- 1Center for Agricultural Resources Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Shijiazhuang, China
- 2National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 3State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 4Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing, China
Rye (Secale cereale L.), a naturally cross-pollinating relative of wheat, is a tertiary gene donor and of substantial value in wheat improvement. Wheat powdery mildew is caused by Blumeria graminis f. sp. tritici (Bgt), which seriously affects yield and quality worldwide. Identifying and transferring new, effective resistance genes against powdery mildew from rye is important for wheat breeding. The current study developed a wheat-rye line YT2 resistant to powdery mildew by crossing, backcrossing, and self-pollination for multiple generations between octoploid triticale 09R2-100 and common wheat cultivar Shixin 616. YT2 was confirmed to be a 6R disomic addition and T1RS⋅1BL translocation line by genomic in situ hybridization (GISH), multicolor fluorescence in situ hybridization (mc-FISH), multicolor-GISH (mc-GISH), and molecular marker analyses. Disease responses to different Bgt isolates and genetic analysis showed that the powdery mildew resistance gene of YT2 was derived from the rye chromosome 6R of 09R2-100, which differed from the previously reported Pm genes from rye including Pm20 on 6RL. Resistance phenotype of different translocation lines and deletion lines derived from YT2 combined with newly developed 6RL-specific markers analysis suggested that the powdery mildew resistance gene of YT2 was localized to the region in chromosome 6RL: 890.09–967.51 Mb and flanked by markers XM189 and X4M19, corresponding to the reference genome of Weining rye. Therefore, YT2 could be used as a promising bridging parent for wheat disease resistance improvement.
Introduction
Common wheat (Triticum aestivum L.) is one of the most important grain crops worldwide ensuring national food security. Powdery mildew, caused by Blumeria graminis (DC.) Speer f. sp. tritici emend. É. J. Marchal (Bgt), can significantly decrease wheat yield and quality worldwide by 5–40% and, in severe cases, up to 62% (Singh et al., 2016; Sun et al., 2018). Using host resistance is considered the most effective and sustainable way of controlling this disease, so a large number of powdery mildew (Pm) resistance genes/alleles continue to be explored, with more than a hundred Pm genes (Pm1–Pm68, noting that Pm8 = Pm17, Pm18 = Pm1c, Pm22 = Pm1e, Pm23 = Pm4c, and Pm31 = Pm21) reported so far (McIntosh et al., 2020; He et al., 2021). However, most of the Pm genes cannot be directly applied in wheat breeding programs due to undesirable linkage drag. In addition, Pm genes are easily overcome after long periods of utilization in practical production by the highly variable pathogen (Mohler and Stadlmeier, 2019). A well-known example is the “boom-bust” of Pm8, which has caused continual severe epidemics in the main wheat production regions in China (He et al., 2011, 2015; An et al., 2019). Therefore, it is a continuously urgent need to explore and utilize sustainable resistance against powdery mildew in new gene resources.
For a long time, wheat breeding has mainly focused on the crossing between cultivars, leading to homogeneous genetic backgrounds and narrowed genetic diversity (Gupta et al., 2010). A growing number of studies have revealed the immense potential of mining useful genes from wheat relatives (Friebe et al., 1996; Colmer et al., 2006; Tester and Langridge, 2010; Chen et al., 2013; Hurni et al., 2015; Li H.H. et al., 2020; Li J.B. et al., 2020). Rye (Secale cereale L.) is a naturally cross-pollinating relative of common wheat, which possesses huge diversity for using as a gene donor in wheat improvement (Targońska et al., 2016; An et al., 2019). Many resistance genes on chromosome 6R of rye have been identified and transferred into wheat, including Pm56 on 6RS from Qinling rye and Pm20 on 6RL from Prolific rye for powdery mildew resistance (Friebe et al., 1994; Hao et al., 2018); Yr83 on 6RL from T-701 triticale derivatives for stripe rust resistance (Li J.B. et al., 2020); CreR on 6RL from T-701 triticale derivatives for cereal cyst nematode (Heterodera avenae Woll.) resistance (Taylor et al., 1998); and H25 on 6RL from Balbo rye for Hessian fly resistance (Friebe et al., 1991, 1996). Apart from these formally designated resistance genes, some wheat-rye 6R derivative lines have also exhibited resistance that was identified to be derived from rye. For instance, 6R from German White rye and 6RL from Kustro rye both conferred resistance to powdery mildew (Fu et al., 2014; An et al., 2015; Du et al., 2018). The chromosome 6R from Kriszta rye appeared to be resistant to stripe rust (Schneider et al., 2016). The recently published reference genome of Weining rye revealed that as many as 287 disease resistance-associated genes were predicted on chromosome 6R (Li et al., 2021). So, widening the variation of 6R to achieve its full potential for novel resistance gene resources is an ongoing and attractive prospect.
Molecular markers specific for alien chromosomes are powerful for easily detecting alien chromatin in the wheat background (Hechanova et al., 2018). To date, many types of molecular markers specific for the chromosome 6R were developed (Korzun et al., 1998; Stojalowski et al., 2009; Milczarski et al., 2011; Xu et al., 2012; Li et al., 2013, 2016; Qiu et al., 2016; Du et al., 2018; Han et al., 2020a; Li J.B. et al., 2020), which has accelerated gene mapping, improved the accuracy of germplasm selection, and promoted germplasm innovation. Nevertheless, the number of markers covering the entire chromosome 6R is still in large demand, especially for those with accurate physical location information and tightly linked to the targeted genes. Fortunately, the recent release of the high-quality rye reference genome sequence can greatly accelerate the development of rye-specific markers and the identification of useful genes (Li et al., 2021; Rabanus-Wallace et al., 2021).
Octoploid triticale (× Triticosecale Wittmack, 2n = 8x = 56, AABBDDRR) is synthesized artificially through distant hybridization between common wheat and rye. It possesses multiple resistance genes to wheat disease and can be more easily crossed with wheat than rye, thus can be used as an alternative source in wheat improvement (Oettler et al., 2005; Li et al., 2018). In the present study, after many years of selection of disease resistance and satisfactory agronomic traits, a stable wheat-rye 6R derivative line YT2 with high resistance to powdery mildew at the adult plant stage was obtained. To better understand YT2 and to facilitate the utilization of its powdery mildew resistance, this study reported the following: (1) its chromosome composition; (2) the assessment and genetic analysis of its powdery mildew resistance; (3) the development of chromosome arm 6RL-specific markers; (4) the physical localization of its powdery mildew resistance gene; and (5) the evaluation of its agronomic performance for breeding.
Materials and Methods
Plant Materials
A wheat-rye derivative line, designated as YT2, was produced by crossing octoploid triticale 09R2-100 (2n = 8x = 56, AABBDDRR) with wheat cultivar Shixin 616, with self-pollination for multiple generations. Wheat cultivar Mingxian 169 susceptible to all the Bgt isolates tested was used as a susceptible control for resistance assessment to powdery mildew at both seedling and adult stages. To evaluate the powdery mildew resistance of YT2, 47 wheat genotypes with the known Pm gene(s) were used as controls, including TAM104/Thatcher with Pm20 from 6RL, Kavkaz with Pm8, and Amigo with Pm17 that both located on 1RS, and CI14189 with Pm7 from 2RL. Each genotype was tested with 22 single-pustule-derived powdery mildew isolates in the same way to compare their reaction patterns. A population of 97 F2 plants from a cross of YT2 and susceptible commercial cultivar Gao 8901 was used to determine the resistance source of powdery mildew resistance.
Wheat cultivars Chinese Spring (CS) and Holdfast, rye cultivars Weining and King II, 6R disomic addition line of Holdfast × King II (DA 6R), 6RS ditelosomic addition line (DtA 6RS), and 6RL ditelosomic addition line (DtA 6RL) were used to verify the specificity of molecular markers.
The complete set of disomic addition lines (DA 1R to 7R) of CS × Imperial, two hexaploid triticale lines 10R2-193-2 and 10R2-194-2 (AABBRR), and two T1BL⋅1RS wheat-rye translocation lines Lovrin 10 and Lovrin 13 (Rabinovich, 1998) were used as controls to characterize YT2 in molecular marker analysis.
Sequential Genomic in situ Hybridization, Multicolor-Fluorescence in situ Hybridization, and Non-denaturing FISH Analyses
Genomic in situ hybridization and FISH were performed to determine the chromosomal composition of YT2. The mitotic metaphase chromosomes from root tips of YT2 were prepared and observed as previously described (An et al., 2013). Total genomic DNA of German White labeled with fluorescein-12-dUTP using nick translation method was used as a probe for GISH. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI), and detection and visualization of rye chromatin were conducted according to An et al. (2013).
After rinsing the GISH hybridization probe signals, the probe pSc119.2 (green) labeled with fluorescein-12-dUTP combined with probe pAs1 (red) labeled with Texas-red-5-dCTP was used in mc-FISH detection. For ND-FISH detection, Oligo-pSc119.2-2 and Oligo-pTa535-2 were 5′-end-labeled with 6-carboxyfluorescein (6-FAM) producing green signals and 6-carboxytetramethylrhodamine (TAMRA) producing red signals, respectively. The FISH patterns were referred to Tang et al. (2014). These oligonucleotide probes were synthesized by Shanghai Invitrogen Biotechnology Co., Ltd. (Shanghai, China). Mc-FISH analysis was performed according to the method of An et al. (2013). ND-FISH analysis was operated according to Fu et al. (2015).
