- 1The Key Laboratory for Silviculture and Conservation of Ministry of Education, College of Forestry, Beijing Forestry University, Beijing, China
- 2Key Laboratory of Forest Protection of National Forestry and Grassland Administration, Ecology and Nature Conservation Institute, Chinese Academy of Forestry, Beijing, China
- 3Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, China
The introduction of the pine wood nematode (Bursaphelenchus xylophilus) to new areas has affected the international forestry industry because this pathogen causes pine wilt disease (PWD). Therefore, methods for the accurate and reliable detection of B. xylophilus are essential for controlling and managing this pest. The PCR and Loop-Mediated Isothermal Amplification (LAMP) techniques developed in this study involve species-specific primer sets targeting B. xylophilus genes encoding potential molecular mimicry proteins (Bx-tlp-1, Bx-tlp-2, and Bx-cpi), which are associated with pathogenicity. The PCR and LAMP results revealed that the primers were specific for B. xylophilus Bx-tlp-1, Bx-tlp-2, and Bx-cpi. Moreover, our LAMP assay targeting Bx-tlp-1 conducted at 63°C detected B. xylophilus within 20 min and B. xylophilus from Monochamus alternatus or M. saltuarius within 30 min. The lower limits of detection for the LAMP and PCR assays were 10 pg and 10 ng genomic DNA, respectively, implying these assays may be useful for the rapid detection of B. xylophilus in pine forests. Designing primers specific for Bx-tlp-1, Bx-tlp-2, and Bx-cpi enabled the relatively rapid detection of B. xylophilus isolates as well as M. alternatus or M. saltuarius carrying B. xylophilus. These primers, which were designed following a thorough functional analysis of key B. xylophilus pathogenicity-related genes, may be useful for developing improved assays for the early diagnosis and prevention of PWD.
Introduction
Pine is among the most popular timber species worldwide. Its ecological, economic, and social benefits are internationally recognized. Additionally, pine, which is an integral component of many products influencing human lifestyles, is a major forestation species in China (Jones et al., 2008; Soliman et al., 2012). However, it is susceptible to pine wilt disease (PWD) caused by the pine wood nematode Bursaphelenchus xylophilus,(Nickle et al., 1981), which poses a major threat to pine forests (Mamiya and Enda, 1972; Mota et al., 1999; Futai, 2013).
As a global forest quarantine disease, PWD can adversely affect forest ecologies and prevent sustainable timber production in the Americas, Europe, and Asia. Bursaphelenchus xylophilus is native to North America, but it was introduced to Japan in 1905, China in 1982, South Korea in 1988, Portugal in 1999, Spain in 2008, and the Madeira islands in 2010. After it was introduced to Europe, it spread to the Mediterranean coast (Futai, 2013). Since the first outbreak of PWD in China in 1982 (Cheng et al., 1983), the disease has been detected in 726 county-level administrative regions in 19 provinces. In the winter of 2017, PWD was detected in Fushun, Liaoning province, where the annual average temperature is 8.5°C referring to China Meteorological Administration. Almost 100,000 hectares were affected by PWD, resulting in the deaths of more than 50 million pine trees and economic losses exceeding 25 billion RMB. Hence, strengthening the quarantine and preventing the spread of PWD are the primary objectives of PWD disease management program.
The beetle species Monochamus alternatus and M. saltuarius are the principal vectors of B. xylophilus in Asia. These insects carry the dispersal fourth-stage juveniles of B. xylophilus and other nematodes (Ryss and Subbotin, 2017; Kanzaki and Giblin-Davis, 2018). Therefore, reliably discriminating B. xylophilus from other nematodes according to their morphological characteristics is a difficult task. Examining M. alternatus and M. saltuarius for the presence of B. xylophilus juveniles is an important part of an effective disease control strategy. More specifically, accurately identifying the juvenile stages is crucial for halting the spread of PWD.
Nematode species are routinely identified on the basis of their morphological characteristics. More specifically, B. xylophilus is typically identified according to the morphological characteristics of the adult-stage nematode; however, this requires a certain level of expertise and experience. Moreover, it is impossible to accurately identify the larvae. The limitations to morphology-based methods for identifying B. xylophilus have necessitated the development of methods relying on immunological, physiological, biochemical, and genetic characteristics. The current accepted methods for detecting and identifying B. xylophilus primarily involve molecular biology techniques. For example, the PCR(Wang et al., 2009) and loop-mediated isothermal amplification (LAMP) are commonly used detection techniques.
Regarding PCR, which enables the rapid amplification of DNA, two primers specific for the target DNA must be designed. The target sequence is then amplified by DNA polymerase (e.g., Taq DNA polymerase) in three steps (denaturation, renaturation, and extension). The amplified sequence serves as a template for the next cycle. Because each cycle takes 2–4 min to complete, the target sequence can be amplified several million times in 2–3 h. Because of it, the PCR technique is mature for clinical diagnoses and quarantine. The LAMP technique, which can be used to rapidly amplify DNA, requires four primers specific for six regions of the target DNA. The target sequence is then amplified by Bst DNA polymerase at a constant temperature between 60 and 65°C. As a rapid, simple, specific, sensitive, and low-cost technique, LAMP has been exploited for research regarding disease detection and gene chip development (Tomita et al., 2008).
In this study, we developed PCR- and LAMP-based methods for the direct detection of B. xylophilus. The proposed PCR and LAMP techniques use the species-specific primer sets targeting B. xylophilus genes encoding potential molecular mimicry proteins, including thaumatin-like protein-1 (Bx-tlp-1; accession number KM063438.1), thaumatin-like protein-2 (Bx-tlp-2; accession number MK000287), and a cysteine proteinase inhibitor (Bx-cpi; accession number MK000288). Shinya et al. (2013) performed a proteomic analysis and identified two putative B. xylophilus thaumatin-like proteins (TLPs) and one cysteine proteinase inhibitor with sequences that were highly similar to those of plant proteins. These proteins were subsequently determined to induce cell death in Nicotiana benthamiana (Kirino et al., 2020). Moreover, the TLP sequences in B. xylophilus and Pinus massoniana are reportedly similar (Wang et al., 2014). Additionally, Meng et al. (2017) cloned the gene encoding a P. massoniana TLP (Pm-tlp) and revealed that its expression is associated with the expression of the B. xylophilus Bx-tlp-1. The relatively high sequence similarity between potential molecular mimicry proteins and plant proteins suggests that they may have similar functions. An analysis of the expression of the potential molecular mimicry proteins in B. xylophilus infecting pine trees indicated that the temporal changes to the α-pinene content in the trees are consistent with the expression levels of the genes encoding a TLP (CPI) in B. xylophilus and P. massoniana. Thus, these genes are likely important for B. xylophilus infections of pine species (Meng et al., 2017, 2020).
Because the potential molecular mimicry proteins are associated with B. xylophilus pathogenicity, we designed primers specific for the B. xylophilus genome, which may be useful for developing new disease prevention and control measures.
