
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 03 June 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.888449
This article is part of the Research Topic Shaping Plant NLR Diversity: Evolutionary Dynamics and Structure-Function Relationships View all 7 articles
Plants employ multi-layered immune system to fight against pathogen infections. Different receptors are able to detect the invasion activities of pathogens, transduce signals to downstream components, and activate defense responses. Among those receptors, nucleotide-binding domain leucine-rich repeat containing proteins (NLRs) are the major intracellular ones. CHILLING SENSITIVE 3 (CHS3) is an Arabidopsis NLR with an additional Lin-11, Isl-1 and Mec-3 (LIM) domain at its C terminus. The gain-of-function mutant, chs3-2D, exhibiting severe dwarfism and constitutively activated defense responses, was selected as a genetic background in this study for a forward genetic screen. A mutant allele of hsp90.2 was isolated as a partial suppressor of chs3-2D, suggesting that HSP90 is required for CHS3-mediated defense signaling. In addition, HSP90 is also required for the autoimmunity of the Dominant Negative (DN)-SNIPER1 and gain-of-function ADR1-L2 D484V transgenic lines, suggesting a broad role for HSP90 in NLR-mediated defense. Overall, our work indicates a larger contribution of HSP90 not only at the sensor, but also the helper NLR levels.
In nature, plants are challenged by various microbial pathogens, including viruses, bacteria, fungi, and oomycetes. They have evolved a complicated immune system in order to combat infections. Both pattern-triggered immunity (PTI) and effector triggered immunity (ETI) which relies on intracellular receptors nucleotide-binding domain leucine-rich repeat containing proteins (NLRs), can be activated during defense (Jones and Dangl, 2006). In some cases, ETI responses require two types of NLR proteins. One acts as the effector-sensing or sensor NLR. The other serves roles in signaling but may not participate in effector recognition. The latter group is termed helper NLRs (hNLRs) (Bonardi et al., 2011). ACTIVATED DISEASE RESISTANCE 1 (ADR1), N REQUIREMENT GENE 1 (NRG1) and NB-LRR REQUIRED FOR HYPERSENSITIVE RESPONSE-ASSOCIATED CELL DEATH1 (NRC1) are the three characterized hNLR families. Both ADR1 and NRG1 are CNLs (coiled coil type NLR) with N terminal regions similar to the Resistance to powdery mildew 8 (RPW8) domain. Different Toll/Interleukin-1-receptor (TIR)-type sensor NLRs (TNLs) seem to differentially employ ADR1s and NRG1s to transduce downstream defense signals (Wu et al., 2019).
NLRs are able to recognize effectors directly or indirectly. CHILLING SENSITIVE 3 (CHS3) is one of the TNLs with an atypical C-terminal Lin11, Isl-1 and Mec-3 (LIM) domain. The single amino acid substitution C1340Y close to the LIM domain leads to the gain-of-function chs3-2D mutant with autoimmune phenotypes (Xu et al., 2015). We previously conducted a chs3-2D forward genetic screen by using the chemical mutagen ETHYL METHANE SULFONATE (EMS) and found that the autoimmunity of chs3-2D fully requires the adjacent TNL gene CONSTITUTIVE SHADE-AVOIDANCE 1 (CSA1), but marginally relies on PAD4, which is a positive regulator downstream of many TNLs (Xu et al., 2015; Lapin et al., 2022).
SNIPER1 (snc1-influencing plant E3 ligase reverse genetic screen, 1) and its homolog SNIPER2 globally control the protein levels of sensor NLRs, but not hNLRs. Upon SNIPER1 overexpression, sensor NLRs’ levels were reduced and resulted in enhanced disease susceptibility to pathogens. In contrast, more sNLR accumulation and enhanced disease resistance were observed when the dominant-negative (DN) form of SNIPER1 was expressed (Wu et al., 2020). However, SNIPER1 has no effect on the autoimmunity mediated by the gain-of-function helper NLRs ADR1-L2 D484V or NRG1A D485V (Wu et al., 2020). The direct E3-substrate relationship between SNIPER1 and the sNLRs seems to be through the well conserved NB domain, which exists in all typical plant sensor NLRs (Wu et al., 2020).
Here, we describe an independent suppressor screen in the chs3-2D background, which was carried out to identify new signaling components of CHS3-mediated defense. Multiple mutants that can suppress the chs3-2D autoimmune phenotypes were identified and characterized. Using mapping-by-sequencing, we found mutations in a number of genes, including novel alleles of known regulators, such as IBR5 (INDOLE-3-BUTYRIC ACID RESPONSE 5) and CSA1 (Liu et al., 2015; Xu et al., 2015). In addition to the known regulators, our study revealed that the autoimmunity of chs3-2D is also partially suppressed by mutation in HSP90.2.
Seeds were sterilized by soaking them in a solution of 15% bleach and 0.1% Tween 20 followed by rinsing twice with sterile water. For soil-grown plants, seeds were stratified at 4°C for two days, sown onto sterile soil and transferred to plant growth rooms with either room temperature (23°C, 16 h light/8 h dark; ∼50% relative humidity) or high temperature (28°C, 16 h light/8 h dark; ∼50% relative humidity) conditions as specified in figure legends. Plate-grown plants were grown on half strength Murashige and Skoog (1/2 MS) medium at 22°C and exposed to a 16 h light and 8 h dark regime.
