was developed to select HapB only.
Evaluation of Nitrogen-Use Efficiency Genes in Germplasm Accessions and Approved Cultivars
In order to facilitate the genetic improvement of NUE in GJ cultivars, we examined the haplotypes of 14 NUE genes in 41 germplasm accessions from 12 countries (Supplementary Table S16) and 71 GJ cultivars approved in China (Supplementary Table S17) using the 18 makers developed above.
We firstly analyzed the distribution of elite NUE haplotypes in 77 germplasm accessions, including the 36 accessions with reference genome (Table 2) and 41 accessions examined with developed markers (Supplementary Table S16). The elite haplotype of each NUE gene was mainly existed in XI accessions, except for that of SBM1 and DEP1 (Figure 3). The number of elite haplotypes carried by an XI accession ranged from 4 to 10, with an average value of 7.56. Three XI accessions, namely, IR64, R527, and YX1, carried 10 elite haplotypes. In contrast, few elite haplotypes were carried by GJ accessions, and 3 accessions carried only one elite haplotype, which were Ballila carrying allele dep1, and IRAT261 and Suyunuo carrying MYB61.
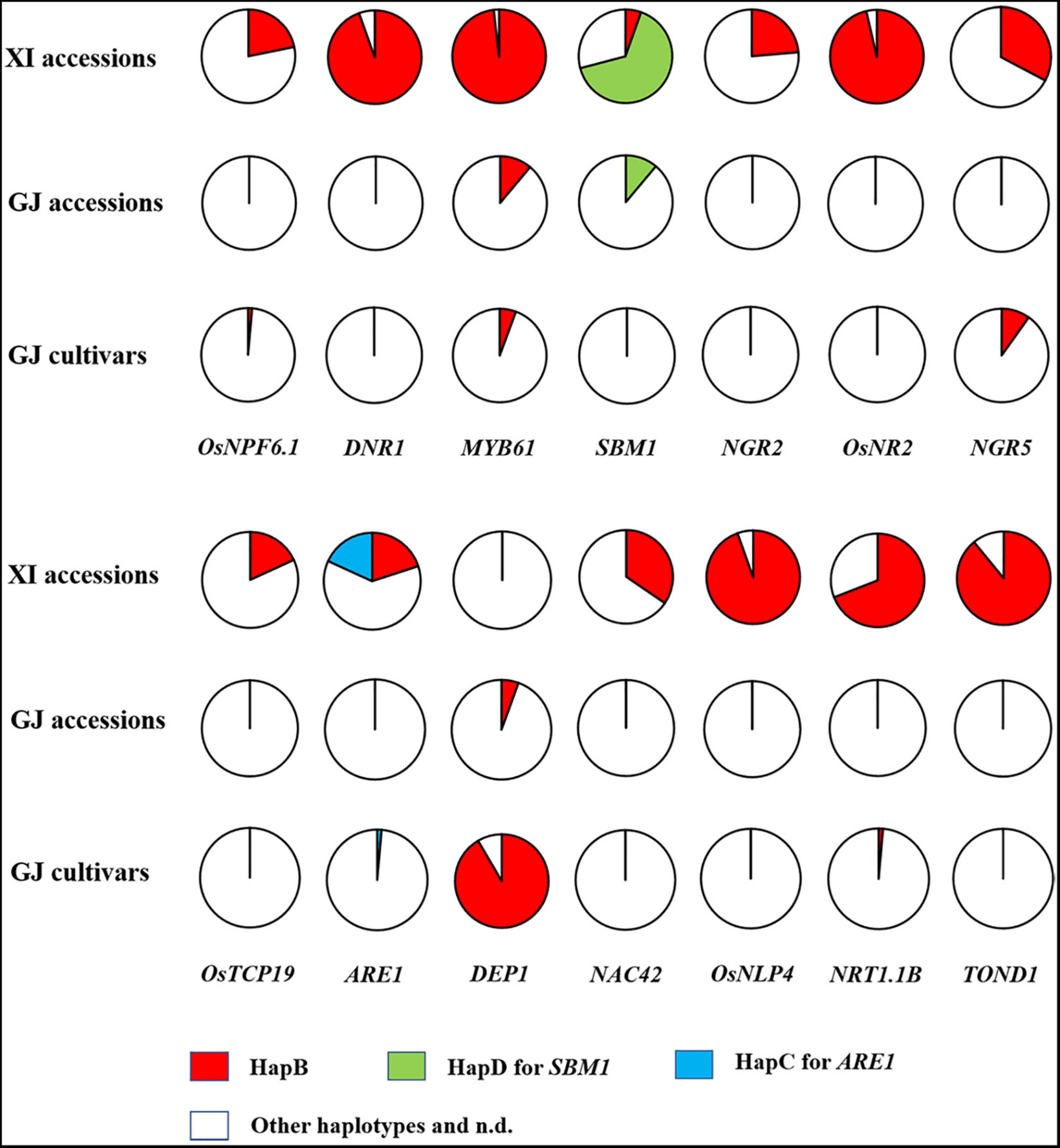
Figure 3. Distribution of elite haplotypes of 14 NUE genes in 55 XI accessions, 18 GJ accessions, and 71 GJ cultivars.