Materials and Methods
Nematodes and Beetles
Both B. xylophilus and Bursaphelenchus mucronatus were raised on a Botrytis cinerea mycelial lawn on potato dextrose agar medium in plates at 25°C. The nematode cultures were stored in the laboratory. Monochamus alternatus and M. saltuarius were collected in different provinces in China, such as Fujian, Anhui, Zhejiang, Shandong, Guangdong, Tianjin, and Liaoning province. For each beetle, the thorax tissues were cut into two equal parts. One-half was used to isolate and observe nematodes by microscope, and the other half was used to extract DNA for detection, which was described as Meng et al. (2018).
Genomic DNA Extraction
Genomic DNA was extracted from the nematode and beetle tissues as described by Meng et al. (2018) with some modifications. Single nematode was isolated from the culture medium, and transferred to a 200-μl Eppendorf tube. After adding 10 μl TE buffer, the tube was maintained in liquid nitrogen for 1 min and then incubated at 85°C for 1 min. This freeze–thaw treatment was performed three times. The supernatant was collected as the template nematode DNA sample. Beetle thorax tissues (0.2 g) were washed in sterilized water, centrifuged to remove as much water as possible, and then transferred to a 2-ml Eppendorf tube. After adding 200 μl TE buffer, the tube was placed in a grinder operated at 60 Hz for 5 min. After a centrifugation at 10,000 g for 1 min, the beetle samples were maintained in liquid nitrogen for 1 min and then incubated at 85°C for 1 min. This freeze–thaw treatment was performed three times, after which the samples were centrifuged at 10,000 g for 1 min. The supernatant was transferred to a 1.5-ml Eppendorf tube, and the freeze–thaw treatment and centrifugation were repeated. The supernatant was collected as the template beetle DNA sample.
DNA Oligonucleotides
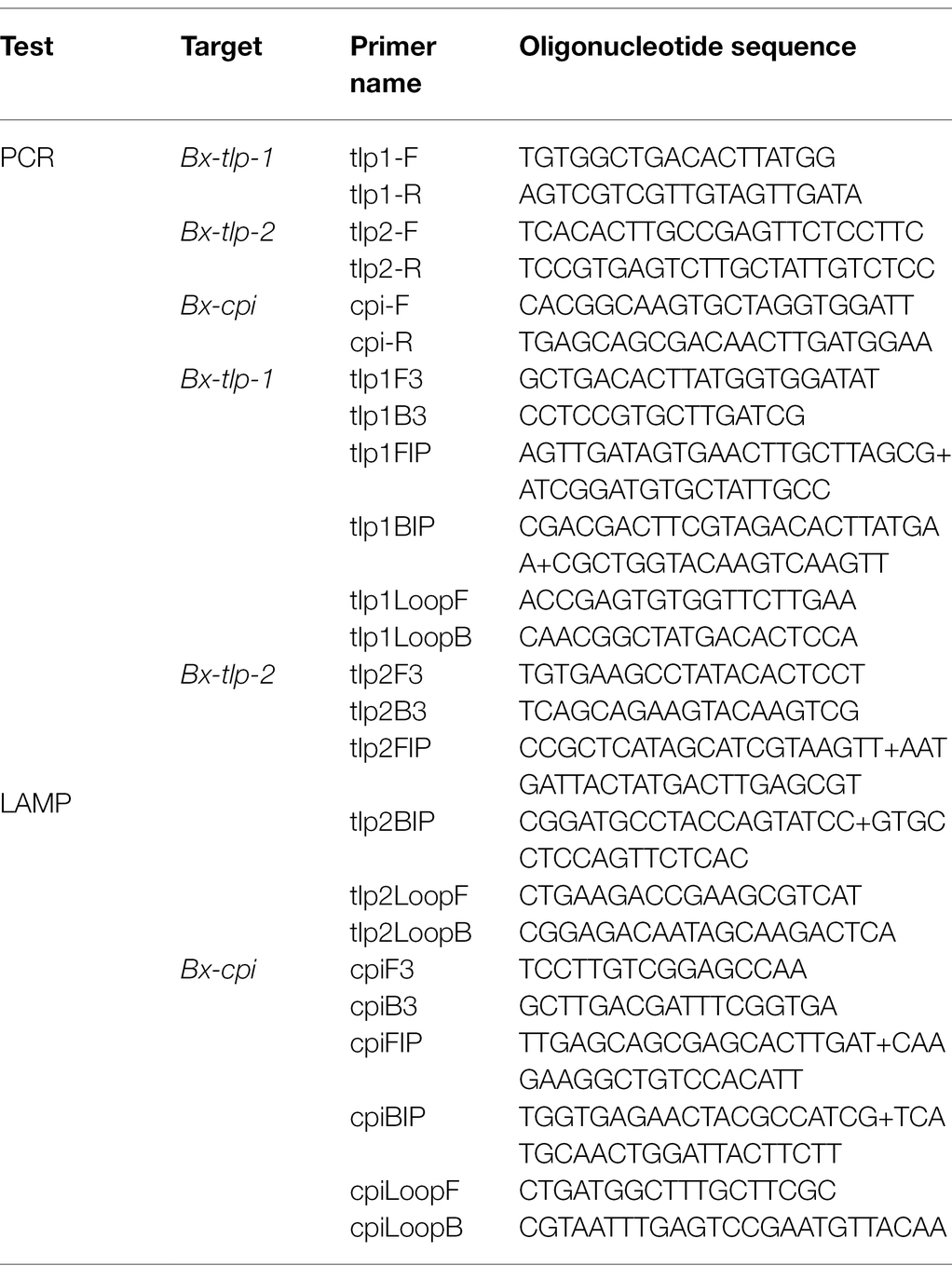
The PCR and LAMP primers designed and used in this study (Table 1) targeted B. xylophilus genes (Bx-tlp-1, Bx-tlp-2, and Bx-cpi) encoding previously identified potential molecular mimicry proteins (Meng et al., 2020). The PCR primers were designed using Primer 6.0. The six LAMP primers were designed using an online LAMP primer design program1 according to previously described criteria (Notomi et al., 2000). The primer set comprised two outer primers (F3 and B3), one forward inner primer (FIP), one reverse inner primer (BIP), and two loop primers (Loop F and Loop B). These primers, which anneal specifically to six distinct regions of the target DNA sequence, were synthesized by the Beijing Genomics Institute (Beijing, China). In addition, the LAMP primers targeting syg-2 designed by Meng et al. (2018) are provided in the Supplementary Material (Supplementary Table S1).
PCR and LAMP
The PCR mixture (25 μl) consisted of the following: PrimeSTAR HS DNA (Premix; TaKaRa, Japan), 12.5 μl; forward and reverse primers, 1 μl each; DNA template, 1 μl; and ddH2O, 9.5 μl. The PCR program was as follows: 94°C for 3 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; 72°C for 10 min. The amplified products were analyzed by agarose gel electrophoresis.
