
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 May 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.881212
This article is part of the Research Topic Shaping Plant NLR Diversity: Evolutionary Dynamics and Structure-Function Relationships View all 7 articles
From a reverse genetic screen using CRISPR/Cas9 gene editing tool, we unintentionally identified an autoimmune mutant. Map-based cloning and whole-genome sequencing revealed that it contains a deletion in SMALL UBIQUITIN-RELATED MODIFIER (SUMO) protease encoding gene EARLY IN SHORT DAYS 4 (ESD4). Previous studies reported that esd4 mutants accumulate elevated levels of plant defense hormone salicylic acid (SA). However, upregulated PATHOGENESIS-RELATED GENE 1 (PR1) expression in esd4 only partly relies on SA level. In this study, we show that plant metabolite N-hydroxypipecolic acid (NHP) biosynthetic genes are upregulated in esd4, and NHP biosynthesis mutant flavin-dependent-monooxygenase 1 (fmo1) partially suppresses the autoimmune phenotypes of esd4, suggestive of a requirement of NHP signaling for the autoimmunity in esd4. As activation of nucleotide-binding leucine-rich repeat immune receptors (NLRs) are associates with the biosynthesis of SA and NHP and lipase-like protein ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) is a key component downstream of many NLRs, we examined the relationship between EDS1 and ESD4 by analyzing the eds1 esd4 double mutant. We found that eds1 largely suppresses esd4 autoimmunity and blocks the elevated expressions of SA and NHP biosynthesis-related genes in esd4. Overall, our study provides evidence supporting the hypothesis that SUMO protease ESD4 likely targets a yet to be identified guardee of NLR by removing its SUMO modification to avoid recognition by the cognate NLR. Loss of ESD4 results in activation of NLR-mediated autoimmunity.
To defend themselves against pathogen infections, plants have evolved a complicated immune system which mainly relies on two types of immune receptors: plasma membrane-localized pattern recognition receptors (PRRs), and intracellular nucleotide-binding leucine-rich repeat immune receptors (NLRs; Dangl and Jones, 2001; Jones and Dangl, 2006). PRRs are receptor-like kinases (RLKs) or receptor-like proteins (RLPs), which recognize conserved microbial features, pathogen-associated molecular patterns (PAMPs), leading to pattern-triggered immunity (PTI). However, pathogens can deliver effectors into intracellular space to inhibit PTI, thereby facilitating pathogenesis. To counteract, intracellular NLRs can directly or indirectly recognize effectors to trigger a stronger defense response termed effector-triggered immunity (ETI), which is often associated with programmed cell death to activate the hypersensitive response (HR; Nishimura and Dangl, 2010; Cui et al., 2015).
Plant NLRs are divided into three classes based on their N-terminal domains: CNLs (CC-NLRs) have N-terminal coiled-coil domains, TNLs (TIR-NLRs) contain N-terminal Toll/interleukin-1 receptor domain (TIR) and RNLs (RPW8-NLRs) have N-terminal coiled-coil domains similar to RESISTANCE TO POWDERY MILDEW 8 (Maruta et al., 2022). TNLs and CNLs function as “sensor” NLRs to sense pathogenic effectors or their modifications on host targets, while RNLs are required for immune signaling and cell death induction mediated by TNLs and CNLs, and thus are considered as “helper” NLRs. ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1), a lipase-like protein, is an important hub downstream of TNLs and a few CNLs (Falk et al., 1999; Lapin et al., 2020). In parallel, a glycosylphosphatidylinositol-anchored plasma membrane-localized protein, NON-RACE-SPECIFIC DISEASE RESISTANCE (NDR1), was shown to be required for the signaling of CNLs (Century et al., 1997; Aarts et al., 1998; Shapiro and Zhang, 2001; Knepper et al., 2011). How EDS1 and NDR1 regulate NLR-mediated signaling is still unclear.
Activation of NLRs induces a series of downstream defense responses including the generations of the phenolic defense hormone salicylic acid (SA) and N-hydroxypipecolic acid (NHP), which are required for both PTI/ETI at local sites and induced immunity in distal tissues. In Arabidopsis thaliana, pathogen-induced SA is mainly synthesized from chorismate by ISOCHORISMATE SYNTHASE 1 (ICS1) and AVRPPHB SUSCEPTIBLE 3 (PBS3; Rekhter et al., 2019; Torrens-Spence et al., 2019). ENHANCED DISEASE SUSCEPTIBILITY 5 (EDS5) is responsible for exporting SA precursor from plastids to the cytosol (Rekhter et al., 2019). Whereas, NHP is biosynthesized from L-lysine via a series of biochemical reactions catalyzed by enzymes including ABERRANT GROWTH AND DEATH2 (AGD2)-LIKE DEFENSE RESPONSE PROTEIN 1(ALD1; Song et al., 2004a,b; Navarova et al., 2012), SAR DEFICIENT 4 (SARD4; Ding et al., 2016; Hartmann et al., 2017) and FLAVIN-DEPENDENT MONOOXYGENASE 1 (FMO1; Chen et al., 2018; Hartmann et al., 2018).
SUMOylation, one of the post-translational protein modifications (PTMs), is conserved in all eukaryotes (Miura et al., 2007). SUMOylation is the process of attaching SUMO (small ubiquitin-like modifier) to target lysine residues of protein via a series of biochemical reactions facilitated by SUMO-activating enzyme (E1), the SUMO-conjugating enzyme (E2), and the SUMO E3 ligase. Firstly, SUMO is activated by E1 in an ATP-dependent manner to form a thioester bond between the carboxyl group of glycine in SUMO and the sulfhydryl group of the catalytic cysteine residue in E1. Activated SUMO is subsequently transferred to a cysteine residue in E2. Finally, E3 ligase aids the transfer of SUMO from E2 to the target substrate. It is worth mentioning that SUMOylation of a target protein can occur without the help of E3 ligases (Wilkinson and Henley, 2010). SUMOylation is a reversible and highly dynamic modification due to the presence of SUMO proteases (Yates et al., 2016). On one hand, SUMO proteases possess activity to release free SUMO from the target protein. On the other hand, they are responsible for the generation of mature SUMO by cleaving off the C-terminus of immature SUMO.
The diverse roles of SUMOylation in plant immune signaling have been emerging during the past few years (He et al., 2017; Verma et al., 2018). For instance, flagellin perception induces the SUMOylation of its receptor FLAGELLIN-SENSITIVE 2 (FLS2) through the degradation of SUMO protease Desi3a, resulting in the release of BOTRYTIS-INDUCED KINASE1 (BIK1) and the activation of downstream immune signaling (Orosa et al., 2018). Recently, it was shown that the SUMOylation of transcriptional factor WRKY33 controlled by two SUMO proteases SPF1/SPF2 facilitates the activation of downstream immune-related genes (Verma et al., 2021).
Arabidopsis SUMO protease ESD4 was identified due to its early flowering phenotype especially under short photoperiods (Reeves et al., 2002; Pineiro et al., 2003). In this study, we show that loss of ESD4 results in autoimmunity including upregulated defense-related genes, spontaneous cell death, highly accumulated H2O2, and enhanced resistance against virulent oomycete pathogen Hyaloperonospora arabidopsidis (H.a.) Noco2. Further genetic analysis showed that mutation in EDS1 largely suppresses the autoimmunity and elevated SA and NHP biosynthesis in esd4, suggesting that loss function of ESD4 leads to EDS1-dependent NLR(s) activation.
