Corrigendum: Integrated analysis of transcriptome and metabolome reveals the mechanism of chlorine dioxide repressed potato (Solanum tuberosum L.) tuber sprouting
- 1Agricultural Product Storage and Processing Research Institute, Gansu Academy of Agricultural Sciences, Lanzhou, China
- 2College of Food Science and Engineering, Gansu Agricultural University, Lanzhou, China
- 3Gansu Innovation Center of Fruit and Vegetable Storage and Processing, Lanzhou, China
Sprouting is an irreversible deterioration of potato quality, which not only causes loss in their commercial value but also produces harmful toxins. As a popular disinfectant, ClO2 can inhibit the sprouting of potato tubers. Using transcriptomic and metabolomic approaches to understand the repressive mechanism of ClO2 in potato sprouting is yet to be reported. Sequencing the transcriptome and metabolome of potatoes treated with ClO2 in this study revealed a total of 3,119 differentially expressed genes, with 1,247 and 1,872 genes showing down- and upregulated expression, respectively. The majority of the downregulated genes were associated with plant hormone signal transduction, whereas upregulated differential genes were associated primarily with biological processes, such as phenylpropanoid biosynthesis and the mitogen-activated protein kinase (MAPK) signaling pathway. Metabonomic assays identified a total of 932 metabolites, with 33 and 52 metabolites being down- and upregulated, respectively. Downregulated metabolites were mostly alkaloids, amino acids, and their derivatives, whereas upregulated metabolites were composed mainly of flavonoids and coumarins. Integrated transcriptomic and metabolomic analyses showed that many different metabolites were regulated by several different genes, forming a complex regulatory network. These results provide new insights for understanding the mechanism of ClO2-mediated repression of potato sprouting.
Introduction
Potato is one of the most important food crops in the world, with a high nutritional value. However, its postharvest storage remains a challenging step to maintaining quality and minimizing market losses (Huang et al., 2014). During the potato storage in the fresh food market, or storage of tubers for seed harvesting and industrial processing, sudden sprouting occurs (Teper-Bamnolker et al., 2010). Tuber sprouting is controlled by several factors, such as plant hormones, genetic factors, signaling molecules, genotype, temperature, and other storage conditions (Wang et al., 2020). Sprouting is also determined by metabolic activities that regulate phytohormones to exert growth control (Sonnewald, 2001).
Potato sprouting causes undesirable tuber weight loss, decay, and overall quality decline in fresh food and seed tubers, leading to significant market losses (Kleinkopf et al., 2003). In addition, sprouting increases the accumulation of solanine in tubers, this might cause mild to acute poisoning when the content exceeds 200 mg/kg (Liu et al., 2020). Thus, the application of sprout inhibitors is necessary during the storage of potato tubers to avoid postharvest losses. Chlorpropham is registered as a commercial product that has been shown to have the best sprout-repressive effect on potato tubers (Corsini et al., 1978; Campbell et al., 2010). However, residual chlorpropham in the treated seed tubers inhibited field sprouting; thus, developing an effective potato sprouting suppressant is an urgent research target (Conte et al., 1995).
Chlorine dioxide (ClO2) has broad and high-biocidal activity because of its strong oxidizing and sanitizing properties (Aday and Caner, 2011). ClO2 was cleared by the Food and Drug Administration (FDA (Food Drug Administration)., 1998) as a safe antibacterial agent for cleaning and treating fruits and vegetables. Chlorine significantly reduces sprouting and effectively controls dry rot, soft rot, and silver scurf of the tubers (Tweddell et al., 2003). For example, cleaning fruit surfaces with ClO2 reduced air-dried Salmonella in tomato and Escherichia coli O157:H7 strain in apples (Du et al., 2003; Pao et al., 2009). In addition, ClO2 treatment maintained quality, reduced rot rate, and extended the shelf life of kiwifruit (Niu et al., 2009), mulberry (Chen et al., 2011), and litchi (Wu et al., 2011), and reduced pericarp browning in longan (Atinut et al., 2019).
Currently, most studies are focused on the use of ClO2 in the control of pathogenic microbes, maintenance of fruit quality, and extending the shelf life of fruit. However, no studies on the repressive mechanism of ClO2 in potato sprouting using transcriptome and metabolome approaches have been reported. This study analyzed the transcriptomic and metabolic profiles of ClO2-treated tubers to uncover the molecular mechanism of ClO2-mediated repression of potato sprouting.
Materials and Methods
Materials
The tubers of the “Atlantic” cultivar were obtained from Kaikai Potato Seed Co., Ltd. (Gansu, China). After discarding the tubers with injuries or infections, potatoes were transferred to the laboratory and stored at 20 ± 3°C until use. A formulation with 12% ClO2 as the active ingredient was purchased from Tianjin Zhangda Technology Development Co., Ltd. (Tianjin, China).
Treatment of Tubers and Sampling
After dormancy, tubers were cleaned and disinfected by immersion in 1% sodium hypochlorite for 3 min, rinsed with sterile water, and air-dried. Tubers were then soaked in 180 mg/L ClO2H3 for 15 min, and a control group (CKH3) was soaked in sterile water. After drying, all treated tubers were stored in the dark at 20 ± 2°C with a relative humidity of 75–85%. Experiments were carried out in three replicates per treatment, with each containing 30 tubers.
Methods
Sampling
Sampling of the ClO2H3-treated tubers with no sprouts was performed after detecting sprouting in the CKH3 treatments. About 5 g of tuber tissue was cut 1 cm deep under the skin at the bud growth site (bud eye), with a 1 cm stainless steel punch. Samples were wrapped in tin foil paper, frozen in liquid nitrogen, and stored at −80°C until use.
RNA Sequencing (RNA-Seq)
Total RNA extraction (three duplicate samples) was performed following the method described by Woolfson et al. (2018). cDNA libraries were developed in three duplicates as described by Foucart et al. (2006). Libraries were sequenced using the Illumina HiSeq 2500 platform (Jisi Huiyuan Biotechnology Co., Ltd., Nanjing, China), with 2 × 150 paired-end reads. The high-quality reads were assembled into unigenes following the methods by Haas et al. (2013). To count the assemblies, TopHat 2 software was used to map RNA sequence data to the Solanum tuberosum reference genome (DM_1-3_516_R44_potato.v6.1), downloaded from the Potato Genomics Resource (http://spuddb.uga.edu/dm_v6_1_download.shtml).
Functional Annotation
The unigenes were queried against the Kyoto Encyclopedia of Genes and Genomes (KEGG), Non-redundant (Nr), Eukaryotic Orthologous Groups of protein (KOG), Uniprot, and Gene Ontology (GO) public databases, with a threshold e-value of <10−5 to obtain the functional annotations. The KEGG pathway analyses and GO functional classifications were visualized using the Web Gene Ontology Annotation Plot (WEGO) and KEGG automatic annotation servers, respectively.