The LAMP mixture (25 μl) comprised the following: primers F3 and B3, 5 pmol each; primers FIP and BIP, 40 pmol each; primers Loop F and Loop B, 20 pmol each; 2 × Reaction Mix (Eiken Chemical, Japan), 12.5 μl; and DNA template, 1 μl. Samples were incubated at 63°C for 60 min and then at 85°C for 3 min to terminate the reaction. A positive control (purified B. xylophilus DNA) and a negative control (distilled water) were included in each run. The LAMP amplicons were detected by fluorescence using LightCycler 480 II system (Roche Diagnostics Ltd., Switzerland) or by a color change in the reaction mixture under natural light.
Analysis of PCR and LAMP Assay Specificity and Sensitivity
To evaluate the specificity of the PCR and LAMP primer sets, the assays were performed using M. alternatus and M. saltuarius genomics DNA as well as the genomic DNA extracted from the following nematode, plant, and fungal species: B. xylophilus, B. mucronatus, Bursaphelenchus fraudulentus, Bursaphelenchus conicaudatus, Bursaphelenchus corneolus, Bursaphelenchus firmae, Bursaphelenchus luxuriosae, Bursaphelenchus sexdentati, Aphelenchoides sp., Meloidogyne incongnita, Caenorhabditis elegans, Pinus thunbergii, Pinus massoniana, Helianthus annuus, Oryza brachyantha, Beauveria bassiana, Pochonia chlamydosporia, Penicillium griseofulvum, Paecilomyces lilacinus, Fusarium oxysporum Schltdl, B. cinerea, and Pestalotia diospyri (Kikuchi et al., 2009; Ye and Giblin-Davis, 2013; Meng et al., 2020). At least three replicates were analyzed for each assay.
The sensitivity for the PCR and LAMP primer sets (i.e., minimal number of copies that could be detected) was assessed by performing the assays using a range of copy numbers (106 to 101) per reaction.
Results
PCR Detection of the Target Genes
Fragments of the B. xylophilus genes encoding potential molecular mimicry proteins (Bx-tlp-1, Bx-tlp-2, and Bx-cpi) were amplified by PCR. The amplified Bx-tlp-1, Bx-tlp-2, and Bx-cpi fragments were 755, 241, and 202 bp, respectively (Figure 1). The sequences of these fragments are provided in the Supplementary Material (Supplementary Figures S1–S3).
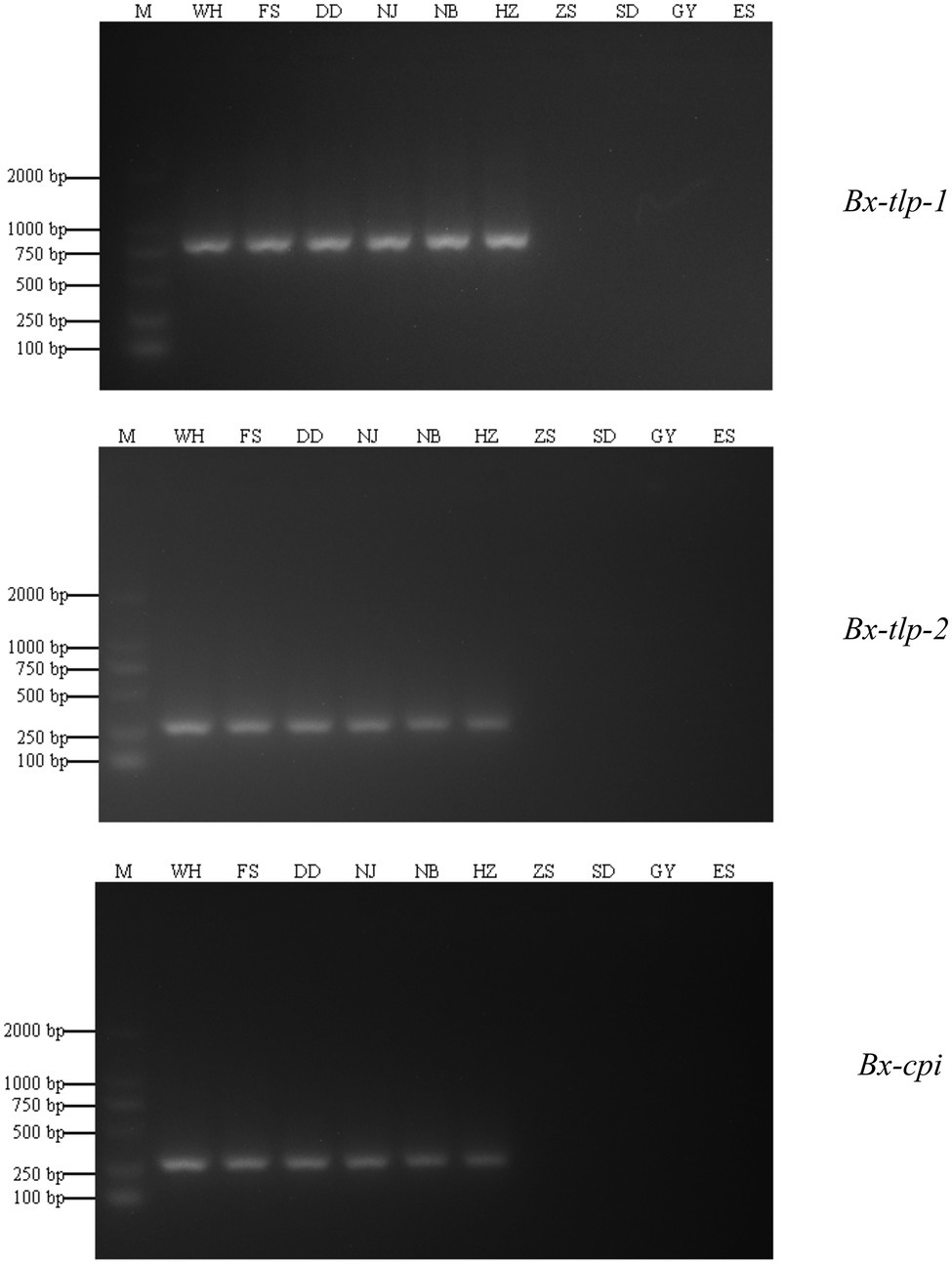
Figure 1. Specificity of the PCR primers for Bursaphelenchus xylophilus and B. mucronatus genes encoding potential molecular mimicry proteins. M, marker; WH, PCR amplification results for B. xylophilus from Shandong; NJ: the PCR amplification results for B. xylophilus from Jiangsu; NB: PCR amplification results for B. xylophilus from Zhejiang; HZ: PCR amplification results for B. xylophilus from Guangdong; FS and DD: PCR amplification results for B. xylophilus from Liaoning; ZS: PCR amplification results for B. mucronatus from Zhejiang; SD: PCR amplification results for B. mucronatus from Hunan; GY: PCR amplification results for B. mucronatus from Sichuan; ES: PCR amplification results for B. mucronatus from Hubei.