The chs3-2D suppressor screen was carried out as previously described using an EMS-mutagenized chs3-2D population (Li and Zhang, 2016). 100 mg (∼5000) chs3-2D seeds were soaked in a solution of 0.1 M sodium phosphate (pH 5), 5% dimethyl sulfoxide, and 0.25% EMS for 16 h with constant shaking. After incubation, the seeds were washed twice in 100 mM sodium thiosulphate for 15 min and three times in distilled water for 15 min. The mutagenized chs3-2D seeds were planted on 1/2 MS plate for 10 days. Then ∼4000 seedlings were transplanted on soil and grown at 28°C as the dwarfism and lethality of chs3-2D can be suppressed at 28°C. M2 seeds from 25 M1 plants were harvested in each pool. For screening, ∼500 M2 seeds from each pool were planted. Mutants were identified in M2 by selecting plants completely or partially reverted to WT morphology in the extremely dwarfed mutant background (Bi et al., 2011). Mutants were backcrossed with chs3-2D; F1s are heterozygous for the mutated gene, while 25% of F2s are homozygotes if the suppression is caused by single nuclear gene mutation. We followed homozygous F2s to F3 generation to confirm their phenotypes, and pooled F3 plants were subjected to the next generation sequencing (NGS) analysis.
For next generation sequencing, total genomic DNA was extracted from 24-day-old candidate suppressors using the CTAB method (Li et al., 2001) followed by DNA purification using Qiagen plant DNA extraction kit (Qiagen, Germany). The library preparation and Illumina sequencing were carried out by the BGI (Beijing Genomic Institute, China).
The genomic fragment of At5G56030 containing its native promoter was PCR-amplified from wildtype (WT) Col-0 genomic DNA. The fragment was then digested and cloned into the vector pCambia1305 to generate HSP90.2:HSP90.2. For Arabidopsis transformation, the above constructed binary vectors were transformed into Agrobacterium tumefaciens GV3101 (pMP90) by electroporation and subsequently transformed into Arabidopsis plants by the floral dip method (Clough and Bent, 1998). Transgenic plants were screened on soil by spraying 100 mg/L BASTA four times with a two-day interval between each treatment.
The mutants hsp90.2 G122S, hsp90.3 S100F (Huang et al., 2014) and sgt1b (Austin et al., 2002) were used to generate double mutants with DN-SNIPER1 (Wu et al., 2020) and ADR1-L2 D484V (Roberts et al., 2013). To identify homozygous double mutants, F1s were planted on BASTA selection plates and genotyped for heterozygous backgrounds. F1s were individually harvested, and F2s were planted on BASTA selection plates to select for ADR1-L2 D484V or DN-SNIPER1 transgenes. Only plants that showed no segregation and were homozygous for the transgenes in F3 were genotyped for verification of homozygosity of the mutated HSP90 chaperone gene.
Three-week-old seedlings were spray-inoculated with Hyaloperonospora arabidopsidis (H.a.) Noco2 at a concentration of 1 × 105 spores per ml. The oomycete was allowed to propagate in a humid growth chamber (12 h light/12 h dark, 18°C) for 7 days before the number of spores on the plant surface was quantified. The 12 plants from each genotype were divided into groups of three and placed in 1 ml of ddH2O in 15 ml test tubes (4 plants per tube). Spores were suspended in solution by vortexing and counted using a hemocytometer. Three independent replicates were performed.
The GraphPad Prism 9.0 software (GraphPad Software, Inc.) and Microsoft Excel were used for the statistical analyses in this study.
The chs3-2D autoimmune mutant exhibits severe dwarfism (Bi et al., 2011). In order to search for components required for CHS3-mediated defense response, a suppressor screen was conducted. Suppression of stunted growth was used as a criterion during the primary screen. The genetic background of each mutant was verified by genotyping the chs3-2D locus. Then, mutants exhibiting morphological suppression of chs3-2D-associated phenotypes were subjected to a secondary screen, where resistance to the virulent oomycete strain Hyaloperonospora arabidopsidis (H.a.) Noco2 was examined. Mutants that displayed enhanced susceptibility to H.a. Noco2 as compared to chs3-2D were selected for further characterization.
