- 1Department of Horticultural Biotechnology, College of Life Science, Kyung Hee University, Yongin, South Korea
- 2Department of Life Sciences, Pohang University of Science and Technology, Pohang, South Korea
- 3School of Interdisciplinary Bioscience and Bioengineering, Pohang University of Science and Technology, Pohang, South Korea
- 4Department of Plant Science, Plant Immunity Research Center, Plant Genomics and Breeding Institute, Research Institute for Agriculture and Life Sciences, Seoul National University, Seoul, South Korea
- 5Graduate School of Biotechnology, Kyung Hee University, Yongin, South Korea
Clavibacter michiganensis, a Gram-positive plant-pathogenic bacterium, utilizes apoplastic effectors for disease development in host plants. Here, we determine the roles of Pat-1Cm (a putative serine protease) in pathogenicity and plant immunity. Pat-1Cm was found to be a genuine secreted protein, and the secreted mature form did not carry the first 33 amino acids predicted to be a signal peptide (SP). The pat-1Cm mutant impaired to cause wilting, but still caused canker symptom in tomato. Moreover, this mutant failed to trigger the hypersensitive response (HR) in a nonhost Nicotiana tabacum. Among orthologs and paralogs of pat-1Cm, only chp-7Cs from Clavibacter sepedonicus, a potato pathogen, successfully complemented pat-1Cm function in pathogenicity in tomato, whereas all failed to complement pat-1Cm function in HR induction in N. tabacum. Based on the structural prediction, Pat-1Cm carried a catalytic triad for putative serine protease, and alanine substitution of any amino acids in the triad abolished both pathogenicity and HR-inducing activities of Pat-1Cm in C. michiganensis. Ectopic expression of pat-1Cm with an SP from tobacco secreted protein triggered HR in N. tabacum, but not in tomato, whereas a catalytic triad mutant failed to induce HR. Inoculation of the pat-1Cm mutant mixed with the mutant of another apoplastic effector CelA (cellulase) caused severe wilting in tomato, indicating that these two apoplastic effectors can functionally cooperate in pathogenicity. Overall, these results indicate that Pat-1Cm is a distinct secreted protein carrying a functional catalytic triad for serine protease and this enzymatic activity might be critical for both pathogenicity and HR-eliciting activities of Pat-1Cm in plants.
Introduction
Plant pathogenic bacteria use diverse effector proteins to manipulate host metabolism and suppress host immunity (Dou and Zhou, 2012; Toruno et al., 2016). Effectors can be divided into intracellular and apoplastic effectors. Intracellular effectors are delivered directly into the host cell cytoplasm through type III or type VI secretion systems (T3SS or T6SS), whereas apoplastic effectors are secreted mostly through a type II secretion system (T2SS) and function in the apoplastic space of plant tissues (Costa et al., 2015; Galan and Waksman, 2018). The types and functions of intracellular effector proteins have been well studied in Gram-negative bacteria such as Pseudomonas syringae and Xanthomonas spp. Generally, these effectors act as virulence factors in susceptible host plants (Cunnac et al., 2009; White et al., 2009; Lindeberg et al., 2012; Timilsina et al., 2020). However, some effectors act as avirulence factors and are recognized by the Nod-like receptor (NLR) proteins to trigger an immune response in nonhost and resistant plants (Lee et al., 2017; van Wersch et al., 2020). In contrast, apoplastic effectors such as cell wall-degrading enzymes, cellulases and pectate lyases, and proteases have been studied in soft rot pathogens (Davidsson et al., 2013) and have been shown only as virulence factors. Unlike soft rot pathogens, Cladosporium fulvum, a fungal pathogen that causes leaf mold disease in tomato, uses apoplastic effectors such as Avr2, Avr4, and Avr9 as virulence factors to suppress pattern-triggered immunity and can trigger immune responses upon recognition by plasma membrane-localized resistance proteins, Cf (de Wit, 2016).
Unlike Gram-negative bacteria, the types and functions of effector proteins have not been well studied in Gram-positive plant-pathogenic bacteria. Nevertheless, some apoplastic effectors have been studied as major virulence factors. As an example, a Gram-positive phytopathogenic bacterium, Clavibacter michiganensis, causes bacterial canker and wilting in tomato and uses a cellulase CelA, a pectate lyase PelA, and serine proteases such as Pat-1Cm and the Chp (Chromosomal homologs of Pat-1) protein family as major virulence factors (Eichenlaub and Gartemann, 2011; Thapa et al., 2017; Hwang et al., 2018, 2019). Meanwhile, other Gram-positive bacteria such as Streptomyces scabies, which causes scab in potato tuber, and Rhodococcus fascians, which causes the leafy gall disease, use the phytotoxin, thaxtomin, and phytohormones as major pathogenicity or virulence factors, respectively (Bignell et al., 2010; Stes et al., 2013; Francis et al., 2016; Liu et al., 2021).
Clavibacter michiganensis Pat-1Cm is a putative serine protease encoded by the pat-1 gene in the pCM2 plasmid. It carries a putative signal peptide (SP) in its N-terminus (Lu et al., 2015), indicating that Pat-1Cm may be secreted. Observations of a C. michiganensis strain lacking pCM2 and complementation of this strain with a DNA fragment carrying the pat-1Cm gene implied that the pat-1Cm gene functions as a pathogenicity factor, although there were no experimental data using deletion or defective mutants of the pat-1Cm gene (Dreier et al., 1997; Burger et al., 2005). Although the enzymatic activity of these proteins has not been demonstrated yet, Pat-1Cm and Chp family proteins have a shared serine residue within a conserved GDSGG motif that might be one of a catalytic triad (together with histidine and aspartate) for protease activity (Ruiz-Perez and Nataro, 2014; Lu et al., 2015). A Pat-1 ortholog in Clavibacter sepedonicus, Chp-7Cs was found to be involved in virulence in potato and hypersensitive response (HR) induction in Nicotiana tabacum (Nissinen et al., 2009), indicating that Pat-1Cm and its orthologs might act as both pathogenicity or virulence factors and immunity elicitors in plants. Moreover, the analysis of whole genome sequence data of Clavibacter species has shown that many genes encoding these putative serine proteases with SP are present in various Clavibacter species, including C. michiganensis, Clavibacter capsici, and C. sepedonicus (Nissinen et al., 2009; Lu et al., 2015; Hwang et al., 2018; Mendez et al., 2020). Overall, these findings suggest that Pat-1Cm and its orthologs are important apoplastic effectors for the interaction of Clavibacter species with plants.
Similar cases have been shown in other pathogenic bacteria. The secreted serine protease, PrtA, of Xylella fastidiosa and a cysteine protease HopN1 of the YopT/AvrPphB effector family in P. syringae pv tomato DC3000 contribute to the virulence and HR induction, respectively (Shao et al., 2002; Gouran et al., 2016). However, unlike human and animal pathogens (Ruiz-Perez and Nataro, 2014), the role of serine proteases in virulence of phytopathogenic bacteria is still elusive, especially in Gram-positive bacterial pathogens such as genus Clavibacter.
In this study, we characterized Pat-1Cm with respect to its protein secretion, pathogenicity, and ability to induce HR in plants. We found that Pat-1Cm is indeed a secreted protein with a functional SP and has roles in pathogenicity and plant immunity. Moreover, the catalytic triad of Pat-1Cm for putative serine protease is critical for both its pathogenicity and HR induction in plants. Our findings provide us a distinct role of apoplastic protease effectors of pathogenic bacteria for interactions with host and nonhost plants.
Materials and Methods
Bacterial Strains, Culture Conditions, and Inoculum Preparation
The C. michiganensis type strain LMG7333, its mutant strains, Tn::pat-1Cm and Tn::celACm generated by transposon mutagenesis, and the complemented strains were used in this study (Hwang et al., 2019). All bacterial strains were grown on KB medium (20 g of protease peptone no. 3, 1.5 g of K2HPO4, 6 ml of 1 M MgSO4, and 16 ml of 50% glycerol per liter) supplemented with appropriate antibiotics: kanamycin (100 μg ml−1), neomycin (100 μg ml−1), and chloramphenicol (10 μg ml−1) at 26°C for 24–48 h. For plant inoculation, single colonies of cultured C. michiganensis strains were incubated in KB broth overnight with shaking at 140 rpm, and then cells were collected by centrifugation and resuspended in 10 mM of MgCl2.
Plant Growth Conditions
Tomato (Solanum lycopersicum L., cv. “Betatini”) and N. tabacum (cv. “Samsun”) plants were grown in a growth chamber at 26°C with a 14:10-h light:dark photoperiod condition. Then, 2- or 3-week-old tomato plants and 6-week-old N. tabacum plants were used for disease and HR assays, respectively.
Selection of Tn::pat-1Cm Mutant
To select a mutant with a transposon insertion in the pat-1Cm gene, we screened approximately 1,400 C. michiganensis strain LMG7333 mutants generated with a transposon, Tn1409Cβ, in the vector pKGT452Cβ (Kirchner et al., 2001; Hwang et al., 2019) by virulence assay in tomato and by PCR using specific primer set (Hwang et al., 2019). The transposon insertion site was determined by whole genome sequencing of the mutant using Illumina sequencing and genome comparison with the genome sequence of the WT C. michiganensis strain LMG7333 (GenBank accession nos. CP080437, CP080438, and CP080439). Furthermore, the single transposon insertion was confirmed by southern hybridization using the chloramphenicol resistance gene as a probe.
Plasmid Curing in Clavibacter michiganensis LMG7333
To remove the large plasmid (pCM2) carrying the pat-1Cm gene, the WT C. michiganensis strain LMG7333 was incubated at 26°C for 2 days and moved to 37–42°C for 3 days. This temperature change was repeated several times in a fresh medium, and then the bacterial culture was spread on KB plates. To check the presence of pCM2, colonies grown on KB plate were screened by PCR using primer sets targeting plasmid backbone genes and the pat-1Cm gene (Supplementary Table 1). The pCM2-cured strain, LMG7333ΔpCM2, was selected and used for this study.
Virulence and HR Assays in Plants
Two-week-old tomato seedlings were inoculated with C. michiganensis strains by the root-dipping inoculation method described by Hwang et al. (2019). Inoculums were prepared from freshly cultured bacteria, and their concentration was adjusted to OD600 = 2.0 [1 × 109 colony forming units (CFU) ml−1] with 10 mM MgCl2. Whole tomato seedlings were pulled out of the soil, and, if necessary, trimmed with sterile scissors. Then, the prepared seedlings were submerged into tubes containing 1 ml of bacterial inoculum for 30 min. As a negative control (mock), seedlings were inoculated with 10 mM MgCl2. Inoculated seedlings were transplanted into soil again in mini pots and grown in a growth chamber at 26°C. Approximately 10–14 days after inoculation (dai), the disease severity of above-ground disease symptoms of all seedlings was evaluated. Wilting symptom severity for each seedling was rated from 0 to 5 scales based on disease index defined by Hwang et al. (2019), which is as follows: 0, no visible symptoms; 1, one or two leaves mildly wilted; 2, more than two leaves mildly wilted, but less than two leaves severely wilted; 3, 25–50% of leaves severely wilted; 4, 50–75% of leaves severely wilted or dead; and 5, all leaves severely wilted or dead. All experiments were performed at least three times with 10 plants per treatment (n = 10). For stem inoculation, the stems of 3-week-old tomato plants were wounded above the cotyledons, the wounds were inoculated with 10 μl of C. michiganensis inoculum, and then the infected plants were transferred to a growth chamber for 3 weeks for canker development.