LAMP Detection of the Target Genes
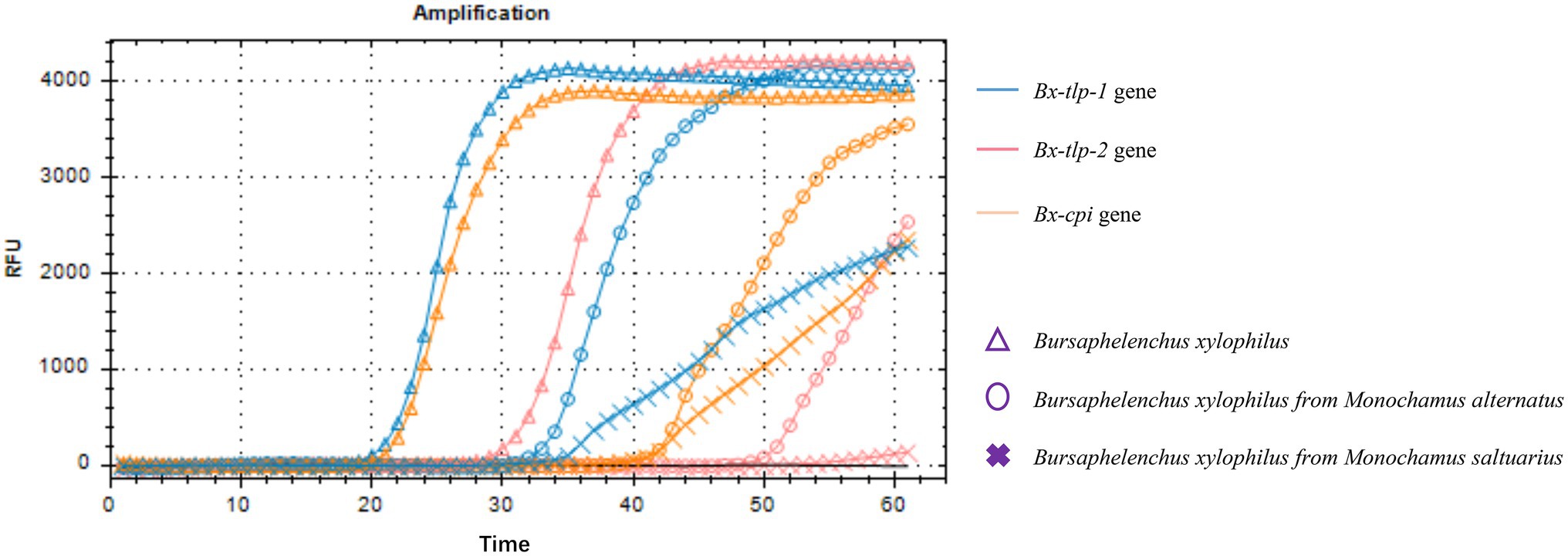
Amplified products are detectable in a LAMP assay within 60 min. In this study, the Bx-tlp-1 and Bx-cpi LAMP amplicons were detectable within 20 min (Figures 2A,C), whereas the Bx-tlp-2 LAMP amplicon was detectable within 30 min (Figure 2B).
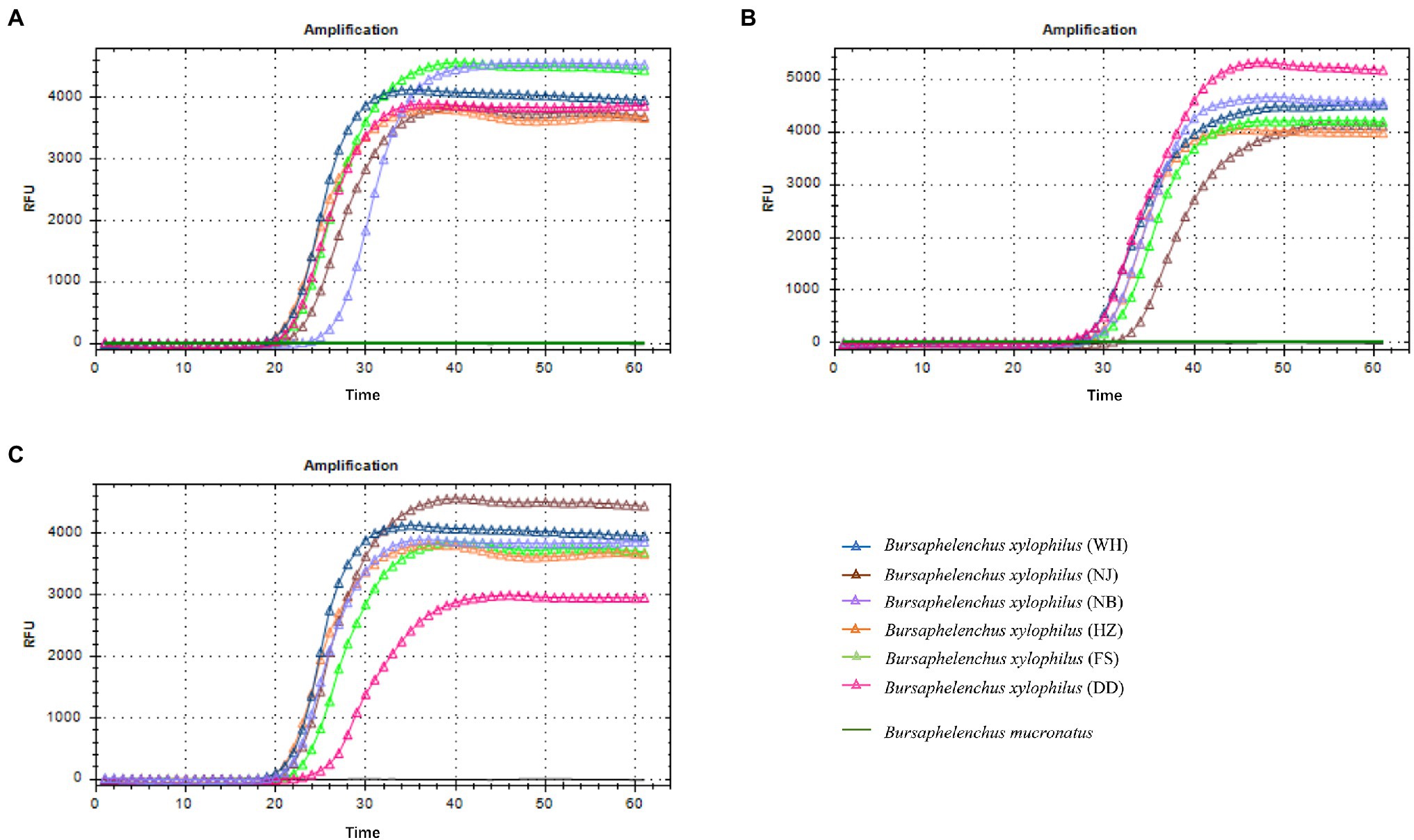
Figure 2. Specificity of the LAMP primers for Bursaphelenchus xylophilus. (A) Bx-tlp-1 gene; (B) Bx-tlp-2 gene; (C) Bx-cpi gene; WH: LAMP results for B. xylophilus from Shandong; NJ: LAMP results for B. xylophilus from Jiangsu; NB: LAMP results for B. xylophilus from Zhejiang; HZ: LAMP results for B. xylophilus from Guangdong; and FS and DD: LAMP results for B. xylophilus from Liaoning.
