
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 29 April 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.884990
This article is part of the Research Topic Physiological, Molecular and Genetic Perspectives of Chilling Tolerance in Horticultural Crops, Volume II View all 9 articles
The ALAD gene encodes an enzyme that is essential for chlorophyll biosynthesis and is involved in many other physiological processes in plants. In this study, the CaALAD gene was cloned from pepper and sequenced. Multiple sequence alignment and phylogenetic analysis of ALAD proteins from nine plant species showed that ALAD is highly conserved, and that CaALAD shows the highest homology with the ALAD protein from eggplant. Subcellular localization indicated that the CaALAD protein is mainly localized to the chloroplasts. After transferring CaALAD into the Arabidopsis thaliana genome, cold tolerance of the transgenic lines improved. Overexpression of CaALAD increased the relative transcription of the AtCBF2, AtICE1, and AtCOR15b genes in transgenic Arabidopsis plants exposed to low temperature (4°C) stress, and the contents of reactive oxygen species decreased due to increased activities of superoxide dismutase, peroxidase, and catalase. Moreover, chlorophyll biosynthesis, as determined by the contents of porphobilinogen, protoporphyrin IX, Mg-protoporphyrin IX, prochlorophyllate, and chlorophyll in the transgenic Arabidopsis plants, increased in response to low temperature stress. In addition, the transgenic lines were more sensitive to exogenous ALA and NaHS, and the H2S content of transgenic line plants increased more rapidly than in the wild-type, suggesting that CaALAD may respond to low temperatures by influencing the content of H2S, a signaling molecule. Our study gives a preliminary indication of the function of CaALAD and will provide a theoretical basis for future molecular breeding of cold tolerance in pepper.
Pepper is an important vegetable crop and condiment, and the normal growth temperature of pepper plants is 20–30°C (Guo, 2013). Growth and development are hindered if the temperature is below 15°C, and plants will be seriously damaged or can die if the temperature falls below 5°C (Wang et al., 2021a). In the winter and spring in northern China, low temperatures are the main abiotic stress that limits the growth, development, and yield of pepper (Tang et al., 2021). Low temperatures will reduce stomatal conductance, change the biological characteristics of thylakoid membranes, reduce chlorophyll content, and limit photosynthetic electron transfer, thus reducing the photosynthetic capacity of plants (Allen and Ort, 2001; Tang et al., 2021). Low temperatures can also lead to the production of reactive oxygen species (ROS), which cause oxidative damage to plants (Wang et al., 2021b).
In order to resist the effects of low temperature, plants have evolved complex response mechanisms to low temperature stress, and cold-resistance genes play a key role. Cold tolerance in plants is regulated by multiple genes, and the low-temperature response transcription network composed of ICE1-CBF-COR is the most widely studied pathway in the plant response to cold. Previous studies have shown that expression of three members of the CBF gene family, CBF1, CBF2, and CBF3, can be induced briefly and rapidly by low temperature stress (Medina et al., 1999; Gilmour et al., 2010). The induced CBF transcription factor proteins can bind to the cis-elements present in the COR gene promoter and activate COR gene expression. ICE is a transcriptional activator that induces the expression of the CBF gene family at low temperatures. ICE can specifically bind to the promoter sequence of CBF at low temperatures to induce the expression of CBF, and the induced CBF protein then binds to the DRE sequence in the promoter of its downstream target gene to induce the expression of COR, thus improving frost tolerance in plants (Tang et al., 2020).
Our previous studies showed that 5-aminolevulinic acid (ALA) and hydrogen sulfide enhanced the photosynthesis and antioxidant capacity of cold-sensitive pepper plants (Wang et al., 2021b). We found that ALA and H2S significantly induced the expression of ALAD in response to chilling stress (Wang et al., 2021b). Therefore, we hypothesized that ALAD might be involved in regulating the resistance to low temperature stress in pepper.
The non-proteinogenic amino acid ALA is a common precursor in the biosynthesis of tetrapyrroles in plants, and ALA is considered to be a new plant growth regulator (Wu et al., 2018b). ALA can regulate plant growth and development, vegetative growth, seed germination, and fruit coloring (Zhang, 2010; Zhao et al., 2013; Kobayashi and Masuda, 2016; Wu et al., 2018a,b). In addition, ALA regulates certain metabolic processes, such as chlorophyll, heme, and siroheme biosynthesis, and alleviates abiotic stress by regulating photosynthesis, nutrient absorption, antioxidant defense, and osmotic regulation (Kim et al., 2014; Wu et al., 2018b). 5-aminolevulinate dehydratase (ALAD) is a necessary enzyme for the biosynthesis of tetrapyrroles, and it catalyzes the condensation of two molecules of ALA to form porphobilinogen (PBG; Yang, 2008). Therefore, the activity of ALAD directly determines the metabolism and accumulation of ALA in plants (Long et al., 2013). Moreover, ALA does not generally accumulate in developing leaves, but is directly transformed into PBG by ALAD (Killiny et al., 2018). At present, the ALAD gene has been successfully cloned from spinach (Berglund and Tegenfeldt, 1992), soybean (Boese et al., 1991), tomato (Polking et al., 1995), pea (Cheung et al., 1997), and some other plant species. Previous studies have shown that the ALAD gene sequence and structure are highly conserved (Yang, 2008; Lu, 2013).
The activity of ALAD is affected by temperature, pH, sugars, and mercaptan (Yang, 2008; Xu, 2014). D-fructose and D-glucose inhibit ALAD activity (Lee et al., 2003; Yang, 2008). Low concentrations of dithiothreitol can improve ALAD activity, but high concentrations can inhibit activity (Xu, 2014). Osmotic stress induced by sorbitol was found to inhibit ALAD activity in maize leaves (Jain et al., 2018). Also, ALAD activity strongly depends on the binding of metal ions (including Zn2+, Mn2+, and Mg2+). In vitro assays of ALAD enzyme activity found that Ca2+, Mn2+, and K+ significantly increase activity, while Cu2+ significantly inhibits the activity of ALAD (Long et al., 2013). In cucumber, it was found that Al3+ can significantly inhibit ALAD activity and growth in cucumber. Al3+ may affect the growth and development of plants by forming complexes with nucleotides, cell walls, and other biomolecules (Pereira et al., 2006).
Some previous studies have shown that the ALAD protein plays important roles in plant growth, development, and stress resistance. Lu (2013) found that in garlic, changes in ALAD activity were consistent with the PBG content and the degree to which the plants turn green through ALAD activity, and also the relative expression of ALAD in different varieties of garlic. The expression level of ALAD was found to be much higher at low temperature than at normal temperature (Lu, 2013). Chai et al. (2017) found that the cotton lesion mimic mutant Ghllm, which results from a mutation in a gene encoding ALAD, causes necrotic spots on cotton leaves (Chai et al., 2017). This mutation leads to the accumulation of ALA, which in turn leads to the production of ROS, and thus induces the expression of GhEDS1, GhPAL, and GhPAD4, leading to an increase in the salicylic acid content (Chai et al., 2017). Also, the expression of SA leads to the increased expression of the PR gene, thus increasing the resistance of cotton to verticillium wilt (Chai et al., 2017). ALAD1 overexpression in wheat improved ALA tolerance in tobacco plants (Yu et al., 2011). In addition, silencing of the ALAD gene can lead to plant stress, resulting in yellow necrotic spots on citrus leaves and stems, decreased levels of chlorophyll, starch, sucrose, trans-violet and cis-violet xanthin, α and β cryptoxanthin, and increased levels of zeaxanthin (Killiny et al., 2018). Studies in Arabidopsis using RNAi gene silencing and HEMB1 (ALAD) mutants showed that the loss or reduction of HEMB1 can seriously affect plant growth and development, resulting in seedling and embryo death (Tang et al., 2012). Wittmann et al. (2018) found that silencing the TRX-M1/M2/M4 thioredoxin (TRX) genes leads to decreased stability of ALAD, and that TRX in chloroplasts can interact with ALAD to improve its activity in Arabidopsis (Wittmann et al., 2018). FHY3/FAR1 binds with the promoter of ALAD to initiate the expression of ALAD and enhance its activity, thus affecting the biosynthesis of chlorophyll (Tang et al., 2012). The results of our previous study also showed that the transcription of ALAD (HEMB) was induced by treatment with exogenous ALA and hydrogen sulfide in pepper seedlings under low temperature stress (Wang et al., 2021b). Therefore, it is necessary to understand the molecular mechanisms that underly cold tolerance mediated by ALAD in pepper.
In this study, the CaALAD gene was cloned from pepper and expressed in Arabidopsis to study its effect on low temperature stress tolerance. The relative cold tolerance of transgenic Arabidopsis plants overexpressing the pepper ALAD gene was determined by phenotypic and physiological indicators, and the results will establish a genetic foundation for the study of cold tolerance in pepper.
Wild type (WT) Col-0 and CaALAD-overexpressing Arabidopsis lines, the pepper cultivar “Hangjiao 2” (Purchased from Shenzhou Lvpeng Company, Tianshui, China) and Nicotiana benthamiana were used in this experiment. Arabidopsis seeds were sown on substrate (a 1:1 mixture of grass charcoal soil and vermiculite) or sterile Murashige and Skoog (MS) medium and grown at 23°C and 65% relative humidity with a 16 h/8 h (light/dark) photoperiod and a light intensity of 200 μmol m–2 s–1 (RT). The pepper seeds were spread on a wet towel and incubated in the dark for 3–5 day at a constant temperature of 28°C. After the seeds had germinated, they were planted in a substrate consisting of a 1:3 mixture of vermiculite and cultivation substrate. N. benthamiana seedlings were grown on the same mixed substrate as pepper under the following conditions: relative humidity 65%; temperature and photoperiod = 28°C/18°C (12 h light/12 h dark); and light intensity = 300 μmol m–2 s–1.
Arabidopsis seeds are rigorously sterilized before sowing on MS medium. WT and transgenic Arabidopsis seeds were surface sterilized with 75% alcohol for 1 min, washed with sterile water once and then treated with 1% sodium hypochlorite for 10 min, washed with sterile water five times, and cultured on MS medium with or without 50 mg/L kanamycin antibiotic. Kanamycin was used to screen genetically modified seeds for the presence of the Kmr gene that is linked to the CaALAD transgene, and the third generation of the transgenic lines (T3) that were screened were used in the cold tolerance tests.