For the HR assay, leaves of 6-week-old N. tabacum, a nonhost plant, were used. Each C. michiganensis strain inoculum, adjusted to a concentration of OD600 = 0.05, was infiltrated by needless syringe into 6-week-old N. tabacum plant leaves. HR development in the infiltrated leaves was observed for 48 h.
Agrobacterium-Mediated Transient Expression Assay in Nicotiana tabacum Leaves
The ORFs of pat-1Cm, chpCCm, chpECm, chpFCm, and chpGCm genes without the stop codon were amplified from genomic DNA of C. michiganensis strain LMG7333 (Supplementary Table 1). The amplified DNA fragments were cloned into the pENTR/SD/D-TOPO vector (Invitrogen, CA, United States) according to the manufacturer’s instructions. After cloning, a gene fragment encoding a signal peptide of tobacco PR1b (GenBank accession no. X03465.1) was inserted into their N-terminal sites to generate in-frame fusion proteins and was verified by DNA sequencing. The resulting entry clones were recombined with the Gateway pGWB417 destination vector by LR reactions (Nakagawa et al., 2007). All cloned genes were also fused with c-Myc tag in their C-termini and expressed under the control of the 35S promoter. Recombinant plasmid constructs were transformed into Agrobacterium tumefaciens strain GV3101 for further assay.
For transient expression assay in N. tabacum leaves, A. tumefaciens strains with target genes were grown overnight in YEP medium (10 g of yeast extract, 1 0 g of peptone, and 5 g of NaCl per liter) with rifampicin (50 μg ml−1) and spectinomycin (50 μg ml−1) at 26°C. Agrobacterium cells were collected by centrifugation, washed with infiltration buffer (10 mM MES and 10 mM MgCl2), and resuspended in the same buffer. Bacterial suspension with 100 μM acetosyringone was incubated at 25°C for more than 3 h and centrifuged again. After adding the infiltration buffer with 100 μM acetosyringone, the bacterial suspension was adjusted to an OD600 of 0.4. Leaves of 6-week-old N. tabacum plants were infiltrated with an Agrobacterium suspension using a needleless syringe, and the plants were kept in the light to dry the leaves and subsequently incubated at 25°C.
Ion Conductivity Measurement
To measure electrolyte leakage to quantify the degree of HR development, a total of six leaf disks (9.2 mm diameter) were harvested from infiltrated leaves at 0 and 24 hai. Detached leaf disks were washed with 20 ml of deionized water for 30 min and then placed in a new tube containing 20 ml of deionized water and shaken at 160 rpm for 2 h. Ion conductivity was measured at the indicated time points with a conductometer (CON6 portable conductivity meter; Oakton, IL, United States).
Generation of pat-1 Gene Constructs
The pat-1Cm gene and its variants, including an SP-deleted mutant and six alanine-substituted mutants with 0.4-kb native promoter region and a FLAG tag on their C-termini were amplified from C. michiganensis strain LMG7333 by PCR and cloned into the pTOP blunt V2 vector (Enzynomics, Daejeon, Korea), named pTOP-pat-1Cm. Moreover, to generate Tn::pat-1Cm strains in which pat-1Cm orthologous genes present in C. capsici and C. sepedonicus were expressed under the native pat-1Cm gene promoter region, pat-1Cc was amplified from C. capsici strain PF008 by PCR. The pat-1Cs and the paralogous chp-7Cs gene from C. sepedonicus were obtained by custom gene synthesis with the promoter region of the pat-1Cm gene (Bioneer, Daejeon, Korea). The DNA fragments of all pat-1 genes digested by SpeI/HindIII restriction enzymes were ligated into the linearized pK2-22 vector with the same enzymes.
To generate Pat-1Cm variants with alanine substitutions at specific amino acid residues, amino acid substitutions of pat-1Cm were performed by site-directed mutagenesis. Using the construct pTOP-pat-1Cm as a template, PCR was performed with two mutagenic primers and then treated with DpnI to digest the methylated template. The final product was transformed into Escherichia coli strain DH5α and then confirmed by PCR and DNA sequencing. These variant genes were cloned into the pK2-22 vector.
Bacterial complementation using the pK2-22 vector was performed as previously described (Hwang et al., 2018). Briefly, the mutant Tn::pat-1Cm was transformed with each gene construct, in which a FLAG tag was fused at the C-terminus, into the pK2-22 vector. After transformation by electroporation with up to 4–5 μg of plasmid DNA, the transformants were grown over 3 days at 26°C, and then the single transformed bacterial colony was selected with neomycin (50 μg ml−1) as a selectable marker.
Western Blotting and Proteomics Analysis
Bacterial strains containing C-terminal FLAG-tagged gene constructs were grown in half-strength KB media with 0.4% CMC. Incubated bacterial cells were harvested by centrifugation, and its supernatant was precipitated with 10% w/v of chilled trichloroacetic acid (TCA)/acetone at a ratio of 4:1 after filtration with a 0.22 μm pore size sterile filter. Proteins collected in the supernatant by centrifugation were washed twice with ice-cold acetone and resuspended in 0.1 ml distilled water. Proteins from the cell pellets were lysed at 25°C for 20 min using B-PER™ Bacterial Protein Extraction Reagent (Thermo Scientific, Rockford, IL, United States) supplemented with 100 μg ml−1 lysozyme. After centrifugation, the supernatant extracted from the cell pellet was used for western blot analysis.
For total protein extraction from N. tabacum leaves, inoculated leaves were harvested at 36 hai and ground into a fine powder in liquid nitrogen. Samples were homogenized in protein extraction buffer [10% glycerol, 150 mM Tris–HCl (pH 7.5), 1 mM EDTA, 150 mM NaCl, 5 mM dithiothreitol, protease inhibitor cocktail (Sigma, St. Louis, MO, United States), and 0.2% Triton X-100 (Sigma)]. The homogenates were centrifuged at 12,000 g for 20 min at 4°C, and supernatants were collected for further study.
For western blot analysis, protein samples in sodium dodecyl sulfate (SDS) sample buffer [100 ml of 1.5 M Tris (pH 6.8), 60 ml of 20% SDS, 300 ml of glycerol, 150 ml of β-mercaptoethanol, and 18 mg of bromophenol blue per liter] were denatured by boiling and separated in a 12% SDS-polyacrylamide gel at 100 V for 2–3 h. After electrophoresis, separated proteins were transferred onto a polyvinylidene difluoride membrane (Millipore, Burlington, MA, United States). Subsequently, blots were blocked for 1 h with 5% w/v nonfat milk in phosphate-buffered saline (PBS) with 0.1% w/v Tween 20 and immunoblotted with horseradish peroxidase-conjugated anti-FLAG antibody (Sigma-Aldrich, St. Louis, MO, United States) or anti-c-Myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA, United States), followed by anti-mouse IgG-HRP secondary antibody (Santa Cruz Biotechnology). Proteins were detected by the HRP activity using enhanced chemiluminescence plus western blotting detection reagent (GE Healthcare, Little Chalfont, United Kingdom).
For 2D gel electrophoresis, proteins were precipitated from cell-free supernatant of C. michiganensis type strain LMG7333 by 10% TCA-acetone precipitation. Isoelectric focusing was performed using 24 cm immobilized pH gradient strips with a non-linear pH 4–10 gradient. 2D gel electrophoresis and MALDI-TOF MS analysis were conducted at GenoMine (Pohang, Gyeongbuk, Korea).
Protein Structure Prediction
A tertiary structure of the Pat-1Cm protein was predicted using phyre21 based on the principles of homology-based modeling (Kelley et al., 2015).
Purification of Mature Pat-1Cm
To obtain a large amount of Pat-1Cm proteins tagged with C-terminal FLAG, C. michiganensis Tn::pat-1Cm expressing pat-1cm or its variants was cultured in 1/5 strength KB media with 0.2% CMC for 2 days at 26°C with constant shaking. The large-scale cultures (500 ml) were centrifuged to collect the supernatant, and then the supernatant was filtered through a 0.22 μm sterile filter (Sartorius Stedim Biotech GmbH, Göttingen, Germany). To immune-precipitate Pat-1Cm protein, 300 μl of washed Pierce Anti-DYKDDDDK Magnetic Agarose (A36797; Thermo Scientific) was added to 50 ml of filtered supernatant and incubated with rotation for 1 h. The tube with the anti-FLAG magnetic beads/protein was placed on a magnetic stand, and the supernatant was removed. This procedure was repeated several times to collect enough protein for the experiment. After removing the supernatant, bound beads were washed twice with PBS (pH 7.4) and once with distilled water. Washed beads were eluted with Pierce IgG Elution buffer (pH 2.8, 21004; Thermo Scientific) or SDS-PAGE sample buffer.
N-Terminal Sequencing of Mature and Secreted Pat-1Cm
To determine the N-terminal amino acids of mature and secreted Pat-1Cm, purified proteins extracted from cell-free supernatant were separated by 1D SDS-PAGE and blotted onto PVDF (Immobilon-P membrane, Merck Millipore, MA, United States). After electro-blotting, the membrane was stained with Coomassie Blue R-250 for 30 min, de-stained multiple times with de-staining solution (10% acetone and 45% methanol–water per liter), and then rinsed with distilled water to remove the high concentration of other buffers, including transfer buffers. This membrane was dried at 25°C, and the targeted protein bands were excised with a blade. For N-terminal sequencing, the cut membrane was subjected to automated Edman degradation using a Procise 492 Protein Sequencing System (Applied Biosystems, CA, United States) at PROTEINWORKS (Daejeon, Korea).
Statistical Analysis
Statistical analysis of disease severity data was done by applying a non-parametric Kruskal-Wallis test with Dunnett’s multiple comparisons (p < 0.01) using the software statistiXL version 2.0 (statistiXL, Broadway, Australia). Duncan’s multiple range test was performed to analyze other results for comparisons between independent groups (p < 0.05).
Results
Clavibacter michiganensis pat-1Cm Gene Is Critical for the Development of Wilting Symptoms, but Not Canker Symptoms, in Tomato
Previously, the curing of plasmid pCM2 and complementation analysis with the pat-1Cm gene in pCM2 showed that the pat-1Cm gene is critical for wilting caused by C. michiganensis strain NCPPB382 in tomato (Meletzus et al., 1993; Dreier et al., 1997). To further study the role of the pat-1Cm gene as a pathogenicity factor, we screened a C. michiganensis type strain LMG7333 mutant library generated by transposon (Tn1409Cβ) insertion (Hwang et al., 2019) and selected the LMG733-Tn::pat-1Cm mutant strain (hereafter Tn::pat-1Cm strain; Supplementary Figure 1). The transposon’s position was determined by whole genome sequencing using Illumina MiSeq. The transposon was located at a distance of 542 bp from the translation start site (ATG) of pat-1Cm (Supplementary Figure 1A), and a single insertion was confirmed by Southern blot analysis (Supplementary Figure 1B). Simultaneously, we generated the pCM2-curred strain (7333ΔpCM2) by plasmid curing. Transposon insertion and pCM2 absence were confirmed by PCR with primer pairs targeting pat-1Cm and the plasmid backbone gene within pCM2 (Supplementary Figure 1C).