Metabolite Extraction
The frozen samples were ground in a mixer mill for 1 min at 45 Hz. Approximately, 100 mg of samples were then mixed with 1.5 mL methanol–water (3:1). The samples were homogenized and sonicated for 15 min at 0°C, extracted with a shaker for 24 h at 4°C, followed by centrifugation with 13,800 × g at 4°C for 15 min. The supernatant was filtered through a membrane, the filtrate was diluted twice with methanol–water (3:1), and then transferred to glass vials; 20 μl of this solution was pooled as QC sample for ultra-high-performance liquid chromatography-mass spectrometry (UHPLC-MS) analyses (Sawada et al., 2009).
UHPLC-MS Analyses
Ultra-high-performance liquid chromatography-mass spectrometry separation was performed using an EXIONLC System, with mobile phases A and B as formic acid (0.1%) and acetonitrile, respectively. The column and the auto-sampler temperatures were set at 40°C. A Sciex QTrap 6500+ (Sciex Technologies) was applied. Ion source parameters were set as follows: temperature, 400°C; curtain gas, 35 psi; ion source gas 2, 60 psi; DP, ±100 V; ion source gas, 1:60 psi; and ion spray voltage, +5,500/−4,500 V (Zhang et al., 2015). The UHPLC-MS measurements were carried out in five replicates per sample.
Data Pre-Processing, Annotation, and Screening of Differentially Accumulated Metabolites
The MRM data were acquired with the SCIEX Analyst Work Station Software (Version 1.6.3), then the raw (.wiff) MS data files were converted to.txt format using the MSconverter. MS peak detection and annotation were performed in the R program (Kuhl et al., 2012). Differentially accumulated metabolites (DAMs) obtained were screened with pairwise comparison using the Variable Importance in the Projection (VIP) with the first principal component of the OPLS-DA model as >1 and p < 0.05, Student's t-test, as the threshold.
qRT-PCR Analyses
qRT-PCR analyses were performed following the methods of Woolfson et al. (2018), with the same RNA and cDNA samples used in the RNA-seq analysis. The accession numbers of selected genes based on the Potato Genomics Resource assembly database and their specific primers are presented in Supplementary Table 1. The primers for the housekeeping gene, ef1, were designed according to Nicot et al. (2005). The RT-qPCR reaction system was prepared as follows: Ultra SYBR mixture, 10 μl; RNase-free water, 8.4 μl; template cDNA, 0.8 μl; and each primer, 0.4 μl. RT-qPCR was performed with the Roche LightCycler 480 (Roche, Bazel, Switzerland) thermocycler, with the program set as follows: initial 95°C for 10 min, followed by 40 reaction cycles of 95°C for 30 s, 72°C for 30 s, one final cycle of 72°C for 10 min, and one cycle of 40°C for 30 s. The relative gene expression was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Results
Quality Assessments of Sequencing Data and Differential Gene Expression Analyses
A total of 161.77 GB quality clean sequencing reads were obtained, with an average of 5.98 GB clean data per sample, and percentages Q20 and Q30 of bases above 97 and 93%, respectively, were observed. The guanine–cytosine (GC) content of each sample was above 42% after filtration (Supplementary Table 2). All clean reads were compared to the potato reference genome (DM_1-3_516_R44_potato.v6.1), and 89.25–89.59% or 89.25–90.10% of sequences from control or ClO2 treatments were mapped to the reference genome, which demonstrated the high reliability of our data (Supplementary Table 3).
To further investigate the gene expression patterns, differentially expressed genes (DEGs) were screened by pairwise comparison with Fold Change (FC) ≥2 and False Discovery Rate (FDR) <0.05 and Fragments per Kilobase of transcript per Million fragments mapped (FPKM) values were calculated for each sample to normalize the gene expression. Hierarchical clustering analysis was performed on the filtered differentially expressed genes, and the genes with the same or similar expression behaviors were clustered, and the results of the clustering of differentially expressed genes are shown in Figure 1A. A total of 3,119 DEGs were detected, with 1,247 and 1,872 as down- and upregulated genes, respectively, after ClO2 treatment (Figure 1B).
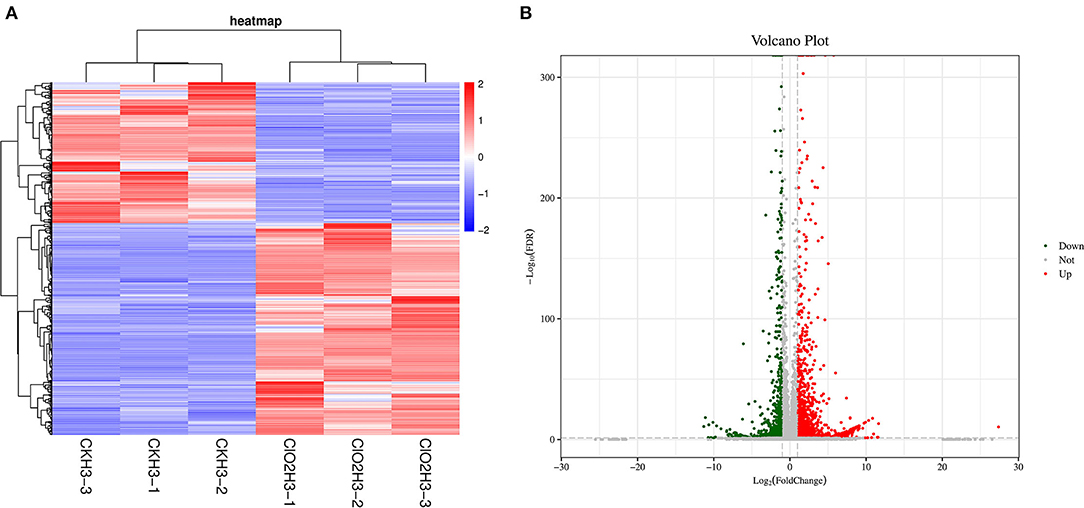
Figure 1. The cluster heat map of the differentially expressed genes (A) and volcanic map (B) in control and ClO2 treated tubers. Three biological repeats were shown in the figure. CKH3 represents the tubers of safflower treated with sterile water; ClO2H3 represents the flowers of safflower treated with ClO2.
GO Annotation and Enrichment
GO classification was used to define the functions of DEGs from the comparisons between control and ClO2. As a result, 43 functional DEG categories were identified, including 12 “cellular components”, 10 “molecular functions”, and 21 “biological processes” (Figure 2A). Most DEGs were classified into the cell (1,728) and cell part (1,724) in the cellular components, whereas 1,288, 1,278, 1,534, and 1,425 DEGs were classified in binding, catalytic activity, molecular functions and cellular processes, and metabolic processes, respectively (Figure 2B).