The seeds of WT and transgenic Arabidopsis were grown on MS medium for 1 week and transferred to square petri dishes. The seeds were cultured vertically at 4°C for another week and the effects of low temperature on the growth of the plants were then observed. The 25-day-old WT and transgenic seedlings which were sown on substrate were transferred to 4°C (LT) for 7 days. Samples were taken at different times during this period to determine the various indicators of cold stress or tolerance. Seedlings grown under regular conditions (23°C, RT) were used as the controls.
To clone CaALAD from C. annuum, the ALAD gene sequence of Arabidopsis was downloaded from the TAIR database1, this sequence was used as a query in BLAST searches of the pepper database; the gene with the highest sequence homology to Arabidopsis ALAD was named CaALAD. A pair of gene-specific primers, TL-F and TL-R, were designed using Primer 5 based on the CaALAD sequence (Table 1). The full-length cDNA of CaALAD was obtained via PCR amplification using C. annuum total RNA, the gene-specific primer pair, and Tks Gflex DNA Polymerase (TaKaRa, Japan). The PCR products were then cloned into the pEASY ® -Blunt Simple Cloning Vector (TransGene, Beijing, China) and transferred into Escherichia coli strain Trans1-T1 (Transgene) for DNA sequencing.
The amplified target DNA fragment was cloned into the pBI121 vector by homologous recombination, and the restriction sites on the vector were Xbal and BamH1. Recombinant pBI121-CaALAD plasmids were introduced into the Agrobacterium strain GV3101. Agrobacterium-mediated transformation was performed to produce T0-generation transgenic lines using the floral dip method (Martins et al., 2015). Transgenic Arabidopsis T1-generation seeds were screened on solid MS medium containing 50 mg/L kanamycin. Homozygous T3-generation lines were used in the experiments.
The homologous ALAD genes of tomato (Solanum lycopersicum), potato (Solanum tuberosum), eggplant (Solanum melongena), tobacco (Nicotiana tabacum), Arabidopsis thaliana, rice (Oryza sativa), wheat (Triticum aestivum), and maize (Zea may) were obtained by BLAST searches of the NCBI database using the deduced protein sequence of CaALAD as the query. The phylogenetic relationships among the predicted ALAD proteins of these nine species were analyzed using DNAMAN 6.0 (Lynnon Biosoft, Montreal, Canada). A phylogenetic analyses was performed using the neighbor-joining method as implemented in MEGA 6.0 with 1,000 bootstrap replicates (Taghizadeh et al., 2020). The functional domains and tertiary structure of CaALAD were predicted using the conserved domain database (CDD)2, and SWISS-MODEL3.
To determine the subcellular localization of CaALAD, we cloned CaALAD (without the termination codon) into the pYBA1132 vector to give an in-frame fusion with the EGFP reporter protein gene. As with the pBI121 vector, the restriction sites on the pYBA1132 vector were also Xbal and BamH1. The p35S:CaALAD-EGFP and 35S:EGFP control plasmids were transformed into Agrobacterium GV3101, and the recombinant strains were infiltrated into 6–8 tobacco (Nicotiana tabacum) leaves with a syringe as described previously (Sparkes et al., 2006). The EGFP signals in leaf epidermal cells were subsequently observed under a fluorescence microscope (Zeiss Vert.A1, Oberkochen, Germany).
Total RNA was extracted from plant tissues using an RNA Extraction Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. First-strand cDNA was synthesized from total RNA using EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgene, Beijing, China). The AtUBQ4 gene was used as the reference for normalization of gene expression. The primer sequences used for amplification of AtUBQ4, CaALAD, and other cold-tolerance genes (AtCBF1, AtCBF2, AtCOR15a, AtCOR15b, AtICE1, and AtICE2) are given in Table 2 (Wang et al., 2021b). All oligonucleotide primers were synthesized by Shenggong (Shanghai, China).
To quantify the expression of CaALAD in transgenic Arabidopsis plants overexpressing the pepper ALAD gene, RNA extracted from the transgenic and wild-type (WT) plants was amplified by PCR using RT-PCR primers specific for CaALAD (HEMB) and reference primers for the Arabidopsis gene AtUBQ4, followed by agarose gel electrophoresis. The annealing temperature was set to 58°C, and 32 cycles of amplification were used.
The contents of Protoporphyrin IX (Proto IX), Mg-protoporphyrin IX (Mg-proto IX), Protochlorophyllide (Pchl), and Chl in WT and transgenic plants were determined at 0 days and 7 days after low temperature (4°C) treatment. The methods used are the same as those used in our previous study (Wang et al., 2021b).
The PBG contents of WT and transgenic plants were determined at 0, 1, and 7 days after low temperature treatment. The determination of PBG content was based on the method of Kayser et al. (2005) with slight modifications. The details are as follows: 1.0 g of leaf blade was ground in an ice bath with 4 mL of 0.05M Tris–HCl (pH = 8.0), and vortex mixed. The slurry was centrifuged at 12,000 rpm at 4°C for 10 min, and the supernatant was absorbed into a new EP tube. One milliliter of ALA buffer (0.15 g ALA dissolved in 50 mL 0.05 mol/L Tris–HCl, pH 8.0) was added, and the mixture was mixed well. One volume of Ehrlich’s reagent (84 mL glacial acetic acid, 16 mL 70% perchloric acid, 2 g dimethylaminobenzaldehyde) was added and the solution was incubated in the dark for 15 min. The tube was then centrifuged at 12,000 rpm for 10 min, and the absorbance was then measured at 555 nm. The concentration was calculated according to the molar extinction coefficient of PBG (6.1 × 104 mol–1 cm–1). PBG concentration = A552/(molar extinction coefficient × diameter).
The H2O2 and O2•– contents were determined by the nitroblue tetrazolium [NBT, 0.1% (w/v)] and 3,3-diaminobenzidine (DAB, 1 mg/ml) uptake methods, respectively, as previously described (Xianyang et al., 2011; Ma et al., 2018). The contents of H2O2 and superoxide anion O2•–, as well as the activities of the antioxidant enzymes superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) were determined before and after 1 day of low temperature stress. These indicators were measured using kits purchased from Comin Biotechnology (Suzhou, China) following the manufacturer’s instructions.
T3-generation transgenic and WT seeds were soaked in solutions containing 100 μmol/L NaHS, 500 μmol/L NaHS, and 1,000 μmol/L NaHS for 12 h. The transgenic and WT seeds were soaked in 1 mg/L ALA, 10 mg/L ALA, and 100 mg/L ALA for 24 h, then washed once with sterile water. The seeds were sterilized with 75% alcohol for 1 min, washed once with sterile water, and sterilized with 1% sodium hypochlorite for 10 min. After washing with sterile water five times, the seeds were sown on MS medium and the germination rate was calculated after 5 days.
The H2S contents of the WT and transgenic plants were determined after 0, 2, 6, 12, and 24 h at 4°C using a kit obtained from Comin Biotechnology (Suzhou, China).
All experiments consisted of three independent replicates, and SPSS 22.0 (SPSS Institute Inc., United States) was used to determine statistical significance at *p < 0.05 and **p < 0.01; Student’s t-test was used to analyze the significance difference of the means. Origin 9 software (OriginLab Institute Inc., United States) was used for drawing the figures.
A phylogenetic tree derived from a multiple protein sequence alignment showed that the ALAD proteins are highly homologous in the nine plant species examined (Figure 1A). As shown in Figure 1B, the pepper ALAD protein is most closely related to the protein from eggplant; the ALAD proteins from five species in the Solanaceae plus Arabidopsis form a well-supported clade, while the proteins from three monocot species form a second well-supported clade, which also indicates that ALAD is a highly evolutionarily conserved protein in higher plants. The domain prediction results show that the CaALAD contains a single ALAD domain, which implies that CaALAD belongs to the ALAD family (Figure 1C). As with the domain prediction result, the predicted tertiary structure also shows that the CaALAD protein consists of one main part, and there are many folding and crimping in the protein (Figure 1C).

Figure 1. Sequence alignment and phylogenetic relationship between CaALAD and its homologous genes from other species. (A) ALAD protein sequences alignment of the pepper (CaALAD), tomato (SlALAD), potato (StALAD), eggplant (SmALAD), tobacco (NaALAD), Arabidopsis (AtALAD), rice (OsALAD), wheat (TaALAD), and maize (ZmALAD). Identical residues are displayed by mazarine. (B) Phylogenetic tree of CaALAD and its homologous proteins from tomato, potato, eggplant, tobacco, Arabidopsis, rice, wheat, and maize. (C) The predicted CaALAD functional domain and tertiary structure.
The subcellular localization analysis showed that the control EGFP fluorescence signal was evenly distributed throughout the N. benthamiana leaf epidermal cells, while the signal from the CaALAD-EGFP fusion protein was localized to the chloroplasts (Figure 2).