We next determined the pathogenicity of the Tn::pat-1Cm strain and LMG7333ΔpCM2 for wilting symptoms in 2-week-old tomato plants via the root-dipping inoculation method and for bacterial canker symptom in 3-week-old tomato plants via the stem-inoculation method. The wild-type (WT) LMG7333 strain caused severe wilting that led to the death of inoculated plants (Supplementary Figure 1D). However, tomato plants inoculated with either Tn::pat-1Cm strain or 7333ΔpCM2 strain exhibited only very mild or no wilting symptoms (Supplementary Figure 1D), although overall plant growth was slightly reduced after infection with both mutant strains. Interestingly, the C. michiganensis WT and its two mutants caused a similar degree of canker symptoms around the inoculation sites (Supplementary Figure 1E). When the stems of the WT-inoculated plants showing canker symptoms were cut lengthwise and examined, the brown discoloration in the vascular tissues and the tissue collapse typical of severe wilting symptoms were observed (Supplementary Figure 2). In contrast, the decrease in discoloration of vascular tissues was only observed in the absence of tissue collapse and of wilting symptoms in plants inoculated with the Tn::pat-1Cm strain (Supplementary Figure 2).
The mutant strains were complemented by transformation with the intact pat-1Cm gene controlled by its native promoter and fused with a FLAG tag at its 3′-terminus, named Tn::pat-1Cm (pat-1Cm) and 7333ΔpCM2 (pat-1Cm). The ability to cause wilting in tomato as much as the WT was restored in both complemented strains (Figure 1; Supplementary Figure 3), indicating that the pat-1Cm gene is a critical factor for wilting in the host plant, tomato. Nevertheless, all strains, including mutants and the complementary strains, were consistently able to cause bacterial canker on plant stems (Supplementary Figures 1E, 2). Overall, these results indicate that pat-1Cm in the plasmid pCM2 is critical for C. michiganensis to cause wilting, but not canker, in tomato.
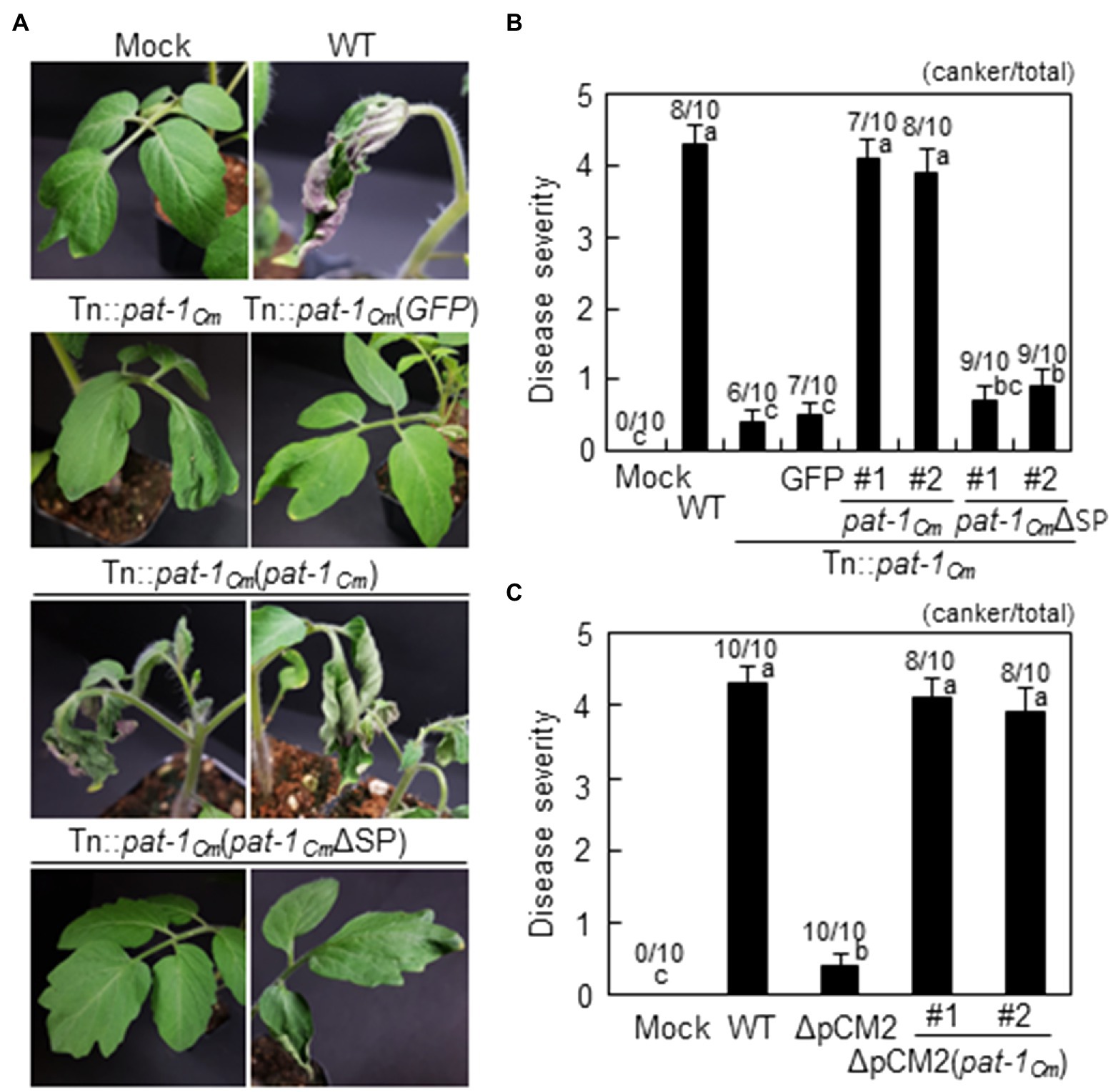
Figure 1. Pathogenicity recovery of Clavibacter michiganensis mutant Tn::pat-1Cm transformed with intact pat-1Cm gene, but not pat-1CmΔSP, in tomato. (A) Disease symptom development in infected tomato plants with indicated strains. Complemented strains was generated by overexpressing either full-length pat-1Cm or signal peptide (SP)-lacking pat-1CmΔSP. Two-week-old plants were used, and disease symptoms were photographed 14 days after inoculation (dai). (B,C) Disease severity of wilting in tomato plants inoculated with the indicated strains at 14 dai calculated based on disease index. Moreover, plants with bacterial canker were marked as the number of plants that displayed canker on the stem (canker/total). Error bars indicate SE (n = 10). Nonparametric Kruskal–Wallis test with Dunnett’s multiple comparisons (p < 0.05) was used to analyze the level of disease severity in tomato plants, and different letters indicate statistically significant differences at p < 0.05. C. Mock, 10 mM MgCl2; WT, C. michiganensis LMG7333 wild–type (WT).
The Pat-1Cm Protein Carries a Functional SP and Is Secreted
The pat-1Cm gene of the C. michiganensis type strain LMG7333 encodes a protein consisting of 280 amino acids that harbors a putative SP in the N-terminal region (Supplementary Figure 4A). Additional protein comparison revealed that Pat-1 proteins from various Clavibacter species contain a conserved protein sequence, including three amino acids for a catalytic triad of putative serine proteases and two cysteines. This holds true even with the Pat-1Cs protein from C. sepedonicus, which lacks an SP. Since Pat-1 was predicted to have an SP in its N-terminal region, we hypothesized that Pat-1Cm is a secreted protein with a functional SP. To ensure the importance of Pat-1Cm secretion for C. michiganensis pathogenicity, the first 33 amino acids, predicted as an SP, were removed, and the resulting truncated form, pat-1CmΔSP was tagged with FLAG at its C-terminus and transformed into the Tn::pat-1Cm strain. This complementary strain did not cause wilting in tomato (Figures 1A,B). Immunoblot with a polyclonal FLAG antibody detected the FLAG-tagged full-length of Pat-1Cm from both the supernatant and pellet after growth of both complemented strains in King’s B (KB) medium with 0.4% carboxymethyl cellulose (CMC) as a substrate (Figure 2A). However, the Pat-1CmΔSP in the Tn::pat-1Cm strain was not detected in either fraction. These results indicate that Pat-1Cm protein is expressed in the complemented strains and that the predicted SP is required for secretion and might be critical for protein stability.
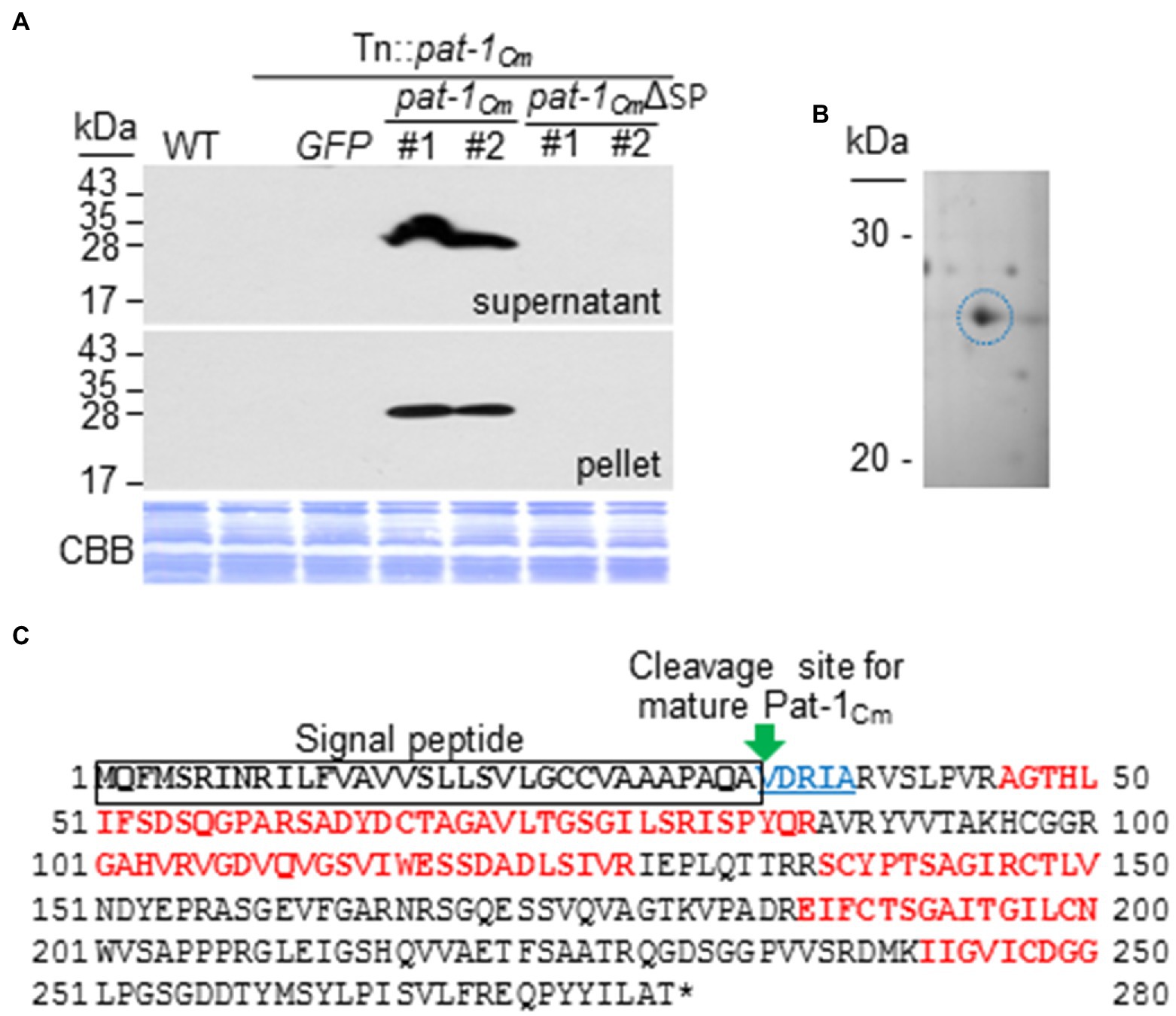
Figure 2. Pat-1Cm proteins are secreted and well-expressed in complementary strains. (A) Expression and secretion of Pat-1Cm proteins in complemented strains by western blotting. Pat-1Cm protein and its derivative were fused to FLAG on their C-termini and expressed in Tn::pat-1Cm strain. Total proteins from the supernatant and pellet were analyzed by immunoblotting using the FLAG antibody. CBB, Coomassie Brilliant Blue. Mock, 10 mM MgCl2; WT, Clavibacter michiganensis LMG7333 wild type. (B) Pat-1Cm proteins secreted in supernatant and separated by 2D gel. Pat-1Cm protein spot (blue circle) was analyzed by mass spectrometry (see Supplementary Figure 2C). (C) Identification of Pat-1Cm proteins by 2D-LC MS/MS. Red bolded letters indicate peptide sequences obtained from mass spectrometry. Black bolded letters indicate predicted SP sequence. Five blue bolded underlined letters indicate N-terminal sequences of mature Pat-1Cm obtained by Edman degradation.