We subsequently analyzed the distribution of elite NUE haplotypes in GJ cultivars approved in the northern provinces of China. A very few elite haplotypes were carried by these cultivars, and the number of elite haplotypes carried by an accession ranged from 0 to 3. Surprisingly, the elite allele dep1 was carried by 65 out of 71 accessions (Figure 3 and Supplementary Table S17). The HapB of NGR5 was carried by six accessions and the HapB of MYB61 was carried by four accessions. In addition, the HapB of OsNPF6.1 and NRT1.1B was carried by an accession, respectively.
Discussion
In this study, haplotype analysis of the 14 NUE genes using high-quality genomic sequences facilitated the identification of some important variations and haplotypes. For example, HapC of ARE1 carried by XI accession 9311 contained several variations in the 5′UTR that was different from reported variations, including a 7,808-bp insertion at position −466 in this study (Supplementary Table S10) and a 6-bp insertion at position −314 in reported study (Wang Q. et al., 2018). HapD of SBM1 was carried by 14 out of 36 accessions in this study (Supplementary Table S5), but was not identified from 1,140 rice accessions of broad genetic diversity (Xu et al., 2021). Therefore, high-quality genomic sequence was of great importance in haplotype analysis. Intragenic or functional variations that resided in the region covering from 2-kb upstream of start codon to 1-kb downstream of stop codon were selected for marker development. The markers developed in this study were ideal markers for its co-segregating with elite alleles. Haplotype analysis of the 14 NUE genes from 77 genetically diverse rice germplasm accessions revealed that the number of elite haplotypes carried by an XI accession ranged from 4 to 10, while that carried by a GJ accession ranged from 0 to 3, which gives a good explanation of the NUE difference between the two subpopulations of Asian rice cultivars (Table 2 and Supplementary Table S16). Finally, developed intragenic markers for NUE genes and evaluated germplasm accessions in this study could be of great use in guiding the improvement of NUE in GJ cultivars in the future.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
PL performed most of the experiments and wrote the manuscript. ZL participated in marker development and materials evaluation. XL, HZ, and QW participated in materials evaluation. NL and HD provided some rice accessions and cultivars. FY designed and supervised the study. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (32101756), the Natural Science Foundation of Shandong Province (ZR2019BC105), the Key Research and Development Project of Shandong Province (2021LZGC025, 2019LZGC017, and 2019LZGC003), the Rice Industry Technology Program of Shandong (SDAIT-17-03), and the Agricultural Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2021B24).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Prof. Guiquan Zhang from South China Agricultural University, Prof. Yuqing He and Master Qinglu Zhang from Huazhong Agricultural University, Researcher Wenfeng Chen from Guangdong Academy of Agricultural Sciences, and Associate Researcher Tianqing Zheng from Chinese Academy of Agricultural Sciences, for providing some rice germplasm accessions. We also thank Researcher Wenyin Zhu and Associate Researcher Feng Chen from our institute, and Director Baizhan Yang from Seed Company of Tancheng County, for providing some rice cultivars approved in Shandong province.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.891860/full#supplementary-material
Footnotes
References
Gao, Y., Xu, Z., Zhang, L., Li, S., Wang, S., Yang, H., et al. (2020). MYB61 is regulated by GRF4 and promotes nitrogen utilization and biomass production in rice. Nat. Commun. 11:5219. doi: 10.1038/s41467-020-19019-x
Gao, Z., Wang, Y., Chen, G., Zhang, A., Yang, S., Shang, L., et al. (2019). The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 10, 5207. doi: 10.1038/s41467-019-13110-8
Hall, T.A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Sump. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838. doi: 10.1038/ng.3337
Huang, X., Qian, Q., Liu, Z., Sun, H., He, S., Luo, D., et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. doi: 10.1038/ng.352
International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436, 793–800. doi: 10.1038/nature03895
Jiang, H., Li, Z., Liu, J., Shen, Z., Gao, G., Zhang, Q., et al. (2019). Development and evaluation of improved lines with broad-spectrum resistance to rice blast using nine resistance genes. Rice (N Y) 12:29. doi: 10.1186/s12284-019-0292-z
Khush, G. S. (1999). Green revolution: preparing for the 21st century. Genome 42, 646–655. doi: 10.1139/g99-044
Li, H., Hu, B., and Chu, C. (2017). Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. J. Exp. Bot. 68, 2477–2488. doi: 10.1093/jxb/erx101
Li, S., Tian, Y., Wu, K., Ye, Y., Yu, J., Zhang, J., et al. (2018). Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560, 595–600. doi: 10.1038/s41586-018-0415-5
Liu, Y., Wang, H., Jiang, Z., Wang, W., Xu, R., Wang, Q., et al. (2021). Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 590, 600–605. doi: 10.1038/s41586-020-03091-w
Mao, T., Zhu, M., Ahmad, S., Ye, G., Sheng, Z., Hu, S., et al. (2021). Superior japonica rice variety YJ144 with improved rice blast resistance, yield, and quality achieved using molecular design and multiple breeding strategies. Mol. Breed 41:65. doi: 10.1007/s11032-021-01259-4
Neff, M. M., Turk, E., and Kalishman, M. (2002). Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 18, 613–615. doi: 10.1016/s0168-9525(02)02820-2
Qin, P., Lu, H., Du, H., Wang, H., Chen, W., Chen, Z., et al. (2021). Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 184, 3542.e–3558.e. doi: 10.1016/j.cell.2021.04.046
Qing, D. J., Liu, K. Q., Yang, Y. Y., Gao, L. J., Huang, J., Gao, J., et al. (2018). Development of molecular marker of rice blast resistance gene Pigm on basis of PARMS technology. Southwest China J. Agric. Sci. 31, 1617–1621.