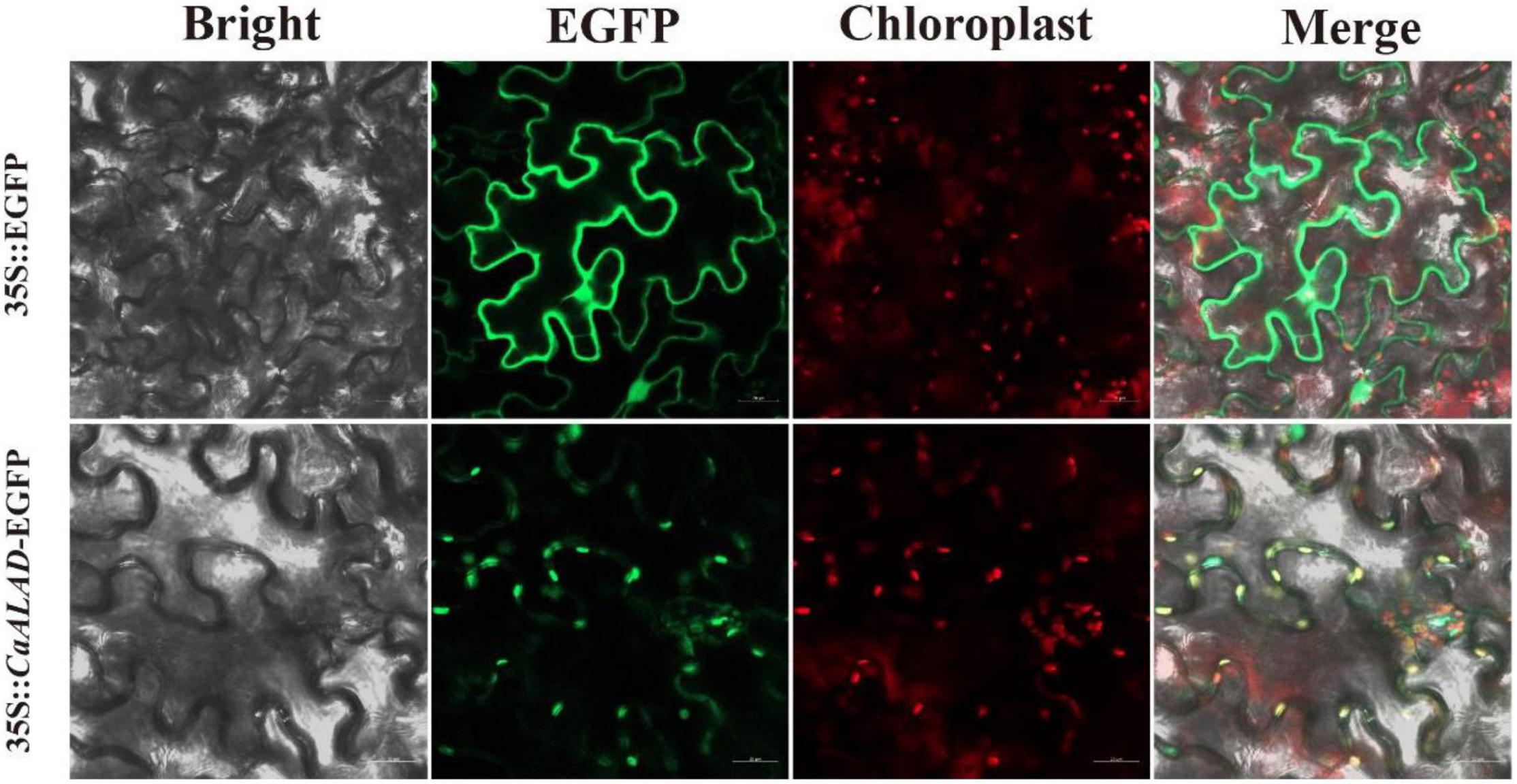
Figure 2. Subcellular localization of CaALAD in N. benthamiana leaf cells. Fluorescence microscopy of the 35S:CaALAD-EGFP and 35S:EGFP (control) plasmids. Scale bars = 20 μm.
In order to examine the expression of CaALAD in Arabidopsis, the reference gene AtUBQ4 and the CaALAD-specific primers were used to conduct semi-quantitative PCR using cDNA of the WT and CaALAD-overexpressing transgenic plants as the templates. The results are shown in Figure 3A; there is no band from the amplification of WT Arabidopsis cDNA with the primer pair specific for CaALAD. The bands amplified with the AtUBQ4 primers were clear, and the CaALAD-overexpressing lines had clear bands amplified with the CaALAD-specific primers. This indicates that the CaALAD gene is successfully transcribed in the transgenic Arabidopsis plants.
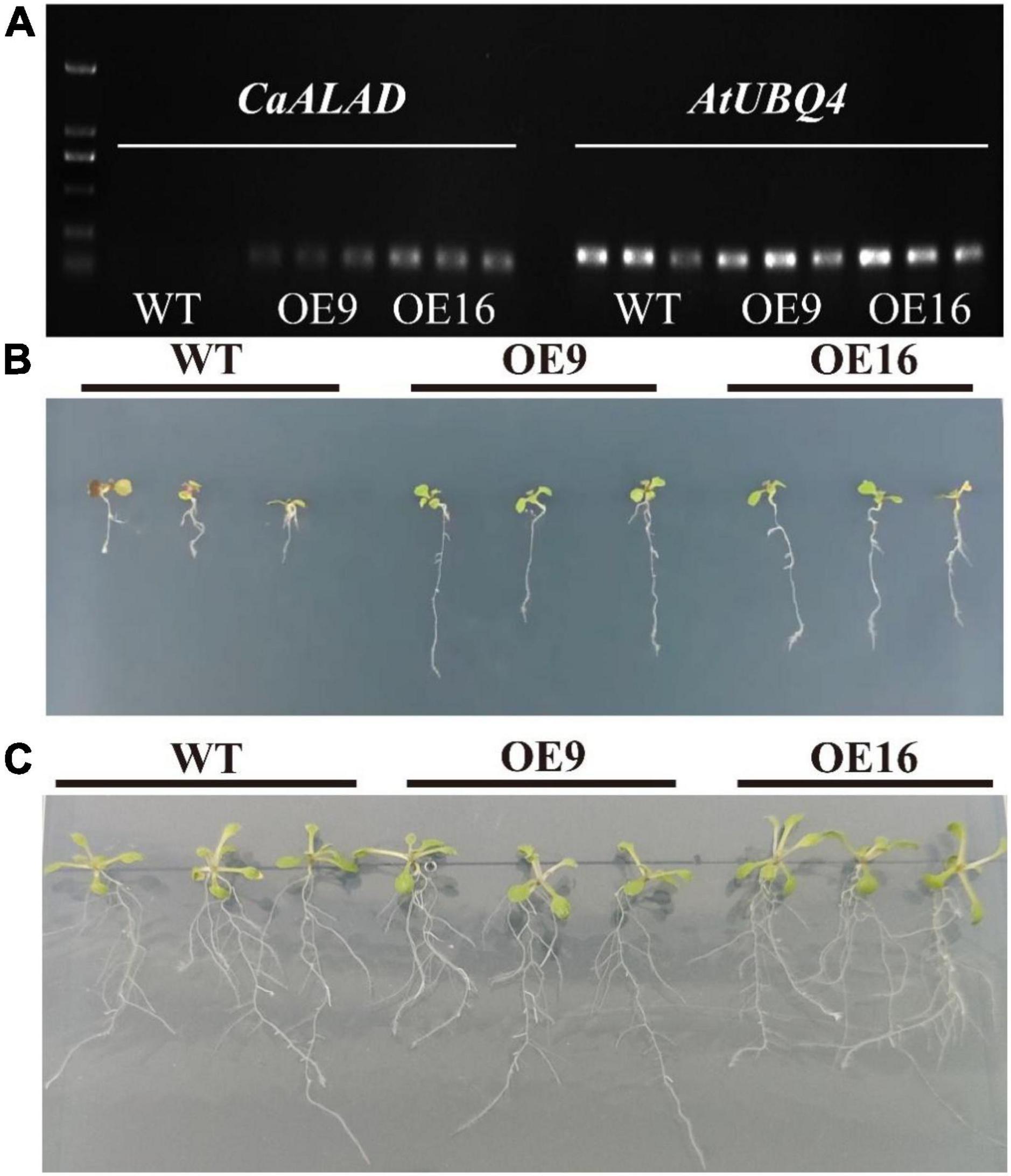
Figure 3. Effects of the overexpression of CaALAD on growth in transgenic Arabidopsis Col-0 plants exposed to low temperature stress for 1 week. WT: wild-type Arabidopsis thaliana Col-0. OE: the CaALAD-overexpressing (OE) lines. (A) Electrophoresis results of CaALAD in A. thaliana by semi-quantitative PCR. (B) The phenotypes of WT and OE plants that were exposed to low temperature stress at 4°C for 1 week. (C) The phenotypes of WT and OE plants grown at 23°C for 1 week.
The growth of transgenic plants after 1 week of exposure to low temperature stress compared to normal culture conditions is shown in Figures 3B,C. However, the growth of CaALAD-overexpressing transgenic plants was significantly better than that of WT plants after low temperature stress, and the transgenic plants were less damaged by exposure to low temperature than were the WT plants.
In order to identify the effect of CaALAD on cold tolerance in transgenic Arabidopsis, the relative expression of six cold stress-related genes, AtCBF1, AtCBF2, AtICE1, AtICE2, AtCOR15a, and AtCOR15b, was measured at different times during low temperature stress (Figure 4). In response to low temperature stress, the transcription of AtCBF1 in both WT and the CaALAD transgenic lines increased sharply at first, then decreased, and reached its highest expression level at 2 h (Figure 4A). At 0 h, the expression level of AtCBF1 in plants of the two transgenic lines was only 25% of that in WT plants (Figure 4A). AtCBF1 expression in the WT decreased rapidly by 97.3% at 12 h under low temperature stress compared with 6 h, but it decreased by 34.1 and 4.8% in OE9 and OE16, respectively, (Figure 4A). The expression profiles of AtCBF2 were similar to those of AtCBF1. The relative expression of AtCBF2 also increased rapidly at 2 h and then gradually decreased from 6 to 48 h (Figure 4B). The initial expression of AtCBF2 in the transgenic plants at 0 h was the 2-fold higher than in WT (Figure 4B). At 2 h, the expression of AtCBF2 in the OE9 and OE16 lines increased by 1.1- and 1.6-fold, respectively, compared with WT (Figure 4B). At 12 h, the expression of AtCBF2 in WT decreased drastically by 96% compared with 6 h, while the expression of AtCBF2 in OE9 and OE16 decreased by only 53.8 and 63%, respectively, (Figure 4B). After low temperature stress, the expression of AtICE1 in WT decreased, while the expression of AtICE1 in the plants of both transgenic lines increased at 2 h, then decreased, and then increased at 24 h (Figure 4C). At 2 h, the expression of AtICE1 in OE9 and OE16 plants increased 11.2- and 10.9-fold, respectively, compared with WT (Figure 4C). During low temperature stress, the relative expression of AtICE2 in the WT increased first and decreased significantly after 6 h (Figure 4D). However, the relative expression of AtICE2 in the transgenic plants showed a more gradual decrease from 6 to 12 h of low temperature stress, then increased by 2- to 3-fold at 24 h, before decreasing again at 48 h (Figure 4D). At 0 h, the relative expression of AtICE2 in OE9 and OE16 plants increased 1.9- and 2.1-fold, respectively, compared with WT (Figure 4D). The relative expression of AtCOR15a in both WT and transgenic plants increased rapidly, beginning at 2 h of low temperature stress, but the expression in the transgenic plants decreased at 48 h while expression in the WT peaked at 48 h (Figure 4E). In response to low temperature stress treatment, the relative expression of AtCOR15b in WT and transgenic plants showed similar profiles in that first expression increased and then decreased (Figure 4F). The expression level of AtCOR15b in the WT peaked at 6 h, while it peaked in the OE lines at 12 h. Relative expression of AtCOR15b was significantly higher in the transgenic plants at 2, 6, 12, and 24 h than in the WT (Figure 4F).