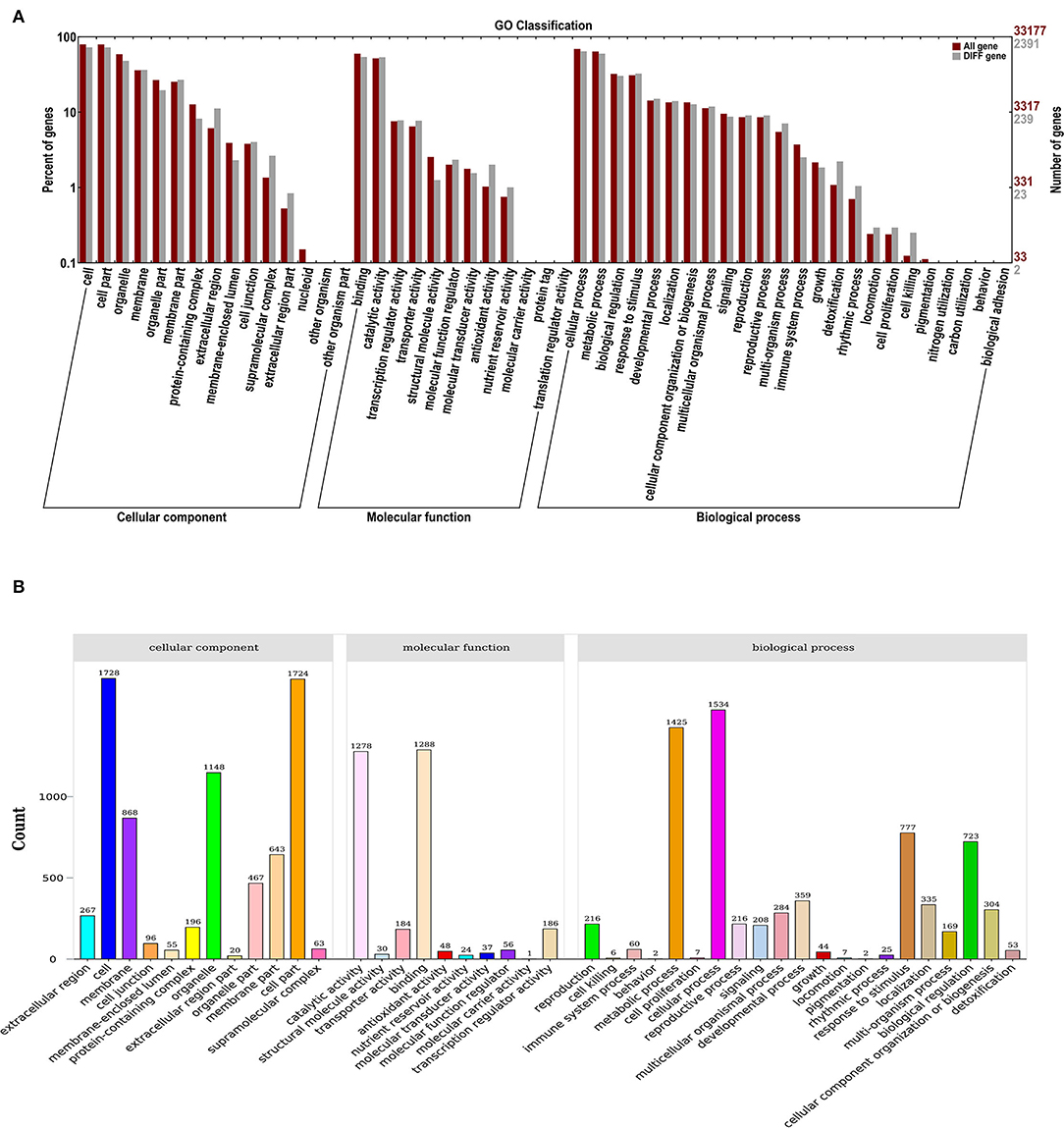
Figure 2. GO enrichment analysis (A) and statistical classification analysis (B) of differentially expressed genes.
KEGG Annotation and Enrichment
DEGs were screened in the KEGG ortholog database for functional enrichment analyses. As a result, the comparison between the control and ClO2-treated tubers assigned 19 pathways to different categories, including organismal systems, metabolism, environmental information processing, genetic information, cellular processes, and human diseases. In addition, carbohydrate metabolism, signal transduction, biosynthesis of other secondary metabolites, amino acid metabolism, and lipid metabolism were significantly enriched in DEGs (Figure 3A).
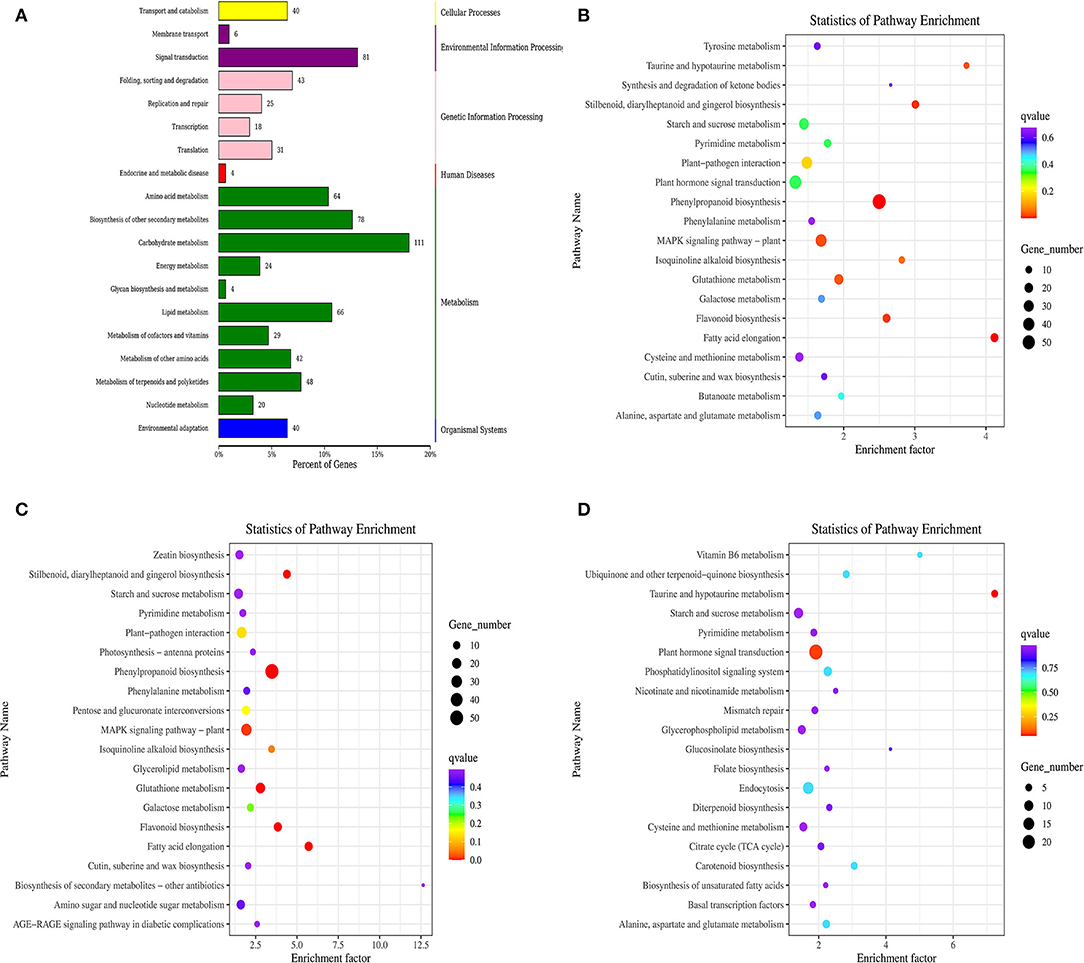
Figure 3. KEGG classification of differentially expressed genes (A) and KEGG pathway enrichment scatter of differentially expressed (B), up-regulated gene (C), and down-regulated gene (D).
