
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 08 September 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.883139
This article is part of the Research Topic The Plant Cell Wall: Advances and Current Perspectives View all 14 articles
 Guillermo Garcia-Gimenez1†
Guillermo Garcia-Gimenez1† Miriam Schreiber2
Miriam Schreiber2 George Dimitroff3
George Dimitroff3 Alan Little3
Alan Little3 Rohan Singh3
Rohan Singh3 Geoffrey B. Fincher3
Geoffrey B. Fincher3 Rachel A. Burton3
Rachel A. Burton3 Robbie Waugh1,2
Robbie Waugh1,2 Matthew R. Tucker3
Matthew R. Tucker3 Kelly Houston1*
Kelly Houston1*(1,3;1,4)-β-Glucan is a non-cellulosic polysaccharide required for correct barley grain fill and plant development, with industrial relevance in the brewing and the functional food sector. Barley grains contain higher levels of (1,3;1,4)-β-glucan compared to other small grain cereals and this influences their end use, having undesirable effects on brewing and distilling and beneficial effects linked to human health. HvCslF6 is the main gene contributing to (1,3;1,4)-β-glucan biosynthesis in the grain. Here, the transcriptional regulation of HvCslF6 was investigated using an in-silico analysis of transcription factor binding sites (TFBS) in its putative promoter, and functional characterization in a barley protoplast transient expression system. Based on TFBS predictions, TF classes AP2/ERF, MYB, and basic helix-loop-helix (bHLH) were over-represented within a 1,000 bp proximal HvCslF6 promoter region. Dual luciferase assays based on multiple HvCslF6 deletion constructs revealed the promoter fragment driving HvCslF6 expression. Highest HvCslF6 promoter activity was narrowed down to a 51 bp region located −331 bp to −382 bp upstream of the start codon. We combined this with TFBS predictions to identify two MYB TFs: HvMYB61 and HvMYB46/83 as putative activators of HvCslF6 expression. Gene network analyses assigned HvMYB61 to the same co-expression module as HvCslF6 and other primary cellulose synthases (HvCesA1, HvCesA2, and HvCesA6), whereas HvMYB46/83 was assigned to a different module. Based on RNA-seq expression during grain development, HvMYB61 was cloned and tested in the protoplast system. The transient over-expression of HvMYB61 in barley protoplasts suggested a positive regulatory effect on HvCslF6 expression.
The primary cell walls of certain members of the Poaceae, including barley, are enriched with (1,3;1,4)-β-glucan, a distinctive non-cellulosic polysaccharide that predominantly accumulates during cell expansion (Carpita et al., 2001). In contrast, secondary walls of the Poaceae are mainly characterized by the presence of heteroxylans as the major non-cellulosic component, and tissue-specific lignin accumulation (Burton and Fincher, 2014). This differs from eudicots where xyloglucan and pectins are the most abundant non-cellulosic polysaccharides found in primary cell walls and secondary walls are characterized by the presence of heteroxylans, predominantly 4-O-methylglucuronoxylans, heteromannans and lignin (Harris, 2006).
The discovery of Cellulose synthase-like (Csl) genes required for (1,3;1,4)-β-glucan biosynthesis, including members of the CslF, H and J families (Burton et al., 2006; Doblin et al., 2009; Little et al., 2018), provided the basis for detailed biochemical and molecular studies of (1,3;1,4)-β-glucan structure, assembly and function. These genetic discoveries complemented initial biochemical and physicochemical studies of (1,3;1,4)-β-glucan carried out predominantly in barley (Woodward et al., 1983), wheat (Bacic and Stone, 1980), and oats (Aaman and Graham, 1987), which reported remarkable differences in relative polysaccharide abundance and structure across cereal species (Bacic and Stone, 1981; Burton and Fincher, 2012). Previous studies identified polymorphisms in the HvCslF6 gene, a key gene involved in grain (1,3;1,4)-β-glucan biosynthesis (Cory et al., 2012; Taketa et al., 2012; Hu et al., 2014), variations in CslF transcript abundance (Burton et al., 2008; Wong et al., 2015), HvCslF6 subcellular location (Wilson et al., 2015) and amino acids contributing to HvCslF6 protein structure and (1,3;1,4)-β-glucan solubility (Jobling, 2015; Dimitroff et al., 2016). Despite this, no clear relationship has been established between barley cultivar-specific polymorphisms in the HvCslF6 upstream region, or the introns of this gene, and the wide range in grain (1,3;1,4)-β-glucan levels observed in barley (Houston et al., 2014; Garcia-Gimenez et al., 2019).
Little is known about regulatory mechanisms affecting primary cell wall formation in grasses, and specifically the biosynthesis of (1,3;1,4)-β-glucan in developing barley grain. In a co-expression analysis of transcribed genes during the cellularization of developing barley endosperm, Zhang et al. (2016a) identified several classes of candidate TF genes that might be involved in cell wall synthesis, but at that stage direct interactions between TF and specific cell wall genes were not investigated. In the model grass Brachypodium distachyon a trihelix transcription factor (THX1) was validated as a positive regulator of BdCslF6 binding to a GT-rich cis-element in the second intron of this gene (Fan et al., 2018). In rice, Zhao et al. (2019) carried out a comprehensive study of transcriptional regulators within the context of cell wall composition. They identified several genes, including members of the MYB gene family, that changed the transcript levels of known cell wall synthesis genes in a transient expression system. One gene, OsMYB61a, was of interest due to its ability within this system to regulate the expression of numerous cell wall genes and the observation that knock out lines had altered cell wall composition (Zhao et al., 2019).
Regulation of cell wall composition has also been studied in Arabidopsis thaliana, particularly in the context of secondary cell walls. This led to the identification of key transcription factors (i.e., secondary wall NAC; SWN) (Taylor-Teeples et al., 2015) that act as regulatory switches enhancing the expression of downstream MYB master regulators, MYB46 and MYB83 (McCarthy et al., 2009). These MYBs interact with a wide range of cell wall-associated transcription factors (Kim et al., 2014) and networks affecting polysaccharide biosynthesis. In rice and maize, functional orthologs of these Arabidopsis secondary cell wall MYB master regulators were identified, suggesting a conserved regulatory mechanism across vascular plants (Zhong et al., 2011; Rao and Dixon, 2018).
Here, our work aimed to explore the transcriptional regulation of HvCslF6 by identifying putative cis-elements in the 5′ promoter region of HvCslF6 and the transcription factor(s) binding to them. Such data would provide another avenue to investigate the regulation of HvCslF6 gene expression and how this affects (1,3;1,4)-β-glucan abundance.
First, the ability of a 3,000 bp HvCslF6 upstream region to drive expression was tested by GFP tagging in transgenic barley lines. To identify regulatory motifs in the HvCslF6 promoter, a series of 5′ deletion constructs generated from the 3,000 bp HvCslF6 upstream region were fused to the dual luciferase reporter system. A barley protoplast-based transient expression system was adapted (Rao, 2007; Yoo et al., 2007) and used to test activity of the HvCslF6 promoter deletion constructs, permitting the analysis of six deletion constructs simultaneously. Our results from the HvCslF6 promoter characterization were combined with in silico predictions for transcription factor binding sites (TFBS), gene network and co-expression analyses, and previous findings from other cereal species, which suggested several candidate transcription factors that may regulate HvCslF6 expression.
HvCslF6 is expressed across a range of tissues including grain and young/developing tissues such as the coleoptile, root and leaves (Burton et al., 2008). To study the transcriptional regulation of this gene in planta, transgenic barley lines carrying endoplasmic reticulum-targeted GFP (mGFP-ER) driven by a 3,000 bp 5′ sequence upstream of HvCslF6 (pHvCslF6) were generated and screened in grain and vegetative tissues. Transverse sections of transgenic mature grain showed strong mGFP-ER expression in the starchy endosperm (Figure 1A) compared to the negative control (empty vector) (Figure 1B). Similarly, increased mGFP-ER expression around vascular bundles was detected in transverse coleoptile sections of transgenic lines (Figure 1C) compared to the negative control (Figure 1D). Therefore, we confirmed that the 3,000 bp 5′ sequence upstream of HvCslF6 is sufficient to drive expression of this GFP gene in both grain and vegetative tissues.
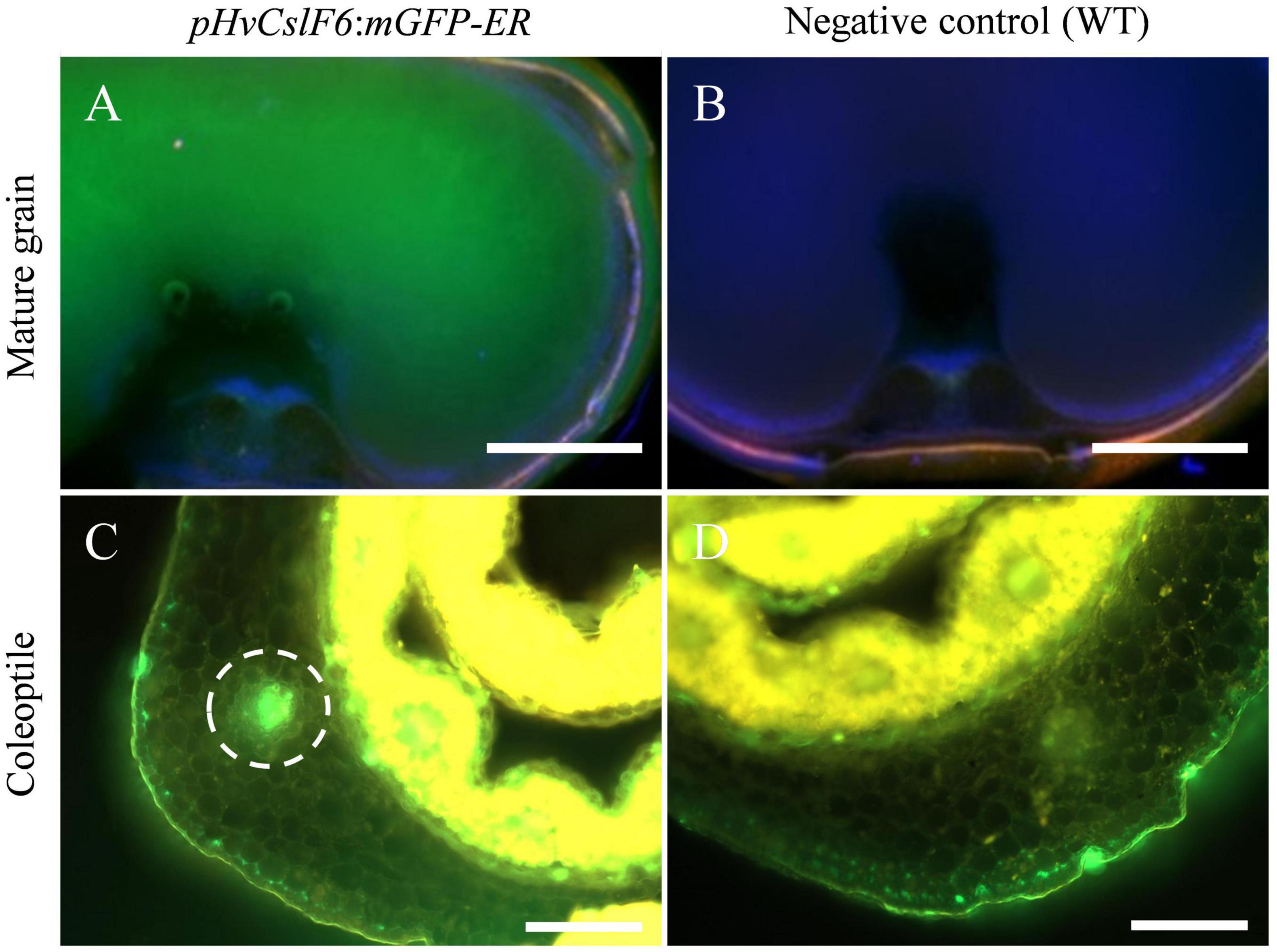
Figure 1. Fluorescent images of pHvCslF6:mGFP-ER (–3,000 bp) barley transverse sections in mature grain and coleoptile tissues (cv. Golden Promise). (A) Mature grain from line transformed with pHvCslF6:mGFP-ER, (B) negative control of wild type grain section, (C) coleoptile from line transformed with pHvCslF6:mGFP-ER, (D) negative control of wild type coleoptile section. The positive control is mGFP-ER driven by the 35S CaMV promoter. The dashed circle indicates the location of a vascular bundle. Images were collected on a Zeiss M2 AxioImager equipped with DIC optics and an Apotome.2 (Zeiss, Germany). GFP was excited at 488 nm and emission collected from 499 to 530 nm, in green. Scale bars are 1 mm.