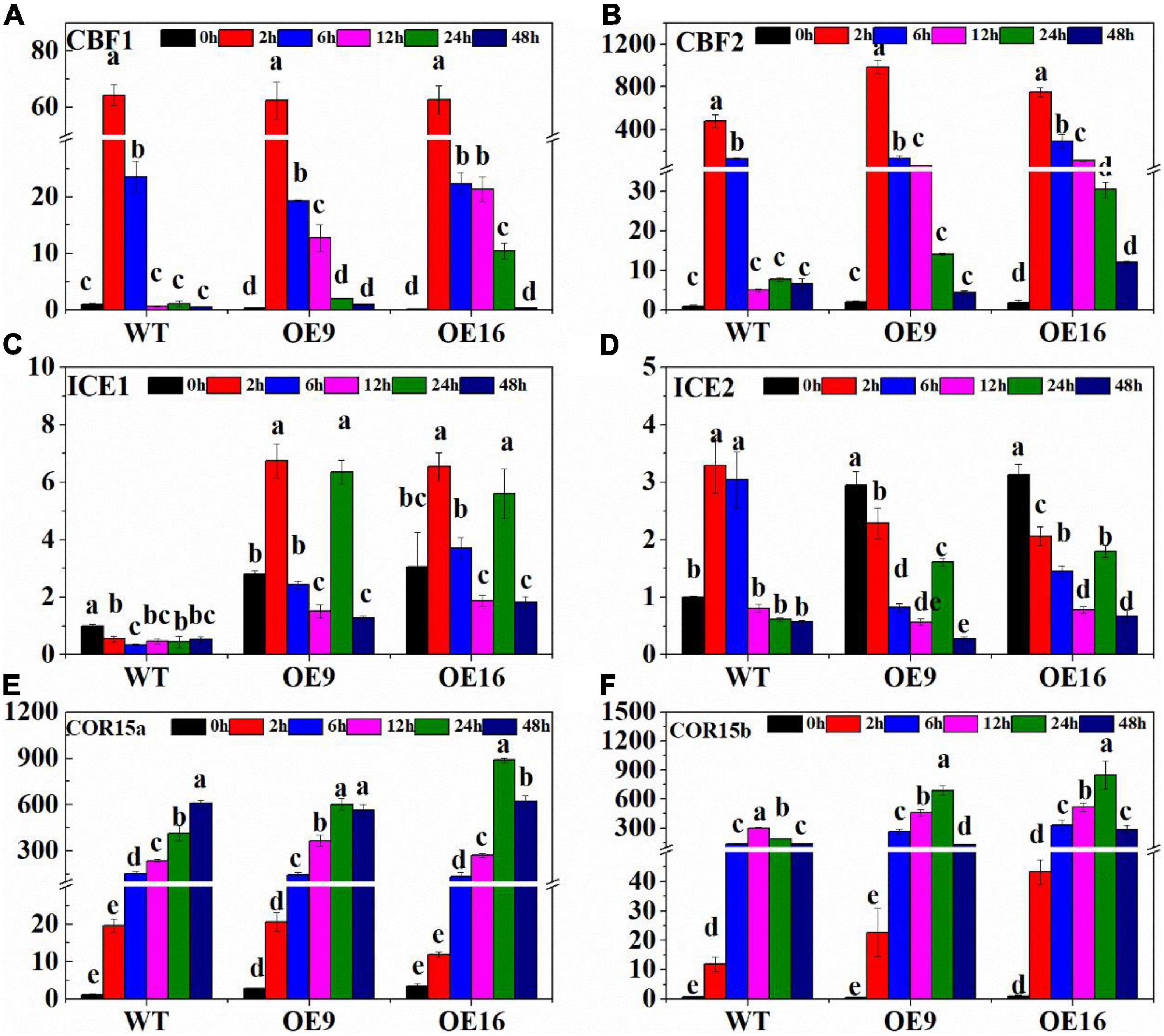
Figure 4. The relative expression of cold stress-related genes including AtCBF1 (A), AtCBF2 (B), AtICE1 (C), AtICE2 (D), AtCOR15a (E), and AtCOR15b (F) in wild-type (WT) and transgenic plants exposed to low temperature stress for 0, 2, 6, 12, and 24 h. OE: the CaALAD-overexpressing (OE) lines. The error bars represent the standard errors for three independent tests. Different letters indicate significant differences (p < 0.05) based on Duncan’s test.
To further determine whether CaALAD is involved in regulating cold tolerance in Arabidopsis, we performed histochemical staining and determined the contents of H2O2 and O2•– and the activities of the antioxidant enzymes SOD, POD, and CAT after 24 h of low temperature stress. The transgenic (OE) plants stained less intensely with both NBT and DAB compared to the WT plants following exposure to low temperature stress (Figures 5A,B). The contents of H2O2 and O2•– and the activities of SOD, POD, and CAT were not significantly different between WT and transgenic lines in the control room temperature treatment (Figures 5, 6). The contents of H2O2 and O2•– and the activities of SOD, POD, and CAT increased in both WT and transgenic plants after 24 h of low temperature treatment (Figures 5, 6). The H2O2 and O2•– contents in WT were significantly higher than in the transgenic plants, and the activities of SOD, POD, and CAT were significantly lower than in the transgenic plants after 1 day at 4oC (Figures 5, 6). After low temperature stress, the O2•– content of the OE9 and OE16 plants decreased by 29.6 and 27.5%, respectively, compared with WT, and the H2O2 content decreased by 22.3 and 17.8%, respectively, compared with WT (Figures 5B,D). In addition, after 24 h at 4oC, the activities of antioxidant enzymes in OE9 and OE16 plants showed increases of 42.4 and 54.0% (SOD), 30.3 and 30.5% (POD), and 14.6 and 18.3% (CAT), respectively, compared with WT plants (Figure 6).
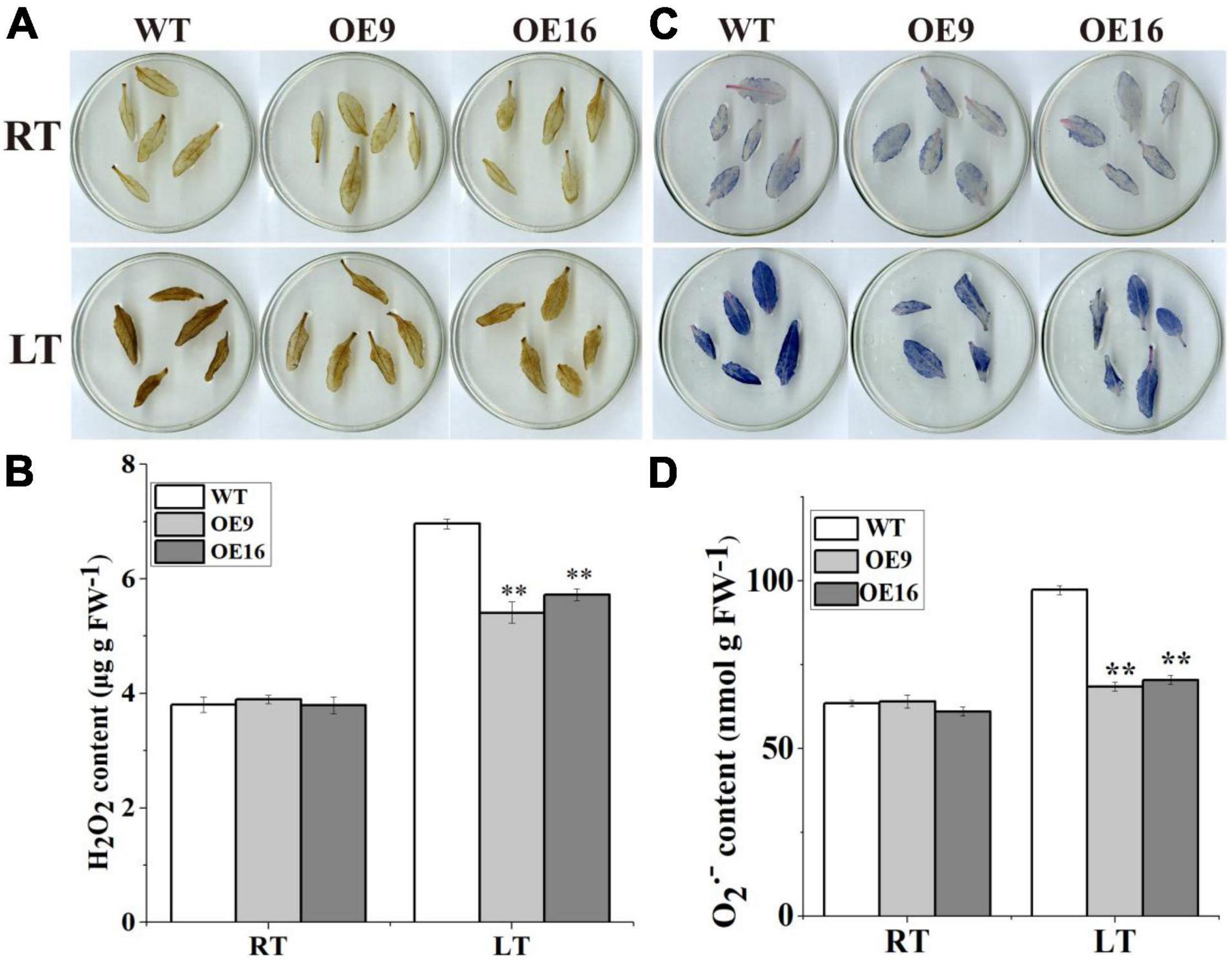
Figure 5. The contents of ROS and the activities of antioxidant enzymes including in the wild-type (WT) and transgenic plants under 4oC for 1 day. (A) DAB staining; (B) the content of H2O2; (C) NBT staining; and (D) the content of O2–. The error bars represent the standard errors for three independent tests, and the asterisks (**) indicate significant differences from the WT at p < 0.01.