From the screen, six independent chs3-2D suppressor lines were isolated. As shown in Figures 1A,B, all six chs3-2D suppressors can either partially or fully suppress the morphology of chs3-2D. Consistent with the morphological suppression, all of them lost the constitutive resistance response of chs3-2D against H.a. Noco2 (Figure 1C). Taken together, these data suggest that all six mutants contain mutations suppressing the autoimmune phenotypes of chs3-2D.
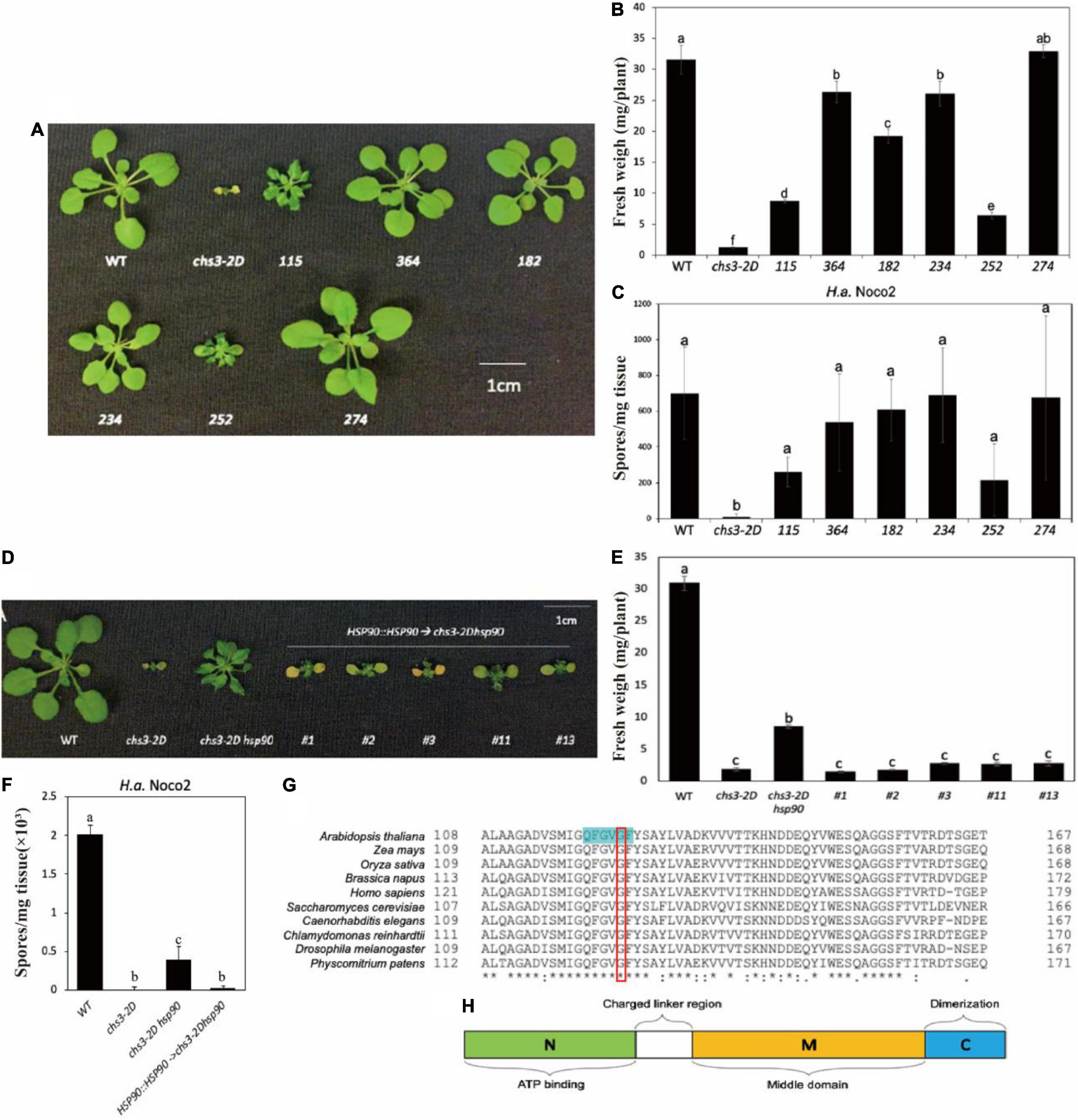
Figure 1. Identification and characterization of chs3-2D suppressors. (A) Morphology of 3-week-old soil-grown plants of chs3-2D suppressors at 18 °C. (B) Weights of 3-week-old soil-grown plants of chs3-2D suppressors at 18 °C. (C) Quantification of H.a. Noco2 sporulation on the leaf surface of chs3-2D suppressors. (D) Morphology of 3-week-old soil-grown plants of the mutant chs3-2D hsp90.2 complementary transgenic lines at 18 °C. (E) Weights of 3-week-old soil-grown plants of the mutant chs3-2D hsp90.2 complementary transgenic lines at 18 °C. (F) Quantification of H.a. Noco2 conidia growth on the leaf surface of the mutant chs3-2D hsp90.2 complementary transgenic lines. (G) Amino acid alignments of selected HSP90 homologs from eukaryotic organisms. The amino acids highlighted in green are the nucleotide binding site. The mutated amino acid is labeled with a red box. For panel (G), List of the organisms included: mouse-ear cress Arabidopsis thaliana HSP90.2, maize Zea mays (C3UZ63), rice Oryza sativa (Q0J4P2), rapeseed Brassica napus (A0A078GRJ1), human Homo sapiens (CAA33259.1), yeast Saccharomyces cerevisiae (AJS65103.1), nematode Caenorhabditis elegans (NP_506626.1), alga Chlamydomonas reinhardtii (XP_001695264.1), fruit fly Drosophila melanogaster (NP_001261362.1), and moss Physcomitrium patens (XP_024385281.1). Protein sequences were aligned using CLUSTAL. (H) Schematic diagram of the HSP90 domains. Hsp90 is comprised of three domains: an N-terminal ATP-binding domain (N, green) that may contain a peptide binding element; a middle domain (M, yellow) that interacts with client proteins and also contains a loop that catalyzes ATP hydrolysis; and a C-terminal dimerization domain (C, blue). A charged region exists between the N and M domains (linker region, white). Data information: For panels (C,F), Two-week-old soil-grown seedlings were sprayed with a spore suspension of H.a. Noco2 at a concentration of 100,000 spores/ml of water. The plants were then covered and incubated for seven days in a high humidity growth chamber. Spores were counted in water suspension using a hemocytometer (bars represent means of n replicates ± SD, n = 3 with 4 plants each). One-way ANOVA followed by Tukey’s post hoc test were performed for panels (A–F). Statistical significance is indicated by different letters (P < 0.05). Error bars represent mean SD (n = 5).
These six independent chs3-2D suppression lines were backcrossed with chs3-2D separately. Mapping-by-sequencing was used on the co-segregating homozygous suppressor plants in F2 and F3, and the NGS data can be found at NCBI GenBank (PRJNA820297). Table 1 summarizes the candidate causal mutations identified in these mutants. Most of them contain mutations in known genes where loss-of-function mutations would suppress chs3-2D (Liu et al., 2015; Xu et al., 2015). Mutant 252 carried a point mutation in the fifth exon of CHS3 (At5g17890), resulting in a P965 to S965 substitution. In addition, the complete suppressor 234-1 had a G to A splice site mutation in the first intron of Indole-3-Butyric Acid Response 5 (IBR5, At2G04550), where the encoded protein is known to physically interact with CHS3 and is required for its defense responses (Liu et al., 2015). Lastly, 364, 274-1, and 182-1 carried mutations in CSA1 (At5G17880) (Xu et al., 2015). One csa1 allele had a point mutation in the third exon, leading to amino acid change from K862 to E862. The other two alleles contained mutations in the second and first exons, respectively, which result in early stop codon in premature protein (Table 1).