Notably, abundant DEGs were involved in phenylpropanoid biosynthesis, MAPK signaling pathway-plant, plant hormone signal transduction, plant–pathogen interaction, starch and sucrose metabolism, and glutathione metabolism(Figure 3B). Interestingly, only the plant hormone signal transduction pathway was enriched with downregulated genes, whereas the other two pathways were enriched with upregulated genes (Figures 3C,D).
Analyses of DEG in Phenylpropanoid Biosynthesis and Plant Hormone Signal Transduction Pathways
Phenylpropanoid biosynthesis and plant hormone signal transduction were the most enriched pathways, with upregulated and downregulated DEGs, respectively. The three differential genes of phenylpropanoid biosynthesis were downregulated, whereas the others were upregulated. RT-qPCR analyses using eight genes associated with the phenylpropanoid pathway and 12 genes of plant hormone signal transduction were used to further confirm the RNA-seq results (Figure 4). As a result, consistent gene expression patterns were observed between qPCR and RNA-seq (Supplementary Figure 1).
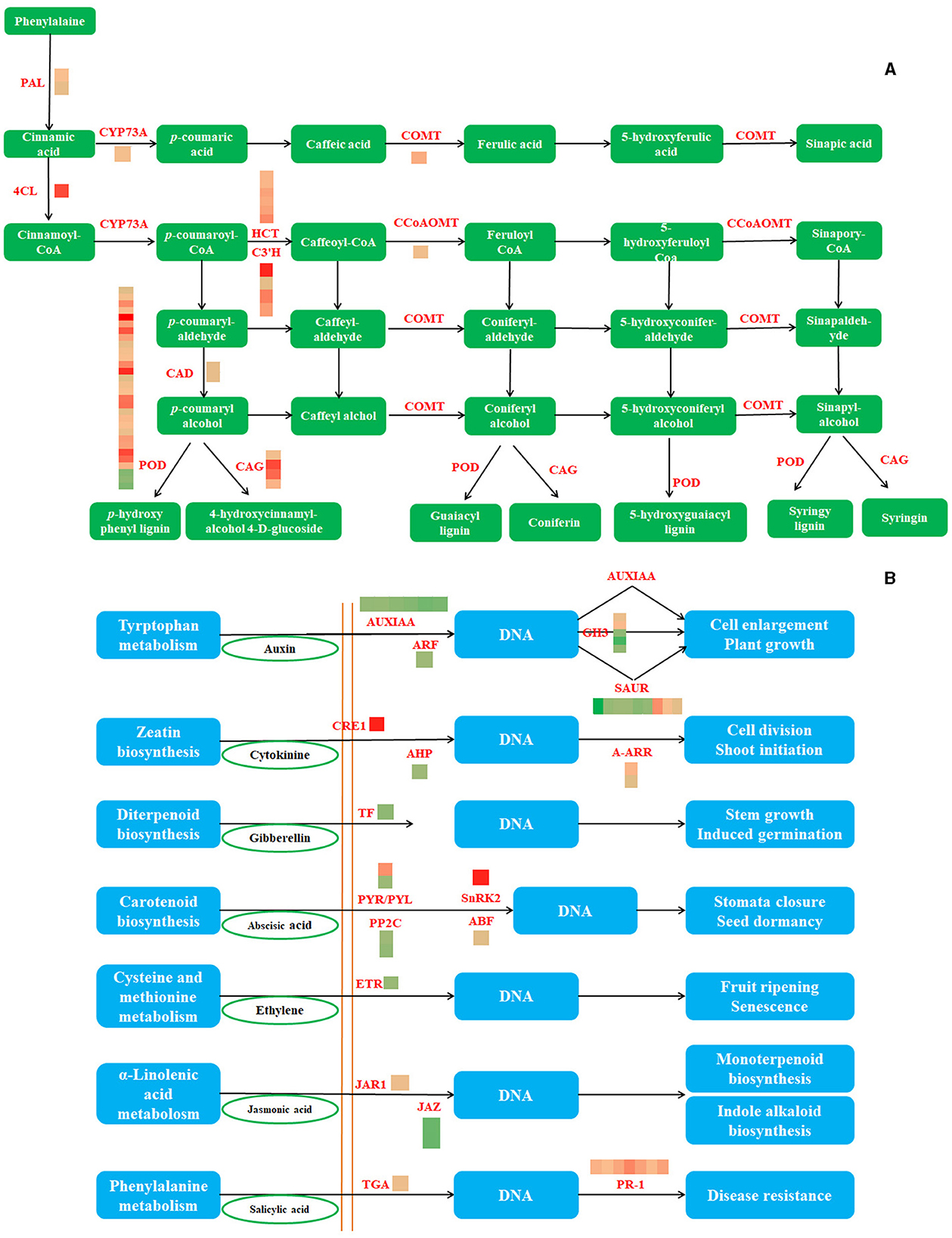
Figure 4. The phenylpropane biosynthesis pathway and the DEGs involved in this pathway (A) (PAL, phenylalanine ammonia-lyase; 4CL, 4-coumarate–CoA ligase; CYP73A, trans-cinnamate 4-monooxygenase; CAD, cinnamyl-alcohol dehydrogenase; COMT, caffeic acid 3-O-methyltransferase; POD, peroxidase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CAG, coniferyl-alcohol glucosyltransferase) and The plant hormone signal transduction and the DEGs involved in this pathway (B) (AUXIAA, auxin-responsive protein IAA; ARF, auxin response factor;GH3, auxin-responsive GH3 gene family; SAUR, SAUR family protein; CRE1, cytokinin receptor; AHP, histidine-containing phosphotransfer protein; A-ARR, two-component response regulator ARR-A family; TF, phytochrome-interacting factor 4; PYR/PYL, abscisic acid receptor PYR/PYL family; PP2C, protein phosphatase 2C; SnRK2, serine/threonine-protein kinase SRK2; ABF, ABA-responsive element binding factor; ETR, ethylene receptor; JAR1, jasmonic acid-amino synthetase; JAZ, jasmonate ZIM domain-containing protein; TGA, transcription factor TGA; PR-1, pathogenesis-related protein 1).