Specificity of the PCR and LAMP Primers for Bursaphelenchus xylophilus
Amplified products were obtained for the PCR analysis of genomic DNA extracted from the B. xylophilus isolates from Shandong, Jiangsu, Zhejiang, Guangdong, and Liaoning provinces. In contrast, PCR products were not generated for the other nematodes, plants, and fungi or for the negative control (i.e., no template). Accordingly, the PCR primers designed to target Bx-tlp-1, Bx-tlp-2, and Bx-cpi were specific for B. xylophilus. The LAMP primers targeting syg-2 designed by Meng et al. (2018) were unable to produce detectable amplicons within 20 min for the B. xylophilus isolates from Liaoning province (FS and DD; Table 2). However, Bx-tlp-1 and Bx-cpi LAMP amplicons were detectable within 20 min for all B. xylophilus isolates, suggesting the targeted Bx-tlp-1 and Bx-cpi sequences may be specific to B. xylophilus (Supplementary Table S2; Supplementary Figure S4).
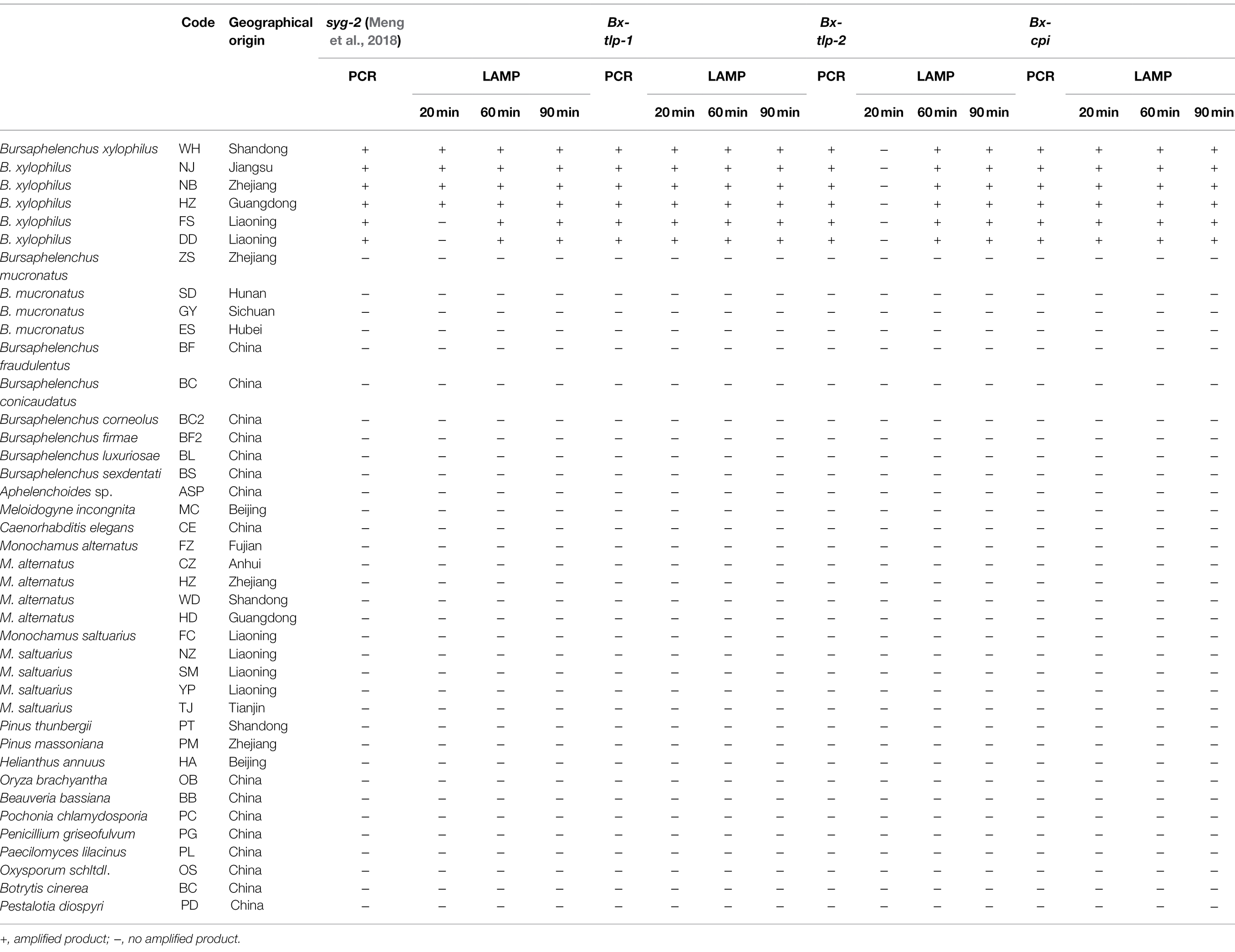
Table 2. Specificity of the PCR and Loop-Mediated Isothermal Amplification (LAMP) assays for Bursaphelenchus xylophilus.
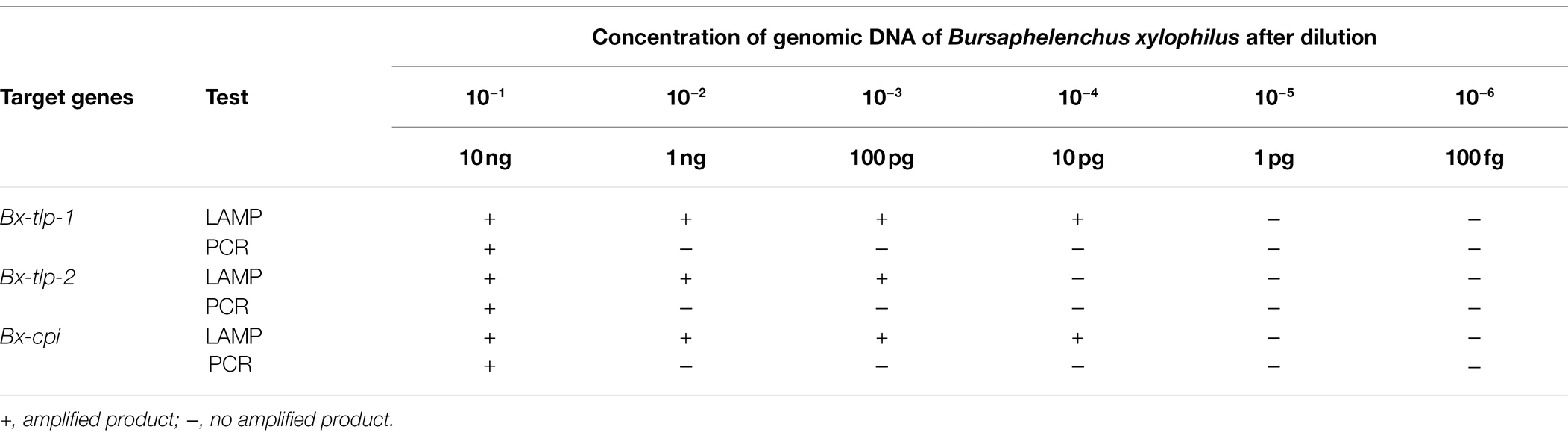
Sensitivity of the PCR and LAMP Assays
On the basis of a comparative analysis of the sensitivities of the two assays, we concluded that the detection limit of the fluorochrome dye used in the LAMP assay was 10 pg for the two positive results (i.e., for Bx-tlp-1 and Bx-cpi), whereas the detection limit of the PCR assay for the same genes was 10 ng (Table 3). The LAMP primers used in this study to target the genes encoding potential molecular mimicry proteins were more sensitive than the primers designed by Meng et al. (2018), which were also associated with B. xylophilus pathogenicity.