As the 3,000 bp 5′ promotor region was sufficient to drive the temporal and spatial expression of HvCslF6 in developing and young tissues, we explored whether known transcription factor binding motifs were either present or enriched within this sequence. First, three TFBS prediction software packages (TRANSFAC® v2014, JASPAR v2020 and PlantPAN v3.0) were compared to identify which produced the most detailed motif predictions with the least number of redundant results. TFBS prediction outputs were compared across the three programs, revealing differences in frequency (number of times a particular motif was predicted within the 3,000 bp pHvCslF6) and annotation (profile summary of each motif comprising TF class and origin species) of predicted TFBSs (Supplementary Figure 1 and Supplementary Table 1). It should be noted that a high rate of false positives in predicted TFBSs has been reported in most of these prediction tools (Shahmuradov et al., 2005). Therefore, choosing a comprehensive and experimentally validated database of TFBSs was key to obtain reliable in silico predictions. TRANSFAC® was discarded due to the relatively low number of predicted motifs that were also identified using JASPAR and PlantPAN. A comparative motif analysis between PlantPAN and JASPAR identified seven classes of TFBSs with similar relative frequencies (allowing a ± 10% variation in motif prediction results across both programs). These were: MYB/SANT (12.5% in JASPAR and 14.0% in PlantPAN, respectively), DNA-binding with one zinc finger DOF (13.3% and 8.9%), APETALA2/Ethylene response factors AP2/ERF (9.3% and 8.4%), basic leucine zipper bZIP (4.7% and 6.5%), WRKY (5.4% and 3.2%), auxin response factors ARF (B3 domain; 3.1% and 5.2%), and NAC (4.6% and 3.6%). Notably, 52% of PlantPAN predicted motifs could not be classified because no information/description was available regarding their hit sequence.
Based on these results, JASPAR was chosen for subsequent in silico analyses of the proximal promoter region of HvCslF6 (1,000 bp upstream of the start codon) and 355 non-redundant motifs were identified. JASPAR TFBS scores are based on position frequency matrices (PFMs), defined as occurrences of each nucleotide at each position in a set of observed TF-DNA interactions (Fornes et al., 2020). We selected JASPAR for further analyses due to the higher number of non-redundant predicted TFBS and motif-associated information compared to TRANSFAC and PlantPAN. In addition, JASPAR is a regularly maintained open-access, manually curated and experimentally defined database for plant-specific TFBSs (Castro-Mondragon et al., 2022).
These 355 predicted motifs were filtered in accordance with an arbitrary prediction score threshold of ≥10 (maximum score was 15.9) to minimize the detection of false positives/redundant motifs in identical promoter locations (unfiltered TFBS predictions are shown in Supplementary Figure 2). By applying this filter, 107 unique predicted motifs with prediction scores between 0.7 and 15.9 were identified and grouped into eight classes of TFBSs including: MYB/SANT, AP2/ERF, basic helix-loop-helix (bHLH), ARF, DOF, Homeodomain-leucine zipper (HD-ZIP), Zinc Finger (ZF; SBP and SWIM type), NAC/NAM and a mixed class of TFBSs (low-represented putative TF classes with high prediction scores) (Figure 2A). Predicted motifs grouped into AP2/ERF, MYB/SANT and bHLH TF classes were the most abundant, hence over-represented within the HvCslF6 proximal promoter region (−1,000 bp) when compared to the total of 107 non-redundant motifs identified. JASPAR prediction scores for the three most abundant TF classes were similar to each other (11.6, MYB/SANT; 11.4, bHLH and 11.7, AP2/ERF; Shown on top of each column in orange, Figure 2A).
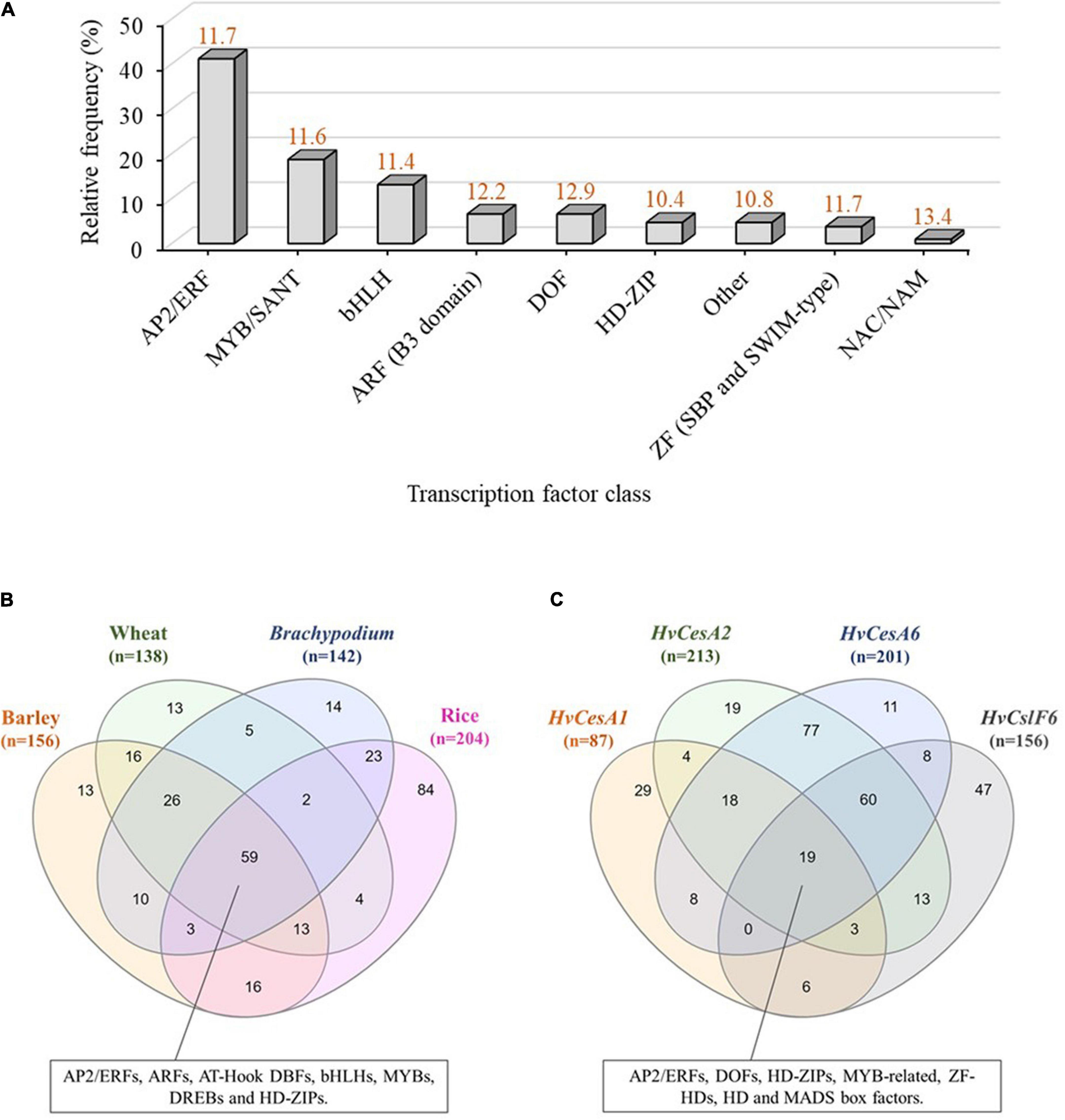
Figure 2. In-silico analysis of transcription factor binding sites (TFBSs) in the CslF6 promoter. (A) TFBS predictions identified within a 1,000 bp upstream region of HvCslF6 start codon using JASPAR (Fornes et al., 2020). TFBS predictions were grouped into TF classes and expressed as relative frequency % of TFBSs corresponding to a particular TF family from the total TFBSs predicted (n = 107, after applying a ≥ 10 matrix score cut-off to maximize TFBS prediction accuracy). Other TF class indicate a mixed class of low-represented TFBSs (frequency within HvCslF6 full length promoter ≤ 2%). Average prediction scores for each TF class are shown in orange on top of each bar and were inferred from position frequency matrices (PFMs) and TF flexible models (TFFMs) in JASPAR. (B) Venn diagram of TFBS predicted within putative CslF6 promoter regions (–1,000 bp) across barley (Hordeum vulgare), wheat (Triticum aestivum), Brachypodium distachyon and rice (Oryza sativa). (C) Venn diagram of TFBSs predicted within HvCesA1, HvCesA2, HvCesA6, and HvCslF6 putative promoter regions (–1,000 bp) with identical filtering as described above. Venn diagrams were created using InteractiVenn (Heberle et al., 2015).
To understand if there was conservation of binding site motifs across grass species, we carried out in-silico analysis of the putative CslF6 promoter sequences in wheat (Triticum aestivum), Brachypodium distachyon and rice (Oryza sativa), and compared the TFBS to those identified in barley (filtered by a ≥ 10 prediction accuracy score cut-off, as previously applied to HvCslF6 full length promoter analysis). This revealed 59 common motifs across all species (Figure 2B and Supplementary Table 2) corresponding to: MYBs/SANTs, ARFs, AP2/ERFs, AT-Hook DNA-binding factors, bHLHs, DREBs, and HD-ZIPs TF classes. The second largest group of common motifs (26) was shared between barley, wheat and Brachypodium CslF6 upstream sequences and corresponded to putative binding sites for MYB-related TFs, (predominantly MYBs/SANTs: MYB55/61, MYB59, MYB111 and MYB113; GARP/G2-LIKE: PHL11, PHL12; MYB-related: UIF1), and other less abundant bHLH and DOF-related motifs. The rice CslF6 upstream sequence contained the highest number of unique TFBS predictions (84) compared to other cereals (13, barley; 13, wheat; and 14, Brachypodium). The frequency, location, and description of predicted TFBS across all species is shown in Supplementary Table 2.
Next, the analysis of TFBSs was expanded to include three more members of the Cellulose synthase gene superfamily, namely HvCesA1, HvCesA2 and HvCesA6, which are involved in cellulose biosynthesis in the primary cell wall (Burn et al., 2002; Burton et al., 2006) and known to be co-expressed with HvCslF6 (Burton et al., 2004; Burton and Fincher, 2009). Common predicted motifs across HvCesA1, HvCesA2, HvCesA6, and HvCslF6 corresponded to MYB-related, AP2/ERF, HD-ZIP, DOF, ZF-HD, Homeo Domain (HD), and MADS box factors. The largest set of common motifs was found across HvCesA2 and HvCesA6 putative promoters (77), followed by HvCesA2 HvCesA6, and HvCslF6 (60) (Figure 2C). As a comparison three HvCesA genes (HvCesA4, HvCesA7, and HvCesA8) which are not co-expressed with HvCslF6 were also analyzed for TFBSs. This analysis identified one common predicted TFBS, a SQUAMOSA promoter-binding protein-like (SPL) motif across all putative promoters (SPL TFs are mainly involved in plant growth and development; Tripathi et al., 2018) and abundant gene-specific, not shared, motif subsets. The largest set of TFBSs was found in the HvCslF6 promoter (57) containing MYBs, AP2/ERFs HD-ZIP among other TF classes. (Supplementary Figure 3 and Supplementary Table 3).
