- Zhejiang Provincial Key Laboratory of Biology of Crop Pathogens and Insects, Ministry of Agriculture and Rural Affairs (MARA) Key Laboratory of Molecular Biology of Crop Pathogens and Insects, College of Agriculture and Biotechnology, Institute of Biotechnology, Zhejiang University, Hangzhou, China
Methyl-CPG-Binding Domain (MBD) proteins play important roles in plant growth, development, and stress responses. The present study characterized the MBD families in watermelon and other cucurbit plants regarding the gene numbers and structures, phylogenetic and syntenic relationships, evolution events, and conserved domain organization of the MBD proteins. The watermelon ClMBD proteins were found to be localized in nucleus, and ClMBD2 and ClMBD3 interacted with ClIDM2 and ClIDM3. ClMBD2 bound to DNA harboring methylated CG sites but not to DNA with methylated CHG and CHH sites in vitro. The ClMBD genes exhibited distinct expression patterns in watermelon plants after SA and MeJA treatment and after infection by fungal pathogens Fusarium oxysporum f.sp. niveum and Didymella bryoniae. Overexpression of ClMBD2, ClMBD3, or ClMBD5 in Arabidopsis resulted in attenuated resistance against Botrytis cinerea, accompanied by down-regulated expression of AtPDF1.2 and increased accumulation of H2O2 upon B. cinerea infection. Overexpression of ClMBD1 and ClMBD2 led to down-regulated expression of AtPR1 and decreased resistance while overexpression of ClMBD5 resulted in up-regulated expression of AtPR1 and increased resistance against Pseudomonas syringae pv. tomato DC3000. Transcriptome analysis revealed that overexpression of ClMBD2 in Arabidopsis up-regulated the expression of a small set of genes that negatively regulate Arabidopsis immunity. These data suggest the importance of some ClMBD genes in plant immunity and provide the possibility to improve plant immunity through modification of specific ClMBD genes.
Introduction
As sessile organisms, plants have to face invasive attacks from diverse pathogenic microorganisms in the environment. To defense these pathogenic invasions, plants have evolved a complicated but fine-tuned innate immune system (Jones and Dangl, 2006; Yuan et al., 2021; Ngou et al., 2022). The first layer of the innate immunity, called PAMP-triggered immunity (PTI), is triggered by the recognition of microbial patterns via cell surface-localized pattern-recognition receptors, while the second layer, called effector-triggered immunity (ETI), is activated by the direct or indirect interaction between predominantly intracellularly localized nucleotide-binding leucine-rich repeat receptors and pathogen effectors. Activation of PTI and/or ETI is fine-tuned by a complicated molecular and genetic network, in which epigenetic regulation including DNA methylation/demethylation play critical roles (Huang and Jin, 2022).
Methylation of DNA, a conserved epigenetic mark, is one of the main mechanisms that play critical roles in epigenetic regulation of various biological processes including plant growth, development, and response to environmental cues (He et al., 2011; Arıkan et al., 2018; Zhang et al., 2018). Among DNA methylation, cytosine methylation (5-mC) is the most common epigenetic phenomenon that regulates the fate of gene expression (Grafi et al., 2007; Zhang et al., 2018). In higher plants, 5-mC occurs in CG dinucleotide regions, and CHG/CHH (H represents A, T, or C) trinucleotide regions (Gruenbaum et al., 1981). DNA methylation is a dynamic process that are achieved by different enzymes (Moore et al., 2013; Zhang et al., 2018) and is involved in many molecular processes, including genome stability, gene regulation, transposon silencing, and chromosome interactions (Zhang et al., 2006; Cokus et al., 2008; Lang et al., 2017). Extensive studies have indicated that DNA methylation plays important roles in plant growth and development, such as vegetable growth, pattern formation, flowering time, seed development, and fruit ripening (Gehring et al., 2009; He et al., 2011; Ibarra et al., 2012; Arıkan et al., 2018; Zhang et al., 2018) as well as in abiotic stress responses (Rambani et al., 2015; Xu et al., 2015; Yong-Villalobos et al., 2015; Hewezi et al., 2017). Importantly, DNA methylation, as one of the epigenetic regulation mechanisms, plays crucial roles in plant immunity (Huang and Jin, 2022); for example, Arabidopsis mutants with DNA hypomethylation are more resistant to Pseudomonas syringae pv. tomato (Pst) DC3000 and exhibit an elevated salicylic acid (SA)-dependent response (Dowen et al., 2012; Yu et al., 2013; Cambiagno et al., 2021).
In epigenetic model, proteins of the Methyl-CpG-binding domain (MBD) family are a group of key interpreters of DNA methylation and are generally associated with transcriptional silencing (Law and Jacobsen, 2010; He et al., 2011). The MBD proteins typically contain an MBD domain, with the ability to bind to 5-mC DNA (Ohki et al., 2001; Zemach and Grafi, 2003). Generally, the MBD proteins recognize 5-mC and recruit histone deacetylases, chromatin remodelers, and histone methyltransferases to repress transcription (Gigek et al., 2016). Genes coding for MBD proteins have been characterized in some plant species including Arabidopsis, rice, maize, poplar, potato, tomato, petunia, common bean, soybean, and rapeseed (Grafi et al., 2007; Parida et al., 2018; Coelho et al., 2022; Shi et al., 2022; Xiao et al., 2022); for example, 13 AtMBDs in Arabidopsis, 17 OsMBDs in rice, 14 ZmMBDs in maize, and 14 PtMBDs in poplar were identified (Grafi et al., 2007). The Arabidopsis AtMBD proteins can be divided into different subclasses (Zemach and Grafi, 2003; Springer and Kaeppler, 2005). AtMBD1, 2, 4, 8, 11 cannot specifically bind to 5-mC DNA, while AtMBD4 and AtMBD11 bind to methylated and unmethylated DNAs with or without 5-mC (Ito et al., 2003; Grafi et al., 2007). AtMBD5, 6, 7 show specific binding ability to mCG sites in vitro (Zemach and Grafi, 2003). Additionally, AtMBD5 also binds to mCHH sites while AtMBD6 binds to non-specific mCHH and mCHG sites (Ito et al., 2003). The binding activity and specificity have not been established for AtMBD3, 9, 10, 12, and 13 (Ito et al., 2003; Scebba et al., 2003; Grafi et al., 2007). Recently, it was demonstrated that AtMBD6 and AtMBD7 are actually readers for methylated DNA (Wu et al., 2022). AtMBD5 and AtMBD6, which are closely related and may have redundant functions (Berg et al., 2003), are recruited to chromatin by recognition of CG methylation to redundantly repress a subset of genes and transposons (Ichino et al., 2021), or participate in the formation of HDAC complexes to modulate the chromatin structure and gene transcription (Zemach et al., 2005). AtMBD6 also functions in RNA-mediated gene silencing (Parida et al., 2017). AtMBD7 interacts with the histone acetyltransferase Increased DNA Methylation 1 (IDM1) and its partners Increased DNA Methylation 2 (IDM2) and Increased DNA Methylation 3 (IDM3), and participates in DNA demethylation (Lang et al., 2015; Wang et al., 2015). Furthermore, AtMBD7 is also required for the H3K18 and H3K23 acetylation (Li Q. et al., 2015). AtMBD9 recognizes histone acetylation marks by IDM1 and functions in H2A.Z deposition (Nie et al., 2019). Furthermore, the biochemical activities of maize ZmMBD101 and tomato SlMBD5 have also been recently established (Li Y. et al., 2015; Questa et al., 2016).
The functions of MBD genes in plant growth, development, and response to abiotic stress have been explored. Mutation in AtMBD8 or knockdown of AtMBD11 led to a delay in flowering time, while the atmbd9 mutant showed a significantly earlier flowering time (Berg et al., 2003; Peng et al., 2006; Stangeland et al., 2009). Overexpression of Salix viminalis L. SvMBD5 led to an early flowering phenotype in transgenic Arabidopsis (Cheng et al., 2020). These observations indicate that the MBD proteins play critical roles in regulation of flowering in plants. The AtMBD11 knockdown mutant also displayed a variety of phenotypic effects, e.g., aerial rosettes, serrated leaves, abnormal position of flowers, and fertility problems (Berg et al., 2003), while the atmbd9 mutants produced more shoot branches (Peng et al., 2006). Overexpression of OsMBD707 leads to larger tiller angles and reduced photoperiod sensitivity in rice (Qu et al., 2021). The atmbd4 mutant exhibited altered root architecture and up-regulated expression of many phosphate transporters and transcription factors, indicating that AtMBD4 negatively regulates the phosphate starvation response (Parida et al., 2019). Some of the wheat TaMBD genes and most of the petunia PhMBD genes were highly induced by abiotic stress and hormones (Hu et al., 2011; Shi et al., 2016, 2022). However, the possible involvement of the MBD genes in plant immunity remains elusive.
Watermelon (Citrullus lanatus L.) is one of important horticultural crops, providing favorite fresh fruits worldwide. Fusarium wilt, caused by Fusarium oxysporum f.sp. niveum (Fon), and gummy stem blight, caused by Didymella bryoniae (Db), are two of the most devasting fungal diseases that lead to significant yield losses in watermelon industry (Michielse and Rep, 2009; Keinath, 2011). However, knowledge on the molecular mechanism of resistance in watermelon against Fon and Db is currently limited, which significantly impedes the breeding for watermelon cultivars with improved resistance against these two fungal diseases. The present study aimed to identify the watermelon ClMBD family by characterization and expression analyses and explore the putative mechanism of the ClMBD family in disease resistance. The transcript levels of the ClMBD genes were changed after treatment with SA and methyl jasmonate (MeJA) and infection by Fon and Db. Functional analyses revealed that ClMBD2, ClMBD3, and ClMBD5 negatively regulate resistance against Botrytis cinerea and that ClMBD1 and ClMBD2 negatively while ClMBD5 positively regulate resistance against Pst DC3000 in Arabidopsis.
Materials and Methods
Plant Materials and Growth Conditions
Watermelon (Citrullus lanatus) cv. Zaojia was used for all experiments. Nicotiana benthamiana plants expressing a known nucleus-localized marker protein RFP-H2B (Chakrabarty et al., 2007) were used for subcellular localization and bimolecular fluorescence complementation (BiFC) assays. Plants were grown in a soil mix (clay: soil = 3:1) in a growth room under fluorescent light (200 μE m2 s–1) at 22–24°C with 70% relative humidity (RH) and a 14 h light/10 h dark cycle. Arabidopsis seeds were surface sterilized in 75% ethanol for 5 min and 4% sodium hypochlorite for 10 min, rinsed with sterile water for three times, sowed on 1/2 MS plates and vernalized for 2 days at 4°C. Arabidopsis seedlings were grown on 1/2 MS plates at 22°C with 75% RH with a 16 h light/8 h dark cycle for 7 days and then transplanted to a soil mix (clay: soil = 1:1) in a growth room at 22°C with 75% humidity under a 16 h light/8 h dark cycle for normal growth or under a 8 h light/16 h dark cycle for disease assays.
Hormone Treatment and Pathogen Inoculation for Gene Expression Analysis in Watermelon
For analysis of tissue-specific expression, leaf, stem and root samples of 4-week-old watermelon plants were collected and stored at −80°C till use. For SA and MeJA treatment, 4-week-old watermelon plants were treated by foliar spraying with 1 mM SA, 100 μM MeJA or an equal volume of solution containing only 0.1% ethanol and 0.02% Tween-20 as controls, and leaf samples were collected at different time points after treatment.
For analysis of gene expression in response to Fon infection, pathogen inoculation was performed according to a previously reported method (Song et al., 2015). Briefly, mycelial plugs from 6-day-old culture of Fon race 1 strain ZJ1 were transferred into 200 mL mung bean liquid broth (mung bean 20 g/L, boiled for 20 min, pH7.0) and incubated with shaking (250 rpm) at 26°C for 2 days. Spores were collected and spore suspension was adjusted to 1 × 107 spores/mL for inoculation. Three-week-old watermelon plants were uprooted, washed in tap water, the main roots were cut up of one-third, and then dipped for 15 min in Fon spore suspension or in mung bean liquid broth as mock-inoculated controls. The inoculated plants were replanted in soil and allowed to grow in the same growth room as described above. Root samples were collected, frozen in liquid nitrogen and stored at −80°C until use.
Db strain DBTL4 was grown at 26°C on PNA (potato 200 g/L, NH4H2PO4 2 g/L, agar 15 g/L, pH7.0) for 6–7 days in dark and then treated with a 12 h UV light/12 h dark cycle for 5 days to induce spore production. After induction, the mycelial plugs were picked into distilled water, spores were collected and the spore suspension was adjusted to 2 × 106 spores/mL. Five-week-old watermelon plants were foliar sprayed with Db spore suspension containing 0.05% Tween-20 or with an equal volume of 0.05% Tween-20 solution as mock controls. The inoculated plants were placed in a 22°C chamber with 100% RH for 48 h. Leaf samples were collected, frozen in liquid nitrogen, and stored at −80°C until use.
Identification of Watermelon ClMBD Genes and Proteins
Arabidopsis AtMBD protein sequences were obtained from TAIR1 and were used as queries to search for putative MBD genes and proteins in watermelon, melon, cucumber, pumpkin, and zucchini genomes at Cucurbit Genomics Databases.2 The obtained nucleotide and protein sequences were examined by domain analysis programs PFAM3 (PF01429) and SMART4 with the default cutoff parameters. The isoelectric points and molecular weights were predicted on the ExPASy Proteomics Server.5 Sequence alignment was carried out by the ClustalX program. Phylogenetic trees were constructed using the neighbor-joining method of the MEGA7 program with the p-distance and complete deletion option parameters.
Synteny Analysis of the ClMBD Genes
The reliability of the obtained trees was tested using a bootstrapping method with 1,000 replicates. The MCScanX algorithm with default parameters (Wang et al., 2012) was used to scan orthologous regions containing the watermelon ClMBD genes. The corresponding plot was created by Dual Synteny Plot for MCscanX in TBtools software (Chen et al., 2020). The chromosomal localization of ClMBDs in the C. lanatus genome was obtained by TBtools software (Chen et al., 2020) according to the annotation data of the C. lanatus genome. The genomic and annotation data of melon, cucumber, zucchini, and pumpkin were downloaded from the Cucurbit Genomics Database (see text footnote 2), and those of Arabidopsis were downloaded from TAIR (see text footnote 1). The synteny relationship of the orthologous MBD genes obtained between watermelon and other selected species was visualized by the Advance Circos package of TBtools (Chen et al., 2020). DnaSP software was used to calculate the non-synonymous (Ka)/synonymous (Ks) values of the duplicated ClMBD gene pairs (Librado and Rozas, 2009).
Cloning of the ClMBD Genes
Total RNA was extracted using RNA Isolater reagent (Vazyme, Nanjing, China) according to the manufacturer’s instructions. RNA was treated with RNase-free DNase and then reverse-transcribed into cDNA using the HiScript QRT SuperMix kit (Vazyme, Nanjing, China). The obtained cDNAs were used for cloning, semi-RT-PCR and qRT-PCR. The coding sequences (CDs) of ClMBDs were amplified using gene-specific primers (Supplementary Table 1) and cloned into pCAMBIA1300s vector, yielding pCAMBIA1300s-ClMBDs-GFP. After confirmation by sequencing, these pCAMBIA1300s-ClMBDs-GFP plasmids were used as templates to amplify the target genes for further experiments.