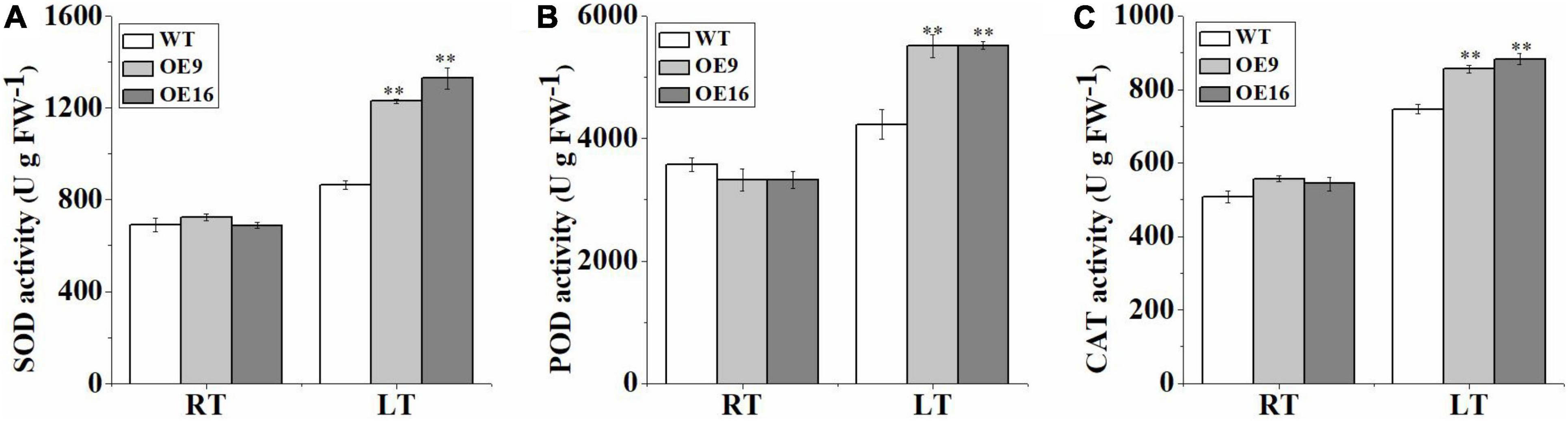
Figure 6. The activities of the antioxidant enzymes SOD (A), POD (B), and CAT (C) in the wild-type (WT) and transgenic plants exposed to 4°C for 1 day. The error bars represent the standard errors of three independent tests and the asterisks (**) indicate significant differences from the WT at p < 0.01.
The ALAD protein catalyzes the condensation of two molecules of ALA into PBG, which can affect the biosynthesis of chlorophyll. In this study, the contents of PBG, Proto IX, Mg-proto IX, Pchl, and Chl in transgenic and WT Arabidopsis plants subjected to low temperature stress were measured. The results showed that the PBG content of the transgenic plants was significantly higher than in WT plants under both normal and low temperature conditions (Figure 7). The PBG contents of WT and transgenic Arabidopsis plants increased significantly after 1 day of low temperature treatment (Figure 7). In addition, the contents of Proto IX, Mg-proto IX, Pchl, and Chl in the transgenic plants under low temperature stress were significantly higher than in WT plants (Figure 8). After 7 days of low temperature stress, Proto IX in the OE9 and OE16 transgenic plants increased by 18.1 and 15.3%, Mg-proto IX increased by 20.9 and 13.2%, Pchl increased by 15 and 16.8%, Chl a increased by 14.6 and 16.1%, Chl b increased by 20.1 and 21.6%, and total Chl increased by 16 and 17.5%, respectively, compared to WT (Figure 8).
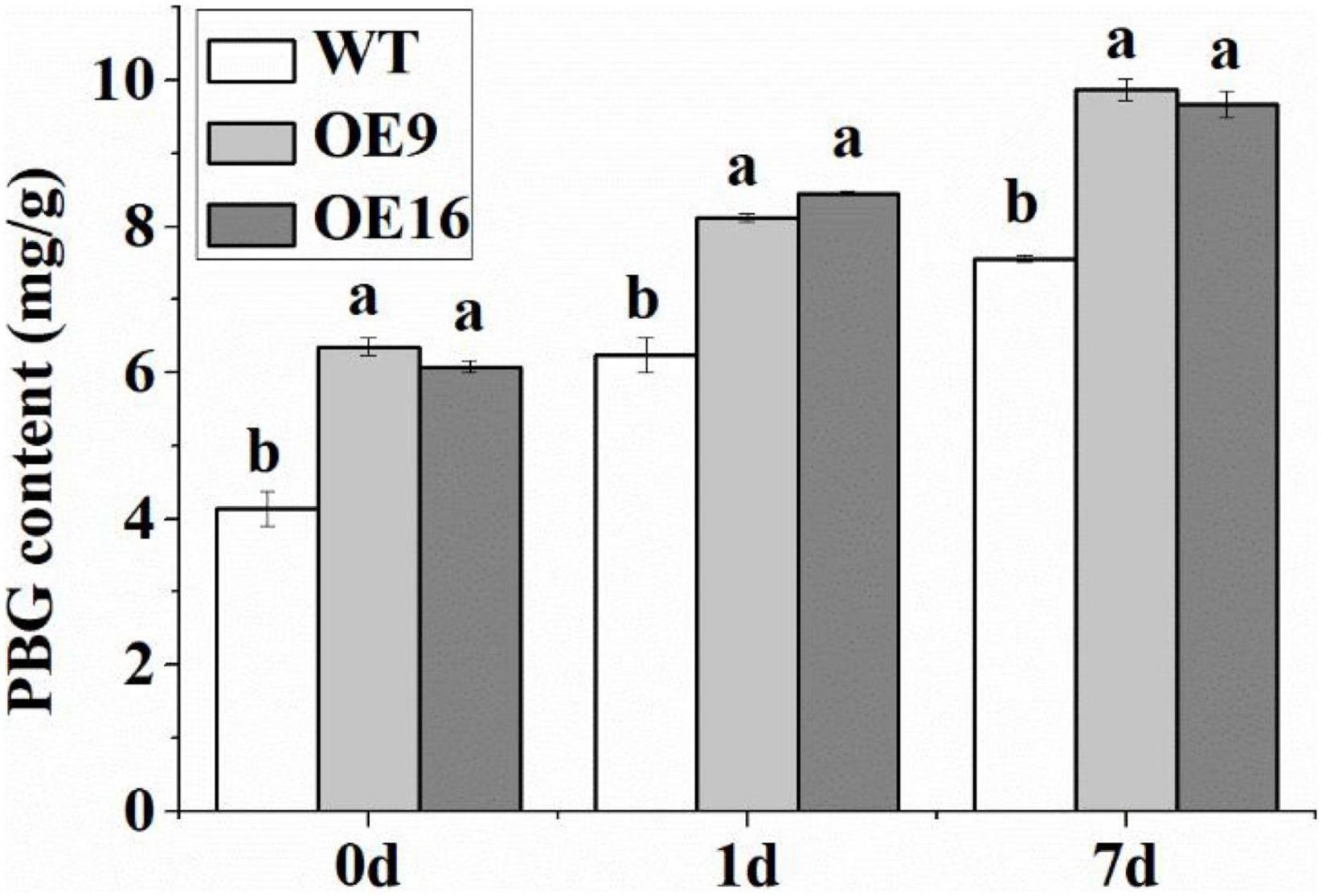
Figure 7. The PBG content of the wild-type (WT) and transgenic plants in response to 4oC treatment for 0, 1, and 7 day. OE: the CaALAD-overexpressing (OE) lines. The error bars represent the standard errors for three independent tests. Different letters show significant differences (p < 0.05) based on Duncan’s test.