Metabolomic Data Analyses
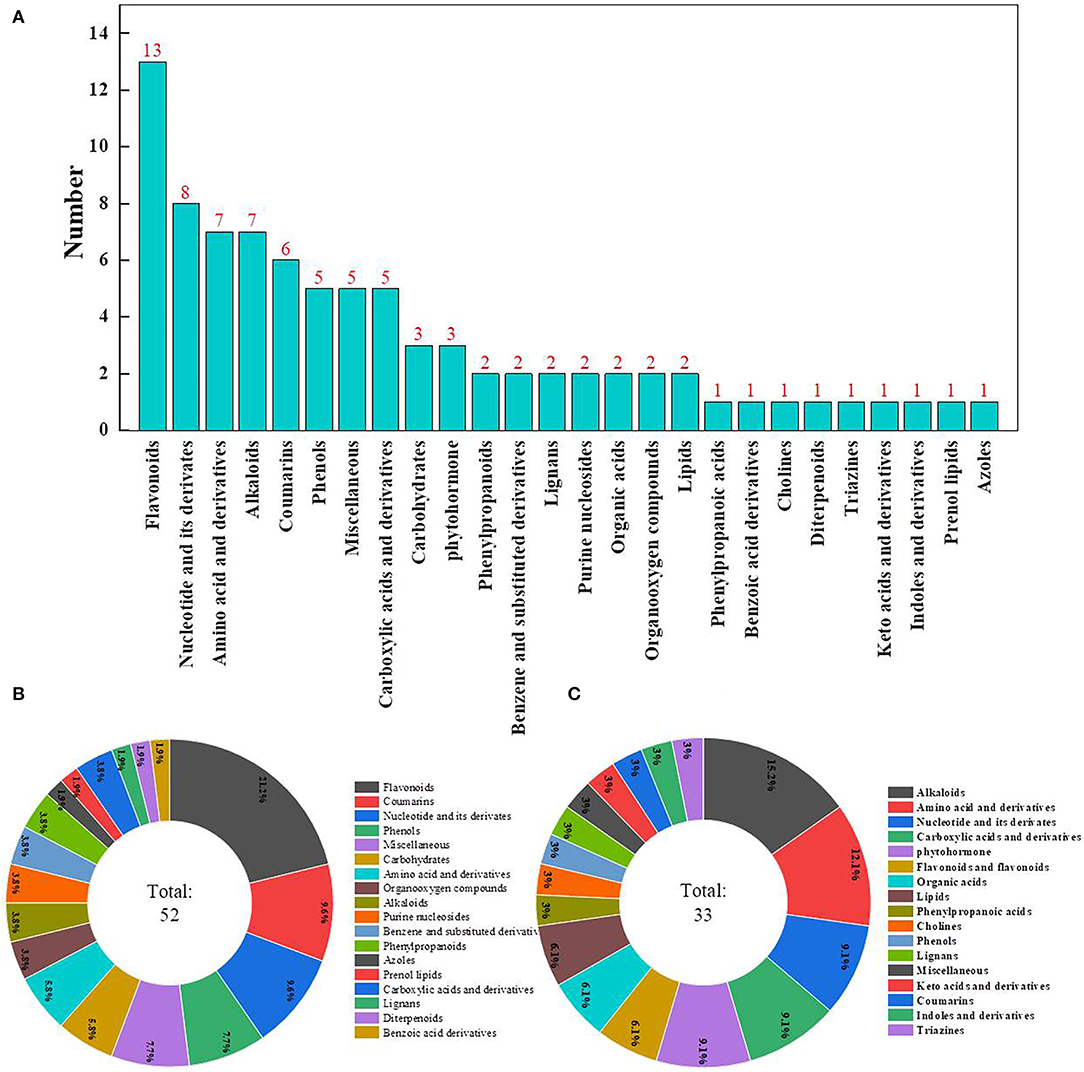
The metabolomic assay identified a total of 932 metabolites. Principal components analysis evidently separated metabolites into treated and control groups, and samples with 95% confidence intervals (Hotelling's t-squared ellipse) were selected for further analyses to screen DAMs (Figure 5A). As a result, 85 DAMs were detected, with 33 and 52 DAMs being down- and upregulated, respectively, after ClO2 treatment (Figure 5B).
Comparison of Metabolites
The most abundant DAMs identified were flavonoids, followed by nucleotides and their derivatives, amino acids and their derivatives, alkaloids, coumarins, phenols, miscellaneous, and carboxylic acids and their derivatives (Figure 6A). A total of 52 DAMs were upregulated, whereas 33 DAMs were downregulated in the pericarp of ClO2-treated tubers relative to that of control tubers. The most decreased metabolites were alkaloids, followed by amino acids and their derivatives, nucleotides and their derivatives, carboxylic acids and their derivatives, and phytohormones (Figure 6B). Most increased metabolites included flavonoids, followed by coumarins, nucleotides and their derivatives, phenols, and miscellaneous compounds (Figure 6C).
Integrated Transcriptomic and Metabolomic Analyses
The regulatory networks were analyzed by correlating DEGs and DAMs from each treatment. Results showed that multiple genes were positively or negatively regulated by multiple metabolites. For example, 17 DAMs were negatively or positively regulated by 20 or 30 DEGs, respectively, whereas 33 DAMs were negatively or positively regulated by 30 or 20 DEGs, respectively (Figure 7A). About three metabolites, 4-methylumbelliferyl acetate, methoxyindoleacetic acid, and DL-tyrosine, were strongly correlated with nine DEGs, with seven and two genes showing positive and negative correlations, respectively. Similarly, bruceine D and L-3-phenyllactic acid were significantly correlated with 10 DEGs, with eight and two genes showing positive and negative correlations, respectively, and 10 DEGs (three positively correlated, seven negatively correlated), respectively. In addition, (S)-2-acetolactate and vasicine were correlated with nine DEGs, with six and three genes showing positive and negative correlations, respectively, and eight DEGs (five positively correlated, three negatively correlated), respectively. Dalbergioidin and D-alpha-aminobutyric acid were markedly correlated with six DEGs, with one and five genes showing positive and negative correlations, respectively (Figure 7B). Notably, additional control networks were identified (Supplementary Figure 2).