Subcellular Localization Assays
The recombinant pCAMBIA1300s-ClMBDs-GFP plasmids were transformed into Agrobacterium tumefaciens strain GV3101. Agrobacteria carrying pCAMBIA1300s-ClMBDs-GFP or pCAMBIA1300s-GFP were separately infiltrated into leaves of Nicotiana benthamiana plants expressing a known nucleus-localized marker protein RFP-H2B (Chakrabarty et al., 2007). At 48 h after agroinfiltration, GFP fluorescence signals were excited at 488 nm and detected under a Zeiss LSM780 confocal laser scanning microscope (Zeiss, Oberkochen, Germany) using a 500–530 nm emission filter.
Yeast Two-Hybrid Assays
Putative interactions between ClMBDs and ClIDM2 or ClIDM3 were examined using the yeast two-hybrid (Y2H) System according to the manufacturer’s instructions (Clontech, Mountain View, CA, United States). The CDs of ClMBDs were amplified using gene-specific primers (Supplementary Table 1) from pCAMBIA1300s-ClMBDs-GFP and cloned into pGBKT7 vector, yielding pGBKT7-ClMBDs. ClIDM2 and ClIDM3 were obtained by homologous searching using Arabidopsis AtIDM2 and AtIDM3 as queries and the CDs of ClIDM2 and ClIDM3 were amplified with gene-specific primers (Supplementary Table 1) and cloned into pGADT7 vector, generating pGADT7-ClIDM2 and pGADT7-ClIDM3. The resultant pGBKT7-ClMBD plasmids were transformed with or without pGADT7-ClIDM2 or pGADT7-ClIDM3 into yeast strain Y2HGold by the LiAc/SS carrier DNA/PEG method and confirmed by colony PCR. The transformed yeasts were cultivated on DDO (SD/-Leu/-Trp) medium (Clontech, Mountain View, CA, United States) at 30°C for 3 days, followed by screening on QDO medium containing 40 μg/mL X-α-Gal (Clontech, Mountain View, CA, United States) and 125 ng/mL Aureobasidin A (Clontech, Mountain View, CA, United States). Interactions between ClMBDs and ClIDM2/3 were evaluated according to the growth performance of the transformed yeast cells on QDO and the production of blue pigments after the addition of X-α-Gal. Co-transformation of pGBKT7-53 or pGBKT7-Lam and pGADT7-T were used as positive and negative controls, respectively.
Bimolecular Fluorescence Complementation Assays
The CDs of ClMBD2 and ClMBD3 were amplified using gene-specific primers (Supplementary Table 1) and inserted into p2YN vector, yielding p2YN-ClMBD2 and p2YN-ClMBD3. Similarly, the CDs of ClIDM2 and ClIDM3 were inserted into p2YC vector, yielding p2YC-ClIDM2 and p2YC-ClIDM3. Agrobacteria harboring different indicated pairs of plasmids were infiltrated into leaves of N. benthamiana plants expressing a red nuclear marker protein RFP-H2B (Chakrabarty et al., 2007). At 48 h after agroinfiltration, YFP and RFP signals were detected and photographed under a Zeiss LSM780 confocal laser scanning microscope (Zeiss, Oberkochen, Germany).
Electrophoretic Mobility Shift Assays
The CDs of the ClMBD genes were amplified using gene-specific primers (Supplementary Table 1) and inserted into pGEX-4T-3 vector, generating pGEX-4T-3-GST-ClMBDs, followed by transforming into Escherichia coli strain BL21 (DE3), a widely used non-T7 expression strain that is suitable for transformation and protein expression (New England BioLabs, Beverly, MA, United States). To induce the expression of GST-ClMBD proteins, isopropyl-D-thiogalactoside was added to the bacterial cultures to a final concentration of 1 mM and incubated at 18°C for 20 h. The recombinant GST-ClMBD fusion proteins were purified using glutathione resin columns (Genscript, Shanghai, China) according to the manufacturer’s protocol. The following double-stranded DNA probes were synthesized and used in EMSA assay: 5mCG (GCTCGTAGCTAACGAGCTCGACTCGTTGACATAGGCCAT GGCGTAGACTC) (methylated nucleotides underlined) and its complementary strand with m5C at symmetrical positions, 5mCHG (GCTCTGAGCTAACAGGCTCAGC TCTGTGACATAGGCCATGGCTGAGACTC) (methylated nucleotides underlined) and its complementary strand with m5C at symmetrical positions, 5mCHH (GCTCTTAGCTAACA AGCTCAACTCTATGACATAGGCCATGGCTTAGACTC) (methylated nucleotides underlined) and its complementary strand (GAGTCTAAGCCATGGCCTA TGTCATAGAGGTGAGCTTGTTAGCTAAGAGC) (Ito et al., 2003). Equal volumes of single-stranded DNAs were mixed in annealing buffer (10 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl, pH7.5) and incubated at 85°C for 5 min to form double-stranded DNAs. EMSA was performed as previously described (Yuan et al., 2019) using LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, Waltham, MA, United States). In brief, binding reactions (10 μL) contained 1 μL 10 × binding buffer, 2 μg GST-ClMBD protein or GST protein (as a negative control) and 1 μL biotin-labeled 5mCG, 5mCHG, or 5mCHH probe. In the competitive reactions, unlabeled 5mCG probe was added in excess of 500 times. The binding reactions were incubated at 28°C for 20 min and separated on 6% native PAGE gels. After electrophoresis, the gels were transferred onto Amersham Hybond-N+ nylon membrane (GE Healthcare, Buckinghamshire, United Kingdom), and signals from the biotin-labeled probes were detected using a Chemiluminescent Biotin-labeled Nucleic Acid Detection Kit (Beyotime Biotechnology, Haimen, China) according to the manufacturer’s recommendations.
Generation and Characterization of ClMBDs-Overexpressing Transgenic Arabidopsis Lines
Arabidopsis transformation was performed using the floral dip method as previously described (Clough and Bent, 1998). In brief, flowers of 5-week-old Arabidopsis plants were dipped in a suspension of agrobacteria carrying pCAMBIA1300s-ClMBD-GFP plasmids for 1 min. The infected plants were placed in dark for 12 h under 100% RH, returned to the growth room with normal conditions and allowed to grow until the silique maturation. T0 seeds were surface sterilized and then plated on 1/2 MS plates containing 50 μg/mL hygromycin. After treatment at 4°C for 2 days, the plates were transferred to 22°C under a 16 h light/8 h dark cycle, seedlings showing hygromycin resistance, regarded as positive transgenic plants, were transferred in the mixed nutrient soil and allowed for growth for 5–6 weeks to collect seeds. Putative single-copy transgenic lines and homozygous lines were obtained by screening for a 3:1 segregation ratio of hygromycin-resistant (HgrR) character and 100% HgrR phenotype in T2 and T3 generations on 1/2 MS medium supplemented with 50 μg/mL hygromycin, respectively. The transcript levels of the ClMBD genes in the transgenic Arabidopsis lines were analyzed by semi-PCR and qRT-PCR. Two homozygous transgenic Arabidopsis lines with single-copy for each of the ClMBD genes (T3 generation) and similar expression levels of the transgenes were chosen for further experiments.
Disease Assays on Transgenic Arabidopsis Plants and Measurement of in planta Pathogen Growth
Disease assays with B. cinerea were performed as previously described (Wang et al., 2009). Briefly, spores were collected from 8∼10-day-old culture of B. cinerea strain BO5.10 grown on 2 × V8 plates and resuspended in 4% maltose and 1% peptone buffer to a final concentration of 2 × 105 spores/mL. Four-week-old Arabidopsis plants were inoculated by foliar spraying with the spore suspension containing 0.05% Tween-20 or with an equal volume of 0.05% Tween-20 solution as mock controls. The inoculated plants were placed in a 22°C chamber with 100% RH for 48 h, and disease development was continuously observed. Measurement of in planta fungal growth was performed by analyzing the transcript level of B. cinerea BcActin gene and comparing with the transcript level of an Arabidopsis Actin gene as an internal control according to a previously reported protocol (Wang et al., 2009).
Disease assays with Pst DC3000 were carried out as previously described (Zhang et al., 2016). Pst DC3000 was grown on King’s B (KB) broth and bacteria were collected and re-suspended in 10 mM MgCl2 solution to OD600 = 0.002. The bacterial inoculation was performed by hand infiltration using 1-mL syringes without needle into rosette leaves of 4-week-old Arabidopsis plants and the inoculated plants were kept in sealed containers 22°C for 72 h. For quantification of in planta bacterial growth, leaf discs from inoculated leaves were collected and homogenized in 10 mM MgCl2. After a series of gradient dilutions, the homogenate was plated on KB plates supplemented with 25 μg/mL rifampicin and bacterial colonies were counted at 3 days after incubation at 28°C.
In situ Detection of H2O2 Accumulation
Detection of H2O2 was performed using the DAB staining method (Thordal-Christensen et al., 1997). Leaf samples were collected from Arabidopsis plants with or without infection of B. cinerea at 24 h post inoculation (hpi) and dipped into DAB solution (1 mg/mL) in 10 mM Na2HPO4 (pH7.0). After incubation for 5 h in dark with shaking (80 rpm) at room temperature, the DAB-treated leaves were transferred into acetic acid/glycerol/ethanol (1:1:1, vol/vol/vol) and boiled for 5 min, followed by several washes with the same solution. The DAB-stained leaves were photographed using a digital camera.
RNA-Seq Analyses
Leaf samples were collected from 4-week-old Col-0 and ClMBD2-OE2 Arabidopsis plants, frozen in liquid nitrogen and stored at −80°C. RNA-seq was performed by BioMarker Technologies (Beijing, China) on Hiseq 2500 platform (Illumina). Raw data were filtered to get clean data, sequence comparison with the GCF_000001735.4_TAIR10.1. FPKM (Fragments Per Kilobase of transcript Per Million Fragments Mapped) was used to analyze the level of gene expression (Florea et al., 2013). The expression changes of differentially expressed genes (DEGs) ≥ 1.5-fold and P-value < 0.05. Gene Ontology (GO) enrichment analysis of DEGs was implemented by the GOseq R packages based Wallenius non-central hyper-geometric distribution (Young et al., 2010). KOBAS (Mao et al., 2005) software were used to test the statistical enrichment of differential expression genes in KEGG pathways (Kanehisa et al., 2008).
Semiquantitative RT-PCR and qRT-PCR Analyses
Extraction and treatment of total RNA were performed as mentioned above. Semiquantitative RT-PCR reactions contained 0.5 μL Phanta Max Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China), 0.5 μL dNTP Mix, 12.5 μL 2 × Phanta Max Buffer, 0.1 μg cDNA, 7.5 pmol of each of gene-specific primers (Supplementary Table 1), and 8.5 μL ddH2O in a final volume of 25 μL. Arabidopsis AtActin was used as the control. Each qPCR reaction contained 10 μL 2 × AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China), 0.1 mg cDNA and 7.5 pmol of each of gene-specific primers (Supplementary Table 1) in a final volume of 20 mL, and had two independent biological replicates. The qPCR was performed in a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, United States). Watermelon ClGAPDH or Arabidopsis AtActin were used as internal controls to normalize the data. Relative gene expression level was calculated using 2–△△CT method as described.
Statistical Analysis
All experiments were independently repeated three times and the obtained data were subjected to statistical analysis according to the Student’s t-test. The probability values of p < 0.05 were considered as significant difference between the treatments and corresponding controls.
Results
Identification and Characterization of the Watermelon ClMBD Family
To identify putative ClMBD genes in watermelon, BLASTp searches were performed against the watermelon genome database using the Arabidopsis AtMBDs as queries and 10 non-redundant sequences that are putative ClMBD genes were identified (Table 1). For convenience, unique identities to each of the identified ClMBD genes were assigned as ClMBD1–10 according to their chromosomal locations (Table 1). The CDs of ClMBD1–10 were confirmed by cloning of the full-length cDNAs using primers designed according to their predicted cDNA sequences. The sizes of the ClMBD open reading frames (ORF) ranged from 798 bp (ClMBD2) to 6,636 bp (ClMBD8) and the sizes of the encoded proteins varied from 265 amino acids (ClMBD2) to 2,211 amino acids (ClMBD8), with molecular weight of 23.34∼244.99 kDa and pI of 4.80∼9.49 (Table 1). Similarly, the MBD families in other cucurbit plants were also characterized and 9, 10, 15, and 16 MBD genes in melon, cucumber, zucchini, and pumpkin, respectively, were identified (Supplementary Table 2).
Structure of ClMBD Genes and Organization of Conserved Domains in ClMBD Proteins
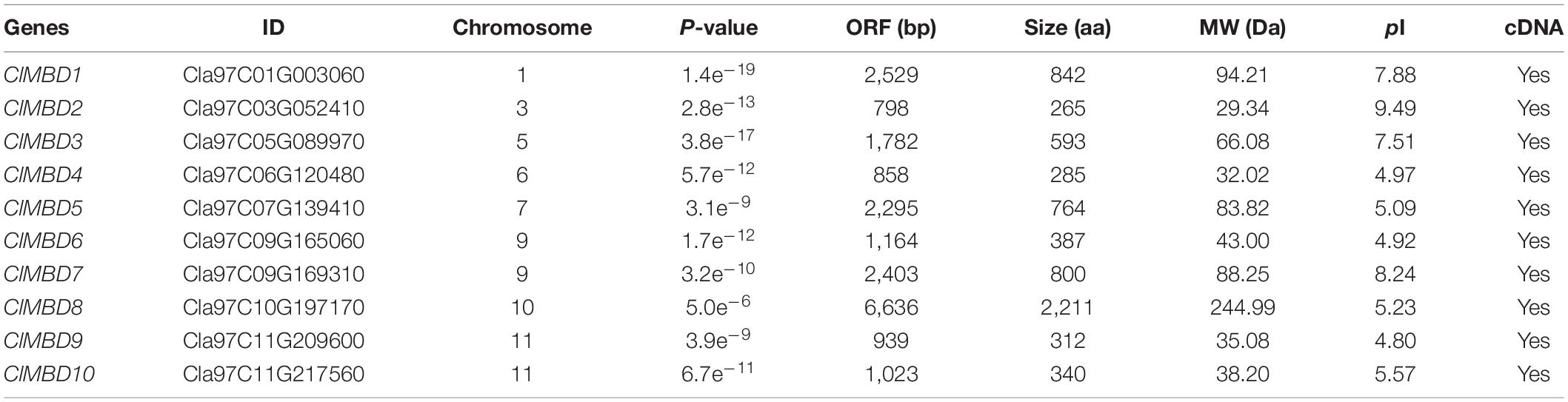
The 10 ClMBD genes are unevenly distributed on eight chromosomes in the watermelon genome and chromosomes 2, 4, and 8 do not host any ClMBD gene (Table 1 and Supplementary Figure 1). Chromosomes 9 and 11 harbor two ClMBD genes while each of the other chromosomes 1, 3, 5, 6, 7, and 10 carry one ClMBD gene (Table 1 and Supplementary Figure 1). Phylogenetic tree analysis revealed that the watermelon ClMBD proteins were divided into two clades: Clade I contained six ClMBD proteins (ClMBD1, 3, 4, 5, 6, and 9) while Clade II contained four ClMBD proteins (ClMBD2, 7, 8, and 10) (Figure 1A). Phylogenetic tree analysis of the MBD proteins from cucurbit plants showed that the MBD proteins from watermelon, melon, cucumber, pumpkin, and zucchini have a high level of similarity in the amino acid sequences (Supplementary Figure 2).