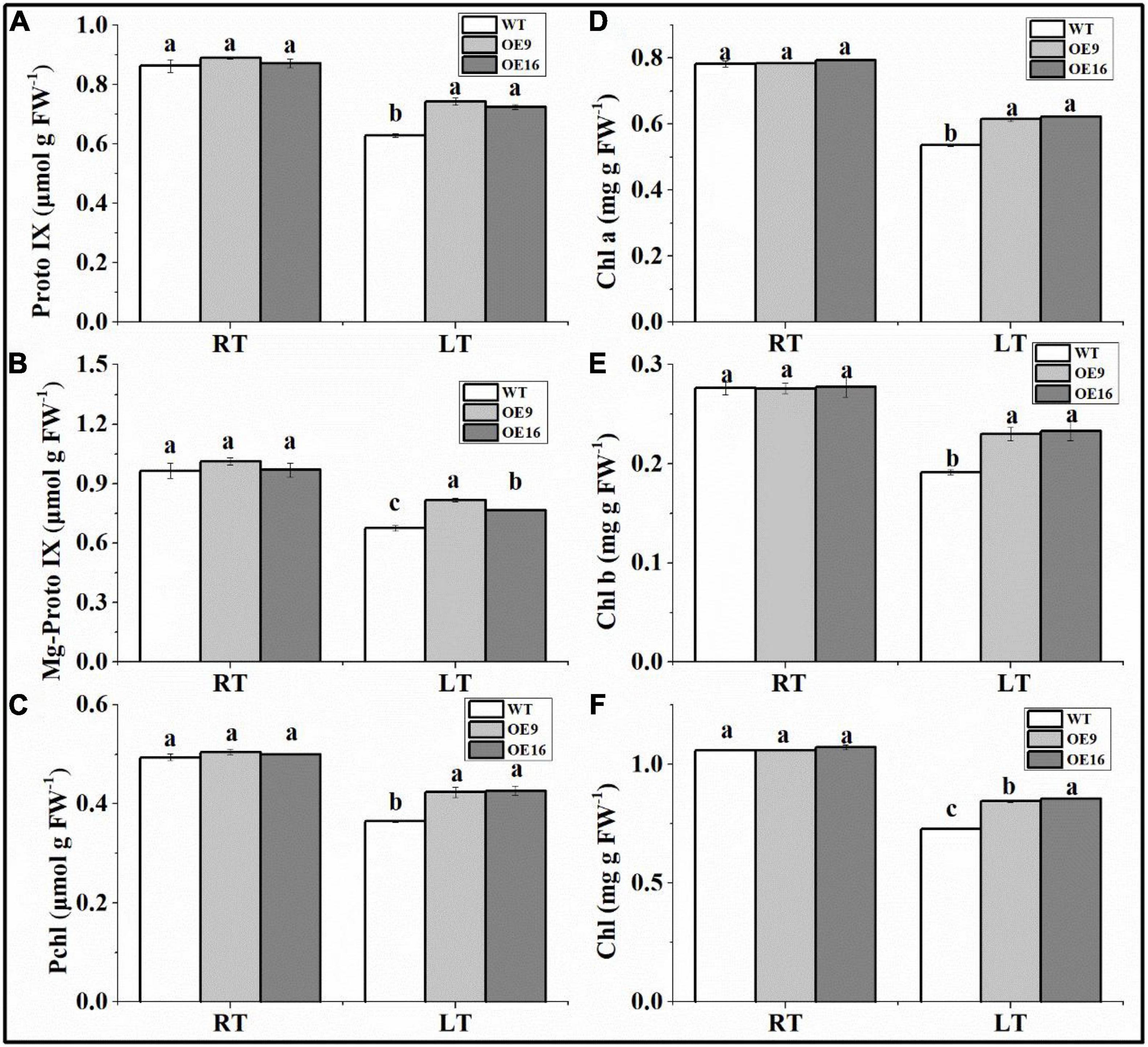
Figure 8. The contents of Proto IX (A), Mg-proto IX (B), Pchl (C), Chl a (D), Chl b (E), and total Chl (F) in the wild-type (WT) and transgenic plants exposed to 4°C for 7 day (LT). OE: the CaALAD-overexpressing (OE) lines. The error bars represent the standard errors for three independent tests. Different letters show significant differences (p < 0.05) based on Duncan’s test.
Wild-type and transgenic Arabidopsis seeds were treated with exogenous ALA and NaHS to determine their sensitivity to ALA and H2S. The results of this experiment suggest that exogenous ALA and NaHS treatment can significantly improve the germination rate of Arabidopsis seeds, while the germination rate of the transgenic seeds was always higher than that of WT seeds (Figure 9). The germination rate of Arabidopsis seeds treated with exogenous ALA was the highest at 100 mg/L (Figure 9A). The germination rates of transgenic seeds treated with ALA at 1, 10, and 100 mg/L increased by 117.3, 94.7, and 66.5% compared with WT seeds, respectively, (Figure 9A). The germination rate of transgenic seeds treated with 10 μmol/L NaHS was the highest (Figure 9B). The germination rates of transgenic seeds treated with 10 μmol/L NaHS, 100 μmol/L NaHS, and 1,000 μmol/L NaHS increased by 68.7, 33.6, and 29.3% compared with WT seeds, respectively, (Figure 9B).
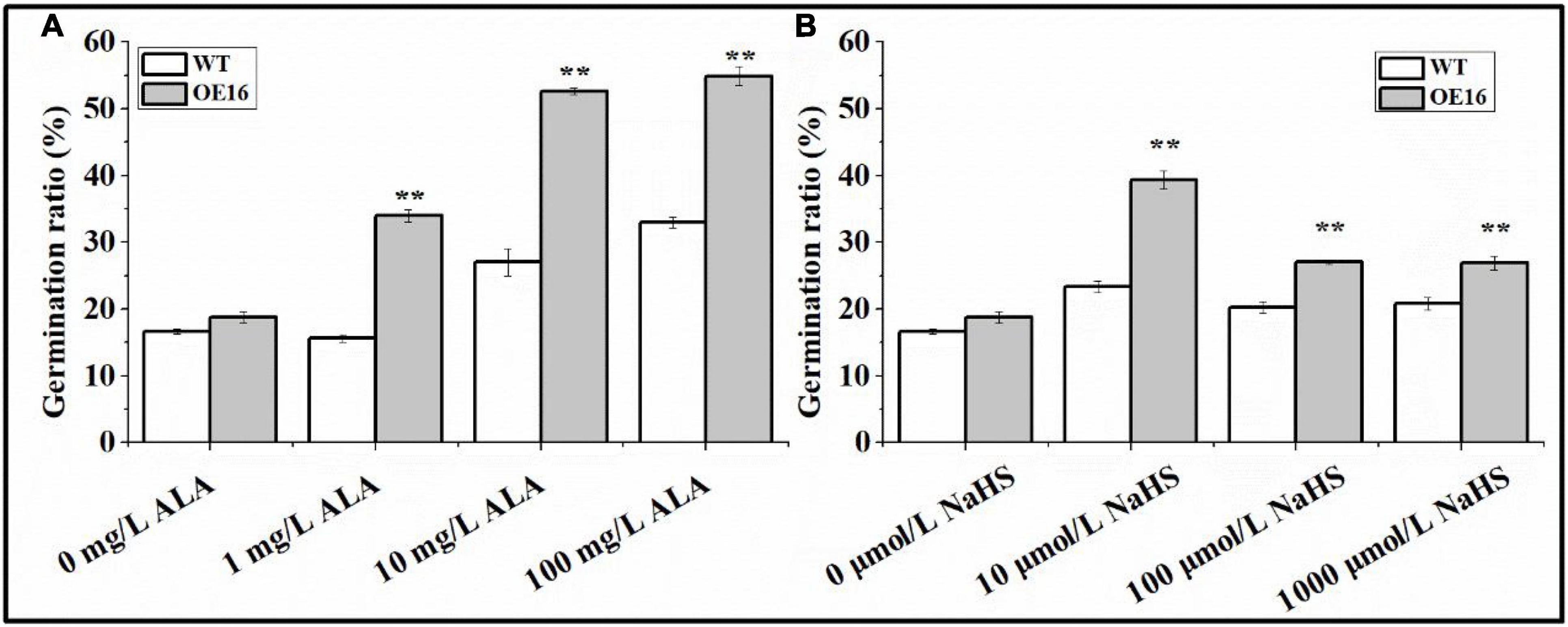
Figure 9. The response of seeds from CaALAD-overexpressing plants to exogenous ALA and NaHS. (A) The germination rates of transgenic seeds treated with 1 mg/L ALA, 10 mg/L ALA, and 100 mg/L ALA. (B) The germination rates of transgenic seeds treated with 10 μmol/L NaHS, 100 μmol/L NaHS, and 1,000 μmol/L NaHS. WT: Arabidopsis Col-0. OE: the CaALAD-overexpressing (OE) lines. NaHS: the donor of hydrogen sulfide. The error bars are the standard errors for three independent tests and asterisks indicate significant differences from the WT at **p < 0.01.
During low temperature treatment at 4oC, the H2S contents of WT and transgenic plants increased rapidly at 2 h, then decreased sharply at 6 h, and reached a low level at 24 h (Figure 10). However, compared with WT, the H2S contents of the transgenic lines were ∼2-fold higher at 2 h than in the WT: the levels in OE9 and OE16 plant increased by 86.7 and 96.6%, respectively, (Figure 10). The H2S content of WT plants then increased to the same level as that of the transgenic lines at 6 h, while the H2S contents of the transgenic OE plants were the same at 2 h and 6 h (Figure 10).

Figure 10. The H2S contents of the wild-type (WT) and transgenic plants on cold stress for 0, 2, 6, 12, and 24 h. OE: the CaALAD-overexpressing (OE) lines. The error bars are the standard errors for three independent tests.
The ALAD protein plays an important role in higher organisms, and the enzymatic activity of ALAD determines the metabolism of ALA in plants, thus affecting the synthesis of tetrapyrrole molecules, especially chlorophyll (Long et al., 2013). Studies have shown that ALA plays an important role in alleviating abiotic stress, including low temperature stress (Wu et al., 2018b). ALAD can catalyze the condensation of ALA into PBG, which is likely to be involved in the plant response to low temperature stress. Our previous study found that ALAD may be induced by ALA and H2S to alleviate the low temperature stress of pepper seedlings (Wang et al., 2021b). In our study, we found that transgenic plants overexpressing CaALAD grew better under low temperature stress, with more and longer roots, indicating that the CaALAD protein alleviates the inhibition of low temperature on the growth of Arabidopsis seedlings (Figure 3). By analyzing the amino acid sequence of CaALAD and comparing it with homologous proteins from other plant species, we found that ALAD is a conserved gene, which is consistent with the results of Long et al. (2013). In addition, we found that it is most similar to the protein from eggplant, a related species in the Solanaceae family. Moreover, the results of subcellular localization experiments and software prediction showed that the CaALAD protein is localized to the chloroplast, which is an organelle that is highly sensitive to low temperature stress. This also provides an indication that CaALAD is involved in the response to chilling stress in pepper.
Abiotic stress causes plants to produce large amounts of ROS, and excessive accumulation of ROS can cause oxidative stress and cell death (Gill and Tuteja, 2010). Over the course of evolution, plants developed an effective scavenging system to remove ROS. Antioxidant enzymes such as SOD, POD, and CAT play an important role in scavenging ROS and enhancing stress resistance in plants (Hu et al., 2012; Wang et al., 2012). In this study, low temperature stress caused increases in the levels of O2•– and H2O2, and the transgenic plants showed reduced accumulation of O2•– and H2O2 compared to WT (Figures 4A,B). In addition, compared with WT, plants of the transgenic Arabidopsis lines showed significantly increased activities of SOD, POD, and CAT in response to low temperature stress (Figures 4C–E). These results indicate that CaALAD mitigates oxidative damage in the transgenic Arabidopsis plants by regulating antioxidant enzyme activity. Chai et al. (2017) found that mutation of the ALAD gene in cotton leads to a large accumulation of ROS, which is consistent with the results of our study, and indicates that the ALAD protein may be involved in maintaining the ROS balance in plant cells (Chai et al., 2017).