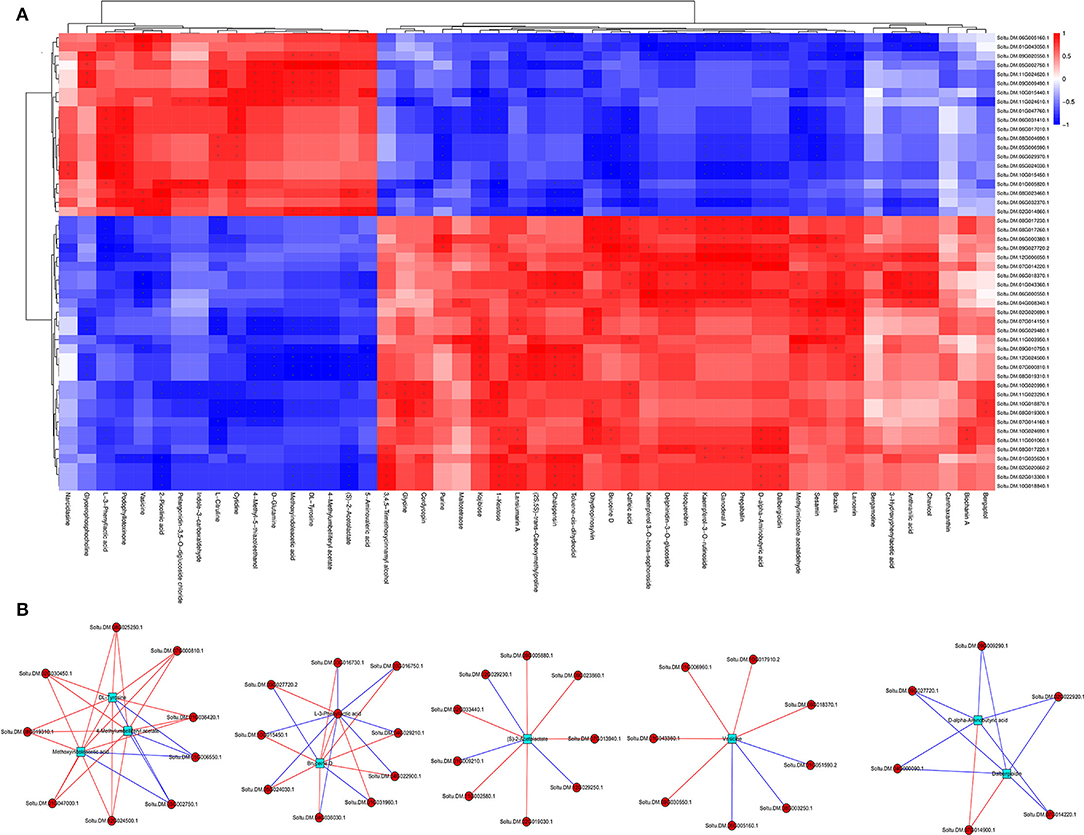
Figure 7. Correlation heat map (A) and network regulation map (B) between different genes and different metabolites.
Discussion
Potato tuber sprouting caused commercial losses and results in increased content of solanine in the tuber, which poses a potential threat to human health (Liu et al., 2020). Transcriptomics and metabolomics can be used to better understand the mechanism of ClO2 -mediated repression of potato tuber sprouting. RNA-seq results identified 3,119 DEGs after ClO2 treatment, with the majority showing enrichment in phenylpropanoid biosynthesis, the MAPK signaling pathway plant, and plant hormone signal transmission pathways. Phenylpropanoid biosynthesis and MAPK signaling pathways were significantly activated, whereas the plant hormone signal transduction pathway was significantly inhibited. Consistently, ClO2 treatment could activate phenylpropane metabolism to improve apple tolerance (Zhao et al., 2011) and promote the accumulation of MAPK, which was shown to regulate browning in longan (Chumyam and Saengnil, 2020). Interestingly, ClO2 treatment hindered signal transduction during potato tuber sprouting. Phytohormones are crucial endogenous regulators of tuber sprouting, and dynamic changes in endogenous hormones related to stages of sprouting were observed in hormone-treated tubers (Aksenova et al., 2013). This suggests that ClO2 repressed potato sprouting by limiting plant hormone signal transduction. In addition, ClO2 could improve the stress tolerance of tubers by activating phenylpropanoid biosynthesis and MAPK signaling pathways. This could be because of the ability of ClO2 to induce the production of reactive oxygen species, which damage cell membranes, eventually leading to the obstruction of signal transduction during tuber sprouting.
Metabolites are important determinants of potato tuber sprouting (Aksenova et al., 2013). During the sprouting process, starch and protein degradation are initiated, and soluble sugars and amino acids are formed (Sonnewald, 2001). Metabolites are the final or intermediate regulatory products of the cell's biological processes (Pichersky and Lewinsohn, 2011), and their accumulation is controlled by numerous endogenous and exogenous factors. Plant growth and development are regulated by metabolites (Luca et al., 2012). Thus, integrated metabolomic and transcriptomic analyses can be used to identify key metabolic pathways and functional genes associated with plant growth and development (Matus, 2016; Wang et al., 2017). Potato sprouting is often accompanied by changes in a large number of metabolites (Sonnewald, 2001). Metabolomic results in this study revealed that most differentially upregulated metabolites associated with the repressing potato tuber sprouting were enriched in flavonoids and coumarins. In contrast, the most differentially downregulated metabolites were alkaloids, amino acids, and their derivatives. Flavonoids have health-promoting benefits because of their antioxidant capacity through cell signaling, pro-oxidative activity, or membrane characteristic mechanism (Mein et al., 2008). Potato tubers accumulate a high concentration of alkaloids, which might potentially compromise food safety because of their toxicity (Friedman, 2006). For example, the accumulation of glycoalkaloids can inhibit acetylcholinesterase activity and damage stomach cell membranes and has been associated with systemic side effects and gastrointestinal disturbance in people (Yamashoji and Matsuda, 2013). Alkaloid intake in small quantities can lead to gastrointestinal symptoms, such as abdominal pain, vomiting, and diarrhea, whereas higher alkaloid doses can cause acute poisoning or severe symptoms, such as coma, neurological disorders, cardiac failure, and paralysis (Mensinga et al., 2005). Our results showed that ClO2 could improve the antioxidant capacity and the safety of potato tubers by promoting the accumulation of flavonoids and inhibiting the synthesis of alkaloids, respectively (Deng et al., 2020; Wang et al., 2020).
Integrated metabolomic and RNA-seq were performed to determine the correlation between identified DEGs and DAMs. The results showed that different metabolites were regulated by different genes. For example, 4-methylumbelliferyl acetate and methoxyindoleacetic acid were regulated by nine genes, with seven and two genes as positive and negative regulators, respectively. The results suggest a complex regulatory mechanism between the changes in metabolite accumulation and gene expression abundance in potato tubers. Thus, we speculated that the mechanism of ClO2-repressed sprouting is achieved by a coordinated complex process involving several metabolic pathways, which inhibit signal transduction, plant hormones, and alkaloid synthesis in potato tubers, leading to repressed sprouting. The activity of ClO2 thus differs from other sprout inhibitors, such as chlorpropham, which is a mitotic inhibitor used to control tuber sprouting during storage (Vaughn and Lehnen, 1991). Essential oils instead act by damaging sprouts but do not really prevent their sprouting (Shukla et al., 2019). However, ClO2 is a safer and inexpensive sprout inhibitor compared to chlorpropham and essential oils, which are environmentally harmful and expensive (Fu et al., 2019). This study provides a basis for understanding the mechanism of ClO2-repressed sprouting; however, its specific regulatory roles still need further clarification.