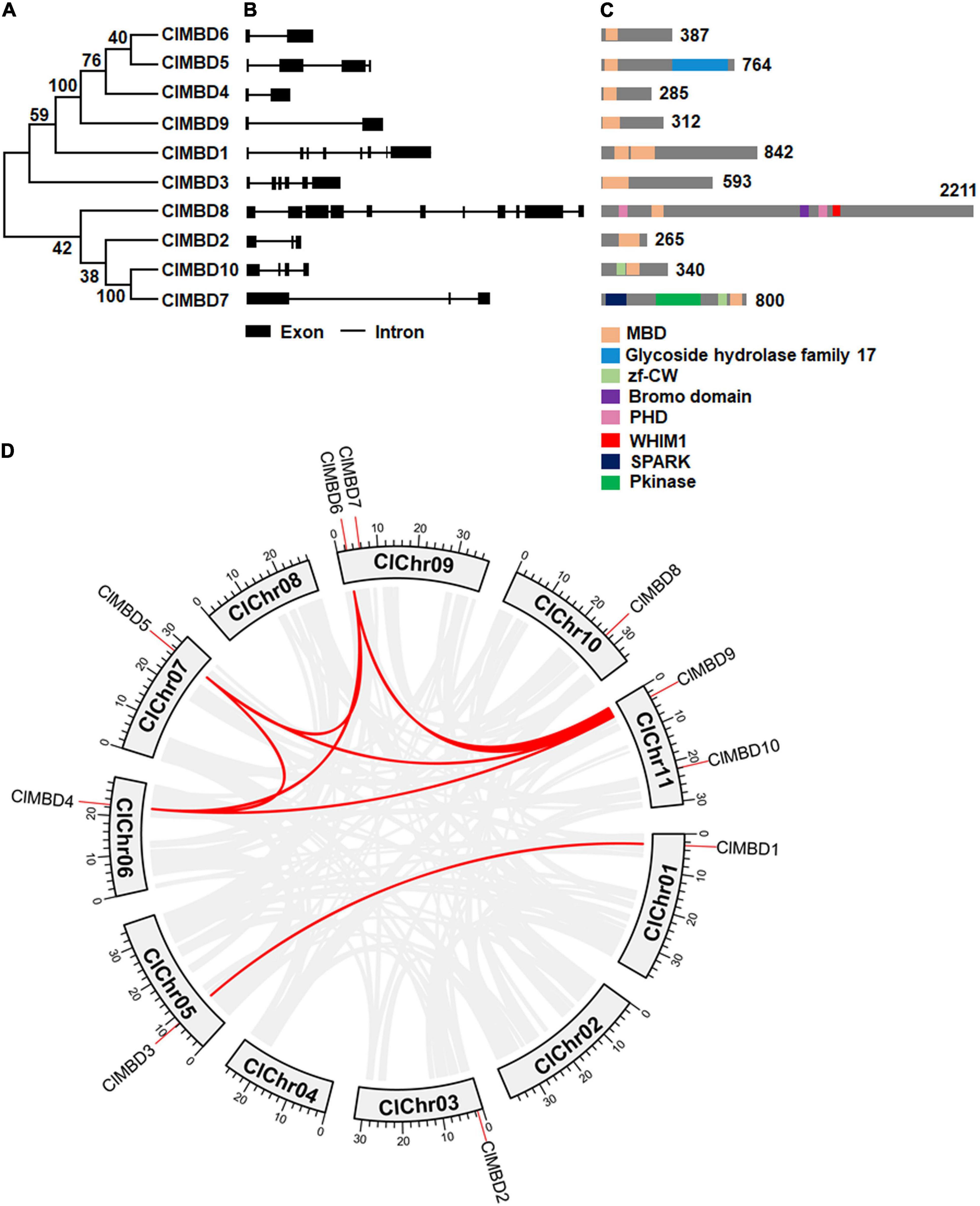
Figure 1. Phylogenetic tree and structure of genes and proteins of the watermelon ClMBD family and the evolutionary relationships among the watermelon ClMBD family members. (A) Phylogenetic relationships of the watermelon ClMBDs. Clustal X2 program was used for multiple sequence alignments and neighbor joining method of MEGA7 was used for constructing the phylogenic tree. (B) Structure of the watermelon ClMBD genes. The exon-intron structure of the coding regions of the ClMBD genes are shown. Filled boxes indicate exons while lines represent introns. (C) Conserved domains and their organization in the watermelon ClMBD proteins. (D) Interchromosomal relationships of the watermelon ClMBD genes. Gray lines indicate the synteny blocks in the genome, and the red lines indicate tandem duplications of the ClMBD gene pairs.
The structure of the ClMBD genes in the CDs is highly divergent in terms of the exon and intron numbers, with two (ClMBD9) to 11 (ClMBD8) exons and one (ClMBD9) to 10 (ClMBD8) introns (Figure 1B). A similar diverse exon-intron structure was also observed in the MBD genes in other cucurbit plants such as melon, cucumber, zucchini, and pumpkin (Supplementary Figure 3). The divergent gene structure may imply that the MBD genes in cucurbit plants possess divergent functions during their evolution.
The ClMBD proteins contain a characteristic conserved MBD domain (Figure 1C), ranging from 60 to 125 aa in size. The MBD domains in ClMBDs show 13∼43% of sequence identity and harbor some conserved amino acids, e.g., 15W/F, 35Y/F, 38P, and 54L/V (Supplementary Figure 4). Notably, ClMBD1 and ClMBD3 have two MBD domains while the other ClMBDs contain a single MBD domain (Figure 1C). Except for ClMBD7 whose MBD domain locates at the C-terminal, the MBD domains are generally located as the N-terminals in the ClMBD proteins (Figure 1C). ClMBD1, 2, 3, 4, 6, and 9 harbor the sole conserved MBD domains, while ClMBD5, 7, 8, and 10 contain other conserved domains in addition to the MBD domain (Figure 1C). For example, ClMBD5 has a Glycoside Hydrolase Family 17 domain; ClMBD10 has a zf-CW domain; ClMBD7 has a SPARK domain, a PKINase domain, a zf-CW domain; ClMBD8 has a Bromo Domain, a WHIM1 domains and 2 PHD domains (Figure 1C). Similar features in the presence of conserved amino acids in MBD domains (Supplementary Figure 4) and of the additional conserved domains in MBD proteins from melon, cucumber, zucchini, and pumpkin (Supplementary Figure 5) were also detected. The divergence of conserved domains between watermelon and other cucurbit plants may result in the diversity of functions and complexity of the biochemical and molecular mechanisms of the MBD proteins in plants.
Evolution and Interspecific Synteny of the Watermelon ClMBD Family
Gene duplication events in ClMBDs in the watermelon genome were detected and seven gene pairs, ClMBD1/3, ClMBD4/5, ClMBD4/6, ClMBD4/9, ClMBD5/6, ClMBD5/9, and ClMBD6/9, were localized in duplicated genomic regions (Figure 1D), implying the occurrence of gene duplication during the evolution of the ClMBD gene family in watermelon. The Ka/Ks ratios of ClMBD1/3, ClMBD4/6, ClMBD4/9, ClMBD5/6, ClMBD5/9, and ClMBD6/9 were < 1 (Supplementary Table 3), indicating that these gene pairs evolved through purifying selection. Interspecific collinearity analyses identified 9, 15, 20, 19, and 26 collinear gene pairs between watermelon and other tested plant species Arabidopsis, melon, cucumber, zucchini, and pumpkin, respectively (Figure 2 and Supplementary Table 4). Some ClMBD genes, e.g., ClMBD4 and ClMBD6, were found to be associated with at least 15 collinear gene pairs identified between watermelon and other tested plant species (Supplementary Table 4), indicating that ClMBD4 and ClMBD6 may play essential roles during evolution of the ClMBD genes. ClMBD2, 4, 6, 8, and 9 showed syntenic relationships with corresponding MBD genes in Arabidopsis and other cucurbit crops (Figure 2 and Supplementary Table 4), implying that these pairs of collinear genes may already exist before the ancestral divergence. Particularly, a total of 19,252 collinear gene pairs were identified between watermelon and melon, and 8 watermelon ClMBD genes on 6 chromosomes (Chr03, Chr06, Chr07, Chr09, Chr10, and Chr11) and 8 melon CmMBD genes on 6 chromosomes (Chr01, Chr02, Chr04, Chr05, Chr07, and Chr12) constituted 15 collinear gene pairs (Figure 2 and Supplementary Table 4). Four colinear gene pairs of watermelon ClMBDs distributed on each of Chr06, Chro9, and Chr11, while one colinear gene pair existed on each of Chr03, Chr07, and Chr10 (Figure 2 and Supplementary Table 4). These genes may originate from the same ancestors. Overall, there are more collinear gene pairs between watermelon and other cucurbit plants, indicating that these species were associated with the phylogenetic relationship and that the ClMBD gene family may be considered as marker genes in plant evolutionary.
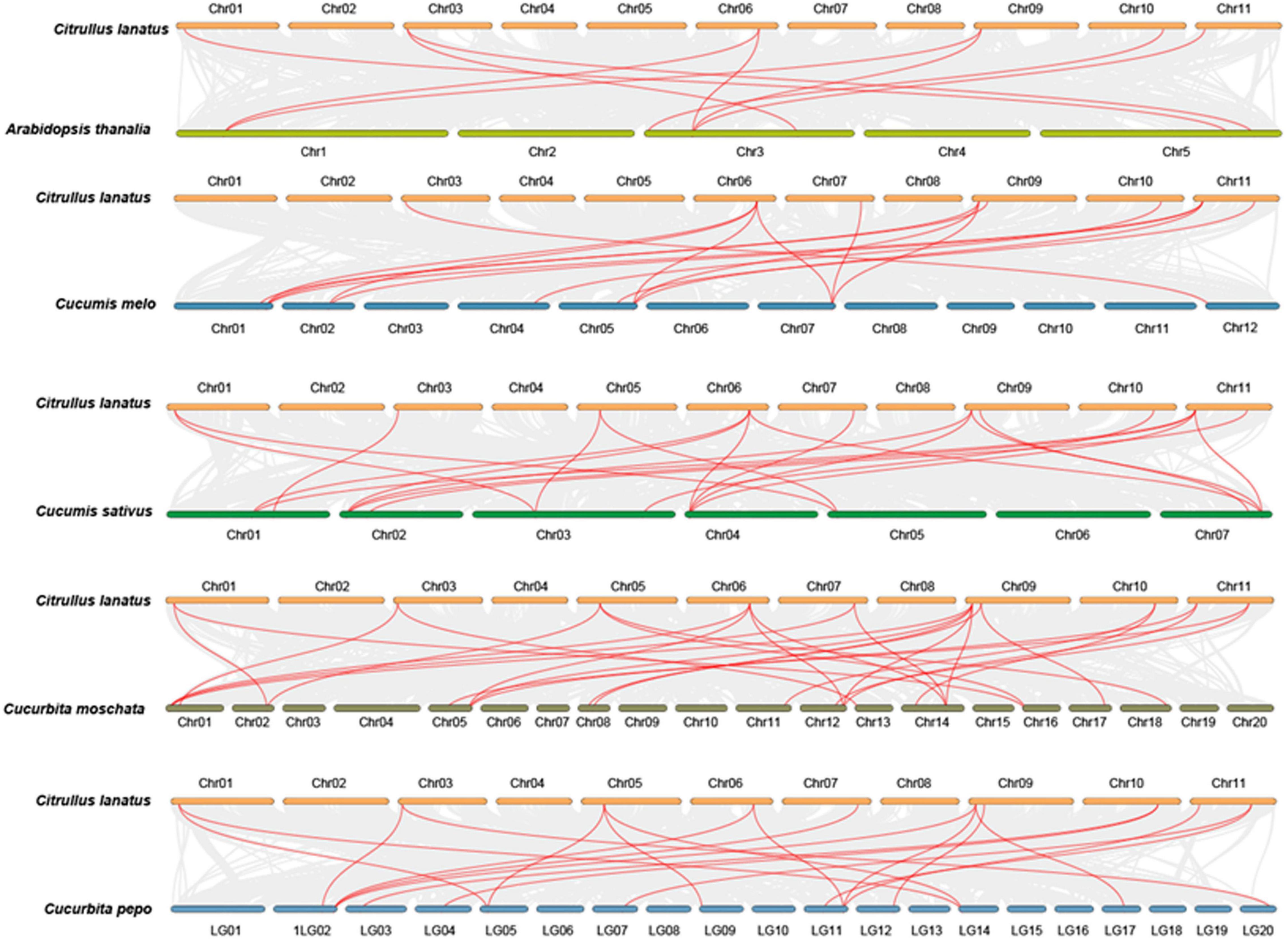
Figure 2. Syntenic relationships of the watermelon ClMBD genes with other MBD genes in representative plant species. Gray lines in the background indicate the collinear blocks within the watermelon and other plant genomes, while the red lines highlight the syntenic MBD gene pairs between watermelon and other plant species.
ClMBDs Are Nucleus-Localized Proteins
To explore the subcellular localization of the ClMBD proteins, agrobacteria carrying ClMBD1–10-GFP or GFP was infiltrated into leaves of N. benthamiana plants expressing a red nuclear marker RFP-H2B protein (Chakrabarty et al., 2007). The ClMBD1–10-GFP protein was solely localized to the nucleus, which was co-localized with the known nucleus marker RFP-H2B protein (Figure 3A). By contrast, GFP alone distributed ubiquitously throughout the cell without specific compartmental localization (Figure 3A). These results indicate that ClMBD1-ClMBD10 are nucleus-localized proteins.
Figure 3. Subcellular localization of the ClMBD proteins and the interactions between ClMBD2/3 and ClIDM2/3. (A) ClMBDs are nucleus-localized proteins. Agrobacteria carrying pCAMBIA1300-ClMBDs-GFP or pCAMBIA1300-GFP were infiltrated into leaves of N. benthamiana plants expressing a known nucleus-localized marker protein RFP-H2B. At 48 h after agroinfiltration, GFP signals were visualized under a confocal laser scanning microscope in dark field for green fluorescence (left), red fluorescence (middle left), white field for cell morphology (middle right) and in combination (right), respectively. (B) Interactions of ClMBD2 and ClMBD3 with ClIDM2 and ClIDM3. Agrobacteria carrying indicated pairs of p2YC and p2YN plasmids were infiltrated into leaves of N. benthamiana plants expressing a red nuclear marker RFP-H2B protein, and YFP and RFP signals were observed at 48 h after infiltration. Images were taken in dark field for green fluorescence (left) and red fluorescence (middle right), white field for cell morphology (middle left) and in combination (right), respectively. Experiments in (A,B) were repeated for three times with similar results.