Plants exposed to low temperatures can improve their cold tolerance, which involves changes in plant gene expression (Ma et al., 2018). ICE-CBF-COR is the most important cold defense mechanism in plants. CBF (C-repeat binding factor) is an important transcription factor in the plant response to low temperature stress and plays a core role in the low temperature stress regulatory network in plants (Lee et al., 2012; Zhou, 2013). When plants are subjected to low temperature stress, CBF transcription factors are rapidly induced and bind to cis-acting elements in the COR (Cold-regulated) gene promoter, thereby activating the expression of downstream genes and ultimately improving plant cold tolerance (Shi et al., 2018; Chen et al., 2019; Ye et al., 2021). In this study, expression of both CBF1 and CBF2 increased rapidly after 2 h of low temperature stress, while the relative increase in the transgenic Arabidopsis plants was significantly higher than in WT. ICE1 and ICE2 are positive regulators of CBF, although they are not directly regulated by low temperature. COR15 is a chloroplast-localized specific cold resistance gene, which can enhance the cold resistance of chloroplasts and reduce the cold-induced chloroplast membrane damage by changing the location of membrane lipids (Zhou, 2013; Wang et al., 2017). In this study, the expression levels of the cold tolerance genes AtCBF1, AtCBF2, AtICE2, AtCOR15a, and AtCOR15b increased rapidly in response to low temperature stress (Figure 4). Compared with WT plants, OE transgenic line plants showed increased transcription of AtCBF2, AtICE1, and AtCOR15b at low temperature (Figure 4). This suggests that the CaALAD protein regulates the expression of cold tolerance genes, and thus participates in the ICE-CBF-COR cold defense mechanism.
Low temperature stress will destroy the photosynthetic system of plants and reduce their photosynthetic capacity. The chloroplast is an essential part of photosynthesis and plays a crucial role in the development and growth of plants through net carbon fixation, biosynthesis of fatty acids, and other physiological processes (Jarvis and López-Juez, 2013). Low temperature is an important environmental factor that affects chloroplast development (Liu et al., 2018). Chlorophyll synthesis is severely reduced under low temperature stress (Tang et al., 2021). ALA is a precursor in the chlorophyll biosynthesis pathway. Lu (2013) concluded that ALAD protein enzyme activity and the PBG content of garlic were significantly increased at low temperature (Lu, 2013). Consistent with this result, we also found that the PBG content increased significantly after 24 h of low temperature stress (Figure 6). The studies of Cui et al. (2001) and He et al. (2008) found that chlorophyll content decreased significantly after low temperature stress, but that the content of PBG, a precursor of chlorophyll synthesis, increased significantly, which was also consistent with the results of our study (Cui et al., 2001; He et al., 2008). Four PBG molecules combine to form a hydroxymethylbilane (HMB), which goes through three steps to form Proto IX (Kobayashi and Masuda, 2016). Mg2+ is inserted into Proto IX to form Mg-proto IX, which forms prochloroimide (Pchl), the key precursor of mature Chl, through a three-step reaction (Wu et al., 2018b). Chlorophyll is eventually formed by the activity of chlorophyll synthase. In this study, low temperature (4oC) significantly reduced the levels of Proto IX, Mg-proto IX, Pchl, and chlorophyll, while the damage to chlorophyll synthesis caused by low temperature was alleviated in CaALAD-OE transgenic plants. Chai et al. (2017) found that mutations in ALAD reduced chlorophyll content and caused necrotic spots on leaves in cotton, suggesting that ALAD plays an important role in maintaining chlorophyll synthesis (Chai et al., 2017).
Zhao et al. (2013) found that treatment with ALA can improve the germination index of tomato seeds (Zhao et al., 2013). Treating seeds with hydrogen sulfide improved the germination rates in rice and maize by regulating the antioxidant system and changing physicochemical properties during seed germination (Liu, 2017; Wang, 2018). Our results also showed that both ALA and H2S could significantly improve seed germination rate in Arabidopsis. Our previous study showed that exogenous application of ALA and NaHS increased the transcription level of ALAD (HEMB) in pepper seedlings under low temperature stress (Wang et al., 2021b). Therefore, transgenic and WT seeds were treated with exogenous ALA and NaHS. The effects of ALA and NaHS on the germination rate were quantified to determine whether CaALAD responds to ALA and NaHS. The results showed that the transgenic seeds showed a higher germination rate. Due to its gaseous properties, H2S can shuttle to all parts of plant cells and is an important signal molecule involved in plant growth and development and the response to stress (Pandey and Gautam, 2020; Zhang et al., 2021). In this study, the content of H2S increased rapidly after 2 h of low temperature stress, which is consistent with the results of previous studies; that is, exposure to low temperatures can rapidly induce a transient rise in H2S levels (Fu et al., 2013; Du et al., 2017; Wang et al., 2021b). Moreover, we found that the H2S content of the transgenic plants changed more rapidly than it did in WT plants at low temperature, suggesting that H2S may be involved with the ALAD protein as an important signaling molecule to regulate the plant response to low temperature stress.
Our study is the first to show that expression of the ALAD gene responds to low temperature stress. ALAD improves cold tolerance by participating in the regulation of the transcription of cold tolerance genes, enhancing antioxidant systems, and enhancing chlorophyll synthesis. In addition, ALAD may also respond to low temperature stress by participating in a hydrogen sulfide-mediated signaling pathway. These results suggest that ALAD is an important gene for the response to low temperature stress in pepper.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
HW, ZL, and JX designed the research. HW and ZL performed the data analysis. HW wrote the manuscript. JL, JZ, JY, LH, and GZ revised the manuscript. All authors have read and agreed to this version of the manuscript.
This work was supported by the National Natural Science Foundation of China (32072657), the Special Fund Project of Leading Science and Technology Innovation Development of Gansu Province, China (2018ZX02), and the Special Fund for Technical System of Melon and Vegetable Industry of Gansu Province, China (GARS-GC-1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.884990/full#supplementary-material
Allen, D. J., and Ort, D. R. (2001). Impacts of chilling temperatures on photosynthesis in warm-climate plants. Trends Plant Sci. 6, 36–42. doi: 10.1016/s1360-1385(00)01808-2
Berglund, B., and Tegenfeldt, J. (1992). Characterization of plastid 5-aminolevulinate dehydratase (ALAD; EC 4.2.1.24) from spinach (Spinacia oleracea L.) by sequencing and comparison with non-plant ALAD enzymes. Zeitschrift Naturforschung C 47, 77–84. doi: 10.1515/znc-1992-1-214
Boese, Q. F., Spano, A. J., Li, J., and Timko, M. P. (1991). Aminolevulinic acid dehydratase in pea (Pisum sativum L.). identification of an unusual metal-binding domain in the plant enzyme. J. Biol. Chem. 266, 17060–17066. doi: 10.1016/s0021-9258(19)47339-0
Chai, Q., Shang, X., Wu, S., Zhu, G., Cheng, C., Cai, C., et al. (2017). 5-aminolevulinic acid dehydratase gene dosage affects programmed cell death and immunity. Plant Physiol. 175, 511–528. doi: 10.1104/pp.17.00816
Chen, T., Zhang, H., and An, L. (2019). Research progress of ICE1-CBF pathway and its function in plant cold tolerance and development. Acta Botanica. Boreal-Occidentalia. Sinica 39, 1513–1520.
Cheung, K. M., Spencer, P., Timko, M. P., and Shoolingin-Jordan, P. M. (1997). Characterization of a recombinant pea 5-aminolevulinic acid dehydratase and comparative inhibition studies with the Escherichia coli dehydratase. Biochemistry 36:1148. doi: 10.1021/bi961215h
Cui, H., Xia, Y., and Gao, M. (2001). Effects of temperature on leaf color and chlorophyll biosynthesis of rice mutant W1. Acta Agriculturae Nucleatae Sinica 15, 269–273.
Du, X., Jin, Z., Liu, D., Yang, G., and Pei, Y. (2017). Hydrogen sulfide alleviates the cold stress through MPK4 in Arabidopsis thaliana. Plant Physiol. Biochem. 120, 112–119. doi: 10.1016/j.plaphy.2017.09.028
Fu, P., Wang, W., Hou, L., and Liu, X. (2013). Hydrogen sulfide is involved in the chilling stress response in Vitis vinifera L. Acta Soc. Botanicorum Poloniae 82, 295–302. doi: 10.5586/asbp.2013.031
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Gilmour, S. J., Zarka, D. G., Stockinger, E. J., Salazar, M. P., Houghton, J. M., and Thomashow, M. F. (2010). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. doi: 10.1046/j.1365-313x.1998.00310.x
Guo, W. (2013). Response to Chilling Stress in Pepper as Well as Characterization and Functional Analysis of Chiling-Related Genes. China: Northwest A&F University.
He, H., Xue, L., Tian, L., and Yuanliang, C. (2008). Evaluation on low temprature and light intensity tolerance of different watermelon varieties at seedling stage. Northern Horticulture 4, 13–16.
Hu, L., Li, H., Pang, H., and Fu, J. (2012). Responses of antioxidant gene, protein and enzymes to salinity stress in two genotypes of perennial ryegrass (Lolium perenne) differing in salt tolerance. J. Plant Physiol. 169, 146–156. doi: 10.1016/j.jplph.2011.08.020
Jain, M., Tiwary, S., and Gadre, R. (2018). Modulation of δ-Aminolevulinic acid dehydratase activity by the sorbitol-induced osmotic stress in maize leaf segments. Biochem. Biokhimiia 83, 32–36. doi: 10.1134/S0006297918010042
Jarvis, P., and López-Juez, E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. doi: 10.1038/nrm3702
Kayser, H. Krull-Savage, U., and Gessel, R. V. (2005). Developmental profiles of 5-aminolevulinate, porphobilinogen and porphobilinogen synthase activity in Pieris brassicae related to the synthesis of the bilin-binding protein. Insect Biochem. Mol. Biol. 35, 165–174. doi: 10.1016/j.ibmb.2004.11.001
Killiny, N., Hijaz, F., Nehela, Y., Hajeri, S., and Gowda, S. (2018). Effects of δ“minolevulinic acid dehydratase silencing on the primary and secondary metabolisms of citrus. Plant Direct. 2:e00072. doi: 10.1002/pld3.72
Kim, J. G., Back, K., Lee, H. Y., Lee, H. J., Phung, T. H., Grimm, B., et al. (2014). Increased expression of Fe-chelatase leads to increased metabolic flux into heme and confers protection against photodynamically induced oxidative stress. Plant Mol. Biol. 86, 271–287. doi: 10.1007/s11103-014-0228-3
Kobayashi, K., and Masuda, T. (2016). Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 7:1811. doi: 10.3389/fpls.2016.01811
Lee, D.-H., Jun, W.-J., Kim, K.-M., and Dong-Hoon, S. (2003). Inhibition of 5-aminolevulinic acid dehydratase in recombinant Escherichia coli using d-glucose. Enzyme Microbial Technol. 32, 27–34. doi: 10.1016/s0141-0229(02)00241-7
Lee, S. C., Lim, M. H., Yu, J. G., Park, B. S., and Yang, T. J. (2012). Genome-wide characterization of the CBF/DREB1 gene family in Brassica rapa. Plant Physiol. Biochem. 61, 142–152. doi: 10.1016/j.plaphy.2012.09.016
Liu, J. (2017). Physiological Effect of Hydrogen Sulfide on Regulating Amylase in Response to Drought Stress in Germinating Rice Seeds. Hainan Agricultural University.