The chs3-2D partial suppressor, mutant 115, carried a mutation in the third exon of HSP90.2 (At5G56030), leading to an amino acid change from G122 to S122 (Table 1), suggesting that HSP90.2 may positively regulate CHS3-mediated defense responses. To confirm that the partial suppression phenotype of the mutant 115 is due to the mutation in HSP90.2, a wild-type copy of HSP90.2 driven by its native promoter was transformed in 115 mutant plants. The 115 mutant lines harboring the HSP90.2 transgene reverted to a smaller size (Figure 1D) and lower weight (Figure 1E), and showed enhanced resistance against H.a Noco2 (Figure 1F) compared with the 115 mutant plants, comparable to chs3-2D. Therefore HSP90.2 can fully complement the partial suppression phenotypes of mutant 115, suggesting that the mutation in HSP90.2 is responsible for partial suppression of the chs3-2D in 115.
BLAST analysis revealed that the mutated amino acid G122 in HSP90.2 is conserved among HSP90 homologs from various eukaryotic organisms (Figure 1G). Furthermore, G122 is located at the central nucleotide binding site (Figure 1H; Xu et al., 2012). Thus, the mutation from G122 to S122 might cause a loss of HSP90.2 function. In Arabidopsis, HSP90.2 is redundant with HSP90.1/3/4. When we compared the current hsp90.2 allele with other previously reported hsp90.1/2/3/4 alleles (Table 2), this hsp90.2 G122S mutation seems to be a novel mutant allele of HSP90.2.
As HSP90 and suppressor of the G2 allele of skp1 (SGT1) are known chaperons for NLRs, and loss-of-function mutations in SGT1b are known to suppress chs3-1, another autoimmune allele of CHS3 (Kadota et al., 2010; Yang et al., 2010), we asked how widely HSP90 and SGT1b contribute to sensor NLRs. From our previous study, SNIPER1/2 can target a large range of sensor NLRs for ubiquitination and degradation. Overexpression of SNIPER1 results in enhanced disease susceptibility, while a dominant-negative form of SNIPER1 (DN SNIPER1) leads to more sensor NLRs accumulation and enhanced disease resistance (Wu et al., 2020). To address whether chaperone proteins HSP90 and SGT1b can regulate the autoimmunity caused by overaccumulation of sensor NLRs, we crossed DN-SNIPER1 with hsp90 or sgt1b. As shown in Figure 2, the dwarfism and enhanced resistance of DN SNIPER1 against H.a Noco2 were largely suppressed by hsp90.2, hsp90.3, or sgt1b, indicating that chaperone proteins HSP90s and SGT1b are generally required by sensor NLRs.
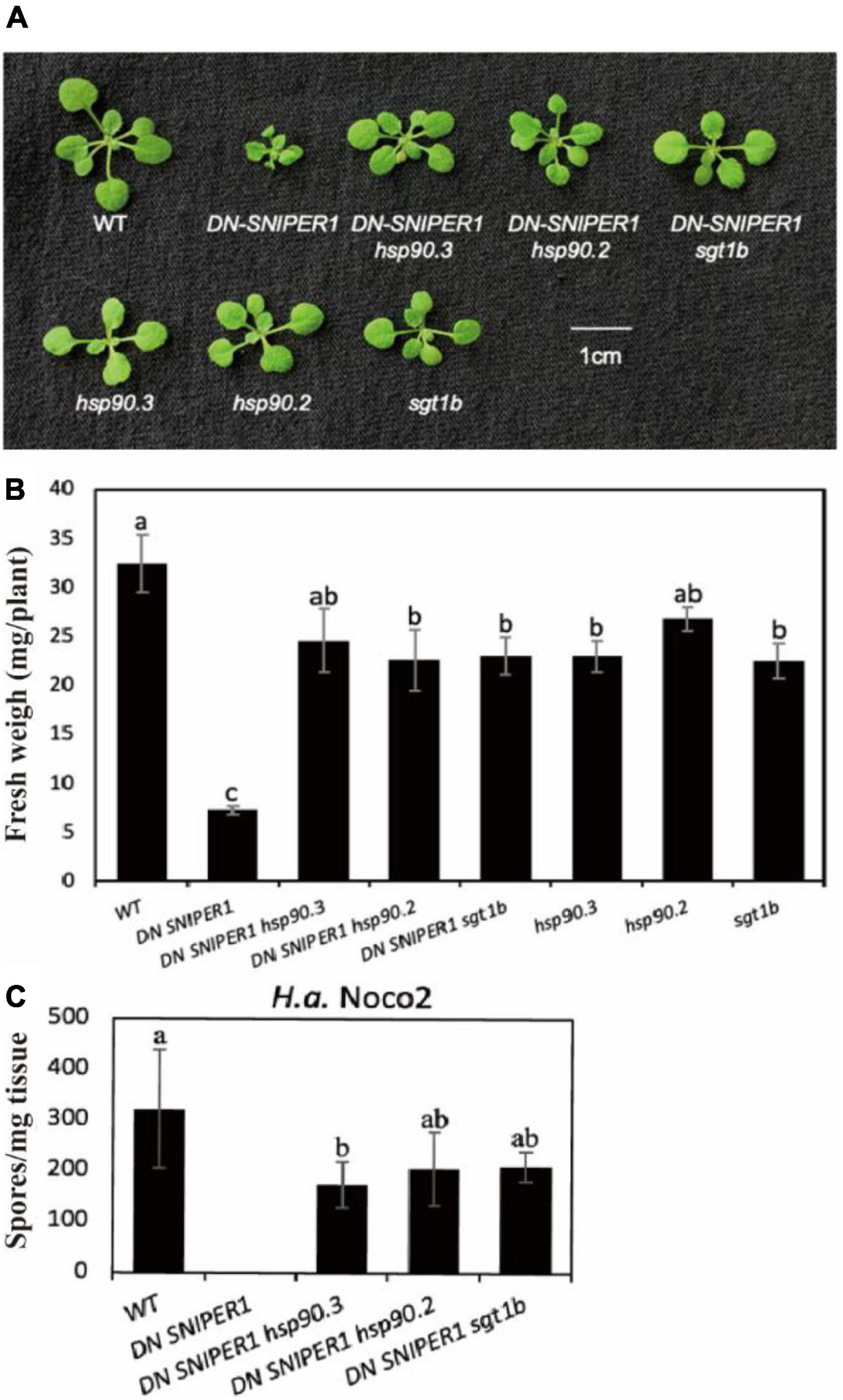
Figure 2. Chaperone protein HSP90s and SGT1b contribute to the autoimmunity caused by overaccumulation of sensor NLRs. (A) Morphology of 3-week-old soil-grown plants of DN-SNIPER1 hsp90, DN-SNIPER1 sgt1b double mutant at 23 °C. (B) Weights of 3-week-old soil-grown plants of DN-SNIPER1 hsp90, DN-SNIPER1 sgt1b double mutant at 23 °C. (C) Quantification of H.a. Noco2 conidia growth on the leaf surface of DN-SNIPER1 hsp90, DN-SNIPER1 sgt1b double mutant. Data information: For panel C, the experimental procedure was carried out as described in Figure 1. One-way ANOVA followed by Tukey’s post hoc test were performed for panels (B,C). Statistical significance is indicated by different letters (P < 0.05). Error bars represent mean SD (n = 5).