To identify the HvCslF6 promoter region necessary for transcriptional activation, seven progressively deleted promoter constructs were generated from the 3,000 bp region upstream of the HvCslF6 start codon (−3,000, −1,846, −1,357, −858, −607, −382, and −199 bp) and tested in a barley leaf-derived protoplast system (previously optimized using GFP/YFP-expressing constructs; Supplementary Figure 4). The region of the promoter that produced the highest relative luciferase assay activity was a −382 bp region upstream of the HvCslF6 start codon (Figure 3A). Additionally, deletion constructs containing a −607 bp or −199 bp HvCslF6 promoter region showed a significant decrease in promoter activity based on the relative luciferase assay compared to the −382 bp region. Therefore, we had delimited a 183 bp sequence within the proximal HvCslF6 promoter between the two smallest deletion constructs (from −382 to −199 bp) that showed a relative increase in luciferase activity and could be responsible for upregulation of HvCslF6 expression. HvCslF6 promoter activity was comparatively low from −607 to −3,000 bp upstream of the HvCslF6 CDS, indicating that repressors of HvCslF6 expression could be binding to this −382 to −607 bp region. Furthermore, no significant differences in promoter activity were observed across constructs carrying the longest HvCslF6 promoter fragments (−3,000, −1,846, and −1,357 bp; Supplementary Figure 5).
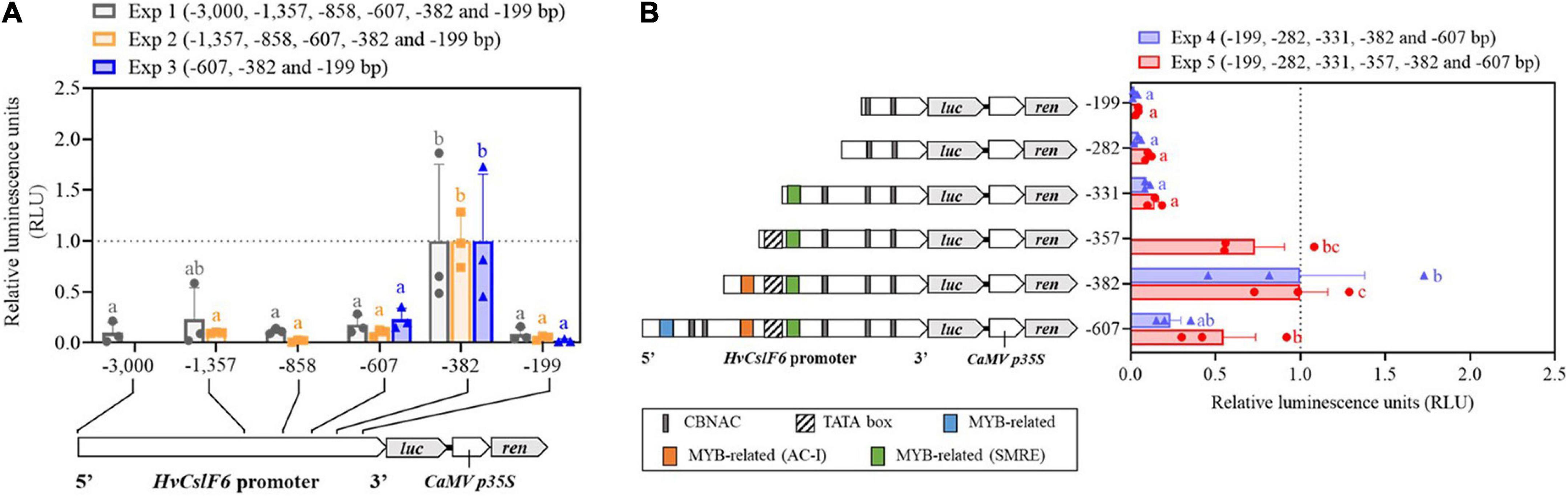
Figure 3. Characterization of the HvCslF6 promoter using dual luciferase assays to identify region(s) response for increase in expression. Promoter activity in panels (A,B) is expressed as relative luciferase activity (luciferase/renilla ratio) and normalized to the –382 construct (with highest promoter activity, dashed gray line) on each experiment, respectively. Three independent protoplasts transfections (biological replicates) were performed per deletion construct and averaged to calculate mean reporter activity 24 h after transfection for each experiment. Negative controls (n = 3 empty vector lacking HvCslF6 promoter) were performed in parallel and luciferase background activity subtracted from tested constructs (n = 3 dual luciferase assays per construct, as technical replicates). (A) HvCslF6 promoter activity analyzed in six deletion constructs (from –3,000 bp full length promoter to –199 bp) using dual luciferase assays across three independent experiments (Exp1, Exp2, and Exp3). Letters above each construct indicate significant differences within each experiment determined by one-way ANOVA (p-value 0.041, Exp1; p-value < 0.01, Exp2 and p-value 0.027, Exp3) followed by Tukey’s multi-comparison test. Error bars represent standard error (n = 3). (B) HvCslF6 promoter activity of 5′ HvCslF6 promoter deletion constructs from –607 bp to –199 bp in barley protoplasts. Gray bar indicates CBNAC calmodulin-binding NAC motif, blue bars indicate MYB-related motifs, yellow bars denote MYB-related AC-I motifs, green bars indicate secondary wall MYB-responsive element, (SMRE) and the diagonally stripped bar denotes predicted location of the TATA box consensus sequence. Different letters above each construct (i.e., a,b) indicate significant differences determined by one-way ANOVA (p-value < 0.01) followed by Tukey’s test. Error bars represent standard error (n = 3).
We retrieved HvCslF6 proximal promoter regions (1,000 bp upstream from start codon) from cv. Morex and cv. Golden Promise and carried out a pairwise sequence alignment. Both promoters were identical based on the barley pangenome sequences available at GrainGenes1 therefore subsequent in silico prediction of TF binding sites could also be extended to cv. Golden Promise.
To further delimit putative TF binding sites within the region that showed increased luciferase activity, three additional deletion constructs were generated. Constructs containing −282 bp, −331 bp, and −357 bp of the HvCslF6 promoter, were tested alongside three previously tested deletion constructs (−199 bp, −382 bp, and −607 bp) to normalize dual luciferase measurements and compare promoter activity across deletion constructs (Figure 3B). A significant increase in promoter activity (p-Value 0.029, Tukey′s test) was detected in the −357 bp deletion construct. In-silico prediction of this promoter fragment indicates that it includes a putative TATA box element (5′–TATAAA–3′). This likely explains the increase in HvCslF6 promoter activity with the inclusion of this region of the promoter, and the low/zero levels of promoter activity in the first three constructs which based on our in-silico prediction lack a TATA box. Highest HvCslF6 promoter activity was narrowed down to a 51 bp region located −331 bp to −382 bp upstream of the start codon, although no significant differences (p-Value 0.321, Tukey’s test) were determined between the −357 bp and −382 bp deletion constructs. Moreover, PlantPAN and JASPAR in-silico motif analysis of the additional 25 bp included in the −382 bp fragment compared to the −357 bp fragment predicted a single MYB-related motif, AC-I, enriched in AC bases. Results also indicated low HvCslF6 promoter activity from −199 bp to −331 bp based on three deletion constructs analyzed (−199 bp, −282 bp, and −331 bp). This region contained two (three in the case of −331 bp construct) predicted calmodulin-binding NAC elements and a MYB-related motif, described as a secondary wall MYB-responsive element (SMRE) based on JASPAR analysis (Figure 3B).
Based on previous studies which suggest a key role for NAC and MYB TF families in SCW polysaccharide biosynthesis (Taylor-Teeples et al., 2015) and the involvement of MYB TFs in grass cell wall synthesis including hemicelluloses (Zhao et al., 2019), we mapped two putative MYB-related cis-elements (among other predicted motifs from our in-silico analysis) onto HvCslF6 promoter regions that conveyed increased promoter activity (Figure 3B and Table 1). A six-nucleotide, AC-I element (5′–ACCTAC–3′) was predicted from −364 to −358 bp upstream from the HvCslF6 start codon. Prouse and Campbell (2013), demonstrated that the R2R3-MYB AtMYB61 can bind to this AC-I element, therefore we were interested in the barley ortholog of this gene as a potential regulator of HvCslF6 expression. A putative binding site in construct −331 (5′–CGTTGGT–3′/3′–ACCAACG–5′) corresponds to another MYB consensus motif, also described as a secondary wall MYB-responsive element (SMRE) or a MYB46-responsive cis-regulatory element, M46RE (Zhong and Ye, 2012). This cis-element was detected in reverse orientation (3′–5′) and in Arabidopsis the same motif can be bound by AtMYB46 and AtMYB83 (Kim et al., 2012; Ko et al., 2014). Additionally, a secondary wall MYB-responsive element (SMRE) motif was predicted from −299 to −292 bp, downstream of the TATA box in the HvCslF6 promoter (see construct −331, Figure 3B). However, this had limited impact on promoter activity, likely due to the absence of the TATA box.
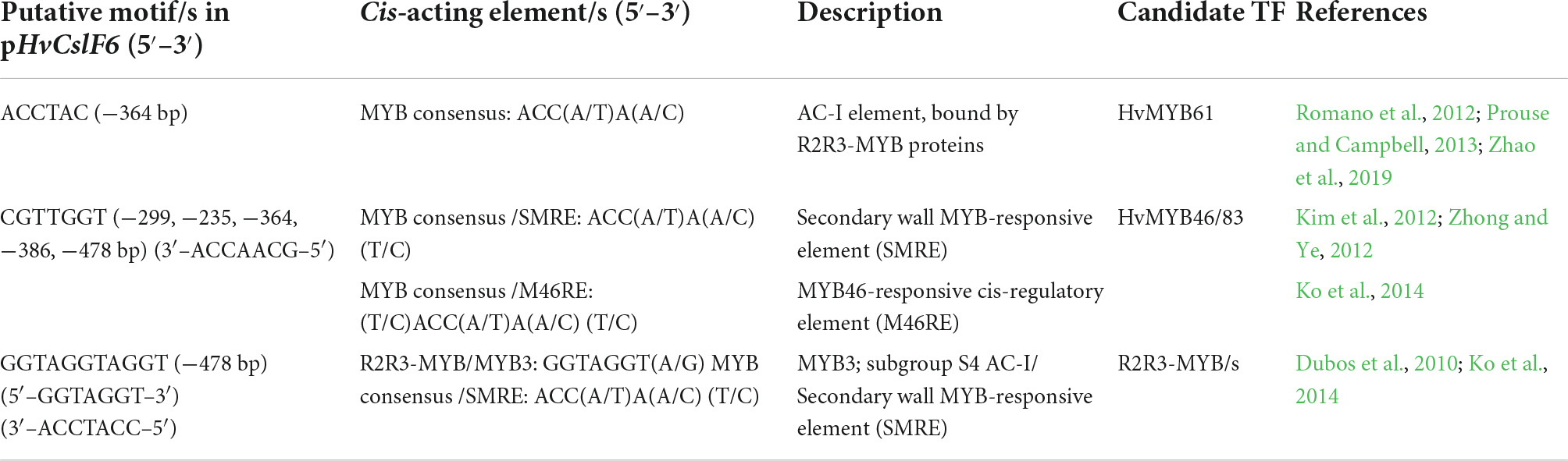
Table 1. Description of putative binding sites found in the HvCslF6 promoter based on dual luciferase results of deletion constructs with differential promoter activity.
To assess protein sequence similarities in MYB46, MYB83, and MYB61 orthologs between barley (Table 2), Arabidopsis (Romano et al., 2012) and rice (Zhong et al., 2011), we constructed an unrooted phylogenetic tree of R2R3-type MYB transcription factors which includes those that could putatively bind to the −382 bp HvCslF6 promoter region (Supplementary Figure 6). The candidate MYB transcription factors that correspond to the predicted binding sites belong to the plant specific R2R3-MYB subfamily, which is characterized by the presence of two highly conserved MYB DNA-binding domains (Pfam PF00249; Bateman et al., 2017) and constitutes the largest MYB subgroup. HvMYB61 was grouped together with their orthologs from Arabidopsis (AtMYB61; Prouse and Campbell, 2013) and rice (OsMYB61a and OsMYB61b; Zhao et al., 2019). The best fitting barley ortholog for AtMYB46 and AtMYB83 was a single gene named HvMYB46/83, following previous studies in rice (OsMYB46/83; Rao and Dixon, 2018; Zhao et al., 2019) and grouped with its orthologs (Supplementary Figure 6).