Conclusion
This study used ClO2-treated and control tubers to analyze the dynamic changes in metabolite accumulation and gene expression levels using UHPLC-MS and RNA-seq methods. The results revealed a total of 3,119 DEGs between the treated and control tubers. Most DEGs were enriched in phenylpropane metabolism, the MAPK signal pathway, and plant signal transduction pathways. ClO2 could inhibit potato tuber sprouting by altering plant signal transduction. Metabolomic analyses identified 85 differential metabolites, with flavonoids and coumarins as the most upregulated differential metabolites, and alkaloids and amino acids and their derivatives as the downregulated differential metabolites. The ClO2-mediated suppression of potato tuber sprouting is controlled by a complex regulatory network involving several different genes and metabolites.
Data Availability Statement
The authors acknowledge that the data presented in this study must be deposited and made publicly available in an acceptable repository, prior to publication. Frontiers cannot accept an article that does not adhere to our open data policies.
Author Contributions
XZ contributed to writing the original draft and writing, reviewing, and editing the manuscript. ML contributed to project administration and funding acquisition. ST contributed to supervision. SL contributed to formal analysis. JC contributed to project administration. XZ investigated the study. XW contributed to methodology. XG contributed to methodology software. JT contributed to data curation. YM contributed to resources. JS conceptualized the study. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflictof interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This research was supported by the National Natural Science Foundation (31760476), Special Project of National Modern Agricultural Industrial Technology System of the Ministry of Finance and the Ministry of Agriculture and Rural Areas (CARS-09-P26), and Innovation Team Construction Project of Gansu Academy of Agricultural Sciences (2017GAAS31).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.887179/full#supplementary-material
Supplementary Figure 1. Verification of some gene expression related to phenylpropane biosynthesis (A) and plant hormone signal transduction (B). Bars indicate standard error (±SE). Asterisks indicate a significant difference (p < 0.05).
Supplementary Figure 2. The network regulation map between different genes and different metabolites.
Supplementary Table 1. Gene primer sequence of genes related to the synthesis of suberin and lignin.
Supplementary Table 2. Statistical sequencing data.
Supplementary Table 3. Statistical comparison efficiency of each sample.
References
Aday, M. S., and Caner, C. (2011). The applications of 'active packaging and chlorine dioxide' for extended shelf life of fresh strawberries. Pack. Technol. Sci. 24, 123–136. doi: 10.1002/pts.918
Aksenova, N. P., Sergeeva, L. I., Konstantinova, T. N., Golyanovskaya, S. A., Kolachevskaya, O. O., and Romanov, G. A. (2013). Regulation of potato tuber dormancy and sprouting. Russ. J. Plant Physiol. 60, 301–312. doi: 10.1134/S1021443713030023
Atinut, J., Jamnong, U., Pathrapol, L., and Kobkiat, S. (2019). Induced expression of NOX and SOD by gaseous sulfur dioxide and chlorine dioxide enhances antioxidant capacity and maintains fruit quality of 'daw' longan fruit during storage through H2O2 signaling-sciencedirect. Postharvest Biol. Technol. 156, 110938–110938. doi: 10.1016/j.postharvbio.2019.110938
Campbell, M., Gleichsner, A., Alsbury, R., Horvath, D., and Suttle, J. (2010). The sprout inhibitors chlorpropham and 1, 4-dimethylnaphthalene elicit different transcriptional profiles and do not suppress growth through a prolongation of the dormant state. Plant Mol. Biol. 73, 181–189. doi: 10.1007/s11103-010-9607-6
Chen, Z., Zhu, C., and Han, Z. (2011). Effects of aqueous chlorine dioxide treatment on nutritional components and shelf-life of mulberry fruit (Morus alba L.). J. Biosci. Bioeng. 111, 675–681. doi: 10.1016/j.jbiosc.2011.01.010
Chumyam, A., and Saengnil, K. (2020). Transient H2O2 induction by ClO2 fumigation alters prx-trx system and causes MAPK accumulation attenuating browning in harvested longan fruit. Chiang Mai Univ. J. Nat. Sci. 20:e2021013. doi: 10.12982/CMUJNS.2021.013
Conte, E., Imbroglini, G., Bertolini, P., and Camoni, I. (1995). Presence of sprout inhibitor residues in potatoes in relation to application techniques. J. Agricult. Food Chem. 43:2985–2987. doi: 10.1021/jf00059a039
Corsini, D., Stallknecht, G., and Sparks, W. (1978). A simplified method for determining sprout inhibiting levels of chlorpropham (CIPC) in potatoes. J. Agricult. Food Chem. 26, 990–991. doi: 10.1021/jf60218a030
Deng, Y. M., He, M. Y., Feng, F., Feng, X. S., Zhang, Y., and Zhang, F. (2020). The distribution and changes of glycoalkaloids in potato tubers under different storage time based on maldi-tof mass spectrometry imaging. Talanta, 221, 121453. doi: 10.1016/j.talanta.2020.121453
Du, J., Han, Y., and Linton, R. H. (2003). Efficacy of chlorine dioxide gas in reducing Escherichia coli O157:H7 on apple surfaces, Food Microbiology. 20, 583–591. doi: 10.1016/S0740-0020(02)00129-6
FDA (Food and Drug Administration). (1998). Bacteriological Analytical Manual, 8th Edition (Revision A) (Chapter 4) (Gaithersburg, MD: AOAC International), 4.20–4.26.