Helper NLRs serve functional roles downstream of sensor NLRs (Jubic et al., 2019; van Wersch et al., 2020). To test whether activation of helper NLRs also require chaperone/co-chaperone proteins HSP90 and SGT1b, we crossed a gain-of-function (g-o-f) variant ADR1 (ADR1-L2 D484V) (Roberts et al., 2013) with chaperone mutants hsp90.2 or sgt1b (Figure 3). The double mutants ADR1-L2 D484V hsp90 and ADR1-L2 D484V sgt1b showed partially suppressed morphological phenotypes (Figures 3A,B), which are consistent with the oomycete infection results (Figure 3C). This suggests the constitutive activation of defense responses in ADR1-L2 D484V also relies on chaperones HSP90 and SGT1b, as both sgt1b and hsp90.2 can largely suppress the autoimmune phenotype of ADR1-L2 D484V plants. Taken together, general chaperone activities are not only required for sensor NLRs, but also helper NLRs.
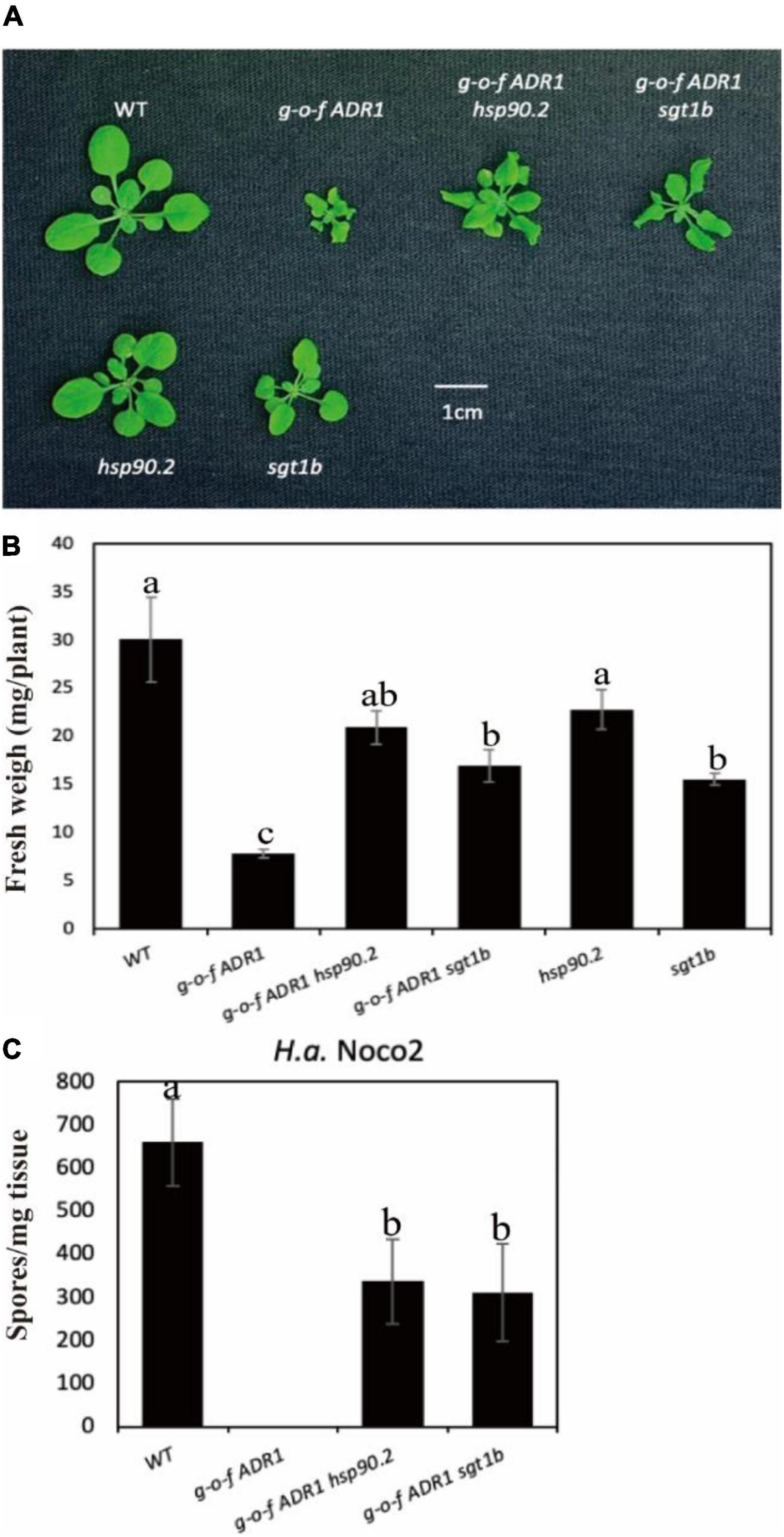
Figure 3. Mutations at HSP90s or SGT1b partially suppress the autoimmunity of ADR1-L2 D484V. (A) Morphology of 3-week-old soil-grown plants of ADR1-L2 D484V hsp90, ADR1-L2 D484V sgt1b double mutant at 23 °C. (B) Weights of 3-week-old soil-grown plants of ADR1-L2 D484V hsp90, ADR1-L2 D484V sgt1b double mutant at 23 °C. (C) Quantification of H.a. Noco2 conidia growth on leaf surface of ADR1-L2 D484V hsp90, ADR1-L2 D484V sgt1b double mutant. The experiment was carried out as described in Figure 1. One-way ANOVA followed by Tukey’s post hoc test were performed for panels (B,C). Statistical significance is indicated by different letters (P < 0.05). Error bars represent mean SD (n = 5).