To reconfirm that Pat-1Cm proteins are secreted, C. michiganensis strain LMG7333 was first grown in KB medium with 0.4% CMC and pelleted, and then the cell-free supernatant was collected. The total protein content of the cell-free supernatant was separated by 2D gel electrophoresis, and distinct protein spots were analyzed by MALDI TOF MS/MS. As a result, Pat-1Cm protein was detected (Figure 2A) and possessed a 100% match to five peptide sequences (Figure 2B).
To determine the SP cleavage site of this protein, we analyzed the N-terminal sequences of mature and secreted Pat-1Cm using Edman degradation. The first five amino acid residues (V-D-R-I-A) were obtained by N-terminal sequencing (Figure 2C; Supplementary Figure 5). These results indicate that Pat-1Cm is a genuine secreted protein with the functional SP consisting of the first 33 amino acids.
The pat-1Cm Gene Is Required for HR Induction in a Nonhost Plant
Previously, it was shown that Clavibacter species induced HR in nonhost plant species, and that the chp-7Cs gene, a homolog of pat-1Cm that encodes a putative serine protease in C. sepedonicus, is involved in HR induction in N. tabacum (Nissinen et al., 2009). To determine if Pat-1Cm plays a role in C. michiganensis LMG7333 HR induction in a nonhost plant, we performed the HR assay in N. tabacum (cv. Samsun) plants. Infiltration of WT C. michiganensis into N. tabacum leaves induced a typical HR within 24 h after infiltration (hai); however, an HR was not induced by the mutant strains, Tn::pat-1Cm (Figure 3A) and 7333ΔpCM2 (Supplementary Figure 6). Moreover, the HR-eliciting ability of both mutants was restored by complementation with an intact pat-1Cm gene. To quantify C. michiganensis-triggered HR, we measured electrolyte leakage during HR induction in tobacco leaves. Compared to the WT, electrolyte leakage after infiltration with the Tn::pat-1Cm strain was significantly and consistently reduced. Furthermore, complementing with the intact pat-1Cm restored electrolyte leakage in tobacco leaves to normal levels (Figure 3B). Our finding that the expression of pat-1Cm is directly associated with HR development in a nonhost plant suggests that the pat-1Cm gene in C. michiganensis is required not only for wilting development in a host plant, but also for HR induction in a nonhost plant.
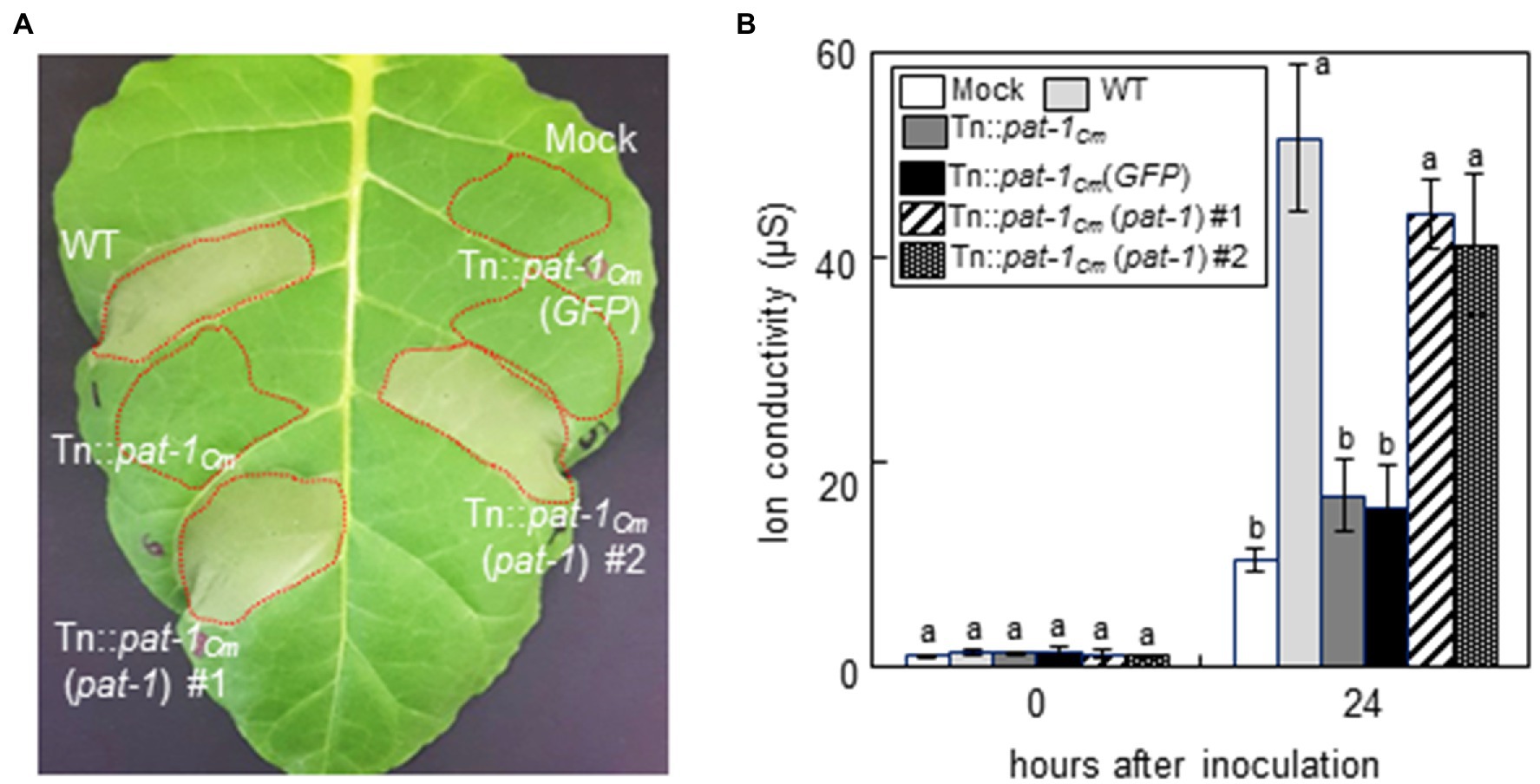
Figure 3. Loss of hypersensitive response (HR)-eliciting activity of Clavibacter michiganensis mutant Tn::pat-1Cm in a nonhost plant, Nicotiana tabacum. (A) HR phenotype in N. tabacum after infiltration with the indicated strains. Mature leaves of 5-week-old plants were infiltrated, and a representative leaf was photographed 36 h after infiltration (hai). Red dotted lines indicate infiltrated regions. (B) Ion conductivity in leaves infiltrated with the indicated strains. At 0 and 24 hai, leaf disks were excised to measure electrolyte leakage. Error bars indicate SD (n = 4). Different letters indicate statistically significant differences as determined by Duncan’s multiple range test (p < 0.05). Mock, 10 mM MgCl2; WT, C. michiganensis LMG7333 wild type.
The chp-7Cs Is Functionally Conserved With pat-1Cm in Pathogenicity
The pat-1Cm orthologs were found in several species, including C. michiganensis, C. capsici, and C. sepedonicus. Those genes, pat-1Cc, pat-1Cs, and chp-7Cs, are conserved in their plasmids with amino acid sequences that are approximately 77% identical to pat-1Cm (Supplementary Figure 4A). The chp-7Cs, which is a chromosomal putative serine protease of C. sepedonicus, was also classified as a pat-1Cm ortholog. A functional SP was predicted in Pat-1Cc and Chp-7Cs, but not in Pat-1Cs (Supplementary Figure 4A).
Subsequently, we determined if the pat-1Cm orthologs confer virulence of by complementing C. michiganensis Tn::pat-1Cm with each of pat-1Cc, pat-1Cs, and chp-7Cs genes under control of a 0.4-kb native promoter of pat-1Cm (Supplementary Figure 4B). The Tn::pat-1Cm strain with chp-7Cs caused wilting in tomato, but disease severity was significantly less than with the WT C. michiganensis (Figures 4A,B). The amount of expressed and secreted Chp-7Cs proteins in Tn::pat-1Cm was significantly lower than that of Pat-1Cm (Figure 4C). In contrast, the strains with either pat-1Cc or pat-1Cs failed to recover virulence activity (Figures 4A,B). When their expression and secretion levels were examined, Pat-1Cc was expressed and secreted, but less than Chp-7Cs, and was only detected after immunoprecipitation with FLAG affinity beads (Figure 4C). As expected, due to the lack of the N-terminal SP on Pat-1Cs, it was not detected. Next, the HR induction was examined with complemented strains. Complementation with pat-1Cm induced a strong HR similar to the WT, but none of the other three complemented strains could induce HR in N. tabacum leaves (Figure 4D). These results indicate that the virulence of only chp-7Cs is conserved with pat-1Cm and that the amount of secreted proteins might be directly correlated with disease severity.