Interactions Between ClMBDs and ClIDM2/3
The Arabidopsis AtMBD7 interacts with AtIDM2 and AtIDM3 to activate ROS1 to function in demethylation (Lang et al., 2015). To examine whether the ClMBD proteins have similar properties, the interactions of ClMBDs with ClIDM2 and ClIDM3 were examined. In Y2H assays, ClMBD2 and ClMBD3 interacted with ClIDM2 and ClIDM3, but the remaining ClMBDs did not (Supplementary Figure 6). Due to self-activation of ClMBD2 and ClIDM3 in Y2H, the interactions of ClMBD2 and ClMBD3 with ClIDM2 and ClIDM3 were further confirmed using the BiFC assays. YFP signal was not detected in N. benthamiana leaves co-infiltrated with agrobacteria harboring p2YN-ClMBD2 + p2YC, p2YN + p2YC-ClIDM2, p2YN-ClMBD3 + p2YC, or p2YN + p2YC-ClIDM3; by contrast, like that in the positive control, significant YFP fluorescence was clearly observed in leaves co-infiltrated with agrobacteria carrying p2YN-ClMBD2 + p2YC-ClIDM2, p2YN-ClMBD3 + p2YC-ClIDM2, p2YN-ClMBD2 + p2YC-ClIDM3, or p2YN-ClMBD3 + p2YC-ClIDM3 (Figure 3B). These results confirmed the interactions of ClMBD2 and ClMBD3 with ClIDM2 and ClIDM3.
ClMBD2 Specifically Binds to Methylated CG DNA
It is well known that MBD proteins have the capability to bind methylated DNA (Ito et al., 2003; Zemach and Grafi, 2003; Grafi et al., 2007). To explore the biochemical activity of the watermelon ClMBDs, recombinant GST-tagged ClMBD1–7, 9, 10 proteins were purified (Supplementary Figure 7) and their binding activity to methylated CG DNA was examined by EMSA. Two complementary single-stranded DNA probes with 5 methylated CG sites (5mCG) were synthesized and the double-stranded 5mCG DNA was generated (Figure 4A). In repeated EMSA, only ClMBD2 bound to labeled double-stranded 5mCG DNA, and the binding of ClMBD2 to labeled double-stranded 5mCG DNAs was specific as this binding was completely suppressed by the excessive unlabeled double-stranded 5mCG DNA in the competition binding assay (Figure 4A). The remaining ClMBDs did not show binding activity to the labeled double-stranded 5mCG DNA (Figure 4A). However, ClMBD2 did not bind to DNA harboring mCHG or mCHH sites (Figure 4B). These results indicate that ClMBD2 specifically binds to mCG DNA, but not to mCHG DNA or mCHH DNA.
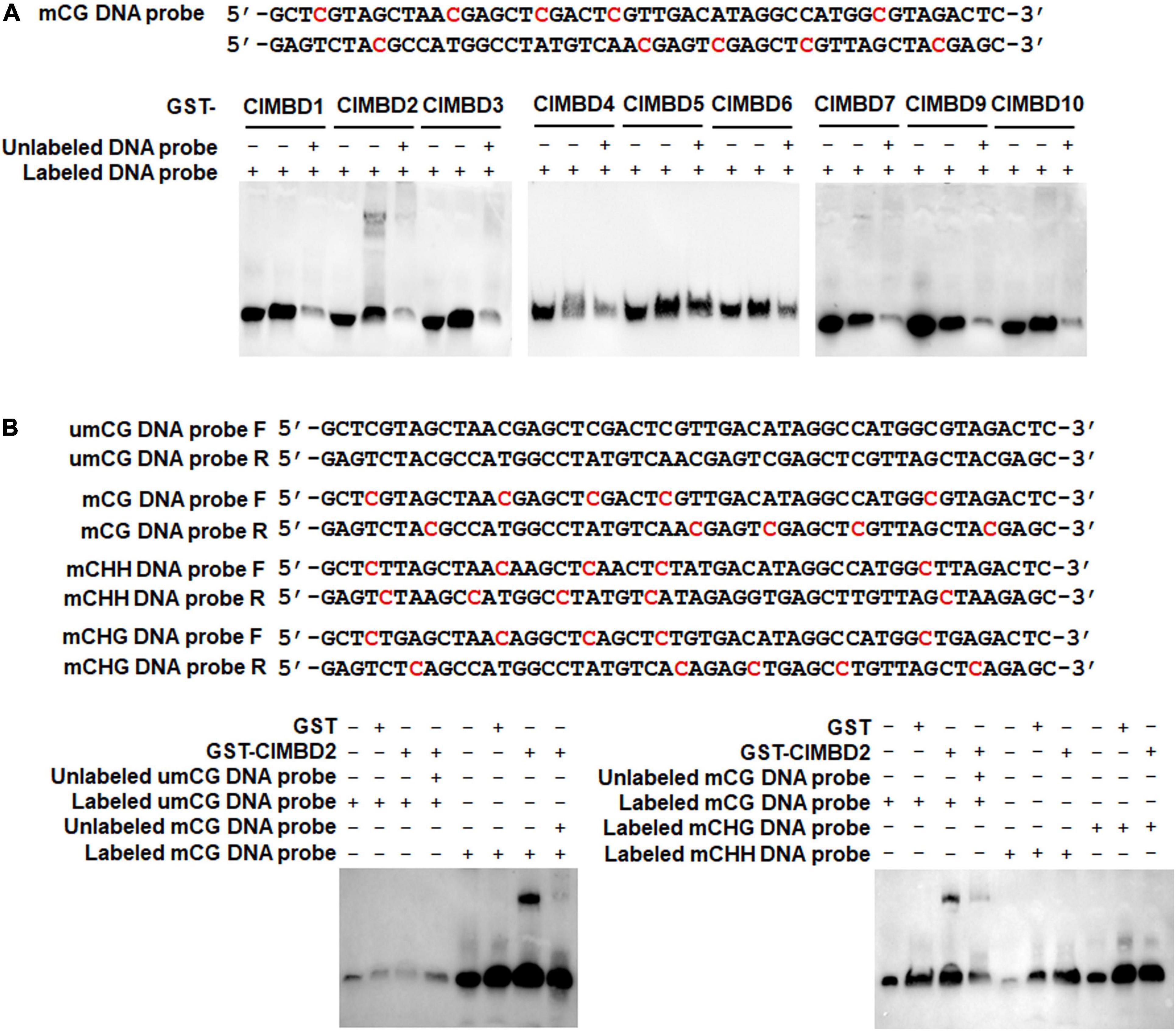
Figure 4. Binding activity of ClMBD2 to methylated CG DNA. (A) Binding of ClMBD proteins to the methylated CG DNA. Biotin-labeled mCG DNA (for binding assays) or biotin-labeled mCG DNA in combination with excessive unlabeled mCG DNA (for competitive assays) were incubated with GST-ClMBD or GST (as a negative control). (B) Binding activity of ClMBD2 to methylated CG DNA, methylated CHG DNA and methylated CHH DNA. Biotin-labeled mCG DNA, biotin-labeled mCHG DNA and biotin-labeled mCHH DNA (for binding assays) or biotin-labeled mCG DNA in combination with excessive unlabeled mCG DNA (for competitive assays) were incubated with GST-ClMBD or GST (as a negative control). The mCG, mCHG, and mCHH DNA sequences are shown and the methylated sites are indicated in red color. Experiments in (A,B) were repeated for three times with similar results.
ClMBDs Have Similar Expression Patterns in Root, Stem, and Leaf Tissues
The expression patterns of the ClMBD gene in root, stem and leaf tissues of 4-week-old watermelon plants were analyzed and the qRT-PCR results showed that the ClMBD genes have similar expression patterns: highest expression in leaves, moderate in stems, and lowest in root (Supplementary Figure 8).
ClMBDs Are Responsive to Defense Hormones Salicylic Acid and Methyl Jasmonate
To explore the possible involvement of the ClMBD genes in disease resistance, expression changes of the ClMBD genes were analyzed in watermelon plants after treatment with SA and MeJA. After foliar spraying with 1 mM SA, the expression of ClNPR1 and ClPR1, the marker genes of SA signaling pathway, was significantly up-regulated. Particularly, the expression of ClPR1 significantly up-regulated at 6 h after treatment and peaked at 12 h, showing a > 59-fold increase, as compared with that in mock control (Figure 5A). After SA treatment, the expression of most of the ClMBD genes were up-regulated with distinct patterns: ClMBD2, 3, 6, 7, 8, 9, and 10 were up-regulated at 6 h; ClMBD1, 4, 5, 7, and 10 were up-regulated at 12 h; while ClMBD1, 6, and 8 were markedly up-regulated at 24 h, as compared with those in the mock controls (Figure 5A). Notably, ClMBD2, 3, 9, and 10 showed similar expression patterns after SA treatment, implying that these ClMBD genes may have similar functions. These data indicate that the ClMBD genes can respond to SA and thus may be involved in disease resistance in watermelon.
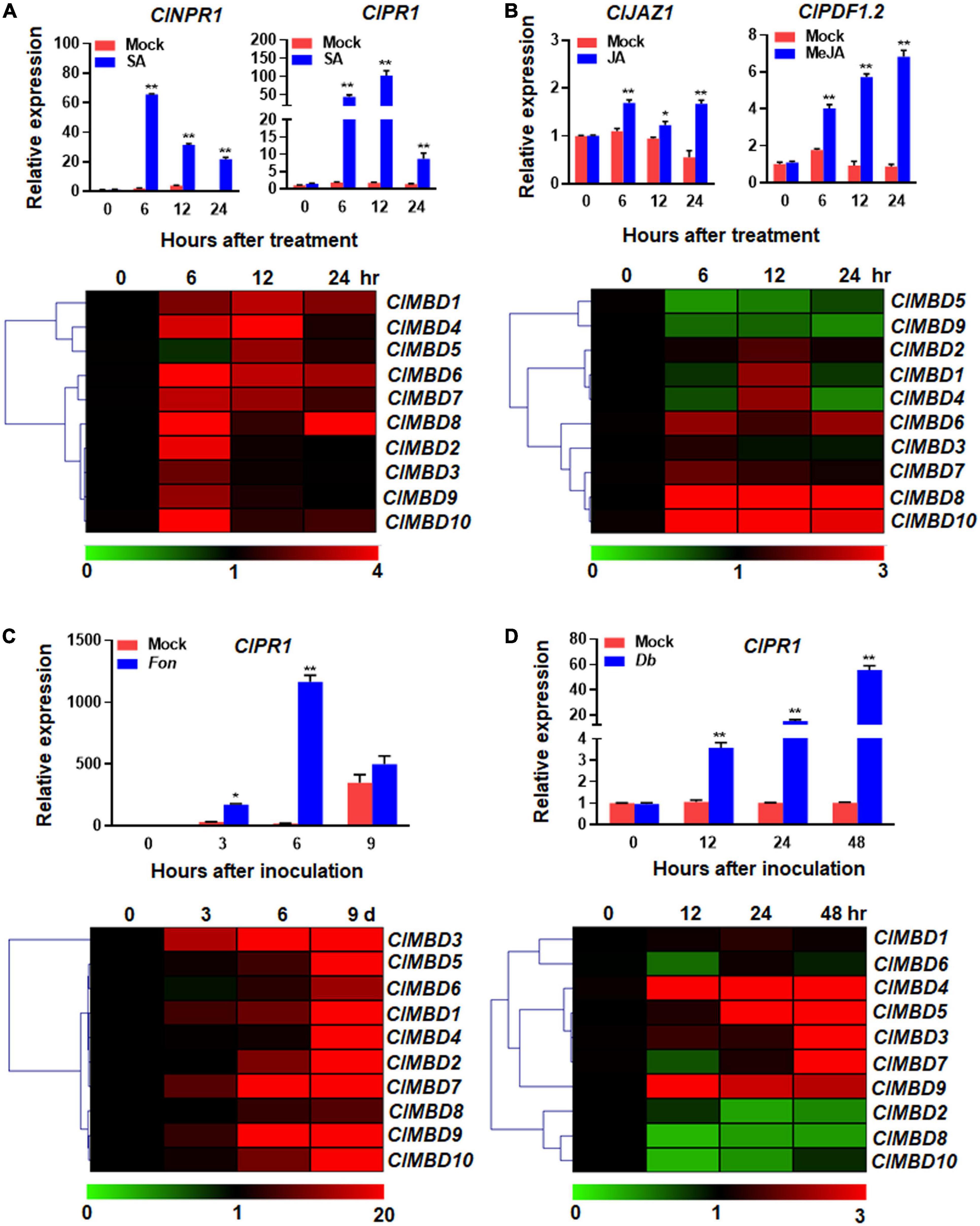
Figure 5. Expression changes of the watermelon ClMBD genes in response to defense hormones and fungal pathogens. (A,B) Expression changes of the ClMBD genes to SA (A) or MeJA (B). Four-week-old watermelon plants were treated by foliar spraying with 1 mM SA, 100 μM MeJA or similar volume of solution (as mock controls) and leaf samples were collected at indicated time points after treatment. (C) Expression changes of the ClMBD genes to Fusarium oxysporum f.sp. niveum. Three-week-old plants were inoculated by dipping the roots in spore suspension (1 × 107 spores/mL) of F. oxysporum f.sp. niveum or in mung bean liquid broth as mock-inoculated controls, and root samples were collected at indicated time points after inoculation. (D) Expression changes of the ClMBD genes to Didymella bryoniae. Five-week-old watermelon plants were inoculated by foliar spraying with D. bryoniae spore suspension (2 × 106 spores/mL) or similar volume of solution as mock controls, and leaf samples were collected at indicated time points after inoculation. qRT-PCR was performed using the watermelon ClGAPDH gene as an internal control. Relative expression was calculated using the 2–ΔΔCT method. Experiments were repeated for three times and the data presented are the means ± SE from three independent experiments. * or ** above the columns indicate significant difference at p < 0.05 or p < 0.01 levels (Student’s t-test), respectively, between treatment/inoculation and mock controls at the same time point.
After foliar spraying with 100 μM MeJA, the expression of ClJAZ1 and ClPDF1.2, the marker genes of the JA signaling pathway (Yang et al., 2019), was significantly up-regulated and peaked at 24 h (Figure 5B). After MeJA treatment, ClMBD2, 6, 7, 8, and 10 were highly up-regulated, while ClMBD5 and ClMBD9 were significantly down-regulated, as compared with those in the mock controls (Figure 5B). ClMBD4 was up-regulated at 12 h but down-regulated at 24 h, as compared with those in the mock controls (Figure 5B). Notably, ClMBD8/ClMBD10 and ClMBD1/ClMBD4 exhibited similar expression patterns in response to exogenous MeJA, indicating these two pairs of the ClMBD genes may have similar biological functions. These data indicate that the ClMBD genes differentially respond to MeJA and thus may play different roles in disease resistance in watermelon.