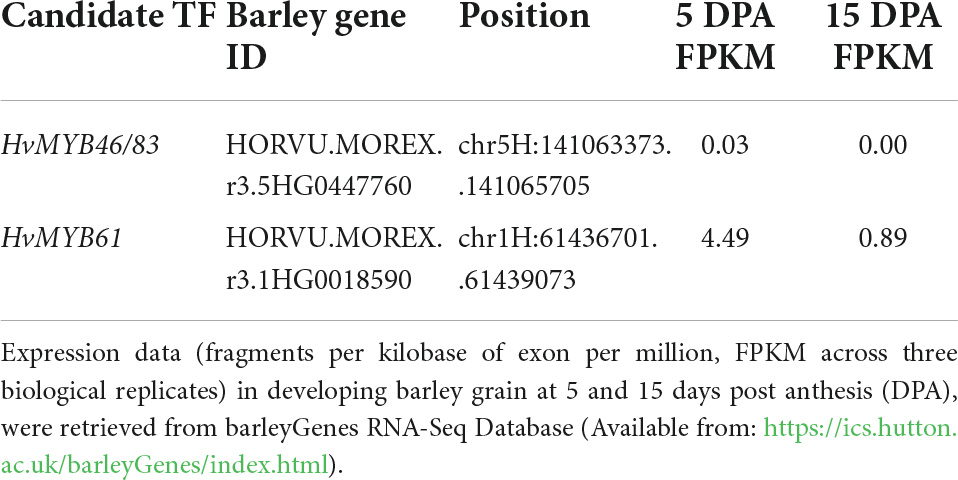
Table 2. Description of candidate transcription factors retrieved from GrainGenes (https://wheat.pw.usda.gov/GG3/) based on cv. MorexV3 (Mascher et al., 2021).
Using gene expression data from 808 individual samples (Supplementary Table 4; Milne et al., 2021), we constructed a co-expression network to identify genes closely connected to HvCslF6. The samples covered a wide range of biotic and abiotic stresses, tissue types and cultivars, resulting in a robust dataset. Low expressed genes were removed (see section “Materials and methods”) and a weighted gene correlation network analysis (WGCNA) performed (Langfelder and Horvath, 2008). We identified 33 modules, with a color assigned to each module, plus one module (gray) which contains the unallocated genes. HvCslF6, along with the primary cell wall cellulose synthases (HvCesA1, HvCesA2, HvCesA6) were assigned to the yellow module (Supplementary Figure 7). HvMYB61 (one of the candidate HvCslF6 regulators) and HvTHX1, the barley ortholog of BdTHX1, which binds to the intronic region of BdCslF6 and acts as a positive regulator of expression of this gene (Fan et al., 2018), were all members of the yellow module. Visualization of the yellow network in Gephi showed a gene network comprising 992 genes and 54,090 gene connections (Supplementary Figure 8). To better investigate the connections, an intermediate stringency threshold of 0.188 (the correlation ranged from 0.15 to 0.335) was applied to define a subnetwork of the top 50 genes connected to HvCslF6.
This subnetwork based around HvCslF6 still included HvMYB61 (BART1_0-p01380 and the three cellulose synthases; HvCesA1, HvCesA2, HvCesA6, BART1_0-p60277, BART1_0-p40943, BART1_0-p44934) (Figure 4). All are either known to be involved in primary cell wall synthesis in barley, or in the case of HvMYB61 the ortholog of OsMYB61 has been shown to influence to cell wall synthesis (Zhao et al., 2019). Additionally, this module contained other proteins that have been linked to cell wall processes (Supplementary Table 5). COBRA-like protein 3 (BART1_0-p39134) located on chromosome 5H was shown to be co-expressed with HvCesA1, HvCesA2, and HvCesA6 (Houston et al., 2015). The other candidate regulator, HvMYB46/83 did not pass the filtering threshold when removing genes with low levels of expression or were assigned to the black instead of yellow module. Hence, this candidate regulator is unlikely to be a global regulator of HvCslF6.
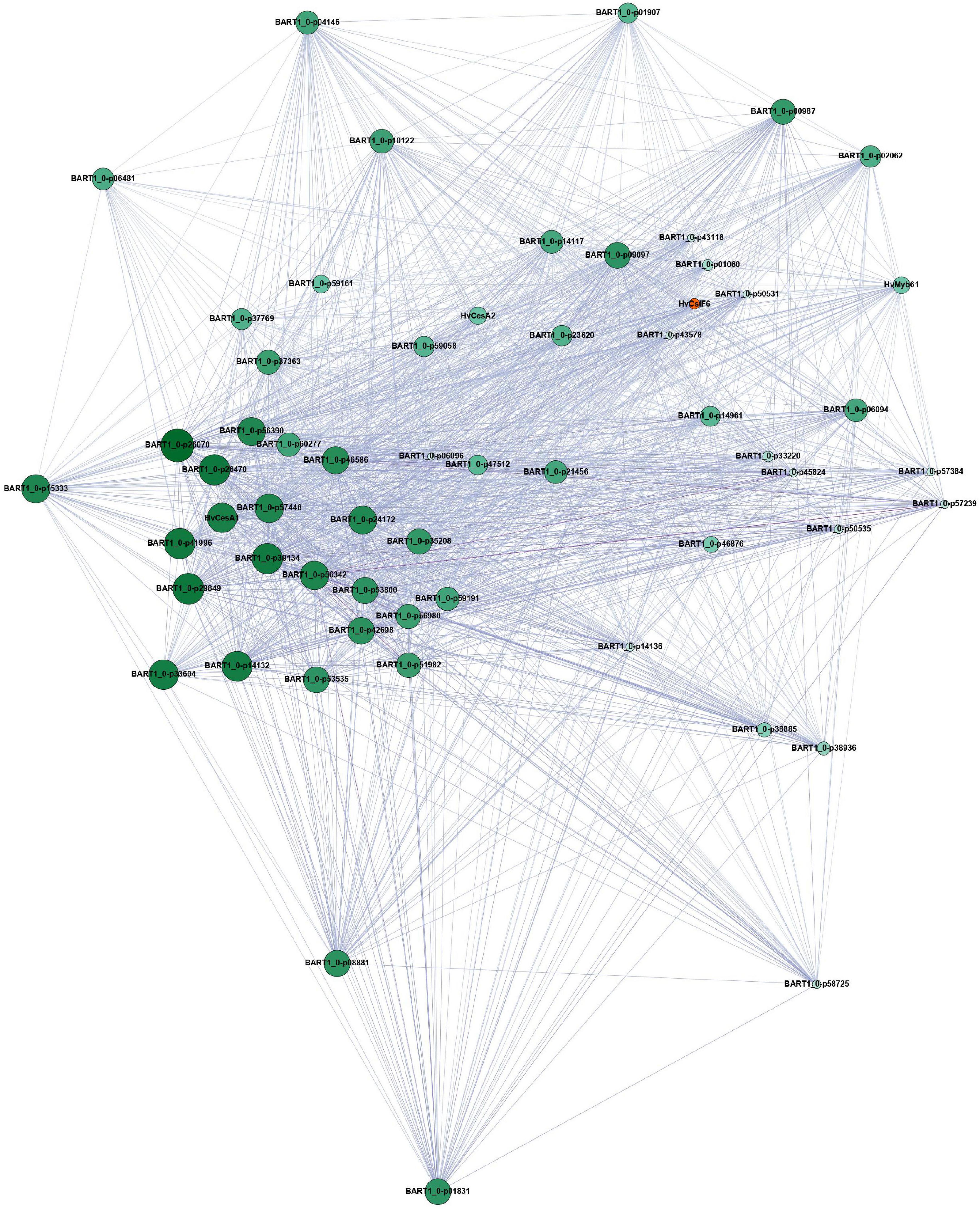
Figure 4. Filtered yellow module network based on genes connected to HvCslF6 (n = 50), visualized using Gephi. This subnetwork was obtained from the publicly available EoRNA database (Milne et al., 2021). Filtering parameters are described in the corresponding methods section. Node size and color correspond to gene degree centrality. Big and dark green nodes have a high number of linked nodes, while small and light green nodes only have a small number of linked nodes. The edges are colored by weight. A dark purple corresponds to a high weight and therefore correlation, while a light purple color corresponds to a lower weight and less strong correlation between the nodes. HvCslF6 is highlighted in orange. Gene names correspond to the description in Supplementary Table 5. Genes not listed in the table do not contain protein annotation and are potential non-coding genes.
Then we carried out a correlation analysis using transcript expression data from the eoRNA database (Milne et al., 2021), which contains 22 publicly available datasets and >800 RNA-seq datasets from a wide range of tissues, development stages and experimental treatments. We focused on two subsets of genes. The first included our candidate TF gene HvMYB61, a selection of genes from the network analysis and others known to be co-expressed with HvCslF6 (i.e., HvCesA1, HvCesA2, and HvCesA6). The second subset was comprised of secondary cell wall genes not co-expressed with HvCslF6 (i.e., HvCesA4, HvCesA7, HvCesA8), and included other members of the HvCslF/H/J gene family, and HvTHX1, the barley ortholog of BdTHX1, a positive regulator of BdCslF6 (Figure 5A and Supplementary Figure 9). We hypothesized that if HvMYB61 shows comparable or greater correlation with HvCslF6 compared to HvCesA1, HvCesA2, HvCesA6, this would provide further confidence in a relationship between these candidates. However, if strong positive correlation is observed between the expression of HvMYB61, our candidate gene, and the group considered to be secondary cell wall HvCesA genes (HvCesA4, HvCesA7, and HvCesA8), which show no correlation with HvCslF6, it might suggest that our candidate gene is unlikely to be involved in the broader regulation HvCslF6 expression.