The chaperones are a large and diverse group of unrelated proteins that control both non-covalent folding and post-translational maintenance of proteins in the cells (Liberek et al., 2008). They participate in diverse biological and cellular processes such as growth, development, disease resistance, and signal transduction (Kadota and Shirasu, 2012). Misfolded or misused sensors are threats to the cell and must be immediately inactivated and discarded to avoid inappropriate activation of downstream pathways. Therefore, chaperones are crucial in maintenance of NLR-type sensors (Shirasu, 2009). Interestingly, a point mutation allele of hsp90.2 was identified from the chs3-2D suppressor screen. Our results show that this new allele of hsp90.2 partially suppressed the autoimmune phenotypes of chs3-2D mutant plants (Figure 1), suggesting that HSP90 contributes to CHS3-mediated defense responses. Combined with previous report where sgt1b and rar1 were found to fully suppress chs3-1 (Yang et al., 2010), HSP90, required for Mla12 resistance (RAR1) and SGT1 seem to be all serving chaperon roles for CHS3.
HSP90 chaperone proteins are highly conserved and abundant in diverse organisms, from bacteria to plants (e.g., moss, alga, rice, and maize) and mammals. In eukaryotes, HSP90 is involved in the assembly, stabilization and maturation of key signaling proteins, including protein kinases and hormone receptors (Kadota and Shirasu, 2012). It contains an N-terminal ATP-binding domain (ND), a middle domain (MD) for binding substrate proteins, and a C-terminal constitutive dimerization domain (CD) (Figure 1G). Plants without functional HSP90s may exhibit increased susceptibility to pathogens and reduced R protein mediated immunity and lower accumulation of immune receptors, suggesting that these chaperones are critical for NLR protein complex assembly, stability, and/or activation (Shirasu, 2009), which is consistent with our observation that a mutation in HSP90 can suppress the autoimmunity caused by a gain-of-function sensor NLR CHS3-2D (Figure 2). Such an observation is also consistent with previous studies suggesting that HSP90 chaperones are broadly involved in serving different sensor NLR client proteins, including RPM1, RPS5, Rx, MLA1, MLA6, and RPS2 (Hubert et al., 2003; Lu et al., 2003; Bieri et al., 2004; Holt et al., 2005; Kadota and Shirasu, 2012). Although we did not test the interactions between CHS3 or ADR1s with HSP90, we would expect that as NLRs, they likely also use the same chaperones for complex assembly during resistosome formation and defense activation.
In Arabidopsis, there are four cytosolic HSP90 genes (HSP90.1-4), all locateed to the same region of chromosome 5 (Donato and Geisler, 2019; Krishna and Gloor, 2001). There is high degree of sequence homology among these paralogs, suggest functional redundancy (Queitsch et al., 2002; Hubert et al., 2003). Although the Arabidopsis HSP90 paralogs are highly related, they exhibit different expression profiles (Prasinos et al., 2005). Moreover, previous studies shown that different point mutations in HSP90s can cause drastically different or even opposite biological consequences (Supplementary Table 1 in Hubert et al., 2009; Table 2), supporting uneven functional redundancy among the family members. Our hsp90.2 allele suppresses chs3-2D, gain-of-function helper NLR (ADR1-L2 D484V) and overaccumulation of sensor NLRs (DN-SNIPER1), indicating that HSP90 contributes broadly to chaperone both sensor and helper NLRs. As both sensor and helper NLRs require oligomerization for activation, HSP90, together with SGT1 and likely RAR1, probably assist in the assembly of resistosomes during NLR oligomerization and defense activation. However, it should be kept in mind that chaperones can not only assist with NLR receptor complex assembly during activation, they can also aid in other protein complexes’ organization during NLR degradation. For example, hsp90.3 mutation affects the SCFCPR1 complex formation, leading to increased SNC1 levels and autoimmunity (Huang et al., 2014). Therefore, sometimes the phenotypes of an hsp90 allele can be a combinatory consequence from both positive and negative effects of the HSP90 in different complexes, which may cause confusion in data interpretation.