Arabidopsis mutant line1-12 (esd4-3) was obtained from Agrobacterium-mediated genetic transformation. esd4-4 and fmo1esd4 were generated by CRISPR-cas9 gene editing system (Wang, 2015). eds1-2 and fmo1 were previously described (McDowell et al., 2000; Mishina and Zeier, 2006). All the mutants used in this study are in Col-0 ecotype. Col-0 was used as wild-type control and Ler ecotype was used for generating a mapping population with line1-12. As esd4 mutants are sterile, the heterozygous esd4 plants were used for producing offspring. The detailed information of mutants used in this study are summarized in Supplementary Table S1. All the primers used for mutant identification are listed in Supplementary Table S2.
For soil-grown plants, seeds were vernalized at 4°C for 2 days, sown onto sterile soil, and transferred to plant growth rooms under 16-h day/8-h night cycle, 22°C, and 65 ± 10% humidity conditions or 16-h day/8-h night cycle, 28°C, and 65 ± 10% humidity conditions. For selecting T1 transgenic plants, sterilized T1 seeds were sown in culture dishes containing 1/2MS medium with hygromycin (40 μg/ml) in growth chamber under 16-h day/8-h night cycle, 22°C and 65 ± 10% humidity conditions.
For map-based cloning, line1-12 homozygote (in Col background) was crossed with Ler (Landsberg-erecta) to generate F1 seeds. Then F1 plants were self-fertilized and the resulting F2 population was used for crude mapping. 64 plants exhibiting dwarfed morphology were picked out from F2 progenies and used for linkage analysis. The markers used for crude mapping were designed using the Monsanto Arabidopsis polymorphism and Ler sequence collections and listed in Supplementary Table S2 (Jander et al., 2002). After the mutation was narrowed down to a region of about 4.51 Mb between markers T4C9 (6.5 Mb) and F9F13 (11.01 Mb) on Chromosome 4, whole-genome resequencing was performed to identify mutations within this region. 1 g tissue from 52 F2 dwarfed plants from a backcross population between line1-12 and Col-0 was collected for genomic DNA extraction following a previously described procedure (Huang et al., 2019). The purified DNA was sequenced using Illumina whole-genome resequencing and the indel mutation in AT4G15880 was confirmed by Sanger sequencing using flanking primers listed in Supplementary Table S2.
The CRISPR/Cas9 constructs for deleting ESD4 were made following the protocol described previously (Wang, 2015). Briefly, ESD4 deletion target primers were designed using the CRISPR-PLANT website.1 The ESD4 CRISPR cassette was generated by PCR amplification using pCBC-DT1T2 as template with designed primer pairs. The PCR products were digested with BsaI and cloned into binary vector pHEE401E, yielding ESD4-pHEE401E. The ESD4-pHEE401E construct was transformed into Col-0 or fmo1 plants by Agrobacterium-mediated floral-dip transformation (Clough and Bent, 1998). Genotyping primers flanking each target site were used to select T1 plants that harbored deletions in ESD4. Sanger sequencing was performed to define the ESD4 deletion. All the primers used are listed in Supplementary Table S2.
For oomycete pathogen Hyaloperonospora arabidopsidis (H.a.) Noco2 infection assays, seeds of the indicated genotypes were planted on soil and grown in a growth room under a 16 h day/8 h night cycle, 22°C and 65 ± 10% humidity. Two-week-old seedlings (3 pots for each genotype and 12 plants in each pot) were spray-inoculated with freshly harvested H.a. Noco2 conidiospores at a concentration of 1×105 spores per mL sterile water. Infected plants were covered with a plastic lid and kept in a growth chamber under 12-h light/12-h dark cycle, 18°C, and 80% humidity. Seven days later, 1 g plant tissue of wild-type or esd4 mutant from each pot was collected and suspended in 2 ml sterile water. After vortexing, the conidiospores were counted using a hemocytometer under microscope.
Trypan Blue staining was performed following a previously described procedure (Zhang et al., 2012). Specifically, true leaves were transferred into 1.5-ml microcentrifuge tubes containing 1 ml lactophenol Trypan Blue solution (10 mg Trypan Blue, 10 g phenol, 10 ml lactic acid, 10 ml glycerol, and 10 ml water) diluted 1:1 in ethanol. After boiling for 2 min, removed staining solution and added 2 ml chloralhydrate solution (2.5 g/ml) on an orbital shaker for 2 h. Samples were further de-stained overnight with new chloralhydrate solution before examination by a light microscope.
DAB staining procedure was performed according to previously described procedures with minor modifications (Daudi and O’Brien, 2012). Briefly, true leaves were transferred into 2.5 ml micro centrifuge tubes containing 2 ml DAB solution (1 mg/ml DAB, pH 3.8). After overnight shaking on an oscillator, DAB solution was removed. 2 ml de-staining solution (70% ethanol) was added for de-colorization. Photos were taken under a light microscope afterwards.
Total RNA was extracted from 3-week-old soil-grown plants (50 mg) using Eastep plant RNA extraction kit (Promega, Ls1040). 2 μg RNA was reverse-transcribed to cDNA using Go Script reverse transcriptase (Promega, A5001). 50 ng cDNA was added as template in a 10 μl reaction on an ABI Step one Real-Time system machine. Real-time PCR was performed using a SYBR-Green PCR HS kit (AG, AG11701). qRT-PCR was carried out as described previously (Zhang et al., 2003). ACTIN1 was used to normalize the expression value. The primers used are listed in Supplementary Table 2.
With the arrival of better CRISPR-Cas9 gene editing methods in different organisms, knocking out redundant genes has become easier than ever. Designing two guide RNAs (gRNAs) targeting the flanking genes in a tandem array would yield a deletion to knock out these duplicated genes for reverse genetic analysis. Here, we tried to use this system with two gRNAs to knock out a tandem array of 20 genes encoding redundant receptor-like kinases (RLKs) from At4g23130 to At4g23320 in the Col-0 ecotype (Wang, 2015). In the T1 generation, among 114 plants tested, we did not detect any deletion products by PCR using primers flanking the two gRNA sites. However, an early flowering dwarfed plant that exhibited abnormal leaf phenotype and sterility was recovered and designated as line1-12. Interestingly, line1-12 resembled mutants with autoimmune defects (Figure 1A). When the defense marker genes Pathogenesis-Related 1 (PR1) and PR2 were examined by quantitative real-time PCR (qRT-PCR) in line1-12, heightened expression was observed (Figures 1B,C). Furthermore, cell death and accumulated H2O2 were observed in line1-12 using trypan blue staining and 3,3′-diaminobenzidine staining (Figures 1D,E), respectively. As Col is susceptible to H.a. Noco2, this model pathogen on Arabidopsis was used to test for enhanced immunity. When challenged with virulent pathogen H.a. Noco2 (Figure 1F), line1-12 showed less oomycete spores growth compared with wild-type plants. All these data indicate the autoimmunity of line1-12. We predicted that the mutation in this mutant should reveal a novel negative regulator of immunity.