Utility of the PCR and LAMP Assays for Detecting Bursaphelenchus xylophilus From Monochamus alternatus or Monochamus saltuarius
The results of the PCR amplification conducted to detect B. xylophilus from M. alternatus or M. saltuarius collected from 10 regions in China are presented in Figure 3. Additionally, the LAMP assay results are presented as an amplification curve visualized using the fluorochrome dye. The results indicated that the primers targeting Bx-tlp-1 and Bx-cpi were able to detect the genomic DNA of B. xylophilus from M. alternatus or M. saltuarius within 40 min, whereas the primers targeting Bx-tlp-2 detected B. xylophilus genomic DNA within 50 min (Figure 4). The primers targeting syg-2 (Meng et al., 2018) detected the genomic DNA of B. xylophilus from M. saltuarius within 50 min (Table 4). Hence, the primers designed for the genes encoding potential molecular mimicry proteins (Bx-tlp-1, Bx-tlp-2, and Bx-cpi) may be more sensitive than the primers targeting syg-2.

Figure 3. Specificity of the PCR primers for the genes encoding potential molecular mimicry proteins in Bursaphelenchus xylophilus from Monochamus alternatus or M. saltuarius. M: DNA marker AL2000; No: no B. xylophilus in M. alternatus; (1): PCR amplification results for B. xylophilus in M. alternatus from Fujian; (2): PCR amplification results for B. xylophilus in M. alternatus from Anhui; (3): PCR amplification results for B. xylophilus in M. alternatus from Zhejiang; (4): PCR amplification results for B. xylophilus in M. alternatus from Shandong; (5): PCR amplification results for B. xylophilus in M. alternatus from Guangdong; (6–9): PCR amplification results for B. xylophilus in M. saltuarius from Liaoning; and (10): PCR amplification results for B. xylophilus in M. saltuarius from Tianjin.
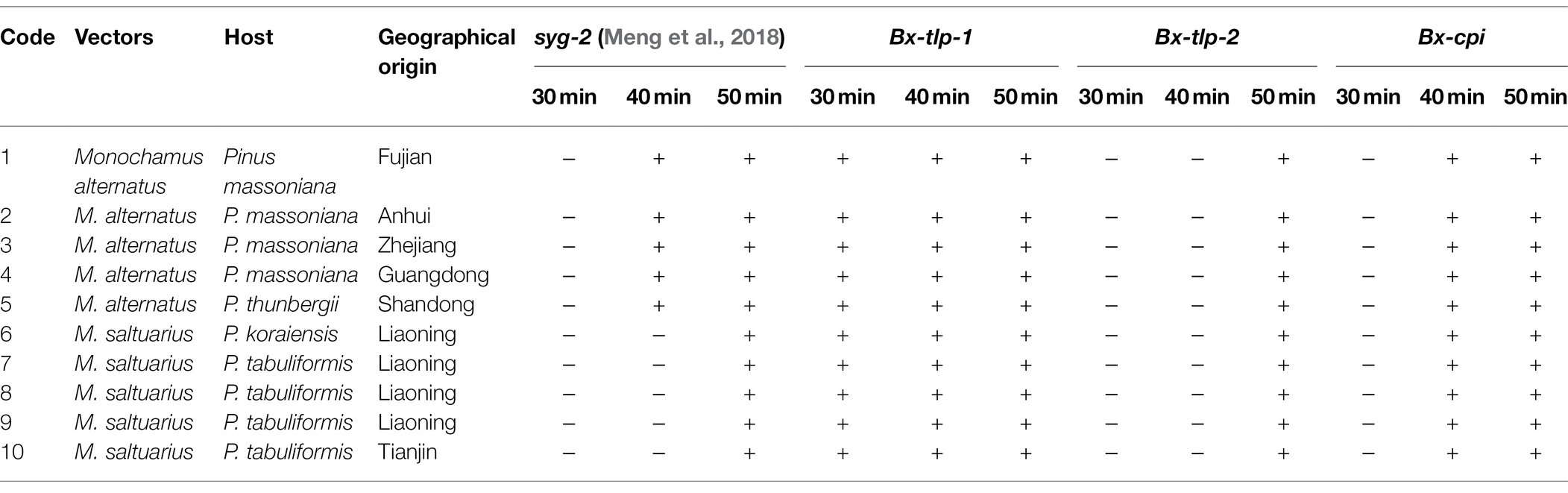
Table 4. LAMP-based detection of Bursaphelenchus xylophilus from Monochamus alternatus or M. saltuarius. +, LAMP amplicon detected; −, LAMP amplicon not detected.
Discussion
The causative agent of PWD, B. xylophilus, is a prevalent organism carried by beetles (i.e., M. alternatus or M. saltuarius). Therefore, the numbers of beetles (M. alternatus or M. saltuarius) in a pine forest is a crucial indicator of the possibility of a PWD outbreak. Unless B. xylophilus from M. alternatus or M. saltuarius is specifically treated in the primary phase, the resulting PWD will often lead to tree death. The efficient and rapid detection of B. xylophilus from M. alternatus or M. saltuarius is necessary for preventing outbreaks via the implementation of specific treatments (Wang et al., 2009; Cardoso et al., 2012). Because death by PWD is very rapid, a novel method for detecting PWD must produce results quickly.
Proteins secreted by pathogens that are structurally or functionally similar to host defense-related proteins are called molecular mimicry proteins. Researchers identified substances released by pathogens that mimic plant defense-related compounds and disrupt physiologically important plant signaling pathways (Winn et al., 2021). Previous studies revealed the relatively high similarity between the Bx-TLP (Bx-CPI) and Pm-TLP (Pm-CPI) sequences, indicative of a similarity in their functions (Wang et al., 2014; Meng et al., 2017, 2020, 2022). Moreover, the expression of B. xylophilus genes encoding potential molecular mimicry proteins (Bx-tlp-1, Bx-tlp-2, and Bx-cpi) is responsive to α-pinene, which may affect terpene metabolism in pine trees and influence the pathogenicity of B. xylophilus (Meng et al., 2019, 2020, 2022).