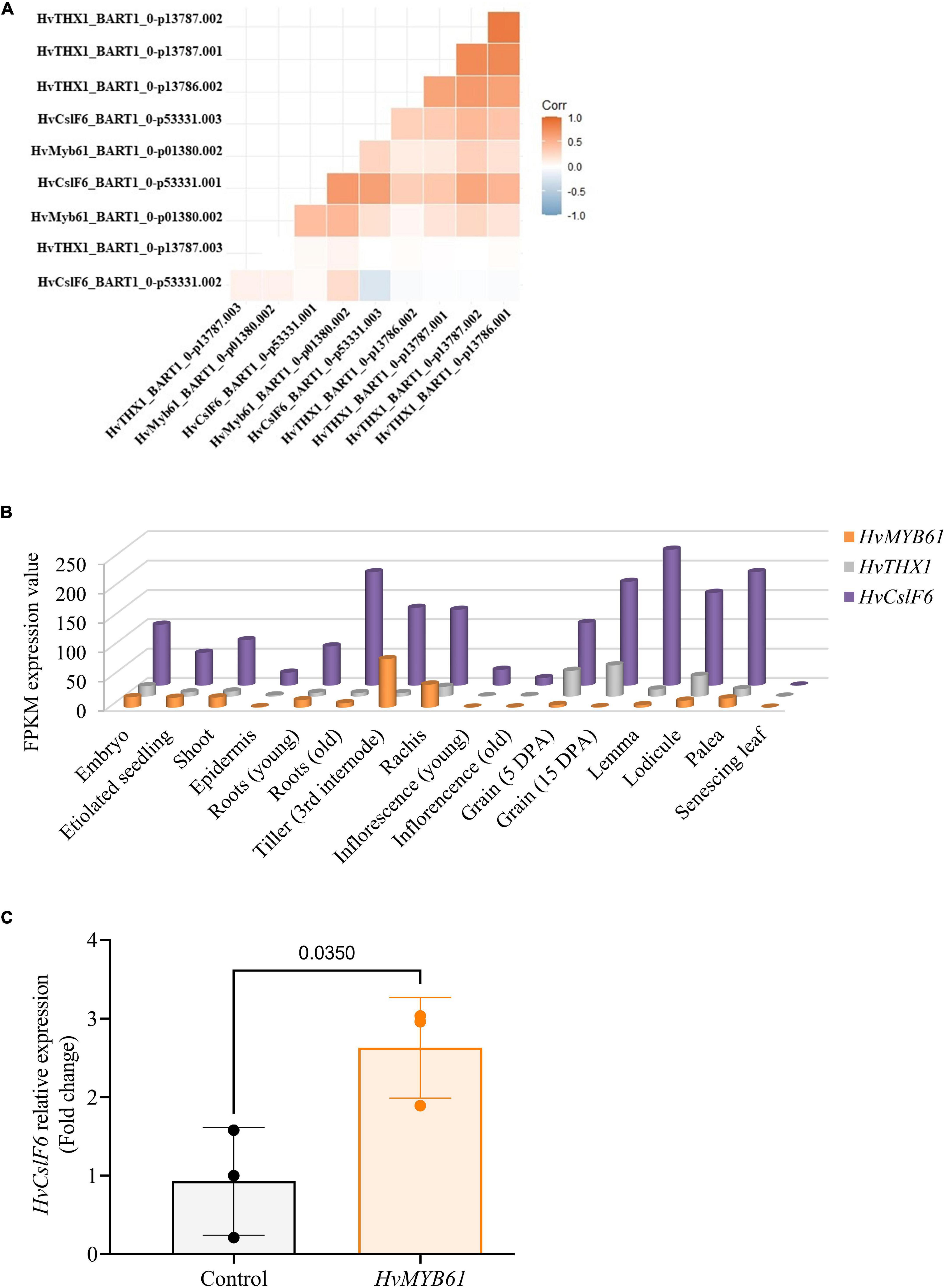
Figure 5. Expression profiles of candidate upregulators of HvCslF6 expression in several datasets. (A) Correlation between TPM values of selected candidate genes from the EoRNA database ordered based on hierarchical clustering. (B) Expression levels of HvMYB61 candidate transcription factor, HvTHX1 and HvCslF6 across 16 barley tissues obtained from barleyGenes RNA-seq database. (Available from: https://ics.hutton.ac.uk/barleyGenes/, The James Hutton Institute), expressed as FPKM (fragments per kilobase of exon per million fragments mapped) across three biological replicates per tissue. (C) Relative expression of HvCslF6 in barley protoplasts transfected with an empty vector and over-expressing HvMYB61, respectively, after 24 h. Internal control genes: α-tubulin, GAPDH and HSP70 were used for normalization of relative expression (2–ΔΔCT method; Vandesompele et al., 2002). Error bars represent standard deviation associated with three independent experiments (transfection assays n = 3). qRT-PCR reactions were performed in triplicate. P-value was calculated based on Student’s t-test (two-tailed).
For the transcripts of genes considered to be primary cell wall related CesA genes and HvCslF6, we observed Pearson correlation coefficients of between r = 0.9 to −0.1, while the secondary cell wall CesA genes had r = 0.4 to −0.2 (Supplementary Figure 9). The range of values is not only due to several genes being compared but also multiple transcripts per gene. For several transcripts there was a high degree of correlation between the expression profile for HvMYB61 and HvCslF6 (Person correlation coefficient r = 0.7 to 0.1) and HvTHX1 and HvCslF6 (Person correlation coefficient r = 0.6 to −0.1). Therefore, this analysis from an extensive range of samples and conditions provides additional evidence supporting the similar expression dynamics of HvMYB61 and HvCslF6.
To further investigate the expression profile of candidate MYB transcription factors we surveyed several smaller transcript datasets from barley that are independent of those described above. An RNA-seq dataset from cv. Morex across 16 barley tissues showed that HvMYB61 is expressed abundantly across the entire barley plant (Figure 5B). HvMYB61 expression was detected in 10-day seedling, shoot (10 cm), tiller (3rd internode), rachis, developing grain at 5 days post anthesis (DPA) and embryo; these are tissues where (1,3;1,4)-β-glucan is present. Similarly, HvTHX1 is also expressed in multiple tissues including developing grain (5 and 15 DPA). Expanding the RNA-seq expression data across barley grain development, an independent expression dataset based on six different spring barley cultivars (cvs. Hopper, Sloop, Extract, Taphouse, Alabama, and Pewter; Aubert et al., 2018; Matros et al., 2021) was used to retrieve the expression profiles of the candidate genes from early to mid-late grain development. HvMYB61 was mainly expressed during early grain development, from 7 to 11 DPA, whereas HvMYB46/83 expression remained almost undetectable from 7 to 20 DPA. In the RNA-seq dataset (developing grain) Pearson correlation coefficients with HvCslF6 were: 0.95 for HvMYB61 and 0.98 for HvTHX1 (showing abundant expression from 7 to 20 DPA; Supplementary Table 6). Our results showed that the positive correlation between HvMYB61 and HvCslF6 across different tissues and datasets, in the context of HvTHX1 (ortholog of BdTHX1, known activator of BdCslF6; Fan et al., 2018) and other HvCesAs (co-expressed and not co-expressed), are consistent with a regulatory effect of HvMYB61.
Based on expression profiles, our luciferase assay results and data from gene network analyses, we focused subsequent assays on the HvMYB61 transcription factor, since evidence suggested this gene may influence HvCslF6 expression. The candidate regulator HvMYB46/83 was ruled out based on low levels of expression from gene co-expression analyses. The candidate transcription factor HvMYB61 was cloned and transfected into barley protoplasts plus an empty vector control, respectively. After 24 h, protoplasts were harvested and HvCslF6 relative transcript abundance was measured. We observed a significant increase in HvCslF6 expression in the presence of HvMYB61, representing a fold change of 2.63 ± 0.64 in HvCslF6 expression compared to protoplasts transfected with an equal amount of empty vector (p-Value 0.035, Student’s t-test; Figure 5C). Taken together, our results from the functional characterization of the HvCslF6 promoter, TF identification and gene co-expression analyses, indicate that the HvMYB61 transcription factor may be a positive regulator of HvCslF6 expression in barley.
The transcriptional regulation of primary cell wall formation and in particular (1,3;1,4)-β-glucan biosynthesis, which is present across grasses including many economically important crop species, remains largely unresolved. The interest in the identification of regulators able to fine-tune HvCslF6 expression and grain (1,3;1,4)-β-glucan content ultimately arises from the beneficial health effects associated with (1,3;1,4)-β-glucan-rich diets (Bai et al., 2019) and its importance for efficient brewing and distilling (Gupta et al., 2010). Several transcription factor families (predominantly NACs and MYBs) are known to regulate polysaccharide biosynthesis in secondary cell walls, some of them with conserved functions between grasses and Arabidopsis (McCarthy et al., 2009; Zhou et al., 2009; Ko et al., 2014).
We delimited a region of the functional promoter of HvCslF6 which contained a binding site motif for MYB61. We observed that when this gene was overexpressed in the presence of HvCslF6 it led to an increase in HvCslF6 expression relative to control samples which lacked HvMYB61. This reflects the results of Zhao et al. (2019) who observed that OsCslF6 and other cell wall related genes were upregulated in rice protoplasts overexpressing OsMYB61. The authors also reported a 31% decrease in leaf (1,3;1,4)-β-glucan content was observed in OsMYB61a knockout mutants compared to WT. Barley and rice have distinct patterns of (1,3;1,4)-β-glucan distribution throughout the plant, with barley having relatively high levels of this polysaccharide in the grain compared to vegetative tissue (Burton and Fincher, 2014); the opposite is true of rice with extremely low levels of (1,3;1,4)-β-glucan present in the grain (Vega-Sanchez et al., 2012). Phenotypic analyses of AtMYB61 loss- and gain-of-function mutants showed similar results to OsMYB61 RNAi lines (Hirano et al., 2013), confirming a role for this gene in secondary cell wall formation in Arabidopsis, in addition to regulating other traits (Penfield et al., 2001; Newman et al., 2004; Romano et al., 2012; Romero-Romero et al., 2018). At present, evidence for a direct interaction between HvMYB61 and the putative binding site in the promoter of HvCslF6 is lacking, as we did not carry out a yeast-1-hybrid experiment. Although Zhao et al. (2019) did not observe any evidence of direct binding between OsMYB61 and OsCslF6, it is important to consider that the levels of (1,3;1,4)-β-glucan are typically 5-fold lower in rice than barley, and hence the expression or function of regulatory genes might be different between these two species.
Unlike rice, which contains two MYB61 genes (OsMYB61a and OsMYB61b; Zhao et al., 2019), HvMYB61 was only present as a single gene on barley chromosome 1H at 47.8 cM. Notably, this co-located with an association peak for grain (1,3;1,4)-β-glucan content (48.4 cM) which was identified in a previous genome wide association scan using elite barley germplasm (Houston et al., 2014). A QTL for malt (1,3;1,4)-β-glucan was detected in the same genomic region using a cv. Steptoe × Morex population (Han et al., 1995). Other genes described in this region of chromosome 1H are HvCslF9, a putative grain (1,3;1,4)-β-glucan synthase based on sequence similarity to other Csl genes, and HvGlbI, a (1,3;1,4)-β-glucan endohydrolase. HvGlbI has been shown to hydrolyse both malt (1,3;1,4)-β-glucan and (1,3;1,4)-β-glucan from germinating grains (Slakeski and Fincher, 1992; Betts et al., 2017) and therefore is a plausible candidate for contributing to variation underlying the QTL’s mentioned above (Han et al., 1995; Houston et al., 2014). However, because knockout mutants for HvCslF9 exhibit similar (1,3;1,4)-β-glucan content to wild-type barley grain, it seems likely that this gene plays a minor role, if any, in determining mature grain (1,3;1,4)-β-glucan content (Garcia-Gimenez et al., 2020). Hence, while HvGlbI and HvCslF9’s contribution to grain (1,3;1,4)-β-glucan content is at least already partially understood,HvMYB61 appears to be an enticing candidate for further investigations of variation in HvCslF6 expression during grain development.
We also identified another R2R3-MYB binding site located from −299 to −292 bp in the HvCslF6 promoter. This was functionally characterized as a secondary cell wall MYB-responsive element, SMRE which can be bound by MYB46 and MYB83 in Arabidopsis thaliana, regulating secondary wall biosynthesis (Kim et al., 2012; Zhong and Ye, 2012). This SMRE motif was also described as a MYB46-responsive cis-regulatory element, M46RE (Ko et al., 2014). However, HvMYB4/83 mRNA levels were barely detectable in most barley tissues, inconsistent with a major role in controlling (1,3;1,4)-β-glucan content. In contrast, HvMYB61 expression was detected across several tissues where (1,3;1,4)-β-glucan is present, (and HvCslF6 is expressed) including early stages of grain development when (1,3;1,4)-β-glucan biosynthesis is occurring. No significant differences in promoter activity were observed for constructs carrying the longer HvCslF6 promoter fragments (−3,000, −1,846 and −1,357 bp; Supplementary Figure 5), with these fragments generating comparatively low levels of promoter activity. One of these fragments encompassed the section of the HvCslF6 promoter (from −1,560 to −1,567 bp of the HvCslF6 start codon) which Wong et al. (2015) had identified an 8 bp insertion in cv. TR251. However, this deletion was absent from all other cultivars in Wong et al. (2015) and the present study.
We observed further support for a relationship between HvMYB61 and HvCslF6 from a comprehensive gene network analysis of HvCslF6 using 807 gene expression sets from different developmental stages and treatments. This analysis assigned HvMYB61 to the same co-expression module as HvCslF6, the primary cellulose synthases (HvCesA1, HvCesA2, and HvCes6), and HvTHX1, the ortholog of a known enhancer of BdCslF6 expression (Fan et al., 2018), consistent with our hypothesis that HvMYB61 could act as positive regulator of HvCslF6 expression. Subsequent analysis of HvMYB61 by transient over-expression in a barley protoplast system led to a significant increase in HvCslF6 expression in the presence of HvMYB61 compared to protoplasts transfected with an equal amount of empty vector across three independent experiments. In line with the study from Zhao et al. (2019) in rice, our results suggest that in cereals, MYB61 may have a conserved role in regulating the expression of grass-specific cell wall biosynthetic enzymes including HvCslF6.