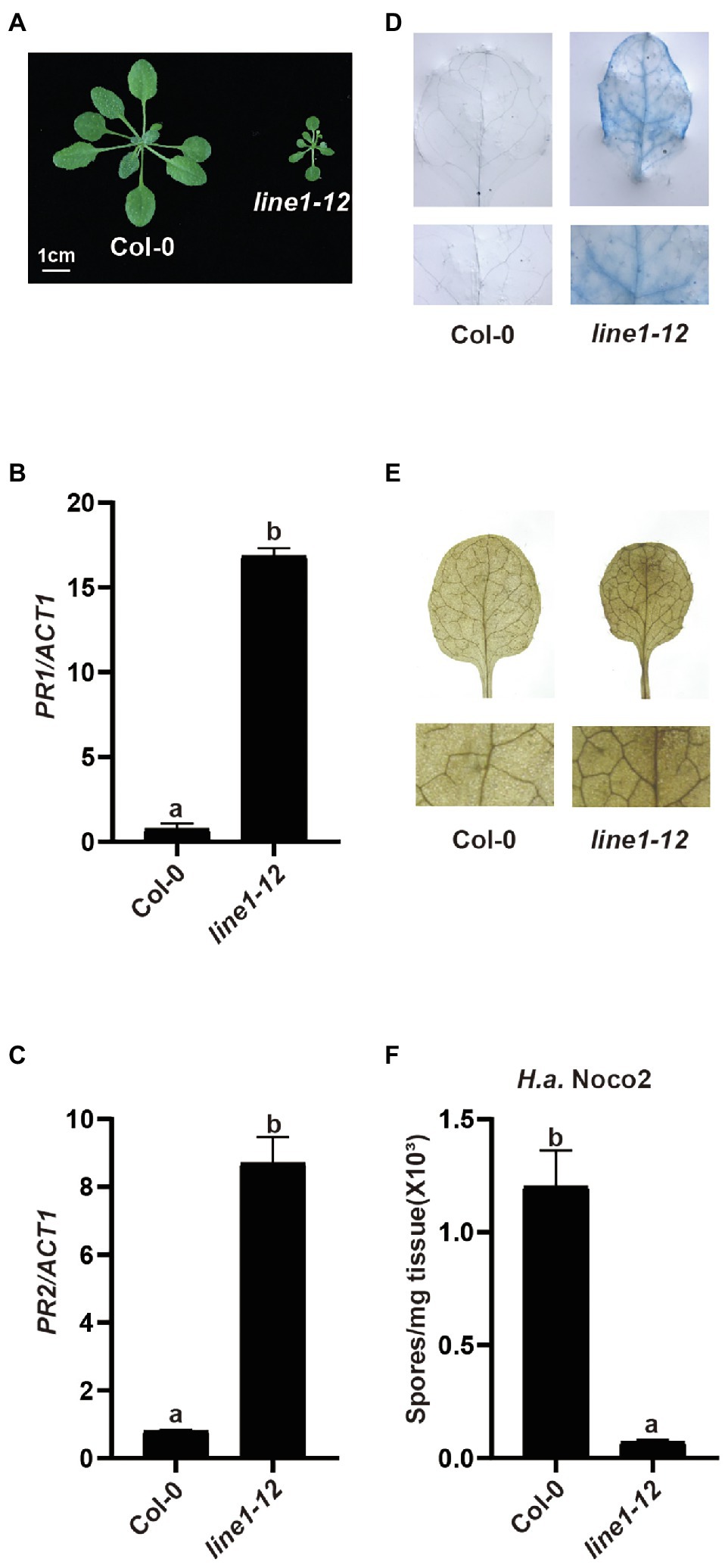
Figure 1. Characterization of line1-12. (A) Morphology of 3-week-old soil-grown plants of wild-type (Col-0) and line1-12 under long-day conditions. (B,C) Expression of PR1 (B) and PR2 (C) in indicated genotypes. ACTIN1 serves as internal control. Total RNA was extracted from 3-week-old seedlings grown on soil. Error bars represent means of 3 biological replicates ± SD. Statistical differences among the samples are labeled with different letters (p < 0.05, one-way ANOVA; n = 3). (D) Trypan blue staining of 3-week-old Col-0 and esd4-3 mutant leaves. (E) DAB staining of 3-week-old Col-0 and esd4-3 mutant leaves. (F) Quantification of H.a. Noco2 conidia spores on indicated genotypes. 1 × 105 spores/ml inoculum were spray-inoculated on 2-week-old soil-grown seedlings. Numbers of spores were counted at 7 days post-inoculation. Error bars represent means of 3 biological replicates ± SD. Statistical differences among the samples are labeled with different letters (p < 0.05, one-way ANOVA; n = 3).
Using map-based cloning, we tested if the mutation responsible for the autoimmunity of line1-12 is linked to the targeted loci by CRISPR-Cas9. When line1-12 was crossed with Ler, 64 dwarfed plants were observed in total 261 F2 population, suggestive of a single recessive mutation. Indeed, the mutation was mapped to chromosome 4 (Figure 2A), where the originally targeted RLK genes are linked to. After failing to find point or small insertion/deletion (Indel) mutations in the target genes through Sanger sequencing, we reasoned that there could be complex rearrangements in the locus. Whole-genome sequencing (WGS) was therefore conducted on the ¼ co-segregants that were dwarfed and flowered earlier in the Col-backcrossed F2 generation. Sequence analysis identified only one mutation, which is an exonic deletion of 49 bp in At4g15880 in the mapped region (Figure 2A).
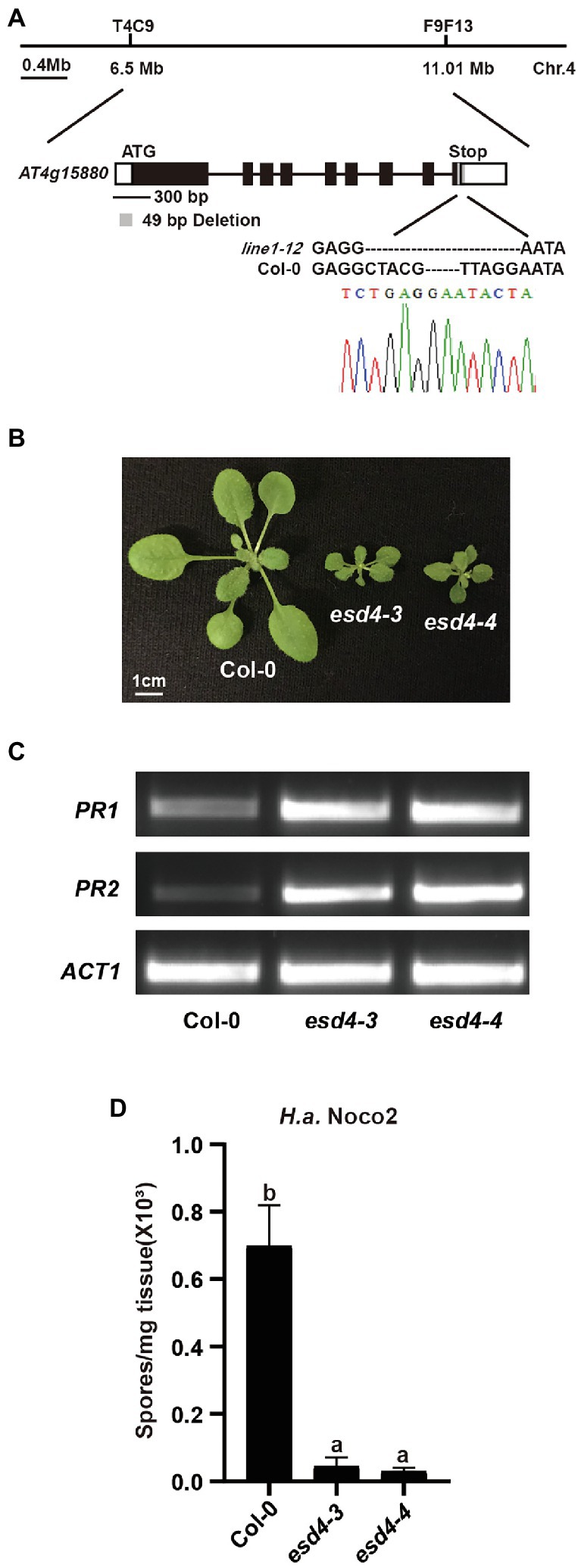
Figure 2. Loss function of ESD4 is responsible for autoimmunity in line1-12. (A) Mapping-based cloning of line1-12. Line1-12 was mapped to a region between marker T4C9 and F9F13 on chromosome 4. Gene structure of ESD4 (AT4G15880) with esd4-3 deletion is highlighted in gray. Black boxes and lines indicate exons and introns, respectively. White boxes represent UTRs. Chromatogram of Sanger sequencing confirmed the deletion in esd4-3. (B) Morphologies of 3-week-old soil-grown plants of the indicated genotypes under long-day conditions. (C) Expression of PR1 and PR2 in the indicated genotypes. ACTIN1 serves as internal control. Experiments were repeated three times with similar results. (D) Growth of H.a. Noco2 on the indicated genotypes. The values indicate averages of replicates ± SD (n = 3). Different letters indicate statistical differences among different samples (p < 0.05, one-way ANOVA, n = 3). Experiments were repeated twice with similar results.