Multicolor-GISH Analysis
Total genomic DNA was isolated from the fresh leaves of Triticum urartu (2n = 2x = 14, AA), Aegilops speltoides (2n = 2x = 14, BB), and Aegilops tauschii (2n = 2x = 14, DD). The total genomic DNA of rye and T. urartu was labeled with fluorescein-12-dUTP, and the total genomic DNA of Ae. tauschii was labeled with Texas-red-5-dCTP, while total genomic DNA of Ae. speltoides was used for blocking (Fu et al., 2012). Detection and visualization of mc-GISH signals were performed as the same as for GISH.
Polymerase Chain Reaction Analysis
Three specific markers SW22057 (F: 5′-GAAGAGGACC GATGCCACTA-3′, R: 5′-TCACACTCCGGACAATGCTA-3′), SW22063 (F: 5′-TCTGCTTGATGATGATCTGCTT, R: 5′-TC CGCAAACCCTAACATTTC-3′), and SW5284 (F: 5′- CCTATC CCTCTGTGGCGAAT -3′, R: 5′- GTCGTGTTCCCCTACA TCCA -3′) were used to detect rye chromosome arms 6RS, 6RL, and 1RS, respectively (Han et al., 2020a). The marker O11B3/O11B5 (F: 5′-GGTACCAACAACAACAACCC-3′, R: 5′- GTTGCTGCTGAGGTTGGTTC-3′) was used as a diagnostic marker to detect wheat chromosome arm 1BS (Van Campenhout et al., 1995).
Assessment of Resistance to Powdery Mildew
Seedling stage reactions of YT2 to 22 single-pustule-derived isolates were separately assessed with three replicates at the greenhouse as described (Qie et al., 2019). These isolates were collected from different wheat production regions of China and were characterized by their individual identifying host (Supplementary Table 1). Mingxian 169 and a set of 47 wheat genotypes with documented Pm gene(s) were used as controls (Supplementary Table 1). For each line, 20 seeds were planted in 128-well rectangular trays (54 cm × 28 cm × 4 cm), with 5 seeds per well (3 cm × 3 cm × 4 cm). After 15 days, when sporulation was observed on the first leaf of Mingxian 169, the plants were scored using a 0–4 scale with infection types (ITs) 0–2 considered resistant, while ITs 3–4 considered susceptible (Si et al., 1992).
Adult plant reactions to powdery mildew were tested on YT2, Shixin 616, 09R2-100, TAM104/Thatcher, CI14189, Kavkaz, and Amigo in field conditions, using three replicates during the 2018–2020 growing seasons at Luancheng Agro-Ecological Experimental Station, Chinese Academy of Sciences, Shijiazhuang, China (37°53′15″N, 114°40′47″E). The wheat cultivar Mingxian 169 was planted as susceptible control and inoculum spreader. In late March, Mingxian 169 was artificially inoculated with a mixture of Bgt isolates which mainly comprised virulent isolates E09, E18, and E20 collected from northern China. In May, when the susceptible control Mingxian 169 showed severe disease symptoms, the disease reaction of the tested plants at the adult stage was assessed using a 0–9 scale, in which 0–4 were considered resistant and 5–9 were considered susceptible (Sheng and Duan, 1991). Each plant was assessed twice for confirmation.
Genetic Analysis of the Resistance to Powdery Mildew
To determine the resistance source of YT2, an F2 population of 97 plants derived from YT2 × Gao 8901 was tested for powdery mildew resistance. YT2, Gao 8901, and Mingxian 169 were also included in the test. At the seeding stage, the F2 plants were inoculated with Bgt isolate E09, a prevalent isolate in northern China (Li et al., 2011), to evaluate their seedling reactions and then transplanted into the field to test the adult plant reactions. Both seedling and adult plant reactions of each plant were assessed twice for confirmation. Three molecular markers, 6RS-specific marker SW22057, 6RL-specific marker SW22063, and 1RS-specific marker SW5284, combined with GISH were used to detect the rye chromatin in YT2.
Development of 6RL-Specific Molecular Markers
To localize the powdery mildew resistance gene in a certain chromosome region, PCR-based markers specific for the 6RL chromosome were developed, based on the different reference sequences between Chinese Spring (International Wheat Genome Sequencing Consortium et al., 2018) and Weining rye (Li et al., 2021). All the primers were synthesized by Shanghai Sangon Biotechnology Co., Ltd. (Shanghai, China).
Chromosomal Localization of Powdery Mildew Resistance Gene of YT2
60Coγ-ray radiation was used to develop progenies of YT2 containing translocations or deletions with different sizes of segments of chromosome 6R. At the flowering stage, the pollens of YT2 plants were treated with 60Co radiation at a dosage of 18 Gy and dosage rate of 1 Gy/min and then crossed with Gao 8901. To physically map the powdery mildew resistance locus, the irradiated F1 plants and their selfing F2 progenies were subject to GISH and molecular analysis and then tested for resistance to powdery mildew at the seedling and adult plant stages.
Evaluation of Agronomic Traits
YT2 and its octoploid triticale parent 09R2-100 and wheat parent Shixin 616 were planted in the field. An evaluation of agronomic traits was conducted from 2018 to 2020 with three replicates in a randomized complete block design. Sowing and assessment methods were performed as described by Han et al. (2020b). At physiological maturity, plant height (PH), spike length (SL), spike number per plant (SNPP), fertile spikelet number per spike (FSS), thousand-kernel weight (TKW), kernel number per spike (KNS), and grain yield per plant (GYPP) of 20 randomly selected plants in the middle of the two internal rows were assessed. The analysis of variance (ANOVA) and least significant difference (LSD) test were performed using software SPSS 22.0 (SPSS Inc., Chicago, IL, United States) to test the significance of differences between YT2 and its parents Shixin 616 and 09R2-100 for each agronomic trait.
Results
Genomic in situ Hybridization Analysis of YT2
The GISH results demonstrated that YT2 included 44 chromosomes in a cell and had one pair of intact chromosomes and two chromosome arms that displayed bright-green hybridization signals. The two chromosome arms contained an obvious secondary constriction and were translocated onto one pair of wheat chromosomes. The other chromosomes showed only the blue signals from counterstaining with DAPI (Figure 1A). The consistent results of GISH analysis for three consecutive generations of selfed YT2 further confirmed that it had high cytological stability.
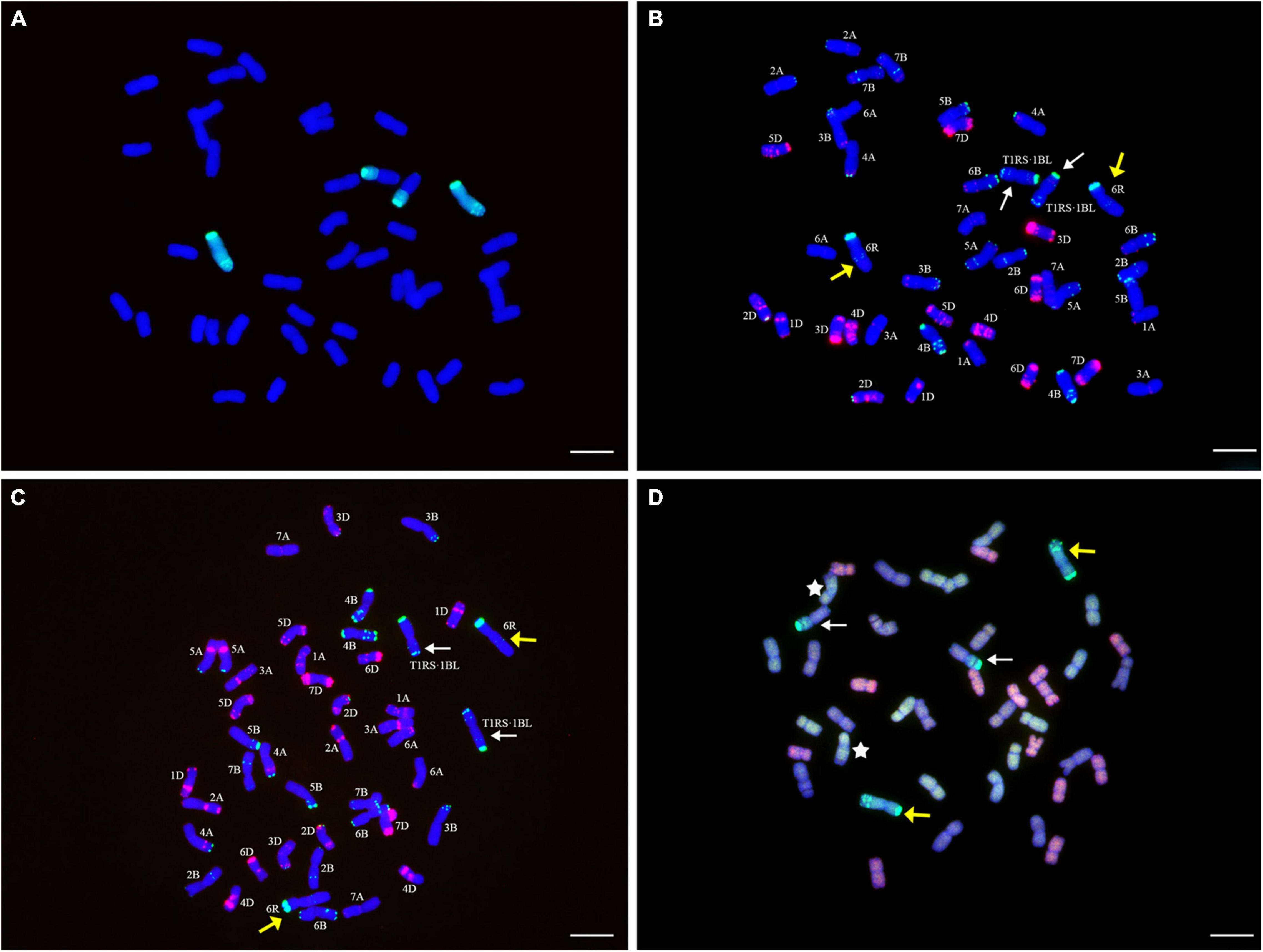
Figure 1. Genomic in situ hybridization (GISH), multicolor fluorescence in situ hybridization (mc-FISH), and multicolor-GISH (mc-GISH) identification of YT2. (A) GISH analysis of YT2 using total genomic DNA of rye cultivar German White (green) as a probe. (B) mc-FISH analysis of the same metaphase cell after GISH analysis (A) with pSc119.2 (green) and pAs1 (red). (C) ND-FISH analysis of YT2 with Oligo-pSc119.2-2 (green) and Oligo-pTa535-2 (red). (D) mc-GISH analysis of YT2, R-genome, A-genome, B-genome, and D-genome chromosomes displayed bright-green, yellow-green, brown, or gray, and pink or red fluorescence signals, respectively. The yellow arrows indicate the rye chromosomes 6R, the white arrows indicate the translocation chromosomes T1RS⋅1BL (B–D), the white stars represent the T7A⋅4B translocations (D), and the bar represents 10 μm.