Liu, X., Lan, J., Huang, Y., Cao, P., Zhou, C., Ren, Y., et al. (2018). WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J. Exp. Bot. 69, 3949–3961. doi: 10.1093/jxb/ery214
Long, F., Li, S., and Li, M. (2013). Bioinformatics analysis of ALAD gene in seven plants. Genom. Appl. Biol. 32:13.
Lu, X. (2013). Cloning and Expression of 5-aminolevulinate Dehydratase Gene and its Relationship to Garlic Greening. China: Shandong Agricultural University.
Ma, X., Chen, C., Yang, M., Dong, X., Lv, W., and Meng, Q. (2018). Cold-regulated protein (SlCOR413IM1) confers chilling stress tolerance in tomato plants. Plant Physiol. Biochem. 124, 29–39. doi: 10.1016/j.plaphy.2018.01.003
Martins, P. K., Nakayama, T. J., Ribeiro, A. P., Cunha, B. A. D. B. D., and Molinari, H. J. B. R. (2015). Setaria viridis floral-dip: a simple and rapid Agrobacterium-mediated transformation method. Biotechnol. Rep. 6, 61–63. doi: 10.1016/j.btre.2015.02.006
Medina, J., Bargues, M., Terol, J., Perez-Alonso, M., and Salinas, J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration 1. Plant Physiol. 119, 463–470. doi: 10.1104/pp.119.2.463
Pandey, A. K., and Gautam, A. (2020). Stress responsive gene regulation in relation to hydrogen sulfide in plants under abiotic stress. Physiol. Plant. 168, 511–525. doi: 10.1111/ppl.13064
Pereira, L. B., Tabaldi, L. A., Goncalves, J. F., Jucoski, G. O., Pauletto, M. M., Weis, S. N., et al. (2006). Effect of aluminum on d-aminolevulinic acid dehydratase (ALA-D) and the development of cucumber (Cucumis sativus). Environ. Exp. Bot. 57, 106–115. doi: 10.1016/j.envexpbot.2005.05.004
Polking, G. F., Hannapel, D. J., and Gladon, R. J. (1995). A cDNA clone for 5-aminolevulinic acid dehydratase from tomato (Lycopersicon esculentum Mill.). Plant Physiol. 107, 366–369. doi: 10.1104/pp.107.3.1033
Shi, Y., Ding, Y., and Yang, S. (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23, 623–637. doi: 10.1016/j.tplants.2018.04.002
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. J. N. P. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Taghizadeh, M., Tarahomi, E., and Fahimi, H. (2020). Quantitative comparison of tree pairs resulted fromgene and protein phylogenetic trees for sulfite reductase flavoprotein alpha-component and 5S rRNA and taxonomic trees in selected bacterial species. J. Health Biomed. Inform. 7, 332–336.
Tang, C., Xie, J., Lv, J., Li, J., Zhang, J., Wang, C., et al. (2021). Alleviating damage of photosystem and oxidative stress from chilling stress with exogenous zeaxanthin in pepper (Capsicum annuum L.) seedlings. Plant Physiol. Biochem. 162, 395–409. doi: 10.1016/j.plaphy.2021.03.010
Tang, K., Zhao, L., Ren, Y., Yang, S., Zhu, J. K., and Zhao, C. (2020). The transcription factor ICE1 functions in cold stress response by binding to the promoters of CBF and COR genes. J. Integr. Plant Biol. 62, 258–263. doi: 10.1111/jipb.12918
Tang, W., Wang, W., Chen, D., Ji, Q., Jing, Y., Wang, H., et al. (2012). Transposase-Derived proteins FHY3/FAR1 interact with phytochrome-interacting factor1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24, 1984–2000. doi: 10.1105/tpc.112.097022
Wang, D.-Z., Jin, Y.-N., Ding, X.-H., Wen-Jia, Z., and Shan-Shan, B. (2017). Gene regulation and signal transduction in the ICE-CBF-COR signaling pathway during cold stress in plants. Biochemistry 82, 1103–1117. doi: 10.1134/S0006297917100030
Wang, C., Zhang, S., Yang, X., Jiang, X., Li, Y., Qizhong, H., et al. (2021a). Research on screening index of chilling and weak light tolerance in pepper seedlings. J. Nuclear Agricultural Sci. 35, 0989–0996.
Wang, H., Liu, Z., Luo, S., Li, J., Zhang, J., Li, L., et al. (2021b). 5-Aminolevulinic acid and hydrogen sulphide alleviate chilling stress in pepper (Capsicum annuum L.) seedlings by enhancing chlorophyll synthesis pathway. Plant Physiol. Biochem. 167, 567–576. doi: 10.1016/j.plaphy.2021.08.031
Wang, Q., Wu, C., Xie, B., Liu, Y., Cui, J., Chen, G., et al. (2012). Model analysing the antioxidant responses of leaves and roots of switchgrass to NaCl-salinity stress. Plant Physiol. Biochem. 58, 288–296. doi: 10.1016/j.plaphy.2012.06.021
Wang, Y. (2018). Study of Repair Mechanism of Hydrogen Sulfide Donor on Seed Vigor of Maize (Zea may.L). China: Henan Agricultural University.
Wittmann, D., Klove, S., Wang, P., and Grimm, B. (2018). Towards initial indications for a thiol-based redox control of Arabidopsis 5-Aminolevulinic acid dehydratase. Antioxidants 7:152. doi: 10.3390/antiox7110152
Wu, Y., Jin, X., Liao, W., Hu, L., Dawuda, M. M., Zhao, X., et al. (2018a). 5-Aminolevulinic Acid (ALA) alleviated salinity stress in cucumber seedlings by enhancing chlorophyll synthesis pathway. Front. Plant Sci. 9:635. doi: 10.3389/fpls.2018.00635
Wu, Y., Liao, W., Dawuda, M. M., Hu, L., and Yu, J. (2018b). 5-Aminolevulinic acid (ALA) biosynthetic and metabolic pathways and its role in higher plants: a review. Plant Growth Regulation 87, 357–374. doi: 10.1007/s10725-018-0463-8
Xianyang, C., Heping, H., Ping, J., Lingling, N., and Hexigeduleng, B. (2011). Transformation of beta-lycopene cyclase genes from Salicornia europaea and Arabidopsis conferred salt tolerance in Arabidopsis and tobacco. Plant Cell Physiol. 52, 909–921. doi: 10.1093/pcp/pcr043
Xu, X. (2014). δ-aminolevulinic Acid Dehydratase from Garlic and its Relationship to Garlic Greening. China: Shandong Agricultural University.
Yang, J. (2008). Cloning and Expression of 5-aminolevulinate Dehydratase from Escherichia coli Rosetta (DE3) and Primary Study on its Enzymatic Properties. China: Zhejiang University.
Ye, Z., Gao, C., Gagoshidze, Z., Shu, H., Bao, W., Zhu, J., et al. (2021). Genome-wide identification and analysis of the cold responsive transcription factor (CBF) in Capsicum Chinense. Mol. Plant Breed. 19, 4903–4910.
Yu, T., Nakajima, H., Kanamaru, K., and Takumi, S. (2011). Characterization of three homoeologous cDNAs encoding chloroplast-targeted aminolevulinic acid dehydratase in common wheat. J. Int. Plant Biol. 53, 942–950. doi: 10.1111/j.1744-7909.2011.01083.x
Zhang, J., Zhou, M., Zhou, H., Zhao, D., Gotor, C., Romero, L. C., et al. (2021). Hydrogen sulfide, a signaling molecule in plant stress responses. J. Int. Plant Biol. 63:15. doi: 10.1111/jipb.13022
Zhang, Z. (2010). Changes of Physiological Characteristics of the Obtained Trangenic YHEM1 Plants with Ability to Over-produce 5-aminolevulinic Acid. China: Nanjing Agricultural University.
Zhao, Y., Hu, X., Zou, Z., and Yan, F. (2013). Effects of seed soaking with different concentrations of 5-aminolevulinic acid on the germination of tomato (Solanum lycopersicum) seeds under NaCl stress. Acta Ecol. Sinica 33, 62–70. doi: 10.5846/stxb201110311628
Keywords: 5-aminolevulinic acid, hydrogen sulfide, reactive oxygen species, low temperature, cold tolerance
Citation: Wang H, Liu Z, Xie J, Li J, Zhang J, Yu J, Hu L and Zhang G (2022) The CaALAD Gene From Pepper (Capsicum annuum L.) Confers Chilling Stress Tolerance in Transgenic Arabidopsis Plants. Front. Plant Sci. 13:884990. doi: 10.3389/fpls.2022.884990
Received: 27 February 2022; Accepted: 11 April 2022;
Published: 29 April 2022.
Edited by:
Shifeng Cao, Zhejiang Wanli University, ChinaReviewed by:
Yang Sheng, Fujian Agriculture and Forestry University, ChinaCopyright © 2022 Wang, Liu, Xie, Li, Zhang, Yu, Hu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianming Xie, eGllamlhbm1pbmdAZ3NhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.