At4g15880 encodes EARLY IN SHORT DAYS 4 (ESD4), which was reported to exhibit dwarfism and early flowering with abnormal leaves and sterility (Reeves et al., 2002; Pineiro et al., 2003; Xu et al., 2007). In addition, esd4 plants also have increased levels of defense hormone salicylic acid (SA; Villajuana-Bonequi et al., 2014), a well-known defense hormone. Thus, we concluded that the mutant line1-12 is an allele of esd4. As two alleles of esd4 were reported in the past, we named line1-12 as esd4-3. When the flanking sequences of the deletion in ESD4 were examined carefully, we did not observe any similarity with the original gRNA sequences, suggesting that the deletion in ESD4 might be due to CRISPR-Cas9 independent causes, likely through Agrobacterium-mediated mutations.
To further confirm the autoimmune phenotypes of esd4-3, we generated another allele of esd4, named esd4-4, by CRIPSPR-Cas9. esd4-4 which contains a 2,772 bp deletion in ESD4 (Supplementary Figure S1A), is also much smaller than wild-type plants and exhibits infertility phenotype under our growth condition (Figure 2B). Semi-qRT-PCR showed that the expression PR1 and PR2 were much higher in esd4-4 compared with that in wild-type (Figure 2C). Similar to esd4-3, esd4-4 showed enhanced resistance to H.a. Noco2 (Figure 2D). Taken together, our data further demonstrated that loss function of ESD4 leads to autoimmunity, indicating a negative role of ESD4 in plant immunity.
Previous study showed esd4 mutants have increased level of defense hormone SA and SA biosynthesis mutant ics1 partially reduced the PR1 expression in esd4 plants (Villajuana-Bonequi et al., 2014), suggesting that the autoimmunity in esd4 partly relies on SA signaling and additional factors contribute to the autoimmune phenotypes in esd4. As plant signal molecules NHP and SA were reported to cooperatively regulate plant immunity (Sun et al., 2020), we examined whether NHP signaling pathway was involved in the autoimmunity of esd4. We generated esd4 fmo1 double mutant by transforming the CRISPR/Cas9 construct for ESD4 deletion into fmo1, a mutant defective in NHP biosynthesis. A new allele of esd4, harboring a deletion of 123 bp, was isolated in the fmo1 background in T1 generation (Supplementary Figure S1B). As shown in Figure 3A, the esd4 fmo1 mutant exhibits intermediate size compared to wild-type and esd4 plant. Upregulated PR1 and PR2 levels in esd4 are also largely suppressed by fmo1 (Figures 3B,C). In addition, more H.a. Noco2 conidiospores grew on the esd4 fmo1 double mutant compared with esd4 single mutant (Figure 3D). Taken together, these data showed that fmo1 partially suppresses the autoimmunity in esd4, suggesting that NHP signaling pathway also contributes to autoimmune responses activated in esd4 mutant.
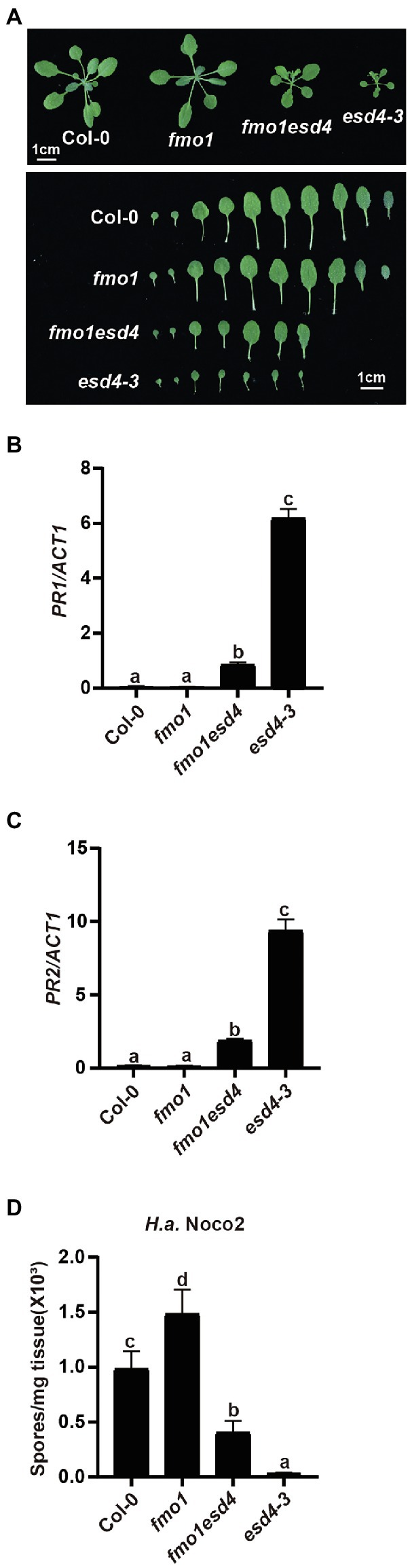
Figure 3. The constitutively activated immunity in esd4 is partially suppressed by fmo1. (A) Morphologies of 3-week-old soil-grown plants of the indicated genotypes under long-day conditions. (B,C) Expression of PR1 (B) and PR2 (C) in indicated genotypes. ACTIN1 serves as internal control. (D) Growth of H.a. Noco2 on the indicated genotypes. The values indicate averages of replicates ± SD (n = 3). Different letters indicate statistical differences among different samples (p < 0.05, one-way ANOVA, n = 3). Experiments were repeated twice with similar results.
To test how NHP signaling pathway contributes to the enhanced disease resistance in esd4 mutant, expressions of NHP biosynthetic genes were examined. As shown in Figures 4A–C, ALD1, SARD4, and FMO1 were all highly upregulated in esd4, suggestive of elevated NHP level in esd4. NHP biosynthetic genes are coordinately regulated by two master transcriptional regulators SARD1 and CBP60g in A. thaliana (Sun et al., 2015). Interestingly, both the expression of SARD1 and CBP60g are greatly induced in esd4 mutant (Figures 4D,E). SARD1 and CBP60g are also the main regulators of SA biosynthetic genes, including ICS1, EDS5, and PBS3 (Zhang et al., 2010; Wang et al., 2011; Sun et al., 2015). Previously, it was shown that esd4 accumulates much higher SA level compared to wild-type plant and ICS1 is upregulated in esd4 mutant under short-day condition (Villajuana-Bonequi et al., 2014). Agreeably, the expressions of other SA biosynthesis genes EDS5 and PBS3 are also upregulated in esd4 mutant (Figures 4F,G). Taken together, these data demonstrate that NHP and SA biosynthesis are upregulated in esd4, likely due to the elevated expression of SARD1 and CBP60g.