In the present study, we designed specific primer sets targeting Bx-tlp-1, Bx-tlp-2, and Bx-cpi for the rapid detection of B. xylophilus and M. alternatus or M. saltuarius carrying B. xylophilus. The PCR and LAMP assays specifically detected all B. xylophilus isolates, with no amplification for all other examined species. The high specificity of the LAMP assay was conferred by the six primers targeting six regions of the Bx-tlp-1 and Bx-cpi sequences, which were specific to B. xylophilus. The specificity of the LAMP primers used in this work was significantly better than that reported by Kikuchi et al. (2009; Supplementary Table S2), who detected B. mucronatus and B. fraudulentus in 90 min. On the other hand, there were no amplified products in other Bursaphelenchus group species, such as B. firmae, B. luxuriosae, and B. sexdentati. Meanwhile, the average concentration of genomic DNA from a single B. xylophilus was about 32 ng/μl (the volume was 10 μl), and the lower limits of detection for the LAMP and PCR assays were 10 pg and 10 ng genomic DNA, respectively. Moreover, LAMP primers targeting Bx-tlp-1 or Bx-cpi were 5-fold more sensitive than the LAMP primers targeting syg-2 (51.4 pg; Meng et al., 2018) and were able to rapidly detect B. xylophilus from M. alternatus or M. saltuarius. Thus, the key genes associated with B. xylophilus pathogenicity may be excellent targets for the rapid detection of the nematode because they may also be related to the fitness of the hosts, insects, and environment.
Considering the high sensitivity of the LAMP assay, the post-amplification procedures should be performed in a separate room (i.e., away from the PCR and LAMP reagents) to minimize the possibility of contamination (Kikuchi et al., 2009; Zhu et al., 2009; Meng et al., 2018). The LAMP assay requires only a regular water bath or a heat block that maintains the temperature at 63°C. Therefore, it is more cost-effective than a conventional PCR assay. Moreover, the two additional loop primers increase the speed and specificity of the amplification, resulting in faster reactions (relative to a conventional PCR). Additionally, the key genes associated with pathogenicity (e.g., those encoding potential molecular mimicry proteins) can be used as the specific targets for detection, which may be significant for preventing or controlling disease outbreaks. Our LAMP-based method detected B. xylophilus Bx-tlp-1 within 20 min and B. xylophilus from M. alternatus or M. saltuarius within 30 min. These results are better than the LAMP assay results obtained by Meng et al. (2018) who detected the B. xylophilus target gene within 25 min and B. xylophilus from M. alternatus within 50 min. Furthermore, in contrast to a conventional PCR assay, the LAMP assay enables the detection of a positive amplification by the naked eye. The reactions and results can be interpreted simply by observing the color change in the reaction mixture. In addition to eliminating the need for the time-consuming electrophoretic analysis, LAMP assays can be performed without the sophisticated equipment needed for PCR (Kang et al., 2009; Huang et al., 2010; Hu et al., 2011; Kanetani et al., 2011; Meng et al., 2018).
In conclusion, M. alternatus is the main insect vector for B. xylophilus. The number of nematodes in M. alternatus is an important factor related to the distribution of B. xylophilus. Therefore, the fast and efficient detection of M. alternatus carrying B. xylophilus is necessary for the monitoring and early detection of PWD (Kikuchi et al., 2009; Hu et al., 2011; Cardoso et al., 2012; Meng et al., 2018). In this study, we designed PCR and LAMP primers specific for the B. xylophilus genes encoding potential molecular mimicry proteins (Bx-tlp-1, Bx-tlp-2, and Bx-cpi). The specific amplified fragments for Bx-tlp-1, Bx-tlp-2, and Bx-cpi were 755, 241, and 202 bp, respectively. However, no specific amplification products were detected for the other analyzed nematodes (e.g., B. mucronatus). These findings indicate that Bx-tlp-1, Bx-tlp-2, and Bx-cpi can be used to specifically detect B. xylophilus as well as M. alternatus or M. saltuarius carrying B. xylophilus. Furthermore, designing primers specific for B. xylophilus following an in-depth analysis of the functions of key pathogenicity genes may have important implications for future attempts at developing reliable methods for the early diagnosis and prevention of PWD.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
FM, YL, and XZ: experimental design. FM and ZL: experimental implementation. ZL: material contribution. FM: data analysis and manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
The present research was funded by the Fundamental Research Funds for the Central Universities (2021ZY03) and China Postdoctoral Science Foundation (2020M680410).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.890949/full#supplementary-material
Footnotes
References
Cardoso, J. M. S., Fonseca, L., and Abrantes, I. (2012). Direct molecular detection of the pine wood nematode, Bursaphelenchus xylophilus, from pine wood, bark and insect vector. Eur. J. Plant Pathol. 133, 419–425. doi: 10.1007/s10658-011-9915-y
Cheng, H., Lin, M., Li, W., and Fang, Z. (1983). The occurrence of a pine wilting disease caused by a nematode found in Nanjing. Forest Pest Dis. 4, 1–5.