ClMBDs Differentially Respond to Fungal Pathogens
To explore the possible involvement of ClMBDs in watermelon disease resistance, the expression changes of the ClMBD genes in watermelon plants after infection with different fungal pathogens were analyzed. Fon is the most important soilborne vascular pathogen that causes Fusarium wilt, one of the most devastating fungal diseases in watermelon (Michielse and Rep, 2009). Fon infects watermelon plants through root system and proliferation within the xylem vessels (Michielse and Rep, 2009). Therefore, the expression changes of the ClMBD genes in root tissues of watermelon plants after Fon infection were analyzed. The qRT-PCR results showed that the expression level of ClPR1 started to increase at 3 days post inoculation (dpi), peaked at 6 days, leading to 65.5-fold higher over that in mock-inoculated plants, and then decreased (Figure 5C). The expression of ClMBD1 and ClMBD10 in root tissues of the Fon-infected watermelon plants were up-regulated, as compared with those in the mock-inoculated plants, at 3 dpi (Figure 5C). As compared with those in the mock-inoculated plants, the expression of ClMBD2, 6, and 9 was up-regulated at 6 dpi, while the expression of the ClMBD genes, except for ClMBD3, 5, and 10, was up-regulated at 9 dpi (Figure 5C). Overall, the expression changes of the ClMBD genes exhibited similar significant up-regulation patterns in root tissue at 3, 6, or 9 dpi; for example, the expression level of ClMBD7 in root tissue was markedly up-regulated with a > 639-fold increase than that in mock-inoculated plants at the 9 dpi (Figure 5C). Notably, the expression changes of ClMBD8, 9, and 10, and of ClMBD1, 2, 4, 5, 6, and 7 showed similar patterns in response to Fon, implying that these two groups of the ClMBD genes may be involved in resistance with similar functions. The results suggest that the ClMBD genes are responsive to Fon infection during the relative late stage of the pathogenesis and thus may play roles in the process of regulating watermelon resistance to vascular Fusarium wilt disease.
Db is another devasting fungal pathogen that infects leaf and stem tissues and causes gummy stem blight, which is a very common fungal disease on cucurbitaceous crops including watermelon (Keinath, 2011). The responsiveness of the ClMBD genes to Db infection was also analyzed in leaf tissues of watermelon plants after foliar spraying with a fungal spore suspension. After Db inoculation, the expression of ClPR1 in leaf tissues started to increase at 12 hpi, gradually increased and peaked at 48 h, leading to a 55-fold increase over that in mock-inoculated plants (Figure 5D). The expression of the ClMBD genes in leaf tissues exhibited distinct patterns in response to Db infection. The expression levels of ClMBD3, 4, 7, and 9 in Db-infected leaves were significantly up-regulated at 12, 24, or 48 hpi, as compared with those in mock-inoculated leaves (Figure 5D). By contrast, the expression levels of ClMBD2, 6, 8, and 10 in Db-infected leaves were highly down-regulated at 12, 24, or 48 hpi, as compared with those in mock-inoculated leaves (Figure 5D). The expression of ClMBD1 and ClMBD5 in leaf tissues was not significantly affected by Db infection during a 48 h period of the experiments (Figure 5D). Notably, the expression changes of ClMBD3, 5, and 7 exhibited similar patterns in response to Db, indicating similar involvement for these three ClMBD genes in Db resistance. These results indicate that the expression of the ClMBD genes in leaf tissues exhibited distinct patterns in response to Db infection and therefore may play different roles in the process of regulating watermelon resistance against Db.
Generation and Characterization of ClMBD-Overexpressing Arabidopsis Lines
To investigate the functions of the ClMBD genes, transgenic Arabidopsis lines with overexpression of an individual ClMBD gene were generated. The ClMBD genes were transcribed normally in their own transgenic Arabidopsis lines (Supplementary Figures 9A,B). The ClMBD-overexpressing Arabidopsis plants showed no significant defect in growth and development, including plant height and size, in comparison to WT plants, when grown in a greenhouse (Supplementary Figure 10).
ClMBD2, ClMBD3, and ClMBD5 Negatively Regulate Arabidopsis Immunity Against Botrytis cinerea
To explore the possible functions of the ClMBD genes in plant immunity, disease resistance phenotype of the ClMBD-overexpressing Arabidopsis lines and the wild-type (WT) Col-0 plants after infection with B. cinerea, a necrotrophic fungus causing grey mold disease, was assessed. In repeated detached leaf punch inoculation assays, B. cinerea-caused necrotic lesions on leaves detached from the ClMBD2-OE, ClMBD3-OE, and ClMBD5-OE plants were significantly larger than those on leaves of WT plants, resulting in increases of approximately 88.9, 55.6, and 66.7% in lesion length, respectively, as compared with those on WT leaves (Supplementary Figure 11). By contrast, B. cinerea-caused necrotic lesions on leaves detached from the ClMBD1-OE, ClMBD4-OE, ClMBD6-OE ClMBD7-OE, ClMBD8-OE, ClMBD9-OE, and ClMBD10-OE plants were comparable to those on leaves of WT plants (Supplementary Figure 11). To confirm these results, the ClMBD-overexpressing plants were inoculated by foliar spraying with B. cinerea spore suspension and disease severity and fungal growth were compared with those in WT plants. After infection, typical B. cinerea-caused disease symptom was seen at 3 dpi. Much severe diseases were observed on leaves of the ClMBD2-OE, ClMBD3-OE, and ClMBD5-OE plants, especially the B. cinerea-infected ClMBD2-OE plants decayed and died at 3 dpi (Figure 6A). By contrast, disease severity on leaves of the B. cinerea-infected ClMBD1-OE, ClMBD4-OE, ClMBD6-OE, ClMBD7-OE, ClMBD8-OE, ClMBD9-OE, and ClMBD10-OE plants were similar to that in WT plants (Supplementary Figure 12A). Accordingly, the ClMBD2-OE, ClMBD3-OE, and ClMBD5-OE plants supported more in planta fungal growth, leading to increases of 57.5–851.3% over that in WT plants (Figure 6B), while the ClMBD1-OE, ClMBD4-OE, ClMBD6-OE, ClMBD7-OE, ClMBD8-OE, ClMBD9-OE, and ClMBD10-OE plants supported similar amounts of in planta fungal growth (Supplementary Figure 12B). These data from detached leaf punch inoculation and whole plant inoculation assays indicate that overexpression of ClMBD2, 3, and 5 attenuates the resistance of transgenic Arabidopsis plants against B. cinerea, while overexpression of each of the remaining ClMBD genes does not affect the resistance against B. cinerea.
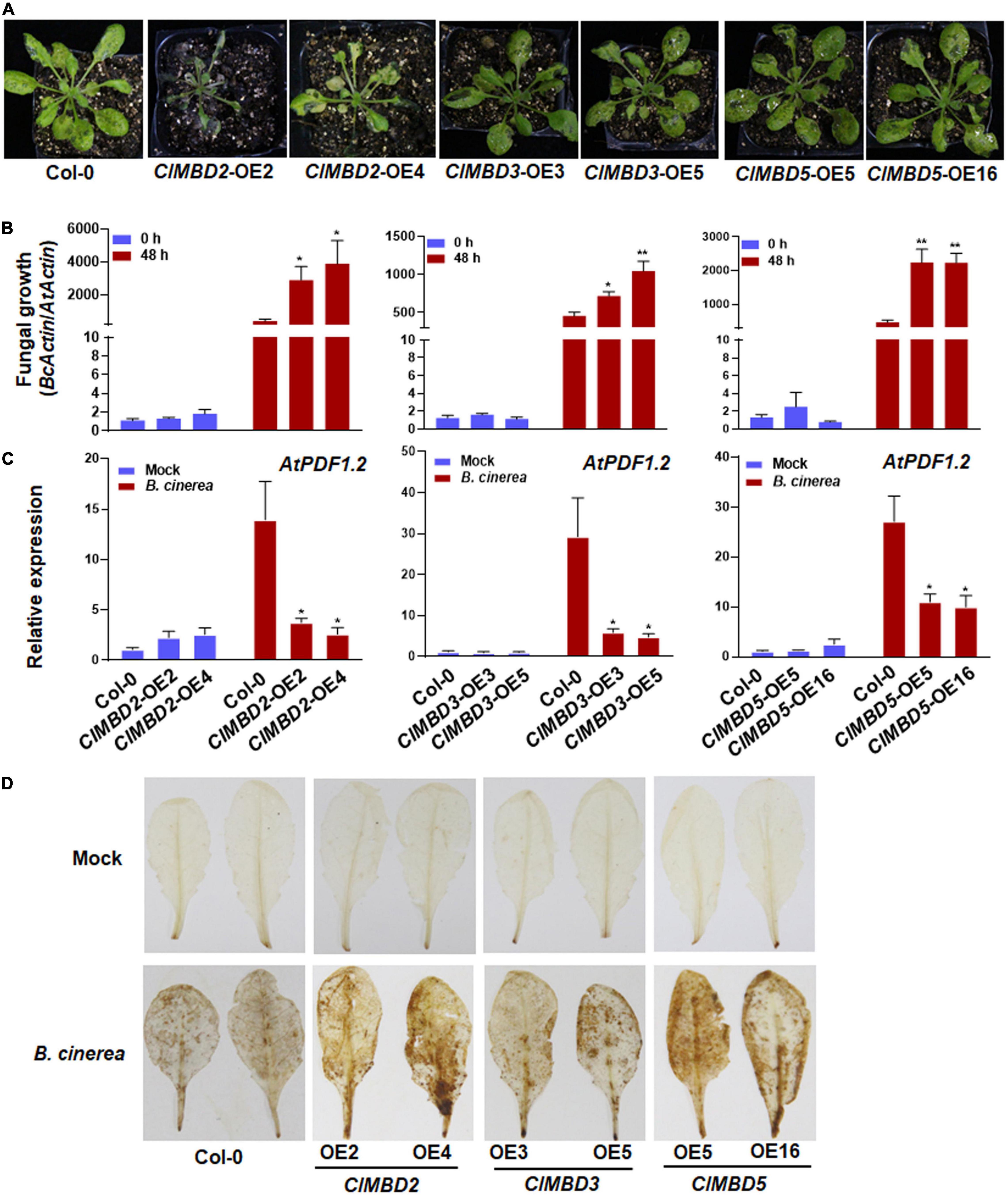
Figure 6. ClMBD2, ClMBD3, and ClMBD5 negatively regulate resistance of the transgenic Arabidopsis plants against Botrytis cinerea. (A) Typical B. cinerea-caused disease on WT, ClMBD2-OE, ClMBD3-OE, and ClMBD5-OE plants. Four-week-old plants were inoculated by foliar spraying with B. cinerea spore suspension (2 × 105 spores/mL) and photographed at 3 dpi. (B) In planta fungal growth in inoculated plants. Fungal growth was shown as ratios of the transcript level of B. cinerea BcActin to that of the Arabidopsis AtActin. (C) Relative expression of AtPDF1.2 in the mock- and B. cinerea-inoculated plants. qRT-RCR was performed using AtActin as an internal control to analyze the expression level of AtPDF1.2. (D) Accumulation of H2O2 in leaves of the in mock- and B. cinerea-inoculated plants, as revealed by DAB staining at 24 hpi. Experiments in (A,D) were repeated for three times with similar results, and results from one representative experiment are shown. Data presented in (B,C) are the means ± SE from three independent experiments and * or ** above the columns indicate significant differences at p < 0.05 or p < 0.01 levels (Student’s t-test), respectively, between the ClMBD2/3/5-OE plants and WT plants at the same time point.
To get insights in the possible mechanism of the attenuated B. cinerea resistance, the expression of a marker defense gene AtPDF1.2 and accumulation of reactive oxygen species (ROS) were analyzed and compared between the ClMBD2-OE, ClMBD3-OE, and ClMBD5-OE plants and WT plants after infection by B. cinerea. In mock-inoculated plants, the expression level of AtPDF1.2 in the ClMBD2-OE, ClMBD3-OE, and ClMBD5-OE plants was similar to that in WT plants (Figure 6C). At 24 hpi with B. cinerea, the expression level of AtPDF1.2 in the ClMBD2-OE, ClMBD3-OE, and ClMBD5-OE plants and WT plants were markedly up-regulated, as compared with those in the mock-inoculated plants; however, the expression level of AtPDF1.2 in the ClMBD2-OE, ClMBD3-OE, and ClMBD5-OE plants were significantly suppressed, resulting in a decrease of 73–81, 80–84, and 59–63%, respectively, as compared with that in Col-0 plants (Figure 6C). Similarly, no difference in accumulation of H2O2, as revealed by in situ DAB staining, was observed among the WT, ClMBD2-OE, ClMBD3-OE, and ClMBD5-OE plants without B. cinerea challenge (Figure 6D). At 24 hpi with B. cinerea, accumulation of H2O2 increased markedly in B. cinerea-infected leaves of the WT, ClMBD2-OE, ClMBD3-OE, and ClMBD5-OE plants, as compared with those in mock-inoculated controls (Figure 6D). However, more staining for H2O2 in B. cinerea-infected leaves of the ClMBD2-OE, ClMBD3-OE, and ClMBD5-OE plants was detected, as compared to that in WT plants (Figure 6D). These data indicate that overexpression of ClMBD2, 3, and 5 in transgenic Arabidopsis plants attenuates the B. cinerea-induced expression of defense genes but promotes the B. cinerea-induced ROS accumulation.
ClMBD1 and ClMBD2 Negatively but ClMBD5 Positively Regulate Arabidopsis Immunity Against Pseudomonas syringae pv. tomato DC3000
The possible involvement of the ClMBD genes in resistance against Pst DC3000, a hemibiotrophic pathogen that causes bacterial spot disease, was also investigated. At 3 dpi, typical Pst DC3000-provoked symptom with chlorotic lesions was seen in WT plants and the ClMBD-overexpressing plants (Figure 7A and Supplementary Figure 13A). The ClMBD1-OE and ClMBD2-OE plants displayed much severe disease with extensive chlorotic lesion while the ClMBD5-OE plants showed less severe disease (Figure 7A). Accordingly, the bacterial growth in the ClMBD1-OE and ClMBD2-OE plants was 0.43–0.69 order of magnitude higher while the growth in the ClMBD5-OE plants was ∼1.0 order of magnitude lower, as compared to that in WT at 3 dpi (Figure 7B). Disease severity and bacterial growth in the ClMBD3-OE, ClMBD4-OE, ClMBD6-OE, ClMBD7-OE, ClMBD8-OE, ClMBD9-OE, and ClMBD10-OE plants were indistinguishable to those in WT plants (Supplementary Figure 13B). These data indicate that overexpression of ClMBD1 and ClMBD2 leads to attenuated resistance while overexpression of ClMBD5 results in increased resistance against Pst DC3000.