Shen, C., Chen, K., Cui, Y., Chen, J., Mi, X., Zhu, S., et al. (2021). QTL mapping and favorable allele mining of nitrogen deficiency tolerance using an interconnected breeding population in rice. Front. Genet. 12:616428. doi: 10.3389/fgene.2021.616428
Song, J. M., Xie, W. Z., Wang, S., Guo, Y. X., Koo, D. H., Kudrna, D., et al. (2021). Two gap-free reference genomes and a global view of the centromere architecture in rice. Mol. Plant 14, 1757–1767. doi: 10.1016/j.molp.2021.06.018
Sun, H., Qian, Q., Wu, K., Luo, J., Wang, S., Zhang, C., et al. (2014). Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 46, 652–656. doi: 10.1038/ng.2958
Tang, W., Ye, J., Yao, X., Zhao, P., Xuan, W., Tian, Y., et al. (2019). Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat. Commun. 10:5279. doi: 10.1038/s41467-019-13187-1
Wang, Q., Nian, J., Xie, X., Yu, H., Zhang, J., Bai, J., et al. (2018). Genetic variations in ARE1 mediate grain yield by modulating nitrogen utilization in rice. Nat. Commun. 9:735. doi: 10.1038/s41467-017-02781-w
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. doi: 10.1038/s41586-018-0063-9
Wang, Y., Jiang, W., Liu, H., Zeng, Y., Du, B., Zhu, L., et al. (2017). Marker assisted pyramiding of Bph6 and Bph9 into elite restorer line 93-11 and development of functional marker for Bph9. Rice (N Y) 10:51. doi: 10.1186/s12284-017-0194-x
Wu, K., Wang, S., Song, W., Zhang, J., Wang, Y., Liu, Q., et al. (2020). Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 367:eaaz2046. doi: 10.1126/science.aaz2046
Xu, J., Shang, L., Wang, J., Chen, M., Fu, X., He, H., et al. (2021). The SEEDLING BIOMASS 1 allele from indica rice enhances yield performance under low-nitrogen environments. Plant Biotechnol. J. 19, 1681–1683. doi: 10.1111/pbi.13642
Yu, J., Xuan, W., Tian, Y., Fan, L., Sun, J., Tang, W., et al. (2021). Enhanced OsNLP4-OsNiR cascade confers nitrogen use efficiency by promoting tiller number in rice. Plant Biotechnol. J. 19, 167–176. doi: 10.1111/pbi.13450
Zhang, H., Wang, Y., Deng, C., Zhao, S., Zhang, P., Feng, J., et al. (2022). High-quality genome assembly of Huazhan and Tianfeng, the parents of an elite rice hybrid Tian-you-hua-zhan. Sci. China Life Sci. 65, 398–411. doi: 10.1007/s11427-020-1940-9
Zhang, S., Zhu, L., Shen, C., Ji, Z., Zhang, H., Zhang, T., et al. (2021). Natural allelic variation in a modulator of auxin homeostasis improves grain yield and nitrogen use efficiency in rice. Plant Cell 33, 566–580. doi: 10.1093/plcell/koaa037
Zhang, Y., Tan, L., Zhu, Z., Yuan, L., Xie, D., and Sun, C. (2015). TOND1 confers tolerance to nitrogen deficiency in rice. Plant J. 81, 367–376. doi: 10.1111/tpj.12736
Keywords: rice, nitrogen-use efficiency, haplotype analysis, intragenic marker, germplasm accession
Citation: Li P, Li Z, Liu X, Zhang H, Wang Q, Li N, Ding H and Yao F (2022) Development and Application of Intragenic Markers for 14 Nitrogen-Use Efficiency Genes in Rice (Oryza sativa L.). Front. Plant Sci. 13:891860. doi: 10.3389/fpls.2022.891860
Received: 08 March 2022; Accepted: 28 March 2022;
Published: 09 May 2022.
Edited by:
Xia Xin, Institute of Crop Sciences (CAAS), ChinaReviewed by:
Weihua Qiao, Institute of Crop Science (CAAS), ChinaJian Sun, Shenyang Agricultural University, China
Copyright © 2022 Li, Li, Liu, Zhang, Wang, Li, Ding and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fangyin Yao, eWFvZnlAMTYzLmNvbQ==
 Pingbo Li
Pingbo Li Zhen Li1
Zhen Li1