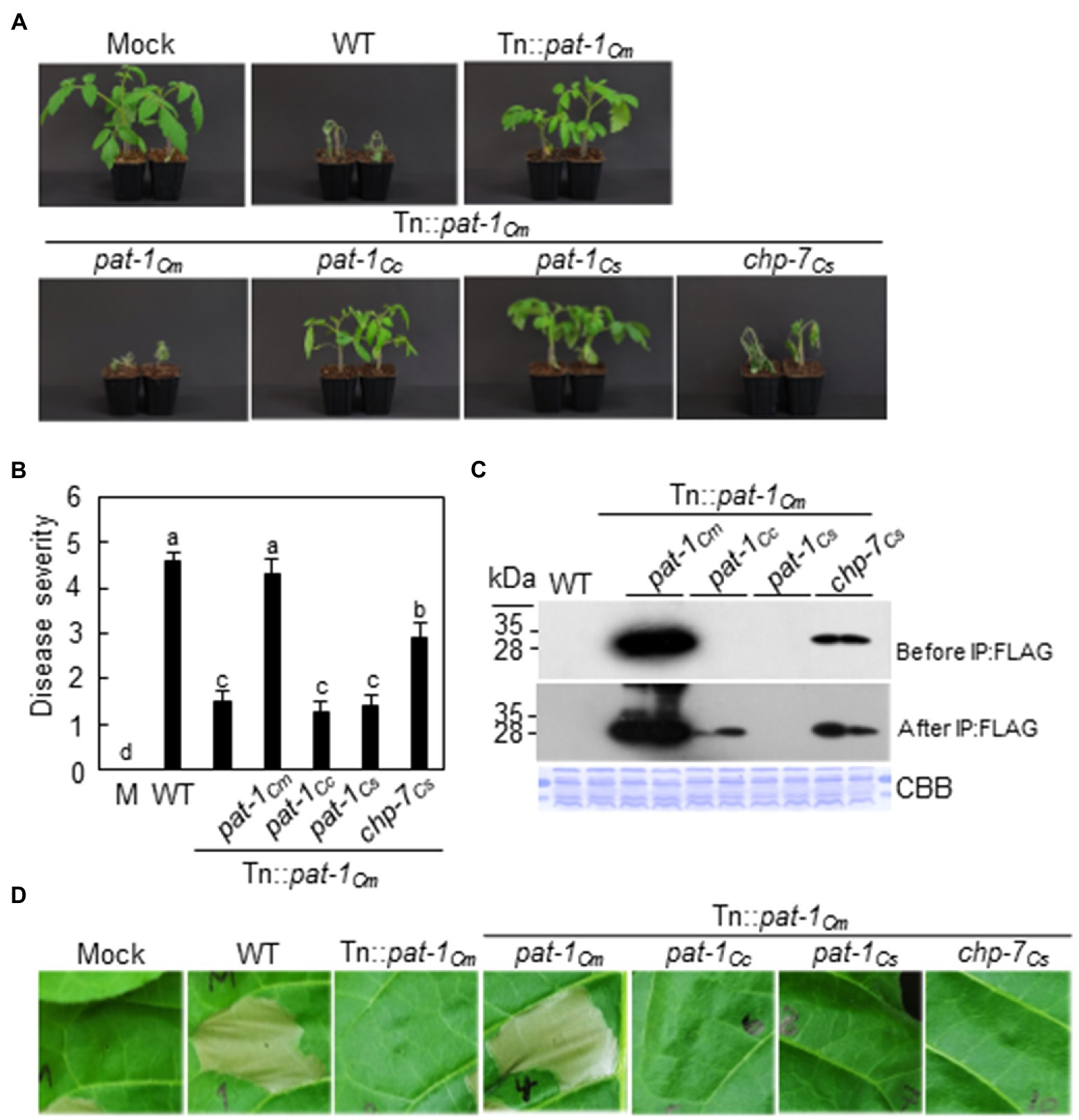
Figure 4. Partial recovery of Clavibacter michiganensis LMG7333 Tn::pat-1Cm mutant virulence by chp-7Cs gene of C. sepedonicus. (A) Wilting symptom development by inoculation of Tn::pat-1Cm strain carrying the pat-1Cm and its orthologs. Inoculated plants were observed for 2 weeks and photographed 14 dai. (B) Disease severity of wilting in tomato plants inoculated with the indicated strains at 14 dai. Error bars indicate SE (n = 10). Nonparametric Kruskal–Wallis test with Dunnett’s multiple comparisons (p < 0.05) was used to analyze the level of disease severity in tomato plants, and different letters indicate statistically significant differences at p < 0.05. (C) Expression and secretion of Pat-1Cm and its orthologs in the complemented strains. All proteins were fused to FLAG on their C-termini and expressed in Tn::pat-1Cm. Total proteins from supernatant before and after immunoprecipitation (IP:FLAG) with anti-FLAG-agarose beads were analyzed by immunoblot using anti-FLAG antibody. CBB, Coomassie Brilliant Blue. (D) HR phenotype in Nicotiana tabacum after infiltration with the indicated strains. Leaves of 5-week-old plants were inoculated, and representative leaves were photographed 36 h after infiltration. Red dotted lines indicate infiltrated regions. Mock, 10 mM MgCl2.
Roles of Pat-1Cm in Pathogenicity and HR Elicitation Are Distinct From Other Chp proteins
In the chromosome of C. michiganensis strain LMG7333, there are seven genes (chpACm to chpGCm) known as chromosomal homology to pat-1 (chp; Stork et al., 2008). Three (chpACm, chpBCm, and chpDCm) are pseudogenes containing internal stop codons, whereas the remaining four genes encode mature forms of putative serine proteases. These latter four proteins have a putative SP sequence and were functionally classified as a serine protease family. However, protein sequence identities between these four proteins and Pat-1Cm were lower than 40% (Supplementary Figures 7A,B). To determine if any of these chp genes could functionally complement pat-1Cm, full length (FL) chpCCm, chpECm, chpGCm, and chpFCm genes tagged with FLAG on their C-termini were transformed into both Tn::pat-1Cm and 7333ΔpCM2, and then strains’ ability to cause wilting in tomato and to elicit HR in N. tabacum was examined. Intriguingly, none of the four chp genes enabled either mutant to cause wilting in tomato (Figures 5A,B; Supplementary Figures 8A,B) or to elicit HR in N. tabacum (Figure 5C; Supplementary Figure 8C), even though they were expressed and secreted similar to Pat-1Cm proteins (Figure 5D). These results indicate that the manner in which Pat-1Cm interacts with plants is distinct from other homologous Chp proteases.
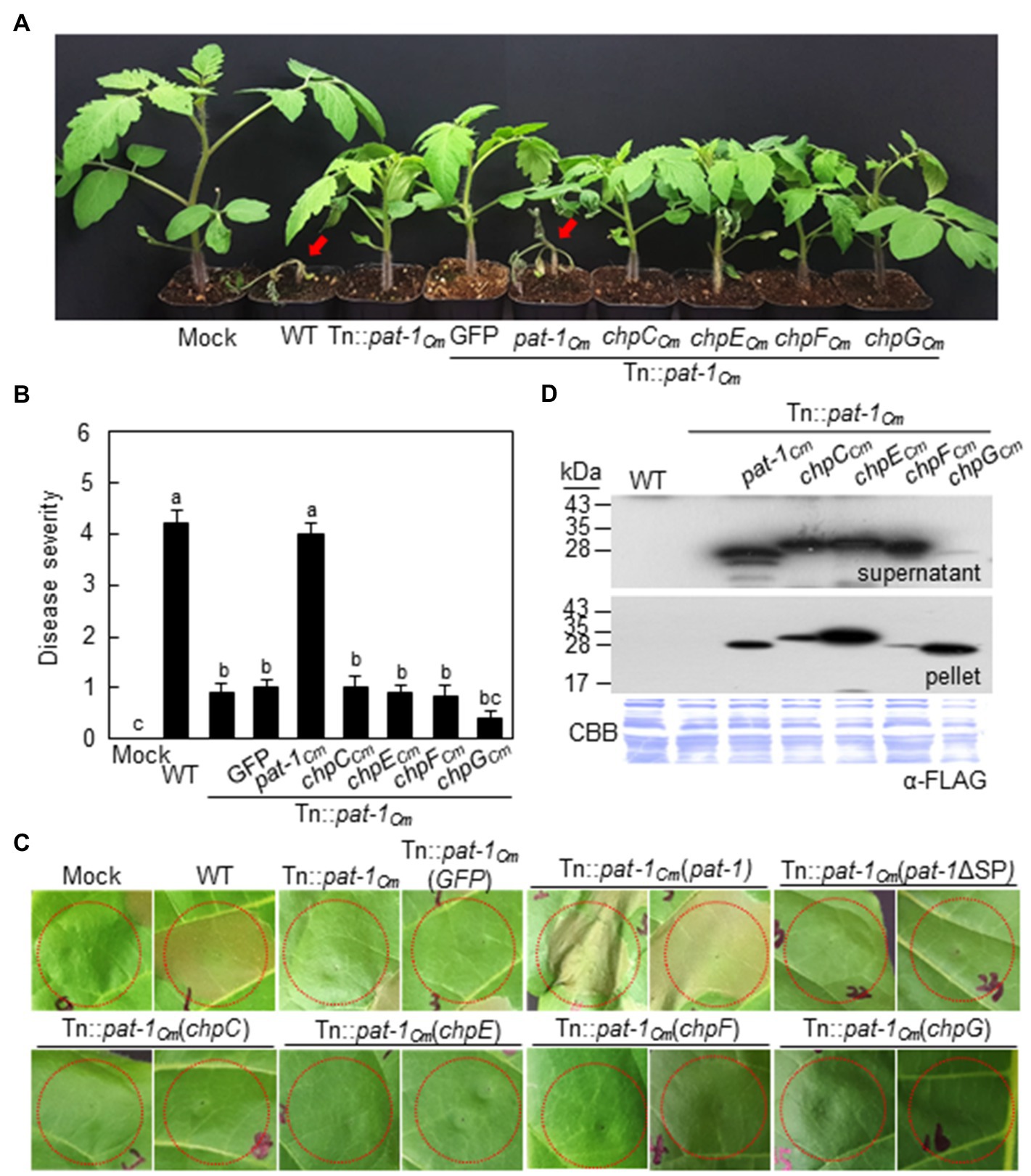
Figure 5. No recovery of either pathogenicity or HR-eliciting activity of Clavibacter michiganensis mutant Tn::pat-1Cm by chromosomal homologs of pat-1Cm (chp) genes. (A) Wilting symptom development after inoculation with Tn::pat-1Cm carrying chpCCm, chpECm, chpFCm, or chpGCm. Inoculated plants were observed for 2 weeks and photographed 14 dai. (B) Disease severity of wilting in tomato plants inoculated with the indicated strains at 14 dai. Error bars indicate SE (n = 10). Nonparametric Kruskal–Wallis test with Dunnett’s multiple comparisons (p < 0.05) was used to analyze the level of disease severity in tomato plants, and different letters indicate statistically significant differences at p < 0.05. (C) HR phenotype in Nicotiana tabacum after infiltration with the indicated strains. Leaves of 5-week-old plants were inoculated, and representative leaves were photographed at 36 h after infiltration. Red dotted lines indicate the infiltrated regions. (D) Expression and secretion of Pat-1Cm and Chp proteins in complemented strains. All proteins were fused to FLAG on their C-termini and expressed in Tn::pat-1Cm. Total proteins from the supernatant and pellet were analyzed by immunoblot using anti-FLAG antibody. CBB, Coomassie Brilliant Blue. Mock, 10 mM MgCl2; WT, C. michiganensis LMG7333 wild type.
The Conserved Catalytic Triad for Putative Serine Protease in Pat-1Cm Is Critical for Disease Development and HR Induction in Plants
Pat-1Cm is considered a putative serine protease due to its C-terminal conserved motif, Gly-ASP-Ser-Gly-Gly (GDSGG; Krem et al., 1999; Burger et al., 2005; Nissinen et al., 2009). Here, Pat-1Cm protein structures were predicted using the Phyre2 program to determine the key functional amino acid residues. Protein modeling showed that Pat-1Cm protein contains three residues in the following order histidine (His, H96), aspartate (Asp, D122), and serine (Ser, S231). This sequence is consistent with the catalytic triad of well-characterized serine proteases (Figure 6A; Supplementary Figure 9). The Ser/His/Asp catalytic triad is highly conserved in all Pat-1 proteins from Clavibacter species (Supplementary Figure 4A) and the Chp protein family in the chromosome of LMG7333 (Supplementary Figure 7A). In addition, protein modeling showed that the Pat-1Cm protein has two cysteine residues at positions 189 and 199 that are connected by disulfide bond to form a proper tertiary structure (Supplementary Figure 9). Similar to the catalytic triad, two cysteine residues are found in Pat-1 proteins and the Chp protein family except for ChpC (Supplementary Figures 4A, 7A).