The dimerized HSP90 usually works in association with co-chaperone proteins that modulate its ATPase activity or recruit substrates (Hoter et al., 2018). Studies in plants revealed that HSP90 and its co-chaperones, SGT1 and RAR1, are major stabilizing factors for NLR proteins (Kadota et al., 2010). HSP90 interacts with the cysteine-and histidine-rich zinc-binding domain (CHORD) of RAR1 as well as the CHORD domain and SGT1 motif of SGT1 (Boter et al., 2007). The Arabidopsis SGT1 (AtSGT1b) gene was identified by loss of Hyaloperonospora arabidopsidis resistance in the sgt1b mutant that would otherwise be provided by RPP5 or RPP7 (Austin et al., 2002; Tor et al., 2002), indicating that SGT1 also plays an important role in NLR activation in plants. In plants, SGT1 is needed for the maintenance of steady-state levels of NLRs including Rx and N (Azevedo et al., 2006; Mestre and Baulcombe, 2006; Boter et al., 2007). Similar to HSP90, SGT1 not only positively regulates plant NLR activity such as CHS3, but also performs a negative role in controlling the turnover of NLRs such as RPM1, RPS5, and SNC1 (SUPPRESSOR OF NPR1, CONSTITUTIVE 1), as these NLR proteins accumulate to higher levels in sgt1b mutant plants (Holt et al., 2005; Li et al., 2010). The contribution of HSP90 and SGT1 in NLR turnover control can be explained by their contributions to the Skp1-Cullin-F-box (SCF) E3 ligase complex assembly for NLR ubiquitination and degradation (Huang et al., 2014; Kapos et al., 2019). The multi-functionality of SGT1 and HSP90 suggests a diverse client base involving different protein complexes assembled during immune responses (Huang et al., 2014). Furthermore, SGT1 is also needed for immune responses mediated by non-NLR-type sensors such as Cf4, Cf9, or RPW8 (Shirasu, 2009). Lastly, as mutations in either SGT1 or HSP90 can suppress autoactivation of helper NLR (Figure 3), both chaperones contribute not only at sensor NLR, but also helper NLR levels. Such broad roles should be taken into consideration for future biochemical studies of these chaperones in NLR biology.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/, PRJNA820297.
JL, WL, NZ, and SW carried out the experiments. JL, NZ, and XL wrote the manuscript with inputs from all authors. XL conceived the original idea and supervised the project.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (http://dx.doi.org/10.13039/501100000038). Research described is supported by funding from CFI-JELF, NSERC-Discovery and NSERC-CREATE-PRoTECT programs. JL, WL, and SW were partly supported by fellowships from China Scholarship Council (CSC) and NSERC.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors cordially thank Jeff Dangl and Jane Parker for seeds of ADR1-L2 D484V and sgt1b.
Austin, M. J., Muskett, P., Kahn, K., Feys, B. J., Jones, J. D., and Parker, J. E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295, 2077–2080. doi: 10.1126/science.1067747
Azevedo, C., Betsuyaku, S., Peart, J., Takahashi, A., Noel, L., Sadanandom, A., et al. (2006). Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 25, 2007–2016. doi: 10.1038/sj.emboj.7601084
Bao, F., Huang, X., Zhu, C., Zhang, X., Li, X., and Yang, S. (2014). Arabidopsis HSP90 protein modulates RPP4-mediated temperature-dependent cell death and defense responses. New Phytol. 202, 1320–1334. doi: 10.1111/nph.12760
Bi, D., Johnson, K. C., Zhu, Z., Huang, Y., Chen, F., Zhang, Y., et al. (2011). Mutations in an atypical TIR-NB-LRR-LIM resistance protein confer autoimmunity. Front. Plant Sci. 2:71. doi: 10.3389/fpls.2011.00071
Bieri, S., Mauch, S., Shen, Q. H., Peart, J., Devoto, A., Casais, C., et al. (2004). RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16, 3480–3495. doi: 10.1105/tpc.104.026682
Bonardi, V., Tang, S., Stallmann, A., Roberts, M., Cherkis, K., and Dangl, J. L. (2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. U.S.A. 108, 16463–16468. doi: 10.1073/pnas.1113726108
Boter, M., Amigues, B., Peart, J., Breuer, C., Kadota, Y., Casais, C., et al. (2007). Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell 19, 3791–3804. doi: 10.1105/tpc.107.050427
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Donato, M., and Geisler, M. (2019). HSP90 and co-chaperones: a multitaskers’ view on plant hormone biology. FEBS Lett. 593, 1415–1430. doi: 10.1002/1873-3468.13499
Holt, B. F. III, Belkhadir, Y., and Dangl, J. L. (2005). Antagonistic control of disease resistance protein stability in the plant immune system. Science 309, 929–932. doi: 10.1126/science.1109977
Hoter, A., El-Sabban, M. E., and Naim, H. Y. (2018). The HSP90 family: structure, regulation, function, and implications in health and disease. Int. J. Mol. Sci. 19:2560. doi: 10.3390/ijms19092560
Huang, S., Monaghan, J., Zhong, X., Lin, L., Sun, T., Dong, O. X., et al. (2014). HSP90s are required for NLR immune receptor accumulation in Arabidopsis. Plant J. 79, 427–439. doi: 10.1111/tpj.12573
Hubert, D. A., He, Y., McNulty, B. C., Tornero, P., and Dangl, J. L. (2009). Specific Arabidopsis HSP90.2 alleles recapitulate RAR1 cochaperone function in plant NB-LRR disease resistance protein regulation. Proc. Natl. Acad. Sci. U.S.A. 16, 9556–9563. doi: 10.1073/pnas.0904877106