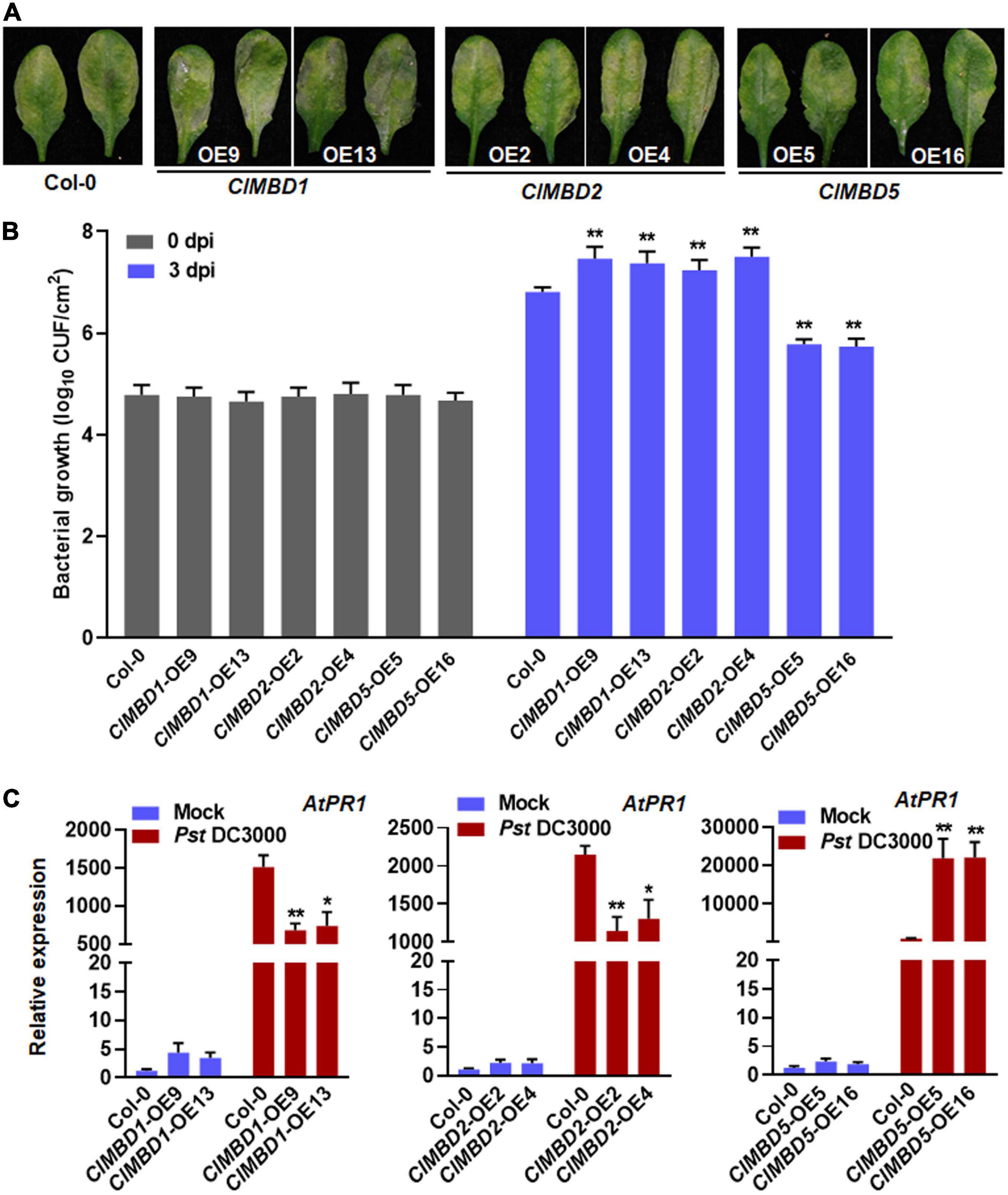
Figure 7. ClMBD1 and ClMBD2 negatively but ClMBD5 positively regulate resistance of the transgenic Arabidopsis plants against Pseudomonas syringae pv. tomato DC3000. (A) Typical P. syringae pv. tomato DC3000-caused disease on WT, ClMBD1-OE, ClMBD2-OE, and ClMBD5-OE plants. Four-week-old plants were inoculated by injecting with P. syringae pv. tomato DC3000 bacterial suspension (OD600 = 0.0002) and photographed at 72 hpi. (B) In planta bacterial growth in inoculated leaves. Leaf samples were collected at 0 and 3 dpi and bacterial growth in CFU/cm2 leaf area are shown. (C) Relative expression of AtPR1 in the mock- and P. syringae pv. tomato DC3000-inoculated plants. qRT-RCR was performed using AtActin as an internal control to analyze the expression level of AtPR1 Experiments in (A,B) were repeated for three times with similar results, and results from one representative experiment are shown. Data presented in (C) are the means ± SE from three independent experiments and * or ** above the columns indicate significant differences at p < 0.05 or p < 0.01 levels (Student’s t-test), respectively, between the ClMBD1/2/5-OE plants and WT plants at the same time point.
To gain insights in the possible mechanism of the altered Pst DC3000 resistance, the expression of a marker defense gene AtPR1 was analyzed and compared between the ClMBD1-OE, ClMBD2-OE, and ClMBD5-OE plants and WT plants after infection by Pst DC3000. In mock-inoculated plants, the expression level of AtPR1 in the ClMBD1-OE, ClMBD2-OE, and ClMBD5-OE plants was not significantly affected, as compared with that in WT plants (Figure 7C). At 24 hpi with Pst DC3000, the expression level of AtPR1 in the ClMBD1-OE, ClMBD2-OE, and ClMBD5-OE plants and WT plants were markedly up-regulated, as compared with those in the mock-inoculated plants (Figure 7C). However, the Pst DC3000-induced expression of AtPR1 in the ClMBD1-OE and ClMBD2-OE plants was significantly suppressed, resulting in a decrease of 51–55 and 39–47%, respectively, while the Pst DC3000-induced expression of AtPR1 in the ClMBD5-OE plants was markedly increased by ∼21-folds (Figure 7C). These data indicate that overexpression of ClMBD1 and ClMBD2 in transgenic Arabidopsis plants attenuates while overexpression of ClMBD5 strengthens the Pst DC3000-induced expression of defense genes.
Identification of Differentially Expressed Genes in ClMBD2-OE Plants
Considering that overexpression of ClMBD2 led to attenuated resistance against B. cinerea and Pst DC3000 (Figures 6, 7), transcriptome profiling of the ClMBD2-OE2 and WT plants grown under normal growth conditions was performed to gain insights into the possible molecular mechanisms of ClMBD2 in regulating resistance against the two pathogens. With criteria of expression change > 1.5-folds and P < 0.05, a total of 70 genes (52 up-regulated and 18 down-regulated) were found to be differentially expressed genes (DEGs) in the ClMBD2-OE2 plants as compared with WT plants (Supplementary Tables 5, 6). The DEGs in the ClMBD2-OE2 plants grown under normal condition were categorized into functional groups based on Gene Ontology (GO). DEGs that were up-regulated (Supplementary Table 5) or down-regulated (Supplementary Table 6) in the ClMBD2-OE2 plants were clustered into 31 (Supplementary Table 7) and 22 categories (Supplementary Table 8), respectively. Some overrepresented categories include genes involved in DNA binding, molecular transducer activity, and transcriptional factor activity in molecular function category, and response to stimulus, immune system process, signaling, and biological regulation in biological processes category (Figures 8A,B), implying that overexpression of ClMBD2 in transgenic Arabidopsis plants may affect the immune signaling and response. Among the DEGs (Supplementary Tables 5, 6), some genes have been previously reported to be involved in Arabidopsis immunity, including AtWRKY18 (Xu et al., 2006), AtWRKY30 (Zou et al., 2019), AtWRKY54 (Chen et al., 2021), AtANAC019 (Zheng et al., 2012), AtMLO6 (Acevedo-Garcia et al., 2017; Kuhn et al., 2017), and AtNATA1 (Lou et al., 2016). The expression patterns of 10 selected genes were further validated by qRT-PCR in ClMBD2-OE and WT plants before and after the infection of B. cinerea and Pst DC3000. In the ClMBD2-OE plants without pathogen infection, the expression levels of AtWRKY18, AtWRKY30, AtANAC019, AtARCK1, AtMLO6, and AtERF54 were significantly up-regulated while the expression levels of AtMAF5, AtBEE1, AtbZIP34, and AT5G52190 were markedly down-regulated (Figure 8C). After infection of B. cinerea, the expression of AtbZIP34 was down-regulated, while the expression of other genes was up-regulated in ClMBD2-OE and WT plants (Figure 8C). The expression levels of AtWRKY18 and AtbZIP34 were significantly increased, while the expression levels of AtANAC019 and AtBEE1 were significantly suppressed in ClMBD2-OE plants after infection of B. cinerea, as compared with those in WT plants (Figure 8C). After infection of Pst DC3000, the expression of AtWRKY18, AtWRKY30, AtANAC019, AtARCK1, AtERF54, AtMLO6, and AtMAF5 was up-regulated, but the expression of AtBEE1, AT5G52190, and AtbZIP34 was down-regulated in ClMBD2-OE and WT plants (Figure 8C). The expression levels of AtWRKY18, AtERF54, AtMAF5, AT5G52190, and AtBEE1 were significantly decreased in ClMBD2-OE plants after infection of Pst DC3000, as compared with those in the WT plants (Figure 8C). These data consistently conformed the results from RNA-seq analysis and indicate that ClMBD2 regulates a small set of defense and signaling genes that are involved in Arabidopsis immunity.
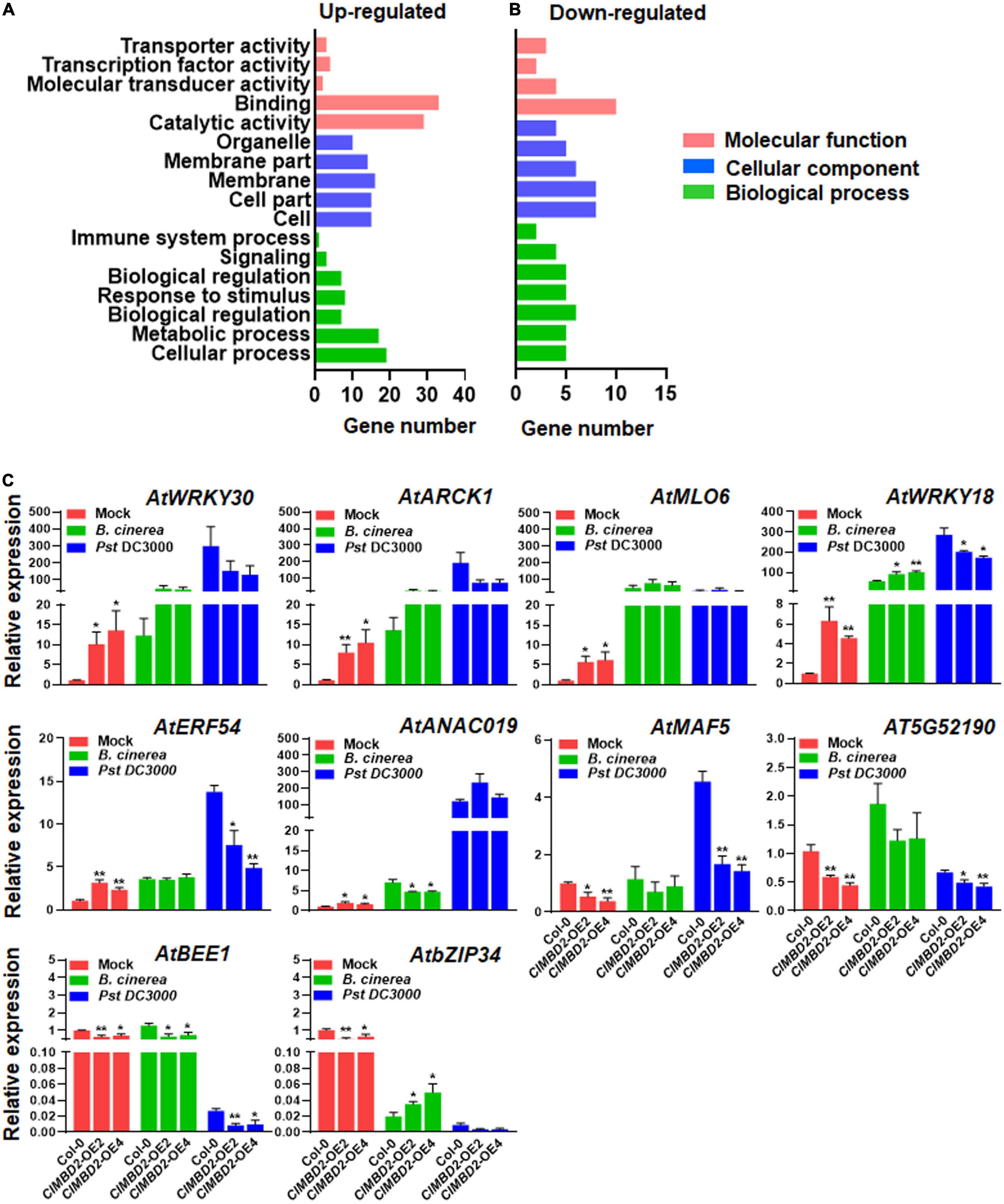
Figure 8. Differentially expressed genes in ClMBD2-OE2 plants. (A,B) Enriched GO terms of up-regulated (A) and down-regulated (B) genes in ClMBD2-OE2 plants, in comparison to WT plants. (C) qRT-PCR validation of expression patterns of selected differentially expressed genes in ClMBD2-OE and WT plants with or without infection of B. cinerea or Pst DC3000 at 24 hpi. Relative expression of the selected genes was analyzed by qRT-RCR using AtActin as an internal control. Data presented in (C) are the means ± SE from three independent experiments and * or ** above the columns indicate significant differences at p < 0.05 or p < 0.01 levels (Student’s t-test), respectively, between the ClMBD2-OE plants and WT plants.
Discussion
It has been documented that the MBD proteins play important roles in plant growth, development, and abiotic stress response; however, the involvement of the MBD proteins in plant immunity has not been established. The present study characterized the MBD families in watermelon and other cucurbit plants, examined the subcellular localization and binding activity of ClMBDs to 5-mC DNAs, analyzed the expression patterns of ClMBDs in response to defense hormones and pathogens, and explored the functions of ClMBDs in disease resistance. The functional analysis in transgenic Arabidopsis revealed that ClMBD1, 2, 3, and 5 play roles in immunity against B. cinerea and Pst DC3000, providing novel insights into the function of the MBD genes in plant immunity and a possibility to improve plant disease resistance through genetic manipulation of specific MBD genes.