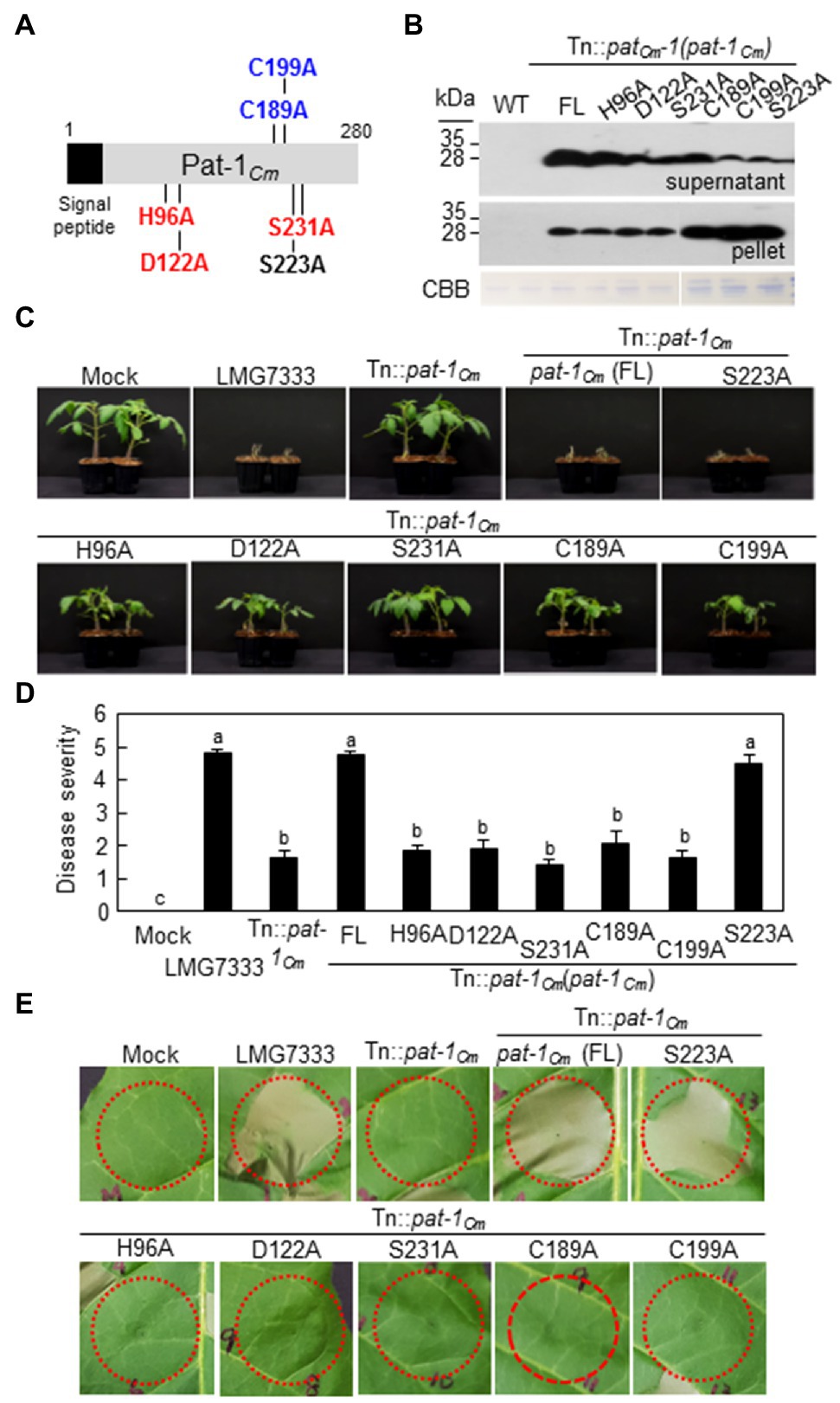
Figure 6. Key amino acids in the Pat-1Cm protein for both pathogenicity and HR-eliciting activities of Clavibacter michiganensis. (A) Schematic of amino acid substitutions in the Pat-1Cm protein. Variants with catalytic triad replacements (H96A, D122A, and S231A) and disulfide bond replacements (C189A and C199A) are shown in red and blue, respectively. Variant S222A was randomly selected as a control. (B) Expression and secretion of Pat-1Cm and its variants in the indicated strains. All proteins were fused to FLAG on their C-termini and expressed in Tn::pat-1Cm strains. Total proteins from supernatant and pellet were analyzed by immunoblot using anti-FLAG antibody. CBB, Coomassie Brilliant Blue. (C) Wilting symptom development in tomato plants inoculated with C. michiganensis Tn::pat-1Cm strain carrying diverse variants of the full length (FL) pat-1Cm gene. Two-week-old plants were inoculated and were photographed 14 dai. (D) Disease severity of wilting in tomato plants inoculated with the indicated strains 14 dai. Error bars indicate SE (n = 10). Nonparametric Kruskal–Wallis test with Dunnett’s multiple comparisons (p < 0.05) was used to analyze the level of disease severity in tomato plants, and different letters indicate statistically significant differences at p < 0.05. (E) HR phenotype in Nicotiana tabacum after infiltration with indicated strains. Five-week-old plant leaves were inoculated, and representative leaves were photographed 36 h after infiltration. Red dotted lines indicate infiltrated regions. Mock, 10 mM MgCl2; FL, full length pat-1Cm.
To investigate if the five residues in the catalytic triad (H96, D122, and S231) and the disulfide bond (C189, C199) play crucial roles in the function of Pat-1Cm, the individual residues were substituted with alanine by site-directed mutagenesis (Figure 6A) and introduced into the mutant strain, Tn:: pat-1Cm. Additionally, the serine residue at position 223 (Ser, S223) located in the un-conserved region was substituted with alanine as an internal control to rule out the possibility that alanine substitution itself affects the protein structure. All alanine substituted proteins were expressed and secreted similar to the WT Pat-1Cm, and the S223A substituent was expressed the least (Figure 6B). The Tn:: pat-1Cm mutant strain carrying all alanine substitutions except for at S223A failed to restore the mutant’s abilities to cause wilting in tomato (Figures 6C,D) or to elicit HR in N. tabacum (Figure 6E). These results indicate that the catalytic triad for putative serine protease and the specific disulfide bond for proper tertiary structure formation is essential for Pat-1Cm functioning in plants.
Ectopic Expression of Secreted Pat-1Cm Proteins Alone Elicits HR in Nicotiana tabacum Leaves
We showed that the mutant Tn::pat-1Cm lost its ability to elicit HR in a nonhost plant (Figure 3). To determine whether the Pat-1Cm protein alone elicits HR, the SP of tobacco PR1a was fused to the N-terminus of full-length Pat-1Cm to mimic Pat-1Cm secretion and translocation to the apoplast of N. tabacum leaves. In addition, a c-Myc tag was fused to the C-terminus of Pat-1Cm to check protein expression. The final construct, 35S-SP::Pat-1Cm under control of the 35S promoter, was transformed into Agrobacterium bacterium. Agrobacterium-mediated transient expression of Pat-1Cm in N. tabacum leaves, but not in a host plant tomato, elicited HR within 48 h, whereas the empty vector (EV) did not (Figure 7A; Supplementary Figure 10). Since the catalytic triad and disulfide bond are important for Pat-1Cm functioning, we generated two more constructs carrying Pat-1Cm–C189A and Pat-1Cm–S231A and expressed in N. tabacum leaves. These two variants failed to elicit HR, unlike the WT Pat-1Cm (Figure 7A), although they were expressed at the same level as the WT Pat-1Cm (Figure 7B). These results indicate that the Pat-1Cm protein alone, when located in plant apoplast, can elicit HR in a nonhost plant. In addition, the results suggest that both the catalytic triad for putative serine protease and a disulfide bond dependent structure in Pat-1Cm are required for HR induction in a nonhost plant.
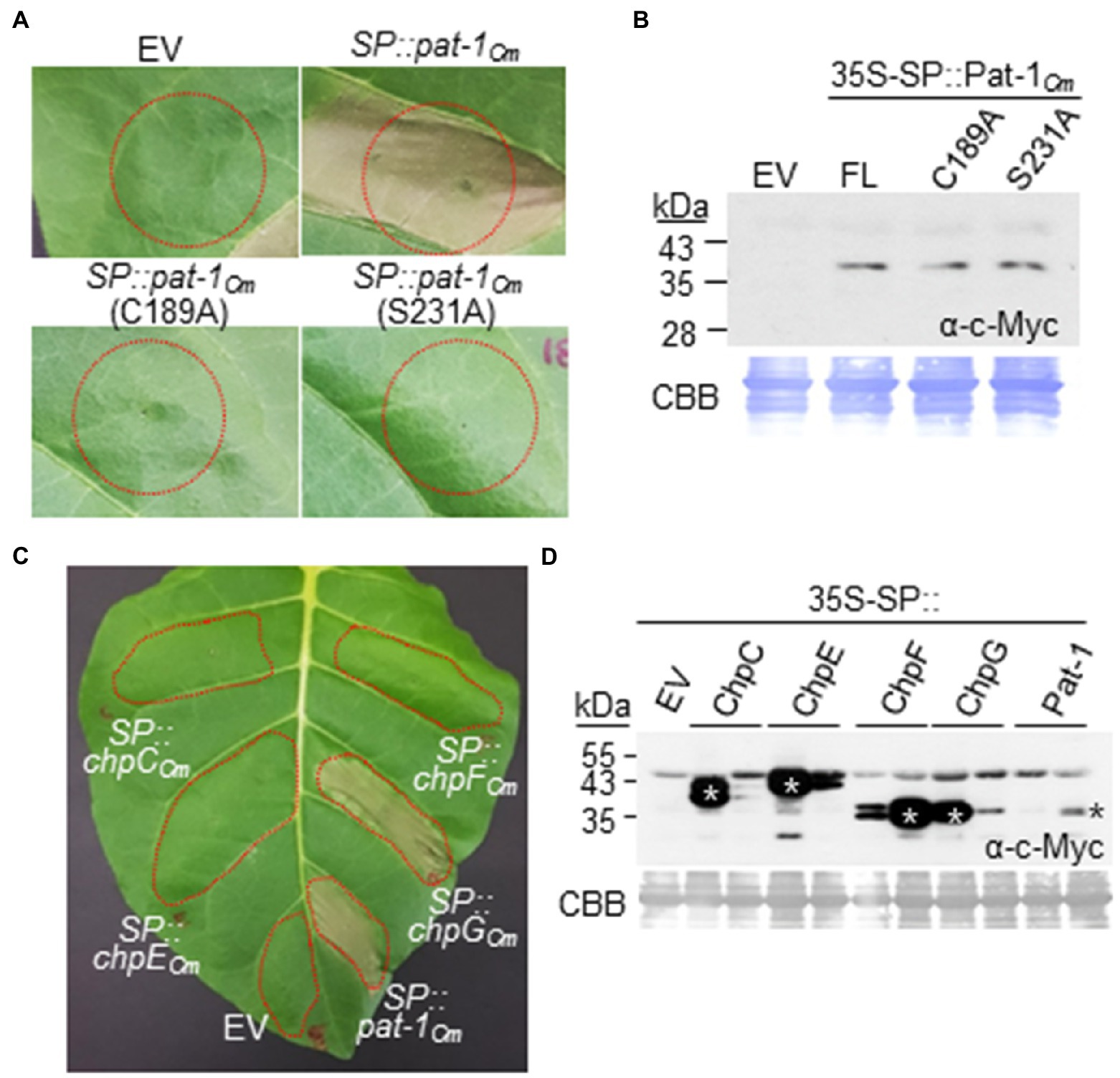
Figure 7. Hypersensitive response-eliciting activity of Pat-1Cm, its variants, and homologous Chp proteins by Agrobacterium-mediated transient expression in Nicotiana tabacum. Five-week-old N. tabacum leaves were infiltrated with Agrobacterium carrying each construct for constitutive expression of the indicated proteins with the SP of the tobacco PR1a protein. HR phenotypes in N. tabacum by expression of Pat-1Cm and its variants (A) and homologous Chp proteins (C) were shown. Representative leaf was photographed 36 h after infiltration (hai). Red dotted lines indicate infiltrated regions. Empty vector (EV) was used as a negative control. (B,D) Western blotting of transiently expressed proteins tagged with c-Myc in their C-termini using c-Myc antibody. Infiltrated leaves were sampled at the time point before HR was fully progressed, and total proteins were extracted. * in (D) indicates the right bands for each protein. CBB, Coomassie Brilliant Blue.