The focus of the current study was to delimit putative cis-elements and transcription factors controlling the expression of HvCslF6, and to use these findings to further refine our knowledge of what determines (1,3;1,4)-β-glucan content and ultimately provide diagnostics to enable selection for variation in this trait. Based on our findings, we speculate that selected MYB transcription factors may act as positive regulators of (1,3;1,4)-β-glucan accumulation in barley primary cell walls. This relationship relates predominantly to HvMYB61 and the proximal promoter region of HvCslF6 and is supported by co-expression in a range of tissues including the young endosperm. However, we anticipate that the regulation of HvCslF6 expression could be tissue- and genotype-specific. We previously demonstrated that HvCslF6 expression is genotype-dependent in a relatively genetically narrow selection of barley cultivars (Garcia-Gimenez et al., 2019). Additionally, the temporal and spatial variation of the expression of this gene has been well characterized previously (Burton et al., 2008). Therefore, expanding this work to include a wider set of germplasm, tissues and generating expression profiles for additional genes such as HvMYB61 would provide additional insight into the relationship between these genes.
The decrease in HvCslF6 promoter activity observed in the −607 bp construct compared to the −382 bp construct requires further investigation. Recent studies also describe the emerging role of MYBs as repressors, negatively affecting secondary cell wall biosynthesis (Zhao and Bartley, 2014; Rao and Dixon, 2018) and cold acclimation (Zhang et al., 2016b), among other traits (Zhou et al., 2017; Ma and Constabel, 2019). It is also possible that repressive cis-elements are present in the HvCslF6 promoter. The dual luciferase system allowed us to screen a more distal promoter region from −382 to −607 bp, which showed comparatively low levels of luciferase activity. This suggests that other potential regulatory factors may bind to the HvCslF6 promoter in the protoplast system, inhibiting the expression of this gene. TFBS predictions in this 225 bp region correspond to several TF families and include additional MYB-related elements. Additionally, it will be intriguing to assess the function of THX1 barley orthologs, and any potential additive effect with HvMYB61.
Barley transgenic lines (cv. Golden Promise) carrying a 3,000 bp upstream region from HvCslF6 start codon fused to a endoplasmic reticulum-targeted GFP (pHvCslF6:mGFP-ER) construct were generated via Agrobacterium-mediated transformation at The University of Adelaide following the protocol outlined in Burton et al. (2011). Transgenic lines (T2) were screened using a Zeiss Axioskop 2 Plus microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) equipped with an external UV light source (HBO 100). mGFP was excited at 488 nm and emission collected at 500–530 nm. For coleoptile screening, lines were grown in a petri dish with filter paper for 3–4 days in the dark and screened 5 days after gemination. Transverse section images were captured with an AxioCam 512. Subsequent to collection, images were processed with Zen Software v6.0, utilizing global adjustment tools only.
A 3,000 bp region upstream of the HvCslF6 start codon (cv. Morex) was retrieved from the barley genome explorer (Mascher et al., 2021) and used for TFBS prediction comparison using TRANSFAC® v2014 (Matys et al., 2006), JASPAR v2020 (Fornes et al., 2020) and PlantPAN v3.0 (Chow et al., 2019). Motif over-representation analysis was carried out using HvCslF6 TFBS predictions from JASPAR and filtered based on ≥ 10.0 prediction score (maximum score = 15) to minimize false positive motif predictions. Upstream CslF6 sequences from wheat (Triticum aestivum), Brachypodium distachyon and rice (Oryza sativa) were retrieved from Ensembl Plants2. TFBS comparisons across species were carried out within a 1,000 bp CslF6 putative promoter region using JASPAR (Fornes et al., 2020) and predicted motifs filtered based on ≥ 10.0 prediction score. The same filter was also applied for the TFBS analysis of HvCesA upstream regions (1,000 bp). Venn diagrams were created using InteractiVenn (Heberle et al., 2015).
Barley plants (cv. Golden Promise) were grown in a growth chamber with a 16-h light/8-h dark photoperiod at the Functional Genomics (FUNGEN) facility, at The James Hutton Institute, United Kingdom. After 3 weeks, 0.25 g of primary leaves were harvested for protoplast assays. Leaves were cut lengthwise and crosswise (∼20 mm × 10 mm) and peeled (epidermis removal from the abaxial side) in sterile conditions using a sharp razor, scalpel and tweezers under a Leica MZ6 stereo microscope.
The barley protoplast isolation protocol was mainly based on the protocol for preparation of Arabidopsis mesophyll protoplasts (Yoo et al., 2007) with modifications described in Supplementary Method 1. Briefly, barley leaves without the epidermis were transferred into 5 mL of the enzyme solution [2% w/v Cellulase Onozuka R-10 (Duchefa, Haarlem, Netherlands), 0.1% w/v Macerozyme R-10 (Duchefa, Haarlem, Netherlands), 0.1% w/v BSA (Sigma-Aldrich, St. Louis, MO, United States), 0.55 M Mannitol (Sigma-Aldrich, St. Louis, MO, United States), pH 5.7]. Leaves were incubated in a 6-well cell culture plate (Sigma-Aldrich, St. Louis, MO, United States) in the dark for 2 h at 28°C. Released protoplasts were mixed with one volume of W5 solution [154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES (Sigma-Aldrich, St. Louis, MO, United States, all), pH 5.7], filtered through a 70 μM gauze (Sigma-Aldrich, St. Louis, MO, United States), washed with 2.5 mL of W5 solution and centrifuged for 3 min at 70 × g, RT (Centrifuge 5810 R, Sigma-Aldrich, St. Louis, MO, United States). Protoplasts were gently re-suspended in 5 mL W5 solution and kept on ice for 30 min to allow cell sedimentation by gravity. Protoplasts were re-suspended to a final concentration of approximately 2 × 105 protoplasts/mL in MMG solution (0.6 M Mannitol, 4 mM MES, 15 mM MgCl2, pH 5.7). Fluorescein diacetate (FDA) was used to determine cell viability under a using a UV-light Zeiss Axioskop 2 Plus microscope and Canon EOS D100 digital camera.
For each transfection assay, 100 μL of protoplasts were mixed with 7 μg of construct DNA and 110 μL of freshly prepared 40% PEG-solution [0.4 M mannitol, 40% w/v PEG-4000 (Sigma-Aldrich, St. Louis, MO, United States), 0.1 M CaCl2 (Sigma-Aldrich, St. Louis, MO, United States), pH 5.7]. After a 10 min incubation in the dark, protoplasts were washed twice with 1.5 mL of W5 solution and centrifuged at 200 × g for 5 min (Centrifuge 5415 D, Sigma-Aldrich, St. Louis, MO, United States). Protoplasts were re-suspended in 80 μL of W5 solution and incubated in the dark for 24 h, at room temperature. Three independent transfection assays were performed per construct and averaged as biological replicates.
The HvCslF6 (HORVU.MOREX.r3.7HG0698110) promoter deletion constructs were generated by PCR amplification of a 3,000 bp region upstream HvCslF6 start codon as follows: 1 μL cv. Golden Promise gDNA, 1.25 μL each primer (forward and reverse listed in Supplementary Table 7) at 10 mM, 5 μL 5X Phusion HF Buffer, 0.2 μL Phusion HF DNA polymerase (Thermo Fisher Scientific, Waltham, MA, United States) and 11.3 μL sdH2O in a total volume of 20 μL. Phusion PCR reaction conditions: 98°C for 30 s, 35 cycles (98°C for 10 s, 55°C for 30 s, 72°C for 30 s/kb), 72°C for 5 min and 4°C hold. Promoter fragments were ligated by NotI restriction enzyme cloning into pGreenII 0800-LUC vector which contains luciferase and renilla reporter genes, enabled to be used in Dual Luciferase Reporter Assays (Promega, Madison, WI, United States). A total of seven PCR amplicons corresponding to HvCslF6 promoter fragments were NotI digested and purified with the QIAquick Gel Extraction Kit (Qiagen GmbH, Hilden, Germany). pGreenII-0800 vector was linearized in a similar digestion reaction using NotI with the addition of 1 μL TSAP (Promega, Madison, WI, United States) and purified as described for the pHvCslF6 fragments. Following this, T4 DNA ligations (Promega, Madison, WI, United States) were set up for each construct and 1 μL of the ligation reaction was used for E. coli transformation. Plasmids were purified with QIAprep Spin Miniprep Kit (Qiagen GmbH, Hilden, Germany) and confirmed by Sanger sequencing using M13F primer at the Genome Technology facility, The James Hutton Institute.
The Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, United States) was used to determine the promoter activity of HvCslF6 deletion constructs in barley protoplasts, 24 h after transfection. Luminescence results were obtained using a Varioskan LUX Multimode Microplate Reader (Thermo Fisher Scientific, Waltham, MA, United States) following the dual luciferase kit instructions. Protoplasts were lysed with 80 μL of a 1X passive lysis buffer and incubated for 15 min at room temperature. In parallel, 100 μL of LAR II (Luciferase Assay Reagent II) were pre-dispensed in a Corning® 96-well opaque flat bottom microplate (Thermo Fisher Scientific, Waltham, MA, United States). For all assays, 20 μL of lysed protoplasts were transferred to the 96-well plate and incubated for 1 min, room temperature. After Firefly luciferase (Fluc) activity was measured (3 s per sample with a 1.5 s delay between wells), 100 μL of Stop and Glo Reagent were added enabling Renilla luciferase (Rluc) detection (1.5 s with a 0.5 s delay between wells). For each construct tested, three technical replicates were performed per transfection assay. A negative control, which included protoplasts transformed with empty vector and dual luciferase reagents, was used in each batch of transfections. Differences in dual luciferase reporter assays across HvCslF6 promoter deletion constructs were determined by one-way ANOVA followed by Tukey’s test using GraphPad Prism 8.4.2 Software (CA, United States). Pairwise comparisons were determined by two-tailed Student’s t-tests.
The protein sequences of cv. Morex v3 genome3 were used to identify R2R3 MYB family members. The sequences were uploaded to a webserver4 for automatic identification of MYB gene family members (Pucker, 2022) using default parameters. Sequences which were not classified as R2R3 MYB were removed for the phylogenetic analysis. Protein sequences of selected Arabidopsis and rice MYB proteins (MYB61, including OsMYB61a and OsMYB61b and MYB46/83) were added and the sequences aligned using ClustalW in MEGA (Kumar et al., 2018). Unreliable positions in the alignment were removed using BMGE (Block Mapping and Gathering with Entropy; Criscuolo and Gribaldo, 2010) v2.0. Model selection for amino acid substitution was done in MEGA5 resulting in LG model plus gamma distribution with invariant sites (LG+G+I) as the best choice. The unrooted phylogenetic tree was build using Maximum Likelihood with 100 bootstrap replications to estimate bootstrap support. Only bootstrap values above 0.5 are displayed.
Gene expression values from 808 individual samples (Supplementary Table 4) were obtained and are available from Milne et al. (2021). In short, the reads were downloaded from the sequence read archive6 and mapped against BaRTv1 (Rapazote-Flores et al., 2019) using Salmon (Patro et al., 2017). Expression of the isoforms were added up to obtain gene expression. For network construction low expressed genes were removed by filtering for a TPM (transcript per million) of above 5 in at least a third of the samples. This condensed the number of expressed genes from 60,444 to 17,678. Weighted gene correlation network analysis (WGCNA; Langfelder and Horvath, 2008) was used for network construction. Euclidean clustering beforehand highlighted one sample from E15.MicrosporeEmbryogenic (SRR6433018) as outlier. Therefore, this sample was removed, leaving 807 samples for further analysis. The soft-thresholding power was set to 10 to achieve scale-free topology. Network analysis was carried out using the blockwise module method (Langfelder and Horvath, 2008), with a power of 10, TOMType = unsigned, corType = bicor, networkType = signed hybrid, minModuleSize = 30, maxPOutliers = 0.05, mergeCutHeight = 0.15, deepSplit = 3. Visualization of the HvCslF6 network was performed in Gephi7. Size and coloring of the nodes was determined by degree centrality (the number or proportion of other nodes linked to a specific node). Edge coloring was based on weight. The subnetwork was extracted by filtering for nodes connected to HvCslF6 (BART1_0-p53331). For this we applied an edge weight which is a threshold, whereby connections between gene represent by values greater or equal to 0.188 are correlated.