The present study identified 10 watermelon ClMBD genes and 9, 10, 15, and 16 MBD genes in melon, cucumber, zucchini, and pumpkin, respectively (Table 1 and Supplementary Table 2). The numbers of ClMBDs in watermelon and MBD genes in other cucurbit plants are comparable to those of Arabidopsis (13), rice (17), maize (14), poplar (14), potato (15), tomato (18), and petunia (11) (Grafi et al., 2007; Parida et al., 2018; Shi et al., 2022). The presence of gene pairs in duplicated genomic regions of the watermelon genome and syntenic collinearity gene pairs between watermelon and other cucurbit plant species (Figures 1, 2 and Supplementary Tables 3, 4) suggests that gene duplication events occurred during the evolution of the ClMBD family. In addition to the typical MBD domain, other conserved domains such as zf-CW domain, SPARK domain, PKINase domain, and Bromo domain were also identified in some of the ClMBD proteins (Figure 1). Similar conserved domains are also present in Arabidopsis AtMBDs, tomato SlMBDs, and potato StMBDs (Grafi et al., 2007; Parida et al., 2018). It is thus likely that some watermelon ClMBD proteins may exert their functions in affecting transcription of target genes through different biochemical mechanisms including protein-protein interactions. Subcellular localization observations revealed that the ClMBD proteins were localized in nucleus when transiently expressed in N. benthamiana (Figure 3). This is consistent with the previous observations that most of the Arabidopsis AtMBD proteins displayed clear localization within the nucleus in onion cells (Berg et al., 2003; Ito et al., 2003) and that ZmMBD101 localized to nucleoplasmic foci (Questa et al., 2016). Another, ClMBD2 and ClMBD3 interacted with ClIDM2 and ClIDM3, and the interactions occurred in nucleus in BiFC assays (Figure 3). This feature is similar to the interaction of the Arabidopsis AtMBD7 with AtIDM2 and AtIDM3 (Lang et al., 2015), and further confirmed the nuclear localization of the ClMBD2 and ClMBD3. ClMBD2 is phylogenetically related to AtMBD5 and AtMBD6 and also shows an evolutionary syntenic relationship with AtMBD5 (At3G46580) and AtMBD6 (At5G59380) (Supplementary Table 4), implying that ClMBD2 may have a similar biochemical activity to AtMBD5 and AtMBD6. In the present study, ClMBD2 showed the ability to bind to mCG DNA (Figure 4), similar to AtMBD5 and AtMBD6, maize ZmMBD101, and tomato SlMBD5, which have the binding ability to mCG DNA (Ito et al., 2003; Scebba et al., 2003; Grafi et al., 2007; Li Y. et al., 2015; Questa et al., 2016). However, ClMBD2 did not bind to mCHH and mCHG DNA (Figure 4), different from AtMBD5 and AtMBD6, which also have the ability to bind to mCHH DNA (Ito et al., 2003; Scebba et al., 2003; Grafi et al., 2007). Surprisingly, the binding activity of the other ClMBD proteins to mCG DNA was not detected in the present study (Figure 4), implying that the ClMBD proteins may have different biochemical activities in recognizing methylated or unmethylated DNA and thus confer specific biological functions.
It was previously observed that the expression of some tomato SlMBD, wheat TaMBD and petunia PhMBD genes were affected by abscisic acid and abiotic stress, e.g., drought, salt, and cold stress (Li et al., 2008; Hu et al., 2011; Parida et al., 2018; Shi et al., 2022). The expression of most of the watermelon ClMBD genes was up-regulated after SA or MeJA treatment, except that ClMBD5 and ClMBD9 were down-regulated by MeJA and that the expression of ClMBD1 and ClMBD3 was not affected by MeJA (Figure 5). In response to Fon, the expression of almost all of the ClMBD genes in root tissues was up-regulated (Figure 5). By contrast, the expression of ClMBD3, 4, 7, and 9 was up-regulated, while the expression of ClMBD2, 6, 8, and 10 was down-regulated in response to Db (Figure 5). Notably, the expression changes of the ClMBD genes exhibited differential but inconsistent patterns in leaf and root tissues of watermelon plants in response to treatment of SA and MeJA and to infection of Fon and Db. However, the expression changes induced by the two defense hormones and the two fungal pathogens imply the involvement of the watermelon ClMBD genes in disease resistance, probably through affecting transcription of a set of genes including those involved in defense response.
The responsiveness of the watermelon ClMBD genes to exogenous SA and JA, two hormones that mediated defense response against (hemi)biotrophic pathogens such as Pst DC3000 and necrotrophic fungi like B. cinerea (Glazebrook, 2005; Grant and Jones, 2009) led to evaluate the disease resistance phenotype of the ClMBD-OE Arabidopsis lines against Pst DC3000 and B. cinerea. In the present study, ectopic overexpression of ClMBD2, 3, and 5 in transgenic Arabidopsis led to increased susceptibility to B. cinerea (Figure 7 and Supplementary Figure 12), suggesting that ClMBD2, 3, and 5 are negative regulators of defense response against B. cinerea. This is further supported by the suppression of pathogen-induced expression of defense gene AtPDF1.2, an indicator gene of defense response against necrotrophic fungal pathogens, and overaccumulation of ROS in ClMBD2-OE, ClMBD3-OE, and ClMBD5-OE plants (Figure 6). This is consistent with the general concept that excessive ROS accumulation during early stage often benefits the infection by the necrotrophic fungi like B. cinerea, but is different from the phenomenon that early ROS accumulation is critical to the activation of immune response against (hemi)biotrophic pathogens (Mengiste, 2012). On the other hand, overexpression of ClMBD1 and ClMBD2 in transgenic Arabidopsis attenuated while overexpression of ClMBD5 strengthened resistance to Pst DC3000 (Figure 7), indicating that ClMBD1 and ClMBD2 are negative regulators while ClMBD5 is a positive regulator of immunity against this bacterial pathogen. This is consistent with the suppression of expression of AtPR1 in ClMBD1-OE and ClMBD2-OE plants but elevation of expression of AtPR1 in ClMBD5-OE plants after infection of Pst DC3000 (Figure 7). This is also indirectly supported by the down-regulated expression of ClMBD2 in leaf tissues of watermelon plants after infection of Db, a heminecrotrophic fungal pathogen (Figure 5). Notably, overexpression of ClMBD2 in transgenic Arabidopsis resulted in attenuated immunity against both B. cinerea and Pst DC3000; however, overexpression of ClMBD5 led to opposite functions in immunity against these two pathogens, e.g., attenuated immunity against B. cinerea but strengthened immunity against Pst DC3000 (Figures 6, 7). It is generally accepted that immune response against (hemi)biotrophic pathogens such as Pst DC3000 is modulated through the SA signaling while the defense response against necrotrophic pathogens like B. cinerea is regulated by the JA/ET signaling (Glazebrook, 2005; Grant and Jones, 2009). Both antagonistic interaction and synergistic cross-talks between the SA and JA/ET signaling pathways occur and allow plants to mount appropriate immune responses against different invading pathogens (Glazebrook, 2005; Koornneef and Pieterse, 2008; Verhage et al., 2010). It is therefore likely that ClMBD2 and ClMBD5 function in immunity through regulating different mechanisms.
Transcriptome profiling identified a limited number of DEGs in the ClMBD2-OE2 plants grown under normal growth conditions (Supplementary Tables 5, 6). The fact that genes involved in DNA binding, transcriptional factor activity, response to stimulus, and immune system process were overrepresented in DEGs in ClMBD2-OE2 plants (Figures 8A,B) further confirms the function of ClMBD2 in immunity of the transgenic Arabidopsis plants against B. cinerea and Pst DC3000. Generally, the MBD proteins recognize the methylated CG sites and recruit chromatin remodelers and histone deacetylases to repress transcription of target genes (Lang et al., 2015). Considering the attenuated immunity against B. cinerea and Pst DC3000 (Figures 6, 7), it is speculated that overexpression of ClMBD2 should lead to down-regulation of a set of genes that are involved in Arabidopsis immunity. Surprisingly, only 18 genes were identified as down-regulated genes (expression change > 1.5-folds and P-value < 0.05) in the ClMBD2-OE2 plants (Supplementary Table 6). Among these down-regulated genes, AtWRKY54 was previously reported to function as a positive regulator of SARD1 and CBP60g expression in immunity against P. syringae pv. maculicola (Chen et al., 2021; Figure 8). No other gene with known function in Arabidopsis immunity was identified in the down-regulated genes in ClMBD2-OE2 plants (Supplementary Table 6). This might be due to the fact that samples from healthy ClMBD2-OE2 plants without pathogen infection were used for RNA-seq analysis. Indeed, the expression of defense genes such as AtPR1 and AtPDF1.2 was suppressed significantly in the ClMBD2-OE plants upon infection with Pst DC3000 and B. cinerea, respectively (Figures 6, 7). If it is the case that ClMBD2, like its closely related AtMBD5 and AtMBD6, acts to repress transcription of target genes, this may imply that the function of ClMBD2 in suppression of transcription of defense genes in transgenic Arabidopsis plants occurs upon pathogen infection. By contrast, some genes that negatively regulate Arabidopsis immunity were found to be up-regulated in the ClMBD2-OE2 plants (Figure 8 and Supplementary Table 5). For example, AtWRKY18 negatively regulates resistance against Pst DC3000 but positively modulates resistance to B. cinerea (Xu et al., 2006), AtANAC019 negatively regulates immune response through repressing AtICS1 and thus inhibiting SA accumulation (Zheng et al., 2012), AtMLO6 is a susceptible gene for powdery mildew disease (Acevedo-Garcia et al., 2017; Kuhn et al., 2017), and AtNATA1 negatively regulates immunity against Pst DC3000 by acetylating putrescine and decreasing ROS accumulation (Lou et al., 2016). It seems that overexpression of ClMBD2 activates an unknown pathway that up-regulates the expression of a subset of genes with negative functions in Arabidopsis immunity.
In summary, the present study characterized the watermelon ClMBD family and the MBD families in other cucurbit plants in terms of gene structures, conserved domain organization, phylogenetic and syntenic relationships, evolution events, subcellular localization, biochemical activity, and expression patterns in response to defense hormones and pathogen infection. The present study also provided the information on the possible involvement of each of the watermelon ClMBD genes in disease resistance when they were ectopically expressed in Arabidopsis. Functional analyses in transgenic Arabidopsis revealed that CMBD2, 3, and 5 negatively regulate Arabidopsis resistance against B. cinerea, and that ClMBD1 and ClMBD2 negatively while ClMBD5 positively regulate Arabidopsis resistance against Pst DC3000. Transcriptome analysis showed that overexpression of ClMBD2 in transgenic Arabidopsis affected the expression of a small set of genes that are involved in Arabidopsis immunity. Further analyzing the DNA methylation levels and characterizing the genome-wide binding sites in the ClMBD2-OE and ClMBD5-OE transgenic Arabidopsis plants will definitely provide detailed molecular mechanisms by which ClMBD2 and ClMBD5 regulate immunity against B. cinerea and Pst DC3000. Due to the divergence of gene functions in immunity between Arabidopsis and watermelon, the functional analysis in the present study performed by ectopic overexpression in Arabidopsis may not reflect the intrinsic functions of the ClMBD gene in watermelon disease resistance. Therefore, further investigations in watermelon through overexpression and CRISPR/Cas9-based knockout approaches will be critical to elucidate the functions and molecular mechanisms of the ClMBD genes, especially the ClMBD1, 2, 3, and 5 in disease resistance against Fon, Db, and other pathogens.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA803007.
Author Contributions
FS and DL conceived the project and designed the experiments. JL generated all material used in this study (cloning, vector, transformations, transgenic plants). JL, XL, YW, and XW performed the experiments and collected the data. FS, JL, and DL analyzed the data. FS and JL drafted the manuscript. All authors commented on the manuscript.