Fluorescence in situ Hybridization Analysis of YT2
After GISH analysis, mc-FISH, using the combination of probes pAs1 and pSc119.2, and ND-FISH, using the combination of probes Oligo-pSc119.2-2 and Oligo-pTa535-2, determined the identity of the individual chromosomes of YT2. The probe pSc119.2 produced strong green hybridization signals in the telomere regions of the short arm and four weak green points in the long arm of the intact chromosome. Meanwhile, strong green hybridization signals in the telomere regions and weak hybridization signals in the subtelomere regions on both the long and short arms of the translocated chromosomes were observed. The two introgressed rye chromosomes and chromosome arms did not bear any hybridization signals of pAs1 (Figure 1B). Using Oligo-pSc119.2-2 and Oligo-pTa535-2 as probes, the detection results were consistent with those by using pSc119.2 and pAs1 as probes (Figure 1C). Thus, the additional rye chromosomes were identified as 6R and the rye chromosome arm 1RS that translocated onto the wheat chromosome arm 1BL.
Multicolor-GISH Analysis of YT2
To further reveal the genomic characteristics of YT2, mc-GISH analysis was performed by using genomic DNA of German White and three diploid progenitors of common wheat as probes or blockers. The results of the cross-hybridization signals made the rye chromosomes display green hybridization signals, whereas the wheat A-, B-, and D-genome chromosomes displayed yellow-green, brown or gray, and pink or red fluorescent signals, respectively (Figure 1D). The results demonstrated that YT2 had seven pairs of genome chromosomes for the A- and D-genome each, six pairs of B-genome chromosomes, one pair of rye chromosomes, and one pair of chromosome arms translocated onto one pair of B-genome chromosomes. The universal T7B⋅4A translocation in the wheat background was also observed in YT2 (Figure 1D).
Polymerase Chain Reaction Analysis of YT2
Four molecular markers were used to confirm the chromosome composition of YT2. The 6RS-specific marker SW22057, 6RL-specific marker SW22063, and 1RS-specific marker SW5284 could amplify their approximate 400 bp targeted fragments in triticale parent 09R2-100 but not in wheat parent Shixin 616. By using markers SW22057 and SW22063, the corresponding DNA fragments were detected in YT2, DA 6R of CS × Imperial, two hexaploid triticale lines 10R2-193-2 and 10R2-194-2, but not in the other addition lines of CS × Imperial and T1BL⋅1RS translocation lines Lovrin 10 and Lovrin 13 (Figures 2A,B). In addition, YT2, 10R2-193-2, 10R2-194-2, Lovrin 10, and Lovrin 13 amplified the diagnostic fragments by using the 1RS-specific marker SW5284, but not in other corresponding genotypes (Figure 2C). Meanwhile, the 1BS-diagnostic marker O11B3/O11B5 amplified approximate 630 bp targeted fragments in all the tested genotypes except for YT2, Lovrin 10, and Lovrin 13 (Figure 2D). Therefore, YT2 was confirmed to bear chromosome 6R and chromosome arm 1RS, but not 1BS. Taking the results from the molecular marker and GISH/FISH analyses together, YT2 is a 6R disomic addition and T1RS⋅1BL translocation line.
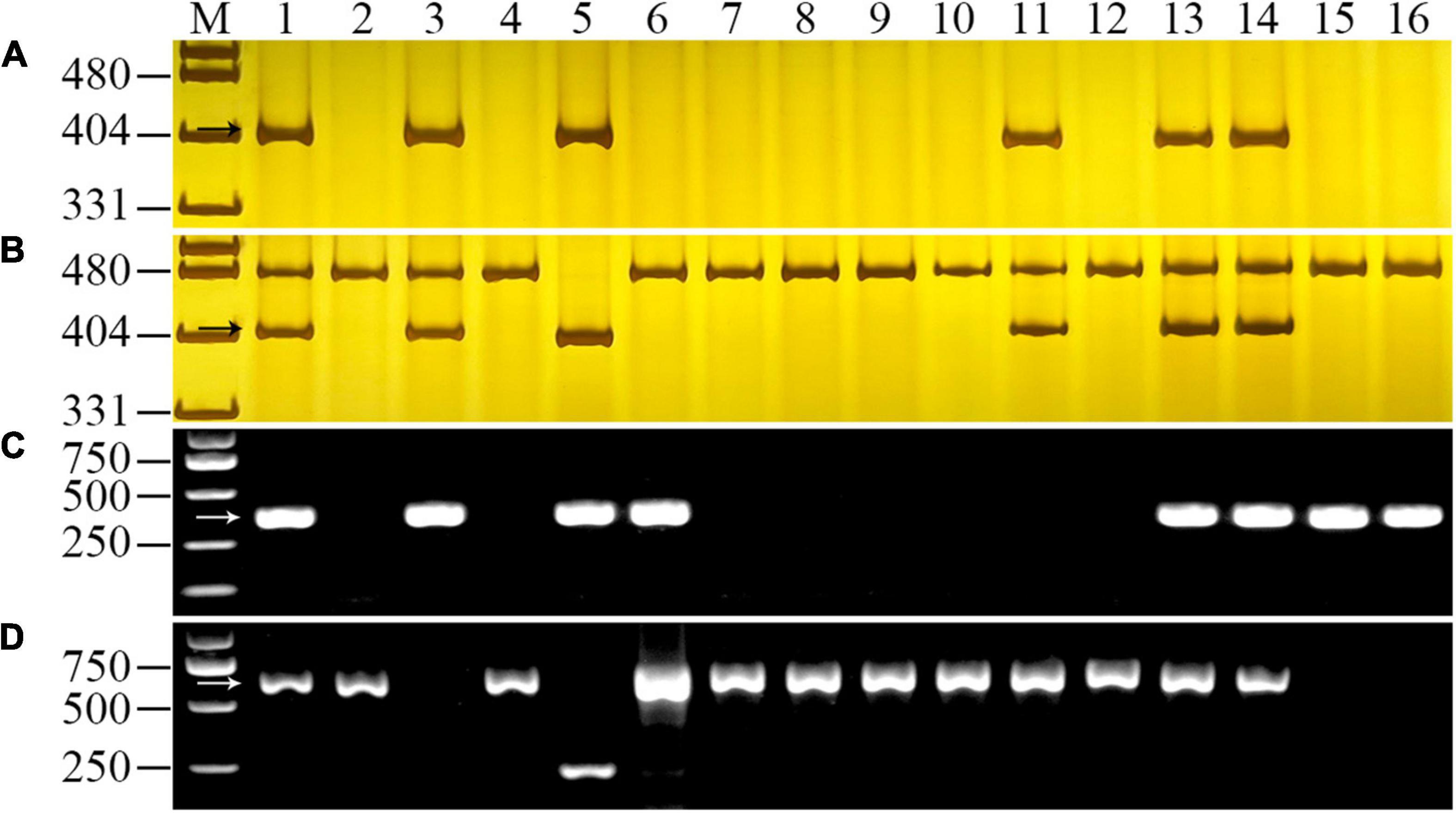
Figure 2. PCR amplification of 6RS-specific marker SW22057 (A), 6RL-specific marker SW22063 (B), 1RS-specific marker SW5284 (C), and 1BS-diagnosed marker O11B3/O11B5 (D) for chromosome composition analysis of YT2. Lanes M, pUC19 MspI in (A,B) and Trans2K Plus in (C,D). 1, octoploid triticale 09R2-100; 2, Shixin 616; 3, YT2; 4, Chinese Spring; 5, Imperial rye; 6–12, 1R-7R disomic addition lines of “Chinese Spring × Imperial”; 13–14, two hexaploid triticale lines 10R2-193-2 and 10R2-194-2; 15–16, two T1BL⋅1RS translocation lines Lovrin 10 and Lovrin 13. The arrows indicated the diagnostic fragments of the markers.
Responses of YT2 to Powdery Mildew
To further determine the resistance to powdery mildew, spectrum analysis to 22 Bgt isolates at the seedling stage was conducted on YT2 along with 47 wheat genotypes. YT2 was resistant to 12 of 22 Bgt isolates tested, showing a significantly different response spectrum in comparison with all the genotypes (Supplementary Table 1). Especially among the four genotypes with known Pm gene(s) from rye, TAM104/Thatcher with Pm20 from 6RL of Prolific rye was resistant to 15 but susceptible to seven Bgt isolates. CI14189 with Pm7 derived from 2RL of Rosen rye was highly susceptible to all the 22 isolates; Kavkaz with Pm8 from 1RS of Petkus rye was resistant to isolates E50 and E60; Amigo with Pm17 from 1RS of Insave rye was resistant to E06 and E13. In addition, compared with WR49-1, a wheat-rye 6R disomic addition line derived from wheat cultivar Xiaoyan 6 and rye cultivar German white that was previously identified by our laboratory, YT2 had a narrower response spectrum but was resistant to the four Bgt isolates E18, E19, E20, and E49, while WR49-1 was susceptible (An et al., 2015). These results suggest that YT2 may carry a new resistance gene.