Although four intact Chp proteins in C. michiganensis, ChpCCm, ChpECm, ChpFCm, and ChpGCm, could not complement the Tn::pat-1Cm mutant to restore its HR-eliciting ability (Figure 5C), we checked whether any of these four Chp proteins alone could elicit HR when ectopically expressed on N. tabacum leaves. An HR was induced by expression of ChpGCm, but not ChpCCm, ChpECm, or ChpFCm (Figure 7C). All four proteins were expressed at higher levels than Pat-1Cm (Figure 7D). These results indicate that, although ChpGCm protein alone can elicit HR, it cannot substitute the ability of Pat-1Cm in C. michiganensis to elicit HR in N. tabacum.
Pat-1Cm Can Cooperate With CelA, a Secreted Cellulase, to Cause Wilting in Tomato
Previously, we reported that celA, another plasmid-borne pathogenicity gene in C. michiganensis that encodes a secreted cellulase, is critical for the development of wilting in tomato (Hwang et al., 2019). Because both Pat-1Cm and CelA encode secreted proteins with different enzymatic activities, we hypothesized that both proteins originated from different mutant bacteria present in the same site can functionally cooperate to cause wilting in tomato. To test this hypothesis, mutants Tn::celA and Tn::pat-1Cm were inoculated individually or co-inoculated on 2-week-old tomato plants by the root-dipping inoculation method. The individually inoculated plants did not show wilting, whereas the co-inoculated plants showed severe wilting, similar to WT-inoculated plants (Figures 8A,B). Moreover, cellulase activity similar to that of the WT strain was observed in Tn::pat-1Cm, whereas Tn::celA lost its ability to produce cellulase (Figure 8C). These results indicate that two different but important pathogenicity proteins, CelA and Pat-1Cm, function cooperatively to cause wilting in a host plant.

Figure 8. Functional cooperation of Pat-1Cm with another major pathogenicity factor, CelA (cellulase), during Clavibacter michiganensis infection of tomato plants. (A) Wilting symptom development in tomato plants inoculated with individual or mixed strains. Two-week-old plants were inoculated, and disease symptoms were photographed 14 dai. (B) Disease severity of wilting in tomato plants inoculated with mixed strains at 14 dai. Error bars indicate SE (n = 10). Nonparametric Kruskal–Wallis test with Dunnett’s multiple comparisons (p < 0.05) was used to analyze the level of disease severity in tomato plants, and different letters indicate statistically significant differences at p < 0.05. (C) Plate assay for detection of cellulase activity in Tn::celA and Tn::pat-1Cm mutant strains using carboxymethyl cellulose (CMC) agar plates strained with Congo Red. Mock, 10 mM MgCl2; WT, C. michiganensis LMG7333 wild type.
Discussion
In this study, we determined that C. michiganensis Pat-1Cm is a genuine secreted protein with a functional SP and acts as both a pathogenicity factor in a host plant tomato and an HR inducer in a nonhost plant N. tabacum. To our knowledge, Pat-1Cm and its close ortholog, Chp-7Cs, are only proteins to perform both activities in Gram-positive plant pathogenic bacteria, similar to the Hrp proteins in Gram-negative bacteria. Hrp proteins function mostly as structural proteins to form T3SS in a bacterial plasma membrane, whereas Pat-1Cm is secreted and very likely localizes to the apoplastic space to function. Based on genomic analyses, C. michiganensis does not have T3SS or T6SS, but probably has a T2SS and a TAT secretion system (Supplementary Table 2). Because the secreted mature form of Pat-1Cm does not have an SP, suggesting that the SP is cleaved during secretion, this protein might be secreted by either T2SS or TAT secretion system, although this inference remains to be confirmed. The SP appears to be crucial for the correct expression and secretion of Pat-1Cm because no Pat-1CmΔSP protein was detected in either the pellet or supernatant, when the protein was expressed in the Tn::pat-1Cm mutant, whereas the SP-cleaved form of mature Pat-1Cm from the WT was well detected.
Pat-1Cm was previously predicted to be a chymotypsin-like serine protease with a catalytic triad for enzymatic activity (Rawlings et al., 2018), although such activity had not been observed yet. We tried to detect the enzymatic activity of mature Pat-1Cm after purification and mixing with casein, a well-known protease substrate to evaluate if casein can be cleaved by Pat-1Cm, but not by Pat-1Cm (S231A). However, we failed to observe the cleavage of casein protein (data not shown). This might indicate that the target(s) of mature Pat-1Cm is specific and present only in host plants. Nevertheless, in this study, we showed that the catalytic triad of Pat-1Cm for predicted serine protease is required for the protein to act as both a pathogenicity factor and an immunity elicitor. This implies that there might be a different target(s) of Pat-1Cm in host and nonhost plants, if so, these targets need to be identified and characterized to fully understand the underlying mechanisms of Pat-1Cm.
Close orthologs of pat-1Cm have been reported in at least two more Clavibacter species, C. capsici (pat-1Cc) and C. sepedonicus (pat-1Cs and chp-7Cs), and are located on a large plasmid or a pathogenicity island in a chromosome (Dreier et al., 1997; Burger et al., 2005; Holtsmark et al., 2008). In addition, many chp family genes including chpC, chpE, chpG, and chpF that are less homologous to pat-1Cm than its close orthologs have been found in Clavibacter species (Eichenlaub and Gartemann, 2011; Hwang et al., 2020). When any of these orthologs or homologs were expressed in the C. michiganensis Tn::pat-1Cm mutant under the pat-1Cm promoter, none of them, except chp-7Cs, successfully replaced pat-1Cm’s pathogenicity function. Although C. michiganensis chpCCm has been shown to be involved in virulence (Stork et al., 2008), it failed to restore pat-1Cm pathogenicity. Based on SP prediction and protein modeling, all of those proteins have an intact SP and a catalytic triad, indicating that these orthologs and homologs are very likely secreted proteins and act as putative serine proteases similar to Pat-1Cm. Therefore, a lack of functional conservation among Pat-1Cm orthologs and homologs indicates that each protein might have a different target(s) in plants during infection.
Interestingly, Chp-7Cs and ChpGCm, which have been shown to trigger HR in N. tabacum leaves (Lu et al., 2015), failed to replace the HR-eliciting activity of Pat-1Cm. Although the amount of secreted Chp-7Cs proteins was significantly less than Pat-1Cm, it was sufficient for replacing Pat-1Cm pathogenicity, but not for HR induction. The reason that the Tn::pat-1Cm mutant carrying the chp-7Cs gene did not induce HR might be related to the amount of Chp-7Cs protein in the supernatant. Overall, Pat-1Cm function in plants appears distinct from its close orthologs and homologs. Functional redundancy among serine proteases participating in pathogenicity or virulence in Gram-negative bacteria has been reported, whereas, in Gram-positive bacteria, it is limited (Xia, 2004; Figaj et al., 2019). Instead, these proteins might have evolved in concert with functional diversifications in Clavibacter species (Sacristan and Garcia-Arenal, 2008; Plissonneau et al., 2017). In the future, identifying the protein(s) targeted by Pat-1Cm in host and nonhost plants will help to clarify the molecular mechanisms of Pat-1Cm functioning beyond pathogenicity.
The Pat-1Cm catalytic triad was necessary for pathogenicity and HR induction. Pat-1Cm alanine substituents were stably expressed and secreted, indicating that the catalytic triad is not necessary for protein stability, but critical for function. This means that the protein’s activity as a serine protease might be critical for pathogenicity and HR induction. Protease families containing a catalytic triad in plant pathogenic bacteria appear structurally similar to those found in human and animal pathogens, implying that these proteases also might play a role in pathogenicity or virulence depending on the proteolytic activity mediated by these residues (Figaj et al., 2019). Additionally, disulfide bond formation between pairs of cysteine residues is involved in bacterial virulence by contributing to protein stability (Reardon-Robinson and Ton-That, 2015; Smith et al., 2016). Dsb (disulfide bond formation) family proteins known to catalyze disulfide bonding are found in many plant-pathogenic bacteria, such as Xanthomonas campestris pv. campestris and Pseudomonas aeruginosa (Braun et al., 2001; Jiang et al., 2008). Disulfide bonding in many bacterial-secreted proteins is directly involved in the protein structural stability by encouraging proper folding (Heras et al., 2009). Two cysteines (C189 and C199) were predicted to form a disulfide bond, and alanine substituents disabled Pat-1Cm pathogenicity and HR induction. Because their protein expression and secretion appeared normal like the WT, this disulfide bond is likely important for maintaining this protein’s proper structure.
Co-inoculation of tomato with C. michiganensis Tn::celA and Tn::pat-1Cm mutants caused almost the same degree of wilting as observed after inoculation like the WT strain. celA encodes a secreted cellulase, which is important for wilting development in tomato (Hwang et al., 2019). After these two proteins are secreted during infection, it is highly likely that they move to the apoplast together and then function cooperatively regardless of their distinct origins. Thus far, the infection route of C. michiganensis is unknown. As shown previously (Hwang et al., 2019) and in this study, both C. michiganensis Tn::celA and Tn::pat-1Cm mutants possessed the ability to cause canker symptom, implying that these two genes are critical for wilting development. If this pathogen infects host plants through their roots, then it should pass several cell layers to reach xylem vessels for wilting development and systemic movement. Where CelA and Pat-1Cm are needed during this process should be dissected in detail.