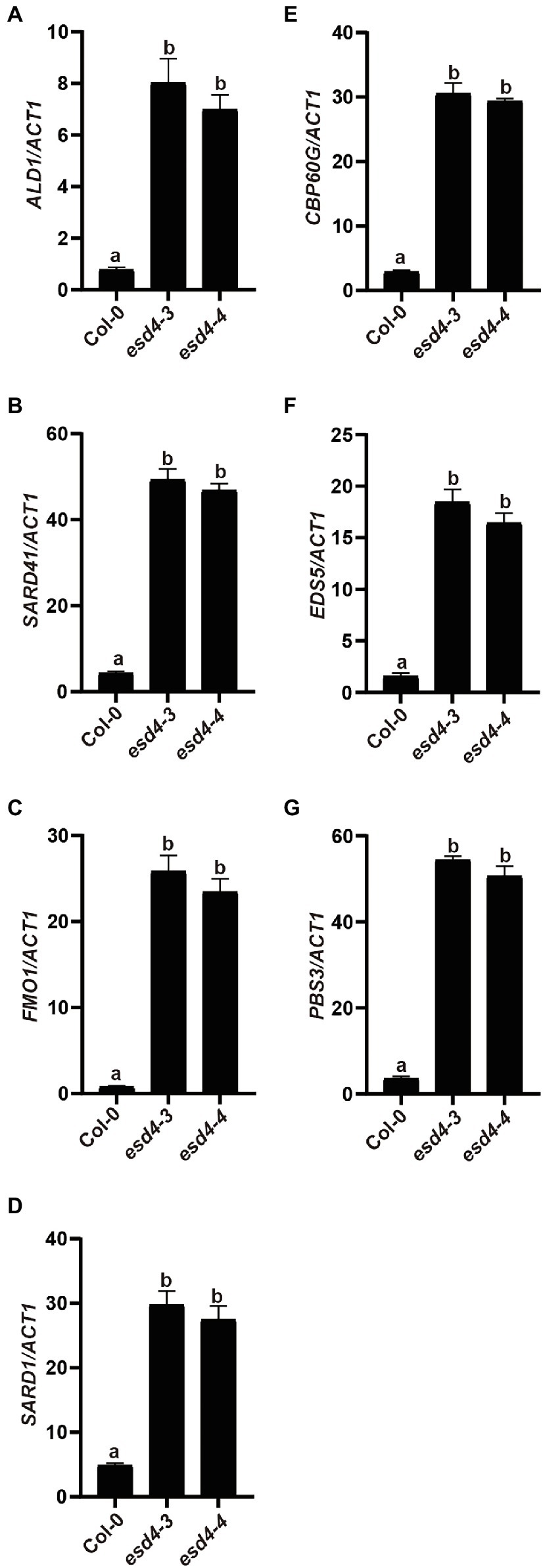
Figure 4. NHP and SA biosynthesis genes are upregulated in esd4 mutant. qRT-PCR was used to examine the NHP biosynthesis genes (A-C), SARD1 (D), CBP60g (E) and SA biosynthesis genes (F,G) on three-week-old Col-0, esd4-3 and esd4-4 plants. ACTIN1 serves as internal control. Error bars represent standard deviations. Letters indicate statistical differences (p < 0.05, one-way ANOVA, n = 3).
EDS1 is a central player downstream of many NLRs and upstream of SA and NHP biosynthesis (Zhang and Li, 2019; Huang et al., 2020). As ESD4 seems to act upstream of SA and NHP biosynthesis, we further examined its relationship with the upstream defense regulator EDS1. esd4-3 was crossed to loss-of-function mutant of EDS1, eds1-2 (Wagner et al., 2013) to generate eds1esd4 double mutant. As shown in Figure 5A, the dwarfism of esd4-3 was largely suppressed by eds1-2. Although autoimmunity often results in reduced fertility, eds1 only mildly restored esd4 infertility (Supplementary Figure S2). qRT-PCR showed that eds1-2 also fully blocked the upregulated PR1 and PR2 in esd4-3 (Figures 5B,C). In addition, analysis of resistance to H.a. Noco2 indicates that enhanced resistance to oomycete in esd4-3 was largely compromised by mutation in EDS1 (Figure 5D). Taken together, these data demonstrate that autoimmunity in esd4 largely relies on EDS1, indicating that loss function of ESD4 activates NLR-mediated immunity.
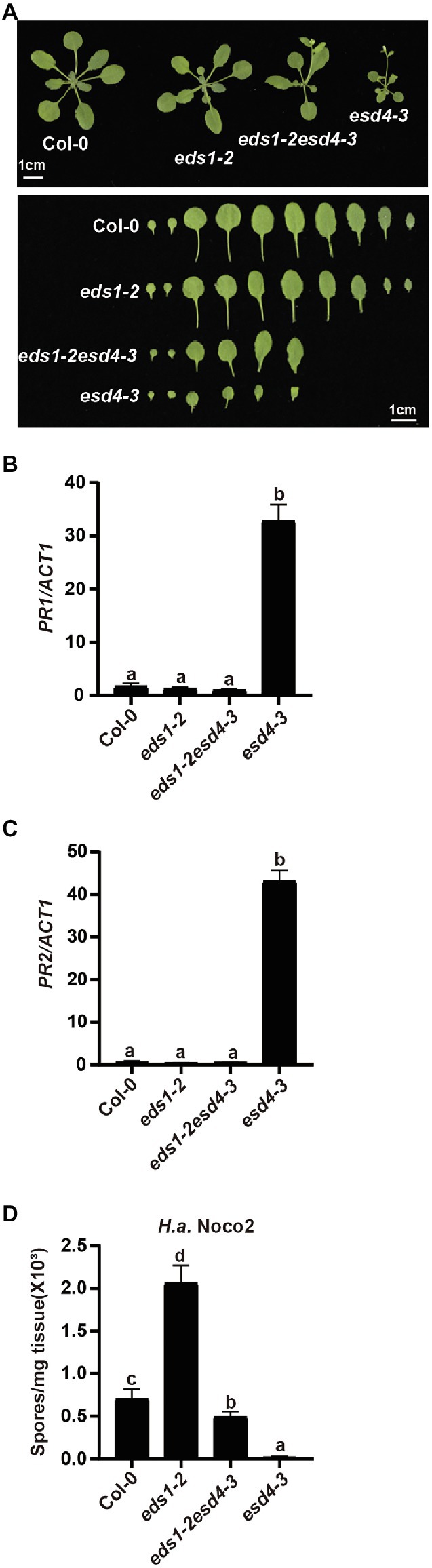
Figure 5. The constitutively activated immunity in esd4 is suppressed by eds1. (A) Morphologies of 4-week-old soil-grown plants of the indicated genotypes under long-day conditions. (B,C) Expression of PR1 (B) and PR2 (C) in indicated genotypes. ACTIN1 serves as internal control. (D) Growth of H.a. Noco2 on the indicated genotypes. The values indicate averages of replicates ± SD (n = 3). Different letters indicate statistical differences among different samples (p < 0.05, one-way ANOVA, n = 3). Experiments were repeated twice with similar results.
Because activation of NLRs can trigger SA and NHP biosynthesis, we then tested whether elevated SA and NHP biosynthesis in esd4 mutants is due to the activation of certain NLR(s). We measured expressions of SA and NHP biosynthetic-related genes in eds1esd4 double mutant. As shown in Figure 6, except for PBS3 and CBP60g, all the other highly expressed genes in esd4 are reduced to wild-type level by eds1, further verifying our hypothesis that esd4 activates NLR-mediated immunity.