During the 2018–2020 wheat-growing seasons, YT2, 09R2-100, and Shixin 616, along with TAM104/Thatcher, Kavkaz, CI14189, and Amigo, were inoculated with the mixture of Bgt isolates including E09, E18, and E20 collected from northern China. The results demonstrated that at the adult stage, YT2, 09R2-100, and TAM104/Thatcher were highly resistant that developed no mildew symptoms with IT 0, while Shixin 616 and the four controls Mingxian 169, Kavkaz, CI14189, and Amigo all showed susceptible reactions with ITs 8–9.
Powdery Mildew Resistance Gene Is Conferred by Chromosome 6R
The F2 population of YT2 × Gao 8901 tested the reactions to powdery mildew resistance at both seedling and adult plant stages. All of the 97 F2 plants resistant or susceptible to Bgt isolate E09 at the seeding stage were identified by specific markers and GISH analysis for the detection of chromosomes 6R and 1RS. The result showed that all of the 50 resistant plants possessed at least one intact rye chromosome 6R, whereas the other 47 susceptible plants lacked any rye chromosome or only possessed the rye chromosome arm 1RS. At the adult plant stage, the F2 plants showed a consistent disease reaction that agreed with that of the seedling resistance test. Thus, co-segregation of the resistance phenotype with chromosome 6R indicated that powdery mildew resistance of YT2 was conferred by rye chromosome 6R (Figure 3).
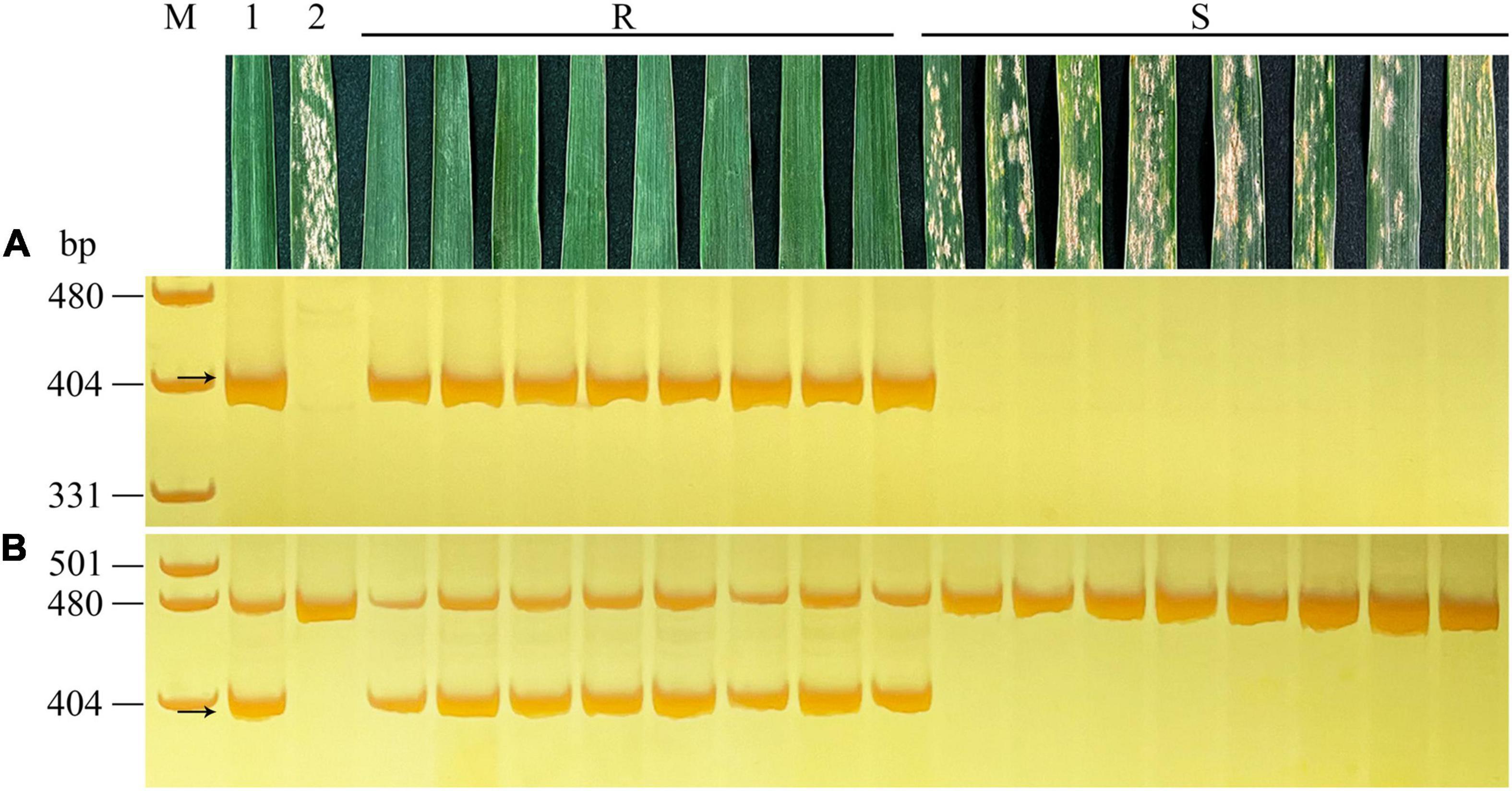
Figure 3. Evaluation of powdery mildew resistance at the adult plant stage and molecular detection of the 6R chromosomes from YT2 using 6RS-specific marker SW22057 (A) and 6RL-specific marker SW22063 (B) in F2 plants of YT2 × Gao 8901. Lanes M, pUC19 MspI. 1, YT2; 2, Gao 8901; and R, resistant; S, susceptible F2 plants. The arrows indicate the diagnosed amplified fragments.
Development and Validation of 6RL-Specific Markers
A series of 6RL-specific markers with approximate 10 Mb apart were designed by comparing the sequences of the 6R chromosome of the Weining rye and CS genome. CS, Weining, Holdfast, King II, and DA 6R, DtA 6RS, DtA 6RL of Holdfast × King II were used to validate the specificity of the markers. For example, the newly developed markers XM84, XM86, XM103, XM104, XM108, XMM20, XM127, and XMM47 were designed based on the locations of approximate 381.87 Mb, 390.02 Mb, 470.03 Mb, 479.87 Mb, 500.34 Mb, 581.48 Mb, 590.06 Mb, and 1023.93 Mb, respectively, corresponding to the 6R chromosome of Weining reference genome. All of these markers can only amplify the diagnostic fragments in Weining, King II, DA 6R, and DtA 6RL, but not in the CS, Holdfast, and DtA 6RS (Figure 4). From the centromere region to the terminal region of the 6RL chromosome, a total of 73 pairs of specific markers with certain locations were finally developed (Supplementary Table 2).
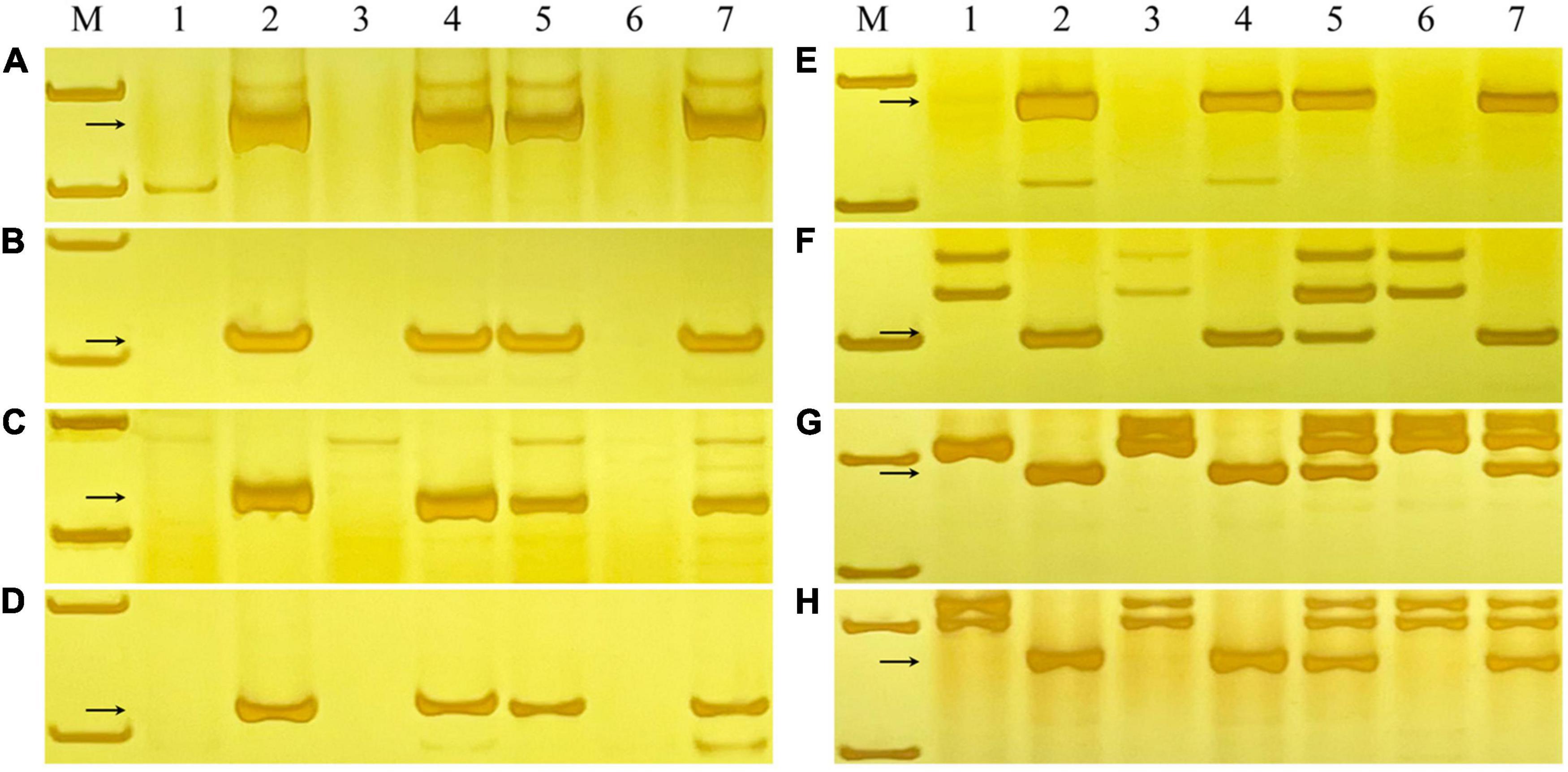
Figure 4. PCR amplification patterns of 6RL-specific markers XM84, XM86, XM103, XM104, XM108, XMM20, XM127, and XMM47 (A–H). Lanes M, pUC19 MspI. 1, Chinese Spring; 2, Weining rye; 3, Holdfast; 4, King II rye; 5, DA 6R of Holdfast × King II; 6, DtA 6RS of Holdfast × King II; 7, DtA 6RL of Holdfast × King II. The arrows indicate the diagnosed amplified fragments.