Overall, we provide a unique apoplastic effector of Gram-positive pathogenic bacterium with dual functions for interaction with host and nonhost plants, and the serine protease activity might be required for both interactions. This is an example of novel roles of serine proteases in plant-pathogen interactions. Moreover, the finding that the functional cooperation of two apoplastic effectors, a serine protease (Pat-1Cm) and a cellulase (CelA), is critical for pathogenicity in a host plant tells us a dynamic functional relationship among apoplastic effectors in plant-pathogen interactions.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
IH, E-JO, ES, DC, and C-SO designed the study, performed the experiments, and drafted the manuscript. YL and KS performed the genome analysis. IH, E-JO, IP, YL, KS, DC, and C-SO substantially revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT; 2019R1A2C2004568 and 2018R1A5A1023599, SRC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.888290/full#supplementary-material
Footnotes
References
Bignell, D. R., Huguet-Tapia, J. C., Joshi, M. V., Pettis, G. S., and Loria, R. (2010). What does it take to be a plant pathogen: genomic insights from Streptomyces species. Antonie Van Leeuwenhoek 98, 179–194. doi: 10.1007/s10482-010-9429-1
Braun, P., Ockhuijsen, C., Eppens, E., Koster, M., Bitter, W., and Tommassen, J. (2001). Maturation of Pseudomonas aeruginosa elastase: formation of the disulfide bonds. J. Biol. Chem. 276, 26030–26035. doi: 10.1074/jbc.M007122200
Burger, A., Grafen, I., Engemann, J., Niermann, E., Pieper, M., Kirchner, O., et al. (2005). Identification of homologues to the pathogenicity factor Pat-1, a putative serine protease of Clavibacter michiganensis subsp. michiganensis. Microbiol. Res. 160, 417–427. doi: 10.1016/j.micres.2005.03.006
Costa, T. R., Felisberto-Rodrigues, C., Meir, A., Prevost, M. S., Redzej, A., Trokter, M., et al. (2015). Secretion systems in gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359. doi: 10.1038/nrmicro3456
Cunnac, S., Lindeberg, M., and Collmer, A. (2009). Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr. Opin. Microbiol. 12, 53–60. doi: 10.1016/j.mib.2008.12.003
Davidsson, P. R., Kariola, T., Niemi, O., and Palva, E. T. (2013). Pathogenicity of and plant immunity to soft rot pectobacteria. Front. Plant Sci. 4:191. doi: 10.3389/fpls.2013.00191
de Wit, P. J. (2016). Cladosporium fulvum effectors: weapons in the arms race with tomato. Annu. Rev. Phytopathol. 54, 1–23. doi: 10.1146/annurev-phyto-011516-040249
Dou, D., and Zhou, J. M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12, 484–495. doi: 10.1016/j.chom.2012.09.003
Dreier, J., Meletzus, D., and Eichenlaub, R. (1997). Characterization of the plasmid encoded virulence region pat-1 of phytopathogenic Clavibacter michiganensis subsp. michiganensis. Mol. Plant-Microbe Interact. 10, 195–206. doi: 10.1094/MPMI.1997.10.2.195
Eichenlaub, R., and Gartemann, K. H. (2011). The Clavibacter michiganensis subspecies: molecular investigation of gram-positive bacterial plant pathogens. Annu. Rev. Phytopathol. 49, 445–464. doi: 10.1146/annurev-phyto-072910-095258
Figaj, D., Ambroziak, P., Przepiora, T., and Skorko-Glonek, J. (2019). The role of proteases in the virulence of plant pathogenic bacteria. Int. J. Mol. Sci. 20:672. doi: 10.3390/ijms20030672
Francis, I. M., Stes, E., Zhang, Y., Rangel, D., Audenaert, K., and Vereecke, D. (2016). Mining the genome of Rhodococcus fascians, a plant growth-promoting bacterium gone astray. New Biotechnol. 33, 706–717. doi: 10.1016/j.nbt.2016.01.009
Galan, J. E., and Waksman, G. (2018). Protein-injection machines in bacteria. Cell 172, 1306–1318. doi: 10.1016/j.cell.2018.01.034
Gouran, H., Gillespie, H., Nascimento, R., Chakraborty, S., Zaini, P. A., Jacobson, A., et al. (2016). The secreted protease PrtA controls cell growth, biofilm formation and pathogenicity in Xylella fastidiosa. Sci. Rep. 6:31098. doi: 10.1038/srep31098
Heras, B., Shouldice, S. R., Totsika, M., Scanlon, M. J., Schembri, M. A., and Martin, J. L. (2009). DSB proteins and bacterial pathogenicity. Nat. Rev. Microbiol. 7, 215–225. doi: 10.1038/nrmicro2087
Holtsmark, I., Takle, G. W., and Brurberg, M. B. (2008). Expression of putative virulence factors in the potato pathogen Clavibacter michiganensis subsp. sepedonicus during infection. Arch. Microbiol. 189, 131–139. doi: 10.1007/s00203-007-0301-2
Hwang, I. S., Lee, H. M., Oh, E. J., Lee, S., Heu, S., and Oh, C. S. (2020). Plasmid composition and the chpG gene determine the virulence level of Clavibacter capsici natural isolates in pepper. Mol. Plant Pathol. 21, 808–819. doi: 10.1111/mpp.12932
Hwang, I. S., Oh, E. J., Kim, D., and Oh, C. S. (2018). Multiple plasmid-borne virulence genes of Clavibacter michiganensis ssp. capsici critical for disease development in pepper. New Phytol. 217, 1177–1189. doi: 10.1111/nph.14896
Hwang, I. S., Oh, E. J., Lee, H. B., and Oh, C. S. (2019). Functional characterization of two cellulase genes in the gram-positive pathogenic bacterium Clavibacter michiganensis for wilting in tomato. Mol. Plant-Microbe Interact. 32, 491–501. doi: 10.1094/MPMI-08-18-0227-R
Jiang, B. L., Liu, J., Chen, L. F., Ge, Y. Y., Hang, X. H., He, Y. Q., et al. (2008). DsbB is required for the pathogenesis process of Xanthomonas campestris pv. campestris. Mol. Plant-Microbe Interact. 21, 1036–1045. doi: 10.1094/MPMI-21-8-1036
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kirchner, O., Gartemann, K. H., Zellermann, E. M., Eichenlaub, R., and Burger, A. (2001). A highly efficient transposon mutagenesis system for the tomato pathogen Clavibacter michiganensis subsp. michiganensis. Mol. Plant-Microbe Interact. 14, 1312–1318. doi: 10.1094/MPMI.2001.14.11.1312
Krem, M. M., Rose, T., and Di Cera, E. (1999). The C-terminal sequence encodes function in serine proteases. J. Biol. Chem. 274, 28063–28066. doi: 10.1074/jbc.274.40.28063
Lee, H. A., Lee, H. Y., Seo, E., Lee, J., Kim, S. B., Oh, S., et al. (2017). Current understandings of plant nonhost resistance. Mol. Plant-Microbe Interact. 30, 5–15. doi: 10.1094/MPMI-10-16-0213-CR
Lindeberg, M., Cunnac, S., and Collmer, A. (2012). Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 20, 199–208. doi: 10.1016/j.tim.2012.01.003
Liu, J., Nothias, L. F., Dorrestein, P. C., Tahlan, K., and Bignell, D. R. D. (2021). Genomic and metabolomic analysis of the potato common scab pathogen Streptomyces scabiei. ACS Omega 6, 11474–11487. doi: 10.1021/acsomega.1c00526
Lu, Y., Hatsugai, N., Katagiri, F., Ishimaru, C. A., and Glazebrook, J. (2015). Putative serine protease effectors of Clavibacter michiganensis induce a hypersensitive response in the apoplast of Nicotiana species. Mol. Plant-Microbe Interact. 28, 1216–1226. doi: 10.1094/MPMI-02-15-0036-R
Meletzus, D., Bermphol, A., Dreier, J., and Eichenlaub, R. (1993). Evidence for plasmid-encoded virulence factors in the phytopathogenic bacterium Clavibacter michiganensis subsp. michiganensis NCPPB382. J. Bacteriol. 175, 2131–2136. doi: 10.1128/jb.175.7.2131-2136.1993
Mendez, V., Valenzuela, M., Salva-Serra, F., Jaen-Luchoro, D., Besoain, X., Moore, E. R. B., et al. (2020). Comparative genomics of pathogenic Clavibacter michiganensis subsp. michiganensis strains from Chile reveals potential virulence features for tomato plants. Microorganisms 8:1679. doi: 10.3390/microorganisms8111679
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. doi: 10.1263/jbb.104.34
Nissinen, R., Xia, Y., Mattinen, L., Ishimaru, C. A., Knudson, D. L., Knudson, S. E., et al. (2009). The putative secreted serine protease Chp-7 is required for full virulence and induction of a nonhost hypersensitive response by Clavibacter michiganensis subsp. sepedonicus. Mol. Plant-Microbe Interact. 22, 809–819. doi: 10.1094/MPMI-22-7-0809
Plissonneau, C., Benevenuto, J., Mohd-Assaad, N., Fouche, S., Hartmann, F. E., and Croll, D. (2017). Using population and comparative genomics to understand the genetic basis of effector-driven fungal pathogen evolution. Front. Plant Sci. 8:119. doi: 10.3389/fpls.2017.00119
Rawlings, N. D., Barrett, A. J., Thomas, P. D., Huang, X., Bateman, A., and Finn, R. D. (2018). The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46, D624–D632. doi: 10.1093/nar/gkx1134
Reardon-Robinson, M. E., and Ton-That, H. (2015). Disulfide-bond-forming pathways in gram-positive bacteria. J. Bacteriol. 198, 746–754. doi: 10.1128/JB.00769-15
Ruiz-Perez, F., and Nataro, J. P. (2014). Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cell. Mol. Life Sci. 71, 745–770. doi: 10.1007/s00018-013-1355-8
Sacristan, S., and Garcia-Arenal, F. (2008). The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9, 369–384. doi: 10.1111/j.1364-3703.2007.00460.x
Shao, F., Merritt, P. M., Bao, Z., Innes, R. W., and Dixon, J. E. (2002). A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109, 575–588. doi: 10.1016/s0092-8674(02)00766-3
Smith, R. P., Paxman, J. J., Scanlon, M. J., and Heras, B. (2016). Targeting bacterial Dsb proteins for the development of anti-virulence agents. Molecules 21:811. doi: 10.3390/molecules21070811
Stes, E., Francis, I., Pertry, I., Dolzblasz, A., Depuydt, S., and Vereecke, D. (2013). The leafy gall syndrome induced by Rhodococcus fascians. FEMS Microbiol. Lett. 342, 187–195. doi: 10.1111/1574-6968.12119
Stork, I., Gartemann, K. H., Burger, A., and Eichenlaub, R. (2008). A family of serine proteases of Clavibacter michiganensis subsp. michiganensis: chpC plays a role in colonization of the host plant tomato. Mol. Plant Pathol. 9, 599–608. doi: 10.1111/j.1364-3703.2008.00484.x
Thapa, S. P., Pattathil, S., Hahn, M. G., Jacques, M. A., Gilbertson, R. L., and Coaker, G. (2017). Genomic analysis of Clavibacter michiganensis reveals insight into virulence strategies and genetic diversity of a gram-positive bacterial pathogen. Mol. Plant-Microbe Interact. 30, 786–802. doi: 10.1094/MPMI-06-17-0146-R
Timilsina, S., Potnis, N., Newberry, E. A., Liyanapathiranage, P., Iruegas-Bocardo, F., White, F. F., et al. (2020). Xanthomonas diversity, virulence and plant-pathogen interactions. Nat. Rev. Microbiol. 18, 415–427. doi: 10.1038/s41579-020-0361-8
Toruno, T. Y., Stergiopoulos, I., and Coaker, G. (2016). Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54, 419–441. doi: 10.1146/annurev-phyto-080615-100204
van Wersch, S., Tian, L., Hoy, R., and Li, X. (2020). Plant NLRs: the whistleblowers of plant immunity. Plant Commun. 1:100016. doi: 10.1016/j.xplc.2019.100016
White, F. F., Potnis, N., Jones, J. B., and Koebnik, R. (2009). The type III effectors of Xanthomonas. Mol. Plant Pathol. 10, 749–766. doi: 10.1111/j.1364-3703.2009.00590.x
Keywords: bacterial canker, catalytic triad, Clavibacter michiganensis, pathogenicity, serine proteases
Citation: Hwang IS, Oh E-J, Song E, Park IW, Lee Y, Sohn KH, Choi D and Oh C-S (2022) An Apoplastic Effector Pat-1Cm of the Gram-Positive Bacterium Clavibacter michiganensis Acts as Both a Pathogenicity Factor and an Immunity Elicitor in Plants. Front. Plant Sci. 13:888290. doi: 10.3389/fpls.2022.888290
Edited by:
Youfu “Frank” Zhao, Washington State University, United StatesReviewed by:
Xiang Sean Li, Canadian Food Inspection Agency, CanadaXiaochen Yuan, Michigan State University, United States
Copyright © 2022 Hwang, Oh, Song, Park, Lee, Sohn, Choi and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang-Sik Oh, Y28zNUBraHUuYWMua3I=
 In Sun Hwang1
In Sun Hwang1 Kee Hoon Sohn
Kee Hoon Sohn Doil Choi
Doil Choi Chang-Sik Oh
Chang-Sik Oh