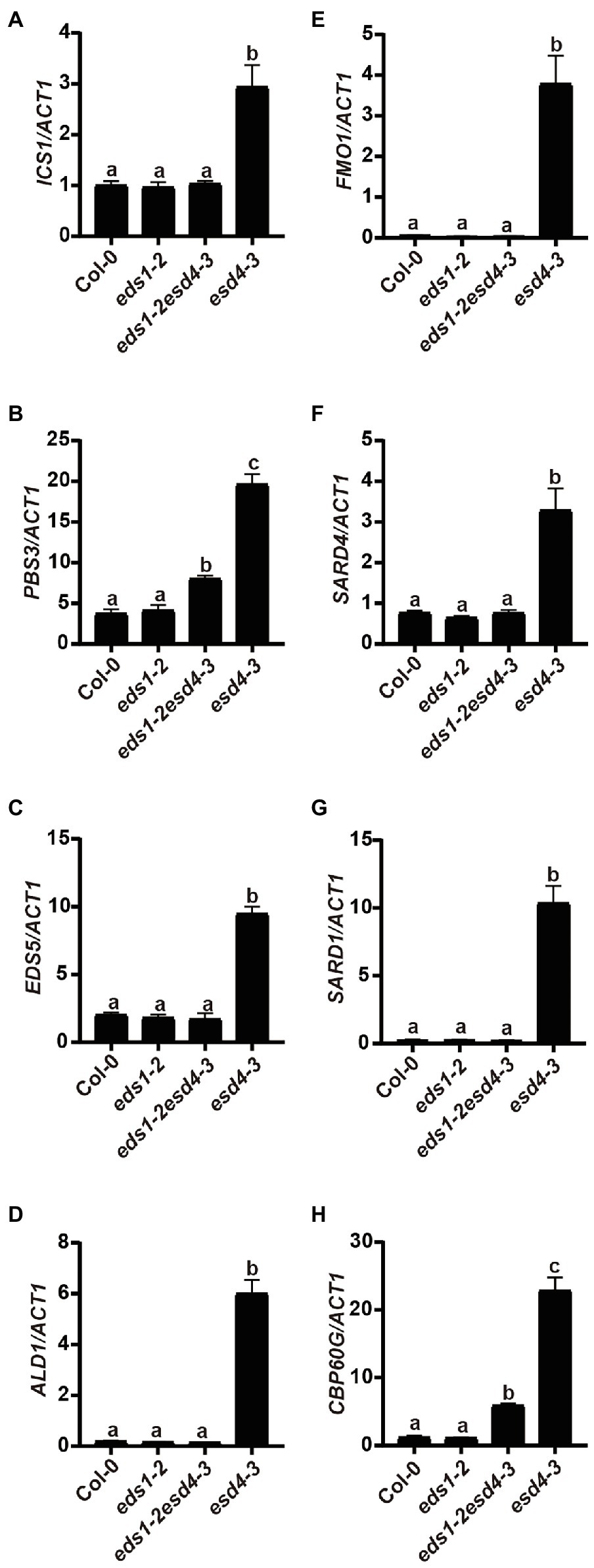
Figure 6. Elevated SA and NHP biosynthetic genes in esd4 are reduced by eds1. qRT-PCR was used to examine the SA biosynthesis genes (A-C), NHP biosynthesis genes (D-F), SARD1 (G) and CBP60g (F) on three-week-old Col-0, eds1-2, eds1-2esd4-3, and esd4-3 mutants. ACTIN1 serves as internal control. Error bars represent standard deviations. Letters indicate statistical differences (p < 0.05, one-way ANOVA, n = 3).
Previous studies showed that the autoimmune phenotypes of many autoimmune mutants are temperature-dependent and high temperature often inhibits NLR-mediated signaling. To test whether the autoimmune phenotype observed in esd4 is temperature-dependent, we grew esd4 under 28°C conditions. As shown in Supplementary Figure S3, the dwarf morphology and the expression of defense marker genes PR1 and PR2 were largely suppressed at 28°C. This is consistent with our finding that activation of NLR(s) leads to autoimmunity in esd4.
The plant immune system is elaborate and must be tightly controlled. Insufficient immune activation can lead to susceptibility against pathogens, whereas over-activation under non-pathogenic conditions may result in autoimmunity that impairs plant growth. The typical autoimmune phenotypes are associated with dwarfism, upregulated defense-related genes, spontaneous cell death, elevated SA, and enhanced disease resistance to pathogens. Autoimmunity could be caused by gain-of-function mutations in positive immune receptors/regulators or loss-of-function mutations in negative immune regulators (Van Wersch et al., 2016). SUMO protease ESD4 was initially identified and characterized due to its early flowering phenotype especially under short-day condition (Reeves et al., 2002; Pineiro et al., 2003). In this study, we reported that loss function of ESD4 leads to autoimmunity, suggesting that ESD4 is a negative regulator of plant immunity.
Forward genetic screen under short-day condition designed to search for genes that contribute to early flowering phenotype in esd4 mutant identified ICS1, a major SA biosynthesis gene (Villajuana-Bonequi et al., 2014). Loss of ICS1 reduces the elevated SA level in esd4 compared to wild-type, but only suppresses part of upregulated PR1 in esd4 mutant under short-day condition. We also analyzed the activated defense response in sid2 esd4 double mutant under long-day condition and obtained similar results (data not shown), indicative of the partial contribution of SA signaling on the autoimmunity in esd4. In parallel of SA, NHP also participates in immune signaling (Bartsch et al., 2006; Liu et al., 2020; Sun et al., 2020). From our analysis of the fmo1 esd4 double mutant, which is deficient in NHP biosynthesis, we showed that NHP signaling is also required for the autoimmunity in esd4.
From our further epistatic analysis between esd4 and eds1, we revealed that the autoimmunity of esd4 is largely dependent on EDS1, the immune regulator immediately downstream of many NLR(s), especially for TNLs. This suggests that the autoimmunity of esd4 is mostly contributed by activation of unknown NLR(s). ESD4, previously reported with SUMO protease activity in vitro and in vivo (Murtas et al., 2003), is mainly responsible for recycling SUMO from SUMOylated targets, as less free SUMO and more SUMO conjugates than wild-type plants are observed in esd4 mutant. It is possible that ESD4 directly targets an NLR to remove its SUMO modification for deactivation. However, as there is no evidence on the requirement of SUMOylation in NLR activation, an alternative hypothesis through a guard model may be more plausible. ESD4 was reported to localize to the nucleus and predominantly to the periphery of the nucleus (Murtas et al., 2003). Here the NLR likely guards a host guardee protein via its SUMOylation status, which is in turn regulated by ESD4. In wild-type plants, ESD4 removes SUMO from the guardee, preventing its perception by the cognate NLR. In esd4 mutant plants, the increased SUMOylation of the guardee can be sensed by this NLR, leading to its activation including the upregulation of downstream immune signaling pathways mediated by SA and NHP (Figure 7).
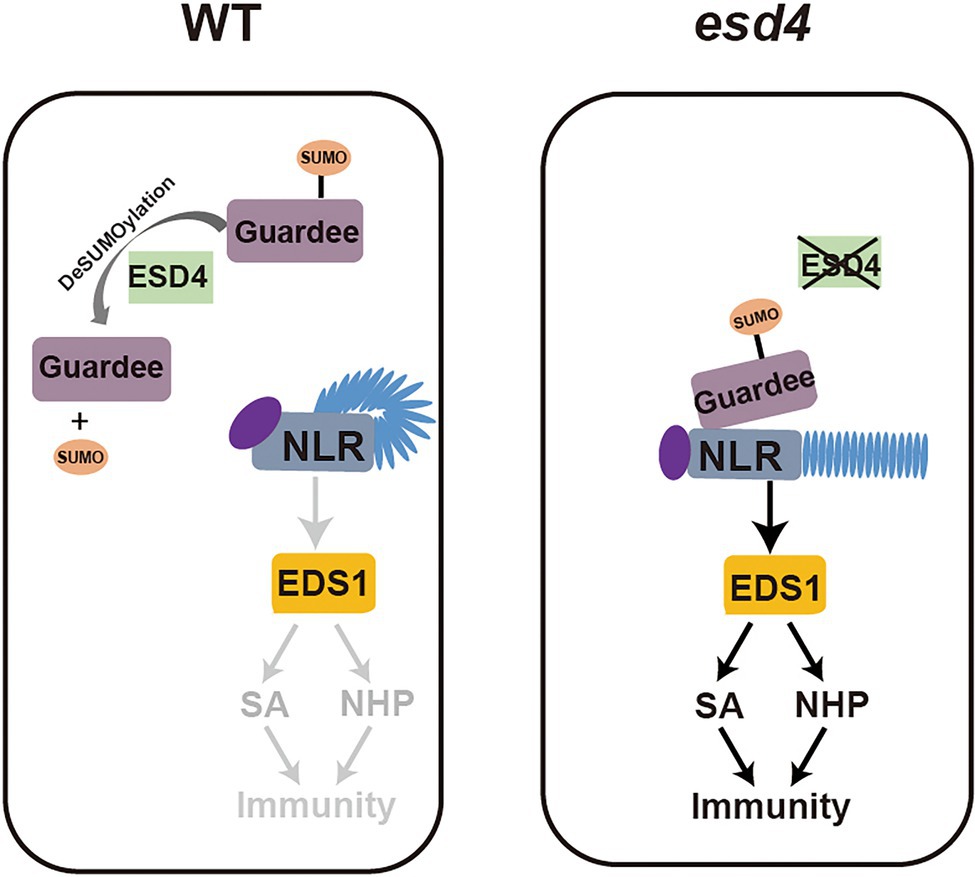
Figure 7. Working model. In wild-type plants, ESD4 deSUMOylates guardee protein to inhibit NLR activation. In esd4 mutants, the SUMOylating status of guardee is sensed by NLR, leading to the activation of downstream immune signaling including SA and NHP biosynthesis.