Physical Localization of the Powdery Mildew Resistance Gene on Chromosome 6R
To confirm the physical location of the powdery mildew resistance gene on chromosome 6R, the irradiated F1 progenies were identified with GISH analysis and assessed for their resistance to powdery mildew at the seedling stage. As shown in Figure 5, nine variants containing different chromosome 6R segments were obtained. The results showed that translocation line G121 possessed intact 6RL chromosomes and was highly resistant to Bgt isolate E09, indicating that the powdery mildew resistance of YT2 was derived from 6RL. Translocation lines G16, G66, and G19 were resistant, while G104, G42, G59, and the deletion line G128 were susceptible. The chromosomal breakpoint positions of these variants were further determined using the 26 of 73 6RL-specific markers newly developed in this study. Combined with their GISH detection, responses to powdery mildew, and molecular marker analyses, the powdery mildew resistance gene of YT2 is mapped to the region in chromosome 6RL: 890.09–967.51 Mb, flanked by markers XM189 and X4M19, corresponding to the reference genome of Weining rye. Disease responses and molecular marker analysis on their selfing F2 progenies also confirmed this result.
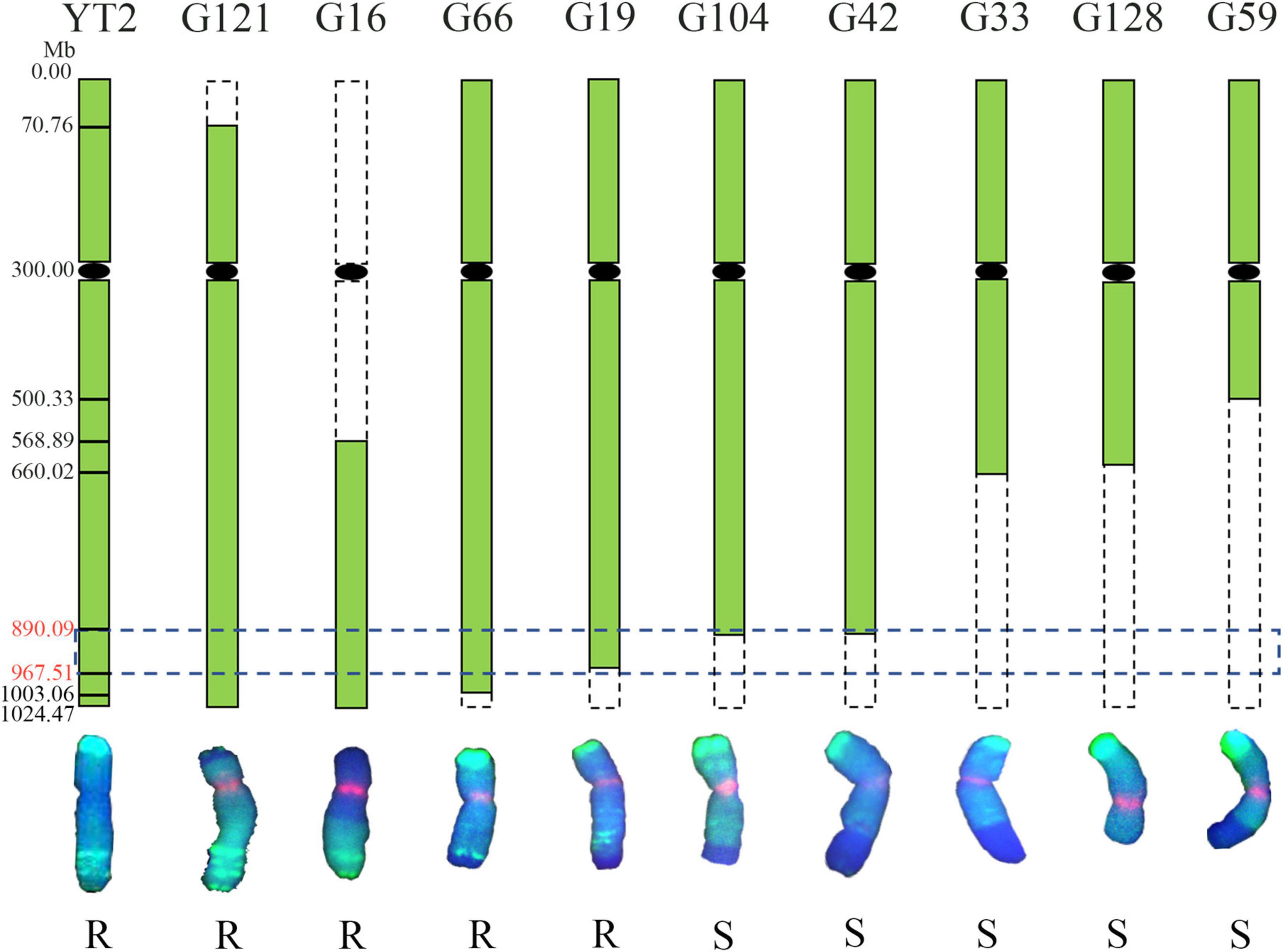
Figure 5. Physical mapping of the powdery mildew resistance gene of YT2. The green boxes represent 6R chromosomes, and the horizontal dotted box indicates that the powdery mildew resistance gene of YT2 was mapped to chromosome 6RL: 890.09–967.51 Mb, corresponding to the reference genome of Weining rye. The vertical dotted boxes represent missing chromosome segments of 6R. The bottom chromosomes were captured in GISH analysis and counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue), and rye chromatin displays green signals. The letters R and S indicate the derivative lines that were resistant and susceptible to powdery mildew.
Agronomic Performance of YT2
After strict bagging self-pollinating for five consecutive generations, neither morphology nor cytology segregation was observed on YT2. Compared to its parents 09R2-100 and Shixin 616, YT2 had a comparable performance on SNPP and KNS, but TKW and GYPP were significantly lower (Table 1).
Discussion
Rye, a naturally cross-pollinating relative of common wheat, which contains extensive genetic variation within and among cultivars, is an important gene donor for wheat improvement (Chikmawati et al., 2012; Targońska et al., 2016). Through distant hybridization and chromosome engineering, many wheat-rye addition, substitution, and translocation lines have been constantly developed as bridging parents in wheat chromosome engineering breeding (An et al., 2006, 2013, 2015, 2019, 2021; Han et al., 2022). In this study, a wheat-rye line YT2 was artificially selected from consecutive self-generations derived from the cross of octoploid triticale 09R2-100 and wheat cultivar Shixin 616, which exhibited high resistance to powdery mildew at both seedling and adult plant stages. Molecular cytogenetic identification, including GISH, mc-FISH, and specific molecular marker analysis, demonstrated that YT2 was a wheat-rye 6R disomic addition and T1RS⋅1BL translocation line. Genetic analysis indicated that the powdery mildew resistance was conferred by chromosome 6R. At the seedling stage, YT2 was resistant to 12 of 22 Bgt isolates, while at the adult plant stage, it exhibited high levels of resistance to a mixture of Bgt isolates collected in northern China, showing a significantly different response spectrum from the cataloged Pm20, Pm7, Pm8, and Pm17 that also derived from rye. To physically map the resistance locus, a total of 73 markers specific for 6RL were developed and the resistance gene was finally localized to the region in chromosome 6RL: 890.09–967.51 Mb, suggesting that YT2 may carry a new powdery mildew resistance gene that differs from previously reported genes from rye.