Futai, K. (2013). Pine wood nematode, Bursaphalenchus xylopholus. Annu. Rev. Phytopathol. 51, 61–83. doi: 10.1146/annurev-phyto-081211-172910
Hu, Y. Q., Kong, X. C., Wang, X. R., Zhong, T. K., Zhu, X. W., Mota, M. M., et al. (2011). Direct PCR-based method for detecting Bursaphelenchus xylophilus, the pine wood nematode in wood tissue of Pinus massoniana. Forest Pathol. 41, 165–168. doi: 10.1111/j.1439-0329.2010.00692.x
Huang, L., Ye, J. R., Wu, X. Q., Xu, X. L., Sheng, J. M., and Zhou, Q. X. (2010). Detection of the pine wood nematode using a real-time PCR assay to target the DNA topoisomerase I gene. Eur. J. Plant Pathol. 127, 89–98. doi: 10.1007/s10658-009-9574-4
Jones, J. T., Moens, M., Mota, M., Li, H. M., and Kikuchi, T. (2008). Bursaphelenchus xylophilus: opportunities in comparative genomics and molecular host-parasite interactions. Mol. Plant Pathol. 9, 357–368. doi: 10.1111/j.1364-3703.2007.00461.x
Kanetani, S., Kikuchi, T., Akiba, M., Nakamura, K., Ikegame, H., and Tetsuka, K. (2011). Detection of Bursaphelenchus xylophilus from old discs of dead Pinus armandii var. amamiana trees using a new detection kit. Forest Pathol. 41, 387–391. doi: 10.1111/j.1439-0329.2010.00695.x
Kang, J. S., Moon, I. S., Lee, S. G., Shin, S. C., and Lee, S. H. (2009). Rapid and accurate prediction of the frequencies of Bursaphelenchus xylophilus and B. mucronatus in mixed nematode samples using real-time species-specific PCR. Nematology 11, 289–299. doi: 10.1163/156854109X429619
Kanzaki, N., and Giblin-Davis, R. M. (2018). Diversity and plant pathogenicity of Bursaphelenchus and related nematodes in relation to their vector bionomics. Curr. Forest. Rep. 4, 85–100. doi: 10.1007/s40725-018-0074-7
Kikuchi, T., Aikawa, T., Oeda, Y., Karim, N., and Kanzali, N. (2009). A rapid and precise diagnostic method for detecting the pinewood nematode Bursaphelenchus xylophilus by loop-mediated isothermal amplification. Phytopathology 99, 1365–1369. doi: 10.1094/PHYTO-99-12-1365
Kirino, H., Yoshimoto, K., and Shinya, R. (2020). Thaumatin-like proteins and a cysteine protease inhibitor secreted by the pine wood nematode Bursaphelenchus xylophilus induce cell death in Nicotiana benthamiana. PLoS One 15:e0241613. doi: 10.1371/journal.pone.0241613
Mamiya, Y., and Enda, N. (1972). Transmission of Bursaphelenchus lignicolus (Nematoda: Aphelenchoididae) by Monochamus alternatus (Coleoptera: Cerambycidae). Nematologica 18, 159–162. doi: 10.1163/187529272X00395
Meng, F. L., Li, Y. X., Liu, Z. K., Feng, Y. Q., Wang, X., and Zhang, X. Y. (2022). Expression of the thaumatin-like protein-1 gene (Bx-tlp-1) from pine wood nematode Bursaphelenchus xylophilus affects terpene metabolism in pine trees. Phytopathology 112, 888–897. doi: 10.1094/PHYTO-07-21-0289-R
Meng, F. L., Li, Y. X., Liu, Z. K., Wang, X., Feng, Y. Q., Wei, Z., et al. (2020). Potential molecular mimicry proteins responsive to α-pinene in Bursaphelenchus xylophilus. Int. J. Mol. Sci. 21:982. doi: 10.3390/ijms21030982
Meng, F. L., Li, Y. X., Wang, X., Feng, Y. Q., Liu, Z. K., Zhang, W., et al. (2019). Thaumatin-like protein-1 gene (Bx-tlp-1) is associated with the pathogenicity of Bursaphelenchus xylophilus. Phytopathology 109, 1949–1956. doi: 10.1094/PHYTO-03-19-0082-R
Meng, F. L., Wang, X. Z., Wang, L. F., Gou, D. P., Liu, H. J., Wang, Y. N., et al. (2018). A loop-mediated isothermal amplification-based method for detecting Bursaphelenchus xylophilus from Monochamus alternatus. For. Pathol. 48:e12404. doi: 10.1111/efp.12404
Meng, F. L., Wang, J., Wang, X., Li, Y. X., and Zhang, X. Y. (2017). Expression analysis of thaumatin-like proteins from Bursaphelenchus xylophilus and Pinus massoniana. Physiol. Mol. Plant Pathol. 100, 178–184. doi: 10.1016/j.pmpp.2017.10.002
Mota, M. M., Braasch, H., Bravo, M. A., Penas, A. C., Burgermeister, W., Metge, K., et al. (1999). First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology 1, 727–734. doi: 10.1163/156854199508757
Nickle, W. R., Golden, A. M., Mamiya, Y., and Wergin, W. P. (1981). On the taxonomy and morphology of the pine wood nematode, Bursaphelenchus xylophilus (Steiner & Buhrer 1934) Nickle 1970. J. Nematol. 13, 385–392.
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. doi: 10.1093/nar/28.12.e63
Ryss, A. Y., and Subbotin, S. A. (2017). Coevolution of wood-inhabiting nematodes of the genus Bursaphelenchus Fuchs, 1937 with their insect vectors and plant hosts. Zh. Obshch. Biol. 78, 13–42.
Shinya, R., Morisaka, H., and Kikuchi, T. (2013). Secretome analysis of the pine wood nematode Bursaphelenchus xylophilus reveals the tangled roots of parasitism and its potential for molecular mimicry. PLoS One 8:e67377. doi: 10.1371/journal.pone.0067377
Soliman, T., Mourits, M. C. M., Vanderwerf, W., Hengeveld, G. M., Robinet, C., and Lansink, A. G. J. (2012). Framework for modeling economic impacts of invasive species, applied to pine wood nematode in Europe. PLoS One 7:e45505. doi: 10.1371/journal.pone.0045505
Tomita, N., Mori, Y., Kanda, H., and Notomi, T. (2008). Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of the products. Nat. Protoc. 3, 877–882. doi: 10.1038/nprot.2008.57
Wang, J., Han, S., Li, Y., Deng, X., and Zhang, X. Y. (2014). Cloning of TLP-1 gene and prediction of TLP-1 protein structure of Bursaphelenchus xylophilus. J. Sichuan Agri. Univ. 32, 305–310. doi: 10.3969/j.issn.1000-2650.2014.03.011
Wang, X. R., Zhu, X. W., Hu, Y. Q., Huang, H. H., Kong, X. C., and Jia, W. H. (2009). A PCR-based method for detecting the pine wood nematodes—Bursaphelenchus xylophilus from Monochamus alternatus. Sci. Silvae Sin. 45, 70–75. doi: 10.11707/j.1001-7488.20090712
Winn, M., Rowlinson, M., Wang, F. H., Bering, L., and Micklefield, J. (2021). Discovery, characterization and engineering of ligases for amide synthesis. Nature 593, 391–398. doi: 10.1038/s41586-021-03447-w
Ye, W. M., and Giblin-Davis, R. M. (2013). Molecular characterization and decelopment of real-time PCR assay for pine-wood nematode Bursaphelenchus xylophilus (Nematoda: parasitaphelenchidae). PLoS One 8:e78804. doi: 10.1371/journal.pone.0078804
Keywords: Bursaphelenchus xylophilus, potential molecular mimicry proteins, specific target, rapid detection, PCR and LAMP
Citation: Meng F, Liu Z, Li Y and Zhang X (2022) Genes Encoding Potential Molecular Mimicry Proteins as the Specific Targets for Detecting Bursaphelenchus xylophilus in PCR and Loop-Mediated Isothermal Amplification Assays. Front. Plant Sci. 13:890949. doi: 10.3389/fpls.2022.890949
Edited by:
Anna Filipiak, Institute of Plant Protection – National Research Institute, PolandReviewed by:
Kai Guo, Zhejiang Agriculture and Forestry University, ChinaAlfonso Navas, CSIC, Spain
Copyright © 2022 Meng, Liu, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongxia Li, bHl4MDIwNDE5QGNhZi5hYy5jbg==
 Fanli Meng
Fanli Meng Zhenkai Liu
Zhenkai Liu Yongxia Li2,3*
Yongxia Li2,3*