Among the 16 putative SUMO proteases identified in Arabidopsis (Morrell and Sadanandom, 2019), OVERLY TOLERANT TO SALT1 (OST1) and OST2 were also reported to redundantly modulate defense responses via SA signaling (Campanaro et al., 2016). Similar to esd4, ots1ots2 double mutants also exhibited autoimmune phenotypes. As highly accumulated SA level and upregulated expression of ICS1 were observed in ots1ots2 plants, the constitutively activated immune responses in ots1ots2 mutants was attributed to the SA signaling. Whether NLR activation leads to the autoimmunity of ots1ots2, as in esd4, remains to be determined.
The regulations of SA and NHP biosynthesis in plant immunity are complicated (Huang et al., 2020). Even though we proved that the upregulation of SA and NHP biosynthesis observed in esd4 is largely due to the activation of NLR, we cannot rule out the possibility that ESD4 may directly regulate SA and NHP levels. By sequencing analysis, we found that most of enzymes involved in SA and NHP biosynthesis genes contain either predicted SUMOylated consensus residues (ψKXE/D,ψis a hydrophobic residue; K is a lysine residue; X is any residue; D/E is an aspartic acid or glutamic acid residue) or SUMO interaction motifs (SIMs; Supplementary Figure S4), indicating that they might be targets of ESD4 and their activity or protein level could be directly modulated by ESD4. In addition, some of SA and NHP biosynthesis regulators like SARD1 and CBP60g also have predicted SUMOylated consensus residues and SIMs. Whether they are targets of ESD4 remains unclear. As ESD4 may simultaneously target multiple proteins at different levels, these models can play in orchestra to fine-tune immunity. Identifying the direct targets of ESD4 in the future will shed light on its exact roles in regulating SA and NHP accumulation. In summary, our study provides sufficient evidence demonstrating that loss of SUMO protease ESD4 leads to the autoimmunity-mediated by NLR(s).
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
XL and SX designed the study. XH performed the experiments and prepared the figures and tables. YL and XH wrote the first draft of the manuscript. JH analyzed the whole-genome sequencing data. WF, XL, and SX discussed the results, supervised, and edited the paper. All authors contributed to the article and approved the submitted version.
This research is supported by grants from National Natural Science Foundation of China (Grants 30970247 and 31971836) to SX; Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery program received by WF and XL; the Dewar Cooper Memorial Fund from the University of British Columbia to XL. YL is supported by the China Postdoctoral International Exchange Program (Talent-Introduction Program).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.881212/full#supplementary-material
Aarts, N., Metz, M., Holub, E., Staskawicz, B. J., Daniels, M. J., and Parker, J. E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 95, 10306–10311. doi: 10.1073/pnas.95.17.10306
Bartsch, M., Gobbato, E., Bednarek, P., Debey, S., Schultze, J. L., Bautor, J., et al. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18, 1038–1051. doi: 10.1105/tpc.105.039982
Campanaro, A., Battaglia, R., Galbiati, M., Sadanandom, A., Tonelli, C., and Conti, L. (2016). SUMO proteases OTS1 and 2 control filament elongation through a DELLA-dependent mechanism. Plant Reprod. 29, 287–290. doi: 10.1007/s00497-016-0292-8
Century, K. S., Shapiro, A. D., Repetti, P. P., Dahlbeck, D., Holub, E., and Staskawicz, B. J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278, 1963–1965. doi: 10.1126/science.278.5345.1963
Chen, Y. C., Holmes, E. C., Rajniak, J., Kim, J. G., Tang, S., Fischer, C. R., et al. (2018). N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 115, E4920–E4929. doi: 10.1073/pnas.1805291115
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cui, H. T., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Dangl, J. L., and Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Daudi, A., and O’Brien, J. A. (2012). Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio Protoc 2:e263. doi: 10.21769/BioProtoc.263
Ding, P., Rekhter, D., Ding, Y., Feussner, K., Busta, L., Haroth, S., et al. (2016). Characterization of a Pipecolic acid biosynthesis pathway required for systemic acquired resistance. Plant Cell 28, 2603–2615. doi: 10.1105/tpc.16.00486
Falk, A., Feys, B. J., Frost, L. N., Jones, J. D., Daniels, M. J., and Parker, J. E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. U. S. A. 96, 3292–3297. doi: 10.1073/pnas.96.6.3292
Hartmann, M., Kim, D., Bernsdorff, F., Ajami-Rashidi, Z., Scholten, N., Schreiber, S., et al. (2017). Biochemical principles and functional aspects of Pipecolic acid biosynthesis in plant immunity. Plant Physiol. 174, 124–153. doi: 10.1104/pp.17.00222
Hartmann, M., Zeier, T., Bernsdorff, F., Reichel-Deland, V., Kim, D., Hohmann, M., et al. (2018). Flavin Monooxygenase-generated N-Hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 173(2), 456–469. doi: 10.1016/j.cell.2018.02.049
He, Z. Q., Huang, T. T., Ao, K., Yan, X. F., and Huang, Y. (2017). Sumoylation, phosphorylation, and acetylation fine-tune the turnover of plant immunity components mediated by Ubiquitination. Front. Plant Sci. 8:1682. doi: 10.3389/fpls.2017.01682
Huang, J., Sun, Y., Orduna, A. R., Jetter, R., and Li, X. (2019). The mediator kinase module serves as a positive regulator of salicylic acid accumulation and systemic acquired resistance. Plant J. 98, 842–852. doi: 10.1111/tpj.14278
Huang, W., Wang, Y., Li, X., and Zhang, Y. (2020). Biosynthesis and regulation of salicylic acid and N-Hydroxypipecolic acid in plant immunity. Mol. Plant 13, 31–41. doi: 10.1016/j.molp.2019.12.008
Jander, G., Norris, S. R., Rounsley, S. D., Bush, D. F., Levin, I. M., and Last, R. L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129, 440–450. doi: 10.1104/pp.003533
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Knepper, C., Savory, E. A., and Day, B. (2011). Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol. 156, 286–300. doi: 10.1104/pp.110.169656
Lapin, D., Bhandari, D. D., and Parker, J. E. (2020). Origins and immunity networking functions of EDS1 family proteins. Annu. Rev. Phytopathol. 58, 253–276. doi: 10.1146/annurev-phyto-010820-012840
Liu, Y., Sun, T., Sun, Y., Zhang, Y., Radojicic, A., Ding, Y., et al. (2020). Diverse roles of the salicylic acid receptors NPR1 and NPR3/NPR4 in plant immunity. Plant Cell 32, 4002–4016. doi: 10.1105/tpc.20.00499