YT2 was resistant to 12 of the 22 Bgt isolates tested at the seedling stage through resistance spectrum analysis, and it showed immunity to all the mixed Bgt isolates at the adult plant stage in the field. Some powdery mildew resistance genes have been reported from chromosome 6RL of rye cultivars including Prolific, Jingzhouheimai, and Kustro, among which the resistance gene from Kustro was localized in the region from the site between 2.3 and 2.5 to the telomere (Friebe et al., 1994; Wang et al., 2010; Li et al., 2016; Du et al., 2018). A powdery mildew APR gene(s) from the wild species Secale africanum was mapped on the long arm of 6Rafr at FL0.85–1.00 (Li G.R. et al., 2020). Our laboratory also reported a wheat-rye 6R addition line carrying powdery mildew resistance gene on its 6R, while it showed significantly different reaction patterns in comparison with YT2 (An et al., 2015). Rye has strong heterogeneity, and many studies have shown different disease reactions of the derivatives even from the same original rye donor (Ren et al., 2011; Li et al., 2016; Schneider et al., 2016). Thus, the powdery mildew resistance gene in YT2 might be different from the previously reported genes from rye and could enrich the gene resources for wheat powdery mildew resistance breeding. Of course, fine mapping and cloning of the key genes should really explain their differences and the evolutionary relationship among those resistance stocks.
The realization of alien gene transfer plays an important role in improving the genetic diversity for wheat improvement (Zhang et al., 2016). Due to the extremely low recombination rate between alien chromosomes and wheat homoeologous counterparts, creating wheat-alien chromosomal translocations lines and deletion lines is one of the main means to determine the physical location of resistance genes derived from wheat relatives (Qi et al., 1998; Lukaszewski et al., 2001; Pu et al., 2015; Song et al., 2016; Zhang et al., 2016; Duan et al., 2017; Liu et al., 2017; He et al., 2018; Li J.B. et al., 2020). Given less linkage drag and regular meiotic behavior, small segment translocation lines are more preferred in wheat breeding practice (Falke et al., 2009; Qi et al., 2016; Ma et al., 2018). Therefore, as increasing resistance genes on rye chromosome 6R are identified, specific markers of high density on 6R appear of particular importance. They can rapidly anchor the targeted resistance regions based on the different phenotypes of different deletions and translocations, thereby promoting fine mapping and utilization of genes. In this study, according to the high-quality reference genome sequence of Weining rye (Li et al., 2021), a total of 73 6RL-specific PCR-based markers with an average density of approximate 10 Mb were developed. Using these developed markers and the deletion and translocation lines created by radiation, the powdery mildew resistance gene of YT2 was mapped to a certain region of the chromosome which is referred to as the reference genome of Weining rye. Based on these results, the creation of smaller segment translocations and the enrichment of interval markers will be more purposeful.
Whether a valuable disease resistance gene could be efficiently applied in resistance breeding largely depends on the agronomic traits of its carrier, but often, disease resistance is at the expense of some agronomic traits and reduced plant adaptation (Deng et al., 2017; Ma et al., 2018; Jia et al., 2020). Compared with the previously reported chromosome addition lines such as WR49-1, YT2 carried an additional T1RS⋅1BL chromosome translocation, which may have an additional positive value on breeding utilization of YT2. Among all of the rye translocation lines, 1RS translocation line was regarded as particularly notable success in wheat breeding because it carried multiple resistance genes against powdery mildew, stripe rust, leaf rust, and stem rust, as well as genes associated with superior agronomic traits and abiotic-stress tolerance (Lukaszewski, 1990; Kumlay et al., 2003; Shearman et al., 2005; Howell et al., 2014, 2019). In spite of the newly variant pathogen virulence circumventing the disease resistance, the T1RS⋅1BL translocations have continued to be widely utilized in wheat breeding because of their yield potential and wide environmental adaptability (Ren et al., 2017; Wang et al., 2017). In this study, the 1RS of YT2 inherited from its triticale parent showed no resistance to powdery mildew, but its role in agronomic traits should not be underestimated in future work. In addition, it is notable that T1RS⋅1BL translocations increased yield in only specific combinations of 1RS and wheat genetic background (Jung and Lelley, 1985; Ren et al., 2017). To clarify the future breeding strategy of YT2, comprehensive evaluations of agronomic traits were conducted on YT2 and their parents 09R2-100 and Shixin 616. Compared to its wheat parent Shixin 616, YT2 had significantly lower TKW and GYYP, probably due to its small and shriveled seeds. Thus, YT2 could be designed to cross with various wheat cultivars with good performance on TKW and GYPP while transferring its powdery mildew resistance.
Conclusion
In this study, we developed and identified a novel wheat-rye 6R addition and T1RS⋅1BL translocation line YT2 with high powdery mildew resistance, developed new rye chromosome-specific markers, and further mapped its powdery mildew resistance locus. This study provides an efficient model from developing germplasm to comprehensively and accurately identifying it. The new germplasm YT2 can serve as a promising genetic stock in wheat improvement.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DA, GH, and HY conceived the research. GH, HY, JW, LC, and SL performed the experiments. LL and XL contributed to the development of the materials. YZ and JF performed phenotypic assessments. GH and HY wrote the manuscript. DA and LL supervised the research. All authors read and approved the final manuscript.
Funding
This research was supported by the National Key Research and Development Program of China (2021YFD1200600), Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24030102), and National Natural Science Foundation of China (31771793).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the following researchers for providing valuable plant materials: Zongxiang Tang (Sichuan Agricultural University, Chengdu, China) for wheat cultivar Holdfast and a set of wheat-rye disomic addition lines of Holdfast × King II, Yiwen Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) for rye cultivar King II, Adrian Turner (John Innes Centre, Norwich, United Kingdom) for wheat-rye 6RS and 6RL ditelosomic addition lines of Holdfast × King II, and John Raupp (Kansas State University, Manhattan, KS, United States) for rye cultivar Imperial and a complete set of disomic addition lines of CS × Imperial.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.889494/full#supplementary-material
References
An, D. G., Han, G. H., Wang, J., Yan, H. W., Zhou, Y. L., Cao, L. J., et al. (2021). Cytological and genetic analyses of a wheat-rye 2RL ditelosomic addition line with adult plant resistance to powdery mildew. Crop J. [Epub ahead of print]. doi: 10.1016/j.cj.2021.08.011
An, D. G., Li, L. H., Li, J. M., Li, H. J., and Zhu, Y. G. (2006). Introgression of resistance to powdery mildew conferred by chromosome 2R by crossing wheat nullisomic 2D with rye. J. Integr. Plant Biol. 48, 838–847. doi: 10.1111/j.1744-7909.2006.00275.x
An, D. G., Ma, P. T., Zheng, Q., Fu, S. L., Li, L. H., Han, F. P., et al. (2019). Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor. Appl. Genet. 132, 257–272. doi: 10.1007/s00122-018-3214-3
An, D. G., Zheng, Q., Luo, Q. L., Ma, P. T., Zhang, H. X., Li, L. H., et al. (2015). Molecular cytogenetic identification of a new wheat-rye 6R chromosome disomic addition line with powdery mildew resistance. PLoS One 10:e0134534. doi: 10.1371/journal.pone.0134534
An, D. G., Zheng, Q., Zhou, Y. L., Ma, P. T., Lv, Z. L., Li, L. H., et al. (2013). Molecular cytogenetic characterization of a new wheat-rye 4R chromosome translocation line resistant to powdery mildew. Chrom. Res. 21, 419–432. doi: 10.1007/s10577-013-9366-8
Chen, P. D., You, C. F., Hu, Y., Chen, S. W., Zhou, B., Cao, A. Z., et al. (2013). Radiation induced translocations with reduced Haynaldia villosa chromatin at the Pm21 locus for powdery mildew resistance in wheat. Mol. Breed. 31, 477–484. doi: 10.1007/s11032-012-9804-x
Chikmawati, T., Miftahudin, M., Skovmand, B., and Gustafson, J. P. (2012). Amplified fragment length polymorphism-based genetic diversity among cultivated and weedy rye (Secale cereale L.) accessions. Genet. Resour. Crop Evol. 59, 1743–1752. doi: 10.1007/s10722-012-9796-8