Hubert, D. A., Tornero, P., Belkhadir, Y., Krishna, P., Takahashi, A., Shirasu, K., et al. (2003). Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22, 5679–5689. doi: 10.1093/emboj/cdg547
Jubic, L. M., Saile, S., Furzer, O. J., El Kasmi, F., and Dangl, J. L. (2019). Help wanted: helper NLRs and plant immune responses. Curr. Opin. Plant Biol. 50, 82–94. doi: 10.1016/j.pbi.2019.03.013
Kadota, Y., and Shirasu, K. (2012). The HSP90 complex of plants. Biochim. Biophys. Acta 1823, 689–697. doi: 10.1016/j.bbamcr.2011.09.016
Kadota, Y., Shirasu, K., and Guerois, R. (2010). NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem. Sci. 35, 199–207. doi: 10.1016/j.tibs.2009.12.005
Kapos, P., Devendrakumar, K. T., and Li, X. (2019). Plant NLRs: from discovery to application. Plant Sci. 279, 3–18. doi: 10.1016/j.plantsci.2018.03.010
Krishna, P., and Gloor, G. (2001). The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6, 238–246. doi: 10.1379/1466-1268(2001)006<0238:thfopi>2.0.co;2
Lapin, D., Johanndrees, O., Wu, Z., Li, X., and Parker, J. E. (2022). Molecular innovations in plant TIR-based immunity signaling. Plant Cell 34, 1479–1496. doi: 10.1093/plcell/koac035
Li, X., Song, Y., Century, K., Straight, S., Ronald, P., Dong, X., et al. (2001). A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J. 27, 235–242. doi: 10.1046/j.1365-313x.2001.01084.x
Li, X., and Zhang, Y. (2016). Suppressor screens in Arabidopsis. Methods Mol. Biol. 1363, 1–8. doi: 10.1007/978-1-4939-3115-6_1
Li, Y., Li, S., Bi, D., Cheng, Y. T., Li, X., and Zhang, Y. (2010). SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog. 6:e1001111. doi: 10.1371/journal.ppat.1001111
Liberek, K., Lewandowska, A., and Zietkiewicz, S. (2008). Chaperones in control of protein disaggregation. EMBO J. 27, 328–335. doi: 10.1038/sj.emboj.7601970
Liu, J., Yang, H., Bao, F., Ao, K., Zhang, X., Zhang, Y., et al. (2015). IBR5 Modulates Temperature-Dependent, R Protein CHS3-mediated defense responses in Arabidopsis. PLoS Genet. 11:e1005584. doi: 10.1371/journal.pgen.1005584
Lu, R., Malcuit, I., Moffett, P., Ruiz, M. T., Peart, J., Wu, A. J., et al. (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. doi: 10.1093/emboj/cdg546
Mestre, P., and Baulcombe, D. C. (2006). Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 18, 491–501. doi: 10.1105/tpc.105.037234
Prasinos, C., Krampis, K., Samakovli, D., and Hatzopoulos, P. (2005). Tight regulation of expression of two Arabidopsis cytosolic Hsp90 genes during embryo development. J. Exp. Bot. 56, 633–644. doi: 10.1093/jxb/eri035
Queitsch, C., Sangster, T. A., and Lindquist, S. (2002). Hsp90 as a capacitor of phenotypic variation. Nature 417, 618–624. doi: 10.1038/nature749
Roberts, M., Tang, S., Stallmann, A., Dangl, J. L., and Bonardi, V. (2013). Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet. 9:e1003465. doi: 10.1371/journal.pgen.1003465
Shirasu, K. (2009). The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60, 139–164. doi: 10.1146/annurev.arplant.59.032607.092906
Takahashi, A., Casais, C., Ichimura, K., and Shirasu, K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 11777–11782. doi: 10.1073/pnas.2033934100
Tor, M., Gordon, P., Cuzick, A., Eulgem, T., Sinapidou, E., Mert-Turk, F., et al. (2002). Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14, 993–1003. doi: 10.1105/tpc.001123
Toumi, I. G., Pagoulatou, M., Margaritopoulou, T., Milioni, D. A., and Roubelakis-Angelakis, K. (2019). Genetically modified heat shock protein90s and polyamine oxidases in Arabidopsis reveal their interaction under heat stress affecting polyamine acetylation, oxidation and homeostasis of reactive oxygen species. Plants 8:323. doi: 10.3390/plants8090323
van Wersch, S., Tian, L., Hoy, R., and Li, X. (2020). Plant NLRs: the Whistleblowers of Plant Immunity. Plant Commun. 1:100016. doi: 10.1016/j.xplc.2019.100016
Wu, Z., Li, M., Dong, O. X., Xia, S., Liang, W., Bao, Y., et al. (2019). Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol. 222, 938–953. doi: 10.1111/nph.15665
Wu, Z., Tong, M., Tian, L., Zhu, C., Liu, X., Zhang, Y., et al. (2020). Plant E3 ligases SNIPER1 and SNIPER2 broadly regulate the homeostasis of sensor NLR immune receptors. EMBO J. 39, e104915. doi: 10.15252/embj.2020104915
Xu, F., Zhu, C., Cevik, V., Johnson, K., Liu, Y., Sohn, K., et al. (2015). Autoimmunity conferred by chs3-2D relies on CSA1, its adjacent TNL-encoding neighbour. Sci. Rep. 5:8792. doi: 10.1038/srep08792
Xu, Z. S., Li, Z. Y., Chen, Y., Chen, M., Li, L. C., and Ma, Y. Z. (2012). Heat shock protein 90 in plants: molecular mechanisms and roles in stress responses. Int. J. Mol. Sci. 13, 15706–15723. doi: 10.3390/ijms131215706
Keywords: plant immunity, HSP90, CHILLING SENSITIVE 3, SNIPER1, sensor NLR, helper NLR, CHS3, CSA1
Citation: Lu J, Liang W, Zhang N, van Wersch S and Li X (2022) HSP90 Contributes to chs3-2D-Mediated Autoimmunity. Front. Plant Sci. 13:888449. doi: 10.3389/fpls.2022.888449
Received: 02 March 2022; Accepted: 04 May 2022;
Published: 03 June 2022.
Edited by:
Andrei-Jose J. Petrescu, Institute of Biochemistry of the Romanian Academy, RomaniaReviewed by:
Xiufang Xin, Institute of Plant Physiology and Ecology (CAS), ChinaCopyright © 2022 Lu, Liang, Zhang, van Wersch and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Li, eGlubGlAbXNsLnViYy5jYQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.