Maruta, N., Burdett, H., Lim, B. Y. J., Hu, X., Desa, S., Manik, M. K., et al. (2022). Structural basis of NLR activation and innate immune signalling in plants. Immunogenetics 74, 5–26. doi: 10.1007/s00251-021-01242-5
McDowell, J. M., Cuzick, A., Can, C., Beynon, J., Dangl, J. L., and Holub, E. B. (2000). Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J. 22, 523–529. doi: 10.1046/j.1365-313x.2000.00771.x
Mishina, T. E., and Zeier, J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141, 1666–1675. doi: 10.1104/pp.106.081257
Miura, K., Jin, J. B., and Hasegawa, P. M. (2007). Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. 10, 495–502. doi: 10.1016/j.pbi.2007.07.002
Morrell, R., and Sadanandom, A. (2019). Dealing With stress: A review of plant SUMO proteases. Front. Plant Sci. 10:1122. doi: 10.3389/fpls.2019.01122
Murtas, G., Reeves, P. H., Fu, Y. F., Bancroft, I., Dean, C., and Coupland, G. (2003). A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant cell 15, 2308–2319. doi: 10.1105/tpc.015487
Navarova, H., Bernsdorff, F., Doring, A. C., and Zeier, J. (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24, 5123–5141. doi: 10.1105/tpc.112.103564
Nishimura, M. T., and Dangl, J. L. (2010). Arabidopsis and the plant immune system. Plant J. 61, 1053–1066. doi: 10.1111/j.1365-313X.2010.04131.x
Orosa, B., Yates, G., Verma, V., Srivastava, A. K., Srivastava, M., Campanaro, A., et al. (2018). SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signalling in plant innate immunity. Nat. Commun. 9:5185. doi: 10.1038/s41467-018-07696-8
Pineiro, M., Gomez-Mena, C., Schaffer, R., Martinez-Zapater, J. M., and Coupland, G. (2003). EARLY BOLTING IN SHORT DAYS is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant Cell 15, 1552–1562. doi: 10.1105/tpc.012153
Reeves, P. H., Murtas, G., Dash, S., and Coupland, G. (2002). Early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 129, 5349–5361. doi: 10.1242/dev.00113
Rekhter, D., Ludke, D., Ding, Y., Feussner, K., Zienkiewicz, K., Lipka, V., et al. (2019). Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 365, 498–502. doi: 10.1126/science.aaw1720
Shapiro, A. D., and Zhang, C. (2001). The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 127, 1089–1101. doi: 10.1104/pp.010096
Song, J. T., Lu, H., and Greenberg, J. T. (2004a). Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, aberrant growth and death2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell 16, 353–366. doi: 10.1105/tpc.019372
Song, J. T., Lu, H., McDowell, J. M., and Greenberg, J. T. (2004b). A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 40, 200–212. doi: 10.1111/j.1365-313X.2004.02200.x
Sun, T., Huang, J., Xu, Y., Verma, V., Jing, B., Sun, Y., et al. (2020). Redundant CAMTA transcription factors negatively regulate the biosynthesis of salicylic acid and N-Hydroxypipecolic acid by modulating the expression of SARD1 and CBP60g. Mol. Plant 13, 144–156. doi: 10.1016/j.molp.2019.10.016
Sun, T., Zhang, Y., Li, Y., Zhang, Q., Ding, Y., and Zhang, Y. (2015). ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 6:10159. doi: 10.1038/ncomms10159
Torrens-Spence, M. P., Bobokalonova, A., Carballo, V., Glinkerman, C. M., Pluskal, T., Shen, A., et al. (2019). PBS3 and EPS1 complete salicylic acid biosynthesis from Isochorismate in Arabidopsis. Mol. Plant 12, 1577–1586. doi: 10.1016/j.molp.2019.11.005
Van Wersch, R., Li, X., and Zhang, Y. (2016). Mighty dwarfs: Arabidopsis autoimmune mutants and their usages in genetic dissection of plant immunity. Front. Plant Sci. 7:1717. doi: 10.3389/fpls.2016.01717
Verma, V., Croley, F., and Sadanandom, A. (2018). Fifty shades of SUMO: its role in immunity and at the fulcrum of the growth-defence balance. Mol. Plant Pathol. 19, 1537–1544. doi: 10.1111/mpp.12625
Verma, V., Srivastava, A. K., Gough, C., Campanaro, A., Srivastava, M., Morrell, R., et al. (2021). SUMO enables substrate selectivity by mitogen-activated protein kinases to regulate immunity in plants. Proc. Natl. Acad Sci. U. S. A. 118:10. doi: 10.1073/pnas.2021351118
Villajuana-Bonequi, M., Elrouby, N., Nordstrom, K., Griebel, T., Bachmair, A., and Coupland, G. (2014). Elevated salicylic acid levels conferred by increased expression of ISOCHORISMATE SYNTHASE 1 contribute to hyperaccumulation of SUMO1 conjugates in the Arabidopsis mutant early in short days 4. Plant J. 79, 206–219. doi: 10.1111/tpj.12549
Wagner, S., Stuttmann, J., Rietz, S., Guerois, R., Brunstein, E., Bautor, J., et al. (2013). Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe. 14, 619–630. doi: 10.1016/j.chom.2013.11.006
Wang, Z. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16:144. doi: 10.1186/s13059-015-0715-0
Wang, L., Tsuda, K., Truman, W., Sato, M., Nguyen le, V., Katagiri, F., et al. (2011). CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J. 67, 1029–1041. doi: 10.1111/j.1365-313X.2011.04655.x
Wilkinson, K. A., and Henley, J. M. (2010). Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428, 133–145. doi: 10.1042/BJ20100158
Xu, X. M., Rose, A., Muthuswamy, S., Jeong, S. Y., Venkatakrishnan, S., Zhao, Q., et al. (2007). NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19, 1537–1548. doi: 10.1105/tpc.106.049239
Yates, G., Srivastava, A. K., and Sadanandom, A. (2016). SUMO proteases: uncovering the roles of deSUMOylation in plants. J. Exp. Bot. 67, 2541–2548. doi: 10.1093/jxb/erw092
Zhang, Y., Goritschnig, S., Dong, X., and Li, X. (2003). A gain-of-function mutation in a plant disease resistance gene lead to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15, 2636–2646. doi: 10.1105/tpc.015842
Zhang, Y., and Li, X. (2019). Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 50, 29–36. doi: 10.1016/j.pbi.2019.02.004
Zhang, Z., Wu, Y., Gao, M., Zhang, J., Kong, Q., Liu, Y., et al. (2012). Disruption of PAMP-induced MAP kinase cascade by a pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11, 253–263. doi: 10.1016/j.chom.2012.01.015
Keywords: ESD4, SUMO modification, NHP signaling, EDS1, NLR, autoimmunity
Citation: Huang X, Liu Y, Huang J, Fernando WGD, Li X and Xia S (2022) Activation of NLR-Mediated Autoimmunity in Arabidopsis Early in Short Days 4 Mutant. Front. Plant Sci. 13:881212. doi: 10.3389/fpls.2022.881212
Received: 22 February 2022; Accepted: 04 May 2022;
Published: 25 May 2022.
Edited by:
Andrei-Jose J. Petrescu, Institute of Biochemistry of the Romanian Academy, RomaniaReviewed by:
Avijit Tarafdar, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), IndiaCopyright © 2022 Huang, Liu, Huang, Fernando, Li and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Li, eGlubGlAbXNsLnViYy5jYQ==; Shitou Xia, eHN0b25lMDUwNUBodW5hdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.