Colmer, T. D., Flowers, T. J., and Munns, R. (2006). Use of wild relatives to improve salt tolerance in wheat. J. Exp. Bot. 57, 1059–1078. doi: 10.1093/jxb/erj124
Deng, Y. W., Zhai, K. R., Xie, Z., Yang, D. Y., Zhu, X. D., Liu, J. Z., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355:eaai8898. doi: 10.1126/science.aai8898
Du, H. M., Tang, Z. X., Duan, Q., Shu, Y., and Fu, S. L. (2018). Using the 6RLKu minichromosome of rye (Secale cereale L.) to create wheat-rye 6D/6RLKu small segment translocation lines with powdery mildew resistance. Int. J. Mol. Sci. 19:3933. doi: 10.3390/ijms19123933
Duan, Q., Yang, W. Y., Qiu, L., Ren, T. H., Zhi, L., Fu, S. L., et al. (2017). Physical location of new PCR-based markers and powdery mildew resistance gene(s) on rye (Secale cereale L.) chromosome 4 using 4R dissection lines. Front. Plant Sci. 8:1716. doi: 10.3389/fpls.2017.01716
Falke, K. C., Susic, Z., Wilde, P., Wortmann, H., Mohring, J., Piepho, H. P., et al. (2009). Testcross performance of rye introgression lines developed by marker-assisted backcrossing using an Iranian accession as donor. Theor. Appl. Genet. 118, 1225–1238. doi: 10.1007/s00122-009-0976-7
Friebe, B., Hatchett, J. H., Gill, B. S., Mukai, Y., and Sebesta, E. E. (1991). Transfer of Hessian fly resistance from rye to wheat via radiation-induced terminal and intercalary chromosomal translocations. Theor. Appl. Genet. 83, 33–40. doi: 10.1007/BF00229223
Friebe, B., Heun, M., Tuleen, N., Zeller, F. J., and Gill, B. S. (1994). Cytogenetically monitored transfer of powdery mildew resistance from rye into wheat. Crop Sci. 34, 621–625. doi: 10.2135/cropsci1994.0011183X003400030003x
Friebe, B., Jiang, J., Raupp, W. J., McIntosh, R. A., and Gill, B. S. (1996). Characterization of wheat alien translocations conferring resistance to diseases and pests: current status. Euphytica 91, 59–87. doi: 10.1007/BF00035277
Fu, S. L., Chen, L., Wang, Y. Y., Li, M., Yang, Z. J., Qiu, L., et al. (2015). Oligonucleotide probes for ND-FISH analysis to identify rye and wheat 435 chromosomes. Sci. Rep. 5:10552. doi: 10.1038/srep10552
Fu, S. L., Lv, Z. L., Qi, B., Guo, X., Li, J., Li, B., et al. (2012). Molecular cytogenetic characterization of wheat-Thinopyrum elongatum addition, substitution and translocation lines with a novel source of resistance to wheat fusarium head blight. J. Genet. Genomics 39, 103–110. doi: 10.1016/j.jgg.2011.11.008
Fu, S. L., Ren, Z. L., Chen, X. M., Yan, B. J., Tan, F. Q., Fu, T. H., et al. (2014). New wheat-rye 5DS-4RS⋅4RL and 4RS-5DS⋅5DL translocation lines with powdery mildew resistance. J. Plant Res. 127, 743–753. doi: 10.1007/s10265-014-0659-6
Gupta, P. K., Langridge, P., and Mir, R. R. (2010). Marker-assisted wheat breeding: present status and future possibilities. Mol. Breed. 26, 145–161. doi: 10.1007/s11032-009-9359-7
Han, G. H., Li, H. W., Cao, L. J., Liu, S. Y., Yan, H. W., Wang, J., et al. (2022). A novel wheat-rye 2R(2D) disomic substitution line pyramids two types of resistance to powdery mildew. Plant Dis. [Epub ahead of print]. doi: 10.1094/PDIS-12-21-2765-RE
Han, G. H., Liu, S. Y., Jin, Y. L., Jia, M. S., Ma, P. T., Liu, H., et al. (2020a). Scale development and utilization of universal PCR-based and high-throughput KASP markers specific for chromosome arms of rye (Secale cereale L.). BMC Genomics 21:206. doi: 10.1186/s12864-020-6624-y
Han, G. H., Liu, S. Y., Wang, J., Jin, Y. L., Zhou, Y. L., Luo, Q. L., et al. (2020b). Identification of an elite wheat-rye T1RS⋅1BL translocation line conferring high resistance to powdery mildew and stripe rust. Plant Dis. 104, 2940–2948. doi: 10.1094/PDIS-02-20-0323-RE
Hao, M., Liu, M., Luo, J. T., Fan, C. L., Yi, Y. J., Zhang, L. Q., et al. (2018). Introgression of powdery mildew resistance gene Pm56 on rye chromosome arm 6RS into wheat. Front. Plant Sci. 9:1040. doi: 10.3389/fpls.2018.01040
He, H. G., Liu, R. K., Ma, P. T., Du, H. N., Zhang, H. H., and Wu, Q. H. (2021). Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor. Appl. Genet. 134, 53–62. doi: 10.1007/s00122-020-03681-2
He, H. G., Zhu, S. Y., Zhao, R. H., Jiang, Z. N., Ji, Y. Y., Ji, J., et al. (2018). Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant. 6, 879–882. doi: 10.1016/j.molp.2018.03.004
He, Z. H., Xia, X. C., Chen, X. M., Zhang, Y., Zhang, Y., Yan, J., et al. (2015). “Application of molecular markers in plant quality and disease resistance breeding,” in Proceedings of the Seventh National Symposium on Wheat Genetics and Breeding, Zhengzhou.
He, Z. H., Xia, X. C., Chen, X. M., and Zhuang, Q. S. (2011). Progress of wheat breeding in China and the future perspective. Acta Agron. Sin. 37, 202–215. doi: 10.3724/SP.J.1006.2011.00202
Hechanova, S. L., Prusty, M. R., Kim, S. R., Ballesfin, L., Ramos, J., Prahalada, G. D., et al. (2018). Monosomic alien addition lines (MAALs) of Oryza rhizomatis in Oryza sativa: production, cytology, alien trait introgression, molecular analysis and breeding application. Theor. Appl. Genet. 131, 2197–2211. doi: 10.1007/s00122-018-3147-x
Howell, T., Hale, I., Jankuloski, L., Bonafede, M., Gilbert, M., and Dubcovsky, J. (2014). Mapping a region within the 1RS⋅1BL translocation in common wheat affecting grain yield and canopy water status. Theor. Appl. Genet. 127, 2695–2709. doi: 10.1007/s00122-014-2408-6
Howell, T., Moriconi, J. I., Zhao, X. Q., Hegarty, J., Fahima, T., Santa-Maria, G. E., et al. (2019). A wheat/rye polymorphism affects seminal root length and yield across different irrigation regimes. J. Exp. Bot. 70, 4027–4037. doi: 10.1093/jxb/erz169
Hurni, S., Brunner, S., Stirnweis, D., Herren, G., Peditto, D., McIntosh, R. A., et al. (2015). The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J. 79, 904–913. doi: 10.1111/tpj.12593
International Wheat Genome Sequencing Consortium [Iwgsc], Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Jia, M. S., Xu, H. X., Liu, C., Mao, R. X., Li, H. S., Liu, J. J., et al. (2020). Characterization of the powdery mildew resistance gene in the elite wheat cultivar Jimai 23 and its application in marker-assisted selection. Front. Genet. 11:241. doi: 10.3389/fgene.2020.00241
Jung, C., and Lelley, T. (1985). Genetic interaction between wheat and rye genomes in triticale. Theor. Appl. Genet. 70, 427–432. doi: 10.1007/BF00273749
Korzun, V., Malyshev, S., Kartel, N., Westermann, T., Weber, W. E., and Borner, A. (1998). A genetic linkage map of rye (Secale cereale L.). Theor. Appl. Genet. 96, 203–208. doi: 10.1007/s001220050728
Kumlay, A. M., Baenziger, P. S., Gill, K. S., Shelton, D. R., Graybosch, R. A., Lukaszewski, A. J., et al. (2003). Understanding the effect of rye chromatin in bread wheat. Crop Sci. 43, 1643–1651. doi: 10.1016/S0261-2194(03)00139-X
Li, F., Li, Y. H., Cao, L. R., Liu, P. Y., Geng, M. M., Zhang, Q., et al. (2018). Simultaneous transfer of leaf rust and powdery mildew resistance genes from hexaploid triticale cultivar Sorento into bread wheat. Front. Plant Sci. 9:85. doi: 10.3389/fpls.2018.00085
Li, G. R., Tang, L. R., Yin, Y., Zhang, A. H., Yu, Z. H., Yang, E. N., et al. (2020). Molecular dissection of Secale africanum chromosome 6Rafr in wheat enabled localization of genes for resistance to powdery mildew and stripe rust. BMC Plant Biol. 20:2351. doi: 10.1186/s12870-020-02351-1
Li, G. W., Wang, L. J., Yang, J. P., He, H., Jin, H. B., Li, X. M., et al. (2021). A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat. Genet. 53, 574–584. doi: 10.1038/s41588-021-00808-z
Li, H. H., Dong, Z. J., Ma, C., Xia, Q., Tian, X. B., Sehgal, S., et al. (2020). A spontaneous wheat-Aegilops longissima translocation carrying Pm66 confers resistance to powdery mildew. Theor. Appl. Genet. 133, 1149–1159. doi: 10.1007/s00122-020-03538-8
Li, H. J., Wang, X. M., Song, F. J., Wu, C. P., Wu, X. F., Zhang, N., et al. (2011). Response to powdery mildew and detection of resistance genes in wheat cultivars from China. Acta Agron. Sin. 37, 943–954. doi: 10.1016/S1875-2780(11)60026-6
Li, J. B., Dundas, I., Dong, C. M., Li, G. R., Trethowan, R., Yang, Z. J., et al. (2020). Identification and characterization of a new stripe rust resistance gene Yr83 on rye chromosome 6R in wheat. Theor. Appl. Genet. 133, 1095–1107. doi: 10.1007/s00122-020-03534-y
Li, J. J., Endo, T. R., Saito, M., Ishikawa, G., Nakamura, T., and Nasuda, S. (2013). Homoeologous relationship of rye chromosome arms as detected with wheat PLUG markers. Chromosoma 122, 555–564. doi: 10.1007/s00412-013-0428-7
Li, M., Tang, Z. X., Qiu, L., Wang, Y. Y., Tang, S. Y., and Fu, S. L. (2016). Identification and physical mapping of new PCR-based markers specific for the long arm of rye (Secale cereale L.) chromosome 6. J. Genet. Genomics 43, 199–206. doi: 10.1016/j.jgg.2015.11.005