A nested PCR was used to amplify HvMYB61 (HORVU.MOREX.r3.1HG0018590) containing: 1 μL of cv. Golden Promise protoplast cDNA (1:10 diluted in sdH2O), 1.25 μL of each primer (forward and reverse listed on Supplementary Table 7) at 10 mM, 5 μL of 5X Phusion HF Buffer, 0.2 μL of Phusion HF DNA polymerase (Thermo Fisher Scientific, Waltham, MA, United States) and 11.3 μL of sdH2O in a total volume of 20 μL. Phusion PCR reaction conditions are described above. A 1 μL aliquot of the PCR mix was used as a template to attach Gateway® attB sites, 8x His-tag and thrombin cleavage site at the C-terminus in a similar Phusion HF PCR reaction and gel purified. HvMYB61 was cloned into pDONR207™, linearized with EcoRI by a BP reaction containing: 3 μL of the purified attB-PCR product (34 ng/μL), 1 μL of linearized pDONR207™ (50 ng/μL), 2 μL 5X BP Clonase™ Reaction Buffer (Thermo Fisher Scientific, Waltham, MA, United States) and 4 μL of TE buffer, pH 8.0 in a total volume of 10 μL. The BP reaction was incubated at RT overnight and stopped by adding 2 μL of Proteinase K and incubated for 10 min at 37°C. 1 μL of the BP reaction was used for E. coli transformation. Positive colonies were transferred to 4 mL of LB medium with gentamycin (100 μg/μL) and incubated overnight, 37°C and shaking at 230 rpm. Plasmids were purified using the QIAprep Spin Miniprep Kit (Qiagen GmbH, Hilden, Germany) and confirmed by Sanger sequencing. HvMYB61 was transferred to pBract214m-HSPT plant expression vector by LR reaction containing: 2 μL of EcoRI linearized pDONR207™-HvMYB61 (40 ng/μL), 5 μL of pBract214m-HSPT (30 ng/μL), 1 μL TE Buffer, pH 8.0 and 2 μL LR Clonase™ II enzyme mix (Thermo Fisher Scientific, Waltham, MA, United States) in a total volume of 10 μL. The reaction was incubated at 25°C for 3 h and stopped by adding 1 μL of the Proteinase K (37°C for 10 min). After E. coli transformation, positive colonies were grown on LB medium with kanamycin (100 μg/μL) for plasmid purification. pBract214m-HSPT-HvMYB61 construct sequence was confirmed as described above using the ZmUbi forward primer (Supplementary Table 7).
Total mRNA was extracted from ∼2 × 105 protoplasts (100 μL) using TRIzol® Reagent (Thermo Fisher Scientific, Waltham, MA, United States) according to the manufacturer’s instructions. RNA concentration and purity were measured using NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, United States). Total mRNA to cDNA conversion was performed using cDNA EcoDry™ Premix (Takara, Kyoto, Japan). For each reaction, 100 ng of total mRNA was added to the lyophilized master mix following the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed in a StepOne Real-Time PCR machine (Thermo Fisher Scientific, Waltham, MA, United States) using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, Waltham, MA, United States) to determine HvCslF6 relative transcript abundance. Three independent experiments were performed, each of them included: HvMYB61 PEG-mediated transfection, RNA extraction, cDNA synthesis and qPCRs, respectively. Three replicate qRT-PCR reactions were performed for each cDNA sample (technical replicates) including a negative control (sdH2O as template). Each qRT-PCR reaction contained: 2 μL protoplast cDNA (1:2 dilution), 5 μL SYBR Green, 1 μL each forward and reverse primer at 4 mM and 1 μL of sdH2O in a total volume of 10 μL. Primer sequences for HvCslF6 and qRT-PCR conditions were used as in Burton et al. (2008) and described in Supplementary Table 7. Relative HvCslF6 gene expression was normalized to α-Tubulin, Gapdh and Hsp70 housekeeping genes and calculated using the 2–ΔΔCT method (Vandesompele et al., 2002) for multiple control genes. Statistical differences in HvCslF6 expression were determined by Student’s t-test compared to WT protoplasts (empty vector) using GraphPad Prism 9.1 Software (CA, United States).
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
GG-G, GD, KH, MT, RB, and RW conceived the work. GD initiated the cloning of HvCslF6 promoter deletion series. GG-G performed the laboratory work, TFBS, and data analyses. KH carried out co-expression analyses. MS performed the gene network construction. RS carried out the barley transformations. GG-G, MS, KH, and MT drafted the manuscript. GD, GF, RB, and RW reviewed the manuscript. All authors have read and approved the manuscript.
This work was supported by the Australian Research Council, the BBSRC, and Rural and Environment Science and Analytical Services Division of the Scottish Government.
We thank Abdellah Barakate for providing pZmUBI:bcoGFP-ER and pBract214m-HSPT constructs. We also thank the Genome Technology team at the James Hutton Institute (United Kingdom) for Sanger sequencing and collaborators at The University of Adelaide (Australia) for barley transformations.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.883139/full#supplementary-material
Aaman, P., and Graham, H. (1987). Analysis of total and insoluble mixed-linked (1,3; 1,4)-β-D-glucans in barley and oats. J. Agric. Food Chem. 35, 704–709. doi: 10.1021/jf00077a016
Aubert, M. K., Coventry, S., Shirley, N. J., Betts, N. S., Würschum, T., Burton, R. A., et al. (2018). Differences in hydrolytic enzyme activity accompany natural variation in mature aleurone morphology in barley (Hordeum vulgare L.). Sci. Rep. 8:11025. doi: 10.1038/s41598-018-29068-29064
Bacic, A., and Stone, B. (1980). A (1→3)- and (1→4)-linked β-D-glucan in the endosperm cell-walls of wheat. Carbohydr. Res. 82, 372–377. doi: 10.1016/S0008-6215(00)85713-85714
Bacic, A., and Stone, B. A. (1981). Chemistry and organization of aleurone cell wall components from wheat and barley. Funct. Plant Biol. 8, 475–495. doi: 10.1071/PP9810475
Bai, J., Ren, Y., Li, Y., Fan, M., Qian, H., Wang, L., et al. (2019). Physiological functionalities and mechanisms of β-glucans. Trends Food Sci. Technol. 88, 57–66. doi: 10.1016/j.tifs.2019.03.023
Bateman, A., Martin, M. J., O’Donovan, C., Magrane, M., Alpi, E., Antunes, R., et al. (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169. doi: 10.1093/nar/gkw1099
Betts, N. S., Wilkinson, L. G., Khor, S. F., Shirley, N. J., Lok, F., Skadhauge, B., et al. (2017). Morphology, carbohydrate distribution, gene expression, and enzymatic activities related to cell wall hydrolysis in four barley varieties during simulated malting. Front. Plant Sci. 8:1872. doi: 10.3389/fpls.2017.01872
Burn, J. E., Hocart, C. H., Birch, R. J., Cork, A. C., and Williamson, R. E. (2002). Functional analysis of the Cellulose synthase genes CesA1, CesA2, CesA3, and Arabidopsis. Plant Physiol. 129, 797–807. doi: 10.1104/pp.010931
Burton, R. A., Collins, H. M., Kibble, N. A. J., Smith, J. A., Shirley, N. J., Jobling, S. A., et al. (2011). Over-expression of specific HvCslF, Cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-β-d-glucans and alters their fine structure. Plant Biotechnol. J. 9, 117–135. doi: 10.1111/j.1467-7652.2010.00532.x
Burton, R. A., and Fincher, G. B. (2009). (1,3;1,4)-β-D-Glucans in cell walls of the poaceae, lower plants, and fungi: a tale of two linkages. Mol. Plant 2, 873–882. doi: 10.1093/mp/ssp063
Burton, R. A., and Fincher, G. B. (2012). Current challenges in cell wall biology in the cereals and grasses. Front. Plant Sci. 3:130. doi: 10.3389/fpls.2012.00130
Burton, R. A., and Fincher, G. B. (2014). Evolution and development of cell walls in cereal grains. Front. Plant Sci. 5:456. doi: 10.3389/fpls.2014.00456
Burton, R. A., Jobling, S. A., Harvey, A. J., Shirley, N. J., Mather, D. E., Bacic, A., et al. (2008). The genetics and transcriptional profiles of the Cellulose synthase-like, HvCslF gene family in barley. Plant Physiol. 146, 1821–1833. doi: 10.1104/pp.107.114694
Burton, R. A., Shirley, N. J., King, B. J., Harvey, A. J., and Fincher, G. B. (2004). The CesA gene family of barley. quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol. 134, 224–236. doi: 10.1104/pp.103.032904
Burton, R. A., Wilson, S. M., Hrmova, M., Harvey, A. J., Shirley, N. J., Medhurst, A., et al. (2006). Cellulose synthase-like, CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science 311, 1940–1942. doi: 10.1126/science.1122975
Carpita, N., Tierney, M., and Campbell, M. (2001). “Molecular biology of the plant cell wall: searching for the genes that define structure, architecture and dynamics,” in Plant Cell Walls, eds N. C. Carpita, M. Campbell, and M. Tierney (Dordrecht: Springer).