Foucart, C., Paux, E., Ladouce, N., Grima-Pettenati, J., and Sivadon, P. (2006). Transcript profiling of a xylem vs phloem cDNA subtractive library identifies new genes expressed during xylogenesis in Eucalyptus. New Phytologist, 170, 739–752. doi: 10.1111/j.1469-8137.2006.01705.x
Friedman. (2006). Potato glycoalkaloids and metabolites: roles in the plant and in the diet. J. Agricult. Food Chem. 54, 8655–8681. doi: 10.1021/jf061471t
Fu, M.R., Zhang, X.M., Jin, T., Li, B.Q., Zhang, Z.Q., and Tian, S.P. (2019). Inhibitory of grey mold on green pepper and winter jujube by chlorine dioxide (ClO2) fumigation and its mechanisms. LWT. 100, 335–340. doi: 10.1016/j.lwt.2018.10.092
Haas, B. J., Papanicolaou, A., Y assour, M., Grabher, M., Blood, P. D., and Bouden, J. (2013). De novotranscript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Huang, Z., Tian, S. L., Ge, X., Zhang, J., Li, S. Q., Li, M., et al. (2014). Complexation of chlorpropham with hydroxypropyl-β-cyclodextrin and its application in potato sprout inhibition. Carbohydrate Polymers, 107, 241–246. doi: 10.1016/j.carbpol.2014.02.072
Kleinkopf, G., Oberg, N., and Olsen, N. (2003). Sprout inhibition in storage: Current status, new chemistries and natural compounds. Am. J. Potato Res. 80, 317–327. doi: 10.1007/BF02854316
Kuhl, C., Tautenhahn, R. B., Ttcher, C., Larson, T. R., and Neumann, S. (2012). Camera: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal.Chem. 84, 283–289. doi: 10.1021/ac202450g
Liu, J, M., Wang, S, S., Zheng, Xu., Jin, N., Lu, J., Huang, Y, T., et al. (2020). Antimicrobial activity against phytopathogens and inhibitory activity on Solanine in potatoes of the Endophytic Bacteria isolated from potato tubers. Front. Microbiol. 17, 570926. doi: 10.3389/fmicb.2020.570926
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using realtime quantitative PCR and the 2–ΔΔCT method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Luca, V. D., Satim, V., Atsumi, S. M., and Fang, Y. (2012). Mining the biodiversity of plants: a revolution in the making. Science. 336, 1658. doi: 10.1126/science.1217410
Matus, J. T. (2016). Transcriptomic and metabolomic networks in the grape berry illustrate that it takes more than flavonoids tofight against ultraviolet radiation. Front. Plant Sci. 7, 1337. doi: 10.3389/fpls.2016.01337
Mein, J.R., Lian, F., and Wang, X.D. (2008). Biological activity of lycopene metabolites: implications for cancer prevention. Nutr. Rev. 66, 667–683. doi: 10.1111/j.1753-4887.2008.00120.x
Mensinga, T. T., Sips, A., Rompelberg, C., Twillert, K. V., Meulenbelt, J., Top, H., et al. (2005). Potato glycoalkaloids and adverse effects in humans: an ascending dose study. Regul. Toxicol. Pharmacol. 41, 66–72. doi: 10.1016/j.yrtph.2004.09.004
Nicot, N., Hausman, J. F, Hoffmann, L., and Danièle, Evers. (2005). Reference gene selection for rt-qpcrnormalization in potato during biotic and abiotic stress. J. Exp. Bot. 56, 2907–2914. doi: 10.1093/jxb/eri285
Niu, R. X., Hui, C. X., Yin-Hua, T. U., and Jin, H. (2009). Effects of chlorine dioxide (ClO2) treatment on fresh-keeping and storage quality of qinmei kiwifruit. Sci. Technol. Food Indus. 1, 289–292. doi: 10.13386/j.issn1002-0306.2009.01.072
Pao, S., Kelsey, D. F., and Long, W. (2009). Spray washing of tomatoes with chlorine dioxide to minimize salmonella on inoculated fruit surfaces and cross-contamination from revolving brushes. J. Food Protect. 72, 2448-2452. doi: 10.4315/0362-028X-72.12.2448
Pichersky, E., and Lewinsohn, E. (2011). Convergent evolution in plant specialized metabolism. Ann. Rev. Plant Biol. 62, 549–566. doi: 10.1146/annurev-arplant-042110-103814
Sawada, Y. J., Kenji, A., Akane, S., Ayuko, K., Hitomi, O., Tetsuya, S., et al. (2009). Widely targeted metabolomics based on large-scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant Cell Physiol. 50, 37–47. doi: 10.1093/pcp/pcn183
Shukla, S., Pandey, S. S., Chandra, M., Pandey, A., Bharti, N., and Barnawal, D. (2019). Application of essential oils as a natural and alternate method for inhibiting and inducing the sprouting of potato tubers. Food Chem. 284, 171–179. doi: 10.1016/j.foodchem.2019.01.079
Sonnewald, U. (2001). Control of potato tuber sprouting. Trends Plant Sci. 6, 333–335. doi: 10.1016/S1360-1385(01)02020-9
Teper-Bamnolker, P., Dudai, N., Fischer, R., Belausov, E., Zemach, H., Shoseyov, O., et al. (2010). Mint essential oil can induce or inhibit potato sprouting by differential alteration of apical meristem. Planta. 232, 179–186. doi: 10.1007/s00425-010-1154-5
Tweddell, R. J., Boulanger, R., and Arul, J. (2003). Effect of chlorine atmospheres on sprouting and development of dry rot, soft rot and silver scurf on potato tubers. Postharvest Biol. Technol. 28, 445–454. doi: 10.1016/S0925-5214(02)00205-3
Vaughn, K. C., and Lehnen, L. P. (1991). Mitotic disrupter herbicides. Weed Sci. 39, 450–457. doi: 10.1017/S0043174500073215
Wang, Z. K., Ma, R., Zhao, M. S., Wang, F. F., Zhang, N., and Si, H. J. (2020). NO and ABA interaction regulates tuber dormancy and sprouting in potato. Front. Plant Sci. 11, 311. doi: 10.3389/fpls.2020.00311
Wang, Z. R., Cui, Y. Y., V ainstein, A., Chen, S. W., and Ma, H. Q. (2017). Regulation of fig (Ficus carica L.) fruit color: metabolomic and transcriptomic analyses of the flavonoid biosynthetic pathway. Front. Plant Sci. 8, 1990. doi: 10.3389/fpls.2017.01990
Woolfson, K. N., Haggitt, M. L., Zhang, Y., Kachura, A., Bjelica, A., Rincon, M. A. R., et al. (2018). Differential induction of polar and non-polar metabolism during wound-induced suberization in potato (Solanum tuberosum L.) tubers. Plant J. Cell Mol. Biol. 93, 931-942. doi: 10.1111/tpj.13820
Wu, B., Li, X., Hu, H., Liu, A., and Chen, W. (2011). Effect of chlorine dioxide on the control of postharvest diseases and quality of litchi fruit. Afr. J. Biotechnol. 10, 6030-6039.
Yamashoji, S., and Matsuda, T. (2013). Synergistic cytotoxicity induced by α-solanine and α-chaconine. Food Chem. 141, 669-674. doi: 10.1016/j.foodchem.2013.03.104
Zhang, Z. M., Tong, X., Peng, Y., Ma, P., Zhang, M. J., Lu, H. M., et al. (2015). Multiscale peak detection in wavelet space. Analyst. 140, 7955-7964. doi: 10.1039/C5AN01816A
Keywords: potato tuber, chlorine dioxide, repression of sprout, transcriptome, metabolome
Citation: Zheng XY, Li M, Tian SL, Li SQ, Chen JC, Zhang XJ, Wu XH, Ge X, Tian JC, Mu YW and Song J (2022) Integrated Analysis of Transcriptome and Metabolome Reveals the Mechanism of Chlorine Dioxide Repressed Potato (Solanum tuberosum L.) Tuber Sprouting. Front. Plant Sci. 13:887179. doi: 10.3389/fpls.2022.887179
Received: 01 March 2022; Accepted: 19 April 2022;
Published: 23 May 2022.
Edited by:
Luigi Lucini, Catholic University of the Sacred Heart, ItalyReviewed by:
Luciana Lanteri, Consejo Nacional de Investigaciones Científicas y Técnicas, ArgentinaMichelle Boivin, Cégep de Trois-Rivières, Canada
Copyright © 2022 Zheng, Li, Tian, Li, Chen, Zhang, Wu, Ge, Tian, Mu and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei Li, 908260866@qq.com
 Xiaoyuan Zheng
Xiaoyuan Zheng Mei Li1,3*
Mei Li1,3* Xia Ge
Xia Ge