Funding
This work was financially supported by the Chinese Agriculture Research System of MOF and MARA of China (Grant No. CARS-25).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.886965/full#supplementary-material
Footnotes
- ^ https://www.arabidopsis.org
- ^ http://www.icugi.org/
- ^ http://pfam.xfam.org
- ^ http://smart.embl-heidelberg.de
- ^ http://expasy.org/
References
Acevedo-Garcia, J., Gruner, K., Reinstädler, A., Kemen, A., Kemen, E., Cao, L., et al. (2017). The powdery mildew-resistant Arabidopsis mlo2 mlo6 mlo12 triple mutant displays altered infection phenotypes with diverse types of phytopathogens. Sci. Rep. 7:9319. doi: 10.1038/s41598-017-07188-7
Arıkan, B., Özden, S., and Turgut-Kara, N. (2018). DNA methylation related gene expression and morphophysiological response to abiotic stresses in Arabidopsis thaliana. Environ. Exp. Bot. 149, 17–26. doi: 10.1016/j.envexpbot.2018.01.011
Berg, A., Meza, T. J., Mahiæ, M., Thorstensen, T., Kristiansen, K., and Aalen, R. B. (2003). Ten members of the Arabidopsis gene family encoding methyl-CpG-binding domain proteins are transcriptionally active and at least one, AtMBD11, is crucial for normal development. Nucleic Acids Res. 31, 5291–5304. doi: 10.1093/nar/gkg735
Cambiagno, D. A., Torres, J. R., and Alvarez, M. E. (2021). Convergent epigenetic mechanisms avoid constitutive expression of immune receptor gene subsets. Front. Plant Sci. 12:703667. doi: 10.3389/fpls.2021.703667
Chakrabarty, R., Banerjee, R., Chung, S. M., Farman, M., Citovsky, V., Hogenhout, S. A., et al. (2007). PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol. Plant-Microb. Interact. 20, 740–750. doi: 10.1094/MPMI-20-7-0740
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, S., Ding, Y., Tian, H., Wang, S., and Zhang, Y. (2021). WRKY54 and WRKY70 positively regulate SARD1 and CBP60g expression in plant immunity. Plant Signal. Behav. 16:1932142. doi: 10.1080/15592324.2021.1932142
Cheng, Y., Cheng, L., Cao, Q., Zou, J., Li, X., Ma, X., et al. (2020). Heterologous expression of SvMBD5 from Salix viminalis L. promotes flowering in Arabidopsis thaliana L. Genes 11, 285–197. doi: 10.3390/genes11030285
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coelho, F. S., Sangi, S., Moraes, J. L., Santos, W., Gamosa, E. A., Fernandes, K., et al. (2022). Methyl-CpG binding proteins (MBD) family evolution and conservation in plants. Gene 824:146404. doi: 10.1016/j.gene.2022.146404
Cokus, S. J., Feng, S., Zhang, X., Chen, Z., Merriman, B., Haudenschild, C. D., et al. (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219. doi: 10.1038/nature06745
Dowen, R. H., Pelizzola, M., Schmitz, R. J., Lister, R., Dowen, J. M., Nery, J. R., et al. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 109, E2183–E2191. doi: 10.1073/pnas.1209329109
Florea, L., Song, L., and Salzberg, S. L. (2013). Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2:188. doi: 10.12688/f1000research.2-188.v2
Gehring, M., Bubb, K. L., and Henikoff, S. (2009). Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324, 1447–1451. doi: 10.1126/science.1171609
Gigek, C. O., Chen, E. S., and Smith, M. A. (2016). Methyl-CpG-Binding Protein (MBD) Family: epigenomic read-outs functions and roles in tumorigenesis and psychiatric diseases. J. Cell. Biochem. 117, 29–38. doi: 10.1002/jcb.25281
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Grafi, G., Zemach, A., and Pitto, L. (2007). Methyl-CpG-binding domain (MBD) proteins in plants. Biochim. Biophys. Acta 1769, 287–294. doi: 10.1016/j.bbaexp.2007.02.004
Grant, M. R., and Jones, J. D. (2009). Hormone (dis)harmony moulds plant health and disease. Science 324, 750–752. doi: 10.1126/science.1173771
Gruenbaum, Y., Naveh-Many, T., Cedar, H., and Razin, A. (1981). Sequence specificity of methylation in higher plant DNA. Nature 292, 860–862. doi: 10.1038/292860a0
He, X. J., Chen, T., and Zhu, J. K. (2011). Regulation and function of DNA methylation in plants and animals. Cell Res. 21, 442–465. doi: 10.1038/cr.2011.23
Hewezi, T., Lane, T., Piya, S., Rambani, A., Rice, J. H., and Staton, M. (2017). Cyst nematode parasitism induces dynamic changes in the root epigenome. Plant Physiol. 174, 405–420. doi: 10.1104/pp.16.01948
Hu, Z., Yu, Y., Wang, R., Yao, Y., Peng, H., Ni, Z., et al. (2011). Expression divergence of TaMBD2 homoeologous genes encoding methyl CpG-binding domain proteins in wheat (Triticum aestivum L.). Gene 471, 13–18. doi: 10.1016/j.gene.2010.10.001
Huang, C. Y., and Jin, H. (2022). Coordinated epigenetic regulation in plants: A potent managerial tool to conquer biotic stress. Front. Plant Sci. 12:795274. doi: 10.3389/fpls.2021.795274
Ibarra, C. A., Feng, X., Schoft, V. K., Hsieh, T. F., Uzawa, R., Rodrigues, J. A., et al. (2012). Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337, 1360–1364. doi: 10.1126/science.1224839
Ichino, L., Boone, B. A., Strauskulage, L., Harris, C. J., Kaur, G., Gladstone, M. A., et al. (2021). MBD5 and MBD6 couple DNA methylation to gene silencing through the J-domain protein SILENZIO. Science 6549, 1434–1439. doi: 10.1126/science.abg6130
Ito, M., Koike, A., Koizumi, N., and Sano, H. (2003). Methylated DNA-binding proteins from Arabidopsis. Plant Physiol. 133, 1747–1754. doi: 10.1104/pp.103.026708
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Keinath, A. P. (2011). From native plants in Central Europe to cultivated crops worldwide: The emergence of Didymella bryoniae as a cucurbit pathogen. HortScience 46, 532–535. doi: 10.21273/HORTSCI.46.4.532
Koornneef, A., and Pieterse, C. M. (2008). Cross talk in defense signaling. Plant Physiol. 146, 839–844. doi: 10.1104/pp.107.112029
Kuhn, H., Lorek, J., Kwaaitaal, M., Consonni, C., Becker, K., Micali, C., et al. (2017). Key components of different plant defense pathways are dispensable for powdery mildew resistance of the Arabidopsis mlo2 mlo6 mlo12 triple mutant. Front. Plant Sci. 8:1006. doi: 10.3389/fpls.2017.01006
Lang, Z., Lei, M., Wang, X., Tang, K., Miki, D., Zhang, H., et al. (2015). The methyl-CpG-binding protein MBD7 facilitates active DNA demethylation to limit DNA hyper-methylation and transcriptional gene silencing. Mol. Cell 57, 971–983. doi: 10.1016/j.molcel.2015.01.009
Lang, Z., Wang, Y., Tang, K., Tang, D., Datsenka, T., Cheng, J., et al. (2017). Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 114, E4511–E4519. doi: 10.1073/pnas.1705233114
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Li, Q., Wang, X., Sun, H., Zeng, J., Cao, Z., Li, Y., et al. (2015). Regulation of active DNA demethylation by a methyl-CpG-binding domain protein in Arabidopsis thaliana. PLoS Genet. 11:e1005210. doi: 10.1371/journal.pgen.1005210
Li, Y., Deng, H., Miao, M., Li, H., Huang, S., Wang, S., et al. (2015). Tomato MBD5, a methyl CpG binding domain protein, physically interacting with UV-damaged DNA binding protein-1, functions in multiple processes. New Phytol. 210, 208–226. doi: 10.1111/nph.13745
Li, Y., Meng, F., Yin, J., Liu, H., Si, Z., Ni, Z., et al. (2008). Isolation and comparative expression analysis of six MBD genes in wheat. Bioch. Biophy. Acta 1779, 90–98. doi: 10.1016/j.bbagrm.2007.09.004
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lou, Y. R., Bor, M., Yan, J., Preuss, A. S., and Jander, G. (2016). Arabidopsis NATA1 acetylates putrescine and decreases defense-related hydrogen peroxide accumulation. Plant Physiol. 171, 1443–1455. doi: 10.1104/pp.16.00446
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Mengiste, T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294. doi: 10.1146/annurev-phyto-081211-172955
Michielse, C. B., and Rep, M. (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10, 311–324. doi: 10.1111/j.1364-3703.2009.00538.x
Moore, L. D., Le, T., and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38. doi: 10.1038/npp.2012.112
Ngou, B. P. M., Jones, J. D. G., and Ding, P. (2022). Plant immune networks. Trends Plant Sci. 27, 255–273. doi: 10.1016/j.tplants.2021.08.012
Nie, W. F., Lei, M., Zhang, M., Tang, K., Huang, H., Zhang, C., et al. (2019). Histone acetylation recruits the SWR1 complex to regulate active DNA demethylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 116, 16641–16650. doi: 10.1073/pnas.1906023116
Ohki, I., Shimotake, N., Fujita, N., Jee, J., Ikegami, T., Nakao, M., et al. (2001). Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell 105, 487–497. doi: 10.1016/s0092-8674(01)00324-5
Parida, A. P., Raghuvanshi, U., Pareek, A., Singh, V., Kumar, R., and Sharma, A. K. (2018). Genome-wide analysis of genes encoding MBD domain-containing proteins from tomato suggest their role in fruit development and abiotic stress responses. Mol. Biol. Rep. 45, 2653–2669. doi: 10.1007/s11033-018-4435-x
Parida, A. P., Sharma, A., and Sharma, A. K. (2017). AtMBD6, a methyl CpG binding domain protein, maintains gene silencing in Arabidopsis by interacting with RNA binding proteins. J. Biosci. 42, 57–68. doi: 10.1007/s12038-016-9658-1
Parida, A. P., Sharma, A., and Sharma, A. K. (2019). AtMBD4: A methylated DNA binding protein negatively regulates a subset of phosphate starvation genes. J. Biosci. 44:14.
Peng, M., Cui, Y., Bi, Y. M., and Rothstein, S. J. (2006). AtMBD9: a protein with a methyl-CpG-binding domain regulates flowering time and shoot branching in Arabidopsis. Plant J. 46, 282–296. doi: 10.1111/j.1365-313X
Qu, M., Zhang, Z., Liang, T., Niu, P., Wu, M., Chi, W., et al. (2021). Overexpression of a methyl-CpG-binding protein gene OsMBD707 leads to larger tiller angles and reduced photoperiod sensitivity in rice. BMC Plant Biol. 21:100. doi: 10.1186/s12870-021-02880-3
Questa, J. I., Rius, S. P., Casadevall, R., and Casati, P. (2016). ZmMBD101 is a DNA-binding protein that maintains Mutator elements chromatin in a repressive state in maize. Plant Cell Environ. 39, 174–184. doi: 10.1111/pce.12604
Rambani, A., Rice, J. H., Liu, J., Lane, T., Ranjan, P., Mazarei, M., et al. (2015). The methylome of soybean roots during the compatible interaction with the soybean cyst nematode. Plant Physiol. 168, 1364–1377. doi: 10.1104/pp.15.00826
Scebba, F., Bernacchia, G., De Bastiani, M., Evangelista, M., Cantoni, R. M., Cella, R., et al. (2003). Arabidopsis MBD proteins show different binding specificities and nuclear localization. Plant Mol. Biol. 53, 715–731. doi: 10.1023/B:PLAN.0000019118.56822.a9
Shi, L., Shen, H., Liu, J., Hu, H., Tan, H., Yang, X., et al. (2022). Exploration of the potential transcriptional regulatory mechanisms of DNA methyltransferases and MBD genes in petunia anther development and multi-stress responses. Genes 13:314. doi: 10.3390/genes13020314
Shi, R., Zhang, J., Li, J., Wang, K., Jia, H., Zhang, L., et al. (2016). Cloning and characterization of TaMBD6 homeologues encoding methyl-CpG-binding domain proteins in wheat. Plant Physiol. Biochem. 109, 1–8. doi: 10.1016/j.plaphy.2016.08.024
Song, Q., Li, D., Dai, Y., Liu, S., Huang, L., Hong, Y., et al. (2015). Characterization, expression patterns and functional analysis of the MAPK and MAPKK genes in watermelon (Citrullus lanatus). BMC Plant Biol. 15:298. doi: 10.1186/s12870-015-0681-4
Springer, N. M., and Kaeppler, S. M. (2005). Evolutionary divergence of monocot and dicot methyl-CpG-binding domain proteins. Plant Physiol. 138, 92–104. doi: 10.1104/pp.105.060566
Stangeland, B., Rosenhave, E. M., Winge, P., Berg, A., Amundsen, S. S., Karabeg, M., et al. (2009). AtMBD8 is involved in control of flowering time in the C24 ecotype of Arabidopsis thaliana. Physiol. Plant 136, 110–126. doi: 10.1111/j.1399-3054
Thordal-Christensen, H., Zhang, Z. G., Wei, Y. D., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1104/pp.123.4.1289
Verhage, A., van Wees, S. C., and Pieterse, C. M. (2010). Plant immunity: it’s the hormones talking, but what do they say? Plant Physiol. 154, 536–540. doi: 10.1104/pp.110.161570
Wang, C., Dong, X., Jin, D., Zhao, Y., Xie, S., Li, X., et al. (2015). Methyl-CpG-binding domain protein MBD7 is required for active DNA demethylation in Arabidopsis. Plant Physiol. 167, 905–914. doi: 10.1104/pp.114.252106
Wang, X., Basnayake, B. M., Zhang, H., Li, G., Li, W., Virk, N., et al. (2009). The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol. Plant-Microb. Interact. 22, 1227–1238. doi: 10.1094/MPMI-22-10-1227
Wang, Y., Tang, H., Tan, X., Li, J., and Wang, X. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wu, Z., Chen, S., Zhou, M., Jia, L., Li, Z., Zhang, X., et al. (2022). Family-wide characterization of methylated DNA binding ability of Arabidopsis MBDs. J. Mol. Biol. 434:167404. doi: 10.1016/j.jmb.2021.167404
Xiao, Y., Li, M., and Wang, J. (2022). The impacts of allopolyploidization on Methyl-CpG-Binding Domain (MBD) gene family in Brassica napus. BMC Plant Biol. 22:103. doi: 10.1186/s12870-022-03485-0
Xu, R., Wang, Y., Zheng, H., Lu, W., Wu, C., Huang, J., et al. (2015). Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J. Exp. Bot. 66, 5997–6008. doi: 10.1093/jxb/erv312
Xu, X., Chen, C., Fan, B., and Chen, Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18, 1310–1326. doi: 10.1105/tpc.105.037523
Yang, Y., Ahammed, G. J., Wan, C., Liu, H., Chen, R., and Zhou, Y. (2019). Comprehensive analysis of TIFY transcription factors and their expression profiles under jasmonic acid and abiotic stresses in watermelon. Int. J. Genom. 2019:6813086. doi: 10.1155/2019/6813086
Yong-Villalobos, L., González-Morales, S. I., Wrobel, K., Gutiérrez-Alanis, D., Cervantes-Peréz, S. A., Hayano-Kanashiro, C., et al. (2015). Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proc. Natl. Acad. Sci. USA 112, E7293–E7302. doi: 10.1073/pnas.1522301112
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Yu, A., Lepere, G., Jay, F., Wang, J., Bapaume, L., Wang, Y., et al. (2013). Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA 110, 2389–2394. doi: 10.1073/pnas.1211757110
Yuan, M., Ngou, B., Ding, P., and Xin, X. F. (2021). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62:102030. doi: 10.1016/j.pbi.2021.102030
Yuan, X., Wang, H., Cai, J. T., Bi, Y., Li, D. Y., and Song, F. M. (2019). Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 19:278. doi: 10.1186/s12870-019-1883-y
Zemach, A., and Grafi, G. (2003). Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J. 34, 565–572. doi: 10.1046/j.1365-313x.2003.01756
Zemach, A., Li, Y., Wayburn, B., Ben-Meir, H., Kiss, V., Avivi, Y., et al. (2005). DDM1 binds Arabidopsis methyl-CpG binding domain proteins and affects their subnuclear localization. Plant Cell 17, 1549–1558. doi: 10.1105/tpc.105.031567
Zhang, H., Hong, Y., Haung, L., Li, D., and Song, F. (2016). Arabidopsis AtERF014 acts as a dual regulator that differentially modulates immunity against. Sci. Rep. 2016:30251. doi: 10.1038/srep30251
Zhang, H., Lang, Z., and Zhu, J. K. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell. Biol. 19, 489–506. doi: 10.1038/s41580-018-0016-z
Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S. W., Chen, H., et al. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126, 1189–1201. doi: 10.1016/j.cell.2006.08.003
Zheng, X. Y., Spivey, N. W., Zeng, W., Liu, P. P., Fu, Z. Q., Klessig, D. F., et al. (2012). Coronatine promotes. Cell Host Microb. 2012, 587–596. doi: 10.1016/j.chom.2012.04.014
Keywords: watermelon (Citrullus lanatus L.), methyl-CPG-binding domain (MBD) protein, ClMBD2, disease resistance, DNA methylation, Arabidopsis
Citation: Liang J, Li X, Wen Y, Wu X, Wang H, Li D and Song F (2022) Genome-Wide Characterization of the Methyl CpG Binding Domain-Containing Proteins in Watermelon and Functional Analysis of Their Roles in Disease Resistance Through Ectopic Overexpression in Arabidopsis thaliana. Front. Plant Sci. 13:886965. doi: 10.3389/fpls.2022.886965
Received: 01 March 2022; Accepted: 20 April 2022;
Published: 09 May 2022.
Edited by:
Ivan Baccelli, Istituto per la Protezione Sostenibile delle Piante, Sede Secondaria Firenze, ItalyReviewed by:
Hui Wei, Nanjing Forestry University, ChinaYuanzheng Yue, Nanjing Forestry University, China
Xiaomei Dong, Shenyang Agricultural University, China
Copyright © 2022 Liang, Li, Wen, Wu, Wang, Li and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dayong Li, ZHlsaUB6anUuZWR1LmNu; Fengming Song, Zm1zb25nQHpqdS5lZHUuY24=
 Jiayu Liang
Jiayu Liang Xiaodan Li
Xiaodan Li Hui Wang
Hui Wang Dayong Li
Dayong Li Fengming Song
Fengming Song