Liu, W. X., Koo, D. H., Xia, Q., Li, C. X., Bai, F. Q., Song, Y. L., et al. (2017). Homoeologous recombination-based transfer and molecular cytogenetic mapping of powdery mildew-resistant gene Pm57 from Aegilops searsii into wheat. Theor. Appl. Genet. 130, 841–848. doi: 10.1007/s00122-017-2855-y
Lukaszewski, A. J. (1990). Frequency of 1RS⋅1AL and 1RS⋅1BL translocations in United States wheats. Crop Sci. 30, 1151–1153. doi: 10.2135/cropsci1990.0011183X003000050041x
Lukaszewski, A. J., Porter, D. R., Baker, C. A., Rybka, K., and Lapinski, B. (2001). Attempts to transfer Russian wheat aphid resistance from a rye chromosome in Russian triticales to wheat. Crop Sci. 41, 1743–1749. doi: 10.2135/cropsci2001.1743
Ma, P. T., Xu, H. X., Xu, Y. F., Song, L. P., Liang, S. S., Sheng, Y., et al. (2018). Characterization of a powdery mildew resistance gene in wheat breeding line 10v-2 and its application in marker-assisted selection. Plant Dis. 102, 925–931. doi: 10.1094/PDIS-02-17-0199-RE
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Xia, X. C., and Raupp, W. J. (2020). Catalogue of Gene Symbols For Wheat 2020 Supplement. Available online at: https://wheat.pw.usda.gov/GG3/WGC
Milczarski, P., Bolibok-Bragoszewska, H., Myskow, B., Stojalowski, S., Heller-Uszynska, K., Goralska, M., et al. (2011). A high density consensus map of rye (Secale cereale L.) based on DarT markers. PLoS One 6:e28495. doi: 10.1371/journal.pone.0028495
Mohler, V., and Stadlmeier, M. (2019). Dynamic QTL for adult plant resistance to powdery mildew in common wheat (Triticum aestivum L.). J. Appl. Genet. 60, 291–300. doi: 10.1007/s13353-019-00518-7
Oettler, G., Tams, S. H., Utz, H. F., Bauer, E., and Melchinger, A. E. (2005). Prospects for hybrid breeding in winter triticale. Crop Sci. 45, 1476–1482. doi: 10.2135/cropsci2004.0462
Pu, J., Wang, Q., Shen, Y. F., Zhuang, L. F., Li, C. X., Tan, M. F., et al. (2015). Physical mapping of chromosome 4J of Thinopyrum bessarabicum using gamma radiation-induced aberrations. Theor. Appl. Genet. 128, 1319–1328. doi: 10.1007/s00122-015-2508-y
Qi, L., Wang, S. L., Chen, P. D., Liu, D. J., and Gill, B. S. (1998). Identification and physical mapping of three Haynaldia villosa chromosome-6V deletion lines. Theor. Appl. Genet. 97, 1042–1046. doi: 10.1007/s001220050989
Qi, W. L., Tang, Y., Zhu, W., Li, D. Y., Diao, C. D., Xu, L. L., et al. (2016). Molecular cytogenetic characterization of a new wheat-rye 1BL⋅1RS translocation line expressing superior stripe rust resistance and enhanced grain yield. Planta 244, 405–416. doi: 10.1007/s00425-016-2517-3
Qie, Y. M., Sheng, Y., Xu, H. X., Jin, Y. L., Ma, F. F., Li, L. H., et al. (2019). Identification of a new powdery mildew resistance gene pmDHT at or closely linked to the Pm5 locus in the Chinese wheat landrace Dahongtou. Plant Dis. 10, 2645–2651. doi: 10.1094/PDIS-02-19-0401-RE
Qiu, L., Tang, Z. X., Li, M., and Fu, S. L. (2016). Development of new PCR-based markers specific for chromosome arms of rye (Secale cereale L.). Genome 59, 159–165. doi: 10.1139/gen-2015-0154
Rabanus-Wallace, M. T., Hackauf, B., Mascher, M., Lux, T., Wicker, T., Gundlach, H., et al. (2021). Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat. Genet. 53, 564–573. doi: 10.1038/s41588-021-00807-0
Rabinovich, S. V. (1998). Importance of wheat-rye translocations for breeding modern cultivars of Triticum aestivum L. (Reprinted from Wheat: prospects for global improvement). Euphytica 100, 323–340. doi: 10.1023/A:1018361819215
Ren, T. H., Chen, F., Yan, B. J., and Zhang, H. Q. (2011). Genetic diversity of wheat-rye 1BL⋅1RS translocation lines derived from different wheat and rye sources. Euphytica 183, 133–146. doi: 10.1007/s10681-011-0412-3
Ren, T. H., Tang, Z. X., Fu, S. L., Yan, B. J., Tan, F. Q., Ren, Z. L., et al. (2017). Molecular cytogenetic characterization of novel wheat-rye T1RS⋅1BL translocation lines with high resistance to diseases and great agronomic traits. Front. Plant Sci. 8:799. doi: 10.3389/fpls.2017.00799
Schneider, A., Rakszegi, M., Molnár-Láng, M., and Szakács, É (2016). Production and cytomolecular identification of new wheat-perennial rye (Secale cereanum) disomic addition lines with yellow rust resistance (6R) and increased arabinoxylan and protein content (1R, 4R, 6R). Theor. Appl. Genet. 129, 1045–1059. doi: 10.1007/s00122-016-2682-6
Shearman, V. J., Sylvester-Bradley, R., Scott, R. K., and Foulkes, M. J. (2005). Physiological processes associated with wheat yield progress in the UK. Crop Sci. 45, 175–185. doi: 10.1016/j.cropro.2004.06.010
Sheng, B. Q., and Duan, X. Y. (1991). Improvement of scale 0-9 method for scoring adult plant resistance to powdery mildew of wheat. Beijing Agr. Sci. 1, 38–39.
Si, Q. M., Zhang, X. X., Duan, X. Y., Sheng, B. Q., and Zhou, Y. L. (1992). On gene analysis and classification of powdery mildew (Erysiphe graminis f. sp. tritici) resistant wheat varieties. Acta Phytopathol. Sin. 22, 349–355.
Singh, R. P., Singh, P. K., Rutkoski, J., Hodson, D. P., He, X. Y., Jørgensen, L. N., et al. (2016). Disease impact on wheat yield potential and prospects of genetic control. Annu. Rev. Phytopathol. 54, 303–322. doi: 10.1146/annurev-phyto-080615-095835
Song, L. Q., Lu, Y. Q., Zhang, J. P., Pan, C. L., Yang, X. M., Li, X. Q., et al. (2016). Physical mapping of Agropyron cristatum chromosome 6P using deletion lines in common wheat background. Theor. Appl. Genet. 129, 1023–1034. doi: 10.1007/s00122-016-2680-8
Stojalowski, S., Mysow, B., Milczarski, P., and Masojc, P. (2009). A consensus map of chromosomes 6R in rye (Secale cereale L.). Cell Mol. Biol. Lett. 14, 190–198. doi: 10.2478/s11658-008-0042-5
Sun, H. G., Hu, J. H., Wei, S., Dan, Q., Cui, L., Wu, P. P., et al. (2018). Pm61: a recessive gene for resistance to powdery mildew in wheat landrace Xuxusanyuehuang identified by comparative genomics analysis. Theor. Appl. Genet. 131, 2085–2097. doi: 10.1007/s00122-018-3135-1
Tang, Z. X., Yang, Z. J., and Fu, S. L. (2014). Oligonucleotides replacing the roles of sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Targońska, M., Bolibok-Brągoszewska, H., and Rakoczy-Trojanowska, M. (2016). Assessment of genetic diversity in Secale cereale based on SSR markers. Plant Mol. Biol. Rep. 34, 37–51. doi: 10.1007/s11105-015-0896-4
Taylor, C., Shepherd, K. W., and Langridge, P. (1998). A molecular genetic map of the long arm of chromosome 6R of rye incorporating the cereal cyst nematode resistance gene, CreR. Theor. Appl. Genet. 97, 1000–1012. doi: 10.1007/s001220050984
Tester, M., and Langridge, P. (2010). Breeding technologies to increase crop production in a changing world. Science 327, 818–822. doi: 10.1126/science.1183700
Van Campenhout, S., Vander Stappen, J., Sagi, L., and Volckaert, G. (1995). Locus specific primers for LMW glutenin genes on each of the group 1 chromosomes of hexaploid wheat. Theor. Appl. Genet. 91, 313–319. doi: 10.1007/bf00220893
Wang, D., Zhuang, L. F., Sun, L., Feng, Y. G., Pei, Z. Y., and Qi, Z. J. (2010). Allocation of a powdery mildew resistance locus to the chromosome arm 6RL of Secale cereale L. cv. ‘Jingzhouheimai’. Euphytica 176, 157–166. doi: 10.1007/s10681-010-0199-7
Wang, J., Liu, Y. L., Su, H. D., Guo, X. R., and Han, F. P. (2017). Centromere structure and function analysis in the wheat-rye translocation lines. Plant J. 91, 199–207. doi: 10.1111/tpj.13554
Xu, H. X., Yin, D. D., Li, L. H., Wang, Q. X., Li, X. Q., Yang, X. M., et al. (2012). Development and application of EST-based markers specific for chromosome arms of rye (Secale cereale L.). Cytogenet. Genome Res. 136, 220–228. doi: 10.1159/000336478
Keywords: powdery mildew, Secale cereale, Triticum aestivum, chromosome-specific marker, 6R
Citation: Han G, Yan H, Wang J, Cao L, Liu S, Li X, Zhou Y, Fan J, Li L and An D (2022) Molecular Cytogenetic Identification of a New Wheat-Rye 6R Addition Line and Physical Localization of Its Powdery Mildew Resistance Gene. Front. Plant Sci. 13:889494. doi: 10.3389/fpls.2022.889494
Received: 04 March 2022; Accepted: 13 April 2022;
Published: 12 May 2022.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Michał Tomasz Kwiatek, Poznań University of Life Sciences, PolandSylwia Okoń, University of Life Sciences in Lublin, Poland
Copyright © 2022 Han, Yan, Wang, Cao, Liu, Li, Zhou, Fan, Li and An. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihui Li, bGlsaWh1aUBjYWFzLmNu; Diaoguo An, ZGdhbkBzanppYW0uYWMuY24=
†These authors have contributed equally to this work
 Guohao Han1†
Guohao Han1† Hanwen Yan
Hanwen Yan Jing Wang
Jing Wang Jieru Fan
Jieru Fan Lihui Li
Lihui Li Diaoguo An
Diaoguo An