Castro-Mondragon, J. A., Riudavets-Puig, R., Rauluseviciute, I., Berhanu Lemma, R., Turchi, L., Blanc-Mathieu, R., et al. (2022). JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50, D165–D173. doi: 10.1093/nar/gkab1113
Chow, C. N., Lee, T. Y., Hung, Y. C., Li, G. Z., Tseng, K. C., Liu, Y. H., et al. (2019). PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from chip-seq experiments in plants. Nucleic Acids Res. 47, D1155–D1163. doi: 10.1093/nar/gky1081
Cory, A. T., Båga, M., Anyia, A., Rossnagel, B. G., and Chibbar, R. N. (2012). Genetic markers for CslF6 gene associated with (1,3;1,4)-β-glucan concentration in barley grain. J. Cereal Sci. 56, 332–339. doi: 10.1016/j.jcs.2012.02.003
Criscuolo, A., and Gribaldo, S. (2010). BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10:210. doi: 10.1186/1471-2148-10-210
Dimitroff, G., Little, A., Lahnstein, J., Schwerdt, J. G., Srivastava, V., Bulone, V., et al. (2016). (1,3;1,4)-β-Glucan biosynthesis by the CSLF6 enzyme: position and flexibility of catalytic residues influence product fine structure. Biochemistry 55, 2054–2061. doi: 10.1021/acs.biochem.5b01384
Doblin, M. S., Pettolino, F. A., Wilson, S. M., Campbell, R., Burton, R. A., Fincher, G. B., et al. (2009). A barley Cellulose synthase-like CslH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. U S A. 106, 5996–6001. doi: 10.1073/pnas.0902019106
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Fan, M., Herburger, K., Jensen, J. K., Zemelis-Durfee, S., Brandizzi, F., Fry, S. C., et al. (2018). A trihelix family transcription factor is associated with key genes in mixed-linkage glucan accumulation. Plant Physiol. 178, 1207–1221. doi: 10.1104/pp.18.00978
Fornes, O., Castro-Mondragon, J. A., Khan, A., Van Der Lee, R., Zhang, X., Richmond, P. A., et al. (2020). JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92. doi: 10.1093/nar/gkz1001
Garcia-Gimenez, G., Russell, J., Aubert, M. K., Fincher, G. B., Burton, R. A., Waugh, R., et al. (2019). Barley grain (1,3;1,4)-β-glucan content: effects of transcript and sequence variation in genes encoding the corresponding synthase and endohydrolase enzymes. Sci. Rep. 9:17250. doi: 10.1038/s41598-019-53798-53798
Garcia-Gimenez, G., Barakate, A., Smith, P., Stephens, J., Khor, S. F., Doblin, M. S., et al. (2020). Targeted mutation of barley (1,3;1,4)−β−glucan synthases reveals complex relationships between the storage and cell wall polysaccharide content. Plant J. 104, 1009–1022. doi: 10.1111/tpj.14977
Gupta, M., Abu-Ghannam, N., and Gallaghar, E. (2010). Barley for brewing: characteristic changes during malting, brewing and applications of its by-products. Compr. Rev. Food Sci. Food Saf. 9, 318–328. doi: 10.1111/j.1541-4337.2010.00112.x
Han, F., Ullrich, S. E., Chirat, S., Menteur, S., Jestin, L., Sarrafi, A., et al. (1995). Mapping of β-glucan content and β-glucanase activity loci in barley grain and malt. Theor. Appl. Genet. 91, 921–927. doi: 10.1007/BF00223901
Harris, P. J. (2006). Primary and secondary plant cell walls: a comparative overview. New Zeal. J. For. Sci. 36, 36–53. doi: 10.1111/j.1365-2621.2006.01470.x
Heberle, H., Meirelles, V. G., da Silva, F. R., Telles, G. P., and Minghim, R. (2015). InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16:169. doi: 10.1186/s12859-015-0611-613
Hirano, K., Kondo, M., Aya, K., Miyao, A., Sato, Y., Antonio, B. A., et al. (2013). Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol. 54, 1791–1802. doi: 10.1093/pcp/pct122
Houston, K., Burton, R. A., Sznajder, B., Rafalski, A. J., Dhugga, K. S., Mather, D. E., et al. (2015). A genome-wide association study for culm cellulose content in barley reveals candidate genes co-expressed with members of the Cellulose synthase A gene family. PLoS One 10:e0130890. doi: 10.1371/journal.pone.0130890
Houston, K., Russell, J., Schreiber, M., Halpin, C., Oakey, H., Washington, J. M., et al. (2014). A genome wide association scan for (1,3;1,4)-β-glucan content in the grain of contemporary 2-row Spring and Winter barleys. BMC Genomics 15:907. doi: 10.1186/1471-2164-15-907
Hu, G., Burton, C., Hong, Z., and Jackson, E. (2014). A mutation of the Cellulose synthase-like, CslF6 gene in barley (Hordeum vulgare L.) partially affects the β-glucan content in grains. J. Cereal Sci. 59, 189–195. doi: 10.1016/j.jcs.2013.12.009
Jobling, S. A. (2015). Membrane pore architecture of the CSLF6 protein controls (1-3,1-4)-β-glucan structure. Sci. Adv. 1:e1500069. doi: 10.1126/sciadv.1500069
Kim, W. C., Ko, J. H., and Han, K. H. (2012). Identification of a cis-acting regulatory motif recognized by MYB46, a master transcriptional regulator of secondary wall biosynthesis. Plant Mol. Biol. 78, 489–501. doi: 10.1007/s11103-012-9880-9887
Kim, W. C., Reca, I. B., Kim, Y. S., Park, S., Thomashow, M. F., Keegstra, K., et al. (2014). Transcription factors that directly regulate the expression of CslA9 encoding mannan synthase in Arabidopsis thaliana. Plant Mol. Biol. 84, 577–587. doi: 10.1007/s11103-013-0154-159
Ko, J. H., Jeon, H. W., Kim, W. C., Kim, J. Y., and Han, K. H. (2014). The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann. Bot. 114, 1099–1107. doi: 10.1093/aob/mcu126
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9:559. doi: 10.1186/1471-2105-9-559
Little, A., Schwerdt, J. G., Shirley, N. J., Khor, S. F., Neumann, K., O’Donovan, L. A., et al. (2018). Revised phylogeny of the Cellulose Synthase gene superfamily: insights into cell wall evolution. Plant Physiol. 177, 1124–1141. doi: 10.1104/pp.17.01718
Ma, D., and Constabel, C. P. (2019). MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 24, 275–289. doi: 10.1016/j.tplants.2018.12.003
Mascher, M., Wicker, T., Jenkins, J., Plott, C., Lux, T., Koh, C. S., et al. (2021). Long-read sequence assembly: a technical evaluation in barley. Plant Cell 33, 1888–1906. doi: 10.1093/plcell/koab077
Matros, A., Houston, K., Tucker, M. R., Schreiber, M., Berger, B., Aubert, M. K., et al. (2021). Genome-wide association study reveals the genetic complexity of fructan accumulation patterns in barley grain. J. Exp. Bot. 72, 2383–2402. doi: 10.1093/jxb/erab002
Matys, V., Kel-Margoulis, O. V., Fricke, E., Liebich, I., Land, S., Barre-Dirrie, A., et al. (2006). TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110. doi: 10.1093/nar/gkj143
McCarthy, R. L., Zhong, R., and Ye, Z. H. (2009). MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964. doi: 10.1093/pcp/pcp139
Milne, L., Bayer, M., Rapazote-Flores, P., Mayer, C. D., Waugh, R., and Simpson, C. G. (2021). EORNA, a barley gene and transcript abundance database. Sci. Data 8:90. doi: 10.1038/s41597-021-00872-874
Newman, L. J., Perazza, D. E., Juda, L., and Campbell, M. M. (2004). Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 37, 239–250. doi: 10.1046/j.1365-313X.2003.01953.x
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Penfield, S., Meissner, R. C., Shoue, D. A., Carpita, N. C., and Bevan, M. W. (2001). MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13, 2777–2791. doi: 10.1105/tpc.13.12.2777
Prouse, M. B., and Campbell, M. M. (2013). Interactions between the R2R3-MYB transcription factor, AtMYB61, and target DNA binding sites. PLoS One 8:e65132. doi: 10.1371/journal.pone.0065132
Pucker, B. (2022). Automatic identification and annotation of MYB gene family members in plants. BMC Genomics 23:220. doi: 10.1186/s12864-022-08452-5
Rao, A. L. N. (2007). Preparation and inoculation of mesophyll protoplasts from monocotyledenous and dicotyledenous hosts. Curr. Protoc. Microbiol. Chapter 16:Unit 16D.2. doi: 10.1002/9780471729259.mc16d02s4.
Rao, X., and Dixon, R. A. (2018). Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front. Plant Sci. 9:399. doi: 10.3389/fpls.2018.00399
Rapazote-Flores, P., Bayer, M., Milne, L., Mayer, C. D., Fuller, J., Guo, W., et al. (2019). BaRTv1.0: an improved barley reference transcript dataset to determine accurate changes in the barley transcriptome using RNA-seq. BMC Genomics 20:968. doi: 10.1186/s12864-019-6243-6247
Romano, J. M., Dubos, C., Prouse, M. B., Wilkins, O., Hong, H., Poole, M., et al. (2012). AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytol. 195, 774–786. doi: 10.1111/j.1469-8137.2012.04201.x
Romero-Romero, J. L., Inostroza-Blancheteaud, C., Orellanaa, D., Aquea, F., Reyes-Díaz, M., Gil, M. P., et al. (2018). Stomata regulation by tissue-specific expression of the Citrus sinensis MYB61 transcription factor improves water-use efficiency in Arabidopsis. Plant Physiol. Biochem. 130, 54–60. doi: 10.1016/j.plaphy.2018.06.034
Shahmuradov, I. A., Solovyev, V. V., and Gammerman, A. J. (2005). Plant promoter prediction with confidence estimation. Nucleic Acids Res. 33, 1069–1076. doi: 10.1093/nar/gki247
Slakeski, N., and Fincher, G. B. (1992). Developmental regulation of (1,3;1,4)-β-glucanase gene expression in barley. Plant Physiol. 99, 1226–1231. doi: 10.1104/pp.99.3.1226
Taketa, S., Yuo, T., Tonooka, T., Tsumuraya, Y., Inagaki, Y., Haruyama, N., et al. (2012). Functional characterization of barley betaglucanless mutants demonstrates a unique role for CslF6 in (1,3;1,4)-β-D-glucan biosynthesis. J. Exp. Bot. 63, 381–392. doi: 10.1093/jxb/err285
Taylor-Teeples, M., Lin, L., De Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Tripathi, R. K., Bregitzer, P., and Singh, J. (2018). Genome-wide analysis of the SPL/miR156 module and its interaction with the AP2/miR172 unit in barley. Sci. Rep. 8:7085. doi: 10.1038/s41598-018-25349-0
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034
Vega-Sanchez, M. E., Verhertbruggen, Y., Christensen, U., Chen, X., Sharma, V., Varanasi, P., et al. (2012). Loss of Cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 159, 56–69. doi: 10.1104/pp.112.195495
Wilson, S. M., Ho, Y. Y., Lampugnani, E. R., Van de Meene, A. M. L., Bain, M. P., Bacic, A., et al. (2015). Determining the subcellular location of synthesis and assembly of the cell wall polysaccharide (1,3; 1,4)-β-D-glucan in grasses. Plant Cell 27, 754–771. doi: 10.1105/tpc.114.135970
Wong, S. C., Shirley, N. J., Little, A., Khoo, K. H. P., Schwerdt, J., Fincher, G. B., et al. (2015). Differential expression of the HvCslF6 gene late in grain development may explain quantitative differences in (1,3;1,4)-β-glucan concentration in barley. Mol. Breed. 35:20. doi: 10.1007/s11032-015-0208-6
Woodward, J. R., Fincher, G. B., and Stone, B. A. (1983). Water-soluble (1→3), (1→4)-β-D-glucans from barley (Hordeum vulgare) endosperm. II. fine structure. Carbohydr. Polym. 3, 207–225. doi: 10.1016/0144-8617(83)90019-X
Yoo, S.-D., Cho, Y.-H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, R., Tucker, M. R., Burton, R. A., Shirley, N. J., Little, A., Morris, J., et al. (2016a). The dynamics of transcript abundance during cellularization of developing barley endosperm. Plant Physiol. 170, 1549–1565. doi: 10.1104/pp.15.01690
Zhang, Z., Hu, X., Zhang, Y., Miao, Z., Xie, C., Meng, X., et al. (2016b). Opposing control by transcription factors MYB61 and MYB3 increases freezing tolerance by relieving c-repeat binding factor suppression. Plant Physiol. 172, 1306–1323. doi: 10.1104/pp.16.00051
Zhao, K., and Bartley, L. E. (2014). Comparative genomic analysis of the R2R3 MYB secondary cell wall regulators of Arabidopsis, poplar, rice, maize, and switchgrass. BMC Plant Biol. 14:135. doi: 10.1186/1471-2229-14-135
Zhao, K., Lin, F., Romero-Gamboa, S. P., Saha, P., Goh, H. J., An, G., et al. (2019). Rice genome-scale network integration reveals transcriptional regulators of grass cell wall synthesis. Front. Plant Sci. 10:1275. doi: 10.3389/fpls.2019.01275
Zhong, R., Lee, C., McCarthy, R. L., Reeves, C. K., Jones, E. G., and Ye, Z. H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 52, 1856–1871. doi: 10.1093/pcp/pcr123
Zhong, R., and Ye, Z. H. (2012). MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 53, 368–380. doi: 10.1093/pcp/pcr185
Zhou, J., Lee, C., Zhong, R., and Ye, Z. H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248–266. doi: 10.1105/tpc.108.063321
Keywords: cell wall, (1,3;1,4)-β-glucan, barley, transcription factors, MYB
Citation: Garcia-Gimenez G, Schreiber M, Dimitroff G, Little A, Singh R, Fincher GB, Burton RA, Waugh R, Tucker MR and Houston K (2022) Identification of candidate MYB transcription factors that influence CslF6 expression in barley grain. Front. Plant Sci. 13:883139. doi: 10.3389/fpls.2022.883139
Received: 24 February 2022; Accepted: 17 August 2022;
Published: 08 September 2022.
Edited by:
Rowan Andrew Craig Mitchell, Rothamsted Research, United KingdomReviewed by:
Paula Del Carmen Fernandez, Instituto Nacional de Tecnología Agropecuaria, ArgentinaCopyright © 2022 Garcia-Gimenez, Schreiber, Dimitroff, Little, Singh, Fincher, Burton, Waugh, Tucker and Houston. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kelly Houston, S2VsbHkuSG91c3RvbkBodXR0b24uYWMudWs=
†Present address: Guillermo Garcia-Gimenez, Department of Animal, Plant and Soil Sciences, School of Life Sciences, La Trobe Institute for Agriculture and Food, La Trobe University, Bundoora, VIC, Australia
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.